Abstract
Protein design and engineering are evolving at an unprecedented pace leveraging the advances in deep learning. Current models nonetheless cannot natively consider non-protein entities within the design process. Here, we introduce a deep learning approach based solely on a geometric transformer of atomic coordinates and element names that predicts protein sequences from backbone scaffolds aware of the restraints imposed by diverse molecular environments. To validate the method, we show that it can produce highly thermostable, catalytically active enzymes with high success rates. This concept is anticipated to improve the versatility of protein design pipelines for crafting desired functions.
Subject terms: Protein design, Protein design, Enzymes
Advances in deep learning are transforming protein design. Here, authors introduce a method using geometric transformers to predict protein sequences, resulting in highly thermostable and catalytically active enzymes with high success rates.
Introduction
Designing proteins de novo to engineer their properties for functional tasks is a grand challenge with direct implications for biology, medicine, biotechnology, and materials science. One key application area is the engineering of protein therapeutics1,2. It involves creating proteins tailored to target specific diseases or conditions with high precision. Such an approach has been shown to be a competitive alternative to small molecule-based medicines3. It opens possibilities to revolutionize the way we treat many health issues, from autoimmune diseases to cancers, offering potentially more effective and personalized treatments compared to conventional drugs.
Additionally, engineering enzymatic functions represent another promising challenging task for protein design. Enzymes, as natural catalysts, play crucial roles in various biological processes. By designing new enzymes, or modifying existing ones, one can create catalysts that facilitate reactions that are rare or non-existent in nature4. This can have profound implications for numerous industries, including pharmaceuticals, where custom enzymes can be used to synthesize complex drug molecules more efficiently5, and in environmental technology, where designed enzymes might break down pollutants or plastics with enhanced efficiency6. Protein engineering, therefore, not only shows tremendous potential for industrial processes and environmental sustainability but also opens new avenues in scientific research and biotechnological innovation.
While physics-based approaches have contributed to the advancements of protein engineering, deep learning methods have recently brought a dramatic acceleration by enhancing the success rates and versatility of protein design pipelines7. Among the most recent and notable examples, ProteinMPNN, based on an encoder-decoder neural network, is able to generate protein sequences experimentally proven to fold as intended8,9. More recently, coupled with denoising diffusion probabilistic models for the generation of protein backbones, ProteinMPNN and RFdiffusion have shown remarkable success10. In addition, ESM-IF1, based on a hybrid protein language model and structural model, is capable of generating highly diverse proteins well outside the known universe of natural sequences11,12. The model has also recently found experimental validation reporting a very high success rate13. More broadly, deep learning approaches are pervasive in the field, finding broad application in several protein design tasks14–17, like for example MaSIF which specializes in the design of protein interactions via learned protein surface fingerprints18,19 or Chroma which is able to generate protein backbones and sequences under arbitrary constraints using diffusion20.
Current protein design models can natively handle multiple protein chains in their inputs, allowing them to design the sequences of interacting proteins. However, they only poorly handle non-protein entities within the design process, which hampers their versatility and limits their scope of applicability. We have recently introduced a deep learning model that can help mitigate these limitations, the Protein Structure Transformer (PeSTo21), a geometric transformer architecture that operates on atom point clouds. It integrates different advances in deep learning such as transformer attention22 and utilizes both a scalar and vector state to represent the atoms23. Representing molecules uniquely by element names and coordinates, PeSTo can be applied to and predict protein interfaces with virtually any kind of molecules, either other proteins, nucleic acids, lipids, ions, small ligands, cofactors or carbohydrates24.
In this work, we leverage the unique features of this model and introduce CARBonAra (Context-aware Amino acid Recovery from Backbone Atoms and heteroatoms), a protein sequence generator model based on PeSTo. Trained uniquely on structural data available in the PDB25, CARBonAra predicts amino acid confidences at all positions of a given backbone scaffold that can be provided alone or complexed to any kind and number of molecules that help to drive sequence design. The model performs excellently against the state of the art in in silico benchmarks and it is validated experimentally for the challenging case of engineering the structure and function of an enzymatic system.
Results
Sequence prediction from backbone scaffolds
Building on the architecture of our previous Protein Structure Transformer – PeSTo Model21, CARBonAra predicts the likelihood of finding a given amino acid at each position of a protein sequence from an input backbone scaffold using a deep learning model composed of geometric transformers (Fig. 1a). CARBonAra takes as input the coordinates and elements of the backbone atoms (Cα, C, N, O) and adds virtual Cβ atoms using ideal bond angles and lengths. The geometry is described using distances and normalized relative displacement vectors between each atom. At its core, CARBonAra is built of geometric transformer operations, each gradually processing the information of a larger local neighborhood from 8 up to 64 nearest neighbors. The geometric information is equivariantly encoded from the vectorial state, while the scalar state represents invariant quantities of the geometry under the global rotation of the atomic point cloud. The geometric transformer operation encodes the interactions of all nearest neighbors and employs a transformer to process the scalar and vectorial information and update the state of each atom. Finally, by pooling the atom states from atomic to residue level, we trained the model to predict amino acid confidences at each position of a protein’s sequence in the form of a position-specific scoring matric (PSSM, Fig. 1a, Supplementary Algorithm 1, Supplementary Fig. 1 and “Methods”). Practically, these confidences can be interpreted as and mapped into probabilities by characterizing the probability of a correct prediction given a prediction confidence for each amino acid type (Supplementary Fig. 2). Like other models such as ProteinMPNN, CARBonAra supports autoregressive predictions by imprinting the prior sequence information of specific amino acids into the backbone atoms using one-hot encoding see “Methods”.
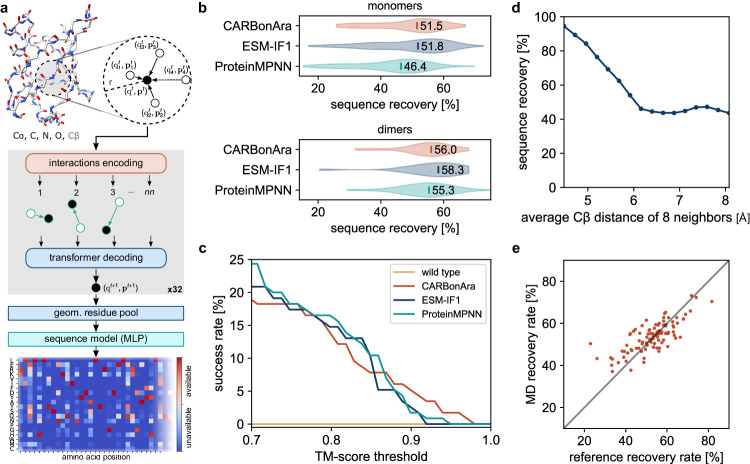
Fig. 1. CARBonAra’s architecture and comparison with state-of-the-art methods.
a The model applies multiple geometric transformer operations to the coordinates and atom element of a backbone scaffold with added virtual Cβ to predict the amino acid confidence at each position in the sequence expressed as a position-specific scoring matrix. b Comparison of the sequence recovery of different methods for monomers and dimers with indicated median sequence recovery. c Percentage of AlphaFold predicted structures, in single-sequence mode, above a TM-score threshold using sequences sampled in the same way for different methods. d Sequence recovery as a function of the average Cβ distance of the 8 nearest neighbors. e Consensus prediction recovery rate against the reference experimental structure recovery rate, derived from 500 frames sampled from 1 μs molecular dynamics simulations for 80 monomers.
Most importantly, CARBonAra inherits PeSTo’s capability to work solely with element names and atomic coordinates, eliminating the need for extensive parametrizations and thus allowing for easy adaptation to various scenarios. As a result, CARBonAra can parse and process any molecular entity near the protein backbone being designed, which includes a range of inputs such as other proteins, small molecules, nucleic acids, lipids, ions, and water molecules. Leveraging this inherent flexibility of CARBonAra, we have been able to incorporate all biological assemblies from the RCSB PDB into our training dataset see “Methods”. This includes proteins in complex with other molecular entities like ions, ligands, nucleic acids, and more. The training dataset is composed of approximately 370,000 subunits, with an additional 100,000 subunits utilized in the validation dataset, all sourced from RCSB PDB biological assemblies annotated as the most likely25. Following a slightly more stringent protocol compared to previously established methods8,11, the testing dataset consists of about 70,000 subunits, distinct from the training set with no shared CATH domains and filtered at less than 30% sequence identity see “Methods”. This selection criterion ensures the robustness of our testing, as it excludes similar folds and sequences present in the training dataset.
For sequence design of isolated proteins or protein complexes from backbone structures without non-protein molecules in the context, CARBonAra performs on par with state-of-the-art methods like ProteinMPNN and ESM-IF1 for sequence prediction (Fig. 1b) at a competitive computational cost (approximately 3 times faster than ProteinMPNN and 10 times faster than ESM-IF1 on GPUs, see Supplementary Fig. 3). Our method achieves a median sequence recovery rate of 51.3% for protein monomer design and 56.0% for dimer design when reconstructing protein sequences from backbone structures. Although having a similar recovery rate, the median sequence identity between the optimal sequence of the three methods ranges from 54 to 58% (Supplementary Fig. 4). Moreover, we observe that CARBonAra can generate high-quality sequences that fold as intended when predicted using AlphaFold in single-sequence mode, with a TM-score above 0.9 (Fig. 1c). We observed that CARBonAra learned the tighter amino acid packing at protein cores, thus resulting in higher recovery rates and reflecting the lower tolerance to substitutions typical of buried amino acids (Fig. 1d and Supplementary Figs. 5a, b) while allowing for higher variability on the protein surface unless additional functional or structural constraints are provided.
Methods for sequence prediction from a backbone scaffold are trained mainly on experimental data with ideal backbone geometry, which can lead to a decrease in performance when applied to the generated backbone. Adding noise to the geometry during the training can mediate this problem8. We characterized the robustness of our method by applying CARBonAra to structural trajectories from molecular dynamics (MD) simulations. We observed no significant decrease in sequence recovery (53 ± 10%) from the consensus prediction (54 ± 7%) due to conformation changes of the backbone and an increase in cases that previously showed low recovery rates (Fig. 1e). Simultaneously, we observed a general reduction in the number of possible amino acids predicted per position (Supplementary Fig. 6) suggesting that exploring the conformational space is limiting the sequence space thus enabling the design of targeted structural conformations.
Sampling the sequence space
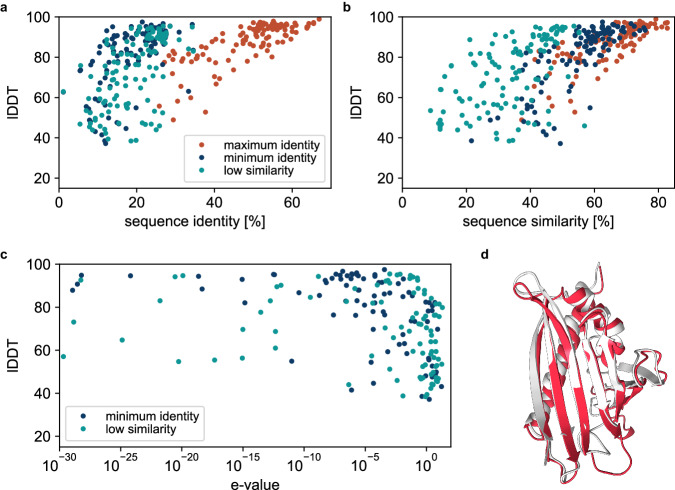
As in other methods for protein sequence prediction, the site-specific amino acid confidences/probabilities need to be somehow sampled to derive sequences that embody actual proteins. ProteinMPNN and ESM-IF1 use the logit output of the model as the energy of a Boltzmann distribution to describe the amino acid probabilities at a user-defined sampling temperature. In contrast, CARBonAra uses multi-class amino acid predictions that generate a space of potential sequences, opening various possibilities for sequence sampling. For example, one can tailor sequences to meet specific objectives, such as achieving minimal sequence identity or low sequence similarity to design unique sequences with a specific fold. We show that we can generate sequences with as low as approximately 10% sequence identity and 20% sequence similarity while still recovering an AlphaFold predicted structure close to the scaffold structure (lDDT > 80) (Fig. 2a, b, see also “Methods”). Some of the generated sequences are not only different from the scaffold protein but also from any known protein (Fig. 2c). As an example, one of these cases uses the birch pollen allergen Bet v 1 protein (PDB ID: 6R3C) as scaffold. We generated a sequence with 7% identity and 13% similarity to the original scaffold, pushing the limits of sequence similarity. A BLAST26 search reveals no significant matches and the AlphaFold predicted structure of the created sequence has a lDDT of 70 (Fig. 2d).
Fig. 2. Sequence sampling.
Local Distance Difference Test (lDDT) of AlphaFold predicted structures to the reference scaffold monomers for sequences generated using CARBonAra with, as objective, maximum sequence identity, minimum sequence identity, and low sequence similarity, against the (a) sequence identity and (b) sequence similarity. c lDDT of the AlphaFold predicted structures as a function of the highest expect value (E-value) of the generated sequences from a BLAST26 search. d Scaffold, in white, from the birch pollen allergen Bet v 1 protein (PDB ID: 6R3C). AlphaFold predicted structure, shown in red, from the generated sequence has a lDDT of 70 from the reference scaffold. The generated sequence has a 7% identity and 13% similarity with the original scaffold protein.
Context-aware sequence prediction
More importantly, leveraging PeSTo’s architecture, CARBonAra has the ability to perform protein sequence prediction conditioned by a specific non-protein molecular context. On a test set composed of structures with folds different than the training set (see “Methods”), we show that the overall structure median sequence recovery increased from 54% to 58% (Supplementary Fig. 7) when an additional molecular context is provided. In particular, CARBonAra achieves median sequence recovery rates of 56% at protein interfaces for protein interacting partners and 55% for interfaces with nucleic acids, i.e., a significant improvement over predictions without context (Fig. 3a). Similarly, recovery rates at protein interfaces improve significantly if small-molecule entities such as ions (67%), lipids (57%), ligands (61%) and glycans (50%) are included. This correlates with the latest developments of PeSTo where we addressed the possibility of re-training specific models addressing carbohydrate-protein interfaces (PeSTo-Carbs24). Including these molecules not only boosts sequence recovery in their surroundings but also reduces the number of amino acid possibilities to sample from (Fig. 3b).
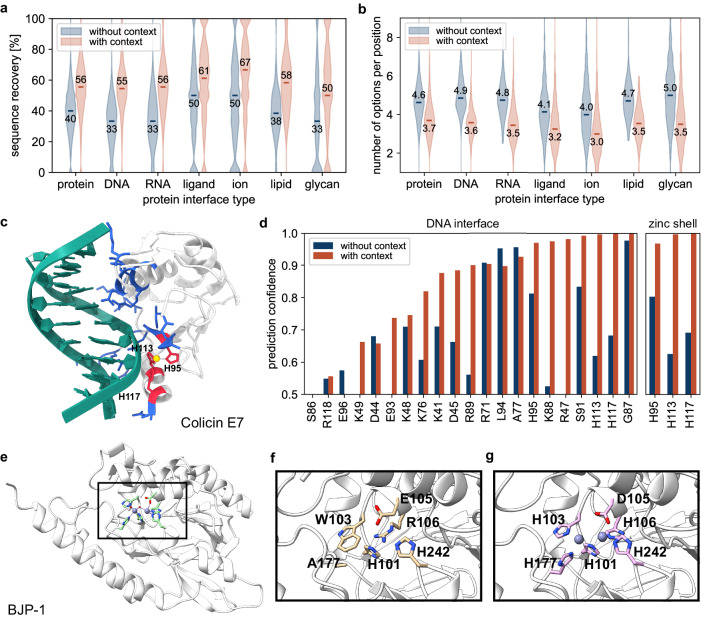
Fig. 3. Context-aware amino acid recovery allows the design of functional proteins.
a Sequence recovery at the interface (residues within 5 Å) without and with proteins, nucleic acids, ligands, ions, lipids, and glycans binders. b Number of predicted possible amino acids per position at the interface (residues within 5 Å) without and with proteins, nucleic acids, ligands, ions, lipids, and glycans binders (considering a confidence prediction threshold of 0.5). c Colicin E7 endonuclease domain in complex with DNA and a zinc ion (PDB ID: 1ZNS). The protein-DNA interface (residues within 4 Å) is highlighted in blue. The protein-zinc shell is highlighted in red (residues within 3 Å). d Estimated accurate prediction probability for the scaffold amino acids at the protein-DNA interface and the protein-zinc shell with and without the presence of DNA and zinc. e Metallo β-lactamase structure of BJP−1 with the catalytic pocket containing two zinc ions (PDB ID: 3LVZ). The pocket of an AlphaFold predicted structure from a CARBonAra-designed sequence applied to the scaffold backbone without zinc ions (f) and containing the original zinc ions (g).
An exemplary case to illustrate the power of this approach is the endonuclease domain of ColE7, which interacts with duplex DNA in a zinc-dependent manner27 (Fig. 3c). The sequence recovery rate obtained by CARBonAra showed a significant increase from 29% to 52% at the metal and DNA interfaces when the zinc ion or the 12-bp DNA duplex was included as resolved in the native structure (Fig. 3d). Thus, imposing the presence of non-protein interacting interfaces can enhance the sequence recovery rate significantly, also with respect to predictions done by ProteinMPNN (24%) and ESM-IF1 (43%) (Supplementary Table 1). Interestingly, when a non-native molecular context is provided, such as a larger ion (e.g., calcium) the sequence recovery rate decreases (Supplementary Fig. 8). Thus, the predicted amino acid confidence of an ion pocket is widely dependent on the given context, as also illustrated for the case of BJP-1, a zinc-dependent metallo β-lactamase (Fig. 3e). In the absence of the zinc ions in BJP-1’s active site, CARBonAra’s prediction does not lead to the complete recovery of the zinc coordinating residues (Fig. 3f). By keeping the zinc ions in the structure, CARBonAra’s context awareness allows the full recovery of the correct zinc coordinating residues in the active site of the Metallo β-lactamase (Fig. 3g). This showcases that the absence or presence of different atoms leads to different predictions indicating a high sensitivity of the context for the outputs from CARBonAra.
Engineering a β-lactamase enzyme
We next sought to test CARBonAra’s predictions by designing variants of an enzyme and studying their structural and functional features in vitro. Relevant for enzyme design is the possibility of designing sequences under the restraints provided by a desired substrate or high-affinity ligand. We thus used the TEM-1 β-lactamase backbone scaffold in complex with a β-lactam substrate (i.e., nitrocefin) to generate potential new sequences holding β-lactamase activity (Fig. 4a). Without context, the catalytic S70 and substrate-binding R244 are not predicted, having low confidences of 0.39 and 0.11, respectively (Fig. 4b). However, when the prediction is done with nitrocefin docked at the catalytic pocket, the catalytic triad S70, K73, and E166, along with key residues necessary for β-lactam binding (i.e., N132, R244) all have a high prediction confidence ( > 0.8) and low ranking (top 2) (Supplementary Fig. 9). Importantly, in this case, the sequence recovery is maximal when also the catalytic water is considered, hinting at a very high sensitivity for the molecular context.
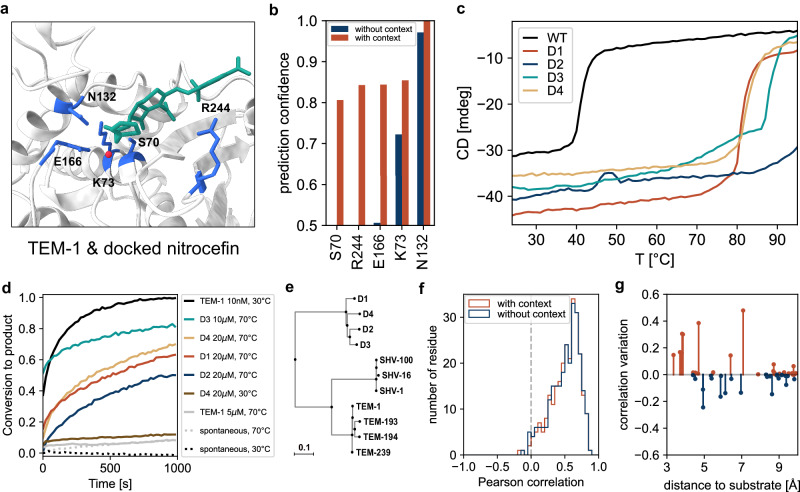
Fig. 4. β-lactamase enzyme engineering and experimental characterization.
a Nitrocefin docked using AutoDock Vina48 at the active site of the serine β-lactamase TEM-1 (PDB ID: 1BT5). Relevant residues for substrate recognition and hydrolysis are shown in blue, nitrocefin in green, and the catalytic water molecule in red. b Prediction confidence with and without the substrate and the catalytic water for the relevant amino acids at the catalytic pocket. (c–d) Experimental characterization of the 4 soluble designs based on the TEM-1 backbone. (c) Thermal denaturation profiles presented as the circular dichroism signal at 222 nm against temperature (see also Supplementary Figs. 10–12 for further structural characterization). d Catalytic activity as fraction of substrate converted to product upon hydrolysis of 200 μM nitrocefin by TEM-1 and the TEM-like lactamase designs, at different temperatures. Proteins were incubated at the indicated concentration (e) Extract of the phylogenetic tree of class A β-lactamases focused on TEM β-lactamases (see Supplementary Fig. 13). f Correlation of the predictions with deep sequencing analysis of TEM-1. g Correlation variation by adding the context (nitrocefin and catalytic water) for the amino acids close (in Cβ distance) to the substrate.
In order to test designed TEM-like enzymes, we sampled CARBonAra’s predictions with docked nitrocefin using imprinting. Imprinting in CARBonAra allows the identification of arbitrary sequence information to any position in the backbone scaffold as prior information for the prediction. By randomly imprinting previously predicted amino acids, this protocol allows the generation of diversity in sampled sequences while using the maximum confidence prediction, ensuring high-quality sequences (see “Methods”). Using this approach, we generated 900 sequences and ranked them using the predicted lDDT provided by AlphaFold (pLDDT) in single-sequence mode (see “Methods”). Single-sequence predictions have been shown to correlate with experimental success, offering a metric for assessing the foldability and functional potential of new designs28.
We tested the top 10 sequences with the highest pLDDT for in vitro validation. While all 10 variants expressed at high yields in E. coli, 4 were also soluble at high concentrations and displayed features of well-folded proteins: far-UV circular dichroism spectra features similar to those of TEM-1 (Supplementary Fig. 10), thermal denaturation profiles indicative of much higher stability (Tm ~80 °C or higher vs. ~ 42 °C for wild-type TEM-1) and of two-state unfolding behavior (Fig. 4c), size exclusion profiles and SEC-MALS-derived molecular weights consistent with monomeric states (Supplementary Fig. 11), and well-spread resonances in NMR spectra (Supplementary Fig. 12). All 4 designed mutants displayed weak to no enzymatic activity against nitrocefin at 30 °C but substantial activity at 70 °C, a temperature at which the natural β-lactamase TEM-1 is totally inactive (Fig. 4d). From the concentrations required to see similar levels of activity for TEM-1 at 30 °C and the designed β-lactamases at 70 °C, we estimate the latter have lower catalytic efficiency at this temperature (estimated kcat/KM ~ 103−104 M−1s−1 vs. 106−107 M−1s−1 for TEM-1 at 30 °C). In this context, it is important to stress that the large excess in thermal stability relative to TEM-1 (40 °C or more) confers the designed mutants ample space for the evolution of their catalytic properties. Comparing the new designs to other class A β-lactamases, they have at most ~55% identity to the TEM-1 β-lactamase and from ~30 to ~50% identity with other proteins of the family such as SHV, KPC, or CTX-M (Supplementary Fig. 13 and Supplementary Table 2). They are more diverse within their own clade than the other families (Fig. 4e), effectively clustering into a new family-level group within class A β-lactamases.
Implications for molecular evolution
Given its features, CARBonAra has promising prospects for the studies of protein evolution, as a means to relate sequence and structure entirely in silico. Specific to the case of TEM β-lactamases tested here experimentally, protein design methods can clearly advance possible routes of mutation by predicting which amino acids are tolerated in the mutational landscape. Notably, the TEM designs picked up substitutions (relative to TEM-1) known to be relevant for substrate profile extension (R164H, A237T, A237S, I127V, H153R, T200S, I173V, M155I, V84I)29,30 and stabilizing mutations like F60Y, S82H, G92D, plus some very strong stabilizing mutations like M182T and R275Q that stand as global suppressors of the destabilization caused by function-enhancing mutations29,31,32.
Along this line, given that tolerance to amino acid substitutions has been widely studied in TEM β-lactamases through full mutational mapping, we took the chance to compare CARBonAra’s residue-wise amino acid probabilities with those measured experimentally through deep sequencing of a library of all single-residue mutants of the TEM-1 β-lactamase33. We observed an average correlation of 0.51 ± 0.21 between the amino acid preferences inferred from CARBonAra and those measured via deep sequencing (Fig. 4f), which is similar to the correlation between the deep sequencing data and the multiple sequence alignment of this enzyme’s family (0.52 ± 0.22). Moreover, we observed that adding the context to the active site of TEM-1 (i.e., docked nitrocefin and the catalytic water) improved the correlation locally (i.e., for amino acids within 5 Å) and also affected the predictions of amino acids further away (up to 10 Å), hinting at the possibility to study the effect of a specific context locally as well as their long-range influences such as allosteric pathways (Fig. 4g). By sampling a large number of sequences, the derived patterns of amino acid variation recover structure-consistent coevolution patterns just as seen in alignments of natural serine β-lactamases (Supplementary Fig. 14). Such kind of predictions could be relevant to inform future biophysics-consistent models of protein evolution34,35.
Discussion
Here, we have presented CARBonAra, a method for sequence prediction from backbone coordinates that is sensitive to the molecular context, allowing the crafting of defined functionalities, accepts imprinting of specific amino acids, requires no atomic parametrization nor surface calculations, and runs design tasks in few seconds. We have also presented in silico examples of CARBonAra’s unique feature, i.e., the capability to design protein sequences that fold as intended under the constraints of a specific interacting molecular environment. We showed experimentally on a workhorse system (i.e., TEM-1 serine β-lactamase) how CARBonAra can approach the challenging case of enzyme engineering, designing actual proteins that fold and remain catalytically active at high temperatures. We furthermore explored focused strategies for sampling protein sequence space from CARBonAra’s output.
Since it is not given that a protein sequence produced from the top-scoring amino acids is functional, sampling strategies are needed to produce proteins that can actually be expressed recombinantly in vitro and are stable and functional. To the best of our knowledge, this has not been tested thoroughly, not even in works reporting successful designs. We have shown here that appropriate sampling strategies can generate rich information not only to produce proteins that work but also to generate synthetic multiple-sequence alignments that reflect the natural variation observed in natural sequences or sampled by mutagenesis and selection experiments. This has implications beyond the niche of protein design itself, particularly opening a window to peek into how proteins evolve within the framework of biophysics-consistent models of protein evolution.
Increasing success rates upon computational design is important on the fundamental side to really achieve mastery in this field and, more practically, to lower costs while attempting actual expression and purification in the lab. Now that AI-based methods are starting to settle, this becomes an important point for discussion7. Different methods and reports present success rates that vary largely, but it is often not clear how each method is evaluated. With a very conservative evaluation, Chroma sets its success rate at around 3%20 while the RoseTTAFold/ProteinMPNN papers report an average success of 15% across several proteins10. From our side, we have one, possibly anecdotal observation hitting at 40% successful proteins with the TEM-1 β-lactamase designs reported in this work. Similarly high success rates hitting 40–55% have been reported for TIM barrels and NTF2 folds, standing out from their previous average 15%10.
Beyond its direct application in designing new proteins and tuning protein functions, CARBonAra seems to be well-suited for improving thermostability, as seen with other protein design methods that also produce robust, highly thermostable proteins. One interesting aspect opened by this observation relates to the intellectual property of designed sequences for stabilizing enzymes for manufacture and industrial processes: often, designed enzymes are protected in a way that covers a small but substantial range of sequence similarity. This has historically been comprehensive enough; however, modern protein design methods, CARBonAra included, can come up with proteins of much lower similarity that preserve function and are highly stable36.
Looking into the future and compared to other methods for protein design, CARBonAra runs with some advantages, mainly related to its inner working based only on element names and coordinates, not requiring any further parametrization or intermediate computation. CARBonAra appears thus more flexible than the alternatives, in that it can intrinsically parse any kind of molecular system and thus can be trained on other kinds of biomolecules (e.g., nucleic acids, small molecules, ions, even water) or molecules not found in biological assemblies, such as materials and surfaces, provided sufficient data are available.
In conclusion, CARBonAra, uniquely based on structural data, stands as a conceptually different approach to protein sequence prediction and design, possessing the additional flexibility required to address the future challenges in molecular design and synthetic biology.
Methods
Datasets
The training dataset is composed of ~370,000 subunits, the validation dataset of ~100,000, all downloaded from RCSB PDB, labeled as the first biological assembly (95% of which are annotated as such by the authors or automatically predicted as such and then confirmed by the authors). The test dataset is composed of ~70,000 subunits (single-chain proteins) with no shared CATH domains with the training set and less than 30% sequence identity with the test set. Within the test dataset, we extracted subunits without any shared CATH domains and maximum 30% sequence identity with any training set of PeSTo ( ~ 370,000 subunits), ProteinMPNN ( ~ 540,000 subunits), or ESM-IF1 ( ~ 18,000 subunits). This comparison dataset is composed of 228 subunits: 76 monomers, 37 dimers, and other 22 multimers. Note that ProteinMPNN and ESM-IF1 both use CATH classification and 40% sequence identity clustering for training and testing.
Features and labels
During the processing phase, we kept only the backbone of proteins (Cα, C, N, O), disregarding the hydrogen atoms, while adding the virtual Cβ using the ideal angle and bond length in the same way as in ProteinMPNN8. The structures we used to train the model can contain any type of molecule, including water, ions, nucleic acids, and any other non-protein molecules. The input scalar state contains the one-hot encoded 30 most frequent atomic elements in the PDB database. The last one-hot channel represents any other or unknown element. The input vector state is initialized randomly drawn from an isotropic normal distribution. We incorporated the geometric features using the pair-wise distance matrices and normalized displacement vector tensor. The output of the model is prediction confidence for each amino acid position among the 20 possible amino acid types (Supplementary Fig. 11). These types are represented as one-hot encoded labels. We optimized the model for multi-class classification of the 20 possible amino acids per position using a binary cross-entropy loss function.
Protein structure transformer architecture
The deep learning architecture of CARBonAra is almost identical to PeSTo21. We first embedded the input features into an input state size (S) of 32 using a three-layer neural network with a hidden layer size of 32. We then applied sequentially four sets of eight geometric transformers (S = 32, Nkey = 3, Nhead = 2), see Supplementary Algorithm 1 and Supplementary Fig. 1. The four sets of eight geometric transformers have a corresponding increasing number of nearest neighbors (nn = 8, 16, 32, 64). In instances where the number of atoms is less than the set number of nearest neighbors (nn), we assigned any additional non-existent interactions to a sink node. We configured this sink node with a constant scalar and vector state of zero. Next, the geometric residue pooling module reduced the atomic-level encoding of the structure into a residue-level description. This aggregation used a local multi-head mask on the atoms that constitute each residue (S = 64, Nhead = 4). Finally, we employed a multi-layer perceptron in the last module, which used three layers of hidden size (S = 64) to decode the state of all residues and computed the prediction, consequently generating a confidence score of the 20 possible amino acids through a sigmoid function ranging from 0 to 1.
Training
We trained our neural network architecture for 16 days on a single NVIDIA V100 (32 GB) GPU. To manage memory usage during training, we limited the subunits to a maximum of 8192 atoms (approximately 100 kDa), excluding hydrogen atoms. Furthermore, subunits containing fewer than 48 amino acids were not considered in the training process. The post-processing effective dataset contains 86610 structures in the training dataset and 24601 structures in the validation dataset.
Sequence sampling
We sampled the optimal sequence by taking the highest confidence amino acid per position from the prediction. To generate sequences with minimum sequence identity to the scaffold, we selected the highest confidence predicted possible amino acid above the positive prediction threshold of 0.5, which is not the original amino acid from the scaffold. The original amino acid is only used in the sequence generated if it is the only possible option within the positive predictions. Our criterion for defining the similarity between two amino acids relies on their BLOSUM37 62 score. We considered them as similar if this score is above zero. We sampled low sequence similarity to the original scaffold by restricting the positively predicted amino acids. When the options were available, we selected the amino acid with the highest BLOSUM 62 score below or equal to zero compared to the reference scaffold. If there are no options with a BLOSUM 62 score below or equal to zero, we sampled the positive predicted amino acid with the lowest BLOSUM 62 score. We noticed that taking only the minimum BLOSUM 62 similarity score generates sequences with a bias towards special amino acids (i.e., cysteine, proline, glycine). We performed a BLAST26,38 analysis to measure the novelty of the generated sequences with minimum identity and low similarity using the non-redundant protein sequences database with an expected value (E-value) cut-off at 100.
Alphafold and alphafold-multimer validation
In the case of the monomers, we sampled the highest confidence sequence from the predictions of CARBonAra for 142 subunits of the testing dataset. We also generated sequences using ProteinMPNN and ESM-IF1, both with a sampling temperature of 1e–6. We modeled the structures from the generated sequences with ColabFold39 (version 1.5.2) using the alphafold2_ptm model, in single-sequence mode and with 3 recycles40. In the case of the dimers, we generated sequences for one subunit, given the sequence of the other subunit. We sampled the sequence with the highest confidence from CARBonAra for the 31 dimers in the testing dataset for a total of 62 complexes with conditioning. We predicted the structures from the generated sequences with ColabFold (version 1.5.2) using the alphafold2_multimer_v2 model, in single-sequence mode and with 5 recycles41. To evaluate the sampling flexibility of CARBonAra, we sampled sequences with maximum identity, minimum identity, and low similarity using CARBonAra’s multi-class predictions. In this case, we used AlphaFold using multiple sequence alignment since a low sequence identity or similarity negates the sequences matching the reference scaffold in the multiple sequence alignment information. We assessed the predicted structures from the generated sequence with the original scaffold using the TM-score42 and Local Distance Difference Test43 (lDDT) on the Cα coordinates.
Molecular dynamics simulations
We selected 20 complexes from the Protein-Protein Docking Benchmark 5.0 dataset44 based on structure resolution and parameterization difficulty. For each complex, we conducted a standard 1 µs-long molecular dynamics (MD) simulation in the NPT ensemble (at 1 atm and 300 K, following a 2 ns NVT equilibration and using settings as per ref. 45) for the bound receptor, unbound receptor, bound ligand, and unbound ligand. We set up all systems using Amber ff14SB46 and its recommended TIP3P water model, running MD simulations with Amber1647. For the 80 (single chain structure) MD, we sampled 500 frames for each simulation and computed the average prediction confidence.
Comparison with deep sequencing
As a case study, we showed that CARBonAra’s residue-wise estimated probabilities (Supplementary Fig. 11) can be reliably correlated with experimentally determined mutations for the class A β-lactamase TEM-1. This widely studied enzyme has been subjected to deep mutagenesis by Deng et al., where the authors analyzed the effect of consecutive triple point mutations along the whole extension of the protein, covering all 20 naturally occurring amino acids per position33. The generated libraries were introduced in E. coli and selected based on ampicillin resistance. These data were used to compute a statistical change in free energy of binding (ΔΔGstat) of mutation of all wild-type residues in the protein. This value was calculated as ΔΔGstat = RT ln(pwt/pmut), where pwt and pmut are the probabilities of finding the wild-type and mutant amino acids, respectively, at the analyzed sequence position. Deng et al. also performed the same calculation on a MSA of 156 sequences of class A β-lactamases, to compare the conservation profile of this family with the requirements imposed by the mutagenesis assays. Aiming at assessing CARBonAra’s ability to recover evolutionary-related residue profiles, we used its residue-wise estimated probabilities to compute the ΔΔGstat per position of TEM-1. We used two structures of TEM-1 as input for the model: TEM-1 in the apo state (PDB ID: 1JTG, removing all non-protein atoms) and TEM-1 retaining its catalytic water and β-lactam nitrocefin at the catalytic pocket. Docking of this ligand to TEM-1 was carried out with AutoDock Vina48 and the analyzed pose was selected based on the proximity of the carbonyl group of the β-lactam ring to the catalytic residue S70. We then calculated Pearson’s correlation coefficient (ρ) of the deep sequencing and CARBonAra’s estimated ΔΔGstat per sequence position.
Nitrocefin docking to TEM-1
This step was necessary because there are no structures currently available of nitrocefin complexed with TEM-1. The docking was carried out with AutoDock Vina48. We obtained the 3D coordinates of nitrocefin from the PubChem database (PubChem CID: 6436140) and used a search space of size 40x40x40 Å centered on the enzyme’s active site (determined by visual inspection). The exhaustiveness parameter was set to 200 and 30 models were generated. The analyzed pose was selected based on the proximity of the carbonyl group of the β-lactam ring to the catalytic residue S70. We also looked for interactions between nitrocefin and residues R244 and N132, known for the stabilization of cephalosporins in TEM-149,50.
Sequence sampling using imprinting
CARBonAra produces matrices (i.e., position-specific scoring matrices, Fig. 1a) that score preferences for each of the 20 amino acids at each position of the designed sequence. The prediction confidence can be converted into a probability (Supplementary Fig. 11). Imprinting in CARBonAra allows the specification of arbitrary sequence information to any position in the backbone scaffold as prior information for the prediction. To efficiently sample the sequence space, we developed an imprinting protocol where first we predict and select the amino acids with the highest confidence score at each sequence position. Afterward, we randomly select between 10 to 90% of these amino acids to imprint on their corresponding backbone positions. We then use the backbone with imprinted information to run a second CARBonAra prediction, from which we select the amino acids with the highest confidence score at each position to build the designed sequence. This strategy promotes sequence diversity while providing high-confidence amino acids per position by scrambling each time the set of amino acids is imprinted after the first prediction. When designing TEM-like enzymes, we used as the input its structure constrained by the presence of the catalytic water and nitrocefin. We predicted a total of 900 sequences within the imprinting range 10-90%, and first filtered for sequences that were able to recover TEM-1’s catalytic triad plus two residues known to accommodate β-lactams in the active site (namely S70, K73, N132, E166, and R244). These sequences were then modeled with AlphaFold in single-sequence mode and ranked based on the highest plDDT. Only the first 10 top-ranked sequences were selected for in vitro validation. The sequences of the 4 that were soluble underwent further experimental validation and are reported in Supplementary Dataset 1.
Material
We purchased HEPES from Chemie Brunschwig AG (Basel, CH), isopropyl β-d-1-thiogalactopyranoside (IPTG) from Huberlab (Aesch, CH) and all other chemicals from Merck (Darmstadt, DE), unless specified.
Protein expression and purification of TEM-like designs
The coding sequences for WT TEM-1 and all the variants were optimized for Escherichia coli (E. coli) expression and cloned into the pET28a(+)-TEV expression vector (Genscript) between NdeI and XhoI, such that the resulting constructs have a N-terminal His-tag followed by a TEV cleavage site. The plasmids were transformed in Rosetta (DE3) cells (Promega). Protein expression was induced by the addition of 1 mM IPTG when the cells reached an optical density of 0.6 and subsequent growth overnight at 20 °C. Cell pellets were resuspended in lysis buffer (20 mM HEPES, pH 7.5, 500 mM NaCl and cOmplete™ Protease Inhibitor Cocktail (Roche)), lysed using sonication, and centrifuged (20000xg for 35 min at 4 °C). The supernatant was applied to a HisTrap HP column (Cytiva) previously equilibrated with lysis buffer. The proteins were eluted with a continuous gradient over 40 column volumes of elution buffer (20 mM HEPES, pH 7.5, 500 mM NaCl, 500 mM Imidazole). Subsequently, pure fractions were additionally purified by Size Exclusion Chromatography (Superdex S200 Increase, Cytiva) in 20 mM HEPES, pH 7.5, 300 mM NaC, 1 mM TCEP. The proteins were flash-frozen in liquid nitrogen and stored at –20 °C.
Size exclusion chromatography coupled to multi-angle light scattering
The molecular weights of the constructs were determined by size exclusion chromatography coupled to multi-angle light scattering (SEC-MALS). The mass measurements were performed on a Dionex UltiMate3000 HPLC system equipped with a 3 angles miniDAWN TREOS static light scattering detector (Wyatt Technology). The sample volumes of 5−10 μl at a concentration of 8 mg/mL, were applied to a Superose 6 Increase 3.2/300 column (Cytiva) previously equilibrated with 20 mM HEPES pH 7.5, 300 mM NaCl at a flow rate of 0.08 mL/min. The data were analyzed using the ASTRA 6.1 software package (Wyatt technology), using the absorbance at 280 nm and the theoretical extinction coefficient for concentration measurements.
Circular dichroism
CD spectra were collected on 10 μM protein solutions in 50 mM Tris pH 7.5, 150 mM NaCl, using a Chirascan CD polarimeter (Applied Photophysics, UK) in 1 mm path cuvettes, at 20 °C. Thermal denaturation curves were acquired by heating the sample from 20 to 98 °C every 1 °C, collecting full spectra and plotting the trace at 222 nm.
NMR spectroscopy
The 1H, 15N HSQC spectrum of TEM design D4 was obtained on a 15N–labeled 250 µM sample prepared in MES pH 6.5 with 200 mM NaCl and 10% 2H2O, at 318 K. It was acquired in a Bruker Avance II 800 MHz (1H frequency) spectrometer equipped with a CPTCIXYZ cryoprobe, using a standard 15N HSQC pulse program with water suppression and sensitivity enhancement, 256 increments in the indirect dimension, and a recycle delay of 1 s.
β-Lactamase activity
TEM−1 and 4 TEM-like designed proteins were incubated at the indicated concentrations (Fig. 2f) with 200 µM nitrocefin in 20 mM HEPES, pH 7.5, 300 mM NaCl at either 30 or 70 °C in a 1 mm path length cuvette while monitoring the absorbance at 485 nm changing over time using a Chirascan CD polarimeter (Applied Photophysics, UK). All temperatures reported involve thermal equilibration of cuvette, buffer, nitrocefin and protein at the indicated temperature for 5 min before mixing.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Source data
Acknowledgements
The Swiss National Science Foundation is acknowledged for supporting this work (grant number 205321_192371 to MDP). We also acknowledge the Swiss National Supercomputing Center (CSCS) for the generous computing time allocation used to run molecular dynamics simulations.
Author contributions
L.F.K., L.A.A., and M.D.P. conceived and designed the research project. L.F.K. designed and implemented the CARBonAra code. F.A.M. designed the TEM-like proteins. L.A.A., M.J.M., J.D., and S.V. characterized in vitro TEM-like designs. L.F.K., F.A.M., L.A.A., M.J.M., and M.D.P. analyzed the data. L.F.K., F.A.M., L.A.A., M.J.M., and M.D.P. wrote the paper.
Peer review
Peer review information
Nature Communications thanks Marco Giulini and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
The data that support this study are available from the corresponding authors upon request. We used publicly available data as described in Methods. The data and code to reproduce the datasets and experiments are available at [https://github.com/LBM-EPFL/CARBonAra]. The molecular dynamics simulations used for the analysis are available at [10.5281/zenodo.12636580]. Previously published structure can be accessed via the accession codes: 6R3C, 1ZNS, 3LVZ, 1BT5 and 1JTG. The source data underlying Figs. 1b-e, 2a-c, 3a-b,d and 4b–d, f, g are provided as a Source Data file. Source data are provided in this paper.
Code availability
The source code is available at [https://github.com/LBM-EPFL/CARBonAra]. An archived version of the code used to produce the results presented in this work is available at [https://github.com/LBM-EPFL/CARBonAra/releases/tag/article].
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-50571-y.
References
- 1.Leader, B., Baca, Q. J. & Golan, D. E. Protein therapeutics: a summary and pharmacological classification. Nat. Rev. Drug Discov.7, 21–39 (2008). 10.1038/nrd2399 [DOI] [PubMed] [Google Scholar]
- 2.Chevalier, A. et al. Massively parallel de novo protein design for targeted therapeutics. Nature550, 74–79 (2017). 10.1038/nature23912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ebrahimi, S. B. & Samanta, D. Engineering protein-based therapeutics through structural and chemical design. Nat. Commun.14, 2411 (2023). 10.1038/s41467-023-38039-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, K. & Arnold, F. H. Engineering new catalytic activities in enzymes. Nat. Catal.3, 203–213 (2020). 10.1038/s41929-019-0385-5 [DOI] [Google Scholar]
- 5.Li, J.-K. et al. Rational enzyme design for enabling biocatalytic Baldwin cyclization and asymmetric synthesis of chiral heterocycles. Nat. Commun.13, 7813 (2022). 10.1038/s41467-022-35468-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu, A., Zhou, J., Blank, L. M. & Jiang, M. Future focuses of enzymatic plastic degradation. Trends Microbiol.31, 668–671 (2023). 10.1016/j.tim.2023.04.002 [DOI] [PubMed] [Google Scholar]
- 7.Khakzad, H. et al. A new age in protein design empowered by deep learning. Cell Syst.14, 925–939 (2023). 10.1016/j.cels.2023.10.006 [DOI] [PubMed] [Google Scholar]
- 8.Dauparas, J. et al. Robust deep learning–based protein sequence design using proteinMPNN. Science378, 49–56 (2022). 10.1126/science.add2187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wicky, B. I. M. et al. Hallucinating symmetric protein assemblies. Science378, 56–61 (2022). 10.1126/science.add1964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Watson, J. L. et al. De novo design of protein structure and function with RFdiffusion. Nature620, 1089–1100 (2023). 10.1038/s41586-023-06415-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hsu, C. et al. Learning inverse folding from millions of predicted structures. In Proc. 39th International Conference on Machine Learning162, 8946–8970 (2022).
- 12.Lin, Z. et al. Evolutionary-scale prediction of atomic-level protein structure with a language model. Science379, 1123–1130 (2023). 10.1126/science.ade2574 [DOI] [PubMed] [Google Scholar]
- 13.Verkuil, R. et al. Language models generalize beyond natural proteins. bioRxiv10.1101/2022.12.21.521521 (2022).
- 14.Ingraham, J., Garg, V., Barzilay, R. & Jaakkola, T. Generative models for graph-based protein design. In Advances in Neural Information Processing Systems 32 (Curran Associates, Inc., 2019).
- 15.Sgarbossa, D., Lupo, U. & Bitbol, A.-F. Generative power of a protein language model trained on multiple sequence alignments. eLife12, e79854 (2023). 10.7554/eLife.79854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou, X. et al. ProRefiner: an entropy-based refining strategy for inverse protein folding with global graph attention. Nat. Commun.14, 7434 (2023). [DOI] [PMC free article] [PubMed]
- 17.Lisanza, S. L. et al. Joint generation of protein sequence and structure with RoseTTAFold sequence space diffusion. bioRxiv10.1101/2023.05.08.539766 (2023).
- 18.Gainza, P. et al. Deciphering interaction fingerprints from protein molecular surfaces using geometric deep learning. Nat. Methods17, 184–192 (2020). 10.1038/s41592-019-0666-6 [DOI] [PubMed] [Google Scholar]
- 19.Gainza, P. et al. De novo design of protein interactions with learned surface fingerprints. Nature617, 176–184 (2023). 10.1038/s41586-023-05993-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ingraham, J. B. et al. Illuminating protein space with a programmable generative model. Nature623,1070–1078 (2023). [DOI] [PMC free article] [PubMed]
- 21.Krapp, L. F., Abriata, L. A., Cortés Rodriguez, F. & Dal Peraro, M. PeSTo: parameter-free geometric deep learning for accurate prediction of protein binding interfaces. Nat. Commun.14, 2175 (2023). 10.1038/s41467-023-37701-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vaswani, A. et al. Attention is all you need. http://arxiv.org/abs/1706.03762 (2017).
- 23.Jing, B., Eismann, S., Suriana, P., Townshend, R. J. L. & Dror, R. Learning from protein structure with geometric vector perceptrons. http://arxiv.org/abs/2009.01411 (2021).
- 24.Bibekar, P., Krapp, L. & Peraro, M. D. PeSTo-Carbs: geometric deep learning for prediction of protein–carbohydrate binding interfaces. J. Chem. Theory Comput20, 2985–2991 (2024). 10.1021/acs.jctc.3c01145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berman, H. M. et al. The protein data bank. Nucleic Acids Res.28, 235–242 (2000). 10.1093/nar/28.1.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol.215, 403–410 (1990). 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 27.Doudeva, L. G. et al. Crystal structural analysis and metal-dependent stability and activity studies of the ColE7 endonuclease domain in complex with DNA/Zn2+ or inhibitor/Ni2+. Protein Sci.15, 269–280 (2006). 10.1110/ps.051903406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bennett, N. R. et al. Improving de novo protein binder design with deep learning. Nat. Commun.14, 2625 (2023). 10.1038/s41467-023-38328-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abriata, L. A., Salverda, M. L. M. & Tomatis, P. E. Sequence-function-stability relationships in proteins from datasets of functionally annotated variants: the case of TEM β-lactamases. FEBS Lett.586, 3330–3335 (2012). 10.1016/j.febslet.2012.07.010 [DOI] [PubMed] [Google Scholar]
- 30.Blázquez, J., Negri, M.-C., Morosini, M.-I., Gómez-Gómez, J. M. & Baquero, F. A237T as a modulating mutation in naturally occurring extended-spectrum tem-type β-lactamases. Antimicrob. Agents Chemother.42, 1042–1044 (1998). 10.1128/AAC.42.5.1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang, W. & Palzkill, T. A natural polymorphism in β-lactamase is a global suppressor. Proc. Natl Acad. Sci. USA94, 8801–8806 (1997). 10.1073/pnas.94.16.8801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brown, N. G., Pennington, J. M., Huang, W., Ayvaz, T. & Palzkill, T. Multiple global suppressors of protein stability defects facilitate the evolution of extended-spectrum TEM β-lactamases. J. Mol. Biol.404, 832–846 (2010). 10.1016/j.jmb.2010.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deng, Z. et al. Deep sequencing of systematic combinatorial libraries reveals β-lactamase sequence constraints at high resolution. J. Mol. Biol.424, 150–167 (2012). 10.1016/j.jmb.2012.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abriata, L. A., Palzkill, T. & Dal Peraro, M. How structural and physicochemical determinants shape sequence constraints in a functional enzyme. PLoS One10, e0118684 (2015). 10.1371/journal.pone.0118684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mayorov, A., Dal Peraro, M. & Abriata, L. A. Active site-induced evolutionary constraints follow fold polarity principles in soluble globular enzymes. Mol. Biol. Evol.36, 1728–1733 (2019). 10.1093/molbev/msz096 [DOI] [PubMed] [Google Scholar]
- 36.Yeh, A. H.-W. et al. De novo design of luciferases using deep learning. Nature614, 774–780 (2023). 10.1038/s41586-023-05696-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Henikoff, S. & Henikoff, J. G. Amino acid substitution matrices from protein blocks. Proc. Natl Acad. Sci. USA89, 10915–10919 (1992). 10.1073/pnas.89.22.10915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Camacho, C. et al. BLAST+: architecture and applications. BMC Bioinforma.10, 421 (2009). 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mirdita, M. et al. Colabfold: making protein folding accessible to all. Nat. Methods19, 679–682 (2022). 10.1038/s41592-022-01488-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jumper, J. et al. Highly accurate protein structure prediction with alphafold. Nature596, 583–589 (2021). 10.1038/s41586-021-03819-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Evans, R. et al. Protein complex prediction with AlphaFold-Multimer. bioRxiv10.1101/2021.10.04.463034 (2021).
- 42.Zhang, Y. & Skolnick, J. Scoring function for automated assessment of protein structure template quality. Proteins: Struct., Funct., Bioinforma.57, 702–710 (2004). 10.1002/prot.20264 [DOI] [PubMed] [Google Scholar]
- 43.Mariani, V., Biasini, M., Barbato, A. & Schwede, T. lDDT: a local superposition-free score for comparing protein structures and models using distance difference tests. Bioinformatics29, 2722–2728 (2013). 10.1093/bioinformatics/btt473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vreven, T. et al. Updates to the integrated protein–protein interaction benchmarks: docking benchmark version 5 and affinity benchmark version 2. J. Mol. Biol.427, 3031–3041 (2015). 10.1016/j.jmb.2015.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abriata, L. A. & Dal Peraro, M. Assessment of transferable forcefields for protein simulations attests improved description of disordered states and secondary structure propensities, and hints at multi-protein systems as the next challenge for optimization. Computational Struct. Biotechnol. J.19, 2626–2636 (2021). 10.1016/j.csbj.2021.04.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maier, J. A. et al. ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput.11, 3696–3713 (2015). 10.1021/acs.jctc.5b00255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Case, D. A. et al. Amber 2016, University of California, San Francisco. 10.13140/RG.2.2.27958.70729 (2016).
- 48.Trott, O. & Olson, A. J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J. Comput Chem.31, 455–461 (2010). 10.1002/jcc.21334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marciano, D. C., Brown, N. G. & Palzkill, T. Analysis of the plasticity of location of the Arg244 positive charge within the active site of the TEM−1 β-lactamase. Protein Sci.18, 2080–2089 (2009). 10.1002/pro.220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cantu, C., Huang, W. & Palzkill, T. Cephalosporin substrate specificity determinants of TEM−1 β-lactamase*. J. Biol. Chem.272, 29144–29150 (1997). 10.1074/jbc.272.46.29144 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support this study are available from the corresponding authors upon request. We used publicly available data as described in Methods. The data and code to reproduce the datasets and experiments are available at [https://github.com/LBM-EPFL/CARBonAra]. The molecular dynamics simulations used for the analysis are available at [10.5281/zenodo.12636580]. Previously published structure can be accessed via the accession codes: 6R3C, 1ZNS, 3LVZ, 1BT5 and 1JTG. The source data underlying Figs. 1b-e, 2a-c, 3a-b,d and 4b–d, f, g are provided as a Source Data file. Source data are provided in this paper.
The source code is available at [https://github.com/LBM-EPFL/CARBonAra]. An archived version of the code used to produce the results presented in this work is available at [https://github.com/LBM-EPFL/CARBonAra/releases/tag/article].