Abstract
Integration of retroviral cDNA involves coupled joining of the two ends of the viral genome at precisely spaced positions in the host cell DNA. Correct coupled joining is essential for viral replication, as shown, for example, by the finding that viral mutants defective in coupled joining are defective in integration and replication. To date, reactions with purified human immunodeficiency virus type 1 (HIV-1) integrase protein in vitro have supported mainly uncoupled joining of single cDNA ends. We have analyzed an activity stimulating coupled joining present in HIV-1 virions, which led to the finding that the HIV-1 nucleocapsid (NC) protein can stimulate coupled joining more than 1,000-fold under some conditions. The requirements for stimulating coupled joining were investigated in assays with mutant NC proteins, revealing that mutations in the zinc finger domains can influence stimulation of integration. These findings (i) provide a means for assembling more authentic integrase complexes for mechanistic studies, (ii) reveal a new activity of NC protein in vitro, (iii) indicate a possible role for NC in vivo, and (iv) provide a possible method for identifying a new class of inhibitors that disrupt coupled joining.
Integration of the reverse-transcribed retroviral cDNA into host cell DNA is a required step in retroviral replication (for reviews, see references 16 and 33). Correct integration requires covalent attachment of viral cDNA to target DNA with a specific number of target base pairs between the cDNA ends. Such coupled joining is probably a consequence of correct assembly of nucleoprotein complexes that hold the cDNA ends in defined relative positions. Studies of retroviruses that generate aberrant integration products indicate that defects in coupled joining are associated with impairments in replication (17, 18, 47). To date, reactions with recombinant human immunodeficiency virus type 1 (HIV-1) integrase protein—the virus-encoded protein responsible for the early steps of integration—have yielded almost exclusively uncoupled products, probably indicating a defect in higher-order assembly in vitro (10, 12, 14, 38).
The DNA cutting and joining reactions that mediate HIV cDNA integration in vivo are well understood (16). Prior to integration, two nucleotides are removed from each 3′ end of the linear cDNA, possibly to remove heterogeneous additional 3′ nucleotides occasionally added by reverse transcriptase (45, 48). The recessed 3′ hydroxyls generated by cleavage then become joined to protruding 5′ ends in the target DNA. Melting of the 5 bp of target DNA between the points of joining yields gaps at each host-virus DNA junction which are then repaired, probably by host-encoded enzymes.
Integration in vivo is carried out by large nucleoprotein complexes named preintegration complexes (PICs) (6, 20, 22). Compositional studies have revealed that the viral integrase (IN), matrix (MA), reverse transcriptase (RT) (7, 23, 27, 45), and a cellular protein, HMG I(Y) (21), cofractionate with PICs of HIV-1. Other proteins, such as viral protein R (Vpr) and nucleocapsid (NC) have been detected in some studies (7, 27). The cDNA ends are bound by proteins (15, 45, 58) that bridge the two cDNA termini (45). Coupled joining by PICs is evident from the finding of correct 5-bp duplications at virus-host DNA junctions after repair of integration intermediates (20, 32, 47a).
Although recombinant HIV-1 integrase carries out coupled joining in vitro only poorly, avian sarcoma-leukosis virus (ASLV) integrase and Moloney murine leukemia virus (MLV) integrase can carry out coupled joining (1, 9, 19, 25, 40, 56, 57). The reason for the difference between HIV integrase and other integrases is unclear but serves to focus interest on the unique determinants in HIV.
Progress in understanding the requirements for coupled joining in the HIV system came from the finding that lysates of virions could direct coupled joining of model cDNA substrates in vitro (28, 29). We have analyzed the virion proteins in such extracts and found that the HIV-1 NC protein can greatly stimulate coupled joining by recombinant HIV-1 integrase. Analysis of the activities of NC mutants revealed that mutations altering the zinc fingers of NC influence the ability to stimulate integration.
MATERIALS AND METHODS
DNA manipulation.
General DNA manipulations were carried out essentially as described previously (3). Oligonucleotides used in this study can be found in Table 1. To construct pTA-LTR, the long terminal repeat (LTR) from NL4-3 was amplified with primers U3NdeI and U5NdeI, giving rise to a complete LTR DNA containing an NdeI site at each end. The PCR product was cloned into the pCR2.1 vector (Invitrogen, Carlsbad, Calif.). Plasmid pTA-LTRsupF, which contains a supF gene within the LTR sequence, was constructed as follows. The supF coding region was obtained from pUCsupF (a gift of Robert Naviaux), in which the supF gene is cloned at bp 763 to 1254 of pUC19. The supF gene was released from plasmid pUCsupF by cleavage with BamHI and ligated into the PpuMI site of pTA-LTR, yielding pTA-LTRsupF. The zeocin-transducing plasmid pZeo+ (3,498 bp) was constructed by digesting pcDNA3.1/Zeo(+) (Invitrogen) with PvuI and EcoRV and reclosing by ligation. This manipulation removed the beta-lactamase (Ap) coding region.
TABLE 1.
DNA oligonucleotides used in this study
| Oligonucleotide | Sequence | Comment |
|---|---|---|
| U3NdeI | 5′ CATATGGAAGGGCTAATTTGGTCCC 3′ | pNL43 primer for preparing LTR substrate |
| U5NdeI | 5′ CATATGCTAGAGATTTTCCACACTG 3′ | pNL43 primer for preparing LTR substrate |
| CH1 | 5′ CAGGGAAGTAGCCTTGTGTGTGGTAG 3′ | Primer for sequencing coupled products |
| HU3A | 5′ CTGGTAACTAGAGATCCCTCA 3′ | Primer for sequencing coupled products |
| T7 20-mer | 5′ TAATACGACTCACTATAGGG 3′ | Sequencing primer (T7 Universal) |
| FB 253 | 5′ CGCCATATGAGTGAGTCGAGCTCGAAGTCC 3′ | Primer for amplifying HMG I(Y) |
| FB 254 | 5′ CTGGATCCTCACAGTTTTTTGGGTCTGCCCCTTGG 3′ | Primer for amplifying HMG I(Y) |
| 4658-333 | 5′ AAGATCTGGACGACGACGACAAGATACAGAAAGGCAATTTTAGGAAC 3′ | Primer |
| 4658-356 | 5′ GTACGTGTCGACCCTAATTAATTAGCCTGTCTCTCAGT 3′ | Antisense primer |
To generate LTR DNAs for use in integration assays, plasmid pTA-LTR or pTA-LTRsupF was cleaved with NdeI and the fragment corresponding to the LTR (1,090 bp) was purified on an agarose gel. LTR DNAs were end labeled by sequential treatment with phosphatase and kinase in the presence of [γ-32P]ATP.
Preparation of virions.
HXB2 virus was obtained from supernatants of the chronic producer line Molt IIIB. R9 and R9ΔIN were obtained from supernatants of cultures of 293T cells transfected with pR9 (53) or pR9ΔIN (26). Virions were purified from cell culture supernatants by centrifugation over a sucrose cushion and size exclusion chromatography on Sephacryl S-1000, as described previously (13).
Integration reactions with viral lysates.
Virions (200 ng of p24 protein per integration reaction) were lysed in a solution of 0.5% Triton X-100, 4 μg of RNase (DNase free [Boehringer Mannheim]) per ml, 20 mM HEPES (pH 7.2), and 150 mM KCl (6-μl final volume) and incubated for 10 min on ice. Subsequently 19 μl of buffer was added to the lysed virus to yield a 25-μl final volume of 10 mM MgCl2, 20 mM HEPES (pH 7.2), 12% dimethyl sulfoxide (DMSO), 6% polyethylene glycol (PEG), 1 mM dithiothreitol (DTT), 40 mM KCl, 0.12% Triton, and 0.96 μg of RNase A per ml. Samples were chilled on ice for another 10 min and then centrifuged for 10 min at 7,000 rpm in an Eppendorf centrifuge at 4°C. To the pellet, a solution of 25 μl of 10 mM MgCl2, 20 mM HEPES (pH 7.2), 12% DMSO, 6% PEG, and 1 mM DTT was added together with 15 ng of LTR and 100 ng of supercoiled plasmid pCR2.1 (3.9 kb; Invitrogen). The samples were incubated for 20 min on ice. The reaction was then carried out by incubating samples for 30 min at 37°C. The reaction was stopped by addition of sodium dodecyl sulfate to 1% and 1 μg of proteinase K, and the samples were incubated at 56°C for 1 h. The proteins were extracted with an equal volume of phenol-chloroform-isoamylalcohol and then chloroform only. The samples were analyzed on 1.2% Tris-acetate-EDTA agarose gels containing ethidium bromide (0.5 μg/ml). Gels were dried and visualized by autoradiography. Proteins from both HIV strains HXB2 and R9 were both found to be active in this assay. Added recombinant integrase (325 nM) was incubated in the viral lysis buffer and centrifuged as above with or without the ΔIN viral lysate.
Purification of proteins used in this study.
Recombinant HIV-1 integrase was purified as described previously (2). His-tagged integrase containing the F185K and C280S substitutions (37) was purified as described previously and used in experiments with NC mutants.
Mutant and wild-type NC proteins were prepared from thioredoxin fusions. The coding regions for the mutant or wild-type NC proteins were obtained by PCR amplification of existing proviral or recombinant NC-encoding clones (30, 31, 59) with the sense primer 4658-333 and the antisense primer 4658-356 (Table 1). Oligonucleotide 4658-333 contains a BglII site and a sequence encoding an enterokinase cleavage site (Asp-Asp-Asp-Asp-Lys), and the remaining 24 nucleotides (3′ end) code for the eight N-terminal amino acid residues of the HIV-1 p7NC protein from the pNL4-3 proviral clone. Oligonucleotide 4658-356 contains a SalI site, the complement of a TAA stop codon, and the complement of the sequence that codes for the six C-terminal amino acid residues of NC. The fragment that codes for the 55-amino-acid NC protein was amplified, digested with BglII and SalI, and ligated into the homologous sites of the pET32a vector (Novagen, Inc., Madison, Wis.). Certain NC mutants were created by altering existing pET32a clones with NC gene inserts and using the Stratagene (La Jolla, Calif.) Quick Change mutagenesis kit.
For NC protein expression, plasmids were transformed into competent Escherichia coli BL21 (DE3) cells and cultures were induced by adding isopropylthio-β-galactoside to 0.4 mM and incubated with shaking for an additional 3 h. Bacteria were pelleted and resuspended in 40 ml of 100 mM CAPS buffer, pH 10, and sonicated for ∼1.5 h. The lysate was centrifuged at 3,600 × g in a Beckman JS-4.2 rotor for 20 min at 4°C. Ten milliliters of supernatant was mixed with 106 μl of 4 M Tris buffer (pH 8.5), 213 μl of 5 M NaCl, 0.7 μl of 30 mM zinc acetate, and 21 μl of 1 M CaCl2. The solution was digested with 7 units of EKMax enterokinase (Invitrogen) for 3 h at room temperature. NC proteins were purified by reverse-phase high-pressure liquid chromatography with a C18 reverse-phase column, as described previously (35, 36). The purified NC proteins were analyzed for the correct molecular mass by MALDI-TOF mass spectrometry. Quantitation of purified NC proteins was performed by amino acid analysis on a Beckman System 6300 amino acid analyzer (Beckman Coulter, Inc., Fullerton, Calif.). Purified proteins were aliquoted, one equivalent of zinc acetate per Zn2+ finger was added (unless otherwise noted), and the samples were lyophilized.
Sources for purified nucleic acid binding proteins were as follows: MA protein was a gift of D. Trono (Geneva, Switzerland); Rev protein was a gift of M. Orsini, D. Rekosh, and M.-L. Hammarskjold (University of Virginia, Charlottesville); Tat protein was a gift of Kathy Jones (Salk Institute, La Jolla, Calif.) and J. Brady; HIV-1 RT heterodimer was a gift of S. LeGrice (Case Western Reserve, Cleveland, Ohio); HMG-1 was a gift of L. Li (Salk Institute); HMG-2 was a gift of R. Johnson (UCLA); histone H1 was purchased from Boehringer Mannheim; HU protein was a gift of A. Segal (SDSU, San Diego, Calif.); BAF protein was a gift of L. Li (Salk Institute); RNase A was from Qiagen; and polylysine was from Sigma Chemical Co. HMG I(Y) amino acids 1 to 90 was purified as described previously for the full-length protein (21).
Integration reactions with recombinant HIV-1 integrase.
A 35 nM concentration of integrase was incubated on ice in a buffer containing 20 mM HEPES (pH 7.2), 1 mM DTT, 12% DMSO, 6% PEG 8000 (buffered at pH 7.2 with HEPES), 0.1 mg of bovine serum albumin (BSA) per ml, and 40 mM NaCl in a volume of 20 μl. Integrase was diluted in a solution of 1 M NaCl, 10 mM DTT, 0.1 mg of BSA per ml, and 50 mM HEPES (pH 7.6). DNA binding proteins to be tested for stimulation were also added at this stage as appropriate. DNA binding proteins were diluted in a solution of 20 mM HEPES (pH 7.2), 150 mM KCl, 1 mM DTT, and 0.1 mg of BSA per ml. After 10 min, 5 μl of a mix containing 40 ng of labeled LTR (20 nM LTR), 200 ng of pZeo+ target, and 50 mM MgCl2 was added and further incubated on ice for 20 min. To carry out reactions, samples were incubated for 1 h at 37°C. Labeled products were quantitated with a Molecular Dynamics PhosphorImager.
Different preparations of HIV-1 integrase and NC were found to support coupled joining to different degrees (unpublished data). For integrase, most active was protein purified according to Allen et al. (2). Next most active was integrase purified by the original method of Sherman and Fyfe (52). Least active, though still sufficient for coupled joining, was His-tagged integrase containing the F185K and C280S mutations (37). Different integrase preparations purified by the same method were not extensively compared. The basis for this difference is unknown. In the case of NC, different batches of NC displayed different activities, and batches with initially high activities sometimes lost activity during storage. This variability probably reflects at least in part the sensitivity of NC protein to oxidation.
Cloning and sequencing of coupled integration products.
In order to clone integration products for sequencing, a standard integration reaction was carried out in the presence of viral lysates (see Table 2) or recombinant integrase and NC (see Table 3) by using LTR-supF as donor and pZeo+ as target. The integration reaction was stopped by the addition of proteinase K-SDS, and DNAs were deproteinized by phenol-chloroform extraction and purified by ethanol precipitation. Integration products were then cleaved with EcoRI, which cleaves within the LTR-supF DNA, and ligated. This step converted coupled joining events involving two LTRs into single-LTR-coupled products. Only events involving one U3 end and one U5 end religated to yield an intact LTR-supF DNA. The ligated DNAs were then transformed into MC1061/p3 cells. The bacteria were plated on Luria-Bertani agar containing zeocin, tetracycline, ampicillin, and kanamycin. Zeocin selected for the integration target; kanamycin selected for the p3 plasmid, containing the ampicillin and tetracycline resistance genes bearing amber mutations; and ampicillin and tetracycline selected for the supF gene. Plasmids were isolated from quadruply resistant colonies, and LTR-target DNA junctions were sequenced. Base numbering in Tables 2 and 3 is as in the original pcDNA3.1+/Zeo plasmid (Invitrogen).
TABLE 2.
Junction sequences from reactions with lysed HXB2 virus
| Product | Duplication sequence | Duplication or deletion | Integration site in pZeo |
|---|---|---|---|
| pI1 | GTGAG | 5-bp duplication | 1991–1995 |
| pI17 | CGCAG | 5-bp duplication | 2224–2228 |
| pEL8 | GTAAT | 5-bp duplication | 2192–2196 |
| pEL9 | GTTTG | 5-bp duplication | 2097–2101 |
| pEL14 | ACTCA | 5-bp duplication | 2181–2185 |
| pEL25 | 55-bp deletion | 2163–2208 | |
| pEL34 | 38-bp duplication | 2141–2279 | |
| pEL38 | 36-bp duplication | 2097–2133 | |
| pEL40 | GGATA | 5-bp duplication | 2218–2222 |
TABLE 3.
Junction sequences from reactions with purified integrase and NC
| Product | Duplication sequence | Duplication or deletion | Integration site in pZeo |
|---|---|---|---|
| pN1 | 50-bp deletion | 1047–1097 | |
| pN2 | CGTTT | 5-bp duplication | 38–42 |
| pN3 | 28-bp deletion | 1889–1917 | |
| pN4 | GGTGG | 5-bp duplication | 215–219 |
| pN5 | 1,300-bp deletion | 878–3064 | |
| pN6 | 250-bp deletion | 3168–3413 | |
| pN7 | 850-bp deletion | 424–3072 | |
| pN8 | CTGTA | 5-bp duplication | 1852–1856 |
| pN11 | 820-bp deletion | 156–973 | |
| pN12 | GTGAG | 5-bp duplication | 1205–1209 |
| pN13 | CAATT | 5-bp duplication | 1932–1936 |
| pN15 | AAGGT | 5-bp duplication | 1202–1206 |
| pN16 | AAGGC | 5-bp duplication | 2258–2262 |
| pN17 | 51-bp deletion | 3034–3085 | |
| pN18 | 37-bp deletion | 1987–2024 |
RESULTS
Integration in vitro directed by extracts from HIV-1 virions.
Our studies of coupled joining by HIV-1 integrase initially employed a viral lysate system based on that used by Goodarzi and coworkers (29) (Fig. 1). As a model of the viral cDNA, a 636-bp DNA molecule matching one HIV LTR was used. LTR DNAs were 32P labeled at each 5′ end to permit detection of integration products. A circular plasmid was used as target. HIV virions were lysed by exposure to 0.5% Triton X-100 and then preincubated with LTR DNA, target DNA, and 10 mM MgCl2 on ice. Reactions were carried out by warming the mixture to 37°C for 30 min. Products were then deproteinized, separated by gel electrophoresis, and visualized by autoradiography.
FIG. 1.
Diagram of the integration reaction and reaction products.
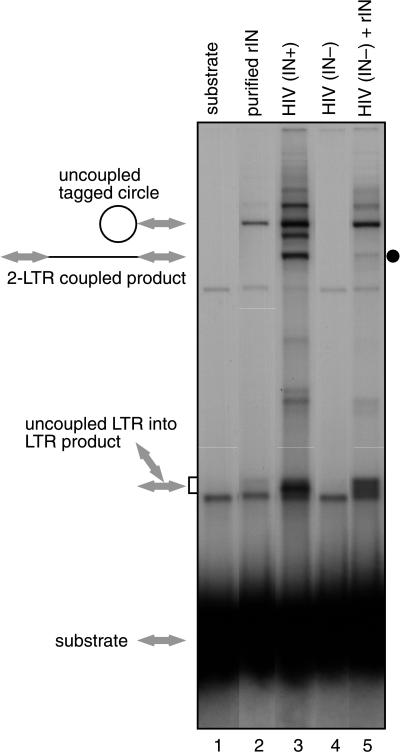
Figure 2 presents a comparison of integration products generated in reactions containing purified recombinant HIV-1 integrase (lane 2) and lysates of HIV-1 virions (lane 3). The structures of integration products are shown beside the gel. In the presence of recombinant integrase, two main DNA products were seen. One corresponded to joining of one end of the labeled LTR substrate to the circular DNA target, yielding a “tagged circle” (illustrated in Fig. 1). The second corresponded to integration of one LTR DNA into a second LTR DNA and, as expected, was seen in reactions without added target (data not shown). The population of these “uncoupled LTR into LTR products” (illustrated in Fig. 1) ran as a broad band, since different molecules in the population had different branch points and hence different electrophoretic mobilities. Both the tagged circle and the uncoupled LTR into LTR products involved only one LTR end and hence are products of uncoupled joining (10, 29).
FIG. 2.
Integration products generated in the presence of lysed HIV-1 virions or purified integrase. Lanes: 1, unreacted substrate; lane 2, 325 nM purified integrase protein; lane 3, lysed HIV-1 (HXB2) virions; lane 4, lysed HIV ΔIN; lane 5, mixture of R9 ΔIN and 325 nM recombinant integrase. Expected structures of integration products are indicated beside the gel; LTR DNAs are indicated by arrows, and target DNA is indicated by thin lines.
In reactions containing lysates from wild-type HIV-1 virions, several further integration products were seen (Fig. 2, lane 3). One new form had the DNA length expected for coupled joining of two LTR molecules to target DNA (“two LTR coupled product,” Fig. 1). This reaction resembled coupled joining in vivo in that one end on each of two LTR molecules was recruited for integration and joined in a coupled fashion, thereby linearizing the target DNA.
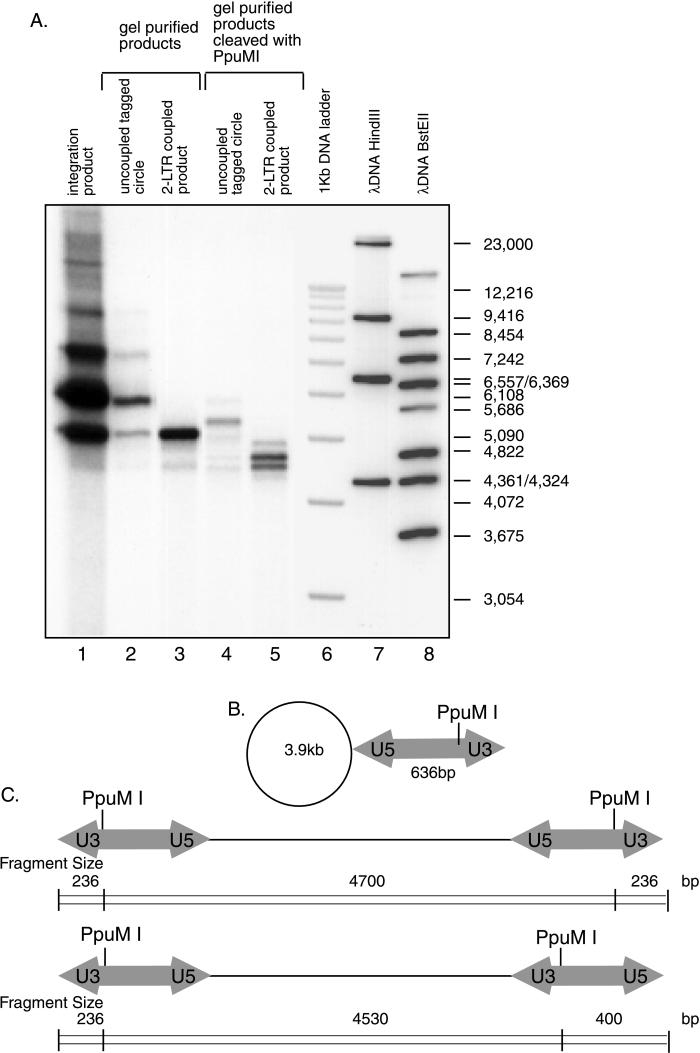
The tagged circle and coupled integration products were characterized by isolation of the product DNA and cleavage with diagnostic restriction enzymes. Cleavage of the purified tagged circle (Fig. 3A, lane 2) or two-LTR product (Fig. 3A, lane 3) with PpuM1, which cleaves in the LTR DNA, yielded fragments of the lengths expected (Fig. 3A, lanes 4 and 5) from the product structure diagrams in Fig. 3B and C. Products involving the U5 LTR end were more abundant, as expected from the previous finding that the U5 end was more reactive than the U3 end (10, 29). Previous product characterization by Goodarzi et al. (29) yielded essentially identical conclusions.
FIG. 3.
Characterization of products of integration reactions with lysed virions by digestion with restriction enzymes. Unintegrated HIV cDNA and solo LTR substrates have U3 sequences at one end and U5 sequences at the other end. (A) Autoradiogram of integration products analyzed by cleavage with restriction enzymes. Lane 1 displays the isolated integration product. Lane 2 presents the partially purified tagged circle form. Lane 3 presents the isolated two-LTR coupled form. Lanes 4 and 5 show these two forms cleaved with PpuMI. Lanes 6 to 8 contain the indicated markers. (B) Expected structure of the predominant tagged circle form. (C) Major coupled two-LTR products. The target DNA used was circular pCR2.1 (3.9 kb); the LTR DNA was 636 bp in length. The major products detected in panel A are as expected from the diagrams in panels B and C. The expected tagged circle form cleaved with PpuM1 runs between the uncleaved form and the relaxed circle form of the target DNA. Cleavage of the two-LTR coupled-joining product yielded two bands that migrated between the 4,822- and 4,361-bp markers; the expected sizes are 4,700 and 4,530 bp. Due to preferential joining of the U5 end, coupled products contained either two U5 ends or one U5 end and one U3 end joined to the target (C).
Products of coupled joining were then characterized by cloning and sequencing (Table 2). The LTR substrate used for this purpose was modified to contain a supF gene between the LTR ends. Integration was carried out into a target containing a zeocin resistance gene. Integration products were recovered after transformation into E. coli by selecting for zeocin resistance and SupF function. Sequencing revealed that six of nine contained the 5-bp duplications of target DNA sequences at integration junctions characteristic of coupled joining (Table 2) (29). Other products contained longer duplications or a deletion, the origins of which were unclear.
As a first step in identifying factors capable of stimulating coupled joining, extracts from integrase-deleted virions were prepared and analyzed. Extracts from HIV-1 ΔIN virions failed to support integration, as expected (Fig. 2, lane 4). However, mixture of extracts from the HIV-1 ΔIN virus with recombinant integrase resulted in the appearance of a new band of the mobility expected for the coupled two-LTR product (Fig. 2, lane 5 [dot]). These data support the hypothesis that extracts of virions contain factors in addition to integrase that stimulate coupled joining.
Stimulation of coupled joining by HIV-1 NC and other proteins.
To identify HIV-1 virion factors capable of stimulating coupled joining, lysed virions were fractionated by differential centrifugation. Analysis by Western blotting revealed that integrase, MA, RT, and NC cofractionated with the integration activity while CA did not (data not shown). The viruses studied were mutant in Vpr, consistent with the idea that Vpr protein is also not important. These fractions proved difficult to purify further but did provide some initial indications of candidate stimulatory proteins.
Purified viral proteins were then tested individually for the ability to stimulate coupled joining by purified integrase (Fig. 4A). Of particular interest was NC protein, which in previous studies has been reported to stimulate uncoupled joining (14, 41) or integration by PICs (20). HIV-1 NC protein (Fig. 4A, lanes 3 to 5), MA protein (lanes 8 to 10), Rev (lanes 11 to 13), Tat (lanes 14 to 16), and RT (p66/p51 heterodimeric form, lanes 17 to 19) were each titrated into reaction mixtures with recombinant HIV-1 integrase. Some stimulation of both coupled and uncoupled joining was seen upon addition MA, Tat, or RT p51 homodimer (data not shown), but the stimulatory effect was greatest by far with NC. NC increased the recovery of coupled products up to 1,000-fold in the presence of low concentrations (35 nM) of purified integrase. Under the conditions in Fig. 4A, lane 5, 26% of the input labeled LTR was converted to the coupled joining product and another 48% was converted to the uncoupled product, corresponding to a 1,200-fold increase in coupled joining and a 230-fold increase in uncoupled joining.
FIG. 4.
Stimulation of coupled joining by various nucleic acid binding proteins. (A) Products generated in the presence of 35 nM purified integrase and the indicated viral proteins. Expected structures of integration products are shown beside the gel. Maximum concentrations for each (right-most lane in each titration) are as follows: NC, 8 μg/ml (1 μM); MA, 16 μg/ml (0.94 μM); Rev, 4 μg/ml (0.2 μM); Tat, 4 μg/ml (0.28 μM); RT, 4 μg/ml (78 nM). Each protein was diluted 1:10 and 1:100 in the two left lanes. (B) Products generated in the presence of 35 nM purified integrase and the indicated cellular DNA binding proteins. Maximum concentrations for each (right-most lane in each titration) are as follows: NC, 8 μg/ml (1 μM); HMG I(Y), 16 μg/ml (1.4 μM); HMG-1, 4 μg/ml (0.16 μM); HMG-2, 8 μg/ml (0.64 μM); histone H1, 8 μg/ml (0.4 μM); Hu, 2.4 μg/ml (0.13 μM); BAF, 2 μg/ml (0.2 μM); RNase A, 4 μg/ml (0.3 μM); polylysine, 4 μg/ml (4 μM). Each protein was diluted 1:10 and 1:100 in the two left lanes.
To assess the specificity of the NC effect, several cellular nucleic acid-binding proteins were also tested for the ability to stimulate coupled joining (Fig. 4B). HMG I(Y) DNA binding domain, HMG-1, and BAF proteins were tested, since they have previously been proposed to be involved in integration (1, 21, 42), together with several other DNA binding proteins for comparison. Slight stimulation was seen with HMG-1 (lanes 8 to 10), HMG-2 (lanes 11 to 13), and polylysine (lanes 26 to 28). No detectable stimulation was seen with HMG I(Y) (lanes 5 to 7), histone H1 (lanes 14 to 16), bacterial HU (lanes 17 to 19), BAF (lanes 20 to 22), or RNase A (lanes 23 to 25). The protein concentrations used were selected based on preliminary titrations to optimize activity. In the presence of high concentrations, BAF and HMG I(Y) inhibited integration (data not shown). None of the cellular proteins stimulated coupled joining as well as did NC protein (Fig. 4B, lane 4).
Optimizing conditions for coupled joining.
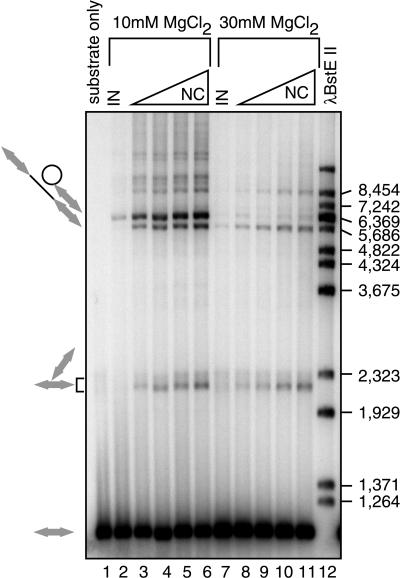
Titrations of reaction components revealed that high concentrations of Mg2+ favored production of coupled joining products over uncoupled products (Fig. 5). Little or no coupled joining is detectable at 10 mM MgCl2 in the absence of NC (Fig. 5, lane 2), while in the presence of NC both the coupled and uncoupled products are greatly increased in abundance (Fig. 5, lanes 3 to 6). At 30 mM MgCl2, the coupled product predominates even in the absence of NC (Fig. 5, lane 7) and is increased in abundance in the presence of NC (Fig. 5, lanes 8 to 11). MnCl2, the divalent metal used in many previous studies, did not support coupled joining under the conditions studied here (unpublished data). Addition of zinc (8, 60) also did not stimulate coupled joining (unpublished data). Six percent PEG was required for coupled joining (unpublished data). In contrast, under conditions reported previously for stimulation of uncoupled joining by NC (i.e., in the presence of 10% glycerol) (14), high concentrations of MgCl2 were inhibitory. These findings highlight the fastidious nature of the requirements for coupled joining.
FIG. 5.
Improvement of the yield of coupled-joining products by optimizing the concentrations of MgCl2 and NC. Concentrations of NC were 100 nM (lanes 3 and 8), 500 nM (lanes 4 and 9), 1 μM (lanes 5 and 10), and 2 μM (lanes 6 and 11). The integrase concentration was 35 nM. For this experiment, the LTR-supF donor was used. Other markings are as in Fig. 2.
The conditions of the coupled-joining reaction strongly influence the magnitude of stimulation by NC. In the presence of 30 mM MgCl2, the degree of stimulation is reduced because the basal level of coupled joining is higher and added NC does not stimulate coupled joining to as high a final level. The greatest stimulation by NC is seen in the presence of 10 mM MgCl2 and minimal concentrations of integrase.
Characterization of products made in the presence of purified integrase and NC.
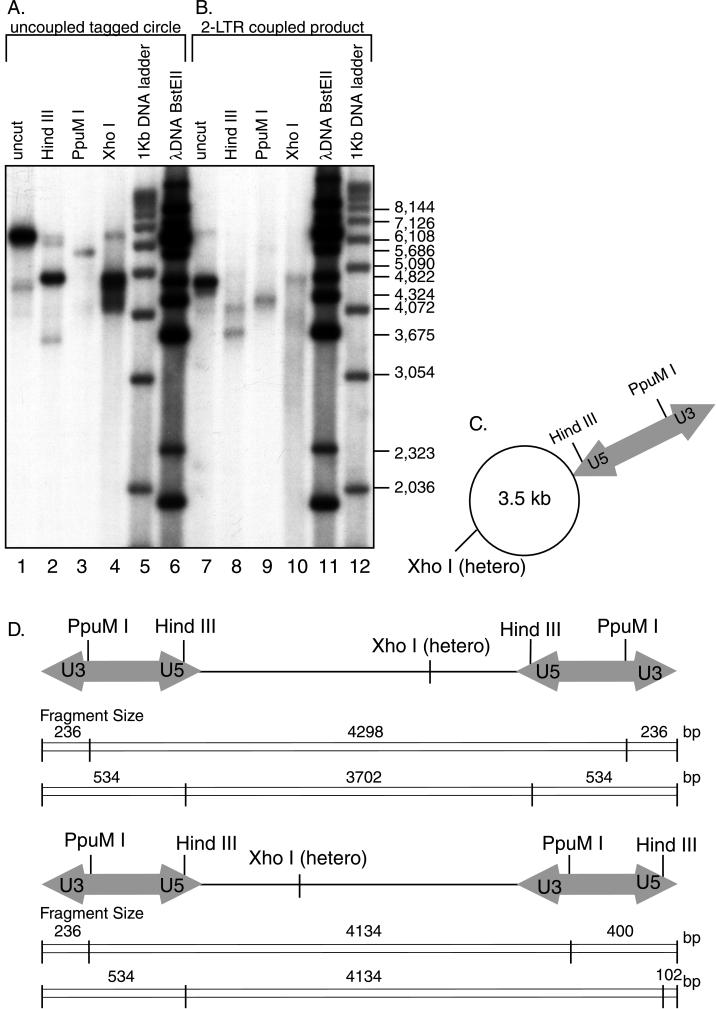
The products generated in reactions with purified recombinant integrase comigrated with those generated in reactions with viral lysates, suggesting that they were identical in structure. Nevertheless, DNA products made with purified integrase were characterized by digestion with restriction enzymes and DNA sequencing. Diagnostic digestions were carried out with PpuM1, as in Fig. 3, and also with HindIII and XhoI (Fig. 6). Cleavage tests were carried out for both the uncoupled tagged circle (Fig. 6A) and the two-LTR coupled product (Fig. 6B). Structures of digestion products were fully consistent with assignment of the DNA forms as tagged circle and two-LTR coupled product (Fig. 6C and D). In this case also, joining of the U5 end was favored over the U3 end. Similar analysis confirmed the structure of the coupled product generated with 30 mM MgCl2 in the absence of NC (data not shown).
FIG. 6.
Characterization of integration products generated in reactions with recombinant integrase and NC. The suspected tagged circle product (A) and coupled two-LTR product (B) were isolated from gels and characterized by digestion with restriction enzymes (diagrammed in panels C and D). The target DNA used was circular pZeo+. Cleavage of the putative tagged circle with HindIII (panel A, lane 2), which cleaves the LTR near the U5 terminus, yields predominantly a shortened labeled form that migrates near the target DNA relaxed circle form (C). Cleavage with PpuMI yielded a longer form, of a size consistent with cleavage near the U3 end (panel A, lane 3). Cleavage with XhoI, which cuts within the target, yields a smear of products (panel A, lane 4), as expected for Y-shaped cleavage products with heterogeneous branch points. The putative two-LTR coupled-joining product (panel B, lane 7) migrates at the molecular weight expected for a linearized target DNA joined in a coupled fashion to two LTRs (4,770 bp [D]). Cleavage with HindIII (panel B, lane 8) yielded a shortened labeled form of the size expected for the joining of two U5 ends (3,702 bp, diagrammed in panel D [top]). A longer form, of the size expected for joining of one U5 end and one U3 end, was also seen (4,134 bp, diagrammed in panel D [bottom]). Cleavage with PpuMI yielded predominantly a form of the size expected for the U5-U5 coupled two-LTR product (4,298 bp [panel B, lane 9]). The second expected PpuMI product, representing the U3-U5 form (expected size, 4,134 bp), migrated just below the U5-U5 form and is not clearly resolved in this gel. Cleavage with XhoI in the target DNA yielded a smear of labeled products, as expected since the points of integration in the target differ from molecule to molecule. The designation “hetero” indicates that this site is heterogeneously positioned in different product molecules due to the use of different integration sites.
Further characterization of coupled-joining products was carried out by cloning and sequencing of junctions between viral and target DNA (Table 3). Integration reactions were carried out with LTR-supF and a pZeo DNA as target. DNAs were transformed into bacteria, and resistance to zeocin and supF function was selected; 517 colonies were recovered from a typical reaction with recombinant integrase and NC. In contrast, reactions without NC or without integrase yielded no colonies (data not shown).
Seven of 15 insertion products sequenced displayed 5-bp duplications of target sequences at host-viral DNA junctions characteristic of HIV-1 integration. The other eight products contained deletions of unclear origin. The larger deletions would not be expected to comigrate with the coupled-joining product seen on gels and presumably arose from other DNA forms. Taken together, these data indicate that coupled-joining products are generated efficiently in reactions with recombinant integrase and NC.
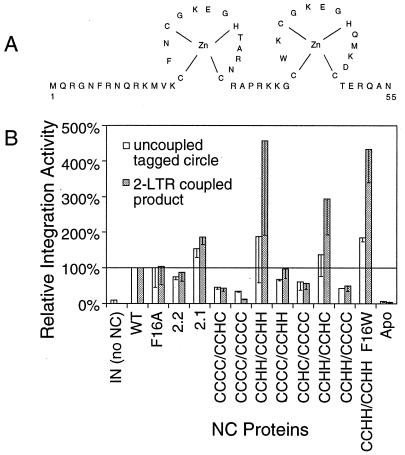
Stimulation of coupled joining by NC mutants.
To investigate the determinants in NC important for stimulating coupled joining, a panel of NC variants was tested (Fig. 7). NC is a basic protein of 55 amino acids, with two Cys-X2-Cys-X4-His-X4-Cys (CCHC) sequences that bind zinc (4, 34). Mutants tested included amino acid substitutions (F16A or F16W) or changes in zinc fingers. The F16A mutant reduced the efficiency of the acceleration of nucleic acid annealing by NC (24), thus allowing the relationship between accelerating of annealing and stimulating of integration to be probed. Mutants in which the types of zinc-binding residues were swapped (Cys to His or vice versa) were also studied. These mutants were chosen in an effort to preserve zinc binding while introducing subtle changes in NC. These NC mutants have also been studied for their effects on viral replication, offering a chance to begin connecting in vitro observations to replication in vivo (31). Zinc finger mutants included ones in which the first zinc finger sequence was replaced with the second (2.2) or in which the order of zinc fingers (2.1) was reversed (30). Several further mutants contained substitutions of the C or H residues, including CCCC/CCHC, CCCC/CCCC, CCHH/CCHH, CCCC/CCHH, CCHC/CCCC, CCHH/CCHC, and CCHH/CCHH F16W (C and H indicate the types of zinc-binding residues in the first or second finger). Wild-type NC lacking zinc was also tested (apo-NC) as was the NC of MLV.
FIG. 7.
Stimulation of integration in vitro by NC mutants. Mutants were tested at a concentration of 1.4 μM and integrase was present at 137.5 nM (His-tagged form), in otherwise standard reactions. The abundances of the two-LTR coupled-joining products (shaded bars) and the tagged circle products (open bars) were quantitated by PhosphorImager and normalized to wild-type NC. Each bar denotes the average of three reactions; half of the standard deviation is shown by the error bar.
Several NC proteins, including CCCC/CCCC, CCCC/CCHC, CCHC/CCCC, CCHH/CCCC, apo-NC, and MLV NC, showed reduced abilities to stimulate coupled joining. Similar though less-pronounced effects were seen for uncoupled joining. Two other mutants, CCHH/CCHH and CCHH/CCHH F16W, consistently stimulated coupled integration to a greater degree than wild-type NC. The other mutants stimulated coupled and uncoupled joining at levels close to wild-type NC.
The NC mutants were then tested for the ability to accelerate annealing of complementary DNA strands, a well-characterized activity of NC protein (data not shown). Annealing assays were carried out in integration buffer to facilitate comparison to integration assays. Wild-type NC and all mutants, with the exception of CCCC/CCCC (inactive) and CCCC/CCHH and CCHH/CCCC (partially active), displayed this activity. The apo-NC protein was active in accelerating annealing though inactive for stimulating coupled joining. Thus, the requirements in vitro for stimulating integration could be separated from the requirements for accelerating annealing. Taken together, the data support the view that intact NC zinc fingers are important for stimulating coupled joining in vitro.
DISCUSSION
Here we present conditions under which purified recombinant HIV-1 integrase carries out correct coupled joining of pairs of cDNA ends in vitro. The previous finding of Goodarzi and coworkers (29) that HIV-1 viral lysates could support coupled joining raised the question of whether recombinant HIV-1 integrase was defective as purified from bacteria. Data presented here indicate that purified recombinant integrase can carry out coupled joining relatively efficiently in the presence of a viral cofactor (NC) and optimized solution conditions.
Studies of HIV-1 viruses containing mutations in NC have the potential to clarify whether or not NC is important for integration in vivo. However, many mutations in NC disrupted steps in replication prior to integration, such as RNA packaging or reverse transcription (31, 54). Thus, it was not possible to determine whether these mutants also impaired integration. The CCHH-containing mutants that hyperstimulated integration, for example, produced little or no viral cDNA after infection, so their effects on integration could not be assessed (31).
The CCCC/CCHC NC mutant, however, may affect integration in vivo at least in part. The mutant protein displayed reduced stimulation of coupled integration in vitro. In vivo, viruses containing this substitution can form cDNA to within 10% of the wild type, but infectivity is reduced by greater than 104-fold (31). Thus, the CCCC/CCHC mutant may be blocking integration in those viruses that successfully carry out reverse transcription. Further studies are needed to support the possible effect of this NC mutant in integration in vivo.
Several proteins in addition to NC have been reported to affect integration in vitro, raising the question of which are most likely to be important biologically. Diverse proteins have been reported to stimulate uncoupled joining, among them NC protein (14, 38, 41, 44). In addition, HMG-1 protein was reported to stimulate coupled joining by ASLV integrase three- to fivefold in vitro, providing another candidate (1). Studies of PICs have also revealed candidates for stimulatory proteins. In a study of HIV-1, PICs were depleted of required factors by exposure to high concentrations of salt. Cellular fractions were used to reconstitute activity, and purification of the activity yielded HMG I(Y) protein. Further studies demonstrated that HMG I(Y) protein is present in PIC fractions prior to depletion and contributes the predominant reconstitution activity in PIC preparations (21, 43). Another protein, BAF, can stimulate integration in these assays (15, 42), though BAF has not yet been found in PICs or linked to integration in vivo. Purified NC protein was also able to stimulate the activity of salt-stripped PICs but was not detectable in the PIC fractions studied (21). However, in one study, NC was detected in association with HIV cDNA in crude PIC fractions (27). NC is likely associated with replication intermediates in target cells, since NC is believed to be important for reverse transcription during infection. Although more data is needed to assess in vivo significance, the presence of NC and HMG I(Y) in probable replication intermediates, together with their in vitro activities, supports their potential importance in vivo.
As yet, the mechanism by which NC stimulates coupled joining is not fully clarified. Models involving a direct interaction between integrase and NC do not seem likely at present, since coimmunoprecipitation studies have not revealed an interaction (unpublished data). NC is known to promote dimerization of viral RNA, packaging of viral RNA, and strand transfer during reverse transcription, all reactions involving nucleic acid condensation (16, 41, 50, 55). In the case of integration, condensation of duplex DNA by NC might facilitate association of the two cDNA ends in an integration complex. However, effects on DNA condensation alone may not be sufficient to explain the stimulation of coupled integration. Several proteins known to condense DNA [Apo-NC, MLV NC, BAF, and HMG I(Y)] did not stimulate coupled joining, suggesting that additional mechanisms may operate. Possibly NC stimulates integration in part by promoting DNA distortion. NC is known to melt DNA (55), and DNA distortion can promote integration under some circumstances (5, 11, 39, 46, 49, 51). Whether any of these candidate mechanisms operate in vivo remains to be determined.
The establishment of coupled joining in vitro by HIV-1 integrase may potentiate a variety of further studies. Mutants of integrase can be analyzed in vitro for their effects on coupled joining, aiding in understanding of the protein-protein interfaces important for higher-order assembly. Integration intermediates formed by coupled joining in vitro may also be useful as substrates for studying the DNA repair reactions involved in completing integration. Inhibitory molecules can now be screened for the ability to block coupled joining, potentially allowing the development of a new class of integrase inhibitors.
ACKNOWLEDGMENTS
We thank members of the Salk Institute Infectious Disease Laboratory for suggestions, materials, and comments on the manuscript; Allison Bocksruker for artwork; Leslie Orgel for comments on the manuscript; and Didier Trono for the HIV ΔIN virus. From the AIDS Vaccine Program, we thank Bradley P. Kane and Donald G. Johnson for assistance in preparing recombinant NC proteins.
This work was supported by grants AI34786 and GM56553 to F.D.B. R.J.G. was supported by the National Cancer Institute, U.S. Department of Health and Human Services, under contract NO1-CO-56000 with SAIC Frederick. S.C. was supported in part by a fellowship from the Rau Foundation. F.D.B. is a scholar of the Leukemia Society of America.
ADDENDUM IN PROOF
C. Deminie has also observed stimulation of coupled joining by NC protein (unpublished data). In addition, Hindmarsh et al. (P. Hindmarsh, T. Ridky, R. Reeves, M. Andrake, A. M. Skalka, and J. Leis, J. Virol. 73:2994–3003, 1999) have also reported conditions for coupled joining by HIV IN.
REFERENCES
- 1.Aiyar A, Hindmarsh P, Skalka A M, Leis J. Concerted integration of linear retroviral DNA by the avian sarcoma virus integrase in vitro: dependence on both long terminal repeat termini. J Virol. 1996;70:3571–3580. doi: 10.1128/jvi.70.6.3571-3580.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen P, Worland S, Gold L. Isolation of high-affinity RNA ligands to HIV-1 integrase from a random pool. Virology. 1995;209:327–336. doi: 10.1006/viro.1995.1264. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons, Inc.; 1987. [Google Scholar]
- 4.Bess J W, Powell P J, Issaq H J, Schumack L J, Grimes M K, Henderson L E, Arthur L O. Tightly bound zinc in human immunodeficiency virus type 1, human T-cell leukemia virus type I, and other retroviruses. J Virol. 1992;66:840–847. doi: 10.1128/jvi.66.2.840-847.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bor Y-C, Bushman F, Orgel L. In vitro integration of human immunodeficiency virus type 1 cDNA into targets containing protein-induced bends. Proc Natl Acad Sci USA. 1995;92:10334–10338. doi: 10.1073/pnas.92.22.10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown P O, Bowerman B, Varmus H E, Bishop J M. Correct integration of retroviral DNA in vitro. Cell. 1987;49:347–356. doi: 10.1016/0092-8674(87)90287-x. [DOI] [PubMed] [Google Scholar]
- 7.Bukrinsky M I, Sharova N, McDonald T L, Pushkarskaya T, Tarpley G W, Stevenson M. Association of integrase, matrix, and reverse transcriptase antigens of human immunodeficiency virus type 1 with viral nucleic acids following acute infection. Proc Natl Acad Sci USA. 1993;90:6125–6129. doi: 10.1073/pnas.90.13.6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burke C J, Sanyal G, Bruner M W, Ryan J A, LaFemina R L, Robbins H L, Zeft A S, Middaugh C R, Cordingley M G. Structural implications of spectroscopic characterization of a putative zinc finger peptide from HIV-1 integrase. J Biol Chem. 1992;267:9639–9644. [PubMed] [Google Scholar]
- 9.Bushman F D, Craigie R. Sequence requirements for integration of Moloney murine leukemia virus DNA in vitro. J Virol. 1990;64:5645–5648. doi: 10.1128/jvi.64.11.5645-5648.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bushman F D, Craigie R. Activities of human immunodeficiency virus (HIV) integration protein in vitro: specific cleavage and integration of HIV DNA. Proc Natl Acad Sci USA. 1991;88:1339–1343. doi: 10.1073/pnas.88.4.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bushman F D, Craigie R. Integration of human immunodeficiency virus DNA: adduct interference analysis of required DNA sites. Proc Natl Acad Sci USA. 1992;89:3458–3462. doi: 10.1073/pnas.89.8.3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bushman F D, Fujiwara T, Craigie R. Retroviral DNA integration directed by HIV integration protein in vitro. Science. 1990;249:1555–1558. doi: 10.1126/science.2171144. [DOI] [PubMed] [Google Scholar]
- 13.Camaur D, Trono D. Characterization of human immunodeficiency virus type 1 Vif particle incorporation. J Virol. 1996;70:6106–6111. doi: 10.1128/jvi.70.9.6106-6111.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carteau S, Batson S C, Poljak L, Mouscadet J-F, Rocquigny H, Darlix J-L, Roques B P, Kas E, Auclair C. Human immunodeficiency virus type 1 nucleocapsid protein specifically stimulates Mg2+-dependent DNA integration in vitro. J Virol. 1997;71:6225–6229. doi: 10.1128/jvi.71.8.6225-6229.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen H, Engelman A. The barrier-to-autointegration protein is a host factor for HIV type 1 integration. Proc Natl Acad Sci USA. 1998;95:15270–15274. doi: 10.1073/pnas.95.26.15270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coffin J M, Hughes S H, Varmus H E. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- 17.Colicelli J, Goff S P. Mutants and pseudorevertants of Moloney murine leukemia virus with alterations at the integration site. Cell. 1985;42:573–580. doi: 10.1016/0092-8674(85)90114-x. [DOI] [PubMed] [Google Scholar]
- 18.Colicelli J, Goff S P. Sequence and spacing requirements of a retrovirus integration site. J Mol Biol. 1988;199:47–59. doi: 10.1016/0022-2836(88)90378-6. [DOI] [PubMed] [Google Scholar]
- 19.Craigie R, Fujiwara T, Bushman F. The IN protein of Moloney murine leukemia virus processes the viral DNA ends and accomplishes their integration in vitro. Cell. 1990;62:829–837. doi: 10.1016/0092-8674(90)90126-y. [DOI] [PubMed] [Google Scholar]
- 20.Ellison V H, Abrams H, Roe T, Lifson J, Brown P O. Human immunodeficiency virus integration in a cell-free system. J Virol. 1990;64:2711–2715. doi: 10.1128/jvi.64.6.2711-2715.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farnet C, Bushman F D. HIV-1 cDNA integration: requirement of HMG I(Y) protein for function of preintegration complexes in vitro. Cell. 1997;88:1–20. doi: 10.1016/s0092-8674(00)81888-7. [DOI] [PubMed] [Google Scholar]
- 22.Farnet C M, Haseltine W A. Integration of human immunodeficiency virus type 1 DNA in vitro. Proc Natl Acad Sci USA. 1990;87:4164–4168. doi: 10.1073/pnas.87.11.4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Farnet C M, Haseltine W A. Determination of viral proteins present in the human immunodeficiency virus type 1 preintegration complex. J Virol. 1991;65:1910–1915. doi: 10.1128/jvi.65.4.1910-1915.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feng Y X, Copeland T D, Henderson L E, Gorelick R J, Bosche W J, Levin J G, Rein A. HIV-1 nucleocapsid protein induces “maturation” of dimeric retroviral RNA in vitro. Proc Natl Acad Sci USA. 1996;93:7577–7581. doi: 10.1073/pnas.93.15.7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fitzgerald M L, Vora A C, Zeh W G, Grandgenett D P. Concerted integration of viral DNA termini by purified avian myeloblastosis virus integrase. J Virol. 1992;66:6257–6263. doi: 10.1128/jvi.66.11.6257-6263.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gallay P, Hope T, Chin D, Trono D. HIV-1 infection of nondividing cells through the recognition of integrase by the importin/karyopherin pathway. Proc Natl Acad Sci USA. 1997;94:9825–9830. doi: 10.1073/pnas.94.18.9825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gallay P, Swingler S, Song J, Bushman F, Trono D. HIV nuclear import is governed by the phosphotyrosine-mediated binding of matrix to the core domain of integrase. Cell. 1995;17:569–576. doi: 10.1016/0092-8674(95)90097-7. [DOI] [PubMed] [Google Scholar]
- 28.Goodarzi G, Chiu R, Brackmann K, Kohn K, Pommier Y, Grandgenett D P. Host site selection for concerted integration by human immunodeficiency virus type-1 virions in vitro. Virology. 1997;231:210–217. doi: 10.1006/viro.1997.8558. [DOI] [PubMed] [Google Scholar]
- 29.Goodarzi G, Im G-J, Brackmann K, Grandgenett D. Concerted integration of retrovirus-like DNA by human immunodeficiency virus type 1 integrase. J Virol. 1995;69:6090–6097. doi: 10.1128/jvi.69.10.6090-6097.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gorelick R J, Chabot D J, Rein A, Henderson L E, Arthur L O. The two zinc fingers in the human immunodeficiency virus type 1 nucleocapsid protein are not functionally equivalent. J Virol. 1993;67:4027–4036. doi: 10.1128/jvi.67.7.4027-4036.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gorelick, R. J., T. D. Gagliardi, W. J. Bosche, T. A. Wiltrout, L. V. Coren, D. J. Chabot, J. D. Lifson, L. E. Henderson, and L. O. Arthur. Strict conservation of the retroviral nucleocapsid (NC) protein zinc-finger is strongly influenced by its role in viral infection processes: characterization of HIV-1 particles containing mutant NC zinc-coordinating sequences. Virology 256:92–104. [DOI] [PubMed]
- 32.Hansen M, Bushman F D. HIV-2 preintegration complexes: activities in vitro and response to inhibitors. J Virol. 1997;71:3351–3356. doi: 10.1128/jvi.71.4.3351-3356.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hansen M S T, Carteau S, Hoffmann C, Li L, Bushman F. Retroviral cDNA integration: mechanism, applications and inhibition. In: Setlow J K, editor. Genetic engineering: principles and methods. New York, N.Y: Plenum Press; 1998. pp. 41–62. [DOI] [PubMed] [Google Scholar]
- 34.Hendersen L E, Bowers M A, Sowder R C, Serabyn S A, Johnson D G, Bess J W, Arthur L O, Bryant D K, Fenselau C. Gag proteins of the highly replicative MN strain of human immunodeficiency virus type 1: posttranslational modifications, proteolytic processings, and complete amino acid sequences. J Virol. 1992;66:1856–1865. doi: 10.1128/jvi.66.4.1856-1865.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Henderson L E, Benveniste R E, Sowder R, Copeland T D, Schultz A M, Oroszlan S. Molecular characterization of gag proteins from simian immunodeficiency virus (SIVMne) J Virol. 1988;62:2587–2595. doi: 10.1128/jvi.62.8.2587-2595.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Henderson L E, Sowder R, Copeland T D, Smythers G, Oroszlan S. Quantitative separation of murine leukemia virus proteins by reversed-phase high-pressure liquid chromatography reveals newly described gag and env cleavage products. J Virol. 1984;52:492–500. doi: 10.1128/jvi.52.2.492-500.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jenkins T M, Engelman A, Ghirlando R, Craigie R. A soluble active mutant of HIV-1 integrase: involvement of both the core and carboxyl-terminal domains in multimerization. J Biol Chem. 1996;271:7712–7718. doi: 10.1074/jbc.271.13.7712. [DOI] [PubMed] [Google Scholar]
- 38.Kalpana G V, Marmon S, Wang W, Crabtree G R, Goff S P. Binding and stimulation of HIV-1 integrase by a human homolog of yeast transcription factor SNF5. Science. 1994;266:2002–2006. doi: 10.1126/science.7801128. [DOI] [PubMed] [Google Scholar]
- 39.Katz R A, Gravuer K, Skalka A M. A preferred target DNA structure for retroviral integrase in vitro. J Biol Chem. 1998;273:24190–24195. doi: 10.1074/jbc.273.37.24190. [DOI] [PubMed] [Google Scholar]
- 40.Katz R A, Merkel G, Kulkosky J, Leis J, Skalka A M. The avian retroviral IN protein is both necessary and sufficient for integrative recombination in vitro. Cell. 1990;63:87–95. doi: 10.1016/0092-8674(90)90290-u. [DOI] [PubMed] [Google Scholar]
- 41.Lapadat-Tapolsky M, De Rocquigny H, Van Gent D, Roques B, Plasterk R, Darlix J-L. Interactions between HIV-1 nucleocapsid protein and viral DNA may have important functions in the viral life cycle. Nucleic Acids Res. 1993;21:831–839. doi: 10.1093/nar/21.4.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee M S, Craigie R. A previously unidentified host protein protects retroviral DNA from autointegration. Proc Natl Acad Sci USA. 1998;95:1528–1533. doi: 10.1073/pnas.95.4.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li L, Farnet C M, Anderson W F, Bushman F D. Modulation of activity of Moloney murine leukemia virus preintegration complexes by several host factors in vitro. J Virol. 1998;72:2125–2131. doi: 10.1128/jvi.72.3.2125-2131.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miller M, Bor Y-C, Bushman F D. Target DNA capture by HIV-1 integration complexes. Curr Biol. 1995;5:1047–1056. doi: 10.1016/s0960-9822(95)00209-0. [DOI] [PubMed] [Google Scholar]
- 45.Miller M D, Farnet C M, Bushman F D. Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. J Virol. 1997;71:5382–5390. doi: 10.1128/jvi.71.7.5382-5390.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Muller H-P, Varmus H E. DNA bending creates favored sites for retroviral integration: an explanation for preferred insertion sites in nucleosomes. EMBO J. 1994;13:4704–4714. doi: 10.1002/j.1460-2075.1994.tb06794.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murphy J E, Goff S P. A mutation at one end of Moloney murine leukemia virus DNA blocks cleavage of both ends by the viral integrase in vivo. J Virol. 1992;66:5092–5095. doi: 10.1128/jvi.66.8.5092-5095.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47a.Myrik, K., and C. Farnet. Unpublished data.
- 48.Patel P H, Preston B D. Marked infidelity of human immunodeficiency virus type 1 reverse transcriptase at RNA and DNA template ends. Proc Natl Acad Sci USA. 1994;91:549–553. doi: 10.1073/pnas.91.2.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pruss D, Bushman F D, Wolffe A P. Human immunodeficiency virus integrase directs integration to sites of severe DNA distortion within the nucleosome core. Proc Natl Acad Sci USA. 1994;91:5913–5917. doi: 10.1073/pnas.91.13.5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rein A, Henderson L E, Levin J G. Nucleic-acid-chaperone activity of retroviral nucleocapsid proteins: significance for viral replication. Trends Biochem Sci. 1998;23:297–301. doi: 10.1016/s0968-0004(98)01256-0. [DOI] [PubMed] [Google Scholar]
- 51.Scottoline B P, Chow S, Ellison V, Brown P O. Disruption of the terminal base pairs of retroviral DNA during integration. Genes Dev. 1997;11:371–382. doi: 10.1101/gad.11.3.371. [DOI] [PubMed] [Google Scholar]
- 52.Sherman P A, Fyfe J A. Human immunodeficiency virus integration protein expressed in Escherichia coli possesses selective DNA cleaving activity. Proc Natl Acad Sci USA. 1990;87:5119–5123. doi: 10.1073/pnas.87.13.5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Swingler S, Gallay P, Camaur D, Song J, Abo A, Trono D. The Nef protein of human immunodeficiency virus type 1 enhances serine phosphorylation of the viral matrix. J Virol. 1997;71:4372–4377. doi: 10.1128/jvi.71.6.4372-4377.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tanchou V, Decimo D, Pechoux C, Lener D, Rogemond V, Berthoux L, Ottmann M, Darlix J-L. Role of the N-terminal zinc finger of human immunodeficiency virus type 1 nucleocapsid protein in virus structure and replication. J Virol. 1998;72:4442–4447. doi: 10.1128/jvi.72.5.4442-4447.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tsuchihashi Z, Brown P O. DNA strand exchange and selective DNA annealing promoted by the human immunodeficiency virus type 1 nucleocapsid protein. J Virol. 1994;68:5863–5870. doi: 10.1128/jvi.68.9.5863-5870.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vora A C, Chiu R, McCord M, Goodarzi G, Stahl S J, Mueser T C, Hyde C C, Grandgenett D P. Avian retrovirus U3 and U5 DNA inverted repeats. Role of nonsymmetrical nucleotides in promoting full-site integration by purified virion and bacterial recombinant integrases. J Biol Chem. 1997;272:23938–23945. doi: 10.1074/jbc.272.38.23938. [DOI] [PubMed] [Google Scholar]
- 57.Vora A C, Grandgenett D P. Assembly and catalytic properties of retrovirus integrase-DNA complexes capable of efficiently performing concerted integration. J Virol. 1995;69:7483–7488. doi: 10.1128/jvi.69.12.7483-7488.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wei S-Q, Mizuuchi K, Craigie R. Footprints of the viral DNA ends in Moloney murine leukemia virus preintegration complexes reflect a specific association with integrase. Proc Natl Acad Sci USA. 1998;95:10535–10540. doi: 10.1073/pnas.95.18.10535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu W, Henderson L E, Copeland T D, Gorelick R J, Bosche W J, Rein A, Levin J G. Human immunodeficiency virus type 1 nucleocapsid protein reduces reverse transcriptase pausing at a secondary structure near the murine leukemia virus polypurine tract. J Virol. 1996;70:132–142. doi: 10.1128/jvi.70.10.7132-7142.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zheng R, Jenkins T M, Craigie R. Zinc folds the N-terminal domain of HIV-1 integrase, promotes multimerization, and enhances catalytic activity. Proc Natl Acad Sci USA. 1996;93:13659–13664. doi: 10.1073/pnas.93.24.13659. [DOI] [PMC free article] [PubMed] [Google Scholar]