Abstract
Full-site integration by recombinant wild-type and mutant simian immunodeficiency virus (SIV) integrase (IN) was investigated with linear retrovirus-like DNA (469 bp) as a donor substrate and circular DNA (2,867 bp) as a target substrate. Under optimized conditions, recombinant SIV IN produced donor-target products consistent with full-site (two donor ends) and half-site (one donor end) reactions with equivalent frequency. Restriction enzyme analysis of the 3.8-kbp full-site reaction products confirmed the concerted insertion of two termini from separate donors into a single target molecule. Donor ends carrying the viral U5 termini were preferred over U3 termini for producing both half-site and full-site products. Bacterial genetic selection was used to isolate individual donor-target recombinants, and the donor-target junctions of the cloned products were characterized by sequencing. Analysis of 149 recombinants demonstrated approximately 84% fidelity for the appropriate simian retrovirus 5-bp host duplication. As seen previously in similar reactions with human immunodeficiency virus type 1 (HIV-1) IN from lysed virions, approximately 8% of the donor-target recombinants generated with recombinant SIV IN incurred specific 17- to 18- or 27- to 29-bp deletions. The efficiency and fidelity of the full-site integration reaction mediated by the purified, recombinant SIV IN is comparable to that of HIV-1 IN from virions. These observations suggest that a purified recombinant lentivirus IN is itself sufficient to recapitulate the full-site integration process.
For all retroviruses, productive infection requires integration of the retroviral DNA genome into the host cellular genome by the viral enzyme integrase (IN). The insertion of the linear viral DNA into the host DNA is a multistep process in which two distinct catalytic reactions are performed at each end of the viral genome (3, 17, 35). In the first of these reactions, 3′ endonucleolytic processing, a dinucleotide is removed from each of the 3′ OH termini on the viral blunt-ended DNA. In the subsequent reaction, strand transfer, the two recessed 3′ OH DNA ends are inserted into the host DNA by a transesterification reaction. The insertion of each end of the viral DNA typically occurs 4 to 6 bp apart on the cellular DNA; thus, the full-site integration reaction generates a small duplication (4 to 6 bp) of cellular DNA sequence subsequent to repair of the resultant gap. The size of the duplication is virus specific, for example, human immunodeficiency virus type 1 (HIV-1) has a 5-bp host duplication.
In the context of virus replication, integration is mediated by IN associated with the viral DNA as a component of stable, high-molecular-weight nucleoprotein complexes called the viral preintegration complexes (PIC). PIC isolated from murine leukemia virus- (4, 26, 40, 41), HIV-1- (14, 15, 31), and Rous sarcoma virus (RSV)-infected cells (28) catalyze full-site integration when provided with a target DNA substrate in vitro. IN is the only protein known to be essential for each of the basic catalytic steps in integration (3′ processing and strand transfer). Cellular factors or other viral proteins, or both, present in the PIC may facilitate integration either directly, by promoting the assembly of appropriate integration intermediates (14), or indirectly, by preventing “autointegration” (26, 40). Several such candidate factors have been identified and shown to stimulate integration reactions performed in vitro with PIC isolated from virus-infected cells.
Efficient full-site integration has been demonstrated by using IN purified from avian myeloblastosis virus (AMV) (38, 39), recombinant RSV PrA IN (37), and IN in nonionic-detergent-lysed HIV-1 virions (18, 19). Efficient full-site integration is defined as 5 to 15% of the input donor substrate being incorporated into full-site products in 10 to 20 min at 37°C. In contrast to IN derived from virions, most recombinant IN are ineffective in catalyzing the full-site, bimolecular donor reaction (Fig. 1), whereas catalysis of the 3′-end processing and strand transfer of a single viral DNA end can be readily demonstrated with these proteins (3, 6, 8, 11, 22, 35, 36). Although in vitro reconstitution systems exhibiting full-site integration activity have been reported with recombinant IN (1, 5, 6, 16, 24), discordance between the activities of the recombinant enzymes and the homologous virus-derived materials remains.
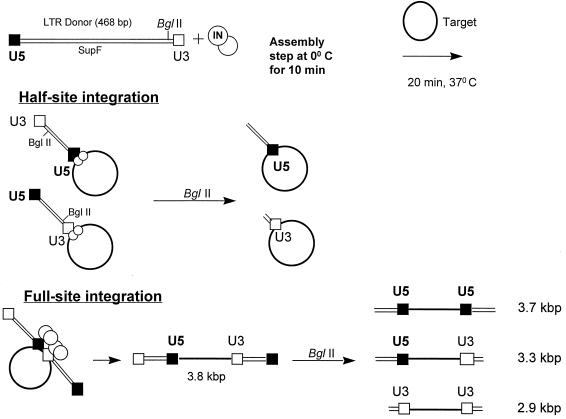
FIG. 1.
Schematic for the full-site integration assay. The 5′ 32P-labeled LTR donor (468 bp) is preincubated with IN (small circles) at 0°C to assemble nucleoprotein complexes. The solid and open boxes on the donor represent the U5 and U3 termini, respectively. Strand transfer is initiated by the addition of a circular target (2,867 bp) (large circles). The nucleoprotein complexes produce both half-site (middle) and full-site (bottom) donor-target products. A unique BglII site is located ∼40 bp from the U3 donor end. By using BglII restriction analysis, it is possible to differentiate the frequency with which LTR termini are used for both the half-site and full-site integration products (bottom right). The products were resolved by agarose gel electrophoresis and analyzed with a Molecular Dynamics PhosphorImager.
In this report, we show that purified, recombinant simian immunodeficiency virus (SIV) IN is capable of efficiently recapitulating full-site integration activity in vitro in the absence of additional cellular or viral proteins. SIV IN catalyzes concerted integration as efficiently as and with a fidelity for the correct host duplication greater than IN associated with nonionic-detergent-lysed HIV-1 virions (18, 19). Approximately 50% of the donor-target products in SIV IN-catalyzed reactions are shown to be the result of two donor substrate insertions producing linear 3.8-kbp products (Fig. 1). Of the 149 sequenced linear donor-target products characterized, 83% were the result of full-site integration events. Eight percent of the linear 3.8-kbp products sequenced contained specific small target deletions of 17 to 18 or 27 to 29 bp. Remarkably, the periodicity of these deletions paralleled those in integration reactions directed by HIV-1 IN from virions (18, 19). Several mutant SIV INs with severely reduced activities in half-site strand transfer assays in vitro were also inactive in full-site integration reactions. These studies demonstrate that retroviral IN can provide a basis for future studies comparing both the enzyme and substrate requirements for full-site and half-site integration activity in vitro.
MATERIALS AND METHODS
Purification of recombinant wt and mutant SIV IN.
Cloning and expression in Escherichia coli of the wild-type (wt) SIV IN from Mac239 and SIV IN mutants were as reported earlier for HIV-1 IN (25). The procedure for purification of SIV IN was as described for recombinant HIV-1 IN (21). Briefly, cells were lysed with a combination of sonication in hypotonic buffer and the addition of lysozyme. After centrifugation, the insoluble fraction was treated with DNase I and IN was solubilized with 1 M NaCl and CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate} (final concentration, 10 mM). Following centrifugation, the supernatant was diluted 1 to 5 (i.e., final NaCl concentration, 250 mM) and loaded onto a heparin-agarose column equilibrated in 50 mM Tris-HCl (pH 7.6), 250 mM NaCl, and 10% glycerol. The column was washed extensively, and IN was eluted in the above-mentioned buffer with a final NaCl concentration of 1.0 M. The purity of IN, as assessed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, was between 85 and 90%. Enzymatic activities were demonstrated by using a gel assay for 3′ processing and strand transfer reactions (21) as well as with the microtiter plate assay for strand transfer activity described below.
Microtiter plate assay for half-site strand transfer.
Assays for IN-catalyzed strand transfer were performed as described previously (21) with the following immobilized oligonucleotides as long terminal repeat (LTR) donor substrates in the reaction as appropriate: HIV-1 U5 (5′-ACCCTTTTAGTCAGTGTGGAAAATCTCTAGCAGT), HIV-1 U3 (5′-ACCCTCTTCTTTCACTGTAAAACATCCCTTCCAGT), SIV U5 (5′-ACCCTTTTAGAAGCAGGAAAATCCCTAGCAGT), and SIV U3 (5′-ACCCTTTTAGACTGGAAGGGATTTATTACAGT) (Table 1). All donor substrates were immobilized with a given directionality by using the unique 5′ phosphate, as on each oligonucleotide pair. Reactions were performed in a final concentration of 10% dimethyl sulfoxide. Representative data from at least two independent experiments are presented. In general, the standard deviation in these assays is ±10%. Note that the U5 LTR termini of HIV-1 and SIV are very similar: in ∼20 nucleotides there is only 1 nucleotide difference in position 9; the SIV and HIV-1 U3 LTR termini diverge at the fifth nucleotide from the end (10).
TABLE 1.
Strand transfer activities of HIV-1 and SIV IN with HIV-1 and SIV oligonucleotide substratesa
| Recombinant IN | Activity
|
|||
|---|---|---|---|---|
| HIV-1 U5 | HIV-1 U3b | SIV U5b | SIV U3b | |
| HIV-1 wt | 1.00 | 0.20 | 0.85 | 0.75 |
| SIV wt | 1.00 | 0.25 | 0.97 | 0.92 |
| SIV H12Ac | 0.02 | ND | ND | ND |
| SIV H16Ac | 0.10 | ND | ND | ND |
| N-terminal His SIVc | 0.85 | ND | ND | ND |
Oligonucleotides are described in Materials and Methods. For each substrate, INs were tested in duplicate at four different protein concentrations (25, 50, 100, and 200 nM). ND, not determined.
Activity relative to HIV-1 U5 substrate.
Activity relative to SIV wt enzyme.
Labeling of donor substrates for full-site strand transfer.
NdeI digestion of a pUC19-based construct yielded a 468-bp restriction fragment that contains wt HIV-1 U5 and U3 LTR termini (designated U5-U3) (18, 19) (Fig. 1). Similarly, a 469-bp fragment that contained two HIV-1 U5 LTR termini (designated U5-U5) was also obtained by NdeI digestion. The donors were 5′ end labeled with [γ-32P]ATP and T4 polynucleotide kinase. The specific activities of the labeled DNA were between ∼5,000 and 10,000 cpm per ng. Supercoiled pGEM (2,867 bp) that lacks a BglII restriction site was used as the target substrate.
Full-site strand transfer assay conditions.
Conditions for full-site integration of the LTR donor substrates into circular DNA for SIV IN are similar to those described for nonionic-detergent-lysed HIV-1 virions (18, 19). The assay conditions for full-site integration with the 469-bp donors were 20 mM HEPES (pH 7.6), 5 mM dithiothreitol, 10 mM MgCl2, 25 μM ZnCl2, 12% dimethyl sulfoxide, 10% polyethylene glycol 6000 (PEG), and 100 mM NaCl. The reaction volume was 20 μl. Preincubation of IN with DNA was performed for 10 min on ice in two different ways (Fig. 1). IN was preincubated with donor substrate prior to the addition of target followed by incubation at 37°C for 20 min, or IN was preincubated with donor and target together prior to incubation at 37°C. The amount of donor was varied between 40 and 60 ng with pGEM set at 100 ng. At 40 ng of pGEM, the donor-to-target molar ratio is 2.6 to 1.
Restriction enzyme analysis and genetic selection of donor-target recombinants.
The donor-target products produced by SIV IN were analyzed by BglII restriction digestion as previously described (18, 19) (Fig. 1). Samples were subjected to agarose gel electrophoresis, and the quantities of the labeled donor-target products were determined with a Molecular Dynamics PhosphorImager. The U5-U5 and U3-U5 donors contained the supF gene that allows for the genetic selection of the donor-target recombinants which are the result of full-site strand transfer reactions. Products of scale-up reactions (done 20 times) with SIV IN were digested with BglII for isolation of the linear 3.3-kbp recombinant DNA used for genetic selection. The isolated restriction fragment was ligated and transformed into CA244 cells that contain amber mutations for Trp biosynthesis and for LacZ expression (2, 29). The donor-target junctions of the plasmids isolated from individual colonies were sequenced. Sequencing was performed as previously described (16) or by the use of a Perkin-Elmer ABI Prism 310 genetic analyzer. The sequence information was analyzed for host site duplications and deletions (18, 19).
RESULTS
Characterization of recombinant SIV IN half-site strand transfer activities.
Wt and SIV IN with mutations in the N-terminal zinc-binding domain were cloned, expressed in E. coli, and purified by heparin-agarose chromatography. The purified SIV IN proteins were evaluated for 3′ processing and strand transfer activities with unprocessed donor substrates in a microtiter plate assay which quantifies the production of strand transfer products.
HIV-1 and SIV U3 and U5 oligonucleotides were evaluated as substrates with recombinant HIV-1 and SIV IN (Table 1). Wt SIV IN was able to use the HIV-1 U5 donor substrate as well as the homologous SIV U5 donor in an equivalent fashion. In addition, the specific activities observed for SIV IN with either the HIV-1 U5 donor or the SIV U5 donor were indistinguishable from that of HIV-1 IN. Both HIV-1 and SIV IN showed a decreased preference (approximately 75%) for the HIV-1 U3 donor relative to U5, while no major difference in activity was detected when the SIV U3 donor was used (Table 1). As shown previously for HIV-1 IN (7, 13, 23, 27, 34, 43), mutations in the N-terminal zinc-binding domain (H12A and H16A) of SIV IN severely reduced the activity of IN in the microtiter plate assay (Table 1).
Optimizing production of linear 3.8-kbp donor-target recombinants by SIV IN.
A schematic defining the general requirements for assembly of IN-LTR donor complexes as well as the reaction products produced is shown in Fig. 1. Substrates and assay conditions for analyzing half-site and full-site strand transfer activities with recombinant SIV IN were similar to conditions previously observed for nonionic-detergent-lysed HIV-1 virions (19). A 469-bp DNA fragment containing two wt HIV-1 U5 LTR termini with recessed 3′-OH ends (designated U5-U5) was used as the donor, and supercoiled pGEM (2,867 bp) was the target (Fig. 2). The donor was labeled at its 5′ ends, and the donor-target products were analyzed by agarose gel electrophoresis. The products were visualized by autoradiography and subjected to PhosphorImager analysis.
FIG. 2.
Effect of varying SIV IN concentrations on full-site and half-site strand transfer reactions under different DNA preincubation conditions. The concentrations of IN (indicated by triangles) in lanes 1 to 5 and lanes 6 to 10 were 0, 40, 80, 160, and 320 nM, respectively. In lanes 1 to 5, IN was preincubated with the U5-U5 donor and the target on ice for 10 min prior to strand transfer. In lanes 6 to 10, IN was preincubated with only the U5-U5 donor prior to the addition of target and subsequent strand transfer. The samples were processed, and equivalent quantities of radioactivity from each sample were subjected to 1% agarose gel electrophoresis. The circular half-site, full-site, and donor-donor products are indicated on the left along with the input donor. The dried gel was exposed to X-ray film at 25°C for 25 h.
As was observed with nonionic-detergent-lysed HIV-1 virions (18, 19), recombinant SIV IN produced three major strand transfer products (Fig. 2). They are (i) the insertion of one donor end into another donor (donor-donor products); (ii) the insertion of a single donor end into a circular target (circular half-site products); and (iii) the concerted insertion of two donor ends into a circular target (linear 3.8-kbp DNA or full-site products) (Fig. 1). Optimal conditions for producing the linear 3.8-kbp product with SIV IN were similar to those determined with HIV-1 IN from nonionic-detergent-lysed virions. With recombinant SIV IN in the presence of 10% PEG, the optimum NaCl concentration for producing either the linear 3.8-kbp product or the circular half-site product was between 80 and 100 mM. Titration of ZnCl2 in the assay also showed that each of the three products generated by SIV IN were increased severalfold in the presence of ZnCl2, with the optimum zinc concentration being ∼25 μM (data not shown). Zinc has previously been shown to stimulate the half-site strand transfer activity of recombinant HIV-1 IN (27, 43).
The quantity of each of the three major products generated by SIV IN under optimized assay conditions were also comparable both in number and in proportion to those produced in reactions with nonionic-detergent-lysed HIV-1 virions (19). Within a range of SIV IN between 40 and 160 nM, approximately 15 to 20% of the input donor DNA was used for strand transfer activities (Fig. 2). Preincubation of IN with the donor (Fig. 2, lanes 6 to 10) did not significantly influence the proportion of donor incorporated into pGEM as half-site or full-site products when compared to preincubation of IN with both donor and target prior to strand transfer (Fig. 2, lanes 1 to 5). Under either condition, the amount of donor incorporated into the pGEM target to produce full-site products was generally equivalent to the amount incorporated into circular half-site products: in each case, approximately 5% of the input donor substrate was incorporated (Fig. 2). However, donor-donor products were slightly more abundant in reactions where IN was preincubated with donor substrate alone (Fig. 2, compare lanes 7 to 9 with lanes 2 to 4) (19). With either preincubation condition, all strand transfer activities were reduced at SIV IN concentrations of ∼160 nM or higher (Fig. 2, lanes 5 and 10, and data not shown).
Although the total yield of strand transfer products was somewhat variable between SIV IN preparations, strand transfer products generally ranged from 10 to 20% of the input donor at 80 nM IN (Fig. 2). In all cases, both half-site and full-site products were produced by recombinant wt SIV IN with equivalent efficiency and the overall strand transfer activity was indistinguishable from that observed with HIV-I virion-derived IN under comparable conditions. A mutant SIV IN (E136K) was similar to wt SIV IN for both reactions at the same protein concentrations as used for Fig. 2 (data not shown). The same mutation in SIV compensated for nucleotide changes introduced into the SIV LTR in vivo (10). SIV IN with mutations in the N-terminal zinc-binding region exhibited severely reduced activity in half-site strand transfer reactions (Table 1) and did not promote full-site integration (data not shown). The N-terminal His tag on wt SIV IN reduced the half-site and full-site strand transfer activities with the U5-U5 donor approximately 50% or more relative to those with wt SIV IN lacking the His tag (data not shown). These results suggest that purified recombinant wt SIV IN can promote the full-site integration reaction without the addition of either viral or cellular proteins associated with SIV virions.
Recombinant SIV IN produces full-site products equivalent to those produced by nonionic-detergent-lysed HIV-1 virions.
Recombinant HIV-1 IN prefers the U5 LTR terminus over the U3 LTR terminus for half-site strand transfer reactions or 3′-OH processing of blunt-ended LTR termini (3, 5, 9, 35). Recombinant SIV IN also exhibited a preference for the HIV-1 U5 sequence over that of HIV-1 U3 in half-site strand transfer reactions (Table 1). To determine whether recombinant SIV IN exhibits a similar preference for the U5 LTR for full-site strand transfer, we used a U3-U5 LTR donor substrate. A unique BglII restriction site in the donor was used to distinguish the donor-target recombinants produced with either U3 or U5 termini based on relative size (Fig. 1). Simple BglII restriction enzyme digestion of the circular half-site and linear 3.8-kbp products followed by agarose gel electrophoresis permits a physical evaluation of the frequency of LTR terminal usage by IN (39).
Donor-target products produced by nonionic-detergent-lysed HIV-1 virions and recombinant SIV IN with the HIV-1 U3-U5 donor (Fig. 3, lanes 2 and 4, respectively) were subjected to BglII digestion. For either IN, digestion of the circular half-site products showed the preferential utilization of the U5 LTR terminus over the U3 LTR terminus for half-site catalysis (Fig. 2, lanes 3 and 5). These results are consistent with the data obtained in the microtiter plate half-site strand transfer reactions with the HIV-1 U3 and U5 oligonucleotides (Table 1). Analysis of full-site products also demonstrated a clear preference for the use of the U5 LTR as a substrate in full-site reactions by both proteins. Digestion of the linear 3.8-kbp full-site products by BglII produced three linear products with sizes of 3.7 (U5-target-U5), 3.3 (U3-target-U5), and 2.9 (U3-target-U3) kbp (Fig. 1 and 3, right side). From PhosphorImager data, the molar ratio of these three fragments is ∼1.2 to 1 to 0.05, respectively. The observation that three discrete products of the appropriate sizes are obtained from the restriction digest is also consistent with the incorporation of LTR termini from two different donors into each target molecule.
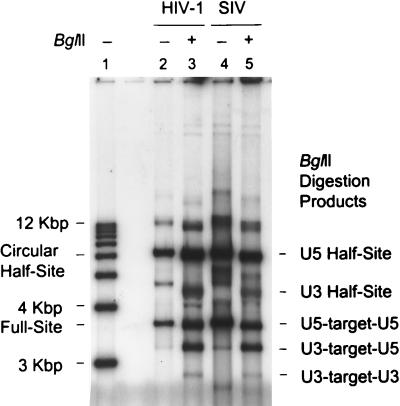
FIG. 3.
BglII restriction analysis reveals preferential usage of U5 over U3 LTR termini by SIV IN for full-site and half-site strand transfer. Standard strand transfer assays were performed with SIV IN and nonionic-detergent-lysed HIV-1 virions with the HIV-1 U5-U3 donor (469 bp). The products were separated on 1.5% agarose gels. The samples were not (−) or were (+) digested with BglII as indicated at the top. Lane 1, molecular size markers; lane 2, HIV-1 IN products; lane 3, BglII digestion of HIV-1 products; lane 4, SIV IN products; lane 5, BglII digestion of SIV IN products. The undigested circular half-site and full-site products of both reactions are marked on the left side along with the sizes of the DNA ladder. The BglII digestion products are shown on the right. The digested full-site products are indicated as LTR donor-target-LTR donor fragments.
To rule out potential artifacts in the analysis due to downstream sequence or context effects, circular half-site and full-site integration products produced by SIV IN with a U5-U5 donor (Fig. 1) were subjected to similar BglII restriction analyses. In contrast to the results obtained with the U3-U5 donor, the U5-U5 substrate produced all three restriction fragments with the appropriate near-molar ratio (1 to 2 to 1, respectively) (37), indicating that both U5 termini were used equivalently (data not shown). The observation that the proportion of products containing two U5 LTR termini is most abundant in the previous analysis (Fig. 3) with the U3-U5 substrate suggests that the U5 sequence is highly preferred for full-site strand transfer in vitro.
High fidelity for producing SIV 5-bp host site duplications with recombinant SIV IN.
When presented with the appropriate LTR donor substrates and assay conditions, recombinant SIV IN is capable of promoting full-site strand transfer reactions with nearly the same efficiency and specificity as IN associated with HIV-1 virions (Fig. 2 and 3). However, to demonstrate that the recombinant SIV IN is capable of faithfully recapitulating full-site integration activity as it occurs during infection, it is necessary to establish the fidelity of the SIV 5-bp host site duplication characteristic of integration in vivo (3, 33).
The fidelity of the SIV IN full-site reaction was evaluated by isolating and sequencing individual recombinants derived from the linear 3.8-kbp donor-target products. Three independent reactions representing different combinations of donors or preincubation conditions were scaled up for this analysis (Table 2). The strand transfer products of these reactions were subjected to BglII digestion, and the linear 3.3-kbp DNA was isolated from each reaction (Fig. 1) (18, 19). The DNAs were ligated, and each was used to transform E. coli CA244 cells. Individual colonies from the three independent reactions were isolated, and the donor-target junctions were sequenced (Table 2).
TABLE 2.
Sequence analysis of donor-target junction sites produced by purified SIV INa
| Characteristic | Size (bp) | No. of recombinants
|
% of total recombinants | |||
|---|---|---|---|---|---|---|
| U5-U3
|
U5-U5
|
Total | ||||
| Donor with target; IN added | Donor with target; IN added | Donor complex to IN; target added | ||||
| Duplication | 5 | 49 | 35 | 34 | 118 | 79.2 |
| 6 | 2 | 1 | 0 | 3 | 2.1 | |
| 4 | 2 | 0 | 1 | 3 | 2.1 | |
| 0 | 6 | 2 | 1 | 9 | 6.0 | |
| Deletion | 17, 18 | 0 | 1 | 2 | 3 | 2.0 |
| 27–29 | 5 | 3 | 1 | 9 | 6.0 | |
| Larger | 3 | 0 | 1 | 4 | 2.7 | |
| Total | 67 | 42 | 40 | 149 | ||
The donor-target junctions of 149 recombinants produced from three independent reactions were sequenced. There were 11 additional clones selected that contained either only one or no LTR termini; some were smaller or had other aberrations suggesting rearrangements or deletions had occurred.
A total of 149 full-site donor-target recombinants were sequenced, and >80% had host site duplications (5 bp) that were characteristic of SIV integration (Table 2). The most abundant duplication, the 5-bp host site duplication, was observed in 79% of the total recombinants. In addition, 2% of the recombinants had duplications of either 4 or 6 bp (Table 2). In the full-site reactions, the fidelity of 5-bp duplications was independent of preincubation conditions or whether the U5-U5 or the U5-U3 donors were used as a substrate.
Specific small deletions were observed in 8% of the sequenced recombinants (Table 2). These size set deletions were never smaller than 17 bp in length (2%) or they appear to represent a turn of the DNA helix for the 27-bp deletion set (6%). The size and periodicity of these small deletions are similar to those observed with HIV-1 virion-derived IN reactions (18, 19). However, the virion-directed reactions produced a larger percentage (35%) of these small deletions than the 8% produced by SIV IN. A small percentage (2.7%) of larger deletions that contained both LTR termini were produced by SIV IN.
Other variations besides the above-mentioned donor-target recombinants were evident. Eleven clones had other major rearrangements or deletions in the LTR region or the plasmid (Table 2, bottom). Nine recombinants lacked any duplication or deletion of host sequence (Table 2, top, with 6% total). Each of these nine recombinants contained two independent donor insertions on adjacent nucleotides but on opposite strands. One of the two LTR termini had a TA dinucleotide inserted between the viral CA dinucleotide and the adjacent host nucleotide, suggesting that the 5′ 2-base TA overhang of the noncatalytic strand was not removed (18). One of the above-mentioned nine recombinants also lacked the terminal A nucleotide of the conserved LTR 3′ CA. The sequences of these recombinant products suggest inadequate or inappropriate repair by the host bacterial cell.
In summary, sequence analysis of the donor-target recombinants isolated from the linear 3.8-kbp DNA products produced by SIV IN revealed a high fidelity for the SIV 5-bp duplication. This fidelity of host site duplications and the production of specific small deletions mirror the catalytic capability of virion-derived HIV-1 IN in vitro, using similar donor substrates and assay conditions.
Efficient half-site and full-site integration by SIV IN is dependent on LTR donor length.
The ability to monitor both half-site and full-site strand transfer activities concurrently allows an evaluation of the substrate and enzymatic requirements for each process and possibly provides some insight into the general lack of success in recapitulating full-site activity in vitro with other recombinant INs. Inverted-repeat sequences at the LTR termini are essential for 3′-OH processing and strand transfer reactions catalyzed by IN both in vivo and in vitro (3, 8, 35). With in vitro half-site reactions, specific sequences within 10 bases proximal to the 3′ end are critical for activity, although downstream sequence has been reported to have varying effects as well (35). Full-site integration activity is sensitive to the proximal sequence effects in vivo (17, 30). We next investigated the effect that the length of the DNA donor substrate has on both the half-site and full-site reactions catalyzed by SIV IN.
The 469-bp U5-U5 donor was labeled with 32P at both ends. The donor was digested with various restriction enzymes to obtain donor substrates of varying lengths containing a single U5 LTR terminus and a nonspecific end. Fragments of 433 to 134 bp were obtained. The total quantity of each isolated fragment was varied in the reaction mixture to maintain a constant IN-to-donor end ratio (5 to 1) while maintaining the IN concentration constant at 80 nM (Fig. 4A, lanes 3 to 10). The target concentration was not varied. IN was incubated together with the donor and target on ice prior to strand transfer.
FIG. 4.
The full-site integration reaction is more sensitive to donor length than the circular half-site reaction under standard assay conditions. (A) A large quantity of the double-ended U5-U5 donor was 5′ end labeled with 32P to produce uniformly labeled DNA. The DNA was independently digested with a variety of restriction enzymes to produce different-size donors that contain only one LTR terminus and a nonspecific end. The end-labeled fragments were separated on agarose gels and purified. A constant IN-donor end molar ratio (5 to 1) was maintained for each fragment. The concentration of IN was 80 nM, and the quantity of each single-ended fragment was varied to maintain this constant ratio. Lane 1 contains no IN with the double-ended U5-U5 donor. Lane 2 is the control U5-U5 reaction. Lanes 3 to 10 contain the single-ended U5 LTR fragments whose sizes (in base pairs) are 433, 375, 331, 290, 242, 187, 175, and 134. The restriction enzymes used to produce these different-size donors were XbaI, DrdI, BsrI, EaeI, HinfI, HinfI, EaeI, and BsrI, respectively. Nearly equivalent quantities (∼34,000 cpm) of each strand transfer reaction were subjected to agarose gel electrophoresis, and the dried gel was exposed to X-ray film. The circular half-site and full-site products along with the various-sized input donor fragments are indicated on the left. The donor-donor products migrated just above each input donor and are not marked on the side. (B) The products shown in panel A were subjected to analysis on a PhosphorImager. The percentage of donor incorporated into each product was determined as indicated on the left. The single open rectangle and triangle represent the control double-ended U5-U5 reaction, while the corresponding solid symbols represent the single-ended U5 reaction for circular half-site and full-site reactions, respectively. The sizes of the single-ended LTR fragments are indicated on the bottom.
The percentages of input donor incorporated into each of the three major DNA products (full site, circular half site, and donor-donor) were determined (Fig. 4B). For the control double-ended U5-U5 donor, the percentages of products produced were 5, 7.5, and 6, respectively (Fig. 4A, lane 2). For the first single-ended U5 donor (433 bp), the percentages of the products identified above were 3.5, 7.5, and 7, respectively (Fig. 4, lane 3). With the 360-, 331-, and 290-bp single-ended U5 donors (Fig. 4A, lanes 4 to 6, respectively), the circular half-site and the donor-donor products were only slightly affected compared to the 433-bp single-ended LTR donor. In contrast, the full-site reaction products with these same donors were decreased to a greater extent (Fig. 4B). Thereafter, both the half-site and full-site products were significantly less than the 433-bp single-ended U5 donor (Fig. 4). Similar results were obtained whether IN (80 nM) was preincubated with the donor substrates alone or the IN-to-donor end ratio was decreased to 2.5 to 1 (data not shown). In each case, the half-site and full-site products by SIV IN were sensitive to the length of the donor substrate, with the full-site products being the most severely affected.
DISCUSSION
We have demonstrated efficient, full-site integration activity by purified recombinant SIV IN. Recombinant SIV IN catalyzes concerted integration with an efficiency and fidelity indistinguishable from that shown for IN associated with nonionic-detergent-disrupted HIV-1 virions. The observation that a purified recombinant IN faithfully promotes the process of full-site integration demonstrates that IN alone is sufficient for the reaction in vitro (6, 24). As we have described, the system is amenable to studies to define the requirements for half-site versus full-integration activities as well as reconstitution studies with host or viral proteins, or both, to identify potential cofactors which may facilitate the integration process.
The fidelity of the 5-bp host site duplication produced by SIV IN is nearly identical to that observed with HIV-1 virions (Table 2) (18, 19). However, the total percentage (83%) of these duplications produced by SIV IN is higher than that observed with HIV-1, which varied between 60 and 70%. In addition, the percentage (8%) of small deletions (17 or 28 bp) produced by SIV IN is significantly lower that the ∼35% observed with HIV-1 virions. The total sequenced donor-target recombinants for SIV IN and for IN associated with HIV-1 virions are 149 and 193, respectively. Therefore, besides the obvious experimental flexibility derived by working with recombinant proteins, the recombinant SIV IN appears to be a better source of enzymatic activity for potential biochemical studies of full-site integration than virion-derived materials.
The origin of the apparently specific small deletions is unknown. But, the similarities in the products produced by the HIV-1 virion enzyme (18, 19) and the recombinant SIV IN in this report suggest that the deletions may be the direct result of IN itself rather than of proteins present in HIV-1 virions or impurities present in the recombinant material. A model demonstrating the potential origin of the HIV-1 and SIV IN deletions is shown in Fig. 5 and is similar to a model previously described for producing deletions by IN in the target DNA (6). Recombinant HIV-1 IN has the ability to form multimers on DNA varying in size from dimers up to structures containing at least ∼10 monomers (12, 13, 32, 42). In our model, an undefined number of IN subunits would multimerize on the target DNA (the 17-bp region in the model) either before or after binding to the LTR donor termini. The next size set of deletions are ∼27 to 29 bp (Table 1) (18, 19), suggesting that both sets of deletions that are produced were contacted on the same face of the DNA by IN. Half-site strand transfer would occur normally, creating the usual free 3′-OH terminus on the target (Fig. 5). The nonspecific endonucleolytic DNA-nicking activity associated with IN (or contaminating endonucleases associated with purified IN preparations and HIV-1 virions) would produce a nick opposite the half-site strand transfer event, thus creating the apparent deletion. Nonspecific DNA endonuclease activities that require Mg2+ are apparent in nonionic-detergent-lysed HIV-1 virions and to a far lesser extent in SIV IN preparations (data not shown); therefore, we cannot rule out their contribution to the observed effect.
FIG. 5.
Model for production of small, 17-bp deletions by SIV IN and IN associated with nonionic-detergent-lysed HIV-1 virions. The LTR donors are shown as U5 and U3 donor substrates (light lines). The 3′-OH termini of two donors are inserted into the target (dark lines), resulting in free 3′-OH target ends. Note that the inserted donor termini must be pointing in opposite directions on the target DNA to produce a deletion. For full-site integration resulting in a 5-bp host site duplication, the donor termini must be pointing toward each other. The 17-bp spacing is located on the target DNA that is occupied by a minimal-size protein structure whose number of IN subunits is unknown. The two thin arrows indicate nicking events by IN or a nonspecific nuclease. The resulting structure is indicated at the bottom. Either full-site integration or the formation of the indicated deletion by IN will produce linear 3.8-kbp donor-target recombinants.
In contrast to the number of host site deletions observed in the HIV-1 and SIV IN full-site integration assays, AMV and RSV PrA IN produce fewer deletions (less than 3% of 141 sequenced) (37–39), and these deletions do not possess the repetitive nature observed in the HIV-1 and SIV systems. In addition, the same genetic selection system was used to isolate the HIV-1–SIV and the avian donor-target recombinants, suggesting again that the host site deletions observed in the HIV-1 and SIV systems are not due to the selection system itself. The similar lack of deletions produced in the target by another avian recombinant IN with a blunt-ended LTR donor substrate has been reported (1). These results suggest that HIV-1 and SIV IN may possess properties different from those of the avian enzymes.
We have observed that both the half-site and full-site integration activities exhibited a sensitivity to the length of the donor substrate, although the full-site reaction appears to be affected more severely. Although the reason for this observation is unknown, there are several potential explanations. As described above, IN can assemble as multimers on DNA. It is possible that additional molecules of IN associated with the longer DNA may facilitate the synapse or juxtaposition of donor DNA via protein-protein interactions. AMV IN is capable of looping DNA by either an intramolecular or an intermolecular mechanism (20). The viral DNA in the PIC isolated from virus-infected cells is held together by a protein bridge (31) encompassing at least ∼200 bases from the LTR termini which appears to be the result of a specific association with IN (41). Alternatively, the requirement for longer DNA to promote full-site strand transfer may merely reflect the molecular crowding events associated with the presence of PEG that are necessary to promote full-site strand transfer (38). Molecular probing studies of reconstituted nucleoprotein complexes capable of full-site strand transfer will be necessary to address the location and number of IN subunits involved in this process.
ACKNOWLEDGMENTS
This work was supported in part by a grant (AI-31334) from the National Institutes of Health to D.G.
We thank R. Chiu for sequencing some of the donor-target recombinants. We thank M. McCord for sharing the information on the donor length effect observed with avian retrovirus full-site integration prior to publication.
REFERENCES
- 1.Aiyar A, Hindmarsh P, Skalka A M, Leis J. Concerted integration of linear retroviral DNA by the avian sarcoma virus integrase in vitro: dependence on both long terminal repeat termini. J Virol. 1996;70:3571–3580. doi: 10.1128/jvi.70.6.3571-3580.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brenner S, Beckwith J R. Oche mutants, a new class of suppressible nonsense mutants. J Mol Biol. 1965;13:629–637. [Google Scholar]
- 3.Brown P O. Retroviruses. In: Coffin J M, Hughes S H, Varmus H E, editors. Integration. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1998. pp. 161–203. [Google Scholar]
- 4.Brown P O, Bowerman B, Varmus H E, Bishop J M. Correct integration of retroviral DNA in vitro. Cell. 1987;49:347–356. doi: 10.1016/0092-8674(87)90287-x. [DOI] [PubMed] [Google Scholar]
- 5.Bushman F D, Craigie R. Sequence requirements for integration of Moloney murine leukemia virus DNA in vitro. J Virol. 1990;64:5645–5648. doi: 10.1128/jvi.64.11.5645-5648.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bushman F D, Fujiwara T, Craigie R. Retroviral DNA integration directed by HIV integration protein in vitro. Science. 1990;249:1555–1558. doi: 10.1126/science.2171144. [DOI] [PubMed] [Google Scholar]
- 7.Cannon P M, Wilson W, Byles E, Kingsman S M, Kingsman A J. Human immunodeficiency virus type 1 integrase: effect on viral replication of mutations at highly conserved residues. J Virol. 1994;68:4768–4775. doi: 10.1128/jvi.68.8.4768-4775.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Craigie R, Fujiwara T, Bushman F D. The IN protein of Moloney murine leukemia virus processes the viral DNA ends and accomplishes their integration in vitro. Cell. 1990;62:829–837. doi: 10.1016/0092-8674(90)90126-y. [DOI] [PubMed] [Google Scholar]
- 9.Craigie R, Hickman A B, Engelman A. Integrase. In: Karn J, editor. HIV. II. A practical approach. Oxford, England: Oxford University Press; 1995. pp. 53–71. [Google Scholar]
- 10.Du Z, Iyinskii P O, Lally K, Desrosiers R C, Engelman A. A mutation in integrase can compensate for mutations in the simian immunodeficiency virus att site. J Virol. 1997;71:8124–8132. doi: 10.1128/jvi.71.11.8124-8132.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ellison V, Abrams H, Roe T, Lifson J, Brown P. Human immunodeficiency virus integration in a cell-free system. J Virol. 1990;64:2711–2715. doi: 10.1128/jvi.64.6.2711-2715.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ellison V, Gerton J, Vincent K A, Brown P. An essential interaction between distinct domains of HIV-1 integrase mediates assembly of the active multimer. J Biol Chem. 1995;270:3320–3326. doi: 10.1074/jbc.270.7.3320. [DOI] [PubMed] [Google Scholar]
- 13.Engelman A, Craigie R. Identification of conserved amino acid residues critical for HIV-1 integrase function in vitro. J Virol. 1992;66:6361–6369. doi: 10.1128/jvi.66.11.6361-6369.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farnet C M, Bushman F D. HIV-1 cDNA integration: requirement of HMG1(Y) protein for function of preintegration complexes in vitro. Cell. 1997;88:483–492. doi: 10.1016/s0092-8674(00)81888-7. [DOI] [PubMed] [Google Scholar]
- 15.Farnet C M, Haseltine H A. Integration of human immunodeficiency virus type 1 DNA in vitro. Proc Natl Acad Sci USA. 1990;87:4164–4168. doi: 10.1073/pnas.87.11.4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fitzgerald M L, Vora A C, Zeh W G, Grandgenett D P. Concerted integration of viral DNA termini by purified avian myeloblastosis virus integrase. J Virol. 1992;66:6257–6263. doi: 10.1128/jvi.66.11.6257-6263.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goff S P. Genetics of retroviral integration. Annu Rev Genet. 1992;26:527–544. doi: 10.1146/annurev.ge.26.120192.002523. [DOI] [PubMed] [Google Scholar]
- 18.Goodarzi G, Chiu R, Brackmann K, Kohn K, Prommier Y, Grandgenett D P. Host site selection for concerted integration by human immunodeficiency virus type-1 virions in vitro. Virology. 1997;231:210–217. doi: 10.1006/viro.1997.8558. [DOI] [PubMed] [Google Scholar]
- 19.Goodarzi G, Im G J, Brackmann K, Grandgenett D P. Concerted integration of retrovirus-like DNA by human immunodeficiency virus type I integrase. J Virol. 1995;69:6090–6097. doi: 10.1128/jvi.69.10.6090-6097.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grandgenett D P, Inman R B, Vora A C, Fitzgerald M. Comparison of DNA binding and integration half-site selection by avian myeloblastosis virus integrase. J Virol. 1993;67:2628–2636. doi: 10.1128/jvi.67.5.2628-2636.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hazuda D J, Hastings J C, Wolfe A L, Emini E A. A novel assay for the DNA strand transfer reaction of HIV-1 integrase. Nucleic Acids Res. 1994;22:1121–1122. doi: 10.1093/nar/22.6.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jenkins T M, Engleman A, Ghirlando R, Craigie R. A soluble mutant of HIV-1 integrase: involvement of both the core and carboxyl-terminal domains in multimerization. J Biol Chem. 1996;271:370–374. doi: 10.1074/jbc.271.13.7712. [DOI] [PubMed] [Google Scholar]
- 23.Jonsson C B, Roth M J. Role of His-Cys finger of MuLV protein in integration and disintegration. J Virol. 1993;67:5562–5571. doi: 10.1128/jvi.67.9.5562-5571.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katz R A, Merkel G, Kulkosky J, Leis J, Skalka A M. The avian retrovirus IN protein is both necessary and sufficient for integrative recombination in vitro. Cell. 1990;63:87–95. doi: 10.1016/0092-8674(90)90290-u. [DOI] [PubMed] [Google Scholar]
- 25.LaFemina R L, Graham P L, LeGrow K, Hastings J C, Wolfe A, Young S D, Emini E A, Hazuda D J. Inhibition of human immunodeficiency virus integrase by bis-catechols. Antimicrob Agents Chemother. 1995;39:320–324. doi: 10.1128/aac.39.2.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee M S, Craigie R. Protection of retroviral DNA from autointegration: involvement of a cellular factor. Proc Natl Acad Sci USA. 1994;91:9823–9827. doi: 10.1073/pnas.91.21.9823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee S P, Xiao J, Knutson J R, Lewis M S, Han M K. Zn2+ promotes the self-association of human immunodeficiency virus type-1 integrase in vitro. Biochemistry. 1997;36:173–180. doi: 10.1021/bi961849o. [DOI] [PubMed] [Google Scholar]
- 28.Lee Y M, Coffin J M. Relationship of avian retrovirus DNA synthesis to integration in vitro. Mol Cell Biol. 1991;11:1419–1430. doi: 10.1128/mcb.11.3.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lutz C T, Hollifield W C, Seed B, Davie J M, Huang H V. Syrinx 2A: an improved λ phage vector designed for screening DNA libraries by recombination in vivo. Proc Natl Acad Sci USA. 1987;84:4379–4383. doi: 10.1073/pnas.84.13.4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Masuda T, Kuroda M J, Harada S. Specific and independent recognition of U3 and U5 att sites by human immunodeficiency virus type 1 integrase in vitro. J Virol. 1998;72:8396–8402. doi: 10.1128/jvi.72.10.8396-8402.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller M D, Farnet C M, Bushman F D. Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. J Virol. 1997;71:5382–5390. doi: 10.1128/jvi.71.7.5382-5390.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pemberton I K, Buckle M, Buc H. The metal ion-induced cooperative binding of HIV-1 integrase to DNA exhibits a marked preference for Mn2+ rather than Mg2+ J Biol Chem. 1996;271:1498–1506. doi: 10.1074/jbc.271.3.1498. [DOI] [PubMed] [Google Scholar]
- 33.Stevens S W, Griffith J D. Sequence analysis of the human DNA flanking site of human immunodeficiency virus type 1 integration. J Virol. 1996;70:6459–6462. doi: 10.1128/jvi.70.9.6459-6462.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vincent K A, Ellison V, Chow S A, Brown P O. Characterization of HIV-1 integrase expressed in Escherichia coli and analysis of amino-terminal mutations. J Virol. 1993;67:425–437. doi: 10.1128/jvi.67.1.425-437.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vink C, Plasterk R H A. The human immunodeficiency virus integrase protein. Trends Genet. 1993;9:433–438. doi: 10.1016/0168-9525(93)90107-s. [DOI] [PubMed] [Google Scholar]
- 36.Vink C, Oude Groeneger A A M, Plasterk R H A. Identification of the catalytic and DNA-binding region of the HIV-1 integrase protein. Nucleic Acids Res. 1993;21:1419–1425. doi: 10.1093/nar/21.6.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vora A C, Chiu R, McCord M, Goodarzi G, Stahl S J, Mueser T C, Hyde C C, Grandgenett D P. Avian retrovirus U3 and U5 DNA inverted repeats: role of nonsymmetrical nucleotides in promoting full-site integration by purified virion and bacterial recombinant integrases. J Biol Chem. 1997;272:23938–23945. doi: 10.1074/jbc.272.38.23938. [DOI] [PubMed] [Google Scholar]
- 38.Vora A C, Grandgenett D P. Assembly and catalytic properties of retrovirus integrase-DNA complexes capable of efficiently performing concerted integration. J Virol. 1995;69:7483–7488. doi: 10.1128/jvi.69.12.7483-7488.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vora A C, McCord M, Fitzgerald M L, Inman R B, Grandgenett D P. Efficient concerted integration of retrovirus-like DNA in vitro by avian myeloblastosis virus integrase. Nucleic Acids Res. 1994;22:4454–4461. doi: 10.1093/nar/22.21.4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wei S Q, Muzuuchi K, Craigie R. A large nucleoprotein assembly at the ends of the viral DNA mediates retroviral DNA integration. EMBO J. 1997;16:7511–7520. doi: 10.1093/emboj/16.24.7511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wei S Q, Mizuuchi K, Craigie R. Footprints on the viral DNA ends in Moloney murine leukemia virus preintegration complexes reflect a specific association with integrase. Proc Natl Acad Sci USA. 1998;95:10535–10540. doi: 10.1073/pnas.95.18.10535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wolfe A L, Felock P J, Hastings J C, Uncapher Blau C, Hazuda D J. The role of manganese in promoting multimerization and assembly of human immunodeficiency virus type 1 integrase as a catalytically active complex on immobilized long terminal repeat substrates. J Virol. 1996;70:1424–1432. doi: 10.1128/jvi.70.3.1424-1432.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zheng R, Jenkins T M, Craigie R. Zinc folds the N-terminal domain of HIV-1 integrase, promotes multimerization, and enhances catalytic activity. Proc Natl Acad Sci USA. 1996;93:13659–13664. doi: 10.1073/pnas.93.24.13659. [DOI] [PMC free article] [PubMed] [Google Scholar]