Abstract
The Barbary macaque (Macaca sylvanus) is the earliest offshoot of the genus Macaca and the only extant African representative, all other species being Asiatic. Once distributed throughout North Africa, M. sylvanus is now restricted to isolated forest fragments in Algeria and Morocco. The species is threatened; the maximum total wild population size is estimated at 10,000 individuals. Relationships among surviving wild subpopulations in Algeria (96 samples) and Morocco (116 samples) were examined by using 468-bp sequences from hypervariable region I of the mitochondrial DNA control region. Twenty-four different haplotypes were identified, differing by 1-26 mutational steps (0.2-5.6%) and 1 insertion. With one exception (attributable to secondary introduction in coastal Morocco), Algerian and Moroccan haplotypes are clearly distinct. However, whereas Moroccan subpopulations show little divergence in hypervariable region I sequences and little correspondence with geographical distribution, there is a deep division between two main subpopulations in Algeria and one marked secondary division, with haplotypes generally matching geographical distribution. Accepting an origin of the genus Macaca of 5.5 million years ago, the Moroccan population and the two main Algerian subpopulations diverged ≈1.6 million years ago. Distinction between Moroccan and Algerian haplotypes permitted analysis of the origin of the Gibraltar colony of Barbary macaques (68 samples; 30% of the population). It is generally held that the present Gibraltar population descended from a dozen individuals imported during World War II. However, the Gibraltar sample was found to include Algerian and Moroccan haplotypes separated by at least 16 mutational steps, revealing a dual origin of the founding females.
Keywords: conservation, fragmentation, haplotype network, molecular clock, mtDNA
Population genetic studies describe genetic diversity within and among populations and contribute to knowledge about evolutionary mechanisms that change natural and artificial populations (1). Previous research has established crucial features of mtDNA, such as its relatively rapid sequence evolution, lack of recombination, and maternal inheritance (2). These properties make mtDNA particularly valuable for phylogenetic and phylogeographic reconstructions of relatively recent cladogenic events in species with female philopatry, such as macaques. Female philopatry involves a social system in which males disperse while females remain in their natal group. It is characterized by local homogeneity and large interpopulational differences in mtDNA (3).
Macaques are widely distributed in southern and eastern Asia as well as in limited areas of western North Africa. They are classified into at least 19 extant species, with the Barbary macaque (Macaca sylvanus) as the only extant African representative (4, 5). Barbary macaques live in multimale, multifemale groups with an approximately balanced adult sex ratio. Natural groups contain an average of ≈30 animals living in a highly promiscuous mating system (6). Because of its small numbers and major habitat reduction, M. sylvanus has been formally recognized as a vulnerable taxon by the World Conservation Union (2002 International Union for Conservation of Nature and Natural Resources Red List of Threatened Species).
After an inferred origin in northern Africa at least 5.5 million years ago (mya), early macaques presumably spread into Eurasia, and one lineage entered southern Europe by the beginning of the Pliocene (7-9). Fossils of Pliocene and Pleistocene macaques have been discovered throughout southerly regions of Europe and reliably identified as representatives of the M. sylvanus lineage (10-13). M. sylvanus apparently flourished in Europe only during the warm phases of the Pliocene and Pleistocene, persisting at least until the last interglacial and then eventually retreating from the continent entirely (7). The species was formerly widely distributed in the once entirely forested Maghreb, inhabiting diverse environments ranging from lowland shrub to high-altitude coniferous forests (14). The Maghreb has been continuously subjected to human influence for at least 4,000 years, and the most significant factor in extensive forest destruction has probably been overgrazing by goats. Two decades ago, the maximum population size of M. sylvanus was estimated at 20,000 individuals in a few refuges in Morocco and Algeria (14, 15) (map in Fig. 2). Since then, the wild population has apparently decreased to ≈10,000 (16). Remaining wild subpopulations are completely isolated in forest fragments separated by extensive intervening built-up areas, such that natural genetic exchange between them is ruled out.
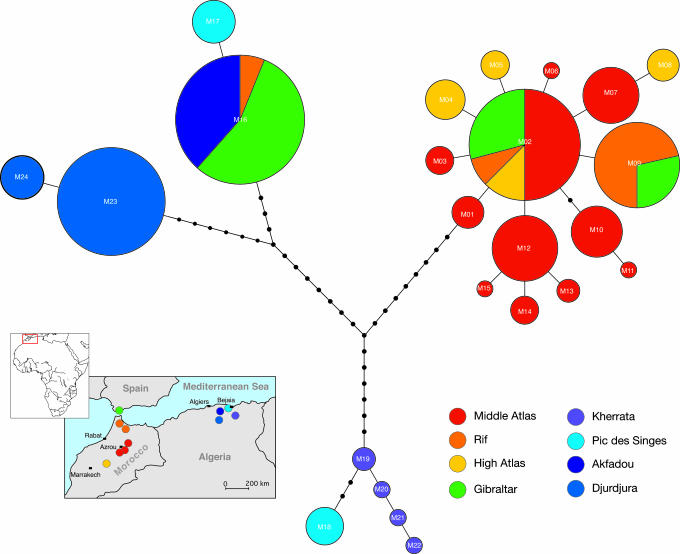
Fig. 2.
Haplotype network generated on the basis of the maximum-likelihood tree. Each line between points represents a single mutational step. A haplotype (numbers refer to Table 1) is represented by a circle whose size is proportional to the number of individuals showing that haplotype. Haplotypes are colored to match the respective population in the map. Note that the only observed insertion occurred between haplotypes M23 and M24.
Although there are now three major captive colonies of Barbary macaques in France (Kintzheim and Rocamadour) and Germany (Salem), the only long-established European population is the provisioned free-ranging colony on Gibraltar. The origin of the Gibraltar colony is unclear; some authors have postulated a remnant European population, whereas other anecdotes suggest that animals were imported from North Africa between 711 and 1492 A.D. However, the first written record of Barbary macaques on Gibraltar dates from 1704 (17). The population was subsequently subjected to artificial intervention to regulate group size over a long period. Numbers were at times kept low by removing individual animals and at others restocked with importations from North Africa. The last introduction of new stock occurred during the second World War, reportedly from Morocco (18).
The pronounced philopatric nature of female macaques has been shown in other macaque species to preserve multiple mitochondrial types and can lead to local fixation of deeply divergent lineages (5, 19). This pattern is reinforced by the ever increasing anthropogenic impacts on M. sylvanus in North Africa. Habitat destruction has not only reduced overall population size to a very low level but has also promoted marked isolation of the remaining subpopulations. Study of the genetic effects of population fragmentation is therefore of central importance for formulation of long-term conservation guidelines.
In this study, we present a phylogenetic and phylogeographic analysis of the Barbary macaques in North Africa, with special emphasis on the origin of the Gibraltar colony. We analyzed the most variable segment of the control region of mtDNA from seven wild-living populations in Algeria and Morocco and the semicaptive colony on Gibraltar. Our primary aim was to elucidate phylogeographic relationships among the individual Barbary macaque populations and to examine the extent to which habitat fragmentation has left its mark on mtDNA variation. Previous studies indicated reduced genetic variability relative to other macaque species (20). A linearized tree approach was applied, based on molecular clock calibrations, to estimate dates for divergence between lineages. We then reconstructed the geographical origin of the Gibraltar population, while recognizing that some lineages may have already become extinct.
Materials and Methods
Sample Collection. This study includes 90 and 96 samples from wild populations of Barbary macaques in Morocco and Algeria, respectively (map in Fig. 2 and Table 1). Samples from 26 additional early-generation individuals from the Moroccan Middle Atlas originated from outdoor enclosures in Germany and France. Sixty-eight samples were collected in Gibraltar from individually known animals living in six different social groups (≈30% of the total Gibraltar population).
Table 1.
Names of sample locations (see Fig. 2), sample sizes (N), and haplotype frequencies
| Population | N | M01 | M02 | M03 | M04 | M05 | M06 | M07 | M08 | M09 | M10 | M11 | M12 | M13 | M14 | M15 | M16 | M17 | M18 | M19 | M20 | M21 | M22 | M23 | M24 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gibraltar | 68 | 24 | 8 | 36 | |||||||||||||||||||||
| Rif | 28 | 4 | 20 | 4 | |||||||||||||||||||||
| Middle Atlas | 69 | 4 | 16 | 3 | 1 | 12 | 10 | 1 | 16 | 2 | 3 | 1 | |||||||||||||
| High Atlas | 19 | 6 | 6 | 3 | 4 | ||||||||||||||||||||
| Akfadou | 25 | 25 | |||||||||||||||||||||||
| Djurdjura | 54 | 45 | 9 | ||||||||||||||||||||||
| Pic d. Singes | 12 | 7 | 5 | ||||||||||||||||||||||
| Kherrata | 5 | 2 | 1 | 1 | 1 |
Collection of blood and hair samples was carried out as far as possible by trapping animals in baited cages and sedating them (Algeria and Gibraltar). When trapping was not possible, fecal samples were collected (Morocco). For the Algerian population, samples were collected from four of the seven remaining wild subpopulations (Akfadou, Djurdjura, Kherrata, and Pic des Singes) and from individual groups at eight different localities (five social groups at Djurdjura and single groups at the other three localities). Samples from Morocco originate from three different areas of distribution, namely from the Rif Mountains, from the Azrou Forest in the Middle Atlas, and from Cascade d'Ouzoud and Ourika Valley in the High Atlas.
Laboratory Methods. DNA was extracted from hair and blood samples by using PCI (25:24:1 mix of phenol, chloroform, and isoamyl-alcohol) and chloroform (21). From fecal samples, DNA was extracted with the QIAamp DNA stool kit (Qiagen) according to the manufacturer's protocol. Initially, a fragment of ≈1,200 bp of the mtDNA was amplified from blood samples and a shorter fragment of 500 bp was amplified from fecal and hair samples. This was then used to sequence 468 nt of hypervariable region I of the control region. PCR amplifications were performed in a 100-μl final volume by using 0.06 M Tris, 0.015 M (NH4)2SO4, 0.0015 M MgCl2, 0.78 M dimethyl sulfoxide (DMSO), 0.025 mM each dNTP, 1 mM each primer, and 2.5 units of Promega Taq polymerase in an Applied Biosystems 9700 thermal cycler. The primers used were M2Pro (5′-TCC ACC ACC AAC ACC CAA AGC-3′), M282 (5′-AAG GCC AGG ACC AAG CCT-3′), and DLR3 (5′-GAA CCA GAT GTC GGA TAC AG-3′) as an internal primer for the sequencing reaction. The reaction cycles consisted of denaturation for 30 s at 95°C, primer annealing for 60 s at 50°C, and extension for 60 s at 72°C. Cycles were repeated 35 times, with the final cycle including an extension reaction of 5 min. The PCR products were purified with the QIAquick PCR purification kit (Qiagen) according to the supplier's instructions. Cycle sequencing reactions were performed by using BigDye Terminator cycle sequencing chemistry (Applied Biosystems). Sequencing products were purified by Sephadex G-50 in CentriSep columns (Princeton Separations), and sequences were analyzed by using an Applied Biosystems 377 sequencing system. All templates were sequenced for both strands.
Phylogenetic and Population Genetic Analyses. Sequences were aligned by using the program sequencher (Gene Codes, Ann Arbor, MI). For phylogenetic analyses, we performed maximum-likelihood, maximum-parsimony, and neighbor-joining analyses in parallel by using the program paup* 4.0b10 (22). modeltest 3.06 (23) was run to determine the appropriate model of molecular evolution in a likelihood ratio test framework. Six representatives of three of the four major Macaca lineages (Macaca tonkeana, M. nigra, M. nemestrina, M. mulatta, M. fascicularis, and M. arctoides) were taken collectively as the outgroup (4, 24). Gaps were treated as a fifth state in parsimony analyses. Bootstrap analyses were performed with 5,000 replicates for maximum parsimony and neighbor-joining; for maximum likelihood, we performed 100 full heuristic bootstrap replicates. For Bayesian inference, we used the program mrbayes v.3.0b4 (25). Four Monte Carlo Markov chains were run for 100,000 generations, sampling every 10 generations; the initial 5% of trees were discarded as burn-in.
To infer timing of major branching events for the Barbary macaques, we performed molecular clock analysis, based on a maximum-likelihood tree in which the molecular clock was enforced. Average uncorrected pairwise distances between and among the four major lineages were calculated to infer absolute time scales, based on an inferred minimum date for the initial divergence of M. sylvanus from other macaque species at ≈5.5 mya (7, 8, 19). To examine relationships between the haplotypes observed in the 286 specimens sequenced, we applied a haplotype network approach. To this end, we translated the maximum-likelihood topology into an unrooted network, describing the optimal maximum-likelihood topology under the maximum parsimony criterion to map the number of mutations assigned to each branch of the network.
Hierarchical analysis of molecular variance (AMOVA) (26) was performed to compare levels of genetic diversity within and among four principal Barbary macaque lineages by using arlequin 2.0 (27) and 1,000 permutations. Both the Gibraltar population and the Rif group at locality 2 (M16) were excluded from this analysis because they involve artificially introduced animals. For AMOVA, the remaining wild populations were arranged according to geographical distribution into seven groups, three from Morocco and four from Algeria (Table 1). A mismatch analysis (28) was then performed with arlequin 2.0 to compare the demographic histories of all lineages according to the phylogenetic analysis. Finally, the moment estimator of time to expansion (τ) was used in the equation τ = 2μt to infer a time scale for the demographic expansion (where t is the coalescent time in generations, which corresponds to the time of the last common ancestor, and μ is the mutation rate for the whole sequence), assuming an average generation time of 5 years (29).
Results
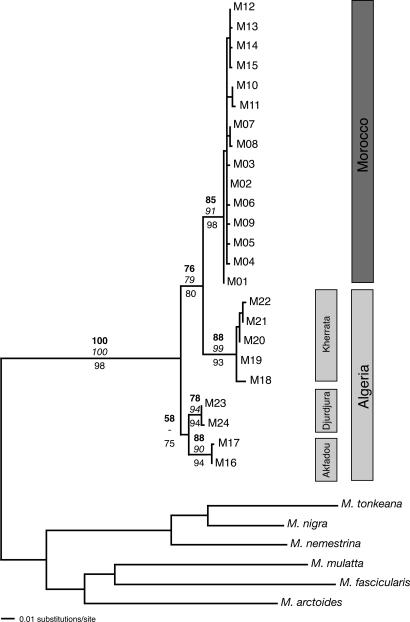
Phylogenetic Analyses. Using collapse 1.1 (30), we identified 24 different haplotypes within the entire sample of Barbary macaques, differing by 1-26 mutational steps (0.21-5.6%). Haplotype M24, shared by nine individuals of a single social group in Djurdjura, showed an insertion at position 155 in our alignment. Of the total 468 nucleotide positions, 116 were parsimony-informative when including the six outgroup taxa and 35 without the outgroup taxa. The HKY+G model (31) was selected by modeltest as the best-fitting model, with a gamma shape correction of 0.21, a transition/transversion ratio of 7.94, and base frequencies of A = 0.308, C = 0.298, and G = 0.129 (the parameters being estimated from the data). All phylogenetic analyses performed resulted in almost identical tree topologies (Fig. 1), identifying four main clusters among the Barbary macaques. The sole Akfadou haplotype (M16) and one aberrant haplotype from Pic des Singes (M17) clustered with the two haplotypes found in Djurdjura (M23/M24). This clade was resolved as sister group to a clade containing haplotypes from Kherrata/Pic des Singes (M18-M22) and Morocco (M1-M15). Generally, there were high bootstrap supports in all analyses, as well as high Bayesian posterior probabilities with respect to the branching order of the lineages.
Fig. 1.
Maximum-likelihood tree for all 24 haplotypes of Barbary macaques and for 6 outgroup taxa. Bootstrap percentages recorded for maximum-likelihood full heuristic search (100 replicates) and maximum parsimony trees (5,000 replicates with fast stepwise addition option) are shown above major branches in bold and italics, respectively. Bayesian posterior probability values are presented below internodes. The haplotype numbers refer to those in Table 1. The population of Pic des Singes is divided between the Akfadou and Kherrata clades.
We calculated average uncorrected pairwise distances between principal lineages in the genus Macaca for a molecular clock calibration, using the minimal initial divergence time of 5.5 mya based on the fossil record (7, 19). The average pairwise distance resulted in a mutation rate μ of 2.70-3.04% (see Table 4, which is published as supporting information on the PNAS web site). According to the average pairwise distances obtained, the initial split between the Morocco/Kherrata clade and the Algerian Akfadou/Djurdjura subclade was dated at 1.24-1.85 mya, and the separation between Morocco and Kherrata at 1.22-1.62 mya. The basal split among individuals assigned to the Akfadou/Djurdjura subclade was estimated at 0.90 mya. Distances within groups were lower, with estimated maximal divergence times of 64-359 thousand years ago (kya) for the Kherrata subpopulation, 110-282 kya for the Moroccan clade, and about 70-78 kya for the Akfadou and Djurdjura subpopulations. Although the absolute timing of divergences may be questionable [notably because direct calibration from the fossil record may lead to serious underestimation (8)], at least the sequence and relative timing depicted here convincingly reflect the evolutionary history of Barbary macaques.
Population Genetic Analyses. In the haplotype network (Fig. 2), three main haplotype clusters emerged. The most abundant haplotype M16, shared by 65 individuals, was found in three different sampling areas, and constitutes [together with one Pic des Singes haplotype found in seven individuals (M17)] a cluster with the Algerian subclade Djurdjura. In the Djurdjura subclade, haplotype M23 was found in all except one social group, which shared haplotype M24. Another Algerian group was resolved including the Kherrata representatives (M19-M22) and a haplotype found in five individuals of Pic des Singes (M18). Within the Moroccan cluster, diversity was largest in the Middle Atlas population, reflected both by the number of haplotypes and the distances between them. The most abundant and widespread Moroccan haplotype M02 is connected by one step to the ancestral haplotype M01, found only in the Middle Atlas. Four haplotypes of the Moroccan clade were restricted to only two geographical populations (with Gibraltar excluded as a nonwild population), namely the Rif Mountains (M09) and the High Atlas (M04-M05/M08), respectively. The Gibraltar population was resolved into three different haplotypes, clearly suggesting a Moroccan (M02/M09) and Algerian (M16) origin. The Algerian haplotype M16 is separated from the Moroccan haplotype M02 by 22 mutational steps and from the other Moroccan haplotype M09 by 23 steps.
AMOVA and Mismatch Analyses. To quantify population genetic structure within and among populations, a hierarchical AMOVA was performed, grouping all but the Gibraltar population and the presumably introduced social group in the Rif (with Algerian haplotype M16) according to geographical distribution (Table 2). AMOVA revealed significant levels of genetic structuring. In total, 61.1% of the genetic variance was partitioned among the four mtDNA phylogroups, whereas 31.4% of the molecular variance resulted from differences among populations within the groups, and only 7.5% accounted for variance within the populations. The mean pairwise value of ΦST was 0.92, indicating a low level of gene flow and genetically isolated populations. On the whole, the high levels of significance show that the inferred genetic structure is representative of the sampled populations. A mismatch analysis was performed to compare the demographic histories of the four major lineages. The distribution differed substantially among the lineages, with the Kherrata population showing the oldest expansion signals (Table 3), followed by the Djurdjura population. The Morocco and Akfadou populations showed the most recent expansion. The values of θ0 and θ1 were similar to each other, and only the sample of Morocco (θ0 = 0.0; θ1 = 3,338) showed instances of sudden population growth at ≈60 kya. The chart of the mismatch distribution for the total sample was multimodal (see Fig. 3, which is published as supporting information on the PNAS web site), with one peak corresponding to differences between the four lineages, one peak corresponding to differences between Akfadou and Djurdjura, and the third peak corresponding to differences within populations.
Table 2. Results of hierarchical AMOVA.
| Hierarchical structure | Source of variation | Sum of squares (df) | Variance component | P | Fixation index | Percent variation |
|---|---|---|---|---|---|---|
| 1 | Among groups | 777.4 (1) | Va = 6.16 | <0.00001 | ΦCT = 0.61 | 61.1 |
| 2 | Among populations | 378.1 (5) | Vb = 3.16 | <0.00001 | ΦSC = 0.81 | 31.4 |
| 3 | Within populations | 150.9 (201) | Vc = 0.75 | <0.05 | ΦST = 0.92 | 7.5 |
Structure 1, two groups, Morocco and Algeria; structures 2 and 3, all seven populations.
Table 3. Results of the mismatch analyses.
| Clade | τ | Obs. | θo | θ1 | Ragged. | texpansion, kyr |
|---|---|---|---|---|---|---|
| Kherrata | 4.79 (1.34-12.29) | 2.69 | 0.0 (0.0-6.3) | 5.1 (1.3-3890) | 0.101 | 194 (54-498) |
| Morocco | 1.56 (0.76-1.89) | 1.43 | 0.0 (0.0-0.9) | 3338 (18.62-6699) | 0.821 | 63 (31-76) |
| Djurdjura | 3.20 (0.45-4.70) | 0.57 | 0.0 (0.0-0.8) | 0.4 (0.0-1749) | 0.674 | 130 (18-191) |
| Akfadou | 0.28 (0.001-0.91) | 0.18 | 0.0 (0.0-0.3) | 0.5 (0.0-2399) | 0.446 | 11 (0.040-37) |
The expansion parameter (τ), the mismatch observed mean (Obs.), the mutation parameter before (θo) and after (θ1) expansion, and the raggedness index (Ragged.) (48) were calculated for all four major groups of haplotypes according to obtained phylogenies. Values in brackets refer to 95% confidence intervals. The timing of the most important demographic expansion (texpansion) was determined by assuming a mutation rate of 2.90% per million years, based on the results of the average uncorrected pairwise distances. See Fig. 3 for graphs.
Discussion
Phylogenetic Reconstruction. mtDNA data revealed significant genetic structuring among Barbary macaques, showing deep genetic divergence between some lineages (Fig. 1). For the Algerian lineages, this finding concords with a previous study based on microsatellites (32). Of particular interest is the initial split occurring at ≈1.6 mya, dividing the Moroccan clade and one major Algerian clade from a second major Algerian clade (Fig. 1). This result is surprising in that Morocco is sister to the Algerian clade (Kherrata/Pic des Singes) that currently shows the greatest geographical separation. This finding might be explained by climatic oscillations influencing patterns of geographic distribution in the past. Otherwise, the tree reconstruction corresponds to geographic relationships, with the single exception of the locality of Pic des Singes. The large mtDNA difference shown by M17/M18 and the haplotypic affiliation to neighboring populations could have resulted from recent isolation of Pic des Singes from previously connected lineages. This is an interesting case, indicating that the single social group in Pic des Singes contains two completely distinct lineages. The results are consistent with the findings that today the small subpopulation of 50 animals inhabits only a relict fragment of a once widespread oak forest (14). Microsatellite data (32) also support a relatively recent separation of the Pic des Singes population from that at Kherrata. Furthermore, the Kherrata population displays the greatest variability despite having the smallest sample size. Similar findings have been reported in a previous study of microsatellites, in which Kherrata was found to have the highest number of private alleles in one social group (32). However, the presence of three markedly divergent mtDNA lineages in Algeria suggests that the observed geographical patterns probably arose through extinction of intermediate populations and physical barriers to dispersal between regions.
Surprisingly, all Moroccan haplotypes are grouped into a fully supported monophyletic clade, in contrast to the paraphyletic Algerian populations. Phylogenetic analyses failed to resolve the Moroccan clade into its three geographical populations, revealing very close relationships among them. The apparently oldest haplotype M01 is exclusively located in the Middle Atlas, from which the central and most abundant haplotype M02 derives. In addition to the Middle Atlas, M02 also occurs in the Rif Mountains and in the High Atlas. These two localities each have one (M09) and three (M04-M05/M08) private haplotypes, respectively, in addition to the central haplotype M02 shared with the Middle Atlas. This presumably indicates that M02 expanded from this region northward into the Rif and southward into the High Atlas.
Phylogeography from the Perspective of Glacial Events. Pleistocene climatic changes have shaped patterns of geographical distribution as well as demographic history and genetic diversification (33). For most species, repeated climatic oscillations resulted in range contractions into separate refugia during climatic cooling and drying (34). The coalescent theory (35) predicts that the most widespread and frequent haplotypes indicate the locations of refugial populations. According to the inferred coalescent time for Kherrata and Morocco, haplotypes diverged in isolation ≈213 kya and 196 kya, before and during the onset of the Riss glaciation. Taking into account the fact that the most frequent and widespread haplotype M02 is present in Morocco, one may speculate that both Morocco and Kherrata (Algeria) were refuges for Barbary macaques during the last glaciation. Barbary macaques from Akfadou and Djurdjura might have diverged from a different lineage ≈74 kya during the interstadials when climatic conditions were still moderate (36). By the mid-Holocene, habitats of M. sylvanus in Morocco and Algeria might have been interconnected through extended forests until the beginning of forest exploitation by human populations, dated ≈2 kya (37). There is reason to believe that the North African forest was heavily depleted in Roman times as a result of exportation of cedars and oaks. The Arab conquest and immigration of nomads intensified the destruction, with the result that by the end of the 17th century most of the lowlands had been cleared (38). The lack of forests probably presented an obstacle to migration, leading to the geographical separation of the mtDNA haplotypes that we see today. These patterns are reinforced by the marked philopatric nature of female macaques, causing sharp geographical clustering of mtDNA haplotypes (3). Low levels of control region diversity have been seen not only in species with pronounced female philopatry, but also in species known to have undergone population bottlenecks in the recent past (39-41). It has also been argued that reduced genetic variability in M. sylvanus compared with other macaques might be the result of successive bottlenecks during withdrawal from a wider Pleistocene expansion to contemporary areas of dispersal (42). The model of sudden expansion for the Moroccan population is supported by the bell-shaped mismatch distribution (see Fig. 3), as well as by the star-like structure of the haplotype network and its short branches (Fig. 2) (43). Moroccan populations perhaps underwent sudden expansion ≈63 kya, starting from one matriline in the Middle Atlas and extending northward and southward (Table 3). This inference fits with the more favorable climate during this interstadial that permitted some expansion of Mediterranean oak forests (36). Akfadou and Djurdjura showed a skewed distribution due to excess of similar or identical haplotypes, which can arise in two ways. First, it is possible that recent expansion of a young population occurred. If so, then the two subpopulations might have suffered a bottleneck in the past or were recolonized only recently. Alternatively, it is possible that sampling was conducted within the geographical boundaries of a single genetic stock. This can be excluded at least for Djurdjura, where sampling took place in distinct geographical areas. For Kherrata, no expansion model can be derived because of the small sample number.
The results for mtDNA found in this study of M. sylvanus are in good agreement with sharp geographical clustering found in M. mulatta, M. maura, and M. tonkeana (3, 44) and with limited mtDNA variation within populations of M. mulatta, M. sinica, and M. fascicularis (3, 45, 46). This conformity could stem both from the social system of female philopatry in macaques and the persistent geographical isolation of some populations in macaque species generally.
The Origin of the Gibraltar Colony. Repeated introduction of animals and the lack of reliable data concerning founders of the present-day population has obscured the origin of the Gibraltar colony. The fact that all extant Gibraltarian mtDNA haplotypes were also found in North Africa, combined with the lack of fossil evidence of M. sylvanus in Gibraltar at the end of the last glaciation, greatly diminishes the possibility that the Gibraltar macaques represent or include any remnant of the original European population. According to haplotype distribution, the most probable sources of the extant Gibraltar colony are populations in both Morocco and Algeria. Because of the large number of intervening mutational steps, the present population must descend from individuals bearing at least one haplotype (M16) originating from Akfadou and two (M02/M09) from the Rif Mountains of Morocco. Another possible origin of M02 could be the Middle Atlas, but this is relatively unlikely because of the much greater distance from Gibraltar. To restrict population size on Gibraltar, removal of animals by culling has long been [and unfortunately still is (47)] the predominant approach. But the unexpected finding of one social group in the Rif with an Algerian haplotype (M16) highlights an alternative possibility. It is conceivable that surplus animals were exported from Gibraltar to the Rif, or were perhaps introduced directly from Algeria, although we cannot exclude the remote possibility of a surviving relict population. Given the fact that more ancient Algerian haplotypes would be expected in the Rif in the latter case, and given that this single social group shares the same haplotype M16 with Gibraltar and Akfadou, it is more likely that animals were reintroduced from Gibraltar or translocated from Algeria.
Conclusion
Analysis of mtDNA reveals overall correspondence with current geographical distribution of Barbary macaques in Algeria and Morocco and clearly reflects the history of the species in these regions. It is notable that the Algerian population shows considerably greater diversity than the Moroccan population. Clear distinction between Algerian and Moroccan haplotypes permits attribution of the Gibraltar colony to founders from both regions. Within recent history, the wild Barbary macaque population in both Algeria and Morocco has declined steeply, becoming fragmented as a result of habitat loss. Given that all extant populations of M. sylvanus are genetically distinct, future guidelines should aim at overall protection of existing populations, but conservation efforts are still in their infancy. It is hoped that this phylogenetic and phylogeographic study will contribute to a sound basis for future management of wild populations of the species.
Supplementary Material
Acknowledgments
We thank F. Botte-von Segesser, N. Ménard, J. Pastorini, W. Scheffrahn, and D. Vallet for providing samples from Algeria; M. Bruford, K. Hodges, J. Pastorini, U. Möckli, U. Möhle, C. Roos, and H. Zischler for help in sample collection and laboratory work; and the Gibraltar Ornithological and Natural History Society and the Gibraltar Veterinary Clinic for research permission and collaboration. This work was supported by a grant from the A. H. Schultz Foundation and by Swiss National Science Foundation Grants 5001-034878 and 3100-045923 (to R.D.M.).
Author contributions: R.D.M. and L.M. designed research; L.M. performed research; L.M. and W.S. analyzed data; and L.M. and R.D.M. wrote the paper.
Abbreviations: AMOVA, analysis of molecular variance; kya, thousand years ago; mya, million years ago.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. DQ001976-DQ002005).
References
- 1.Page, R. D. M. & Holmes, E. C. (1998) Molecular Evolution: A Phylogenetic Approach (Blackwell Scientific, London).
- 2.Brown, W. M., George, M. & Wilson, A. C. (1979) Proc. Natl. Acad. Sci. USA 76, 1967-1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Melnick, D. J. & Hoelzer, G. A. (1992) Int. J. Primatol. 13, 379-393. [Google Scholar]
- 4.Fooden, J. (1980) in The Macaques: Studies in Ecology, Behavior, and Evolution, ed. Lindburg, D. G. (Van Nostrand Reinhold, New York), pp. 1-9.
- 5.Hayasaka, K., Fujii, K. & Horai, S. (1996) Mol. Biol. Evol. 13, 1044-1053. [DOI] [PubMed] [Google Scholar]
- 6.Ménard, N., Scheffrahn, W., Vallet, D., Zidane, C. & Reber, C. (1992) in Paternity in Primates: Genetic Tests and Theories, eds. Martin, R. D., Dixson, A. F. & Wickings, E. J. (Karger, Basel), pp. 155-174.
- 7.Delson, E. (1980) in The Macaques: Studies in Ecology, Behavior, and Evolution, ed. Lindburg, D. G. (Van Nostrand Reinhold, New York), pp. 10-30.
- 8.Tavaré, S., Marshall, C. R., Will, O., Soligo, C. & Martin, R. D. (2002) Nature 416, 726-729. [DOI] [PubMed] [Google Scholar]
- 9.Martin, R. D. (1990) Primate Origins and Evolution (Chapman & Hall/Princeton Univ. Press, London/Princeton).
- 10.Koehler, M., Moyà-Solà, S. & Alba, D. M. (2000) J. Hum. Evol. 38, 447-452. [DOI] [PubMed] [Google Scholar]
- 11.Fladerer, F. A. (1991) Z. Saugetierkd. 56, 272-283. [Google Scholar]
- 12.Singer, R., Wolff, R. G., Gladfelter, B. G. & Wymer, J. J. (1982) Folia Primatol. 37, 141-152. [DOI] [PubMed] [Google Scholar]
- 13.Rook, L., Mottura, A. & Gentili, S. (2001) J. Hum. Evol. 40, 187-202. [DOI] [PubMed] [Google Scholar]
- 14.Taub, D. M. (1977) Folia Primatol. 27, 108-133. [DOI] [PubMed] [Google Scholar]
- 15.Taub, D. M. (1984) in The Barbary Macaque: A Case Study in Conservation, ed. Fa, J. E. (Plenum, New York), pp. 71-78.
- 16.Camperio Ciani, A. (2003) Antropol. Mediterran. 1, 57-68. [Google Scholar]
- 17.Fa, J. E. (1981) Oryx 16, 43-46. [Google Scholar]
- 18.Fa, J. E. (1984) in The Barbary Macaque: A Case Study in Conservation, ed. Fa, J. E. (Plenum, New York), pp. 263-306.
- 19.Tosi, A. J., Morales, J. C. & Melnick, D. J. (2003) Evolution (Lawrence, Kans.) 57, 1419-1435. [DOI] [PubMed] [Google Scholar]
- 20.Pastorini, J., Epplen, J. T. & Martin, R. D. (2000) Folia Primatol. 71, 161-168. [DOI] [PubMed] [Google Scholar]
- 21.Sambrook, J., Fritsch, E. & Maniatis, T. (1989) Molecular Cloning, A Laboratory Manual (Cold Spring Harbor Lab. Press, Cold Spring Harbor, NY).
- 22.Swofford, D. L. (2002) paup*: Phylogenetic Analysis Using Parsimony (*and Other Methods) (Sinauer, Sunderland, MA), Version 4.0b10.
- 23.Posada, D. & Crandall, K. A. (1998) Bioinformatics 14, 817-818. [DOI] [PubMed] [Google Scholar]
- 24.Tosi, A. J. (2000) Mol. Phylogenet. Evol. 17, 133-144. [DOI] [PubMed] [Google Scholar]
- 25.Huelsenbeck, J. P. & Ronquist, F. (2001) Bioinformatics 17, 754-755. [DOI] [PubMed] [Google Scholar]
- 26.Excoffier, L., Smouse, P. E. & Quattro, J. M. (1992) Genetics 131, 479-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schneider, S., Roessli, D. & Excoffier, L. (1999) arlequin (Genetics and Biometry Laboratory, Univ. of Geneva, Geneva), Version 2.000.
- 28.Schneider, S. & Excoffier, L. (1999) Genetics 152, 1079-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ménard, N. & Vallet, D. (1993) Int. J. Primatol. 14, 479-500. [Google Scholar]
- 30.Posada, D. (1999) collapse (Department of Zoology, Brigham Young Univ., Provo, UT), Version 1.1.
- 31.Hasegawa, M., Kishino, H. & Yano, T. (1985) J. Mol. Evol. 22, 160-174. [DOI] [PubMed] [Google Scholar]
- 32.Von Segesser, F., Ménard, N., Gaçi, B. & Martin, R. D. (1999) Mol. Ecol. 8, 433-442. [DOI] [PubMed] [Google Scholar]
- 33.Avise, J. C. (2000) Phylogeography (Harvard Univ. Press, Cambridge, MA).
- 34.Hewitt, G. M. (1996) Biol. J. Linn. Soc. 58, 247-276. [Google Scholar]
- 35.Crandall, K. A. & Templeton, A. R. (1993) Genetics 134, 959-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hooghiemstra, H., Stalling, H., Agwu, C. O. C. & Dupont, L. M. (1992) Rev. Palaeobot. Palyno. 74, 1-53. [Google Scholar]
- 37.Lamb, H. F., Eicher, U. & Switsur, V. R. (1989) J. Biogeogr. 16, 65-74. [Google Scholar]
- 38.Mikesell, M. W. (1960) Science 132, 441-448. [DOI] [PubMed] [Google Scholar]
- 39.Barrat, E. M., Gurnell, J., Malarky, G., Deaville, R. & Bruford, M. W. (1999) Mol. Ecol. 8, S55-S63. [DOI] [PubMed] [Google Scholar]
- 40.Li, H., Meng, S.-J., Men, Z.-M., Fu, Y. & Zhang, Y.-P. (2003) Genetics 164, 269-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Randi, E., Tabaroni, C., Rimondi, S., Luchini, V. & Sfougaris, A. (2003) Mol. Ecol. 12, 2201-2214. [DOI] [PubMed] [Google Scholar]
- 42.Scheffrahn, W., Ménard, N., Vallet, D. & Gaçi, B. (1993) Primates 34, 381-394. [Google Scholar]
- 43.Tajima, F. (1989) Genetics 123, 585-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Evans, B. J., Supriatna, J. & Melnick, D. J. (2001) Evolution (Lawrence, Kans.) 55, 1686-1702. [DOI] [PubMed] [Google Scholar]
- 45.Hoelzer, G. A., Dittus, W. P. J., Ashley, M. V. & Melnick, D. J. (1994) Mol. Ecol. 3, 451-458. [DOI] [PubMed] [Google Scholar]
- 46.Perwitasari-Farajallah, D., Kawamoto, Y., Kyes, R. C., Lelana, R. P. A. & Sajuthi, D. (2001) Primates 42, 141-153. [Google Scholar]
- 47.Schiermeier, Q. (2003) Nature 426, 111. [DOI] [PubMed] [Google Scholar]
- 48.Harpending, H. (1994) Hum. Biol. 66, 591-600. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.