Mutations in the chromatin reader ENL drive AML by altering chromatin modifications and gene expression through condensate formation, the disruption of which via mutagenesis or with a small molecule blocks leukemogenesis, revealing a novel therapeutic strategy.
Abstract
Gain-of-function mutations in the histone acetylation “reader” eleven-nineteen-leukemia (ENL), found in acute myeloid leukemia (AML) and Wilms tumor, are known to drive condensate formation and gene activation in cellular systems. However, their role in tumorigenesis remains unclear. Using a conditional knock-in mouse model, we show that mutant ENL perturbs normal hematopoiesis, induces aberrant expansion of myeloid progenitors, and triggers rapid onset of aggressive AML. Mutant ENL alters developmental and inflammatory gene programs in part by remodeling histone modifications. Mutant ENL forms condensates in hematopoietic stem/progenitor cells at key leukemogenic genes, and disrupting condensate formation via mutagenesis impairs its chromatin and oncogenic function. Moreover, treatment with an acetyl-binding inhibitor of the mutant ENL displaces these condensates from target loci, inhibits mutant ENL-induced chromatin changes, and delays AML initiation and progression in vivo. Our study elucidates the function of ENL mutations in chromatin regulation and tumorigenesis and demonstrates the potential of targeting pathogenic condensates in cancer treatment.
Significance: A direct link between ENL mutations, condensate formation, and tumorigenesis is lacking. This study elucidates the function and mechanism of ENL mutations in leukemogenesis, establishing these mutations as bona fide oncogenic drivers. Our results also support the role of condensate dysregulation in cancer and reveal strategies to target pathogenic condensates.
Introduction
Precise control of gene expression is critical for normal development and tissue homeostasis; accordingly, disruptions to this process can drive various diseases, notably cancer (1). Achieving proper gene expression requires the coordinated assembly and function of numerous proteins at specific genomic loci. While still not fully understood, one emerging mechanism underlying this coordination is through the formation of dynamic, locally concentrated assemblies known as transcriptional condensates or hubs (2–8). These assemblies form through a combination of high-affinity “lock and key” interactions and multivalent, nonstochiometric weak interactions involving proteins and nucleic acids (8–11). Here, we use “condensates” to refer to such assemblies, making no assumption regarding the process by which they form or their biophysical properties (12). It has been proposed that multivalent interactions underlying condensate formation can enhance transcriptional activation in a phase separation–dependent (2, 3, 7, 13, 14) or –independent manner (4, 15–17). Although research has unveiled an increasing number of gene regulatory proteins with the potential to form condensates in reconstitution and cellular assays, the biological function of transcriptional condensates in physiologically relevant, in vivo contexts remains less clear.
Accumulating evidence has suggested that disease-associated mutations in gene regulatory proteins could perturb the formation or properties of transcriptional condensates, raising the exciting possibility that condensate dysregulation contributes to disease phenotypes (4, 18–31). In the context of cancer, recent studies have linked dysregulation of transcriptional condensates to various malignancies, including Ewing sarcoma and AML. Oncogenic fusion proteins, such as EWS–FLI1 and NUP98–HOXA9, often result from the fusion of a phase separation prone intrinsically disordered region (IDR) to a transcription factor DNA-binding domain, enabling them to form condensates in both biochemical assays and in cells. The transformation ability of EWS–FLI1 and NUP98–HOXA9 is compromised when their IDRs are mutated or deleted (4, 20–23). Additionally, cancer-associated mutations in the histone demethylase UTX has been shown to disrupt condensate formation and abolish its tumor suppressive function (25). While these and other findings highlight potential links between condensate formation and cancer, further investigation is needed to understand how cancer-associated mutations that modulate condensate behaviors function to drive tumorigenesis in clinically relevant models. Furthermore, exploring the therapeutic targeting of these mechanisms may unveil new avenues for cancer treatment.
In this study, we explore these important questions using gain-of-function mutations found in the chromatin regulator, ENL. ENL, also known as MLLT1, is a chromatin reader protein that recognizes histone acetylation at actively transcribed gene promoters through its well-conserved YEATS (Yaf9, ENL, AF9, Taf14, Sas5) domain (32, 33). Upon binding to acetylation marks, ENL recruits co-factors such as the super elongation complex (SEC/P-TEFb) and DOT1L (34–38) to promote productive transcription elongation (32, 33). ENL has been implicated in cancers through various mechanisms. It is frequently fused with the mixed lineage leukemia protein (MLL1 or KMT2A) through chromosomal translocation, resulting in MLL–ENL fusion proteins that are potent drivers of leukemias associated with poor prognosis (39, 40). Moreover, wild-type (WT) ENL supports oncogenic gene expression programs necessary for the maintenance of a subset of AMLs, particularly those harboring MLL fusions or NPM1 mutations (32, 33, 41). A small-molecule inhibitor targeting the acetyl-binding YEATS domain of ENL has recently been shown to exhibit anticancer efficacy in preclinical models for these subsets of AML (41). More recently, a series of hotspot mutations in the ENL YEATS domain, characterized by small insertions or deletions within the same region of the protein, have been found in Wilms tumor and AML (19, 42–46). Our previous studies in HEK293 cells examined the function of eight of these mutations, and the results showed that each mutation enhances the chromatin occupancy and transcriptional activation activity of ENL on selected target genes (19, 31). ENL mutants, but not WT, form discrete condensates at endogenous genomic targets, and disrupting the formation or properties of these condensates abolishes ENL mutant-induced gene activation in cellular systems (31). The high specificity and gain-of-function nature of these mutations make them a powerful model for exploring how condensate dysregulation contributes to cancer development.
Currently, it is unknown how ENL mutations drive tumorigenesis in vivo and whether their condensate formation property contributes to this function. By creating a conditional knock-in mouse model for the most prevalent ENL YEATS domain mutation found in cancer (42, 43), referred to as ENL-T1, we show that this mutant ENL is a bona fide oncogenic driver for AML. Heterozygous expression of mutant ENL in the hematopoietic system gives rise to highly aggressive AML in mice, and it does so by perturbing the normal hematopoietic hierarchy and causing aberrant expansion of myeloid progenitors. The mutant ENL induces dynamic changes of H3K27ac and H3K27me3 histone modifications during cell differentiation and drives leukemic gene expression programs implicated in human AML. Notably, physiologically relevant levels of mutant ENL forms condensates at key leukemogenic gene loci, including Meis1 and Hoxa cluster genes, in hematopoietic stem and progenitor cells (HSPC). Disrupting condensate formation through targeted mutagenesis impairs both the chromatin function and oncogenic activity of mutant ENL. Finally, blocking the acetyl-binding activity of mutant ENL via a small-molecule inhibitor displaces these condensates from their target loci, dampens their effects on histone modifications and gene activation, and delays the initiation and progression of AML in vivo. This study provides new insights into AML pathogenesis by revealing the biological function and therapeutic potential of ENL mutations. It also offers crucial in vivo evidence that supports a role of condensate dysregulation in cancer.
Results
Heterozygous Enl Mutation Drives Aggressive AML in Mice
Using HEK293 as a cellular model, our previous study has shown that eight hotspot mutations clustered in the ENL YEATS domain (referred to as T1–T8; Supplementary Fig. S1A) identified in Wilms tumor and AML (19, 42–46) enhance the transcriptional activation activity of ENL. Moreover, these ENL mutants but not the WT form submicron-sized condensates at select genomic targets, and perturbing these condensates via targeted mutagenesis abolishes ENL mutation-induced gene activation (19, 31). These findings establish ENL mutations as a pioneer example in which gain-of-function mutations in a transcriptional regulator drive gene hyperactivation through condensate formation. However, whether and how such a property drives tumorigenesis in vivo remains unknown.
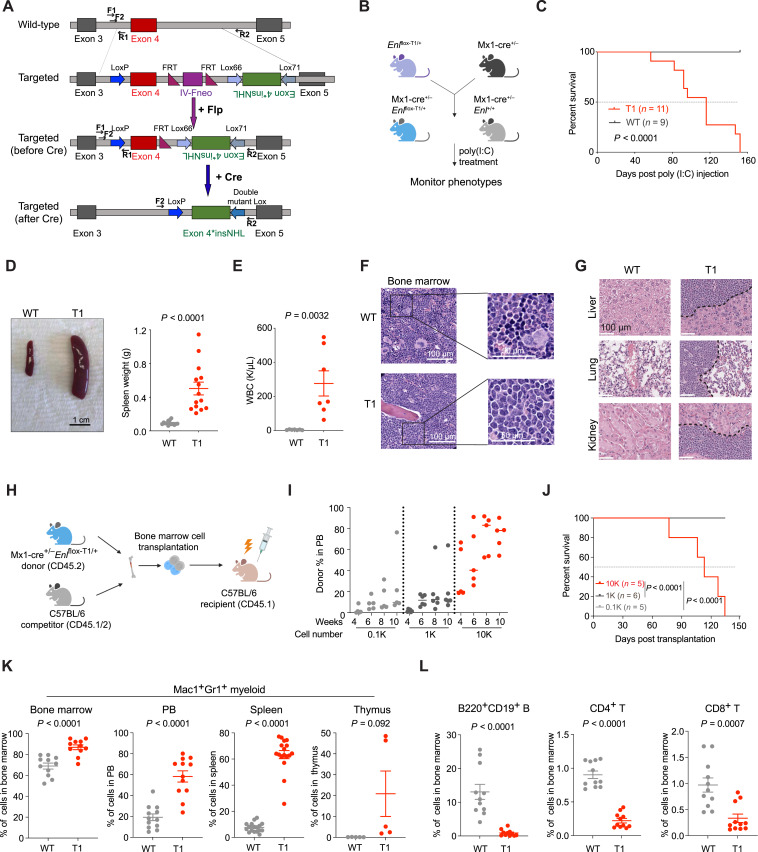
In this study, we set out to employ ENL mutations as a model to explore how condensate dysregulation contributes to tumorigenesis. We generated a conditional knock-in mouse model for ENL-T1, the most frequent ENL mutation found in cancer which involves an insertion of three amino acids (p.117_118insNHL; Supplementary Fig. S1A; refs. 19, 42, 43). The targeted allele was designed using an inversion strategy (Fig. 1A). Before induction of cyclization recombinase (cre) activity, the targeted allele is expressed as Enl-WT. After two steps of cre-mediated recombination, the inverted exon 4 containing the T1 mutation is flipped to the correct direction and expressed, whereas WT exon 4 is excised, thus leading to expression of the targeted allele as Enl-T1 (Fig. 1A). PCR genotyping was used to successfully distinguish the WT and T1 Enl alleles (Supplementary Fig. S1B). To explore the function of Enl-T1 in leukemogenesis, we crossed mice heterozygous for the Enl-T1 allele with the Mx1-cre strain to obtain Mx1-cre/Enlflox-T1/+ (hereafter referred to as Enl-T1) and Mx1-cre/Enl+/+ mice (hereafter referred to as Enl-WT; Fig. 1B). PCR analysis showed that the recombination efficiency of the Enl-T1 allele reached nearly 100% in bone marrow (BM) cells 2 weeks after poly (I:C) treatment, whereas no recombination was observed before the treatment (Supplementary Fig. S1C).
Figure 1.
Heterozygous Enl-T1 mutation drives aggressive AML in mice. A, Schematic design of a conditional knock-in mouse model for Enl-T1. Two pairs of primers (F1/R1 and F2/R2) used for genotyping shown in Supplementary Fig. S1 are depicted as black lines with arrowheads; Flp, flippase; FRT, flippase recombinase target; Lox66, Lox71, engineered recombination target sites derived from the WT loxP site. B, Breeding strategies to obtain Mx1-cre/Enlflox-T1/+ mice for experiments. poly (I:C), polyinosinic:polycytidylic acid. C, Kaplan–Meier survival curves of Enl-WT (n = 9) or Enl-T1 (n = 11) mice. Enl-WT, Mx1-cre/Enl+/+; Enl-T1, Mx1-cre/Enlflox-T1/+. P value using log-rank test. D, Representative images (left) and weight quantification (right) of spleen harvested from Enl-WT or T1 mice. Scale bar, 1 cm; bars represent the median (n = 14). P value using unpaired, two-tailed Student t test. E, WBC count of PB samples harvested from Enl-WT or T1 mice. Bars represent the median (n = 7). P value using unpaired, two-tailed Student t test. F, Representative hematoxylin and eosin (H&E) staining of BM harvested from Enl-WT or T1 mice. Scale bar, 100 μm (left, zoomed out); 50 μm (right, zoomed in). G, Representative H&E staining of liver (top), lung (middle), kidney (bottom) harvested from Enl-WT or T1 mice. Scale bar, 100 μm. H, BM transplantation experiment workflow. I, Flow cytometric quantification of Enl-T1 BM cells (CD45.2+CD45.1−) in the PB harvested at indicated time points from C57BL/6 recipient mice. Bars represent the median (0.1K, n = 5; 1K, n = 6; 10K, n = 5). J, Kaplan–Meier survival curves of C57BL/6 recipient mice that received 0.1K, 1K, 10K Enl-T1 BM cells. 0.1K, n = 5; 1K, n = 6; 10K, n = 5. P value using Log-rank test. K, Percentage of Mac1+Gr1+ myeloid cell population in the BM, PB, spleen, and thymus samples harvested from Enl-WT or T1 mice. Bars represent the median (BM, n = 11; PB, n = 12; spleen, n = 17; thymus, n = 5). P value using unpaired, two-tailed Student t test. L, Percentage of B220+CD19+ B, CD4+ T, and CD8+ T cells in BM samples harvested from Enl-WT or T1 mice. Bars represent the median (n = 11). P value using unpaired, two-tailed Student t test.
Heterozygous Enl-T1 expression in mice resulted in mortality as early as 2 months post poly (I:C) treatment, and all Enl-T1 mice died within 5 months (Fig. 1C). Upon necropsy, Enl-T1 mice exhibited splenomegaly (Fig. 1D) and significantly elevated white blood cell (WBC) counts (Fig. 1E). A peripheral blood (PB) smear corroborated the occurrence of leukocytosis in these mice (Supplementary Fig. S2A). Further histologic analysis revealed that blast cells had replaced the normal trilineage hematopoiesis in the BM (Fig. 1F). There was also notable blast cell infiltration in the spleen (Supplementary Fig. S2B), as well as other organs including the liver, lungs, and kidneys (Fig. 1G). To confirm the expression of the Enl-T1 allele, we extracted mRNA from BM cells of Mx1-cre/Enlflox-T1/+ mice 3 months post poly (I:C) treatment and reversed transcribed them into cDNA. We then performed PCR amplification of the Enl cDNA followed by deep sequencing (Supplementary Fig. S3A). Analysis of results from two mice consistently showed an almost equal expression of the Enl-WT and Enl-T1 alleles in these cells (Supplementary Fig. S3B). Furthermore, to determine if there were any cooperating mutations in Enl-T1-driven leukemia cells, we performed targeted genomic DNA sequencing on a panel of 611 cancer-related genes (47–49). Apart from the expected heterozygous Enl-T1 mutation found in nearly all cells, only two other mutations were detected in a minority of the cells: one in the Kit gene (p.V562_P580dup, VAF ∼ 0.1) and one in the Ptpn11 gene (p.E76 K, VAF ∼ 0.1; Supplementary Fig. S3C). The identified Kit mutation is not known to play a role in leukemogenesis. Although the Ptpn11 mutation has been shown to promote leukemia development in mouse models, it leads to less-severe phenotypes than Enl-T1 in the same Mx1-cre model (50). These, together with the low frequency of these mutations, suggest that the Enl-T1 mutation is likely the primary driver of the disease phenotypes observed in our mouse model.
To validate our findings in a different model, we crossed Enlflox-T1/+ mice with the UBC-cre-ERT2 strain to obtain UBC-cre-ERT2/Enlflox-T1/+ and UBC-cre-ERT2/Enl+/+ progeny (Supplementary Fig. S4A). Following tamoxifen treatment, all UBC-cre-ERT2/Enlflox-T1/+ mice succumbed to the disease within 5 months (Supplementary Fig. S4B). Histopathological examination of these mice revealed marked splenomegaly (Supplementary Fig. S4C) and extensive infiltration of blast cells in the BM and other organs (Supplementary Fig. S4D–S4F). These phenotypes were highly consistent with those observed in the Mx1-cre model. Furthermore, we performed BM transplantation experiments with BM cells taken from Mx1-cre/Enlflox-T1/+ leukemic mice to assess their ability to give rise to leukemia in recipient mice (Fig. 1H). We injected increasing numbers of these cells into sublethally irradiated syngeneic recipient mice. Monitoring the engraftment over time, we observed a dosage-dependent phenotype. Mice receiving 100 (0.1K) or 1,000 (1K) Enl-T1 cells showed a low but steady increase in Enl-T1 cell abundance in the PB, and they all remained alive 5 months post-transplantation. In contrast, mice that received 10,000 (10K) cells exhibited a much higher level of engraftment starting 2 weeks post transplantation and rapidly developed terminal disease, resulting in 100% mortality within 5 months post transplantation (Fig. 1I and J). Therefore, our in vivo studies indicate that heterozygous expression of the Enl-T1 mutant drives the development of aggressive acute leukemia in mice with a short latency and 100% penetrance, thus establishing it as a bona fide and potent oncogenic driver in vivo.
To identify the specific subtype of leukemia induced by Enl-T1, we profiled various hematopoietic cell compartments in Mx1-cre/Enlflox-T1/+ mice using flow cytometric analysis. We observed a consistent increase in the proportion and/or absolute numbers of myeloid cells (Mac1+Gr1+) in the BM, PB, spleen, and thymus of Enl-T1 leukemic mice (Fig. 1K; Supplementary Fig. S5A–S5C). Moreover, imaging analysis revealed that the BM cells from these mice exhibited marked myeloid hyperplasia and an aberrantly high myeloid to erythroid ratio (Supplementary Fig. S5D). In contrast, there was a marked decrease in the number and/or the percentage of B cells (B220+CD19+) as well as CD4+ and CD8+ T cells in these same tissues in Enl-T1 leukemic mice (Fig. 1L; Supplementary Fig. S6A–S6L). The same analysis on Enl-T1 mice at 4 weeks post poly (I:C) treatment—prior to the overt onset of leukemia—revealed a similar trend in the expansion in the myeloid compartment and a decrease in other cell lineages at the pre-leukemic stage (Supplementary Fig. S7A–S7M). Taken together, these results suggest that Enl-T1 drives the development of AML-like disease in mice.
Enl Mutation Perturbs the Normal Hematopoietic Hierarchy and Leads to Abnormal Expansion of Myeloid Progenitors with Increased Self-renewal Properties
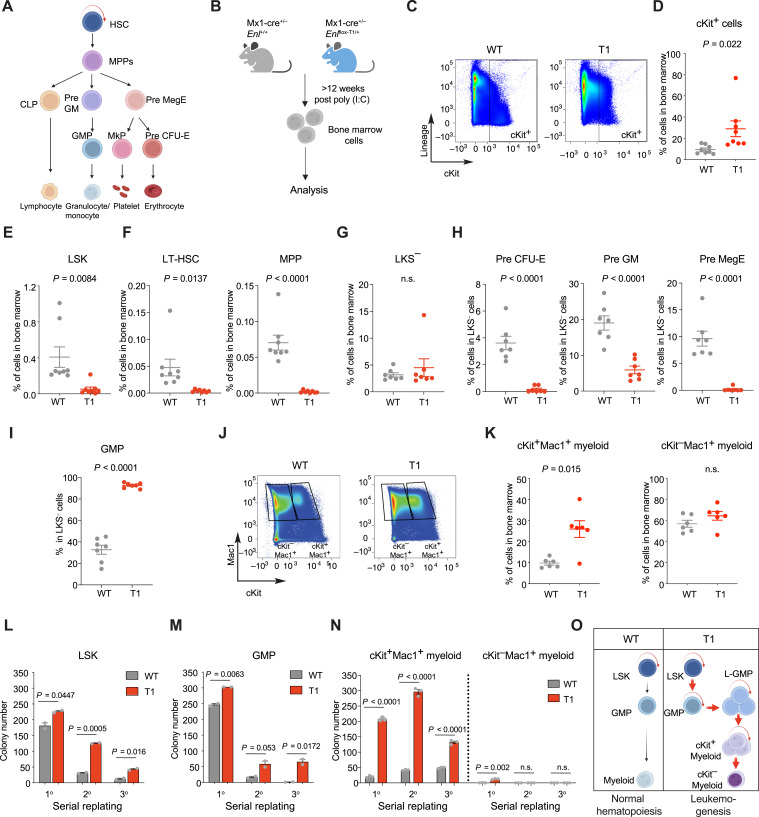
During normal hematopoietic development, hematopoietic stem cells (HSC) possess self-renewal properties and differentiate into multipotent progenitors (MPP) and downstream hematopoietic progenitor cells, including common lymphoid progenitors, pre-megakaryocyte/erythrocyte progenitors (Pre MegE), and granulocyte/monocyte progenitors (GMP). These progenitors further differentiate into all types of blood lineage cells, such as myeloid cells, T cells, and B cells (Fig. 2A; refs. 51–53). Our data show that the expression of Enl-T1 resulted in the expansion of the myeloid compartment and aggressive AML in vivo (Fig. 1). To further identify specific cell populations affected by Enl-T1, we performed comprehensive immunophenotype analysis of the BM cells from Enl-T1 leukemic mice to determine the distribution of different HSPCs (Fig. 2B). The percentage and absolute numbers of cells expressing cKit, a common marker for HSPCs (54, 55), were higher in Enl-T1 leukemic mice than in age-matched WT mice (Fig. 2C and D; Supplementary Fig. S8A). Interestingly, while cKit+ cells in WT mice were mostly negative for lineage markers, cKit+ cells in Enl-T1 leukemic mice expressed high levels of lineage markers (Fig. 2C), suggesting that the immunophenotype of cKit+ cells was altered by Enl-T1. Moreover, Enl-T1 mice had a decrease in HSPC-enriched LSK (Lin−Sca-1+c-Kit+) cells in the BM (Fig. 2E; Supplementary Fig. S8B and S8C). Furthermore, within the LSK population we observed a decrease in multiple sub-populations, including the long-term HSCs (LT-HSC, Lin−Sca-1+c-Kit+CD150+CD48−) and MPP (Lin−Sca-1+c-Kit+CD150−CD48−; Fig. 2F; Supplementary Fig. S8B and S8C). Next, we focused on the myelo-erythroid progenitor-enriched LKS− (Lin−c-Kit+Sca-1−) population, which contains multiple types of committed progenitors. We found that while the percentage and absolute numbers of LKS− cells in the BM were mostly unchanged by Enl-T1 (Fig. 2G; Supplementary Fig. S8D and S8E), their composition was altered. The percentage and absolute numbers of Pre MegE, pre-granulocyte/macrophage progenitor (Pre GM), and pre-colony-forming unit-erythroid progenitor (Pre CFU-E) cells within the LKS− population and whole BM were decreased in Enl-T1 leukemic mice (Fig. 2H; Supplementary Fig. S8D, S8F, and S8G), whereas those of the GMP population were increased (Fig. 2I; Supplementary Fig. S8D, S8F, and S8G).
Figure 2.
Enl mutation perturbs the normal hematopoietic hierarchy and leads to abnormal expansion of myeloid progenitors with increased self-renewal properties. A, Schematic representation of hematopoietic development from HSCs to differentiated blood cells. B, Schematic workflow of flow cytometric analysis for BM cells harvested from Enl-T1 leukemia mice and age-matched control mice (data shown in C–K). C and D, Representative flow cytometric plots (C) and the percentage (D) of cKit+ cells in BM samples. n = 8. E and F, Percentage of LSK (E), LT-HSC (F, left), and MPP (F, right) populations in BM. n = 8. G, Percentage of LKS− population in BM samples. n = 7. H and I, Percentage of Pre CFU-E (H, left), Pre GM (H, middle), Pre MegE (H, right), and GMP (I) cells in LKS− population from BM samples. n = 7. J and K, Representative flow cytometric gating plots (J) and the percentage (K) of cKit+Mac1+ (left) and cKit−Mac1+ (right) cells in BM. n = 6. L–N, Quantification of colonies formed by Enl-WT or T1 LSK (L), GMP (M), cKit+Mac1+ (N) and cKit−Mac1+ (N) cells. Error bars represent mean ± SEM (LSK, n = 2; GMP, n = 2; cKit+Mac1+, n = 3; cKit−Mac1+, n = 3). P values using unpaired, two-tailed Student t test. O, Schematic depicting how the Enl mutation perturbs the normal hematopoietic hierarchy and leads to the abnormal expansion of myeloid progenitors with increased self-renewal properties. D–I, K, Bars represent the median; P values using unpaired, two-tailed Student t test. n.s., not significant.
To ask whether Enl-T1 impacts the distribution of HSPCs at the pre-leukemic phase, we profiled the BM of Enl-T1 mice 4 weeks after poly (I:C) treatment, a time point before any overt disease phenotype was observed (Supplementary Fig. S9A). We found no significant differences in the proportion of cKit+ cells between pre-leukemic Enl-T1 mice and their control counterparts (Supplementary Fig. S9B–S9D). Although there was a noticeable trend toward a decrease in the percentage and absolute numbers of various HSPC compartments such as LSK, MPP, LKS−, Pre MegE, Pre GM, and Pre CFU-E (Supplementary Fig. S9E–S9J), the GMP population within the LKS− population as well as in the whole BM did not show an increase in the pre-leukemic Enl-T1 mice (Supplementary Fig. S9H, S9K, and S9L).
Previous studies have suggested that some subtypes of AML exhibit a leukemic cell hierarchy. Leukemia stem cells (LSC) in these models are not distinguished by a specific immunophenotype. Rather, some LSCs express normal HSPC markers (such as cKit+) and are negative for lineage markers, whereas others represent aberrant committed myeloid progenitors that express cKit as well as myeloid differentiation markers such as Mac1 and Gr1 (56, 57). In Enl-T1-induced leukemia, most cKit+ cells had high expression of lineage markers (Fig. 2C). Flow cytometric analysis further revealed an increase in cKit+Mac1+ committed myeloid progenitor cells but no changes in the cKit−Mac1+ differentiated myeloid cells in the BM of Enl-T1 leukemic mice when compared to age-matched Enl-WT mice (Fig. 2J and K). Together, our results suggest that during the development of leukemia, expression of Enl-T1 suppresses normal hematopoiesis and negatively impacts HSCs and most committed progenitors while inducing the expansion of GMP and abnormal committed myeloid progenitors (cKit+Mac1+).
AML is thought to arise from the accumulation of somatic mutations in HSPCs, which often results in aberrant self-renewal and/or impaired differentiation (58, 59). Thus, we examined the self-renewal properties of LSK and GMP cells with serial replating assays. Compared with cells from Enl-WT mice, freshly sorted LSK and GMP cells from pre-leukemic Enl-T1 mice exhibited an enhanced colony-forming ability in vitro (Fig. 2L and M). To test whether this phenotype is due to an intrinsic effect of Enl-T1 on HSPCs, we sorted LSK and GMP cells from the BM of normal C57BL/6 mice and transduced them with FLAG-tagged human WT or mutant ENL transgenes ex vivo followed by colony formation assays (Supplementary Fig. S10A). We tested two out of the eight ENL mutations identified in cancer, ENL-T1 and ENL-T4, as T1 is the most frequent ENL mutation found in Wilms tumor and T4 is found in AML (19, 42–44). Consistent with results from the knock-in mouse model, ectopic introduction of ENL-T1 or T4 ex vivo enhanced the serial replating potential of both LSK and GMP cells (Supplementary Fig. S10B and S10C). Furthermore, while cKit+Mac1+ cells from Enl-WT mice had a much lower potential to form colonies compared with LSK and GMP cells, cKit+Mac1+ cells from Enl-T1 leukemic mice exhibited a strong colony formation ability (Fig. 2N). In contrast, cKit−Mac1+ cells from both Enl-WT and Enl-T1 mice exhibited minimal colony formation potential (Fig. 2N). These results suggest that Enl-T1 enhances the self-renewal properties of HSPCs as well as cKit+Mac1+ committed myeloid progenitors, raising the possibility that these populations serve as LSCs that fuel the development of leukemia in Enl-T1 mice (56, 57). Collectively, our data suggest that Enl-T1 perturbs the normal hematopoietic hierarchy and establishes a leukemia cell hierarchy which leads to the aberrant expansion of myeloid progenitors with increased self-renewal properties (Fig. 2O).
Enl Mutation Induces Developmental and Inflammatory Gene Signatures in HSPCs
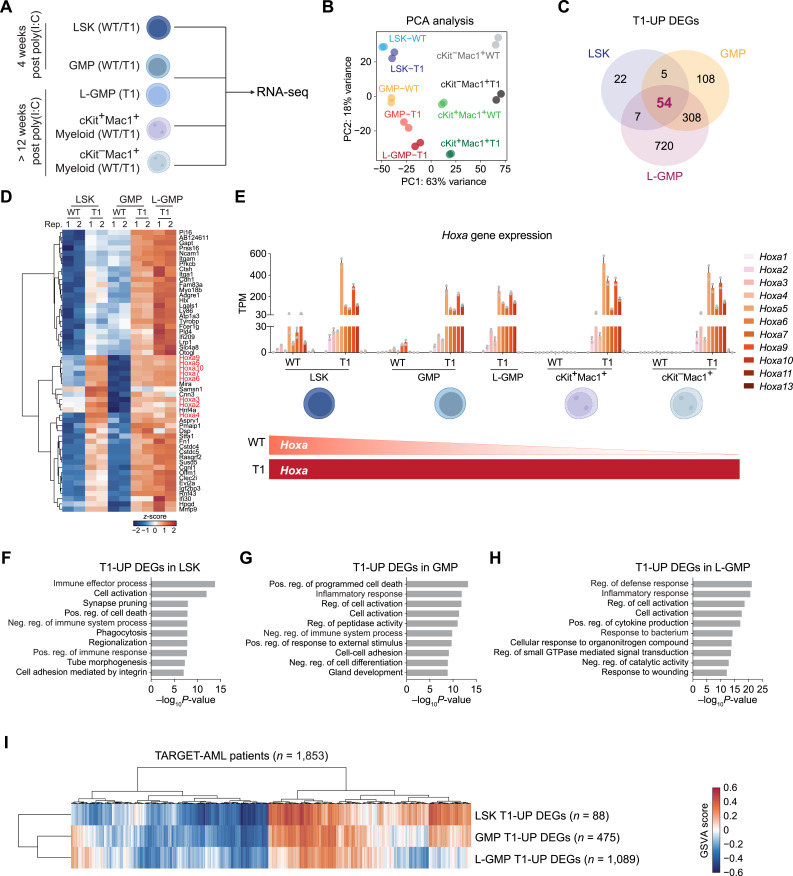
To understand the molecular mechanisms underlying Enl mutation-induced leukemogenesis, we performed RNA sequencing (RNA-seq) on LSK and GMP cells from pre-leukemic Enl-T1 mice [4 weeks post poly (I:C) treatment], as well as leukemic GMP (L-GMP), cKit+Mac1+, and cKit−Mac1+ myeloid cells from leukemic Enl-T1 mice [>12 weeks post poly (I:C) treatment]. For comparison, we included cells from age-matched Enl-WT mice that also received poly (I:C) treatment (Fig. 3A). We first assessed endogenous Enl expression in these cell populations from WT mice. The expression level of Enl was highest in HSPCs (LSK and GMP) and gradually decreased during differentiation (Supplementary Fig. S11A), suggesting a potentially critical role of Enl in HSPCs. Principal component analysis (PCA) and pairwise comparisons demonstrated that LSK cells from Enl-WT and Enl-T1 mice were more similar to each other than to other cell populations (Fig. 3B). Similar observations were obtained for GMP, cKit+Mac1+, and cKit−Mac1+ populations. These data indicate that Enl-T1 does not lead to drastic transcriptional changes that would alter hematopoietic cell identity. Differential expression analysis revealed that there were more genes with upregulated (LSK, n = 88; GMP, n = 475; 1.5-fold change, P. adj < 0.01) than downregulated expression (LSK, n = 28; GMP, n = 276) in Enl-T1 LSK and GMP when compared with Enl-WT counterparts, whereas Enl-T1 led to similar numbers of upregulated and downregulated transcripts in L-GMP, cKit+Mac1+, and cKit−Mac1+ cells (Supplementary Fig. S11B; Supplementary Tables S1–S5).
Figure 3.
Enl mutation induces developmental and inflammatory gene signatures in HSPCs. A, Schematic representation of RNA-seq assay for different hematopoietic populations. B, PCA of all the hematopoietic populations listed in A, including LSK, GMP, L-GMP, cKit+Mac1+, and cKit−Mac1+ cells in BM samples harvested from Enl-WT or T1 mice. Biological replicates are highlighted with the same color. C, Venn diagram showing the overlap of T1 upregulated (T1-UP) DEGs among LSK, GMP, and L-GMP (leukemia GMP) cells in BM samples harvested from Enl-WT or T1 mice. See Supplementary Tables S1–S3. D, Heatmap showing the expression of the 54 triple-overlapped T1-UP DEGs highlighted in C. Gene expression is normalized by z-score. Hoxa genes are highlighted in red. See Supplementary Table S6. E, Bar plots showing the gene expression of all Hoxa genes in LSK, GMP, L-GMP, cKit+Mac1+, and cKit−Mac1+ populations in BM samples harvested from Enl-WT or T1 mice. The bottom schematic summarizes expression trends during hematopoietic differentiation under Enl-WT and T1 conditions. Error bars represent mean ± SEM (n = 2). See Supplementary Table S7. F–H, Bar plots showing GO term analysis of T1-UP DEGs for LSK (F), GMP (G) and L-GMP (H), respectively. Neg., negative; Pos., positive; Reg., regulation. I, Clustered heatmap showing sample-wise gene set enrichment scores calculated by GSVA. The three gene sets are T1-UP DEGs of LSK, GMP, and LGMP, respectively. Each column represents one sample from TARGET-AML dataset (n = 1,853). See Supplementary Table S9.
We next characterized the transcriptional changes induced by Enl-T1 in HSPCs (LSK, GMP, L-GMP). There were 54 shared genes upregulated by Enl-T1 in the three HSPC subsets (Fig. 3C; Supplementary Table S6). Notably, this list included multiple Hoxa genes (Fig. 3D and E; Supplementary Table S6), which are known to promote the self-renewal of normal and malignant HSPCs and are overexpressed in 50% to 70% of AML cases (60–62). In WT mice, Hoxa genes were highly expressed in LSK cells and became gradually silenced during differentiation (Fig. 3E; Supplementary Table S7), consistent with previous reports (63). In contrast, the presence of Enl-T1 resulted in much higher expression levels of Hoxa genes in LSK, which were aberrantly maintained in progenitors (GMP, L-GMP, cKit+Mac1+) as well as terminally differentiated myeloid cells (cKit−Mac1+; Fig. 3E). These results suggest that Enl-T1 not only induces hyper-activation but also impairs differentiation-associated repression of the Hoxa cluster genes. Interestingly, though Enl-T1 induced hyperactivation of Hoxa genes across all five populations we have examined, only stem and progenitor populations (LSK, GMP, L-GMP, cKit+Mac1+), but not terminally differentiated myeloid cells (cKit−Mac1+), showed increased self-renewal abilities (Fig. 2L–N). These data suggest that Enl-T1 promotes self-renewal of HSPCs in part through hyper-activation of Hoxa genes, but Hoxa genes alone are not sufficient to grant self-renewal properties once hematopoietic cells enter a “point of no return” commitment to differentiation (64).
To categorize functional pathways induced by Enl-T1, we performed gene ontology (GO) term analysis on identified DEGs in each of the five cell populations. Interestingly, we found that gene signatures related to inflammatory and immune pathways were highly enriched in Enl-T1-upregulated genes in LSK, GMP, and L-GMP cells (Fig. 3F–H). These signatures contain genes that are associated with inflammation and are known to alter the BM microenvironment during leukemogenesis, including genes encoding Toll-like receptors (TLR) and Interleukins (Fig. 3F–H). GO term analysis of Enl-T1 downregulated genes in LSK, GMP, and L-GMP cells revealed enrichment of protein folding and homeostasis-associated pathways (Supplementary Fig. S11C–S11E). When hematopoietic cells enter the maturation process, they depend on the upregulation of metabolic pathways to support rapid expansion and function (65). Of note, genes regulated by Enl-T1 in cKit+Mac1+ and cKit−Mac1+ myeloid populations showed strong enrichment for metabolic and cell cycle–related pathways, suggesting that Enl-T1, directly or indirectly, alters metabolic pathways to facilitate the expansion and function of these myeloid populations (Supplementary Fig. S11F–S11I). Furthermore, cell activation pathways, which are involved in both immune response and cell differentiation, were highly enriched in Enl-T1-upregulated genes across all the populations (Fig. 3F–H; Supplementary Fig. S11F and S11G). These data indicate that Enl-T1 might regulate selected differentiation events. Given that Enl-T1 led to the expansion of the myeloid compartment and the development of AML, we performed gene set enrichment analysis (GSEA) to test whether Enl-T1 influences myeloid differentiation. Interestingly, Enl-T1 led to a gain of myeloid differentiation signatures in HSPCs (LSK, GMP, and L-GMP) but not in cKit+Mac1+ or cKit−Mac1+ cells when compared with WT counterparts (Supplementary Fig. S12A and S12B; Supplementary Table S8). Furthermore, Enl-T1 LSK cells gained expression of GMP-like signature (Supplementary Fig. S12C; Supplementary Table S8). These results are intriguing as they suggest that Enl-T1 induces transcriptional programs that promote self-renewal (e.g., through Hoxa genes) as well as myeloid commitment of HSPCs, both of which may contribute to the development of AML. In support of this concept, a recent study using MLL–AF9 and MOZ–TIF2 AML models have shown that myeloid differentiation to GMP is critical for the transformation of HSPCs (66).
To investigate the clinical relevance of Enl-T1-regulated genes in HSPCs identified in our mouse model, we examined their expression in samples obtained from patients with AML. We downloaded the transcriptomic data for 1,853 samples from patients with AML from the TARGET-AML database (46) and performed gene set variation analysis (GSVA) to calculate the sample-wise enrichment scores for T1-upregulated genes identified in LSK, GMP, and L-GMP cells. From the samples from patients with AML, we found that sample-wise enrichment scores of all three signatures highly correlated with each other and that ∼30% of AML samples had high expression scores for these signatures (Fig. 3I; Supplementary Table S9), supporting the implication of Enl-T1-regulated genes identified in the mouse model in a significant portion of human AML.
Enl Mutation Leads to Increases in H3K27ac and p300 Occupancy at Subsets of Genes in HSPCs
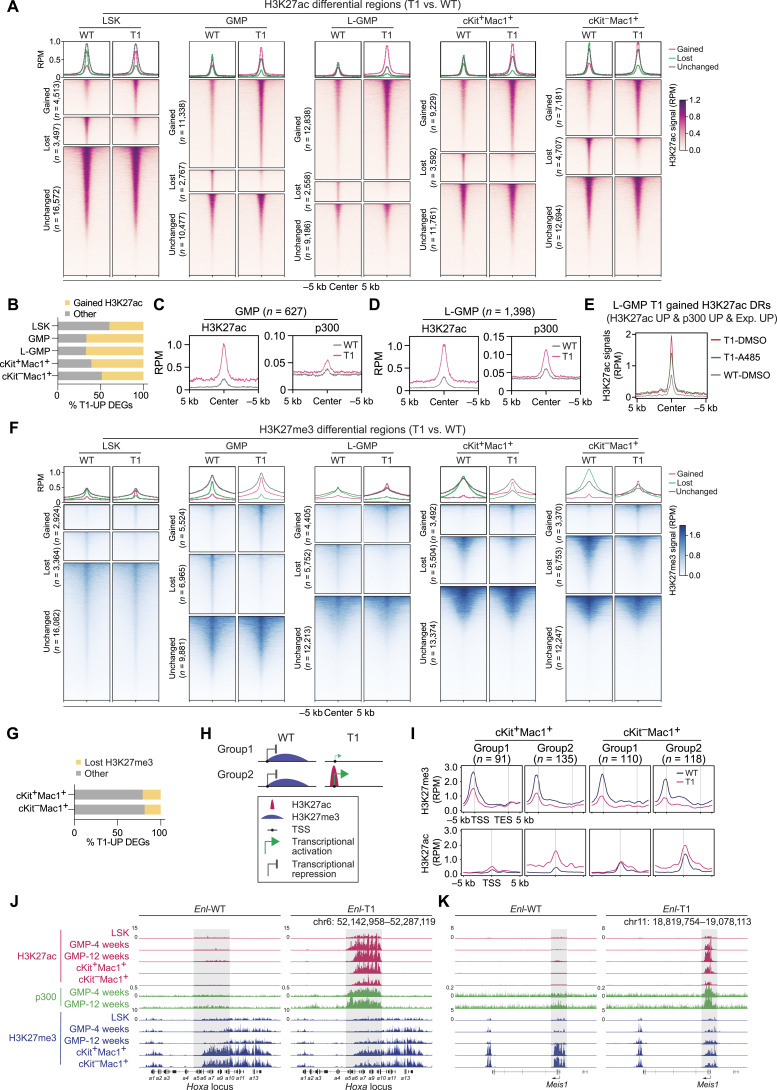
Having determined that most Enl-T1-upregulated genes in HSPCs were involved in leukemia-associated signatures, we asked whether alterations in the chromatin state correlate with the observed transcriptional changes. We first profiled genome-wide distribution of histone 3 Lysine 27 acetylation (H3K27ac), a chromatin mark associated with gene activation, via CUT&Tag (Cleavage Under Targets & Tagmentation; ref. 67) in Enl-WT and Enl-T1 LSK, GMP, L-GMP, cKit+Mac1+, and cKit−Mac1+ cells. We compared H3K27ac patterns between WT and T1 conditions in each cell population and identified H3K27ac peaks that were gained, lost, or unchanged (1.5-fold cut-off) by Enl-T1 (Fig. 4A). Overall, around 30% of H3K27ac peaks were altered by Enl-T1 in LSK, and more than half of them were gained peaks. Furthermore, Enl-T1 impacted about 50% of H3K27ac peaks in GMP, L-GMP, cKit+Mac1+, and cKit−Mac1+ cells, and the majority of changed peaks were gained in T1, especially in GMP and L-GMP populations (Fig. 4A; Supplementary Fig. S13A; Supplementary Tables S10–S15). We identified genes associated with T1-gained H3K27ac peaks and observed that about half of T1-upregulated genes in each of the five cell populations were associated with increased H3K27ac (Fig. 4B; Supplementary Table S16). These results suggest the possibility that Enl-T1 regulates transcription in hematopoietic cells in part by, directly or indirectly, remodeling the H3K27ac landscape.
Figure 4.
Enl mutation results in altered H3K27ac and H3K27me3 landscapes in hematopoietic cells. A, H3K27ac CUT&Tag data in LSK, GMP, L-GMP, cKit+Mac1+ and cKit−Mac1+ populations were plotted as average occupancies (top) and heatmap (bottom) across the respective H3K27ac differential regions (DR) between WT and T1. The CUT&Tag signals are normalized by reads per million (RPM). All regions are defined as gained (upregulated in T1, red), lost (downregulated in T1, green) and unchanged (gray) regions, and the corresponding numbers are shown on the left, respectively. See Supplementary Tables S11–S15. B, Bar plots showing the percentage of T1-UP DEGs that are associated with T1-gained H3K27ac DRs in indicated cell populations. Supplementary Table S16. C and D, Average occupancies of H3K27ac and p300 at genomic regions that exhibit T1-induced increase in H3K27ac and are associated with T1-UP DEGs in GMP (C) or L-GMP (D) from Enl-WT (gray) or T1 (red) mice. The CUT&Tag or ChIP-seq signals are normalized by RPM. See Supplementary Table S17. E, Average occupancies of H3K27ac at genomic regions that exhibit T1-induced increase in H3K27ac and p300 and are associated with T1-UP DEGs in GMP (WT) or L-GMP (T1) cells treated with DMSO or A-485 (1 µmol/L) for 24 hours. The CUT&Tag signals are normalized by RPM. See Supplementary Table S20. F, H3K27me3 CUT&Tag signals in indicated cell populations were plotted as average occupancies (top) and heatmap (bottom) across H3K27me3 differential regions (DR) between WT and T1. See Supplementary Tables S24–S28. G, Bar plots showing the percentage of T1-UP DEGs that are associated with T1-lost H3K27me3 DRs in cKit+Mac1+ and cKit−Mac1+ populations. See Supplementary Table S29. H, Schematic showing the two groups of T1-UP DEGs that are associated with lost H3K27me3 DRs. I, Average occupancies of H3K27me3 (top) and H3K27ac (bottom) on group1 and two genes along the transcription unit in cKit+Mac1+ and cKit−Mac1+ from Enl-WT (blue) or T1 (red) mice. The CUT&Tag signals are normalized by RPM. See Supplementary Table S30. J and K, Genome browser view of H3K27ac, p300, and H3K27me3 CUT&Tag or ChIP-seq signals at Hoxa (J) and Meis1 (K) gene loci in indicated hematopoietic populations from Enl-WT (left) or T1 (right) mice.
As a histone acetylation reader, ENL predominantly localizes to the transcriptional start site and gene body of subsets of actively transcribed genes that are highly enriched for H3K9ac and H3K27ac, and ENL cancer mutants (e.g., T1/T2/T3) occupy largely similar genomic loci as WT ENL (19, 32, 33). Although the regulation of histone acetylation by ENL or its cancer mutants has not been described, previous studies have suggested read–write feedback loops as a general mechanism contributing to the deposition of histone modifications (68). We therefore performed chromatin immunoprecipitation assays followed by sequencing (ChIP-seq) for p300, a histone acetyltransferase that catalyzes H3K27ac, in GMP and L-GMP cells which exhibited the most prominent T1-induced increase in H3K27ac (Fig. 4A). We focused on T1-gained H3K27ac peaks that were associated with T1-upregulated genes and compared p300 levels at these peak regions (GMP, n = 627; L-GMP, n = 1,398) between WT and T1 conditions. p300 colocalized with H3K27ac at these regions and exhibited an increase in signal in Enl-T1 GMP and L-GMP cells compared with WT counterparts (Fig. 4C and D; Supplementary Fig. S13B and S13C; Supplementary Tables S17 and S18). Notably, among Enl-T1 upregulated genes associated with increased H3K27ac, 30% to 50% were also associated with increased p300 signals at H3K27ac peaks (Supplementary Fig. S13D; Supplementary Table S19). Furthermore, the majority of p300-gained peaks that were correlated with increased H3K27ac signals and target gene expression were predominantly located at TSS regions (Supplementary Fig. S13E), consistent with ENL localization at target genes (19, 32, 33). GREAT analysis of genomic regions with gained p300 signals revealed strong enrichment of immune and development related pathways (Supplementary Fig. S13F and S13G). These results suggest that increased recruitment of p300 may contribute to ENL-T1-mediated increases in H3K27ac at a subset of genomic regions in HSPCs. To test this hypothesis, we sorted L-GMP cells from the BM of Enl-T1 leukemic mice and treated them with the p300 inhibitor A-485 (69) ex vivo for 3, 6, and 24 hours, followed by H3K27ac CUT&Tag experiments (Supplementary Fig. S13H). At genomic regions that exhibited ENL-T1-induced increases in p300 and H3K27ac enrichment and were also associated with T1-upregulated genes, A-485 treatment resulted in a reduction in H3K27ac across all three time points tested (Fig. 4E; Supplementary Fig. S13I; Supplementary Table S20). A-485 treatment also resulted in a decrease in the expression of key ENL-T1 target genes, such as Meis1 and Hoxa cluster genes (Supplementary Fig. S13J). These findings suggest that the ENL-T1-mediated increase in H3K27ac in HSPCs is mediated in part through the recruitment of p300, a mechanism that likely contributes to transcriptional changes induced by ENL-T1.
Differentiation-Associated Gain of H3K27me3 is Impaired in Enl-Mutated Hematopoietic Cells
The execution of developmental processes requires dynamic activation and repression of differentiation-related genes. In addition to H3K27ac, Polycomb-mediated methylation of H3K27 (H3K27me3) has been shown to play important roles in development (70–73). Currently, the dynamic changes of H3K27me3 during normal hematopoietic development remain incompletely understood. Thus, we determined the genome-wide H3K27me3 distribution in LSK, GMP, cKit+Mac1+, and cKit−Mac1+ cells freshly sorted from WT mice via CUT&Tag. Interestingly, we found that in WT mice, the overall signal for H3K27me3 was lowest in LSK cells and highest in cKit+Mac+ and cKit−Mac+ myeloid cells (Supplementary Fig. S14A; Supplementary Table S21). We identified 3,397 H3K27me3 peaks whose signals gradually increased during hematopoietic differentiation, and most of these peaks were located at gene body or intergenic regions (Supplementary Fig. S14B and S14C; Supplementary Table S22). GREAT analysis on these peaks revealed significant association with developmental processes such as pattern specification and cell fate commitment (Supplementary Fig. S14D). Consistent with H3K27me3 being a repressive mark, the expression levels of genes associated with increasing H3K27me3 during differentiation were significantly higher in LSK cells compared with GMP, cKit+Mac1+, and cKit−Mac+ cells (Supplementary Fig. S14E and S14F), and these genes were highly enriched for developmental pathways (Supplementary Fig. S14G). These results highlight a potential role of H3K27me3-mediated repression of developmental genes during hematopoietic differentiation, prompting us to ask whether Enl-T1 may perturb the H3K27me3 landscape during leukemogenesis.
We performed H3K27me3 CUT&Tag in different cell populations from WT and T1 BM. Interestingly, Enl-T1 induced modest changes at regions with low H3K27me3 signals in HSPCs (LSK, GMP, and L-GMP). However, in cKit+Mac1+ and cKit−Mac1+ cells, Enl-T1 led to a marked decrease in signal at subsets of broad H3K27me3 peaks (Fig. 4F; Supplementary Tables S23–S28) which are functionally involved in developmental pathways (Supplementary Fig. S15A and S15B). Furthermore, about 20% to 30% of Enl-T1-induced H3K27me3 lost regions overlapped with (Supplementary Fig. S15C) and accounted for, 40% of regions that naturally gain H3K27me3 during normal hematopoietic differentiation (Supplementary Fig. S15D–S15F), suggesting that H3K27me3-mediated repression of developmental genes may be disrupted in Enl-T1 mice. Indeed, about 20% of Enl-T1 upregulated genes in cKit+Mac1+ or cKit−Mac1+ cells were associated with loss in H3K27me3 signals (Fig. 4G; Supplementary Table S29). Of note, among genes that were associated with loss of H3K27me3, one subset (group 1) exhibited loss of H3K27me3 without significant changes of H3K27ac, and the other (group 2, including Hoxa and Meis1) exhibited simultaneous loss of H3K27me3 and gain of H3K27ac (Fig. 4H and I; Supplementary Table S30). Interestingly, genes in group 2 were expressed at higher levels than those in group 1 (Supplementary Fig. S15G and S15H) and were enriched for immune response and differentiation pathways (Supplementary Fig. S15I and S15J).
Taken together, our epigenomic analyses reveal dynamic changes in active and repressive histone modifications in Enl-T1 mice that are associated with altered expression at subsets of development related genes. These include genes that are important for normal hematopoietic development and/or leukemogenesis, such as the Hoxa cluster, Meis1, and Igf2bp3 (Fig. 4J and K; Supplementary Fig. S15K; refs. 61, 62, 74–78). When zooming in on the Hoxa and Meis1 loci, we observed a decrease in H3K27ac and an increase in H3K27me3 during normal differentiation (Fig. 4J and K; Supplementary Fig. S15L). At these loci, the presence of Enl-T1 induced a marked increase in H3K27ac in LSK and a further increase in myeloid progenitors (Fig. 4J and K). Notably, the levels of H3K27ac at Hoxa and Meis1 in differentiated cKit−Mac1+ cells from Enl-T1 mice were even higher than that in WT LSK cells (Fig. 4J and K). Concurrently, differentiation-associated gain of H3K27me3 at these loci were impaired in Enl-T1 mice (Fig. 4J and K). Additionally, some other gene loci exhibited changes in either H3K27ac (Supplementary Fig. S15M) or H3K27me3 (Supplementary Fig. S15N), but not both. While further studies are needed to fully understand the mechanisms by which ENL mutations directly or indirectly affect these histone modifications, their alterations likely play a role in the transcriptional changes induced by ENL-T1 in hematopoietic cells that contribute to leukemia development.
Condensate Formation Ability Correlates with Mutant ENL’s Oncogenic Function in HSPCs
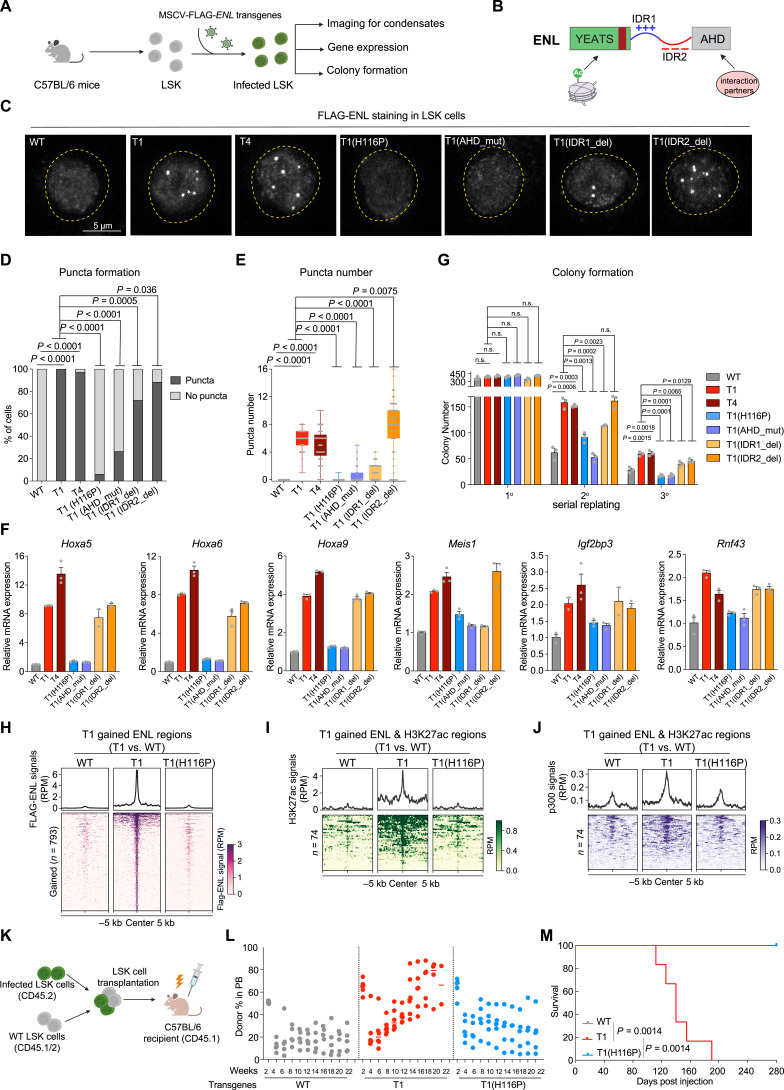
Our previous study in HEK293 cells showed that cancer-associated mutations enable ENL to form condensates at select genomic target loci, and these condensates are required for ENL mutation-induced gene activation (31). Therefore, we set out to ask whether ENL mutants form condensates in HSPCs. Due to the lack of a reliable antibody for detecting mouse ENL protein via immunofluorescence (IF) staining, we introduced various FLAG-tagged human ENL transgenes into LSK cells from normal mice and performed IF staining using an anti-FLAG antibody (Fig. 5A). This strategy also enabled a direct comparison of different ENL proteins. We found that ENL-T1 and T4, but not WT, consistently formed distinct nuclear puncta in ∼100% of LSK cells (Fig. 5B–E; Supplementary Fig. S16A–S16E), and this was evident even when the transgenes were expressed at significantly lower levels than the endogenous mouse Enl (Supplementary Fig. S16A). Importantly, FLAG-ENL staining coupled with DNA fluorescence in situ hybridization (FISH) revealed that at least one Hoxa9 allele colocalized with an ENL-T1 puncta in the majority of LSK cells (Supplementary Fig. S16F and S16G). Furthermore, the expression of ENL-T1 and T4 variants in LSK cells resulted in increased expression of key target genes, including the Hoxa cluster (Supplementary Fig. S16H). To ask whether similar phenotypes occur in human hematopoietic cells, we transduced primary human CD34+ HSPCs with WT and T1 FLAG-ENL transgenes (Supplementary Fig. S17A). In line with findings from mouse HSPCs, ENL-T1 expression resulted in puncta formation (Supplementary Fig. S17B–S17E), an upregulation of HOXA genes (Supplementary Fig. S17F), and enhanced colony formation in human HSPCs (Supplementary Fig. S17G).
Figure 5.
Mutant ENL forms condensates at key target genes, and disrupting condensate formation impairs its oncogenic activity in HSPCs. A, Schematic representation of the transduction of various FLAG-tagged ENL transgenes in LSK cells and subsequent experiments (shown in C–G). B, Schematic depiction of domain structure and features of the ENL mutant protein. C, Representative images of anti-FLAG IF staining in LSK cells expressing the indicated FLAG-ENL transgenes. WT, ENL-WT; T1, ENL-T1; T4, ENL-T4; T1(H116P), H116P mutation in the YEATS domain of ENL-T1; T1(AHD_mut), DRD538RFR mutations in the AHD domain of ENL-T1; T1(IDR1_del), deletion of IDR1 in ENL-T1; T1(IDR2_del), deletion of IDR2 in ENL-T1. D and E, Percentage of nuclei with or without FLAG-ENL puncta (D) and the number of FLAG-ENL puncta (E) in each nucleus in LSK cells. Error bars represent mean ± SEM. P values using unpaired, two-tailed Student t test. F, RT-qPCR analysis showing mRNA expression of Hoxa5/6/9, Meis1, Igf2bp3, and Rnf43 in LSK cells expressing indicated FLAG-ENL transgenes. Error bars represent mean ± SEM (n = 3). G, Quantification of colonies formed by LSK cells expressing indicated FLAG-ENL transgenes. Error bars represent mean ± SEM (n = 3). P value using unpaired, two-tailed Student t test. H, Average occupancies (top) and heatmap (bottom) representation of FLAG-ENL-bound peak regions that show increased occupancy by T1 compared with WT ENL in LSK cells expressing the indicated FLAG-ENL transgenes. The CUT&Tag signals are normalized by reads per million (RPM). See Supplementary Table S32. I and J, Average occupancies (top) and heatmap (bottom) showing H3K27ac (I) and p300 (J) at genomic regions that exhibit T1-induced increase in H3K27ac and FLAG-ENL in LSK cells expressing the indicated FLAG-ENL transgenes. The CUT&Tag or ChIP-seq signal are normalized by RPM. See Supplementary Table S33. K, Schematic workflow of LSK cells transplantation experiment. L, Flow cytometric quantification of FLAG-ENL transgenes expressing cells (CD45.2+CD45.1−) in the PB harvested at indicated time points from C57BL/6 recipient mice. Bars represent the median (WT, n = 5; T1, n = 6; T1(H116P), n = 5). P value using unpaired, two-tailed Student t test. M, Kaplan–Meier survival curves of C57BL/6 recipient mice that received LSK cells expressing the indicated FLAG-ENL transgenes. WT, n = 5; T1, n = 6; T1(H116P), n = 5. The gray line (WT) overlapped with the blue line [T1 (H116P)]. P value using log-rank test.
To further investigate the relationship between ENL-T1’s condensate formation ability and its oncogenic function, we engineered a series of FLAG-ENL-T1 variants with specific mutations or deletions within different domains of the ENL-T1 protein, including the YEATS domain, IDR1, IDR2, and the AHD domain (Fig. 5B). These variants were previously characterized in HEK293 cells, where they exerted differential impacts on condensate formation via different biochemical mechanisms (31). We introduced these variants into freshly sorted LSK cells at similar levels (Supplementary Fig. S18A and S18B) and assessed their ability to form condensates. The H116P substitution, which reverts specific structural changes in the YEATS domain caused by the T1 mutation (31), abolished ENL-T1’s ability to form condensates in LSK cells. Similar results were observed in GMP cells (Supplementary Fig. S19A–S19E). Moreover, mutations within the AHD domain (DRD538RFR), designed to disrupt ENL’s interactions with its partners SEC/P-TEFb and DOT1L, also markedly impaired condensate formation (Fig. 5C–E). In contrast, the deletion of IDR1 only partially reduced the number of condensates, while IDR2 deletion exerted a negligible effect (Fig. 5C–E). Importantly, we found that alterations that resulted in condensate formation deficiency, namely H116P and AHD_mut, abolished T1-induced increases in target gene expression (Fig. 5F) and colony formation (Fig. 5G). Similar results were observed in GMP cells (Supplementary Fig. S19F and S19G). IDR1 deletion led to a reduced expression of certain target genes, such as Meis1, indicating a partial functional impairment. IDR2 deletion, which did not affect condensate formation, had minimal impact on ENL-T1’s ability to promote gene expression and colony formation in LSK cells (Fig. 5F and G).
We next asked whether disrupting the condensate formation ability of ENL-T1 affects its chromatin function. We first determined the genome-wide occupancies of FLAG-ENL, H3K27ac, and p300 in LSK cells expressing WT, T1, and the condensate-deficient T1(H116P) variant. Overall, WT, T1, and T1(H116P) ENL proteins localize to largely similar genomic locations (Supplementary Fig. S20A; Supplementary Table S31). By comparing FLAG-ENL occupancy patterns between WT and T1, we identified a list of T1-enhanced ENL-bound regions (n = 793), and quite remarkably, this increased occupancy was diminished by the H116P mutation (Fig. 5H; Supplementary Fig. S20B and S20C; Supplementary Table S32). Furthermore, we identified regions that showed augmented FLAG-ENL and H3K27ac signals in T1-expressing cells (Fig. 5I; Supplementary Fig. S20B and S20C; Supplementary Table S33) and observed an increase in p300 occupancy at these same regions (Fig. 5J; Supplementary Fig. S20B and S20C; Supplementary Table S33). The H116P mutation significantly reduced T1-induced increases in H3K27ac and p300 occupancy at these sites (Fig. 5I and J; Supplementary Fig. S20B and S20C; Supplementary Table S33). These findings further support the role of ENL-T1 in p300/H3K27ac regulation and suggest that its condensate formation ability contributes to this process.
Furthermore, we compared the leukemogenic ability of ENL-WT, T1, and T1(H116P) via BM transplantation (Fig. 5K). Mice receiving ENL-T1-expressing LSK cells showed the highest levels of engraftment in the PB and rapidly developed lethal leukemia, resulting in 100% mortality within 6 months post-transplantation (Fig. 5L and M). In contrast, mice that received WT or T1(H116P)-expressing LSK cells displayed low engraftment and remained healthy and alive even at 9 months post transplantation (Fig. 5L and M). At necropsy, mice transplanted with ENL-T1 cells exhibited splenomegaly (Supplementary Fig. S21A) and high BM engraftment (Supplementary Fig. S21B). FLAG-ENL staining further confirmed that nearly all leukemic BM cells taken from these mice harbored distinct nuclear puncta (Supplementary Fig. S21C–S21E). Taken together, our results reveal that mutant ENL forms condensates at key leukemogenic genes and this condensate formation ability strongly correlates with its chromatin and gene regulatory function as well as oncogenic activity in disease-relevant models.
Small-Molecule Inhibition of the Acetyl-Binding Activity of Mutant ENL Displaces Its Condensates from Target Loci and Impairs Its Chromatin Function in HSPCs
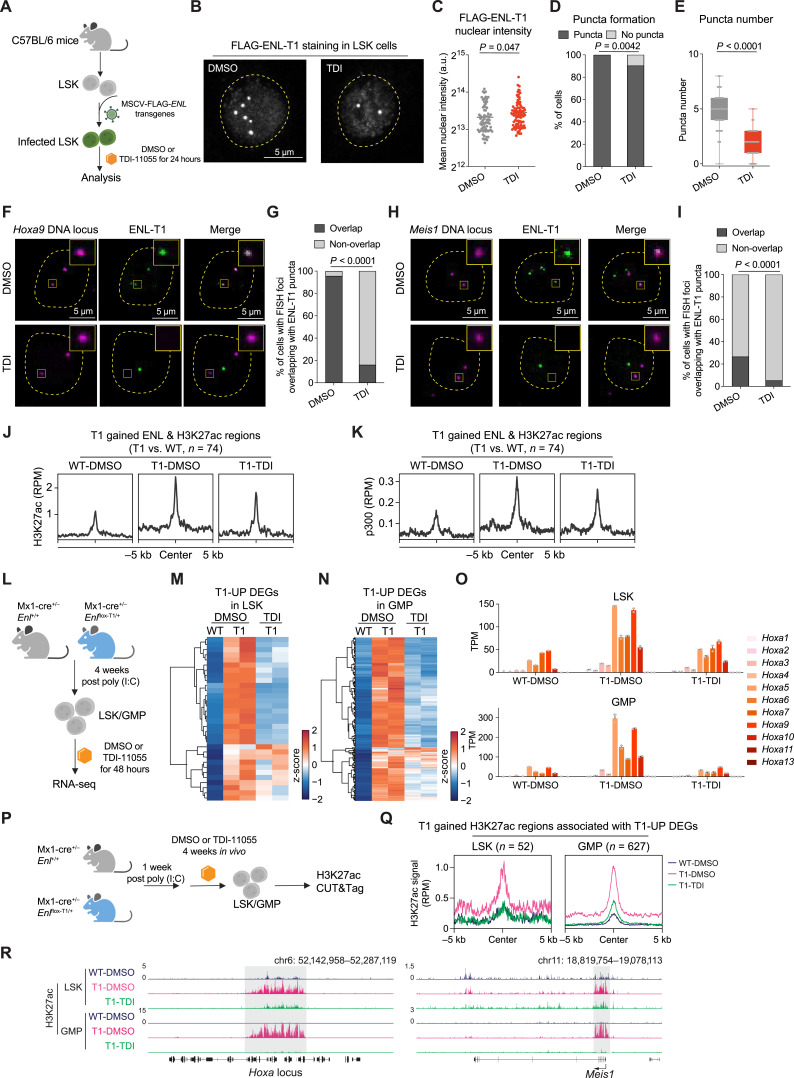
ENL recognizes histone acetylation at actively transcribed gene promoters through its YEATS domain (32, 33). Our previous studies in HEK293 cells showed that introducing the Y78A mutation, known to disrupt acetyl-binding activity, decreases chromatin occupancy of ENL cancer mutants (T1/T2/T3) and abolishes target gene hyperactivation (19, 31). Notably, the Y78A mutation did not abolish puncta formation; instead, it resulted in nuclear puncta that no longer localized to H3K27ac-marked chromatin, indicating a critical role for acetyl-binding in the proper localization of ENL mutant condensates (19, 31). Based on these findings, we hypothesized that blocking the acetyl-binding activity could be an effective strategy to inhibit mutant ENL’s function. Recently, we reported on TDI-11055, a potent, in vivo bioactive ENL inhibitor that displaces WT ENL from chromatin by blocking the interaction of its YEATS domain with acetylated histones (41). As cancer-associated mutations do not significantly alter ENL’s acetyl-binding pocket (19, 31), we speculated that TDI-11055 could also target ENL mutants. Thus, we set out to evaluate the efficacy of TDI-11055 against ENL mutation-induced condensate formation and oncogenic functions in HSPCs.
We introduced the FLAG-ENL-T1 transgene into LSK cells isolated from WT mice, treated these cells with either DMSO or TDI-11055 for 24 hours, and then assessed condensate formation and chromatin changes (Fig. 6A). Similar to the Y78A mutation, TDI-11055 did not abrogate condensate formation in HSPCs but did reduce their numbers (Fig. 6A–E). FLAG-ENL staining coupled with DNA FISH for two key target genes, Hoxa9 and Meis1, revealed that TDI-11055 treatment significantly reduced the colocalization of ENL-T1 condensates with these genomic loci (Fig. 6F–I). The treatment also decreased ENL-T1-induced enrichment of H3K27ac and p300 at a subset of genomic regions, including the Hoxa cluster and Meis1 (Fig. 6J and K; Supplementary Fig. S22A–S22C; Supplementary Table S33) and led to reduced expression of these genes (Supplementary Fig. S22D).
Figure 6.
Small-molecule inhibition of the acetyl-binding activity of mutant ENL displaces its condensates from target loci and impairs its chromatin function in HSPCs. A, Schematic representation of LSK cells expressing FLAG-ENL-T1 transgene under DMSO or 10 µmol/L TDI-11055 (TDI for short) treatment for 24 hours and subsequent experiments (shown in A–K). B, Representative images of anti-FLAG IF staining in LSK cells treated with DMSO or TDI-11055. C–E, Nuclear intensity (C), percentage of nuclei with or without FLAG-ENL puncta (D), and the number of FLAG-ENL puncta (E) in each nucleus in LSK cells treated with DMSO or TDI-11055 (TDI). Error bars represent mean ± SEM. P values using unpaired, two-tailed Student t test. F and G, Representative images (F) and quantification (G) showing percentage of DMSO or TDI-11055 treated cells containing at least one Hoxa9 DNA locus overlapping with an ENL-T1 puncta. DMSO, n = 89; TDI-11055, n = 75. P value using unpaired, two-tailed Student t test. H and I, Representative images (H) and quantification (I) showing percentage of DMSO or TDI-11055 treated cells containing at least one Meis1 DNA locus overlapping with an ENL-T1 puncta. DMSO, n = 79; TDI-11055, n = 76. P value using unpaired, two-tailed Student t test. J and K, Average occupancies of H3K27ac (J) and p300 (K) at genomic regions that exhibit T1-induced increase in FLAG-ENL and H3K27ac occupancies in LSK cells expressing FLAG-ENL-T1 transgenes under DMSO or TDI-11055 treatment. The CUT&Tag or ChIP-seq signals are normalized by RPM. See Supplementary Table S33. L, Schematic showing ex vivo treatment of LSK/GMP cells sorted from Enl-WT or T1 mice and following RNA-seq analysis (shown in M–O). M and N, Heatmap showing gene expression of T1-UP DEGs (identified in Supplementary Fig. S23A and S23B) in LSK (M) and GMP (N) treated with DMSO or TDI-11055. Gene expression is scaled by z-score. See Supplementary Tables S34 and S35. O, Bar plots showing the gene expression of Hoxa genes in WT or T1 LSK (top) and GMP (bottom) treated with DMSO or TDI-11055. Gene expression is obtained from the RNA-seq data and normalized by TPM. Dots represent different biological replicates. Bars represent the median. See Supplementary Table S36. P, Schematic showing in vivo treatment of Enl-WT or T1 mice and subsequent H3K27ac profiling in LSK/GMP cells. Q, Average occupancy of H3K27ac on T1-induced H3K27ac DRs that are associated with T1-UP DEGs in LSK (left) and GMP (right) treated with DMSO or TDI-11055. See Supplementary Table S17. R, Genome browser view of H3K27ac CUT&Tag signals at select genes under indicated treatment conditions in LSK and GMP.
Having explored the effect of TDI-11055 using the transgene-based system, we then examined its impact on cells obtained directly from mouse models. To determine its impact on gene expression, we performed RNA-seq analysis on LSK and GMP cells sorted from Enl-WT or T1 mice and treated with DMSO or TDI-11055 ex vivo for 48 hours (Fig. 6L). We identified 32 and 220 genes with upregulated expression by Enl-T1 in LSK and GMP cells, respectively (1.5-fold change, P. adj < 0.05; Supplementary Fig. S23A and S23B; Supplementary Tables S34 and S35), most of which showed a decrease in expression upon TDI-11055 treatment (Fig. 6M and N; Supplementary Fig. S23C and S23D; Supplementary Tables S34 and S35). Remarkably, TDI-11055 treatment in Enl-T1 LSK and GMP cells was sufficient to bring expression of Hoxa genes back to that of their Enl-WT counterparts (Fig. 6O; Supplementary Table S36). We next assessed whether TDI-11055 treatment could revert Enl-T1-induced chromatin changes in HSPCs from the mouse model. Enl-WT and T1 mice were treated with DMSO or TDI-11055 daily for 28 consecutive days. Subsequently, LSK and GMP cells were sorted from the BM and subjected to H3K27ac CUT&Tag analysis (Fig. 6P). TDI-11055 treatment in vivo abolished T1-enhanced H3K27ac signals that are associated with genes showing T1-induced upregulated expression in both LSK and GMP cells (Fig. 6Q and R; Supplementary Fig. S23E–S23G; Supplementary Table S17). Taken together, our results suggest that TDI-11055 treatment can displace ENL mutant condensates from specific genomic loci and inhibit chromatin and transcription changes induced by mutant ENL in HSPCs.
Small-Molecule Inhibition of the Acetyl-Binding Activity of Mutant ENL Impairs the Onset and Progression of AML In Vivo
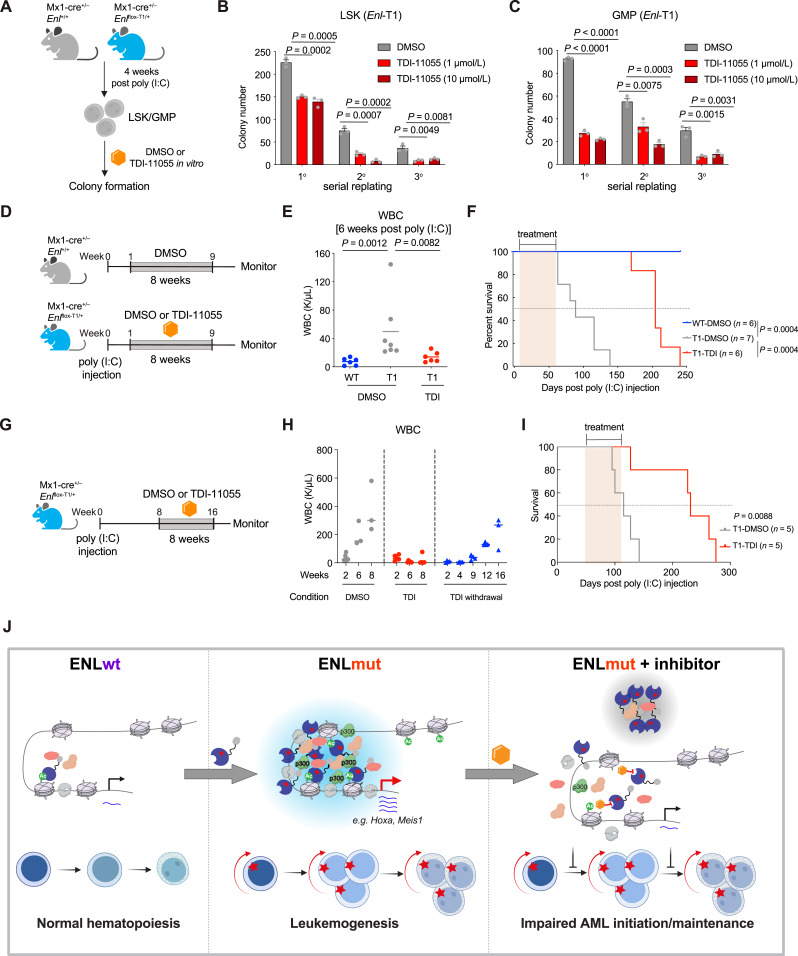
To examine the effects of TDI-11055 on ENL mutant’s oncogenic function, we first performed a colony formation assay on LSK and GMP cells sorted from preleukemic Enl-T1 mice under DMSO or TDI-11055 treatment conditions (Fig. 7A). We observed a marked decrease in colony number with TDI-11055 treatment (Fig. 7B and C). Similar results were obtained in separate experiments in which ENL-T1 or T4 was ectopically introduced into LSK and GMP cells (Supplementary Fig. S24A–S24C).
Figure 7.
Small-molecule inhibition of the acetyl-binding activity of mutant ENL impairs the onset and progression of AML in vivo. A, Schematic experimental plan with LSK/GMP cells sorted from Enl-WT or T1 mice. B and C, Quantification of colonies formed by Enl-T1 LSK (B) or GMP cells (C) treated with DMSO or TDI (1 or 10 µmol/L). Error bars represent mean ± SEM (n = 3). P values using unpaired, two-tailed Student t test. D, Schematic of in vivo treatment of Enl-WT or T1 mice to examine the impact of TDI-11055 on leukemia onset. E, WBC count of PB samples harvested from Enl-WT or T1 mice under DMSO or TDI-11055 treatment conditions. Bars represent the median (WT-DMSO, n = 6; T1-DMSO, n = 7; T1-TDI, n = 6). P values using unpaired, two-tailed Student t test. F, Kaplan–Meier survival curves of Enl-WT or Enl-T1 mice under DMSO or TDI-11055 treatment conditions. WT-DMSO, n = 6; T1-DMSO, n = 7; T1-TDI, n = 6. P values using log-rank test. G, Schematic of in vivo treatment of Enl-WT or T1 mice to examine the impact of TDI-11055 on leukemia maintenance. H, WBC count of PB samples harvested from Enl-T1 mice at indicated time points under DMSO or TDI-11055 treatment. For DMSO and TDI groups, numbers on the X-axis indicate weeks of treatment; for the TDI withdrawal group, numbers on the X-axis indicates weeks after the cessation of treatment. Bars represent the median (T1-DMSO, n = 5; T1-TDI, n = 5). I, Kaplan–Meier survival curves of Enl-T1 mice under DMSO or TDI-11055 treatment conditions. T1-DMSO, n = 5; T1-TDI, n = 5. P values using log-rank test. J, Schematic showing mutant ENL forms discrete condensates at critical gene loci and alters transcriptional and histone modification changes in HSPCs, resulting in the aberrant expansion of myeloid progenitors and rapid onset of aggressive AML. Small-molecule inhibition of acetyl-binding displaces ENL-mutant condensates from target loci, dampens their impact on histone modification and gene activation, and effectively delays the initiation and progression of AML in vivo.
We next explored the therapeutic potential of TDI-11055 in Enl-T1-driven leukemia in vivo. To determine impacts on the onset of leukemia, treatment with either DMSO or TDI-11055 commenced immediately following poly (I:C) administration in Enl-T1 mice and continued once daily for eight consecutive weeks (Fig. 7D). By the 6th week of treatment, Enl-T1 mice treated with DMSO exhibited a significant increase in WBC counts compared to WT mice. This increase was suppressed in the TDI-11055-treated Enl-T1 mice, indicating a delay in leukemia onset (Fig. 7E). Furthermore, while all DMSO-treated Enl-T1 mice succumbed to AML within 5 months, those treated with TDI-11055 remained alive at the 5-month mark (Fig. 7F). Notably, short-term treatment with TDI-11055 during the early stages of leukemogenesis had a prolonged effect, as these mice remained active more than 100 days after discontinuing treatment (Fig. 7F). The overall survival of Enl-T1 mice in the TDI-11055 group was significantly improved compared with mice in the vehicle group, with a 127% increase in median survival (91 vs. 207 days).
Additionally, we initiated TDI-11055 treatment in Enl-T1 mice already in the leukemic phase to evaluate its efficacy in halting leukemia progression (Fig. 7G). DMSO-treated Enl-T1 mice exhibited elevated WBC counts over time and 100% mortality within 5 months post poly (I:C) treatment (Fig. 7H and I). In contrast, TDI-11055 treatment suppressed the rise in WBC counts in Enl-T1 leukemic mice, and these mice maintained normal levels throughout the treatment period. Remarkably, an increase in WBC counts was not observed until 9 weeks following the cessation of TDI-11055 treatment (Fig. 7H), indicating a lasting effect in controlling leukemia progression. Furthermore, the overall survival of Enl-T1 mice in the TDI-11055 group significantly improved, doubling the median survival time (115 vs. 231 days) compared to the vehicle group (Fig. 7I). Collectively, these results demonstrate that small-molecule inhibition of mutant ENL’s acetyl-binding activity effectively impairs the onset and progression of leukemia.
Discussion
Despite extensive biochemical investigations into the molecular basis of condensate formation, the functional significance of transcriptional condensates and their dysregulation in physiologically relevant in vivo contexts are only beginning to be addressed. Recent studies on cancer-associated ENL mutations (19, 31) revealed their ability to trigger condensate formation via a network of multivalent interactions and induce hyper-activation of target genes in HEK293 cells, but these studies did not explore the role of these mutations in tumorigenesis and the contribution of condensate formation to this process.
In this study, we show that heterozygous knock-in of a specific ENL mutation (T1) into the hematopoietic system leads to the rapid onset of lethal AML with 100% penetrance, thus establishing the mutant ENL as a highly potent oncogene in spontaneous mouse models. Mechanistically, mutant ENL perturbs the normal hematopoietic hierarchy, promotes myeloid differentiation from HSPCs, and results in abnormal expansion of myeloid progenitors with increased self-renewal properties by altering histone modifications and transcription programs across various hematopoietic populations. Notably, we demonstrate that ENL cancer mutants (T1 and T4) expressed at physiological levels form discrete condensates at key target genomic loci (e.g., Hoxa and Meis1) in HSPCs. Disrupting condensate formation through targeted mutagenesis impairs mutant ENL’s ability to regulate histone modifications, activate target genes, and drive tumorigenesis in vivo. Finally, we show that inhibiting the acetyl-binding activity of mutant ENL with a small molecule displaces its condensates from target genes, impairs its chromatin function, and blocks leukemogenesis driven by mutant ENL in vivo (Fig. 7J). While our study primarily focused on the ENL-T1 mutation, our previous work has established that eight ENL mutations (T1–T8, Supplementary Fig. S1A), found in Wilms tumor and AML, induce similar structural changes in the YEATS domain and activate target genes via condensate formation (31). Notably, through further analysis of whole-genome sequencing projects for AML (45, 46), we identified three new ENL mutations involving three-amino acid insertions (p.118_119insTTC, p.118_119insTCA, and p.C119SPAR), similar to the T1 and T4 mutations we characterized in this study. Interestingly, these mutation patterns have been observed in pediatric AML and Wilms tumor but not in adult AML, suggesting that specific developmental stages are more susceptible to transformation by ENL mutations—a hypothesis that warrants further investigation. Future research is also needed to validate our findings in ENL-mutated patient samples as they become available. Our study advances the understanding of AML pathogenesis by elucidating the biological function and therapeutic potential of ENL mutations. It also provides concrete in vivo evidence that supports a role of condensate dysregulation in cancer.
AML typically arises from dysregulated transcriptional programs that result in the malignant self-renewal and/or impaired differentiation of HSPCs. Our study reveals that mutant ENL induces the aberrant expression of developmental and inflammatory gene signatures in HSPCs that are implicated in human AML. Notably, among the genes most significantly upregulated by mutant ENL are the Hoxa cluster genes, which are crucial for the self-renewal of normal and malignant HSPCs and are overexpressed in a significant portion of AML cases (62, 74–76). Thus, ENL mutants join a list of oncogenes, such as NUP98-fusions (20, 22, 23, 79–83), MLL/KMT2A fusions (84, 85), MN1 fusion (86, 87), and mutant NPM1 (24, 88, 89), that drive leukemogenesis in part by elevating Hoxa expression. Intriguingly, similar to ENL mutants, NUP98 fusions and mutant NPM1 are also known to form nuclear condensates in leukemia cells (20, 22–24). ENL can also drive leukemogenesis in the form of MLL–ENL fusion; however, whether MLL–ENL functions through the formation of condensates remains to be investigated. Nevertheless, our comprehensive functional and mechanistic investigation of ENL mutants and their condensates could offer valuable insights and motivate future studies to explore condensate dysregulation as a potential mechanism underlying the hyperactivation cancer-promoting genes such as Hoxa cluster in cancer (20, 22–24).
To our surprise, we show that mutant ENL not only promotes self-renewal but also induces a gain of myeloid differentiation signatures in HSPCs. This observation suggests that the commitment of HSPCs to myeloid lineage might be critical for AML development. Supporting this idea, previous studies have shown that blocking myeloid differentiation of HSCs and common myeloid progenitors (CMP) by deleting CEBPA blunts MLL-AF9- and MOZ-TIF2-driven leukemogenesis in murine models (66, 90–92). Conversely, mutant ENL appears to block the differentiation of committed myeloid progenitors (cKit+Mac1+) and sustain them in an aberrant state with self-renewal properties. Thus, we propose that a delicate balance in self-renewal and differentiation is important for the transformation of HSPCs and AML pathogenesis.
In previous studies using HEK293 cells, we showed that ENL mutants drive gene activation through enriching co-factors (e.g., SEC/P-TEFb and DOT1L) into condensates and promote RNA Pol II elongation (19). Our current study adds another layer of mechanisms by which ENL mutants influence transcription. We show that mutant ENL expression results in increased occupancies of H3K27ac and the acetyltransferase p300 at key developmental genes in HSPCs. Inhibiting the enzymatic activity of p300 can partially reverse mutant ENL-induced increases in H3K27ac and gene expression at these genes. This partial suppression of H3K27ac suggests either the contribution of a non-enzymatic activity of p300 or the involvement of other acetyltransferases in mutant ENL-mediated increases in H3K27ac. Given p300’s known interactions with certain condensate-forming proteins (14, 93, 94), we speculate that mutant ENL may promote p300 recruitment by incorporating it into condensates. This hypothesis is supported by our observation that disrupting the condensate-forming ability of mutant ENL leads to reduced p300 recruitment and lower H3K27ac levels at a subset of target genes. It is important to note, however, that this regulation of p300/H3K27ac by mutant ENL seems context-dependent, as it is not observed in HEK293 cells. In addition to H3K27ac, our study reveals that many development-related genes gain H3K27me3 during hematopoietic cell differentiation. This process is partially hindered in the Enl-T1 mice, correlating with the incomplete repression of key development genes like Hoxa and Meis1 even in terminally differentiated cells. Thus, we propose that ENL mutants drive extended hyper-activation and impair differentiation-associated repression of key developmental genes in HSPCs in part through dynamic remodeling of histone H3K27ac and H3K27me3 landscapes. While our study focused on H3K27ac and H3K27me3, it remains to be determined whether ENL mutants may impact other chromatin features to regulate oncogenic gene expression and the disease phenotypes.
We have provided several lines of evidence supporting the role of condensate formation in mediating mutant ENL’s oncogenic function in HSPCs. First, DNA FISH experiments confirmed the formation of ENL condensates at key leukemogenic targets, notably the Hoxa cluster genes and Meis1. Second, our mutagenesis studies, targeting four distinct regions of the ENL-T1 protein, revealed a strong correlation between its condensate formation ability and its role in oncogenic transformation in HSPCs. Among the engineered mutations, the H116P point mutation disrupts condensate formation by reversing T1-induced structural changes in the ENL YEATS domain (31). To our knowledge, this occurs without affecting other known ENL functions. We show that this point mutation significantly reduces T1-induced increases in chromatin occupancies of ENL, H3K27ac, and p300 in HSPCs. It also inhibits T1-mediated leukemogenesis in vivo. Lastly, treatment with the acetyl-binding inhibitor TDI-11055 leads to the displacement of ENL-T1 condensates from target genes, accompanied by reduced H3K27ac and p300 occupancies, decreased target gene activation, and impaired leukemogenesis in vivo. These results establish a strong link between mutant ENL’s condensate formation ability and its oncogenic function.
Interestingly, mutant ENL forms larger and fewer puncta compared to many other gene regulators known to form condensates. The number of condensates formed by the ENL mutant is noticeably smaller than the number of genes it occupies and upregulates in HSPCs. Therefore, while our results indicate that condensate formation underlies the hyperactivation of key oncogenic targets like Hoxa genes and Meis1 by mutant ENL, mutant ENL might regulate some other genes through the formation of smaller clusters not easily detectable in current assays or through other mechanisms. Previous studies have shown that condensates can vary significantly in size and composition, and transcriptional activation may occur through multivalent interactions among cofactors without forming discrete condensates (95). Future research employing super-resolution and single-molecule imaging will enhance our understanding of how oncogenic ENL mutants cluster on chromatin and their impact on gene expression. Nevertheless, our findings suggest that pathogenic condensates, including those formed by ENL mutants and possibly other oncogenic proteins, might selectively target key cancer-driving genomic loci. Future studies are needed to explore the mechanisms underlying this specificity.
The dysregulation of biomolecular condensates has emerged as a pathogenic mechanism in various diseases, sparking interest in targeting them for therapeutic interventions. However, such efforts are hindered by a limited understanding of the molecular interactions governing their formation and regulation, and the lack of biologically relevant readouts for most condensates (96, 97). Recent high-throughput drug screenings have identified small molecules that can modulate stress granules associated with neurodegenerative diseases, showing potential benefits in motor neuron pathology (98–100). Although the therapeutic targeting of transcriptional condensates remains underexplored, our study demonstrates its potential effectiveness. We show that small-molecule inhibition of the acetyl-binding activity displaces ENL mutant condensates from critical oncogenic targets, leading to reduced H3K27ac and p300 enrichment as well as mRNA expression at these loci in HSPCs. Moreover, in vivo treatment with this inhibitor significantly impairs the onset and progression of leukemia driven by mutant ENL in mice. These findings suggest that targeting the ENL YEATS domain represents a promising strategy for treating ENL-mutated cancers. Our results offer proof-of-concept for the mechanism-based targeting of oncogenic condensates as a viable approach in cancer therapy.
Methods
Mouse Model
We generated a conditional knock-in mouse model for Enl-T1 which involves an insertion of three amino acids (p.117_118insNHL) following the steps described below.
Targeting Vector for Conditional Activation of the Enl/Mllt1 Mutations
An 8.9 kb genomic DNA sequence used to construct the targeting vector was first subcloned from a positively identified C57BL/6 fosmid clone (WI1-2250I3). The region was designed such that the long homology arm extends ∼6.1 kb 5′ to the 5′ LoxP cassette, and the short homology arm extends about 2.1 kb 3′ to the insertion of the inversion cassette. The inversion cassette is flanked by two mutant Lox sites (Lox71/66) and contains mutant exon 4 (AACCACCTG duplication) and the flanking genomic sequences for correct splicing (Inv.saE4*Sd). This cassette was inserted in the reverse direction downstream of exon 4. The FRT-flanked Neo cassette was inserted immediately upstream of the inversion cassette and is 175 bp away from WT exon 4. The targeting region containing exon four is 628 bp.
Screening and Reconfirmation of Recombinant Clones
A total of 10 μg of the targeting vector was linearized and then transfected by electroporation of HF4 (129/SvEv x C57Bl/6; FLP Hybrid) embryonic stem cells. After selection with the G418 antibiotic, surviving clones were expanded for PCR analysis to identify recombinant ES clones. The Neo cassette in the targeting vector was removed during ES clone expansion. The recombinant clones were further confirmed by PCR and DNA sequencing.
Generation of Enl-T1 Knock-in Mice
The correctly targeted ES clone was injected into blastocysts. Injected embryos were transferred to pseudo-pregnant recipient females. The chimeras were evaluated for germline transmission and bred to establish the knock-in mouse line. Knock-in mice were further confirmed by PCR and DNA sequencing.
Flow Cytometric Analysis of Different Hematopoietic Cell Compartments in the BM, Spleen, Thymus, and PB
Enl-WT or T1 mice were sacrificed, and BM cells were extracted from femurs, coxae, and vertebral columns; spleen and thymus cells were collected by gently smashing the organ and filtering through a 40 µm filter; PB was collected via orbital bleeding. BM cells were stained with murine cKit, Sca1, CD150, CD48, CD41, CD105, CD16/32, lineage antibodies, and Zombie Aqua fixable viability kit (BioLegend, #423102); cells were run through the FACS Aria Flow Cytometer (BD) to perform hematopoietic stem/progenitor cell compartments analysis. BM, spleen, thymus, and PB cells were stained with murine TCRb, B220, CD19, CD4, CD8, Mac1, Gr1, TruStain FcX, and Zombie Aqua fixable viability kit (BioLegend, #423102); cells were run through the FACS Aria Flow Cytometer (BD) to collect mature hematopoietic cell compartments.
Hematoxylin and Eosin Staining
Paraffin-embedded tissues were sectioned, stained in Mayers Hematoxylin for 1 minute, washed with PBS, and stained in Alcoholic-Eosin for 1 minute. Then the slides were dehydrated until clear and then samples were mounted with resinous mounting medium. Images were captured using the EVOS M5000 Imaging system (Thermo Fisher Scientific).
PB and BM Smear
PB and BM were harvested from Enl-WT or Enl-T1 mice. A small drop of blood or BM cells was placed on one end of slide. Holding the edge of another slide at an approximately 45° angle on the first glass slide, the top slide was rapidly but gently pushed forward through the blood or BM drop to spread the cells on the first slide. Slides were stained with Wright’s-Giemsa Stain Kit (Thermo Fisher Scientific, #9990701). Images were captured using a Leica DM2500 LED Optical microscope.
BM or LSK Cells Transplantation Assay
Eight-week-old CD45.1 recipient mice (The Jackson Laboratory, #002014) were sublethally irradiated (a split dose of 11Gy). For the BM transplantation assay, 0.1K, 1K, and 10K of total BM cells from Enl-T1 leukemic mice (CD45.2) were mixed with 300K of total BM cells from normal C57BL/6 mice (CD45.1/45.2), respectively. For the LSK cell transplantation assay, LSK cells from Enl-WT mice (CD45.2) were transduced with GFP-linked FLAG-tagged ENL constructs. GFP+ cells were sorted and then mixed with 300K of total BM cells from normal C57BL/6 mice (CD45.1/45.2). The mixed cells were transplanted by retro-orbital injection into sublethally irradiated CD45.1 recipient mice 3 hours post irradiation. Animals were monitored daily, and body weights were measured every 2 days throughout the treatment period. To assess the engraftment, PB was extracted from mice via retro-orbital bleeding, stained for mouse CD45.1−CD45.2+ %, and subjected to flow cytometry analysis. Kaplan–Meier survival curves were created using the GraphPad Prism (v9) software.
Cell Sorting
Enl-WT or Enl-T1 mice were sacrificed, and BM cells were extracted from femurs, coxae, and vertebral columns. BM cells underwent cKit positive cell enrichment using the EasySep Mouse CD117 (cKit) positive selection kit (Stemcell Technologies, #18757). The cKit enriched BM cells were stained with murine cKit, Sca1, CD150, CD41, CD16/32, lineage antibodies, and Zombie Aqua fixable viability kit (BioLegend, #423102). The total BM cells were stained with murine cKit, Mac1, TruStain FcX, and Zombie Aqua fixable viability kit (BioLegend, #423102). Cells were sorted using FACS Aria Flow Cytometer (BD) to obtain the Lin−Sca-1+cKit+ (LSK), Lin−Sca-1−cKit+CD41−CD150−CD16/32+ (GMP), cKit+Mac1+, cKit−Mac1+ cells.
Plasmids
FLAG-tagged WT human ENL was cloned into a MSCV_Puro_IRES_GFP vector (Addgene, 18751). ENL-T1, ENL-T4, and ENL-T1 (H116P) were introduced using a site-mutagenesis approach.
LSK and GMP Cell Culture and Virus Transduction
Mouse LSK and GMP cells were cultured in SFEM (Stemcell Technologies, #09600) supplemented with 10% FBS (HyClone, #SH30910.03), 20 ng/mL mFlt3-Ligand (PeproTech, #250-31L), 20 ng/mL mIL-6 (PeproTech, #216-16), 100 ng/mL mSCF (PeproTech, #250-03), 20 ng/mL mTPO (PeproTech, #315-14), 10−4 mol/L 2-mercaptoethanol (Thermo Fisher Scientific, #21985023), 100 U/mL penicillin-streptomycin (Thermo Fisher Scientific, #15140122), and L-glutamine (Thermo Fisher Scientific, #MT25005CI).
The tissue culture plates were precoated with RetroNectin (TaKaRa, #T100B) and incubated overnight at 4°C. The RetroNectin was removed the next day, and the plates were blocked with 2% BSA for 30 minutes at room temperature, then washed with HBSS once. The plates were then loaded with retrovirus and spun at 3,000 rpm at 10°C for 1 hour. Then, after removing the supernatants, LKS/GMP cells were added to the plate and spin-infected at 1,500 rpm for 90 minutes at 37°C with 10 μg/mL polybrene (EMD Millipore, #TR-1003-G). The infected cells were incubated at 37°C overnight, and the medium was changed the next day.
Colony Formation
A total of 6,000 LSK, GMP, cKit+Mac1+, or cKit−Mac1+ cells were resuspended in 600 μL IMDM media, respectively. The indicated cells were added to 4.5 mL Methyl Cellulose media (StemCell Technologies, #M3234) supplemented with 10 ng/mL IL-3, 20 ng/mL SCF, and 10 ng/mL IL-6 (PeproTech, #231-13, 250-03, 216-16), then indicated cells were plated as triplicate (2K cells/replicate). The number of colonies was determined after incubation in 37°C incubator for ∼1 week.
Immunofluorescence
Coverslips were transferred to a 24-well plate and coated with Poly-D-Lysine (Thermo Fisher Scientific, #A3890401) for 1 hour at room temperature. Coverslips were washed with H2O twice. HSPCs were then plated on coverslips for 15 minutes at 37°C, then spun at 1,200 rpm at room temperature for 4 minutes to allow 200,000 to 300,000 cells to attach to coverslips. Cells were fixed with 4% paraformaldehyde at room temperature for 10 minutes and washed in PBS three times, 5 minutes for each wash. To permeabilize nuclei, cells were incubated with 0.1% Triton X-100 at room temperature for 9 minutes, and then cells were washed in PBS three times, 5 minutes for each wash. Cells were incubated with 10% goat serum at room temperature for 30 minutes, and then incubated overnight at 4°C with anti-Flag antibody. The next day, cells were washed in PBST (PBS with 0.1% Tween 20) three times, 5 minutes for each wash, and then the cells were incubated with secondary antibody at room temperature for 1 hour. Finally, cells were washed in PBS three times, 5 minutes for each wash, and coverslips were mounted on slides with resinous mounting medium. Images were captured using Zeiss LSM 880 Confocal.
DNA FISH
Slides were placed on gaskets and coated with Poly-L-Lysine (Electron Microscopy Sciences, #63478-AS) at room temperature for 10 minutes. Coated slides were dried at room temperature overnight. 200,000 HSPCs were plated to each gasket well and incubated at 37°C for 30 minutes. Cells were fixed with 4% paraformaldehyde at room temperature for 10 minutes and washed in PBS three times, 10 minutes for each wash. To permeabilize nuclei, cells were incubated with 0.5% Triton X-100 at room temperature for 15 minutes, then washed in PBS three times, 5 minutes for each wash. Cells were incubated with 70% ethanol for 2 minutes, 90% ethanol for 2 minutes, and 100% ethanol for 2 minutes. Then, cells were treated with 2× SSCT (2× saline-sodium citrate with 0.1% Tween 20) at room temperature for 5 minutes, and 70% formamide buffer at room temperature for 5 minutes followed with 70% formamide buffer at 37°C for 1 hour. Hoxa9 or Meis1 DNA probes were mixed with dNTPs, formamide buffer, and hybridization buffer. DNA probe master mix was added into the cells and denatured at 83°C for 30 minutes. Then, cells were hybridized with denatured DNA probe master mix at 37°C overnight. The next day, cells were washed with 2× SSCT at 60°C for 15 minutes, 2× SSCT at room temperature for 10 minutes, and 0.2× SSC at room temperature for 10 minutes. Secondary oligonucleotide was mixed with formamide and hybridization buffer. Cells were incubated with secondary hybridization master mix at room temperature for 2 hours followed with 2× SSCT at 60°C for 15 minutes, 2× SSCT at room temperature for 10 minutes, and 0.2× SSC at room temperature for 10 minutes. Finally, cells were incubated with DAPI buffer at room temperature for 5 minutes, washed in 2× SSC for 5 minutes, and slides were mounted on coverslips with mounting medium (Thermo Fisher Scientific, S36936). Images were captured using Zeiss LSM 880 Confocal.
In Vivo Drug Treatment
All animal experiments related to this study were approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Pennsylvania. To prepare TDI-11055 for in vivo treatment, the compound was resuspended in DMSO at 400 mg/mL and diluted with 10% hydroxypropyl β-cyclodextrin (HPB; KLEPTOSE, #346112). The solution was adjusted for pH to 4.0 with 2 mol/L HCl, sonicated at 4°C for 2 minutes to totally dissolve the TDI-11055, and further diluted 1:20 with 10% HPB to the final concentration of 20 mg/mL. DMSO solution was prepared with the same protocol but without the addition of TDI-11055. Enl-WT and Enl-T1 mice were treated with DMSO or 200 mg/kg TDI-11055 once daily via oral gavage for 8 consecutive weeks starting from 7 days or 8 weeks post poly (I:C) treatment. Animals were monitored daily, and body weights were measured every 2 days throughout the treatment period. To assess leukemia progression, PB was extracted from the mice via retro-orbital bleeding, stained for the percentage of CD45.1−CD45.2+ cells, and subjected to flow cytometry analysis every 2 weeks throughout the study. Kaplan–Meier survival curves were created using the GraphPad Prism (v9) software.
RT-qPCR Analyses
Total RNA was isolated using RNeasy UCP Micro kit (Qiagen, #73934) and reverse transcribed with the high-capacity cDNA reverse transcription kit (Thermo Fisher, #4368814) in accordance with the manufacturer’s instructions. Quantitative PCR was performed using the SYBR Green PCR Master Mix (Fisher Scientific, #A25778) with the ViiA 7 Real-time PCR system (Thermo Fisher).
MSK Mouse-IMPACT Capture-Based Next-Generation Sequencing
Genomic DNA was isolated from BM cells of Enl-WT or Enl-T1 mice using the Puregene Cell kit (Qiagen, #158043). The genomic DNA was then sheared and prepared for DNA sequencing library using the MSK Mouse-IMPACT platform. The samples were sequenced using Illumina Hiseq2500.
Data analysis was performed as previously described (47–49). Briefly, adapter sequences at the end of the reads were removed using cutadapt (v1.6). Reads were mapped to the mouse reference genome (mm10) using BWA mapper (bwa mem v0.7.12). The mapped SAM files were sorted and read group tags were added using the PICARD tools. The sorted and grouped BAM files were processed with PICARD MarkDuplicates, followed with GATK toolkit (v3.2) analysis according to the best practices for tumor and normal pairs. The tumor and normal pairs were realigned using ABRA (v 0.92), and then the base quality values were recalibrated with the BaseQRecalibrator. Somatic variants were then called in the processed BAMs using muTect (v1.1.7) for SNV and the Haplotype caller from GATK with a custom post-processing script to call somatic indels.
RNA Sequencing
Total RNA was extracted using RNeasy UCP Micro kit (Qiagen, #73934), and 500 ng total RNA was used for library preparation using the polyA mRNA magnetic isolation module (NEB, #E7490) and RNA library prep kit (NEB, #7770) following the manufacturer’s instructions. RNA samples were sequenced using Illumina NextSeq 500.
Reads were aligned to the mouse reference genome (mm10) using HISAT2 v 2.2.1 with default parameters (101). The package featureCounts v 2.0.2 was used for counting the mapped reads in each mouse gene (102). Transcript per million (TPM) was used for normalizing the gene expression. Differentially expressed genes between conditions were statistically determined by R package DESeq2 v1.38.3 (103). Genes with adjusted P value < 0.05 and fold change ≥ 1.5 were reported as differential genes. Volcano plots and heatmap were generated directly in R.
Principal Component Analysis
R package FactoMineR v 2.8 was used to perform PCA. Briefly, gene expression profiles, normalized by TPM for all the samples, were provided as input for the FactoMineR package module PCA. By employing the main dimensions PC1 and PC2, we generated a dot plot for visualization.
Gene Set Enrichment Analysis
GSEA was performed using the GSEA v4.1.0 software with 1,000 gene set permutations. Gene sets used were manually curated from published or our own datasets. A detailed description of GSEA methodology and interpretation can be found at http://www.broadinstitute.org/gsea/doc/GSEAUserGuideFrame.html. Gene rank lists were generated by ordering the expression fold change (T1 vs. WT) from largest to smallest in indicated cells. Enrichment score reflects the degree to which a gene set is overrepresented at the top or bottom of a ranked list of genes. The normalized enrichment score is the enrichment score with normalization across analyzed gene sets. The false discovery rate q value is the estimated probability that a gene set with a given normalized enrichment score represents a false positive finding. All gene sets used in this study are provided in Supplementary Table S8.
CUT&Tag
Hematopoietic cells were collected and nuclei were extracted. The nuclei were crosslinked with 0.1% formaldehyde in PBS at room temperature for 1 minute and stopped with 125 mmol/L glycine for 5 minutes. Then, 40K nuclei were incubated with 10 μL activated ConA beads (Bangs Laboratories, #BP531) at room temperature for 10 minutes. Next, 50 μL cold antibody150 buffer (20 mmol/L HEPES, PH 7.5; 150 mmol/L NaCl; 0.5 mmol/L Spermidine; 1× EDTA-free protease inhibitor) was added, with 1 μg indicated antibodies, mixed and incubated overnight at 4°C. The beads were washed, and then incubated with secondary antibody at room temperature for 30 minutes. Samples were incubated with pAG-Tn5 (Epicypher, #15-1017) and chromatin tagmentation was performed. The CUT&Tag DNA was amplified using CUTANA High Fidelity 2× PCR Master Mix (Epicypher, #15-1018), and purified with AMPure beads (Beckman Coulter, #A63881).
CUT&Tag samples were sequenced using the Illumina NextSeq2000. Reads were aligned to the mouse reference genome (mm10) by the package Bowtie2 v 2.2.5 with the default parameters (104). The package samtools v 1.6 was used for converting to BAM format, sorting, removing duplicates, and indexing (105). Peak calling was performed by the package MACS2 v 2.2.8 with the parameters “-f BAMPE −nomodel -q 1E-2 −broad −keep-dup all” for both H3K27ac and H3K27me3 data (106). The chromatin distribution of the CUT&Tag peaks was determined by HOMER module annotatePeaks.pl v 4.11 (107). To make the heatmaps, the filtered, sorted, and indexed BAM files were converted to bigwig files using the bamCoverage from deeptools v3.5.0 and then normalized by counts per million (CPM; ref. 108). Heatmaps were generated using the module computeMatrix and plotHeatmap from deeptools v3.5.0. The coverage plots were generated by either the module plotCoverage from deeptools v3.5.0 or the package ngs.plot.r v2.63 (109).
ChIP-Seq
GMP or L-GMP cells were collected, washed, and crosslinked with 1% formaldehyde in PBS at room temperature for 10 minutes and stopped with 125 mmol/L glycine for 5 minutes. Cells were resuspended and sonicated in RIPA 0.3 buffer (0.1% SDS, 1% Triton X-100, 10 mmol/L Tris-HCl (pH 7.4), 1 mmol/L EDTA (pH 8.0), 0.1% NaDOC, 0.3 mol/L NaCl, 0.25% sarkosyl, 1 mmol/L DTT, and protease inhibitors) using a Covaris Ultrasonicator. 10 μg antibody was pre-incubated with 75 μL Protein A Dynabeads at 4°C for >4 hours. The beads were washed three times with PBS plus 0.01% tween 20 which was then added to and incubated with the samples overnight at 4°C. The immunoprecipitates were washed twice with low salt wash buffer (50 mmol/L Tris pH 8.0, 150 mmol/L NaCl, 1 mmol/L EDTA, 1% Triton X-100, and 0.1% SDS), twice with high salt wash buffer (50 mmol/L Tris pH 8.0, 500 mmol/L NaCl, 1 mmol/L EDTA, 1% Triton X-100, and 0.1% SDS), twice with LiCl wash buffer (50 mmol/L Tris pH 8.0, 150 mmol/L LiCl, 1 mmol/L EDTA, 1% NP-40, and 0.5% Na-Deoxycholate, 0.1% SDS), and once with TE buffer (1 mmol/L EDTA, 10 mmol/L Tris-HCl pH8.0) plus 50 mmol/L NaCl. Bound DNA was eluted using 200 μL ChIP elution buffer (1% SDS, 50 mmol/L Tris-HCl, pH 8.0, 10 mmol/L EDTA, 200 mmol/L NaCl), reverse crosslinked, and then purified using a PCR purification kit (Qiagen, #28106) to a final volume of 50 μL.
ChIP-seq sample libraries were prepared using the NEBNext Ultra II DNA Library Prep Kit (NEB, #E7645L) following the manufacturer’s instructions. The samples were sequenced using an Illumina NextSeq2000. The analysis procedures for ChIP-seq data are almost identical to those for CUT&Tag data, except for the peak calling step. In ChIP-seq analysis, we utilized the corresponding input data as a control during peak calling; other parameters are the same as what we used for CUT&Tag.
Clinical Patient Data Analysis
The patient RNA-seq data were obtained from the TARGET-AML database. GSVA was employed for determining the gene signature score for each patient with AML. The R package GSVA v 1.46.0 was utilized to perform the analysis (110).
Functional Analysis
The predicted biological function of certain gene list was determined by GO analysis, which was performed by the Metascape website (https://metascape.org; ref. 111). The prediction of the function of cis-regulatory regions were performed by GREAT analysis (http://bejerano.stanford.edu/great/public/html/) with the whole genome as the background regions (112).
Statistical Analyses
No statistical methods were used to predetermine sample size. Experimental data are presented as mean ± SEM, unless stated otherwise. Statistical significance was calculated by two-tailed, unpaired t test on two experimental conditions with P < 0.05 considered statistically significant unless stated otherwise.
Oligos
All oligos used are provided in Supplementary Table S37.
Antibodies
All antibodies used are provided in Supplementary Table S38.
Data Availability
The CUT&Tag, ChIP–seq, and RNA-seq data have been deposited in the Gene Expression Omnibus database under accession numbers GSE239470. All other raw data generated or analyzed during this study are included in this published article (and its Supplementary Files). Codes used for data analysis are available upon request.
Supplementary Material
Supplementary Table S1. Genes differentially expressed between Enl-T1 and Enl-WT LSK cells. Supplementary Table S2. Genes differentially expressed between Enl-T1 and Enl-WT GMP cells. Supplementary Table S3. Genes differentially expressed between Enl-T1 and Enl-WT L-GMP cells. Supplementary Table S4. Genes differentially expressed between Enl-T1 and Enl-WT cKit + Mac1+ cells. Supplementary Table S5. Genes differentially expressed between Enl-T1 and Enl-WT cKit-Mac1+ cells. Supplementary Table S6. Shared Enl-T1 up-regulated DEGs in LSK, GMP, and L-GMP cells. Supplementary Table S7. Expression of Hoxa genes in Enl-WT and Enl-T1 hematopoietic populations. Supplementary Table S8. GSEA gene sets used in Supplementary Figure S12. Supplementary Table S9. GSVA score of patients from TARGET-AML database. Supplementary Table S10. H3K27ac peaks in all hematopoietic populations. Supplementary Table S11. H3K27ac differential regions of ENL-T1 versus ENL-WT in LSK cells. Supplementary Table S12. H3K27ac differential regions of ENL-T1 versus ENL-WT in GMP cells. Supplementary Table S13. H3K27ac differential regions of ENL-T1 versus ENL-WT in L-GMP cells. Supplementary Table S14. H3K27ac differential regions of ENL-T1 versus ENL-WT in cKit + Mac1+ cells. Supplementary Table S15. H3K27ac differential regions of ENL-T1 versus ENL-WT in cKit-Mac1+ cells. Supplementary Table S16. T1-UP DEGs associated with H3K27ac T1 gained DRs in all hematopoietic populations. Supplementary Table S17. H3K27ac T1 gained differential regions associated with T1-UP DEGs in all hematopoietic populations. Supplementary Table S18. p300 ChIP-seq normalized signal at T1 gained H3K27ac differential regions in Enl-WT and Enl-T1 GMP, L-GMP cells. Supplementary Table S19. T1 gained H3K27ac differential regions with both p300 UP and associated gene expression UP in Enl-T1 cells for GMP and L-GMP. Supplementary Table S20. T1 gained H3K27ac differential regions with p300 UP and associated gene expression UP in Enl-T1 cells for L-GMP under A-485 treatment. Supplementary Table S21. H3K27me3 peaks in wildtype hematopoietic populations. Supplementary Table S22. Hematopoietic differentiation associated-H3K27me3 peaks. Supplementary Table S23. H3K27me3 peaks in all hematopoietic populations. Supplementary Table S24. H3K27me3 differential regions of ENL-T1 versus ENL-WT in LSK cells. Supplementary Table S25. H3K27me3 differential regions of ENL-T1 versus ENL-WT in GMP cells. Supplementary Table S26. H3K27me3 differential regions of ENL-T1 versus ENL-WT in L-GMP cells. Supplementary Table S27. H3K27me3 differential regions of ENL-T1 versus ENL-WT in cKit + Mac1+ cells. Supplementary Table S28. H3K27me3 differential regions of ENL-T1 versus ENL-WT in cKit-Mac1+ cells. Supplementary Table S29. T1-UP DEGs associated with H3K27me3 lost differential regions in all hematopoietic populations. Supplementary Table S30. Group1 and group2 gene list in cKit + Mac1+ and cKit-Mac1+ cells. Supplementary Table S31. FLAG-ENL peaks in LSK cells expressing the indicated FLAG-ENL transgenes. Supplementary Table S32. FLAG-ENL gained regions of T1 versus WT in LSK cells expressing the indicated FLAG-ENL transgenes. Supplementary Table S33. H3K27ac T1 gained differential regions associated with T1 gained FLAG-ENL DRs in LSK cells expressing the indicated FLAG-ENL transgenes. Supplementary Table S34. Genes differentially expressed between Enl-T1-DMSO and Enl-WT-DMSO LSK cells. Supplementary Table S35. Genes differentially expressed between Enl-T1-DMSO and Enl-WT-DMSO in GMP cells. Supplementary Table S36. Expression of Hoxa genes under Enl-WT-DMSO, Enl-T1-DMSO and Enl-T1-TDI conditions in LSK and GMP cells. Supplementary Table S37. Oligos used in this study. Supplementary Table S38. Antibodies used in this study.
Supplementary Figure S1. Generation of a conditional knock-in mouse model for the Enl-T1 mutation. Supplementary Figure S2. Impact of Enl mutation on the peripheral blood and spleen. Supplementary Figure S3. Characterization of Enl-T1 allele expression and concurrent mutations in heterozygous knock-in Enl-T1 mouse model. Supplementary Figure S4. UBC-cre-ERT2/Enl flox-T1/+ mice develop aggressive acute leukemia following tamoxifen treatment. Supplementary Figure S5. Enl mutation leads to expansion of myeloid cells in mice in the leukemic phase. Supplementary Figure S6. Enl mutation leads to the decrease of B220+CD19+ B, CD4+T, CD8+ T cells in mice in the leukemic phase. Supplementary Figure S7. Impact of the Enl mutation on the bone marrow, peripheral blood, spleen, and thymus in mice in the pre-leukemic phase. Supplementary Figure S8. Enl mutation perturbs the normal hematopoietic hierarchy and leads to abnormal expansion of myeloid progenitors in mice in the leukemic phase. Supplementary Figure S9. Enl mutation does not lead to the expansion of myeloid progenitors in mice in the pre-leukemic phase. Supplementary Figure S10. Enl mutation promotes self-renewal properties of HSPCs. Supplementary Figure S11. Enl mutation-induced up- and down-regulated genes are related to distinct biological functions. Supplementary Figure S12. Enl mutation leads to a gain of myeloid differentiation signatures in HSPCs. Supplementary Figure S13. Mutant ENL-induced H3K27ac signals correlate with upregulation of development and inflammation associated transcriptional programs. Supplementary Figure S14. HSPCs gain H3K27me3 during differentiation in wildtype mice. Supplementary Figure S15. Differentiation-associated gain of H3K27me3 is impaired in Enl-mutated hematopoietic cells. Supplementary Figure S16. ENL mutants form condensate at key target genes and increase gene expression in HSPCs. Supplementary Figure S17. Condensate formation property correlates with mutant ENL’s oncogenic function in human CD34+ HSPCs. Supplementary Figure S18. Expression levels of different FLAG-ENL transgenes in LSK cells. Supplementary Figure S19. Condensate formation property correlates with mutant ENL’s oncogenic function in GMP cells. Supplementary Figure S20. Disrupting condensate formation by the H116P mutation reduces ENL-T1-induced increases in chromatin occupancy of FLAG-ENL, H3K27ac, and p300 at a subset of target genes. Supplementary Figure S21. Impact of mutant ENL on leukemia development and condensate formation. Supplementary Figure S22. Small molecule inhibition of the acetyl-binding activity of mutant ENL suppresses chromatin function and Hoxa cluster gene activation in LSK cells. Supplementary Figure S23. Small molecule inhibition of the acetyl-binding activity of mutant ENL impairs its chromatin and transcriptional function in HSPCs. Supplementary Figure S24. Small molecule inhibition of the acetyl-binding activity of mutant ENL inhibits its impact on the self-renewal property in HSPCs.
Acknowledgments
The authors would like to thank members of the Wan laboratory for technical support and scientific input throughout the study; Gerd Blobel and Kathrin Bernt for critical reading of the manuscript; the flow cytometry and cell sorting resource laboratories at the Children’s Hospital of Philadelphia and University of Pennsylvania for technical support; animal facility at the University of Pennsylvania for assistance with housing and maintenance of mice. The authors appreciate the support from C. David Allis (The Rockefeller University) and the St. Jude Children’s Research Hospital Collaborative Initiative on Chromatin Regulation in Pediatric Cancer during the initial stages of mouse model generation. The research was supported by a NIH Director’s New Innovator Award (1DP2HG012443 to L. Wan), a NIH pathway to Independence Award (R00CA226399 to L. Wan), a Pew-Stewart Scholar Award (L. Wan), a V Foundation Scholar Award (L. Wan), an ASH Scholar Award (L. Wan), a LLS Scholar Award (L. Wan), an ACS Scholar Award (L. Wan), an AFCRI Postdoctoral Fellowship (Y. Liu), a LLS Special Fellow Award (Y. Liu), a NIH R01 award (R01AI091627 to I. Maillard), and a NIH fellowship award (F30AI161873 to A. Vanderbeck).
Footnotes
NoteSupplementary data for this article are available at Cancer Discovery Online (http://cancerdiscovery.aacrjournals.org/).
Authors’ Disclosures
I. Maillard reports grants from NIAID and grants from NIAID during the conduct of the study; other support from Genentech and Regeneron; and personal fees from Garuda Therapeutics outside the submitted work. L. Wan reports personal fees from Bridge Medicines outside the submitted work; in addition, L. Wan has a patent for US No. 62/949,160 issued. No disclosures were reported by the other authors.
Authors’ Contributions
Y. Liu: Data curation, formal analysis, investigation, visualization, methodology, writing–original draft, writing–review and editing. Q. Li: Data curation, formal analysis, investigation, visualization, methodology, writing–review and editing. L. Song: Data curation, investigation. C. Gong: Data curation, investigation. S. Tang: Data curation, investigation, writing–review and editing. K.A. Budinich: Data curation, investigation, writing-review and editing. A. Vanderbeck: Investigation, methodology. K.M. Mathias: Investigation. G.B. Wertheim: Investigation, writing–review and editing. S.C. Nguyen: Investigation, methodology. R. Outen: Resources. E.F. Joyce: Investigation, methodology. I. Maillard: Supervision, investigation, methodology, writing–review and editing. L. Wan: Conceptualization, data curation, formal analysis, supervision, investigation, visualization, methodology, writing-original draft, writing–review and editing.
References
- 1. Bradner JE, Hnisz D, Young RA. Transcriptional addiction in cancer. Cell 2017;168:629–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cho W-K, Spille J-H, Hecht M, Lee C, Li C, Grube V, et al. Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science 2018;361:412–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sabari BR, Dall’Agnese A, Boija A, Klein IA, Coffey EL, Shrinivas K, et al. Coactivator condensation at super-enhancers links phase separation and gene control. Science 2018;361:eaar3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chong S, Dugast-Darzacq C, Liu Z, Dong P, Dailey GM, Cattoglio C, et al. Imaging dynamic and selective low-complexity domain interactions that control gene transcription. Science 2018;361:eaar2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mir M, Stadler MR, Ortiz SA, Hannon CE, Harrison MM, Darzacq X, et al. Dynamic multifactor hubs interact transiently with sites of active transcription in Drosophila embryos. Elife 2018;7:e40497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cisse II, Izeddin I, Causse SZ, Boudarene L, Senecal A, Muresan L, et al. Real-time dynamics of RNA polymerase II clustering in live human cells. Science 2013;341:664–7. [DOI] [PubMed] [Google Scholar]
- 7. Hnisz D, Shrinivas K, Young RA, Chakraborty AK, Sharp PA. A phase separation model for transcriptional control. Cell 2017;169:13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bhat P, Honson D, Guttman M. Nuclear compartmentalization as a mechanism of quantitative control of gene expression. Nat Rev Mol Cell Biol 2021;22:653–70. [DOI] [PubMed] [Google Scholar]
- 9. Banani SF, Lee HO, Hyman AA, Rosen MK. Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol 2017;18:285–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Alberti S, Gladfelter A, Mittag T. Considerations and challenges in studying liquid-liquid phase separation and biomolecular condensates. Cell 2019;176:419–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shin Y, Brangwynne CP. Liquid phase condensation in cell physiology and disease. Science 2017;357:eaaf4382. [DOI] [PubMed] [Google Scholar]
- 12. Mittag T, Pappu RV. A conceptual framework for understanding phase separation and addressing open questions and challenges. Mol Cell 2022;82:2201–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Han X, Yu D, Gu R, Jia Y, Wang Q, Jaganathan A, et al. Roles of the BRD4 short isoform in phase separation and active gene transcription. Nat Struct Mol Biol 2020;27:333–41. [DOI] [PubMed] [Google Scholar]
- 14. Ma L, Gao Z, Wu J, Zhong B, Xie Y, Huang W, et al. Co-condensation between transcription factor and coactivator p300 modulates transcriptional bursting kinetics. Mol Cell 2021;81:1682–97.e7. [DOI] [PubMed] [Google Scholar]
- 15. Chong S, Graham TG, Dugast-Darzacq C, Dailey GM, Darzacq X, Tjian R. Tuning levels of low-complexity domain interactions to modulate endogenous oncogenic transcription. Mol Cell 2022;82:2084–97.e5. [DOI] [PubMed] [Google Scholar]
- 16. Trojanowski J, Frank L, Rademacher A, Mücke N, Grigaitis P, Rippe K. Transcription activation is enhanced by multivalent interactions independent of phase separation. Mol Cell 2022;82:1878–93.e10. [DOI] [PubMed] [Google Scholar]
- 17. McSwiggen DT, Mir M, Darzacq X, Tjian R. Evaluating phase separation in live cells: diagnosis, caveats, and functional consequences. Genes Dev 2019;33:1619–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Basu S, Mackowiak SD, Niskanen H, Knezevic D, Asimi V, Grosswendt S, et al. Unblending of transcriptional condensates in human repeat expansion disease. Cell 2020;181:1062–79.e30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wan L, Chong S, Xuan F, Liang A, Cui X, Gates L, et al. Impaired cell fate through gain-of-function mutations in a chromatin reader. Nature 2020;577:121–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ahn JH, Davis ES, Daugird TA, Zhao S, Quiroga IY, Uryu H, et al. Phase separation drives aberrant chromatin looping and cancer development. Nature 2021;595:591–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Boulay G, Sandoval GJ, Riggi N, Iyer S, Buisson R, Naigles B, et al. Cancer-specific retargeting of BAF complexes by a prion-like domain. Cell 2017;171:163–78.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chandra B, Michmerhuizen NL, Shirnekhi HK, Tripathi S, Pioso BJ, Baggett DW, et al. Phase separation mediates NUP98 fusion oncoprotein leukemic transformation. Cancer Discov 2022;12:1152–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Terlecki-Zaniewicz S, Humer T, Eder T, Schmoellerl J, Heyes E, Manhart G, et al. Biomolecular condensation of NUP98 fusion proteins drives leukemogenic gene expression. Nat Struct Mol Biol 2021;28:190–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang XQD, Fan D, Han Q, Liu Y, Miao H, Wang X, et al. Mutant NPM1 hijacks transcriptional hubs to maintain pathogenic gene programs in acute myeloid leukemia. Cancer Discov 2023;13:724–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shi B, Li W, Song Y, Wang Z, Ju R, Ulman A, et al. UTX condensation underlies its tumour-suppressive activity. Nature 2021;597:726–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Alberti S, Dormann D. Liquid–liquid phase separation in disease. Annu Rev Genet 2019;53:171–94. [DOI] [PubMed] [Google Scholar]
- 27. Tsang B, Pritišanac I, Scherer SW, Moses AM, Forman-Kay JD. Phase separation as a missing mechanism for interpretation of disease mutations. Cell 2020;183:1742–56. [DOI] [PubMed] [Google Scholar]
- 28. Li CH, Coffey EL, Dall’Agnese A, Hannett NM, Tang X, Henninger JE, et al. MeCP2 links heterochromatin condensates and neurodevelopmental disease. Nature 2020;586:440–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fan C, Zhang H, Fu L, Li Y, Du Y, Qiu Z, et al. Rett mutations attenuate phase separation of MeCP2. Cell Discov 2020;6:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang L, Hu M, Zuo MQ, Zhao J, Wu D, Huang L, et al. Rett syndrome-causing mutations compromise MeCP2-mediated liquid-liquid phase separation of chromatin. Cell Res 2020;30:393–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Song L, Yao X, Li H, Peng B, Boka AP, Liu Y, et al. Hotspot mutations in the structured ENL YEATS domain link aberrant transcriptional condensates and cancer. Mol Cell 2022;82:4080–98.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wan L, Wen H, Li Y, Lyu J, Xi Y, Hoshii T, et al. ENL links histone acetylation to oncogenic gene expression in acute myeloid leukaemia. Nature 2017;543:265–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Erb MA, Scott TG, Li BE, Xie H, Paulk J, Seo H-S, et al. Transcription control by the ENL YEATS domain in acute leukaemia. Nature 2017;543:270–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lin C, Smith ER, Takahashi H, Lai KC, Martin-Brown S, Florens L, et al. AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol Cell 2010;37:429–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yokoyama A, Lin M, Naresh A, Kitabayashi I, Cleary ML. A higher-order complex containing AF4 and ENL family proteins with P-TEFb facilitates oncogenic and physiologic MLL-dependent transcription. Cancer Cell 2010;17:198–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Biswas D, Milne TA, Basrur V, Kim J, Elenitoba-Johnson KS, Allis CD, et al. Function of leukemogenic mixed lineage leukemia 1 (MLL) fusion proteins through distinct partner protein complexes. Proc Natl Acad Sci U S A 2011;108:15751–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chou S, Upton H, Bao K, Schulze-Gahmen U, Samelson AJ, He N, et al. HIV-1 Tat recruits transcription elongation factors dispersed along a flexible AFF4 scaffold. Proc Natl Acad Sci U S A 2013;110:E123–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mueller D, Bach C, Zeisig D, Garcia-Cuellar M-P, Monroe S, Sreekumar A, et al. A role for the MLL fusion partner ENL in transcriptional elongation and chromatin modification. Blood J Am Soc Hematol 2007;110:4445–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Winters AC, Bernt KM. MLL-rearranged leukemias—an update on science and clinical approaches. Front Pediatr 2017;5:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer 2007;7:823–33. [DOI] [PubMed] [Google Scholar]
- 41. Liu Y, Li Q, Alikarami F, Barrett DR, Mahdavi L, Li H, et al. Small-molecule inhibition of the acyl-lysine reader ENL as a strategy against acute myeloid leukemia. Cancer Discov 2022;12:2684–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Perlman EJ, Gadd S, Arold ST, Radhakrishnan A, Gerhard DS, Jennings L, et al. MLLT1 YEATS domain mutations in clinically distinctive Favourable Histology Wilms tumours. Nat Commun 2015;6:10013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gadd S, Huff V, Walz AL, Ooms AH, Armstrong AE, Gerhard DS, et al. A Children’s Oncology Group and TARGET initiative exploring the genetic landscape of Wilms tumor. Nat Genet 2017;49:1487–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hetzner K, Garcia-Cuellar M-P, Büttner C, Slany RK. The interaction of ENL with PAF1 mitigates polycomb silencing and facilitates murine leukemogenesis. Blood J Am Soc Hematol 2018;131:662–73. [DOI] [PubMed] [Google Scholar]
- 45. Umeda M, Ma J, Westover T, Ni Y, Song G, Maciaszek JL, et al. A new genomic framework to categorize pediatric acute myeloid leukemia. Nat Genet 2024;56:281–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bolouri H, Farrar JE, Triche T, Ries RE, Lim EL, Alonzo TA, et al. The molecular landscape of pediatric acute myeloid leukemia reveals recurrent structural alterations and age-specific mutational interactions. Nat Med 2018;24:103–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Loberg MA, Bell RK, Goodwin LO, Eudy E, Miles LA, SanMiguel JM, et al. Sequentially inducible mouse models reveal that Npm1 mutation causes malignant transformation of Dnmt3a-mutant clonal hematopoiesis. Leukemia 2019;33:1635–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Leibold J, Ruscetti M, Cao Z, Ho YJ, Baslan T, Zou M, et al. Somatic tissue engineering in mouse models reveals an actionable role for WNT pathway alterations in prostate cancer metastasis. Cancer Discov 2020;10:1038–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pronier E, Bowman RL, Ahn J, Glass J, Kandoth C, Merlinsky TR, et al. Genetic and epigenetic evolution as a contributor to WT1-mutant leukemogenesis. Blood 2018;132:1265–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Dong L, Yu WM, Zheng H, Loh ML, Bunting ST, Pauly M, et al. Leukaemogenic effects of Ptpn11 activating mutations in the stem cell microenvironment. Nature 2016;539:304–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Oguro H, Ding L, Morrison SJ. SLAM family markers resolve functionally distinct subpopulations of hematopoietic stem cells and multipotent progenitors. Cell Stem Cell 2013;13:102–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pronk CJ, Rossi DJ, Månsson R, Attema JL, Norddahl GL, Chan CKF, et al. Elucidation of the phenotypic, functional, and molecular topography of a myeloerythroid progenitor cell hierarchy. Cell Stem Cell 2007;1:428–42. [DOI] [PubMed] [Google Scholar]
- 53. Cheng H, Zheng Z, Cheng T. New paradigms on hematopoietic stem cell differentiation. Protein Cell 2020;11:34–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lennartsson J, Rönnstrand L. Stem cell factor receptor/c-Kit: from basic science to clinical implications. Physiol Rev 2012;92:1619–49. [DOI] [PubMed] [Google Scholar]
- 55. Ikuta K, Weissman IL. Evidence that hematopoietic stem cells express mouse c-kit but do not depend on steel factor for their generation. Proc Natl Acad Sci U S A 1992;89:1502–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Somervaille TC, Cleary ML. Identification and characterization of leukemia stem cells in murine MLL-AF9 acute myeloid leukemia. Cancer Cell 2006;10:257–68. [DOI] [PubMed] [Google Scholar]
- 57. Kirstetter P, Schuster MB, Bereshchenko O, Moore S, Dvinge H, Kurz E, et al. Modeling of C/EBPalpha mutant acute myeloid leukemia reveals a common expression signature of committed myeloid leukemia-initiating cells. Cancer Cell 2008;13:299–310. [DOI] [PubMed] [Google Scholar]
- 58. Vetrie D, Helgason GV, Copland M. The leukaemia stem cell: similarities, differences and clinical prospects in CML and AML. Nat Rev Cancer 2020;20:158–73. [DOI] [PubMed] [Google Scholar]
- 59. Yamashita M, Dellorusso PV, Olson OC, Passegué E. Dysregulated haematopoietic stem cell behaviour in myeloid leukaemogenesis. Nat Rev Cancer 2020;20:365–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Argiropoulos B, Humphries RK. Hox genes in hematopoiesis and leukemogenesis. Oncogene 2007;26:6766–76. [DOI] [PubMed] [Google Scholar]
- 61. Collins CT, Hess JL. Deregulation of the HOXA9/MEIS1 axis in acute leukemia. Curr Opin Hematol 2016;23:354–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Drabkin HA, Parsy C, Ferguson K, Guilhot F, Lacotte L, Roy L, et al. Quantitative HOX expression in chromosomally defined subsets of acute myelogenous leukemia. Leukemia 2002;16:186–95. [DOI] [PubMed] [Google Scholar]
- 63. Magli MC, Largman C, Lawrence HJ. Effects of HOX homeobox genes in blood cell differentiation. J Cell Physiol 1997;173:168–77. [DOI] [PubMed] [Google Scholar]
- 64. Blanco MA, Sykes DB, Gu L, Wu M, Petroni R, Karnik R, et al. Chromatin-state barriers enforce an irreversible mammalian cell fate decision. Cell Rep 2021;37:109967. [DOI] [PubMed] [Google Scholar]
- 65. Rattigan KM, Zarou MM, Helgason GV. Metabolism in stem cell–driven leukemia: parallels between hematopoiesis and immunity. Blood J Am Soc Hematol 2023;141:2553–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ye M, Zhang H, Yang H, Koche R, Staber PB, Cusan M, et al. Hematopoietic differentiation is required for initiation of acute myeloid leukemia. Cell Stem Cell 2015;17:611–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kaya-Okur HS, Wu SJ, Codomo CA, Pledger ES, Bryson TD, Henikoff JG, et al. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat Commun 2019;10:1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zhang T, Cooper S, Brockdorff N. The interplay of histone modifications–writers that read. EMBO Rep 2015;16:1467–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Lasko LM, Jakob CG, Edalji RP, Qiu W, Montgomery D, Digiammarino EL, et al. Discovery of a selective catalytic p300/CBP inhibitor that targets lineage-specific tumours. Nature 2017;550:128–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Sankar A, Mohammad F, Sundaramurthy AK, Wang H, Lerdrup M, Tatar T, et al. Histone editing elucidates the functional roles of H3K27 methylation and acetylation in mammals. Nat Genet 2022;54:754–60. [DOI] [PubMed] [Google Scholar]
- 71. Bogliotti YS, Ross PJ. Mechanisms of histone H3 lysine 27 trimethylation remodeling during early mammalian development. Epigenetics 2012;7:976–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev 2006;20:1123–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 2006;441:349–53. [DOI] [PubMed] [Google Scholar]
- 74. Bach C, Buhl S, Mueller D, García-Cuéllar M-P, Maethner E, Slany RK. Leukemogenic transformation by HOXA cluster genes. Blood J Am Soc Hematol 2010;115:2910–8. [DOI] [PubMed] [Google Scholar]
- 75. Chen S-L, Qin Z-Y, Hu F, Wang Y, Dai Y-J, Liang Y. The role of the HOXA gene family in acute myeloid leukemia. Genes 2019;10:621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Andreeff M, Ruvolo V, Gadgil S, Zeng C, Coombes K, Chen W, et al. HOX expression patterns identify a common signature for favorable AML. Leukemia 2008;22:2041–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Argiropoulos B, Yung E, Humphries RK. Unraveling the crucial roles of Meis1 in leukemogenesis and normal hematopoiesis. Genes Dev 2007;21:2845–9. [DOI] [PubMed] [Google Scholar]
- 78. Tran TM, Philipp J, Bassi JS, Nibber N, Draper JM, Lin TL, et al. The RNA-binding protein IGF2BP3 is critical for MLL-AF4-mediated leukemogenesis. Leukemia 2022;36:68–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Kaltenbach S, Soler G, Barin C, Gervais C, Bernard OA, Penard-Lacronique V, et al. NUP98-MLL fusion in human acute myeloblastic leukemia. Blood 2010;116:2332–5. [DOI] [PubMed] [Google Scholar]
- 80. Gough SM, Slape CI, Aplan PD. NUP98 gene fusions and hematopoietic malignancies: common themes and new biologic insights. Blood 2011;118:6247–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Gough SM, Lee F, Yang F, Walker RL, Zhu YJ, Pineda M, et al. NUP98-PHF23 is a chromatin-modifying oncoprotein that causes a wide array of leukemias sensitive to inhibition of PHD histone reader function. Cancer Discov 2014;4:564–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. de Rooij JD, Hollink IH, Arentsen-Peters ST, van Galen JF, Berna Beverloo H, Baruchel A, et al. NUP98/JARID1A is a novel recurrent abnormality in pediatric acute megakaryoblastic leukemia with a distinct HOX gene expression pattern. Leukemia 2013;27:2280–8. [DOI] [PubMed] [Google Scholar]
- 83. Wang GG, Cai L, Pasillas MP, Kamps MP. NUP98-NSD1 links H3K36 methylation to Hox-A gene activation and leukaemogenesis. Nat Cell Biol 2007;9:804–12. [DOI] [PubMed] [Google Scholar]
- 84. Krivtsov AV, Twomey D, Feng Z, Stubbs MC, Wang Y, Faber J, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature 2006;442:818–22. [DOI] [PubMed] [Google Scholar]
- 85. Ayton PM, Cleary ML. Transformation of myeloid progenitors by MLL oncoproteins is dependent on Hoxa7 and Hoxa9. Genes Dev 2003;17:2298–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Heuser M, Yun H, Berg T, Yung E, Argiropoulos B, Kuchenbauer F, et al. Cell of origin in AML: susceptibility to MN1-induced transformation is regulated by the MEIS1/AbdB-like HOX protein complex. Cancer Cell 2011;20:39–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Riedel SS, Lu C, Xie HM, Nestler K, Vermunt MW, Lenard A, et al. Intrinsically disordered Meningioma-1 stabilizes the BAF complex to cause AML. Mol Cell 2021;81:2332–48.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Uckelmann HJ, Haarer EL, Takeda R, Wong EM, Hatton C, Marinaccio C, et al. Mutant NPM1 directly regulates oncogenic transcription in acute myeloid leukemia. Cancer Discov 2023;13:746–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Brunetti L, Gundry MC, Sorcini D, Guzman AG, Huang YH, Ramabadran R, et al. Mutant NPM1 maintains the leukemic state through HOX expression. Cancer Cell 2018;34:499–512.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Goardon N, Marchi E, Atzberger A, Quek L, Schuh A, Soneji S, et al. Coexistence of LMPP-like and GMP-like leukemia stem cells in acute myeloid leukemia. Cancer Cell 2011;19:138–52. [DOI] [PubMed] [Google Scholar]
- 91. Ye M, Zhang H, Amabile G, Yang H, Staber PB, Zhang P, et al. C/EBPa controls acquisition and maintenance of adult haematopoietic stem cell quiescence. Nat Cell Biol 2013;15:385–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Ohlsson E, Hasemann MS, Willer A, Lauridsen FK, Rapin N, Jendholm J, et al. Initiation of MLL-rearranged AML is dependent on C/EBPα. J Exp Med 2014;211:5–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Zhang Y, Brown K, Yu Y, Ibrahim Z, Zandian M, Xuan H, et al. Nuclear condensates of p300 formed though the structured catalytic core can act as a storage pool of p300 with reduced HAT activity. Nat Commun 2021;12:4618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Kasper LH, Brindle PK, Schnabel CA, Pritchard CE, Cleary ML, van Deursen JM. CREB binding protein interacts with nucleoporin-specific FG repeats that activate transcription and mediate NUP98–HOXA9 oncogenicity. Mol Cell Biol 1999;19:764–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Lyon AS, Peeples WB, Rosen MK. A framework for understanding the functions of biomolecular condensates across scales. Nat Rev Mol Cell Biol 2021;22:215–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Mitrea DM, Mittasch M, Gomes BF, Klein IA, Murcko MA. Modulating biomolecular condensates: a novel approach to drug discovery. Nat Rev Drug Discov 2022;21:841–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Conti BA, Oppikofer M. Biomolecular condensates: new opportunities for drug discovery and RNA therapeutics. Trends Pharmacol Sci 2022;43:820–37. [DOI] [PubMed] [Google Scholar]
- 98. Fang MY, Markmiller S, Vu AQ, Javaherian A, Dowdle WE, Jolivet P, et al. Small-molecule modulation of TDP-43 recruitment to stress granules prevents persistent TDP-43 accumulation in ALS/FTD. Neuron 2019;103:802–19.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Babinchak WM, Dumm BK, Venus S, Boyko S, Putnam AA, Jankowsky E, et al. Small molecules as potent biphasic modulators of protein liquid-liquid phase separation. Nat Commun 2020;11:5574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Simone R, Balendra R, Moens TG, Preza E, Wilson KM, Heslegrave A, et al. G-quadruplex-binding small molecules ameliorate C9orf72 FTD/ALS pathology in vitro and in vivo. EMBO Mol Med 2018;10:22–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 2015;12:357–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014;30:923–30. [DOI] [PubMed] [Google Scholar]
- 103. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods 2012;9:357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment/map format and SAMtools. Bioinformatics 2009;25:2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, et al. Model-based analysis of ChIP-seq (MACS). Genome Biol 2008;9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 2010;38:576–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res 2016;44:W160–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Shen L, Shao N, Liu X, Nestler E. ngs.plot: quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC Genomics 2014;15:284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 2013;14:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun 2019;10:1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol 2010;28:495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table S1. Genes differentially expressed between Enl-T1 and Enl-WT LSK cells. Supplementary Table S2. Genes differentially expressed between Enl-T1 and Enl-WT GMP cells. Supplementary Table S3. Genes differentially expressed between Enl-T1 and Enl-WT L-GMP cells. Supplementary Table S4. Genes differentially expressed between Enl-T1 and Enl-WT cKit + Mac1+ cells. Supplementary Table S5. Genes differentially expressed between Enl-T1 and Enl-WT cKit-Mac1+ cells. Supplementary Table S6. Shared Enl-T1 up-regulated DEGs in LSK, GMP, and L-GMP cells. Supplementary Table S7. Expression of Hoxa genes in Enl-WT and Enl-T1 hematopoietic populations. Supplementary Table S8. GSEA gene sets used in Supplementary Figure S12. Supplementary Table S9. GSVA score of patients from TARGET-AML database. Supplementary Table S10. H3K27ac peaks in all hematopoietic populations. Supplementary Table S11. H3K27ac differential regions of ENL-T1 versus ENL-WT in LSK cells. Supplementary Table S12. H3K27ac differential regions of ENL-T1 versus ENL-WT in GMP cells. Supplementary Table S13. H3K27ac differential regions of ENL-T1 versus ENL-WT in L-GMP cells. Supplementary Table S14. H3K27ac differential regions of ENL-T1 versus ENL-WT in cKit + Mac1+ cells. Supplementary Table S15. H3K27ac differential regions of ENL-T1 versus ENL-WT in cKit-Mac1+ cells. Supplementary Table S16. T1-UP DEGs associated with H3K27ac T1 gained DRs in all hematopoietic populations. Supplementary Table S17. H3K27ac T1 gained differential regions associated with T1-UP DEGs in all hematopoietic populations. Supplementary Table S18. p300 ChIP-seq normalized signal at T1 gained H3K27ac differential regions in Enl-WT and Enl-T1 GMP, L-GMP cells. Supplementary Table S19. T1 gained H3K27ac differential regions with both p300 UP and associated gene expression UP in Enl-T1 cells for GMP and L-GMP. Supplementary Table S20. T1 gained H3K27ac differential regions with p300 UP and associated gene expression UP in Enl-T1 cells for L-GMP under A-485 treatment. Supplementary Table S21. H3K27me3 peaks in wildtype hematopoietic populations. Supplementary Table S22. Hematopoietic differentiation associated-H3K27me3 peaks. Supplementary Table S23. H3K27me3 peaks in all hematopoietic populations. Supplementary Table S24. H3K27me3 differential regions of ENL-T1 versus ENL-WT in LSK cells. Supplementary Table S25. H3K27me3 differential regions of ENL-T1 versus ENL-WT in GMP cells. Supplementary Table S26. H3K27me3 differential regions of ENL-T1 versus ENL-WT in L-GMP cells. Supplementary Table S27. H3K27me3 differential regions of ENL-T1 versus ENL-WT in cKit + Mac1+ cells. Supplementary Table S28. H3K27me3 differential regions of ENL-T1 versus ENL-WT in cKit-Mac1+ cells. Supplementary Table S29. T1-UP DEGs associated with H3K27me3 lost differential regions in all hematopoietic populations. Supplementary Table S30. Group1 and group2 gene list in cKit + Mac1+ and cKit-Mac1+ cells. Supplementary Table S31. FLAG-ENL peaks in LSK cells expressing the indicated FLAG-ENL transgenes. Supplementary Table S32. FLAG-ENL gained regions of T1 versus WT in LSK cells expressing the indicated FLAG-ENL transgenes. Supplementary Table S33. H3K27ac T1 gained differential regions associated with T1 gained FLAG-ENL DRs in LSK cells expressing the indicated FLAG-ENL transgenes. Supplementary Table S34. Genes differentially expressed between Enl-T1-DMSO and Enl-WT-DMSO LSK cells. Supplementary Table S35. Genes differentially expressed between Enl-T1-DMSO and Enl-WT-DMSO in GMP cells. Supplementary Table S36. Expression of Hoxa genes under Enl-WT-DMSO, Enl-T1-DMSO and Enl-T1-TDI conditions in LSK and GMP cells. Supplementary Table S37. Oligos used in this study. Supplementary Table S38. Antibodies used in this study.
Supplementary Figure S1. Generation of a conditional knock-in mouse model for the Enl-T1 mutation. Supplementary Figure S2. Impact of Enl mutation on the peripheral blood and spleen. Supplementary Figure S3. Characterization of Enl-T1 allele expression and concurrent mutations in heterozygous knock-in Enl-T1 mouse model. Supplementary Figure S4. UBC-cre-ERT2/Enl flox-T1/+ mice develop aggressive acute leukemia following tamoxifen treatment. Supplementary Figure S5. Enl mutation leads to expansion of myeloid cells in mice in the leukemic phase. Supplementary Figure S6. Enl mutation leads to the decrease of B220+CD19+ B, CD4+T, CD8+ T cells in mice in the leukemic phase. Supplementary Figure S7. Impact of the Enl mutation on the bone marrow, peripheral blood, spleen, and thymus in mice in the pre-leukemic phase. Supplementary Figure S8. Enl mutation perturbs the normal hematopoietic hierarchy and leads to abnormal expansion of myeloid progenitors in mice in the leukemic phase. Supplementary Figure S9. Enl mutation does not lead to the expansion of myeloid progenitors in mice in the pre-leukemic phase. Supplementary Figure S10. Enl mutation promotes self-renewal properties of HSPCs. Supplementary Figure S11. Enl mutation-induced up- and down-regulated genes are related to distinct biological functions. Supplementary Figure S12. Enl mutation leads to a gain of myeloid differentiation signatures in HSPCs. Supplementary Figure S13. Mutant ENL-induced H3K27ac signals correlate with upregulation of development and inflammation associated transcriptional programs. Supplementary Figure S14. HSPCs gain H3K27me3 during differentiation in wildtype mice. Supplementary Figure S15. Differentiation-associated gain of H3K27me3 is impaired in Enl-mutated hematopoietic cells. Supplementary Figure S16. ENL mutants form condensate at key target genes and increase gene expression in HSPCs. Supplementary Figure S17. Condensate formation property correlates with mutant ENL’s oncogenic function in human CD34+ HSPCs. Supplementary Figure S18. Expression levels of different FLAG-ENL transgenes in LSK cells. Supplementary Figure S19. Condensate formation property correlates with mutant ENL’s oncogenic function in GMP cells. Supplementary Figure S20. Disrupting condensate formation by the H116P mutation reduces ENL-T1-induced increases in chromatin occupancy of FLAG-ENL, H3K27ac, and p300 at a subset of target genes. Supplementary Figure S21. Impact of mutant ENL on leukemia development and condensate formation. Supplementary Figure S22. Small molecule inhibition of the acetyl-binding activity of mutant ENL suppresses chromatin function and Hoxa cluster gene activation in LSK cells. Supplementary Figure S23. Small molecule inhibition of the acetyl-binding activity of mutant ENL impairs its chromatin and transcriptional function in HSPCs. Supplementary Figure S24. Small molecule inhibition of the acetyl-binding activity of mutant ENL inhibits its impact on the self-renewal property in HSPCs.
Data Availability Statement
The CUT&Tag, ChIP–seq, and RNA-seq data have been deposited in the Gene Expression Omnibus database under accession numbers GSE239470. All other raw data generated or analyzed during this study are included in this published article (and its Supplementary Files). Codes used for data analysis are available upon request.