Abstract
Ascoschoengastia indica is one of the dominant chigger species in Southeast Asia and a potential carrier of scrub typhus, due in part to its cosmopolitan nature. This study explored the possible biological significance of the observed dimorphism in the shape of its scutum sensilla. Sensilla are specialized structures that are generally adapted to perform specific functions related to sensory capabilities, so their shape and sizes are expected to vary between taxa. We describe morphological variation of the sensilla of A. indica in Thailand. The sensilla had either a round or an ovoid, club-shaped form, which was not dependent on the particularly locality or host. Ignoring the precise function of the sensilla and their morphological variation, our study attempted to answer the following single question: Do the distinct forms of the sensilla indicate possible heterogeneity of the A. indica species? The two forms, named S1 and S2, were compared by genetic and morphometric techniques. The genetic analysis was based on the COI sequences, while the morphometric comparison used the scutum, an organ shown to be of taxonomic value for chigger mites. Neither morphometric nor genetic data revealed any evidence of a speciation process underlying the morphological variation in sensillum types.
Keywords: Ascoschoengastia indica, Sensilla dimorphism, Geometric morphometrics, COI, Urban parks, Bangkok
Highlights
-
•
The scutum sensilla of A. indica in Thailand are round or ovoid, club-shaped.
-
•
Geometric morphometry of the sensilla contour confirmed the visual observation.
-
•
Geometric analyses of the scutum contour or landmarks revealed no notable variation.
-
•
The distinct sensilla forms showed no genetic divergence suggestive of speciation.
1. Introduction
In Thailand, scrub typhus continues to be a leading cause of acute febrile illnesses, with an average of 6,394 cases reported annually in 2018–2022 [1]. It is a vector-borne disease transmitted by the bite of the infected larval stage of Trombiculid mites, which is the stage considered in this study. A gram-negative bacterium, Orientia tsutsugamushi, is the causative agent of this disease [2,3]. More than 3,000 species of chiggers have been identified worldwide [4]. In 2016, 99 species of Trombiculid mites were confirmed and recorded in the revised checklist of chigger mites in Thailand [5]. Recently, the number of chigger species recorded as currently being present in Thailand has increased to 156 [6]. Trombiculid mites are common not only in rural habitats but also in urban areas where cosmopolitan rats are present [7].
The Trombiculid mite Ascoschoengastia indica (Hirst) (Prostigmata, Trombiculidae) is one of the dominant chigger species in Southeast Asia and a potential carrier of scrub typhus, due in part to its cosmopolitan nature [8,9]. Identification of chiggers to the species level is crucial to identify which of them act as vectors of scrub typhus. Medical entomologists and also acarologists are expected to identify species, especially cryptic species. Limitation in the available information on DNA sequences of chigger mites is a critical issue that needs to be overcome in order to aid the accurate classification of various chigger mite species [10]. The current taxonomic classification mainly relies on external morphological traits, but such traits can sometimes be confusing. In addition to environmentally induced morphological variation, the existence of hybridization [11] or the presence of undetected biological species [12] might introduce taxonomic uncertainties when relying on external morphological traits. Against this background, geometric morphometrics can be useful [13]. This morphological approach is based on quantitative rather than qualitative traits, and its relevance has been demonstrated on numerous occasions, including in chigger mites [14] or ticks [15,16].
In the original description by Hirst [17], sensilla of Schoengastia indica (a synonym of Ascoschoengastia indica) were described as having an elongated oval shape. A similar club-shaped aspect was later reported by Womersley [18] and more recently by Fernandes and Kulkarni [19]. The main aim of the current study on A. indica is to understand the possible evolutionary significance of locally observed sensilla dimorphism in this species in Thailand, named here S1 and S2.
The sensilla are specialized structures usually related to sensory abilities [20], so their shape and size are expected to vary from species to species, but this is actually only the case between quite distantly related species. Against this background, this study used the scutum to check for possible species heterogeneity in our sample, which should correspond to the observed sensilla variation. The scutum shape has indeed been identified as a characteristic that can distinguish chigger species, as confirmed by molecular techniques [14].
Molecular approaches including COI gene sequencing [21] have been applied to analyses of a great variety of arthropods [[22], [23], [24]] and Acari [14,15,25]. Evaluation of the COI gene sequence is a convenient way of performing not only species identification but also molecular phylogenetic analysis [26].
The aim of this study was to detect a hidden speciation event as possibly suggested by dimorphism of the sensilla observed in two forms of A. indica using morphometric and genetic approaches in Thailand.
2. Materials and methods
2.1. Chigger collection
In this study, we used chiggers attached to rodent ears previously collected in earlier research (Survey of Rodent-borne Disease in Public Park of Bangkok Metropolitan and the Faculty of Tropical Medicine, Mahidol University) [9]. The ears were collected from rodents captured from September to December 2018 in seven public parks in Metropolitan Bangkok. Our morphometric study was limited to 124 samples of chigger mites collected from two public parks: Suan Wachirabenjatas (WB) and Suan Vareepirom (VP). This study was conducted following the guidelines of animal care and use of Mahidol University, Thailand. The Faculty of Tropical Medicine - Animal Care and Use Committee (FTM-ACUC), Mahidol University, Bangkok, Thailand (Certificate no. FTM-ACUC 015/2020E), approved the animal care and all experimental procedures. Small mammals’ ears were scraped under a stereomicroscope on a Petri dish with a fine needle to detach the chiggers.
2.2. Chigger identification and preparation
Berlese fluid (TCS Biosciences Ltd., Buckingham, UK) was used for the clearing process and for making permanent slides for morphological identification and geometric morphometric analysis of the chiggers. Slide-mounted specimens were examined under a stereomicroscope and identified according to available taxonomic keys [5,27,28]. Each identified individual A. indica was temporarily mounted on a microscope slide with a double coverslip using distilled water. Dorsal and ventral sides of the chiggers were observed under a compound microscope by a flip sandwich coverslip, as previously described [10]. The specimens on these temporarily mounted slides were identified using a ZEISS LSM 700 laser scanning confocal microscope with a fluorescein isothiocyanate (FITC) filter and bright-field mode without capturing images.
Intact bodies of A. indica at the same larval stage were mounted on permanent slides and sorted according to the shape of the sensilla: elongated (S1) or rounded (S2). A total of 124 scutum samples of A. indica, corresponding to different shapes of scutal sensilla (S1 and S2), were selected. Chigger samples were isolated either from a “single host” (only A. indica infesting the host) or a “shared host” (A. indica sharing the host with other mites such as Leptotrombidium deliense).
2.3. Geometric morphometric analysis
In our study, we used two distinct morphometric approaches to increase the possibility of detecting differences between the S1 and S2 morphotypes. The first method (the landmark-based one) uses the relative positions of distinct anatomical landmarks, while the second method (the outline-based one) focuses on the contour of the organ housing these anatomical landmarks.
The scutum presents both a contour and visible landmarks, so it was explored using both techniques. However, the sensilla, owing to their lack of visible landmarks, were explored only by the contour method. For both techniques, the precision of the digitization of images was tested by estimating the repeatability of two digitizing sessions performed on the same specimens [29].
2.3.1. Landmark-based approach
The landmark-based method uses a set of anatomical landmarks to describe shape [30,31]. The autofluorescent properties of the scutum allowed clear identification of the fluorescent insertions of setae and of sensilla, that served as specific and identifiable morphological features of chiggers [10]. Here, to increase the quality of the landmark collection, we used only type I landmarks. These sorts of landmarks are easily relocatable from one individual to another, reducing the digitization error [32]. The landmarks were thus based on the insertion of scutal setae and of sensilla on the scutum. These features were easily recognized under the ZEISS LSM 700 laser scanning confocal microscope in the fluorescein isothiocyanate (FITC) filter mode.
The “centroid size” (CS) was computed for estimation of the scutum size; it is defined as the square root of the sum of the squared distances between the center of the configuration of landmarks and each individual landmark [32]. Its variation was illustrated by quantile plots. The statistical significance of mean CS difference among populations was estimated using nonparametric, permutation-based tests (1,000 cycles, with Bonferroni correction).
The variables relating to scutum shape were computed as principal components (PCs) of the residual coordinates after Procrustes superimposition to the consensus form (“Procrustes residuals”). In this principal component approach, these variables are sometimes called “Procrustes components.” The shape variation was illustrated by the factor maps of the first two Procrustes components. In the same way as performed for the size comparisons, the statistical significance of shape differences was tested using nonparametric methods (1,000 permutations, with Bonferroni correction).
2.3.2. Outline-based approach
The outline-based analyses used the contours of scutums and sensilla. After digitization, the coordinates were subjected to an elliptic Fourier analysis [33]. The normalization of the outlines (coefficients corrected for orientation and size) was performed using the semimajor axis (SMA) of the first ellipse of the Fourier decomposition. Because the perimeter is easier to determine than the SMA and is generally highly correlated to it, we used the perimeter to compare the sizes of scutum and of sensilla between A. indica S1 and S2.
The normalized elliptic Fourier coefficients (NEF) were used for subsequent statistical analyses of shape, including the fourth coefficient of the first ellipse, the one related to the width-on-length ratio of the outline [34]. The number of harmonics retained for the comparisons was the one allowing a power of 99.9 % (the power to reconstitute the shape), and the number of first PCs of the corresponding coefficients was selected to represent at least 95 % of the total variance.
The statistical significance of the size or the shape comparisons was based on nonparametric, permutation-based tests (1,000 runs), with Bonferroni correction at a p-value of <0.05.
The offline application, CLIC (accessed at http://xyom-clic.eu/), was used to digitize the images [35]. Landmark-based and outline-based statistical analyses used the same online application, XYOM (version 2), which is freely available online (http://xyom.io) [36]. The random forest analysis used the ml-random-forest.js software (https://www.npmjs.com/package/ml-random-forest). For the two-factor ANOVA on shape, R software was used (https://www.r-project.org/).
2.4. Reclassification
2.4.1. Unsupervised reclassification
The K-means algorithm is a blind classification technique allowing the identification of possible natural groupings within the data without using the individual labels [37]. This analysis is based mainly on interindividual distances; we used the Euclidean distance computed from shape variables. In the reclassification process, the K parameter needed by the algorithm is the number of predefined clusters, which is 2 here (A. indica S1 and S2).
The composition of the two clusters defined by the K-means algorithm was compared with the pre-established ones (A. indica S1 and S2), allowing us to compute the rate of correct classification. The initialization of the algorithm was based on the “naive sharding centroid” method (https://www.kdnuggets.com/2017/03/naive-sharding-centroid-initialization-method.html).
Meanwhile, the optimum number of clusters was estimated using the same algorithm for a large range of K entries. The optimal K was the one beyond which no more significant reduction of within-group variance was obtained, which is generally visible using the “elbow graph” method.
2.4.2. Supervised reclassification
Two methods were applied for supervised reclassification: one parametric and the other nonparametric. Both techniques produced rankings that were then validated and adjusted for prior probabilities, i.e., the probabilities of being correct by chance [38]. As input, we used shape variables only, derived from seven landmarks or from the outline of the scutum of A. indica S1 and S2. We also submitted the sensilla to a geometric morphometric analysis, based on outlines only because of their lack of visible landmarks.
The parametric procedure was based on the shortest Mahalanobis distance of each individual to the center of each group. The reclassification was submitted to external cross-validation using the leave-one-out method. The final total accuracy was adjusted for prior probabilities. With two groups and almost the same number of individuals by group, the prior probability for each one to be correctly assigned at random was close to 50 % [38,39]. The adjusted accuracy gives a standardized measure of improvement over random assignment as estimated by group sample sizes [40].
The powerful Mahalanobis distance-based reclassification method requires intense statistical assumptions such as multinormality, homoscedasticity, and linearity. To confirm its final result, we also applied a nonparametric approach: the random forest (RF) algorithm [41]. As a machine learning approach, the RF algorithm does not require assumptions such as linearity or normality of the variables. By using an out-of-the-bag (OOB) procedure, the RF algorithm provides an alternative to the cross-validation: in this procedure, the reclassification error was calculated on instances (OOB) not included in the training of the corresponding trees, a process analogous to the cross-validated classification. As for the parametric estimation described above, we subjected the final total accuracy to adjustment for prior probabilities.
2.5. ANOVA
Since the material subjected to geometric morphometric analysis could be subdivided by either the type of sensilla (rounded, elongated) or the public park of origin in Bangkok (WB, VP), we explored the influences of each of these factors on the scutum shape by two-factor ANOVA.
2.6. Molecular identification
DNA was extracted from 12 individual samples of A. indica (8 S1 and 4 S2) following a modified version of the protocol for DNA extraction from the gSYNC™ DNA Extraction Kit Quick (Geneaid Biotech Ltd., Taipei, Taiwan). Prior to DNA extraction, chiggers were placed in a 1.5 mL Eppendorf tube and dried by removing and evaporating the remaining ethanol. Chiggers were submerged in 20 μL of GT lysis buffer, crushed with a sterile 21G needle under a microscope, and homogenized by the addition of 180 μL of GT lysis buffer. The homogenate was then mixed with 20 μL of proteinase K and incubated for 5 h at 60 °C. Subsequently, 200 μL of GSB buffer was added to the samples and vortexed vigorously for 10 s. Two hundred microliters of cold absolute ethanol was added to the sample lysate and then immediately mixed by vortexing for 10 s. The samples were transferred to GS columns placed in 2 mL collection tubes and applied to the GS column and DNA was eluted with 30 μL of elution buffer and stored at −20 °C for the next amplification.
Twelve samples were subjected to polymerase chain reaction (PCR) to amplify a region of the mitochondrial cytochrome oxidase 1 (COI) gene using the universal primers, LCO1490: 5′-GGT CAA CAA ATC ATA AAG ATA TTG G-3′ and HCO2198: 5′-TAA ACT TCA GGG TGA CCA AAA AAT CA-3′, which generate a control amplicon of approximately 710 bp [42]. Mitochondrial cytochrome oxidase 1 is a suitable molecular marker to classify mites at a lower taxonomic level [43] as mitochondrial DNA analysis is one of the most effective phylogenetic tools in mites [44]. PCR amplification was performed in a reaction volume of 50 μL that comprised 5 μL of DNA template, OnePCR Master Mix (GeneDireX, Taiwan), and 2 μL of both forward and reverse primers (final concentration: 0.4 μM). Subsequently, PCR was run for 40 cycles: initial denaturation at 94 °C for 1 min; 5 cycles of 94 °C for 1 min, 45 °C for 90 s, and 72 °C for 90 s; 35 cycles of 94 °C for 1 min, 50 °C for 90 s, and 72 °C for 1 min; and then final extension at 72 °C for 5 min. Post-PCR amplicons were visualized by 1.2 % agarose gel electrophoresis with the gel comprising SYBR Safe dye (Invitrogen, USA) at 100 V for 30 min. PCR products were sent for purification and BT sequencing processes at U2Bio (Thailand) Sequencing Service.
The neighbor joining (NJ) method was used [45] to construct a phylogenetic tree [46] based on the pairwise distance (p-distance) between sequences. The p-distance is computed as the number of nucleotide differences between each pair of sequences divided by the length of the sequence. The resulting NJ tree was resampled with replacement (bootstrap) 1,000 times and the proportion of replicated trees that clustered together was shown on the branch nodes.
In addition to the 8 S1 and 4 S2 sequences from Bangkok (Thailand), the composition of the NJ tree was as follows: (i) 2 A. indica from Nan Province (Thailand) with GenBank accession no. MG728110 and MG728111; (ii) 4 A. indica from Laos GenBank accession no. KY930728, KY930729, KY930730, and KY930732; (iii) 1 genus Ascoschoengastia sp. from Japan accession no. AB300501; and finally (iv) 1 Leptotrombidium deliense (HQ324975) and 1 Walchia ewingi lupella (KY930719) sequences as outgroups to generate the phylogenetic tree.
Bioedit version 7.2.5 software was used to check and edit the DNA base sequence [47]. ClustalX 2.1 software was used to align the DNA base sequences [48]. MEGA-X software [49] was used to compute mean distances within and between groups using the p-distance method, as well as to build the corresponding NJ tree.
3. Results
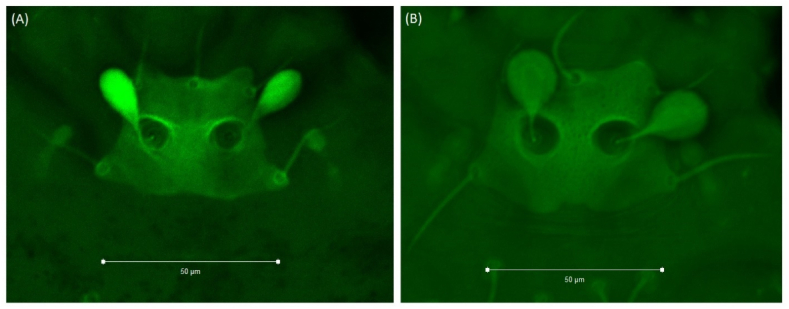
We observed clear-cut dimorphism regarding the morphology of sensilla: club-shaped sensilla (S1, Fig. 1A), typical of A. indica, and the newly observed rounded sensilla (S2, Fig. 1B). The numbers of both morphotypes used for geometric morphometric analysis are shown in Table 1. In the two public parks used as sites in this study, both shapes (S1 and S2) were found together irrespective of the animal host or collection site (Table 2).
Fig. 1.
Ascoschoengastia indica's scutum S1 as club-shaped sensilla (A) and S2 as rounded sensilla (B).
Table 1.
Number of A. indica belonging to the S1 (elongated shape) and to the S2 (rounded shape) sensilla morphotypes used for geometric morphometric analysis.
| Data (Organ) | A. indica S1 | A. indica S2 |
|---|---|---|
| 7 Landmarks (Scutum) | 43 VR + 19 WB = 62 | 42 VR + 20 WB = 62 |
| Outline (Scutum) | 41 VR + 19 WB = 60 | 44 VR + 20 WB = 64 |
| Outline (Sensilla) | 41 VR + 19 WB = 60 | 44 VR + 20 WB = 64 |
Legend of Table 1. For the morphometric analyses, S1 and S2 morphotypes, samples came from two collecting sites: Suan Wachirabenjatas (WB) and Suan Vareepirom (VP).
Table 2.
A. indica specimens used for the NJ tree of Fig. 7.
| ID | Sensillum type | GenBank accession number | Site | Rodent ID | Rodent species |
|---|---|---|---|---|---|
| GM 1 | S1 | OQ291245 | WB | R7775 | R. tanezumi |
| GM 2 | S1 | OQ291246 | TB | R7879 | R. rattus complex |
| GM 3 | S1 | OQ291247 | WB | R7775 | R. tanezumi |
| GM 4 | S1 | OQ291248 | RM | R7807 | R. tanezumi |
| GM 5 | S1 | OQ291249 | ST | R7838 | R. tanezumi |
| GM 6 | S2 | OQ291250 | RM | R7807 | R. tanezumi |
| GM 7 | S2 | OQ291251 | VP | R7870 | R. tanezumi |
| GM 8 | S2 | OQ291252 | RM | R7807 | R. tanezumi |
| GM 9 | S2 | OQ291253 | VP | R7870 | R. tanezumi |
| GM 21 | S1 | OQ291254 | TB | R7879 | R. rattus complex |
| GM 29 | S1 | OQ291255 | TB | R7879 | R. rattus complex |
| GM 31 | S1 | OQ291256 | TB | R7879 | R. rattus complex |
Legend of Table 2. The ID (column 1) and the accession numbers (column 3) are indicated in the tree (Fig. 7). The sensillum types (column 2) are either an elongated type (S1) or a rounded type (S2). R., Rattus; WB, Suan Wachirabenjatas park; TB, Suan Thonburirom park; RM, Suan Luang Rama IX park; ST, Suan Serithai park and VP, Suan Vareepirom park.
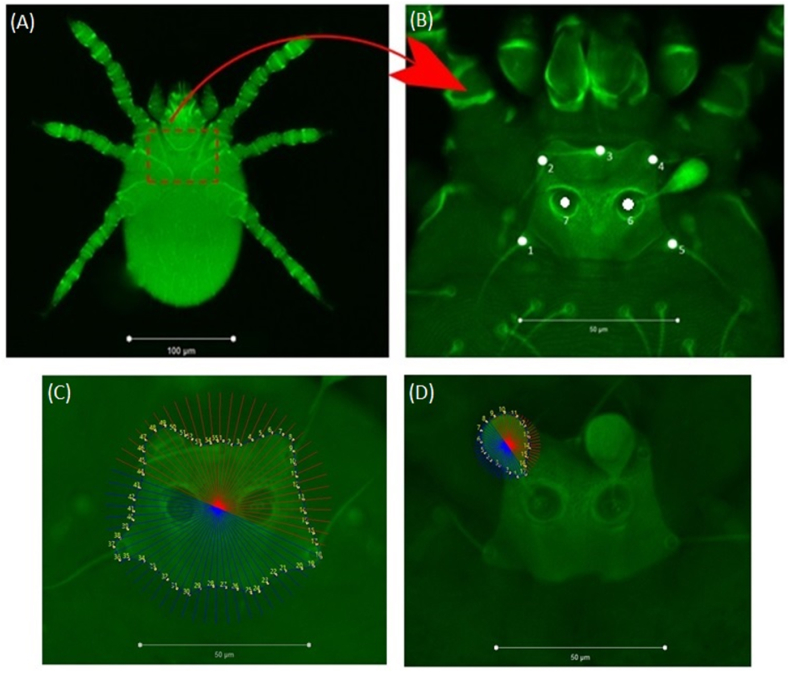
Fig. 2 shows the scutal seven-landmark configuration (Fig. 2A and B) used for landmark-based geometric morphometric analysis, and digitization dots describing the contours of the scutum (Fig. 2C) and the sensilla (Fig. 2D) for outline-based analysis.
Fig. 2.
Scutal seven-landmark configuration (A and B) used for landmark-based geometric morphometric analysis, and digitization dots describing the contours of the scutum (C) and the sensilla (D) for outline-based analysis.
3.1. Morphometric study (specimens from Bangkok)
The CS and perimeter repeatability score were high, at 0.99 and 0.97, respectively. The repeatability score for shape was 0.90 for the landmark-based analysis and 0.95 for the outline-based one.
3.1.1. Scutum and sensilla size variation
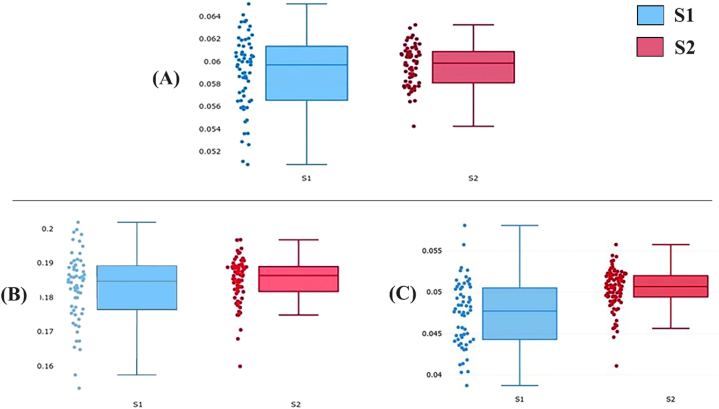
There was no significant difference in the (seven-landmark) CS of S1 (0.059 mm) and S2 (0.060 mm) (P = 0.2058), and also no significant difference in the scutum perimeter of S1 (0.182 mm) and S2 (0.184 mm) (P = 0.2058), but there was a highly significant difference in size (P < 0.0001) between the perimeters of sensilla S1 (0.047 mm) and S2 (0.050 mm) (Fig. 3; Table 3).
Fig. 3.
Quantile boxes, horizontal line in the box is the median. Dots represent individuals. Comparison between A. indica S1 (blue boxes) and S2 (red boxes) scutum CS (A), perimeter of scutum (B), and perimeter of sensilla (C). S1 refers to the club-shaped sensilla and S2 refers to the rounded sensilla.
Table 3.
Mean size of scutum and sensilla of A. indica S1 and S2 based on landmark and outline-based analyses.
| Organ |
Scutum |
Sensilla |
||||
|---|---|---|---|---|---|---|
| Size | Centroid size | Perimeter | Perimetera | |||
| Type (No. used) | S1 (62) | S2 (62) | S1 (60) | S2 (64) | S1 (60) | S2 (64) |
| Mean size | 0.059 | 0.060 | 0.182 | 0.184 | 0.047 | 0.050 |
| S.D. | 0.003 | 0.002 | 0.010 | 0.007 | 0.004 | 0.002 |
Legend of Table 3. Units are millimeters. S.D. Standard deviation. S1 refers to the A. indica with club-shaped sensilla and S2 refers to the rounded sensilla.
Means statistically different between S1 and S2 at P < 0.0001.
3.1.2. Scutum and sensilla shape variation
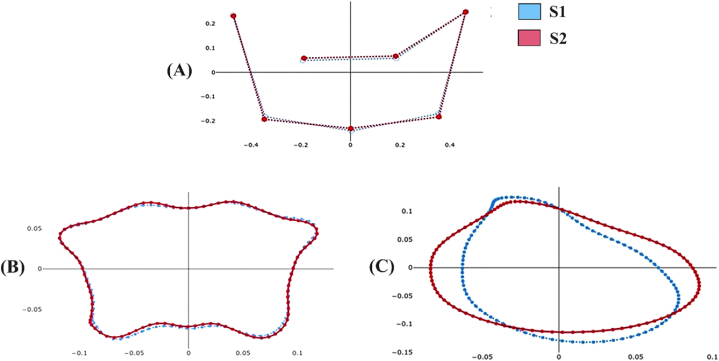
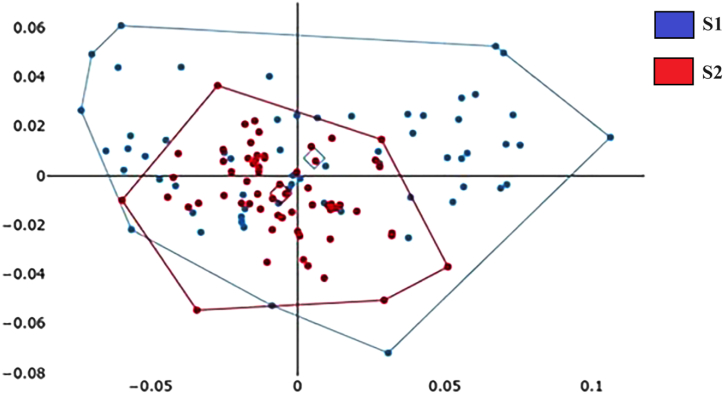
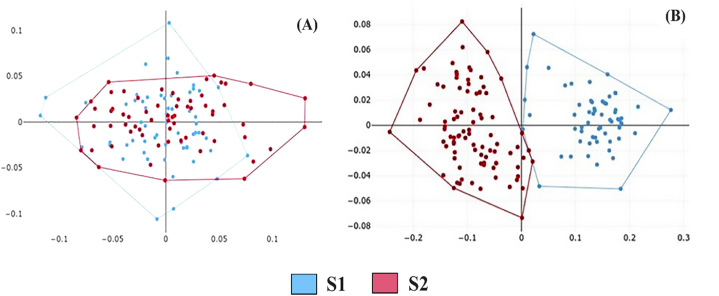
The polygon superimposition of mean landmark (Fig. 4A) or pseudolandmark (Fig. 4B) configurations of the scutum showed slight shape variation between A. indica S1 and S2; however, the factor map of the first two PCs of shape showed complete overlap of the two forms (Fig. 5, Fig. 6A).
Fig. 4.
Procrustes superposition of the mean shape of the landmark configurations of the scutum (A), of the mean scutum (B) and sensilla (C) contours of A. indica S1 (club-shaped sensilla) and S2 (rounded sensilla).
Fig. 5.
Factor map of the principal component analysis (PCA) of the scutal seven-landmark residual coordinates after Procrustes superposition. A posteriori convex hulls were drawn to compare S1 (club-shaped sensilla) and S2 (rounded sensilla). The horizontal axis is the first principal component (PC1) and the vertical axis is the second principal component (PC2), with 42 % and 16 % their respective contributions to the total variation of shape.
Fig. 6.
Factor map of the principal component analysis (PCA) computed from the shape variables describing the contour of the scutum (A) and the sensilla (B) of A. indica. A posteriori convex hulls were drawn to compare S1 (club-shaped sensilla) and S2 (rounded sensilla). The horizontal axis is the first principal component (PC1) and the vertical axis is the second principal component (PC2), with 36 % and 19 % their respective contributions to the total variation of shape.
In contrast, the mean shape of the sensilla showed a clear difference between S1 and S2 (Fig. 5, Fig. 6B). The sensilla validated reclassification scores confirmed the separation of the S1 and S2 sensilla. Both supervised and unsupervised reclassification scores ranged from 96 % to 99 % (Table 4). For the scutum, all reclassification tests were unsatisfactory: supervised or not, parametric or not, and irrespective of the method used, namely, the landmark-based or outline-based one (Table 4).
Table 4.
Validated reclassification scores of A. indica S1 and S2 after correction for prior probabilities, as based on the scutum shape (from either landmarks or outline) and on the sensilla shape (outline).
| Organ and method used | No used (n1, n2) | Clustering |
Validated supervised reclassification |
|
|---|---|---|---|---|
| K-Means (K = 2) | Discriminant analysis | Random Forest | ||
| Sensilla outline | 60, 64 | 96 % (optK = 2) | 99 % (17PCs) | 97 % (17PCs) |
| Scutum outline | 60, 64 | 53 % (optK = 7) | 43 % (17PCs) | 40 % (17PCs) |
| Scutum landmarks | 62, 62 | 64 % (optK = 4) | 42 % (10PCs) | 42 % (10PCs) |
Legend of Table 4. Percentages of correctly assigned individuals to either S1 (club-shaped sensilla) or S2 (rounded sensilla) morphotypes (Fig. 1), according to unsupervised and supervised reclassification and after adjustment for prior probabilities (see Materials and Methods). The K-means analysis was performed with K = 2, using the total number of shape variables; opt K, the optimum number of groups (see Materials and Methods for more details). The random forest algorithm used 400 bootstraps. The set of first principal components (PCs) used as input are indicated between brackets: they were PCs of shape variables (excluding size) and represented together at least 95 % of the total shape variation.
The results of two-factor ANOVA, which only displays the statistical significance associated with each factor (sensilla and parks), are presented in Table 5.
Table 5.
Two-factors ANOVA, sensilla and parks.
| Methods | Outline-based method | Landmark-based method | ||
|---|---|---|---|---|
| Factors | PC1 (36 %) | PC2 (19 %) | PC1 (42 %) | PC2 (16 %) |
| Sensilla | NS | NS | NS | 0.001 |
| Parks | NS | 0.05 | 0.010 | NS |
| Interaction | NS | NS | 0.001 | NS |
Legend of Table 5. Partial output of two-factor ANOVA, showing only the statistical significance associated with each factor (sensilla, parks) after ANOVA performed on first (PC1) and second (PC2) principal components of shape, as derived from either outlines or landmarks. Between brackets, the contribution of the PC to the total variation. NS = not significant.
3.1.3. Scutum shape variation, sensilla, and parks
The two-factor ANOVA performed on the first (PC1) and second (PC2) components of scutum shape, as derived from either outlines or landmarks, suggested that the geographical origin (public parks of Bangkok) had more of an effect than the type of sensilla (S1, S2). This suggests that variation in scutum shape was not related to sensilla dimorphism, but perhaps influenced by the environment. For the outline-based approach, the PC1 variation appeared to be unaffected by either a possible spatial effect (parks WB, VB) or the sensilla type; only the second principal component (PC2) appeared to change according to the park, but there was no effect at all of the sensilla type (Table 5). For the landmark-based scutum shape, the first component (PC1) showed a significant effect of geographical origin, not of sensilla type. For the landmark shape only, a significant interaction of sensilla and parks was identified (Table 5).
3.2. Genetic analyses
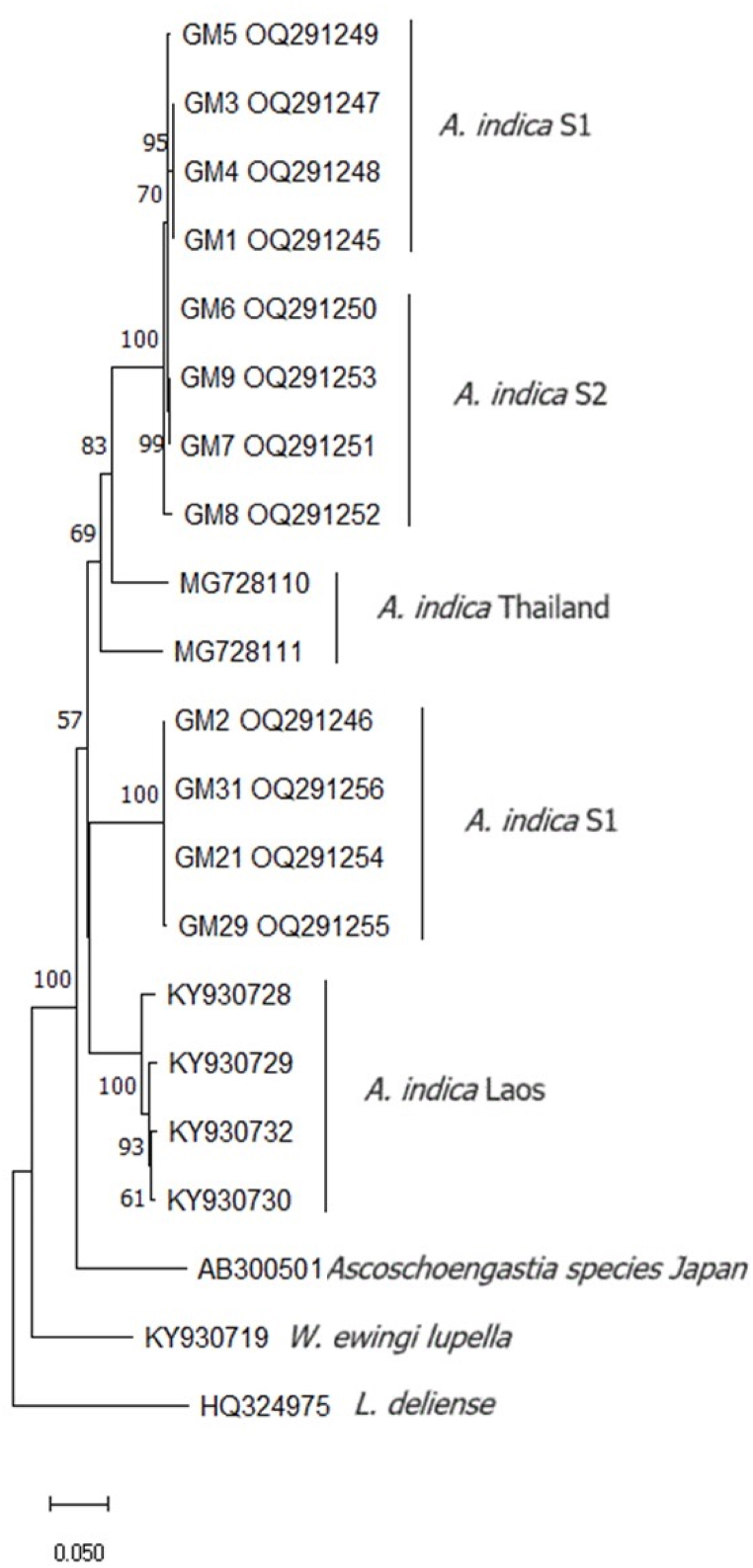
Amplification of the mitochondrial cytochrome oxidase 1 (COI) gene was conducted on 12 selected individuals (8 individuals of A. indica S1 and 4 of S2 from various study sites). Using L. deliense and W. ewingi lupella as outgroups, the NJ phylogenetic reconstruction illustrated the evolutionary relationships between sequences of A. indica (S1, S2, Thailand, and Laos) and one sequence of an unknown species of the genus Ascoschoengastia sp. (Fig. 7).
Fig. 7.
Phylogenetic NJ tree based on the COI sequences of A. indica S1 and S2. S1 refers to the club-shaped sensilla and S2 refers to the rounded sensilla. Bootstrap values (1,000 replicates) of bifurcations are shown at the nodes.
The nucleotide sequences of chiggers established in this study were deposited in the GenBank database under accession numbers OQ291245–OQ291256.
The nucleotide contents were similar between the A. indica individuals. The AT richness of Ascoschoengastia sp. from Japan showed a consistently lower value than for A. indica, even lower than the one of the outgroups L. deliense and W. ewingi lupella (Table 6).
Table 6.
Nucleotide contents of Ascoschoengastia sp. materials.
| Species | Group | AT content (%) | GC content (%) |
|---|---|---|---|
| A. indica | S1 | 66.9 | 33.1 |
| A. indica | S2 | 67.0 | 33.0 |
| A. indica | Thailand | 68.6 | 31.4 |
| A. indica | Laos | 67.1 | 32.9 |
| Ascoschoengastia sp. | Japan | 64.8 | 35.2 |
| W. ewingi lupella | Thailand | 68.1 | 31.9 |
| L. deliense | Thailand | 63.1 | 36.9 |
Legend of Table 6. W., Walchia; L., Leptotrombidium; AT, adenine-thymine; GC, guanine-cytosine; S1 refers to the A. indica with club-shaped sensilla and S2 refers to the A. indica with rounded sensilla.
The 18 sequences of A. indica (8 S1, 4 S2, 2 Thailand, and 4 Laos) always showed low within-group p-distance values, except when the comparison related to geographically distant localities, like between Bangkok and the northern province of Nan, or between Thailand and Laos.
The mean genetic distance within S1 (0.080), as well as that within S2 (0.008), was of the same order of magnitude as the mean distance within A. indica from Thailand (0.057) or within A. indica from Laos (0.042). Meanwhile, the genetic distances of S1, or of S2, with other A. indica sequences from Thailand were also very low, ranging from 0.036 to 0.075 (Table 7). Thus, the distances among S1, S2, and other A. indica sequences were of the same order of magnitude, which was illustrated by the phylogenetic tree. Another noteworthy point is that S1 and S2 specimens did not constitute separate clusters (Fig. 7). The between-species values of p-distances, like between A. indica and Ascoschoengastia sp. from Japan, or A. indica and L. deliense, were larger or much larger, ranging from 0.167 to 0.295 (Table 7).
Table 7.
Mean p-distances between groups of COI sequences between Ascoshoengastia indica.
| A. indica S 1 | A. indica S 2 | A. indica Thailand | A. indica Laos | Ascoshoengastia sp. Japan | W. ewingi lupella | L. deliense | |
|---|---|---|---|---|---|---|---|
| A. indica S 1 (n = 8) | 0.080 | ||||||
| A. indica S 2 (n = 4) | 0.074 | 0.008 | |||||
| A. indica Thailand (n = 10) | 0.075 | 0.036 | 0.057 | ||||
| A. indica Laos (n = 6) | 0.134 | 0.138 | 0.135 | 0.042 | |||
| Ascoshoengastia sp. Japan (n = 1) | 0.180 | 0.176 | 0.177 | 0.167 | 0.000 | ||
| W. ewingi lupella (n = 1) | 0.207 | 0.208 | 0.208 | 0.201 | 0.228 | 0.000 | |
| L. deliense (n = 1) | 0.276 | 0.295 | 0.286 | 0.279 | 0.276 | 0.028 | 0.000 |
Legend of Table 7. Mean intraspecific p-distance is shown in bold. W., Walchia; L., Leptotrombidium. S1 refers to the A. indica with club-shaped sensilla and S2 refers to the A. indica with rounded sensilla.
4. Discussion
To the best of our knowledge, this is the first time that clear dimorphism of the sensilla has been observed in the same larval stage of A. indica. Morphometric analyses of the sensilla shapes, even without using the labels indicating S1 or S2, clearly confirmed our visual observation: there were two distinct morphotypes of sensilla, S1 and S2, which differed in both size and shape. The K-means algorithm accuracy score was close to perfection (97 %). This almost-perfect score was repeated in the supervised classifications used here: both parametric (Mahalanobis distances) and nonparametric (RF algorithm) ones, with accuracy scores close to perfection even after correction for prior probabilities (from 96 % to 99 %).
4.1. The hypothesis of sexual dimorphism
Could the S1 and S2 morphotypes correspond to different sexes? Trombiculidae larvae do not present morphological characteristics allowing their future sex to be recognized [50]. After development of the larval stage to three nymph stages (protonymph, deutonymph, and tritonymph), active deutonymphs become identical to small adult stages, but do not complete sexual development [51]. Most previous knowledge on the sexual differentiation and sex ratio of chiggers comes from laboratory rearing experiments of the genus Leptotrombidium [52,53], with possible sexual dimorphism of the larvae not being mentioned. In the absence of any reference in the scientific literature on A. indica, we cannot definitively rule out the hypothesis that the shape of the sensilla represents a sexual character.
4.2. Host or environment hypothesis
In the present study, the presence of A. indica S1 and of S2 specimens (all of them collected in Bangkok) did not exhibit an obvious link to the host origin or to the spatial location. Indeed, most of the two morphotypes of sensilla were found in the same species, Rattus tanezumi, which does not suggest an influence of the host [54,55]. However, R. tanezumi is part of the Rattus rattus complex, and some cryptic species in our sample could have gone unnoticed. To bring to the fore some unexpected host effect, more accurate rodent species identification and potentially a larger spectrum of hosts for A. indica should be considered.
The effect of microgeographic variation was also difficult to invoke since the two forms S1 and S2 were found in the same parks of the same city. Therefore, in our data, there is weak support for a host effect or structuring based on geographical location.
4.3. The taxonomic question
Sensilla are sensory organs that play a crucial role in detecting various stimuli from the environment [56]. As a consequence, they can have some taxonomic importance and it was reasonable to question the species homogeneity of the morphospecies A. indica given its possession of differently shaped sensilla [20]. We explored the possibility that using both molecular and geometric morphometric techniques and showed that S1 and S2 is not a single species.
4.4. Genetic variation
The similar nucleotide contents of S1, S2, and other A. indica (Table 6) were consistent with conspecificity, while the sequence of the unknown Ascoschoengastia sp. from Japan showed rather different values, as would be expected for a probably different species. However, the nucleotide contents of our outgroups belonging to other genera (Leptotrombidium and Walchia) did not depart too much from the A. indica one, suggesting the need for complementary genetic measurements. We used the p-distance i.e., the number of nucleotide differences between each pair of sequences divided by the length of the sequence. This method assumes an equal substitution rate among nucleotides, treating all substitutions as equally likely, which is compatible with sequences of the same or closely related species and applicable to scenarios involving low genetic divergence.
A total of twenty-eight COI sequences were calculated for p-distances. The p-distances among the three species included among our material (Ascoschoengastia sp. from Japan, L. deliense, and A. indica) ranged from 0.167 to 0.295; these values were much larger than those between the two morphotypes S1 and S2 (0.074 on average, see Table 7). The mean p-distance between A. indica S1 and S2 was of the same order of magnitude as the one within S1 or within S2, and as the one found between the other A. indica sequences in our material, coming from either Thailand (0.057) or Laos (0.042). Thus, our data perfectly fitted the hypothesis of these two forms belonging to a single species, actually the A. indica species (Table 7). Interestingly, some genetic differentiation was apparent between A. indica of different geographical origins, namely, Thailand and Laos, exhibiting a mean p-distance of 0.135, which is intermediate between conspecific (<0.13) and interspecific taxa (>0.19). These observations were confirmed and clearly illustrated by the phylogenetic approach (Fig. 7).
4.5. Morphometric variation
In the vast majority of cases, different species present more or less marked variations in geometric shape at various parts of the body, even in the case of sibling species [13,35]. To detect possible speciation revealed by the dimorphism of the sensilla, we explored the shape variation of another part of the body. Here, the scutum was selected because of its recognized taxonomic importance [14]. By using two geometric morphometric approaches, landmark-based [30,32] and outline-based ones [33,57], our idea was to reduce the risk of rejecting a difference because of an incomplete or deficient capture of shape.
The estimation of repeatability produced highly satisfactory results, suggesting that our measurements of metric properties were reliable, and confirming the quality of the photography protocol and data recording. However, statistically significant differences of the scutum size, either CS or perimeter, could not be identified between S1 and S2. The very large overlap of sizes between these two morphotypes (Figs. 3A and 4A) made it unlikely that scutum size could be used as a distinctive trait between them.
Visual comparison of the scutum shapes by superimposing them, either the outline or the LM configuration, did not provide any obvious clue to recognize the forms “S1” and “S2”: Slight differences were scattered across the body with apparently no particular localization (Fig. 4A and B). The unsupervised classification based on the K-means algorithm was not satisfactory, suggesting that the variation in shape of the scutum did not reflect that of the sensilla (Table 4). The same conclusion also arose from the shape supervised classification based on either a parametric or a nonparametric method: the scutum shape could not be used satisfactorily to separate the S1 and S2 types (Table 4). Moreover, the variation in scutum shape showed a stronger association with the geographical location (two different public parks) than with the sensilla variation (Table 5).
5. Conclusion
Our study was designed to detect a possible speciation process, but both morphometric and genetic analyses provided results supportive of conspecificity. Thus, in the absence of a hidden speciation process, we considered other hypotheses explaining the variation in sensilla shape as either a sexual character or an adaptation to some environmental factor. The hypothesis regarding sensilla as a sexual character is very speculative since, at present, the larval stage of Trombiculidae is not known to harbor a morphological feature allowing the sex to be recognized.
Meanwhile, if there is an environmental factor that affects the shape of the sensilla, it would probably not differ from one local geographical context to another. Indeed, the S1 and S2 types were found in the same public parks of the same locality (Bangkok). An adaptation to different host species is contradicted by the presence of the two morphotypes in the same species (R. tanezumi), but it cannot be completely ruled out because the R. rattus complex hides cryptic species not resolved in our material. The only robust conclusion that can currently be drawn is the lack of evidence for any speciation process explaining the sensilla dimorphism of A. indica.
Ethical statement
This study was conducted following the guidelines of animal care and use of Mahidol University, Thailand. The Faculty of Tropical Medicine - Animal Care and Use Committee (FTM-ACUC), Mahidol University, Bangkok, Thailand (Certificate no. FTM-ACUC 015/2020E), approved the animal care and all experimental procedures.
Funding statement
Financial support for the project was granted by the Thailand Research Fund (Grant no. DBG6180016) and the Office of the Higher Education Commission and the Thailand Research Fund (Grant no. MRG6180023). Additionally, funding for M.Sc. in Tropical Medicine of Shobiechah Aldillah Wulandhari from the Malaria Consortium's Dr. Sylvia Meek Scholarship for Entomology.
Data availability statement
Data included in article/supplementary material/referenced in article.
Additional information
No additional information is available for this paper.
CRediT authorship contribution statement
Shobiechah Aldillah Wulandhari: Writing – review & editing, Writing – original draft, Formal analysis, Data curation. Vachirapong Charoennitiwat: Writing – review & editing, Data curation. Yudthana Samung: Writing – review & editing, Data curation. Piengchan Sonthayanon: Writing – review & editing, Formal analysis, Conceptualization. Rawadee Kumlert: Writing – review & editing, Data curation. Serge Morand: Writing – review & editing, Formal analysis, Conceptualization. Kittipong Chaisiri: Writing – review & editing, Supervision, Formal analysis, Data curation, Conceptualization. Tanawat Chaiphongpachara: Writing – review & editing, Data curation. Jean-Pierre Dujardin: Writing – review & editing, Validation, Supervision, Software, Formal analysis. Suchada Sumruayphol: Writing – review & editing, Supervision, Funding acquisition, Formal analysis, Data curation, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We would like to gratefully thank the Environment Department, Bangkok Metropolitan Administration (Director of the Environment Department, Mrs. Wullaya Wattanarat and the Director of Bangkok Public Park Office, Mrs. Arom Wongmaha) for granting a permission to work in the area of Bangkok public parks. We would like to deliver special appreciation also to the staff from the seven public parks for their kind cooperation and facilitation during the fieldworks conducted at their properties.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e33908.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Bureau of Epidemiology . Scrub typhus; 2023. Department of Disease Control, Ministry of Public Health, Thailand. Annual Epidemiologic Surveillance Report.http://doe.moph.go.th/surdata/disease.php?ds=44 Available online: [Google Scholar]
- 2.Chaisiri K., Gill A.C., Stekolnikov A.A., et al. Ecological and microbiological diversity of chigger mites, including vectors of scrub typhus, on small mammals across stratified habitats in Thailand. Anim. Microbiome. 2019;1:18. doi: 10.1186/s42523-019-0019-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wangrangsimakul T., Elliott I., Nedsuwan S., et al. The estimated burden of scrub typhus in Thailand from national surveillance data (2003-2018) PLoS Neglected Trop. Dis. 2020;14(4) doi: 10.1371/journal.pntd.0008233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elliott I., Pearson I., Dahal P., Thomas N.V., Roberts T., Newton P.N. Scrub typhus ecology: a systematic review of Orientia in vectors and hosts. Parasites Vectors. 2019;12:513. doi: 10.1186/s13071-019-3751-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chaisiri K., Stekolnikov A.A., Makepeace B.L., Morand S. A revised checklist of chigger mites (Acari: Trombiculidae) from Thailand, with the description of three new species. J. Med. Entomol. 2016;53(2):321–342. doi: 10.1093/jme/tjv244. PMID: 26744466. [DOI] [PubMed] [Google Scholar]
- 6.Stekolnikov A.A. A checklist of chigger mites (Acariformes: Trombiculidae) of Southeast Asia. Zootaxa. 2021;19:4913. doi: 10.11646/zootaxa.4913.1.1. [DOI] [PubMed] [Google Scholar]
- 7.Traub R., Wisseman C.L., Jr. Ecological considerations in scrub typhus: 2. Vector species. Bull. World Health Organ. 1968;39(2):219–230. [PMC free article] [PubMed] [Google Scholar]
- 8.Nadchatram M. Correlation of habitat, environment and color of chiggers, and their potential significance in the epidemiology of scrub typhus in Malaya (Prostigmata: Trombiculidae) J. Med. Entomol. 1970;7(2):131–144. doi: 10.1093/jmedent/7.2.131. [DOI] [PubMed] [Google Scholar]
- 9.Wulandhari S., Paladsing Y., Saesim W., et al. High prevalence and low diversity of chigger infestation in small mammals found in Bangkok Metropolitan parks. Med. Vet. Entomol. 2021;35(4):534–546. doi: 10.1111/mve.12531. [DOI] [PubMed] [Google Scholar]
- 10.Kumlert R., Chaisiri K., Anantatat T., et al. Autofluorescence microscopy for paired-matched morphological and molecular identification of individual chigger mites (Acari: Trombiculidae), the vectors of scrub typhus. PLoS One. 2018;13 doi: 10.1371/journal.pone.0193163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lv Y., Guo X.G., Jin D.C. Research progress on Leptotrombidium deliense. Kor. J. Parasitol. 2018;56(4):313–324. doi: 10.3347/kjp.2018.56.4.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu E.S., Wu H.Y. Study on morphological and biological differences between two types of Trombicula deliensis Walch. Acta Entomol. Sin. 1959;9(1):66–74. [Google Scholar]
- 13.Dujardin J.P. Morphometrics applied to medical entomology. Infect. Genet. Evol. 2008;8(6):875–890. doi: 10.1016/j.meegid.2008.07.011. [DOI] [PubMed] [Google Scholar]
- 14.Sungvornyothin S., Kumlert R., Paris D.H., et al. Geometric morphometrics of the scutum for differentiation of trombiculid mites within the genus Walchia (Acariformes: Prostigmata: Trombiculidae), a probable vector of scrub typhus. Ticks Tick Borne Dis. 2019;10(2):495–503. doi: 10.1016/j.ttbdis.2018.11.013. [DOI] [PubMed] [Google Scholar]
- 15.Dupraz M., Toty C., Noël V., et al. Linking morphometric and genetic divergence with host use in the tick complex, Ornithodoros capensis sensu lato. Infect. Genet. Evol. 2016;46:12–22. doi: 10.1016/j.meegid.2016.10.005. [DOI] [PubMed] [Google Scholar]
- 16.Diaha-Kouame A.C.A., Tian-Bi T.Y.N., Yao K.P., et al. Contribution of geometric morphometry in the control of Rhipicephalus (Boophilus) microplus (Canestrini, 1888) on the transhumance corridor between Côte d'Ivoire and Burkina Faso. Int. J. Biol. Chem. Sci. 2017;11(6):2630–2648. doi: 10.4314/ijbcs.v11i6.7. [DOI] [Google Scholar]
- 17.Hirst S. On some new Acarine parasites of rats. Bull. Entomol. Res. 1915;6(2):183–190. [Google Scholar]
- 18.Womersley H. The scrub-typhus and scrub-itch mites (Trombiculidae, Acarina) of the Asiatic-Pacific. Rec. South Aust. Mus. 1952;10:1–435. [Google Scholar]
- 19.Fernandes S.J.S., Kulkarni S.M. Studies on the trombiculid mite fauna of India. Record Zool. Surv. India Occas. Pap. 2003;212:1–539. [Google Scholar]
- 20.Hallberg E., Hansson B.S. Arthropod sensilla: morphology and phylogenetic considerations. Microsc. Res. Tech. 1999;47(6):428–439. doi: 10.1002/(SICI)1097-0029(19991215)47:6<428::AID-JEMT6>3.0.CO. 2. [DOI] [PubMed] [Google Scholar]
- 21.Hebert P.D.N., Cywinska A., Ball S.L., DeWaard J.R. Biological identifications through DNA barcodes. Proc. Biol. Sci. 2003;270(1512):313–321. doi: 10.1098/rspb.2002.2218. 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sumruayphol S., Apiwathnasorn C., Ruangsittichai J., et al. DNA barcoding and wing morphometrics to distinguish three Aedes vectors in Thailand. Acta Trop. 2016;159:1–10. doi: 10.1016/j.actatropica.2016.03.010. [DOI] [PubMed] [Google Scholar]
- 23.Weeraratne T.C., Surendran S.N., N S., Parakrama Karunaratne S.H.P. DNA barcoding of morphologically characterized mosquitoes belonging to the subfamily Culicinae from Sri Lanka. Parasites Vectors. 2018;11:266. doi: 10.1186/s13071-018-2810-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rodrigues B.L., Galati E.A.B. Molecular taxonomy of phlebotomine sand flies (Diptera, Psychodidae) with emphasis on DNA barcoding: a review. Acta Trop. 2023;238 doi: 10.1016/j.actatropica.2022.106778. [DOI] [PubMed] [Google Scholar]
- 25.Krčmar S., Klobučar A., Vucelja M., et al. DNA barcoding of hard ticks (Ixodidae), notes on distribution of vector species and new faunal record for Croatia. Ticks Tick Borne Dis. 2022;13(3) doi: 10.1016/j.ttbdis.2022.101920. [DOI] [PubMed] [Google Scholar]
- 26.Hebert P.D., Gregory T.R. The promise of DNA barcoding for taxonomy. Syst. Biol. 2005;54(5):852–859. doi: 10.1080/10635150500354886. [DOI] [PubMed] [Google Scholar]
- 27.Nadchatram M., Dohany A.L. Institute Penyelidikan Perubatan, Kuala Lumpur; 1974. A Pictorial Key to the Subfamilies, Genera and Subgenera of Southeast Asian Chiggers (Acari, Prostigmata, Trombiculidae) [Google Scholar]
- 28.Stekolnikov A.A. Leptotrombidium (Acari: Trombiculidae) of the world. Zootaxa. 2013;3728:1–173. doi: 10.11646/zootaxa.3728.1.1. [DOI] [PubMed] [Google Scholar]
- 29.Arnqvist G., Mårtensson T. Measurement error in geometric morphometrics: empirical strategies to assess and reduce its impact on measure of shape. Acta zool. Acad. Sci. Hung. 1998;44(1–2):73–96. [Google Scholar]
- 30.Rohlf F.J. In: Proceedings of the Michigan Morphometrics Workshop. Rohlf F.J., Bookstein F.L., editors. University of Michigan Museum of Zoology; Ann Arbor, Mich: 1990. Rotational fit (Procrustes) methods; pp. 227–236. 1990. [Google Scholar]
- 31.Rohlf F.J., Slice D.E. Extensions of the Procrustes method for the optimal superimposition of landmarks. Syst. Zool. 1990;39(1):40–59. doi: 10.2307/2992207. [DOI] [Google Scholar]
- 32.Bookstein F.L. Cambridge University Press; NY: 1991. Morphometric Tools for Landmark Data. Geometry and Biology. [Google Scholar]
- 33.Kuhl F.P., Giardina C.R. Elliptic Fourier features of a closed contour. Comput. Graph. Image Process. 1982;18(3):236–258. doi: 10.1016/0146-664X(82)90034-X. 1982. [DOI] [Google Scholar]
- 34.Lestrel P.E. Cambridge University Press; Cambridge: 1997. Fourier Descriptors and Their Applications in Biology; p. 18. [Google Scholar]
- 35.Dujardin J.P., Slice D. In: Encyclopedia of Infectious Diseases: Modern Methodologies. Tibayrenc M., editor. John Wiley & Sons Inc; Hoboken, NJ: 2007. Contributions of morphometrics to medical entomology; pp. 435–447. [Google Scholar]
- 36.Dujardin S., Dujardin J.P. Geometric morphometrics in the cloud. Infect. Genet. Evol. 2019;70:189–196. doi: 10.1016/j.meegid.2019.02.018. [DOI] [PubMed] [Google Scholar]
- 37.Pérez-Ortega J., Almanza-Ortega N.N., Vega-Villalobos A., Pazos-Rangel R., Zavala-Díaz C., Martínez-Rebollar A. IntechOpen; 2020. The K-Means Algorithm Evolution. 2020. [DOI] [Google Scholar]
- 38.McGarigal K., Cushman S., Stafford S. Springer-Verlag; New York: 2000. Multivariate Statistics for Wildlife and Ecology Research; p. 283. [Google Scholar]
- 39.Kovarovic K., Aiello L.C., Cardini A., Lockwood C.A. Discriminant function analyses in archaeology: are classification rates too good to be true? J. Archaeol. Sci. 2011;38(11):3006–3018. doi: 10.1016/j.jas.2011.06.028. [DOI] [Google Scholar]
- 40.Klecka W.R., analysis Discriminant. Sage Publications; Beverly Hills, CA: 1980. Sage University Paper Series on Quantitative Applications in the Social Sciences, Series No. 07–019. [Google Scholar]
- 41.Breiman L., Random Forests L. Mach. Learn. 2001;45:5–32. [Google Scholar]
- 42.Folmer O., Black M., Hoeh W., et al. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994;3(5):294–299. [PubMed] [Google Scholar]
- 43.Cruickshank R. Molecular markers for the phylogenetics of mites and ticks. Syst. Appl. Acarol. 2002;7(1):3–14. doi: 10.11158/saa.7.1.1. [DOI] [Google Scholar]
- 44.Dabert M. DNA markers in the phylogenetics of the Acari. Biol. Lett. 2006;43(2):97–107. [Google Scholar]
- 45.Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 46.Nei M., Kumar S. Oxford University Press; New York: 2000. Molecular Evolution and Phylogenetics. [Google Scholar]
- 47.Hall T.A., editor. Nucleic Acids Symposium Series. Information Retrieval Ltd; 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT; pp. c1979–c2000. London. 1999. [Google Scholar]
- 48.Larkin M.A., Blackshields G., Brown N.P., et al. Clustal W and clustal X version 2.0. Bioinformation. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 49.Kumar S., Stecher G., Li M., Knyaz C., Tamura K. Mega X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35(6):1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tuegel M.A., Wrenn W.J. Sexual dimorphism in morphology and development of the pest chigger, Eutrombicula cinnabaris (Acari: Trombiculidae) Int. J. Acarol. 1998;24(3):199–211. doi: 10.1080/01647959808683585. [DOI] [Google Scholar]
- 51.Shatrov A.B., Kudryashova N.I. Micromammals and Macroparasites: from Evolutionary Ecology to Management. Springer; 2006. Taxonomy, life cycles and the origin of parasitism in trombiculid mites; pp. 119–140. [Google Scholar]
- 52.Roberts L.W., Garrison R., Francis G.G. Sex ratios in Rickettsia tsutsugamushi-infected and noninfected colonies of Leptotrombidium (Acari: Trombiculidae) J. Med. Entomol. 1977;14(1):89–92. doi: 10.1093/jmedent/14.1.89. [DOI] [PubMed] [Google Scholar]
- 53.Phasomkusolsil S., Tanskul P., Ratanatham S., et al. Influence of Orientia tsutsugamushi infection on the developmental biology of Leptotrombidium imphalum and Leptotrombidium chiangraiensis (Acari: Trombiculidae) J. Med. Entomol. 2012;49(6):1270–1275. doi: 10.1603/me12100. [DOI] [PubMed] [Google Scholar]
- 54.Egan M.E., Barth R.H., Hanson F.E. Chemically-mediated host selection in a parasitic mite. Nature. 1975;257:788–790. doi: 10.1038/257788a0. [DOI] [PubMed] [Google Scholar]
- 55.Egan M.E. The chemosensory bases of host discrimination in a parasitic mite. J. Comp. Physiol. 1976;109:69–89. [Google Scholar]
- 56.Foelix R.F. In: Neurobiology of Arachnids. Barth F.G., editor. Springer; Berlin, Heidelberg: 1985. Mechano- and chemoreceptive sensilla. [Google Scholar]
- 57.Dujardin J.P., Kaba D., Solano P., et al. Outline-based morphometrics, an overlooked method in arthropod studies? Infect. Genet. Evol. 2014;28:704–714. doi: 10.1016/j.meegid.2014.07.035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data included in article/supplementary material/referenced in article.