Abstract
Healthy skin reflects a healthy microbiome and vice versa. The contemporary society, marked by a sharp increase in skin irritation cases, has compelled researchers, dermatologists, and the cosmetics industry to investigate the correlation between skin microbiomes and the use of skincare products. Different cosmetics can change skin's normal flora to a varying degree –some changes can be detrimental, there are also instances where these alterations aid in restoring the skin microbiome. Previous studies using artificial skin models, metagenomic analysis, and culture-based approaches have suggested that skincare products play an important role in skin microbial alteration. This article assessed current knowledge on microbial shifts from daily use of various personal and skincare products. We have also introduced a readily applicable framework, synthesized from various observations, which can be employed to identify the normal skin microbiome and evaluate the impact of personal care and skincare products on it. We also discussed how lifestyle choice remake skin microbial makeup. Future studies are warranted to examine the effect of personal and skincare product usage on skin microbiome across various age groups, genders, and body sites with a multi-study approach.
Keywords: Active ingredients, Metagenomic, Microbiome, Microbiota changes, Skincare products
1. Introduction
Beauty is a cherished aspect for everyone, recognizing inner intelligence and external beauty propels toward motivation and refinement. Opting for cosmetics or skincare products regularly—daily, weekly, or monthly—instead of pills or supplements is a convenient, hassle-free way to enhance our appearance. This preference contributes to the exponential growth of the global cosmetics market, expected to reach $463.5 billion by 2027 with a 5.3 % CAGR (compound annual growth rate) [1]. So, skin, the most versatile interface organ, comprising an average 30 m2 surface area in adults, is the center of interest here [2]. Along with diversified chemicals and structural regimes, one thing that boosts skin's anisotropic heterogeneous nature is the composition of skin-dwelling microbiota. This is an absolute habitat for a diverse group of microorganisms, including bacteria, fungi, micro-eukaryotes (dust mites, Demodex mites), archaea, viruses, and phages [3]. A newborn baby gets colonized hugely after birth [4]. Gradually, due to topographical and site-specific diversity, the skin microbiota turns into a highly variable composition of an extremely versatile community depending on the areas of the body, between individuals, and over time. Contrary to a popular myth, not all skin microorganisms are inherently dangerous; pathogenicity only happens when the ecosystem's equilibrium is interrupted and variety is diminished. This issue of disruption has stirred the curiosity of health-conscious individuals.
Over the past 5–10 years, there has been a significant rise in skin damage rates. While there could be multiple environmental factors contributing to this issue, researchers have identified the increased usage of synthetic chemical components in modern-day cosmetics as the primary culprit posing a significant threat [5]. Skin care products are highly appreciated until they attract some uninvited guests. Within the personal care industry, the skin microbiota represents a promising yet often overlooked area, as evidenced by the increasing use of formulations that may impact the balance of the skin microbiota. Numerous instances of skin diseases have been linked to this dysbiosis or perturbation of the skin microbiome [6]. The imbalance can often be traced back to excessive product usage. Understanding the intricate relationship between skin chemistry, microbes, and their variations due to cosmetic application is essential. Gaining a better understanding of the correlation between microorganisms and skin care routines could significantly expand our knowledge of the relationship between the skin microbiome, skin diseases, and overall skin health. This, in turn, might pave the way for a more sustainable and healthier human existence. This review aims to highlight the latest findings on how different skin care products affect the skin's microbiome, as well as the potential consequences associated with these alterations. It also provides insights into various methods for assessing the skin microbiome and screening the effects of skin care products on skin inhabitants.
2. Conducting literature review
The literature search strategy entailed a comprehensive review of scholarly articles published up to November 2023, focusing on the contemporary discussion surrounding microbial shifts arising from the habitual use of various personal and skincare products. Here we reviewed more than 100 research articles to compile regarding the whole knowledge. We searched online databases including Web of Science, Science Direct, Scopus, and Google Scholar. Keyword selection was guided by the thematic structures and content of the review that included skin microbiome, cosmetics impacts, microbial dysbiosis, dermatological research, microbial alteration, active ingredients of skin care products and so on. Moreover, the search criteria were strictly limited to English-language publications.
3. Skin microbiome: composition and diversity
Skin microbiota, also known as skin flora, refers to microorganisms that live on the skin. It is the second-largest microbiota of the human body in mass [7]. Skin provides vital nutrients for the development of this microbiota through sweat or stratum corneum, which is composed of 75–80 % proteins (primarily keratins and membrane proteins), 5–15 % lipids (ceramides, cholesterol mainly), 5–10 % unknown compounds, and water (15–20 % of the total tissue dry weight) [8]. Within 2- and 3-mm thickness of the skin, microbiome has the most variation over time than the gut's and the mouth's microbiomes [9]. The skin harbors a vast array of bacterial species, with over 1200 identified. Among them, more than 90 % belong to four phyla: Actinobacteria (52 %), including Micrococcus, Propionibacteria, and Corynebacteria genera; Firmicutes (24 %), comprising Staphylococcus, Lactobacillus, and Streptococcus genera; Proteobacteria (16 %), which includes Paracoccus, Haematobacter, and Sphingomonas genera; and Bacteroidetes (6 %), represented by the genera Porphyromonas, Prevotella, and Flavobacterium [10,11]. Estimates suggest that Cutibacterium, Staphylococcus, and Corynebacterium genera, found throughout various sites on the skin, constitute approximately 45–80 % of the skin microbiome [12]. Additionally, up to 4.2 % of the prokaryotic skin microbiome is composed of archaea, with the dominant group being Thaumarchaeota phyla [13].
Various fungi have been notably found on different body regions, including Malassezia spp., Aspergillus, Cladosporium, Epicoccum, Phoma, Cryptococcus, Rhodotorula, Microsporum, Trichophyton, Saccharomyces, Candida, and Epidermophyton [14].
Regarding viruses or phages, the skin can harbor Molluscum contagiosum virus, Merkel cell polyomavirus (MCPyV), Alphapapillomavirus, Simian virus, Acheta domestica densovirus (AdDNV), Actinomyces phage, Propionibacterium phage, Polyomavirus HPyV7, Streptococcus phage, Stenotrophomonas phage, Enterobacteria phage, and more [7].
The resident microflora plays an advantageous role in modulating the immune system, occupying niches, and preventing the entry of harmful and contagious microorganisms into the skin. While skin microorganisms have been broadly identified and categorized [15], quantitative measurements are still necessary to compare investigations carried out by different researchers using various methodologies to consolidate their findings.
4. Interplay between cosmetics and skin microbiome
A vast array of skin care products, including body washes, gels, lotions, exfoliants, moisturizers, toners, and sunscreens, is available to address various beauty conditions, with a primary focus on nourishing the skin from within. However, researchers must acknowledge that cosmetic components can have both positive and negative impacts on the skin's microbiota [16]. Additionally, under specific physiochemical conditions, cosmetic compounds can serve as nutrient sources, promoting the growth of opportunistic pathogenic microorganisms that may lead to significant infections and diseases [17].
Certain research points to “microbiome resilience,” suggesting that there may be no negative effects on microorganisms due to the use of skin wash products. According to these studies, the concentrations of microorganisms are dramatically reduced by 5–10 times during product application [18]. However, the durability of these products varies depending on the application site, and their effects can last for weeks with highly individual reactions. These reactions may include changes in steroid and pheromone levels, as well as alterations in the structure and dynamics of bacterial and archaeal ecosystems, leading to a modified scenario [18].
As richness in microbial diversity has been proven to be linked with healthy skin [19], any kind of effect upon them destines changes leading to threats. But due to the broader range, studying their variations over skin has always been a challenge. These effects are not only occupied in the applied sites, but also other parts of the body [20]. For launching cosmetics, in depth studies are needed in context of microbiome. It is a fact of optimism that such practices are being encouraged nowadays in modern cosmetics. The first “Microbiome-friendly” certification for a cosmetic product was premiered in 2019 and was established by ‘MyMicrobiome’. Here the product's purity, the targeted area's specific bacteria's safety, the preservation of the microbiome diversity, and the preservation of the skin's natural balance are all confirmed (neither through the induction of harmful microorganisms nor by the repression of commensals) [21].
5. Assessing skin microbiome and screening the response of cosmetics
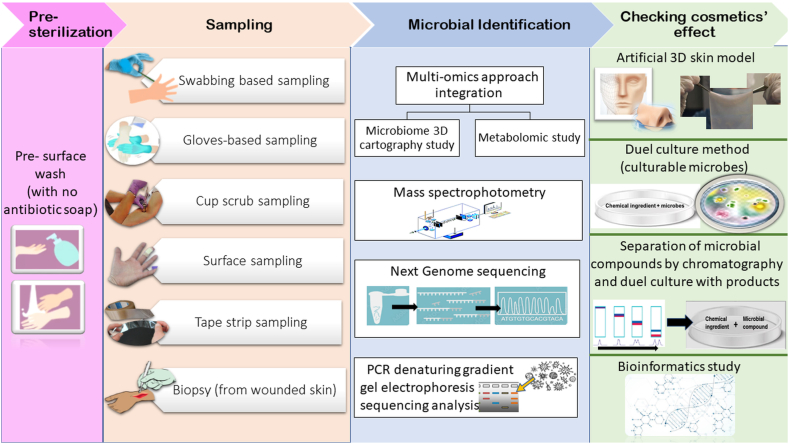
There is a need to explore effective approaches for comparing the skin microbiome after using skin care products to determine any deviations from normal skin conditions. This initial step is crucial in establishing a connection between the amount of synthetic ingredients in a product and its impact on skin health. In the 1950s, American dermatologist Albert Kligman pioneered human microbiota research in dermatology, employing advanced cell culture methods [22,23]. Today, modern research techniques allow us to identify non-culturable, newer forms of microbes residing in the surface and deeper layers of human skin. Overcoming the limitations of culture-dependent studies, molecular methods, such as multi-omics approach integration [20], 18S and 16S rRNA gene sequencing, DNA barcoding, PCR-denaturing gradient gel electrophoresis sequencing [24], enable the detection of microbes present in low quantities. Additionally, studying the chemicals produced by microbes through mass spectrometry visualization and molecular networking provides a wide range of microbial studies [16]. To identify the microbes, various methods like swabbing, biopsy, or tape stripping [25] are used, with the selection depending on the categories of microbes to be identified [26]. For assessing the ultimate outcomes resulting from the application of different products, the use of artificial 3D skin models and leather skin models has become popular [27,28]. Other methods for screening include the duel culture method, Mass spectrophotometry, FTIR spectro microscopy (Fourier-transform infrared spectroscopy), or magnetic resonance imaging [29,30]. Incorporating bioinformatics studies can also shed light on the effects of chemicals and bioactive products from microbes. Fig. 1 illustrates a workflow for identifying skin microbes and outlines possible approaches for screening cosmetics in relation to changes in the skin microbiome. Molecular approaches are commendable, but they do raise some counter-thoughts, such as their inability to differentiate between the genes of living and dead organisms, which can lead to confusing results. Therefore, to gain a comprehensive understanding of skin microbes, it is essential to combine whole genome sequencing with skin models and cultural approaches.
Fig. 1.
A workflow of assessing skin microbe and cosmetics' response.
6. Effect of cosmetics on the skin's microbiome
6.1. Effect of common cosmetic preservatives on skin microbe dynamics
In cosmetics, ingredients such as water, oils, peptides, and carbohydrates create an environment conducive to the growth of microorganisms. To prevent contamination, preservatives are commonly added, which remain active on the skin upon application. However, their blind use solely for preservation with antimicrobial properties might not serve their intended purpose [31].
A study conducted by Pinto, Ciardiello [32] revealed that various tested preservatives had an impact on the dynamics of targeted bacteria, including Cutibacterium acnes (previously known as Propionibacterium acnes), Staphylococcus epidermidis, and Staphylococcus aureus. While the combination of different preservatives in one bacterial strain (S. aureus) showed somewhat positive results, it proved to be both beneficial and harmful for the other two bacteria.
The most effective mixtures for reestablishing a pre-existing dysbiosis were found to contain hydroxyacetophenone, phenylpropanol, propanediol, caprylyl glycol, tocopherol, and tetrasodium glutamate diacetate because they act to moderately inhibit C. acnes and strongly inhibit S. aureus without simultaneously inhibiting S. aureus growth. The finest combinations to use in topical solutions for the skin and scalp where it is vital to maintain the eubiosis of the microbiota were proposed to be C1 (Sodium benzoate phenoxyethanol, ethylhexylglycerin), C4 (Sodium anisate, 1,2-hexanediol, ammonium acryloyldimethyltaurate/vp copolymer), C6 (Hydroxyacetophenone, phenylpropanol, propanediol, caprylyl glycol, tocopherol, disodium EDTA), and C7 (Benzyl alcohol, benzoic acid, dehydroacetic acid, ammonium acryloyldimethyltaurate/vp copolymer).
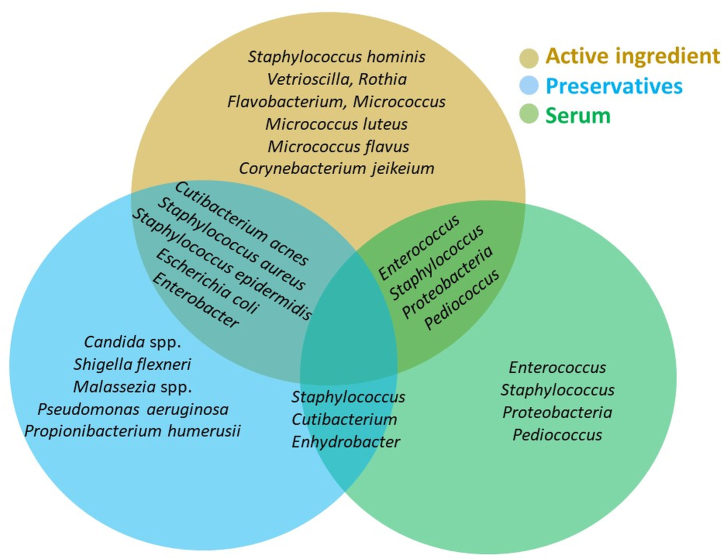
Toner, emulsion, cream, and baby cream contain common preservatives like parabens, 1, 2-hexanediol, phenoxyethanol. They exhibited potent antibacterial effects against S. aureus, Escherichia coli, Pseudomonas aeruginosa as well as other skin-resident bacteria such as S. epidermidis, Shigella flexneri, Enterobacter aerogenes and so on that are not the target at all [33]. Paraben, used in common cream had shown to have robust effects against skin filamentous fungi and yeasts like Candida spp. and Malassezia spp. [34]. C. acnes and S. epidermis were significantly inhibited by the short-term usage of 0.5–5 % methylparaben in cosmetics [35]. Methylisothiazolinone was seen to inhibit S. epidermidis [36]. The use of phenoxyethanol disturbed the skin microbiota conferring the change both at the phylum level (Proteobacteria increased) and at species level (Propionibacterium humerusii, S. epidermidis decreased) [33,36]. The influence of preservatives may go beyond the parameters of the product composition and may have a potentially negative impact on the skin microbiota, according to a theory put up in light of the expanding understanding of the significance of the human microbiome. The specified and some common microbes changed by basic ingredients that are used in skin care products are illustrated in Fig. 2.
Fig. 2.
A Venn diagram showing the specified and some common microbes changed by basic ingredients that are used in skin care products. Different color code represents different groups (Bronze- Active ingredient (eg.Kojic Acid, Isotretinoin etc.); Blue- Preservative (eg.Parabens, Methylparaben, Ethylparaben, Phenoxyethanol, Methylisothiazolinone etc.); Green- Serum (Galacto-oligosaccharide, Hyaluronic Acid etc.).
6.2. Changes in hand and palm microbiome by cosmetics
Hand and palm microbiome seems quite interesting with 83 % difference between the left and right hands of same individual, 87 % differences in difference in individuals and a greater overall diversity in female hands than males [37]. The amount of exposure to environmental elements, how often hands are washed all likely play a role in the variation in the skin microbiota of the hands. It is obviously affirmative to have a health care setting strategies for cleaning hands in but chronic washing is potential for dysbiosis in some individuals [38]. Healthcare personnel who routinely washed their hands have more pathogenic bacteria on them than those who didn't [39] indicating less microbial diversity led to the potentiality to increase pathogenic species including S. aureus, Enterococcus spp., Candida albicans [40]. The alteration is most in the prominent skin taxa than others in antiseptic treatments [41]. The alcohol-based hand sanitizer and ethanol showed a reduction of the levels of viable aerobic-anaerobic bacteria [42]. Antimicrobial soap had impacts on epidermal antibacterial defense system and upon skin microbes alike Streptococcus [43]. Antibacterial wipes resulted in a decrease in the number of S. aureus [44].
Many cleansers use surfactants that not only take away the key defensive components of our protective skin barrier exposing it to outside aggressors, but also misbalances the pH of skin [42]. Skin pH is fundamental to maintaining a healthy microbial community. Higher concentration of NaOH in cleansers, soaps, makeup, creams or lotions leads to higher pH level, causing Candida albicans, a commensal microbe turning into a fungal infection [45]. Extreme amount of citric acid and lactic acid in cleanser or exfoliants lowered normal skin pH causing decreased C. acnes. Similarly, with the increase in pH, the activity and population had been seen to be boosted for C. acnes and S. aureus [46].
6.3. Microbiome alterations induced by the active ingredients in cosmetics
Active ingredients in skin care products are specifically formulated for various purposes such as brightening, addressing dryness, combating aging, treating sunburn, and managing acne. In addition to different extracts and chemicals, pre-pro-postbiotics are also included in this list. Collectively, all these groups can have both positive and negative impacts on the skin's microbiome. Plant extracts, fruit extracts, and seaweed extracts used in cosmetics have shown incredible potential in modifying C. acnes populations, effectively combating skin pathogenesis like acne vulgaris [47,48]. However, certain ingredients, like kojic acid found in skin lightning creams and lotions, have been found to be capable of traveling into the bloodstream [49]. Moreover, pure kojic acid, when exposed to UV light, has the potential to induce gene mutations in E. coli strains [50]. As E. coli strains naturally occur on the skin in small amounts, such mutations might lead to harmful consequences [51]. Table 1 provides an overview of research findings highlighting the changes in skin microbes caused by the active ingredients of cosmetics and Fig. 2 shows microbial changes caused by active ingredients along with preservatives and serum. In contrast to synthetic substances, which humans have only encountered in the last 60 years of their 200 000-year history, natural components, in the quantities found in nature, are not perceived as “foreign” to the skin's natural condition [52]. This is why branding products as “natural or organic” easily captures attention. Consumers often prefer using products labelled as “natural cosmetics” due to concerns about their skin. However, it's essential to recognize that natural products may not be as pure as they seem in reality. For instance, Methylisothiazolinone (MI), a synthetic compound used in some ‘natural’ labelled products, has been associated with potential harm, including neurotoxicity [53], allergic reactions [54], and changes in microorganisms [55]. Alarmingly, this MI preservative is not limited to adult products like makeup, eyeliners, makeup removers, blush, face powder, hair care products, nail and waxing products, moisturizing creams, and sunscreen; it is also used in baby wipes and bath products. Even in toddlers, their diapers, skin pH, and dermatitis are linked with Candida albicans and S. aureus [56]. The research conducted by Wallen-Russell [57] investigated the microbial community shifts caused by using three different cosmetic conditions: a 100 % truly natural product (JooMo's face wash), a product labelled as natural but containing 70 % synthetic ingredients, and a synthetic product with 75 % synthetic components. Microbiome sampling was done before product use (T1), after two weeks (T2), and after four weeks (T3) of product use. During the transition from T1 to T2 and T1 to T3, JooMo showed the fastest rise in Chao1 diversity and species richness. This suggests that cosmetics companies should conduct studies on skin microbiota when launching new active ingredients to ensure their products maintain, improve, or restore a healthy skin-microbiome balance, especially in cases of a disrupted microbiome.
Table 1.
The Alteration in skin microbes caused by the active ingredients used in cosmetics.
| Active ingredients/A mixture of active ingredients | Alteration in skin microbes | References |
|---|---|---|
| Maltodextrin, Zymomonas ferment extract, honey extract, aqua |
Corynebacterium jeikeium
|
[58] |
Micrococcus luteus
| ||
Staphylococcus aureus
| ||
Micrococci
| ||
Staphylococcus epidermidis
| ||
Staphylococcus hominis
| ||
Micrococcus flavus
| ||
Cutibacterium avidum
| ||
| Glycerin, Laminaria digitata extract, Chlorella vulgaris extract, saccharide isomerate, phenoxyethanol, ethylhexylglycerin, aqua, seawater |
Staphylococcus aureus
|
[58] |
Staphylococcus hominis Micrococcus luteus
| ||
Cutibacterium avidum
| ||
S. epidermidis
| ||
Micrococcus flavus
| ||
| Fermented oil |
Staphylococcus
|
[59] |
Proteobacteria
| ||
| ExpoZenfi by GREENTECH | Bacterial diversity
|
[60] |
Staphylococcus epidermidi
| ||
| Isotretinoin | Rothia
|
[61] |
| Flavobacterium | ||
| Enterobacter | ||
| Micrococcus | ||
Cutibacterium acnes
| ||
| Halymenia durvillei (red algae) extract | Proteobacteria phyla
|
[60] |
Bacteriodetes phyla
| ||
Actinobacteria phyla
| ||
Firmicutes phyla
| ||
Corynebacterium kroppendenstedtii
|
 indicates increase,
indicates increase,  indicates decrease.
indicates decrease.
6.4. Alternation of skin microbiome by different types of cosmetics
The topical application of personal hygiene products can alter the skin's lipid film, affecting microbial diversity and the overall richness of bacterial species on the skin. For instance, active body wash products have demonstrated potential advantages, as they can eliminate harmful bacteria such as Brevibacterium casei and Rhodotorula mucilaginosa, while promoting the growth of beneficial bacteria like C. acnes [62].
Furthermore, a basic skin care routine consisting of skin softener, lotion, essence, and cream with moisturizing compounds was found to impact the abundance of two common skin phyla: Actinobacteria (decreased) and Proteobacteria (increased) on facial cheeks [63]. The cause of these changes could be attributed to other skin bacteria growing and utilizing ingredients from the basic cosmetics, or the cosmetics may be hindering the growth of normal skin bacterial groups, or even altering the skin environment itself. The following Table 2 provides an overview of product-based variations that influence skin microbes.
Table 2.
Some common skin care products causing microbial alteration in skin.
| Product category | Alteration in skin microbes | References |
|---|---|---|
| Lotion application in lower foot (against xerosis, extreme dryness) |
Staphylococcus epidermidis
|
[64] |
Xanthomonas campestris
| ||
Xanthomonas spp.
| ||
| Short chain fructo-oligosaccharides (A prebiotic used in powder etc.) |
Staphylococcus epidermidis at lower concentration (0.5–5 %) Staphylococcus aureu lower concentration (0.5–5 %) Staphylococcus aureu s s
|
[65] |
Staphylococcus epidermidis at higher concentration (10–15 %) C. acnes (complete halt) higher concentration (10–15 %) C. acnes (complete halt)
| ||
| Spermidine used in lotion, cream |
Staphylococcus pneumonia
|
[66] |
Staphylococcus infantis
| ||
Staphylococcus thermophiles
| ||
| Ceramides in moisturizers |
Streptococcus spp.
|
[67] |
| Serum cosmetics containing galacto-oligosaccharides |
Burkholderia
|
[68] |
Bifidobacteria
| ||
Lactobacilli
| ||
Lactococcus
| ||
Sphingomonas
| ||
Thermoanaerobacterium
| ||
Staphylococcus aureus
| ||
Staphylococcus
| ||
Proteobacteria
| ||
Cutibacterium
| ||
Pediococcus
| ||
Enhydrobacter
| ||
Enterobacteriaceae family
| ||
| Foot powder use |
Micrococcus
|
[20] |
Anaerococcus
| ||
Streptococcus
| ||
Brevibacterium Acinetobacter
| ||
Moraxellaceae family
| ||
| Selenium in lotion, sunscreen, creams |
Staphylococcus aureus
|
[69,70] |
| Moisturizers containing lipids |
Staphylococcus
|
[16,71,72] |
Propionibacterium
|
 indicates increase,
indicates increase,  indicates decrease.
indicates decrease.
Sebocytes create sebum, a natural moisturizer that is essential for maintaining healthy skin. Excessive sebum production can lead to issues like acne, greasy skin, and clogged pores. To prevent such problems, many individuals use different skin care products, such as lotions [73], which have proven to be effective in reducing sebum production. However, decreased sebum can also result in a reduced food source for microbes, leading to a decline in the number of Staphylococcus bacteria and causing an imbalance in the skin microbiome. A comparison between makeup users and non-users revealed significantly higher microbial diversity on the forehead skin of makeup users, including an increase in the genera Selenomonas, Aggregatibacter, and Aquicella [74].
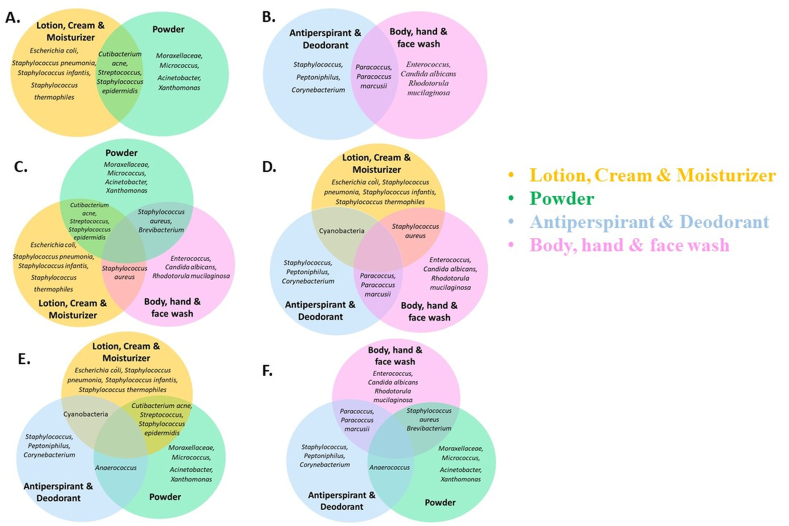
The changes in skin microbes are intricate and interconnected. Fig. 3A–F depicts the range of products (lotion, cream and moisture; powder, antiperspirant and deodorant; body, hand and face wash) that commonly influence bacterial or fungal populations, detailing those that have individual and combined effects on these species as evidenced by multiple research studies.
Fig. 3.
Venn diagrams illustrating the specific and common microbial changes influenced by skincare products. A: Alteration of microbes by Lotion, Cream & Moisturizer + Powder. B: Alteration of microbes by Antiperspirant & Deodorant + Body, hand & face wash. C: Alteration of microbes by Powder + Lotion, Cream & Moisturizer + Body, hand & face wash. D: Alteration of microbes by Lotion, Cream & Moisturizer + Antiperspirant & Deodorant + Body, hand & face wash. E: Alteration of microbes by Lotion, Cream & Moisturizer + Antiperspirant & Deodorant + Powder. F: Alteration of microbes by Body, hand & face wash + Antiperspirant & Deodorant + Powder.
6.5. Microplastic
Microplastics and nanoplastics are ubiquitous constituents of human daily consumption, serving as pervasive pollutants that appear in many forms such as microbeads and fibers. The impact of microplastic load is terrifying for flora-fauna, terrestrial, ocean organisms [75,76] and soil microbiota [77], covering all sort of environmental microbiomes [78]. They can adsorb organic and inorganic contaminants on their surface [79]. In addition, biofilms can form on their surface and act as carriers of pathogenic vectors and antimicrobial resistance [80], career for pollutants [81], microorganisms and resistance genes [82]. Their effects on different animals’ microbiome had been notified.
Polystyrene reduced of the relative abundance of phyla Firmicutes, Bacteroidetes and Verrucomicrobia, increased Actinobacteria with 0.5 μm particles size in the gastrointestinal tract of mice [83]. Polystyrene microplastics fibres and fragments reduced Actinobacteria; fibers affected specific bacteria genera while fragments caused a decrease in Pseudomonas and Aeromonas genus and all three shapes increased the relative abundance of Gordonia [84]. It's 5 μm increased Staphylococcus and 200 μm boosted the genera Vibrio, Acinetobacter, Porphyromonas, Haemophilus, Neisseria and Lactococcus in zebra fish [85]. Phyla Cyanobacteria, Chloroflexi, Fusobacteria and Proteobacteria were increased, while Nitrospirae, Bacteroidetes, and Firmicutes were reduced in freshwater crabs [86] via microplastic intervention. In Human, exposure to Polystyrene MPs induced gut microbial shifts increasing α-diversity and abundance of potentially harmful pathobionts, such as Dethiosulfovibrionaceae and Enterobacteriaceae family [87]. Polyethylene alters Firmicutes, Bacteroidetes, Proteobacteria, and Verrucomicrobia phyla [88]. Reductions in Bifidobacterium spp., Clostridium spp., Staphylococcus spp., total aerobic and anaerobic bacteria, and increase in Escherichia or Shigella, Cloacibacillus, Bilophila, Eisenbergiella, Megasphaera and Oscillibacter genus including rise in Firmicutes phylum [89] had been shown in different chambers of gut. Polypropylene microplastics caused significant changes in fish gut microbiome, reducing beneficial lactic acid bacteria and increasing potential pathogenic microorganism Proteobacteria and Vibrionales [90].
Regarding human microbiomes, mammal microbiomes are more similar to that of humans than non-mammal microbiomes. Microbead is the primary source of Microplastics used as cleansing or exfoliating agents in a diversity of personal care and cosmetic products such as shower gels, toothpaste, nail polishes, or eye shadows, amongst many others. From the above-mentioned studies, we can predict there is highest possible chance of those microplastic altering our normal skin microbiome. In a research conducted in Macao showed 100 skin care products (facial skin, body skin, cosmetics) out of 144 are with microplastic [91]. A significant quantity of personal care and cosmetics products are found to include microplastics such as polyethylene, polyethylene terephthalate, polypropylene, and other similar substances. The aforementioned alterations in the environment and gut microbiota may lead us to speculate that they could potentially impact the composition of the skin microbiome. Unfortunately, there is a lack of relevant data regarding this particular matter. A comprehensive investigation is necessary to ascertain the impact of microplastic exposure, either alone or in conjunction with skin-care products, on the host microbiome.
7. Consequences of altered microbiome
In contrast to individual variances, it is a well-known fact that most adult human microbiomes remain stable without intervention [9,92,93]. However, shifts can occur due to both intrinsic and extrinsic factors, leading to the opposite phenomenon. Thus far, it has been established that the use of various skincare products can trigger microbial alterations. Whether this shifting will be named after positive or negative, is dependent upon the species of microbes being changed. Depending upon the types of products use, the consequences might vary. For sure, many of the skin care products offer us a variety of favors in skin commensals. One of the consequences that terrifies all is microbes misbalance causes their shifting from friend to foe for instance C. acnes [94]. The same statement is justified as commensals S. epidermidis would be turned into pathogen if its number is increased creating serious skin irritation by boosting atopic dermatitis [95]. But in normal skin, S. epidermidis aids in skin homeostasis and lowers the pathogenic inflammation triggered by C. acnes as it drops off the TLR2 protein (Toll like receptor 2 protein) production, which reasons skin inflammation [96]. Dysbiosis and altered microbial biodiversity of the skin microbiome has been linked with many diseases [97]. Acne was modulated by Proteobacteria, Firmicutes and Actinobacteria [98] and also by many species like C. acnes [99], Streptococcus [100], S. epidermidis [101], Malassezia species [102,103] etc. Atopic dermatitis, a chronic skin inflammatory disease impacts roughly 15 %–20 % of children and 1 %–3 % of adults worldwide and has skyrocketed 2- to 3-fold [104]. It had seen to have relation with significant skin microbial changes such as S. aureus, S. epidermidis and Staphylococcus haemolyticus [105,106], Propiniumbacterium, Corynebctarium and Streptococcus [105,107]. Some common skin dwellers Staphylococcus, Corynebacterium, Streptococcus and Propionibacterium were related to skin diseases like psoriatic lesion [108]. Malassezia fungal species linking with folliculitis [109], Staphylococcus with diabetic skin [110], S. aureus with allergies [111], Streptococcus, Staphylococcus, Fusobacterium with leishmaniasis [112], Acinetobacter lwoffii with allergic sensitization and inflammation [113] had been reported. Apart from the common one (Malassezia fungi) [114] some low abundant genera like Gordonia and Geobacillus also had seen linking with skin disease-rosacea [115]. Regarding bodies defense mechanism, some ardently serious issues are involved. Shifting of normal microbiome of Staphylococcus, and in particular S. epidermidis results in the disruption of skin immune system causing the natural antimicrobial bacteriocins production hampered [116]. Changes in Porphyromonas, Streptococcus, Peptostreptococcus, Sphingomonas, Stenotrophomonas, Anaerococcus, Staphylococcus, Corynebacterium etc. were related to wound healing process [117]. So even minor shift may result some catastrophe. These findings hold significance from an immunological perspective, as they imply a direct communication link between host health and microbial cells in the skin.
While individual microorganisms alone may or may not directly cause cancer, the overall composition and diversity of the skin microbiome can have an impact. Cosmetics can alter the skin microbiome by reducing its diversity and balance, resulting in microbial imbalance (dysbiosis). This, in turn, can lead to inflammation, immune suppression, oxidative stress, and infections, all of which can contribute to the promotion of skin cancer [118]. The studies mentioned above indicate varying levels of microbial increase and decrease in the skin microbial community. Some of these common microbes have been shown to be linked to cancer. For instance, S. aureus is associated with the carcinogenic progression from actinic keratosis to squamous cell carcinoma (SCC), a type of skin cancer that develops in the flat squamous cells located in the outer layer of the skin.” [119,120]. Moreover, it can influence the expression of Human beta-defensin-2, thereby causing SCC proliferation [121]. Cutaneous T cell lymphoma, another type of skin cancer had been linked with S. aureus [122]. Some research showed that Corynebacterium species may influence the development of malignant melanoma, the deadliest form of skin cancer, accounting for 75 % of all skin cancer-related deaths [123,124]. Skin cancers might be boosted as a result of skin microbiome alteration demonstrating skin barrier disruption [125] for Staphylococcus epidermidis [126].
Coffee aqueous extract is being used in many skins care formula that had demonstrated links with Colorectal cancer [127]. Tryptophan, a microbial metabolite detected on skin, also used in cosmetics [128] has effect on aryl hydrocarbon receptor. Imbalanced regulation of aryl hydrocarbon receptor expression or activity promotes cancer development alike breast cancer [129], lung cancer [130], oesophageal cancer [131] etc. Malassezia sp. affects skin cancer [132]. It increased aryl hydrocarbon receptor activity conferring increased tumorigenesis [133]. Candida is related with an increased risk of numerous malignancies, including hematologic, head and neck, pancreatic, skin, and thyroid cancer [134]. Viruses are also included in this list [135]. More study is needed to better understand how various cosmetics products promoter different forms of cancer and how they can be managed to prevent or treat cancer.
8. Conclusion
A diverse and thriving skin microbiome is essential for human health, making it a novel objective for skin care products to focus on maintaining or restoring the skin's microbiota. The application of additional skin care products can significantly impact the microbial diversity on the skin, resulting in both positive and negative differences. Each type of product used on the skin has its own individual and combined effects. Common skin microbes like Staphylococcus aureus, Staphylococcus epidermidis, Corynebacterium, Cutibacterium acnes, and Malassezia spp. are frequently altered by diversified arrays of such products application. As microbial alteration is linked with lots of diseases ranging from skin irritation to cancers not only in skin but also of other organs, precise studies are needed. A combination of sampling methods, culture-based strategies, and modernized research tools, such as artificial skin models, spectrophotometry, bioinformatics, and metagenomic analysis, is necessary. To gain a comprehensive understanding of the skin's microbiome and its complex interactions when using various cosmetics, further investigations considering factors like age, location, and specific body sites are essential. Ultimately, these studies aim to ensure a healthy human skin microbiome. No definitive conclusion has been reached to universally categorize any product as entirely positive or negative. The question arises whether the negative impacts outweigh the positive ones.
Data availability statement
No data was used for the research described in the article.
CRediT authorship contribution statement
Mahjabin Ferdaous Mim: Writing – original draft, Visualization, Data curation, Conceptualization. Mahmudul Hasan Sikder: Writing – review & editing. Md. Zahid Hasan Chowdhury: Visualization, Data curation. Ashkar-Ul-Alam Bhuiyan: Visualization. Nayeematul Zinan: Data curation. Shah Mohammad Naimul Islam: Writing – review & editing, Conceptualization.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:Shah Mohammad Naimul Islam reports administrative support was provided by Bangabandhu Sheikh Mujibur Rahman Agricultural University.
References
- 1.Shankar Cosmetics market by category, gender and distribution channel: opportunity analysis and industry forecast, 2021–2027. Allied market research. 2021 https://www.alliedmarketresearch.com/press-release/cosmetics-market.html [Google Scholar]
- 2.Gallo R.L. Human skin is the largest epithelial surface for interaction with microbes. J. Invest. Dermatol. 2017;137:1213–1214. doi: 10.1016/j.jid.2016.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Verbanic S., Kim C.Y., Deacon J.M., Chen I.A. Improved single-swab sample preparation for recovering bacterial and phage DNA from human skin and wound microbiomes. BMC Microbiol. 2019;19:1–13. doi: 10.1186/s12866-019-1586-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oh J., Byrd A.L., Park M., Kong H.H., Segre J.A. Temporal stability of the human skin microbiome. Cell. 2016;165:854–866. doi: 10.1016/j.cell.2016.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wallen-Russell C., Wallen-Russell S. Meta analysis of skin microbiome: new link between skin microbiota diversity and skin health with proposal to use this as a future mechanism to determine whether cosmetic products damage the skin. Cosmet. Toilet. 2017;4:14. [Google Scholar]
- 6.Myles I.A., Williams K.W., Reckhow J.D., Jammeh M.L., Pincus N.B., Sastalla I., Saleem D., Stone K.D., Datta S.K. Transplantation of human skin microbiota in models of atopic dermatitis. JCI Insight. 2016;1 doi: 10.1172/jci.insight.86955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Byrd A.L., Belkaid Y., Segre J.A. The human skin microbiome. Nat. Rev. Microbiol. 2018;16:143–155. doi: 10.1038/nrmicro.2017.157. [DOI] [PubMed] [Google Scholar]
- 8.Barbieri J.S., Wanat K., Seykora J. 2014. Skin: Basic Structure and Function. [Google Scholar]
- 9.Costello E.K., Lauber C.L., Hamady M., Fierer N., Gordon J.I., Knight R. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grice E.A., Kong H.H., Renaud G., Young A.C., NISC Comparative Sequencing Program, Bouffard G.G., Blakesley R.W., Wolfsberg T.G., Turner M.L., Segre J.A. A diversity profile of the human skin microbiota. Genome Res. 2008;18:1043–1050. doi: 10.1101/gr.075549.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grice E.A., Kong H.H., Conlan S., Deming C.B., Davis J., Young A.C., NISC Comparative Sequencing Program, Bouffard G.G., Blakesley R.W., Murray P.R. Topographical and temporal diversity of the human skin microbiome. Science. 2009;324:1190–1192. doi: 10.1126/science.1171700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Samaras S., Hoptroff M. 2020. The Microbiome of Healthy Skin, Skin Microbiome Handbook: from Basic Research to Product Development; pp. 1–32. [Google Scholar]
- 13.Probst A.J., Auerbach A.K., Moissl-Eichinger C. Archaea on human skin. PLoS One. 2013;8 doi: 10.1371/journal.pone.0065388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li H., Goh B.N., Teh W.K., Jiang Z., Goh J.P.Z., Goh A., Wu G., Hoon S.S., Raida M., Camattari A. Skin commensal Malassezia globosa secreted protease attenuates Staphylococcus aureus biofilm formation. J. Invest. Dermatol. 2018;138:1137–1145. doi: 10.1016/j.jid.2017.11.034. [DOI] [PubMed] [Google Scholar]
- 15.Sfriso R., Egert M., Gempeler M., Voegeli R., Campiche R. Revealing the secret life of skin‐with the microbiome you never walk alone. Int. J. Cosmet. Sci. 2020;42:116–126. doi: 10.1111/ics.12594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bouslimani A., Porto C., Rath C.M., Wang M., Guo Y., Gonzalez A., Berg-Lyon D., Ackermann G., Moeller Christensen G.J., Nakatsuji T. vol. 112. Proceedings of the National Academy of Sciences; 2015. pp. E2120–E2129. (Molecular Cartography of the Human Skin Surface in 3D). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neza E., Centini M. Microbiologically contaminated and over-preserved cosmetic products according Rapex 2008–2014. Cosmet. Toilet. 2016;3:3. [Google Scholar]
- 18.Murphy B., Hoptroff M., Arnold D., Eccles R., Campbell-Lee S. In-vivo impact of common cosmetic preservative systems in full formulation on the skin microbiome. PLoS One. 2021;16 doi: 10.1371/journal.pone.0254172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Finlay B.B., Arrietta M.-C. Greystone Books; 2016. Let Them Eat Dirt. [Google Scholar]
- 20.Bouslimani A., da Silva R., Kosciolek T., Janssen S., Callewaert C., Amir A., Dorrestein K., Melnik A.V., Zaramela L.S., Kim J.-N. The impact of skin care products on skin chemistry and microbiome dynamics. BMC Biol. 2019;17:1–20. doi: 10.1186/s12915-019-0660-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.MyMicrobiome. Retrieved. February 3, 2023. https://microbiome-friendly.com/en/ from.
- 22.Dréno B., Araviiskaia E., Berardesca E., Gontijo G., Sanchez Viera M., Xiang L.F., Martin R., Bieber T. Microbiome in healthy skin, update for dermatologists. J. Eur. Acad. Dermatol. Venereol. 2016;30:2038–2047. doi: 10.1111/jdv.13965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pillsbury D.M. Manual OF dermatology. AJN The American Journal of Nursing. 1943;43:791. [Google Scholar]
- 24.Li W., Han L., Yu P., Ma C., Wu X., Xu J. Nested PCR-denaturing gradient gel electrophoresis analysis of human skin microbial diversity with age. Microbiol. Res. 2014;169:686–692. doi: 10.1016/j.micres.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 25.Kong H.H., Andersson B., Clavel T., Common J.E., Jackson S.A., Olson N.D., Segre J.A., Traidl-Hoffmann C. Performing skin microbiome research: a method to the madness. J. Invest. Dermatol. 2017;137:561–568. doi: 10.1016/j.jid.2016.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Verbanic S., Kim C.Y., Deacon J.M., Chen I.A. Improved single-swab sample preparation for recovering bacterial and phage DNA from human skin and wound microbiomes. BMC Microbiol. 2019;19:1–13. doi: 10.1186/s12866-019-1586-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Niehues H., Bouwstra J.A., El Ghalbzouri A., Brandner J.M., Zeeuwen P.L., Van Den Bogaard E.H. 3D skin models for 3R research: the potential of 3D reconstructed skin models to study skin barrier function. Exp. Dermatol. 2018;27:501–511. doi: 10.1111/exd.13531. [DOI] [PubMed] [Google Scholar]
- 28.Pinto D., Ciardiello T., Franzoni M., Pasini F., Giuliani G., Rinaldi F. Effect of commonly used cosmetic preservatives on skin resident microflora dynamics. Sci. Rep. 2021;11:8695. doi: 10.1038/s41598-021-88072-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coenye T., Nelis H.J. In vitro and in vivo model systems to study microbial biofilm formation. J. Microbiol. Methods. 2010;83:89–105. doi: 10.1016/j.mimet.2010.08.018. [DOI] [PubMed] [Google Scholar]
- 30.Gannesen A.V., Zdorovenko E.L., Botchkova E.A., Hardouin J., Massier S., Kopitsyn D.S., Gorbachevskii M.V., Kadykova A.A., Shashkov A.S., Zhurina M.V. Composition of the biofilm matrix of Cutibacterium acnes acneic strain RT5. Front. Microbiol. 2019;10:1284. doi: 10.3389/fmicb.2019.01284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cao M.C., Feng T.T., Zhang X.C. Investigation on preservatives use in commercial cosmetics. J Environ Hyg. 2017;7:296–300. [Google Scholar]
- 32.Pinto D., Ciardiello T., Franzoni M., Pasini F., Giuliani G., Rinaldi F. Effect of commonly used cosmetic preservatives on skin resident microflora dynamics. Sci. Rep. 2021;11:8695. doi: 10.1038/s41598-021-88072-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jeong J.-J., Kim D.-H. Effects of cosmetics and their preservatives on the growth and composition of human skin microbiota. Journal of the Society of Cosmetic Scientists of Korea. 2015;41:127–134. [Google Scholar]
- 34.Nasrollahi S.A., Fattahi M., Khamesipoor A., Amiri F., Ahmadi M., Kavkani M.S., Lotfali E., Ayatollahi A., Skandari S.E., Firooz A. Effects of cosmetic preservatives on healthy facial skin microflora. J. Clin. Aesthet. Dermatol. 2022;15:34. [PMC free article] [PubMed] [Google Scholar]
- 35.Halla N., Fernandes I.P., Heleno S.A., Costa P., Boucherit-Otmani Z., Boucherit K., Rodrigues A.E., Ferreira I.C., Barreiro M.F. Cosmetics preservation: a review on present strategies. Molecules. 2018;23:1571. doi: 10.3390/molecules23071571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Q., Cui S., Zhou L., He K., Song L., Liang H., He C. Effect of cosmetic chemical preservatives on resident flora isolated from healthy facial skin. J. Cosmet. Dermatol. 2019;18:652–658. doi: 10.1111/jocd.12822. [DOI] [PubMed] [Google Scholar]
- 37.Fierer N., Hamady M., Lauber C.L., Knight R. vol. 105. Proceedings of the National Academy of Sciences; 2008. pp. 17994–17999. (The Influence of Sex, Handedness, and Washing on the Diversity of Hand Surface Bacteria). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nash A.A., Dalziel R.G., Fitzgerald J.R. Mims' Pathogenesis of Infectious Disease. Elsevier; Amsterdam, The Netherlands: 2015. Attachment to and entry of microorganisms into the body; pp. 9–49. [Google Scholar]
- 39.Larson E. Skin hygiene and infection prevention: more of the same or different approaches? Clin. Infect. Dis. 1999;29:1287–1294. doi: 10.1086/313468. [DOI] [PubMed] [Google Scholar]
- 40.Rosenthal M., Aiello A., Larson E., Chenoweth C., Foxman B. Healthcare workers' hand microbiome may mediate carriage of hospital pathogens. Pathogens. 2014;3:1–13. doi: 10.3390/pathogens3010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.SanMiguel A.J., Meisel J.S., Horwinski J., Zheng Q., Bradley C.W., Grice E.A. Antiseptic agents elicit short-term, personalized, and body site–specific shifts in resident skin bacterial communities. J. Invest. Dermatol. 2018;138:2234–2243. doi: 10.1016/j.jid.2018.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zapka C., Leff J., Henley J., Tittl J., De Nardo E., Butler M., Griggs R., Fierer N., Edmonds-Wilson S. Comparison of standard culture-based method to culture-independent method for evaluation of hygiene effects on the hand microbiome. mBio. 2017;8 doi: 10.1128/mbio.00093–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Two A.M., Nakatsuji T., Kotol P.F., Arvanitidou E., Du-Thumm L., Hata T.R., Gallo R.L. The cutaneous microbiome and aspects of skin antimicrobial defense system resist acute treatment with topical skin cleansers. J. Invest. Dermatol. 2016;136:1950–1954. doi: 10.1016/j.jid.2016.06.612. [DOI] [PubMed] [Google Scholar]
- 44.Ameri S., Ahmad Nasrollahi S., Samadi A., Amiri F., Ahmadvand S., Yadangi S., Fattahi M., Ehsani M., Firooz A. Assessment of skin microbiota and biometric parameters: a comprehensive comparison of four types of hand cleansers. Iranian Journal of Dermatology. 2021;24:306–314. [Google Scholar]
- 45.Rippke F., Berardesca E., Weber T.M. vol. 54. 2018. pp. 87–94. (pH and microbial infections, pH of the Skin: Issues and Challenges). [DOI] [PubMed] [Google Scholar]
- 46.Korting H.C., Hübner K., Greiner K., Hamm G., Braun-Falco O. Differences in the skin surface pH and bacterial microflora due to the long-term application of synthetic detergent preparations of pH 5.5 and pH 7.0. Results of a crossover trial in healthy volunteers. Acta Derm. Venereol. 1990;70:429–431. [PubMed] [Google Scholar]
- 47.Gervason S., Metton I., Gemrot E., Ranouille E., Skorski G., Cabannes M., Berthon J.-Y., Filaire E. Rhodomyrtus tomentosa fruit extract and skin microbiota: a focus on C. acnes phylotypes in acne subjects. Cosmet. Toilet. 2020;7:53. [Google Scholar]
- 48.Lee J.-H., Eom S.-H., Lee E.-H., Jung Y.-J., Kim H.-J., Jo M.-R., Son K.-T., Lee H.-J., Kim J.H., Lee M.-S. In vitro antibacterial and synergistic effect of phlorotannins isolated from edible brown seaweed Eisenia bicyclis against acne-related bacteria. ALGAE. 2014;29:47–55. [Google Scholar]
- 49.Fukase H. 2005. Percutaneous Absorption Study of Kojic Acid in Humans, CPC Clinic, Medical Facility. Kagoshima, Japan. [Google Scholar]
- 50.Wollny H.E. 1998. Salmonella typhimurium and Escherichia coli Reverse Muation Assay with Kojic Acid. RCC-CCR Project Number 612701, Unpublished Data. [Google Scholar]
- 51.Petkovsek Z., Elersic K., Gubina M., Zgur-Bertok D., Starcic Erjavec M. Virulence potential of Escherichia coli isolates from skin and soft tissue infections. J. Clin. Microbiol. 2009;47:1811–1817. doi: 10.1128/JCM.01421-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Blaser M.J., Falkow S. What are the consequences of the disappearing human microbiota? Nat. Rev. Microbiol. 2009;7:887–894. doi: 10.1038/nrmicro2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Burnett C.L., Bergfeld W.F., Belsito D.V., Klaassen C.D., Marks J.G., Shank R.C., Slaga T.J., Snyder P.W., Andersen F.A. Final report of the safety assessment of methylisothiazolinone. Int. J. Toxicol. 2010;29:187S–213S. doi: 10.1177/1091581810374651. [DOI] [PubMed] [Google Scholar]
- 54.Scherrer M.A.R., Rocha V.B., Andrade A.R.C. Contact dermatitis to methylisothiazolinone. An. Bras. Dermatol. 2015;90:912–914. doi: 10.1590/abd1806-4841.20153992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fournière M., Latire T., Souak D., Feuilloley M.G.J., Bedoux G. Staphylococcus epidermidis and Cutibacterium acnes: two major sentinels of skin microbiota and the influence of cosmetics. Microorganisms. 2020;8:1752. doi: 10.3390/microorganisms8111752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Šikić Pogačar M., Maver U., Marčun Varda N., Mičetić‐Turk D. Diagnosis and management of diaper dermatitis in infants with emphasis on skin microbiota in the diaper area. Int. J. Dermatol. 2018;57:265–275. doi: 10.1111/ijd.13748. [DOI] [PubMed] [Google Scholar]
- 57.Wallen-Russell C. The role of every-day cosmetics in altering the skin microbiome: a study using biodiversity. Cosmet. Toilet. 2018;6:2. [Google Scholar]
- 58.Rademacher M., Zinn M.-K., Beinio R., Bockmühl D.P. A new model to investigate the effects of cosmetics on skin microorganisms in vitro. Cosmet. Toilet. 2022;9:88. [Google Scholar]
- 59.Ciardiello T., Pinto D., Marotta L., Giuliani G., Rinaldi F. Effects of fermented oils on alpha-biodiversity and relative abundance of cheek resident skin microbiota. Cosmet. Toilet. 2020;7:34. [Google Scholar]
- 60.Filaire E., Vialleix C., Cadoret J.-P., Guénard S., Muller C., Dreux-Zigha A., Berthon J.-Y. Characterization of reactive and sensitive skin microbiota: effect of Halymenia durvillei (HD) extract treatment. Cosmet. Toilet. 2019;6:69. [Google Scholar]
- 61.McCoy W.H., Otchere E., Rosa B.A., Martin J., Mann C.M., Mitreva M. Skin ecology during sebaceous drought–how skin microbes respond to isotretinoin. J. Invest. Dermatol. 2019;139:732. doi: 10.1016/j.jid.2018.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sfriso R., Claypool J. Microbial reference frames reveal distinct shifts in the skin microbiota after cleansing. Microorganisms. 2020;8:1634. doi: 10.3390/microorganisms8111634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee H.J., Jeong S.E., Lee S., Kim S., Han H., Jeon C.O. Effects of cosmetics on the skin microbiome of facial cheeks with different hydration levels. Microbiologyopen. 2018;7 doi: 10.1002/mbo3.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Murphy B., Grimshaw S., Hoptroff M., Paterson S., Arnold D., Cawley A., Adams S.E., Falciani F., Dadd T., Eccles R. Alteration of barrier properties, stratum corneum ceramides and microbiome composition in response to lotion application on cosmetic dry skin. Sci. Rep. 2022;12:5223. doi: 10.1038/s41598-022-09231-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Le Bourgot C., Meunier C., Gaio E., Murat V., Micheletto M., Tedesco E., Benetti F. Effects of short chain fructo-oligosaccharides on selected skin bacteria. Sci. Rep. 2022;12:9702. doi: 10.1038/s41598-022-13093-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kim G., Kim M., Kim M., Park C., Yoon Y., Lim D.-H., Yeo H., Kang S., Lee Y.-G., Beak N.-I. Spermidine-induced recovery of human dermal structure and barrier function by skin microbiome. Commun. Biol. 2021;4:231. doi: 10.1038/s42003-020-01619-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Howard B., Bascom C.C., Hu P., Binder R.L., Fadayel G., Huggins T.G., Jarrold B.B., Osborne R., Rocchetta H.L., Swift D. Aging-associated changes in the adult human skin microbiome and the host factors that affect skin microbiome composition. J. Invest. Dermatol. 2022;142:1934–1946. e21. doi: 10.1016/j.jid.2021.11.029. [DOI] [PubMed] [Google Scholar]
- 68.Hong K.-B., Hong Y.H., Jung E.Y., Jo K., Suh H.J. Changes in the diversity of human skin microbiota to cosmetic serum containing prebiotics: results from a randomized controlled trial. J. Personalized Med. 2020;10:91. doi: 10.3390/jpm10030091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kaur K., Rath G. Formulation and evaluation of UV protective synbiotic skin care topical formulation. J. Cosmet. Laser Ther. 2019;21:332–342. doi: 10.1080/14764172.2019.1658878. [DOI] [PubMed] [Google Scholar]
- 70.Stolzoff M., Wang S., Webster T. 2016. Efficacy and Mechanism of Selenium Nanoparticles as Antibacterial Agents. [Google Scholar]
- 71.Unno M., Cho O., Sugita T. Inhibition of Propionibacterium acnes lipase activity by the antifungal agent ketoconazole. Microbiol. Immunol. 2017;61:42–44. doi: 10.1111/1348-0421.12464. [DOI] [PubMed] [Google Scholar]
- 72.Holland C., Mak T.N., Zimny-Arndt U., Schmid M., Meyer T.F., Jungblut P.R., Brüggemann H. Proteomic identification of secreted proteins of Propionibacterium acnes. BMC Microbiol. 2010;10:1–11. doi: 10.1186/1471-2180-10-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Weber N., Schwabe K., Schempp C.M., Wölfle U. Effect of a botanical cleansing lotion on skin sebum and erythema of the face: a randomized controlled blinded half‐side comparison. J. Cosmet. Dermatol. 2019;18:821–826. doi: 10.1111/jocd.12680. [DOI] [PubMed] [Google Scholar]
- 74.Staudinger T., Pipal A., Redl B. Molecular analysis of the prevalent microbiota of human male and female forehead skin compared to forearm skin and the influence of make‐up. J. Appl. Microbiol. 2011;110:1381–1389. doi: 10.1111/j.1365-2672.2011.04991.x. [DOI] [PubMed] [Google Scholar]
- 75.Jin Y., Xia J., Pan Z., Yang J., Wang W., Fu Z. Polystyrene microplastics induce microbiota dysbiosis and inflammation in the gut of adult zebrafish. Environ. Pollut. 2018;235:322–329. doi: 10.1016/j.envpol.2017.12.088. [DOI] [PubMed] [Google Scholar]
- 76.van Raamsdonk L.W.D., van der Zande M., Koelmans A.A., Hoogenboom R.L., Peters R.J.B., Groot M.J., Peijnenburg A.A., Weesepoel Y.J.A. Current insights into monitoring, bioaccumulation, and potential health effects of microplastics present in the food chain. Foods. 2020;9:72. doi: 10.3390/foods9010072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Guo J.-J., Huang X.-P., Xiang L., Wang Y.-Z., Li Y.-W., Li H., Cai Q.-Y., Mo C.-H., Wong M.-H. Source, migration and toxicology of microplastics in soil. Environ. Int. 2020;137 doi: 10.1016/j.envint.2019.105263. [DOI] [PubMed] [Google Scholar]
- 78.Wei H., Wu L., Liu Z., Saleem M., Chen X., Xie J., Zhang J. Meta-analysis reveals differential impacts of microplastics on soil biota. Ecotoxicol. Environ. Saf. 2022;230 doi: 10.1016/j.ecoenv.2021.113150. [DOI] [PubMed] [Google Scholar]
- 79.Dissanayake P.D., Kim S., Sarkar B., Oleszczuk P., Sang M.K., Haque M.N., Ahn J.H., Bank M.S., Ok Y.S. Effects of microplastics on the terrestrial environment: a critical review. Environ. Res. 2022;209 doi: 10.1016/j.envres.2022.112734. [DOI] [PubMed] [Google Scholar]
- 80.Kaur K., Reddy S., Barathe P., Oak U., Shriram V., Kharat S.S., Govarthanan M., Kumar V. Microplastic-associated pathogens and antimicrobial resistance in environment. Chemosphere. 2022;291 doi: 10.1016/j.chemosphere.2021.133005. [DOI] [PubMed] [Google Scholar]
- 81.Amelia T.S.M., Khalik W.M.A.W.M., Ong M.C., Shao Y.T., Pan H.-J., Bhubalan K. Marine microplastics as vectors of major ocean pollutants and its hazards to the marine ecosystem and humans. Prog. Earth Planet. Sci. 2021;8:1–26. [Google Scholar]
- 82.Stenger K.S., Wikmark O.G., Bezuidenhout C.C., Molale-Tom L.G. Microplastics pollution in the ocean: potential carrier of resistant bacteria and resistance genes. Environ. Pollut. 2021;291 doi: 10.1016/j.envpol.2021.118130. [DOI] [PubMed] [Google Scholar]
- 83.Lu L., Wan Z., Luo T., Fu Z., Jin Y. Polystyrene microplastics induce gut microbiota dysbiosis and hepatic lipid metabolism disorder in mice. Sci. Total Environ. 2018;631:449–458. doi: 10.1016/j.scitotenv.2018.03.051. [DOI] [PubMed] [Google Scholar]
- 84.Qiao R., Deng Y., Zhang S., Wolosker M.B., Zhu Q., Ren H., Zhang Y. Accumulation of different shapes of microplastics initiates intestinal injury and gut microbiota dysbiosis in the gut of zebrafish. Chemosphere. 2019;236 doi: 10.1016/j.chemosphere.2019.07.065. [DOI] [PubMed] [Google Scholar]
- 85.Gu W., Liu S., Chen L., Liu Y., Gu C., Ren H.-Q., Wu B. Single-cell RNA sequencing reveals size-dependent effects of polystyrene microplastics on immune and secretory cell populations from zebrafish intestines. Environ. Sci. Technol. 2020;54:3417–3427. doi: 10.1021/acs.est.9b06386. [DOI] [PubMed] [Google Scholar]
- 86.Liu Z., Yu P., Cai M., Wu D., Zhang M., Chen M., Zhao Y. Effects of microplastics on the innate immunity and intestinal microflora of juvenile Eriocheir sinensis. Sci. Total Environ. 2019;685:836–846. doi: 10.1016/j.scitotenv.2019.06.265. [DOI] [PubMed] [Google Scholar]
- 87.Fournier E., Ratel J., Denis S., Leveque M., Ruiz P., Mazal C., Amiard F., Edely M., Bezirard V., Gaultier E. Exposure to polyethylene microplastics alters immature gut microbiome in an infant in vitro gut model. J. Hazard Mater. 2023;443 doi: 10.1016/j.jhazmat.2022.130383. [DOI] [PubMed] [Google Scholar]
- 88.Zhao Y., Qin Z., Huang Z., Bao Z., Luo T., Jin Y. Effects of polyethylene microplastics on the microbiome and metabolism in larval zebrafish. Environ. Pollut. 2021;282 doi: 10.1016/j.envpol.2021.117039. [DOI] [PubMed] [Google Scholar]
- 89.Tamargo A., Molinero N., Reinosa J.J., Alcolea-Rodriguez V., Portela R., Bañares M.A., Fernández J.F., Moreno-Arribas M.V. PET microplastics affect human gut microbiota communities during simulated gastrointestinal digestion, first evidence of plausible polymer biodegradation during human digestion. Sci. Rep. 2022;12:528. doi: 10.1038/s41598-021-04489-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Montero D., Rimoldi S., Torrecillas S., Rapp J., Moroni F., Herrera A., Gómez M., Fernández-Montero Á., Terova G. Impact of polypropylene microplastics and chemical pollutants on European sea bass (Dicentrarchus labrax) gut microbiota and health. Sci. Total Environ. 2022;805 doi: 10.1016/j.scitotenv.2021.150402. [DOI] [PubMed] [Google Scholar]
- 91.Bashir S.M., Kimiko S., Mak C.-W., Fang J.K.-H., Gonçalves D. Personal care and cosmetic products as a potential source of environmental contamination by microplastics in a densely populated Asian city. Front. Mar. Sci. 2021;8 [Google Scholar]
- 92.Flores G.E., Caporaso J.G., Henley J.B., Rideout J.R., Domogala D., Chase J., Leff J.W., Vázquez-Baeza Y., Gonzalez A., Knight R. Temporal variability is a personalized feature of the human microbiome. Genome Biol. 2014;15:1–13. doi: 10.1186/s13059-014-0531-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Oh J., Byrd A.L., Park M., Kong H.H., Segre J.A. Temporal stability of the human skin microbiome. Cell. 2016;165:854–866. doi: 10.1016/j.cell.2016.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dessinioti C., Katsambas A.D. The role of Propionibacterium acnes in acne pathogenesis: facts and controversies. Clin. Dermatol. 2010;28:2–7. doi: 10.1016/j.clindermatol.2009.03.012. [DOI] [PubMed] [Google Scholar]
- 95.Brown M.M., Horswill A.R. Staphylococcus epidermidis—skin friend or foe? PLoS Pathog. 2020;16 doi: 10.1371/journal.ppat.1009026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Claudel J.-P., Auffret N., Leccia M.-T., Poli F., Corvec S., Dréno B. Staphylococcus epidermidis: a potential new player in the physiopathology of acne? Dermatology. 2019;235:287–294. doi: 10.1159/000499858. [DOI] [PubMed] [Google Scholar]
- 97.McDonald D., Ackermann G., Khailova L., Baird C., Heyland D., Kozar R., Lemieux M., Derenski K., King J., Vis-Kampen C. Extreme dysbiosis of the microbiome in critical illness. mSphere. 2016;1 doi: 10.1128/msphere.00199–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Dreno B., Martin R., Moyal D., Henley J.B., Khammari A., Seité S. Skin microbiome and acne vulgaris: Staphylococcus, a new actor in acne. Exp. Dermatol. 2017;26:798–803. doi: 10.1111/exd.13296. [DOI] [PubMed] [Google Scholar]
- 99.Balato A., Cacciapuoti S., Di Caprio R., Marasca C., Masarà A., Raimondo A., Fabbrocini G. Human microbiome: composition and role in inflammatory skin diseases. Arch. Immunol. Ther. Exp. 2019;67:1–18. doi: 10.1007/s00005-018-0528-4. [DOI] [PubMed] [Google Scholar]
- 100.Coughlin C.C., Swink S.M., Horwinski J., Sfyroera G., Bugayev J., Grice E.A., Yan A.C. The preadolescent acne microbiome: a prospective, randomized, pilot study investigating characterization and effects of acne therapy. Pediatr. Dermatol. 2017;34:661–664. doi: 10.1111/pde.13261. [DOI] [PubMed] [Google Scholar]
- 101.Wang Y., Kuo S., Shu M., Yu J., Huang S., Dai A., Two A., Gallo R.L., Huang C.-M. Staphylococcus epidermidis in the human skin microbiome mediates fermentation to inhibit the growth of Propionibacterium acnes: implications of probiotics in acne vulgaris. Appl. Microbiol. Biotechnol. 2014;98:411–424. doi: 10.1007/s00253-013-5394-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Findley K., Oh J., Yang J., Conlan S., Deming C., Meyer J.A., Schoenfeld D., Nomicos E., Park M. Topographic diversity of fungal and bacterial communities in human skin. Nature. 2013;498:367–370. doi: 10.1038/nature12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Grice E.A. 2014. The Skin Microbiome: Potential for Novel Diagnostic and Therapeutic Approaches to Cutaneous Disease, NIH Public Access. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kim K.H. Overview of atopic dermatitis, asia pac. Allergy. 2013;3:79–87. doi: 10.5415/apallergy.2013.3.2.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kong H.H., Oh J., Deming C., Conlan S., Grice E.A., Beatson M.A., Nomicos E., Polley E.C., Komarow H.D., Murray P.R. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res. 2012;22:850–859. doi: 10.1101/gr.131029.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Henley J.B., Ing R.M.M., Zelenkova H. Microbiome of affected and unaffected skin of patients with atopic dermatitis before and after emollient treatment. J. Drugs Dermatol. 2014;13:1365–1372. [PubMed] [Google Scholar]
- 107.Gonzalez M.E., Schaffer J.V., Orlow S.J., Gao Z., Li H., Alekseyenko A.V., Blaser M.J. Cutaneous microbiome effects of fluticasone propionate cream and adjunctive bleach baths in childhood atopic dermatitis. J. Am. Acad. Dermatol. 2016;75:481–493. e8. doi: 10.1016/j.jaad.2016.04.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yerushalmi M., Elalouf O., Anderson M., Chandran V. The skin microbiome in psoriatic disease: a systematic review and critical appraisal. Journal of Translational Autoimmunity. 2019;2 doi: 10.1016/j.jtauto.2019.100009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Velegraki A., Cafarchia C., Gaitanis G., Iatta R., Boekhout T. Malassezia infections in humans and animals: pathophysiology, detection, and treatment. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1004523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gardiner M., Vicaretti M., Sparks J., Bansal S., Bush S., Liu M., Darling A., Harry E., Burke C.M. A longitudinal study of the diabetic skin and wound microbiome. PeerJ. 2017;5:e3543. doi: 10.7717/peerj.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Tsilochristou O., du Toit G., Sayre P.H., Roberts G., Lawson K., Sever M.L., Bahnson H.T., Radulovic S., Basting M., Plaut M. Association of Staphylococcus aureus colonization with food allergy occurs independently of eczema severity. J. Allergy Clin. Immunol. 2019;144:494–503. doi: 10.1016/j.jaci.2019.04.025. [DOI] [PubMed] [Google Scholar]
- 112.Salgado V.R., de Queiroz A.T.L., Sanabani S.S., de Oliveira C.I., Carvalho E.M., Costa J.M.L., Barral-Netto M., Barral A. The microbiological signature of human cutaneous leishmaniasis lesions exhibits restricted bacterial diversity compared to healthy skin. Mem. Inst. Oswaldo Cruz. 2016;111:241–251. doi: 10.1590/0074-02760150436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Fyhrquist N., Ruokolainen L., Suomalainen A., Lehtimäki S., Veckman V., Vendelin J., Karisola P., Lehto M., Savinko T., Jarva H. Acinetobacter species in the skin microbiota protect against allergic sensitization and inflammation. J. Allergy Clin. Immunol. 2014;134:1301–1309. e11. doi: 10.1016/j.jaci.2014.07.059. [DOI] [PubMed] [Google Scholar]
- 114.Schommer N.N., Gallo R.L. Structure and function of the human skin microbiome. Trends Microbiol. 2013;21:660–668. doi: 10.1016/j.tim.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zaidi A.K., Spaunhurst K., Sprockett D., Thomason Y., Mann M.W., Fu P., Ammons C., Gerstenblith M., Tuttle M.S., Popkin D.L. Characterization of the facial microbiome in twins discordant for rosacea. Exp. Dermatol. 2018;27:295–298. doi: 10.1111/exd.13491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Christensen G.J.M., Brüggemann H. Bacterial skin commensals and their role as host guardians. Benef. Microbes. 2014;5:201–215. doi: 10.3920/BM2012.0062. [DOI] [PubMed] [Google Scholar]
- 117.Gardner S.E., Hillis S.L., Heilmann K., Segre J.A., Grice E.A. The neuropathic diabetic foot ulcer microbiome is associated with clinical factors. Diabetes. 2013;62:923–930. doi: 10.2337/db12-0771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Woo Y.R., Cho S.H., Lee J.D., Kim H.S. The human microbiota and skin cancer. Int. J. Mol. Sci. 2022;23:1813. doi: 10.3390/ijms23031813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kullander J., Forslund O., Dillner J. Staphylococcus aureus and squamous cell carcinoma of the skin. Cancer Epidemiol. Biomarkers Prev. 2009;18:472–478. doi: 10.1158/1055-9965.EPI-08-0905. [DOI] [PubMed] [Google Scholar]
- 120.Wood D.L.A., Lachner N., Tan J.-M., Tang S., Angel N., Laino A., Linedale R., Lê Cao K.-A., Morrison M., Frazer I.H. A natural history of actinic keratosis and cutaneous squamous cell carcinoma microbiomes. mBio. 2018;9 doi: 10.1128/mbio.01432–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Madhusudhan N., Pausan M.R., Halwachs B., Durdević M., Windisch M., Kehrmann J., Patra V., Wolf P., Boukamp P., Moissl-Eichinger C. Molecular profiling of keratinocyte skin tumors links Staphylococcus aureus overabundance and increased human β-defensin-2 expression to growth promotion of squamous cell carcinoma. Cancers. 2020;12:541. doi: 10.3390/cancers12030541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Jackow J.C.M., Cather J.C., Hearne V., Asano A.T., Musser J.M., Duvic M. Association of erythrodermic cutaneous T-cell lymphoma, superantigen-positive Staphylococcus aureus, and oligoclonal T-cell receptor Vβ gene expansion, Blood. Am. J. Hematol. 1997;89:32–40. [PubMed] [Google Scholar]
- 123.Glud M., Gniadecki R. MicroRNAs in the pathogenesis of malignant melanoma. J. Eur. Acad. Dermatol. Venereol. 2013;27:142–150. doi: 10.1111/j.1468-3083.2012.04579.x. [DOI] [PubMed] [Google Scholar]
- 124.Mizuhashi S., Kajihara I., Sawamura S., Kanemaru H., Makino K., Aoi J., Makino T., Masuguchi S., Fukushima S., Ihn H. Skin microbiome in acral melanoma: Corynebacterium is associated with advanced melanoma. J. Dermatol. 2021;48:e15–e16. doi: 10.1111/1346-8138.15633. [DOI] [PubMed] [Google Scholar]
- 125.Zoschke C., Ulrich M., Sochorová M., Wolff C., Vávrová K., Ma N., Ulrich C., Brandner J.M., Schäfer-Korting M. The barrier function of organotypic non-melanoma skin cancer models. J. Contr. Release. 2016;233:10–18. doi: 10.1016/j.jconrel.2016.04.037. [DOI] [PubMed] [Google Scholar]
- 126.Williams M.R., Cau L., Wang Y., Kaul D., Sanford J.A., Zaramela L.S., Khalil S., Butcher A.M., Zengler K., Horswill A.R. Interplay of staphylococcal and host proteases promotes skin barrier disruption in Netherton syndrome. Cell Rep. 2020;30:2923–2933. e7. doi: 10.1016/j.celrep.2020.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Chapkin R.S., Davidson L.A., Park H., Jin U., Fan Y., Cheng Y., Hensel M.E., Landrock K.K., Allred C., Menon R. Role of the aryl hydrocarbon receptor (AhR) in mediating the effects of coffee in the colon. Mol. Nutr. Food Res. 2021;65 doi: 10.1002/mnfr.202100539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Qiao P., Zhang C., Yu J., Shao S., Zhang J., Fang H., Chen J., Luo Y., Zhi D., Li Q. Quinolinic acid, a tryptophan metabolite of the skin microbiota, negatively regulates NLRP3 inflammasome through AhR in psoriasis. J. Invest. Dermatol. 2022;142:2184–2193. e6. doi: 10.1016/j.jid.2022.01.010. [DOI] [PubMed] [Google Scholar]
- 129.Gearhart-Serna L.M., Davis J.B., Jolly M.K., Jayasundara N., Sauer S.J., Di Giulio R.T., Devi G.R. A polycyclic aromatic hydrocarbon-enriched environmental chemical mixture enhances AhR, antiapoptotic signaling and a proliferative phenotype in breast cancer cells. Carcinogenesis. 2020;41:1648–1659. doi: 10.1093/carcin/bgaa047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Portal-Nuñez S., Shankavaram U.T., Rao M., Datrice N., Atay S., Aparicio M., Camphausen K.A., Fernández-Salguero P.M., Chang H., Lin P. Aryl hydrocarbon receptor-induced adrenomedullin mediates cigarette smoke carcinogenicity in humans and mice. Cancer Res. 2012;72:5790–5800. doi: 10.1158/0008-5472.CAN-12-0818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Roth M.J., Wei W.-Q., Baer J., Abnet C.C., Wang G.-Q., Sternberg L.R., Warner A.C., Johnson L.L., Lu N., Giffen C.A. Aryl hydrocarbon receptor expression is associated with a family history of upper gastrointestinal tract cancer in a high-risk population exposed to aromatic hydrocarbons. Cancer Epidemiol. Biomarkers Prev. 2009;18:2391–2396. doi: 10.1158/1055-9965.EPI-08-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Gaitanis G., Magiatis P., Hantschke M., Bassukas I.D., Velegraki A. The Malassezia genus in skin and systemic diseases. Clin. Microbiol. Rev. 2012;25:106–141. doi: 10.1128/CMR.00021-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sato Y., Fujimura T., Tanita K., Chunbing L., Matsushita S., Fujisawa Y., Otsuka A., Yamamoto Y., Hidaka T., Aiba S. Malassezia‐derived aryl hydrocarbon receptor ligands enhance the CCL20/Th17/soluble CD163 pathogenic axis in extra‐mammary Paget's disease. Exp. Dermatol. 2019;28:933–939. doi: 10.1111/exd.13944. [DOI] [PubMed] [Google Scholar]
- 134.Chung L.-M., Liang J.-A., Lin C.-L., Sun L.-M., Kao C.-H. Cancer risk in patients with candidiasis: a nationwide population-based cohort study. Oncotarget. 2017;8 doi: 10.18632/oncotarget.18855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Spurgeon M.E., Lambert P.F. Merkel cell polyomavirus: a newly discovered human virus with oncogenic potential. Virology. 2013;435:118–130. doi: 10.1016/j.virol.2012.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.