SUMMARY
Pancreatic ductal adenocarcinoma (PDAC) is an aggressive disease with a low 5-year survival rate and is associated with poor response to therapy. Elevated expression of the myeloid-specific hematopoietic cell kinase (HCK) is observed in PDAC and correlates with reduced patient survival. To determine whether aberrant HCK signaling in myeloid cells is involved in PDAC growth and metastasis, we established orthotopic and intrasplenic PDAC tumors in wild-type and HCK knockout mice. Genetic ablation of HCK impaired PDAC growth and metastasis by inducing an immune-stimulatory endotype in myeloid cells, which in turn reduced the desmoplastic microenvironment and enhanced cytotoxic effector cell infiltration. Consequently, genetic ablation or therapeutic inhibition of HCK minimized metastatic spread, enhanced the efficacy of chemotherapy, and overcame resistance to anti-PD1, anti-CTLA4, or stimulatory anti-CD40 immunotherapy. Our results provide strong rationale for HCK to be developed as a therapeutic target to improve the response of PDAC to chemo- and immunotherapy.
In brief
Poh et al. show that ablation of host HCK, or inhibition of its kinase, reduces pancreatic adenocarcinoma by stimulating myeloid cells, which limits desmoplasia and promotes effector cell recruitment/activation. Accordingly, HCK inhibition overcomes resistance to immunotherapy and improves response to chemotherapy, thereby establishing HCK as a drug target for PDAC.
Graphical abstract
INTRODUCTION
Pancreatic ductal adenocarcinoma (PDAC) is an aggressive disease with a 5-year survival rate of less than 10% (Raimondi et al., 2009). While chemotherapy confers transient tumor regression in 30% of patients, 9 out of 10 patients that undergo surgery still die of the disease due to local recurrence and/or metastasis (Conroy et al., 2011). Likewise, immune checkpoint therapies including anti-(α)PD1 or αCTLA4 have failed to translate into meaningful improvements in a majority of PDAC patients (Diamond et al., 2021; Galon and Bruni, 2019).
The poor response of PDAC to immune cell-related therapies can be accounted for by two major obstacles. The first tumor-intrinsic barrier relates to insufficient immune activation due to limited immunogenic mutations and presentation of cancer neo-epitopes, resulting in the current clinical recommendation for αPD1 to be limited as the second-line therapy for <1% of PDAC patients with DNA-mismatch repair deficient disease (Bailey et al., 2016; Rojas and Balachandran, 2021; Tempero et al., 2019). The second tumor-extrinsic barrier arises from an immunosuppressive and desmoplastic microenvironment characterized by an influx of cancer-associated fibroblasts (CAFs), myeloid-derived suppressor cells (MDSCs), and tumor-associated macrophages (TAMs), which collectively promote the exclusion of cytotoxic T cells and natural killer (NK) effector cells from tumors (Ho et al., 2020). Thus, therapies that can simultaneously activate tumor immunity and relieve immune suppression represent promising adjuvant strategies to better control PDAC progression and metastasis.
Elevated expression of the myeloid SRC family kinase hematopoietic cell kinase (HCK) is observed in most human solid malignancies including PDAC, where more than 95% of all HCK expression occurs in immune cells and correlates with poor patient survival (Bailey et al., 2016; Crnogorac-Jurcevic et al., 2005; Heidenblad et al., 2004; Isella et al., 2015; Poh et al., 2015; Zhu et al., 2021). We have previously demonstrated a tumor-extrinsic role for myeloid HCK signaling in gastric and colon cancer by promoting an immunosuppressive tumor microenvironment (Poh et al., 2017, 2020). Conversely, genetic ablation or pharmacologic inhibition of HCK reduced tumor growth (Poh et al., 2017, 2020). Here, we establish the therapeutic benefit of targeting HCK in PDAC to reduce immune suppression, attenuate the desmoplastic response, and re-invigorate adaptive anti-tumor immunity.
RESULTS
Genetic ablation of HCK in hosts reduces PDAC growth and metastasis
To explore a potential tumor-promoting role for HCK in pancreatic cancer, we interrogated the expression level of HCK in PDAC patients (Tang et al., 2017) and observed elevated expression in tumor samples compared with matched normal tissues (Figure 1A). We also analyzed single-cell RNA sequencing (scRNA-seq) datasets of human and mouse PDAC tumors (Elyada et al., 2019), and we confirmed that HCK was most prominently expressed in tumor-associated myeloid cells (Figures 1B and 1C).
Figure 1. Genetic ablation of HCK in myeloid cells impairs PDAC tumor growth and metastasis.
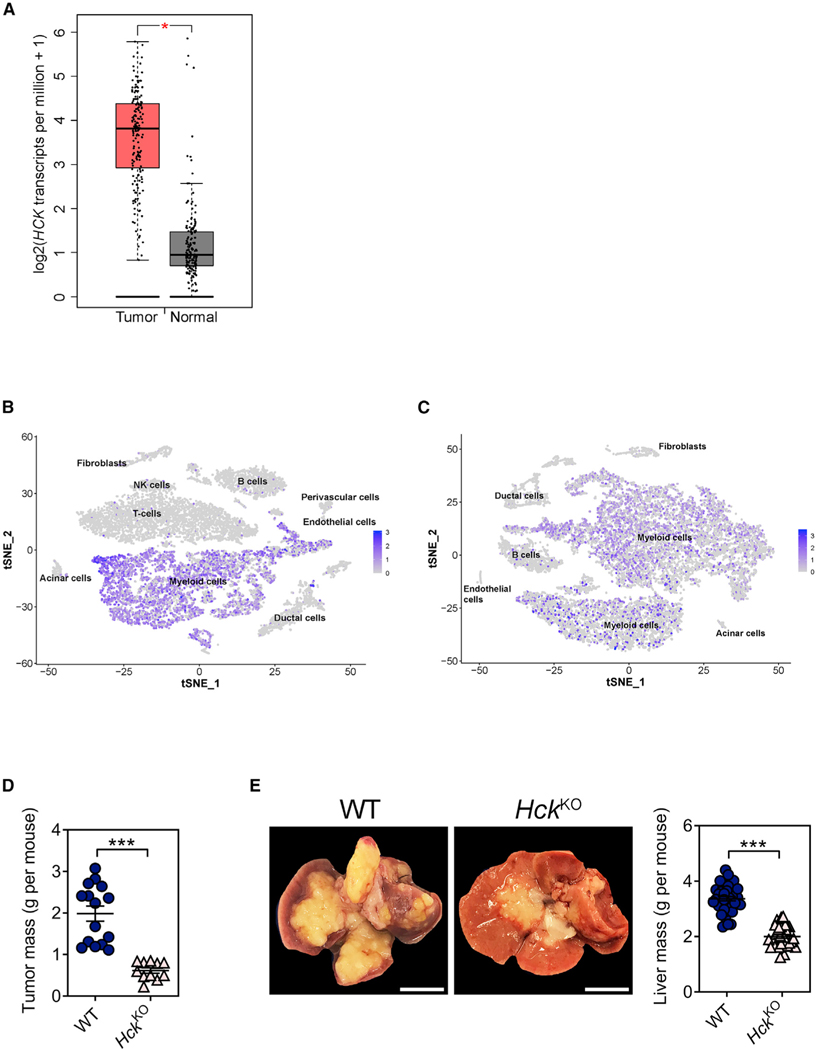
(A) HCK gene expression in tumors and matched normal tissue samples of human pancreatic adenocarcinoma patients (n = 179) using the GEPIA online tool (Tang et al., 2017).
(B) tSNE plot depicting HCK gene expression in human PDAC tumors using primary data from Elyada et al. (2019).
(C) tSNE plot depicting Hck gene expression in mouse KPC PDAC tumors using primary data from Elyada et al. (2019).
(D) Mass of primary PDAC tumors from WT and HckKO hosts 5 weeks following orthotopic injection of KPC tumor cells. Each symbol represents an individual mouse. n ≥ 11 mice per group.
(E) Representative whole mounts and corresponding liver weights of WT and HckKO hosts 3 weeks after intrasplenic injection of KPC tumor cells. Scale bar: 1 cm. Each symbol represents an individual mouse. n ≥ 34 mice per group. Data represent mean ± SEM; ***p < 0.001, with statistical significance determined by an unpaired Student’s t test for comparison between two means. See also Figure S1.
To determine whether aberrant myeloid HCK signaling in the host is involved in PDAC growth and metastasis, we orthotopically engrafted syngeneic KPC pancreatic tumor cells into the distal pancreas of wild-type (WT) and HCK knockout (HckKO) hosts, and we observed significantly smaller pancreatic tumors in HckKO hosts compared with their WT counterparts (Figures 1D and S1A). Moreover, HckKO hosts did not develop metastatic lesions, which we consistently observed in WT mice, including in the liver (n = 15/15), spleen (15/15), intestine (7/15), peritoneum (4/15), and kidneys (4/15) (Figure S1B). To obtain better insights into the contribution of HCK during PDAC metastasis, we injected KPC tumor cells into the spleen followed by splenectomy to protect hosts against premature death arising from overgrowth of the primary tumor. Again, we observed reduced incidence of liver metastasis in HckKO hosts (Figures 1E and S1A), and we confirmed in both orthotopic and intrasplenic PDAC models that Hck expression was restricted to the myeloid cell compartment of KPC tumors (Figure S1C). To formally prove that the enhanced anti-tumor response in HckKO hosts was an intrinsic consequence of hematopoietic cells lacking Hck expression, we generated reciprocal bone marrow chimeras and subjected these mice to the intrasplenic tumor model. We observed improved survival of WT←KO (Recipient←Donor) bone marrow chimeras compared with WT←WT hosts, and reduced survival of KO←WT hosts compared with KO←KO hosts (Figure S1D and Table S1). We also extended these observations to a therapeutic setting by treating WT mice with the HCK-specific small molecule inhibitor RK20449 (Saito et al., 2013) after orthotopic or intrasplenic injection of KPC tumor cells. Compared with vehicle-treated mice, RK20449 treatment impaired the growth of primary tumors and liver metastases (Figures S1E and S1F).
Genetic ablation of HCK enhances the immune-stimulatory endotype of myeloid cells and promotes an influx of activated cytotoxic effector cells into tumors
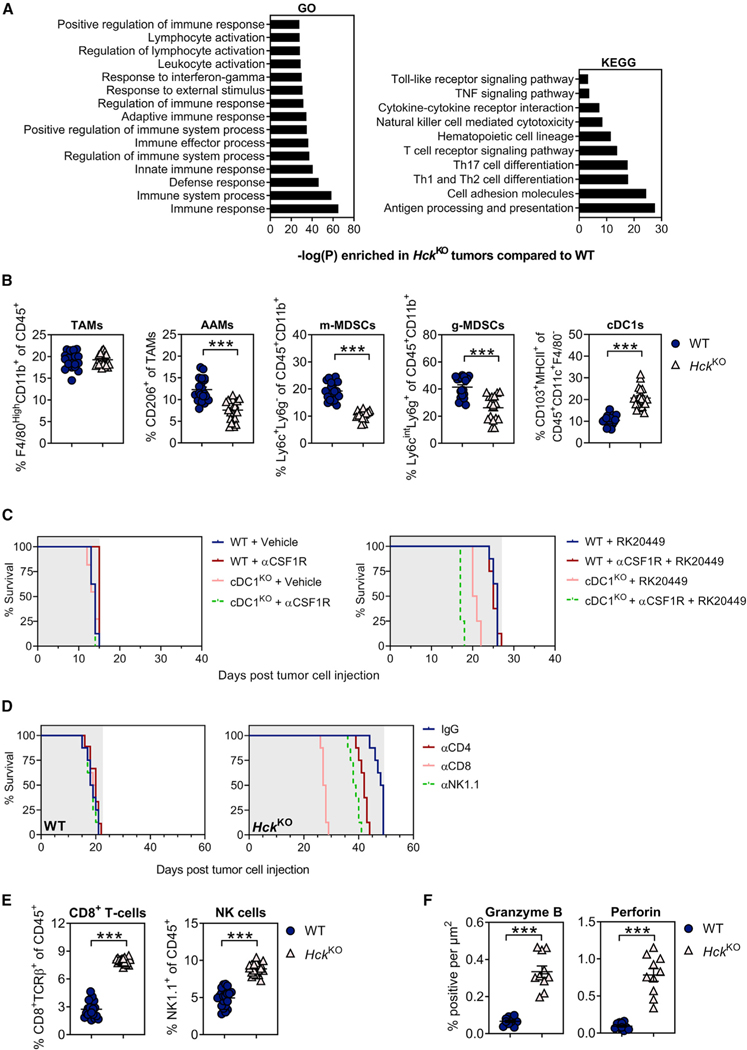
To functionally link reduced Hck expression in myeloid cells to an improved anti-tumor immune response, we performed Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) pathway analysis on bulk RNA sequenced KPC liver metastases recovered from WT and HckKO hosts. In tumors of HckKO hosts, we observed a significant enrichment of pathways associated with innate and adaptive immune responses, including myeloid cell activation and effector cell-mediated cytotoxicity (Figure 2A).
Figure 2. Genetic ablation of HCK in myeloid cells enhances cytotoxic effector cell recruitment and activation.
(A) Enriched KEGG and GO signaling pathways in KPC liver metastases of HckKO hosts compared with WT mice. n = 5 mice per group.
(B) Flow cytometry quantification of myeloid cells in KPC liver metastases of WT and HckKO hosts. Each symbol represents an individual mouse. n = 20 mice per group.
(C) Kaplan-Meier survival analysis of WT bone marrow chimeras reconstituted with cDC1-deficient (cDC1KO) or cDC1-proficient (WT) bone marrow. To deplete TAMs, half of each cohort were treated with αCSF1R prior to intrasplenic KPC tumor cell injection and continued until clinical endpoint. Following establishment of intrasplenic KPC tumors, mice were treated with the small molecule HCK inhibitor RK20449 or Captisol vehicle control. Shaded area indicates treatment period. n = 8 mice per group. A Mantel-Cox log rank test was used to evaluate statistical significance (see Table S3).
(D) Kaplan-Meier survival analysis of tumor-bearing WT and HckKO hosts following NK cell, CD4+ T cell, or CD8+ T cell depletion. Shaded area indicates treatment period. n = 8 mice per group. A Mantel-Cox log rank test was used to evaluate statistical significance (see Table S5).
(E) Flow cytometry quantification of CD8+ T cells and NK cells in KPC liver metastases of WT and HckKO hosts. Each symbol represents an individual mouse. n = 20 mice per group.
(F) Quantification of granzyme B and perforin immunohistochemical staining in KPC liver metastases of WT and HckKO hosts. Each symbol represents an individual mouse. n = 10 mice per group. Data represent mean ± SEM; ***p < 0.001, with statistical significance determined by an unpaired Student’s t test for comparison between two means. See also Figure S2 and Tables S3 and S5.
Given the contribution of immunosuppressive myeloid cells to PDAC growth and metastasis (Poh and Ernst, 2021), we next profiled myeloid cells in KPC liver metastases of WT and HckKO hosts and found that HCK deficiency did not affect the overall abundance of TAMs. Instead, we observed a reduction of both CD206+ alternatively activated macrophages and MDSCs, and an increase in CD103+ conventional type 1 dendritic cells (cDC1s) in tumors of HckKO hosts (Figure 2B). Importantly, DCs and TAMs from tumors of HckKO hosts displayed increased expression of immune-stimulatory factors (i.e., Il12α, Ifnγ, Tnf, Cxcl9, and Cxcl10) and a concomitant downregulation of genes associated with immune suppression (i.e., Il4, Il10, Il13, Tgfβ, Arg1) and matrix remodeling (i.e, Mmp7, Mmp9) (Figure S2A). Given that myeloid cells are the primary source of IL12 and CXCL9/CXCL10 (Arnold et al., 2019; Chow et al., 2019; Garris et al., 2018; House et al., 2020; Reschke and Gajewski, 2022), we next determined the contribution of these molecules to HCK-dependent suppression of anti-tumor immunity using neutralizing antibodies against either IL12 or CXCR3 (cognate receptor for CXCL9/CXCL10). Blockade of IL12 or CXCR3 abrogated the survival benefit of HckKO hosts, while a difference in survival was not observed in WT hosts treated with these antibodies (Figure S2B and Table S2). Together, our findings suggest that genetic ablation of HCK promotes a shift of DCs and TAMs toward an activated endotype.
We next assessed the contribution of cDC1s and TAMs to the enhanced anti-tumor response in HckKO hosts. Following reconstitution of lethally irradiated WT hosts with bone marrow from either cDC1-deficient (ItgaxCreIrf8fl/fl; Caton et al., 2007; Chopin et al., 2013; Feng et al., 2011; referred to as cDC1KO) or cDC1-proficient (WT) mice, we treated half of each cohort with a neutralizing antibody against CSF1R to also deplete TAMs prior to intrasplenic injection of KPC tumor cells. Following establishment of intrasplenic KPC tumors, mice were treated with the small molecule HCK inhibitor RK20449 or Captisol vehicle until clinical endpoint. While neither cDC1 nor TAM depletion affected the overall survival of vehicle-treated hosts compared with their immune cell-proficient controls, cDC1 depletion reduced the overall survival of RK20449-treated hosts, which was further reduced when TAMs were also simultaneously depleted (Figure 2C and Table S3).
We then clarified the contribution of adaptive immunity to the enhanced anti-tumor response observed in HckKO hosts by exploiting the intrasplenic KPC model to establish liver metastasis in WT, HckKO, and lymphocyte-deficient Rag1KO and HckKO; Rag1KO compound mutant hosts. We observed enhanced liver tumor burden and reduced overall survival in HckKO;Rag1KO hosts compared with HckKO mice, as well as extended survival in HckKO hosts compared with all other cohorts (Figure S2C and Table S4). To delineate the role of NK and T cells in HckKO hosts, we individually depleted NK cells, CD4+ T cells, or CD8+ T cells, and we observed that CD8+ T cell depletion abrogated the tumor-suppressive effects conferred in the absence of HCK expression more effectively than either NK or CD4+ T cell depletion (Figure 2D and Table S5). These observations were consistent with an increased proportion of CD8+ T cells and NK cells in tumors of HckKO hosts compared with WT mice (Figures 2E and S2D). Furthermore, CD8+ T cells and NK cells isolated from tumors of HckKO hosts also showed elevated expression of genes encoding cytotoxic activities (i.e., Ifnγ, Tnf, GzmB, Prf1) (Figure S2E), and this in turn correlated with more abundant staining for granzyme B and perforin in tumors of HckKO hosts (Figure 2F). We surmised that these effects are primarily mediated by IL12 and CXCR3 signaling by myeloid cells, since administration of neutralizing antibodies against IL12 or CXCR3 significantly reduced CD8+ T cell recruitment and cytotoxicity in tumors of HckKO hosts (Figures S2F and S2G). Collectively, our findings suggest that CD8+ T cells are a major effector cell population that mediates the enhanced anti-tumor response in HckKO hosts.
Genetic ablation of HCK in myeloid cells reduces the desmoplastic response of PDAC
CAFs produce and remodel most of the extracellular matrix (ECM) in PDAC (Geng et al., 2021) and interact with myeloid cells to collectively amplify and sustain the immune-suppressive and fibrotic tumor microenvironment (Poh and Ernst, 2021). Consistent with these observations, KPC liver metastases of HckKO hosts displayed significantly less ECM including collagen and fibronectin compared with their WT counterparts (Figure 3A). We therefore examined whether CAFs associated with tumors of HckKO hosts differed quantitatively and qualitatively from those of tumors from WT hosts. Immunohistochemistry and flow cytometry revealed that tumors of HckKO hosts contained fewer inflammatory, myofibroblastic, and antigen-presenting CAFs (Figures 3B and 3C). Likewise, CAFs from HckKO hosts also displayed reduced expression of genes associated with immune suppression (i.e., Tgfβ, Il10), fibrosis (i.e., Il11), and ECM remodeling (i.e., Mmp3, Mmp7, Mmp9, Col1a1) than CAFs from tumors of WT hosts (Figure 3D).
Figure 3. Genetic ablation of HCK in myeloid cells reduces the desmoplastic tumor microenvironment.
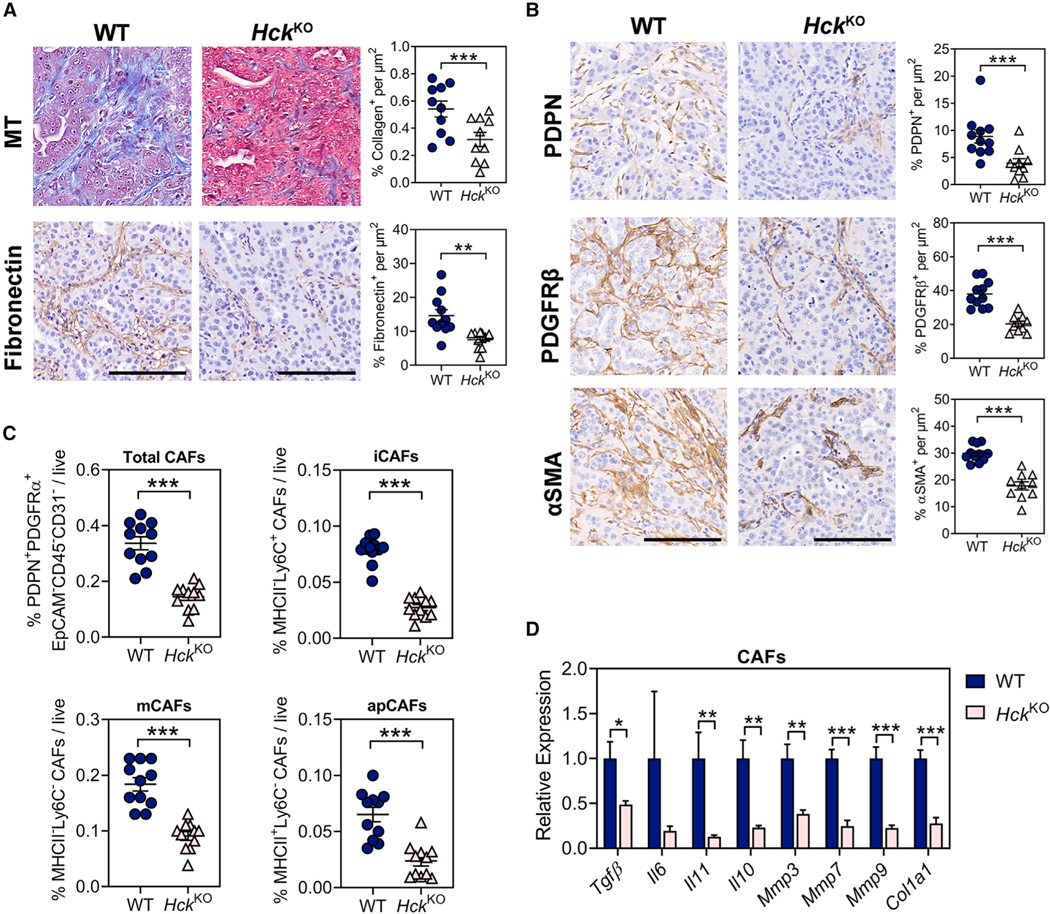
(A) Representative immunohistochemical staining and quantification of ECM proteins in KPC liver metastases of WT and HckKO hosts. Collagen was visualized by Masson’s Trichrome (MT) staining. Scale bar: 100 μm. Each symbol represents an individual mouse. n ≥ 10 mice per group.
(B) Representative immunohistochemical staining and quantification of pan-CAF markers in KPC liver metastases of WT and HckKO hosts. Scale bar: 100 μm. Each symbol represents an individual mouse. n ≥ 10 mice per group.
(C) Flow cytometry quantification of CAFs in KPC liver metastases of WT and HckKO mice. iCAFs: inflammatory CAFs; mCAFs: myofibroblasts; apCAFs: antigen-presenting CAFs. Each symbol represents an individual mouse. n = 11 mice per group.
(D) qPCR analysis on CD45−EpCAM−CD31−PDPN+PDGFRα+ CAFs isolated from KPC liver metastases of WT and HckKO mice. n = 5 mice per group. Data represent mean ± SEM; *p < 0.05, **p < 0.01, ***p < 0.001, with statistical significance determined by an unpaired Student’s t test.
Inhibition of HCK improves therapeutic response to chemotherapy and immunotherapy
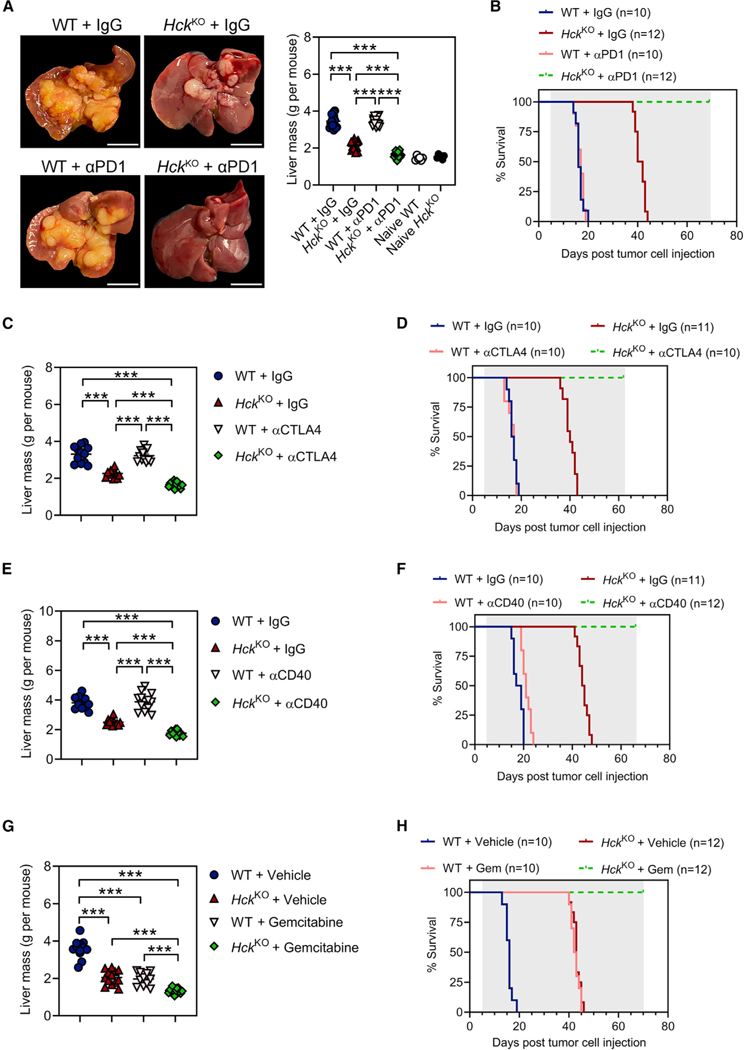
The immune cell-excluded and fibrotic transcriptional profile of human PDAC correlates with poor clinical outcomes and resistance to immunotherapy (Bagaev et al., 2021). Given that durable responses to αPD1 therapy are limited by the exhaustion of cytotoxic effector cells and an immunosuppressive and desmoplastic stroma, we first examined whether HCK deficiency sensitizes treatment-refractory PDAC tumors to αPD1. We treated tumor-bearing WT or HckKO hosts with αPD1, and we observed that genetic ablation of HCK in hosts enabled response to αPD1 immunotherapy and blocked the outgrowth of liver metastases (Figures 4A and S3A). This correlated with prolonged survival of all corresponding αPD1-treated HckKO mice well beyond that of all other treatment cohorts (Figure 4B and Table S6).
Figure 4. Genetic ablation of HCK in myeloid cells sensitizes PDAC tumors to immunotherapy and augments the efficacy of chemotherapy.
(A) Representative whole mounts and corresponding liver weights of WT and HckKO hosts treated once every 3 days with αPD1 or a matched IgG isotype control. Treatment commenced on the 5th day after intrasplenic KPC tumor cell injection and continued for 2 weeks. Scale bar: 1 cm. Each symbol represents an individual mouse. n ≥ 15 mice per treatment group, n = 6 mice per treatment naive group.
(B) Kaplan-Meier survival analysis of WT and HckKO hosts treated as described in Figure 4A until clinical endpoint. Shaded area indicates treatment period. n ≥ 10 mice per group. A Mantel-Cox log rank test was used to evaluate statistical significance (see Table S6).
(C and E) Liver weights of WT and HckKO hosts treated once every 3 days with (C) αCTLA4, (E) αCD40, or a matched IgG isotype control. Treatment commenced on the 5th day after intrasplenic KPC tumor cell injection and continued for 2 weeks. Each symbol represents an individual mouse. n ≥ 11 mice per group.
(D and F) Kaplan-Meier survival analysis of WT and HckKO hosts treated as described above with (D) αCTLA4, (F) αCD40, or a matched IgG isotype control until clinical endpoint. Shaded area indicates treatment period. n ≥ 10 mice per group. A Mantel-Cox log rank test was used to evaluate statistical significance (see Tables S7 and S8).
(G) Liver weights of WT and HckKO hosts treated weekly with gemcitabine or PBS vehicle. Treatment commenced on the 5th day after intrasplenic KPC tumor cell injection and continued for 2 weeks. Each symbol represents an individual mouse. n ≥ 11 mice group.
(H) Kaplan-Meier survival analysis of WT and HckKO mice treated as described in Figure 4G until clinical endpoint. Shaded area indicates treatment period. n ≥ 10 mice per group. A Mantel-Cox log rank test was used to evaluate statistical significance (see Table S9). Data represent mean ± SEM; ***p < 0.001, with statistical significance determined by one-way ANOVA followed by Tukey’s multiple comparison test or Mantel-Cox log rank test for Kaplan-Meier analysis. See also Figures S3 and S4 and Tables S6–S9.
To determine whether HCK inhibition not only alleviated αPD1-mediated local immunosuppression but also stimulated antigenic priming, we next assessed the contribution of HCK ablation in the host to therapeutic CTLA4 inhibition or CD40 stimulation, respectively. Unlike the PD1 checkpoint, which suppresses T cell activation in the tumor microenvironment, CTLA4 signaling controls T cell priming in lymph nodes (Mellman et al., 2011). Meanwhile, stimulation of CD40 enhances cross-presentation by cDC1s and broadens T cell response through epitope spreading (Diamond et al., 2021). We observed that treatment with either antagonistic αCTLA4 or agonistic αCD40 antibodies protected HckKO hosts from liver metastases and extended their survival over that of WT hosts treated with these immunotherapies (Figures 4C–4F and Tables S7–S8). Moreover, we observed that the standard-of-care chemotherapy gemcitabine extended the survival of HckKO hosts with no histological evidence of liver metastases in tissue sections (Figures 4G, 4H, and S3B and Table S9).
Finally, we expanded our findings to a therapeutic setting by treating tumor-bearing WT hosts with RK20449 in combination with immunotherapy or chemotherapy. RK20449 treatment of WT hosts reduced tumor growth by enabling response of PDAC tumors to αPD1 therapy (Figure S4A), and it enhanced CD8+ T cell recruitment and activation (Figures S4B and S4C). RK20449 treatment also improved the efficacy of αCTLA4, αCD40, and gemcitabine (Figures S4D–S4F) and significantly extended survival in tumor-bearing WT hosts compared with hosts from monotherapy-treated groups (Figures S4G and S4H and Tables S10 and S11). Collectively, our data suggest that inhibition of HCK activity in hosts with established tumors licenses immune checkpoint-dependent anti-tumor immunity and provides a compelling rationale for targeting HCK as an adjuvant therapy to boost response of PDAC to chemo- and immunotherapy.
DISCUSSION
Myeloid cells are one of the earliest infiltrating cells in PDAC tumors and are associated with disease progression, recurrence, metastasis, and reduced overall survival (Poh and Ernst, 2021). Accordingly, therapeutic strategies aimed at eliminating myeloid cells, inhibiting their infiltration, and/or reprogramming them toward an immune-stimulatory endotype have shown potential in both pre-clinical PDAC models (Kaneda et al., 2016; Stromnes et al., 2014; Zhu et al., 2014) and clinical trials (NCT01413022, NCT02345408, NCT00711191, NCT03214250).
Here, we have identified HCK as a critical promotor of myeloid-mediated immune suppression, metastasis, and desmoplasia in PDAC. Mechanistically, genetic ablation of HCK in myeloid cells suppressed the growth of primary and metastatic tumors by skewing DCs and TAMs toward an immune-stimulatory endotype and by reducing the abundance of MDSCs. These changes were associated with increased NK and CD8+ T cell recruitment into tumors and a dramatic shift toward a tumor microenvironment enriched for non-exhausted effector cells. While our observations demonstrate thatexcessive HCK activity in myeloid cells promotes tumor progression in part through a T cell-dependent mechanism, our findings here also expand on our previously described T cell-independent mechanisms (Poh et al., 2017, 2020). Here, we attribute the latter to increased immune suppression and matrix remodeling associated with alternative macrophage polarization as a consequence of excessive HCK activity. Meanwhile, high HCK expression is associated with increased abundance of immune-suppressive macrophage and exhausted T cell signatures that correlate with reduced overall survival in pancreatic (Bailey et al., 2016), gastric (Poh et al., 2020), and colon cancer patients (Poh et al., 2017). Thus, we surmise that inhibition of HCK signaling offers an opportunity to simultaneously reprogram immune-suppressive myeloid cells and stimulate anti-tumor immunity.
Our findings reinforce a pivotal role for myeloid cells in promoting and sustaining a desmoplastic tumor microenvironment. During PDAC progression, TAMs undergo significant expansion and directly shape fibrotic responses through the production of signaling molecules that stimulate ECM deposition and remodeling (Xue et al., 2015; Zhu et al., 2017). TAMs also indirectly contribute to desmoplasia through the secretion of immune-suppressive growth factors and cytokines that activate CAFs and perpetuate a feed-forward loop to sustain fibrosis (Candido et al., 2018; Kaneda et al., 2016; Nielsen et al., 2016). Our observations imply that the concerted reduction of TGFβ, IL11 (Cook and Schafer, 2020), and other promoters of tissue fibrosis observed in tumors of HckKO hosts may reduce the ECM remodeling and the immunosuppressive endotype of both TAMs and CAFs. Indeed, studies in pancreatic cancer (Creeden et al., 2020), atherosclerosis (Smolinska et al., 2011), and renal and pulmonary fibrosis (Ernst et al., 2002; Smolinska et al., 2011) suggest a link between excessive HCK activity and desmoplasia. However, our observation that tumors of HckKO mice contained fewer antigen-presenting CAFs suggests their limited contribution to the improved anti-tumor immune response elicited by inhibition of HCK signaling.
Our observations suggest that the limited response of PDAC to immunotherapy may arise from an immune-suppressive tumor environment rather than insufficient de novo antigenicity of tumor cells, supporting findings that high tumor antigenicity is insufficient to elicit T cell-mediated tumor control in the pancreas (Diamond et al., 2021; Galon and Bruni, 2019). Indeed, our data complements observations that CD40 activation enables priming of T cells through DC activation independent of pattern recognition receptors (Morrison et al., 2020) and that escape of antigenic tumors can be antagonized by enhancing DC function through administration of FLT3 ligands (Hegde et al., 2020; Vonderheide, 2018). Thus, targeting the catalytic activity of HCK not only overcomes major immunological barriers that limit therapeutic response of fibrotic tumors with minimal infiltration of effector cells, but it also enables durable anti-tumor responses when combined with chemo- or immunotherapy.
Limitations of the study
Limitations of the results presented here are that the KPC model does not evaluate which subtypes of human PDAC likely to benefit most from HCK inhibition, and whether HCK is an effective therapeutic target in PDACs that harbor mutations in genes other than KRAS and TP53. The latter include PDACs with loss of function mutations in SMAD4 that occur in approximately 30% of human pancreatic cancers. Likewise, we are aware that insights from the orthotopic KPC allograft model have provided limited prediction for the outcomes of clinical trials targeting CSF1R, CCR2, IDO, and some other immune modulatory molecules.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Matthias Ernst (matthias.ernst@onjcri.org.au).
Materials availability
This study did not generate new unique reagents.
Data and code availability
Bulk RNA sequencing data generated from KPC liver metastases of WT and HckKO mice are publicly available at the Gene Expression Omnibus (GEO) under accession number GSE185540.
All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
C57BL/6 WT, HckKO (Lowell et al., 1994), Rag1KO, HckKO;Rag1KO and ItgaxCreIrf8fl/fl (Chopin et al., 2013) mice were bred and maintained in specific pathogen-free facilities at La Trobe University, The Austin Hospital, and The Walter and Eliza Hall Institute, Australia. 10-week-old male and female littermates were randomly assigned to experimental groups. No sex differences were observed. All animal studies were approved and conducted in accordance with the Animal Ethics Committee at La Trobe University, the Olivia Newton-John Cancer Research Institute/Austin Hospital and The Walter and Eliza Hall Institute.
Cell line
Luciferase labelled KPC cells derived from pancreatic tumors of KRasG12D;Trp53R172H;Pdx-1Cre (KPC) mice (Vennin et al., 2017) were maintained in DMEM/F-12 media (Gibco Cat#11320033) supplemented with 10% FCS at 37°C with 10% CO2. The cell line was confirmed mycoplasma negative.
METHOD DETAILS
Tumor models
To establish orthotopic PDAC tumors, KPC pancreatic cancer cells were re-suspended as a single cell solution in PBS. A cell viability of >95% was confirmed using trypan blue exclusion. Prior to surgery, Carprofen analgesic (Zoetis, 5 mg/kg) was administered subcutaneously. Mice were anesthetized using Ketamine (Baxter, 100 mg/kg i.p.) and Xylazine (Ilium, 10 mg/kg i.p.). A left subcostal incision was made to access the peritoneal cavity and expose the tail of the pancreas. 5×103 KPC cells were drawn up into a pre-cooled Hamilton Syringe (Sigma Aldrich Cat#20702) and injected directly into the pancreas tail using a 27-gauge needle over a period of 15 seconds (Nikfarjam et al., 2013). A cotton tip was applied to the injection site for an additional 15 seconds to minimize leakage. The abdominal muscle and skin were closed separately using 5–0 coated Vicryl sutures, and 0.12% Bupivacaine analgesic (AstraZeneca) was applied to the incision site. Normal saline was instilled into the peritoneal cavity by i.p. injection. Mice were sacrificed 5 weeks after tumor cell injection. To confirm the absence of micro-metastases in HckKO mice, we obtained at least six sections from each organ at a depth of 200 μM apart. Sections were stained with H&E and analyzed with Aperio ImageScope v11.2.0.780 software for the presence of metastases.
To establish PDAC liver metastasis, KPC pancreatic cancer cells (>95% viability) were re-suspended as a single cell solution in PBS. Prior to surgery, Carprofen analgesic (Zoetis, 5 mg/kg) was administered via subcutaneous injection, and mice were anesthetized via 2% isoflurane inhalation. A left subcostal incision was made to access the peritoneal cavity and expose the spleen. 8–10×105 KPC tumor cells were drawn up into a pre-cooled Hamilton Syringe (Sigma Aldrich Cat#20702) and injected into the spleen using a 27-gauge needle over a period of 40 seconds to allow cells to perfuse into the liver (Nielsen et al., 2016). Cotton gauze was applied to the injection site for an additional minute to minimize leakage before a splenectomy was performed by cautery. The abdominal muscle and skin were closed separately using 5–0 coated Vicryl sutures and wound clips, respectively. Bupivacaine analgesic (AstraZeneca, 0.12%) was applied to the incision site, and normal saline was instilled into the peritoneal cavity by i.p. injection.
Drug treatments
For the orthotopic PDAC model, mice were either treated with RK20449 (synthesized by Reagency, 30 mg/kg, dissolved in 12% Captisol, twice daily i.p.) or Captisol vehicle control (Cat#RC-0C7-K01) commencing one week after KPC tumor cell injection for 4 consecutive weeks. For the intrasplenic PDAC model, mice were treated with αCD40 (Clone FGK45, JPP Biologics, 100 μg once every 3 days i.p.), αPD1 (Clone RMP1–14, JPP Biologics, 200 μg once every 3 days i.p.), αCTLA4 (Clone 4F10, JPP Biologics, 200 μg once every 3 days i.p.), isotype-matched IgG (JPP Biologics, 200 μg once every 3 days i.p.), Gemcitabine (Pfizer, 120 mg/kg, weekly, i.p.) or PBS vehicle (weekly, i.p.) on the 5th day after KPC tumor cell injection for 2 consecutive weeks. In some cases, mice were also treated with RK20449 (30 mg/kg, dissolved in 12% Captisol, twice daily i.p.) or Captisol vehicle control (Cat#RC-0C7-K01).
For Kaplan-Meier survival analysis, treatment commenced on the 5th day following intrasplenic KPC tumor cell injection and continued until mice reached clinical endpoint (i.e., lost ≥15% of their body weight or developed jaundice or other clinical signs of metastasis). To confirm the absence of micro-metastases in chemo- or immunotherapy treated HckKO mice, we obtained at least six sections from the liver at a depth of 200 μM apart. Sections were stained with H&E and analyzed with Aperio ImageScope v11.2.0.780 software for the presence of metastases.
For antibody-mediated depletion/neutralization experiments, mice were pre-treated with αCD4 (Clone GK1.5, JPP Biologics, 200 μg), αCD8 (Clone YTS169, JPP Biologics, 200 μg), αNK1.1 (Clone PK136, JPP Biologics, 200 μg), αCSF1R (Clone AFS98, BioxCell Cat#BP0213, 500 μg), αIL12 (Clone R2–9A5, BioxCell Cat#BE0233, 500 μg), or αCXCR3 (Clone CXCR3–173, BioxCell #BE0249, 500 μg) once every 3 days (total 3 treatments, i.p.) prior to intrasplenic KPC tumor cell injection, and continued until mice reached clinical endpoint.
Generation of bone marrow chimeras
Bone marrow was harvested from the femurs and tibias of donor mice by flushing with sterile PBS (Poh et al., 2017). Cell suspensions were filtered, washed twice in PBS, and kept on ice in PBS until ready for use. Recipient mice were lethally irradiated with 2 doses of 5.5 Gy γ-irradiation 3 hours apart, before receiving 5×106 donor bone marrow cells via tail-vein injection using a 27-gauge needle. Mice were maintained on neomycin-supplemented water (ThermoFisher Cat#21810031, 2 mg/mL) for 3 weeks, and aged for an additional 8 weeks to allow for complete bone marrow reconstitution.
Flow cytometry
Tumors were minced into 1 mm pieces and digested in collagenase/dispase (Roche Cat#11097113001) and DNase I (Roche Cat#10104159001) diluted in Ca2+/Mg2+-free HBSS media (Gibco Cat#14170112) plus 10% FCS (Poh et al., 2017, 2020). Samples were incubated at 37°C for 20 min under continuous rotation, then vortexed for 30 seconds to dissociate immune cells. Cell suspensions were filtered and washed in PBS plus 10% FCS, and incubated with Fc block (eBioscience Cat#14–9161-73) on ice for 10 min. Samples were then stained with fluorophore-conjugated primary antibodies for 20 min on ice in the dark, washed twice, and re-suspended in PBS plus 10% FCS.
Fluorophore-conjugated primary antibodies were directed against mouse CD45 (Clone 30-F11, BioLegend Cat#103116 or Cat#103131), NK1.1 (Clone PK136, eBioscience Cat#17–5941-82), TCRβ (Clone H57–597, BioLegend Cat#109208), CD8a (Clone 53–6.7, BioLegend #100722), F4/80 (Clone BM8, BioLegend Cat#123108 or eBioscience Cat#25–4801-82), CD11b (Clone M1/70, BD Biosciences Cat#553311), Ly6G (Clone 1A8, BD Biosciences Cat#560602), Ly6C (Clone HK1.4, eBioscience Cat#48–5932-82 or BioLegend Cat#128016), CD206 (Clone C068C2, BioLegend Cat#141708), CD11c (Clone 3.9, eBioscience Cat#11–0116-42), MHC II (Clone M5/114.15.2, eBioscience Cat#48–5321-82 or BioLegend Cat#107645), CD103 (Clone 2E7, eBioscience Cat#12–1031-82), PDGFRα (Clone APA5, BioLegend Cat#135906), CD31 (Clone 390, BioLegend Cat#102418), PDPN (Clone 8.1.1, BioLegend Cat#127418) and EpCAM (Clone G8.8, BioLegend Cat#118208).
Flow cytometry was performed and analyzed on the BD FACSCanto and Aria cell sorter. Background fluorescence was estimated by substituting the primary antibodies with their specific isotype controls, and/or fluorescent-minus-one controls, as well as unstained controls. Dead cells were identified by Sytox Blue (Invitrogen Cat#S34857) and excluded from analysis. Analysis was performed using compensated data with FlowJo software (Version 10).
RNA extraction and qPCR
RNA extraction on FACS purified cells was performed using the RN-easy Micro Plus kit (Qiagen Cat#74034) and cDNA was generated with the SuperScript™ IV First-Strand Synthesis System (Invitrogen Cat#18091050). RNA extraction on tumor samples was performed using the RN-easy Mini Plus kit (Qiagen Cat#74134).
qPCR analysis on each biological sample was performed using technical replicates with TaqMan Fast Universal PCR Master Mix (Applied Biosystems Cat#4352042) and probes (Table S12) using the Viia7 Real-Time PCR System for 40 cycles (95°C for 15 seconds, 60°C for 1 min) and following an initial holding stage (50°C for 2 min, 95°C for 10 min). The cDNA concentration of target genes was normalized by amplification of 18S rRNA or Gapdh and fold changes in gene expression were obtained using the 2-ΔΔCT method (Livak and Schmittgen, 2001).
Taqman probes used were mouse 18s (Mm04277571_s1), Gapdh (Mm99999915_g1), Hck (Mm01241463_m1), Il4 (Mm004452 59_m1), Il6 (Mm00446190_m1), Il10 (Mm01288386_m1), Il11 (Mm00434162_m1), Il13 (Mm00434204_m1), Arg1 (Mm004759 88_m1), Tgfβ (Mm01227699_m1), Il12α (Mm00434169_m1), Ifnγ (Mm01168134_m1), Cxcl9 (Mm00434946_m1), Cxcl10 (Mm00445235_m1), Tnf (Mm00443258_m1), Prf1 (Mm00812512_m1), GzmB (Mm00442837_m1), Mmp3 (Mm00440295_m1), Mmp7 (Mm00487724_m1), Mmp9 (Mm00442991_m1), and Col1a1 (Mm00801666_g1).
Immunohistochemistry
Paraffin-embedded formalin-fixed sections were dewaxed in xylene and rehydrated in ethanol. Antigen retrieval was performed by boiling slides in citrate buffer (pH 6) for 15 min using a microwavable pressure cooker (Poh et al., 2017, 2020). Sections were immersed in 3% H2O2 for 20 min at room temperature to inhibit endogenous peroxidase activity, washed in TBS, then blocked in 10% normal goat serum (Gibco Cat#PCN5000) for 1 hour at room temperature. Sections were stained with primary antibodies (diluted in 10% normal goat serum) at 4°C in a humidified chamber overnight.
Primary antibodies used were Fibronectin (Abcam Cat#ab2143), PDPN (Abcam Cat#ab11936), αSMA (Abcam Cat#ab5694), PDGFRβ (Abcam Cat#ab32570), CD8a (Clone 4SM15, eBioscience #14–0808-82), Granzyme B (Clone D6E9W, Cell Signaling Technology Cat#44153), and Perforin (Clone E3W4I, Cell Signaling Technology Cat#31647). Biotinylated secondary antibodies from the Avidin Biotin Complex kit (Vector Laboratories Cat#BA-4001 or Cat#BA-1000) were used according to the manufacturer’s instructions.
Antigen visualization was achieved using 3,3-Diaminobenzine (DAKO). Sections were counterstained with Mayer’s haematoxylin, developed in Scott’s tap water, and dehydrated in ethanol and xylene before cover-slipping. Images were collected and analyzed with Aperio ImageScope v11.2.0.780 software. Quantification of positive staining per μm2 was performed using an automated cell counter script in FIJI (ImageJ) (Schneider et al., 2012).
Bulk RNA sequencing and analysis
RNA extracted from whole tumors were submitted to the Australian Genome Research Facility (AGRF) for sequencing on the Illumina NovaSeq 6000 platform with v1 200 cycle chemistry (100 bp paired ends). The Illumina TruSeq Stranded Total RNA Library Prep workflow with Ribo-Zero Gold was used to process the samples as per the manufacturer’s instructions. Quality control was evaluated by the AGRF, and reads were also screened for the presence of any Illumina adapter/overrepresented sequences and cross-species contamination.
Paired-end RNA-seq reads were aligned to the mouse reference genome GRCm38/mm10 using the Subread aligner (Rsubread version 2.2.6) (Liao et al., 2013). Gene-level read counts were obtained by running featureCounts, (Liao et al., 2014) a read count summarisation program within the Rsubread package (Liao et al., 2019) and the inbuilt Rsubread annotation that is a modified version of NCBI RefSeq mouse (mm10) genome annotation build 38.1. Pseudo genes or genes that did not meet a CPM (counts per million) read cut-off of 0.5 in at least 5 libraries were excluded from further analysis. Read counts were converted to log2-CPM, quantile normalized, and precision weighted with the voom function of the limma package (Law et al., 2014; Ritchie et al., 2015). A linear model was fitted to each gene, and empirical Bayes moderated t-statistic was used to assess differences in expression (McCarthy and Smyth, 2009; Smyth, 2004). Genes were called differentially expressed (DE) if they achieved a False Discovery Rate (FDR) of 5% or less. Gene Ontology (GO) terms enrichment analysis on the differentially expressed genes was performed using the goana function within the limma package (Young et al., 2010). Pathway enrichment against the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways for differentially expressed genes was performed using the kegga function also implemented in the limma package. A p-value cut-off of 0.001 was applied when determining enriched GO terms or KEGG pathways. RNA sequencing data were submitted to the GEO repository under the accession number GSE185540.
Analysis of scRNA-seq datasets
scRNA-seq gene expression data together with metadata of tumor specimens from human PDAC patients which has been previously published (Elyada et al., 2019) was obtained from the authors. Raw counts were imported into R and processed using the Seurat pipeline (version 4) (Hao et al., 2021). Briefly, cells with a mitochondrial content of >20%, or ribosomal content >40%, or <200 or >4,000 detected genes were excluded from analysis. Furthermore, genes that failed to express (an expressed gene has at least 1 UMI count) in at least 3 cells in at least 1 sample together with Mitochondrial and Ribosomal genes were excluded from analysis. Data was then normalized using a global-scaling normalization method “LogNormalize” that normalizes the feature expression measurements for each cell by the total expression, multiplies this by a scale factor of 10,000, and log-transforms the result. A subset of highly variable genes between cells were identified using the ‘FindVariableFeatures’ function within Seurat. A linear transformation was applied to the normalized data prior to dimensional reduction. Principal component analysis was performed using the 2000 most variable genes and dimension reduction to identify clusters was performed using 10 principal components and a resolution of 0.5. Cell type annotation of clusters was performed by computing the mean expression of marker genes identified in the paper (Elyada et al., 2019) and by manual curation. The gene expression profile of HCK in each cell type was computed and overlaid on tSNE plots.
Furthermore, raw scRNA-seq data of KPC mouse tumors consisting of viable cells were downloaded from Sequence Read Archive (SRA) with accession number SRP191615. The raw data was processed by cellCounts, a function within Rsubread (Liao et al., 2019) for quantifying 10x scRNA-seq data. Sequence reads were mapped to the mouse genome (GRCm38) based on the align function (Liao et al., 2013) and UMI counts were generated for each gene in each cell based on featureCounts (Liao et al., 2014). The inbuilt mouse (mm10) annotation in Rsubread was used for quantification. Similarly, the generated counts were processed, normalized and dimension reduction performed using the steps discussed above. Clusters were also annotated based on marker genes identified in the paper as above. The gene expression profile of Hck was computed and visualized on tSNE plots to highlight differences Hck expression across different cell types.
QUANTIFICATION AND STATISTICAL ANALYSIS
All experiments were performed at least twice with a minimum of four age- and sex-matched mice per group. The specific n (number of animals) used per cohort is indicated in the respective figure legends and shown as individual data points. No data was excluded from analysis. Tumor and liver weights were recorded by an independent assessor who was blinded to the experimental conditions. Statistical analysis was conducted using GraphPad Prism Software (Version 8). For comparison between multiple groups, a one-way ANOVA followed by Tukey’s multiple comparison test was performed as appropriate. Comparisons between two mean values were performed with a 2-tailed Student’s t-test. For survival studies, a Mantel-Cox log-rank test was used to evaluate statistical significance in Kaplan-Meier analysis. A p value of less than 0.05 was considered statistically significant.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| αCD40 (Clone FGK45) | JPP Biologics | N/A |
| αPD1 (Clone RMP1–14) | JPP Biologics | N/A |
| αCTLA4 (Clone 4F10) | JPP Biologics | N/A |
| αCD4 (Clone GK1.5) | JPP Biologics | N/A |
| αCD8 (Clone YTS169) | JPP Biologics | N/A |
| αNK1.1 (Clone PK136) | JPP Biologics | N/A |
| αCSF1R (Clone AFS98) | BioxCell | Cat#BP0213 |
| αIL12 (Clone R2–9A5) | BioxCell | Cat#BE0233 |
| αCXCR3 (Clone CXCR3–173) | BioxCell | Cat#BE0249 |
| Fc block | eBioscience | Cat#14–9161-73 |
| APC/Cyanine7 rat anti-mouse CD45 (Clone 30-F11) | BioLegend | Cat#103116 |
| PerCP/Cyanine5.5 rat anti-mouse CD45 Antibody (Clone 30-F11) | BioLegend | Cat#103131 |
| APC mouse anti-mouse NK1.1 (Clone PK136) | eBioscience | Cat#17–5941-82 |
| PE rat anti-mouse TCR β chain (Clone H57–597) | BioLegend | Cat#109208 |
| CD8a (Clone 53–6.7) | BioLegend | Cat#100722 |
| FITC rat anti-mouse F4/80 (Clone BM8) | BioLegend | Cat#123108 |
| PE-Cy7 rat anti-mouse F4/80 (Clone BM8) | eBioScience | Cat#25–4801-82 |
| PE rat anti-mouse CD11b (Clone M1/70 | BD Biosciences | Cat#553311 |
| PerCP-Cyanine5.5 rat anti-mouse Ly6G (Clone 1A8) | BD Biosciences | Cat#560602 |
| eFluor 450 rat anti-mouse Ly6C (Clone HK1.4) | eBioscience | Cat#48–5932-82 |
| APC rat anti-mouse Ly6C (Clone HK1.4) | BioLegend | Cat#128016 |
| APC rat anti-mouse CD206 (Clone C068C2) | BioLegend | Cat#141708 |
| FITC rat anti-mouse CD11c (Clone 3.9) | eBioscience | Cat#11–0116-42 |
| eFluor 450 rat anti-mouse MHC II (Clone M5/114.15.2) | eBioscience | Cat#48–5321-82 |
| Brilliant Violet 785 rat anti-mouse MHC II (Clone M5/114.15.2) | BioLegend | Cat#107645 |
| PE hamster anti-mouse CD103 (Clone 2E7) | eBioscience | Cat#12–1031-82 |
| PE rat anti-mouse PDGFRα (Clone APA5) | BioLegend | Cat#135906 |
| Pe-Cy7 rat anti-mouse CD31 (Clone 390) | BioLegend | Cat#102418 |
| APC-Cy7 hamster anti-mouse PDPN (Clone 8.1.1) | BioLegend | Cat#127418 |
| FITC rat anti-mouse EpCAM (Clone G8.8) | BioLegend | Cat#118208 |
| Rabbit anti-mouse Fibronectin | Abcam | Cat#ab2143 |
| Rabbit anti-mouse PDPN | Abcam | Cat#ab11936 |
| Rabbit anti-mouse αSMA | Abcam | Cat#ab5694 |
| Rabbit anti-mouse PDGFRβ | Abcam | Cat#ab32570 |
| Rat anti-mouse CD8a (Clone 4SM15) | eBioscience | Cat#14–0808-82 |
| Rabbit anti-mouse Granzyme B (Clone D6E9W) | Cell Signaling Technology | Cat#44153 |
| Rabbit anti-mouse Perforin (Clone E3W4I) | Cell Signaling Technology | Cat#31647 |
| Rabbit anti-rat biotinylated IgG (Avidin Biotin Complex kit) | Vector Laboratories | Cat#BA-4001 |
| Goat anti-rabbit biotinylated IgG (Avidin Biotin Complex kit) | Vector Laboratories | Cat#BA-1000 |
|
| ||
| Critical commercial assays | ||
|
| ||
| RN-easy Micro Plus kit | Qiagen | Cat#74034 |
| RN-easy Mini Plus kit | Qiagen | Cat#74134 |
| SuperScript™ IV First-Strand Synthesis System | Invitrogen | Cat#18091050 |
| TaqMan™ Fast Universal PCR Master Mix (2X), no AmpErase™ UNG | Applied Biosystems | Cat#4352042 |
|
| ||
| Deposited data | ||
|
| ||
| Bulk RNA sequencing data of KPC liver metastases from WT and HckKO mice | This paper | GEO repository GSE185540 |
| Single cell RNA sequencing of human pancreatic cancer and mouse KPC tumors | Elyada et al., 2019 |
Elyada et al., 2019 PMID: 31197017 |
|
| ||
| Experimental models: Cell lines | ||
|
| ||
| Mouse KPC pancreatic cancer cells | A/Prof Marina Pajic, The Garvan Institute of Medical Research, Australia. |
Vennin et al., 2017 PMID: 28381539 |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| Mouse: Wild-type C57BL/6 | In-house (Prof Matthias Ernst), The Olivia Newton-John Cancer Research Institute. | N/A |
| Mouse: HckKO | In-house (Prof Clifford Lowell), The Olivia Newton-John Cancer Research Institute. |
Lowell et al. (1994) PMID: 8125254 |
| Mouse: Rag1KO | In-house (Prof Matthias Ernst), The Olivia Newton-John Cancer Research Institute. | N/A |
| Mouse: HckKO;Rag1KO | In-house (Prof Matthias Ernst), The Olivia Newton-John Cancer Research Institute. | N/A |
| Mouse: ItgaxCreIrf8fl/fl | In-house (Dr Michael Chopin), The Walter and Eliza Hall Institute, Australia. |
Chopin et al. (2013) PMID: 24249112 |
|
| ||
| Oligonucleotides | ||
|
| ||
| See Table S12 for Taqman Probes | N/A | |
|
| ||
| Software and algorithms | ||
|
| ||
| Aperio ImageScope v11.2.0.780 | Leica Biosystems | https://www.leicabiosystems.com/en-au/digital-pathology/manage/aperio-imagescope/ |
| FIJI (ImageJ) |
Schneider et al. (2012) PMID: 22930834 |
https://imagej.nih.gov/ij/ |
| FlowJo (Version 10) | Flowjo | https://www.flowjo.com/ |
| GraphPad Prism Software (Version 8) | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| Subread aligner (Rsubread version 2.2.6) |
Liao et al., 2013, 2019 PMIDs: 23558742 & 30783653 |
https://bioconductor.org/packages/release/bioc/html/Rsubread.html |
| featureCounts (Rsubread version 2.2.6) |
Liao et al. 2014, 2019 PMIDs: 24227677 & 30783653 |
https://bioconductor.org/packages/release/bioc/html/Rsubread.html |
| limma (3.44.3) |
Law et al. (2014) PMID: 24485249 Ritchie et al. (2015) PMID: 25605792 |
https://bioconductor.org/packages/release/bioc/html/limma.html |
| Seurat (version 4) |
Hao et al. (2021) PMID: 34062119 |
https://cran.r-project.org/web/packages/Seurat/index.html |
| R version 4.0.3 | CRAN | https://cran.csiro.au/src/base/R-4/R-4.0.3.tar.gz |
| cellCounts (Rsubread version 2.2.6) |
Liao et al., 2013, 2019 PMIDs: 23558742 & 30783653 |
https://bioconductor.org/packages/release/bioc/html/Rsubread.html |
|
| ||
| Other | ||
|
| ||
| DMEM/F-12 media | Gibco | Cat#11320033 |
| HBSS media | Gibco | Cat#14170112 |
| Ketamine | Baxter | N/A |
| Xylazine | Ilium | N/A |
| Isoflurane | Baxter | N/A |
| Carprofen | Zoetis | N/A |
| Bupivacaine | AstraZeneca | N/A |
| Gemcitabine | Pfizer | N/A |
| Neomycin | ThermoFisher | Cat#21810031 |
| RK20449 | Reagency | Saito et al. (2013) PMID: 23596204 |
| Captisol | Captisol | Cat#RC-0C7-K01 |
| Collagenase / Dispase | Roche | Cat#11097113001 |
| DNase I | Roche | Cat#10104159001 |
| Sytox Blue | Invitrogen | Cat#S34857 |
| Normal goat serum | Gibco | Cat#PCN5000 |
Highlights.
Myeloid HCK expression is elevated in human pancreatic ductal adenocarcinoma
HCK inhibition, or its ablation in hosts, limits primary/metastatic PDAC growth
HCK ablation prevents an immune-suppressive and desmoplastic PDAC stroma
HCK inhibition enables and improves PDAC response to immunotherapy/chemotherapy
ACKNOWLEDGMENTS
This work was supported in parts through the Victorian State Government Operational Infrastructure Support and the National Health and Medical Research Council (NHMRC) of Australia project and development grants 1081373, 1092788, and 2014063. C.A.L. received funding from the UC Cancer Research Coordinating Committee (CRR-20-636450). M.E. received funding from Ludwig Cancer Research and is a NHMRC Investigator grant recipient (1173814). A.R.P. received support from 2018 Priority-driven Collaborative Cancer Research Scheme Grant (1157894) co-funded by Cancer Australia, Cure Cancer and Pancare Foundation; a Tour de Cure Early Career Seed Grant (RSP-060-18/19); and an Avner Collaboration Grant from PanKind, The Australian Pancreatic Cancer Foundation in collaboration with Tour de Cure and Woolworths Limited through Woolies on Wheels and Walks. A.R.P. is an NHMRC Peter Doherty Early Career Fellow (1166447). M.P. is supported by an NHMRC career development fellowship (1162556) and NHMRC project grant (1162860). We acknowledge The Collie Foundation for providing funds to purchase the Leica Aperio slide scanner. We also acknowledge the use of the services and facilities of the AGRF.
DECLARATION OF INTERESTS
A.R.P. and M.E. are inventors on a patent application related to this work filed by The Olivia Newton-John Cancer Research Institute (PCT/AU2021/051073) on the 16th of September 2021.
INCLUSION AND DIVERSITY
We support inclusive, diverse, and equitable conduct of research.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.111479.
REFERENCES
- Arnold IC, Zhang X, Artola-Boran M, Fallegger A, Sander P, Johansen P, and Müller A. (2019). BATF3-dependent dendritic cells drive both effector and regulatory T-cell responses in bacterially infected tissues. PLoS Pathog. 15, e1007866. 10.1371/journal.ppat.1007866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagaev A, Kotlov N, Nomie K, Svekolkin V, Gafurov A, Isaeva O, Osokin N, Kozlov I, Frenkel F, Gancharova O, et al. (2021). Conserved pan-cancer microenvironment subtypes predict response to immunotherapy. Cancer Cell 39, 845–865.e7. 10.1016/j.ccell.2021.04.014. [DOI] [PubMed] [Google Scholar]
- Bailey P, Chang DK, Nones K, Johns AL, Patch AM, Gingras MC, Miller DK, Christ AN, Bruxner TJC, Quinn MC, et al. (2016). Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 531, 47–52. 10.1038/nature16965. [DOI] [PubMed] [Google Scholar]
- Candido JB, Morton JP, Bailey P, Campbell AD, Karim SA, Jamieson T, Lapienyte L, Gopinathan A, Clark W, McGhee EJ, et al. (2018). CSF1R+ macrophages sustain pancreatic tumor growth through T cell suppression and maintenance of key gene programs that define the squamous subtype. Cell Rep. 23, 1448–1460. 10.1016/j.cel-rep.2018.03.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caton ML, Smith-Raska MR, and Reizis B. (2007). Notch-RBP-J signaling controls the homeostasis of CD8- dendritic cells in the spleen. J. Exp. Med 204, 1653–1664. 10.1084/jem.20062648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopin M, Seillet C, Chevrier S, Wu L, Wang H, Morse HC III, Belz GT, and Nutt SL (2013). Langerhans cells are generated by two distinct PU.1-dependent transcriptional networks. J. Exp. Med 210, 2967–2980. 10.1084/jem.20130930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow MT, Ozga AJ, Servis RL, Frederick DT, Lo JA, Fisher DE, Freeman GJ, Boland GM, and Luster AD (2019). Intratumoral activity of the CXCR3 chemokine System is required for the efficacy of anti-PD-1 therapy. Immunity 50, 1498–1512.e5. 10.1016/j.immuni.2019.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conroy T, Desseigne F, Ychou M, Bouché O, Guimbaud R, Bécouarn Y, Adenis A, Raoul JL, Gourgou-Bourgade S, de la Fouchardière C, et al. (2011). FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N. Engl. J. Med 364, 1817–1825. 10.1056/NEJ-Moa1011923. [DOI] [PubMed] [Google Scholar]
- Cook SA, and Schafer S. (2020). Hiding in plain sight: interleukin-11 emerges as a master regulator of fibrosis, tissue integrity, and stromal inflammation. Annu. Rev. Med 71, 263–276. 10.1146/annurev-med-041818-011649. [DOI] [PubMed] [Google Scholar]
- Creeden JF, Alganem K, Imami AS, Brunicardi FC, Liu S-H, Shukla R, Tomar T, Naji F, and McCullumsmith RE (2020). Kinome array profiling of patient-derived pancreatic ductal adenocarcinoma identifies differentially active protein tyrosine kinases. Int. J. Mol. Sci 21, 8679. 10.3390/ijms21228679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crnogorac-Jurcevic T, Gangeswaran R, Bhakta V, Capurso G, Lattimore S, Akada M, Sunamura M, Prime W, Campbell F, Brentnall TA, et al. (2005). Proteomic analysis of chronic pancreatitis and pancreatic adenocarcinoma. Gastroenterology 129, 1454–1463. 10.1053/j.gastro.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Diamond MS, Lin JH, and Vonderheide RH (2021). Site-dependent immune escape due to impaired dendritic cell cross-priming. Cancer Immunol. Res 9, 877–890. 10.1158/2326-6066.Cir-20-0785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elyada E, Bolisetty M, Laise P, Flynn WF, Courtois ET, Burkhart RA, Teinor JA, Belleau P, Biffi G, Lucito MS, et al. (2019). Cross-species single-cell analysis of pancreatic ductal adenocarcinoma reveals antigen-presenting cancer-associated fibroblasts. Cancer Discov. 9, 1102–1123. 10.1158/2159-8290.CD-19-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst M, Inglese M, Scholz GM, Harder KW, Clay FJ, Bozinovski S, Waring P, Darwiche R, Kay T, Sly P, et al. (2002). Constitutive activation of the SRC family kinase Hck results in spontaneous pulmonary inflammation and an enhanced innate immune response. J. Exp. Med 196, 589–604. 10.1084/jem.20020873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Wang H, Shin DM, Masiuk M, Qi CF, and Morse HC 3rd. (2011). IFN regulatory factor 8 restricts the size of the marginal zone and follicular B cell pools. J. Immunol 186, 1458–1466. 10.4049/jimmunol.1001950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galon J, and Bruni D. (2019). Approaches to treat immune hot, altered and cold tumours with combination immunotherapies. Nat. Rev. Drug Discov 18, 197–218. 10.1038/s41573-018-0007-y. [DOI] [PubMed] [Google Scholar]
- Garris CS, Arlauckas SP, Kohler RH, Trefny MP, Garren S, Piot C, Engblom C, Pfirschke C, Siwicki M, Gungabeesoon J, et al. (2018). Successful anti-PD-1 cancer immunotherapy requires T cell-dendritic cell cross-talk involving the cytokines IFN-g and IL-12. Immunity 49, 1148–1161.e7. 10.1016/j.immuni.2018.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng X, Chen H, Zhao L, Hu J, Yang W, Li G, Cheng C, Zhao Z, Zhang T, Li L, and Sun B. (2021). Cancer-associated fibroblast (CAF) heterogeneity and targeting therapy of CAFs in pancreatic cancer. Front. Cell Dev. Biol 9, 655152. 10.3389/fcell.2021.655152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y, Hao S, Andersen-Nissen E, Mauck WM 3rd, Zheng S, Butler A, Lee MJ, Wilk AJ, Darby C, Zager M, et al. (2021). Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29. 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde S, Krisnawan VE, Herzog BH, Zuo C, Breden MA, Knolhoff BL, Hogg GD, Tang JP, Baer JM, Mpoy C, et al. (2020). Dendritic cell paucity leads to dysfunctional immune surveillance in pancreatic cancer. Cancer Cell 37, 289–307.e9. 10.1016/j.ccell.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidenblad M, Schoenmakers EFPM, Jonson T, Gorunova L, Veltman JA, van Kessel AG, and Höglund M. (2004). Genome-wide array-based comparative genomic hybridization reveals multiple amplification targets and novel homozygous deletions in pancreatic carcinoma cell lines. Cancer Res. 64, 3052–3059. 10.1158/0008-5472.can-03-3159. [DOI] [PubMed] [Google Scholar]
- Ho WJ, Jaffee EM, and Zheng L. (2020). The tumour microenvironment in pancreatic cancer — clinical challenges and opportunities. Nat. Rev. Clin. Oncol 17, 527–540. 10.1038/s41571-020-0363-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- House IG, Savas P, Lai J, Chen AXY, Oliver AJ, Teo ZL, Todd KL, Henderson MA, Giuffrida L, Petley EV, et al. (2020). Macrophage-derived CXCL9 and CXCL10 are required for antitumor immune responses following immune checkpoint blockade. Clin. Cancer Res 26, 487–504. 10.1158/1078-0432.Ccr-19-1868. [DOI] [PubMed] [Google Scholar]
- Isella C, Terrasi A, Bellomo SE, Petti C, Galatola G, Muratore A, Mellano A, Senetta R, Cassenti A, Sonetto C, et al. (2015). Stromal contribution to the colorectal cancer transcriptome. Nat. Genet 47, 312–319. 10.1038/ng.3224. [DOI] [PubMed] [Google Scholar]
- Kaneda MM, Cappello P, Nguyen AV, Ralainirina N, Hardamon CR, Foubert P, Schmid MC, Sun P, Mose E, Bouvet M, et al. (2016). Macrophage PI3Kγ drives pancreatic ductal adenocarcinoma progression. Cancer Discov. 6, 870–885. 10.1158/2159-8290.CD-15-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law CW, Chen Y, Shi W, and Smyth GK (2014). voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15, R29. 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, and Shi W. (2013). The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 41, e108. 10.1093/nar/gkt214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, and Shi W. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, and Shi W. (2019). The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res. 47, e47. 10.1093/nar/gkz114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, and Schmittgen TD (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Lowell CA, Soriano P, and Varmus HE (1994). Functional overlap in the src gene family: inactivation of hck and fgr impairs natural immunity. Genes Dev. 8, 387–398. [DOI] [PubMed] [Google Scholar]
- McCarthy DJ, and Smyth GK (2009). Testing significance relative to a fold-change threshold is a TREAT. Bioinformatics 25, 765–771. 10.1093/bioinformatics/btp053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellman I, Coukos G, and Dranoff G. (2011). Cancer immunotherapy comes of age. Nature 480, 480–489. 10.1038/nature10673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison AH, Diamond MS, Hay CA, Byrne KT, and Vonderheide RH (2020). Sufficiency of CD40 activation and immune checkpoint blockade for T cell priming and tumor immunity. Proc. Natl. Acad. Sci. USA 117, 8022–8031. 10.1073/pnas.1918971117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen SR, Quaranta V, Linford A, Emeagi P, Rainer C, Santos A, Ireland L, Sakai T, Sakai K, Kim Y-S, et al. (2016). Macrophage-secreted granulin supports pancreatic cancer metastasis by inducing liver fibrosis. Nat. Cell Biol 18, 549–560. 10.1038/ncb3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikfarjam M, Yeo D, He H, Baldwin G, Fifis T, Costa P, Tan B, Yang E, Wen S.w., and Christophi C. (2013). Comparison of two syngeneic orthotopic murine models of pancreatic adenocarcinoma. J. Invest. Surg 26, 352–359. 10.3109/08941939.2013.797057. [DOI] [PubMed] [Google Scholar]
- Poh AR, and Ernst M. (2021). Tumor-associated macrophages in pancreatic ductal adenocarcinoma: therapeutic opportunities and clinical challenges. Cancers 13, 2860. 10.3390/cancers13122860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poh AR, Dwyer AR, Eissmann MF, Chand AL, Baloyan D, Boon L, Murrey MW, Whitehead L, O’Brien M, Lowell CA, et al. (2020). Inhibition of the SRC kinase HCK impairs STAT3-dependent gastric tumor growth in mice. Cancer Immunol. Res 8, 428–435. 10.1158/2326-6066.CIR-19-0623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poh AR, Love CG, Masson F, Preaudet A, Tsui C, Whitehead L, Monard S, Khakham Y, Burstroem L, Lessene G, et al. (2017). Inhibition of hematopoietic cell kinase activity suppresses myeloid cell-mediated colon cancer progression. Cancer Cell 31, 563–575.e5. 10.1016/j.ccell.2017.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poh AR, O’Donoghue RJJ, and Ernst M. (2015). Hematopoietic cell kinase (HCK) as a therapeutic target in immune and cancer cells. Oncotarget 6, 15752–15771. 10.18632/oncotarget.4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raimondi S, Maisonneuve P, and Lowenfels AB (2009). Epidemiology of pancreatic cancer: an overview. Nat. Rev. Gastroenterol. Hepatol 6, 699–708. 10.1038/nrgastro.2009.177. [DOI] [PubMed] [Google Scholar]
- Reschke R, and Gajewski TF (2022). CXCL9 and CXCL10 bring the heat to tumors. Sci. Immunol 7, eabq6509. 10.1126/sciimmunol.abq6509. [DOI] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, and Smyth GK (2015). Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47. 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas LA, and Balachandran VP (2021). Scaling the immune incline in PDAC. Nat. Rev. Gastroenterol. Hepatol 18, 453–454. 10.1038/s41575-021-00475-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito Y, Yuki H, Kuratani M, Hashizume Y, Takagi S, Honma T, Tanaka A, Shirouzu M, Mikuni J, Handa N, et al. (2013). A pyrrolo-pyrimidine derivative targets human primary AML stem cells in vivo. Sci. Transl. Med 5, 181ra52. 10.1126/scitranslmed.3004387. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, and Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolinska MJ, Page TH, Urbaniak AM, Mutch BE, and Horwood NJ (2011). Hck tyrosine kinase regulates TLR4-induced TNF and IL-6 production via AP-1. J. Immunol 187, 6043–6051. 10.4049/jimmunol.1100967. [DOI] [PubMed] [Google Scholar]
- Smyth GK (2004). Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol 3, Article3. 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- Stromnes IM, Brockenbrough JS, Izeradjene K, Carlson MA, Cuevas C, Simmons RM, Greenberg PD, and Hingorani SR (2014). Targeted depletion of an MDSC subset unmasks pancreatic ductal adenocarcinoma to adaptive immunity. Gut 63, 1769–1781. 10.1136/gutjnl-2013-306271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z, Li C, Kang B, Gao G, Li C, and Zhang Z. (2017). GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 45, 98–102. 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tempero MA, Malafa MP, Chiorean EG, Czito B, Scaife C, Narang AK, Fountzilas C, Wolpin BM, Al-Hawary M, Asbun H, et al. (2019). Pancreatic adenocarcinoma, version 1.2019. J. Natl. Compr. Canc. Netw 17, 202–210. 10.6004/jnccn.2019.0014. [DOI] [PubMed] [Google Scholar]
- Vennin C, Chin VT, Warren SC, Lucas MC, Herrmann D, Magenau A, Melenec P, Walters SN, Del Monte-Nieto G, Conway JRW, et al. (2017). Transient tissue priming via ROCK inhibition uncouples pancreatic cancer progression, sensitivity to chemotherapy, and metastasis. Sci. Transl. Med 9, eaai8504. 10.1126/scitranslmed.aai8504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonderheide RH (2018). The immune revolution: a case for priming, not checkpoint. Cancer Cell 33, 563–569. 10.1016/j.ccell.2018.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue J, Sharma V, Hsieh MH, Chawla A, Murali R, Pandol SJ, and Habtezion A. (2015). Alternatively activated macrophages promote pancreatic fibrosis in chronic pancreatitis. Nat. Commun 6, 7158. 10.1038/ncomms8158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young MD, Wakefield MJ, Smyth GK, and Oshlack A. (2010). Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 11, R14. 10.1186/gb-2010-11-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Herndon JM, Sojka DK, Kim K-W, Knolhoff BL, Zuo C, Cullinan DR, Luo J, Bearden AR, Lavine KJ, et al. (2017). Tissue-resident macrophages in pancreatic ductal adenocarcinoma originate from embryonic hematopoiesis and promote tumor progression. Immunity 47, 323–338.e6. 10.1016/j.immuni.2017.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Knolhoff BL, Meyer MA, Nywening TM, West BL, Luo J, Wang-Gillam A, Goedegebuure SP, Linehan DC, and DeNardo DG (2014). CSF1/CSF1R blockade reprograms tumor-infiltrating macrophages and improves response to T-cell checkpoint immunotherapy in pancreatic cancer models. Cancer Res. 74, 5057–5069. 10.1158/0008-5472.Can-13-3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Tang C, Xu T, and Zhao Z. (2021). Molecular analysis of prognosis and immune pathways of pancreatic cancer based on TNF family members. J. Oncol 2021, 2676996. 10.1155/2021/2676996. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Bulk RNA sequencing data generated from KPC liver metastases of WT and HckKO mice are publicly available at the Gene Expression Omnibus (GEO) under accession number GSE185540.
All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.