Abstract
Genetically encoded biosensors are crucial for enhancing our understanding of how molecules regulate biological systems. Small molecule biosensors, in particular, help us understand the interaction between chemicals and biological processes. They also accelerate metabolic engineering by increasing screening throughput and eliminating the need for sample preparation through traditional chemical analysis. Additionally, they offer significantly higher spatial and temporal resolution in cellular analyte measurements. In this review, we discuss recent progress in biosensors and control systems—biosensor-based controllers—in metabolic engineering. We emphasize cutting-edge protein-based biosensor development. Furthermore, we explore engineered protein-based biosensors that utilize less commonly exploited signaling mechanisms, such as protein stability and induced degradation, compared to the prevalent transcription factor binding and allosteric regulation. Lastly, we propose that these lesser-used mechanisms will be significant for engineering eukaryotic systems and slower-growing prokaryotic systems where protein turnover may facilitate a more rapid and reliable measurement and regulation of the current cellular state.
Keywords: Biosensors, metabolic engineering, synthetic biology, rational design, directed evolution, metabolic controllers, protein degradation
Introduction
Metabolic engineering has gained significant attention due to its potential to impact human and environmental health as well as the global economy through enhancing industrial production of biochemicals, agrochemicals, biofuels, pharmaceuticals, nutraceuticals, other chemicals, and biomolecules. Through turning an organism into a cell factory and rewiring metabolic pathways, biomanufacturing and metabolic engineering offer an alternative and environmentally friendly method to produce natural active compounds, metabolites, antibiotics, fatty acids, or de novo synthetic compounds[3–11]. An organism that has been specifically modified to serve as a platform on which new genetic functions and specific metabolic pathways can be designed and assembled to produce such useful compounds is typically called a chassis organism[12–16]. Heterologous gene expression and editing of native genes in the chassis result in optimization of the biochemical reactions, signaling, and metabolic pathways, for production of compounds of interest via the chassis’ metabolism instead of traditional chemical synthesis or extraction from plants[15, 17–19]. Moreover, chassis organisms can also be engineered to make use of low value feedstocks such as food waste or waste plant biomass to generate beneficial, high value compounds[20–24]. Another impactful application of metabolic engineering is the engineering of chassis organisms to degrade undesired chemical compounds in the environment[25–27].
Metabolic engineering is a complex, difficult, and often unpredictable process. The production yield of desired molecule by the chassis is often initially low and limited due to low heterologous expression levels, undesired catalytic properties, incorrect folding, feedback inhibition, and environmental triggers[3, 28, 29]. Further iteration of chassis development, or perhaps total redesign, is required to improve yield. Engineering biological systems is generally a sophisticated process that requires costly Design-Build-Test-Learn (DBTL) cycles to iteratively seek the optimal system (Figure 1)[30, 31]. The test phase, which evaluates the performance of the constructed biological system for example, determining the amount of the desired chemical produced, is often particularly laborious, costly, and time-consuming[32, 33]. Conventional assays are often used, based on liquid or gas chromatography sometimes paired with mass spectrometry or immunoassays, have limited capacity for processing samples and are intricate and time-consuming for optimizing the production of valuable compounds of interest[34–37]. The DBTL cycle is repeated, often with many designs being built and tested in parallel, until the desired specific function is achieved, such as a strain that can convert a specific substrate into the desired product at an economically viable rate[30, 31]. Although many functional genes and biosynthesis pathways in many organisms have been identified, it is still challenging to identify suitable enzymes and pathways in specific hosts. Factors such as metabolic flux balance, host strain choices, and chassis tolerance to heterologous expression, feedstocks, intermediates and products are important metabolic engineering design parameters[11, 38–41]. Due to the complexity of metabolic pathways, it is difficult to explore, reveal, and engineer every single component. The above bottlenecks impede the development of cell factories for producing compounds of interest at high yields. Genetically encoded biosensors are essential to resolving bottlenecks in chassis selection and metabolic engineering as high-throughput and efficient tools for identifying pathway components, monitoring and regulating biosynthesis pathways, and facilitating screening and selection of host cells with desired phenotypes[42–45]. Biosensors have the potential to significantly improve the throughput of Design-Build-Test-Learn (DBTL) processes and reduce the number of conventional assays needed for identifying key components and fluxes within the metabolic pathway.[46–48]. They detect and quantify small molecules, using biological macromolecules like proteins, DNA, or RNA as recognition units to sense specific target molecules[42, 44, 49, 50]. These units connect to measurable reporter signals, such as changes in spectrophotometric, chemical, or electrical properties. Biosensors can be engineered to function outside of cells, as in vitro biochemical reactions, or engineered to be encoded in the genome, or delivered to cells by some other means, and function in vivo, entirely within host cells.
Figure 1.
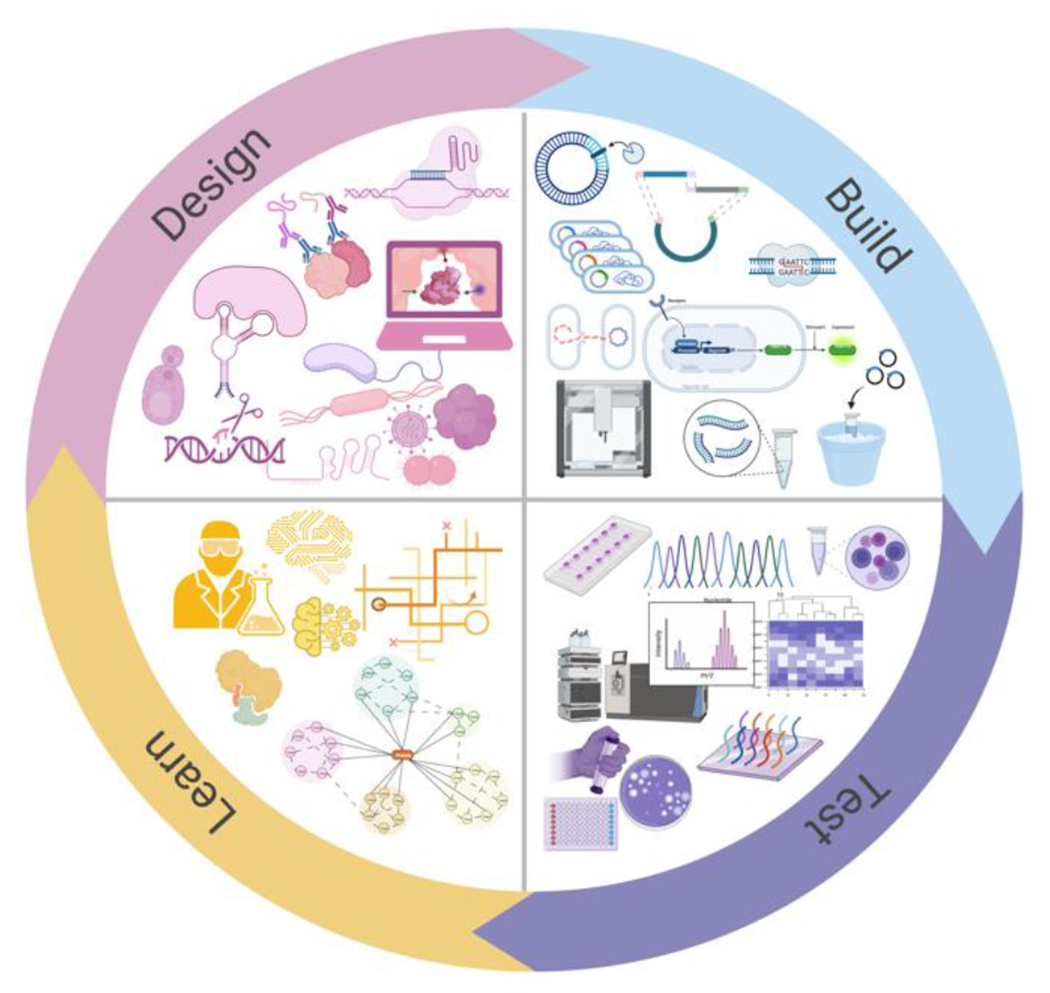
A diagram representing a “design-build-test-learn” (DBTL) cycle in metabolic engineering. The DBTL framework is a systematic and iterative process for the development of optimal components and systems. The Design phase identifies the biological components such as genes, proteins, enzymes, and metabolic pathways, including host cells, and combinatorial arrangements of these proposed to give rise to desired outcomes. The Build phase involves the synthesis, and assembly of various parts to be expressed in the host. The Test phase validates the engineered strains for target molecule production, efficiency and function of enzymes, and optimization of the metabolic pathway, among other possible outcomes. The Learn phase analyzes the data from the Test phase to improve the initial design in the next iteration.
This review centers on in vivo genetically encoded biosensors that expedite chassis organism development and optimization in metabolic engineering[51–54]. We explore their use as metabolic controllers, and other applications. We also highlight the protein-based biosensor development, the potential of protein-based controller that control cellular protein abundance for metabolic engineering, and future perspectives.
Biosensors accelerate metabolic engineering
Biosensors have proven to be valuable tools in metabolic engineering (Table 1), facilitating rapid and efficient screening of intracellular target compounds produced by recombinant chasses[46, 50, 55–57]. They provide high temporal and spatial resolution in reporting the cell’s metabolic state[55–57], thus boosting throughput (Figure 2)[58–60]. This high-throughput approach enables the identification of optimal enzymes and regulatory genes of chasses[61, 62], discovery of novel biosynthesis pathways[43, 46, 63, 64], and monitoring metabolite production[51], ultimately leading to the enhancement of the production of a target molecule of interest.
Table 1.
Examples of the successful integration of genetically encoded biosensors into assisted cell factory engineering in metabolic engineering application.
| Target small molecule | Biorecognition or sensing element | Reporter/output | Host/Chassis | Purposes |
|---|---|---|---|---|
| Caprolactam, valerolactam[69] | OplR transcription factor | RFP | P. putida | Monitoring target molecule production |
| Resveratrol, flavonoid naringenin[62] | TtgR transcription repressor | GFP | E. coli | Screening enzyme activity and monitoring production |
| D-glucaric acid[70] | cdaR transcription factor | GFP | S. cerevisiae | Screening and selection for optimal chassis |
| Resveratrol, naringenin[62] | P. putida/TtgT transcription repressor | RFP | E. coli | Screening enzyme activity for the optimal genetic variants |
| N-acetylneuraminate (NeuAc) [71] | Self-cleavage aptazyme | GFP | E. coli | Screening for optimal enzyme activity of NeuAc synthase |
| Vanillate[72] | Caulobacter crescentus VanR-VanO | YFP | E. coli | Screening and selection for optimal chassis |
| Anthranilic acid[73] | NahR regulatory protein | tetA gene | E. coli | Screening and selection for optimal system and chassis |
| l-phenylalanine (l-Phe)[74] | pTF-TyrR1 | YFP | E. coli | Screening and selection for optimal chassis |
| Salicylate[75] | AraC transcription factor | GFP | E. coli | Screening for regulatory gene expression patterns |
| Naringenin[76] | FdeR transcription factor | GFP | S. cerevisiae | Screening and selection for optimal chassis |
| Malonyl-CoA[77] | Type III polyketide synthase RppA | flaviolin | E. coli, P. putida, C. glutamicum | Screening and selection for optimal chassis |
| Macrolides[78] | MphR transcription factor | GFP | E. coli | Screening and selection for optimal chassis |
| Malate[79] | Malate response regulator MalR | GFP | B. licheniformis | Screening and selection the optimal pathway |
| ε-caprolactam, δ-valerolactam, butyrolactam[80] | ChnR/Pb transcription factor-promoter pair | mCherry | E. coli | Discriminate against lactam biosynthetic intermediates during biomanufacturing |
| Muconic acid[81] | benM transcription factor | GFP | S. cerevisiae | Screening and selection for optimal chassis |
| N-acetylglucosamine (GlcNAc) [82] | glmS ribozyme | glmM mRNA | B. subtilis | Regulating and screening the optimal genetic variants |
| 3-Dehydroshikimate [83] | CusR transcriptional regulator | GFP | E. coli | Screening and selection for optimal chassis |
| Alkanes[84] | AlkS transcriptional factor | GFP | E. coli | Screening and selection for optimal chassis |
| Bicyclic Monoterpenes[85] | Camphor-responsive TetR-family regulator CamR | GFP | E. coli | Screening and selection for optimal chassis |
| p-Coumaroyl-CoA[86] | CouR transcriptional repressor | GFP | S. cerevisiae | Dynamically regulate naringenin synthetic pathway |
| Pinene[41] | MexR transcriptional repressor | acrAB gene | E. coli | Screening and selection for optimal chassis |
| D-psicose[87] | PsiR transcription factor | GFP | E. coli | Monitoring and regulating chemical production |
| Erythromycin[88] | MphR transcriptional repressor | GFP | E. coli | Screening and selection for optimal chassis |
| Lignin[89] | EmrR transcriptional regulator | GFP | E. coli | Screening and selection for optimal chassis |
| Nitrilase, amidase, and NHase[90] | BsNadR transcriptional repressor | GFP | E. coli | Screening nitrile metabolism-related enzymes |
| Mannitol 1-phosphate dehydrogenases/phosphatases[91] | AraC transcription factor | RFP | C. necator | Screening genetic variants, monitoring chemical production and growth conditions |
| shikimic acid[92] | LysR-type transcriptional regulator (ShiR) | GFP | C. glutamicum | Screening and selection for optimal chassis |
| D-allulose[93] | PsiR transcription factor | mCherry, LacZ gene | E. coli | Screening function enzyme mutants |
| L-arginine[94] | ArgP transcriptional regulator | GFP | E. coli | Screening genetic variants and optimal chassis |
| Phytase, Laccase, β-Casein and β-Lactoglobulin[95] | Split GFP construct | GFP-TEV | P. pastoris | Monitoring the recombinant protein production and screening for optimal chassis |
| L-DOPA[37] | DOPA dioxygenase (DOD) | RFP | S. cerevisiae | Monitoring and optimizing enzyme activity |
| S-Adenosyl-l-homocysteine[96] | Riboswitch and aptamer fusion | GFP | E. coli | Monitoring intracellular levels of the metabolite |
Figure 2.
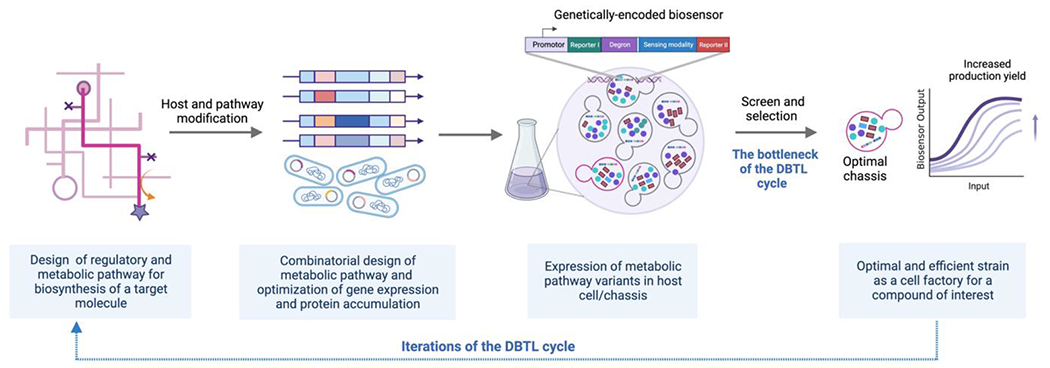
A schematic illustrating an example of biosensor-facilitated chassis development aimed at enhancing the production of a desired molecule. In the Test phase, a genetically encoded biosensor facilitates the screening and selection of optimal strains.
Biosensors can also be used to screen large libraries of genetic variants or metabolic pathways, which allow further optimization of the production of desired compounds by adjusting the gene expressions and altering metabolic pathways[44, 60, 65–68]. This helps to accelerate the development and optimization of the productivity of a chassis organism. While the concept of cellular biosensors is not a new innovation, as all living organisms naturally possess intrinsic biosensors to regulate and balance their metabolisms, these genetically encoded biosensors provide a more efficient and alternative mechanism to quantify a molecule of interest than the classical assays of quantitative chemistry. Therefore, harnessing and engineering molecular biosensors for high-throughput screening of desired compounds or metabolic pathways can aid the understanding and selecting valuable genetic variants and discovering metabolic pathways. This, in turn, can expedite development of efficient biomanufacturing platforms. Here, we summarized the studies that utilized genetically-encoded biosensors for metabolic engineering.
Genetically encoded biosensors offer a valuable tool for monitoring metabolic states by detecting specific target molecules and generating signals corresponding to their concentrations. These biosensors also facilitate high-throughput screening and selection, significantly reducing the time required for analyzing optimal enzyme, biosynthetic pathway, as well as high-producing strains from a library of natural or engineered strains in the production of various chemicals.
Biosensors as metabolic controllers
The quantification and regulation of metabolites within pathways is critical to enhance microbial production bioprocesses. In metabolic engineering, it is well known that accumulation of certain intermediate compounds in metabolic pathways can exhibit toxicity, imposing a burden on the host chassis, and exemplifying the necessity of balancing flux through metabolic pathways by controlling the levels of biosynthetic enzymes. To address this issue, the regulation of specific components, such as transcription factors or gene regulators, can be employed to control gene expression. By exploiting the underlying mechanism of biosensors, which involves sensing a target molecule and triggering transcriptional activation or altering protein stability as an output, these biosensors can be utilized as controllers to actively regulate metabolism and cellular functions. This rewiring of in vivo cellular processes enables the creation of novel signaling pathways within cells.
The utilization of various controllers, functioning as cellular switches, has revolutionized research involving bacteria, yeast, and animals by effectively regulating cellular mechanisms and processes. Significantly, numerous metabolite-responsive transcription factor-based biosensors have been extensively explored to regulate genes of interest and control cellular functions, including metabolic flux. For instance, an example of such a biosensor is FapR, a responsive transcription factor that negatively regulates fatty acid and phospholipid biosynthesis in Bacillus subtilis. By engineering the regulatory circuit sensor for malonyl-CoA in E. coli, the group showed that the malonyl-CoA sensor–actuator biosensor could help alleviate the toxicity in the chassis, therefore improve fatty acid titer and productivity. When excess malonyl-CoA is accumulated, the biosensor will turn on the expression of specific gene lacI, which then downregulates the expression acetyl-CoA carboxylase, alleviating toxicity caused by acetyl-CoA carboxylase gene overexpression. On the other hand, when the intracellular malonyl-CoA concentration is low, the biosensor is capable to up-regulate acetyl-CoA carboxylase expression. Therefore, the acetyl-CoA carboxylase can convert acyl-CoA the desired product of fatty acid. This system effectively prevents metabolic overflow, the production of unnecessary proteins during the early stages of fermentation, thereby increasing the production of fatty acid[63]. Similarly, a regulatory sensor based on FadR was developed to facilitate the production of fatty acid–based products in E. coli. The fatty acid/acyl-CoA biosensors were able to response to both exogenous and endogenously produced fatty acids. In this case, when there is sufficient fatty acid, FadR promotes production of acyl-CoA. The fatty acid would be first activated to acyl-CoA which then antagonize the DNA-binding activity of FadR resulting in expression of genes that encode enzymes to produce ethanol, activate more free fatty acid to fatty acyl-CoA, and convert ethanol and acyl-CoA to fatty acid ethyl ester[64]. Another study has developed and demonstrated a malonyl-CoA-responsive biosensor to dynamically regulate fatty acid biosynthesis, optimizing the expression and accumulation of the acetyl-CoA carboxylase gene. This approach alleviates the toxicity and cell growth inhibition associated with gene overexpression, while maintaining high malonyl-CoA production[97]. In a recent study by Peng et al. (2022), novel genetic circuits were effectively employed to regulate GAL promoter-driven heterologous pathways in S. cerevisiae. These engineered circuits enable precise activation and repression of the gene of interest under the control of the endogenous yeast galactose-inducible (GAL) promoter. Through the implementation of these synthetic regulatory circuits, a significant increase in the production of the terpenoid nerolidol was achieved, even in flask cultivation conditions[98]. These successful demonstrations highlight the efficacy of genetic circuitry in enhancing bioproduction. Furthermore, the engineering of promoters represents a global strategy for metabolic control, offering a promising approach to facilitate efficient bioproduction processes[99–102].
Biosensors also facilitates drug-discovery and drug development pipelines
Biosensors have demonstrated their value not only in detection and metabolic engineering but also in diverse research and applications. Over decades, biosensors are used along the drug-discovery pipeline in pharmaceutical and diagnostic industry. The application includes drug target identification, ligand fishing, lead molecule screening and selection, and pharmacokinetic study. In a recent study, Scott and colleagues (2022) conducted research involving the engineering of a biosensor designed to detect HIF-1α/HIF-1β dimerization inhibition, which represents a significant target in anticancer therapies. The biosensor was utilized within a yeast two-hybrid system as a drug discovery platform. The utilization of fluorescence and auxotrophy-based selections enabled the identification of outcomes indicating the occurrence of HIF-1α/HIF-1β dimerization inhibition. Additionally, the research group successfully applied the biosensor to monitor cellular activity and assess potential off-target effects of drugs. The biosensor introduced in this study demonstrates its utility as a valuable drug discovery platform, particularly for the development of protein-based new modality drugs[52]. Wehr and coworkers (2016) reported on the utilization of genetically encoded biosensors to accurately and robustly monitor protein-protein interactions within living cells. The biosensor mechanism relies on protein fragment complementation, wherein the readouts are delivered through fluorescent proteins or luciferases fused to each protein when the split proteins interact. Furthermore, Dong et al. (2021) developed a genetically encoded fluorescent biosensor based on the 5-hydroxytryptamine 2A receptor (5-HT2AR) for serotonin. This biosensor facilitated in vivo detection of endogenous serotonin dynamics and enabled the identification of potential therapeutics[103]. Moreover, Prével and colleagues (2014) also developed the fluorescent biosensors to investigate the behavior of Cyclin-dependent kinases (CDK/Cyclins) both in vitro and in vivo. These biosensors permit the detection and monitoring of CDK/Cyclin abundance and activity, which are central to the regulation of cell cycle progression and other critical biological processes. The CDK/Cyclin biosensors were additionally employed to identify new classes of inhibitors for cancer therapeutics[104]. Additionally, Sato et al. (2009) utilized genetically encoded fluorescent biosensors to detect extracellular signal-regulated kinase (ERK). Through this engineered biosensor, the spatiotemporal dynamics of protein phosphorylation by ERK in the cytosol and nucleus were revealed[54]. The high-throughput nature of biosensors proves advantageous for the screening and selection of functional drug variants, rendering these biosensors efficient tools in the field of drug development. Collectively, these studies underscore the potential of biosensors in drug discovery and emphasize the importance of developing innovative approaches to enhance the efficacy and efficiency of drug development pipelines.
Types of Biosensors
Genetically encoded biosensors can be classified based on the biological material used as a receptor scaffold and the mechanism of sensing. For example, transcription factor-based biosensors typically utilize a transcriptional activator or repressor protein that regulates the expression of reporter genes under a specific promoter. This transcription factor responds to the small molecule of interest through direct or indirect interaction with a ligand-binding domain for the molecule. This interaction leads to changes in the conformation and/or stabilization of the transcriptional complex, promoting or restricting accessibility of RNA polymerase and the transcription process. Many natural transcription factors contain ligand-binding domains that bind to small molecules or metabolites and affect allosteric or other mechanisms of transcriptional regulation. Many of these ligand binding domains have been successfully engineered to bind novel ligands and act as biosensors, regulating gene expression in both prokaryotic and eukaryotic cells[46]. The dynamic response and output of this biosensor can be fine-tuned by changing the position and affinity of the ligand-binding site[105] or by replacing the native promoter with a proximal promoter that is minimally affected by endogenous molecules[55]. Transcription-based biosensors have a wide range of applications, including optimizing and boosting the production of small molecules in eukaryotes, regulating the activity of the Cas9 protein in human cells[106], optimizing chemical biosynthesis and pathway variants in prokaryotes[61], and controlling gene expression in heterogeneous living cells[107].
Nucleic-acid-based biosensors refer to either natural or synthetic nucleotides that can identify complementary DNA or RNA sequences of an analyte, or act as a receptor for a specific biomolecule or target chemical substance. Nucleic-acid-based binding modalities are termed aptamers. The aptamer can selectively bind to a specific target, including proteins, peptides, small molecules, toxins, and living cells. Upon target binding, it changes its tertiary structure such as hairpin shape or a hybridization form delivering the output from target recognition. Aptamers have been applied widely in diagnosis of diseases[108–111], detection of small molecules[112, 113], and monitoring environmental contamination[114, 115]. Furthermore, the aptamers can also be engineered to regulate the formation of mRNA synthesis by binding to a specific molecule. For example, an aptamer specific to tetracycline can be placed at the 5′-splice of RNA to prevents slicing and protein expression in the presence of tetracycline[116, 117]. Moreover, aptamers can be integrated with an expression platform known as a riboswitch. The aptamer acts as a receptor for a ligand or analyte, while the expression platform acts directly on gene expression through its ability to switch between two distinct secondary structures in response to ligand binding[118]. Many riboswitch classes with ability to sense diverse metabolites have been discovered[119] such as guanidine[110], flavin mononucleotide (FMN)[120], S-adenosylmethionine[121], lysine[122], etc.
Immuno-based biosensors, on the other hand, rely on recombinant antibodies to detect an antigen of interest. The antibodies are typically immobilized on a surface that incorporates a reporter for detection and quantification. Numerous studies have shown that the antigen-binding activity and sensitivity of this type of biosensor depend on the orientation of the immobilized antibody[123–126]. Engineered antibodies hold great promise as a biosensor for medical diagnosis[123, 127], as well as therapeutics for diseases[128–130].
Membrane proteins are proteins that naturally reside on the cell membrane. They play a central role in sensing various stimuli. Taking inspiration from this natural sensing mechanism, membrane protein-based biosensors have been developed for numerous applications. These biosensors can be created by either incorporating the membrane protein into a lipid bilayer or expressing it within a cell. The membrane-expressing platform is then linked to a sensing device for output signals, which include fluorescence expression and electrical alterations[131]. An example of such a lipid bilayer-based biosensor is the biological nanopore. These membrane proteins are readily producible, form pores, and can be inserted into a membrane. They have been extensively investigated and utilized for detecting a wide range of organic and inorganic molecules, including nucleic acids. Membrane proteins, such as E. coli outer membrane protein G (OmpG), E. coli curli transport channel (CsgG), E. coli ferric hydroxamate uptake component A (FhuA), and S. aureus α-hemolysin (α-HL), can be engineered as sensing receptors[132]. The sensing mechanism relies on the distinct conductance of ions resulting from the translocation of analytes through the pore[133]. For instance, an α-hemolysin nanopore, combined with an antigen-binding fragment (Fab) of the HED10 antibody, has been developed for nucleic acid detection[134, 135]. Furthermore, membrane protein-based biosensors can also be constructed using G protein-coupled receptors (GPCRs), olfactory receptors (ORs), or OR co-receptors (Orco) embedded within a cell. In the presence of specific analytes, these receptors bind to the analytes and subsequently generate a signal response. This response can manifest as fluorescence protein expression, cellular division in a suitable cellular environment, or the initiation of a signaling cascade within the cell[132].
Similar to membrane proteins, transmembrane channel proteins also reside on the cell membrane. They play an essential role in various physiological responses and multiple intracellular functions across bacteria, archaea, fungi, plants, and animals[136–138]. This sensing process involves controlling the expressed channel receptors on the cell membrane, which can stretch open directly in response to mechanical forces from membrane tension. As a result, molecules and ions are transported through these channel receptors upon stimulation, initiating cellular responses[136–141]. Drawing inspiration from natural mechanosensitive sensing mechanisms, researchers have developed mechanosensitive channel-based biosensors by incorporating circularly permuted green fluorescence protein (cpGFP) into mechanosensitive channel from E. coli. This channel experiences significant conformational changes upon activation by mechanical forces, converting them into intracellular biochemical signals[141]. Moreover, the mechanosensitive channel from E. coli has been adapted to sense changes in osmotic pressure. Furthermore, the mechanosensitive biosensors are successfully engineered and programmed to respond to the external stimulus by inducing gene expression within a synthetic cell[142, 143], controlling drug release from matrices or carriers, and releasing desired contents from vesicle systems[144]. These mechanosensitive biosensors, equipped with functional actuation, provide an invaluable platform and tools for studying and engineering cell signaling, regulating gene expression, and influencing cell behaviors[140].
Protein/enzyme-based biosensors mimic the mechanism of how a protein interacts with its substrate to regulate biochemical reactions, metabolism, and homeostasis. Proteins are the fundamental building block that construct the physiological body of every species. Most proteins or enzymes, as a receptor, bind to specific substrates, namely ions, small ligands, or macromolecules, to maintain dynamic activities within living cells. The diverse function and activities of proteins rely on genetic information and associated three-dimensional structures[145]. Protein-based biosensors can be obtained from a wide range of proteins by rewiring their recognition domains and employed into a variety of host cells[146, 147]. They are often designed to fit a target ligand or metabolite, which subsequently induces a structural or conformational change, or promotes changes of another relevant protein. Protein-based biosensors are versatile tools for many applications and research fields, such as monitoring molecules of interest in the environment, detection and diagnosis tool[148–150], quantifying target molecules[151], monitor intracellular metabolites in microbial hosts[77, 152], and studying biological processes and dynamics within living cells[53, 153, 154].
Protein-based biosensors offer several advantages over other types of biosensors, particularly for metabolic engineering. Firstly, both the sensing and detecting mechanisms of protein-based biosensors occur within the cytosol rather than the nucleus. As a result, genetic information within the nucleus is unlikely to be interfered with. Secondly, protein-based biosensors also act independently of transcription factor activation in nucleus, which leads to a faster response and output signal. For example, changes in the fluorescence yield of protein biosensors through changes in structure or stability usually occur within minutes, while a transcriptional-driven reporters may take hours for transcription and translation to affect a new steady state. Therefore, biosensors that are independent of transcription for their sensing mechanism can allow more quick, accurate and robust observations of changes in metabolism and quantifies of molecules of interest. Thirdly, engineering a protein-based biosensor to sense a specific molecule can be achieved through several approaches for desired properties and suitable performance, such as rational design, directed evolution, de novo design, mutagenesis, DNA shuffling, and recombinant DNA techniques[155, 156]. Therefore, we can improve the sensitivity and selectivity of the protein recognition units for defined target molecules. Lastly, protein-based biosensors can be expressed and employed in many living cells as a heterologous system for simultaneous and continuous sensing in dynamic, living environments. For instance, protein-based biosensors can detect biomarkers in cancer cells[157–159], study protein structural changes[160], and dynamic protein–protein interactions in living cells[161].
Herein we focus on the current state-of-the-art in protein-based biosensor engineering for the detection of small molecules primarily in vivo in a non-destructive manner, utilizing two major approaches: rational protein engineering and evolution-guided engineering (Figure 3).
Figure 3.
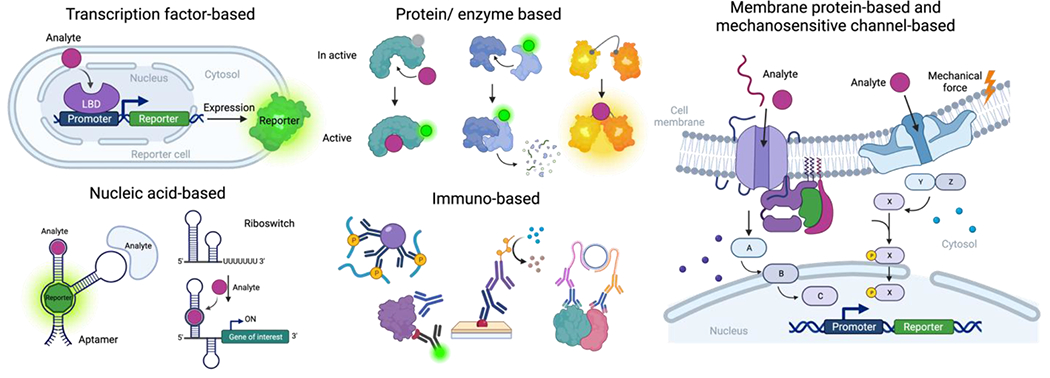
Types of biosensors are classified based on the biological material used as a receptor scaffold and the mechanism of sensing. These classifications include transcription factor-based, nucleic acid-based, protein-based, immuno-based, membrane protein-based, and mechanosensitive channel-based biosensors.
Development of protein-based biosensors
The development of protein-based biosensors for small molecules requires careful consideration of several key elements and attributes. Protein-based biosensors are generally composed of a ligand-binding domain (LBD), a linker, and a reporter domain that provides detection and quantification. The biosensor-analyte coupling generates a signal perceived in the ligand-binding domain that is transmitted through linker and affects a change in the reporter domain. The signal from the biosensor-analyte coupling can be a conformational change of molecule, physiochemical change, the generation of an intermediate, or a change in stability or quantity of the reporter. The reporter domain in addition to this potential for change must also be measurable by optical, electrochemical, or other means. In this review, we will focus on non-destructive in vivo optical reporters that produce signals based on chemical gradients proportional to the intracellular concentration of target molecule. This type of reporters includes fluorescence and chemiluminescence. Fluorescent proteins, such as RFP, GFP, CFP, etc., as well as luciferases are widely used in the genetically encoded biosensor constructs and provide high spatial and temporal resolution. Using the optical modality in the biosensor construct offer several advantages such as direct, and real-time detection of many target molecules, small size and low cost[162].
Several critical factors must be carefully considered when designing protein-based biosensors for small molecules. Ligand specificity, binding affinity, sensitivity, robustness, and biosensor architecture all play crucial roles in biosensor design. Specificity and sensitivity depend primarily on the recognition unit or receptor, which must be carefully engineered to recognize and couple with a particular molecule accurately. Other attributes such as reproducibility, robustness, and stability must also be optimized to obtain high biosensor performance. Reproducibility is the ability of a biosensor to perform consistently under the same operating conditions. The changes of energy and mass, bonding, or interaction between biosensors and analytes are critical to maintain their reproducibility. Furthermore, the stability of the biosensor, specifically its degree of stability to ambient disturbances, especially in the test-case biological system, needs to be considered. In addition, background-level detection, including light reflection from chromophore-containing molecules, organelle autofluorescence, and cellular heterogeneity may also require adjustments to the biosensor design to ensure sensitivity and accuracy of measurement. In summary, designing an efficient protein-based biosensor for small molecules demands thoughtful consideration and optimization of multiple factors. This is necessary to ensure excellent performance and functionality across various systems and use-cases. The following outlines two approaches that can be employed in the engineering of protein-based biosensors:
Rational protein design and engineering
Rational design is a targeted and knowledge-driven approach to constructing protein-based biosensors for small molecules or other protein engineering goals. This approach relies on a deep understanding of the structure-function relationship, including the three-dimensional structure of the protein, its catalytic and/or ligand-binding domains, and other critical regions. Rational design involves the intentional modification of a protein’s sequence/structure based on a detailed understanding of its function, with the goal of improving or altering the protein’s properties. Computational simulations, mathematical models, and molecular modeling techniques, such as maps of electrostatic potential, solvent-accessible surface area, ligand-receptor binding mode prediction, and bonding free energy calculations, are frequently employed to predict key residues or functionally allosteric regions that could enhance the characteristics and functionality of biosensors constructed from proteins[163–166]. In order to implement rational design for protein-based biosensor development, the initial step involves identifying the key residues or regions that modulate the functions of the sensor and reporter domains. Although domain insertions can be advantageous for biosensor construction, they either require a comprehensive understanding of protein structures or demand significant effort to generate all possible insertion sites[167]. Nonetheless, this identification process helps narrow down the possibilities and pinpoint the hotspots for potential protein variants to be examined, effectively reducing the amount of experimental work required. Once the identification of key residues or regions is completed, site-directed mutagenesis techniques, involving the deletion, insertion, or replacement of specific amino acids, can be employed to induce conformational and structural changes in the active binding site, linker, and/or reporter domains. These modifications can enhance the specificity and sensitivity of the biosensor. Following mutagenesis, expression and functional characterization experiments are conducted to evaluate the potential of the protein biosensor. Overall, rational design represents a powerful tool for constructing protein-based biosensors for small molecules, leading to more precise and sensitive detection of specific molecules. This approach combines computational predictions, targeted mutagenesis, and experimental characterization to optimize the performance of protein biosensors.
Numerous studies have demonstrated the effectiveness of rational design strategies in protein engineering. For instance, Solscheid et al. (2015) demonstrated successful engineering of the PstS phosphate binding protein from E. coli for detection of inorganic phosphate (Pi). This was achieved through mutational approaches between the protein’s two lobes, as well as the insertion of glycine in the hinge region to increase protein flexibility, thereby widening the range of substrate detection. Binding of Pi to the protein resulted in lobe rotation, conformational changes, and a response to Pi concentration. Tetramethylrhodamine fluorescence attached to the mutated PstS phosphate receptor serves as a reporter modality sending a readout signals proportional to the Pi concentration (Figure 4A). However, because of the necessity for fluorophore conjugation this biosensor is limited to primarily in vitro use. Furthermore, Chen and colleagues (2015) demonstrated successful rational design of homoserine dehydrogenase (HSDH) of Corynebacterium glutamicum to recognize an unnatural inhibitor L-lysine[168]. HSDH is a natural allosteric receptor for threonine and isoleucine. The binding site of the HSDH protein was modified by mutagenesis to respond to L-lysine using various point mutations and their combinations. These mutations resulted in a larger binding pocket and a more negatively charged environment for L-lysine binding and preventing threonine binding, thereby enabling a novel allosteric regulation with an unnatural molecule. This work demonstrated the re-engineering of protein could be used to detect L-lysine production. Interestingly, Cormann and colleagues (2018) employed rational design techniques to create novel biosensors capable of detecting L-malate, ethylmalonate, and phthalate, which are aromatic compounds. To achieve this, the researchers replaced key ligand-binding residues in the sensory unit of the CitA citrate-binding histidine kinase from Geobacillus thermoleovorans with corresponding residues found in similar histidine kinases that bind different ligands. For large families of ligand binding domains, this study demonstrates that the specificity of these sensing units can be modified through pocket grafting to recognize non-native ligands. Additionally, the research suggests that the flexibility of the protein receptor scaffold in the absence of a ligand reduces entropy costs during ligand binding, allowing the protein to adapt to bind non-native ligands with micromolar affinity[169].
Figure 4.
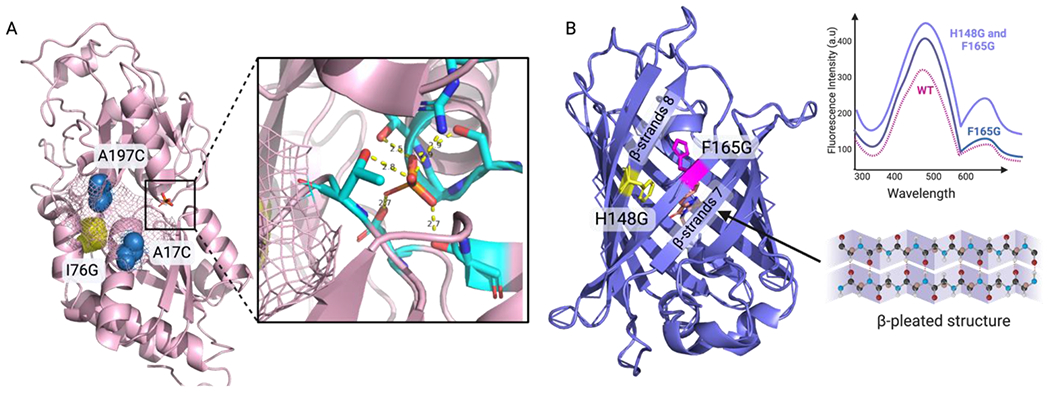
Rational design approach for protein engineering. A) The mutation site in the hinge region (I76G) is presented in yellow and the sites for tetramethylrhodamine fluorescence attachment (A17C, A197C) are labeled in blue. The binding site residues and the interactions with Pi are shown as yellow dashed lines (PDB:1A55). B) The double mutant H148G/F165G in the GFP structure results in a larger cavity, leading to an increase in fluorescence intensity compared to that of WT GFP (PDB:1B9C).
Small molecule biosensors often undergo a dynamic change at the molecular level upon recognizing the ligand within the ligand binding domain that is then translated to the reporter domain. To achieve optimal detection output from protein-ligand interaction, the concept of induced protein stabilization can be utilized [106]. For example, a protein biosensor can be engineered by fusing to a ligand-stabilized ligand binding domain with fluorescence genes or other reporter genes, resulting in a stable and detectable protein when it binds to the target small molecule. Consequently, the binding of the target ligand to the protein biosensor stabilizes the protein structure, enabling the detection of signals from the target ligand[106]. Additionally, mutations in the fluorescent protein structure can induce fluorescence quenching, thereby enhancing the fluorescent intensity or output signal of the biosensor. To achieve a reliable output signal, a fluorescent protein, which serves as a versatile transducer generating observable output, should be strategically placed in a spot or region that is highly sensitive to the ligand of interest[170]. Chesterfield et al (2020) also demonstrated that inserting fluorescent protein into a biosensor protein’s flexible loop without altering the ligand-binding site makes it highly sensitive to local conformational changes and can increase the signal output of the biosensors[171]. Moreover, a library of randomly inserted fluorescent proteins in the ligand-binding domain of a protein can be generated to identify allosteric hotspots and explore the optimal residue for inserting the fluorescent-based biosensor. Additionally, Tansila et al. (2007) presented another strategy to enhance the sensitivity to the ligand of interest and the output signal. They achieved this by by introducing mutations to residues within the inter-strand space of GFP, a region potentially formed during the folding process. This region can be targeted for engineering residues for ligand binding sites without adversely affecting the intrinsic fluorescence and structural integrity of the fluorescent protein[2] (Figure 4). Site-directed mutagenesis, based on rational design, has also led to the production of high-affinity enhanced green fluorescent protein (EGFP) variants targeting endotoxin binding motifs in gram-negative bacteria[170]. Through engineering their barrel structures, a wide range of fluorescence proteins spanning from blue to red wavelengths have been extensively employed in various applications[172, 173].
The rational design approach is a valuable tool for predicting and comprehending the molecular structure, as well as the interactions between ligands and protein receptors acting as biosensors. These cost-effective designs, facilitated by computational aids and mathematical models, enable the identification of protein domains, allosteric hotspots, and the simulation and prediction of molecular dynamics. Predicted variants, via substitution, insertion, or deletion, can enhance the ligand specificity, sensitivity, and overall function of a protein biosensor. However, changes in amino acid residues in the protein structure may sometimes make it challenging to predict the protein’s overall function. Therefore, identifying allosteric binding sites or critical residues requires careful consideration. It is crucial to experimentally investigate the effect of predicted mutations using methods such as site-directed mutagenesis and appropriate screening techniques. In doing so, a structure-function relationship of protein variants can be established. By leveraging existing structural knowledge and functional insights gained through rational engineering approaches, a proof-of-concept for further engineering biosensors for molecules of interest can be established.
Evolution-guided engineering
The advancements in synthetic biology and metabolic engineering has made it possible to reprogram organisms, creating new metabolic pathways that allow for efficient chemical synthesis using the modified chassis. Directed evolution aims to generate massively diverse populations of chassis organisms and apply artificial selective pressure to this population to select for the desired organismal phenotypes, metabolite characteristics, and pathway dynamics. Directed evolution is also a bottom-up approach for accelerating the development of protein biosensors. Directed evolution has been demonstrated to achieve the desired phenotype based on the collaboration between design choices and the proteins’ evolved nature, limiting the design decisions and introducing significant random chance into the development process, as opposed to rational design in which computational algorithms make pointed choices about specific variants to test[174, 175]. Frances Arnold won the Nobel Prize in Chemistry for successfully using directed evolution to obtain a better version of an enzyme. Specifically, mutations were introduced into the gene encoding the peptide-cleaving enzyme subtilisin E. Those gene variants were then introduced into bacteria, Bacillus subtilis, which are naturally not able to cleave peptide bonds in organic solvents. The cells carrying randomly mutated versions of the subtilisin E coding sequence were cultured on solid media containing organic solvents and milk proteins. This enables the identification of colonies capable of cleaving milk proteins, leaving clear spots on the milky plates, acting as the artificial selective pressure. With several rounds of in vitro evolution, mimicking natural evolution by generating genetic variation, and applying a selective pressure for the most fit, highest function variants (in terms of ability to cleave milk proteins in the presence of organic solvent), an enzyme was generated that worked 256 times better in organic solvents [176–179].
The strategy inspired by natural evolution involves creating a library of biosensor variants and carefully selecting the desired molecular characteristics. Directed evolution for protein-based biosensors involves mutating a native receptor, mainly at the ligand binding site, using methods like error-prone PCR, site-directed mutagenesis, and DNA shuffling, to generate a diverse variant pool[180]. Additionally, we can link the functions of these two domains by randomly inserting a coding sequence for a reporter domain into the receptor domain or vice versa. Circular permutation of one or both coding sequences is another strategy that can be employed for the same purpose.[181, 182]. The library of variants of a candidate biosensor design is then introduced into living cells. Living cells expressing the protein variants are grown in the presence and absence of the small molecule of interest and cells demonstrating the desired output behavior are enriched. Although the variations are introduced randomly, the selection process directs the evolution of the protein towards the desired outcome. With rounds of diversification and selection, typically through a rapid high-throughput screening or growth selection, the variant population that interacts properly with its ligand to induce changes in reporter signal is enriched, while the variants that do not properly bind to the ligand or do not couple the binding event with output signal are reduced. The selected variants can then be subjected to further structural and functional analysis as well as additional rounds of directed evolution. Molecular docking can be performed to identify the binding modes and understand the structural properties that govern specific interaction between the small molecule and biosensor. In this way, directed evolution can inform future rational design approaches, and likewise rational design can inform variant library design for directed evolution.
Directed evolution strategies may uncover non-intuitive mutations or other variation in genes to improve the function and performance of the encoded protein, in terms of binding affinity, selectivity, catalytic activity, operational range, tolerance to the selective pressure in a new environment, or other functions. This becomes a starting point of adaptive and novel functions. Although this evolutionary method is based on a different perspective theory and methodology from rational design, it may actually prove to be an efficient alternative. This is particularly true if there are specific and sensitive screening and selection methods available for the desired functions. Successful directed evolution for engineering a protein-based biosensor requires careful consideration of several factors: selecting appropriate mutagenesis methods, determining strategic locations for changes (e.g., throughout the protein, in the active site region, or at flexible sites), choosing a suitable host for gene expression, and employing sensitive methods to screen and select the desired protein.
In recent years, directed evolution has emerged as a promising strategy particularly for enhancing the substrate/ligand specificity of biological molecules, as evidenced by numerous successful applications of protein engineering. For instance, Sneok et al. (2019) presented a high-throughput method for evolving allosteric transcription factors of E. coli and transferring the output functions to a eukaryote, Saccharomyces cerevisiae. The transcription factor, BenM, was evolved to change its ligand specificity, dynamic range, inversion-of-function, and operational range, resulting in a ligand-specific biosensor for muconic acid that showed great promise for the development of biosensors[183] (Figure 5). Similarly, Ambrosio et al. (2020) successfully employed directed evolution to develop the VanR transcription factor, which exhibited improved responsiveness to vanillin and reduced affinity for the native ligand effector, vanillic acid[105]. In another example, TtgR transcriptional repressor from Pseudomonas putida coupled to a green fluorescent reporter was engineered by directed evolution to detect wide range of antibiotics and other organic compounds[184]. Ogawa et al. (2019) similarly utilized random mutagenesis library construction and directed evolution techniques to engineer a transcriptional activator, XylS, derived from Pseudomonas putida. The aim was to create variants that could more efficiently recognize p-toluic acid while displaying minimal response to m-toluic acid. These efforts demonstrate the potential of XylS variants as highly specific biosensors capable of accurately detecting desired ligands[185]. Likewise, Brandsen et al. (2018) have developed biosensors in E. coli that act through ligand-dependent stabilization of a transcriptional repressor, LacI. Using error-prone PCR mutagenesis of lacI and subsequent selection, the researchers acquired a biosensor with multiple mutations that allowed for the detection of isopropyl β-D-1-thiogalactopyranoside (IPTG) as well as D-fucose. The study further demonstrated that a single mutation in the LacI biosensor is sufficient for achieving selectivity and biosensor activity. Overall, this research highlights the potential of mutagenesis and selection strategies for the development of biosensors with enhanced specificity and sensitivity[186]. Furthermore, Tang Rui-Qi et al. (2020) utilized directed evolution techniques to engineer a novel biosensor capable of detecting a wide range of xylose concentrations. The team focused on XylR, a transcriptional activator derived from E. coli, and successfully created a series of mutant xylose-responsive biosensors and showed that the biosensors containing the mutant XylR exhibits an increase operational range by nearly 10-fold comparing with the control. These biosensors were also employed to regulate the expression of various genes, including those involved in lycopene biosynthesis, by utilizing xylose as an inexpensive inducer. As a result, the biosensor system demonstrated a marked increase in lycopene production in E. coli compared to the wild-type XylR[187]. Bali et al. (2018) successfully identified and obtained mutant libraries of nicotinamide riboside transporters, PnuC, that accept thiamine as a new substrate using directed evolution. This finding highlights that identifying a single specific residue could yield a larger change in specificity and function of the target protein biosensor[188]. Moreover, Machado et al. (2019) demonstrated directed evolution of the PcaV allosteric transcription factor, which changed ligand specificity toward vanillin and other aromatic aldehyde effectors. Their study resulted in the design of a biosensor derived from a directed evolution strategy[189]. Similarly, Kasey and colleagues (2017) utilized directed evolution to engineer the transcription factor MphR for the detection of macrolides produced by different mutant enzymes, pathways, and strains. The resulting MphR biosensor variants showed improved detection capabilities for in-cell macrolide production, demonstrating the potential of directed evolution for drug discovery and development applications[78]. In a recent study conducted by Flachbart and their colleagues (2021), a series of highly specific biosensors capable of detecting different target molecules, including 4-hydroxybenzoic acid, p-coumaric acid, 5-bromoferulic acid, and 6-methyl salicylic acid, were developed. The team employed directed evolution techniques to evolve the transcriptional repressor HcaR towards a more relaxed-ligand specificity. As a result, several HcaR variants were successfully engineered, each with unique ligand specificities. This approach has the potential to expand the repertoire of biosensors available for detecting a diverse range of target molecules[190]. Liu et al (2017) carried out successful random mutagenesis on the TyrR1 transcription factor and generated a biosensor capable of detecting intracellular L-phenylalanine (L-Phe). This biosensor facilitated the directed evolution of L-Phe hyperproducing strains[74]. Additionally, d’Oelsnitz et al. (2022) have developed genetically encoded, generalist monoterpene biosensors using directed evolution techniques. The sensory unit of these biosensors is based on the camphor-responsive TetR-family transcriptional regulator CamR from Pseudomonas putida. By subjecting the CamR biosensors to directed evolution, the effector specificity of the sensors was expanded to recognize four distinct monoterpenes: borneol, fenchol, eucalyptol, and camphene[85]. Since many monoterpenes are toxic to microbes, the developed biosensors can also aid in balancing expression levels to reduce the metabolic load of heterologous pathways. The above repertoire of biosensors provides a valuable toolbox for facilitating metabolic engineering applications. Overall, this research demonstrates the potential of directed evolution in expanding the range of biosensors for applications in metabolic engineering. However, the above biosensors are all based on re-engineered transcription factors, while providing signal amplification through transcription and translations, are also limited to the associated hour-long timescale and cellular resolution.
Figure 5. Evolution-guided engineering of transcriptional factor as small molecule biosensors.
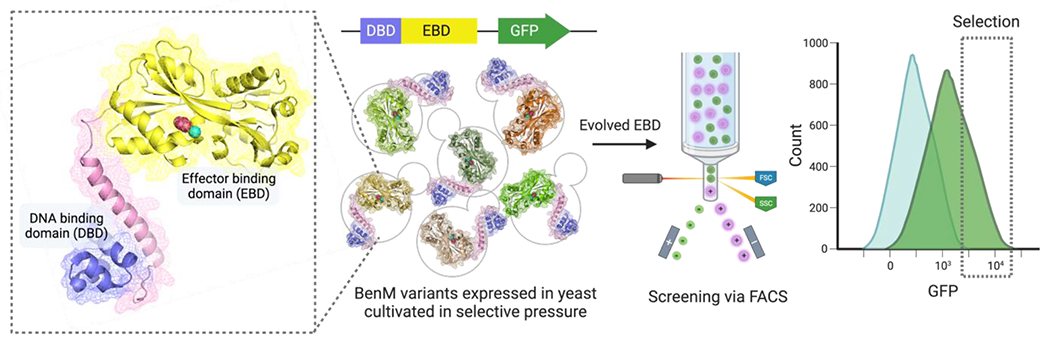
The DNA-binding domain of the BenM transcription factor is evolved to have a broader operational range for cis,cis-muconic acid and the new ligand, adipic acid. Yeasts expressing these BenM variants are cultivated in the absence of ligands, with muconic acid, and adipic acid as inducers or stressors. FACS is employed in the screening and selection of variants with specificity towards both muconic acid and adipic acid.
For environmental research and applications, coarse-grain time and space resolution are often sufficient. Towards environmental applications, Cai Y and colleagues (2022) have demonstrated the potential of using cadmium resistance transcriptional regulatory (CadR) as a sensing unit by expressing it in bacteria. To improve the performance and sensitivity of the CadR biosensor for the detection of cadmium in environmental samples, the researchers generated mutant libraries using error-prone PCR and subjected them to five rounds of directed evolution facilitated by fluorescence-activated cell sorting (FACS). The resulting biosensor derived from directed evolution exhibited superior performance compared to the initial construct, thereby providing a valuable tool for sensitive and specific detection of cadmium in real-world applications[191]. Zimran et al. (2022) have developed a biosensor for detecting a diverse range of herbicide ligands by utilizing abscisic acid receptors as a potential scaffold. The research team employed directed evolution techniques to screen hundreds of receptor variants for activation by various groups of herbicides, leading to the identification of improved herbicide receptors. This study further demonstrated the utility of expressing the receptor variants in a yeast two-hybrid platform, which enabled functional selection by FACS. The isolated receptors exhibited a low nanomolar sensitivity and a broad dynamic range in detecting a ubiquitous group of chloroacetamide herbicides[192]. This research highlights the potential of directed evolution in improving biosensor performance and its application in detecting environmental contaminants.
Overall, directed evolution has shown great potential in enhancing the substrate specificity and function of biological molecules, which can have significant implications in the development of biosensors and other biotechnological applications. The knowledge and logic of directed evolution is a versatile alternative to rational design that can be effectively used to solve complex systems related to substrate specificity of a biological molecule. Directed evolution allows enhancing key enzyme activities in a biosynthetic pathway, and biosensors specific for the target product aid in high-throughput screening/selection and improving biosynthesis of target molecules. Directed evolution often allows rapid identification of several variants with greater activity/function than the parental sequence. It also allows us to discover novel protein structures to accommodate a variety of molecules.
A collaborative approach is the best approach
Current advancements in knowledge and technology have enabled the utilization of both rational design and directed evolution as complementary strategies for protein design, including the creation of biosensors (Table 1). Rational design has emerged as a powerful tool for designing novel protein structures; however, incomplete knowledge of underlying protein structure, hidden factors, and mechanisms of action can lead to unsuccessful attempts. Additionally, predicting the higher-ordered structure of the protein or focusing only on specific amino acids or domains can cause rational design methods to overlook important considerations that lead to protein function failure. On the other hand, directed evolution, which relies on the theory of evolution by natural selection, can enhance the selectivity and sensitivity of biosensors without requiring a comprehensive understanding of protein structure and how it is related to function. While some amino acids may change through the evolutionary process, significant changes in protein structure for improved function may come from alterations in the backbone or core structure. However, the astronomical size of possible amino acid changes and rearrangements of the possible backbones of protein biosensors are limitations of directed evolution along with the need for extreme-throughput screening and selection methods to identify the desired properties in a large pool of evolved protein sequences. To address this challenge, smaller more cost-effective variant libraries guided by rational design can be created based on knowledge of protein structure and function and subjected to artificial selections. These more targeted libraries contain fewer variants, reducing the need for many high-throughput screening and selection experiments compared to traditional random mutagenesis. Therefore, to achieve an effective protein-based biosensor most efficiently, it is advisable to carry out rational design and directed evolution approaches in parallel (Figure 6).
Figure 6.
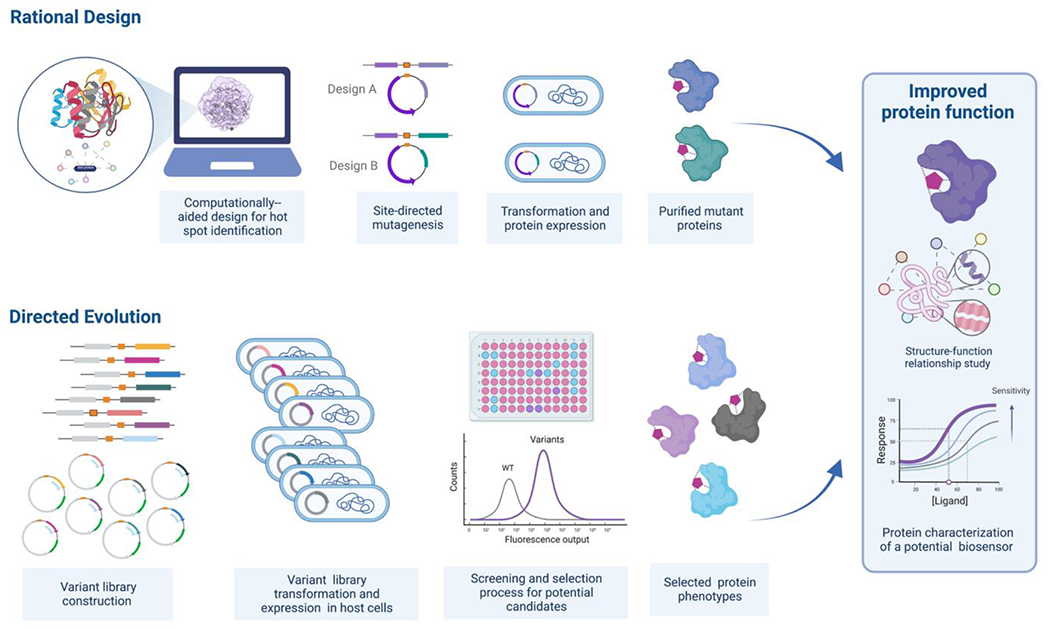
A schematic illustrating the engineering of a protein biosensor, employing both rational design and directed evolution approaches. Rational design relies largely on crystal structure datasets, along with biophysical information, protein structure-function relationships, ligand-receptor interactions, and other measurements and predictions, to guide design, in this case, of site-directed mutagenesis. This approach primarily involves computational modeling for hypothesis generation. Conversely, evolution-guided engineering mimics the natural selection process. The gene of interest undergoes iterative rounds of random or potentially targeted mutagenesis to generate variant libraries. The resulting variants are then screened and selected for specific functionality using screening and selection methods to measure and/or select for greatest desired protein function.
Several examples of the complementary application of rational design and directed evolution now exist in biosensor design. Taylor et al (2016) successfully engineered an allosteric transcriptional factor, LacI from E.coli, to respond to a new molecule using both computational protein design and directed evolution approaches[198]. Overall, the complementary use of these strategies is essential to develop highly efficient biosensors. Moreover, Chi and colleagues (2022) have developed a biosensor for chloride detection based on the proton-pumping rhodopsin wtGR from Gloeobacter violaceus. The researchers first introduced a single point mutation (D121V) at the Schiff counterion position of wtGR fused to a fluorescent protein (CFP). They then explored and engineered the rhodopsin sequence space to improve its function through coevolutionary analysis with directed evolution. The iterative selection process resulted in additional mutations to the rhodopsin receptor domain resulting in an improved dynamic range for extracellular chloride[199]. In their recent study, Li and colleagues (2022) developed a biosensor for detecting phthalic acid (PA) and terephthalic acid (TPA), which are the primary hydrolysis products of plastics and plasticizers. The biosensor utilized the organic acid responsive transcription factor XylS, which was subjected to rational design and directed evolution to generate novel variants. Site-directed saturation mutagenesis was employed to target residues located around the active site, resulting in XylS mutants with enhanced specificity and responsiveness towards TPA and PA ligands[200]. These studies highlight the effectiveness of the rational design and directed evolution framework as an efficient methodology for engineering biosensors with desired properties.
Protein design and engineering hold significant importance in the realm of metabolic engineering and various research areas. We can utilize rational design for site-specific mutagenesis and/or learning from nature through directed evolution to enhance protein function or craft novel protein traits[201]. Nevertheless, both approaches depend on the pool of generated and analyzed protein mutants. The ultimate protein is acquired via screening and selection from the available choices within the library. A larger and more specific library increases the likelihood of obtaining the desired protein compared to a smaller, more random library. However, it is time-consuming for screening or selection of a large mutational sequence space. Recently, computational and machine learning algorithms have played a crucial role in narrowing down, identifying, and prioritizing hotspots from libraries. They also predict the potential effects of numerous mutations during the process of customizing proteins for various applications. Several commercially available software can aid in rational protein design, particularly in predicting 3D protein models; these include Rosetta and Modeller. Additionally, many protein-ligand docking programs are available, such as AutoDock, LeDock, MOE-Dock, GOLD, and FlexAID, each with different algorithms for calculating and predicting interactions. Moreover, GROMACS and CHARMM are valuable tools for conducting molecular dynamics simulations and free energy calculations. Furthermore, FireProt, FoldX, UniRep, and PredictSNP offer essential features for predicting protein stability and the impact of mutations. Notably, computer software such as ScaffoldSeq, Proseeker, and SCHEMA facilitates directed evolution by simulating natural selection processes on a screen, notably diminishing experiment duration from months to mere days. Recent research had utilized machine learning algorithms to predict the optimal design of variant libraries from sequence-function datasets. This approach eliminates the requirement for a profound understanding of deep structure-function relationships, thereby accelerating the process of directed protein evolution[202, 203] and the development of biosensors[204] as reviewed by Chu et al in 2021[205]. Collectively, research demonstrated the potential of combining these approaches with computational tools to create biosensors with enhanced functionality and specificity. Overall, employing software and in silico tools as a data-driven approach to design and reengineer proteins enables us to effectively achieve the desired protein characteristics.
Protein-based biosensor to control cellular protein abundance and metabolism through degradation
Previous dynamic metabolic controllers mostly based on transcriptional regulation have shown increased yields of engineered biosynthetic pathways.[206, 207]. However, they are limited by several-minute delay and cellular spatial scale of transcription and translation[208]. Interestingly, protein-based biosensors, which couple a specific molecule and subsequently induce protein degradation, present an attractive solution for regulating and fine-tuning protein levels, as well as for studying the effects of loss-of-function characteristics[209–211]. This protein degradation acts as a metabolic controller, providing tight and precise control of signal transduction and protein levels. Although many protein-based biosensors as metabolism controllers have not been extensively exploited, they have the potential to regulate cellular metabolism and overcome the limitations of transcriptional-based controllers.
In bacteria, the protein degradation system plays a crucial role in naturally regulating and maintaining protein homeostasis. The degradation system involves bacterial proteases and specific amino acids called degrons that are attached to the target protein. The degrons act as recognition domains, enabling the degron-protein complex to be recognized by proteases and subsequently degraded by the protein degradation machinery. Bacterial proteases typically regulate intracellular protein levels through their diverse binding affinities for different degrons [43]. Taking inspiration from this natural system, a protease-degron-induced degradation system has been employed to regulate protein levels in both prokaryotes [13,14] and eukaryotes [15–17]. By tagging the protein of interest with a degron, it provides a strategy to control and fine-tune the degradation rates of the protein.
In metabolic engineering, degron-induced degradation systems are often employed by tagging degrons to target proteins that exhibit high expression levels in host cells to reduce the burden of excessive protein accumulation, with the goal of maximizing production yield. These synthetic circuits alleviate the cellular burden caused by the accumulation of diverse proteins and address imbalances in mass-energy equilibrium within the cell [28–31]. However, it is important to acknowledge that the effectiveness of this synthetic degradation system may vary depending on factors such as strains, cell growth rate, protease saturation, and protein turnover rate. For example, the impact on protein levels by the degradation system will be more pronounced for highly expressed proteins as compared to those with slower expression and lower abundance in the cell.
Degrons can either exist inherently within the protein sequences [43] or be added to the C-terminal [20] or N-terminus of a target protein, allowing degradation by various protease proteolytic complexes. Among the native degrons, the ones derived from E.coli SsrA (ec-ssrA) and its variants are the most commonly exploited ones to date[212–214]. SsrA has been utilized in studying protein kinetics, such as protein degradation and turnover rate[215–217]. The SsrA degron is attached to the C-terminal of a defective protein resulting from translational errors, which is then recognized and degraded by the ClpXP protease[218–220]. The selectivity of the target protein for degradation is determined by the efficiency of the initial binding affinity, which can be adjusted by an adaptor protein. One such adaptor protein is SspB, a chaperone that has been engineered to connect target proteins to ClpXP. The target protein is subsequently transported to the ClpXP proteolytic complex for degradation[221–223].
Apart from E. coli, the degron tagging system has been employed in other bacterial species[209, 211, 224–227]. However, the specificity to the ClpXP protease is low in bacteria, resulting in recognition by other proteases, which hampers the ability to tightly control degradation. Additionally, the SsrA tag does not provide inducible control over degradation. Therefore, more robust circuits are required for applications in bacterial or other living systems. Various approaches have been developed to enhance robustness and specificity in degradation, such as utilizing hybrid tags with cleavage sites identified by proteases[228] and utilizing natural SsrA variants[214, 229]. Cameron and Collins (2014) developed a tunable control of protein degradation based on components of the Mesoplasma florum tmRNA system. The M. florum ssrA tag (mf-ssrA) is specifically degraded by its protease called mf-Lon, but not by E. coli ClpXP. The authors demonstrated that mf-Lon-mediated protein degradation and the mf-ssrA variant tags exhibited temporal degradation dynamics. The study emphasized that targeted protein degradation depends not only on the target protein and the mf-ssrA variant tag, but also on mf-Lon expression levels[214]. Therefore, expanding the repertoire of degron sequences and inducible proteolytic complexes that can be integrated into the genome will provide a diverse orthogonal targeted degradation system that is tunable and robust in controlling intracellular protein levels. The protein degradation system enables the regulation of protein abundance in a cell without interfering with transcriptional and translational regulation. In the field of metabolic engineering research, the protein degradation system has been applied to control the abundance of specific enzymes in biosynthesis pathways. Brockman and Prather (2015) demonstrated inducible degradation of phosphofructokinase-I (Pfk-I) in E. coli by tagging the SsrA degron to the coding sequence of Pfk-I and knocking out the native copy of the adaptor protein chaperones SspB. This strategy enables rapid changes in the steady-state level of Pfk-I, resulting in increased myo-inositol product yield[230].
Apart from C-terminal tagged degrons, N-terminal fusions have also been developed to expand options for inducible protein degradation. In this approach, a cleavable degron is fused N-terminally to the target protein, resulting in the degradation of the target once the N-degron is cleaved. Genetically encoded N-terminal modifications, along with ClpP protease, have shown high degradation rates for proteins such as mCherry and beta-galactosidase[231]. This strategy increases the range of proteins that can be targeted since some proteins require a free C-terminus for activity[232]. Moreover, Liu et al. (2017) introduced the prokaryotic N-terminal targeted proteolysis system. The degrons were exploited and obtained from the LexA-like regulator HdiR of Streptococcus mutans. The N-degron tags showed highly efficient constitutive proteolysis of the target proteins. The N-terminal degron tag could also have its activity modulated by fusing the degron to the ubiquitin-like protein NEDD8 (NEDD8-degron tag). This study provided evidence that the degron can be effectively regulated or activated through fusion with a third-party protein. Additionally, the system exhibited a high degree of engineering flexibility, enabling open manipulation and modification of the system [233].
Another approach with great potential for targeted intracellular protein degradation involves the use of adaptor proteins to specifically target substrates. Davis et al. (2011) developed an engineered rapamycin-dependent split-adaptor system in E. coli. In this system, the assembly and delivery of protein targets to the ClpXP protease by the adaptor protein are induced by the small molecule rapamycin. The ssrA degron tag was mutated from LAA to DAS and tagged to the target protein. Rapamycin drives the assembly of a functional SspB adaptor, which delivers the DAS-tagged substrates to ClpXP for degradation (Figure 4A). It was demonstrated that the target protein (GFP) degraded rapidly within a timeframe of seconds to minutes. Furthermore, in the presence of rapamycin, the LacI repressor and the essential cell-division protein FtsA were degraded by ClpXP, resulting in pronounced filamentation. However, the normal morphology was restored after rapamycin removal. This split-adaptor system provides controllable and reversible protein degradation in a rapamycin-dependent manner[213]. To our knowledge, this rapamycin-induced proteolysis is the only means of engineering small-molecule-induced direct (transcription-independent) proteolysis in prokaryotes, although there may be some advances beyond transcription-independent systems in prokaryotes in the near future[234, 235].
In a somewhat related application, induced protein degradation has recently been used to relieve catabolite repression and induce expression from GAL promoters in yeast, while still allow growth on glucose[236]. This study used auxin-induced protein degradation via the ubiquitin–proteasome pathway to induce degradation of the Mig1p glucose-dependent repressor. Auxin-induced protein degradation, also known as AID, has been shown to function in many protein contexts and many eukaryotic systems, from yeast and animal cells[237], to worms[238], mice[239] and various types of cell line[237, 240]. AID is an example of a chemically activated ubiquitin ligase where the chemical auxin, or indole-3-acetic acid, acts as a molecular glue facilitating the recruitment of degron-tagged proteins to the auxin-receptor-containing SCF ubiquitin ligase, which polyubiquitinates the degron-tagged proteins, targeting them for degradation (Figure 4B). The AID system is considered a reversible protein depletion system in the presence of auxin, where the degradation of the target protein can be reverted by removing auxin. This system allows us to efficiently, specifically, and rapidly control the level of the target protein[240]. Engineered variants of auxin and its receptor have been developed to avoid the wide-ranging nature of auxin[241].
The implementation of a chemically activated ubiquitin ligase as a controller offers a promising solution for achieving real-time, cell-specific balancing of metabolic pathways. This idea is inspired by plants, which contain several chemically activated ubiquitin ligases to control various aspects of growth, development and behavior[242]. Engineered chemically activated ubiquitin ligases could interact with target metabolites and associated enzymes and transcriptional regulators to mitigate the production and accumulation of toxic intermediates in the host cells. This would require engineering novel chemically activated ubiquitin ligases and degrons that exhibit the desired chemical specificity through rational design or directed evolution techniques, as mentioned earlier. Induced protein degradation is a precise control mechanism enabling efficient modulation of the target enzymes, without the delay associated with transcriptional repression. Moreover, degrons could also be fused to transcriptional activators or repressors and employed to regulate the expression of other enzymes in the pathway, facilitating improved conversion of metabolites into the desired end product. This dynamic metabolic controller thus enables simultaneous negative and positive regulation. By leveraging biosensors based on the ubiquitin-proteasome system, more sophisticated synthetic regulatory networks can be established within living cells, enabling precise degradation of transcription factors, triggering the degradation of unwanted proteins or enzymes in various applications. However, most current systems for chemically induced protein degradation are limited to the ubiquitin–proteasome system of eukaryotic chasses.
By utilizing protein degradation as a control modality, it is possible to exert control over cellular proteasome and metabolism on a sub-minute timescale and with sub-cellular spatial resolution. Metabolic controllers based on induced protein degradation have the potential to overcome issues related to the accumulation of inhibitory or toxic intermediates and the heterogeneity of cellular states and environments by preventing accumulation of causal enzymes through negative feedback control. Protein degradation systems offer the advantage of transcriptional independence over other exogenous approaches for modulating protein activity, such as transcriptional repression[243], RNAi[244, 245], CRISPR/Cas9 and/or dCas9[246, 247], toggle switches[244, 248]. Knocking out, silencing, or turning off protein-encoding genes often disrupts cognate genes, leading to slow growth phenotypes and deleterious effects on cell metabolism[240]. Genetically encoded, inducible protein degradation system provides an alternative approach to effectively controlling intracellular proteins, balancing metabolic flux, studying gene and protein function, and is applicable in wide-ranging research fields.
Conclusion
Genetically encoded biosensors allow precise and timely assessment and control of intracellular metabolism. While rational design and evolution-guided engineering both offer effective strategies for biosensor development, the combination of both approaches proves essential for optimizing the functionality of biosensors. These two distinct methodologies, operating towards a shared objective, provide a cost-effective means of constructing proteins with desired functions. The Test phase within the Design-Build-Test-Learn cycle of engineering biological systems can be executed more expeditiously and efficiently through the utilization of biosensors. Biosensors not only serve as tools for detection and quantification in biotechnology but also can be implemented as controllers of metabolism and cellular states. Consequently, biosensors are critical aids in maximizing the production of metabolites or chemicals by living organisms, thereby promoting green alternative approaches for industrial applications. In the near future, biosensors are anticipated to play an increasingly essential and widespread role across various fields. To fully capitalize on biotechnology’s potential for enhancing manufacturing sustainability, it is crucial to explore novel designs and modalities for biosensors.
Figure 7.
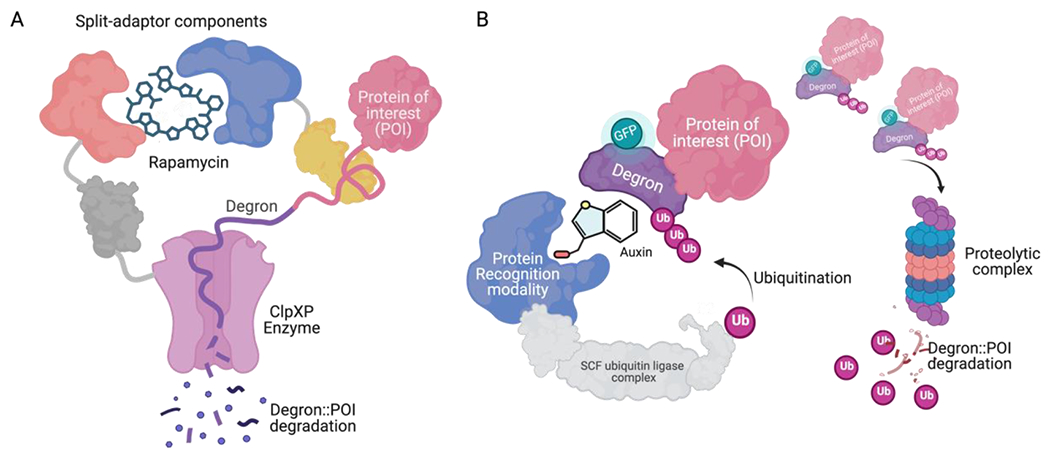
Examples of a small molecule-induced protein degradation system. The systems offer an efficient, specific, and rapid strategy to regulate the level of the target protein within the cell. A) Rapamycin-induced protein degradation: The split-adaptor system is engineered to assemble and deliver degron-tagged substrates to the protease in a rapamycin-dependent manner. B) Auxin-induced protein degradation: In this system, the degron-tagged protein is recruited by the auxin-receptor-containing SCF ubiquitin ligase, leading to ubiquitination and degradation of the IAA degron-tagged protein of interest (POI) by the proteasome. As a result, the intracellular level of POI is depleted in an auxin-dependent manner.
Table 2.
Comparison table of rational design and directed evolution strategies for protein engineering as a biosensor[156, 180, 193–197]
| Comparison | Rational design | Directed evolution |
|---|---|---|
| Process | Top-down | Bottom-up |
| Comprehensive knowledge of protein structure related to the function | Required | Not required |
| Mutagenesis/ mutation pattern | Site-directed | Random |
| Method/platform used to create the mutation | Molecular simulation/Computational design, site directed mutagenesis | Error-prone PCR, Saturation mutagenesis, genetic drift libraries, amino acid replacement, insertion or deletion of residues, etc. |
| Time required for mutation and obtaining desired protein | Minutes to hours | Hours to days |
| Specific domain engineering | Feasible | Less feasible |
| Higher-ordered structure modification | Not feasible | Feasible |
| Optimization of desired properties | Very much required | Required |
| Cost | Less | More |
Acknowledgments
Research in the Wright Plant Synthetic Biology Laboratory: the United States Department of Agriculture National Institute of Food and Agriculture (USDA-NIFA), Agriculture and Food Research Initiative (AFRI) Plant Breeding for Agricultural Production Grant No. 2022-67013-36293 and Hatch Project [VA-1021738]; Virginia Space Grant Consortium; Virginia Tech Institute for Critical Technologies and Sciences Junior Faculty Award; and Virginia Tech College of Agriculture and Life Sciences Strategic Plan Advancement 2021 Integrated Internal Competitive Grants through the Center for Advanced Innovation in Agriculture. Additional support for P.C. was provided by the Ministry of Science and Technology, Royal Thai Government. BioRender was used to create all figures.
Footnotes
Disclosure of competing interests
All authors declare that they have no known competing financial, personal or profession interests that might influence the work reported in this paper.
References
- [1].Solscheid C, Kunzelmann S, Davis CT, Hunter JL, Nofer A, Webb MR. Development of a reagentless biosensor for inorganic phosphate, applicable over a wide concentration range. Biochemistry. 2015;54:5054–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Tansila N, Tantimongcolwat T, Isarankura-Na-Ayudhya C, Nantasenamat C, Prachayasittikul V. Rational design of analyte channels of the green fluorescent protein for biosensor applications. International journal of biological sciences. 2007;3:463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Nielsen J, Keasling JD. Engineering cellular metabolism. Cell. 2016;164:1185–97. [DOI] [PubMed] [Google Scholar]
- [4].Budin I, Keasling JD. Synthetic biology for fundamental biochemical discovery. Biochemistry. 2018;58:1464–9. [DOI] [PubMed] [Google Scholar]
- [5].Ward VC, Chatzivasileiou AO, Stephanopoulos G. Metabolic engineering of Escherichia coli for the production of isoprenoids. FEMS microbiology letters. 2018;365:fny079. [DOI] [PubMed] [Google Scholar]
- [6].Das M, Patra P, Ghosh A. Metabolic engineering for enhancing microbial biosynthesis of advanced biofuels. Renewable and Sustainable Energy Reviews. 2020;119:109562. [Google Scholar]
- [7].Yuan S-F, Alper HS. Metabolic engineering of microbial cell factories for production of nutraceuticals. Microbial cell factories. 2019;18:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Blazeck J, Miller J, Pan A, Gengler J, Holden C, Jamoussi M, et al. Metabolic engineering of Saccharomyces cerevisiae for itaconic acid production. Applied microbiology and biotechnology. 2014;98:8155–64. [DOI] [PubMed] [Google Scholar]
- [9].Tariq H, Asif S, Andleeb A, Hano C, Abbasi BH. Flavonoid Production: Current Trends in Plant Metabolic Engineering and De Novo Microbial Production. Metabolites. 2023;13:124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Gu Y, Jiao X, Ye L, Yu H. Metabolic engineering strategies for de novo biosynthesis of sterols and steroids in yeast. Bioresources and Bioprocessing. 2021;8:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Stephanopoulos G, Aristidou AA, Nielsen J. Metabolic engineering: principles and methodologies. 1998. [Google Scholar]
- [12].Liu J, Wu X, Yao M, Xiao W, Zha J. Chassis engineering for microbial production of chemicals: from natural microbes to synthetic organisms. Current Opinion in Biotechnology. 2020;66:105–12. [DOI] [PubMed] [Google Scholar]
- [13].Ke J, Yoshikuni Y. Multi-chassis engineering for heterologous production of microbial natural products. Current opinion in biotechnology. 2020;62:88–97. [DOI] [PubMed] [Google Scholar]
- [14].Voigt CA. Synthetic Biology: Methods for part/device characterization and chassis engineering: Academic Press; 2011. [Google Scholar]
- [15].Calero P, Nikel PI. Chasing bacterial chassis for metabolic engineering: a perspective review from classical to non-traditional microorganisms. Microbial Biotechnology. 2019;12:98–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Nielsen J, Larsson C, van Maris A, Pronk J. Metabolic engineering of yeast for production of fuels and chemicals. Current opinion in biotechnology. 2013;24:398–404. [DOI] [PubMed] [Google Scholar]
- [17].Lee SY, Kim HU. Systems strategies for developing industrial microbial strains. Nature biotechnology. 2015;33:1061–72. [DOI] [PubMed] [Google Scholar]
- [18].Keasling JD. Synthetic biology for synthetic chemistry. ACS chemical biology. 2008;3:64–76. [DOI] [PubMed] [Google Scholar]
- [19].Liu J, Wang X, Dai G, Zhang Y, Bian X. Microbial chassis engineering drives heterologous production of complex secondary metabolites. Biotechnology Advances. 2022;59:107966. [DOI] [PubMed] [Google Scholar]
- [20].Tiwari P, Dufossé L. Focus and Insights into the Synthetic Biology-Mediated Chassis of Economically Important Fungi for the Production of High-Value Metabolites. Microorganisms. 2023;11:1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Anthony WE, Carr RR, DeLorenzo DM, Campbell TP, Shang Z, Foston M, et al. Development of Rhodococcus opacus as a chassis for lignin valorization and bioproduction of high-value compounds. Biotechnology for biofuels. 2019;12:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Li Y, Wang H, Zhang Y, Martin C. Can the world’s favorite fruit, tomato, provide an effective biosynthetic chassis for high-value metabolites? Plant Cell Reports. 2018;37:1443–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Xu X, Liu Y, Du G, Ledesma-Amaro R, Liu L. Microbial chassis development for natural product biosynthesis. Trends in biotechnology. 2020;38:779–96. [DOI] [PubMed] [Google Scholar]
- [24].Hong K-K, Nielsen J Metabolic engineering of Saccharomyces cerevisiae: a key cell factory platform for future biorefineries. Cellular and Molecular Life Sciences. 2012;69:2671–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Ruomeng B, Meihao O, Siru Z, Shichen G, Yixian Z, Junhong C, et al. Degradation strategies of pesticide residue: From chemicals to synthetic biology. Synthetic and Systems Biotechnology. 2023;8:302–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Qi X, Yan W, Cao Z, Ding M, Yuan Y. Current advances in the biodegradation and bioconversion of polyethylene terephthalate. Microorganisms. 2021;10:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Bugg TD, Williamson JJ, Alberti F. Microbial hosts for metabolic engineering of lignin bioconversion to renewable chemicals. Renewable and Sustainable Energy Reviews. 2021;152:111674. [Google Scholar]
- [28].Sun J, Alper HS. Metabolic engineering of strains: from industrial-scale to lab-scale chemical production. Journal of Industrial Microbiology and Biotechnology. 2015;42:423–36. [DOI] [PubMed] [Google Scholar]
- [29].Keasling JD. Synthetic biology and the development of tools for metabolic engineering. Metabolic engineering. 2012;14:189–95. [DOI] [PubMed] [Google Scholar]
- [30].Whitford CM, Cruz-Morales P, Keasling JD, Weber T. The design-build-test-learn cycle for metabolic engineering of streptomycetes. Essays in biochemistry. 2021;65:261–75. [DOI] [PubMed] [Google Scholar]
- [31].Li X, Lan C, Li X, Hu Z, Jia B. A review on design-build-test-learn cycle to potentiate progress in isoprenoid engineering of photosynthetic microalgae. Bioresource Technology. 2022:127981. [DOI] [PubMed] [Google Scholar]
- [32].Tellechea-Luzardo J, Otero-Muras I, Goñi-Moreno A, Carbonell P. Fast biofoundries: coping with the challenges of biomanufacturing. Trends in biotechnology. 2022. [DOI] [PubMed] [Google Scholar]
- [33].Kamminga T, Slagman S-J, Dos Santos VAM, Bijlsma JJ, Schaap PJ. Risk-based bioengineering strategies for reliable bacterial vaccine production. Trends in biotechnology. 2019;37:805–16. [DOI] [PubMed] [Google Scholar]
- [34].Guomiao Z, Xin Y, Yuan ZHANG JW, Jian T, Chao W, Nana Z, et al. Biofoundry and its industrial application. Synthetic Biology Journal. 2023:1. [Google Scholar]
- [35].Petzold CJ, Chan LJG, Nhan M, Adams PD. Analytics for metabolic engineering. Frontiers in bioengineering and biotechnology. 2015;3:135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Harris DC. Quantitative chemical analysis: Macmillan; 2010. [Google Scholar]
- [37].DeLoache WC, Russ ZN, Narcross L, Gonzales AM, Martin VJ, Dueber JE. An enzyme-coupled biosensor enables (S)-reticuline production in yeast from glucose. Nature chemical biology. 2015;11:465–71. [DOI] [PubMed] [Google Scholar]
- [38].Antoniewicz MR. A guide to metabolic flux analysis in metabolic engineering: Methods, tools and applications. Metabolic engineering. 2021;63:2–12. [DOI] [PubMed] [Google Scholar]
- [39].Kim HU, Kim TY, Lee SY. Metabolic flux analysis and metabolic engineering of microorganisms. Molecular BioSystems. 2008;4:113–20. [DOI] [PubMed] [Google Scholar]
- [40].Varma A, Palsson BO. Metabolic flux balancing: basic concepts, scientific and practical use. Bio/technology. 1994;12:994–8. [Google Scholar]
- [41].Siu Y, Fenno J, Lindle JM, Dunlop MJ. Design and selection of a synthetic feedback loop for optimizing biofuel tolerance. ACS synthetic biology. 2018;7:16–23. [DOI] [PubMed] [Google Scholar]
- [42].Zhang J, Jensen MK, Keasling JD. Development of biosensors and their application in metabolic engineering. Current opinion in chemical biology. 2015;28:1–8. [DOI] [PubMed] [Google Scholar]
- [43].Teng Y, Zhang J, Jiang T, Zou Y, Gong X, Yan Y. Biosensor-enabled pathway optimization in metabolic engineering. Current opinion in biotechnology. 2022;75:102696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Qin L, Liu X, Xu K, Li C. Mining and design of biosensors for engineering microbial cell factory. Current Opinion in Biotechnology. 2022;75:102694. [DOI] [PubMed] [Google Scholar]
- [45].Hossain GS, Saini M, Miyake R, Ling H, Chang MW. Genetic biosensor design for natural product biosynthesis in microorganisms. Trends in biotechnology. 2020;38:797–810. [DOI] [PubMed] [Google Scholar]
- [46].Rogers JK, Taylor ND, Church GM. Biosensor-based engineering of biosynthetic pathways. Current opinion in biotechnology. 2016;42:84–91. [DOI] [PubMed] [Google Scholar]
- [47].Kim SK, Woo SG, Kim TH, Park SH, Lee JJ, Park AY, et al. Transcription Factor-Based Biosensors and Their Application in Microbiome Engineering. Principles in Microbiome Engineering. 2022:277–304. [Google Scholar]
- [48].Liu Z, Wang J, Nielsen J. Yeast synthetic biology advances biofuel production. Current Opinion in Microbiology. 2022;65:33–9. [DOI] [PubMed] [Google Scholar]
- [49].Mehrotra P Biosensors and their applications–A review. Journal of oral biology and craniofacial research. 2016;6:153–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Yan Q, Fong SS. Biosensors for metabolic engineering. Systems Biology Application in Synthetic Biology: Springer; 2016. p. 53–70. [Google Scholar]
- [51].Rogers JK, Church GM. Genetically encoded sensors enable real-time observation of metabolite production. Proceedings of the National Academy of Sciences. 2016;113:2388–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Scott LH, Wigglesworth MJ, Siewers V, Davis AM, David F. Genetically Encoded Whole Cell Biosensor for Drug Discovery of HIF-1 Interaction Inhibitors. ACS Synthetic Biology. 2022;11:3182–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Oldach L, Zhang J. Genetically encoded fluorescent biosensors for live-cell visualization of protein phosphorylation. Chemistry & biology. 2014;21:186–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Sato M, Kawai Y, Umezawa Y. Genetically encoded fluorescent indicators to visualize protein phosphorylation by extracellular signal-regulated kinase in single living cells. Analytical chemistry. 2007;79:2570–5. [DOI] [PubMed] [Google Scholar]
- [55].Wan X, Marsafari M, Xu P. Engineering metabolite-responsive transcriptional factors to sense small molecules in eukaryotes: current state and perspectives. Microbial Cell Factories. 2019;18:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Liu Y, Liu Y, Wang M. Design, optimization and application of small molecule biosensor in metabolic engineering. Frontiers in microbiology. 2017;8:2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Liu D, Evans T, Zhang F. Applications and advances of metabolite biosensors for metabolic engineering. Metabolic engineering. 2015;31:35–43. [DOI] [PubMed] [Google Scholar]
- [58].Lim HG, Jang S, Jang S, Seo SW, Jung GY. Design and optimization of genetically encoded biosensors for high-throughput screening of chemicals. Current opinion in biotechnology. 2018;54:18–25. [DOI] [PubMed] [Google Scholar]
- [59].Schallmey M, Frunzke J, Eggeling L, Marienhagen J. Looking for the pick of the bunch: high-throughput screening of producing microorganisms with biosensors. Current opinion in biotechnology. 2014;26:148–54. [DOI] [PubMed] [Google Scholar]
- [60].Kaczmarek JA, Prather KL. Effective use of biosensors for high-throughput library screening for metabolite production. Journal of Industrial Microbiology and Biotechnology. 2021;48:kuab049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Dietrich JA, Shis DL, Alikhani A, Keasling JD. Transcription factor-based screens and synthetic selections for microbial small-molecule biosynthesis. ACS synthetic biology. 2013;2:47–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Xiong D, Lu S, Wu J, Liang C, Wang W, Wang W, et al. Improving key enzyme activity in phenylpropanoid pathway with a designed biosensor. Metabolic engineering. 2017;40:115–23. [DOI] [PubMed] [Google Scholar]
- [63].Liu D, Xiao Y, Evans BS, Zhang F. Negative feedback regulation of fatty acid production based on a malonyl-CoA sensor–actuator. ACS synthetic biology. 2015;4:132–40. [DOI] [PubMed] [Google Scholar]
- [64].Zhang F, Carothers JM, Keasling JD. Design of a dynamic sensor-regulator system for production of chemicals and fuels derived from fatty acids. Nature biotechnology. 2012;30:354–9. [DOI] [PubMed] [Google Scholar]
- [65].Tellechea-Luzardo J, Stiebritz MT, Carbonell P. Transcription factor-based biosensors for screening and dynamic regulation. Frontiers in Bioengineering and Biotechnology. 2023;11:1118702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Tay SB, Ang EL. In Vivo Biosensors for Directed Protein Evolution. Protein Engineering: Tools and Applications. 2021:29–55. [Google Scholar]
- [67].Chen C, Liu J, Yao G, Bao S, Wan X, Wang F, et al. A novel, genetically encoded whole-cell biosensor for directed evolution of myrcene synthase in Escherichia coli. Biosensors and Bioelectronics. 2023;228:115176. [DOI] [PubMed] [Google Scholar]
- [68].Marsafari M, Samizadeh H, Rabiei B, Mehrabi A, Koffas M, Xu P. Biotechnological production of flavonoids: an update on plant metabolic engineering, microbial host selection, and genetically encoded biosensors. Biotechnology Journal. 2020;15:1900432. [DOI] [PubMed] [Google Scholar]
- [69].Thompson MG, Pearson AN, Barajas JF, Cruz-Morales P, Sedaghatian N, Costello Z, et al. Identification, characterization, and application of a highly sensitive lactam biosensor from Pseudomonas putida. ACS synthetic biology. 2019;9:53–62. [DOI] [PubMed] [Google Scholar]
- [70].Zheng S, Hou J, Zhou Y, Fang H, Wang T-T, Liu F, et al. One-pot two-strain system based on glucaric acid biosensor for rapid screening of myo-inositol oxygenase mutations and glucaric acid production in recombinant cells. Metabolic engineering. 2018;49:212–9. [DOI] [PubMed] [Google Scholar]
- [71].Yang P, Wang J, Pang Q, Zhang F, Wang J, Wang Q, et al. Pathway optimization and key enzyme evolution of N-acetylneuraminate biosynthesis using an in vivo aptazyme-based biosensor. Metabolic engineering. 2017;43:21–8. [DOI] [PubMed] [Google Scholar]
- [72].Kunjapur AM, Prather KL. Development of a vanillate biosensor for the vanillin biosynthesis pathway in E. coli. ACS synthetic biology. 2019;8:1958–67. [DOI] [PubMed] [Google Scholar]
- [73].Li Z, Lu Y, Wang X, Vekaria A, Jiang M, Zhang H. Enhancing anthranilic acid biosynthesis using biosensor-assisted cell selection and in situ product removal. Biochemical Engineering Journal. 2020;162:107722. [Google Scholar]
- [74].Liu Y, Zhuang Y, Ding D, Xu Y, Sun J, Zhang D. Biosensor-based evolution and elucidation of a biosynthetic pathway in Escherichia coli. ACS synthetic biology. 2017;6:837–48. [DOI] [PubMed] [Google Scholar]
- [75].Qian S, Li Y, Cirino PC. Biosensor-guided improvements in salicylate production by recombinant Escherichia coli. Microbial Cell Factories. 2019;18:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Wang R, Cress BF, Yang Z, Hordines JC III, Zhao S, Jung GY, et al. Design and characterization of biosensors for the screening of modular assembled naringenin biosynthetic library in Saccharomyces cerevisiae. ACS Synthetic Biology. 2019;8:2121–30. [DOI] [PubMed] [Google Scholar]
- [77].Yang D, Kim WJ, Yoo SM, Choi JH, Ha SH, Lee MH, et al. Repurposing type III polyketide synthase as a malonyl-CoA biosensor for metabolic engineering in bacteria. Proceedings of the National Academy of Sciences. 2018;115:9835–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Kasey CM, Zerrad M, Li Y, Cropp TA, Williams GJ. Development of transcription factor-based designer macrolide biosensors for metabolic engineering and synthetic biology. ACS synthetic biology. 2018;7:227–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Zhang Y, Li Y, Xiao F, Wang H, Zhang L, Ding Z, et al. Engineering of a biosensor in response to malate in Bacillus licheniformis. ACS Synthetic Biology. 2021;10:1775–84. [DOI] [PubMed] [Google Scholar]
- [80].Zhang J, Barajas JF, Burdu M, Ruegg TL, Dias B, Keasling JD. Development of a transcription factor-based lactam biosensor. ACS synthetic biology. 2017;6:439–45. [DOI] [PubMed] [Google Scholar]
- [81].Wang G, Øzmerih Sl, Guerreiro R, Meireles AC, Carolas A, Milne N, et al. Improvement of cis, cis-muconic acid production in Saccharomyces cerevisiae through biosensor-aided genome engineering. ACS synthetic biology. 2020;9:634–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Niu T, Liu Y, Li J, Koffas M, Du G, Alper HS, et al. Engineering a glucosamine-6-phosphate responsive glmS ribozyme switch enables dynamic control of metabolic flux in Bacillus subtilis for overproduction of N-acetylglucosamine. ACS Synthetic Biology. 2018;7:2423–35. [DOI] [PubMed] [Google Scholar]
- [83].Li L, Tu R, Song G, Cheng J, Chen W, Li L, et al. Development of a synthetic 3-dehydroshikimate biosensor in Escherichia coli for metabolite monitoring and genetic screening. ACS Synthetic Biology. 2019;8:297–306. [DOI] [PubMed] [Google Scholar]
- [84].Chen D, Xu S, Li S, Tao S, Li L, Chen S, et al. Directly Evolved AlkS-Based Biosensor Platform for Monitoring and High-Throughput Screening of Alkane Production. ACS Synthetic Biology. 2023;12:832–41. [DOI] [PubMed] [Google Scholar]
- [85].d’Oelsnitz S, Nguyen V, Alper HS, Ellington AD. Evolving a generalist biosensor for bicyclic monoterpenes. ACS Synthetic Biology. 2022;11:265–72. [DOI] [PubMed] [Google Scholar]
- [86].Liu D, Sica MS, Mao J, Chao LF-I, Siewers V. A p-Coumaroyl-CoA Biosensor for Dynamic Regulation of Naringenin Biosynthesis in Saccharomyces cerevisiae. ACS Synthetic Biology. 2022;11:3228–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Pandi A, Grigoras I, Borkowski O, Faulon J-L. Optimizing cell-free biosensors to monitor enzymatic production. ACS synthetic biology. 2019;8:1952–7. [DOI] [PubMed] [Google Scholar]
- [88].Wang Y, Li S, Xue N, Wang L, Zhang X, Zhao L, et al. Modulating Sensitivity of an Erythromycin Biosensor for Precise High-Throughput Screening of Strains with Different Characteristics. ACS Synthetic Biology. 2023. [DOI] [PubMed] [Google Scholar]
- [89].Ho JC, Pawar SV, Hallam SJ, Yadav VG. An improved whole-cell biosensor for the discovery of lignin-transforming enzymes in functional metagenomic screens. ACS synthetic biology. 2018;7:392–8. [DOI] [PubMed] [Google Scholar]
- [90].Han L, Liu X, Cheng Z, Cui W, Guo J, Yin J, et al. Construction and application of a high-throughput in vivo screening platform for the evolution of nitrile metabolism-related enzymes based on a desensitized repressive biosensor. ACS Synthetic Biology. 2022;11:1577–87. [DOI] [PubMed] [Google Scholar]
- [91].Hanko EK, Sherlock G, Minton NP, Malys N. Biosensor-informed engineering of Cupriavidus necator H16 for autotrophic D-mannitol production. Metabolic Engineering. 2022;72:24–34. [DOI] [PubMed] [Google Scholar]
- [92].Liu C, Zhang B, Liu Y-M, Yang K-Q, Liu S-J. New intracellular shikimic acid biosensor for monitoring shikimate synthesis in Corynebacterium glutamicum. ACS Synthetic Biology. 2018;7:591–601. [DOI] [PubMed] [Google Scholar]
- [93].Armetta J, Berthome R, Cros A, Pophillat C, Colombo BM, Pandi A, et al. Biosensor-based enzyme engineering approach applied to psicose biosynthesis. Synthetic Biology. 2019;4:ysz028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Jiang S, Wang R, Wang D, Zhao C, Ma Q, Wu H, et al. Metabolic reprogramming and biosensor-assisted mutagenesis screening for high-level production of L-arginine in Escherichia coli. Metabolic Engineering. 2023;76:146–57. [DOI] [PubMed] [Google Scholar]
- [95].Navone L, Moffitt K, Behrendorff J, Sadowski P, Hartley C, Speight R. Biosensor-guided rapid screening for improved recombinant protein secretion in Pichia pastoris. Microbial Cell Factories. 2023;22:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Su Y, Hickey SF, Keyser SG, Hammond MC. In vitro and in vivo enzyme activity screening via RNA-based fluorescent biosensors for S-adenosyl-l-homocysteine (SAH). Journal of the American Chemical Society. 2016;138:7040–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Xu P, Wang W, Li L, Bhan N, Zhang F, Koffas MA. Design and kinetic analysis of a hybrid promoter–regulator system for malonyl-CoA sensing in Escherichia coli. ACS chemical biology. 2014;9:451–8. [DOI] [PubMed] [Google Scholar]
- [98].Peng B, Bandari NC, Lu Z, Howard CB, Scott C, Trau M, et al. Engineering eukaryote-like regulatory circuits to expand artificial control mechanisms for metabolic engineering in Saccharomyces cerevisiae. Communications Biology. 2022;5:135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].He S, Zhang Z, Lu W. Natural promoters and promoter engineering strategies for metabolic regulation in Saccharomyces cerevisiae. Journal of Industrial Microbiology and Biotechnology. 2023;50:kuac029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Alper H, Fischer C, Nevoigt E, Stephanopoulos G. Tuning genetic control through promoter engineering. Proceedings of the National Academy of Sciences. 2005;102:12678–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Blazeck J, Alper HS. Promoter engineering: recent advances in controlling transcription at the most fundamental level. Biotechnology journal. 2013;8:46–58. [DOI] [PubMed] [Google Scholar]
- [102].Muñoz-Fernández G, Montero-Bullón J-F, Revuelta JL, Jiménez A. New promoters for metabolic engineering of Ashbya gossypii. Journal of Fungi. 2021;7:906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Dong C, Ly C, Dunlap LE, Vargas MV, Sun J, Hwang I-W, et al. Psychedelic-inspired drug discovery using an engineered biosensor. Cell. 2021;184:2779–92.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Prével C, Kurzawa L, Van TNN, Morris MC. Fluorescent biosensors for drug discovery new tools for old targets–Screening for inhibitors of cyclin-dependent kinases. European Journal of Medicinal Chemistry. 2014;88:74–88. [DOI] [PubMed] [Google Scholar]
- [105].D’Ambrosio V, Pramanik S, Goroncy K, Jakočiūnas T, Schönauer D, Davari MD, et al. Directed evolution of VanR biosensor specificity in yeast. Biotechnology notes. 2020;1:9–15. [Google Scholar]
- [106].Feng J, Jester BW, Tinberg CE, Mandell DJ, Antunes MS, Chari R, et al. A general strategy to construct small molecule biosensors in eukaryotes. Elife. 2015;4:e10606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Singh S Systems biology application in synthetic biology: Springer; 2016. [Google Scholar]
- [108].Lee J-H, Jucker F, Pardi A. Imino proton exchange rates imply an induced-fit binding mechanism for the VEGF165-targeting aptamer, Macugen. FEBS letters. 2008;582:1835–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Liu G, Mao X, Phillips JA, Xu H, Tan W, Zeng L. Aptamer– nanoparticle strip biosensor for sensitive detection of cancer cells. Analytical chemistry. 2009;81:10013–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Wang P, Yang Y, Hong H, Zhang Y, Cai W, Fang D. Aptamers as therapeutics in cardiovascular diseases. Current medicinal chemistry. 2011;18:4169–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Lupold SE, Hicke BJ, Lin Y, Coffey DS. Identification and characterization of nuclease-stabilized RNA molecules that bind human prostate cancer cells via the prostate-specific membrane antigen. Cancer research. 2002;62:4029–33. [PubMed] [Google Scholar]
- [112].Ruscito A, DeRosa MC. Small-molecule binding aptamers: Selection strategies, characterization, and applications. Frontiers in chemistry. 2016;4:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Alkhamis O, Canoura J, Yu H, Liu Y, Xiao Y. Innovative engineering and sensing strategies for aptamer-based small-molecule detection. TrAC Trends in Analytical Chemistry. 2019;121:115699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Nguyen V-T, Kwon YS, Gu MB. Aptamer-based environmental biosensors for small molecule contaminants. Current opinion in biotechnology. 2017;45:15–23. [DOI] [PubMed] [Google Scholar]
- [115].Park J, Yang K-A, Choi Y, Choe JK. Novel ssDNA aptamer-based fluorescence sensor for perfluorooctanoic acid detection in water. Environment International. 2022;158:107000. [DOI] [PubMed] [Google Scholar]
- [116].Finke M, Brecht D, Stifel J, Gense K, Gamerdinger M, Hartig JS. Efficient splicing-based RNA regulators for tetracycline-inducible gene expression in human cell culture and C. elegans. Nucleic Acids Research. 2021;49:e71–e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Weigand JE, Suess B. Tetracycline aptamer-controlled regulation of pre-mRNA splicing in yeast. Nucleic acids research. 2007;35:4179–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Kavita K, Breaker RR. Discovering riboswitches: the past and the future. Trends in biochemical sciences. 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Barrick JE, Breaker RR. The distributions, mechanisms, and structures of metabolite-binding riboswitches. Genome biology. 2007;8:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Winkler WC, Cohen-Chalamish S, Breaker RR. An mRNA structure that controls gene expression by binding FMN. Proceedings of the National Academy of Sciences. 2002;99:15908–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Winkler WC, Nahvi A, Sudarsan N, Barrick JE, Breaker RR. An mRNA structure that controls gene expression by binding S-adenosylmethionine. Nature Structural & Molecular Biology. 2003;10:701–7. [DOI] [PubMed] [Google Scholar]
- [122].Sudarsan N, Wickiser JK, Nakamura S, Ebert MS, Breaker RR. An mRNA structure in bacteria that controls gene expression by binding lysine. Genes & development. 2003;17:2688–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Asal M, Özen Ö, Şahinler M, Polatoğlu İ. Recent developments in enzyme, DNA and immuno-based biosensors. Sensors. 2018;18:1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Batalla P, Mateo C, Grazu V, Fernandez-Lafuente R, Guisan JM. Immobilization of antibodies through the surface regions having the highest density in lysine groups on finally inert support surfaces. Process Biochemistry. 2009;44:365–8. [Google Scholar]
- [125].Shen M, Rusling JF, Dixit CK. Site-selective orientated immobilization of antibodies and conjugates for immunodiagnostics development. Methods. 2017;116:95–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Ngernpimai S, Srijampa S, Thongmee P, Teerasong S, Puangmali T, Maleewong W, et al. Insight into the covalently oriented immobilization of antibodies on gold nanoparticle probes to improve sensitivity in the colorimetric detection of listeria monocytogenes. Bioconjugate Chemistry. 2022;33:2103–12. [DOI] [PubMed] [Google Scholar]
- [127].Aloraij Y, Alsheikh A, Alyousef RA, Alhamlan F, Suaifan GA, Muthana S, et al. Development of a Rapid Immuno-Based Screening Assay for the Detection of Adenovirus in Eye Infections. ACS omega. 2022;7:17555–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Beck A, Goetsch L, Dumontet C, Corvaïa N. Strategies and challenges for the next generation of antibody–drug conjugates. Nature reviews Drug discovery. 2017;16:315–37. [DOI] [PubMed] [Google Scholar]
- [129].Carter PJ, Lazar GA. Next generation antibody drugs: pursuit of the’high-hanging fruit’. Nature Reviews Drug Discovery. 2018;17:197–223. [DOI] [PubMed] [Google Scholar]
- [130].Schrama D, Reisfeld RA, Becker JC. Antibody targeted drugs as cancer therapeutics. Nature reviews Drug discovery. 2006;5:147–59. [DOI] [PubMed] [Google Scholar]
- [131].Misawa N, Osaki T, Takeuchi S. Membrane protein-based biosensors. Journal of the Royal Society Interface. 2018;15:20170952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Crnković A, Srnko M, Anderluh G. Biological nanopores: Engineering on demand. Life. 2021;11:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Cao C, Long Y-T. Biological nanopores: confined spaces for electrochemical single-molecule analysis. Accounts of chemical research. 2018;51:331–41. [DOI] [PubMed] [Google Scholar]
- [134].Kasianowicz JJ, Brandin E, Branton D, Deamer DW. Characterization of individual polynucleotide molecules using a membrane channel. Proceedings of the National Academy of Sciences. 1996;93:13770–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Ying Y-L, Li D-W, Li Y, Lee JS, Long Y-T. Enhanced translocation of poly (dt) 45 through an α-hemolysin nanopore by binding with antibody. Chemical Communications. 2011;47:5690–2. [DOI] [PubMed] [Google Scholar]
- [136].Kloda A, Martinac B. Mechanosensitive channels in archaea. Cell biochemistry and biophysics. 2001;34:349–81. [DOI] [PubMed] [Google Scholar]
- [137].Kung C, Martinac B, Sukharev S. Mechanosensitive channels in microbes. Annual review of microbiology. 2010;64:313–29. [DOI] [PubMed] [Google Scholar]
- [138].Kung C, Saimi Y, Martinac B. Mechano-sensitive ion channels in microbes and the early evolutionary origin of solvent sensing. Current Topics in Membranes and Transport: Elsevier; 1990. p. 145–53. [Google Scholar]
- [139].Martinac B Mechanosensitive ion channels: molecules of mechanotransduction. Journal of cell science. 2004;117:2449–60. [DOI] [PubMed] [Google Scholar]
- [140].Haswell ES, Phillips R, Rees DC. Mechanosensitive channels: what can they do and how do they do it? Structure. 2011;19:1356–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Hsu Y-Y, Resto Irizarry AM, Fu J, Liu AP. Mechanosensitive Channel-Based Optical Membrane Tension Reporter. ACS sensors. 2023;8:12–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Garamella J, Majumder S, Liu AP, Noireaux V. An adaptive synthetic cell based on mechanosensing, biosensing, and inducible gene circuits. ACS Synthetic Biology. 2019;8:1913–20. [DOI] [PubMed] [Google Scholar]
- [143].Majumder S, Garamella J, Wang Y-L, DeNies M, Noireaux V, Liu AP. Cell-sized mechanosensitive and biosensing compartment programmed with DNA. Chemical Communications. 2017;53:7349–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Hsu Y-Y, Hwang S-W, Chen SJ, Alsberg E, Liu AP. Development of mechanosensitive synthetic cells for biomedical applications. SLAS technology. 2023. [DOI] [PubMed] [Google Scholar]
- [145].Petsko GA, Ringe D. Protein structure and function: New Science Press; 2004. [Google Scholar]
- [146].Alexandrov K, Vickers CE. In vivo protein-based biosensors: seeing metabolism in real time. Trends in biotechnology. 2022. [DOI] [PubMed] [Google Scholar]
- [147].Ibraheem A, Campbell RE. Designs and applications of fluorescent protein-based biosensors. Current opinion in chemical biology. 2010;14:30–6. [DOI] [PubMed] [Google Scholar]
- [148].Barbosa AJ, Oliveira AR, Roque AC. Protein-and peptide-based biosensors in artificial olfaction. Trends in biotechnology. 2018;36:1244–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Guo Z, Murphy L, Stein V, Johnston WA, Alcala-Perez S, Alexandrov K. Engineered PQQ-glucose dehydrogenase as a universal biosensor platform. Journal of the American Chemical Society. 2016;138:10108–11. [DOI] [PubMed] [Google Scholar]
- [150].Adamson H, Jeuken LJ. Engineering protein switches for rapid diagnostic tests. ACS sensors. 2020;5:3001–12. [DOI] [PubMed] [Google Scholar]
- [151].Cook A, Hari-Gupta Y, Toseland CP. Application of the SSB biosensor to study in vitro transcription. Biochemical and biophysical research communications. 2018;496:820–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Goers L, Ainsworth C, Goey CH, Kontoravdi C, Freemont PS, Polizzi KM. Whole-cell Escherichia coli lactate biosensor for monitoring mammalian cell cultures during biopharmaceutical production. Biotechnology and bioengineering. 2017;114:1290–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Randriamampita C, Lellouch AC. Imaging early signaling events in T lymphocytes with fluorescent biosensors. Biotechnology journal. 2014;9:203–12. [DOI] [PubMed] [Google Scholar]
- [154].Kunzelmann S, Solscheid C, Webb MR. Fluorescent biosensors: design and application to motor proteins. Fluorescent Methods for Molecular Motors. 2014:25–47. [DOI] [PubMed] [Google Scholar]
- [155].Quijano-Rubio A, Yeh H-W, Park J, Lee H, Langan RA, Boyken SE, et al. De novo design of modular and tunable protein biosensors. Nature. 2021;591:482–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].K Singh R, Lee J-K, Selvaraj C, Singh R, Li J, Kim S-Y, et al. Protein engineering approaches in the post-genomic era. Current Protein and Peptide Science. 2018;19:5–15. [DOI] [PubMed] [Google Scholar]
- [157].Jayanthi VSA, Das AB, Saxena U. Recent advances in biosensor development for the detection of cancer biomarkers. Biosensors and Bioelectronics. 2017;91:15–23. [DOI] [PubMed] [Google Scholar]
- [158].Yang G, Xiao Z, Tang C, Deng Y, Huang H, He Z. Recent advances in biosensor for detection of lung cancer biomarkers. Biosensors and Bioelectronics. 2019;141:111416. [DOI] [PubMed] [Google Scholar]
- [159].Bohunicky B, Mousa SA. Biosensors: the new wave in cancer diagnosis. Nanotechnology, science and applications. 2010:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].de la Cova C, Townley R, Regot S, Greenwald I. A real-time biosensor for ERK activity reveals signaling dynamics during C. elegans cell fate specification. Developmental cell. 2017;42:542–53.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Wehr MC, Rossner MJ. Split protein biosensor assays in molecular pharmacological studies. Drug Discovery Today. 2016;21:415–29. [DOI] [PubMed] [Google Scholar]
- [162].Aydin EB, Aydin M, Sezginturk MK. Biosensors in drug discovery and drug analysis. Current Analytical Chemistry. 2019;15:467–84. [Google Scholar]
- [163].Khoshbin Z, Housaindokht MR, Izadyar M, Bozorgmehr MR, Verdian A. Recent advances in computational methods for biosensor design. Biotechnology and Bioengineering. 2021;118:555–78. [DOI] [PubMed] [Google Scholar]
- [164].Vongsouthi V, Whitfield JH, Unichenko P, Mitchell JA, Breithausen Br, Khersonsky O, et al. A rationally and computationally designed fluorescent biosensor for d-serine. ACS sensors. 2021;6:4193–205. [DOI] [PubMed] [Google Scholar]
- [165].Marvin JS, Hellinga HW. Conversion of a maltose receptor into a zinc biosensor by computational design. Proceedings of the National Academy of Sciences. 2001;98:4955–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Yang W, Lai L. Computational design of ligand-binding proteins. Current opinion in structural biology. 2017;45:67–73. [DOI] [PubMed] [Google Scholar]
- [167].Kanwar M, Wright RC, Date A, Tullman J, Ostermeier M. Protein switch engineering by domain insertion. Methods in enzymology: Elsevier; 2013. p. 369–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Chen Z, Rappert S, Zeng A-P. Rational design of allosteric regulation of homoserine dehydrogenase by a nonnatural inhibitor L-lysine. ACS synthetic biology. 2015;4:126–31. [DOI] [PubMed] [Google Scholar]
- [169].Cormann KU, Baumgart M, Bott M. Structure-Based Design of Versatile Biosensors for Small Molecules Based on the PAS Domain of a Thermophilic Histidine Kinase. ACS synthetic biology. 2018;7:2888–97. [DOI] [PubMed] [Google Scholar]
- [170].Goh YY, Ho B, Ding JL. A novel fluorescent protein-based biosensor for gram-negative bacteria. Applied and Environmental Microbiology. 2002;68:6343–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Chesterfield RJ, Whitfield JH, Pouvreau B, Cao D, Beveridge CA, Vickers CE. Rational design of novel fluorescent enzyme biosensors for direct detection of strigolactones. bioRxiv. 2020. [DOI] [PubMed] [Google Scholar]
- [172].Campbell RE. Fluorescent-protein-based biosensors: modulation of energy transfer as a design principle. ACS Publications; 2009. [DOI] [PubMed] [Google Scholar]
- [173].Shaner NC, Patterson GH, Davidson MW. Advances in fluorescent protein technology. Journal of cell science. 2007;120:4247–60. [DOI] [PubMed] [Google Scholar]
- [174].Szymanski E, Calvert J. Designing with living systems in the synthetic yeast project. Nature Communications. 2018;9:2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Szymanski EA, Henriksen J. Reconfiguring the Challenge of Biological Complexity as a Resource for Biodesign. Msphere. 2022;7:e00547–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Romero PA, Arnold FH. Exploring protein fitness landscapes by directed evolution. Nature reviews Molecular cell biology. 2009;10:866–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Arnold FH. Design by directed evolution. Accounts of chemical research. 1998;31:125–31. [Google Scholar]
- [178].Arnold FH, Wintrode PL, Miyazaki K, Gershenson A. How enzymes adapt: lessons from directed evolution. Trends in biochemical sciences. 2001;26:100–6. [DOI] [PubMed] [Google Scholar]
- [179].Arnold FH. Innovation by evolution: bringing new chemistry to life (Nobel Lecture). Angewandte Chemie International Edition. 2019;58:14420–6. [DOI] [PubMed] [Google Scholar]
- [180].Kazlauskas RJ, Bornscheuer UT. Finding better protein engineering strategies. Nature chemical biology. 2009;5:526–9. [DOI] [PubMed] [Google Scholar]
- [181].Guntas G, Kanwar M, Ostermeier M. Circular permutation in the Ω-loop of TEM-1 β-lactamase results in improved activity and altered substrate specificity. PLoS One. 2012;7:e35998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Bliven S, Prlić A. Circular permutation in proteins. PLoS computational biology. 2012;8:e1002445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Snoek T, Chaberski EK, Ambri F, Kol S, Bjørn SP, Pang B, et al. Evolution-guided engineering of small-molecule biosensors. Nucleic acids research. 2020;48:e3–e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Espinosa-Urgel M, Serrano L, Ramos JL, Fernández-Escamilla AM. Engineering biological approaches for detection of toxic compounds: a new microbial biosensor based on the Pseudomonas putida TtgR repressor. Molecular Biotechnology. 2015;57:558–64. [DOI] [PubMed] [Google Scholar]
- [185].Ogawa Y, Katsuyama Y, Ueno K, Ohnishi Y. Switching the ligand specificity of the biosensor XylS from meta to para-toluic acid through directed evolution exploiting a dual selection system. ACS Synthetic Biology. 2019;8:2679–89. [DOI] [PubMed] [Google Scholar]
- [186].Brandsen BM, Mattheisen JM, Noel T, Fields S. A biosensor strategy for E. coli based on ligand-dependent stabilization. ACS synthetic biology. 2018;7:1990–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Tang R-Q, Wagner JM, Alper HS, Zhao X-Q, Bai F-W. Design, evolution, and characterization of a xylose biosensor in Escherichia coli using the XylR/xylO system with an expanded operating range. ACS Synthetic Biology. 2020;9:2714–22. [DOI] [PubMed] [Google Scholar]
- [188].Bali AP, Genee HJ, Sommer MO. Directed evolution of membrane transport using synthetic selections. ACS synthetic biology. 2018;7:789–93. [DOI] [PubMed] [Google Scholar]
- [189].FM Machado L, Currin A, Dixon N. Directed evolution of the PcaV allosteric transcription factor to generate a biosensor for aromatic aldehydes. Journal of biological engineering. 2019;13:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [190].Flachbart LK, Gertzen CGW, Gohlke H, Marienhagen J. Development of a biosensor platform for phenolic compounds using a transition ligand strategy. ACS synthetic biology. 2021;10:2002–14. [DOI] [PubMed] [Google Scholar]
- [191].Cai Y, Zhu K, Shen L, Ma J, Bao L, Chen D, et al. Evolved biosensor with high sensitivity and specificity for measuring cadmium in actual environmental samples. Environmental Science & Technology. 2022;56:10062–71. [DOI] [PubMed] [Google Scholar]
- [192].Zimran G, Feuer E, Pri-Tal O, Shpilman M, Mosquna A. Directed Evolution of Herbicide Biosensors in a Fluorescence-Activated Cell-Sorting-Compatible Yeast Two-Hybrid Platform. ACS Synthetic Biology. 2022;11:2880–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [193].Lane MD, Seelig B. Advances in the directed evolution of proteins. Current opinion in chemical biology. 2014;22:129–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [194].Cobb RE, Chao R, Zhao H. Directed evolution: past, present, and future. AIChE Journal. 2013;59:1432–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [195].Wang Y, Xue P, Cao M, Yu T, Lane ST, Zhao H. Directed evolution: methodologies and applications. Chemical reviews. 2021;121:12384–444. [DOI] [PubMed] [Google Scholar]
- [196].Eriksen DT, Lian J, Zhao H. Protein design for pathway engineering. Journal of structural biology. 2014;185:234–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [197].Lutz S Beyond directed evolution—semi-rational protein engineering and design. Current opinion in biotechnology. 2010;21:734–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [198].Taylor ND, Garruss AS, Moretti R, Chan S, Arbing MA, Cascio D, et al. Engineering an allosteric transcription factor to respond to new ligands. Nature methods. 2016;13:177–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [199].Chi H, Zhou Q, Tutol JN, Phelps SM, Lee J, Kapadia P, et al. Coupling a Live Cell Directed Evolution Assay with Coevolutionary Landscapes to Engineer an Improved Fluorescent Rhodopsin Chloride Sensor. ACS Synthetic Biology. 2022;11:1627–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [200].Li J, Nina MRH, Zhang X, Bai Y. Engineering transcription factor XylS for sensing phthalic acid and terephthalic acid: an application for enzyme evolution. ACS Synthetic Biology. 2022;11:1106–13. [DOI] [PubMed] [Google Scholar]
- [201].Bornscheuer UT, Hohne M. Protein Engineering: Springer; 2018. [Google Scholar]
- [202].Yang KK, Wu Z, Arnold FH. Machine-learning-guided directed evolution for protein engineering. Nature methods. 2019;16:687–94. [DOI] [PubMed] [Google Scholar]
- [203].Wu Z, Kan SJ, Lewis RD, Wittmann BJ, Arnold FH. Machine learning-assisted directed protein evolution with combinatorial libraries. Proceedings of the National Academy of Sciences. 2019;116:8852–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [204].Unger EK, Keller JP, Altermatt M, Liang R, Matsui A, Dong C, et al. Directed evolution of a selective and sensitive serotonin sensor via machine learning. Cell. 2020;183:1986–2002. e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [205].Chu HY, Wong AS. Facilitating Machine Learning-Guided Protein Engineering with Smart Library Design and Massively Parallel Assays. Advanced Genetics. 2021;2:2100038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [206].Liu D, Mannan AA, Han Y, Oyarzún DA, Zhang F. Dynamic metabolic control: towards precision engineering of metabolism. Journal of Industrial Microbiology and Biotechnology. 2018;45:535–43. [DOI] [PubMed] [Google Scholar]
- [207].Dinh CV, Prather KL. Development of an autonomous and bifunctional quorum-sensing circuit for metabolic flux control in engineered Escherichia coli. Proceedings of the National Academy of Sciences. 2019;116:25562–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [208].Milo R, Phillips R. Cell biology by the numbers: Garland Science; 2015. [Google Scholar]
- [209].Griffith KL, Grossman AD. Inducible protein degradation in Bacillus subtilis using heterologous peptide tags and adaptor proteins to target substrates to the protease ClpXP. Molecular microbiology. 2008;70:1012–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [210].Sakamoto KM, Kim KB, Kumagai A, Mercurio F, Crews CM, Deshaies RJ. Protacs: Chimeric molecules that target proteins to the Skp1–Cullin–F box complex for ubiquitination and degradation. Proceedings of the National Academy of Sciences. 2001;98:8554–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [211].Kim J-H, Wei J-R, Wallach JB, Robbins RS, Rubin EJ, Schnappinger D. Protein inactivation in mycobacteria by controlled proteolysis and its application to deplete the beta subunit of RNA polymerase. Nucleic acids research. 2011;39:2210–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [212].McGinness KE, Baker TA, Sauer RT. Engineering controllable protein degradation. Molecular cell. 2006;22:701–7. [DOI] [PubMed] [Google Scholar]
- [213].Davis JH, Baker TA, Sauer RT. Small-molecule control of protein degradation using split adaptors. ACS chemical biology. 2011;6:1205–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [214].Cameron DE, Collins JJ. Tunable protein degradation in bacteria. Nature biotechnology. 2014;32:1276–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [215].Wong WW, Tsai TY, Liao JC. Single-cell zeroth-order protein degradation enhances the robustness of synthetic oscillator. Molecular systems biology. 2007;3:130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [216].Lei Y, Chen W, Xiang L, Wu J, Zhen Z, Jin J-M, et al. Engineering an SspB-mediated degron for novel controllable protein degradation. Metabolic Engineering. 2022;74:150–9. [DOI] [PubMed] [Google Scholar]
- [217].Farrell CM, Baker TA, Sauer RT. Altered specificity of a AAA+ protease. Molecular cell. 2007;25:161–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [218].Gottesman S, Roche E, Zhou Y, Sauer RT. The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes & development. 1998;12:1338–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [219].Fritze J, Zhang M, Luo Q, Lu X. An overview of the bacterial SsrA system modulating intracellular protein levels and activities. Applied microbiology and biotechnology. 2020;104:5229–41.32342145 [Google Scholar]
- [220].Fei X, Bell TA, Jenni S, Stinson BM, Baker TA, Harrison SC, et al. Structures of the ATP-fueled ClpXP proteolytic machine bound to protein substrate. Elife. 2020;9:e52774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [221].Levchenko I, Seidel M, Sauer RT, Baker TA. A specificity-enhancing factor for the ClpXP degradation machine. Science. 2000;289:2354–6. [DOI] [PubMed] [Google Scholar]
- [222].Flynn JM, Levchenko I, Sauer RT, Baker TA. Modulating substrate choice: the SspB adaptor delivers a regulator of the extracytoplasmic-stress response to the AAA+ protease ClpXP for degradation. Genes & development. 2004;18:2292–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [223].Wah DA, Levchenko I, Baker TA, Sauer RT. Characterization of a specificity factor for an AAA+ ATPase: assembly of SspB dimers with ssrA-tagged proteins and the ClpX hexamer. Chemistry & biology. 2002;9:1237–45. [DOI] [PubMed] [Google Scholar]
- [224].Guiziou S, Sauveplane V, Chang H-J, Clerté C, Declerck N, Jules M, et al. A part toolbox to tune genetic expression in Bacillus subtilis. Nucleic acids research. 2016;44:7495–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [225].Volke DC, Turlin J, Mol V, Nikel PI. Physical decoupling of XylS/Pm regulatory elements and conditional proteolysis enable precise control of gene expression in Pseudomonas putida. Microbial Biotechnology. 2020;13:222–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [226].Prindle A, Selimkhanov J, Danino T, Samayoa P, Goldberg A, Bhatia SN, et al. Genetic circuits in Salmonella typhimurium. ACS synthetic biology. 2012;1:458–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [227].Zhang M, Luo Q, Sun H, Fritze J, Luan G, Lu X. Engineering a controllable targeted protein degradation system and a derived OR-GATE-type inducible gene expression system in Synechococcus elongatus PCC 7942. ACS Synthetic Biology. 2021;11:125–34. [DOI] [PubMed] [Google Scholar]
- [228].Fernandez-Rodriguez J, Voigt CA. Post-translational control of genetic circuits using Potyvirus proteases. Nucleic acids research. 2016;44:6493–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [229].Gur E, Sauer RT. Evolution of the ssrA degradation tag in Mycoplasma: specificity switch to a different protease. Proceedings of the National Academy of Sciences. 2008;105:16113–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [230].Brockman IM, Prather KL. Dynamic knockdown of E. coli central metabolism for redirecting fluxes of primary metabolites. Metabolic engineering. 2015;28:104–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [231].Sekar K, Gentile AM, Bostick JW, Tyo KE. N-terminal-based targeted, inducible protein degradation in Escherichia coli. PLoS One. 2016;11:e0149746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [232].Zorzini V, Buts L, Schrank E, Sterckx YG, Respondek M, Engelberg-Kulka H, et al. Escherichia coli antitoxin MazE as transcription factor: insights into MazE-DNA binding. Nucleic acids research. 2015;43:1241–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [233].Liu N, Chaudhry MT, Xie Z, Kreth J, Merritt J. Identification of new degrons in Streptococcus mutans reveals a novel strategy for engineering targeted, controllable proteolysis. Frontiers in Microbiology. 2017;8:2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [234].Izert MA, Klimecka MM, Górna MW. Applications of bacterial degrons and degraders—toward targeted protein degradation in bacteria. Frontiers in Molecular Biosciences. 2021;8:669762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [235].Gopal P, Sarathy JP, Yee M, Ragunathan P, Shin J, Bhushan S, et al. Pyrazinamide triggers degradation of its target aspartate decarboxylase. Nature communications. 2020;11:1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [236].Hayat IF, Plan M, Ebert BE, Dumsday G, Vickers CE, Peng B. Auxin-mediated induction of GAL promoters by conditional degradation of Mig1p improves sesquiterpene production in Saccharomyces cerevisiae with engineered acetyl-CoA synthesis. Microbial Biotechnology. 2021;14:2627–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [237].Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nature methods. 2009;6:917–22. [DOI] [PubMed] [Google Scholar]
- [238].Zhang L, Ward JD, Cheng Z, Dernburg AF. The auxin-inducible degradation (AID) system enables versatile conditional protein depletion in C. elegans. Development. 2015;142:4374–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [239].Macdonald L, Taylor GC, Brisbane JM, Christodoulou E, Scott L, Von Kriegsheim A, et al. Rapid and specific degradation of endogenous proteins in mouse models using auxin-inducible degrons. Elife. 2022;11:e77987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [240].Snyder NA, Kim A, Kester L, Gale AN, Studer C, Hoepfner D, et al. Auxin-inducible depletion of the essentialome suggests inhibition of TORC1 by auxins and inhibition of Vrg4 by SDZ 90–215, a natural antifungal cyclopeptide. G3: Genes, Genomes, Genetics. 2019;9:829–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [241].Yesbolatova A, Saito Y, Kitamoto N, Makino-Itou H, Ajima R, Nakano R, et al. The auxin-inducible degron 2 technology provides sharp degradation control in yeast, mammalian cells, and mice. Nature communications. 2020;11:5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [242].Shabek N, Zheng N. Plant ubiquitin ligases as signaling hubs. Nature structural & molecular biology. 2014;21:293–6. [DOI] [PubMed] [Google Scholar]
- [243].Skerra A Use of the tetracycline promoter for the tightly regulated production of a murine antibody fragment in Escherichia coli. Gene. 1994;151:131–5. [DOI] [PubMed] [Google Scholar]
- [244].Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403:339–42. [DOI] [PubMed] [Google Scholar]
- [245].Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. cell. 2004;116:281–97. [DOI] [PubMed] [Google Scholar]
- [246].Perez-Pinera P, Kocak DD, Vockley CM, Adler AF, Kabadi AM, Polstein LR, et al. RNA-guided gene activation by CRISPR-Cas9–based transcription factors. Nature methods. 2013;10:973–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [247].Alzhanuly B, Mukhatayev ZY, Botbayev DM, Ashirbekov Y, Katkenov ND, Dzhaynakbaev NT, et al. Modulation of Insulin Gene Expression with CRISPR/Cas9-based Transcription Factors. Open Access Macedonian Journal of Medical Sciences. 2021;9:876–81. [Google Scholar]
- [248].Atkinson MR, Savageau MA, Myers JT, Ninfa AJ. Development of genetic circuitry exhibiting toggle switch or oscillatory behavior in Escherichia coli. Cell. 2003;113:597–607. [DOI] [PubMed] [Google Scholar]


