Summary
Background
The search for factors beyond the radiotherapy dose that could identify patients more at risk of developing radio-induced toxicity is essential to establish personalised treatment protocols for improving the quality-of-life of survivors. To investigate the role of the intestinal microbiota in the development of radiotherapy-induced gastrointestinal toxicity, the MicroLearner observational cohort study characterised the intestinal microbiota of 136 (discovery) and 79 (validation) consecutive prostate cancer patients at baseline radiotherapy.
Methods
Gastrointestinal toxicity was assessed weekly during RT using CTCAE. An average grade >1.3 over time points was used to identify patients suffering from persistent acute toxicity (endpoint). The microbiota of patients was quantified from the baseline faecal samples using 16S rRNA gene sequencing technology and the Ion Reporter metagenomic pipeline. Statistical techniques and computational and machine learning tools were used to extract, functionally characterise, and predict core features of the bacterial communities of patients who developed acute gastrointestinal toxicity.
Findings
Analysis of the core bacterial composition in the discovery cohort revealed a cluster of patients significantly enriched for toxicity, displaying a toxicity rate of 60%. Based on selected high-risk microbiota compositional features, we developed a clinical decision tree that could effectively predict the risk of toxicity based on the relative abundance of genera Faecalibacterium, Bacteroides, Parabacteroides, Alistipes, Prevotella and Phascolarctobacterium both in internal and external validation cohorts.
Interpretation
We provide evidence showing that intestinal bacteria profiling from baseline faecal samples can be effectively used in the clinic to improve the pre-radiotherapy assessment of gastrointestinal toxicity risk in prostate cancer patients.
Funding
Italian Ministry of Health (Promotion of Institutional Research INT-year 2016, 5 × 1000, Ricerca Corrente funds). Fondazione Regionale per la Ricerca Biomedica (ID 2721017). AIRC (IG 21479).
Keywords: Radiation toxicity, Intestinal microbiota, Machine learning, Prostate cancer
Research in context.
Evidence before this study
From a search in PubMed for articles including in their title or abstract the words (“radiotherapy” OR “radiation therapy”) AND “microbiota” AND “toxicity” AND “prostate cancer” we found 9 articles published between 2019 and January 2024 including 3 reviews.
Evidence from studies conducted over the last decade suggests that the microbiota composition of the gastrointestinal tract could play a role in the development of radiotherapy-induced gastrointestinal toxicity.
These studies have shown that patients displaying symptoms of gastrointestinal toxicity following radiotherapy have faecal microbiota profiles exhibiting a lower species diversity and distinguishable compositional features compared to those without. Processes including dysbiosis-induced inflammation and reduction in short-chain fatty acids production are believed to link radiation-induced toxicity and an altered host–microbiota equilibrium. Despite the evidence, no microbial signature predictive of toxicity has been identified and established by previous studies.
Added value of this study
Here we identified a set of commensal bacterial genera characteristic of the intestinal microbiota that are predictive of the development of acute gastrointestinal toxicity in prostate cancer patients during radiotherapy. To characterise the microbiome, we used 16S rRNA sequencing technology, which offers taxonomic resolution up to the scale of bacterial genera and is cost effective.
Implications of all the available evidence
The study opens up concrete prospects for the use of baseline faecal samples in the clinical practice to improve pre-radiotherapy toxicity risk assessment, and it is the starting point for any research looking for ways to act directly on the intestinal microbiome of prostate cancer patients, for example by use of probiotics or faecal transplant to reduce treatment side effects. Analysis of the bacterial communities of patients with higher rates of toxicity pointed to a selection for genera more adapted for coping with oxidative stress and phosphate scarcity, suggesting that the microbiota composition reflects a host environment characterised by increased levels of reactive oxygen species that, in turn, predispose to toxicity from radiation oxidative effects.
Introduction
Radiotherapy (RT) is often used as a curative treatment for prostate cancer (PCa) either definitively or as adjuvant/salvage (post-prostatectomy) treatment with excellent efficacy outcomes. However, many patients experience significant early and/or long-term gastrointestinal (GI) side effects due to the unavoidable irradiation of healthy gut tissues surrounding the tumour target. About 10–50% of treated patients suffer from moderate to severe acute GI side effects (manifesting within 90 days after the end of RT), including proctitis, diarrhoea, rectal bleeding and abdominal pain.1 The onset of acute side effects during RT is particularly relevant, as late intestinal side effects (manifesting more than 90 days after the end of RT) show a sequential behaviour, and patients with more severe and particularly long-lasting non-healing acute toxicity are at increased risk of long-term toxicity.2,3
The incidence of radio-induced GI toxicity can be reduced through radiation dose-volume constraints that limit the volume of intestinal/rectal tissue irradiated to significant dose levels.4 However, the heterogeneity of toxicity levels observed after treatment is challenging to model only based on the dose distribution in organs at risk. The dose-toxicity relationship is modulated by many patient-specific factors that affect the tissue response to radiation, such as genetic background and expression patterns, premorbid conditions and use of drugs, as well as the intestinal microenvironment.5 In addition, different causes can contribute to the same symptom, e.g., diarrhoea can be caused by epithelial injury, small bowel bacterial overgrowth or bile acid malabsorption.4
The intestinal microbiota plays an essential role in maintaining intestinal homeostasis and is affected by radiotherapy in the pelvic district. Specific patterns of bacterial communities in the gut are associated with developing various intestinal disorders, including Crohn's disease, ulcerative colitis and pseudomembranous colitis. These conditions result in symptoms that are often similar to those observed in radio-induced GI toxicity.5
Small cohort studies (n < 12) observed a peculiar intestinal microbiota composition and, more specifically, a significantly lower microbial diversity before RT in cancer patients who developed diarrhoea during pelvic RT.6, 7, 8
The most compelling evidence that the intestinal microbiota might be predictive of the development of RT-induced GI toxicity in PCa patients came from the MARS study.9 MARS investigated the associations of the intestinal microbiota with both acute and late radiation-induced toxicity. In a cohort representative of early acute toxicity (n = 32), where sequential faecal samples were obtained longitudinally over early time points to identify patterns and dynamics of the microbiome, the authors observed that a microbiota characterised by a comparatively reduced bacterial diversity over-time was associated with a significantly higher incidence of acute GI toxicity. These changes were mirrored in two other cohorts (n = 103) evaluated for late GI toxicity. In addition, an association of higher relative abundance of some short-chain fatty acid (SCFA) producers with both acute and late toxicity was found. With the patients having received homogeneous treatment and no cytotoxic systemic therapies, and reporting on the largest cohorts at the time, MARS was a cornerstone in the demonstration that alterations in the intestinal microbiota associated with both early and late radiation enteropathy.
These studies suggest that the intestinal microbiome presents opportunities for predicting and treating radio-induced GI toxicity. However, further research is needed to understand whether the information on the microbiome can be used in clinical practice to assess the risk of toxicity development, allowing clinicians to modulate treatment or use other intervention strategies to mitigate that risk.10
We herein report the results of the MicroLearner PCa study, where we obtained pre-RT intestinal microbiome data from faecal samples of 215 PCa patients and applied machine learning techniques to elucidate whether the intestinal microbiome can predict the risk of development of acute GI toxicity.
Methods
Ethics
The MicroLearner observational cohort study was approved by the local Ethical Committee (ID: INT 11/17) and prospectively registered on ClinicalTrials.gov (ID: NCT03294122). It enrolled 244 PCa patients who provided written Informed Consent and agreed that incidental findings would not be disclosed to them or any clinician.
Participants and setting
All patients were treated with high-dose RT (curative intent, exclusive RT or post-prostatectomy RT) at the Fondazione IRCCS Istituto Nazionale dei Tumori di Milano. All patients underwent RT over 5 (hypofractionation) or 7–8 (conventional fractionation) weeks according to the standard of practice at Fondazione IRCCS Istituto Nazionale dei Tumori di Milano. Detailed information on patient- and treatment-related features were prospectively registered using standardised Case Report Forms. Details on participation criteria can be found on ClinicalTrials.gov: https://clinicaltrials.gov/study/NCT03294122.
Study design
According to the study design, PCa patients were divided into two consecutive cohorts, namely MicroLearner discovery and MicroLearner validation. The size of the MicroLearner study was designed to have sufficient statistical power to detect the effect of risk factors with OR ≥2.5 (the protocol is available on ClinicalTrials.gov: https://cdn.clinicaltrials.gov/large-docs/22/NCT03294122/Prot_SAP_000.pdf). Therefore, differences in toxicity rates associated with smaller ORs cannot be statistically significant. Faecal samples were collected before RT initiation to profile the baseline intestinal microbiota of the patients using 16S rRNA amplicon sequencing. Clinical data were available for all the patients, and data on the use of antibiotics and probiotics were also recorded, along with data on diet and physical activity. Blood samples were collected before RT initiation from a fraction of patients in the discovery cohort to investigate the level of serum pro-inflammatory cytokines CCL2, PDGF-BB, TGF-β1, TNF-α and TNFR1, previously reported to directly associate with the development of radio-induced GI toxicity.11
Information on radio-induced side-effects was prospectively collected before, during treatment (once a week) and every 6 months till five-year follow-up by using health professionals’ assessment Common Terminology Criteria for Adverse Events (CTCAE) v4.0. Patient-reported questionnaires (Expanded Prostate Cancer Index Composite-26, International Prostatic Symptom Score, International Consultation on Continence Questionnaire, International Index of Erectile Function-5, EORTC Core Quality of Life Questionnaire) were collected before RT, at RT end and every 6 months in five-year follow-up. Data on coping with cancer (Mini-Mental Adjustment to Cancer) and with anxiety (Memorial Anxiety Scale—Prostate Cancer) are at baseline.
Before RT, patients also completed a questionnaire on lifestyle with information on diet habits and physical activity. The diet and physical activity questionnaire template, including the food frequency questionnaire (FFQ) section, is reported in Supplementary Table S3.
Toxicity endpoint definition
For every GI symptom included in the CTCAE, we calculated the average grade across all the early time points (≤8 weeks from RT initiation). Patients with at least one symptom with average grade >1.3 were defined as having developed a persistent form of acute GI toxicity (hereafter termed ‘with toxicity’ or ‘in the toxicity group’) which was the primary endpoint. This threshold allowed the capture of sustained forms of early acute toxicity for which at least one grade 2 is observed in one week and an average grade >1 is measured across the remaining weeks.
Collection and processing of biological samples
Faecal sample collection and processing
Faecal specimens were collected using the OMNIgene•GUT stool devices (DNA Genotek Inc. Ottawa, ON, Canada) consisting of a tube with a preservation buffer to stabilise microbial DNA and a bearing steel bead. Patients were instructed to collect the faeces into the tube, avoid contaminations, and homogenise the sample by shaking. The samples were stored at room temperature and delivered to the centralised laboratory for metagenomics analyses.
Upon arrival, after mechanical lysis with silica beads, DNA was extracted by QIAsymphony DSP Virus/Pathogen Midi kit (Qiagen) on an automated QIAsymphony station (Qiagen). Microbial DNA was quantified using a Qubit fluorometer (ThermoFisher), and quality was assessed using a 4200 TapeStation (Agilent). Samples reaching good quality (DNA integrity number >7) were used for metagenomics profiling.
The NGS libraries were prepared using 16S Metagenomics kit (ThermoFisher) following the manufacturer's instructions. The 16S region was amplified with primer sets recognising V2, V4, V8 and V3, V6-7, and V9 hypervariable regions in 2 separate PCR reactions. Fifty nanograms of amplicons were combined and processed for library prep using the Ion Plus Fragment Library Kit and Ion Xpress Barcodes Adapters (ThermoFisher). After PCR amplification (1 cycle of 95 °C for 5 min; 5 cycles of 95 °C for 15 sec, 58 °C for 15 s, 70 °C for 1 min) and purification using 1.4 volumes of Agencourt AMPure beads (Beckman Coulter), libraries were eluted and their size and quantity were assessed with TapeStation.
16S rRNA sequencing and bioinformatic analysis
Sequencing was performed for 16S libraries by Ion S5 XL, whereas base calling and demultiplexing was performed by Torrent Suite (ThermoFisher). The ThermoFisher Ion Reporter Software metagenomics 16S analysis pipeline was used to generate operational taxonomic unit abundances from the 16S rRNA reads and to assign taxonomy at the genus level by clustering sequences at a 97% similarity threshold.
Cytokines measurement
Ten millilitres of EDTA blood samples (BD Vacutainer™ K2 EDTA-367525) were obtained at baseline. Samples were kept at 4 °C for a maximum of 1 h after sampling and then centrifuged for 20 min at 2200g at 4 °C. Plasma was collected (∼4 mL) and centrifugated for 10 min at 2200g at 4 °C. Supernatant was immediately stored at ≤−80 °C until analysis (stored in Nalgene CryoBox-50260909).
All analyses were carried out blind to patient and therapy factors. The concentrations of CCL2, PDGF-BB, TGF-β1, TNF-α and TNFR1 were determined using commercially available ELISA kits (Quantikine®ELISA R&D Systems Inc., Minneapolis, MN, USA), according to manufacturer's protocols: Quantikine®ELISA Human TGF-β1 cod. DB100B R&D System, Quantikine®ELISA Human CCL2/MCP-1 cod. DCP00 R&D System, Quantikine®ELISA Human PDGF-BB cod. DBB00 R&D System, Quantikine®ELISA Human TNF-α cod. DTA00C R&D System and Quantikine®ELISA Human TNF RI cod. DRT100 R&D System.
MARS external data
MARS microbiome data9 and clinical data from the early cohort (n = 32), were used as external validation data. To homogenise the endpoint between the studies at best, we employed a robust GI toxicity endpoint for MARS patients defined as the consensus of grade ≤2 toxicity between patient-reported outcomes and clinician-reported outcomes. We used the data from the MARS early cohort and the MARS late cohort (n = 87) to assess microbiome-based predictors of toxicity (Supplementary Fig. S15).
Statistics
The computational methodology developed in this paper aimed to construct a model for assessing the risk of toxicity based on the profiled intestinal microbiota from baseline faecal samples. The analysis of the microbiota of the patients in the discovery cohort resulted in the identification of risk classes for the development of acute GI toxicity. Machine learning was used to develop a decision tree that could accurately predict patients at high risk of developing toxicity. The decision tree was trained on the discovery cohort and validated both on the validation cohort and on external data from the MARS study.9
Analysis of microbiota diversity
We measured the alpha diversity from the samples according to the Shannon's index and to the inverse Simpson's index12(hereafter termed ‘Simpson's index’ for simplicity) because they showed no correlation with the total number of mapped reads (Supplementary Fig. S6).
Microbiota-based clustering of patients
We performed hierarchical clustering using the Euclidean distance and the Ward linkage method on the core centre-log ratio (clr) of the abundance profiles standardised across the discovery population (z-scores).
The core included all genera having relative abundance ≥2% in ≥10% of the discovery patients, and the count data of the core sub-composition were clr-transformed after imputation of zeros via Geometric Bayesian Multiplicative replacement method previous to clustering.
We defined the optimal number of clusters as the one maximising the Jaccard similarity index between the partition of toxicity events obtained by using core bacterial genera and the one obtained by using core bacterial families, defined as the families having relative abundance ≥2% in ≥10% of the discovery patients (Supplementary Figs. S8 and S9).
A microbiota risk class for acute GI toxicity (high risk/moderate risk/low risk) was assigned to each microbiota-based cluster of patients based on a Fisher's exact test on the toxicity rate observed inside that cluster (‘Acute GI toxicity’ enrichment/‘No acute GI toxicity’ enrichment).
Polycytokinic risk score
We developed a logistic stepwise backward regression model to predict the classes of ‘Acute GI toxicity’ (positive cases) and ‘No acute GI toxicity’ (negative cases) using as predictors the log-transformed values of concentration of the plasma cytokines measured. We used the stepAIC function from the R package MASS version 7.3–5713 to identify the best predictors. The logit model based on the selected predictors was used to obtain a risk probability for each patient, and the polycytokinic risk score was defined by standardising the probabilities across the patient population.
Statistical analysis
Continuous variables, including age, BMI, the log-transformed concentrations of the plasma cytokines, doses, alpha diversity indices, and discrete variables (such as the number of genera) were tested for their statistical association with the toxicity endpoint using Wilcoxon's rank test.
Categorical variables, including comorbidities, probiotic use, drug use, surgeries previous to RT, diet-related groups and physical activity class, were tested for enrichment in the toxicity or no toxicity groups using Fisher's exact test.
Differential abundance analysis of genera between toxicity groups was performed using the Wilcoxon's rank test. To account for the compositional nature of the data,14 count data were transformed using the centred log-ratio transformation (clr) after imputation of zeros via Geometric Bayesian Multiplicative replacement method using R package zCompositions version 1.4.0.15 p values were adjusted using Benjamini–Hochberg method to control the false discovery rate at the level of 5%. Microbiota-based clusters of patients were tested for enrichment in toxicity and other categorical factors using Fisher's exact test. Statistical differences in the polycytokinic risk score, mean rectal dose, alpha diversity indices, number of core genera, age and BMI between microbiota-based risk classes were performed using Wilcoxon's rank test.
Multivariable logistic regression models of acute GI toxicity
We developed logistic multivariable regression models using the function lrm from the R package rms.16 We considered different combinations of predictors among the set including the level of pro-inflammatory cytokines CCL2, PDGF-BB, TGF-β1, TNF-α and TNFR1, BMI, age, mean rectal dose and the Shannon's index measured on the core microbiota.
Machine learning methods for clinical decision tree construction
We developed the clinical decision tree to categorise patients in the high-risk/moderate-risk/low-risk microbiota classes by using R package rpart version 4.1.1617 while aiming at reducing the sensitivity of the prediction to potential factors specific to the training data set, including the geography of the cohort and the sequencing platform.
Accordingly, we developed the tree algorithm with the following requirements:
-
1.
base the decision on the relative abundance of genera profiled from a single sample;
-
2.
having final predictors that allowed external testing on the MARS cohort data;
-
3.
obtaining after training a reduced depth of the branches for clinical interpretability.
As a feature pre-selection process before the training, we considered the logarithm of the relative abundances of the core genera that were significantly differentially abundant (package limma version 3.50.0,18 adjusted p < 0.05, |logFC|>1.5) in the high-risk microbiota cluster compared to any other cluster in the discovery cohort (Supplementary Table S13).
We then filtered out the pre-selected genera that were not profiled in the MARS early cohort, resulting in retaining the relative abundance of six genera, namely Faecalibacterium, Bacteroides, Parabacteroides, Alistipes, Prevotella, Phascolarctobacterium. The count distribution of these genera is shown in Supplementary Fig. S15 for the MicroLearner discovery and validation cohorts and for the MARS early and late cohorts.
To improve prediction robustness, we trained the decision tree model using the relative abundances of the retained genera rounded to the closest per cent point to predict the microbiota risk class of patients in the discovery cohort. To limit the complexity of the tree, we pruned it at the minimum number of nodes and splits necessary to reach 80% prediction accuracy on the high-risk class. This process kept all six retained core genera in use as effective predictors in the final tree model.
The model was tested by assigning a predicted microbiota risk class to patients in our validation cohort (n = 79) and the MARS early cohort (n = 32) and measuring the toxicity rate within the predicted classes. The tree model training and validation details are reported in Supplementary Tables S14 and S15.
Functional imputed metagenomics
We performed the functional imputed metagenomic analysis of the core microbiota profiles of the patients in the discovery cohort using PICRUSt version 1.1.4.19 We referenced the core metagenome profiles to the most updated reference collection of OTUs from Greengenes (gg_13_5_otus).20 Because the taxonomy assignment by Ion Reporter is based on ThermoFisher-curated Greengenes references, this choice maximised the coherence between inferred functional profiles and taxonomic abundance profiles.
Profiles were normalised by the known or predicted 16S copy number abundance, and a final metagenome was predicted and mapped to KEGG Orthologs (KOs). Inferred KEGG Orthologs (KO) IDs annotations based on the Integrated Microbial Genomes and Microbiomes21 were manually verified and curated against the most recent version of KEGG (v.104, 2022/10) and of the Transport Classification database22 and annotated by KEGG modules or Brites.
We checked the quality of the inferred metagenome by calculating the average Nearest Sequenced Taxon Index (NSTI)19 across samples, which quantifies the average over the samples of the average substitutions per site separating each OTU from the reference bacterial genome. We obtained NSTI = 0.071+−0.028, consistent with values expected from faecal samples.19
Functional analysis of microbiota risk classes
We identified a first set of KOs of interest by selecting KO terms with average standardised relative abundance >1.5, in absolute value, across the high-risk microbiota patients.
A second set of KOs of interest was identified by selecting KO terms significantly different in terms of log-transformed relative abundance between the low-risk microbiota patients and the high-risk microbiota patients, by using R package limma version 3.50.018 with significance threshold given by |log(FC)|>1.5 and adjusted p < 0.05 (Supplementary Table S12).
Functional profiles (KOs relative abundances) were clustered using hierarchical clustering with Euclidean distance and Ward's linkage method. The relative contribution of individual genera to the abundance of the KO of interest was also extracted using PICRUSt function metagenome_contributions.py (Supplementary Figs. S13 and S14).
Role of funders
The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Results
Patient population, RT treatment and acute toxicity rates
Of 244 PCa patients enrolled, 9 dropped before completing RT, 4 lacked toxicity data, and 16 had samples with low DNA integrity number and were not profiled. Information from a patient-compiled diet and lifestyle survey was available for 135 patients in the discovery cohort, and the levels of serum pro-inflammatory cytokines were measured in 99 patients of the discovery cohort.
To investigate the association between the intestinal microbiota and the development of GI toxicity during the RT treatment, we analysed faecal samples from n = 136 MicroLearner discovery patients and n = 79 MicroLearner validation patients whose characteristics are reported in Supplementary Table S1.
All patients received Volumetric Modulated Arc RT (VMAT). One hundred and forty-five patients received conventionally fractionated RT. Fifty-seven were post-prostatectomy patients (adjuvant setting 11 patients, salvage 46 patients) treated with a prescribed dose of 70–72 Gy. Eighty-eight radical patients had a prescribed dose of 74–78 Gy. Seventy radical patients received moderately hypofractionated RT (2.6 Gy/day) with a prescribed dose of 65–67.6 Gy. One hundred and seventy-two patients received whole-pelvis RT (50 Gy) and 142 irradiation of seminal vesicles (66–70 Gy). Treatment details are reported in Supplementary Table S2.
Twenty-four patients were scored with an average grade >1.3 for acute GI toxicity, of which 16 were in the discovery cohort (12% of the cohort), and 8 were in the validation cohort (10% of the cohort).
Association of acute GI toxicity with dosimetric and patient-specific features
The mean dose to the rectum stratified by toxicity group and RT intention is shown across the pooled cohort in Fig. 1a. We did not find statistically significant associations with acute GI toxicity. No significant dose-toxicity association was found in the populations who received whole-pelvis or prostate-only RT (Supplementary Fig. S2).
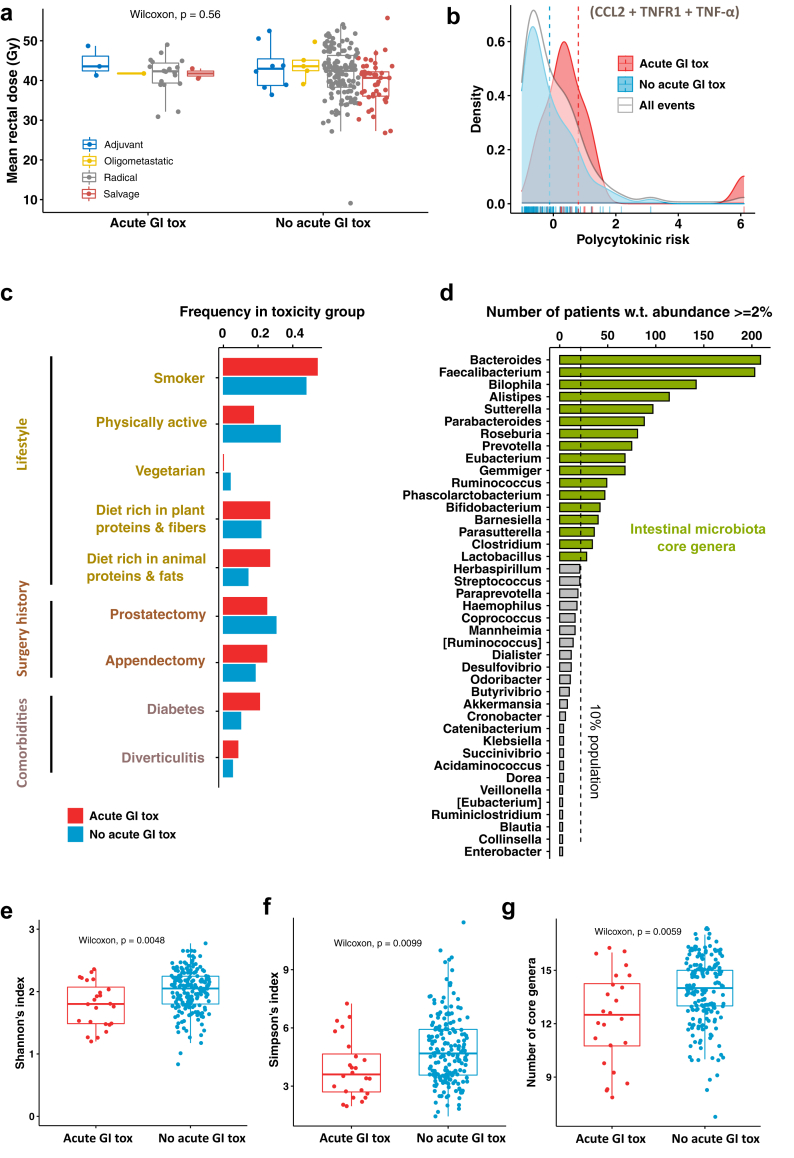
Fig. 1.
Analysis of the risk factors for the development of early acute GI toxicity. (a) Values of the mean rectal dose across the patient population, stratified by RT intent (n = 215) and by toxicity group. (b) Density plots of the risk score associated with RT-related pro-inflammatory cytokines (polycytokinic risk) across the overall population (grey) and in the toxicity groups (blue, red). (c) Frequency of potential protective and risk factors measured within the toxicity groups. (d) Statistic of the intestinal microbiota bacterial genera detected in the RT-baseline faecal samples of the patient population with a relative abundance of at least 2%; bacterial genera measured with at least 2% relative abundance in at least 10% of the sample population define the core intestinal microbiota. (e) Shannon's index, (f) Simpson's index and (g) number of core genera measured from the microbiota profiles (genus level) and stratified by toxicity group (a small vertical jitter is applied to dots to improve visualisation).
To explore the possible influence of the inflammatory environment in modulating the response to radiation, we assigned to each patient a measure of inflammatory-driven radio-sensitivity, the polycytokinic risk score, based on the plasma concentration of CCL2, TNFR1 and TNF-α. In Fig. 1b the distributions of the score in toxicity (red) and no toxicity (blue) patients are shown (overall distribution in grey). Despite an Odds Ratio (OR) of 2.04 (95% CI = 1.29–2.79, p = 0.02) for a 1-point increase in the polycytokinic risk score, and a 5% toxicity rate among patients with score ≤0 compared to 25% in patients with score >0 (Fisher's exact test p = 0.005; Supplementary Table S8), the score distributions overlapped significantly (Welch's t-test p = 0.075), denoting a limited discrimination power of the cytokines (details of the polycytokinic risk model are reported in Supplementary Table S8).
No significant associations between BMI or age and acute GI toxicity were found (Supplementary Fig. S7a and b). We developed a multivariable logistic model to check whether the interplay of these two factors with cytokines and mean rectal dose could reveal any predictive potential for toxicity when used in combination as predictors and found that no regression coefficient was significantly different from zero (see Supplementary Table S16).
For what concerns diet and lifestyle, specific food classes from the FFQ were aggregated to analyse the diet composition (Supplementary Fig. S1). Patient categories related to diet and physical activity were defined according to the criteria reported in Supplementary Table S5. Supplementary Table S4 reports the statistics of these categories in the patient population. We compared potential risk/beneficial factors for GI toxicity (Fig. 1c) that were represented in ≥3% of the patients (n ≥ 7). Specifically, smoking history, physical activity, vegetarian diet, plant proteins and fibres-rich diet, animal fats and protein-rich diet, previous prostatectomy, previous appendectomy, diabetes and diverticulitis, and found no statistically significant differences (Supplementary Table S7). However, a trend was observed for physical activity (protective factor, 6.3% toxicity rate in active patients vs 13.7% in sedentary patients, p = 0.1), diabetes (risk factor, 20.8% toxicity rate in patients with diabetes vs 9.9% in patients without, p = 0.11), and a diet rich in animal fats and proteins (risk factor, 19% toxicity rate in patients declaring high intake of animal fats and proteins vs 9.6% for patients with low intake of animal fats and proteins, p = 0.18) (Fig. 1c and Supplementary Table S7). Also, we observed the sub-group of patients taking statins as regular medication to have a significantly lower risk of toxicity (rate 3.4% vs 14% for patients not taking statins, p = 0.019) and a trend for ACE inhibitors to be beneficial (4.7% vs 12.8%, p = 0.1) (details in Supplementary Table S7).
Association of acute GI toxicity with intestinal microbiota diversity
We analysed the average relative abundance of the bacterial genera and their occurrence (Supplementary Figs. S3 and S4). The core microbiota composition at the genus level (Fig. 1d) included 17 out of 115 genera sequenced in both cohorts of validation and discovery (∼15%).
When we measured the alpha-diversity of microbiota at the genus level (Shannon's index Fig. 1e, Simpson's index Fig. 1f) we observed that a lower community diversity before the radiotherapy was significantly associated with acute GI toxicity development (Wilcoxon's rank test; Shannon's index: p = 0.0048; Simpson's index: p = 0.0099). The presence of a lower number of core genera at baseline was also associated with acute GI toxicity (p = 0.0059; Fig. 1g). Interestingly, the significance held when remeasuring the alpha-diversity on the restricted core microbiota sub-composition (Supplementary Fig. S7c and d), suggesting that the core microbiota can capture alone the observed gap in alpha-diversity between groups (Shannon's index: p = 0.0028; Simpson's index: p = 0.0083).
When we included the Shannon's index of alpha diversity measured on the core microbiota among the predictors of multivariable logistic models of acute GI toxicity including combinations of clinical risk factors (BMI, age and mean rectal dose) and cytokines, we found that the microbiota diversity was the only feature with a coefficient significantly different from zero (see Supplementary Tables S17–S19).
Microbiota-based clustering of patients
On univariate analysis, no core genus differed significantly in abundance between patients with/without acute GI toxicity in the discovery cohort. Among non-core genera, Flavonifractor tested significantly differentially abundant in the discovery cohort (Wilcoxon's rank test adjusted p = 0.042) but was not significantly different in the validation cohort (Supplementary Table S9, Supplementary Fig. S5).
To further test whether microbial clusters rather than individual features predicted GI toxicity, we performed a multivariate analysis of the core microbiota composition via hierarchically clustering the abundance core profiles in the discovery cohort (n = 136). We found 8 clusters of patients (Fig. 2a) with toxicity rates ranging from 0% to 60% (Fig. 2b). The cluster with the highest toxicity rate (60%, termed high-risk microbiota class) included 10 patients (7% of the cohort, 38% of patients with toxicity) with significantly higher toxicity rates (Fisher's exact test p = 0.00016). A separate cluster with 26 patients had a 0% toxicity rate and significantly less toxicity than other clusters (Fisher's exact test p = 0.027); we termed it low-risk microbiota class. Other clusters were not significantly different in terms of toxicity rates range (3%–25%) and were grouped to define a moderate-risk microbiota class.
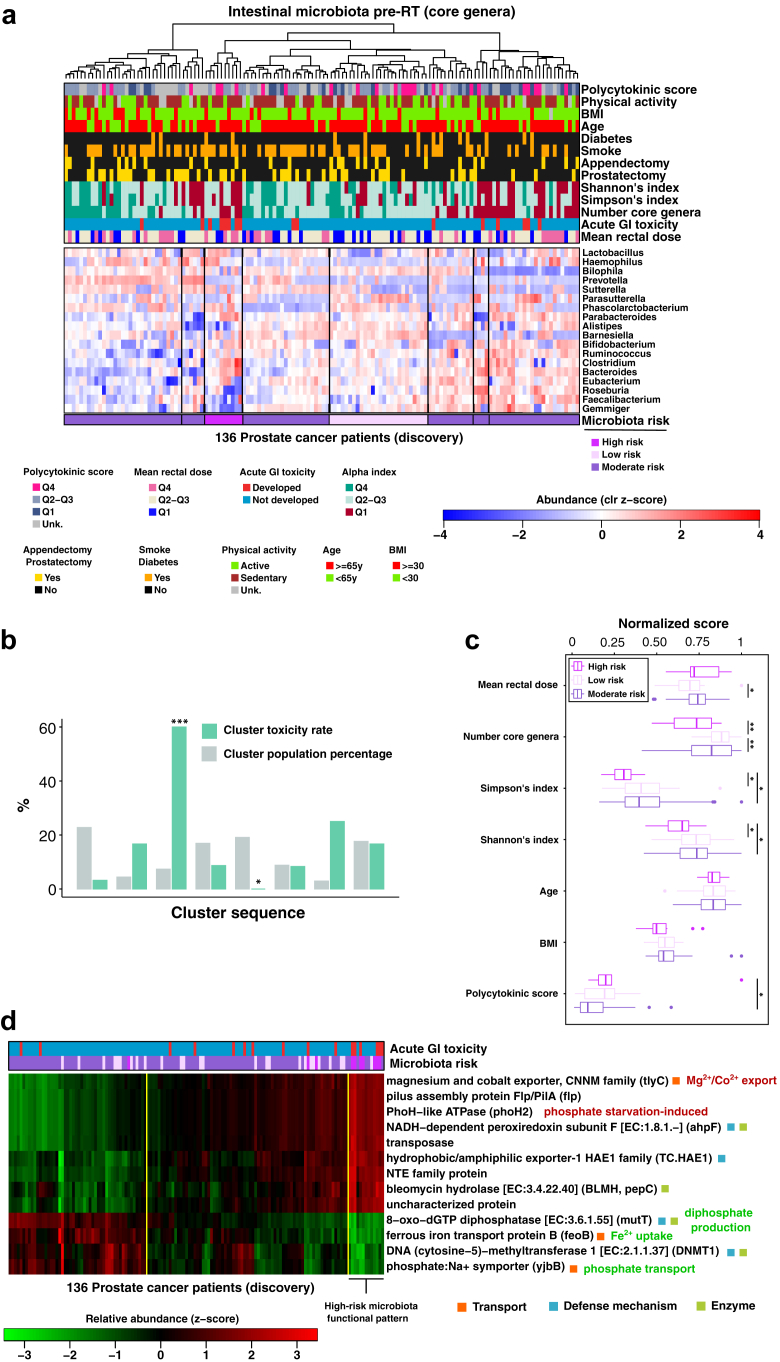
Fig. 2.
Association of the intestinal microbiota with RT-induced GI toxicity. (a) Heatmap showing the normalised abundance profiles (standardised clr-transformed relative abundance values) of the core microbiota genera (rows) from patients in the MicroLearner PCa discovery cohort (columns); core genera were defined as those genera found in at least 10% of the cohort with relative abundance ≥2%; hierarchical clustering was used to discover 8 clusters of patients (black vertical lines) with similar core compositions to which we assigned a microbiota class for the risk for developing acute GI toxicity during RT. (b) Bar plot showing the toxicity rate observed in each cluster of patients and the cluster size in unit of population percentage (cluster sequence same as in the heatmap); cluster number three was significantly enriched for toxicity (60% toxicity rate, high microbiota risk) while cluster five was significantly enriched for no toxicity (0% toxicity rate, low microbiota risk); other clusters show toxicity rate in the range 3–25% (moderate microbiota risk). (c) Statistic of normalised indices of toxicity risk factors visualised by microbiota risk class. (d) Heatmap showing the microbiota functional profiles (standardised relative abundance values) of 13 selected KEGG Orthologs (KOs, rows) imputed from the core microbiota abundance profiles of the patients in the MicroLearner PCa discovery cohort (columns); KOs selected have an average within-group absolute z-score abundance value >1 in the high-risk microbiota patients; hierarchical clustering was used to define 3 clusters of patients (yellow vertical lines) and to reveal the functional pattern associated with the microbiota composition at high-risk for toxicity.
Stratification of alpha-diversity scores by microbiota risk class (Fig. 2c) showed statistically significant gaps in the average diversity passing from low-to moderate-to high-risk, while age, BMI, mean rectal dose and polycytokinic risk score did not differ significantly on average between the low-risk class and the high-risk class, indicating that other risk factors did not confound the microbiota risk classes.
Functional imputed metagenomics and functional analysis
Patients in the high-risk microbiota class showed an unbalanced core microbiota, with genus abundances deviating from the average cohort levels significantly more than in the low-risk microbiota class (Wilcoxon's rank test on the L1-norm of the profiles, high-risk vs low-risk, p = 0.000059, Supplementary Fig. S10a). Also, the average within-cluster unbalance showed a positive trend with increased toxicity rate (R2 = 0.64, p = 0.085; Supplementary Fig. S10b).
We thus investigated functional traits characteristic of the core microbiota of the patients at higher risk of toxicity by performing a functional imputation analysis. Fig. 2d shows the relative abundance of 13 selected KEGG Orthologs (KOs) imputed from the core genera abundance profiles of the discovery cohort.
The functional pattern characteristic of the high-risk microbiota class can be visualised by re-clustering patients using functional profiles into three functional groups. This pattern clustered together 7 of the 10 patients with a high-risk microbiota (5/7, 71%, with observed toxicity), suggesting that combining functional information with data on the community composition might capture the risk of developing toxicity more accurately.
Several identified genes were either related to transport or to defence mechanisms. TlyC (over-represented in the high-risk microbiota) is associated with the efflux of intracellular magnesium and cobalt in Proteobacteria and Firmicutes. YjbB and feoB (both under-represented in the high-risk microbiota) are involved in phosphate and iron uptake, respectively. The enzyme 8-oxo-dGTP diphosphatase (under-represented in the high-risk microbiota) prevents the incorporation of oxidised purine nucleoside triphosphates into DNA, in a reaction catalysed by magnesium that produces hydrogen diphosphate. The enzyme ahpF (over-represented in the high-risk microbiota) has a defensive function and protects the cell against DNA damage by alkyl peroxide.
To investigate whether some peculiar intestinal microbiota composition protects from radiation-induced effects, we performed a differential abundance analysis of the inferred core metagenomes between low-risk and high-risk microbiota groups. We found 29 significantly different KOs (Supplementary Table S12) related to different aspects of metabolism, including carbon metabolism and biosynthesis and transport of amino acids (Supplementary Fig. S13).
Hierarchical clustering of the functional profiles into three groups (Supplementary Fig. S11a) identified patterns associated with high-risk and low-risk microbiota patients and, in turn, identified three biological pathways containing metagenes consistently over-represented (butanoate metabolism, biosynthesis of cofactors) or under-represented (histidine metabolism) in the low-risk pattern (Supplementary Fig. S11b).
A cross-comparison of the functional clusters obtained with the sets of 13 KOs and 29 KOs (Supplementary Fig. S12) revealed an overlap of 100% between the high-risk functional pattern groups and 79% overlap between the low-risk functional pattern groups, indicating that the sets of features are both characteristic of the high-risk microbiota and that the set of 29 KOs is more distinctive with respect to the low-risk microbiota.
Development of a microbiota-based clinical decision tree to predict the risk of acute GI toxicity
We built an interpretable machine learning model that could assess the microbiota-related risk of the individual patient to develop toxicity during treatment starting from his baseline faecal sample.
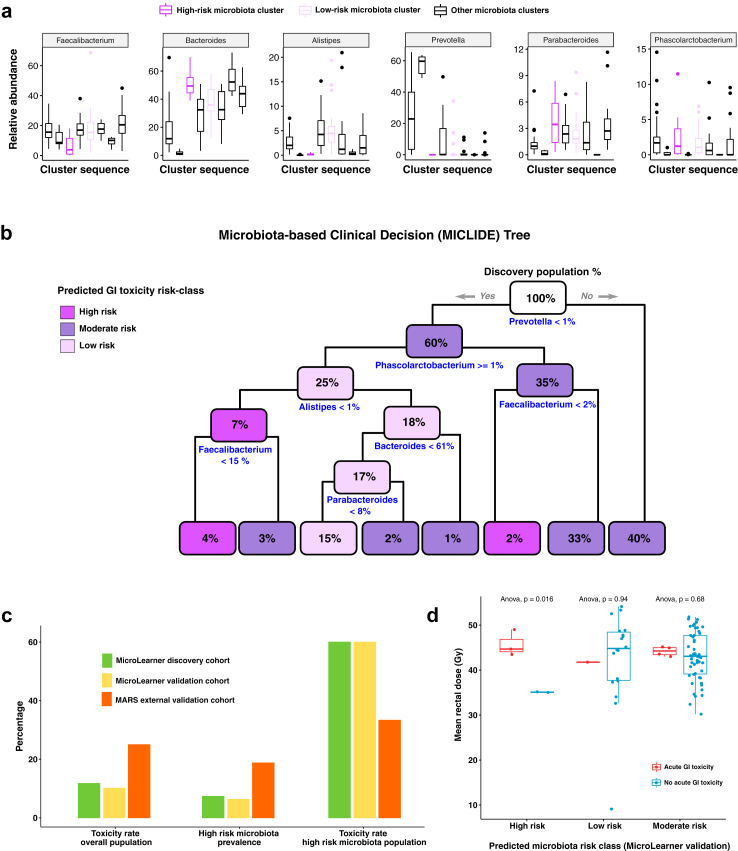
In Fig. 3a, the distribution of the selected features evaluated by the clinical decision tree (the MICLIDE Tree, MIcrobiota-based CLInical DEcision Tree) is shown across the discovery cohort, stratified by clusters of microbiota composition. They include the relative abundance of the core genera Faecalibacterium, Bacteroides, Parabacteroides, Alistipes, Prevotella and Phascolarctobacterium. Fig. 3b gives an overview of the set of logical rules used by the clinical decision tree to assess the microbiota-associated risk of an individual microbiota profile. Fig. 3c reports the measures used to evaluate the model performance, including the rates of acute GI toxicity evaluated in the cohorts used to train (discovery cohort) and test the model (validation cohorts), the prevalence of patients at high risk for toxicity as determined by microbiota classes that were measured in the discovery cohort (established via unsupervised clustering) and predicted (by the decision tree model) in the validation cohorts, and the rate of acute GI toxicity in the subgroup of patients found (discovery cohort) or predicted (validation cohorts) to have high-risk microbiota.
Fig. 3.
Prediction of acute GI toxicity from RT-baseline faecal samples. (a) Statistics of the relative abundance of the six core genera used as predictors of acute GI toxicity, visualised across the eight clusters of the discovery cohort, including the high- and the low-risk microbiota clusters. (b) Schematic of the clinical decision tree algorithm (MICLIDE) developed to predict the risk of developing RT-induced acute GI toxicity from individual faecal samples at treatment baseline; the prediction rules to assign patients to risk classes based on the proportion of selected genera are shown in blue under each node of the tree; starting from the root node (top), the decision rules are evaluated for any individual patient microbiota profile (if yes, follow left branch otherwise follow right branch); each node reports the percentage of the discovery population classified to that level of the tree hierarchy and it is coloured according to the prevalence of microbiota risk class within. (c) Bar plots showing the performance of the MICLIDE tree predicting acute GI toxicity; the plot on the left shows the rates of acute GI toxicity measured in the populations used to train the model (MicroLearner discovery cohort) and to test the model (MicroLearner validation cohort and MARS cohort); the plot in the centre compares the prevalence of patients at high-risk microbiota found in the MicroLearner discovery cohort with the same prevalence predicted by the model in the MicroLearner validation cohort and in the MARS cohort; the plot on the right shows the rate of acute GI toxicity measured in the subgroup of patients with high-risk microbiota across the discovery and the validation cohorts. (d) Statistic of mean rectal dose received by patients in the MicroLearner validation cohort stratified by microbiota risk class predicted by the MICLIDE tree and by toxicity group.
In the MicroLearner validation cohort, the model predicted 5 patients with high-risk microbiota (6% of the population), with a 60% toxicity rate (3/5), consistent with the incidence and characteristic toxicity rate observed in high-risk microbiota patients of the discovery cohort (Supplementary Tables S14 and S15).
In the MARS cohort, a higher rate of toxicity in the high-risk class predicted by the model was also observed, with the model separating high-risk (toxicity rate: 33%) and moderate-risk (23%) classes, with no patients in the low-risk class (Supplementary Table S15).
Additionally, when we stratified the mean rectal dose received by patients in the MicroLearner validation cohort by predicted microbiota risk class and by toxicity group (Fig. 3d), we observed that a significantly lower dose was associated with high-risk microbiota patients that did not develop toxicity compared to high-risk patients that did develop acute GI toxicity and to patients in other microbiota risk classes (Anova, p = 0.0016).
Discussion
To our knowledge, we report the most extensive study investigating whether the intestinal microbiome at RT starts associates with the development of acute GI toxicity in a clinical setting, with data from 215 consecutive PCa patients enrolled between 2017 and 2019 who received high-dose exclusive or post-prostatectomy RT.
We show that a low diversity of the intestinal microbiota is a clinical risk indicator of the onset of GI side effects induced by RT during treatment. Bacterial diversity is a high-level measure of a healthy microbiota, and reduced diversity is associated with multiple conditions, including inflammatory bowel diseases, diabetes and obesity, and is often associated with radio-induced GI toxicity in cohort studies.5
We also found that a lower alpha-diversity of the core intestinal microbiota (made of 18 genera having a relative abundance ≥2% in ≥10% of the discovery cohort) was significantly associated with toxicity, suggesting that diversity can be robustly estimated from a subset of the microbiome and this measure can then be used for prediction purposes. This observation also suggested that core microbiota composition may predict a patient-specific risk of radiation-induced intestinal side effects. We did not find, however, a specific bacterial genus in the core associated with toxicity. This is consistent with previous studies and illustrates the functional redundancy of many bacteria, suggesting that the microbiome function is at least as relevant as taxonomy for prediction.9
An unsupervised clustering analysis of the core microbiota composition identified a subset of patients with higher toxicity rates. The functional characterisation of the bacterial communities of these high-risk patients pointed to a selection of species that are more adapted for coping with oxidative stress and phosphate scarcity. Specifically, we found a higher relative abundance of Bacteroides and Sutterella (enhanced ahpF and phosphate-deficiency induced ATPase PhoH2) and, conversely, under-abundance of oxygen-sensitive Roseburia and phosphate-metabolising Faecalibacterium.
This suggests that the microbiota composition might reflect a host environment characterised by increased reactive oxygen species (ROS) levels. Such an environment can be expected to predispose to more severe damage of the intestinal epithelium and prolonged pro-inflammatory responses following radiation oxidative effects23,24 and, consequently, to radiation-induced GI toxicity.
Bacterial communities might also directly contribute to altered levels of ROS and oxygen gradients in the intestinal environment through the exchange of cations, such as Mg2+, Fe2+ and Na+, and the release of signalling bacterial metabolites, including short-chain fatty acids. Short-chain fatty acids (SCFAs), including butyrate (butanoate), are bacterial-produced metabolites essential for intestinal homeostasis and have been suggested to protect against radiation-induced GI toxicity by consolidating the mucus layer and acting as molecular triggers for the recruitment of enterocytes and mucosal Treg cells.24,25 Moreover, butyrate utilisation by enterocytes increases the consumption of oxygen and activates hypoxia-inducible factors, contributing to the basal hypoxic tone of the intestine.23 Roseburia and Faecalibacterium showed the highest contribution to butanoate metabolism (Supplementary Fig. S13) and were more abundant in the low-risk compared to the high-risk class, consistent with the fact that R. intestinalis and F. prausnitzii are the most abundant butyrate-producing bacteria found in human faeces.
Consistently, our analysis highlighted the benefit of butyrate synthesis, which was found to be enhanced in the intestinal microbiota of patients at low risk for toxicity. Interestingly, Guo et al., reporting on the radioprotective role of SCFAs, observed that butyrate- and propionate-treated mice presented significantly reduced levels of intracellular ROS in bone marrow stem cells after whole-body irradiation.26
Besides, these characteristic microbiota traits associated with the risk of developing toxicity share similarities with dysbiosis observed in pro-inflammatory intestinal states in general. For example, a higher abundance of bacteria capable of withstanding a highly oxidative environment27 is also associated with intestinal inflammation in Crohn's disease, and a profoundly decreased abundance of butyrate-producing genera was observed in patients following conditioning chemotherapy.28 Interestingly, the monotypic genus Flavonifractor, which we found significantly overabundant in the toxicity patients among non-core genera, was reported to associate with colorectal cancer in an Indian cohort with a potential explanation in its ability to degrade beneficial anticarcinogenic flavonoids, such as quercetin, that could function as antioxidant molecules.29
We detected an influence of the inflammatory environment in modulating the response to radiation, with an OR of 2 for a one-point increase in the polycitokinic risk score based on the plasma concentration of CCL2, TNFR1 and TNF-α. Despite this association, the polycitokinic score distributions for patients with and without toxicity overlapped significantly, and the polycitokinic risk score had limited discrimination power.
We also investigated the association between GI toxicity and dosimetric/patient-specific risk factors. We did not find statistically significant associations between rectal doses and acute GI morbidity. The mean dose to the rectum was already optimised in the planning phase following guidelines (maximum mean rectal dose 55.7 Gy; interquartile range [38–45] Gy), and the limited variation of this parameter intrinsically limits its potential as a toxicity predictor. We found no statistically significant association between toxicity and patient factors (age, BMI, comorbidities, diet, smoking), even after including them in a multivariable logistic regression together with Shannon's index for the alpha diversity of the patients' core microbiota. We observed trends for physical activity (6.2% vs 13.7% toxicity rate in active and sedentary patients, respectively, p = 0.1), diabetes (20.8% vs 9.9% in diabetic and non-diabetic patients, p = 0.11), and consumption of animal fats and proteins (19% vs 9.6% toxicity rate in patients with high vs low intake). Plant-based diets are associated with healthy and diverse intestinal microbiota,23 while diabetes is known to increase the risk of RT-induced side effects, likely due to microvasculature dysfunction and impairment in tissue repair mechanisms.30,31 Our study was limited by design in the possibility of investigating how dietary habits could modulate the response to radiation and how the microbiota composition of high-risk patients could be associated with dietary features. Indeed, much longer times are required to enrol patients on diverse non-prevalent diets, such as vegetarian or vegan diets.
When considering drugs, patients taking statins regularly exhibited a significantly lower risk of toxicity (3.4% vs 14% for statin and non-statin users, p = 0.019), consistent with previously reported protective effects of this class of drugs.32
We delineated a methodology to construct a clinical model predicting a microbiota-based risk of developing toxicity for single patients. After using unsupervised clustering to discover patterns within the microbiota composition, we computed the toxicity rate in each cluster to distinguish clusters in those with a significantly higher toxicity rate (high risk), those with a significantly lower toxicity rate (low risk) and those with intermediate non-significantly different toxicity rates (moderate or intermediate risk). Clustering is more effective than building signatures using multivariable regression when the feature set is highly dimensional and when the endpoint of interest is associated with anomaly detection (patients with radio-induced toxicity in well-optimised radiotherapy plans can be considered anomalies/outliers). Accordingly, we used machine learning to develop a decision tree (the MICLIDE Tree) to categorise patients in the high-/moderate-/low-risk classes, and we tested the model on the independent populations by checking that patients classified as high-risk still exhibited a significantly higher toxicity rate.
Our clinical decision tree algorithm was implemented and validated using microbiota data from the MicroLearner/Italy (Thermofisher sequencing) and the MARS/UK cohorts (Illumina sequencing). Despite the limitations due to the mono-institutional nature of the MicroLearner study, the results in external validation show that the predictive features identified, consisting of the relative abundance of the genera Faecalibacterium, Bacteroides, Parabacteroides, Alistipes, Prevotella and Phascolarctobacterium, appear to generalise prediction beyond sequencing technology and platforms and minimise biases associated with geographic origin of patients. In the MicroLearner validation cohort, the model predictions were consistent with the incidence and characteristic toxicity rate observed in high-risk microbiota patients of the discovery cohort. In the MARS cohort, a higher rate of acute side effects was expected compared to the other cohorts because all patients received whole pelvis RT, rather than mixed whole pelvis and prostate-only RT. Also, different criteria to score toxicity (see methods) might have contributed to capturing more punctual, less-sustained forms of acute GI toxicity. Despite these differences, a higher toxicity rate in the high-risk class predicted by the model was also observed, with the model separating high-risk (toxicity rate: 33%) and moderate-risk (23%) classes, with no patients in the low-risk class. We acknowledge potential biases of 16S rRNA marker gene sequencing for bacterial community profiling due to uneven PCR amplification of gene regions. However, in terms of costs and time, it remains the fastest and most affordable way to implement large-scale microbiota profiling of patients in the clinics.
The MICLIDE Tree can be straightforwardly translated into clinical practice to assess pre-radiotherapy toxicity risk and optimise radiotherapy planning by, for example, tightening radiation dose constraints in patients at higher risk of toxicity or using rectum spacers selectively in patients at higher risk of GI side effects. It may also aid the design of interventional measures to manipulate bacterial composition to improve early and late side effects, particularly by selecting patients more likely to benefit from those interventions. Given that 68% of the MicroLearner patient population received irradiation to the whole pelvis, the analytical approach and methodology developed here and the clinical decision tree model obtained might have broader applicability to other types of cancer whose treatment is also based on the irradiation of the abdomen or pelvis.
Interestingly, there exists a consequential behaviour between acute and late toxicity, with patients developing acute gastrointestinal toxicity having higher chances of developing also late gastrointestinal side effects. This known clinical implication also indirectly makes the results presented in this manuscript relevant for understanding late toxicity in the context of prostate cancer radiotherapy.
Our findings are based on statistical associations and patterns discovered by the use of machine learning techniques. To better elucidate the consequences that an altered community functionality might have on the host and open to interventional bacteriotherapy in patients at high risk for toxicity, we envisage that future research by means of different omics technologies, including shotgun metagenomics and metabolomics, will be the optimal strategy.
Contributors
Conceptualisation: T.R., R.V., E.O., L.D.C., F.P.; Data Curation: F.B., J.I., M.S.S., A.C., A.D., E.G., L.P.; Formal analysis: J.I., A.D.; Funding Acquisition: T.R., R.V., E.O.; Investigation: M.S.S., B.A., V.D., T.G., M.D., E.M., B.N.C., S.V.; Methodology: J.I., T.R., N.Z., L.D.C.; Project administration: T.R.; Resources: J.I., A.D., T.G.; Software: J.I.; Supervision: T.R., R.V., B.A., N.Z., L.D.C.; Validation: M.S.S., J.I., M.R.F., N.Z., L.D.C.; Visualisation: J.I., T.R., M.R.F.; Writing (original draft): J.I., T.R., M.R.F.; Writing (review and editing): all the authors. All authors read and approved the final version of the manuscript. T.R., L.D.C. accessed and verified the underlying data.
Data sharing statement
All sequencing data are deposited into the GEO (Gene Expression Omnibus) repository of NCBI (National Center for Biotechnology Expression) (http://www.ncbi.nlm.nih.gov/geo/), accession number GSE268869. IonReporter-generated OTU tables and associated metadata that can be used to reproduce the main findings of the study have also been deposited in the same repository.
Declaration of interests
The authors declare no conflict of interest.
Acknowledgements
MicroLearner was funded under the Call for the Promotion of Institutional Research INT-year 2016, 5 x 1000 by the Italian Ministry of Health. This work was founded by the Italian Ministry of Health – Ricerca Corrente funds. J.I. acknowledges funding from Fondazione Regionale per la Ricerca Biomedica, grant ID 2721017 (MicBioRadio). L.P. acknowledges funding from AIRC IG 21479.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2024.105246.
Appendix A. Supplementary data
References
- 1.Budäus L., Bolla M., Bossi A., et al. Functional outcomes and complications following radiation therapy for prostate cancer: a critical analysis of the literature. Eur Urol. 2012;61(1):112–127. doi: 10.1016/j.eururo.2011.09.027. [DOI] [PubMed] [Google Scholar]
- 2.Pinkawa M., Holy R., Piroth M.D., et al. Consequential late effects after radiotherapy for prostate cancer - a prospective longitudinal quality of life study. Radiat Oncol. 2010;5:27. doi: 10.1186/1748-717X-5-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heemsbergen W.D., Peeters S.T.H., Koper P.C.M., Hoogeman M.S., Lebesque J.V. Acute and late gastrointestinal toxicity after radiotherapy in prostate cancer patients: consequential late damage. Int J Radiat Oncol Biol Phys. 2006;66(1):3–10. doi: 10.1016/j.ijrobp.2006.03.055. [DOI] [PubMed] [Google Scholar]
- 4.Hauer-Jensen M., Denham J.W., Andreyev H.J.N. Radiation enteropathy--pathogenesis, treatment and prevention. Nat Rev Gastroenterol Hepatol. 2014;11(8):470–479. doi: 10.1038/nrgastro.2014.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferreira M.R., Muls A., Dearnaley D.P., Andreyev H.J.N. Microbiota and radiation-induced bowel toxicity: lessons from inflammatory bowel disease for the radiation oncologist. Lancet Oncol. 2014;15(3):e139–e147. doi: 10.1016/S1470-2045(13)70504-7. [DOI] [PubMed] [Google Scholar]
- 6.Wang A., Ling Z., Yang Z., et al. Gut microbial dysbiosis may predict diarrhea and fatigue in patients undergoing pelvic cancer radiotherapy: a pilot study. PLoS One. 2015;10(5) doi: 10.1371/journal.pone.0126312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Manichanh C., Varela E., Martinez C., et al. The gut microbiota predispose to the pathophysiology of acute postradiotherapy diarrhea. Am J Gastroenterol. 2008;103(7):1754–1761. doi: 10.1111/j.1572-0241.2008.01868.x. [DOI] [PubMed] [Google Scholar]
- 8.Jang B.S., Chung M.G., Lee D.S. Association between gut microbial change and acute gastrointestinal toxicity in patients with prostate cancer receiving definitive radiation therapy. Cancer Med. 2023;12(22):20727–20735. doi: 10.1002/cam4.6636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reis Ferreira M., Andreyev H.J.N., Mohammed K., et al. Microbiota- and radiotherapy-induced gastrointestinal side-effects (MARS) study: a large pilot study of the microbiome in acute and late-radiation enteropathy. Clin Cancer Res. 2019;25(21):6487–6500. doi: 10.1158/1078-0432.CCR-19-0960. [DOI] [PubMed] [Google Scholar]
- 10.Roy S., Trinchieri G. Microbiota: a key orchestrator of cancer therapy. Nat Rev Cancer. 2017;17(5):271–285. doi: 10.1038/nrc.2017.13. [DOI] [PubMed] [Google Scholar]
- 11.Bedini N., Cicchetti A., Palorini F., et al. Evaluation of mediators associated with the inflammatory response in prostate cancer patients undergoing radiotherapy. Dis Markers. 2018;2018 doi: 10.1155/2018/9128128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Madonsela S., Cho M.A., Ramoelo A., Mutanga O. Remote sensing of species diversity using Landsat 8 spectral variables. ISPRS J Photogrammetry Remote Sens. 2017;133:116–127. [Google Scholar]
- 13.Venables W.N., Ripley B.D. Springer; New York: 2002. Modern applied statistics with S. Fourth. [Google Scholar]
- 14.Gloor G.B., Macklaim J.M., Pawlowsky-Glahn V., Egozcue J.J. Microbiome datasets are compositional: and this is not optional. Front Microbiol. 2017;8:2224. doi: 10.3389/fmicb.2017.02224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Palarea-Albaladejo J., Martín-Fernández J.A. zCompositions — R package for multivariate imputation of left-censored data under a compositional approach. Chemometr Intell Lab Syst. 2015;143:85–96. [Google Scholar]
- 16.Harrell Jr FE. R package version; 2016. rms: regression modeling strategie; pp. 1–263. [Google Scholar]
- 17.Therneau T and Ab. B. A, Ripley B. R package version 4.1-3; 2013. Rpart: recursive partitioning. [Google Scholar]
- 18.Ritchie M.E., Phipson B., Wu D., et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Langille M.G.I., Zaneveld J., Caporaso J.G., et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31(9):814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DeSantis T.Z., Hugenholtz P., Larsen N., et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72(7):5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen I.M.A., Chu K., Palaniappan K., et al. The IMG/M data management and analysis system v.7: content updates and new features. Nucleic Acids Res. 2023;51(D1):D723–D732. doi: 10.1093/nar/gkac976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saier M.H., Reddy V.S., Moreno-Hagelsieb G., et al. The transporter classification database (TCDB): 2021 update. Nucleic Acids Res. 2021;49(D1):D461–D467. doi: 10.1093/nar/gkaa1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Singhal R., Shah Y.M. Oxygen battle in the gut: hypoxia and hypoxia-inducible factors in metabolic and inflammatory responses in the intestine. J Biol Chem. 2020;295(30):10493–10505. doi: 10.1074/jbc.REV120.011188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ney L.M., Wipplinger M., Grossmann M., Engert N., Wegner V.D., Mosig A.S. Short chain fatty acids: key regulators of the local and systemic immune response in inflammatory diseases and infections. Open Biol. 2023;13(3) doi: 10.1098/rsob.230014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tian T., Zhao Y., Yang Y., et al. The protective role of short-chain fatty acids acting as signal molecules in chemotherapy- or radiation-induced intestinal inflammation. Am J Cancer Res. 2020;10(11):3508–3531. [PMC free article] [PubMed] [Google Scholar]
- 26.Guo H., Chou W.C., Lai Y., et al. Multi-omics analyses of radiation survivors identify radioprotective microbes and metabolites. Science. 2020;370(6516) doi: 10.1126/science.aay9097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kostic A.D., Xavier R.J., Gevers D. The microbiome in inflammatory bowel disease: current status and the future ahead. Gastroenterology. 2014;146(6):1489–1499. doi: 10.1053/j.gastro.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Touchefeu Y., Montassier E., Nieman K., et al. Systematic review: the role of the gut microbiota in chemotherapy- or radiation-induced gastrointestinal mucositis - current evidence and potential clinical applications. Aliment Pharmacol Ther. 2014;40(5):409–421. doi: 10.1111/apt.12878. [DOI] [PubMed] [Google Scholar]
- 29.Gupta A., Dhakan D.B., Maji A., et al. Association of flavonifractor plautii, a flavonoid-degrading bacterium, with the gut microbiome of colorectal cancer patients in India. mSystems. 2019;4(6) doi: 10.1128/mSystems.00438-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alashkham A., Paterson C., Hubbard S., Nabi G. What is the impact of diabetes mellitus on radiation induced acute proctitis after radical radiotherapy for adenocarcinoma prostate? A prospective longitudinal study. Clin Transl Radiat Oncol. 2019;14:59–63. doi: 10.1016/j.ctro.2017.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Herold D.M., Hanlon A.L., Hanks G.E. Diabetes mellitus: a predictor for late radiation morbidity. Int J Radiat Oncol Biol Phys. 1999;43(3):475–479. doi: 10.1016/s0360-3016(98)00460-x. [DOI] [PubMed] [Google Scholar]
- 32.Berliner C. Are the solutions to radiotherapy side effects on the gastrointestinal tract right at our doorstep? eBioMedicine. 2021;74 doi: 10.1016/j.ebiom.2021.103687. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.