Abstract
The genetic factors contributing to complex trait variation may reside in regulatory, rather than protein-coding portions of the genome. Within noncoding regions, SNPs in regulatory elements are more likely to contribute to phenotypic variation than those in nonregulatory regions. Thus, it is important to be able to identify and annotate noncoding regulatory elements. DNA conservation among diverged species successfully identifies noncoding regulatory regions. However, because rapidly evolving regulatory regions will not generally be conserved across species, these will not detected by using purely conservation-based methods. Here we describe additional approaches that can be used to identify putative regulatory elements via signatures of nonneutral evolution. An examination of the pattern of polymorphism both within and between populations of Drosophila melanogaster, as well as divergence with its sibling species Drosophila simulans, across 24.2 kb of noncoding DNA identifies several nonneutrally evolving regions not identified by conservation. Because different methods tag different regions, it appears that the methods are complementary. Patterns of variation at different elements are consistent with the action of selective sweeps, balancing selection, or population differentiation. Together with regions conserved between D. melanogaster and Drosophila pseudoobscura, we tag 5.3 kb of noncoding DNA as potentially regulatory. Ninety-seven of the 408 common noncoding SNPs surveyed are within putatively regulatory regions. If these methods collectively identify the majority of functional noncoding polymorphisms, genotyping only these SNPs in an association mapping framework would reduce genotyping effort for noncoding regions 4-fold.
A major goal of modern biological research is to understand the relationship between genotype and phenotype. The search for genetic variation contributing to differences among individuals is exemplified by association studies that aim to identify those segregating genetic polymorphisms that confer risk to common polygenic, or complex diseases in humans (1). Association mapping involves genotyping a dense set of SNPs in a large population of individuals, and asking whether there is evidence of an association between the genotype at each SNP and the phenotype. A significant association suggests that the genotyped SNP is either itself responsible for conferring disease risk or strongly correlated, i.e., is in linkage disequilibrium, with the causal site. We refer to SNPs contributing to phenotypic variation as functional SNPs (fSNPs).
The human genome harbors 4.6-7.1 million common SNPs (minor allele frequency above 5%; refs. 2 and 3), with the vast majority presumed to be nonfunctional. Unfortunately, it is not yet cost-effective to exhaustively test every SNP for an association with a disease phenotype. Despite a great deal of academic and private research, genotyping technology remains unable to efficiently genotype millions of SNPs in thousands of individuals at reasonable cost (4). Thus, some intelligent way of reducing the genotyping effort is needed.
One such method, the HapMap project (5), seeks to take advantage of the level of linkage disequilibrium (LD) across the genome and choose a subset of SNPs to genotype that explain the majority of haplotype information. This approach is favored for humans, with the recent suggestion that the genome exhibits a block-like LD structure (6-8). Under the HapMap plan, between 200,000 and 1 million SNPs need to be genotyped to achieve complete genome coverage (7-10). However, this plan is critically dependent on the degree to which available SNPs capture human haplotype diversity, which is hotly debated (9, 11), and on the reliability of the block definitions across different populations, which is also unclear (8). Perhaps a more fundamental difficulty with this methodology is that haplotypes do not cause disease. Finding an association to a haplotype block is not an endpoint, it merely delimits the search, and further genotyping is required to finally identify the causal mutation.
An alternative strategy to reduce total genotyping effort is to genotype the subset of SNPs most likely to contribute to the examined phenotype. In a seminal paper, Risch and Merikangas (12) showed that association studies for complex traits have higher power than linkage mapping approaches, and the paper is widely cited as supporting the use of association mapping. However, an important aspect of the theoretical treatment put forward by Risch and Merikangas (12) is often overlooked: the actual disease-causing site must be one of the sites genotyped. The power of association studies is greatly reduced if the causative site is not among those genotyped (13, 14).
Based on data acquired from analyses of Mendelian diseases, Botstein and Risch (15) have suggested that causal polymorphisms may generally be coding, which immediately suggests a strategy for selecting putatively disease-causing SNPs on which to focus: identify and genotype all SNPs in coding regions. This approach would ensure a large reduction in total genotyping effort, and provided complex traits are somewhat similar to Mendelian traits in their genetic architecture is likely to uncover some fraction of phenotypically relevant genetic variation. Nevertheless, some clear examples of genetic factors underlying complex trait variation suggest that the responsible polymorphisms may reside in regulatory regions (16-18). The strategy suggested by Botstein and Risch (15) will be undermined if variation in complex traits is generally determined by regulatory genetic variants.
Methods that allow us to identify functional noncoding regulatory domains, such as promoters or enhancers capable of modulating spatial and temporal gene expression, would enable SNPs to be classified based on their position relative to these domains. Genotyping only those SNPs present within regulatory domains would allow for a reduction in the total genotyping effort in association studies. Such a strategy is simple in principal, but it is a major challenge to sift through the ocean of noncoding DNA to find those polymorphisms that are truly cis-regulatory in function. In Drosophila melanogaster, the amount of noncoding DNA is 95.9 megabases (Mb), or ≈80% of the euchromatic genome (19). In humans, the disparity between coding and noncoding DNA is more extreme, with 2,817 Mb of noncoding DNA representing 98.8% of the genome (20). It is possible that statistical tests coopted from the fields of population genetics and molecular evolution can be adapted to identify regulatory regions of the noncoding genome: if a region can be shown to have evolved in a nonneutral manner, presumably there are functional elements buried in these regions. Statistical tests are available to explicitly test for evidence of selection in coding regions (e.g., refs. 21 and 22), but these tests cannot be applied to noncoding DNA because they rely on the ability to parse the sequence into synonymous and nonsynonymous sites.
Here we examine several statistics that can be applied to noncoding DNA to detect regions of sequence subject to the action of past natural selection: conservation between phylogenetically diverged species, the ratio of polymorphism to divergence between sibling species, the polymorphism frequency spectrum, and the level of population structure. We explore graphical sliding window presentations (23) of these statistics, because our goal is to suggest regions likely to harbor fSNPs rather than to apply rigorous statistical tests. Such graphical, sliding-window tests are also more easily generalized to genome-scale data. Compared to SNPs in regions that do not show evidence for past natural selection, those in regions showing departures from neutral expectation are stronger candidates for fSNPs. Because nonneutrally evolving regions are likely to be enriched for fSNPs, a reduction in genotyping effort could be achieved in association studies by preferentially genotyping SNPs from nonneutrally evolving regions.
We select 26 ≈1-kb fragments of primarily noncoding DNA near known genes distributed across the D. melanogaster genome. We examine the rate at which the various proposed tools are capable of “tagging” potential regulatory elements in these regions, and also determine the degree to which the statistics tag the same or different areas as nonneutrally evolving. Because we chose the noncoding regions for this study randomly with respect to cis-regulatory annotation, the rate at which we tag potential regulatory elements is likely typical of the genome as a whole. The tests we propose could be applied to any noncoding sequence. Proving that tagged regions are cis-regulatory elements harboring fSNPs is a more difficult problem that remains to be addressed.
Materials and Methods
Sequenced Regions. We chose 26 loci distributed evenly with respect to genetic location along the five major chromosome arms of D. melanogaster (Table 1). These loci fall into three categories: those known to interact with the Notch signaling pathway (seven genes), those thought to affect development of the peripheral nervous system or that have been shown to have quantitative effects on bristle number (16 genes), and finally three genes selected to ensure coverage of the genome. Primers were developed for an ≈1-kb amplicon at each locus using Primer3 (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi; sequences are provided in Table 2, which is published as supporting information on the PNAS web site). None of the developed amplicons appear to encompass known regulatory elements. Fig. 1 highlights the annotated regions sequenced for each amplicon (taken from Release 4.0 of the D. melanogaster genome sequence), Table 1 documents the position of the amplicon relative to the start codon of the gene, and Fig. 3, which is published as supporting information on the PNAS web site, details the exact positions of the amplicons relative to the structure of the loci.
Table 1. Details of the loci examined.
| Gene name | Gene symbol | Gene position | Amplicon position | Functional category (ref.) |
|---|---|---|---|---|
| deltex | dx | X, 17.0 | −553 | Notch |
| cut | ct | X, 20.0 | +3870 | PNS |
| dishevelled | dsh | X, 34.5 | −385 | Notch |
| scalloped | sd | X, 51.5 | −2441 | PNS |
| Beadex | Bx | X, 59.4 | −30244 | PNS (24) |
| split ends | spen | 2L, 0.5 | −2652 | PNS (25) |
| friend of echinoid | fred | 2L, 11.5 | −730 | PNS |
| wingless | wg | 2L, 21.9 | +34125 | PNS (26) |
| numb | numb | 2L, 35.5 | −13229 | Notch |
| daughterless | da | 2L, 41.3 | −967 | PNS |
| deadpan | dpn | 2R, 57.5 | −1709 | PNS (24, 27) |
| scabrous | sca | 2R, 66.7 | −768 | PNS |
| mastermind | mam | 2R, 70.3 | −18725 | Notch |
| cousin of atonal | cato | 2R, 79.5 | −635 | PNS |
| smooth | sm | 2R, 91.5 | +3796 | PNS (28) |
| Distal-less | DII | 2R, 107.8 | −808 | — |
| extra macrochaetae | emc | 3L, 0.0 | −407 | PNS |
| vein | vn | 3L, 16.2 | −2767 | — |
| quemao | qm | 3L, 23.0 | +254 | PNS (29) |
| Bearded | Brd | 3L, 42.0 | +392 | Notch |
| neuralized | neur | 3R, 48.5 | +1381 | PNS |
| Actin 88F | Act88F | 3R, 57.1 | −1062 | — |
| Hairless | H | 3R, 69.5 | −499 | Notch |
| pointed | pnt | 3R, 79.0 | −4659 | PNS |
| Serrate | Ser | 3R, 92.0 | −722 | Notch |
| tramtrack | ttk | 3R, 102.0 | −2030 | PNS |
Gene position is the chromosome arm on which the gene resides, followed by its genetic position. Amplicon position is the distance between the midpoint of the amplicon and the gene start codon in bp (−, base pair is upstream of the start codon; +, base pair is downstream). Functional categories were determined by using the Gene Ontology (www.geneontology.org) unless references are provided. Notch, these genes functionally interact with the Notch signaling pathway (Gene Ontology terms GO:0007219, GO:0030179, and GO:0005112); PNS, these genes are involved in peripheral nervous system and sensory organ development, or bristle morphogenesis (GO:0007422, GO:0007423, and GO:0008407). - indicates that genes are unlikely to have any involvement in neurogenesis.
Fig. 1.
The type of DNA sequence surveyed. Each of the 26 amplicons is referred to by the symbol for the closest known gene, and amplicons are grouped according to functional category (see Table 1 for full gene names and a description of the categorization). The amplicons are each represented by a bar, scaled to the length of the D. melanogaster alignment, and shaded to reflect the D. melanogaster release 4.0 genome annotation.
D. melanogaster Stocks. All 26 amplicons were sequenced for 16 wild-type lines representing a worldwide sample. The stock numbers for these lines are: B1 (Canton, OH), B3839 (Bermuda), B3841 (Bogata, Colombia), B3844 (Barcelona, Spain), B3846 (Capetown, South Africa), B3852 (Koriba Dam, South Africa), B3853 (Koriba Dam, South Africa), B3864 (Israel), B3870 (Riverside, CA), B3875 (Athens), B3886 (Red Top Mountain, GA), T14021-0231.0 (Oahu, Hawaii), T14021-0231.1 (Ica, Peru), T14021-0231.4 (Kuala Lumpur, Malaysia), T14021-0231.6 (Mysore, India), and T14021-0231.7 (Ken-ting, Taiwan), where “B” and “T” refer to the Bloomington and Tucson Drosophila stock centers, respectively. Before sequencing, the 16 lines were propagated by using single male-female pairs for between 2 and 12 generations to reduce heterozygosity.
In addition, for each amplicon, we sequenced eight strains from a single population. For the X- and third-chromosome amplicons, we sequenced eight chromosomal extraction strains, where the natural alleles were derived from Napa Valley, CA, whereas for amplicons on the second chromosome, we sequenced eight inbred lines derived from North Carolina (kindly provided by C. H. Langley, Center for Population Biology, University of California, Davis).
Outgroup Sequences. Using shotgun sequencing assemblies provided by the Genome Sequencing Center, Washington University Medical School (http://genome.wustl.edu/projects/simulans), we obtained the homologous region in Drosophila simulans from one of the strains, sim4, sim6 or w501 with blastn, for each amplicon. We used a similar procedure to identify the homologous region for each amplicon from the Drosophila pseudoobscura genome assembly (release 1.03), taken as a 4-kb window centered on the position of the best blastn hit. Details of the regions extracted from these outgroup species are provided in Table 3, which is published as supporting information on the PNAS web site.
Sequence and Population Genetics Analyses. Sequence traces for each D. melanogaster strain/amplicon combination were assembled by using seqmanii (version 5.01, DNASTAR), and for each amplicon, the D. melanogaster and D. simulans sequences were manually aligned by using bioedit (www.mbio.ncsu.edu/BioEdit/bioedit.html). All D. melanogaster sequences were deposited in the GenBank database (accession nos. AY863438-AY864021).
After alignment, each amplicon was represented by at least six within-population D. melanogaster lines, and at least 13 worldwide D. melanogaster lines. Missing sequences were due largely to repeated PCR failures, but were also due to ambiguous sequence reads caused by heterozygous insertion/deletion polymorphisms that remained in some of the worldwide and North Carolina lines despite inbreeding. Twelve of the 26 amplicons showed between one and four sequences harboring at least one heterozygous nucleotide, and before analysis, on a per amplicon basis, each heterozygous sequence was arbitrarily split into a pair of pseudohaplotypes. This split is justified because PCR was performed on DNA extracted from single males, so the heterozygous sequence reflects the presence of two alleles. None of the diversity measures we estimate are affected by the phase of the polymorphism data.
Using a sliding window approach with a window size of 250 bp, stepping through each sequence alignment in 1-bp increments, we estimated (i) nucleotide diversity (π) across the D. melanogaster sequences, (ii) divergence (K) between D. melanogaster and D. simulans, (iii) Tajima's D (30), which provides a measure of the polymorphism frequency spectrum, (iv) πw estimated from the alleles obtained from the single D. melanogaster population (either Napa Valley or North Carolina), and (v) πb estimated from the worldwide D. melanogaster samples. A comparison of πw and πb serves as a proxy for population structure in that differences between within- and among-population nucleotide diversity can be assessed. Sites segregating for more than two alleles were ignored for all calculations, with window size kept constant with respect to the remaining informative sites. Missing data and gaps were treated as a reduction in the sample size, and values were weighted accordingly. All analyses were performed by using custom scripts in the statistical programming language r (www.r-project.org).
Finally, we extracted the consensus sequence for each D. melanogaster alignment and used a sliding-window approach to blast 31-bp sections against the homologous region of the D. pseudoobscura genome, stepping through the consensus sequence in 1-bp increments. For each D. melanogaster query sequence, we recorded the position, orientation, and score of the highest blast hit in D. pseudoobscura, and considered only hits with a score >45 in further analyses.
Results
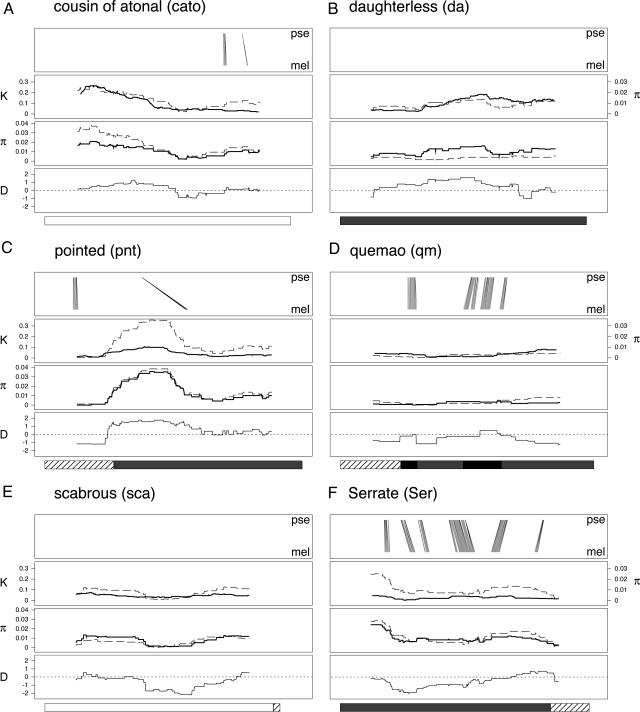
We sequenced 26 ≈1-kb amplicons in D. melanogaster, primarily from noncoding regions in or near genes involved in peripheral nervous system development and/or regulation of Notch signaling. For each amplicon, we also identified the homologous region from the closely related D. simulans species, and from D. pseudoobscura, which is thought to have diverged from D. melanogaster ≈25 million years ago (31). The degree to which studied amplicons harbor cis-regulatory elements is unknown. These data allowed us to examine a set of sequence attributes across each of the amplicons to examine for regions exhibiting nonneutral evolution: (i) the level of sequence conservation between D. melanogaster and D. pseudoobscura, (ii) the amount of nucleotide polymorphism within D. melanogaster relative to the level of divergence between D. melanogaster and its sibling species D. simulans, (iii) the polymorphism frequency spectrum in D. melanogaster, and (iv) the amount of population structure within D. melanogaster, by comparing the nucleotide diversity within a single D. melanogaster population to the diversity observed in a worldwide panel. Because the footprint of selection may be small, a sliding-window framework is likely to be more informative than examining the average values of the statistics for each amplicon (see Table 4, which is published as supporting information on the PNAS web site). Fig. 2 shows the sliding-window analyses for six selected amplicons, and Fig. 4, which is published as supporting information on the PNAS web site, presents analyses for all amplicons. Below we document those sequenced noncoding regions that have patterns in the sliding-window plots suggesting deviation from neutral expectation, and also note the number of SNPs present within such regions.
Fig. 2.
Signatures of selection across sequenced amplicons. Six of the 26 amplicons are detailed, and each is composed as follows. (Top) Conservation between D. melanogaster and D. pseudoobscura. Each line represents a blast hit (with a score >45) between a 31-bp subsection of the D. melanogaster consensus sequence and the homologous sequence from D. pseudoobscura, with the endpoints of each line showing the position of the hit in each genome. All blast hits are between subsequences in the same orientation. (Upper Middle) Nucleotide diversity (π) within D. melanogaster (dashed line), and divergence (K) between D. melanogaster and D. simulans (solid line). (Lower Middle) Nucleotide diversity within the lines derived from the single D. melanogaster population (dashed line), and within the worldwide panel of D. melanogaster strains (solid line). (Bottom) Tajima's D statistic (30). Below each figure is an annotation bar describing the type of sequence surveyed for each amplicon, with the shading as described in Fig. 1.
Deep Sequence Conservation. Random neutral mutation will tend to erode similarity between neutrally evolving sequences in independent lineages. Thus, conservation of DNA sequence across taxa diverged by many millions of years is taken as evidence of function, as such regions are presumed to be subject to negative, or purifying selection to preserve sequence. This has become a guiding principle in the detection of functional noncoding DNA (32-35).
Nine of the 26 amplicons show no fine-scale conservation using our blast approach [Bx, da (Fig. 2B), dsh, mam, sca (Fig. 2E), sd, sm, ttk, and vn], 10 show low conservation [Brd, cato (Fig. 2A), dpn, dx, fred, H, neur, numb, pnt (Fig. 2C), and spen; defined as showing three or fewer short (<60-bp) stretches of conservation], and 7 show high conservation [Act88F, ct, Dll, emc, qm (Fig. 2D), Ser (Fig. 2F), and wg]. In two of the amplicons with high conservation, qm (Fig. 2D) and emc, the regions of conservation map to known exons. Overall, of the 24.2 kb of sequenced noncoding DNA in D. melanogaster, 2.1 kb (8.6%) is highly conserved between D. melanogaster and D. pseudoobscura, suggesting that it may have regulatory significance. There are 408 common (>5% minor allele frequency) biallelic SNPs in the 24.2 kb of noncoding sequence, and 14 (3.4%) are present within the detected conserved regions.
Our blast approach reveals that D. melanogaster and D. pseudoobscura do not appear to differ by any conserved micro-inversions, as all strong blast hits are between subsequences in the same orientation, or by any conserved local rearrangements, because none of the hit lines cross. This finding is in accordance with previous results at the Enhancer of split locus (36). However, we did observe at least three cases where there appears to have been a large insertion/deletion in one of the two genomes [Act88F, ct, and pnt (Fig. 2C)].
Patterns of Neutral Evolution. For the remaining analyses not involving D. pseudoobscura, sliding-window plots for 13 amplicons show no noticeable departure from the pattern of diversity predicted by neutral theory [Brd, Bx, ct, Dll, dpn, dsh, emc, H, mam, qm (Fig. 2D), sm, spen, and ttk]. Several criteria suggest the absence of recent, detectable selective forces acting on these regions. Within-species diversity (π) and between-species divergence (K) generally track each other, indicating no change in mutational processes between species. There are also no obvious differences between the nucleotide diversity within the single D. melanogaster population, and diversity across the worldwide sample of D. melanogaster lines, implying that no population-specific forces are at work. Finally, for these 13 amplicons we see no clear departure from the allele frequency distribution predicted under neutrality as measured by Tajima's D statistic (30).
Positive Selection. A low level of diversity coupled with a frequency spectrum skewed toward an excess of rare variants (i.e., negative Tajima's D) is generally taken as evidence for a selective sweep or positive selection (37, 38). A selective sweep removes variation around the advantageous mutation, and observed polymorphisms are rare having arisen since the sweep. We identify three amplicons, neur, sca (Fig. 2E), and sd, showing patterns of diversity and divergence consistent with the action of a weak selective sweep.
The amplicon upstream of the sca gene represents a particularly clear example (Fig. 2E). In a central 200-bp section of this amplicon there is a marked dip both in the level of nucleotide diversity and in Tajima's D, suggesting that a site within this short region has swept to fixation in D. melanogaster. A similar pattern is observed for the amplicon in an intronic region of the neur gene: reduced π and negative D for the second half of the amplicon. In contrast, the entire amplicon upstream of the sd gene shows very low nucleotide polymorphism (just four polymorphisms exist, three of which are singletons), whereas interspecific divergence is normal, suggesting that the entire sequenced region has been impacted by a positive selection event. We estimate that 1.4 kb (5.8%) of the sequenced noncoding DNA in D. melanogaster has been impacted by positive selection, and these regions collectively harbor two common biallelic SNPs (0.5% of the total common SNPs discovered).
Balancing Selection. Balanced polymorphisms are segregating sites maintained in a population at intermediate frequency due to heterozygote advantage, frequency-dependent selection, by selection on alternate alleles in different environments, or by antagonistic pleiotropy. A balanced polymorphism can theoretically be maintained indefinitely and will enhance the level of neutral polymorphism surrounding it, with the size of the affected region dependent on the local recombination rate. Thus, the presence of a balanced polymorphism will generate a high level of diversity compared to divergence, and a greater number of frequent polymorphisms (i.e., positive Tajima's D). Three amplicons, Act88F, dx, and pnt (Fig. 2C), exhibit patterns suggestive of balancing selection.
The best example is provided by the amplicon in a 5′ UTR/intronic region of the pnt gene (Fig. 2C), where starting at the transition between 5′ UTR and intron, and continuing within the intron for ≈300 bp, the level of nucleotide diversity is very high, and D is positive. It is of interest that the affected region may represent a previously uncharacterized insertion relative to D. pseudoobscura. The amplicon about the dx gene is around one-third 5′ UTR, and for about 200 bp upstream of the 5′ UTR the level of nucleotide diversity is high relative to divergence, and D is positive. Soon after the start of the transcribed region, diversity returns to lower values, and D falls to its neutral expectation of zero.
The amplicon upstream of the Act88F gene also exhibits a pattern consistent with balancing selection for the first ≈300 bp. However, this amplicon is also noteworthy for a single sequence from the Napa Valley D. melanogaster population that has a unique haplotype. The presence of this sequence in the D. melanogaster-D. simulans alignment generates 48 singleton polymorphic sites and substantial insertion/deletion variation, such that particularly in the central portion of the Act88F amplicon, nucleotide diversity is high, and Tajima's D is negative (see Fig. 4). In comparison, analyses based on an alignment lacking the aberrant Act88F sequence show a Tajima's D and nucleotide diversity not inconsistent with neutrality, although the signature of balancing selection at the start of the amplicon remains. Because this unique D. melanogaster haplotype is not similar to the D. simulans sequence, it is unclear whether it represents a single event or the aftermath of a series of mutational events. Rare, extremely diverged haplotypes perhaps deserve special treatment.
The amount of noncoding DNA sequenced in D. melanogaster that shows a pattern of nucleotide diversity consistent with balancing selection is 0.8 kb (3.4%), and these regions harbor 38 common biallelic SNPs (9.3% of the common SNPs identified in the survey).
Population Structure. Two types of D. melanogaster population-specific effects are evident from our sequenced amplicons. The first type is when the single population shows lower sequence variation than does the worldwide panel. This observation is indicative of a geographically localized reduction in diversity, possibly via local adaptation. Two of the amplicons show this pattern, the central 300 bp of the intronic region sequenced for the da gene (Fig. 2B), and the end of the amplicon upstream of the fred gene. The second pattern is the reverse, where there is less variation in the worldwide sample than would be predicted based on variation within the single population. The maintenance of higher variation within a single population than across multiple populations is potentially the result of balancing selection. This pattern is apparent for the 300 bp at the end of the amplicon upstream of the vn gene, and for the 400 bp at the start of the amplicon upstream of the cato gene (Fig. 2A). Together the two patterns highlighting population structure within D. melanogaster encompass 1.0 kb (3.9%) of the noncoding sequence and hold 43 (10.5%) of the common biallelic SNPs uncovered in our survey.
Unexpected Patterns. Finally, two 5′ UTR/intronic amplicons, within the genes Ser (Fig. 2F) and numb, and a single amplicon downstream of the wg gene, show higher nucleotide polymorphism than expected given the level of sequence divergence between D. melanogaster and D. simulans. However, in no case is this accompanied by a coordinated skew in the polymorphism frequency spectrum. These three amplicons imply that, as we collect larger DNA sequence data sets from a range of sequence types, we are likely to see patterns of polymorphism that neither conform to neutral expectation nor neatly fit with our current ideas about the expected result of selective events.
Discussion
There is considerable interest in developing methods to identify functional domains from primary sequence data. One goal is to detect regions likely to harbor fSNPs that contribute to intraspecific phenotypic variation in complex traits. To identify such regions, we propose employing a series of tests based on population genetics theory, which should complement approaches based purely on deep phylogenetic conservation.
Conservation. Over evolutionary time, separately evolving taxa will accumulate random neutral mutations, and only regions under functional constraint will be conserved. Comparative genome sequencing has proved quite useful for both gene prediction and for identifying conserved noncoding regions (34), which in some instances have been shown to exhibit regulatory activity (32, 33, 39). In the present study, 8.6% of the noncoding sequence we surveyed was conserved between the diverged species D. melanogaster and D. pseudoobscura, distributed in short sections across 17 of the 26 amplicons. The 14 common SNPs located within these identified regions are candidate fSNPs.
We have previously demonstrated that regulatory elements within the Enhancer of split locus in D. melanogaster are often conserved in D. pseudoobscura (36), suggesting that they retain a similar regulatory function in this species. However, a recent analysis of 142 bona fide regulatory elements showed that they were only 4-8% more conserved between D. melanogaster and D. pseudoobscura than were control regions (40). These results imply that the signal of function in deep pairwise species comparisons may be both weak and heterogeneous across the genome. A further difficulty with relying completely on a conservation approach to functionally annotate a genome and identify fSNPs is that, although sequence conservation may imply function, a lack of conservation does not imply the absence of function. This was elegantly shown by Ludwig et al. (41) for the even-skipped stripe 2 embryonic expression pattern in Drosophila. Here, the expression pattern itself is strongly conserved between the species D. melanogaster and D. pseudoobscura, whereas the regulatory region giving rise to the pattern is very different in sequence between the two species. Thus, true regulatory regions can be missed by using phylogenetic conservation. It is entirely possible that those cis-regulatory elements that contribute to within species variation for complex traits are fast evolving, and as a result are unlikely to be conserved in wide phylogenetic comparisons. The 8.6% of noncoding D. melanogaster DNA tagged by conservation with D. pseudoobscura harbors just 3.4% of the common SNP variation in D. melanogaster. This lack of polymorphism might imply that highly conserved regions are too constrained to tolerate variation, and may actually be less likely to harbor fSNPs contributing to within-species phenotypic variation than less conserved regions identified by other means.
These concerns, coupled with the fact that conservation implies the action of a single form of selection (purifying), suggests that other methods of locating noncoding regulatory domains may be helpful.
Polymorphism and Divergence. The neutral theory of molecular evolution states that, for neutrally evolving DNA, the expected ratio of polymorphism within a species to divergence between species should be constant throughout the genome (42). This expectation has a large variance, because of both the sampling and the particular genealogy of the tested region, but departures from neutrality can be detected, for instance with the widely applied HKA test (43).
A few clear cases of candidate regions associated with selective sweeps have been identified in Drosophila, for example the Sdic gene that encodes a subunit of the sperm axoneme (44), and the cytochrome P450 gene Cyp6g1 associated with DDT resistance (45, 46). Also, cases of balanced polymorphisms have been shown, such as that centered on the Adh fast/slow polymorphism in D. melanogaster (23). However, in these instances, the magnitude of the population genetic signature was greater than those observed in our survey.
In this study, our goal is not to test for rigorous statistical significance, but instead to suggest regions that are likely to harbor fSNPs. We made use of a graphical approach (23) allowing visual inspection of departures from neutrality across each of the 26 amplicons. Six amplicons exhibit patterns indicative of nonneutral evolution, with three suggesting past positive selection (selective sweeps) and three implicating a balanced polymorphism. It is possible that, despite modest power to detect nonneutral events, the magnitude of departure from neutrality based on the ratio of polymorphism to divergence is predictive of the likelihood of a region being regulatory in function. In this regard, we note that within known enhancer regions in the Drosophila locus Enhancer of split, using a test adapted from the McDonald-Kreitman (21) and HKA tests (43), the ratio of polymorphism to divergence differs significantly between transcription factor binding sites and adjacent nonbinding sites (P = 0.004, ref. 36).
Population Structure. Wright's F statistics (47) seek to partition allelic variation into within individual, within population, and between population components, and the FST statistic represents the degree of population differentiation. Under neutrality, the same level of population subdivision should be seen across the genome, but local adaptation can result in regional departures from this genome-wide expectation. In Drosophila, regions showing a strong departure can be quite small, from a single site to short regions of a few hundred base pairs. For instance, the Adh fast/slow polymorphism and  insertion/deletion polymorphism are both functional, and both show much stronger clinal variation across D. melanogaster populations than do neighboring polymorphisms in the same gene (48, 49).
insertion/deletion polymorphism are both functional, and both show much stronger clinal variation across D. melanogaster populations than do neighboring polymorphisms in the same gene (48, 49).
Typically, FST is based on allele frequency estimates obtained from several subpopulations each consisting of number of individuals. Such an approach may be of limited use in genome-wide scans for fSNPs, as the economics of sequencing favors generating complete sequence data from a much more limited sample. Here, we use a proxy for standard measures of FST based on comparing the nucleotide diversity in a single population sample (within-population variance) to that across a set of lines of worldwide distribution (among-population variance). In the context of genome-wide scans, this approach may have greater utility than traditional measures of FST because it requires characterizing just 24 alleles. Using this approach, regions in four amplicons showed greater or lesser worldwide variation than expected based on the variation in a single population. We hypothesize that such regions are more likely to include fSNPs than regions showing no population subdivision.
Prospects for in Silico Functional Annotation. We surveyed 24.2 kb of noncoding DNA in D. melanogaster, encompassing 408 common SNPs, and identified putative regulatory regions using deep phylogenetic conservation (8.6% of the sequence, 3.4% of common SNPs), the pattern of positive selection (5.8% of sequence, 0.5% of SNPs), the pattern of balancing selection (3.4% of sequence, 9.3% of SNPs), and evidence for population structure (3.9% of sequence, 10.5% of SNPs). It is clear that the different tests identify different regions as potentially harboring fSNPs. This finding suggests that any one method may fail to annotate many functionally important areas, and at present it is premature to rely on a single method (such as deep phylogenetic conservation) at the expense of the others. It is also of note that, although deep conservation and positive selection tag 14.4% of the DNA sequenced, the tagged regions collectively harbor only 3.9% of the common SNPs. In contrast, balancing selection and population structure demarcate a much smaller portion of the sequence (7.3%) but many more SNPs (19.8%). That is, tests based on diversity-reducing forces of selection identify large regions containing few SNPs, whereas diversity-enhancing forces identify smaller regions with higher SNP density.
Collectively, we tag 5.3 kb (21.8%) of the surveyed noncoding DNA as potentially regulatory, and the identified regions harbor 97 of the 408 common biallelic SNPs discovered. Assuming that the subset of SNPs we identify includes the majority of fSNPs, if we adopted an association study approach genotyping only these 97 sites, our genotyping effort would be reduced 4-fold over genotyping all common noncoding SNPs. This value is not inconsistent with the reduction in genotyping effort for the HapMap proposal implied by some studies (7, 10), although the actual level of reduction possible under the HapMap plan remains unclear. We note that the reduction in genotyping effort we propose assumes coding variants contribute little to phenotypic variation.
The major remaining question is how often each of the annotation methods identifies functional regions that influence complex phenotypes. An obvious reverse approach to answering this question is to assess the ability of the sequence-based methods we propose to identify known regulatory elements. We have previously examined this for the Enhancer of split locus in Drosophila (36), and as more regulatory elements are identified by using molecular and developmental techniques, the ability of sequence-based methods alone to detect them can be assessed. Several forward approaches are also possible. For highly conserved regions, tests of function have taken two forms, the ability to drive gene expression in promoter-reporter constructs (33, 39) and the ability to bind transcription factors (32). The population genetic approaches we present likely identify more quickly evolving regions, which may harbor regulatory elements that influence only a subset of tissues and/or developmental times. Such elements may make important contributions to complex traits, but their functional role may be difficult to confirm with promoter-reporter assays. An alternative approach may be to identify a set of putative regulatory regions using an array of methods, and exhaustively test all polymorphisms in these regions for an association with phenotype. The degree of association at the sites could then be used to assess the rate at which each annotation method falsely classifies a DNA region as harboring an fSNP. Clearly, this experiment lacks finesse, but it has the advantage of directly providing an estimate of the phenotypic effect associated with each SNP identified on the basis of primary sequence data.
A model system that is probably most amenable to this test is the classic D. melanogaster bristle number quantitative trait, shown to be under stabilizing selection (50), and its associated set of candidate genes (24, 51). Many of the proteins encoded by these genes are members of the Notch signaling pathway, regulate members of this pathway, or are involved in the development of the peripheral nervous system in Drosophila. For our sequencing survey, 23 of 26 amplicons were developed in or near genes involved in these processes (Table 1). Thus, we have a strong a priori prediction that fSNPs in regions visible to selection at these candidate loci are likely to contribute to natural variation for bristle number. Clearly, SNPs in nonneutrally evolving regions around these genes do not necessarily have to affect bristle number, but mutant alleles associated with these genes regularly have pleiotropic effects on bristle number and patterning (www.flybase.org, ref. 24). So, although regions of these loci experiencing recent selection are not expected to directly map to those fSNPs contributing to bristle number variation, we do expect the two sets of regions to overlap to some extent.
It is important to understand the ability of different methods of genome annotation to uncover functional regulatory variation to direct future genome sequencing studies. The current model for genome annotation employs a comparative approach, whereby annotation of a focal genome is aided by sequence comparisons to one or a set of diverged species genomes. However, depending on the performance of other annotation methods, it may be extremely valuable to sequence multiple individuals from a single species in addition to single individuals from multiple species.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grant GM58564 (to A.D.L.).
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Systematics and the Origin of Species: On Ernst Mayr's 100th Anniversary,” held December 16-18, 2004, at the Arnold and Mabel Beckman Center of the National Academies of Science and Engineering in Irvine, CA.
Abbreviation: fSNP, functional SNP.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY863438-AY864021).
References
- 1.Carlson, C. S., Eberle, M. A., Kruglyak, L. & Nickerson, D. A. (2004) Nature 429, 446-452. [DOI] [PubMed] [Google Scholar]
- 2.Kruglyak, L. & Nickerson, D. A. (2001) Nat. Genet. 27, 234-236. [DOI] [PubMed] [Google Scholar]
- 3.Stephens, J. C., Schneider, J. A., Tanguay, D. A., Choi, J., Acharya, T., Stanley, S. E., Jiang, R., Messer, C. J., Chew, A., Han, J.-H., et al. (2001) Science 293, 489-493. [DOI] [PubMed] [Google Scholar]
- 4.Syvänen, A.-C. (2001) Nat. Rev. Genet. 2, 930-942. [DOI] [PubMed] [Google Scholar]
- 5.The International HapMap Consortium (2003) Nature 426, 789-796. [DOI] [PubMed] [Google Scholar]
- 6.Daly, M. J., Rioux, J. D., Schaffner, S. F., Hudson, T. J. & Lander, E. S. (2001) Nat. Genet. 29, 229-232. [DOI] [PubMed] [Google Scholar]
- 7.Patil, N., Berno, A. J., Hinds, D. A., Barrett, W. A., Doshi, J. M., Hacker, C. R., Kautzer, C. R., Lee, D. H., Marjoribanks, C., McDonough, D. P., et al. (2001) Science 294, 1719-1723. [DOI] [PubMed] [Google Scholar]
- 8.Gabriel, S. B., Schaffner, S. F., Nguyen, H., Moore, J. M., Roy, J., Blumenstiel, B., Higgins, J., DeFelice, M., Lochner, A., Faggart, M., et al. (2002) Science 296, 2225-2229. [DOI] [PubMed] [Google Scholar]
- 9.Carlson, C. S., Eberle, M. A., Rieder, M. J., Smith, J. D., Kruglyak, D. & Nickerson, D. A. (2003) Nat. Genet. 33, 518-521. [DOI] [PubMed] [Google Scholar]
- 10.Goldstein, D. B., Ahmadi, K. R., Weale, M. E. & Wood, N. W. (2003) Trends Genet. 19, 615-622. [DOI] [PubMed] [Google Scholar]
- 11.Reich, D. E., Gabriel, S. B. & Altshuler, D. (2003) Nat. Genet. 33, 457-458. [DOI] [PubMed] [Google Scholar]
- 12.Risch, N. & Merikangas, K. (1996) Science 273, 1516-1517. [DOI] [PubMed] [Google Scholar]
- 13.Kruglyak, L. (1999) Nat. Genet. 22, 139-144. [DOI] [PubMed] [Google Scholar]
- 14.Long, A. D. & Langley, C. H. (1999) Genome. Res. 9, 720-731. [PMC free article] [PubMed] [Google Scholar]
- 15.Botstein, D. & Risch, N. (2003) Nat. Genet. 33, Suppl., 228-237. [DOI] [PubMed] [Google Scholar]
- 16.Robin, C., Lyman, R. F., Long, A. D., Langley, C. H. & Mackay, T. F. (2002) Genetics 162, 155-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ueda, H., Howson, J. M. M., Esposito, L., Heward, J., Snook, H., Chamberlain, G., Rainbow, D. B., Hunter, K. M. D., Smith, A. N., Di Genova, G., et al. (2003) Nature 423, 506-511. [DOI] [PubMed] [Google Scholar]
- 18.Shapiro, M. D., Marks, M. E., Peichel, C. L., Blackman, B. K., Nereng, K. S., Jónsson, B., Schluter, D. & Kingsley, D. M. (2004) Nature 428, 717-723. [DOI] [PubMed] [Google Scholar]
- 19.Adams, M. D., Celniker, S. E., Holt, R. A., Evans, C. A., Gocayne, J. D., Amanatides, P. G., Scherer, S. E., Li, P. W., Hoskins, R. A., Galle, R. F., et al. (2000) Science 287, 2185-2195. [DOI] [PubMed] [Google Scholar]
- 20.International Human Genome Sequencing Consortium (2004) Nature 431, 931-945. [DOI] [PubMed] [Google Scholar]
- 21.McDonald, J. H. & Kreitman, M. (1991) Nature 351, 652-654. [DOI] [PubMed] [Google Scholar]
- 22.Nielsen, R. & Yang, Z. (1998) Genetics 148, 929-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kreitman, M. & Hudson, R. R. (1991) Genetics 127, 565-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Norga, K. K., Gurganus, M. C., Dilda, C. L., Yamamoto, A., Lyman, R. F., Patel, P. H., Rubin, G. M., Hoskins, R. A., Mackay, T. F. & Bellen, H. J. (2003) Curr. Biol. 13, 1388-1397. [DOI] [PubMed] [Google Scholar]
- 25.Kuang, B., Wu, S. C., Shin, Y., Luo, L. & Kolodziej, P. (2000) Development (Cambridge, U.K.) 127, 1517-1529. [DOI] [PubMed] [Google Scholar]
- 26.Ramain, P., Khechumian, K., Seugnet, L., Arbogast, N., Ackermann, C. & Heitzler, P. (2001) Curr. Biol. 11, 1729-1738. [DOI] [PubMed] [Google Scholar]
- 27.Bier, E., Vaessin, H., Younger-Shepherd, S., Jan, L. Y. & Jan, Y. N. (1992) Genes Dev. 6, 2137-2151. [DOI] [PubMed] [Google Scholar]
- 28.Lage, P. Z., Shrimpton, A. D., Flavell, A. J., Mackay, T. F. C. & Brown, A. J. L. (1997) Genetics 146, 607-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lai, C., McMahon, R., Young, C., Mackay, T. F. C. & Langley, C. H. (1998) Genetics 149, 1051-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tajima, F. (1989) Genetics 123, 585-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Russo, C. A. M., Takezaki, N. & Nei, M. (1995) Mol. Biol. Evol. 12, 391-404. [DOI] [PubMed] [Google Scholar]
- 32.Boffelli, D., McAuliffe, J., Ovcharenko, D., Lewis, K. D., Ovcharenko, I., Pachter, L. & Rubin, E. M. (2003) Science 299, 1391-1394. [DOI] [PubMed] [Google Scholar]
- 33.Hong, R. L., Hamaguchi, L., Busch, M. A. & Weigel, D. (2003) Plant Cell 15, 1296-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kellis, M., Patterson, N., Endrizzi, M., Birren, B. & Lander, E. S. (2003) Nature 423, 241-254. [DOI] [PubMed] [Google Scholar]
- 35.Berman, B. P., Pfeiffer, B. D., Laverty, T. R., Salzberg, S. L., Rubin, G. M., Eisen, M. B. & Celniker, S. E. (2004) Genome Biol. 5, R61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Macdonald, S. J. & Long, A. D. (2005) Mol. Biol. Evol. 22, 1-13. [DOI] [PubMed] [Google Scholar]
- 37.Andolfatto, P. & Przeworski, M. (2001) Genetics 158, 657-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim, Y. & Stephan, W. (2002) Genetics 160, 765-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson, D. S., Davidson, B., Brown, C. D., Smith, W. C. & Sidow, A. (2004) Genome Res. 14, 2448-2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Richards, S., Liu, Y., Bettencourt, B. R., Hradecky, P., Letovsky, S., Nielsen, R., Thornton, K., Hubisz, M. J., Chen, R., Meisel, R. P., et al. (2005) Genome Res. 15, 1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ludwig, M. Z., Bergman, C., Patel, N. H. & Kreitman, M. (2000) Nature 403, 564-567. [DOI] [PubMed] [Google Scholar]
- 42.Kimura, M. (1983) The Neutral Theory of Molecular Evolution (Cambridge Univ. Press, Cambridge, U.K.).
- 43.Hudson, R. R., Kreitman, M. & Aguadé, M. (1987) Genetics 116, 153-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nurminsky, D. I., Nurminskaya, M. V., De Aguiar, D. & Hartl, D. L. (1998) Nature 396, 572-575. [DOI] [PubMed] [Google Scholar]
- 45.Daborn, P. J., Yen, J. L., Bogwitz, M. R., Le Goff, G., Feil, E., Jeffers, S., Tijet, N., Perry, T., Heckel, D., Batterham, P., et al. (2002) Science 297, 2253-2256. [DOI] [PubMed] [Google Scholar]
- 46.Schlenke, T. A. & Begun, D. J. (2004) Proc. Natl. Acad. Sci. USA 101, 1626-1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wright, S. (1951) Ann. Eugen. 15, 323-354. [DOI] [PubMed] [Google Scholar]
- 48.Berry, A. & Kreitman, M. (1993) Genetics 134, 869-893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stam, L. F. & Laurie, C. C. (1996) Genetics 144, 1559-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.García-Dorado, A. & González, J. A. (1996) Evolution (Lawrence, Kans.) 50, 1573-1578. [DOI] [PubMed] [Google Scholar]
- 51.Mackay, T. F. (1995) Trends Genet. 11, 464-470. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.