Abstract
Phospholipids and sphingolipids play critical roles in signal transduction, intracellular membrane trafficking, and control of cell growth and survival. We discuss recent progress in the identification and characterization of a family of integral membrane proteins with central roles in bioactive lipid metabolism and signalling. These five groups of homologous proteins, which we collectively term LPTs (lipid phosphatases/phosphotransferases), are characterized by a core domain containing six transmembrane-spanning α-helices connected by extramembrane loops, two of which interact to form the catalytic site. LPT family members are localized to all major membrane compartments of the cell. The transmembrane topology of these proteins places their active site facing the lumen of endomembrane compartments or the extracellular face of the plasma membrane. Sequence conservation between the active site of the LPPs (lipid phosphate phosphatases), SPPs (sphingosine phosphate phosphatases) and the recently identified SMSs (sphingomyelin synthases) with vanadium-dependent fungal oxidases provides a framework for understanding their common catalytic mechanism. LPPs hydrolyse LPA (lysophosphatidic acid), S1P (sphingosine 1-phosphate) and structurally-related substrates. Although LPPs can dephosphorylate intracellularly generated substrates to control intracellular lipid metabolism and signalling, their best understood function is to regulate cell surface receptor-mediated signalling by LPA and S1P by inactivating these lipids at the plasma membrane or in the extracellular space. SPPs are intracellularly localized S1P-selective phosphatases, with key roles in the pathways of sphingolipid metabolism linked to control of cell growth and survival. The SMS enzymes catalyse the interconversion of phosphatidylcholine and ceramide with sphingomyelin and diacylglycerol, suggesting a pivotal role in both housekeeping lipid synthesis and regulation of bioactive lipid mediators. The remaining members of the LPT family, the LPR/PRGs (lipid phosphatase-related proteins/plasticity-related genes) and CSS2s (type 2 candidate sphingomyelin synthases), are presently much less well studied. These two groups include proteins that lack critical amino acids within the catalytic site, and could therefore not use the conserved LPT reaction mechanism to catalyse lipid phosphatase or phosphotransferase reactions. In this review, we discuss recent ideas about their possible biological activities and functions, which appear to involve regulation of cellular morphology and, possibly, lipid metabolism and signalling in the nuclear envelope.
Keywords: lipid phosphatase, lysophosphatidic acid, phospholipid metabolism, sphingolipid metabolism, sphingomyelin synthase, sphingosine 1-phosphate
Abbreviations: C1P, ceramide 1-phosphate; CPO, chloroperoxidase; CSS2, type 2 candidate sphingomyelin synthase; DG, diacylglycerol; DPP, diacylglycerolpyrosphosphate phosphatase; EST, expressed sequence tag; G6P, glucose 6-phosphatase; LPA, lysophosphatidic acid; LPP, lipid phosphate phosphatase; LPR/PRG, lipid phosphatase-related protein/plasticity-related gene; LPT, lipid phosphatase/phosphotransferase; PA, phosphatidic acid; PC, phosphatidylcholine; PLD, phospholipase D; S1P, sphingosine 1-phosphate; SM, sphingomyelin; SMS, sphingomyelin synthase; SPP, sphingosine phosphate phosphatase
INTRODUCTION
The realization that membrane lipids play critical roles in many aspects of cell regulation is one of the most important advances in biomedical research in the past 30 years. The identification of many of the enzymes responsible for catalysing critical reactions in pathways of bioactive lipid metabolism has often lagged behind what continues to be steady and impressive progress in the definition of reactions and pathways responsible for the synthesis and inactivation of bioactive lipids and their metabolites. This is largely because the proteins responsible are often tightly associated with cell membranes, making them difficult to isolate and work with biochemically. Activity of these enzymes is characteristically dependent on the physical form of their substrates, which impedes the design and interpretation of in vitro assays to detect and quantify them. This is a particular issue with assays using detergent-solubilized substrates, which are a necessity when working with enzymes that require membrane solubilization. The identification of several important families of these lipid-metabolizing enzymes has therefore most often resulted from a combination of hard work, serendipity and ingenuity, rather than the ‘traditional’ approach of protein sequencing, cDNA cloning and, more recently, bioinformatics. The progress discussed in this review began in the 1960s with the identification of membrane-associated PA (phosphatidic acid) phosphatase activities [1,2], and has culminated in the recent recognition that the enzymes responsible for this activity, which are collectively termed LPPs (lipid phosphate phosphatases) [3–6], belong to a larger family of proteins that includes the SPPs (sphingosine phosphate phosphatases) [7,8], the long-sought animal SMSs (sphingomyelin synthases) [9], as well as two groups of proteins, the LPR/PRGs (lipid phosphatase-related proteins/plasticity related genes) [10–13] and CSS2s (type 2 candidate sphingomyelin synthases) [9], with less well-characterized enzymatic activities and functions.
IDENTIFICATION OF THE LPT (LIPID PHOSPHATASE/PHOSPHOTRANSFERASE) FAMILY
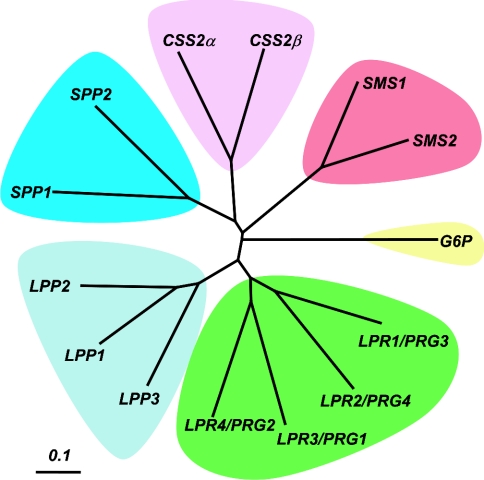
Although the important role of LPPs in glycerophospholipid metabolism and signalling had been long recognized and their activities characterized biochemically in some detail [14,15], difficulties in purifying the proteins responsible meant that these enzymes were not characterized at a molecular level until 1996 [3,4]. Recognition of homologies between two Saccharomyces cerevisiae open reading frames and a bacterial integral membrane lipid phosphatase that dephosphorylates phosphatidylglycerol phosphate [16], coupled with success in purification and sequencing of a mammalian LPP, revealed the existence of a family of homologous enzymes in yeast and mammals with broad specificity for lipid phosphates, including LPA (lysophosphatidic acid), S1P (sphingosine 1-phosphate), PA, C1P (ceramide 1-phosphate) and DGPP (diacylglycerol pyrophosphate) [3,4,6,17,18]. A genetic screen in yeast subsequently identified two further homologous proteins that were selective S1P phosphatases [19], and mammalian homologues of these enzymes were identified by both a complementation cloning approach and genome-wide sequence analysis [8,20,21]. While progress in understanding the functions of these mammalian LPP and SPP enzymes has continued steadily, exciting advances in the past year have led to the identification of three further groups of proteins that share the overall transmembrane topology of the LPPs and SPPs, but are characterized by variations in the core catalytic domain sequence motifs that define this family of proteins, and in the length and primary sequence of the N- and C-terminal portions of the proteins that flank the catalytic core. In this review, we collectively refer to this multigene family of integral membrane enzymes as LPTs. One group of these newly identified proteins, the SMSs, are mammalian homologues of the S. cerevisiae Aur1 gene product that catalyse the reversible interconversion of PC (phosphatidylcholine) and ceramide with DG (diacylglycerol) and SM (sphingomyelin) [9]. The other two groups of LPT family proteins are presently much less well described. The first of these have been termed LPR/PRGs. Two of the four LPR/PRG proteins have a novel and unexpected role in regulation of cellular morphology [10–12]. The final homology group of LPT family proteins, provisionally termed CSS2s, have presently uncharacterized enzymatic activities and functions [9]. Figure 1 shows a dendrogram that illustrates the overall sequence similarity relationships between the five classes of LPT proteins, noting a more distant relationship of these proteins to G6P (glucose 6-phosphatase), another integral membrane phosphatase enzyme with a similar transmembrane topology and, as discussed in more detail below, partial active site homology with the LPT family proteins. Although the tissue distribution of RNA and protein has been reported for some LPT family members [3,4,10,11,20,22], the expression of these genes in mammalian cells and tissues has not yet been examined in a comprehensive and systematic manner. Analysis of the tissue distribution and relative abundance of ESTs (expressed sequence tags) provides a useful and quantitative way to evaluate gene expression patterns. Table 1 summarizes extensive information regarding the expression pattern of all members of the LPT family in adult human tissues compiled from EST abundance data (http://www.ncbi.nlm.nih.gov/UniGene/). Expression patterns of the individual genes are discussed below in more detail in the corresponding sections of the article.
Figure 1. Primary sequence relationships between LPT family members.
A dendrogram representing primary sequence homology relationships between members of the LPT family is shown. The dendrogram was generated from a TCoffee alignment of the relevant sequences using TreeView [94].
Table 1. Expression of LPT family proteins in adult human tissues.
Expression profiles for the indicated genes were compiled from data available online at http://www.ncbi.nlm.nih.gov/UniGene/. The abundance of EST transcripts corresponding to each gene is expressed as transcripts per million.
| EST abundance (transcripts per million) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LPP | SPP | LPR/PRG | SMS | CSS2 | |||||||||
| LPP1 | LPP2 | LPP3 | SPP1 | SPP2 | LPR1/PRG3 | LPR2/PRG4 | LPR3/PRG2 | LPR4/PRG1 | SMS1 | SMS2 | CSS2α | CSS2β | |
| Bladder | 191 | 0 | 47 | 191 | 47 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 47 |
| Bone | 35 | 17 | 143 | 0 | 0 | 0 | 53 | 0 | 0 | 0 | 17 | 17 | 35 |
| Bone marrow | 27 | 0 | 136 | 54 | 0 | 0 | 0 | 27 | 0 | 109 | 27 | 0 | 27 |
| Brain | 107 | 30 | 166 | 35 | 19 | 48 | 181 | 17 | 124 | 48 | 13 | 45 | 24 |
| Cervix | 72 | 48 | 24 | 0 | 0 | 0 | 24 | 0 | 0 | 24 | 0 | 0 | 0 |
| Colon | 64 | 347 | 58 | 0 | 88 | 0 | 35 | 11 | 0 | 17 | 5 | 0 | 17 |
| Eye | 80 | 43 | 222 | 0 | 12 | 18 | 92 | 30 | 18 | 55 | 0 | 37 | 6 |
| Heart | 161 | 0 | 287 | 0 | 0 | 0 | 53 | 0 | 0 | 71 | 35 | 17 | 53 |
| Kidney | 172 | 44 | 202 | 37 | 112 | 29 | 7 | 0 | 0 | 44 | 14 | 7 | 37 |
| Larynx | 41 | 0 | 292 | 0 | 0 | 0 | 0 | 0 | 0 | 251 | 0 | 0 | 0 |
| Liver | 22 | 7 | 197 | 38 | 7 | 0 | 0 | 7 | 0 | 68 | 15 | 0 | 76 |
| Lung | 67 | 56 | 106 | 14 | 81 | 7 | 24 | 3 | 10 | 35 | 38 | 3 | 10 |
| Lymph node | 39 | 0 | 724 | 31 | 7 | 0 | 0 | 23 | 0 | 85 | 0 | 0 | 15 |
| Mammary gland | 132 | 24 | 490 | 0 | 33 | 0 | 41 | 0 | 0 | 57 | 33 | 0 | 8 |
| Muscle | 119 | 0 | 82 | 9 | 18 | 0 | 0 | 0 | 0 | 36 | 73 | 18 | 9 |
| Ovary | 63 | 116 | 21 | 0 | 31 | 10 | 116 | 10 | 0 | 31 | 10 | 21 | 21 |
| Pancreas | 74 | 211 | 161 | 0 | 12 | 0 | 87 | 12 | 0 | 12 | 37 | 0 | 0 |
| Peripheral nervous system | 318 | 0 | 238 | 0 | 39 | 0 | 159 | 0 | 0 | 119 | 0 | 39 | 0 |
| Placenta | 108 | 4 | 535 | 77 | 4 | 0 | 21 | 0 | 8 | 82 | 38 | 0 | 30 |
| Pituitary | – | – | – | – | – | 0 | 38 | 0 | 0 | 46 | 7 | – | – |
| Prostate | 483 | 38 | 576 | 7 | 15 | 0 | 42 | 0 | – | 12 | 0 | 31 | 23 |
| Skin | 30 | 12 | 6 | 24 | 24 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6 |
| Soft tissue | 102 | 0 | 153 | 0 | 0 | 51 | 0 | 0 | 0 | 0 | 102 | 0 | 0 |
| Spleen | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 120 | 0 | 0 | 0 |
| Stomach | 77 | 58 | 48 | 48 | 77 | 0 | 48 | 0 | 0 | 29 | 67 | 0 | 58 |
| Tongue | 36 | 0 | 36 | 0 | 145 | 0 | 0 | 0 | 72 | 109 | 0 | 0 | 0 |
| Testis | 60 | 7 | 68 | 7 | 15 | 7 | 7 | 15 | 15 | 83 | 30 | 0 | 30 |
| Thymus | 0 | 0 | 0 | 222 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 666 |
| Uterus | 196 | 156 | 214 | 0 | 17 | 0 | 11 | 5 | 0 | 57 | 34 | 0 | 17 |
| Vascular system | 154 | 0 | 656 | 0 | 38 | 0 | 0 | 0 | 0 | 308 | 38 | 0 | 0 |
| Whole blood | 105 | 0 | 39 | 13 | 13 | 0 | 52 | 26 | 0 | 66 | 0 | 0 | 0 |
PRIMARY SEQUENCE AND TRANSMEMBRANE ORIENTATION
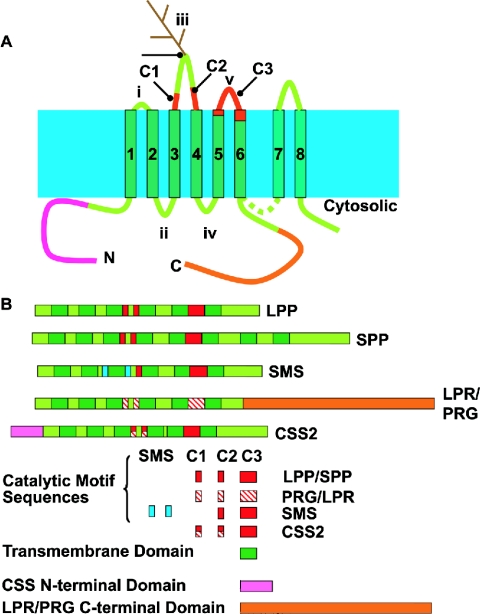
Figure 2 shows the proposed transmembrane orientation, topology and structural organization of members of the LPT family. These are all single polypeptide enzymes. Hydropathy analysis identifies six regions of hydrophobic sequence that could form transmembrane α-helices which are numbered 1–6. These helices are linked by extramembrane loops numbered i–v. The lengths of these loops vary between family members. This structurally conserved core domain is flanked by N- and C-terminal extensions. These extensions are also of varying lengths, and differences in the size and sequence composition of the N- and C-termini are defining features of particular members of the five classes of LPT proteins. This proposed transmembrane topology has only been confirmed experimentally for LPP3 [23]. A protease-susceptibility study of the SMS enzymes indicates a cytoplasmic orientation of the C-terminus, which is also consistent with this predicted topology [9]. The yeast and mammalian SPPs and two of the LPR/PRG proteins have much longer C-termini than all other LPT family members. Whereas the C-termini of the LPR/PRG proteins are highly hydrophilic and are unlikely to form additional membrane spans, the C-termini of the SPPs contain additional hydrophobic sequences that, by hydropathy analysis, are predicted to form two additional membrane-spanning α-helices, denoted 7 and 8 in Figure 2 [8,19,21]. A recent examination of the protease susceptibility of a series of epitope-tagged variants of one of the yeast SPP enzymes demonstrated the presence of these additional membrane helices [24], and it will be interesting to determine if this topology is conserved in the mammalian SPPs. Two other lines of evidence support the proposed transmembrane topology of the common core domain of these enzymes. Firstly, some, but not all, family members contain a functional glycosylation sequence in loop iii, implying that this region of the proteins has access to the lumen of the endoplasmic reticulum and Gogi apparatus as it traverses the secretory pathway [4,22,23]. Secondly, as discussed in detail in the following section, the catalytic site of the enzymes is formed from three sequence motifs, located in loops iii and v, and transmembrane sequences that immediately precede these loop regions [16,25,26]. The fact that these regions must be adjacent to each other to interact to form the catalytic site of the enzymes also requires that transmembrane helices 4–6 are oriented as shown in Figure 2. A detailed study of the transmembrane topology and active site organization of the integral membrane G6P is also consistent with a functional interaction between catalytic residues located in loops iii and v [27].
Figure 2. Structural organization of LPT family members.
(A) Diagram illustrating the transmembrane topology of members of the LPT family. (B) Diagram illustrating the structural organization of members of the LPT family.
ACTIVE SITE STRUCTURE AND CATALYTIC MECHANISM
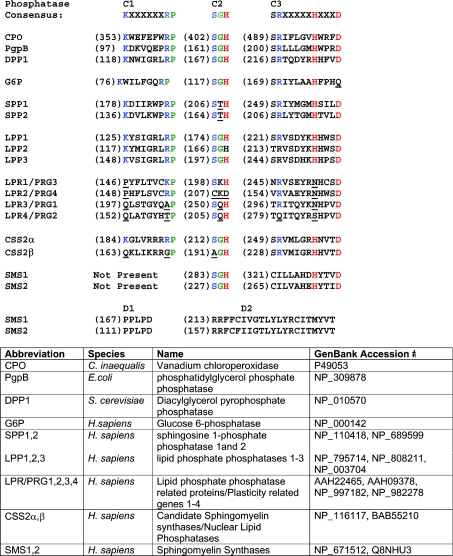
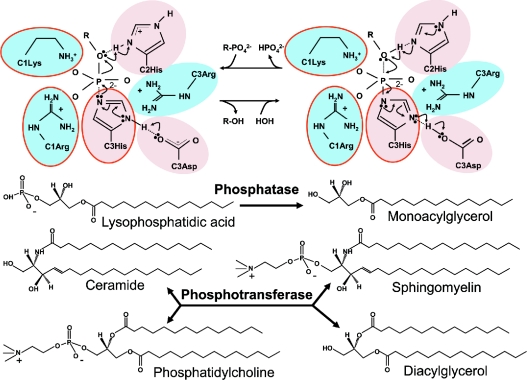
Shortly following the identification of the mammalian LPP and yeast DPP (diacylglycerolpyrosphosphate phosphatase) genes, three groups of workers noted that LPPs belong to a phosphatase superfamily that is defined by a shared motif comprising three separate sequences, denoted C1, C2 and C3, located in the third and fifth extramembrane loops and proximal transmembrane sequences [16,25,26]. This motif is conserved in the S. cerevisiae DPPs, Escherichia coli phosphatidylglycerol phosphatase, and mammalian G6Ps and SPPs. Figure 3 shows alignments of the relevant catalytic domain sequences of all known mammalian members of the LPT family. A critical insight, independently noted by these investigators, was that this phosphatase motif is also found in the active site of fungal oxyanion-dependent haloperoxidases. These enzymes share a common active site configuration and reaction chemistry that has been best studied for the vanadium-dependent CPO (chloroperoxidase) from the fungus Curvularia inaequalis. This enzyme uses hydrogen peroxide to catalyse the oxidation of halides to their corresponding hypohalous acids. Its three-dimensional structure has been solved at atomic scale resolution [26]. The active site of CPO is formed from four α-helices linked, on one face, by short polypeptides containing the phosphatase-consensus motif sequences. Prompted by the observation that the active site of CPO is homologous with that of the integral membrane phosphatases, CPO was also shown to be a broad specificity phosphatase with activity against p-nitrophenol phosphate and several other synthetic substrates [28]. High-resolution structural information, coupled with mutagenesis and measurements of enzymatic activity, as well as spectroscopic studies of oxyanion binding to CPO isoforms from several fungi, indicated that the trigonal bipyrimidal vanadate cofactor required for the halide oxidation reaction interacts with the enzyme in the same manner as the transition state phosphate intermediate of the phosphatase reaction. The C3 histidine and aspartic acid residues operate as a charge-relay system in which the histidine residues acts as a nucleophile, driving formation of a phosphohistidine intermediate. The C2 histidine facilitates phosphate bond cleavage and participates in the second step of the reaction in which the phosphohistidine intermediate is hydrolysed, freeing the active site for another round of catalysis. The invariant lysine, arginine, serine and glycine residues of the C1, C2 and C3 regions donate hydrogen bonds to the phosphate oxygen molecules and stabilize the transition state of the reaction (Figure 4) [26,28–30]. Although no structures are presently available for any of the integral membrane phosphatase/phosphotransferases, this detailed information about the catalytic mechanism of CPO provides an experimentally validated framework for understanding how the LPP and SPP enzymes catalyse hydrolysis of their substrates [31–33], and indicates how the SMS enzymes have probably adapted this reaction chemistry to perform a phosphotransferase reaction [9]. The incomplete conservation of the phosphatase active site in the LPR/PRG proteins and in one of the CSS2 proteins implies that they could not catalyse a phosphatase reaction using this mechanism [9–12].
Figure 3. Catalytic domain sequence conservation among LPT family members.
Alignment of catalytic domain sequences of members of the LPT family with cognate sequences from a fungal haloperoxidase (CPO), bacterial phosphatidylglycerol phosphatase (PgpB), yeast DPP1 and human G6P. H. sapiens, Homo sapiens.
Figure 4. LPT reaction mechanism.
A representation of the catalytic mechanism of LPT family members, highlighting functions for the conserved residues of the C1, C2 and C3 sequences of conserved catalytic motif in the reaction chemistry.
LPPs
Structural organization and enzymology
LPPs are the prototypic members of the LPT family. The three mammalian enzymes, LPP1–LPP3, catalyse divalent cation ion-independent hydrolysis of lipid phosphomonoesters [3,4,6] and can also perform complete dephosphorylation of diacylglycerolpyrophosphate, indicating that they have phosphodiesterase activity against a pyrophosphate bond [34]. However, they will not cleave the phosphodiester bond of glycerophospholipids. Although there are modest differences in Vmax and interfacial Km values when analysed using a mixed micellar assay system, LPPs hydrolyse a range of lipid phosphomonoesters, which includes LPA, PA, S1P and C1P, with broadly similar activities [6,14]. They do not hydrolyse the phosphomonoester groups of the phosphoinositides PtdIns(4)P and PtdIns(4,5)P2, and will not hydrolyse soluble substrates such as glycerol 3-phosphate. Although LPPs can hydrolyse substrates in complex with BSA [6] and are clearly enzymatically active in biological membranes and when reconstituted into artificial lipid bilayers [35], they are markedly more active against substrates dispersed in non-ionic detergents, particularly Triton X-100 [6,14]. The invariant amino acids of the C1, C2 and C3 phosphatase motif are completely conserved in these mammalian LPP enzymes, and mutagenesis studies of S. cerevisiae DPP1, LPP1 and LPP3 indicate that their catalytic mechanism is identical with that demonstrated for CPO [32,33,36]. The issue of what makes these enzymes broadly specific for lipid phosphate substrates in comparison with the SPP and SMS enzymes, which are highly selective (see below), is presently unresolved. As discussed below, the SPPs also contain a completely conserved C1-C2-C3 phosphatase motif with a minor substitution in the C2 sequence (Ser-Thr-His instead of Ser-Gly-His, Figure 3), yet these enzymes appear to be absolutely selective for S1P [37]. The possibility that this simple substitution in the C2 sequence accounts for this dramatic difference in substrate specificity is an interesting, but, as yet, untested idea.
Expression, localization and functions
LPP1 and LPP3 are widely expressed in human tissues, whereas levels of LPP2 transcripts are somewhat lower and more restricted. However, the majority of tissues examined express all three genes (Table 1), and these results are broadly consistent with the published analyses of LPP RNA distribution and, in cases where appropriate antibodies are available, protein expression [3,4,17,22]. cDNAs encoding apparent splice variants of LPP1 and LPP2 with alternative N-terminal sequences and truncated C-termini have been deposited in GenBank® [38]. Two of these cDNAs encode proteins without a complete catalytic domain. It is presently not known if proteins corresponding to these variant cDNAs are actually expressed, so the significance of these findings remains unclear. The subcellular localization of LPPs has been examined in a number of different cell types by overexpression of epitope-tagged variants, or, in a smaller number of cases, endogenously expressed LPPs have been visualized by indirect immunofluorescence using selective antibodies [18,22,39,40]. The proteins predominantly localize to the endoplasmic reticulum and plasma membrane, with possible localization to other endomembrane compartments, including the Golgi apparatus and endosomes also being noted. Increases in intact cell lipid phosphatase activity against exogenously applied substrates observed in cells overexpressing LPP1 and LPP3 are also consistent with localization to the plasma membrane [18,35,41]. Biochemical fractionation experiments indicate that LPP1 and LPP3 exhibit cell-specific localization to detergent-resistant membrane domains, which are considered to represent lipid rafts or caveolae [22,42]. Recruitment of LPPs to these membrane domains may be an important way to compartmentalize them with LPA or S1P receptors or with lipid signalling enzymes, particularly PC-specific PLD (phospholipase D) enzymes which are also found in caveolae and raft domain membranes [43].
Two major functions have been proposed for the LPPs. Firstly, they have been suggested to have roles in regulation of intracellular lipid metabolism by controlling the balance between PA and DG with obvious consequences for both cell signalling and influence on the synthesis of choline- and inositol-containing phospholipids. Although overexpression of LPP1 [39,44–46] and overexpression [22] or knock-out [36] of LPP3 clearly have effects on intracellular lipid metabolism in some, but not all, cells, the mechanism and significance of these observations remain largely unexplored. In this regard, perhaps the most interesting possible function for LPPs in intracellular lipid metabolism and signalling involves hydrolysis of PA generated by PLD. In this situation, LPP activity would terminate PLD signalling by hydrolysis of PLD-generated PA, concurrently forming DG which could activate lipid-responsive protein kinase C isoforms. In support of this idea, overexpression of LPP3 was shown to enhance PLD-dependent production of DG, possibly as a result of co-localization of LPP3 with a specific PLD isoenzyme, PLD2, in detergent-insoluble lipid raft domains [22]. Similarly, the Raf protein kinase appears to be a signalling target of PLD-generated PA and overexpression of LPP1 and LPP2 attenuates activation of the downstream Raf effector MAPK (mitogen-activated protein kinase) by stimulation of G-protein-coupled cell surface receptors [39]. Secondly, by virtue of their localization to the plasma membrane in many cell types [6,18,22,35,40,47] and their demonstrated ability to function as ecto-enzymes that dephosphorylate bioactive lipids with receptor-directed signalling actions, such as LPA and S1P [6,18,35,48], LPPs have been proposed to function as negative regulators of receptor-directed signalling by these bioactive lipid mediators. Experimental data, based in large part on overexpression studies and to a lesser extent on loss of function experiments using antisense RNA and pharmacological inhibitors, support this idea. The role of LPPs in regulation of signalling by cell surface lysophospholipid receptors has been reviewed extensively by others [13,38,49,50] and will not be discussed further here.
LPP3 knock-out mice exhibit defects in patterning during early development
It is perhaps more interesting that attempts to ascribe functions to the mammalian LPPs at a cellular and organismal level are beginning to reveal unexpected functions, some of which may not directly involve their catalytic activities. The broad and overlapping expression patterns of the three LPPs (Table 1) suggests that their functions may be non-redundant and consistent with this idea; mice lacking LPP2 are viable and were reported to exhibit no overt phenotype [51]. Inactivation of LPP1 in mice has not yet been reported. However, transgenic overexpression of LPP1 under control of a ubiquitously active actin promoter produced a runted phenotype with defects in spermatogenesis and fur growth. Circulating levels of LPA in the blood of these animals were not different from those measured in wild-type mice, and their phenotype was suggested to result from effects on the intracellular accumulation of DG which was observed in LPP1-overexpressing cell lines derived from these animals [46]. In marked contrast with the relatively benign effects of manipulating LPP1 and LPP2 expression in mice, homozygous inactivation of the murine LPP3 gene results in early embryonic lethality and a complex phenotype characterized by defects in both vasculogenesis and patterning during early development [36]. The duplication of axis symmetry observed in embryos from LPP3 null mice is similar to that observed when the Wnt signalling pathway is activated, for example, by ectopic embryonic overexpression of Wnts [52]. This phenotype also results from inactivation of axin which is a scaffolding protein functioning as a negative regulator of Wnt signalling by binding activated β-catenin [53]. Experiments using cells derived from LPP3 null mice reveal a novel role for LPP3 as a negative regulator of the Wnt signalling pathway. Expression of a Wnt reporter gene construct was higher in LPP3 null cells than in wild-type cells, whereas ectopic expression of LPP3 in LPP3 null cells reduced Wnt signalling activity to wild-type levels and concurrently reduced nuclear translocation of β-catenin [36]. The mechanism by which LPP3 inhibits Wnt signalling is currently not known. In addition to the ‘cannonical’ Wnt signalling pathway, Wnts also activate a phospholipase C-coupled pathway, leading to protein kinase C activation [54]. Overactivation of this ‘Wnt/Ca2+’ pathway results in attenuation of the cannonical Wnt signalling pathway and a ventralizing phenotype [55], which is very similar to that seen when mammalian LPP3 was ectopically expressed in Xenopus embryos [36]. As noted above, overexpression of LPP3 has been shown to result in increased levels of DG in cells [22], and DG levels were decreased in LPP3 null cells with a concurrent decrease in activated protein kinase C, which might, in turn, result in a stimulation of the canonical Wnt signalling pathway. On the other hand, studies using catalytically inactive LPP3 mutants indicate that the ability of overexpressed LPP3 to suppress Wnt signalling in LPP3 null cells is, at least partially, independent of enzymatic activity, implying the involvement of an alternative, as yet unidentified, mechanism [36]. Although overexpression of a catalytically inactive mutant of LPP1 has been shown to attenuate LPA-stimulated cytokine secretion in bronchial airway cells [41], the only ‘non-enzymatic’ function suggested for LPP3 is regulation of cell–cell interactions by an integrin-binding RGD (Arg-Gly-Asp) sequence present in the third extramembrane loop [56]. Although this finding is provocative in light of the involvement of LPP3 in Wnt signalling, which can also be regulated through another cell surface-adhesion molecule, p120 catenin, its broad relevance for mammalian LPP3 function is questionable, because the cognate sequence in murine LPP3 is RGE (Arg-Gly-Glu), which would not be expected to bind integrins.
Drosophila LPPs regulate germ cell migration and survival during early development
Studies in Drosophila have identified roles for two LPP homologues, the products of the wunen genes wun and wun2, in regulation of embryonic germ cell migration and survival [57–59]. Drosophila germ cells form at the posterior pole of the developing embryo and are swept into the hindgut and posterior midgut during gastrulation. The germ cells then move across the midgut, re-orient dorsally and migrate into the mesoderm to associate with the somatic gonadal precursor cells [59–61]. wun and wun2 act redundantly as repellant factors to guide migrating germ cells in the Drosophila embryo. These genes are normally expressed in somatic tissues that the germ cells avoid. However, in wunen mutants, in which both genes are disrupted, the germ cells scatter throughout the embryo, failing to reach the mesoderm and eventually dying. By contrast, overexpression of wun or wun2 in somatic tissues, such as the midgut and mesoderm, that normally attract germ cells, results in germ cell repulsion and death [57,59]. The repulsive effect of these LPPs on germ cell migration has been suggested to be a result of their ability to degrade a lipid factor that both guides germ cells and serves as a germ cell survival factor during their migration in the developing gut. However, whereas both wun and wun2 are functional LPPs whose enzymatic activity is necessary for their ability to repel germ cells [59,62], the chemical identity of this lipid signal has not yet been determined. The finding that mammalian LPP3 can also repel germ cells when expressed in the somatic cells of Drosophila embryos indicates a conserved function between insect and mammalian LPPs, although surprisingly mammalian LPP1 cannot repel germ cells in this assay [62,63]. The basis for this difference in biological activity between LPP3 and LPP1 is not known. Although their substrate specificities and enzymatic activites are very similar in vitro, these two LPPs have been shown to localize selectively to the apical and basolateral surface of polarized cells [40], and LPP3 has also been reported to localize to detergent-resistant membrane domains [22], so it is possible that differences in subcellular localization or, perhaps, regulation of these LPPs, rather than in their intrinsic enzymatic activity or substrate selectivity, accounts for their distinct effects on germ cell migration when expressed in Drosophila embryos. Interestingly, wun and mammalian LPP1 and LPP3 were recently shown to form stable homodimers through interactions requiring the C-termini of the proteins in overexpression experiments conducted using epitope-tagged proteins [63a]. Although, at least in the case of wun, this interaction did not appear to have functional relevance, these findings raise the possibility that interactions between LPPs may modulate their localization or activities in some settings [62].
A new study identifies a novel and surprising additional role for wun2 as a germ cell-specific factor required for proper germ cell migration and survival [63]. wun2 is specifically expressed in germ cells of the developing Drosophila embryo from maternally inherited RNA. Expression of catalytically active wun2 in germ cells is required for germ cell survival in otherwise wild-type embryos and, as in the germ cell repulsion assays described above, when expressed in germ cells, LPP3 can substitute for wun2. Germ cell-specific expression of wild-type, but not catalytically inactive alleles of Wun2 or of mammalian LPP3, could suppress germ cell death resulting from somatic overexpression of wun2. wun2 therefore exhibits paradoxical cell- and tissue-specific effects on germ cell survival. When expressed in germ cells, wun2 promotes their survival, whereas expression of either wun2 or wun in somatic cells repels migrating germ cells and promotes germ cell death. Experiments in which expression levels of wun2 in germ cells or somatic cells were separately regulated revealed a reciprocal relationship between the actions of wun2 in germ cells and somatic cells. Germ cell death induced by somatic overexpression of wun2 could be suppressed by overexpression of wun2 in germ cells, whereas germ cell death resulting from a lack of maternally expressed germ cell wun2 could be rescued by reducing somatic wun2 expression. These results suggest a model in which somatic wun and wun2 compete with germ cell wun2 for a common lipid substrate. While hydrolysis of this substrate in the soma results in decreased germ cell survival, hydrolysis of the substrate by germ cells promotes their survival. As noted above, both wun and wun2 are active LPPs with substrate specificities and catalytic properties that are very similar to those of the mammalian LPPs [62,63]. As was observed for mammalian LPP1 [35], overexpression of wun2 in cultured insect cells results in a significant enhancement of cellular accumulation of lipid dephosphorylation products when the cells are incubated with fluorescent analogues of PA [63]. Hydrolysis-coupled intracellular accumulation of PA dephosphorylation products has been described in mammalian cells [64,65]. This phenomenon may involve partitioning of the lipid substrate into the plasma membrane where LPP-catalysed hydrolysis generates a non-polar lipid product, DG, that can enter the cell by endocytosis where it becomes susceptible to further metabolism. It is therefore plausible that, although the activities of wun and wun2 in the soma may be linked to degradation of a pool of a lipid phosphate substrate that is required for germ cell guidance and survival, the germ cell-specific action of maternally inherited wun2, which is necessary for germ cell survival, is linked to wun2-facilitated lipid uptake and the intracellular accumulation of a bioative lipid signalling molecule. In light of the apparent absence of G-protein-coupled lysophospholipid receptors from the Drosophila genome [66,67], it is possible that the ability to facilitate lipid uptake might represent a primary signalling function for wun and/or wun2. Clearly, the identification of the relevant lipid substrate hydrolysed by wun/wun2 in the somatic and germ cells of Drosophila embryos remains a major challenge. The possibility that mammalian LPPs also have signalling functions that are coupled to the intracellular accumulation of lipid dephosphorylation products is also an area worth examining further.
SPPs
Structural organization and enzymology
The identification of the mammalian SPPs was preceded by studies of yeast genes that encode enzymes that dephosphorylate phosphorylated sphingoid bases and are named YSR3/LBP2 and YSR2/LBP1/LCB3 [19]. These yeast enzymes are non-redundant and both exhibit high phosphatase activity against long-chain sphingoid base phosphates. Although SPP1 has been characterized in most detail, in contrast with the broad specificity of LPP1–LPP3 for lipid phosphomonoester substrates, SPP1 and 2 are highly selective for S1P [20,37]. Another notable difference between the LPP and SPP enzymes is that, whereas the LPPs are most active against detergent-solubilized substrates, the SPPs are inhibited by detergents and show a marked preference for substrates that are bound to a protein carrier [8,68]. As noted above, SPPs contain a consensus phosphatase/phosphotransferase catalytic motif with a minor substitution in the C2 sequence, but, in contrast with the LPPs, their C-terminus is much longer and may contain an additional two transmembrane spans.
Expression, localization and functions
SPP1 and SPP2 have broad and partially overlapping expression patterns in mammalian tissues. Expression of SPP2 appears more widespread than SPP1 [20] (Table 1). Like the LPPs, it has proven difficult to generate effective antibodies against mammalian SPPs, so overexpression studies using epitope-tagged proteins have been used to investigate their subcellular localization. In contrast with the LPPs which are broadly localized to both endomembrane compartments, primarily the endoplasmic reticulum, and to the plasma membrane, SPP1 and SPP2 are restricted to the endoplasmic reticulum [20,37], which is similar to the subcellular localization of their yeast counterparts [69]. Ideas about the function of these enzymes therefore emphasize the possibility that they have roles in intracellular sphingolipid metabolism, in particular generation of sphingosine for ceramide synthesis, rather than a primary direct role in the inactivation of S1P at the cell surface and consequent termination of its signalling actions, which, as discussed above, is a more likely potential function for the LPPs. The most detailed information about SPP function comes from studies in yeast, which are now being extrapolated to mammalian cells. Yeast sphingolipids are structurally analogous to mammalian sphinoglipids, except that the polar head group is phosphoinositol, whereas phytosphingosine is the predominant sphingoid base, as opposed to choline and sphingosine respectively. Yeast sphingolipids, in particular ceramide, have well-defined functions in protection from heat, osmotic and low pH stresses [70]. Deletion of LBP1 and LBP2 results in a marked accumulation of phosphorylated sphingoid bases, a concomitant reduction in ceramide levels and a dramatic increase in resistance to these stresses. Conversely, overexpression of LBP1 results in ceramide accumulation and increased sensitivity to stress [19]. Mutational analysis and suppressor screens demonstrate that LBP1p functions to control the balance between sphingoid bases, which are used for sphingolipid synthesis, and phosphorylated sphingoid bases, which are recycled for phospholipid synthesis. Interestingly, LBP1 mutants are also unable to incorporate exogenously supplied sphingosine into sphingolipids, which suggests that phosphorylation and dephosphorylation of these lipids is somehow required for their use as precursors for ceramide synthesis [70,71]. Studies in mammalian cells reveal an analogous role for SPP1 in control of ceramide levels and apoptosis, but suggest additional roles for the mammalian SPPs in control of sphingolipid metabolism and signalling.
SPPs regulate survival, apoptosis and migration in mammalian cells
Forced overexpression of SPP1 in HEK-293 cells results in significantly increased levels of intracellular ceramide. This SPP1-dependent increase in ceramide levels can be enhanced further by exogenous application of S1P that presumably enters the cells and is dephosphorylated by SPP1 at the endoplasmic reticulum to generate sphingosine, which in turn serves as a substrate for ceramide biosynthesis [72]. In mammalian cells, a substantial body of evidence identifies ceramide as an intracellular signal for stress-induced apoptosis [73]. The increased levels of ceramide observed in SPP1-overexpressing cells were associated with reduced cell survival and increased apoptosis [8,72]. A curious observation from these experiments was that, although an equally good substrate for SPP1, exogenous application of dihydroS1P had no effect on intracellular ceramide levels or apoptosis. These interesting findings imply different roles for ceramide and dihydroceramide in sphingolipid synthesis and regulation of apoptosis, possibly related to trafficking of ceramide and dihydroceramide between different endomembrane compartments that serve as sites for the synthesis and actions of these lipids [68,72]. Although mouse knock-out models of SPP1 have not yet been reported, RNA interference studies conducted in mammalian cells reveal a role for SPP1 in control of levels of S1P, both within cells and extracellularly, and suggest an addition mechanism by which SPP1 could regulate apoptosis. Knock-down of SPP1 resulted in significant accumulation of both intra- and extra-cellular S1P, suggesting that SPP1 activity normally opposes the synthesis of S1P by sphingosine kinase. Suppression of SPP1 expression in these cells rendered them resistant to apoptosis induced by TNF (tumour necrosis factor) and chemotherapy drugs [21]. Through actions mediated by specific G-protein-coupled cell surface receptors, S1P is a potent and effective survival factor for many mammalian cells [67], so it is reasonable to postulate that the resistance to apoptosis observed when SPP1 expression is suppressed results from an enhancement of S1P signalling [21]. These results raise the possibility that the increases in apoptosis associated with overexpression of SPP1 may result from a decrease in S1P levels, rather than increases in ceramide levels as suggested above. These competing ideas are not mutually exclusive and clearly this issue will require further experimentation to resolve. For example, one critical consideration is that the extracellular levels of S1P that result in increased intracellular accumulation of ceramide are much higher than the levels required to promote cell signalling responses through actions mediated by S1P receptors. It is also unclear if these effects on intracellular sphingolipid metabolism are restricted to the SPPs or if overexpression of one or more of the LPPs, which can both dephosphorylate S1P and localize to the endoplasmic reticulum in many cells, can also increase intracellular levels of ceramide or regulate levels of S1P inside and outside of cells. It is also important to note that all of these studies have focused on SPP1 and, although it is clearly an active S1P phosphatase in vitro, the cellular functions of SPP2 also need to be examined.
SMSs
Structure and enzymology
The sole pathway for SM synthesis in mammalian cells involves the enzymatic transfer of the phosphocholine group of PC to ceramide, generating SM and DG [74]. The enzyme responsible, SMS, therefore occupies a central position at a crossroads of sphingolipid and phospholipid metabolism. The actions of this enzyme not only generate sphingomyelin, but also regulate cellular levels of the signalling lipids DG and ceramide [73,75]. However, despite the pivotal role of this membrane-associated activity in both phospho- and sphingo-lipid metabolism, the enzymes responsible for this reaction in mammalian cells were only identified very recently. Studies published in the past year have described a family of two animal SMSs termed SMS1 and SMS2 [9]. These are integral membrane proteins that, as discussed in detail above, have the common six transmembrane-spanning core domain topology that is common to the LPT family. SMS1 and SMS2 contain the C2 and C3 catalytic motifs which are likely to be responsible for the phosphotransferase step of catalysis, whereas the C1 motif is absent and replaced by unique SMS-specific sequence motifs that are likely to be responsible for substrate recognition and orientation in the active site [9] (Figures 2–4). SMS1 and SMS2 were identified using a candidate gene expression cloning strategy. The essential S. cerevisiae Aur 1 gene product catalyses the transfer of the headgroup of phosphatidylinositol to phytoceramide, generating inositol-phosphorylceramide and DG [76,77]. BLAST searches for novel sequences encoding integral membrane proteins containing the C2 and C3 domain common to Aur1p and LPPs without homologues in yeast or previously characterized biochemical functions identified three groups of candidate genes with homologues in multiple animal species. Epitope-tagged variants of these proteins were expressed in yeast, and SMS activity was determined in vitro using membrane extracts as a source of activity by monitoring the incorporation of fluorescent ceramide into SM which proceeds using endogenous membrane PC as a substrate. Two of the human proteins tested, SMS1 and SMS2, were active in these assays. SMS activity could be observed using exogenously provided PC, but not lyso-PC, choline, phosphorylcholine or nonphosphocholine lipids, including phosphatidylethanolamine, suggesting that the enzyme recognizes the two acyl chains attached to the phosphocholine headgroup. Interestingly, in these experiments, SM itself could also serve as a headgroup donor, indicating that, as observed previously using membrane preparations as a source of enzyme activity [78], the SMS reaction is reversible. A subsequent publication reported expression cloning of SMS1 using an equally elegant complementation strategy employing mammalian cell lines with previously described defects in SMS activity. This study is of importantance as it demonstrated that SMS1 is functional when expressed in intact cells [79]. As noted above and consistent with the presence of SM in many organisms, SMS homologues are found in many species. Drosophila, which lacks SM, but instead synthesizes ethanolamine phosphoceramide, is a notable exception. Although the Drosophila genome does not contain SMS homologues, a single SMS-related gene was identified that might encode an ethanolamine phosphotransferase responsible for ethanolamine phosphoceramide synthesis [9].
Localization and function
Prior to the identification of the SMS genes, fractionation studies suggested that SMS activity was associated with both the Gogi apparatus and plasma membrane [80,81]. In agreement with these findings, indirect immunofluorescence analysis of epitope-tagged SMS1 and SMS2 in HeLa cells revealed two distinct localization patterns. SMS1 was mainly found at the Golgi apparatus, whereas SMS2 displayed a predominantly plasma membrane localization and only partial co-localization with the Golgi marker sialyltransferase. The C-terminally appended V5 epitope tag of SMS2 was insensitive to proteolysis when trypsin was added to intact cells, but could be readily degraded when the plasma membrane was permeabilized with detergent. This finding implies that, as is the case with other LPT family members, the C-termini of SMSs are oriented towards the cytosol, whereas the active sites are directed towards the lumen of the Golgi apparatus or the extracellular space [9]. The orientation and localization of SMSs suggest that SMS1 may have a primary ‘housekeeping’ role in SM synthesis within the cell, whereas SMS2 could have a lipid signalling function at the plasma membrane, possibly working in conjunction with sphingomyelinases to regulate plasma membrane levels of the pro-apoptotic signalling lipid ceramide. Nothing is presently known about how SMS1 and SMS2 activity is regulated. In light of the reversibility of the SMS reaction, it is possible that the relative concentrations of DG and ceramide resulting from the actions of phospholipases C and sphingomyelinases in the vicinity of SMS are a relevant determinant of the direction and rate of the SMS reaction. In this regard it is interesting to note that up- and down-regulation of SMS activity has been linked to mitogenic and pro-apoptotic signalling in several mammalian cell types [73]. The identification of SMS1 and SMS2 provides unique tools to investigate the regulation and functions of these long-sought enzymes.
LPR/PRGs
Structure and enzymology
In comparison with other members of the LPT family, the four members of the LPR/PRG family are characterized by incomplete conservation of the C1, C2 and C3 catalytic motif sequences. As mentioned above and illustrated in Figure 3, these proteins share a non-conservative substitution of the C3 nucleophilic histidine residue and of the C1 lysine and arginine residues, and there are additional non-conservative substitutions of the C2 motif residues in some LPR/PRG family proteins. The proposal that these genes are named LPR proteins [12,13] was intended to reflect this incomplete structural relationship to the LPPs and other LPT family members. One of these genes, LPR3, was cloned from a rat brain cDNA library and given the name PRG1 to reflect the regulation of its expression during brain development and response to experimentally induced hippocampal lesions [10]. In agreement with this nomenclature, the other members of this gene family have been termed PRG2, PRG3 and PRG4 [11]. The LPR nomenclature has been adopted by GenBank®, because of the pre-existing use of the acronym ‘PRG’ for an unrelated family of secreted proteoglycan genes [82]. For clarity and consistency, we refer to LPR/PRG proteins in the present article. Ideally, this imprecise terminology will soon be replaced by a nomenclature that reflects the biological functions of these proteins. The incomplete conservation of the catalytic motif in all of the LPR/PRG proteins, and, in particular, the non-conservative substitution of amino acid residues that are critical for catalysis in other LPT family members, implies that none of the LPR/PRG proteins could catalyse lipid phosphatase or phosphotransferase reactions using the mechanism discussed in detail above (Figures 3 and 4). Consistent with this prediction, the LPR/PRG proteins do not have SMS activity when expressed in yeast [9], and two of these proteins, LPR1/PRG3 and LPR3/PRG1 do not have LPA phosphatase activity when expressed in HEK-293 cells and assayed using either intact cells or cell membranes as the source of enzyme activity [12]. These experiments were conducted using a highly sensitive ‘phosphate-release’ assay with 32P-labelled substrates, or a less sensitive assay in which the formation of radiolabelled monoacylglycerol by hydrolysis of acyl chain labelled LPA substrate was monitored under conditions where activity of a co-expressed LPP could be readily detected. Both the LPR/PRG proteins and the control LPP protein were expressed to similar levels, as shown by Western blotting using antibodies against an appended C-terminal tag. In accordance with these findings, overexpression of LPR1/PRG3 in a cultured neuronal cell line did not result in increases in the rate of hydrolysis of exogenously added LPA by intact cells [11]. Paradoxically, however, overexpression of LPR3/PRG1 has been reported to result in variable increases in rates of LPA hydrolysis (either 5-fold or 2-fold compared with the control) in the same neuronal cell line [10,11]. Given the inactivity of LPR3/PRG1 in more carefully controlled experiments, if these observations can be substantiated by others, one explanation may be that overexpression of LPR3/PRG1 in some cells results in an upregulation of the activity of endogenously expressed LPPs at the cell surface. As discussed in more detail below, resolution of this issue is critical to our understanding of the biological activities of these LPR/PRG proteins. The other noteworthy structural feature of the LPR/PRG proteins is the very long C-terminus (approx. 400 amino acids) found in LPR3/PRG1 and LPR4/PRG2, which is unique among the entire LPT family. This region of the proteins is highly enriched in charged amino acid residues and contains PEST (Pro-, Glu-, Ser- and Thr-containing) sequences that, in other proteins, function as signals for ubiqutin-dependent targeting for proteolytic degradation [83]. Multiple species of C-terminally tagged LPR3/PRG1 are detected when the protein is overexpressed in mammalian cells, suggesting that proteolysis may be a highly relevant mechanism for regulation of the turnover and expression levels of the protein [12]. Inspection of both EST sequences and genomic sequences for LPR3/PRG1 and LPR4/PRG2 provides evidence for the existence of splice variants with alternative C-termini, which may provide an additional mechanism for regulating the biological activities of these proteins.
Expression, localization and functions
In contrast with the LPPs and SPPs, the individual members of the LPR/PRG family exhibit much more restricted expression patterns. Northern blot analysis showed that LPR3/PRG1 was most strongly expressed in the brain, although weaker expression in peripheral tissues was also apparent, which is consistent with the tissue distribution of corresponding ESTs with transcripts found in the eye, kidney and ovary [10]. LPR4/PRG2 is more widely expressed than LPR3/PRG1. Similarly, LPR1/PRG3 exhibits a more restricted expression pattern than LPR2/PRG4 (Table 1). The distinct and, in many cases, non-overlapping expression patterns of the LPR/PRG genes suggests that they may not function redundantly. The characterization of the LPR/PRG proteins is at an early stage. Overexpression of wild-type LPR3/PRG1, but not of an LPR3/PRG1 mutant containing a non-conservative substitution of one amino acid residue that is conserved within the LPT C2 catalytic motif and known to be critical for catalysis in the LPPs, was reported to protect cultured neuronal cells from the neurite-collapsing actions of exogenously applied LPA. In conjunction with the finding that neuronal expression of LPR3/PRG1 is increased during brain development, and regenerative sprouting of axons and neurites induced in response to hippocampal injury, these observations were suggested to identify a normal role for LPR3/PRG1 as a regulator of these kinds of neuronal membrane protrusions during brain development and responses to injury through the localized attenuation of LPA signalling [10]. Clearly identification of the mechanism by which overexpression of LPR3/PRG1 apparently increases rates of dephosphorylation of exogenously provided LPA is of critical importance to understanding this phenomenon. More importantly, this suggestion implies that endogenously-formed LPA has a normal role in regulation of neuronal plasticity, which is an interesting, but as yet unproven, idea [84,85]. Further testing of these interesting ideas regarding LPR3/PRG1 function will require loss-of-function approaches and the identification of the LPA receptors responsible for these putatively LPR3/PRG1-regulated signalling pathways. In apparent contrast with these effects of the overexpression of LPR3/PRG1 on neurite outgrowth, overexpression of LPR1/PRG3 was reported to cause the spontaneous outgrowth of structures suggested to be ‘neurites’ by cultured neuronal cells and COS-7 cells [11]. The true identity of these membrane protrusions, a description of their composition, dynamics and relationship to neurites, which are formed in cells overexpressing LPR3/PRG1 through apparently LPA-dependent processes, needs to be established. Because overexpression of LPR1/PRG3 does not result in increases in rates of dephosphorylation of exogenously provided LPA [11,12], these observations imply that overexpression of LPR1/PRG3 can induce changes in cell morphology through processes that do not involve attenuation of LPA signalling, as was suggested for LPR3/PRG1. Taken together, these studies show that two members of the LPR/PRG family have a role in regulation of cellular morphology. However, the mechanistic basis for this surprising activity is presently not known. In light of their primary sequence and inactivity in properly controlled experiments, the most parsimonious conclusion is that none of the LPR/PRG proteins have intrinsic lipid phosphatase or phosphotransferase activity which raises the question of whether they are enzymes at all. As has been found for other classes of protein and lipid phosphatases, an intriguing possibility is that the LPR/PRG proteins are ‘non-enzymes’ – catalytically incompetent homologues of active enzymes lacking residues critical for catalysis, but retaining residues required for the capacity to interact non-productively with their lipid substrates [86]. For example, enzymatically inactive homologues of the phosphoinositide phosphatase, myotubularin, can still bind D-3 phosphorylated inositol lipids and function as adapter proteins [87]. Although the identified G-protein-coupled LPA receptors clearly account for may of the identified signalling actions of LPA, genetic and pharmacological data imply that additional mechanisms of LPA signalling exist [88]. Perhaps the LPR/PRG proteins can interact with LPA, S1P or a related lipid-signalling molecule and have ‘receptor-like’ activities that are coupled to pathways that regulate cell morphology? Whatever the case, these fascinating new proteins are a very attractive area for future studies.
CSS2s
Structure and enzymology
The final group of LPT family proteins contains two members. These proteins have been provisionally termed CSS2s [9], but, aside from a report that they do not exhibit SMS activity when overexpressed in yeast, their enzymatic activities are unexplored. One of these proteins, CSS2α contains a completely conserved phosphatase motif that is identical with that found in the LPPs, which strongly suggests that it may be a functional phosphatase. CSS2β contains a variant phosphatase motif that is, in some respects, similar to the apparently inactive phosphatase motif found in the LPR/PRG proteins. Clearly, the potential enzymatic activities of these proteins need to be investigated directly. In comparison with other LPT family members, the defining shared structural characteristic of the CSS2 proteins is an extended N-terminus preceding the first transmembrane domain. This region is approx. 80 amino acids long and quite divergent between the two proteins, although a short motif close to the transmembrane domain is well conserved. Hydropathy analysis of these proteins also predicts the presence of six transmembrane α-helices, although it is noteworthy that the putative extramembrane loop sequence between helices 4 and 5 is very short, suggesting that these two helices may not span the plasma membrane completely, perhaps forming a single ‘half-membrane-spanning’ helix, as has been observed in some aquaporin family channel proteins [89].
Expression, localization and functions
Analysis of EST abundance reveals that CSS2β is widely expressed in mammalian tissues, whereas CSS2α has a more restricted expression pattern (Table 1). Intriguingly, a subtractive proteomic screen suggests that CSS2β localizes, at least partially, to the nuclear envelope [90]. The strategy used to isolate proteins for sequencing in this study implies that CSS2β is at least partially resistant to extraction of the nuclear membrane with non-ionic detergents, suggesting that nuclear envelope localization and retention of the protein involves an interaction with a component of the nuclear lamina or matrix. The nuclear envelope proteins sequenced in this screen were obtained from liver which appears to only express CSS2β (Table 1), therefore clearly the subcellular localization of both CSS2α and CSS2β needs to be evaluated directly. The apparent nuclear envelope localization of at least one of the CSS2 proteins is provocative, because, although LPP activity has not been explored explicitly in isolated nuclei, both direct and indirect biochemical observations of enzyme activities that can generate DG from PA in nuclei have been reported [91,92]. A growing body of work identifies roles for glycerophospholipids in both the nuclear matrix and surrounding membrane as regulators of aspects of nuclear function that include signal transduction and regulation of nuclear RNA export [93]. The possibility that one or both of the CSS2 proteins functions in nuclear phospholipid metabolism linked to control of these, and possibly other nuclear functions, is worth considering.
CONCLUDING COMMENTS
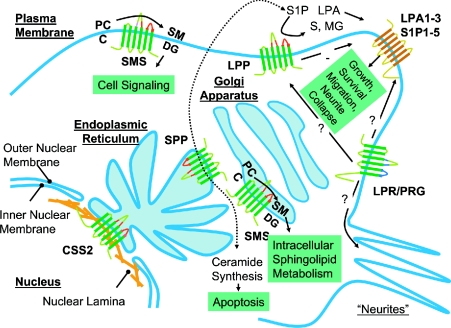
Figure 5 summarizes ideas presented in this review about the subcellular localization and possible functions of the different classes of LPT proteins. It is important to stress that the information presented in Figure 5 comes from a relatively small number of studies conducted using divergent cell types and relying to a great extent on overexpression approaches. Moreover, some of the ideas presented, in particular about the localization and functions of the LPR/PRG and CSS2 proteins are admittedly speculative, but incorporate what limited information is presently available. Nevertheless, the observations and ideas brought together in this Figure suggest roles for members of the LPT family in aspects of lipid metabolism and signalling that involve all major endomembrane systems of the cell. The advances discussed in this review promise to stimulate the application of more sophisticated approaches to the analysis of the function of the LPT family proteins in cells and animals. Investigators now have molecular tools to examine the phenotypic consequences of down-regulation and overexpression of these proteins, and can employ genetic and mutational approaches to dissect the structural basis for their activities. As integral membrane proteins, segregation and sorting of LPT family members to different membrane compartments is clearly of great importance for understanding their cellular functions, and this issue can now be explored by the construction and analysis of epitope-tagged variants of the proteins. In particular, members of the LPP, SPP and SMS families are highly amenable to C-terminal tagging with green fluorescent protein, which can be used to investigate their localization and trafficking in live cells. Although many important questions can now be addressed using cell culture systems, analysis of LPT family gene function at the organismal level is now possible. The use of knock-out technology to probe the function of LPT family members in multicultural organisms is at a very early stage. The lessons learned from the work on murine LPP2 and LPP3 and the Drosophila wunen genes indicate that, although there is may be some functional redundancy between members within a particular class of LPT proteins, this is likely to continue to be a very fruitful and productive approach. These gene inactivation approaches would benefit from the identification of homologues in simpler genetically manipulatable organisms, and this is would be another fruitful avenue for functional analysis of members of the LPT family. As noted in the relevant sections of this review, budding yeast has been a very important system for studies of the LPP and SPP enzymes and for the identification of animal SMSs, whereas studies in Drosophila continue to provide the most valuable insights into LPP function during early development. In this regard, we note that homologues of the two most interesting and underexplored members of the LPT family, the LPR/PRG and CSS2 proteins, are restricted to multicellular organisms, including Drosophila and Caenorhabditis elegans, which might indicate a role in some aspect of cellular communication. Finally, as noted at the very beginning of this article, enzymatic analysis of these enzymes necessarily involves studies using exogenously provided substrates presented in unnatural physical forms. The development of assay systems, for example, using fluorescent substrates as reporters, to monitor enzyme activity in intact cells or tissues would allow researchers to evaluate LPT activity in situ.
Figure 5. Subcellular localization and functions of LPT family members.
The localization of the CSS2 proteins to the nuclear envelope is speculative and on the basis of results obtained from a subtractive proteomic screen conducted using rat and mouse liver-derived subcellular fractions. The subcellular localizations of other LPT family members are based on published reports that, for the most part, involve studies using overexpressed epitope-tagged proteins. C, ceramide; S, sphingosine.
Acknowledgments
Work in our laboratory is supported by grants from the National Institutes of Health. Y.J.S. is a pre-doctoral fellow of the American Heart Association. We are grateful to Joost Holthius, Colin Stewart, Andrew Renault and Ruth Lehman for useful discussions.
References
- 1.Hokin L. E., Hokin M. R. Diglyceride kinase and phosphatidic acid phosphatase in erythrocyte membranes. Nature (London) 1961;189:836–837. doi: 10.1038/189836a0. [DOI] [PubMed] [Google Scholar]
- 2.Coleman R., Huebscher G. Metabolism of phospholipids. V. Studies of phosphatidic acid phosphatase. Biochim. Biophys. Acta. 1962;56:479–490. doi: 10.1016/0006-3002(62)90600-5. [DOI] [PubMed] [Google Scholar]
- 3.Kai M., Wada I., Imai S., Sakane F., Kanoh H. Cloning and characterization of two human isozymes of Mg2+-independent phosphatidic acid phosphatase. J. Biol. Chem. 1997;272:24572–24578. doi: 10.1074/jbc.272.39.24572. [DOI] [PubMed] [Google Scholar]
- 4.Kai M., Wada I., Imai S., Sakane F., Kanoh H. Identification and cDNA cloning of 35-kDa phosphatidic acid phosphatase (type 2) bound to plasma membranes. Polymerase chain reaction amplification of mouse H2O2-inducible hic53 clone yielded the cDNA encoding phosphatidic acid phosphatase. J. Biol. Chem. 1996;271:18931–18938. doi: 10.1074/jbc.271.31.18931. [DOI] [PubMed] [Google Scholar]
- 5.Brindley D. N., Waggoner D. W. Mammalian lipid phosphate phosphohydrolases. J. Biol. Chem. 1998;273:24281–24284. doi: 10.1074/jbc.273.38.24281. [DOI] [PubMed] [Google Scholar]
- 6.Roberts R., Sciorra V. A., Morris A. J. Human type 2 phosphatidic acid phosphohydrolases. Substrate specificity of the type 2a, 2b, and 2c enzymes and cell surface activity of the 2a isoform. J. Biol. Chem. 1998;273:22059–22067. doi: 10.1074/jbc.273.34.22059. [DOI] [PubMed] [Google Scholar]
- 7.Mandala S. M. Sphingosine-1-phosphate phosphatases. Prostaglandins Other Lipid Mediat. 2001;64:143–156. doi: 10.1016/s0090-6980(01)00111-3. [DOI] [PubMed] [Google Scholar]
- 8.Mandala S. M., Thornton R., Galve-Roperh I., Poulton S., Peterson C., Olivera A., Bergstrom J., Kurtz M. B., Spiegel S. Molecular cloning and characterization of a lipid phosphohydrolase that degrades sphingosine-1-phosphate and induces cell death. Proc. Natl. Acad. Sci. U.S.A. 2000;97:7859–7864. doi: 10.1073/pnas.120146897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huitema K., van den D. J., Brouwers J. F., Holthuis J. C. Identification of a family of animal sphingomyelin synthases. EMBO J. 2004;23:33–44. doi: 10.1038/sj.emboj.7600034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brauer A. U., Savaskan N. E., Kuhn H., Prehn S., Ninnemann O., Nitsch R. A new phospholipid phosphatase, PRG-1, is involved in axon growth and regenerative sprouting. Nat. Neurosci. 2003;6:572–578. doi: 10.1038/nn1052. [DOI] [PubMed] [Google Scholar]
- 11.Savaskan N. E., Brauer A. U., Nitsch R. Molecular cloning and expression regulation of PRG-3, a new member of the plasticity-related gene family. Eur. J. Neurosci. 2004;19:212–220. doi: 10.1046/j.1460-9568.2003.03078.x. [DOI] [PubMed] [Google Scholar]
- 12.McDermott M. I., Sigal Y. J., Sciorra V. A., Morris A. J. Is PRG-1 a new lipid phosphatase? Nat. Neurosci. 2004;7:789–790. doi: 10.1038/nn0804-789a. [DOI] [PubMed] [Google Scholar]
- 13.Brindley D. N. Lipid phosphate phosphatases and related proteins: signaling functions in development, cell division, and cancer. J. Cell Biochem. 2004;92:900–912. doi: 10.1002/jcb.20126. [DOI] [PubMed] [Google Scholar]
- 14.Waggoner D. W., Gomez-Munoz A., Dewald J., Brindley D. N. Phosphatidate phosphohydrolase catalyzes the hydrolysis of ceramide 1-phosphate, lysophosphatidate, and sphingosine 1-phosphate. J. Biol. Chem. 1996;271:16506–16509. doi: 10.1074/jbc.271.28.16506. [DOI] [PubMed] [Google Scholar]
- 15.Waggoner D. W., Martin A., Dewald J., Gomez-Munoz A., Brindley D. N. Purification and characterization of novel plasma membrane phosphatidate phosphohydrolase from rat liver. J. Biol. Chem. 1995;270:19422–19429. doi: 10.1074/jbc.270.33.19422. [DOI] [PubMed] [Google Scholar]
- 16.Stukey J., Carman G. M. Identification of a novel phosphatase sequence motif. Protein Sci. 1997;6:469–472. doi: 10.1002/pro.5560060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hooks S. B., Ragan S. P., Lynch K. R. Identification of a novel human phosphatidic acid phosphatase type 2 isoform. FEBS Lett. 1998;427:188–192. doi: 10.1016/s0014-5793(98)00421-9. [DOI] [PubMed] [Google Scholar]
- 18.Jasinska R., Zhang Q. X., Pilquil C., Singh I., Xu J., Dewald J., Dillon D. A., Berthiaume L. G., Carman G. M., Waggoner D. W., Brindley D. N. Lipid phosphate phosphohydrolase-1 degrades exogenous glycerolipid and sphingolipid phosphate esters. Biochem. J. 1999;340:677–686. [PMC free article] [PubMed] [Google Scholar]
- 19.Mao C., Wadleigh M., Jenkins G. M., Hannun Y. A., Obeid L. M. Identification and characterization of Saccharomyces cerevisiae dihydrosphingosine-1-phosphate phosphatase. J. Biol. Chem. 1997;272:28690–28694. doi: 10.1074/jbc.272.45.28690. [DOI] [PubMed] [Google Scholar]
- 20.Ogawa C., Kihara A., Gokoh M., Igarashi Y. Identification and Characterization of a novel human sphingosine-1-phosphate phosphohydrolase, hSPP2. J. Biol. Chem. 2003;278:1268–1272. doi: 10.1074/jbc.M209514200. [DOI] [PubMed] [Google Scholar]
- 21.Johnson K. R., Johnson K. Y., Becker K. P., Bielawski J., Mao C., Obeid L. M. Role of human sphingosine-1-phosphate phosphatase 1 in the regulation of intra- and extracellular sphingosine-1-phosphate levels and cell viability. J. Biol. Chem. 2003;278:34541–34547. doi: 10.1074/jbc.M301741200. [DOI] [PubMed] [Google Scholar]
- 22.Sciorra V. A., Morris A. J. Sequential actions of phospholipase D and phosphatidic acid phosphohydrolase 2b generate diglyceride in mammalian cells. Mol. Biol. Cell. 1999;10:3863–3876. doi: 10.1091/mbc.10.11.3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barila D., Plateroti M., Nobili F., Muda A. O., Xie Y., Morimoto T., Perozzi G. The Dri 42 gene, whose expression is up-regulated during epithelial differentiation, encodes a novel endoplasmic reticulum resident transmembrane protein. J. Biol. Chem. 1996;271:29928–29936. doi: 10.1074/jbc.271.47.29928. [DOI] [PubMed] [Google Scholar]
- 24.Kihara A., Sano T., Iwaki S., Igarashi Y. Transmembrane topology of sphingoid long-chain base-1-phosphate phosphatase, Lcb3p. Genes Cells. 2003;8:525–535. doi: 10.1046/j.1365-2443.2003.00653.x. [DOI] [PubMed] [Google Scholar]
- 25.Neuwald A. F. An unexpected structural relationship between integral membrane phosphatases and soluble haloperoxidases. Protein Sci. 1997;6:1764–1767. doi: 10.1002/pro.5560060817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hemrika W., Renirie R., Dekker H. L., Barnett P., Wever R. From phosphatases to vanadium peroxidases: a similar architecture of the active site. Proc. Natl. Acad. Sci. U.S.A. 1997;94:2145–2149. doi: 10.1073/pnas.94.6.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pan C. J., Lei K. J., Annabi B., Hemrika W., Chou J. Y. Transmembrane topology of glucose-6-phosphatase. J. Biol. Chem. 1998;273:6144–6148. doi: 10.1074/jbc.273.11.6144. [DOI] [PubMed] [Google Scholar]
- 28.Renirie R., Hemrika W., Wever R. Peroxidase and phosphatase activity of active-site mutants of vanadium chloroperoxidase from the fungus Curvularia inaequalis. Implications for the catalytic mechanisms. J. Biol. Chem. 2000;275:11650–11657. doi: 10.1074/jbc.275.16.11650. [DOI] [PubMed] [Google Scholar]
- 29.Macedo-Ribeiro S., Hemrika W., Renirie R., Wever R., Messerschmidt A. X-ray crystal structures of active site mutants of the vanadium-containing chloroperoxidase from the fungus Curvularia inaequalis. J. Biol. Inorg. Chem. 1999;4:209–219. doi: 10.1007/s007750050306. [DOI] [PubMed] [Google Scholar]
- 30.Renirie R., Hemrika W., Piersma S. R., Wever R. Cofactor and substrate binding to vanadium chloroperoxidase determined by UV-VIS spectroscopy and evidence for high affinity for pervanadate. Biochemistry. 2000;39:1133–1141. doi: 10.1021/bi9921790. [DOI] [PubMed] [Google Scholar]
- 31.Furneisen J. M., Carman G. M. Enzymological properties of the LPP1-encoded lipid phosphatase from Saccharomyces cerevisiae. Biochim. Biophys. Acta. 2000;1484:71–82. doi: 10.1016/s1388-1981(99)00202-4. [DOI] [PubMed] [Google Scholar]
- 32.Toke D. A., McClintick M. L., Carman G. M. Mutagenesis of the phosphatase sequence motif in diacylglycerol pyrophosphate phosphatase from Saccharomyces cerevisiae. Biochemistry. 1999;38:14606–14613. doi: 10.1021/bi991472x. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Q. X., Pilquil C. S., Dewald J., Berthiaume L. G., Brindley D. N. Identification of structurally important domains of lipid phosphate phosphatase-1: implications for its sites of action. Biochem. J. 2000;345:181–184. [PMC free article] [PubMed] [Google Scholar]
- 34.Dillon D. A., Chen X., Zeimetz G. M., Wu W. I., Waggoner D. W., Dewald J., Brindley D. N., Carman G. M. Mammalian Mg2+-independent phosphatidate phosphatase (PAP2) displays diacylglycerol pyrophosphate phosphatase activity. J. Biol. Chem. 1997;272:10361–10366. doi: 10.1074/jbc.272.16.10361. [DOI] [PubMed] [Google Scholar]
- 35.Roberts R. Z., Morris A. J. Role of phosphatidic acid phosphatase 2a in uptake of extracellular lipid phosphate mediators. Biochim. Biophys. Acta. 2000;1487:33–49. doi: 10.1016/s1388-1981(00)00081-0. [DOI] [PubMed] [Google Scholar]
- 36.Escalante-Alcalde D., Hernandez L., Le Stunff H., Maeda R., Lee H. S., Jr-Gang-Cheng, Sciorra V. A., Daar I., Spiegel S., Morris A. J., Stewart C. L. The lipid phosphatase LPP3 regulates extra-embryonic vasculogenesis and axis patterning. Development. 2003;130:4623–4637. doi: 10.1242/dev.00635. [DOI] [PubMed] [Google Scholar]
- 37.Le Stunff H., Peterson C., Thornton R., Milstien S., Mandala S. M., Spiegel S. Characterization of murine sphingosine-1-phosphate phosphohydrolase. J. Biol. Chem. 2002;277:8920–8927. doi: 10.1074/jbc.M109968200. [DOI] [PubMed] [Google Scholar]
- 38.Nanjundan M., Possmayer F. Pulmonary phosphatidic acid phosphatase and lipid phosphate phosphohydrolase. Am. J. Physiol. Lung Cell Mol. Physiol. 2003;284:L1–L23. doi: 10.1152/ajplung.00029.2002. [DOI] [PubMed] [Google Scholar]
- 39.Alderton F., Darroch P., Sambi B., McKie A., Ahmed I. S., Pyne N., Pyne S. G-protein-coupled receptor stimulation of the p42/p44 mitogen-activated protein kinase pathway is attenuated by lipid phosphate phosphatases 1, 1a, and 2 in human embryonic kidney 293 cells. J. Biol. Chem. 2001;276:13452–13460. doi: 10.1074/jbc.M006582200. [DOI] [PubMed] [Google Scholar]
- 40.Jia Y. J., Kai M., Wada I., Sakane F., Kanoh H. Differential localization of lipid phosphate phosphatases 1 and 3 to cell surface subdomains in polarized MDCK cells. FEBS Lett. 2003;552:240–246. doi: 10.1016/s0014-5793(03)00931-1. [DOI] [PubMed] [Google Scholar]
- 41.Zhao Y., Usatyuk P. V., Cummings R., Saatian B., He D., Watkins T., Morris A., Spannhake E. W., Brindley D. N., Natarajan V. Lipid phosphate phosphatase-1 regulates lysophosphatidic acid-induced calcium release, NF-κB activation and IL-8 secretion in human bronchial epithelial cells. Biochem. J. 2004;385:493–502. doi: 10.1042/BJ20041160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nanjundan M., Possmayer F. Pulmonary lipid phosphate phosphohydrolase in plasma membrane signalling platforms. Biochem. J. 2001;358:637–646. doi: 10.1042/0264-6021:3580637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Czarny M., Lavie Y., Fiucci G., Liscovitch M. Localization of phospholipase D in detergent-insoluble, caveolin-rich membrane domains. Modulation by caveolin-1 expression and caveolin-182-101. J. Biol. Chem. 1999;274:2717–2724. doi: 10.1074/jbc.274.5.2717. [DOI] [PubMed] [Google Scholar]
- 44.Leung D. W., Tompkins C. K., White T. Characterization of two spliced variants of human phosphatidic acid phosphatase cDNAs that are differentially expressed in normal and tumor cells. Adv. Exp. Med. Biol. 1999;469:639–646. doi: 10.1007/978-1-4615-4793-8_92. [DOI] [PubMed] [Google Scholar]
- 45.Leung D. W., Tompkins C. K., White T. Molecular cloning of two alternatively spliced forms of human phosphatidic acid phosphatase cDNAs that are differentially expressed in normal and tumor cells. DNA Cell Biol. 1998;17:377–385. doi: 10.1089/dna.1998.17.377. [DOI] [PubMed] [Google Scholar]
- 46.Yue J., Yokoyama K., Balazs L., Baker D. L., Smalley D., Pilquil C., Brindley D. N., Tigyi G. Mice with transgenic overexpression of lipid phosphate phosphatase-1 display multiple organotypic deficits without alteration in circulating lysophosphatidate level. Cell. Signalling. 2004;16:385–399. doi: 10.1016/j.cellsig.2003.08.012. [DOI] [PubMed] [Google Scholar]
- 47.Alderton F., Sambi B., Tate R., Pyne N. J., Pyne S. Assessment of agonism at G-protein coupled receptors by phosphatidic acid and lysophosphatidic acid in human embryonic kidney 293 cells. Br. J. Pharmacol. 2001;134:6–9. doi: 10.1038/sj.bjp.0704278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smyth S. S., Sciorra V. A., Sigal Y. J., Pamuklar Z., Wang Z., Xu Y., Prestwich G. D., Morris A. J. Lipid phosphate phosphatases regulate lysophosphatidic acid production and signaling in platelets. J. Biol. Chem. 2003;278:43214–43223. doi: 10.1074/jbc.M306709200. [DOI] [PubMed] [Google Scholar]
- 49.Brindley D. N., English D., Pilquil C., Buri K., Ling Z. C. Lipid phosphate phosphatases regulate signal transduction through glycerolipids and sphingolipids. Biochim. Biophys. Acta. 2002;1582:33–44. doi: 10.1016/s1388-1981(02)00135-x. [DOI] [PubMed] [Google Scholar]
- 50.Sciorra V. A., Morris A. J. Roles for lipid phosphate phosphatases in regulation of cellular signaling. Biochim. Biophys. Acta. 2002;1582:45–51. doi: 10.1016/s1388-1981(02)00136-1. [DOI] [PubMed] [Google Scholar]
- 51.Zhang N., Sundberg J. P., Gridley T. Mice mutant for Ppap2c, a homolog of the germ cell migration regulator wunen, are viable and fertile. Genesis. 2000;27:137–140. doi: 10.1002/1526-968x(200008)27:4<137::aid-gene10>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 52.Popperl H., Schmidt C., Wilson V., Hume C. R., Dodd J., Krumlauf R., Beddington R. S. Misexpression of Cwnt8C in the mouse induces an ectopic embryonic axis and causes a truncation of the anterior neuroectoderm. Development. 1997;124:2997–3005. doi: 10.1242/dev.124.15.2997. [DOI] [PubMed] [Google Scholar]
- 53.Zeng L., Fagotto F., Zhang T., Hsu W., Vasicek T. J., Perry W. L., III, Lee J. J., Tilghman S. M., Gumbiner B. M., Costantini F. The mouse Fused locus encodes Axin, an inhibitor of the Wnt signaling pathway that regulates embryonic axis formation. Cell. 1997;90:181–192. doi: 10.1016/s0092-8674(00)80324-4. [DOI] [PubMed] [Google Scholar]
- 54.Kuhl M., Sheldahl L. C., Park M., Miller J. R., Moon R. T. The Wnt/Ca2+ pathway: a new vertebrate Wnt signaling pathway takes shape. Trends Genet. 2000;16:279–283. doi: 10.1016/s0168-9525(00)02028-x. [DOI] [PubMed] [Google Scholar]
- 55.Kuhl M., Sheldahl L. C., Malbon C. C., Moon R. T. Ca2+/calmodulin-dependent protein kinase II is stimulated by Wnt and Frizzled homologs and promotes ventral cell fates in Xenopus. J. Biol. Chem. 2000;275:12701–12711. doi: 10.1074/jbc.275.17.12701. [DOI] [PubMed] [Google Scholar]
- 56.Humtsoe J. O., Feng S., Thakker G. D., Yang J., Hong J., Wary K. K. Regulation of cell–cell interactions by phosphatidic acid phosphatase 2b/VCIP. EMBO J. 2003;22:1539–1554. doi: 10.1093/emboj/cdg165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang N., Zhang J., Purcell K. J., Cheng Y., Howard K. The Drosophila protein Wunen repels migrating germ cells. Nature (London) 1997;385:64–67. doi: 10.1038/385064a0. [DOI] [PubMed] [Google Scholar]
- 58.Zhang N., Zhang J., Cheng Y., Howard K. Identification and genetic analysis of wunen, a gene guiding Drosophila melanogaster germ cell migration. Genetics. 1996;143:1231–1241. doi: 10.1093/genetics/143.3.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Starz-Gaiano M., Cho N. K., Forbes A., Lehmann R. Spatially restricted activity of a Drosophila lipid phosphatase guides migrating germ cells. Development. 2001;128:983–991. doi: 10.1242/dev.128.6.983. [DOI] [PubMed] [Google Scholar]
- 60.Forbes A., Lehmann R. Cell migration in Drosophila. Curr. Opin. Genet. Dev. 1999;9:473–478. doi: 10.1016/s0959-437x(99)80072-0. [DOI] [PubMed] [Google Scholar]
- 61.Starz-Gaiano M., Lehmann R. Moving towards the next generation. Mech. Dev. 2001;105:5–18. doi: 10.1016/s0925-4773(01)00392-6. [DOI] [PubMed] [Google Scholar]
- 62.Burnett C., Howard K. Fly and mammalian lipid phosphate phosphatase isoforms differ in activity both in vitro and in vivo. EMBO Rep. 2003;4:793–799. doi: 10.1038/sj.embor.embor900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Renault A. D., Sigal Y. J., Morris A. J., Lehmann R. Soma-germ line competition for lipid phosphate uptake regulates germ cell migration and survival. Science. 2004;305:1963–1966. doi: 10.1126/science.1102421. [DOI] [PubMed] [Google Scholar]
- 63a.Burnett C., Makridou P., Hewlett L., Howard K. Lipid phosphate phosphatases dimerise, but this interaction is not required for in vivo activity. BMC Biochem. 2004;5:2. doi: 10.1186/1471-2091-5-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Perry D. K., Stevens V. L., Widlanski T. S., Lambeth J. D. A novel ecto-phosphatidic acid phosphohydrolase activity mediates activation of neutrophil superoxide generation by exogenous phosphatidic acid. J. Biol. Chem. 1993;268:25302–25310. [PubMed] [Google Scholar]
- 65.Pagano R. E., Longmuir K. J., Martin O. C., Struck D. K. Metabolism and intracellular localization of a fluorescently labeled intermediate in lipid biosynthesis within cultured fibroblasts. J. Cell Biol. 1981;91:872–877. doi: 10.1083/jcb.91.3.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Renault A. D., Starz-Gaiano M., Lehmann R. Metabolism of sphingosine 1-phosphate and lysophosphatidic acid: a genome wide analysis of gene expression in Drosophila. Gene Expression Patterns. 2002;2:337–345. doi: 10.1016/s0925-4773(02)00389-1. [DOI] [PubMed] [Google Scholar]
- 67.Hla T., Lee M. J., Ancellin N., Paik J. H., Kluk M. J. Lysophospholipids-receptor revelations. Science. 2001;294:1875–1878. doi: 10.1126/science.1065323. [DOI] [PubMed] [Google Scholar]
- 68.Le Stunff H., Peterson C., Liu H., Milstien S., Spiegel S. Sphingosine-1-phosphate and lipid phosphohydrolases. Biochim. Biophys. Acta. 2002;1582:8–17. doi: 10.1016/s1388-1981(02)00132-4. [DOI] [PubMed] [Google Scholar]
- 69.Mao C., Obeid L. M. Yeast sphingosine-1-phosphate phosphatases: assay, expression, deletion, purification, and cellular localization by GFP tagging. Methods Enzymol. 2000;311:223–232. doi: 10.1016/s0076-6879(00)11085-7. [DOI] [PubMed] [Google Scholar]
- 70.Obeid L. M., Okamoto Y., Mao C. Yeast sphingolipids: metabolism and biology. Biochim. Biophys. Acta. 2002;1585:163–171. doi: 10.1016/s1388-1981(02)00337-2. [DOI] [PubMed] [Google Scholar]
- 71.Mao C., Saba J. D., Obeid L. M. The dihydrosphingosine-1-phosphate phosphatases of Saccharomyces cerevisiae are important regulators of cell proliferation and heat stress responses. Biochem. J. 1999;342:667–675. [PMC free article] [PubMed] [Google Scholar]
- 72.Le Stunff H., Galve-Roperh I., Peterson C., Milstien S., Spiegel S. Sphingosine-1-phosphate phosphohydrolase in regulation of sphingolipid metabolism and apoptosis. J. Cell Biol. 2002;158:1039–1049. doi: 10.1083/jcb.200203123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hannun Y. A., Obeid L. M. The ceramide-centric universe of lipid-mediated cell regulation: stress encounters of the lipid kind. J. Biol. Chem. 2002;277:25847–25850. doi: 10.1074/jbc.R200008200. [DOI] [PubMed] [Google Scholar]
- 74.Voelker D. R., Kennedy E. P. Cellular and enzymic synthesis of sphingomyelin. Biochemistry. 1982;21:2753–2759. doi: 10.1021/bi00540a027. [DOI] [PubMed] [Google Scholar]
- 75.Luberto C., Hannun Y. A. Sphingomyelin synthase, a potential regulator of intracellular levels of ceramide and diacylglycerol during SV40 transformation. Does sphingomyelin synthase account for the putative phosphatidylcholine-specific phospholipase C? J. Biol. Chem. 1998;273:14550–14559. doi: 10.1074/jbc.273.23.14550. [DOI] [PubMed] [Google Scholar]
- 76.Nagiec M. M., Nagiec E. E., Baltisberger J. A., Wells G. B., Lester R. L., Dickson R. C. Sphingolipid synthesis as a target for antifungal drugs. Complementation of the inositol phosphorylceramide synthase defect in a mutant strain of Saccharomyces cerevisiae by the AUR1 gene. J. Biol. Chem. 1997;272:9809–9817. doi: 10.1074/jbc.272.15.9809. [DOI] [PubMed] [Google Scholar]
- 77.Dickson R. C., Lester R. L. Metabolism and selected functions of sphingolipids in the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta. 1999;1438:305–321. doi: 10.1016/s1388-1981(99)00068-2. [DOI] [PubMed] [Google Scholar]
- 78.Marggraf W. D., Kanfer J. N. The phosphorylcholine acceptor in the phosphatidylcholine:ceramide cholinephosphotransferase reaction. Is the enzyme a transferase or a hydrolase? Biochim. Biophys. Acta. 1984;793:346–353. doi: 10.1016/0005-2760(84)90248-0. [DOI] [PubMed] [Google Scholar]
- 79.Yamaoka S., Miyaji M., Kitano T., Umehara H., Okazaki T. Expression cloning of a human cDNA restoring sphingomyelin synthesis and cell growth in sphingomyelin synthase-defective lymphoid cells. J. Biol. Chem. 2004;279:18688–18693. doi: 10.1074/jbc.M401205200. [DOI] [PubMed] [Google Scholar]
- 80.Futerman A. H., Stieger B., Hubbard A. L., Pagano R. E. Sphingomyelin synthesis in rat liver occurs predominantly at the cis and medial cisternae of the Golgi apparatus. J. Biol. Chem. 1990;265:8650–8657. [PubMed] [Google Scholar]
- 81.van Helvoort A., van't Hof W., Ritsema T., Sandra A., van Meer G. Conversion of diacylglycerol to phosphatidylcholine on the basolateral surface of epithelial (Madin–Darby canine kidney) cells. Evidence for the reverse action of a sphingomyelin synthase. J. Biol. Chem. 1994;269:1763–1769. [PubMed] [Google Scholar]
- 82.Avraham S., Stevens R. L., Gartner M. C., Austen K. F., Lalley P. A., Weis J. H. Isolation of a cDNA that encodes the peptide core of the secretory granule proteoglycan of rat basophilic leukemia-1 cells and assessment of its homology to the human analogue. J. Biol. Chem. 1988;263:7292–7296. [PubMed] [Google Scholar]
- 83.Rechsteiner M., Rogers S. W. PEST sequences and regulation by proteolysis. Trends Biochem. Sci. 1996;21:267–271. [PubMed] [Google Scholar]
- 84.Ishii I., Fukushima N., Ye X., Chun J. Lysophospholipid receptors: signaling and biology. Annu. Rev. Biochem. 2004;73:321–354. doi: 10.1146/annurev.biochem.73.011303.073731. [DOI] [PubMed] [Google Scholar]
- 85.Ye X., Fukushima N., Kingsbury M. A., Chun J. Lysophosphatidic acid in neural signaling. NeuroReport. 2002;13:2169–2175. doi: 10.1097/00001756-200212030-00002. [DOI] [PubMed] [Google Scholar]
- 86.Todd A. E., Orengo C. A., Thornton J. M. Sequence and structural differences between enzyme and nonenzyme homologs. Structure. 2002;10:1435–1451. doi: 10.1016/s0969-2126(02)00861-4. [DOI] [PubMed] [Google Scholar]
- 87.Nandurkar H. H., Caldwell K. K., Whisstock J. C., Layton M. J., Gaudet E. A., Norris F. A., Majerus P. W., Mitchell C. A. Characterization of an adapter subunit to a phosphatidylinositol (3)P 3-phosphatase: identification of a myotubularin-related protein lacking catalytic activity. Proc. Natl. Acad. Sci. U.S.A. 2001;98:9499–9504. doi: 10.1073/pnas.171306098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Luquain C., Sciorra V. A., Morris A. J. Lysophosphatidic acid signaling: how a small lipid does big things. Trends Biochem. Sci. 2003;28:377–383. doi: 10.1016/S0968-0004(03)00139-7. [DOI] [PubMed] [Google Scholar]
- 89.Stroud R. M., Miercke L. J., O'Connell J., Khademi S., Lee J. K., Remis J., Harries W., Robles Y., Akhavan D. Glycerol facilitator GlpF and the associated aquaporin family of channels. Curr. Opin. Struct. Biol. 2003;13:424–431. doi: 10.1016/s0959-440x(03)00114-3. [DOI] [PubMed] [Google Scholar]
- 90.Schirmer E. C., Florens L., Guan T., Yates J. R., III, Gerace L. Nuclear membrane proteins with potential disease links found by subtractive proteomics. Science. 2003;301:1380–1382. doi: 10.1126/science.1088176. [DOI] [PubMed] [Google Scholar]
- 91.D'santos C. S., Clarke J. H., Irvine R. F., Divecha N. Nuclei contain two differentially regulated pools of diacylglycerol. Curr. Biol. 1999;9:437–440. doi: 10.1016/s0960-9822(99)80193-6. [DOI] [PubMed] [Google Scholar]
- 92.Vann L. R., Wooding F. B., Irvine R. F., Divecha N. Metabolism and possible compartmentalization of inositol lipids in isolated rat-liver nuclei. Biochem. J. 1997;327:569–576. doi: 10.1042/bj3270569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Irvine R. F. Nuclear lipid signalling. Nat. Rev. Mol. Cell Biol. 2003;4:349–360. doi: 10.1038/nrm1100. [DOI] [PubMed] [Google Scholar]
- 94.Page R. D. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]