Summary
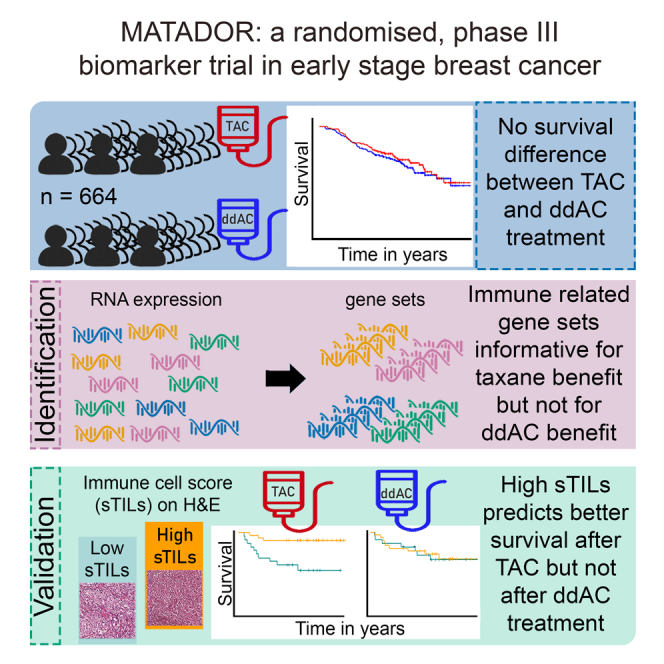
The primary objective of the prospective, randomized, multicenter, phase 3 biomarker Microarray Analysis in breast cancer to Taylor Adjuvant Drugs Or Regimens trial (MATADOR: ISRCTN61893718) is to generate a gene expression profile that can predict benefit from either docetaxel, doxorubicin, and cyclophosphamide (TAC) or dose-dense scheduled doxorubicin and cyclophosphamide (ddAC). Patients with a pT1-3, pN0-3 tumor were randomized 1:1 between ddAC and TAC. The primary endpoint was a gene profile-treatment interaction for recurrence-free survival (RFS). We observed 117 RFS events in 664 patients with a median follow-up of 7 years. Hallmark gene set analyses showed significant association between enrichment in immune-related gene expression and favorable outcome after TAC in hormone receptor-negative, human epidermal growth factor receptor 2 (HER2)-negative breast cancer (BC) (triple-negative breast cancer [TNBC]). We validated this association in TNBC patients treated with TAC on H&E slides; stromal tumor-infiltrating lymphocytes (sTILs) ≥20% was associated with longer RFS (hazard ratio 0.18, p = 0.01), while in patients treated with ddAC no difference in RFS was seen (hazard ratio 0.92, p = 0.86, pinteraction = 0.02).
Subject areas: Cancer, Clinical genetics, Oncology
Graphical abstract
Highlights
-
•
This proof-of-concept randomized biomarker trial design demonstrates scientific utility
-
•
High sTILs can guide chemotherapy choice in high-risk triple-negative breast cancer
-
•
Gene expression analyses identify biological processes informative for taxane benefit
-
•
Immune processes interact differentially with outcome after TAC vs. ddAC chemotherapy
Cancer; Clinical genetics; Oncology
Introduction
For patients with early-stage breast cancer, both the addition of taxanes and dose-dense scheduling of adjuvant chemotherapy reduced the risk of early breast cancer relapse and death.1,2 The eight-year survival improves from 83.3% to 86.5% with the addition of taxanes to a (neo)adjuvant anthracycline-based chemotherapy. Current state-of-the-art medicine is mainly based on results obtained from randomized-controlled trials in all comers, and predictive biomarkers are generally not the primary objective. Therefore, we can only determine the best treatment for the whole group of breast cancer patients, while it remains unclear whether subgroups of patients exist that would benefit more from the experimental or control treatment in the setting of a randomized clinical trial. In order to discover a biomarker, we hypothesize that the following clinical trial may have a higher likelihood to yield a clinically useful test: (1) randomize patients between two regimens that have been found to be equally effective based on meta-analysis,3 (2) a prespecified plan of in-depth characterization of the tumor of each patient enrolled, and (3) in a prespecified training sample, use this information to determine whether subgroups exist that would benefit significantly more from one particular adjuvant chemotherapy regimen and validate findings in a test set within the same trial. Several gene expression profiles that might predict sensitivity to chemotherapy have been identified.4,5,6,7,8,9 However, early investigations were based on single-arm studies resulting in a profile predictive of response to that particular treatment, without information on what the benefit to an alternative regimen could be. Such studies produce gene expression profiles that can have both prognostic as well as predictive value. More recent studies were well designed but did not yield definitive results.9,10 A neo-adjuvant design accelerates assessment of a biomarker/treatment interaction by using pathological complete remission (pCR) as a surrogate endpoint, especially in an adaptive clinical trial design like the I-SPY (Investigation of Serial studies to Predict Your Therapeutic Response with Imaging and Molecular AnaLysis) studies.11 The disadvantage of pCR as endpoint, however, is that it cannot reliably be used to read out improvements in long-term recurrence-free survival when comparing novel treatments to the standard of care.12
In the randomized phase 3 MATADOR study we tested, as primary objective, the hypothesis that a gene expression profile can be defined that predicts advantage for either docetaxel, doxorubicin, and cyclophosphamide (TAC) or dose-dense scheduled doxorubicin and cyclophosphamide (ddAC) in early-stage breast cancer. Such a profile enables us to predict which of the treatment regimens, which are equally effective in the whole study population,13 will most benefit subgroups of patients. To our knowledge, this is the first trial with a primary objective testing whether a gene expression profile can be generated that estimates the recurrence-free survival (RFS) benefit of one chemotherapy regimen over another regimen in subgroups of patients, while both regimens are considered equally effective in the whole patient group.
Results
Between August 2004 and November 2012, 664 patients were enrolled (Figure S1). Sixty-five patients had a clinically low risk of recurrence according to Adjuvant! Online14 and would not receive adjuvant chemotherapy according to current guidelines. Since for these patients there is no need to develop a predictive test, these patients were excluded from the analyses. Furthermore, patients were excluded because of not being treated according to protocol (n = 18), ineligibility to be included in the study (n = 3), or no follow-up data (n = 1); more details are reported by Van Rossum.13 For the analysis of the primary objective, 81 samples failed the RNA quality control and are left out (Mendeley bioinformatics file section 1.5).15 In the whole cohort, we observed 117 RFS events with a median follow-up of 7 years (Table S1). Treatment groups were not significantly different regarding clinicopathologic characteristics (shown before).13
No predictive biomarker with gene expression profile using single genes
Unsupervised hierarchical clustering grouped patients showed a correlation between Prediction Analysis of Microarray 50 (PAM50) and immunohistochemistry (IHC) subtypes (Figure S2, Mendeley bioinformatics file section 1.7).15,16 The association between the PAM50 classification and RFS is shown in Figure S3. As expected, in the first years after diagnosis, the RFS events occurred mostly in the basal-like breast cancers and the human epidermal growth factor receptor 2 (HER2)-enriched subgroups, while the events in the luminal A and the luminal B subgroup were more or less constant over time (Figure S3). We found that expression levels of individual genes displayed interaction with treatment in a way that seems distinct from mere noise for triple-negative breast cancer TNBC (Mendeley bioinformatics file section 2.3).15 However, because of multiple testing we cannot conclude that the results are statistically significant.
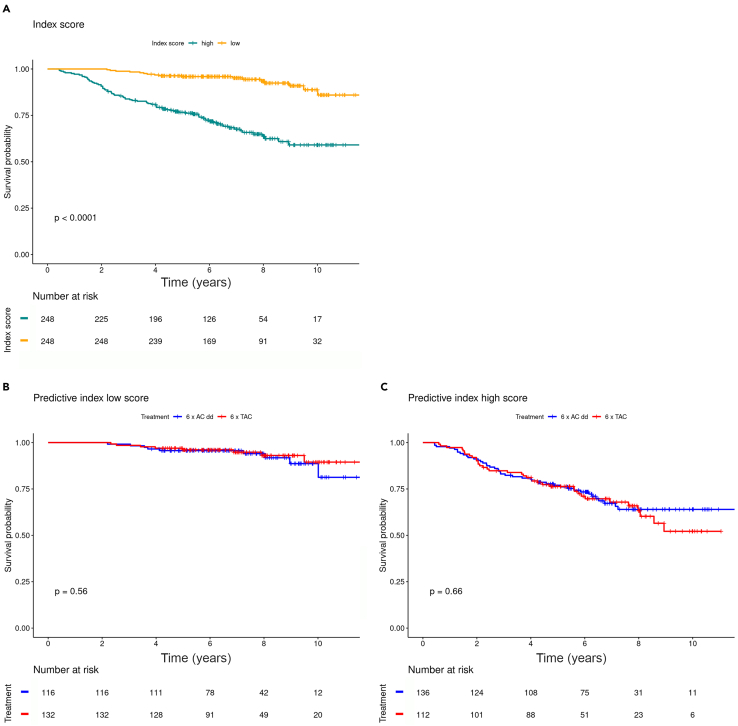
We therefore decided to fit a Least Absolute Shrinkage and Selection Operator (LASSO) regression model using all genes to generate a profile index score (Mendeley bioinformatics file section 3).15 Besides the gene expression, the model was fitted using the treatment effect and the interactions of each gene’s expression and treatment as explanatory variables. Each patient was classified below or above the median profile index score. This binary profile index score had a significant association with grade, subtype (Table 1), and RFS (adjusted hazard ratio 5.64, 95% confidence interval [CI] 3.22–9.86, p < 0.001), with a significantly longer RFS for patients with a low-profile index score compared with those with a high-profile index score (Figure 1A). When considering the low- and high-profile index score groups per treatment arm, no significant association with RFS was observed, indicating that the profile index score had no predictive value for outcome after ddAC or TAC (Figures 1B and 1C). The genes that were included in this profile are listed in Table S2.
Table 1.
Characteristics of patients for stratified profile index score
| Variable | Profile index score | Low | n = 248 | High | n = 248 | p valuea |
|---|---|---|---|---|---|---|
| Treatment | ddAC | 116 | (47%) | 136 | (55%) | 0.09 |
| TAC | 132 | (53%) | 112 | (45%) | ||
| Age groups | <50 years | 119 | (48%) | 105 | (42%) | 0.24 |
| ≥50 years | 129 | (52%) | 143 | (58%) | ||
| Surgery | breast | 128 | (52%) | 135 | (54%) | 0.59 |
| mastectomy | 120 | (48%) | 113 | (46%) | ||
| T stage | T1 | 114 | (46%) | 103 | (42%) | 0.25 |
| T2 | 124 | (50%) | 124 | (50%) | ||
| T3 | 9 | (4%) | 19 | (8%) | ||
| T4 | 1 | (0.4%) | 1 | (0.4%) | ||
| N stage | N0 | 37 | (15%) | 42 | (17%) | 0.33 |
| N1 | 165 | (67%) | 154 | (62%) | ||
| N2 | 38 | (15%) | 36 | (15%) | ||
| N3 | 8 | (3%) | 16 | (6%) | ||
| Grade | good | 19 | (8%) | 4 | (2%) | <0.001 |
| intermediate | 135 | (57%) | 88 | (37%) | ||
| poor | 82 | (35%) | 145 | (61%) | ||
| Histology | ductal | 196 | (80%) | 208 | (85%) | 0.31 |
| lobular | 41 | (17%) | 31 | (13%) | ||
| other | 9 | (4%) | 6 | (2%) | ||
| Subtype | HR-positive HER2-negative | 231 | (93%) | 171 | (69%) | <0.001 |
| HER2-positive | 1 | (0.4%) | 14 | (6%) | ||
| Triple negative | 16 | (6%) | 63 | (25%) |
A, doxorubicin; C, cyclophosphamide; T, docetaxel; dd, dose-dense.
Pearson chi-squared test, missing values excluded; subtypes were defined as (1) hormone receptor (HR)-positive, human epidermal growth factor receptor 2 (HER2)-negative; (2) HER2-positive, regardless of ER or PR status; (3) triple (ER, PR, HER2) negative.
Figure 1.
Association between validation profile index score and recurrence-free survival
(A–C) Profile obtained using a model with LASSO penalty17 on 496 patients with high quality of RNA reads (A). Low-profiled index score stratified for treatment (B). High-profile index score stratified for treatment (C). p value of the log rank test is shown.
Hallmark gene sets appear predictive in taxane-treated subgroup
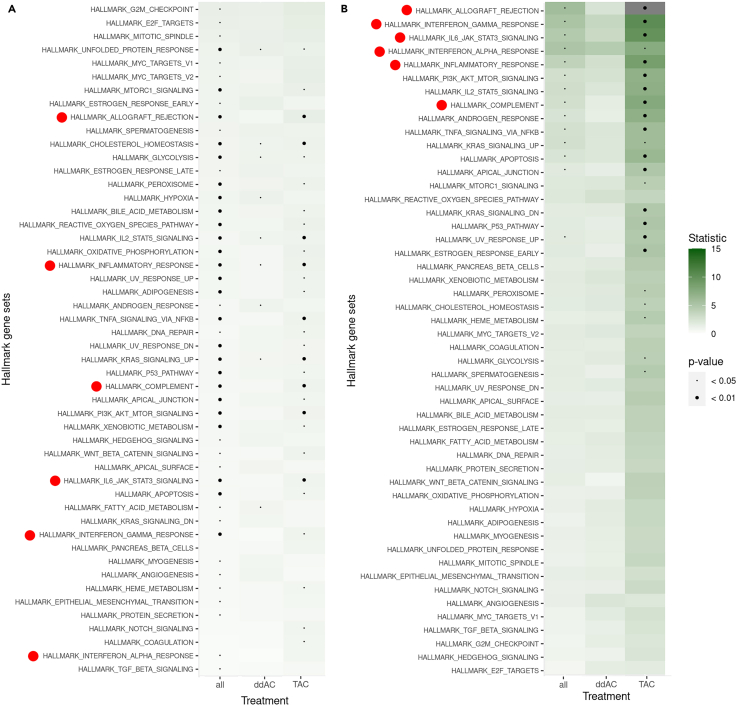
Next, we tested the associations between well-described biological processes represented in Hallmark gene sets18 and RFS (Figure 2 and Mendeley bioinformatics file section 4).15,19 Whereas none of the gene sets had a significant association with RFS in the ddAC-treated patients, 13 gene sets were significantly associated with RFS in the TAC-treated subgroup (Mendeley bioinformatics file section 4.1).15 In the subgroup of TNBC, we observed a more profound difference in the associations with RFS between the treatment arms. Whereas no gene sets were significantly associated with RFS in the ddAC-treated TNBC, 16 gene sets had a significant association with RFS in the TAC-treated subgroup (Mendeley bioinformatics file section 4.3).15 Interestingly, high expression of immune-related gene sets was associated with favorable outcome in the TAC-treated subgroup, while this was not observed in ddAC-treated patients (Figure 2B).
Figure 2.
Strength of associations of Hallmark gene sets with recurrence-free survival
(A and B) Shown for all patients and split by treatment arm (A), and in triple-negative breast cancer only (B). The gene sets are ordered according to Goeman’s globaltest statistic,19 and p values are represented by the size of the black dot. Immune-related processes are depicted by a red dot. A, doxorubicin; C, cyclophosphamide; T, docetaxel; dd, dose-dense.
The associations between the individual genes of the top 3 immune gene sets (allograft rejection, interferon gamma response, and interleukin-6 (IL-6) JAK STAT3 signaling) and RFS split by treatment subgroup are given in Table S3 and Mendeley bioinformatics file section 5.15
Tumor-infiltrating lymphocytes as validation
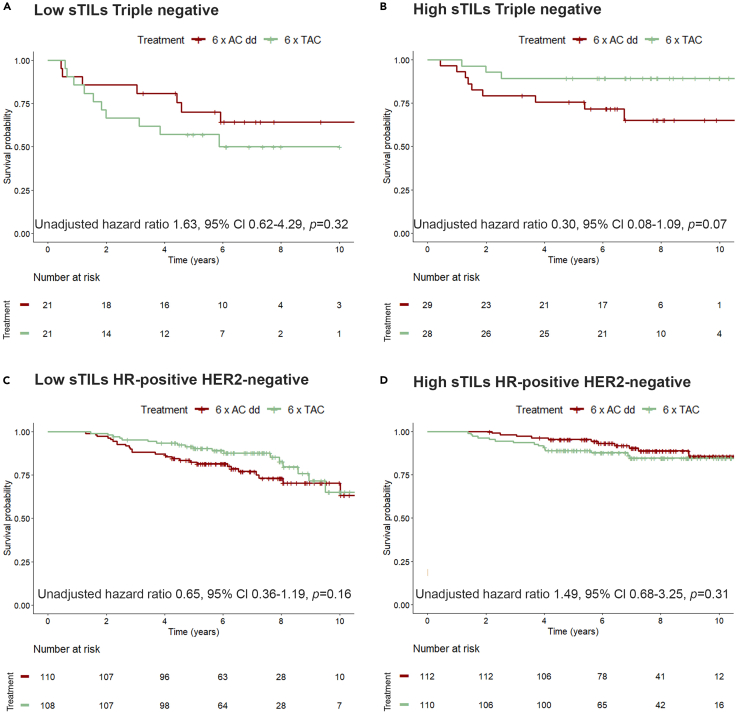
A standardized scoring method for stromal tumor-infiltrating lymphocytes (sTILs) based on H&E slides has been developed that is close to clinical application in TNBC, making it an ideal candidate biomarker to assess endogenous immune responses in breast cancer.20 We used standardized sTILs scoring to assess whether we could validate the association between immune-related gene sets and RFS in relation to chemotherapeutic regimen received in 455 patients with hormone receptor(HR)-positive human epidermal growth factor receptor 2 (HER2)-negative and 99 patients with TNBC (Figure S1). When considering the sTILs percentage as continuous variable in all clinical high-risk patients, a significant interaction between sTILs and treatment is observed (Table S4). When the TNBCs are analyzed separately, this resulted in a median sTILs score of 20% (interquartile range [IQR] 10–50) (Table 2). Patients were divided in two groups according to the median: low sTILs (<20%) and high sTILs (≥20%). Abundance of sTILs was significantly associated with RFS in TNBC patients treated with TAC (hazard ratio 0.18, 95% CI 0.05–0.64, p = 0.01), but not in patients treated with ddAC (hazard ratio 0.92, 95% CI 0.34–2.46, p = 0.86; Figures S4A and S4B), with significant interaction between sTILs and treatment (adjusted pinteraction = 0.02; Table 3; Figures 3A and 3B). For the HR-positive HER2-negative subgroup, the median sTILs was 10% (IQR 5–15; Table 2). In this subgroup sTILs were correlated with improved RFS in the ddAC-treated patients (hazard ratio 0.34, 95% CI 0.20–0.77, p < 0.01), but not in the TAC-treated patients (hazard ratio 0.77, 95% CI 0.39–1.56 p = 0.46; Figures S4C and S4D) and there was no significant interaction between sTILs and treatment (Figures 3C and 3D; Table 4).
Table 2.
Characteristics of clinical high-risk patients for whom stromal tumor-infiltrating lymphocytes (sTILs) score was available, shown for patients with triple-negative breast cancer and with hormone receptor (HR)-positive human epidermal growth factor receptor 2 (HER2)-negative, stratified in low and high sTILs score
| Variable | TNBC | n = 99 | HR-positive HER2-negative | n = 455 | |
|---|---|---|---|---|---|
| sTILs | median [Q1–Q3] | 20 [10–50] | 10 [5–15] | ||
| sTILs | sTILs | ||||
| <20% | ≥ 20% | <10% | ≥ 10% | ||
| Treatment | ddAC | 21 (50%) | 29 (51%) | 110 (50%) | 112 (50%) |
| TAC | 21 (50%) | 28 (49%) | 108 (50%) | 110 (50%) | |
| Age groups | <50 years | 24 (57%) | 27 (47%) | 83 (53%) | 108 (49%) |
| ≥50 years | 18 (43%) | 30 (53%) | 135 (47%) | 114 (51%) | |
| Surgery | breast-conserving surgery | 18 (43%) | 38 (67%) | 116 (46%) | 110 (50%) |
| mastectomy | 24 (57%) | 19 (33%) | 102 (54%) | 112 (50%) | |
| T stage | T1 | 17 (40%) | 24 (42%) | 94 (43%) | 95 (43%) |
| T2-4 | 25 (60%) | 33 (58%) | 123 (57%) | 127 (57%) | |
| N stage | N0 | 11 (26%) | 18 (32%) | 29 (13%) | 27 (12%) |
| N+ | 31 (74%) | 39 (68%) | 189 (87%) | 195 (88%) | |
| Grade | good | 0 (0%) | 1 (2%) | 16 (8%) | 10 (5%) |
| intermediate | 3 (7%) | 4 (7%) | 125 (62%) | 112 (53%) | |
| poor | 37 (93%) | 52 (91%) | 62 (31%) | 90 (43%) | |
| Histology | ductal | 38 (95%) | 50 (91%) | 168 (77%) | 172 (78%) |
| lobular | 1 (2%) | 0 (0%) | 42 (19%) | 41 (19%) | |
| other | 1 (2%) | 5 (9%) | 7 (3%) | 7 (3%) |
Cutoff defined at the median of each subtype. A, doxorubicin; C, cyclophosphamide; T, docetaxel; dd, dose-dense.
Table 3.
Multivariate Cox regression model of the association between stromal tumor-infiltrating lymphocytes (sTILs) in an interaction with treatment and recurrence-free survival (RFS) in clinically high-risk triple-negative patients
| Variable | n | adjusted hazard ratio | 95% CI | p value | |
|---|---|---|---|---|---|
| Treatment | ddAC | 49 | reference | ||
| TAC | 48 | 0.77 | 0.36–1.64 | 0.49 | |
| Age | <50 years | 49 | reference | ||
| ≥50 years | 48 | 1.07 | 0.50–2.30 | 0.85 | |
| T stage | T1 | 39 | reference | ||
| T2-4 | 58 | 1.29 | 0.60–2.77 | 0.51 | |
| N stage | N0 | 28 | reference | ||
| N+ | 69 | 14.16 | 1.91–105.0 | 0.009 | |
| Histologic grade | good/intermediate | 8 | reference | ||
| poor | 89 | 1.78 | 0.41–7.66 | 0.44 | |
| Type of surgery | breast-conserving surgery | 56 | reference | ||
| mastectomy | 41 | 0.87 | 0.41–1.87 | 0.72 | |
| sTILs | <20% | 40 | reference | ||
| ≥20% | 57 | 0.46 | 0.21–0.98 | 0.04 | |
| sTILs ∗ treatment | 0.14 | 0.03–0.74 | 0.02 |
Test for interaction between sTILs and treatment is shown in the last row. A, doxorubicin; C, cyclophosphamide; T, docetaxel; dd, dose-dense; sTILs, stromal tumor-infiltrating lymphocytes; CI, confidence interval.
Figure 3.
Recurrence-free survival stratified by treatment, shown for patients with low and high number of stromal tumor-infiltrating lymphocytes (sTILs)
(A–D) Kaplan-Meier curve for clinical high-risk patients. Triple-negative breast cancer and sTILs abundance labeled as low (<20%; A) and high (≥20%; B). Test for interaction (p 0.02) is calculated in Table 3. Patients with hormone receptor (HR)-positive human epidermal growth factor receptor 2 (HER2)-negative (HR+HER2−)tumors, low sTILs <10%: C), and high sTILs (≥10%; D). Test for interaction (p 0.16) is calculated in Table 4. Unadjusted cox proportional hazard ratio with a 95% confidence interval (CI) and corresponding p value is shown. A, doxorubicin; C, cyclophosphamide; T, docetaxel; dd, dose-dense.
Table 4.
Multivariate Cox regression model of the association between stromal tumor-infiltrating lymphocytes (sTILs; cut-off at 10%) in an interaction with treatment and recurrence-free survival (RFS) in clinically high-risk HR-positive HER2-negative patients
| Variable | n | adjusted hazard ratio | 95% CI | p value | |
|---|---|---|---|---|---|
| Treatment | ddAC | 216 | reference | ||
| TAC | 203 | 0.88 | 0.54–1.43 | 0.61 | |
| Age | <50 years | 179 | reference | ||
| ≥50 years | 240 | 1.11 | 0.67–1.84 | 0.68 | |
| T stage | T1 | 182 | reference | ||
| T2-4 | 237 | 2.13 | 1.24–3.66 | 0.006 | |
| N stage | N0 | 52 | reference | ||
| N+ | 367 | 2.12 | 0.76–5.92 | 0.15 | |
| Histologic grade | good/intermediate | 266 | reference | ||
| poor | 153 | 1.70 | 1.04–2.79 | 0.03 | |
| Type of surgery | breast-conserving surgery | 216 | reference | ||
| mastectomy | 203 | 0.75 | 0.46–1.24 | 0.26 | |
| sTILs | <10% | 205 | reference | ||
| ≥10% | 214 | 0.47 | 0.28–0.78 | 0.003 | |
| sTILs ∗ treatment | 2.08 | 0.76–5.71 | 0.16 |
A, doxorubicin; C, cyclophosphamide; T, docetaxel; dd, dose-dense; sTILs, stromal tumor-infiltrating lymphocytes; CI, confidence interval.
Discussion
The primary objective of the randomized, phase 3 MATADOR study was to identify a gene expression profile that can predict RFS benefit for one of the treatments based on the hypothesis that, while on average RFS is similar after ddAC or TAC, some patients derive more benefit from ddAC and others from TAC. The primary endpoint was not met, since the objective was to define a gene expression profile using single genes. Interestingly, when using well-defined gene sets, we were able to reveal an enrichment in immune-related processes that predicted differential benefit from TAC versus ddAC, which was restricted to the TNBC subtype. Hence, co-development of a biomarker during a randomized clinical trial using tumor RNA sequencing (RNA-seq) data in combination with a gene sets-centered bioinformatics approach seems possible. Such an approach may be considered in future registration studies to help restrict the indication to a subset of patients that derives substantial benefit from the novel agent. In addition, it might even “rescue” randomized clinical trials that turn out negative for the primary objective of superiority.
Here, our findings suggest that TNBC tumors with a stronger endogenous immune response are more susceptible to the addition of a taxane. This finding is supported by data from a mouse model for TNBC where docetaxel was able to deplete myeloid-derived immune-suppressive cells, a subset of sTILs, in a specific manner.21 Also, high expression of immune-related genes has been linked to high likelihood of achieving a pCR in women with TNBC treated with taxane-containing regimens.22,23 In addition, an 8-gene tumor-infiltrating lymphocyte signature has been described that suggested a similar relationship for anthracycline-based schedules,24 although the authors wrote that they could not exclude a role for other aspects of the miscellaneous regimens studied (i.e., other agents like taxanes and timing of administration). Many preclinical studies have addressed the interaction of chemotherapeutics with the immune system (reviewed in Galluzzi, Cancer Cell 2015; Coffelt, Trends Immunol 2015; Fridman, Nat Rev Clin Oncol. 2017).25,26,27 Recently, preclinical data have revealed a novel T lymphocyte-mediated, T cell receptor-independent mode of action of taxanes important for cancer cell killing.28 Further functional studies in model systems are needed to dissect the differential effects of chemotherapy combinations, also addressing timing of administration, on various components of the immune system. Breast cancer subtype should also be considered in these experiments.26,29 Wimmer et al. nicely show that the combination of epirubicin and cyclophosphamide reduces the number of circulating B and T cells, while no effect on immune cells is seen after docetaxel treatment in a selection of patients of the ABCSG-34 trial. This reduction in circulating B and T cells aligns with our results that the putative favorable prognostic effect of high sTILs in TNBC is negated by 6 cycles of ddAC.30
Importantly, a simple H&E-based score of the immune infiltrate confirmed the signal from gene expression data. In patients with TNBC, high abundance of sTILs favored TAC over ddAC, while better outcome after ddAC was observed in the low sTILs group, with a significant interaction between abundance of sTILs and treatment. These results are in line with previous reports on sTILs, overexpression of immune-response genes and pCR after taxane-containing chemotherapy,31,32,33 and a recent report on the predictive value of high tumor expression of immune-related genes and improved disease-free and overall survival after dose-dense, taxane-based chemotherapy when compared to standard chemotherapy without taxanes.34 Remarkably, Yam et al. found the same Hallmark immune-associated pathways that were associated with higher T cell activity resulting in a high chance of achieving a pCR on AC-taxane-based chemotherapy (allograft rejection, interferon gamma, and interferon alpha).35 However, in two other studies no significant interaction was observed between sTILs and adjuvant anthracycline-based or anthracycline/docetaxel-containing chemotherapy for (invasive) disease-free survival in estrogen receptor (ER)-negative, HER2-negative breast cancer patients.36,37 This may be explained by a substantial difference in chemotherapy schedules including cumulative doses of anthracyclines and docetaxel administered. Taking together all the evidence, low sTILs deserves further study as a relatively cheap and simple biomarker to select especially TNBC patients who can forego a taxane and receive ddAC only, thereby reducing the risk of peripheral neuropathy, neutropenia, muscle and joint pain, mucositis, and rash.38 On the other hand, these data raise the question whether TNBC patients selected on the basis of high sTILs, eventually complemented by T cell-to-macrophage ratio and tumor cell-T lymphocyte distance,35 could forego doxorubicin/epirubicin and only receive a taxane, eventually combined with cyclophosphamide or carboplatin.39 This hypothesis requires further studies.
In the HR-positive HER2-negative breast cancer subgroup, the median sTILs abundance was lower than that in the TNBC subgroup, which is in accordance with the current literature.40 Interestingly, higher sTILs (≥10%) in HR-positive HER2-negative breast cancer patients were associated with an improved outcome. This signal was more pronounced in patients treated with ddAC than in patients treated with TAC. The test for interaction however was not significant. It should be noted that, in the MATADOR study analysis plan, only clinically high-risk HR-positive HER2-negative breast cancer patients are considered. This patient selection might drive the finding here that sTILs ≥10% have a better RFS compared to patients with sTILs<10%.40,41
To identify a predictive gene expression profile by utilizing the expression levels of thousands of single genes in a dataset with 117 events results in statistically high penalties precluding success. Although previous groups could define a putative predictive profile,4,5,6,7,8,9 reports on the validation of these classifications are lacking, indicating the difficulty of generating a robust predictive gene expression-based classification using single genes. A biological explanation for the lack of success is the presence of a variety of resistance mechanisms in the tumors analyzed in these studies.42 If a resistance mechanism is not shared by a large fraction of the tumors, finding a predictive gene expression profile will be complicated. Also, the heterogeneity of the subclones within a tumor might influence the process.43
Strengths of our phase 3 study include the randomized setup, with finding a predictive gene expression profile as the primary objective, choosing between two regimens that are equally effective for all patients. An additional strength is that we could recapitulate the regimen-specific transcriptomic chemo-immune interactions on the morphological level (sTILs).
Limitations of the study
Although in line with the high-risk patients of the Microarray In Node-negative and 1 to 3 positive lymph node Disease may Avoid ChemoTherapy (MINDACT) study,14 the survival of our patients was better than anticipated at the start of the trial, which compromised power due to a lower number of events. In addition, when this trial was started in 2004, both treatments arms were deemed suitable for all included patients. Nowadays both treatment regimens have largely been replaced by newer schedules. During the trial it became clear that HER2-positive patients required anti-HER2 directed therapy as an add-on and should not have been part of the study population. During follow-up, MINDACT taught that clinically low-risk patients are overtreated with chemotherapy and hence should not have been part of the study population.14 Nowadays, we know that HR-positiveHER2-negative and TNBC are biologically different subtypes and should be studied separately. These factors partially explain why finding a gene expression profile predictive for differential outcome after ddAC or TAC of certain patients within the total study population was not successful. By stratifying for subtype, we try to overcome part of this limitation. Finally, bulk RNA derived from the tumor will only reflect the most prevalent tumor cell types. Further research will elucidate whether disentangling the bulk signal into contributions of individual cellular components derived from single-cell RNA-seq can have predictive value.44
Conclusion
Using whole-transcriptome RNA-seq data, we failed to meet the primary endpoint of our randomized biomarker study. However, analyses using well-established gene sets revealed immune-related processes as important predictors of RFS after either ddAC or TAC, especially in patients with TNBC. Furthermore, high abundance of sTILs appeared to be a significant predictor of RFS benefit from docetaxel-based adjuvant chemotherapy in TNBC. If the taxane interaction with immune cells can be validated in an independent cohort, the abundance of sTILs in the primary tumor may help us to further personalize adjuvant chemotherapy, especially in patients with TNBC. Notably, our prespecified analyses revealed a biomarker for which clinical implementation could be relatively easy (H&E slide), highlighting the value of the concept of our biomarker trial that can serve as a template for future randomized clinical trials with biomarker co-development in the oncology field and beyond.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| ER (SP1) | Roche/Ventana | RRID:AB_2857956 |
| PR (1E2) | Roche/Ventana | 5277990001 |
| HER2 (4B5) | Roche/Ventana | Roche RRID:AB_2921204 |
| HER2 DNA Probe | Roche/Ventana | 5273439001 |
| Deposited data | ||
| Clinical data per patient | Mendeley data | Opdam et al.15 |
| RNA-sequencing data | Gene Expression Omnibus | https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE167977 |
| Software and algorithms | ||
| ALEA system | FormsVision | https://www.aleaclinical.eu/ |
| R version 4.3.1 | Comprehensive R Archive Network | https://cran.r-project.org/bin/windows/base/old/ |
| globaltest version 5.54.0 | Goeman JJ | https://www.bioconductor.org/packages/release/bioc/html/globaltest.html |
| edgeR 3.42.2 | Robinson MD | https://bioconductor.org/packages/release/bioc/html/edgeR.html |
| glmnet version 4.1-717 | Friedman J | https://cran.r-project.org/web/packages/glmnet/index.html |
| survminer version 0.4.945 | Kassambara A | https://www.rdocumentation.org/packages/survminer/versions/0.4.9 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Sabine Linn (s.linn@nki.nl).
Materials availability
This study did not generate new unique reagents.
Data and code availability
Anonymised patient data that support the finding of this study is shared in a Mendeley repository and RNA sequencing data on GEO (GSE167977).15 In the Mendeley bioinformatics file the code for the quality control steps and gene expression profiling is described in detail.15 Any additional information required to reanalyse the data reported in this work paper is available from the lead contact upon request.
Experimental model and study participant details
The MATADOR (Microarray Analysis in breast cancer to Tailor Adjuvant Drugs Or Regimens, ISRCTN61893718) study is an open-label, randomized clinical trial. Six hundred sixty-four female patients (average age 51) with pT1-3, N0-3, M0 breast cancer were recruited onto the trial. The inclusion criteria were described in detail elsewhere.13,46
Method details
Study design
The MATADOR study is an open-label, randomised clinical trial conducted in 29 centres in the Netherlands. The study protocol and amendments were approved by the ethical committee of the Netherlands Cancer Institute.15 At trial start, trastuzumab was not part of standard adjuvant treatment for patients with HER2-positive breast cancer yet. Therefore, these patients were initially enrolled in the MATADOR study. With emerging evidence that trastuzumab, especially when given concurrently with chemotherapy, improved survival in HER2-positive breast cancer patients, these patients became ineligible to participate in the trial. The study was conducted in agreement with Good Clinical Practice guidelines and with the Declaration of Helsinki. All patients provided written informed consent to participate in the trial and to use the tumour tissue removed at surgery for translational research. The REMARK (Reporting Recommendations for Tumor Marker Prognostic Studies) criteria were used to report this study.47
Participants
Six hundred sixty-four female patients with pT1-3, N0-3, M0 breast cancer were recruited onto the trial (Figure S1). The inclusion criteria were described in detail elsewhere and in the protocol.13,15,46 In short, all female breast cancer patients of at least 18 years old with good performance status (WHO ≤1) and all ethnicity could participate in this study, but was not recorded. Based on the incidence of various ethnicities in the Netherlands it can be assumed that most patients were of Dutch origin.48
Randomisation and masking
Patients were randomly assigned (1:1) by means of the automated ALEA system (FormsVision BV, Abcoude, the Netherlands). Patients were stratified using Pocock's minimization technique.49 Stratification factors included treatment centre, menopausal status (pre vs. post), type of surgery (mastectomy vs. lumpectomy), hormone receptor status (ER and/or PR+ vs. both negative), HER2 receptor status, nodal status (pN0, pN1(sn), pN1, pN2/3 (AJCC staging manual sixth edition 2002)), tumour size (pT1 vs. pT2 vs. pT3), and sequence of chemotherapy-radiotherapy (chemotherapy followed by radiotherapy or vice versa). Randomisation was performed centrally at the Netherlands Cancer Institute. Radiation therapy and endocrine therapy were given according to the contemporary Dutch guidelines.50
Procedures
Intravenous administration of 6 cycles of doxorubicin 60 mg/m2 and cyclophosphamide 600 mg/m2 every 2 weeks (ddAC) with 6 mg granulocyte-colony stimulating factor (pegfilgrastim) support OR intravenous docetaxel 75 mg/m2, doxorubicin 50 mg/m2 and cyclophosphamide 500 mg/m2 every 3 weeks (TAC) with 6 mg pegfilgrastim support.
Tumour histology and immunohistochemistry (IHC)
Histologic grade was assessed according to the modified Bloom-Richardson classification.51 Tumours were scored centrally for expression of the oestrogen receptor (ER), progesterone receptor (PR) and human epidermal growth factor receptor 2 (HER2) as previously described.46 ER and PR were considered positive if 10% or mofre cells showed nucleic staining. HER2 score of 3+, or 2+ with confirmation by in situ hybridization (HER2 DNA probe), were classified as positive. IHC-based breast cancer subtypes were defined as 1. ER and/or PR-positive, HER2-negative (HR-positive HER2-negative); 2. HER2-positive, regardless of ER and PR status (HER2-positive) or 3. ER, PR and HER2 negative (TNBC).
RNA isolation and sequencing
RNA was isolated from formalin-fixed, paraffin-embedded tissue with a tumour cell percentage of at least 40% using the AllPrep DNA/RNA mini kit according to the manufacturer’s instructions (Qiagen, Hilden, Germany). Quantification and purity were measured using the NanoDrop 2000 spectrophotometer (Thermofisher Scientific, Waltham, Massachusetts, USA) and the 2100 Bioanalyzer (Agilent Technologies, Santa Clara, California, USA). cDNA libraries were constructed with the TruSeq RNA Access Library Prep Kit (Illumina, San Diego, California, USA) and single-end sequenced using the HiSeq 2500 (Illumina). Reads were aligned to the reference genome (hg38) using TopHat.52 The number of uniquely assigned reads per gene was calculated with HTSeq.53 Gene expression was quantified either as total reads with trimmed-mean of median (TMM) normalizing factors, or as normalised counts per million, both computed using edgeR (Mendeley bioinformatics file).15,54
Outcomes
The primary objective was to identify a gene expression profile that can predict recurrence-free survival (RFS) benefit of either ddAC chemotherapy or a taxane-containing regimen based on the hypothesis that, while on average RFS is similar after ddAC or TAC, some patients derive more benefit from ddAC while others from TAC. RFS is defined as the time from randomisation to locoregional recurrence, distant metastasis or death by any cause, whichever occurred first. Second primary cancers and contra-lateral breast cancers will not be considered as events and will not be censored.55 The clinical risk of recurrence was assessed using the modified Adjuvant!Online classification (based on ER and HER2-status, grade, nodal-status and tumour size) in line with the classification used in the MINDACT trial.14 Patients with a clinically low risk of recurrence would not receive adjuvant chemotherapy nowadays according to current guidelines. For these patients, there is no clinical need for a predictive test guiding the decision of which chemotherapy regimen will be most effective. Therefore, these patients were excluded from the analysis of the primary objective, as defined in the statistical analysis plan.15 The secondary objective was to directly compare RFS, overall survival and toxicity of the two treatment arms and has been reported by Van Rossum.13,46
Molecular subtypes
Patients were grouped in five molecular subtypes using the PAM50 gene expression-based classifier.16 Gene expression data was visualised using unsupervised hierarchical clustering with subtypes, received treatment and RFS event as labels.
Gene expression single genes
The impact of expression levels of each individual gene, as well as of these gene expressions and their interaction with treatment, was studied separately for patients with HR-positive HER2-negative disease and patients with TNBC. A profile index score was constructed as follows. Applying 10-fold cross-validation, 10 separate penalised-likelihood Cox proportional hazards regression models were fitted which included treatment, the main effect of each gene and all pairwise treatment-gene interactions as variables for each patient. Note that the coefficient corresponding to a gene’s main effect is equivalent to the log-hazard ratio for that gene. A LASSO penalty was used on the main effect of the genes and the treatment-gene interaction effects. The scheme used means that the model fit was optimised on 9/10 of the patients, and profile scores were computed for the remaining (independent) patients. This was repeated until all patients were assigned a profile index score. Patients were then split using their dichotomised profile scores: high-profile for patients with profile score above the median score, all others in the low-profile group. RFS of these subgroups were compared using the log-rank test. Subsequently a multivariate Cox regression model was used to compare the RFS of the subgroups, while correcting for the main effects of tumour size, lymph node status, histologic grade, age and type of surgery. For the high- and low-profile score subgroups, the association between treatment and RFS was tested using the Kaplan-Meier method and compared using the log-rank test.
Hallmark gene sets
Tests for association between well-defined biological processes (Hallmark gene sets: Broad Institute, Cambridge, MA, USA)18 and RFS were performed using Goeman’s global test. Essentially, this test is for the association between the RFS and the expression profile of all genes in a gene set at once, leading to a clear conclusion per gene set. We tested the association between the Hallmark gene sets and RFS in all patients, as well as within the TNBC and the HR-positive HER2-negative subgroups, according to chemotherapy regimen received. For each individual gene set, test results were further decomposed for the genes involved, yielding a selection of the most important genes driving the test result formed by those with a family-wise error-rate (FWER) corrected pvalue below 0.05 (Meinshausen method).56
Tumour-infiltrating lymphocytes
Stromal tumour-infiltrating lymphocytes (sTILs) were scored by HH (pathologist) for the patients with triple negative breast cancer (TNBC) and by MO (supervised by HH). Scoring of sTILs was performed on digitalised hematoxylin and eosin-stained (H&E) whole slides according to previously published recommendations57 with high inter-observer concordance in an online environment.58,59 The association between sTILs and RFS was assessed using the Kaplan-Meier method and compared using the log-rank test. The interaction between sTILs and treatment was tested in a multivariate Cox regression model while correcting for the main effect of tumour size, lymph node status, histologic grade, age and type of surgery. These analyses were performed within the patients with TNBC as well as in the HR-positive HER2-negative subgroup with the median sTILs per subgroup used as cut-off point.
Quantification and statistical analysis
A gain was defined as the improvement in RFS at 5 years with the treatment strategy using the profile, over the strategy in which all patients would get the same treatment (either ddAC or TAC), assuming that both chemotherapy regimens would be equally effective for the whole group. It was assumed that, if the profile were derived using data from 400 patients, the standard error (SE) of the estimate of the gain would be less than 2.5%. The SE was calculated by propagation of error (delta-method). In this calculation, the variance resulting from the randomness of the treatment used as reference in the calculation of the gain, was considered negligible. The sample size of the study was set at 660 so that 1/3 of the data could be used as a validation cohort, allowing for 10% early dropout. During the course of the study, it became clear that the event rate was lower than expected. Therefore, an amendment was made to the protocol to use a cross-validation method instead of a separation into a training and a validation cohort.
Acknowledgments
First, we would like to thank all the patients and their families. Also, we would like to express our gratitude to the study teams of the participating centers (see also supplemental file), the Data Centre of the Netherlands Cancer Institute for collecting the data, the Core Facility – Molecular Pathology and Biobanking of the Netherlands Cancer Institute for their help with the immunohistochemistry stainings, the Genomics Core Facility of the Netherlands Cancer Institute for their work on the RNA sequencing, and the Dutch Breast Cancer Research Group (BOOG) for their role in coordinating the study. Finally, we thank the Dutch Cancer Society and the Dutch Ministry of Health, Welfare and Sport for their funding and Sanofi and Amgen for their unrestricted research grants.
This study was supported by the Dutch Cancer Society (CKTO 2004-04) and unrestricted research grants from Sanofi and Amgen. The funders had no role in the study design; in the collection, analysis, and interpretation of data; in the writing of the paper; or in the decision to submit the paper for publication.
Author contributions
S.C.L., H.M.O., E.J.R., and H.v.T. were involved in the conception and design of the trial. S.C.L., H.M.O., E.J.R., A.L.T.I., J.E.A.P., M.E.M.M.B., A.v.B., I.A.M.M., and all members of the MATADOR trialists’ group contributed to the recruitment of patients onto the trial, acquisition of data, or both. M.O., J.W., and H.M.H. were involved in the revision of the IHC data and sTIL scoring. R.X.d.M., A.G.J.v.R., M.H., G.B., E.v.W., M.O., L.K., J.v.R., S.C., and L.W. performed the statistical analyses. A.G.J.v.R., M.H., G.B., E.v.W., M.O., S.C., L.W., M.K., S.C.L., and H.M.O. interpreted the data. M.O., A.G.J.v.R., G.B., R.X.d.M., and S.C.L. drafted the manuscript. All authors were involved in the manuscript development and approved the final version.
Declaration of interests
S.C.L. and H.M.O. received an institutional unrestricted research grant from Sanofi and Amgen to conduct the MATADOR study (ISRCTN61893718); H.M.O. received institutional research support funding from Roche; M.K. received institutional research support funding from Roche, BMS, and AZ; S.C.L. received institutional research support funding from Agendia, Amgen, AstraZeneca, Eurocept, Genentech, Immunomedics (now Gilead), Roche, Sanofi, and TESARO (now GSK). S.C.L. is an advisory board member for AstraZeneca for which the institute receives compensation, and for Cergentis (pro bono). S.C.L. receives funding for educational activities from Daiichi Sankyo, paid to the institute. H.M.O. is an advisory board member for Roche, Pfizer, and Novartis. M.K. is an advisory board member for AZ, BMS, Roche, MSD, and Daiichi for which the institute receives compensation.
Published: June 29, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.110425.
Contributor Information
Sabine C. Linn, Email: s.linn@nki.nl.
the MATADOR trialists’ group for the Dutch Breast Cancer Research Group (BOOG):
Sabine C. Linn, Marcel Soesan, Rianne M. Oosterkamp, Frank Jeurissen, Nir Weijl, Alex L.T. Imholz, Johanneke E.A. Portielje, Karin J. Beelen, Monique E.M.M. Bos, Aart van Bochove, Gerty de Klerk, Suzan Vrijaldenhoven, Annette van der Velden, Hiltje de Graaf, Marielle Smeets, Jetske Meerum Terwogt, Jolanda Schrama, Philomeen Kuijer, Hanneke Wilmink, Ronald Hoekstra, Judith Kroep, Hans F.M. Pruijt, Leander van Gerven, Allert H. Vos, Frans Erdkamp, Willemien G. van Leeuwen-Breuk, and Alexander de Graeff
Supplemental information
References
- 1.Early Breast Cancer Trialists' Collaborative Group EBCTCG. Peto R., Davies C., Godwin J., Gray R., Pan H.C., Clarke M., Cutter D., Darby S., McGale P., et al. Comparisons between different polychemotherapy regimens for early breast cancer: meta-analyses of long-term outcome among 100,000 women in 123 randomised trials. Lancet. 2012;379:432–444. doi: 10.1016/s0140-6736(11)61625-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Early Breast Cancer Trialists' Collaborative Group EBCTCG Increasing the dose intensity of chemotherapy by more frequent administration or sequential scheduling: a patient-level meta-analysis of 37 298 women with early breast cancer in 26 randomised trials. Lancet. 2019;393:1440–1452. doi: 10.1016/s0140-6736(18)33137-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Simon R. Biomarker based clinical trial design. Chin. Clin. Oncol. 2014;3:39. doi: 10.3978/j.issn.2304-3865.2014.02.03. [DOI] [PubMed] [Google Scholar]
- 4.Hess K.R., Anderson K., Symmans W.F., Valero V., Ibrahim N., Mejia J.A., Booser D., Theriault R.L., Buzdar A.U., Dempsey P.J., et al. Pharmacogenomic predictor of sensitivity to preoperative chemotherapy with paclitaxel and fluorouracil, doxorubicin, and cyclophosphamide in breast cancer. J. Clin. Oncol. 2006;24:4236–4244. doi: 10.1200/JCO.2006.05.6861. [DOI] [PubMed] [Google Scholar]
- 5.Gianni L., Zambetti M., Clark K., Baker J., Cronin M., Wu J., Mariani G., Rodriguez J., Carcangiu M., Watson D., et al. Gene expression profiles in paraffin-embedded core biopsy tissue predict response to chemotherapy in women with locally advanced breast cancer. J. Clin. Oncol. 2005;23:7265–7277. doi: 10.1200/JCO.2005.02.0818. [DOI] [PubMed] [Google Scholar]
- 6.Hatzis C., Pusztai L., Valero V., Booser D.J., Esserman L., Lluch A., Vidaurre T., Holmes F., Souchon E., Wang H., et al. A genomic predictor of response and survival following taxane-anthracycline chemotherapy for invasive breast cancer. JAMA. 2011;305:1873–1881. doi: 10.1001/jama.2011.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang J.C., Wooten E.C., Tsimelzon A., Hilsenbeck S.G., Gutierrez M.C., Elledge R., Mohsin S., Osborne C.K., Chamness G.C., Allred D.C., O'Connell P. Gene expression profiling for the prediction of therapeutic response to docetaxel in patients with breast cancer. Lancet. 2003;362:362–369. doi: 10.1016/S0140-6736(03)14023-8. [DOI] [PubMed] [Google Scholar]
- 8.Okuma H.S., Koizumi F., Hirakawa A., Nakatochi M., Komori O., Hashimoto J., Kodaira M., Yunokawa M., Yamamoto H., Yonemori K., et al. Clinical and microarray analysis of breast cancers of all subtypes from two prospective preoperative chemotherapy studies. Br. J. Cancer. 2016;115:411–419. doi: 10.1038/bjc.2016.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bertheau P., Turpin E., Rickman D.S., Espié M., de Reyniès A., Feugeas J.P., Plassa L.F., Soliman H., Varna M., de Roquancourt A., et al. Exquisite sensitivity of TP53 mutant and basal breast cancers to a dose-dense epirubicin-cyclophosphamide regimen. PLoS Med. 2007;4 doi: 10.1371/journal.pmed.0040090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tabchy A., Valero V., Vidaurre T., Lluch A., Gomez H., Martin M., Qi Y., Barajas-Figueroa L.J., Souchon E., Coutant C., et al. Evaluation of a 30-gene paclitaxel, fluorouracil, doxorubicin, and cyclophosphamide chemotherapy response predictor in a multicenter randomized trial in breast cancer. Clin. Cancer Res. 2010;16:5351–5361. doi: 10.1158/1078-0432.CCR-10-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang H., Yee D. I-SPY 2: a Neoadjuvant Adaptive Clinical Trial Designed to Improve Outcomes in High-Risk Breast Cancer. Curr. Breast Cancer Rep. 2019;11:303–310. doi: 10.1007/s12609-019-00334-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cortazar P., Zhang L., Untch M., Mehta K., Costantino J.P., Wolmark N., Bonnefoi H., Cameron D., Gianni L., Valagussa P., et al. Pathological complete response and long-term clinical benefit in breast cancer: the CTNeoBC pooled analysis. Lancet. 2014;384:164–172. doi: 10.1016/S0140-6736(13)62422-8. [DOI] [PubMed] [Google Scholar]
- 13.van Rossum A.G.J., Kok M., van Werkhoven E., Opdam M., Mandjes I.A.M., van Leeuwen-Stok A.E., van Tinteren H., Imholz A.L.T., Portielje J.E.A., Bos M.M.E.M., et al. Adjuvant dose-dense doxorubicin-cyclophosphamide versus docetaxel-doxorubicin-cyclophosphamide for high-risk breast cancer: First results of the randomised MATADOR trial (BOOG 2004-04) Eur. J. Cancer. 2018;102:40–48. doi: 10.1016/j.ejca.2018.07.013. [DOI] [PubMed] [Google Scholar]
- 14.Cardoso F., van't Veer L.J., Bogaerts J., Slaets L., Viale G., Delaloge S., Pierga J.Y., Brain E., Causeret S., DeLorenzi M., et al. 70-Gene Signature as an Aid to Treatment Decisions in Early-Stage Breast Cancer. N. Engl. J. Med. 2016;375:717–729. doi: 10.1056/NEJMoa1602253. [DOI] [PubMed] [Google Scholar]
- 15.Opdam M., van Rossum A., Menezes R., Kok M., Oosterkamp R., Linn S. MATADOR: A prospective, randomised, phase III biomarker trial in breast cancer patients. Mendeley Data. 2024;V2 doi: 10.17632/n24jkp487r.2. [DOI] [Google Scholar]
- 16.Parker J.S., Mullins M., Cheang M.C.U., Leung S., Voduc D., Vickery T., Davies S., Fauron C., He X., Hu Z., et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J. Clin. Oncol. 2009;27:1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedman J., Hastie T., Tibshirani R. Regularization Paths for Generalized Linear Models via Coordinate Descent. J. Stat. Software. 2010;33:1–22. [PMC free article] [PubMed] [Google Scholar]
- 18.Liberzon A., Birger C., Thorvaldsdóttir H., Ghandi M., Mesirov J.P., Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goeman J.J., van de Geer S.A., de Kort F., van Houwelingen H.C. A global test for groups of genes: testing association with a clinical outcome. Bioinformatics. 2004;20:93–99. doi: 10.1093/bioinformatics/btg382. [DOI] [PubMed] [Google Scholar]
- 20.Burstein H.J., Curigliano G., Loibl S., Dubsky P., Gnant M., Poortmans P., Colleoni M., Denkert C., Piccart-Gebhart M., Regan M., et al. Estimating the benefits of therapy for early-stage breast cancer: the St. Gallen International Consensus Guidelines for the primary therapy of early breast cancer 2019. Ann. Oncol. 2019;30:1541–1557. doi: 10.1093/annonc/mdz235. [DOI] [PubMed] [Google Scholar]
- 21.Kodumudi K.N., Woan K., Gilvary D.L., Sahakian E., Wei S., Djeu J.Y. A novel chemoimmunomodulating property of docetaxel: suppression of myeloid-derived suppressor cells in tumor bearers. Clin. Cancer Res. 2010;16:4583–4594. doi: 10.1158/1078-0432.CCR-10-0733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee H.J., Lee J.J., Song I.H., Park I.A., Kang J., Yu J.H., Ahn J.H., Gong G. Prognostic and predictive value of NanoString-based immune-related gene signatures in a neoadjuvant setting of triple-negative breast cancer: relationship to tumor-infiltrating lymphocytes. Breast Cancer Res. Treat. 2015;151:619–627. doi: 10.1007/s10549-015-3438-8. [DOI] [PubMed] [Google Scholar]
- 23.Denkert C., von Minckwitz G., Brase J.C., Sinn B.V., Gade S., Kronenwett R., Pfitzner B.M., Salat C., Loi S., Schmitt W.D., et al. Tumor-Infiltrating Lymphocytes and Response to Neoadjuvant Chemotherapy With or Without Carboplatin in Human Epidermal Growth Factor Receptor 2–Positive and Triple-Negative Primary Breast Cancers. J. Clin. Oncol. 2015;33:983–991. doi: 10.1200/jco.2014.58.1967. [DOI] [PubMed] [Google Scholar]
- 24.West N.R., Milne K., Truong P.T., Macpherson N., Nelson B.H., Watson P.H. Tumor-infiltrating lymphocytes predict response to anthracycline-based chemotherapy in estrogen receptor-negative breast cancer. Breast Cancer Res. 2011;13 doi: 10.1186/bcr3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Galluzzi L., Buqué A., Kepp O., Zitvogel L., Kroemer G. Immunological Effects of Conventional Chemotherapy and Targeted Anticancer Agents. Cancer Cell. 2015;28:690–714. doi: 10.1016/j.ccell.2015.10.012. [DOI] [PubMed] [Google Scholar]
- 26.Coffelt S.B., de Visser K.E. Immune-mediated mechanisms influencing the efficacy of anticancer therapies. Trends Immunol. 2015;36:198–216. doi: 10.1016/j.it.2015.02.006. [DOI] [PubMed] [Google Scholar]
- 27.Fridman W.H., Zitvogel L., Sautès-Fridman C., Kroemer G. The immune contexture in cancer prognosis and treatment. Nat. Rev. Clin. Oncol. 2017;14:717–734. doi: 10.1038/nrclinonc.2017.101. [DOI] [PubMed] [Google Scholar]
- 28.Vennin C., Cattaneo C.M., Bosch L., Vegna S., Ma X., Damstra H.G.J., Martinovic M., Tsouri E., Ilic M., Azarang L., et al. Taxanes trigger cancer cell killing in vivo by inducing non-canonical T cell cytotoxicity. Cancer Cell. 2023;41:1170–1185.e12. doi: 10.1016/j.ccell.2023.05.009. [DOI] [PubMed] [Google Scholar]
- 29.Gil Del Alcazar C.R., Alečković M., Polyak K. Immune Escape during Breast Tumor Progression. Cancer Immunol. Res. 2020;8:422–427. doi: 10.1158/2326-6066.Cir-19-0786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wimmer K., Sachet M., Ramos C., Frantal S., Birnleitner H., Brostjan C., Exner R., Filipits M., Bago-Horvath Z., Rudas M., et al. Differential immunomodulatory effects of epirubicin/cyclophosphamide and docetaxel in breast cancer patients. J. Exp. Clin. Cancer Res. 2023;42:300. doi: 10.1186/s13046-023-02876-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Denkert C., Loibl S., Noske A., Roller M., Müller B.M., Komor M., Budczies J., Darb-Esfahani S., Kronenwett R., Hanusch C., et al. Tumor-associated lymphocytes as an independent predictor of response to neoadjuvant chemotherapy in breast cancer. J. Clin. Oncol. 2010;28:105–113. doi: 10.1200/JCO.2009.23.7370. [DOI] [PubMed] [Google Scholar]
- 32.Yamaguchi R., Tanaka M., Yano A., Tse G.M., Yamaguchi M., Koura K., Kanomata N., Kawaguchi A., Akiba J., Naito Y., et al. Tumor-infiltrating lymphocytes are important pathologic predictors for neoadjuvant chemotherapy in patients with breast cancer. Hum. Pathol. 2012;43:1688–1694. doi: 10.1016/j.humpath.2011.12.013. [DOI] [PubMed] [Google Scholar]
- 33.Fasching P.A., Szeto C., Denkert C., Benz S., Weber K., Spilman P., Budczies J., Schneeweiss A., Stickeler E., Schmatloch S., et al. Inferred Immune-Cell Activity Is an Independent Predictor of HER2-Negative Breast Cancer Prognosis and Response to Paclitaxel-Based Therapy in the GeparSepto Trial. Clin. Cancer Res. 2023;29:2456–2465. doi: 10.1158/1078-0432.CCR-22-2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reinisch M., Bruzas S., Gluz O., Ataseven B., Schmid P., Cortés J., Blohmer J.U., Shenoy S., Dyson M.H., Dittmer-Grabowski C., et al. Prognostic and predictive impact of gene expression in node-positive early breast cancer patients receiving dose-dense versus standard-dose adjuvant chemotherapy. Mol. Oncol. 2023;17:1060–1075. doi: 10.1002/1878-0261.13435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yam C., Yen E.-Y., Chang J.T., Bassett R.L., Jr., Alatrash G., Garber H., Huo L., Yang F., Philips A.V., Ding Q.-Q., et al. Immune Phenotype and Response to Neoadjuvant Therapy in Triple-Negative Breast Cancer. Clin. Cancer Res. 2021;27:5365–5375. doi: 10.1158/1078-0432.Ccr-21-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Loi S., Sirtaine N., Piette F., Salgado R., Viale G., Van Eenoo F., Rouas G., Francis P., Crown J.P.A., Hitre E., et al. Prognostic and predictive value of tumor-infiltrating lymphocytes in a phase III randomized adjuvant breast cancer trial in node-positive breast cancer comparing the addition of docetaxel to doxorubicin with doxorubicin-based chemotherapy: BIG 02-98. J. Clin. Oncol. 2013;31:860–867. doi: 10.1200/JCO.2011.41.0902. [DOI] [PubMed] [Google Scholar]
- 37.Loi S., Drubay D., Adams S., Pruneri G., Francis P.A., Lacroix-Triki M., Joensuu H., Dieci M.V., Badve S., Demaria S., et al. Tumor-Infiltrating Lymphocytes and Prognosis: A Pooled Individual Patient Analysis of Early-Stage Triple-Negative Breast Cancers. J. Clin. Oncol. 2019;37:559–569. doi: 10.1200/JCO.18.01010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eckhoff L., Nielsen M., Moeller S., Knoop A. TAXTOX - a retrospective study regarding the side effects of docetaxel given as part of the adjuvant treatment to patients with primary breast cancer in Denmark from 2007 to 2009. Acta Oncol. 2011;50:1075–1082. doi: 10.3109/0284186x.2011.602111. [DOI] [PubMed] [Google Scholar]
- 39.Gluz O., Nitz U., Kolberg-Liedtke C., Prat A., Christgen M., Kuemmel S., Mohammadian M.P., Gebauer D., Kates R., Paré L., et al. De-escalated Neoadjuvant Chemotherapy in Early Triple-Negative Breast Cancer (TNBC): Impact of Molecular Markers and Final Survival Analysis of the WSG-ADAPT-TN Trial. Clin. Cancer Res. 2022;28:4995–5003. doi: 10.1158/1078-0432.Ccr-22-0482. [DOI] [PubMed] [Google Scholar]
- 40.Denkert C., von Minckwitz G., Darb-Esfahani S., Lederer B., Heppner B.I., Weber K.E., Budczies J., Huober J., Klauschen F., Furlanetto J., et al. Tumour-infiltrating lymphocytes and prognosis in different subtypes of breast cancer: a pooled analysis of 3771 patients treated with neoadjuvant therapy. Lancet Oncol. 2018;19:40–50. doi: 10.1016/s1470-2045(17)30904-x. [DOI] [PubMed] [Google Scholar]
- 41.Fujimoto Y., Watanabe T., Hida A.I., Higuchi T., Miyagawa Y., Ozawa H., Bun A., Fukui R., Sata A., Imamura M., et al. Prognostic significance of tumor-infiltrating lymphocytes may differ depending on Ki67 expression levels in estrogen receptor-positive/HER2-negative operated breast cancers. Breast Cancer. 2019;26:738–747. doi: 10.1007/s12282-019-00977-0. [DOI] [PubMed] [Google Scholar]
- 42.Borst P., Wessels L. Do predictive signatures really predict response to cancer chemotherapy? Cell Cycle. 2010;9:4836–4840. doi: 10.4161/cc.9.24.14326. [DOI] [PubMed] [Google Scholar]
- 43.Swanton C. Intratumor Heterogeneity: Evolution through Space and Time. Cancer Res. 2012;72:4875–4882. doi: 10.1158/0008-5472.Can-12-2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Azizi E., Carr A.J., Plitas G., Cornish A.E., Konopacki C., Prabhakaran S., Nainys J., Wu K., Kiseliovas V., Setty M., et al. Single-Cell Map of Diverse Immune Phenotypes in the Breast Tumor Microenvironment. Cell. 2018;174:1293–1308.e36. doi: 10.1016/j.cell.2018.05.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alboukadel Kassambara M.K., Biecek P., Fabian S. survminer: Drawing Survival Curves using 'ggplot2'. 2021. https://cran.r-project.org/web/packages/survminer/index.html
- 46.van Rossum A.G.J., Kok M., McCool D., Opdam M., Miltenburg N.C., Mandjes I.A.M., van Leeuwen-Stok E., Imholz A.L.T., Portielje J.E.A., Bos M.M.E.M., et al. Independent replication of polymorphisms predicting toxicity in breast cancer patients randomized between dose-dense and docetaxel-containing adjuvant chemotherapy. Oncotarget. 2017;8:113531–113542. doi: 10.18632/oncotarget.22697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McShane L.M., Altman D.G., Sauerbrei W., Taube S.E., Gion M., Clark G.M., Statistics Subcommittee of the NCI-EORTC Working Group on Cancer Diagnostics REporting recommendations for tumor MARKer prognostic studies (REMARK). Nature clinical practice. Oncology. 2005;2:416–422. [PubMed] [Google Scholar]
- 48.Kuijer A., Verloop J., Visser O., Sonke G., Jager A., van Gils C.H., van Dalen T., Elias S.G. The influence of socioeconomic status and ethnicity on adjuvant systemic treatment guideline adherence for early-stage breast cancer in the Netherlands. Ann. Oncol. 2017;28:1970–1978. doi: 10.1093/annonc/mdx204. [DOI] [PubMed] [Google Scholar]
- 49.Pocock S.J., Simon R. Sequential treatment assignment with balancing for prognostic factors in the controlled clinical trial. Biometrics. 1975;31:103–115. [PubMed] [Google Scholar]
- 50.Rutgers E.J.T., Nortier J.W.R., Tuut M.K., van Tienhoven G., Struikmans H., Bontenbal M., von Meyenfeldt M.F., Vreugdenhil G., Benraadt T., Garssen B., et al. Nationaal Borstkanker Overleg Nederland; Kwaliteitsinstituut voor de Gezondheidszorg.[Dutch Institute for Healthcare Improvement guideline,‘Treatment of breast cancer’] Ned. Tijdschr. Geneeskd. 2002;146:2144–2151. [PubMed] [Google Scholar]
- 51.Elston C.W., Ellis I.O. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology. 1991;19:403–410. doi: 10.1111/j.1365-2559.1991.tb00229.x. [DOI] [PubMed] [Google Scholar]
- 52.Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Anders S., Pyl P.T., Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hudis C.A., Barlow W.E., Costantino J.P., Gray R.J., Pritchard K.I., Chapman J.A.W., Sparano J.A., Hunsberger S., Enos R.A., Gelber R.D., Zujewski J.A. Proposal for standardized definitions for efficacy end points in adjuvant breast cancer trials: the STEEP system. J. Clin. Oncol. 2007;25:2127–2132. doi: 10.1200/jco.2006.10.3523. [DOI] [PubMed] [Google Scholar]
- 56.Meinshausen N. Hierarchical testing of variable importance. Biometrika. 2008;95:265–278. doi: 10.1093/biomet/asn007. [DOI] [Google Scholar]
- 57.Salgado R., Denkert C., Demaria S., Sirtaine N., Klauschen F., Pruneri G., Wienert S., Van den Eynden G., Baehner F.L., Penault-Llorca F., et al. The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: recommendations by an International TILs Working Group 2014. Ann. Oncol. 2015;26:259–271. doi: 10.1093/annonc/mdu450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Buisseret L., Desmedt C., Garaud S., Fornili M., Wang X., Van den Eyden G., de Wind A., Duquenne S., Boisson A., Naveaux C., et al. Reliability of tumor-infiltrating lymphocyte and tertiary lymphoid structure assessment in human breast cancer. Mod. Pathol. 2017;30:1204–1212. doi: 10.1038/modpathol.2017.43. [DOI] [PubMed] [Google Scholar]
- 59.Slide Score. http://www.slidescore.com.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Anonymised patient data that support the finding of this study is shared in a Mendeley repository and RNA sequencing data on GEO (GSE167977).15 In the Mendeley bioinformatics file the code for the quality control steps and gene expression profiling is described in detail.15 Any additional information required to reanalyse the data reported in this work paper is available from the lead contact upon request.