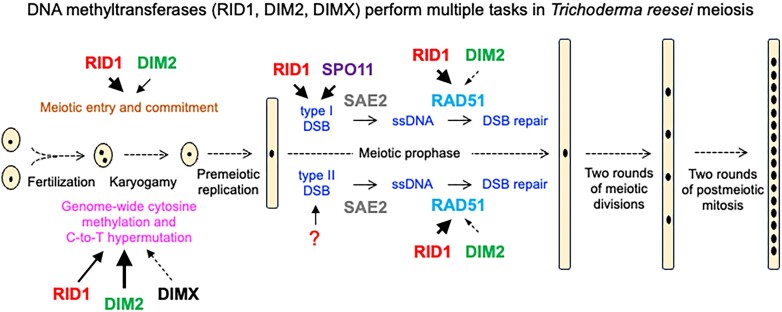
Abstract
Trichoderma reesei is an economically important enzyme producer with several unique meiotic features. spo11, the initiator of meiotic double-strand breaks (DSBs) in most sexual eukaryotes, is dispensable for T. reesei meiosis. T. reesei lacks the meiosis-specific recombinase Dmc1. Rad51 and Sae2, the activator of the Mre11 endonuclease complex, promote DSB repair and chromosome synapsis in wild-type and spo11Δ meiosis. DNA methyltransferases (DNMTs) perform multiple tasks in meiosis. Three DNMT genes (rid1, dim2 and dimX) differentially regulate genome-wide cytosine methylation and C:G-to-T:A hypermutations in different chromosomal regions. We have identified two types of DSBs: type I DSBs require spo11 or rid1 for initiation, whereas type II DSBs do not rely on spo11 and rid1 for initiation. rid1 (but not dim2) is essential for Rad51-mediated DSB repair and normal meiosis. rid1 and rad51 exhibit a locus heterogeneity (LH) relationship, in which LH-associated proteins often regulate interconnectivity in protein interaction networks. This LH relationship can be suppressed by deleting dim2 in a haploid rid1Δ (but not rad51Δ) parental strain, indicating that dim2 and rid1 share a redundant function that acts earlier than rad51 during early meiosis. In conclusion, our studies provide the first evidence of the involvement of DNMTs during meiotic initiation and recombination.
Graphical Abstract
Graphical Abstract.
Introduction
5-Methylcytosine (5mC), an important epigenetic mark, has been implicated in a variety of biological processes and human diseases. This heritable DNA modification is performed by DNA-methyltransferases (DNMTs) that catalyze transfer of a methyl group from S-adenosyl-l-methionine to cytosine bases (see reviews (1,2)). DNA cytosine methylation and demethylation also occur during meiosis, a specialized germ cell cycle that generates haploid gametes via one round of DNA replication and two rounds of meiotic nuclear divisions. For example, loss of male-lineage-specific RNA-directed DNA methylation in Arabidopsis thalianadisrupts meiosis due to mis-splicing of the Multipolar Spindle 1 (MPS1) gene, also known as Putative Recombination initiating Defects 2 (PRD2) (3). The genomes of mouse spermatocytes undergo DNA replication-dependent DNA demethylation. Mouse spermatocytes gradually but unevenly regain DNMT3-dependent DNA methylation after DNA replication, suggesting that key meiotic prophase I events might occur in the context of hemi-methylated genomes (4).
A hallmark of meiotic prophase I is that nonsister chromatids of homologous chromosomes undergo DNA recombination and chromosome synapsis. In most studied sexual eukaryotes, meiotic DNA recombination is initiated by Spo11-induced double strand breaks (DSBs). Spo11 is a meiosis-specific topoisomerase VI subunit A endonuclease (5,6). The covalently linked Spo11-oligo complexes are then released from the DSB ends by the Mre11-Rad50-Xrs2/Nbs1 endonuclease complexes with the assistance of Sae2 (also called Com1 or CtIP) (7–9). The processed DSB ends undergo 5′-to-3′ resection to create 3′ single-strand DNA (ssDNA) tails. Eukaryotic RecA-like recombinases (ubiquitous Rad51 and meiosis-specific Dmc1) cooperate to repair these ssDNA-associated DSBs (ssDSBs), preferentially using a homologous non-sister chromatid (interhomolog or IH repair) rather than a sister chromatid (intersister or IS repair) as template. Rad51 and/or Dmc1 first polymerize on these ssDSBs to form right-handed helical nucleoprotein filaments to initiate homologous recombination (HR) by promoting homologous pairing and strand exchange reactions. The nucleoprotein filaments, often referred to as the presynaptic complex, can pair with double-strand DNA (dsDNA) with homologous or homeologous sequences to yield synaptic complexes consisting of triplex-helical DNA pairing intermediates. Intriguingly, several sexual eukaryotes lack Dmc1, including filamentous ascomycetes (e.g. Neurospora crassa, Sordaria macrospora and Trichoderma reesei), Drosophila melanogaster and Caenorhabditis elegans. The Rad51 proteins in these Rad51-only organisms have acquired Dmc1’s capability to tolerate mismatched sequences during homologous pairing and strand exchange activity (10,11).
The IH–HR pathway ultimately results in crossover (CO) and noncrossover (NCO) recombination products. COs create new genetic diversity in gametes, whereas NCO or gene conversion (GC) involves the unidirectional transfer of genetic information (12). NCOs form early during meiosis via a process termed synthesis-dependent strand-annealing (SDSA). Early NCO formation is abolished in S. cerevisiae mutants lacking Sgs1, the ortholog of mammalian Bloom RecQ-like helicase (13). Two subclasses of COs co-exist in many sexual eukaryotes (14–17). The ‘CO obligatory rule’ reflects the fact that the class I CO pathway ensures that each pair of homologous chromosomes receives at least one obligatory chiasma during the diplotene stage of meiosis prophase I. The absence of a chiasma generally results in improper chromosomal segregation and aneuploidy. Class I COs are interference-sensitive, so the formation of COs near each other appears to be suppressed. Class II COs are non-interfering. The ‘CO interference rule’ reflects the fact that superfluous IH-COs lead to incorrect chromosome segregation at the first meiotic nuclear division (MI). In S. cerevisiae, the class I CO pathway accounts for 70–85% of COs and requires a group of meiosis-specific ZMM proteins. In contrast, class II COs depend on several nucleases, including Mms4–Mus81, Yen1 and Slx1–Slx4 (18,19). ZMM proteins also mediate the assembly of the synaptonemal complex (SC), a protein lattice that connects paired homologous chromosomes in meiotic prophase (15). For most studied sexual eukaryotes, SC assembly is coupled to the formation of recombination intermediates, such as in S. cerevisiae, S. macrospora and Mus musculus (20). The ZMM family in S. cerevisiae includes eight functionally collaborating proteins: the SC transverse filament protein Zip1, the SUMO E3 ligase Zip3, the Zip2–Zip4–Spo16 (ZZS) complex, the DNA helicase Mer3 and the Msh4-Msh5 (MutSγ) complex (15,21–31). The SC transverse filament proteins in different eukaryotes (e.g. S. cerevisiae Zip1, S. macrospora Sme4, Arabidopsis thaliana ZYP1 and mammalian SYCP1) share no sequence similarity but strong structural homology; all comprise long internal coiled-coil domains (with sizes correlated with the width of the SC; ∼100 nm) flanked by globular N- and C-terminal domains (20,32). In contrast, the MutSγ complex not only shares strong sequence similarity but is also functionally relevant to the Rad51/Dmc1 protein interaction network in several sexual eukaryotes. For instance, the S. cerevisiae Msh4/5 complex binds to Spo11-induced DSBs and stabilizes the single-end invasion intermediates that form during early stages of recombination to ensure CO formation (33–35). C. elegans MSH4/5 colocalizes with RAD51 on meiotic chromosomes and is required for timely disappearance of RAD51 foci (36). A. thaliana MSH4 and RAD51 foci appear almost simultaneously and exhibit strong colocalization along early meiotic chromosomes (37). Human MSH4 protein physically interacts with both RAD51 and DMC1 (38). Mouse MSH4 colocalizes with DMC1/RAD51 complexes on meiotic chromosomes (38).
DNA cytosine methylation also plays important roles in the Pezizomycotina (‘sac’) fungi, including the filamentous fungal model organism Neurospora crassa, Ascobolus immersus (39), pathogenic fungi (e.g. Aspergillus fumigatus, Magnaporthe oryzae, Fusarium graminearum and Leptosphaeria maculans) (40,41), as well as the industrial enzyme producer T. reesei (this study). These fungi possess two evolutionarily conserved DNMT genes, such as dim-2 (DNA methyltransferase 2) and rid-1 (RIP defective 1) in N. crassa (42) or dim2 and rid1 in T. reesei. N. crassa dim-2 is responsible for most known DNA cytosine methylation during vegetative growth (43), whereas rid-1 is involved more profoundly than dim-2 in cytosine(C):guanine(G)-to-thymine(T):adenine(A) or C-to-T mutations during the operation of repeat-induced point mutation (RIP) (44,45). RIP is a fungus-specific genome defense mechanism that mitigates the deleterious consequences of repeated genomic regions and transposable elements (TEs) during meiosis (44,45). Notably, closely positioned repeats can also induce dim-2 and heterochromatin-dependent C-to-T mutations of the adjoining nonrepetitive regions in N. crassa meiocytes (46). The first member of the rid1 gene family to be identified was Ascobolus immersus masc1 (encoding methyltransferase from Ascobolus 1), which mediates a gene-silencing process termed ‘methylation induced premeiotically (MIP)’ (47–49).
The molecular mechanisms and physiological impacts of DNA cytosine methylation in N. crassa are distinct from those in other filamentous fungi. Neither dim-2 nor rid-1 is essential for N. crassa sexual development or meiosis (42). In contrast, the rid1 orthologs in other filamentous fungi are essential for meiosis and sexual development, including A. immersus masc1 (39), Aspergillus nidulans dmtA (50), Podospora anserina rid (51), as well as T. reesei rid1 (this study). Pierre Grognet and Fabienne Malagnac have provided important insights into the roles of P. anserina rid during sexual development (51). First, sexual development is blocked in homozygous rid1Δ crosses just before individualization of the dikaryotic cells to meiocytes. This phenotype is similar to those of A. immersus masc1Δ (39), A. nidulans dmtAΔ (50) and T. reesei rid1Δ (this study). Second, the putative catalytic residues of P. anserina RID methylase are essential for its role during sexual development. Third, although P. anserina rid is not essential for the formation of maternal tissues (i.e. peridium or envelope), it is required for female fertility because the presence of RID in the female reproductive structures is necessary for normal sexual development. Finally, the rid-controlled genetic network might overlap with the well-known mating-type gene developmental pathway common to many filamentous fungi (51). The exact roles of these rid/masc1 genes in regulating meiosis are still unclear.
T. reesei is a cellulolytic filamentous fungus widely used for industrial enzyme production. The natural isolate QM6a was originally isolated from the Solomon Islands during World War II. A serial mutagenesis program was employed during the 1980s to generate high-enzyme-producing strains, including RUT-C30 and QM9414 (52). QM6a has a MAT1-2 mating type locus, but it is female sterile due to mutations in ham5. ham5, also known as idc1, is a Pezizomycotina-specific gene essential for vegetative hyphal fusion (anastomosis) and for the formation of the immature female mating structures (protoperithecia) (53). Accordingly, RIP has not operated in QM6a for >75 years, as QM6a was vegetatively or asexually propagated at least until Hypocrea jecorina was demonstrated as being the teleomorph (the sexual reproductive stage) of QM6a (54,55). H. jecorina CBS999.97 fruiting bodies were originally collected from French Guiana. CBS999.97(MAT1-1) and CBS999.97(MAT1-2) are two sexually-competent haploid strains that were derived from two ascospores (sexual spores) of a CBS999.97 fruiting body (FB). Sexual crossing of CBS999.97(MAT1-1) with either QM6a or CBS999.97(MAT1-2) can produce FBs with many asci, which are sac-like structures each containing a rosette of 16 linear ascospores (55).The 16 ascospores in the same ascus are generated from meiosis followed by two rounds of post-meiotic mitosis (11,56). Due to geographical isolation, QM6a and CBS999.97(MAT1-1) exhibit high levels of sequence heterogeneity, including 1138579 single nucleotide polymorphisms (SNPs) and 5019401 base pairs (bps) of insertions and deletions (InDels). Thus, the QM6a/CBS999.97(MAT1-1) zygotes are ideal models for studying the molecular mechanisms of hybrid meiosis (11,57,58).
Notably, compared to those of QM6a and CBS999.97(MAT1-2), the genome of CBS999.97(MAT1-1) displays reciprocal exchange (re) between the L segment (∼30 kb) at the right terminus of the second chromosome and the D segment (∼0.5 Mb) at the right terminus of the fourth chromosome. Accordingly, the wild-type (wt) natural isolates of CBS999.97(MAT1-1), CBS999.97(MAT1-2), and QM6a are referred to as CBS999.97(MAT1-1, re), CBS999.97(MAT1-2, wt) and QM6a (MAT1-2, wt), respectively (11,56) (Table 1A). We have shown previously that all 16 ascospores generated from sexual crossing of CBS999.97(MAT1-1, wt) with CBS999.97(MAT1-2, wt) or CBS999.97(MAT1-1, re) with CBS999.97(MAT1-2, re) are viable, as they can germinate and form vegetative mycelia with green conidia (asexual spores). In contrast, sexual crosses of CBS999.97(MAT1-1, re) with QM6a (MAT1-2, wt) or CBS999.97(MAT1-2, wt) only generate 65–80% viable ascospores; 70–80% of the asci contain 16 euploid ascospores and 20–30% of the asci display two different types of segmentally aneuploid (SAN) ascospores (Table 1A). The SAN1 ascospores cannot germinate due to loss of the D segment (∼0.5 Mb). The SAN2 ascospores possess two D segments but lack the L segment (∼30 kb). The SAN2 ascospores germinate and form vegetative mycelia with white conidia because the missing L segment harbors polyketide synthase 4 (pks4), which is responsible for the formation of green conidial pigmentation. The euploid ascospores germinate and form vegetative mycelia with green conidia (11,56,57). We have also shown previously that QM6a/CBS999.97 (MAT1-1) hybrid meiosis not only produces IH-HR and ‘canonical’ RIP products, but also considerable C-to-T mutations at chromosomal regions with low AT content or lacking duplicated sequences (11,57–60). However, it has not been clear which DNMT genes play important roles in promoting IH-HR, RIP and genome-wide C-to-T hypermutations during T. reesei meiosis.
Table 1.
Summary of the phenotypes of different sexual crosses
| Sexual crosses | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MAT1-1 | MAT1-2 | ||||||||||||
| WTH strain number and genetic background | Genotype | ham5 a | WTH strain number and genetic background | Genotype | ham5 a | Fruiting bodies | Mature asciib | Mature sporesc | No. of dissected ascii/spores | No. of viable sexual spores | Sexual pore viability (%) | ||
| A | |||||||||||||
| 0011 | C(re)d | wild type (wt) | + | 0015 | Qd | wt | − | + | +++ | +++ | 30/480 | 356 | 74 |
| 0011 | C (re) | wt | + | 0013 | C | wt | + | + | +++ | +++ | 30/480 | 332 | 69 |
| 0011 | C (re) | wt | + | 0282 | C (re) | wt | + | + | +++ | +++ | 20/320 | 320 | 100 |
| 0270 | C | wt | + | 0013 | C | wt | + | + | +++ | +++ | 20/320 | 320 | 100 |
| 7205 | H | spo11Δ | + | 7217 | H (re) | spo11Δ | + | + | +++ | +++ | 39/624 | 480 | 77 |
| 7205 | H | spo11Δ | + | 10586 | H | spo11Δ | – | + | +++ | +++ | 26/416 | 416 | 100 |
| 10610 | H (re) | spo11Δ | + | 10586 | H | spo11Δ | – | + | +++ | +++ | 5/80 | 57 | 71 |
| 12794 | H | rad51Δ | − | 12867 | H (re) | rad51Δ | + | + | +++ | − | − | − | − |
| 0011 | C (re) | wt | + | 12867 | H (re) | rad51Δ | + | + | +++ | +++ | 5/80 | 56 | 70 |
| 12902 | H (re) | spo11Δrad51Δ | − | 12901 | H (re) | spo11Δrad51Δ | + | + | +++ | − | − | − | − |
| 13049 | H | sae2Δ | + | 13040 | Q | sae2Δ | + | + | +++ | + | 26/232 | 23 | 10 |
| 13049 | H | sae2Δ | + | 12867 | H (re) | rad51Δ | + | + | +++ | +++ | 3/48 | 19 | 40 |
| 13053 | H | spo11Δsae2Δ | − | 13054 | H | spo11Δsae2Δ | + | + | +++ | + | 26/230 | 34 | 15 |
| B | |||||||||||||
| 0011 | C (re) | wt | + | 13058 | Q | rid1Δ | − | + | +++ | +++ | 3/48 | 34 | 71 |
| 0011 | C (re) | wt | + | 13060 | Q | dim2Δ | − | + | +++ | +++ | 5/80 | 60 | 80 |
| 0011 | C (re) | wt | + | 13215 | Hd | dim2 rid1Δ | + | + | +++ | +++ | 5/80 | 60 | 80 |
| 13059 | C (re) | rid1Δ | + | 0015 | Q | wt | − | + | +++ | +++ | 2/32 | 24 | 75 |
| 13059 | C (re) | rid1Δ | + | 13058 | Q | rid1Δ | − | + | + | − | − | − | − |
| 13059 | C (re) | rid1Δ | + | 13060 | Q | dim2Δ | − | + | +++ | +++ | 4/64 | 52 | 81 |
| 13059 | C (re) | rid1Δ | + | 13215 | H | dim2Δrid1Δ | + | + | + | − | − | − | − |
| 13079 | H | dim2Δ | + | 0015 | Q | wt | − | + | +++ | +++ | 5/80 | 76 | 95 |
| 13079 | H | dim2Δ | + | 13058 | Q | rid1Δ | − | + | +++ | +++ | 5/80 | 63 | 79 |
| 13079 | H | dim2Δ | + | 13060 | Q | dim2Δ | − | + | +++ | +++ | 5/80 | 73 | 91 |
| 13079 | H | dim2Δ | + | 13215 | H | dim2Δrid1Δ | + | + | +++ | +++ | 3/48 | 48 | 100 |
| 13147 | H (re) | dim2Δrid1Δ | + | 0015 | Q | wt | − | + | +++ | +++ | 2/32 | 23 | 72 |
| 13147 | H (re) | dim2Δrid1Δ | + | 13058 | Q | rid1Δ | − | + | + | − | − | − | − |
| 13147 | H (re) | dim2Δrid1Δ | + | 13060 | Q | dim2Δ | − | + | +++ | +++ | 10/160 | 123 | 77 |
| 13147 | H (re) | dim2Δrid1Δ | + | 13215 | H | dim2Δrid1Δ | + | + | − | − | − | − | − |
| 13059 | C (re) | rid1Δ | + | 10586 | H | spo11Δ | − | + | +++ | +++ | 10/160 | 104 | 65 |
| 13079 | H | dim2Δ | + | 10586 | H | spo11Δ | − | + | +++ | +++ | 4/64 | 46 | 72 |
| 13289 | H | spo11Δrid1Δ | + | 13290 | H | spo11Δrid1Δ | + | + | +++ | − | − | − | − |
| 13269 | H (re) | spo11Δdim2Δ | + | 13268 | H (re) | spo11Δdim2Δ | − | + | +++ | +++ | 3/48 | 43 | 90 |
| C | |||||||||||||
| 13059 | C (re) | rid1Δ | + | 12867 | H (re) | rad51Δ | + | + | +++ | +/− | − | − | − |
| 13059 | C (re) | rid1Δ | + | 12901 | H (re) | spo11Δrad51Δ | + | + | +++ | − | − | − | − |
| 12794 | H | rad51Δ | − | 13290 | H | spo11Δrid1Δ | + | + | + | − | − | − | − |
| 13297 | H | rad51Δ | + | 13058 | Q | rid1Δ | − | + | +++ | +/− | − | − | − |
| 13289 | H | spo11Δrid1Δ | + | 12867 | H (re) | rad51Δ | + | + | +++ | +/− | − | − | − |
| 13289 | H | spo11Δrid1Δ | + | 12901 | H (re) | spo11Δrad51Δ | + | + | +++ | + | − | − | − |
| 13299 | H | rad51Δrid1Δ | + | 13315 | H | rad51Δrid1Δ | + | + | +++ | − | − | − | − |
| 13287 | H | rad51Δdim2Δ | + | 13286 | H | rad51Δdim2Δ | + | + | +++ | − | − | − | − |
| 13059 | C (re) | rid1Δ | + | 13040 | H | sae2Δ | + | + | +++ | + | 7/58 | 38 | 66 |
| 13079 | H | dim2Δ | + | 12867 | H (re) | rad51Δ | + | + | +++ | +++ | 4/64 | 42 | 66 |
| 13269 | H (re) | spo11Δdim2Δ | + | 12867 | H (re) | rad51Δ | + | + | +++ | +++ | 4/64 | 52 | 81 |
| 13147 | H (re) | dim2Δrid1Δ | + | 12867 | H (re) | rad51Δ | + | + | +++ | +++ | 6/96 | 92 | 96 |
| 13059 | C (re) | rid1Δ | + | 13286 | H | rad51Δdim2Δ | + | + | +++ | +/- | − | − | − |
| 12794 | H | rad51Δ | − | 13215 | H | rid1Δdim2Δ | + | + | +++ | +++ | 3/48 | 45 | 94 |
| 13287 | H | rad51Δ dim2Δ | + | 13058 | Q | rid1Δ | + | + | +++ | +/- | 3/48 | 46 | 96 |
| 13287 | H | rad51Δdim2Δ | + | 13215 | H | rid1Δdim2Δ | + | + | +++ | +++ | 4/64 | 64 | 100 |
| 13147 | H (re) | dim2Δrid1Δ | + | 13286 | H | rad51Δdim2Δ | + | + | +++ | +/- | 3/47 | 14 | 30 |
| D | |||||||||||||
| 0270 | C | wt | + | 13352 | Q | msh4Δ | - | + | +++ | +++ | 11/176 | 101 | 68 |
| 13297 | H | rad51Δ | + | 13352 | Q | msh4Δ | - | + | +++ | +++ | 8/128 | 82 | 64 |
| 13359 | H | msh4Δ | - | 12867 | C (re) | rad51Δ | + | + | +++ | - | - | - | - |
| 13359 | H | msh4Δ | - | 13354 | H | msh4Δ | + | + | +++ | +++ | 8/128 | 97 | 76 |
a ham5+ encodes a wild-type HAM5 protein, whereas ham5− encodes a truncated protein of only 151 amino acids due to a point mutation (G531 → T) at the first position of the second intron, thus changing the invariant G+1 of the donor splicing site (53).
b+++: the majority of asci were >50 μm in length; +: <10% asci were >50 μm; −: no elongated asci.
c+++: the majority of asci contain 16 mature ascospores; +: the majority of asci (e.g. sae2Δ/sae2Δ or spo11Δsae2Δ/spo11Δsae2Δ) formed <16 mature ascospores; +/−: very few (<1%) asci formed mature ascospores; −: the elongated asci hardly formed any mature ascospores.
dQ represents QM6a (MAT1-2) and the gene deletion mutants that are directly generated from this wild isolate by homologous recombination.
eC represents CBS999.97(MAT1-1, re), CBS999.97(MAT1-1, wt), and the gene deletion mutants directly generated from these two wild isolates by homologous recombination, with ‘re’ and ‘wt’ representing reciprocal exchange and wild type, respectively (11,56).
In this report, we characterized the roles of DNMT genes and their functional relationships with spo11, sae2, rad51 and msh4 during T. reesei meiosis. Our results indicate that T. reesei rid1 exerts multiple functions during meiosis, explaining why it is essential for meiosis and sporulation.
Materials and methods
Miscellaneous
Fungal growth, culture media, sexual crossing, single ascospore isolation, PCR genotyping and Southern hybridization were conducted as described previously (11,56,57). The DNeasy Plant Mini Kit (cat. no. 69104, Qiagene, USA) was applied to isolate genomic DNA (gDNA) from vegetative mycelia and FBs (11). All buffers and RNase A were provided by the vendor. Vegetative mycelia or FBs (600 mg) were mixed with liquid nitrogen and disrupted by a mortar (7 cm in diameter) and pestle (2.5 cm in diameter). The frozen lysate was collected in a 50 ml FALCON centrifuge tube, mixed with 2400 μl Buffer AP1 and 24 μl RNase A (100 mg/mL), briefly vortexed, and incubated at 65°C for 10 min. The lysate was centrifuged for 10 min at 20 000 × g, mixed with 780 μl Buffer P3, and then incubated on ice for 5 min. One-third of the lysate was aliquoted into a QIAshredder spin column placed in a 2 ml collection tube, and then centrifuged for 2 min at 20 000 × g. All flow-through from three QIAshredder spin columns was collected and transferred into a 15 ml centrifuge tube without disturbing the pellet if present. Then, 4800 μl Buffer AW1 was added and mixed by gently inverting the tube. Next, we transferred 650 μl of the mixture into a DNeasy Mini spin column placed in a 2 ml collection tube. All flow-through from three DNeasy Mini spin column was collected into a 15 ml centrifuge tube and centrifuged for 30 s at 20 000 × g. The flow-through was discarded. The DNeasy Mini spin column was placed into a new 2 ml collection tube, 500 μl Buffer AW2 was aliquoted in, and centrifuged for 30 s at 20 000 × g. The flow-through was discarded. The DNeasy Mini spin column was placed into yet another new 2 ml collection tube, 500 μl Buffer AW2 was aliquoted in, and centrifuged for 30 s at 20 000 × g. The DNeasy Mini spin column was then transferred to a new 1.5 ml microcentrifuge tube, 50 μl Buffer AE was aliquoted in for elution, incubated at room temperature (15–25°C) for 5 min, and then centrifuged for 2 min at 20 000 × g. All eluted samples (150 μl) from three DNeasy Mini spin columns were collected into a new 1.5 ml microcentrifuge tube. The gDNA was then collected by ethanol precipitation.
For cytological analysis, asci were stained with 4′,6-diamidino-2-phenylindole (DAPI) or SYTOX™ Green Nucleic Acid Stain (Invitrogen, USA) and then visualized using a DeltaVision Core Imaging System (Applied Precision, LLC, USA) or utilizing AxioImager Z1 fluorescence microscopy (Zeiss, German) (11,56,57,60,61).
All T. reesei gene deletion mutants used in this report (Table 1 and Supplementary Dataset DS1) were generated by homologous recombination, followed by sexual crossing, and then confirmed by gDNA polymerase chain reaction (gPCR) and Southern hybridization (Supplementary Figures S1 and S2).
Reverse-transcription quantitative polymerase chain reaction (RT-qPCR) was performed as described previously (61), with transcripts of the ribosome protein gene rpl6e being used for normalization of the RT-gPCR data. All quantitative data are shown as means plus/minus standard error of the mean (SEM), as indicated in each figure legend. Graphs were plotted using GraphPad Prism 10.1.0 (GraphPad software). Statistical analyses were performed using two-way analysis of variance (ANOVA) with Bonferroni's post-test correction. All statistical analyses were performed using Sigmastat 3.5 (SigmaPlot software). P values ≥ 0.05 were considered nonsignificant. #P value < 0.05; **P value < 0.01; ***P value < 0.001.
For physical analysis of meiotic DSBs, genomic DNA (gDNA) was isolated from the vegetative mycelia of parental haploid strains, from the developing FBs harvested at three, six, or eight days (D3, D6, D8) after the initiation of sexual crosses, as well as from the ascospores released from the mature FBs at D8. The gDNA was then restriction-digested using either AgeI or BglII, separated by electrophoresis in a 0.8% agarose gel (UltraPure Agarose, Invitrogen, USA), and subjected to Southern hybridization. Visualization/quantification of Southern hybridization band intensity was performed using a BAS-IP MS204 phosphorimaging plate (Cytiva, Japan) either with a Typhoon 5 biomolecular imager (Cytiva, Japan) or a TyphoonFLA 9000 biomolecular imager (Cytiva, Japan). The pixel resolution was 25 μm, whereas the PMT voltage value was 1000 (11,62). An act1 or smp3 DNA probe was used as the loading control for Southern hybridization. Since the emergence timing of T. reesei white primordial stromata after the induction of sexual crosses was asynchronous, it was important to harvest FBs of similar size at the indicated time points during sexual development. The PCR primers used in RT-qPCR and Southern hybridization are listed in Supplementary Table S1.
Transmission electron microscopy (TEM) tomography
Developing FBs were collected and dissected into thin sections (0.2 mm thick) using a Vibratome 1000 PLUS tissue section system. Pre-fixation was performed in 2.5% glutaraldehyde/0.1 M sodium cacodylate (pH 7.2) for 3 h at room temperature, and then shifted to 4°C overnight. The specimens were washed three times in 0.1 M sodium cacodylate (pH 7.2) for 15 min for each wash. Post-fixation was performed by 2% OsO4 solution/0.1 M sodium cacodylate buffer at room temperature for 1 h. Next, the specimens were washed three times with 0.1 M sodium cacodylate for 15 min each wash. We used 2% uranyl acetate for in-block staining at room temperature for 1 h. For dehydration, a series of ethanol solutions at 10% increments (from 10% to 100%) was used. The specimens were immersed twice in 100% 1,2-propylene oxide (PO) for 20 min. Infiltration was first performed for 4 h in a mixture of PO with 10% Spurr's resin. We then increased the concentration of Spurr's resin by 10% for 8–12 h each change until we reached 100%. The specimens were immersed in Spurr's resin for 12 hours twice, and then a further 12 h in Spurr's resin with the accelerator dimethylaminoethanol (DMAE), before being transferred to the embedding molds. Polymerization proceeded at 70°C for 12 h. Serial 200 nm-thick sections were collected and scanned with a Thermo Scientific™ Talos L120C TEM system (4 × 4k CMOS Ceta camera) operating at 120 kV for 0 and 90°. The TEM tomography images were taken at 6700× magnification. The Inspect 3D 4.3 program was applied for image stack alignment, reconstruction and axis registration for each section. Amira Software 6.0 was used to align and concatenate all image sections. Three-dimensional models were generated by inputting the concatenated images into Imaris9.1.2 software.
Whole genome sequencing
PacBio RSII or Sequel technology, Oxford Nanopore technology (ONT) and Illumina next-generation sequencing (NGS) technology were applied for gDNA or RNA sequencing as described previously (57,59,60,63). Genome-wide mapping of interhomolog recombination products was performed using the same approaches described recently for recombination analysis of QM6a/CBS999.97(MAT1-1) hybrid meiosis (11,59).
Genome-wide 5mC profiling during vegetative growth and sexual development
The EZ DNA Methylation-Gold™ Kit (Zymo Research, Orange, CA) was used for NGS-based bisulfite sequencing (BS+ NGS-seq) to map and visualize 5mC in the genomic DNA isolated from vegetative mycelia, as well as at different stages of sexual development. Unmethylated cl857 Sam7 Lambda DNA (catalog # D1521; Promega, US) was spiked-in (0.5% w/w) to estimate the BS conversion efficiency. The genomic DNA with BS+ treatment was then subjected to library preparation using the Accel-NGS Methyl-Seq DNA library kit (catalog # 30024; Swift Bioscience, USA). The libraries were sequenced using a NextSeq 500 sequencing system (Illumina, USA), as described previously (64). BatMeth2, an open source software program (https://github.com/GuoliangLi-HZAU/BatMeth2/) (64), was used for genome-wide DNA methylation calling. Notably, the BS conversion efficiency of the unmethylated Lambda DNA was 97.5–98.7% for all BS+ samples we examined herein. The genome-wide ratios of 5mC and C-to-T mutations were revealed by BS+ and BS− NGS-seq, respectively, and then visualized using TSETA (Third-generation Sequencing to Enable Tetrad Analysis), a bioinformatic software package for aligning, mapping and visualizing multiple genome sequences at the scale of the chromosomal landscape to individual nucleotides (59). The AT-rich regions are indicated by their GC contents (GC%).
Meiotic ssDNA enrichment on benzoylated naphthoylated DEAE (BND) cellulose
T. reesei FBs were generated and harvested on the fourth to sixth day (D4-D6) after sexually crossing two corresponding haploid strains, before isolating intact genomic DNA as described previously (60). The same method for enriching ssDNA in the S. cerevisiae dmc1Δ mutant (65) was applied to isolate meiotic ssDNA from the FBs generated from QM6a x CBS999.97(MAT1-1), sae2Δ × sae2Δ, rad51Δ × rad51Δ and rad51Δ spo11Δ × rad51Δ spo11Δ crosses. The vegetative mycelia of QM6a and CBS999.97(MAT1-1) were used as negative controls for meiotic ssDNA enrichment experiments. All the sequencing raw data are publicly available at NCBI (Supplementary Dataset DS1). All experiments were performed in triplicate. The enriched ssDNA was quality checked using both Agilent 2100 Bioanalyzer and the 2200 TapeStation system, as described previously (60). The raw sequencing reads from each sample were mapped to the QM6a genome using the BWA (v0.7.17) alignment tool (66). We used the package BAMscale (v1.0) to quantify the sequencing peaks and normalize the coverage tracks across all samples (67). With the ‘scale’ function, peak coverages were calculated based on the sum of per-base coverage of reads and then scaled to 1× genome coverage. The resulting values are referred to as the ‘normalized coverage index (NCI)’ (Supplementary Dataset DS2).
Additionally, the sequencing coverage tracks were scaled with a bin size of 10 bps using the parameter ‘–binsize 10’. After checking for correlation across replicate samples, we combined the replicate tracks by calculating the average scaled coverage tracks for three replicates. To identify the differential peaks between conditions, we subtracted the average scaled coverage tracks between two conditions and the regions with differences >2 were defined as differential regions. Significant differential regions were determined by using a Kolmogorov–Smirnov test (R function ks.test) with a P-value of <0.05 (Supplementary Datasets DS5-DS7).
Identification of rid1-dependent differentially expressed genes (DEGs)
Raw RNA-seq reads generated by Illumina were initially processed for quality filtering and adaptor trimming in ‘Cutadapt’ (v1.12) (68). The trimmed reads were aligned and mapped to the near-complete QM6a genome sequence (57) using ‘STAR’ (v2.5.3a) (69). Paired aligned reads were recovered from each of these RNA-seq libraries. The expected read counts of each gene were estimated using ‘RSEM’ (v1.2.31) (70), before adjusting the counts per million (CPM) with the effective library size using the calcNormFactors function implemented in the R package ‘edgeR’ (v.3.26.8) (71). Differentially expressed genes (DEGs) were identified by using the exactTest function in ‘edgeR’. Genes with a false discovery rate (FDR) of fold-change >2 (or <0.5) were considered as significant DEGs. Next, we conducted Gene Ontology and KEGG pathway enrichment analysis on each set of DEGs using the R package ‘clusterProfiler’ (v 3.18.1) (72) (Supplementary Dataset DS3). The putative function of each DEG was revealed by referring to the genome-wide annotation datasets of QM6a and CBS999.97(MAT1-1) (11,57). All the sequencing raw data are publicly available at NCBI (Supplementary Dataset DS1).
Results
spo11 is dispensable for T. reesei sexual development, meiosis, and sporulation
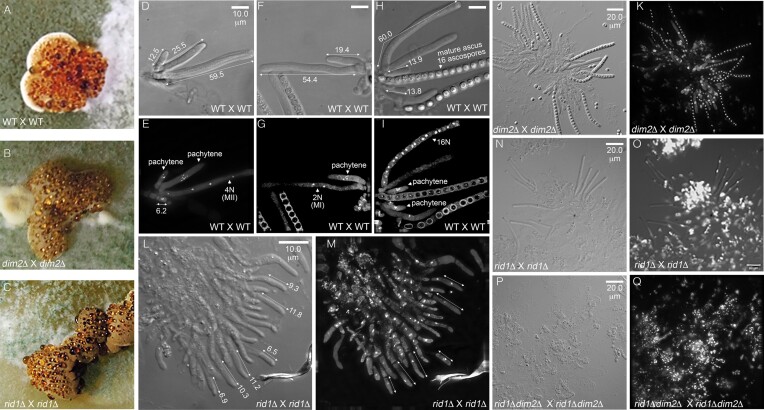
To monitor the process of sexual development, we established a protocol for relatively synchronous crosses under conditions of 25°C and a 12-h light/dark cycle (60,61). In brief, the vegetative ham5+ mycelia [e.g. CBS999.97(MAT1-1, re)] were grown for 4 days on a maltose extract agar (MEA) plate to serve as the female in the subsequent cross. To induce sexual crossing, conidial (asexual spores) suspensions of ham5− (e.g. QM6a) or ham5+ [e.g. CBS999.97(MAT1-2, wt)] were taken as the male and dropped onto the ham5+ female mycelia [e.g. CBS999.97(MAT1-1, re)] at day zero (D0). White primordial stromata began to emerge at day one (D1) after inducing sexual mating. Young and pale-brown stromata first emerged between the first two days (i.e. D1 and D2) after inducing sexual development. Multiple spherical chambers (called perithecia) gradually developed just under the dark-colored stromatal surface (Figure 1A–C), and eventually became flask-shaped cavities hosting an apical opening (60). Asci were observed at the base or on the sides of the perithecium from D3 to D4. Before the first meiotic nuclear division (MI), the zygotes or meiocytes began to develop into long cylindrical asci (50–65 μm in length) (Figure 1D–I). The mature asci with 16 liner ascospores gradually emerged after D5 or D6 and then began to disintegrate after D9 or D10. Eventually, the mature ascospores were forcibly discharged from perithecia via the apical pore or ostiole (60).
Figure 1.
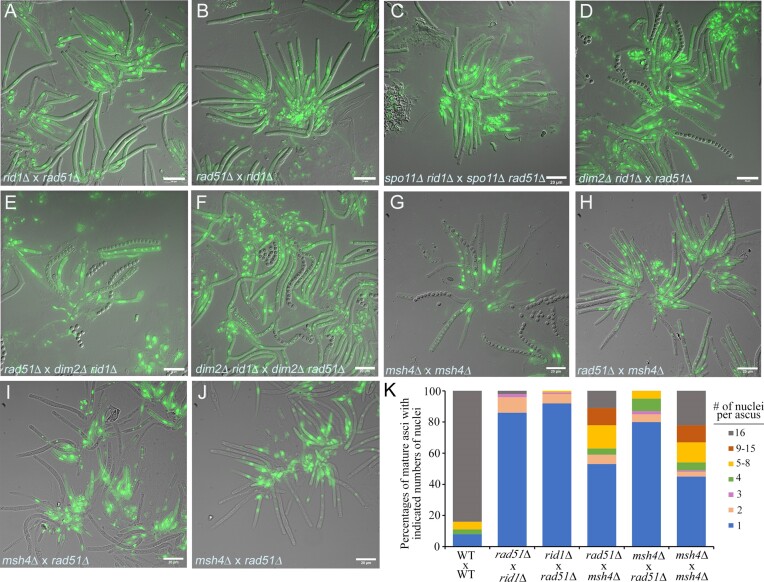
rid1 (but not dim2) is required for normal meiosis. (A–C) rid1 and dim2 are dispensable for sexual mating and the formation of mature stromata. (D–Q) rid1 (but not dim2) is required for normal development of mature ascospores. Rosettes of asci were manually dissected from perithecia, stained with either DAPI or SYTOX™, and then visualized by a DeltaVision Core Imaging System. Differential interference contrast (DIC; D, F, H, L, J, N and P) and DAPI/SYTOX fluorescence images (E, G, H, I, K, M, O and P) are shown. Wild-type (WT; QM6a X CBS999.97(MAT1-1)) (D–I) and dim2Δ (J, K) can generate mature asci (>50 μm in length) with 16 ascospores. Some rid1Δ asci can elongate into mature asci, but they contain only one or two nuclei (L–O). The rid1Δ dim2Δ meiocytes hardly elongate into mature asci (P, Q). Bar = 10 or 20 μm.
spo11 is dispensable for interhomolog recombination and chromosome synapsis in T. reesei meiosis
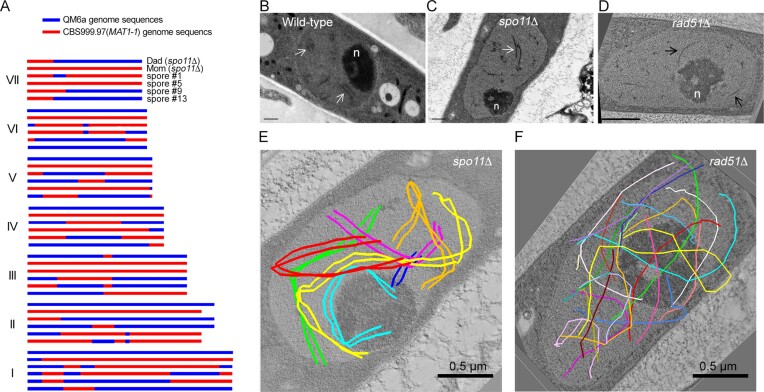
The reference genomes of QM6a(MAT1-1, wt), CBS999.97(MAT1-1, re) and CBS999.97(MAT1-2, wt) all encode a copy of spo11 (Supplementary Figure S1), rad51 and sae2/com1, respectively (57,60). Next, we generated spo11Δ, rad51Δ and sae2Δ haploid mutants by homologous recombination and sexual crossing, which were confirmed by Southern hybridization (Supplementary Figure S2). Herein, we show that spo11 is dispensable for sexual development and meiosis, as homozygous spo11Δ crosses result in the formation of FBs, perithecia, asci, and viable ascospores (Table 1A, Supplementary Figures S3A, S3B and S4). We also show by PacBio Sequel and a fungus-centric software pipeline TSETA (58,59) that a representative spo11Δ meiotic event generated 26 interhomolog meiotic products (19 COs and 7 NCOs) (Figure 2A). The same method was used previously to map 24 COs and 10 NCOs in a representative QM6a/CBS999.97(MAT1-1) meiotic event (11). T. reesei also possesses a sme4 gene that encodes a conserved SC traverse filament protein (11,57,60). Through three-dimensional transmission electron microscopy (3D TEM) imaging experiments, we show that both wild-type (WT) (Figure 2B) and spo11Δ (Figure 2C and E, and Supplementary Movie S1) homozygous meiotic cells can generate fully developed SC.
Figure 2.
spo11 is dispensable for T. reesei interhomolog recombination and chromosome synapsis. (A) Genome-wide mapping of meiotic recombination products in the absence of spo11 using PacBio Sequel technology and the TSETA software tool (59,60). The seven horizontal rows of sequence data represent the seven full-length chromosomes (I to VII) in two parental spo11Δ strains (Dad and Mom) and the four representative F1 progeny. Nucleotide sequences identical to those of the QM6a and CBS999.97(MAT1-1) reference genomes are indicated in blue and red. COs are located where 2:2 markers undergo a reciprocal genotype change. (B–D) Representative TEM images of wild-type (B), spo11Δ (C) and rad51Δ (D) meiotic cells at the pachytene stages. The SCs in WT and spo11Δ are marked by white arrows, whereas axial elements in rad51Δ are marked by black arrows. The enlarged nucleolus (n) is a hallmark of meiotic prophase nuclei. (E, F) 3D-TEM tomography. The seven pairs of lateral elements in spo11Δ (E) and the fourteen axial elements in rad51Δ (F) are highlighted as colored lines. Black bar: 0.5 μm.
rad51 is required for interhomolog recombination and chromosome synapsis in T. reesei meiosis
We observed previously that the majority of rad51Δ asci (>65%) were arrested at meiotic prophase and possessed only one nucleus, even 10 days after mating QM6a rad51Δ with CBS999.97(MAT1-1) rad51Δ. Moreover, approximately one-third of the rad51Δ asci advanced to MI and/or MII, but none of these asci underwent sporulation or formed ascospores (11) (Supplementary Figure S4). Here, we further demonstrate that only unsynapsed axial elements were detected in all 16 rad51Δ meiotic nuclei we examined (Figures 2F, S5–S9, and Supplementary Movie S2). Thus, normal repair of meiotic DSBs is needed for SC assembly in T. reesei, as is the case in S. cerevisiae and S. macrospora (20,32). Notably, in terms of sexual development and meiosis, the spo11Δ rad51Δ mutant is phenotypically similar to rad51Δ (11), as the majority (>60%) of long cylindrical spo11Δ rad51Δ asci (>50 μm in length) have only one nucleus. Both the spo11Δ rad51Δ and rad51Δ mutants failed to generate any mature ascospores (Table 1A and Supplementary Figures S3 and S4).
sae2, like rad51, is required for normal T. reesei meiosis
In most sexual eukaryotes, meiotic DSBs first undergo 5′-to-3′ resection to create 3′ ssDNA tails. The unprocessed DSB precursors (e.g. the covalent Spo11-DSB adducts) accumulate in certain DSB processing-defective and repair-defective mutants, including mre11S, rad50S, as well as the gene deletion mutants sae2Δ/com1Δ (Saccharomyces cerevisiae), ctp1Δ (Schizosaccharomyces pombe) and com1 (plants) (7,8,73). The Mre11 complex and Sae2 also play important roles in modulating the DNA damage checkpoint response during meiosis. In S. cerevisiae, the mre11S, rad50S or sae2Δ/com1Δ mutants display activation of Tel1 (TELomere maintenance 1) (74–80). Tel1 and Mec1 (Mitotic Entry Checkpoint 1) are generally regarded as the budding yeast homologs of ATM (Ataxia Telangiectasia Mutated) and ATR (ATM and RAD3-related), respectively. Mec1 performs most of the kinase-linked functions of both ATM and ATR, and deletion of TEL1 only elicits modest checkpoint phenotypes (81–84). It was reported previously that≥50% of S. cerevisiae sae2/com1 null mutant cells, like rad50S, could complete two rounds of meiotic nuclear divisions, but hardly generated any viable spores (7,8). Since Spo11 is responsible for generating meiotic DSBs in S. cerevisiae, it was reported that the sae2-1 spo11Δ double mutant generates >100-fold more ascospores, a frequency similar to that of the spo11Δ mutant alone (7).
By staining T. reesei asci with the green fluorescent nuclear and chromosome dye SYTOX, we found that the majority of T. reesei sae2Δ and spo11Δ sae2Δ asci underwent meiotic nuclear divisions or even completed one or two rounds of post-meiotic mitosis. Some sae2Δ and spo11Δ sae2Δ asci even formed mature ascospores, but these asci displayed fewer than 16 ascospores (Supplementary Figures S3 and S4). We then applied yeast tetrad dissection technology to analyze 26 sae2Δ asci (232 mature ascospores) and 26 spo11Δ sae2Δ asci (230 mature ascospores). The percentages of viable ascospores in those asci were 9.9% (sae2Δ) and 14.8% (spo11Δ sae2Δ) (Table 1A). These results indicate that T. reesei sae2Δ and spo11Δ sae2Δ exhibited less severe meiotic defects than T. reesei rad51Δ or spo11Δ rad51Δ. The sporulation phenotypes of T. reesei sae2Δ and spo11Δ sae2Δ are similar to that of S. cerevisiae sae2Δ (but not spo11Δ sae2Δ), which accumulates unprocessed DSBs and exhibits a mild meiotic arrest phenotype. For two reasons, T. reesei sae2Δ and spo11Δ sae2Δ mutants can generate more viable spores than S. cerevisiae sae2Δ and rad50S mutants, which hardly produce viable spores (7,8,85). First, S. cerevisiae and T. reesei zygotes possess 16 or 7 pairs of homologous chromosomes, respectively. Thus, T. reesei zygotes may have a much higher probability of euploidy through random chromosome segregation than S. cerevisiae zygotes. Consistent with that notion, >6% viable spores were generated by the rad50S mutants of the fission yeast S. pombe (86), which possesses three pairs of homologous chromosomes. Second, as described above, two rounds of postmeiotic mitosis occur after meiosis in T. reesei but not in S. cerevisiae. Accordingly, the true percentages of viable ascospores generated from meiosis ought to be <2.5% and <3.7% for these sae2Δ and spo11Δ sae2Δ asci, respectively.
In conclusion, T. reesei rad51Δ and rad51Δ spo11Δ mutants are DSB processing-capable but repair-defective, whereas T. reesei sae2Δ and sae2Δ spo11Δ mutants are DSB processing-defective (also see Figures 7 and 8).
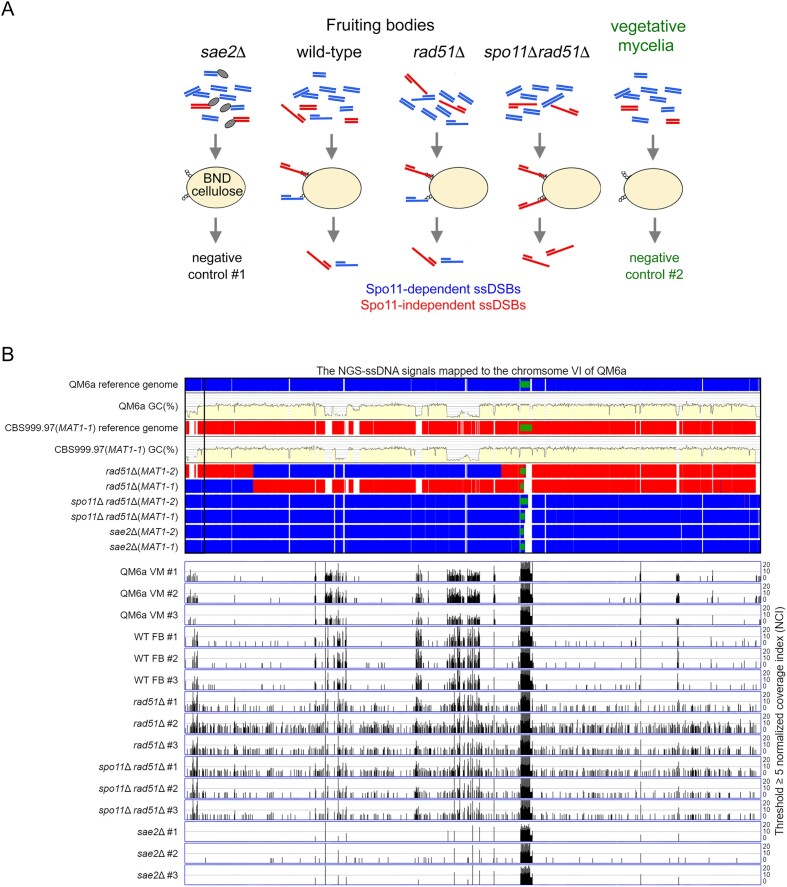
Figure 7.
Genome-wide mapping of ssDNA-associated DSBs (ssDSBs) in wt, rad51Δ and rad51Δ spo11Δ fruiting bodies (FB). (A) Schematic illustrating the ssDNA enrichment procedures using BND cellulose, as described previously (65). The ssDNAs formed in QM6a (WHY00015) × CBS999.97(MAT1-1) (WHY00011), rad51Δ (WTH12867) × rad51Δ (WTH12794) and spo11Δ rad51Δ (WTH12901) × spo11Δ rad51Δ (WTH12902) FBs, as well as those from QM6a (WHY00015) and CBS999.97(MAT1-1) vegetative mycelia, were enriched employing BND cellulose. The ssDNAs isolated from the sae2Δ (WTH13049) x sae2Δ (WTH13040) FBs and from QM6a and CBS999.97(MAT1-1) vegetative mycelia were used as negative controls. All experiments were performed in triplicate(see Materials and methods). (B) The enriched ssDNA peaks along the sixth chromosome were identified using a threshold of ≥ 5 normalized coverage index (NCI) (see Materials and methods). The reference genome sequences of QM6a (WTH0011) and CBS999.97(MAT1-1) (WTH0015) determined by PacBio RSII technology (11) are indicated in red and blue, respectively. The GC contents (window size 500 bp) for the telomere-to-telomere sequence of the sixth chromosome of QM6a are shown in yellow. The near-complete genome sequences of six haploid parental strains were determined by Oxford Nanopore Technology, and their nucleotide sequences identical to the two reference genomes are visualized in blue and red, respectively. The two telomeric sequences and each 45S rDNA array are indicated by black empty and green solid rectangles, respectively. The raw sequencing reads from each sample were mapped to the QM6a genome using the BWA (v0.7.17) alignment tool (66). We used the package BAMscale (v1.0) to quantify the sequencing peaks and normalize the coverage tracks across all samples (67). Using its ‘scale’ function, peak coverages were calculated based on the sum of per-base coverage of reads and then scaled to 1× genome coverage. The resulting values of ‘normalized coverage index’ are referred to as NCI. QM6a and CBS999.97(MAT1-1) vegetative mycelia (VM) and the three sae2 FBs were used as negative controls. All experiments were performed in triplicate.
Figure 8.
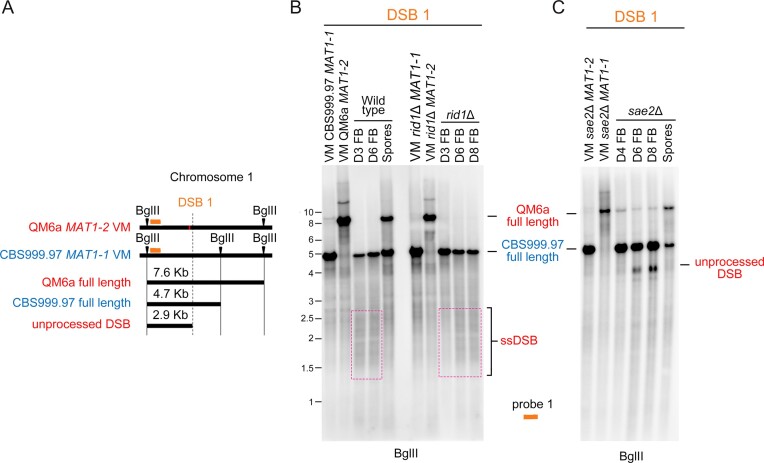
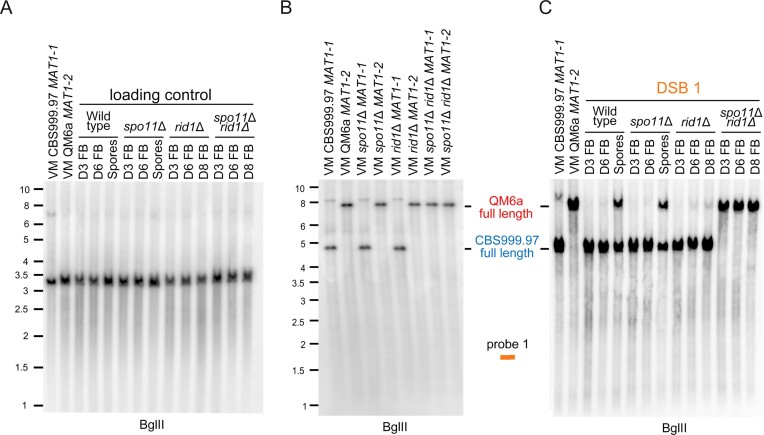
The roles of rid1 and sae2 in initiation and repair of DSB1. (A) Restriction map of DSB1 in the first chromosome of QM6a and CBS999.97(MAT1-1). Polymorphic DNA sequences in QM6a and CBS999.97(MAT11-1) are indicated in red and blue, respectively. The BglII restriction enzyme sites and the two break sites revealed by our BND ssDNA enrichment experiment are indicated by green arrowheads and ‘X’, respectively. After BglII digestion, the expected fragment sizes for the full-length bands, unprocessed DSB, and ssDSB are designated. The location of the DNA probe (red bar) used for Southern hybridization is shown above the chromosomes. (B, C) Southern hybridization of gDNA isolated from two haploid maternal and paternal vegetative mycelia (VM) and the corresponding fruiting bodies (FBs) at indicated days after the initiation of sexual crosses, as well as the mature ascospores released from wild-type and sae2Δ FBs. Visualization/quantification of Southern hybridization band intensity was performed using a BAS-IP MS204 phosphorimaging plate (Cytiva, Japan) and a TyphoonFLA 9000 biomolecular imager (Cytiva, Japan).
rid1 (but not dim2) is essential for T. reesei meiosis
The protein products of T. reesei dim2 and rid1 display highly similar amino acid sequences to their homologs in N. crassa, F. fumigatus, and P. anserina, respectively (Supplementary Figures S10 and S11, Supplementary Table S2). Unlike T. reesei rad51 (11) and sae2 (this study), neither rid1 nor dim2 is required for DNA damage repair during vegetative growth. Specifically, we subjected the conidia of the corresponding haploid mycelia to spot assays with five-fold serial dilutions on malt extract agar (MEA) plates containing no or 0.015% (wt/vol) of the DNA damage agent methyl methanesulfonate (MMS). Only rad51Δ and sae2Δ (but not rid1Δ, dim2Δ or rid1Δ dim2Δ) haploid mutants exhibited slow vegetative growth in the presence of 0.015% MMS (Supplementary Figure S12).
We also found that T. reesei dim2 is dispensable for normal sexual development and meiosis, as homozygous dim2Δ x dim2Δ crosses resulted in well-developed FBs (Figure 1C) that contained many mature asci with viable ascospores (Figure 1J, K and Table 1B). Heterozygous dim2Δ x dim2Δ ridΔ crosses are phenotypically identical to homozygous dim2Δ x dim2Δ crosses (Table 1B). T. reesei rid1 is also dispensable for the formation of well-developed FBs, perithecia, and asci (Figure 1C). However, as revealed by staining with SYTOX, homozygous rid1Δ x rid1Δ crosses often resulted in oblong asci (6–12 μm in length) with one nucleus before D5 (Figure 1I and J). Like homozygous rad51Δ x rad51Δ asci, some homozygous rid1Δ x rid1Δ asci developed into mature asci (>50 μm in length) and underwent one or two rounds of meiotic nuclear divisions (Figure 1L–O). Thus, rid1 (but not dim2) is essential for normal meiosis. Notably, homozygous rid1Δ dim2Δ × rid1Δ dim2Δ crosses resulted in FBs without any elongated asci (Figure 1P and Q), indicating that rid1 is functionally linked to dim2 either before or during early meiosis (also see the Discussion section). Lastly, unlike P. anserina rid (51), T. reesei rid1 is not essential for female fertility, as heterozygous wt × rid1Δ or rid1Δ × wt crosses resulted in no apparent defects in sexual development or meiosis (Table 1B).
dim2 and rid1 differentially mediate genome-wide DNA cytosine methylation and C-to-T hypermutation during vegetative growth and sexual development
Although RIP operates in T. reesei (57), it was previously unknown if T. reesei dim2 and rid1 can mediate DNA cytosine methylation and/or C-to-T mutation during vegetative growth and sexual development. We isolated genomic DNA (gDNA) on the indicated days after sexual crossing [i.e. CBS999.97(MAT1-1; re) × QM6a(MAT1-2; wt), dim2Δ × rid1Δ dim2Δ, rid1Δ × rid1Δ, rid1Δ dim2Δ × rid1Δ dim2Δ], as well as from the vegetative mycelia of seven haploid parental strains, respectively (Table 2 and Supplementary Dataset DS1). Their genome-wide 5mC profiles were revealed by means of genome-wide mapping of the BS+ NGS-seq reads. Genome-wide C-to-T mutations were uncovered by mapping the BS– NGS-seq reads to the indicated parental haploid genomes. To ensure proper mapping of BS− and BS+ NGS-seq reads, we applied ONT to sequence and de novo assemble all corresponding haploid parental genomes. All the near-complete genome sequences and NGS datasets are publicly available at NCBI GenBank (Supplementary Dataset DS1). The genome-wide sequence alignment tool TSETA (58,59) was applied to compare all newly determined haploid genome sequences with those of the QM6a (57) and CBS999.97(MAT1-1) (11) reference genomes, respectively. These two reference genomes were previously determined using PacBio RSII technology and are also publicly available at NCBI GenBank (Supplementary Dataset DS1). Our results reveal that the T. reesei genomes are stable during vegetative growth because the genome sequences determined by PacBio RSII and ONT contain very few allelic variations (Figures 3–5). By cross-referencing against the QM6a reference genome (57), all C-to-T allelic variations in the reference CBS999.97(MAT1-1) genome and the seven haploid parental genomes were excluded for subsequent identification of newly generated C-to-T mutations (Figures 3–5). The genome-wide transcriptional profiles of wild-type and rid1Δ samples were revealed by mapping the RNA NGS-seq reads to the QM6a reference genome (Supplementary Figures S13–S15 and Supplementary Dataset DS1).
Table 2.
Summary of 5mCs, C-to-T mutations and transcriptional profiles in three representative chromosomal segments
| Vegetative growtha | Sexual developmentb | ||||||
|---|---|---|---|---|---|---|---|
| Representative chromosomal segment (RCS) | Chromosomal location in QM6a | Transcription profiles | Long (>500 bps) AT-rich DNA | 5mCs | C-to-T mutations | 5mCs | C-to-T mutations |
| RCS1-GTX | ChXI GTX-BGC ±10 000 bp | Genes upregulated during sexual development | 0 | rare | rare | dim2 | dim2 |
| RCS2-SOR | ChV usk1-SOR-BGC-cel61a ±10 000 bp | Genes downregulated during sexual development | 0 | rare | rare | dim2 | dim2 |
| RCS3-AT containing five QM6a-specific, gene-free, AT-rich sequences | ChV 298991–447710 ±100 000 bp | QM6a-specific, gene-free, AT-rich DNA | 5 | dim2 | rare | dim2 rid1 dimX | dim2 rid1 dimX |
| Non-AT-rich Genes up- or down-regulated during sexual development | 0 | dim2 | rare | dim2 rid1 dimX | dim2 rid1 dimX | ||
aDim2 is responsible for 5mCs at RSC3-AT in wild-type and rid1Δ vegetative mycelia, whereas C-to-T mutations were hardly detected at RCS1-GTX, RCS2-SOR and RCS3-AT in vegetative mycelia.
bOnly Dim2 is responsible for 5mCs at RCS1-GTX and RCS2-SOR in the wild-type and rid1Δ fruiting bodies, whereas Dim2, Rid1 and DimX act redundantly to mediate 5mCs and C-to-T mutations at RCS3-AT in the fruiting bodies.
Figure 3.
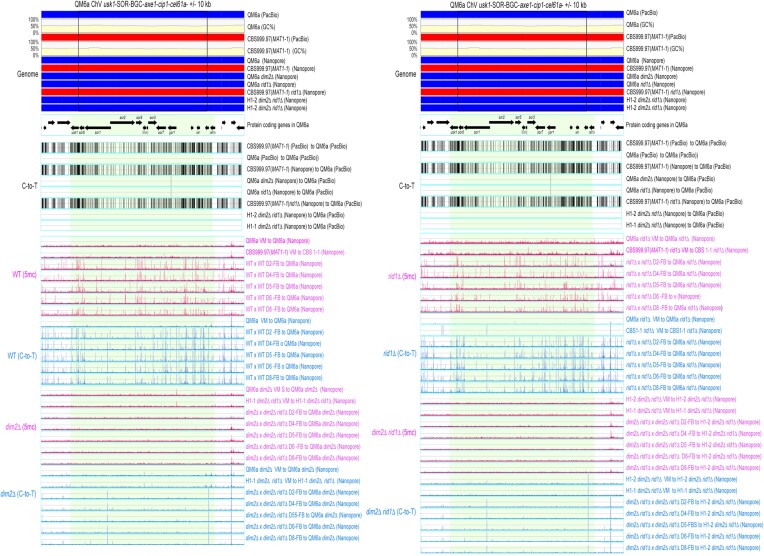
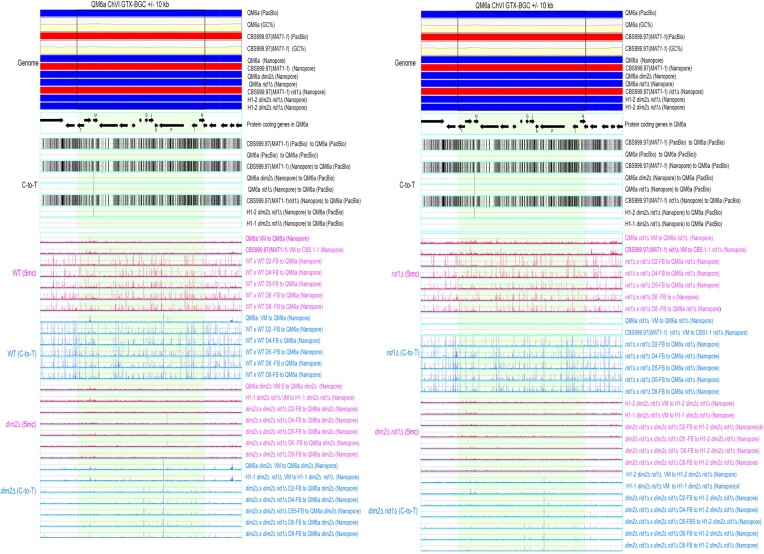
The 5mC (in blue; BS+/NGS) and C-to-T mutation (in pink; BS−/NGS) profiles in RCS1-GTX. RCS1-GTX contains the GTX-BGC chromosomal region and its upstream 10 kb and downstream 10 kb sequences. The two boundaries of GTX-BGC are indicated by two vertical black lines. Genomic DNA was isolated from indicated vegetative mycelia (VM) of the two parental haploid strains, as well as the fruiting bodies (FB) at different days (D2-D8) after initiating sexual crosses of WT (WTH0011) × WT (WTH0015), dim2Δ (WTH13060) × rid1Δ dim2Δ (WTH13147), rid1Δ (WTH13058) × rid1Δ (WTH13059) and rid1Δ dim2Δ (WTH13215) × rid1Δ dim2Δ (WTH13147), respectively (Table 1). The results of BS− and BS+ NGS gDNA-seq were analyzed and visualized using ‘TSETA’ (58,59). The reference genome sequences of QM6a (WTH0011) and CBS999.97(MAT1-1) (WTH0015) determined by PacBio RSII technology (11) are indicated in red and blue, respectively. The GC contents (window size = 500 bp) for the telomere-to-telomere sequence of the sixth chromosome of QM6a are shown in yellow. The protein-coding genes in QM6a were visualized by arrows. The near-complete genome sequences of seven haploid parental strains were determined by Oxford Nanopore Technology, and their nucleotide sequences identical to the two reference genomes are visualized in blue and red, respectively. Compared to the genome sequence of QM6a, all C-to-T allelic variants in the genomes of CBS999.97(MAT1-1) and the seven parental haploid genomes are indicated by vertical black lines and were excluded to enable identification of newly generated C-to-T mutations.
Figure 5.
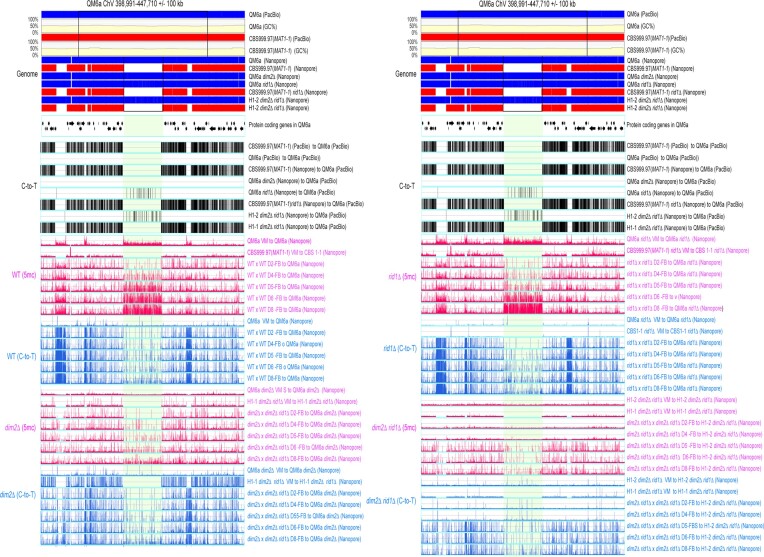
The 5mC (in blue; BS+/NGS) and C-to-T mutation (in pink; BS−/NGS) profiles in RCS3-AT. RCS3-AT contains a long QM6a chromosomal fragment (ChV: 398991–447710 bp) and its upstream 10 kb and downstream 10 kb sequences. The two boundaries of this long QM6a chromosomal fragment are indicated by two vertical black lines. All results were analyzed and visualized as described in Figure 3. The five QM6a-specific, AT-rich, and protein-free DNA sequences are indicated by 1–5.
To illustrate the global impacts of rid1 and dim2 on 5mC, C-to-T hypermutation, and transcription, respectively, the results of three representative chromosomal segments (RCSs)—namely RCS1-GTX, RCS2-SOR and RCS3-AT—are depicted in Figures 3–5 and Supplementary Figures S13–S15, and summarized in Table 1. RCS1-GTX contains the gliotoxin (GTX)-like (gli) biosynthetic gene cluster (BGC) and its upstream and downstream 10-kilobase flanking regions. GTX-BGC in QM6a comprises 13 gli genes (Figure 3) (87) that are transcriptionally up-regulated relative to vegetative mycelia in wild-type and rid1Δ during sexual development (Supplementary Figure S13). RCS2-SOR includes usk1 (unique SOR cluster kinase 1) (88), a T. reesei-specific sorbicillinoid (SOR)-BGC (89), and a well-characterized carbohydrate-active enzyme (CAZ) gene cluster harboring three CAZyme genes: axe1 (acetyl xylan esterase), cip1 (a CBM-containing auxiliary factor), and cel61a (endoglucanase-4) (90) (Figure 4). Unlike those in RSC1-GTX, most of the protein-encoding genes (e.g. sor1, sor2, sor5, ypr1, axe1, cip1 and cel61a) in RCS2-SOR are transcriptionally down-regulated at one or two days after induction of sexual crossing (Supplementary Figure S14). There is no long (>500 bps) AT-rich DNA or strain-specific sequence in RSC1-GTX or RSC2-SOR (Figure 3 and 4). In contrast, the third chromosomal region (i.e. RSC3-AT) harbors five QM6a-specific sequences (referred to as QS1-QS5, respectively) that are highly AT-rich and gene-free (Supplementary Figure S15). Their lengths and nucleotide coordinates in QM6a are QS1 (13096 bp; ChV:17125–330220), QS2 (2934 bp; ChV: 351545–354487), QS3 (1084 bp; ChV: 358556–359639), QS4 (48720 bp; ChV: 398991–447710) and QS5 (6077 bp; ChV: 477058–483134), respectively (Figure 5). Pairwise sequence comparisons using the NCBI BLASTN software tool (word size = 5) revealed that there are more repetitive sequences in QS1 and QS5 than in QS2, QS3 or QS4 (Supplementary Figure S16). We postulate that intensive RIP activity might have occurred in QS2, QS3 and QS4 before QM6a was isolated during WWII. Thus, QS1 and QS5 represent better targets than QS2, QS3 and QS4 for canonical RIP. Unlike those in RCS1-GTX or RCS2-SOR, the protein-encoding genes in RSC3-AT either are constitutively expressed or they are down- or up-regulated after the induction of sexual crosses (Supplementary Figure S15).
Figure 4.
The 5mC (in blue; BS+/NGS) and C-to-T mutation (in pink; BS−/NGS) profiles in RCS2-SOR. RCS2-SOR contains the usk1-SOR-BGC-axe1-cip1-cel61a chromosomal region and its upstream 10 kb and downstream 10 kb sequences. The two boundaries of usk1-SOR-BGC-axe1-cip1-cel61a are indicated by two vertical black lines. All results were analyzed and visualized as described in Figure 3.
These findings provide several important insights (Table 2). First, generally speaking, 5mC marks occur more frequently during sexual development than in vegetative growth. Compared to all non-AT-rich DNA in the three RSCs, the five QM6a-specific DNA sequences (QS1–QS5) possess more 5mC marks during vegetative growth and sexual development. Notably, in vegetative mycelia, C-to-T mutations hardly occurred in all three RSCs regardless of their AT contents, duplicated sequences, and the presence or absence of protein-coding genes (Figures 3–5). These results support the notion that genome-wide C-to-T mutations preferentially occur during sexual development rather than during vegetative growth. Secondly, dim2 (but not rid1) mediates 5mC marking at highly AT-rich chromosomal regions in vegetative mycelia, as well as genome-wide 5mC marks and C-to-T hypermutations in RSC1-TX and RSC2-SOR during sexual development (Figures 3 and 4). Thirdly, dim2 and rid1 act redundantly to mediate 5mC marks and C-to-T hypermutations in RSC3-AT, including nucleotide sequences within, near or distal to QS1–QS5. Residual 5mC marks and C-to-T mutations still occur in rid1Δ dim2Δ × rid1Δ dim2Δ FBs, indicating that T. reesei has at least one novel DNMT gene (which we refer to as ‘dimX’; see also the Discussion section). The hierarchical contribution of these three DNMTs to the formation of 5mC marks and C-to-T hypermutations in RSC3-AT is dim2 > rid1 >> dimX (Figure 5). Finally, loss of rid1 hardly causes any significant changes to the transcriptional profiles of RSC1-GTX, RSC2-SOR or RSC3-AT (Supplementary Figures S13–S15). Thus, neither DNA cytosine methylation nor C-to-T mutations can explicitly explain why rid1 is essential for T. reesei meiosis.
rid1 exhibits locus heterogeneity (LH) with rad51 in meiosis
Since rid1Δ and rad51Δ cause similar meiotic phenotypes (Figure 1), we investigated the genetic relationships between spo11, rad51, sae2, rid1 and dim2 by performing a panel of heterozygous crosses between the corresponding single and double null mutants, respectively (Table 1C). LH describes the scenario whereby mutations in different genes can generate the same disorder (91). Genes displaying LH often encode proteins that mediate or regulate interconnectivity in the human protein interaction network, such as genes linked to hypertrophic cardiomyopathy (92), retinitis pigmentosa (93), Cornelia de Lange syndrome and Bardet–Biedl syndrome (94).
We found that rid1 and rad51 exhibit LH, regardless of the presence or absence of spo11, given that rid1Δ x rad51Δ (Figure 6A), rad51Δ × rid1Δ (Figure 6B), and spo11Δ rid1Δ × spo11Δ rad51Δ (Figure 6C) heterozygous asci all exhibited meiotic defects similar to those of rid1Δ, rad51Δ or rid1Δ rad51Δ homozygous asci (Figure 1). In contrast, LH does not occur in the heterozygous asci of the rid1Δ × spo11Δ, rid1Δ × sae2Δ, dim2Δ × spo11Δ, dim2Δ × rad51Δ or dim2Δ × sae2Δ crosses, respectively (Table 1C). Notably, the LH relationship between rid1 and rad51 can be (partly) suppressed by deleting dim2 from the haploid rid1Δ parental mutants (Figure 6D–F and Table 1), but not by deleting dim2 alone from the haploid rad51Δ parental mutant. Since the rid1Δ dim2Δ zygotes exhibit more severe early meiotic defects than the rid1Δ zygotes and the rid1Δ rad51Δ zygotes (Figure 1P and Q), our results indicate that dim2 and rid1 also share a redundant function that acts earlier than rad51 during meiosis.
Figure 6.
The LH relationship between rad51Δ × rid1Δ or rid1Δ × rad51Δ heterozygous zygotes is regulated by dim2 and partly mediated by msh4. (A–J) Rosettes of asci from indicated sexual crosses were manually dissected from perithecia, stained with SYTOX™, and then visualized by a DeltaVision Core Imaging System. Differential interference contrast (DIC) and DAPI/SYTOX fluorescence images (in green) are shown. Bar = 10 or 20 μm. (K) Percentages of cylindrical asci (≥50 μm in length) with the indicated number of nuclei are shown.
Identification of rid1-dependent differentially expressed genes (DEGs)
The near-complete QM6a genome has >10500 protein-encoding genes (57,60). We identified 199 rid1-dependent DEGs that exhibit statistically significant changes in NGS RNA-seq read counts and/or expression levels/indexes during wild-type and rid1Δ sexual development (see Materials and methods) (Supplementary Dataset DS3). We found that 10 DEGs encode homologs in N. crassa or other filamentous ascomycetes that are involved in meiosis, sexual development or DNA repair. The names and gene identities of their respective N. crassa homologs are msh-4 (NCU10895), rec-8 (NCU03190), rsp (round spore; NCU02764), ts (tan spore; NCU01459), mlh-2 (NCU09373), msh-3 (NCU8115), qde-2 (quelling-defective 2; NCU04730), stk-21 (NCU03242), stk-53 (NCU09064) and per-1 (perithecial-1; NCU03584). Meiotic cohesin subunit Rec8 replaces a mitotic cohesin subunit Scc1 to promote assembly of axial filaments and it constitutes part of the axial filaments generated by almost all eukaryotic species analyzed to date. S. cerevisiae Rec8 is required for both reciprocal recombination and for preventing hyperresection of DSBs during meiosis (95). N. crassa qde-2 regulates an RNA silencing pathway (quelling) during vegetative growth (96). Notably, sms-2, the qde-2 homolog in the cereal pathogen Fusarium graminearum, controls meiosis and subsequent developmental pathways (97). stk-21Δ mutants are defective in the late stages of ascospore development and often produce morphologically abnormal ascospores and fewer than eight ascospores per ascus (98). gin4 is the homolog of stk-53 in the filamentous fungus Aspergillus nidulans, and it is required for early sexual development (99). N. crassa per-1 encodes a polyketide synthetase required for female development (100).
The LH relationship between rid1 and rad51 is not solely regulated by msh4
Among the 192 down-regulated and 7 up-regulated DEGs (Supplementary Dataset DS3), msh4 is the most relevant to the Rad51 protein interaction network (33–38). The msh4 transcriptional profiles in wild-type and rid1Δ FBs (Supplementary Figure S18) are not well correlated with the msh4 DNA methylation profiles revealed by our BS+ NGS-seq experiments (data not shown). Next, we performed RT-qPCR (61) to compare the steady-state levels of msh4 transcripts in the FBs generated by different sexual crosses. As revealed in Supplementary Figure S19 and Supplementary Dataset DS4, the msh4 transcripts were induced after D4 in all six sexual crosses, regardless of the absence of one or both rid1 alleles. The order of the steady-state levels of msh4 transcripts in FBs at D8 is WT × WT > rad51Δ dim2Δ × rid1Δ dim2Δ > rid1Δ dim2Δ × rid1Δ dim2Δ > rid1Δ × rid1Δ > rad51Δ × rid1Δ. These results are partially consistent with the notion that msh4 expression is rid1-dependent and that additional dim2 deletion can rescue msh4 expression.
Next, we generated msh4Δ(MAT1-1) and msh4Δ(MAT1-2) mutants by homologous recombination and sexual crossing, which were confirmed by Southern hybridization (Supplementary Figure S2). We found that sexual crosses of msh4Δ(MAT1-1) with msh4Δ(MAT1-2) or rad51Δ(MAT1-1) with msh4Δ(MAT1-2) resulted in mature asci and viable ascospores (Figure 6G, H and Table 1D). In contrast, sexual crosses of msh4Δ(MAT1-1) with rad51Δ(MAT1-2) resulted in full-elongated asci but hardly any mature ascospores (Figures 6I, J, and Table 1D). Although T. reesei msh4 is dispensable for meiosis and sporulation, it exhibits a moderate LH relationship with rad51. We conclude that rid1-dependent induction of msh4 transcripts alone cannot explain the LH relationship of rid1 and rad51 (see also the Discussion section).
Genome-wide mapping of T. reesei meiotic DSBs
Next, we performed genome-wide mapping of ssDNA-associated DSBs (ssDSBs) using BND cellulose, which selectively binds ssDNA (101) (Figure 7A). This BND-ssDNA methodology has been applied previously to enrich meiotic ssDSBs in S. cerevisiae dmc1Δ and dmc1Δ rad51Δ mutants (65). We applied Illumina NGS technology to sequence the enriched ssDSBs from three replicate experiments of different biological samples, including the vegetative mycelia of QM6a and CBS999.97(MAT1-1), as well as the D4-D6 FBs generated from sexual crosses of QM6a x CBS999.97(MAT1-1), sae2Δ × sae2Δ, rad51Δ × rad51Δ and spo11Δ rad51Δ × spo11Δ rad51Δ, respectively. To ensure proper mapping of NGS reads, we applied ONT to sequence and assemble the near-complete genome sequences of six corresponding haploid mutants (Supplementary Figure S19). These near-complete genome sequences are publicly available in the NCBI databases (Supplementary Dataset DS1). Next, we designed a JavaScript software program, AS-ssDSB-NGS-BAMscale (publicly available on Github: https://github.com/labASIMBTFWang/AS-ssDSB-NGS-BAMScale), to conduct genome-wide normalization and mapping of the NGS reads generated from the BND-ssDNA enrichment experiments (Supplementary Dataset DS2). BAMscale (https://github.com/ncbi/BAMscale) is a versatile bioinformatic tool for accurate quantification of NGS peaks and for generating scaled coverage tracks from several genome-wide datasets, including RNA-seq, chromatin immunoprecipitation sequencing (ChIP-seq) and DNA DSB mapping sequencing (END-seq) (67).
To accurately determine the correct threshold for meiotic ssDSB mapping, normalized NGS-ssDNA signals from both QM6a and CBS999.97(MAT1-1) vegetative mycelia, as well as the sae2Δ FBs, were used as negative controls because meiotic DSBs supposedly do not occur during vegetative growth or are not nucleolytically processed into shorter ssDSBs in the absence of sae2 (see below) (7–9). As revealed in Supplementary Figures S20–S23, we found that a threshold of ≥5 normalized coverage index (NCI) was suitable for genome-wide identification of meiotic ssDSB peaks. Accordingly, we observed that there are more ssDSB peaks in three rad51Δ FBs and three spo11Δ rad51Δ FBs than in three WT FBs, three sae2Δ FBs or six vegetative mycelia (Figures 7 and S19–S22). The ssDSB peaks exist not only within chromosomal regions containing protein-encoding genes (DS5) and intergenic regions (DS6) but those within AT-rich blocks (DS7).
Notably, in all 18 biological samples we examined here, highly concentrated ssDNA signals were observed within the large ribosomal DNA (rDNA) locus, as well as in many highly AT-rich chromosomal regions (with the exception of the three sae2Δ FBs) (Figure 7 and Supplementary Figures S20–S23). The large rDNA locus of QM6a and CBS999.97(MAT1-1) is located on the right arm of their respective sixth chromosome and contains 9 and 11 tandem ‘head-to-tail’ 45S rDNA arrays, respectively. Each 45S rDNA array (∼7.8 kb) contains an 18S–5.8S–26S rRNA gene cluster and an untranscribed intergenic spacer (IGS) (58). The large rDNA locus is an intrinsically unstable genomic region, with high rRNA transcriptional levels often eliciting transcription-replication conflicts known to cause DNA damage and chromosomal rearrangements (102). Our results in Figure 7 and Supplementary Figures S20-S23 reveal that, during both vegetative growth and sexual development, bulk ssDNA signals occur in a sae2-dependent manner on highly AT-rich chromosomal sequences, whereas those located at the large rDNA locus are sae2-independent (see also the Discussion section). Moreover, regardless of the GC contents of chromosomal sequences other than the large rDNA locus, sae2 is required for processing DSBs into ssDNA during meiosis and DNA damage repair during vegetative growth because we detected much fewer ssDSB signals in the three sae2Δ FBs than in the other 15 biological samples. This scenario is also consistent with our results showing that sae2Δ vegetative mycelia are more sensitive to the DNA-damaging agent MMS than wild-type vegetative mycelia (Supplementary Figure S12).
Loss of rid1 impairs meiotic DSB initiation and repair
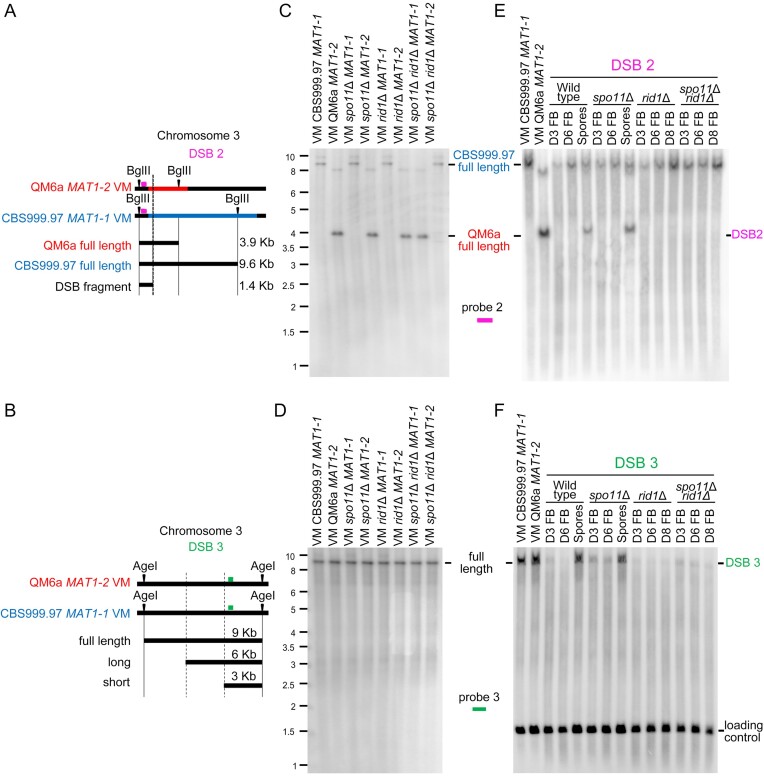
Next, we confirmed three true meiotic DSBs (DSB1–DSB3) by Southern hybridization (Figures 8–10). First, Southern hybridization revealed both haplotype specificity and restriction fragment length polymorphism in DNA sequences containing DSB1 (Figure 8A) and DSB2 (Figure 10A), but not DSB3 (Figure 10B). The full-length DNA fragment containing DSB1 is present in QM6a (MAT1-2), rid1Δ(MAT1-2) (Figure 8B), sae2Δ(MAT1-1) (Figure 8C), spo11Δ(MAT1-2), spo11Δ rid1Δ(MAT1-1) and spo11Δ rid1Δ(MAT1-2) (Figure 9B and C), but not in CBS999.97(MAT1-1), rid1Δ(MAT1-1) (Figure 8B), sae2Δ(MAT1-2) (Figure 8C), or spo11Δ(MAT1-1) (Figure 9B and C). The full-length DNA fragment containing DSB2 exists in QM6a (MAT1-2), spo11Δ(MAT1-2), rid1Δ(MAT1-2) and spo11Δ rid1Δ(MAT1-1), but not in CBS999.97(MAT1-1), spo11Δ(MAT1-1), rid1Δ(MAT1-1) or spo11Δ rid1Δ(MAT1-2) (Figure 10C and E). In contrast, the full-length DNA fragments containing DSB3 in all eight parental haploid strains exhibited the same restriction fragment length (Figure 10D and F).
Figure 10.
The roles of rid1 and spo11 in initiation and repair of DSB2 and DSB3. (A) Restriction map of DSB2 on the third chromosome. Polymorphic DNA sequences in QM6a and CBS999.97(MAT11-1) are indicated in red and blue, respectively. The BglII restriction enzyme sites and the putative break site revealed by BND ssDNA enrichment experiments are indicated by black arrowheads and ‘X’, respectively. (B) Restriction map of DSB1 in the first chromosome of QM6a and CBS999.97(MAT1-1). The AgeI restriction enzyme sites and the two break sites revealed by our BND ssDNA enrichment experiment are indicated by two vertical lines, respectively. After AgeI digestion, the expected fragment sizes for the full-length band, ssDSB-1 (long), and ssDSB-2 (short) are designated. The location of the DNA probe (in green) used for Southern hybridization is shown above the chromosome (C–F) Southern hybridization. After BglII or AgeI digestion, the expected fragment sizes for the full-length DNA bands, DSB2 and DSB3 are designated. The location of the two DNA probes (in pink and orange) used for Southern hybridization are shown above the chromosome. (C, D) Southern hybridization of gDNA isolated from eight haploid maternal and paternal vegetative mycelia (VM). (E, F) Southern hybridization of gDNA isolated from the corresponding fruiting bodies (FBs) harvested at indicated days after the initiation of sexual crosses, as well as the mature ascospores released from the indicated fruiting bodies. After BglII digestion, the smp3 DNA was used as the loading control for Southern hybridization (Supplementary Figure S24A). After AgeI digestion, the act1 DNA was used as the loading control for Southern hybridization (F). Visualization/quantification of Southern hybridization band intensity was performed using a BAS-IP MS204 phosphorimaging plate (Cytiva, Japan) with a Typhoon 5 biomolecular imager (Cytiva, Japan).
Figure 9.
The roles of rid1 and spo11 in initiation and repair of DSB1 (Figure 8A). (A) The smp3 DNA was used as the loading control for Southern hybridization. (B) Southern hybridization of gDNA isolated from eight haploid maternal and paternal vegetative mycelia (VM). (C) Southern hybridization of gDNA isolated from the corresponding fruiting bodies (FBs) at indicated days after the initiation of sexual crosses, as well as the mature ascospores released from the indicated fruiting bodies. Visualization/quantification of Southern hybridization band intensity was performed using a BAS-IP MS204 phosphorimaging plate (Cytiva, Japan) with a Typhoon 5 biomolecular imager (Cytiva, Japan).
Second, the WT, spo11Δ and sae2Δ FBs (but not rid1Δ or spo11Δ rid1Δ FBs) form mature ascospores and eventually discharge mature ascospores after D6. After the gDNA of corresponding samples were digested by BglII, the full-length DNA fragments and the expected length of unprocessed of DSB1 are ∼7.6 and ∼2.9 kb, respectively (Figure 8A). The full-length DNA fragments containing DSB1 were detected in the corresponding WT, rid1Δ and sae2Δ parental vegetative mycelia (Figure 8B, C). DSB1s were initiated and nuclealytically processed into many shorter ssDSBs (shown as weak smear signals with apparent lengths ≤2.9 kb) in the WT FBs at D3 and D6, as well as in the rid1Δ FBs at D6 and D8, but the full-length fragments reappeared only in the ascospores of the WT FBs (Figure 8B). Together with our findings that rid1Δ and rad51Δ display similar meiotic arrest phenotypes and that rid1 exhibits LH with rad51 in meiosis (Figure 1 and Table 1), we conclude that rid1 is indispensable for repair of ssDSBs.
Notably, unprocessed DSB1s were detected in the sae2Δ FBs at D6 and D8, but as ‘bands’ with stronger signals. Because Spo11 or the Spo11-independent DSB inducing enzyme(s) covalently bind to unprocessed DSB1, the apparent length of unprocessed DSB1 (∼4.0 kb) (Figure 8C) is longer than the expected length of unprocessed DSB1 (∼2.9 kb) (Figure 8A). The full-length fragments containing DSB1 reappeared in sae2Δ ascospores (Figure 8C). Since DSB1 is not nucleolytically processed into shorter ssDSBs in the sae2Δ FBs, it is unlikely to be repaired by Rad51-dependent homologous recombination. Instead, the unprocessed DSB1s might be repaired via nonhomologous end joining (NHEJ) or alternative DSB repair pathways in the absence of sae2, explaining why 10–15% viable ascospores were produced in the sae2Δ and sae2Δ spo11Δ FBs, respectively (Table 1).
Third, the full-length DNA fragments containing DSB1, DSB2, and DSB3 detected in the corresponding parental vegetative mycelia, were lacking (i.e. absent due to nucleotlytic processing into many shorter ssDSBs) from the WT and spo11Δ FBs at D3 and D6, as well as the spo11Δ rid1Δ FBs at D3, D6 and D8, but they reappeared in the WT and spo11Δ ascospores (Figures 9 and 10). All three DSBs we examined here were fully repaired in the WT and spo11Δ FBs, but not in the rid1Δ or rid1Δ spo11Δ FBs (Figures 9C, 10E, 10F). Most interestingly, DSB1 was produced in WT, spo11Δ and rid1Δ FBs, but not in those of spo11Δ rid1Δ (Figure 9C).
Lastly, we also observed by Southern hybridization that rid1Δ and rad51Δ elicit the same defects in DSB repair because DSB1 was initiated and then absent from (due to nucleolytic processing) the rad51Δ FBs (Supplementary Figure S24), i.e. as determined for the rid1Δ FBs (Figures 8B and 9C).
In conclusion, all three DSBs are meiosis-specific because they are not detected during vegetative growth and can only be induced through sexual crossing. Our results also reveal at least two distinct types of meiotic DSBs. Initiation of DSB1 requires either spo11 or rid1, whereas initiation of DSB2 and DSB3 is independent of spo11 and rid1. rid1 is also required for Rad51-dependent DSB repair, explaining why rid1, like rad51, is indispensable for meiosis. Therefore, initiation and repair of meiotic DSBs in T. reesei are both epigenetically regulated.
Discussion
In this study, we report that T. reesei DNMTs exert multiple roles during meiosis. First, rid1 is required for Rad51-mediated repair of all three DSBs (DSB1-DSB3) we identified and characterized in this study. Notably, rid1 acts redundantly with spo11 to initiate DSB1 during meiosis. We identified ten rid1-dependent DEGs whose gene products may be involved in meiosis, sexual development, or DNA repair. Our genetic, NGS mRNA sequencing, and RT-qPCR data (Table 1C, Supplementary Figures S18, and S19) reveal that rid1-dependent induction of msh4 transcripts cannot fully explain the LH relationship of rid1 and rad51. Further investigations will reveal whether other rid1-dependent DEG(s) act alone or cooperate with msh4 to facilitate rid1’s essential roles in meiosis.
We also present two lines of evidence supporting that rid1 and dim2 are functionally linked before or during early meiosis (Figures 1F–G, 6D–F, and Table 1C). Since the LH relationship between rid1 and rad51 can only be suppressed by deleting dim2 from the rid1Δ parental mutants, but not by deleting dim2 ‘alone’ from the rad51Δ parental mutants, we theorize that the underlying mechanism(s) may be similar to how RIP operates in N. crassa. Previous studies have revealed that RIP-dependent C-to-T mutations during meiosis of filamentous fungi (e.g. N. crassa) only occur within or close to repetitive sequences. Accordingly, it has been suggested that RIP might be mediated by an efficient and global homology search mechanism because repetitive sequences are recognized irrespective of their sequence, coding capacity, or genomic positions (46,103–105). RIP occurs between fertilization and karyogamy or premeiotic S phase (106,107). Previously, we demonstrated empirically that RIP operates in T. reesei just as it does in N. crassa (57,108). N. crassa rid-1 and dim-2 (but not mei-3/rad51) are involved in the operation of RIP (44,45).
Second, we have shown herein that rid1, dim2 and dimX differentially control genome-wide 5mC and C-to-T mutations during T. reesei sexual development. Methylation-dependent C-to-T mutations are the most abundant single-base changes observed in human cancer cells and also contribute significantly to the appearance of internal mutations in microorganisms (109–113). Our demonstration of ‘genome-wide’ C-to-T hypermutations in T. reesei meiosis but not in its vegetative mycelia provides intriguing perspectives not only for basic research (e.g. genome diversity and evolution), but also for improving economically important strains via sexual crossing. Nevertheless, several important questions still need to be addressed: (a) how does dim2 promote DNA cytosine methylation and C-to-T hypermutation in chromosomal regions [e.g. RCS-GTX (Figure 3) and RCS-SOR (Figure 4)] with low AT contents and far away from repetitive or duplicated sequences; (b) how do C-to-T hypermutations preferentially occur during T. reesei meiosis but not in its vegetative mycelia and do C-to-T mutations occur spontaneously or are they developmentally controlled during meiosis; (c) do 5mC marks or C-to-T hypermutations participate in homology-directed repair of DSBs in the context of hemimethylated, methylated or mismatched genome sequences during meiotic prophase I and (d) what are the evolutionary impacts of meiotic-driven C-to-T hypermutations? Taking a long-term perspective, continuous sexual crosses might induce excessive and wide-ranging C-to-T hypermutations that are deleterious to the genomes. Thus, it would be illuminating to investigate if specialized protective mechanism(s) might have evolved to maintain genome stability or to avoid sexual mating and development. Intriguingly, in this latter case, many Trichoderma spp. natural isolates are known to have become male or female sterile (e.g. T. reesei QM6a) or it is difficult to find compatible mating partners for them in natural environments (54,87,114). Another fundamental question raised by our findings is if T. reesei possesses a third DNMT gene. From the genome-wide annotation results that we reported previously (11,57,60), the T. reesei genome encodes a short polypeptide of 185 amino acids (Supplementary Table S2) that structurally resembles the catalytic domain (amino acid residues 1–179) of human O6-Alklyguanine-DNA alkyltransferase (AGT) or 6-O-methylguanine DNA methyltransferase (MGMT). AGT is an important DNA repair protein that protects mammalian cells from mutagenesis and toxicity arising from alkylating agents. The X-ray crystal structure of the human AGT catalytic domain bound to dsDNA with a chemically modified cytosine base has already been determined (115). Retention of full methyltransferase activity had also been shown for a more extreme truncation variant (residues 1–175) of human AGT (116). We conducted a BLASTP search of the NCBI database and observed that homologs of this polypeptide can be found in S. cerevisiae (Mgt1), N. crassa (NCU11088), Podospora comata (NCBI accession: VBB78373.1), Sordaria sp. MPI-SDFR-AT-0083 (NCBI accession: KAH7625286.1), and several other Trichoderma species (including T. virens, T. harzianum, T. atroviride, T. asperellum, and T. longibrachiatum), but not in S. macrospora or P. anserina (Supplementary Table S2).
Why is T. reesei msh4 dispensable for meiosis and sporulation? It was reported previously that Msh4 and Msh5 belong to the ZMM family of proteins and they form a MutSγ complex that promotes DSB repair and proper SC assembly in budding yeast and several other sexual eukaryotes (e.g. C. elegans, higher plants and mammals) (33–38). Like T. reesei msh4Δ mutants, S. cerevisiae msh4Δ and msh5Δ zygotes can generate some viable spores (spore viability > 30%) (34). Notably, some sexual organisms lack Msh4 and Msh5 homologs. Despite it being a fully sexual species and displaying the ability to form SC, D. melanogaster does not possess several meiosis-specific protein homologs, e.g. Dmc1, Hop2, Mnd1, Hop1, Zip4 and Mer3. The fission yeast S. pombe lacks SC and the entire ZMM machinery (Zip4, Mer3, Msh4 and Msh5 homologs), yet still undergoes normal meiosis. The absence of homologs of these meiosis-specific proteins in these two model organisms can be explained by their lack of the class I CO pathway. Instead, obligatory COs in fly and fission yeast are resolved by alternative pathways; for example, S. pombe relies on the class II (non-interfering) pathway for CO resolution (117,118). From this perspective, T. reesei msh4Δ may also utilize alternative pathways to facilitate the production and/or resolution of obligatory COs. Since T. reesei also possesses several other ZMM-like genes, including sme4, msh5, zip4 and mer3 (Supplementary Table S2), further investigations will likely reveal if their protein products are required for interhomolog recombination and SC assembly in T. reesei.
In this study, we show that meiotic DSBs are initiated via at least two distinct modes (Figures 8 and 9). It will be important to study how rid1 acts redundantly with spo11 to initiate DSB1, and to determine which gene(s) is responsible for initiating spo11- and rid1-independent DSBs (e.g. DSB2 and DSB3) during T. reesei meiosis. It is also imperative to reveal the roles of T. reesei spo11 in regulating the timing of ascus rupture or ascospore release given that spo11Δ homozygous asci undergo rapid disintegration at D9 and D10 after induction of sexual crossing (Supplementary Figure S3B, lower panels).
Spo11 and TopoVIBL form a catalytic complex that is the main initiator of meiotic recombination in most studied sexual eukaryotes (6,119). In S. cerevisiae, Spo11, TopoVIBL (i.e. the Rec102–Rec104 heterodimer), and the antiviral protein Ski8 form the DSB catalytic core complex. The Rec114–Mei4 heterodimer and the Mer2 tetramer bind DNA complexes in a highly cooperative manner and assemble into condensates that might recruit the catalytic core complex to DNA based on interactions observed by yeast two-hybrid assays (120,121). SKI8, Mer2 (ASY2) and REC114 (ASY3), as well as two SPO11 proteins, have been identified and/or characterized in S. macrospora (122–124), but the near-complete genome of T. reesei only possesses a spo11 gene (this study) and a ski8-like gene (Supplementary Table S2). Notably, Spo11-independent DSBs have been identified during meiosis of several eukaryotes. Social amoebae (or Dictyostelids) lack a spo11 gene, but meiotic recombination still occurs at high frequencies through unknown mechanisms (125). Moreover, residual Spo11-independent DSBs and IH-HR proceed in N. crassa wild-type and spo11Δ meiosis (126). Post-meiotic DSBs occur in the unicellular ciliated eukaryote Tetrahymena thermophila and they require DNA topoisomerase II and Spo11 (127). SPO11-induced DSBs are important for the initiation of meiotic sex chromosome inactivation (MSCI) and meiotic silencing of unsynapsed chromatin (MSUC) in mouse meiocytes (128). Transcription of a long noncoding RNA (lncRNA) can induce DSBs and initiate meiotic recombination via local chromatin remodeling in the fission yeast S. pombe (129). During transcription, the nascent RNA and the template dsDNA form an R loop, a peculiar three-stranded RNA–dsDNA hybrid structure. R-loops are well known sources of genome instability from yeast to human because they induce DSBs and/or single-strand breaks (SSBs) (see review in (130)). Studies on mouse have indicated that defective formation of an R loop can cause transcriptional and transcription-coupled DNA damage during meiosis (131). It was reported recently that high levels of active demethylation induce oxidation of 5mCs by ten-eleven translocation enzymes (TETs), with subsequent excision by thymidine DNA glycosylases (TDGs) then enhancing the number of SSBs (132). These SSBs can consequently be converted into DSBs during S phase, i.e. when the broken DNA is unwound by the replication fork. Since our genome-wide BS+ NGS results reveal that active demethylation occurs after the induction of sexual crossing (Figure 3–5), it will be important to determine if TETs and TDGs are involved in generating meiotic DSBs during T. reesi meiosis.
Another intriguing observation arising from the current T. reesei study is that abundant NGS-ssDNA signals at the rDNA locus exist in a sae2-independent manner during vegetative growth and sexual development. In contrast, most (if not all) NGS-ssDNA signals that we detected at other chromosomal regions during vegetative growth and meiosis require sae2 (Figure 7 and Supplementary Figures S20–S23). These results are (at least partly) consistent with a previous study (133) reporting that all 45S rDNA arrays in Arabidopsis thaliana become transcriptionally active and are recruited into the nucleous early in meiosis. The nucleolar rDNA in Arabidopsis thaliana is protected from Spo11-mediated DSBs (133). During meiotic prophase, DSB repair in the 45S rDNA locus requires DNA ligase IV (Lig4) but not Sae2 (Com1) or Rad51. Lig4 is used for canonical nonhomologous end joining (C-NHEJ) but not HR and microhomology-mediated end joining (MMEJ), whereas Sae2 and Rad51 are employed in HR but not C-NHEJ or MMEJ (133). Notably, due to its high transcriptional activity, the 45S rDNA locus is a conserved hotspot from yeast to humans for accumulations of RNA–DNA hybrids (134). Accordingly, we suggest that the abundant ssDNA signals at the 45S rDNA locus that we observed in T. reesei vegetative mycelia and FBs may be due to our use of RNase A during the process of isolating gDNA. RNase A cleaves single-stranded and double-stranded RNA as well as the RNA strand in RNA–DNA hybrids. Taken together, our results presented herein support the notion that T. reesei sae2 is required for genome-wide DSB repair during vegetative growth and sexual development.
In conclusion, given its small genome size and the possibility of isolating and characterizing all 16 ascospores in the asci generated from inbred and hybrid meiosis (56–59,63), our results from this study demonstrate that T. reesei is an ideal model organism to unravel the molecular mechanism of Spo11-independent meiosis and the epigenetic control on meiosis. Most importantly, our discovery of genome-wide 5mC and C-to-T hypermutations in T. reesei meiosis provides unprecedented insights into how sexual reproduction promotes ultrafast and extensive divergence of fungal genomes.
Supplementary Material
Acknowledgements
We thank Shu-Yun Tung (IMB Genomics Core) for NGS sequencing service, Kun-Hai Ye and Yi-Ning Chen (IMB Bioinformatics Core) for statistical help and bioinformatics consulting service, John O′Brien for English editing, Sue-Ping Li (IMB Imaging Core) for help with 3D TEM tomography, and Yu-Tang Huang (IMB Computer Room) for maintaining the computer workstation.
Authors’ contributions: J.S.C., C.C.C., I.C.L., H.C.L., W.C.L., Y.C.C., L.T., C.H.Y., H.N.L., C.I.Y., C.Y.S., W.L.P., R.S.C., Y.P.C., Y.P.H., H.Y. and T.F.W. performed the experiments and analyzed the data. T.F.W. conceived and designed the experiments and wrote the paper. Y.P.H., H.Y. and T.F.W. funding and supervision. All of the authors read and approved the manuscript.
Contributor Information
Lavernchy Jovanska, Institute of Molecular Biology, Academia Sinica, Taipei 11529, Taiwan.
I-Chen Lin, Institute of Molecular Biology, Academia Sinica, Taipei 11529, Taiwan; Chi-Mei Medical Center, Tainan 71004, Taiwan.
Jhong-Syuan Yao, Institute of Molecular Biology, Academia Sinica, Taipei 11529, Taiwan.
Chia-Ling Chen, Institute of Molecular Biology, Academia Sinica, Taipei 11529, Taiwan.
Hou-Cheng Liu, Institute of Molecular Biology, Academia Sinica, Taipei 11529, Taiwan.
Wan-Chen Li, Institute of Molecular Biology, Academia Sinica, Taipei 11529, Taiwan.
Yu-Chien Chuang, Institute of Molecular Biology, Academia Sinica, Taipei 11529, Taiwan.
Chi-Ning Chuang, Institute of Molecular Biology, Academia Sinica, Taipei 11529, Taiwan.
Albert Chen-Hsin Yu, Institute of Molecular Biology, Academia Sinica, Taipei 11529, Taiwan.
Hsin-Nan Lin, Institute of Molecular Biology, Academia Sinica, Taipei 11529, Taiwan.
Wen-Li Pong, Institute of Molecular Biology, Academia Sinica, Taipei 11529, Taiwan.
Chang-I Yu, Institute of Molecular Biology, Academia Sinica, Taipei 11529, Taiwan.
Ching-Yuan Su, Institute of Molecular Biology, Academia Sinica, Taipei 11529, Taiwan.
Yi-Ping Chen, Institute of Molecular Biology, Academia Sinica, Taipei 11529, Taiwan.
Ruey-Shyang Chen, Department of Biochemical Science and Technology, National Chiayi University, Chiayi, Taiwan.
Yi-Ping Hsueh, Institute of Molecular Biology, Academia Sinica, Taipei 11529, Taiwan.
Hanna S Yuan, Institute of Molecular Biology, Academia Sinica, Taipei 11529, Taiwan.
Ljudmilla Timofejeva, Institute of Molecular Biology, Academia Sinica, Taipei 11529, Taiwan; Centre of Estonian Rural Research and Knowledge, J. Aamisepa 1, Jõgeva 48309, Estonia.
Ting-Fang Wang, Institute of Molecular Biology, Academia Sinica, Taipei 11529, Taiwan; Department of Biochemical Science and Technology, National Chiayi University, Chiayi, Taiwan.
Data availability
The raw datasets from PacBio Sequel, Oxford Nanopore and Illumina NGS sequencing approaches, as well as the assembled near-complete haploid genomes, have been deposited in the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/bioproject/) (Dataset DS1). TSETA-2023 (The software program used in Figures 3-7 and S13-S15) is available on Github (https://github.com/tfwangasimb/TSETA-2023) and (https://doi.org/10.5281/zenodo.12561256).
Supplementary data
Supplementary Data are available at NAR Online.
Funding
Institute of Molecular Biology, Academia Sinica, Taiwan, Republic of China (Intramural fund); National Science and Technology Council, Taiwan, Republic of China [NSTC 111-2311-B-001-MY3 to T.F.W.]; Lavernchy Jovanska was supported by a postdoctoral fellowship from National Science and Technology Council, Taiwan, Republic of China [NSTC 112-2811-B-001-021 to T.F.W.]. Funding for open access charge: Academia Sinica.
Conflict of interest statement. None declared.
References
- 1. Lyko F. The DNA methyltransferase family: a versatile toolkit for epigenetic regulation. Nat. Rev. Genet. 2018; 19:81–92. [DOI] [PubMed] [Google Scholar]
- 2. Nai Y.S., Huang Y.C., Yen M.R., Chen P.Y.. Diversity of fungal DNA methyltransferases and their association with DNA methylation patterns. Front. Microbiol. 2021; 11:616922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Walker J., Gao H., Zhang J., Aldridge B., Vickers M., Higgins J.D., Feng X.. Sexual-lineage-specific DNA methylation regulates meiosis in Arabidopsis. Nat. Genet. 2018; 50:130–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gaysinskaya V., Miller B.F., De Luca C., van der Heijden G.W., Hansen K.D., Bortvin A.. Transient reduction of DNA methylation at the onset of meiosis in male mice. Epigenetics Chromatin. 2018; 11:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Keeney S., Giroux C.N., Kleckner N.. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997; 88:375–384. [DOI] [PubMed] [Google Scholar]
- 6. de Massy B. Initiation of meiotic recombination: how and where? Conservation and specificities among eukaryotes. Annu. Rev. Genet. 2013; 47:563–599. [DOI] [PubMed] [Google Scholar]
- 7. McKee A.H., Kleckner N.. A general method for identifying recessive diploid-specific mutations in Saccharomyces cerevisiae, its application to the isolation of mutants blocked at intermediate stages of meiotic prophase and characterization of a new gene SAE2. Genetics. 1997; 146:797–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Prinz S., Amon A., Klein F.. Isolation of COM1, a new gene required to complete meiotic double-strand break-induced recombination in Saccharomyces cerevisiae. Genetics. 1997; 146:781–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nicola Y., Bret H., Cannavo E., Acharya A., Cejka P., Borde V., Guerois R.. Molecular insights into the activation of Mre11-Rad50 endonuclease activity by Sae2/CtIP. Mol. Cell. 2024; 84:2223–2237. [DOI] [PubMed] [Google Scholar]
- 10. Steinfeld J.B., Belan O., Kwon Y., Terakawa T., Al-Zain A., Smith M.J., Crickard J.B., Qi Z., Zhao W., Rothstein R.et al.. Defining the influence of Rad51 and Dmc1 lineage-specific amino acids on genetic recombination. Genes Dev. 2019; 33:1191–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li W.C., Lee C.Y., Lan W.H., Woo T.T., Liu H.C., Yeh H.Y., Chang H.Y., Chuang Y.C., Chen C.Y., Chuang C.N.et al.. Trichoderma reesei Rad51 tolerates mismatches in hybrid meiosis with diverse genome sequences. Proc. Natl Acad. Sci. U.S.A. 2021; 118:e2007192118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Storlazzi A., Xu L., Cao L., Kleckner N.. Crossover and noncrossover recombination during meiosis: timing and pathway relationships. Proc. Natl. Acad. Sci. U.S.A. 1995; 92:8512–8516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. De Muyt A., Jessop L., Kolar E., Sourirajan A., Chen J., Dayani Y., Lichten M.. BLM helicase ortholog Sgs1 is a central regulator of meiotic recombination intermediate metabolism. Mol. Cell. 2012; 46:43–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kleckner N. Meiosis: how could it work. Proc. Natl. Acad. Sci. U.S.A. 1996; 93:8167–8174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lynn A., Soucek R., Borner G.V.. ZMM proteins during meiosis: crossover artists at work. Chromosome Res. 2007; 15:591–605. [DOI] [PubMed] [Google Scholar]
- 16. Berchowitz L.E., Copenhaver G.P.. Genetic interference: don’t stand so close to me. Curr. Genomics. 2010; 11:91–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hunter N. Meiotic recombination: the essence of heredity. Cold Spring Harb. Perspect. Biol. 2015; 7:a016618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. de los Santos T., Hunter N., Lee C., Larkin B., Loidl J., Hollingsworth N.M.. The Mus81/Mms4 endonuclease acts independently of double-Holliday junction resolution to promote a distinct subset of crossovers during meiosis in budding yeast. Genetics. 2003; 164:81–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Holloway J.K., Booth J., Edelmann W., McGowan C.H., Cohen P.E.. MUS81 generates a subset of MLH1-MLH3-independent crossovers in mammalian meiosis. PLoS Genet. 2008; 4:e1000186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zickler D. From early homologue recognition to synaptonemal complex formation. Chromosoma. 2006; 115:158–174. [DOI] [PubMed] [Google Scholar]
- 21. Sym M., Engebrecht J.A., Roeder G.S.. Zip1 is a synaptonemal complex protein required for meiotic chromosome synapsis. Cell. 1993; 72:365–378. [DOI] [PubMed] [Google Scholar]
- 22. Ross-Macdonald P., Roeder G.S.. Mutation of a meiosis-specific MutS homolog decreases crossing over but not mismatch correction. Cell. 1994; 79:1069–1080. [DOI] [PubMed] [Google Scholar]
- 23. Hollingsworth N.M., Ponte L., Halsey C.. MSH5, a novel MutS homolog, facilitates meiotic reciprocal recombination between homologs in Saccharomyces cerevisiae but not mismatch repair. Genes Dev. 1995; 9:1728–1739. [DOI] [PubMed] [Google Scholar]
- 24. Nakagawa T., Ogawa H.. The Saccharomyces cerevisiae MER3 gene, encoding a novel helicase-like protein, is required for crossover control in meiosis. EMBO J. 1999; 18:5714–5723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang T.F., Kleckner N., Hunter N.. Functional specificity of MutL homologs in yeast: evidence for three Mlh1-based heterocomplexes with distinct roles during meiosis in recombination and mismatch correction. Proc. Natl. Acad. Sci. U.S.A. 1999; 96:13914–13919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cheng C.H., Lo Y.H., Liang S.S., Ti S.C., Lin F.M., Yeh C.H., Huang H.Y., Wang T.F.. SUMO modifications control assembly of synaptonemal complex and polycomplex in meiosis of Saccharomyces cerevisiae. Genes Dev. 2006; 20:2067–2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shinohara M., Hayashihara K., Grubb J.T., Bishop D.K., Shinohara A.. DNA damage response clamp 9-1-1 promotes assembly of ZMM proteins for formation of crossovers and synaptonemal complex. J. Cell Sci. 2015; 128:1494–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. De Muyt A., Pyatnitskaya A., Andreani J., Ranjha L., Ramus C., Laureau R., Fernandez-Vega A., Holoch D., Girard E., Govin J.et al.. A meiotic XPF-ERCC1-like complex recognizes joint molecule recombination intermediates to promote crossover formation. Genes Dev. 2018; 32:283–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pyatnitskaya A., Borde V., De Muyt A.. Crossing and zipping: molecular duties of the ZMM proteins in meiosis. Chromosoma. 2019; 128:181–198. [DOI] [PubMed] [Google Scholar]
- 30. Grey C., de Massy B.. Coupling crossover and synaptonemal complex in meiosis. Genes Dev. 2022; 36:4–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pyatnitskaya A., Andreani J., Guerois R., De Muyt A., Borde V.. The Zip4 protein directly couples meiotic crossover formation to synaptonemal complex assembly. Genes Dev. 2022; 36:53–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Espagne E., Vasnier C., Storlazzi A., Kleckner N.E., Silar P., Zickler D., Malagnac F.. Sme4 coiled-coil protein mediates synaptonemal complex assembly, recombinosome relocalization, and spindle pole body morphogenesis. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:10614–10619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Borner G.V., Kleckner N., Hunter N.. Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell. 2004; 117:29–45. [DOI] [PubMed] [Google Scholar]
- 34. Nishant K.T., Chen C., Shinohara M., Shinohara A., Alani E.. Genetic analysis of baker's yeast Msh4-Msh5 reveals a threshold crossover level for meiotic viability. PLoS Genet. 2010; 6:e1001083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nandanan K.G., Salim S., Pankajam A.V., Shinohara M., Lin G., Chakraborty P., Farnaz A., Steinmetz L.M., Shinohara A., Nishant K.T.. Regulation of Msh4-Msh5 association with meiotic chromosomes in budding yeast. Genetics. 2021; 219:iyab102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Colaiacovo M.P., MacQueen A.J., Martinez-Perez E., McDonald K., Adamo A., La Volpe A., Villeneuve A.M.. Synaptonemal complex assembly in C. elegans is dispensable for loading strand-exchange proteins but critical for proper completion of recombination. Dev. Cell. 2003; 5:463–474. [DOI] [PubMed] [Google Scholar]
- 37. Higgins J.D., Armstrong S.J., Franklin F.C., Jones G.H.. The Arabidopsis MutS homolog AtMSH4 functions at an early step in recombination: evidence for two classes of recombination in Arabidopsis. Genes Dev. 2004; 18:2557–2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Neyton S., Lespinasse F., Moens P.B., Paul R., Gaudray P., Paquis-Flucklinger V., Santucci-Darmanin S.. Association between MSH4 (MutS homologue 4) and the DNA strand-exchange RAD51 and DMC1 proteins during mammalian meiosis. Mol. Hum. Reprod. 2004; 10:917–924. [DOI] [PubMed] [Google Scholar]
- 39. Malagnac F., Wendel B., Goyon C., Faugeron G., Zickler D., Rossignol J.L., Noyer-Weidner M., Vollmayr P., Trautner T.A., Walter J.. A gene essential for de novo methylation and development in Ascobolus reveals a novel type of eukaryotic DNA methyltransferase structure. Cell. 1997; 91:281–290. [DOI] [PubMed] [Google Scholar]
- 40. Novikova O.S., Fet V., Blinov A.G.. Homology-dependent inactivation of LTR retrotransposons in genomes of Aspergillus fumigatus and A. nidulans. Mol. Biol. 2007; 41:973–981. [PubMed] [Google Scholar]
- 41. He C., Zhang Z., Li B., Tian S.. The pattern and function of DNA methylation in fungal plant pathogens. Microorganisms. 2020; 8:227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Galagan J.E., Calvo S.E., Borkovich K.A., Selker E.U., Read N.D., Jaffe D., FitzHugh W., Ma L.J., Smirnov S., Purcell S.et al.. The genome sequence of the filamentous fungus neurospora crassa. Nature. 2003; 422:859–868. [DOI] [PubMed] [Google Scholar]
- 43. Kouzminova E., Selker E.U.. dim-2 encodes a DNA methyltransferase responsible for all known cytosine methylation in Neurospora. EMBO J. 2001; 20:4309–4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Freitag M., Williams R.L., Kothe G.O., Selker E.U.. A cytosine methyltransferase homologue is essential for repeat-induced point mutation in Neurospora crassa. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:8802–8807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gladyshev E., Kleckner N.. DNA sequence homology induces cytosine-to-thymine mutation by a heterochromatin-related pathway in Neurospora. Nat. Genet. 2017; 49:887–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Carlier F., Nguyen T.S., Mazur A.K., Gladyshev E.. Modulation of C-to-T mutation by recombination-independent pairing of closely positioned DNA repeats. Biophys. J. 2021; 120:4325–4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Barry C., Faugeron G., Rossignol J.L.. Methylation induced premeiotically in Ascobolus: coextension with DNA repeat lengths and effect on transcript elongation. Proc. Natl. Acad. Sci. U.S.A. 1993; 90:4557–4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rossignol J.L., Faugeron G.. MIP: an epigenetic gene silencing process in Ascobolus immersus. Curr. Top. Microbiol. Immunol. 1995; 197:179–191. [DOI] [PubMed] [Google Scholar]
- 49. Goyon C., Faugeron G.. Targeted transformation of Ascobolus immersus and de novo methylation of the resulting duplicated DNA sequences. Mol. Cell. Biol. 1989; 9:2818–2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lee D.W., Freitag M., Selker E.U., Aramayo R.. A cytosine methyltransferase homologue is essential for sexual development in Aspergillus nidulans. PLoS One. 2008; 3:e2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Grognet P., Timpano H., Carlier F., Ait-Benkhali J., Berteaux-Lecellier V., Debuchy R., Bidard F., Malagnac F.. A RID-like putative cytosine methyltransferase homologue controls sexual development in the fungus Podospora anserina. PLoS Genet. 2019; 15:e1008086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kubicek C.P. Systems biological approaches towards understanding cellulase production by Trichoderma reesei. J. Biotechnol. 2013; 163:133–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Linke R., Thallinger G.G., Haarmann T., Eidner J., Schreiter M., Lorenz P., Seiboth B., Kubicek C.P.. Restoration of female fertility in Trichoderma reesei QM6a provides the basis for inbreeding in this industrial cellulase producing fungus. Biotechnol. Biofuels. 2015; 8:155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Samuels G.J. Trichoderma: systematics, the sexual state, and ecology. Phytopathology. 2006; 96:195–206. [DOI] [PubMed] [Google Scholar]
- 55. Seidl V., Seibel C., Kubicek C.P., Schmoll M.. Sexual development in the industrial workhorse Trichoderma reesei. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:13909–13914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chuang Y.C., Li W.C., Chen C.L., Hsu P.W., Tung S.Y., Kuo H.C., Schmoll M., Wang T.F.. Trichoderma reesei meiosis generates segmentally aneuploid progeny with higher xylanase-producing capability. Biotechnol. Biofuels. 2015; 8:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Li W.C., Huang C.H., Chen C.L., Chuang Y.C., Tung S.Y., Wang T.F.. Trichoderma reesei complete genome sequence, repeat-induced point mutation, and partitioning of CAZyme gene clusters. Biotechnol. Biofuels. 2017; 10:170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Li W.C., Liu H.C., Lin Y.J., Tung S.Y., Wang T.F.. Third-generation sequencing-based mapping and visualization of single nucleotide polymorphism, meiotic recombination, illegitimate mutation and repeat-induced point mutation. NAR Genomics Bioinformatics. 2020; 2:lqaa056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Liu H.C., Li W.C., Wang T.F.. TSETA: a third-generation sequencing-based computational tool for mapping and visualization of SNPs, meiotic recombination products, and RIP mutations. Methods Mol. Biol. 2021; 2234:331–361. [DOI] [PubMed] [Google Scholar]
- 60. Li W.C., Wang T.F.. PacBio long-read sequencing, assembly, and funannotate reannotation of the complete genome of Trichoderma reesei QM6a. Methods Mol. Biol. 2021; 2234:311–329. [DOI] [PubMed] [Google Scholar]
- 61. Chen C.L., Kuo H.C., Tung S.Y., Hsu P.W., Wang C.L., Seibel C., Schmoll M., Chen R.S., Wang T.F.. Blue light acts as a double-edged sword in regulating sexual development of Hypocrea jecorina (Trichoderma reesei). PLoS One. 2012; 7:e44969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Woo T.T., Chuang C.N., Higashide M., Shinohara A., Wang T.F.. Dual roles of yeast Rad51 N-terminal domain in repairing DNA double-strand breaks. Nucleic Acids Res. 2020; 48:8474–8489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Chen C.L., Li W.C., Chuang Y.C., Liu H.C., Huang C.H., Lo K.Y., Chen C.Y., Chang F.M., Chang G.A., Lin Y.L.et al.. Sexual crossing, chromosome-level genome sequences, and comparative genomic analyses for the medicinal mushroom Taiwanofungus Camphoratus (Syn. Antrodia Cinnamomea, Antrodia Camphorata). Microbiol. Spectr. 2022; 10:e0203221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zhou Q., Lim J.Q., Sung W.K., Li G.. An integrated package for bisulfite DNA methylation data analysis with Indel-sensitive mapping. BMC Bioinf. 2019; 20:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Buhler C., Borde V., Lichten M.. Mapping meiotic single-strand DNA reveals a new landscape of DNA double-strand breaks in Saccharomyces cerevisiae. PLoS Biol. 2007; 5:e324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Li H., Durbin R.. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009; 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Pongor L.S., Gross J.M., Vera Alvarez R., Murai J., Jang S.M., Zhang H., Redon C., Fu H., Huang S.Y., Thakur B.et al.. BAMscale: quantification of next-generation sequencing peaks and generation of scaled coverage tracks. Epigenetics Chromatin. 2020; 13:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011; 17:10–12. [Google Scholar]
- 69. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Li B., Dewey C.N.. RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinf. 2011; 12:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Robinson M.D., McCarthy D.J., Smyth G.K.. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010; 26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Yu G., Wang L.G., Han Y., He Q.Y.. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012; 16:284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Keeney S. Mechanism and control of meiotic recombination initiation. Curr. Top. Dev. Biol. 2001; 52:1–53. [DOI] [PubMed] [Google Scholar]
- 74. Usui T., Ogawa H., Petrini J.H.. A DNA damage response pathway controlled by Tel1 and the Mre11 complex. Mol. Cell. 2001; 7:1255–1266. [DOI] [PubMed] [Google Scholar]
- 75. Nakada D., Matsumoto K., Sugimoto K.. ATM-related Tel1 associates with double-strand breaks through an Xrs2-dependent mechanism. Genes Dev. 2003; 17:1957–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. You Z., Chahwan C., Bailis J., Hunter T., Russell P.. ATM activation and its recruitment to damaged DNA require binding to the C terminus of Nbs1. Mol. Cell. Biol. 2005; 25:5363–5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Clerici M., Mantiero D., Lucchini G., Longhese M.P.. The Saccharomyces cerevisiae Sae2 protein negatively regulates DNA damage checkpoint signalling. EMBO Rep. 2006; 7:212–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Carballo J.A., Johnson A.L., Sedgwick S.G., Cha R.S.. Phosphorylation of the axial element protein Hop1 by Mec1/Tel1 ensures meiotic interhomolog recombination. Cell. 2008; 132:758–770. [DOI] [PubMed] [Google Scholar]
- 79. Chuang C.N., Cheng Y.H., Wang T.F.. Mek1 stabilizes Hop1-Thr318 phosphorylation to promote interhomolog recombination and checkpoint responses during yeast meiosis. Nucleic Acids Res. 2012; 40:11416–11427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Cheng Y.H., Chuang C.N., Shen H.J., Lin F.M., Wang T.F.. Three distinct modes of Mec1/ATR and Tel1/ATM activation illustrate differential checkpoint targeting during budding yeast early meiosis. Mol. Cell. Biol. 2013; 33:3365–3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Weinert T.A., Kiser G.L., Hartwell L.H.. Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes Dev. 1994; 8:652–665. [DOI] [PubMed] [Google Scholar]
- 82. Lydall D., Nikolsky Y., Bishop D.K., Weinert T.. A meiotic recombination checkpoint controlled by mitotic checkpoint genes. Nature. 1996; 383:840–843. [DOI] [PubMed] [Google Scholar]
- 83. Mallory J.C., Petes T.D.. Protein kinase activity of Tel1p and Mec1p, two Saccharomyces cerevisiae proteins related to the human ATM protein kinase. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:13749–13754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Corcoles-Saez I., Dong K., Johnson A.L., Waskiewicz E., Costanzo M., Boone C., Cha R.S.. Essential function of Mec1, the budding yeast ATM/ATR checkpoint-response kinase, in protein homeostasis. Dev. Cell. 2018; 46:495–503. [DOI] [PubMed] [Google Scholar]
- 85. Alani E., Padmore R., Kleckner N.. Analysis of wild-type and rad50 mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell. 1990; 61:419–436. [DOI] [PubMed] [Google Scholar]
- 86. Rothenberg M., Kohli J., Ludin K.. Ctp1 and the MRN-complex are required for endonucleolytic Rec12 removal with release of a single class of oligonucleotides in fission yeast. PLoS Genet. 2009; 5:e1000722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Li W.C., Lin T.C., Chen C.L., Liu H.C., Lin H.N., Chao J.L., Hsieh C.H., Ni H.F., Chen R.S., Wang T.F.. Complete genome sequences and genome-wide characterization of Trichoderma biocontrol agents provide new insights into their evolution and variation in genome organization, sexual development, and fungal-plant interactions. Microbiol. Spectr. 2021; 9:e0066321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Beier S., Hinterdobler W., Monroy A.A., Bazafkan H., Schmoll M.. The kinase USK1 regulates cellulase gene expression and secondary metabolite biosynthesis in Trichoderma reesei. Front. Microbiol. 2020; 11:974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Druzhinina I.S., Kopchinskiy A.G., Kubicek E.M., Kubicek C.P.. A complete annotation of the chromosomes of the cellulase producer Trichoderma reesei provides insights in gene clusters, their expression and reveals genes required for fitness. Biotechnol. Biofuels. 2016; 9:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Martinez D., Berka R.M., Henrissat B., Saloheimo M., Arvas M., Baker S.E., Chapman J., Chertkov O., Coutinho P.M., Cullen D.et al.. Genome sequencing and analysis of the biomass-degrading fungus trichoderma reesei (syn. Hypocrea jecorina). Nat. Biotechnol. 2008; 26:553–560. [DOI] [PubMed] [Google Scholar]
- 91. McClellan J., King M.C.. Genetic heterogeneity in human disease. Cell. 2010; 141:210–217. [DOI] [PubMed] [Google Scholar]
- 92. Solomon S.D., Jarcho J.A., McKenna W., Geisterfer-Lowrance A., Germain R., Salerni R., Seidman J.G., Seidman C.E.. Familial hypertrophic cardiomyopathy is a genetically heterogeneous disease. J. Clin. Invest. 1990; 86:993–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Daiger S.P., Sullivan L.S., Bowne S.J.. Genes and mutations causing retinitis pigmentosa. Clin. Genet. 2013; 84:132–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Keith B.P., Robertson D.L., Hentges K.E.. Locus heterogeneity disease genes encode proteins with high interconnectivity in the human protein interaction network. Front. Genet. 2014; 5:434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Klein F., Mahr P., Galova M., Buonomo S.B., Michaelis C., Nairz K., Nasmyth K.. A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell. 1999; 98:91–103. [DOI] [PubMed] [Google Scholar]
- 96. Fagard M., Boutet S., Morel J.B., Bellini C., Vaucheret H.. AGO1, QDE-2, and RDE-1 are related proteins required for post-transcriptional gene silencing in plants, quelling in fungi, and RNA interference in animals. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:11650–11654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Kim H.K., Jo S.M., Kim G.Y., Kim D.W., Kim Y.K., Yun S.H.. A large-scale functional analysis of putative target genes of mating-type loci provides insight into the regulation of sexual development of the cereal pathogen Fusarium graminearum. PLoS Genet. 2015; 11:e1005486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Liu H., Li Y., Chen D., Qi Z., Wang Q., Wang J., Jiang C., Xu J.R.. A-to-I RNA editing is developmentally regulated and generally adaptive for sexual reproduction in Neurospora crassa. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:E7756–E7765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. De Souza C.P., Hashmi S.B., Osmani A.H., Andrews P., Ringelberg C.S., Dunlap J.C., Osmani S.A.. Functional analysis of the Aspergillus nidulans kinome. PLoS One. 2013; 8:e58008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Chinnici J.L., Fu C., Caccamise L.M., Arnold J.W., Free S.J.. Neurospora crassa female development requires the PACC and other signal transduction pathways, transcription factors, chromatin remodeling, cell-to-cell fusion, and autophagy. PLoS One. 2014; 9:e110603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Feng W., Collingwood D., Boeck M.E., Fox L.A., Alvino G.M., Fangman W.L., Raghuraman M.K., Brewer B.J.. Genomic mapping of single-stranded DNA in hydroxyurea-challenged yeasts identifies origins of replication. Nat. Cell Biol. 2006; 8:148–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Korsholm L.M., Gal Z., Nieto B., Quevedo O., Boukoura S., Lund C.C., Larsen D.H.. Recent advances in the nucleolar responses to DNA double-strand breaks. Nucleic Acids Res. 2020; 48:9449–9461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Selker E.U. DNA methylation and chromatin structure: a view from below. Trends Biochem. Sci. 1990; 15:103–107. [DOI] [PubMed] [Google Scholar]
- 104. Gladyshev E., Kleckner N.. Direct recognition of homology between double helices of DNA in Neurospora crassa. Nat. Commun. 2014; 5:3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Gladyshev E., Kleckner N.. Recombination-independent recognition of DNA homology for repeat-induced point mutation. Curr. Genet. 2017; 63:389–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Aramayo R., Selker E.U.. Neurospora crassa, a model system for epigenetics research. Cold Spring Harb. Perspect. Biol. 2013; 5:a017921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Gladyshev E. Repeat-induced point mutation and other genome defense mechanisms in fungi. Microbiol. Spectr. 2017; 5: 10.1128/microbiolspec.FUNK-0042-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Li W.C., Chen C.L., Wang T.F.. Repeat-induced point (RIP) mutation in the industrial workhorse fungus trichoderma reesei. Appl. Microbiol. Biotechnol. 2018; 102:1567–1574. [DOI] [PubMed] [Google Scholar]
- 109. Hollstein M., Sidransky D., Vogelstein B., Harris C.C.. p53 mutations in human cancers. Science. 1991; 253:49–53. [DOI] [PubMed] [Google Scholar]
- 110. Denissenko M.F., Chen J.X., Tang M.S., Pfeifer G.P.. Cytosine methylation determines hot spots of DNA damage in the human P53 gene. Proc. Natl. Acad. Sci. U.S.A. 1997; 94:3893–3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Lutsenko E., Bhagwat A.S.. Principal causes of hot spots for cytosine to thymine mutations at sites of cytosine methylation in growing cells. A model, its experimental support and implications. Mutat. Res. 1999; 437:11–20. [DOI] [PubMed] [Google Scholar]
- 112. Harris R.S. Cancer mutation signatures, DNA damage mechanisms, and potential clinical implications. Genome Med. 2013; 5:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Lewis C.A. Jr, Crayle J., Zhou S., Swanstrom R., Wolfenden R. Cytosine deamination and the precipitous decline of spontaneous mutation during Earth's history. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:8194–8199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Druzhinina I., Kubicek C.P.. Species concepts and biodiversity in Trichoderma and Hypocrea: from aggregate species to species clusters?. J. Zhejiang Univ. Sci. B. 2005; 6:100–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Duguid E.M., Rice P.A., He C.. The structure of the human AGT protein bound to DNA and its implications for damage detection. J. Mol. Biol. 2005; 350:657–666. [DOI] [PubMed] [Google Scholar]
- 116. Daniels D.S., Mol C.D., Arvai A.S., Kanugula S., Pegg A.E., Tainer J.A.. Active and alkylated human AGT structures: a novel zinc site, inhibitor and extrahelical base binding. EMBO J. 2000; 19:1719–1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Sekelsky J.J., Brodsky M.H., Burtis K.C.. DNA repair in Drosophila: insights from the Drosophila genome sequence. J. Cell Biol. 2000; 150:F31–F36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Hofstatter P.G., Ribeiro G.M., Porfirio-Sousa A.L., Lahr D.J.G.. The sexual ancestor of all eukaryotes: a defense of the “Meiosis Toolkit”: a rigorous survey supports the obligate link between meiosis machinery and sexual recombination. Bioessays. 2020; 42:e2000037. [DOI] [PubMed] [Google Scholar]
- 119. Lam I., Keeney S.. Mechanism and regulation of meiotic recombination initiation. Cold Spring Harb. Perspect. Biol. 2014; 7:a016634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Arora C., Kee K., Maleki S., Keeney S.. Antiviral protein Ski8 is a direct partner of Spo11 in meiotic DNA break formation, independent of its cytoplasmic role in RNA metabolism. Mol. Cell. 2004; 13:549–559. [DOI] [PubMed] [Google Scholar]
- 121. Yadav V.K., Claeys Bouuaert C.. Mechanism and control of meiotic DNA double-strand break formation in S. cerevisiae. Front. Cell Dev. Biol. 2021; 9:642737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Storlazzi A., Tesse S., Gargano S., James F., Kleckner N., Zickler D. Meiotic double-strand breaks at the interface of chromosome movement, chromosome remodeling, and reductional division. Genes Dev. 2003; 17:2675–2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Tesse S., Storlazzi A., Kleckner N., Gargano S., Zickler D. Localization and roles of Ski8p protein in Sordaria meiosis and delineation of three mechanistically distinct steps of meiotic homolog juxtaposition. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:12865–12870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Tesse S., Bourbon H.M., Debuchy R., Budin K., Dubois E., Liangran Z., Antoine R., Piolot T., Kleckner N., Zickler D.et al.. Asy2/Mer2: an evolutionarily conserved mediator of meiotic recombination, pairing, and global chromosome compaction. Genes Dev. 2017; 31:1880–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Bloomfield G. Spo11-independent meiosis in social amoebae. Annu. Rev. Microbiol. 2018; 72:293–307. [DOI] [PubMed] [Google Scholar]
- 126. Bowring F.J., Yeadon P.J., Catcheside D.E.. Residual recombination in Neurospora crassa spo11 deletion homozygotes occurs during meiosis. Mol. Genet. Genomics. 2013; 288:437–444. [DOI] [PubMed] [Google Scholar]
- 127. Akematsu T., Fukuda Y., Garg J., Fillingham J.S., Pearlman R.E., Loidl J.. Post-meiotic DNA double-strand breaks occur in Tetrahymena, and require Topoisomerase II and Spo11. eLife. 2017; 6:e261176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Carofiglio F., Inagaki A., de Vries S., Wassenaar E., Schoenmakers S., Vermeulen C., van Cappellen W.A., Sleddens-Linkels E., Grootegoed J.A., Te Riele H.P.et al.. SPO11-independent DNA repair foci and their role in meiotic silencing. PLoS Genet. 2013; 9:e1003538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Senmatsu S., Asada R., Oda A., Hoffman C.S., Ohta K., Hirota K.. 2021) lncRNA transcription induces meiotic recombination through chromatin remodelling in fission yeast. Commun. Biol. 4:295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Rinaldi C., Pizzul P., Longhese M.P., Bonetti D. Sensing R-loop-associated DNA damage to safeguard genome stability. Front. Cell Dev. Biol. 2020; 8:618157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Fujiwara Y., Handel M.A., Okada Y.. 2022) R-loop formation in meiosis: roles in meiotic transcription-associated DNA damage. Epigenomes. 6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Wang D., Wu W., Callen E., Pavani R., Zolnerowich N., Kodali S., Zong D., Wong N., Noriega S., Nathan W.J.et al.. Active DNA demethylation promotes cell fate specification and the DNA damage response. Science. 2022; 378:983–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Sims J., Copenhaver G.P., Schlogelhofer P.. Meiotic DNA repair in the nucleolus employs a nonhomologous end-joining mechanism. Plant Cell. 2019; 31:2259–2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Vydzhak O., Luke B., Schindler N.. Non-coding RNAs at the eukaryotic rDNA locus: RNA-DNA hybrids and beyond. J. Mol. Biol. 2020; 432:4287–4304. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw datasets from PacBio Sequel, Oxford Nanopore and Illumina NGS sequencing approaches, as well as the assembled near-complete haploid genomes, have been deposited in the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/bioproject/) (Dataset DS1). TSETA-2023 (The software program used in Figures 3-7 and S13-S15) is available on Github (https://github.com/tfwangasimb/TSETA-2023) and (https://doi.org/10.5281/zenodo.12561256).