Abstract
Telomere length (TL) is increasingly recognized as a molecular marker that reflects how reproductive aging affects intergenerational transmissions. Here, we investigated the effects of parental age on offspring survival and the regulation of TL by examining the telomere-elongating activity of telomerase in the Pacific oyster. We assessed the classical hallmarks of aging in parents at three age classes (young, middle-aged, and old) and crossbred them using a split-brood design to examine the consequences of the nine maternal-by-paternal age combinations on their offspring. Reproductive aging leads to increased larval mortality and accelerated telomere shortening in spats, rendering them more susceptible to infection by the Ostreid herpesvirus. Viral exposure stimulates telomerase activity, a response that we identified as adaptive, but weakened by parental aging. While telomerase lengthens a spat’s telomere, paradoxically, longer individual TL predicts higher mortality in adults. The telomerase-telomere complex appeared as a conservative biomarker for distinguishing survivors and losers upon exposure to polymicrobial diseases.
Elderly oysters limit growth and life chance of their offspring by altering the cellular machinery of aging.
INTRODUCTION
The aging capital of an organism is determined by its genetic makeup, yet it is largely influenced by complex interactions between epigenetics and environmental factors, which can either accelerate or shorten the pace of development and life expectancy. Among the diversity of cellular factors dictating an individual’s aging rate, cytogenetic abnormalities and metabolic changes shape the accumulation of cellular damage, mitochondrial dysfunctions, loss of proteostasis, and telomere shortening (1–3). Over time, these aging-related processes may compromise an individual’s fitness and survival prospects, alongside what is known as reproductive senescence (4, 5). In most species, advancing age correlates with a decline in gamete quality (sperm motility, DNA fragmentation, and aneuploidy), which may have detrimental trans-generational effects on offspring fitness (6). However, characterizing the influence of parental age on offspring traits remains challenging due to the inherent complexity of species biology. Factors such as sex-specific responses between daughters and sons, variations in the degree of parental care, and multi-pairing reproductive systems often confound clear generalizations regarding alterations encoded by parental age at conception (7). Addressing these limitations requires the use of simple model organisms (e.g., sexually undetermined without parental care) to interrogate factors affecting the influence of reproductive aging on offspring survival.
In recent years, studies on telomeres in evolutionary ecology have provided valuable insights into such intergenerational transmissions (8–10), particularly in quantifying the heritability of aging rate among populations (11). Telomeres, located at the ends of eukaryotic chromosomes, consist of nucleoproteins that encapsulate and protect coding DNA from oxidative damage (12). The inevitable shortening of telomeres with each cell division makes telomere length (TL) a well-established hallmark of aging (1–3, 12). Moreover, telomeres can further shrink due to intrinsic oxidative stress (13, 14) and extrinsic environmental factors (15, 16). The rate of individual telomere attrition over time serves as an indicator of cellular aging and can potentially predict remaining life expectancy (17) and the lifetime of an organism’s reproductive success (18). Early in life, offspring inherit TL phenotypes directly from their parents, but the heritability of TL within populations varies between species (19). Recent investigations on invertebrates have shown high values of TL heritability in field crickets (20). This questions the potential of TL to evolve under selection. Here, we investigated whether telomeres may carry parental effects to the next generation of offspring and underscore the heritable role of telomeres in reproductive aging (21).
The telomeres lost throughout life can be replenished by the regenerating action of telomerase (12, 22), a unique ribonucleoprotein complex composed of a reverse transcriptase, a noncoding RNA template, and multiple associated proteins (12). Telomerase acts by aligning on telomere motifs (5′-TTAGGG-3′)n at the end of chromosomes, which catalyzes their regeneration (23), particularly during gamete formation and early embryonic development. While telomerase activity can be highly active in these contexts, it is also observed in the vast majority of cancers (24, 25), where it facilitates cell immortalization and indefinite cancer cell replication (12, 26, 27). In addition, telomerase activity represents a homeostatic cellular response to multiple oncogenic virus strains (28, 29). However, characterization of this cellular response remains largely restricted to in vitro studies, emphasizing the need for investigating it in controlled in vivo systems.
In this study, we investigated the in vivo impact of reproductive aging on the telomerase-telomere complex in the Pacific oyster (Crassostrea gigas) and explored their narrow-sense heritability and association with offspring resistance to the Ostreid herpesvirus 1 microvariant (OsHV-1). OsHV-1 infection, which has caused extensive mortality in juvenile oyster populations worldwide since 2008, results in hemocyte immune exhaustion leading to opportunistic bacteremia and death (30). Previous research has shed light on drivers that can aggravate this polymicrobial disease (31, 32), including sea water temperature ranging between 16° and 24°C and high food concentrations used to maximize metabolism (33). Notably, susceptibility to OsHV-1 exhibits high levels of narrow-sense heritability (h2), ranging from h2 = 0.65 to h2 = 0.99 (34), and is largely influenced by epigenetic variations, genetic differences and their interactions (35). Given that parental age modulates epigenetic control of gene expression, we hypothesized that increasing parental age would negatively affect reproductive performance and offspring survival. To test this hypothesis, we crossbred adult oysters of varying age categories (fig. S1) and performed whole-organism proteomic profiling to map common hallmarks of aging. Subsequently, we assessed the offspring life history traits and survival in relation to parental age during successive life stages (larval, spat and young adults; fig. S2). This included developmental success, growth rate, survival, and resistance to OsHV-1. In addition, we explored whether these offspring phenotypic traits were heritable and potentially modulated by telomere dynamics and telomerase activity. Last, we conducted a data mining analysis of gene expression related to telomerase activity to elucidate family-wide susceptibility to OsHV-1 experimental infection. Our data reveals that reproductive aging has a notable effect on larval mortality and telomere erosion in spats, making them more susceptible to OsHV-1 infection. These results underscore the potential for using heritable telomerase-telomere complex as a biomarker for predicting outcomes of polymicrobial infection.
RESULTS
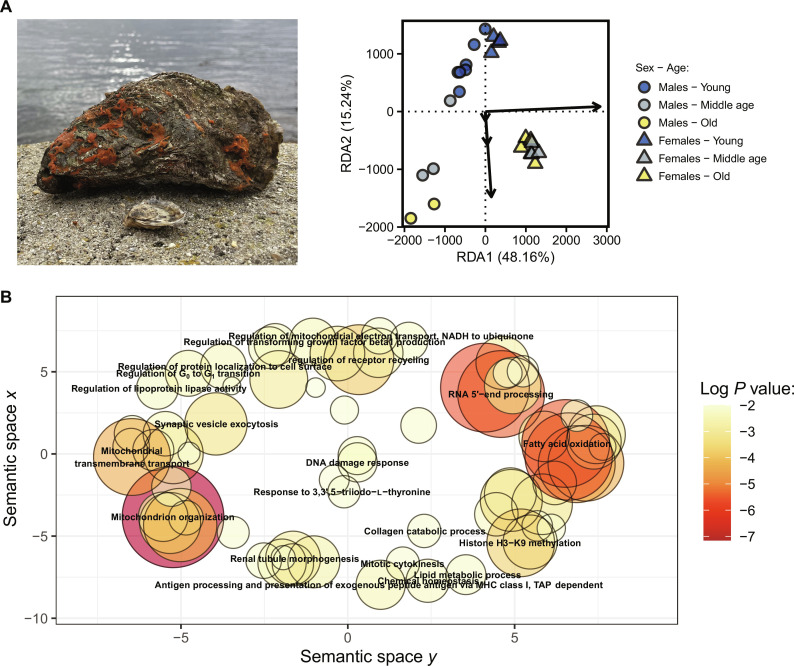
Comparative proteomic confirms hallmarks of parental aging
To assess the impact of parental aging on reproductive success and offspring survival, we crossbred 24 adult oysters (12 females and 12 males) across three age classes (young, middle-aged, or old; fig. S1) to produce 36 full-sibling and 72 half-sibling family combinations (fig. S1). We then performed whole-individual parental proteome profiling. Factors sex and age explained 59.90% of the total variation in oyster proteomic profiles (Fig. 1A). Sex-specific proteomes explained most of the variance, but the clusters of co-expressed proteins (hereafter called “modules”) associated with sex effect were not significantly associated with age class effect, except for a visible trend for module “Brown” (fig. S3). This suggests that aging effects on proteome pathways are conserved across sexes. Neither TL nor telomerase activity differed between young, middle-aged, and old parents (all, P > 0.471). This is consistent with previous cross-sectional comparisons in the ocean quahog clam (Arctica islandica), which is one of the longest-lived animals able to reach ages of ~500 years (36). However, gene ontologies (GOs) that are associated to telomere regulation and telomerase (fig. S4A) or to DNA damage and oxidative stress (fig. S4B) were identified to be within the same modules. This confirms that there are functional interconnections between these biological processes (13, 14). Although functionally related, the use of TL alone as an index of chronological age is increasingly criticized due to several factors. These include high intra-and inter-species variation in TL, methodological issues in telomere assay protocol or storage conditions, and publication biases (37). These issues, however, were not completely circumvented by our sampling design where we used cross-sectional measures of individuals at different ages. That is, we could not avoid previous selective disappearance of individuals with short TL. Besides, contrary to most evidences in vertebrates, TL may not decrease during aging in adult bivalves. In support of this alternative hypothesis, we found that distribution of TL among adults were slightly biased to high TL as compared to those measured in spats of less than 1 year old (fig. S5). However, we cannot draw any firm conclusions regarding the TL-age relationship given the paucity of studies on the age dependence of TL among adult shellfishes [but see (36)].
Fig. 1. Global parental proteomic profiles of Pacific oyster compared by sex and age class.
(A) Protein expression differs between sex and age in this species (n = 24 oysters) with sex and age class accounting for 70.07 and 14.18% of the inter-individual variance explained by the model [permutation analysis of variance (ANOVA); P < 0.001], respectively. (B) Scatterplot of the most significant biological processes increasing in oyster growing old, regardless of their sex. NADH, reduced form of nicotinamide adenine dinucleotide (oxidized form). TAP, antigen peptide transporter.
Here, our main goal was to focus on the intergenerational impacts of reproductive aging and to use comparative proteomics to confirm that old oysters endured the hallmarks of aging. As expected, aging-related phenotypes were confirmed by the enrichment of gene ontologies (176 GOs in old oysters), which included mitochondrial dysfunction, increased lipid oxidation, histone methylation, DNA damage response, and loss of perceptive receptors (Fig. 1B). Most of these functions can be attributed to the classical hallmarks of aging among species (3). Collectively, these proteomic analyses confirm that aging alters multiple traits of reproductive parents.
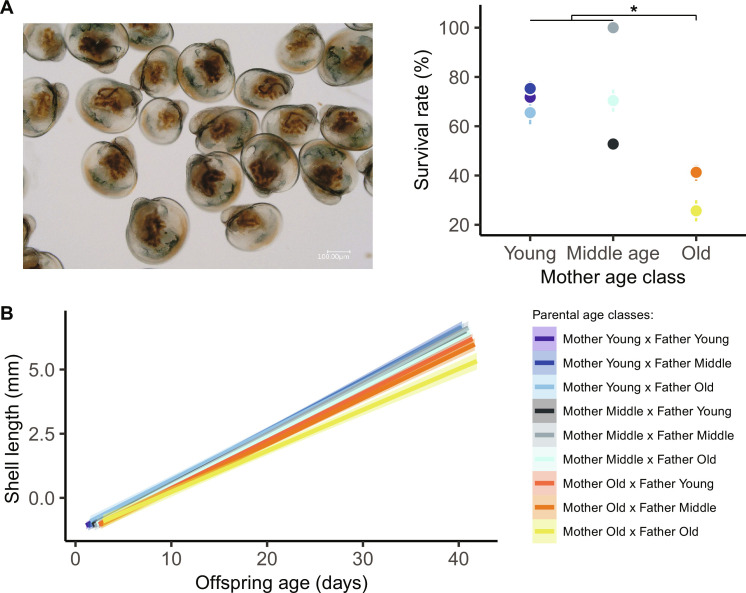
Parental aging diminishes offspring survival in early life
We next sought to investigate the impact of parental aging on offspring survival. To this end, we surveyed the developmental success of larvae and spats, including their survival and shell growth rates. Maternal aging significantly affected larval survival at 2 weeks [Generalized Linear Mixed Model (GLMM): χ2 = 11.8, df = 2, P = 0.003], regardless of paternal age (GLMM: χ2 = 4.7, df = 2, P = 0.097) and interaction term (GLMM: χ2 = 5.7, df = 4, P = 0.226), when pediveliger larvae metamorphose and become sessile (fig. S2). Larvae born from old mothers exhibited lower survival rates (39%) than those from young (72%) and middle-aged (71%) dams (all pairwise post hoc tests: z > 2.5, P < 0.026; Fig. 2A). From 2 to 40 days after fertilization, growth rates were slower among offspring born from old mothers (GLMM: χ2 = 9.8, df = 2, P = 0.007). This trend was exacerbated when larvae were spawned from two old parents (GLMM interaction term: χ2 = 42.2, df = 4, P < 0.001; Fig. 2B); however, this trait was not heritable [GLMMs using Markov chain Monte Carlo (MCMCglmm): h2 = 0.00, 95% confidence interval (CI): 0.00 to 0.01; Table 1]. Similar trends in oyster growth occurred during the spat life stage (from 40 to 365 days old) where the negative effects of maternal and paternal age on body size changes were evident and not heritable (fig. S6A). During their first winter and spring, spat survival remained above 95% (fig. S6B), which was independent of parental age (P > 0.173). In agreement with our hypothesis, these findings indicate that parental aging, especially when two parents were old, negatively alters offspring developmental success in the earliest developmental stages.
Fig. 2. Reproductive aging alters offspring survival and growth rates during larval development.
(A) At the time of metamorphosis (14 days old), survival rate falls in pediveliger larvae (n = 7217) born from old mothers compared to offspring from young or middle-aged mothers (means ± SE, all, *P < 0.026) regardless of paternal age. (B) Using high-resolution microscopy, we highlighted that larval growth rate (n = 6806) is negatively influenced by the interactive effect of maternal and paternal age [solid and color lines, model predictions surrounded by 95% confidence intervals (CIs)].
Table 1. Impacts of parental aging and estimation of narrow-sense heritability on offspring traits in the Pacific oyster (C. gigas).
Table summarizes the main effect of maternal age, paternal age, and their interaction (−, negative effect; n.s., nonsignificant effect) on the F1 generation at multiple life stages, in larvae (2 to 40 days old), spats (40 to 365 days old), and young adults (>365 old; see timelines of sampling in fig. S2). It also reports the narrow-sense heritability (h2) values surrounded by the 95% confidence interval (CI).
| Offspring life stage | Offspring response | n | Maternal age | Paternal age | Interaction between parental ages | h 2 | 95% CI |
|---|---|---|---|---|---|---|---|
| Larvae | Growth rate | 6806 | − | − | − | 0.00 | 0.00–0.01 |
| Spat | Growth rate | 5042 | n.s. | − | n.s. | 0.00 | 0.00–0.01 |
| Larvae | Survival rate | 7217 | − | n.s. | n.s. | 0.10 | 0.00–0.28 |
| Spats | Resistance to infectious disease (OsHV-1) | 4325 | − | − | n.s. | 0.63 | 0.37–0.86 |
| Adults | Resistance to infectious disease (Vibriosis) | 357 | n.s. | n.s. | n.s. | 0.11 | 0.00–0.31 |
| Spats | Intrinsic ΔTL | 443 | − | − | n.s. | 0.53 | 0.35–0.72 |
| Extrinsic ΔTL | 501 | n.s. | n.s. | n.s. | 0.55 | 0.30–0.85 | |
| Intrinsic ΔTelomerase | 72 | n.s. | n.s. | n.s. | 0.09 | 0.00–0.31 | |
| Extrinsic ΔTelomerase | 72 | − | − | n.s. | 0.07 | 0.00–0.25 |
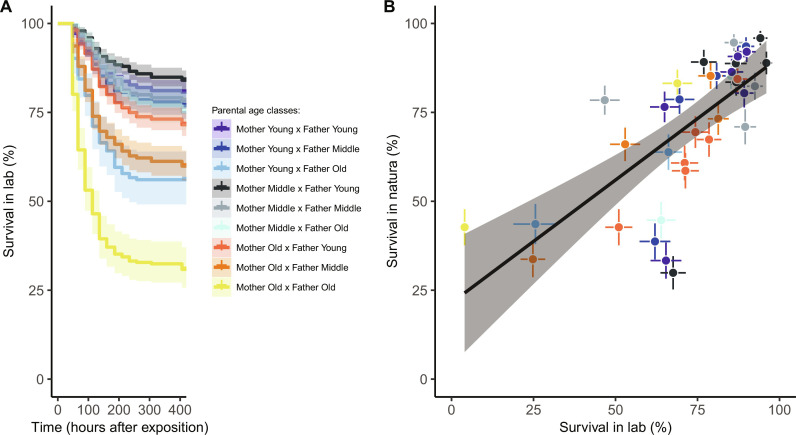
Under controlled laboratory conditions (i.e., seawater at 21°C and addition of food as ad libitum), maternal and paternal ages influenced offspring resistance to OsHV-1 equally [Cox proportional hazards models containing mixed effects (COXME): maternal age: χ2 = 10.1, df = 2, P = 0.006; paternal age: χ2 = 10.8, df = 2, P = 0.005]. The presence of one old parent decreased survival rates in spats, and, with an old-old parent combination, mortality rates were maximized (Fig. 3A). We detected DNA of OsHV-1 in 100% of moribund oysters from the 36 familial pools, with a viral load range from 2.5 × 104 to 3.5 × 105 viral DNA copies. When replicating this experiment in wild conditions, we confirmed that family differences surpassed environmental context, as the survival rates to OsHV-1 in this setting were positively correlated with laboratory results [Linear Model (LM): F1,34 = 36.1, P < 0.001, coefficient of determination (r2) = 0.50; Fig. 3B]. Of note is the susceptibility to OsHV-1, which was moderately heritable with h2 = 0.63 (MCMCglmm: 95% CI, 0.37 to 0.86; Table 1) falls within the range of previous estimations for this host-pathogen interaction (34).
Fig. 3. Negative impacts of parental age on spat oyster survival to OsHV-1 challenge.
(A) In the laboratory, we optimized conditions for viral replication (seawater at 21°C, food ad libitum) and found lower survival chance in offspring (n = 4325) from both old parents. (B) Offspring exposed to natural conditions (n = 3407) in Bay of Brest had similar mortality risks correlated with laboratory measures (dot colors, means ± SE of familial average; solid line, model predictions surrounded by 95% CIs).
Dynamics of offspring telomerase-telomere complex with and without pathogenic pressure
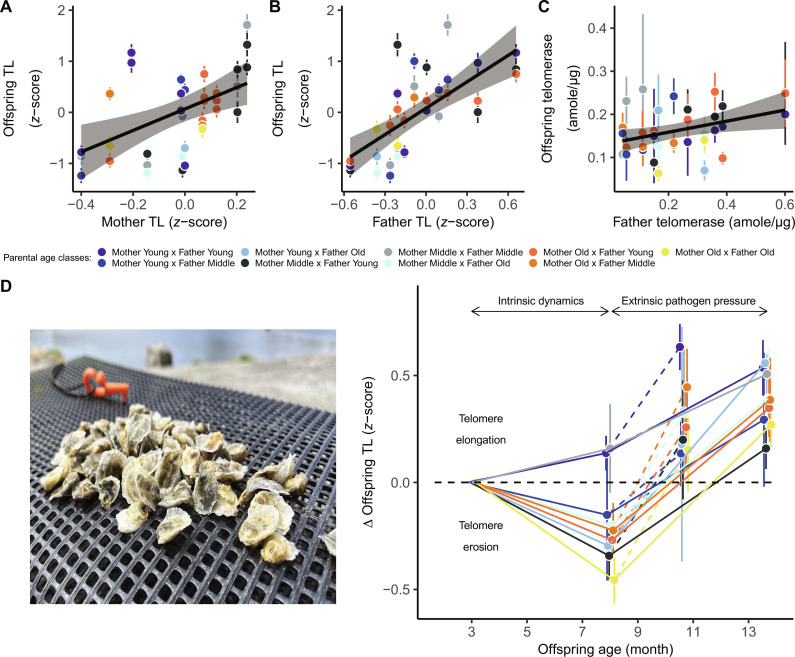
We tested the impact of reproductive aging on offspring telomerase-telomere complex by checking inter-familial changes in TL and telomerase activity. During the first 8 months of life without pathogenic selective pressure, we found that spat TL was moderately heritable (MCMCglmm: h2 = 0.53; 95% CI, 0.35 to 0.72; Table 1) and telomerase activity was poorly heritable (MCMCglmm: h2 = 0.09; 95% CI, 0.00 to 0.32; Table 1). Offspring TL was highly correlated with both maternal TL (GLMM: χ2 = 11.8, β = 1.78 ± 0.52, P < 0.001, r2 = 0.25; Fig. 4A) and paternal TL (GLMM: χ2 = 23.6, β = 1.30 ± 0.27, P < 0.001, r2 = 0.43; Fig. 4B), whereas offspring telomerase was slightly correlated with father telomerase (GLMM: χ2 = 5.3, β = 0.12 ± 0.06, P = 0.022, r2 = 0.07; Fig. 4C). TL was positively correlated with offspring shell length (GLMM: χ2 = 5.6, P = 0.018), a pattern also seen in other bivalves and interpreted as marker of individual quality. Over time, offspring TL covaried in interaction between chronological age and both maternal (GLMM: χ2 = 7.1, df = 2, P = 0.029) and paternal age (GLMM: χ2 = 6.1, df = 2, P = 0.047), reflecting greater telomere erosion as a function of reproductive aging (Fig. 4D). Thus, offspring experienced significant telomere erosion from families composed from either old mothers and/or old fathers (3 versus 8 months old; all pairwise post hoc tests: z > 3.1, P < 0.002; Fig. 4D). The greater influence of parental age on dynamic telomere changes (ΔTL), rather than a static estimates (baseline TL), is not restricted to our study as this phenomenon has been previously observed in vertebrates (38). This further confirms a parental TL inheritance, as offspring ΔTL (again not baseline TL) was positively correlated with two modules of parental aging (“Salmon” and “Darkred,” fig. S3), confirming that proteins involved in telomere regulation among parents may also influence telomere regulation of offspring.
Fig. 4. Impact of reproductive aging on the offspring telomerase-telomere complex.
We distinguished two phases related to intrinsic aging rate and extrinsic pathogenic pressure. Without pathogens, spat TL (n = 443) positively correlated with both (A) maternal TL, and (B) paternal TL (dot colors, means ± SE of familial average; solid lines, model predictions surrounded by 95% CIs). (C) Offspring telomerase (n = 72 familial pools of 10 individuals per pool) slightly correlated with father telomerase. (D) TL erosion with age was then greater in offspring from old parents, as shown by interaction between offspring age and maternal and paternal age. After extrinsic pathogen exposure, offspring TL (n = 501) increased in all families, in the lab (dotted lines) and in two natural environments (solid lines).
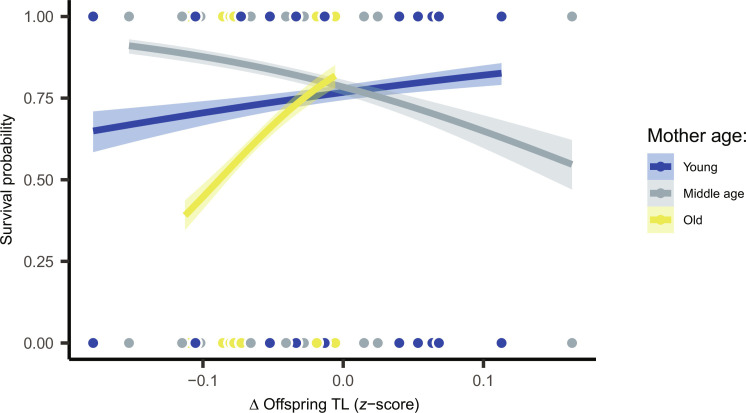
Preemptive dynamics of ΔTL correlated with inter-familial chances of survival to OsHV-1; however, there was an interaction with maternal age (COXME: χ2 = 9.1, df = 2, P = 0.011). The expected relationship between survival and TL was restricted to families born from old mothers (Fig. 5). This is probably because it adds to the other uncontrolled maternal effects. In this particular case, telomere shortening preceded, rather than resulting from viral infection. When considering two nonexclusive theories that link pathogen susceptibility to host telomeres, our results appear consistent with the immune-senescence hypothesis (i.e., shorter telomeres predict greater susceptibility to infection), rather than the aging cost of infection hypothesis (i.e., infection shortens telomeres), as proposed by Giraudeau et al. (39). The interaction effect with condition of mothers, as seen here, has been previously documented in birds (40) and reinforces the assumption that relationships between mortality and early life TL remain complex and context dependent (41).
Fig. 5. Dynamic changes in offspring TL (ΔTL) predicted their resistance to OsHV-1 challenge.
However, relationships depended upon maternal age. Inter-familial survival–ΔTL relationship was positive in offspring from old mothers (z = −2.06, P = 0.039), negative in those from middle-aged mothers (z = 2.11, P = 0.035), and nonsignificant among families from young mothers (z = −0.89, P = 0.370).
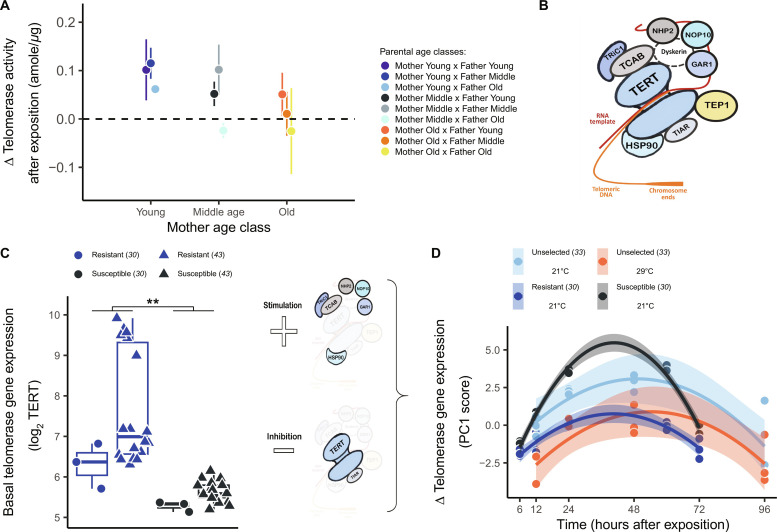
We then questioned the consequences of viral challenges on the telomerase-telomere complex. Interaction with OsHV-1, which induced marked changes in TL compared to the unperturbed baseline conditions, is concomitant to enhanced telomerase activity. Upon exposure of subjects to OsHV-1 between 10 and 14 months old, offspring TL remained positively linked to shell length (P < 0.001). However, TL elongated in both the laboratory and wild conditions at the end of the summer period (after versus before infection; all pairwise post hoc tests: z > 5.0, P < 0.016; Fig. 4D). In the laboratory, surviving oysters exhibited greater telomerase activity (GLMM: χ2 = 13.5, df = 1, P < 0.001), a response mitigated by maternal and paternal reproductive aging (maternal age: GLMM: χ2 = 15.7, df = 2, P < 0.001; paternal age GLMM: χ2 = 8.0, df = 2, P = 0.018; Fig. 6A). Together, our in vivo findings demonstrate that viral stimulation of telomerase activity is consistent with previous in vitro observations in human cells exposed to oncogenic viruses (28, 29).
Fig. 6. Adaptive stimulation of telomerase activity by the virus OsHV-1.
(A) Telomerase activity increased in offspring surviving viral challenge (n = 72 familial pools of 10 individuals per pool), and this response is mitigated in those from old parents. (B) We used genome annotation of C. gigas (42) to explore telomerase gene expression in response to OsHV-1, obtained from previously published RNA-seq projects (30, 33, 43) including the constitutive telomerase reverse transcriptase (TERT) and the accessory proteins [Tric1, TCAB, NPH2, NOP10, GAR1, TEP1, TIAR, and HSP90; see (25)]. (C) At the baseline state, TERT expression discriminates oyster lineages experimentally selected for their resistance or susceptibility to OsHV-1 (30, 43) (**P = 0.007). (D) In response to viral exposition, all telomerase genes but one (TEP 1) covaried together along a main axis of a principal components analysis (PCA) and inversely with TERT expression (fig. S7). Extracting normalized scores (PC1) for each experimental replicate, we identified a quadratic trend with response peaking at 24 to 48 hours after infection (time: F1,48 = 46.0, P < 0.001; time2: F1,48 = 50.8, P < 0.001). Peak intensity was specific both to family types (susceptible and resistant at 21°C) and acclimation temperature (unselected families at 21° or 29°C). Gene expression trends paralleled the associated mortality rates. In other words, inhibition of TERT gene expression was similar between families selected for susceptibility and unselected families acclimated 21°C (pairwise post hoc: t = −0.02, P > 0.999). Inhibition of TERT was lower in surviving than susceptible oysters (all pairwise post hoc: |t| > 4.2, P < 0.001) and similar between in resistant and among oysters conditioned to 29°C (pairwise post hoc: t = −0.1, P > 0.999).
Conserved telomerase response to the virus OsHV-1
Telomerase is a holoenzyme composed of a reverse transcriptase (TERT) and multiple accessory proteins binding to an RNA template to align on telomeric DNA (12). We identified nine genes annotated in the C. gigas genome (42) that coded for TERT or accessory proteins (Fig. 6B). To investigate a potential association between telomerase response and viral exposure, we mined publicly available transcriptomic datasets of OsHV-1 in Pacific oysters (30, 33, 43) and tested whether baseline and virus-induced telomerase gene expression differed between genetic oysters lineages (table S1). These oyster lineages were previously obtained by artificial selection over four generations with either high or low susceptibility to OsHV-1 (44).
At the baseline state, we compared the main constitutive telomerase TERT gene expression in oysters acclimated to 21°C between four families with high resistance to OsHV-1 [mortality < 40%; (30, 43)] against four families exhibiting high viral susceptibility [mortality > 90%; (30, 43)]. Families selected for high susceptibility to OsHV-1 exhibited significantly lower baseline TERT expression than those selected for high resistance (GLMM: χ2 = 7.4, df = 1, P = 0.007; Fig. 6C). Among 21,462 genes identified by RNA sequencing (RNA-seq) (30, 43), 355 genes (1.7%) were differentially expressed between resistant and susceptible families, which included the TERT gene (ranked in the top 50 genes). This result indicates that basal telomerase expression adds to other physiological pathways (e.g., immune responses, DNA repair, and stress response) that genetically predispose oysters to survive OsHV-1 challenge (43).
We further investigated the dynamics of telomerase-related genes in response to virus exposure under controlled conditions (fig. S7). We compared selected families (30) versus nonselected oysters [individuals born from a pooled of genitors; (31)] that were acclimated to either 21° or 29°C (33). In this temperature experiment, oysters acclimated at 29°C for 10 days exhibited a protective effect against the disease (mortality rate, 14%) compared to those acclimated at 21°C (mortality rate, 48%). Susceptible families showed greater stimulation of regulatory proteins but a stronger inhibition of TERT expression, when compared to resistant families (Fig. 6D). Acclimating nonselected oysters to 29°C resulted in a similar gene expression response to those experimentally selected for resistance (Fig. 6D). Together, this suggests that telomerase response is crucial for survival against OsHV-1 challenge and is highly influenced by the thermal environment. Future research will need to determine whether the host, the virus, or both modulate greater telomerase gene expression (28).
Adaptive telomerase response comes at delayed survival costs in adulthood
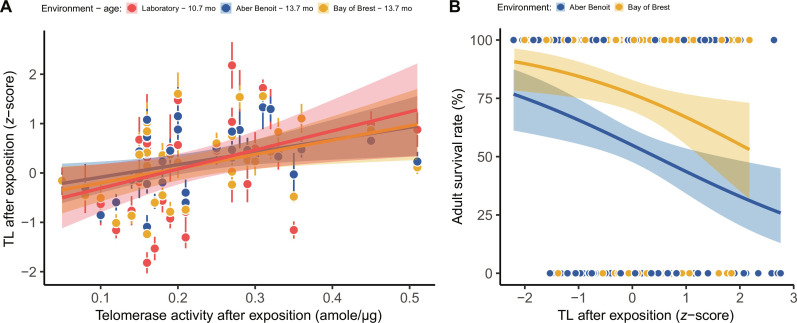
TL was positively correlated with enhanced telomerase activity both in laboratory settings and 3 months later, following exposure at two different field sites (GLMM: χ2 = 7.5, df = 1, P = 0.006; Fig. 7A). This confirms that telomerase activity contributes to elongate TL at the whole-organism level. However and unexpectedly, longer TL was associated with a greater risk of mortality among adults (GLMM: χ2 = 5.1, df = 1, P = 0.023; Fig. 7B). This result challenges conventional evidence in vertebrates, where longer telomeres usually predict greater life expectancies (17). This suggests that there is a need to explore the relationship between TL and the resistance to polymicrobial infectious diseases (39).
Fig. 7. Elongation of telomere by telomerase conducted to greater adult mortality over the next 6 months.
(A) After exposition to OsHV-1 challenge, lengthening TL positively correlated with inter-familial stimulation telomerase activity regardless of the context (laboratory and field sites). (B) Individuals TL in early fall (n = 357) negatively predicted survival rate the following months, again irrespective of field site (dot colors, means ± SE of familial average in each environment; solid and color lines, model predictions by environment surrounded by 95% CIs).
We identified the pathogenic bacterium Vibrio aestuarianus by quantitative polymerase chain reaction (qPCR) (34) as the primary pathogen strain causing adult mortality in 89% of individuals. Resistance to this bacterial strain was poorly heritable (MCMCglmm: h2 = 0.11; 95% CI, 0.00 to 0.31; Table 1), which is consistent with previous investigations (34). These delayed survival costs associated to telomerase response might reflect selective pressure that mitigates overexpression of this enzyme in somatic tissue as a way to avoid abnormal cell immortalization and tumor risks.
DISCUSSION
In this study, we characterized the intergenerational impacts of reproductive aging and the cellular vehicles linking parental age to offspring traits. Our findings demonstrate that offspring from elderly parents exhibited lower developmental success, which is concomitant with intrinsic telomere erosion and extrinsic telomere elongation due to a telomerase response to Ostreid herpesvirus 1. Although non-informative with the chronological age of adults, the complex of telomerase and telomeres emerges as a potential mechanism linking offspring traits to parental age at conception.
Our studies reveal a high number of enriched biological functions in parent oysters growing old. Across both sexes, multiple age-related protein modules identified among adults correlated with offspring phenotypes, particularly traits related to telomere dynamics and telomerase response to infection. The strong correlation with parental TL and substantial telomere heritability (h2 = 0.53) suggests that the variation in offspring TL results from parental inheritance acquired through germline TL, as well as genetically based differences in TL regulation. This later process may reflect genetic differences in line with the father-offspring telomerase relationship. By further exploring the parental proteomic modules and correlations of offspring traits, our study identifies relevant pathways that may regulate these intergenerational effects. Notably, the two parental age-decreasing modules that correlate with offspring telomere changes revealed a strong enrichment of the WNT signaling pathway and β-catenin (fig. S3), two important regulatory factors of telomerase in stem cells and cancer (45). We hypothesized that declining WNT/β-catenin levels in older parents was likely passed on to the next generation and potentially mitigated the reparation of offspring telomeres. However, this assumption requires further testing at the individual scale.
In many species, and especially early in life, telomere dynamics predict higher reproductive success and survival rates (10, 17, 41, 46). Telomere dynamics also influences the susceptibility to viral infection [i.e., as observed in over 200 diseases in humans; (47)]. Our results confirm the predictive power of telomere dynamics regarding survival to pathogens but demonstrate the complexity of this relationship. At 10 months old, the inter-familial differences in the erosion of telomeres positively predicted mortality to OsHV-1, only among offspring from old dams. Additional studies are now required to clearly establish, at the individual scale, whether shorter TL predicts greater susceptibility to OsHV-1. Between the age of 14 and 20 months, longer individual telomeres predicted a higher subsequent adult mortality risk to vibriosis, an unexpected finding that was confirmed in two independent environments that contrasted bacterial prevalence (31). We speculate that longer TL may contribute to mortality from vibriosis through indirect effects of telomerase on cytogenetic alterations. Shellfish can tolerate a relatively high proportions of aneuploidy (up to 30%) (48), and this proportion of abnormal chromosomes increases during OsHV-1 infection, but the functional effects remain unknown (49). We propose that telomerase is one such mechanism, similar to that in human cancer (50). If this assumption holds true, then longer TL would have been indirectly associated with chromosome instability and to a greater oyster susceptibility to bacterial infection (51).
OsHV-1 stimulated telomerase activity in vivo, a response that, to our knowledge, had only been documented in human in vitro cell culture (28, 29). Mining into the C. gigas transcriptomic datasets, we found that telomerase level predetermines oyster susceptibility and that telomerase response to OsHV-1 is initiated within a few hours after exposure to the virus (30, 33, 43). Baseline TERT activity was shown to discriminate between susceptible and resistant genotypes and with resistant oysters characterized by reduced TERT inhibition upon viral exposure. The role of telomerase as an adaptive response to discern whether individuals may survive or die from infectious disease is further supported by single-nucleotide polymorphism–based studies, which showed that telomerase genes appear as an encoding differential resistance to virus (35, 52). Consistently, parental aging inhibited this adaptive telomerase response, a process that we see as an underlying factor associated with a greater risk of mortality to OsHV-1. Besides, we confirmed that telomerase response can be regulated by the environment, because the acclimation of unselected oysters to high temperatures (29°C) displayed the same viral protection (33) and telomerase response, rather than being a result of the genotypes selected for viral resistance. However, the adaptive telomerase response to OsHV-1 in juveniles was counterbalanced by increased mortality in adults. Overall, we unraveled that telomerase represents a central and adaptive mechanism in oyster response to OsHV-1, and, like many adaptive traits in ectotherms, its regulation is influenced by environmental temperature (53).
This study provides insights into how reproductive aging affects oyster developmental success, telomere shortening, and resistance to polymicrobial disease. Already in larval life, elderly parents produced offspring with lower survival. We further highlight the importance of the telomere-telomerase system as a molecular indicator of parental age, which can predict the outcomes of OsHV-1 infection. The abnormal expression of telomerase in adult somatic cells can have oncogenic effects (12, 26, 27), similar to observations in human tumors of viral etiology (50). Mimicking human tumors, transcriptomic profiles of mussels confirmed that telomerase expression strongly increases in neoplastic cells (54). We argue that oyster telomerase response to OsHV-1 in Pacific oysters will thereby offer an unparalleled setting to understand the in vivo interactions between virus, telomerase activation, and cytogenetic regulations.
MATERIALS AND METHODS
Parent origin and crossbreeding design
Beginning in 2012, we collected 1-month-old spats of the same oyster cultch along the coastline of the Aix Island, France (46.0121; −1.1609), each year and transferred them to a Britain tideway, the “Aber Benoit” (48.5751; −4.6051). We separated oyster cohorts into mesh bags (100 cm by 50 cm by 10 cm) attached to iron tables in the middle bathymetry of the foreshore level. At this bathymetric height, animals experience natural tidal cycles, food conditions, and suitable temperature for growth and are able to reach sexual maturity and an age up to 10 years. We sampled 50 individuals of the different cohorts on 11 August 2022 and maintained them at the laboratory facility of Argenton in 300-liter tank with ad libitum food conditions (Tisochrysis lutea and Chaetoceros sp.) and seawater controlled at 17°C (55). On 16 August, we isolated the gametes of 12 mothers [young (2 years old), n = 4; middle-aged (6 years old), n = 4; and old (10 years old), n = 4; Fig. 1] and 12 fathers [young (2 years old), n = 6; middle-aged (6 years old), n = 4; and old (8 to 9 years old), n = 2; Fig. 1] by incising the animal gonads with a scalpel in purified seawater and sieving the milt of females (diameter, 20 μm) and males (diameter, 60 μm) in 5- and 2-liter beakers, respectively. Each parent contributed to produce three families to obtain half-sibling relatedness (fig. S1). We did not mate reproductive pairs randomly because we intended to crossbreed each female with three males of the three age classes, and vice versa (fig. S1). We then realized 36 fecundations (700,000 oocytes per family) targeting a 1:10 ratio of ovocyte:sperms in 36 beakers (5 liters) and ensured that, at 2 hours, fecundation rate (bourgeoning larvae) was above 95%. No animal experimentation permit was required for the experiments conducted here.
Offspring early life development
We monitored offspring developmental performance for 1 year by sampling different individuals over time (fig. S2). First, larval development between 2-hour-old embryos and 40 day-old “spats” followed the zootechnical protocol as regularly used in this species (31, 55). That is, when larvae are mobile (until ~14 days old), they are reared in 5-liter cylindrical Plexiglas tubes (one tube per family) with a seawater flow rate of 75 ml/min, a bubbling system, food ad libitum, and water temperature set at 25°C. We counted larvae at day 2 (D-larvae stage), a critical life stage in this species and again at day 14, which marks the end of the mobile phase, before metamorphosis. At days 2, 6, 9, and 13, we fixed ~100 larvae in a 5% formaldehyde solution. We measured shell length as the maximal distance between the umbo and the ventral edge of the shell using a high-resolution microscope (Keyence VHX 2000E, 1-μm resolution images). At day 14, we transferred surviving larvae to 36 riddles filled with sand so they could find suitable support to metamorphose. At days 28, 33, and 41, we non-lethally scanned (Epson Perfection 2480, HDR images) ~50 larvae per family to extract shell length using ImageJ software. Then, we sampled spat oysters (life stage between 40 and 365 days old) regularly to depict the impact of parental aging on the dynamics of the telomerase-telomere complex (fig. S2).
Parent proteomic profiles
Immediately after gamete extraction, the whole parents were flash frozen in liquid nitrogen. We grounded frozen individuals using an MM400 homogenizer (Retsch, Eragny, France) to smooth intra-individual tissue variation. We extracted the total proteins content from frozen oyster powder using a protocol previously optimized for C. gigas (56). The powder (~200 mg) was incubated on ice in a lysis buffer for 30 min and centrifuged at 12,000g. Total protein content in each lysate was analyzed using the detergent compatible (DC) protein assay (Bio-Rad), in 96-well microplates (Nunc) using a microplate reader (Bio-TekSynergyTM HT, Thermo Fisher Scientific, Les Ulis, France). Total protein concentrations were obtained using Gen5 version 2.03 software (Bio-Tek). Proteomic profiles of each oyster were determined with mass spectrometry using 5 μg of protein on a thermal ionization mass spectrometry–time-of-flight pro mass spectrometer (Bruker Daltonics) with a modified nano-electrospray ion source (CaptiveSpray, Bruker Daltonics). We followed the same protocol as detailed previously (57), fitted onto an updated UniProt C. gigas database. We quantified the relative levels of protein abundance between samples using the differential enrichment analysis of proteomics data (DEP) package from R software (58). We checked and imputed missing patterns of protein levels using the “imputePCA” function and regularized method implemented in missMDA R package.
We assessed the relative contribution of sex and age on the protein levels using a redundancy discriminant analysis (RDA). First, we computed a Euclidian distance matrix on proteomic data and performed a principal coordinates analysis (PCoA). A distance-based RDA (db-RDA) was then produced with the retained PCoA factors as the response matrix and the variables sex and age as the explanatory matrix. A total of four PCoA axes were retained based on a Gower dissimilarity index, explaining 59.90% of the total variance. Then, two partial db-RDAs and an analysis of variance [analysis of variance (ANOVA), 999 permutations] were used to validate the effect of each factor while controlling for the other factor.
We compared adult proteomic profiles using a signed co-expression network, built on the imputed protein level matrix using the R package WGCNA following the protocol detailed previously (59). We scored oyster sex (male, 1; female, 2) and age class (young, 1; middle-aged, 2; and old, 3). Soft threshold power was fixed to 12 using the scale-free topology criterion to reach a model fit (|r|) of 0.90. The modules were defined using the “cutreeDynamic” function (minimum of 20 genes by module and default cutting-height of 0.99) based on the topological overlap matrix and a module eigengene distance threshold of 0.25 that was used to merge highly similar modules. For each module, we defined the module membership (kME) and correlation between module eigengene value and condition traits. Only modules with a significant correlation with one of the condition factors (sex or age; P < 0.001) were conserved for a downstream functional analysis. Functional enrichment was conducted with a rank-based gene ontology approach with adaptive clustering using a Mann-Whitney U test for each independent module (kME value when belonging to the module, 0 otherwise), which was then implemented into the GO_MWU R tools (60) (go.obo database, downloaded October 2023, https://geneontology.org/docs/download-ontology/). We used REviGO to reduce redundancy and to visualize the most significant GO (P < 0.01) (61).
Parent and offspring TL
We measured TL in parents (~15-mg gills, n = 24) and their offspring (~15 mg of a mix of tissues, n = 980) at different times (n = 184 at 90 day-old, n = 259 at 240 day-old, n = 144 at 320 day-old, and n = 357 at 410 day-old; fig. S2) using a similar protocol that has been previously described for this species (62). We extracted high–molecular weight DNA using a DNA isolation kit for cells and tissue (Sigma-Aldrich, Roche). We used a NanoDrop 2000 (Thermo Fisher Scientific) to check that absorbance ratios of extracted DNA met the optimal criteria (means ± SD: A260/A280, 1.88 ± 0.05; and A260/A230, 2.22 ± 0.30). We ran qPCR from 5 ng of DNA by measuring real-time fluorescence as a means to amplify telomere regions using Sybr Green Supermix (Bio-Rad, California, USA) and universal telomere primers (Tel1 and Tel2). These were corrected by the amplification of a reference gene [glyceraldehyde-3-phosphate dehydrogenase (GAPDH)]. We used 30 and 40 amplification cycles at an annealing and extension temperature of 55° and 60°C, with primer concentrations of 0.1 and 0.2 μM, respectively. We assessed cycle threshold (Cq) of telomeres and GAPDH on a Bio-Rad CFX96 Touch (Bio-Rad, USA) as detailed previously (62). Samples were measured in duplicates with high repeatability [telomeric DNA: correlation coefficient (r) = 0.98, P < 0.001; GAPDH: r = 0.99, P < 0.001]. We used a DNA positive control (pool of 30 samples) on each plate to smooth interplate variation and a non-template control to verify the absence of contamination. Relative TL was determined using gene efficiency (ETelo = 99.3% and EGAPDH = 105.4%) obtained from the serial dilution of the positive control and Cq differential for each gene as
On each plate, we measured three samples three times to assay intraplate repeatability of TL measure (r = 0.90, P < 0.001), and we used four individuals on all plates to determine interplate repeatability of TL (r = 0.85, P < 0.001). Individual TL was transformed to a z-score (mean = 0 and SD = 1) to ease model convergence and inter-study comparisons (63).
Parent and offspring telomerase activity
We measured whole-organism telomerase activity in parents and their offspring (pools of 10 offspring sampled for each family) at three times (fig. S2). Offspring were sampled at day 40 at the end of larval life stage to determine parent-offspring relationship of telomerase activity. We also sampled offspring at days 290 and 320, respectively, before and after OsHV-1 challenge (see below). We determined telomerase activity using telomeric repeat amplification protocol assays (TRAPeze Kit RT Telomerase Detection Kit, S7710, Merck Millipore). We extracted proteins from the frozen oyster powder (~20 to 30 mg) using a CHAPS lysis buffer (200 μl) and then incubated for 30 min on ice. Samples were centrifuged for 20 min at 12,000g. Protein lysates were collected and diluted at a 1:10 ratio. We assessed the total protein content using the DC protein assay (Bio-Rad; see above). Telomerase activity was assayed from FAM carboxyfluorescein with Bio-Rad CFX96 Touch (Bio-Rad, USA) following the manufacturer’s protocol. During the initial phase (30 min at 30°C), telomerase elongates telomere from a telomeric DNA template. Then, telomere products were quantified by qPCR on the basis of real time fluorescence emission over 40 cycles. We used a standard curve from a serial dilution of known telomerase quantity (40 to 0.04 amoles) on each plate to obtain a slope of the Cq against telomerase. Each sample was measured in duplicate with a high level of repeatability (r = 0.95, P < 0.001). We also used three samples among all plates to calculate interplate repeatability (r = 0.94, P < 0.001).
OsHV-1 viral challenge
We combined two approaches to study the response of oysters to OsHV-1 infection: laboratory (~120 spats per family) or in the field (~100 spats per family) (31, 32, 34, 35). In the laboratory, we controlled food availability (ad libitum) and seawater temperature (21°C) to maximize OsHV-1 virulence (31, 32). We exposed receiving oysters (n = 4325) from 36 families to oyster donors (~2000 spat oysters) impregnated during a field mortality event in the Bay of Brest (48.3355; −4.3175). This occurred in mid-June, a time known as the annual peak of OsHV-1 replication and high mortality risk (64). We exposed them for 48 hours by cohabitation with natural viral load before bringing them back to the laboratory. Contact between the receivers and donors occurred on 16 June at 17:00. We counted alive and dead oysters daily for 2 weeks. We verified that OsHV-1 was the pathogen responsible of oyster mortalities by quantifying viral presence through qPCR, in seawater (fig. S8) and in the moribund oysters as described previously (34). Concomitantly, we deployed 3600 spats (100 per family) to the same field site in the Bay of Brest to obtain mortality risk in natura. Families were isolated using nets within two mesh bags (18 families per bag) beginning on 7 June. We recorded spat mortality daily during the first 2 weeks and again monthly over the remaining summer.
Statistical analyses
We analyzed the impacts of parental age on offspring traits using R software (58). We built generalized linear mixed models [package “lme4” (65)] to determine the fixed effects of maternal and paternal age and the interaction on offspring traits by setting offspring family as a random intercept. We used Akaike information criterion (AIC)–based model comparisons [package “MuMIn” (66)] to identify the most parsimonious explanatory variables. Depending on whether or not offspring response was continuous (shell growth, TL, and telomerase activity) or binary (mortality rate: alive, 0; or dead, 1), we considered Gaussian or Binomial (loglink function) family to describe error distribution, respectively. We analyzed the kinetic of survival probability (larval intrinsic survival, 3 time sessions; susceptibility to OsHV-1, 13 time sessions) with cox mixed models [package “coxme” (67)] containing fixed effects of parental age class, interaction effects, and random intercepts of family and tank. We produced Kaplan-Meier survival curves using the survfit function of the package “survival,” with a log transformation to compute 95% CIs from SEs (68).
We built animal models by fitting a Bayesian algorithm of the Markov Chain Monte Carlo model [package “MCMCglmm” (69)] used to quantify narrow-sense heritability. These models decompose the variance explained by a combination of fixed and random factors. We considered the final models selected by frequentists analyses (described above), which categorized the fixed and random structure of each model. In addition, all models included the random factor of “animal,” which referred to the offspring relatedness matrix (r = 0.5 between full siblings, r = 0.25 between half-siblings, and r = 0 between unrelated). Following recommendations from de Villemereuil (70), we considered non-informative priors for both the Gaussian (residuals: V = 1, nu = 0.02; random terms: nu = 1, alpha.mu = 0, and alpha.V = 1000) and threshold models (residuals: V = 1 and fix = 1; random terms: V = 1, nu = 1000, alpha.mu = 0, and alpha.V = 1). We set simulations to 5 × 105 iterations, with a 5 × 104 burn-in period and a thin interval of 50 to record iterations. We ensured appropriate model convergence by graphically inspecting the MCMC traces and verifying that random and fixed terms passed the Heidelberg stationarity tests (all P > 0.05). The model on larval growth necessitated an increase of iterations by 1 × 106 and a burn-in period to 2 × 105 to pass the Heidelberg test. We estimated h2 for offspring traits and 95% CIs while accounting for the ratio between additive genetic variance (VA) obtained from the random “animal” factor against total phenotypic variance (VP). VP was calculated from the sum of variances explained by both random and fixed effects (70).
RNA-seq meta-analysis
We explored the conserved response of telomerase related genes expression in C. gigas. We selected transcriptomic datasets previously published by studies that investigated oyster resistance/susceptibility to OsHV-1. Three transcriptomic studies representing n = 102 samples (each sample composed by a pool of n = 9 to 10 oysters) were retrieved from National Center for Biotechnology Information server (table S1).
First, we assessed whether family resistance phenotype was associated with differential TERT gene expression in the basal state. We subset the datasets to keep only individuals with prior exposure or without experimental viral exposition [n = 42 pools of 10 individuals per pool, from two studies (30, 43)]. Raw data were trimmed with Trimmomatic v0.36 (71) and mapped to the C. gigas reference genome (42) using STAR v2.7.10b (72). The default options were selected, but specificity was applied depending on the data type for each study (reported in table S1). Gene counts were computed with HTseq-count v0.9.1 (73). All count data were normalized with median of ratio implemented in DESeq2 R package (74). Batch effect (i.e., different experiments) was controlled using RUVseq and a k value of 6, following previously published recommendations (75). Differentially expressed genes across phenotypes (resistant versus susceptible) were detected using the likelihood ratio test with k vectors as covariates. We applied a conservative approach by considering genes as differentially expressed when the absolute log2 fold change (|log2FC|) was >2 and with a false discovery rate < 0.01. We tracked telomerase related genes in different temperature regimes or across phenotypes following experimental infection [n = 70 pools of 10 individuals per pool, from two studies (30, 33)]. We ran a principal components analysis (PCA) on the log2-normalized counts to explore how the multiple genes were co-expressed at different times after infection (fig. S7). We extracted the main axis score for each replicate as an integrative measure of telomerase gene expression (see Fig. 5).
Acknowledgments
We thank M. Diagne for producing phytoplankton used to feed larvae and oysters during laboratory experiments. We also thank M.-A. Travers and E. Bouchereau who produced the positive control for bacterial assays. We thank I. Queau, V. Leroy, T. Lemaitre, R. Quillien, and C. Cotté for helping in fieldwork survey. We are grateful to L. Madec, V. Quillien, C. Quéré, C. Dubreuil, and F. Pernet for helpful contribution and recommendations during viral challenge. We lastly thank J. Carmona (LifeScience editor) and C. Miller for scientific editing and English checks and F. Pernet and M.-A. Travers for constructive feedbacks on early draft manuscript.
Funding: This work was supported by Ifremer and ISblue project [Interdisciplinary graduate school for the blue planet (ANR-17-EURE-0015) under “TeloRepro” and “TeloTabo” projects].
Author contributions: Conceptualization: A.D., H.K., M.H., and J.L.L. Methodology: A.D., H.K., M.H., P.d.V., B.M., and J.L.L. Investigation: A.D., H.K., M.H., P.M., J.L.G., C.C., E.F., B.B., and J.L.L. Visualization: A.D. and J.L.L. Supervision: A.D. Writing—original draft: A.D. Writing—review and editing: A.D., H.K., M.H., P.M., J.L.G., C.C., E.F., B.B., P.d.V., B.M., and J.L.L.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Raw data and codes used here are available at Zenodo repository (DOI: 10.5281/zenodo.11954194).
Supplementary Materials
This PDF file includes:
Figs. S1 to S8
Table S1
REFERENCES AND NOTES
- 1.Chakravarti D., LaBella K. A., DePinho R. A., Telomeres: History, health, and hallmarks of aging. Cell 184, 306–322 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lopez-Otin C., Blasco M. A., Partridge L., Serrano M., Kroemer G., The hallmarks of aging. Cell 153, 1194–1217 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lopez-Otin C., Blasco M. A., Partridge L., Serrano M., Kroemer G., Hallmarks of aging: An expanding universe. Cell 186, 243–278 (2023). [DOI] [PubMed] [Google Scholar]
- 4.Lemaître J.-F., Gaillard J.-M., Reproductive senescence: New perspectives in the wild. Biol. Rev. 92, 2182–2199 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Lemaître J.-F., Gaillard J.-M., Editorial: The evolutionary roots of reproductive ageing and reproductive health across the tree of life. Front. Ecol. Evol. 11, 1349845 (2023). [Google Scholar]
- 6.Monaghan P., Maklakov A. A., Metcalfe N. B., Intergenerational transfer of ageing: Parental age and offspring lifespan. Trends Ecol. Evol. 35, 927–937 (2020). [DOI] [PubMed] [Google Scholar]
- 7.Ivimey-Cook E. R., Shorr S., Moorad J. A., The distribution of the Lansing Effect across animal species. Evolution 77, 608–615 (2023). [DOI] [PubMed] [Google Scholar]
- 8.Haussmann M. F., Heidinger B. J., Telomere dynamics may link stress exposure and ageing across generations. Biol. Lett. 11, 20150396 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heidinger B. J., Young R. L., cross-generational effects of parental age on offspring longevity: Are telomeres an important underlying mechanism? Bioessays 42, e1900227 (2020). [DOI] [PubMed] [Google Scholar]
- 10.Meillère A., Buchanan K. L., Eastwood J. R., Mariette M. M., Pre- and postnatal noise directly impairs avian development, with fitness consequences. Science 384, 475–480 (2024). [DOI] [PubMed] [Google Scholar]
- 11.Dugdale H. L., Richardson D. S., Heritability of telomere variation: It is all about the environment!. Philos. Trans. R. Soc. Lond. B Biol. Sci. 373, 20160450 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shay J. W., Wright W. E., Telomeres and telomerase: Three decades of progress. Nat. Rev. Genet. 20, 299–309 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Armstrong E., Boonekamp J., Does oxidative stress shorten telomeres in vivo? A meta-analysis. Ageing Res. Rev. 85, 101854 (2023). [DOI] [PubMed] [Google Scholar]
- 14.Reichert S., Stier A., Does oxidative stress shorten telomeres in vivo? A review. Biol. Lett. 13, 20170463 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dupoué A., Blaimont P., Angelier F., Ribout C., Rozen-Rechels D., Richard M., Miles D., de Villemereuil P., Rutschmann A., Badiane A., Aubret F., Lourdais O., Meylan S., Cote J., Clobert J., Le Galliard J.-F., Lizards from warm and declining populations are born with extremely short telomeres. Proc. Natl. Acad. Sci. U.S.A. 119, e2201371119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rouan A., Pousse M., Djerbi N., Porro B., Bourdin G., Carradec Q., Hume B. C. C., Poulain J., Lê-Hoang J., Armstrong E., Agostini S., Salazar G., Ruscheweyh H.-J., Aury J.-M., Paz-García D. A., McMinds R., Giraud-Panis M.-J., Deshuraud R., Ottaviani A., Morini L. D., Leone C., Wurzer L., Tran J., Zoccola D., Pey A., Moulin C., Boissin E., Iwankow G., Romac S., de Vargas C., Banaigs B., Boss E., Bowler C., Douville E., Flores M., Reynaud S., Thomas O. P., Troublé R., Thurber R. V., Planes S., Allemand D., Pesant S., Galand P. E., Wincker P., Sunagawa S., Röttinger E., Furla P., Voolstra C. R., Forcioli D., Lombard F., Gilson E., Telomere DNA length regulation is influenced by seasonal temperature differences in short-lived but not in long-lived reef-building corals. Nat. Commun. 14, 3038 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilbourn R. V., Moatt J. P., Froy H., Walling C. A., Nussey D. H., Boonekamp J. J., The relationship between telomere length and mortality risk in non-model vertebrate systems: A meta-analysis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 373, 20160447 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eastwood J. R., Hall M. L., Teunissen N., Kingma S. A., Hidalgo Aranzamendi N., Fan M., Roast M., Verhulst S., Peters A., Early-life telomere length predicts lifespan and lifetime reproductive success in a wild bird. Mol. Ecol. 28, 1127–1137 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Chik H. Y. J., Sparks A. M., Schroeder J., Dugdale H. L., A meta-analysis on the heritability of vertebrate telomere length. J. Evol. Biol. 35, 1283–1295 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boonekamp J., Rodríguez-Muñoz R., Hopwood P., Zuidersma E., Mulder E., Wilson A., Verhulst S., Tregenza T., Telomere length is highly heritable and independent of growth rate manipulated by temperature in field crickets. Mol. Ecol. 31, 6128–6140 (2022). [DOI] [PubMed] [Google Scholar]
- 21.Marasco V., Boner W., Griffiths K., Heidinger B., Monaghan P., Intergenerational effects on offspring telomere length: Interactions among maternal age, stress exposure and offspring sex. Proc. Biol. Sci. 286, 20191845 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith S., Hoelzl F., Zahn S., Criscuolo F., Telomerase activity in ecological studies: What are its consequences for individual physiology and is there evidence for effects and trade-offs in wild populations. Mol. Ecol. 31, 6239–6251 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He Y., Wang Y., Liu B., Helmling C., Sušac L., Cheng R., Hong Zhou Z., Feigon J., Structures of telomerase at several steps of telomere repeat synthesis. Nature 593, 454–459 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim N. W., Piatyszek M. A., Prowse K. R., Harley C. B., West M. D., Ho P. L. C., Coviello G. M., Wright W. E., Weinrich S. L., Shay J. W., Specific association of human telomerase activity with immortal cells and cancer. Science 266, 2011–2015 (1994). [DOI] [PubMed] [Google Scholar]
- 25.Roake C. M., Artandi S. E., Regulation of human telomerase in homeostasis and disease. Nat. Rev. Mol. Cell Biol. 21, 384–397 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harley C. B., Telomerase and cancer therapeutics. Nat. Rev. Cancer 8, 167–179 (2008). [DOI] [PubMed] [Google Scholar]
- 27.Shay J. W., Role of telomeres and telomerase in aging and cancer. Cancer Discov. 6, 584–593 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bellon M., Nicot C., Regulation of telomerase and telomeres: Human tumor viruses take control. J. Natl. Cancer Inst. 100, 98–108 (2008). [DOI] [PubMed] [Google Scholar]
- 29.Yuan X., Larsson C., Xu D., Mechanisms underlying the activation of TERT transcription and telomerase activity in human cancer: Old actors and new players. Oncogene 38, 6172–6183 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Lorgeril J., Lucasson A., Petton B., Toulza E., Montagnani C., Clerissi C., Vidal-Dupiol J., Chaparro C., Galinier R., Escoubas J. M., Haffner P., Dégremont L., Charrière G. M., Lafont M., Delort A., Vergnes A., Chiarello M., Faury N., Rubio T., Leroy M. A., Pérignon A., Régler D., Morga B., Alunno-Bruscia M., Boudry P., Le Roux F., Destoumieux-Garzόn D., Gueguen Y., Mitta G., Immune-suppression by OsHV-1 viral infection causes fatal bacteraemia in Pacific oysters. Nat. Commun. 9, 4215–4215 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petton B., Boudry P., Alunno-Bruscia M., Pernet F., Factors influencing disease-induced mortality of Pacific oysters Crassostrea gigas. Aquac. Environ. Interact. 6, 205–222 (2015). [Google Scholar]
- 32.Petton B., Alunno-Bruscia M., Mitta G., Pernet F., Increased growth metabolism promotes viral infection in a susceptible oyster population. Aquac. Environ. Interact. 15, 19–33 (2023). [Google Scholar]
- 33.Delisle L., Pauletto M., Vidal-Dupiol J., Petton B., Bargelloni L., Montagnani C., Pernet F., Corporeau C., Fleury E., High temperature induces transcriptomic changes in Crassostrea gigas that hinders progress of Ostreid herpesvirus (OsHV-1) and promotes survival. J. Exp. Biol. 223, jeb226233 (2020). [DOI] [PubMed] [Google Scholar]
- 34.Azèma P., Lamy J.-B., Boudry P., Renault T., Travers M.-A., Dégremont L., Genetic parameters of resistance to Vibrio aestuarianus, and OsHV-1 infections in the Pacific oyster, Crassostrea gigas, at three different life stages. Genet. Sel. Evol. 49, 23 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gawra J., Valdivieso A., Roux F., Laporte M., de Lorgeril J., Gueguen Y., Saccas M., Escoubas J.-M., Montagnani C., Destoumieux-Garzόn D., Lagarde F., Leroy M. A., Haffner P., Petton B., Cosseau C., Morga B., Dégremont L., Mitta G., Grunau C., Vidal-Dupiol J., Epigenetic variations are more substantial than genetic variations in rapid adaptation of oyster to Pacific oyster mortality syndrome. Sci. Adv. 9, eadh8990 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gruber H., Schaible R., Ridgway I. D., Chow T. T., Held C., Philipp E. E. R., Telomere-independent ageing in the longest-lived non-colonial animal, Arctica islandica. Exp. Gerontol. 51, 38–45 (2014). [DOI] [PubMed] [Google Scholar]
- 37.Remot F., Ronget V., Froy H., Rey B., Gaillard J.-M., Nussey D. H., Lemaitre J.-F., Decline in telomere length with increasing age across nonhuman vertebrates: A meta-analysis. Mol. Ecol. 31, 5917–5932 (2022). [DOI] [PubMed] [Google Scholar]
- 38.Heidinger B. J., Herborn K. A., Granroth-Wilding H. M. V., Boner W., Burthe S., Newell M., Wanless S., Daunt F., Monaghan P., Parental age influences offspring telomere loss. Funct. Ecol. 30, 1531–1538 (2015). [Google Scholar]
- 39.Giraudeau M., Heidinger B., Bonneaud C., Sepp T., Telomere shortening as a mechanism of long-term cost of infectious diseases in natural animal populations. Biol. Lett. 15, 20190190 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Asghar M., Hasselquist D., Hansson B., Zehtindjiev P., Westerdahl H., Bensch S., Hidden costs of infection: Chronic malaria accelerates telomere degradation and senescence in wild birds. Science 347, 436–438 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Eastwood J. R., Dupoué A., Delhey K., Verhulst S., Cockburn A., Peters A., When does early-life telomere length predict survival? A case study and meta-analysis. Mol. Ecol. 32, 3000–3013 (2023). [DOI] [PubMed] [Google Scholar]
- 42.Penaloza C., Gutierrez A. P., Eöry L., Wang S., Guo X., Archibald A. L., Bean T. P., Houston R. D., A chromosome-level genome assembly for the Pacific oyster Crassostrea gigas. Gigascience 10, giab020 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.de Lorgeril J., Petton B., Lucasson A., Perez V., Stenger P.-L., Dégremont L., Montagnani C., Escoubas J.-M., Haffner P., Allienne J.-F., Leroy M. A., Lagarde F., Vidal-Dupiol J., Gueguen Y., Mitta G., Differential basal expression of immune genes confers Crassostrea gigas resistance to Pacific oyster mortality syndrome. BMC Genomics 21, 63 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dégremont L., Nourry M., Maurouard E., Mass selection for survival and resistance to OsHV-1 infection in Crassostrea gigas spat in field conditions: Response to selection after four generations. Aquaculture 446, 111–121 (2015). [Google Scholar]
- 45.Hoffmeyer K., Raggioli A., Rudloff S., Anton R., Hierholzer A., Del Valle I., Hein K., Vogt R., Kemler R., Wnt/β-catenin signaling regulates telomerase in stem cells and cancer cells. Science 336, 1549–1554 (2012). [DOI] [PubMed] [Google Scholar]
- 46.Dupoué A., Angelier F., Ribout C., Meylan S., Rozen-Rechels D., Decencière B., Agostini S., Le Galliard J.-F., Chronic water restriction triggers sex-specific oxidative stress and telomere shortening in lizards. Biol. Lett. 16, 20190889 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schneider C. V., Schneider K. M., Teumer A., Rudolph K. L., Hartmann D., Rader D. J., Strnad P., Association of telomere length with risk of disease and mortality. JAMA Intern. Med. 182, 291–300 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leitao A., Boudry P., Thiriot-Quievreux C., Negative correlation between aneuploidy and growth in the Pacific oyster, Crassostrea gigas: Ten years of evidence. Aquaculture 193, 39–48 (2001). [Google Scholar]
- 49.Batista F. M., Lo M., Ruano F., Renault T., Fonseca V. G., Leitao A., Insights on the association between somatic aneuploidy and ostreid herpesvirus 1 detection in the oysters Crassostrea gigas, C. angulata and their F1 hybrids. Aquacult. Res. 47, 1530–1536 (2016). [Google Scholar]
- 50.Christodoulidou A., Raftopoulou C., Chiourea M., Papaioannou G. K., Hoshiyama H., Wright W. E., Shay J. W., Gagos S., The roles of telomerase in the generation of polyploidy during neoplastic cell growth. Neoplasia 15, 156–168 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Azèma P., Travers M.-A., Benabdelmouna A., Dégremont L., Single or dual experimental infections with Vibrio aestuarianus and OsHV-1 in diploid and triploid Crassostrea gigas at the spat, juvenile and adult stages. J. Invertebr. Pathol. 139, 92–101 (2016). [DOI] [PubMed] [Google Scholar]
- 52.Yao S., Li L., Guan X., He Y., Jouaux A., Xu F., Guo X., Zhang G., Zhang L., Pooled resequencing of larvae and adults reveals genomic variations associated with Ostreid herpesvirus 1 resistance in the Pacific oyster Crassostrea gigas. Front. Immunol. 13, 928628 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Friesen C. R., Wapstra E., Olsson M., Of telomeres and temperature: Measuring thermal effects on telomeres in ectothermic animals. Mol. Ecol. 31, 6069–6086 (2022). [DOI] [PubMed] [Google Scholar]
- 54.Burioli E. A. V., Hammel M., Vignal E., Vidal-Dupiol J., Mitta G., Thomas F., Bierne N., Destoumieux-Garzón D., Charrière G. M., Transcriptomics of mussel transmissible cancer MtrBTN2 suggests accumulation of multiple cancer traits and oncogenic pathways shared among bilaterians. Open Biol. 13, 230259 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Di Poi C., Brodu N., Gazeau F., Pernet F., Life-history traits in the Pacific oyster Crassostrea gigas are robust to ocean acidification under two thermal regimes. ICES J. Mar. Sci. 79, 2614–2629 (2022). [Google Scholar]
- 56.Corporeau C., Vanderplancke G., Boulais M., Suquet M., Quéré C., Boudry P., Huvet A., Madec S., Proteomic identification of quality factors for oocytes in the Pacific oyster Crassostrea gigas. J. Proteomics 75, 5554–5563 (2012). [DOI] [PubMed] [Google Scholar]
- 57.Fantin J., Toutain J., Pérès E. A., Bernay B., Mehani S. M., Helaine C., Bourgeois M., Brunaud C., Chazalviel L., Pontin J., Corroyer-Dulmont A., Valable S., Cherel M., Bernaudin M., Assessment of hypoxia and oxidative-related changes in a lung-derived brain metastasis model by [64Cu][Cu(ATSM)] PET and proteomic studies. EJNMMI Res. 13, 102 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.R Core Team, R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna, Austria, 2022).
- 59.Langfelder P., Horvath S., WGCNA: An R package for weighted correlation network analysis. Bioinformatics 9, 559 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wright R. M., Aglyamova G. V., Meyer E., Matz M. V., Gene expression associated with white syndromes in a reef building coral, Acropora hyacinthus. BMC Genomics 16, 371 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Supek F., Bosnjak M., Skunca N., Smuc T., REVIGO summarizes and visualizes long lists of gene ontology terms. PLOS ONE 6, e21800 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dupoué A., Mello D. F., Trevisan R., Dubreuil C., Queau I., Petton S., Huvet A., Guével B., Com E., Pernet F., Salin K., Fleury E., Corporeau C., Intertidal limits shape covariation between metabolic plasticity, oxidative stress and telomere dynamics in Pacific oyster (Crassostrea gigas). Mar. Environ. Res. 191, 106149 (2023). [DOI] [PubMed] [Google Scholar]
- 63.Verhulst S., Improving comparability between qPCR-based telomere studies. Mol. Ecol. Resour. 20, 11–13 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mazaleyrat A., Normand J., Dubroca L., Fleury E., A 26-year time series of mortality and growth of the Pacific oyster C. gigas recorded along French coasts. Sci. Data 9, 392 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bates D., Maechler M., Bolker B., Walker S., Fitting linear mixed-effects models using lme4. J. Stat. Softw. 67, 1–48 (2015). [Google Scholar]
- 66.K. Barton, MuMIn: Multi-Model Inference, R package version 1436 (2019).
- 67.T. M. Therneau, coxme: Mixed Effects Cox Models (2015).
- 68.T. M. Therneau, A Package for Survival Analysis, R package version 3.4-0 (2022); https://CRAN.R-project.org/package=survival.
- 69.Hadfield J. D., MCMC methods for multi-response generalized linear mixed models: The MCMCglmm R package. J. Stat. Softw. 33, 1–22 (2010).20808728 [Google Scholar]
- 70.de Villemereuil P., Quantitative genetic methods depending on the nature of the phenotypic trait. Ann. N. Y. Acad. Sci. 1422, 29–47 (2018). [DOI] [PubMed] [Google Scholar]
- 71.Bolger A. M., Lohse M., Usadel B., Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T. R., STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Anders S., Pyl P. T., Huber W., HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Risso D., Ngai J., Speed T. P., Dudoit S., Normalization of RNA-seq data using factor analysis of control genes or samples. Nat. Biotechnol. 32, 896–902 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S8
Table S1