Summary
Background
Medications prescribed for chronic diseases can lead to short-term neuropsychiatric symptoms, but their long-term effects on brain structures and psychiatric conditions remain unclear.
Methods
We comprehensively analyzed the FDA Adverse Event Reporting System database and conducted drug target Mendelian Randomization (MR) studies on six categories of common drugs, 477 brain imaging-derived phenotypes (IDPs) and eight psychiatric disorders. Genetic instruments were extracted from expression quantitative trait loci (eQTLs) in blood, brain, and other target tissues, protein quantitative trait loci (pQTLs) in blood, and genome-wide association studies (GWAS) of hemoglobin and cholesterol. Summary statistics for brain IDPs, psychiatric disorders, and gut microbiome were obtained from the BIG40, Psychiatric Genomics Consortium, and MiBioGen. A two-step MR and mediation analysis were employed to screen possible mediators of drug-IDP effects from 119 gut microbiota genera and identify their mediation proportions.
Findings
Among 19 drug classes, six drugs were found to be associated with higher risks of psychiatric adverse events, while 11 drugs were associated with higher risks of gastrointestinal adverse events in the FAERS analysis. We identified ten drug-psychiatric disorder associations, 202 drug-IDP associations, 16 drug-microbiota associations, and four drug-microbiota-IDP causal links. For example, PPARG activation mediated HbA1c reduction caused a higher risk of bipolar disorder (BD) II. Genetically proxied GLP-1R agonists were significantly associated with an increase in the volume of the CA3-head of the right hippocampus and the area of the left precuneus cortex, both of which have been shown to correlate with cognition in previous studies.
Interpretation
Common drugs may affect brain structure and risk of psychiatric disorder. Oral medications in particular may exert some of these effects by influencing gut microbiota. This study calls for greater attention to be paid to the neuropsychiatric adverse effects of drugs and encourages drug repurposing.
Funding
National Natural Science Foundation of China (grant No. 82330035, 82130043, 82172685, and 82001223), National Natural Science Foundation of Hunan Province (grant No. 2021SK1010), and the Science Foundation for Distinguished Young Scholars of Changsha (grant No. kq2209006).
Keywords: Pharmacovigilance, Mendelian randomization, Common drugs, Psychiatric disorders, Brain structure, Gut microbiome
Research in context.
Evidence before this study
Previous studies and data from pharmacovigilance databases suggested that some drugs used for chronic diseases were associated with short-term neuropsychiatric symptoms.
Added value of this study
Through drug target Mendelian Randomization, we discovered ten drug-psychiatric disorder associations, 202 drug-IDP associations, 16 drug-microbiota associations, and four drug-microbiota-IDP causal links. Some of the effects were also observed in real-world data from FAERS, a pharmacovigilance database. This study allows a better understanding of how medications can affect our brains and mental health in the long term. It also showed how the gut microbiome can play a role in this process, especially when many common drugs are administrated orally.
Implications of all the available evidence
This study is a great reminder to pay more attention to the effects of common drugs on our brains and hopefully encourages the repurposing of old drugs for new psychiatric conditions.
Introduction
Chronic diseases, such as diabetes, cardiovascular diseases, and lung diseases, require prolonged medication. Many drugs used for these diseases have diverse degrees of neurological adverse reactions,1,2 while a few medications may have potential protective effects for neuropsychiatric conditions.3, 4, 5, 6, 7, 8, 9 For instance, treatment with liraglutide, a glucagon-like peptide-1 receptor (GLP-1R) agonist, enhanced associative learning and memory in individuals with obesity.10 Nevertheless, pre-marketing clinical trials of drugs often evaluate only short-term neuropsychiatric symptoms, leaving the long-term effects of drugs on brain structures and associated psychiatric conditions unknown. Addressing these gaps can significantly enhance clinical practice by improving patient care, facilitating early screening for drug-related neuropsychiatric adverse effects, aiding in decision-making when prescribing medications, and increasing public awareness of the long-term impacts of common drugs.
Mendelian randomization (MR) is an approach to uncover causal relationships by employing outputs from genome-wide association studies (GWAS) devoid of confounding variables. In essence, genetic variants, usually single nucleotide polymorphism (SNP), that are strongly associated with the exposure are selected as instrumental variables (IVs), and the causal effect of the exposure on the outcome is thus estimated by the effect of IVs on the outcome.11 By utilizing cis-expression quantitative trait loci (cis-eQTL) or SNPs linked with the drug target effect as IVs, it is also feasible to evaluate the impact of drugs on diseases or phenotypes, usually referred to as drug target MR. Following the general concepts of Mendelian randomization, drug target MR selected cis-variants that are associated with the drug target effect to proxy a certain drug class, allowing researchers to assess the potential effects of drugs on various outcomes.
Further, how may oral drugs impact brain structures and diseases? In addition to direct ways such as crossing the blood–brain barrier, affecting target gene expression, and activating signalling pathways in the neurons,12 the gut microbiota may serve as a key mediator linking oral medication use with brain structure and disease. Firstly, pharmacomicrobiomics has revealed that drugs can impact the composition of gut microbiota through direct microbial killing, modulation of host immune reaction, and alterations in intestinal pH.13, 14, 15 Secondly, gut microbiota can influence brain function via the microbiota-gut-brain axis, producing tryptophan metabolites and other neurotransmitters.16,17 Based on these established relationships, we hypothesize that common drugs, most of which are orally administrated, can affect brain structure partially by altering the gut microbiome, ultimately resulting in altered risks of psychiatric disorders.
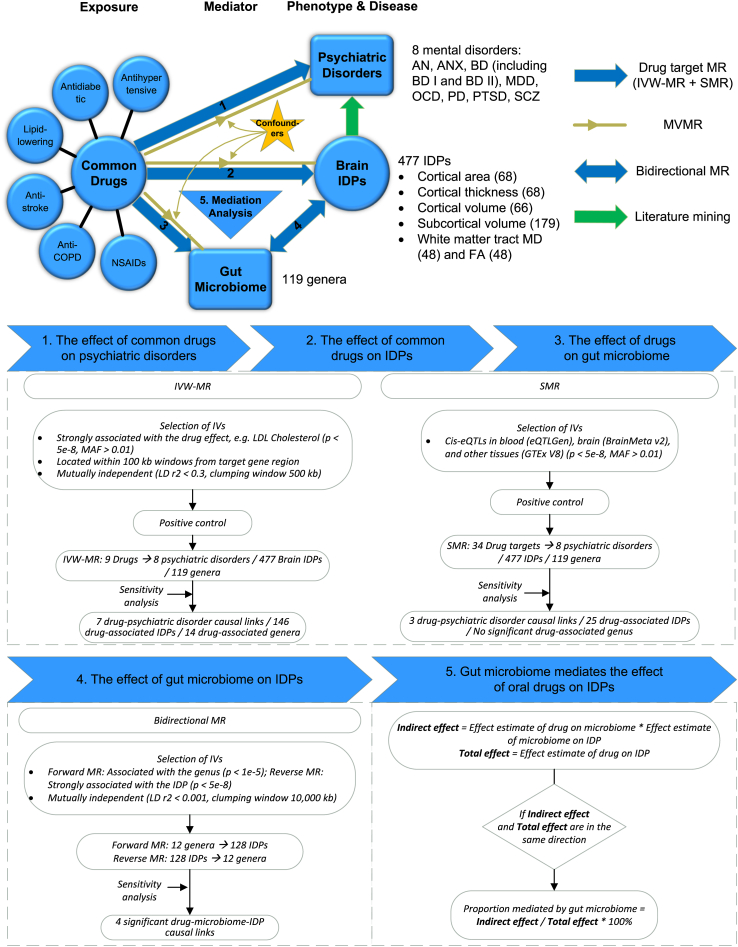
The primary goal of this study is to investigate the causal effect of six categories of common drugs on 477 brain imaging-derived phenotypes (IDPs) and eight psychiatric disorders using MR. Pharmacovigilance analysis of the FDA Adverse Event Reporting System (FAERS) database was conducted to support the MR findings. Through a two-step MR technique, 119 genera of gut microbiota were screened to identify possible mediators, followed by mediation analysis to determine the proportion mediated by a specific genus. A brief overview of this study is provided in Fig. 1. This study can uncover long-term neuropsychiatric effects of common medications, provide guidance on the appropriate usage of medication for patients afflicted by both chronic disease and poor mental health, and explore repurposing of marketed drugs for the treatment of psychiatric disorders.
Fig. 1.
Study workflow. Abbreviations: AN, anorexia nervosa; ANX, anxiety disorders; BD, bipolar disorder; COPD, chronic obstructive pulmonary disease; eQTL, expression quantitative trait loci; FA, fractional anisotropy; IDP, imaging-derived phenotype; IV, instrumental variable; IVW, inverse variance-weighted; LD, linkage disequilibrium; LDL, low-density lipoprotein; MAF, minor allele frequency; MD, mean diffusivity; MDD, major depressive disorder; MR, Mendelian randomization; MVMR, multivariable MR; NSAIDs, nonsteroidal anti-inflammatory drugs; OCD, obsessive-compulsive disorder; PD, panic disorder; PTSD, post-traumatic stress disorder; SCZ, schizophrenia; SMR, summary-data-based Mendelian randomization.
Methods
Study overview and data sources
An overview of the study workflow is provided in Fig. 1. The study was a two-sample drug target MR study supplemented by pharmacovigilance analysis of the FDA Adverse Event Reporting System (FAERS) database. We incorporated six categories of common drugs used for chronic diseases, including antidiabetic drugs, antihypertensive agents, lipid-lowering drugs, anti-stroke agents (anticoagulants and antiplatelet agents), chronic obstructive pulmonary disease (COPD) medications, and nonsteroidal anti-inflammatory drugs (NSAIDs). Detailed drug target information was listed in eTable 1. This study is reported according to the Strengthening the reporting of observational studies in epidemiology using mendelian randomisation (STROBE-MR) guidelines.18
Detailed information on all GWAS data sources is listed in eTable 2a. IDP GWAS summary statistics were obtained from the Oxford Brain Imaging Genetics Server (BIG40, https://open.win.ox.ac.uk/ukbiobank/big40/). The original study analyzed brain imaging data of 39,691 participants from UK biobank.19 We selected 477 IDPs from the original set of 3935, including 68 cortical area metrics, 68 cortical thickness metrics, 66 cortical volume metrics, 179 subcortical volume measures, and 96 white matter tract measures, with detailed information provided in eTable 2b. GWAS summary data of eight psychiatric disorders were acquired mainly through the Psychiatric Genomics Consortium (PGC, https://pgc.unc.edu/), with sample sizes ranging from ∼10,000 to ∼500,000. Microbiome quantitative trait loci (mbQTLs) were obtained from MiBioGen (https://mibiogen.gcc.rug.nl/). The original study involved an analysis of genotypes and fecal 16 S microbiome data from 18,340 participants across 24 cohorts.20 We only included 119 known genera for MR analysis as listed in eTable 2c. GWAS data from other studies such as the Megastroke project21 were used for positive control analysis.
Psychiatric, and digestive adverse events of common drugs: a pharmacovigilance analysis of the FAERS database
We conducted a pharmacovigilance investigation on the psychiatric and digestive adverse effects of the drugs of interest based on the FAERS database, a large public database of AEs reported by healthcare workers, pharmacists and consumers worldwide. We selected 130 FDA-approved drugs with oral products available as drugs of interest, with detailed information listed in eTable 3a and b. We analyzed adverse events (AEs) that listed the drugs of interest as PS (Primary Suspect Drug) and were reported by MD (Physician), PH (Pharmacist), or OT (Other health-professional) from January 2004 to March 2024. Duplicate reports were removed according to the following procedure: 1) sort the reports in the order of CASEID, FDA_DT, and PRIMARYID; 2) retain the reports with the largest FDA_DT value for reports with the same CASEID; 3) retain the reports with the largest PRIMARYID value for reports with the same CASEID and FDA_DT. We deleted all the irrelevant events including product issues, medication errors (administration, confusion, monitoring, etc.), and social circumstances, as listed in eTable 4. Demographic and clinical information of all the analyzed reports was summarized by each drug target. To compare the risk of AEs between the drug of interest and other drugs in the database, a two-by-two contingency table was constructed (eTable 5) and a disproportionality analysis was conducted using the reporting odds ratio (ROR) and proportional reporting ratio (PRR). ROR, PRR and their 95% confidence intervals (CIs) were calculated using the following formula:
AE signals were regarded as valid when the number of reports was no less than three and the lower threshold of the 95% CI of the ROR exceeded one.22 Robust signals were defined as valid signals meeting additional criteria of a PRR of at least 2 and a chi-squared value of at least 4.23 For each drug target, we reported the ROR and PRR of reports with SOC (System Organ Classes) names of ‘Psychiatric disorders’ and ‘Gastrointestinal disorders’. Additionally, we reported five PTs (Preferred Terms) with the highest RORs in the SOC of ‘Psychiatric disorders’ and ‘Gastrointestinal disorders’.
Within the dataset analyzed, over 80% of cases had at least one missing value of clinical characteristics. Direct deletion of all these cases could potentially reduce statistical power and introduce new biases, such as reports from developed countries being more standardized and complete compared to those from developing countries. To address this, we performed sensitivity analyses by sequentially deleting cases with missing values for each clinical characteristic. Specifically, sensitivity analyses were conducted for sex, age, weight, and country to determine if the cases with missing values will greatly affect the results.
The effect of common drugs on IDP, psychiatric disorders, and gut microbiome
Selection of IVs
MR analysis relies on three basic assumptions to make the causal inference: 1) the genetic variant is strongly associated with the exposure; 2) the genetic variant is not associated with the outcome via a confounding pathway; and 3) the genetic variant does not directly affect the outcome.24 Based on these assumptions, we used two methods to select IVs to proxy the drug effect, as described in previous MR studies.25
For the first method, we selected SNPs that meet the following requirements: 1) associated with the drug effect: SNPs associated with glycated hemoglobin (HbA1c) [false discovery rate (FDR) < 0.05] for antidiabetic drugs; SNPs associated with low-density lipoprotein cholesterol (LDL-C) (P-value < 5e-8) for lipid-lowering drugs; protein quantitative trait loci (pQTLs), i.e., SNPs associated with plasma protein level of F10 (P-value < 1e-5) and F2 (FDR < 0.05) for anticoagulants; pQTLs of PDE4A (P-value < 1e-5) for COPD medications; 2) located within ±100 kb windows from the drug target gene region; 3) minor allele frequency (MAF) > 0.01. As for the first two requirements, we intend to obtain SNPs that best mimic the effects of drugs on the human body as drug target IVs. Therefore, we preferentially select SNPs that are associated with the target effects (e.g., blood glucose reduction for antidiabetic drugs), and are located within certain range around the target gene region. No SNPs meeting the above criteria for antihypertensive agents, anticoagulants, and NSAIDs were identified. However, inspired by the work of Harshfield et al.,26 we realized that coagulation Factor II and Factor Xa protein expression could be used as a proxy for coagulation function, which is the target effect of thrombin inhibitors and Factor Xa inhibitors. In other words, the effects of thrombin inhibitors and Factor Xa inhibitors, are more direct without involving other mediating mechanisms. Under such circumstances, we used cis-pQTLs of F2, F10 and PDE4A as drug target IVs. As for the P value threshold, we start from the most stringent threshold (P < 5e-8) and gradually loosen the criteria to false discovery rate (FDR) < 0.05 or P < 1e-5, which were both used in previous publications.27,28 Our purpose was to obtain an adequate number of IVs so that the MR results will be more statistically powerful. Subsequent linkage disequilibrium (LD) clumping was conducted based on LD r2 < 0.3 and a clumping window of 500 kb, utilizing the 1000 genomes reference panel (“EUR”). The clumping threshold was chosen to balance the statistical power and the stability of drug target MR. Specifically, a loose LD r2 threshold reduces the reliability of MR results, while a stringent LD r2 threshold reduces the statistical power.29 Therefore, we adopted the clumping threshold of LD r2 < 0.3, a widely used threshold in drug target MR.25,30,31 Exposure and outcome data were harmonized and palindromic SNPs with intermediate allele frequencies were eliminated. To confirm the validity of IVs, positive control analyses were performed for each drug class: for antidiabetic drugs, we included glucose and type 2 diabetes mellitus (T2D) as the outcome; for lipid-lowering drugs, we incorporated coronary artery disease (CAD) as the outcome; for anti-stroke agents, we included any stroke (AS), any ischemic stroke (AIS), large artery stroke (LAS), cardioembolic stroke (CES), and small vessel stroke (SVS) as the outcome; for COPD medications, we included forced expiratory volume in 1 s/forced vital capacity (FEV1/FVC) ratio and percentage of predicted forced expiratory volume in the first second of expiration (FEV1% predicted) as the outcome.
For the second approach, we extracted common (MAF >0.01) expression quantitative trait loci (eQTLs) associated with the expression of drug target genes (P-value < 5e-8) from eQTLGen Consortium, BrainMeta v2, and GTEx V8, with detailed information listed in eTable 2a. To reduce potential pleiotropy, we only included cis-eQTLs as IVs, defined as eQTLs located within ±1 Mb range from the encoded gene. The prioritization order of different database sources for selecting IVs is determined as follows. Generally, since we are interested in neuropsychiatric effects of common drugs and gene expressions in the brain seems to more directly affect psychiatric disorders and brain structures than gene expressions in the blood or other tissues, using brain eQTLs as IVs may be more appropriate. However, the sample size of BrainMeta v2 is relatively small (around 2400) compared with eQTLGen (around 31,000) and relevant studies have demonstrated a high correlation of cis-eQTLs across different tissues, especially between brain and blood.32 Therefore, we preferentially select blood eQTLs from eQTLGen. For other drug targets without cis-eQTLs that meet the requirements in the eQTLGen Consortium, we extract eQTLs in brain (from BrainMeta v2), and subsequently in corresponding target tissues (from GTEx V8) if there are no qualified cis-eQTLs in BrainMeta v2, since these eQTLs can reflect the major target effects of common drugs. Specifically, we extracted eQTLs in skeletal muscle for CACNG1, eQTLs in tibial artery for CACNA1S, and eQTLs in subcutaneous adipose for NPC1L1. Although these drugs have more than one target tissues, the other target tissues have no qualified eQTLs. We investigated genetic associations between selected IVs and drug indications or drug effects as positive control analyses. Specifically, we incorporated Fasting glucose (FG), 2 h-glucose post–challenge (2hGlu), Glycated hemoglobin (HbA1c), and Fasting insulin (FI) as the outcome for antidiabetic drugs; systolic blood pressure (SBP) and diastolic blood pressure (DBP) as the outcome for antihypertensive agents; five stroke types (AS, AIS, LAS, CES, and SVS) as the outcome for anti-stroke agents; FEV1/FVC ratio as the outcome for COPD medications; pain in joint and low back pain as the outcome for NSAIDs.
To validate the effectiveness of the IVs, the F-statistic was computed for each SNP using the provided formula33:
where n = sample size, k = number of IVs, R2 = explained variance of genetic instruments on exposure, β = effect size of SNPs, and se = standard error of effect size.
We ensured that the F-statistics of all the IVs used for the MR analysis exceeded 10 to prevent any violations of Assumption 1 of mendelian randomization analysis.34
Primary MR analysis
The inverse variance-weighted (IVW) method with a multiplicative random-effects model was applied for the first set of IVs. This method meta-analyzes the Wald ratios of each IVs to obtain an overall estimate, which has the highest statistical power compared with other MR methods.35 Causal estimates were reported per 1% increase of HbA1c for antidiabetic drugs, per 1 mmol/L increase of LDL cholesterol for lipid-lowering drugs, and per one standard deviation (SD) increase of protein expression for anti-stroke agents and COPD medications.
Summary-data-based MR (SMR) was employed for the second set of IVs (eQTLs extracted from different sources). This method first identifies the ‘top SNP’ that has the most significant associations with mRNA expression of exposure genes, and then uses the top SNP to analyze the genetic associations between mRNA expressions and outcome traits. SMR is usually combined with heterogeneity in dependent instruments (HEIDI) analysis to eliminate false positive results due to linkage disequilibrium (LD) between the Top SNP and other SNPs associated with the outcome, which will be discussed later.36 Causal estimates were reported per one SD increase of mRNA expression of target genes.
FDR correction was utilized to account for multiple tests. Findings with FDR values less than 0.05 were viewed as significant, while those greater than 0.05 but with P-values less than 0.05 were considered nominally significant.
Assessment of the MR assumptions and sensitivity analysis
For IVW-MR method, we computed the F-statistic as previously stated to assess the relevance assumption. To examine the robustness of the IVW results, we utilized the MR-Egger method for sensitivity analysis. The MR-Egger method remains efficacious even when all IVs are invalid, but is based on a weaker assumption termed InSIDE (Instrument Strength Independent of Direct Effect).37 The slope coefficient and the intercept estimate obtained through the MR-Egger regression were utilized to estimate the causal effects and identify potential pleiotropy, respectively. Results with an MR-Egger intercept significantly differing from zero (P-value <0.05) were deemed unreliable and biased due to pleiotropy. Additionally, we drew funnel plots to assess heterogeneity across genetic variants visually and calculated Cochran's Q statistics from the IVW. I2 was calculated using the formula I2 = (Q-df)/Q × 100%, where Q = Cochran's Q statistics and df = degree of freedom; if I2 < 0, it was set to 0. To verify if the IVW results were affected by a genetic variant, a leave-one-out analysis was performed. To identify potential outlier SNPs, MR pleiotropy residual sum and outlier (MR-PRESSO) were conducted.38 In cases where the MR-PRESSO global test P-value was less than 0.05, IVW was re-run after excluding outlier SNPs.
Additionally, several other approaches were leveraged to test the MR assumptions. MR Steiger test was performed to determine whether the IVs were more associated with the outcome than with the exposure.39 False directionality was considered present when ‘steiger_dir’ is FALSE and ‘steiger_pval’ is less than 0.05. Under such circumstances, we re-performed IVW-MR after removing IVs with false directionality. Moreover, to test the independence assumption, we searched for evidences that suggest the associations between the IVs and potential confounders. Confounders are any factors that are linked with both the exposure and the outcome, which may bias the causal inference. However, it is difficult to identify all potential confounders, and hence we chose four major confounders that have been proven to be associated with psychiatric disorders based on established work, including smoking, alcohol consumption, education attainment, and income.40, 41, 42, 43 We queried three databases for associations regarding all the IVs used in the IVW-MR analysis with a P value threshold of < 5e-8, including the Genome-Wide Repository of Associations Between SNPs and Phenotypes (GRASP)44 (https://grasp.nhlbi.nih.gov/Search.aspx), the MRC IEU OpenGWAS database45,46 (https://gwas.mrcieu.ac.uk/), and the GWAS Catalog47 (https://www.ebi.ac.uk/gwas/). Queries in the GRASP were manually done while searches in the other two databases were through their REST API, with resources provided at the end of Methods. Subsequently, we re-performed IVW-MR after removing IVs associated with the four confounders. Multivariable MR (MVMR) is a MR method that can investigate the effects of multiple exposures on the outcome simultaneously, which can be used to adjust for potential confounders, or calculating direct effects in mediation analysis.48, 49, 50 Eventually, multivariable MR (MVMR) were conducted for significant associations between drugs and psychiatric disorder/IDP/gut microbiota to adjust for the four confounders, with similar processes as univariate MR. We retrieved summary statistics of the four confounders using IVs selected in the former univariate MR, with detailed GWAS information listed in eTable 2a. Then exposure data were combined and harmonized with outcome data, and MVMR was performed using the IVW method. Furthermore, since we used GWAS summary statistics of HbA1c to proxy the target effects of antidiabetic drugs, and HbA1c is also related to red blood cell (RBC) count, RBC count is likely to be a confounding factor due to its relationship with psychiatric disorders.51 Therefore, for antidiabetic drugs we performed additional MVMR adjusting for RBC count. Although the lipid-lowering drugs included in our study primarily reduce LDL-C, some of them may affect other lipid components like HDL-C and triglycerides (TG) as well.52, 53, 54 To explore potential pathways other than reducing LDL-C, we performed MVMR for formerly identified significant associations between lipid-lowering drugs and psychiatric disorder/IDP/gut microbiota, incorporating LDL-C, HDL-C, and TG as exposures. In MVMR analysis, we considered results with a P value of <0.05 as significant.
For the SMR method, the F-statistics of the top SNP were calculated using the above formula. HEIDI analysis was performed, which incorporates significant SNPs other than the top SNP in the cis-eQTL region to test whether genetic associations were due to linkage disequilibrium (LD). Results with HEIDI P-value <0.05 were considered due to linkage rather than functional association.
Bidirectional MR testing the effect of gut microbiome on IDPs
Selection of IVs
In the forward MR, gut microbiome was included as exposures and IDPs were included as outcomes, while in the reverse MR, IDPs were seen as exposures and gut microbiome was seen as outcomes. SNPs that meet the following requirements were selected as IVs: 1) associated with the gut microbiome for forward MR (P-value < 1e-5) or IDPs for reverse MR (P-value < 5e-8); 2) mutually independent (LD r2 < 0.001) and clumping window of 10 Mb using the “EUR” panel; and 3) MAF >0.01. Exposure and outcome data were harmonized and palindromic SNPs with intermediate allele frequencies were eliminated. The F-statistic was computed for each SNP using the formula described above.
Primary MR analysis
The IVW method with a multiplicative random-effects model was applied for the primary MR analysis. Causal estimates were reported per one SD increase in bacterial abundance for the forward MR, and per one SD increase in the phenotype for the reverse MR.
Assessment of the MR assumptions and sensitivity analysis
Similar to the former process, we computed the F-statistics and incorporated the weighted median, weighted mode, and MR-Egger methods for sensitivity analysis. The weighted median method can generate a reliable estimate when less than half of the IVs are invalid.55 The weighted mode method is useful when over half of the IVs are invalid, and depends on the zero-modal pleiotropy assumption (ZEMPA). This assumption implies that, among all the causal estimates obtained from each instrument, the most frequent estimate is a robust estimate of the true causal effect.56 For forward MR, results with IVW P-value <0.05 and one of the additional three methods P-value <0.05 were considered significant. For reverse MR, results with IVW P-value <0.05 were considered significant. Results with an Egger intercept P-value <0.05 were excluded from further analysis due to pleiotropy bias. To assess heterogeneity, we computed Cochran's Q statistics and I2 from IVW and used funnel and leave-one-out plots for visual examination. MR-PRESSO was conducted and if the global P-value of the MR-PRESSO test was less than 0.05, IVW was repeated after excluding outlier SNPs. Additional sensitivity analysis was performed by removing IVs with false directionality identified by the Steiger test and subsequently removing IVs associated with confounders, as described earlier.
Mediation analysis
We then performed a mediation analysis to evaluate the proportion of drug-IDP effect mediated by gut microbiota. The direct effect was the IVW estimate of drug-IDP associations. The indirect effect was calculated by the product of drug-microbiome IVW causal estimate and microbiome-IDP IVW causal estimate. The mediation proportion was then derived by dividing the indirect effect by the direct effect. The point estimate and 95% CI were obtained using the Monte Carlo method.
Replication analyses based on UK Biobank research
We replicated the IVW-MR analysis of drugs on psychiatric disorders using different GWAS data sources listed in eTable 2a, applying a stringent P-value threshold for IV selection. For antidiabetic drugs, we used a recent GWAS study of HbA1c involving 338,919 UK Biobank participants.57 For anticoagulants, we utilized the pQTL data from the UK Biobank Pharma Proteomics Project (UKB-PPP) including 34,557 European participants.58 The P-value threshold was set to 5e-8. Other processes, including subsequent IV filtering, positive control analysis, and IVW-MR analysis were conducted as previously described.
Software and packages
All the MR analyses, including sensitivity analysis, were performed via either ‘TwoSampleMR’ package (v0.5.6) in R software 4.2.3 (https://www.r-project.org/) or SMR software (v1.3.1)36 (https://yanglab.westlake.edu.cn/software/smr/#Download). Queries for the associations of IVs with confounders were performed using the ‘tophits’ function implemented in the ‘ieugwasr’ package (v1.0.0) and the ‘fromJSON’ function in the ‘jsonlite’ package (v1.8.8) to access the REST API of the IEU OpenGWAS and the GWAS Catalog, respectively. Cortical structures were visualized with the BrainNet Viewer59 (http://www.nitrc.org/projects/bnv/). Subcortical structures were drawn using the ‘ggseg’ package (v1.6.6). White matter tracts were visualized via the ‘ggsegICBM’ package (v1.0.1) and ‘ggseg3d’ package (v1.6.3). FDR values were calculated using the Benjamini-Hochberg method via the ‘p.adjust’ function implemented in the ‘stats’ package (v4.2.3). The Monte Carlo method for the calculation of the point estimate and 95% CI in mediation analysis was implemented by the ‘ci’ function of the ‘RMediation’ package (v1.2.2).
Ethics
All MR analyses were conducted using publicly available GWAS datasets, with detailed information provided in eTable 2a. All these GWAS studies have received ethical approval and the participants have provided informed consent. No individual-level data were utilized in this study, thus eliminating the need for new ethical review board approval.
Role of funders
The funders had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Results
Discovery of psychiatric adverse effects of common drugs through analysis of the FAERS database
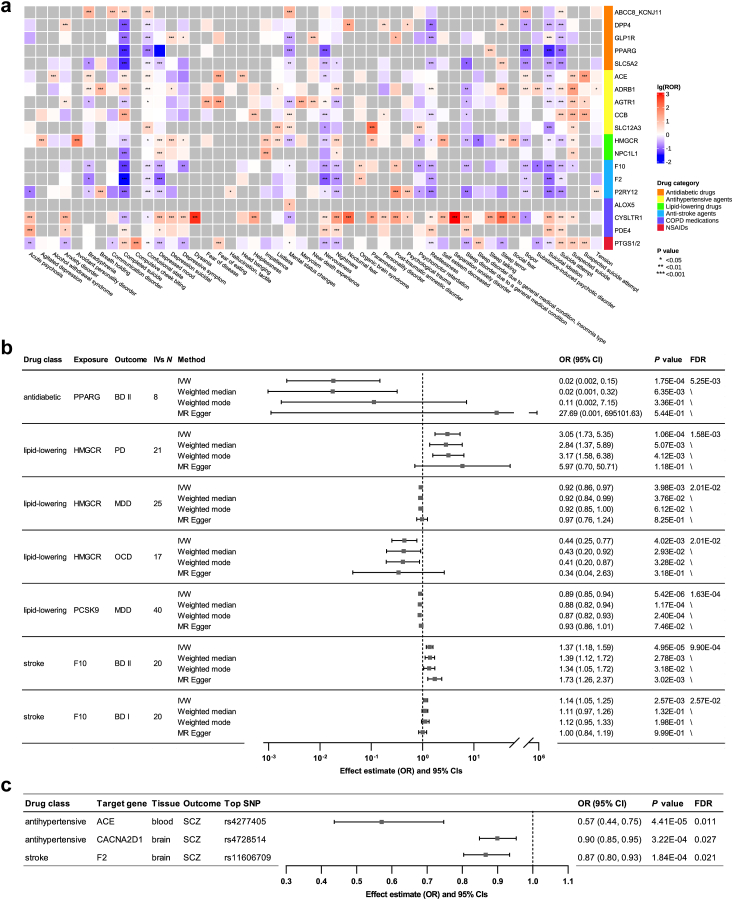
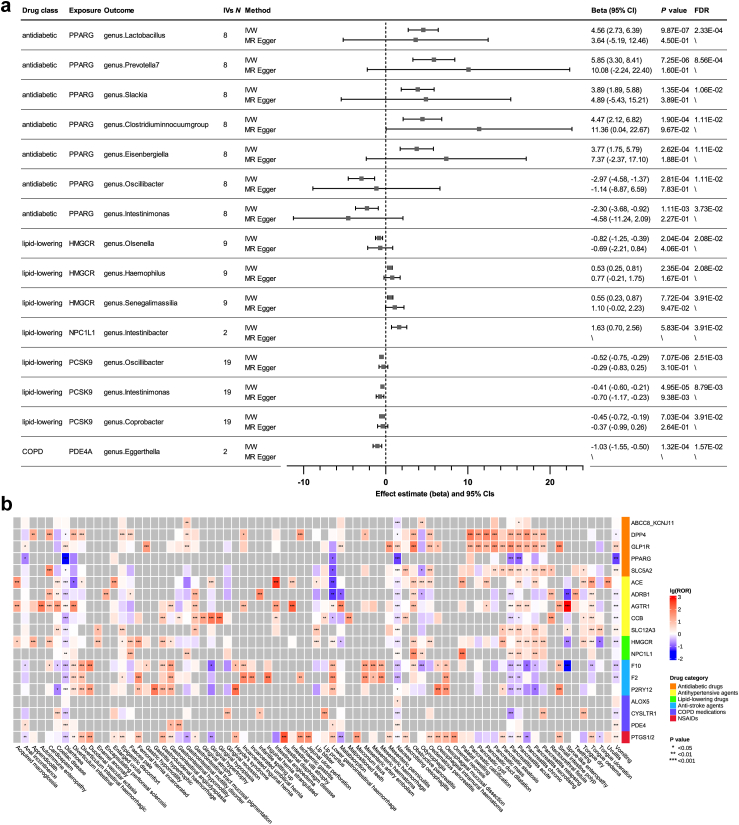
We first carried out a retrospective analysis of the psychiatric adverse effects of common drugs using the FAERS database. We searched for adverse event (AE) records concerning the drugs under investigation and found that most products documented in the database were administered orally. Therefore, only orally administered medications were included to eliminate adverse reactions caused by the modes of administration, such as skin reactions from injections. Clinical characteristics of psychiatric AE reports, including sex, age, weight, country, reporter type, and patient outcome were summarized in eTable 6. Among 19 drug classes analyzed, six drugs were significantly associated with higher psychiatric AEs based on the reporting odds ratio (ROR) criteria, with a robust signal detected for leukotriene receptor antagonists (Table 1). Leukotriene receptor antagonists have the highest effects on psychiatric AEs (ROR [95% CI] = 7.37 [7.16, 7.6]). For each drug class, five PTs (Preferred Terms) belonging to SOCs (System Organ Classes) of ‘Psychiatric disorders’ with the highest ROR were listed in eTable 8. As shown in the ROR heatmap (Fig. 2a), leukotriene receptor antagonists were associated with an increased risk of multiple psychiatric PTs, with the highest effects on ‘Separation anxiety disorder’ (ROR = 1019.07 [432.75, 2399.79]).
Table 1.
ROR and PRR of psychiatric AEs among 19 drugs.
| Drug category | Drug target | Number of psychiatric AEs (a) | Total number of AEs (a+b) | Percentage of psychiatric AEs [a/(a+b)] | ROR (95% CI) | P_value_ROR | PRR (95% CI) | χ2 | P_value_PRR |
|---|---|---|---|---|---|---|---|---|---|
| Antidiabetic drugs | ABCC8+KCNJ11∗ | 522 | 6940 | 7.52% | 1.51 (1.38, 1.65) | 1.56E-19 | 1.47 (1.39, 1.56) | 83.8 | 3.81E-39 |
| Antidiabetic drugs | DPP4 | 647 | 26,378 | 2.45% | 0.47 (0.43, 0.51) | 2.11E-67 | 0.48 (0.4, 0.56) | 383.3 | 1.22E-17 |
| Antidiabetic drugs | GLP1R | 1218 | 50,432 | 2.42% | 0.46 (0.43, 0.49) | 4.02E-120 | 0.47 (0.42, 0.53) | 754.7 | 4.40E-37 |
| Antidiabetic drugs | PPARG | 197 | 53,836 | 0.37% | 0.07 (0.06, 0.08) | 1.68E-287 | 0.07 (0, 0.21) | 2502.0 | 1 |
| Antidiabetic drugs | SLC5A2 | 1057 | 67,339 | 1.57% | 0.3 (0.28, 0.31) | 0 | 0.31 (0.25, 0.37) | 1742.5 | 1.13E-31 |
| Antihypertensive agents | ACE | 3457 | 87,128 | 3.97% | 0.77 (0.74, 0.79) | 2.42E-55 | 0.78 (0.74, 0.81) | 233.2 | 4.47E-27 |
| Antihypertensive agents | ADRB1∗ | 7455 | 105,052 | 7.10% | 1.42 (1.39, 1.46) | 3.11E-172 | 1.39 (1.37, 1.42) | 865.9 | 5.54E-284 |
| Antihypertensive agents | AGTR1 | 2578 | 79,569 | 3.24% | 0.62 (0.6, 0.65) | 3.28E-121 | 0.63 (0.6, 0.67) | 573.1 | 1.54E-60 |
| Antihypertensive agents | CCB∗ | 9489 | 100,768 | 9.42% | 1.94 (1.9, 1.98) | 0 | 1.85 (1.83, 1.87) | 3889.9 | 0 |
| Antihypertensive agents | SLC12A3 | 796 | 18,701 | 4.26% | 0.83 (0.77, 0.89) | 4.58E-07 | 0.83 (0.77, 0.9) | 27.8 | 2.84E-06 |
| Lipid-lowering drugs | HMGCR | 5766 | 137,391 | 4.20% | 0.81 (0.79, 0.84) | 2.69E-41 | 0.82 (0.8, 0.85) | 235.0 | 1.09E-37 |
| Lipid-lowering drugs | NPC1L1 | 271 | 8643 | 3.14% | 0.6 (0.53, 0.68) | 9.36E-16 | 0.61 (0.5, 0.73) | 69.2 | 3.05E-07 |
| Anti-stroke agents | F10 | 2062 | 170,257 | 1.21% | 0.23 (0.22, 0.24) | 0 | 0.24 (0.19, 0.28) | 5373.3 | 3.50E-47 |
| Anti-stroke agents | F2 | 473 | 48,321 | 0.98% | 0.18 (0.17, 0.2) | 0 | 0.19 (0.1, 0.28) | 1701.8 | 2.57E-10 |
| Anti-stroke agents | P2RY12 | 1687 | 106,795 | 1.58% | 0.3 (0.28, 0.31) | 0 | 0.31 (0.26, 0.36) | 2753.5 | 3.39E-45 |
| COPD medications | ALOX5 | 6 | 145 | 4.14% | 0.8 (0.35, 1.82) | 5.96E-01 | 0.81 (0.03, 1.59) | 0.3 | 8.35E-01 |
| COPD medications | CYSLTR1∗∗ | 6007 | 21,230 | 28.29% | 7.37 (7.16, 7.6) | 0 | 5.57 (5.55, 5.59) | 23600.1 | 0 |
| COPD medications | PDE4∗ | 599 | 6018 | 9.95% | 2.06 (1.89, 2.24) | 2.00E-62 | 1.95 (1.87, 2.03) | 292.3 | 3.96E-223 |
| NSAIDs | PTGS1+PTGS2∗ | 16,523 | 305,619 | 5.41% | 1.06 (1.05, 1.08) | 5.14E-16 | 1.06 (1.05, 1.08) | 58.7 | 5.14E-16 |
Disproportionality analysis were performed by calculating the ROR and PRR. Significant adverse reaction signals (the number of reports was not less than three and the lower limit of the 95% CI of the ROR exceeds one) were marked with an asterisk (∗). Robust signals (meeting additional criteria of a PRR of at least 2 and a chi-squared value of at least 4) were marked with two asterisks (∗∗).
Fig. 2.
Psychiatric effects of common drugs. a, ROR heatmap for psychiatric PTs of common drugs from the disproportionality analysis on the FAERS database. The data are expressed as log-transformed ROR values with unadjusted P values (disproportionality analysis). ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001. b, Significant results (FDR <0.05, RE-IVW) from IVW-MR on the effects of common drugs on psychiatric disorders. Data were analyzed using IVW as the primary method, and other three methods (weighted median, weighted mode, and MR Egger) as sensitivity analysis. The causal estimates are expressed as odds ratio values with 95% CI values. The width of the lines extending from the midpoint represent the 95% CI (the scale has been log-transformed). Both unadjusted P values (RE-IVW, weighted median, weighted mode, and MR Egger) and FDR values (RE-IVW) are given. c, Significant results (FDR <0.05, SMR) from SMR on the effects of common drugs on psychiatric disorders. The causal estimates are expressed as odds ratio values with 95% CI values. The width of the lines extending from the midpoint represent the 95% CI. Both unadjusted P values (SMR) and FDR values (SMR) are given.
In sensitivity analyses, most results still remain stable after deleting cases with missing age, country or sex, respectively. For example, all of the valid signals in the SOC level still remained significant, and 76.1%, 98.5%, and 89.6% of the top five PTs still remained in the top five list after deleting the cases missing age, country or sex, respectively. However, since the number of cases missing weight is relatively large, only 50% of the valid signals in the SOC level and 38.8% of the top five PTs still remained after deleting the cases missing weight.
The effect of common drugs on psychiatric disorders
Since all drugs under investigation have specific target genes, it's feasible to proxy the drug effects using cis-genetical variants. To further decipher the long-term effect of common drugs on psychiatric disorders, we conducted drug target MR using two different methods, inverse variance-weighted MR (IVW-MR) and summary-data-based MR (SMR).
IV selection and validation
As shown in eTable 9, the minimum F-statistic of the IVs for IVW-MR was 13, suggesting a low probability of weak instrument bias. In positive control analyses (eTable 10a), most IVs yielded significant results, indicating the reliability of the IVs. For instance, GLP1R-mediated HbA1c increase (per one percent increase) was significantly associated with a higher risk of type 2 diabetes mellitus (T2D) (odds ratio [OR] = 78.18 [9.24, 661.74], P = 6.33e-5, random-effect inverse variance-weighted [RE-IVW]).
For the SMR method, the minimum F-statistic of the top SNP was 31, indicating strong associations between the IVs and gene expression (eTable 10b). However, in the positive control analysis (eTable 10b), only 18.7% (17/91) results yielded significant results (p_SMR <0.05 and p_HEIDI >0.05), among which nine results yielded estimates that were directionally correct.
Main findings
IVW-MR identified seven significant associations between four drugs and four psychiatric disorders (Fig. 2b and eTable 11). PPARG-mediated HbA1c increase was associated with a significantly lower risk of bipolar disorder Ⅱ (BD Ⅱ) (OR = 0.02 [0.00, 0.15], P = 1.75e-04, RE-IVW). HMGCR-mediated LDL cholesterol increase was associated with a lower risk of major depressive disorder (MDD) (OR = 0.92 [0.86, 0.97], P = 3.98e-03, RE-IVW), a lower risk of obsessive-compulsive disorder (OCD) (OR = 0.44 [0.25, 0.77], P = 4.02e-03, RE-IVW), and a higher risk of panic disorder (PD) (OR = 3.05 [1.73, 5.35], P = 1.06e-04, RE-IVW). PCSK9-mediated LDL cholesterol increase was associated with a lower risk of MDD (OR = 0.89 [0.85, 0.94], P = 5.42e-06, RE-IVW). Genetically determined F10 protein expression was associated with a higher risk of BD Ⅰ (OR = 1.14 [1.05, 1.25], P = 2.57e-03, RE-IVW) and BD Ⅱ (OR = 1.37 [1.18, 1.59], P = 4.95e-05, RE-IVW).
In sensitivity analyses, MR Egger produced one significant result (P < 0.05, MR Egger) out of the seven associations identified by IVW, which was in a consistent direction as that from IVW-MR. Other sensitivity analyses revealed no abnormalities (eTable 11). The scatter plots of the seven significant results are shown in eFig. 1. We did not observe significant asymmetry in funnel plots (eFig. 2). Leave-one-out analysis revealed that the combined effect estimate was not driven by any single SNP (eFig. 3). MR Steiger test did not detect any IVs with wrong directionality. After removing IVs associated with the four confounders (i.e., smoking, drinking, education, and income) as listed in eTable 12, the IVW-MR results did not change significantly, as shown in eTable 13. In the MVMR analysis adjusting for the four confounders as shown in eTable 14a, although relatively large mediation effects were observed in the effects of HMGCR on OCD by smoking heaviness, and of PPARG on BD II by drinking and income, 77.8% of direct effects remained significant after adjustment. Among the three antidiabetic drugs, only PPARG agonists have more than one SNP as drug target IVs, hence we performed MVMR for PPARG agonists adjusting for red blood cell (RBC) count. As shown in eTable 14b, there are no significant changes after adjusting for RBC count, suggesting the robustness of our results. To explore potential mechanisms behind these associations, we performed MVMR for lipid-lowering drugs incorporating LDL-C, HDL-C, and triglycerides (TG). As demonstrated in eTable 14c, the effect of HMGCR mediated LDL decrease on OCD became insignificant after adjustment while the effect of HDL on OCD obtained significant results, suggesting that the effect of HMGCR inhibitors on OCD may be mediated through TG reduction rather than LDL reduction.
SMR analyses revealed three significant associations between three drugs and schizophrenia (SCZ), as shown in Fig. 2c and eTable 15. Genetically determined ACE expression was associated with a lower risk of SCZ (OR = 0.57 [0.44, 0.75], P = 4.41e-05, SMR). Genetically determined CACNA2D1 expression was associated with a lower risk of SCZ (OR = 0.90 [0.85, 0.95], P = 3.22e-04, SMR). Genetically determined F2 expression was associated with a lower risk of SCZ (OR = 0.87 [0.80, 0.93], P = 1.84e-04, SMR). HEIDI tests did not show any significant findings that could be attributed to linkage disequilibrium.
The effect of common drugs on IDPs
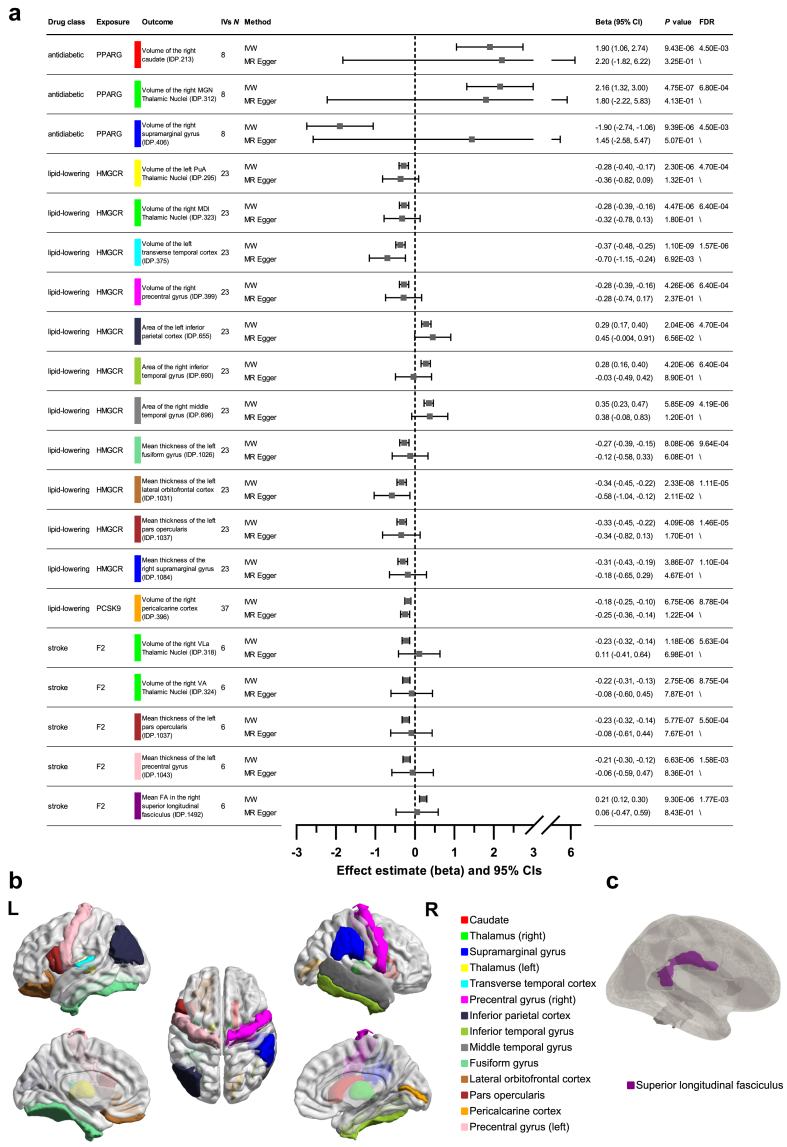
IVs selection and validation were already described above. IVW-MR analyses identified 174 significant (false discovery rate [FDR] < 0.05, RE-IVW) drug-IDP associations between eight drugs (except PDE4A) and 143 IDPs (including 22 cortical volume metrics, 16 cortical area metrics, 28 cortical thickness metrics, 11 subcortical volume metrics, 37 subregions of Amygdala Nuclei, Hippocampal Subfields, and Thalamic Nuclei, 29 white matter tract measures), as shown in eTable 16. The top 20 significant results, which also passed Bonferroni correction, are shown in Fig. 3a, with the schematic diagram of the relevant brain regions presented in Fig. 3b and c. Interestingly, both adverse effects and positive effects of common drugs on brain structures were found. Among all the drugs, genetically proxied HMGCR inhibitors had the most extensive impact on brain structures, which were linked with 92 IDPs with the majority being positive associations. It was followed by PCSK9 inhibitors and F2 inhibitors, which were associated with 27 and 21 IDPs respectively, also with the majority of them being positive associations. Notably, PPARG agonists, SGLT2 inhibitors and F10 inhibitors mainly had adverse effects on brain structures. Additionally, genetically proxied GLP1R agonists were significantly associated with a substantial increase in the volume of the CA3-head of the right hippocampus (|beta| = 4.64) and the area of the left precuneus cortex (|beta| = 4.40).
Fig. 3.
Effects of common drugs on IDPs identified by IVW-MR. a, Top 20 significant results, also those passing Bonferroni correction. Data were analyzed using IVW as the primary method and MR Egger as sensitivity analysis. The causal estimates are expressed as β values with 95% CI values. The width of the lines extending from the midpoint represent the 95% CI. Both unadjusted P values (RE-IVW and MR Egger) and FDR values (RE-IVW) are given. b, Schematic diagram of the brain regions in lateral, medial, and dorsal views. c, Schematic diagram of the white matter tract in right lateral view.
In sensitivity analyses, MR Egger yielded 29 significant results (P < 0.05, MR Egger) among 174 associations identified by IVW, which were all in the same direction as those from IVW-MR. MR-Egger intercept analysis indicated no significant horizontal pleiotropy. Cochran's Q test revealed significant heterogeneity between the genetic variants for F2-IDP.328 (P = 0.034, Cochran's Q test, I2 = 58%). However, this heterogeneity will not significantly affect our results since we used the IVW method with a multiplicative random-effects model. MR-PRESSO revealed no significant outlier SNPs. All the above results are displayed in eTable 16. The scatter plots of the top 20 significant results are shown in eFig. 4. Asymmetry in funnel plots was observed for PCSK9 inhibitors on IDP.396 (eFig. 5). Leave-one-out analysis revealed that the combined effect estimate was not affected by the removal of any single SNP (eFig. 6). As presented in eTable 17a, MR Steiger test detected five SNPs with wrong directionality, but after removing those SNPs, the MR results did not change significantly. After removing IVs associated with the four confounders as listed in eTable 12, 97.2% of the IVW-MR results did not change significantly, as shown in eTable 17b. However, four associations previously identified as significant became insignificant, and nine associations previously identified as insignificant became significant after removing confounder-associated SNPs. In the MVMR analysis adjusting for the four confounders as shown in eTable 18a, although relatively large mediation effects were observed in some cases such as the effects of PPARG on Volume of 5th-Ventricle by the four confounders, 78.2% of direct effects remained significant after adjustment. As shown in eTable 18b, 78.2% of the results remained significant or insignificant after adjusting for RBC count. As demonstrated in eTable 18c, 47.3% of the direct effects remained significant after adjusting for HDL and TG, suggesting that half of associations between lipid-lowering drugs and IDPs may be mediated through other mechanisms other than LDL.
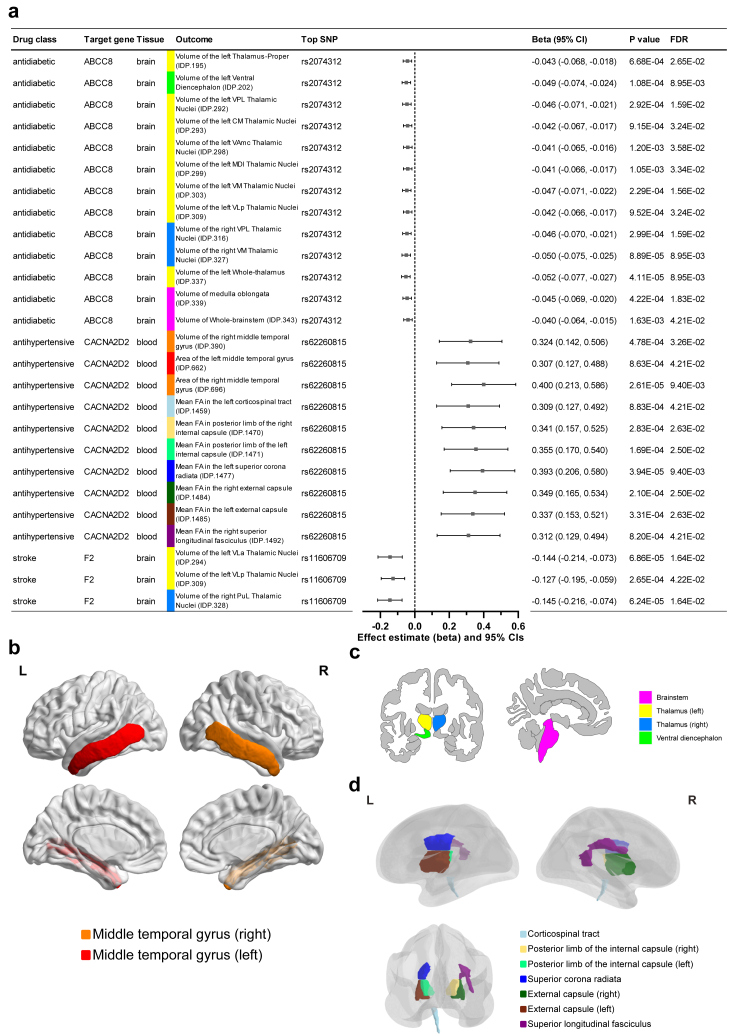
SMR analyses revealed 26 significant gene-IDP associations (FDR <0.05, SMR) between three drug target genes (F2, ABCC8, and CACNA2D2) and 25 IDPs (including 3 cortical measures, 15 subcortical measures, and 7 white matter tract measures), as shown in Fig. 4 and eTable 19. Only positive drug-IDP associations were identified for genetically determined F2 and ABCC8 expressions, while genetically determined CACNA2D2 expression only had negative drug-IDP associations. Specifically, genetically proxied CACNA2D2 inhibition led to a reduction in the area and volume of the middle temporal gyrus, and a decrease in mean FA in the left corticospinal tract, right superior longitudinal fasciculus, external capsule, posterior limb of internal capsule, and left superior corona radiata. HEIDI tests revealed no significant results that were due to linkage disequilibrium.
Fig. 4.
Effects of common drugs on IDPs identified by SMR. a, Significant results with FDR <0.05 (SMR). The causal estimates are expressed as β values with 95% CI values. The width of the lines extending from the midpoint represent the 95% CI. Both unadjusted P values (SMR) and FDR (SMR) values are given. b, Schematic diagram of the brain cortical structures in lateral and medial views. c, Schematic diagram of the subcortical structures in coronal and sagittal section views. d, Schematic diagram of the white matter tracts in lateral and rear views.
The effect of common drugs on gut microbiota and gastrointestinal AEs of common drugs
IVs selection and validation were already described above. IVW-MR identified 15 drug-microbiota associations between five drugs (PPARG agonists, HMGCR inhibitors, NPC1L1 inhibitors, PCSK9 inhibitors, and PDE4 inhibitors) and 13 genera, as shown in Fig. 5a and eTable 20.
Fig. 5.
Causal effects of drugs on gut microbiota and ROR heatmap for gastrointestinal AEs of common drugs. a, Significant results (FDR <0.05, RE-IVW) from IVW-MR on the effects of common drugs on gut microbiota. Data were analyzed using IVW as the primary method and MR Egger as sensitivity analysis. The causal estimates are expressed as β values with 95% CI values. The width of the lines extending from the midpoint represent the 95% CI. Both unadjusted P values (RE-IVW and MR Egger) and FDR values (RE-IVW) are given. b, ROR heatmap for gastrointestinal PTs of common drugs from the disproportionality analysis on the FAERS database. The data are expressed as log-transformed ROR values with unadjusted P values (disproportionality analysis). ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001.
In sensitivity analyses, MR Egger produced one significant result (P < 0.05, MR Egger) out of the 15 associations identified by IVW, which was in a consistent direction as that from IVW-MR. Other sensitivity analyses revealed no abnormalities (eTable 20). The scatter plots of the 15 significant results are shown in eFig. 7. Asymmetry was observed in some funnel plots, probably due to the insufficient number of SNPs (eFig. 8). Leave-one-out analysis revealed that the combined effect estimate was not driven by any single SNP (eFig. 9). As presented in eTable 21a, MR Steiger test detected ten SNPs with wrong directionality, but after removing those SNPs, the MR results did not change significantly. After removing IVs associated with the four confounders as listed in eTable 12, 99.1% of the IVW-MR results did not change significantly, as shown in eTable 21b. The effect of F2 inhibitors on genus.Romboutsia was previously insignificant but became significant after removing confounder-associated SNPs. In the MVMR analysis adjusting for the four confounders as shown in eTable 22a, 36.9% of direct effects remained significant after adjustment. As shown in eTable 22b, 64.7% of the results remained significant or insignificant after adjusting for RBC count. As demonstrated in eTable 22c, 83.3% of the direct effects of lipid-lowering drugs on gut microbiota remained significant after adjusting for HDL and TG.
SMR analyses did not produce significant drug-microbiota associations (eTable 23).
Gut microbiota is a critical component of the intestinal ecosystem and its disturbance can contribute to the development of many gastrointestinal diseases.60,61 Hence, we next investigated the gastrointestinal adverse effects of common drugs using the FAERS database to verify the MR findings. Clinical characteristics of gastrointestinal adverse event (AE) reports were summarized in eTable 24. Among 19 drug classes analyzed, 11 drugs were significantly associated with higher gastrointestinal AEs based on the ROR criteria, with two robust signals detected for GLP-1R agonists and angiotensin receptor blockers (eTable 25). GLP-1R agonists have the highest effects on gastrointestinal AEs (ROR = 4.51 [4.43, 4.6]). For each drug class, five PTs (Preferred Terms) belonging to SOCs (System Organ Classes) of ‘Gastrointestinal disorders’ with the highest ROR were listed in eTable 26. As shown in Fig. 5b, DPP-IV inhibitors, GLP-1R agonists, and angiotensin receptor blockers were associated with increased risk of multiple gastrointestinal PTs, with angiotensin receptor blockers having the greatest effect on ‘Sprue-like enteropathy’ (ROR = 802.17 [733.72, 877.01]).
In sensitivity analyses, most results still remain stable after deleting cases with missing age, country or sex, respectively. For example, 90.9%, 90.9%, 100%, and 90.9% of the valid signals in the SOC level still remained significant after deleting the cases missing age, country, sex or weight, respectively. Additionally, 73.5%, 96.4%, and 81.9% of the top five PTs still remained in the top five list after excluding the cases missing age, country or sex, respectively. However, since the number of cases missing weight is relatively large, only 50.6% of the top five PTs still remained after deleting the cases missing weight.
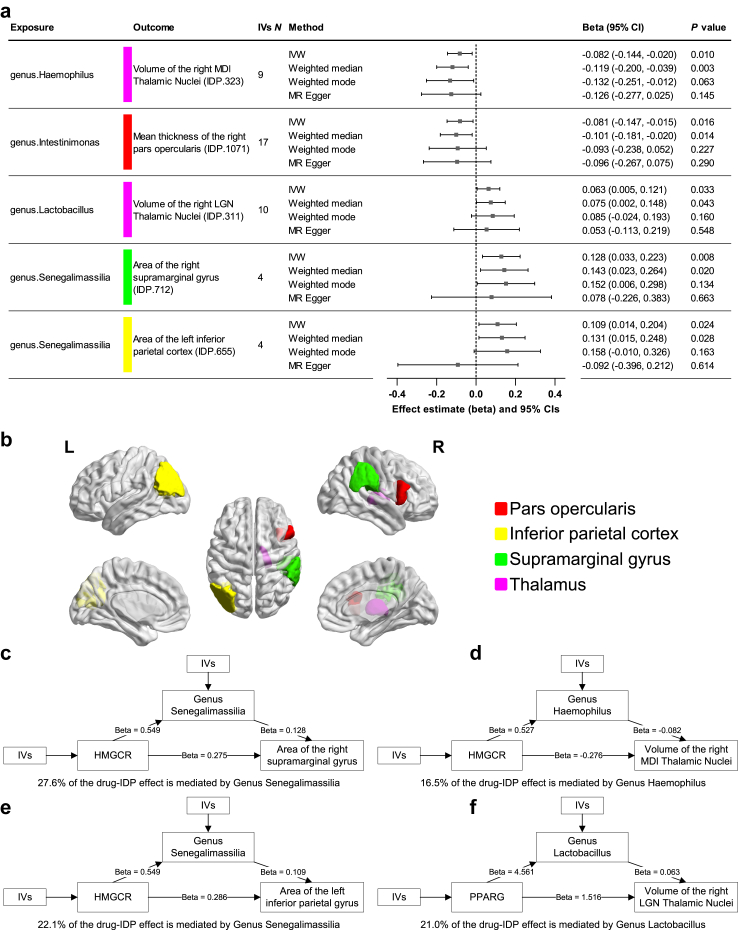
Causal associations between gut microbiota and IDPs
IVs used for forward and reverse MR were given in eTable 27a and b, respectively. The minimum F-statistic was 19, suggesting a low risk of weak instrumental bias.
We identified five results with IVW P value < 0.05 plus one of the additional three methods P value < 0.05, as shown in Fig. 6a and b and eTable 28. In sensitivity analyses, weighted median also produced significant results (P < 0.05, weighted median), which was in a consistent direction as that from IVW-MR, suggesting that the results are reliable when less than half of the IVs are invalid. Other sensitivity analyses revealed no abnormalities (eTable 28). The scatter plots of the five significant results are shown in eFig. 10. Asymmetry was not observed in funnel plots (eFig. 11). The leave-one-out analysis did not yield favorable results (eFig. 12). MR Steiger test did not detect any SNPs with wrong directionality. In the reverse MR, IVW did not identify significant reverse causations for the five microbiota-IDP pairs (eTable 29). In the MR of IDPs on gut microbiota, 96.7% of the results remained to be significant or insignificant after removing IVs associated with the confounders (eTable 30).
Fig. 6.
Effects of gut microbiota on IDPs identified by IVW-MR and mediation analysis. a, Significant results with IVW P-value <0.05 (RE-IVW) and one of the additional three methods P-value <0.05 (weighted median, weighted mode, and MR Egger). Data were analyzed using IVW as the primary method and other three methods (weighted median, weighted mode, and MR Egger) as sensitivity analysis. The causal estimates are expressed as β values with 95% CI values. The width of the lines extending from the midpoint represent the 95% CI. Unadjusted P values (RE-IVW, weighted median, weighted mode, and MR Egger) are given. b, Schematic diagram of the brain regions in lateral, medial, and dorsal views. c-f, Significant results from the mediation analysis.
Mediation analysis
Combining the findings above, we identify five possible drug→microbiota→IDP causal links (eTable 31). We conducted a mediation analysis to determine the extent to which the gut microbiota mediates the effect of common drugs on IDPs (Fig. 6c–f and eTable 31). The proportions mediated by gut microbiota vary between 16.5% and 27.6%. We did not calculate the mediation proportion of genus Intestinimonas since the total effect and the product of drug-microbiota causal estimate and microbiota-IDP causal estimate were in opposite directions.
Replication analyses based on UK Biobank research
We further performed replication analysis for antidiabetic drugs and anticoagulants on psychiatric disorders using UK Biobank data as the source of IV selection, applying a P-value threshold of 5e-8. As shown in eTable 32, the minimum F-statistic of the IVs for IVW-MR was 31, which was larger than that in the previous analysis. In positive control analyses (eTable 33), most IVs yielded significant results. As presented in eTable 34, 66.7% (2/3) of the original significant results remained significant in the replication analysis. Besides, we identified five new associations that were not discovered in the original analysis. For instance, SLC5A2-mediated HbA1c increase was associated with higher risk of OCD (OR = 0.02 [0.003, 0.20], P = 3.29e-4, RE-IVW).
Discussion
We explore the long-term effects of common medications for chronic conditions on the central nervous system (CNS) through genetic methods and retrospective analysis of public database. A variety of positive and negative genetic associations between common drugs and brain structures or psychiatric disorders were identified. The results from the retrospective analyses of the adverse event records in the FAERS database generally support the findings from MR.
Among all categories of common drugs, genetically proxied HMGCR inhibition and its representative drug statins showed concordant results between MR and FAERS analysis. Statins are lipid-lowering agents that function by inhibiting 3-hydroxy-3-methyglutaryl coenzyme A (HMG-CoA) reductase (HMGCR), a rate-limiting enzyme in the cholesterol synthesis pathway. We identified that genetically proxied HMGCR inhibition was associated with higher risks of MDD through MR. Our analysis of FAERS database revealed that statins were associated with higher risks of ‘Self esteem decreased’ (ROR = 18.92 [14.09, 25.42]), ‘Agitated depression’ (ROR = 9.76 [4.54, 20.99]) compared with other drugs (eTable 8), which is consistent with the MR results. Likewise, a recent cross-sectional study using medical claims data (n = 7,481,168) showed that the association between statin use and depression may be dose-dependent.62 However, previous meta-analyses on observational studies proposed that statins were not associated with risks of MDD or severity of depressive symptoms,63, 64, 65 emphasizing the necessity of rigorous randomized controlled trials to solve the problems. Furthermore, we discovered that statins were associated with significant changes in various brain structures, suggesting the wide impact of statins on the CNS. Emerging evidence supports that statins exert active functions in the CNS mainly through inhibiting isoprenylation of small GTPases and play a neuroprotective role in neurodegenerative disorders, including AD and Parkinson's disease, and neurodevelopmental disorders like Rett syndrome, fragile X syndrome (FXS), and tuberous sclerosis.66,67 Notably, the three lipid-lowering drugs have opposite effects on several IDPs, as identified by the IVW-MR (eTable 16), including the area of the left inferior parietal cortex, the area of the right middle temporal gyrus, and the volume of the right middle temporal gyrus. This can be explained by different mediation effects of other lipids, as shown in eTable 18c, and possible pleiotropic effects.68, 69, 70
Besides, GLP-1R agonists, a highly recommended class of glucose-lowering agents for the treatment of T2D, have accumulated evidence of improving cognitive functions, which was indicated by the significant increase in the volume of the CA3-head of the right hippocampus and the area of the left precuneus cortex in our investigation. Proximal CA3 interacts with the dentate gyrus (DG) to facilitate pattern separation, the ability to make two similar inputs more distinct, while distal CA3 forms an auto-associative network to perform pattern completion, the ability to retrieve stored full pattern from incomplete inputs.71 The precuneus is an important region involved in complex cognitive functions including episodic memory, theory of mind and self-referential processes, etc.72 Therefore, GLP-1R agonists may improve cognitive functions like episodic memory, which has also been proved by clinical studies.3,10,73, 74, 75 For example, a 16-week randomized controlled trial (RCT) demonstrated that liraglutide improved cognitive functions of delayed memory, attention, and executive function, in part through a direct effect on left hippocampal activation.76 A recent study revealed that GLP-1R agonists exert anti-inflammatory effects depending on central neuronal GLP-1Rs, further indicating the significance of the CNS effects of this drug class.77
Moreover, combining our MR results with previous MR studies can help discover potential causal links. Thiazolidinediones are a class of glucose-lowering agents that act by activating the peroxisome proliferator-activated receptor-γ and thus improving tissue sensitivity to insulin. We have identified that it was significantly associated with a higher risk of BD Ⅱ. Combining our findings with previous MR of IDPs on psychiatric disorders,33 we identified that thiazolidinediones may increase the risk of BD by decreasing the volume of the left accumbens. An RCT also discovered that pioglitazone treatment was not superior, or even worse than placebo, in terms of antidepressant efficacy.78
Combining the idea of pharmacomicrobiomics13 and the microbiota-gut-brain axis,16,17,79 we speculate that gut microbiota may function as an important mediator in the causal effects of common drugs on brain structures. Through step-by-step screening, we successfully identified five significant drug-microbiota-IDP causal links and calculated the proportion mediated by the microbiota. Previous studies have proved that statin therapy can modulate the gut microbiome composition and reduce the risk of microbiota dysbiosis in obese participants80 and patients with acute coronary syndrome.81 An animal study also suggested that long-term atorvastatin treatment led to improved cognitive function via modulating the gut microbiome, affecting retinoic acid metabolism and altering immune cells in naturally aging rats.82 Zhang et al. reported that atorvastatin treatment reduced neuroinflammation and improved gut barrier function via restoring changed gut microbiome composition in mice with middle cerebral artery occlusion (MCAO). Fecal microbiota transplantation (FMT) from atorvastatin-treated mice significantly improved cognitive function and inhibited neuroinflammation in the MCAO group, indicating that altered microbiota in atorvastatin-treated mice played a crucial role in the neuroprotective effect.83 Research findings on the effects of PPARG agonists (thiazolidinediones) on gut microbiota composition are inconsistent. Madsen et al. found no significant effect of rosiglitazone treatment on gut microbiota composition in diabetic db/db mice.84 In contrast, another study revealed that pioglitazone treatment resulted in significant alterations in the gut microbiome of high-fat diet (HFD)-fed obese mice, including an increase in the abundance of genus Lactobacillus, which was also identified in our MR.85 Further studies are warranted to confirm the mediation model identified in this study.
One interesting point is that IVW-MR appears to yield more positive results than SMR. This may be attributed to the disparity in the two methodologies; IVW meta-analyzes effect estimates from multiple genetic variants, while SMR only leverages the top SNP to derive the effect estimate. Prior studies have illustrated that SMR utilizing summary statistics of the top SNP yielded less power than multi-SNP prediction when identifying gene expression-trait relationships.86 Considering the undesirable performance in the positive control analysis, the results of SMR need to be interpreted with caution.
These findings have extensive implications for the appraisal of drug side effects, the clinical use of medications and drug repurposing. For example, genetically proxied HMGCR inhibition has wide impacts on multiple IDPs and is linked with higher risks of MDD and OCD. Therefore, long-term statin users may require regular mental status assessments and patients with MDD or OCD should use statins with caution. Since genus Senegalimassilia mediates the effect of statins on the area of the right supramarginal gyrus and the left inferior parietal cortex, proper supplementation of this genus may reduce the neuropsychiatric side effects of statins. Additionally, genetically proxied Factor Xa inhibitors were associated with lower risks of BD Ⅰ and BD Ⅱ, suggesting that this drug class may be repurposed for the treatment of affective disorders. We have only touched on some of the potential applications of our study, with the intention of encouraging further thought and discussion.
Our study has the following strengths. Firstly, in terms of research content, we incorporated multiple common drugs and systematically investigated their effects on various brain structures and psychiatric disorders. Secondly, the implementation of two MR methodologies, IVW and SMR, allows us to assess the long-term neuropsychiatric effects of common drugs without interference from confounders. Supplementation with retrospective analysis of the FAERS database also enriched this study, making the findings more realistic. Thirdly, we hypothesized that the gut microbiome acts as a mediator in drug-IDP associations and identified four drug-microbiota-IDP causal links.
However, several limitations also exist. Firstly, this study leverages GWAS summary statistics of European populations, and thus further analyses remain needed for other populations. Secondly, the FAERS database is a self-reporting system with some inherent downsides. For example, most reports were from the United States with the majority of ethnicity being European and not all AEs of drugs that happened were recorded. Thirdly, our study faced limitations in accessing GWAS data with large sample sizes for enough genome-wide significant IV selection. To achieve better statistical efficacy, the IVs selected for simulating antidiabetic drugs, anticoagulants, and COPD medications did not meet the genome-wide significance threshold (P < 5e-8). Despite this limitation, the minimum F-statistic for the drug target IVs in our study was 13, exceeding the conventional threshold of F > 10,87,88 which indicates a low risk of weak instruments.89, 90, 91 To further validate our results, we conducted a replication analysis using the large-scale UKB data, employing a stringent P-value threshold (<5e-8). This replication was only employed in simulating drug effects on psychiatric disorders, since exploring other mediation effects using IVs from UKB data will violate the two-sample MR assumption. The outcomes of this replication were largely consistent with our original findings, thereby reinforcing the robustness of our results.
In conclusion, this study unraveled the long-term effects of common drugs on brain IDPs and psychiatric disorders through drug target MR. Both positive and negative effects were found, implicating clinical drug use and drug repurposing. Further MR analysis identified four drug-IDP associations that were partially mediated by gut microbiota with mediation proportions of around 20%. Retrospective analysis of the FAERS database identified psychiatric and gastrointestinal side effects of common drugs, supporting the MR findings. These findings have wide applications and may hopefully stimulate the utilization of genetic methods in drug effect prediction.
Contributors
All authors read and approved the final version of the manuscript.
Conceptualization, Z.C, X.W, Z.L, F.L, and K.X; methodology, Z.C, X.W, Z.T, and J.H; investigation, Z.C, X.W, Z.T, J.H, J.M, C.Q, and Y.W; formal analysis, Z.C, X.W, and Z.T; visualization, Z.C and X.W; writing—original draft, Z.C and X.W; writing—review & editing, Z.T, J.H, J.M, C.Q, and Y.W; supervision, Z.L, F.L, and K.X; resources, Z.L, F.L, and K.X; project administration, Z.L, F.L, and K.X; funding acquisition, J.H, Z.L, F.L, and K.X.
Z.L, F.L, and K.X verified the underlying data.
Data sharing statement
All GWAS summary statistics involved in this study are publicly available on the corresponding websites listed in eTable 2a. Adverse event reports of drugs are publicly available and can be retrieved from the FAERS database (https://fis.fda.gov/extensions/FPD-QDE-FAERS/FPD-QDE-FAERS.html). R codes for relevant analysis in this study will be shared upon reasonable request to the corresponding authors.
Declaration of interests
The authors declare no competing interests.
Acknowledgements
Data on glycaemic traits have been contributed by MAGIC investigators and have been downloaded from www.magicinvestigators.org. Data on coronary artery disease have been contributed by CARDIoGRAMplusC4D investigators and have been downloaded from http://www.cardiogramplusc4d.org/. The MEGASTROKE project received funding from sources specified at http://www.megastroke.org/acknowledgments.html. The OCD summary data was supported by grants from the Judah Foundation, the Tourette Association of America, NIH grants MH079489 and MH073250, American Recovery and Re-investment Act (ARRA) awards NS40024-07S1, NS16648-29S1, MH071507, MH079489, MH079487, MH079488 and MH079494. Other data sources are listed and described in eTable 2a.
This work was supported by the National Natural Science Foundation of China (grant No. 82330035, 82130043, 82172685, and 82001223), National Natural Science Foundation of Hunan Province (grant No. 2021SK1010), and the Science Foundation for Distinguished Young Scholars of Changsha (grant No. kq2209006).
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2024.105314.
Contributor Information
Zhixiong Liu, Email: zhixiongliu@csu.edu.cn.
Fangkun Liu, Email: liufangkun@csu.edu.cn.
Kun Xia, Email: xiakun@sklmg.edu.cn.
Appendix A. Supplementary data
Scatter plots for MR of common drugs on psychiatric disorders.
Funnel plots for MR of common drugs on psychiatric disorders.
Leave-one-out plots for MR of common drugs on psychiatric disorders.
Scatter plots for MR of common drugs on IDPs.
Funnel plots for MR of common drugs on IDPs.
Leave-one-out plots for MR of common drugs on IDPs.
Leave-one-out plots for MR of common drugs on IDPs.
Funnel plots for MR of common drugs on gut microbiome.
Leave-one-out plots for MR of common drugs on gut microbiome.
Scatter plots for MR of gut microbiome on IDPs.
Funnel plots for MR of gut microbiome on IDPs.
Leave-one-out plots for MR of gut microbiome on IDPs.
References
- 1.Cham S., Koslik H.J., Golomb B.A. Mood, personality, and behavior changes during treatment with statins: a case series. Drug Saf Case Rep. 2016;3(1):1. doi: 10.1007/s40800-015-0024-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feng Z., Zhao Q., Wu J., et al. Nonselective beta-adrenoceptor blocker use and risk of Parkinson's disease: from multiple real-world evidence. BMC Med. 2023;21(1):437. doi: 10.1186/s12916-023-03122-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Horska K., Ruda-Kucerova J., Skrede S. GLP-1 agonists: superior for mind and body in antipsychotic-treated patients? Trends Endocrinol Metab. 2022;33(9):628–638. doi: 10.1016/j.tem.2022.06.005. [DOI] [PubMed] [Google Scholar]
- 4.Nowell J., Blunt E., Gupta D., Edison P. Antidiabetic agents as a novel treatment for Alzheimer's and Parkinson's disease. Ageing Res Rev. 2023;89 doi: 10.1016/j.arr.2023.101979. [DOI] [PubMed] [Google Scholar]
- 5.Kosowski M., Smolarczyk-Kosowska J., Hachuła M., et al. The effects of statins on neurotransmission and their neuroprotective role in neurological and psychiatric disorders. Molecules. 2021;26(10) doi: 10.3390/molecules26102838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adesuyan M., Jani Y.H., Alsugeir D., et al. Antihypertensive agents and incident Alzheimer's disease: a systematic review and meta-analysis of observational studies. J Prev Alzheimers Dis. 2022;9(4):715–724. doi: 10.14283/jpad.2022.77. [DOI] [PubMed] [Google Scholar]
- 7.Fenger-Grøn M., Vestergaard C.H., Ribe A.R., et al. Association between bipolar disorder or schizophrenia and oral anticoagulation use in Danish adults with incident or prevalent atrial fibrillation. JAMA Netw Open. 2021;4(5) doi: 10.1001/jamanetworkopen.2021.10096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delhaye S., Bardoni B. Role of phosphodiesterases in the pathophysiology of neurodevelopmental disorders. Mol Psychiatry. 2021;26(9):4570–4582. doi: 10.1038/s41380-020-00997-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaduševičius E. Novel applications of NSAIDs: insight and future perspectives in cardiovascular, neurodegenerative, diabetes and cancer disease therapy. Int J Mol Sci. 2021;22(12) doi: 10.3390/ijms22126637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vadini F., Simeone P.G., Boccatonda A., et al. Liraglutide improves memory in obese patients with prediabetes or early type 2 diabetes: a randomized, controlled study. Int J Obes. 2020;44(6):1254–1263. doi: 10.1038/s41366-020-0535-5. [DOI] [PubMed] [Google Scholar]
- 11.Smith G.D., Ebrahim S. 'Mendelian randomization': can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol. 2003;32(1):1–22. doi: 10.1093/ije/dyg070. [DOI] [PubMed] [Google Scholar]
- 12.Dehnavi S., Kiani A., Sadeghi M., et al. Targeting AMPK by statins: a potential therapeutic approach. Drugs. 2021;81(8):923–933. doi: 10.1007/s40265-021-01510-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao Q., Chen Y., Huang W., Zhou H., Zhang W. Drug-microbiota interactions: an emerging priority for precision medicine. Signal Transduct Target Ther. 2023;8(1):386. doi: 10.1038/s41392-023-01619-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jackson M.A., Verdi S., Maxan M.E., et al. Gut microbiota associations with common diseases and prescription medications in a population-based cohort. Nat Commun. 2018;9(1):2655. doi: 10.1038/s41467-018-05184-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jackson M.A., Goodrich J.K., Maxan M.E., et al. Proton pump inhibitors alter the composition of the gut microbiota. Gut. 2016;65(5):749–756. doi: 10.1136/gutjnl-2015-310861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Socała K., Doboszewska U., Szopa A., et al. The role of microbiota-gut-brain axis in neuropsychiatric and neurological disorders. Pharmacol Res. 2021;172 doi: 10.1016/j.phrs.2021.105840. [DOI] [PubMed] [Google Scholar]
- 17.Mayer E.A., Nance K., Chen S. The gut-brain axis. Annu Rev Med. 2022;73:439–453. doi: 10.1146/annurev-med-042320-014032. [DOI] [PubMed] [Google Scholar]
- 18.Skrivankova V.W., Richmond R.C., Woolf B.A.R., et al. Strengthening the reporting of observational studies in epidemiology using mendelian randomization: the STROBE-MR statement. JAMA. 2021;326(16):1614–1621. doi: 10.1001/jama.2021.18236. [DOI] [PubMed] [Google Scholar]
- 19.Smith S.M., Douaud G., Chen W., et al. An expanded set of genome-wide association studies of brain imaging phenotypes in UK Biobank. Nat Neurosci. 2021;24(5):737–745. doi: 10.1038/s41593-021-00826-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kurilshikov A., Medina-Gomez C., Bacigalupe R., et al. Large-scale association analyses identify host factors influencing human gut microbiome composition. Nat Genet. 2021;53(2):156–165. doi: 10.1038/s41588-020-00763-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malik R., Chauhan G., Traylor M., et al. Multiancestry genome-wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nat Genet. 2018;50(4):524–537. doi: 10.1038/s41588-018-0058-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou C., Peng S., Lin A., et al. Psychiatric disorders associated with immune checkpoint inhibitors: a pharmacovigilance analysis of the FDA Adverse Event Reporting System (FAERS) database. eClinicalMedicine. 2023;59 doi: 10.1016/j.eclinm.2023.101967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Evans S.J.W., Waller P.C., Davis S. Use of proportional reporting ratios (PRRs) for signal generation from spontaneous adverse drug reaction reports. Pharmacoepidemiol Drug Saf. 2001;10(6):483–486. doi: 10.1002/pds.677. [DOI] [PubMed] [Google Scholar]
- 24.Didelez V., Sheehan N. Mendelian randomization as an instrumental variable approach to causal inference. Stat Methods Med Res. 2007;16(4):309–330. doi: 10.1177/0962280206077743. [DOI] [PubMed] [Google Scholar]
- 25.Huang W., Xiao J., Ji J., Chen L. Association of lipid-lowering drugs with COVID-19 outcomes from a Mendelian randomization study. Elife. 2021;10 doi: 10.7554/eLife.73873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harshfield E.L., Sims M.C., Traylor M., Ouwehand W.H., Markus H.S. The role of haematological traits in risk of ischaemic stroke and its subtypes. Brain. 2020;143(1):210–221. doi: 10.1093/brain/awz362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang J., He X., Qian L., et al. Association between plasma proteome and childhood neurodevelopmental disorders: a two-sample Mendelian randomization analysis. eBioMedicine. 2022;78 doi: 10.1016/j.ebiom.2022.103948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tang B., Wang Y., Jiang X., et al. Genetic variation in targets of antidiabetic drugs and alzheimer disease risk: a mendelian randomization study. Neurology. 2022;99(7):e650–e659. doi: 10.1212/WNL.0000000000200771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Woolf B., Rajasundaram S., Cronjé H.T., Yarmolinsky J., Burgess S., Gill D. A drug target for erectile dysfunction to help improve fertility, sexual activity, and wellbeing: mendelian randomisation study. BMJ. 2023;383 doi: 10.1136/bmj-2023-076197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ference B.A., Ray K.K., Catapano A.L., et al. Mendelian randomization study of ACLY and cardiovascular disease. N Engl J Med. 2019;380(11):1033–1042. doi: 10.1056/NEJMoa1806747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rosoff D.B., Bell A.S., Jung J., Wagner J., Mavromatis L.A., Lohoff F.W. Mendelian randomization study of PCSK9 and HMG-CoA reductase inhibition and cognitive function. J Am Coll Cardiol. 2022;80(7):653–662. doi: 10.1016/j.jacc.2022.05.041. [DOI] [PubMed] [Google Scholar]
- 32.Qi T., Wu Y., Zeng J., et al. Identifying gene targets for brain-related traits using transcriptomic and methylomic data from blood. Nat Commun. 2018;9(1):2282. doi: 10.1038/s41467-018-04558-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo J., Yu K., Dong S.-S., et al. Mendelian randomization analyses support causal relationships between brain imaging-derived phenotypes and risk of psychiatric disorders. Nat Neurosci. 2022;25(11):1519–1527. doi: 10.1038/s41593-022-01174-7. [DOI] [PubMed] [Google Scholar]
- 34.Burgess S., Thompson S.G. Avoiding bias from weak instruments in Mendelian randomization studies. Int J Epidemiol. 2011;40(3):755–764. doi: 10.1093/ije/dyr036. [DOI] [PubMed] [Google Scholar]
- 35.Burgess S., Butterworth A., Thompson S.G. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol. 2013;37(7):658–665. doi: 10.1002/gepi.21758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu Z., Zhang F., Hu H., et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat Genet. 2016;48(5):481–487. doi: 10.1038/ng.3538. [DOI] [PubMed] [Google Scholar]
- 37.Bowden J., Davey Smith G., Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44(2):512–525. doi: 10.1093/ije/dyv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Verbanck M., Chen C.Y., Neale B., Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. 2018;50(5):693–698. doi: 10.1038/s41588-018-0099-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hemani G., Tilling K., Davey Smith G. Orienting the causal relationship between imprecisely measured traits using GWAS summary data. PLoS Genet. 2017;13(11) doi: 10.1371/journal.pgen.1007081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prochaska J.J., Das S., Young-Wolff K.C. Smoking, mental illness, and public health. Annu Rev Public Health. 2017;38:165–185. doi: 10.1146/annurev-publhealth-031816-044618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hjorthøj C., Østergaard M.L., Benros M.E., et al. Association between alcohol and substance use disorders and all-cause and cause-specific mortality in schizophrenia, bipolar disorder, and unipolar depression: a nationwide, prospective, register-based study. Lancet Psychiatr. 2015;2(9):801–808. doi: 10.1016/S2215-0366(15)00207-2. [DOI] [PubMed] [Google Scholar]
- 42.McCloud T., Kamenov S., Callender C., Lewis G., Lewis G. The association between higher education attendance and common mental health problems among young people in England: evidence from two population-based cohorts. Lancet Public Health. 2023;8(10):e811–e819. doi: 10.1016/S2468-2667(23)00188-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ridley M., Rao G., Schilbach F., Patel V. Poverty, depression, and anxiety: causal evidence and mechanisms. Science. 2020;370(6522) doi: 10.1126/science.aay0214. [DOI] [PubMed] [Google Scholar]
- 44.Leslie R., O'Donnell C.J., Johnson A.D. GRASP: analysis of genotype-phenotype results from 1390 genome-wide association studies and corresponding open access database. Bioinformatics. 2014;30(12):i185–i194. doi: 10.1093/bioinformatics/btu273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Elsworth B., Lyon M., Alexander T., et al. The MRC IEU OpenGWAS data infrastructure. bioRxiv. 2020 doi: 10.1101/2020.08.10.244293. [DOI] [Google Scholar]
- 46.Hemani G., Zheng J., Elsworth B., et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife. 2018;7 doi: 10.7554/eLife.34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sollis E., Mosaku A., Abid A., et al. The NHGRI-EBI GWAS Catalog: knowledgebase and deposition resource. Nucleic Acids Res. 2023;51(D1):D977–D985. doi: 10.1093/nar/gkac1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Burgess S., Thompson S.G. Multivariable Mendelian randomization: the use of pleiotropic genetic variants to estimate causal effects. Am J Epidemiol. 2015;181(4):251–260. doi: 10.1093/aje/kwu283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Carter A.R., Sanderson E., Hammerton G., et al. Mendelian randomisation for mediation analysis: current methods and challenges for implementation. Eur J Epidemiol. 2021;36(5):465–478. doi: 10.1007/s10654-021-00757-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sanderson E., Davey Smith G., Windmeijer F., Bowden J. An examination of multivariable Mendelian randomization in the single-sample and two-sample summary data settings. Int J Epidemiol. 2019;48(3):713–727. doi: 10.1093/ije/dyy262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lindqvist D., Mellon S.H., Dhabhar F.S., et al. Increased circulating blood cell counts in combat-related PTSD: associations with inflammation and PTSD severity. Psychiatry Res. 2017;258:330–336. doi: 10.1016/j.psychres.2017.08.052. [DOI] [PubMed] [Google Scholar]
- 52.Fruchart J.C., Duriez P. HDL and triglyceride as therapeutic targets. Curr Opin Lipidol. 2002;13(6):605–616. doi: 10.1097/00041433-200212000-00003. [DOI] [PubMed] [Google Scholar]
- 53.Arsenault B.J., Boekholdt S.M. Clinical and biological relevance of statin-mediated changes in HDL metabolism. Curr Atheroscler Rep. 2014;16(1):379. doi: 10.1007/s11883-013-0379-8. [DOI] [PubMed] [Google Scholar]
- 54.Schoch L., Sutelman P., Suades R., et al. Hypercholesterolemia-induced HDL dysfunction can Be reversed: the impact of diet and statin treatment in a preclinical animal model. Int J Mol Sci. 2022;23(15) doi: 10.3390/ijms23158596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bowden J., Davey Smith G., Haycock P.C., Burgess S. Consistent estimation in mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. 2016;40(4):304–314. doi: 10.1002/gepi.21965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hartwig F.P., Davey Smith G., Bowden J. Robust inference in summary data Mendelian randomization via the zero modal pleiotropy assumption. Int J Epidemiol. 2017;46(6):1985–1998. doi: 10.1093/ije/dyx102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sinnott-Armstrong N., Tanigawa Y., Amar D., et al. Genetics of 35 blood and urine biomarkers in the UK Biobank. Nat Genet. 2021;53(2):185–194. doi: 10.1038/s41588-020-00757-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sun B.B., Chiou J., Traylor M., et al. Plasma proteomic associations with genetics and health in the UK Biobank. Nature. 2023;622(7982):329–338. doi: 10.1038/s41586-023-06592-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xia M., Wang J., He Y. BrainNet viewer: a network visualization tool for human brain connectomics. PLoS One. 2013;8(7) doi: 10.1371/journal.pone.0068910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jones R.M., Neish A.S. Gut microbiota in intestinal and liver disease. Annu Rev Pathol. 2021;16:251–275. doi: 10.1146/annurev-pathol-030320-095722. [DOI] [PubMed] [Google Scholar]
- 61.Cruz M.S., Tintelnot J., Gagliani N. Roles of microbiota in pancreatic cancer development and treatment. Gut Microb. 2024;16(1) doi: 10.1080/19490976.2024.2320280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Leutner M., Matzhold C., Kautzky A., et al. Major depressive disorder (MDD) and antidepressant medication are overrepresented in high-dose statin treatment. Front Med. 2021;8 doi: 10.3389/fmed.2021.608083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Köhler-Forsberg O., Otte C., Gold S.M., Østergaard S.D. Statins in the treatment of depression: Hype or hope? Pharmacol Ther. 2020;215 doi: 10.1016/j.pharmthera.2020.107625. [DOI] [PubMed] [Google Scholar]
- 64.Lee M.C., Peng T.R., Lee C.H., et al. Statin use and depression risk: a systematic review and meta-analysis. J Affect Disord. 2021;282:308–315. doi: 10.1016/j.jad.2020.12.164. [DOI] [PubMed] [Google Scholar]
- 65.De Giorgi R., Waters S., Pesci N.R., Rosso G., Cowen P.J., Harmer C.J. The effects of statin monotherapy on depressive symptoms: a systematic review and meta-analysis. J Affect Disord. 2022;311:336–343. doi: 10.1016/j.jad.2022.05.113. [DOI] [PubMed] [Google Scholar]
- 66.Ling Q., Tejada-Simon M.V. Statins and the brain: new perspective for old drugs. Prog Neuro Psychopharmacol Biol Psychiatr. 2016;66:80–86. doi: 10.1016/j.pnpbp.2015.11.013. [DOI] [PubMed] [Google Scholar]
- 67.Fracassi A., Marangoni M., Rosso P., et al. Statins and the brain: more than lipid lowering agents? Curr Neuropharmacol. 2018;17(1):59–83. doi: 10.2174/1570159X15666170703101816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Basiak M., Kosowski M., Cyrnek M., et al. Pleiotropic effects of PCSK-9 inhibitors. Int J Mol Sci. 2021;22(6) doi: 10.3390/ijms22063144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.German C.A., Liao J.K. Understanding the molecular mechanisms of statin pleiotropic effects. Arch Toxicol. 2023;97(6):1529–1545. doi: 10.1007/s00204-023-03492-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Oesterle A., Laufs U., Liao J.K. Pleiotropic effects of statins on the cardiovascular system. Circ Res. 2017;120(1):229–243. doi: 10.1161/CIRCRESAHA.116.308537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee H., GoodSmith D., Knierim J.J. Parallel processing streams in the hippocampus. Curr Opin Neurobiol. 2020;64:127–134. doi: 10.1016/j.conb.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dadario N.B., Sughrue M.E. The functional role of the precuneus. Brain. 2023;146(9):3598–3607. doi: 10.1093/brain/awad181. [DOI] [PubMed] [Google Scholar]
- 73.Mansur R.B., Ahmed J., Cha D.S., et al. Liraglutide promotes improvements in objective measures of cognitive dysfunction in individuals with mood disorders: a pilot, open-label study. J Affect Disord. 2017;207:114–120. doi: 10.1016/j.jad.2016.09.056. [DOI] [PubMed] [Google Scholar]
- 74.Yaribeygi H., Rashidy-Pour A., Atkin S.L., Jamialahmadi T., Sahebkar A. GLP-1 mimetics and cognition. Life Sci. 2021;264 doi: 10.1016/j.lfs.2020.118645. [DOI] [PubMed] [Google Scholar]
- 75.Monney M., Jornayvaz F.R., Gariani K. GLP-1 receptor agonists effect on cognitive function in patients with and without type 2 diabetes. Diabetes Metabol. 2023;49(5) doi: 10.1016/j.diabet.2023.101470. [DOI] [PubMed] [Google Scholar]
- 76.Cheng H., Zhang Z., Zhang B., et al. Enhancement of Impaired Olfactory neural activation and cognitive capacity by liraglutide, but not Dapagliflozin or Acarbose, in patients with type 2 diabetes: a 16-week randomized parallel comparative study. Diabetes Care. 2022;45(5):1201–1210. doi: 10.2337/dc21-2064. [DOI] [PubMed] [Google Scholar]
- 77.Wong C.K., McLean B.A., Baggio L.L., et al. Central glucagon-like peptide 1 receptor activation inhibits Toll-like receptor agonist-induced inflammation. Cell Metab. 2023;36:130. doi: 10.1016/j.cmet.2023.11.009. [DOI] [PubMed] [Google Scholar]
- 78.Aftab A., Kemp D.E., Ganocy S.J., et al. Double-blind, placebo-controlled trial of pioglitazone for bipolar depression. J Affect Disord. 2019;245:957–964. doi: 10.1016/j.jad.2018.11.090. [DOI] [PubMed] [Google Scholar]
- 79.Góralczyk-Bińkowska A., Szmajda-Krygier D., Kozłowska E. The microbiota–gut–brain Axis in psychiatric disorders. Int J Mol Sci. 2022;23(19) doi: 10.3390/ijms231911245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vieira-Silva S., Falony G., Belda E., et al. Statin therapy is associated with lower prevalence of gut microbiota dysbiosis. Nature. 2020;581(7808):310–315. doi: 10.1038/s41586-020-2269-x. [DOI] [PubMed] [Google Scholar]
- 81.Hu X., Li H., Zhao X., et al. Multi-omics study reveals that statin therapy is associated with restoration of gut microbiota homeostasis and improvement in outcomes in patients with acute coronary syndrome. Theranostics. 2021;11(12):5778–5793. doi: 10.7150/thno.55946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xu T.-C., Lv Y., Liu Q.-Y., Chen H.-S. Long-term atorvastatin improves cognitive decline by regulating gut function in naturally ageing rats. Immun Ageing. 2022;19(1) doi: 10.1186/s12979-022-00311-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang P., Zhang X., Huang Y., et al. Atorvastatin alleviates microglia-mediated neuroinflammation via modulating the microbial composition and the intestinal barrier function in ischemic stroke mice. Free Radic Biol Med. 2021;162:104–117. doi: 10.1016/j.freeradbiomed.2020.11.032. [DOI] [PubMed] [Google Scholar]
- 84.Madsen M.S.A., Grønlund R.V., Eid J., et al. Characterization of local gut microbiome and intestinal transcriptome responses to rosiglitazone treatment in diabetic db/db mice. Biomed Pharmacother. 2021;133 doi: 10.1016/j.biopha.2020.110966. [DOI] [PubMed] [Google Scholar]
- 85.Wang D., Liu J., Zhong L., et al. Potential benefits of metformin and pioglitazone combination therapy via gut microbiota and metabolites in high-fat diet-fed mice. Front Pharmacol. 2022;13 doi: 10.3389/fphar.2022.1004617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Veturi Y., Ritchie M.D. How powerful are summary-based methods for identifying expression-trait associations under different genetic architectures? Pac Symp Biocomput. 2018;23:228–239. [PMC free article] [PubMed] [Google Scholar]
- 87.Skrivankova V.W., Richmond R.C., Woolf B.A.R., et al. Strengthening the reporting of observational studies in epidemiology using mendelian randomisation (STROBE-MR): explanation and elaboration. BMJ. 2021;375 doi: 10.1136/bmj.n2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Davies N.M., Holmes M.V., Davey Smith G. Reading Mendelian randomisation studies: a guide, glossary, and checklist for clinicians. BMJ. 2018;362 doi: 10.1136/bmj.k601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lawlor D.A., Harbord R.M., Sterne J.A., Timpson N., Davey Smith G. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat Med. 2008;27(8):1133–1163. doi: 10.1002/sim.3034. [DOI] [PubMed] [Google Scholar]
- 90.Staiger D., Stock J.H. Instrumental variables regression with weak instruments. Econometrica. 1997;65(3):557–586. [Google Scholar]
- 91.Sekula P., Del Greco M.F., Pattaro C., Köttgen A. Mendelian randomization as an approach to assess Causality using observational data. J Am Soc Nephrol. 2016;27(11):3253–3265. doi: 10.1681/ASN.2016010098. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Scatter plots for MR of common drugs on psychiatric disorders.
Funnel plots for MR of common drugs on psychiatric disorders.
Leave-one-out plots for MR of common drugs on psychiatric disorders.
Scatter plots for MR of common drugs on IDPs.
Funnel plots for MR of common drugs on IDPs.
Leave-one-out plots for MR of common drugs on IDPs.
Leave-one-out plots for MR of common drugs on IDPs.
Funnel plots for MR of common drugs on gut microbiome.
Leave-one-out plots for MR of common drugs on gut microbiome.
Scatter plots for MR of gut microbiome on IDPs.
Funnel plots for MR of gut microbiome on IDPs.
Leave-one-out plots for MR of gut microbiome on IDPs.