Abstract
Rift Valley fever virus (RVFV), a phlebovirus of the family Bunyaviridae, is a major public health threat in Egypt and sub-Saharan Africa. The viral and host cellular factors that contribute to RVFV virulence and pathogenicity are still poorly understood. All pathogenic RVFV strains direct the synthesis of a nonstructural phosphoprotein (NSs) that is encoded by the smallest (S) segment of the tripartite genome and has an undefined accessory function. In this report, we show that MP12 and clone 13, two attenuated RVFV strains with mutations in the NSs gene, were highly virulent in IFNAR−/− mice lacking the alpha/beta interferon (IFN-α/β) receptor but remained attenuated in IFN-γ receptor-deficient mice. Both attenuated strains proved to be excellent inducers of early IFN-α/β production. In contrast, the virulent strain ZH548 failed to induce detectable amounts of IFN-α/β and replicated extensively in both IFN-competent and IFN-deficient mice. Clone 13 has a defective NSs gene with a large in-frame deletion. This defect in the NSs gene results in expression of a truncated protein which is rapidly degraded. To investigate whether the presence of the wild-type NSs gene correlated with inhibition of IFN-α/β production, we infected susceptible IFNAR−/− mice with S gene reassortant viruses. When the S segment of ZH548 was replaced by that of clone 13, the resulting reassortants became strong IFN inducers. When the defective S segment of clone 13 was exchanged with the wild-type S segment of ZH548, the reassortant virus lost the capacity to stimulate IFN-α/β production. These results demonstrate that the ability of RVFV to inhibit IFN-α/β production correlates with viral virulence and suggest that the accessory protein NSs is an IFN antagonist.
Alpha/beta interferons (IFNs-α/β) are key components of the innate immune mechanisms that protect the host against invading viruses (23, 31, 42). The extraordinary power of the IFN system has prompted many viruses to adopt strategies that inhibit IFN production or action (for a review, see reference 13). We therefore considered the possibility that virulent strains of Rift Valley fever virus (RVFV) differ from attenuated strains in their capacity to actively antagonize the IFN response of the host. RVFV is a mosquito-borne virus which belongs to the Bunyaviridae family (Phlebovirus genus). Periodically, the virus causes epidemics and epizootics in sub-Saharan countries of Africa and in Egypt. In humans, infection leads to a wide spectrum of clinical symptoms that range from a benign fever to severe encephalitis, retinitis, and fatal hepatitis with hemorrhagic fever (27). Among animals, sheep and goats are severely affected.
Like all members of the family, RVFV possesses a single-stranded segmented RNA genome composed of a large (L), a medium (M), and a small (S) segment (for reviews, see references 9, 11, and 40). The L and M segments are of negative polarity. The L segment codes for the RNA-dependent RNA polymerase. The M segment codes for a polyprotein which is the precursor to the glycoproteins G1 and G2 and two nonstructural proteins, 14K and 78K. The S segment has two open reading frames that do not overlap. They code for the nucleoprotein N and the nonstructural protein NSs in an ambisense coding strategy. The roles of the nonstructural proteins, in particular the NSs protein, are still unknown.
Safe and efficient vaccines for both human and veterinary use are urgently needed. In the past, efforts were undertaken to develop live attenuated vaccine strains by natural selection from wild-type strains. Thus, the Smithburn neurotropic strain of RVFV was derived from the virulent Entebbe strain after numerous intracerebral passages in mice (41). It is still in use as a veterinary live virus vaccine, irrespective of its considerable neurotropism, abortogenicity, and teratogenicity. More recently, two additional attenuated strains, MP12 and clone 13, were obtained. The MP12 strain was derived from the virulent Egyptian strain ZH548 (6). Strain ZH548 was originally isolated from the serum of an uncomplicated human febrile case of RVF that occurred during the Egyptian outbreak of 1977 (26). The virus was then propagated for 12 serial tissue culture passages in the presence of the mutagenic agent 5-fluorouracil, generating strain MP12. It was later shown that MP12 carries attenuating mutations in each genomic segment and was considered a safe vaccine (39). Clone 13 represents an avirulent virus variant that has a large deletion in the S segment and is naturally attenuated. It was biologically cloned by plaque purification from a field isolate obtained from a nonfatal human case during the 1974 RVF outbreak in Bangui, Central African Republic (30). Clone 13 is unique in that it grows well in Vero cells but is avirulent in vivo. It has no pathogenicity for mice or hamsters, in that these animals survive large infectious doses of up to 106 PFU without developing any signs of disease. In addition, clone 13 is highly immunogenic, leading to long-lasting immunity (30).
Recent work with reassortant viruses demonstrated that the attenuation phenotype is mediated by the S segment of clone 13, which contains a defective NSs gene (45). The wild-type NSs gene codes for a nonstructural protein of 265 amino acids that is abundantly expressed in infected cells. The NSs gene of clone 13 has a large internal in-frame deletion of 549 nucleotides which removes 70% of the open reading frame but conserves the N and C termini of the protein. As a consequence, a truncated NSs protein of 82 amino acids is produced but is rapidly degraded in infected cells by the proteasome pathway (45). The NSs protein of RVFV is unique among members of the Bunyaviridae family, as it is phosphorylated and found in the nucleus of infected cells, where it forms large filamentous structures (21, 47). This nuclear localization is surprising because RVFV, like all members of the family, replicates exclusively in the cytoplasm. All attempts to define a role for NSs have failed so far. In experimental reconstitution systems, the protein has neither a stimulatory nor an inhibitory effect on transcription or replication of RVFV (24, 36). The NSs-deficient clone 13 is able to multiply normally in IFN-deficient Vero cells as well as in mosquito cells or whole Culex pipiens mosquitoes (30). This may suggest that the NSs gene has evolved during adaptation of RVFV to the mammalian host and that an important role of the NSs protein was to provide a mechanism to circumvent the IFN response of vertebrate cells. Using mice which are unable to respond to IFN-α/β or IFN-γ, we demonstrate that the S segment of RVFV determines IFN-α/β production in the infected host. The present results suggest that the NSs protein is an important virulence factor that prevents IFNs-α/β from being induced early during the course of RVFV infection.
MATERIALS AND METHODS
Viruses.
Virus strains ZH548, MP12 (6), clone 13 (30), and reassortants between ZH548 and clone 13 (45) were propagated in Vero cells using a multiplicity of infection (MOI) of 0.001. Virus present in the supernatant was usually harvested 3 days after infection. Virus titers were determined by plaque assay in Vero cells.
Animal studies.
IFN-competent mice of inbred strain 129/SvPasIco were obtained from Iffa Credo, Les Oncins, France. Transgenic mice with targeted disruptions of the β-subunit of the IFN-α/β receptor (IFNAR−/−) or the IFN-γ receptor (IFNGR−/−) on an inbred 129SV/Ev genetic background were bred and raised as described (16, 19, 31). Age- and sex-matched animals were inoculated intraperitoneally with 104 PFU of RVFV. Mortality was recorded twice a day. Animals were observed for a maximum of 21 days.
IFN assay.
At various times after infection, venous blood was collected from each group of mice and pooled. The pooled sera were treated at pH 2 overnight, readjusted to pH 7.2, and stored frozen (−72°C) until use. Serum IFN titers were determined in a biological assay, using mouse L929 cells and vesicular stomatitis virus, as described previously (15). IFN titers are expressed as units per milliliter of serum, standardized against a standard preparation of recombinant human IFN-αB/D (Novartis, Basel, Switzerland) known to be active on mouse cells (18).
Histopathological examination.
Specimens including liver and spleen were fixed in formalin and embedded in paraffin. Serial 5-μm-thick tissue sections were stained with hematoxylin and eosin, periodic acid Schiff, and Gordon sweet stains according to the protocols described by Bancroft et al. (2). Immunohistochemical analysis was performed as described (14), using a mouse polyclonal antibody directed against baculovirus-expressed nucleoprotein N of RVFV strain MP12. Briefly, tissue sections were immersed in 200 ml of citrate and incubated three times for 5 min in a microwave oven at 650 W before staining. The alkaline phosphatase method with fast red as a chromogen (LSAB 2 universal alkaline phosphatase kit; Dako, Trappes, France) was used to detect the secondary antibody. Slides were counterstained with Meyer's hematoxylin. Negative controls included tissue samples obtained from uninfected mice.
In situ hybridization.
A 35S probe was prepared by labeling the EcoRI-cleaved DNA insert of plasmid pBS-N in the presence of [35S]dATP using the random-primed DNA labeling kit (Boehringer, Mannheim, Germany). The pBS-N plasmid contains the N protein sequence of the MP12 strain from nucleotides 1008 to 1691 (according to the sequence of the genome strand reported previously [12]) inserted at the unique EcoRI site of the BlueScript polylinker (Stratagene). In situ hybridization was performed as previously described (32). Briefly, deparaffinized slides were dehydrated in a blocking solution to inhibit nonspecific binding of the probe. The slides were acetylated in 0.1 M triethanolamine (pH 8) and washed in a solution composed of formamide and 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). The probe containing 5 × 104 cpm/ml was denatured for 2 min at 80°C and incubated for 16 h. The slides were then washed in SSC solution, followed by incubation in a photographic emulsion. Slides were counterstained in Harris hematoxylin and mounted in Eukitt medium.
RESULTS
Attenuated RVFV strains are fully pathogenic for mice lacking a functional IFN-α/β system.
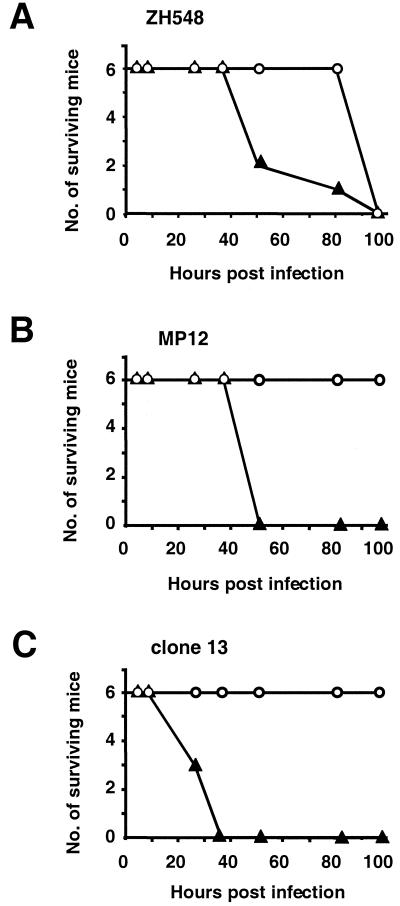
It has previously been shown that the virulent strain ZH548 is pathogenic for laboratory mice after intraperitoneal or subcutaneous inoculation, causing acute hepatitis and death of the animals within a few days after infection with a dose as low as 10 PFU (45). In contrast, strains MP12 and clone 13 are highly attenuated, as indicated by the fact that mice survive experimental infections with virus doses of up to 105 to 106 PFU (6). The mechanisms responsible for this astonishing difference in pathogenicity have not been elucidated. To assess a possible role of virus-induced IFN-α/β, we infected genetically modified mice defective for the IFN-α/β receptor (IFNAR−/− mice) and appropriate control mice with either the attenuated strain MP12, the attenuated strain clone 13, or the virulent strain ZH548. Six mice in each group were infected intraperitoneally with 104 PFU of virus and observed for 21 days. Blood samples were obtained at various times after infection to monitor viremia and serum IFN-α/β levels. Figure 1 shows that strain ZH548 killed both IFNAR−/− mice and wild-type mice within 4 to 5 days, as expected. IFNAR−/− mice showed an accelerated death rate, indicating that they were somewhat more susceptible than wild-type mice (Fig. 1A). In contrast, wild-type mice infected with MP12 or clone 13 survived for 21 days without any signs of disease, as previously reported (45) (Fig. 1B and C). However, none of the IFNAR−/− mice survived. In fact, all the mice died from the infection within 2 to 3 days. This rapid death was unexpected, as it resembled that of mice infected with virulent strain ZH548. Therefore, additional experiments with MP12 and clone 13 were performed. They gave the same results and showed that IFNAR−/− mice died very rapidly after infection with virus doses as low as 10 PFU (data not shown). The 50% lethal dose (LD50) of clone 13 was found to be approximately 1 to 5 PFU for IFNAR−/− mice. It thus resembles the LD50 of virulent strain ZH548 for wild-type mice (45).
FIG. 1.
High virulence of attenuated RVFV strains in IFN-deficient mice. IFNAR−/− mice lacking the receptor for IFN-α/β (solid triangles) and genotype-matched normal mice (open circles) were infected intraperitoneally with 104 PFU of either wild-type virus ZH548 (A), attenuated strain MP12 (B), or attenuated strain clone 13 (C). Survival was assessed daily for up to 21 days.
To investigate whether IFN-γ plays a comparable role in mediating in vivo attenuation of RVFV strains, mice having a targeted mutation inactivating the IFN-γ receptor were infected with 104 PFU of attenuated virus strain MP12 or clone 13. IFNAR−/− mice and wild-type mice were included in these experiments as susceptible and resistant controls, respectively. All six IFNGR−/− mice survived infection without showing disease symptoms, as did the six wild-type mice, whereas all six of the IFNAR−/− mice succumbed to infection with either strain. These results clearly demonstrate that IFN-γ plays at best a negligible role in RVFV attenuation but that IFN-α/β is absolutely required for survival.
Attenuated strains cause fulminant hepatitis in IFNAR−/− mice.
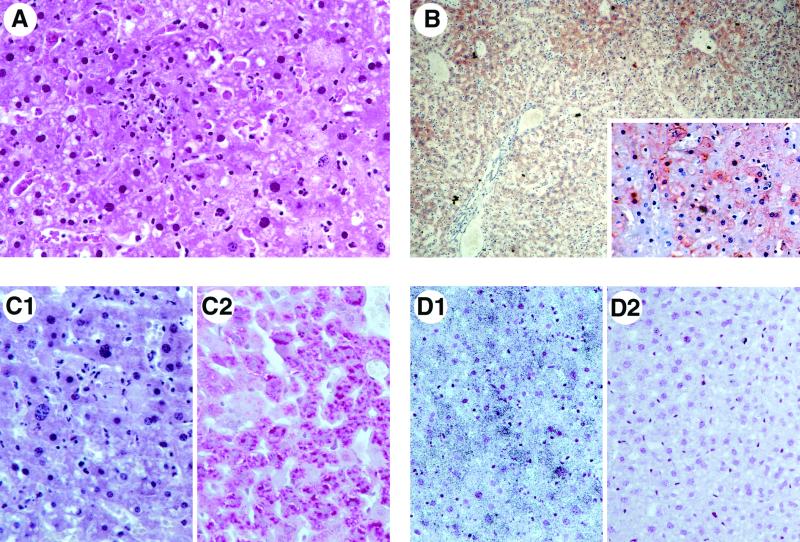
To assess the cause of death due to infection with the attenuated viruses in IFNAR−/− mice, histopathological examinations were performed. On post mortem examination of mice inoculated with clone 13, a swollen and congested liver was most prominent. Microscopically, the livers exhibited the salient features associated with RVFV infection (25, 29). The mice showed signs of a fulminant hepatitis with centrolobular, mostly perivascular coagulative necrosis and massive destruction of the whole lobules except for a layer of hepatocytes around the portal area. In these areas, numerous apoptotic nuclei were detectable (Fig. 2A). Polymorphonuclear or lymphocytic infiltrates were absent, indicating a rapid onset and fulminant course of the destructive process. Staining for glycogen revealed a loss of glycogen from infected livers (Fig. 2C1) compared to control livers (Fig. 2C2), indicating fresh lesions of acute hepatitis.
FIG. 2.
Fulminant hepatitis caused by attenuated clone 13 in IFN-deficient mice. Histology, immunostaining, and in situ hybridization of post mortem liver sections from IFNAR−/− mice inoculated with 104 PFU of clone 13. (A) Hematoxylin-eosin staining showing perivascular coagulative necrosis and numerous apoptotic nuclei around the portal area. (B) Immunostaining for viral N protein. (C) Loss of glycogen as revealed by periodic acid Schiff staining. (D) In situ hybridization detecting virus-specific nucleic acids in infected (D1) or uninfected (D2) hepatocytes. Magnifications: (A and C) ×360; (B) ×90; (inset) ×225; (D) ×225.
In addition, immunohistochemistry with a monospecific antibody directed against the viral N protein was performed. Infected hepatocytes were most prominently detected in the perivascular areas but extended into the whole lobule, as demonstrated by strong immunostaining (Fig. 2B). These hepatocytes were heavily infected, as revealed by the presence of large amounts of viral antigen in the cytoplasm (insert in Fig. 2B). Finally, in situ hybridization with an S segment-specific probe demonstrated that numerous hepatocytes contained virus-specific nucleic acids (RNA) in infected (Fig. 2D1) but not in uninfected (Fig. 2D2) livers. In the spleen, immunohistochemistry and in situ hybridization confirmed the presence of infected cells and demonstrated that the macrophages of the perifollicular areas are the main target of the virus (data not shown). These histopathological features were identical to those observed in livers and spleens of wild-type mice inoculated with virulent strain ZH548 (data not shown).
Rapid growth of attenuated RVFV strains in IFNAR−/− mice.
Next, we determined the kinetics and extent of virus growth in wild-type and IFNAR−/− mice infected intraperitoneally with 104 PFU of RVFV strain ZH548, MP12, or clone 13 (Fig. 3). The development and degree of viremia were assessed by titrating infectious virus in serum samples obtained at various times after infection. ZH548 caused a high level of viremia in both normal and IFN-deficient mice (Fig. 3A). Clearly, virus growth was faster and reached 100-fold-higher titers in IFNAR−/− mice compared to control mice. In contrast, both attenuated virus strains were unable to cause viremia in the IFN-competent wild-type mice (Fig. 3B and C). Obviously, virus multiplication was severely restricted, and the infecting virus was unable to reach the bloodstream in detectable amounts. This lack of viral spread corresponded well with the absence of overt clinical disease and gross pathological changes in these animals.
FIG. 3.
Attenuated RVFV strains cause viremia in IFN-deficient mice. Growth of virulent virus ZH548 (A), attenuated strain MP12 (B), and attenuated strain clone 13 (C) in IFNAR−/− mice (solid triangles) and control mice (open circles). Serum samples were collected from six mice per group at the indicated times after infection with 104 PFU and pooled before plaque titration on Vero cells.
Interestingly, both MP12 and clone 13 showed a completely different behavior than wild-type virus in the IFNAR−/− mice. High virus titers were detectable with MP12 in sera collected 48 h after infection (Fig. 3B). Surprisingly, clone 13 virus grew extremely fast and reached titers of 107 PFU/ml of serum within 28 h (Fig. 3C). In IFN-deficient mice, this otherwise attenuated virus seemed to be even more virulent than strain ZH548. The rapid viral growth correlated well with the rapid progression of disease and the early death observed in these animals (Fig. 1C).
Virulent strains are poor inducers of IFN-α/β.
Given these findings, it was clear that IFN-α/β was the key player responsible for the attenuation phenotype observed with MP12 and clone 13 in normal mice. We investigated IFN-α/β production by determining the amount of acid-stable IFN-α/β in aliquots of the serum samples used for virus titrations. Figure 4A shows that IFN-α/β was barely detectable in serum samples from mice infected with the virulent strain ZH548. This was unexpected, given the extensive virus multiplication in these animals (Fig. 3A). Interestingly, the MP12 and clone 13 viruses induced large amounts of IFN-α/β in IFNAR−/− mice that were easily detectable in late serum samples (Fig. 4B and C). The time course of IFN production correlated well with the rapid virus growth in these IFN-nonresponsive mice, indicating that both viruses are good inducers of IFN-α/β. Significantly, serum IFN was also detectable in normal wild-type mice early in the course of infection with clone 13 (Fig. 4C). This is remarkable because no viremia was present, again indicating that the attenuated strain was able to induce an early production of IFN-α/β that was restricting further virus growth in the IFN-competent animals. This early IFN peak is likely to be responsible for the attenuation phenotype of clone 13 and possibly MP12 in normal mice. Remarkably, it is not detectable in mice infected with the virulent strain ZH548 (Fig. 4A), despite extensive virus replication (Fig. 3A).
FIG. 4.
Virulent RVFV strain ZH548 is a poor inducer of IFN-α/β. Aliquots of the pooled serum samples described in Fig. 3 were inactivated at low pH and tested for acid-stable IFN-α/β in a standard bioassay. Sera were from IFNAR−/− mice (solid triangles) or control mice (open circles) infected with 104 PFU of either wild-type virus ZH548 (A), attenuated strain MP12 (B), or attenuated strain clone 13 (C).
S segment of RVFV controls IFN induction.
Clone 13 carries a large in-frame deletion of the NSs gene, which is encoded by the S segment of the tripartite genome. This deletion leads to a truncated and abnormal NSs protein (45). Genetic reassortment between virulent ZH548 and attenuated clone 13 demonstrated that the altered S segment carries a major virulence/attenuation determinant (45). It was conceivable that this genetic alteration was also responsible for the increased IFN-α/β production. To investigate this possibility, we used these reassortant viruses and their parental strains to infect IFNAR−/− mice. The reassortant virus carrying the L and M segments of strain ZH548 and the defective S segment of clone 13 was named Z/Z/C. Conversely, the reassortant carrying the intact S segment of ZH548 in a clone 13 genetic background was called C/C/Z. Groups of six IFNAR−/− mice were infected with 104 PFU of either wild-type virus ZH548 (Z/Z/Z), mutant clone 13 (C/C/C), or the reassortant virus Z/Z/C or C/C/Z. Individual blood samples were collected at the indicated times after infection. Sera were pooled within each group, and aliquots were assayed in parallel for infectious virus and IFN-α/β. Figure 5 shows that the virulent Z/Z/Z virus grew to high titers and killed the animals within 48 h of infection, but it did not induce detectable levels of IFN-α/β. When the S segment of ZH548 was replaced by that of clone 13, the resulting Z/Z/C virus also grew to high titers, but it strongly induced IFN synthesis. Likewise, clone 13 (C/C/C) grew well and induced large amounts of IFN-α/β. In contrast, reassortant C/C/Z had lost its capacity to induce IFN-α/β, although it maintained the growth characteristics of clone 13. These results formally demonstrate that the S segment of RVFV carries determinants that influence IFN-α/β production in the infected host. They further suggest that the NSs protein targets the IFN-α/β system of the host but is otherwise not required for efficient virus growth. Taken together, the present results strongly suggest that the IFN response governed by NSs determines viral virulence or attenuation in normal hosts.
FIG. 5.
S segment of RVFV determines IFN inducibility. IFNAR−/− mice were infected with 104 PFU of either wild-type virus ZH548 (Z/Z/Z), attenuated clone 13 (C/C/C), or the reassortant virus Z/Z/C or C/C/Z. Individual blood samples were collected at the indicated times after infection. Sera from six mice per group were pooled, and aliquots were assayed in parallel for infectious virus in a plaque assay (solid triangles) and for IFN-α/β in a standard bioassay (open circles).
DISCUSSION
The function of the nonstructural protein NSs of RVFV during infection is still unknown. It is a matter of great interest, because the phosphoprotein accumulates in large amounts in the nucleus of infected cells, whereas virus replication takes place exclusively in the cytoplasm. The nuclear localization of NSs is compatible with the view that NSs is not directly involved in virus growth but may serve an accessory function. Here we provide evidence suggesting that the main role of the NSs protein of RVFV is to inhibit the IFN response of the host by blocking virus-induced IFN-α/β production. The evidence is based on a classical genetic approach, combining genetic modification of the virus with genetic manipulation of the host.
We show that the NSs-expressing ZH548 virus is highly virulent and grows to high titers in laboratory mice without inducing detectable amounts of IFN-α/β, whereas the NSs-defective clone 13 virus is an excellent IFN inducer and is highly attenuated in IFN-responsive mice. The mutant virus shows restored growth and wild-type virulence in genetically altered mice that are nonresponsive to IFN-α/β. The growth and virulence of the NSs-defective virus are not restored in genetically altered mice that have a defect in the IFN-γ system. This specific phenotypic restoration clearly identifies the IFN-α/β system as the target of NSs. Finally, we show that a reassortant ZH548 virus with the NSs-defective S segment of clone 13 becomes a phenocopy of clone 13, and vice versa: a reassortant clone 13 virus containing the intact S segment of the virulent strain is now a poor IFN inducer but replicates efficiently in IFN-competent mice. These results demonstrate for the first time that the S segment of RVFV carries determinants that regulate IFN-α/β production in the infected host. However, these results do not formally prove that NSs is the responsible factor. Comparisons of the S segment sequences of clone 13 and ZH548 show that there are a few changes in addition to the large deletion in the NSs gene, namely, one amino acid change in the N protein sequence (glycine versus glutamic acid at position 159) and six nucleotide changes in the intergenic region (30, 45). It is highly unlikely that these differences rather than the loss of NSs function are responsible for the different phenotypes described here. However, a definitive answer will have to await the availability of recombinant viruses with targeted deletions in the NSs gene. A reverse genetics system exists for Bunyamwera virus, another member of the Bunyaviridae family (5). A recent report describes a recombinant Bunyamwera virus that lacks NSs and exhibits an attenuation phenotype in mice (A. Bridgen, J. K. Fazakerley, and R. M. Elliott, Abstr. 11th Int. Cong. Virol. abstr. VW47.06, 1999). Interestingly, this NSs-minus virus seems to have gained IFN-inducing properties, indicating that the NSs protein of Bunyamwera virus may also have IFN-antagonistic functions (F. Weber, A. Bridgen, and R. M. Elliott, Abstr. 11th Int. Conf. Negative-Strand Viruses, abstr. 117, 2000).
The MP12 strain of RVFV was derived from strain ZH548 by serial tissue culture passages and cloning in the presence of 5-fluorouracil, a mutagenic substance. The close genetic relationship between the two virus strains allows a direct comparison of their sequences (46). The S segment of MP12 has acquired one nucleotide exchange in the intergenic region and three exchanges in the NSs coding region, leading to a single-amino-acid substitution from valine to alanine at position 513. This point mutation occurred early in the course of passaging, when reduced virulence and reduction in plaque size of the resulting virus was first observed (39, 46). It is therefore conceivable that this mutation is responsible for the attenuated phenotype observed. However, MP12 acquired additional attenuating mutations in the other gene segments during subsequent passages in tissue culture (43). Unlike its parental strain, the attenuated MP12 proved to be a good IFN inducer in the present experiments. Moreover, it exhibited virulence in IFNAR−/− mice, exactly like clone 13. It can therefore be assumed that the point mutation in NSs acquired during attenuation of MP12 inactivates the anti-IFN function of the protein. Alternatively, some of the other attenuating mutations may contribute to the phenotype.
The present results clearly show that both attenuated RVFV strains are fully competent to replicate and to display their inherent pathogenic potential in IFN-α/β-insensitive mice, leading to severe disease and death within a few days after infection. In fact, clone 13 was even more virulent than wild-type strain ZH548, indicating that a functional NSs protein is totally dispensable for virus growth and pathogenicity in these animals. However, in the presence of a competent IFN response, these viruses grew poorly and were apathogenic. It was conceivable that the attenuated viruses were not only better IFN inducers, as shown here, but were also more sensitive to the antiviral action of IFNs than the pathogenic strains. We tested this hypothesis using virus-permissive Vero cells and recombinant human IFN-α, but were unable to detect significant differences in IFN sensitivity between MP12, clone 13, and ZH548 (data not shown). This is in agreement with a previous study reporting that RVFV strains that differed in virulence for rats were equally sensitive to the antiviral effect of recombinant human IFN-α (1). We conclude that the virulence of a given RVFV strain is not dictated by its IFN sensitivity but resides in its capacity to efficiently block the production of IFN-α/β.
The present findings are not without precedent. An early study by Higashihara and coworkers found that less virulent RVFV strains were generally more potent IFN-α/β inducers in mice than the corresponding more virulent strains (17). Avirulent strains were able to induce a first peak of IFN at a very early stage of infection, whereas virulent strains were unable to do so but had circulating IFN later on shortly before death, when the animals had a high viremia (17). Likewise, Peters and coworkers concluded from extensive studies that the outcome of RVFV infections in rodents and monkeys is controlled by early IFN-α/β (1, 28, 29, 33, 34).
Most viruses must multiply and spread in the presence of IFNs that activate innate host defense mechanisms. Many viruses seem therefore to have evolved specific means that circumvent the IFN response of the host (for a review, see reference 13). Such strategies are especially important for RNA viruses. During their life cycle, RNA viruses produce large amounts of double-stranded RNA (dsRNA) which, by itself, is an excellent inducer of IFN-α/β (20). It is therefore not surprising that most negative-strand RNA viruses seem to code for accessory proteins that subvert IFN production and/or action (13). A major strategy for blocking IFN-α/β production is to provide a virally encoded dsRNA-binding protein that sequesters dsRNA molecules and prevents them from inducing IFN-α/β (10). The sequestration of dsRNA is expected to reduce the dsRNA-dependent activation of a number of antiviral proteins, including PKR (4, 7, 22), which is also involved in efficient induction of IFN-α/β (8). A variety of additional strategies have been found to operate in different viruses. For example, some viruses encode proteins inhibiting the activities of cellular transcription factors mediating IFN-α/β induction, such as NF-κB (35, 37) and members of the IFN-regulatory factor family (3, 37, 38, 44). Given the plethora of mechanisms through which NSs could work, it will be interesting to determine which strategy is used by RVFV.
ACKNOWLEDGMENTS
We thank Agnes Billecocq, Jason Paragas, Michael Frese, Georg Kochs, Peter Staeheli, and Adolfo Garcia-Sastre for stimulating discussions and critically reading the manuscript.
This work was supported in part by grant HA 1582 from the Deutsche Forschungsgemeinschaft.
REFERENCES
- 1.Anderson G W, Jr, Peters C J. Viral determinants of virulence for Rift Valley fever (RVF) in rats. Microb Pathog. 1988;5:241–250. doi: 10.1016/0882-4010(88)90096-4. [DOI] [PubMed] [Google Scholar]
- 2.Bancroft J D, Cook H C, Stirling R W, Turner D R. Manual of histological technics and their diagnostic applications. London, U.K: Churchill Livingstone; 1994. [Google Scholar]
- 3.Barnard P, McMillan N A. The human papillomavirus E7 oncoprotein abrogates signaling mediated by interferon-alpha. Virology. 1999;259:305–313. doi: 10.1006/viro.1999.9771. [DOI] [PubMed] [Google Scholar]
- 4.Bergmann M, Garcia-Sastre A, Carnero E, Pehamberger H, Wolff K, Palese P, Muster T. Influenza virus NS1 protein counteracts PKR-mediated inhibition of replication. J Virol. 2000;74:6203–6206. doi: 10.1128/jvi.74.13.6203-6206.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bridgen A, Elliot R M. Rescue of a segmented negative-stranded RNA virus entirely from cloned complementary DNAs. Proc Natl Acad Sci USA. 1996;93:15400–15404. doi: 10.1073/pnas.93.26.15400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caplen H, Peters C J, Bishop D H L. Mutagen-directed attenuation of Rift Valley fever virus as a method for vaccine development. J Gen Virol. 1985;66:2271–2277. doi: 10.1099/0022-1317-66-10-2271. [DOI] [PubMed] [Google Scholar]
- 7.Chang H W, Watson J C, Jacobs B L. The E3L gene of vaccinia virus encodes an inhibitor of the interferon-induced, double-stranded RNA-dependent protein kinase. Proc Natl Acad Sci USA. 1992;89:4825–4829. doi: 10.1073/pnas.89.11.4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chu W M, Ostertag D, Li Z W, Chang L, Chen Y, Hu Y, Williams B, Perrault J, Karin M. JNK2 and IKKbeta are required for activating the innate response to viral infection. Immunity. 1999;11:721–731. doi: 10.1016/s1074-7613(00)80146-6. [DOI] [PubMed] [Google Scholar]
- 9.Elliott R M, editor. The bunyaviridae. New York, N.Y: Plenum Press; 1996. [Google Scholar]
- 10.Garcia-Sastre A, Egorov A, Matassov D, Brandt S, Levy D E, Durbin J E, Palese P, Muster T. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology. 1998;252:324–330. doi: 10.1006/viro.1998.9508. [DOI] [PubMed] [Google Scholar]
- 11.Giorgi C. Molecular biology of phleboviruses. In: Elliott R M, editor. The bunyaviridae. New York, N.Y: Plenum Press; 1996. pp. 105–128. [Google Scholar]
- 12.Giorgi C, Accardi L, Nicoletti L, Gro M C, Takehara K, Hilditch C, Morikawa S, Bishop D H. Sequences and coding strategies of the S RNAs of Toscana and Rift Valley fever viruses compared to those of Punta Toro, Sicilian sandfly fever, and Uukuniemi viruses. Virology. 1991;180:738–753. doi: 10.1016/0042-6822(91)90087-r. [DOI] [PubMed] [Google Scholar]
- 13.Goodbourn S, Didcock L, Randall R E. Interferons: cell signalling, immune modulation, antiviral response and virus countermeasures. J Gen Virol. 2000;81:2341–2364. doi: 10.1099/0022-1317-81-10-2341. [DOI] [PubMed] [Google Scholar]
- 14.Hall W C, Crowell T P, Watts D M, Barros V L, Kruger H, Pinheiro F, Peters C J. Demonstration of yellow fever and dengue antigens in formalin-fixed paraffin-embedded human liver by immunohistochemical analysis. Am J Trop Med Hyg. 1991;45:408–417. doi: 10.4269/ajtmh.1991.45.408. [DOI] [PubMed] [Google Scholar]
- 15.Haller O, Arnheiter H, Gresser I, Lindenmann J. Genetically determined, interferon-dependent resistance to influenza virus in mice. J Exp Med. 1979;149:601–612. doi: 10.1084/jem.149.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hefti H P, Frese M, Landis H, Di Paolo C, Aguzzi A, Haller O, Pavlovic J. Human MxA protein protects mice lacking a functional alpha/beta interferon system against La Crosse virus and other lethal viral infections. J Virol. 1999;73:6984–6991. doi: 10.1128/jvi.73.8.6984-6991.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Higashihara M, Koyama H, Igarashi Y. Induction of virus-inhibiting factor or interferon in mice by strains with different virulence of Rift Valley fever virus. Kitasato Arch Exp Med. 1972;45:33–43. [PubMed] [Google Scholar]
- 18.Horisberger M A, de Staritzky K. A recombinant human interferon-alpha B/D hybrid with a broad host-range. J Gen Virol. 1987;68:945–948. doi: 10.1099/0022-1317-68-3-945. [DOI] [PubMed] [Google Scholar]
- 19.Huang S, Hendriks W, Althage A, Hemmi S, Bluethmann H, Kamijo R, Vilcek J, Zinkernagel R M, Aguet M. Immune response in mice that lack the interferon-gamma receptor. Science. 1993;259:1742–1745. doi: 10.1126/science.8456301. [DOI] [PubMed] [Google Scholar]
- 20.Jacobs B L, Langland J O. When two strands are better than one: the mediators and modulators of the cellular responses to double-stranded RNA. Virology. 1996;219:339–349. doi: 10.1006/viro.1996.0259. [DOI] [PubMed] [Google Scholar]
- 21.Kohl A, di Bartolo V, Bouloy M. The Rift Valley fever virus nonstructural protein NSs is phosphorylated at serine residues located in casein kinase consensus motifs in the carboxy-terminus. Virology. 1999;263:517–525. doi: 10.1006/viro.1999.9978. [DOI] [PubMed] [Google Scholar]
- 22.Langland J O, Pettiford S, Jiang B, Jacobs B L. Products of the porcine group C rotavirus NSP3 gene bind specifically to double-stranded RNA and inhibit activation of the interferon-induced protein kinase PKR. J Virol. 1994;68:3821–3829. doi: 10.1128/jvi.68.6.3821-3829.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leib D A, Machalek M A, Williams B R, Silverman R H, Virgin H W. Specific phenotypic restoration of an attenuated virus by knockout of a host resistance gene. Proc Natl Acad Sci USA. 2000;97:6097–6101. doi: 10.1073/pnas.100415697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lopez N, Muller R, Prehaud C, Bouloy M. The L protein of Rift Valley fever virus can rescue viral ribonucleoproteins and transcribe synthetic genome-like RNA molecules. J Virol. 1995;69:3972–3979. doi: 10.1128/jvi.69.7.3972-3979.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McGavran M H, Easterday B C. Rift Valley fever virus hepatitis light and electron microscopic studies in the mouse. Am J Pathol. 1963;42:587–607. doi: 10.21236/ad0410394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meegan J M. The Rift Valley fever epizootic in Egypt 1977–78: description of the epizootic and virological studies. Trans R Soc Trop Med Hyg. 1979;73:618–623. doi: 10.1016/0035-9203(79)90004-x. [DOI] [PubMed] [Google Scholar]
- 27.Meegan J M, Bailey C J. Rift Valley fever. In: Monath T P, editor. The arboviruses: epidemiology and ecology. IV. Boca Raton, Fla: CRC Press Inc.; 1988. pp. 51–76. [Google Scholar]
- 28.Morrill J C, Jennings G B, Cosgriff T M, Gibbs P H, Peters C J. Prevention of Rift Valley fever in rhesus monkeys with interferon-alpha. Rev Infect Dis. 1989;11(Suppl. 4):815–825. doi: 10.1093/clinids/11.supplement_4.s815. [DOI] [PubMed] [Google Scholar]
- 29.Morrill J C, Jennings G B, Johnson A J, Cosgriff T M, Gibbs P H, Peters C J. Pathogenesis of Rift Valley fever in rhesus monkeys: role of interferon response. Arch Virol. 1990;110:195–212. doi: 10.1007/BF01311288. [DOI] [PubMed] [Google Scholar]
- 30.Muller R, Saluzzo J F, Lopez N, Dreier T, Turell M, Smith J, Bouloy M. Characterization of clone 13, a naturally attenuated avirulent isolate of Rift Valley fever virus, which is altered in the small segment. Am J Trop Med Hyg. 1995;53:405–411. doi: 10.4269/ajtmh.1995.53.405. [DOI] [PubMed] [Google Scholar]
- 31.Muller U, Steinhoff U, Reis L F, Hemmi S, Pavlovic J, Zinkernagel R M, Aguet M. Functional role of type I and type II interferons in antiviral defense. Science. 1994;264:1918–1921. doi: 10.1126/science.8009221. [DOI] [PubMed] [Google Scholar]
- 32.Ozden S, Aubert C, Gonzalez-Dunia D, Brahic M. In situ analysis of proteolipid protein gene transcripts during persistent Theiler's virus infection. J Histochem Cytochem. 1991;39:1305–1309. doi: 10.1177/39.10.1940303. [DOI] [PubMed] [Google Scholar]
- 33.Peters C J, Anderson G W J. Pathogenesis of Rift Valley fever. Contrib Epidemiol Biostat. 1981;3:21–41. [Google Scholar]
- 34.Peters C J, Reynolds J A, Slone T W, Jones D E, Stephen E L. Prophylaxis of Rift Valley fever with antiviral drugs, immune serum, an interferon inducer, and a macrophage activator. Antiviral Res. 1986;6:285–297. doi: 10.1016/0166-3542(86)90024-0. [DOI] [PubMed] [Google Scholar]
- 35.Powell P P, Dixon L K, Parkhouse R M. An IκB homolog encoded by African swine fever virus provides a novel mechanism for downregulation of proinflammatory cytokine responses in host macrophages. J Virol. 1996;70:8527–8533. doi: 10.1128/jvi.70.12.8527-8533.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prehaud C, Lopez N, Blok M J, Obry V, Bouloy M. Analysis of the 3′ terminal sequence recognized by the Rift Valley fever virus transcription complex in its ambisense S segment. Virology. 1997;227:189–197. doi: 10.1006/viro.1996.8324. [DOI] [PubMed] [Google Scholar]
- 37.Revilla Y, Callejo M, Rodriguez J M, Culebras E, Nogal M L, Salas M L, Vinuela E, Fresno M. Inhibition of nuclear factor kappaB activation by a virus-encoded IkappaB-like protein. J Biol Chem. 1998;273:5405–5411. doi: 10.1074/jbc.273.9.5405. [DOI] [PubMed] [Google Scholar]
- 38.Ronco L V, Karpova A Y, Vidal M, Howley P M. Human papillomavirus 16 E6 oncoprotein binds to interferon regulatory factor-3 and inhibits its transcriptional activity. Genes Dev. 1998;12:2061–2072. doi: 10.1101/gad.12.13.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saluzzo J F, Smith J F. Use of reassortant viruses to map attenuating and temperature-sensitive mutations of the Rift Valley fever virus MP-12 vaccine. Vaccine. 1990;8:369–375. doi: 10.1016/0264-410x(90)90096-5. [DOI] [PubMed] [Google Scholar]
- 40.Schmaljohn C S. Bunyaviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley D M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 1447–1471. [Google Scholar]
- 41.Smithburn K C. Rift valley fever: the neurotropic adaption of the virus and the experimental use of this modified virus as a vaccine. Br J Exp Pathol. 1949;30:1–16. [PMC free article] [PubMed] [Google Scholar]
- 42.Stark G R, Kerr I M, Williams B R, Silverman R H, Schreiber R D. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 43.Takehara K, Min M K, Battles J K, Sugiyama K, Emery V C, Dalrymple J M, Bishop D H L. Identification of mutations in the mRNA of a candidate vaccine strain of Rift Valley fever virus. Virology. 1989;169:452–457. doi: 10.1016/0042-6822(89)90171-2. [DOI] [PubMed] [Google Scholar]
- 44.Talon J, Horvath C M, Polley R, Basler C F, Muster T, Palese P, Garcia-Sastre A. Activation of interferon regulatory factor 3 is inhibited by the influenza A virus NS1 protein. J Virol. 2000;74:7989–7996. doi: 10.1128/jvi.74.17.7989-7996.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vialat P, Billecocq A, Kohl A, Bouloy M. The S segment of Rift valley fever Phlebovirus (Bunyaviridae) carries determinants for attenuation and virulence in mice. J Virol. 2000;74:1538–1543. doi: 10.1128/jvi.74.3.1538-1543.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vialat P, Muller R, Vu T H, Prehaud C, Bouloy M. Mapping of the mutations present in the genome of the Rift Valley fever virus attenuated MP12 strain and their putative role in attenuation. Virus Res. 1997;52:43–50. doi: 10.1016/s0168-1702(97)00097-x. [DOI] [PubMed] [Google Scholar]
- 47.Yadani F-Z, Kohl A, Préhaud C, Billecocq A, Bouloy M. The carboxy-terminal acidic domain of Rift Valley fever virus NSs protein is essential for the formation of filamentous structures but not for the nuclear localization of the protein. J Virol. 1999;73:5018–5025. doi: 10.1128/jvi.73.6.5018-5025.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]