Abstract
When an organism encounters a pathogen, the host innate immune system activates to defend against pathogen colonization and toxic xenobiotics produced. C. elegans employ multiple defense systems to ensure survival when exposed to Pseudomonas aeruginosa including activation of the cytoprotective transcription factor SKN-1/NRF2. Although wildtype C. elegans quickly learn to avoid pathogens, here we describe a peculiar apathy-like behavior towards PA14 in animals with constitutive activation of SKN-1, whereby animals choose not to leave and continue to feed on the pathogen even when a non-pathogenic and healthspan-promoting food option is available. Although lacking the urgency to escape the infectious environment, animals with constitutive SKN-1 activity are not oblivious to the presence of the pathogen and display the typical pathogen-induced intestinal distension and eventual demise. SKN-1 activation, specifically in neurons and intestinal tissues, orchestrates a unique transcriptional program which leads to defects in serotonin signaling that is required from both neurons and non-neuronal tissues. Serotonin depletion from SKN-1 activation limits pathogen defenses capacity, drives the pathogen-associated apathy behaviors and induces a synthetic sensitivity to selective serotonin reuptake inhibitors. Taken together, our work reveals interesting insights into how animals perceive environmental pathogens and subsequently alter behavior and cellular programs to promote survival.
Subject terms: Behavioural genetics, Ageing, Pathogens, Innate immunity
Here, Nair et al show that constitutive activation of the cytoprotective transcription factor SKN-1 within the digestive and nervous system of Caenorhabditis elegans impedes serotonin signalling and leads to pathogen-apathy behaviour.
Introduction
Avoidance of a potential pathogen is one of the earliest defenses an organism employs to ensure survival. In its natural environment, Caenorhabditis elegans feeds upon a variety of bacterial diets, with varying dietary benefits and pathogenic quality, which can affect organismal health1,2. When exposed to a pathogenic microbe, C. elegans must initiate a proper immune response to defend against infection3. Even though C. elegans lack a classical adaptive immune system, they initiate a potent innate immune response, through the upregulation of genes like c-type lectins, anti-microbial peptides, and other defense mechanisms in order to fight off pathogenic infection4.
Of the various bacteria that C. elegans encounter, Pseudomonas aeruginosa (PA14) is also a human opportunistic pathogen that is lethal as a diet to C. elegans with prolonged exposure. When C. elegans are infected by PA14, they consequently activate a conserved innate immune pathway, including the p38 mitogen-activated protein kinase (MAPK) that plays a critical role in survival to pathogen exposure5,6 by controlling the expression of downstream immune effector molecules (e.g., lysozymes, antimicrobial peptides, and C-type lectins)7.
SKN-1/NRF2 is a multifaceted cytoprotective transcription factor that regulates phase 2 detoxification and redox balance8,9. Recently, it has been studied for its essential role in the immune response downstream of the p38 MAPK pathway10–12. Upon activation, SKN-1 activates the expression of the phase II cellular detoxification system and multiple pathogen response genes10,11,13,14 that help protect the organism from pathogen exposure. Relatedly, animals with constitutive activation of SKN-1 display enhanced resistance to killing by pathogen exposure11. However, the enhanced innate immune response to pathogens occurs at the expense of organismal lipid homeostasis and results in a rapid redistribution of lipid stores from the soma to the germline that leads to impaired organismal health, reduced oxidative stress resistance, and a shortened lifespan11,15. Transcriptional redirection of activated SKN-1 away from pathogen resistance and immune response genes towards oxidative stress and xenobiotic detoxification genes restores metabolic homeostasis and prevents the depletion of somatic fat from the intestines, thereby improving healthspan and lifespan11. As such, although SKN-1 activation plays a critical role in balancing cytoprotection, failure to turn off this response is pleiotropic.
Another mechanism C. elegans utilizes to prevent lasting harm and death from pathogens is an avoidance phenotype16,17. Initially, naïve worms that have never experienced a pathogenic infection will preferentially move toward PA14 over E. coli through stimulation of olfactory behavioral responses16,18. However, this preference does not persist as sophisticated intracellular and cell non-autonomous signaling pathways are initiated to switch the behavior of the host toward avoiding the pathogenic bacteria19–21. This brief exposure to PA14 functions as an important learning experience for the animal to avoid future exposure to the pathogen22,23, and this learned behavior to avoid pathogens can be transmitted to progeny as well24.
The ability to avoid pathogenic bacteria, even after a brief exposure, is essential for the health of the organism, as the inability to leave can lead to a more rapid death. The signaling pathways driving aversion behaviors are complex but clearly require neuropeptide signaling and downstream neurogenic targets20,23,25. Although intestinal bloating has been shown to drive leaving behaviors, several of the molecular details that influence the decision to leave pathogenic bacteria for other food sources are not fully understood. Among the array of neurometabolic signaling molecules that mediate homeostasis, serotonin plays a critical role in the modulation of gut-neuronal signaling in response to dietary inputs and regulating immune responses26. Serotonin synthesis, in response to environmental and nutritional cues, regulates fat mobilization, which may be utilized to elicit immune responses27. Here, we demonstrate that animals with constitutive activation of SKN-1, without the capacity to turn it off, have an activated innate immune system yet fail to initiate leaving behaviors from PA14, despite ultimately dying from exposure. This apathetic early response to the pathogen is driven by cell non-autonomous communication between the digestive and nervous systems stemming from defective serotonin signaling. Understanding why constitutive SKN-1 activation leads to deficiencies in pathogen-leaving behavior will enhance our understanding of the molecular drivers of pathogen avoidance and the role that SKN-1 transcriptional activation plays in this essential survival response to environmental pathogens.
Results
Constitutive activation of SKN-1 drives pathogen apathy
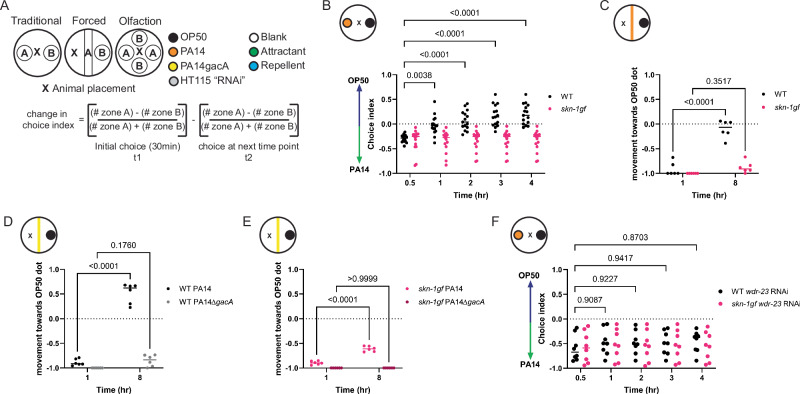
Studies of host responses to pathogenic diets traditionally use preference plates where animals are placed equidistant from two different food options (Fig. 1A and Source Data file 1), and the initial choice between the two options is recorded. Beyond this initial choice, animals can choose to remain on one food source or explore other options, but the decision matrix used in this complex behavior requires additional study. Despite the numerous reports on pathogen food choice, there has not been a consistent experimental design for these studies, and as such, we tested several paradigms of PA14 culturing to standardize our studies of the choice index. Ultimately, our studies showed a very distinct and reproducible pathogen leaving response when the PA14 pathogen was cultured at 37 °C28 (Supplementary Fig. S1A). We compared the choice dynamics of WT and skn-1gf mutants with constitutive SKN-1 activity and discovered that although initial choice was not significantly different, skn-1gf mutants that first choose the PA14 bacteria failed to leave the pathogen environment like age-matched WT animals (Fig. 1B and Supplementary Fig. S1B–D); behaving similar to animals given a choice of E. coli or a non-virulent PA14 strain harboring a mutation in the gacA gene (Supplementary Fig. S1E, F)29. This difference in choice was not influenced by animal movement speed, as all animals reached the first bacteria at similar rates (Supplementary Fig. S1G), nor made minor differences in developmental timing in the skn-1gf mutant influence choice kinetics (Supplementary Fig. S1H).
Fig. 1. Activation of SKN-1 drives pathogen apathy.
A Graphical representation of the variations in food choice assays performed. B, C Apathy displayed by skn-1gf worms in the (B) traditional choice assay or (C) forced choice assays. The pathogen leaving response of WT worms is absent in response to the non-pathogenic strain PA14∆gacA (D) and resembles the apathy behavior of skn-1gf animals (E). Both WT and skn-1gf mutant animals display apathy for PA14 following depletion of wdr-23 by RNAi (F). Each of the food choice assays comprised of N ≥ 3 (with ≥ 150 worms per biological replicate per strain/condition) and analyzed via two-way ANOVA test; **(p < 0.01) ***(p < 0.001) ****(p < 0.0001). Individual “p” value numbers for the comparisons within each graph are placed above the respective comparison bars in the graph. Source data for all behavioral responses are provided as a Source Data file 1.
A confounding factor of traditional food choice assays is the variation in the initial choice at the start of the experiment, which cannot be overcome by changing the distance between the two diets (Supplementary Fig. S1I) and limits the power to study leaving behavior after the initial choice. To overcome this limitation, we developed a “forced” choice assay (Fig. 1A) where animals must interact with one diet first, as a line on the plate, and can then choose to leave that food source for a second food option. In this experimental setup, skn-1gf mutant animals reach the PA14 line within 30 min (Supplementary Fig. S1J), like WT animals, but remain on the pathogen, whereas most WT animals move toward the E. coli option over an 8-hour time course, which ensured ample amounts of both food options were available (Fig. 1C and Supplementary Fig. S1K). As expected, pathogen leaving behavior is sensitive to the pathogenicity of PA14 as neither genotype chooses to leave the attenuated PA14ΔgacA mutant (Fig. 1D, E). We tested whether other mechanisms of SKN-1 activation could impede pathogen leaving and confirmed that RNAi knockdown of the wdr-23 gene, a negative regulator of SKN-1 activity that results in potent activation of SKN-1 when impaired30,31, was sufficient to delay WT animals from leaving the pathogen food source (Fig. 1F) and was not additive with skn-1gf mutation (Figure SL-M). We next measured the pathogen leaving response of daf-2lf and eat-2lf animals to test if these long-lived and enhanced pathogen resistance mutants might display indifference toward the pathogen. However, neither displayed behaviors like the skn-1gf mutants (Supplementary Fig. S1N, O). Lastly, we tested skn-1lf mutant animals in the forced food choice assay, which revealed a normal leaving behavior (Supplementary Fig. S1P). This result supports a model where constitutive SKN-1 activation impedes leaving behavior but also uncouples the behavioral response from the SKN-1-dependent and pathogen-related phenotype of somatic lipid depletion that is suppressed by the E. coli/K-12 “HT115” diet used for RNAi11. Taken together, we coin this failure to leave a pathogenic environment as a form of “pathogen apathy”.
Pathogen apathy is a specific ASI neuronal response
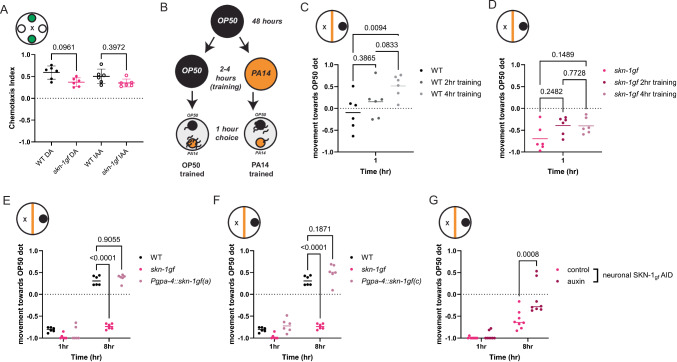
SKN-1 has established roles in ASI neurons, and most recently, we discovered that the phenotypes associated with the skn-1gf allele begin in the nervous system to initiate systemic responses cells non-autonomously32. With this in mind, and considering the sensory role of the ASI neurons, we first tested whether skn-1gf mutants are generally defective in olfaction and found that neither chemoattraction to diacetyl or isoamyl alcohol (Fig. 2A) nor chemorepulsion from octanol or nonanone (Supplementary Fig. S2A) was impaired, but a modest defect in the chemotaxis response toward the repellant 1-undecene was observed (Supplementary Fig. S2B). These data match the observation that skn-1gf mutants reach the first food source with the same kinetics. Behaviors associated with a pathogenic environment are learned and informed by experience, so we tested if animals with constitutive activation of SKN-1 that are previously exposed to PA14 would restore normal pathogen-leaving behaviors. As previously reported, WT animals that are trained with short exposures to the pathogen (2–4 h) display an enhanced leaving response to PA14 (Fig. 2B, C and Supplementary Fig. S2C, D)33,34. In contrast, neither 2 h nor 4 h of training was sufficient to change the apathy behavior of the skn-1gf mutants (Fig. 2D and Supplementary Fig. S2E, F). Taken together, these findings suggest that the behavior defect that induces apathetic leaving behaviors is a specific choice, but not clearly associated with the general perception of volatile odorants within the diet.
Fig. 2. Pathogen apathy requires neuronal SKN-1.
Chemotaxis towards (A) attractants [diacetyl (DA) and isoamyl alcohol (IAA)] are similar for both WT and skn-1gf worms. B Graphical representation of pathogen training and subsequent food choice assay. Differences in pathogen leaving response in (C) WT and (D) skn-1gf worms following pathogen training for 0, 2, and 4 hr of exposure. Specific expression of SKN-1gf isoform a (E) or isoform c (F) in ASI neurons does not elicit failed pathogen leaving behaviors as observed in the skn-1gf mutant (G) Pan-neuronal degradation of SKN-1gf restores pathogen leaving behavior in the skn-1gf mutant. Each of the chemotaxis assays comprised of N ≥ 3 (with ≥ 150 worms per biological replicate per strain/condition) and analyzed via one-way ANOVA test data represented as mean values +/− SEM for the. Each of the food choice assays comprised of N ≥ 3 (with ≥ 150 worms per biological replicate per strain/condition) and analyzed via two-way ANOVA test; **(p < 0.01) ***(p < 0.001) ****(p < 0.0001). Individual “p” value numbers for the comparisons within each graph are placed above the respective comparison bars in the graph. Source data for all behavioral responses are provided as a Source Data file 1.
Our previous work has demonstrated that constitutive SKN-1 activation, specifically in ASI neurons, is sufficient to drive cell non-autonomous effects throughout the organism32. We next expressed the two skn-1 isoforms impacted by the skn-1gf mutation exclusively in ASI neurons. Surprisingly, the expression of skn-1a (gpa-4p::skn-1gf(a)) or skn-1c (gpa-4p::skn-1gf(c)) in the ASI could not replicate the failed pathogen leaving behavior observed in the skn-1gf mutants (Fig. 2E, F). We next examined if SKN-1 activity in neurons, in general, was necessary for the apathy behavior observed in the skn-1gf mutant by degrading the SKN-1gf protein, specifically in the nervous system, by an auxin-inducible degradation (AID) system35. Pan-neuronal degradation of SKN-1gf was sufficient to restore pathogen-leaving behavior in the skn-1gf mutant (Fig. 2G) and had no effect on the pathogen-leaving behavior of WT animals with loss of SKN-1wt-AID in the nervous system (Supplementary Fig. S2G). Based on the necessity of SKN-1gf activity in the nervous system, but perhaps outside of the ASI neuron pair, we modified the expression of the skn-1gf isoforms pan-neuronally, but like ASI-specific expression, pan-neuronal expression also could not phenocopy the skn-1gf mutant failure to leave the PA14 food source (Supplementary Fig. S2H, I). Collectively, these data reveal that skn-1gf activity in neurons is necessary, but not sufficient, to drive apathetic leaving behavior in response to pathogens and indicate that additional determinants, beyond neuronal expression, are important.
Dynamics of nuclear localization of intestinal SKN-1 does not influence luminal distention in response to Pseudomonas infection
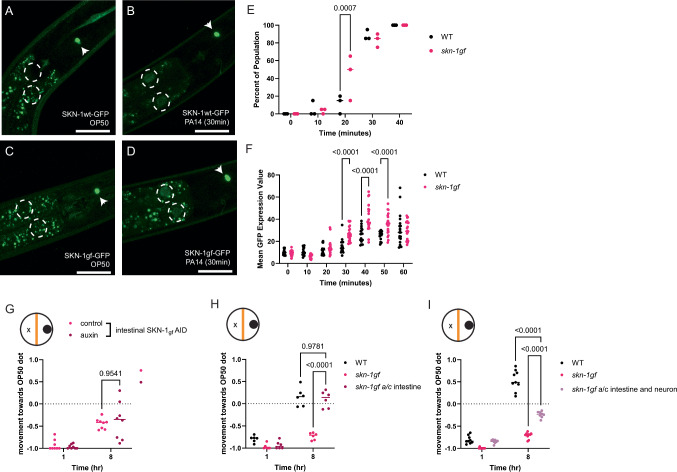
In response to environmental stressors, SKN-1 is stabilized and accumulates within the nucleus, particularly in the intestine9,32, where it can promote the transcription of cytoprotective genes that function to alleviate the stressful condition36–38. Although skn-1gf mutants have constitutive activation of the SKN-1 protein, the polypeptide is still rapidly turned over in the absence of exogenous stress32. To assess how pathogen exposure might differentially impact SKN-1 stabilization, we exposed animals harboring a CRISPR/Cas9-edited GFP at the C-terminal end of either SKN-1wt or SKN-1gf with exposure to PA14 to assess protein stabilization and nuclear localization. Outside of the constitutive expression of both SKN-1wt and SKN-1gf in the ASI neurons32, when we exposed these animals to PA14, SKN-1gf-GFP accumulated in the first two intestinal nuclei earlier than SKN-1wt-GFP (Fig. 3A–E, Supplementary Fig. S3A–F and Source Data file 2) and the abundance of nuclear-localized protein was marginally greater in SKN-1gf-GFP mutants (Fig. 3F). We next examined the loss of SKN-1 nuclear accumulation post-PA14 exposure but observed no major differences between WT and skn-1gf worms (Supplementary Fig. S3G–P and Source Data file 2). Collectively, these data suggest that the failure to leave a pathogenic environment is not a result of impaired stabilization and subcellular localization of SKN-1.
Fig. 3. Precocious activation of intestinal SKN-1 in responses to PA14.
A–D Intestinal accumulation and stabilization of SKN-1-gf-GFP and SKN-1wt-GFP upon PA14 exposure in comparison to OP50 (time point: 30 mins); the first two intestinal nuclei of each worm are outlined by white-dotted circles and the constitutive expression of SKN-1 in the ASI neurons is marked with white arrows. The scale bar represents 25 µm. Quantification of (E) percent of the population and (F) the intensity of SKN-1 nuclear localization. G Intestine-specific degradation of SKN-1gf does not restore pathogen-leaving behavior in skn-1gf mutant. H Co-expression of the a and c isoforms of SKN-1gf in the intestine does not elicit apathy behavior, as observed in the skn-1gf mutant, while (I) dual expression in both neurons and the intestine induces a modest apathy response that resembles skn-1gf mutants. For the population studies, a total of 60 (N = 3; n = 20) animals were counted per time point per condition. For quantification studies, a minimum of three animals per time point per condition was taken into consideration. Each of the nuclear accumulation time points comprised of N ≥ 3 (with n ≥ 3 per biological replicate per strain/condition) and analyzed via two-way ANOVA test; **(p < 0.01) ***(p < 0.001) ****(p < 0.0001). Each of the food choice assay comprised of N ≥ 3 (with ≥ 150 worms per biological replicate per strain/condition) and analyzed via two-way ANOVA test; **(p < 0.01) ***(p < 0.001) ****(p < 0.0001). Individual “p” value numbers for the comparisons within each graph is placed above the respective comparison bars in the graph. Source data for all nuclear activation responses are provided as a Source Data file 2. Source data for all behavioral responses are provided as a Source Data file 1.
Intestinal bloating in response to pathogen colonization in the gut is an important driver of pathogen-leaving behaviors19. With this in mind, we measured intestinal lumen distention following exposure to PA14 and discovered that bloating was unremarkable between WT and skn-1gf mutant animals (Supplementary Fig. S3Q–T). As such, the failure to leave the PA14 diet is not due to the absence of the intestinal bloating trigger, and noting that WT and skn-1gf mutant animals display normal luminal morphology in the presence of E. coli OP50 bacteria. RNAi of aex-5 or egl-8 have previously been demonstrated to induce intestinal bloating and pathogen-leaving responses in WT animals19. However, only egl-8 RNAi was capable of marginally inducing the leaving behavior of the skn-1gf mutant animals, albeit not to the level of WT animals with normal egl-8 expression and far worse than WT animals subjected to egl-8 RNAi (Supplementary Fig. S3U, V). These data confirm that the distention of the intestinal lumen is unremarkable in skn-1gf mutant animals, and the failure to leave the pathogen environment following the ingestion of PA14 is a defect in a downstream response.
We next tested if the intestinal expression of the skn-1gf allele was important for the failed pathogen-leaving behavior of skn-1gf mutant by specifically degrading SKN-1gf-AID in the intestine. Degradation of SKN-1 in the intestine had minimal capacity to restore pathogen-leaving behavior in the skn-1gf mutant (Fig. 3G) and no effect on SKN-1wt-AID animals (Supplementary Fig. S3W). Moreover, intestinal-specific expression of the individual gain-of-function variants of skn-1a (vha-6p::skn-1gf(a) or skn-1c (vha-6p::skn-1gf(c) (Supplementary Fig. S3X, Y) nor intestinal-specific co-expression of both i.e., vha-6p::skn-1gf(a&c) could elicit a pathogen apathy response like skn-1gf mutants (Fig. 3H). Intriguingly, co-expression of skn-1gf in both neurons and the intestine could elicit a modest apathy-like response, albeit not to the same magnitude as the skn-1gf mutant (Fig. 3I). Taken together, these data reveal that although the SKN-1gf protein can be stabilized more rapidly within the intestinal nuclei of PA14-exposed animals, unlike the nervous system where SKN-1gf is necessary for pathogen apathy behaviors, expression within the intestine is neither necessary nor sufficient to influence pathogen leaving behavior.
Constitutive SKN-1 activation induces a unique and context-specific transcriptional signature
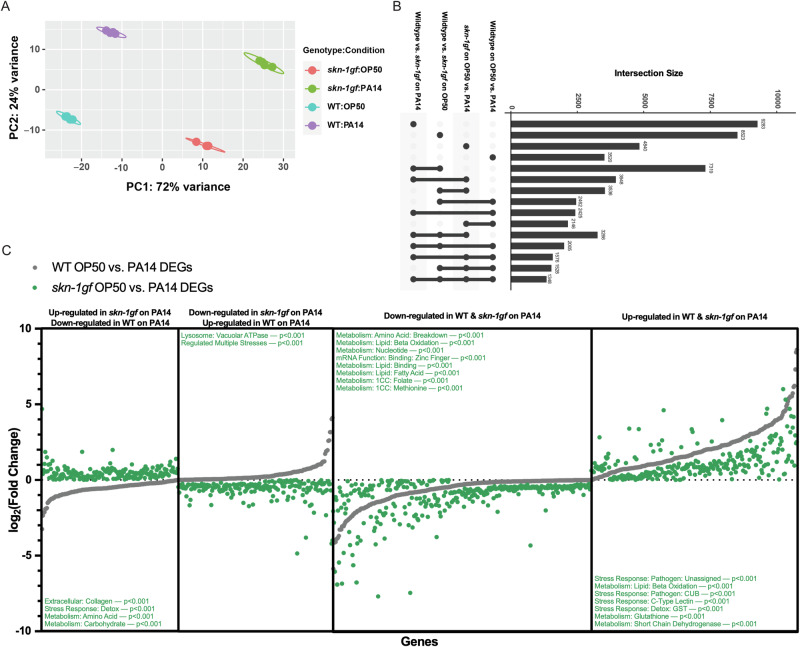
The precocious nuclear localization of SKN-1gf protein upon exposure to PA14 suggested to us that the pathogen apathy observed is a failure in responses following the initial exposure to the pathogen. As such, to elucidate the molecular basis underlying the difference in pathogen leaving between WT and skn-1gf animals, we employed RNAseq to define differential transcriptional responses to PA14 exposure in animals with and without constitutively activated SKN-1 (Supplementary Fig. S4A and Source Data file 3). We used the DESeq2 package in R to compare changes in transcriptional response across genotypes (WT or skn-1gf) and conditions (OP50 or PA14 exposure). Principal component analysis of the four sample groups (WT - OP50, WT - PA14, skn-1gf - OP50, and skn-1gf - PA14) clearly demonstrates that each sample is unique due to the lack of overlaps between groups (Fig. 4A). Pairwise analysis of the four sample groups identified a significant number of genes both down- and up-regulated, along with several specific pathways altered, most of which were associated with metabolism and stress responses (Supplementary Fig. S4A). Although distinctly different, the analysis revealed similarities when conducting pairwise comparisons and three-way comparisons, along with identifying 1348 genes that are differentially expressed across all comparisons (Fig. 4B).
Fig. 4. Context dependent transcriptional signature of PA14 exposure.
A Principal component analysis of the four sample groups (WT - OP50, WT - PA14, skn-1gf - OP50, and skn-1gf - PA14) indicates unique and context-dependent transcriptional responses. B Analyses of the various classes of genes with unique transcriptional responses revealed similarities upon pairwise comparisons and three-way comparisons, along with identifying 1348 genes that are differentially expressed across comparisons. C Genotype-specific responses to PA14 identified 363 genes with reversed directionality of transcriptional response between WT and skn-1gf animals (170 down-regulated and 193 up-regulated in WT in comparison to skn-1gf). 579 genes with similar directionality but the differential magnitude of the transcriptional response between WT and skn-1gf mutants (322 down-regulated and 257 up-regulated). RNAseq data is available at the NIH (GEO) Gene Expression Omnibus (GSE251677). Source data for all Genotype-specific transcriptional responses are provided as a Source Data file 3.
When examining genotype-specific responses to PA14 at the L4 stage, we identified 363 genes where the directionality of the transcriptional response was reversed; 193 that are up in WT but down in skn-1gf (Fig. 4C and Source Data file 3) and 170 down in WT but up in skn-1gf (Fig. 4C and Source Data file 3). Moreover, we identified 579 genes where the directionality was the same, but the magnitude of the transcriptional response was significantly different between WT and skn-1gf mutants: 322 down-regulated (Fig. 4C and Source Data file 3) and 257 up-regulated (Fig. 4C and Source Data file 3). Intriguingly, gene set enrichment analysis39 revealed reproductive systems, the pharynx, and socket cells, which make the pore where sensory dendrites extend into the cuticle and exterior, were over-represented in the context-dependent transcriptional response (Supplementary Fig. S4B). The phenotypes associated with the differentially regulated genes included intestinal function, shortened lifespan, and behavior (Supplementary Fig. S4C), and gene ontology terms that were enriched included adherens junctions, proton transport across membranes, and biosynthetic and catabolic processes (Supplementary Fig. S4D). Taken together, these data reveal the unique and context-specific transcriptional signatures of animals with constitutive SKN-1 activation when exposed to different bacterial environments.
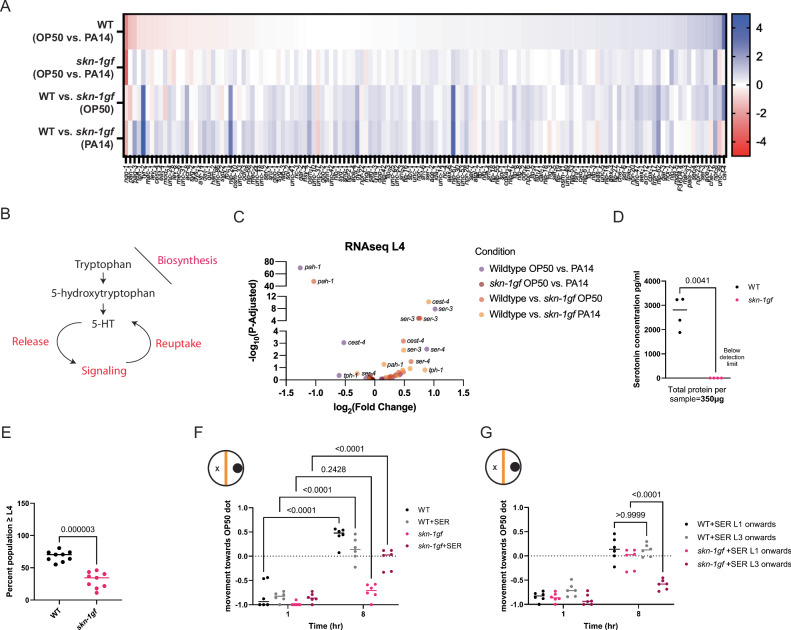
Constitutive activation of SKN-1 results in a serotonin-limiting state
The GO-term enrichment among the genes that differentially respond to PA14 based on the presence of the skn-1gf allele was intriguing, and we noted a distinct difference in the genotype-dependent response of genes with roles in neuronal function (Fig. 5A). Moreover, genes involved in serotonin signaling (Fig. 5B) were particularly sensitive to both genetic and dietary conditions (Fig. 5C and Supplementary Fig. S5A). As such, we measured whole animal serotonin levels by ELISA and found that serotonin levels were remarkably low in skn-1gf mutants compared to WT; in fact, serotonin was below the levels of detection in skn-1gf mutant animals (Fig. 5D). Serotonin availability is regulated at multiple levels (Fig. 5B) including reuptake, which effectively recycles serotonin previously released and as such, we tested what would happen in animals treated with fluoxetine, a selective serotonin reuptake inhibitor (SSRI) and found that skn-1gf mutants displayed enhanced sensitivity to fluoxetine, resulting in a significant delay in the development into adulthood as compared to WT animals (Fig. 5E) and this physiological readout confirms our finding that serotonin bioavailability is diminished in skn-1gf mutant animals.
Fig. 5. Constitutive SKN-1 Activation results in serotonin limitations.
A Heat map of neuronal enrichment genes differentially expressed upon PA14 exposure between WT and skn-1gf animals, in comparison to OP50. B Representation of serotonin signaling pathways, including biosynthesis, uptake, and reuptake. C Selective representation of differentially expressed genes involved in serotonin signaling between WT and skn-1gf upon PA14 and OP50 exposure. D ELISA measurement of serotonin levels in whole animal extracts from WT and skn-1gf. E skn-1gf animals display enhanced sensitivity to treatment with the selective serotonin reuptake inhibitor fluoxetine. F Serotonin supplementation alleviates the apathy response of skn-1gf animals but only with continuous treatment from the earliest larval stage (G). Each of the food choice assay comprised of N ≥ 3 (with ≥ 150 worms per biological replicate per strain/condition) and analyzed via two-way ANOVA test; **(p < 0.01) ***(p < 0.001) ****(p < 0.0001). For serotonin ELISA (N = 2; n = 2) were analyzed via a two-tailed/sided paired t test; **(p < 0.01) ***(p < 0.001) ****(p < 0.0001). Individual “p” value numbers for the comparisons within each graph are placed above the respective comparison bars in the graph. All experiments were conducted at 25 °C. RNAseq data is available at the NIH (GEO) Gene Expression Omnibus (GSE251677). Source data for all Genotype-specific transcriptional responses are provided as a Source Data file 3. Source data for all behavioral responses are provided as a Source Data file 1.
With this in mind, we next tested whether treatment with the monoamine neurotransmitter serotonin (5-hydroxytryptamine, 5-HT) could restore pathogen-leaving behaviors. Strikingly, treatment of skn-1gf animals with serotonin from the earliest larval stage (L1) significantly reversed the pathogen leaving behavior without changing the leaving behavior of WT animals, as compared to vehicle-treated controls (Fig. 5F). This result was specific to serotonin (5-HT), as treatment with 5-hydroxytryptophan (5-HTP), the rate-limiting precursor of serotonin biosynthesis (Supplementary Fig. S5B) did not reverse apathy for leaving the pathogen environment observed in mock-treated skn-1gf animals. Similarly, treatment with other neurotransmitters, including dopamine (Supplementary Fig. S5C), octopamine (Supplementary Fig. S5D), and acetylcholine (Supplementary Fig. S5E), was unable to alter pathogen-leaving behaviors, which suggests that serotonin is specifically limiting in skn-1gf mutant animals. Lastly, we note the importance of the timing of 5-HT treatment as acute treatment at the L3 larval stage onwards was unable to restore normal pathogen behavior as compared to the sustained treatment beginning at the L1 larval stage (Fig. 5G). Collectively, these data suggest that skn-1gf mutants have diminished serotonin availability that influences behavioral responses to PA14.
Pathogen apathy is a result of limited systemic serotonin bioavailability
In C. elegans, the tph-1 gene encodes a tryptophan hydroxylase that catalyzes the first and rate-limiting step of neuronal serotonin biosynthesis40,41. To further confirm that the apathy-like behavioral response to PA14 was due to a lack of serotonin and not another outcome of constitutive SKN-1 activation, we next tested tph-1lf mutant animals for changes in pathogen leaving responses. Surprisingly, tph-1 mutants displayed an unremarkable leaving response from PA14 as compared to WT (Fig. 6A). However, a recent discovery describes the phenylalanine hydroxylase, pah-1 in the enzymatic conversion of phenylalanine to serotonin in non-neuronal tissues42. Although two alleles of pah-1lf (Fig. 6B and Supplementary Fig. S5F) failed to significantly alter pathogen-leaving behaviors, we recalled that neither neuronal- nor intestinal-specific expression of skn-1gf could recapitulate the pathogen-leaving behaviors of the skn-1gf mutant. As such, we next tested a tph-1lf pah-1lf double mutant animals and found that these animals with systemic loss of serotonin biosynthesis were could phenotype the pathogen apathy behavior of the skn-1gf mutant (Fig. 6C). We confirmed by qRT-PCR that the context-dependent changes in expression of both tph-1 and pah-1 observed in our RNAseq analyses from skn-1gf mutant animals (Supplementary Fig. S5G, H), and like animals with constitutive activation of SKN-1, this failure of tph-1lf pah-1lf double mutant animals to leave a pathogenic environment is reversed with 5-HT treatment (Fig. 6D). 5-HT treatment failed to significantly alter the nuclear localization of SKN-1wt or SKN-1gf protein following PA14 exposure (Supplementary Fig. S6A–F and Source Data file 2) nor the turnover after removal from the pathogen (Supplementary Fig. S6G–L and Source Data file 2). Taken together, these data reveal the serotonin-dependent influence of pathogen apathy has contributions from both neuronal and non-neuronal tissues and that the impact of this limited serotonin bioavailability occurs after SKN-1 stabilization and nuclear localization.
Fig. 6. Pathogen apathy stems from systemic serotonin depletion.
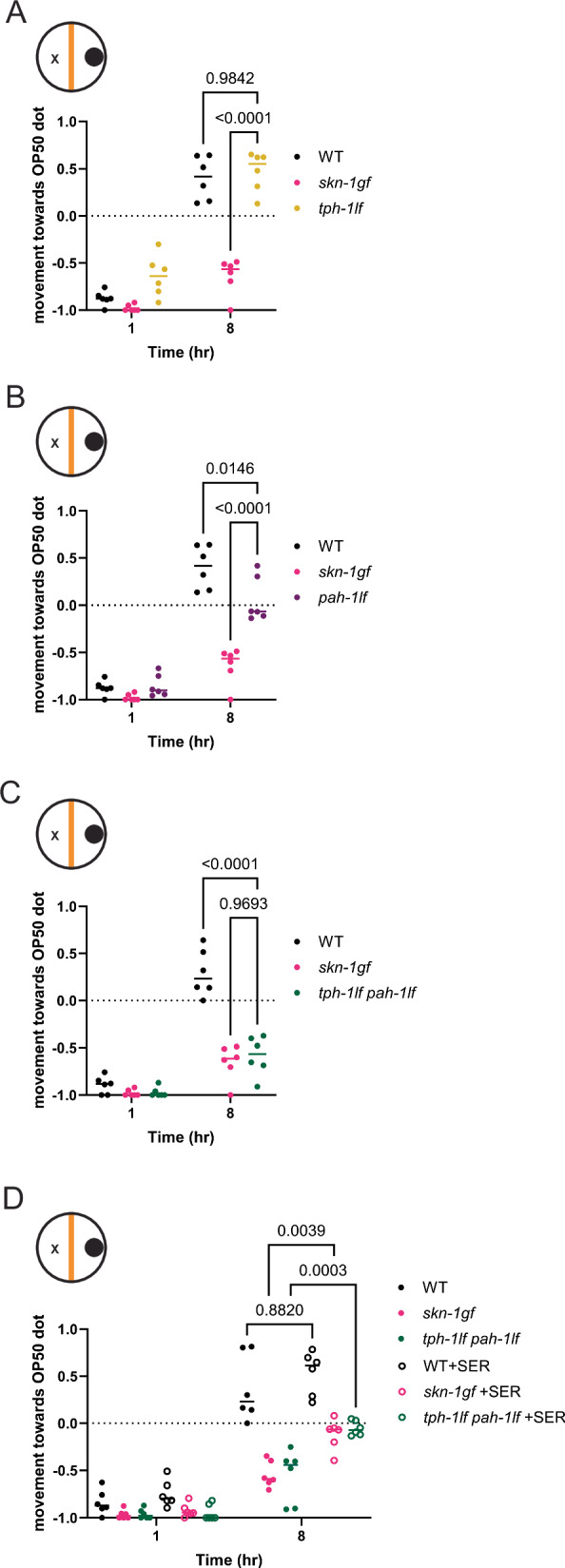
Pathogen leaving behavior of (A) tph-1lf and (B) pah-1lf animals. C tph-1lf pah-1lf double mutants display pathogen apathy like skn-1gf mutants which is alleviated with supplementation of serotonin (D). Each of the food choice assays comprised of N ≥ 3 (with ≥ 150 worms per biological replicate per strain/condition) and analyzed via two-way ANOVA test; **(p < 0.01) ***(p < 0.001) ****(p < 0.0001). Individual “p” value numbers for the comparisons within each graph is placed above the respective comparison bars in the graph. Source data for all behavioral responses are provided as a Source Data file 1.
Serotonin depletion limits pathogen resistance with SKN-1 activation
The impaired pathogen leaving behavior observed in the skn-1gf mutants is a failure in one of the earliest responses to PA14 exposure that normally occurs within one hour of encountering toxins produced by the bacteria (Fig. 1). Although serotonin signaling from the nervous system is important for host-pathogen behaviors16, a role for non-neuronal serotonin42 in response to SKN-1 activity in ASI neurons is not known. As such, we examined whether conditions that influence pathogen apathy behavior also influence pathogen resistance in the “fast kill” assay, which is a response dependent on the presence of virulence factors that also contribute to pathogen avoidance behaviors. We previously reported that skn-1gf mutants display enhanced pathogen resistance (Epr) when compared to WT animals11, and surprisingly treatment with 5-HT resulted in a further enhancement of the Epr phenotype in the skn-1gf mutant but had negligible effects on WT animals survival in the fast kill paradigm (Fig. 7A, B and Source Data file 4). We also examined the serotonin biosynthesis mutants in the fast kill assay and discovered that the tph-1lf pah-1lf double mutant animals display increased sensitivity to PA14 exposure (Fig. 7C and Source Data file 4), as compared to either single mutant, which further connects the enhanced survival phenotype of skn-1gf mutants with serotonin availability but only in the context of SKN-1 activation. These data indicate a role for both neuronal and non-neuronal serotonin signaling in host behavioral responses to pathogens and survival from pathogen infection. As such, pathogen leaving behavior and pathogen resistance are serotonin-sensitive physiological responses that are influenced by SKN-1 activity in both neuronal and non-neuronal tissues.
Fig. 7. Serotonin depletion limits pathogen resistance.
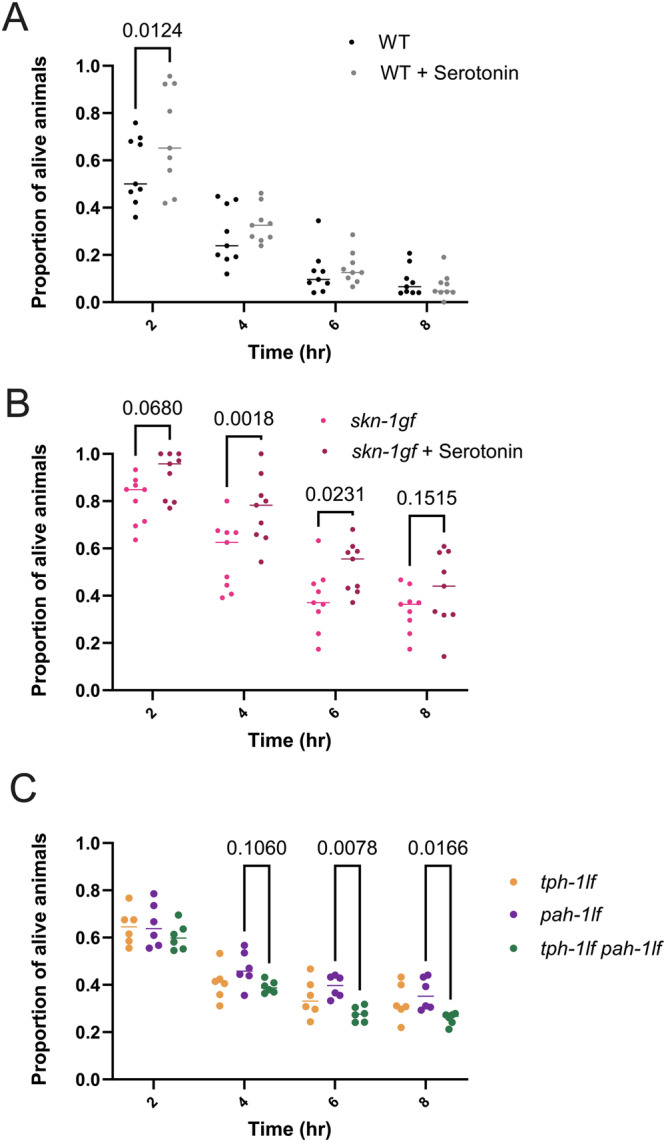
Survival analysis of (A) WT and (B) skn-1gf animals with and without serotonin supplementation on PA14 fast kill assay plates. C Survival analysis of tph-1lf, pah-1lf, and tph-1lf pah-1lf on PA14-seeded fast kill assay plates. Each of the fast kill assays comprised N ≥ 3 (with ≥ 150 worms per biological replicate per strain/condition) and analyzed via two-way ANOVA test; **(p < 0.01) ***(p < 0.001) ****(p < 0.0001). Individual “p” value numbers for the comparisons within each graph are placed above the respective comparison bars in the graph. Source data for pathogen survival data are provided as a Source Data file 4.
Taken together, these data reveal a consequence of constitutive SKN-1 activation is the depletion of available serotonin that leads to a defect in pathogen leaving behavior. This phenotype is critical for survival in response to exposure to a bacterial pathogen and connects SKN-1 functions and serotonin signaling in cytoprotective defenses to animal decisions on food-dependent behaviors like whether to forage, avoid, or dwell (Fig. 8). Collectively, our study enhances our understanding of why the inability to turnoff of cytoprotective pathways results in pleiotropic consequences that are relevant to health across the lifespan.
Fig. 8. Model of pathogen apathy responses influenced by SKN-1 activity and serotonin.
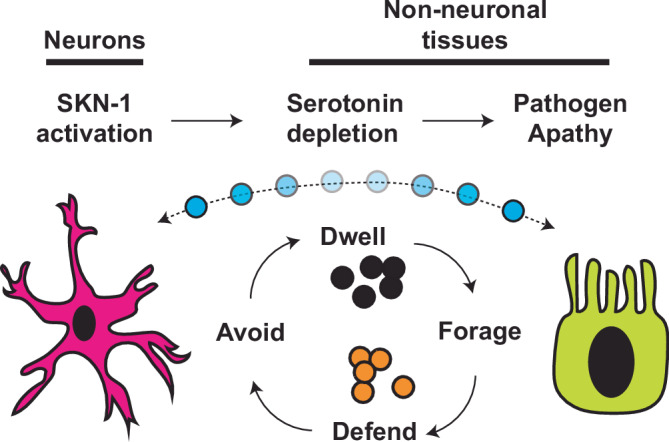
SKN-1 activation results in limited serotonin bioavailability that drives pathogen apathy; a complex phenotype influenced by animal behavioral responses to microorganisms and regulated by both neurons and non-neuronal tissues.
Discussion
Host-pathogen interactions are complex and an animal’s ability to sense and respond to potentially toxic components in the diet is essential for long-term health, and often, survival. In their natural environment, C. elegans are exposed to numerous bacteria, some of which are potentially pathogenic43,44. A proper immune response is required to survive the immediate toxins produced by pathogenic bacteria45, but the worm must also be able to escape and avoid the pathogen or potential pathogen colonization in the gut, which otherwise risks further long-term damage and eventual death. Our work demonstrates how SKN-1 activity is critical for the earliest behaviors post-exposure to a pathogen, and as such, have developed a model to test how the gut and brain coordinate to ensure an appropriate organism-level response is executed when an animal encounters a potentially toxic environment.
Although not fully understood, several pathogens, like PA14, stimulate attraction behaviors through the olfactory system in C. elegans, leading to a preferential association with these microbes over the standard OP50 E. coli/B diet16,22. Although skn-1gf mutants behave similarly to WT animals in their initial choice of PA14 over OP50, they fail to evoke normal pathogen-leaving behaviors, thus appearing apathetic to the pathogenic environment. Two prominent models of leaving behavior link a sophisticated coordination of gut-brain signaling initiated from sensory neurons17,46 or intestinal bloating caused by pathogen infection that sends cues to the nervous system to drive pathogen avoidance19. Our endogenous SKN-1-GFP reporters confirm the steady-state presence of SKN-1 exclusively in ASI neurons32, which is an essential neuron that is required for proper transgenerational inheritance of pathogen avoidance47, however, expression of skn-1gf only in ASI was not sufficient to influence pathogen leaving behavior, indicating the necessity of other neurons to elicit this response. This idea is supported by specifically degrading activated SKN-1 throughout the nervous system to restore pathogen-leaving behaviors. Previous expression profiling reveals that the endogenous expression of skn-1 is lower in each of the tissues and cell types examined as compared to the expression induced by the intestinal-specific and neuronal-specific drivers utilized48–51, and as such, that lack of sufficiency to drive pathogen apathy when skn-1gf is specifically expressed in these isolated compartments is unlikely due to transcription. Moreover, our discovery of the precocious nuclear accumulation of SKN-1 within intestinal nuclei, upon exposure to PA14, further connects the brain-gut axis model of SKN-1 activity to pathogen-related behaviors and survival however, the lack of sufficiency or necessity of skn-1gf expression in the intestine to induce apathy, suggests a correlation rather than causation between this behavior and SKN-1 nuclear dynamics.
Our first evidence that constitutive SKN-1 activation resulted in a differential response to pathogens, as compared to animals that maintain the capacity to turn SKN-1 off, was in the distinct and context-dependent transcriptional response to PA14 which included changes in multiple serotonin signaling pathway genes. 5-HT can potently stimulate fatty acid B-oxidation, which results in fat loss in the intestine, and animals with constitutive activation of SKN-1 deplete intestinal lipids in an age-dependent manner11,52. Our discovery that animals lacking serotonin biosynthesis through TPH-1 and PAH-1 also display pathogen apathy provides additional dimensions to previous work that demonstrates how serotonin signaling can suppress the innate immune response, limit the rate of pathogen clearance, and modulate the immune system in response to changes in external stimuli53. SKN-1 is a key regulator of environmental stress and constitutive activation by multiple mechanisms, including activation by RNAi of wdr-23 in WT (Fig. 1F) and in animals lacking serotonin biosynthesis (Supplementary Fig. S5I) display a clear apathy response. Our finding that serotonin supplementation, in the context of constitutively active SKN-1, results in enhanced pathogen resistance is significant because it indicates that the loss of available serotonin resulting from activated SKN-1 limits organismal capacity to fight off infection, but perhaps more importantly, it suggests that the overall impact of SKN-1/NRF2 cytoprotection can be realized with pharmacological interventions.
Our finding that the apathy behaviors observed in the skn-1gf mutants can be reversed by sustained treatment with serotonin is intriguing as serotonin signaling is associated with food responses across animals, but also apathy behaviors in humans54,55. The apathy observed in skn-1gf mutants is phenocopied only when serotonin synthesis is disrupted in both neuronal and non-neuronal cell types, suggesting that at least for basal levels of activity, a certain degree of compensation can occur. In many patients taking selective serotonin reuptake inhibitors (SSRIs), a common treatment for depression, outcomes associated with loss of motivation, energy, and lack of curiosity (collectively referred to as apathy) are commonly experienced55. Our discovery reveals that skn-1gf mutant animals exhibit a synthetic interaction with serotonin depletion by SSRI treatment that results in developmental arrest and mimics the worsened outcomes of individuals starting or increasing SSRI treatment, especially if they have apathy - which is often misdiagnosed for depression56. Our work suggests that NRF2 activation could be a facile biomarker to measure when considering treatments that manipulate serotonin signaling.
Nuclear factor erythroid 2-related factor 2 (Nrf2), the human homolog of SKN-1, is known for its pivotal role in inducing antioxidant stress and regulating inflammatory responses. It plays a crucial role in the progression of intestinal fibrosis and carcinogenesis associated with inflammatory bowel diseases (IBD)57. The inflammatory response exhibited upon the onset of Ulcerative colitis (a form of IBD) leads to the dysregulation of the immune system, and constitutive activation of Nrf2 has been demonstrated to exacerbate cases of acute colitis to carcinoma57,58. Serotonin synthesized mainly in the GI tract is involved in a vast array of biological processes revolving around the gut-brain axis. It also plays a role in regulating inflammation, as increased inflammation in IBD lowers serotonin reuptake59. Further, apathy is associated with polymorphisms in serotonin uptake transporter genes that affect their functionality60,61, and serotonin reuptake inhibitor (SSRI) treatments offered to patients with depression often display higher cases of apathy62. As such, physiological responses revealed in this study of C. elegans could inform and model human disease states at the intersection of diet, behaviors, and immunity axes.
Methods
C. elegans strains and maintenance
All worms were grown at 20 °C on 6 cm Nematode Growth Medium (NGM) agar plates with streptomycin and seeded with 250 μL E. coli strain OP50-1 unless otherwise noted. The following strains were used: WT – N2 Bristol (reference strain), MT15434 - tph-1(mg280) II, MT7988 - bas-1(ad446) III, PS8116 - C17H12.4(sy1192) IV, LC74 - pah-1(ok687) II, PHX3596 - tph-1(mg280) pah-1(syb3596) II, PHX3601 - pah-1(syb3601) II, CB1370 - daf-2(e1370), DA465 - eat-2(ad465)II, VC1772 - skn-1(ok2315) IV/nT1 [qIs51] (IV;V) were ordered from CGC (Caenorhabditis Genetics Center). SPC227 - skn-1gf(lax188), SPC2002 - skn-1wt-GFP, SPC2003 - skn-1gf-GFP, SPC602 - skn-1wt-AID; rgef-1p::TIR1, SPC603 - skn-1wt-AID; ges-1p::TIR1, SPC2048 - skn-1gf-AID; rgef-1p::TIR1, SPC2047 - skn-1gf-AID; ges-1p::TIR1, SPC2065 - rgef-1p::skn-1gf-isoformA, SPC2064 - rgef-1p::skn-1gf-isoformC, SPC2058 - vha-6p::skn-1gf-isoformA, SPC2057 - vha-6p::skn-1gf-isoformC, SPC2108 - vha-6p::skn-1gf-isoformA;vha-6p::skn-1gf-isoformC, SPC2068 - gpa-4p::skn-1gf-isoformA, SPC2067 - gpa-4p::skn-1gf-isoformC were ordered from SunyBiotech.
For all experiments, eggs were collected from day 1 adult animals by hypochlorite treatment and allowed to hatch overnight for L1 synchronization. For testing on day 3 adults, worms had to be washed to new plates every day beginning at day 1 of adulthood, to separate worms from progeny. M9 + 0.01% Triton X-100 (M9T) was used to wash the worms from the plate into microcentrifuge tubes. Worms were allowed to gravity settle and the supernatant containing the progeny was removed without disturbing the adult pellet. This was repeated three times, and worms were dropped onto fresh NGM plates.
Bacterial strain and maintenance
E. coli, strains OP50-1 and HT115 and P. aeruginosa, strain PA14 and PA14ΔgacA were used. All strains were streaked onto LB agar plates, with different antibiotic conditions for each strain. OP50 was streaked on plates with streptomycin, HT115 on plates with ampicillin, PA14, and PA14ΔgacA on plates without antibiotics. OP50, which was used for preference plates, was streaked on LB plates without antibiotics to match PA14. Following streaking, plates were grown overnight at 37 °C and either used the next day or stored at 4 °C for no longer than a week.
RNAi experiments
RNAi treatment was performed as previously mentioned63. Briefly, E. coli HT115 bacteria carrying specific double-stranded RNA-expression plasmids were seeded on NGM plates containing 5 mM isopropyl-b-D-thiogalactoside (IPTG) and 50 mg ml-1 carbenicillin. RNAi were induced at room temperature for 24 h. Synchronized L1 animals were added onto RNAi plates to knock down indicated genes. L4 animals were subjected to food choice assays, and the choice index was measured.
Fast Kill Assays
Fast Kill Assay was performed in 3.5 cm peptone glucose media plates (1% Bacto-Peptone; 1% NaCl; 1% Glucose; 1.7% Bacto-Agar) containing 0.15 M sorbitol. PA14 cultures were grown overnight at 37 °C for 14-15 h. 5ul of the overnight culture was spread over the PGM + sorbitol plates using a spreader made from an open loop-tipped glass pasture pipette. The plates were incubated at 37 °C for 24 h and then at 25 °C overnight. 30–40 L4 animals were placed on each plate. The assay was performed at 25 °C. Survival of animals was plotted over a period of 8 hrs with intervals of 2 hrs (0, 2, 4, 6, 8 hr)64,65.
Behavioral analyses
Two-choice preference assay
Preference plates were made by using the same NGM plates for growth, except 0.25% peptone is replaced by 0.35% peptone, and no antibiotics are used. The night before seeding, a single colony from each bacteria that is to be used in the preference assay was picked into 3 mL of LB and grown in a 37 °C shaker overnight. The following day, the OD600 was measured by diluting 100 μL of bacteria with 900 μL of LB and multiplying the OD600 readout by ten. The bacteria were then diluted to an OD600 of 1.0. Two spots were marked equidistant from the center, yet not too close to the edge, and 15 μL of diluted bacteria were placed on the marked spots. Plates were allowed to dry and then transferred to the 37 °C for 24 h, followed by the 25 °C incubator for 48 h unless otherwise noted.
On the day of the assay, the plates were removed from the 25 °C incubator to reach room temperature. Worms were collected from NGM growth plates in the M9 buffer and further subjected to three washes in the M9T buffer. Finally, the worms were placed onto the assay plate and recorded.
Forced choice assay
Plates were made of a similar composition as the two-choice preference assay plates. A single colony from each Pseudomonas aeruginosa and E. coli bacteria were inoculated into 3 mL of LB for overnight primary culture at 37 °C. The OD600 of each of the primary cultures was measured, following which the cultures were diluted to an OD600 of 1.0. A drop of 15 μL bacterial culture was seeded onto NGM plates (without streptomycin) equidistant from the center to the point where the animals (worms) were being dropped (not too close to the edge). The complimentary bacterial culture was stamped on the plates diametrically using an “L” tipped glass pasture pipette. The plates were dried and transferred to the 37 °C for 24 h, followed by the 25 °C incubation for 48 h, unless noted otherwise.
On the day of the assay, the plates were removed from the 25 °C incubator to reach room temperature. Worms were collected from NGM growth plates in the M9 buffer and further subjected to three washes in the M9T buffer. Finally, the worms were placed onto the assay plate and recorded.
Chemoattraction and chemorepulsion
Chemotaxis assays were performed as previously described66. Briefly, the underside of 5 cm unseeded NGM plates was divided into 4 equal quadrants. A circle of 0.5 cm radius was marked around the center. Diagonally opposite quadrants were marked either for the test odorant or as the vector control, ethanol. The sites where the odorants or Ethanol were to be spotted were equidistant from the center and each other. Equal volumes of the test odorant and 0.5 M azide or control and 0.5 M azide were mixed to make working solutions. Young adult worms were washed and resuspended in 100 μl of M9 buffer. 50–100 worms were added to the center of the plate. Immediately after, 2 μl of the test solution and control solution were added to the respective sites in each quadrant. Once the odorant drops were absorbed in the agar, the lids were placed, and the plates were inverted. After 60 min, the assay plates were placed at 4 °C. Only the assay plates required to be assessed and counted were removed from 4 °C. The number of worms in each quadrant that crossed the center was recorded. For each odorant set, replicates were in an order of (n = 3; N = 3). The chemotaxis index was measured as follows: Chemotaxis Index = (# Worms in Both Test Quadrants - # Worms in Both Control Quadrants) / (Total # of Scored Worms). + 1.0 score indicated maximal attraction towards the compound, and − 1 indicated maximal repulsion.
Pharmacological treatments
Serotonin hydrochloride: 5-HT (Sigma, H9523) powder was dissolved in MilliQ to a working stock concentration of 0.1 M and added to NGM plates seeded with E. coli OP50 to a final plate concentration of 5 mM67. Plates were allowed to equilibrate, and synchronized L1-stage animals were added onto 5-HT-treated plates. At the L4 stage, worms were then subjected to the indicated food choice assays and physiological assays like fast kill and subjected to the visualization of pathogen-mediated localization of SKN-1.
Dopamine Hydrochloride: DOPA (Sigma, H8502) powder was dissolved in MilliQ to a working stock concentration of 1 M and added to NGM plates seeded with E. coli OP50 to a final plate concentration of 10 mM and 20 mM68. Plates were allowed to equilibrate, and synchronized L1-stage animals were added to dopamine-treated plates. At the L4 stage, worms were then subjected to the indicated food choice assays.
5-hydroxytryptophan; 5-HTP (Sigma, H9772) powder was dissolved into a working stock concentration of 0.1 M and added to NGM plates seeded with E. coli OP50 to a final plate concentration of 5 mM69. The plates were allowed to equilibrate, and synchronized L1-stage animals were added onto 5-HTP-treated plates. At the L4 stage, worms were then subjected to the indicated food choice assays.
Octopamine Hydrochloride; OCT (Sigma, O0250) powder was dissolved in MilliQ and added to NGM plates to make the final concentrations of 10 mM and 5 mM before plates were seeded with E. coli OP5070. Synchronized L1-stage animals were added onto Octopamine-treated plates. At the L4 stage, worms were then subjected to the indicated food choice assays.
Acetylcholine chloride: AcC (Sigma, PHR1546) powder was dissolved in MilliQ and added to NGM plates to make the final concentrations of 5 mM and 15 mM prior to plates being seeded with E. coli OP50. Synchronized L1-stage animals were added onto acetylcholine-treated plates. At the L4 stage, worms were then subjected to the indicated food choice assays.
Fluoxetine Hydrochloride (Sigma, PHR1394) powder was dissolved in MilliQ and added to NGM plates seeded with E. coli OP50 to make the final concentrations of 145 µM (0.5 mg/ml)71. Synchronized L4-stage animals were added to Fluoxetine-treated plates to assess survival percentages.
Crawling assays
Worms were egg-prepped, and eggs were allowed to hatch overnight for a synchronous L1 population. The next day, worms were dropped onto plates seeded with OP50. Worms were then allowed to grow until each time point (48 h post-drop for L4s). Once worms were at the required stage of development, 30–50 worms were washed off a plate in 50 uL of M9 with a cut and M9 + triton-coated P1000 tip and dropped onto PA14 plates (as described above).
Pseudomonas (PA14) was prepared with an overnight culture of LB incubating at 37 °C. A 15 µL drop of PA14 was placed on one end of a standard NGM plate with no streptomycin. Plates were incubated at 37 °C for 24 h, followed by 25 °C for 48 h. L4 worms were placed directly opposite from the bacteria culture. The M9 was allowed to dissipate, and the time to reach the PA14 food was recorded. Crawling was imaged with the MBF Bioscience WormLab microscope and analysis was performed with WormLab version 2022. Worm crawling on the plate was imaged for 1 min for each condition at frame rate 7.5 ms. Values were analyzed under the “Speed” tab. Mean speed values of each individual animal captured the overall movement speed for the time interval. Worm crawling was analyzed with the software, and only worms that moved for at least 90% of the time were included in the analysis. Irregular speed values were discarded from the dataset using normalization to the overall average of the speed values. Statistical analysis of crawling speed was done via one-way ANOVA with multiple t test.
Serotonin ELISA
The competitive serotonin ELISA kit was adapted from the Abcam protocol (ab133053). Samples comprising ~ 20000 worms synchronized to L4 development stage was prepared using fast freezing facilitated by liquid nitrogen and FA lysis buffer [EDTA (1 mM, pH 8.0), HEPES-KOH (50 mM, pH 7.5) NaCl (140 mM) with protease inhibitors (100X), diluted to 1X final concentration, Sodium deoxycholate (0.1%, w/v), Triton X-100 (1%, v/v)]. Frozen samples were sonicated (30 sec ON and 30 sec OFF, 30 cycles) in a water bath-based sonicator (Diagenode, USA). The protein concentration in the supernatant was estimated by using a Bradford reagent prior to starting the assay.
RNAseq and analyses
RNAseq analysis was conducted as outlined44,72. Worms were egg-prepped, and eggs were allowed to hatch overnight for a synchronous L1 population. The next day, L1s were dropped onto seeded NGM plates and allowed to grow 48 h (L4 stage), then exposed to PA14 before collection. Animals were washed 3 times with M9 buffer and frozen in TRI reagent at − 80 °C until use. Animals were homogenized, and RNA extraction was performed via the Zymo Direct-zol RNA Miniprep kit (Cat. #R2052). Qubit™ RNA BR Assay Kit was used to determine RNA concentration. The RNA samples were sequenced and read counts were reported by Novogene. Read counts were then used for differential expression (DE) analysis using the R package DESeq2 created using R version 3.5.2. Statistically significant genes were chosen based on the adjusted p-values that were calculated with the DESeq2 package (p < 0.01). Gene Ontology was analyzed using the most recent version of WormCat 2.073.
QRT-PCR analysis
Approximately 1 μg of RNA was converted to cDNA with the help of Superscript
III Reverse Transcriptase enzyme and poly-T primers (Invitrogen, USA). To determine
the relative gene expression levels, QRT-PCR analysis was performed using the 2X
Quanta PerfeCTa® SYBR® FastMix® and BioRad Real-time PCR System (.C1000 Touch Thermal Cycler; CFX96 Real-Time System) Primers used from IDT (Integrated DNA Technologies) include:
tph-1 FP: 5’ CGTGACAGTCAAGATGGAAGT 3’
tph-1 RP: 5’ CTCATGAACATCAAGCCCATTAAG 3’
pah-1 FP: 5’ GAATTTGCTGAAGCTGAAGACC 3’
pah-1 RP: 5’ TCCAGTCTTGAACAAGAACCTT 3’
Auxin-inducible degradation
For tissue-specific degradation experiments, worms were egg-prepped and allowed to hatch overnight for a synchronous L1 population. The next day, worms were dropped onto plates with vehicle 4 mM ethanol or 4 mM auxin (Sigma Aldrich, I3750) raised to L4 stage, and then moved to experiment plates.
Imaging
Fluorescence
For confocal microscopy (SKN-1-GFP, 491 nm), imaging was performed on a Stellaris 5 confocal microscope equipped with a white light laser source and spectral filters, HyD detectors, 63x/1.4 Plan ApoChromat Oil objective, and run on LAS X 4.4.0.24861 software.
Intestinal distension
The NGM plates (without streptomycin) were seeded with an overnight culture of Pseudomonas Aeruginosa PA14 and allowed to dry. Further, the plates were incubated at 37 °C for 24 h, followed by the 25 °C incubation for 48 h. Perfect L4 staged worms (45–48 hrs post L1 starvation) were placed onto the PA14 seeded plates and observed over a period of 24 hrs. The images of intestinal distension were taken with LAS X software and Leica Thunder Imager flexacam C3 color camera (Magnification- 63X).
Statistics
All statistical analyses were performed using GraphPad Prism version 10.0. The respective figure legends provide information on the specific statistical tests used for each assay.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Source data
Acknowledgements
We thank S. Ledgerwood for technical assistance and C.M. Ramos for critical reading of the manuscript. This work was funded by the NIH R01AG058610 and Hevolution Foundation award HF AGE-004 to SPC, T32AG000037 to BAW, T32AG052374, and F31AG077873 to N.L.S. We also thank the USC School of Gerontology Imaging Core, which is funded in part by the Nathan Shock Center of Excellence P30AG068345. Some strains were provided by the CGC, which is funded by the NIH Office of Research Infrastructure Programs (P40 OD010440). We thank WormBase for database curation and data access.
Author contributions
Conceptualization: S.P.C.; Methodology: T.N., B.A.W., N.L.S., J.D.N., and S.P.C.; Investigation: T.N., B.A.W., N.L.S., J.D.N., and S.P.C.; Visualization: T.N., B.A.W., N.L.S., J.D.N., and S.P.C.; Supervision: S.P.C.; Writing (original draft): S.P.C.; Writing (reviewing & editing): T.N., B.A.W., N.L.S., and S.P.C.
Peer review
Peer review information
Nature Communications thanks Jonathan Lalsiamthara and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
All data are available in the main text or the supplementary materials. RNAseq data is available at the NIH Gene Expression Omnibus (GEO) https://www.ncbi.nlm.nih.gov/geo (GSE251677), Source data are provided with this paper.
Competing interests
All authors declare that they have no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Brandy A. Weathers, Nicole L. Stuhr.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-52233-5.
References
- 1.Zhang, J., Holdorf, A. D. & Walhout, A. J. C. elegans and its bacterial diet as a model for systems-level understanding of host-microbiota interactions. Curr. Opin. Biotechnol.46, 74–80 (2017). 10.1016/j.copbio.2017.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu, Y., Samuel, B. S., Breen, P. C. & Ruvkun, G. Caenorhabditis elegans pathways that surveil and defend mitochondria. Nature508, 406–410 (2014). 10.1038/nature13204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Head, B. P., Olaitan, A. O. & Aballay, A. Role of GATA transcription factor ELT-2 and p38 MAPK PMK-1 in recovery from acute P. aeruginosa infection in C. elegans. Virulence8, 261–274 (2017). 10.1080/21505594.2016.1222334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martineau, C. N., Kirienko, N. V. & Pujol, N. Innate immunity in C. elegans. Curr. Top. Dev. Biol.144, 309–351 (2021). 10.1016/bs.ctdb.2020.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim, D. H. et al. A conserved p38 MAP kinase pathway in Caenorhabditis elegans innate immunity. Science297, 623–626 (2002). 10.1126/science.1073759 [DOI] [PubMed] [Google Scholar]
- 6.Garsin, D. A. et al. Long-lived C. elegans daf-2 mutants are resistant to bacterial pathogens. Science300, 1921 (2003). 10.1126/science.1080147 [DOI] [PubMed] [Google Scholar]
- 7.Troemel, E. R. et al. p38 MAPK regulates expression of immune response genes and contributes to longevity in C. elegans. PLoS Genet.2, e183 (2006). 10.1371/journal.pgen.0020183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.An, J. H. & Blackwell, T. K. SKN-1 links C. elegans mesendodermal specification to a conserved oxidative stress response. Genes Dev.17, 1882–1893 (2003). 10.1101/gad.1107803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Inoue, H. et al. The C. elegans p38 MAPK pathway regulates nuclear localization of the transcription factor SKN-1 in oxidative stress response. Genes Dev.19, 2278–2283 (2005). 10.1101/gad.1324805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li, L. et al. Glucose negatively affects Nrf2/SKN-1-mediated innate immunity in C. elegans. Aging10, 3089–3103 (2018). 10.18632/aging.101610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nhan, J. D. et al. Redirection of SKN-1 abates the negative metabolic outcomes of a perceived pathogen infection. Proc. Natl. Acad. Sci. USA116, 22322–22330 (2019). 10.1073/pnas.1909666116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoeven, R., McCallum, K. C., Cruz, M. R. & Garsin, D. A. Ce-Duox1/BLI-3 generated reactive oxygen species trigger protective SKN-1 activity via p38 MAPK signaling during infection in C. elegans. PLoS Pathog.7, e1002453 (2011). 10.1371/journal.ppat.1002453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li, W.-H., Chang, C.-H., Huang, C.-W., Wei, C.-C. & Liao, V. H.-C. Selenite enhances immune response against Pseudomonas aeruginosa PA14 via SKN-1 in Caenorhabditis elegans. PLOS ONE9, e105810 (2014). 10.1371/journal.pone.0105810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Papp, D., Csermely, P. & Sőti, C. A role for SKN-1/Nrf in pathogen resistance and immunosenescence in Caenorhabditis elegans. PloS Pathog.8, e1002673 (2012). [DOI] [PMC free article] [PubMed]
- 15.Ramos, C. M. & Curran, S. P. Comparative analysis of the molecular and physiological consequences of constitutive SKN-1 activation. Geroscience45, 3359–3370 (2023). 10.1007/s11357-023-00937-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang, Y., Lu, H. & Bargmann, C. I. Pathogenic bacteria induce aversive olfactory learning in Caenorhabditis elegans. Nature438, 179–184 (2005). 10.1038/nature04216 [DOI] [PubMed] [Google Scholar]
- 17.Meisel, J. D. & Kim, D. H. Behavioral avoidance of pathogenic bacteria by Caenorhabditis elegans. Trends Immunol.35, 465–470 (2014). 10.1016/j.it.2014.08.008 [DOI] [PubMed] [Google Scholar]
- 18.Singh, J. & Aballay, A. Neural control of behavioral and molecular defenses in C. elegans. Curr. Opin. Neurobiol.62, 34–40 (2020). 10.1016/j.conb.2019.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Singh, J. & Aballay, A. Intestinal infection regulates behavior and learning via neuroendocrine signaling. ELife8, e50033 (2019). 10.7554/eLife.50033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harris, G. et al. Dissecting the signaling mechanisms underlying recognition and preference of food odors. J. Neurosci.34, 9389–9403 (2014). 10.1523/JNEUROSCI.0012-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Styer, K. L. et al. Innate immunity in Caenorhabditis elegans is regulated by neurons expressing NPR-1/GPCR. Science322, 460–464 (2008). 10.1126/science.1163673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jin, X., Pokala, N. & Bargmann, C. I. Distinct circuits for the formation and retrieval of an imprinted olfactory memory. Cell164, 632–643 (2016). 10.1016/j.cell.2016.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ha, H.-i et al. Functional organization of a neural network for aversive olfactory learning in Caenorhabditis elegans. Neuron68, 1173–1186 (2010). 10.1016/j.neuron.2010.11.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moore, R. S., Kaletsky, R. & Murphy, C. T. Piwi/PRG-1 Argonaute and TGF-beta mediate transgenerational learned pathogenic avoidance. Cell177, 1827–1841 (2019). 10.1016/j.cell.2019.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen, Z. et al. Two insulin-like peptides antagonistically regulate aversive olfactory learning in C. elegans. Neuron77, 572–585 (2013). 10.1016/j.neuron.2012.11.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dag, U. et al. Dissecting the functional organization of the C. elegans serotonergic system at whole-brain scale. Cell186, 2574–2592 (2023). 10.1016/j.cell.2023.04.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Srinivasan, S. et al. Serotonin regulates C. elegans fat and feeding through independent molecular mechanisms. Cell Metab.7, 533–544 (2008). 10.1016/j.cmet.2008.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tan, M. W., Mahajan-Miklos, S. & Ausubel, F. M. Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proc. Natl. Acad. Sci. USA96, 715–720 (1999). 10.1073/pnas.96.2.715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parkins, M. D., Ceri, H. & Storey, D. G. Pseudomonas aeruginosa GacA, a factor in multihost virulence, is also essential for biofilm formation. Mol. Microbiol.40, 1215–1226 (2001). 10.1046/j.1365-2958.2001.02469.x [DOI] [PubMed] [Google Scholar]
- 30.Lo, J. Y., Spatola, B. N. & Curran, S. P. WDR23 regulates NRF2 independently of KEAP1. PLoS Genet.13, e1006762 (2017). 10.1371/journal.pgen.1006762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spatola, B. N., Lo, J. Y., Wang, B. & Curran, S. P. Nuclear and cytoplasmic WDR-23 isoforms mediate differential effects on GEN-1 and SKN-1 substrates. Sci. Rep.9, 11783 (2019). 10.1038/s41598-019-48286-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Turner, C. D., Stuhr, N. L., Ramos, C. M., Van Camp, B. T. & Curran, S. P. A dicer-related helicase opposes the age-related pathology from SKN-1 activation in ASI neurons. Proc. Natl. Acad. Sci. USA120, e2308565120 (2023) [DOI] [PMC free article] [PubMed]
- 33.Filipowicz, A., Lalsiamthara, J. & Aballay, A. TRPM channels mediate learned pathogen avoidance following intestinal distention. Elife10, 10.7554/eLife.65935 (2021). [DOI] [PMC free article] [PubMed]
- 34.Kaletsky, R. et al. C. elegans interprets bacterial non-coding RNAs to learn pathogenic avoidance. Nature586, 445–451 (2020). 10.1038/s41586-020-2699-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang, L., Ward, J. D., Cheng, Z. & Dernburg, A. F. The auxin-inducible degradation (AID) system enables versatile conditional protein depletion in C. elegans. Development142, 4374–4384 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ewe, C. K., Alok, G. & Rothman, J. H. Stressful development: integrating endoderm development, stress, and longevity. Dev. Biol.471, 34–48 (2021). 10.1016/j.ydbio.2020.12.002 [DOI] [PubMed] [Google Scholar]
- 37.Fukushige, T., Smith, H. E., Miwa, J., Krause, M. W. & Hanover, J. A. A genetic analysis of the. Genetics206, 939–952 (2017). 10.1534/genetics.117.202515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oliveira, R. P. et al. Condition-adapted stress and longevity gene regulation by Caenorhabditis elegans SKN-1/Nrf. Aging Cell8, 524–541 (2009). 10.1111/j.1474-9726.2009.00501.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Angeles-Albores, D., Lee, R., Chan, J. & Sternberg, P. Two new functions in the WormBase Enrichment Suite. MicroPubl. Biol.10.17912/W25Q2N (2018). [DOI] [PMC free article] [PubMed]
- 40.Sze, J. Y., Victor, M., Loer, C., Shi, Y. & Ruvkun, G. Food and metabolic signalling defects in a Caenorhabditis elegans serotonin-synthesis mutant. Nature403, 560–564 (2000). 10.1038/35000609 [DOI] [PubMed] [Google Scholar]
- 41.Sze, J. Y., Zhang, S., Li, J. & Ruvkun, G. The C. elegans POU-domain transcription factor UNC-86 regulates the tph-1 tryptophan hydroxylase gene and neurite outgrowth in specific serotonergic neurons. Development129, 3901–3911 (2002). 10.1242/dev.129.16.3901 [DOI] [PubMed] [Google Scholar]
- 42.Yu, J. et al. Parallel pathways for serotonin biosynthesis and metabolism in C. elegans. Nat. Chem. Biol.19, 141–150 (2023). 10.1038/s41589-022-01148-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Couillault, C. & Ewbank, J. J. Diverse bacteria are pathogens of Caenorhabditis elegans. Infect. Immun.70, 4705–4707 (2002). 10.1128/IAI.70.8.4705-4707.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stuhr, N. L. & Curran, S. P. Bacterial diets differentially alter lifespan and healthspan trajectories in C. elegans. Commun. Biol.3, 653 (2020). 10.1038/s42003-020-01379-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Irazoqui, J. E. et al. Distinct pathogenesis and host responses during infection of C. elegans by P. aeruginosa and S. aureus. PLoS Pathog.6, e1000982 (2010). 10.1371/journal.ppat.1000982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meisel, J. D., Panda, O., Mahanti, P., Schroeder, F. C. & Kim, D. H. Chemosensation of bacterial secondary metabolites modulates neuroendocrine signaling and behavior of C. elegans. Cell159, 267–280 (2014). 10.1016/j.cell.2014.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moore, R. S. et al. The role of the Cer1 transposon in horizontal transfer of transgenerational memory. Cell184, 4697–4712 (2021). 10.1016/j.cell.2021.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wormbase. http://www.wormbase.orgWS293 (2024).
- 49.Hammarlund, M., Hobert, O., Miller, D. M. 3rd & Sestan, N. The CeNGEN Project: The complete gene expression map of an entire nervous system. Neuron99, 430–433 (2018). 10.1016/j.neuron.2018.07.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mundade, R., Ozer, H. G., Wei, H., Prabhu, L. & Lu, T. Role of ChIP-seq in the discovery of transcription factor binding sites, differential gene regulation mechanism, epigenetic marks and beyond. Cell Cycle13, 2847–2852 (2014). 10.4161/15384101.2014.949201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gerstein, M. B. et al. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science330, 1775–1787 (2010). 10.1126/science.1196914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lynn, D. A. et al. Omega-3 and −6 fatty acids allocate somatic and germline lipids to ensure fitness during nutrient and oxidative stress in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA112, 15378–15383 (2015). 10.1073/pnas.1514012112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Anderson, A., Laurenson-Schafer, H., Partridge, F. A., Hodgkin, J. & McMullan, R. Serotonergic chemosensory neurons modify the C. elegans immune response by regulating G-protein signaling in epithelial cells. PLoS Pathog.9, e1003787 (2013). 10.1371/journal.ppat.1003787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Devanand, D. P. Management of neuropsychiatric symptoms in dementia. Curr. Opin. Neurol.36, 498–503 (2023). 10.1097/WCO.0000000000001199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Padala, P. R. et al. Selective serotonin reuptake inhibitors-associated apathy syndrome: A cross sectional study. Med. Baltim.99, e21497 (2020). 10.1097/MD.0000000000021497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Klionsky, D. J. et al. Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition)(1). Autophagy17, 1–382 (2021). 10.1080/15548627.2020.1797280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Peng, S. et al. The role of Nrf2 in the pathogenesis and treatment of ulcerative colitis. Front. Immunol.14, 1200111 (2023). 10.3389/fimmu.2023.1200111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Geertsema, S. et al. The NRF2/Keap1 pathway as a therapeutic target in inflammatory bowel disease. Trends Mol. Med.29, 830–842 (2023). 10.1016/j.molmed.2023.07.008 [DOI] [PubMed] [Google Scholar]
- 59.Jørandli, J. W. et al. The serotonin reuptake transporter is reduced in the epithelium of active Crohn’s disease and ulcerative colitis. Am. J. Physiol. Gastrointest. Liver Physiol.319, G761–g768 (2020). 10.1152/ajpgi.00244.2020 [DOI] [PubMed] [Google Scholar]
- 60.Tang, L. et al. The Association between 5HT2A T102C and behavioral and psychological symptoms of dementia in Alzheimer’s disease: A meta-analysis. Biomed. Res. Int.2017, 5320135 (2017). 10.1155/2017/5320135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chakraborty, S. et al. Serotonergic system, cognition, and BPSD in Alzheimer’s disease. Neurosci. Lett.704, 36–44 (2019). 10.1016/j.neulet.2019.03.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wongpakaran, N., van Reekum, R., Wongpakaran, T. & Clarke, D. Selective serotonin reuptake inhibitor use associates with apathy among depressed elderly: a case-control study. Ann. Gen. Psychiatry6, 7 (2007). 10.1186/1744-859X-6-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dalton, H. M. & Curran, S. P. Hypodermal responses to protein synthesis inhibition induce systemic developmental arrest and AMPK-dependent survival in Caenorhabditis elegans. PLOS Genet.14, e1007520 (2018). 10.1371/journal.pgen.1007520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mahajan-Miklos, S., Tan, M. W., Rahme, L. G. & Ausubel, F. M. Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa-Caenorhabditis elegans pathogenesis model. Cell96, 47–56 (1999). 10.1016/S0092-8674(00)80958-7 [DOI] [PubMed] [Google Scholar]
- 65.Cezairliyan, B. et al. Identification of Pseudomonas aeruginosa phenazines that kill Caenorhabditis elegans. PLoS Pathog.9, e1003101 (2013). 10.1371/journal.ppat.1003101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Matsuura, T., Izumi, J., Hioki, M., Nagaya, H. & Kobayashi, Y. Sensory interaction between attractant diacetyl and repellent 2-nonanone in the nematode Caenorhabditis elegans. J. Exp. Zool. A Ecol. Genet Physiol.319, 285–295 (2013). 10.1002/jez.1795 [DOI] [PubMed] [Google Scholar]
- 67.Sawin, E. R., Ranganathan, R. & Horvitz, H. R. C. elegans locomotory rate is modulated by the environment through a dopaminergic pathway and by experience through a serotonergic pathway. Neuron26, 619–631 (2000). 10.1016/S0896-6273(00)81199-X [DOI] [PubMed] [Google Scholar]
- 68.Barros, A. G. et al. Dopamine signaling regulates fat content through β-oxidation in Caenorhabditis elegans. PLoS ONE9, e85874 (2014). 10.1371/journal.pone.0085874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jafari, G., Xie, Y., Kullyev, A., Liang, B. & Sze, J. Y. Regulation of extrasynaptic 5-HT by serotonin reuptake transporter function in 5-HT-absorbing neurons underscores adaptation behavior in Caenorhabditis elegans. J. Neurosci.31, 8948–8957 (2011). 10.1523/JNEUROSCI.1692-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.O’Donnell, M. P., Fox, B. W., Chao, P. H., Schroeder, F. C. & Sengupta, P. A neurotransmitter produced by gut bacteria modulates host sensory behaviour. Nature583, 415–420 (2020). 10.1038/s41586-020-2395-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kullyev, A. et al. A genetic survey of fluoxetine action on synaptic transmission in Caenorhabditis elegans. Genetics186, 929–941 (2010). 10.1534/genetics.110.118877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cedillo, L. et al. Ether lipid biosynthesis promotes lifespan extension and enables diverse pro-longevity paradigms in Caenorhabditis elegans. Elife12, 10.7554/eLife.82210 (2023). [DOI] [PMC free article] [PubMed]
- 73.Holdorf, A. D. et al. WormCat: An online tool for annotation and visualization of Caenorhabditis elegans Genome-Scale Data.Genetics214, 279–294 (2020). 10.1534/genetics.119.302919 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in the main text or the supplementary materials. RNAseq data is available at the NIH Gene Expression Omnibus (GEO) https://www.ncbi.nlm.nih.gov/geo (GSE251677), Source data are provided with this paper.