Abstract
Diaporthe species are known as endophytes, saprobes and pathogens infecting a wide range of plants and resulting in important crop diseases. In the present study, four strains of Diaporthe were obtained from diseased leaves of Bauhiniavariegata in Guangdong Province, China. Phylogenetic analyses were conducted to identify these strains using five gene regions: internal transcribed spacer (ITS), calmodulin (cal), histone H3 (his3), translation elongation factor 1-α (tef1) and β-tubulin (tub2). The results combined with morphology revealed two new species of Diaporthe named D.bauhiniicola in D.arecae species complex and D.guangzhouensis in D.sojae species complex.
Key words: Diaporthales, morphology, multi-gene phylogeny, taxonomy, two new taxa
Introduction
Diaporthe (syn. Phomopsis) is the type genus of Diaporthaceae in Diaporthales (Hyde et al. 2014; Maharachchikumbura et al. 2016). Before the implementation of “one fungus, one name”, it has been a common practice to use two names for the fungal species with pleomorphic life cycles (Taylor 2011). The genus Diaporthe established in 1870 predates Phomopsis established in 1905, thus Diaporthe is recommended for use (Rossman et al. 2015). More than 1200 epithets for Diaporthe have been listed in Index Fungorum with names often based on host association (http://www.indexfungorum.org/, accessed June 2024).
The teleomorph of Diaporthe is characterized by aggregated spherical ascomata with tapering necks, unitunicate, 8-spored, elongate to clavate asci, and septate or aseptate, elongated to elliptical, hyaline ascospores with larger guttules at center and smaller ones at the ends (Senanayake et al. 2018; Yang et al. 2020). The anamorph is characterized by black, ostiolate pycnidia containing cylindrical phialides often producing three types of hyaline, aseptate conidia called α-conidia, β-conidia and γ-conidia (Udayanga et al. 2012a; Dissanayake et al. 2017; Fan et al. 2018). The α-conidia and β-conidia are produced frequently, but the γ-conidia are rarely observed (Gomes et al. 2013).
Diaporthe species are associated with a wide range of plant hosts as pathogens, endophytes and saprobes of crops, forest trees and ornamentals (Farr et al. 2002a; Crous 2005; Udayanga et al. 2012b, 2014a, 2014b, 2015; Jiang et al. 2021; Zhu et al. 2023). As plant pathogens, Diaporthe species cause severe diseases, e.g., leaf spots, blights, dieback, scab, decay, stem end rots and wilt of many economically important plants including species of Citrus (Guarnaccia and Crous 2017), Macadamia (Wrona et al. 2020), Rosa (Caio et al. 2021), Vaccinium (Farr et al. 2002b), Vitis (Manawasinghe et al. 2019) and many more (Yang et al. 2018, 2021; Guarnaccia et al. 2020; Guo et al. 2020; Ariyawansa et al. 2021). In addition, Diaporthe species can live inside the healthy host tissues as endophytes (Huang et al. 2015; Dong et al. 2021). In addition, species of Diaporthe have been also reported as saprobes from different woody hosts (Dissanayake et al. 2020).
Species identification of Diaporthe has traditionally been based on host as well as morphological characters such as the size and shape of fruiting bodies and spores (Mostert et al. 2001; Santos and Phillips 2009). However, recent studies have shown that many species of Diaporthe are not host-specific i.e., one species may infect more than one host species (Vrandecic et al. 2011; Bai et al. 2015; Zhang et al. 2018; Huang et al. 2021; Sun et al. 2021; Cao et al. 2022). Moreover, many Diaporthe species that are morphologically similar have proven to be genetically distinct (van Rensburg et al. 2006; Yang et al. 2018). Phylogenetic analysis using a five-locus dataset (ITS-tef1-tub2-cal-his3) has been widely used to identify species of Diaporthe species (Santos et al. 2017; Marin-Felix et al. 2019; Hilário et al. 2021b; Norphanphoun et al. 2022). Diaporthe was clustered into 13 groups, namely D.arecae, D.biconispora, D.carpini, D.decedens, D.eres, D.oncostoma, D.pustulata, D.rudis, D.scobina, D.sojae, D.toxica, D.varians and D.vawdreyi species complexes and nine singletons as D.acerina, D.acutispora, D.crataegi, D.multiguttulata, D.ocoteae, D.perjuncta, D.pseudoalnea, D.spartinicola and D.undulata based on multilocus phylogeny (Norphanphoun et al. 2022; Hongsanan et al. 2023).
Bauhiniavariegata is a flowering plant species belonging to Fabaceae. It is native to China and cultivated as an ornamental tree in subtropical and tropical climate for its scented flowers. The aim of the present study was to identify new isolates collected from diseased leaves of Bauhiniavariegata in China following the combined approaches of morphology and phylogeny in the genus Diaporthe.
Materials and methods
Isolation and morphological characterization
In 2022, a plant disease investigation was conducted in Guangdong Province, China. Small and irregular leaf spots were observed on the leaves of Bauhiniavariegata, and 14 leaves were collected for isolation. The leaves were firstly surface-sterilized for 1 min in 75% ethanol, 3 min in 1.25% sodium hypochlorite and 1 min in 75% ethanol, rinsed for 2 min in distilled water and blotted on dry sterile filter paper. Then, the discolored areas were cut into 0.5 × 0.5 cm pieces and transferred to the surface of potato dextrose agar plates (PDA; 200 g potatoes, 20 g dextrose, 20 g agar per litre), incubated at 25 °C to obtain pure cultures. The cultures were deposited in the China Forestry Culture Collection Center (CFCC; http://cfcc.caf.ac.cn/) and the specimen was deposited in the Herbarium of the Chinese Academy of Forestry (CAF; http://museum.caf.ac.cn/).
The isolates were grown on PDA, MEA and SNA plates, incubated at 25 °C under a 12 h near-ultraviolet light/12 h dark cycle to induce sporulation. Colony characters and pigment production on PDA, MEA and SNA were noted for the 10-day culture. Microscopic structures of the fungi growing on medium were mounted in water and examined under an Axio Imager 2 microscope (Zeiss, Oberkochen, Germany). At least 30 measurements were made for each structure examined.
DNA extraction, amplification and sequencing
The genomic DNA was extracted from the fresh mycelium harvested from PDA plates after seven days using a cetyltrimethylammonium bromide (CTAB) method (Doyle and Doyle 1990). For initial genus confirmation, the internal transcribed spacer (ITS) region was sequenced. After confirmation of Diaporthe species, four additional gene regions coding for translation elongation factor 1-alpha (tef1), beta-tubulin (tub2), calmodulin (cal) and his-tone H3 (his3) were sequenced. The primer pairs and amplification conditions for each of the above-mentioned gene regions are provided in Table 1.
Table 1.
Loci assessed in this study with used PCR primers and program.
| Loci | Primers | PCR: Thermal Cycles: (Annealing Temp. in Bold) | Reference |
|---|---|---|---|
| ITS | ITS1f/ITS4 | (95 °C: 30 s, 48 °C: 30 s, 72 °C: 1 min) × 35 cycles | White et al. 1990 |
| cal | CAL228F/CAL737R | (95 °C: 15 s, 54 °C: 20 s, 72 °C: 1 min) × 35 cycles | Carbone and Kohn 1999 |
| his3 | CYLH3F/H3-1b | (95 °C: 30 s, 57 °C: 30 s, 72 °C: 1 min) × 35 cycles | Crous et al. 2004; Glass and Donaldson 1995 |
| tef1 | EF1-728F/EF1-986R | (95 °C: 15 s, 54 °C: 20 s, 72 °C: 1 min) × 35 cycles | Carbone and Kohn 1999 |
| tub2 | T1(Bt2a)/Bt2b | (95 °C: 30 s, 55 °C: 30 s, 72 °C: 1 min) × 35 cycles | Glass and Donaldson 1995; O’Donnell and Cigelnik 1997 |
A PCR reaction was conducted in a 20 µL reaction volume, and the components were as follows: 1 µL DNA template (20 ng/μL), 1 µL forward 10 µM primer, 1 µL reverse 10 µM primer, 10 µL T5 Super PCR Mix (containing Taq polymerase, dNTP and Mg2+, Beijing Tisingke Biotech Co., Ltd., Beijing, China), and 7 µL sterile water. Amplifications were performed using a T100 Thermal Cycler (Bio-Rad, Hercules, CA, USA). All amplified PCR products were evaluated visually with 1.4% agarose gels stained with ethidium bromide and PCR positive products sent to Sangon Biotech (Shanghai) Co., Ltd., (Beijing, China) for sequencing. Strands were sequenced in both directions using PCR primers. The new sequences generated in this study, as well as the reference sequences of all isolates used in the present study, are listed in Table 2.
Table 2.
Isolates and GenBank accession numbers used in the phylogenetic analyses of Diaporthe.
| Species | Location | Host | Strain | GenBank Accession Number | ||||
|---|---|---|---|---|---|---|---|---|
| ITS | tef1 | tub2 | cal | his3 | ||||
| Diaportheabsenteum | China | Camelliasinensis | LC3429* | KP267897 | KP267971 | KP293477 | NA | KP293547 |
| D.absenteum | China | Camelliasinensis | LC3564 | KP267912 | KP267986 | KP293492 | NA | KP293559 |
| D.acaciarum | Tanzania | Acaciatortilis | CBS 138862* | KP004460 | NA | KP004509 | NA | KP004504 |
| D.acericola | Italy | Acernegundo | MFLUCC 17-0956* | KY964224 | KY964180 | KY964074 | KY964137 | NA |
| D.aceris | Japan | Acer sp. | LC8112 | KY491547 | KY491557 | KY491567 | KY491575 | NA |
| D.actinidiae | New Zealand | Actinidiadeliciosa | ICMP 13683* | KC145886 | KC145941 | NA | NA | NA |
| D.acuta | China | Pyruspyrifolia | CGMCC 3.19600* | MK626957 | MK654802 | MK691225 | MK691124 | MK726161 |
| D.alangii | China | Alangiumkurzii | CFCC 52556* | MH121491 | MH121533 | MH121573 | MH121415 | MH121451 |
| D.alangii | China | Alangiumkurzii | CFCC 52557 | MH121492 | MH121534 | MH121574 | MH121416 | MH121452 |
| D.alnea | Netherlands | Alnus sp. | CBS 146.46 | KC343008 | KC343734 | KC343976 | KC343250 | KC343492 |
| D.amaranthophila | Japan | Amaranthustricolor | MAFF 246900 | LC459575 | LC459577 | LC459579 | LC459583 | LC459581 |
| D.ambigua | South Africa | Pyruscommunis | CBS 114015* | KC343010 | KC343736 | KC343978 | KC343252 | KC343494 |
| D.angelicae | Austria | Heracleumsphondylium | CBS 111592* | KC343027 | KC343753 | KC343995 | KC343269 | KC343511 |
| D.anhuiensis | China | Cunninghamialanceolata | CNUCC 201901* | MN219718 | MN224668 | MN227008 | MN224549 | MN224556 |
| D.arctii | Austria | Arctiumlappa | CBS 139280* | KJ590736 | KJ590776 | KJ610891 | KJ612133 | KJ659218 |
| D.arecae | India | Arecacatechu | CBS 161.64* | KC343032 | KC343758 | KC344000 | KC343274 | KC343516 |
| D.arengae | Hong Kong | Arengaengleri | CBS 114979* | KC343034 | KC343760 | KC344002 | KC343276 | KC343518 |
| D.arezzoensis | Italy | Cytisus sp. | MFLUCC 15-0127 | MT185503 | NA | NA | NA | NA |
| D.aseana | Thailand | Unidentified dead leaf | MFLUCC 12-0299a* | KT459414 | KT459448 | KT459432 | KT459464 | NA |
| D.australiana | Australia | Macadamia | CBS 146457 | MN708222 | MN696522 | MN696530 | NA | NA |
| D.bauhiniicola | China | Bauhiniavariegata | CFCC 58154* | PP864723 | PP938599 | PP938603 | PP938607 | PP938611 |
| D.bauhiniicola | China | Bauhiniavariegata | GZ13B | PP864724 | PP938600 | PP938604 | PP938608 | PP938612 |
| D.batatas | USA | Ipomoeabatatas | CBS 122.21* | KC343040 | KC343766 | KC344008 | KC343282 | KC343524 |
| D.beilharziae | Australia | Indigoferaaustralis | BRIP 54792* | JX862529 | JX862535 | KF170921 | NA | NA |
| D.biconispora | China | Citrusgrandis | ZJUD62 | KJ490597 | KJ490476 | KJ490418 | MT227578 | KJ490539 |
| D.biguttulata | China | Citruslimon | ZJUD47* | KJ490582 | KJ490461 | KJ490403 | NA | KJ490524 |
| D.brasiliensis | Brazil | Aspidosperma sp. | CBS 133183* | KC343042 | KC343768 | KC344010 | KC343284 | KC343526 |
| D.caatingaensis | Brazil | Tacingainamoena | CBS 141542* | KY085927 | KY115603 | KY115600 | NA | KY115605 |
| D.camelliae-oleiferae | China | Camelliaoleifera | HNZZ027* | MZ509555 | MZ504707 | MZ504718 | MZ504685 | MZ504696 |
| D.caryae | China | Caryaillinoensis | CFCC 52563* | MH121498 | MH121540 | MH121580 | MH121422 | MH121458 |
| D.caryae | China | Caryaillinoensis | CFCC 52564 | MH121499 | MH121541 | MH121581 | MH121423 | MH121459 |
| D.cercidis | China | Cercischinensis | CFCC 52565* | MH121500 | MH121542 | MH121582 | MH121424 | MH121460 |
| D.cercidis | China | Cercischinensis | CFCC 52566 | MH121501 | MH121543 | MH121583 | MH121425 | MH121461 |
| D.chiangraiensis | Thailand | Bauhinia sp. | MFLUCC 17-1669* | MF190119 | MF377598 | NA | NA | NA |
| D.chrysalidocarpi | China | Chrysalidocarpuslutescens | SAUCC194.35 | MT822563 | MT855760 | MT855876 | MT855646 | MT855532 |
| D.cichorii | Italy | Cichoriumintybus | MFLUCC 17-1023* | KY964220 | KY964176 | KY964104 | KY964133 | NA |
| D.cinmomi | China | Cinnamomum sp. | CFCC 52569* | MH121504 | MH121546 | MH121586 | NA | MH121464 |
| D.cinmomi | China | Cinnamomum sp. | CFCC 52570 | MH121505 | MH121547 | MH121587 | NA | MH121465 |
| D.citriasiana | China | Citrusunshiu | CGMCC 3.15224* | JQ954645 | JQ954663 | KC357459 | KC357491 | KJ490515 |
| D.columnaris | USA | Vacciniumvitisidaea | AR3612* | AF439625 | NA | NA | NA | NA |
| D.compacta | China | Camelliasinensis | CGMCC 3.17536* | KP267854 | KP267928 | KP293434 | NA | KP293508 |
| D.convolvuli | Turkey | Convolvulusarvensis | CBS 124654* | KC343054 | KC343780 | KC344022 | KC343296 | KC343538 |
| D.cucurbitae | Canada | Cucumis sp. | DAOM 42078* | KM453210 | KM453211 | KP118848 | NA | KM453212 |
| D.cuppatea | South Africa | Aspalathuslinearis | CBS 117499* | KC343057 | KC343783 | KC344025 | KC343299 | KC343541 |
| D.cyatheae | Taiwan | Cyathealepifera | YMJ 1364* | JX570889 | KC465406 | KC465403 | KC465410 | NA |
| D.discoidispora | China | Citrusunshiu | ZJUD89* | KJ490624 | KJ490503 | KJ490445 | NA | KJ490566 |
| D.drenthii | Australia | Macadamia | CBS 146453 | MN708229 | MN696526 | MN696537 | NA | NA |
| D.durionigena | Vietnam | Duriozibethinus | VTCC 930005 | MN453530 | MT276157 | MT276159 | NA | NA |
| D.endocitricola | China | Citrusmaxima | ZHKUCC20-0012* | MT355682 | MT409336 | MT409290 | MT409312 | NA |
| D.endophytica | Brazil | Schinusterebinthifolius | CBS 133811* | KC343065 | KC343791 | KC344033 | KC343307 | KC343549 |
| D.eucalyptorum | China | Eucalyptus | CBS 132525* | MH305525 | NA | NA | NA | NA |
| D.eugeniae | Indonesia | Eugeniaaromatica | CBS 444.82* | KC343098 | KC343824 | KC344066 | KC343340 | KC343582 |
| D.fraxini-angustifoliae | Australia | Fraxinusangustifolia | BRIP 54781* | JX862528 | JX862534 | KF170920 | NA | NA |
| D.fructicola | Japan | Passifloraedulis × P. edulis | MAFF 246408* | LC342734 | LC342735 | LC342736 | LC342738 | LC342737 |
| D.fulvicolor | China | Pyruspyrifolia | CGMCC 3.19601* | MK626859 | MK654806 | MK691236 | MK691132 | MK726163 |
| D.ganjae | USA | Cannabissativa | CBS 180.91* | KC343112 | KC343838 | KC344080 | KC343354 | KC343596 |
| D.goulteri | Australia | Helianthusannuus | BRIP 55657a* | KJ197290 | KJ197252 | KJ197270 | NA | NA |
| D.guangdongensis | China | Citrusmaxima | ZHKUCC20-0014* | MT355684 | MT409338 | MT409292 | MT409314 | NA |
| D.guangxiensis | China | Vitisvinifera | JZB320094* | MK335772 | MK523566 | MK500168 | MK736727 | NA |
| D.guangzhouensis | China | Bauhiniavariegataa | CFCC 58151* | PP864725 | PP938601 | PP938605 | PP938609 | PP938613 |
| D.guangzhouensis | China | Bauhiniavariegata | GZ13E | PP864726 | PP938602 | PP938606 | PP938610 | PP938614 |
| D.gulyae | Australia | Helianthusannuus | BRIP 54025* | JF431299 | JN645803 | KJ197271 | NA | NA |
| D.guttulata | China | Unknown | CGMCC 3.20100 | MT385950 | MT424685 | MT424705 | MW022470 | MW022491 |
| D.helianthi | Serbia | Helianthusannuus | CBS 592.81* | KC343115 | KC343841 | KC344083 | KC343357 | KC343599 |
| D.heterostemmatis | China | Heterostemmagrandiflorum | SAUCC194.85* | MT822613 | MT855925 | MT855810 | MT855692 | MT855581 |
| D.hongkongensis | China | Dichroafebrífuga | CBS 115448* | KC343119 | KC343845 | KC344087 | KC343361 | KC343603 |
| D.hordei | Norway | Hordeumvulgare | CBS 481.92* | KC343120 | KC343846 | KC344088 | KC343362 | KC343604 |
| D.huangshanensis | China | Camelliaoleifera | CNUCC 201903* | MN219729 | MN224670 | MN227010 | NA | MN224558 |
| D.hubeiensis | China | Vitisvinifera | JZB320123 | MK335809 | MK523570 | MK500148 | MK500235 | NA |
| D.hunanensis | China | Camelliaoleifera | HNZZ023* | MZ509550 | MZ504702 | MZ504713 | MZ504680 | MZ504691 |
| D.infecunda | Brazil | Schinus sp. | CBS 133812* | KC343126 | KC343852 | KC344094 | KC343368 | KC343610 |
| D.infertilis | Suriname | Camelliasinensis | CBS 230.52* | KC343052 | KC343778 | KC344020 | KC343294 | KC343536 |
| D.kochmanii | Australia | Helianthusannuus | BRIP 54033* | JF431295 | JN645809 | NA | NA | NA |
| D.kongii | Australia | Portulacagrandifla | BRIP 54031* | JF431301 | JN645797 | KJ197272 | NA | NA |
| D.krabiensis | Thailand | marine based habitats | MFLUCC 17-2481* | MN047101 | MN433215 | MN431495 | NA | NA |
| D.leucospermi | Australia | Leucospermum sp. | CBS 111980* | JN712460 | KY435632 | KY435673 | KY435663 | KY435653 |
| D.limonicola | Malta | Citruslimon | CPC 28200* | NR_154980 | MF418501 | MF418582 | MF418256 | MF418342 |
| D.litchiicola | Australia | Litchichinensis | BRIP 54900* | JX862533 | JX862539 | KF170925 | NA | NA |
| D.lithocarpi | China | Lithocarpusglabra | CGMCC 3.15175* | KC153104 | KC153095 | KF576311 | KF576235 | NA |
| D.longicolla | USA | Glycinemax | FAU599* | KJ590728 | KJ590767 | KJ610883 | KJ612124 | KJ659188 |
| D.longispora | Canada | Ribes sp. | CBS 194.36* | KC343135 | KC343861 | KC344103 | KC343377 | KC343619 |
| D.lusitanicae | Portugal | Foeniculumvulgare | CBS 123212 | KC343136 | KC343862 | KC344104 | KC343378 | KC343620 |
| D.lusitanicae | Portugal | Foeniculumvulgare | CBS 123213* | MH863280 | KC343863 | KC344105 | KC343379 | KC343621 |
| D.malorum | Portugal | Malusdomestica | CAA 734* | KY435638 | KY435627 | KY435668 | KY435658 | KY435648 |
| D.manihotia | Rwanda | Manihotutilissima | CBS 505.76 | KC343138 | KC343864 | KC344106 | KC343380 | KC343622 |
| D.masirevicii | Australia | Helianthusannuus | BRIP 57892a* | KJ197276 | KJ197239 | KJ197257 | NA | NA |
| D.mayteni | Brazil | Maytenusilicifolia | CBS 133185 | KC343139 | KC343865 | KC344107 | KC343381 | KC343623 |
| D.megalospora | Not stated | Sambucuscanadensis | CBS 143.27 | KC343140 | KC343866 | KC344108 | KC343382 | KC343624 |
| D.melitensis | Malta | Citruslimon | CPC 27873* | MF418424 | MF418503 | MF418584 | MF418258 | MF418344 |
| D.melonis | USA | Cucumismelo | CBS 507.78* | KC343142 | KC343868 | KC344110 | KC343384 | KC343626 |
| D.melonis | Indonesia | Glycinesoja | CBS 435.87 | KC343141 | KC343867 | KC344109 | KC343383 | KC343625 |
| D.middletonii | Australia | Rapistrumrugostrum | BRIP 54884e* | KJ197286 | KJ197248 | KJ197266 | NA | NA |
| D.millettiae | China | Millettiareticulata | GUCC9167* | MK398674 | MK480609 | MK502089 | MK502086 | NA |
| D.minusculata | China | saprobic on decaying wood | CGMCC 3.20098* | MT385957 | MT424692 | MT424712 | MW022475 | MW022499 |
| D.miriciae | Australia | Helianthusannuus | BRIP 54736j* | KJ197282 | KJ197244 | KJ197262 | NA | NA |
| D.musigena | Australia | Musa sp. | CBS 129519* | KC343143 | KC343869 | KC344111 | KC343385 | KC343267 |
| D.myracrodruonis | Brazil | Astroniumurundeuva | URM 7972* | MK205289 | MK213408 | MK205291 | MK205290 | 17 |
| D.nelumbonis | Taiwan | Nelumbonucifera | R. Kirschner 4114* | KT821501 | NA | LC086652 | NA | NA |
| D.neoarctii | USA | Ambrosiatrifi | CBS 109490* | KC343145 | KC343871 | KC344113 | KC343387 | KC343629 |
| D.neoraonikayaporum | Thailand | Tectonagrandis | MFLUCC 14-1136* | KU712449 | KU749369 | KU743988 | KU749356 | NA |
| D.oculi | Japan | Homosapiens | HHUF 30565* | LC373514 | LC373516 | LC373518 | NA | NA |
| D.osmanthi | China | Osmanthusfragrans | GUCC9165* | MK398675 | MK480610 | MK502091 | MK502087 | NA |
| D.ovalispora | China | Citruslimon | CGMCC 3.17256* | KJ490628 | KJ490507 | KJ490449 | NA | KJ490570 |
| D.oxe | Brazil | Maytenusilicifolia | CBS 133186* | KC343164 | KC343890 | KC344132 | KC343406 | KC343648 |
| D.pandanicola | Thailand | Pandanus sp. | MFLUCC 17-0607* | MG646974 | NA | MG646930 | NA | NA |
| D.paranensis | Brazil | Maytenusilicifolia | CBS 133184* | KC343171 | KC343897 | KC344139 | KC343413 | KC343655 |
| D.pascoei | Australia | Perseaamericana | BRIP 54847* | JX862532 | JX862538 | KF170924 | NA | NA |
| D.passiflorae | South America | Passiflaedulis | CBS 132527* | JX069860 | KY435633 | KY435674 | KY435664 | KY435654 |
| D.passifloricola | Malaysia | Passiflorafoetida | CBS 141329* | KX228292 | NA | KX228387 | NA | KX228367 |
| D.perseae | Netherlands | Perseagratissima | CBS 151.73* | KC343173 | KC343899 | KC343141 | KC343415 | KC343657 |
| D.pescicola | China | Prunuspersica | MFLUCC 16-0105* | KU557555 | KU557623 | KU557579 | KU557603 | NA |
| D.phaseolorum | USA | Phaseolusvulgaris | AR4203* | KJ590738 | KJ590739 | KJ610893 | KJ612135 | KJ659220 |
| D.phoenicicola | India | Arecacatechu | CBS 161.64* | MH858400 | GQ250349 | JX275440 | JX197432 | NA |
| D.podocarpi-macrophylli | China | Podocarpusmacrophyllus | CGMCC 3.18281* | KX986774 | KX999167 | KX999207 | KX999278 | KX999246 |
| D.pseudobauhiniae | Thailand | Bauhinia sp. | MFLU 17-1670 | MF190118 | MF377599 | NA | NA | NA |
| D.pseudobauhiniae | Thailand | Bauhinia sp. | MFLUCC 17-1669* | MF190119 | MF377598 | NA | NA | NA |
| D.pseudolongicolla | Serbia | Glycinemax | PL42* | JQ697843 | JQ697856 | NA | NA | NA |
| D.pseudolongicolla | Croatia | Glycinemax | CBS 127269 | KC343155 | KC343881 | KC344123 | KC343397 | KC343639 |
| D.pseudomangiferae | Dominican Republic | Mangiferaindica | CBS 101339* | KC343181 | KC343907 | KC344149 | KC343423 | KC343665 |
| D.pseudooculi | Japan | Homosapiens | HHUF 30617* | NR_161019 | LC373517 | LC373519 | NA | NA |
| D.pseudophoenicicola | Spain | Phoenixdactylifera | CBS 462.69* | KC343184 | KC343910 | KC344152 | KC343426 | KC343668 |
| D.pseudophoenicicola | Iraq | Mangiferaindica | CBS 176.77 | KC343183 | KC343909 | KC344151 | KC343425 | KC343667 |
| D.pterocarpicola | Thailand | Pterocarpusindicus | MFLUCC 10-0580a* | JQ619887 | JX275403 | JX275441 | JX197433 | NA |
| D.pyracanthae | Portugal | Pyracanthacoccinea | CBS 142384* | KY435635 | KY435625 | KY435666 | KY435656 | KY435646 |
| D.racemosae | South Africa | Euclearacemosa | CPC 26646* | MG600223 | MG600225 | MG600227 | MG600219 | MG600221 |
| D.raonikayaporum | Brazil | Spondiasmombin | CBS 133182* | KC343188 | KC343914 | KC344156 | KC343430 | KC343672 |
| D.rhodomyrti | China | Rhodomyrtustomentosa | CFCC 53101 | MK432643 | MK578119 | MK578046 | MK442965 | MK442990 |
| D.rhodomyrti | China | Rhodomyrtustomentosa | CFCC 53102 | MK432644 | MK578120 | MK578047 | MK442966 | MK442991 |
| D.rosae | Thailand | Rosa sp. | MFLUCC 17-2658* | MG828894 | NA | MG843878 | MG829273 | NA |
| D.rosiphthora | Brazil | Rosa sp. | COAD 2914* | MT311197 | MT313693 | NA | MT313691 | NA |
| D.rossmaniae | Portugal | Vacciniumcorymbosum | CAA762* | MK792290 | MK828063 | MK837914 | MK883822 | MK871432 |
| D.sackstonii | Australia | Helianthusannuus | BRIP 54669b* | KJ197287 | KJ197249 | KJ197267 | NA | NA |
| D.salinicola | Thailand | Xylocarpus sp. | MFLU 18-0553* | MN047098 | MN077073 | NA | NA | NA |
| D.sambucusii | China | Sambucuswilliamsii | CFCC 51986* | KY852495 | KY852507 | KY852511 | KY852499 | KY852503 |
| D.sambucusii | China | Sambucuswilliamsii | CFCC 51987 | KY852496 | KY852508 | KY852512 | KY852500 | KY852504 |
| D.schimae | China | Schimasuperba | CFCC 53103* | MK432640 | MK578116 | MK578043 | MK442962 | MK442987 |
| D.schimae | China | Schimasuperba | CFCC 53104 | MK432641 | MK578117 | MK578044 | MK442963 | MK442988 |
| D.schini | Brazil | Schinusterebinthifolius | CBS 133181* | KC343191 | KC343917 | KC344159 | KC343433 | KC343675 |
| D.schoeni | Italy | Schoenusnigricans | MFLU 15-1279* | KY964226 | KY964182 | KY964109 | KY964139 | |
| D.sclerotioides | Netherlands | Cucumissativus | CBS 296.67* | KC343193 | KC343919 | KC344161 | KC343435 | KC343677 |
| D.searlei | Australia | Macadamia | CBS 146456* | MN708231 | NA | MN696540 | NA | NA |
| D.sennae | China | Sennabicapsularis | CFCC 51636* | KY203724 | KY228885 | KY228891 | KY228875 | NA |
| D.sennae | China | Sennabicapsularis | CFCC 51637 | KY203725 | KY228886 | KY228892 | KY228876 | NA |
| D.serafiniae | Australia | Helianthusannuus | BRIP 55665a* | KJ197274 | KJ197236 | KJ197254 | NA | NA |
| D.siamensis | Thailand | Dasymaschalon sp. | MFLUCC 10-0573a* | JQ619879 | JX275393 | JX275429 | JX197423 | NA |
| D.sinensis | China | Amaranthus sp. | ZJUP0033-4* | MK637451 | MK660449 | MK660447 | NA | MK660451 |
| D.sojae | USA | Glycinemax | FAU635* | KJ590719 | KJ590762 | KJ610875 | KJ612116 | KJ659208 |
| D.spinosa | China | Pyruspyrifolia | CGMCC 3.19602* | MK626849 | MK654811 | MK691234 | MK691129 | MK726156 |
| D.stewartii | Not stated | Cosmosbipinnatus | CBS 193.36* | MH867279 | GQ250324 | JX275421 | JX197415 | NA |
| D.subellipicola | China | On dead wood | KUMCC 17-0153* | MG746632 | MG746633 | MG746634 | NA | NA |
| D.subordinaria | New Zealand | Plantagolanceolata | CBS 464.90* | KC343214 | KC343940 | KC344182 | KC343456 | KC343698 |
| D.taiwanensis | Taiwan | Ixorachinensis | NTUCC 18-105-1* | MT241257 | MT251199 | MT251202 | MT251196 | NA |
| D.taoicola | China | Prunuspersica | MFLUCC 16-0117* | KU557567 | KU557635 | KU557591 | NA | NA |
| D.tarchonanthi | South Africa | Tarchonanthuslittoralis | CBS 146073* | MT223794 | NA | MT223733 | NA | MT223759 |
| D.tecomae | Brazil | Tabebuia sp. | CBS 100547* | KC343215 | KC343941 | KC344183 | KC343457 | KC343699 |
| D.tectonae | Thailand | Tectonagrandis | MFLUCC 12-0777* | KU712430 | KU749359 | KU743977 | KU749345 | NA |
| D.tectonendophytica | Thailand | Tectonagrandis | MFLUCC 13-0471* | KU712439 | KU749367 | KU743986 | KU749354 | NA |
| D.tectonigena | China | Tectonagrandis | MFLUCC 12-0767* | KU712429 | KU749371 | KU743976 | KU749358 | NA |
| D.tectonigena | China | Camelliasinensis | LC6512 | KX986782 | KX999174 | KX999214 | KX999284 | KX999254 |
| D.terebinthifolii | Brazil | Schinusterebinthifolius | CBS 133180* | KC343216 | KC343942 | KC344184 | KC343458 | KC343700 |
| D.thunbergiicola | Thailand | Thunbergialaurifolia | MFLUCC 12-0033* | KP715097 | KP715098 | NA | NA | NA |
| D.tulliensis | Australia | Theobromacacao | BRIP 62248a* | KR936130 | KR936133 | KR936132 | NA | NA |
| D.ueckeri | USA | Cucumismelo | FAU656* | KJ590726 | KJ590747 | KJ610881 | KJ612122 | KJ659215 |
| D.unshiuensis | China | Fortunellamargarita | CGMCC 3.17566* | KJ490584 | KJ490463 | KJ490405 | NA | KJ490526 |
| D.unshiuensis | China | Caryaillinoensis | CFCC 52594 | MH121529 | MH121571 | MH121606 | MH121447 | MH121487 |
| D.unshiuensis | China | Caryaillinoensis | CFCC 52595 | MH121530 | MH121572 | MH121607 | MH121448 | MH121488 |
| D.vawdreyi | Australia | Psidiumguajava | BRIP 57887a | KR936126 | KR936129 | KR936128 | NA | NA |
| D.vexans | USA | Solanummelongena | CBS 127.14 | KC343229 | KC343955 | KC344197 | KC343471 | KC343713 |
| D.viniferae | China | Vitisvinifera | JZB320071* | MK341550 | MK500107 | MK500112 | MK500119 | NA |
| D.vochysiae | Brazil | Vochysiadivergens | LGMF1583* | MG976391 | MK007526 | MK007527 | MK007528 | MK033323 |
| D.xishuangbanica | China | Camelliasinensis | CGMCC 3.18283* | KX986784 | KX999176 | KX999217 | NA | NA |
| D.xishuangbanica | China | Camelliasinensis | LC6707 | KX986783 | KX999175 | KX999216 | NA | KX999255 |
Notes: NA, not applicable. * ex-type strains.
Phylogeny
For the phylogenetic analysis, sequences of reference Diaporthe species and related taxa were downloaded from NCBI GenBank based on recent publications on the genus Diaporthe (Norphanphoun et al. 2022) (Table 2). Downloaded sequences were aligned together with the sequences obtained in the present study using MAFFT version 7.526 (Katoh and Standley 2013) and manually corrected using Bioedit 7.0.9.0 (Hall 1999). The phylogenetic analyses of the combined gene regions were performed using Maximum Likelihood (ML) and Bayesian Inference (BI) methods. ML was conducted using PhyML v. 3.0 (Guindon et al. 2010), with 1000 bootstrap replicates while BI was performed using a Markov Chain Monte Carlo (MCMC) algorithm in MrBayes v. 3.0 (Ronquist and Huelsenbeck 2003). Two MCMC chains, started from random trees for 1,000,000 generations and trees, were sampled every 100th generation, resulting in a total of 10,000 trees. The first 25% of trees were discarded as burn-in of each analysis. Branches with significant Bayesian Posterior Probabilities (BPP) were estimated in the remaining 7500 trees. Phylogenetic trees were visualized with FigTree v.1.3.1 (Rambaut and Drummond 2010) and processed by Adobe Illustrator CS5. The nucleotide sequence data of the new taxa were deposited in GenBank (Table 2)
Results
Phylogenetic analyses
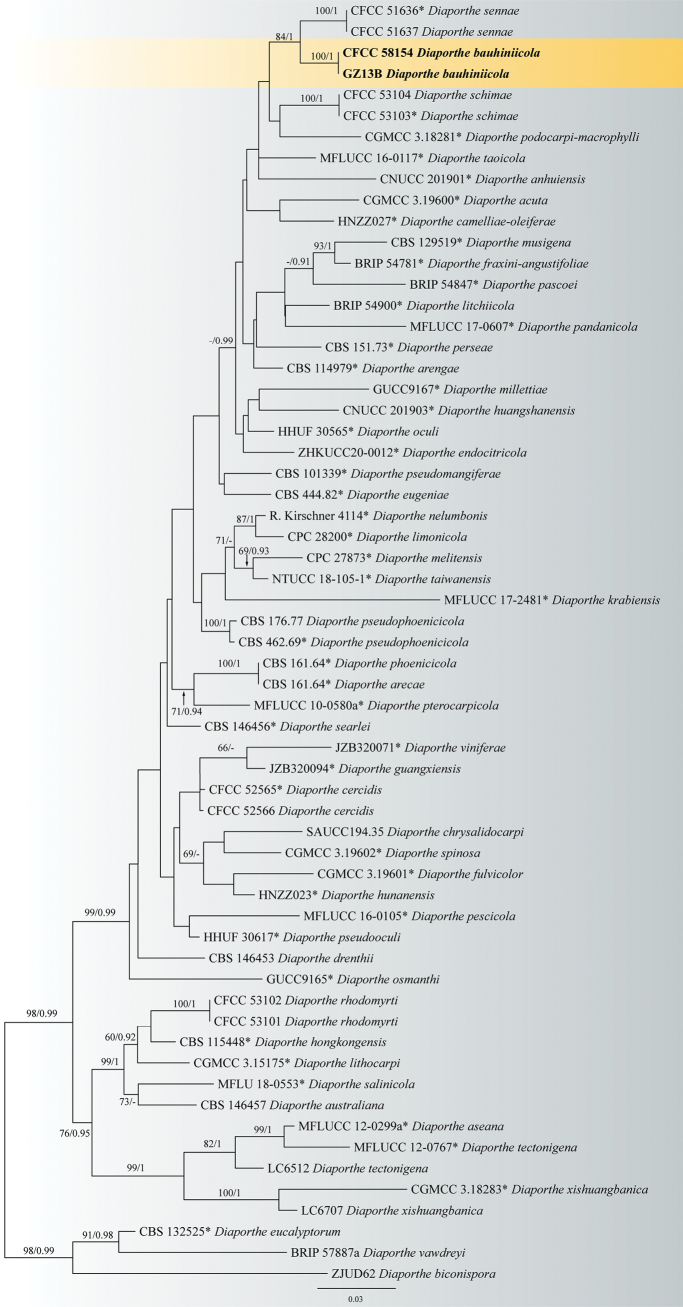
In the present study, we inferred a genus tree of Diaporthe covering a large proportion of sequence data available as last summarized by Norphanphoun et al. (2022). Two strains CFCC 58154 and GZ13B formed a clade in the D.arecae species complex, and the other strains CFCC 58151 and GZ13E in the D.sojae species complex.
In the D.arecae species complex, the combined sequence alignments comprised 61 strains, with D.eucalyptorum (CBS 13252), D.biconispora (ZJUD62) and D.vawdreyi (BRIP 57887a) as the outgroup taxa. The dataset comprised 2662 characters including alignment gaps (590 for ITS, 499 for cal, 485 for his3, 375 for tef1 and 713 for tub2). CFCC 58154 and GZ13B from Bauhiniavariegata formed a distinct clade close to D.sennae (Fig. 1). In the D.sojae species complex, the combined sequence alignments comprised 166 strains (Fig. 2), with D.aceris (LC8112) and D.alnea (CBS 146.46) as the outgroup taxa. The dataset comprised 3025 characters including alignment gaps (602 for ITS, 592 for cal, 521 for his3, 483 for tef1 and 827 for tub2). CFCC 58151 and GZ13E from B.variegata clustered in a distinct clade close to D.tulliensis (Fig. 2).
Figure 1.
Phylogram of Diaporthearecae species complex resulting from a maximum likelihood analysis based on a combined matrix of ITS, cal, his3, tef1 and tub2 loci. Numbers above the branches indicate ML bootstrap values (left, ML BS ≥ 50%) and Bayesian posterior probabilities (right, BPP ≥ 0.9). Isolates from the present study are in bold and ex-type strains are marked with *.
Figure 2.
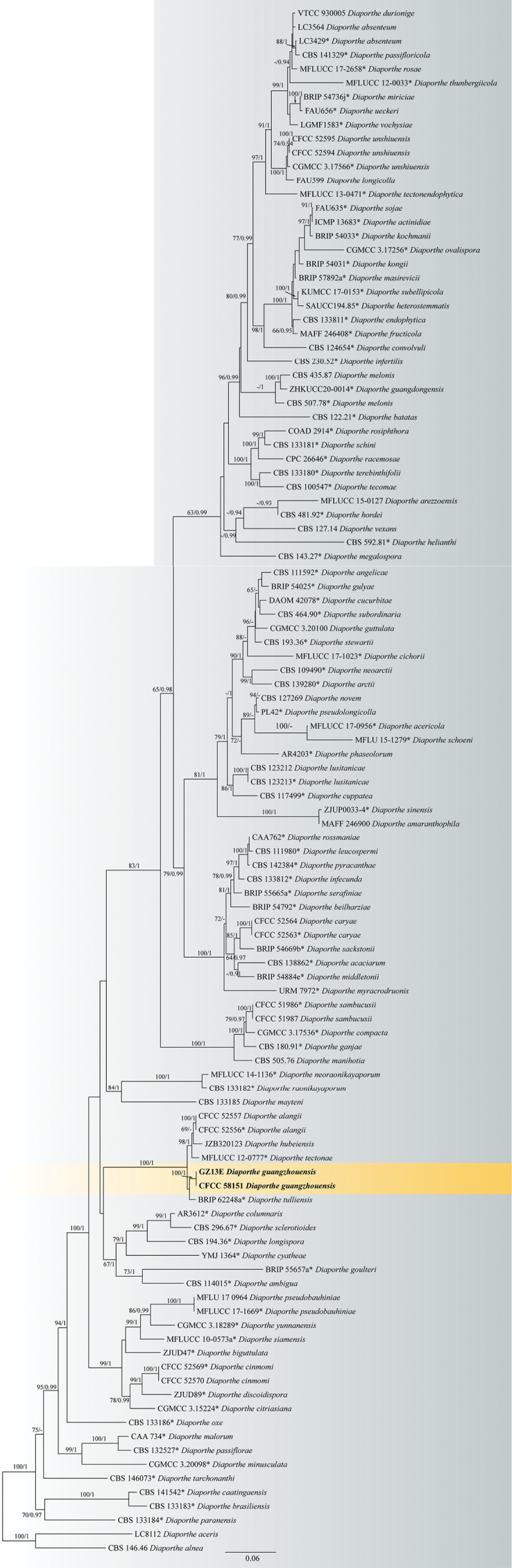
Phylogram of Diaporthesojae species complex resulting from a maximum likelihood analysis based on a combined matrix of ITS, cal, his3, tef1 and tub2 loci. Numbers above the branches indicate ML bootstrap values (left, ML BS ≥ 50%) and Bayesian posterior probabilities (right, BPP ≥ 0.9). Isolates from the present study are in bold and ex-type strains are marked with *.
Taxonomy
. Diaporthe bauhiniicola
Ning Jiang & Y.Q. Zhu sp. nov.
2D6AD0F1-0758-54ED-B0EB-B5BDDB93E656
854183
Figure 3.
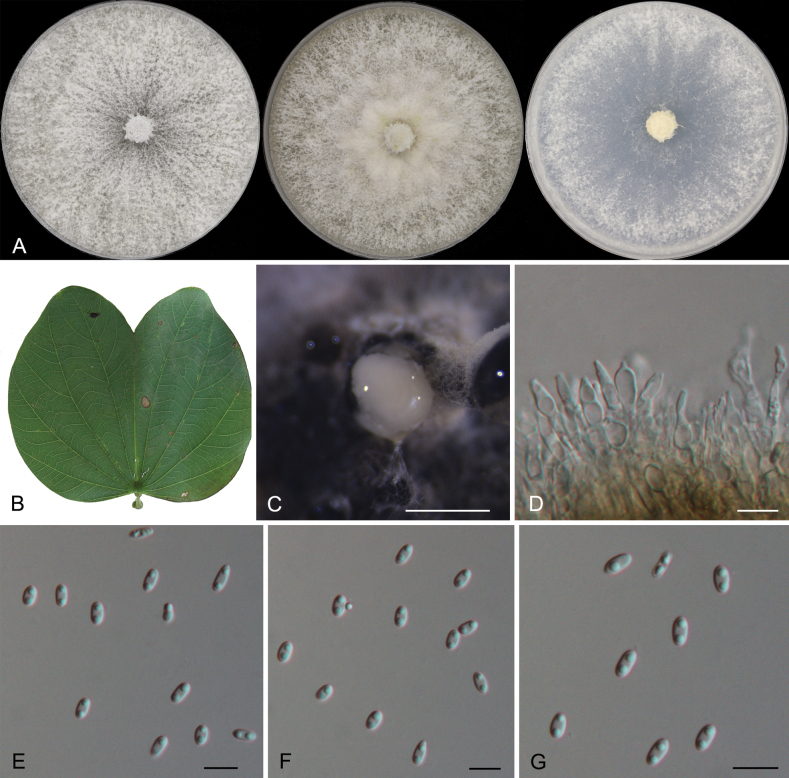
Morphology of DiaporthebauhiniicolaA colonies on PDA, MEA and SNA at 25 °C after 2 weeks B a diseased leaf of BauhiniavariegataC conidioma formed on PDA after 30 days D conidiogenous cells with attached alpha conidia E–G alpha conidia. Scale bars: 200 µm (C); 10 µm (D–J).
Holotype.
China • Guangdong Province, Guangzhou City, Luhu Park, 23°9'11.15"N, 113°16'46.01"E, 92 m asl, on diseased leaves of Bauhiniavariegata, 8 Aug 2022, Yong Li, Chengbin Wang & Yaquan Zhu, (holotype: CAF800094; ex-type culture: CFCC 58154).
Etymology.
Named after the host genus, Bauhinia.
Description.
Conidiomata formed on PDA pycnidial, scattered to aggregated, black, erumpent, raising above surface of culture medium, subglobose, 150–300 μm diam., exuding white or yellowish creamy conidial droplets from central ostioles after 30 days at 25 °C. Conidiophores reduced to conidiogenous cells. Conidiogenous cells hyaline, unbranched, septate, straight, slightly tapering towards the apex, 6.0–15.0 × 1.5–4.0 μm. Alpha conidia hyaline, aseptate, ellipsoidal to spindle-shaped, biguttulate or with one guttulate, 4.5–7.0 × 2.0–3.0 μm. Beta conidia and gamma conidia not observed. Teleomorph not observed.
Culture characteristics.
Colonies covering entire plate after 2 weeks. On PDA with profuse aerial mycelium, white surface, reverse fulvous. On MEA with fluffy aerial mycelium, dirty white surface, reverse ochreous. On SNA white sparse aerial mycelium, surface and reverse white.
Additional material examined.
China • Guangdong Province, Guangzhou City, Luhu Park, 23°9'11.15"N, 113°16'46.01"E, 92 m asl, on diseased leaves of Bauhiniavariegata, 8 Aug 2022, Yong Li, Chengbin Wang & Yaquan Zhu, living culture GZ13B.
Notes.
Two strains representing Diaporthebauhiniicola clustered in a clade distinct from its closest phylogenetic neighbour, D.sennae (Fig. 1). D.sennae has been reported from the host Sennabicapsularis in China (Yang et al. 2017). D.bauhiniicola differs from D.sennae by wider alpha conidia (4.5–7.0 × 2.0–3.0 μm in D.bauhiniicola vs. 5.0–6.5 × 1.5–1.8 μm in D.sennae) (Yang et al. 2017). Diaporthebauhiniicola differs from D.sennae in nucleotide sequence data (18/529 in ITS, 5/490 in cal, 15/351 in tef1, 14/677 in tub2) (Yang et al. 2017).
. Diaporthe guangzhouensis
Ning Jiang & Y.Q. Zhu sp. nov.
2A0959D8-088A-5FA5-A7A2-765C956730CF
854184
Figure 4.
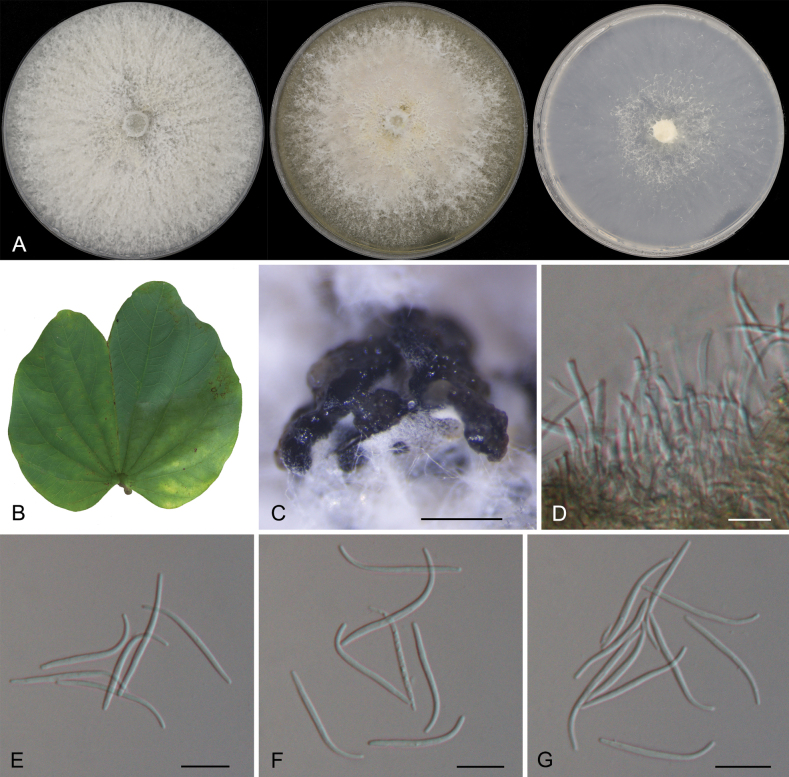
Morphology of DiaportheguangzhouensisA colonies on PDA, MEA and SNA at 25 °C after 2 weeks B the leaf of BauhiniavariegataC conidiomata D conidiogenous cells with attached beta conidia E–G beta conidia. Scale bars: 200 µm (C); 10 µm (D–J).
Etymology.
Named after the collection site of the type specimen, Guangzhou City.
Holotype.
China • Guangdong Province, Guangzhou City, Longdong straight street, 23°11'41.02"N, 113°22'8.33"E, 46 m asl, on diseased leaves of Bauhiniavariegata, 8 Aug 2022, Yong Li, Chengbin Wang & Yaquan Zhu, (holotype: CAF800095; ex-type culture: CFCC 58151).
Description.
Conidiomata pycnidial, scattered to aggregated, black, erumpent, raising above surface of culture medium, subglobose, 150–450 µm diam, exuding white or yellowish creamy conidial droplets from central ostioles after 30 days at 25 °C. Conidiophores 12.5–24.5 × 1–2.5 μm, cylindrical, hyaline, unbranched, straight to sinuous. Conidiogenous cells densely aggregated, phiailidic, unbranched, straight or slightly curved, 5.5–10 × 2.0–7.5 μm. Beta conidia filiform, hyaline, straight or slightly curved, aseptate, 17.0–29.5 × 1.0–2.0 μm. Alpha conidia and gamma conidia not observed. Teleomorph not observed.
Culture characteristics.
Colonies covering entire plate after 2 weeks. On PDA with profuse aerial mycelium, white surface, reverse amber. On MEA with fluffy aerial mycelium, dirty white surface, reverse ochreous. On SNA white sparse aerial mycelium, surface and reverse white.
Additional material examined.
China • Guangdong Province, Guangzhou City, Longdong straight street, 23°11'41.02"N, 113°22'8.33"E, 46 m asl, on diseased leaves of Bauhiniavariegata, 8 Aug 2022, Yong Li, Chengbin Wang & Yaquan Zhu, living culture GZ13E.
Notes.
Diaportheguangzhouensis from the present study is phylogenetically close to D.tulliensis (Fig. 2). Diaportheguangzhouensis differs from D.tulliensis in nucleotide sequence data (5/526 in ITS, 9/347 in tef1, 13/711 in tub2) (Crous et al. 2015). In addition, host and distribution data are vital for species identification (D.guangzhouensis inhabiting Bauhiniavariegata in China vs. D.tulliensis inhabiting Theobromacacao in Australia) (Crous et al. 2015).
Discussion
In the current study, phylogenetic analyses based on five combined loci (ITS, cal, his3, tef1 and tub2), as well as morphological characters of the anamorph obtained in culture, revealed D.bauhiniicola and D.guangzhouensis spp. nov. from Bauhiniavariegata, which contributed to our knowledge of the diversity of Diaporthe species in China.
Diaporthepseudobauhiniae (syn. D.chiangraiensis, Chiangraiomycesbauhiniae) was described as a saprobic fungus on branches of Bauhinia sp. in Thailand (Senanayake et al. 2017). D.bauhiniae was introduced from branches of B.purpurea in China (Yang et al. 2021). Hence, a total of four species of Diaporthe have been recorded from the host genus Bauhinia. Phylogenetically, D.bauhiniae belongs to D.varians species complex; D.bauhiniicola belongs to D.arecae species complex; D.guangzhouensis and D.pseudobauhiniae belong to D.sojae species complex (Figs 1, 2) (Norphanphoun et al. 2022). Furthermore, D.guangzhouensis and D.pseudobauhiniae formed different clades in D.sojae species complex (Fig. 2). Morphologically, D.bauhiniicola has larger alpha conidia than D.pseudobauhiniae, but longer alpha conidia than D.bauhiniae (4.5–7.0 × 2.0–3.0 μm in D.bauhiniicola vs. 3–5 × 2–4 μm in D.pseudobauhiniae vs. 7.5–14 × 1.5–3 μm in D.bauhiniae) (Senanayake et al. 2017; Yang et al. 2021). D.guangzhouensis shares similar beta conidia size with D.pseudobauhiniae that are shorter and wider than D.bauhiniae (17.0–29.5 × 1.0–2.0 μm in D.guangzhouensis vs. 18–38 × 1.5–2 μm in D.pseudobauhiniae vs. 25–43 × 1 µm in D.bauhiniae) (Senanayake et al. 2017; Yang et al. 2021). Another species named Phomopsisbauhiniae was recorded on the branches of Bauhiniavariegata in Spain, however, this species was only studied in morphology and has not been combined in Diaporthe (Uecker 1988). Diaporthebauhiniicola has shorter but wider alpha conidia than P.bauhiniae morphologically (Uecker 1988). The molecular analyses are necessary for the species P.bauhiniae based on the ex-type culture in the future.
The initial species concept of Diaporthe based on the assumption of host-specificity, resulted in the introduction of more than 1000 taxa (http://www.indexfungorum.org/). However, more than one species of Diaporthe have been often discovered from the same host (Gomes et al. 2013; Guarnaccia and Crous 2017; Guarnaccia et al. 2020; Guo et al. 2020). For example, D.caryae and an additional 18 Diaporthe species are associated with pear shoot canker in China (Guo et al. 2020); D.sennae and D.sennicola inhabit branches of Sennabicapsularis causing canker diseases (Yang et al. 2017). The current study further supports this phenomenon.
Diaporthe is considered as a species-rich genus. Nevertheless, an emerging perspective posits that the quantity of recognized Diaporthe species may have been substantially overestimated. The D.amygdali species complex has been proven a single species evidenced from the genealogical concordance phylogenetic species recognition principle (GCPSR) and coalescence-based models: general mixed yule-coalescent (GMYC) and poisson tree processes (PTP), with several species becoming synonyms (Hilário et al. 2021b). Similarly, several species in the D.eres species complex such as D.betulae and D.padina were treated as synonyms (Hilário et al. 2021a). A comprehensive study is necessary to clarify species boundaries of Diaporthe in the future. This will help improve our understanding of the species concept within this genus.
Supplementary Material
Citation
Zhu Y, Ma L, Xue H, Li Y, Jiang N (2024) New species of Diaporthe (Diaporthaceae, Diaporthales) from Bauhinia variegata in China. MycoKeys 108: 317–335. https://doi.org/10.3897/mycokeys.108.128983
Funding Statement
This study was supported by Fundamental Research Funds of CAF (CAFYBB2023PA002), and the National Microbial Resource Center of the Ministry of Science and Technology of the People’s Republic of China (NMRC-2023-7).
Additional information
Conflict of interest
The authors have declared that no competing interests exist.
Ethical statement
No ethical statement was reported.
Funding
This study was supported by Fundamental Research Funds of CAF (CAFYBB2023PA002).
Author contributions
Conceptualization: LM, YL, NJ. Methodology: YZ. Formal analysis: HX. Investigation: YL. Data Curation: LM, HX. Writing - Original draft: YZ. Writing - Review and Editing: NJ. Visualization: NJ.
Author ORCIDs
Yaquan Zhu https://orcid.org/0000-0002-3296-239X
Han Xue https://orcid.org/0000-0003-0414-6237
Yong Li https://orcid.org/0000-0002-4406-1329
Ning Jiang https://orcid.org/0000-0002-9656-8500
Data availability
All of the data that support the findings of this study are available in the main text.
Reference
- Ariyawansa HA, Tsai I, Wang JY, Withee P, Tanjira M, Lin SR, Suwannarach N, Kumla J, Elgorban AM, Cheewangkoon R. (2021) Molecular phylogenetic diversity and biological characterization of Diaporthe species associated with leaf spots of Camelliasinensis in Taiwan. Plants 10(7): 1434. 10.3390/plants10071434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Q, Zhai LF, Chen XR, Hong N, Xu WX, Wang GP. (2015) Biological and molecular characterization of five Phomopsis species associated with pear shoot canker in China. Plant Disease 99(12): 1704–1712. 10.1094/PDIS-03-15-0259-RE [DOI] [PubMed] [Google Scholar]
- Caio P, Bruno F, Carlos AP, Robert B. (2021) Diaportherosiphthora sp. nov.: Yet another rose dieback fungus. Crop Protection (Guildford, Surrey) 139: 105365. 10.1016/j.cropro.2020.105365 [DOI]
- Cao L, Luo D, Lin W, Yang Q, Deng X. (2022) Four new species of Diaporthe (Diaporthaceae, Diaporthales) from forest plants in China. MycoKeys 91: 25–47. 10.3897/mycokeys.91.84970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone I, Kohn LM. (1999) A Method for designing primer sets for speciation studies in filamentous Ascomycetes. Mycologia 91(3): 553–556. 10.1080/00275514.1999.12061051 [DOI] [Google Scholar]
- Crous PW. (2005) Impact of molecular phylogenetics on the taxonomy and diagnostics of fungi. Bulletin OEPP. EPPO Bulletin. European and Mediterranean Plant Protection Organisation 35(1): 47–51. 10.1111/j.1365-2338.2005.00811.x [DOI] [Google Scholar]
- Crous PW, Groenewald JZ, Risède JM, Simoneau P, Hywel-Jones NL. (2004) Calonectria species and their Cylindrocladium anamorphs: Species with sphaeropedunculate vesicles. Studies in Mycology 50: 415–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Roux JJ, Richardson DM, Strasberg D, Shivas RG, Alvarado P, Edwards J, Moreno G, Sharma R, Sonawane MS, Tan YP, Altés A, Barasubiye T, Barnes CW, Blanchette RA, Boertmann D, Bogo A, Carlavilla JR, Cheewangkoon R, Daniel R, de Beer ZW, Yáñez-Morales MJ, Duong TA, Fernández-Vicente J, Geering ADW, Guest DI, Held BW, Heykoop M, Hubka V, Ismail AM, Kajale SC, Khemmuk W, Kolařík M, Kurli R, Lebeuf R, Lévesque CA, Lombard L, Magista D, Manjón JL, Marincowitz S, Mohedano JM, Nováková A, Oberlies NH, Otto EC, Paguigan ND, Pascoe IG, Pérez-Butrón JL, Perrone G, Rahi P, Raja HA, Rintoul T, Sanhueza RMV, Scarlett K, Shouche YS, Shuttleworth LA, Taylor PWJ, Thorn RG, Vawdrey LL, Solano-Vidal R, Voitk A, Wong PTW, Wood AR, Zamora JC, Groenewald JZ. (2015) Fungal Planet description sheets: 371–399. Persoonia 35(1): 264–327. 10.3767/003158515X690269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dissanayake AJ, Phillips AJL, Hyde KD, Yan JY, Li XH. (2017) The current status of species in Diaporthe. Mycosphere : Journal of Fungal Biology 8(5): 1106–1156. 10.5943/mycosphere/8/5/5 [DOI]
- Dissanayake AJ, Chen YY, Liu JK. (2020) Unravelling Diaporthe species associated with woody hosts from karst formations (Guizhou) in China. Journal of Fungi (Basel, Switzerland) 6(4): 251. 10.3390/jof6040251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Z, Manawasinghe IS, Huang Y, Shu Y, Phillips AJL, Dissanayake AJ, Hyde KD, Xiang M, Luo M. (2021) Endophytic Diaporthe associated with Citrusgrandis cv. tomentosa in China. Frontiers in Microbiology 11: e3621. 10.3389/fmicb.2020.609387 [DOI] [PMC free article] [PubMed]
- Doyle JJ, Doyle JL. (1990) Isolation of plant DNA from fresh tissue. Focus 12: 13–15. [Google Scholar]
- Fan XL, Yang Q, Bezerra JDP, Alvarez LV, Tian CM. (2018) Diaporthe from walnut tree (Juglansregia) in China, with insight of Diaportheeres complex. Mycological Progress 17(7): 1–13. 10.1007/s11557-018-1395-4 [DOI] [Google Scholar]
- Farr DF, Castlebury LA, Rossman AY. (2002a) Morphological and molecular characterization of Phomopsisvaccinii and additional isolates of Phomopsis from blueberry and cranberry in the eastern United States. Mycologia 94(3): 494–504. 10.1080/15572536.2003.11833214 [DOI] [PubMed] [Google Scholar]
- Farr DF, Castlebury LA, Rossman AY, Putnam ML. (2002b) A new species of Phomopsis causing twig dieback of Vacciniumvitisidaea (lingonberry). Mycological Research 106(6): 745–752. 10.1017/S095375620200583X [DOI] [Google Scholar]
- Glass NL, Donaldson GC. (1995) Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Applied and Environmental Microbiology 61(4): 1323–1330. 10.1128/aem.61.4.1323-1330.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes RR, Glienke C, Videira SIR, Lombard L, Groenewald JZ, Crous PW. (2013) Diaporthe: A genus of endophytic, saprobic and plant pathogenic fungi. Persoonia 31(1): 1–41. 10.3767/003158513X666844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarnaccia V, Crous PW. (2017) Emerging citrus diseases in Europe caused by Diaporthe spp. IMA Fungus 8(2): 317–334. 10.5598/imafungus.2017.08.02.07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarnaccia V, Martino I, Tabone G, Brondino L, Gullino ML. (2020) Fungal pathogens associated with stem blight and dieback of blueberry in northern Italy. Phytopathologia Mediterranea 59(2): 229–245. 10.14601/Phyto-11278 [DOI] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Systematic Biology 59(3): 307–321. 10.1093/sysbio/syq010 [DOI] [PubMed] [Google Scholar]
- Guo YS, Crous PW, Bai Q, Fu M, Yang MM, Wang XH, Du YM, Hong N, Xu WX, Wang GP. (2020) High diversity of Diaporthe species associated with pear shoot canker in China. Persoonia 45(1): 132–162. 10.3767/persoonia.2020.45.05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall T. (1999) BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41: 95–98. [Google Scholar]
- Hilário S, Micael FM, Artur A. (2021a) Using genealogical concordance and coalescent-based species delimitation to assess species boundaries in the Diaportheeres complex. Journal of Fungi 7(7): 507. 10.3390/jof7070507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilário S, Santos L, Alves A. (2021b) Diaportheamygdali, a species complex or a complex species? Fungal Biology 125(7): 505–518. 10.1016/j.funbio.2021.01.006 [DOI] [PubMed]
- Hongsanan S, Norphanphoun C, Senanayake IC, Jayawardena RS, Manawasinghe IS, Abeywickrama PD, Khuna S, Suwannarach N, Senwanna C, Monkai J, Hyde KD, Gentekaki E, Bhunjun CS. (2023) Annotated notes on Diaporthe species. Mycosphere 14(1): 918–1189. 10.5943/mycosphere/14/1/12 [DOI] [Google Scholar]
- Huang F, Udayanga D, Wang X, Hou X, Mei X, Fu Y, Hyde KD, Li HY. (2015) Endophytic Diaporthe associated with Citrus: A phylogenetic reassessment with seven new species from China. Fungal Biology 119(5): 331–347. 10.1016/j.funbio.2015.02.006 [DOI] [PubMed] [Google Scholar]
- Huang ST, Xia JW, Zhang XG, Sun WX. (2021) Morphological and phylogenetic analyses reveal three new species of Diaporthe from Yunnan, China. MycoKeys 78: 49–77. 10.3897/mycokeys.78.60878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde KD, Nilsson RH, Alias SA, Ariyawansa HA, Blair JE, Cai L, De Cock AWAM, Dissanayake AJ, Glockling SL, Goonasekara ID, Gorczak M, Hahn M, Jayawardena RS, Van Kan JAL, Laurence MH, Lévesque CA, Li XH, Liu JK, Maharachchikumbura SSN, Manamgoda DS, Martin FN, McKenzie EHC, McTaggart AR, Mortimer PE, Nair PVR, Pawłowska J, Rintoul TL, Shivas RG, Spies CFJ, Summerell BA, Taylor PWJ, Terhem RB, Udayanga D, Vaghefi N, Walther G, Wilk M, Wrzosek M, Xu JX, Yan JY, Zhou N. (2014) One stop shop: backbones trees for important phytopathogenic genera: I. Fungal Diversity 67(1): 21–125. 10.1007/s13225-014-0298-1 [DOI] [Google Scholar]
- Jiang N, Voglmayr H, Piao CG, Li Y. (2021) Two new species of Diaporthe (Diaporthaceae, Diaporthales) associated with tree cankers in the Netherlands. MycoKeys 85: 31–56. 10.3897/mycokeys.85.73107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. (2013) MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Molecular Biology and Evolution 30(4): 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maharachchikumbura SSN, Hyde KD, Jones EBG, McKenzie EHC, Bhat JD, Dayarathne MC, Huang SK, Norphanphoun C, Senanayake IC, Perera RH, Shang QJ, D’souza MJ, Hongsanan S, Jayawardena RS, Daranagama DA, Konta S, Goonasekara ID, Zhuang WY, Jeewon R, Phillips AJL, Abdel-Wahab MA, Al-Sadi AM, Bahkali AH, Boonmee S, Boonyuen N, Cheewangkoon R, Dissanayake AJ, Kang J, Li QR, Liu JK, Liu XZ, Liu ZY, Luangsa-ard JJ, Pang KL, Phookamsak R, Promputtha I, Suetrong S, Stadler M, Wen T, Wijayawardene NN. (2016) Families of Sordariomycetes. Fungal Diversity 79(1): 1–317. 10.1007/s13225-016-0369-6 [DOI] [Google Scholar]
- Manawasinghe IS, Dissanayake AJ, Li X, Liu M, Wanasinghe DN, Xu J, Zhao W, Zhang W, Zhou Y, Hyde KD, Brooks S, Yan J. (2019) High genetic diversity and species complexity of Diaporthe associated with grapevine dieback in China. Frontiers in Microbiology 10: 1936. 10.3389/fmicb.2019.01936 [DOI] [PMC free article] [PubMed]
- Marin-Felix Y, Hernandez-Restrepo M, Wingfield MJ, Akulov A, Carnegie AJ, Cheewangkoon R, Gramaje D, Groenewald JZ, Guarnaccia V, Halleen F, Lombard L, Luangsa-ard J, Marincowitz S, Moslemi A, Mostert L, Quaedvlieg W, Schumacher RK, Spies CFJ, Thangavel R, Taylor PWJ, Wilson AM, Wingfield BD, Wood AR, Crous PW. (2019) Genera of phytopathogenic fungi: GOPHY 2. Studies in Mycology 92(1): 47–133. 10.1016/j.simyco.2018.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostert L, Crous PW, Kang J-C, Phillips AJL. (2001) Species of Phomopsis and a Libertella sp. occurring on grapevines with specific reference to South Africa: Morphological, cultural, molecular and pathological characterization. Mycologia 93(1): 146–167. 10.1080/00275514.2001.12061286 [DOI] [Google Scholar]
- Norphanphoun C, Gentekaki E, Hongsanan S, Jayawardena R, Senanayake C, Manawasinghe I, Abeywickrama P, Bhunjun CS, Hyde KD. (2022) Diaporthe: Formalizing species-group concepts. Mycosphere 13(1): 752–819. 10.5943/mycosphere/13/1/9 [DOI] [Google Scholar]
- O’Donnell K, Cigelnik E. (1997) Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Molecular Phylogenetics and Evolution 7(1): 103–116. 10.1006/mpev.1996.0376 [DOI] [PubMed] [Google Scholar]
- Rambaut A, Drummond A. (2010) FigTree v.1.3.1. Institute of Evolutionary Biology, University of Edinburgh, Edinburgh.
- Ronquist F, Huelsenbeck JP. (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19(12): 1572–1574. 10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Rossman AY, Adams GC, Cannon PF, Castlebury LA, Crous PW, Gryzenhout M, Jaklitsch WM, Mejia LC, Stoykov D, Udayanga D, Voglmayr H, Walker DM. (2015) Recommendations of generic names in Diaporthales competing for protection or use. IMA Fungus 6(1): 145–154. 10.5598/imafungus.2015.06.01.09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos JM, Phillips AJL. (2009) Resolving the complex of Diaporthe (Phomopsis) species occurring on Foeniculumvulgare in Portugal. Fungal Diversity 34(11): 111–125. [Google Scholar]
- Santos L, Alves A, Alves R. (2017) Evaluating multi-locus phylogenies for species boundaries determination in the genus Diaporthe. PeerJ 5: e3120. 10.7717/peerj.3120 [DOI] [PMC free article] [PubMed]
- Senanayake IC, Crous PW, Groenewald JZ, Maharachchikumbura SSN, Jeewon R, Phillips AJL, Bhat DJ, Perera RH, Li QR, Li WJ, Tangthirasunun N, Norphanphoun C, Karunarathna SC, Camporesi E, Manawasighe IS, Al-Sadi AM, Hyde KD. (2017) Families of Diaporthales based on morphological and phylogenetic evidence. Studies in Mycology 86(1): 217–296. 10.1016/j.simyco.2017.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senanayake IC, Jeewon R, Chomnunti P, Wanasinghe DN, Norphanphoun C, Karunarathna A, Pem D, Perera RH, Camporesi E, McKenzie EHC, Hyde KD, Karunarathna SC. (2018) Taxonomic circumscription of Diaporthales based on multigene phylogeny and morphology. Fungal Diversity 93(1): 241–443. 10.1007/s13225-018-0410-z [DOI] [Google Scholar]
- Sun W, Huang S, Xia J, Zhang X, Li Z. (2021) Morphological and molecular identification of Diaporthe species in south-western China, with description of eight new species. MycoKeys 77: 65–95. 10.3897/mycokeys.77.59852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JW. (2011) One Fungus = One Name: DNA and fungal nomenclature twenty years after PCR. IMA Fungus 2(2): 113–120. 10.5598/imafungus.2011.02.02.01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udayanga D, Liu X, Crous PW, McKenzie EH, Chukeatirote E, Chukeatirote E, Hyde KD. (2012a) A multi-locus phylogenetic evaluation of Diaporthe (Phomopsis). Fungal Diversity 56(1): 157–171. 10.1007/s13225-012-0190-9 [DOI] [Google Scholar]
- Udayanga D, Liu X, McKenzie EH, Chukeatirote E, Hyde KD. (2012b) Multi-locus phylogeny reveals three new species of Diaporthe from Thailand. Cryptogamie. Mycologie 33(3): 295–309. 10.7872/crym.v33.iss3.2012.295 [DOI] [Google Scholar]
- Udayanga D, Castlebury LA, Rossman AY, Hyde KD. (2014a) Species limits in Diaporthe: Molecular re-assessment of D.citri, D.cytosporella, D.foeniculina and D.rudis. Persoonia 32(1): 83–101. 10.3767/003158514X679984 [DOI] [PMC free article] [PubMed]
- Udayanga D, Castlebury LA, Rossman AY, Chukeatirote E, Hyde KD. (2014b) Insights into the genus Diaporthe: Phylogenetic species delimitation in the D.eres species complex. Fungal Diversity 67(1): 203–229. 10.1007/s13225-014-0297-2 [DOI] [Google Scholar]
- Udayanga D, Castlebury LA, Rossman AY, Chukeatirote E, Hyde KD. (2015) The Diaporthesojae species complex: Phylogenetic re-assessment of pathogens associated with soybean, cucurbits and other field crops. Fungal Biology 119(5): 383–407. 10.1016/j.funbio.2014.10.009 [DOI] [PubMed] [Google Scholar]
- Uecker FA. (1988) A world list of Phomopsis names with notes on nomenclature, morphology and biology. Morphology and Biology 13: 1–231. [Google Scholar]
- van Rensburg JCJ, Lamprecht SC, Groenewald JZ, Castlebury LA, Crous PW. (2006) Characterization of Phomopsis spp. associated with dieback of rooibos (Aspalathuslinearis) in South Africa. Studies in Mycology 55: 65–74. 10.3114/sim.55.1.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrandecic K, Jurkovic D, Cosic J, Postic J, Riccioni L. (2011) First report of cane blight on blackberry caused by Diaportheeres in Croatia. Plant Disease 95(5): 612–612. 10.1094/PDIS-11-10-0860 [DOI] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protocols: A Guide to Methods and Applications 18: 315–322. 10.1016/B978-0-12-372180-8.50042-1 [DOI] [Google Scholar]
- Wrona CJ, Mohankumar V, Schoeman MH, Tan YP, Shivas RG, Jeff‐Ego OS, Akinsanmi OA. (2020) Phomopsis husk rot of macadamia in Australia and South Africa caused by novel Diaporthe species. Plant Pathology 69(5): 911–921. 10.1111/ppa.13170 [DOI] [Google Scholar]
- Yang Q, Fan XL, Du Z, Tian CM. (2017) Diaporthe species occurring on Sennabicapsularis in southern China, with descriptions of two new species. Phytotaxa 302(2): 145–155. 10.11646/phytotaxa.302.2.4 [DOI] [Google Scholar]
- Yang Q, Fan XL, Guarnaccia V, Tian CM. (2018) High diversity of Diaporthe species associated with dieback diseases in China, with twelve new species described. MycoKeys 39: 97–149. 10.3897/mycokeys.39.26914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Jiang N, Tian CM. (2020) Three new Diaporthe species from Shaanxi Province, China. MycoKeys 67: 1–18. 10.3897/mycokeys.67.49483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Jiang N, Tian CM. (2021) New species and records of Diaporthe from Jiangxi Province, China. MycoKeys 77: 41–64. 10.3897/mycokeys.77.59999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang QM, Yu CL, Li GF, Wang CX. (2018) First report of Diaportheeres causing twig canker on Zizyphusjujuba (Jujube) in China. Plant Disease 102(7): e1458. 10.1094/PDIS-12-17-1910-PDN [DOI]
- Zhu YQ, Ma CY, Xue H, Piao CG, Li Y, Jiang N. (2023) Two new species of Diaporthe (Diaporthaceae, Diaporthales) in China. MycoKeys 95: 209–228. 10.3897/mycokeys.95.98969 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All of the data that support the findings of this study are available in the main text.