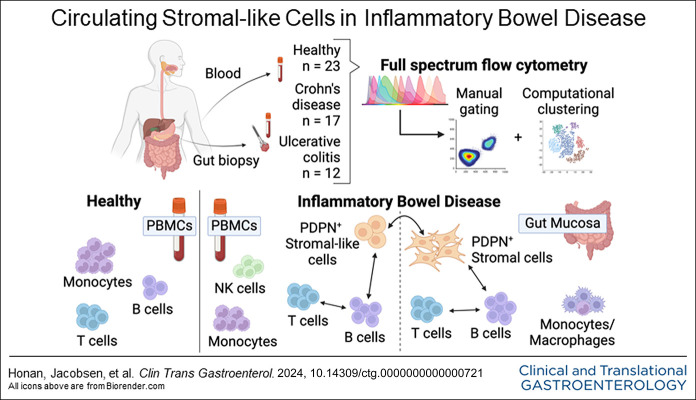
Abstract
INTRODUCTION:
Recent studies have identified a critical role of stromal-immune cell interactions in immunity and immune tolerance. Transcriptomic profiling has implicated stromal cells in immune-mediated disorders including the 2 common forms of inflammatory bowel disease (IBD), Crohn's disease (CD), and ulcerative colitis (UC). Stromal-immune interactions may edify inflammatory state and the development of IBD-related complications such as fibrosis, yet the lack of protein markers has hampered studying stromal-immune perturbation.
METHODS:
In this study, we designed a 40-color spectral flow cytometry assay to characterize hematopoietic and nonhematopoietic cells in intestinal biopsies and matched blood samples from patients with CD or UC.
RESULTS:
We identified circulating stromal-like cells that are significantly more abundant in IBD blood samples than in healthy controls. Those cells expressed podoplanin (PDPN), a commonly used marker for fibroblasts, and they were associated with activated and memory T and B cells and altered natural killer cell, monocyte, and macrophage populations. PDPN+ cells in the blood correlated with PDPN+ cells in the colon. Principal component analysis distinctly separated healthy blood samples from IBD blood samples, with stromal-like cells and B-cell subtypes dominating the IBD signature; Pearson correlation detected an association between PDPN+ stromal-like cells and B-cell populations in IBD blood and gut biopsies.
DISCUSSION:
These observations suggest that PDPN+ cells in the blood may serve as a biomarker of IBD. Understanding the relationship between stromal cells and immune cells in the intestine and the blood may provide a window into disease pathogenesis and insight into therapeutic targets for IBD.
KEYWORDS: inflammatory bowel disease, stromal cells, immune cells, spectral flow cytometry
INTRODUCTION
Inflammatory bowel disease (IBD) is an immune-mediated intestinal disorder characterized by chronic inflammation in the absence of a known pathogen. The 2 most common forms of IBD are Crohn's disease (CD), which manifests as patchy, often transmural lesions anywhere along the digestive tract, and ulcerative colitis (UC), consisting of continuous mucosal inflammation limited to the colon. The exact cause of IBD is not fully understood, but a number of contributing factors are implicated including genetic predisposition, mucosal barrier dysfunction, disturbance in the gastrointestinal microbiota, and lifestyle that interplay with innate and adaptive immune dysregulation (1,2) as well as disruption of stromal compartment (3).
Recently, researchers have explored a new dimension of immune regulation by focusing on the role of the stromal compartment in controlling or perpetuating immune-mediated diseases (4,5). Stromal cells are known for their role in providing structural and nutrient support to lymphoid and nonlymphoid tissues. They have also been shown to play a role in shaping the organ microenvironment through interactions with immune cells. Stromal cells shape T-cell maturation and differentiation in central and peripheral lymphoid organs and thus, control autoimmune and other immune-mediated diseases (6,7). In the clinical setting, recent studies have highlighted a critical role of stromal cells, such as fibroblasts, in mediating rheumatoid arthritis (RA) (8). It has been suggested that they may also be involved in pathogenesis of other immune-mediated diseases, including IBD (9).
Most IBD therapies have focused on decreasing inflammation in the gut by manipulating the immune system, specifically targeting cytokine pathways and T cell homing to the intestine (10). However, accumulating evidence also highlights the important role of the nonhematopoietic compartment in IBD pathogenesis. Mesenchymal stem cells, which constitute the majority of stromal cells in the bone marrow supporting hematopoiesis, are being used to treat refractory perianal fistulas in CD (3). Therapeutically introduced mesenchymal stem cells may promote tissue repair while potentially modulating immune responses. Furthermore, single-cell RNA sequencing studies of CD and UC gut tissue samples have implicated stromal cell lineages within lesions, including podoplanin (PDPN)-expressing fibroblasts (11–13). These stromal cells are also believed to contribute to refractoriness to anti-TNF therapy in both CD and UC (12,13). Thus, stromal cell pathways may serve as targets or biomarkers for therapeutic interventions (3).
For our study, we reasoned that a deeper investigation of stromal and immune cells in matched blood and tissue could yield important information about the contribution of stromal cells to the pathobiology of CD and UC. By directly comparing gut mucosal cells and peripheral blood cells collected simultaneously, we aimed to uncover circulating cellular or molecular markers that reflect inflammatory stromal-immune interactions within the gut tissue microenvironment. We also aimed to characterize immune cell subsets in the blood matched with intestinal samples of patients with IBD using advanced full spectral flow cytometry to uncover novel cell populations. We used spectral flow cytometry to facilitate high throughput characterization of 40+ parameters at the single-cell level. We characterized the phenotypes of major stromal and immune cell subtypes and compared populations by disease state (CD, UC, or healthy) and inflammatory state (inflamed or uninflamed). We uncovered evidence of stromal-immune interactions in the gut mucosa and intriguing populations of stromal-like cells in the blood circulation of patients with IBD that correlate with immune perturbation.
METHODS
Sample collection
The use of human samples for this study was approved by the University of Miami Institutional Review Board (IRB ID: 20081100). Samples were obtained from patients with CD and UC who provided informed consent for the IBD Clinical Phenotype Database and Specimen Collection. Demographic characteristics of patients with IBD can be found in Table 1. Peripheral blood was collected by venipuncture into 1 heparin tube per patient. Six biopsies of approximately 3 mm diameter were obtained from the terminal ileum, ascending colon, and/or sigmoid colon of each patient. Cold single-use radial jaw jumbo forceps with needle (Boston Scientific, Marlborough, MA) were used to retrieve 2 biopsies at a time from adjacent locations. Mucosal biopsies were placed in hypothermosol solution (MilliporeSigma, St. Louis, MO) at 4°C for preservation and processed within 6 hours of collection. Adjacent biopsies from each location were sent to pathologists as part of standard patient care; the subsequent pathology reports were used to characterize biopsies. Biopsy samples were designated inflamed or uninflamed based on a combination of endoscopic signs and histologic findings, such as increased lymphocytes and other reactive changes. Separately from the IRB protocol, deidentified healthy control blood samples (buffy coat) were obtained from a local blood bank located in the same geographic area for which the University hospital serves and the donors were screened for health status and tested for infectious agents per the standard blood bank policy. Healthy donors were recruited by the blood blank without any input from the researchers. The healthy donor and IBD patient groups had a similar median age ∼50 years and similar age ranges ∼20–∼80 years. The sex distribution of the healthy blood donor, 57% men and 43% women was similar to that of the IBD patient group, 55% men and 45% women.
Table 1.
IBD patient demographics and clinical characteristics
| Total | N = 29 |
| Sex, n (%) | |
| Male | 16 (55) |
| Female | 13 (45) |
| Ethnicity, n (%) | |
| Hispanic | 12 (41) |
| Non-Hispanic | 17 (59) |
| Race, n (%) | |
| White | 24 (83) |
| Black | 3 (10) |
| Asian | 1 (3) |
| Other | 1 (3) |
| Current BMI, median | 25.3 |
| Current age, yr, median | 47 |
| Years of disease, yr, median | 17 |
| Current inflammation, n (%) | 12 (41) |
| Inflamed total SES-CD, median | 4.5 |
| Inflamed total Mayo, median | 2 |
| CD, n (%) | 17 (58) |
| Disease location, n (%) | |
| Ileum only | 7 (24) |
| Ileum and colon | 9 (31) |
| Colon only | 1 (3) |
| Disease behavior, n (%) | |
| Inflammatory | 12 (41) |
| Fibrostenotic | 7 (24) |
| Penetrating/perforating | 4 (14) |
| UC, n (%) | 12 (41) |
| Disease location, n (%) | |
| Pancolitis | 5 (17) |
| Left sided | 7 (24) |
| Disease severity, n (%) | |
| Mild | 3 (10) |
| Moderate | 9 (31) |
| Current medications, n (%) | |
| None | 2 (7) |
| Aminosalicylates | 6 (21) |
| Steroids | 0 (0) |
| Immunomodulators | 3 (10) |
| Biologics | 23 (79) |
| Anti-TNF | 12 (41) |
| Anti-integrin | 5 (17) |
| Anti-IL-12/23 | 5 (17) |
| JAK inhibitor | 1 (3) |
BMI, body mass index; CD, Crohn's disease; IBD, inflammatory bowel disease; IL, interleukin; JAK, Janus kinase; SES-CD, Simple Endoscopic Score for CD; TNF, tumor necrosis factor.
Biopsy and blood sample processing into single-cell suspensions
Mucosal biopsies, about 3 mm each, were shaken in Dulbecco's Modified Eagle Medium with 10 mM dithiothreitol and 0.5 mM ethylenediaminetetraacetic acid for 20 minutes to remove the majority of intestinal epithelial cells. Lamina propria cells were then dissociated by digesting tissue in Dulbecco's Modified Eagle Medium with 250 μg/mL Liberase (MilliporeSigma) and 10 μg/mL DNase I (Lucigen Corporation, Middleton, WI) at 37°C for 30 minutes. Digested tissue was further mechanically dissociated by pipetting and filtered using a 70 μm strainer to obtain a single-cell suspension. Peripheral blood mononuclear cells (PBMCs) were isolated from IBD and healthy control blood samples using the same standard laboratory protocol with the same type of Ficoll gradient. Both IBD and healthy control PBMCs were cryopreserved in RPMI with 10% human AB serum and 10% dimethylsulfoxide and thawed in batches for flow cytometry.
Full spectrum flow cytometry
We developed a 40-color flow cytometry panel for comprehensive immunophenotyping using Cytek Aurora. Single-cell preparation and flow cytometry staining were performed using a standard procedure (14). Cells were blocked from nonspecific binding using a cocktail of anti-CD16/32 and normal mouse serum. The cells were then stained with fluorescent-antibody conjugates to determine cell phenotype. For intracellular staining, the Transcription Factor Buffer Set (BD Bioscience) was used for fixation and permeabilization of cells after staining for cell surface markers. Zombie NIR (Biolegend) was used to exclude dead cells from analyses. Doublets were gated out before further analysis of marker expression by single cells. Isotype controls were used to assess nonspecific binding of fixed and permeabilized cells by the antibody reagents for intracellular markers. Fluorescent minus one (FMO) controls were implemented by staining a sample with all fluorochromes except one. This allows for the upper boundary of background signal to be determined for the omitted fluorochrome. FMO controls also assisted in determining potential spillover due to fluorochromes with similar spectra. All manual gates were set based on FMO controls before the analysis of samples. Gating and analysis were performed using FCS Express 7 software.
viSNE and SPADE clustering analysis
Spectral flow cytometry data were analyzed using the Cytobank platform. Hematopoietic and nonhematopoietic cell population FCS files were pregated using FCS Express software. Hematopoietic cell analysis was conducted on live/dead− CD45+ cells, and nonhematopoietic cell analysis was conducted on live/dead− CD45−CD3− CD19− cells. Data were first analyzed using viSNE dimensionality reducing analysis on the Cytobank software. viSNE analysis transformed FCS data into 2 dimensions using the Barnes-Hut implementation of the t-distributed stochastic neighbor embedding (tSNE) algorithm. Analysis was performed on the samples using proportional sampling, with 7,500 iterations, a perplexity of 30, and a θ of 0.5, following Cytobank user guides and similar procedures reported in a previous study (15). The following markers were used as channels for hematopoietic cell viSNE analysis: CCR7, CD103, CD11b, CD11c, CD127, CD14, CD15, CD16, CD19, CD200, CD206, CD25, CD27, CD3, CD4, CD45RA, CD56, CD64, CD66b, CD8, CD86, cytotoxic T-lymphocyte-associated protein 4 (CTLA4), forkhead box P3 (FOXP3), GATA-binding protein 3, human leukocyte antigen DR (HLA-DR), IgD, PD-1, PD-L1, RAR-related orphan receptor gamma, T-BET, TCRαβ, and TCRγδ. The following markers were used as channels for nonhematopoietic cell viSNE analysis CD31, fibroblast activation protein (FAP), HLA-DR, PD-L1, PDPN1, and smooth muscle actin alpha (SMAα) as clustering markers. Spanning-tree Progression Analysis of Density-normalized Events (SPADE) clustering (16) was done on the viSNE analysis. Hematopoietic cell populations were clustered based on dimensionality reduction channels (tSNE1 and tSNE2) followed with marker identification in the clusters of the resulting viSNE analysis tSNE plots. Nonhematopoietic SPADE analysis was conducted using FAP, HLA-DR, SMAα, PD-1, CCR7, PDPN, PD-L1, CD31, and epithelial cell adhesion molecule (EPCAM) as clustering channels. SPADE trees were manually analyzed, and nodes were gated based on marker expression. Gates were then overlaid on the viSNE plots for visualization.
Statistical analyses
All statistical calculations and graphs were made using GraphPad Prism (GraphPad Software, San Diego, CA) including the Pearson correlation tests for correlations between stromal cell, T-cell, and B-cell subsets for potential evidence of stromal-immune interaction. Two-way comparisons were made using unpaired t test for parametric and Mann-Whitney test for nonparametric comparisons. Welch correction was added to the unpaired t test for samples with significantly different standard deviations. Three-way comparisons were made using ordinary 1-way ANOVA for parametric and the Kruskal-Wallis test for nonparametric comparisons. Principal component analysis (PCA) was calculated using normalized cell counts for 110 PBMC cell subtypes and 101 tissue cell subtypes as variables. Only cell subtypes with >0 counts in at least 5 samples were included as variables. Simple linear regression was used for correlation analyses. *P < 0.05 **, P < 0.01 ***, P < 0.001, ****P < 0.0001.
RESULTS
Patient characteristics
Twenty-three (n = 23) healthy control blood samples were deidentified of all demographic data before use in the study. Twenty-nine (n = 29) patients with IBD were enrolled in the study (Table 1); all 29 provided gut mucosal biopsies and 26 provided blood for PBMC isolation. Notably, a large proportion (n = 12, 41%) of the patients were Hispanic, half of whom were foreign born. Patients were considered inflamed through a combination of endoscopic scoring (total Simple Endoscopic Score for CD >2 or total Mayo ≥2) and pathologist impression of histologic findings (e.g., increased lymphoplasmacytic infiltrate). Almost half of the patients (n = 12, 41%) had inflammation at the time of sample collection. Of these 12 inflamed patients, 10 were on medication, indicating some level of resistance to treatment. Seven of 17 patients with CD also had fibrostenotic disease. The current patient demographic and IBD disease phenotype reflects the spectrum of IBD and the unmet need in treating IBD.
Increased PDPN+ stromal-like cells in peripheral blood of patients with IBD and the relationship to intestinal stromal cell populations
CD45 is a broad marker of hematopoietic cells and was used for initial gating of cell populations. First, we analyzed CD45– cells (nonhematopoietic lineages) in both blood and intestinal tissue. In blood, the abundance of total CD45– cells was similar between CD, UC, and healthy PBMC samples (Figure 1a and b). We then characterized these cells based on PDPN and CD31 expression to distinguish fibroblast-like and endothelial cells, respectively (Figure 1a). PDPN is a mucin-like transmembrane protein and a commonly used marker to identify fibroblast populations within nonhematopoietic lineage (17–20), while CD31 is constitutively expressed by endothelial cells (21). We found that patients with IBD (both CD and UC) had increased numbers of circulating PDPN+CD31– stromal-like cells compared with healthy controls (Figure 1a and b).
Figure 1.
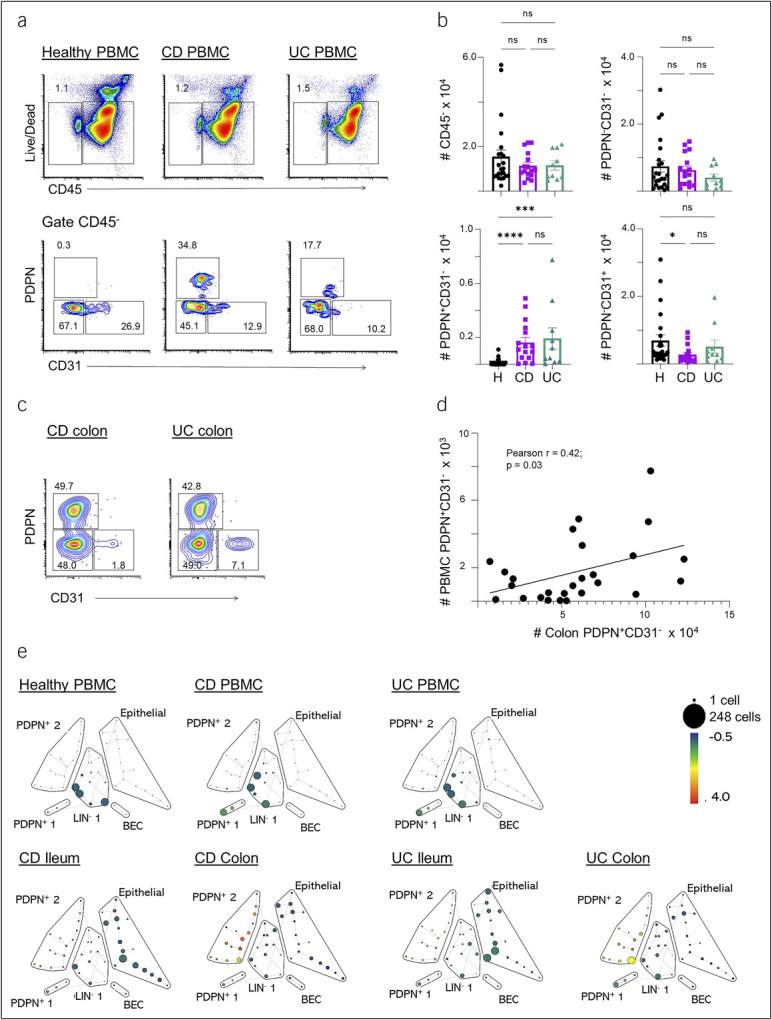
Characterization of stromal cells in the blood, ileum, and colon of patients with IBD. (a) Representative flow cytometry plots of the CD45– cell population and its subsets based on PDPN and CD31 markers in healthy control (left), CD (middle), and UC (right) blood samples. (b) Summary cell counts for total CD45– population and the PDPN+CD31–, PDPN−CD31– and PDPN–CD31+ subsets. (c) Representative flow cytometry plots of the stromal cell populations (gated single cells, live CD45–CD3–CD19–) from the colon tissue from patients with CD and UC. (d) Correlation analysis of PDPN+CD31– cell numbers in PBMCs and colon biopsies. Cell counts are normalized to counts per million. The number in the flow cytometry plots is the gated percentage. Each data point represents 1 sample (mean ± SEM, n = 10–26), *P < 0.05, ***P < 0.001, ****P < 0.0001. (e) SPADE tree derived from a 40-marker spectral flow cytometry panel depicting the nonhematopoietic compartment from healthy controls or IBD patient samples. The tree is colored by the signal intensity for PDPN. Cell counts are scaled by size of the filled circles (“nodes”). Representative plots of the nonhematopoietic cells found in PBMCs (top row) and in the colon and ileum of patients with CD and UC (bottom row) are shown. BEC: blood endothelial cells (PDPN–CD31+); LIN–: (PDPN–CD31–EpCAM–). CD, Crohn's disease; IBD, inflammatory bowel disease; PBMC, peripheral blood mononuclear cell; PDPN, podoplanin; SEM, standard error of mean; SPADE, Spanning-tree Progression Analysis of Density-normalized Events; UC, ulcerative colitis.
We further examined if treatment with biologics affected the PDPN+CD31– stromal-like cell population. The vast majority of the patients had received biologics (n = 21), and only 5 patients received nonbiologics or no treatment. The PDPN+CD31– stromal-like cell number in these 2 groups were 2,662 ± 413 and 1,496 ± 800 (mean ± standard error of mean), respectively, P = 0.105. Therefore, the fact that the majority of patients were being treated with biologics limits our statistical power to determine whether the treatment with biologics affected the PDPN+ stromal-like cell population.
We next characterized CD45– populations in gut mucosal samples using the same markers. We identified both fibroblast (PDPN+CD31–) and endothelial cell (PDPN–CD31+) populations in the ileum and colon of patients with CD and UC (see Supplementary Figure 1A, Supplementary Digital Content 1, http://links.lww.com/CTG/B138). We also found an increased fibroblast (PDPN+CD31–) population in CD ileum compared with UC ileum (see Supplementary Figure 1B and C, Supplementary Digital Content 1, http://links.lww.com/CTG/B138). We compared the gut tissue samples with or without active inflammation determined by both endoscopic and histologic examination. However, we found no significant differences in the number of fibroblasts or endothelial cells between the tissue samples with or without active inflammation (see Supplementary Figure 1A, Supplementary Digital Content 1, http://links.lww.com/CTG/B138). We also did not find any correlations between the number of fibroblasts and disease phenotype, such as fibrostenotic CD. We next examined the relationship between PDPN+ cells in the blood compared with their presence in ileum or colon samples from the same patient. We found that there was a numerical correlation between PDPN+ cells in the blood with PDPN+ cells in distal colon biopsies (Figure 1c and d). The same relationship was not seen in the ileum (Figure 1 or see Supplementary Figure 1D, Supplementary Digital Content 1, http://links.lww.com/CTG/B138).
To extend our findings, we performed a high-dimensional single-cell analysis of the nonhematopoietic population using the SPADE algorithm. This allowed us to corroborate our manual gating with an unbiased computational method that clusters cell populations based on expression of defined markers (FAP, HLA-DR, SMAa, programmed cell death protein 1 [PD1], CCR7, PDPN, PDL1, CD31, EPCAM). Our SPADE analysis identified 2 clusters of PDPN+ cells: PDPN+ 1 and PDPN+ 2. Both CD and UC PBMCs had higher frequencies of the PDPN+ 1 cells than healthy controls (Figure 1e). By contrast, the PDPN+ 2 cells were more abundant in intestinal biopsies compared with blood samples. To assess the differences between PDPN+1 and PDPN+ 2, we determined the expression levels for SMAα, FAP, EPCAM, and HLA-DR (a class II major histocompatibility complex molecule specialized in antigen presentation). This analysis revealed that the PDPN+ 1 group expressed a lower level of PDPN and SMAα and a higher level of FAP, EPCAM, and HLA-DR compared with the PDPN+ 2 group (PDPNloFAPhiEPCAM+SMAα–HLADR+ and PDPNhiFAP−/loEPCAM–/loSMAα+HLADR–/lo, respectively) (see Supplementary Figure 2A–D, Supplementary Digital Content 2, http://links.lww.com/CTG/B139). These phenotypic differences could imply that the PDPN+ 1 group might represent a pathogenic fibroblast subset (22), whereas the PDPN+ 2 group might be related to myofibroblasts.
To corroborate our spectral flow cytometry results, we asked if PDPN transcripts were also differentially expressed in IBD vs healthy samples. We used the IBD Transcriptome and Metatranscriptome Meta-Analysis platform to search publicly available RNA sequencing data pooled from multiple studies (23) (see Supplementary Figure 3, Supplementary Digital Content 3, http://links.lww.com/CTG/B140). This included 1,850 ileum samples, 448 colon samples, and 209 blood samples. In the ileum, PDPN expression was significantly higher in CD compared with UC or control samples. There was no difference in expression in UC ileum compared with controls. In the colon, PDPN expression was significantly higher in both CD and UC compared with controls, as well as in UC compared with CD. These results complement what we observed in our spectral flow cytometry data and support that PDPN expression correlates with disease state, although we do not know which subset(s) of the cells contributed to the increase of PDPN expression uncovered in the analysis of the bulk RNA-seq data. While there were no significant differences in PDPN expression seen in the blood sample comparisons, we believe this is due to the relatively small number of PDPN+ cells in the blood and therefore, a low PDPN transcript count in whole blood. These data highlight that PDPN+ cells are found in peripheral blood, ileum, and colon of patients with IBD although the phenotype of these cells in the specific compartments may be distinct.
Peripheral blood T-cell profiling demonstrates a TH17 shift and increased regulatory T cells in IBD
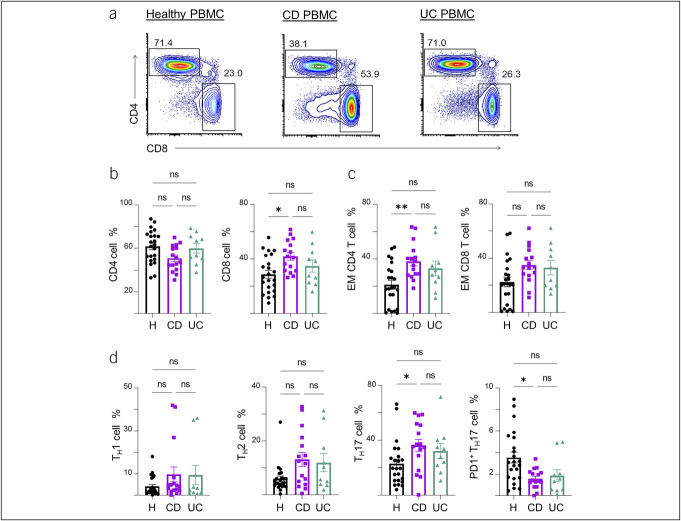
We next focused on characterizing adaptive immune cell types in IBD peripheral blood compared with healthy samples to understand the utility of spectral flow in identifying populations that may serve as biomarkers of disease state. CD45+ hematopoietic cell populations in PBMCs were examined for T-cell subsets in CD, UC, and healthy subjects. Patients with CD had a higher proportion of CD8+ cells in the blood compared with healthy controls (Figure 2a and b). We characterized T-cell memory phenotypes according to commonly used markers: naïve, CCR7+CD45RA+; central memory, CCR7+CD45RA−CD27+; effector (EFF), CCR7−CD27−; and effector memory (EM), CCR7−CD27+. For EFF and EM cells, although the majority were CD45RA−, CD45RA+ cells are included based on literature evidence that CD45RA can be expressed in EFF and EM cells (24–26). We found an increase in CD4 EM cells in CD vs controls but no significant differences in CD8 EM cells (Figure 2c).
Figure 2.
Profiles of αβ T-cell subsets and markers of memory formation and effector differentiation in blood samples from patients with IBD. (a) Representative flow cytometry plots of the CD4 and CD8 T-cell populations in the PBMCs of healthy control (left), CD (middle), and UC (right) blood samples (gated on CD3+TCRαβ+ populations), followed by (b) summary of the subset percentages. (c) Summary percentages of CD4 and CD8 EM T cells in the PBMC of healthy controls, CD, and UC samples. (d) Summary percentages of TH1, TH2, and TH17 effector subsets including a further subset of PD1+ TH17 cells. The number in the flow cytometry plots is the gated percentage. Each data point represents 1 sample from a patient (mean ± SEM), *P < 0.05, **P < 0.01. Cell counts are shown in Supplementary Figure 5 (see Supplementary Digital Content 5, http://links.lww.com/CTG/B142). CD, Crohn's disease; EM, effector memory; IBD, inflammatory bowel disease; PBMC, peripheral blood mononuclear cell; SEM, standard error of mean; UC, ulcerative colitis.
TH17-mediated inflammation has been implicated in mucosal inflammation in IBD (27). We therefore profiled our samples for TH-cell subsets with intracellular staining for the T-box transcription factor TBX21, GATA-binding protein 3, and RORγt, which are required for the development of TH1, TH2, and TH17 cells, respectively. We found an increased frequency of TH17 populations in CD vs healthy controls and a similar trend in UC samples (Figure 2d). We further used the PD1 marker to examine T-cell checkpoint inhibition. We found that the PD1+ subset of TH17 cells was decreased in CD compared with healthy control blood, suggesting suggests defective checkpoint regulation in the TH17 subsets in CD. There were similar trends of TH17 and PD1+ TH17 population differences in UC PBMCs vs healthy controls, but these were not significant. We did not detect significant differences in TH1 or TH2 frequencies between CD, UC, or healthy blood samples. Taken together, the results demonstrate that peripheral blood of patients with IBD, especially CD, harbor a higher proportion of TH17 cells and further suggest that these cells are more active because of their decreased expression of PD1, which is an inhibitory checkpoint molecule.
We then assessed peripheral T-cell regulation in IBD by analyzing 2 key regulatory molecules, FOXP3 and CTLA4. FOXP3 is highly expressed by Treg cells, while CTLA4 is an immunosuppressive checkpoint marker expressed by both Treg cells and activated conventional T cells (28). Both CD and UC PBMC samples had increased populations of CD4+FOXP3+ Treg cells and its CTLA4+ subset cells compared with healthy controls (Figure 3a). They also had increased CTLA4+ conventional CD4 and CD8 T cells, suggesting heightened regulation, extrinsically through the Treg cell suppression or intrinsically by intracellular CTLA4 signaling in conventional T cells, for both CD4 and CD8 T-cell subsets (Figure 3b). These data demonstrate that regulatory T-cell pathways are increased in IBD patient samples and are reflected in peripheral blood.
Figure 3.
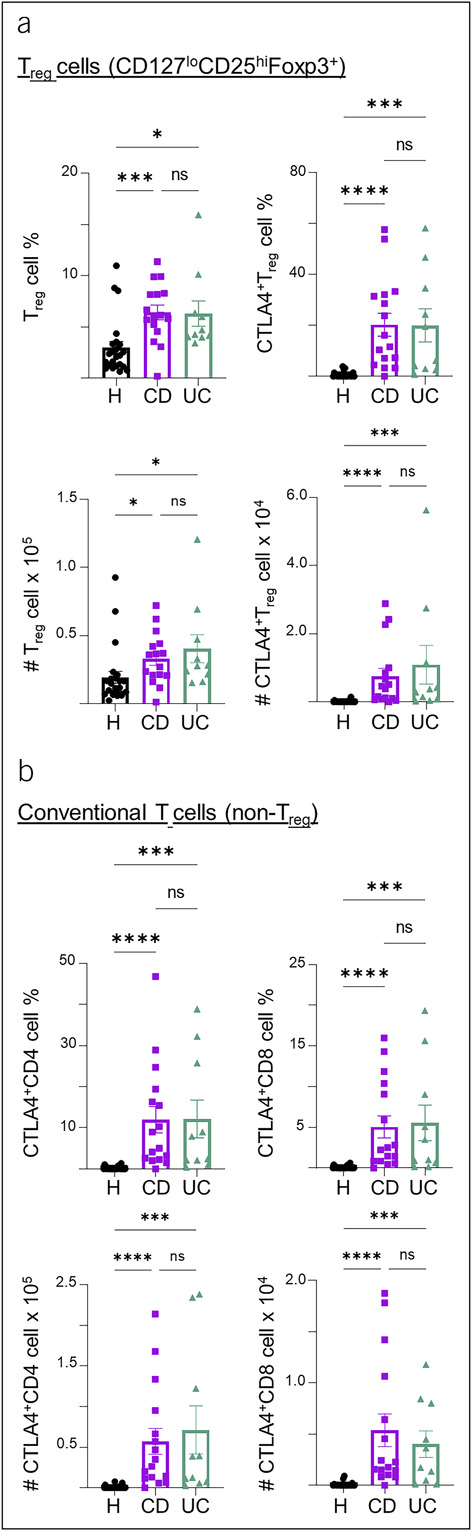
Profiles of αβ T-cell regulation and checkpoints in blood samples from patients with IBD. (a) Summary percentages and cell counts of CD4+CD127loCD25hiFoxp3+ Treg cells and the CTLA4+ subset of Treg cells in the PBMC of healthy control, CD, and UC samples. (b) Summary percentages and cell counts of conventional (non-Treg) CD4 or CD8 T cells expressing CTLA4 in healthy control, CD, and UC PBMC samples. Cell counts are normalized to counts per million. Each data point represents 1 sample from a patient (mean ± SEM), *P < 0.05, ***P < 0.001, ****P < 0.0001. CD, Crohn's disease; IBD, inflammatory bowel disease; PBMC, peripheral blood mononuclear cell; SEM, standard error of mean; UC, ulcerative colitis.
Spectral flow cytometry coupled with bioinformatic analysis of high dimensional data can facilitate identification of small populations of cells among various subsets of cells. We performed a SPADE on viSNE analysis of the PBMC spectral flow cytometry data (see Supplementary Figure 4, Supplementary Digital Content 4, http://links.lww.com/CTG/B141). Our analysis highlighted a clear γδ T-cell cluster in CD and UC but not in healthy blood (see Supplementary Figure 4A, Supplementary Digital Content 4, http://links.lww.com/CTG/B141). Furthermore, we identified an elevated percentage of EM γδ T-cell frequencies in CD and UC samples compared with healthy controls (see Supplementary Figure 5A, Supplementary Digital Content 5, http://links.lww.com/CTG/B142). By contrast, we found a higher proportion of naïve γδ T cells in healthy samples. Healthy samples also had significantly higher percentages of naïve CD4 and CD8 αβ T cells than CD samples but not UC samples (see Supplementary Figure 5B, Supplementary Digital Content 5, http://links.lww.com/CTG/B142). The variation in the proportions of naïve vs memory subsets may explain some of the heterogeneous results in previous studies on peripheral γδ T cells in IBD, where early studies found increased peripheral γδ T cells in CD compared with controls, but later studies found a deficit (29). These data taken together demonstrate the utility of deep flow cytometric characterization of peripheral blood cell populations in IBD as a window into mucosal pathology.
Switch memory B cells and markers of B-T cell interaction are increased in peripheral blood and tissue of patients with IBD
Recent studies have highlighted a role for B cells in IBD (30,31). We next analyzed B cells from the blood of patients with IBD and healthy controls for naïve and memory cell populations based on the differential expression of IgD and CD27. Naïve B cells were identified as IgD+CD27–, nonswitch memory (SM) B cells were identified as IgD+CD27+, SM B cells were identified as IgD–CD27+, and double negative memory B cells were identified as IgD–CD27– (Figure 4a). Both CD and UC PBMCs had increases in SM B cells over heathy controls (Figure 4b). In the ileum, we also observed an increase in the number of SM B cells in inflamed tissue compared with uninflamed tissue (Figure 4c). However, there were no differences in the number of naïve, non-SM, or double negative B cells in inflamed compared with uninflamed tissue (data not shown). SM B cells, which have undergone class switching and affinity maturation in germinal centers, are known to maintain high diversity in the gut to quickly respond to commensal bacteria (32). Our results suggest a role of this memory B-cell population in IBD, particularly in active ileal CD.
Figure 4.
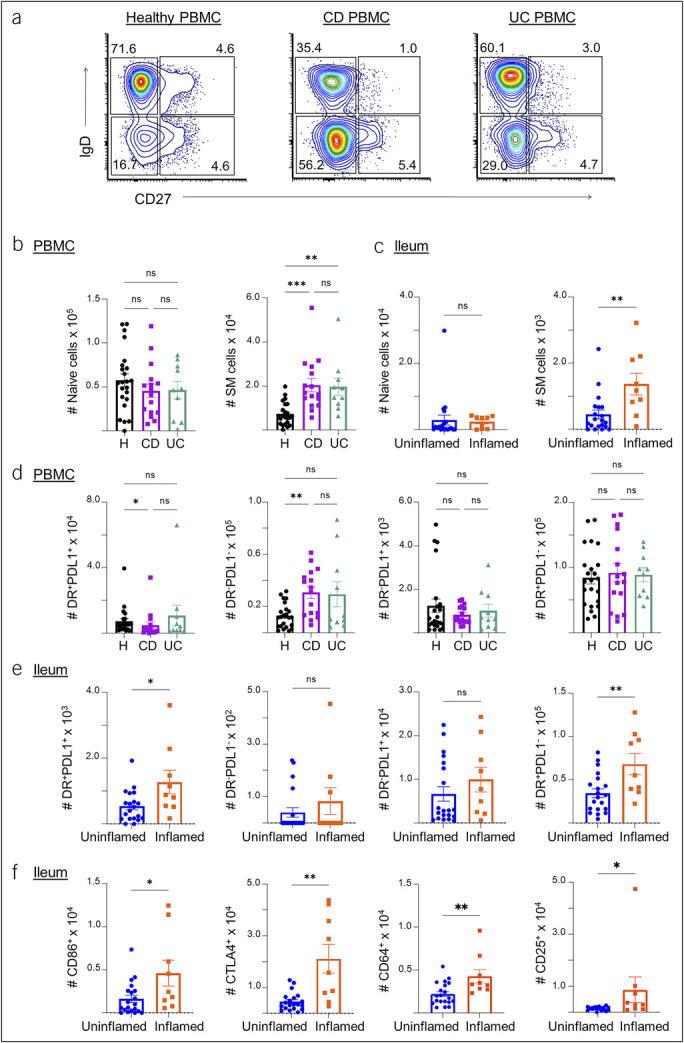
Characterization of B cell subsets in the blood, ileum, and colon of patients with IBD. (a) Representative flow cytometry plots for the naïve and memory B-cell populations in healthy control, CD, and UC PBMC samples, followed by (b) summary cell counts of naïve and SM B cells. (c) Summary counts of naïve and SM B cells in uninflamed vs inflamed ileum tissue samples from patients with IBD. (d) Summary cell counts of HLA-DR+PDL1+, HLA-DR–PDL1–, HLA-DR−PDL1+, and HLA-DR+PDL1− populations in healthy, CD, and UC PBMC samples. (e) Summary cell counts of HLA-DR+PDL1+, HLA-DR–PDL1–, HLA-DR−PDL1+, and HLA-DR+PDL1− B-cell populations in uninflamed and inflamed ileum samples of patients with IBD. (f) Summary cell counts of B cells with CD86 or CTLA4 markers in uninflamed vs inflamed ileum samples of patients with IBD. Each data point represents 1 sample from a patient (mean ± SEM), *P < 0.05, **P < 0.01, ***P < 0.001. CD, Crohn's disease; IBD, inflammatory bowel disease; PBMC, peripheral blood mononuclear cell; SEM, standard error of mean; SM, switch memory; UC, ulcerative colitis.
We then analyzed HLA-DR and PDL1 checkpoint expression patterns in B cells. We observed decreased numbers of HLA-DR+PDL1+ B cells in CD PBMC samples (but not in UC) and increased numbers of HLA-DR+PDL1− B cells in CD when compared with healthy control PBMC samples (Figure 4d). When comparing inflamed and uninflamed tissue samples, we also observed increases in HLA-DR+PDL1+ and HLA-DR+PDL1− B cells in the inflamed ileum (Figure 4e). Furthermore, in inflamed samples, we observed increases in B cells expressing CTLA4, CD86, a costimulatory molecule for T-cell activation, CD64 (FcγRI), the high affinity IgG receptor, and CD25, a subunit of the high affinity IL2 receptor (Figure 4f). Overall, increased expression of these immune checkpoint and T-cell activation markers suggests increased B-T cell interactions and immunoregulation in IBD, particularly in ileal CD.
NK cells and CD206+/CD64+ monocyte and macrophage lineages are increased in IBD PBMCs or gut tissue
Finally, we characterized innate immune cells in both PBMCs and gut tissue samples. Phagocytic innate immune cells are involved in IBD pathology as they interact with dysbiotic gut microbiota and serve as professional antigen presenting cells (33). Neutrophilic infiltrate is also considered a hallmark of acute inflammation in IBD. We included markers for monocytes, macrophages, dendritic cells (DCs), natural killer (NK) cells, and neutrophils, as well as various subtypes of these cells in our flow panel. We found that NK cells (CD56+) were significantly increased in IBD PBMC samples compared with healthy control samples (Figure 5a). This was true of both immature (CD16lo) and mature NK cells (CD16hi), although mature NK cells had a greater increase. SPADE clustering of PBMC samples similarly showed an increase in NK cells and reduction in monocyte populations in IBD compared with healthy control PBMCs (see Supplementary Figure 4, Supplementary Digital Content 4, http://links.lww.com/CTG/B141).
Figure 5.
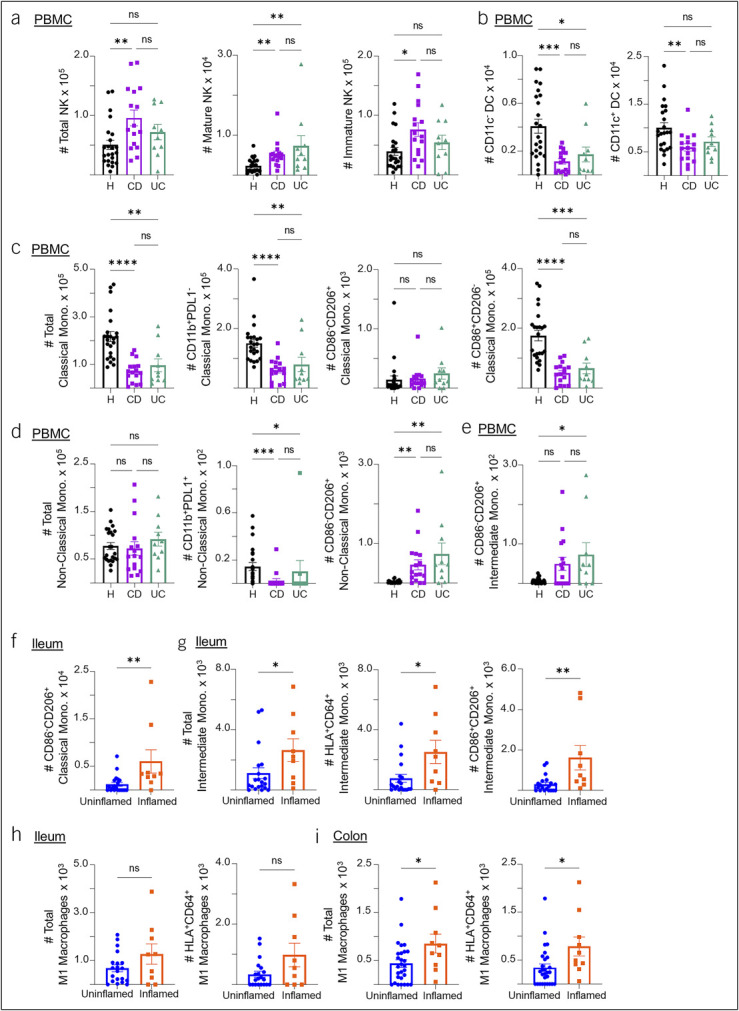
Characterizations of innate immune populations in the blood, ileum, and colon of patients with IBD. (a) Summary cell counts of NK cells and their mature (CD56hiCD16hi) and immature (CD56hi CD16lo) subsets in healthy control, CD, and UC PBMC samples. (b) Summary cell counts of CD11c– and CD11c+ dendritic cells (DC) in healthy control, CD, and UC PBMC samples. (c) Summary cell counts of classical monocyte (CD14+CD15–CD16–) total population followed by counts of the CD11b+PDL1–, CD86–CD206+ and CD86+CD206– subsets in healthy control, CD, and UC PBMC samples. (d) Summary cell counts of the nonclassical monocyte (CD14–CD16+) total population, followed by counts of the CD11b+PDL1+ and CD86−CD206+ subsets in healthy control, CD, and UC PBMC samples. (e) Summary cell counts of the CD86−CD206+ subset of intermediate monocyte (CD14+CD16+) in healthy, CD, and UC PBMC samples. (f) Summary cell counts of the CD86–CD206+ classical monocyte subset in uninflamed vs inflamed ileum samples of patients with IBD. (g) Summary cell counts of the intermediate monocyte total population counts followed by counts of the HLA+CD64+ and CD86+CD206+ subsets in uninflamed vs inflamed ileum samples of patients with IBD. (h, i) Summary cell counts of total M1 macrophages (CD86+, CD206–) and its HLA-DR+CD64+ subset in the ileum (h) and colon (i) of patients with IBD. Cell counts are normalized to counts per million. Each data point represents 1 sample from a patient (mean ± SEM), *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. CD, Crohn's disease; IBD, inflammatory bowel disease; NK, natural killer; PBMC, peripheral blood mononuclear cell; SEM, standard error of mean; SM, switch memory; UC, ulcerative colitis.
We also examined a number of DC subsets in our samples. We included markers CD11b and CD11c, integrin molecules that aid in cell migration and phagocytosis. Both are expressed by innate immune cells, but the highest expression of CD11c is on DCs. We found that both CD11c+ and CD11c− DC populations were significantly decreased in IBD PBMCs compared with healthy PBMCs (Figure 5b) but observed no differences at the tissue level.
We next characterized an array of monocyte and macrophage subsets. For this, we included the PDL1 checkpoint marker, the CD86 costimulatory molecule, and the CD206 innate pattern-recognition molecule. CD206 is also known as the macrophage mannose receptor; it is upregulated during inflammation and is a marker of active phagocytosis in cells such as M2 macrophages and DCs. CD86 is classically associated with M1 macrophages, while CD206 is associated with M2 macrophages (34). We found that CD and UC PBMCs had smaller populations of classical monocytes and their CD11b+PDL1– and CD86+CD206– subsets compared with healthy controls (Figure 5c). In addition, IBD PBMCs had an increase in the CD11b+PDL1+ subset and a decrease in the CD86–CD206+ subset of nonclassical monocytes compared with healthy samples (Figure 5d). By contrast, there was an increase in the CD86–CD206+ subset of intermediate monocytes in IBD blood samples compared with healthy controls (Figure 5e). We next observed similar trends in the ileum, where we saw a significant increase in CD86–CD206+ classical monocytes, as well as total intermediate monocytes and their HLA-DR+CD64+ and CD86+CD206+ subsets (Figure 5f and g). The increased CD206 expression in IBD monocytes may be due to increased phagocytosis or differentiation into M2 macrophages. These differences in monocyte populations can also be observed in our SPADE analysis, with decreased expression of monocyte markers such as CD11b, CD14, CD16, and HLA-DR in CD and UC PBMCs (see Supplementary Figure 4, Supplementary Digital Content 4, http://links.lww.com/CTG/B141).
In the colon, we did not observe the same monocyte trends as in the blood and ileum. However, total M1 macrophages (defined as CD86+ and CD206–) and HLA-DR+CD64+ M1 macrophages were significantly increased in inflamed colon samples and had a similar but not significant trend in the ileum (Figure 5h and i). HLA-DR and CD64 expression on macrophages implies interactions between these macrophages and adaptive immune cells. Overall, this suggests that different innate immune cell populations participate in inflammation in the ileum vs the colon in IBD.
Finally, we were able to identify neutrophils, a population that is often absent in transcriptomic studies, in the gut tissue samples based on CD15 expression. We found that neutrophils were numerically increased in the inflamed ileum but did not reach significance (data not shown). While neutrophils are the primary indicator of acute inflammation in IBD, recent studies found that they might also have an important role in chronic inflammation (35), even in a setting dominated by a lymphocytic infiltrate.
IBD PBMC samples and CD ileal samples cluster separately with differences driven by stromal-like cells and B cells
Owing to the distinct differences between healthy and IBD samples in various cell populations, we aimed to determine if the overall cellular signatures of PBMC samples or tissue samples were different between groups. We performed PCA using over 100 cell subtypes determined by spectral flow cytometry as variables. PBMC, ileum, and colon samples were analyzed separately. We plotted samples by their PC scores and looked for clustering based on sample characteristics such as healthy vs IBD, CD vs UC, inflamed vs uninflamed, or current IBD medications. We then looked at the top loadings for each quadrant to determine which variables drove the sample clustering in each quadrant.
Remarkably, when plotting PC1 and PC3 for PBMC samples, the majority of healthy samples and none of the IBD samples clustered in quadrant III (Figure 6a). The top loadings in this quadrant were classical monocyte subtypes and total innate immune cells, meaning the healthy PBMC cellular signature was dominated by these cell types. Meanwhile, stromal-like cell subsets made up the top loadings in quadrant IV, which contained about half of the inflamed IBD samples. These samples also included 4 of 5 patients not currently on biologic drugs (see Supplementary Figure 6A, Supplementary Digital Content 6, http://links.lww.com/CTG/B143). Quadrant I contained IBD samples and was dominated by B cell and intermediate monocyte loadings, while quadrant II, which contained both healthy and IBD samples, was dominated by central memory CD4 T cells, B-cell subsets, and nonclassical monocytes. Altogether, the IBD PBMC cellular signature can be distinguished from a healthy signature primarily due to an increase in stromal-like cells and B cells.
Figure 6.
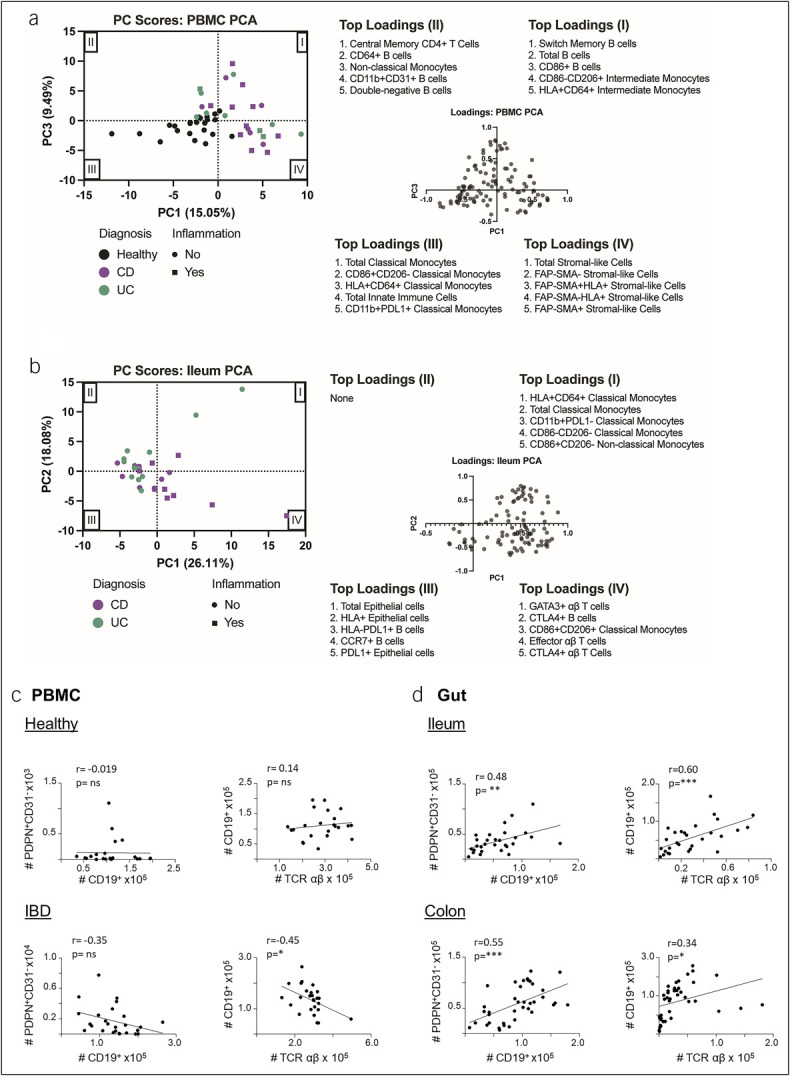
PCA and Pearson correlation tests using the cell subtypes identified by spectral flow cytometry as variables. Unsupervised clustering through principal component analysis was performed on (a) PBMC samples (n = 49) and (b) ileum samples (n = 29). Cell counts determined through manual gating were used as variables (110 cell subtypes for PBMC samples and 98 for ileum samples). Each sample is represented by a dot, where color indicates diagnosis and shape indicates active inflammation. The loadings plot and top 5 loadings of each quadrant are shown. (c) Correlation analyses between the number of CD19+ B cells and PDPN+CD31– or αβ T cells and CD19+ B cells in PBMC samples from healthy controls (top) or patients with IBD (bottom). (d) Correlation analyses between the number of CD19+ B cells and PDPN+CD31– or αβ T cells and CD19+ B cells in ileum (top) and colon (bottom) biopsies from patients with IBD. *P < 0.05, **P < 0.01, ***P < 0.001. IBD, inflammatory bowel disease; PBMC, peripheral blood mononuclear cell; PCA, principal component analysis.
We next examined clustering of ileal samples from CD and UC samples in this agnostic way. Ileal samples in UC are considered normal, as UC does not affect the ileum. A subset of CD samples, including 5 of the 9 inflamed samples, clustered in quadrant IV and were completely separate from UC samples (Figure 6b). This quadrant was dominated by activated T-cell and B-cell loadings. Stromal cell and neutrophil loadings also fell into quadrant IV but with lower PC scores (data not shown). The other quadrants (I, II, and III), which contained the majority of uninflamed CD and UC samples, were dominated by classical monocytes and epithelial cell loadings. Samples from patients on anti-TNF medications almost all clustered in quadrants II and III (see Supplementary Figure 6B, Supplementary Digital Content 6, http://links.lww.com/CTG/B143). PCA of colon samples did not show any differential clustering in any of the PC score graphs (see Supplementary Figure 6C, Supplementary Digital Content 6, http://links.lww.com/CTG/B143). These data demonstrate that unbiased computer-driven clustering of cellular characteristics can separate healthy blood samples from IBD samples and actively inflamed CD samples from uninflamed ileal samples.
Stromal-like cell and immune cell populations correlate in IBD blood and colon
To gain a clue on potential relationships between stromal cell subsets and immune cell lineages in IBD, we analyzed multiple correlations of cell numbers between individual cell types. We detected significant although modest inverse correlations between PDPN+CD31− fibroblast-like cell numbers and B-cell numbers in blood from patients with IBD but not healthy controls. The number of B cells also inversely correlated with the number of T cells in PBMCs from patients with IBD but not in healthy control blood (Figure 6c). In the gut tissue, positive correlations were detected between B cell and fibroblast numbers as well as between B-cell and T-cell numbers in both ileum and colon (Figure 6d). Although we did not observe a correlation between PDPN+CD31− fibroblast-like cell numbers and total αβ T-cell numbers in IBD blood samples (not shown), both CTLA4+CD4+ and CTLA4+CD8+ subsets of conventional αβ T cells correlated with PDPN+CD31− cells or their FAP−SMAα+ or FAP−SMAα− subsets in IBD PBMCs (see Supplementary Figure 7A, Supplementary Digital Content 7, http://links.lww.com/CTG/B144). Again, no correlations were detected in healthy control PBMCs between these cell types. Furthermore, the numbers of B cells inversely correlated with the number of FAP−SMAα− or FAP−SMAα+ subsets in IBD PBMCs, but positive correlations were detected between B cells and the stromal cell subsets in ileum and colon samples (see Supplementary Figure 7B–D, Supplementary Digital Content 7, http://links.lww.com/CTG/B144). Overall, these correlations between stromal cells, T cells, and B cells in IBD samples but not healthy controls suggest a complex network of stromal-immune interactions in IBD, potentially contributing to disease pathology and tissue inflammation which is reflected in peripheral blood.
DISCUSSION
The root cause of IBD remains elusive. Extensive studies on immune pathogenesis have led to several immune-based treatments that have significantly improved patients' quality of life and led to healing of the intestinal mucosa—at least while on therapy. Nevertheless, a substantial portion of patients with IBD either do not respond to these therapies or become refractory after an initial response (2). There is also an unmet need for peripheral blood biomarkers in IBD. Until recently, research showed that the pathologic state was best represented in the intestine and had very few identifiable markers in the blood (36). Past characterization of blood and tissue by flow cytometry was limited in depth—yet had the advantage over RNA-based techniques of confirming protein expression and allowing for single-cell characterization.
In this study, we took advantage of full spectral flow cytometry to characterize a diverse cohort of patients with IBD using matched blood and intestinal tissue collected on the same day. Previous studies have used cytometry by time of flight to obtain similarly large protein expression profiles of immune cells (37–40). In our studies, in addition to using matched blood and tissue samples, we captured stromal markers along with traditional adaptive and innate immune markers. This allowed us to characterize nonhematopoietic cells such as epithelial cells, endothelial cells, and fibroblast-like cells alongside a wide array of immune cell subsets.
Our first notable finding is that peripheral blood of patients with IBD has a small population of circulating PDPN-expressing cells that are further characterized by expression of SMAα, FAP, EPCAM, and HLA-DR to varying degrees. This cell type was not seen in healthy samples. Indeed, in unbiased clustering analysis, peripheral PDPN+ stromal-like cells distinguish IBD PBMCs from healthy samples. This provocative finding suggests that chronic intestinal inflammation may induce expression of PDPN and/or systemic availability of stromal-like cells. This is reminiscent of recent findings in RA research, where fibroblast-like synoviocytes are the main mediators of inflammation. PDPN is upregulated in these cells when compared with healthy controls (41), and in vitro treatment of synoviocytes with proinflammatory cytokines leads to the upregulation of PDPN (42). Furthermore, a recent study with RNA profiling found circulating PDPN+ nonhematopoietic cells similar to the fibroblast-like synoviocytes, and those cells could serve as a biomarker for RA flares (8).
We also found PDPN+ cells with similar phenotypes present in both ileum and colon. We classified the PDPN+CD31− cells as fibroblasts based on their generally recognized phenotype and found several fibroblast subsets increased in CD ileum compared with UC ileum. A limitation of our study was that we did not have healthy intestinal biopsies for comparison. However, in silico analysis of publicly available RNA data does show increased expression of PDPN in ileum and colon biopsies of patients with IBD compared with healthy controls. Furthermore, although we could not prove that these tissue-resident PDPN+ cells are the same as the ones in the blood, we found a statistically significant correlation between the number of PDPN+ cells in peripheral blood and matched colon PDPN+ cells, suggesting that the former may be somehow related to the latter. Along this line, we should note that we designed the experiments and data analyses in such a way that the lack of healthy intestinal samples would not substantially affect the data interpretation. Indeed, the key conclusions from this study did not rely on the comparison of IBD and healthy intestinal biopsies. Altogether, this suggests that these stromal-like cells in the peripheral blood of patients with IBD may come from chronic inflammation of the colon and may serve as a biomarker of IBD.
Part of the interest in this finding of a PDPN signature stems from work suggesting that stromal cells contribute to IBD inflammation and refractoriness to therapy (12). It is hypothesized that activated or pathogenic fibroblast populations contribute to the proinflammatory environment through multiple mechanisms including production of proinflammatory cytokines and downregulation of immune inhibitory molecules (22). A previous study also found that fibroblast molecular signatures contributed to matched biopsy molecular inflammation scores (36). In addition, myofibroblast-T-cell interactions have recently been implicated in CD (9). However, these discoveries were largely made using RNA sequencing, which may not necessarily translate into changes in protein expression and are harder to quantify as a clinical test. Our study confirms the PDPN protein expression. Furthermore, while our study was not large enough to determine if the presence of these peripheral PDPN+ cells correlated with refractory disease, most of the patients with active inflammation while on anti-TNF medication had a strong PDPN+ signature.
In addition to profiling PDPN+ cells, our study captured activation, effector, and memory profiles of B and T cells in the setting of IBD. Enrichment of TH17 cells and Treg cells has been found in IBD mucosa in a previous study (40). Our study not only observed the increased frequency of TH17 cells and Treg cells but also detected reduced expression of PD1 inhibitory checkpoint in TH17 cells and increased expression of CTLA4 checkpoint in Treg cells in the peripheral blood of patients with IBD. Future larger studies can determine whether these cells are gut homing or relate to response to specific types of therapy, such as in a previous study which found that the activation state of Treg cells was an important predictor of response to vedolizumab, a biologic that targets integrin α4β7 (43). We also found an enrichment of SM B cells in IBD peripheral blood and inflamed ileum. There is a renewed interest in targeting B cells in IBD (31). Conceivably, combinatorial immune therapies targeting inflammatory T cells and B cells may improve controlling gut inflammation and promoting gut tissue repair and regeneration.
In addition to possible involvement of direct or indirect T-B-cell interactions, we also found evidence of B-stromal cell interactions within the intestinal tissue. The positive correlation between total B cells and fibroblasts in ileum and colon, but lack of significant correlation between total T cells and fibroblast populations (although some T-cell subsets correlated with fibroblasts), was somewhat surprising given the experimental evidence of major histocompatibility complex-based interactions between T cells and fibroblasts from a recent study (44). On the other hand, the relationship between B cells and stromal cells has been well documented in lymphoid organs (45). While we have no evidence for the functional effect of the B cell-fibroblast correlation in IBD, fibroblasts could play a role in retention or expansion of tissue B cells. Conversely, B cells in the tissue could also promote the differentiation of fibroblasts and thus stimulate a process of wound healing, or fibrosis if there are chronically dysregulated interactions. Overall, our results suggest that B cells may not only play a role in directly or indirectly affecting T-cell activation within the intestine but also perhaps fibroblast activation and subsequent aberrant wound healing or fibrosis, particularly in CD.
Regarding innate immune cells, we found an increase in NK cells in IBD blood. Previous studies on peripheral NK cells in IBD have been heterogeneous, with some demonstrating an increase in IBD (46) and others showing a decrease (38). In intestinal tissue, a previous work found that lamina propria-resident CD11b+ phagocytic cells, including monocytes and macrophages, differed greatly in gene expression between ileum and colon populations (33). In this study, we found more classical monocytes in inflamed ileum expressing CD206, a marker of active phagocytosis in M2 macrophages and DCs. Meanwhile, in the colon, M1 macrophages were higher in inflamed tissue, in particular HLA-DR+CD64+ M1 macrophages. Macrophage phenotypes are thought to exist on a spectrum, where M1 macrophages are classically defined as proinflammatory and involved in acute inflammation, and M2 macrophages are anti-inflammatory and promote wound healing (34). However, M2 macrophages have also been associated with fibrosis in organs such as the kidney and may similarly contribute to fibrosis in CD (47). CD64 was also previously found to be expressed by a unique population of macrophages associated with inflamed colon in patients with CD (48). Our findings therefore corroborate this study.
In summary, our study fills the gap in knowledge on IBD protein markers at a single-cell level with spectral flow profiling of stromal and immune cells in the gut mucosa and circulation simultaneously. We identified a new subset(s) of nonhematopoietic PDPN+ cells in the blood of patients with IBD which showed corresponding phenotypes to stromal cells within the gut mucosa. Our spectral flow cytometry panel could distinguish IBD blood samples from healthy controls driven by hematopoietic and nonhematopoietic cells, particularly B cells and PDPN+ cells. We believe this may reflect what is happening in tissue and that the interplay might critically shape the inflammatory milieu toward pathogenic destruction vs regeneration and repair of damaged tissue. Further studies are needed to examine the function of these circulating stromal-like cells associated with IBD. Integrating stromal and immune cell biology will likely lead to a better understanding of IBD pathogenesis, as well as potential clinical utility of stromal-immune cell signatures as biomarkers of IBD and its responsiveness to treatment.
CONFLICTS OF INTEREST
Guarantors of the article: Zhibin Chen, MD, PhD, and Maria T. Abreu, MD.
Specific author contributions: A.M.H., G.E.J., M.T.A., and Z.C.: contributed to the design of the study and initial drafting of manuscript. H.D., A.M.H., G.E.J., E.N.V., and K.V.d.J.: contributed to the statistical and bioinformatic analysis. A.M.H., G.E.J., M.T.A., and Z.C.: had full access to all the data in the study and took responsibility for the integrity of the data and the accuracy of the data analysis. M.T.A. and Z.C.: supervised the study. All authors contributed to acquisition, analysis or interpretation of the clinical data or laboratory analysis data, drafting the article or revising it critically for important intellectual content. All authors approved the final version of the manuscript.
Financial support: This work was supported in part by grants from National Institutes of Health/National Institute of Diabetes and Digestive and Kidney Diseases (R01DK099076 to M.T.A.) and National Institutes of Health/National Institute of Allergy and Infectious Diseases (T32AI162624 to G.E.J.; R01AI134903 to Z.C.).
Potential competing interests: Dr. Abreu has received research funding from the National Institute of Health Research, DOD and charities including The Leona M. and Harry B. Helmsley Charitable Trust, Crohn’s and Colitis Foundation, as well as relationships with industry: AbbVie Inc, Consultant/Advisory Board; Alimentiv, teaching, lecturing or speaker; Amgen Inc, Advisor; Bristol Myers Squibb, Consultant/Advisory Board; Celsius Therapeutics, Consultant/Advisory Board; Eli Lilly and Company, Consultant/Advisory Board; Gilead Sciences Inc, Consultant/Advisory Board; Janssen Pharmaceuticals, Consultant/Advisory Board, teaching; Materia Prima, Consultant; Pfizer Pharmaceutical, Consultant/Advisory Board; Takeda Pharmaceuticals, teaching, lecturing or speaker. The other authors report no competing interests.
IRB approval: The use of human samples for this study was approved by the University of Miami Institutional Review Board (IRB ID: 20081100).
Study Highlights.
WHAT IS KNOWN
✓ Single-cell RNA-sequencing studies have revealed dynamic mRNA expression profiles in both stromal and immune cells in patients with inflammatory bowel disease (IBD).
WHAT IS NEW HERE
✓ We designed a forty-color flow cytometry panel for single-cell characterization of protein markers in hematopoietic and nonhematopoietic lineages.
✓ PDPN+ fibroblast-like cells are more abundant in IBD blood than healthy controls.
✓ These cells correlated with fibroblasts in IBD colon biopsies and were associated with innate and adaptive immune activation and dysregulation in IBD.
Supplementary Material
Footnotes
SUPPLEMENTARY MATERIAL accompanies this paper at http://links.lww.com/CTG/B138, http://links.lww.com/CTG/B139, http://links.lww.com/CTG/B140, http://links.lww.com/CTG/B141, http://links.lww.com/CTG/B142, http://links.lww.com/CTG/B143, and http://links.lww.com/CTG/B144
Amanda M. Honan and Gillian E. Jacobsen contributed equally to this work.
Contributor Information
Amanda M. Honan, Email: amh367@miami.edu.
Gillian E. Jacobsen, Email: gjacobsen@miami.edu.
Hannah Drum, Email: hbd24@miami.edu.
Emily N. Vazquez, Email: env6@miami.edu.
Maria A. Quintero, Email: mxq155@miami.edu.
Amar R. Deshpande, Email: adeshpande@miami.edu.
Daniel A. Sussman, Email: dsussman@miami.edu.
David H. Kerman, Email: dkerman@miami.edu.
Oriana M. Damas, Email: omazorra@miami.edu.
Siobhan Proksell, Email: sproksell@miami.edu.
Kevin Van der Jeught, Email: kxv560@miami.edu.
Maria T. Abreu, Email: mabreu1@miami.edu.
REFERENCES
- 1.Burgueno JF, Abreu MT. Epithelial Toll-like receptors and their role in gut homeostasis and disease. Nat Rev Gastroenterol Hepatol 2020;17(5):263–78. [DOI] [PubMed] [Google Scholar]
- 2.Chang JT. Pathophysiology of inflammatory bowel diseases. N Engl J Med 2020;383(27):2652–64. [DOI] [PubMed] [Google Scholar]
- 3.Che Z, Ye Z, Zhang X, et al. Mesenchymal stem/stromal cells in the pathogenesis and regenerative therapy of inflammatory bowel diseases. Front Immunol 2022;13:952071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oltz EM, Schwab S. Stromal immunology: Frameworks for development and response. J Immunol 2021;206(2):241–2. [DOI] [PubMed] [Google Scholar]
- 5.Krishnamurty AT, Turley SJ. Lymph node stromal cells: Cartographers of the immune system. Nat Immunol 2020;21(4):369–80. [DOI] [PubMed] [Google Scholar]
- 6.Anderson MS, Venanzi ES, Klein L, et al. Projection of an immunological self shadow within the thymus by the aire protein. Science 2002;298(5597):1395–401. [DOI] [PubMed] [Google Scholar]
- 7.Shipman WD, Sandoval MJ, Veiga K, et al. Fibroblast subtypes in tissues affected by autoimmunity: With lessons from lymph node fibroblasts. Curr Opin Immunol 2020;64:63–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Orange DE, Yao V, Sawicka K, et al. RNA identification of PRIME cells predicting rheumatoid arthritis flares. N Engl J Med 2020;383(3):218–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Filidou E, Valatas V, Drygiannakis I, et al. Cytokine receptor profiling in human colonic subepithelial myofibroblasts: A differential effect of Th polarization-associated cytokines in intestinal fibrosis. Inflamm Bowel Dis 2018;24(10):2224–41. [DOI] [PubMed] [Google Scholar]
- 10.Salas A, Hernandez-Rocha C, Duijvestein M, et al. JAK-STAT pathway targeting for the treatment of inflammatory bowel disease. Nat Rev Gastroenterol Hepatol 2020;17(6):323–37. [DOI] [PubMed] [Google Scholar]
- 11.Kinchen J, Chen HH, Parikh K, et al. Structural remodeling of the human colonic mesenchyme in inflammatory bowel disease. Cell 2018;175(2):372–86.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin JC, Chang C, Boschetti G, et al. Single-cell analysis of Crohn's disease lesions identifies a pathogenic cellular module associated with resistance to anti-TNF therapy. Cell 2019;178(6):1493–508.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smillie CS, Biton M, Ordovas-Montanes J, et al. Intra- and inter-cellular rewiring of the human colon during ulcerative colitis. Cell 2019;178(3):714–30.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miska J, Lui JB, Toomer KH, et al. Initiation of inflammatory tumorigenesis by CTLA4 insufficiency due to type 2 cytokines. J Exp Med 2018;215(3):841–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van der Jeught K, Sun Y, Fang Y, et al. ST2 as checkpoint target for colorectal cancer immunotherapy. JCI Insight 2020;5(9):e136073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qiu P, Simonds EF, Bendall SC, et al. Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nat Biotechnol 2011;29(10):886–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farr AG, Berry ML, Kim A, et al. Characterization and cloning of a novel glycoprotein expressed by stromal cells in T-dependent areas of peripheral lymphoid tissues. J Exp Med 1992;176(5):1477–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mizoguchi F, Slowikowski K, Wei K, et al. Functionally distinct disease-associated fibroblast subsets in rheumatoid arthritis. Nat Commun 2018;9(1):789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Friedrich M, Pohin M, Jackson MA, et al. IL-1-driven stromal-neutrophil interactions define a subset of patients with inflammatory bowel disease that does not respond to therapies. Nat Med 2021;27(11):1970–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brugger MD, Basler K. The diverse nature of intestinal fibroblasts in development, homeostasis, and disease. Trends Cell Biol 2023;33(10):834–49. [DOI] [PubMed] [Google Scholar]
- 21.Imhof BA, Dunon D. Leukocyte migration and adhesion. Adv Immunol 1995;58:345–416. [DOI] [PubMed] [Google Scholar]
- 22.Wei K, Nguyen HN, Brenner MB. Fibroblast pathology in inflammatory diseases. J Clin Invest 2021;131(20):e149538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Massimino L, Lamparelli LA, Houshyar Y, et al. The inflammatory bowel disease transcriptome and metatranscriptome meta-analysis (IBD TaMMA) framework. Nat Comput Sci 2021;1(8):511–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoflich C, Docke WD, Busch A, et al. CD45RA(bright)/CD11a(bright) CD8+ T cells: effector T cells. Int Immunol 1998;10(12):1837–45. [DOI] [PubMed] [Google Scholar]
- 25.Tian Y, Babor M, Lane J, et al. Unique phenotypes and clonal expansions of human CD4 effector memory T cells re-expressing CD45RA. Nat Commun 2017;8(1):1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu H, Galdos FX, Lee D, et al. Identification of pathogenic immune cell subsets associated with checkpoint inhibitor-induced myocarditis. Circulation 2022;146(4):316–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cao H, Diao J, Liu H, et al. The pathogenicity and synergistic action of Th1 and Th17 cells in inflammatory bowel diseases. Inflamm Bowel Dis 2023;29(5):818–29. [DOI] [PubMed] [Google Scholar]
- 28.Chambers CA, Kuhns MS, Egen JG, et al. CTLA-4-mediated inhibition in regulation of T cell responses: Mechanisms and manipulation in tumor immunotherapy. Annu Rev Immunol 2001;19:565–94. [DOI] [PubMed] [Google Scholar]
- 29.Catalan-Serra I, Sandvik AK, Bruland T, et al. Gammadelta T cells in Crohn's disease: A new player in the disease pathogenesis? J Crohns Colitis 2017;11(9):1135–45. [DOI] [PubMed] [Google Scholar]
- 30.Uzzan M, Martin JC, Mesin L, et al. Ulcerative colitis is characterized by a plasmablast-skewed humoral response associated with disease activity. Nat Med 2022;28(4):766–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Castro-Dopico T, Colombel JF, Mehandru S. Targeting B cells for inflammatory bowel disease treatment: Back to the future. Curr Opin Pharmacol 2020;55:90–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lindner C, Thomsen I, Wahl B, et al. Diversification of memory B cells drives the continuous adaptation of secretory antibodies to gut microbiota. Nat Immunol 2015;16(8):880–8. [DOI] [PubMed] [Google Scholar]
- 33.Jacobsen GE, Fernandez I, Quintero MA, et al. Lamina propria phagocyte profiling reveals targetable signaling pathways in refractory inflammatory bowel disease. Gastro Hep Adv 2022;1(3):380–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Orecchioni M, Ghosheh Y, Pramod AB, et al. Macrophage polarization: Different gene signatures in M1(LPS+) vs. Classically and M2(LPS-) vs. Alternatively activated macrophages. Front Immunol 2019;10:1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Herrero-Cervera A, Soehnlein O, Kenne E. Neutrophils in chronic inflammatory diseases. Cell Mol Immunol 2022;19(2):177–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Argmann C, Hou R, Ungaro RC, et al. Biopsy and blood-based molecular biomarker of inflammation in IBD. Gut 2023;72(7):1271–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rubin SJS, Bai L, Haileselassie Y, et al. Mass cytometry reveals systemic and local immune signatures that distinguish inflammatory bowel diseases. Nat Commun 2019;10(1):2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kosoy R, Kim-Schulze S, Rahman A, et al. Deep analysis of the peripheral immune system in IBD reveals new insight in disease subtyping and response to monotherapy or combination therapy. Cell Mol Gastroenterol Hepatol 2021;12(2):599–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Corridoni D, Chapman T, Antanaviciute A, et al. Inflammatory bowel disease through the lens of single-cell RNA-seq technologies. Inflamm Bowel Dis 2020;26(11):1658–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mitsialis V, Wall S, Liu P, et al. Single-cell analyses of colon and blood reveal distinct immune cell signatures of ulcerative colitis and Crohn's disease. Gastroenterology 2020;159(2):591–608.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ekwall AK, Eisler T, Anderberg C, et al. The tumour-associated glycoprotein podoplanin is expressed in fibroblast-like synoviocytes of the hyperplastic synovial lining layer in rheumatoid arthritis. Arthritis Res Ther 2011;13(2):R40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Honma M, Minami-Hori M, Takahashi H, et al. Podoplanin expression in wound and hyperproliferative psoriatic epidermis: Regulation by TGF-beta and STAT-3 activating cytokines, IFN-gamma, IL-6, and IL-22. J Dermatol Sci 2012;65(2):134–40. [DOI] [PubMed] [Google Scholar]
- 43.Abreu MT, Davies JM, Quintero MA, et al. Transcriptional behavior of regulatory T cells predicts IBD patient responses to vedolizumab therapy. Inflamm Bowel Dis 2022;28(12):1800–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Honan AM, Vazquez EN, Chen Z. Lymph node stromal cell-intrinsic MHC class II expression promotes MHC class I-restricted CD8 T cell lineage conversion to regulatory CD4 T cells. J Immunol 2021;207(6):1530–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lamaison C, Tarte K. B cell/stromal cell crosstalk in health, disease, and treatment: Follicular lymphoma as a paradigm. Immunol Rev 2021;302(1):273–85. [DOI] [PubMed] [Google Scholar]
- 46.Samarani S, Sagala P, Jantchou P, et al. Phenotypic and functional changes in peripheral blood natural killer cells in Crohn disease patients. Mediators Inflamm 2020;2020:6401969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tang PM, Nikolic-Paterson DJ, Lan HY. Macrophages: Versatile players in renal inflammation and fibrosis. Nat Rev Nephrol 2019;15(3):144–58. [DOI] [PubMed] [Google Scholar]
- 48.Chapuy L, Bsat M, Sarkizova S, et al. Two distinct colonic CD14(+) subsets characterized by single-cell RNA profiling in Crohn's disease. Mucosal Immunol 2019;12(3):703–19. [DOI] [PubMed] [Google Scholar]