Abstract
In the transmissible spongiform encephalopathies, disease is closely associated with the conversion of the normal proteinase K-sensitive host prion protein (PrP-sen) to the abnormal proteinase K-resistant form (PrP-res). Amino acid sequence homology between PrP-res and PrP-sen is important in the formation of new PrP-res and thus in the efficient transmission of infectivity across species barriers. It was previously shown that the generation of mouse PrP-res was strongly influenced by homology between PrP-sen and PrP-res at amino acid residue 138, a residue located in a region of loop structure common to PrP molecules from many different species. In order to determine if homology at residue 138 also affected the formation of PrP-res in a different animal species, we assayed the ability of hamster PrP-res to convert a panel of recombinant PrP-sen molecules to protease-resistant PrP in a cell-free conversion system. Homology at amino acid residue 138 was not critical for the formation of protease-resistant hamster PrP. Rather, homology between PrP-sen and hamster PrP-res at amino acid residue 155 determined the efficiency of formation of a protease-resistant product induced by hamster PrP-res. Structurally, residue 155 resides in a turn at the end of the first alpha helix in hamster PrP-sen; this feature is not present in mouse PrP-sen. Thus, our data suggest that PrP-res molecules isolated from scrapie-infected brains of different animal species have different PrP-sen structural requirements for the efficient formation of protease-resistant PrP.
The transmissible spongiform encephalopathy (TSE) diseases include scrapie in sheep and goats, bovine spongiform encephalopathy (BSE) in cattle, and Creutzfeldt-Jakob disease in humans. A central pathogenic event in the TSE diseases involves the mammalian prion protein (PrP). PrP is a glycophosphatidylinositol (GPI)-anchored cell surface glycoprotein present in many different tissues (1, 11, 30) but present at particularly high levels in the brain (1). During the course of TSE infection, normal host PrP-sen, a protein which is both sensitive to digestion with proteinase K (PK) and detergent soluble, is converted to an abnormal, detergent-insoluble form which is partially resistant to PK digestion. This PK-resistant form of PrP, PrP-res, accumulates to high levels in the lymphoreticular and central nervous systems of the infected host. PrP-sen expression and PrP-res accumulation are both believed to be involved in the neurodegeneration which leads to the characteristic spongiform changes in the brains of infected animals (8). The close association of PrP-res with infectivity and the lack of any well-documented bacterial or viral association with TSE diseases have led to the hypothesis that PrP-res itself is the infectious agent (35). Although this hypothesis has yet to be proven, PrP-res and PrP-sen clearly play important roles in disease pathogenesis (4, 8, 9).
With the TSE diseases, there can be a strong barrier to infection of one animal species with the TSE agent of a different species. This resistance is manifested either as a long disease incubation time upon primary passage in the host animal or as a lack of clinical disease altogether. Species barriers in TSE diseases are of particular importance given the probability that BSE has crossed species barriers to cause variant Creutzfeldt-Jakob disease in humans in the United Kingdom (50). In the United States, the possibility exists that chronic wasting disease, a TSE identified for wild and captive populations of deer and elk in several western states (51, 52), could cross species barriers to infect range cattle and potentially expose the human population to a new TSE infection. Thus, it is important to understand the mechanisms underlying species barriers to infection with the TSE diseases and to determine how to prevent cross-species transmission of TSE infection.
Studies with transgenic mice have shown that the sequence of PrP influences the interspecies transmission of TSE infection between mice and Syrian hamsters (45, 46) and between humans and mice (49). In these studies, amino acid sequence homology between host PrP-sen and PrP-res associated with the incoming TSE agent appeared to be necessary for the efficient transmission of TSE infection across species (36, 45). Homology in the middle portion of the PrP molecule was particularly important (27, 34, 46, 47). Therefore, at the molecular level, TSE species barriers can be at least partly explained by the dependence of PrP-res formation on PrP amino acid sequence homology.
In vitro studies with mouse neuroblastoma cells persistently infected with mouse scrapie (Sc+-MNB cells) have demonstrated that protease-resistant PrP formation can be extremely sensitive to even minor differences between the PrP-sen and the PrP-res amino acid sequences (24, 32, 34, 46). In Sc+-MNB cells, substitution of the mouse-specific isoleucine with a hamster-specific methionine at residue 138 in mouse PrP-sen significantly inhibited the species-specific formation of mouse PrP-res (34). Interestingly, an isoleucine-to-methionine substitution occurs naturally at the equivalent residue (position 142) in goat PrP and is associated with resistance to both sheep scrapie and BSE infection, as indicated by a significant increase in disease incubation time (18). Thus, the same polymorphism at a single amino acid residue has been shown to have an effect on PrP-res formation in vitro and on cross-species transmission of TSE infection in vivo. This finding suggests that, for some animal models of scrapie, a mismatch at this amino acid residue between host PrP-sen and TSE-associated PrP-res could interfere with the transmission of TSE infection across species barriers.
In order to determine if homology at amino acid 138 also mediated PrP-res formation in other species, both Sc+-MNB cells and a cell-free model of protease-resistant PrP formation were used to determine the amino acid residues necessary for the formation of hamster PrP-res. Our data show that homology at amino acid residue 155, not at the hamster equivalent of mouse PrP residue 138 (i.e., hamster PrP residue 139), is necessary for the efficient formation of protease-resistant hamster PrP. Residue 155 is located in a structural region of PrP different from that in which residue 139 is located. Therefore, our data suggest that PrP-res molecules isolated from different species of TSE-infected animals have different PrP-sen structural requirements for inducing the formation of more PrP-res. Consequently, there is no single amino acid position which acts similarly in all species to allow the cross-species formation of PrP-res.
MATERIALS AND METHODS
Cells.
Sc+-MNB cells have been described previously (38, 39). These cells express mouse PrP-sen, accumulate mouse PrP-res, and replicate the mouse scrapie agent (38). The retrovirus packaging cell lines PA317 and Ψ2 have been described previously (43).
Antibodies.
The anti-hamster PrP-specific mouse monoclonal antibody 3F4 recognizes within hamster PrP an epitope which includes the hamster-specific methionines at positions 109 and 112 (5, 25). Normal mouse PrP-sen is not recognized by the antibody 3F4 (25). Substitution of the leucine and valine residues at the equivalent mouse positions (residues 108 and 111) with methionine results in the expression of the 3F4 epitope in mouse PrP (16). All of the recombinant hamster and mouse PrP-sen molecules used in this study expressed this antibody epitope. The anti-PrP peptide rabbit polyclonal antibody R.30 was raised to a PrP peptide encompassing residues 89 to 103 and recognizes both mouse PrP and hamster PrP (15).
Clones.
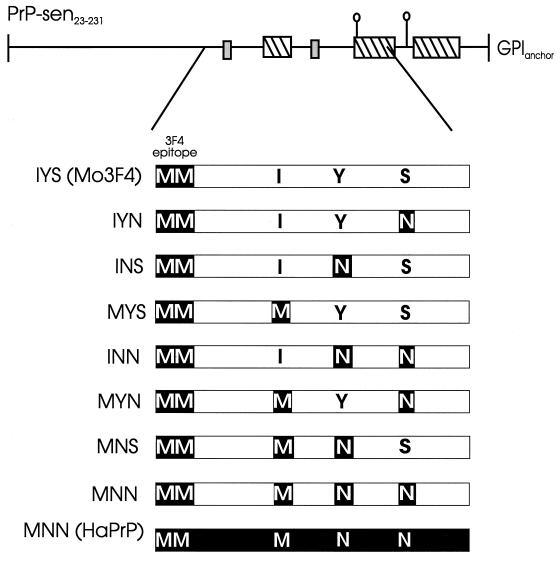
Mouse PrP-sen mutated to contain the 34F antibody epitope and a unique NaeI restriction endonuclease site (Mo3F4) and normal hamster PrP with a unique BstEII restriction endonuclease site have been described previously (16, 34). Mo3F4 clones containing amino acid mutations at residues 138, 154, and 169 were derived using a series of 10 overlapping oligonucleotides containing the desired mutations (34). These oligonucleotides spanned the region of Mo3F4 from the NaeI site to the BstEII site (nucleotides 436 to 660). For Mo3F4 without the GPI anchor [Mo3F4(GPINEG)], the GPI anchor addition site was removed by inserting a stop codon at residue 231 and deleting C-terminal amino acid residues 232 to 254 (26). Clones with the Mo3F4(GPINEG) background are designated by a three-letter code. Each letter represents the amino acid present at residue 138, 154, or 169, respectively (see Fig. 2).
FIG. 2.
Structures of recombinant PrP-sen (GPINEG) molecules. The top line shows the secondary structure of processed mouse PrP-sen (PrP-sen23–231) (23). The line designates turns and loops or disordered structure, the small gray boxes indicate areas of beta strands, and the hatched boxes indicate alpha helices. The N-linked glycosylation sites are indicated by lollipops. The GPI anchor is indicated on the right. The region of PrP from mouse residues 108 to 188, associated with the species-specific formation of PrP-res (27, 34, 47), is expanded below. White bars represent mouse PrP sequence, and black bars represent hamster PrP sequence. The two methionines at mouse and hamster PrP positions 108 and 109 and positions 111 and 112, respectively, which comprise the 3F4 antibody epitope, are indicated. The three variant amino acid residues between mouse PrP and hamster PrP at mouse and hamster positions 138 and 139, 154 and 155, and 169 and 170, respectively, are indicated. Residues shaded in black are specific to hamster PrP, while nonshaded residues are specific to mouse PrP. Clone designations are shown on the left.
Hamster PrP mutated at position 155 was derived by subcloning the NaeI-BstEII fragment of the mutant Mo3F4(GPINEG) construct MYN (see Fig. 2) into hamster PrP containing a unique BstEII site (34). All recombinant PrP-sen molecules were subcloned into the retrovirus expression vector pSFF and transfected into a 1:1 mixture of the retrovirus packaging cell lines PA317 and Ψ2 (10, 32). These cells were used both as a source of recombinant PrP-sen and as a source of infectious retrovirus encoding recombinant PrP-sen.
Analysis of recombinant PrP-sen and PrP-res in Sc+-MNB cells.
Sc+-MNB cells were transduced with PA317 and Ψ2 tissue culture supernatants containing infectious retroviruses encoding recombinant PrP-sen molecules (10, 32). Following transduction, cell surface immunofluorescence with antibody 3F4 showed that 80 to 100% of the Sc+-MNB cells expressed the transduced PrP-sen mutants (data not shown). Cells were also analyzed for recombinant PrP expression by radiolabeling with 35S-methionine-cysteine (Tran35S; NEN) followed by immunoprecipitation using the hamster-specific 3F4 antibody epitope as previously described (13, 32). In Sc+-MNB cells, the 3F4 antibody epitope allows exogenous recombinant PrP-sen to be distinguished from endogenous mouse PrP-sen and mouse PrP-res molecules, which do not contain the 3F4 epitope (32, 47). Following PK treatment, PrP-res derived from the exogenous PrP-sen mutants was detected by Western blotting with antibody 3F4, while PrP-res derived from both endogenous and exogenous PrP-sen was detected with the anti-PrP peptide rabbit polyclonal antibody R.30 (26, 32). Western blots were developed with the enhanced chemiluminescence system (Amersham) as specified by the manufacturer.
Cell-free conversion assays.
The contents of flasks (25 cm2) of a confluent Ψ2-PA317 cell culture expressing the desired recombinant PrP-sen were labeled with 1.5 mCi of Tran35S as described previously (10, 26). PrP-res was purified from brains of Syrian hamsters infected with the hamster scrapie strain 263K or VM/DK mice infected with the mouse scrapie strain 87V (10, 20). The in vitro conversion of PrP-sen to protease-resistant PrP has been described elsewhere (10, 26). Briefly, 200 ng of guanidine hydrochloride-treated PrP-res was mixed with 20,000 cpm (∼2 ng) of radiolabeled, immunoprecipitated PrP-sen. The reaction mixture was incubated at 37°C for 2 days. After incubation, 10% of the reaction mixture was precipitated in methanol (total PrP). The remaining 90% was treated with 12 μg of PK/ml for 1 h at 37°C. PK was inactivated by the addition of protease inhibitors, and the protein was methanol precipitated (PrP-res). Radiolabeled protease-resistant products were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The amounts of protease-resistant and protease-sensitive proteins were determined using a Molecular Dynamics Storm PhosphorImager system. Bands were quantified in terms of the integrated peak volume, and the percent conversion was calculated using the following formula: [(volume of PrP-res/volume of total PrP)(10)] × 100. The percent relative conversion was determined by comparing the level of conversion of hamster PrP-sen to that of mutant PrP-sen using the following formula: (percentage of mutant PrP-sen converted/percentage of hamster PrP-sen converted) × 100.
RESULTS
Homology at amino acid residue 139 is not sufficient for hamster PrP-res formation in Sc+-MNB cells.
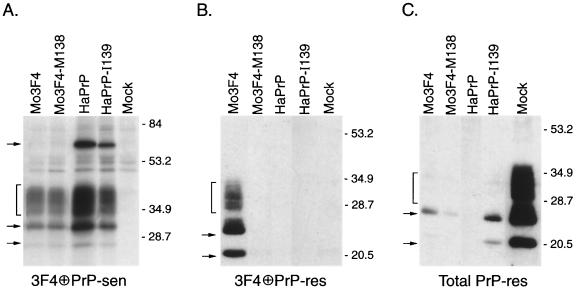
It was previously shown that the conversion of Mo3F4 PrP-sen by mouse PrP-res in Sc+-MNB cells was strongly dependent upon amino acid homology at position 138 (34). In order to determine if homology at the same position in hamster PrP-sen would allow its conversion by mouse PrP-res, the hamster-specific methionine at the equivalent hamster residue 139 was replaced with the mouse-specific isoleucine. The resultant mutant, HaPrP-I139, was expressed in Sc+-MNB cells and assayed for its ability to convert to PrP-res. When expressed at similar levels (Fig. 1A), neither HaPrP-I139 nor wild-type hamster PrP-sen was converted to PrP-res (Fig. 1B). Therefore, a mouse-specific isoleucine at position 139 was not sufficient to allow the cross-species conversion of hamster PrP-sen by mouse PrP-res.
FIG. 1.
Hamster PrP with a mouse-specific isoleucine at position 139 is not converted to PrP-res in Sc+-MNB cells. (A) Analysis of PrP-sen expression in Sc+-MNB cells transduced with the designated PrP-sen construct by radioimmunoprecipitation with antibody 3F4 (13, 32). The exposure time was 5 days. HaPrP, hamster PrP. (B) Analysis of PrP-res derived from the indicated 3F4 antibody epitope-positive recombinant PrP-sen constructs by Western blotting with antibody 3F4. Of the transduced constructs, only Mo3F4 PrP-sen was converted to PrP-res. (C) Analysis of overall PrP-res levels from endogenous and mutant PrP-res molecules in Sc+-MNB cells by Western blotting with the rabbit anti-PrP polyclonal antibody R.30. For all panels, the brackets and arrows on the left indicate PrP-specific bands and molecular mass markers in kilodaltons are shown on the right. The upper arrow in panel A designates the PrP dimer expressed by hamster PrP-sen (33). All data are from the same experiment, which was repeated five times. Extra lanes were excised from the gels for the purpose of data presentation. Mock, Sc+-MNB cells not expressing any exogenous 3F4-reactive PrP-sen.
Previous experiments have shown that introduction of the 3F4 epitope into mouse PrP-sen (i.e., a methionine at amino acid residues 108 and 111) can interfere with the formation of PrP-res from endogenous mouse PrP-sen in Sc+-MNB cells (32). If the level of expression of recombinant 3F4 epitope-containing PrP-sen is high enough, no PrP-res can be detected (31, 32). Since all of the constructs tested contained the 3F4 epitope, the lack of conversion of HaPrP-I139 to PrP-res in Sc+-MNB cells could be explained simply by a shutdown in PrP-res formation. In order to determine if Sc+-MNB cells expressing HaPrP-I139 still accumulated PrP-res, overall PrP-res levels were assayed by Western blotting using the rabbit polyclonal antibody R.30. This antibody detects both endogenous and recombinant PrP molecules. Endogenous mouse PrP-res was still detectable, albeit at low levels, in Sc+-MNB cells expressing HaPrP-I139 (Fig. 1C). However, even in experiments where little or no interference with endogenous mouse PrP-res formation was observed, protease-resistant HaPrP-I139 was never detected (data not shown). Thus, the inability of HaPrP-I139 to be converted to protease resistance was not a consequence of a shutdown in total PrP-res formation in Sc+-MNB cells. Overall, our results suggest that homology at amino acid residues other than 138 is necessary for hamster PrP-sen to be converted to PrP-res.
Homology at amino acid residue 155 influences the formation of protease-resistant hamster PrP.
In the region of PrP which has been shown to influence the species-specific formation of PrP-res (amino acids 112 to 188) (27, 34, 47), there are three differences between mouse PrP and hamster PrP, at mouse and hamster residues 138 and 139, 154 and 155, and 169 and 170, respectively (29). The formation of hamster PrP-res therefore could be dependent upon homology at a residue(s) other than position 139. Since tissue culture cells persistently infected with hamster scrapie are not available, we used a cell-free conversion system (26) to assay the ability of PrP-res derived from the brains of scrapie strain 263K-infected hamsters to convert a panel of radiolabeled PrP-sen molecules which had been mutated at the three variant residues (Fig. 2).
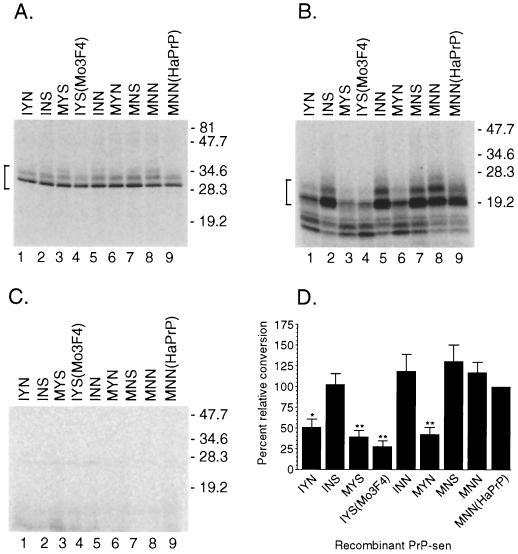
Since N-linked glycans are not required for PrP-res formation in the cell-free reaction (26, 41), recombinant PrP-sen molecules from which the GPI membrane anchor had been removed (GPINEG) were initially used in our mapping studies (26). Glycosylation is drastically reduced in GPINEG clones. As a result, these PrP molecules yield species-specific, appropriately sized, and easily identified conversion products in the cell-free conversion assay (26). All GPINEG clones contained the mouse PrP amino acid sequence, except that hamster-specific residues were present at positions 108 and 111 (the 3F4 epitope) and/or at positions 138, 154, and 169 (Fig. 2). Amino acid homology in regions of PrP-sen other than at residues 112 to 188 has been shown to have little or no influence on the species-specific formation of PrP-res (27, 34, 47). Thus, Mo3F4(GPINEG) PrP-sen with hamster PrP-specific residues from 108 to 188 (clone MNN) (Fig. 2) was converted to protease-resistant PrP by hamster PrP-res as efficiently as the homologous GPINEG hamster clone [clone MNN(HaPrP)] (Fig. 3B, compare lanes 8 and 9). Therefore, we initially used mutations in the Mo3F4(GPINEG) background to map the critical amino acid residues in hamster PrP-res-mediated conversions.
FIG. 3.
Homology at mouse PrP-sen amino acid residue 154 is important for hamster PrP-res-induced formation of protease-resistant PrP. (A to C) Lanes 1 to 8, formation of protease-resistant PrP from mutant Mo3F4(GPINEG) PrP-sen molecules; lane 9, GPINEG recombinant hamster PrP-sen. The name of the clone is indicated above each lane. Molecular mass markers in kilodaltons are indicated on the right. In panel A, 10% of the total reaction mixture without PK treatment but in the presence of 263K-derived hamster PrP-res was used. The input amount of radiolabeled recombinant PrP was equivalent for each reaction. The PrP bands which were quantified are indicated by the bracket on the left. In panel B, the remaining 90% of the reaction mixture from panel A following digestion with PK was used. The correctly sized protease-resistant PrP bands (26) which were quantified are indicated by the bracket on the left. In panel C, in the absence of 263K-derived hamster PrP-res, no protease-resistant products remained following PK digestion. (D) Percent relative conversion of the recombinant PrP-sen molecules from panel B. The graph represents data from 11 to 13 samples from several independent experiments. The percentage of homologous GPINEG hamster PrP converted to protease-resistant PrP by hamster PrP-res was set to 100% and compared to the amount of protease-resistant product formed by the heterologous PrP-sen mutants. This value, designated the percent relative conversion, as detailed in Materials and Methods, is shown on the left. The constructs tested are indicated below the graph. Error bars represent the standard error of the mean. Constructs for which results were significantly different from the homologous conversion of GPINEG hamster PrP at a P value of ≤0.01 are denoted by double asterisks, while a P value of ≤0.05 is denoted by a single asterisk. Statistical analysis was performed using a one-way repeated-measures analysis of variance with Dunnett's post test.
For all of the Mo3F4(GPINEG) mutants tested (Fig. 3A), the formation of protease-resistant PrP was always dependent upon the addition of 263K-derived hamster PrP-res (Fig. 3C). Mouse-specific amino acid residues at position 138 or 169 did not affect the hamster PrP-res-mediated conversion of PrP-sen (Fig. 3B, compare lanes 5 and 7 to lane 8). However, when a mouse-specific tyrosine was substituted for the hamster-specific asparagine at position 154, the level of protease-resistant PrP generated dropped significantly (Fig. 3B, compare lanes 6 and 8). Although the formation of protease-resistant PrP was never completely abolished, the effect of a tyrosine at residue 154 reproducibly reduced the formation of protease-resistant PrP by two- to fourfold regardless of homology at residues 138 and 169 (Fig. 3D). For example, Mo3F4(GPINEG) PrP-sen with a hamster-specific asparagine at position 154 but mouse-specific residues at positions 138 and 169 was converted to protease-resistant PrP as efficiently as GPINEG hamster PrP-sen (Fig. 3B, compare lanes 2 and 9, and Fig. 3D). Hamster PrP-res therefore requires homology at residue 154 (residue 155 in hamster PrP-sen) to efficiently convert PrP-sen to protease-resistant products.
Amino acid residue 155 affects protease-resistant PrP formation in wild-type hamster PrP-sen.
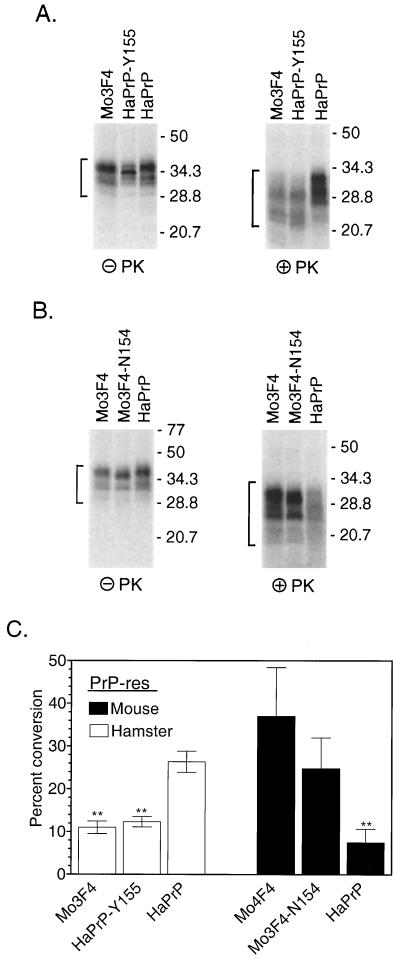
Normal PrP-sen is located at the cell surface and is inserted into the cell membrane via a GPI membrane anchor (12, 48) which may influence PrP-res formation. In order to determine if homology at residue 154 also affected protease-resistant PrP formation from GPI anchor-positive (GPIPOS) hamster PrP-sen (i.e., wild-type hamster PrP), GPIPOS hamster PrP-sen with a mouse-specific tyrosine at hamster residue 155 (HaPrP-Y155) was tested with the cell-free conversion assay. The efficiency of conversion of HaPrP-Y155 induced by hamster PrP-res was significantly lower than that of wild-type hamster PrP-sen (Fig. 4A, right panel). Furthermore, the pattern of the protease-resistant products was different from that of GPIPOS hamster PrP-sen and indistinguishable from that of Mo3F4(GPIPOS) PrP-sen (Fig. 4A, right panel). Thus, a mismatch at position 155 is important both qualitatively and quantitatively (Fig. 4C) in the formation of protease-resistant PrP from wild-type hamster PrP.
FIG. 4.
Efficient conversion of GPIPOS PrP-sen mutated at amino acid residue 154 or 155 is dependent upon the species of PrP-res. (A and B) Cell-free conversion of mutant (GPIPOS) mouse or hamster PrP-sen by PrP-res derived from scrapie-infected hamster (A) or mouse (B) brains. In each panel, the names of the radiolabeled PrP-sen molecules are indicated above the lanes. HaPrP, hamster PrP. The left panels show 10% of the total reaction mixture without PK treatment but in the presence of the indicated PrP-res constructs and demonstrate that the input amounts of radiolabeled PrP in the reactions were equivalent. The PrP bands which were quantified are indicated by the brackets on the left. The right panels show the remaining 90% of the reaction mixture following digestion with PK. The PrP-res bands which were quantified are indicated by the brackets on the left. The formation of protease-resistant PrP was dependent upon the addition of PrP-res (data not shown). Molecular mass markers in kilodaltons are indicated on the right. Extra lanes were excised from the gels for the purpose of data presentation. (C) Data from four independent repeats of the experiments in panels A and B. Error bars indicate the standard error of the mean. Statistical analysis was performed using a one-way repeated-measures analysis of variance with Dunnett's post test; double asterisks indicate a P value of <0.001.
Homology at amino acid residue 154 does not influence mouse PrP-res-mediated cell-free conversions.
In Sc+-MNB cells, amino acid residue 154 did not appear to significantly influence mouse PrP-res formation (34). This finding suggested that the strong negative effect of a mismatch at this residue was specific to conversions induced by 263K-derived hamster PrP-res. In order to determine if this was the case, a hamster-specific asparagine was substituted for the mouse-specific tyrosine in GPIPOS Mo3F4 PrP-sen (Mo3F4-N154). PrP-res derived from the brains of mice infected with mouse strain 87V converted Mo3F4-N154 almost as efficiently as wild-type Mo3F4 (Fig. 4B, right panel) but converted GPIPOS hamster PrP-sen poorly (Fig. 4B, right panel, and Fig. 4C). Thus, consistent with the results for Sc+-MNB cells (34), the formation of protease-resistant PrP by mouse PrP-res was not influenced by homology at residue 154 (residue 155 in hamster PrP).
DISCUSSION
We have shown that amino acid residue 155 in hamster PrP-sen is critically involved in the species-specific induction of protease-resistant PrP formation by hamster PrP-res. This residue is different from those which have been identified in species barriers to infection for other animal TSE diseases, including those of mice (34), goats (18), humans (41), and sheep (6, 7, 41). Our data suggest that different species of PrP-res have different amino acid sequence homology requirements for the efficient formation of protease-resistant PrP. Thus, it is likely that there is no universal amino acid residue in PrP which acts similarly in all PrP species as a “switch” to allow cross-species formation of PrP-res.
Several different studies have shown that efficient cross-species transmission of TSE infection between mice and Syrian hamsters can be affected by homology at certain amino acid residues between the PrP-sen of the mouse and the PrP-res associated with the incoming hamster scrapie agent (36, 45, 46). In our studies, the negative effect of a mismatch at position 155 did not lead to the complete abolition of protease-resistant PrP formation. This result suggests that in vivo, a mismatch at residue 155 between mouse PrP-sen and hamster PrP-res would not be absolutely protective but might simply delay disease development. Consistent with this prediction, recent studies have shown that mice infected with hamster scrapie propagate the scrapie agent (37), eventually begin to accumulate PrP-res, and develop clinical disease after extremely long incubation times (19). Our data provide an explanation at the molecular level for these results. Inefficient conversion of mouse PrP-sen to mouse PrP-res by the incoming hamster agent is strongly influenced by homology at amino acid residue 155, which could be responsible, at least in part, for the strong, but not absolute, TSE species barrier between mice and Syrian hamsters.
The precise mechanism by which position 155 affects protease-resistant PrP formation is unclear. At least two broadly defined and sequential events occur during the formation of PrP-res: (i) PrP-PrP binding and (ii) conversion (2, 17, 21). One analysis of the hamster PrP-sen nuclear magnetic resonance structure suggested that the difference between a mouse-specific tyrosine and a hamster-specific asparagine at position 155 would have little structural effect (28), while another predicted that changes at position 155 might modify the specificity of intermolecular interactions (3). Thus, a mismatch at position 155 could affect protease-resistant PrP formation by interfering with the specific binding of PrP-sen to PrP-res. Although our data are consistent with this possibility, residue 155 is outside of the region of PrP recently implicated in the initial interaction of PrP-sen with PrP-res (21). Furthermore, heterologous PrP-sen and PrP-res molecules appear to bind as efficiently as homologous PrP molecules (22; S. Priola, unpublished data), suggesting that a mismatch at residue 155 would have little effect. Therefore, if amino acid residue 155 has an influence on PrP-PrP binding, it would occur at an as-yet-unidentified inter- or intramolecular PrP-PrP interaction which follows the initial binding event.
Unlike the initial binding between PrP-sen and PrP-res, the conversion of PrP to protease resistance is very sensitive to changes in the PrP amino acid sequence (6, 7, 40, 41). Amino acid residue 155 is variable between mice and hamsters, a difference which is probably responsible for one minor change in secondary structure between the two molecules. In mouse PrP-sen, the tyrosine at 154 is at the C-terminal end of the first alpha helix (23, 42), while in hamster PrP-sen, residue 155 is an asparagine which forms a hydrogen-bonded turn at the end of the first alpha helix (28). If residue 155 influences a conversion event which follows the initial binding of PrP-sen to PrP-res, our data would suggest that this hydrogen-bonded turn is important in the efficient conversion of PrP to protease resistance induced by hamster PrP-res but not mouse PrP-res. This requirement for a hydrogen-bonded turn is different from the structural requirement for mouse PrP-res formation in which loop structures appear to be important (31, 34). That PrP-res molecules isolated from mouse or hamster scrapie-infected animals have different PrP-sen structural requirements is consistent with the different PrP-res conformations associated with these animal models of scrapie (14, 44).
ACKNOWLEDGMENTS
We thank Bruce Chesebro for making the GPINEG versions of our mutant mouse PrP molecules. We also thank Byron Caughey, Ina Vorberg, Bruce Chesebro, Kim Hasenkrug, and Karin Peterson for suggestions on and critiques of the manuscript.
REFERENCES
- 1.Bendheim P E, Brown H R, Rudelli R D, Scala L J, Goller N L, Wen G Y, Kascsak R J, Cashman N R, Bolton D C. Nearly ubiquitous tissue distribution of the scrapie agent precursor protein. Neurology. 1992;42:149–156. doi: 10.1212/wnl.42.1.149. [DOI] [PubMed] [Google Scholar]
- 2.Bessen R A, Kocisko D A, Raymond G J, Nandan S, Lansbury P T, Jr, Caughey B. Nongenetic propagation of strain-specific phenotypes of scrapie prion protein. Nature. 1995;375:698–700. doi: 10.1038/375698a0. [DOI] [PubMed] [Google Scholar]
- 3.Billeter M, Riek R, Wider G, Hornemann S, Glockshuber R, Wuthrich K. Prion protein NMR structure and species barrier for prion diseases. Proc Natl Acad Sci USA. 1997;94:7281–7285. doi: 10.1073/pnas.94.14.7281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blattler T, Brandner S, Raeber A J, Klein M A, Voigtlander T, Weissmann C, Aguzzi A. PrP-expressing tissue required for transfer of scrapie infectivity from spleen to brain. Nature. 1997;389:69–73. doi: 10.1038/37981. [DOI] [PubMed] [Google Scholar]
- 5.Bolton D C, Seligman S J, Bablanian G, Windsor D, Scala L J, Kim K S, Chen C M J, Kascsak R J, Bendheim P E. Molecular location of a species-specific epitope on the hamster scrapie agent protein. J Virol. 1991;65:3667–3675. doi: 10.1128/jvi.65.7.3667-3675.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bossers A, Belt P B G M, Raymond G J, Caughey B, de Vries R, Smits M A. Scrapie susceptibility-linked polymorphisms modulate the in vitro conversion of sheep prion protein to protease-resistant forms. Proc Natl Acad Sci USA. 1997;94:4931–4936. doi: 10.1073/pnas.94.10.4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bossers A, de Vries R, Smits M A. Susceptibility of sheep for scrapie as assessed by in vitro conversion of nine naturally occurring variants of PrP. J Virol. 2000;74:1407–1414. doi: 10.1128/jvi.74.3.1407-1414.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brandner S, Isenmann S, Raeber A, Fischer M, Sailer A, Kobayashi Y, Marino S, Weissmann C, Aguzzi A. Normal host prion protein necessary for scrapie-induced neurotoxicity. Nature. 1996;379:339–343. doi: 10.1038/379339a0. [DOI] [PubMed] [Google Scholar]
- 9.Bueler H, Aguzzi A, Sailer A, Greiner R-A, Autenried P, Aguet M, Weissmann C. Mice devoid of PrP are resistant to scrapie. Cell. 1993;73:1339–1347. doi: 10.1016/0092-8674(93)90360-3. [DOI] [PubMed] [Google Scholar]
- 10.Caughey B, Kocisko D A, Priola S A, Raymond G J, Race R E, Bessen R A, Lansbury P T, Jr, Chesebro B. Methods for studying prion protein (PrP) metabolism and the formation of protease-resistant PrP in cell culture and cell-free systems. In: Baker H F, Ridley R M, editors. Prion diseases. Totowa, N.J: Humana Press; 1996. pp. 285–300. [DOI] [PubMed] [Google Scholar]
- 11.Caughey B, Race R E, Chesebro B. Detection of prion protein mRNA in normal and scrapie-infected tissues and cell lines. J Gen Virol. 1988;69:711–716. doi: 10.1099/0022-1317-69-3-711. [DOI] [PubMed] [Google Scholar]
- 12.Caughey B, Race R E, Ernst D, Buchmeier M J, Chesebro B. Prion protein (PrP) biosynthesis in scrapie-infected and uninfected neuroblastoma cells. J Virol. 1989;63:175–181. doi: 10.1128/jvi.63.1.175-181.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caughey B, Raymond G J. The scrapie-associated form of PrP is made from a cell surface precursor that is both protease- and phospholipase-sensitive. J Biol Chem. 1991;266:18217–18223. [PubMed] [Google Scholar]
- 14.Caughey B, Raymond G J, Bessen R A. Strain-dependent differences in beta-sheet conformations of abnormal prion protein. J Biol Chem. 1998;273:32230–32235. doi: 10.1074/jbc.273.48.32230. [DOI] [PubMed] [Google Scholar]
- 15.Caughey B, Raymond G J, Ernst D, Race R E. N-terminal truncation of the scrapie-associated form of PrP by lysosomal protease(s): implications regarding the site of conversion of PrP to the protease-resistant state. J Virol. 1991;65:6597–6603. doi: 10.1128/jvi.65.12.6597-6603.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chesebro B, Wehrly K, Caughey B, Nishio J, Ernst D, Race R. Foreign PrP expression and scrapie infection in tissue culture cell lines. Dev Biol Stand. 1993;80:131–140. [PubMed] [Google Scholar]
- 17.DebBurman S K, Raymond G J, Caughey B, Lindquist S. Chaperone-supervised conversion of prion protein to its protease-resistant form. Proc Natl Acad Sci USA. 1997;94:13938–13943. doi: 10.1073/pnas.94.25.13938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goldmann W, Martin T, Foster J, Hughes S, Smith G, Hughes K, Dawson M, Hunter N. Novel polymorphisms in the caprine PrP gene: a codon 142 mutation associated with scrapie incubation period. J Gen Virol. 1996;77:2885–2891. doi: 10.1099/0022-1317-77-11-2885. [DOI] [PubMed] [Google Scholar]
- 19.Hill A F, Joiner S, Linehan J, Desbruslais M, Lantos P L, Collinge J. Species-barrier-independent prion replication in apparently resistant species. Proc Natl Acad Sci USA. 2000;97:10248–10253. doi: 10.1073/pnas.97.18.10248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hope J, Morton L J D, Farquhar C F, Multhaup G, Beyreuther K, Kimberlin R H. The major polypeptide of scrapie-associated fibrils (SAF) has the same size, charge distribution and N-terminal protein sequence as predicted for the normal brain protein (PrP) EMBO J. 1986;5:2591–2597. doi: 10.1002/j.1460-2075.1986.tb04539.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horiuchi M, Chabry J, Caughey B. Specific binding of normal prion protein to the scrapie form via a localized domain initiates its conversion to the protease-resistant state. EMBO J. 1999;18:3193–3203. doi: 10.1093/emboj/18.12.3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horiuchi M, Priola S A, Chabry J, Caughey B. Interactions between heterologous forms of prion protein: binding, inhibition of conversion, and species barriers. Proc Natl Acad Sci USA. 2000;97:5836–5841. doi: 10.1073/pnas.110523897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hornemann S, Korth C, Oesch B, Riek R, Wider G, Wuthrich K, Glockshuber R. Recombinant full-length murine prion protein, mPrP(23–231): purification and spectroscopic characterization. FEBS Lett. 1997;413:277–281. doi: 10.1016/s0014-5793(97)00921-6. [DOI] [PubMed] [Google Scholar]
- 24.Kaneko K, Zulianello L, Scott M, Cooper C M, Wallace A C, James T L, Cohen F E, Prusiner S B. Evidence for protein X binding to a discontinuous epitope on the cellular prion protein during scrapie prion propagation. Proc Natl Acad Sci USA. 1997;94:10069–10074. doi: 10.1073/pnas.94.19.10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kascsak R J, Rubenstein R, Merz P A, Tonna-DeMasi M, Fersko R, Carp R I, Wisniewski H M, Diringer H. Mouse polyclonal and monoclonal antibody to scrapie-associated fibril proteins. J Virol. 1987;61:3688–3693. doi: 10.1128/jvi.61.12.3688-3693.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kocisko D A, Come J H, Priola S A, Chesebro B, Raymond G J, Lansbury P T, Caughey B. Cell-free formation of protease-resistant prion protein. Nature. 1994;370:471–474. doi: 10.1038/370471a0. [DOI] [PubMed] [Google Scholar]
- 27.Kocisko D A, Priola S A, Raymond G J, Chesebro B, Lansbury P T, Jr, Caughey B. Species specificity in the cell-free conversion of prion protein to protease-resistant forms: a model for the scrapie species barrier. Proc Natl Acad Sci USA. 1995;92:3923–3927. doi: 10.1073/pnas.92.9.3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu H, Farr-Jones S, Ulyanov N B, Llinas M, Marqusee S, Groth D, Cohen F E, Prusiner S B, James T L. Solution structure of Syrian hamster prion protein rPrP(90–231) Biochemistry. 1999;38:5362–5377. doi: 10.1021/bi982878x. [DOI] [PubMed] [Google Scholar]
- 29.Locht C, Chesebro B, Race R, Keith J M. Molecular cloning and complete sequence of prion protein cDNA from mouse brain infected with the scrapie agent. Proc Natl Acad Sci USA. 1986;83:6372–6376. doi: 10.1073/pnas.83.17.6372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oesch B, Westaway D, Walchli M, McKinley M P, Kent S B H, Aebersold R, Barry R A, Tempst P, Teplow D B, Hood L E, Prusiner S B, Weissmann C. A cellular gene encodes scrapie PrP 27 to 30 protein. Cell. 1985;40:735–746. doi: 10.1016/0092-8674(85)90333-2. [DOI] [PubMed] [Google Scholar]
- 31.Priola S A. Prion protein and species barriers in the transmissible spongiform encephalopathies. Biomed Pharmacother. 1999;53:27–33. doi: 10.1016/s0753-3322(99)80057-2. [DOI] [PubMed] [Google Scholar]
- 32.Priola S A, Caughey B, Race R E, Chesebro B. Heterologous PrP molecules interfere with accumulation of protease-resistant PrP in scrapie-infected murine neuroblastoma cells. J Virol. 1994;68:4873–4878. doi: 10.1128/jvi.68.8.4873-4878.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Priola S A, Caughey B, Wehrly K, Chesebro B. A 60-kDa prion protein (PrP) with properties of both the normal and scrapie-associated forms of PrP. J Biol Chem. 1995;270:3299–3305. doi: 10.1074/jbc.270.7.3299. [DOI] [PubMed] [Google Scholar]
- 34.Priola S A, Chesebro B. A single hamster amino acid blocks conversion to protease-resistant PrP in scrapie-infected mouse neuroblastoma cells. J Virol. 1995;69:7754–7758. doi: 10.1128/jvi.69.12.7754-7758.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prusiner S B. Novel proteinaceous infectious particles cause scrapie. Science. 1982;216:136–144. doi: 10.1126/science.6801762. [DOI] [PubMed] [Google Scholar]
- 36.Prusiner S B, Scott M, Foster D, Pan K M, Groth D, Mirenda C, Torchia M, Yang S L, Serban D, Carlson G A, Hoppe P C, Westaway D, DeArmond S J. Transgenetic studies implicate interactions between homologous PrP isoforms in scrapie prion replication. Cell. 1990;63:673–686. doi: 10.1016/0092-8674(90)90134-z. [DOI] [PubMed] [Google Scholar]
- 37.Race R, Chesebro B. Scrapie infectivity found in resistant species. Nature. 1998;392:770. doi: 10.1038/33834. [DOI] [PubMed] [Google Scholar]
- 38.Race R E, Caughey B, Graham K, Ernst D, Chesebro B. Analyses of frequency of infection, specific infectivity, and prion protein biosynthesis in scrapie-infected neuroblastoma cell clones. J Virol. 1988;62:2845–2849. doi: 10.1128/jvi.62.8.2845-2849.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Race R E, Fadness L H, Chesebro B. Characterization of scrapie infection in mouse neuroblastoma cells. J Gen Virol. 1987;68:1391–1399. doi: 10.1099/0022-1317-68-5-1391. [DOI] [PubMed] [Google Scholar]
- 40.Raymond G J, Bossers A, Raymond L D, O'Rourke K I, McHolland L E, Bryant III P K, Miller M W, Williams E S, Smits M, Caughey B. Evidence of a molecular barrier limiting susceptibility of humans, cattle and sheep to chronic wasting disease. EMBO J. 2000;19:4425–4430. doi: 10.1093/emboj/19.17.4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raymond G J, Hope J, Kocisko D A, Priola S A, Raymond L D, Bossers A, Ironside J, Will R G, Chen S G, Petersen R B, Gambetti P, Rubenstein R, Smits M A, Lansbury P T, Jr, Caughey B. Molecular assessment of the transmissibilities of BSE and scrapie to humans. Nature. 1997;388:285–288. doi: 10.1038/40876. [DOI] [PubMed] [Google Scholar]
- 42.Riek R, Hornemann S, Wider G, Billeter M, Glockshuber R, Wuthrich K. NMR structure of the mouse prion protein domain PrP(121–231) Nature. 1996;382:180–182. doi: 10.1038/382180a0. [DOI] [PubMed] [Google Scholar]
- 43.Robertson N M, Spangrude G J, Hasenkrug K, Perry L, Nishio J, Wehrly K, Chesebro B. Role and specificity of T-cell subsets in spontaneous recovery from Friend virus-induced leukemia in mice. J Virol. 1992;66:3271–3277. doi: 10.1128/jvi.66.6.3271-3277.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Safar J, Wille H, Itri V, Groth D, Serban H, Torchia M, Cohen F E, Prusiner S B. Eight prion strains have PrP(Sc) molecules with different conformations. Nat Med. 1998;4:1157–1165. doi: 10.1038/2654. [DOI] [PubMed] [Google Scholar]
- 45.Scott M, Foster D, Mirenda C, Serban D, Coufal F, Walchli M, Torchia M, Groth D, Carlson G, DeArmond S J, Westaway D, Prusiner S B. Transgenic mice expressing hamster prion protein produce species-specific scrapie infectivity and amyloid plaques. Cell. 1989;59:847–857. doi: 10.1016/0092-8674(89)90608-9. [DOI] [PubMed] [Google Scholar]
- 46.Scott M, Groth D, Foster D, Torchia M, Yang S L, DeArmond S J, Prusiner S B. Propagation of prions with artificial properties in transgenic mice expressing chimeric PrP genes. Cell. 1993;73:979–988. doi: 10.1016/0092-8674(93)90275-u. [DOI] [PubMed] [Google Scholar]
- 47.Scott M R, Kohler R, Foster D, Prusiner S B. Chimeric prion protein expression in cultured cells and transgenic mice. Protein Sci. 1992;1:986–997. doi: 10.1002/pro.5560010804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stahl N, Borchelt D R, Hsiao K, Prusiner S B. Scrapie prion protein contains a phosphatidylinositol glycolipid. Cell. 1987;51:229–240. doi: 10.1016/0092-8674(87)90150-4. [DOI] [PubMed] [Google Scholar]
- 49.Telling G C, Scott M, Hsiao K K, Foster D, Yang S L, Torchia M, Sidle K C, Collinge J, DeArmond S J, Prusiner S B. Transmission of Creutzfeldt-Jakob disease from humans to transgenic mice expressing chimeric human-mouse prion protein. Proc Natl Acad Sci USA. 1994;91:9936–9940. doi: 10.1073/pnas.91.21.9936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Will R G, Ironside J W, Zeidler M, Cousens S N, Estibeiro K, Alperovitch A, Poser S, Pocchiari M, Hofman A, Smith P G. A new variant of Creutzfeldt-Jakob disease in the UK. Lancet. 1996;347:921–925. doi: 10.1016/s0140-6736(96)91412-9. [DOI] [PubMed] [Google Scholar]
- 51.Williams E S, Young S. Chronic wasting disease of captive mule deer: a spongiform encephalopathy. J Wildl Dis. 1980;16:89–98. doi: 10.7589/0090-3558-16.1.89. [DOI] [PubMed] [Google Scholar]
- 52.Williams E S, Young S. Spongiform encephalopathy of Rocky Mountain elk. J Wildl Dis. 1982;18:465–471. doi: 10.7589/0090-3558-18.4.465. [DOI] [PubMed] [Google Scholar]