Abstract
Intronic elements flanking the splice-site consensus sequences are thought to play a role in pre-mRNA splicing. However, the generality of this role, the catalog of effective sequences, and the mechanisms involved are still lacking. Using molecular genetic tests, we first showed that the ∼50-nt intronic flanking sequences of exons beyond the splice-site consensus are generally important for splicing. We then went on to characterize exon flank sequences on a genomic scale. The G+C content of flanks displayed a bimodal distribution reflecting an exaggeration of this base composition in flanks relative to the gene as a whole. We divided all exons into two classes according to their flank G+C content and used computational and statistical methods to define pentamers of high relative abundance and phylogenetic conservation in exon flanks. Upstream pentamers were often common to the two classes, whereas downstream pentamers were totally different. Upstream and downstream pentamers were often identical around low G+C exons, and in contrast, were often complementary around high G+C exons. In agreement with this complementarity, predicted base pairing was more frequent between the flanks of high G+C exons. Pseudo exons did not exhibit this behavior, but rather tended to form base pairs between flanks and exon bodies. We conclude that most exons require signals in their immediate flanks for efficient splicing. G+C content is a sequence feature correlated with many genetic and genomic attributes. We speculate that there may be different mechanisms for splice site recognition depending on G+C content.
Pre-mRNA splicing is a fundamental step in gene expression. During this process, splice sites must be recognized within large introns before subsequent steps occur. How the splicing machinery manages to recognize splice sites remains a central problem in the study of splicing and its regulation. Three conserved sequence motifs, the 5′ splice site, the 3′ splice site and the branch-point (BP), are known to be necessary for the two sequential transesterification reactions by which introns are removed and exons are joined (Adams et al. 1996; Jurica and Moore 2003). However, these sequences are quite degenerate; one can find about two orders of magnitude more intronic sequences that match either splice-site consensus, as well as or better than the sites that are actually used (Sun and Chasin 2000; Zhang et al. 2003). The BP consensus is even more degenerate (Harris and Senapathy 1990; Senapathy et al. 1990).
There is strong genetic and biochemical evidence for the idea that the exon is the unit of initial recognition, and that there is coupling between the recognition of 5′ and 3′ splice sites across the exon (Robberson et al. 1990; Carothers et al. 1993; Berget 1995). However, even if pairing of 3' and 5' splice site-like sequences is stipulated and the distance between them constrained to a range typical for real exons, these “pseudo exons” still out-number the real exons by at least an order of magnitude (Sun and Chasin 2000). Thus, the information conveyed by splice-site sequences alone is obviously insufficient for exon recognition.
One source of this additional information resides in the bodies of the exons themselves in the form of exonic splicing-enhancer (ESE) sequence elements. Most knowledge about ESEs has come from studies of their role in stimulating the use of particular alternatively used splice sites (Manley and Tacke 1996; Graveley 2000; Black 2003). However, ESEs also function in constitutive splicing (Xu et al. 1993; Yeakley et al. 1996; Mayeda et al. 1999; Schaal and Maniatis 1999a; Zhang and Chasin 2004). A wide variety of ESE sequences have been identified by systematic searches using iterative selection experiments (Tacke and Manley 1995; Coulter et al. 1997; Liu et al. 1998; Cavaloc et al. 1999; Schaal and Maniatis 1999b; Liu et al. 2000; Tian and Kole 2001; Cartegni et al. 2003) and by computational approaches (Fairbrother et al. 2002; Zhang and Chasin 2004). The sequences found are quite degenerate and appear to be very abundant in exons. In a similar manner, exonic splicing silencers (ESS) have been found, albeit at a lower prevalence, in alternatively and constitutively spliced exons (Ladd and Cooper 2002; Black 2003; Wang et al. 2004; Zhang and Chasin 2004).
The analogous regulatory elements in introns, intronic-splicing enhancers (ISEs), and intronic-splicing silencers (ISSs) have been less systematically studied and will be the subject of this report. Many ISEs and ISSs have been identified through the study of individual alternatively spliced genes and have been compiled in several recent reviews (Ladd and Cooper 2002; Black 2003; Zheng 2004). Most of these elements are located near the 5′ or 3′ ends of introns, but some can be found hundreds of nucleotides (nt) from an exon border. In many cases, these elements have been shown to bind hnRNP proteins, in particular hnRNP A1 (Chabot et al. 2003) or PTB (Wagner and Garcia-Blanco 2001). Iterative functional selections have not yet been applied toward a functional definition of ISE or ISS elements, although they have been used to define the binding sites of specific splicing inhibitory factors, e.g., PSI, PTB, and hnRNP A1 (Burd and Dreyfuss 1994; Singh et al. 1995; Amarasinghe et al. 2001). Iterative selections have been used to define functional polypyrimidine tracts and BP sequences (Buvoli et al. 1997; Lund et al. 2000). Although these are intronic sequences necessary for splicing, they are considered elements integral to the splicing process rather than enhancers.
Computational implication of ISE sequence motifs have been made by searching for sequences that are overrepresented in exon flanks. We define exon flanks here as intronic sequences beyond the customary limits of the consensus splice sites, upstream of -14 and downstream of +6 with respect to the exon borders. Note that this definition puts the BP within what we are calling the exon flank. Since the BP occupies a variable position (usually between 18 and 40 nt upstream of the exon) it could be intermingled with ISEs and ISSs (Reed and Maniatis 1985). Moreover, since BP sequences display such a wide latitude, they themselves could constitute part of an ISE. Another element in the upstream flank that may be distinct from an ISE is an anchoring site of 20 nt just upstream of the BP. This region has been implicated in sequence-independent binding of U2 snRNP proteins (Gozani et al. 1996), but its role in splicing can be sequence dependent (Ast et al. 2001). Finally, taking the extent of the polypyrimidine tract to be -14 is a bit arbitrary, as the overrepresentation of pyrimidines extends at a lower level beyond that distal limit (Penotti 1991; Stephens and Schneider 1992; Zhang et al. 2003).
Studies of small introns (McCullough and Berget 2000), as well as introns in general (Nussinov 1989; Engelbrecht et al. 1992; Louie et al. 2003; Zhang et al. 2003; Yeo et al. 2004) have identified C-rich and G-rich motifs as potential ISEs. This role was supported by their preferred association with weak splice sites (Yeo et al. 2004) and by empirical tests in the case of GGG triplets (McCullough and Berget 2000). Sorek and Ast reported that the flanks of alternative exons are more conserved than those of constitutive exons, suggesting a functional role for these flanking sequences (Sorek and Ast 2003).
Using machine learning in the form of a support vector machine (SVM) classifier, we previously showed that the flanks of constitutive exons can be distinguished from those of pseudo exons by the overrepresentation and underrepresentation of several classes of pentamers (Zhang et al. 2003). Candidates for positive motifs included G-rich, C-rich, and TG-rich pentamers, as well as pentamers that resemble the BP consensus. In examining these sequences, we were struck by the preponderance of G and C among the downstream winning pentamers. To see whether such a disproportion was true in general, we examined the global distribution of G+C content (GC%) in 50-nt exon flanks. To our surprise, these regions displayed a striking bimodality in GC%. This finding raised the possibility that exons are divided into two classes that presented highly divergent splicing signals. On the other hand, GC% has been correlated with many aspects of gene structure and behavior (Versteeg et al. 2003); thus, it is possible that the high and low GC% exon flanks were serving some purpose other than splicing. Thus, before embarking on a detailed statistical characterization of exon flank sequences based on this partition, we wanted to test the hypothesis that an exon generally requires compatible flanks in order to be spliced. In support of this idea are the numerous reports of particular ISEs and ISSs mentioned above. However, since these reports are almost always based on a priori positive results, they may not reflect the general state of affairs. We therefore carried out a limited prospective survey of arbitrarily chosen exons to ask whether their splicing efficiency depended on the inclusion of their natural flanks. In most cases, that proved to be the case, and so we proceeded with a genomic analysis of the flanks of GC-rich and AT-rich exons aimed at identifying candidate ISE motifs for each class, and comparing their organization.
We used straightforward statistical methods and phylogenetic conservation to identify pentamers that are overrepresented in each set of exon flanks. Our separate treatment of GC-rich and AT-rich exons allowed us to detect motifs that were missed when examining exons as a whole, particularly among the AT-rich exons. The pentamer sequences from these two classes of exons present quite different targets for splicing factors. Further analysis suggested secondary structure may play a role in the recognition of GC-rich exons, but not AT-rich exons. Overall, these results raise the possibility that exons in GC-rich and AT-rich genes are spliced by distinguishable mechanisms.
Results
The intronic flanking regions of exons are functionally important in splicing
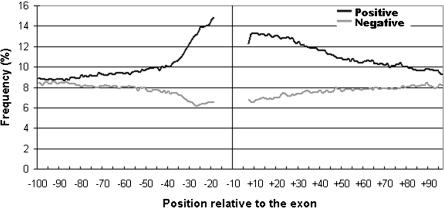
In a previous study, we identified groups of pentamers in intronic regions upstream of the polypyrimidine tract of exons (<-14) and downstream of the consensus 5′ splice site (>+6) that helped distinguish real exons from intronic pseudo exons. We defined these pentamers as “positive” or “negative”, depending on whether they were preferentially associated with real exons or pseudo exons, respectively. The frequencies of these four pentamer classes around exons are plotted in Figure 1. The positive pentamers are up to 50% more frequent in the 50-nt flanks than in deeper intronic regions. The prevalence of these pentamers was limited to about 50 nt beyond the splice site consensus sequences and, unless noted otherwise, our reference to “flanks” will be to the regions from -64 to -15 upstream and +7 to +56 downstream of exons. We concluded that exon flanks are highly nonrandom and proposed that they may be generally important in splicing (Zhang et al. 2003).
Figure 1.
Distribution around exons of positive and negative flanking pentamers characteristic of exons found by SVM analysis (Zhang et al. 2003). Raw frequencies of all 64 positive (black line) and 62 negative (gray line) pentamers are plotted against the positions relative to the exon. Upstream and downstream pentamers are plotted only in the corresponding flanks.
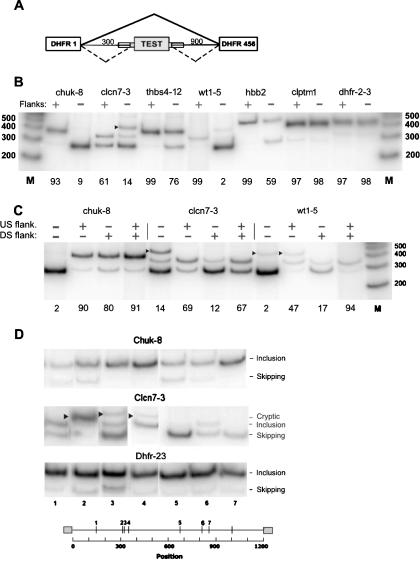
Before proceeding with a more detailed analysis of flank sequences, we wanted to test the proposed role of these regions in splicing. We thus measured the splicing of seven internal exons from seven genes in the presence or absence of their ∼50-nt flanks. Six of these exons (chuk-8, clcn7-3, thbs4-12, clptm1-13, hbb-2, and wt1-5) were human. The seventh was a hamster exon, dhfr-2-3, the fused exons 2 and 3 of the 6-exon dhfr gene. Among the six human exons, five are constitutively spliced and one is a well-known alternative exon, the fifth exon of the wt-1 gene (Natoli et al. 2002). Each exon was inserted as the central exon in a 3-exon hamster dhfr minigene (Fig. 2A). The minigene was derived from a construct originally comprised of dhfr exon 1, exon 2+3, and exons 4+5+6, each separated by an intron. Exon 2+3 with its splice consensus sequences was replaced by a NotI site to serve as an insertion site for heterologous exons. Thus, the sequences surrounding the NotI insertion site in the test exon retain the original dhfr flanks, including the branch point of intron 1 at -24 (Chen and Chasin 1993). The experiment was designed in this way to increase the chance of revealing sequences or sequence relationships that influence splicing over and above the presence of a functional branch site; a similar consideration applies to the downstream flank; the flank deletion experiments here are testing for effects that cannot be fulfilled by any ISEs that might be present in the original dhfr flank. In the special case of dhfr-2-3, when we inserted the exon without its flanks back into the NotI site, we considered this construct to be “with flanks”, since the original flanks were restored. We then inserted a 25-nt bacterial sequence (from transposon Tn5, see Supplemental methods) on each side of this exon, thus distancing each splice site from its original flank by this amount on each side. Although not strictly the case, we put this version of dhfr-2-3 into the “flankless” category.
Figure 2.
Effect of flanks on splicing. (A) Schematic diagram of the test construct used. PCR was used to insert the test exon with both, neither, or one flank of ∼50 nt beyond the splice-site consensus. (Thin lines) Introns; (large rectangles) exons; (small rectangles superimposed on lines) flanks. (B) Effect of inclusion of both flanks on splicing. Cloned plasmids were transfected into 293 cells and subjected to RT-PCR using incorporation of [32P]dATP and polyacrylamide gel electrophoresis; the intensity of the radioactive bands was quantified with a PhosphorImager. Markers are from an end-labeled 100-bp DNA ladder (Invitrogen). (chuk) Conserved helix-loop-helix ubiquitous kinase; (clcn7) chloride channel 7; (thbs4) thrombospondin 4; (wt1) Wilms tumor 1; (hbb) human β-globin; (clptm1) cleft lip and palate transmembrane protein 1; (dhfr-2-3) fused exons 2 and 3 of the Chinese hamster dhfr gene. The number after the hyphen denotes the exon number. An arrowhead indicates use of a cryptic donor splice site found by sequencing the PCR product to be gaa|gtaagt at +83 in the downstream dhfr intron 3; otherwise, the upper band position represents exon inclusion, and the lower band position represents exon skipping. The percent inclusion (radioactivity in the included band representing exon splicing divided by the total of all bands) is indicated below each panel. (C) Testing the effect of individual flanks in the case of 3 exons as in B above. (D) Splicing of the indicated flankless exons inserted at random locations within the 1200-nt intronic sequence of the test construct. In this experiment, each exon was bounded on each side by the same 25-nt bacterial sequence used for transposition. Cloned plasmids were transfected into 293 cells and analyzed by RT-PCR as in B. (Arrowheads) Splicing at unidentified cryptic sites. The seven insertion positions were at the following distances from the 5′ end of the intron: 143, 310, 321, 339, 674, 809, and 864, respectively. The insertion point for the experiments shown in B and C was 304, the natural end of intron 1 of the hamster dhfr gene.
Splicing of these transcripts was assayed by transient transfection of human 293 cells. For five of the six human exons, the flankless version was spliced much less efficiently than its flanked counterpart (Fig. 2B), confirming a functional role of flanks in splicing. This test does not strictly apply to the dhfr exon, as its natural flanks are restored in the insertion, except for the introduction of a few nucleotides comprised by the engineered NotI site. We also tested the upstream and downstream flanks separately for three of these exons (Fig. 2C). For chuk-8, the two flank effects were redundant; either flank was adequate to increase splicing from 10% to at least 80%. For clcn7-3, the upstream flank alone sufficed. For wt1-5, each flank alone had a partial positive effect; only the two together produced efficient splicing. In two of these cases (wt1-5 and clcn7-3), removal of a flank also partially activated a cryptic 5′ splice site downstream of the inserted exons in the host sequence.
In a second type of experiment, we tested the effect of varying the sequence context of a given exon by placing it at different locations within an intron. We used a Tn5 transposon system to insert an exon at random positions within the dhfr intron (a composite of intron 1 and an abbreviated intron 3). Three exons (dhfr-2-3, chuk-8, and clcn7-3) were inserted as the central exon into seven different positions within the 1200-nt intron in the test minigene. Each exon included its splice-site sequence from -14 to +1 on the upstream side and -3 to +6 on the downstream side, but no additional flanking sequence. Splicing of the 21 constructs was then assayed by transient transfection. Each of the exons displayed a different splicing profile for position effects (Fig. 2D). One exon, dhfr-2-3, was fairly insensitive to the change of intronic context, whereas the other two exons displayed different degrees of context dependence. In particular, splicing of clcn7-3 was highly dependent on its environment, in that it was almost entirely skipped at certain positions (e.g., positions 3 and 5 in Fig. 2D), and a cryptic site was activated at some other positions (e.g., position 2). Note that the distance between positions 2 and 3 is only 11 nt. These results are consistent with the data showing that splicing of chuk-8 and clcn7-3 is compromised when flanks are removed (Fig. 2B). All these data support the idea that particular flanking sequences are important for the splicing of many, if not most exons.
Having confirmed that the flanking sequences of exons are often required for splicing, we proceeded to carry out a detailed computational analyses of exon flanks on a genomic scale. We reasoned that knowledge gained from computational analyses would suggest hypotheses and guide more pointed experiments in the future.
GC% of the 50-nt flanks of exons display a bimodal distribution
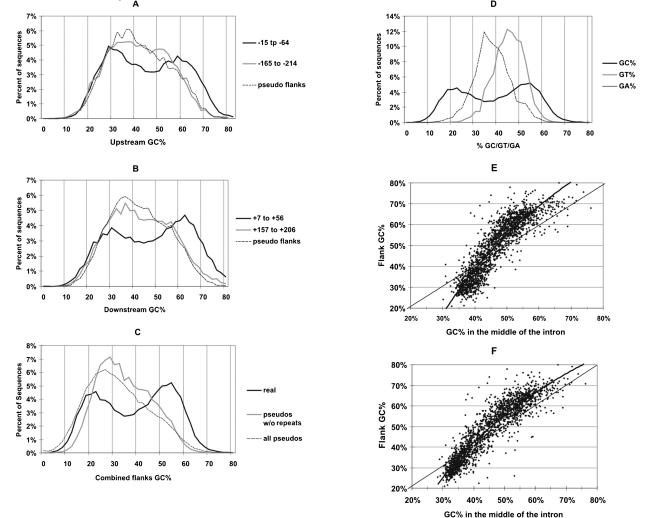
We noticed that the pentamers defined as positive signals in downstream flanks in our previous study (Zhang et al. 2003) had an unusually high GC% (G+C content) of 68%. In contrast, the GC% of the negative pentamers was only 39%. This observation led us to examine the overall GC% distribution in exon flanks. In examining over 16,000 exons, we found that the GC% of both the upstream flanks and the downstream flanks displayed a bimodal distribution (Fig. 3, A and B, respectively). Upstream flanks had a lower GC% in general than downstream flanks (the bimodal curve is shifted leftward in Fig. 3A compared with that in Fig. 3B), probably because T is more abundant than C in the extended polypyrimidine tracts in the upstream flanks and G-triplets are common in downstream flanks (see the following results). When the GC% of the upstream and downstream flank across each exon were averaged, the bimodality was even more dramatic (Fig. 3C). The same distribution does not result when deeper intronic fragments of the same size are examined (as shown by the thick gray curves in Fig. 3A,B). Thus, the bimodality is specific to the immediate flanks of exons. For comparison, we repeated this analysis for pseudo exons rather than real exons; pseudo exon flanks do not have this property (Fig. 3A,B, the lightest gray curves). To see whether this unusual distribution is particular to the G+C base combination we analyzed distributions of GA% and GT%; neither of these combinations showed a bimodal distribution (Fig. 3D). The peak of pseudo exons can be seen to be skewed toward low GC% due to the fact that pseudo exons are derived from introns and introns are generally AT-rich in the human genome.
Figure 3.
GC% distributions of flanks. (A,B) GC% distributions of 50-nt flank regions upstream and downstream of exons, respectively. The solid curves show the GC% distribution of sequence windows at the indicated distances (black curves, -15 to -64; gray curves, -165 to -214) from the 3′ or 5′ end of exons. The dashed curve shows the GC% distribution of the closer 50-nt windows in the flanks of pseudo exons. (C) GC% distributions of averaged GC% of both closest 50-nt windows (i.e., -15 to -64 and +7 to +56) upstream and downstream of exons, compared with the same distribution of pseudo exons with or without repeats. (D) GC% distribution for real exon flanks as in C compared with their GT% and GA% distributions. (E,F) Flanks tend to exaggerate GC%. (E) GC% of flanks is plotted against GC% of the remainder of the introns. (F) The same as A, but with repeats masked.
It is generally accepted that most human genes lie entirely within high GC% regions or within low GC% regions of the genome (Bernardi 2000; Versteeg et al. 2003). If large-scale variations in G+C content alone were responsible for the bimodality of GC% in exon flanks, one would expect that pseudo exon flanks and deeper intron segments would have a similar GC% distribution. However, these regions do not exhibit the same GC% bimodality (Fig. 3A,B). The insertion of highly repeated sequences during the course of evolution can dilute GC% variation (Smit 1999; Duret and Hurst 2001), and it is known that repeated sequences are relatively scarce in exon flanks (Majewski and Ott 2002) compared with deeper regions of introns. We therefore considered the possibility that the restriction of the bimodal distribution to exon flanks was the indirect result of a difference in repeat distribution. However, masking all the repeats in our data set did not reveal any hidden bimodal GC% distribution in the flanks of pseudo exons (Fig. 3C).
Since upstream flanks and downstream flanks displayed very similar GC% distributions (Fig. 3A,B), we asked whether there was a correlation of GC% between the flanks that bound an exon. Because GC% is not homogeneously distributed in the human genome as a whole, we expected to see such a correlation between any two 50-nt sequences within a given gene. We measured the correlation across the control pseudo exons as a measure of this background, finding a correlation of r2 = 0.01. If pseudo exons that overlap highly repeated sequences are eliminated, then the correlation increases to r2 = 0.08, reflecting the damping effect of repeat insertions mentioned above (Smit 1999; Duret and Hurst 2001). The correlation across real exons is much stronger, r2 = 0.35. The difference between 0.35 and 0.08 is highly significant (P <10-100). We also saw a significant correlation between the flanks that bound an intron (r2 = 0.20); this correlation is also significantly different from our background measurement of 0.08 (P <10-100).
Flanks tend to exaggerate the GC% of the gene in which they reside
In comparing the GC% of flanks with that of the rest of the intron sequence, we found that there is a substantial exaggeration of GC% in flanks. That is, if the introns are rich in GC%, the flanks will be even richer in GC%, and if the introns are AT-rich, the flanks will be even more AT-rich (Fig. 3E). To address the argument mentioned in the previous paragraph, we also plotted the data after removal of repeats (Fig. 3F). Although somewhat diminished, the exaggeration is still obvious and remains highly significant (P < 10-35 for GC% < 45% and P < 10-60 for GC% ≥ 45%, one tail paired t-test).
Downstream flanks of high and low GC% exons have different potential splicing signals
That exon flanks exhibit a dichotomous GC% distribution, yet contain functional sequences, raises an interesting question; Do GC-rich and GC-poor flanks nevertheless harbor similar splicing signals? To identify potential splicing signals, we sought sequence motifs that were overrepresented in exon flanks compared with other intronic regions. To address the possibility that such signals would be different according to the GC% of flanks, we divided all of the real exons into two classes, those with a flank GC% > 55% (the high GC% class or HGC) and those with a flank GC% < 45% (the low GC% class or LGC). Flank GC% here is defined as the average GC% of the upstream (-64 to -15) and downstream (+7 to +56) flanks combined. The two classes comprised about 80% of all exons. The upstream flanks and downstream flanks were treated separately in this search, resulting in four different data sets as follows: HGC upstream flanks, HGC downstream flanks, LGC upstream flanks, and LGC downstream flanks. As a basis for comparison, we generated four sets of randomly chosen repeat-free 50-nt intronic fragments that had exactly the same GC% distribution as each of the four experimental sets. We determined the frequencies of all 1024 possible pentamers in each flank set and its background set, and calculated a Z-score for each pentamer. The Z-score is a measure of the degree to which the frequency of a given pentamer differs between the test set and the background set. To estimate an empirical significance of the Z-scores, we carried out simulations comparing a portion of each background set to itself (10% vs. 90%, see Supplemental Methods). The maximum Z-score found in these simulations was 3.9, and so we chose a Z-score of +4 as our threshold for significance (P = 3.2 × 10-5 or 0.033 after a Bonferroni correction).
As a second criterion of potential functional significance, we demanded phylogenetic conservation of an overrepresented pentamer. We compared orthologous human and mouse genes by devising a novel word-based method that obviates the need for an accurate alignment, allowing cis-acting splicing elements to be located anywhere within the 50-nt flanks (see Supplemental Methods for details). We used this criterion to eliminate pentamers that did not exhibit a level of conservation greater than that shown by deeper intronic regions (201-250 nt from the exon border). This filter removed about half the overrepresented pentamers, leaving 120 winning pentamers (3%) of 4096 total candidates (1024 × 2 flanks × 2 GC% classes). These winning pentamers are shown in Figure 4.
Figure 4.
Pentamer winners selected by comparing exon flanks to intron sequences with exactly the same GC% range and distributions. Winners were classified into four categories according to the GC% of their flanks (HGC or LGC) and location (upstream or downstream). Winners in italics have reverse complementary sequences as winners in the opposite flanks in the same class. Winners in bold are common to both upstream and downstream flanks in the same class. The underlined winners were also identified in a previous study (Zhang et al. 2003). The winners with asterisks overlap the branch-site consensus YTRAY.
Most of the winning pentamers in upstream flanks were comprised of motifs related (four of five match) to the BP consensus YTRAY or polypyrimidines residing in an extended polypyrimidine tract. This resemblance was true for both the HGC exons (41/48) and the LGC exons (28/29). Indeed, eight of these upstream winners were common to both the HGC and LGC sets, a proportion six times higher than that expected by chance. Eight pentamers did not resemble either of these motifs and are candidates for novel signals. The situation was quite different for the downstream flanks, where none of the 43 winners (27 HGC and 16 LGC) were in common. We concluded that the two classes include overlapping upstream winners, but highly distinct downstream winners.
We then examined the positional distribution of these winners within the exon flank. To avoid circularity, this positional information was extracted from a new data set of ∼100,000 exons that did not include the exons that were used to identify these winners. These exons were also divided into HGC and LGC classes. As expected, distributions of pentamers resembling the branch site YTRAY were dramatically overrepresented in the region of -37 to -15, with a peak at about -28 (the bulged A would be at -25), whereas the frequencies of polypyrimidines decreased monotonically from a peak at -15 abutting the canonical polypyrimidine tract (Supplemental Fig. S1). These patterns were nearly identical in the HGC and LGC classes. As controls, pseudo exons and scrambled real exon flanks were examined in the same way; neither showed similar distribution profiles. That these known elements were found at their expected positions demonstrates that this approach is effective in finding splicing related motifs.
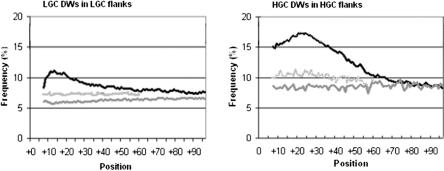
A different result was seen in the downstream flanks. Compared with more distal regions, LGC winners were as much as 30% more frequent in the region downstream of LGC exons from +7 to +27 with a maximum at position +13 (Fig. 5, left). The HGC winners were ∼100% overrepresented in a broader window downstream of HGC exons, from +7 to +57, with a peak at position +22 (Fig. 5, right). Again, pseudo exons and scrambled flanks did not display any of these patterns. Moreover, in these same restricted regions, the winning pentamers of the HGC class were underrepresented in the flanks of LGC exons and vice versa (Supplemental Fig. S1). Overall, these different sequences and their different distribution results suggest that HGC and LGC genes use different intronic signals in splicing.
Figure 5.
Distributions of downstream winning pentamers (DW) in the flanks of HGC and LGC exons. The distribution around real exons (black curve) is compared with the distribution around pseudo exons (dark-gray curve) and after scrambling the flanks of real exons (light-gray curve). Total frequencies of each class of winners are plotted as percent. (Left) LGC winners; (right) HGC winners.
The HGC winners are GC-rich (GC% = 78.5), and the LGC winners are AT-rich (GC% = 12.5). Thus, one might argue that it should not be surprising to see more AT-rich pentamers in AT-rich flanks and vice versa. On the other hand, if the splicing signals were acting as targets for splicing factors without regard to GC%, then we would expect to see them emerge as such despite the GC% context, or even display a contrasting GC%. This situation did result, in large part, for the upstream flanks. It should be remembered that the winners were extracted by comparing flanks to sets of intronic fragments with exactly the same GC% range and distribution. Therefore, our extraction should not necessarily yield winners with strong GC% biases. The fact that we did get such winners in downstream flanks means that these pentamers are distinctive for other reasons (i.e., functional importance). Furthermore, when we scrambled the region downstream of HGC or LGC exons (keeping the GC% unchanged), the peaks in frequency disappeared (Fig. 5). These results suggest that the differences we have observed are not simply due to the similarity of the flanks and the pentamer winners in GC%.
HGC and LGC classes may use distinct modes of interaction between their flanks
Comparing winners in downstream versus upstream flanks within each class led to some intriguing findings. Of 75 winners in the HGC class, 22 are pentamers whose reverse complementary sequences are also winners in the opposite flank (Fig. 4), a number much greater than expected by chance (P <10-10, see Supplemental Methods). In sharp contrast, there are no complementary pairs between upstream and downstream winners among the 45 winners in the LGC class. Indeed, this situation is avoided in the LGC class; the complements of the majority of downstream winners are vastly underrepresented in the upstream flank (∼100,000 exons, Z-scores lower than -4).
Additionally, if we consider pentamer sequences that are identical in both flanks, the HGC and LGC classes again show differences. In the LGC class, 20 of the 45 winning pentamers are represented in both flanks (44%), 22 times more than expectation (P <10-10, see Supplemental Methods). If the BP-like upstream pentamers are ignored, this fraction rises to 61%; and if, additionally, one mismatch is allowed, 100% of the pentamers are represented in both flanks. In contrast, in the HGC class, only 10 of 75 winners have this property, with corresponding fractions being 13%, 16%, and 35%, respectively (Fig. 4). These features suggest that different types of cross-exon interactions may be taking place in the two classes. The complementarity between upstream and downstream flanks in the HGC class suggests that secondary structures may be formed across exons or introns, whereas the identity of many winners in upstream and downstream flanks in the LGC class hints at interactions between identical proteins or protein subunits. We explore these possibilities further in the next two sections.
HGC flanks are predicted to have secondary structures
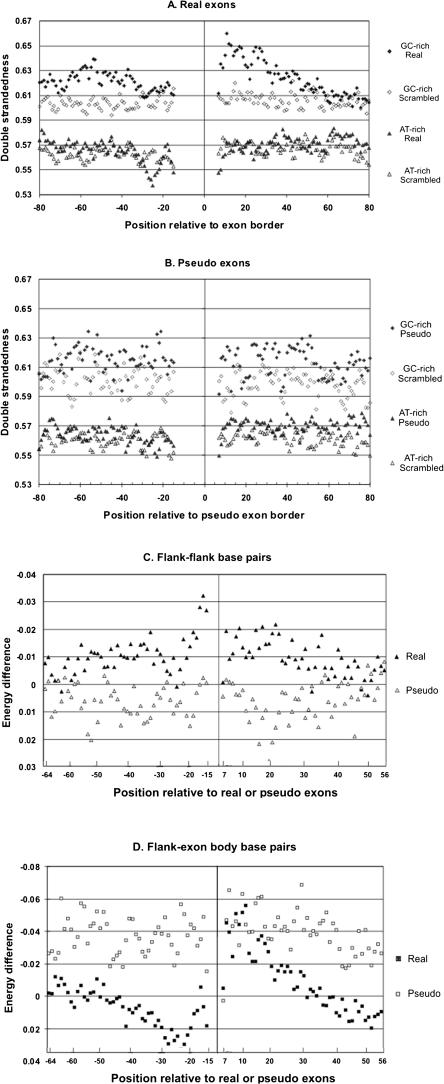
To investigate potential secondary structures, we used the Mfold program to predict the occurrence of base pairing in the region from ∼ -100 to +100 and including the exons. Each base was allowed to pair with any other base within this region. As a control, we scrambled the two flanks separately, reconnected them to the corresponding exon, and repeated the folding. We examined the likelihood that each individual base would be in a stem (paired), and designated this feature as double strandedness (DS). DS is calculated by summarizing the information from all predicted structures whose energies lie within 5% of the most stable predicted structure.
We folded real HGC and LGC exons (5000 each), as well as HGC and LGC pseudo exons (1000 and 5000, respectively), and calculated the DS value for each flank position from -100 to -15 and +7 to +100. Base pairing between flanks and exons, within flanks, and across exons, were all calculated, but DS values for positions within the exons and splice sites were not examined further. The results are shown in Figure 6A. From the average positions of the points on the vertical axis, it can be seen that GC-rich sequences formed more stems than AT-rich sequences, as expected from the greater hydrogen bonding capacity of the former. A comparison of DS in the actual versus the scrambled sequences reveals predicted secondary structure in flanks over and above that dictated by GC% itself. The data in the upper part of Figure 6A show excess DS at most positions in the region from -67 to -33 upstream and from +7 to +40 downstream of HGC splice sites. In contrast, there was little difference between the LGC flanks and their scrambled controls. The one noticeable distinction was a valley of DS centered at position -25 (Fig. 6A, left lower curves). Interestingly, this is the predicted peak position for the bulged A of the branch point. However, the very fact that A is overrepresented at this position could account for a reduced probability of participating in a stem.
Figure 6.
Secondary structure analysis. (A) Double strandedness in real exon flanks. Exons with their flanks were folded using Mfold. As a control, each upstream flank was scrambled and each downstream flank was scrambled, and the scrambled flanks were reconnected to the original exon body and then folded using Mfold. Both the original and scrambled versions of the sequences were divided into a HGC class (GC% of the most proximal 50-nt flanks >55) and an LGC class (GC% of the most proximal 50-nt flanks <45), leading to four different data sets as follows: original HGC (♦), original LGC (▴), scrambled HGC (⋄), and scrambled LGC (▵). We then plotted the double strandedness as a function of positions in flanks. Double strandedness reflects the frequency of all predicted base pairing at each position (see Results and Methods). (B) Double strandedness in pseudo exon flanks. Exactly the same analysis of pseudo exons as a control. (C) Flank-flank base pairing around HGC exons. The incremental contribution of each interflank base pair to the energy of each predicted stable structure was extracted from the Mfold output after folding both original exon plus flank sequences and again after scrambling the flanks. The difference between these two values (original sequence energies—scrambled sequence energies) is plotted as an indication of the excess secondary structure contributed at each position (filled symbols). A negative value represents more base pairing. For comparison, the same process was carried out on pseudo exons (open symbols). (D) Flank-exon base pairing around HGC exons. Differences in the free energy contributions of individual base pairs were calculated and displayed as in C, except only base pairs between flank and exon positions were chosen. (Real exons) Filled symbols; (pseudo exons) open symbols.
In contrast, pseudo exons flanks displayed little or no excess DS (Fig. 6B). The flanks of HGC pseudo exons did have slightly higher DS values compared with their scrambled counterparts, but the difference was not position correlated (Fig. 6B) and was barely significant. For instance, for the region +7 to +31, the area between the natural and scrambled curves for the real HGC exons was 0.84 ± 0.18 (s.d.), but only 0.33 ± 0.30 for the pseudo HGC exons. Moreover, folding of randomly chosen intronic HGC and LGC sequences 250 to 450 long yielded no DS differences between native and scrambled versions (data not shown). Based on these data, we concluded the HGC flanks, but not LGC flanks, have a propensity to form stems. We examined here only secondary structures formed within and across exons; it is possible that excess DS can be predicted to form across introns as well.
We next asked whether there was a particular tendency for base pairing between flanks, i.e., across the HGC exons. Mfold was run on 20,000 HGC real exons and on 7700 HGC pseudo exons. In this case, to provide a weighted differentiation among base pairs, we extracted the individual free energy contribution of each base pair to each winning structure. Although the entire region was allowed to fold, only base pairs between opposite flanks were considered; intra-flank base pairs and base pairs between flanks and exons were ignored. Once again, we combined the data from all structures within 5% of the most stable structure. We then summed the energies of all class members at each position to obtain a measure of the likelihood that a given position is involved in flank-flank base pairing. We repeated this calculation for the data set in which the flank sequences had been scrambled. We then subtracted the energies of the scrambled versions from those of the actual sequences to obtain a ΔΔG at each position. The same free energy differences were calculated for pseudo exons. The results are shown in Figure 6C. Unlike the pseudo exon controls, flanking regions upstream and downstream of real exons exhibit a tendency to base pair with each other (solid symbols, negative ΔΔG values). Once again, this tendency disappears in the region of the branch point.
With the intention of providing another contrasting control, we extracted the data for base pairing between the flanks and the exon, expecting to see no base pairing beyond that exhibited by pseudo exons. We found that not only was there no tendency to base pair, but that there was some tendency not to base pair (Fig. 6D). Except for the region from +8 to +27, real exons tend to avoid forming predicted stems between flanks and exon bodies. In contrast, pseudo exons have a tendency to do just that, displaying negative free energy differences compared with the scrambled control (Fig. 6D, open symbols).
The occurrence of LGC winners in one flank is independent of that in the other flank
Most of the downstream LGC winners were also winners in the upstream flank (Fig. 4), raising the possibility that the binding of the same factor on both sides of the exon is a required feature in the splicing of these exons. If so, then there would be strong dependence for the occurrence of these pentamers in both flanks of a given exon. Alternatively, if such a protein can bind to either side of an exon in order to function, these motifs should occur relatively independently in both flanks. Using a χ2 test for each pentamer, we failed to see any significant correlations between the occurrences of the LGC winners in opposite flanks (data not shown). We also did not find any such correlations across introns (data not shown). The fact that these motifs occur independently in the two LGC flanks is consistent with the idea that the same protein can carry out its function from either side of an exon.
Discussion
The importance of exon flanks
From the results of our previous genomic analysis, we proposed that exons flanks would be important for splicing and that exons might fall into different classes based on GC%. To a large extent, both of these predictions have been borne out. Regarding the first hypothesis, the empirical experiments not only showed a dependence on one or more flanks for efficient splicing, but pointed to a specificity in this requirement. The majority of the flankless exons required their own flanks rather than those provided at the host insertion site. This requirement did not correlate with the strength of the splice sites. With respect to the second hypothesis, most human exons were found to possess either GC-rich or AT-rich flanks, and the two classes harbor noticeably different candidates for ISEs. These ISEs may be involved in an early recognition step for exons or may enhance a later step in splicing.
The importance of the classification by GC%
Does classification of exons by GC% aid in identifying novel signals and in gaining a better understanding of the role of exon flanks? One way to examine this question is to compare the search for signals in downstream flanks using unclassified data (Fig. 1) with that using the classified sets (Fig. 5). If the results of the LGC (Fig. 5, left) and HGC (Fig. 5, right) classes were combined, one might naively expect an additive effect, but the result is just the opposite (subtractive). With both data sets combined, winning pentamer incidence in the downstream flanks was ∼3% over background (13%-10% background at deeper intron positions in Fig. 1), whereas it was 8%, nearly three times as great, when only HGC exons were considered (17%-9% background in Fig. 5, right).
Another way to ask this question is to compare the winning pentamers found here with those found in our previous study using a SVM on unclassified exons. In upstream flanks, both studies yielded extended polypyrimidine tract and branch-point sequences. In downstream flanks, both studies yielded G-rich and C-rich sequences. However, the T-rich sequences found here in the LGC downstream flanks (Fig. 4, right-most column) were missed by the SVM methodology, which treated all exons as a single class. Moreover, it was essential that the classification be by GC%; classification by GT% failed to identify the T-rich signals (data not shown). T-rich sequences have also been found using a different method, which relies on the overrepresentation of motifs compared with a local background (Louie et al. 2003).
A third indication of the usefulness of the GC% dichotomy came from repeating the SVM analysis using classified data. Using unclassified exons and a combination of features that include flanking pentamers, we could distinguish real from pseudo exons with a sensitivity of 0.82 and a specificity of 0.80. When we repeated this procedure after first classifying the exons by GC%, these numbers increased to 0.95 and 0.89, respectively (average of very similar values for an HGC and an LGC data set, data not shown).
The dichotomy of GC% in exon flanks goes beyond that seen in the genome
It has been known for almost 30 years that the human genome has large-scale GC% variations, designated by some authors as isochores (Bernardi 2000; Li et al. 2003; Versteeg et al. 2003). As shown in Figure 3, pseudo exon flanks and deeper intron segments do not display the bimodal GC% distribution of exon flanks, indicating that genomic GC% variation alone is not responsible for the bimodality in real exon flanks. The distribution cannot be explained by the location of highly repeated sequences within introns (see Supplemental material for a more detailed discussion).
What is the role of secondary structure in the flanks of HGC exons?
Sequences in and around LGC exons showed no particular tendency to form predicted secondary structures over and above that of scrambled sequences of the same base composition. In contrast, HGC exons did show such a tendency. Analysis of the pairwise base-pairing combinations predicted that, except for bases +8 to +27 downstream of HGC exons, flanks would pair more with each other than with exon body sequences (Fig. 6C,D). This result suggests that there has been a selection against flank-exon pairing in many cases, perhaps to allow access of protein factors to exonic regulatory sequences. There are many individual cases in which secondary structure has been suggested to play a role in splicing (for review, see Buratti and Baralle 2004); our results are consistent with the common use of such a mechanism for HGC exons.
Another negative correlation was the avoidance of base pairing of the branch sites. A straightforward interpretation of these results is that, like the exon, the branch sites need to keep their sequences accessible, in this case for binding SF1 and U2 snRNA. This accessibility has previously been suggested by the relative nuclease sensitivity of the branch-site sequence (Munroe and Duthie 1986). However, we cannot rule out the possibility that this effect is simply the consequence of the branch-site consensus sequence (TRAY) being relatively AT-rich and, therefore, being at a disadvantage for Watson-Crick hydrogen bonding.
We have used pseudo exons as a basis of comparison to highlight the features of real exons. However, our results also bear on the properties of pseudo exons themselves. The rather uniform tendency of pseudo exons flanks to pair with exon bodies (negative-free energies in Fig. 6D, open symbols) rather than the opposite flank (Fig. 6C) suggests that secondary structure could also play a role in keeping pseudo exons from being spliced.
The difference in folding between real exons and their pseudo exons counterparts is limited to the exon and about 50 nt of surrounding flank, which is just the region found by SVM, and by the statistical analysis presented here to distinguish these two classes. With regard to folding, a 50-nt distance is also consistent with the finding that the introduction of sequences complementary to the 3′ splice site inhibited splicing only when placed within 50 nt of their target (Eperon et al. 1988). These data suggest that local pre-mRNA folding may most often be restricted to this distance in vivo.
The average free-energy difference between the top 5% of the structures predicted for the 20,000 HGC exon-flank regions and their flank-scrambled counterparts was -3.9 kcal/mol, representing 3.1% of the average total free energy of the original sequences (126 kcal/mol). While this difference is highly significant (P <10-100), we point out that the original structures exhibit a rather wide range of free energies with a standard deviation of 6.9 kcal/mol, and that about 25% of the predicted structures actually yield a positive difference (less stability) compared with their scrambled versions (note that only the flanks were scrambled in these experiments). Additional statistical tests for a general role of secondary structure can be formulated, but they unfortunately require rather intensive computation (it took several weeks of constant computation on a modern desktop computer to generate the data in Fig. 6). In the end, it will take empirical experiments based on a priori hypotheses to provide a compelling test of these ideas.
The experimentally tested exons contain HGC and LGC winning pentamers
Although the exons we tested for flank-dependent splicing were few, it is still of some interest to examine the HGC or LGC winners in their flanks. As can be seen in Figure 7, A-G, winning pentamers usually comprise a substantial proportion of the flanks of exons that exhibited flank-dependent splicing (see the shaded sequences), about five times that expected by chance (assuming that each pentamer has an equal probability of occurrence, P <10-10). Noticeably, the same winning pentamer occurs multiple times for a given exon (e.g., CCCTG thrice in the clcn7-3 downstream flank, and tandem TCCCCAC sequences in the upstream flanks of wt1-5). This reiteration is suggestive of cooperative protein binding within a flank. Some of these particular sequences resemble binding sites of known factors, e.g., CTCTCT in clcn7-3 has been seen in core polypyrimidine-binding protein-binding sites (Southby et al. 1999; Carstens et al. 2000), AGGG in thbs4-12 is part of several hnRNP A1-binding sites (Burd and Dreyfuss 1994; Boerkoel et al. 1995; Abdul-Manan et al. 1996; Expert-Bezancon et al. 2004), GGGA in thbs4-12 is the core element of the hnRNP H family (Caputi and Zahler 2001) and sequences bearing G-triplets can potentially bind U1 snRNP (McCullough and Berget 2000). We also compared the effect of flank deletions on winner content and phenotype. In the five cases in which flank deletion produced 20% to 100% exon skipping the number of winning pentamers located in both flanks, dropped from an average of 14.4 to 4.8 (P < 0.005), in agreement with a role of these pentamers as ISEs.
Figure 7.
LGC and HGC winning pentamers residing in the flanks of the seven tested exons. (A-G) Exonic sequences are shown by uppercase letters and the splice site consensus sequences are in bold. Appropriate winners (i.e., of the cognate location and GC% class) are shaded; those that are inappropriate (of the same location, but opposite class) are underlined. Winners that overlap with exons or splice-site sequences (-14 to +1 and -3 to +6 relative to exon borders) are not shown. In G, the known branch point is indicated by an arrow and the consensus branch site sequence is italicized. Note the frequent overlap of winning pentamers in clusters.
For comparison, we also show in Figure 7 the winners from the opposite GC% class in each sequence (underlined). One can see that some winners from the opposite GC% class appear in upstream flanks, but rarely in downstream flanks. The coincidence of upstream winners probably reflects their use as branch sites, regardless of GC% class. We also found several pentamer winners in an exon that showed no flank dependence in our splicing test (Fig. 7E). In this case, either the winners are redundant with other positive signals (e.g., ESEs), or surrogate flank signals were provided by the minigene host. In a previous study, we showed that insertion of exonic splicing silencers reversed the positive effect of flank signals, and insertion of a variety of ESEs counteracted a deficiency in flanks in chuk-8 (Zhang and Chasin 2004). These results indicate that intronic and exonic splicing signals can compensate for one another to dictate splice-site recognition. Exon dhfr-2-3 (Fig. 7G), contained no winners of either class in its downstream flank and showed noteworthy flank independence when placed at several locations within an intron (Fig. 2D); its autonomous behavior could be explained by the presence of numerous splicing enhancers within the exon body and splice sites with close agreement to the consensus. The absence of winners in the flank of dhfr-2-3 may be significant with respect to the experimental design used here. These flanks constitute the sequences that surround all test exons inserted into the NotI site in the test vector. The lack of pentamer winners in this region may have fortuitously allowed splicing signals in the test exon flanks to be manifested.
Is there more than one exon-recognition mechanism?
The exaggerated dichotomy seen in exon flanks raises the possibility that exons of GC-rich genes are recognized by a different mechanism than exons of AT-rich genes. Certainly, this need not be the case, since the very same machinery could have evolved to accommodate the different GC% of different genes. For instance, the splice-site consensus themselves vary according to the GC% of a gene (Clark and Thanaraj 2002), yet they presumably are acted upon by the same snRNPs. Moreover, ESEs selected for responsiveness to a particular SR protein display a considerable range of GC%, e.g., the core hexamers of 24 sequences responding to SRp55 range from 33% to 84% in GC% (Liu et al. 1998). Even if distinct machinery were used for GC-rich and AT-rich transcripts, the mechanisms could be exactly the same. An example of such a case is translation, where different adapters (tRNAs) are used for synonymous codons of different GC% within the same mechanistic framework (e.g., AGA and GGC for arginine). In an analogous way, splicing factors could be specialized for high or low GC% elements, yet work through a common mechanism.
Nonetheless, there are reasons to think that the exons of GC-rich and AT-rich genes may be recognized by distinct mechanisms. (1) GC-rich and AT-rich genes are dichotomous for several other characteristics, i.e., surrounding gene density, intron length, expression level, codon usage bias, mutation rate, and recombination rate (Duret et al. 1995; Gardiner 1996; Karlin and Mrazek 1996; Zoubak et al. 1996; Castillo-Davis et al. 2002; D'Onofrio 2002; Versteeg et al. 2003). In particular, GC-rich genes have shorter introns (Lander et al. 2001) and are expressed at higher rates (Versteeg et al. 2003) than AT-rich genes; both of these features can influence splicing (Talerico and Berget 1994; Cramer et al. 1997). (2) The exon flanks differ in the way winning pentamers are grouped; HGC flanks include many complementary pentamers, while LGC flanks include many identical pentamers. (3) HGC flanks are predicted to have excess secondary structure, while this is not the case for LGC flanks. (4) Finally, and most intriguingly, it has been noted that GC-rich chromosome regions preferentially associate with nuclear speckles rich in the splicing factor SC35, presumably via an interaction with nascent RNA (Smith et al. 1999; Shopland et al. 2003). This observation is consistent with the idea that the splicing machinery is specialized for transcripts of different GC%.
Methods
Plasmid constructs, the random transposition of exons, splicing assays, the sources of data sets, the definition of pseudo exons, and other statistical methods are described in Supplemental materials.
Secondary structure analyses
We folded exons or pseudo exons with their flanks from -99 to +91 using Mfold 3.1, with the command “nafold” and the following parameter settings: number of tracebacks: 200, window size: 8, percentage of sort: 5, and default energy files. Detailed information about these parameters can be found in the Mfold manual. Among the output files, those that have the suffix .det contain complete information about the positions and the incremental energies of the base pairs. We took into account all of the structures whose free energy was within 5% of the best prediction and first calculated the average proportion of sequences predicted to be base paired at each position. A probability of being involved in stems (called double strandedness, DS) was then calculated based on each nucleotide position in all sequences. As a control, we scrambled the flanks (not the exons) and performed the same analyses. Approximately 5000 exons were examined for all classes except HGC pseudo exons, for which 1000 pseudo exons were used (Fig. 6A,B).
We also specifically examined the predicted base pairs between the upstream and downstream flanks, (Fig. 6C) or between flanks and exon bodies (Fig. 6D). In these cases, we calculated the average incremental free energy of every base pair of interest in the top 5% structures of each sequence. We made the same calculations for the flank-scrambled sequences and subtracted that incremental energy from the corresponding energy of the actual sequences at each position. The differences (ΔΔG) are plotted as a function of position in Figure 6, C and D. Approximately 20,000 HGC examples were examined for real exons and 7700 for pseudo exons.
χ2 analysis of coincidence of pentamers in upstream vs. downstream flanks
For each pentamer of interest, we asked how many times it was present at least once in both flanks, present in upstream flanks but not in downstream flanks, present in downstream flanks but not in upstream flanks, and present in neither flank. These four numbers constituted a 2 × 2 table, from which a χ2 was calculated and the independence of pentamer occurrence in the two flanks was tested.
Acknowledgments
We thank Dennis Weiss for useful discussions, James Lee for help with exon classification, and an anonymous reviewer for helpful suggestions. We thank Michael Zucker for the Mfold program and Arian Smit for RepeatMasker. L.C. was supported by funds from Columbia University. X.Z. is a predoctoral Faculty Fellow of Columbia University. C.L. was supported by a grant from the PhRMA Foundation and NIH grant LM07276.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.3217705.
References
- Abdul-Manan, N., O'Malley, S.M., and Williams, K.R. 1996. Origins of binding specificity of the A1 heterogeneous nuclear ribonucleoprotein. Biochemistry 35: 3545-3554. [DOI] [PubMed] [Google Scholar]
- Adams, M.D., Rudner, D.Z., and Rio, D.C. 1996. Biochemistry and regulation of pre-mRNA splicing. Curr. Opin. Cell. Biol. 8: 331-339. [DOI] [PubMed] [Google Scholar]
- Amarasinghe, A.K., MacDiarmid, R., Adams, M.D., and Rio, D.C. 2001. An in vitro-selected RNA-binding site for the KH domain protein PSI acts as a splicing inhibitor element. RNA 7: 1239-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ast, G., Pavelitz, T., and Weiner, A.M. 2001. Sequences upstream of the branch site are required to form helix II between U2 and U6 snRNA in a trans-splicing reaction. Nucleic Acids Res. 29: 1741-1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berget, S.M. 1995. Exon recognition in vertebrate splicing. J. Biol. Chem. 270: 2411-2414. [DOI] [PubMed] [Google Scholar]
- Bernardi, G. 2000. Isochores and the evolutionary genomics of vertebrates. Gene 241: 3-17. [DOI] [PubMed] [Google Scholar]
- Black, D.L. 2003. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 72: 291-336. [DOI] [PubMed] [Google Scholar]
- Boerkoel, C.F., Exelbert, R., Nicastri, C., Nichols, R.C., Miller, F.W., Plotz, P.H., and Raben, N. 1995. Leaky splicing mutation in the acid maltase gene is associated with delayed onset of glycogenosis type II. Am. J. Hum. Genet. 56: 887-897. [PMC free article] [PubMed] [Google Scholar]
- Buratti, E. and Baralle, F.E. 2004. Influence of RNA secondary structure on the pre-mRNA splicing process. Mol. Cell. Biol. 24: 10505-10514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd, C.G. and Dreyfuss, G. 1994. RNA binding specificity of hnRNP A1: Significance of hnRNP A1 high-affinity binding sites in pre-mRNA splicing. EMBO J. 13: 1197-1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buvoli, M., Mayer, S.A., and Patton, J.G. 1997. Functional crosstalk between exon enhancers, polypyrimidine tracts and branchpoint sequences. EMBO J. 16: 7174-7183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caputi, M. and Zahler, A.M. 2001. Determination of the RNA binding specificity of the heterogeneous nuclear ribonucleoprotein (hnRNP) H/H′/F/2H9 family. J. Biol. Chem. 276: 43850-43859. [DOI] [PubMed] [Google Scholar]
- Carothers, A.M., Urlaub, G., Grunberger, D., and Chasin, L.A. 1993. Splicing mutants and their second-site suppressors at the dihydrofolate reductase locus in Chinese hamster ovary cells. Mol. Cell. Biol. 13: 5085-5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carstens, R.P., Wagner, E.J., and Garcia-Blanco, M.A. 2000. An intronic splicing silencer causes skipping of the IIIb exon of fibroblast growth factor receptor 2 through involvement of polypyrimidine tract binding protein. Mol. Cell. Biol. 20: 7388-7400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartegni, L., Wang, J., Zhu, Z., Zhang, M.Q., and Krainer, A.R. 2003. ESEfinder: A web resource to identify exonic splicing enhancers. Nucleic Acids Res. 31: 3568-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo-Davis, C.I., Mekhedov, S.L., Hartl, D.L., Koonin, E.V., and Kondrashov, F.A. 2002. Selection for short introns in highly expressed genes. Nat. Genet. 31: 415-418. [DOI] [PubMed] [Google Scholar]
- Cavaloc, Y., Bourgeois, C.F., Kister, L., and Stevenin, J. 1999. The splicing factors 9G8 and SRp20 transactivate splicing through different and specific enhancers. RNA 5: 468-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabot, B., LeBel, C., Hutchison, S., Nasim, F.H., and Simard, M.J. 2003. Heterogeneous nuclear ribonucleoprotein particle A/B proteins and the control of alternative splicing of the mammalian heterogeneous nuclear ribonucleoprotein particle A1 pre-mRNA. Prog. Mol. Subcell. Biol. 31: 59-88. [DOI] [PubMed] [Google Scholar]
- Chen, I.T. and Chasin, L.A. 1993. Direct selection for mutations affecting specific splice sites in a hamster dihydrofolate reductase minigene. Mol. Cell. Biol. 13: 289-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, F. and Thanaraj, T.A. 2002. Categorization and characterization of transcript-confirmed constitutively and alternatively spliced introns and exons from human. Hum. Mol. Genet. 11: 451-464. [DOI] [PubMed] [Google Scholar]
- Coulter, L.R., Landree, M.A., and Cooper, T.A. 1997. Identification of a new class of exonic splicing enhancers by in vivo selection. Mol. Cell. Biol. 17: 2143-2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer, P., Pesce, C.G., Baralle, F.E., and Kornblihtt, A.R. 1997. Functional association between promoter structure and transcript alternative splicing. Proc. Natl. Acad. Sci. 94: 11456-11460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Onofrio, G. 2002. Expression patterns and gene distribution in the human genome. Gene 300: 155-160. [DOI] [PubMed] [Google Scholar]
- Duret, L. and Hurst, L.D. 2001. The elevated GC content at exonic third sites is not evidence against neutralist models of isochore evolution. Mol. Biol. Evol. 18: 757-762. [DOI] [PubMed] [Google Scholar]
- Duret, L., Mouchiroud, D., and Gautier, C. 1995. Statistical analysis of vertebrate sequences reveals that long genes are scarce in GC-rich isochores. J. Mol. Evol. 40: 308-317. [DOI] [PubMed] [Google Scholar]
- Engelbrecht, J., Knudsen, S., and Brunak, S. 1992. G+C-rich tract in 5′ end of human introns. J. Mol. Biol. 227: 108-113. [DOI] [PubMed] [Google Scholar]
- Eperon, L.P., Graham, I.R., Griffiths, A.D., and Eperon, I.C. 1988. Effects of RNA secondary structure on alternative splicing of pre-mRNA: Is folding limited to a region behind the transcribing RNA polymerase? Cell 54: 393-401. [DOI] [PubMed] [Google Scholar]
- Expert-Bezancon, A., Sureau, A., Durosay, P., Salesse, R., Groeneveld, H., Lecaer, J.P., and Marie, J. 2004. HnRNP A1 and SR proteins, ASF/SF2 and SC35 have antagonistic functions in splicing of β -tropomyosin exon 6B. J. Biol. Chem. 279: 38249-38259. [DOI] [PubMed] [Google Scholar]
- Fairbrother, W.G., Yeh, R.F., Sharp, P.A., and Burge, C.B. 2002. Predictive identification of exonic splicing enhancers in human genes. Science 297: 1007-1013. [DOI] [PubMed] [Google Scholar]
- Gardiner, K. 1996. Base composition and gene distribution: Critical patterns in mammalian genome organization. Trends Genet. 12: 519-524. [DOI] [PubMed] [Google Scholar]
- Gozani, O., Feld, R., and Reed, R. 1996. Evidence that sequence-independent binding of highly conserved U2 snRNP proteins upstream of the branch site is required for assembly of spliceosomal complex A. Genes & Dev. 10: 233-243. [DOI] [PubMed] [Google Scholar]
- Graveley, B.R. 2000. Sorting out the complexity of SR protein functions. RNA 6: 1197-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, N.L. and Senapathy, P. 1990. Distribution and consensus of branch point signals in eukaryotic genes: A computerized statistical analysis. Nucleic Acids Res. 18: 3015-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurica, M.S. and Moore, M.J. 2003. Pre-mRNA splicing: Awash in a sea of proteins. Mol. Cell 12: 5-14. [DOI] [PubMed] [Google Scholar]
- Karlin, S. and Mrazek, J. 1996. What drives codon choices in human genes? J. Mol. Biol. 262: 459-472. [DOI] [PubMed] [Google Scholar]
- Ladd, A.N. and Cooper, T.A. 2002. Finding signals that regulate alternative splicing in the post-genomic era. Genome Biol. 3: reviews0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander, E.S., Linton, L.M., Birren, B., Nusbaum, C., Zody, M.C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860-921. [DOI] [PubMed] [Google Scholar]
- Li, W., Bernaola-Galvan, P., Carpena, P., and Oliver, J.L. 2003. Isochores merit the prefix `iso'. Comput. Biol. Chem. 27: 5-10. [DOI] [PubMed] [Google Scholar]
- Liu, H.X., Zhang, M., and Krainer, A.R. 1998. Identification of functional exonic splicing enhancer motifs recognized by individual SR proteins. Genes & Dev. 12: 1998-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H.X., Chew, S.L., Cartegni, L., Zhang, M.Q., and Krainer, A.R. 2000. Exonic splicing enhancer motif recognized by human SC35 under splicing conditions. Mol. Cell. Biol. 20: 1063-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louie, E., Ott, J., and Majewski, J. 2003. Nucleotide frequency variation across human genes. Genome Res. 13: 2594-2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund, M., Tange, T.O., Dyhr-Mikkelsen, H., Hansen, J., and Kjems, J. 2000. Characterization of human RNA splice signals by iterative functional selection of splice sites. RNA 6: 528-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewski, J. and Ott, J. 2002. Distribution and characterization of regulatory elements in the human genome. Genome Res. 12: 1827-1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manley, J.L. and Tacke, R. 1996. SR proteins and splicing control. Genes & Dev. 10: 1569-1579. [DOI] [PubMed] [Google Scholar]
- Mayeda, A., Screaton, G.R., Chandler, S.D., Fu, X.D., and Krainer, A.R. 1999. Substrate specificities of SR proteins in constitutive splicing are determined by their RNA recognition motifs and composite pre-mRNA exonic elements. Mol. Cell. Biol. 19: 1853-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough, A.J. and Berget, S.M. 2000. An intronic splicing enhancer binds U1 snRNPs to enhance splicing and select 5′ splice sites. Mol. Cell. Biol. 20: 9225-9235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munroe, S.H. and Duthie, R.S. 1986. Splice site consensus sequences are preferentially accessible to nucleases in isolated adenovirus RNA. Nucleic Acids Res. 14: 8447-8465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natoli, T.A., McDonald, A., Alberta, J.A., Taglienti, M.E., Housman, D.E., and Kreidberg, J.A. 2002. A mammal-specific exon of WT1 is not required for development or fertility. Mol. Cell. Biol. 22: 4433-4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussinov, R. 1989. Conserved signals around the 5′ splice sites in eukaryotic nuclear precursor mRNAs: G-runs are frequent in the introns and C in the exons near both 5′ and 3′ splice sites. J. Biomol. Struct. Dyn. 6: 985-1000. [DOI] [PubMed] [Google Scholar]
- Penotti, F.E. 1991. Human pre-mRNA splicing signals. J. Theor. Biol. 150: 385-420. [DOI] [PubMed] [Google Scholar]
- Reed, R. and Maniatis, T. 1985. Intron sequences involved in lariat formation during pre-mRNA splicing. Cell 41: 95-105. [DOI] [PubMed] [Google Scholar]
- Robberson, B.L., Cote, G.J., and Berget, S.M. 1990. Exon definition may facilitate splice site selection in RNAs with multiple exons. Mol. Cell. Biol. 10: 84-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaal, T.D. and Maniatis, T. 1999a. Multiple distinct splicing enhancers in the protein-coding sequences of a constitutively spliced pre-mRNA. Mol. Cell. Biol. 19: 261-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 1999b. Selection and characterization of pre-mRNA splicing enhancers: Identification of novel SR protein-specific enhancer sequences. Mol. Cell. Biol. 19: 1705-1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senapathy, P., Shapiro, M.B., and Harris, N.L. 1990. Splice junctions, branch point sites, and exons: Sequence statistics, identification, and applications to genome project. Methods Enzymol. 183: 252-278. [DOI] [PubMed] [Google Scholar]
- Shopland, L.S., Johnson, C.V., Byron, M., McNeil, J., and Lawrence, J.B. 2003. Clustering of multiple specific genes and gene-rich R-bands around SC-35 domains: Evidence for local euchromatic neighborhoods. J. Cell Biol. 162: 981-990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, R., Valcarcel, J., and Green, M.R. 1995. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science 268: 1173-1176. [DOI] [PubMed] [Google Scholar]
- Smit, A.F. 1999. Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr. Opin. Genet. Dev. 9: 657-663. [DOI] [PubMed] [Google Scholar]
- Smith, K.P., Moen Jr., P.T., Wydner, K.L., Coleman, J.R., and Lawrence, J.B. 1999. Processing of endogenous Pre-mRNAs in association with SC-35 domains is gene specific. J. Cell Biol. 144: 617-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek, R. and Ast, G. 2003. Intronic sequences flanking alternatively spliced exons are conserved between human and mouse. Genome Res. 13: 1631-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southby, J., Gooding, C., and Smith, C.W. 1999. Polypyrimidine tract binding protein functions as a repressor to regulate alternative splicing of α-actinin mutally exclusive exons. Mol. Cell. Biol. 19: 2699-2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens, R.M. and Schneider, T.D. 1992. Features of spliceosome evolution and function inferred from an analysis of the information at human splice sites. J. Mol. Biol. 228: 1124-1136. [DOI] [PubMed] [Google Scholar]
- Sun, H. and Chasin, L.A. 2000. Multiple splicing defects in an intronic false exon. Mol. Cell. Biol. 20: 6414-6425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tacke, R. and Manley, J.L. 1995. The human splicing factors ASF/SF2 and SC35 possess distinct, functionally significant RNA binding specificities. EMBO J. 14: 3540-3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talerico, M. and Berget, S.M. 1994. Intron definition in splicing of small Drosophila introns. Mol. Cell. Biol. 14: 3434-3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, H. and Kole, R. 2001. Strong RNA splicing enhancers identified by a modified method of cycled selection interact with SR protein. J. Biol. Chem. 276: 33833-33839. [DOI] [PubMed] [Google Scholar]
- Versteeg, R., van Schaik, B.D., van Batenburg, M.F., Roos, M., Monajemi, R., Caron, H., Bussemaker, H.J., and van Kampen, A.H.C. 2003. The human transcriptome map reveals extremes in gene density, intron length, GC content, and repeat pattern for domains of highly and weakly expressed genes. Genome Res. 13: 1998-2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, E.J. and Garcia-Blanco, M.A. 2001. Polypyrimidine tract binding protein antagonizes exon definition. Mol. Cell. Biol. 21: 3281-3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z., Rolish, M.E., Yeo, G., Tung, V., Mawson, M., and Burge, C.B. 2004. Systematic identification and analysis of exonic splicing silencers. Cell 119: 831-845. [DOI] [PubMed] [Google Scholar]
- Xu, R., Teng, J., and Cooper, T.A. 1993. The cardiac troponin T alternative exon contains a novel purine-rich positive splicing element. Mol. Cell. Biol. 13: 3660-3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeakley, J.M., Morfin, J.P., Rosenfeld, M.G., and Fu, X.D. 1996. A complex of nuclear proteins mediates SR protein binding to a purine-rich splicing enhancer. Proc. Natl. Acad. Sci. 93: 7582-7587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo, G., Hoon, S., Venkatesh, B., and Burge, C.B. 2004. Variation in sequence and organization of splicing regulatory elements in vertebrate genes. Proc. Natl. Acad. Sci. 101: 15700-15705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X.H. and Chasin, L.A. 2004. Computational definition of sequence motifs governing constitutive exon splicing. Genes & Dev. 18: 1241-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X.H., Heller, K.A., Hefter, I., Leslie, C.S., and Chasin, L.A. 2003. Sequence information for the splicing of human pre-mRNA identified by support vector machine classification. Genome Res. 13: 2637-2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, Z.M. 2004. Regulation of alternative RNA splicing by exon definition and exon sequences in viral and mammalian gene expression. J. Biomed. Sci. 11: 278-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoubak, S., Clay, O., and Bernardi, G. 1996. The gene distribution of the human genome. Gene 174: 95-102. [DOI] [PubMed] [Google Scholar]