Abstract
Motivation
We are now in the era of being able to routinely generate highly contiguous (near telomere-to-telomere) genome assemblies of human and nonhuman species. Complex structural variation and regions of rapid evolutionary turnover are being discovered for the first time. Thus, efficient and informative visualization tools are needed to evaluate and directly observe structural differences between two or more genomes.
Results
We developed SVbyEye, an open-source R package to visualize and annotate sequence-to-sequence alignments along with various functionalities to process alignments in PAF format. The tool facilitates the characterization of complex structural variants in the context of sequence homology helping resolve the mechanisms underlying their formation.
Availability and implementation
SVbyEye is available at https://github.com/daewoooo/SVbyEye.
Introduction:
Informative and efficient visualization of genomic structural variation (SV) is an important step to evaluate the validity of the most complex regions of the genome, helping us to develop new hypotheses and draw biological conclusions. With advances in long-read sequencing technologies, such as HiFi (high-fidelity) PacBio (Wenger et al. 2019) and ONT (Oxford Nanopore Technologies) (Deamer and Branton 2002), we are now able to fully assemble even the most complex regions of the genome, such as segmental duplications (Vollger et al. 2022), acrocentric regions (Guarracino et al. 2023), and centromeres (Logsdon et al. 2024) into continuous, highly accurate linear assemblies—also known as telomere-to-telomere (T2T) assemblies (Jarvis et al. 2022; Nurk et al. 2022). A large part of our understanding of the evolution of complex biological systems comes from comparative analyses, including direct visual observations (Paparella et al. 2023; Yoo et al. 2024).
The challenge with these analyses is that many large-scale structural changes between genomes are mediated by large, highly identical repeat sequences that are not readily annotated by existing software. This necessitates the development of visualization tools to complement T2T comparative studies. We developed SVbyEye for three purposes: 1) to directly characterize structurally complex regions, including insertions, duplications, deletions and inversions, by comparison to a linear genome reference; 2) to place these changes in the context of sequence homology by characterizing associated sequence identity; and 3) to define the breakpoints, including the length and orientation of homologous sequence mediating the rearrangement. SVbyEye is inspired by the previously developed tool called Miropeats (Parsons 1995) and brings its visuals to the popular scripting language R and visualization paradigm using ggplot2 (Wickham 2009).
Materials and Methods:
SVbyEye uses as input DNA sequence alignments in PAF (Pairwise mApping Format) format, which can be easily generated with minimap2 (Li 2018). In principle, however, any sequence-to-sequence aligner that can export alignments in standard PAF format should be sufficient. We note, however, that we tested our tool only using minimap2 alignments. Such alignments can be read using the ‘readPaf’ function. Subsequently, imported alignments can be filtered and flipped into the desired orientation using ‘filterPaf’ and ‘flipPaf’ functions, respectively,
Visualization modes
There are four visualization modes offered by SVbyEye: visualization of pairwise alignments, alignments between more than two sequences, alignments within a single sequence, and whole-genome alignments.
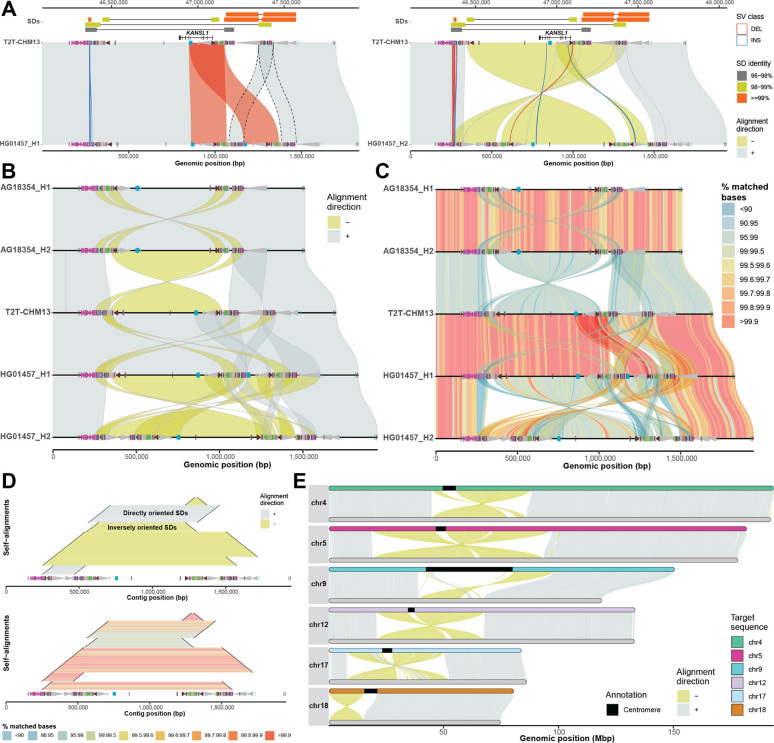
The main function of this package, called ‘plotMiro’, serves to visualize pairwise sequence alignments in a horizontal layout with the target (reference) sequence at the top and the query at the bottom (Fig. 1A). The user has control over a number of visual and alignment processing features. For instance, users can color sequence alignments by their orientation or percentage of matched bases (Supplementary Notes).
Figure 1: Example of SVbyEye visualization modes.
A) The plot depicts a minimap alignment of a 1.7 Mbp region from chromosome 17q21.31 of two human sequences: HG01457 haplotypes (query) vs. T2T-CHM13 reference (target). Segmental duplication (SD) pairwise alignments are shown (top) (connected by horizontal line) colored by their sequence identity with gene annotation (KANSL1 exons) depicted below as annotated in the UCSC Genome Browser. Minimap2 alignments are shown as gray (forward ‘+’ orientation) and yellow (reverse ‘−’ orientation) polygons between query (bottom) and target (top) sequences. Duplicon annotations as defined by DupMasker (Jiang et al. 2008) are indicated for both query and target sequences by colored arrowheads pointing forward or backward based on their orientation. An SV embedded within the SDs between query and target sequences (≥1 kbp) is highlighted as blue (insertion) and red (deletion) outlines facilitating breakpoint definition. B) A “stacked” SVbyEye plot depicting the 17q21.31 region for two chimpanzee haplotypes followed by three human haplotypes from T2T-CHM13 and HG01457. Each sequence is compared to the sequence immediately above and clearly defines a 750 kbp inversion between chimpanzee and human flanked by inverted repeats. A larger 900 kbp inversion polymorphism is also identified in human mediated by inverted SDs. C) The plot shows the same alignments as in B but with a “% identity grid” colored by the percentage of matched bases per 10-kbp-long sequence bin. Human inversion shows significant divergence indicating a deeper coalescence of the 17q21.31 region (Zody et al. 2008). D) A ‘horizontal dotplot’ visualization that shows self-alignments of HG01457 (haplotype 2) indicating the size (black line), the orientation (inverted=yellow and gray=direct; top panel), and the pairwise identity (colored grid; bottom panel). The largest and most identical segments are preferred sites for non-allelic homologous recombination (NAHR) breakpoints. E) A T2T view of six chimpanzee chromosomes (query, bottom) aligned to human syntenic chromosomes (T2T-CHM13, target, top). This view readily defines the extent of paracentric and pericentric inversions.
SVbyEye also allows visualization of alignments between more than two sequences. This can be done by aligning multiple sequences to each other using so-called all-versus-all (AVA) or stacked alignments and submitting them to the ‘plotAVA’ function. In this way, alignments are visualized in subsequent order where the alignment of the first sequence is shown with respect to the second and then second sequence to the third and so on (Fig. 1B). Many of the same parameters from ‘plotMiro’ also apply to ‘plotAVA’ as well. We illustrate a use of binning PAF alignments into defined bins (by setting a parameter ‘binsize’) and coloring them by the percentage of matched bases—a useful feature to reflect regional or pairwise differences in sequence identity (Fig. 1C).
To accommodate visualization of regions that are homologous to each other within a single sequence, we implemented the ‘plotSelf’ function. This function takes PAF alignments of a sequence to itself and visualizes them in a so-called horizontal dotplot (Fig. 1D). Such visualization can tell us a relative orientation, identity, and size of intrachromosomally aligned regions, an important feature of segmental duplications that predispose intervening sequence to recurrent rearrangements (Itsara et al. 2012; Coe et al. 2014; Porubsky et al. 2022). We note that self-alignments can also be visualized as arcs or arrowed rectangles connecting aligned regions (Supplementary Notes).
To allow for a full overview of whole-genome assembly with respect to a reference, SVbyEye offers ‘plotGenome’ function. With this function whole-genome alignments can be visualized to observe large structural rearrangements, such as large para- and pericentromeric inversions between the chimpanzee and human genomes (Fig. 1E).
Alignment processing and annotation functionalities
SVbyEye has the ability to break PAF alignments at the positions of insertions and deletions and thereby delineate their breakpoints. This is done by parsing alignment CIGAR strings if reported in the PAF file. Thus, by setting the minimum size of insertions and deletions to be reported, one can visualize SVs as red (deletions) and blue (insertions) outlines (Fig. 1A). For further interrogation users can also opt to report alignments embedded insertions and/or deletions in a data table format using the ‘breakPaf’ function.
An important feature of SVbyEye is its capability to annotate query and target sequences with genomic ranges such as gene position, position of segmental duplications, or other DNA functional elements. This is done by adding extra annotation layers on top of the target and/or query alignments using the ‘addAnnotation’ function (Supplementary Notes). Annotation ranges are visualized as either a rectangle or an arrowhead. Arrowheads are especially useful for conveying an orientation of a genomic range. Similar to PAF alignments, annotation ranges can also be colored by a user defined color scheme (Fig. 1A). If there is a need to highlight specific PAF alignments between a query and a target, one can do so with the ‘addAlignments’ function that adds selected alignment(s) over the original plot highlighted by a unique outline and/or color (Fig. 1A).
There are several other useful functionalities that come with SVbyEye, for instance, lifting coordinates from target to query and vice versa provided by the ‘liftRangesToAlignment’ function. Users can also subset alignments from a desired region on a target sequence using the ‘subsetPaf’ function. Lastly, there is a possibility to disjoin PAF alignments at regions where two and more alignments overlap each other with the ‘disjoinPafAlignments’ function to provide exact boundaries of duplicated regions (Fig. 1A).
Conclusion:
We developed SVbyEye, a data visualization R package, to facilitate direct observation of structural differences between two or more sequences. SVbyEye provides several visualization modes depending on the desired application. It offers ample ways to annotate both query and target sequences along with many functionalities to process alignments in PAF format. A more detailed package documentation along with code examples can be found at: https://htmlpreview.github.io/?https://github.com/daewoooo/SVbyEye/blob/master/man/doc/SVbyEye.html.
Supplementary Material
Acknowledgements:
This article is subject to HHMI’s Open Access to Publications policy. HHMI lab heads have previously granted a non-exclusive CC BY 4.0 license to the public and a sublicensable license to HHMI in their research articles. Pursuant to those licenses, the author-accepted manuscript of this article can be made freely available under a CC BY 4.0 license immediately upon publication.
Funding:
This research was supported, in part, by funding from the National Institutes of Health (NIH) grants R01 HG002385 and R01 HG010169 (to E.E.E.). E.E.E. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Conflict of Interest: E.E.E. is a scientific advisory board (SAB) member of Variant Bio, Inc.
References:
- Coe Bradley P., Witherspoon Kali, Rosenfeld Jill A., van Bon Bregje W. M., Vulto-van Silfhout Anneke T., Bosco Paolo, Friend Kathryn L., et al. 2014. “Refining Analyses of Copy Number Variation Identifies Specific Genes Associated with Developmental Delay.” Nature Genetics 46 (10): 1063–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deamer David W., and Branton Daniel. 2002. “Characterization of Nucleic Acids by Nanopore Analysis.” Accounts of Chemical Research 35 (10): 817–25. [DOI] [PubMed] [Google Scholar]
- Guarracino Andrea, Buonaiuto Silvia, de Lima Leonardo Gomes, Potapova Tamara, Rhie Arang, Koren Sergey, Rubinstein Boris, et al. 2023. “Recombination between Heterologous Human Acrocentric Chromosomes.” Nature 617 (7960): 335–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itsara Andy, Vissers Lisenka E. L. M., Steinberg Karyn Meltz, Meyer Kevin J., Zody Michael C., Koolen David A., de Ligt Joep, et al. 2012. “Resolving the Breakpoints of the 17q21.31 Microdeletion Syndrome with next-Generation Sequencing.” American Journal of Human Genetics 90 (4): 599–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis Erich D., Formenti Giulio, Rhie Arang, Guarracino Andrea, Yang Chentao, Wood Jonathan, Tracey Alan, et al. 2022. “Semi-Automated Assembly of High-Quality Diploid Human Reference Genomes.” Nature 611 (7936): 519–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Zhaoshi, Hubley Robert, Smit Arian, and Eichler Evan E.. 2008. “DupMasker: A Tool for Annotating Primate Segmental Duplications.” Genome Research 18 (8): 1362–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Heng. 2018. “Minimap2: Pairwise Alignment for Nucleotide Sequences.” Bioinformatics 34 (18): 3094–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logsdon Glennis A., Rozanski Allison N., Ryabov Fedor, Potapova Tamara, Shepelev Valery A., Catacchio Claudia R., Porubsky David, et al. 2024. “The Variation and Evolution of Complete Human Centromeres.” Nature 629 (8010): 136–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurk Sergey, Koren Sergey, Rhie Arang, Rautiainen Mikko, Bzikadze Andrey V., Mikheenko Alla, Vollger Mitchell R., et al. 2022. “The Complete Sequence of a Human Genome.” Science 376 (6588): 44–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paparella Annalisa, L’Abbate Alberto, Palmisano Donato, Chirico Gerardina, Porubsky David, Catacchio Claudia R., Ventura Mario, Eichler Evan E., Maggiolini Flavia A. M., and Antonacci Francesca. 2023. “Structural Variation Evolution at the 15q11-q13 Disease-Associated Locus.” International Journal of Molecular Sciences 24 (21): 15818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons J. D. 1995. “Miropeats: Graphical DNA Sequence Comparisons.” Bioinformatics 11 (6): 615–19. [DOI] [PubMed] [Google Scholar]
- Porubsky David, Wolfram Höps Hufsah Ashraf, Hsieh Pinghsun, Rodriguez-Martin Bernardo, Yilmaz Feyza, Ebler Jana, et al. 2022. “Recurrent Inversion Polymorphisms in Humans Associate with Genetic Instability and Genomic Disorders.” Cell 185 (11): 1986–2005.e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollger Mitchell R., Guitart Xavi, Dishuck Philip C., Mercuri Ludovica, Harvey William T., Gershman Ariel, Diekhans Mark, et al. 2022. “Segmental Duplications and Their Variation in a Complete Human Genome.” Science 376 (6588): eabj6965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenger Aaron M., Peluso Paul, Rowell William J., Chang Pi-Chuan, Hall Richard J., Concepcion Gregory T., Ebler Jana, et al. 2019. “Accurate Circular Consensus Long-Read Sequencing Improves Variant Detection and Assembly of a Human Genome.” Nature Biotechnology 37 (10): 1155–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham Hadley. 2009. Ggplot2: Elegant Graphics for Data Analysis. New York, NY: Springer. [Google Scholar]
- Yoo Dongahn, Rhie Arang, Hebbar Prajna, Antonacci Francesca, Logsdon Glennis A., Solar Steven J., Antipov Dmitry, et al. 2024. “Complete Sequencing of Ape Genomes.” bioRxiv.org: The Preprint Server for Biology, July. 10.1101/2024.07.31.605654. [DOI] [Google Scholar]
- Zody Michael C., Jiang Zhaoshi, Fung Hon-Chung, Antonacci Francesca, Hillier Ladeana W., Cardone Maria Francesca, Graves Tina A., et al. 2008. “Evolutionary Toggling of the MAPT 17q21.31 Inversion Region.” Nature Genetics 40 (9): 1076–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.