Significance
Acinetobacter baumannii poses a significant health threat due to its extensive antibiotic resistance. Antibiotic resistance is conferred by a diverse family of chromosomal islands, but their cost on the bacterial fitness is inconsistent with their high prevalence in the species. We show that the islands consistently disrupt a gene involved in natural transformation, a mechanism proposed to allow genetic diversification but also to cure the genome of genetic parasites. This gene disruption causes specific alterations in natural transformation which, under modeled contexts of antibiotic exposure, favors the propagation of the costly islands in bacterial populations. This illustrates a strategy by which genomic islands manipulate natural transformation to prevent their elimination, thereby driving the prevalence of antibiotic resistance genes.
Keywords: Acinetobacter baumannii, natural transformation, antibiotic resistance, mobile genetic element, recombination
Abstract
The opportunistic pathogen Acinetobacter baumannii, carries variants of A. baumannii resistance islands (AbaR)-type genomic islands conferring multidrug resistance. Their pervasiveness in the species has remained enigmatic. The dissemination of AbaRs is intricately linked to their horizontal transfer via natural transformation, a process through which bacteria can import and recombine exogenous DNA, effecting allelic recombination, genetic acquisition, and deletion. In experimental populations of the closely related pathogenic Acinetobacter nosocomialis, we quantified the rates at which these natural transformation events occur between individuals. When integrated into a model of population dynamics, they lead to the swift removal of AbaRs from the population, contrasting with the high prevalence of AbaRs in genomes. Yet, genomic analyses show that nearly all AbaRs specifically disrupt comM, a gene encoding a helicase critical for natural transformation. We found that such disruption impedes gene acquisition, and deletion, while moderately impacting acquisition of single nucleotide polymorphism. A mathematical evolutionary model demonstrates that AbaRs inserted into comM gain a selective advantage over AbaRs inserted in sites that do not inhibit or completely inhibit transformation, in line with the genomic observations. The persistence of AbaRs can be ascribed to their integration into a specific gene, diminishing the likelihood of their removal from the bacterial genome. This integration preserves the acquisition and elimination of alleles, enabling the host bacterium—and thus its AbaR—to adapt to unpredictable environments and persist over the long term. This work underscores how manipulation of natural transformation by mobile genetic elements can drive the prevalence of multidrug resistance.
Acinetobacter baumannii is responsible for a wide range of nosocomial infections (1). Due to the constant worldwide increase in its multidrug resistance (MDR), including to antibiotics of last resort (carbapenems, colistin) (2), A. baumannii has been declared critical for research efforts by the WHO (3) and is considered a major threat to human health by the CDC (4). A. baumannii is thought to be a rapidly evolving species, showing high rates of recombination and horizontal gene transfer (HGT) (5–7). Indeed, clinically relevant resistance levels are largely the result of horizontally acquired antibiotic resistance genes (ARGs) which are hitchhiking on mobile genetic elements (MGEs) (8, 9). Frequent carriers of ARGs are the transposon-derived and closely related genomic islands AbaR-type and AbGRI-type (hereafter collectively called A. baumannii resistance islands, AbaRs) (10–12). AbaRs are widespread in A. baumannii but are also found in closely related pathogenic Acinetobacter species from the A. baumannii-Acinetobacter calcoaceticus complex, namely, Acinetobacter nosocomialis, Acinetobacter haemolyticus, Acinetobacter pittii, and Acinetobacter seifertii (10, 13, 14). Examination of AbaRs shows that they exist in as many as 53 genetic configurations, resulting from a complex history of multiple successive transposition events taking place within an original Tn7-like backbone (11, 15, 16). AbaRs often contain a class 1 integron (16, 17) and additional modules carrying ARGs conferring resistance to aminoglycosides, tetracycline and sulfonamide and beta-lactams (12). The intense transposition activity driving AbaRs plasticity suggests that they represent a safe harbor and reservoir for diverse TEs and ARGs (12). This plasticity could, for example, be behind the recent emergence of the AbaR4 variant, carrying the carbapenem resistance gene blaOXA-23, which is a major concern for the evolution of antibiotic resistance in Acinetobacter pathogenic species (18, 19).
AbaRs can be considered highly successful genomic islands, as they can be found in most clinical isolates of A. baumannii. This could be due to their ability to transfer across bacteria, by hitchhiking on plasmids (10, 12) but also by direct transfer through homologous recombination (20, 21). The genomic signature of these transfer events, and the fact that most A. baumannii isolates are competent for natural transformation (22–24) hint at a major role of this process in their HGT. Natural transformation (hereafter called “transformation”) is the process by which bacteria actively import exogenous DNA to recombine it in their chromosome (25). Confirming genomic evidence, transformation was found to allow the transfer of AbaRs at high rates between individual cells of mixed populations (26). Although AbaRs could use transformation for their dissemination, the contribution of transformation in the dynamics of MGEs, and more broadly in genome evolution, is currently questioned (27–29). It is well accepted that transformation can serve as a means for genetic diversification, allowing the acquisition by recombination of genetic polymorphisms (allelic transfer) and the efficient adaptation to fluctuating environments (30–32). Because transformation allows import of large DNA fragments, it also permits the acquisition of large nonhomologous DNA sequences (gene transfer) if these are flanked by sequences homologous between donor and recipient genomes (33–36). A consequence is that transformation opens the door to the acquisition of MGEs, such as transposon and integrons, prophages, and genomic islands (37–39) which often encode adaptive traits (antibiotic resistance). Yet, MGEs are associated with a fitness cost and, when not conferring an adaptive advantage, bacteria could use transformation as a means to delete these MGEs from the chromosome (29). Alternating the acquisition and removal of MGEs, and thus the transient expression of MGEs-carried traits, would maximize fitness by buffering against unpredictable environmental fluctuations (40).
Although potentially beneficial in the long term for bacteria, frequent acquisition and removal of adaptive MGEs is not compatible with the propagation and persistence of specific MGEs. Thus, a form of genetic conflict may exist between bacteria and MGEs. In that sense, a growing list of MGEs are found to counteract transformation through diverse strategies (41–47), suggesting an evolutionary response of MGEs against the cleansing activity of transformation in the host bacteria. Although critical to the persistence and dynamics of ARGs, the consequences of transformation inhibition on the evolutionary success of MGEs remains underappreciated. In this study, we examine the strategy of AbaRs islands of A. baumannii, which are commonly discovered inserted within the comM gene (10). This gene encodes a helicase that plays a specific role in the recombination of DNA obtained through transformation (48). As a result, AbaRs could impede the process of transformation. We here test the hypothesis that the interference of AbaR with transformation, mediated by their insertion site, is a strategy that gives them a selective advantage. We first established the strong preference of AbaR for the comM site among a large number of potential sites. Using an AbaR-naive Acinetobacter nosocomialis strain, closely related to A. baumannii, we then quantified the cost of AbaR acquisition in terms of growth of the bacterial host population. In addition, we quantified the effects of inserting AbaR into different genes on the efficiency of transformation (acquisition and elimination of heterologous sequences) naturally occurring in experimental populations. This work was used to develop and parameterize an evolutionary model in which AbaRs that differ in their strategies (insertion site) compete in exploiting the bacteria.
Results
AbaR Prevalence Is Associated with Insertional Inactivation of comM.
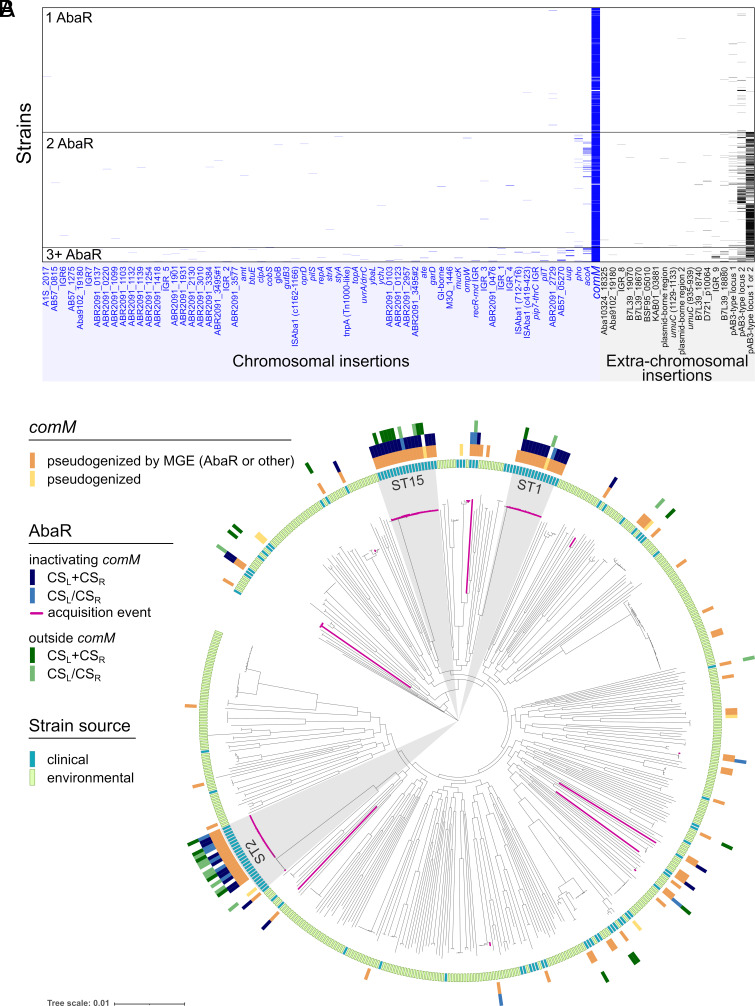
AbaRs islands share a Tn7-like backbone structure with conserved sequence at their left (CSL) and right (CSR) boundaries. An extensive query of these sequences was previously conducted on publicly available genomes, the majority of which are from clinical strains belonging to the lineages of sequence type 1 (ST) and 2 (ST2) (10). AbaRs were found in 66% of strains (10). Mainly present in the chromosome (85%), they were found inserted at 51 different core genome loci and in 12 loci within other MGEs (prophage, IS). Considering that over half of the strains carry more than one AbaR, we used this dataset to analyze the distribution of insertion sites in the genomes. We found that comM is interrupted by an AbaR in 96% of the genomes (Fig. 1A). Other occurrences of AbaR are found almost only when one AbaR is already inserted in comM and are typically in plasmids (Fig. 1A). Interestingly, insertional inactivation of other genes essential for natural transformation (pilT, pilS) is rare. AbaR elements are almost ubiquitous in the genomes of the strains, mostly of clinical origin, making it difficult to date and assess the independence of their acquisition. Hence, we searched the CSL and CSR in a smaller but much more diverse collection of 496 A. baumannii genomes including many environmental isolates that are expected to lack resistance islands (49) (Fig. 1B). We identified 60 strains from 19 different STs with comM disrupted by an AbaR. We conducted an ancestral reconstruction of the presence of AbaRs in the species phylogenetic tree and found 15 independent AbaR acquisitions (Fig. 1B and SI Appendix, Table S1). This indicates that AbaRs frequently inactivate comM. As before, there were 35 strains in which AbaR islands could be detected in sites other than comM. Out of these, 20 have comM readily interrupted by another AbaR, 3 strains display a pseudogenized comM and 7 have a comM gene interrupted by an MGE other than AbaR. These results show that the highly frequent inactivation of comM by AbaR in the clinical dataset is not the result of one single random event. Interestingly, in nonclinical isolates, the comM gene is often pseudogenized by the insertion of other large sequences. In two specific cases of pairs of closely related strains from clinical and environmental origins, comM is interrupted by an AbaR in the clinical strains, and by a CBASS defense system in the environmental ones. Overall, this indicates that A. baumannii’s MGEs frequently target and inactivate the comM gene, suggesting that this favors the propagation of AbaRs in clinical strains.
Fig. 1.
AbaR are preferentially inserted in the comM gene. (A) Distribution of AbaR islands insertion sites (x axis) in the genome of 3,148 A. baumannii strains, mainly of clinical lineages (ST2, n = 1,908; ST1, n = 125; ST25, n = 53). Strains are sorted on the y axis by the number of AbaR insertions. Data were retrieved from work by Bi et al. (10). Names of some insertion sites were abbreviated (Material and Methods). (B) Phylogenetic distribution of AbaR islands in a collection of 496 diverse A. baumannii strains of clinical (blue) and environmental sources (green). AbaRs were detected by searching genomes for their left and right boundaries (CSL and CSR) as displayed when inactivating comM (blue) or inserted elsewhere in the genome (green). The branches of the clades descending from the nodes where acquisitions of AbaR islands disrupting comM occurred were colored in dark pink.
AbaR Imposes a Fitness Cost on Their Host.
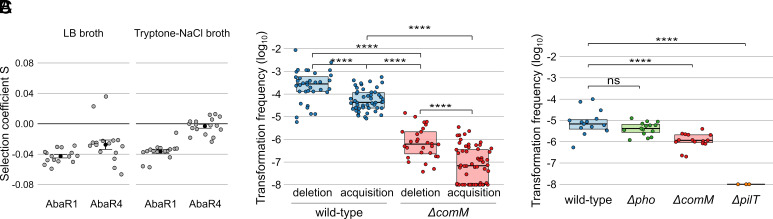
Acquisition of a single AbaR confers host with resistance to multiple antibiotics (26), providing a strong selective advantage under potentially diverse conditions of antibiotic stress. However, fixation of MGEs in bacterial genomes depends on their fitness effect on the bacterial host in absence of antibiotics (50). We therefore assessed the cost of AbaR acquisition by a naive host using an isogenic bacterial competition assay. As a native host, we chose the strain M2 of the A. nosocomialis species. This species is closely related to A. baumannii, and harbors similar AbaRs (10, 14), yet the M2 strain (previously reported as an A. baumannii) does not carry any AbaR (51). We selected two distinct AbaRs, AbaR1, and AbaR4. AbaR4 is an emerging, 16-kbp long AbaR, that contains the Tn2006-blaOXA-23 transposon conferring resistance to carbapenems (13, 52, 53). In contrast, AbaR1 is an 86-Kb long AbaR including 25 putative resistance genes (54), conferring resistance to several old-generation antibiotics (aminoglycosides, cephalosporins, tetracycline, and sulfonamide) (26), and which is not found in modern isolates. AbaR1 and AbaR4 are found inserted in the comM gene in the chromosome of strains AYE and 40288, respectively. To disentangle the effect on fitness of the insertion site and of the island, we first quantified the fitness cost of the inactivation of comM by running competitions between M2 and M2 ΔcomM strains, alternatively labeled by the introduction of the Hu-sfGFP marker (marker swap to deduce its cost on fitness; see Material and Methods). The selection coefficient of the M2 ΔcomM, corrected from marker cost, appeared weak and negative (mean = −0.011, SE = 0.005). Thus, inactivation of comM does not confer a fitness advantage that could have simply explained the high occurrence of AbaRs at this site. Next, to assess the effect on fitness of the selected AbaR, they were horizontally transferred to the M2 recipient to perform competition experiments between parental and AbaR-carrying derived strains. Competitions were conducted in rich and low-nutrient media (LB broth and tryptone-NaCl media, respectively). Acquisition of AbaR1 was deleterious to their host cells with selection coefficients of −0.043 and −0.037 in LB and tryptone-NaCl, respectively (Fig. 2A). The acquisition of AbaR4 seems less deleterious than that of AbaR1, with a selection coefficient of −0.027 in rich LB medium. Similarly to AbaR1, this cost is lower in the tryptone-NaCl condition. In both media, AbaR1 showed a higher fitness cost than AbaR4. This could be due to its large size (86 kb), and is consistent with the higher prevalence of shorter AbaR in A. baumannii genomes (10 to 20 kb) (10). Overall, the data suggest that in absence of antibiotic stress, AbaRs tend to incur a fitness cost. However, these fitness costs can be influenced by factors such as the content and size of the islands, as well as the prevailing growth conditions.
Fig. 2.
Fate of AbaRs and natural transformation events within mixed populations. (A) Experimental determination of the cost on fitness of AbaR4 and AbaR1. The effect on fitness of acquisition of AbaR4 and AbaR1 by the M2 strain was measured using bacterial competitions with their parental M2 strain in LB medium or in tryptone-NaCl medium. Selection coefficient (S) of the AbaR-carrying strains was determined relative to the parental M2 strain. Independent measures of selection coefficients are represented by the gray dots, black dots represent mean values, and error bars represent the SE. (B) Frequency of transformation events resulting in the deletion or acquisition of inserts of length from 10 to 16 kb in a wild-type strain and ΔcomM mutant. Equal parts of strains carrying or not the insert are mixed. Following incubation, the frequency of cells not previously carrying the insert and which acquired the insert is determined (acquisition). Similarly, the frequency of cells which have lost the insert (deletion) is determined (Materials and Methods). (C) Frequency of transformation events resulting in the acquisition of a SNP allele conferring resistance to rifampicin. As in A, equal parts of strains carrying or not the allele are mixed. The fraction of cells which acquired the SNP from the other cells is determined by plating (Materials and Methods). (ns: P > 0.05; *: P ≤ 0.05; **: P ≤ 0.01; ***: P ≤ 0.001; ****: P ≤ 0.0001). The detection limit of transformation events is 1 × 10−8. Boxplots represent the 25th percentile, median, 75th percentile.
Within-Population Natural Transformation Favors Deletion of AbaR.
Individual bacteria of a population may occasionally acquire an AbaR through horizontal gene transfer. The transfer of AbaR from AbaR-carrying individuals to noncarrying individuals spontaneously occurs in an experimental mixed population (26). These within-population HGT are attributed to the natural release of DNA by some cells, which can then be acquired through transformation by other cells within the population. Genomic analyses showed that within-population acquisition of AbaR results from the import of large, two-digit kb-long DNA fragments (26). However, such fragments released by AbaR-free cells could also promote the deletion of the island in AbaR-carrying cells, a phenomenon which has not been experimentally evaluated.
In order to determine the fate of AbaR in populations, we designed experimental populations of AbaR-carrying and AbaR-free cells that were genetically modified to determine the rate at which AbaR-carrying and AbaR-free cells respectively delete or acquire AbaR from one another. Importantly, deletion and acquisition events can only result from DNA released by the cells, as no other DNA source is provided. In addition, to test the importance of the size of AbaR elements on their acquisition/deletion rates, we varied the length of AbaR-derived elements from 2 to 16 Kb. To do so, we constructed a set of mutants, called M2 AbaR4-Lj and which carry an AbaR4-derived insert (j) of variable length (L) (SI Appendix, Fig. S1A). Another set of mutants, called M2 AbaR4-ΔLj was produced by deleting the insert while keeping the flanking regions (SI Appendix, Fig. S1A). The pairwise combination of these two sets of mutants in a mixed population makes it possible to measure the rate at which the AbaR4-derived insert is acquired or deleted. Indeed, AbaR4-ΔLj cells which acquired the insert using DNA released by AbaR-Lj could be detected by the gain of the kanamycin resistance conferred by the aphA gene (SI Appendix, Fig. S2A). In contrast, AbaR-Lj cells which took up DNA released by AbaR4-ΔLj cells will have the insert deleted, reconstituting the aacC4 gene conferring resistance to apramycin (SI Appendix, Fig. S2B). In each population consisting of a mix of M2 AbaR4-Lj and M2 AbaR4-ΔLj, either strain could act as donor or recipient. To distinguish between these events, we alternatively deleted the comEC gene in one genotype to make it a donor-only (transformation is inactivated by loss of comEC) while introducing the selectable rpoB(rif R) allele conferring resistance to rifampicin in the recipient genotype.
We observed that the acquisition rate of the insert only slightly decreases with its length (SI Appendix, Fig. S2C), confirming previous results that the full-length, 16.6 kb-long AbaR4 island is acquired 4-times less efficiently than the 4.8 kb-long Tn2006 (26). Yet, while statistically significant (linear regression test; n = 101, t = −2.467, P-value = 0.0153), the size effect remained limited (slope of −0.027 kb−1 and r2 = 0.048). In contrast, deletion rates do not vary significantly with the length of the insert (linear regression test; n = 69, t = 1.865, P = 0.066) indicating that small and large islands are deleted with similar efficiencies (SI Appendix, Fig. S2D). Importantly, in the range of 10 to 16 kb, deletion is more efficient than acquisition (Fig. 2B) (Student’s t test; df = 84, t = −4.89, P = 4.76E-6), an asymmetry that is in line with previous results (55). This indicates that, in the absence of selective pressure to maintain AbaR, natural transformation would be effective at purging them from the population.
Within-Population Natural Transformation Events Are Differentially Altered by comM Inactivation.
Depending on their insertion site, the acquisition of AbaR could alter the rate of subsequent within-population gene and allelic transfer events. Indeed, AbaRs seem to target comM, encoding a DNA helicase implicated in homologous recombination of transforming DNA. When tested using exogenously added DNA carrying a single (and selectable) nucleotide polymorphism (SNP), inactivation of comM was previously shown to reduce the transformation frequency SNP by ~sevenfold in Acinetobacter baylyi (48). We thus next tested the impact of comM inactivation on allelic transfer spontaneously occurring in the mixed-population setup, which here consisted in equal parts of the M2 cells carrying or not a rpoB(rifR) allele in which a single SNP confers resistance to rifampicin. To detect the acquisition by rifampicin-sensitive recipient M2 cells, we used strains carrying a kanamycin resistance marker at a neutral site (attTn7) in order to distinguish them from rpoB(rifR) allele donor cells. Also, to avoid the confounding effect of the rpoB(rifR) allele donor cells acquiring the kanamycin resistance marker of recipients, the comEC gene of donors was inactivated. We found that recombinants displaying both kanamycin and rifampicin resistance were produced at a frequency of 1E-5 (Fig. 2C). Inactivation of pho, a gene of unknown function and in which AbaRs are also found, had no effect. Confirming that HGT in these conditions is due to natural transformation, inactivation of pilT encoding the retraction ATPase of Type IV pilus required for transformation, abrogated transfer of the rpoB(rifR) allele. Inactivation of comM resulted in an eightfold reduction in the acquisition of the rifampicin resistance from neighboring cells (Fig. 2C). Thus, the interindividual transfer of SNP alleles is marginally affected by the inactivation of comM. Given the role of ComM in integration of heterologous DNA (48), we next tested the impact of comM inactivation on within-population acquisition and deletion of large heterologous DNA sequences such as AbaRs. Bacteria lacking the comM gene displayed decreased acquisition rates of the AbaR4-derived insert (j) by around 100-fold (Fig. 2B). Interestingly, the absence of functional ComM did not cause any additional defect in acquisition of inserts of increasing length with a slope of −0.069 kb−1, similar to that observed in wild-type cells (SI Appendix, Fig. S1C). Deletion rates of inserts follow the same trend, being strongly reduced by the inactivation of comM with little dependence on the length of the deleted sequence (SI Appendix, Fig. S1D). Like in the wild-type situation, deletion rates remain higher than acquisition rates. Importantly, deletion rates in comM-defective individuals are ~50 times lower than acquisition rates in wild-type cells (Student’s t test; df = 76; t = 14.382, P < 2.2E-16) (Fig. 2B). This suggests that AbaR islands targeting comM may not be purged by natural transformation within a population.
Transformation-Inhibiting AbaRs Gain a Competitive Advantage in a Fluctuating Environment.
The asymmetry of natural transformation supports a role in removing parasitic MGEs from the chromosome of bacteria [genome curing effect, (29)]. It could be hypothesized that the specific insertional inactivation of the comM gene would represent an evolutionary response of AbaRs. We found that AbaR insertion in comM globally inhibits the transformation rate but this inhibition appears heterogeneous between acquisition or removal of large DNA fragments (gene transfer events, >500-fold reduction) and SNPs (allelic transfer, ~eightfold reduction) (Fig. 2 B and C). To disentangle the effect of comM inactivation on the dynamic of AbaRs, we developed a computational model simulating bacterial populations exposed to AbaR elements (Material and Methods and SI Appendix, Fig. S3A). We considered three distinct AbaRs variants that compete for the exploitation of bacteria and differing in their insertion strategy, i.e., in the genes targeted for their insertion. AbaR inserting in comM (comM::AbaR) which partially inhibits transformation of their host, AbaR inserting in pilT (pilT::AbaR) representing AbaR which completely prevents transformation, and AbaR inserting in pho (pho::AbaR) considered as a generic neutral site with no incidence on natural transformation. Simulated bacterial cells are allowed to divide following a logistic growth model. Cells can thus naturally die at a basal rate, and at an elevated rate when encountering environmental stress. Consequently, their DNA is released, becoming available for other organisms to undergo natural transformation. The capacity for natural transformation of individuals is dictated by their genotype (wild-type, pilT, pho, comM) and calibrated from empirical data (Figs. 1 and 2, summarized in SI Appendix, Tables S3 and S4). AbaR could confer resistance to antibiotic stress but would reduce the growth rate of their host cell (fitness cost) according to experimental values (Fig. 2A).
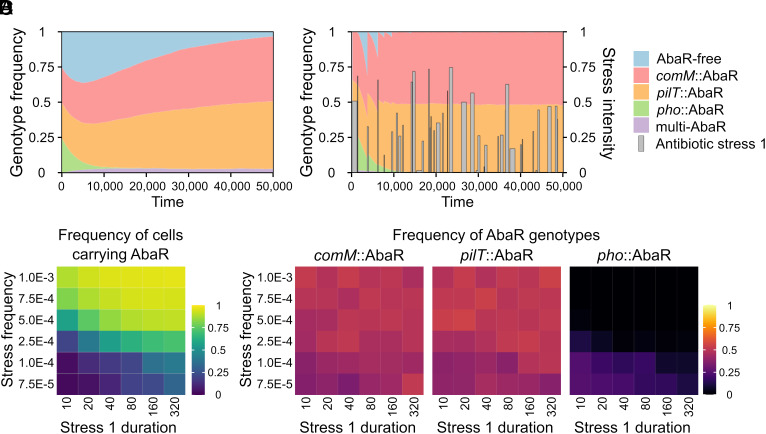
To examine the impact of transformation asymmetry in MGE removal, we initially observed the dynamics of AbaR under a hypothetical scenario where they incur no costs on fitness and do not provide any selective advantage to their bacterial host. We found that when bacteria carrying AbaR at the neutral site (pho::AbaR) are equally mixed with AbaR-free wild-type cells, AbaR are purged from the population (SI Appendix, Fig. S3B, row 1). Thus, the transformation asymmetry in our system of within-population transformation is effective to cure a genomic island (here AbaR) from a population. In contrast, comM and pilT-inactivating AbaRs are not purged and even spread to the entire population. This is due to the fact that the wild-type cells always acquire AbaR at a high rate, while the genotypes comM::AbaR clear their AbaR at a much lower rate or fail to clear them in the case of pilT::AbaR. These results confirm the genome-curing function of natural transformation (SI Appendix, Fig. S3B). Furthermore, still without incurring costs or facing antibiotic-induced stress, when all four genotypes are mixed in the initial population, transformation-inhibiting AbaRs (comM::AbaR or pilT::AbaR) coexist quasi-neutrally and outcompete the AbaR that does not (pho::AbaR) (Fig. 3A; see also SI Appendix, Figs. S3 and S5A). We then considered the more realistic situation where AbaRs are costly and confer antibiotic resistance. In the absence of stress, wild-type cells expectedly gain a competitive advantage over any AbaR-carrying cells (SI Appendix, Fig. S3B, row 2). Single occurrence of stress then strongly favors AbaR-carrying cells, which take over the population under constant or stochastic stress (SI Appendix, Fig. S3B, rows 3 and 4). In simulations initialized with only the wild-type genotype, while AbaRs are added to the extracellular compartment at a residual rate, short and rare exposures to antibiotic stress are sufficient to select and maintain AbaRs (Fig. 3 B and C). If stress is scarce (mean frequency below 1E-4) and short (mean duration inferior to 80), the wild-type genotype is favored, with a low proportion of cells carrying AbaRs (Fig. 3C). In this situation, all AbaR-carrying genotypes perform equally well. In contrast, under frequent and/or longer stress duration, which increase the proportion of AbaR-carrying cells (Fig. 3C), pho::AbaR appear less competitive and can even become extinct, while the transformation-inhibiting AbaRs (comM or pilT::AbaR) coexist in a quasi-neutral way (Fig. 3D and SI Appendix, Fig. S5B). This stems from the genome curing effect: the inevitable removal of AbaR from the transformation-proficient pho::AbaR cells, while transformation-inhibiting AbaR (comM and pilT) are themselves protected from this effect.
Fig. 3.
Transformation-inhibiting AbaRs gain a competitive advantage in a fluctuating environment. (A and B) Simulated temporal dynamics of bacterial genotypes carrying hypothetically costless AbaR in the absence of antibiotic stress (A) or in presence of a stochastic stress (mean peak duration = 320, mean peak frequency = 1E-3) and AbaR imposing a fitness cost (0.01) (B). Simulations were initiated with equal number of cells of each genotype (2.5E5, which is 1/40 of the population carrying capacity) and with initial number of each eDNA type (1E7). (C) Mean frequency of cells carrying AbaR between times 30,000 and 50,000 as a function of stress duration and stress frequency (averaged from 50 simulations each). (D) Mean frequencies of each AbaR (among all AbaR genotypes) as a function of stress duration (d) and stress frequency (f). For each simulation (initialized with AbaR-free cells only = 1E6, initial number of each eDNA type = 1E7), we calculate the mean frequency of genotypes between times 30,000 and 50,000, and then calculate the mean of these frequencies from 50 independent simulations. The number of actual generations per simulation depends on the stress regime which reduces the population density, but also depends on the genotype of bacteria. In 50,000 simulated time units, the number of generations can thus vary from ~5,000 [when, under low stress, the population at its carrying capacity (K) at each time] to up to ~50,000 (if there were theoretically no density dependency). Model parameters are listed in SI Appendix, Table S2.
Altogether, these findings demonstrate the potency of transformation asymmetry in purging AbaRs (or any other MGEs) from the bacterial chromosome. When AbaR provides a selective advantage during stochastic exposure to antibiotic stress, the AbaR variants that hinder transformation gain a competitive advantage over those that do not. Interestingly, comM::AbaR and pilT::AbaR genotypes show similar dynamics, suggesting that partial inhibition of transformation linked to insertion in the comM gene is nearly as effective as complete inhibition.
Transformation-Modulating AbaRs Targeting comM Maintain the Adaptability of Their Hosts.
A role of natural transformation in removing parasitic MGEs is still consistent with its documented involvement in adaptive recombination of beneficial alleles (28). The high rates of recombination in A. baumannii indicate a major role of natural transformation in acquisition of beneficial alleles. This is exemplified by the acquisition of the highly selective SNPs in gyrA and parC genes conferring resistance to fluoroquinolones (20). We then implemented allelic variations in the computational model by introducing a SNP providing resistance to a specific stress 2. Stress 1 (to which AbaR are conferring resistance) and stress 2 occur stochastically and independently.
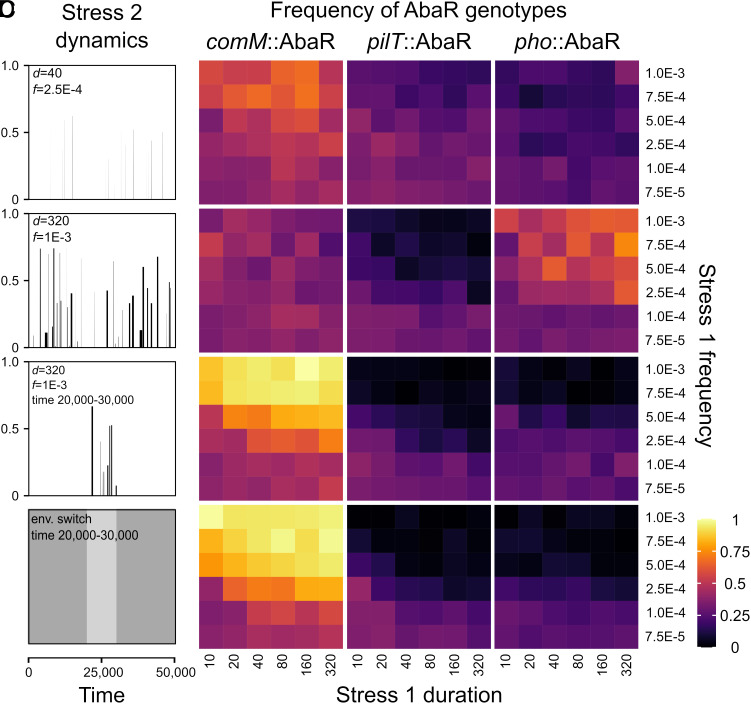
When stress 2 is rare and short (Fig. 4A), we detect an advantage of comM::AbaR over pilT::AbaR and pho::AbaR for numerous stress 1 exposure regimes. comM::AbaR, which maintains a high level of transformation for short sequence changes, allows their host bacteria to acquire the SNP. In contrast, pilT::AbaR that completely inhibits transformation cannot respond to changes in exposure to stress 2 and are at a disadvantage. It should be noted that pilT::AbaR are not completely counterselected because wild-type cells (carrying or not the SNP), which grow during stress 1-free periods, can subsequently acquire pilT::AbaR by transformation, resulting in the persistence of pilT::AbaR in the population (SI Appendix, Fig. S5B). When stress 2 is more frequent and longer (Fig. 4B) and exposure to stress 1 is sustained (frequent and long-lasting), pho::AbaR becomes fixed in the majority of simulations. The SNP persists in the population (SI Appendix, Fig. S4) and maximum transformation activity is selectively advantageous: This ensures great genomic plasticity, i.e., acquisition of the SNP when exposed to stress 2, but also its rapid elimination when it is no longer needed. Bacteria carrying pho::AbaR can acquire and eliminate SNPs 8-times faster than bacteria carrying comM::AbaR and as a result, pho::AbaR has a selective advantage. Note that pho::AbaR does not escape genome curing but as it becomes the only AbaR capable of conferring stress 1 resistance in the majority of simulations, it is regularly recruited/reintegrated by transformation. These results could be interpreted as a form of cooperation (mutualism) between pho::AbaR and their bacterial host, but this does not seem robust from an evolutionary point of view (see below).
Fig. 4.
Frequency of AbaR islands in fluctuating environments favoring adaptation by allelic transfer. Results of simulations of the competition between the three types of AbaR islands, which differ in their insertion strategy. Environmental fluctuations have two components: the bacterial population is exposed to stochastic stress 1 (to which resistance is conferred by AbaR) and another environmental change 2 (to which advantage is conferred by a SNP acquired by allelic transfer). The gray boxes show the dynamics of environmental change 2. In panels (A–C), environmental change 2 corresponds to stochastic exposure to a new stress (stress 2) which from Top to Bottom are (A) rare and short (mean peak duration d = 40, mean peak frequency f = 2.5E-4), (B) frequent and long (mean peak duration d = 320, mean peak frequency f = 1E-3), and (C) frequent and long (mean peak duration d = 320, mean peak frequency f = 1E-3) over a short period (20,000 to 30,000 time units). Last, (D) environmental change 2 is not a stress but a change in resource availability occurring between 20,000 and 30,000 time units. The ratio of bacterial genotypes carrying AbaR was assessed as a function of stress 1 frequency and duration (panel A–D). The cost on fitness of the SNP conferring resistance to stress 2 was set at 0.1 (see SI Appendix, Fig. S4 for similar result with a fitness cost of 0.01). For each simulation (initial number of cells: AbaR-free = 1E6, initial number of each eDNA type = 1E7), we calculate the mean frequency of genotypes between times 30,000 and 50,000, and then calculate the mean of these frequencies from 50 independent simulations. Model parameters are listed in SI Appendix, Table S2.
We then tested situations in which the environmental change occurs over a relatively short period during the modeled trajectories (Fig. 4 C and D) or a longer period (SI Appendix, Fig. S4Q). In the first scenario, the change is characterized by exposure to stress 2 (which increases the mortality of bacteria not resistant to this stress) to which the SNP confers resistance (Fig. 4C). In the second scenario, this change corresponds to a shift in the continuously available resource, which has no effect on bacterial mortality. However, the resource will be better exploited by bacteria carrying the appropriate SNP, resulting in a restored replication rate (Fig. 4D and SI Appendix, Fig. S4Q). In all these situations, the insertion in comM clearly confers a strong selective advantage. During long periods without stress 2, pho::AbaR is cleaned out in favor of comM::AbaR and pilT::AbaR (situation similar to Fig. 3). As soon as environmental change occurs, pilT::AbaR is counterselected because, unlike comM::AbaR, it cannot acquire the SNP by transformation. The success of comM::AbaR is therefore explained by the fact that it maximizes its probability of persistence (reducing the probability of being eliminated by transformation to almost 0) while maintaining a nonnegligible level of transformation activity for allelic changes allowing the host bacterium to respond quickly to the change in its environment. It should be noted that pilT::AbaR could persist if wild-type cells which managed to acquire the SNP subsequently acquire pilT::AbaR during the short environmental change. However, the newly formed pilT::AbaR bacteria would also be rapidly counterselected once the initial environment was restored, in favor of comM::AbaR bacteria that were able to eliminate the SNP by transformation. The results are qualitatively similar when SNPs with a weak (10-times lower) cost are tested (SI Appendix, Fig. S4).
In conclusion, we found that if any allelic change is necessary for survival or adaptation to a reversible environmental shift, then even a single occurrence of such is sufficient to fix in the population the AbaR targeting comM.
Discussion
We investigated the insertion sites of AbaRs in A. baumannii genomes, quantified transformation rates in experimental populations, and performed modeling work to understand the insertion strategy of AbaR islands into the chromosome of bacteria. We found that insertion into the comM gene confers a selective advantage in many contexts, surpassing other insertion strategies that either do not inhibit (such as insertion into the pho gene) or completely inhibit transformation (such as insertion into the pilT gene). This comM insertion strategy should be selected due to two combined effects on transformation. First, inserting in comM allows AbaR to circumvent the genome curing effect of transformation. This curing effect, proposed on the basis of the transformation properties of Streptococcus pneumoniae, requires that deletion rates be greater than acquisition rate (referred to asymmetry of transformation) (29). Such strong asymmetry was verified in S. pneumoniae, with experimental models involving the addition of exogenous DNA (55). We here confirm this phenomenon in A. baumannii, yet in a unique and more realistic model in which transformation occurs spontaneously within a population, not through the addition of exogenous DNA. In this system, the degree of transformation asymmetry is less pronounced which is due to the rates of acquiring naturally released DNA being far less affected by the size of the acquired DNA (2.7E-2 kb−1) than in the S. pneumoniae model (1E-6 kb−1). This is possibly due to the fact that naturally released DNA presents fewer transformation-terminating nicks than purified DNA. Simulations indicate that even a limited asymmetry between acquisition and deletion rates is effective to cure AbaR—or any other chromosome-integrated MGE—from a population (SI Appendix, Fig. S3B, row 1). Through the inactivation of comM, AbaR significantly diminish the deletion rate of their host, leading to a substantial 50-fold asymmetry between the rate at which wild-type cells can acquire AbaR and the rate at which they can subsequently remove it. By counteracting the curing effect of natural transformation, AbaR inserting in the comM gene can invade a population (SI Appendix, Fig. S3B, row 1).
Second, the success of AbaRs is explained by the fact that inserting in comM differentially inhibits allelic and gene transfer events. While AbaRs inserting in comM can oppose the genome curing effect of transformation, their host cell would still allow the transformation-mediated recombination events which can shuffle mutations, break linkage disequilibrium, acquire beneficial mutations, or purge deleterious ones. We observed that AbaR inserting in comM outcompete AbaR inserting in any other site, whether allelic transfer confers a strong selective advantage (resistance to antibiotic) or a milder adaptive advantage linked to the exploitation of a new resource in the environment (Fig. 4). This differential inhibition of allelic and gene transfer events is linked to the role of ComM as the helicase dedicated to natural transformation. Inactivation of any other genes of the transformation pathway, and involved in initial binding (type IV pilus genes, pilS, pilT, pilA, etc), import (comEA, comEC, comFC), protection (dprA), or recombination (recA) of DNA would not result in such differential inhibition (25). Inactivation of these genes would rather cause a complete inhibition, and similarly to the pilT example of the model, AbaR inserting in them would be strongly disadvantaged. In addition, inactivation of some of the genes required for transformation may also be counterselected because these genes are also needed for other important functions. For instance, inactivation of RecA would cause major deficiencies in DNA repair, and loss of Type IV pili would affect the cells capacity to attach and move on surfaces. Even without considering these effects, our model indicates that complete loss of transformation would be detrimental. Thus, among the genes involved in natural transformation, comM represents a unique target because it is specific to this pathway, and its inactivation blocks genome curing yet still allows evolution by acquisition and recombination of new alleles. The results are therefore consistent with the recent demonstration that these Tn7-like elements have evolved to specifically insert in the comM gene (56).
Our simulations demonstrate that the comM insertion strategy is advantageous when the model incorporates some degree of environmental complexity, such as different types of stress, or stress combined with changes in environmental resources. It would thus seem essential to consider the complexity of the environment that may favor AbaR targeting the comM gene of A. baumannii. In A. baumannii isolates from nonclinical lineages, and presumably less exposed to antibiotics (57), comM is often pseudogenized by large insertions unrelated to AbaRs and not carrying ARGs (Fig. 1B and SI Appendix, Table S1). The comM gene has also been found disrupted by distinct MGEs in Mannheimia succiniciproducens (58), Mannheimia haemolytica (58), Francisella philomiragia, and the two species known to be naturally transformable Pseudomonas syringae (29) and Aggregatibacter actinomycetemcomitans (59). Inserting in comM may be a successful strategy for many other MGEs or genomic islands of transformable species evolving in contrasting environments. However, the comM insertion strategy, altering transformation in a peculiar way, may benefit only certain types of MGEs. Some integrative conjugative elements (ICEs) have been reported to completely inhibit transformation (41–43, 45) and this is also expected for prophages inserting into genes essential for transformation (29). Such total inhibition of transformation is not a successful strategy in our model. But, in contrast to genomic islands such as AbaRs, these types of MGEs are autonomous in their horizontal transfer and do not depend on natural transformation to access a new genetic context. Therefore, they can completely inhibit transformation while escaping extinction in the event of environmental change by controlling their own horizontal transfer. On the contrary, nonautonomous MGEs (such as AbaRs) depend on natural transformation to change their genetic context and may have an interest in maintaining basal transformation activity. Interactions between MGEs should also be considered to understand the strategies of MGEs in inhibiting transformation. For example, autonomous MGEs could still benefit from the inhibition of transformation conferred by AbaRs inserted into the comM gene. These MGEs could then dispense with carrying transformation inhibition mechanisms. Understanding the evolution of MGEs strategies for manipulating natural transformation and the diversity of these strategies will require the development of experimental approaches which, combined with modeling work, would provide a better understanding of the combination of traits carried by the MGEs and the interactions between MGEs.
The insertion strategy into the comM gene should be a key element in the dynamics of ARGs, at least in A. baumannii. This may occur in two distinct ways not explored here: First, AbaRs inserted in comM can persist for a long time in bacteria genomes and therefore accumulate transposons or undergo rearrangements, easily leading to the evolution of accessory functions such as antibiotic resistance. Second, AbaRs can have their own intracellular dynamics and change vehicles (genetic support). AbaRs, which may have conserved a transposition activity, can jump to plasmids, thus facilitating new pathways for the spread of antibiotic resistance and new evolutionary trajectories of AbaRs on these new genetic vehicles. In this sense, our genomic analysis reveals that when an AbaR is present on a plasmid, it is also almost always present in comM - although the reverse is not true -, suggesting intracellular movements (Fig. 1A).
In conclusion, studying the strategies of MGEs and their interactions with the natural transformation mechanism of bacteria is crucial for understanding the persistence and evolution of antibiotic resistance. Our findings highlight the importance of considering environmental complexity when investigating the prevalence of specific transformation inhibition strategies, such as the insertion into the comM gene. By uncovering the mechanisms explaining the successful establishment and long-term persistence of MGEs, we could better develop strategies to combat the spread of antibiotic resistance and preserve the effectiveness of antimicrobial interventions.
Materials and Methods
AbaRs Insertion Sites in Bacterial Genomes.
AbaR-type insertion sites identified from 3,148 genomes were retrieved from Bi et al. (10). Insertion sites belonging to the same gene (or intergenic region, IGR) were combined. Each insertion site is attributed to a chromosomal or extrachromosomal category. AbaRs inserted into tetA sites, which correspond to an AbaR inserted within AbaR were removed from the data. AbaR-type insertions were detected by the presence of either the conserved sequence left or right (CSL, CSR) of AbaR-type elements (10). For the AbaRs inserted in the pAB3-type locus 1 and pAB3-type locus 2 sites, we checked that both boundaries were present. Finally, we represent the insertion sites of the different AbaRs for each bacterial strain, by grouping the strains according to the number of AbaRs they carried (analysis and graphical representation: R4.3.1 and ggplot2). The name of some insertion sites were abbreviated as follows: IGR_1, ABR2091_3231-tRNAArg; IGR_2, ABR2091_3550-ABR2091_3551; IGR_3, ABR2091_0006-ABR2091_0007; IGR_4, CBI29_04511-CBI29_04512; IGR_5, ABR2091_1794-ABR2091_1795; IGR_6, AB57_0986-AB57_0987; IGR_7, ABR2091_0111-ABR2091_0112; IGR_8, ACX61_19590-ACX61_19595; IGR_9, CBI29_04511-CBI29_04512
Phylogenetic Distribution of AbaRs in Diverse A. baumannii Genomes.
A diverse collection of 496 A. baumannii strains from clinical and environmental origins (49) was queried for the presence of AbaRs. CSL and CSR were considered present when the combination of tniC and tniA and orf4, respectively, was present. TniC (WP_000736404.1), TniA (WP_000573062.1), and Orf4 (WP_001144958.1) were searched using tblastn (blast+/2.10.1). Each gene was deemed present if the alignment had more than 80% identity and 80% coverage with the query protein. AbaR are deemed present if at least 1 CS is identified. Pseudogenization of comM (tblastn alignment covered between 20% and 80%) was observed in 110 strains. comM was interrupted by large nucleotidic insertions (MGEs, AMR genes, or defense systems) in 101 of them. To verify whether those interruptions were due to an integration of an AbaR in the gene, we confronted the previously identified AbaR hits and their positions with comM region of interruption. comM was considered disrupted by an AbaR when an AbaR hit fell in between the bounds of its region of interruption.
We reconstructed the ancestral states of presence/absence of the gene families that constitute the AbaRs interrupting comM. The reconstruction was made along the recombination-free rooted phylogenetic tree (49) with the MMPA prediction method and the F81 evolutionary model of PastML v.1.9.34 (60). Since the MPPA algorithm can keep several ancestral states per node if they have similar and high probabilities, only the events where both ancestral and descendant nodes had one single distinct state either presence or absence were kept. Hence, at each node, a gene family could only be gained or lost. In one genome a single gene family can have several gene representatives. In that case, with our inference, the acquisition node of the gene family would correspond to the acquisition node of the first gene acquired and not necessarily of the gene inside the AbaR. To avoid this, only the AbaR gene families that had a single representative gene in the corresponding genome were considered. The node where all the gene families composing an AbaR and verifying these criteria were gained was considered to be the node where the AbaR was acquired.
Bacterial Strains, Plasmids, and Genetic Constructs.
Bacterial strains with genotypes and plasmids are detailed in SI Appendix, Table S7. All the oligonucleotides used in this study for genetic modification are listed in SI Appendix, Table S8.
Chromosomal Modifications in Acinetobacter.
Genetic constructions were performed using overlap extension PCR to create assembly PCR fragments carrying the genetic construction with a selection marker flanked by 2 kbp-long fragments that are homologous to the insertion site. A scarless genetic strategy was used to create the mutations without antibiotic resistance markers (SI Appendix, Fig. S1B). All PCRs for genetic modifications were performed with a high-fidelity DNA polymerase (PrimeStarMax, Takara) and chromosomal modifications were confirmed by colony PCR (Dream Taq, Thermo Fisher).
Bacterial Strains for Fitness Cost Determination.
The M2 hu-sfgfp aacC4 (Hu-sfGFP) strain was obtained from a previous work (23). The M2 comM::AbaR4 and M2 comM::AbaR1 strains were generated by naturally transforming the M2 comM::sacB aacC4 with genomic DNA from A. baumannii strains 40288 (AbaR4) (26) AYE (AbaR1) (54). Fluorescent variants were generated by naturally transforming bacteria with a PCR product of the hu-sfgfp aacC4 construct.
Bacterial Strains for SNP Acquisition.
The M2 rpoB(RifR) rifampicin-resistant variant of the A. nosocomialis M2 wild-type (WT) strain was obtained by plating an overnight bacterial culture on a LB agar plate supplemented with rifampicin. Spontaneous resistant mutants were then purified and the rpoB gene from each one was further sequenced. The selected isolate displayed normal growth on LB plates supplemented with rifampicin and the nonsynonymous mutation A1565T (G522L) in the rpoB gene. The nontransformable derivative M2 rpoB(RifR) comEC::aacC4 strain was obtained by naturally transforming the M2 rpoB(RifR) strain with a PCR product of the comEC::aacC4 construct. The M2 ΔcomM strain has a deletion of 231 bp in the coding sequence of the comM gene and the M2 Δpho strain has a deletion of 825 bp in the coding sequence of the pho gene. Both mutants were generated using the scarless genetic strategy. The M2 pilT::aphA was obtained from Gottfried Wilharm (22). To test acquisition of the SNP in mixed culture with the M2 rpoB(RifR) comEC::aacC4 strain, recipient strains were made kanamycin resistant by inserting the marker at the AttTn7 site.
Bacterial Strains for Acquisition and Deletion of Inserts in Mixed Culture.
A. nosocomialis M2 mutants carrying heterologous DNA chromosomal inserts with variable length were generated following the steps described in (SI Appendix, Fig. S1B). Rifampicin-resistant derivatives for selection of recipient strains were obtained by transforming bacteria with a PCR product of the rpoB gene amplified from genomic DNA of the rifampicin-resistant M2 rpoB(RifR) strain. Nontransformable derivatives for donor strains were obtained by transforming bacteria with a PCR product of the comEC::tetA chromosomal modification.
Fitness Cost of AbaR.
Bacterial competitions were performed by competing an A. nosocomialis M2 strain carrying an AbaR island inserted in the comM gene (M2 comM::AbaR4 or M2 comM::AbaR1 against the isogenic M2 with no AbaR. A single chromosomal fluorescent Hu-sfGFP marker was alternately expressed in each genetic background, as this marker discriminates fluorescent cells from the nonfluorescent cells using flow cytometry. The fitness cost associated with the expression of the Hu-sfGFP marker was determined by conducting marker swap experiments and subtracted from the fitness data points. Bacterial competitions were performed as previously described (61). Briefly, bacteria from the competing strains were grown in the competition medium (LB broth or Tryptone-NaCl medium) for one night. The next day, overnight cultures were mixed at an equal ratio in 24-well plates and were preincubated at 37 °C for 1 h. Competitions were then performed in 12-well plates by diluting 1,000-fold each day the competing culture in a fresh liquid medium and incubated for 24 h (10 generations per day). The ratio of AbaR carrying cells to wild-type cells was measured by counting 105 total cells using flow cytometry (Attune acoustic flow cytometer) at 0 and 10 generations. Selection coefficients were determined using the model s = [ln R(t)/R(0)]/t where R is the ratio of mutant to wild-type cells (62).
Within-Population Transformation in Mixed Cultures.
Mixed cultures were setup as previously described (26). Strains were grown overnight on LB agar plates, then inoculated and grown in 2 mL of LB until the cultures reached an optical density (OD, 600 nm) of 1. The bacterial broths were then diluted to an OD of 0.01 in PBS. Then equal volumes of bacterial suspensions were mixed by pairs. The mixture (2.5 µL) was deposited on the surface of 1 mL of tryptone-NaCl medium solidified with 2% agarose D3 (Euromedex) poured in 2 mL microtubes or in wells of 24-well plates and incubated overnight at 37 °C. Bacteria resuspended in PBS were plated on LB agar plates without antibiotics and LB agar plates containing appropriate antibiotics. Recombinant frequencies were determined through calculation of the ratio of the number of CFUs on antibiotic selection to the total number of CFUs on plates without antibiotics.
Transformation Assays with Purified DNA.
Bacteria were grown approximately 5 to 6 h in LB liquid medium at 37 °C and were then diluted in PBS to obtain bacterial suspensions of 107 cells/mL. Then, an equal volume of the bacterial suspension and the DNA substrate solution (at 100 ng/µL) were mixed together and 2.5 µL of this bacteria/DNA mix was spotted on the surface of a 1 mL transformation medium (tryptone 5 g/L, NaCl 2.5 g/L, agarose type D3 2%) freshly prepared in a 2-mL Eppendorf tube. After 20 h at 37 °C, bacteria were harvested by adding 200 µL of PBS in the tube and vortexing. Transformants were selected after plating the bacteria in selective agar media containing various antibiotics (apramycin 30 µg/µL, kanamycin 15 µg/µL, rifampicin 100 µg/mL, tetracycline 15 µg/mL, nalidixic acid 60 µg/mL). Transformation frequencies were determined by calculating the ratio of the number of transformant CFUs (counted in selective media) to the total number of CFU (counted in nonselective media).
Modeling the Dynamics of AbaRs.
We used a stochastic numeric model to generate the dynamics of AbaRs, which have different insertion strategies and compete in the exploitation of a bacterial population. The model comprises two compartments, one composed of bacterial cells, the other of extracellular DNA (eDNA) containing wild-type alleles, AbaRs carrying resistance and SNP (SNPs are only used when modeling a second stress or a change in environmental resources (Fig. 4 and SI Appendix, Fig. S4). Bacteria can uptake eDNA from the extracellular compartment and replace their own using transformation. AbaRs inserted into comM, pilT, and pho inhibit transformation partially, totally or not at all, respectively. We examine the performance of these different AbaR insertion strategies when the bacterial population is exposed to different environmental fluctuation patterns. The model is detailed (SI Appendix) and its main components are described below.
In the first situation modeled, the bacteria are exposed to a single type of stress (stress 1), with no other environmental changes (Fig. 3). The bacterial cells have three AbaR insertion sites in their chromosome (comM, pilT, pho) which can be occupied by wild-type allele (comM, pilT, pho) or AbaR (comM::AbaR, pilT::AbaR, pho::AbaR). Considering all combinations of DNA type, there are a total of 23 = 8 possible genotypes i. Bacterial population growth follows a logistic model and the number of replicating cells per genotype i and per time step dt, is determined using a binomial distribution:
where is the replication rate and is the total number of cells with genotype i at time t. Cells carrying an AbaR have a replication rate lower than that of AbaR-free bacteria.
Bacteria release DNA upon cell lysis, the number of lysed cells per genotype i and time step dt also follows a binomial distribution:
where is the lysis rate per time step at time t. Bacterial populations are faced with stochastic stresses of random duration, frequency, and intensity. In the absence of stress, cells are lysed at a basal rate, and under stress exposure, the lysis rate of AbaR-free cells increases but remains unchanged for cells with an AbaR carrying resistance.
Bacteria can uptake eDNA from the extracellular compartment and replace their own using transformation, the number of competent cells undergoing a transformation event during a time step dt is determined using a binomial distribution:
where is the transformation rate of genotype i at time t.
AbaRs inserted into comM, pilT, and pho inhibit transformation partially, totally or not at all, respectively (Fig. 2 and SI Appendix, Table S3). For each run, the transformation probability values were randomly drawn from experimental data (SI Appendix, Table S5). Then, the transforming cells are distributed thanks to a multinomial law among the different genotypes according to transformation probabilities (SI Appendix, Table S6).
The extracellular compartment is supplied by eDNA from lysed cells, each lysed cell releasing DNA molecules corresponding to their DNA composition. In addition, eDNA is added at a residual rate simulating arrival from neighboring populations (open system). In the extracellular compartment, eDNA is degraded at a constant rate , the number of degraded eDNA molecules j per time step is determined using a binomial distribution:
where corresponds to the number of eDNA of type j.
In the following modeled situations, more complex environmental fluctuations were simulated by either introducing a new stress (stress 2, Fig. 4 A–C) or by modeling a change in environmental resource (Fig. 4D and SI Appendix, Fig. S4 row 5). In both situations, adaptation to the new environment is conferred by a SNP that can be acquired or removed by transformation. In the case of stress 2, the rate of cell lysis increases for cells lacking the SNP, which replenishes the extracellular compartment (Fig. 4C). When there is a change in resource availability, bacteria lacking the appropriate SNP experience a decrease in their replication rate (Fig. 4D and SI Appendix, Fig. S4 row 5). Because of the new insertion site, the number of genotypes is increased and becomes 24 = 16. The previous equations remain true.
The model was implemented in C++ (gcc version 9.4.0), using Code::Blocks IDE (20.03). The code is available on Zenodo (https://doi.org/10.5281/zenodo.11108981).
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
Portions of the paper were developed from the thesis of Anne-Sophie Godeux. This work was supported by the LABEX ECOFECT (ANR-11-LABX-0048) of Université de Lyon, within the program “Investissements d’Avenir” (ANR-11-IDEX-0007) operated by the French National Research Agency (ANR) and by the RESPOND program of the Université de Lyon. PhD funding of A.-S.G. was provided by VetAgro Sup. PhD funding of R.T. was provided the CNRS 80|Prime program. This work was also supported by the French ANR, under grant ANR-20-CE12-0004 (TransfoConflict), and by the Fondation pour la Recherche Médicale, grant number EQU202303016268 to X.C. This work was performed using the computing facilities of the CC LBBE/PRABI. We thank all three referees for their valuable and constructive comments.
Author contributions
R.T., G.C., A.-S.G., F.M., E.P.C.R., M.-H.L., S.V., and X.C. designed research; R.T., G.C., A.-S.G., and F.M. performed research; R.T., G.C., A.-S.G., F.M., E.P.C.R., M.-H.L., S.V., and X.C. analyzed data; and R.T., G.C., A.-S.G., F.M., E.P.C.R., M.-H.L., S.V., and X.C. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Contributor Information
Maria-Halima Laaberki, Email: maria-halima.laaberki@vetagro-sup.fr.
Samuel Venner, Email: samuel.venner@univ-lyon1.fr.
Xavier Charpentier, Email: xavier.charpentier@univ-lyon1.fr.
Data, Materials, and Software Availability
Program of the evolutionary model data have been deposited in Zenodo (https://doi.org/10.5281/zenodo.11108981). Previously published data were used for this work (10). All other data are included in the manuscript and/or SI Appendix.
Supporting Information
References
- 1.Wong D., et al. , Clinical and pathophysiological overview of acinetobacter infections: A century of challenges. Clin. Microbiol. Rev. 30, 409–447 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ayobami O., et al. , The incidence and prevalence of hospital-acquired (carbapenem-resistant) Acinetobacter baumannii in Europe, Eastern Mediterranean and Africa: A systematic review and meta-analysis. Emerg. Microbes Infect. 8, 1747–1759 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.WHO, WHO publishes the WHO medically important antimicrobials list for human medicine. https://www.who.int/news/item/08-02-2024-who-medically-important-antimicrobial-list-2024. Accessed 13 February 2024.
- 4.CDC, The biggest antibiotic-resistant threats in the U.S. (Centers for Disease Control and Prevention, 2022). https://www.cdc.gov/drugresistance/biggest-threats.html. Accessed 25 March 2022. [Google Scholar]
- 5.Snitkin E. S., et al. , Genome-wide recombination drives diversification of epidemic strains of Acinetobacter baumannii. Proc. Natl. Acad. Sci. U.S.A. 108, 13758–13763 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wright M. S., et al. , New insights into dissemination and variation of the health care-associated pathogen Acinetobacter baumannii from genomic analysis. mBio 5, e00963-13 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dougan G., et al. , Five decades of genome evolution in the globally distributed, extensively antibiotic-resistant Acinetobacter baumannii global clone 1. Microb. Genom. 2, e000052 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cruz-López F., et al. , Acquired genetic elements that contribute to antimicrobial resistance in frequent gram-negative causative agents of healthcare-associated infections. Am. J. Med. Sci. 360, 631–640 (2020), 10.1016/j.amjms.2020.06.028. [DOI] [PubMed] [Google Scholar]
- 9.Pagano M., Martins A. F., Barth A. L., Mobile genetic elements related to carbapenem resistance in Acinetobacter baumannii. Braz. J. Microbiol. 47, 785–792 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bi D., et al. , Large-scale identification of AbaR-type genomic islands in Acinetobacter baumannii reveals diverse insertion sites and clonal lineage-specific antimicrobial resistance gene profiles. Antimicrob. Agents Chemother. 63, e02526-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamidian M., Hall R. M., The AbaR antibiotic resistance islands found in Acinetobacter baumannii global clone 1—Structure, origin and evolution. Drug Resist. Updat. 41, 26–39 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Bi D., et al. , Comparative analysis of AbaR-type genomic islands reveals distinct patterns of genetic features in elements with different backbones. mSphere 5, e00349-20 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim D. H., et al. , AbaR4-type resistance island including the blaOXA-23 gene in Acinetobacter nosocomialis isolates. Antimicrob. Agents Chemother. 56, 4548–4549 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim D. H., Ko K. S., AbaR-type genomic islands in non-baumannii Acinetobacter species isolates from South Korea. Antimicrob. Agents Chemother. 59, 5824–5826 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Šeputienė V., Povilonis J., Sužiedėlienė E., Novel variants of AbaR resistance islands with a common backbone in Acinetobacter baumannii isolates of European Clone II. Antimicrob. Agents Chemother. 56, 1969–1973 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krizova L., Dijkshoorn L., Nemec A., Diversity and evolution of AbaR genomic resistance islands in Acinetobacter baumannii strains of European clone I. Antimicrob. Agents Chemother. 55, 3201–3206 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blackwell G. A., Nigro S. J., Hall R. M., Evolution of AbGRI2-0, the progenitor of the AbGRI2 resistance island in global clone 2 of Acinetobacter baumannii. Antimicrob. Agents Chemother. 60, 1421–1429 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoon E.-J., et al. , The blaOXA-23-associated transposons in the genome of Acinetobacter spp. represent an epidemiological situation of the species encountering carbapenems. J. Antimicrob. Chemother. 72, 2708–2714 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Mugnier P. D., Poirel L., Naas T., Nordmann P., Worldwide dissemination of the blaOXA-23 carbapenemase gene of Acinetobacter baumannii. Emerg. Infect. Dis. 16, 35–40 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hamidian M., Hawkey J., Wick R., Holt K. E., Hall R. M., Evolution of a clade of Acinetobacter baumannii global clone 1, lineage 1 via acquisition of carbapenem- and aminoglycoside-resistance genes and dispersion of ISAba1. Microb. Genom. 5, e000242 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hua X., et al. , Acquisition of a genomic resistance island (AbGRI5) from global clone 2 through homologous recombination in a clinical Acinetobacter baumannii isolate. J. Antimicrob. Chemother. 76, 65–69 (2021). [DOI] [PubMed] [Google Scholar]
- 22.Wilharm G., Piesker J., Laue M., Skiebe E., DNA uptake by the nosocomial pathogen Acinetobacter baumannii occurs during movement along wet surfaces. J. Bacteriol. 195, 4146–4153 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Godeux A.-S., et al. , Fluorescence-based detection of natural transformation in drug-resistant Acinetobacter baumannii. J. Bacteriol. 200, e00181-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu Y., He L., Tao X., Meng F., Zhang J., High DNA uptake capacity of international clone II Acinetobacter baumannii detected by a novel planktonic natural transformation assay. Front. Microbiol. 10, 2165 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnston C., Martin B., Fichant G., Polard P., Claverys J.-P., Bacterial transformation: Distribution, shared mechanisms and divergent control. Nat. Rev. Microbiol. 12, 181–196 (2014). [DOI] [PubMed] [Google Scholar]
- 26.Godeux A.-S., et al. , Interbacterial transfer of carbapenem resistance and large antibiotic resistance islands by natural transformation in pathogenic Acinetobacter. mBio 13, e02631-21 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blokesch M., In and out-contribution of natural transformation to the shuffling of large genomic regions. Curr. Opin. Microbiol. 38, 22–29 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Ambur O. H., Engelstädter J., Johnsen P. J., Miller E. L., Rozen D. E., Steady at the wheel: Conservative sex and the benefits of bacterial transformation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 371, 20150528 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Croucher N. J., et al. , Horizontal DNA transfer mechanisms of bacteria as weapons of intragenomic conflict. PLoS Biol. 14, e1002394 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takeuchi N., Kaneko K., Koonin E. V., Horizontal gene transfer can rescue prokaryotes from Muller’s ratchet: Benefit of DNA from dead cells and population subdivision. G3 (Bethesda) 4, 325–339 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moradigaravand D., Engelstädter J., The evolution of natural competence: Disentangling costs and benefits of sex in bacteria. Am. Nat. 182, E112–E126 (2013). [DOI] [PubMed] [Google Scholar]
- 32.Levin B. R., Cornejo O. E., The population and evolutionary dynamics of homologous gene recombination in bacteria. PLoS Genet. 5, e1000601 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mell J. C., Lee J. Y., Firme M., Sinha S., Redfield R. J., Extensive cotransformation of natural variation into chromosomes of naturally competent Haemophilus influenzae. G3 (Bethesda) 4, 717–731 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Croucher N. J., Harris S. R., Barquist L., Parkhill J., Bentley S. D., A high-resolution view of genome-wide pneumococcal transformation. PLoS Pathog. 8, e1002745 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bubendorfer S., et al. , Genome-wide analysis of chromosomal import patterns after natural transformation of Helicobacter pylori. Nat. Commun. 7, 11995 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alfsnes K., et al. , A genomic view of experimental intraspecies and interspecies transformation of a rifampicin-resistance allele into Neisseria meningitidis. Microb. Genom. 4, e000222 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Domingues S., et al. , Natural transformation facilitates transfer of transposons, integrons and gene cassettes between bacterial species. PLoS Pathog. 8, e1002837 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Udden S. M. N., et al. , Acquisition of classical CTX prophage from Vibrio cholerae O141 by El Tor strains aided by lytic phages and chitin-induced competence. Proc. Natl. Acad. Sci. U.S.A. 105, 11951–11956 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Al Suwayyid B. A., et al. , Meningococcal disease-associated prophage-like elements are present in Neisseria gonorrhoeae and some commensal Neisseria species. Genome Biol. Evol. 12, 3938–3950 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carvalho G., et al. , Bacterial transformation buffers environmental fluctuations through the reversible integration of mobile genetic elements. mBio 11, e02443-19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gaasbeek E. J., et al. , A DNase encoded by integrated element CJIE1 inhibits natural transformation of Campylobacter jejuni. J. Bacteriol. 191, 2296–2306 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gaasbeek E. J., et al. , Nucleases encoded by the integrated elements CJIE2 and CJIE4 inhibit natural transformation of Campylobacter jejuni. J. Bacteriol. 192, 936–941 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dalia A. B., Seed K. D., Calderwood S. B., Camilli A., A globally distributed mobile genetic element inhibits natural transformation of Vibrio cholerae. Proc. Natl. Acad. Sci. U.S.A. 112, 10485–10490 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Singh P. K., et al. , Inhibition of Bacillus subtilis natural competence by a native, conjugative plasmid-encoded comK repressor protein. Environ. Microbiol. 14, 2812–2825 (2012). [DOI] [PubMed] [Google Scholar]
- 45.Durieux I., et al. , Diverse conjugative elements silence natural transformation in Legionella species. Proc. Natl. Acad. Sci. U.S.A. 116, 18613–18618 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mashburn-Warren L., Goodman S. D., Federle M. J., Prehna G., The conserved mosaic prophage protein paratox inhibits the natural competence regulator ComR in Streptococcus. Sci. Rep. 8, 16535 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fléchard M., Lucchetti-Miganeh C., Hallet B., Hols P., Gilot P., Intensive targeting of regulatory competence genes by transposable elements in streptococci. Mol. Genet. Genomics 294, 531–548 (2018), 10.1007/s00438-018-1507-5. [DOI] [PubMed] [Google Scholar]
- 48.Nero T. M., et al. , ComM is a hexameric helicase that promotes branch migration during natural transformation in diverse Gram-negative species. Nucleic Acids Res. 46, 6099–6111 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mazzamurro F., et al. , Intragenomic conflicts with plasmids and chromosomal mobile genetic elements drive the evolution of natural transformation within species. bioRxiv [Preprint] (2024). 10.1101/2023.11.06.565790 (Accessed 10 May 2024). [DOI]
- 50.Baltrus D. A., Exploring the costs of horizontal gene transfer. Trends Ecol. Evol. 28, 489–495 (2013). [DOI] [PubMed] [Google Scholar]
- 51.Harding C. M., et al. , Acinetobacter baumannii strain M2 produces type IV Pili which play a role in natural transformation and twitching motility but not surface-associated motility. mBio 4, e00360-13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Adams M. D., Chan E. R., Molyneaux N. D., Bonomo R. A., Genomewide analysis of divergence of antibiotic resistance determinants in closely related isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother. 54, 3569–3577 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hamidian M., Hall R. M., AbaR4 replaces AbaR3 in a carbapenem-resistant Acinetobacter baumannii isolate belonging to global clone 1 from an Australian hospital. J. Antimicrob. Chemother. 66, 2484–2491 (2011). [DOI] [PubMed] [Google Scholar]
- 54.Fournier P.-E., et al. , Comparative genomics of multidrug resistance in Acinetobacter baumannii. PLoS Genet. 2, e7 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Apagyi K. J., Fraser C., Croucher N. J., Transformation asymmetry and the evolution of the bacterial accessory genome. Mol. Biol. Evol. 35, 575–581 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Correa A. III, et al. , Novel mechanisms of diversity generation in Acinetobacter baumannii resistance islands driven by Tn7-like elements. Nucleic Acids Res. 52, 3180–3198 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wilharm G., et al. , On the ecology of Acinetobacter baumannii–Jet stream rider and opportunist by nature. bioRxiv [Preprint] (2024). 10.1101/2024.01.15.572815 (Accessed 10 May 2024). [DOI]
- 58.Redfield R. J., et al. , Evolution of competence and DNA uptake specificity in the Pasteurellaceae. BMC Evol. Biol. 6, 82 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jorth P., Whiteley M., An evolutionary link between natural transformation and CRISPR adaptive immunity. mBio 3, e00309-12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ishikawa S. A., Zhukova A., Iwasaki W., Gascuel O., A fast likelihood method to reconstruct and visualize ancestral scenarios. Mol. Biol. Evol. 36, 2069–2085 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Knöppel A., Lind P. A., Lustig U., Näsvall J., Andersson D. I., Minor fitness costs in an experimental model of horizontal gene transfer in bacteria. Mol. Biol. Evol. 31, 1220–1227 (2014). [DOI] [PubMed] [Google Scholar]
- 62.Dykhuizen D. E., Experimental studies of natural selection in bacteria. Annu. Rev. Ecol. Syst. 21, 373–398 (1990). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
Program of the evolutionary model data have been deposited in Zenodo (https://doi.org/10.5281/zenodo.11108981). Previously published data were used for this work (10). All other data are included in the manuscript and/or SI Appendix.