Abstract
DNA-stabilized silver nanoclusters (AgN-DNAs) have sequence-tuned compositions and fluorescence colors. High-throughput experiments together with supervised machine learning models have recently enabled design of DNA templates that select for AgN-DNA properties, including near-infrared (NIR) emission that holds promise for deep tissue bioimaging. However, these existing models do not enable simultaneous selection of multiple AgN-DNA properties, and require significant expert input for feature engineering and class definitions. This work presents a model for multiobjective, continuous-property design of AgN-DNAs with automatic feature extraction, based on variational autoencoders (VAEs). This model is generative, i.e., it learns both the forward mapping from DNA sequence to AgN-DNA properties and the inverse mapping from properties to sequence, and is trained on an experimental data set of DNA sequences paired with AgN-DNA fluorescence properties. Experimental testing shows that the model enables effective design of AgN-DNA emission, including bright NIR AgN-DNAs with 4-fold greater abundance compared to training data. In addition, Shapley analysis is employed to discern learned nucleobase patterns that correspond to fluorescence color and brightness. This generative model can be adapted for a range of biomolecular systems with sequence-dependent properties, enabling precise design of emerging biomolecular nanomaterials.
Keywords: variational autoencoder, silver nanocluster, DNA, interpretable machine learning, fluorescence, near-infrared
Metal nanoclusters are promising emitters for a range of photonic applications.1 DNA-stabilized silver nanoclusters (AgN-DNAs) are fluorophores with sequence-defined properties and significant promise for applications in bioimaging and biosensing. These emitters range in size from about 10 to 30 silver atoms and are stabilized by single-stranded DNA oligomers.2 AgN-DNAs exhibit favorable fluorescence properties, including high quantum yields,3,4 large Stokes shifts,5,6 and diverse fluorescence colors ranging from blue/green to near-infrared (NIR).2 Because of the sequence-dependent interactions between DNA and silver,7 the specific size and shape of an AgN-DNA depends strongly on the sequence of its templating DNA oligomer.8 This results in the templating DNA sequence influencing AgN-DNA properties such as absorbance and fluorescence spectra, quantum yield, extinction coefficient, and chemical stability (Figure 1a).9−11 Recent efforts have focused on developing AgN-DNAs with bright NIR emission in the tissue transparency window (700–1400 nm) and with sufficient stability for in vivo deep tissue imaging.12−14 Several studies have reported AgN-DNAs with low toxicities to mammalian cells,15,16 making these tunable fluorophores attractive for applications in bioimaging and biosensing.
Figure 1.
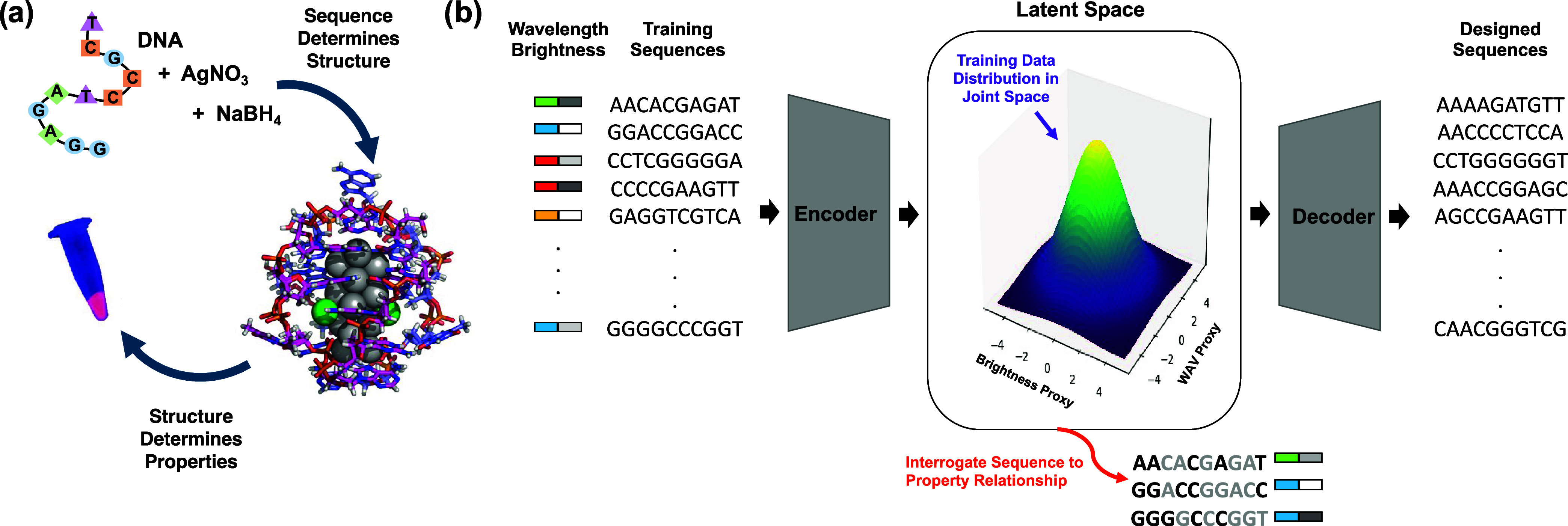
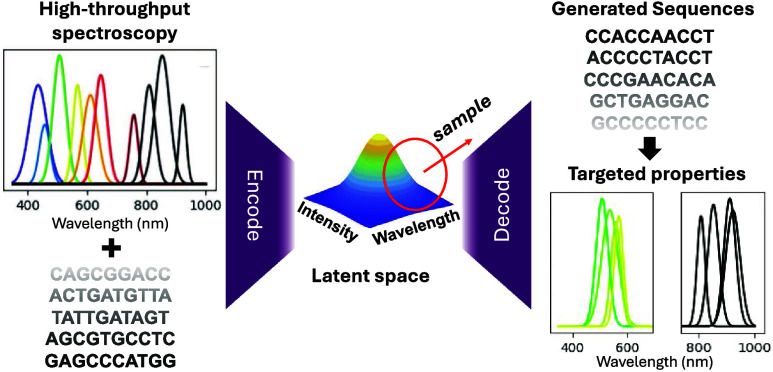
(a) Schematic of relationship between DNA sequence, AgN-DNA structure (PDB accession code 6M2P, adapted with permission from ref (10)., copyright 2023 American Chemical Society), and resulting emission properties. (b) Schematic of generative VAE model for multiobjective design of AgN-DNAs. The VAE is trained on input DNA sequences paired with corresponding nanocluster properties: peak emission wavelength and brightness. Latent space is normally distributed and serves two key purposes: (i) design of new sequences with desired properties by sampling proxy property dimensions for desired ranges of peak wavelength (WAV Proxy) and brightness (LII Proxy) and (ii) interrogation by Shapley value analysis of important DNA features automatically extracted by the VAE.
However, the DNA sequence space is expansive, with 4L possible L-base sequences, making the design of DNA templates for AgN-DNAs a major challenge and hindering the development of suitable fluorophores. Moreover, the sequence-to-color rules that govern AgN-DNAs are highly complex. Together, these factors have slowed progress to develop AgN-DNA fluorophores that are tailored for specific applications. Such a design challenge is general to sequence-encoded biomolecules and materials derived from nucleic acids, peptides, and proteins.17−20
Supervised machine learning (ML) has shown success as a design tool for AgN-DNAs with targeted fluorescence properties. Simple ML classifiers and classifier ensembles have been trained using experimental data libraries of about 700 to 2700 DNA sequences and their corresponding AgN-DNA emission peak wavelength and brightness.21−23 AgN-DNA-based sensors called NanoCluster Beacons have also been designed by this approach.24 Most recently, a chemistry-informed ML model composed of an ensemble of support vector machine (SVM) classifiers was used to design DNA templates that stabilize AgN-DNAs with NIR peak emission >800 nm, achieving 12-fold greater likelihood of selecting DNA sequences for AgN-DNAs in this color window.25 This model’s success relied on the use of chemistry-motivated features inspired by the first X-ray crystal structures of AgN-DNAs.26,27 The model’s simplicity and chemically relevant features also enabled the use of feature analysis tools to interpret the sequence-to-color rules learned by the model. A brief review of ML-guided design of AgN-DNAs, as well as an overview of ML concepts for biomolecular materials, can be found in recent work.28
Despite the success of past ML-guided discovery of AgN-DNAs, existing approaches have several major limitations. First, the models only map sequence onto a single AgN-DNA property, e.g., emission color, and are not well-suited for simultaneous control over multiple important properties such as emission color, brightness, chemical stability, sensitivity to analytes, etc. This limits their utility for designing AgN-DNAs that are well-tailored for imaging and sensing applications, which require such control over multiple properties. Second, the necessity of chemistry-informed features (as in ref (25)) is a major limitation when there is insufficient known information about the fundamental properties of a materials system. At present, very few AgN-DNAs have solved crystal structures,26,27,29,30 and first-principles computational methods to predict AgN-DNA structures are still in development.10,31,32
The lack of fundamental knowledge is a general challenge for developing ML-guided design methods for emerging materials systems. In such cases, structure–property relationships can be too poorly understood to inform ML feature engineering, and first-principles computational models may still be in development.33−35 Thus, progress in materials discovery would be substantially accelerated by the development of artificial intelligence models that enable multiobjective design and automatically perform feature extraction.
Here, we present a variational autoencoder (VAE) model for AgN-DNA design that (1) learns to map DNA sequence directly onto multiple AgN-DNA properties without requiring a class-based approach, (2) automatically extracts features during the learning process, (3) can be used as a generative model for multiobjective design of AgN-DNAs, and (4) can be interpreted to gain insights into how DNA sequence selects AgN-DNA properties. We train the VAE to encode both emission peak wavelength (“color”) and brightness as independent latent dimensions. The model is then employed to generate new template sequences for brightly emissive AgN-DNAs in the green and NIR spectral regions, which are experimentally validated to test the efficacy of the model (Figure 1b). Finally, Shapley analysis is implemented to interpret nucleobase-to-property patterns, providing insights into how DNA sequence selects AgN-DNA properties. This work presents a versatile workflow for developing ML techniques that could be broadly applied to sequence-based biomaterials. By avoiding the need for user-performed featurization and instead directly using biomolecular sequence as input, and by enabling design for multiple properties, models of this type can be applied to a wide variety of design problems, particularly those where there is little fundamental knowledge about detailed materials properties.
Results and Discussion
Training Data Preparation and Statistics
This study focuses on AgN-DNAs stabilized by 10-base DNA sequences, which are by far the most widely studied and can produce AgN-DNA products with a diversity of spectral properties and peak emission ranging from ca. 400 to 1200 nm.6,8,21−23,25,36 (Past work showed that sequence-to-color rules for AgN-DNAs are general across a range of DNA oligomers, and that ML models trained on 10-base sequences can also design AgN-DNA templates of other lengths23). To date, 3003 10-base sequences and the properties of their associated AgN-DNA products have been reported.25 However, only 105 of these sequences have AgN-DNA spectral peaks reported for peak wavelengths greater than 800 nm.25,36 While recent ML-guided design showed success for predicting AgN-DNAs using incomplete training information in the NIR spectral range,25 this incomplete spectral information for 72% of the training sequences is likely to limit the effectiveness of ML models for designing NIR AgN-DNAs.
Therefore, we experimentally synthesized AgN-DNAs for all of the prior studied 3003 DNA sequences and gathered complete emission spectral information from 400 to 1400 nm. High-throughput AgN-DNA synthesis was performed using uniform conditions (see Methods section). Rapid parallel fluorimetry was performed under universal ultraviolet (UV) excitation37 on a commercial multimode fluorimeter sensitive up to ca. 800 nm and a custom plate reader sensitive to 700–1400 nm emission.38 Data curation takes into account the different spectral responsivities of the two fluorimeters. Details of automated synthesis, fluorimetry, and spectral data analysis procedures are provided in past works,8,22,25Methods section, and Supporting Information.
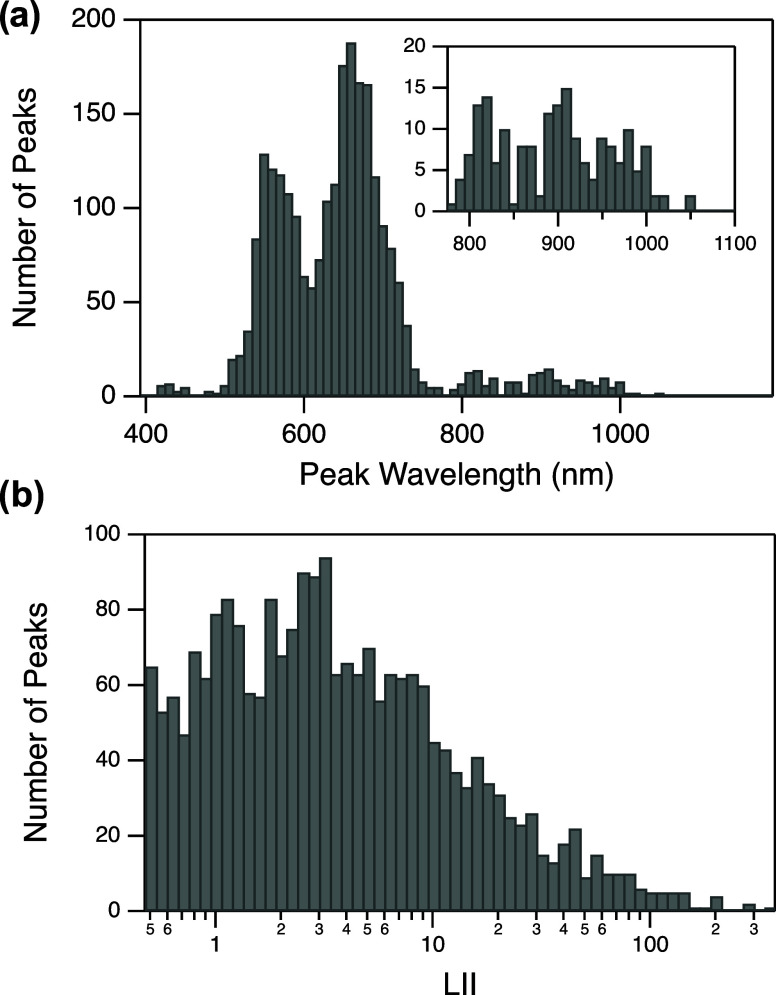
Figure 2 shows the distributions of peak wavelength and peak brightness for all AgN-DNA products stabilized by the 3003 DNA templates. Peak brightness is measured by normalized local integrated intensity (LII) of the Gaussian fit, i.e., the area under the curve of the Gaussian peak.22 (Peaks with LII values <0.5 were excluded from Figure 2 and training data due to consideration of the commercial plate reader’s signal-to-noise ratio. Sequences whose associated spectra exhibit >3 spectral peaks were also omitted from the training data. See details in Methods section and Supporting Information.) The multimodal wavelength distribution in Figure 2a is expected due to the so-called “magic number” properties of AgN-DNAs.6,8 This complete experimental screening of all 3003 10-base sequences identified 157 emissive AgN-DNA sequences with λp > 800 nm (Figure 2a, inset), which is more than double the number of training instances in this long-wavelength range in the previous available incomplete data library.
Figure 2.
Distributions of (a) peak wavelength λp (inset: high-wavelength range) and (b) normalized local integrated intensity (LII) for all AgN-DNA products produced by 10-base DNA sequences in the training data set.
Finally, each DNA sequence is labeled by the peak wavelength, λp, and LII of the brightest spectral peak in its associated emission spectrum. The relative frequency of DNA sequences that yield a single emissive AgN-DNA versus sequences that can stabilize multiple distinct AgN-DNA products with distinct sizes and emission peaks has been reported previously.8,22 While prior ML-guided studies have avoided “multicolored” DNA sequences because they likely contain nucleobase patterns shared by multiple sizes of AgN-DNAs, which could challenge effective learning, here we do not exclude these sequences from the training data library. This provides a greater learning challenge, in addition to the challenge of multiobjective design for λp and LII.
Training and Tuning the VAE
Next, we trained a deep learning model to map DNA sequence onto λp and LII. Specifically, we employed the property-regularized VAE introduced by Moomtaheen et al.39 As shown schematically in Figures 1b and S1, the VAE encodes DNA sequences into a low-dimensional latent space. Constraints were placed on two dimensions of the latent space during training to ensure that the model learns to order sequences in latent space similar to their corresponding values of λp and LII, i.e., the two latent space dimensions are property-regularized. These two latent dimensions are referred to as “wavelength (WAV) proxy” and “LII proxy” respectively. Simultaneously, the model also learns to reverse this latent space encoding by mapping latent space locations back to the original sequences. A detailed description of the model can be found in Supporting Information Section 1.1.
Intuitively, the latent space serves as a map, encoding both properties and sequences, with regions in latent space corresponding to DNA sequences that produce AgN-DNAs with similar spectral properties.40−43 The encoder and decoder of the VAE allow both forward and backward transformations. In the forward direction, the model maps DNA sequences onto a continuous distribution of proxy properties. The backward transformation can be used to sample points from a latent space region and decode these points into sequences, thus generating new DNA templates whose properties are likely to be similar to those in the sampled region. As a generative ML model, the VAE enables direct design of AgN-DNAs by eliminating the need to screen prospective candidate sequences through a discriminative ML model, which can be computationally costly and time-consuming.
The VAE is trained using a standard iterative process, batch gradient descent. Batches of 32 sequences are provided to the model, which learns to encode and decode sequences while also ordering sequences in WAV proxy and LII proxy latent space dimensions according to the properties of their corresponding sequence.39 Training a VAE also requires selection of various hyperparameters that control how the model learns. This step is essential to avoid pitfalls such as overfitting, which can diminish a model’s utility for making predictions about data that is not included in the training data set. To configure the VAE model’s hyperparameters, we employed a training/validation split of 90:10% of the training data, respectively. Hyper-parameters were selected based on the optimal ordering of the latent dimensions for WAV proxy and LII proxy for the validation data set, as well as accurate reconstruction of the original sequences by the decoder (details provided in Supporting Information Section 1.2, including accuracy and breakdown of loss plots in Figure S2a,b). To assess the ordering of WAV proxy and LII proxy values during the course of training, we monitored average WAV proxy and LII proxy values for sequences in four λp ranges and four LII ranges during training. (Recall that these proxy values correspond to the two regularized dimensions of the latent space vector, which is calculated by passing the sequence through the encoder.) The four λp ranges were defined based on known chemical differences among AgN-DNAs, specifically, magic numbers that correlate λp to nanocluster composition and shape8 and are responsible for the multimodal distribution in Figure 2a. These ranges were defined as follows: Green, λp > 590 nm; Red, 590 nm < λp < 660 nm; Far Red, 660 nm < λp < 680 nm; NIR, λp > 800 nm. (Note: here, we use “NIR” to refer to λp > 800 nm, as consistent with past work25). Details for LII proxy are provided in the Supporting Information Figure S2.
It is important to note that the VAE is not a class-based model. Training sequences are labeled by the scalar values of λp and LII, not by categories of these parameters, such as the Green, Red, Far Red, and NIR ranges defined above. The VAE is not provided any information about λp and LII ranges; rather, we simply used these ranges to monitor how well the VAE learns to map sequences with similar structure–property relationships into similar regions of latent space, thereby selecting hyperparameters and the number of training epochs accordingly. This level of care is important for training ML models for materials design to ensure that models are appropriately configured for meaningful prediction.
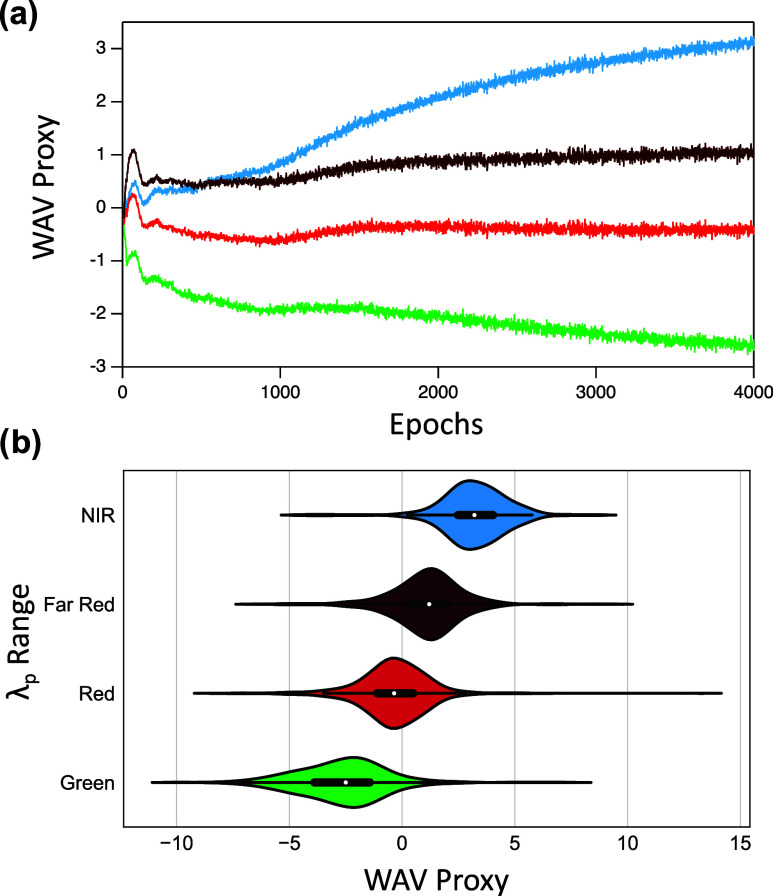
Figure 3 shows that the selected hyperparameters ensure that mean WAV proxy of the four λp ranges are ordered such that Green < Red < Far Red < NIR. Figure S2f shows the corresponding correct ordering of LII proxy. Green, Red, and Far Red mean WAV proxy are correctly ordered after very few epochs (Figure 3a), as are LII proxy values (Figure S2f) However, nearly 3000 epochs were required for the VAE to learn the correct ordering of NIR mean WAV proxy. Moreover, the distributions of WAV proxy values of Far Red and NIR ranges for the trained VAE show a greater degree of overlap than between other consecutive wavelength ranges (Figure 3b). Such increased overlap of NIR and Far Red WAV proxy values may result from the limited number of NIR training sequences, as well as the increased complexity of AgN-DNAs that emit above about 700 nm. Recent studies have shown that the structure–property relationships of longer-wavelength AgN-DNAs are much more diverse than visibly emissive AgN-DNAs. While a clear distinction exists between the valence electron counts of green-emissive AgN-DNAs (with 4 valence electrons) and red-emissive AgN-DNAs (with 6 valence electrons), AgN-DNAs with λp < 700 nm have been reported with varying valence electron counts of 6, 8, 10, and 12.36,44 AgN-DNAs with λp < 700 nm can also exhibit either rod-like or spheroidal nanocluster core geometries, as well as nanosecond-lived fluorescence or microsecond-lived luminescence.2,10,45 Finally, dual-emissive AgN-DNAs have recently been reported, exhibiting both fluorescence at green or red wavelengths and also microsecond-lived emission at longer NIR wavelengths.31,46−48 These distinct nanocluster properties—composition, shape, and emission lifetime—which cannot be distinguished solely by high-throughput fluorimetry, may be correlated with distinct sequence-to-property mappings. Together, these factors are expected to significantly challenge the task of learning to accurately map DNA sequence onto NIR emission properties.
Figure 3.
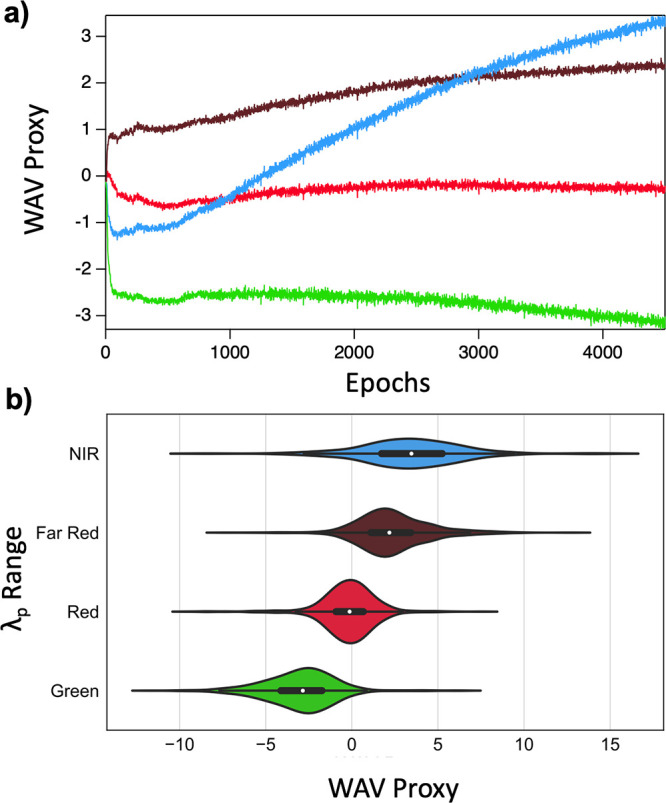
(a) Average wavelength proxy (WAV proxy) for Green (green), Red (red), Far Red (dark red) and NIR (blue) λp ranges as a function of training epoch. (b) Violin plots of WAV proxy values for λp ranges. White points indicate mean WAVE proxy for each λp range.
Experimental Validation of the VAE
We experimentally validated the VAE model’s utility for AgN-DNA design by generating sequences for brightly fluorescent AgN-DNAs in two different color ranges: λp > 590 nm (Green), and λp > 800 nm (NIR). These two spectral windows were selected because they present the greatest design challenges due to having the least amount of training data and because of known complexity of NIR AgN-DNAs, and the historic difficulty in designing brightly emissive green clusters.28 Following hyperparameter tuning, the VAE was trained using all training DNA sequences without a separate validation set, employing 4500 training epochs. Then, we sampled the latent space of the VAE to generate new template sequences that select for AgN-DNA brightness and λp, using an efficient method called truncated normal sampling to target latent subspace regions of desired property values.39
New NIR sequences were generated by sampling the region of latent space corresponding to the upper range of WAV proxy values (associated with high λp values, see Figure 3b) and the upper range of LII proxy values. Specifically, we employ truncated sampling49 where the target sampled WAV proxy values exceed the mean of the NIR range (WAV proxy >3.39) and target LII proxy values exceeding the NIR range mean (LII proxy >0.56). Similarly, Green sequences were generated by sampling the latent space in the region corresponding to the lower range of WAV proxy values (associated with low λp, see Figure 3b) and the upper range of the LII proxy. The truncation bounds were selected based on the Green range, sampling sequences below the mean WAV proxy (WAV proxy < −3.01) and above the mean Green range LII proxy (LII proxy > −1.17).
Next, sampled latent space points were converted to sequences. First, latent space points were passed through the VAE decoder. This step does not itself produce a sequence, with discrete encoding values corresponding to each nucleobase. Rather, decoding produces continuous weights for position-dependent nucleobase. A one-hot encoding approximation was then obtained by selecting the nucleobase with highest decoded weight in each sequence position. Because this process introduces error, candidate sequences were re-encoded to verify that WAV proxy and LII proxy values of candidates fall in the correct range of proxy distributions (details of the re-encoding process available in ref (39)). Figure S6 shows that the distributions of sampled sequences in latent space are shifted toward the preferred WAV proxy regions. Sequences with re-encoded WAV proxy in the target threshold range were retained in the candidate test set for experimental AgN-DNA synthesis.
Finally, we ranked the generated Green and NIR sequences in the candidate test set and selected the top 100 Green, and the top 200 NIR generated sequences for experimental validation. NIR candidate sequences with LII proxy values that were greater than the mean LII proxy of NIR training sequences were then ranked in descending order by re-encoded WAV proxy, with a higher proxy value corresponding to higher ranking. Similarly, Green candidate sequences with LII proxy values that were greater than the mean LII proxy of Green training sequences were ranked in ascending order by re-encoded WAV proxy, with a lower proxy value corresponding to higher ranking. This ranking ensures that we test sequences likely to yield both high LII values and λp values in the target wavelength range.
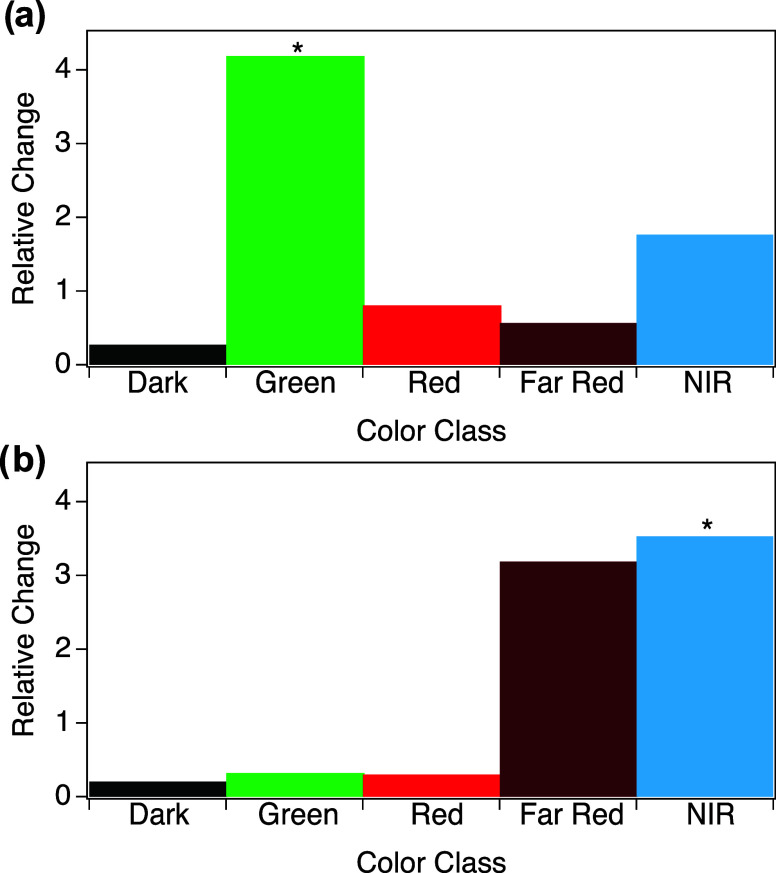
Experimental AgN-DNA synthesis and spectroscopic characterization was performed for the 300 generated sequences using the same experimental and spectral fitting procedures used to generate training data. Figure 4a shows that the VAE can effectively generate Green sequences, increasing the relative abundance of Green products by 4.2 times. Similarly, VAE-guided design increased the abundance of NIR peaks by 3.5 times (Figure 4b), which was the greatest relative change in size as compared to other λp ranges. However, NIR-generated sequences also produced a large number of Far Red products (Figure 4b), with nearly equal relative change in the numbers of Far Red and NIR sequences. This lack of specificity may result from the large degree of overlap of WAV proxy values between the NIR and Far Red ranges (Figure 3b), as well as the relatively few NIR sequences for training and the aforedescribed complexity of Far Red to NIR emitting AgN-DNAs.
Figure 4.
Relative change in the number of sequences in each λp range with high LII values >1 and the number of sequences with “Dark” LII < 0.5, for sequences generated to be (a) Green and (b) NIR.
Improving Performance by Stratification
We hypothesized that design of NIR AgN-DNAs can be further improved by developing a model that better separates the distributions of WAV proxy values for Far Red and NIR sequences. Therefore, we modified the VAE to address the challenge of the significant imbalance in observed λp values in the training data. For each training epoch, random selection of 32 sequences from the training library, whose λp distribution is shown in Figure 2a, is unlikely to contain NIR sequences in most batches. This will limit the VAE’s ability to learn sequence-to-NIR mapping because it is possible that each randomly sampled batch of 32 sequences is not sufficiently diverse in properties. This issue can be addressed by extra care to ensure that batches are stratified, i.e., include sequences stabilizing AgN-DNAs across the entire λp spectrum. Past work on classification demonstrated that batch stratification (ensuring that each batch contains classes in similar proportion to their frequency in the whole training set) are advantageous for model accuracy.50 While our model concerns continuous-valued properties and not classification, we employ a similar stratification approach to ensure the diversity of DNA sequences in training batches. Specifically, our method organizes batches to include a broad spectrum of sequences characterized by varying λp and LII values. To this end, we discretize the continuous property values into bins by employing quantile-based variable discretization and then create batches that match the range distribution of the overall data set (Figures S3 and4). Detailed description is provided in Supporting Information Section 1.3.
Next, we again monitored WAV proxy and LII proxy during training for the previously defined parameter ranges to determine whether stratification enables the VAE to better distinguish Far Red and NIR sequences (Figure S5). Figure 5a shows average WAV proxy for the Green, Red, Far Red, and NIR λp ranges for the stratified model. Compared to the unstratified VAE (Figure 3a), the stratified VAE learns to appropriately order sequences in the NIR range in latent space in far fewer epochs; the average NIR proxy exceeds average Far Red proxy in less than 1000 epochs. Figure 5b illustrates that the stratified model also exhibits significantly less overlap between the distributions of WAV proxy for Far Red and NIR ranges.
Figure 5.
Stratified VAE latent space ordering of wavelength proxy. (a) Average WAV proxy for the Green (green), Red (red), Far Red (dark red), and NIR (blue) spectral windows as a function of training epochs for stratified batch composition. (b) Distributions of WAV proxy values for λp ranges. White points indicate mean WAV proxy for each λp range.
We experimentally tested the efficacy of the stratified VAE model for generating NIR and Green sequences using the same approach as for the unstratified model. Using the previously described sampling strategy, we again generated 100 Green and 200 NIR candidate sequences for experimental validation. For both wavelength ranges, VAE-guided design increased the abundance of AgN-DNAs within the target spectral window. Unlike in the unstratified model, the increase for both Green and NIR using the stratified VAE was markedly greatest for the intended spectral range as compared to other λp ranges (Figure 6a,c). The relative abundance of Green sequences increased by 3.7 times (Figure 6a), and 58% of all sequences exhibited the desired λp > 590 nm (Figure 6c). Furthermore, the stratified model was especially successful at multiobjective design of “bright” Green sequences; the majority of designed Green sequences yielded LII values above the previously defined threshold of LII > 1.0, and average LII values of designed Green products shifted higher than the training data (Figure 6c). Past work reported that design of green-emissive AgN-DNA with high brightness was especially challenging due to sequence motifs shared by Green AgN-DNAs and “dark” DNA sequences that do not stabilize emissive products.22 The stratified VAE model overcomes this challenge, designing the brightest Green-emissive AgN-DNA identified to date.
Figure 6.
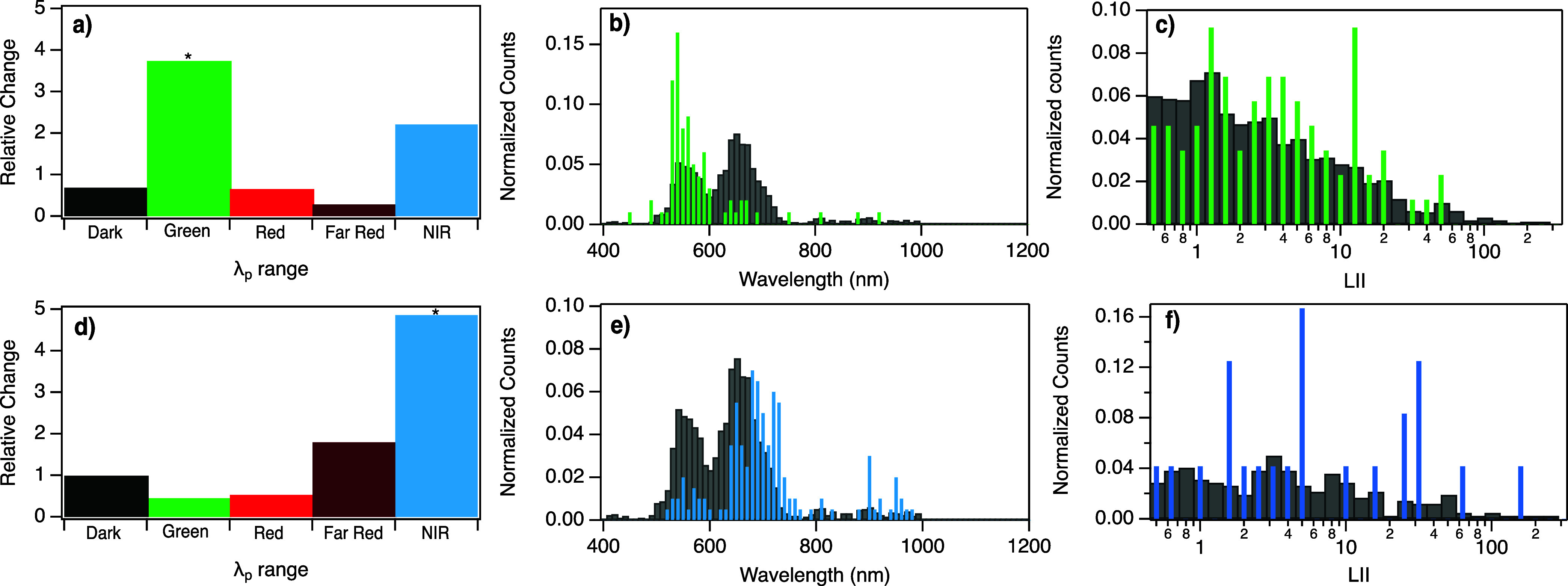
Relative change in the number of sequences in each λp range with high LII value >1 and the number of sequences with “Dark” LII < 0.5, for sequences generated using the stratified VAE to be (a) Green and (d) NIR. All fluorescent peaks with LII > 1 for (b) Green designed template sequences (green bars) and (e) NIR designed template sequences (blue bars), as compared to training data (gray bars). LII values for (c) all Green products, compared to Green products in the training data set, and (f) all NIR products, compared to NIR products in training data.
As hypothesized, the stratified VAE was notably more successful at design of NIR AgN-DNAs than the unstratified model, increasing the relative abundance of NIR AgN-DNAs by 4.9 times (Figure 6b,d). Moreover, 10 of these had λp > 900 nm. Most significantly, the LII values of NIR AgN-DNAs designed using the stratified VAE was shifted significantly higher as compared to the training data (Figure 6f). This finding shows that even with limited training data, generative models can be designed and configured to be effective tools for AgN-DNA design.
Model Interpretation
The complexity of VAE models makes it nontrivial to interpret what the model has learned about the mapping of DNA sequence onto AgN-DNA properties. In general, the lack of interpretability of deep learning models for chemical and materials design can limit their wider adoption because it is scientifically unsettling to use black box algorithms.51 To address this challenge, and to advance fundamental understanding of the sequence-structure–property relationships of AgN-DNAs, we adapted SHAP (SHapley Additive exPlanations) analysis52 to interpret the VAE’s model predictions.
Shapley value analysis is an approach from game theory, where the goal is to attribute team success to individual subteam contributions. In this study, the “team” is the complete DNA sequence, “team members” are nucleobases in specific positions on the template, and “subteams” are subsequences of nucleobases, where a subsequence is a subset of positions of the entire 10-nucleobase sequence. Success is quantified by the VAE’s ability to order templates by WAV proxy and LII proxy in latent space. We seek to quantify which DNA subsequences contribute the most to the correct AgN-DNA property mapping by the VAE. Subsequences are scored using Shapley values based on their contributions to this goal. Intuitively, if the nucleobases in a subsequence with a high importance score are altered, the VAE’s prediction of the AgN-DNA properties will be significantly changed. Details of Shapley value formulation are provided in Supporting Information Section 2.
Table 1 illustrates the top 10 scored subsequences identified by this method for four example sequences, two for the property λp > 590 nm and two for the property λp > 800 nm. Subsequences are ordered in Table 1 by importance score, which is a measure of the subsequence’s influence on the VAE’s mapping of sequences to the property of interest. Note that subsequences are scored by importance for each sequence. The important subsequences listed in Table 1 are thus specific to the four example sequences listed at the top of the table and do not necessarily represent the average effects of such subsequences on λp. Moreover, it is important to note that in this study, subsequences are position-dependent and therefore contain information about both nucleobase type(s) and nucleobase position(s) within the 10-base sequence. This information is distinct from the positionally invariant motifs reported by prior AgN-DNA studies.22,25
Table 1. Top 10 Sub-sequences Derived from the Primary Sequences at the Top of the Table and Ranked by Shapley Scores that Reflect Proximity to the Mean of the λp Rangea.
| λp < 590 nm |
λp < 800 nm |
|||
|---|---|---|---|---|
| index | AAAATCCCTA | AGAGTCCAAC | GGGGACCTAA | CGAGAACTCA |
| 1 | A – – – – C – – – − | A – – – – – – – – – | – G – – – – C – – – | – G – – – – – – – – |
| 2 | – – – – – – – – T – | – – A – – – – – – – | – G – – – – C – – A | – G – – – – – – – A |
| 3 | –A – – – − C – – − | A – A – – – – – – – | – G – – – – – – – A | – G – – – – C – – – |
| 4 | A – – – – C – – – A | A – – – – C – – – − | G G – – – – C – – – | – G – – – – – – C – |
| 5 | A – – – – C C – – A | A – A – – – – – – C | G – – – – – C – – A | – – – – – – – – – A |
| 6 | – – A – T – – – – − | A – – – – – C – – – | – G – – – C – – – A | – G – – – – C – C – |
| 7 | –A– −T C – – – – | – – A – – C– – – – | G – – – – – – – A | – G – – – – C – – A |
| 8 | A – – – – C C – – – | – – A – – – C – – – | – G – – – C C – – A | – G – – – – – – C A |
| 9 | – – A – T – C – – − | A – A – T – – – – − | G – – – – C – – – A | – – – – – – C – – – |
| 10 | – A – – – − C C T – | – – A – T – – – – − | G G – – – C C – – – | – G – G – – – – – A |
Nucleobase positions that are not included in the sub-sequence are marked with “–”. This does not imply that any nucleobase can occupy “–” positions; rather, it indicates that the nucleobases at “–” positions are less critical for the model’s predictions.
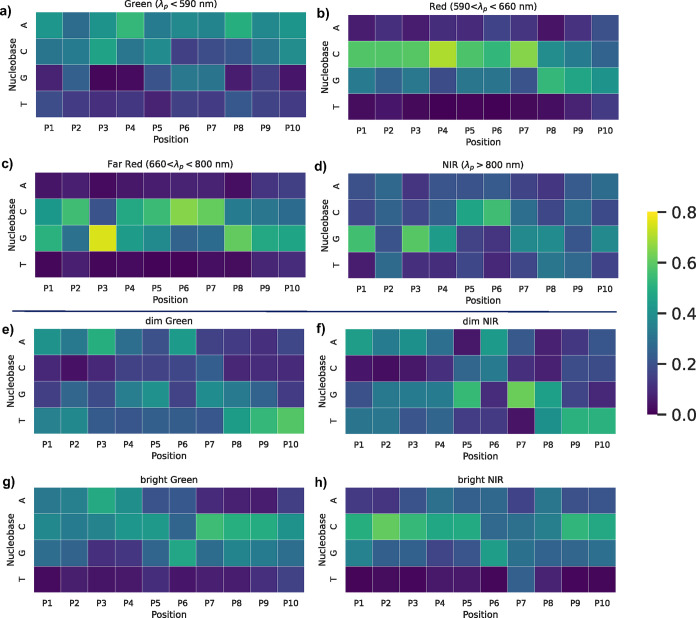
We next analyze the highest scored subsequences from all DNA sequences in the training data to understand general trends about nucleobase patterns that the VAE has learned to be important for mapping sequence onto properties. Figure 7 summarizes the highest scored nucleobase patterns for selecting λp, for each λp range. We find that Green sequences with λp > 590 nm dominantly feature adenines, with cytosines playing secondary role (Figure 7a). Adenines have high scores at nearly every sequence position, with especially high scores for positions 4 (P4) and 8 (P8). Cytosine is the most prominent nucleobase in positions 3 (P3) and 5 (P5), and has the second-highest score in P10. Guanines also have moderately high scores in P6 and P7. These findings agree with past reports of the importance of adenines in particular for Green AgN-DNAs,22,25 and this analysis provides more information about where these adenines prominently feature within Green-selecting DNA sequences.
Figure 7.
Top 20 subsequences derived from primary sequences, ranked by Shapley scores that reflect proximity to the mean of the λp range for (a) Green, (b) Red, (c) Far Red, and (d) NIR sequences. Heat map illustrating the top 20 subsequences from Shapley value analysis for (e) dim Green, (f) dim NIR, (g) bright Green, and (h) bright NIR sequences. “Dim” was defined as sequences with the bottom 30% of LII values; “bright” was defined as the top 30% of LII values. Color displays the probability of A, C, G, T at each position. All probabilities are shown as line graphs in Figure S7.
Figure 7b,c show that cytosine- and guanine-abundant subsequences are both important for Red (590 nm < λp < 660 nm) and Far Red (660 nm < λp < 800 nm). However, distinctions exist in the relative positions of these cytosines and guanines between Red and Far Red. Red sequences favor long uninterrupted runs of cytosines from P1 through P7, guanine scores becoming dominant at the 3′ end. Compared to Red, Far Red sequences have relatively similar abundance of cytosines and prevalence of guanines in P8 through P10, but Far Red much more strongly favors guanine in P1 and P3 as compared to Red sequences, with moderate guanine scores in P4 and P5, as well.
Finally, NIR sequences (λp > 800 nm) predominantly feature guanine-rich subsequences in P1 and P3 at the 5′-end and lesser guanine content at the 3′ end as compared to Red and Far Red (Figure 7d). Cytosines in P5 and P6 are also important for NIR sequences. Comparison of the patterns for Far Red and NIR shows some marked similarities, which likely contributes to the unintended selection of Far Red sequences during NIR sequence sampling.
We also performed Shapley value analysis on Green and NIR sequences with high and low values of LII to evaluate the impact of nucleobase patterns on emission peak brightness. For each λp range, sequences with the top and bottom 30% of LII values from the training data were defined as “bright” and “dim,” respectively. We then analyzed their top 20 subsequences. Figure 7e,g show the nucleobase content for subsequences for bright and dim Green sequences. While adenine positions are similar for bright and dim Green sequences, bright Green sequences show a strong preference for cytosines in P7 through P9, while dim Green prominently feature thymines at the 3′ terminus. Subtler differences in guanine content are also apparent between dim and bright Green.
Shapley value analysis for bright and dim NIR sequences yields especially interesting insights. Bright NIR sequences (Figure 7h) exhibit strong preference for cytosines in P1 through P5 and P9, P10, with moderate preference for guanines throughout the sequence. These patterns share commonalities with Red and Far Red sequences (Figure 7c), and such commonalities could contribute to the challenge of discriminating sequences among these wavelength ranges. In contrast, dim NIR sequences (Figure 7f) have significantly diminished preference for cytosines and an increased preference for adenines, as well as specific preferences for thymines at the 5′ end and guanines at central sequence positions. These patterns of nucleobase importance are similar to dim Green sequences in Figure 7e. Interestingly, sequences generated by both the unstratified and stratified VAEs for Green λp values sometimes yielded unintended NIR products (Figures 4a and 6a). Because of recent reports of dual-emissive AgN-DNAs that exhibit both green fluorescence and microsecond-lived far red to NIR emission,31,46−48 we hypothesize that there exist two “classes” of NIR AgN-DNAs, one whose NIR emission results from a primary fluorescence process and one whose NIR emission results from the less efficient and therefore dimmer microsecond-lived process. Very recently reported experimental methods may be able to separate these two classes of emitters.53
Mass spectrometry has shown that AgN-DNAs stabilized by 10-base oligomers contain two to three oligomer copies per AgN nanocluster core.44 Thus, the training DNA template sequences in this study are almost certainly associated with similar behavior, i.e., two to three identical DNA strands encapsulate a single AgN. As stated above, the VAE performs automatic feature extraction without any prior knowledge of this chemical behavior. It is not clear from Figure 7 whether the average subsequence behavior reflects the fact that the AgN-DNAs are stabilized by multiple oligomers. Future research may address this, as well as on other possible structure–property relationships that may be learned by models with automatic feature extraction.
The sequence patterns identified by Shapley value analysis for the stratified VAE are consistent with nucleobase “staple motifs” scored as important for AgN-DNA color by Mastracco et al.25 However, the previously reported staple motifs were position-invariant and did not provide information about the importance of nucleobase locations in the sequence. The information presented here about nucleobase importance for AgN-DNA color and brightness could enable computational design of AgN-DNA model systems for first-principles modeling. We hope that these findings inspire work by groups who have begun to work on these topics.31,54
Lessons Learned
The VAE model presented here for AgN-DNA requires no expert knowledge for feature engineering or problem formulation, unlike in past works. Our results show that despite the complexity of deep learning models, strategies can and should be implemented to assess their fitness for making predictions regarding chemical and materials design. For example, by monitoring proxy values for peak wavelength and brightness, we identified that the unstratified VAE struggled to capture sequence-to-color trends for the least abundant wavelength range in the training data, λp > 800 nm. By implementing stratification to address issues that arise due to significant data imbalance, we significantly increased the specific selection of NIR sequences by the VAE.
Experiments confirmed that the VAE model can effectively generate sequences that select for two distinct AgN-DNA properties: emission intensity and peak emission wavelength. This multiobjective design approach is particularly important for the design of bright Green AgN-DNAs, which have posed challenges to past models.22,23,25 The model also enabled the discovery of two NIR AgN-DNAs with brighter emission than any similar products in the training data.
Implementation of Shapley value analysis can be used to interrogate the VAE, learning the important nucleobase patterns that select for AgN-DNA properties. This supports that VAE models can be interpretable and that deep learning applied to chemical and materials design does not necessarily require sacrificing the level of interpretability that can be achieved with simpler models.
Conclusions
This work presents the first model for multiobjective AgN-DNA design and with automatic feature extraction. The generative model, a regularized VAE, can effectively learn from highly imbalanced training data and requires no domain expertise for featurization. The model was challenged with the task of generating DNA template sequences for two kinds of AgN-DNAs: bright Green emitters and relatively rare bright NIR emitters, which represent 28 and 7% of the training instances, respectively. Experimental validation showed that the stratified VAE effectively guides discovery of AgN-DNAs with these target properties, increasing the relative abundance of sequences that select for AgN-DNAs with Green and NIR emission by 3.7 and 4.9, respectively. The designed sequences also significantly yield AgN-DNAs with higher emission brightness as compared to training data, demonstrating utility for multiobjective design.
The VAE model can also be interpreted using our implementation of Shapley value analysis. This approach provided insights into the importance of DNA sequence patterns for mapping of DNA sequence onto AgN-DNA emission color and brightness, including the first information about the importance of nucleobase patterns at specific locations in DNA sequences for selecting AgN-DNA properties. These findings may guide modeling efforts for these emerging nanomaterials.31,54
Finally, we note that the model presented here can be adapted for a range of sequence-based biomolecules and their derived materials. This model would be well-suited for designing nucleic acid nanomaterials whose sequence-structure–property relationships are not completely understood, such as metal-mediated DNA complexes.55 This approach could also generalize to protein and peptide-based materials,19 with especial utility for systems with intrinsic disorder,56 where existing methods have less utility, and for emerging peptide- and protein-stabilized metal nanoclusters.57
Methods
High-throughput AgN-DNA Synthesis
AgN-DNA synthesis was performed in 384 well microplates using robotic liquid handling. DNA oligomers (Integrated DNA Technologies, standard desalting) were mixed with an aqueous solution of AgNO3 and NH4OAc (Sigma-Aldrich), pH 7. After 18 min, AgN-DNA solutions were reduced by a freshly prepared solution of NaBH4 (Sigma-Aldrich), at 0.5 molar ratio of NaBH4 to AgNO3. Final DNA concentration was 20 μM, and final NH4OAc concentration was 10 mM. AgNO3 concentrations were selected to match conditions at which training data were collected in previous work,22,25,36 corresponding to a 5 Ag+/DNA stoichiometry for measurements in the visible spectrum, and a 7 Ag+/DNA stoichiometry for measurements in the NIR. Microplates were then centrifuged at low speed for <60 s to remove any small bubbles. Samples were stored in the dark at 4 °C and measured 7 days after synthesis. Additional details are provided in the SI, and full experimental details are provided in past publications.25
Spectroscopy and Data Processing
Fluorescence emission spectra were collected using two microplate readers. A Tecan Spark was used to acquire emission in the visible range (400–850 nm). NIR emission (675–1425 nm) was measured in a Tecan Infinity 200 Pro with a custom-built InGaAs photodetector,38 using 50 nm bandpass filters and posteriorly correcting for detector spectral responsivity. For both instruments, 280 nm light was used to universally excite all AgN-DNAs.37
Custom spectral fitting routines were used to extract peak wavelength, λp and peak brightness, LII. For visible emission spectra (400–850 nm), spectra were fitted to the sum of one to three Gaussians as a function of energy (in eV). LII values were assigned as integrated intensity of the fitted Gaussian peak and then normalized using a control AgN-DNA58 that is included in all well plates to allow LII values to be compared across different experiments. For data collected on the custom NIR plate reader, peak λp was assigned as the intensity-weighted average of the wavelength corresponding to maximum measured intensity its two neighboring points to right and left. Spectra with >3 peaks or with normalized LII < 0.5 were excluded from training data passed on to the VAE. The resulting training data set comprises 2204 10-base DNA sequences and their peak wavelength(s) and brightness properties of the stabilized AgN-DNAs. Details on spectroscopy and data processing are provided in the SI and in past work.25
VAE Model
We employed the generative model introduced by Moomtaheen et al.,39 with modifications including batch stratification for balanced training. This model utilizes a bidirectional long short-term memory based β-VAE framework (Figure S1). The framework comprises two distinct encoder and decoder neural networks. The encoder is trained to learn the posterior distribution qϕ(z|x) by passing through the one-hot encoded feature vectors and mapping their associated λp and LII scores into a lower dimensional space, z, creating distributions within the lower dimensional space. The decoder, represented as pθ(x|z), reconstructs the latent space z back to the original samples by learning likelihood distribution.
To ensure a decoupled and distinct latent space and to achieve a precise reconstruction of the input DNA sequence, we utilize a loss function consisting of three elements: reconstruction LREC, a Kullback–Leibler (KL)-divergence LKL, and a third term corresponding to regularization:
La is given by eq S2. The initial component in the loss function, LREC, encourages the decoder to reconstruct the original samples using the latent representations z effectively. The second component penalizes the Kullback–Leibler (KL) divergence between the approximated distribution qϕ(z|xi) and a prior distribution P(z), a standard multivariate normal distribution. The final component introduces property regularization, which is governed by the hyperparameter γ. This ensures the lower dimensional space captures the desired properties of the AgN-DNAs (specifically, peak wavelength and brightness) in a joint manner. Additionally, the VAE is trained to order training data for λp with a latent dimension serving as WAV proxy, and similarly for LII in z, for a corresponding LII proxy dimension in latent space.
VAE model hyperparameters were selected using a grid search with a 90:10 training/test split. Optimal hyperparameters for the stratified model were α = 0.007, β = 0.007, γ = 1, δ = 1, |z| = 15, h = 13, with a single LSTM layer, and dropout not utilized. Optimal hyperparameters for the unstratified model were α = 0.003, β = 0.007, γ = 2, δ = 1, |z| = 17, h = 15, a single LSTM layer, and dropout not utilized. For experimental validation, the VAE was trained using all training data without a separate validation set. Expanded details about VAE architecture, training, and sampling are provided in the Supporting Information.
Acknowledgments
The authors gratefully acknowledge support from NSF CBET award #2025793 and from the UC multicampus research programs and initiatives (MRPI) under award no. M23PL5990, as well as partial support from the National Science Foundation Materials Research Science and Engineering Center program through the UC Irvine Center for Complex and Active Materials (DMR-2011967). This project has also been made possible in part by grant 2024-337801 from the Chan Zuckerberg Initiative DAF, an advised fund of the Silicon Valley Community Foundation. A.G.-R. acknowledges the support of a fellowship from “la Caixa” foundation (ID LCF/BQ/EU22/11930078).
Data Availability Statement
Code, training data, and experimental validation for the regularized variational autoencoder (VAE) model are available for download at https://github.com/copplab/VAE-Ag-DNA-design.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsnano.4c09640.
Detailed description of the VAE model; grid search and hyper parameter tuning, model validation metrics; batch stratification method; description of Shapley analysis; experimental methods and data analysis (PDF)
Author Contributions
# E.S. and P.M. contributed equally to this work.
The authors declare no competing financial interest.
Supplementary Material
References
- Kang X.; Zhu M. Tailoring the photoluminescence of atomically precise nanoclusters. Chem. Soc. Rev. 2019, 48, 2422–2457. 10.1039/C8CS00800K. [DOI] [PubMed] [Google Scholar]
- Gonzàlez-Rosell A.; Cerretani C.; Mastracco P.; Vosch T.; Copp S. M. Structure and luminescence of DNA-templated silver clusters. Nanoscale Adv. 2021, 3, 1230–1260. 10.1039/D0NA01005G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz D.; Gardner K.; Oemrawsingh S. S. R.; Markešević N.; Olsson K.; Debord M.; Bouwmeester D.; Gwinn E. Evidence for Rod-Shaped DNA-Stabilized Silver Nanocluster Emitters. Adv. Mater. 2013, 25, 2797–2803. 10.1002/adma.201204624. [DOI] [PubMed] [Google Scholar]
- Neacşu V. A.; Cerretani C.; Liisberg M. B.; Swasey S. M.; Gwinn E. G.; Copp S. M.; Vosch T. Unusually large fluorescence quantum yield for a near-infrared emitting DNA-stabilized silver nanocluster. Chem. Commun. 2020, 56, 6384–6387. 10.1039/D0CC01849J. [DOI] [PubMed] [Google Scholar]
- Bogh S. A.; Carro-Temboury M. R.; Cerretani C.; Swasey S. M.; Copp S. M.; Gwinn E. G.; Vosch T. Unusually large Stokes shift for a near-infrared emitting DNA-stabilized silver nanocluster. Methods Appl. Fluoresc. 2018, 6, 024004 10.1088/2050-6120/aaa8bc. [DOI] [PubMed] [Google Scholar]
- Copp S. M.; Gonzàlez-Rosell A. Large-scale investigation of the effects of nucleobase sequence on fluorescence excitation and Stokes shifts of DNA-stabilized silver clusters. Nanoscale 2021, 13, 4602–4613. 10.1039/D0NR08300C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swasey S. M.; Leal L. E.; Lopez-Acevedo O.; Pavlovich J.; Gwinn E. G. Silver (I) as DNA glue: Ag+-mediated guanine pairing revealed by removing Watson-Crick constraints. Sci. Rep. 2015, 5, 10163 10.1038/srep10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copp S. M.; Schultz D.; Swasey S.; Pavlovich J.; Debord M.; Chiu A.; Olsson K.; Gwinn E. Magic Numbers in DNA-Stabilized Fluorescent Silver Clusters Lead to Magic Colors. J. Phys. Chem. Lett. 2014, 5, 959–963. 10.1021/jz500146q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guha R.; Rafik M.; Gonzàlez-Rosell A.; Copp S. M. Heat, pH, and salt: synthesis strategies to favor formation of near-infrared emissive DNA-stabilized silver nanoclusters. Chem. Commun. 2023, 59, 10488–10491. 10.1039/D3CC02896H. [DOI] [PubMed] [Google Scholar]
- Gonzàlez-Rosell A.; Malola S.; Guha R.; Arevalos N. R.; Matus íF.; Goulet M. E.; Haapaniemi E.; Katz B. B.; Vosch T.; Kondo J.; Häkkinen H.; Copp S. M. Chloride Ligands on DNA-Stabilized Silver Nanoclusters. J. Am. Chem. Soc. 2023, 145, 10721–10729. 10.1021/jacs.3c01366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwinn E. G.; ONeill P.; Guerrero A. J.; Bouwmeester D.; Fygenson D. K. Sequence-dependent fluorescence of DNA-hosted silver nanoclusters. Adv. Mater. 2008, 20, 279–283. 10.1002/adma.200702380. [DOI] [Google Scholar]
- Wang X.; Liisberg M. B.; Nolt G. L.; Fu X.; Cerretani C.; Li L.; Johnson L. A.; Vosch T.; Richards C. I. DNA-AgNC Loaded Liposomes for Measuring Cerebral Blood Flow Using Two-Photon Fluorescence Correlation Spectroscopy. ACS Nano 2023, 17, 12862–12874. 10.1021/acsnano.3c04489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J.; Li N.; Yao Y.; Tang D.; Yang D.; Ong’achwa Machuki J.; Li J.; Yu Y.; Gao F. DNA-stabilized silver nanoclusters for label-free fluorescence imaging of cell surface glycans and fluorescence guided photothermal therapy. Anal. Chem. 2018, 90, 14368–14375. 10.1021/acs.analchem.8b03837. [DOI] [PubMed] [Google Scholar]
- Xu J.; Zhu X.; Zhou X.; Khusbu F. Y.; Ma C. Recent advances in the bioanalytical and biomedical applications of DNA-templated silver nanoclusters. TrAC, Trends Anal. Chem. 2020, 124, 115786 10.1016/j.trac.2019.115786. [DOI] [Google Scholar]
- Danai L.; Rolband L. A.; Perdomo V. A.; Skelly E.; Kim T.; Afonin K. A. Optical, structural and antibacterial properties of silver nanoparticles and DNA-templated silver nanoclusters. Nanomedicine 2023, 18, 769–782. 10.2217/nnm-2023-0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S.; Yan Q.; Cao S.; Wang L.; Luo S.-H.; Lv M. Inhibition of bacteria in vitro and in vivo by self-assembled DNA-silver nanocluster structures. ACS Appl. Mater. Interfaces 2022, 14, 41809–41818. 10.1021/acsami.2c13805. [DOI] [PubMed] [Google Scholar]
- Golinski A. W.; Schmitz Z. D.; Nielsen G. H.; Johnson B.; Saha D.; Appiah S.; Hackel B. J.; Martiniani S. Predicting and Interpreting Protein Developability via Transfer of Convolutional Sequence Representation. ACS Synth. Biol. 2023, 12, 2600–2615. 10.1021/acssynbio.3c00196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emami P.; Perreault A.; Law J.; Biagioni D.; John P. S. Plug & play directed evolution of proteins with gradient-based discrete MCMC. Mach. Learn.: Sci. Technol. 2023, 4, 025014 10.1088/2632-2153/accacd. [DOI] [Google Scholar]
- Krishnaji S. T.; Bratzel G.; Kinahan M. E.; Kluge J. A.; Staii C.; Wong J. Y.; Buehler M. J.; Kaplan D. L. Sequence-structure-property relationships of recombinant spider silk proteins: integration of biopolymer design, processing, and modeling. Adv. Funct. Mater. 2013, 23, 241–253. 10.1002/adfm.201200510. [DOI] [Google Scholar]
- Kuang Y.; Yao Z.-F.; Lim S.; Ngo C.; Rocha M. A.; Fishman D. A.; Ardoña H. A. M. Biomimetic Sequence-Templating Approach toward a Multiscale Modulation of Chromogenic Polymer Properties. Macromolecules 2023, 56, 4526–4540. 10.1021/acs.macromol.3c00403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copp S. M.; Bogdanov P.; Debord M.; Singh A.; Gwinn E. Base Motif Recognition and Design of DNA Templates for Fluorescent Silver Clusters by Machine Learning. Adv. Mater. 2014, 26, 5839–5845. 10.1002/adma.201401402. [DOI] [PubMed] [Google Scholar]
- Copp S. M.; Gorovits A.; Swasey S. M.; Gudibandi S.; Bogdanov P.; Gwinn E. G. Fluorescence color by data-driven design of genomic silver clusters. ACS Nano 2018, 12, 8240–8247. 10.1021/acsnano.8b03404. [DOI] [PubMed] [Google Scholar]
- Copp S. M.; Swasey S. M.; Gorovits A.; Bogdanov P.; Gwinn E. G. General Approach for Machine Learning-Aided Design of DNA-Stabilized Silver Clusters. Chem. Mater. 2020, 32, 430–437. 10.1021/acs.chemmater.9b04040. [DOI] [Google Scholar]
- Kuo Y.-A.; Jung C.; Chen Y.-A.; Kuo H.-C.; Zhao O. S.; Nguyen T. D.; Rybarski J. R.; Hong S.; Chen Y.-I.; Wylie D. C.; et al. Massively parallel selection of nanocluster beacons. Adv. Mater. 2022, 34, 2204957 10.1002/adma.202204957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastracco P.; Gonzàlez-Rosell A.; Evans J.; Bogdanov P.; Copp S. M. Chemistry-informed machine learning enables discovery of DNA-stabilized silver nanoclusters with near-infrared fluorescence. ACS Nano 2022, 16, 16322–16331. 10.1021/acsnano.2c05390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huard D. J. E.; Demissie A.; Kim D.; Lewis D.; Dickson R. M.; Petty J. T.; Lieberman R. L. Atomic structure of a fluorescent Ag8 cluster templated by a multistranded DNA scaffold. J. Am. Chem. Soc. 2019, 141, 11465–11470. 10.1021/jacs.8b12203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerretani C.; Kanazawa H.; Vosch T.; Kondo J. Crystal structure of a NIR-Emitting DNA-Stabilized Ag16 Nanocluster. Angew. Chem., Int. Ed. 2019, 58, 17153–17157. 10.1002/anie.201906766. [DOI] [PubMed] [Google Scholar]
- Mastracco P.; Copp S. M. Beyond nature’s base pairs: machine learning-enabled design of DNA-stabilized silver nanoclusters. Chem. Commun. 2023, 59, 10360–10375. 10.1039/D3CC02890A. [DOI] [PubMed] [Google Scholar]
- Cerretani C.; Kondo J.; Vosch T. Mutation of position 5 as a crystal engineering tool for a NIR-emitting DNA-stabilized Ag 16 nanocluster. CrystEngComm 2020, 22, 8136–8141. 10.1039/D0CE01225D. [DOI] [Google Scholar]
- Rück V.; Neacsu V. A.; Liisberg M. B.; M?llerup C. B.; Ju P. H.; Vosch T.; Kondo J.; Cerretani C. Atomic Structure of a DNA-Stabilized Ag11 Nanocluster with Four Valence Electrons. Adv. Opt. Mater. 2024, 12, 2301928 10.1002/adom.202301928. [DOI] [Google Scholar]
- Malola S.; Häkkinen H. On transient absorption and dual emission of the atomically precise, DNA-stabilized silver nanocluster Ag 16 Cl 2. Chem. Commun. 2024, 60, 3315–3318. 10.1039/D3CC06085C. [DOI] [PubMed] [Google Scholar]
- Chen X.; Boero M.; Lopez-Acevedo O. Atomic structure and origin of chirality of DNA-stabilized silver clusters. Phys. Rev. Mater. 2020, 4, 065601 10.1103/PhysRevMaterials.4.065601. [DOI] [Google Scholar]
- Patel R. A.; Webb M. A. Data-driven design of polymer-based biomaterials: high-throughput simulation, experimentation, and machine learning. ACS Appl. Bio Mater. 2024, 7, 510–527. 10.1021/acsabm.2c00962. [DOI] [PubMed] [Google Scholar]
- Stuart S.; Watchorn J.; Gu F. X. Sizing up feature descriptors for macromolecular machine learning with polymeric biomaterials. npj Comput. Mater. 2023, 9, 102 10.1038/s41524-023-01040-5. [DOI] [Google Scholar]
- McDonald S. M.; Augustine E. K.; Lanners Q.; Rudin C.; Brinson L. C.; Becker M. L. Applied machine learning as a driver for polymeric biomaterials design. Nat. Commun. 2023, 14, 4838 10.1038/s41467-023-40459-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swasey S. M.; Copp S. M.; Nicholson H. C.; Gorovits A.; Bogdanov P.; Gwinn E. G. High throughput near infrared screening discovers DNA-templated silver clusters with peak fluorescence beyond 950 nm. Nanoscale 2018, 10, 19701–19705. 10.1039/C8NR05781H. [DOI] [PubMed] [Google Scholar]
- O’Neill P. R.; Gwinn E. G.; Fygenson D. K. UV excitation of DNA stabilized Ag cluster fluorescence via the DNA bases. J. Phys. Chem. C 2011, 115, 24061–24066. 10.1021/jp206110r. [DOI] [Google Scholar]
- Swasey S. M.; Nicholson H. C.; Copp S. M.; Bogdanov P.; Gorovits A.; Gwinn E. G. Adaptation of a visible wavelength fluorescence microplate reader for discovery of near-infrared fluorescent probes. Rev. Sci. Instrum. 2018, 89, 095111 10.1063/1.5023258. [DOI] [PubMed] [Google Scholar]
- Moomtaheen F.; Killeen M.; Oswald J.; Gonzàlez-Rosell A.; Mastracco P.; Gorovits A.; Copp S. M.; Bogdanov P.. et al. In DNA-Stabilized Silver Nanocluster Design via Regularized Variational Autoencoders, Proceedings of the 28th ACM SIGKDD Conference on Knowledge Discovery and Data Mining; ACM, 2022; pp 3593–3602.
- Ochiai T.; Inukai T.; Akiyama M.; Furui K.; Ohue M.; Matsumori N.; Inuki S.; Uesugi M.; Sunazuka T.; Kikuchi K.; et al. Variational autoencoder-based chemical latent space for large molecular structures with 3D complexity. Commun. Chem. 2023, 6, 249 10.1038/s42004-023-01054-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansoor S.; Baek M.; Park H.; Lee G. R.; Baker D. Protein Ensemble Generation through Variational Autoencoder Latent Space Sampling. J. Chem. Theory Comput. 2024, 20, 2689–2695. 10.1021/acs.jctc.3c01057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew A. J.; Buehler M. J. Encoding and exploring latent design space of optimal material structures via a VAE-LSTM model. Forces Mech. 2021, 5, 100054 10.1016/j.finmec.2021.100054. [DOI] [Google Scholar]
- Iovanac N. C.; Savoie B. M. Improved chemical prediction from scarce data sets via latent space enrichment. J. Phys. Chem. A 2019, 123, 4295–4302. 10.1021/acs.jpca.9b01398. [DOI] [PubMed] [Google Scholar]
- Guha R.; Gonzàlez-Rosell A.; Rafik M.; Arevalos N.; Katz B. B.; Copp S. M. Electron count and ligand composition influence the optical and chiroptical signatures of far-red and NIR-emissive DNA-stabilized silver nanoclusters. Chem. Sci. 2023, 14, 11340–11350. 10.1039/D3SC02931J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzàlez-Rosell A.; Guha R.; Cerretani C.; Rück V.; Liisberg M. B.; Katz B. B.; Vosch T.; Copp S. M. DNA stabilizes eight-electron superatom silver nanoclusters with broadband downconversion and microsecond-lived luminescence. J. Phys. Chem. Lett. 2022, 13, 8305–8311. 10.1021/acs.jpclett.2c02207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rück V.; Cerretani C.; Neacşu V. A.; Liisberg M. B.; Vosch T. Observation of microsecond luminescence while studying two DNA-stabilized silver nanoclusters emitting in the 800–900 nm range. Phys. Chem. Chem. Phys. 2021, 23, 13483–13489. 10.1039/D1CP01731D. [DOI] [PubMed] [Google Scholar]
- Petty J. T.; Carnahan S.; Kim D.; Lewis D. Long-lived Ag106+ luminescence and a split DNA scaffold. J. Chem. Phys. 2021, 154, 244302 10.1063/5.0056214. [DOI] [PubMed] [Google Scholar]
- Liisberg M. B.; Rück V.; Romolini G.; Cerretani C.; Vosch T. Hydration Sensitive Orthogonal Dual Emission of a DNA-Stabilized Silver Nanocluster. Adv. Opt. Mater. 2024, 12, 2400345 10.1002/adom.202400345. [DOI] [Google Scholar]
- Li Y.; Ghosh S. K. Efficient sampling methods for truncated multivariate normal and student-t distributions subject to linear inequality constraints. J. Stat. Theory Pract. 2015, 9, 712–732. 10.1080/15598608.2014.996690. [DOI] [Google Scholar]
- Peng D.; Gu T.; Hu X.; Liu C. Addressing the multi-label imbalance for neural networks: An approach based on stratified mini-batches. Neurocomputing 2021, 435, 91–102. 10.1016/j.neucom.2020.12.122. [DOI] [Google Scholar]
- Loecher A.; Bruyns-Haylett M.; Ballester P. J.; Borros S.; Oliva N. A machine learning approach to predict cellular uptake of pBAE polyplexes. Biomater. Sci. 2023, 11, 5797–5808. 10.1039/D3BM00741C. [DOI] [PubMed] [Google Scholar]
- Lundberg S. M.; Lee S.-I.. A unified approach to interpreting model predictions Adv. Neural Inf. Process. Syst. 2017; Vol. 30.
- Liisberg M. B.; Vosch T. Fluorescence Screening of DNA-AgNCs with Pulsed White Light Excitation. Nano Lett. 2024, 24, 7987–7991. 10.1021/acs.nanolett.4c01561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malola S.; Matus M. F.; Häkkinen H. Theoretical Analysis of the Electronic Structure and Optical Properties of DNA-Stabilized Silver Cluster Ag16Cl2 in Aqueous Solvent. J. Phys. Chem. C 2023, 127, 16553–16559. 10.1021/acs.jpcc.3c04103. [DOI] [Google Scholar]
- Vecchioni S.; Lu B.; Livernois W.; Ohayon Y. P.; Yoder J. B.; Yang C.-F.; Woloszyn K.; Bernfeld W.; Anantram M.; Canary J. W.; et al. Metal-Mediated DNA Nanotechnology in 3D: Structural Library by Templated Diffraction. Adv. Mater. 2023, 35, 2210938 10.1002/adma.202210938. [DOI] [PubMed] [Google Scholar]
- Strader R. L.; Shmidov Y.; Chilkoti A. Encoding Structure in Intrinsically Disordered Protein Biomaterials. Acc. Chem. Res. 2024, 57, 302–311. 10.1021/acs.accounts.3c00624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Martinez E.; Gianolio D.; Garcia-Orrit S.; Vega-Mayoral V.; Cabanillas-Gonzalez J.; Sanchez-Cano C.; Cortajarena A. L. Tuning the optical properties of Au nanoclusters by designed proteins. Adv. Opt. Mater. 2022, 10, 2101332 10.1002/adom.202101332. [DOI] [Google Scholar]
- Cerretani C.; Vosch T. Switchable dual-emissive DNA-stabilized silver nanoclusters. ACS Omega 2019, 4, 7895–7902. 10.1021/acsomega.9b00614. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Code, training data, and experimental validation for the regularized variational autoencoder (VAE) model are available for download at https://github.com/copplab/VAE-Ag-DNA-design.