Tumor cell-derived signals induce stromal IL33 expression in pancreatic cancer and stromal IL33 promotes pancreatic cancer growth by supporting immunosuppression.
Abstract
Pancreatic cancer is characterized by an extensive fibroinflammatory microenvironment. During carcinogenesis, normal stromal cells are converted to cytokine-high cancer-associated fibroblasts (CAF). The mechanisms underlying this conversion, including the regulation and function of fibroblast-derived cytokines, are poorly understood. Thus, efforts to therapeutically target CAFs have so far failed. Herein, we show that signals from epithelial cells expressing oncogenic KRAS—a hallmark pancreatic cancer mutation—activate fibroblast autocrine signaling, which drives the expression of the cytokine IL33. Stromal IL33 expression remains high and dependent on epithelial KRAS throughout carcinogenesis; in turn, environmental stress induces interleukin-33 (IL33) secretion. Using compartment-specific IL33 knockout mice, we observed that lack of stromal IL33 leads to profound reprogramming of multiple components of the pancreatic tumor microenvironment, including CAFs, myeloid cells, and lymphocytes. Notably, loss of stromal IL33 leads to an increase in CD8+ T-cell infiltration and activation and, ultimately, reduced tumor growth.
Significance: This study provides new insights into the mechanisms underlying the programming of CAFs and shows that during this process, expression of the cytokine IL33 is induced. CAF-derived IL33 has pleiotropic effects on the tumor microenvironment, supporting its potential as a therapeutic target.
Introduction
The 5-year survival rate for pancreatic ductal adenocarcinoma (PDA) is currently 13%, making it one of the most fatal human malignancies; to date, PDA is the third leading cause of cancer-related death in the United States (1). Oncogenic mutations in the small GTPase KRAS, most frequently KRASG12D, occur in more than 95% of PDA cases (2–4). In mice, pancreas-specific expression of KRASG12D drives the transformation of healthy tissue into premalignant precursor lesions known as pancreatic intraepithelial neoplasia (PanIN), which progress over time into invasive PDA (5, 6). Furthermore, proof of principle studies in mouse models have shown that inactivation of oncogenic KRAS induces tumor regression at all stages of carcinogenesis (7, 8).
Formation of PanIN and progression to PDA is paralleled by the accumulation of a fibroinflammatory stroma that includes cellular components, such as fibroblasts and immune cells (the latter mostly immunosuppressive in nature), and abundant extracellular matrix (9, 10). We and others have shown that oncogenic KRASG12D activity drives accumulation of the stroma; in turn, inactivation of oncogenic KRAS reverses some, but not all, stromal changes (8, 11–16). Indeed, it is telling that an altered microenvironment has been linked to resistance to oncogenic KRAS inhibition in mouse models and human patients (14, 17).
Fibroblasts are a prevalent component of the precursor lesion microenvironment (PME) and the tumor microenvironment (TME), defined in these contexts as PanIN-associated fibroblasts (PAF) and cancer-associated fibroblasts (CAF), respectively. The healthy pancreas includes heterogenous populations of resident fibroblasts and fibroblast-like pancreatic stellate cells (PSC); lineage tracing experiments completed by our group and others have demonstrated that resident fibroblasts and PSCs contribute to PAFs and CAFs (18, 19). Other sources of CAFs have been described, including mesothelial cells, pericytes, endothelial cells, and adipocytes (20). The overarching role of PAFs and CAFs remains poorly understood, with evidence supporting pro- and anti-tumorigenic functions (for review see refs. 16, 21). Likewise, the mechanism through which normal fibroblasts and other cells of the healthy pancreas are converted to PAFs and CAFs also remains unclear.
Furthermore, pancreatic CAFs are heterogenous—although their classification continues to evolve, a general framework for CAF heterogeneity describes three groups: “myofibroblastic CAFs” (myCAF), “inflammatory CAFs” (iCAF), and “antigen-presenting CAFs” (apCAF; refs. 22, 23). myCAFs exist in close proximity to tumor cells and are marked by high expression of α-smooth muscle actin (α-SMA); these cells are major sources of ECM and ECM-remodeling proteins (22). Conversely, iCAFs express little-to-no α-SMA and instead produce and secrete a wide variety of signaling molecules, including many immunoregulatory chemokines and growth factors (22). Of note, these classifications are largely based on single-cell RNA sequencing (scRNA-seq) and are not fully reflected at the protein level (e.g., most pancreatic CAFs have some level of α-SMA protein expression). Epithelial cells expressing oncogenic KRAS activate a secretory phenotype in CAFs (12, 22); as a result, CAF-derived cytokines promote immunosuppression and tumor progression (24, 25). The mechanisms underlying CAF subtype polarization seem intrinsic and extrinsic. For instance, a subset of CAFs expresses the surface marker CD105—this population is stable and retains its status in vitro and in vivo (26). Additionally, apCAFs, a specialized population of CAFs expressing MHC-II components (23), distinctly originate from mesothelial cells (27), yet in the case of the myCAF and iCAF phenotypes, CAF polarization is driven by extracellular signals, with TGFβ signaling promoting the myCAF phenotype and NF-κB-JAK1/2-STAT3 activation promoting the iCAF phenotype (22). Although the myCAF/iCAF distinction is commonly used in the field, ex vivo polarization studies highlight the plasticity of these two groups, suggesting that these classifications may be nonbinary and instead exist on a spectrum, similar to macrophage polarization (22, 28).
CAFs are the main source of tumor-promoting, immunosuppressive cytokines, such as IL6 and SAA3 (25, 29, 30); we have recently shown that these cytokines are activated in PAFs during the earliest stages of carcinogenesis through signals from KRAS-mutant epithelial cells (12). Another cytokine activated in PAFs at this juncture is IL33, an IL1 family member. IL33 is an unusual cytokine—under homeostatic conditions, it is sequestered in the nucleus of cells expressing it, such as endothelial cells, barrier epithelial cells, and fibroblast-like cells. Upon tissue damage, IL33 is released into the extracellular space (31, 32). Released IL33 binds to its receptor, ST2, which may be expressed by a number of cell types, including mast cells, group 2 innate lymphoid cells (ILC2), CD4+ T helper 2 cells (Th2), regulatory T cells (Tregs), NK cells, and macrophages (31). IL33 has been linked to tumor promotion and restriction across various solid tumor types (33). In PDA, epithelial IL33 has been described in multiple studies, whereas stromal IL33 has remained unexplored. Epithelial IL33 is activated downstream of oncogenic KRAS in premalignant (34) and tumor cells (35). Inhibition of epithelial IL33 slows PanIN progression (36), whereas administration of recombinant IL33 promotes PanIN formation (34). In advanced disease, the effect of IL33 has been studied with controversial results: administration of recombinant IL33 into orthotopic tumor-bearing mice has revealed an antitumor role for this cytokine (37, 38); however, orthotopic injection of IL33 deficient tumor cells also results in less tumor growth (35). Beyond this characterization of epithelial IL33 (34–39), the role of stromal IL33 has been largely ignored, despite previous descriptions of IL33 in this compartment in mouse and human PDA and in other pancreas diseases, such as pancreatitis (12, 40, 41). Furthermore, the mechanisms underlying the activation of stromal IL33 during carcinogenesis remain unknown.
Here, we show that pancreatic PAFs/CAFs are a dominant source of IL33 in PanIN and PDA. We show that deletion of compartment-specific stromal IL33 in an orthotopic model of PDA alleviates immunosuppression and suppresses tumor growth. Additionally, we investigate the mechanisms of IL33 induction and show that its expression by PAFs/CAFs is dependent on tumor and fibroblast-derived signaling factors. In summary, this work explores the role of stromal-derived IL33 in the PME and TME and provides new insight into the mechanisms through which KRAS-mutant cells reprogram their surroundings; at the same time, it highlights a new node of intercellular signaling between tumor cells, fibroblasts, and immune cells.
Results
IL33+ Stromal Cells are Abundant in Human and Mouse PDA
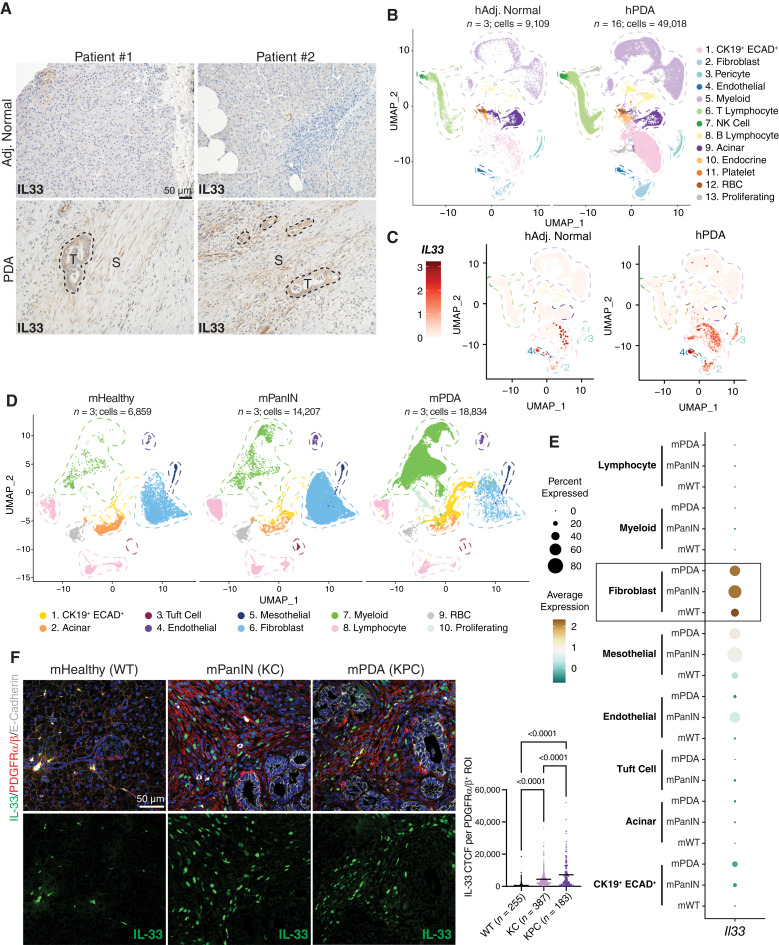
We first assessed the abundance and compartmentalization of IL33 within the PDA microenvironment by performing IL33 IHC on surgically resected human tumors and matched adjacent normal regions (Fig. 1A). We observed robust IL33 staining in tumor cells and disease-associated stromal regions, in contrast with low protein levels in the adjacent normal tissues. To interrogate which stromal cells expressed IL33, we analyzed a human scRNA-seq dataset comprising 16 PDA patient samples and three adjacent normal samples previously created by our group (Fig. 1B; Supplementary Fig. S1A; ref. 42). Notably, overall IL33 transcript abundance was higher in the PDA dataset than in the adjacent normal (Fig. 1C). Furthermore, we found IL33 expression in fibroblasts (cluster 2), pericytes (cluster 3), and endothelial cells (cluster 4) within the TME, in addition to expression by the CK19+ (Cytokeratin-19, KRT19) ECAD+ (E-Cadherin, CDH1) ductal/ductal-like malignant cells (cluster 1).
Figure 1.
IL33+ stromal cells are abundant in human and mouse PDA. A, Human IHC staining of IL33 in matched adjacent normal (“Adj. Normal”) and PDA regions. S, stromal area; T, tumor area. B, UMAP visualization of human scRNA-seq dataset split into adjacent normal and PDA groups. n = number of patients in each dataset. C, Feature plot of IL33 transcription levels in human scRNA-seq. D, UMAP visualization of murine scRNA-seq dataset split into healthy, PanIN, and PDA groups. E, Dot plot representation of Il33 transcription levels across cell types in the murine scRNA-seq dataset. F, Co-IF staining of murine tissues [healthy (wildtype) aka WT, PanIN aka KC (Ptf1a-Cre; LSL-KrasG12D), and PDA aka KPC (Ptf1a-Cre; Trp53R172H/+;LSL-KrasG12D)]. IL33 (green), PDGFRα/β (red), E-Cadherin (white), DAPI (blue). IL33 CTCF was quantified per individual ROI; each ROI encompasses one PDGFRα/β+ cell. N = 3 mice were quantified per group. N in the figure represents the number of ROIs measured per group. P values represent one-way ANOVA testing between groups. Line = mean CTCF.
Next, we examined the compartmentalization of IL33 expression across commonly used genetically engineered mouse models (GEMM) of PDA progression. We curated a murine scRNA-seq dataset of healthy pancreas, PanIN, and PDA tissues to query Il33 gene expression (Fig. 1D; Supplementary Fig. S1B and S1C; refs. 23, 43–45). As in the human samples, Il33 expression was present in CK19+ ECAD+ cells and stromal cells, but it was most highly expressed in fibroblasts across each tissue type, followed by mesothelial and endothelial cells (Fig. 1E). We then performed coimmunofluorescence (co-IF) staining of healthy murine pancreata (WT), spontaneous PanIN (Ptf1a-Cre;LSL-KrasG12D, hereafter KC), and spontaneous pancreatic tumors (Ptf1a-Cre;Trp53R172H/+;LSL-KrasG12D, hereafter KPC) with IL33, the epithelial marker ECAD, and an antibody that recognizes the C-terminal intracellular domains of PDGFRα and PDGFRβ. PDGFRα and PDGFRβ are expressed by fibroblasts (including PAFs/CAFs) and mesothelial cells (Supplementary Fig. S1B), as well as by PSCs (41, 46), whereas PDGFRβ is expressed by pericytes (47)—importantly, these cell types may all give rise to PAFs/CAFs in the pancreatic disease context. We observed an abundance of IL33 in ECAD− PDGFRα/β+ cells in the PanIN and PDA microenvironments, as well as occasional expression in the healthy pancreas (Fig. 1F). Taken together, our findings show that IL33 is robustly expressed in transformed pancreatic tissues and that the stroma is a major source of IL33 in the TME.
Stromal IL33 Promotes PDA Growth
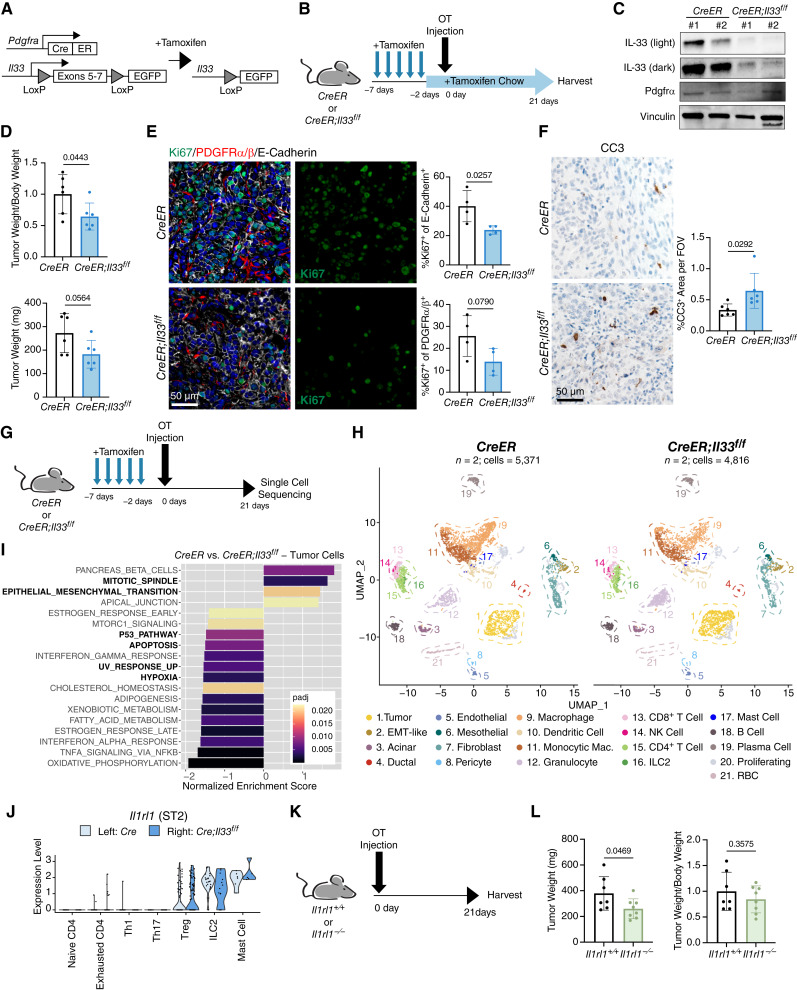
Our observations revealed that the PAF/CAF compartment is the highest expressor of stromal IL33 in pancreatic disease. Therefore, to decipher the impact of stromal IL33 in PDA, we utilized the Pdgfra-CreERT2/+ mouse, which targets fibroblasts, PSCs, and mesothelial cells (the cell types that ultimately give rise to the majority of the PAF/CAF compartment; refs. 18–20, 27). We crossed these mice with Il33f/f-eGFP mice to create Pdgfra-CreERT2/+;Il33f/f-eGFP GEMMs (CreER;Il33f/f) whereby exons 5 to 7 of Il33 are excised upon administration of tamoxifen (Fig. 2A). We induced recombination in these mice via tamoxifen by oral gavage once a day for 5 days prior to orthotopic injection of syngeneic PDA cells (cell line 7940b; ref. 48) and maintained mice on tamoxifen chow throughout the experiment (Fig. 2B). Tumors were harvested 3 weeks after implantation. Using this treatment strategy, we induced targeted deletion of Il33 from PDGFRA+ cells prior to the establishment of disease and continued to induce recombination throughout tumor growth, possibly targeting nascent CAFs derived originally from PDGFRA− sources. PDGFRα+ cells were sorted from the resulting tumors and evaluated for recombination efficiency via western blot; we found drastically reduced IL33 levels in the experimental groups using this method (Fig. 2C). Overall, tumors in CreER;Il33f/f mice were ∼40% smaller than controls, demonstrating that CAF IL33 has a protumorigenic role in PDA (Fig. 2D).
Figure 2.
Stromal IL33 promotes PDA growth. A, Genetic scheme of Pdgfra-CreERT2/+;Il33f/f murine model. Tamoxifen induces activation of the Cre-ERT2 fusion protein, allowing recombination to occur. B, Experimental design for the Pdgfra-CreERT2/+;Il33f/f orthotopic tumor model. OT, orthotopic, CreER = Pdgfra-CreERT2/+, CreER;Il33f/f = Pdgfra-CreERT2/+;Il33f/f. C, Western blot of PDGFRα+ cells sorted from CreER and CreER;Il33f/f orthotopic tumors. Two tumors/mice were pooled in each lane. D, Relative and absolute tumor sizes from CreER and CreER;Il33f/f orthotopic tumors. (E + F) Immunostainings of CreER and CreER;Il33f/f tumors: E, = Co-IF staining of Ki67 (green), PDGFRα/β (red), E-Cadherin (white), and DAPI (blue), F, = IHC staining of Cleaved Caspase-3 (CC3). In quantification, each dot represents one animal. G, Treatment schedule for the Pdgfra-CreERT2/+;Il33f/f orthotopic tumor model adapted for scRNA-seq. H, UMAP visualization of orthotopic scRNA-seq dataset split into CreER and CreER;Il33f/f groups. I, Waterfall plot depicting differential pathway enrichment in tumor cells based on the Hallmark collection of annotations. Positive normalized enrichment scores are enriched in the control group. Pathways of interest are bolded. padj = Bonferroni-corrected P value. J, Violin plot depicting expression of Il1rl1 (ST2) in select leukocytes from scRNA-seq. K, Experimental design for Il1rl1+/+ and Il1rl1−/− orthotopic tumor experiment. L, Relative and absolute tumor sizes from Il1rl1+/+ and Il1rl1−/− orthotopic tumors. Tumor weight/body weight ratios are relative to the control group. Histogram data are mean ± standard deviation. Experiments with two conditions were compared using a two-tailed Student t test.
To characterize changes that might explain tumor size suppression, we performed immunostaining for markers of cell proliferation and cell death (Fig. 2E and F; Supplementary Fig. S2A and S2B). For the former, tumor cells (E-Cadherin+), CAFs (PDGFRα/β+ intratumoral cells), and the proliferation marker Ki67 were measured by co-IF. We saw a clear reduction in overall proliferation in the CreER;Il33f/f model (Supplementary Fig. S2A), including a strong reduction in the fraction of proliferating tumor cells and a trending decrease in proliferating CAFs (Fig. 2E). We also observed increased levels of the cell death marker Cleaved Caspase 3 (CC3) in experimental tissues (Fig. 2F). Interestingly, although we observed an increase in overall CC3 in CreER;Il33f/f tumors, the relative fraction of CC3 colocalizing with E-Cadherin and PDGFRα/β were slightly reduced in the experimental context (69.5% vs. 60.1% and 5.2% vs. 4.5% of total CC3 staining, respectively; Supplementary Fig. S2B). Consequently, these data show an increase in CC3 in an E-Cadherin− PDGFRα/β− population from 25.3% of total CC3 staining in control tumors to 35.4% in CreER;Il33f/f tumors, possibly indicating cell death in E-Cadherin− tumor cells or a PDGFRα/β− stromal compartment. Together these data link the decreased tumor size to loss of proliferation and reduced cell survival.
To delineate the impact of CAF IL33 loss on the TME, we repeated our orthotopic experiment and harvested tissues for scRNA-seq. In this experiment, we adapted our initial orthotopic treatment schedule whereby we gavaged mice once a day for 5 days prior to surgery, but then let mice rest for 2 days before tumor implantation, withholding tamoxifen chow for the entirety of the experiment (Fig. 2G). We took this approach to prevent systemic toxicity of prolonged tamoxifen treatment and to assess the relative contribution of PDGFRA+ healthy cells in total CAF IL33 expression. The tumors from this treatment model exhibited a comparable trend in tumor size reduction between groups, as seen in our tamoxifen chow model (Supplementary Fig. S2C). Two experimental and two control tumors (one male and one female per group) were pooled and submitted for scRNA-seq. The resulting data revealed a complex TME that included tumor cells as well as fibroblasts, immune cells, and other stromal compartments (Fig. 2H; Supplementary Fig. S2D and S2E). Despite withholding tamoxifen chow, we saw a drastic reduction in Il33 transcription in the fibroblast cluster of the experimental group, implying that these CAFs are mostly derived from cells expressing Pdgfra during homeostatic conditions (Supplementary Fig. S2F).
In addition, we observed a population of EMT-like cells expressing the mesenchymal markers Pdgfra, Pdgfrb, Pdpn (Podoplanin), and Col1a1 (Collagen Type 1 Alpha 1), relatively high Krt19 compared with other stromal groups, and high EMT markers such as Ero1l (endoplasmic reticulum oxidoreductase 1 alpha), Vegfa (vascular endothelial growth factor A), Twist1 (twist family BHLH transcription factor 1), and Snai1 (snail family transcriptional repressor 1), that simultaneously lacked expression of the mesothelial markers Lrrn4 (Leucine Rich Repeat Neuronal 4) and Msln (Mesothelin) and the pericyte marker Cspg4 (Chondroitin Sulfate Proteoglycan 4; Supplementary Fig. S2D). Because our implanted tumor cells are derived from a male mouse, yet we used male and female host mice in our orthotopic transplantation study, we investigated whether this EMT-like cell population was of host or tumor cell origin by querying expression of the X-inactivation lncRNA Xist, which is highly expressed in the cells of females but not of males (49, 50). We saw that our tumor cell population had no meaningful Xist expression, but our EMT-like cells exhibited robust Xist expression, similar to that of other host populations such as ductal cells, mesothelial cells, fibroblasts, and pericytes (Supplementary Fig. S2G). This provided evidence that the EMT-like cells are a unique host-derived Krt19high mesenchymal population, possibly arising in response to tissue disruption in areas adjacent to invasive disease. When we examined the expression of Il33 across all cell populations in our scRNA-seq dataset, we detected Il33 in tumor cells and, to a greater extent, in the Pdgfra-expressing stromal cell types, that is, fibroblasts, mesothelial cells, and EMT-like cells; these stromal cells all displayed reduced Il33 levels in the CreER;Il33f/f mice (further supporting the idea that the EMT-like population originates from the host, as the transplanted cancer cells carry neither the CreER nor the Il33 floxed allele; Supplementary Fig. S2F). In contrast, we observed no change in Il33 expression within the tumor cells (Supplementary Fig. S2F).
We next dissected changes in the tumor cell population in the CreER;Il33f/f mice. We first compared the tumor cell transcriptome to the established “classical” and “basal” gene signatures; these molecular subtypes have been associated with improved and worse survival in PDA, respectively (51). We found that the tumor cells of CreER;Il33f/f mice had an enrichment for the “classical” gene signature, whereas the basal signature score was unchanged across the two groups (Supplementary Fig. S2H). We then took an unbiased approach and assessed differential pathway activation between tumor cells in the control and experimental animals using the Hallmark collection of molecular signatures (Fig. 2I). In accordance with our staining result (Fig. 2E and F; Supplementary Fig. S2B), we found enrichment for the “MITOTIC_SPINDLE” signature in the control group and “P53_PATHWAY” and “APOPTOSIS” in the experimental group. We additionally detected enrichment for the “EPITHELIAL_MESENCHYMAL_TRANSITION” signature in the control tumor cells, indicating that tumor cells in the CreER;Il33f/f TME have diminished EMT capabilities. Notably, the experimental tumor cells also displayed enrichment for the cellular stress signatures “HYPOXIA” and “UV_RESPONSE_UP.” Furthermore, CreER;Il33f/f tumor cells were enriched for multiple indicators of a proinflammatory TME, including “TNFA_SIGNALING_VIA_NFKB,” “INTERFERON_ALPHA_RESPONSE,” and “INTERFERON_GAMMA_RESPONSE,” suggesting that the CreER;Il33f/f TME is more immune-permissive.
We next explored the mechanism by which the loss of stromal IL33 induces a tumor-restrictive phenotype. Although canonically IL33 is thought to be constitutively maintained in the nucleus (31), it can also be secreted from living cells through stress-induced mechanisms (35, 41, 52–54). Of particular importance to this work, a recent study in pancreatitis demonstrated that PSCs can secrete IL33 in response to oxidative stress (41). To assess whether this was also true in our cancer model, we grew normal murine pancreatic fibroblasts in vitro (cell line CD1WT; ref. 25) and either cultured them in normal DMEM or polarized them to CAFs by providing murine PDA (cell line 9805; ref. 55) tumor conditioned media (CM) for 24 hours. We then spiked in hydrogen peroxide (H2O2) to induce oxidative stress for an additional 24 hours, collected the resulting CAF CM, and profiled the level of extracellular IL33 via ELISA (Supplementary Fig. S3A). Fibroblasts grown in DMEM did not secrete IL33 (Supplementary Fig. S3B), fitting with the observation in vivo that normal pancreatic fibroblasts express minimal IL33 at baseline (Fig. 1E and F). CAFs treated with PDA CM also showed little-to-no IL33 secretion (Supplementary Fig. S3B). However, CAFs treated with PDA CM + H2O2 did secrete IL33 into the extracellular space (Supplementary Fig. S3B). Oxidative stress is a well-characterized hallmark of the PDA TME (56–59); it is therefore likely that this secretion mechanism also exists in vivo. In fact, when we compared enrichment of the Hallmark reactive oxygen species pathway across cell types in our tumorigenesis scRNA-seq dataset, we detected increasing enrichment for oxidative stress during disease progression in all cell populations but found that fibroblasts were unique in displaying a strong enrichment for reactive oxygen species activation even in healthy tissues (Supplementary Fig. S3C).
The notion that IL33 is secreted from CAFs into the TME prompted us to check for changes in the activation profiles of cells expressing the IL33 receptor, ST2, in our CreER;Il33f/f tumors. Expression of ST2 (Il1rl1) was restricted to mast cells, ILC2s, and a small subset of CD4+ T cells (Supplementary Fig. S2D). To parse out which CD4+ T cells expressed ST2, we subclustered them and discovered that Tregs, but not other helper T-cell populations present in the tumors, were ST2-positive (Fig. 2J; Supplementary Fig. S3D). Interestingly, IL33 activation of mast cells, ILC2s, and Tregs has been linked to disease progression in solid tumors, including IL33-ILC2/Treg activation in pancreatic cancer (35, 36, 39, 60). Given the presence of ST2+ immune cell populations in the tumors, we performed an additional orthotopic PDA implantation experiment using Il1rl1+/+ and Il1rl1−/− mice (Fig. 2K). Tumors in Il1rl1−/− mice were ∼30% smaller than controls, further implicating IL33-ST2 signaling as protumorigenic in PDA (Fig. 2L).
Overall, these data suggest that CAF IL33 supports PDA. The loss of IL33 from PDGFRA+ stromal cells correlates with a decrease in tumor cell proliferation and an increase in tissue apoptosis. Furthermore, the tumor-promoting effect of IL33 is at least in part mediated by secreted IL33 activating signaling in ST2-expressing cells.
Loss of Stromal IL33 Alters the ST2+ Immune Cell Secretome, Resulting in a Shift in CAF Differentiation
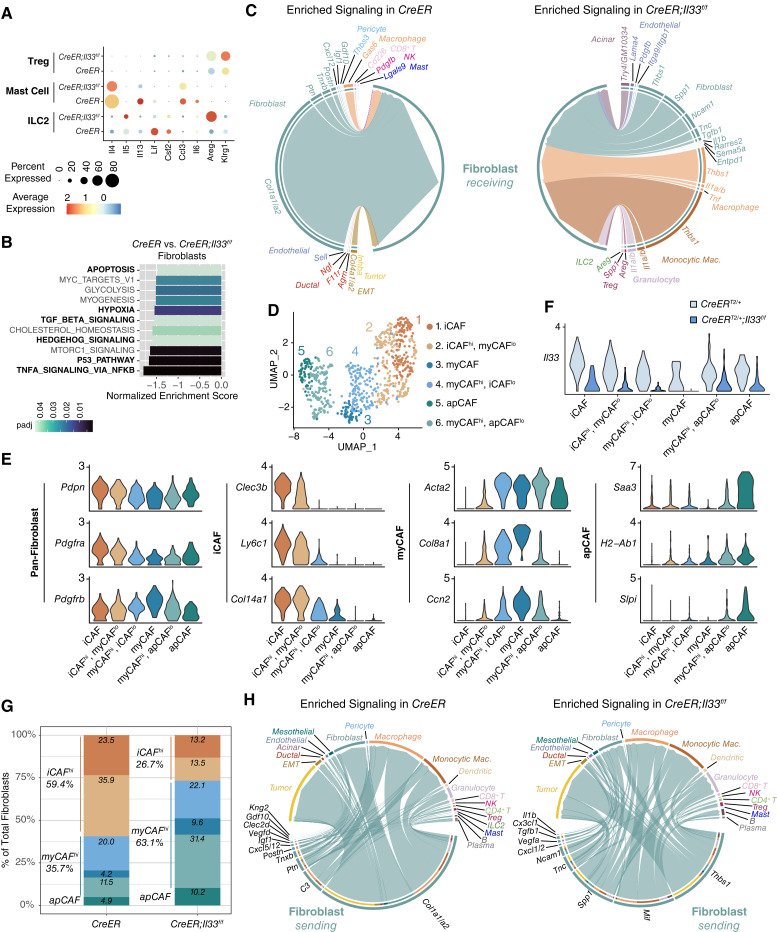
We next investigated gene expression changes in ST2+ cell populations (ILC2s, mast cells, and Tregs) to assess the effect of stromal IL33 loss (Fig. 3A). Mast cells in CreER;IL33f/f tumors expressed less Ccl3 (C–C motif chemokine ligand 3) and Il6, whereas ILC2s showed a decrease in Csf2 (colony-stimulating factor 2, GM-CSF); this reduction was expected, as these factors are linked to IL33 signaling in gastric cancer (Fig. 3A; ref. 60). IL33-ST2 signaling also induces ILC2s to produce IL4, IL5, and IL13 in PDA, as well as LIF (leukemia inhibitory factor) in pancreatitis (35, 41). Although the expressions of Il4, Il13, and Lif were decreased in mast cells and ILC2s in CreER;Il33f/f tumors, the level of ILC2 Il5 was unexpectedly increased (Fig. 3A). Furthermore, we detected upregulation of the EGFR ligand Areg (Amphiregulin) by ILC2s and Tregs in CreER;Il33f/f tumors (Fig. 3A). This result was surprising as IL33 positively regulates ILC2 Areg expression in pancreatitis and other instances of intestinal inflammation (41, 61), and Il1rl1 expression positively correlates with an immunosuppressive AREG+ KLRG1+ Treg phenotype in a murine model of KRASG12D-driven lung cancer (62). Thus, these data possibly highlight tissue-specific functions for IL33. The increase in Areg was particularly interesting as AREG directly stimulates myofibroblast activation and fibrosis in pancreatitis (41) and has recently been shown to contribute to heterogeneity within the myCAF population (63). In our tumors, we found that ILC2s and Tregs were the highest expressors of Areg across all cell types in CreER;Il33f/f mice, whereas CAFs expressed the AREG receptor Egfr (Supplementary Fig. S4A). Given this upregulation of Areg in CreER;Il33f/f tumors, we next looked for changes in CAF activation and polarization. We performed immunostainings to check for differences in total CAFs (intratumoral PDGFRα/β+ cells), activated myofibroblasts (α-SMA and PDGFRα/β colocalization), and total collagen deposition (Gomori trichrome); we detected no substantial change between the two groups based on these parameters (Supplementary Fig. S4B). We then assessed transcriptomic changes within the fibroblast population using scRNA-seq data (note this and subsequent downstream scRNA-seq analyses (Fig. 3B–H) include only the “fibroblast” population and not the “mesothelial” or “EMT-like” cells). Interestingly, the top enriched pathway in CreER;Il33f/f fibroblasts was “TNFA_SIGNALING_VIA_NFKB,” which is downstream of AREG activation (Fig. 3B). Other pathways differentially activated in the IL33-deficient fibroblasts included “HEDGEHOG_SIGNALING” and “TGF_BETA_SIGNALING” (both linked to myCAF differentiation; refs. 11, 22), as well as “P53_PATHWAY,” “HYPOXIA,” and “APOPTOSIS” (the latter of which complements our CC3 IHC staining; Fig. 2F). To understand the drivers of these changes in fibroblasts, we used our scRNA-seq data to plot an unbiased differential predicted interaction analysis between cells across the TME and fibroblasts in the control and CreER;Il33f/f context (Fig. 3C). Our analysis highlighted increased signaling between ILC2- and Treg-derived AREG and fibroblasts in CreER;Il33f/f tumors; at the same time, it revealed strong interaction potential between fibroblasts and myeloid-derived Il1a, Il1b, and Tnf (Tumor necrosis factor), as well as increases in autocrine fibroblast Il1b and Tgfb1 (transforming growth factor beta 1) signaling, possibly contributing to the NF-κB and TGFβ pathway activation seen in the pathway analysis (Fig. 3B).
Figure 3.
Loss of stromal IL33 alters the ST2+ immune cell secretome, resulting in a shift in CAF differentiation. A, Gene expression of activation markers split by CreER and CreER;Il33f/f from scRNA-seq. B, Waterfall plot depicting differential pathway enrichment in fibroblasts based on the Hallmark collection of annotations. Negative normalized enrichment scores are enriched in the experimental group. Pathways of interest are bolded. No genesets were enriched in the control group with Bonferroni-corrected P value (padj) of < 0.05. C, Chord diagram visualizing differentially enriched (Bonferroni-corrected P value < 0.05 and fold-change ≥0.25) predicted to interact with fibroblasts. Edge widths are proportional to predicted interaction strength. D, UMAP visualization of fibroblasts from the CreER and CreER;Il33f/f scRNA-seq datasets. E, Gene expression of markers representing CAF subtypes. F,Il33 expression in each CAF population split by experimental group. G, Histogram depicting the frequency of each CAF population across the CreER and CreER;Il33f/f scRNA-seq datasets. H, Chord diagram visualizing differentially enriched (Bonferroni-corrected P value < 0.05 and fold-change ≥0.25) fibroblast-derived ligands and their predicted interaction partners. Edge widths are proportional to predicted interaction strength.
Given that the NF-κB and TGFβ pathways work in opposition to regulate the iCAF/myCAF/apCAF phenotypes in PDA, we hypothesized that CreER;Il33f/f CAFs may exhibit a modified phenotypic landscape. To investigate this, we subclustered fibroblasts and identified six unique groups, all present in control and experimental samples (Fig. 3D). Although some of these clusters aligned similarly to the iCAF, myCAF, and apCAF molecular signatures (Fig. 3E; refs. 22, 23) others displayed hybrid expression of multiple markers (“iCAFhi, myCAFlo,” “myCAFhi, iCAFlo,” and “myCAFhi, apCAFlo”), consistent with the plastic nature of fibroblast populations. Notably, each CAF cluster displayed some level of Il33 expression in control tumors, with higher expression in iCAF- and apCAF-high cells than in myCAF-high cells; in CreER;Il33f/f fibroblasts, expression was diminished, as expected, across populations (Fig. 3F). Looking broadly at these subtypes, a shift in CAF phenotype between control and experimental tumors emerged. Although fibroblasts in the control tumors mainly exhibited an immunosuppressive secretory phenotype (iCAF/iCAFhi; 59.4% of total fibroblasts), CreER;Il33f/f tumors exhibited an opposite profile, with 63.1% of fibroblasts displaying a myofibroblastic phenotype (myCAF/myCAFhi; Fig. 3G).
IL33 is a nuclear cytokine with a DNA binding motif; as such, it can operate intracellularly to affect the transcriptome (33, 64, 65). This intrinsic function of IL33 is highly context-dependent (31, 66, 67), and whether it factors in pancreatic CAFs is unknown. To query the cell-autonomous effect of IL33 as it may relate to CAF phenotype regulation, we harvested pancreatic fibroblasts from a healthy Il33f/f-eGFP mouse and clonally generated two IL33 KO cell lines (“KO #14” and “KO #16”) and two IL33 WT cell lines (“WT #1” and “WT #20”; Supplementary Fig. S4C). We treated these fibroblasts with murine PDA CM to induce CAF polarization for 24 hours before harvesting for protein or RNA. As expected, CM treatment induced IL33 accumulation in wildtype pancreatic fibroblasts but not in IL33 KO cell lines, confirming efficient gene inactivation (Supplementary Fig. S4D). We then performed bulk RNA-seq. We observed no considerable changes in NF-κB signaling or TGFβ signaling between the IL33 WT and KO cells, either at baseline or upon treatment with PDA CM (Supplementary Fig. S4E). As these pathways define myCAF/iCAF/apCAF differentiation in vivo, it seems unlikely that the nuclear activity of IL33 plays a role in regulating CAF polarization. A more likely explanation is that a change in factors secreted by ST2-expressing immune cells affects fibroblast differentiation status. Importantly, we also measured proliferation in these cell lines and found no differences between the IL33 WT or KO groups, under DMEM or PDA CM treatment (Supplementary Fig. S4F). These data suggest that intrinsic IL33 is dispensable to CAF survival and cell growth.
As an important function of PDA CAFs is to activate protumorigenic intercellular signaling across the TME (12, 21, 22, 24, 29), we next performed the scRNA-seq interaction technique to predict how fibroblast-derived signaling was altered in the context of IL33 loss (Fig. 3H). In control fibroblasts, a variety of ECM-remodeling proteins including Collagen Type 1 (Col1a1 and Col1a2), the glycoprotein Tenascin XB (Tnxb), and the heparin-binding protein Pleiotrophin (Ptn) were differentially enriched compared with the experimental group. Several fibroblast-immune interactions were also higher in control fibroblasts, with the strongest predicted interaction stemming from Complement C3 (C3) signaling to macrophages, which has been shown to activate an immunosuppressive, protumorigenic myeloid phenotype in cancer (68). We also saw expression of Cxcl12 (C–X–C motif chemokine ligand 12), a cytokine linked to PDA cell immune evasion, in control CAFs, which was lost in the CreER;Il33f/f model (24). Conversely, upon fibroblast IL33 loss, we observed an increase in regulators of cell adhesion, plasticity, and vascularization, including Thbs1 (thrombospondin 1; ref. 69), Tgfb1 (70), Spp1 (osteopontin; ref. 71), Tnc (tenascin-C; ref. 72), and Vegfa (Fig. 3H; ref. 73). Furthermore, we observed an increase in genes encoding for immune-regulatory proteins acting on myeloid cells. These include the chemoattractants Cxcl1 and Cxcl2 (potentially recruiting granulocytes), as well as Cx3cl1 (potentially recruiting macrophages). We also saw enrichment for signaling between fibroblast Mif (macrophage migration inhibitory factor) and macrophages and granulocytes; this is notable, as Mif activates a proinflammatory phenotype in myeloid cells (74). To understand the causes of this shift in the fibroblast secretome, we compared the expression levels of the differentially expressed ligands across our CAF compartments (Supplementary Fig. S4G). We found that the different secretome signatures between control and CreER;Il33f/f fibroblasts could be attributed to the shift from an immunosuppressive secretory (iCAF/iCAFhi) phenotype to a myofibroblast-like (myCAF/myCAFhi) phenotype, although we also observed transcriptional upregulation of Mif, Cxcl1, Cxcl2, and Vegfa within the apCAF-like population upon Il33 loss.
Altogether, our data show that loss of IL33 from PDGFRA+ stromal cells profoundly reprograms the TME, including changes in cytokine expression in ST2+ immune cells. In turn, we observed a shift in CAF populations, resulting in differences in their secretome. The result of this phenomenon is the loss of expression of tumor-promoting and immunosuppressive cytokines such as Cxcl12, and an increase in cytokines that potentially support a proinflammatory immune composition.
Inactivation of Stromal IL33 Enables Cytotoxic T-cell Activity
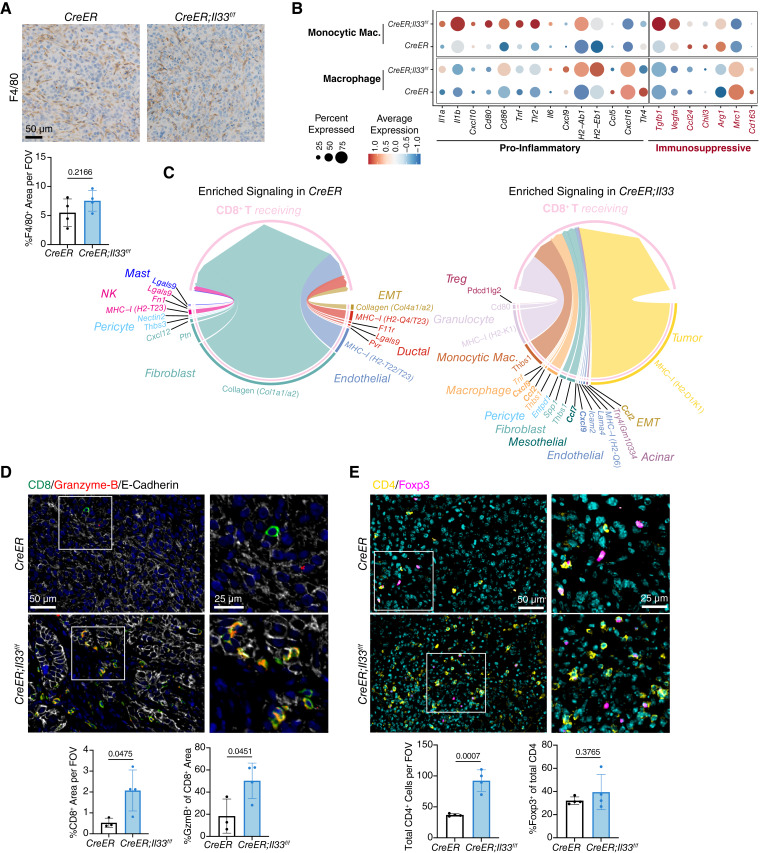
Tumor-infiltrating myeloid cells play a central role in the initiation, progression, and maintenance of PDA through their secretion of tumor-promoting factors and their suppression of CD8+ T cells (28). Given the changes in multiple immune-regulatory factors in CreER;Il33f/f tumors, we sought to determine whether loss of PDGFRA+ stromal cell IL33 caused changes in myeloid populations within the TME. We first stained tumors for macrophages (F4/80) and granulocytes (myeloperoxidase) to measure cell abundance (Fig. 4A; Supplementary Fig. S5A). Although macrophage levels were unchanged, granulocyte infiltration exhibited a dramatic increase; the latter was predicted by the upregulation of fibroblast Cxcl1 and Cxcl2 upon Il33 loss (Fig. 3H; Supplementary Fig. S4G). Notably, increased neutrophil infiltration via CXCL1-CXCR2 signaling is tied to CD8+ T-cell exclusion and pancreatic tumor growth (75).
Figure 4.
Inactivation of stromal IL33 enables cytotoxic T-cell activity. A, IHC staining of F4/80 in CreER and CreER; Il33f/f tumors. B, scRNA-seq gene expression of curated proinflammatory and immunosuppressive markers, grouped by cell type and split by experimental group. C, Chord diagram visualizing ligands differentially enriched (Bonferroni-corrected P value < 0.05 and fold-change ≥0.25) in CreER and CreER;Il33f/f tumors that interact with CD8+ T cells. Edge widths are proportional to predicted interaction strength. Chemokines are bolded. D and E, Co-IF staining of CreER and CreER;Il33f/f tumors: (D) = CD8 (green), Granzyme-B (red), E-Cadherin (white) and DAPI (blue), (E) = CD4 (yellow), Foxp3 (magenta), and DAPI (cyan). For staining quantification, each dot represents one animal, and values were compared using a two-tailed Student t test. Histogram data are mean ± standard deviation.
We next looked for functional changes within myeloid populations using scRNA-seq data. In our dataset, we identified traditional macrophages (Adgre1hi, Mrc1hi, C1qchi, Cd14lo, and Ccr2lo) as well as a distinct population of monocytic macrophages (Adgre1lo, Mrc1lo, C1qclo, Cd14hi, and Ccr2hi; Fig. 2H; Supplementary Fig. S2D). We examined the macrophage populations by comparing their transcriptomes to a curated list of proinflammatory or immunosuppressive markers (Fig. 4B). Both cell types showed upregulation of multiple proinflammatory markers when stromal IL33 was deleted, including but not limited to Il1a, Tnf, Il6, Cxcl9, and MHC2 complex genes H2-Ab1 and H2-Eb1. We also detected downregulation of immunosuppressive markers, including Chil3 (Chitinase-like 3, aka Ym1) and Mrc1 (CD206). Interestingly, Tgfb1 and Vegfa, two immunosuppressive markers, were upregulated in the CreER;Il33f/f group but only by monocytic macrophages. We then evaluated changes in granulocytes by performing differential expression analysis (Supplementary Fig. S5B). Although the CreER;Il33f/f tumors had many more granulocytes than the control tumors, the gene expression between the two groups was mostly unchanged. However, we noted upregulation of Il1b in the CreER;Il33f/f-infiltrating granulocytes, whereas the control granulocytes expressed more Apoe (apolipoprotein E); both genes are linked to NF-κB activation and tumor promotion in pancreatic cancer (44, 76). Furthermore, differential pathway analysis showed enrichment for indicators of activation in CreER;Il33f/f granulocytes, including “TNFA_SIGNALING_VIA_NFKB,” “ALLOGRAFT_REJECTION,” “INTERFERON_ALPHA_RESPONSE,” and “INTERFERON_GAMMA_RESPONSE” (Supplementary Fig. S5C).
Altogether, our data indicate multiple avenues of potential direct and indirect CD8+ T-cell modulation. These include not only supportive phenotypes such as increased proinflammatory polarization in macrophages (Fig. 4B) and a reduced immunosuppressive profile in CAFs (Fig. 3G), but also increased granulocyte recruitment and activation (Supplementary Fig. S5A–S5C), which is potentially CD8+ T-cell suppressive. Given this multifaceted phenotype, we performed an unbiased differential enrichment analysis of all signals from cells across the TME received by CD8+ T cells in control and CreER;Il33f/f tumors to gain a comprehensive understanding of possible changes in CD8+ T-cell function (Fig. 4C). We found that the strongest predicted interactions in control tumors were between CD8+ T cells and collagens originating from fibroblasts and EMT-like cells; in KRASG12D-driven murine models of lung cancer, collagen interactions directly induce CD8+ T-cell exhaustion and correlate with low CD8+ T-cell infiltration (77). Conversely, the strongest signal received by CD8+ T cells in the CreER;Il33f/f model was MHC-I presentation from tumor cells (Fig. 4C), suggesting a potential for increased tumor cell killing and correlating with our increase in CC3 staining (Fig. 2F). We also observed multiple CD8+ T-cell recruiting factors enriched in CreER;Il33f/f tumors, including Cxcl9, Ccl7, and Ccl2 from EMT-like cells, endothelial cells, mesothelial cells, and macrophages (Fig. 4C). Beyond the interaction analysis, the overall gene expression of these chemokines also trended upward in other cell types in CreER;Il33f/f tumors, including Cxcl9 in tumor cells, fibroblasts, monocytic macrophages, and dendritic cells, Ccl7 and Ccl2 in acinar cells, and Ccl2 in fibroblasts, dendritic cells, granulocytes, and mast cells (Supplementary Fig. S5D). Furthermore, CD4+ T cells also expressed the corresponding receptors for these chemokines, Cxcr3, Ccr2, Ccr5, and Ccr4, in the CreER;Il33f/f model (Supplementary Fig. S5D).
To determine whether these transcriptional changes indeed affect T-cell function, we costained tumors for CD8 and the cytotoxicity marker Granzyme-B (Fig. 4D). We measured an increase in the number of CD8+ and Granzyme-B+ CD8+ cells, indicating a heightened cytotoxic T-cell response in tumors lacking stromal IL33. We then costained CD8 and Ki67 (to measure CD8+ T-cell proliferation) and CD44 (which is enriched on memory T cells; ref. 78). We detected a modest trend in CD8+ T-cell proliferation increasing in CreER;Il33f/f tumors as well as an increase in the expression of CD44 on CD8+ T cells, suggesting a potentially heightened memory phenotype, although further markers are needed to evaluate the memory status of this cell population (Supplementary Fig. S5E). We also stained for CD4 and the Treg marker Foxp3 and found an increase in overall CD4+ T cells in CreER;Il33f/f tumors (Fig. 4E), as predicted by our sequencing data (Supplementary Fig. S5D). This included increases in Foxp3+ and Foxp3− CD4+ T cells. Notably, the ratio of Foxp3+ to Foxp3− cells remained unchanged between the CreER and CreER;Il33f/f groups, suggesting that the observed increase in Tregs was driven by an overall increase in CD4+ T-cell recruitment and not by changes in Treg polarization.
In summary, we observed that removing stromal-derived IL33 shifted the abundance and immunosuppressive potential of myeloid cells and that signaling changes across the CreER;Il33f/f TME ultimately resulted in an increase in the recruitment and activation of CD8+ T cells, along with an increase in helper T cells.
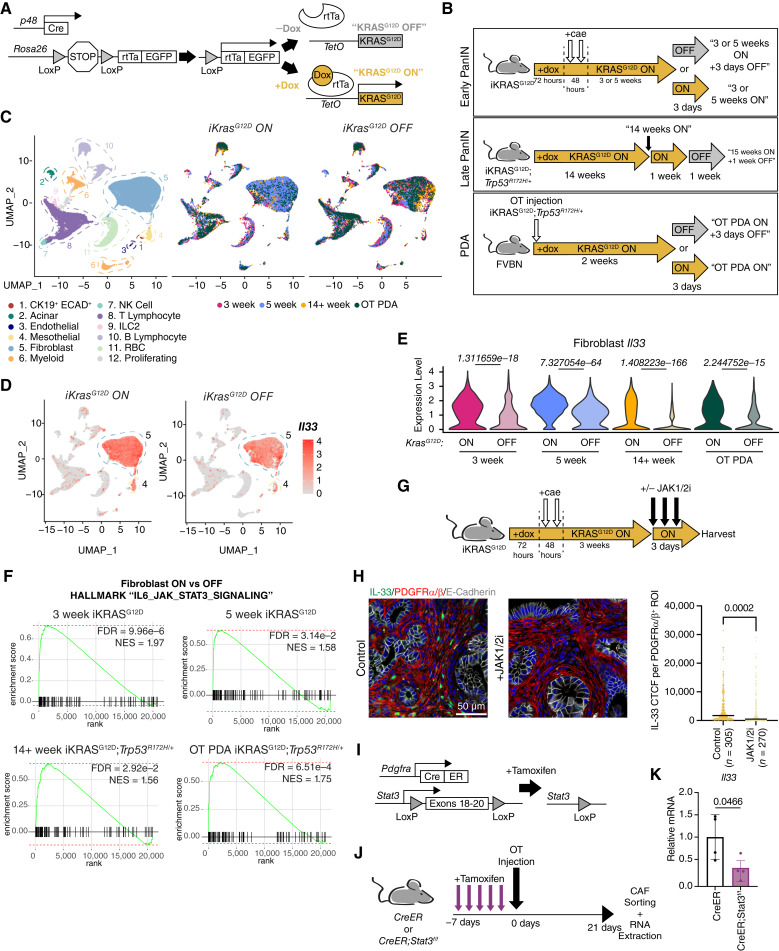
Expression of Fibroblast IL33 is Extrinsically Induced by Epithelial KRASG12D and Requires JAK1/2-STAT3 Activation Throughout Tumorigenesis
Given the pleiotropic effect of stromal IL33 in the TME, we sought to better understand its mechanism of regulation in order to identify potential therapeutic vulnerabilities. We thus utilized the iKRASG12D model, whereby pancreatic epithelial cells express KrasG12D in a doxycycline-dependent, inducible and reversible manner (referred to as “KRASG12D ON” and “KRASG12D OFF,” respectively; Fig. 5A). Using the iKRASG12D model, we have shown that expression of PAF Il33 is dependent on epithelial oncogenic KRAS at the onset of tumor initiation (12), although the mechanism by which this occurs remains unknown. As such, we first endeavored to understand whether stromal IL33 expression remains dependent on oncogenic KRAS throughout tumorigenesis. We generated an “atlas” of previously published and newly generated iKRASG12D scRNA-seq data representing distinct stages of PDA development. This included two “Early PanIN” timepoints (3 and 5 weeks postpancreatitis; refs. 7, 12, 79), a “Late PanIN” timepoint wherein mice also have a full-body Trp53R172H/+ knock-in mutation to accelerate tumorigenesis (55), and a “PDA” model of syngeneic orthotopically injected iKRASG12D;Trp53R172H/+ tumor cells (Fig. 5B; refs. 55, 79). We also included matched “KRASG12D OFF” groups for comparison at each timepoint (7, 12, 55). These datasets were batch-corrected and analyzed collectively. The resulting Uniform Manifold Approximation and Projection for Dimension Reduction (UMAP) visualization revealed a diverse cellular landscape including CK19+ ECAD+ cells (encompassing ductal/malignant-ductal cells), stromal cells including fibroblasts and mesothelial cells, and immune cells at each timepoint (Fig. 5C; Supplementary Fig. S6A and S6B). Notably, the fibroblast compartment (cluster 5) remained the highest expressor of Il33 across cell types in the “KRASG12D ON” and “KRASG12D OFF” contexts, followed by the mesothelial cluster (cluster 4; Fig. 5D). When broken down by timepoint, fibroblast Il33 expression was substantially reduced in each model when KRASG12D was turned “OFF” (Fig. 5E). This pattern was not true for all genes enriched in PME/TME fibroblasts: for example, Igf1 (insulin-like growth factor 1) and Timp2 (TIMP metallopeptidase inhibitor 2) were initially KRASG12D sensitive but became KRASG12D independent as disease progressed (Supplementary Fig. S6C). Interestingly, the expression of Il33 in mesothelial cells (cluster 4) did not display meaningful dependence on KRASG12D (Fig. 5D; Supplementary Fig. S6D). Unfortunately, we did not detect an EMT-like cell population in this dataset comparable with the one found in our orthotopic CreER and CreER;Il33f/f tumors, and as such, we were unable to draw any conclusions about the regulation of Il33 in this compartment. Presently, our findings build off existing knowledge to show that fibroblast Il33 upregulation remains dependent on the activity of epithelial KRASG12D throughout PanIN development and in PDA, whereas mesothelial cells, despite upregulating Il33 during pancreatic disease (Fig. 1E), may regulate Il33 through a KRASG12D-independent mechanism.
Figure 5.
Expression of fibroblast IL33 is extrinsically induced by epithelial KRASG12D and requires JAK1/2-STAT3 activation throughout tumorigenesis. A, Genetic scheme of the iKRASG12D mouse. Doxycycline induces reversible expression of KRASG12D in pancreatic epithelial cells. B, Diagram representing the various iKRASG12D treatment models and collection points across tumorigenesis. Cae, caerulein; OT, orthotopic. C, UMAP visualization of iKRASG12D scRNA-seq dataset. Projection on the left is colored by cell type (all datasets merged). Projections on the right are split by iKRASG12D “ON” and “OFF” status and are colored by timepoint. D, Feature plot representation of Il33 expression levels split by iKRASG12D “ON” and “OFF” status (all timepoints merged). E, Violin plots depicting fibroblast Il33 expression level per timepoint and split by iKRASG12D “ON” and “OFF” status. Wilcoxon rank sum tests were performed between iKRASG12D “ON” and “OFF” pairings per each timepoint, and Bonferroni adjusted P values are displayed above violins. F, GSEA enrichment plots of the Hallmark “IL6_JAK_STAT3_SIGNALING” pathway based on fibroblast iKRASG12D “ON” and “OFF” differential gene expression analysis within each timepoint. G, Treatment scheme for iKRASG12D “ON” model + JAK1/2 inhibitor. H, Co-IF staining of IL33 (green), PDGFRα/β (red), E-Cadherin (white), DAPI (blue). IL33 CTCF was quantified per individual ROI; each ROI encompasses one PDGFRα/β+ cell. N = 3 mice were quantified per group. N in the figure represents the number of ROIs measured per group. P values represent a two-tailed Student t test. Line = Mean CTCF. I, Genetic scheme of Pdgfra-CreERT2/+;Stat3f/f (CreER;Stat3f/f) murine model. Tamoxifen induces activation of the Cre-ERT2 fusion protein, allowing recombination to occur. J, Diagram representing the treatment schedule for the CreER;Stat3f/f orthotopic tumor model. K, Expression levels of Il33 in CAFs from J as measured by RT-qPCR. Values are normalized to Ppia (Cyclophilin A) and relative to the CreER group. Two-tailed Student t test was performed to compare groups; data are mean ± standard deviation.
We next investigated the mechanism by which the expression of fibroblast IL33 is modulated in PanIN and PDA. Our group has previously shown that Il33 upregulation in pancreatic fibroblasts in vitro requires JAK1/2-STAT3 signaling (12). We therefore sought to assess the in vivo relationship between stromal IL33 and JAK1/2-STAT3 activation. First, we performed differential gene expression profiling between fibroblasts in each KRASG12D ON/OFF pairing and found that the Hallmark “IL6_JAK_STAT3_SIGNALING” signature was enriched in the KRASG12D ON group at each timepoint (Fig. 5F). We then performed co-IF staining of our 3-week PanIN tissues and saw decreases in pSTAT3 and IL33 protein levels within PDGFRα/β+ cells at the 3-day “OFF” timepoint (Supplementary Fig. S6E). Thus, high JAK1/2-STAT3 signaling positively correlates with stromal IL33 mRNA and protein expression. Subsequently, we treated our 3-week PanIN-bearing iKRASG12D mice with the JAK1/2 inhibitor ruxolitinib for 3 days prior to harvesting tissue (Fig. 5G). PDGFRα/β+ cells from the treated mice had decreased pSTAT3 and IL33 expression (Fig. 5H; Supplementary Fig. S6F), consistent with the notion that JAK1/2-STAT3 activity is required for IL33 expression even in the presence of oncogenic KRAS. Next, to conditionally disrupt stromal JAK1/2-STAT3 signaling, we bred Pdgfra-CreERT2/+;Stat3f/f (CreER;Stat3f/f) mice, which lose exons 18 to 20 of Stat3 upon activation of recombination by tamoxifen (Fig. 5I). We gavaged mice with tamoxifen once a day for 5 days to suppress Stat3 in Pdgfra+ cells and then implanted syngeneic tumor cells orthotopically to model mature PDA (Fig. 5J; Supplementary Fig. S6G). The resulting tumors also displayed a notable decrease in growth (Supplementary Fig. S6H), in accordance with our CreER;Il33f/f and Il1rl1−/− orthotopic models. We sorted PDGFRα+ cells from these tumors, extracted RNA, and assessed via RT-qPCR to reveal a reduction in Il33 mRNA levels in the CreER;Stat3f/f model (Fig. 5K). Collectively, these findings suggest that KRASG12D-dependent activation of JAK1/2-STAT3 in fibroblasts is required to induce and maintain fibroblast IL33 expression throughout tumorigenesis.
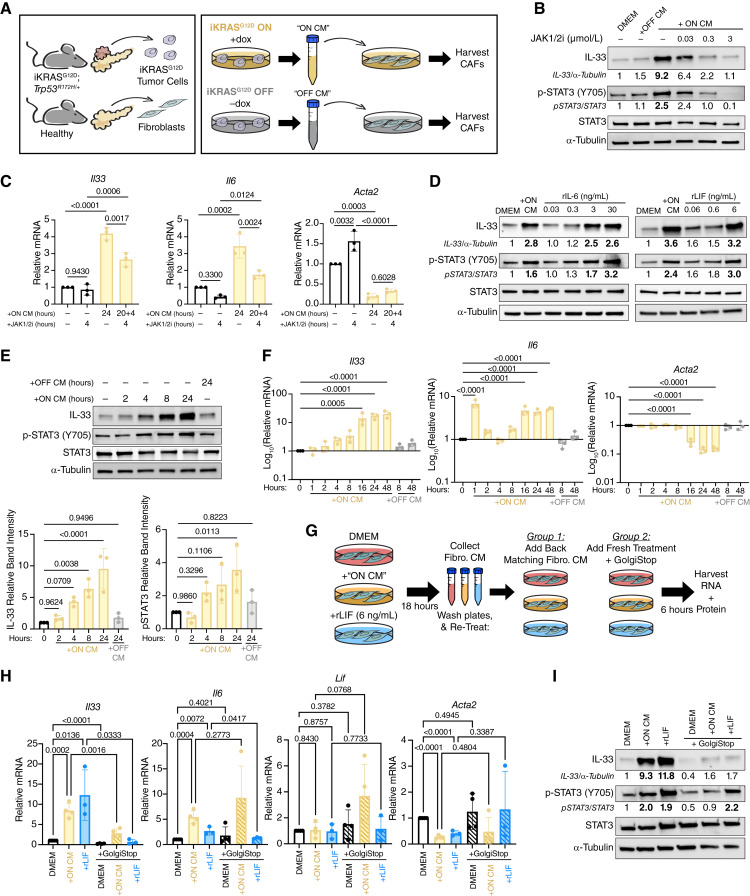
Tumor Cell-Initiated Autocrine Signaling Drives IL33 Upregulation in Pancreatic Fibroblasts
Because KRASG12D expression in vivo inevitably alters multiple compartments throughout the microenvironment, we previously developed an in vitro system to determine whether tumor cell signals directly activate fibroblast JAK1/2-STAT3. To accomplish this, we utilized a previously established iKRASG12D;Trp53R172H/+ tumor cell line (9805; ref. 55). Low-passage 9805 cells (≤p17) maintain doxycycline dependence in vitro and continue to modulate “KRASG12D ON” and “KRASG12D OFF.” We collected CM from “KRASG12D ON” and “KRASG12D OFF” 9805 cultures and provided it to normal pancreatic fibroblasts (CD1WT; Fig. 6A; ref. 25). We have demonstrated already that 24-hour treatment of CD1WT with “KRASG12D ON” CM, but not “KRASG12D OFF” CM instigates Il33 transcriptional upregulation via a heat-liable signaling factor(s) and that this upregulation is blocked by concurrent treatment of CD1WT with a JAK1/2i (12). We repeated this assay and also found that 24 hours of “KRASG12D ON” CM exposure caused accumulation of IL33 protein and Il33 mRNA upregulation, whereas the “KRASG12D OFF” CM did not (Fig. 6B; Supplementary Fig. S7A). We further validated this phenomenon using a pharmacologic approach by treating 9805 cells with the KRASG12D inhibitor MRTX1133 or the MEK1/2 inhibitor Trametinib, generating CM, and providing it to CD1WT fibroblasts (Supplementary Fig. S7B). As expected, MRTX1133- and Trametinib-treated tumor CM resulted in reduced Il33 expression in fibroblasts compared with fibroblasts treated with CM alone. As a negative control, we treated tumor cells with the KRASG12C inhibitor Sotorasib, which had no effect. We also saw that 24 hours of “KRASG12D ON” CM exposure induced pSTAT3 activation, and that upregulation of IL33 and activation of pSTAT3 were blocked by concurrent treatment with a JAK1/2i in a dose-dependent manner (Fig. 6B; Supplementary Fig. S7A). The “KRASG12D OFF” CM failed to induce pSTAT3, indicating that the JAK1/2-activating signaling factor(s) of interest coming from cancer cells is KRASG12D-dependent. Notably, “KRASG12D ON” CM also upregulated the iCAF marker Il6 and downregulated the myCAF marker Acta2 (α-SMA), suggesting a phenotypic switch to a more iCAF-like polarization at this 24-hour interval (Supplementary Fig. S7A). Although the expression of Il6 was also JAK1/2-dependent, Acta2 was not. We then queried whether JAK1/2 was also required to maintain IL33 expression after its initial induction and subsequent CAF polarization. We tested this by pretreating CD1WT with “KRASG12D ON” CM for 20 hours and then supplementing cultures with JAK1/2i for an additional 4 hours (Fig. 6C). By RT-qPCR, we found that this acute spike-in of the JAK1/2i suppressed Il33 and Il6 expression, corroborating our in vivo findings that JAK1/2 activation is continuously required to maintain IL33 upregulation in PDA CAFs (Fig. 5H–K). Next, we investigated the relationship between CAF polarization, JAK1/2 activation, and IL33 expression in a human model. We generated CM from primary patient-derived PDA organoids and provided it to primary fibroblasts collected from either adjacent normal (normal-like fibroblasts) or tumor regions (CAFs). PDA organoid CM induced upregulation of IL33, as well as IL6 and CXCL1, in all three fibroblast lines (Supplementary Fig. S7C). Furthermore, upregulation of IL33 and IL6 was suppressed in the presence of JAK1/2i. Thus, a cancer cell-derived factor(s) induces gene expression of IL33 and IL6 in a JAK1/2-dependent manner in human and mouse fibroblasts.
Figure 6.
Tumor cell-initiated autocrine signaling drives IL33 upregulation in pancreatic fibroblasts. A,Ex vivo culture scheme for iKRASG12D; Trp53R172H/+ (cell line 9805) CM generation and healthy pancreatic fibroblasts (cell line CD1WT). B, Western blot of CD1WT whole cell lysates after 24 hours of treatment with DMEM, iKRASG12D “OFF” CM, iKRASG12D “ON” CM, or concurrent iKRASG12D “ON” CM and JAK1/2i. C, RT-qPCR of CD1WT after treatment with DMEM, JAK1/2i (4 hours, 0.3 μmol/L), iKRASG12D “ON” CM (24 hours), or pretreatment of iKRASG12D “ON” CM for 20 hours followed by spike-in of JAK1/2i (0.3 μmol/L) for an additional 4 hours (24 hours total iKRASG12D “ON” CM treatment). Groups were compared with ordinary one-way ANOVA. D, Western blot of CD1WT whole cell lysates after 24 hours of treatment with DMEM, iKRASG12D “ON” CM, rIL6 (left) or rLIF (right). E, Representative western blot of CD1WT whole cell lysates after treatment with DMEM, iKRASG12D “OFF” CM, or iKRASG12D “ON” CM for increasing intervals of time. Densitometry quantification for IL33 normalized to loading control (α-tubulin) and pSTAT3 normalized to total STAT3 are shown. Quantification is relative to the 0-hour timepoint. Ordinary one-way ANOVA was performed to compare each timepoint to the control. F, RT-qPCR of CD1WT after treatment with DMEM, iKRASG12D “OFF” CM, or iKRASG12D “ON” CM for increasing intervals of time. Values are log10 transformed to better visualize large changes in gene expression level. Ordinary one-way ANOVA was performed to compare each timepoint to the 0-hour timepoint. Only comparisons with P value < 0.05 are shown. G, Experimental scheme to block autocrine signaling in CD1WT. CD1WT were treated with DMEM, iKRASG12D “ON” CM, or rLIF for 18 hours, and then, the resulting CM was set aside. Cells were washed with PBS and then given back their original 18-hour CM or given GolgiStop (1.3 μL/2 mL) + fresh DMEM, iKRASG12D “ON” CM, or rLIF media. Cells were incubated for an additional 6 hours before harvesting CD1WT RNA and protein. H, RT-qPCR of CD1WT after autocrine blocking experiment. Two-tailed Student t test was performed to compare groups of interest (all tested comparisons shown). I, Western blot of CD1WT whole cell lysates after autocrine blocking experiment. In all experiments with iKRASG12D CM, doxycycline is used as a vehicle control. In all experiments with JAK1/2i (ruxolitinib), DMSO was used as a vehicle control. All replicates represent complete, independent experiments. RT-qPCR values are normalized to Ppia (Cyclophilin A) and relative to the untreated DMEM group. Histogram data are mean ± standard deviation.
To identify cancer cell-secreted proteins that are KRAS-dependent, we performed a multiplex ELISA on “KRASG12D ON” and “KRASG12D OFF” CM. We detected low amounts of IL1α, TNFα, and IL6, as well as abundant LIF, in the “KRASG12D ON” CM, all of which were reduced or undetectable in “KRASG12D OFF” CM (IL1β was also probed for, but not detected, in either condition; Supplementary Fig. S7D). IL1α and TNFα activate NF-κB: in pancreatic fibroblasts, it has been shown that NF-κB activation induces the production and secretion of IL6 and LIF, which in turn autocrine activate JAK1/2 signaling and induce an iCAF phenotype (22). Therefore, we sought to determine whether any of the detected KRASG12D-dependent cancer cell-derived factors were sufficient to induce IL33 expression. For this purpose, we treated CD1WT fibroblasts with recombinant IL6 (rIL6) and rLIF (activating JAK1/2) or rIL1α and rTNFα (activating NF-κB). Within 24 hours of treatment with rIL6 and rLIF, both cytokines were able to activate STAT3 phosphorylation and IL33 expression to levels that mirrored the “KRASG12D ON” CM treatment group (Fig. 6D). We then treated CD1WTs with rIL1α or rTNFα for 1 and 24 hours and collected RNA. However, neither cytokine activated Il33 expression; rather, expression of Il33 was reduced below baseline with each (Supplementary Fig. S7E). Interestingly, 1 hour of either rIL1α or rTNFα upregulated Il6 and Lif in CD1WTs, but only rIL1α-induced Il6 remained high at the 24-hour timepoint, whereas Lif expression was either lost or suppressed below baseline expression at that interval (Supplementary Fig. S7E). rIL1α and rTNFα also suppressed Acta2 at 24 hours (Supplementary Fig. S7E), suggesting that each factor was individually sufficient to push the fibroblasts to a “less myCAF-like” state—similar to treatment with full tumor cell CM—despite not upregulating Il33.
It has also been reported that TGFβ signaling can induce Il6 expression in fibroblasts (80) and PSCs (81) and Lif expression in other cell types (82, 83); furthermore, this signaling directly induces upregulation of IL33 in tumor-initiating cells (TIC) in squamous cell carcinoma (SCC; ref. 54). To investigate whether TGFβ could also lead to Il33 upregulation in pancreatic fibroblasts, we treated CD1WT for 24 hours with rTGFβ and harvested the resulting RNA (Supplementary Fig. S7F). Interestingly, we found that TGFβ induced heightened expression of the myCAF marker Acta2 (as predicted given the relationship between TGFβ and the myCAF phenotype; ref. 22) and moderate upregulation of Il6 but suppressed Il33 expression. Lif was also unaffected by rTGFβ at this timepoint. We further investigated the relationship between TGFβ and IL33 by combining the treatment of CD1WT with “KRASG12D ON” CM and a TGFβ receptor inhibitor (Supplementary Fig. S7G). The addition of the inhibitor did not impact the ability of “KRASG12D ON” CM to upregulate IL33, despite dose-dependent loss of pSMAD2/3 in this assay. Therefore, our data indicate that TGFβ is not involved in the upregulation of fibroblast IL33. In fact, Il33 expression may be antagonized by high TGFβ activity, similar to the generation of the iCAF phenotype.
Overall, our findings presented a conundrum: whereas rIL1α, rTNFα, and rTGFβ all induce expression of Lif and/or Il6, but not Il33, LIF and IL6 induce Il33 expression. We considered that the timing of activation might explain this disconnect. We thus investigated the kinetics of STAT3 phosphorylation and IL33 protein accumulation following the “KRASG12D ON” CM treatment. Interestingly, we found that activation of pSTAT3 did not occur until between 2 and 4 hours following CM treatment and continued to increase until 24 hours (Fig. 6E). IL33 protein accumulation followed a comparable pattern. We also assessed our fibroblasts at the RNA level and found that Il33 mRNA gradually increased from 2 hours to peak at ∼16 hours post-“KRASG12D ON” CM exposure (Fig. 6F). Of note, we also observed sustained increases in Il6 and decreases in Acta2 at the 16 hours timepoint, suggesting a major phenotypic CAF switch at this juncture (Fig. 6F). As treatment with “KRASG12D ON” CM did not immediately initiate STAT3 phosphorylation—despite CM containing some IL6 and LIF, which are JAK1/2 activating ligands—our data support a model by which tumor cells may secrete an insufficient concentration of ligands or lack secretion of other factors required to activate JAK1/2 signaling in fibroblasts. We then tested whether a secondary message by fibroblasts was necessary to induce JAK1/2 signaling via a feedforward loop. We designed an in vitro assay in which we treated CD1WT with DMEM alone, “KRASG12D ON” CM, or rLIF for 18 hours to initiate IL33 upregulation, pSTAT3 activation, and CAF polarization. We collected the resulting CD1WT CM, washed cells with PBS, and treated them for an additional 6 hours with either (i) their matching CD1WT CM (i.e., giving the CM back to the originating cells) or (ii) a Golgi blocking agent (GolgiStop) plus fresh treatment (DMEM, “KRASG12D ON” CM, or rLIF) matched to the original treatment condition (Fig. 6G). This design resulted in fibroblasts receiving either (i) up to 24 hours of potential autocrine signaling factors or (ii) withdrawal of any autocrine signaling factors for the last 6 hours of treatment. We then collected RNA and protein from these fibroblasts. Our results showed that “KRASG12D ON” CM-dependent Il33 upregulation was lost when autocrine signaling was blocked (Fig. 6H and I). Importantly, the Golgi blocking agent also prevented the “KRASG12D ON” CM from activating pSTAT3, indicating that tumor-derived factors alone are indeed not sufficient to activate this pathway (Fig. 6I). Furthermore, rLIF did not rescue IL33 upregulation when autocrine signaling was blocked, despite successfully activating pSTAT3 (Fig. 6H and I). Interestingly, upregulation of Il6 was not affected by treatment with the Golgi blocker with or without CM; in contrast, the Golgi blocker reversed the downregulation of Lif previously seen after 24 hours of “KRASG12D ON” CM (Fig. 6H; Supplementary Fig. S7E). The Golgi blocker also prevented the downregulation of Acta2 in response to rLIF treatment (Fig. 6H).
Taken together, our data demonstrate that IL33 upregulation in PAFs/CAFs is dependent on at least two autocrine signaling events. In the first loop, fibroblast pSTAT3 activation is dependent on tumor cell-initiated fibroblast autocrine signaling (Fig. 6I). In the second loop, pSTAT3 activation alone (without tumor CM) is sufficient to drive IL33 upregulation in healthy pancreatic fibroblasts (Fig. 6D) but only in the context of active fibroblast autocrine signaling (Fig. 6I). Additionally, it is possible that this second loop is dependent on pSTAT3, despite being agnostic of tumor CM treatment (Fig. 6I). Overall, our observations shed light on a complex signaling network involving cancer cell-derived signaling factors and autocrine activation that evolves over time as healthy fibroblasts are reprogrammed to CAFs. These data corroborate the preexisting model whereby pancreatic tumor cell CM promotes the iCAF phenotype and suppresses the myCAF phenotype in culture (22); at the same time, they demonstrate that IL33 expression is uniquely regulated compared with the PDA-promoting iCAF cytokines IL6 and LIF (25, 84).
Discussion
Pancreatic cancer is characterized by a complex network of cancer, stromal, and immune cell crosstalk (85) that requires further unraveling to improve therapeutic interventions. CAFs are an abundant population within the stroma, but their role during cancer progression, whether pro- or anti-tumor, remains controversial (86). This lack of understanding likely stems from the complex biologic functions of CAFs: beyond providing structural support, they secrete extracellular matrix and serve as a signaling hub, secreting multiple cytokines and growth factors (12, 15, 21). The function of specific CAF-derived cytokines and mechanisms through which epithelial neoplastic cells drive cytokine expression programs in CAFs remain unclear. One of the cytokines activated in pancreatic fibroblasts in response to epithelial oncogenic KRAS is IL33 (12). Although IL33 expression in cancer-associated stromal cells has been previously noted (40), its function and regulation have remained unexplored.
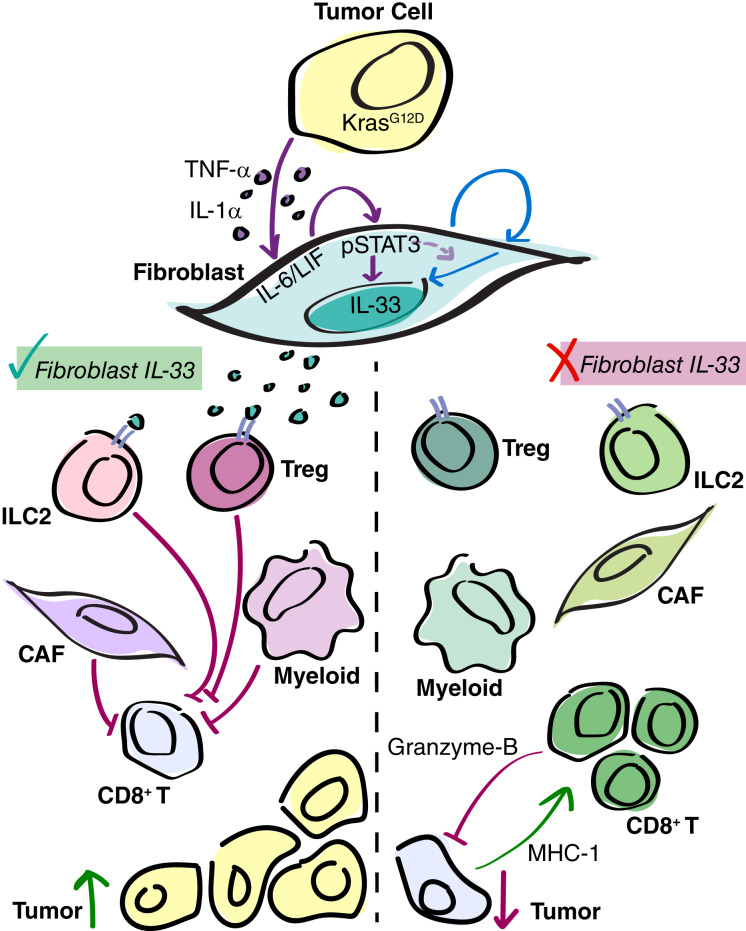
Here, we demonstrate that stromal IL33 is activated early during carcinogenesis and that PAF/CAF IL33 remains dependent on epithelial KRAS at all stages of cancer progression, from low-grade PanIN to malignant disease. Furthermore, we show that stromal IL33 supports tumor progression (Fig. 7).
Figure 7.
Tumor cell KRASG12D initiates upregulation of fibroblast IL33, promoting immunosuppression in PDA. Working model. During PanIN and PDA, KRASG12D-dependent tumor cell-derived signaling factors initiate fibroblast autocrine signaling, including the JAK1/2-STAT3 pathway. This fibroblast reprogramming results in the upregulation of IL33. Furthermore, at least one additional autocrine loop is required for IL33 upregulation (possibly also dependent on pSTAT3). CAF IL33 is secreted in response to oxidative stress, in which it signals to ST2+ immune cells ILC2s and Tregs, promoting an immunosuppressive TME and tumor growth. When stromal IL33 is removed, ILC2s and Tregs exhibit an altered secretory gene signature, and a shift in CAF and myeloid cell polarization is seen. This ultimately results in the recruitment and activation of CD8+ T cells, and the suppression of tumor growth.
Our study complements previous literature addressing the role of epithelial IL33 in cancer. IL33 expression is elevated in many solid tumor types; its regulation is cell type-dependent, whereas its functional role is largely determined by the nature of the cells within the tumor expressing the IL33 receptor, ST2 (33, 87). In a murine model of SCC, TICs upregulate IL33 in response to TGFβ signaling; in turn, secreted IL33 drives a TIC-macrophage positive feedback loop that promotes cancer invasion and drug resistance (54). Conversely, ST2+ NK cells have been identified in multiple human and murine tumors including breast cancer; these cells activate an antitumoral inflammatory response upon IL33 stimulation (bioRxiv 2023.02.14.528486). In pancreatic cancer, the role of IL33 may vary depending on the stage of the disease. Injection of recombinant IL33 into mice bearing KRAS–mutant pancreatic epithelia accelerates PanIN formation, and inactivation of IL33 in KRASG12D-expressing epithelial cells hinders it, despite robust expression of IL33 in other stromal cell populations (12, 34, 36). In invasive diseases, the relationship between IL33 and ST2+ ILC2s has been increasingly explored; the downstream effect is controversial, with studies supporting pro- and anti-tumoral effects (35, 37). Concurrently, IL33 to ST2+ Treg signaling promotes CD8+ T-cell exclusion and increases tumor growth (39).
Importantly, in addition to differences in the function of IL33 during PanIN and PDA, there are likely species-dependent differences in the regulation and role of epithelial and stromal IL33 that should be emphasized. Outside of the disease context, homeostatic IL33 compartmentalization can differ between humans and mice (e.g., although nearly all human endothelial cells express IL33, only a subset of murine endothelial cells show IL33 expression; ref. 31). This observation may also be reflected in our scRNA-seq data: although murine endothelial cells have little-to-no Il33, our human endothelial cells have substantial IL33 expression in adjacent normal and PDA tissues, suggesting that IL33 expression in this compartment may be disease agnostic (it should be noted, however, that some level of dysplasia is expected in adjacent normal tissues due to their close localization with the tumor, making them a limited control for such a comparison). Although we found that the fibroblast compartment was the highest expressor of IL33 across multiple GEMMs of PanIN and PDA, with minimal expression of epithelial IL33 by RNA and protein staining, our human PDA data show robust expression of IL33 in tumor and stromal cells, suggesting that tumor cell IL33 may play a larger role in human disease than in our murine models.
Indeed, it is further possible that the type of cell expressing IL33 plays an important role in its impact on the TME and tumor growth. Nuclear IL33 may have a unique cell-intrinsic transcriptomic effect that promotes or hinders tumor progression. Another consideration is the regulation of IL33 secretion; regarding the relative contribution of each cell type’s IL33 in the extracellular space, it is possible that different extracellular cues are necessary to stimulate active IL33 secretion from each population, and that some cues are more common than others. Here, we show that tumor CM-induced IL33 upregulation does not automatically instigate IL33 secretion from pancreatic fibroblasts; instead, our work corroborates a mechanism by which oxidative stress causes fibroblast IL33 secretion (41)—a relationship that has also been shown in TICs (54). Other stress stimuli that have been shown to cause IL33 release in epithelial cells include fungus (35, 52) and extracellular ATP (53). Overall, the cell-intrinsic and extracellular impacts of IL33 warrant further investigation about cells of origin, with emphasis on the interspecies differences between human and murine biology, in order to fully understand the scope of IL33 function in PanIN and PDA and apply these findings therapeutically.
To focus on the role of stromal IL33 in PDA, we deleted IL33 from PDGFRα+ cells and found a suppression of tumor growth, despite intact IL33 expression in cancer cells. We detected ST2 expression in ILC2s, mast cells, and Tregs, and found fewer Il4+ and Il13+ ILC2s and mast cells in CreER;Il33f/f tumors, supporting previous observations whereby IL33 induces expression of these cytokines (37, 41). We observed upregulation of AREG—an EGFR ligand—by Tregs and ILC2s upon inactivation of IL33. This was unexpected, given that IL33 stimulation elicits AREG upregulation by ILC2s in the gut and during pancreatitis (41, 61), and again, indicates tissue-dependent mechanisms of action.
Thus, the loss of stromal IL33 induced elevated expression of AREG, which was in turn predicted to act on CAFs, which express a high degree of EGFR. Consistent with this notion, CAFs displayed elevated NF-κB pathway enrichment, a canonical downstream mediator of AREG-EGFR signaling. A direct link between AREG and CAF differentiation has been recently described (63), with fibroblast-derived AREG shifting myCAFs to a tumor-promoting subtype. However, we did not detect AREG expression in fibroblasts, rather, we found a potential immune cell-fibroblast paracrine signaling axis. Beyond its effect on fibroblasts, AREG might act on the pancreatic epithelium to reinforce the KRAS-MAPK signaling axis, which is important in the onset of pancreatic carcinogenesis even in the presence of oncogenic KRAS (88, 89). CAFs in CreER;Il33f/f tumors also displayed increased activation of Hedgehog and TGFβ signaling, both of which are linked to myCAF differentiation (11, 22). In addition to an increase in Areg, stromal IL33 deletion results in increased NF-κB ligands, namely Il1a, Il1b, and Tnf from myeloid cells and fibroblasts, as well as increased fibroblast Tgfb1, all of which may act on the CAF compartment. The overarching result of these pleiotropic changes was a shift in fibroblasts toward a myofibroblastic phenotype and a reduction in immunosuppressive cytokine production; these findings support previous literature showing that the myofibroblast-activating TGFβ pathway overrides NF-κB activation and the subsequent iCAF phenotype when the pathways are in competition (22). It should be emphasized that despite this transcriptomic analysis, we did not detect a change in the abundance of the myofibroblast marker α-SMA by immunostaining in our tissues. Indeed, our data depict a nuanced molecular landscape in which there is much overlap between secretory and myofibroblastic CAFs in vivo [to this point, of the six CAF subtypes we detected, five display moderate-to-high Acta2 (α-SMA) gene expression]. Thus, our data highlight the need to continually address the concept of CAF heterogeneity in the context of cell plasticity.
Interestingly, nuclear IL33 has also been implicated in the regulation of NF-κB and TGFβ signaling (64, 65), suggesting that there may be cell-intrinsic effects of IL33 loss driving CAF repolarization, in addition to cues from the microenvironment. However, our in vitro assessment of IL33 WT and IL33 KO fibroblasts with and without tumor CM treatment showed no change in NF-κB or TGFβ signaling, fibroblast proliferation, or fibroblast survival, thus providing no evidence of a cell-intrinsic effect.
Ablation of stromal IL33 caused loss of cancer cell proliferation, increased apoptosis, prompted a transcriptional shift toward the more differentiated “Classical” molecular subtyping, and resulted in an overall reduction in tumor size while profoundly altering the microenvironment. We observed a similar reduction in tumor size when syngeneic orthotopic tumors were implanted in mice lacking the IL33 receptor, ST2. This indicates that the effects of IL33 are largely dependent on its secreted form, rather than the DNA binding, intracellular functionality of IL33. Notably, the effects we observed in the CreER;Il33f/f model occur in the presence of epithelial IL33, supporting the notion that the stroma is the main source of secreted IL33 in this system. We observed direct and indirect effects of IL33 on the TME; ST2+ cells such as ILC2s, Tregs and mast cells had altered cytokine production upon fibroblast IL33 loss. At the same time, in macrophages, we observed a loss of immunosuppressive markers such as Arg1 (Arginase-1) and Mrc1 (CD206) in addition to an increase in proinflammatory genes including Cd80, Cd86, Tnf, Il1a, and Il1b, and MHC2 presentation machinery (H2-Ab1 and H2-Eb1). Tumors lacking stromal IL33 also had increased recruitment and activation of granulocytes which, although generally thought to be immunosuppressive (90), also expressed the T–cell activating component Cd80 in the CreER;Il33f/f context, adding to the proinflammatory milieux of the TME. Given that we did not detect ST2 expression in these myeloid cell populations by scRNA-seq, these changes are likely due to indirect effects of IL33 loss, potentially stemming from changes in the production of other cytokines from ST2+ cells.
In the context of stromal IL33 loss, tumor cell/TME to T-cell interactions were altered in such a way that the overall result was an increase in CD8+ T-cell recruitment and activation, with likely cytotoxic activity, as evidenced by increased CC3 and Granzyme-B. To this point, we also found increased infiltration by helper T cells, which are necessary for driving an antitumoral cytotoxic response (91). We were surprised to find no change in the relative abundance of Th2s or ILC2s by scRNA-seq, as the recruitment of these cell types has been previously linked to extracellular IL33 activity; however, this was likely due to the compounded changes in recruitment factors seen across cell populations when stromal IL33 was removed. Overall, our data indicate that extracellular stromal IL33 has tumor-promoting activity through modulation of the immune microenvironment.
Given the functional importance of stromal IL33, we then sought to address the mechanism through which oncogenic KRAS-expressing cells drive IL33 expression in pancreatic fibroblasts. We found that PAF/CAF IL33 expression in murine models of PanIN and PDA is dependent on fibroblast pSTAT3 and epithelial oncogenic KRAS activity, consistent with our previous observations in vitro (12). Similarly, secreted factors derived from human PDA organoids induce IL33 upregulation in human pancreatic fibroblasts and CAFs with dependence on fibroblast JAK1/2 activation. Using a multicytokine ELISA platform, we showed that pancreatic cancer cells secrete the NF-κB-activating ligands IL1α and TNFα as well as the JAK1/2-STAT3-activating ligands IL6 and LIF in an oncogenic KRAS-dependent manner. Treatment of healthy pancreatic fibroblasts with these cytokines revealed a complex mechanism whereby gene expressions of many cytokines (including Il33) are upregulated or downregulated with different temporal dynamics. Il33 and Il6, but not Lif, are gradually induced by recombinant LIF over the course of 16 hours, which in turn suppresses the expression of Acta2, encoding for α-SMA. Conversely, Il6 and Lif, but not Il33, are induced by recombinant IL1α and TNFα, although these activation relationships are sustained differently over time. Furthermore, despite being a direct activator of IL33 in SCC TICs, TGFβ induces Il6 but not Il33 upregulation in pancreatic fibroblasts (54). It is also notable that we detected the highest levels of IL33 expression in vivo in iCAFs, which are canonically polarized by activation of NF-κB and JAK1/2-STAT3 and suppressed by TGFβ signaling (22). Interestingly, tumor CM-induced upregulation of Il33 is almost completely ablated when combined with GolgiStop, which prevents fibroblast autocrine signaling; thus, a paracrine and an autocrine component are required for Il33 expression. We also found that tumor-instigated fibroblast STAT3 phosphorylation was dependent on fibroblast autocrine signaling, in accordance with existing reports in the literature (22). Importantly, direct STAT3 activation in fibroblasts treated with GolgiStop did not rescue IL33 accumulation, indicating that additional autocrine pathways are required to activate IL33 upregulation. In other cell types, focal adhesion kinase activation has been implicated in IL33 expression (92, 93), and in PSCs, PDGF-BB and IFNγ can both cause IL33 upregulation (40).
Together, our findings shed light on the mechanisms through which cancer cells reprogram the stroma, particularly CAFs—mechanisms that could be targeted therapeutically to “normalize” fibroblasts in the setting of pancreatic cancer, as proposed (94). Our data demonstrate the rapid nature by which CAF polarization can shift due to the addition or loss of extracellular stimuli, further complicating potential efforts to establish sustained CAF reprograming as a therapeutic approach. Importantly, our use of the pharmacologic KRASG12D inhibitor, MRTX1133, showed a reduction of CAF IL33 expression that mirrored the genetic ablation of oncogenic KRAS, highlighting a potential synergistic effect whereby the TME may be reprogrammed in response to targeting cancer cells—a highly relevant observation, as KRASG12D inhibitors are currently entering the clinic (95, 96). Ultimately, targeting IL33 might provide a new avenue for combination therapy in pancreatic cancer.
Methods
Human Samples
Surgical specimens were obtained from patients referred for Whipple or distal pancreatectomy according to IRB HUM000025339. Written informed consent forms were obtained from the patients. Studies were approved by the Institutional Review Boards of the University of Michigan Medical School and were conducted in accordance with recognized ethical guidelines.
Mice
Mice were housed in the specific pathogen-free animal facility at the Rogel Cancer Center at the University of Michigan. Animals were overseen by the University of Michigan Unit for Laboratory Animal Medicine. Male and female age-matched mice were utilized for experiments, and littermates were used whenever possible. Mouse studies were approved by the Institutional Animal Care and Use Committee at the University of Michigan (protocol #PRO00011612).
iKRASG12D [Ptf1a (aka p48)-Cre;TetO-KrasG12D;Rosa26rtTa/+] mice were generated as previously described (7, 12). KRASG12D expression in the pancreatic epithelium was induced in adult mice (8–14 weeks of age) by supplementing 5% sucrose water with doxycycline (0.2 g/L, #D9891, Sigma-Aldrich). After maintaining mice on doxycycline water for 3 to 4 days, acute pancreatitis was induced via intraperitoneal injection of caerulein (75 μg/kg, #C9026, Sigma-Aldrich) 8 times a day for 2 days. Mice were then aged for 3 or 5 weeks after the last day of caerulein, at which point a cohort of mice was randomly selected to switch from doxycycline water to regular water, thus terminating the expression of KRASG12D. After 3 additional days of doxycycline (“3- or 5-week KRASG12D ON”) or regular water (“3- or 5-week KRASG12D ON + 3-day KRASG12D OFF”), animals were sacrificed and pancreata were collected.
iKRASG12D;Trp53R172H/+ (Ptf1a-Cre;TetO-KrasG12D;Rosa26rtTa/+;Trp53R172H/+) mice were generated as previously described (55). Expression of KRASG12D in this model was induced by providing doxycycline chow (1.0 gm/kg, #F3949, Bio-Serv). Mice were aged to 14 weeks and then harvested (“14-week KRASG12D ON”) or remained on doxycycline chow to 15 weeks before being switched back to a regular diet for an additional week before harvesting pancreata (“15-week KRASG12D ON + 1-week KRASG12D OFF”).
Pdgfra-CreER T2/+ (#032770, Jackson Laboratory, mixed background at time of utilization) mice were crossed with either Il33f/f-eGFP mice (#030619, Jackson Laboratory, C57BL/6J) or Stat3flox mice (#016923, Jackson Laboratory, C57BL/6J) to generate Pdgfra-CreERT2/+;Il33f/f-eGFP mice and Pdgfra-CreERT2/+;Stat3f/f mice, respectively. These mice were aged 8 to 12 weeks and then treated with tamoxifen (4 mg/day, #T5648, Sigma-Aldrich) dissolved in corn oil via oral gavage once a day for 5 days. Two days after their last gavage, mice underwent orthotopic tumor implantation surgery. Experimental and control animals used in Pdgfra-CreERT2/+;Il33f/f-eGFP experiments were additionally switched to tamoxifen chow (400 mg/kg, #TD.130860, Teklad Custom Diets) immediately following the last administration of tamoxifen gavage and remained on tamoxifen chow until experiment end (except in the case of animals intended for scRNA-seq, which received standard chow diet).
Il1rl1 −/− mice were originally generated by Dr. Shizuo Akira’s lab and backcrossed to C57BL/6 by Dr. Stefan Writz’s group (97, 98). Mice were aged to 8 weeks old before undergoing orthotopic tumor implantation surgery. Il1rl1+/+ C57BL/6 mice were used as controls.
Murine Cell Lines
All cell lines were maintained in DMEM (#11965, Gibco) + 10% heat-inactivated FBS (#A3840202, Gibco) + 1% penicillin–streptomycin (PS; #15070063, Gibco) at 37°C and 5% CO2. Cells were routinely tested for mycoplasma using the MycoAlert PLUS Mycoplasma Detection Kit (#LT07-710, Lonza), and cell lines used in orthotopic surgeries tested negatively prior to injection.
The iKRASG12D;Trp53R172H/+ cell lines A9993 (aka “iKRASG12DF1,” FVB/N, female) and 9805 (mixed background, male) were previously derived from spontaneous pancreatic tumors (7, 55). Both cell lines were maintained in media supplemented with doxycycline (1 μg/mL, #D9891, Sigma-Aldrich). Only passage 17 and below were utilized to ensure doxycycline dependence in these cell lines. The male KPC (Pdx-Cre;LSL-KrasG12D;LSL-Trp53R172H) cell line 7940b (C57BL/6J) was provided by Dr. Gregory Beatty (48). The fibroblast cell line CD1WT was previously derived from a healthy murine pancreas (25).
To generate IL33 wildtype (“IL33 WT”) and knockout (“IL33 KO”) cell lines, primary fibroblasts were isolated under sterile conditions from the pancreas of a healthy male Il33f/f-eGFP mouse using the outgrowth method (99). Fibroblasts were established and infected with adenovirus (Ad5 CMV-eGFP for IL33 WT lines or Ad5 CMV-eGFP and Ad5 CMV-Cre for IL33 KO lines) as previously described (100). Successfully infected cells were selected by flow cytometry and assessed at the genomic and protein levels for recombination efficiency.
Orthotopic Surgeries
To establish orthotopic KPC tumors, 7940b cells (passage 16 or lower) were injected into the pancreata of adult mice (7.5 × 104 cells/animal for the Pdgfra-CreERT2/+;Il33f/f-eGFP with tamoxifen chow model and 5 × 104 cells/animal for all other orthotopic experiments). Equivalent injections were given to control animals; controls used were C57BL/6 mice for the Il1rl1−/− experiment and Pdgfra-CreERT2/+ mice for the Pdgfra-CreERT2/+;Il33f/f-eGFP and Pdgfra-CreERT2/+;Stat3f/f experiments. All Pdgfra-CreERT2/+ controls were also given tamoxifen oral gavage/chow alongside experimental animals in accordance with the experimental design. 7940b cells were resuspended in a 1:1 ratio of RPMI 1640 medium (#11875, Gibco) supplemented with 10% FBS and Matrigel matrix basement membrane (#354234, Corning) at a concentration of 1,875 cells/μL (for 7.5 × 104 cells/animal; 40 μL/injection) or 1,000 cells/μL (for 5 × 104 cells/animal; 50 μL/injection; for full surgery procedure, see ref. 101). Tumors were allowed to grow for 3 weeks prior to harvest.
To establish the iKRASG12D orthotopic model, 5 × 104 A9993 cells were implanted into syngeneic FVB/N mice (1,000 cells/μL in a 1:1 ratio of RPMI 1640 medium supplemented with 10% FBS and Matrigel matrix basement membrane; ref. 55). Mice were provided doxycycline chow for 2 weeks to maintain KRASG12D expression, and then randomly selected to return to regular diet for an additional 3 days (KRASG12D “OFF”) before tumor harvest.
Histology and Immunohistochemistry
Murine tissues were incubated in 10 mL of 10% neutral-buffered formalin at room temperature (RT) for 24 hours before being submitted to the University of Michigan Tissue & Molecular Pathology Shared Resource core facility for paraffin embedding and sectioning. Gomori trichrome staining was performed following the manufacturer’s guidelines (#87021, Epredia).
For IHC staining, slides were deparaffinized and rehydrated by serial incubation in Xylene (5 minutes, two times), 100% ethanol (5 minutes, two times), 95% ethanol (1 minute, two times), and diH2O (5 minutes). Slides were then subjected to antigen retrieval with Antigen Retrieval Citra Solution (#HK086-9K, BioGenex) and microwaving as per the manufacturer’s instructions. Upon cooling, slides were quenched in 0.3% H2O2 in methanol for 15 minutes before blocking in 1% BSA (#BP1600-100, Fisher Scientific) in PBS for 30 minutes at RT. After blocking, slides were incubated in primary antibody diluted in PBS in a humidified chamber overnight at 4°C. Primary antibodies were used at the following concentrations: human IL33 (1:50, #AF3625, R&D Systems), Cleaved Caspase 3 (1:100, #9664L, Cell Signaling), F4/80 (1:200, #70076, Cell Signaling), and myeloperoxidase (1:200, #AF3667, R&D Systems). The next day, slides were incubated in a biotinylated secondary antibody diluted 1:300 in PBS for 45 minutes at RT. Vectastain Elite ABC Kit, Peroxidase (PK6100, Vector Laboratories) complex was then added following the manufacturer’s protocol, and slides were developed using the DAB (3,3′-diaminobenzidine) Substrate Kit, horseradish peroxidase without nickel (#SK4100, Vector Laboratories). Next, slides were counterstained in hematoxylin (#26381-02, Electron Microscopy Services) for 2 minutes and then dehydrated by serial incubation in 95% ethanol (10 seconds, two times), 100% ethanol (2 minutes, two times), and Xylene (2 minutes, two times). The resulting slides were mounted with Cytoseal (#23-244256, Epredia).
Immunofluorescence
For immunofluorescent (IF) stains, slides were deparaffinized and rehydrated as for IHC. Antigen retrieval was done by microwaving and Antigen Retrieval Citra Solution as previously described, or EDTA Unmasking Solution (#14747, Cell Signaling, following manufacturer’s recommendations) when staining for pSTAT3 and/or IL33 primary antibodies. Upon cooling, slides were blocked in 1% BSA in PBS for 30 minutes (or 10% goat serum if IL33 primary antibody was used) and then incubated in primary antibody overnight in a humidified chamber at 4°C. Primary antibodies were used at the following concentrations: IL33 (1:50, #AF3625, R&D Systems), PDGFRα/β (1:100, #AB32570, Abcam), E-Cadherin (1:50, #14472S, Cell Signaling), pSTAT3 (Y705; 1:100, #9145S, Cell Signaling), Ki67 (1:100, #AB15580, Abcam), α-SMA (1:1,000, #A2547, Sigma-Aldrich), CD8 (1:400, #98941, Cell Signaling), Granzyme-B (1:500, #17215, Cell Signaling), CD4 (1:100, #25229, Cell Signaling), CD44 (1:100, #MA1-10225, Invitrogen), and Foxp3 (1:100, #12653, Cell Signaling). The following day, nuclei were stained by incubating slides in DAPI (4′,6-diamidino-2-phenylindole, dihydrochloride; 1:30,000 in PBS, #D1306, Invitrogen) for 10 minutes in the dark at RT. Secondary antibody incubation was subsequently done at 1:300 in PBS for 45 minutes at RT in the dark. Slides were mounted with Prolong Gold Antifade Mountant (#P36934, Invitrogen). The Tyramide SuperBoost Kit (#B40922, #B40912, or #B40923, Invitrogen) was used when staining with multiple primary antibodies raised in the same host species.
Imaging and Quantification
IHC and IF slides were imaged on an Olympus BX53F microscope outfitted with an Olympus DP80 digital camera and CellSens Standard software (Olympus). Select IF stains (those that included IL33 or pSTAT3 primary antibody) were imaged on the Leica Stellaris 8 Falcon Confocal Microscopy System using LAS X software (Leica Microsystems). Investigators were blind to the experimental condition of the slide when taking images, and at least three mice per group were assessed.
IHC and IF images were quantified using Fiji (102). To quantify % positive staining per field of view (FOV), three to eight images per slide (depending on the size of the tissue) were taken on the Olympus system at 20× magnification with care to capture only tissue area within the FOV. The fraction of stained area for each image was averaged within each mouse, and the resulting % positive scores are reported in the text (each datapoint is a single mouse). To quantify the % positive cells, E-Cadherin+ or PDGFRα/β+ cells (∼100–150 cells each per FOV) or CD4 cells (all cells per FOV) were manually categorized as Ki67/Foxp3 positive or negative; at least three images per mouse were quantified, and total % positive scores were averaged within each mouse and represented in the figure. To quantify the fluorescence intensity of IL33 and/or pSTAT3 within fibroblasts, three to five images were taken of the tissues on the Leica system using the 40× objective. Then, ≥30 regions of interest (ROI) composed of individual PDGFRα/β+ cells were hand-selected for each image (for images with fewer than 30 PDGFRα/β+ cells, all available PDGFRα/β+ cells were selected). Using the single channel image for the protein of interest, the integrated density was recorded for each ROI. The mean gray value of three background ROIs was also recorded. Corrected total cell fluorescence (CTCF) was then calculated for each ROI as CTCF = Integrated Density − (Area of ROI × Average Mean Gray Value of Background ROIs). The CTCF for each ROI is represented in the results section to profile the range and distribution of CTCF scores in each group. Colocalization of CD8/Granzyme-B and PDGFRα/β/α-SMA were measured using Just Another Colocalization Plugin in ImageJ (103).
Generation of Single Cell RNA Sequencing Datasets
We analyzed a number of scRNA-seq datasets previously published by our group and our collaborators, which are detailed in Supplementary Table S1. Here, we present a number of new murine scRNA-seq datasets including three KC PanIN datasets, seven iKRASG12D datasets, and the Pdgfra-CreERT2/+;Il33f/f orthotopic datasets [NCBI Gene Expression Omnibus (GEO) collection GSE269888]. The detailed GEO accession numbers for these datasets can also be found in Supplementary Table S1.
To prepare tissues for sequencing, samples were disrupted to single-cell suspension by mechanically mincing the tissue, followed by enzymatic digestion with Collagenase V (1 mg/mL in RPMI, #C9263, Sigma-Aldrich) at 37°C for 30 minutes under constant shaking. After digestion, suspensions underwent serial filtration through 500-, 100-, and 40-μm mesh cell strainers. We then enriched live cells using the MACS Dead Cell Removal Kit (#130-090-101, Miltenyi Biotec). The resulting single-cell suspensions were brought to the University of Michigan Advanced Genomics Core for cDNA library preparation and sequencing. Samples were run using 50-cycle paired-end reads on the HiSeq 4000 (Illumina) or NovaSeq 6000 (Illumina) to a depth of 100,000 reads. The University of Michigan Advanced Genomics Core also performed processing and alignment of the raw data using Cell Ranger (versions 2.2.0–6.1.2 depending on the date of sequencing) with default settings and an initial expected cell count of 10,000. Details on the sequencing and alignment method for each sample can be found on NCBI GEO.
Single cell RNA Sequencing Analysis
Downstream analysis of the scRNA-seq data was done using R version 4.3.0 “Already Tomorrow,” R Studio version 2023.03.1 + 446, and R package Seurat version 4.3.0.1 (104). We excluded genes that appeared in <3 cells, as well as cells with <100 genes, <1,000 or >100,000 transcripts, or >15% mitochondrial gene composition. Genes were normalized using NormalizeData with the “LogNormalize” method and a scale factor of 10,000. Variable genes were identified using the FindVariableFeatures function. When scRNA-seq datasets from differing run dates were merged, batch correction was performed using the IntegrateData workflow in Seurat (105). All genes were scaled and centered using ScaleData. Principal component analysis (PCA) was performed using RunPCA. Cell clusters were identified using FindNeighbors and FindClusters with dimensions that captured ∼90% variance as defined by the PCA result. UMAP clustering was completed using RunUMAP. To compare gene expression profiles between groups, we used FindMarkers and Wilcoxon rank sum test with Bonferroni correction. In addition to Seurat, the R package scCustomize version 1.1.1 (RRID:SCR_024675) was used for visualization. Pathway analysis was performed with the R package fgsea version 1.26.0 (bioRxiv 060012) and the MSigDB mouse-ortholog hallmark gene sets were downloaded using the R package msigdbr version 7.5.1 (RRID:SCR_022870). Pathway enrichment scores were also calculated using Seurat’s AddModuleScore functionality.
To infer ligand–receptor interactions, we used the R package CellChat version 1.6.1 (106) following their standard workflow. Differential gene expression via Wilcoxon rank sum test with Bonferroni correction was performed using identifyOverExpressedGenes with thresh.pc = 0.1 (minimum 10% of cells expressing the given gene in the cluster), tresh.fc = 0.1 (log fold-change minimum), and thresh. P = 0.05 (P value threshold). For final visualization, ligands with less than 0.25% fold-change were removed. Interactions between the ligand-expressing cluster and the receptor-expressing cell populations were visualized using netVisual_chord_gene with slot.name = “netP” and then manually annotated to include the relevant gene names found in slot.name = “net.”
The complete R script encompassing the analysis and visualization for each dataset presented in the manuscript is publicly available on GitHub (https://github.com/PascaDiMagliano-Lab/).
Tumor Conditioned Media Generation
The 9805 cell line was plated at 1 × 106 cells per 10-cm dish in DMEM + 10% FBS + 1% PS + doxycycline (1 μg/mL). The cells were allowed to adhere overnight, and then, the plates were washed with PBS and replenished with DMEM + 10% FBS + 1% PS with or without doxycycline (1 μg/mL; establishing “KRASG12D ON” and “KRASG12D OFF” conditions, respectively). The cells were again left overnight to transition between “KRASG12D ON” and “KRASG12D OFF” status. The following day, the cells were washed with PBS and replenished with low-serum DMEM + 1% FBS + 1% PS with or without doxycycline. After 48 hours, the resulting CM was centrifuged at 1,000 g for 10 minutes at 4°C to pellet debris. The supernatant was decanted, and then, the CM was stored at either 4°C (for up to 1 week) or −80°C (for up to 3 months) before use. Samples of “KRASG12D ON” and “KRASG12D OFF” CM were submitted to the Michigan Diabetes Research Center Clinical Core to measure extracellular LIF, IL6, IL1α, TNFα, and IL1β using MILLIPLEX (MilliporeSigma) and the Luminex 200 Instrument System.
iKRASG12D JAK1/2i In vivo
For JAK1/2i treatment, iKRASG12D mice 8 to 14 weeks of age were given doxycycline chow for 4 days and then received caerulein-induced acute pancreatitis as previously described. Three weeks following the last dose of caerulein, mice were randomized and received either vehicle or the JAK1/2i ruxolitinib (#INCB18424, MedChemExpress) via oral gavage once a day for 4 days (180 mg/kg in 100 μL of 10% DMSO in corn oil). Mice were harvested the same day following the last dose of ruxolitinib.
Pharmacologic Inhibition of Pancreatic Fibroblasts
CD1WT were plated at 3 × 105 cells per well in six-well dishes with DMEM + 10% FBS + 1% PS. The next day, cells were washed with PBS and treated with 2 mL of fresh, low-serum media (DMEM + 1% FBS + 1% PS), 1 mL of “KRASG12D ON” or “KRASG12D OFF” CM + 1 mL of low-serum media, or a combination of 1 mL of “KRASG12D ON” CM + 1 mL of low-serum media + either JAK1/2i (ruxolitinib) or TGFβRi (Galunisertib, #HY13226, MedChemExpress) at 0.03, 0.3, or 3 μmol/L. DMSO and doxycycline were given as vehicle controls for JAK1/2i/TGFβRi and “KRASG12D ON” CM, respectively. Cells were harvested 24 hours after treatment.
In the shortened adaptation of the JAK1/2i experiment, CD1WT were plated as described and then treated with 2 mL of low-serum DMEM + doxycycline vehicle or 1 mL of “KRASG12D ON” CM + 1 mL of low-serum media. 20 hours following treatment, JAK1/2i or an equivalent volume of DMSO was added directly to the well to reach a total concentration of 0.3 μmol/L. CD1WT were harvested 4 hours following JAK1/2i treatment.
rIL6, rLIF, rIL1α, rTNFα, and rTGF-β Treatment
CD1WT were plated at 3 × 105 cells per well in six-well dishes with DMEM + 10% FBS + 1% PS. The following day, cells were washed with PBS and treated with either 2 mL of low-serum DMEM + doxycycline vehicle, 1 mL of “KRASG12D ON” CM + 1 mL of low-serum DMEM, or 2 mL of low-serum DMEM + doxycycline vehicle + rIL6 (0.03–30 ng/mL, #406-ML, R&D Systems), rLIF (0.06–6 ng/mL, #8878-LF, R&D Systems), or rTGFβ (2 or 20 ng/mL, #7666-MB, R&D Systems). Cells were harvested 24 hours after treatment. For rIL1α (20 ng/mL, #400-ML, R&D Systems) and rTNFα (20 ng/mL, #410-MT, R&D Systems) experiments, three additional wells were given 2 mL of low-serum DMEM + doxycycline vehicle in parallel with the untreated control group to serve as the 1-hour treatment wells. One hour before harvest, these wells were given 1 mL of “KRASG12D ON” CM + 1 mL of low-serum media or 2 mL of rIL1α (20 ng/mL) or rTNFα (20 ng/mL) + doxycycline vehicle and collected simultaneously with the 2-hour treatment group.
Tumor Conditioned Media Time Course
CD1WT were plated at 1.5 × 105 cells per well in six-well dishes with DMEM + 10% FBS + 1% PS. The following day, cells were washed with PBS, then one well was switched to 1 mL of “KRASG12D ON” CM + 1 mL of low-serum DMEM and another was switched to 1 mL of “KRASG12D OFF” CM + 1 mL of low-serum DMEM (this was the 24-hour timepoint for western blot collection series and the 48-hour timepoint for the RNA collection series). All other wells were given 2 mL of low-serum DMEM + doxycycline. Cells were then treated in descending order over time to allow all cells to be harvested simultaneously. The 0-hour timepoint never received CM.
Autocrine Blocking of Pancreatic Fibroblasts
CD1WT were plated at 3 × 105 cells per well in six-well dishes with DMEM + 10% FBS + 1% PS. The next day, cells were washed with PBS and treated with either 2 mL of fresh, low-serum DMEM, 1 mL of “KRASG12D ON” CM + 1 mL of low-serum DMEM, or 2 mL of low-serum DMEM supplemented with 6-ng/mL rLIF (doxycycline was given as a vehicle control to all wells not receiving “KRASG12D ON” CM). Two identical sets of CD1WT were treated under these conditions in parallel (groups 1 and 2). Cells were incubated for 18 hours, at which point the resulting CD1WT CM was removed by pipetting and set aside, and plates were washed with PBS. Immediately following the wash, the fibroblast CM was added back to the well it originated from for the wells in group 1. For the wells in group 2, we instead added fresh aliquots of low-serum DMEM + doxycycline (2 mL), 1 mL of “KRASG12D ON” CM + 1 mL of low-serum DMEM, or 2 mL of low-serum DMEM + doxycycline supplemented with 6-ng/mL rLIF. Group 2 cells also received the protein transport inhibitor GolgiStop (1.3 μL/2 mL, #554724, BD Biosciences). All cells were incubated for an additional 6 hours and then harvested for RNA or protein.
IL33 Secretion
CD1WT were plated 2 × 105 cells per well into four wells of a six-well plate in triplicate and left overnight. The following day, two wells were provided 2 mL of fresh, low-serum DMEM + doxycycline and two wells received 1 mL of “KRASG12D ON” CM + 1 mL of low-serum DMEM. After 24 hours of incubation, we administered 100-μmol/L H2O2 to one control and one “KRASG12D ON” CM well by pipetting directly onto the plate. The cells were incubated for an additional 24 hours, and then, media was collected. Media from three wells were pooled (∼6 mL at the time of collection), centrifuged at 1,000 g for 10 minutes at 4°C to pellet any debris, and then concentrated using Amicon Ultra 4 Centrifugal Filters (#UFC801024, MilliporeSigma) as per the manufacturer’s directions. The resulting concentrated CD1WT CM was submitted for IL33 ELISA at the Rogel Cancer Center Immunology Core, alongside samples of plain “KRASG12D ON” CM, and low-serum DMEM to serve as controls.
Isolation and Culture of Primary Human PDA Organoids and Fibroblasts
Adjacent normal fibroblasts, tumor fibroblasts, and tumor organoids were generated from resected patient tumors. Tissue was digested as described previously (107). To culture organoids, cells were plated in growth factor-reduced Matrigel matrix basement membrane and grown in human complete feeding media [50% L-WRN CM (108) and 50% DMEM/F12 (#12634010, ThermoFisher) supplemented with B27 (#17504044, ThermoFisher) to 1× final concentration, 10-mmol/L nicotinamide (#N0636, Sigma-Aldrich), 1.25-mmol/L N-acetyl-cysteine (#A9165, Sigma-Aldrich), 50-ng/mL hEGF (#236-EG, R&D Systems), 100-ng/mL hFGF (#345-FG, R&D Systems), 10-nmol/L hGastrin1 (#3006, Tocris Bioscience), 10.5-μmol/L Y27632 (#1254, Tocris Bioscience), and 2× PS (107, 109)]. To culture fibroblasts, cells were grown in fibroblast media (DMEM-F12 + 10% FBS + 1× PS). All primary cell cultures were characterized by whole exome sequencing as described previously (107).
Human Fibroblast Treatment with Organoid CM and JAKi
Tumor organoids (cell line 61620400) were seeded at 50% confluence and cultured in human complete feeding media; organoid CM was collected after 72 hours. Adjacent normal (cell line 19809) and tumor fibroblasts (cell lines 1249 and 21498) were cultured with 100% fibroblast media or 50% organoid CM and 50% fibroblast media in the presence or absence of 0.5-μmol/L JAK1/2i (ruxolitinib) for 24 hours before harvesting total RNA. DMSO was used as a vehicle control when JAK1/2i was not provided.
Cell Proliferation Assay
“IL33 WT” and “IL33 KO” cell lines were seeded at 3 × 103 cells per well in DMEM + 10% FBS + 1% PS in four opaque-walled 96-well plates and left to adhere overnight. The following day, cell density was measured on 1 plate (day 0); the remaining plates were aspirated and washed with PBS, and cells were treated with low-serum DMEM or a 1:1 ratio of low-serum DMEM and 9805 “KRASG12D ON” tumor CM. Conditions without tumor CM were given doxycycline as a control. Plates were collected and read every 24 hours for 3 days. Proliferation was measured using CellTiter-Glo Luminescent Cell Viability Assay (#G7570, Promega) following the manufacturer’s instructions. Two wells per condition were averaged for each measurement.
Pharmacologic Inhibition of KRAS in Murine Tumor Cells
The 9805 tumor cell line was grown as described above for the generation of tumor CM; briefly, cells were plated in doxycycline-containing DMEM + 10% FBS + 1% PS in 10 cm dishes (day 1; d1) and then, the following day, washed with PBS and switched to doxycycline-deficient low-serum DMEM (for “KRASG12D OFF” CM) or doxycycline-proficient low-serum DMEM (for “KRASG12D ON” CM; d2). On d3, cells were again washed and replenished with low-serum DMEM (for “KRASG12D OFF” CM), low-serum DMEM + doxycycline (for “KRASG12D ON” CM), or low-serum DMEM + doxycycline + small molecule inhibitor: MRTX1133 (0.5 μmol/L, #HY134813, MedChemExpress), Sotorasib (0.5 μmol/L, #HY114277, MedChemExpress), or Trametinib (50 nmol/L, #S2673, Selleck Chemicals). On d4, inhibitors were again spiked into the media at equivalent concentrations to ensure continued inhibition of KRAS. Tumor CM was collected as described above on d5. DMSO was used as a vehicle control for the untreated conditions.
CD1WT cells were plated at 3 × 105 cells per well in six-well dishes with DMEM + 10% FBS + 1% PS. The next day, cells were washed with PBS and treated with 2 mL of fresh, low-serum media (DMEM + 1% FBS + 1% PS) or 1 mL of low-serum media + 1 mL of the various tumor CMs. Doxycycline was given as vehicle control for “KRASG12D ON” CM. Cells were harvested for RNA 24 hours after treatment.
Western Blotting
Cells were lysed in their culture dish in RIPA buffer (#R0278, Sigma-Aldrich) supplemented with protease and phosphatase inhibitor cocktail (#87785, Sigma-Aldrich). Protein concentration was quantified using the Pierce BCA Protein Assay Kit (#23225, ThermoScientific) following the manufacturer’s protocol. Equal concentrations of protein were separated via SDS-PAGE and transferred to polyvinylidene difluoride membranes using a wet tank system (Bio-Rad). Membranes were rinsed in water and then allowed to dry completely before being reactivated in methanol and stained using Ponceau-S (#P7170, Sigma-Aldrich). Membranes were then blocked at RT with gentle shaking for 1 hour in 5% milk in TBST [1× TBS (#1706435, Bio-Rad) + 0.1% Tween-20 (#P7949, Sigma-Aldrich)]. After blocking, membranes were incubated in primary antibody diluted either in 5% milk in TBST (for total protein targets) or 5% BSA in TBST (for phosphorylation site targets) at 4°C with gentle rocking. Primary antibodies were used at the following concentrations: mouse IL33 (1:100, #AF3625, R&D Systems), PDGFRα (1:500, #3164S, Cell Signaling), pSTAT3 (Y705; 1:1,000, #9145S, Cell Signaling), STAT3 (1:1,000, #9139S, Cell Signaling), p-SMAD2 (S465/467)/SMAD3 (S423/425; 1:500, #8828S, Cell Signaling), SMAD2/3 (1:1,000, #8685S, Cell Signaling), α-tubulin (1:2,000, #3873S, Cell Signaling), vinculin (1:2,000, #13901S, Cell Signaling). Membranes were incubated in primary antibody for 24 to 48 hours, then washed with TBST, and incubated in HRP-conjugated secondary antibody diluted in 5% milk in TBST for 2 hours at RT. Bands were visualized using Clarity Western ECL Substrate (#1705060, Bio-Rad) and the ChemiDoc Imaging System (Bio-Rad). Densitometry was performed in Fiji.
Selection of PDGFRα+ Cells
To harvest PDGFRα+ cells from Pdgfra-CreERT2/+;Il33f/f orthotopic tumors for protein, single-cell suspensions were created as described above, and cells were labeled and subjected to CD140a (PDGFRα) magnetic selection as per the manufacturer’s instructions (#130-101-502, Miltenyi Biotec). The resulting PDGFRα+ cells were pelleted and lysed in RIPA + protease and phosphatase inhibitor cocktail for western blot.
To harvest PDGFRα+ cells from Pdgfra-CreERT2/+;Stat3f/f orthotopic tumors for RNA, tumors were processed into single-cell suspensions and cells were stained for 30 minutes with antimouse PE-CD140a (PDGFRa; #135905, BioLegend), FITC-CD31 (#102405, BioLegend), FITC-CD326 (#1182010, BioLegend), and FITC-CD45 (#157214, BioLegend). Fibroblasts were identified as PDGFRa+, CD31−, CD326−, and CD45−; sorted using Sony SH800 Cell Sorter; and processed for RNA isolation.
RNA Extraction and RT-qPCR
RNA was extracted using the RNeasy Plus Mini Kit (#74134, QIAGEN) following the manufacturer’s protocol. RNA levels and quality were assessed via nanodrop, and cDNA was generated using the High-capacity cDNA Reverse Transcription Kit (#4368814, Applied Biosystems) supplemented with RNAse Inhibitor (#N8080119, Applied Biosystems). Samples were prepared for qPCR using Fast SYBR Green Master Mix (#4385612, Applied Biosystems). Primers are listed in the Supplementary Table S2. Reactions were run on a QuantStudio 6 Pro (Applied Biosystems). Cyclophilin A (Ppia) and GAPDH were used as the housekeeping genes in all mouse and human RT-qPCR experiments, respectively.
Bulk RNA Sequencing
Extracted RNA was submitted to the University of Michigan Advanced Genomics Core for bulk RNA-seq and preprocessing. Samples were subjected to 151-bp paired-end sequencing according to the manufacturer’s protocol (Illumina NovaSeq 6000). BCL Convert Conversion Software version 3.9.3 (Illumina) was used to generate demultiplexed Fastq files. Quality control was performed using FastQC version 0.11.8 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Reads were mapped to the reference genome GRCm38 (ENSEMBL) using STAR version 2.7.8a (110) and assigned count estimated to genes with RSEM version 1.3.3 (111). Alignment options followed ENCODE standards for RNA-seq. Downstream analysis was performed using DESeq2 version 1.40.2 (112). Differentially expressed genes were defined by an FDR of 0.05 and fold changes of 0.5 or more. The MSigDB mouse-ortholog hallmark gene sets were downloaded using the R package msigdbr version 7.5.1 (RRID:SCR_022870). Pathway analysis was performed using the R package fgsea version 1.26.0 (bioRxiv 060012). The raw data from this experiment are available on NCBI GEO accession number GSE269889.
Quantification and Statistical Analysis
GraphPad Prism 10 software was used for all statistical analyses outside of scRNA-seq experiments. Histograms are presented as mean ± standard deviation. Two-tailed Student t test and one-way ANOVA with Tukey test were performed as described in the figure legend. All in vitro assays were performed at least three times with cells of different passage numbers to ensure biologic variability. Outliers were detected using Grubbs’ test by GraphPad Prism and excluded where appropriate. To compare gene expression profiles between groups using scRNA-seq data, we used a Wilcoxon rank sum test with Bonferroni correction. Exact P values for each comparison can be found within the figures or figure legends, with P < 0.05 considered significant.
Data availability
The scRNA-seq datasets generated in this study are publicly available at NCBI GEO accession number GSE269888. Preexisting scRNA-seq data analyzed in this study are available at the accession numbers listed in Supplementary Table S1. Bulk RNA-seq data generated in this study are also available at NCBI GEO GSE269889.
Supplementary Material
Table S1. Single cell RNA sequencing datasets.
Table S2. Primer sequences used in qRT-PCR
Figure S1: IL-33+ stromal cells are abundant in human and mouse PDA.
Figure S2: Stromal IL-33 promotes PDA growth.
Figure S3: Pancreatic fibroblasts secrete IL-33 in response to oxidative stress.
Figure S4: Loss of stromal IL-33 alters the ST2+ immune cell secretome, resulting in a shift in CAF differentiation.
Figure S5: Inactivation of stromal IL-33 enables cytotoxic T cell activity.
Figure S6: Expression of fibroblast IL-33 is extrinsically induced by epithelial KrasG12D and requires JAK1/2-STAT3 activation throughout tumorigenesis.
Figure S7: Tumor cell-initiated autocrine signaling drives IL-33 upregulation in pancreatic fibroblasts.
Acknowledgments
The authors would like to thank Dr. Gregory Beatty (University of Pennsylvania) for generously sharing the murine pancreatic cancer cell line 7940b. We would like to thank Dr. Nicholas Lukacs (University of Michigan) for providing the Il1rl1−/− mouse model. The Ptf1a-Cre mouse was generously gifted by Dr. Chris Wright (Vanderbilt University). We thank Lee Olsen for her editing services as well as Michael Mattea and Christopher Strayhorn for providing histopathology support. We thank Matthew Felten for his assistance with the submission of sequencing data to NCBI GEO. Moreover, we would like to acknowledge technical support from the University of Michigan Advanced Genomics Core, Michigan Diabetes Research Center Clinical Core, Rogel Cancer Center Immunology Core, Rogel Cancer Center Tissue and Molecular Pathology Shared Resource, and University of Michigan Microscopy Core. Funding: K.L. Donahue was supported by NIH/NCI F31-CA265085. K.L. Donahue and E.L. Lasse Opsahl were funded by the NIH/NCI Cancer Biology Training Program T32-CA009676. Work in the M. Pasca di Magliano laboratory was supported by NIH/NCI grant U54CA274371. M. Pasca di Magliano and F. Bednar were supported by NIH/NCI R01-CA271510. Dr. M. Pasca di Magliano and Dr. B. Allen were also supported by NIH/NCI R01-CA275182. M. Pasca di Magliano and T.L. Frankel were supported by NIH/NCI R01-CA268426. T.L. Frankel was also supported by VA BLR&D Merit Award 5I01BX005777, NIH/National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) 5R01DK128102, and NIH/NCI U01CA274154. Moreover, F. Bednar was supported by the Association of Academic Surgery, Association of VA Surgeons, and American Surgical Association Foundation. P. Kadiyala was supported by the NIH/National Institute of Allergy and Infectious Diseases Research Training Grant in Experimental Immunology T32-AI007413 and NIH/National Institute of General Medical Sciences (NIGMS) Training Program in Translational Research T32-GM113900. W. Du, M.K. Scales, and R.E. Menjivar were supported by the NIH/Eunice Kennedy Shriver National Institute of Child Health & Human Development Training Program in Organogenesis T32-HD007505. Furthermore, M.K. Scales was supported by NIH/NCI F31-CA232655 and the Bradley Merrill Patten Memorial Scholarship. R.E. Menjivar was supported by the NIH/NIGMS Cellular and Molecular Biology Training Grant T32-GM007315 and the NIH/NCI F31-CA257533. A. Velez-Delgado was supported by the NIH/NIGMS Cellular Biotechnology Training Program T32-GM008353 and the NIH/NCI F31-CA247037. A.M. Elhossiny was supported by the Rackham International Student Fellowship. P.I. Medina-Cabrera, A. Velez-Delgado, and R.E. Menjivar were supported by the University of Michigan Rackham Merit Fellowship. P.I. Medina-Cabrera was supported by the Cancer Biology Graduate Program at the University of Michigan. B.D. Griffith was supported by the NIH/NCI Surgical Oncology Research Training Program T32-CA009672 and Society of University Surgeons Resident Research Award. A. Rao was supported by the NIH/NCI R37-CA214955-01A1 and CCSG Bioinformatics Shared Resource 5 P30 CA046592. J. Shi was supported by NIH/NCI R37CA262209. Finally, Y. Zhang was funded by NIH/NCI R50CA232985. E.S. Carpenter was supported by the VA BLR&D CDA IK2BX005875, American College of Gastroenterology CDA, and NIH/NIDDK T32-DK094775.
Footnotes
Note Supplementary data for this article are available at Cancer Discovery Online (http://cancerdiscovery.aacrjournals.org/).
Authors’ Disclosures
E.L. Lasse Opsahl reports grants from NIH/NCI during the conduct of the study. A. Rao reports grants from NIH, ACS, DoD, UM MIDAS, and UM MICDE; grants and other support from Agilent; and other support from ASI during the conduct of the study, as well as other support from Telperian, Inc., Tempus Inc., Voxel Analytics, LLC, and TCS Ltd. outside the submitted work. B. Allen reports grants from NIH during the conduct of the study. J. Shi reports grants from NIH during the conduct of the study. E.S. Carpenter reports grants from Cornerstone Pharmaceutical and nonfinancial support from Beigene outside the submitted work. No disclosures were reported by the other authors.
References
- 1. Siegel RL, Giaquinto AN, Jemal A. Cancer statistics, 2024. CA Cancer J Clin 2024;74:12–49. [DOI] [PubMed] [Google Scholar]
- 2. Sarantis P, Koustas E, Papadimitropoulou A, Papavassiliou AG, Karamouzis MV. Pancreatic ductal adenocarcinoma: treatment hurdles, tumor microenvironment and immunotherapy. World J Gastrointest Oncol 2020;12:173–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Witkiewicz AK, McMillan EA, Balaji U, Baek G, Lin WC, Mansour J, et al. Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nat Commun 2015;6:6744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Buscail L, Bournet B, Cordelier P. Role of oncogenic KRAS in the diagnosis, prognosis and treatment of pancreatic cancer. Nat Rev Gastroenterol Hepatol 2020;17:153–68. [DOI] [PubMed] [Google Scholar]
- 5. Aguirre AJ, Bardeesy N, Sinha M, Lopez L, Tuveson DA, Horner J, et al. Activated Kras and Ink4a/Arf deficiency cooperate to produce metastatic pancreatic ductal adenocarcinoma. Genes Dev 2003;17:3112–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hingorani SR, Petricoin EF, Maitra A, Rajapakse V, King C, Jacobetz MA., et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell 2003;4:437–50. [DOI] [PubMed] [Google Scholar]
- 7. Collins MA, Bednar F, Zhang Y, Brisset JC, Galban S, Galban CJ, et al. Oncogenic Kras is required for both the initiation and maintenance of pancreatic cancer in mice. J Clin Invest 2012;122:639–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ying H, Kimmelman AC, Lyssiotis CA, Hua S, Chu GC, Fletcher-Sananikone E, et al. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell 2012;149:656–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sherman MH, Beatty GL. Tumor microenvironment in pancreatic cancer pathogenesis and therapeutic resistance. Annu Rev Pathol 2023;18:123–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ying H, Dey P, Yao W, Kimmelman AC, Draetta GF, Maitra A., et al. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev 2016;30:355–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Steele NG, Biffi G, Kemp SB, Zhang Y, Drouillard D, Syu L, et al. Inhibition of Hedgehog signaling alters fibroblast composition in pancreatic cancer. Clin Cancer Res 2021;27:2023–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Velez-Delgado A, Donahue KL, Brown KL, Du W, Irizarry-Negron V, Menjivar RE, et al. Extrinsic KRAS signaling shapes the pancreatic microenvironment through fibroblast reprogramming. Cell Mol Gastroenterol Hepatol 2022;13:1673–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang Y, Yan W, Mathew E, Kane KT, Brannon A III, Adoumie M, et al. Epithelial-myeloid cell crosstalk regulates acinar cell plasticity and pancreatic remodeling in mice. Elife 2017;6:e27388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hou P, Kapoor A, Zhang Q, Li J, Wu CJ, Li J, et al. Tumor microenvironment remodeling enables bypass of oncogenic KRAS dependency in pancreatic cancer. Cancer Discov 2020;10:1058–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dey P, Li J, Zhang J, Chaurasiya S, Strom A, Wang H, et al. Oncogenic KRAS-driven metabolic reprogramming in pancreatic cancer cells utilizes cytokines from the tumor microenvironment. Cancer Discov 2020;10:608–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Biffi G, Tuveson DA. Diversity and biology of cancer-associated fibroblasts. Physiol Rev 2021;101:147–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ning W, Marti TM, Dorn P, Peng RW. Non-genetic adaptive resistance to KRASG12C inhibition: EMT is not the only culprit. Front Oncol 2022;12:1004669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Garcia PE, Adoumie M, Kim EC, Zhang Y, Scales MK, El-Tawil YS, et al. Differential contribution of pancreatic fibroblast subsets to the pancreatic cancer stroma. Cell Mol Gastroenterol Hepatol 2020;10:581–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Helms EJ, Berry MW, Chaw RC, DuFort CC, Sun D, Onate MK, et al. Mesenchymal lineage heterogeneity underlies nonredundant functions of pancreatic cancer-associated fibroblasts. Cancer Discov 2022;12:484–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang T, Ren Y, Yang P, Wang J, Zhou H. Cancer-associated fibroblasts in pancreatic ductal adenocarcinoma. Cell Death Dis 2022;13:897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sherman MH, Magliano MPD. Cancer-associated fibroblasts: lessons from pancreatic cancer. Annu Rev Cancer Biol 2023;7:43–55. [Google Scholar]
- 22. Biffi G, Oni TE, Spielman B, Hao Y, Elyada E, Park Y, et al. IL1-Induced JAK/STAT signaling is antagonized by TGFβ to shape CAF heterogeneity in pancreatic ductal adenocarcinoma. Cancer Discov 2019;9:282–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Elyada E, Bolisetty M, Laise P, Flynn WF, Courtois ET, Burkhart RA, et al. Cross-species single-cell analysis of pancreatic ductal adenocarcinoma reveals antigen-presenting cancer-associated fibroblasts. Cancer Discov 2019;9:1102–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Feig C, Jones JO, Kraman M, Wells RJ, Deonarine A, Chan DS, et al. Targeting CXCL12 from FAP-expressing carcinoma-associated fibroblasts synergizes with anti-PD-L1 immunotherapy in pancreatic cancer. Proc Natl Acad Sci U S A 2013;110:20212–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang Y, Yan W, Collins MA, Bednar F, Rakshit S, Zetter BR, et al. Interleukin-6 is required for pancreatic cancer progression by promoting MAPK signaling activation and oxidative stress resistance. Cancer Res 2013;73:6359–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hutton C, Heider F, Blanco-Gomez A, Banyard A, Kononov A, Zhang X, et al. Single-cell analysis defines a pancreatic fibroblast lineage that supports anti-tumor immunity. Cancer Cell 2021;39:1227–44.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Huang H, Wang Z, Zhang Y, Pradhan R, Ganguly D, Chandra R, et al. Mesothelial cell-derived antigen-presenting cancer-associated fibroblasts induce expansion of regulatory T cells in pancreatic cancer. Cancer Cell 2022;40:656–73.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. DeNardo DG, Ruffell B. Macrophages as regulators of tumour immunity and immunotherapy. Nat Rev Immunol 2019;19:369–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Djurec M, Graña O, Lee A, Troulé K, Espinet E, Cabras L, et al. Saa3 is a key mediator of the protumorigenic properties of cancer-associated fibroblasts in pancreatic tumors. Proc Natl Acad Sci U S A 2018;115:E1147–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fukuda A, Wang SC, Morris JPT, Folias AE, Liou A, Kim GE, et al. Stat3 and MMP7 contribute to pancreatic ductal adenocarcinoma initiation and progression. Cancer Cell 2011;19:441–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cayrol C, Girard J-P. Interleukin-33 (IL-33): a critical review of its biology and the mechanisms involved in its release as a potent extracellular cytokine. Cytokine 2022;156:155891. [DOI] [PubMed] [Google Scholar]
- 32. Kakkar R, Lee RT. The IL-33/ST2 pathway: therapeutic target and novel biomarker. Nat Rev Drug Discov 2008;7:827–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yeoh WJ, Vu VP, Krebs P. IL-33 biology in cancer: an update and future perspectives. Cytokine 2022;157:155961. [DOI] [PubMed] [Google Scholar]
- 34. Alonso-Curbelo D, Ho Y-J, Burdziak C, Maag JLV, Morris JPT IV, Chandwani R, et al. A gene-environment-induced epigenetic program initiates tumorigenesis. Nature 2021;590:642–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Alam A, Levanduski E, Denz P, Villavicencio HS, Bhatta M, Alhorebi L, et al. Fungal mycobiome drives IL-33 secretion and type 2 immunity in pancreatic cancer. Cancer Cell 2022;40:153–67.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Burdziak C, Alonso-Curbelo D, Walle T, Reyes J, Barriga FM, Haviv D, et al. Epigenetic plasticity cooperates with cell-cell interactions to direct pancreatic tumorigenesis. Science 2023;380:eadd5327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Moral JA, Leung J, Rojas LA, Ruan J, Zhao J, Sethna Z, et al. ILC2s amplify PD-1 blockade by activating tissue-specific cancer immunity. Nature 2020;579:130–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dixit A, Sarver A, Zettervall J, Huang H, Zheng K, Brekken RA, et al. Targeting TNF-α-producing macrophages activates antitumor immunity in pancreatic cancer via IL-33 signaling. JCI Insight 2022;7:e153242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zeng Q, Saghafinia S, Chryplewicz A, Fournier N, Christe L, Xie YQ, et al. Aberrant hyperexpression of the RNA binding protein FMRP in tumors mediates immune evasion. Science 2022;378:eabl7207. [DOI] [PubMed] [Google Scholar]
- 40. Masamune A, Watanabe T, Kikuta K, Satoh K, Kanno A, Shimosegawa T. Nuclear expression of interleukin-33 in pancreatic stellate cells. Am J Physiol Gastrointest Liver Physiol 2010;299:G821–32. [DOI] [PubMed] [Google Scholar]
- 41. Yang X, Chen J, Wang J, Ma S, Feng W, Wu Z, et al. Very-low-density lipoprotein receptor-enhanced lipid metabolism in pancreatic stellate cells promotes pancreatic fibrosis. Immunity 2022;55:1185–99.e8. [DOI] [PubMed] [Google Scholar]
- 42. Steele NG, Carpenter ES, Kemp SB, Sirihorachai VR, The S, Delrosario L, et al. Multimodal mapping of the tumor and peripheral blood immune landscape in human pancreatic cancer. Nat Cancer 2020;1:1097–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hosein AN, Huang H, Wang Z, Parmar K, Du W, Huang J, et al. Cellular heterogeneity during mouse pancreatic ductal adenocarcinoma progression at single-cell resolution. JCI Insight 2019;5:e129212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kemp SB, Carpenter ES, Steele NG, Donahue KL, Nwosu ZC, Pacheco A, et al. Apolipoprotein E promotes immune suppression in pancreatic cancer through NF-κB-Mediated production of CXCL1. Cancer Res 2021;81:4305–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Du W, Menjivar RE, Donahue KL, Kadiyala P, Velez-Delgado A, Brown KL, et al. WNT signaling in the tumor microenvironment promotes immunosuppression in murine pancreatic cancer. J Exp Med 2023;220:e20220503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jin G, Hong W, Guo Y, Bai Y, Chen B. Molecular mechanism of pancreatic stellate cells activation in chronic pancreatitis and pancreatic cancer. J Cancer 2020;11:1505–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Winkler EA, Bell RD, Zlokovic BV. Pericyte-specific expression of PDGF beta receptor in mouse models with normal and deficient PDGF beta receptor signaling. Mol Neurodegener 2010;5:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Long KB, Gladney WL, Tooker GM, Graham K, Fraietta JA, Beatty GL. IFNγ and CCL2 cooperate to redirect tumor-infiltrating monocytes to degrade fibrosis and enhance chemotherapy efficacy in pancreatic carcinoma. Cancer Discov 2016;6:400–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Loda A, Heard E. Xist RNA in action: past, present, and future. PLoS Genet 2019;15:e1008333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Brown CJ, Ballabio A, Rupert JL, Lafreniere RG, Grompe M, Tonlorenzi R, et al. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature 1991;349:38–44. [DOI] [PubMed] [Google Scholar]
- 51. Moffitt RA, Marayati R, Flate EL, Volmar KE, Loeza SGH, Hoadley KA, et al. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat Genet 2015;47:1168–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Katz-Kiriakos E, Steinberg DF, Kluender CE, Osorio OA, Newsom-Stewart C, Baronia A, et al. Epithelial IL-33 appropriates exosome trafficking for secretion in chronic airway disease. JCI Insight 2021;6:e136166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kouzaki H, Iijima K, Kobayashi T, O’Grady SM, Kita H. The danger signal, extracellular ATP, is a sensor for an airborne allergen and triggers IL-33 release and innate Th2-type responses. J Immunol 2011;186:4375–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Taniguchi S, Elhance A, Van Duzer A, Kumar S, Leitenberger JJ, Oshimori N. Tumor-initiating cells establish an IL-33-TGF-β niche signaling loop to promote cancer progression. Science 2020;369:eaay1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Collins MA, Brisset J-C, Zhang Y, Bednar F, Pierre J, Heist KA, et al. Metastatic pancreatic cancer is dependent on oncogenic Kras in mice. PLoS One 2012;7:e49707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mello AM, Ngodup T, Lee Y, Donahue KL, Li J, Rao A, et al. Hypoxia promotes an inflammatory phenotype of fibroblasts in pancreatic cancer. Oncogenesis 2022;11:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Martinez-Useros J, Li W, Cabeza-Morales M, Garcia-Foncillas J. Oxidative stress: a new target for pancreatic cancer prognosis and treatment. J Clin Med 2017;6:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hao X, Ren Y, Feng M, Wang Q, Wang Y. Metabolic reprogramming due to hypoxia in pancreatic cancer: implications for tumor formation, immunity, and more. Biomed Pharmacother 2021;141:111798. [DOI] [PubMed] [Google Scholar]
- 59. Abou Khouzam R, Lehn JM, Mayr H, Clavien PA, Wallace MB, Ducreux M, et al. Hypoxia, a targetable culprit to counter pancreatic cancer resistance to therapy. Cancers (Basel) 2023;15:1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Eissmann MF, Dijkstra C, Jarnicki A, Phesse T, Brunnberg J, Poh AR, et al. IL-33-mediated mast cell activation promotes gastric cancer through macrophage mobilization. Nat Commun 2019;10:2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Monticelli LA, Osborne LC, Noti M, Tran SV, Zaiss DM, Artis D. IL-33 promotes an innate immune pathway of intestinal tissue protection dependent on amphiregulin-EGFR interactions. Proc Natl Acad Sci U S A 2015;112:10762–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Li A, Herbst RH, Canner D, Schenkel JM, Smith OC, Kim JY, et al. IL-33 signaling alters regulatory T cell diversity in support of tumor development. Cell Rep 2019;29:2998–3008.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Mucciolo G, Araos Henríquez J, Jihad M, Pinto Teles S, Manansala JS, Li W, et al. EGFR-activated myofibroblasts promote metastasis of pancreatic cancer. Cancer Cell 2024;42:101–18.e11. [DOI] [PubMed] [Google Scholar]
- 64. Ali S, Mohs A, Thomas M, Klare J, Ross R, Schmitz ML, et al. The dual function cytokine IL-33 interacts with the transcription factor NF-κB to dampen NF-κB–Stimulated gene transcription. J Immunol 2011;187:1609–16. [DOI] [PubMed] [Google Scholar]
- 65. Park JH, Ameri AH, Dempsey KE, Conrad DN, Kem M, Mino-Kenudson M, et al. Nuclear IL-33/SMAD signaling axis promotes cancer development in chronic inflammation. EMBO J 2021;40:e106151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Gautier V, Cayrol C, Farache D, Roga S, Monsarrat B, Burlet-Schiltz O, et al. Extracellular IL-33 cytokine, but not endogenous nuclear IL-33, regulates protein expression in endothelial cells. Sci Rep 2016;6:34255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Travers J, Rochman M, Miracle CE, Habel JE, Brusilovsky M, Caldwell JM, et al. Chromatin regulates IL-33 release and extracellular cytokine activity. Nat Commun 2018;9:3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zha H, Wang X, Zhu Y, Chen D, Han X, Yang F, et al. Intracellular activation of complement C3 leads to PD-L1 antibody treatment resistance by modulating tumor-associated macrophages. Cancer Immunol Res 2019;7:193–207. [DOI] [PubMed] [Google Scholar]
- 69. Daubon T, Léon C, Clarke K, Andrique L, Salabert L, Darbo E, et al. Deciphering the complex role of thrombospondin-1 in glioblastoma development. Nat Commun 2019;10:1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Derynck R, Turley SJ, Akhurst RJ. TGFβ biology in cancer progression and immunotherapy. Nat Rev Clin Oncol 2021;18:9–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Zhao H, Chen Q, Alam A, Cui J, Suen KC, Soo AP, et al. The role of osteopontin in the progression of solid organ tumour. Cell Death Dis 2018;9:356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Sun Z, Schwenzer A, Rupp T, Murdamoothoo D, Vegliante R, Lefebvre O, et al. Tenascin-C promotes tumor cell migration and metastasis through integrin α9β1-mediated YAP inhibition. Cancer Res 2018;78:950–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Yang Y, Cao Y. The impact of VEGF on cancer metastasis and systemic disease. Semin Cancer Biol 2022;86:251–61. [DOI] [PubMed] [Google Scholar]
- 74. Calandra T, Roger T. Macrophage migration inhibitory factor: a regulator of innate immunity. Nat Rev Immunol 2003;3:791–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Bianchi A, De Castro Silva I, Deshpande NU, Singh S, Mehra S, Garrido VT, et al. Cell-autonomous Cxcl1 sustains tolerogenic circuitries and stromal inflammation via neutrophil-derived TNF in pancreatic cancer. Cancer Discov 2023;13:1428–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Das S, Shapiro B, Vucic EA, Vogt S, Bar-Sagi D. Tumor cell-derived IL1β promotes desmoplasia and immune suppression in pancreatic cancer. Cancer Res 2020;80:1088–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Peng DH, Rodriguez BL, Diao L, Chen L, Wang J, Byers LA, et al. Collagen promotes anti-PD-1/PD-L1 resistance in cancer through LAIR1-dependent CD8+ T cell exhaustion. Nat Commun 2020;11:4520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Tietze JK, Wilkins DE, Sckisel GD, Bouchlaka MN, Alderson KL, Weiss JM, et al. Delineation of antigen-specific and antigen-nonspecific CD8+ memory T-cell responses after cytokine-based cancer immunotherapy. Blood 2012;119:3073–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Zhang Y, Lazarus J, Steele NG, Yan W, Lee HJ, Nwosu ZC, et al. Regulatory T-cell depletion alters the tumor microenvironment and accelerates pancreatic carcinogenesis. Cancer Discov 2020;10:422–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Eickelberg O, Pansky A, Mussmann R, Bihl M, Tamm M, Hildebrand P, et al. Transforming growth factor-beta1 induces interleukin-6 expression via activating protein-1 consisting of JunD homodimers in primary human lung fibroblasts. J Biol Chem 1999;274:12933–8. [DOI] [PubMed] [Google Scholar]
- 81. Aoki H, Ohnishi H, Hama K, Shinozaki S, Kita H, Yamamoto H, et al. Existence of autocrine loop between interleukin-6 and transforming growth factor-beta1 in activated rat pancreatic stellate cells. J Cell Biochem 2006;99:221–8. [DOI] [PubMed] [Google Scholar]
- 82. Peñuelas S, Anido J, Prieto-Sánchez RM, Folch G, Barba I, Cuartas I, et al. TGF-beta increases glioma-initiating cell self-renewal through the induction of LIF in human glioblastoma. Cancer Cell 2009;15:315–27. [DOI] [PubMed] [Google Scholar]
- 83. Zhang F, Yan Y, Cao X, Guo C, Wang K, Lv S. TGF-β-driven LIF expression influences neutrophil extracellular traps (NETs) and contributes to peritoneal metastasis in gastric cancer. Cell Death Dis 2024;15:218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Shi Y, Gao W, Lytle NK, Huang P, Yuan X, Dann AM, et al. Targeting LIF-mediated paracrine interaction for pancreatic cancer therapy and monitoring. Nature 2019;569:131–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Halbrook CJ, Lyssiotis CA, Pasca di Magliano M, Maitra A. Pancreatic cancer: advances and challenges. Cell 2023;186:1729–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Helms E, Onate MK, Sherman MH. Fibroblast heterogeneity in the pancreatic tumor microenvironment. Cancer Discov 2020;10:648–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Fournié JJ, Poupot M. The pro-tumorigenic IL-33 involved in antitumor immunity: a yin and yang cytokine. Front Immunol 2018;9:2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Ardito CM, Grüner BM, Takeuchi KK, Lubeseder-Martellato C, Teichmann N, Mazur PK, et al. EGF receptor is required for KRAS-induced pancreatic tumorigenesis. Cancer Cell 2012;22:304–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Blasco MT, Navas C, Martín-Serrano G, Graña-Castro O, Lechuga CG, Martín-Díaz L, et al. Complete regression of advanced pancreatic ductal adenocarcinomas upon combined inhibition of EGFR and C-raf. Cancer Cell 2019;35:573–87.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Jiang W, Li X, Xiang C, Zhou W. Neutrophils in pancreatic cancer: potential therapeutic targets. Front Oncol 2022;12:1025805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Tay RE, Richardson EK, Toh HC. Revisiting the role of CD4+ T cells in cancer immunotherapy–new insights into old paradigms. Cancer Gene Ther 2021;28:5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Griffith BGC, Upstill-Goddard R, Brunton H, Grimes GR, Biankin AV, Serrels B, et al. FAK regulates IL-33 expression by controlling chromatin accessibility at c-Jun motifs. Sci Rep 2021;11:229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Serrels B, McGivern N, Canel M, Byron A, Johnson SC, McSorley HJ, et al. IL-33 and ST2 mediate FAK-dependent antitumor immune evasion through transcriptional networks. Sci Signal 2017;10:eaan8355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Sherman MH, Yu RT, Engle DD, Ding N, Atkins AR, Tiriac H, et al. Vitamin D receptor-mediated stromal reprogramming suppresses pancreatitis and enhances pancreatic cancer therapy. Cell 2014;159:80–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Kemp SB, Cheng N, Markosyan N, Sor R, Kim IK, Hallin J, et al. Efficacy of a small-molecule inhibitor of KRASG12D in immunocompetent models of pancreatic cancer. Cancer Discov 2023;13:298–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Wang X, Allen S, Blake JF, Bowcut V, Briere DM, Calinisan A, et al. Identification of MRTX1133, a noncovalent, potent, and selective KRASG12D inhibitor. J Med Chem 2022;65:3123–33. [DOI] [PubMed] [Google Scholar]
- 97. Hoshino K, Kashiwamura S, Kuribayashi K, Kodama T, Tsujimura T, Nakanishi K, et al. The absence of interleukin 1 receptor-related T1/ST2 does not affect T helper cell type 2 development and its effector function. J Exp Med 1999;190:1541–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. McHedlidze T, Waldner M, Zopf S, Walker J, Rankin AL, Schuchmann M, et al. Interleukin-33-dependent innate lymphoid cells mediate hepatic fibrosis. Immunity 2013;39:357–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Todaro GJ, Green H. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J Cell Biol 1963;17:299–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Scales MK, Velez-Delgado A, Steele NG, Schrader HE, Stabnick AM, Yan W, et al. Combinatorial Gli activity directs immune infiltration and tumor growth in pancreatic cancer. PLoS Genet 2022;18:e1010315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Aiello NM, Rhim AD, Stanger BZ, Orthotopic injection of pancreatic cancer cells .Cold Spring Harb Protoc 2016;2016:pdb.prot078360. [DOI] [PubMed] [Google Scholar]
- 102. Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods 2012;9:676–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Bolte S, Cordelières FP. A guided tour into subcellular colocalization analysis in light microscopy. J Microsc 2006;224:213–32. [DOI] [PubMed] [Google Scholar]
- 104. Hao Y, Hao S, Andersen-Nissen E, Mauck WM III, Zheng S, Butler A, et al. Integrated analysis of multimodal single-cell data. Cell 2021;184:3573–87.e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM III, et al. Comprehensive integration of single-cell data. Cell 2019;177:1888–902.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Jin S, Guerrero-Juarez CF, Zhang L, Chang I, Ramos R, Kuan C-H, et al. Inference and analysis of cell-cell communication using CellChat. Nat Commun 2021;12:1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Carpenter ES, Kadiyala P, Elhossiny AM, Kemp SB, Li J, Steele NG, et al. KRT17high/CXCL8+ tumor cells display both classical and basal features and regulate myeloid infiltration in the pancreatic cancer microenvironment. Clin Cancer Res 2024;30:2497–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Miyoshi H, Stappenbeck TS. In vitro expansion and genetic modification of gastrointestinal stem cells in spheroid culture. Nat Protoc 2013;8:2471–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Boj SF, Hwang CI, Baker LA, Chio II, Engle DD, Corbo V, et al. Organoid models of human and mouse ductal pancreatic cancer. Cell 2015;160:324–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 2011;12:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Single cell RNA sequencing datasets.
Table S2. Primer sequences used in qRT-PCR
Figure S1: IL-33+ stromal cells are abundant in human and mouse PDA.
Figure S2: Stromal IL-33 promotes PDA growth.
Figure S3: Pancreatic fibroblasts secrete IL-33 in response to oxidative stress.
Figure S4: Loss of stromal IL-33 alters the ST2+ immune cell secretome, resulting in a shift in CAF differentiation.
Figure S5: Inactivation of stromal IL-33 enables cytotoxic T cell activity.
Figure S6: Expression of fibroblast IL-33 is extrinsically induced by epithelial KrasG12D and requires JAK1/2-STAT3 activation throughout tumorigenesis.
Figure S7: Tumor cell-initiated autocrine signaling drives IL-33 upregulation in pancreatic fibroblasts.
Data Availability Statement
The scRNA-seq datasets generated in this study are publicly available at NCBI GEO accession number GSE269888. Preexisting scRNA-seq data analyzed in this study are available at the accession numbers listed in Supplementary Table S1. Bulk RNA-seq data generated in this study are also available at NCBI GEO GSE269889.