Abstract
Bacterial extracellular vesicles (BEVs) have emerged as pivotal mediators between bacteria and host. In addition to being crucial players in host homeostasis, they have recently been implicated in disease pathologies such as cancer. Hence, the study of BEVs represents an intriguing and rapidly evolving field with substantial translational potential. In this review, we briefly introduce the fundamentals of BEV characteristics, cargo and biogenesis. We emphatically summarize the current relationship between BEVs across various cancer types, illustrating their role in tumorigenesis, treatment responses and patient survival. We further discuss the inherent advantages of BEVs, such as stability, abundance and specific cargo profiles, that make them attractive candidates for non‐invasive diagnostic and prognostic approaches. The review also explores the potential of BEVs as a strategy for cancer therapy, considering their ability to deliver therapeutic agents, modulate the tumour microenvironment (TME) and elicit immunomodulatory responses. Understanding the clinical significance of BEVs may lead to the development of better‐targeted and personalized treatment strategies. This comprehensive review evaluates the current progress surrounding BEVs and poses questions to encourage further research in this emerging field to harness the benefits of BEVs for their full potential in clinical applications against cancer.
Keywords: bacterial extracellular vesicles, biogenesis, biomarkers, cancer treatment, inter‐kingdom communication
The graphical abstract illustrates the streamlined process by which bacterial extracellular vesicles (BEVs) traverse the intestinal epithelial barrier and enter the bloodstream. These BEVs play a role in carcinogenesis by enhancing mutagenesis, modulating oncogenes or tumor suppressor genes, and impacting immune activity. Cancers associated with BEVs include those of the oral cavity, liver, stomach, pancreas, and others.
1. INTRODUCTION
In 2022, Hanahan expanded the hallmarks of cancer to include 14 distinct characteristics, one of which is the ‘polymorphic microbiome’. This addition highlights the growing understanding of the microbiome's role in regulating polymorphism within tumours and its potential significance in cancer research (Hanahan, 2022). The human body contains a vast number of bacteria, outnumbering host cells by a ratio of almost 3:1, with 38 trillion bacteria and 4500 species (Almeida et al., 2021; Sender et al., 2016). While the majority (around 97%) reside in the colon, the rest can be found in the proximal intestine, skin and lung tissue (Garrett, 2015). These bacteria contribute a significant number of genes to the host genome expanding human genetic diversity (Gilbert et al., 2018). The structure of the bacterial community is influenced by various factors, such as age, race, sex, geographical location, dietary habits and lifestyle. Dysbiosis, or an imbalance in the microbiota, can lead to several pathologies, highlighting the importance of the microbiome beyond genetic factors alone (Tang et al., 2019). The individualized rate of change in normal microbiota composition suggests a potential clinical feature, although the sources of this variation and its impact on human health are not yet well understood (Gilbert et al., 2018). Currently, it is known that approximately 20% of human cancers are associated with the microbiome (Garrett, 2015). Among these ‘oncomicrobes’, only one bacterium has been identified as a human carcinogen, indicating that there is still much to be discovered in this field (Garrett, 2015). It is likely that there are many more bacteria with direct and indirect carcinogenic effects that have not yet been thoroughly studied and identified based on current laboratory and epidemiological evidence (for a comprehensive review of microbiome and human cancer, please refer to Sepich‐Poore et al. (2021)).
The relationship between the host and gut microbiome is multifaceted, necessitating the exchange of substances and signalling pathways for various functions such as immune regulation, stress response and cell proliferation or apoptosis (Hughes & Sperandio, 2008; Koeppen et al., 2016). However, microbiota faces physical and chemical barriers when accessing the epithelial barrier, relying on factors like proteins, metabolites and extracellular vesicles (EVs) to communicate with the host (Koeppen et al., 2016; Kuehn & Kesty, 2005). Recent evidence suggests that intercellular communication is primarily mediated by EVs released by both the gut microbiota and host intestinal cells (Diaz‐Garrido et al., 2021). Bacterial extracellular vesicles (BEVs), a specific type of nanoscale lipid bilayer vesicles secreted by bacteria, play a crucial role in mediating interactions between the host and the environment due to their unique composition of bioactive proteins, nucleic acids, lipids and metabolites. While BEVs have been extensively studied in microbial communities for nutrient transfer, transportation of virulence factors and toxins, horizontal gene transfer, and regulation of host immunity, their potential implications in cancer have been largely overlooked (Kulp & Kuehn, 2010; Riley et al., 2013; Wei et al., 2022; Zhang et al., 2020). However, BEVs may be the effector linking the immune system, bacteria and cancer through dysbiosis, alteration of metabolite/cellular signalling and immunomodulation. The aim of this review is to introduce BEVs and comprehensively discuss their roles in modulating tumour development across various cancer types, as well as their subsequent clinical significance. Throughout this review, the term BEV will be used to refer to all bacterial‐derived vesicles indiscriminately.
2. FUNDAMENTALS OF BEVS
Pioneers have already delved into research on the biogenesis, structure and cargo of BEVs, laying the groundwork for future studies. In the following, we will provide a concise overview for newcomers and those with some experience in the field.
2.1. Biogenesis and structure of BEVs
BEV components normally consist of membrane proteins, lipopolysaccharides (LPS) or lipoteichoic acid (LTA), peptidoglycans (PG), enzymes related to vesicle formation and cargos. The unique architecture of BEVs of gram‐positive and gram‐negative bacteria gives rise to distinct subtypes, which determine their biological function. Excellent summaries of the biogenesis mechanism and taxonomy of BEVs, which were classified into six main categories, were previously done by other groups (for a comprehensive review on this topic, please refer to Toyofuku et al. (2015, 2019, 2023)).
The membrane of gram‐negative bacteria consists of an inner membrane (IM) and outer membrane (OM), with a PG layer in the periplasmic space between them (Bos et al., 2007; Kulp & Kuehn, 2010). The OM is composed of two leaflets, an outer one consisting of LPS, and the inner one primarily made up of phospholipids (Kulp & Kuehn, 2010). Conversely, both leaflets of the IM consist solely of phospholipids. Gram‐negative bacteria are known to produce two main types of BEVs: outer membrane vesicles (OMVs) and outer‐inner membrane vesicles (OIMVs).
OMVs are spherical structures with a diameter ranging from 20 to 250 nm (Figure 1). They are primarily composed of OM and periplasmic proteins while lacking IM and cytoplasmic components (Toyofuku et al., 2019). On the other hand, OIMVs contain both outer and IM components, including proteins from the periplasmic space. Various mechanisms have been proposed to explain the production of OMVs. One of the earliest models suggested that the bulging or blebbing of the OM is due to the loss of OM and PG cross‐linking (Schwechheimer & Kuehn, 2015). Subsequently, the intercalation model was proposed, which suggests that the aggregation of embedded membrane proteins, such as outer membrane protein A (OmpA), Pseudomonas quinolone signals, unsaturated branched‐chain fatty acids, or Lipid A, drives membrane budding and pinching (Elhenawy et al., 2016; Florez et al., 2017; Naradasu et al., 2021; Song et al., 2008). The misfolded protein theory also supports a similar idea, where the aggregation of such proteins in the periplasmic space triggers membrane blebbing as a means to relieve stress by eliminating them (McBroom & Kuehn, 2007; Song et al., 2008). OIMVs, on the other hand, are formed through autolysin‐mediated weakening or inaccurate synthesis of the PG layer. The IM protrudes into the periplasm and pinches off from the cell surface along with the surrounding OM, forming a vesicle (Aktar et al., 2021; Hayashi et al., 2002; Pérez‐Cruz et al., 2013). Both OMVs and OIMVs can further be classified as explosive OMVs/OIMVs (EOMVs/EOIMVs) if they are produced through a burst‐like process (Aktar et al., 2021; Kulp & Kuehn, 2010; Turnbull et al., 2016). Genetic, phototoxic or antibody‐induced stress can lead to endolysin‐mediated degradation of the PG layer, resulting in explosive cell lysis and the generation of EOIMVs and EOMVs (Toyofuku et al., 2023). The irregular shape of these vesicles, stemming from their haphazard release, can cause the inclusion of both OM and IM components, explaining why they may contain cytoplasmic content or DNA (Toyofuku et al., 2023).
FIGURE 1.
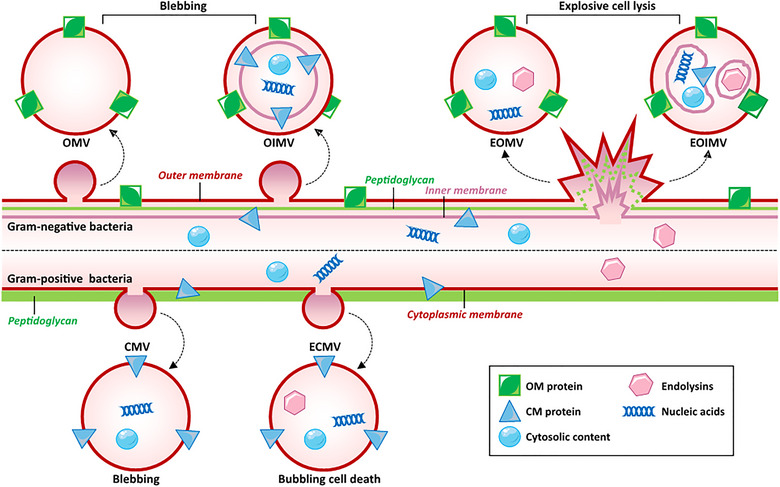
Biogenesis and types of BEVs. Gram‐negative BEVs have two main pathways of formation, namely, blebbing and explosive cell lysis. Blebbing results in the formation of OMV and OIMV, respectively. When the curvature of the membrane changes due to various reasons, the outer membrane protrudes and then falls off, forming OMV. Therefore, OMV contains outer membrane proteins and periplasmic contents but does not include substances from the inner membrane and cytoplasm. On the other hand, OIMV is another blebbing mechanism where the inner membrane protrudes into the periplasm after the weakening of PG layer by autolysins, followed by detachment along with the outer membrane. Therefore, OIMV can contain substances from both the inner membrane and cytoplasm. Explosive cell lysis is triggered by endolysins causing cell protrusion and explosion to form EOMVs. In addition, self‐assembly of the inner membrane with cytoplasmic contents can lead to irregular vesicles within the outer membrane which then form EOIMV. Gram‐positive bacteria also have two pathways for vesicle formation: Blebbing caused by loosening of PG layer allows small vesicles formed by cytoplasmic membrane protrusions to pass through thick PG layers to form CMVs; additionally, endolysins also can induce bubbling cell death resulting in ECMVs which differ from CMVs. BEVs, bacteria extracellular vesicles; CMVs, cytoplasmic membrane‐derived vesicles; ECMVs, explosive cytoplasmic membrane vesicles; EOIMV, explosive outer‐inner membrane vesicle; EOMVs, explosive outer membrane vesicles; OIMV, outer‐inner membrane vesicle; OMV, outer membrane vesicles; PG, peptidoglycan.
In comparison to gram‐negative bacteria, the presence of a thick PG layer surrounding the cytoplasmic membrane creates an obstacle for gram‐positive BEVs release. Nevertheless, the study of gram‐positive BEVs, known as cytoplasmic membrane‐derived vesicles (CMVs), has gained considerable attention in recent years (Brown et al., 2015; Cao & Lin, 2021; Lee et al., 2009). Three non‐exclusive hypotheses have been proposed to explain their production (Brown et al., 2015). The first hypothesis suggests that CMVs are released from the cytoplasm and traverse the cell wall under turgor pressure, with the rate of release regulated by pore size and cell wall thickness (Brown et al., 2015). The second model proposes that cell wall‐modifying enzymes loosen the membrane to facilitate CMVs release (Lee et al., 2009). Finally, the third hypothesis suggests that CMVs undergo dynamic deformation to squeeze through small pores when CMVs precursors generate a push force against a loosened PG wall or degraded cell wall (Jeong et al., 2022). This mechanism is believed to instigate PG encapsulation during CMVs release (Toyofuku et al., 2023). Under stress conditions, gram‐positive bacteria can produce explosive CMVs (ECMVs) through a phenomenon coined ‘bubbling cell death’. Unlike their gram‐negative counterparts, due to the presence of a thick PG layer outside the cytoplasmic membrane, they can form intracellular MVs and ghost cells without complete membrane hydrolysis (Andreoni et al., 2019; da Silva Barreira et al., 2022).
2.2. Cargo composition of BEVs
The composition of cargo within BEVs serves as a unique identifier of their origin and provides valuable insights into their function and formation. Several factors such as growth conditions, biogenesis routes and membrane composition give rise to such heterogeneity.
Nucleic acids, particularly DNA, have been extensively studied as cargo within BEVs. It has been observed that DNA can be carried or embedded on the surface of BEVs, playing a role in biofilm stabilisation, bacterial motility and modulation of pathogenesis or host colonisation (Gloag et al., 2013; Whitchurch et al., 2002). Horizontal gene transfer between the microbiome and host has also been reported, including in tumour tissues, which is believed to contribute to genetic defects and tumour progression (Deatherage & Cookson, 2012). Similarly, various types of RNA, such as mRNA, tRNA, small non‐coding RNA and rRNA, are carried by BEVs. There is sufficient evidence pointing to their involvement in downstream cellular dysregulation (Dauros‐Singorenko et al., 2018; Fan et al., 2023; Koeppen et al., 2016; Liu, Liu, et al., 2021; Zhang et al., 2020).
BEVs are also known to carry a diverse range of proteins, including enzymes, toxins and virulence factors. These proteins play crucial roles in inter‐microbial communication, modulation of host immune responses, and pathogenicity, and can impact various cellular processes such as cell proliferation, resistance, quorum sensing, nucleotide repair and cytokine secretion (Hussein et al., 2023; Wei et al., 2022). Similar to DNA, proteins can be found along the BEV membrane or encapsulated within the vesicle (Won et al., 2023). Other macromolecules, such as metabolites and lipids, can also play a role in BEV‐mediated signalling and communication. Although the collective knowledge of lipids is limited, specific enrichment of certain lipid species in OMVs compared to the OM suggests that formation occurs at membrane‐specific regions (Naradasu et al., 2021). Interestingly, BEVs can carry an array of metabolites, such as amino acids, nucleotides, sugars and organic acids. The complete expression of metabolic pathways within OMVs suggests their ability to function as a self‐sustaining biological system. Both protein and metabolite cargo are predicted to play a substantial role in BEV‐mediated functions (Faddetta et al., 2022; Zakharzhevskaya et al., 2017).
3. CANCER ASSOCIATED WITH BEVS
Maintaining microbial homeostasis is crucial for preserving the physiological equilibrium of the human ecosystem. Commensal bacteria occupy binding sites, establish colonization resistance, and impede the growth of pathogens. However, environmental changes or imbalances can disrupt this balance, resulting in the overgrowth of opportunistic pathogens amidst the depletion of commensal bacteria. In disease conditions, the influence of each microbial niche on cancer promotion is mediated by community‐level interactions facilitated by altered configurations of microbial communities, interaction between individual bacteria, or through secretion and modulation of metabolites (Cullin et al., 2021). Analysis of 1500 samples revealed that seven different tumour types had a specific microbial composition, with the microbiome of breast cancer exhibiting higher α‐ and β‐diversity than the others (Nejman et al., 2020). Certain bacteria with metabolic properties associated with specific cancers were found enriched in lung tumours of current smokers due to their ability to degrade chemicals in tobacco (Nejman et al., 2020). Significant differences in multiple bacterial taxa were observed between melanoma patients who responded positively or negatively to immune checkpoint inhibitors (Nejman et al., 2020).
The relationship between tumour‐associated microbiota and tumorigenesis involves several primary mechanisms. These include promoting tumorigenesis by increasing mutagenesis, regulating oncogenes or tumour suppressor genes, and regulating immune activity within the tumour microenvironment (TME) (Kim et al., 2017; Sepich‐Poore et al., 2021; Won et al., 2023; Wong‐Rolle et al., 2021). BEVs possess virulence factors that allow them to penetrate the epithelial barrier resulting in inflammation in the submucosa. Prolonged bacterial occupation increases BEV exposure of the mucosa leading to chronic inflammation, a notorious start to the pro‐oncogenic environment. Components of BEVs act as microbial‐associated molecular patterns (MAMPs), interacting with toll‐like receptors (TLRs), NOD‐like receptors (NLRs), AIM2 and so forth to kickstart a pro‐inflammatory immune response (Bielaszewska et al., 2018; Chen, Lei, et al., 2023; Han et al., 2019). For example, Fusobacterium nucleatum‐EVs induced F4/80+ iNOS+ M1‐like macrophages and upregulated cytokines such as TNF‐α, IL‐6, IL‐1β and iNOS, while also driving barrier leakage via receptor‐interacting serine/threonine‐protein kinase 1 (RIPK1), a regulator participating in chronic NF‐κB activation (Liu, Liang, et al., 2021; Wong et al., 2020). However, certain BEVs can also induce cellular apoptosis, inhibit cell proliferation, downregulate angiogenesis, mobilize immune cells and trigger anti‐tumour responses (Aly et al., 2021; Kim et al., 2017; Won et al., 2023). Understanding the strain‐specific role of bacteria in tumorigenesis is primordial to leveraging anti‐tumour benefits and circumventing pro‐tumour signalling.
BEVs can exert this widespread influence on the body's remote organs and tissues through both the lymphatic and circulatory systems. Kinetic analysis confirmed that intraperitoneal injection of BEVs in mice can spread to the liver, lungs, spleen, kidneys, pancreas and small intestine (Jang et al., 2015). To mimic the physical condition, oral administration of BEVs showed that they reach the lamina propria blood vessels through dynamin‐dependent endocytosis by the intestinal epithelium and primarily accumulate in the liver (Jones et al., 2020; Schaack et al., 2024). This suggests that BEVs may not only play a role in the formation of local TME but also the establishment of premetastatic niches in distant organs (Amieva et al., 2003; Shimoda et al., 2016; Turkina et al., 2015; Zoaiter et al., 2021). However, comprehensive reviews exploring the specific role of BEVs in various types of cancers are lacking. Therefore, the aim of this section is to delve into the role of BEVs in colon, stomach, liver, oral and other commonly associated tumours (Table 1).
TABLE 1.
The role of bacterial‐derived extracellular vesicles in human cancers.
| Bacteria | Cancer | Cargo | Function | Signalling | References |
|---|---|---|---|---|---|
| Helicobacter pylori | Gastric cancers | VacA |
1. Induce intracellular vacuoles, accompanied by altered iron metabolism and glutathione loss 2. Induce inflammatory responses |
|
(Chitcholtan et al., 2008; Choi et al., 2017) |
| CagA | 1. Induce inflammatory responses | 1. N.D. | (Choi et al., 2017) | ||
| N.D. |
1. Induce the burst of anti‐inflammatory cytokines 2. Impede human T cell responses 3. Induce Th2 and M2‐biased immune responses |
|
(Ahmed et al., 2021; Hock et al., 2017; Liu et al., 2019b) | ||
| sRNA (sR‐2509025, sR‐989262) | Reduce IL‐8 cytokine secretion | N.D. | (Zhang et al., 2020) | ||
| OSM | Promote the development of gastric cancer | JAK/STAT | (Zoaiter et al., 2021) | ||
| Liver cancer | N.D. | Induce expression of hepatic fibrosis markers | N.D. | (Bolori et al., 2023) | |
| Fusobacterium nucleatum | Colorectal cancer | N.D. |
|
|
(Engevik et al., 2021, Lamprinaki et al., 2021, Lin et al., 2021, Liu et al., 2021, Wei et al., 2023, Wu et al., 2023) |
| Breast cancer | N.D. | Promotes the proliferation, migration, and invasion | TLR4 | (Li, Sun et al., 2023) | |
| FomA | Trigger innate immunity of intestinal epithelial cells | Tlr2‐dependent NF‐κB pathway | (Martin‐Gallausiaux et al., 2020) | ||
| Oral cancer | N.D. | Alter expression levels of EMT‐related proteins | N.D. | (Chen et al., 2023a) | |
| Escherichia coli | Colorectal cancer | N.D. |
|
|
(Alvarez et al., 2019, Marzoog et al., 2023, Tyrer et al., 2014) |
| Liver cancer | N.D. |
|
|
(Natsui et al., 2023, Shi et al., 2023) | |
| Breast/Leukaemia cancer | N.D. | Induce a broad inflammatory response | N.D. | (Firth et al., 2023) | |
| Neuroblastoma | N.D. |
|
|
(Jin et al., 2022, Tang et al., 2018) | |
| Bacteroides fragilis | Colorectal cancer | Virulence factors | 1. Promote colon cancer development | N.D. | (Zakharzhevskaya et al., 2017) |
| N.D. | 2. Increase NPC1L1 gene expression | N.D. | (Ahmadi Badi et al., 2020, Badi et al., 2020) | ||
| Bacteroides thetaiotaomicron | Colorectal cancer | N.D. | Increase NPC1L1 gene expression | N.D. | (Ahmadi Badi et al., 2020, Badi et al., 2020) |
| Lacticaseibacillus paracase | Colorectal cancer | N.D. |
|
|
(An & Ha, 2022; Choi et al., 2020; Shi et al., 2021) |
| Limosilactobacillus johnsoni and Limosilactobacillus mucosae | Colorectal cancer | N.D. |
|
ZO‐1 and occludin | (Li, Feng, et al., 2023) |
| Lacticaseibacillus rhamnosus GG | Colorectal cancer | N.D. |
|
|
(Pang et al., 2022) |
| Liver cancer | N.D. |
|
|
(Behzadi et al., 2017) | |
| Limosilactobacillus reuteri | Colorectal cancer | N.D. | Protect the integrity of intestinal barrier |
|
(Pang et al., 2022) |
| Lentilactobacillus buchneri | Colorectal cancer and Gastric cancers | N.D. |
|
|
(Abedi et al., 2024) |
| Lactobacillus plantarum | Colorectal cancer | N.D. |
|
|
(An & Han, 2022) |
| Lactobacillus crispatus | Gastric cancers | N.D. |
|
|
(Fakharian et al., 2024) |
| Porphyromonas gingivalis | Oral cancer | sRNA45033 | Target CBX5 to regulates apoptosis through the methylation of p53 DNA | p53/Bcl‐2 | (Fan et al., 2023) |
| sRNA23392 | Promote the invasion and migration of Cancer cells by targeting DSC2 | N.D. | (Liu et al., 2021a) | ||
| PG | Induce PD‐L1 expression | NOD1‐RIP2 | (Groeger et al., 2020) | ||
| Clostridioides difficile | Liver cancer | N.D. | Induce mitochondrial dysfunction and increased intracellular ROS | N.D. | (Caballano‐Infantes et al., 2023) |
| Akkermansia muciniphila | Prostate cancer | N.D. | Inhibit the development and metastasis of cancer | N.D. | (Luo et al., 2021) |
| Colorectal cancer | N.D. |
|
N.D. | (Chelakkot et al., 2018, Kang et al., 2013, Wang et al., 2023) | |
| Amuc_2172 | Promote CTL‐related immune response | Hsp70 | (Jiang, Xu, et al., 2023) | ||
| Bifidobacterium longum | Breast cancer | N.D. | Induce apoptosis | Bax/Bcl‐2 | (Jiang, Wang, et al., 2023) |
| Staphylococcus aureus | Breast cancer | N.D. | Enhance tamoxifen efficacy | AKT ‐ERK | (An, Kwon, et al., 2022) |
| Bacillus licheniformis | Breast/lung cancer | N.D. | Inhibited cell viability and proliferation by increasing ROS and decreasing glutathione | p53, p21, caspase‐9/3, Bax, Bcl‐2 | (Gurunathan et al., 2023) |
Abbreviations: 5‐FU, 5‐flu‐orouracil; CagA; cytotoxin‐associated gene A; CBX5, chromobox 5; Clec4e, C‐type lectin domain family 4 member E; COX‐2, cyclo‐oxygenase‐2; CTL, cytotoxic T lymphocyte; DSC2, Desmocollin‐2; EMT, epithelial‐mesenchymal transition; LPS, lipopolysaccharide; MET, mesenchymal‐epithelial transition factor; N.D., not determined; NO, nitric oxide; NPC1L1, niemann‐Pick C1‐Like 1; OSM, Oncostatin M; PD‐L1, Programmed death‐ligand 1; PG, peptidoglycan; RIP2, receptor‐interacting serine/threonine‐protein kinase 2; ROS, reactive oxygen species; Siglec‐7, sialic acid‐binding immunoglobulin‐like lectins‐7; TLR4, toll‐like receptor 4; VacA, vacuolar cytotoxin A.
3.1. Colorectal cancer
Colorectal cancer (CRC) is currently the third most prevalent cancer and the second leading cause of cancer mortality worldwide (Sung et al., 2021). The colon harbours the most diverse types of bacteria in the body, with quantities reaching approximately 1013–1014 (Sender et al., 2016). A study on the shotgun metagenome of 526 faecal samples from five countries has shown consistent enrichment of a pro‐tumorigenic ‘core microbiome,’ including Bacteroides fragilis, F. nucleatum, Porphyromonas asaccharolytica, Parvimonas micra, Prevotella intermedia, Alistipes finegoldii and Thermanaerovibrio acidaminovorans (Dai et al., 2018).
F. nucleatum, a key member of the ‘core microbiome’ in CRC, has been strongly associated with the pathogenesis of CRC in the past decade as shown in numerous studies (Castellarin et al., 2012; Kostic et al., 2012, 2013; McCoy et al., 2013). Proteomic analysis of BEVs from F. nucleatum (FnEVs) revealed the presence of virulence factors, including FadA, MORN2, and YadA‐like domain proteins, each with multiple exposed epitope sites (Liu, Hsieh, et al., 2019; Zhang et al., 2023). MORN domain‐containing proteins and other virulence factors are significantly upregulated under acidic conditions (Zhang et al., 2023). FnEVs have been found to promote mitochondrial fusion and cell invasion of CRC cells, and improve the ability of superoxide dismutase enzyme (SOD) and reduce reactive oxygen species (ROS) levels, thus showing the protective effect of CRC cells (Lin et al., 2021; Wu et al., 2023). Furthermore, transcriptome profiling revealed that FnEVs‐treated cells exhibited increased expression of CRC‐related oncogenes (ID2, CCNA1, EGR2, WNT4, RIPK4, PDK4, IL‐11 and MAPK4), along with decreased expression of p53 (Wu et al., 2023). Additionally, a novel oncogene, CCL2, was identified (Wu et al., 2023).
CRC induced by inflammatory bowel disease represents a quintessential model of inflammation‐induced carcinogenesis (Shah & Itzkowitz, 2022). Chronic inflammation generates a significant amount of ROS and reactive nitrogen species, which can cause mutations and double‐strand DNA breaks (Shah & Itzkowitz, 2022). Additionally, elevated levels of pro‐inflammatory cytokines are observed, which promote cancer cell proliferation and survival (Shah & Itzkowitz, 2022). Subsequent studies explored the mechanistic functions of FnEVs in shaping the chronic inflammatory environment and promoting intestinal mucosal barrier damage. Engevik et al. confirmed that FnEVs can stimulate the production of interleukin (IL)‐8 and tumour necrosis factor (TNF) through activation of the TLR4‐MyD88 signalling pathway in colonic epithelial cells (Engevik et al., 2021). This effect was diminished by pharmacological inhibition of TLR4 (Engevik et al., 2021). FnEVs have even been reported to interact with sialic acid‐binding immunoglobulin‐like lectins‐7 (Siglec‐7) on innate immune cells (Lamprinaki et al., 2021). Siglec‐7 has been found to increase PD‐L1 and promote monocyte polarization towards a tumour‐associated macrophage phenotype. Lamprinaki et al. found that FnEVs induce the expression of TNF‐α in dendritic cells and upregulate IL‐10 in macrophages (Lamprinaki et al., 2021). For certain antibiotic‐resistant CRC patients, employing glycan intervention targeting FnEVs and Siglec‐7 could serve as adjunctive therapy for CRC. Further studies confirmed that FnEVs upregulate IL‐1β, IL‐6 and TNF‐α, while downregulating IL‐10 and intercellular tight junction proteins ZO‐1 and occludin in colonic epithelial cells via miR‐574‐5p/CARD3‐dependent autophagy activation (Wei et al., 2023). Indeed, FnEVs significantly promote epithelial barrier dysfunctions and oxidative stress damage via FADD‐RIPK1‐caspase‐3 axis‐mediated apoptosis in macrophage/CRC cell co‐cultures (Liu, Liang, et al., 2021). However, Liu et al. found that FnEVs decreased the level of IL‐10 in macrophages, which contrasts with the findings of Lamprinaki's study. This discrepancy may be due to differences in the doses of FnEVs used in treating the cells. Further experiments are needed to confirm these controversial results. Chen and colleagues recently achieved selective eradication of F. nucleatum at tumour sites by designing a nanomedicine that mimics F. nucleatum and is fused with Colistin‐loaded liposomes (Chen, Zhao, et al., 2023). Furthermore, this anti‐F. nucleatum therapy was able to restore the efficacy of anti‐PD‐1 therapy in F. nucleatum‐infected mice. Therefore, appropriately combined anti‐F. nucleatum therapy is necessary for CRC patients with high F. nucleatum levels (Yu et al., 2017).
Compared with the oncogenic role of F. nucleatum, the strains of Escherichia coli exhibit a wide spectrum of roles, encompassing both probiotics and pathogens. For example, EcEVs derived from probiotic strains E. coli were found to enhance the expression of ZO‐1, occludin and claudin in colonic epithelial cells that were infected with enteropathogenic E. coli O127:H6, thus reversing the cellular barrier damage (Alvarez et al., 2019). EcEVs from the laboratory strain also induce apoptosis in CRC by decreasing the Bcl‐2/Bax ratio (Jiang et al., 2024). Recent research found that EcEVs from standard strain upregulate ROS and induce mitophagy via the Akt/mTOR pathways in CRC cells, resulting in the selective killing of CRC cells (Marzoog et al., 2023).
Interestingly, studies have demonstrated that EVs secreted by probiotics can have tumour‐suppressive effects. For example, BEVs derived from Limosilactobacillus johnsoni and Limosilactobacillus mucosae have been shown to effectively mitigate LPS‐induced damage to the intestinal barrier by upregulating the expression levels of ZO‐1 and occludin, and reverse intestinal inflammatory damage (Li, Feng, et al., 2023). BEVs derived from Limosilactobacillus reuteri also have the ability to decrease barrier leakage caused by enterotoxigenic E. coli (Pang et al., 2022). Lacticaseibacillus paracasei‐EVs (LpEVs) regulate intestinal inflammation by reducing pro‐inflammatory cytokines IL‐1α, IL‐1β, IL‐2 and TNF‐α, while increasing anti‐inflammatory cytokines IL‐10 and TGF‐β. This is followed by the downregulation of inflammatory‐associated proteins cyclo‐oxygenase‐2 (COX‐2), iNOS, and NF‐κB to maintain intestinal homeostasis. Another study demonstrated that LpEVs suppress the proliferation, mobility and survival of CRC cells both in vitro and in vivo by mediating the PDK1/AKT/Bcl‐2 axis (Shi et al., 2021). BEVs derived from Lacticaseibacillus rhamnosus GG (LGG‐EVs) have been shown to effectively suppress the proliferation of CRC cells (Keyhani et al., 2022). LGG‐EVs also downregulated various pro‐inflammatory factors (TNF‐α, IL‐1β, IL‐6, IL‐2) by inhibiting the TLR4/NF‐κB/NLRP3 pathway (Tong et al., 2021). Additionally, oral administration of LGG‐EVs synergistically enhanced the efficacy of anti‐PD‐1 immunotherapy in CRC (Lu et al., 2023). The underlying mechanisms included an increased CD8+ T/CD4+ T cell ratio in mesenteric lymph nodes and enhanced proportions of MHC II+ dendritic cells, CD4+ T cells, and CD8+ T cells within TME (Lu et al., 2023). Lentilactobacillus buchneri‐derived EVs (LbEVs) significantly increased the apoptosis rate in CRC and gastric cancer (GC) by enhancing the expression of BAX, CASP and CASP9 genes. LbEVs induced a cell cycle arrest and reduced the migration of these two cancer cells (Abedi et al., 2024). BEV derived from Lactobacillus plantarum overcame 5‐FU resistance by downregulating PDK2 expression in the p53‐p21 glycolysis signal (An & Han, 2022). These findings suggest that probiotics from the Lactobacillaceae family have demonstrated anti‐tumour effects in CRC. It is worth exploring whether these probiotics could exhibit synergistic effects during cancer treatment, particularly for drug‐resistant patients.
Certain commensal bacteria also have specialized roles. For example, Akkermansia muciniphila‐derived BEVs (AmEVs) have been shown to decrease gut permeability in LPS‐induced Caco‐2 cells by increasing the expression of tight junction protein, occludin (Chelakkot et al., 2018). The bioactive compound Amuc_2172 within AmEVs was found to reshape the TME by enhancing HSP70 secretion and promoting cytotoxic T lymphocyte (CTL)‐mediated immune responses during tumorigenesis (Jiang, Xu, et al., 2023). AmEVs have also been shown to alleviate the production of pro‐inflammatory cytokines such as IL‐6, IFN‐γ and C‐reactive protein in colonic epithelial cells, while increasing the levels of anti‐inflammatory cytokines such as IL‐10, IL‐13 and angiotensin‐converting enzyme 2 (Kang et al., 2013, Wang et al., 2023). Oral administration of AmEVs protected against dextran sulphate sodium‐induced colitis by enhancing the expression of tight junctions ZO‐1 and occludin (Kang et al., 2013, Wang et al., 2023). The combination of AmEVs and anti‐programmed death‐ligand 1 (PD‐L1) optimized the therapy against CRC (Wang et al., 2023). These findings suggest their potential as therapeutic candidates for CRC treatment.
3.2. Gastric cancer (GC)
GC is a significant global health concern, ranking fifth in terms of incidence and fourth in terms of mortality worldwide (Sung et al., 2021). Chronic infection with Helicobacter pylori, which affects at least 50% of the global population (Hooi et al., 2017), is considered the main contributor to GC development.
Many studies have explored the functional impact of HpEVs on GC. In particular, two major virulence factors of H. pylori, cytotoxin‐associated gene A (CagA) and vacuolar cytotoxin A (VacA), have been identified within HpEVs. VacA within HpEVs has been shown to induce intracellular vacuoles, along with altered iron metabolism and loss of glutathione, potentially promoting gastric carcinogenesis (Chitcholtan et al., 2008). HpEVs‐CagA and ‐VacA can stimulate the production of TNF‐α, IL‐6 and IL‐1β by macrophages, as well as IL‐8 by gastric epithelial cells (Choi et al., 2017). HpEVs also upregulate oncostatin M (OSM), an IL‐6 family cytokine, and its type II receptor OSMRβ in GC cells (Zoaiter et al., 2021). Interestingly, OSM expression is upregulated in early GC, potentially influenced by HpEVs originating from H. pylori infection (Shi et al., 2019). Overall, GC patients exhibit higher levels of HpEVs compared to healthy individuals. Both gastric juices and serum HpEVs can induce inflammatory cytokines, and prolonged stimulation may potentially induce malignant features.
However, the coevolution of bacteria and host may result in the induction of an anti‐inflammatory response to evade the host's immune attack. HpEVs strongly induce secretion of IL‐10 and IL‐6 by peripheral blood mononuclear cells (PBMCs), leading to T cell apoptosis (Winter et al., 2014) Furthermore, the addition of HpEVs to PBMCs significantly suppressed T cell proliferation via COX‐2‐expression in monocytes, leading to increased production of PGE2 and IL‐10 (Hock et al., 2017). Ahmed et al. confirmed that HpEVs promoted a Th2 immune response by inducing high levels of IL‐10 and IL‐4 production, and stimulated the M2 phenotype by increasing the ratio of CD206/CD86 in macrophages (Ahmed et al., 2021). Indeed, orally administered HpEVs predominantly induce a Th2‐biased immune response (Liu, Li, et al., 2019). In addition to proteins, HpEVs enriched with certain small noncoding RNAs (sR‐2509025 and sR‐989262) have been shown to reduce the secretion of IL‐8 by GC cells (Zhang et al., 2020). Overall, under the dual influence of long‐term inflammation stimulation and evasion from immune cell attacks, the stomach gradually trends towards malignancy. Recent study showed that Lactobacillus crispatus‐derived EV can reverse the inflammatory environment in gastric cells infected with H. pylori, demonstrating their potential anti‐cancer effects (Fakharian et al., 2024).
3.3. Liver cancer
Liver cancer, specifically hepatocellular carcinoma (HCC), accounts for more than 90% of the total burden and ranks as the sixth most prevalent malignancy worldwide (Sung et al., 2021). However, most current studies on BEVs in liver cancer focus on non‐alcoholic fatty liver disease, which has emerged as a significant contributor to HCC incidence in the past decade, along with chronic hepatitis B virus infection. The gut‐liver axis, formed by a natural anatomical connection, the hepatic portal vein, allows the translocation of gut microbiota and their derived EVs/components into the liver microenvironment (Bertocchi et al., 2021; Fizanne et al., 2023). Analysis of faeces‐derived EVs (fEVs) in patients with non‐alcoholic steatohepatitis (fEVs‐NS) has revealed a reduction in the expression of occludin and ZO‐1 through a pathway dependent on non‐muscular myosin light chain kinase (Fizanne et al., 2023). Additionally, fEVs‐NS were found to enhance endothelial cell permeability via the TL4‐LPS pathway and induce the secretion of multiple chemokine/cytokine molecules in endothelial cells (Fizanne et al., 2023). fEVs‐NS also induced the secretion of pro‐fibrotic and pro‐inflammatory proteins in hepatic stellate cells by activating cGAS/STING (Fizanne et al., 2023; Luo et al., 2022).
Considering the colon and liver are connected, there are ongoing studies investigating the impact of EcEVs on the liver. Conditions such as leaky gut syndrome enhance gut permeability, as more EcEVs enter the liver, inflammation is observed through upregulation of C‐type lectin domain family 4 member E (Clec4e), worsening fibrosis and the reduced albumin production (Natsui et al., 2023). However, oral gavage of a probiotic strain producing EcEVs showed simultaneous improvements in the metabolism of the intestinal and hepatic systems, leading to a reduction in mouse body weight and blood glucose levels (Shi et al., 2023).
In another study, treatment of hepatic stellate cells with HpEVs increased the level of α‐SMA and induced the expression of hepatic fibrosis markers, leading to liver fibrosis (Bolori et al., 2023). Clostridioides difficile‐derived EVs could decrease mitochondrial membrane potential and increase intracellular ROS production in HepG2 cells and impact their metabolic activity (Caballano‐Infantes et al., 2023). LGG‐EVs have been shown to promote apoptosis of HepG2 cells, similar to their effects in CRC cells (Behzadi et al., 2017).
3.4. Oral cancer
Oral cancer, predominantly squamous cell carcinomas, including cancers of the lip and oral cavity, currently ranks as the 18th most common malignant neoplasm worldwide (Sung et al., 2021). While the oral microbiome exhibits the second highest level of alpha diversity after the gut, until recently, no correlation between cancer development and oral microbiota‐derived EVs had been reported. It has been established that persistent periodontitis is a risk factor for oral cancer. Recent research indicates that individuals with periodontitis show higher levels of LPS‐positive BEVs in their saliva compared to the healthy populations (Han et al., 2021). Current studies regarding the association between BEVs and oral cancer primarily focus on the periodontal pathogen Porphyromonas gingivalis. PgEVs have been found to promote apoptosis and inflammatory cytokine secretion in human periodontal ligament cells through sRNA45033, which targets chromobox 5 (CBX5); CBX5 regulates cell apoptosis through p53/Bcl‐2 axis (Fan et al., 2023). PG of PgEVs have also been shown to activate cytosolic receptors, inducing PD‐L1 expression in a receptor‐interacting serine/threonine‐protein kinase 2 (RIP2)‐dependent fashion in human oral carcinoma cells, contributing to immunosuppressive functions (Groeger et al., 2020). However, this study did not specifically characterize BEVs. Another study revealed that sRNA23392 within PgEVs promoted the invasion and migration of oral squamous cell carcinoma by targeting desmocollin‐2 (DSC2), a member of the desmosomal cadherin family associated with highly aggressive tumours and poor survival when suppressed (Liu, Liu et al., 2021). Overall, PgEVs have a negative effect on oral health, promoting tumour progression and metastasis. Recent investigations show FnEVs mediate cancer progression in several ways such as promoting autophagy (Chen, Gao, et al., 2023), upregulating Vimentin and N‐cadherin while downregulating E‐cadherin (Chen, Gao, et al., 2023).
3.5. Other cancers
Studies have investigated the relationship between microbiota and various cancers, including pancreatic cancer, prostate cancer, breast cancer, neuroblastoma, skin cancer and lung cancer.
In pancreatic cancer, 16s rRNA gene sequencing of EVs isolated from paired tumour and normal tissues revealed that Tepidimonas was more abundant in tumours (Jeong et al., 2020). Additionally, larger tumours were associated with lower levels of Leuconostoc and Sutterella, while lymph node metastasis was found to be associated with increased Comamonas and Turicibacter in tumour tissues. Abundancy of Streptococcus and Akkermansia decreased in the case of tumour recurrence (Jeong et al., 2020). Notably, supernatant from Tepidimonas fonticaldi could stimulate cell proliferation and migration and enhance EMT and tricarboxylic acid cycle‐related metabolites (Jeong et al., 2020). Interestingly, gut dysbiosis‐derived microbial DNA‐containing EVs deliver microbial DNAs into beta cells, resulting in elevated inflammation and impaired insulin release by activating the cGAS/STING pathway (Gao et al., 2022). This phenomenon could be blunted through the removal of microbial DNA‐containing EVs by Vsig4+ macrophages (Gao et al., 2022). Given the challenge of detecting pancreatic cancer, a promising approach is to employ a non‐invasive strategy that analyses alterations in gut microbiota, depicted in BEVs, as a means to discover potential diagnostic markers.
Prostate cancer is one of the most prevalent malignancies among men and presents significant treatment challenges. In male mice, intravenous injection of AmEVs reduced the tumour burden of prostate cancer in vivo with no obvious toxicity to normal tissues. This was achieved by an increase in the proportion of GZMB+ and IFN‐γ+ lymphocytes in CD8+ T cells and causing recruitment of tumour‐killing M1 macrophages to suppress the proliferation and invasion of prostate cancer cells (Luo et al., 2021). AmEVs may be a promising treatment to improve the therapeutic efficacy of refractory prostate cancer.
Breast cancer has the highest proportion of new cancer cases worldwide (Siegel et al., 2023). FnEVs exhibited their carcinogenic potential in breast cancer by promoting proliferation, migration and invasion through the activation of TLR4 (Li, Sun, et al., 2023). Another study showed that Bifidobacterium‐derived BEVs induced apoptosis in triple‐negative breast cancer (TNBC) cells by increasing the expression of Bax and decreasing the expression of Bcl‐2, both in vivo and in vitro (Jiang, Wang, et al., 2023). Compared to healthy controls, breast cancer patients exhibit a significant reduction in Staphylococcus aureus. Additionally, S. aureus‐derived BEVs enhance the efficacy of tamoxifen against breast cancer by increasing TNF‐α and decreasing cyclin E2 (An, Kwon, et al., 2022). The combination treatment with tamoxifen and S. aureus EVs resulted in downregulated expression of p‐AKT and p‐ERK and upregulated TNF‐α in ER‐positive breast cancer cells compared to treatment with tamoxifen alone (An, Kwon, et al., 2022). Furthermore, S. aureus EVs reduced the expression of Cyclin E2, which is positively correlated with tamoxifen resistance (An, Kwon, et al., 2022). Additionally, attenuated EcEVs induced a broad inflammatory response with cytolytic Vγ9Vδ2 T cells that combat both breast and leukaemia cancer cells (Firth et al., 2023). The study, however, did not report any common features of BEVs (such as TEM, NTA, etc.). Overall, in TNBC, BEVs derived from S. aureus, E. coli and Bifidobacterium exhibit an anti‐tumour effect and could be utilized as adjunctive therapy.
In neuroblastoma, EcEVs have been found to have an anti‐tumour effect both in vivo and in vitro. This effect is achieved by phosphorylating p53 to downregulate Bcl‐2 and upregulate Bax, thereby promoting apoptosis (Jin et al., 2022). Another study discovered that EcEVs can reduce the migratory ability of neuroblastoma cells, with the inhibition rate of migration showing a dose‐dependent relationship (Tang et al., 2018).
The skin acts as the body's natural barrier to the external environment and serves as a habitat for various bacteria. Numerous studies have explored the connection between BEVs and inflammatory skin diseases (Cros et al., 2023; Kim et al., 2018; Zhou et al., 2023). However, there is currently no research evidence establishing a link between BEVs and skin‐related cancers. Nevertheless, researchers have attempted to utilize engineered BEVs for the treatment of melanoma to prevent metastasis (Cheng et al., 2021).
Similarly, the lungs are directly exposed to the external air. Despite the purifying action of airway cilia, microorganisms can still reach the lungs. Lung immune dysregulation and pathological damage have been associated with BEVs in dust (Kim et al., 2015; Meganathan et al., 2020). Analysis of circulating microbial EVs using 16s rRNA sequencing revealed a significant increase in species richness in lung cancer patients compared to the healthy population (McDowell et al., 2022). One study found that BEVs derived from Bacillus licheniformis (BlEVs) inhibit proliferation and promote apoptosis in lung cancer and breast cancer cells (Gurunathan et al., 2023). BlEVs increased ROS and decreased glutathione, thereby showing their cytotoxicity in lung and breast cancer (Gurunathan et al., 2023). Co‐administration with doxorubicin (DOX) enhances therapeutic efficacy (Gurunathan et al., 2023).
3.6. Methodology used in cancer studies
The lack of a standardized workflow for BEV research has led to variations in quality and inconsistent reporting in research papers, especially for those isolated from tissue and faecal samples. While EV isolation from single bacterial strains has been improved, the functional studies done with in vitro monocultures do not reflect the heterogeneity or lower working concentration of BEVs from the gut microbiome in vivo. Herein, we aim to explore the strengths and weaknesses of the current research status (for a summary of the methodology used for isolating BEVs, please refer to Tables 2, 3, 4, 5, 6, which were organized according to the subdivisions of Section 3). For pre‐processing, due to the diversity of bacterial culture (such as temperature, humidity, oxygen levels, culture composition, agitation conditions and growth phases), researchers should provide detailed reports on their culture methods (Welsh et al., 2024). For separation, many studies have used a combination of ultracentrifugation with 0.22 µm filtration or direct ultrafiltration. Additionally, it is important to consider that different separation methods may enrich specific subtypes of BEVs or aggregate non‐BEV materials, such as pili, flagella, phage, proteins, lipoproteins and nucleoprotein complexes (Welsh et al., 2024). For characterization, the majority of functional studies have reported both microscopic and particle analyses of BEVs. However, a small portion of studies have only shown one of these aspects, which compromises the credibility of the findings. Although there is currently no universally recognized specific protein marker for BEVs, some studies have attempted to use LTA (for gram‐positive bacteria), LPS (for gram‐negative bacteria) and OmpA as indicative proteins for BEVs. In the realm of H. pylori research, prevalent markers of HpEVs include VacA, CagA and urease subunit A (UreA), and some studies have reported one or more of these marker proteins. Additionally, certain studies have shown the protein profile of BEVs.
TABLE 2.
Methodology of functional reported papers on colorectal cancer.
| First author | Year | Bacteria/sample | Method specifics | Particle analysis | Microscopy | Protein analysis | Antibody specifics |
|---|---|---|---|---|---|---|---|
| Lin, LT | 2021 | Fusobacterium nucleatum |
3000 × g for 15 min 35,000 × g for 60 min 200,000 × g for 60 min 0.22‐µm filter |
Y | Y | N | NA |
| Wu, X | 2023 | F. nucleatum |
3000 × g for 10 min 0.45‐µm filter centrifuge for 20h 10,000 × g for 1 h 12,000 × g for 2 min exosome purification filter 3000 × g for 10 min |
Y | Y | Y | N, protein profile |
| Engevik, MA | 2021 | F. nucleatum |
7000 × g for 10 min mixed with 120 g of ammonium sulfate 10,000 × g for 20 min 10,000‐molecular‐weight cutoff dialysis tubing 292,700 × g for 3 h (70 Ti rotor; Beckman Coulter Inc.) 38,400 × g for 3 h |
Y | Y | N | NA |
| Wei, S | 2023 | F. nucleatum |
2500 × g for 10 min 0.22‐µm filter 100,000 × g for 70 min Two wash steps |
Y | Y | N | NA |
| Liu, L | 2021 | F. nucleatum |
10,000 × g for 20 min 0.22‐µm filter 100 kDa ultrafiltration 60,000 × g for 30 min Two wash steps 0.22‐µm filter 150,000 × g for 2 h using a sucrose density× gradient Detoxi‐Gel Endotoxin Removing Column |
Y | Y | N | NA |
| Lamprinaki, D | 2021 | F. nucleatum |
8500 × g for 15 min 0.22‐µm filter 100,000 molecular weight cut‐off filter unit density gradient ultra‐centrifugation 135,000 × g for 16h 200,500 × g for 2 h at 4°C using a Type 45 Ti rotor (Beckman Coulter) 0.22‐µm filter |
Y | N | Y (GC‐MS) | LPS, Siglec‐7 |
| Martin‐Gallausiaux, C | 2020 | F. nucleatum |
4700 × g for 20 min 10,000 × g for 20 min 0.22‐µm filter 100‐kDA cutoff (Amicon) centrifugal filter units 4000 × g Iodixanol × gradient (Optiprep, Sigma) using Beckman ultracentrifuge with a swinging bucket rotor (SW40 Ti) during 16 h at 100,000 × g |
Y | Y | Y | N protein profile |
| Marzoog, TR | 2023 | Escherichia coli |
0.22‐µm filter 100,000 rpm for 5h |
Y | Y | Y | OmpA |
| Ahmadi Badi, S | 2020 | Bacteroides fragilis and Bacteroides thetaiotaomicron |
2900 × g for 1 h 20,000 × g for 60 min 20,000 × g for 120 min ×2 0.22‐µm filter |
N | Y (SEM) | Y | N protein profile |
| Choi, JH | 2020 | Lacticaseibacillus paracasei |
10,000 × g for 20 min 0.22‐µm bottle‐top filter 100‐kDa Pellicon 2 Cassette filter membrane 0.22‐µm bottle‐top filter 150,000 × g for 3 h |
Y | Y | N | N |
| Shi, Y | 2021 | L. paracasei |
4500 × g for 15 min 0.45‐µm filter 120,000 × g for 1 h |
Y | Y | N | N |
| Keyhani, G | 2022 | Lactobacillus rhamnosus |
10,000 × g for 10 min 0.22 µm filter 100,000 × g for 60 min |
N | Y (Low reliability) | N | N |
| Tong, L | 2021 | L. rhamnosus |
8000 × g for 30 min 0.22‐µm PVDF filter Centricon Plus‐70 Centrifugal Filter Units 100,000 × g for 2 h 100,000 × g for 60 min |
Y | Y | Y | TSG101 |
| Li, J | 2023 | Limosilactobacillus johnsoni and Limosilactobacillus mucosae |
8000 × g for 30 min 20,000 × g for 45 min 0.22‐µm filter 120,000 × g for 2 h with an SW 32 Ti rotor ×2 |
Y | Y | N | N |
| Pang, Y | 2022 | Limosilactobacillus reuteri |
5000 × g for 10 min 10,000 × g for 10 min 0.45‐µm pore filter Amicon 100 kDa MWCO (molecular weight cutoff) filter 118,000 × g at 4°C for 3 h ×2 |
Y | Y | Y (Dot‐blot assay) | LTA |
| Alvarez, CS | 2019 | E. coli |
10,000 × g, 4 °C, 20 min 0.22‐µm filter Centricon® Plus‐70 filter device 150,000 × g,1 h |
N | N | N | NA |
| Chelakkot, C | 2018 | Akkermansia muciniphila |
10,000 × g for 20 min 0.45‐µm vacuum filter 0.22‐µm bottle‐top filter 150,000 × g for 2 h |
Y | Y | Y | N, protein profile |
| Jiang, Y | 2023 | A. muciniphila | bottomup optiprep density gradient centrifugation | N | Y | N | NA |
| Kang, CS | 2013 | A. muciniphila | 200,000 × g for 2 h | Y | Y | N | NA |
| Wang, X | 2023 | A. muciniphila |
15,000 × g for 30 min 0.45‐µm filter 170,000 × g for 60 min 0.45‐µm filter |
Y | N | N | NA |
| Montanari, M | 2022 | Campylobacter jejuni |
1000 × g for 15 min 12,000 × g for 20 min 18,000−20,000 × g for 20 min 0.22‐µm filter 110,000 × g for 70 min |
Y | N | N | NA |
| Abedi, A | 2024 | Lentilactobacillus buchneri |
13,000 rpm for 10 min 0.22‐µm‐pore size filter 100,000 × g for 1 h |
N | Y | Y | N, protein profile |
Abbreviations: NA, not available; Omp, outer membrane protein; SEM, scanning electron microscopy; Siglec‐7, sialic acid‐binding immunoglobulin‐like lectins‐7.
TABLE 3.
Methodology of functional reported papers on gastric cancer.
| First author | Year | Bacteria/sample | Method specifics | Particle analysis | Microscopy | Protein analysis | Antibody specifics |
|---|---|---|---|---|---|---|---|
| Chitcholtan, K | 2008 | Helicobacter pylori |
10,000 × g, 15 min ×2 100,000 × g for 2 h ×2 |
N | N | N | N |
| Choi, H I | 2017 | H. pylori |
10,000 × g for 20 min Tan × gential flow filtration system (EMD Millipore, Billerica, MA, USA) with a 100‐kDa hollow‐fibre membrane filter 0.22‐µm filter 150,000× g for 3 h Density gradient ultracentrifugation at 100,000× g for 2 h |
Y | Y | Y | VacA CagA |
| Hock, B. D. | 2017 | H. pylori |
12,000 × g for 15 min ×2 200,000 × g for 2 h ×3 |
N | N | N | NA |
| Winter, J. | 2014 | H. pylori |
6000 × g for 10 min 0.45‐µm filter 0.20‐µm syringe filters Precipitated at room temperature using 40% (wt/vol) ammonium sulphate for 1 h 10,000 × g for 15 min 100,000 × g for 2 h |
N | Y | Y | VacA |
| Ahmed, A. A. Q. | 2021 | H. pylori |
5000 ×g for 20 min × 2 0.45‐µm filter 0.22‐µm filter OMV binding resin Amicon ultra centrifugal filter device with a 3 K MWCO |
Y | Y | Y | N proteins profile |
| Liu, Q. | 2019 | H. pylori |
4500 × g for 1 h 0.45‐µm Steritop bottle‐top filter 20,000 × g for 2 h OptiPrep density gradient 100,000 × g for 24 h |
N | Y | Y | N proteins profile |
| Fakharian, F. | 2024 | Lactobacillus crispatus |
10,000 × g for 20 min 0.22‐µm filter 150,000 × g for 5 h at 4°C |
Y | Y | Y | N Protein profile |
Abbreviations: CagA; cytotoxin‐associated gene A; NA, not available; SEM, scanning electron microscopy; UreA, urease subunit A; VacA, vacuolar cytotoxin A.
TABLE 4.
Methodology of functional reported papers on liver cancer.
| First author | Year | Bacteria/sample | Method specifics | Particle analysis | Microscopy | Protein analysis | Antibody specifics |
|---|---|---|---|---|---|---|---|
| Shi, J | 2023 | Escherichia coli |
5000 × g for 30 min 10,000 × g for 30 min 0.45‐µm filter 0.22‐µm filter 150,000 × g for 3 h |
Y | Y | Y | OmpA OmpC |
| Caballano‐Infantes, E. | 2023 | Clostridioides difficile |
EV original size exclusion columns of 70 nm Vivaspin® 20 100 kDa |
N | Y | N | NA |
| Natsui, K | 2023 | E. coli |
2000 × g for 30 min 10,000 × g at 4°C for 30 min 0.2‐µm filter 100,000 × g at 4°C for 3 h 100,000 × g at 4°C for 2 h |
Y | Y | Y |
OmpA OmpC |
| Bolori, S | 2023 | Helicobacter pylori |
10,000 × g, 20 min 0.45‐µm cellulose filter 200,000 × g for 3 h |
Y | Y (SEM) | N | NA |
| Behzadi, E | 2017 | Lactobacillus rhamnosus |
2000 × g for 10 min 10,000 × g for 30 min 0.22‐µm filter Centricon Plus‐70 TL‐100 rotor was utilized for an ultracentrifugation achievement at 100,000 × g within 60 min |
N | Y (Low reliability) | N | NA |
Abbreviations: NA, not available; Omp, outer membrane protein; SEM, scanning electron microscopy.
TABLE 5.
Methodology of functional reported papers on oral cancer.
| First author | Year | Bacteria/sample | Method specifics | Particle analysis | Microscopy | Protein analysis | Antibody specifics |
|---|---|---|---|---|---|---|---|
| Groeger, S | 2020 | Porphyromonas gingivalis |
10,000 × g for 10 min 0.4‐µm filter 0.2‐µm filter 100,000 × g for 60 min 0.22‐µm Ultrafree spin filters 100,000 × g for 60 min |
N | N | N | NA |
| Liu, D | 2021 | P. gingivalis |
5000 × g for 20 min ExoBacteria OMV isolation kit |
Y | Y | N | NA |
| Fan, R | 2023 | P. gingivalis |
8000 × g for 15 min 0.22‐µm‐pore syringe filter 100,000 × g for 2 h |
Y | Y | Y | N, protein profile |
| Chen, G | 2023 | Fusobacterium nucleatum |
0.22‐µm filter 5000 × g for 30 min 100,000 × g for 120 min 0.22‐µm filter Density gradient ultracentrifugation using 45 Ti rotor 100,000 × g for 2 h |
Y | Y | N | NA |
Abbreviation: NA, not available.
TABLE 6.
Methodology of functional reported papers on other cancers.
| First author | Year | Bacteria/sample | Method specifics | Particle analysis | Microscopy | Protein analysis | Antibody specifics |
|---|---|---|---|---|---|---|---|
| Luo, ZW | 2021 | Akkermansia muciniphila |
6000 × g for 30 min 0.45‐µm filter Amicon Ultra‐15 Centrifugal Filter OptiPrep solution (60% w/v iodixanol; Sigma–Aldrich) at 100,000 × g for 18 h 0.22‐µm filter |
Y | Y | N | NA |
| Li, G | 2023 | Fusobacterium nucleatum |
200,000 × g for 60 min 0.22‐µm syringe filter |
Y | Y | N | NA |
| Jiang, Y | 2023 | Bifdobacterium |
5000 × g for 20 min 0.45‐µm filter 0.22‐µm filter 100 KDa filter ultrafiltration 3900 × g for 20 min 160,000 × g for 2 h |
Y | Y | N | NA |
| An, J | 2022 | Staphylococcus aureus |
A top‐bottom vacuum filter (Corning, NY, USA) with a pore size of 0.45 µm A bottle top vacuum 0.22‐µm filter 150,000 × g for 3 h on a type 45 Ti rotor |
Y | Y | N | NA |
| Firth, J | 2023 | Escherichia coli | 235,000 × g for 2 h with 45 Ti rotor | N | N | N | NA |
| Jin, L | 2022 | E. coli |
6000 × g for 20 min Sterile membrane filters (0.45‐µm pore size × 2 2500 × g/min, 10 min with 100‐kDa Centrifugal Filter Unit 110,000 × g for 70 min 0.45‐µm filter |
Y | Y | N | NA |
| Tang, B | 2018 | E. coli |
10,000 × g for 10 min 0.45‐µm filter 38,400 × g for 1 h 38,000 × g for 1–2 h 0.45‐µm filter |
Y | Y | N | NA |
| Gurunathan, S | 2023 | Bacillus licheniformis |
10,000 × g for 10 min 0.45‐ and 0.22 µm filters 100,000 × g for 3 h, 100 kDa filter |
Y | Y (TEM & SEM) | N | NA |
Abbreviations: NA, not available; SEM, scanning electron microscopy; TEM, transmission electron microscopy.
4. CLINICAL APPLICATIONS OF BEV
Bacteria are present throughout the body, and their BEVs can accurately reflect subtle changes in their state. This makes BEVs a promising tool for diagnosing and monitoring cancer prognosis in various body parts, as they preserve the original bacteria microecology during extraction. There were a few high‐quality review papers that provided comprehensive summaries (Aytar Çelik et al., 2022; Hosseini‐Giv et al., 2022; Li, Zhu, et al., 2023; Xie et al., 2022). In the following section, we expand on existing research by presenting novel findings and outlining the challenges encountered in clinical translation.
4.1. Diagnosis and prognosis
Several studies have explored the use of BEVs as potential non‐invasive, sensitive and specific biomarkers for cancer diagnosis and prognosis. A study comparing the BEVs derived from 227 faecal samples of CRC patients and healthy subjects found a significant reduction in microbial evenness and diversity in the CRC group (Park, Kim, et al., 2021). At the genus level, Catenibacterium, Erysipelotrichaceae, Faecalibacterium, Ruminococcus 2, Blautia, [Eubacterium] hallii group, [Ruminococcus] torques group, Oribacterium, Dorea and Collinsella were enriched in CRC group (Park, Kim, et al., 2021). Interestingly, Ruminococcus 2‐derived EVs exhibited a significant decrease in distal CRC cases compared to proximal CRC (Park, Kim, et al., 2021). Furthermore, Alistipes‐derived EVs have been proposed as novel biomarkers for CRC diagnosis and prognosis. Another study combined data on metabolites (leucine and oxalic acid) and bacterial genera (Collinsella and Solanum melongena) from faecal microbe‐derived EVs of CRC patients and healthy controls to construct an optimal model, achieving an AUC of 1 (Kim, Yang, et al., 2020). However, a sequencing analysis of 16S rRNA in urinary BEVs from CRC patients and healthy individuals yielded different findings, with no apparent disparity in the composition of gut microbiome between early‐ and late‐stage CRC patients, but late‐stage CRC exhibiting even higher α‐diversity compared to early‐stage cases (Yoon et al., 2023). In another study, a potential diagnostic model for distinguishing between GC and a healthy population was developed. The biomarkers, including Peptoniphilus, Diaphorobacter, Neisseria, Staphylococcus, Bifidobacterium, Corynebacterium 1, Actinomyces, Acinetobacter and Sphingomonas, based on urine‐derived BEVs achieved an AUC of 0.823, surpassing the performance of models based on circulating BEVs and faecal BEVs (Park, Kang, et al., 2021). Macro‐genomic analysis of circulating BEVs from HCC and healthy controls revealed that a model based on the five genera (Pseudomonas, Streptococcus, Staphylococcus, Bifidobacterium and Trabulsiella) achieved an AUC of 0.879 (Cho et al., 2019). Furthermore, by utilizing blood‐derived biomarker‐enriched BEVs, compositional differences in the microbiota between pancreatic cancer patients and healthy controls were identified, leading to the discovery of potential biomarkers (Kim et al., 2021). The selected markers of the optimal model at the phylum level were Verrucomicrobia and Actinomycetes, with an AUC of 0.966; 10 species were selected as markers at the genus level, with an AUC of 1. Serum BEVs were also sequenced in cases of benign ovarian tumours and ovarian cancer, which showed a higher abundance of Acinetobacter in ovarian cancer (Kim, Kang, et al., 2020). When combined with age and serum CA‐125 levels, the under area of AUC reached 0.846. It is worth noting that the methods used above are overly simplistic. All researchers claimed they harvested BEVs after centrifugation at 10,000 × g for 30 min (Table 7). However, free DNA may still remain in the supernatant. Therefore, subsequent researchers should use more appropriate methods to isolate and characterize BEVs to ensure accuracy and reliability. In summary, these results suggest that BEVs have great potential as biomarkers for cancer diagnosis and prognosis, but their efficiency and cost‐effectiveness compared to other models need to be demonstrated in clinical practice.
TABLE 7.
BEV isolated from biological samples in this review.
| First author | Year | Disease | Source | Method specifics | Characterization |
|---|---|---|---|---|---|
| Park, J | 2021 | Colorectal cancer | Faeces |
10,000 × g for 10 min at 4°C 0.22‐µm filter Boiled for 40 min at 100 °C 13,000 rpm for 30 min at 4°C DNA isolation kit (DNeasy PowerSoil Kit, QIAGEN, Germany) |
None |
| Kim DJ | 2020 | Colorectal cancer | Faeces |
10,000 × g for 10 min at 4°C 0.22‐µm filter Boiled for 40 min at 100 °C 13,000 rpm for 30 min at 4°C DNA isolation kit (DNeasy PowerSoil Kit, QIAGEN, Germany) |
None |
| Yoon H | 2023 | Colorectal cancer | Urine |
10,000 × g for 10 min at 4°C 0.22‐µm filter Boiled for 40 min at 100 °C 13,000 rpm for 30 min at 4°C DNA isolation kit (DNeasy PowerSoil Kit, QIAGEN, Germany) |
None |
| Park, JY | 2021 | Gastric cancer | Urine, serum, faeces |
10,000 × g for 10 min at 4°C 0.22‐µm filter Boiled for 40 min at 100 °C 13,000 rpm for 30 min at 4°C DNA isolation kit (DNeasy PowerSoil Kit, QIAGEN, Germany) |
None |
| Cho EJ | 2019 | Hepatocellular carcinoma | Serum |
2000 × g for 15 min at 4 °C proteinase K at 56 °C for 30 min Boiled for 40 min at 100 °C 10,000 × g for 30 min at 4°C DNA isolation kit (DNeasy Blood & Tissue Kit, QIAGEN, Germany) |
None |
| Kim JR | 2021 | Pancreatic cancer | Serum |
2000 × g for 15 min at 4 °C Proteinase K at 56 °C for 30 min Boiled for 40 min at 100 °C 10,000 × g for 30 min at 4°C DNA isolation kit (DNeasy Blood & Tissue Kit, QIAGEN, Germany) |
None |
| Kim SL | 2020 | Ovarian cancer | Serum |
3000 rpm for 15 min at 4°C 10,000 × g for 1 min at 4°C 0.22‐um filter Boiled for 40 min at 100 °C 13,000 rpm for 30 min at 4°C DNA isolation kit (PowerSoil DNA Isolation Kit, MO BIO, Carlsbad, CA, USA) |
None |
| Fizanne, L | 2023 | Non‐alcoholic fatty diseases non‐alcoholic steatohepatitis | Faeces, blood |
700 × g for 15 min 3600 × g for 15 min 7800 × g for 15 min 10,000 × g for 30 min 0.45‐µm filter 0.22‐µm filter 150,000 × g for 2 h |
TEM; NTA; WB (LTA) |
Abbreviations: LTA, lipoteichoic acid; NTA, nanoparticle tracking analysis; TEM, transmission electron microscopy; WB, western blot.
4.2. Cancer therapy
Researchers are exploring the use of bioengineered BEVs as drug delivery systems and immunotherapy agents for cancer treatment. Various technologies are being used to assemble or load chemotherapy drugs, photosensitizers, biotoxins and other therapeutic agents into targeted BEVs (Figure 2) (Feng et al., 2022; Li, Wu, et al., 2023; Qing et al., 2020; Ren et al., 2023, Zanella et al., 2021). Additionally, genetically engineered bacteria can curate specific cargo within BEVs. Compared to other nanomaterials, BEVs possess several advantages, including potent immune‐stimulating effects, non‐replicating nature, ease of modification, cargo protection, targeted delivery to tumours through the permeation and retention effect, and enhanced safety. However, there are also drawbacks, such as uncontrollable composition, the presence of residual bacterial toxins, susceptibility to immune clearance, and a lack of standardized large‐scale production methods.
FIGURE 2.
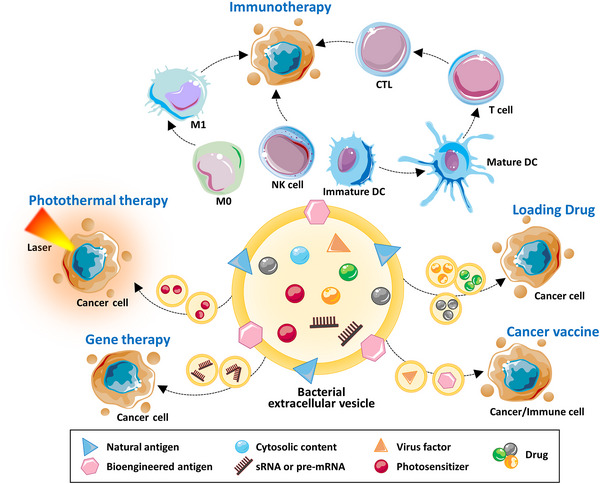
Multiple applications of BEV in cancer therapy. BEVs exhibit potent immunotherapeutic potential by modulating the immune microenvironment of tumours through induction of M1 polarization, augmentation of NK cell activity, and maturation of immature DCs to facilitate T cell differentiation into CTLs. The combination of BEV with photothermal therapy enables the activation of photosensitizers by external laser irradiation, thereby promoting tumour cell death. BEV also can serve as a carrier for various chemotherapy drugs to enhance their efficacy. When combined with gene therapy, BEV penetrates tumour cells and regulates the expression of crucial proteins to induce tumour cell death. Incorporating viral factors can be used to create tumour vaccines. The synergistic integration of these therapeutic modalities often yields optimal treatment outcomes. BEVs, bacterial extracellular vesicles; CTLs, cytotoxic T lymphocyte; DCs, dendritic cells.
BEVs act as immune adjuvants by utilizing their pathogen‐associated molecular patterns to activate anti‐cancer immunity through binding with host pattern recognition receptors. Nano‐vaccines based on BEV tumour antigens are currently being studied in cancer therapy (Wang et al., 2019). Kim et al. demonstrated that BEVs derived from both gram‐negative (E. coli) and gram‐positive (S. aureus and L. acidophilus) exhibit potential anti‐tumour effects via an active IFN‐γ response. This response endures multiple tumour challenges, establishing long‐term anti‐tumour immunity (Kim et al., 2017). Moreover, Carvalho and colleagues found that the intranasal route exhibited higher levels of IgA and IgG compared to oral gavage (Carvalho et al., 2019). This may be due to the upper respiratory tract providing a relatively milder environment, which facilitates the uptake of OMVs by host cells and makes them more accessible to the abundant immune cells in the submucosa. Indeed, studies used detoxified synthetic BEVs as an immunoadjuvant targeted to tumours (Carvalho et al., 2019; Park, Svennerholm, et al., 2021). Park et al. revealed that synthetic bacterial vesicles slowed down melanoma growth by inducing Th1‐type polarization, with the strongest inhibitory effect observed when combined with anti‐PD‐L1 treatment (Park, Svennerholm, et al., 2021). The BEVs were modified by inserting the OM domain of PD1, allowing them, to specifically bind to PD‐L1 on tumour cells and protect T cells from the PD1/PD‐L1 immunosuppressive mechanism (Li et al., 2020). BEVs can also be used for in situ vaccination optimization (Caproni et al., 2023). Salmonella Typhimurium‐derived BEVs (StEVs) act as an immunoadjuvant to increase the infiltration of CD49b‐expressing NKs (Aly et al., 2021). Genetic modification of StEVs’ flagella components significantly enhances the anti‐cancer immune response (Felgner et al., 2020). Surface engineering techniques have been employed to attach CD47 onto the BEV membrane, reducing its immunogenicity. Moreover, CD47‐BEVs remodelled the TME through induction of M1 polarization and blockade of immunosuppressive pathways, even showing long‐term immune memory when challenged by tumours again (Feng et al., 2022). BEVs derived from Neisseria meningitidis combined with ganglioside NAcGM3 also promote M1 polarization (Khan et al., 2022). Gut dysbiosis can hijack and alter the immune microenvironment, such as inducing T cell exhaustion, immunosuppression, thereby, increasing tumour susceptibility (Behary et al., 2021; Fan et al., 2021; Levy et al., 2017). However, immune checkpoint therapy often faces challenges of resistance and lack of durability. Therefore, BEV can serve as an essential immune adjuvant to overcome this dilemma.
Biological engineering systems often utilize BEVs as drug carriers to combine drugs with the immune capabilities of BEVs for more effective drug delivery and targeted therapy. For example, StEVs incorporate a tumour‐targeting ligand Arg‐Gly‐Asp while loading Tegafur, resulting in enhanced susceptibility of cancer cells to CTLs and efficient eradication of cancer cells (Chen et al., 2020). Attenuated Klebsiella pneumoniae‐derived BEVs loaded with DOX show high uptake efficiency by small cell lung cancer cells and exhibit a potent inhibitory effect on tumour growth (Kuerban et al., 2020). EcEVs loaded with perhexiline effectively shift the phenotypes of the TME from M2 to M1 (Jiang, Fu, et al., 2023). Loading MSN‐5‐FU within EcEVs significantly decreases the cumulative drug release rate, thereby prolonging the duration of drug action and mitigating toxic side effects in the treatment of oral squamous cell carcinoma (Huang et al., 2022).
Incorporating viral factors into BEVs can be employed to create tailored tumour vaccines for specific malignancies. For example, the tumoral antigen human papillomavirus type 16 early protein E7 has been successfully incorporated into EcEVs to prepare a potent tumour vaccine (Wang et al., 2017). BEV‐based vaccines also facilitate dendritic cell maturation, inducing the differentiation in CTLs (Wang et al., 2017). Encapsulating autophagy‐inducing virus AD within BEVs effectively triggers autophagy at the tumour site and promotes an immunologically favourable TME (Ban et al., 2023). FnEVs encapsulated with HSV‐1 oncolytic viruses enhanced ZBP1‐mediated PANoptosis in tumour cells (Wang et al., 2024). Additionally, FnEVs improved the efficacy of anti‐PD‐L1 therapy and established long‐term immune memory (Wang et al., 2024).
The non‐replicative nature of BEVs is advantageous in the context of gene therapy. Loading siRNA into BEVs is a viable approach for modulating gene expression in tumour cells. Genetically engineered BEVs loaded with siRNA targeting kinesin spindle protein effectively silenced the human epidermal growth factor receptor 2, resulting in significant regression of tumour growth (Gujrati et al., 2014). EcEVs have also been engineered to carry pre‐microRNAs that can be internalized by cancer cells and suppress the expression of the oncogene CXCR4 (Cui et al., 2022). Furthermore, surface expression of PD1 on BEVs and encapsulation of miRNA‐34a within BEVs synergistically activate immune responses and enhance the therapeutic efficacy of immune checkpoint blockade (Cui et al., 2023).
Combining BEVs with photothermal therapy (PTT) has been shown to enhance tumour antigen recognition and overcome the immunosuppressive microenvironment (Li et al., 2022; Zhang et al., 2022; Zhuang et al., 2021, 2022). In breast cancer, BEVs loaded with photosensitizers and DOX can be phagocytosed by macrophages, inducing cancer cells pyroptosis (Li, Wu, et al., 2023). Additionally, BEVs loaded with photosensitizers and coated with PD‐L1 antibody can activate DCs and CD8+ T cells (Zhang et al., 2022). Furthermore, EcEVs combined with PTT and αvβ3 integrin peptide targeting ligand show potential for complete eradication of melanoma (Gu et al., 2020; Peng et al., 2020). Modification of EcEVs enables targeted delivery of Fe3O4‐MnO2 to the tumour site, where they undergo reactive decomposition and exert a photothermal therapeutic effect (Liu et al., 2023). These ions induce immunogenic cell death and regulate the hypoxic environment of the tumour by supplying oxygen (Liu et al., 2023). Another promising approach involves the use of siRNA‐OMVs incorporating photothermal‐sensitive liposomes and anti‐PD‐1 therapy, which exhibit a pronounced tumour inhibitory effect (Zhai et al., 2021).
In summary, BEVs exhibit high manoeuvrability and multifaceted anticancer properties, allowing for their integration with various treatment modalities and offering alternative options for patients with refractory cancers and resistance. As mentioned earlier, BEVs can typically be combined with various therapeutic approaches to achieve optimal results. The immunotherapeutic potential of BEVs sets them apart from other EVs, making them promising candidates for clinical translation.
5. FUTURE PERSPECTIVES
The field of BEV involvement in pathologies, particularly cancer, is rapidly advancing with new evidence. This suggests a promising avenue for understanding the disease and potentially improving diagnostics, therapeutics and treatment outcomes. However, further research is necessary to address key questions and fill knowledge gaps in order to refine the science.
5.1. BEV cargo
The advancement of multi‐omics technology has revealed that BEVs are more than just carriers of harmful substances – they act as ‘trojan horses’ with a diverse cargo profile and complex purposes. While their involvement in processes like virulence factor transfer and colonization facilitation is evident, their direct relation to carcinogenesis is still underdeveloped. To better understand this, it is necessary to investigate cargo sorting mechanisms and assembly.
Despite emerging evidence, there is currently no universally accepted model for the biogenesis of prokaryotic EVs. The existing knowledge on packaging, sorting and biogenesis remains controversial. Additionally, the exact mechanism of interaction with host processes and the disruptions caused need to be characterized to identify therapeutic targets. The idea that specific enzymes dissolve host cell membranes, for example, has been disputed in existing literature. Furthermore, the precise alteration of the genetic and proteomic landscape in recipient cells for tumorigenesis is not well understood. For instance, the validation of how BEV‐derived RBA silences genes in human cells is still lacking. It is unclear whether it functions as an RNAi system or utilizes ribosomes to enable bacterial gene translation. Additionally, the roles of proximal or internally localized nucleic acids and whether both populations play unique roles in host‐genome immunogenicity interference system need to be explored. The study of RNA secretory mechanisms for BEVs packaging is also imperative. In terms of DNA, the causal relationship must be supplemented with quantification of the efficiency of genetic transfer, especially in the context of BEV vaccine or treatment development. Meanwhile, the investigation of membrane proteins, which are notoriously difficult to study, is crucial for understanding downstream signalling effects.
The lack of consensus on sample preparation compounds the difficulty of distinguishing cargo from structural proteins, and the current use of LPS and LTA for probing membrane proteins via western blot has been highly controversial. Similar challenges are faced in studying lipids, as extraction methods may introduce population or isomer bias, making it difficult to preserve native bilayer membrane lipids and separate them from lipid cargo. Understanding lipid composition is crucial for studying how it mediates host‐microbial interactions during pathogenesis versus commensalism. Overall, the effects of growth conditions, strains, and other permutations should also be studied to gain a holistic view of metabolites, nucleic acids and all macromolecular cargo. Standardizing preparation methods for fairer comparison across studies and conducting a comprehensive meta‐analysis of BEV cargo populations is key. The ability to selectively package or engineer specific contents into BEVs holds great potential as targeted delivery platforms for various diseases.
5.2. BEV research methodology
We can employ 16s rRNA sequencing to differentiate the types of bacteria. Whole‐genome shotgun metagenomic sequencing could offer more precise identification down to the species level, along with their functional characteristics. However, completely eradicating contamination from the environment and reagents remains a challenge, so it is important to include blank controls to assess external contamination. Additionally, the genetic material loaded in BEVs may not be sufficient for sequencing. To address this issue, a strategy based on Raman spectroscopy and deep learning algorithms has been introduced to provide molecular information for identifying bacterial strains (Qin et al., 2022). Combining this approach with a single‐particle automated Raman trapping system allows for analysis at the individual particle level, improving diagnostics and disease monitoring (Penders et al., 2021).
Isolation and purification of BEVs are critical steps in relevant studies. Various methodologies have been reported, establishing a standard, straightforward and cost‐effective method for isolating and purifying BEVs would greatly benefit the field. Researchers have attempted to develop a method based on orthogonal size and density for isolating BEVs from stool and blood plasma (Tulkens, De Wever, et al., 2020). Particularly for diagnostic purposes, it is important to isolate BEVs from eukaryotic EVs. Wu and his colleagues were among the first to focus on optimizing BEVs isolation from faecal samples of mice and humans (Wu et al., 2019). Recently, Erttmann and Gekara described BEVs isolation protocols from mouse blood and colon, but there is a lack of diverse published methods to compare with when establishing an industry gold‐standard (Erttmann & Gekara, 2023). Some studies have utilized immunomagnetic isolation techniques using antibody‐labelled magnetic beads capable of selectively capturing EV surface markers (Koliha et al., 2016). By applying a magnetic field, captured EVs can be separated from other substances (Oksvold et al., 2015). There is promising potential to utilize specific bacterial markers (e.g., LTA, LPS, flagellin) for isolating BEVs from eukaryotic EVs (Schaack et al., 2022). Establishing a unanimous definition of BEV markers would significantly advance research in this area. Currently, most articles rely on NTA for particle analysis and TEM for particle morphology. However, there are no established protein markers that reliably indicate BEV quality. To bridge this gap, some researchers have explored using LPS, LTA and flagellin as potential markers for BEVs (Li, Hsu, et al., 2023; Piroli et al., 2023). Certain BEVs, such as EcEVs, prefer using OmpA as a marker protein, while HpEVs favour VacA. Implementing a standard reporting system for BEV research would be valuable in avoiding misinterpretation of findings and ensuring the repeatability and reproducibility of studies (Watson et al., 2021). Following the guidelines of the EV‐TRACK knowledgebase, we compiled methodology tables of current functional BEV research in the field of cancer (Tables 2, 3, 4, 5, 6). It is recommended that any researcher wishing to conduct studies on BEVs should refer to the guidelines provided by MISEV2023 (Welsh et al., 2024).
Another significant challenge in studying the functional role of BEVs in inter‐kingdom communication lies in the physiological relevance of experimental methods. In laboratory settings, BEVs are often produced in large volumes and concentrated before undergoing functional characterization. However, the amount of BEVs used in these studies may not be physiologically relevant to the levels present in the human circulatory system. Moreover, the composition of BEVs produced in culture media can differ significantly from those produced in vivo. Therefore, it is essential to carefully report, examine and interpret these findings to ensure accurate information and conclusions.
5.3. BEV in homeostasis and pathology
In recent years, research on gut microbiota has shifted from a focus on resident bacteria within the gut to a consideration of BEVs that can enter the systemic circulation in the human body (Bittel et al., 2021; Jones et al., 2020; Park et al., 2017; Poore et al., 2020; Stentz et al., 2018; Tulkens, De Wever, et al., 2020, Tulkens, Vergauwen, et al., 2020). These BEVs have demonstrated significant potential for various multifunctional applications, particularly in the field of cancer‐related diagnosis and treatment. However, despite their immense potential value, a key challenge faced by BEVs is the limited amount of relevant research available (Castillo‐Romero et al., 2023). The evidence of BEVs crossing the altered epithelial barrier has brought them into the spotlight as distant mediators of inter‐kingdom communication and potential biomarkers for monitoring human diseases. While there is a consensus that these BEVs likely originate from gut bacteria, further studies are required to ascertain the contributions from microbial niches in different organ sites. Moreover, further investigation is necessary to determine the precise mechanisms by which BEVs enter the circulatory system, whether they are involved in compromising the gut epithelial barrier for this purpose, or if it is a consequence of gut dysbiosis. The direction of causation and further positive feedback loops during disease onset should further be investigated.
5.4. BEV related therapeutics
Indeed, BEVs offer intrinsic advantages. The pharmacokinetic profile of BEVs demonstrates a peak at 3 h post‐administration, followed by a gradual decline within 12 h, indicating their effectiveness as mediators of long‐distance communication in vivo (Jang et al., 2015). Furthermore, engineered OMVs can be designed for oral administration, improving patient compliance (Yue et al., 2022). Additionally, faecal microbiota transplantation (FMT) shows promise as a potential therapeutic approach for targeting gut microbiota‐associated cancer. Clinical trials have demonstrated its efficacy in overcoming resistance to anti‐PD‐1 treatment in melanoma (Baruch et al., 2021; Davar et al., 2021). It is conceivable that by administering BEVs derived from a healthy host, ‘good’ bacteria or engineered strains, similar or even better efficacy could be achieved in individuals receiving FMT treatments. Moreover, the utilization of natural or engineered BEVs for targeted delivery to antigen‐presenting cells is considered a hot research topic in the development of cancer vaccines.
Rigorous evaluation and the establishment of regulatory guidelines will be essential to establish the necessary standards for clinical translation. As our understanding deepens and breakthroughs emerge, BEVs have the potential to revolutionize the field of medicine and significantly impact patient care.
AUTHOR CONTRIBUTIONS
Aijun Liang: Writing – original draft (lead); writing – review and editing (equal). Lavisha Korani: Writing – review and editing (equal). Cherlie Lot Sum Yeung: Writing – original draft (Supporting). Sze Keong Tey: Conceptualization (equal); writing – review and editing (equal). Judy Wai Ping Yam: Conceptualization (equal); funding acquisition (lead); writing – review and editing (equal).
CONFLICT OF INTEREST STATEMENT
The authors declare no conflicts of interest.
ACKNOWLEDGEMENTS
This work was supported by National Natural Science Foundation of China (Grant No. 81872340 and 82072626).
Liang, A. , Korani, L. , Yeung, C. L. S. , Tey, S. K. , & Yam, J. W. P. (2024). The emerging role of bacterial extracellular vesicles in human cancers. Journal of Extracellular Vesicles, 13, e12521. 10.1002/jev2.12521
Contributor Information
Sze Keong Tey, Email: szekeong@hku.hk.
Judy Wai Ping Yam, Email: judyyam@pathology.hku.hk.
REFERENCES
- Abedi, A. , Tafvizi, F. , Jafari, P. , & Akbari, N. (2024). The inhibition effects of Lentilactobacillus buchneri‐derived membrane vesicles on AGS and HT‐29 cancer cells by inducing cell apoptosis. Scientific Reports, 14(1), 3100. 10.1038/s41598-024-53773-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmadi Badi, S. , Moshiri, A. , Ettehad Marvasti, F. , Mojtahedzadeh, M. , Kazemi, V. , & Siadat, S. D. (2020). Extraction and evaluation of outer membrane vesicles from two important gut microbiota members, Bacteroides fragilis and bacteroides thetaiotaomicron. Cell Journal, 22(3), 344–349. 10.22074/cellj.2020.6499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed, A. A. Q. , Qi, F. , Zheng, R. , Xiao, L. , Abdalla, A. M. E. , Mao, L. , Bakadia, B. M. , Liu, L. , Atta, O. M. , Li, X. , Shi, Z. , & Yang, G. (2021). The impact of ExHp‐CD (outer membrane vesicles) released from Helicobacter pylori SS1 on macrophage RAW 264.7 cells and their immunogenic potential. Life Sciences, 279, 119644. 10.1016/j.lfs.2021.119644 [DOI] [PubMed] [Google Scholar]
- Aktar, S. , Okamoto, Y. , Ueno, S. , Tahara, Y. O. , Imaizumi, M. , Shintani, M. , Miyata, M. , Futamata, H. , Nojiri, H. , & Tashiro, Y. (2021). Incorporation of plasmid DNA into bacterial membrane vesicles by peptidoglycan defects in Escherichia coli . Frontiers in Microbiology, 12, 747606. 10.3389/fmicb.2021.747606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida, A. , Nayfach, S. , Boland, M. , Strozzi, F. , Beracochea, M. , Shi, Z. J. , Pollard, K. S. , Sakharova, E. , Parks, D. H. , Hugenholtz, P. , Segata, N. , Kyrpides, N. C. , & Finn, R. D. (2021). A unified catalog of 204,938 reference genomes from the human gut microbiome. Nature Biotechnology, 39(1), 105–114. 10.1038/s41587-020-0603-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez, C. S. , Giménez, R. , Cañas, M. A. , Vera, R. , Díaz‐Garrido, N. , Badia, J. , & Baldomà, L. (2019). Extracellular vesicles and soluble factors secreted by Escherichia coli Nissle 1917 and ECOR63 protect against enteropathogenic E. coli‐induced intestinal epithelial barrier dysfunction. BMC Microbiology, 19(1), 166. 10.1186/s12866-019-1534-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aly, R. G. , El‐Enbaawy, M. I. , Abd El‐Rahman, S. S. , & Ata, N. S. (2021). Antineoplastic activity of Salmonella Typhimurium outer membrane nanovesicles. Experimental Cell Research, 399(1), 112423. 10.1016/j.yexcr.2020.112423 [DOI] [PubMed] [Google Scholar]
- Amieva, M. R. , Vogelmann, R. , Covacci, A. , Tompkins, L. S. , Nelson, W. J. , & Falkow, S. (2003). Disruption of the epithelial apical‐junctional complex by Helicobacter pylori CagA. Science, 300(5624), 1430–1434. 10.1126/science.1081919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An, J. , & Ha, E. M. (2022a). Extracellular vesicles derived from Lactobacillus plantarum restore chemosensitivity through the PDK2‐mediated glucose metabolic pathway in 5‐FU‐resistant colorectal cancer cells. Journal of Microbiology (Seoul, Korea), 60(7), 735–745. 10.1007/s12275-022-2201-1 [DOI] [PubMed] [Google Scholar]
- An, J. , Kwon, H. , Lim, W. , & Moon, B. I. (2022b). Staphylococcus aureus‐derived extracellular vesicles enhance the efficacy of endocrine therapy in breast cancer cells. Journal of Clinical Medicine, 11(7), 2030. 10.3390/jcm11072030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreoni, F. , Toyofuku, M. , Menzi, C. , Kalawong, R. , Mairpady Shambat, S. , François, P. , Zinkernagel, A. S. , & Eberl, L. (2019). Antibiotics stimulate formation of vesicles in Staphylococcus aureus in both phage‐dependent and ‐independent fashions and via different routes. Antimicrobial Agents and Chemotherapy, 63(2), e01439. 18. 10.1128/aac.01439-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aytar Çelik, P. , Derkuş, B. , Erdoğan, K. , Barut, D. , Blaise Manga, E. , Yıldırım, Y. , Pecha, S. , & Çabuk, A. (2022). Bacterial membrane vesicle functions, laboratory methods, and applications. Biotechnology Advances, 54, 107869. 10.1016/j.biotechadv.2021.107869 [DOI] [PubMed] [Google Scholar]
- Badi, S. A. , Motahhary, A. , Bahramali, G. , Masoumi, M. , Khalili, S. F. S. , Ebrahimzadeh, N. , Nouri, P. , Rahimi, A. , Masotti, A. , Moshiri, A. , & Siadat, S. D. (2020). The regulation of Niemann‐Pick C1‐like 1 (NPC1L1) gene expression in opposite direction by Bacteroides spp. and related outer membrane vesicles in Caco‐2 cell line. Journal of Diabetes & Metabolic Disorders, 19(1), 415–422. 10.1007/s40200-020-00522-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban, W. , Sun, M. , Huang, H. , Huang, W. , Pan, S. , Liu, P. , Li, B. , Cheng, Z. , He, Z. , Liu, F. , & Sun, J. (2023). Engineered bacterial outer membrane vesicles encapsulating oncolytic adenoviruses enhance the efficacy of cancer virotherapy by augmenting tumor cell autophagy. Nature Communications, 14(1), 2933. 10.1038/s41467-023-38679-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baruch, E. N. , Youngster, I. , Ben‐Betzalel, G. , Ortenberg, R. , Lahat, A. , Katz, L. , Adler, K. , Dick‐Necula, D. , Raskin, S. , Bloch, N. , Rotin, D. , Anafi, L. , Avivi, C. , Melnichenko, J. , Steinberg‐Silman, Y. , Mamtani, R. , Harati, H. , Asher, N. , Shapira‐Frommer, R. , … Boursi, B. (2021). Fecal microbiota transplant promotes response in immunotherapy‐refractory melanoma patients. Science, 371(6529), 602–609. 10.1126/science.abb5920 [DOI] [PubMed] [Google Scholar]
- Behary, J. , Raposo, A. E. , Amorim, N. M. L. , Zheng, H. , Gong, L. , McGovern, E. , Chen, J. , Liu, K. , Beretov, J. , Theocharous, C. , Jackson, M. T. , Seet‐Lee, J. , McCaughan, G. W. , El‐Omar, E. M. , & Zekry, A. (2021). Defining the temporal evolution of gut dysbiosis and inflammatory responses leading to hepatocellular carcinoma in Mdr2 ‐/‐ mouse model. BMC Microbiology, 21(1), 113. 10.1186/s12866-021-02171-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behzadi, E. , Mahmoodzadeh Hosseini, H. , & Imani Fooladi, A. A. (2017). The inhibitory impacts of Lactobacillus rhamnosus GG‐derived extracellular vesicles on the growth of hepatic cancer cells. Microbial Pathogenesis, 110, 1–6. 10.1016/j.micpath.2017.06.016 [DOI] [PubMed] [Google Scholar]
- Bertocchi, A. , Carloni, S. , Ravenda, P. S. , Bertalot, G. , Spadoni, I. , Lo Cascio, A. , Gandini, S. , Lizier, M. , Braga, D. , Asnicar, F. , Segata, N. , Klaver, C. , Brescia, P. , Rossi, E. , Anselmo, A. , Guglietta, S. , Maroli, A. , Spaggiari, P. , Tarazona, N. , … Rescigno, M. (2021). Gut vascular barrier impairment leads to intestinal bacteria dissemination and colorectal cancer metastasis to liver. Cancer Cell, 39(5), 708–724. e11. 10.1016/j.ccell.2021.03.004 [DOI] [PubMed] [Google Scholar]
- Bielaszewska, M. , Marejková, M. , Bauwens, A. , Kunsmann‐Prokscha, L. , Mellmann, A. , & Karch, H. (2018). Enterohemorrhagic Escherichia coli O157 outer membrane vesicles induce interleukin 8 production in human intestinal epithelial cells by signaling via Toll‐like receptors TLR4 and TLR5 and activation of the nuclear factor NF‐κB. International Journal of Medical Microbiology, 308(7), 882–889. 10.1016/j.ijmm.2018.06.004 [DOI] [PubMed] [Google Scholar]
- Bittel, M. , Reichert, P. , Sarfati, I. , Dressel, A. , Leikam, S. , Uderhardt, S. , Stolzer, I. , Phu, T. A. , Ng, M. , Vu, N. K. , Tenzer, S. , Distler, U. , Wirtz, S. , Rothhammer, V. , Neurath, M. F. , Raffai, R. L. , Günther, C. , & Momma, S. (2021). Visualizing transfer of microbial biomolecules by outer membrane vesicles in microbe‐host‐communication in vivo. Journal of Extracellular Vesicles, 10(12), e12159. 10.1002/jev2.12159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolori, S. , Shegefti, S. , Baghaei, K. , Yadegar, A. , Moon, K. M. , Foster, L. J. , Nasiri, M. J. , & Dabiri, H. (2023). The effects of Helicobacter pylori‐derived outer membrane vesicles on hepatic stellate cell activation and liver fibrosis in vitro. BioMed Research International, 2023, 4848643. 10.1155/2023/4848643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos, M. P. , Robert, V. , & Tommassen, J. (2007). Biogenesis of the gram‐negative bacterial outer membrane. Annual Review of Microbiology, 61, 191–214. 10.1146/annurev.micro.61.080706.093245 [DOI] [PubMed] [Google Scholar]
- Brown, L. , Wolf, J. M. , Prados‐Rosales, R. , & Casadevall, A. (2015). Through the wall: Extracellular vesicles in gram‐positive bacteria, mycobacteria and fungi. Nature Reviews Microbiology, 13(10), 620–30. 10.1038/nrmicro3480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caballano‐Infantes, E. , Ho‐Plagaro, A. , Lopez‐Gomez, C. , Martin‐Reyes, F. , Rodriguez‐Pacheco, F. , Taminiau, B. , Daube, G. , Garrido‐Sanchez, L. , Alcain‐Martinez, G. , Andrade, R. J. , Garcia‐Cortes, M. , Lucena, M. I. , Garcia‐Fuentes, E. , & Rodriguez‐Diaz, C. (2023). Membrane vesicles of toxigenic Clostridioides difficile affect the metabolism of liver HepG2 cells. Antioxidants (Basel), 12(4), 818. 10.3390/antiox12040818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, Y. , & Lin, H. (2021). Characterization and function of membrane vesicles in Gram‐positive bacteria. Applied Microbiology and Biotechnology, 105(5), 1795–1801. 10.1007/s00253-021-11140-1 [DOI] [PubMed] [Google Scholar]
- Caproni, E. , Corbellari, R. , Tomasi, M. , Isaac, S. J. , Tamburini, S. , Zanella, I. , Grigolato, M. , Gagliardi, A. , Benedet, M. , Baraldi, C. , Croia, L. , Di Lascio, G. , Berti, A. , Valensin, S. , Bellini, E. , Parri, M. , Grandi, A. , & Grandi, G. (2023). Anti‐tumor efficacy of in situ vaccination using bacterial outer membrane vesicles. Cancers (Basel), 15(13), 3328. 10.3390/cancers15133328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho, A. L. , Fonseca, S. , Miquel‐Clopés, A. , Cross, K. , Kok, K. S. , Wegmann, U. , Gil‐Cordoso, K. , Bentley, E. G. , Al Katy, S. H. M. , Coombes, J. L. , Kipar, A. , Stentz, R. , Stewart, J. P. , & Carding, S. R. (2019). Bioengineering commensal bacteria‐derived outer membrane vesicles for delivery of biologics to the gastrointestinal and respiratory tract. Journal of Extracellular Vesicles, 8(1), 1632100. 10.1080/20013078.2019.1632100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellarin, M. , Warren, R. L. , Freeman, J. D. , Dreolini, L. , Krzywinski, M. , Strauss, J. , Barnes, R. , Watson, P. , Allen‐Vercoe, E. , Moore, R. A. , & Holt, R. A. (2012). Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Research, 22(2), 299–306. 10.1101/gr.126516.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo‐Romero, K. F. , Santacruz, A. , & Gonzalez‐Valdez, J. (2023). Production and purification of bacterial membrane vesicles for biotechnology applications: Challenges and opportunities. Electrophoresis, 44(1‐2), 107–124. 10.1002/elps.202200133 [DOI] [PubMed] [Google Scholar]
- Chelakkot, C. , Choi, Y. , Kim, D. K. , Park, H. T. , Ghim, J. , Kwon, Y. , Jeon, J. , Kim, M. S. , Jee, Y. K. , Gho, Y. S. , Park, H. S. , Kim, Y. K. , & Ryu, S. H. (2018). Akkermansia muciniphila‐derived extracellular vesicles influence gut permeability through the regulation of tight junctions. Experimental & Molecular Medicine, 50(2), e450. 10.1038/emm.2017.282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, G. , Gao, C. , Jiang, S. , Cai, Q. , Li, R. , Sun, Q. , Xiao, C. , Xu, Y. , Wu, B. , & Zhou, H. (2023a). Fusobacterium nucleatum outer membrane vesicles activate autophagy to promote oral cancer metastasis. Journal of Advanced Research, 56, 167–179. 10.1016/j.jare.2023.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L. , Zhao, R. , Shen, J. , Liu, N. , Zheng, Z. , Miao, Y. , Zhu, J. , Zhang, L. , Wang, Y. , Fang, H. , Zhou, J. , Li, M. , Yang, Y. , Liu, Z. , & Chen, Q. (2023b). Antibacterial Fusobacterium nucleatum‐mimicking nanomedicine to selectively eliminate tumor‐colonized bacteria and enhance immunotherapy against colorectal cancer. Advanced Materials, 35(45), e2306281. 10.1002/adma.202306281 [DOI] [PubMed] [Google Scholar]
- Chen, Q. , Bai, H. , Wu, W. , Huang, G. , Li, Y. , Wu, M. , Tang, G. , & Ping, Y. (2020). Bioengineering bacterial vesicle‐coated polymeric nanomedicine for enhanced cancer immunotherapy and metastasis prevention. Nano Letters, 20(1), 11–21. 10.1021/acs.nanolett.9b02182 [DOI] [PubMed] [Google Scholar]
- Chen, S. , Lei, Q. , Zou, X. , & Ma, D. (2023c). The role and mechanisms of gram‐negative bacterial outer membrane vesicles in inflammatory diseases. Frontiers in Immunology, 14, 1157813. 10.3389/fimmu.2023.1157813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, K. , Zhao, R. , Li, Y. , Qi, Y. , Wang, Y. , Zhang, Y. , Qin, H. , Qin, Y. , Chen, L. , Li, C. , Liang, J. , Li, Y. , Xu, J. , Han, X. , Anderson, G. J. , Shi, J. , Ren, L. , Zhao, X. , & Nie, G. (2021). Bioengineered bacteria‐derived outer membrane vesicles as a versatile antigen display platform for tumor vaccination via plug‐and‐display technology. Nature Communications, 12(1), 2041. 10.1038/s41467-021-22308-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitcholtan, K. , Hampton, M. B. , & Keenan, J. I. (2008). Outer membrane vesicles enhance the carcinogenic potential of Helicobacter pylori . Carcinogenesis, 29(12), 2400–24005. 10.1093/carcin/bgn218 [DOI] [PubMed] [Google Scholar]
- Cho, E. J. , Leem, S. , Kim, S. A. , Yang, J. , Lee, Y. B. , Kim, S. S. , Cheong, J. Y. , Cho, S. W. , Kim, J. W. , Kim, S. M. , Yoon, J. H. , & Park, T. (2019). Circulating microbiota‐based metagenomic signature for detection of hepatocellular carcinoma. Scientific Reports, 9(1), 7536. 10.1038/s41598-019-44012-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, H. I. , Choi, J. P. , Seo, J. , Kim, B. J. , Rho, M. , Han, J. K. , & Kim, J. G. (2017). Helicobacter pylori‐derived extracellular vesicles increased in the gastric juices of gastric adenocarcinoma patients and induced inflammation mainly via specific targeting of gastric epithelial cells. Experimental & Molecular Medicine, 49(5), e330. 10.1038/emm.2017.47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, J. H. , Moon, C. M. , Shin, T. S. , Kim, E. K. , McDowell, A. , Jo, M. K. , Joo, Y. H. , Kim, S. E. , Jung, H. K. , Shim, K. N. , Jung, S. A. , & Kim, Y. K. (2020). Lactobacillus paracasei‐derived extracellular vesicles attenuate the intestinal inflammatory response by augmenting the endoplasmic reticulum stress pathway. Experimental & Molecular Medicine, 52(3), 423–437. 10.1038/s12276-019-0359-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cros, M. P. , Mir‐Pedrol, J. , Toloza, L. , Knödlseder, N. , Maruotti, J. , Zouboulis, C. C. , Güell, M. , & Fábrega, M. J. (2023). New insights into the role of Cutibacterium acnes‐derived extracellular vesicles in inflammatory skin disorders. Scientific Reports, 13(1), 16058. 10.1038/s41598-023-43354-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, C. , Guo, T. , Zhang, S. , Yang, M. , Cheng, J. , Wang, J. , Kang, J. , Ma, W. , Nian, Y. , Sun, Z. , & Weng, H. (2022). Bacteria‐derived outer membrane vesicles engineered with over‐expressed pre‐miRNA as delivery nanocarriers for cancer therapy. Nanomedicine, 45, 102585. 10.1016/j.nano.2022.102585 [DOI] [PubMed] [Google Scholar]
- Cui, C. , He, Q. , Wang, J. , Kang, J. , Ma, W. , Nian, Y. , Sun, Z. , & Weng, H. (2023). Targeted miR‐34a delivery with PD1 displayed bacterial outer membrane vesicles‐coated zeolitic imidazolate framework nanoparticles for enhanced tumor therapy. International Journal of Biological Macromolecules, 247, 125692. 10.1016/j.ijbiomac.2023.125692 [DOI] [PubMed] [Google Scholar]
- Cullin, N. , Azevedo Antunes, C. , Straussman, R. , Stein‐Thoeringer, C. K. , & Elinav, E. (2021). Microbiome and cancer. Cancer Cell, 39(10), 1317–1341. 10.1016/j.ccell.2021.08.006 [DOI] [PubMed] [Google Scholar]
- da Silva Barreira, D. , Lapaquette, P. , Novion Ducassou, J. , Couté, Y. , Guzzo, J. , & Rieu, A. (2022). Spontaneous prophage induction contributes to the production of membrane vesicles by the gram‐positive bacterium Lacticaseibacillus casei BL23. mBio, 13(5), e0237522. 10.1128/mbio.02375-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, Z. , Coker, O. O. , Nakatsu, G. , Wu, W. K. K. , Zhao, L. , Chen, Z. , Chan, F. K. L. , Kristiansen, K. , Sung, J. J. Y. , Wong, S. H. , & Yu, J. (2018). Multi‐cohort analysis of colorectal cancer metagenome identified altered bacteria across populations and universal bacterial markers. Microbiome, 6(1), 70. 10.1186/s40168-018-0451-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauros‐Singorenko, P. , Blenkiron, C. , Phillips, A. , & Swift, S. (2018). The functional RNA cargo of bacterial membrane vesicles. FEMS Microbiology Letters, 365(5). 10.1093/femsle/fny023 [DOI] [PubMed] [Google Scholar]
- Davar, D. , Dzutsev, A. K. , McCulloch, J. A. , Rodrigues, R. R. , Chauvin, J. M. , Morrison, R. M. , Deblasio, R. N. , Menna, C. , Ding, Q. , Pagliano, O. , Zidi, B. , Zhang, S. , Badger, J. H. , Vetizou, M. , Cole, A. M. , Fernandes, M. R. , Prescott, S. , Costa, R. G. F. , Balaji, A. K. , … Zarour, H. M. (2021). Fecal microbiota transplant overcomes resistance to anti‐PD‐1 therapy in melanoma patients. Science, 371(6529), 595–602. 10.1126/science.abf3363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deatherage, B. L. , & Cookson, B. T. (2012). Membrane vesicle release in bacteria, eukaryotes, and archaea: a conserved yet underappreciated aspect of microbial life. Infection and Immunity, 80(6), 1948–57. 10.1128/iai.06014-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz‐Garrido, N. , Badia, J. , & Baldoma, L. (2021). Microbiota‐derived extracellular vesicles in interkingdom communication in the gut. Journal of Extracellular Vesicles, 10(13), e12161. 10.1002/jev2.12161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elhenawy, W. , Bording‐Jorgensen, M. , Valguarnera, E. , Haurat, M. F. , Wine, E. , & Feldman, M. F. (2016). LPS remodeling triggers formation of outer membrane vesicles in salmonella. mBio, 7(4), e00940. 16. 10.1128/mBio.00940-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engevik, M. A. , Danhof, H. A. , Ruan, W. , Engevik, A. C. , Chang‐Graham, A. L. , Engevik, K. A. , Shi, Z. , Zhao, Y. , Brand, C. K. , Krystofiak, E. S. , Venable, S. , Liu, X. , Hirschi, K. D. , Hyser, J. M. , Spinler, J. K. , Britton, R. A. , & Versalovic, J. (2021). Fusobacterium nucleatum secretes outer membrane vesicles and promotes intestinal inflammation. mBio, 12(2), e02706. 20. 10.1128/mBio.02706-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erttmann, S. F. , & Gekara, N. O. (2023). Protocol for isolation of microbiota‐derived membrane vesicles from mouse blood and colon. STAR Protocols, 4(1), 102046. 10.1016/j.xpro.2023.102046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faddetta, T. , Renzone, G. , Vassallo, A. , Rimini, E. , Nasillo, G. , Buscarino, G. , Agnello, S. , Licciardi, M. , Botta, L. , Scaloni, A. , Palumbo Piccionello, A. , Puglia, A. M. , & Gallo, G. (2022). Streptomyces coelicolor vesicles: Many molecules to be delivered. Applied and Environmental Microbiology, 88(1), e0188121. 10.1128/aem.01881-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakharian, F. , Sadeghi, A. , Pouresmaeili, F. , Soleimani, N. , & Yadegar, A. (2024). Anti‐inflammatory effects of extracellular vesicles and cell‐free supernatant derived from Lactobacillus crispatus strain RIGLD‐1 on Helicobacter pylori‐induced inflammatory response in gastric epithelial cells in vitro. Folia Microbiologica, 69(4), 927–939. 10.1007/s12223-024-01138-3 [DOI] [PubMed] [Google Scholar]
- Fan, R. , Zhou, Y. , Chen, X. , Zhong, X. , He, F. , Peng, W. , Li, L. , Wang, X. , & Xu, Y. (2023). Porphyromonas gingivalis outer membrane vesicles promote apoptosis via msRNA‐regulated DNA methylation in periodontitis. Microbiology Spectrum, 11(1), e0328822. 10.1128/spectrum.03288-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, X. , Jin, Y. , Chen, G. , Ma, X. , & Zhang, L. (2021). Gut microbiota dysbiosis drives the development of colorectal cancer. Digestion, 102(4), 508–515. 10.1159/000508328 [DOI] [PubMed] [Google Scholar]
- Felgner, S. , Spöring, I. , Pawar, V. , Kocijancic, D. , Preusse, M. , Falk, C. , Rohde, M. , Häussler, S. , Weiss, S. , & Erhardt, M. (2020). The immunogenic potential of bacterial flagella for Salmonella‐mediated tumor therapy. International Journal of Cancer, 147(2), 448–460. 10.1002/ijc.32807 [DOI] [PubMed] [Google Scholar]
- Feng, Q. , Ma, X. , Cheng, K. , Liu, G. , Li, Y. , Yue, Y. , Liang, J. , Zhang, L. , Zhang, T. , Wang, X. , Gao, X. , Nie, G. , & Zhao, X. (2022). Engineered bacterial outer membrane vesicles as controllable two‐way adaptors to activate macrophage phagocytosis for improved tumor immunotherapy. Advanced Materials, 34(40), e2206200. 10.1002/adma.202206200 [DOI] [PubMed] [Google Scholar]
- Firth, J. , Sun, J. , George, V. , Huang, J. D. , Bajaj‐Elliott, M. , & Gustafsson, K. (2023). Bacterial outer‐membrane vesicles promote Vgamma9Vdelta2 T cell oncolytic activity. Frontiers in Immunology, 14, 1198996. 10.3389/fimmu.2023.1198996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fizanne, L. , Villard, A. , Benabbou, N. , Recoquillon, S. , Soleti, R. , Delage, E. , Wertheimer, M. , Vidal‐Gómez, X. , Oullier, T. , Chaffron, S. , Martínez, M. C. , Neunlist, M. , Boursier, J. , & Andriantsitohaina, R. (2023). Faeces‐derived extracellular vesicles participate in the onset of barrier dysfunction leading to liver diseases. Journal of Extracellular Vesicles, 12(2), e12303. 10.1002/jev2.12303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florez, C. , Raab, J. E. , Cooke, A. C. , & Schertzer, J. W. (2017). Membrane distribution of the Pseudomonas quinolone signal modulates outer membrane vesicle production in Pseudomonas aeruginosa . mBio, 8(4), e01034. 17. 10.1128/mBio.01034-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, H. , Luo, Z. , Ji, Y. , Tang, K. , Jin, Z. , Ly, C. , Sears, D. D. , Mahata, S. , & Ying, W. (2022). Accumulation of microbial DNAs promotes to islet inflammation and beta cell abnormalities in obesity in mice. Nature Communications, 13(1), 565. 10.1038/s41467-022-28239-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett, W. S. (2015). Cancer and the microbiota. Science, 348(6230), 80–6. 10.1126/science.aaa4972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert, J. A. , Blaser, M. J. , Caporaso, J. G. , Jansson, J. K. , Lynch, S. V. , & Knight, R. (2018). Current understanding of the human microbiome. Nature Medicine, 24(4), 392–400. 10.1038/nm.4517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloag, E. S. , Turnbull, L. , Huang, A. , Vallotton, P. , Wang, H. , Nolan, L. M. , Mililli, L. , Hunt, C. , Lu, J. , Osvath, S. R. , Monahan, L. G. , Cavaliere, R. , Charles, I. G. , Wand, M. P. , Gee, M. L. , Prabhakar, R. , & Whitchurch, C. B. (2013). Self‐organization of bacterial biofilms is facilitated by extracellular DNA. PNAS, 110(28), 11541–6. 10.1073/pnas.1218898110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groeger, S. , Denter, F. , Lochnit, G. , Schmitz, M. L. , & Meyle, J. (2020). Porphyromonas gingivalis cell wall components induce programmed death ligand 1 (PD‐L1) expression on human oral carcinoma cells by a receptor‐interacting protein kinase 2 (RIP2)‐dependent mechanism. Infection and Immunity, 88(5), e00051. 20. 10.1128/IAI.00051-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, T. W. , Wang, M. Z. , Niu, J. , Chu, Y. , Guo, K. R. , & Peng, L. H. (2020). Outer membrane vesicles derived from E. coli as novel vehicles for transdermal and tumor targeting delivery. Nanoscale, 12(36), 18965–18977. 10.1039/d0nr03698f [DOI] [PubMed] [Google Scholar]
- Gujrati, V. , Kim, S. , Kim, S. H. , Min, J. J. , Choy, H. E. , Kim, S. C. , & Jon, S. (2014). Bioengineered bacterial outer membrane vesicles as cell‐specific drug‐delivery vehicles for cancer therapy. ACS Nano, 8(2), 1525–37. 10.1021/nn405724x [DOI] [PubMed] [Google Scholar]
- Gurunathan, S. , Ajmani, A. , & Kim, J. H. (2023). Extracellular nanovesicles produced by Bacillus licheniformis: A potential anticancer agent for breast and lung cancer. Microbial Pathogenesis, 185, 106396. 10.1016/j.micpath.2023.106396 [DOI] [PubMed] [Google Scholar]
- Han, E. C. , Choi, S. Y. , Lee, Y. , Park, J. W. , Hong, S. H. , & Lee, H. J. (2019). Extracellular RNAs in periodontopathogenic outer membrane vesicles promote TNF‐α production in human macrophages and cross the blood‐brain barrier in mice. FASEB Journal, 33(12), 13412–13422. 10.1096/fj.201901575R [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, P. , Bartold, P. M. , Salomon, C. , & Ivanovski, S. (2021). Salivary outer membrane vesicles and DNA methylation of small extracellular vesicles as biomarkers for periodontal status: A pilot study. International Journal of Molecular Sciences, 22(5), 2423. 10.3390/ijms22052423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan, D. (2022). Hallmarks of cancer: New dimensions. Cancer Discovery, 12(1), 31–46. 10.1158/2159-8290.CD-21-1059 [DOI] [PubMed] [Google Scholar]
- Hayashi, J. , Hamada, N. , & Kuramitsu, H. K. (2002). The autolysin of Porphyromonas gingivalis is involved in outer membrane vesicle release. FEMS Microbiology Letters, 216(2), 217–22. 10.1111/j.1574-6968.2002.tb11438.x [DOI] [PubMed] [Google Scholar]
- Hock, B. D. , McKenzie, J. L. , & Keenan, J. I. (2017). Helicobacter pylori outer membrane vesicles inhibit human T cell responses via induction of monocyte COX‐2 expression. Pathogens and Disease, 75(4). 10.1093/femspd/ftx034 [DOI] [PubMed] [Google Scholar]
- Hooi, J. K. Y. , Lai, W. Y. , Ng, W. K. , Suen, M. M. Y. , Underwood, F. E. , Tanyingoh, D. , Malfertheiner, P. , Graham, D. Y. , Wong, V. W. S. , Wu, J. C. Y. , Chan, F. K. L. , Sung, J. J. Y. , Kaplan, G. G. , & Ng, S. C. (2017). Global prevalence of Helicobacter pylori infection: Systematic review and meta‐analysis. Gastroenterology, 153(2), 420–429. 10.1053/j.gastro.2017.04.022 [DOI] [PubMed] [Google Scholar]
- Hosseini‐Giv, N. , Basas, A. , Hicks, C. , El‐Omar, E. , El‐Assaad, F. , & Hosseini‐Beheshti, E. (2022). Bacterial extracellular vesicles and their novel therapeutic applications in health and cancer. Frontiers in Cellular and Infection Microbiology, 12, 962216. 10.3389/fcimb.2022.962216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, J. , Wu, Z. , & Xu, J. (2022). Effects of biofilm nano‐composite drugs OMVs‐MSN‐5‐FU on cervical lymph node metastases from oral squamous cell carcinoma. Frontiers in Oncology, 12, 881910. 10.3389/fonc.2022.881910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, D. T. , & Sperandio, V. (2008). Inter‐kingdom signalling: Communication between bacteria and their hosts. Nature Reviews Microbiology, 6(2), 111–20. 10.1038/nrmicro1836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussein, M. , Jasim, R. , Gocol, H. , Baker, M. , Thombare, V. J. , Ziogas, J. , Purohit, A. , Rao, G. G. , Li, J. , & Velkov, T. (2023). Comparative proteomics of outer membrane vesicles from polymyxin‐susceptible and extremely drug‐resistant Klebsiella pneumoniae . mSphere, 8(1), e0053722. 10.1128/msphere.00537-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang, S. C. , Kim, S. R. , Yoon, Y. J. , Park, K. S. , Kim, J. H. , Lee, J. , Kim, O. Y. , Choi, E. J. , Kim, D. K. , Choi, D. S. , Kim, Y. K. , Park, J. , Di Vizio, D. , & Gho, Y. S. (2015). In vivo kinetic biodistribution of nano‐sized outer membrane vesicles derived from bacteria. Small, 11(4), 456–61. 10.1002/smll.201401803 [DOI] [PubMed] [Google Scholar]
- Jeong, D. , Kim, M. J. , Park, Y. , Chung, J. , Kweon, H. S. , Kang, N. G. , Hwang, S. J. , Youn, S. H. , Hwang, B. K. , & Kim, D. (2022). Visualizing extracellular vesicle biogenesis in gram‐positive bacteria using super‐resolution microscopy. BMC Biology, 20(1), 270. 10.1186/s12915-022-01472-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong, J. Y. , Kim, T. B. , Kim, J. , Choi, H. W. , Kim, E. J. , Yoo, H. J. , Lee, S. , Jun, H. R. , Yoo, W. , Kim, S. , Kim, S. C. , & Jun, E. (2020). Diversity in the extracellular vesicle‐derived microbiome of tissues according to tumor progression in pancreatic cancer. Cancers (Basel), 12(9), 2346. 10.3390/cancers12092346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, S. , Fu, W. , Wang, S. , Zhu, G. , Wang, J. , & Ma, Y. (2023a). Bacterial outer membrane vesicles loaded with perhexiline suppress tumor development by regulating tumor‐associated macrophages repolarization in a synergistic way. International Journal of Molecular Sciences, 24(13), 11222. 10.3390/ijms241311222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, Y. , Xu, Y. , Zheng, C. , Ye, L. , Jiang, P. , Malik, S. , Xu, G. , Zhou, Q. , & Zhang, M. (2023b). Acetyltransferase from Akkermansia muciniphila blunts colorectal tumourigenesis by reprogramming tumour microenvironment. Gut, 72(7), 1308–1318. 10.1136/gutjnl-2022-327853 [DOI] [PubMed] [Google Scholar]
- Jiang, Y. , Wang, L. , Yang, B. , Ma, G. , Chen, Z. , Ma, J. , Chang, X. , Fang, L. , & Wang, Z. (2023c). Bifidobacterium‐derived membrane vesicles inhibit triple‐negative breast cancer growth by inducing tumor cell apoptosis. Molecular Biology Reports, 50(9), 7547–7556. 10.1007/s11033-023-08702-z [DOI] [PubMed] [Google Scholar]
- Jiang, Y. , Ma, J. , Long, Y. , Dan, Y. , Fang, L. , & Wang, Z. (2024). Extracellular membrane vesicles of Escherichia coli induce apoptosis of CT26 colon carcinoma cells. Microorganisms, 12(7), 1446. 10.3390/microorganisms12071446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, L. , Zhang, Z. , Tan, X. , Wang, Z. , Tang, B. , Wang, Z. , Li, M. , Mi, T. , Shen, L. , Long, C. , Wei, G. , & He, D. (2022). Antitumor effect of Escherichia coli‐derived outer membrane vesicles on neuroblastoma in vitro and in vivo. Acta Biochimica et Biophysica Sinica (Shanghai), 54(9), 1301–1313. 10.3724/abbs.2022127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, E. J. , Booth, C. , Fonseca, S. , Parker, A. , Cross, K. , Miquel‐Clopes, A. , Hautefort, I. , Mayer, U. , Wileman, T. , Stentz, R. , & Carding, S. R. (2020). The uptake, trafficking, and biodistribution of Bacteroides thetaiotaomicron generated outer membrane vesicles. Frontiers in Microbiology, 11, 57. 10.3389/fmicb.2020.00057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, C. S. , Ban, M. , Choi, E. J. , Moon, H. G. , Jeon, J. S. , Kim, D. K. , Park, S. K. , Jeon, S. G. , Roh, T. Y. , Myung, S. J. , Gho, Y. S. , Kim, J. G. , & Kim, Y. K. (2013). Extracellular vesicles derived from gut microbiota, especially Akkermansia muciniphila, protect the progression of dextran sulfate sodium‐induced colitis. PLoS ONE, 8(10), e76520. 10.1371/journal.pone.0076520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyhani, G. , Mahmoodzadeh Hosseini, H. , & Salimi, A. (2022). Effect of extracellular vesicles of Lactobacillus rhamnosus GG on the expression of CEA gene and protein released by colorectal cancer cells. Iranian Journal of Microbiology, 14(1), 90–96. 10.18502/ijm.v14i1.8809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan, A. N. H. , Emmons, T. R. , Magner, W. J. , Alqassim, E. , Singel, K. L. , Ricciuti, J. , Eng, K. H. , Odunsi, K. , Tomasi, T. B. , Lee, K. , Abrams, S. I. , Mesa, C. , & Segal, B. H. (2022). VSSP abrogates murine ovarian tumor‐associated myeloid cell‐driven immune suppression and induces M1 polarization in tumor‐associated macrophages from ovarian cancer patients. Cancer Immunology, Immunotherapy, 71(10), 2355–2369. 10.1007/s00262-022-03156-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D. J. , Yang, J. , Seo, H. , Lee, W. H. , Lee, D. H. , Kym, S. , Park, Y. S. , Kim, J. G. , Jang, I.‐J. , Kim, Y.‐K. , & Cho, J.‐Y. (2020a). Colorectal cancer diagnostic model utilizing metagenomic and metabolomic data of stool microbial extracellular vesicles. Scientific Reports, 10(1), 2860. 10.1038/s41598-020-59529-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J. R. , Han, K. , Han, Y. , Kang, N. , Shin, T.‐S. , Park, H. J. , Kim, H. , Kwon, W. , Lee, S. , Kim, Y.‐K. , Park, T. , & Jang, J.‐Y. (2021). Microbiome markers of pancreatic cancer based on bacteria‐derived extracellular vesicles acquired from blood samples: A retrospective propensity score matching analysis. Biology‐Basel, 10(3), 219. 10.3390/biology10030219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, M. H. , Choi, S. J. , Choi, H. I. , Choi, J. P. , Park, H. K. , Kim, E. K. , Kim, M. J. , Moon, B. S. , Min, T. K. , Rho, M. , Cho, Y. J. , Yang, S. , Kim, Y. K. , Kim, Y. Y. , & Pyun, B. Y. (2018). Lactobacillus plantarum‐derived extracellular vesicles protect atopic dermatitis induced by Staphylococcus aureus‐derived extracellular vesicles. Allergy, Asthma & Immunology Research, 10(5), 516–532. 10.4168/aair.2018.10.5.516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, O. Y. , Park, H. T. , Dinh, N. T. H. , Choi, S. J. , Lee, J. , Kim, J. H. , Lee, S. W. , & Gho, Y. S. (2017). Bacterial outer membrane vesicles suppress tumor by interferon‐γ‐mediated antitumor response. Nature Communications, 8(1), 626. 10.1038/s41467-017-00729-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S. I. , Kang, N. , Leem, S. , Yang, J. , Jo, H. , Lee, M. , Kim, H. S. , Dhanasekaran, D. N. , Kim, Y. K. , Park, T. , & Song, Y. S. (2020b). Metagenomic analysis of serum microbe‐derived extracellular vesicles and diagnostic models to differentiate ovarian cancer and benign ovarian tumor. Cancers (Basel), 12(5), 1309. 10.3390/cancers12051309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, Y. S. , Lee, W. H. , Choi, E. J. , Choi, J. P. , Heo, Y. J. , Gho, Y. S. , Jee, Y. K. , Oh, Y. M. , & Kim, Y. K. (2015). Extracellular vesicles derived from gram‐negative bacteria, such as Escherichia coli, induce emphysema mainly via IL‐17A‐mediated neutrophilic inflammation. Journal of Immunology, 194(7), 3361–8. 10.4049/jimmunol.1402268 [DOI] [PubMed] [Google Scholar]
- Koeppen, K. , Hampton, T. H. , Jarek, M. , Scharfe, M. , Gerber, S. A. , Mielcarz, D. W. , Demers, E. G. , Dolben, E. L. , Hammond, J. H. , Hogan, D. A. , & Stanton, B. A. (2016). A novel mechanism of host‐pathogen interaction through sRNA in bacterial outer membrane vesicles. PLoS Pathogens, 12(6), e1005672. 10.1371/journal.ppat.1005672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koliha, N. , Wiencek, Y. , Heider, U. , Jüngst, C. , Kladt, N. , Krauthäuser, S. , Johnston, I. C. , Bosio, A. , Schauss, A. , & Wild, S. (2016). A novel multiplex bead‐based platform highlights the diversity of extracellular vesicles. Journal of Extracellular Vesicles, 5, 29975. 10.3402/jev.v5.29975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostic, A. D. , Gevers, D. , Pedamallu, C. S. , Michaud, M. , Duke, F. , Earl, A. M. , Ojesina, A. I. , Jung, J. , Bass, A. J. , Tabernero, J. , Baselga, J. , Liu, C. , Shivdasani, R. A. , Ogino, S. , Birren, B. W. , Huttenhower, C. , Garrett, W. S. , & Meyerson, M. (2012). Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Research, 22(2), 292–8. 10.1101/gr.126573.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostic, A. D. , Chun, E. , Robertson, L. , Glickman, J. N. , Gallini, C. A. , Michaud, M. , Clancy, T. E. , Chung, D. C. , Lochhead, P. , Hold, G. L. , El‐Omar, E. M. , Brenner, D. , Fuchs, C. S. , Meyerson, M. , & Garrett, W. S. (2013). Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor‐immune microenvironment. Cell Host & Microbe, 14(2), 207–15. 10.1016/j.chom.2013.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehn, M. J. , & Kesty, N. C. (2005). Bacterial outer membrane vesicles and the host‐pathogen interaction. Genes & Development, 19(22), 2645–55. 10.1101/gad.1299905 [DOI] [PubMed] [Google Scholar]
- Kuerban, K. , Gao, X. , Zhang, H. , Liu, J. , Dong, M. , Wu, L. , Ye, R. , Feng, M. , & Ye, L. (2020). Doxorubicin‐loaded bacterial outer‐membrane vesicles exert enhanced anti‐tumor efficacy in non‐small‐cell lung cancer. Acta Pharmaceutica Sinica B, 10(8), 1534–1548. 10.1016/j.apsb.2020.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulp, A. , & Kuehn, M. J. (2010). Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annual Review of Microbiology, 64(1), 163–184. 10.1146/annurev.micro.091208.073413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamprinaki, D. , Garcia‐Vello, P. , Marchetti, R. , Hellmich, C. , McCord, K. A. , Bowles, K. M. , Macauley, M. S. , Silipo, A. , De Castro, C. , Crocker, P. R. , & Juge, N. (2021). Siglec‐7 mediates immunomodulation by colorectal cancer‐associated Fusobacterium nucleatum ssp. animalis . Frontiers in Immunology, 12, 744184. 10.3389/fimmu.2021.744184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, E. Y. , Choi, D. Y. , Kim, D. K. , Kim, J. W. , Park, J. O. , Kim, S. , Kim, S. H. , Desiderio, D. M. , Kim, Y. K. , Kim, K. P. , & Gho, Y. S. (2009). Gram‐positive bacteria produce membrane vesicles: Proteomics‐based characterization of Staphylococcus aureus‐derived membrane vesicles. Proteomics, 9(24), 5425–36. 10.1002/pmic.200900338 [DOI] [PubMed] [Google Scholar]
- Levy, M. , Kolodziejczyk, A. A. , Thaiss, C. A. , & Elinav, E. (2017). Dysbiosis and the immune system. Nature Reviews: Immunology, 17(4), 219–232. 10.1038/nri.2017.7 [DOI] [PubMed] [Google Scholar]
- Li, C. C. , Hsu, W. F. , Chiang, P. C. , Kuo, M. C. , Wo, A. M. , & Tseng, Y. J. (2023a). Characterization of markers, functional properties, and microbiome composition in human gut‐derived bacterial extracellular vesicles. Gut Microbes, 15(2), 2288200. 10.1080/19490976.2023.2288200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, D. , Zhu, L. , Wang, Y. , Zhou, X. , & Li, Y. (2023b). Bacterial outer membrane vesicles in cancer: Biogenesis, pathogenesis, and clinical application. Biomedicine and Pharmacotherapy, 165, 115120. 10.1016/j.biopha.2023.115120 [DOI] [PubMed] [Google Scholar]
- Li, G. , Sun, Y. , Huang, Y. , Lian, J. , Wu, S. , Luo, D. , & Gong, H. (2023c). Fusobacterium nucleatum‐derived small extracellular vesicles facilitate tumor growth and metastasis via TLR4 in breast cancer. BMC Cancer, 23(1), 473. 10.1186/s12885-023-10844-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J. , Feng, S. , Pi, Y. , Jiang, X. , Li, X. , Zhou, Z. , Liu, X. , Wei, H. , & Tao, S. (2023d). Limosilactobacillus johnsoni and Limosilactobacillus mucosae and their extracellular vesicles alleviate gut inflammatory injury by mediating macrophage polarization in a lipopolysaccharide‐challenged Piglet model. Journal of Nutrition, 153(8), 2497–2511. 10.1016/j.tjnut.2023.06.009 [DOI] [PubMed] [Google Scholar]
- Li, Y. , Zhao, R. , Cheng, K. , Zhang, K. , Wang, Y. , Zhang, Y. , Li, Y. , Liu, G. , Xu, J. , Xu, J. , Anderson, G. J. , Shi, J. , Ren, L. , Zhao, X. , & Nie, G. (2020). Bacterial outer membrane vesicles presenting programmed death 1 for improved cancer immunotherapy via immune activation and checkpoint inhibition. ACS Nano, 14(12), 16698–16711. 10.1021/acsnano.0c03776 [DOI] [PubMed] [Google Scholar]
- Li, Y. , Zhang, K. , Wu, Y. , Yue, Y. , Cheng, K. , Feng, Q. , Ma, X. , Liang, J. , Ma, N. , Liu, G. , Nie, G. , Ren, L. , & Zhao, X. (2022). Antigen capture and immune modulation by bacterial outer membrane vesicles as in situ vaccine for cancer immunotherapy post‐photothermal therapy. Small, 18(14), e2107461. 10.1002/smll.202107461 [DOI] [PubMed] [Google Scholar]
- Li, Y. , Wu, J. , Qiu, X. , Dong, S. , He, J. , Liu, J. , Xu, W. , Huang, S. , Hu, X. , & Xiang, D. X. (2023e). Bacterial outer membrane vesicles‐based therapeutic platform eradicates triple‐negative breast tumor by combinational photodynamic/chemo‐/immunotherapy. Bioactive Materials, 20, 548–560. 10.1016/j.bioactmat.2022.05.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, L.‐T. , Shi, Y.‐C. , Choong, C.‐Y. , & Tai, C.‐J. (2021). The fruits of Paris polyphylla inhibit colorectal cancer cell migration induced by Fusobacterium nucleatum‐derived extracellular vesicles. Molecules (Basel, Switzerland), 26(13), 4081. 10.3390/molecules26134081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, D. , Liu, S. , Liu, J. , Miao, L. , Zhang, S. , & Pan, Y. (2021a). sRNA23392 packaged by Porphyromonas gingivalis outer membrane vesicles promotes oral squamous cell carcinomas migration and invasion by targeting desmocollin‐2. Molecular Oral Microbiology, 36(3), 182–191. 10.1111/omi.12334 [DOI] [PubMed] [Google Scholar]
- Liu, J. , Hsieh, C. L. , Gelincik, O. , Devolder, B. , Sei, S. , Zhang, S. , Lipkin, S. M. , & Chang, Y. F. (2019a). Proteomic characterization of outer membrane vesicles from gut mucosa‐derived Fusobacterium nucleatum . Journal of Proteomics, 195, 125–137. 10.1016/j.jprot.2018.12.029 [DOI] [PubMed] [Google Scholar]
- Liu, L. , Liang, L. , Yang, C. , Zhou, Y. , & Chen, Y. (2021b). Extracellular vesicles of Fusobacterium nucleatum compromise intestinal barrier through targeting RIPK1‐mediated cell death pathway. Gut Microbes, 13(1), 1–20. 10.1080/19490976.2021.1902718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Q. , Li, X. , Zhang, Y. , Song, Z. , Li, R. , Ruan, H. , & Huang, X. (2019b). Orally‐administered outer‐membrane vesicles from Helicobacter pylori reduce H. pylori infection via Th2‐biased immune responses in mice. Pathogens and Disease, 77(5), ftz050. 10.1093/femspd/ftz050 [DOI] [PubMed] [Google Scholar]
- Liu, X. Z. , Wen, Z. J. , Li, Y. M. , Sun, W. R. , Hu, X. Q. , Zhu, J. Z. , Li, X. Y. , Wang, P. Y. , Pedraz, J. L. , Lee, J. H. , Kim, H. W. , Ramalingam, M. , Xie, S. , & Wang, R. (2023). Bioengineered bacterial membrane vesicles with multifunctional nanoparticles as a versatile platform for cancer immunotherapy. ACS Applied Materials & Interfaces, 15(3), 3744–3759. 10.1021/acsami.2c18244 [DOI] [PubMed] [Google Scholar]
- Lu, S. , Xu, J. , Zhao, Z. , Guo, Y. , Zhang, H. , Jurutka, P. W. , Huang, D. , Cao, C. , & Cheng, S. (2023). Dietary Lactobacillus rhamnosus GG extracellular vesicles enhance antiprogrammed cell death 1 (anti‐PD‐1) immunotherapy efficacy against colorectal cancer. Food & Function, 14(23), 10314–10328. 10.1039/d3fo02018e [DOI] [PubMed] [Google Scholar]
- Luo, Z. , Ji, Y. , Zhang, D. , Gao, H. , Jin, Z. , Yang, M. , & Ying, W. (2022). Microbial DNA enrichment promotes liver steatosis and fibrosis in the course of non‐alcoholic steatohepatitis. Acta Physiologica (Oxford, England), 235(3), e13827. 10.1111/apha.13827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, Z. W. , Xia, K. , Liu, Y. W. , Liu, J. H. , Rao, S. S. , Hu, X. K. , Chen, C. Y. , Xu, R. , Wang, Z. X. , & Xie, H. (2021). Extracellular vesicles from Akkermansia muciniphila elicit antitumor immunity against prostate cancer via modulation of CD8(+) T cells and macrophages. International Journal of Nanomedicine, 16, 2949–2963. 10.2147/IJN.S304515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin‐Gallausiaux, C. , Malabirade, A. , Habier, J. , & Wilmes, P. (2020). Fusobacterium nucleatum extracellular vesicles modulate gut epithelial cell innate immunity via FomA and TLR2. Frontiers in Immunology, 11, 583644. 10.3389/fimmu.2020.583644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzoog, T. R. , Jabir, M. S. , Ibraheem, S. , Jawad, S. F. , Hamzah, S. S. , Sulaiman, G. M. , Mohammed, H. A. , & Khan, R. A. (2023). Bacterial extracellular vesicles induced oxidative stress and mitophagy through mTOR pathways in colon cancer cells, HT‐29: Implications for bioactivity. Biochimica et Biophysica Acta (BBA) – Molecular Cell Research, 1870(6), 119486. 10.1016/j.bbamcr.2023.119486 [DOI] [PubMed] [Google Scholar]
- McBroom, A. J. , & Kuehn, M. J. (2007). Release of outer membrane vesicles by gram‐negative bacteria is a novel envelope stress response. Molecular Microbiology, 63(2), 545–58. 10.1111/j.1365-2958.2006.05522.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy, A. N. , Araujo‐Perez, F. , Azcarate‐Peril, A. , Yeh, J. J. , Sandler, R. S. , & Keku, T. O. (2013). Fusobacterium is associated with colorectal adenomas. PLoS ONE, 8(1), e53653. 10.1371/journal.pone.0053653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell, A. , Kang, J. , Yang, J. , Jung, J. , Oh, Y. M. , Kym, S. M. , Shin, T. S. , Kim, T. B. , Jee, Y. K. , & Kim, Y. K. (2022). Machine‐learning algorithms for asthma, COPD, and lung cancer risk assessment using circulating microbial extracellular vesicle data and their application to assess dietary effects. Experimental & Molecular Medicine, 54(9), 1586–1595. 10.1038/s12276-022-00846-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meganathan, V. , Moyana, R. , Natarajan, K. , Kujur, W. , Kusampudi, S. , Mulik, S. , & Boggaram, V. (2020). Bacterial extracellular vesicles isolated from organic dust induce neutrophilic inflammation in the lung. American Journal of Physiology, Lung Cellular and Molecular Physiology, 319(6), L893. l907. 10.1152/ajplung.00107.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naradasu, D. , Miran, W. , Sharma, S. , Takenawa, S. , Soma, T. , Nomura, N. , Toyofuku, M. , & Okamoto, A. (2021). Biogenesis of outer membrane vesicles concentrates the unsaturated fatty acid of phosphatidylinositol in Capnocytophaga ochracea . Frontiers in Microbiology, 12, 682685. 10.3389/fmicb.2021.682685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natsui, K. , Tsuchiya, A. , Imamiya, R. , Osada‐Oka, M. , Ishii, Y. , Koseki, Y. , Takeda, N. , Tomiyoshi, K. , Yamazaki, F. , Yoshida, Y. , Ohashi, R. , Ling, Y. , Ueda, K. , Moritoki, N. , Sato, K. , Nakajima, T. , Hasegawa, Y. , Okuda, S. , Shibata, S. , … Terai, S. (2023). Escherichia coli‐derived outer‐membrane vesicles induce immune activation and progression of cirrhosis in mice and humans. Liver International, 43(5), 1126–1140. 10.1111/liv.15539 [DOI] [PubMed] [Google Scholar]
- Nejman, D. , Livyatan, I. , Fuks, G. , Gavert, N. , Zwang, Y. , Geller, L. T. , Rotter‐Maskowitz, A. , Weiser, R. , Mallel, G. , Gigi, E. , Meltser, A. , Douglas, G. M. , Kamer, I. , Gopalakrishnan, V. , Dadosh, T. , Levin‐Zaidman, S. , Avnet, S. , Atlan, T. , Cooper, Z. A. , … Straussman, R. (2020). The human tumor microbiome is composed of tumor type‐specific intracellular bacteria. Science, 368(6494), 973–980. 10.1126/science.aay9189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksvold, M. P. , Neurauter, A. , & Pedersen, K. W. (2015). Magnetic bead‐based isolation of exosomes. Methods in Molecular Biology, 1218, 465–81. 10.1007/978-1-4939-1538-5_27 [DOI] [PubMed] [Google Scholar]
- Pang, Y. , Ermann Lundberg, L. , Mata Forsberg, M. , Ahl, D. , Bysell, H. , Pallin, A. , Sverremark‐Ekström, E. , Karlsson, R. , Jonsson, H. , & Roos, S. (2022). Extracellular membrane vesicles from Limosilactobacillus reuteri strengthen the intestinal epithelial integrity, modulate cytokine responses and antagonize activation of TRPV1. Frontiers in Microbiology, 13, 1032202. 10.3389/fmicb.2022.1032202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, J. , Kim, N. E. , Yoon, H. , Shin, C. M. , Kim, N. , Lee, D. H. , Park, J. Y. , Choi, C. H. , Kim, J. G. , Kim, Y. K. , Shin, T. S. , Yang, J. , & Park, Y. S. (2021a). Fecal microbiota and gut microbe‐derived extracellular vesicles in colorectal cancer. Frontiers in oncology, 11, 650026. 10.3389/fonc.2021.650026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, J. Y. , Choi, J. , Lee, Y. , Lee, J. E. , Lee, E. H. , Kwon, H. J. , Yang, J. , Jeong, B. R. , Kim, Y. K. , & Han, P. L. (2017). Metagenome analysis of bodily microbiota in a mouse model of Alzheimer disease using bacteria‐derived membrane vesicles in blood. Experimental Neurobiology, 26(6), 369–379. 10.5607/en.2017.26.6.369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, J. Y. , Kang, C. S. , Seo, H. C. , Shin, J. C. , Kym, S. M. , Park, Y. S. , Shin, T. S. , Kim, J. G. , & Kim, Y. K. (2021b). Bacteria‐derived extracellular vesicles in urine as a novel biomarker for gastric cancer: Integration of liquid biopsy and metagenome analysis. Cancers (Basel), 13(18), 4687. 10.3390/cancers13184687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, K. S. , Svennerholm, K. , Crescitelli, R. , Lasser, C. , Gribonika, I. , & Lotvall, J. (2021c). Synthetic bacterial vesicles combined with tumour extracellular vesicles as cancer immunotherapy. Journal of Extracellular Vesicles, 10(9), e12120. 10.1002/jev2.12120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penders, J. , Nagelkerke, A. , Cunnane, E. M. , Pedersen, S. V. , Pence, I. J. , Coombes, R. C. , & Stevens, M. M. (2021). Single particle automated raman trapping analysis of breast cancer cell‐derived extracellular vesicles as cancer biomarkers. ACS Nano, 15(11), 18192–18205. 10.1021/acsnano.1c07075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, L. H. , Wang, M. Z. , Chu, Y. , Zhang, L. , Niu, J. , Shao, H. T. , Yuan, T. J. , Jiang, Z. H. , Gao, J. Q. , & Ning, X. H. (2020). Engineering bacterial outer membrane vesicles as transdermal nanoplatforms for photo‐TRAIL‐programmed therapy against melanoma. Science Advances, 6(27), eaba2735. 10.1126/sciadv.aba2735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez‐Cruz, C. , Carrión, O. , Delgado, L. , Martinez, G. , López‐Iglesias, C. , & Mercade, E. (2013). New type of outer membrane vesicle produced by the Gram‐negative bacterium Shewanella vesiculosa M7T: Implications for DNA content. Applied and Environmental Microbiology, 79(6), 1874–81. 10.1128/aem.03657-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piroli, N. H. , Reus, L. S. C. , Mamczarz, Z. , Khan, S. , Bentley, W. E. , & Jay, S. M. (2023). High performance anion exchange chromatography purification of probiotic bacterial extracellular vesicles enhances purity and anti‐inflammatory efficacy. BioRxiv, 10.1101/2023.05.01.538917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poore, G. D. , Kopylova, E. , Zhu, Q. , Carpenter, C. , Fraraccio, S. , Wandro, S. , Kosciolek, T. , Janssen, S. , Metcalf, J. , Song, S. J. , Kanbar, J. , Miller‐Montgomery, S. , Heaton, R. , McKay, R. , Patel, S. P. , Swafford, A. D. , & Knight, R. (2020). Microbiome analyses of blood and tissues suggest cancer diagnostic approach. Nature, 579(7800), 567–574. 10.1038/s41586-020-2095-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin, Y. F. , Lu, X. Y. , Shi, Z. , Huang, Q. S. , Wang, X. , Ren, B. , & Cui, L. (2022). Deep learning‐enabled raman spectroscopic identification of pathogen‐derived extracellular vesicles and the biogenesis process. Analytical Chemistry, 94(36), 12416–12426. 10.1021/acs.analchem.2c02226 [DOI] [PubMed] [Google Scholar]
- Qing, S. , Lyu, C. , Zhu, L. , Pan, C. , Wang, S. , Li, F. , Wang, J. , Yue, H. , Gao, X. , Jia, R. , Wei, W. , & Ma, G. (2020). Biomineralized bacterial outer membrane vesicles potentiate safe and efficient tumor microenvironment reprogramming for anticancer therapy. Advanced Materials, 32(47), e2002085. 10.1002/adma.202002085 [DOI] [PubMed] [Google Scholar]
- Ren, C. , Li, Y. , Cong, Z. , Li, Z. , Xie, L. , & Wu, S. (2023). Bioengineered bacterial outer membrane vesicles encapsulated Polybia‐mastoparan I fusion peptide as a promising nanoplatform for bladder cancer immune‐modulatory chemotherapy. Frontiers in Immunology, 14, 1129771. 10.3389/fimmu.2023.1129771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley, D. R. , Sieber, K. B. , Robinson, K. M. , White, J. R. , Ganesan, A. , Nourbakhsh, S. , & Dunning Hotopp, J. C. (2013). Bacteria‐human somatic cell lateral gene transfer is enriched in cancer samples. PLoS Computational Biology, 9(6), e1003107. 10.1371/journal.pcbi.1003107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaack, B. , Hindré, T. , Quansah, N. , Hannani, D. , Mercier, C. , & Laurin, D. (2022). Microbiota‐derived extracellular vesicles detected in human blood from healthy donors. International Journal of Molecular Sciences, 23(22), 13787. 10.3390/ijms232213787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaack, B. , Mercier, C. , Katby, M. , Hannani, D. , Vollaire, J. , Robert, J. S. , Caffaratti, C. , Blanquet, F. , Nicoud, O. , Josserand, V. , & Laurin, D. (2024). Rapid biodistribution of fluorescent outer‐membrane vesicles from the intestine to distant organs via the blood in mice. International Journal of Molecular Sciences, 25(3), 1821. 10.3390/ijms25031821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwechheimer, C. , & Kuehn, M. J. (2015). Outer‐membrane vesicles from gram‐negative bacteria: Biogenesis and functions. Nature Reviews Microbiology, 13(10), 605–619. 10.1038/nrmicro3525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sender, R. , Fuchs, S. , & Milo, R. (2016). Revised estimates for the number of human and bacteria cells in the body. PLoS Biology, 14(8), e1002533. 10.1371/journal.pbio.1002533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepich‐Poore, G. D. , Zitvogel, L. , Straussman, R. , Hasty, J. , Wargo, J. A. , & Knight, R. (2021). The microbiome and human cancer. Science, 371(6536), eabc4552. 10.1126/science.abc4552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah, S. C. , & Itzkowitz, S. H. (2022). Colorectal cancer in inflammatory bowel disease: Mechanisms and management. Gastroenterology, 162(3), 715–730. e3. 10.1053/j.gastro.2021.10.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, J. , Xu, X. , Du, J. , Cui, H. , & Luo, Q. (2019). Expression of Oncostatin M in early gastric cancer and precancerous lesions. Gastroenterology Research and Practice, 2019, 3616140. 10.1155/2019/3616140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, J. , Ma, D. , Gao, S. , Long, F. , Wang, X. , Pu, X. , Cannon, R. D. , & Han, T. L. (2023). Probiotic Escherichia coli Nissle 1917‐derived outer membrane vesicles modulate the intestinal microbiome and host gut‐liver metabolome in obese and diabetic mice. Frontiers in Microbiology, 14, 1219763. 10.3389/fmicb.2023.1219763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, Y. , Meng, L. , Zhang, C. , Zhang, F. , & Fang, Y. (2021). Extracellular vesicles of Lacticaseibacillus paracasei PC‐H1 induce colorectal cancer cells apoptosis via PDK1/AKT/Bcl‐2 signaling pathway. Microbiological Research, 255, 126921. 10.1016/j.micres.2021.126921 [DOI] [PubMed] [Google Scholar]
- Shimoda, A. , Ueda, K. , Nishiumi, S. , Murata‐Kamiya, N. , Mukai, S. A. , Sawada, S. , Azuma, T. , Hatakeyama, M. , & Akiyoshi, K. (2016). Exosomes as nanocarriers for systemic delivery of the Helicobacter pylori virulence factor CagA. Scientific Reports, 6, 18346. 10.1038/srep18346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel, R. L. , Miller, K. D. , Wagle, N. S. , & Jemal, A. (2023). Cancer statistics. CA: A Cancer Journal for Clinicians, 73(1), 17–48. 10.3322/caac.21763 [DOI] [PubMed] [Google Scholar]
- Song, T. , Mika, F. , Lindmark, B. , Liu, Z. , Schild, S. , Bishop, A. , Zhu, J. , Camilli, A. , Johansson, J. , Vogel, J. , & Wai, S. N. (2008). A new Vibrio cholerae sRNA modulates colonization and affects release of outer membrane vesicles. Molecular Microbiology, 70(1), 100–11. 10.1111/j.1365-2958.2008.06392.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stentz, R. , Carvalho, A. L. , Jones, E. J. , & Carding, S. R. (2018). Fantastic voyage: The journey of intestinal microbiota‐derived microvesicles through the body. Biochemical Society Transactions, 46(5), 1021–1027. 10.1042/BST20180114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung, H. , Ferlay, J. , Siegel, R. L. , Laversanne, M. , Soerjomataram, I. , Jemal, A. , & Bray, F. (2021). Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer Journal for Clinicians, 71(3), 209–249. 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- Tang, B. , He, D. W. , Li, D. , Guo, W. H. , Zhang, D. , & Wei, G. H. (2018). Effect of outer membrane vesicles derived from Escherichia coli on proliferation, apoptosis and migration of human neuroblastoma SK‐N‐SH cells in vitro. Nan Fang Yi Ke Da Xue Xue Bao, 38(3), 334–339. 10.3969/j.issn.1673-4254.2018.03.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, W. H. W. , Bäckhed, F. , Landmesser, U. , & Hazen, S. L. (2019). Intestinal microbiota in cardiovascular health and disease: JACC state‐of‐the‐art review. Journal of the American College of Cardiology, 73(16), 2089–2105. 10.1016/j.jacc.2019.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong, L. , Zhang, X. , Hao, H. , Liu, Q. , Zhou, Z. , Liang, X. , Liu, T. , Gong, P. , Zhang, L. , Zhai, Z. , Hao, Y. , & Yi, H. (2021). Lactobacillus rhamnosus GG derived extracellular vesicles modulate gut microbiota and attenuate inflammatory in DSS‐induced colitis mice. Nutrients, 13(10), 3319. 10.3390/nu13103319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyofuku, M. , Tashiro, Y. , Hasegawa, Y. , Kurosawa, M. , & Nomura, N. (2015). Bacterial membrane vesicles, an overlooked environmental colloid: Biology, environmental perspectives and applications. Advances in Colloid and Interface Science, 226(Pt A), 65–77. 10.1016/j.cis.2015.08.013 [DOI] [PubMed] [Google Scholar]
- Toyofuku, M. , Nomura, N. , & Eberl, L. (2019). Types and origins of bacterial membrane vesicles. Nature Reviews Microbiology, 17(1), 13–24. 10.1038/s41579-018-0112-2 [DOI] [PubMed] [Google Scholar]
- Toyofuku, M. , Schild, S. , Kaparakis‐Liaskos, M. , & Eberl, L. (2023). Composition and functions of bacterial membrane vesicles. Nature Reviews Microbiology, 21(7), 415–430. 10.1038/s41579-023-00875-5 [DOI] [PubMed] [Google Scholar]
- Tulkens, J. , De Wever, O. , & Hendrix, A. (2020a). Analyzing bacterial extracellular vesicles in human body fluids by orthogonal biophysical separation and biochemical characterization. Nature Protocols, 15(1), 40–67. 10.1038/s41596-019-0236-5 [DOI] [PubMed] [Google Scholar]
- Tulkens, J. , Vergauwen, G. , Van Deun, J. , Geeurickx, E. , Dhondt, B. , Lippens, L. , De Scheerder, M. A. , Miinalainen, I. , Rappu, P. , De Geest, B. G. , Vandecasteele, K. , Laukens, D. , Vandekerckhove, L. , Denys, H. , Vandesompele, J. , De Wever, O. , & Hendrix, A. (2020b). Increased levels of systemic LPS‐positive bacterial extracellular vesicles in patients with intestinal barrier dysfunction. Gut, 69(1), 191–193. 10.1136/gutjnl-2018-317726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turkina, M. V. , Olofsson, A. , Magnusson, K. E. , Arnqvist, A. , & Vikstrom, E. (2015). Helicobacter pylori vesicles carrying CagA localize in the vicinity of cell‐cell contacts and induce histone H1 binding to ATP in epithelial cells. FEMS Microbiology Letters, 362(11), fnv076. 10.1093/femsle/fnv076 [DOI] [PubMed] [Google Scholar]
- Turnbull, L. , Toyofuku, M. , Hynen, A. L. , Kurosawa, M. , Pessi, G. , Petty, N. K. , Osvath, S. R. , Carcamo‐Oyarce, G. , Gloag, E. S. , Shimoni, R. , Omasits, U. , Ito, S. , Yap, X. , Monahan, L. G. , Cavaliere, R. , Ahrens, C. H. , Charles, I. G. , Nomura, N. , Eberl, L. , … Whitchurch, C. B. (2016). Explosive cell lysis as a mechanism for the biogenesis of bacterial membrane vesicles and biofilms. Nature Communications, 7, 11220. 10.1038/ncomms11220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyrer, P. C. , Frizelle, F. A. , & Keenan, J. I. (2014). Escherichia coli‐derived outer membrane vesicles are genotoxic to human enterocyte‐like cells. Infectious Agents and Cancer, 9(1), 2. 10.1186/1750-9378-9-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S. , Huang, W. , Li, K. , Yao, Y. , Yang, X. , Bai, H. , Sun, W. , Liu, C. , & Ma, Y. (2017). Engineered outer membrane vesicle is potent to elicit HPV16E7‐specific cellular immunity in a mouse model of TC‐1 graft tumor. International Journal of Nanomedicine, 12, 6813–6825. 10.2147/ijn.S143264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S. , Gao, J. , & Wang, Z. (2019). Outer membrane vesicles for vaccination and targeted drug delivery. Wiley Interdisciplinary Reviews: Nanomedicine and Nanobiotechnology, 11(2), e1523. 10.1002/wnan.1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S. , Song, A. , Xie, J. , Wang, Y. Y. , Wang, W. D. , Zhang, M. J. , Wu, Z. Z. , Yang, Q. C. , Li, H. , Zhang, J. , & Sun, Z. J. (2024). Fn‐OMV potentiates ZBP1‐mediated PANoptosis triggered by oncolytic HSV‐1 to fuel antitumor immunity. Nature Communications, 15(1), 3669. 10.1038/s41467-024-48032-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X. , Lin, S. , Wang, L. , Cao, Z. , Zhang, M. , Zhang, Y. , Liu, R. , & Liu, J. (2023). Versatility of bacterial outer membrane vesicles in regulating intestinal homeostasis. Science Advances, 9(11), eade5079. 10.1126/sciadv.ade5079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson, D. C. , Johnson, S. , Santos, A. , Yin, M. , Bayik, D. , Lathia, J. D. , & Dwidar, M. (2021). Scalable isolation and purification of extracellular vesicles from Escherichia coli and other bacteria. Journal of Visualized Experiments: JoVE, 00(176), 0.3791/63155. 10.3791/63155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, S. , Li, X. , Wang, J. , Wang, Y. , Zhang, C. , Dai, S. , Wang, X. , Deng, X. , Zhao, L. , & Shan, B. (2022). Outer membrane vesicles secreted by Helicobacter pylori transmitting gastric pathogenic virulence factors. ACS Omega, 7(1), 240–258. 10.1021/acsomega.1c04549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, S. , Zhang, J. , Wu, X. , Chen, M. , Huang, H. , Zeng, S. , Xiang, Z. , Li, X. , & Dong, W. (2023). Fusobacterium nucleatum extracellular vesicles promote experimental colitis by modulating autophagy via the miR‐574‐5p/CARD3 axis. Inflammatory Bowel Diseases, 29(1), 9–26. 10.1093/ibd/izac177 [DOI] [PubMed] [Google Scholar]
- Welsh, J. A. , Goberdhan, D. C. I. , O'Driscoll, L. , Buzas, E. I. , Blenkiron, C. , Bussolati, B. , Cai, H. , Di Vizio, D. , Driedonks, T. A. P. , Erdbrügger, U. , Falcon‐Perez, J. M. , Fu, Q. L. , Hill, A. F. , Lenassi, M. , Lim, S. K. , Mahoney, M. G. , Mohanty, S. , Möller, A. , Nieuwland, R. , & Witwer, K. W. (2024). Minimal information for studies of extracellular vesicles (MISEV2023): From basic to advanced approaches. Journal of Extracellular Vesicles, 13(2), e12404. 10.1002/jev2.12404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitchurch, C. B. , Tolker‐Nielsen, T. , Ragas, P. C. , & Mattick, J. S. (2002). Extracellular DNA required for bacterial biofilm formation. Science, 295(5559), 1487. 10.1126/science.295.5559.1487 [DOI] [PubMed] [Google Scholar]
- Winter, J. , Letley, D. , Rhead, J. , Atherton, J. , & Robinson, K. (2014). Helicobacter pylori membrane vesicles stimulate innate pro‐ and anti‐inflammatory responses and induce apoptosis in Jurkat T cells. Infection and Immunity, 82(4), 1372–81. 10.1128/IAI.01443-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won, S. , Lee, C. , Bae, S. , Lee, J. , Choi, D. , Kim, M. G. , Song, S. , Lee, J. , Kim, E. , Shin, H. , Basukala, A. , Lee, T. R. , Lee, D. S. , & Gho, Y. S. (2023). Mass‐produced gram‐negative bacterial outer membrane vesicles activate cancer antigen‐specific stem‐like CD8(+) T cells which enables an effective combination immunotherapy with anti‐PD‐1. Journal of Extracellular Vesicles, 12(8), e12357. 10.1002/jev2.12357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong‐Rolle, A. , Wei, H. K. , Zhao, C. , & Jin, C. (2021). Unexpected guests in the tumor microenvironment: Microbiome in cancer. Protein Cell, 12(5), 426–435. 10.1007/s13238-020-00813-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, J. , Garcia‐Carbonell, R. , Zelic, M. , Ho, S. B. , Boland, B. S. , Yao, S. J. , Desai, S. A. , Das, S. , Planell, N. , Harris, P. A. , Font‐Burgada, J. , Taniguchi, K. , Bertin, J. , Salas, A. , Pasparakis, M. , Gough, P. J. , Kelliher, M. , Karin, M. , & Guma, M. (2020). RIPK1 mediates TNF‐induced intestinal crypt apoptosis during chronic NF‐κB activation. Cellular and Molecular Gastroenterology and Hepatology, 9(2), 295–312. 10.1016/j.jcmgh.2019.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, J. , An, M. , Zhu, J. , Tan, Z. , Chen, G. Y. , Stidham, R. W. , & Lubman, D. M. (2019). A method for isolation and proteomic analysis of outer membrane vesicles from fecal samples by LC‐MS/MS. Journal of Proteomics & Bioinformatics, 12(2), 38–42. 10.4172/0974-276x.1000494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, X. , Xu, J. , Yang, X. , Wang, D. , & Xu, X. (2023). Integrating transcriptomics and metabolomics to explore the novel pathway of Fusobacterium nucleatum invading colon cancer cells. Pathogens, 12(2), 201. 10.3390/pathogens12020201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, J. , Li, Q. , Haesebrouck, F. , Van Hoecke, L. , & Vandenbroucke, R. E. (2022). The tremendous biomedical potential of bacterial extracellular vesicles. Trends in Biotechnology, 40(10), 1173–1194. 10.1016/j.tibtech.2022.03.005 [DOI] [PubMed] [Google Scholar]
- Yoon, H. , Kim, N. E. , Park, J. , Shin, C. M. , Kim, N. , Lee, D. H. , Park, J. Y. , Choi, C. H. , Kim, J. G. , & Park, Y. S. (2023). Analysis of the gut microbiome using extracellular vesicles in the urine of patients with colorectal cancer. Korean Journal of Internal Medicine, 38(1), 27–38. 10.3904/kjim.2022.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, T. , Guo, F. , Yu, Y. , Sun, T. , Ma, D. , Han, J. , Qian, Y. , Kryczek, I. , Sun, D. , Nagarsheth, N. , Chen, Y. , Chen, H. , Hong, J. , Zou, W. , & Fang, J. Y. (2017). Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell, 170(3), 548–563. e16. 10.1016/j.cell.2017.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue, Y. , Xu, J. , Li, Y. , Cheng, K. , Feng, Q. , Ma, X. , Ma, N. , Zhang, T. , Wang, X. , Zhao, X. , & Nie, G. (2022). Antigen‐bearing outer membrane vesicles as tumour vaccines produced in situ by ingested genetically engineered bacteria. Nature Biomedical Engineering, 6(7), 898–909. 10.1038/s41551-022-00886-2 [DOI] [PubMed] [Google Scholar]
- Zakharzhevskaya, N. B. , Vanyushkina, A. A. , Altukhov, I. A. , Shavarda, A. L. , Butenko, I. O. , Rakitina, D. V. , Nikitina, A. S. , Manolov, A. I. , Egorova, A. N. , Kulikov, E. E. , Vishnyakov, I. E. , Fisunov, G. Y. , & Govorun, V. M. (2017). Outer membrane vesicles secreted by pathogenic and nonpathogenic Bacteroides fragilis represent different metabolic activities. Scientific Reports, 7(1), 5008. 10.1038/s41598-017-05264-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanella, I. , König, E. , Tomasi, M. , Gagliardi, A. , Frattini, L. , Fantappiè, L. , Irene, C. , Zerbini, F. , Caproni, E. , Isaac, S. J. , Grigolato, M. , Corbellari, R. , Valensin, S. , Ferlenghi, I. , Giusti, F. , Bini, L. , Ashhab, Y. , Grandi, A. , & Grandi, G. (2021). Proteome‐minimized outer membrane vesicles from Escherichia coli as a generalized vaccine platform. Journal of Extracellular Vesicles, 10(4), e12066. 10.1002/jev2.12066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai, Y. , Ma, Y. , Pang, B. , Zhang, J. , Li, Y. , Rui, Y. , Xu, T. , Zhao, Y. , Qian, Z. , Gu, Y. , & Li, S. (2021). A cascade targeting strategy based on modified bacterial vesicles for enhancing cancer immunotherapy. Journal of Nanobiotechnology, 19(1), 434. 10.1186/s12951-021-01193-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H. , Zhang, Y. , Song, Z. , Li, R. , Ruan, H. , Liu, Q. , & Huang, X. (2020). sncRNAs packaged by Helicobacter pylori outer membrane vesicles attenuate IL‐8 secretion in human cells. International Journal of Medical Microbiology, 310(1), 151356. 10.1016/j.ijmm.2019.151356 [DOI] [PubMed] [Google Scholar]
- Zhang, J. , Li, Z. , Liu, L. , Li, L. , Zhang, L. , Wang, Y. , & Zhao, J. (2022). Self‐assembly catalase nanocomplex conveyed by bacterial vesicles for oxygenated photodynamic therapy and tumor immunotherapy. International Journal of Nanomedicine, 17, 1971–1985. 10.2147/ijn.S353330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X. , Wang, Y. , Fan, R. , Zhang, L. , Li, Z. , Zhang, Y. , Zheng, W. , Wang, L. , Liu, B. , & Quan, C. (2023). Quantitative proteomic analysis of outer membrane vesicles from Fusobacterium nucleatum cultivated in the mimic cancer environment. Microbiology Spectrum, e0039423. 10.1128/spectrum.00394-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, H. , Tan, X. , Chen, G. , Liu, X. , Feng, A. , Liu, Z. , & Liu, W. (2023). Extracellular vesicles of commensal skin microbiota alleviate cutaneous inflammation in atopic dermatitis mouse model by re‐establishing skin homeostasis. Journal of Investigative Dermatology, S0022‐202X(23)00169‐0. Advance online publication. 10.1016/j.jid.2023.02.023 [DOI] [PubMed] [Google Scholar]
- Zhuang, Q. , Xu, J. , Deng, D. , Chao, T. , Li, J. , Zhang, R. , Peng, R. , & Liu, Z. (2021). Bacteria‐derived membrane vesicles to advance targeted photothermal tumor ablation. Biomaterials, 268, 120550. 10.1016/j.biomaterials.2020.120550 [DOI] [PubMed] [Google Scholar]
- Zhuang, W. R. , Wang, Y. , Lei, Y. , Zuo, L. , Jiang, A. , Wu, G. , Nie, W. , Huang, L. L. , & Xie, H. Y. (2022). Phytochemical engineered bacterial outer membrane vesicles for photodynamic effects promoted immunotherapy. Nano Letters, 22(11), 4491–4500. 10.1021/acs.nanolett.2c01280 [DOI] [PubMed] [Google Scholar]
- Zoaiter, M. , Nasser, R. , Hage‐Sleiman, R. , Abdel‐Sater, F. , Badran, B. , & Zeaiter, Z. (2021). Helicobacter pylori outer membrane vesicles induce expression and secretion of oncostatin M in AGS gastric cancer cells. Brazilian Journal of Microbiology, 52(3), 1057–1066. 10.1007/s42770-021-00490-7 [DOI] [PMC free article] [PubMed] [Google Scholar]


