Abstract
Although a substantial number of hormones and drugs increase cellular cAMP levels, the global impact of cAMP and its major effector mechanism, protein kinase A (PKA), on gene expression is not known. Here we show that treatment of murine wild-type S49 lymphoma cells for 24 h with 8-(4-chlorophenylthio)-cAMP (8-CPT-cAMP), a PKA-selective cAMP analog, alters the expression of ≈4,500 of ≈13,600 unique genes. By contrast, gene expression was unaltered in Kin- S49 cells (that lack PKA) incubated with 8-CPT-cAMP. Changes in mRNA and protein expression of several cell-cycle regulators accompanied cAMP-induced G1-phase cell-cycle arrest of wild-type S49 cells. Within 2 h, 8-CPT-cAMP altered expression of 152 genes that contain evolutionarily conserved cAMP-response elements within 5 kb of transcriptional start sites, including the circadian clock gene Per1. Thus, cAMP through its activation of PKA produces extensive transcriptional regulation in eukaryotic cells. These transcriptional networks include a primary group of cAMP-response element-containing genes and secondary networks that include the circadian clock.
Keywords: microarray, cAMP-response element (CRE), S49 cells, circadian clock
The evolutionarily conserved cAMP-signaling system is an important regulator of general and cell-type-specific effects (e.g., metabolic regulation, cell proliferation, cell death, learning, and memory) (1). Elevation of cAMP levels and subsequent activation of cAMP-dependent protein kinase (PKA) result in the proliferation of certain cell types but induce cell-cycle arrest and apoptosis in others (2–6). Increases in cAMP can also alter the sensitivity of cells to killing and growth arrest by other classes of drugs (3, 5, 6). Aberrant regulation of the cAMP-signaling pathway has been found in human cancer cell lines, primary tumors, and transformed cells (7), and targeting of this pathway shows potential for cancer therapy (2, 7, 8).
Cellular cAMP levels are controlled by G protein-coupled receptors that couple to heterotrimeric G proteins that regulate adenylyl cyclases, which catalyze cAMP formation, and by cyclic nucleotide phosphodiesterases, which hydrolyze cAMP. Several key characteristics of this important signaling system were identified in the CD4+ CD8+ S49 T lymphoma cell line derived from a BALB/c mouse tumor (9). When treated with cAMP analogs or agents that increase cAMP levels, wild-type (WT) S49 cells undergo G1-phase cell-cycle arrest and then apoptosis (3, 4). The latter response, which facilitated the isolation of cAMP-resistant clones, has made the S49 cell system a powerful model for investigating signal transduction (9).
Kin- S49 cells, a clonal S49 variant, lack PKA activity and are resistant to cAMP-promoted G1 cell-cycle arrest and apoptosis (4, 9, 10). Despite expressing a functional PKA catalytic transcript, these cells lack detectable catalytic protein (11). The absence of PKA activity allows the identification of cAMP/PKA-dependent events, including enzyme regulation, cell-cycle arrest, and apoptosis (9, 10, 12).
PKA-promoted protein phosphorylation has both transcriptional and posttranscriptional effects. The former occur pre-dominantly by the action of cAMP-response element (CRE) binding protein (CREB) and cAMP-response element modulator (CREM), which bind to cis-acting CREs, short DNA consensus elements, of PKA target genes (13, 14). Several aspects of cAMP/PKA-dependent gene regulation remain unclear, especially in view of recent evidence that cAMP stimulates PKA-independent target proteins (e.g., Epac1, Epac2, and PDZ-GEF1/RA-GEF1) that can regulate gene transcription by activating the Rap1 protein (15). The precise identity and mechanisms of regulation of genes responsible for cAMP-promoted G1/S-phase cell-cycle arrest are also not well defined. Genes such as p21, p27, and the cyclin D3 gene have been implicated (16), but cell-cycle regulators have not been comprehensively examined.
Here, we describe results of gene expression profiling in WT and Kin- S49 cells, analyzed with a model of the cell-cycle pathway to identify G1/S cell-cycle checkpoint genes regulated at the mRNA and protein levels by cAMP/PKA. To distinguish between cAMP/PKA-regulated primary and secondary transcripts, we tested the hypothesis that primary transcripts are regulated early (i.e., within2hofcAMP treatment) and enriched in evolutionarily conserved CRE sites near transcriptional start sites.
Methods
Cell Lines and Treatment with cAMP Analog. WT and Kin- S49 cells were cultured in suspension with minimal changes in media or physical perturbation in Dulbecco's modified Eagles' medium supplemented with 10% heat-inactivated horse serum and 10 mM Hepes (pH 7.4) in a humidified atmosphere containing 10% CO2 at 37°C. Cells were incubated with 100 μM 8-(4-chlorophenylthio)-cAMP (8-CPT-cAMP) (Sigma), a cAMP analog highly specific for PKA activation (17) for 2, 6, and 24 h. Stock cells were subcultured and maintained at a density not greater than 2 × 106 cells per ml in T75 flasks throughout experiments. Cultures were initiated at a density of 4–5 × 105 cells per ml for RNA extraction and at 2.5 × 105 cells per ml for cell-growth experiments and protein extraction.
Sample Preparation for Expression Array Analysis. Total cellular RNA was isolated from cells by using an RNeasy minicolumn (Qiagen, Santa Clara, CA). Total RNA was reverse transcribed with superscript II reverse transcriptase and an oligo(dT) primer containing a T7 RNA polymerase promoter. Single-stranded cDNA was converted into ds cDNA and then purified (MessageAMP aRNA Kit, Ambion, Austin, TX). Biotinylated cRNA was generated from ds cDNA by in vitro transcription (MessageAMP aRNA Kit). After a further round of purification, in vitro transcription reactions yielded 30–70 μg of biotinylated cRNA, which was fragmented to ≈100-bp fragments before hybridization.
Analysis of Transcript Levels and Unique Gene Counts with the 430 A Microarray. Each 430 A chip, which contains probes for 22,626 transcripts (representing 13,642 unique Ensembl or Unigene genes based on annotations from www.affymetrix.com and www.ensembl.org/Multi/martview), was hybridized to 10 μg of fragmented cRNA. Arrays were hybridized and scanned with a GeneArray Scanner (Hewlett–Packard; and Affymetrix, Santa Clara, CA). For each array, the .cel files were generated with the Affymetrix Microarray Suite 5.0 and analyzed with gc rma (18). All of the data are available at the Gene Expression Omnibus (www.ncbi.nlm.nih.gov/projects/geo/) under the accession no. GSE2413.
Comparisons, Statistical Analysis, and Hierarchical Ordered Partitioning and Collapsing Hybrid (HOPACH) Clustering. To identify transcripts that exhibited variability during the study, we generated seven groups of ratios for clustering. Four groups of ratios were generated by subtracting the average of the log2 signal values of 0-h WT cells from the corresponding probe sets of the following four groups: 2-, 6-, and 24-h cAMP-treated WT cells, and 0-h Kin- cells. Three other groups of ratios were generated by subtracting the 0-h Kin- log2 signal values from each of the Kin- cAMP-treated cells (2, 6, and 24 h). The 6,339 most robustly changed probe sets (permutation F test statistic of P < 0.05 and fold change >50% in at least one group) were clustered with the HOPACH clustering algorithm (19). HOPACH clustering produced 6 first-level clusters and 42 second-level clusters (denoted, for example, as 13 indicating first-level cluster 1, second-level cluster 3). The 6,339 robustly changed probe sets were filtered again to 4,947 probe sets that represented unique genes within first-level clusters. These relationships provided a unique gene count in a given comparison (Fig. 1) and a method to exclude redundant probe sets in the first-level HOPACH clusters (Fig. 2). For pairwise comparisons, we computed permutation T test statistics and fold calculations with the log2 gc rma signal values. Because we had three replicates per time point, the permutation tests resulted in the smallest P values of either 0.02 or 0.057; therefore, we used a P < 0.06 to include genes with permutation P values of 0.057. To determine the percentage of genes in the genome changed by cAMP/PKA, we computed a permutation F test statistic with the four WT log2 expression groups (0, 2, 6, and 24 h).
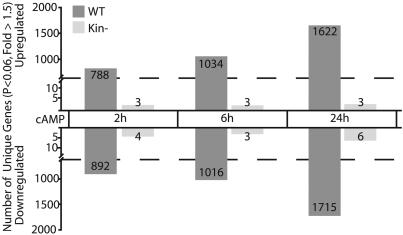
Fig. 1.
Requirement for PKA in 8-CPT-cAMP-regulated gene expression in S49 cells. Treatments of WT and Kin- cells were compared with the 0-h WT and Kin- baselines, respectively, at the indicated times. Pairwise comparisons were made by using the log2 expression values and removing data for redundant probe sets.
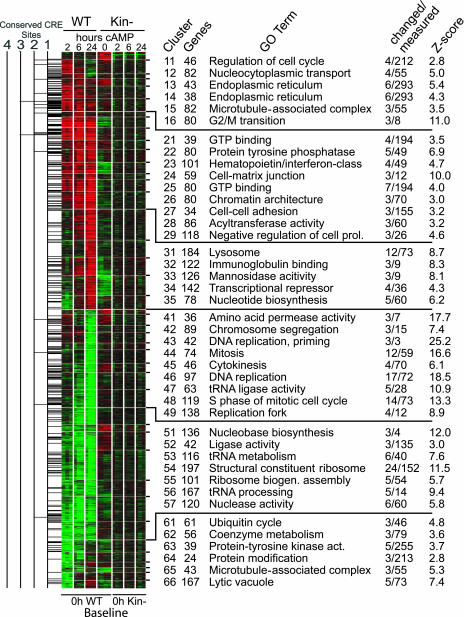
Fig. 2.
HOPACH clustering of cAMP-regulated genes annotated by Gene Ontology (GO) terms and frequencies of conserved CRE sites. HOPACH cluster analysis of 4,947 genes that had a permuted F test (P < 0.05) and at least a 1.5-fold change in any of the seven groups are represented on the cluster column titles. Red, up-regulation; green, down-regulation; black, no change. The number of conserved CRE sites is displayed as a histogram adjacent to each gene. Changed/measured, the number of genes that are in the cluster/number of genes represented on the array; Z-score calculations are described in ref. 20.
mappfinder Results. First-level and second-level clusters were annotated with Gene Ontology (GO) information (www.GeneOntology.org) with the mappfinder program (20). The raw output was sorted for mappfinder terms that had at least three genes changed with a Z-score >2 (approximately equivalent to a P < 0.05). For first-level cluster results (Table 1, which is published as supporting information on the PNAS web site), only the most significant term from a group of “parent–child” relationships is shown, based on a gene number weighted Z-score, thus significantly simplifying the results. For the second-level cluster, mappfinder results yielding the GO term with the highest Z-score are represented (Fig. 2). Z-scores were calculated by subtracting the number of genes expected to be randomly changed in a GO term from the observed number of changed genes in that GO term. This value was then divided by the standard deviation of the observed number of genes under a hypergeometric distribution (20).
CRE Site Mapping. Locus Link IDs were used to determine the genomic loci of the 4,947 genes in the six main HOPACH clusters. Five-kilobase upstream sequences from each mouse transcriptional start site along with orthologous human sequences were extracted from the whole genome alignment of mouse and human (21). CRE sites were identified with the regulatory vista (rvista) program (22), based on transfac Professional 7.4 library (23) to identify CRE (transfac matrix ID: V$CREB_01) sites in each of the aligned sequences, and to determine which of the predicted sites were aligned and conserved between the species in the alignment.
Real-Time Quantitative RT-PCR. As a validation of the microarray results, we used real-time RT-PCR analysis of 15 transcripts at three time points and found a high degree of correlation (R2 = 0.872, slope = 0.997, n = 45; see also Fig. 6, which is published as supporting information on the PNAS web site). Real-time RT-PCR was conducted as described (3). Total RNA, prepared as described above, was used to generate cDNA templates with Reverse Transcriptase Super Script II (Invitrogen), and cDNA amplification was quantitatively analyzed by incorporation of SYBR green (Qiagen, Santa Clara, CA) into dsDNA. Samples were compared by the relative (comparative) threshold cycle (Ct) method. Fold induction or repression was measured relative to controls and calculated after adjusting for GADPH by using --ΔΔCt, where ΔCt = Ct for gene of interest - Ct GADPH and ΔΔCt =ΔCt treatment -ΔCt control. Sequences for primers are listed in Table 2, which is published as supporting information on the PNAS web site.
Immunoblot Analysis. Cells treated or not with 8-CPT-cAMP for 24 h were counted, washed once with ice-cold PBS, sonicated in 1× Nu-Page LDS Sample Buffer (Invitrogen), and the volume was adjusted for cell number. Lysates (10–30 μg per lane) were electrophoresed on 12% precast Nupage gels and transferred to nitrocellulose membranes for incubation with antibodies (Santa Cruz Biotechnology): p27, sc-1641; GADD45α, sc-796; actin, sc-8432; goat anti-rabbit, sc-2030; and goat anti-mouse, sc-2031. Blots were developed with Lumigen TMA-6 (Amersham Pharmacia Biosciences); chemiluminescent signals were captured, quantitated with a Bio-Imager (Ultraviolet Products), and normalized to actin expression. Graphing and statistical analysis were done with prism software (GraphPad, San Diego).
Results and Discussion
PKA Is Required for cAMP-Mediated Gene Expression in S49 Cells. We profiled the expression patterns of 22,626 mouse gene transcripts (13,642 unique Ensembl or Unigene genes) in WT and Kin- S49 lymphoma cells that were treated with 8-CPT-cAMP, a PKA-specific cAMP analog (17), for 2, 6, and 24 h. The fold-changes calculated from the microarray data correlated well with RT-PCR results (R2 = 0.872, slope = 0.997, n = 45; see also Fig. 6). WT and Kin- cells have essentially identical growth kinetics (Fig. 7A, which is published as supporting information on the PNAS web site) unless incubated with 8-CPT-cAMP, which arrests ≈65% of WT, but not Kin-, cells in G1 by 18 h (Fig. 7B).
Incubation of WT S49 cells with 8-CPT-cAMP significantly changed (P < 0.06, fold change >1.5) the expression of 1,680, 2,050, and 3,337 genes at 2, 6, and 24 h, respectively, with a similar number of increases and decreases at each time point (Fig. 1). Overall, this represents a change in 4,475 of the 13,642 transcripts on the microarrays. In similarly treated Kin- cells, only 7, 6, and 9 genes were changed at 2, 6, and 24 h, respectively. Increasing the filtering stringency from 1.5- to 2.0-fold resulted in 714, 890, and 1,861 gene expression changes (P < 0.06) in the WT cells at 2, 6, and 24 h, respectively, but only one alteration in Kin- cells (at 2 h). PKA-independent cAMP signaling can occur by means of Epac, a protein activated by cAMP (15); we assessed for and found expression of this gene (Rapgef3). Thus, although this alternative cAMP signaling system appears to be present in S49 cells, PKA is required for cAMP-mediated gene regulation in these cells and mediates a ≥50% change in expression of ≈12% of genes within 2 h with ≈33% of the genes showing gene expression changes over 24 h.
HOPACH Clustering and GO Analysis of PKA-Mediated Gene Expression Patterns. To characterize gene expression patterns, we used HOPACH (19) to cluster the 4,947 genes with the most prominent changes during 8-CPT-cAMP treatment (Fig. 2). Six large first-level clusters that could be subdivided into smaller second-level clusters were identified: early-induction (cluster 1), sustained-induction (cluster 2), late-induction (cluster 3), late-repression (cluster 4), sustained-repression (cluster 5), and early-repression (cluster 6).
Using mappfinder (www.GenMAPP.org) (20), we annotated the first- (Table 1) and second-level (Fig. 2) clusters with GO (www.GeneOntology.org) terms to obtain a functional description of the gene expression patterns. The early-induction cluster, which we hypothesized would include transcripts regulated preferentially and directly by PKA, contained genes involved in the endoplasmic reticulum (n = 23), kinase activities (n = 30), cell differentiation (n = 12), and others (Table 1). Conversely, late-induction genes were involved in the lysosome (n = 22), hydrolase activity (n = 103), intracellular signaling (n = 43), and cell death (n = 26). The late-repression group included genes involved in the cell cycle (n = 70), RNA binding (n = 54), and protein metabolism (n = 112). By 24 h, 70 genes involved in the mitotic cell cycle were down-regulated, consistent with the idea that transcriptional regulation mediates cAMP/PKA-promoted G1-phase cell-cycle arrest. RNA synthetases and translation factors (Fig. 8, which is published as supporting information on the PNAS web site) were coordinately down-regulated by 24-h treatment of WT S49 cells with 8-CPT-cAMP. This shutdown in protein synthesis accompanied the down-regulation of mitotic cell-cycle genes.
Circadian Rhythm Clock Genes Are Induced by cAMP. Several circadian rhythm clock genes (Table 1) were identified in early-induction cluster 1. Of these clock genes identified by mappfinder in cluster 1, only Per1 was significantly induced by 2 h (4-fold induction; P < 0.05), consistent with a previous report (24). Per1 is under circadian regulation in human and mouse peripheral tissues (25, 26) and is responsible, in part, for maintaining core clock rhythm. Per1 forms heterodimers with other proteins to inhibit the transcriptional activity of the circadian clock transcription factors Clock/Bmal1. Transcriptional regulation controlled by the core circadian rhythm clock is estimated to involve ≈8–10% of peripherally expressed genes (27).
Several lines of evidence support the concept that the cAMP-mediated alteration of circadian-regulated genes in S49 cells contributes to cell synchronization and G1-phase cell-cycle arrest. We tested cAMP-regulated genes against three groups of genes under circadian regulation in mouse liver (n = 426), heart (n = 345), and suprachiasmatic nucleus (n = 343) (28). The circadian gene lists overlapped significantly with cAMP-regulated genes whose expression changed significantly (1.5-fold; P < 0.06) (Table 3, which is published as supporting information on the PNAS web site). These results support the idea that cAMP/PKA alters the regulation of a major secondary transcriptional network controlled by the circadian clock within 2 h. Alterations in circadian clock genes affect cell-cycle-controlled phenotypes in cancer (29) and hepatic regeneration (30).
Mapping of Conserved Human and Mouse CRE Sites. To define mechanisms underlying the transcriptional effects of cAMP and PKA, we assessed all transcripts on the microarrays for evolutionarily conserved CRE sites, DNA consensus elements [TGACGT(C/A)A] that initiate PKA-mediated transcription (13, 14). Using human/mouse genomic alignments to scan 5 kb of upstream conserved sequence(s) from the transcriptional start site, we identified evolutionarily conserved CRE sites with a matrix based on the CRE consensus element. Alignment of CRE sites in human and mouse sequences defined a conserved 21-bp interval with >80% conservation. This approach allowed the identification of ≈90% of experimentally verified binding sites over a 1-Mb gene-rich, well-annotated genomic interval (22).
We hypothesized that target genes of PKA-controlled transcription factors would be regulated within 2 h of cAMP treatment (early-induction cluster 1) and enriched in evolutionarily conserved CRE sites near their transcriptional start sites. Indeed, the genes up-regulated at 2 h included more CRE sites than other clusters or than all genes on the array (Fig. 2, clusters 12, 15, and 16; see also Fig. 9, which is published as supporting information on the PNAS web site). Genome-wide analysis of ≈15,000 aligned promoter regions revealed that the majority of CRE sites were within 250 bp of the transcriptional start site (Fig. 10, which is published as supporting information on the PNAS web site), implicating them as functional CRE sites (31). Of the 1,680 genes that showed 1.5-fold change within 2 h, 111 of the up-regulated genes and 41 of the down-regulated genes contained at least one conserved CRE site (Table 4, which is published as supporting information on the PNAS web site), suggesting that these are likely primary targets of cAMP/PKA. Other genes transcriptionally regulated by cAMP at 2 h may be regulated by other mechanisms, perhaps as secondary transcriptional networks (i.e., the circadian clock as discussed above). This hypothesis is supported by analysis using mappfinder, which indicated that 41 transcription regulator activity genes (P = 0.004) are in the early-induction cluster (Table 1). Another possible explanation for the small percentage of early-induction genes that contain CRE sites is that the mouse transcripts that we measured may contain functional CRE sites that are not evolutionarily conserved or not within 5 kb upstream of the transcriptional start site. Other clusters (sustained-induction clusters 22 and 23, and late-repression clusters 45, 47, and 49) were also enriched in CRE sites, perhaps indicative of differences in properties of CRE-binding protein/CRE modulator and DNA interaction.
The complexity and cell specificity of PKA regulation are underscored by comparison of these findings to the 78 top-scoring “hits” of Conkright et al. (31), who identified conserved CRE sites in a human and mouse genome alignment. Probe sets for 77 of those transcripts were measured by our microarrays but only 7 transcripts were significantly (P < 0.06) up-regulated, whereas 3 transcripts were down-regulated 1.5-fold at 2 h by cAMP/PKA (Fig. 11, which is published as supporting information on the PNAS web site). These results imply that one cannot currently predict transcriptional regulation by cAMP/CRE solely by comparing human and mouse genomes. The data also indicate that substantial CRE silencing may occur in particular tissues and suggest that only a fraction of CRE-containing genes are transcriptionally active in a specific cell type (32).
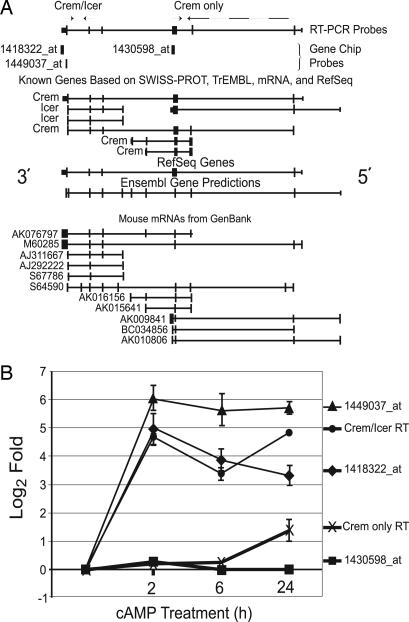
Induction of the Inducible cAMP Early Repressor (Icer) Gene by 2 h. Our data indicated that in 8-CPT-cAMP-treated WT S49 cells, CRE-mediated transcription was inhibited within 2 h, as evidenced by the strong induction of the inhibitory CRE-modulator (CREM) isoform, ICER, which binds to CRE and decreases PKA-mediated transcription (13, 14). Expression of Crem was up-regulated at all three time points (Fig. 3). Two up-regulated probe sets that target the 3′ end of the Crem gene detect both Crem and Icer, and a third probe set, for which there was no change in expression, targets the middle of the gene. Quantitative RT-PCR studies confirmed the unchanged expression of the middle portion of the Crem gene. Thus, Icer was highly induced in S49 cells by cAMP/PKA within 2 h of treatment with 8-CPT-cAMP, suggesting that by this time an autoregulatory PKA feedback loop decreases CRE-mediated transcription. Indeed, Icer induction was detectable between 30 min and 1 h (Fig. 12, which is published as supporting information on the PNAS web site). The induction of Icer is consistent with the conclusion that most gene expression changes in response to 8-CPT-cAMP after 2 h occur by CRE-independent secondary transcriptional networks.
Fig. 3.
Induction of the inducible cAMP early repressor. (A) The target sequences of the three Crem gene probe sets were aligned to the October 2003 build of the mouse genome by blat searching at the University of California Santa Cruz genome browser (http://genome.ucsc.edu/). Arrows, RT-PCR primers; dashed arrow, Crem only reverse primer that spanned two exons. (B) log2 folds of Crem and Icer probes. RT, real-time RT-PCR.
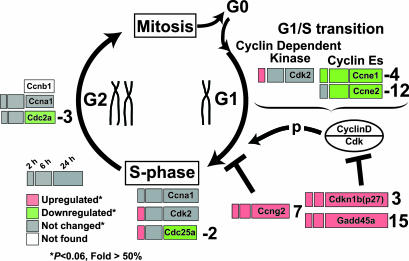
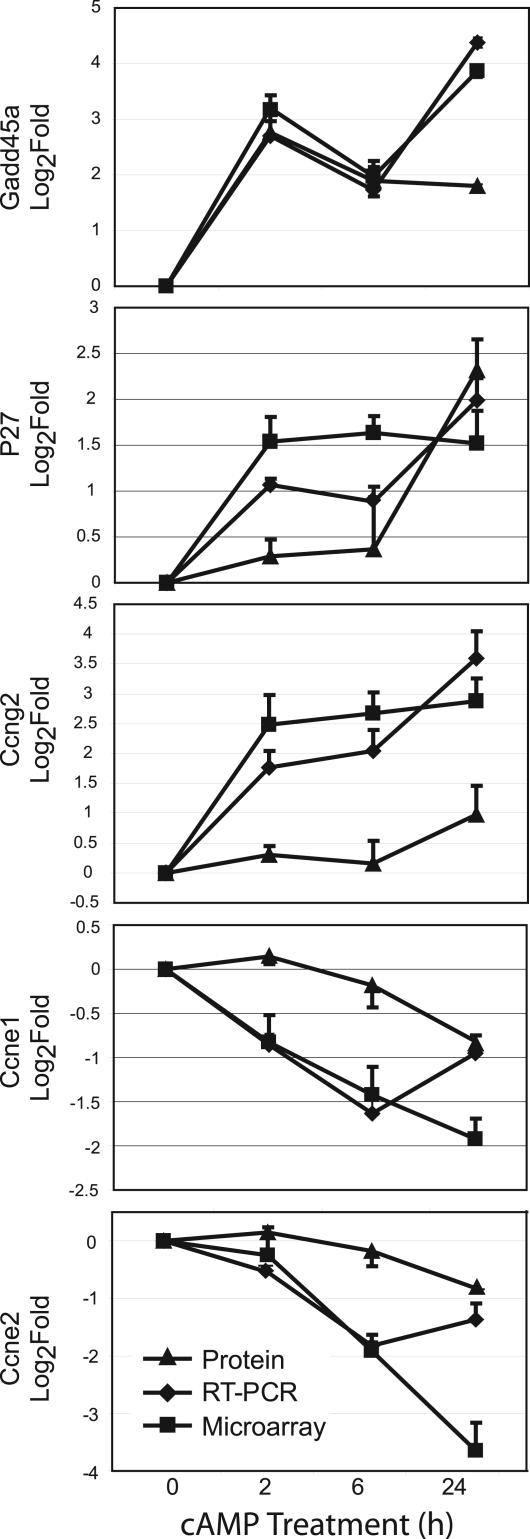
Changes in Cell Cycle G1/S-Phase Mediators and Checkpoints. The expression of several genes involved in the cell cycle was changed in response to cAMP (Fig. 4; see also Fig. 13, which is published as supporting information on the PNAS web site, for expanded pathway). We observed substantial up-regulation of genes that inhibit G1–S progression [e.g., Gadd45a, p27, Ccng2 (cyclin G2)] and, in parallel, a down-regulation of genes that induce the G1–S transition (e.g., Cdk2, Ccne1, and Ccne2). Using quantitative RT-PCR and immunoblotting, we confirmed the changes at the mRNA and protein levels (Fig. 5). The increase in Gadd45a mRNA and protein levels at 2 h was particularly striking, independent of changes in p53 mRNA expression and preceded changes in protein expression of the other G1/S-related transcripts; however, Gadd45a does not have conserved CRE sites <5 kb from its initiation site. We analyzed transcript levels for Gadd45a, p27, Ccng2, cyclin E1, cyclin E2, and p27 at earlier time-points (0.5 and 1 h) by real-time RT-PCR, and only Gadd45a showed substantial regulation at 30 min (8-fold) and 1 h (16-fold) (Fig. 12). Induction of p27 by increasing cAMP levels has been shown (16), but we know of no prior report that PKA activation strongly induces mRNA for Gadd45a (22-fold by 2 h) and Ccng2 (5.5-fold by 2 h). Other evidence implicates changes in Gadd45a expression, perhaps in a p53-independent manner, in G1 cell-cycle arrest, although a role in G2/M has also been noted (33, 34).
Fig. 4.
G1/S cell-cycle control. Boxes, gene transcripts; numbers, microarray-derived fold changes at 24 h compared with 0 h. Pathway was adapted from Reactome (35) and refs. 34 and 36. [See also Fig. 13 for an expanded GenMAPP (37) pathway.]
Fig. 5.
Real-time RT-PCR and immunoblot analysis of cell-cycle targets compared to microarray data. Targets assayed for RNA by DNA microarrays (▪), real time PCR (♦), and for protein by immunoblot analysis (▴). Values were normalized to baseline expression (0 h).
Summary and Discussion. These studies in WT and Kin- S49 cells define a transcriptional “topography” of gene expression regulated by cAMP acting by means of PKA. The results show that PKA activation of WT S49 cells by cAMP for 24 h leads to changes in ≈33% of the genome. Other cells types that have less dramatic phenotypic responses than do S49 cells (i.e., G1 cell-cycle arrest and cytolysis) to cAMP/PKA activation will likely show different and potentially fewer numbers of differentially regulated transcripts (32). Mapping of evolutionarily conserved PKA-responsive transcriptional CRE sites showed that clusters of early-induced genes were enriched in these elements, implying that they are primary PKA-regulated transcripts (Table 4). Of these early-induced genes, only 152 of 1,680 that changed within 2 h of cAMP treatment contained conserved CRE sites, suggesting that a substantial number of secondary transcriptional events occur within 2 h of cAMP treatment. This hypothesis is supported by three observations: (i) cAMP regulates the circadian clock, which can regulate ≈8–10% of peripherally expressed genes; (ii) 41 genes in the early-induction cluster are classified by GO as having transcription regulator activity (P = 0.004); and (iii) CRE-mediated transcription is blunted by the large induction of the inhibitor Icer (Fig. 3).
Using GO classifications, we have defined this transcriptional topography in terms of known biological annotation (Fig. 2; see also Table 1) and have posted these results (web site available from the authors on request) so as to allow visualization of all of the genes represented by the GO terms in Table 1.
We observed early induction (2 h) of several G1-phase cell-cycle checkpoint genes, but when combined with protein analysis, Gadd45a emerged as a potential primary mediator of cell-cycle regulation by cAMP/PKA. Later changes in gene expression, which include down-regulation of genes involved in the G1/S-phase transition (Fig. 4), S-phase DNA replication (Fig. 12), and translation (Fig. 8) likely contribute to secondary aspects of cell-cycle control produced by cAMP and PKA. It will be of interest to determine whether the changes we observed in S49 cells are found in other cell types that undergo cAMP-promoted cell-cycle arrest in the G1 phase and how such changes contrast with those in cells that, unlike S49 cells, have enhanced replication in response to cAMP (2, 5, 7).
Supplementary Material
Acknowledgments
We thank J. Corbeil for advice and the use of the University of California at San Diego Genomic Research Center and Core Laboratory. This study was supported by National Institutes of Health Grant GM 61774 (to L.Z., J.R.K., and P.A.I.); American Heart Association Postdoctoral Fellowship 0425278Y (to A.C.Z.); National Heart, Lung, and Blood Institute, National Institutes of Health Grant U1HL66681B (to S.M., S.P., and I.D.); and U.S. Department of Energy's Office of Science, Biological, and Environmental Research Program, Lawrence Berkeley National Laboratory Contract DE-AC03-76SF00098 (to S.M., S.P., and I.D.).
Author contributions: A.C.Z., L.Z., B.R.C., and P.A.I. designed research; L.Z. and J.R.K. performed research; A.C.Z., S.M., S.P., N.S., and I.D. contributed new reagents/analytic tools; A.C.Z., S.M., S.P., N.S., K.V., and I.D. analyzed data; and A.C.Z. and P.A.I. wrote the paper.
Abbreviations: PKA, protein kinase A; 8-CPT-cAMP, 8-(4-chlorophenylthio)-cAMP; CRE, cAMP-response element; HOPACH, hierarchical ordered partitioning and collapsing hybrid; GO, Gene Ontology; Ct, threshold cycle.
Data deposition: The microarray data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database (accession no. GSE2413).
© 2005 by The National Academy of Sciences of the USA
References
- 1.Skalhegg, B. S. & Tasken, K. (2000) Front. Biosci. 5, D678-D693. [DOI] [PubMed] [Google Scholar]
- 2.Roger, P. P., Reuse, S., Maenhaut, C. & Dumont, J. E. (1995) Vitam. Horm. 51, 59-191. [DOI] [PubMed] [Google Scholar]
- 3.Zhang, L. & Insel, P. A. (2004) J. Biol. Chem. 279, 20858-20865. [DOI] [PubMed] [Google Scholar]
- 4.Yan, L., Herrmann, V., Hofer, J. K. & Insel, P. A. (2000) Am. J. Physiol. 279, C1665-C1674. [DOI] [PubMed] [Google Scholar]
- 5.Thompson, E. B., Medh, R. D., Zhou, F., Ayala-Torres, S., Ansari, N., Zhang, W. & Johnson, B. H. (1999) J. Steroid Biochem. Mol. Biol. 69, 453-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abraham, I., Hunter, R. J., Sampson, K. E., Smith, S., Gottesman, M. M. & Mayo, J. K. (1987) Mol. Cell. Biol. 7, 3098-3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho-Chung, Y. S., Nesterova, M., Becker, K. G., Srivastava, R., Park, Y. G., Lee, Y. N., Cho, Y. S., Kim, M. K., Neary, C. & Cheadle, C. (2002) Ann. N.Y. Acad. Sci. 968, 22-36. [DOI] [PubMed] [Google Scholar]
- 8.Weissinger, E. M., Oettrich, K., Evans, C., Genieser, H. G., Schwede, F., Dangers, M., Dammann, E., Kolb, H. J., Mischak, H., Ganser, A. & Kolch, W. (2004) Br. J. Cancer 91, 186-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Daalen Wetters, T. & Coffino, P. (1987) Methods Enzymol. 151, 9-19. [DOI] [PubMed] [Google Scholar]
- 10.Insel, P. A., Bourne, H. R., Coffino, P. & Tomkins, G. M. (1975) Science 190, 896-898. [DOI] [PubMed] [Google Scholar]
- 11.Orellana, S. A. & McKnight, G. S. (1990) J. Biol. Chem. 265, 3048-3053. [PubMed] [Google Scholar]
- 12.Steinberg, R. A., O'Farrell, P. H., Friedrich, U. & Coffino, P. (1977) Cell 10, 381-391. [DOI] [PubMed] [Google Scholar]
- 13.Mayr, B. & Montminy, M. (2001) Nat. Rev. Mol. Cell Biol. 2, 599-609. [DOI] [PubMed] [Google Scholar]
- 14.Servillo, G., Della Fazia, M. A. & Sassone-Corsi, P. (2002) Exp. Cell Res. 275, 143-154. [DOI] [PubMed] [Google Scholar]
- 15.Kopperud, R., Krakstad, C., Selheim, F. & Doskeland, S. O. (2003) FEBS Lett. 546, 121-126. [DOI] [PubMed] [Google Scholar]
- 16.Stork, P. J. & Schmitt, J. M. (2002) Trends Cell Biol. 12, 258-266. [DOI] [PubMed] [Google Scholar]
- 17.Lamb, D. & Steinberg, R. A. (2002) J. Cell. Physiol. 192, 216-224. [DOI] [PubMed] [Google Scholar]
- 18.Wu, Z. & Irizarry, R. A. (2004) Nat. Biotechnol. 22, 656-658. [DOI] [PubMed] [Google Scholar]
- 19.van der Laan, M. J. & Pollard, K. S. (2001) A New Algorithm for Hybrid Hierarchical Clustering with Visualization and the Bootstrap (Division of Bio-statistics, Univ. of California, Berkeley), Technical Report No. 93. Available at www.stat.berkeley.edu/~laan/Research/Research_subpages/Papers/hopach.pdf.
- 20.Doniger, S. W., Salomonis, N., Dahlquist, K. D., Vranizan, K., Lawler, S. C. & Conklin, B. R. (2003) Genome Biol. 4, R7. Available at http://genomebiology.com/2003/4/1/R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brudno, M., Poliakov, A., Salamov, A., Cooper, G. M., Sidow, A., Rubin, E. M., Solovyev, V., Batzoglou, S. & Dubchak, I. (2004) Genome Res. 14, 685-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loots, G. G., Ovcharenko, I., Pachter, L., Dubchak, I. & Rubin, E. M. (2002) Genome Res. 12, 832-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wingender, E., Chen, X., Fricke, E., Geffers, R., Hehl, R., Liebich, I., Krull, M., Matys, V., Michael, H., Ohnhauser, R., et al. (2001) Nucleic Acids Res. 29, 281-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tischkau, S. A., Mitchell, J. W., Tyan, S. H., Buchanan, G. F. & Gillette, M. U. (2003) J. Biol. Chem. 278, 718-723. [DOI] [PubMed] [Google Scholar]
- 25.King, D. P. & Takahashi, J. S. (2000) Annu. Rev. Neurosci. 23, 713-742. [DOI] [PubMed] [Google Scholar]
- 26.Zambon, A. C., McDearmon, E. L., Salomonis, N., Vranizan, K. M., Johansen, K. L., Adey, D., Takahashi, J. S., Schambelan, M. & Conklin, B. R. (2003) Genome Biol. 4, R61. Available at http://genomebiology.com/2003/4/10/R61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Storch, K. F., Lipan, O., Leykin, I., Viswanathan, N., Davis, F. C., Wong, W. H. & Weitz, C. J. (2002) Nature 417, 78-83. [DOI] [PubMed] [Google Scholar]
- 28.Panda, S., Antoch, M. P., Miller, B. H., Su, A. I., Schook, A. B., Straume, M., Schultz, P. G., Kay, S. A., Takahashi, J. S. & Hogenesch, J. B. (2002) Cell 109, 307-320. [DOI] [PubMed] [Google Scholar]
- 29.Fu, L., Pelicano, H., Liu, J., Huang, P. & Lee, C. (2002) Cell 111, 41-50. [DOI] [PubMed] [Google Scholar]
- 30.Matsuo, T., Yamaguchi, S., Mitsui, S., Emi, A., Shimoda, F. & Okamura, H. (2003) Science 302, 255-259. [DOI] [PubMed] [Google Scholar]
- 31.Conkright, M. D., Guzman, E., Flechner, L., Su, A. I., Hogenesch, J. B. & Montminy, M. (2003) Mol. Cell 11, 1101-1108. [DOI] [PubMed] [Google Scholar]
- 32.Zhang, X., Odom, D. T., Koo, S. H., Conkright, M. D., Canettieri, G., Best, J., Chen, H., Jenner, R., Herbolsheimer, E., Jacobsen, E., et al. (2005) Proc. Natl. Acad. Sci. USA 102, 4459-4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taylor, W. R. & Stark, G. R. (2001) Oncogene 20, 1803-1815. [DOI] [PubMed] [Google Scholar]
- 34.Zhang, X., Ma, L., Enkemann, S. A. & Pledger, W. J. (2003) Oncogene 22, 4166-4174. [DOI] [PubMed] [Google Scholar]
- 35.Joshi-Tope, G., Vastrik, I., Gopinathrao, G., Matthews, L., Schmidt, E., Gillespie, M., D'Eustachio, P., Jassal, B., Lewis, S., Wu, G., et al. (2003) Cold Spring Harbor Symp. Quant. Biol. 68, 237-243. [DOI] [PubMed] [Google Scholar]
- 36.Bennin, D. A., Don, A. S., Brake, T., McKenzie, J. L., Rosenbaum, H., Ortiz, L., DePaoli-Roach, A. A. & Horne, M. C. (2002) J. Biol. Chem. 277, 27449-27467. [DOI] [PubMed] [Google Scholar]
- 37.Dahlquist, K. D., Salomonis, N., Vranizan, K., Lawlor, S. C. & Conkling, B. R. (2002) Nat. Genet. 31, 19-20. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.