Abstract
The synthesis of human immunodeficiency virus type 1 (HIV-1) mRNAs is a complex process by which more than 30 different mRNA species are produced by alternative splicing of a single primary RNA transcript. HIV-1 splice sites are used with significantly different efficiencies, resulting in different levels of mRNA species in infected cells. Splicing of Tat mRNA, which is present at relatively low levels in infected cells, is repressed by the presence of exonic splicing silencers (ESS) within the two tat coding exons (ESS2 and ESS3). These ESS elements contain the consensus sequence PyUAG. Here we show that the efficiency of splicing at 3′ splice site A2, which is used to generate Vpr mRNA, is also regulated by the presence of an ESS (ESSV), which has sequence homology to ESS2 and ESS3. Mutagenesis of the three PyUAG motifs within ESSV increases splicing at splice site A2, resulting in increased Vpr mRNA levels and reduced skipping of the noncoding exon flanked by A2 and D3. The increase in Vpr mRNA levels and the reduced skipping also occur when splice site D3 is mutated toward the consensus sequence. By in vitro splicing assays, we show that ESSV represses splicing when placed downstream of a heterologous splice site. A1, A1B, A2, and B1 hnRNPs preferentially bind to ESSV RNA compared to ESSV mutant RNA. Each of these proteins, when added back to HeLa cell nuclear extracts depleted of ESSV-binding factors, is able to restore splicing repression. The results suggest that coordinate repression of HIV-1 RNA splicing is mediated by members of the hnRNP A/B protein family.
Both simple and complex retroviruses require splicing of a single primary RNA transcript in order to generate mRNA for the viral envelope protein (Env). Complex retroviruses, such as human immunodeficiency virus type 1 (HIV-1), require the production of additional mRNAs for regulatory and accessory proteins. For HIV-1 these include mRNAs for Tat, Rev, Vif, Vpr, and Nef (6, 19, 35, 37, 39). The Rev protein binds to RNAs containing the Rev-responsive element in the env gene sequence. This interaction facilitates nuclear export of unspliced and partially spliced RNAs required for translation and for packaging into progeny virions (16, 17, 20, 21, 30; for a recent review, see reference 12). Early in infection of cells with HIV-1 and prior to the accumulation of Rev, multiply spliced mRNAs predominate in the cytoplasm. Later in infection, the production of Rev allows the cytoplasmic accumulation of unspliced and partially spliced RNAs (24, 25)
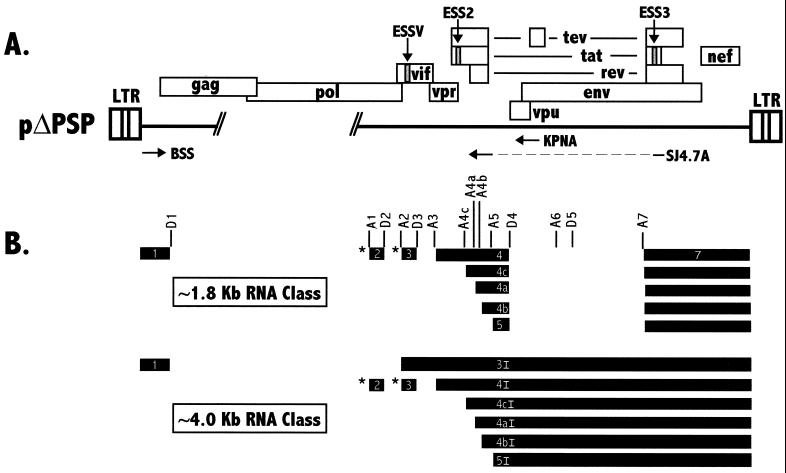
In order to generate mRNAs required for the synthesis of viral proteins, HIV-1 primary RNA transcripts undergo a complex splicing process (Fig. 1). The viral RNA contains both constitutive and alternative 5′ and 3′ splice sites. All spliced mRNAs contain 5′-terminal noncoding exon 1, which is flanked by consensus 5′ splice site D1. Selection of the alternative 3′ splice sites near the middle of the genome determines which proteins are encoded by the mRNAs. Two size classes of spliced RNAs are produced, depending on the removal of the intron spanning D4 to A7 (∼1.8 kb for the small size class and ∼4 kb for the intermediate size class). For instance, splicing at A3 coupled with splicing at D4 to A7 generates ∼1.8-kb Tat mRNA. Similarly, splicing at A4a, A4b, or A4c coupled with splicing at D4 to A7 generates ∼1.8-kb Rev mRNA; splicing at A5 coupled with splicing at D4 to A7 generates ∼1.8-kb Nef mRNA. Splicing at A3 generates an ∼4-kb mRNA encoding a single-exon form of Tat. Splicing of mRNAs at A4a, A4b, A4c, and A5 generates ∼4-kb mRNAs encoding Env. Splicing at A1 and A2 generates ∼4-kb mRNAs encoding Vif and Vpr, respectively. As a further complexity, some mRNAs of both size classes include one or both of two alternative noncoding exons (Fig. 1B): exon 2, which is flanked by A1 and D2, and exon 3, which is flanked by A2 and D3 (18, 35, 39). Finally, some virus strains contain within the env gene cryptic splice sites (D5 and A6) whose usage results in the synthesis of an mRNA encoding a hybrid protein, Tev (8, 38).
FIG. 1.
(A) Structure of the HIV-1 NL4-3 genome. Boxes indicate open reading frames. Hash marks represent endpoints of gag-pol deletion in pΔPSP. ESS sequences are shown by shaded boxes. Oligonucleotide primers used are indicated by arrows designating position and orientation. LTR, long terminal repeat. (B) Structures of the small (∼1.8-kb) and intermediate (∼4.0-kb) size classes of HIV-1 transcripts. Exons are indicated as black bars. The exons within the different RNA species are designated by numbers (and sometimes letters) within the boxes according to the nomenclature of Purcell and Martin (35). The exon designations with the letter I indicate exons present only in the intermediate-size HIV-1 mRNA species. The alternative noncoding exons 2 and 3 are indicated with asterisks. Locations of 5′ (D) and 3′ (A) splice sites are shown.
Different spliced HIV-1 mRNAs are generated with very different efficiencies. For example, ∼1.8-kb mRNAs encoding Tat are present at low levels compared to mRNAs encoding Rev or Nef. Similarly, ∼4-kb mRNAs encoding single-exon Tat are present at low levels compared to mRNAs encoding Env (35). It was previously shown that splice site A3 is repressed by ESS2, an exonic splicing silencer (ESS) within the first tat coding exon (exon 4 in Fig. 1). Mutations within the ESS2 element result in a selective increase in splicing at A3 (3, 4). A second ESS (ESS3) was identified within the second tat/rev coding exon downstream of A7 (exon 7 in Fig. 1). In this case, an adjacent upstream exonic splicing enhancer is juxtaposed to the ESS (4, 43). Both ESS2 and ESS3 appear to bind to a common cellular factor or factors that act to repress splicing (42). It has been reported that ESS2 selectively binds to members of the A/B hnRNP family (hnRNPs A1, A1B, A2, and B1) (11, 14). The addition of hnRNP A1 and other members of the hnRNP A/B protein family restores specific splicing repression in HeLa cell nuclear extracts depleted of ESS2-binding proteins (11). A third potential ESS is present in the env gene, where it may prevent the activation of cryptic exon 6D, which is bordered by splice sites A6 and D5 (46).
The levels of Vpr mRNA singly spliced at 3′ splice site A2 also have been shown to be low in cells infected with HIV-1, indicating that splicing at A2 is inefficient. Furthermore, noncoding exon 3 (Fig. 1) is skipped in the majority of the mRNAs (35). This exon skipping also suggests that splice site A2 or D3 or both of these splice sites are used inefficiently. It has been shown that the branch point used for splicing at splice site A2 is a G rather than the consensus A that is used for most 3′ splice sites. However, replacing the nonconsensus wild-type branch-point sequence with a consensus sequence did not significantly affect splicing efficiency in an in vitro splicing system (13). In this report, we describe additional elements downstream of 3′ splice site A2 that act to repress splicing at this splice site.
MATERIALS AND METHODS
Plasmids.
Infectious HIV-1 plasmid pNL4-3 (GenBank accession no. M19921) was constructed by Adachi et al. (1) and was obtained from the National Institutes of Health AIDS Research and Reference Reagent Program. Plasmid pΔPSP is a derivative of pNL4-3 with a deletion between the SpeI site at nucleotide (nt) 1511 and the BalI site at nt 4551 (23). Mutant plasmid pSPRS, with base changes within noncoding exon 3, was constructed by PCR mutagenesis using a Quickchange mutagenesis kit (Stratagene, La Jolla, Calif.). The mutagenic primers were NL43PSMTF (5′GAAATACCATATTCTGACGTATAGTTCTTCCTCTGTGTGAATATCAAGC) and NL43PSMTR (5′GCTTGATATTCACACAGAGGAAGAACTATACGTCAGAATATGGTATTTC); the changed nucleotides are underlined. Mutant plasmid pSPD3up, with an A-to-T change at position +6 of the D3 splice site, was generated by PCR mutagenesis using the Quickchange mutagenesis kit. The mutagenic primers were D3ATF (5′GGACATAACAAGGTAGGTTCTCTACAGTACTTGG3′) and D3ATR (5′CCAAGTACTGTAGAGAACCTACCTTGTTATGTCC3′). Plasmid pHS1-X, used as a template to synthesize substrates for the splicing assays, has been described previously (3). Plasmid pHS3-ESSV was constructed by replacing the EcoRI-ScaI fragment of pHS1-X with nt 5322 to nt 5479 of pNL4-3. Mutant plasmid pHS3-ESSVx was created by replacing the region between the EcoRI and XhoI sites of pHS3-ESSV with mutated PCR products. The mutated PCR products were synthesized by using a modified megaprimer technique (2). The mutagenic primers were PSALLF (5′CCATATTCTGACGTATAGTTCTTCCTCTGTGTGAA) and PSALLR (5′TTCACACAGAGGAAGAACTATACGTCAGAATATGG). pHS1-ESSV and pHS1-ESSVx are derivatives of pHS1-X which have wild-type and mutant noncoding exon 3 silencers inserted in place of the tat exon 2 ESS2, respectively. pHS1-ESSV and pHS1-ESSVx were generated by PCR mutagenesis using the primers PSWTS (5′TTAGGACGTATAGTTAGTCCTAGGGGAAGCATCCAGGAAGTC) and PSWTA (5′CCTAGGACTAACTATACGTCCTAAACTGGCTCCATTTCTTGC) for pHS1-ESSV and the primers PSMTS (5′TTCTGACGTATAGTTCTTCCTCTGGGAAGCATCCAGGAAGTC) and PSMTA (5′CAGAGGAAGAACTATACGTCAGAAACTGGCTCCATTTCTTGC) for pHS1-ESSVx. The resulting PCR products were used as primers for the synthesis of a larger product by a modified megaprimer technique (2). This PCR product was ligated into pHS1-X cleaved with EcoRI and KpnI. The competitor RNAs used for depleting nuclear extracts were transcribed from linearized plasmids pESS2, pESS2x, pESSV, and pESSVx. pESS2 and pESS2x were created by insertion of 109-bp AccI-RsaI fragments from pHS1 and pΔESS10 (4) into pBluescript SK(+) (Stratagene) cleaved with AccI and EcoRI, respectively. pESSV and pESSVx were created similarly using AccI-RsaI fragments from pHS1-ESSV and pHS1-ESSVx, respectively.
RNA isolation, reverse transcription, and PCR.
Total cellular RNA was isolated from transfected HeLa cells 48 h posttransfection by extraction with Tri-Reagent (Molecular Research Center, Inc.) according to procedures supplied by the manufacturer. Three micrograms of RNA was reverse transcribed for 1 h in a 30-μl total volume containing 20 mM each deoxynucleoside triphosphate, 20 U of RNasin (Promega, Madison, Wis.), 100 pmol of random hexamer (Pharmacia, Piscataway, N.J.), 6 μg of bovine serum albumin, and 200 U of Moloney murine leukemia virus reverse transcriptase (RT) (Life Technologies/Gibco/BRL, Rockville, Md.).
For the semiquantitative analysis of ∼1.8-kb HIV-1 mRNAs, PCR of cDNA was performed with forward oligonucleotide primer BSS (5′GGCTTGCTGAAGCGCGCACGGCAAGAGG; nt 700 to nt 727) and reverse primer SJ4.7A, which spans splice sites D5 and A7 (5′TTGGGAGGTGGGTTGCTTTGATAGAG; nt 8381 to nt 8369 and nt 6044 to nt 6032). Reverse primer KPNA (5′AGAGTGGTGGTTGCTTCCTTCCACACAG) was used with forward primer BSS for the analysis of ∼4.0-kb HIV-1 mRNAs (34). PCR amplification was performed essentially as previously described (10). Thirty cycles of PCR (94°C for 30 s, 60°C for 1 min, and 72°C for 2 min) were completed with a total reaction volume of 50 μl containing 75 mM MgCl2, 10 mM each deoxynucleoside triphosphate, 25 pmol of each primer, and 0.1 U of Perkin-Elmer Amplitaq Gold polymerase. Prior to PCR, the reaction mixture was denatured for 5 min at 94°C. After confirmation of the amplified spliced product by polyacrylamide gel electrophoresis (PAGE) and ethidium bromide staining, products (100 ng) were radiolabeled by performing a single round of PCR with the addition of 10 μCi of [32P]dCTP. Radiolabeled products were analyzed by denaturing electrophoresis on 6% polyacrylamide–7 M urea gels.
RNA substrate synthesis.
To prepare transcription templates, all DNA constructs were linearized with XhoI. In vitro transcription of runoff RNA transcripts labeled with [32P]UTP (NEN, Boston, Mass.) was carried out as previously described (3).
Immobilization of RNA and depletion of HeLa cell nuclear extracts.
Substrate RNAs for bead immobilization were synthesized by in vitro transcription using T7 RNA polymerase (Ambion, Austin, Tex.) and biotin-14 CTP (Life Technologies/Gibco/BRL) with a ratio of modified nucleotide to standard nucleotide of 1:2. RNAs were noncovalently linked to Dynabeads M-280–streptavidin (6.7 × 108 beads/ml) (Dynal, Lake Success, N.Y.) as suggested by the manufacturer. Two micrograms of RNA was incubated with 40 μl of Dynabeads in 1 M NaCl–10 mM Tris HCl (pH 7.5)–1 mM EDTA at room temperature for 15 min with gentle agitation. The Dynabeads with immobilized RNA were washed two times with Dignam's buffer D (15) and then incubated with 15 μl of HeLa cell nuclear extract for 15 min at 30°C with gentle agitation. Nuclear extract depleted of bound factors was separated from the Dynabeads-RNA complex by collecting the complex with a Dynal magnetic particle concentrator for 1 min.
In vitro splicing.
Splicing reactions were carried out essentially as previously described (3). In brief, approximately 8 fmol of 32P-labeled RNA was incubated for 2 h at 30°C in a solution containing 60% (vol/vol) nuclear extract in Dignam's buffer D, 20 mM creatine phosphate, 3 mM MgCl2, 0.8 mM ATP, and 2.6% (wt/vol) polyvinyl alcohol. The final volume of the splicing reaction mixture was 25 μl.
Protein analysis.
Proteins were separated by sodium dodecyl sulfate (SDS)–10% PAGE and visualized by Coomassie blue staining or transferred by electroblotting to nitrocellulose for immunoblot analysis. Monoclonal antibody 4B10 against hnRNP A1, which also detects hnRNP A1B, was provided by G. Dreyfuss (University of Pennsylvania) and used at a concentration of 1:3,000. Monoclonal antibody 2B2 against hnRNP B1 was provided by H. Kamma (University of Tsukuba, Ibaraki, Japan) and used at a concentration of 1:3,000. Rabbit polyclonal anti-A2 antiserum, which also cross-reacts with hnRNP A1, was provided by S. Riva (Istituto di Genetica Biochimica and Evoluzionistica, Pavia, Italy). It was used at a concentration of 1:1,000. Immunoblots were developed using an alkaline phosphatase staining kit (Vector Labs, Burlingame, Calif.).
Preparation of A/B hnRNPs and adding back to depleted extracts.
Recombinant hnRNPs A1, A1B, A2, and B1 were expressed in Escherichia coli and purified as described previously (32, 33). Glutathione S-transferase (GST)–UP1 and GST plasmids (obtained from X. Zhang, University of Arkansas) were expressed in E. coli and lysed by sonication, and the proteins were purified by binding to and elution with glutathione-Sepharose beads (Pharmacia). Proteins were added to depleted nuclear extracts, and splicing was carried out for 2 h at 30°C.
RESULTS
Mutagenesis of 5′ splice site D3 toward the consensus 5′ splice site sequence increases splicing at 3′ splice site A2.
We first investigated elements affecting the efficiency of splicing at HIV-1 splice site A2 in HeLa cell cultures transfected with an HIV-1 genomic deletion construct (pΔPSP; Fig. 1). It has been shown that the splicing of pΔPSP RNA transcripts does not differ significantly from that of wild-type HIV-1 (23). Also, it has been shown that HIV-1 RNAs are spliced identically in transfected HeLa cells and infected peripheral blood mononuclear cells (35). Splice site D3 (AG/GUAGGA) differs at positions +4 and +6 from the mammalian consensus 5′ splice site sequence.
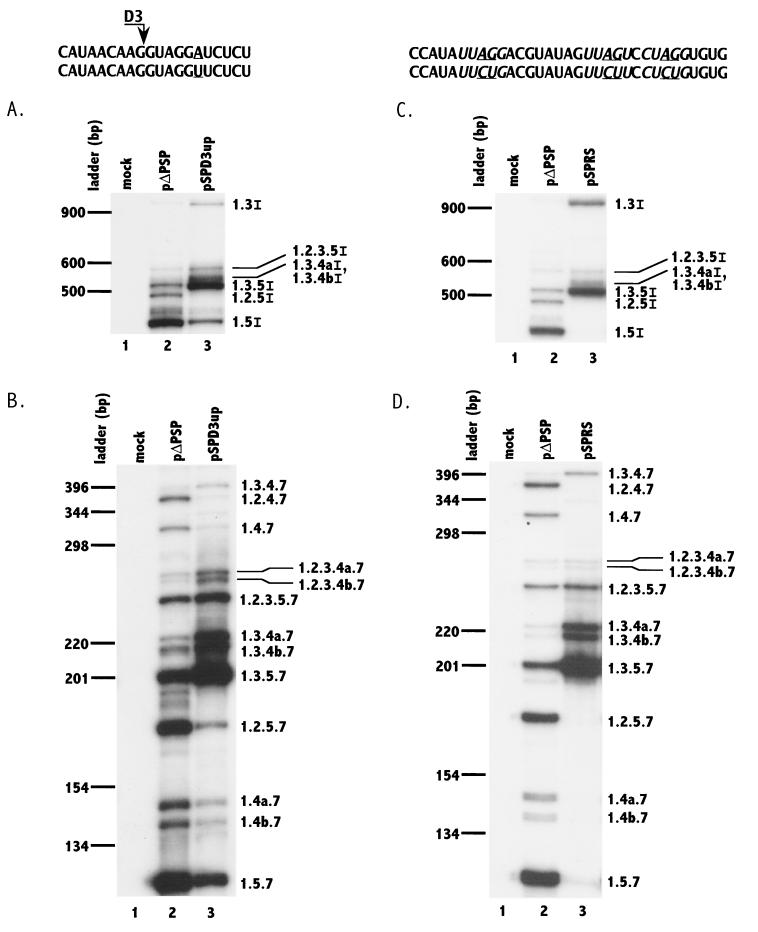
We first tested whether improving this 5′ splice site would increase the efficiency of splicing at splice site A2. Thus, position +6 of splice site D3 was changed from A to U to improve the match to the consensus 5′ splice site sequence. HeLa cells were transfected with wild-type (pΔPSP) and mutant (pSPD3up) constructs, and total RNA was isolated from the cells at 48 h after transfection. Using appropriate oligonucleotide primers, RNA was analyzed by RT-PCR for the relative levels of individual mRNAs of both intermediate (∼4.0 kb) and small (∼1.8 kb) sizes (Fig. 2). The nomenclature for the mRNAs denotes which exons are present in the particular mRNA species (Fig. 1B). The results for the ∼4.0-kb RNA size class indicated that there was a significant increase in the level of singly spliced Vpr mRNA (1.3I) when 5′ splice site D3 was improved (compare lanes 2 and 3 of Fig. 2A). In addition, there were dramatic increases in the levels of ∼4.0-kb Env mRNA species that include noncoding exon 3 (1.3.5I, 1.3.4aI/1.3.4bI, and 1.2.3.5I) and a relative decrease in the major singly spliced species, 1.5I, as well as species 1.2.5I, which includes only noncoding exon 2.
FIG. 2.
Mutagenesis of 5′ splice site D3 toward the consensus 5′ splice site sequence (panels A and B) and putative silencers (panels C and D) increases splicing at 3′ splice site A2 and inclusion of noncoding exon 2. RT-PCR analyses of ∼4.0-kb (A and C) and ∼1.8-kb (B and D) mRNAs from cells transfected with wild-type and mutant plasmids (pΔPSP and pSPD3up in panels A and B; pΔPSP and pSPRS in panels C and D) were performed. Mock-transfected cell mRNA was analyzed in parallel. Denaturing PAGE of 32P-labeled PCR products was performed as described in Materials and Methods. The mutated sequences in each case are shown below the wild-type NL4-3 sequence, and the changes are underlined. The PyUAG sequences are shown in italic type. RNA species are designated with exon numbers (and letters) as in Fig. 1. Note in panels A and C that RNA species 1.3.4aI and 1.3.4bI were not separated. The locations on the gel of DNA ladder bands are shown on the left.
This shift to mRNA species that include exon 3 was also observed with the mutant in the ∼1.8-kb mRNA size class (compare lanes 2 and 3 of Fig. 2B). Increases in the levels of 1.3.5.7, 1.3.4b.7, 1.3.4a.7, 1.2.3.5.7, 1.2.3.4b.7, 1.2.3.4a.7, and 1.3.4.7 mRNAs, all of which include exon 3, were observed. These increases were concomitant with relative decreases in the levels of 1.5.7, 1.4b.7, and 1.4a.7, which exclude both noncoding exons, and 1.2.5.7, which includes only exon 2. These results indicate that in wild-type HIV-1 RNA, the nonconsensus 5′ splice site D3 bordering the 3′ end of noncoding exon 3 affects the splicing efficiency of splice site A2 bordering the 5′ end of this exon. The presence of nonconsensus 5′ splice site D3 also results in the skipping of noncoding exon 3 in the majority of the HIV-1 mRNAs.
Mutations within a putative ESS element in exon 3 increase splicing at 3′ splice site A2.
Since it appeared from the above data that the optimization of 5′ splice site D3 resulted in only partial relief of exon 3 skipping, we investigated whether there were other cis elements repressing splicing at A2. Inspection of noncoding exon 3 revealed that there were three motifs with the general consensus sequence PyUAG. These motifs have sequence homology with previously described ESS elements in the two tat coding exons (42). Previous data indicated that mutations of the AG within this consensus sequence inhibited ESS activity in the tat coding exons (41, 42). To test for a similar effect in noncoding exon 3, we mutated all three of the AGs within the PyUAG motifs to CU. The wild-type and mutated plasmids were transfected into HeLa cells, and the singly and multiply spliced mRNA species were analyzed by RT-PCR as described above (Fig. 2C and D). For the ∼4-kb mRNA class, there was an increase in the level of Vpr mRNA (1.3I) in the mutant-transfected cells, and almost all of the ∼4.0-kb env mRNA species contained noncoding exon 3, indicating that exon inclusion was almost complete (Fig. 2C, lane 3). As shown in Fig. 2D, lane 3, similar results were obtained with the ∼1.8-kb spliced mRNA species; almost all of these mRNAs in mutant-transfected cells contained noncoding exon 3. These results strongly suggested that, in addition to the flanking nonconsensus 5′ splice site D3, exon 3 contains an ESS element whose repressive effect on splice site A2 is relieved by mutagenesis.
HIV-1 noncoding exon 3 contains an ESS element.
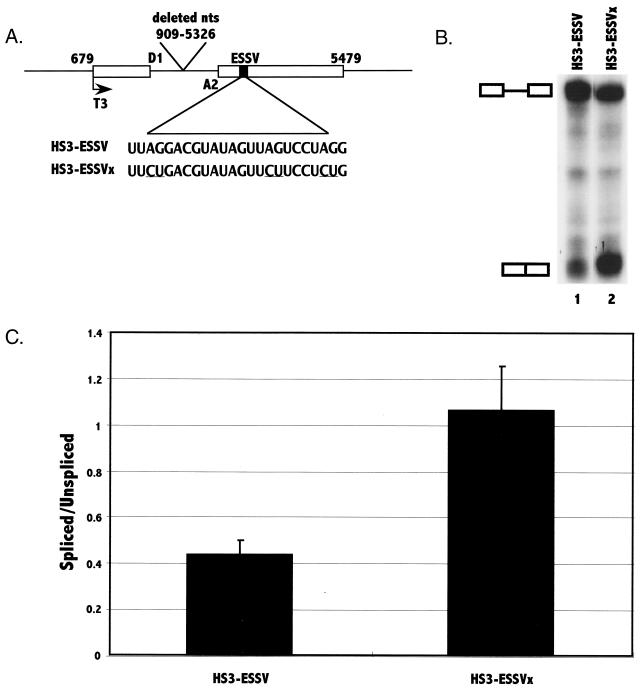
To further establish that the element in noncoding exon 3 was indeed an ESS, we performed additional experiments using in vitro splicing assays with HeLa cell nuclear extracts. We first created wild-type and mutant minigene constructs containing exon 3 (pHS3-ESSV and pHS3-ESSVx, respectively) (Fig 3A). These minigene templates were transcribed with phage T3 polymerase, and the RNA transcripts were used as substrates for in vitro splicing assays. Mutagenesis of all three of the exon 3 AG sequences, as in the pΔPSRS construct used in the in vivo transfection experiments described above, resulted in two- to threefold increases in the ratio of spliced to unspliced RNAs (Fig. 3B and C). These results were consistent with the hypothesis that exon 3 contains an ESS and that mutagenesis of the element results in increased splicing at 3′ splice site A2.
FIG. 3.
Analysis of the presence of an ESS in exon 3 by in vitro splicing assays. (A) The pHS3-ESSV template construct contains the indicated regions of pNL4-3. Shown are 5′ splice site D1 and 3′ splice site A2. The location of the T3 phage polymerase promoter is also shown. The location and the sequence of the putative ESS element are shown with the locations of the mutations underlined. RNA substrates were synthesized from the template as described in Materials and Methods. (B) In vitro splicing of 32P-labeled HIV-1 HS3-ESSV and HS3-ESSVx substrates was analyzed by denaturing PAGE. The positions of the RNA precursor and the spliced product are marked. (C) Ratios of radioactivity in the spliced product compared to that in the unspliced RNA precursor were determined for mutant and wild-type substrates. The results are based on six independent experiments. Standard deviations are shown by error bars.
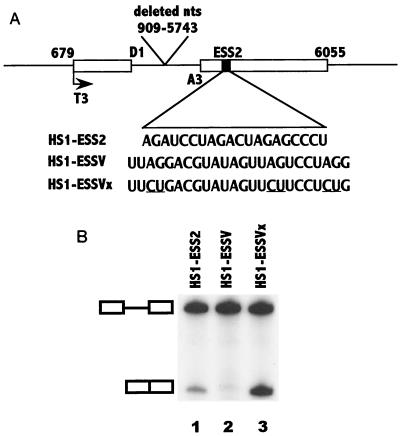
If the sequence in noncoding exon 3 is an ESS, it should act to inhibit splicing when placed into a heterologous context. To this end, we replaced ESS2 in the first HIV-1 tat coding exon with a 24-nt noncoding exon 3 sequence containing the three PyUAG motifs. The resulting plasmid minigene construct, shown in Fig. 4A, was used as a template for the synthesis of in vitro splicing substrates. The data shown in Fig. 4B, lane 2, indicated that splicing at 3′ splice site A3 was indeed inhibited. We found that splicing at splice site A3 of substrate HS1-ESSV with the exon 3 insertion was consistently lower than that of the wild-type HS1-ESS2 substrate containing the wild-type ESS2 element (compare lanes 1 and 2 of Fig. 4B). From these results, we concluded that the noncoding exon 3 sequence was an ESS, which we named ESSV, and that it had a stronger silencing effect on splicing at splice site A3 than did ESS2. This silencing was dependent on the sequence of ESSV, since mutagenesis of all three AG sequences abrogated the splicing inhibition (compare lanes 2 and 3 of Fig. 4B).
FIG. 4.
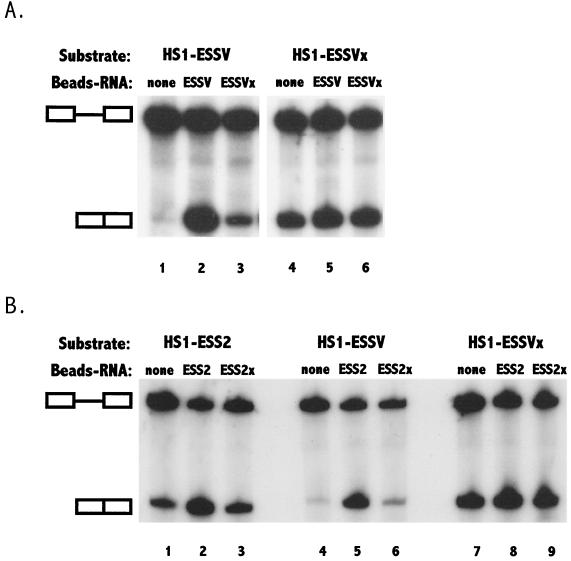
The ESSV sequence acts to inhibit splicing in a heterologous context. (A) Schematic representation of minigene template constructs containing 5′ splice site D1 and 3′ splice site A3 (3). These constructs contain either ESS2, ESSV, or ESSVx. (B) 32P-labeled RNA substrates HS1-ESS2, HS1-ESSV, and HS1-ESSVx were synthesized and RNA splicing was performed as described in Materials and Methods. Products of in vitro splicing of substrates were analyzed on denaturing polyacrylamide gels. The positions of the precursor and the spliced product are marked.
Evidence that ESSV and ESS2 bind to common cellular factors.
If ESSV is an authentic splicing silencer, then it would be expected to bind to cellular factors, as do other ESS elements. We have previously shown that preincubation with excess competitor RNA containing ESS2 increases splicing at the tat 3′ splice site (A3) of RNA splicing substrates containing this element (4). In data not shown, we found that preincubation with RNA containing wild-type ESSV caused relief of splicing inhibition of substrate HS1-ESSV, whereas preincubation with the same amount of mutant ESSV-containing RNA did not significantly affect splicing. Interestingly, preincubation with competitor RNA containing ESS2 also resulted in increased splicing of HS1-ESSV RNA. These results supported the hypothesis that ESSV binds a cellular factor(s) and that ESS2 and ESSV share this factor(s), necessary for this inhibition.
To further test this hypothesis, we performed experiments in which we depleted nuclear extracts of the putative factors by binding to RNAs containing splicing silencers. Competitor ESS RNAs were biotinylated and coupled to streptavidin-coated paramagnetic beads. HeLa cell nuclear extracts were incubated with the RNA-beads, the beads containing bound factors were removed, and the treated extracts were used for splicing reactions. We first compared the splicing of RNA substrates containing ESSV (HS1-ESSV) in nuclear extracts which had been preincubated with wild-type and mutant ESSV RNAs coupled to beads. As shown in Fig. 4B, there was little splicing of HS1-ESSV in untreated extracts (Fig. 5A, lane 1). In extracts preincubated with wild-type ESSV RNA coupled to beads, there was a striking increase in the amount of splicing of HS1-ESSV, indicating that a factor or factors inhibiting splicing at 3′ splice site A3 had been removed (Fig. 5A, lane 2). In contrast, there was only a small increase in the amount of splicing when extracts were preincubated with mutant RNA coupled to beads (Fig. 5A, lane 3). As expected, the mutagenesis of ESSV (substrate HS1-ESSVx) relieved the inhibition of splicing in untreated extracts (Fig. 5A, lane 4), and splicing of this substrate was similar in extracts preincubated with either wild-type or mutant ESSV RNA coupled to beads (Fig. 5A, lanes 5 and 6).
FIG. 5.
Depletion of HeLa cell nuclear extracts with ESSV and ESS2 RNAs removes a common inhibitory cellular factor or factors. In vitro splicing of HS1-ESSV, HS1-ESSVx, or HS1-ESS2 substrates was carried out by use of nondepleted nuclear extracts and nuclear extracts depleted with ESSV- or ESSVx-biotinylated RNAs (A) or ESS2- or ESS2x-biotinylated RNAs (B) immobilized on paramagnetic beads (Beads-RNA) as described in Materials and Methods. The positions of the precursor and the spliced product are marked.
To confirm that ESS2 and ESSV bind to common factors, we treated HeLa cell nuclear extracts with beads coupled to ESS2 and mutant ESS2 RNAs. As expected, there was relief of splicing inhibition of substrates containing ESS2 (Fig. 5B, compare lanes 1 and 2). The results indicated that there was also relief of splicing inhibition of substrates containing ESSV (compare lanes 4 and 5 of Fig. 5B). In extracts that had been preincubated with beads coupled to mutated ESS2 RNA, there was only a small increase in the splicing of substrates containing ESS2 or ESSV (Fig. 5B, lane 3 or 6, respectively). In contrast, the splicing of substrates containing mutated ESSV (HS1- ESSVx) was similar in both treated and untreated extracts (Fig. 5B, lanes 7 to 9). These results reinforced the hypothesis that the splicing silencers in tat exon 2 and noncoding exon 3 bind to common factors.
Specific depletion of cellular hnRNP A/B proteins by ESSV RNA-beads.
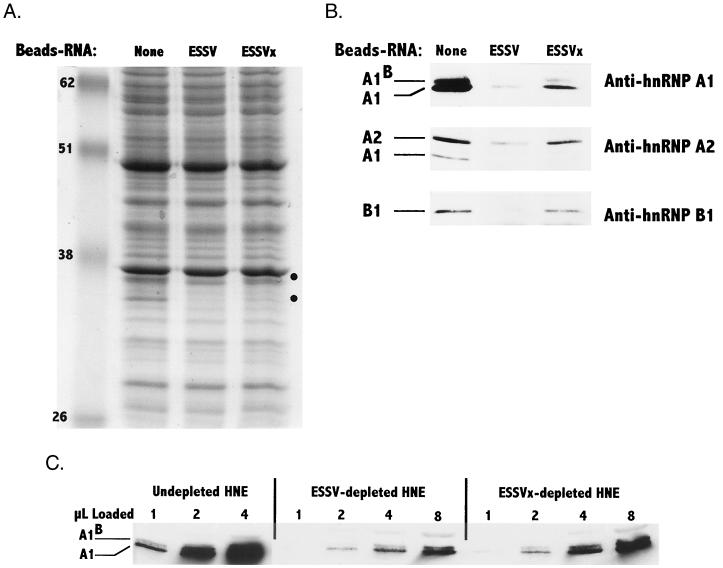
Previous studies have indicated that members of the hnRNP A/B protein family mediate splicing inhibition of HIV-1 substrates containing ESS2 (11). We examined proteins in untreated extracts and in extracts treated with wild-type or mutated ESSV RNA-beads to test for selective depletion of proteins in the size range of the hnRNP A/B proteins. As shown in Fig. 6A, several differences in the 30- to 40-kDa molecular mass range between the ESSV and ESSVx lanes were seen in the Coomassie blue-stained SDS-PAGE patterns. Extracts treated with either wild-type or mutant RNA-beads were depleted of these proteins, but the effect was selectively greater when the wild-type RNA was used.
FIG. 6.
Selective depletion of inhibitory proteins in HeLa cell nuclear extracts with ESSV RNA-beads. (A) Samples (50 μg) from nondepleted nuclear extracts and nuclear extracts depleted with either ESSV or ESSVx paramagnetic bead-immobilized RNAs were separated by SDS–10% PAGE and stained with Coomassie blue. Circles on the right indicate differences between the ESSV RNA-beads and the ESSVx RNA-beads. Apparent molecular weights (in thousands) are shown on the left. (B) Western blot analyses of proteins in depleted and nondepleted extracts were carried out with the indicated anti-hnRNP antibodies as described in Materials and Methods. For the analyses with anti-hnRNP A1 and anti-hnRNP B1 antibodies, 20 μg of protein from the depleted or nondepleted extracts was analyzed. For the analysis with anti-hnRNP A2 antibody, 50 μg of protein from the depleted or nondepleted extracts was analyzed. The anti-hnRNP A2 antiserum also detects hnRNP A1. (C) Aliquots (1 to 8 μl) from HeLa cell nuclear extracts (HNE) nondepleted or depleted with either wild-type (ESSV) or mutant (ESSVx) RNA bound to beads were electrophoresed, and Western blot analysis was carried out using anti-hnRNP A1 antibody.
To identify these proteins, we performed Western blotting using three antibodies directed against hnRNPs A1 and A1B, hnRNP A2, and hnRNP B1 (Fig. 6B). In extracts treated with either wild-type or mutant RNA-beads, there were reductions in the amounts of hnRNP A/B proteins relative to those in untreated extracts. However, in each case, there were further reductions in the amounts of hnRNPs detected in extracts treated with wild-type ESSV RNA-beads compared with the mutant ESSV RNA-beads. These results imply that both wild-type and mutant RNAs bind the hnRNPs but that the affinity for the wild-type ESSV is greater than that for the mutant ESSV. The binding data correlate with the data shown in Fig. 5A, which showed small increases in the splicing of ESSV-containing substrates in extracts treated with mutant ESSV RNA-beads but significantly greater increases in extracts treated with wild-type ESSV RNA-beads.
In order to estimate the concentrations of hnRNP A1 remaining in the extracts treated with wild-type and mutant ESSV RNA-beads, aliquots from nondepleted extracts and proteins from extracts depleted with either ESSV or ESSVx RNA-beads were separated by SDS-PAGE. The amounts of hnRNP A1 were then estimated based on a comparison of the intensities of Western blot staining using anti-A1 antibody and the intensities using known amounts of purified hnRNP A1 (Fig. 6C). The results indicated the following approximate hnRNP A1 concentrations in the nuclear extracts: nondepleted, 7 μM; ESSV RNA-bead depleted, 1 μM; and ESSVx RNA-bead depleted, 2 μM. These results reinforced the conclusion that hnRNP A/B proteins bind to both wild-type and mutant ESSV RNAs. However, there appeared to be an approximate twofold increase in binding to the wild-type sequence, resulting in a preferential depletion of A/B hnRNPs from the nuclear extracts treated with the wild-type RNA-beads.
Addition of A/B hnRNPs to depleted extracts restores specific splicing inhibition.
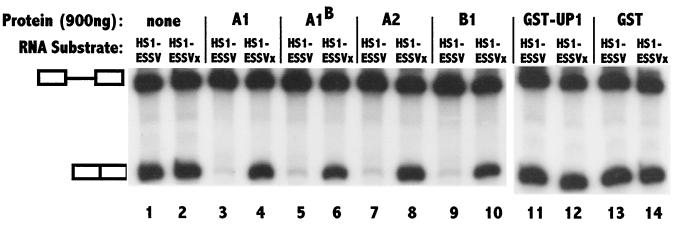
To confirm the hypothesis that splicing inhibition by ESSV is mediated by preferential binding of hnRNP A/B proteins, we added back exogenous hnRNP A/B proteins to extracts depleted with wild-type ESSV RNA and tested these extracts for the restoration of splicing inhibition (Fig. 7). The amounts of hnRNPs added back to the wild-type ESSV RNA-depleted nuclear extracts restored the final concentration in the splicing reactions to approximately the level of the protein in extracts treated with mutant ESSV RNA-beads. As expected, substrates containing either wild-type or mutant ESSV silencers were spliced similarly in the depleted extracts (compare lanes 1 and 2 of Fig. 7). When hnRNP A/B proteins (A1, A1B, A2, and B1) were individually added back to the depleted extracts, repression of splicing was restored to the substrates with the wild-type but not the mutant silencer. The addition of control proteins GST-UP1 (hnRNP A1 containing its two RNA recognition motifs but lacking the C-terminal glycine-rich domain) and GST alone did not affect splicing. We also added back untagged UP1 and showed that this protein also has no effect on the splicing of substrates with the wild-type or the mutant silencers (data not shown). These results indicated that hnRNP A/B proteins are necessary and sufficient to restore the specific splicing inhibition by ESSV. These data also indicate that this inhibition occurs at physiological protein concentrations.
FIG. 7.
Reconstitution of splicing inhibition by addition of hnRNP A/B proteins to depleted extracts. HS1-ESSV and HS1-ESSVx RNA substrates were spliced in ESSV RNA-bead-depleted HeLa cell nuclear extracts in the presence of 900 ng of the indicated hnRNPs. Splicing was also carried out after the addition of the same amounts of control proteins: UP1-GST (two RNA recognition motifs of hnRNP A1 fused to GST) in lanes 11 and 12 and GST alone in lanes 13 and 14. The positions of the precursor and the spliced product are marked.
DISCUSSION
Two elements act in concert to regulate splicing at HIV-1 3′ splice site A2, which defines the 5′ border of noncoding exon 3. The first element is 5′ splice site D3, which defines the 3′ boundary of the exon and is a weak splice site. The second element consists of silencer sequences within the exon. When we mutated splice site D3 to change it to a consensus 5′ splice site, there were increases in both inclusion of noncoding exon 3 in multiply spliced mRNAs and usage of 3′ splice site A2 in singly spliced mRNAs. These results can be explained by the exon bridging hypothesis, which proposes that U1 snRNP binding to the downstream 5′ splice site acts to increase splicing efficiency at the upstream flanking 3′ splice site (9, 22, 36). According to this hypothesis, changing splice site D3 to a consensus 5′ splice site would increase the affinity of U1 snRNP for this splice site and thus increase the efficiency of splicing at 3′ splice site A2. This prediction is consistent with what our data showed. The weak effect of nonconsensus 5′ splice site D3 on splicing at 3′ splice site A2 may contribute to the relatively low singly spliced Vpr mRNA levels and to skipping of exon 3. Alternatively, skipping of exon 3 may result from cis competition, in which consensus 5′ splice site D1 is normally favored over nonconsensus 5′ splice site D3 for the alternative 3′ splice sites A3, A4a, A4b, A4c, and A5 (Fig. 1A). Consistent with the importance of this nonconsensus 5′ splice site, the sequence of splice site D3 is conserved in all sequenced strains of HIV-1 (27). A similar mechanism may contribute to the low efficiency of splicing at 3′ splice site A1, which flanks noncoding exon 2. Consistent with this hypothesis, we have found that improvement of nonconsensus 5′ splice site D2 immediately downstream of noncoding exon 2 to a consensus splice site (AG/GUGAAG to AG/GUGAGU) results in increased inclusion of this exon (P. Bilodeau and C. M. Stoltzfus, unpublished data).
Splicing efficiency at 3′ splice site A2 is also regulated by ESS elements. We showed that mutagenesis of the three PyUAG motifs within noncoding exon 3 abrogated the silencing activity of ESSV. Using in vitro splicing assays, we found that mutagenesis of each motif individually resulted in only small increases in splicing that were not significantly different from the results seen with the wild type (Bilodeau and Stoltzfus, unpublished). Significant differences were seen only when all three AGs within the 24-nt ESSV element were mutated. In this regard, it is of interest that the three PyUAG sequences in noncoding exon 3 are conserved in most sequenced HIV-1 strains belonging to the major HIV group (group M) (27).
Other HIV-1 ESS elements also contain the consensus sequence UAG or PyUAG. We have previously shown that HIV-1 tat exon 2 contains ESS2, whose core sequence is CUAGACUAGA, and tat exon 3 contains ESS3, which contains the sequence UUAG. In each case, mutagenesis of the AG dinucleotides results in abrogation of the silencing effect (41, 42). HIV-1 cryptic exon 6D inclusion has been shown to be activated by a U-to-C mutation at the underlined base in the sequence CAAUAGUAGUAG (46). This latter sequence may also be an ESS element.
The evidence presented here also indicates that the ESS elements downstream of both the Vpr and the Tat splice sites compete for the same cellular factors. We have shown above that these shared factors are members of the A/B hnRNP family. Thus, our data agree with previous reports implicating A/B hnRNP family members as mediators of splicing inhibition by the ESS in tat exon 2 (ESS2) (11, 14). hnRNP A1 has also been shown to bind to UAGG in the K-SAM exon of fibroblast growth factor receptor 2 and to mediate splicing silencing of this alternative exon (14). Recent data have indicated that the binding of A/B hnRNPs to a splicing silencer within alternative exon 16 of protein 4.1R pre-mRNA is necessary for splicing repression of the 3′ splice site bordering exon 16 (J. Conboy, personal communication).
These previous results and the results reported here suggest that members of the hnRNP A/B protein family binding to ESS elements coordinately repress HIV-1 splicing. Thus, HIV-1 exploits these proteins as a means to limit the amount of splicing at both Tat 3′ splice sites and Vpr 3′ splice sites. hnRNP A/B proteins are ubiquitous, and this fact may allow the repression of HIV-1 splicing in a variety of cell types. Our data and those of Caputi et al. (11) indicate that, as determined by in vitro splicing assays, all members of the hnRNP A/B protein family, i.e., hnRNPs A1, A1B, A2, and B1, are able to mediate HIV-1 ESS splicing inhibition. It is of interest that hnRNP A1B, which is an alternatively spliced isoform of hnRNP A1, has equivalent silencer activity in the alternative splicing of HIV-1 pre-mRNAs (11; this study), whereas it has very limited activity for 5′ splice site switching (33). In preliminary experiments, we have found that the splicing of HIV-1 Tat mRNA is repressed in the mouse erythroleukemia cell line CB3 despite the lack of detectable hnRNP A1 and A1B expression in these cells (7, 47). Mutagenesis of ESS2 relieved this repression (J. Domsic and C. M. Stoltzfus, unpublished data). These data suggest that other members of the hnRNP A/B protein family (i.e., hnRNPs A2 and B1) are able to substitute for hnRNPs A1 and A1B in vivo as well as in vitro.
Recent data have indicated that hnRNP A1 binds to splicing silencer elements within human CD44 exon v6 and downregulates the splicing of this exon (26, 31). Interestingly, this repression was relieved by the expression of either oncogenic Ras or oncogenic proteins that are in the Ras effector pathway (31). Thus, in this case, oncogenic signaling appears to interfere with hnRNP A1-mediated silencing. Inclusion of variant exons in CD44 mRNAs is also increased upon activation of normal lymphocytic and dendritic cells (5, 29, 45). It is possible that interference with hnRNP A/B protein silencing upon cell activation is a potential mechanism for increasing the efficiency of HIV-1 splicing, resulting in increased Tat and Vpr mRNA levels in some cell types and at different times after infection. However, CD44 exon v5 does not contain consensus PyUAG sequences, and the splicing silencing activity appears to be spread throughout the 118-nt exon rather than localized, as in the HIV-1 genome (26, 31). Thus, the mechanism by which hnRNP A1 inhibits CD44 splicing may differ from that used in HIV-1 splicing.
ESS elements and members of the hnRNP A/B protein family also play important roles in the replication of other RNA viruses. Borna disease virus is a nonsegmented negative-strand RNA virus which replicates in the nucleus and whose RNA undergoes splicing. It has been shown that the utilization of one of the Borna disease virus 3′ splice sites (SA3) is regulated by an ESS element downstream of this splice site. This ESS contains two PyUAG motifs, and deletion of these motifs abrogates the silencer activity (44). Thus, this ESS is also likely to bind to members of the hnRNP A/B protein family. hnRNP A1 has been shown to specifically bind to UUAG sequences within the RNA of the coronavirus mouse hepatitis virus, the replication of which occurs in the cytoplasm. This binding is required for the regulation of viral RNA transcription and replication (28, 40). The various roles played by hnRNP A/B proteins in cells and during the replication of viruses suggest that these proteins are components of several different complexes that act in a regulatory fashion at different steps of viral and cellular RNA processing as well as viral RNA synthesis.
ACKNOWLEDGMENTS
We thank G. Dreyfuss, H. Kamma, and S. Riva for generously providing the hnRNP antibodies used in this study. We also thank X. Zhang for GST-UP1 and GST plasmids. For review of the manuscript, we thank S. Perlman and W. Maury. We thank Sandrine Jacquenet and Christiane Branlant for helpful discussions and sharing unpublished data early in this study.
This research was supported by PHS grant AI36073 from the National Institute of Allergy and Infectious Diseases to C.M.S. A.R.K. and A.M. were supported by PHS grant CA13106 from the National Cancer Institute. A.M. is a member of the Sylvester Comprehensive Cancer Center and was also supported by funds awarded by the Lucille P. Markey Trust. HeLa cells were obtained from the Cell Culture Center, which is sponsored by the National Center for Research Resources of the NIH.
REFERENCES
- 1.Adachi A, Gendelman H E, Koenig S, Folks T, Willey R, Rabson A, Martin M A. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aiyar A, Leis J. Modification of the megaprimer method of PCR mutagenesis: improved amplification of the final product. BioTechniques. 1993;14:366–369. [PubMed] [Google Scholar]
- 3.Amendt B A, Hesslein D, Chang L-J, Stoltzfus C M. Presence of negative and positive cis-acting RNA splicing elements within and flanking the first tat coding exon of the human immunodeficiency virus type 1. Mol Cell Biol. 1994;14:3960–3970. doi: 10.1128/mcb.14.6.3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amendt B A, Si Z-H, Stoltzfus C M. Presence of exon splicing silencers within HIV-1 tat exon 2 and tat/rev exon 3: evidence for inhibition mediated by cellular factors. Mol Cell Biol. 1995;15:4606–4615. doi: 10.1128/mcb.15.8.4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arch R, Wirth K, Hoffman M, Ponta H, Matzku S, Herrlich P, Zoller M. Participation in normal immune response of a splice variant of CD44 that encodes a metastasis-inducing domain. Science. 1992;257:682–685. doi: 10.1126/science.1496383. [DOI] [PubMed] [Google Scholar]
- 6.Arrigo S, Weitsman S, Zack J A, Chen I S. Characterization and expression of novel singly spliced RNA species of human immunodeficiency virus type 1. J Virol. 1990;64:4585–4588. doi: 10.1128/jvi.64.9.4585-4588.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ben-David Y, Bani M R, Chabot B, De Koven A, Bernstein A. Retroviral insertions downstream of the heterogeneous nuclear ribonucleoprotein A1 gene in erythroleukemia cells: evidence that A1 is not essential for cell growth. Mol Cell Biol. 1992;12:4449–4455. doi: 10.1128/mcb.12.10.4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benko D M, Schwartz S, Pavlakis G N, Felber B K. A novel human immunodeficiency virus type 1 protein, tev, shares sequences with tat, env, and rev proteins. J Virol. 1990;64:2505–2518. doi: 10.1128/jvi.64.6.2505-2518.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berget S M. Exon recognition in vertebrate splicing. J Biol Chem. 1995;270:2411–2414. doi: 10.1074/jbc.270.6.2411. [DOI] [PubMed] [Google Scholar]
- 10.Bilodeau P S, Domsic J K, Stoltzfus C M. Splicing regulatory elements within tat exon 2 of human immunodeficiency virus type 1 (HIV-1) are characteristic of group M but not group O HIV-1 strains. J Virol. 1999;73:9764–9772. doi: 10.1128/jvi.73.12.9764-9772.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caputi M, Mayeda A, Krainer A R, Zahler A M. hnRNP A/B proteins are required for inhibition of HIV-1 pre-mRNA splicing. EMBO J. 1999;18:4060–4067. doi: 10.1093/emboj/18.14.4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cullen B R. Nuclear export pathways. Mol Cell Biol. 2000;20:4181–4187. doi: 10.1128/mcb.20.12.4181-4187.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Damier L, Domenjoud L, Branlant C. The D1–A2 and D2–A2 pairs of splice sites from human immunodeficiency virus type 1 are highly efficient in vitro, in spite of an unusual branch site. Biochem Biophys Res Commun. 1997;237:182–187. doi: 10.1006/bbrc.1997.7091. [DOI] [PubMed] [Google Scholar]
- 14.delGatto-Konczak F, Olive M, Gesnel M-C, Breathnach R. hnRNP A1 recruited to an exon in vivo can function as an exon splicing silencer. Mol Cell Biol. 1999;19:251–260. doi: 10.1128/mcb.19.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Emerman M, Vazeus R, Peden K. The rev gene product of the human immunodeficiency virus affects envelope-specific RNA localization. Cell. 1989;57:1155–1165. doi: 10.1016/0092-8674(89)90053-6. [DOI] [PubMed] [Google Scholar]
- 17.Felber B K, Hadzopoulou-Cladaras M, Cladaras C, Copeland T, Pavlakis G N. Rev protein of human immunodeficiency virus type 1 affects the stability and transport of the viral mRNA. Proc Natl Acad Sci USA. 1989;86:1495–1499. doi: 10.1073/pnas.86.5.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Furtado M R, Balachandran R, Gupta P, Wolinsky S M. Analysis of alternatively spliced human immunodeficiency virus type 1 mRNA species, one of which encodes a novel TAT-ENV fusion protein. Virology. 1991;185:258–270. doi: 10.1016/0042-6822(91)90773-5. [DOI] [PubMed] [Google Scholar]
- 19.Guatelli J C, Gingeras T R, Richman D D. Alternative splice acceptor utilization during human immunodeficiency virus type 1 infection of cultured cells. J Virol. 1990;64:4093–4098. doi: 10.1128/jvi.64.9.4093-4098.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hadzopoulos-Cladaras M, Felber B K, Cladaras C, Athanassopoulos A, Tse A, Pavlakis G. The rev (trs/art) protein of human immunodeficiency virus type 1 affects viral mRNA and protein expression via a cis-acting sequence in the env region. J Virol. 1989;63:1265–1274. doi: 10.1128/jvi.63.3.1265-1274.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hammarskjold M-L, Heimer J, Hammarskjold B, Sangwan I, Albert L, Rekosh D. Regulation of human immunodeficiency virus env expression by the rev gene product. J Virol. 1989;63:1959–1966. doi: 10.1128/jvi.63.5.1959-1966.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoffman B E, Grabowski P J. U1 snRNP targets an essential splicing factor, U2AF65, to the 3′ splice site by a network of interactions spanning the exon. Genes Dev. 1992;6:2554–2568. doi: 10.1101/gad.6.12b.2554. [DOI] [PubMed] [Google Scholar]
- 23.Jacquenet S, Ropers D, Bilodeau P S, Damier L, Mougin A, Stoltzfus C M, Branlant C. Conserved stem-loop structures in the HIV-1 RNA region containing the A3 3′ splice site and its cis-regulatory element: possible involvement in RNA splicing. Nucleic Acids Res. 2001;29:464–478. doi: 10.1093/nar/29.2.464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim S, Byrn R, Groopman J, Baltimore D. Temporal aspects of DNA and RNA synthesis during human immunodeficiency virus infection: evidence for differential gene expression. J Virol. 1989;63:3708–3713. doi: 10.1128/jvi.63.9.3708-3713.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klotman M E, Kim S, Buchbinder A, DeRossi A, Baltimore D, Wong-Staal F. Kinetics of expression of multiply spliced RNA in early human immunodeficiency virus type 1 infection of lymphocytes and monocytes. Proc Natl Acad Sci USA. 1991;88:5011–5015. doi: 10.1073/pnas.88.11.5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Konig H, Ponta H, Herrlich P. Coupling of signal transduction to alternative pre-mRNA splicing by a composite splice regulator. EMBO J. 1998;17:2904–2913. doi: 10.1093/emboj/17.10.2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuiken C, Foley B, Hahn B, Marx P, McCutchan F, Mellors J W, Mullins J, Wolinsky S, Korber B. Human retroviruses and AIDS 1999. Los Alamos, N.Mex: Los Alamos National Laboratory; 1999. [Google Scholar]
- 28.Li H P, Zhang X, Duncan R, Comai L, Lai M M C. Heterogeneous nuclear ribonucleoprotein A1 binds to the transcription-regulatory region of mouse hepatitis virus RNA. Proc Natl Acad Sci USA. 1997;94:9544–9549. doi: 10.1073/pnas.94.18.9544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mackay C R, Terpe H-J, Stauder R, Martson W L, Stark H, Gunthert U. Expression and modulation of CD44 variant isoforms in humans. J Cell Biol. 1994;124:71–82. doi: 10.1083/jcb.124.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malim M H, Hauber J, Le S-Y, Maizel J V, Cullen B R. The HIV-1 rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature. 1989;338:254–257. doi: 10.1038/338254a0. [DOI] [PubMed] [Google Scholar]
- 31.Matter N, Marx M, Weg-Remers S, Ponta H, Herrlich P, Konig H. Heterogeneous ribonucleoprotein A1 is part of an exon-specific splice-silencing complex controlled by oncogenic signaling pathways. J Biol Chem. 2000;275:35353–35360. doi: 10.1074/jbc.M004692200. [DOI] [PubMed] [Google Scholar]
- 32.Mayeda A, Krainer A R. Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell. 1992;68:365–375. doi: 10.1016/0092-8674(92)90477-t. [DOI] [PubMed] [Google Scholar]
- 33.Mayeda A, Munroe S H, Caceres J F, Krainer A R. Function of conserved domains of hnRNP A1 and other hnRNP A/B proteins. EMBO J. 1994;13:5483–5495. doi: 10.1002/j.1460-2075.1994.tb06883.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neumann M, Harrison J, Saltarelli M, Hadziyannis E, Erfle V, Felber B K, Pavlakis G N. Splicing variability in HIV type 1 revealed by quantitative RNA polymerase chain reaction. AIDS Res Hum Retrovir. 1994;10:1531–1542. doi: 10.1089/aid.1994.10.1531. [DOI] [PubMed] [Google Scholar]
- 35.Purcell D F J, Martin M A. Alternative splicing of human immunodeficiency virus type 1 mRNA modulates viral protein expression, replication, and infectivity. J Virol. 1993;67:6365–6378. doi: 10.1128/jvi.67.11.6365-6378.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robberson B L, Cote G J, Berget S M. Exon definition may facilitate splice site selection in RNAs with multiple exons. Mol Cell Biol. 1990;10:84–94. doi: 10.1128/mcb.10.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robert-Guroff M, Popovic M, Gartner S, Markham P, Gallo R C, Reitz M S. Structure and expression of tat-, rev-, and nef-specific transcripts of human immunodeficiency virus type 1 in infected lymphocytes and macrophages. J Virol. 1990;64:3391–3398. doi: 10.1128/jvi.64.7.3391-3398.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salfeld J, Gottlinger H, Sia R, Park R, Sodroski J, Haseltine W. A tripartite HIV-1 tat-env-rev fusion protein. EMBO J. 1990;9:965–970. doi: 10.1002/j.1460-2075.1990.tb08195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schwartz S, Felber B K, Benko D M, Fenyo E-M, Pavlakis G N. Cloning and functional analysis of multiply spliced mRNA species of human immunodeficiency virus type 1. J Virol. 1990;64:2519–2529. doi: 10.1128/jvi.64.6.2519-2529.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shi S T, Huang P, Li H-P, Lai M C. Heterogeneous nuclear ribonucleoprotein A1 regulates RNA synthesis of a cytoplasmic virus. EMBO J. 2000;19:4701–4711. doi: 10.1093/emboj/19.17.4701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Si Z-H, Amendt B A, Stoltzfus C M. Splicing efficiency of human immunodeficiency virus type 1 Tat RNA is determined by both a suboptimal 3′ splice site and a 10 nucleotide exon splicing silencer element located within tat exon 2. Nucleic Acids Res. 1997;25:861–867. doi: 10.1093/nar/25.4.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Si Z-H, Rauch D, Stoltzfus C M. The exon splicing silencer in human immunodeficiency virus type 1 Tat exon 3 is bipartite and acts early in spliceosome assembly. Mol Cell Biol. 1998;18:5404–5413. doi: 10.1128/mcb.18.9.5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Staffa A, Cochrane A. Identification of positive and negative splicing regulatory elements within the terminal tat/rev exon of human immunodeficiency virus type 1. Mol Cell Biol. 1995;15:4597–4605. doi: 10.1128/mcb.15.8.4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tomonaga K, Kobayashi T, Lee B-J, Watanabe M, Kamitani W, Ikuta K. Identification of alternative splicing and negative splicing activity of a nonsegmented negative-strand RNA virus, Borna disease virus. Proc Natl Acad Sci USA. 2000;97:12788–12793. doi: 10.1073/pnas.97.23.12788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weiss J M, Sleeman J, Renkl A C, Dittmar H, Termeer C C, Taxis S, Howells N, Hofmann M, Kohler G, Schopf E, Ponta H, Herrlich P, Simon J C. An essential role for CD44 variant isoforms in epidermal Langerhans cell and blood dendritic cell function. J Cell Biol. 1997;137:1137–1147. doi: 10.1083/jcb.137.5.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wentz M P, Moore B E, Cloyd M W, Berget S M, Donehower L A. A naturally arising mutation of a potential silencer of exon splicing in human immunodeficiency virus type 1 induces dominant aberrant splicing and arrests virus production. J Virol. 1997;71:8542–8551. doi: 10.1128/jvi.71.11.8542-8551.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang X, Bani M-R, Lu S-J, Rowan S, Ben-David Y, Chabot B. The A1 and A1B proteins of heterogeneous nuclear ribonucleoproteins modulate 5′ splice site selection in vivo. Proc Natl Acad Sci USA. 1994;91:6924–6928. doi: 10.1073/pnas.91.15.6924. [DOI] [PMC free article] [PubMed] [Google Scholar]