Abstract
Polycyclic aromatic hydrocarbons (PAHs) are a class of organic compounds frequently detected in the environment with widely varying toxicities. Many PAHs activate the aryl hydrocarbon receptor (AHR), inducing the expression of a battery of genes, including xenobiotic metabolizing enzymes like cytochrome P450s (CYPs); however, not all PAHs act via this mechanism. We screened several parent and substituted PAHs in in vitro AHR activation assays to classify their unique activity. Retene (1-methyl-7-isopropylphenanthrene) displays Ahr2-dependent teratogenicity in zebrafish, but did not activate human AHR or zebrafish Ahr2, suggesting a retene metabolite activates Ahr2 in zebrafish to induce developmental toxicity. To investigate the role of metabolism in retene toxicity, studies were performed to determine the functional role of cyp1a, cyp1b1, and the microbiome in retene toxicity, identify the zebrafish window of susceptibility, and measure retene uptake, loss, and metabolite formation in vivo. Cyp1a-null fish were generated using CRISPR-Cas9. Cyp1a-null fish showed increased sensitivity to retene toxicity, whereas Cyp1b1-null fish were less susceptible, and microbiome elimination had no significant effect. Zebrafish required exposure to retene between 24 and 48 hours post fertilization (hpf) to exhibit toxicity. After static exposure, retene concentrations in zebrafish embryos increased until 24 hpf, peaked between 24 and 36 hpf, and decreased rapidly thereafter. We detected retene metabolites at 36 and 48 hpf, indicating metabolic onset preceding toxicity. This study highlights the value of combining molecular and systems biology approaches with mechanistic and predictive toxicology to interrogate the role of biotransformation in AHR-dependent toxicity.
Keywords: polycyclic aromatic hydrocarbons, aryl hydrocarbon receptor, biotransformation, cytochrome p450, developmental toxicity, zebrafish
PAHs and AHR pathway
Polycyclic aromatic hydrocarbons (PAHs) are a class of organic compounds characterized by multiple fused benzene rings. Exposure to PAHs is a public health concern due to their many sources of production, frequency of detection in the environment, and known and suspected toxicities. The toxicity of different PAHs vary widely with forms of toxicity including carcinogenesis (Siddens et al. 2012; Boada et al. 2015; Stading et al. 2021; Vogeley et al. 2022), immunotoxicity (Yu et al. 2022), cardiac and vascular toxicities (Marris et al. 2020; Mallah et al. 2021; Yu et al. 2021; Ho et al. 2022; Ma et al. 2022), neurotoxicity (Peiffer et al. 2016; Knecht et al. 2017; Das et al. 2019; Tian et al. 2020; Olasehinde and Olaniran 2022), and teratogenesis (Barbieri et al. 1986; Incardona et al. 2004; Geier et al. 2018a). The toxicity of some PAHs is mediated by aryl hydrocarbon receptor (AHR), a ligand-activated transcription factor which regulates the expression of a battery of genes including xenobiotic metabolizing enzymes such as the cytochrome P450s (CYPs) (van Delft et al. 2010; Nebert 2017; Shi et al. 2017; Shankar et al. 2019); however, it is clear that this mechanism is not universal (Incardona et al. 2005; Knecht et al. 2013; Goodale et al. 2015; Brette et al. 2017; Geier et al. 2018a; Shankar et al. 2019; Ainerua et al. 2020).
Cytochrome P450s and PAH metabolism
PAH toxicity mechanisms can diverge through xenobiotic metabolism. CYPs are monooxygenase enzymes involved in phase I metabolism of xenobiotics (Shimada 2006). In humans, there are 57 genes encoding CYPs, 32 of which are conserved in zebrafish (Nelson 2009; Goldstone et al. 2010). CYP families 1 to 3 are most known for biotransformation, together metabolizing an estimated 70% to 80% of xenobiotics (Williams et al. 2004; Zanger and Schwab 2013; Rendic and Guengerich 2015). Many PAHs require bioactivation by CYPs to exert some forms of toxicity, however, inhibition of CYP activity can cause increased toxicity in some instances (Hawkins et al. 2002; Shimada 2006; Fleming and Di Giulio 2011; Xie et al. 2023).
Zebrafish as a toxicological model
The zebrafish (Danio rerio) is a highly sensitive model excelling in whole animal high-throughput screening for chemical hazard assessment and prioritization (Zon and Peterson 2010; Truong et al. 2014; Wiley et al. 2017; Yoganantharjah and Gibert 2017; Zhang et al. 2017; Vranic et al. 2019). Their high fecundity, small size, and optically clear tissues during early development allow for rapid assessment of developmental toxicity. By 48 hours post-fertilization (hpf), larval zebrafish contain many integrated organ systems including a primordial liver, heart, kidneys, and central nervous system (Kimmel 1993; Chu and Sadler 2009; Bakkers 2011; Gerlach and Wingert 2013). Zebrafish express many metabolizing genes early in development, including most cyp1 isoforms by 4 hpf and most cyp2 and cyp3 isoforms by 48 hpf (Goldstone et al. 2010; Nawaji et al. 2020). Zebrafish have been used extensively to study PAH toxicity (Incardona et al. 2006; Gao et al. 2017; Geier et al. 2018a, 2018b; Kompella et al. 2021; Gentile et al. 2023). Of note, zebrafish have 3 Ahr homologs, Ahr1a, Ahr1b, and Ahr2 (Andreasen et al. 2002; Karchner et al. 2005; Hahn et al. 2017). The 3 homologs overlap in their roles, however, Ahr2 is primarily responsible for binding dioxin-like ligands in zebrafish to cause cardiotoxicity and acts most similarly to mammalian AHR in this context (Prasch et al. 2003; Incardona et al. 2006; Shankar et al. 2020).
Retene sources and bioactivity
Retene (1-methyl-7-isopropyl phenanthrene) is a 3-ring alkylated PAH frequently detected in environmental samples. Retene was originally associated with incomplete combustion of resinous softwoods and paper mill effluent, however, it has since been shown that retene is also produced during the combustion of most carbonaceous materials (Ramdahl 1983; Koistinen et al. 1998; Leppänen and Oikari 2001; Shen et al. 2012). Retene is an abundant PAH in environmental sites affected by creosote and crude oil and is the most abundant PAH measured in wildfire smoke (Allan et al. 2011, 2012; de Oliveira Alves et al. 2017; Minick and Anderson 2017; Ghetu et al. 2022). Given its petrogenic and pyrogenic sourcing, retene is likely to present in most PAH-impacted sites, although it is often excluded from analysis because it is not an EPA priority PAH (Hussar et al. 2012). It is similarly excluded from many analyses of human exposure to PAHs, yet it is clear from personal passive sampling and indoor and outdoor air sampling that people are widely exposed to retene at levels comparable with or greater than other considerably more studied PAHs such as benzo[a]pyrene and pyrene (Gustafson et al. 2008; Dixon et al. 2022; Samon et al. 2022; McLarnan et al. 2024).
Despite widespread human exposure, most studies on the toxicity of retene have been in aquatic models. Similar to many PAHs, it causes oxidative stress, mutagenicity, and developmental toxicity in many fish models (Gravato and Santos 2002; Brinkworth et al. 2003; Maria et al. 2005; Scott et al. 2011; da Silva Junior et al. 2021). To date, there are no studies examining retene toxicity in intact mammalian models. A few studies have examined retene toxicity in human A549 lung epithelial cells, Hep G2 liver cells, and SK-W-SH neuroblastoma cells, demonstrating retene induced oxidative stress, genotoxicity, and cell death (de Oliveira Alves et al. 2017; Sarma et al. 2017; Peixoto et al. 2019; Scaramboni et al. 2023). Retene causes increased expression of AHR-associated genes such as Cyp1a in primary human bronchial epithelial cells (Colvin et al. 2024).
Retene is enigmatic because it acts like a strong AHR activator in vivo yet in vitro assays suggest it is a poor ligand. Retene toxicity is absent in Ahr2 knockout zebrafish (Scott et al. 2011; Wilson et al. 2022), and exposure to retene induces cyp1a expression in multiple fish models (Scott et al. 2011; Mu et al. 2016; Vehniäinen et al. 2016; Wilson et al. 2022). Concerning the broader transcriptome, larval zebrafish and larval Oncorhynus mykiss (rainbow trout) exposed to retene display similar transcriptomic signatures compared with larval zebrafish exposed to strong Ahr2 ligands like benzo[k]fluoranthene, benzo[j]fluoranthene, and TCDD (Vehniäinen et al. 2016; Shankar et al. 2019). Conversely, retene is a weak ligand for Ahr2 as determined by the displacement of TCDD in PLHC-1 hepatocarcinoma cells from Poeciliopsis lucida and recombinantly expressed Ahr2a and Ahr2b from rainbow trout (Billiard et al. 2002). Our first experiment in this study aligns with these findings, showing little to no retene activation of Ahr2 in a luciferase-based reporter system in ZF4 zebrafish fibroblast cells. We note the same finding in human HuH-7 epithelial-like hepatoma cells, indicating the phenomena may be relevant in humans. The apparent disconnect between the in vivo and in vitro results would be explained if a retene metabolite activates Ahr2. Hodson et al. (2007) proposed this after observing that high concentrations of the CYP inhibitor α-naphthoflavone eliminated retene toxicity in larval rainbow trout. This would be a tidy solution; however, α-naphthoflavone is also a mild AHR antagonist, which complicates the interpretation of its effect on retene toxicity (Merchant et al. 1990; Santostefano et al. 1993). Therefore, the toxic metabolite hypothesis remains plausible but requires verification.
This study evaluates the role of metabolism in retene toxicity using a combination of in vitro and in vivo tools. We show retene is a weak activator of human AHR and zebrafish Ahr2 in in vitro luciferase reporter-based assays. Through timed exposures and analysis of retene metabolites we show the window of susceptibility for retene teratogenicity to be 24 to 48 hpf during which time at least 4 retene metabolites are formed. Finally, we investigate Cyp1a, Cyp1b1, and the microbiome as potential mediators of retene toxicity through exposures with transgenic knock-out lines and germ-free fish.
Materials and methods
Chemical sourcing
Retene (97% purity) and xanthone (99% purity) were sourced from Santa Cruz Biotechnology (Dallas, Texas). Benzo[j]fluoranthene (BjF) (98.1% purity) was sourced from AccuStandard, Inc (Newhaven, Connecticut). Pyrene (98% purity) was sourced from Thermo Scientific Chemicals (Waltham, Massachusetts). All above chemicals were analytically verified by the Chemical Standards Core at Oregon State University and dissolved in 100% DMSO to make stock solutions. Hydroxyphenanthrene standard mixes were made from 1-, 2-, 3-, 4-, and 9-hydroxyphenanthrene standards acquired from Toronto Research Chemicals (North York, Ontario, Canada). HPLC grade Methanol (MeOH) and DMSO were sourced from VWR (Radnor, Pennsylvania). HPLC grade ethyl acetate (ETOAC) was sourced from Thermo-Scientific Chemicals (Waltham, Massachusetts).
Zebrafish husbandry and developmental toxicity screening
Specific pathogen-free 5D Tropical and CRISPR-Cas9 generated mutant zebrafish were reared at Sinnhuber Aquatic Research Laboratory (Corvallis, Oregon) in accordance with Institutional Animal Care and Use Committee protocols at Oregon State University. Stock fish were housed on a recirculating water system in 50-gallon brood stock tanks under a 14 h light: 10 h dark cycle. Water was supplemented with Instant Ocean salts (Spectrum Brands, Blacksburg, Virginia) and sodium bicarbonate as needed to achieve pH 7.4 and maintained at 28°C ± 1°C. Fish were fed Gemma Micro (Skretting, Inc., Fontaine-Lès-Vervins, France) twice daily across all life stages.
Embryos were collected as previously described and sorted by developmental stage (Westerfield 2007). Sorted embryos were kept in E2 embryo medium (EM) (15 mM NaCl, 0.5 mM KCl, 1 mM CaCl2, 1 mM MgSO4, 0.15 mM KH2PO4, 0.05 mM Na2HPO4, and 0.7 mM NaHCO3 buffered with 1 M NaOH to pH 7.2). At 4 hpf, embryos used for all screening studies except those for TCDD were enzymatically dechorionated using pronase (Fluka, St Louis, Missouri) (Mandrell et al. 2012). Briefly, embryos were placed in glass petri dishes containing 25 ml EM with 50 µl of 50 mg/ml pronase on a modified shaker for 6.5 min with constant agitation then rinsed with EM. Embryos were sorted to remove any damaged during dechorionation then robotically loaded into individual wells of 96-well plates pre-loaded with 100 µl EM (Mandrell et al. 2012).
PAHs were dispensed into 96-well plates using an HP D300e Digital Dispenser and plates were sealed with pressure-sensitive silicone adhesive-backed polyolefin plastic PCR film (ThermalSeal RTS) to reduce chemical volatilization. For all experiments except for determining the window of susceptibility, chemical administration occurred at 6 hpf. For each PAH exposure, concentrations ranged from 0 to 100 µM. Because chemicals were solubilized in DMSO, the DMSO concentration was normalized across treatments by adding dry DMSO to achieve a concentration of 1% in embryo media by volume. Vehicle controls received 1% DMSO which has been shown to be safe for developmental zebrafish toxicity assays (Hoyberghs et al. 2021). Plates were placed on an orbital shaker at 235 RPM overnight under dark conditions at 28°C ± 1°C then kept under the same conditions without shaking until tissue collections or screening.
TCDD exposure methodology was adapted from previously described studies (Garcia et al. 2018; Shankar et al. 2022). Morphologically normal 6 to 8 hpf embryos were exposed to vehicle control (1% DMSO by volume) or TCDD at 0.025, 0.05, 0625, 0.125, 0.25, 0.5, 1.0 ng/ml in capped glass vials in batches of 40 embryos per vial in 4,000 μl of exposure solution. Exposures continued for 1 h on a rocking table. Vials were mixed by inversion at the start of exposure and at 15 min intervals. After exposure, embryos were rinsed 3 times in clean EM, and transferred to individual wells of a 96-well plate, utilizing 36 fish per concentration per phenotype.
For all developmental toxicity screening studies, zebrafish were visually screened for 13 morphological endpoints at 24 and 120 hpf (Table 1). The percent incidence of each endpoint was calculated across replicate plates and the percent incidence of any endpoint occurring at a given concentration was reported as “any effect,” which was used to derive concentration-response curves and identify the concentration which elicited 50% effect above controls through benchmark dose modeling (BMC50).
Table 1.
Developmental toxicity screening endpoints measured at 24 and 120 hpf.
| Zebrafish morphological endpoints | |
|---|---|
| 24 hpf | Mortality, delayed progression, spontaneous movement |
| 48 hpf | Mortality, edemas, bent axis, touch response, and craniofacial, muscular/cardiovascular, lower trunk, brain skin, notochord malformations |
AHR activation assays
Ten PAHs were screened in human (Huh7) and zebrafish (ZF4) cell lines to measure activation of AHR and Ahr2, respectively, by INDIGO Biosciences, Inc. (State College, Pennsylvania). The cells used express native human AHR or zebrafish Ahr2 and are modified with a firefly luciferase reporter with a promotor region containing multiple tandem xenobiotic response elements. For each line, a reporter cell suspension was prepared in cell media and pre-incubated for 4 to 6 h at 28°C, 5% CO2, 85% humidity in 96-well plates. Compound stocks were diluted using DMSO to achieve 1,000× stocks, then diluted with medium to achieve 2× concentrations. After pre-incubation, culture media was discarded and 100 µl of the 2× compound stocks were added to each well in triplicate for final exposure concentrations of 1.52, 4.57, 13.7, 41.2, 123, 370, 1,111, 3,333, 10,000, and 30,000 nM. DMSO was used as a vehicle control and the strong AHR agonist (2′'Z, 3'E)-6-Bromo-1-methylindirubin-3'-oxime (MeBIO) was used as a positive control and to determine fold activation. Plates were incubated for 22 to 24 h in a cell incubator under the same conditions listed above.
After incubation, treatment media was discarded and 100 µl of luciferase detection reagent was added to each well and relative light units (RLU) were measured using a Tecan Spark Microplate Reader in luminescence mode. The plate reader was programmed to perform a single 5 s “plate shake” prior to reading the first assay well. The read time was set to 0.5 s per well. Average RLU values, standard deviation, fold activation (Equation 1), and % coefficient of variation (%CV) (Equation 2) were calculated for each concentration. Relative fold activation was calculated according to Equation (3) and used for dose-response modeling and comparison between chemicals.
| (1) |
| (2) |
| (3) |
Bioactivation of retene in the AHR reporter assay was also tested with a preactivation method and a concurrent activation method in Huh7 reporter cells. Pooled human liver microsomes (Xtreme 200, H2610) and S9 extracts (Xtreme 200, H2610.S9) were acquired from Xenotech (Kansas City, Kansas). For the preactivation experiment, retene was incubated with human liver microsomes (2 mg/ml) in a core buffer (3 mM NADPH, 3 mM MgCl2, 100 mM KPO4, pH 7.4) at 37ºC for 30 min in 10 ml reactions. Vehicle controls were generated by incubating DMSO with liver microsomes, and metabolism negative controls were generated with acid denatured liver microsomes. Reactions were quenched by acidification, and extracted into an equal volume of EtOAc. Extracts were evaporated under a stream of nitrogen and reconstituted in DMSO to yield a solution concentrated to 750 µM retene equivalent assuming mass balance. Samples were tested in the Huh7 agonist reporter system at 3 µM retene equivalence with four biological replicates and three technical replicates using the protocols described above. For the concurrent activation experiment, reporter cells lines were coincubated with S9 or with liver microsomes for the duration of the retene exposure. For S9 coincubation, cells were exposed to 30 µM retene, 0.33 mg/ml S9 fraction, and 5% core buffer. Microsome coincubations were conducted under the same conditions with 0.125 mg/ml microsomal fraction instead of S9 to normalize for CYP concentrations. Treatments included vehicle controls, positive controls with MeBIO, and inactivated S9/Microsome controls. In both bioactivation experiments, reporter signal was measured and analyzed as described above.
PAH toxicity, Ahr2 dependence, and Cyp1a induction in developing zebrafish
Zebrafish developmental toxicity for each PAH was readily available from publications indicated in Table 2. Ahr2 dependence was mined from the literature when available which is also indicated in Table 2. When previous Ahr2 dependence was unavailable, as was the case for benzo[j]fluoranthene and xanthone, Ahr2 dependence was determined by exposing wild-type 5D fish and Ahr2 knockout (ahr2hu3335) fish to a nominal concentration causing morphologic malformations in 80% of WT zebrafish (EC80). EC80 concentrations were determined by exposing and evaluating fish according to the methods described in “Zebrafish husbandry and developmental toxicity screening” section. The EC80 for Xanthone was 52 μM. An EC80 for benzo[j]fluoranthene could not be determined, so 80 μM was used for Ahr2 dependence testing. Both compounds were tested with 48 embryos per genotype. Cyp1a expression is reported from Geier et al. (2018a) in which the authors used immunohistochemistry (IHC) to determine the presence and localization of Cyp1a protein.
Table 2.
Chemicals included in AHR activation study with literature sources for Ahr2 dependence.
| Chemical | CASRN | Abbreviation | Dev. Tox. Lit. source | Ahr2 Dep. Lit. source(s) |
|---|---|---|---|---|
| Benzanthrone | 82-05-3 | BEZO | Geier et al. 2018a | Goodale et al. 2015 |
| Benzo[j]fluoranthene | 205-82-3 | BjF | Geier et al. 2018a | This study |
| Retene | 483-65-8 | Retene | Geier et al. 2018a | Scott et al. 2011; Wilson et al. 2022 |
| Benzo(k)fluoranthene | 207-08-9 | BkF | Shankar et al. 2019 | Garland et al. 2020 |
| Benzanthraquinone | 2498-66-0 | BAAQ | Knecht et al. 2013 | Goodale et al. 2015 |
| Pyrene | 129-00-0 | Pyrene | Geier et al. 2018a | Incardona et al. 2005; Incardona et al. 2006, this study |
| Xanthone | 90-47-1 | XAN | Geier et al. 2018a | This study |
| Benzo[a]pyrene | 50-32-8 | BAP | Geier et al. 2018a | NA |
| Benzo[b]fluoranthene | 205-99-2 | BbF | Geier et al. 2018a | NA |
| Fluoranthene | 206-44-0 | FLO | NA |
Body burden
To investigate retene uptake and loss in zebrafish tissue, fish were exposed to 30 µM retene or 1% DMSO beginning at 6 hpf as described above. At 12, 24, 36, and 48 hpf, fish were collected in four replicate pools of 40 fish each into 1.5 ml Eppendorf SafeLock tubes and rinsed 3× with EM. Fish were anesthetized on ice and flash frozen in liquid nitrogen and kept at −20°C until tissue extraction.
Once all samples were collected, 100 µl of 1 mm glass beads were added to each tube and tubes were placed on ice for 10 min. 250 mg Na2SO4 and 200 µl ethyl acetate was added to each tube and vortexed. Tubes were closed and homogenized in a Bullet Blender tissue homogenizer (Next Advance, New York). An additional 300 µl ethyl acetate was added then tubes were vortexed and the homogenization step was repeated. Homogenate was centrifuged at 4°C for 10 min at speed 16,000 × g and supernatant was removed into a 1 ml amber vial and stored at 4°C until chemical analysis.
Analysis was performed using an analytical method for quantitation and identification of 65 parent and substituted PAHs (Anderson et al. 2015). The method was performed using an Agilent: 7890 gas chromatograph (GC) with a 7000C triple quadrupole mass spectrometer (MS/MS) with an Agilent J&W PAH select column (30 m × 250 µm × 0.15 µm). Internal calibration with at least a 4-point (4 to 7) calibration with correlations ≥0.99 was employed. Specific instrument conditions are detailed in the original method. GC-MS/MS data was analyzed using MassHunter Quantitative Analysis v. B.06.00 SP1 build 6.0.388.1 (Agilent Corp. Wilmington, Delaware) software. Average tissue burdens constituted 1.2% to 4.5% of the total retene added to media during exposures.
Larval metabolite analysis
Metabolite sample acquisition
Larval fish were dechorionated and exposed to 26 µM retene following protocols stated above. At 48 hpf, plates were unsealed, 36 fish from control or exposure groups were pooled, rinsed 3× with cold EM, and flash frozen in liquid nitrogen. For extractions, fish tissue was homogenized in a bullet blender with 1 mm glass beads, acidified in 500 μl of 0.45 M sulfuric acid, and extracted with an equal volume of EtOAC. Metabolites were solvent exchanged to MeOH before analysis.
Metabolite detection by LC-IM-MS
Chromatography separation was performed with an Agilent Ultrahigh performance liquid chromatography (UHPLC) 1290 Infinity II system. A 20 μl aliquot of the reconstituted sample was injected onto an Ascentis 25 cm × 4.6 mm, 5 μm C18 column. The gradient used was water: Acetonitrile starting at 45:55 and going to 20:80 in 2 min, then to 15:85 in 6 min, then to 0:100 in 2 min and holding it at 100% acetonitrile until 22 min, with a constant flow of 0.95 ml/min. The UHPLC system was coupled to an Agilent 6560 Ion Mobility quadrupole TOF MS system (Agilent Technologies, Santa Clara, California) equipped with an electrospray ionization (ESI) source for metabolite detection and with an atmospheric pressure photoionization (APPI) source for retene detection. Samples were analyzed in negative ESI with metabolites detected as deprotonated molecules ([M—H]-), and in positive APPI with retene detected as radical (M•+). Data was acquired in the mass range from m/z 50 to 1700.
LC-IMS-MS data were converted from vendor format (.d) to an open standard (mzML) for processing by DEIMoS (Data Processing for Integrated Multidimensional Spectrometry), an open-source Python package amenable to high dimensional mass spectrometry acquisitions, such as the liquid chromatography—ion mobility spectrometry—tandem mass spectrometry utilized here (Colby et al. 2022). Smoothing was performed only in the drift time dimension with radius = 1 scan (approx. 12 ms) and repeated for three iterations.
Feature detection utilized persistent homology, a topological data analysis algorithm, to identify local maxima in the acquired signal. Although we initially explored feature detection in native dimensionality (i.e. m/z, drift time, and retention time simultaneously), we observed greater sensitivity when detecting features in m/z and drift time for each retention time frame. Putative features were then thresholded to 500 intensity.
By detecting features in each retention time frame, we introduced the potential for duplicate (m/z, drift time) features across frames. To ameliorate, we combined features within 20 ppm m/z, 0.4% drift time, and 1 s retention time, keeping only the most intense feature, according to sensitivity analysis performed on mixtures of 1-, 2-, 3-, 4-, and 9-hydroxyphenanthrene standards.
Study samples were compared against matched control samples from unexposed fish to exclude features independent of the treatment. Although the ion mobility dimension affords potential to separate isomers, only putative formula assignments were made from unique m/z values detected in study samples. This was due to a lack of standards available to confirm putative isomer assignment.
Cyp1a GFP time course
Transgenic zebrafish line cms2TG/+ (Zfin ID: ZDB-GENO-151012-1) were plated in 96-well plates and exposed to 26 μM retene, 8.9 μM benzo[k]fluoranthene, or vehicle control (1% DMSO) at 8 HPF to determine Cyp1a localization over time. We refer to Cms2TG/+ fish as cyp1acms-gfp. Cyp1acms-gfp contain TgBAC(cyp1a: NLS-EGFP) (zfin ID: ZDB-TGCONSTRCT-150928-7), an enhanced green fluorescent protein (EGFP) construct within 8 kb of upstream sequence and 30 kb of downstream sequence of cyp1a. They are sensitive to Ahr2 ligands, expressing EGFP where native Cyp1a is produced (Kim et al. 2013). At 23 hpf, four fish from each treatment were transferred to the same well of a glass bottomed 96-well plate in 300 μl of exposure media. From 23 hpf to 48 hpf brightfield and green fluorescence images were acquired for each group of 4 fish using an Andor BC43 benchtop confocal microscope. Brightfield images were acquired at 20% power with 120 ms exposures. Green fluorescent images were acquired in EPI mode measuring at emission wavelength 529 nm, with excitation wave length at 488 nm, 25% laser intensity, and 200 ms exposures. The experiment was repeated twice, each with four fish per chemical.
Window of susceptibility
To determine the window of time during which zebrafish embryos are most susceptible to retene-induced developmental toxicity, fish were exposed during four developmental windows: 6 to 48, 6 to 120, 24 to 120, and 48 to 120 hpf. These windows were selected to capture various points of organogenesis. For the 6 to 48 hpf exposure, at 48 hpf, fish were washed with a series of repeated 1:1 dilution with EM using a 96-well Rainin Liquidator. 100 µl fresh EM was added to the wells, solution was gently pumped up and down to mix, then 100 µl was removed and discarded. This was repeated 6× for seven total washes to reach <1% the original retene concentration. Exposures were administrated and screening was performed exactly as described above.
Development of a functional Cyp1a mutant zebrafish line
Single-guide RNA design and injection
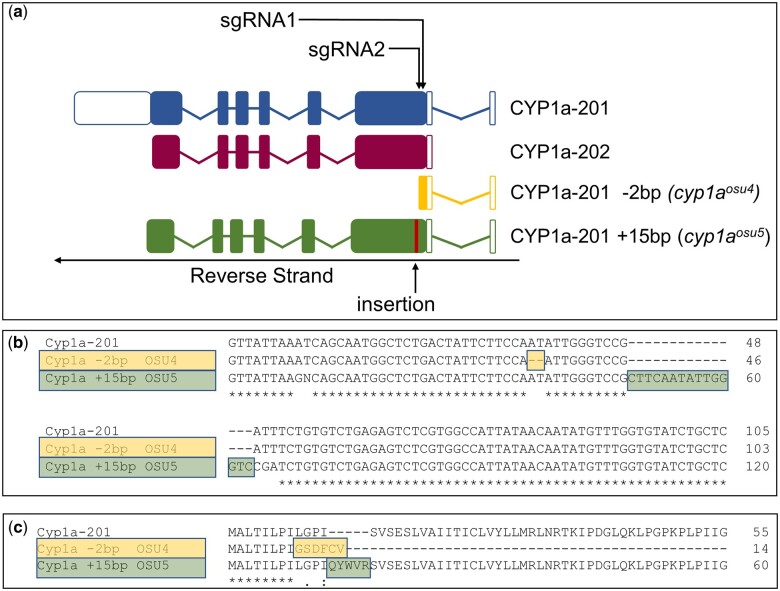
Two Cyp1a mutant lines were developed using CRISPR-Cas9 to investigate the role of Cyp1a in retene toxicity and to serve as a tool for mechanistic investigation in further studies. There are two cyp1a splice variants in zebrafish: cyp1a_201 and cyp1a_202 (Martin et al. 2023). Three single-guide RNA oligos (sgRNA) were synthesized and provided by Synthego all of which targeted zebrafish cyp1a near the 5′ end, in cyp1a_201 exon 2 and cyp1a_202 exon 1. Throughout the rest of the manuscript we utilize cyp1a_201 to identify locations in the gene. Two of the heritable mutations were produced from two of the sgRNAs. sgRNA1 treatment produced a 2 bp deletion and sgRNA2 treatment resulted in a single nucleotide polymorphism and 15 bp addition. The lines from these mutations are cyp1aosu4 and cyp1aosu5, respectively. The sequences of all sgRNA, qPCR and TAQMAN reagents are included in Table S2.
CAS9 administration and F0 generation selection
One-cell stage 5D Tropical wild-type embryos were injected with 1.5 to 2 nl of sgRNA and Cas9 protein mixture resulting in an administered dose of 150 to 200 pg sgRNA and 225 to 300 pg Cas9 into the yolk stream. Embryos were reared in EM and monitored for normal development. At 1 day post fertilization (dpf), a subselection of injected embryos were disassociated with NAOH and screened for mutational efficiency via DNA melt curve analysis. The remaining embryos from sgRNA batches inducing a shift in melt curve peak for >70% of embryos were raised to adulthood to comprise the F0 generation.
Founder screening and progression to stable lines
F0 fish with heritable mutations were identified by outcrossing with wild-type 5D and screening the resulting embryos for mutations by melt curve analysis with whole embryo gDNA. Once identified, F0 founders were outcrossed and the F1 generation was grown to adulthood. Adult F1 fish were genotyped with melt-curve analysis or a Taqman Assay utilizing gDNA isolated from fin clippings. The DNA from fin clippings in confirmed heterozygotes was amplified and sequenced via Sanger sequencing in an ABI 3730 sequencer at the Oregon State University Center for Quantitative Life Sciences. Mixed base reads in heterozygotes were interpreted with Poly Peak Parser to identify indels (Hill et al. 2014). Two mutations were identified: an in-frame 15 BP insertion (cyp1aosu5) and a 2 BP deletion resulting in a premature stop codon (cyp1aosu4). Fish identified with the same mutation were in-crossed to form the F2 generation and homozygous fish from F2 were identified at adulthood by sequencing of gDNA from fin clippings.
gDNA isolation and melt curve analysis
Embryos were placed individually into wells of a 96-well PCR plate (VWR Thermocycling plate 89049-178) and euthanized on ice. Excess embryo media was removed and tissue was homogenized in 20 μl of 50 mM NaOH heated to 95°C for 10 min and vortexed. After tissue dissociation, plates were centrifuged briefly, allowed to cool, and neutralized with 4 μl of 1 M Tris-HCl (pH 8). After dilution with 120 μl of ultrapure water, debris from the mixture was pelletized via centrifugation at 4680 RPM for 10 min. The resulting DNA was utilized for target sequence amplification and melt curve analysis using Power Sybr Green Master Mix (Applied Biosystems, Foster City California, ref 4367659) on a StepOnePlus Realtime PCR System (Applied Biosystems, Waltham, Massachusetts). Reactions consisted of 10 μl 2× master mix, 0.4 μl of forward and reverse primers, and 2 μl of isolated DNA diluted to 20 μl in RNase free water. Temperature protocol consisted of initial activation (95°C held 10 min), 40 amplification cycles (95°C for 15 s, 58°C for 1 min), and melt curve analysis (70°C for 2 min, 0.075°C * s−1 ramp rate, 95°C). NaOH homogenizations for fin clippings were extended to 15 min.
TaqMan assay
The 2 bp deletion in cyp1aosu4 proved difficult to confirm via melt curve analysis alone. To overcome this, a TaqMan assay was developed with primer pairs amplifying a 78 bp region around the expected mutation and with TaqMan probes targeting the mutated or WT sequences. The reaction was performed using TaqMan Universal PCR Mastermix (Applied Biosystems, ref 4304437) under manufacture recommended conditions with a StepOnePlus Realtime PCR System. The TaqMAN probes are included in Table S2.
Whole-mount immunohistochemistry
To validate loss of Cyp1a protein in mutant zebrafish, whole-mount IHC was performed as previously described (Anderson et al. 2022) with minor modifications. Briefly, dechorionated wild type and cyp1aosu4 and cyp1aosu5 mutant embryos were exposed to the potent AHR activator BkF at 8.9 µM or 1% DMSO beginning at 6 hpf as described above. At 3 dpf, fish were transferred to 1.5 ml microcentrifuge tubes in pools of 10 and anesthetized on ice. Once no movement was observed, buffered Tricaine was added to euthanize embryos. Tricaine solution was immediately removed and 500 μl of 4% paraformaldehyde in phosphate-buffered saline (PBS) was added. Tubes were placed at 4°C on a rocker overnight to fix fish tissues. Fish were rinsed with PBS 3 times and stored in 0.02% NaN3 in PBS (PBS-NaN3) preservative at 4°C until proceeding with IHC.
Fixed fish were placed into a 24-well polystyrene plate the following morning and preservative solution was removed. For all IHC immersion and washing steps, solution was added, plates were kept on an orbital shaker at low speed for the indicated duration, then solution was removed. All steps were performed at room temperature unless otherwise stated. Fish were washed 3× for 15 min each in PBS with 0.1% Tween 20 detergent (PBST). Fish tissue was permeabilized using freshly made ice-cold 0.005% trypsin in PBS for 8 min on ice followed by 3× 5 min washes in PBST at room temperature. Fish were post-fixed with 4% PFA for 10 min then washed twice for 5 min with PBST. To block nonspecific protein binding, PBST was removed and 10% normal goat serum in PBST with 0.01% triton X-100 (PBSTx) was added for 10 min. Fish were then transferred to 1.5 ml microcentrifuge tubes and solution was removed. The primary monoclonal Cyp1a antibody (C10-7, mouse anti-fish Cyp1a, Biosense Laboratories, Bergen, Norway) was added at a 1:500 dilution in NGS-PBSTx and incubated overnight at 4°C. An acetylated tubulin (AT) primary antibody (mouse anti-fish AT T6793, Sigma-Aldrich, St Louis, Missouri) was used as a procedural control at 1:4000 NGS-PBSTx.
The following morning, primary antibodies were removed and fish were transferred to a new 24-well plate then rinsed in PBST with 2× 5 min washes followed by 4× 30 min washes. Fish were then incubated in secondary antibodies: Alexa Fluor 594 goat anti-mouse, IgG3 for Cyp1a, Alexa Fluor 594 goat anti-mouse, IgG for acetylated tubulin (Invitrogen, Eugene, Oregon) at 1:2,000 in NGS-PBSTx for 2 h. Fish were rinsed in PBST with 2× 5 min washes followed by 4× 30 min washes then transferred to 1.5 ml microcentrifuge tubes and stored in PBS-NaN3 at 4°C until imaging.
To assess localization of Cyp1a protein, fish were placed in a 12-well glass-bottom plate and imaged using a BZ-X710 fluorescence microscope (Keyence, Osaka, Japan) using a Texas Red filter cube. Upon determining that antibody detection was consistent within each treatment, representative fish from each group were mounted in low-melt agar on 35 mm glass bottom dishes (Matsunami Glass, Bellingham, Washington) and imaged as described above. Imaging exposure time was optimized on BkF exposed control fish and held constant between treatments.
EROD activity assay
To assess Cyp1a mutant fish for spatiotemporal Cyp1a enzymatic activity, an in vivo ethoxyresorufin-o-deethylase (EROD) activity assay was performed. Wild type and cyp1aosu4 and cyp1aosu5 mutant zebrafish embryos were exposed to 50 µM retene or 1% DMSO beginning at 6 hpf as described above. At 72 hpf, 50 µl of exposure solution was removed and 50 µl 0.8 µg/ml 7-ethoxyresorufin (7-ER) dissolved in EM was added to each well under dark conditions. Fish were incubated in 7-ER for 10 min. 0.2 g/l buffered tricaine was added to anesthetize fish prior to imaging. Four fish from each treatment were mounted in low-melt agarose containing 0.2 g/l tricaine and imaged for production of fluorescent resorufin on a BZ-X710 fluorescence microscope (Keyence, Osaka, Japan) using a Texas Red filter cube to capture the target excitation and emission wavelengths of 562 nm and ≥590 nm, respectively. Exposure time for imaging each treatment was performed using the exposure time which produced sufficient resorufin detection in retene-exposed wild type fish, 1/2.5 s.
Mutant line developmental toxicity screening
Cyp1aosu4 and cyp1aosu5 mutant fish were screened for developmental retene toxicity exactly as described in “Zebrafish husbandry and developmental toxicity screening” section. To assess the role of a constitutively expressed enzyme, Cyp1b1, in retene toxicity, Cyp1b1wh4 zebrafish (Zfin ID: ZDB-ALT-240703-14) were procured from Woods Hole Oceanographic Institute. A manuscript on the development and validation of this line is in review (Perone 2024). Cyp1b1wh4 fish contain a cyp1b1 with three deletion mutations of lengths 37, 21, and 8 bp. The first mutation causes a frame shift that results in an early stop codon in exon 2. The truncated protein no longer contains a heme binding domain, rendering it inactive. These fish, which were derived in an AB wild-type background and were outcrossed with 5D wild-type fish twice to reduce background malformations. Wild type AB fish were crossed the same way, resulting in 5D-AB wild type fish to serve as control in chemical screening. Cyp1b1wh4 and wild type fish were screened exactly as described in “Zebrafish husbandry and developmental toxicity screening” section.
Investigation of microbial metabolism
Biotransformer metabolite prediction
Biotransformer 3.0 was used to identify predicted retene metabolites (Djoumbou-Feunang et al. 2019). The knowledge- and machine learning-based approach considered metabolites formed through EC, CYP450, phase II enzymes and human gut microbiome metabolism. The resulting metabolite list was then filtered to remove nonprimary metabolites and large, conjugated molecules unlikely to dock in the Ahr2 ligand binding site. Predicted metabolites can be found in Table S6.
Germ-free derivation and exposure
Metabolite prediction identified metabolites derived from the microbiome. To assess the role of microbiome in retene metabolism, germ-free (GF) zebrafish were derived. Adult zebrafish were spawned as above and embryos were collected into either EM or antibiotic EM (EM with 100 µg/ml ampicillin, 250 ng/ml amphotericin B, 10 µg/ml gentamycin, 1 µg/ml tetracycline, 1 µg/ml chloramphenicol, filter sterilized). Under a laminar flow hood surface-sterilized with 100% ethanol, germ-free embryos were poured into 50 ml beakers and rinsed 3× with filter-sterilized EM. Embryos were immersed in 0.1% PVP-I solution for 2 min then sterile EM was added to dilute the solution and poured off. Embryos were rinsed 3× with sterile EM and kept in sterile EM for 5 min. Embryos were transferred to a new beaker, sterile EM removed, and 0.003% bleach solution added. Embryos were submerged in bleach solution for 10 min, then solution was poured off and embryos were rinsed 3× with sterile EM. Embryos were transferred to a sterile glass petri dish and 1 ml sterile EM was removed and plated on an LB agar plate. 1 ml nonsterile EM from the dish with conventional embryos was plated and plates were incubated at 37°C then visually checked after 24 h, confirming no bacterial growth in germ-free media.
GF embryos were plated by hand using sterilized glassware into sterile 96-well plates pre-filled with 50 µl sterile EM. Working retene stocks were made at 2× concentrations in sterile EM and 50 µl was added to each well for a total of three plates with 36 fish per concentration. Plates were sealed with sterile PCR seals and stored as described above. Conventional embryos were plated and stored exactly the same way, but with non-sterile EM, 96-well plates, and seals.
Statistical analysis
For dose response experiments with luciferase reporter assays, luciferase reporter relative fold changes were fit to log-logistic regression models in GraphPad for each chemical. Log-logistic models were used to determine EC50 values, the concentration causing 50% of the maximal luciferase induction for the compound. EC50 values were used to compare potencies. Chemicals with successfully fit log-logistic models were considered “active” AHR or Ahr2 ligands. If the data would not support a log-logistic model, then the highest concentration was tested for significance by determining a 95% confidence interval for the fold change of luciferase induction compared with controls. If the fold change was greater than 2 and the 95% confidence interval did not include 1, then the induction was considered significant and the compound was marked with an asterisk in Fig. 1.
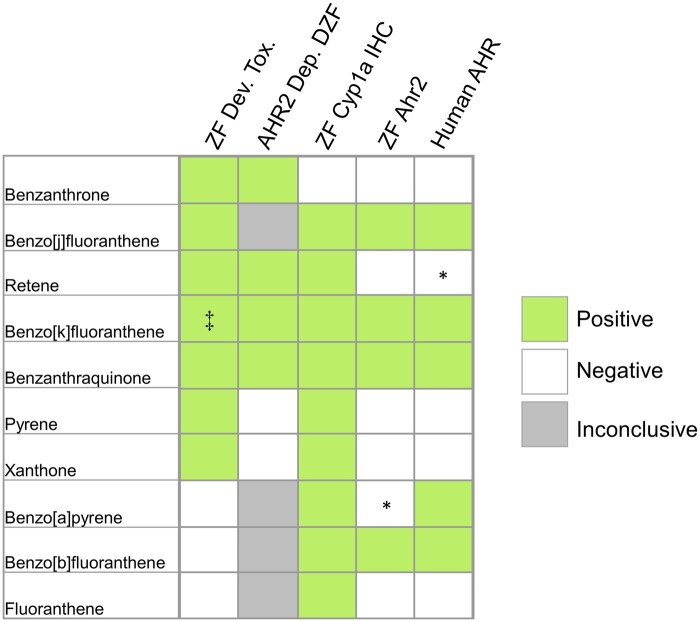
Fig. 1.
Summary of developmental zebrafish toxicity assay (ZF Dev. Tox.), Ahr2 dependence in developmental zebrafish toxicity assay (AHR2 Dep. DZF), Cyp1a induction (ZF Cyp1a IHC), and AHR activation in the AHR reporter assay for zebrafish Ahr2 (ZF Ahr2) and human AHR. Positive Ahr2 dependence indicates attenuated toxicity in Ahr2-null fish. Positive Cyp1a induction indicates detection of Cyp1a via IHC after exposure. Positive results in either reporter assay indicates robust expression of luciferase sufficient to estimate an EC50 with exposures up to 30 µM using a 4 parameter log-logistic model. Asterisk (*) indicates some significant activation at the highest concentrations, but for which a log-logistic model was not significant. Significance of highest concentrations in these cases were determined by fold changes from controls greater than 2 and 95% confidence intervals not including 1. Dagger (‡) indicates Ahr2 dependence was determined by morpholino knockdown.
Metabolic activation experiments were tested for significance in R with 2-way ANOVA models utilizing chemical treatment and metabolic status as factors. Significance between individual treatments was determined by nonoverlap between 95% confidence intervals.
Dose response curve analysis of chemical toxicity in zebrafish embryos was carried out in R using the DRC package. Experimental models were fit to 2 parameter, 3 parameter, and 4 parameter log-logistic models utilizing exposure windows, genotype, or germ status as factors when appropriate. We used the simplest model not significantly different (P-value < 0.05) from the model with one more parameter. The selected models were used to calculate a BMC50 for each test condition. The BMC50 is the nominal concentration of the chemical in the media causing an effect 50% greater than control animals. Differences between treatments of genotypes were determined by computing the 95% confidence interval for BMC50 values. Nonoverlapping BMC50 confidence intervals between factor groups indicated significance. Dose response figures were generated in GraphPad Prism 10 using the models generated in R. Each point indicates the proportion of effected fish with 95% confidence intervals calculated using the Wilson/Brown method in GraphPad Prism 10.
Results
Retene does not activate AHR: zebrafish and human luciferase-based AHR reporter assays
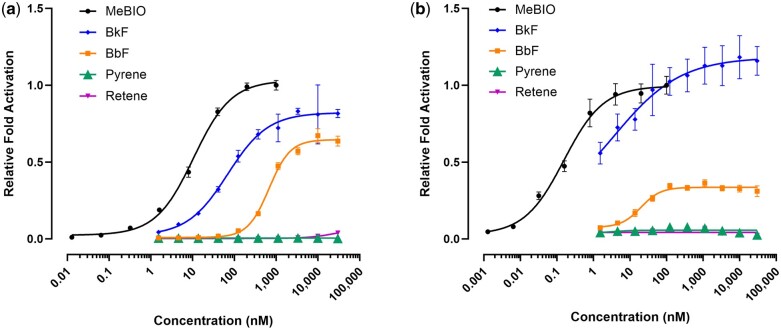
As part of a large-scale effort to classify and bin PAHs by their bioactivity, 10 PAHs with known Ahr2 activity in in vivo zebrafish assays were screened for transcriptional Ahr2 activation in ZF-4 zebrafish luciferase reporter cells. Screening was also performed in HuH-7 human hepatoma luciferase reporter cells to compare transcription activation between zebrafish Ahr2 and human AHR. The results of these assays are displayed in Fig. 1, which also assembles the toxicity in developing zebrafish, Ahr2 dependence, and Cyp1a induction in zebrafish compiled from other publications indicated in Table 2. The presence of Cyp1a as determined by IHC occurs with more chemical exposures than either of the reporter lines would suggest, although this is likely because Cyp1a protein abundance is also sensitive to Ahr1a and Ahr1b activity (Geier et al. 2018a). Benzo[k]fluoranthene, benzanthraquinone, benzo[j]fluoranthene, and benzo[b]fluoranthene were identified as direct AHR activators in both human and zebrafish cell lines (Fig. 2, Tables S3 and S4). Benzo[a]pyrene was an activator in human cells and zebrafish cell, but the slope was too broad to fit a log-logistic model over the concentration range. Retene exhibited minimal reporter activity in human cells with small but significant fold change differences at 10 and 30 μM and no significant activation in zebrafish cells. Retene is the only chemical among these 10 PAHs that displays Ahr2 dependent toxicity and Cyp1a induction, but little to no luciferase activity in the reporter cell lines.
Fig. 2.
Concentration–response curves for luciferase production representative of AHR activation in (a) Human Huh-7 and (b) Zebrafish ZF-4 cells. Each point is the result of 3 replicate wells. Error bars indicate 95% confidence intervals. Fold activation values for all compounds tested, including those not shown here, are in Table S4.
The AHR activation assay in human HuH-7 reporter cells was supplemented with external metabolism in 2 different experiments to test if metabolic activation of retene would activate AHR. Exposure of reporter cells to human liver microsome-reacted retene extracts at 3 μM did not elicit statistically significant luciferase reporter activity (Fig. S1). HuH-7 reporter cells exposed to 30 μM retene had significant induction of AHR activity, but supplementation with human liver microsomes or s9 did not lead to an increase in reporter activity (Figs. S2 and S3). Metabolism was confirmed in the concurrent metabolism study by a decrease in response to a positive control.
Retene is rapidly metabolized between 36 and 48 hpf
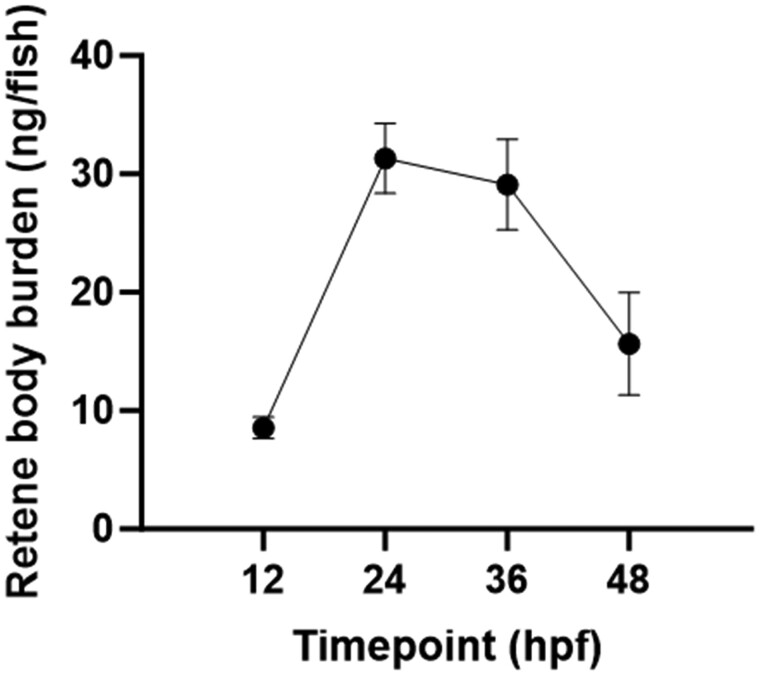
To confirm retene metabolism occurs in larval zebrafish by 36 hpf, retene body burden was assessed at four separate timepoints in zebrafish exposed to 30 μM retene. Fish were exposed at 6 hpf and tissue extracts were collected at 12, 24, 36, and 48 hpf for retene quantification via GC-MS analysis. The mass balance of retene recovered in fish tissue extracts ranged from 1.1% to 4.9%. Retene was detected and measured at each timepoint, with a greater than 3-fold increase in body burden from 12 to 24 hpf. Tissue burden did not change from 24 to 36 hpf, then decreased to near-baseline at 48 hpf (Fig. 3). The leveling off of retene between 24 and 36 hpf indicates a potential onset of xenobiotic metabolic activity. The rapid depletion of retene in fish tissue from 36 to 48 hpf indicates rapid metabolism developing in zebrafish after 36 hpf.
Fig. 3.
Retene mass detected in zebrafish tissue extracts at 4 developmental timepoints after static exposure to 30 μM retene in 96 well plates beginning at 6 hpf. Each point represents the average mass of retene per fish determined from 4 replicates each consisting of extracts from 40 exposed embryos. Error bars indicate the 95% confidence intervals.
In silico prediction of metabolites with Biotransformer
To investigate potential retene metabolites and routes of formation in vivo, a list of predicted metabolites was generated using Biotransformer 3.0 (Djoumbou-Feunang et al. 2019). A table of Biotransformer results is included in Table S6. Over 30,000 total metabolites were initially predicted, with many metabolites predicted repeatedly and derived from multiple biotransformation mechanisms. Upon filtering for unique structures, and removing bulky and polar phase II metabolites that were unlikely to activate AHR, 558 candidate metabolites remained. From this list, 21.5% were predicted to be produced microbially while the remaining 78.5% were predicted to be produced by phase I metabolism.
Retene metabolite identification by LC-MS
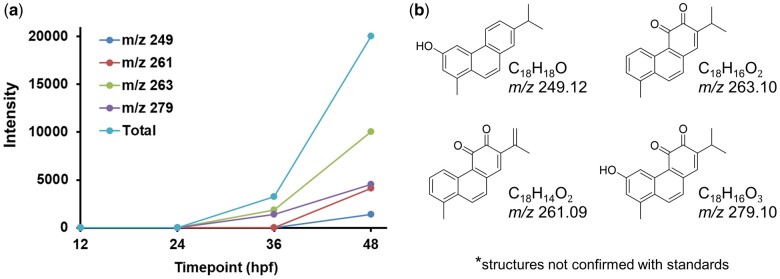
To identify important in vivo retene metabolites, we altered our extraction protocol for metabolite efficiency and scanned tissue extracts from 12, 24, 36, and 48 hpf fish using LC-IM-MS with ESI in negative ion mode for metabolites and with an APPI in positive ion mode for retene detection. The LC-IM-MS data were processed with DEIMoS and features also detected in control samples were excluded. The trend for parent retene ion intensity in metabolite extracts differed somewhat from the quantified retene by GC-MS, with abundance increasing from 12 to 24 hpf, decreasing from 24 to 36 hpf, and remaining relatively constant from 36 to 48 hpf (Fig. S4). Retene metabolites followed an inverse trend from the retene parent, with metabolites appearing at 36 hpf and increasing to 48 hpf. The list of possible retene metabolites generated with Biotransformer was matched by m/z (50 ppm tolerance) to ions uniquely detected in the samples. Proposed structures of the detected metabolites are included in Fig. 4b. We note each m/z matched to multiple Biotransformer indicated isomers and these structures are not validated to analytical standards. No retene metabolites were detected at 12 and 24 hpf, confirming that retene metabolism is not significant up to 24 hpf. Two ions at m/z 263.10 (C18H16O2) and m/z 279.10 (C18H16O3) were detected at 36 hpf indicating the onset of metabolism before 36 hpf. These ions matched with Biotranformer generated metabolites. The intensity of the 2 metabolites at m/z 263.10 and m/z 279.10 increased 5 and 3-fold, respectively, from 36 to 48 hpf. Two other ions at m/z 249.12 (C18H18O) and m/z 261.09 (C18H14O2) were also detected and matched with Biotransformer generated retene metabolites at 48 hpf (Fig. 4). The detection of metabolites at 36 hpf confirms metabolic onset before 36 hpf, and the increase in metabolite by 48 hpf indicates increasing metabolism from 36 to 48 hpf.
Fig. 4.
a) Signal intensity of metabolites identified by LC-MS in extracts from retene exposed embryos at 12, 24, 36, and 48 hpf. Each point represents average ion intensity from at least 3 replicate extracts from 36 fish each. b) Potential structures for metabolites corresponding to observed m/z ions.
Cyp1a-gfp expression in retene exposed fish is delayed to 28 hpf
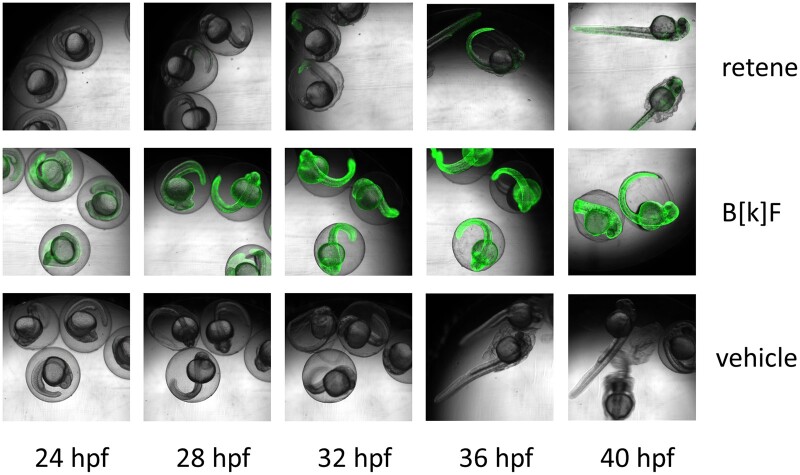
Cyp1acms-gfp Transgenic fish expressing EGFP under a cyp1a promotor region were exposed to retene, the strong AHR ligand benzo[k]fluoranthene, or vehicle control from 6 to 48 hpf and imaged for EGFP expression every 30 min from 23 hpf to 48 hpf to identify Cyp1a protein localization over time (Fig. 5). Control fish exhibited minimal EGFP fluorescence confined to the otic vesicles, skin, urogenital pore, and pectoral fin buds. Benzo[k]fluoranthene exposed fish displayed strong widespread EGFP expression at 23 hpf, the earliest imaged time point, which continued through the time course. Retene exposed fish display control-like EGFP expression in early hours which became noticeably elevated between 26.5 and 28 hpf and increased throughout the exposure. At 26.5 hpf, a single retene-exposed embryo was observed with an increase in fluorescence in the skin of its tail. By 28 hpf, increased fluorescence was observable in all retene exposed embryos in the tail and otic vesicles. Fluorescence increased in retene exposed embryos to include the cardiac region and most of the skin by 33 hpf, and increased in intensity throughout the duration of the exposure. In summary, whereas benzo[k]fluoranthene causes strong, early EGFP fluorescence consistent with a direct agonist, EGFP signal in retene exposed fish is delayed and increases over the same time period as metabolism.
Fig. 5.
Representative overlay images of cyp1acms-gfp containing zebrafish imaged in brightfield and for green fluorescence. Fish were exposed to retene (26 μM), benzo[k]fluoranthene (8.9 μM), or vehicle control (1% DMSO) and imaged every 30 min for green fluorescence and under bright field from 23 to 48 hpf. There were eight replicate fish per treatment and time point.
Teratogenicity requires exposure between 24 and 48 hpf
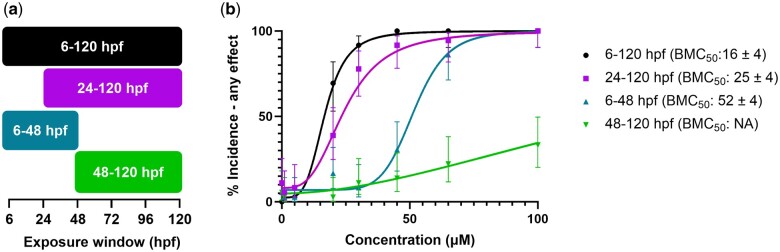
To determine a window of susceptibility to retene-induced developmental toxicity, fish were exposed to the same concentration range during four developmental windows and then screened at 120 hpf (Fig. 6a). Fish exposed to retene 24 to 120 hpf displayed slightly decreased toxicity from fish exposed to 120 hpf (Fig. 6b). Fish exposed from 6 to 48 hpf exhibited reduced toxicity with an BMC50 value of 52 ± 4 µM while fish exposed from 48 to 120 hpf exhibited little response, suggesting exposure during the 24 to 48 hpf time period drives retene-induced developmental toxicity.
Fig. 6.
a) Windows of exposure to retene. b) Concentration-response curves for developmental toxicity screening of retene at four developmental windows. Percent incidence of any effect values are the percent effect in 36 replicate fish and displayed with 95% confidence intervals. BMC50 values include the 95% confidence intervals.
CRISPR-Cas9 produced 2 functional Cyp1a knockouts
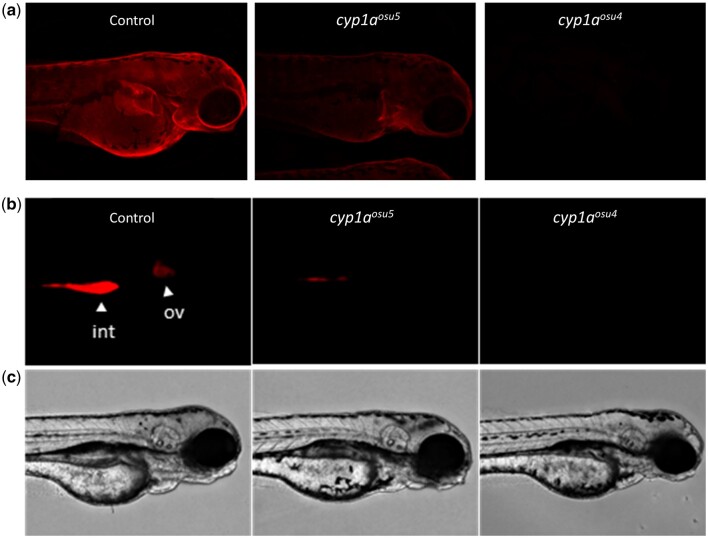
We began investigating in vivo metabolism by constructing a functional Cyp1a KO fish line. Two of the three sgRNA treatments produced heritable mutations early in the cyp1a gene. SgRNA1 produced a 2-bp deletion and frameshift mutation in Exon 2 that resulted in a premature stop codon. SgRNA2 induced 2 consecutive SNPs followed by a 15 bp insertion in Exon 2 leading to an in-frame mutation predicted to produce an elongated protein (Fig. 7a). These were confirmed by SANGER sequencing (Fig. 7b), melt curve analysis, and TAQMAN analysis (data not shown). The line produced from sgRNA1 was submitted to ZFIN as cyp1aosu4 (Zfin ID: ZDB-ALT-240215-1). The line produced with sgRNA2 was submitted to ZFIN as cyp1aosu5 (Zfin ID: ZDB-ALT-240215-2). Homozygous mutant embryos from each line were assessed for Cyp1a protein production and activity. Whole-mount IHC revealed reduced expression of Cyp1a in cyp1aosu5 and no observable expression in cyp1aosu4 (Fig. 8a). Similarly, EROD assays showed reduced Cyp1a metabolic activity in vivo in cyp1aosu5 mutants and no observable activity in cyp1aosu4 mutants (Fig. 8b).
Fig. 7.
Construct description. a) Diagram of Cyp1a transcripts and mutants. b) cDNA of mutated regions. c) Amino Acid Sequence of mutated regions.
Fig. 8.
a) Whole mount IHC shows Cyp1a protein localization in 3 dpf zebrafish exposed to benzo[k]fluoranthene, a potent Cyp1a inducer. Fluorescence signal was normalized to AB background control fish. b) In vivo EROD activity in 72 hpf zebrafish upon retene exposure indicated by red fluorescent signal. Int = intestine; ov = otic vesicle. c) Brightfield images of AB control fish, cyp1aosu5 fish, and cyp1aosu4 fish from row (b).
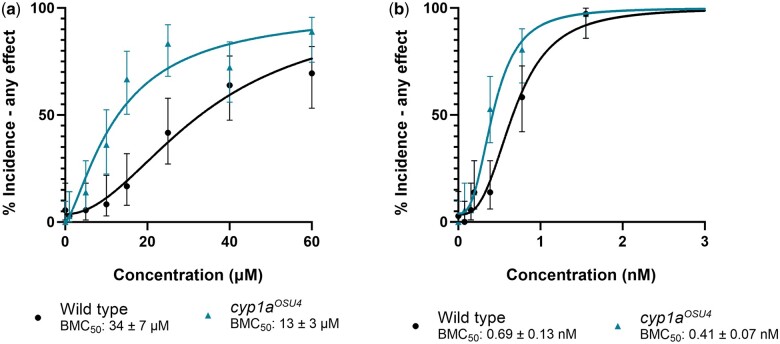
Cyp1aosu4 exhibits increased susceptibility to retene toxicity and slightly increased susceptibility to TCDD
To further validate a functional knockout in these lines, homozygous fish were screened for bioactivity upon exposure to retene, which is biotransformed in fish, and to 2, 3, 7, 8-Tetrachlorodibenzo-p-dioxin, which resists metabolism in zebrafish (Hawkins et al. 2002; Hodson et al. 2007). Cyp1aosu4 fish were more susceptible to retene toxicity having a BMC50 of 13 ± 3 µM compared with the wild type fish which had a BMC50 of 34 ± 7 µM (Fig. 9a). Cyp1aosu4 fish were also more susceptible to TCDD than their wild type counterparts although the effect is less pronounced (BMC50 = 0.41 ± 0.07 nM and 0.69 ± 0.13 nM, respectively) (Fig. 9b). Cyp1aosu5 fish were more susceptible to retene toxicity (BMC50 = 11. ± 7 μM, Fig. S5a), but not significantly more susceptible to TCDD toxicity (BMC = 0.56 ± 0.13, Fig. S5b). Increased retene toxicity in the absence of Cyp1a suggests detoxification by the enzyme overshadows any role it might play in producing an Ahr2 active metabolite.
Fig. 9.
Concentration-response curves for percent incidence of any mortality or morphological effect intervals observed after exposure to (a) retene and (b) TCDD in wild-type and cyp1aosu4 fish. Percent incidence of any effect values are the percent effect in 36 replicate fish and displayed with 95% confidence intervals. BMC50 values include the 95% confidence intervals.
Cyp1b1 and gut microbiome knock out exposures
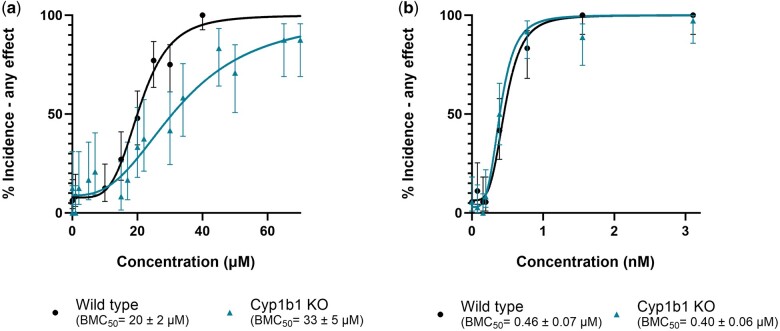
We further investigated in vivo metabolism through developmental toxicity assays in the absence of microbiome metabolism and the important phase I metabolizer Cyp1b1. To investigate the potential role of gut microbiome in retene metabolism, zebrafish embryos were exposed to a range of retene concentrations under conventional and germ-free conditions (Fig. 10). Germ-free zebrafish exhibited a slight decrease in BMC50, although this was largely driven by exposure at 1 concentration and was not statistically significant. We also performed a concentration response in Cyp1b1wh4 fish to test the importance of Cyp1b1 metabolism in retene toxicity and used TCDD as a no-metabolism control (Fig. 11). Retene is less potent in Cyp1b1wh4 fish than in control fish, with a less steep slope in the fitted model causing a BMC50 of 33 ± 5 µM as opposed to the control value of 20 ± 2 µM. Conversely, Cyp1b1wh4 fish exposed to TCDD, which is not metabolized by Cyp1b1, had no change in toxicity from 5D fish exposed to TCDD.
Fig. 10.
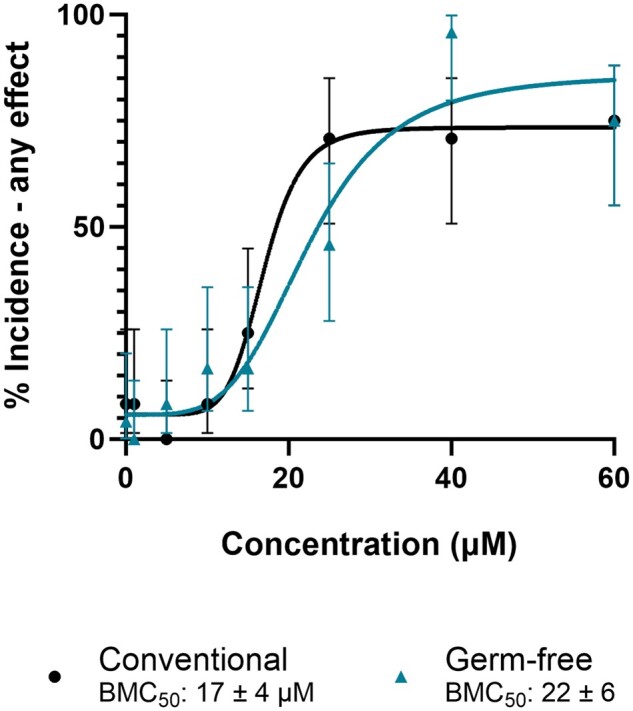
Concentration–response curve for germ free and conventionally reared 5D zebrafish exposed to retene. Percent incidence of any effect values are the percent effect in 24 replicate fish and displayed with 95% confidence intervals. BMC50 values include the 95% confidence intervals.
Fig. 11.
Concentration–response curves for percent incidence of any morphological effect observed after exposure to (a) retene and (b) TCDD in 5D and Cyp1b1wh4 fish. Percent incidence of any effect values are the percent effect in 36 replicate fish for TCDD exposure and 24 replicate fish for retene exposure. Observed effect is displayed with 95% confidence intervals. BMC50 values include the 95% confidence intervals.
Discussion
In vitro and in vivo lack of concordance
Zebrafish ZF-4 luciferase based Ahr2 reporter cells do not respond up to retene to 30 μM, and human HuH-7 AHR reporter cells show a weak response at 30 μM. False negatives due to lack of bioavailability are unlikely. If retene were not sufficiently bioavailable in cellular assays to cause AHR activation, lung cell models exposed to retene would not express AHR-linked genes and 101-L luciferase based AHR reporters would not be sensitive to retene (Jones et al. 2001; Colvin et al. 2024). Instead, the primary difference between the aforementioned cells and the reporter cell lines used in this study is their lack of metabolic capacity. Poor activation of Ahr2 by retene is conserved across teleosts and is likely conserved for human AHR. This observation is corroborated by weak displacement of TCDD from recombinantly expressed rainbow trout Ahr2α and Ahr2β and Ahr2 in PLHC-1 cells (Billiard et al. 2002). Luciferase induction in ZF-4 reporter cells and Ahr2 binding affinity measured by Bisson et al. (2009) show the same rank order and similar magnitude of response in both assays indicating highly conserved binding among teleost Ahr2s for PAHs. Comparison of luciferase induction in zebrafish Ahr2 reporters and human AHR reporters again shows high concordance, suggesting that conserved binding also extends to humans and other mammals. This is consistent with previous homology modeling of binding of TCDD to AHR homologs in mouse, humans, and zebrafish. Interestingly, another reporter in 101-L human hepatoma cells expressing luciferase behind a cyp1a promotor demonstrated moderate induction of luciferase by retene similar to that of benzo[a]pyrene (Jones et al. 2001). The difference in responses observed may indicate the cyp1a promotor is sensitive to other retene-responding transcription factors, or might be due to metabolism within the hepatoma cell line used by Jones et al. (2000) Because the cellular reporter assays in ZF-4 and HuH-7 cells feature minimal metabolism while the 101-L reporters have been shown to actively metabolize PAHs, bioactivation of retene into an AHR-active metabolite is a compelling hypothesis to investigate.
Retene’s potent Ahr2 dependent toxicity in the face of relatively weak binding makes metabolism an even more compelling path of investigation. Retene is 73× and 3× less strong at binding Ahr2 than benzo[b]fluoranthene and 1-methylphenanthrene (Billiard et al. 2002), yet causes severe developmental teratogenicity in 50% of zebrafish embryos by 16.5 μM while benzo[b]fluoranthene and 1-methylphenathrene are not toxic in zebrafish up to 100 uM (data not shown). Potent retene toxicity could be the result of metabolism: the production of a metabolite or metabolites that are strong Ahr2 ligands or which are particularly toxic by another mechanism.
Initial attempts to bioactivate retene and demonstrate AHR activity via concurrent or pre-metabolism by human liver S9 or microsomes did not increase reporter activity (Figs. S1 to S3). Although strong AHR reporter signal with an external metabolizing system would confirm that a retene metabolite activates Ahr2, the negative results of these experiments do not rule out metabolism completely. The bioactivation experiments utilized different metabolic systems and feature different chemical distribution than exposure in a developing zebrafish. We used S9 and microsomes from human liver. Species or tissue specific differences in metabolism may lead to the production of metabolites with different binding affinity between human liver S9, microsomes, and developing zebrafish. Additionally, chemical distribution of metabolites is different in the cellular reporter systems than in the developing zebrafish. The pre-metabolism assay had potential for metabolite loss during extractions and solvent exchange. In both metabolic activation assays, metabolites originate in the media and must accumulate in cells at a concentration high enough to activate AHR. In developing zebrafish, the hydrophobic nature of retene causes high internal exposures. For instance, the 24 hpf body burden of retene of 31 ng/fish equates to roughly 500 μM total internal concentration. If metabolites are formed in the cells of a target tissue, they are available to activate Ahr2; however, if metabolic activation occurs in a tissue other than the target tissue, retene metabolites would have to overcome the limits of cellular uptake. In order to conclusively determine if retene metabolites, rather than the parent compound, activate Ahr2, analytical confirmation of metabolites produced in vivo combined with their exposure in an Ahr2-activation assay is necessary.
Retene metabolism, window of susceptibility, and Ahr2 activation coincide
Exposure to retene between 24 and 48 hpf drives toxicity in developing zebrafish. This window of susceptibility is consistent with the first signs of cardiac toxicity noted by Scott et al. (2011) who found Ahr2 dependent decreased cardiac output, atrial dilation, and reduced cardiac jelly in retene exposed zebrafish arising between 30 and 36 hpf.
Body burdens of retene and detection of retene metabolites in zebrafish indicate metabolism of retene by zebrafish initiates between 24 and 36 hpf. Concentrations of retene in zebrafish increased from 12 to 24 hpf, remained relatively constant from 24 to 36 hpf, and decreased thereafter. Without kinetic modeling, it is difficult to assess the relative contribution of metabolism to the stable body burden of retene between 24 and 36 hpf. Never the less, by 36 hpf two retene metabolites appear in the tissue extracts of retene exposed fish, directly confirming the onset of metabolism beginning between 24 and 36 hpf. This result aligns with the first measurable EROD activity in unexposed zebrafish at 24 hpf in previous studies (Bräunig et al. 2015). A steep decline in retene body burden and increase in number and abundance of retene metabolites between 36 and 48 hpf indicates more extensive retene metabolism after 36 hpf. Metabolism of retene arises during the window of susceptibility, before or during the first indications of toxicity in developing zebrafish.
EGFP expression is delayed in retene exposed cyp1acms-gfp fish. Benzo[k]fluoranthene elicited strong EGFP fluorescence at 23 hpf, the earliest observed time point. Increases in EGFP fluorescence do not appear in retene exposed fish until 26.5 to 28 hpf, after which it increases gradually to a strong signal by 48 hpf. This is consistent with RNA sequencing results that show retene exposure induces cyp1a at levels similar to direct Ahr2 ligands like benzo[k]fluoranthene and benz[a]anthracene by 48 hpf (Shankar et al. 2019). Benzo[k]fluoranthene shows strong AHR activation before the onset of metabolism, whereas retene Ahr2 activation appears to increase gradually with the onset of metabolism. This suggests metabolism must be present for retene's Ahr2 mediated response.
Besides overall retene metabolism and possible bioactivation in the zebrafish embryo, localized, tissue-specific retene metabolism could drive the observed toxicity. For example, Scott et al. (2011) noted Cyp1a in cardiac cells at 48 but not 72 hpf despite static renewal of retene every 24 hpf and attributed this to the fish’s metabolism having developed sufficiently to protect the heart from Ahr2 activation by retene. The decline in retene body burden we note from 36 to 48 hpf, and the decreased susceptibility to retene after 48 hpf corroborate these findings and suggest that extensive metabolism after 48 hpf protects the fish from overt toxicity.
Sources of metabolism
Given the onset of toxicity between 30 and 36 hpf, and window of susceptibility between 24 and 48 hpf, any metabolizing enzymes producing toxic retene metabolites must be present between 24 and 36 hpf. Cyp1a and cyp1b are among the CYPs expressed in zebrafish by 24 hpf (Bräunig et al. 2015; Nawaji et al. 2020). Little is known about the microbiome before gut development in zebrafish, but emerging evidence shows that the microbiome can alter PAH bioactivity (Van de Wiele et al. 2005; Quintanilla-Mena et al. 2021; Stagaman et al. 2024).
We eliminated the microbiome, Cyp1a, and Cyp1b1 to assess the effect of their metabolism on retene toxicity. In this study, elimination of the microbiome did not significantly change retene toxicity, indicating it is not a mediator of retene teratogenicity. Cyp1b1wh4 fish were slightly less susceptible than wild type fish to retene toxicity. This indicates that Cyp1b1 metabolism may contribute to retene toxicity; however, given the modest effect, it is unlikely that Cyp1b1 is the only mediator. Knockout of Cyp1a led to increased retene toxicity in both Cyp1a knockout lines developed in this study. Contrary to inducing a toxic metabolite, it appears that Cyp1a is crucial for the detoxification of retene in larval zebrafish. The role of Cyp1a in retene and broader PAH toxicity is conflicting in the literature. Scott et al. (2011) noted no change in retene toxicity with morpholino knock down of Cyp1a, but their experiment confirmed continued toxicity in exposures at a single high exposure concentration. Hodson et al. (2007) studied retene toxicity in the context of Cyp1 inhibition by alpha naphthoflavone (ANF) in rainbow trout. ANF is a nonspecific inhibitor of multiple Cyp1 isoforms and a weak AHR antagonist (Juvonen et al. 2020). Retene toxicity in juvenile rainbow trout increases with co-exposure to ANF at 32 and 100 μg/L and is attenuated with co-exposure to 320 μg/L ANF (Hodson et al. 2007). Hodson et al. (2007) conclude that retene metabolites drive retene toxicity because near-complete inhibition of apparent metabolism with 320 μg/l prevents retene toxicity in rainbow trout. Their analysis is complicated some by ANF being a weak AHR agonist and inhibiting AHR function in the presence of strong AHR agonists (Merchant et al. 1990; Santostefano et al. 1993). Decreased retene toxicity at the highest concentrations of ANF might instead arise from Ahr2 inhibition instead of elimination of retene metabolites. Although the metabolite explanation for retene toxicity remains possible, it has yet to be proven. Additionally cyp1c1, cyp1c2, cyp1d1, cyp2y3, and cyp3c1, are all expressed to some degree by 24 hpf in larval zebrafish and have yet to be investigated for their role in retene toxicity (Nawaji et al. 2020).
Metabolite analysis
An alternative route to investigating the retene metabolite hypothesis exists in identifying retene metabolite structures during early zebrafish development. Analysis revealed two distinct retene metabolites appearing by 36 hpf, and an additional two by 48 hpf. The highest signal intensity at both 36 and 48 hpf had a m/z of 263.10 (C18H16O2) and possibly corresponds retene-dione (Fig. 4b). PAHs are metabolized to polynuclear diones, which have carbonyls on multiple rings, or orthoquinones in both mammals and teleosts (Smithgall et al. 1988; Staretz et al. 1997; Palackal et al. 2002; Shappell et al. 2003; Scornaienchi et al. 2010). Polynuclear diones are produced by the radical cation pathway, but this typically occurs in larger PAHS with lower ionization energies, making this pathway less likely in retene metabolism (Kuroda 1964; Cavalieri and Rogan 1985). Retene is metabolize to orthoquinones in human HepG2 cells (Huang et al. 2017b). Formation of orthoquinones has been demonstrated for chrysene using rainbow trout liver microsomes, indicating it is a relevant pathway for teleosts (Shappell et al. 2003). Dehydrogenation of the isopropyl group from m/z 263.10 may have formed the metabolite at m/z 261.09 (C18H14O2Fig. 4b). The retene metabolite at m/z 249.12 (C18H18O) may be a retene epoxide, a phenol, or side chain alcohol. Phenols and side chain alcohols are often the dominant phase I metabolites of PAHs, with the latter typically being more abundant (Miranda et al. 1997; Shappell et al. 2003; Huang et al. 2017a; Wang et al. 2022a, 2022b). Interestingly, Huang et al found no evidence of side chain alcohols in their analysis of retene metabolism, indicating retene may act differently than other alkylated phenanthrene (Huang et al. 2017b). Determining whether this is also true in developing zebrafish will likely require confirmation with synthetic standards. Finally, the second most abundant metabolite which occurs at m/z 279.1 (C18H2O3) likely includes both the dione and phenol/alcohol functionalities.
If orthoquinones are the dominant metabolite of retene metabolism, this could account for the high toxicity of retene compared with other PAHs. Retene is mutagenic and causes oxidative stress in fish and in human cells (Maria et al. 2005; Peixoto et al. 2019; da Silva Junior et al. 2021; Scaramboni et al. 2023). Redox cycling of orthoquinones with catechols produces damaging reactive oxygen species (Flowers-Geary et al. 1996; Yu et al. 2002; Park et al. 2008) and orthoquinones are strong electrophiles capable of reacting with guanine nucleotides in DNA (Shou et al. 1993; McCoull et al. 1999). Formation of quinones in target tissues may lead to its potent developmental toxicity.
Future directions
This study presents compelling evidence that retene is not a direct Ahr2 ligand in zebrafish. We show that metabolic onset overlaps with the window of susceptibility, Cyp1a expression, and Ahr2 dependent cardiotoxicity observed by Scott et al. (2011) but were not able to confirm a metabolic source or toxic retene metabolites. Future analysis will focus on in silico docking of retene metabolites to mammalian AHR and zebrafish Ahr2 to produce candidates for analytical verification, in vivo toxicity testing, and confirmation of Ahr2 binding.
Conclusions
Retene presents a yet unresolved case study in which our knowledge of adverse outcome pathways and mechanism based in vitro data does not predict the in vivo response. Retene is a potent Ahr2 dependent toxicant in zebrafish. Apparent AHR mediated retene toxicity extends to other fish and human cellular models, yet it is a weak ligand for both human AHR and teleost Ahr2. In the case of retene, ZF-4 and Huh-7 reporter systems lacking metabolic competence fail to predict AHR activation, whereas systems with metabolic competence such as zebrafish, primary human bronchial epithelial cells, and human hepatoma 101-L cells show clear AHR activation. Further analysis may identify the precise role of metabolism in mediating the toxicity of retene; however, it is unlikely retene is the only chemical, nor AHR the only target, for which this type of disconnect exists. As new approach methodologies seek to better protect human and environmental health via hazard-based mechanistic in vitro screening, it will be necessary to incorporate more complex integrated systems to protect for blind spots and uncover new and important pathways of toxicity.
Supplementary Material
Acknowledgments
The authors would like to thank the screening staff at Sinnhuber Aquatic Research Laboratory for fish husbandry and screening support. Thank you to the Oregon State University/Pacific Northwest National Laboratory Superfund Research Center Chemical Mixtures Core for providing test compounds and Richard Scott and the Laboratory of Dr Kim Anderson for analytical chemistry support. Pacific Northwest National Laboratory is a multi-program national laboratory operated by Battelle for the U.S. Department of Energy under Contract DE-AC05-76RL01830.
Contributor Information
Christian I Rude, Environmental and Molecular Toxicology Department, Oregon State University, Corvallis, OR 97333, United States.
Lindsay B Wilson, Environmental and Molecular Toxicology Department, Oregon State University, Corvallis, OR 97333, United States.
Jane La Du, Environmental and Molecular Toxicology Department, Oregon State University, Corvallis, OR 97333, United States.
Priscila M Lalli, Biological Sciences Division, Pacific Northwest National Laboratory, Richland, WA 99352, United States.
Sean M Colby, Biological Sciences Division, Pacific Northwest National Laboratory, Richland, WA 99352, United States.
Katherine J Schultz, Biological Sciences Division, Pacific Northwest National Laboratory, Richland, WA 99352, United States.
Jordan N Smith, Environmental and Molecular Toxicology Department, Oregon State University, Corvallis, OR 97333, United States; Biological Sciences Division, Pacific Northwest National Laboratory, Richland, WA 99352, United States.
Katrina M Waters, Environmental and Molecular Toxicology Department, Oregon State University, Corvallis, OR 97333, United States; Biological Sciences Division, Pacific Northwest National Laboratory, Richland, WA 99352, United States.
Robyn L Tanguay, Environmental and Molecular Toxicology Department, Oregon State University, Corvallis, OR 97333, United States.
Supplementary material
Supplementary material is available at Toxicological Sciences online.
Funding
This research reported in this manuscript was supported by the National Institute of Environmental Health Sciences of the National Institutes of Health under Award Numbers P42 ES016465, T32 ES007060, and P30 ES030287. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflicts of interest. The authors declare no conflict of interest.
References
- Ainerua MO, Tinwell J, Kompella SN, Sørhus E, White KN, van Dongen BE, Shiels HA.. 2020. Understanding the cardiac toxicity of the anthropogenic pollutant phenanthrene on the freshwater indicator species, the brown trout (Salmo trutta): from whole heart to cardiomyocytes. Chemosphere. 239:124608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan SE, Smith BW, Anderson KA.. 2012. Impact of the deepwater horizon oil spill on bioavailable polycyclic aromatic hydrocarbons in Gulf of Mexico coastal waters. Environ Sci Technol. 46(4):2033–2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan SE, Sower GJ, Anderson KA.. 2011. Estimating risk at a superfund site using passive sampling devices as biological surrogates in human health risk models. Chemosphere. 85(6):920–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson AL, Dubanksy BD, Wilson LB, Tanguay RL, Rice CD.. 2022. Development and applications of a zebrafish (Danio rerio) CYP1A-targeted monoclonal antibody (CRC4) with reactivity across vertebrate taxa: evidence for a conserved CYP1A epitope. Toxics. 10(7):404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson KA, Szelewski MJ, Wilson G, Quimby BD, Hoffman PD.. 2015. Modified ion source triple quadrupole mass spectrometer gas chromatograph for polycyclic aromatic hydrocarbon analyses. J Chromatogr A. 1419:89–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreasen EA, Hahn ME, Heideman W, Peterson RE, Tanguay RL.. 2002. The zebrafish (Danio rerio) aryl hydrocarbon receptor Type 1 is a novel vertebrate receptor. Mol Pharmacol. 62(2):234–249. [DOI] [PubMed] [Google Scholar]
- Bakkers J. 2011. Zebrafish as a model to study cardiac development and human cardiac disease. Cardiovasc Res. 91(2):279–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri O, Ognio E, Rossi O, Astigiano S, Rossi L.. 1986. Embryotoxicity of benzo(a)pyrene and some of its synthetic derivatives in Swiss mice. Cancer Res. 46(1):94–98. [PubMed] [Google Scholar]
- Billiard SM, Hahn ME, Franks DG, Peterson RE, Bols NC, Hodson PV.. 2002. Binding of polycyclic aromatic hydrocarbons (PAHs) to teleost aryl hydrocarbon receptors (AHRs). Comp Biochem Physiol B Biochem Mol Biol. 133(1):55–68. [DOI] [PubMed] [Google Scholar]
- Bisson WH, Koch DC, O'Donnell EF, Khalil SM, Kerkvliet NI, Tanguay RL, Abagyan R, Kolluri SK.. 2009. Modeling of the aryl hydrocarbon receptor (AHR) ligand binding domain and its utility in virtual ligand screening to predict new AHR ligands. J Med Chem. 52(18):5635–5641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boada LD, Henríquez-Hernández LA, Navarro P, Zumbado M, Almeida-González M, Camacho M, Álvarez-León EE, Valencia-Santana JA, Luzardo OP.. 2015. Exposure to polycyclic aromatic hydrocarbons (PAHs) and bladder cancer: evaluation from a gene-environment perspective in a hospital-based case-control study in the canary islands (Spain). Int J Occup Environ Health. 21(1):23–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bräunig J, Schiwy S, Broedel O, Müller Y, Frohme M, Hollert H, Keiter SH.. 2015. Time-dependent expression and activity of cytochrome P450 1s in early life-stages of the zebrafish (Danio rerio). Environ Sci Pollut Res Int. 22(21):16319–16328. [DOI] [PubMed] [Google Scholar]
- Brette F, Shiels HA, Galli GLJ, Cros C, Incardona JP, Scholz NL, Block BA.. 2017. A novel cardiotoxic mechanism for a pervasive global pollutant. Sci Rep. 7(1):41476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkworth LC, Hodson PV, Tabash S, Lee P.. 2003. CYP1A induction and blue sac disease in early developmental stages of rainbow trout (Oncorhynchus mykiss) exposed to retene. J Toxicol Environ Health A. 66(7):627–646. [DOI] [PubMed] [Google Scholar]
- Cavalieri E, Rogan E.. 1985. Role of radical cations in aromatic hydrocarbon carcinogenesis. Environ Health Perspect. 64:69–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu J, Sadler KC.. 2009. New school in liver development: lessons from zebrafish. Hepatology. 50(5):1656–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colby SM, Chang CH, Bade JL, Nunez JR, Blumer MR, Orton DJ, Bloodsworth KJ, Nakayasu ES, Smith RD, Ibrahim YM, et al. 2022. Deimos: an open-source tool for processing high-dimensional mass spectrometry data. Anal Chem. 94(16):6130–6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colvin VC, Bramer LM, Rivera BN, Pennington JM, Waters KM, Tilton SC.. 2024. Modeling PAH mixture interactions in a human in vitro organotypic respiratory model. Int J Mol Sci. 25(8):4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva Junior FC, Agues-Barbosa T, Luchiari AC, Batistuzzo de Medeiros SR.. 2021. Genotoxicity and behavioral alterations induced by retene in adult zebrafish. J Environ Chem Eng. 9(6):106518. [Google Scholar]
- Das L, Patel B, Patri M.. 2019. Adolescence benzo[a]pyrene treatment induces learning and memory impairment and anxiolytic like behavioral response altering neuronal morphology of hippocampus in adult male wistar rats. Toxicol Rep. 6:1104–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira Alves N, Vessoni AT, Quinet A, Fortunato RS, Kajitani GS, Peixoto MS, Hacon SS, Artaxo P, Saldiva P, Menck CFM, et al. 2017. Biomass burning in the amazon region causes DNA damage and cell death in human lung cells. Sci Rep. 7(1):10937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon HM, Bramer LM, Scott RP, Calero L, Holmes D, Gibson EA, Cavalier HM, Rohlman D, Miller RL, Calafat AM, et al. 2022. Evaluating predictive relationships between wristbands and urine for assessment of personal PAH exposure. Environ Int. 163:107226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djoumbou-Feunang Y, Fiamoncini J, Gil-de-la-Fuente A, Greiner R, Manach C, Wishart DS.. 2019. Biotransformer: a comprehensive computational tool for small molecule metabolism prediction and metabolite identification. J Cheminform. 11(1):2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming CR, Di Giulio RT.. 2011. The role of CYP1A inhibition in the embryotoxic interactions between hypoxia and polycyclic aromatic hydrocarbons (PAHs) and PAH mixtures in zebrafish (Danio rerio). Ecotoxicology. 20(6):1300–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flowers-Geary L, Bleczinki W, Harvey RG, Penning TM.. 1996. Cytotoxicity and mutagenicity of polycyclic aromatic hydrocarbon ortho-quinones produced by dihydrodiol dehydrogenase. Chem Biol Interact. 99(1–3):55–72. [DOI] [PubMed] [Google Scholar]
- Gao D, Wang C, Xi Z, Zhou Y, Wang Y, Zuo Z.. 2017. Early-life benzo[a]pyrene exposure causes neurodegenerative syndromes in adult zebrafish (Danio rerio) and the mechanism involved. Toxicol Sci. 157(1):74–84. [DOI] [PubMed] [Google Scholar]
- Garcia GR, Bugel SM, Truong L, Spagnoli S, Tanguay RL.. 2018. AHR2 required for normal behavioral responses and proper development of the skeletal and reproductive systems in zebrafish. PLoS One. 13(3):e0193484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garland MA, Geier MC, Bugel SM, Shankar P, Dunham CL, Brown JM, Tilton SC, Tanguay RL.. 2020. Aryl hydrocarbon receptor mediates larval zebrafish fin duplication following exposure to benzofluoranthenes. Toxicol Sci. 176(1):46–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geier MC, Chlebowski AC, Truong L, Massey Simonich SL, Anderson KA, Tanguay RL.. 2018a. Comparative developmental toxicity of a comprehensive suite of polycyclic aromatic hydrocarbons. Arch Toxicol. 92(2):571–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geier MC, James Minick D, Truong L, Tilton S, Pande P, Anderson KA, Teeguardan J, Tanguay RL.. 2018b. Systematic developmental neurotoxicity assessment of a representative PAH superfund mixture using zebrafish. Toxicol Appl Pharmacol. 354:115–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentile I, Vezzoli V, Martone S, Totaro MG, Bonomi M, Persani L, Marelli F.. 2023. Short-term exposure to benzo(a)pyrene causes disruption of GnRH network in zebrafish embryos. Int J Mol Sci. 24(8):6913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach GF, Wingert RA.. 2013. Kidney organogenesis in the zebrafish: insights into vertebrate nephrogenesis and regeneration. Wiley Interdiscip Rev Dev Biol. 2(5):559–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghetu CC, Rohlman D, Smith BW, Scott RP, Adams KA, Hoffman PD, Anderson KA.. 2022. Wildfire impact on indoor and outdoor PAH air quality. Environ Sci Technol. 56(14):10042–10052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstone J, McArthur A, Kubota A, Zanette J, Parente T, Jonsson M, Nelson D, Stegeman J.. 2010. Identification and developmental expression of the full complement of cytochrome P450 genes in zebrafish. BMC Genomics. 11(1):643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodale BC, La Du J, Tilton SC, Sullivan CM, Bisson WH, Waters KM, Tanguay RL.. 2015. Ligand-specific transcriptional mechanisms underlie aryl hydrocarbon receptor-mediated developmental toxicity of oxygenated PAHs. Toxicol Sci. 147(2):397–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravato C, Santos MA.. 2002. Juvenile sea bass liver biotransformation and erythrocytic genotoxic responses to pulp mill contaminants. Ecotoxicol Environ Saf. 53(1):104–112. [DOI] [PubMed] [Google Scholar]
- Gustafson P, Ostman C, Sällsten G.. 2008. Indoor levels of polycyclic aromatic hydrocarbons in homes with or without wood burning for heating. Environ Sci Technol. 42(14):5074–5080. [DOI] [PubMed] [Google Scholar]
- Hahn ME, Karchner SI, Merson RR.. 2017. Diversity as opportunity: insights from 600 million years of AHR evolution. Curr Opin Toxicol. 2:58–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins SA, Billiard SM, Tabash SP, Brown RS, Hodson PV.. 2002. Altering cytochrome P4501a activity affects polycyclic aromatic hydrocarbon metabolism and toxicity in rainbow trout (Oncorhynchus mykiss). Environ Toxicol Chem. 21(9):1845–1853. [PubMed] [Google Scholar]
- Hill JT, Demarest BL, Bisgrove BW, Su YC, Smith M, Yost HJ.. 2014. Poly peak parser: method and software for identification of unknown indels using Sanger sequencing of polymerase chain reaction products. Dev Dyn. 243(12):1632–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho CC, Wu WT, Lin YJ, Weng CY, Tsai MH, Tsai HT, Chen YC, Yet SF, Lin P.. 2022. Aryl hydrocarbon receptor activation-mediated vascular toxicity of ambient fine particulate matter: contribution of polycyclic aromatic hydrocarbons and osteopontin as a biomarker. Part Fibre Toxicol. 19(1):43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodson PV, Qureshi K, Noble CA, Akhtar P, Brown RS.. 2007. Inhibition of CYP1A enzymes by alpha-naphthoflavone causes both synergism and antagonism of retene toxicity to rainbow trout (Oncorhynchus mykiss). Aquat Toxicol. 81(3):275–285. [DOI] [PubMed] [Google Scholar]
- Hoyberghs J, Bars C, Ayuso M, Van Ginneken C, Foubert K, Van Cruchten S.. 2021. DMSO concentrations up to 1% are safe to be used in the zebrafish embryo developmental toxicity assay. Front Toxicol. 3:804033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M, Mesaros C, Hackfeld LC, Hodge RP, Blair IA, Penning TM.. 2017a. Potential metabolic activation of representative alkylated polycyclic aromatic hydrocarbons 1-methylphenanthrene and 9-ethylphenanthrene associated with the Deepwater Horizon oil spill in human hepatoma (HepG2) cells. Chem Res Toxicol. 30(12):2140–2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M, Mesaros C, Hackfeld LC, Hodge RP, Zang T, Blair IA, Penning TM.. 2017b. Potential metabolic activation of a representative c4-alkylated polycyclic aromatic hydrocarbon retene (1-methyl-7-isopropyl-phenanthrene) associated with the deepwater horizon oil spill in human hepatoma (HepG2) cells. Chem Res Toxicol. 30(4):1093–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussar E, Richards S, Lin ZQ, Dixon RP, Johnson KA.. 2012. Human health risk assessment of 16 priority polycyclic aromatic hydrocarbons in soils of Chattanooga, Tennessee, USA. Water Air Soil Pollut. 223(9):5535–5548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Incardona JP, Carls MG, Teraoka H, Sloan CA, Collier TK, Scholz NL.. 2005. Aryl hydrocarbon receptor-independent toxicity of weathered crude oil during fish development. Environ Health Perspect. 113(12):1755–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Incardona JP, Collier TK, Scholz NL.. 2004. Defects in cardiac function precede morphological abnormalities in fish embryos exposed to polycyclic aromatic hydrocarbons. Toxicol Appl Pharmacol. 196(2):191–205. [DOI] [PubMed] [Google Scholar]
- Incardona JP, Day HL, Collier TK, Scholz NL.. 2006. Developmental toxicity of 4-ring polycyclic aromatic hydrocarbons in zebrafish is differentially dependent on AH receptor isoforms and hepatic cytochrome P4501A metabolism. Toxicol Appl Pharmacol. 217(3):308–321. [DOI] [PubMed] [Google Scholar]
- Jones JM, Anderson JW, Tukey RH.. 2000. Using the metabolism of PAHs in a human cell line to characterize environmental samples. Environ Toxicol Pharmacol. 8(2):119–126. [DOI] [PubMed] [Google Scholar]
- Jones JM, Anderson JW, Wiegel JV, Tukey RH.. 2001. Application of P450 reporter gene system (RGS) in the analysis of sediments near pulp and paper mills. Biomarkers. 6(6):406–416. [DOI] [PubMed] [Google Scholar]
- Juvonen RO, Jokinen EM, Javaid A, Lehtonen M, Raunio H, Pentikäinen OT.. 2020. Inhibition of human cyp1 enzymes by a classical inhibitor α-naphthoflavone and a novel inhibitor n-(3, 5-dichlorophenyl)cyclopropanecarboxamide: an in vitro and in silico study. Chem Biol Drug Des. 95(5):520–533. [DOI] [PubMed] [Google Scholar]
- Karchner SI, Franks DG, Hahn ME.. 2005. AHR1B, a new functional aryl hydrocarbon receptor in zebrafish: tandem arrangement of AHR1B and AHR2 genes. Biochem J. 392(Pt 1):153–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KH, Park HJ, Kim JH, Kim S, Williams DR, Kim MK, Jung YD, Teraoka H, Park HC, Choy HE, et al. 2013. CYP1A reporter zebrafish reveals target tissues for dioxin. Aquat Toxicol. 134–135:57–65. [DOI] [PubMed] [Google Scholar]
- Kimmel CB. 1993. Patterning the brain of the zebrafish embryo. Annu Rev Neurosci. 16:707–732. [DOI] [PubMed] [Google Scholar]
- Knecht AL, Goodale BC, Truong L, Simonich MT, Swanson AJ, Matzke MM, Anderson KA, Waters KM, Tanguay RL.. 2013. Comparative developmental toxicity of environmentally relevant oxygenated PAHs. Toxicol Appl Pharmacol. 271(2):266–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knecht AL, Truong L, Simonich MT, Tanguay RL.. 2017. Developmental benzo[a]pyrene (b[a]p) exposure impacts larval behavior and impairs adult learning in zebrafish. Neurotoxicol Teratol. 59:27–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koistinen J, Lehtonen M, Tukia K, Soimasuo M, Lahtipera M, Oikari A.. 1998. Identification of lipophilic pollutants discharged from a Finnish pulp and paper mill. Chemosphere. 37(2):219–235. [DOI] [PubMed] [Google Scholar]
- Kompella SN, Brette F, Hancox JC, Shiels HA.. 2021. Phenanthrene impacts zebrafish cardiomyocyte excitability by inhibiting IKr and shortening action potential duration. J Gen Physiol. 153(2):e202012733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda H. 1964. Ionization potentials of polycyclic aromatic hydrocarbons. Nature. 201(4925):1214–1215. [Google Scholar]
- Leppänen H, Oikari A.. 2001. Retene and resin acid concentrations in sediment profiles of a lake recovering from exposure to pulp mill effluents. J Paleolimnol. 25(3):367–374. [Google Scholar]
- Ma J, Tan Q, Nie X, Zhou M, Wang B, Wang X, Cheng M, Ye Z, Xie Y, Wang D, et al. 2022. Longitudinal relationships between polycyclic aromatic hydrocarbons exposure and heart rate variability: exploring the role of transforming growth factor-beta in a general Chinese population. J Hazard Mater. 425:127770. [DOI] [PubMed] [Google Scholar]
- Mallah MA, Mallah MA, Liu Y, Xi H, Wang W, Feng F, Zhang Q.. 2021. Relationship between polycyclic aromatic hydrocarbons and cardiovascular diseases: a systematic review. Front Public Health. 9:763706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandrell D, Truong L, Jephson C, Sarker MR, Moore A, Lang C, Simonich MT, Tanguay RL.. 2012. Automated zebrafish chorion removal and single embryo placement: optimizing throughput of zebrafish developmental toxicity screens. J Lab Autom. 17(1):66–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maria VL, Correia AC, Santos MA.. 2005. Anguilla L. Liver EROD induction and genotoxic responses after retene exposure. Ecotoxicol Environ Saf. 61(2):230–238. [DOI] [PubMed] [Google Scholar]
- Marris CR, Kompella SN, Miller MR, Incardona JP, Brette F, Hancox JC, Sorhus E, Shiels HA.. 2020. Polyaromatic hydrocarbons in pollution: a heart-breaking matter. J Physiol. 598(2):227–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin FJ, Amode MR, Aneja A, Austine-Orimoloye O, Azov AG, Barnes I, Becker A, Bennett R, Berry A, Bhai J, et al. 2023. Ensembl 2023. Nucleic Acids Res. 51(D1):D933–D941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoull KD, Rindgen D, Blair IA, Penning TM.. 1999. Synthesis and characterization of polycyclic aromatic hydrocarbon o-quinone depurinating N7-guanine adducts. Chem Res Toxicol. 12(3):237–246. [DOI] [PubMed] [Google Scholar]
- McLarnan SM, Bramer LM, Dixon HM, Scott RP, Calero L, Holmes D, Gibson EA, Cavalier HM, Rohlman D, Miller RL, et al. 2024. Predicting personal PAH exposure using high dimensional questionnaire and wristband data. J Expo Sci Environ Epidemiol. 34(4):679–687. [DOI] [PubMed] [Google Scholar]
- Merchant M, Arellano L, Safe S.. 1990. The mechanism of action of alpha-naphthoflavone as an inhibitor of 2,3,7,8-tetrachlorodibenzo-p-dioxin-induced CYP1A1 gene expression. Arch Biochem Biophys. 281(1):84–89. [DOI] [PubMed] [Google Scholar]
- Minick DJ, Anderson KA.. 2017. Diffusive flux of PAHs across sediment-water and water-air interfaces at urban superfund sites. Environ Toxicol Chem. 36(9):2281–2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda CL, Henderson MC, Williams DE, Buhler DR.. 1997. In vitro metabolism of 7,12-dimethylbenz[a]anthracene by rainbow trout liver microsomes and trout P450 isoforms. Toxicol Appl Pharmacol. 142(1):123–132. [DOI] [PubMed] [Google Scholar]
- Mu J, Jin F, Wang J, Wang Y, Cong Y.. 2016. The effects of CYP1A inhibition on alkyl-phenanthrene metabolism and embryotoxicity in marine medaka (Oryzias melastigma). Environ Sci Pollut Res Int. 23(11):11289–11297. [DOI] [PubMed] [Google Scholar]
- Nawaji T, Yamashita N, Umeda H, Zhang S, Mizoguchi N, Seki M, Kitazawa T, Teraoka H.. 2020. Cytochrome P450 expression and chemical metabolic activity before full liver development in zebrafish. Pharmaceuticals (Basel). 13(12):456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebert DW. 2017. Aryl hydrocarbon receptor (AHR): “pioneer member” of the basic-helix/loop/helix per-Arnt-sim (bHLH/PAS) family of “sensors” of foreign and endogenous signals. Prog Lipid Res. 67:38–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DR. 2009. The cytochrome P450 homepage. Hum Genomics. 4(1):59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olasehinde TA, Olaniran AO.. 2022. Neurotoxicity of polycyclic aromatic hydrocarbons: a systematic mapping and review of neuropathological mechanisms. Toxics. 10(8):417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palackal NT, Lee SH, Harvey RG, Blair IA, Penning TM.. 2002. Activation of polycyclic aromatic hydrocarbon trans-dihydrodiol proximate carcinogens by human aldo-keto reductase (AKR1C) enzymes and their functional overexpression in human lung carcinoma (A549) cells. J Biol Chem. 277(27):24799–24808. [DOI] [PubMed] [Google Scholar]
- Park JH, Mangal D, Tacka KA, Quinn AM, Harvey RG, Blair IA, Penning TM.. 2008. Evidence for the aldo-keto reductase pathway of polycyclic aromatic trans-dihydrodiol activation in human lung A549 cells. Proc Natl Acad Sci USA. 105(19):6846–6851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiffer J, Grova N, Hidalgo S, Salquèbre G, Rychen G, Bisson JF, Appenzeller BMR, Schroeder H.. 2016. Behavioral toxicity and physiological changes from repeated exposure to fluorene administered orally or intraperitoneally to adult male wistar rats: a dose-response study. Neurotoxicology. 53:321–333. [DOI] [PubMed] [Google Scholar]
- Peixoto MS, da Silva Junior FC, de Oliveira Galvao MF, Roubicek DA, de Oliveira Alves N, Batistuzzo de Medeiros SR.. 2019. Oxidative stress, mutagenic effects, and cell death induced by retene. Chemosphere. 231:518–527. [DOI] [PubMed] [Google Scholar]
- Perone DM. 2024. Knocking out the zebrafish cyp1b1 gene alters metabolomic profiles and neurobehavioral functions. In preparation.
- Prasch AL, Teraoka H, Carney SA, Dong W, Hiraga T, Stegeman JJ, Heideman W, Peterson RE.. 2003. Aryl hydrocarbon receptor 2 mediates 2,3,7,8-tetrachlorodibenzo-p-dioxin developmental toxicity in zebrafish. Toxicol Sci. 76(1):138–150. [DOI] [PubMed] [Google Scholar]
- Quintanilla-Mena M, Vega-Arreguin J, Del Rio-Garcia M, Patino-Suarez V, Peraza-Echeverria S, Puch-Hau C.. 2021. The effect of benzo[a]pyrene on the gut microbiota of Nile tilapia (Oreochromis niloticus). Appl Microbiol Biotechnol. 105(20):7935–7947. [DOI] [PubMed] [Google Scholar]
- Ramdahl T. 1983. Retene—a molecular marker of wood combustion in ambient air. Nature. 306(5943):580–582. [Google Scholar]
- Rendic S, Guengerich FP.. 2015. Survey of human oxidoreductases and cytochrome P450 enzymes involved in the metabolism of xenobiotic and natural chemicals. Chem Res Toxicol. 28(1):38–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samon SM, Rohlman D, Tidwell LG, Hoffman PD, Oluyomi AO, Anderson KA.. 2022. Associating increased chemical exposure to hurricane Harvey in a longitudinal panel using silicone wristbands. Int J Environ Res Public Health. 19(11):6670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santostefano M, Merchant M, Arellano L, Morrison V, Denison MS, Safe S.. 1993. Alpha-naphthoflavone-induced CYP1A1 gene expression and cytosolic aryl hydrocarbon receptor transformation. Mol Pharmacol. 43(2):200–206. [PubMed] [Google Scholar]
- Sarma SN, Blais JM, Chan HM.. 2017. Neurotoxicity of alkylated polycyclic aromatic compounds in human neuroblastoma cells. J Toxicol Environ Health A. 80(5):285–300. [DOI] [PubMed] [Google Scholar]
- Scaramboni C, Arruda Moura Campos ML, Junqueira Dorta D, Palma de Oliveira D, Batistuzzo de Medeiros SR, de Oliveira Galvão MF, Dreij K.. 2023. Reactive oxygen species-dependent transient induction of genotoxicity by retene in human liver HepG2 cells. Toxicol In Vitro. 91:105628. [DOI] [PubMed] [Google Scholar]
- Scornaienchi ML, Thornton C, Willett KL, Wilson JY.. 2010. Functional differences in the cytochrome P450 1 family enzymes from zebrafish (Danio rerio) using heterologously expressed proteins. Arch Biochem Biophys. 502(1):17–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott JA, Incardona JP, Pelkki K, Shepardson S, Hodson PV.. 2011. AHR2-mediated, CYP1A-independent cardiovascular toxicity in zebrafish (Danio rerio) embryos exposed to retene. Aquat Toxicol. 101(1):165–174. [DOI] [PubMed] [Google Scholar]
- Shankar P, Dasgupta S, Hahn ME, Tanguay RL.. 2020. A review of the functional roles of the zebrafish aryl hydrocarbon receptors. Toxicol Sci. 178(2):215–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankar P, Garcia GR, La Du JK, Sullivan CM, Dunham CL, Goodale BC, Waters KM, Stanisheuski S, Maier CS, Thunga P, et al. 2022. The AHR2-dependent wfikkn1 gene influences zebrafish transcriptome, proteome, and behavior. Toxicol Sci. 187(2):325–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankar P, Geier MC, Truong L, McClure RS, Pande P, Waters KM, Tanguay RL.. 2019. Coupling genome-wide transcriptomics and developmental toxicity profiles in zebrafish to characterize polycyclic aromatic hydrocarbon (PAH) hazard. Int J Mol Sci. 20(10):2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shappell NW, Carlino-MacDonald U, Amin S, Kumar S, Sikka HC.. 2003. Comparative metabolism of chrysene and 5-methylchrysene by rat and rainbow trout liver microsomes. Toxicol Sci. 72(2):260–266. [DOI] [PubMed] [Google Scholar]
- Shen G, Tao S, Wei S, Zhang Y, Wang R, Wang B, Li W, Shen H, Huang Y, Yang Y, et al. 2012. Retene emission from residential solid fuels in China and evaluation of retene as a unique marker for soft wood combustion. Environ Sci Technol. 46(8):4666–4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Q, Fijten RR, Spina D, Riffo Vasquez Y, Arlt VM, Godschalk RW, Van Schooten FJ.. 2017. Altered gene expression profiles in the lungs of benzo[a]pyrene-exposed mice in the presence of lipopolysaccharide-induced pulmonary inflammation. Toxicol Appl Pharmacol. 336:8–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T. 2006. Xenobiotic-metabolizing enzymes involved in activation and detoxification of carcinogenic polycyclic aromatic hydrocarbons. Drug Metab Pharmacokinet. 21(4):257–276. [DOI] [PubMed] [Google Scholar]
- Shou M, Harvey RG, Penning TM.. 1993. Reactivity of benzo[a]pyrene-7,8-dione with DNA. Evidence for the formation of deoxyguanosine adducts. Carcinogenesis. 14(3):475–482. [DOI] [PubMed] [Google Scholar]
- Siddens LK, Larkin A, Krueger SK, Bradfield CA, Waters KM, Tilton SC, Pereira CB, Löhr CV, Arlt VM, Phillips DH, et al. 2012. Polycyclic aromatic hydrocarbons as skin carcinogens: comparison of benzo[a]pyrene, dibenzo[def,p]chrysene and three environmental mixtures in the FVB/N mouse. Toxicol Appl Pharmacol. 264(3):377–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smithgall TE, Harvey RG, Penning TM.. 1988. Oxidation of the trans-3,4-dihydrodiol metabolites of the potent carcinogen 7,12-dimethylbenz(a)anthracene and other benz(a)anthracene derivatives by 3 alpha-hydroxysteroid-dihydrodiol dehydrogenase: effects of methyl substitution on velocity and stereochemical course of trans-dihydrodiol oxidation. Cancer Res. 48(5):1227–1232. [PubMed] [Google Scholar]
- Stading R, Gastelum G, Chu C, Jiang W, Moorthy B.. 2021. Molecular mechanisms of pulmonary carcinogenesis by polycyclic aromatic hydrocarbons (PAHs): implications for human lung cancer. Semin Cancer Biol. 76:3–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stagaman K, Alexiev A, Sieler MJ, Hammer A, Kasschau KD, Truong L, Tanguay RL, Sharpton TJ.. 2024. The zebrafish gut microbiome influences benzo[a]pyrene developmental neurobehavioral toxicity. Sci Rep. 14(1):14618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staretz ME, Murphy SE, Patten CJ, Nunes MG, Koehl W, Amin S, Koenig LA, Guengerich FP, Hecht SS.. 1997. Comparative metabolism of the tobacco-related carcinogens benzo[a]pyrene, 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone, 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol, and n'- nitrosonornicotine in human hepatic microsomes. Drug Metab Dispos. 25(2):154–162. [PubMed] [Google Scholar]
- Tian FJ, Li WX, Lyu Y, Zhang P, Mu JB, Pei QL, Zheng JP.. 2020. Heat-shock protein 70 (HSP70) polymorphisms affect the risk of coke-oven emission-induced neurobehavioral damage. Neurotoxicology. 76:174–182. [DOI] [PubMed] [Google Scholar]
- Truong L, Reif DM, St Mary L, Geier MC, Truong HD, Tanguay RL.. 2014. Multidimensional in vivo hazard assessment using zebrafish. Toxicol Sci. 137(1):212–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Wiele T, Vanhaecke L, Boeckaert C, Peru K, Headley J, Verstraete W, Siciliano S.. 2005. Human colon microbiota transform polycyclic aromatic hydrocarbons to estrogenic metabolites. Environ Health Perspect. 113(1):6–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Delft JHM, Mathijs K, Staal YCM, van Herwijnen MHM, Brauers KJJ, Boorsma A, Kleinjans JCS.. 2010. Time series analysis of benzo[a]pyrene-induced transcriptome changes suggests that a network of transcription factors regulates the effects on functional gene sets. Toxicol Sci. 117(2):381–392. [DOI] [PubMed] [Google Scholar]
- Vehniäinen ER, Bremer K, Scott JA, Junttila S, Laiho A, Gyenesei A, Hodson PV, Oikari AO.. 2016. Retene causes multifunctional transcriptomic changes in the heart of rainbow trout (Oncorhynchus mykiss) embryos. Environ Toxicol Pharmacol. 41:95–102. [DOI] [PubMed] [Google Scholar]
- Vogeley C, Rolfes KM, Krutmann J, Haarmann-Stemmann T.. 2022. The aryl hydrocarbon receptor in the pathogenesis of environmentally-induced squamous cell carcinomas of the skin. Front Oncol. 12:841721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vranic S, Shimada Y, Ichihara S, Kimata M, Wu W, Tanaka T, Boland S, Tran L, Ichihara G.. 2019. Toxicological evaluation of SiO(2) nanoparticles by zebrafish embryo toxicity test. Int J Mol Sci. 20(4):882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Groot A, Seidel A, Wang L, Kiachaki E, Boogaard PJ, Rietjens I.. 2022a. The influence of alkyl substitution on the in vitro metabolism and mutagenicity of benzo[a]pyrene. Chem Biol Interact. 363:110007. [DOI] [PubMed] [Google Scholar]
- Wang D, Schramm V, Pool J, Pardali E, Brandenburg A, Rietjens I, Boogaard PJ.. 2022b. The effect of alkyl substitution on the oxidative metabolism and mutagenicity of phenanthrene. Arch Toxicol. 96(4):1109–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerfield M. 2007. The zebrafish book. A guide for the laboratory use of zebrafish (Danio rerio). Eugene (OR: ): University of Oregon Press. [Google Scholar]
- Wiley DS, Redfield SE, Zon LI.. 2017. Chemical screening in zebrafish for novel biological and therapeutic discovery. Methods Cell Biol. 138:651–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JA, Hyland R, Jones BC, Smith DA, Hurst S, Goosen TC, Peterkin V, Koup JR, Ball SE.. 2004. Drug-drug interactions for UDP-glucuronosyltransferase substrates: a pharmacokinetic explanation for typically observed low exposure (AUCI/AUC) ratios. Drug Metab Dispos. 32(11):1201–1208. [DOI] [PubMed] [Google Scholar]
- Wilson LB, McClure RS, Waters KM, Simonich MT, Tanguay RL.. 2022. Concentration-response gene expression analysis in zebrafish reveals phenotypically-anchored transcriptional responses to retene. Front Toxicol. 4:950503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie S, Feng Y, Zhou A, Lu Z, JixingZou. 2023. Comparative analysis of two new zebrafish models: the CYP1A low-expression line and cyp1a knockout line under PAHs exposure. Gene. 869:147391. [DOI] [PubMed] [Google Scholar]
- Yoganantharjah P, Gibert Y.. 2017. The use of the zebrafish model to aid in drug discovery and target validation. Curr Top Med Chem. 17(18):2041–2055. [DOI] [PubMed] [Google Scholar]
- Yu D, Berlin JA, Penning TM, Field J.. 2002. Reactive oxygen species generated by PAH o-quinones cause change-in-function mutations in p53. Chem Res Toxicol. 15(6):832–842. [DOI] [PubMed] [Google Scholar]
- Yu J, Fang Q, Liu M, Zhang X.. 2021. Polycyclic aromatic hydrocarbons associated long non-coding RNAs and heart rate variability in coke oven workers. Environ Sci Pollut Res Int. 28(34):47035–47045. [DOI] [PubMed] [Google Scholar]
- Yu YY, Jin H, Lu Q.. 2022. Effect of polycyclic aromatic hydrocarbons on immunity. J Transl Autoimmun. 5:100177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanger UM, Schwab M.. 2013. Cytochrome p450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol Ther. 138(1):103–141. [DOI] [PubMed] [Google Scholar]
- Zhang G, Truong L, Tanguay RL, Reif DM.. 2017. A new statistical approach to characterize chemical-elicited behavioral effects in high-throughput studies using zebrafish. PLoS One. 12(1):e0169408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zon LI, Peterson R.. 2010. The new age of chemical screening in zebrafish. Zebrafish. 7(1):1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.