Abstract
The rapid release of high-performing computer vision models offers new potential to study the impact of different inductive biases on the emergent brain alignment of learned representations. Here, we perform controlled comparisons among a curated set of 224 diverse models to test the impact of specific model properties on visual brain predictivity – a process requiring over 1.8 billion regressions and 50.3 thousand representational similarity analyses. We find that models with qualitatively different architectures (e.g. CNNs versus Transformers) and task objectives (e.g. purely visual contrastive learning versus vision- language alignment) achieve near equivalent brain predictivity, when other factors are held constant. Instead, variation across visual training diets yields the largest, most consistent effect on brain predictivity. Many models achieve similarly high brain predictivity, despite clear variation in their underlying representations – suggesting that standard methods used to link models to brains may be too flexible. Broadly, these findings challenge common assumptions about the factors underlying emergent brain alignment, and outline how we can leverage controlled model comparison to probe the common computational principles underlying biological and artificial visual systems.
Subject terms: Neural encoding, Object vision
Through controlled model-to-brain comparisons across a large-scale survey of deep neural networks, the authors show the data models are trained on matters far more for downstream brain prediction than design factors such as architecture and training task.
Introduction
The biological visual system transforms patterned light along a hierarchical series of processing stages into a useful visual format, capable of supporting object recognition1. Visual neuroscientists have made significant progress in understanding the nature of the tuning in early areas like V12, as well as in crafting normative computational accounts of the ecological and biological conditions under which such tuning might emerge [e.g. sparse coding of natural image statistics]3. However, that same computational clarity has been lacking with respect to the nature of the representation in later stages of the ventral stream supporting object representation, including human object-responsive occipitotemporal cortex (OTC), and the analogue monkey inferotemporal (IT) cortex. In the past decade, this landscape has dramatically changed with the introduction of goal-optimized deep neural networks [DNNs]4,5. These models have revolutionized our methodological capacity to explore the format underlying these late-stage visual representations, and have provided new traction for empirically testing the impact of different pressures guiding high-level visual representation formation6–14.
Landmark findings have demonstrated that deep convolutional neural networks—trained on a rich natural image diet, with the task of object categorization—learn features that predict neural tuning along the ventral visual stream5–7,15,16. For example, the responses of single neurons in monkey IT cortex to different natural images can be captured by weighted combinations of internal units in DNNs with greater accuracy than handcrafted features5. The predictive capacity of these single-neuron encoding models has been further validated in experiments that use these models to synthesize visual stimuli capable of driving neural activity beyond the range evoked by handpicked natural images17,18. The same encoding model procedures carried out using functional magnetic resonance imaging (fMRI) data in humans have shown similarly strong voxel-wise encoding and population-level geometry modeling7,19–25, providing further evidence of the emergent correspondence between the structure of biological visual system responses and the internals of visual DNN models.
However, where there was once a paucity of performant, image-computable visual representations to study, there is now an overabundance. New models with increasingly powerful visual representational competencies, and with variable architectures, objectives, image diets, and learning parameters, are now produced almost weekly. More often than not, these models are designed to optimize performance on canonical computer vision tasks, typically with no reference to brain function nor intent to directly reverse engineer brain mechanisms per se. This has changed the nature of the problem faced by computational neuroscientists trying to understand high-level visual representation, raising new questions for how to proceed. For example, if these DNN models are to be considered direct models of the brain, is there one model neuroscientists should be using until a better one comes along? Or, might there be another way to leverage the model diversity itself for insight into how more general inductive biases, shared among sets of models, lead to more or less “brain-like” representation?
Neural benchmarking platforms such as Brain-Score, Algonauts, and Sensorium directly operationalize the research effort to find the most brain-like model of biological vision, and do so with impressive scale and generality26–29. Crowd-sourcing across neuroscientists and applied machine learning researchers alike, these platforms collect many brain datasets that sample responses from multiple visual areas to a variety of stimuli, allowing users to upload and enter any candidate model for scoring. An automated pipeline fits unit-wise encoding models (under prespecified linking assumptions) to each candidate DNN, and computes an aggregated score across all probe datasets. These neural benchmarking endeavors seem aimed predominantly at identifying the single best predictive model of the target neural system (e.g. a mouse primary visual area, the primate ventral stream), a goal that that is reflected in the leaderboards of top-ranking models that have become the standard outputs of the pipeline.
Here, we take an alternative but complementary methodological approach, wherein we explicitly leverage the diversity and quantity of open-source DNNs as a source of insight into representation formation. Specifically, we conceptualize each of these DNNs as a different model organism—a unique artificial visual system—with performant, human-relevant visual capacities. As such, each DNN is worthy of study, regardless of whether its properties seem to match the biology or depart from it. We take as our next premise that different DNNs can learn different high-level visual representations, based on their architectures, task objectives, learning rules, and visual “diets”. By comparing sets of models that vary only in one of these factors, while holding other factors constant, we can begin to experimentally examine which inductive biases lead to learned representations that are more or less brain-predictive. In our framework, models are not competing to be the best in-silico model of the brain. Instead, we think of them as powerful visual representation learners, with controlled comparisons among them providing empirical traction to study the pressures guiding visual representation formation.
In the current work, we harness sources of controlled variation already present among pre-existing, open-source models to ask broad questions about their emergent brain-predictive capacity. For instance, about meso-scale architectural motifs: do convolutional or transformer encoders learn features that better capture high-level visual responses to natural images, holding task and visual experience constant? Or, about the goal of high-level vision: when visual representations are aligned with language representations, does this provide a better fit to brain responses than purely visual self-supervised objectives, controlling for architecture and visual experience? Or, about visual experience: do certain training datasets (such as faces alone or places alone) lead to more a brain-predictive representational format than others? We supplement these experiments with additional analyses that test the explanatory power of more general factors (e.g. effective dimensionality) that have been proposed to underlie increased brain predictivity, but that do not fit as neatly into the framework of inductive bias. Broadly, the goals of this work are to reveal the relationships between model variations and emergent brain-predictive visual representation, to provide insight into the principles shaping high-level visual representation in both biological and artificial visual systems, and to articulate the next steps for the neuroconnectionist enterprise13.
Results
Our approach involves first searching through pre-trained model repositories and curating distinct sets of models that have performant visual capacities, and which provide meaningful controlled variation in key inductive biases such as architecture, task objective, and visual diet (i.e. training dataset). Each of these analyses involves insolating models that vary along only one of these dimensions, while holding the others constant. In total, we examine the degree to which the representations of 224 distinct DNNs can predict the responses to natural images across human occipitotemporal cortex (OTC) in the 7T Natural Scenes Dataset [NSD]30, using two different model-to-brain linking methods. Details on all aspects of our procedure are available in the Methods Section.
The full set of all included models and their most relevant metadata is described in Supplementary Information Table 1. Trained models (N = 160) were sourced from a variety of online repositories (Fig. 1B). We also collected the randomly-initialized variants (N = 64) of all ImageNet-1K-trained architectures, using the default random initialization procedure provided by the model repository or original authors. We then grouped these models into controlled comparison sets, to enable ‘opportunistic experiments’ between models that differ in only one inductive bias, holding the others constant.
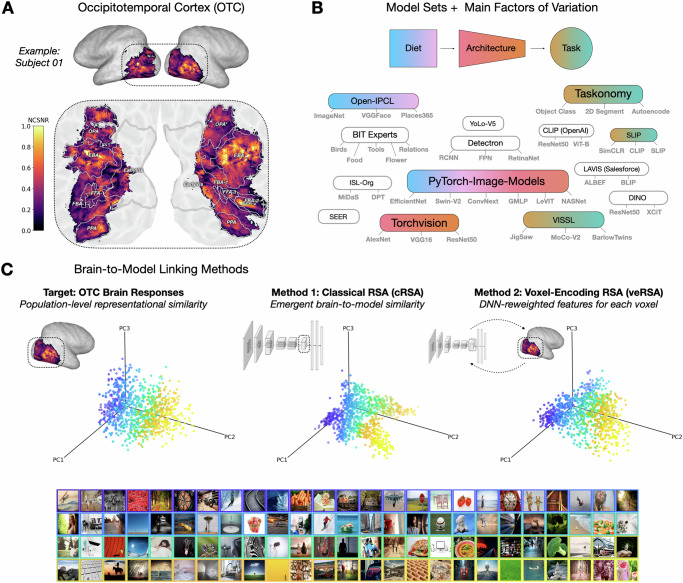
Fig. 1. Overview of our approach.
A The brain region of focus is occipitotemporal cortex (OTC), here shown for an example subject. The voxel-wise noise-ceiling signal-to-noise ratio (NCSNR) is indicated in color. B A large set of models were gathered, schematized here by repository, and colored here by the main experiments to which they contribute. C Brain-linking methods. The left plot depicts the target representational geometry of OTC for 1000 COCO images, plotted along the first three principal components of the voxel space. Each dot reflects the encoding of a natural image, a subset of which are depicted below in a corresponding color outline. The middle panel shows a DNN representational geometry (here the final embedding of a CLIP-ResNet50), plotted along its top 3 principal components. Classical RSA involves directly estimating the emergent similarity between the brain target and the model layer representational geometries. The right plot shows the same DNN layer representation, but after the voxel-wise encoding procedure (veRSA), which involves first re-weighting the DNN features to maximize voxel-wise encoding accuracy, and then estimating the similarity between the target voxel representations and the model-predicted voxel representations. (Note: Images in C are copyright-free images gathered from Pixabay.com using query terms from the COCO captions for 100 of the original NSD1000 images. We are grateful to the original creators for the use of these images).
The main brain targets of our analyses in this work were OTC responses to 1000 natural images in voxels sampled from 4 subjects in the NSD (Fig. 1A). We define the OTC sector individually in each participant using a combination of reliability-based (SNR) and functional metrics. Our key outcome measure was the representational geometry of the OTC brain region, captured by the set of 124,750 pairwise distances between 500 test images. The representational geometry in this dataset was highly reliable (noise ceiling mean across subjects: rPearson = 0.8, 95% CI [0.74, 0.85] across subjects), providing a strong target for arbitrating the relative predictivities of our surveyed models.
We considered two different linking methods to relate the representations learned in models to the response structure measured in brains. Both of these methods predict each individual subject’s OTC representational geometry, using representational similarity analysis (RSA)31. The first method, classical RSA (cRSA), is the more strict linking hypothesis, estimating the degree of correspondence between the brain’s population geometry and the best-fitting model layer’s population geometry, without any feature re-weighting procedures. This measure probes for a fully emergent correspondence between model and brain, making the clear (and reasonable) assumption that as a whole, all units of the DNN layer must contribute equally to capture the population level geometry.
The second method, voxel-encoding RSA (veRSA), tests for the same correspondence, but allows for guided re-weighting of the DNN features32–34. This method first makes the (also reasonable) assumption that different voxels are likely tuned to different features, and thus, that each voxel’s response profile should be modeled as a weighted combination of the units in a layer, using independent brain data for fitting the encoding model. After fitting, each voxel-wise encoding model is used to predict responses to the test images, for which we compare the predicted population geometry to the observed population geometry of neural responses. Thus, these two mapping procedures provide two distinct measures of the degree to which each model’s internal representations are able to predict population geometry of OTC, with either more strict or more flexible linking assumptions.
All results reported are from the most brain-predictive layer of each model. This layer is selected through a nested cross-validation procedure using a training set of 500 images, and the final reported brain prediction scores are assessed using a held-out test set of 500 test images. This process ensures independence between the selection of the most predictive layer and its subsequent evaluation. (For results as a function of model layer depth, see Supplementary Fig. 3).
Architecture comparison
While differences in architecture across models can be operationalized in many different ways (total number of parameters, total number of layers, average width of layers, et cetera), here we focus on a distinct mesoscale architectural motif: the presence (or absence) of an explicit convolutional bias, which is present in convolutional neural networks (CNNs) but absent in vision transformers.
CNN architectures applied to image processing were arguably the central drivers of the last decade’s reinvigorated interest in artificial intelligence4,35. CNNs are considered to be naturally optimized for visual processing applications that benefit from a sliding window (weight sharing) strategy, applying learned local features over an entire image. These models are known for their efficiency, stability, and shift equivariance36,37. Transformers, originally developed for natural language processing, have since emerged as major challengers to the centrality of CNNs in AI research, even in vision. Transformers operate over patched image inputs, using multihead attention modules instead of convolutions38,39. They are designed to better capture long-range dependencies, and are considered in some cases to be a more powerful alternative to CNNs precisely for this reason40. While many variants of CNNs and some kinds of transformers have appeared in benchmarking competitions27–29, comparisons between these models have been largely uncontrolled. Which of these models’ starkly divergent architectural inductive biases lead to learned representations that are more predictive of human ventral visual system responses, controlling for visual input diet and task?
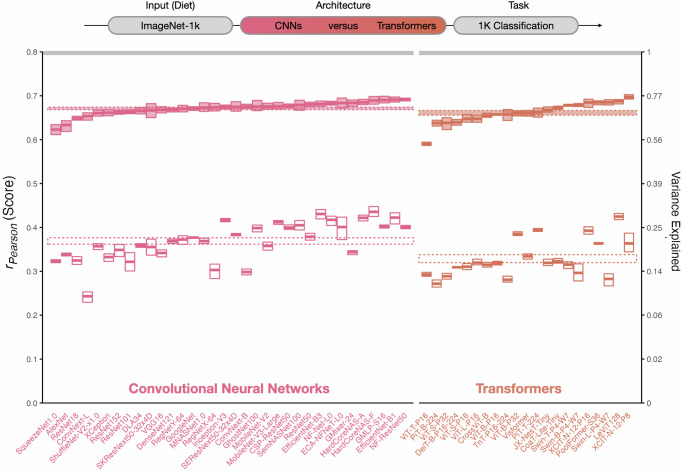
We compared the brain-predictivity scores of 34 CNNs against 21 transformers. Critically, all of these models were trained with the same dataset (ImageNet1K) and the same task objective (1000-way image classification). The results are shown in Fig. 2. Surprisingly, we find that both the CNN and transformer architectures account for the structure of OTC responses almost equally well: in the veRSA comparison, the brain predictivity on average was rPearson = 0.67 [0.67, 0.68] for convolutional models and rPearson = 0.66 [0.65, 0.67] for transformer models. We did find, however, that the aggregate differences between these architectures, while small, were statistically significant (Wald t-distribution statistics, see Methods). Specifically, the transformers were on average less predictive than the CNNs, in both the classical and voxel-encoding RSA metrics (cRSA: β = −0.04 [−0.05, −0.03], p < 0.001; veRSA: β = −0.01 [−0.02, −0.00] p < 0.001). Thus, the CNNs as a class may introduce an inductive bias that leads to a slightly more brain-aligned late-stage visual representation, on average—holding task and visual input diet constant. However, note that the prediction ranges among the surveyed CNN and transformer models were substantially overlapping, so this statistical effect should not be interpreted as a categorical claim that all convolutional models have greater emergent brain predictivity than all transformer models.
Fig. 2. Architecture variation.
Degree of brain predictivity (rPearson) is plotted for the controlled set of convolutional neural networks (CNNs) and transformer models in our survey. Each small box corresponds an individual model. The horizontal midline of each box indicates the mean score of each model’s most brain-predictive layer (selected by cross-validation) across the 4 subjects, with the height of the box indicating the grand-mean-centered 95% bootstrapped confidence intervals (CIs)137 of the model’s score across subjects. The cRSA score is plotted in open boxes, and the veRSA score is plotted in filled boxes. For each class of model architecture (convolutional, transformer) the class mean is plotted as a striped horizontal ribbon. The width of this ribbon reflects the 95% grand-mean-centered bootstrapped 95% CIs over the mean score for all models in a given set. The noise ceiling of the occipitotemporal brain data is plotted in the gray horizontal ribbon at the top of the plot, and reflects the mean of the noise ceilings computed for each individual subject. The secondary y-axis shows explainable variance explained (the squared model score, divided by the squared noise ceiling). Source data are provided as a Source Data file.
Given the dramatic differences between these architectural encoders, we found it surprising how similarly they predicted the structure of the brain responses in high-level visual cortex, which suggests that these models are converging on the same representational format. Worth noting, though, is that the cRSA brain-predictivity scores were both much lower and more variable than the veRSA scores. This implies that feature re-weighting is playing a substantial role in the degree to which these models capture the representational geometry of the high-level visual system. This observation is also consistent with the possibility that the learned representations of these models all capture similar representational sub-spaces after feature re-weighting. We return to this possibility analytically in the “Model-to-Model Comparison” Section.
Task variation
Next, we examined the impact of different task objectives on the emergent capacity of a model to predict the similarity structure of brain responses. Our case studies here probe the effect of different canonical computer vision tasks41, the effect of different self-supervised algorithms42, and the effect of visual representation learning with or without language alignment43. Results from these experiments are summarized in Fig. 3.
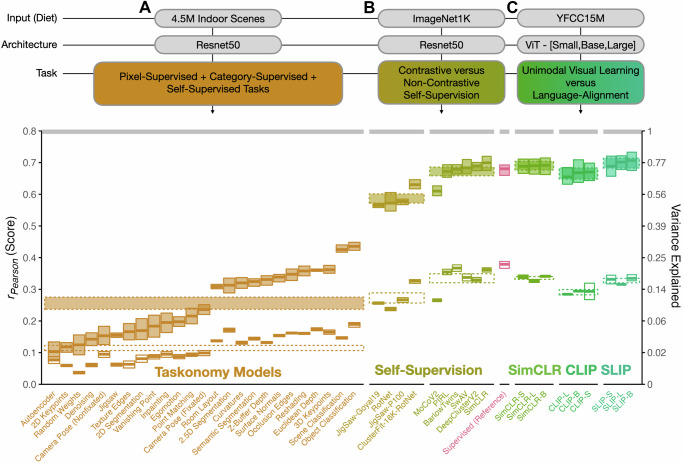
Fig. 3. Task variation.
Degree of brain predictivity (rPearson) is plotted for the sets of models with controlled variation in task. A The first set of models shows scores across the ResNet50 encoders from Taskonomy, trained on a custom dataset of 4.5 million indoor scenes. B The second set of models shows the difference between contrastive and non-contrastive self-supervised learning ResNet50 models (with a category-supervised ResNet50 for reference), trained on ImageNet1K. C The third set of models shows the scores across the vision-only and vision-language contrastive learning ViT-[Small,Base,Large] models from FaceBook’s SLIP Project, trained on the images (or image-text pairs) of YFCC15M. Each small box corresponds an individual model. In all subplots, the horizontal midline of each box indicates the mean score of each model’s most brain-predictive layer (selected by cross-validation) across the 4 subjects, with the height of the box indicating the grand-mean-centered 95% bootstrapped confidence intervals (CIs) of the model’s score across subjects. The cRSA score is plotted in open boxes, and the veRSA score is plotted in filled boxes. The class mean for each distinct set of models is plotted in striped horizontal ribbons across the individual models. The width of this ribbon reflects the 95% grand-mean-centered bootstrapped 95% CIs over the mean score for all models in this set. The noise ceiling of the occipitotemporal brain data is plotted in the gray horizontal ribbon at the top of the plot, and reflects the mean of the noise ceilings computed for each individual subject. The secondary y-axis shows explainable variance explained (the squared model score, divided by the squared noise ceiling). Source data are provided as a Source Data file.
Task variation: the Taskonomy models
First, we examined the Taskonomy models41,44—an early example of controlled model rearing. The Taskonomy models were originally designed to test how well learned representations trained with one task objective transfer to other tasks. Each of these 24 models were trained on different tasks spanning a range of unsupervised and supervised objectives (e.g. autoencoding, depth prediction, scene classification, surface normals, edge detection), some requiring pixel-level labeling and others requiring a single label for the whole image. In all cases, the base encoder architecture is a ResNet50, modified with a specialized projection head to fit the task-specific output. In this analysis, we consider only the feature spaces of the base ResNet50 encoders. The dataset on which the Taskonomy models are trained is a large dataset in terms of raw images, consisting of 4.5 million images, but depicts only images of indoor scenes, with any images of people excluded.
Comparing the brain-predictivity scores of the Taskonomy models (Fig. 3A), we make two key observations. The first is that across the different task objectives, there was indeed a large range of scores. The least brain-aligned task—autoencoding—yielded a rPearson = 0.077 [0.066, 0.085] in cRSA and rPearson = 0.103 [0.096, 0.11] in veRSA, while the most brain-aligned task––object classification––yielded a rPearson = 0.189 [0.178, 0.201] in cRSA and rPearson = 0.436 [0.419, 0.454] in veRSA).
The second key observation is that the overall range of brain-predictivity scores among these models was relatively low—even for the highest-scoring tasks: for object classification, the veRSA score was only rPearson = 0.44 [0.42, 0.45]. For reference, a standard ResNet50 architecture also trained on image classification, but over the ImageNet dataset, shows an average brain predictivity of rPearson = 0.68 [0.63, 0.72] (tStudent(3) = −14.3, p < 0.001). Note that this difference manifests in spite of Taskonomy’s larger training set (∼4.5 M images), nearly thrice that of the ImageNet1K (∼1.2 M images). This observation leads to the hypothesis that the relatively weaker overall brain-predictivity scores for Taskonomy models are related to insufficient diversity of the Taskonomy images. Indeed, the Taskonomy authors estimate that only 100 of the 1000 ImageNet classes are present across the ‘scenes’ of the Taskonomy dataset41.
Task variation: self-supervised algorithms
Early variants of self-supervised objectives involved learning representations by predicting image rotations (RotNet) or unscrambling images (JigSaw). More modern variants of self-supervised objectives operate by learning to represent individual images distinctly from one another in an embedding space (SimCLR, BarlowTwins). In particular, contrastive learning objectives typically build a high-level embedding of images by representing samples (augmentations) of the same image nearby in feature space, and far from the representations of other images. Critically, when these learned representations are probed on the canonical computer vision task of image classification, emergent categorization capacity is nearly comparable to that of models trained with category supervision45,46. Additionally, these contrastive learning models have also been shown to predict brain activity on par with category-supervised models in mice, humans, and non-human primates34,47–49. However, there has not been a systematic comparison of brain predictivity for models trained with these different kinds of self-supervised objectives.
In this experiment (Fig. 3B), we examine the different brain predictivities of contrastive versus non-constrastive self-supervised learning methods using a suite of 10 models from the VISSL model zoo42. Each of these models are trained using a different method of self-supervision, but all with a ResNet50 architectural backbone, and a training dataset that consists of the images (but not the labels) of the ImageNet1K dataset. We divide the models of this set into two groups: models that employ instance-level contrastive learning (N = 6: PIRL, DeepClusterV2, MoCoV2, SwaV, SimCLR, BarlowTwins) and models that do not (N = 4: RotNet, two Jigsaw variants, ClusterFit).
These two different classes of self-supervised learning objectives yield significantly different brain predictivities. Instance-level contrastive learning objectives lead to more brain-predictive representations than non-contrastive objectives by a significant, midsize margin (cRSA: β = −0.06 [−0.04, −0.09], p < 0.001; veRSA: β = −0.09 [−0.07, −0.11] p < 0.001). And, consistent with prior work34,49, most instance-level contrastive objectives provide comparable brain predictivity to the matched category-supervised model, holding architecture and visual input diet constant. (For example, the average predictivity of VISSL’s ResNet50-BarlowTwins was 0.367 [0.353, 0.381] in cRSA and 0.679 [0.637, 0.720] in veRSA; the predictivity of Torchvision’s ImageNet1K-trained category-supervised ResNet50 was rPearson = 0.379 [0.363, 0.39] in cRSA and 0.680 [0.640, 0.718] in veRSA. These models share identical architectures in PyTorch.) Broadly, these results highlight the potential for understanding biological visual representation through deeper exploration of instance-level contrastive objectives, which focus on learning invariances over samples from the same image, while also learning features that discriminate distinct individual images.
Task variation: language alignment (the SLIP models)
Another recent development in self-supervised contrastive learning involves leveraging the structure of another modality—language—to influence representations learned in vision. The preeminent example of this ‘language-aligned’ learning is OpenAI’s CLIP model (Contrastive Language-Image-Pretraining50), which builds representations designed to maximize the cosine similarity between the latent representation of an image and the latent representation of that image’s caption. In computer vision research, CLIP has demonstrated remarkable zero-shot generalization to image classification task variants without further retraining. Indeed, across all the models in our set, the model with the highest brain predictivity is OpenAI’s CLIP-ResNet50 model, with 4 other CLIP-trained models from OpenAI also in the top 10 (see Supplementary Information SI.1). If the constraint of language alignment is key in building more brain-aligned representations, this constitutes a development of deep theoretical import when considering the pressures guiding late stage representations in the ventral visual hierarchy.
A critical issue in using OpenAI’s CLIP model for brain prediction, however, is the fact that OpenAI not only introduced a new objective (image-text alignment via contrastive learning), but also a massive new training dataset (a highly curated and proprietary dataset of 400 million image-text pairs, which has yet to be made available for public research). This makes the direct comparison of CLIP to other models empirically dubious, as it leaves open whether gains in predictivity are attributable to language alignment per se, to massive dataset differences, or the interaction of the two.
Fortunately, Meta AI has since released a set of models—the SLIP models43—that compare self-supervised contrastive learning models with and without language alignment, controlling for training dataset and architecture. These models are trained with one of 3 learning objectives: pure self-supervision (SimCLR), pure language alignment (CLIP), or a combination of self-supervision and language alignment (SLIP). All models consist of a Vision Transformer backbone (three sizes: Small [ViT-S], Base [ViT-B], & Large [ViT-L]), and are all trained on the YFCC15M dataset (15 million images or image-text pairs). Thus, any differences in brain predictivity across these models reflect the impact of language alignment per se, holding image diet and architecture constant.
Comparing OTC predictivity across these models (Fig. 3C), we find that all models have relatively high brain-predictivity scores, with no substantial differences between objectives with and without language alignment. If anything, we see a slight but significant decrease in accuracy for the pure language-aligned CLIP objective. Specifically, in a linear model regressing brain predictivity on the interaction of model size (ViT-[S,B,L]) and task (SimCLR, CLIP, SLIP) plus an additive effect of subject ID, the only significant experimental effect is a slight decrease in the predictive accuracy of the pure CLIP model relative to SimCLR in both metrics (cRSA: β = −0.05 [−0.06, −0.03], p < 0.001; veRSA: β = −0.02 [−0.04, −0.01] p = 0.005). We find no effect of the model size, nor an interaction with the task objective.
These results thus lead to the somewhat surprising conclusion that, in terms of brain predictivity, the superior performance of OpenAI’s CLIP models is likely due to the expanded and proprietary image database, rather than the influence of language alignment per se (though it is possible the benefits of language alignment only emerge at datasets of this more massive scale). Likewise, emerging insights in the machine learning community are also finding that the visual representational robustness of OpenAI’s CLIP is almost exclusively conferred by image dataset51. Our conclusions about the negligible impact of language alignment on brain predictivity also directly contrast with those of recent papers that conclude the opposite, based on uncontrolled comparisons of CLIP-ResNet50 against ImageNet-1K-trained ResNet5052. Thus, our work also strongly underscores the need for more empirically controlled model sets (like those of SLIP) that could better arbitrate questions about the emergent properties of language-aligned models trained on massive datasets.
Input variation
We next directly examined the impact of a model’s input diet on brain predictivity. Here, we define a model’s input diet as the images used to train the model, irrespective of whether their labels factor into the training procedure. While there are many different datasets used across the 160 models in our model set, there were actually relatively few subsets that enabled controlled comparison of the impact of dataset while holding architecture and objective constant. In the two controlled experiments possible, we examined the relative brain predictivity of architecture-matched models trained on ImageNet1K versus ImageNet21K, and of instance-prototype contrastive learning (IPCL34) models trained on datasets of faces, places, objects. The results of these experiments are summarized in Fig. 4.
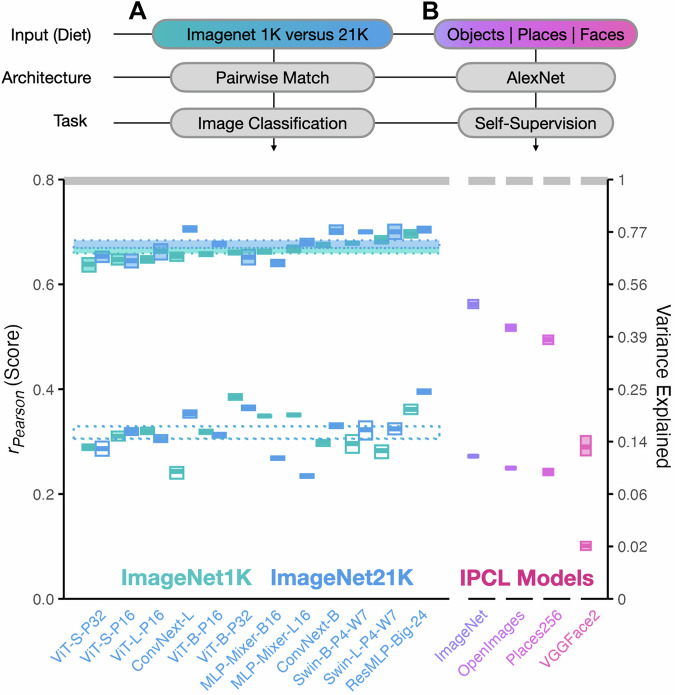
Fig. 4. Input variation.
Degree of brain predictivity (rPearson) is plotted for the sets of models with controlled variation in input diet. A The first set of models shows scores across paired model architectures trained either on ImageNet1K or ImageNet21K (a ∼13× increase in number of training images). B The second set of models shows scores across 4 variants of a self-supervised IPCL-AlexNet model trained on different image datasets. Each small box corresponds an individual model. In all subplots, the horizontal midline of each box indicates the mean score of each model’s most brain-predictive layer (selected by cross-validation) across the 4 subjects, with the height of the box indicating the grand-mean-centered 95% bootstrapped confidence intervals (CIs) of the model’s score across subjects. The cRSA score is plotted in open boxes, and the veRSA score is plotted in filled boxes. The class mean for each distinct set of models is plotted in striped horizontal ribbons across the individual models. The width of this ribbon reflects the 95% grand-mean-centered bootstrapped 95% CIs over the mean score for all models in this set. The noise ceiling of the occipitotemporal brain data is plotted in the gray horizontal ribbon at the top of the plot, and reflects the mean of the noise ceilings computed for each individual subject. The secondary y-axis shows explainable variance explained (the squared model score, divided by the squared noise ceiling). Source data are provided as a Source Data file.
Input variation: Imagenet1K versus Imagenet21K
The first controlled dataset comparison we performed was between models trained either on ImageNet1K or ImageNet21K. Deep learning is notoriously data-intensive, and most if not all DNNs are known to benefit from more training samples when it comes to overall classification accuracy53,54. ImageNet21K is a dataset of ∼14.2 million images across 21,843 classes (many of which are hierarchically labeled). The more popular version of ImageNet1K (with 1000 classes) is a ∼1.2 million image subset of this larger dataset. ImageNet21K is considered by some to be a more diverse dataset55,56, though exact quantification of this diversity has proven difficult. Does training on this larger and purportedly more diverse dataset lead to increased brain predictivity?
We considered 14 pairs of models from the PyTorch-Image-Models57 repository with matched architecture and task-objective, trained either on the ImageNet1K or the ImageNet21K dataset (Fig. 4A). Performing paired statistical comparisons between their prediction levels, we find no effect of input diet on resulting brain predictivity, in either metric (cRSA β = 0.0 [−0.03, 0.03], p = 0.957; veRSA β = 0.01 [0.00, 0.03], p = 0.147). This result highlights first that the raw quantity of training images does not necessarily lead to increased brain-predictivity. This result also highlights that whatever the increased diversity of ImageNet21K, it does not manifest in this particular comparison to high-level visual representations in human OTC.
Input variation: objects versus places versus faces
The second controlled dataset experiment we perform is based on the set of IPCL-Alexnet models, trained on 4 different datasets: object-focused image sets (ImageNet-1K, OpenImages), scene-focused image sets (Places365), or face-focused image sets (VGGFace2). This model set uses the same architecture and the same self-supervised, instance prototype contrastive learning objective34. Notably, these models are trained without labels—further deconfounding task and visual input diet.
In this model set (Fig. 4B), we find that the ImageNet diet leads to the highest overall brain predictivity: relative to the ImageNet-trained model, the OpenImages-trained model yields a decrease in score of β = −0.02 [−0.03,−0.01], p = 0.002 in cRSA, and β = −0.04 [−0.07, −0.02], p < 0.001 in veRSA). The Places-365 trained model yields a decrease in score of β = −0.03 [−0.04,−0.02], p < 0.001 in cRSA, and β = −0.07 [−0.09, −0.04], p < 0.001 in veRSA. Finally, the VGGFace2-trained model is significantly worse than the ImageNet-trained model by a substantial margin in both metrics (cRSA β = −0.17 [−0.18,−0.16], p < 0.001; veRSA β = −0.27 [−0.30, −0.25], p < 0.001). These results highlight that—at least for this model architecture and objective—the visual diet leads to substantial variation in brain predictivity.
Finally, we note that in each of these cases, the VGGFace2 and Places-365 dataset actually contain more images than ImageNet (∼2.75× and ∼1.5×, respectively), again underscoring that the quantity of images is not the relevant factor here. Broadly, these results hint yet again at the importance of a latent dataset diversity factor that has yet to be quantified. A speculative but intriguing reverse inference suggests that ImageNet, OpenImages, Places-365, and VGGFace2 may be ranked in terms of diversity from greatest to least, based on their ability to capture the representational structure of OTC in response to hundreds of natural images.
Impact of training
Research in network initialization techniques has in recent years led to the development of models with randomized, hierarchical representations (effectively, multiscale hierarchical random projections) that are sometimes powerful enough to serve as functional substitutes for trained features in a variety of tasks58. Past experiments with brain-to-model mappings, for example, have suggested that randomly initialized models may, in some cases, be as predictive of the brain as trained models (25,59, see also refs. 60,61 for further studies of untrained models). However, subsequent work has largely converged on the result that trained models have better predictive capacity of visual brain responses25,47,62–64.
Here, we include another such test: for every trained model architecture we test from the TorchVision and PyTorch-Image-Models repositories, we also test one randomly initialized counterpart (N = 64). This paired statistical test confirms that training has a resounding effect on brain predictivity in both metrics (cRSA β = 0.30 [0.29, 0.31], p < 0.001; veRSA β = 0.56 [0.55, 0.75], p < 0.001; see also Supplementary Fig. 3)—the single largest and most robust effect across all of our analyses. In short, training matters.
Overall variation across models
In all analyses to this point, we have focused on understanding variation in brain-predictivity scores through targeted model comparisons, with controlled differences in inductive biases defined by their architecture, task, or input diet. We next focus on understanding variation in brain predictivity across all models in our survey.
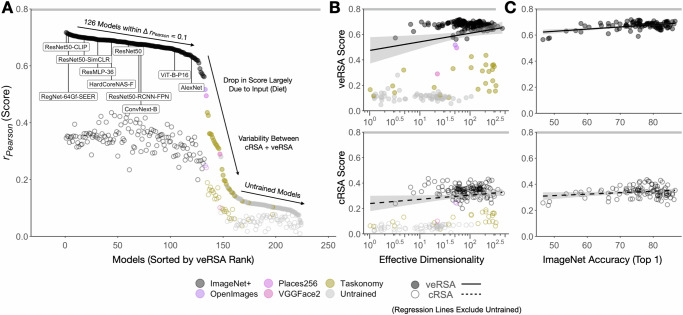
Across the full set of 224 models we tested (including randomly-initialized variants) and both metrics, we observe predictivity scores that span nearly the full range possible between 0 and the noise ceiling (rPearson = 0.8 [0.741, 0.847]). Figure 5A shows the brain predictivity of all 224 models. From this graph, it is clear that a large number of models perform comparably well, with 126 models yielding veRSA brain-predictivity scores that differ by less than rPearson = 0.1. (A bootstrapped segmented regression analysis over these scores indicate a break point at rank 124 [123.6, 124.7] / 224, corresponding to scores of rPearson = 0.623 and lower). The ranks beyond this elbow are populated almost entirely by models trained on image diets less diverse than ImageNet (e.g. Taskonomy), and untrained models. See Supplementary Information for additional analyses on the stability and variation of these scores across subjects (SI.3 and Supplementary Fig. 2.)
Fig. 5. Overall Model Variation.
A Brain predictivity is plotted for all models in this survey (N = 224), sorted by veRSA score. Each point is the score from the most brain-predictive layer (selected by cross-validation) of a single model, plotted for both cRSA (open) and veRSA (filled) metrics. Models trained on different image sets are labeled in color. B Brain predictivity is plotted as a function of the effective dimensionality of the most predictive layer, with veRSA scores in the top panel and cRSA scores in the bottom panel. The regression (± 95% CIs) line is fit only on trained variants of the models (excluding untrained variants). C Brain predictivity is plotted as a function of the top-1 ImageNet1K-categorization accuracy for the models (N = 108) whose metadata includes this measure (veRSA, top panel; cRSA bottom panel). The noise ceiling of the OTC brain data is shown as the gray horizontal bar at the top of each plot. Source data are provided as a Source Data file.
Given this degree of variation, we next focused on three more general factors that have been hypothesized to account for a model’s emergent brain predictivity beyond differences in inductive bias: effective dimensionality, classification accuracy, and parameter count.
Overall variation: effective dimensionality
Recent work in DNN modeling of high-level visual cortical responses in both human and non-human pri- mates has suggested a general principle, where model representations with a higher ‘latent’ or ‘effective’ dimensionality65 are more predictive of high-level visual cortex66. Effective dimensionality (ED), in this case, is a property of manifold geometry defined as the “continuous measurement of the number of principal components needed to explain most of the variance in a dataset”66.
In this analysis, we sought to test whether the relationship between ED and brain predictivity holds across the full set of models in our survey. To do so, we computed the ED of the most OTC-aligned layer representations from each model (as measured by our veRSA metric), using the same 1000 COCO images from our main analysis (see Methods and Supplementary Information for details). The relationship between model layer effective dimensionality and its corresponding brain predictivity is shown in Fig. 5B.
We first considered variation in ED across all models—both trained and randomly-initialized, akin to prior work66. From this perspective, ED appears to be a significant, moderately high predictor of each model’s veRSA score: rSpearman = 0.489 [0.381, 0.580], p < 0.001, across 1000 bootstraps of the sampled models. However, in our data, this relationship seems to be driven almost entirely by the predictivity differences between trained and untrained models. When we computed the relationship between ED and prediction scores for trained and randomly-initialized models separately, variation in the ED of trained models showed no correlation with the veRSA score (rSpearman = −0.063 [−0.31, 0.099], p = 0.692). Similarly, variation in ED among randomly-initialized model layers produced a non-significant, slightly negative correlation with OTC prediction (rSpearman = −0.142 [−0.307, 0.118], p = 0.077). See Supplementary Information SI.2 for a more detailed exploration of the impact of ED in our data in relation to Elmoznino and Bonner66, and the different analytical choices that may underlie the divergence in our results.
Overall, our analyses suggest that this particular effective dimensionality metric is not a general principle explaining emergent brain predictivity. This is by no means a rejection of the more general hypothesis that the geometric and statistical properties of neural manifolds might well transcend inductive bias as predictors of brain similarity, but it does suggest that we may need different metrics (e.g.,67,68) to unveil these underlying principles moving forward.
Overall variation: classification accuracy
Early studies of brain-predictive DNNs provided evidence of a link between a model’s ability to accurately perform object categorization and its capacity to predict the responses to neurons along the primate ventral visual stream5. However, more recent work suggests that across modern neural network architectures, ImageNet top-1 performance acts as a weaker or even null predictor of brain similarity26,27,34,69. We next examined this relationship in our human OTC data, considering the N = 99 trained models for which we have the ImageNet-1K top-1 categorization accuracy available. We focused on trained models here, because we reasoned that if gradations in top-1 accuracy are reliable indicators of emergent brain predictivity, then this relationship should also hold among trained models alone (and not solely rely on the gap between untrained and trained models).
Fig. 5C plots a model’s top-1 accuracy against the brain predictivity of that model’s best-predicting layer. We find little to no relationship between classification accuracy and brain predictivity across these trained models, for either cRSA or veRSA (rSpearman = −0.0057, p = 0.96 in cRSA; rSpearman = 0.17, p = 0.088 in veRSA; rank-order correlation is appropriate given the non-normality of top-1 accuracies). Our results also do not show an obvious plateau in accuracies above 70% (c.f.26); rather, the models in our comparison set have a relatively wide range of top-1 accuracies with a relatively restricted range of brain-predictivity scores. Thus, among trained models, variations in brain predictivity do not seem to be well captured by the fine-grained metric of top-1 object recognition accuracy that was once the primary indicator of this particular competence.
Overall variation: number of trainable parameters
Model size is another attribute commonly suspected to influence many DNN outcome metrics, including emergent predictivity of human brain and behavioral data27,53,70–72, and is typically measured via quantification of depth, width, or total number of parameters. Here, we test for differences in brain predictivity as a function of total parameter count for all models, excluding the Taskonomy and VISSL ResNet50s, as well as IPCL-AlexNet models, which (sharing the same architectures) do not vary in their total number of parameters. We find a somewhat irregular set of patterns. As parameter count increases in trained models, there is a significant, midsize decrease in cRSA brain-predictivity (rSpearman = −0.45, p = 0.026e−7, and a non-significant increase in veRSA brain-predictivity (rSpearman = 0.14, p = 0.129). As parameter count increases in untrained models, there is a significant, small-to-midsize increase in cRSA score (0.28, p = 0.0225), coupled with another non-significant increase in veRSA score (0.083, p = 0.513). In short, there is no consistent influence of total trainable parameter count on subsequent brain predictivity.
Model-to-Model comparison (RSA)
Considered in the aggregate, the opportunistic experiments and overarching statistics in this work are underwritten by over 1.8 billion regression fits and 50,300 representational similarity analyses. Given the scale of our analyses and the surprisingly modest set of significant relationships that arise from them, it would not be entirely unreasonable at this point to arrive at a somewhat deflationary conclusion—that almost none of the factors hypothesized as central to the better modeling of brains actually translate to more brain-aligned representations in practice. Convolutions, language alignment, effective dimensionality—all concepts of deep theoretical import—make almost no difference when it comes to predicting how well a model will ultimately explain high-level visual cortical representations in the largest such fMRI dataset gathered to date. 126 models—many of which appear to differ fundamentally in their design—have veRSA brain-predictivity scores within rPearson = 0.1 of a notably high noise ceiling.
A key question for this research enterprise, then, is to understand whether the different architectures, objectives, and diets actually lead to different representations in the first place. For example, are all of the most brain-aligned models converging on effectively the same representational structure?
To explore this question, we performed a direct model-to-model similarity analysis. Specifically, we computed the pairwise similarities of the most brain-aligned feature spaces from each model, and compared each of these model representations to those from all other models. Critically, we did this using two methods. The first method operates over the classical representational similarity matrices (cRSMs) from each model (i.e. the unweighted RSMs), assessing the pairwise similarity of each model’s cRSM to every other model’s cRSM. The second method operates over the voxel-wise encoding RSMs from each model (i.e. the RSM that is produced after feature reweighting), assessing the pairwise similarity of each model’s veRSM to each other model’s veRSM. Taken together, the output of this analysis is effectively two model-to-model meta-RSMs whose constituent pairwise similarities allow us to assess how similar different models’ most brain-aligned layer representations are, with and without being linearly reweighted to predict fMRI responses.
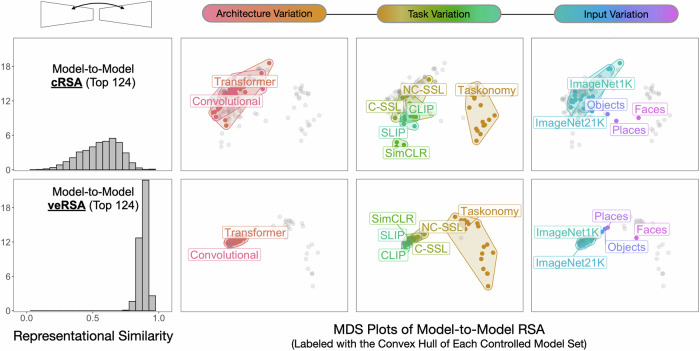
The results are shown in Fig. 6. Considering only the top 124 most brain-predictive models, we find that a direct pairwise comparison of their most brain-aligned layers yields substantial variation in representational similarity, with a range that extends from rPearson = −0.107 to 0.983 (mean = 0.448, SD = 0.148). Thus, these model layers express substantially different representational structure in response to the 500 natural image probe set we use in this analysis. However, the feature-reweighted model representational structure showed a much tighter distribution (mean = 0.881, SD = 0.0313). Thus, the linear reweighting of DNN features in veRSA seems to reveal a remarkably brain-aligned, shared subspace in almost all trained models (or at least, those models trained on a sufficiently diverse image diet).
Fig. 6. Model-to-Model comparison.
Leftmost Panel: Histogram of the pairwise model-to-model representational similarity for the 124 highest-ranking trained models in our survey. The top panel indicates direct layer-to-layer comparisons, while the bottom panel reflects the feature-reweighted layer-to-layer comparisons. Rightward Panels: Results of a multidimensional scaling (MDS) analysis of the model-to-model comparisons, where models whose most brain-predictive layers (selected by cross-validation) share greater representational structure appear in closer proximity. The 3 plots in each row show datapoints output from the same MDS procedure (cRSA, top row; veRSA, bottom row), and the columns show different colored convex hulls that highlight the different model sets from the opportunistic experiments. Note the scale of the MDS plots is the same across all panels. NC-SSL and C-SSL correspond to Non-Contrastive and Contrastive Self-Supervised Learning, respectively. Objects, Faces, and Places correspond to the IPCL models trained on ImageNet1K / OpenImages, Places256, and VGGFace2, respectively. Source data are provided as a Source Data file.
The adjacent subplots in Fig. 6 show a multidimensional scaling plot of the model-to-model comparisons (using all N = 160 trained models), with the axis scales held constant across all facets. Model layers with more similar representational structure are displayed nearby. These plots make clear that there is a substantial degree of raw representational variation amongst these models, which is dramatically compressed when these feature spaces are mapped onto responses in human OTC.
These findings suggest that the somewhat hidden factor of model-to-brain mapping method (and the linking assumptions inherent to these methods) is at least as consequential, if not more consequential, than differences in inductive bias. Indeed, if we treat our metrics themselves (cRSA versus veRSA) as a factor in the same kind of linear regression model we use for our controlled model comparisons, we find that the difference between the two constitutes one of the most substantial effects on brain predictivity of any we assess (second only to that of trained versus random weights): β = 0.23 [0.21, 0.25], p < 0.001 for all models, trained and random; β = 0.30 [0.29, 0.31], p < 0.001 for trained models only.
Discussion
As performant image-computable representation learners, with accessible internal parameters, deep neural networks offer tools for directly operationalizing and testing hypotheses about the formation of high-level visual representations in the brain. This is arguably the core tenet of theoretical frameworks proposed in recent years to unify deep learning and experimental neuroscience (e.g.,11), and encapsulated most strongly in what has been called the “neuroconnectionist research programme”13. According to these frameworks, controlled manipulation across DNNs that learn with different inductive biases, combined with appropriate linking methods and predictivity metrics, has the potential to unveil the pressures that have shaped the representation we measure in the brain, answering questions about “why” these representations appear as they do (14, see also refs. 73,74).
Our work operates within this framework to explore the prediction of representations in human occipitotemporal cortex, using a diverse array of open-source DNN models and a large-scale sampling of brain responses to natural images, related using model-to-brain linking methods that have become the de facto standard in the field. Surprisingly, many of the controlled comparisons among qualitatively different models yield only very small differences in their prediction of high-level visual cortical responses. For example, networks with qualitatively different computational architectures (e.g. convolutional neural networks and vision transformers) yield almost indistinguishable brain-predictivity scores. Furthermore, networks trained to represent natural images using vision-language contrastive learning versus purely visual contrastive learning show remarkably similar capacity to predict OTC responses.
Instead, our analyses point to the importance of a model’s visual experience (i.e. input diet) as a key determinant of downstream brain predictivity that is particularly evident in the poor performance of the indoor-only-trained taskonomy models, and the high performance of OpenAI models trained on their 400 M proprietary image-text database. These results indirectly reveal a currently unquantified factor of dataset diversity as an important predictor of more brain-like visual representation. In addition, our work highlights a critical need to re-examine our standard linking assumptions and model-to-brain pipelines, potentially reducing their flexibility in order to better draw out the representational differences. We next discuss each of these results in turn, and highlight the limitations of the current approach alongside directions for future work in metrics of model-brain representational alignment.
The importance of visual experience
A number of our analyses point to the impact of visual input—not in the size of the image database, but in the diversity of image content—on a model’s emergent brain predictivity. First, we found that the single biggest effect was that of training: untrained models with no visual experience were unable to capture the rich representational structure evident in the late stages of the visual system. Second, impoverished diets (e.g. only faces) yield substantially lower capacity to predict brain responses than richer diets. Taskonomy models showed uniformly lower brain predictivity across the board, which we attribute to an image diet consisting of only indoor scenes. Indeed, many of the tasks in Taskonomy that seem to predict visual cortex rather poorly (e.g. semantic segmentation) seem perfectly capable of producing brain-predictive representations when trained on more diverse image sets (e.g. as is the case with the Detectron models, which rank among the most predictive models in our broader survey). While there has been significant interest in the Taskonomy models for controlled model comparisons (including by us47,75–77), a direct implication of these results is that these models should not be used in future research to make arguments about which brain regions are best fit to which tasks.
The effects of increasing dataset diversity or richness beyond ImageNet1K on brain predictivity are relatively small and difficult to isolate given currently available models and datasets, but are perhaps still evident in our study. For example, we did not find clear improvements between ImageNet1K and 21K, but did find that OpenAI’s 400 million image set seems to be an important factor in positioning the CLIP models as the most predictive of all the models we surveyed (see also51). As a field, computational cognitive neuroscience currently has no single satisfying measure of dataset diversity or richness. Structured image similarity metrics (e.g. SSIM78,79) and perceptual losses (such as those behind neural style-transfer80,81) are in some sense early attempts to address the problem of capturing image similarity in latent representational spaces more complex than pixel statistics, but these algorithms have not been applied directly to the problem of characterizing the intrinsic richness of a visual diet. Key to solving this issue may actually be recent attempts in the computer vision community to distill smaller, less redundant, and more efficient training data from larger image sets by way of “semantic deduplication”—the removal of images from large corpora that image-aligned models like CLIP embed as effectively the same point in their latent spaces82,83—or, similarly, through targeted pruning84. The success of semantic deduplication suggests cognitive neuroscience may be able to leverage comparable measures for understanding the minimal set of visual inputs or experiences necessary for recapitulating the overall patterns of brain responses to natural images.
Finally, another hidden factor of variation related to the input diet is not just the images themselves, but also the suite of data augmentations and related hyperparameters applied over these images during training85. For example, some models experience the ImageNet1K input with a ‘standard’ set of augmentations
which crop and rescale an image each time, with flipped left-right variation and color variation. Other models experience these same images, but with additional augmentations, leveraging some of the newer techniques that blend images and interpolate their labels86,87, increasing performance over standard augmentation schemes36,85. Relatedly, some training regimes use progressive resizing, beginning with small blurred images that ramp upward in resolution over training88,89. These hidden hyperparameters influence what kind of image data is fed to the model at different points in training, and may by implication influence the emergent brain predictivity of learned representations. More generally, there is a need to further curate (or build) sets of DNNs with controlled variation in image diet, image augmentation scheme, and training recipe to explore these factors further.
Model-to-Brain linking methods
In the present work we considered two different model-to-brain linking pipelines. Considered purely in isolation, our cRSA metric might lead us to conclude that our most predictive model layers explain less than a third (∼31%) of the explainable variance in the Natural Scenes Dataset; conversely, our veRSA metric would suggest we have explained the vast majority of that variance (∼79.5%). Currently, it is somewhat unclear which of these is the more correct, not least because we have yet to cohere as a field on the principles of mechanistic intepretability in modeling that logically favor one over the other8,12,90. Relatedly, our model-to-model comparisons highlight that models trained with different inductive biases are indeed learning different representational formats, as revealed by classical RSA. However, the feature re-weighting procedure effectively solves for a similar subspace present in all of the learned feature spaces. This difference demonstrates that there is meaningful diversity in the learned representations of models that our metrics are failing to translate into significantly different brain-predictivity scores. These observations lead to several possible directions for stronger, more diagnostic model-to-brain comparisons.
Analytically, one possible way forward is to develop deeper theoretical commitments to the relationship between single units in the model and single neurons or voxels. For example, adding a sparse positive regularization term to our linear encoding models might better capture the functional role of model unit tuning and require more aligned tuning curves62. Relatedly, we could consider different commitments on the coverage between a given set of model units and brain targets: Perhaps we should allow for the selection of units from across multiple model layers to better account for differences in representational hierarchy63, or maybe even explore one-to-one mappings that require single units or features in models to directly predict single units in the brain91,92? Finally, we could expand the brain target to include not just prediction of the regional geometry and single unit or voxel tuning, but also its topographic organization93–96. Indeed, the right choice of metric (or set of metrics) to evaluate representational alignment between two systems remains an active research frontier (92,97; see ref. 98 for review).
Empirically, another possible route forward is to more carefully select the set of images from which both brain data and model responses are measured. Here, we leveraged the Natural Scenes Dataset30, which contains reliably measured brain responses to a wide variety of natural images—a reasonable and ecologically rich target to try to predict. However, our results highlight that, with this widely sampled natural image set, basically all performant models can capture the major large-scale representational distinctions present in the visual system responses. It is possible—even likely—that this widely sampled image set may actually be obscuring finer-scale representational differences among models and human brain responses that would be revealed with more targeted stimulus comparisons (e.g. the texture-shape cue conflict stimuli of 99; see also refs. 100,101). Exposing models to a targeted array of artificial stimuli (such as line drawings or geometric shapes), or conducting diagnostic psychophysical comparisons (for instance, comparing responses to upright versus inverted or scrambled faces), could help further differentiate models in their ability not only to predict visual brain responses, but visual behavior102–104. Another promising direction would be to pre-select and measure responses to ‘controversial stimuli’, which are novel synthetic images generated to actively differentiate one model from another105,106. A related idea that could be applied to existing datasets is ‘controversial selection’—selecting a subset of images to draw out the differences in model representational geometries, rather than using the whole set.
In sum, it is critical to understand that the results reported here, including the magnitude of RSA scores and the patterns of predictivity across model factors, do not represent absolute truths regarding how “brain-like” these artificial models are across all possible stimuli. All of our conclusions must be understood in the context of both our empirical and analytical choices, which are scoped to assess the brain predictivity of model sets within the class of natural scenes, using two currently standard linking approaches. Relatedly, this means that our method of curating these model sets also matters for the scope of our claims. For example, we intentionally selected models for our comparison of convolutional and transformer architectures from well-known and high-performing model repositories favored by the NeuroAI community. As such, all conclusions about the average effectiveness of convolutional models versus transformer models must be understood with this sampling in mind, and not be interpreted as claims about these classes writ large.
Relationship to prior work
Our approach has some similarities to existing neural benchmarking endeavors, aimed at identifying the most ‘brain-like’ model of the visual system, whether in primates or mice26–29. This approach typically focuses on an aggregate ‘brain-score’, which involves scoring models on data from multiple hierarchical regions, across multiple datasets, and ranking models on a leaderboard according to these aggregate scores. The key addition required of these platforms or pipelines to allow for analyses like ours would be to add a comprehensive collection of model metadata, to serve as the basis of statistical grouping operations. Notably, benchmarking approaches and controlled model comparisons tend to have different theoretical goals. Leaderboards are typically aimed at identifying the single best in-silico model of the biological system. Controlled model comparison is aimed at understanding how higher-order principles govern visual representation formation and emergent brain alignment.
A number of our findings are presaged by previous work that probed individual aspects of the neuroconnectionist research programme, though not necessarily at scale. Regarding architecture, early model-to-brain comparisons found no substantive differences among a set of 9 classic convolutional neural networks (e.g. AlexNet, VGG16, ResNet18) following feature re-mixing and re-weighting25. Regarding input diet, researchers attempting to create a less erroneously labeled alternative to ImageNet found that a more targeted selection of ‘ecologically’ realistic images (‘Eco-Set’) produced a modest but significant effect on downstream predictivity of the human ventral visual system (107; see also ref. 108 for similar findings in mouse visual brain predictivity). More recently and perhaps most relevantly, researchers studying alignment of neural network representations with human similarity judgments (i.e. visual behavior) found also that models trained on ‘larger, more diverse datasets’ (not necessarily in terms of image count alone) were the best predictors of these judgments71.
A number of our findings also contrast with some of the emerging concurrent work in this domain, e.g. arguing for effective dimensionality66, or the primacy of language-aligned features52 as sources of increased brain predictivity. For example, our work does not provide immediate support for effective dimensionality as a model-agnostic predictor of higher brain predictivity. Importantly, however, we do not consider this a claim on the importance of similar model-agnostic metrics more generally: the intersection of manifold statistics and neural coding schemes is an emerging field68,109–111, which we believe will be fruitful for the neuroconnectionist research program, providing deeper insights into representation learning in brains and models alike. Relatedly, another divergent result from our work suggests that language alignment is not in fact the key pressure governing the superlative performance of the OpenAI-CLIP models. Here, too, however, we note that there remains substantial room for more targeted analyses at finer-grained neural resolution and with targeted stimuli that emphasize vision-only versus language-aligned models. These kinds of analyses will be crucial for elucidating what are hypothesized to be significant interactions between vision and language at the most anterior portions of the ventral stream112,113. Indeed, one limitation of our analysis in its current form is that differences in predictivity in smaller (sub)regions of occipitotemporal cortex may potentially be obscured in our more general mask (for example, see ref. 114).
Final considerations
More broadly, even with the 1.8 billion regressions and 50.3 thousand representational similarity analyses underlying the results reported in this paper, we note that our survey approach reflects only a small slice of the possibility space for model-to-brain comparisons with this dataset. For example, we did not try to link hierarchical model layers with hierarchical brain regions (e.g.63), or focus in on category-selective regions (e.g62,64,115) and early visual cortex (though see Supplementary Information SI.5 for initial analyses). To facilitate future work on these fronts, we have open-sourced our codebase (see Data and Code Availability Section), which can be used to conduct these model-to-model, and model-to-brain analyses at scale.
Our aim here was to provide a broad ‘lay-of-the-land’ for relating deep neural network models to the high-level visual system—leveraging controlled variation present in the diversity of available models to conduct opportunistic experiments that isolate factors of architecture, task, and dataset13,14,71,116. Taken together, our results call for a deeper investigation into the impact of visual diet diversity, and highlight the need for conceptual advances in developing theoretically-constrained linking procedures that relate models to brains, along with more diagnostic image sets to further differentiate these highly performant computer vision models.
Methods
Model selection
We collected a set of 224 distinct models (160 trained; 64 randomly-initialized), sourced from the following repositories: the Torchvision (PyTorch) model zoo117; the Pytorch-Image-Models (timm) library57; the VISSL (self-supervised) model zoo42; the OpenAI CLIP collection50; the PyTorch Taskonomy (visualpriors) project41,44,118; the Detectron2 model zoo119; and Harvard Vision Sciences Laboratory’s Open-IPCL project34.
This set of models was collected with a focus on high-level visual representation, and was explicitly intended to span different architectural types, training objectives, and other available variations. Our 64 randomly-initialized models consist of the untrained variants of each ImageNet-1K-trained architecture from the Torchvision and Pytorch-Image-Models repository. (These models were initialized using the default parameters provided by the package, and in most cases are the recommended defaults specified by the contributing authors). Where possible, we extracted the relevant metadata for each model using automatic parsing of web data from the associated repositories; where this automatic parsing was not possible, we manually annotated each model with respect to its associated publication. A list of all included models and their most significant metadata (architecture, task, training data) is included in SI Table 1.
Human fMRI data
The Natural Scenes Dataset30 contains measurements of over 70,000 unique stimuli from the Microsoft Common Objects in Context (COCO) dataset120 at high resolution (7 T field strength, 1.6-s TR, 1.8 mm3 voxel size). In this analysis, we focus on the brain responses to 1000 COCO stimuli that overlapped between subjects, and limit analyses to the 4 subjects (subjects 01, 02, 05, 07) that saw these images in each of their 3 repetitions across scans. The 3 image repetitions were averaged to yield the final voxel-level response values in response to each stimulus. All responses were estimated using a new, publicly available GLM toolbox [GLMsingle121], which implements optimized denoising and regularization procedures to accurately measure changes in brain activity evoked by experimental stimuli.
Voxel selection procedure
To obtain a reasonable signal-to-noise ratio (SNR) in our target data, we implement a reliability-based voxel selection procedure122 to subselect voxels containing stable structure in their responses. Specifically, we use the NCSNR (“noise ceiling signal-to-noise ratio”) metric computed for each voxel as part of the NSD metadata30 as our reliability metric. In this analysis, we include only those voxels with NCSNR > 0.2.
After filtering voxels based on their NCSNR, we then filtered voxels based on region-of-interest (ROI). In our main analyses, we focus on voxels within occipitotemporal cortex (OTC; also referred to as human IT). Our goal was to identify a sector of cortex beyond early visual cortex that covers the ventral and lateral object-responsive cortex, including category-selective regions. To do so, we first considered voxels within a liberal mask of the visual system (“nsdgeneral” ROI). Next we isolated the subset within either the mid-to-high ventral or mid-to-high lateral ROIs (“streams” ROIs). Then, we included all voxels from 11 category-selective ROIs (face, body, word, and scene ROIs, excluding RSC) with a t-contrast statistic > 1; while many of these voxels were already contained in the streams ROIs, this ensures that these regions were included in the larger scale OTC sector. The number of OTC voxels included were 8088 for subject 01, 7528 for subject 02, 8015 for subject 05, and 5849 for subject 07, for a combined total of 29,480 voxels.
Noise ceilings
To contextualize model performance results, we estimated noise ceilings for each of the target brain ROIs. These noise ceilings indicate the maximum possible performance that can be achieved given the level of measurement noise in the data. Importantly, in the present context, noise ceiling estimates refer to within-subject representational similarity matrices (RSMs), where noise reflects trial-to-trial variability in a given subject. This stands in contrast to more conventional group-level representational dissimilarity matrices31, where noise reflects variability across subjects. To estimate within-subject noise ceilings, we developed a novel method based on generative modeling of signal and noise, which we term GSN123. This method estimates, for a given ROI, multivariate Gaussian distributions characterizing the signal and the noise under the assumption that observed responses can be characterized as sums of samples from the signal and noise distributions. A post-hoc scaling is then applied to the signal distribution such that the signal and noise distributions generate accurate matches to the empirically observed reliability of RSMs across independent splits of the experimental data. Noise ceilings are estimated using Monte Carlo simulations in which a noiseless RSM (generated from the estimated signal distribution) is correlated with RSMs constructed from noisy measurements (generated from the estimated signal and noise distributions). All noise ceiling calculations were performed on independent data outside the main analysis.
Feature mapping methods
Feature extraction procedure
For each of our candidate DNN models, we first transform each of our probe images into tensors using the evaluation (“test-time”) image transforms provided with a given model. These image transforms typically involve a resizing operation, followed by pixel normalization using the mean and standard deviation of images within the model’s training dataset. For randomly initialized models, we exclude this normalization step. For the few models whose image transforms are not explicitly defined in the source code, we reconstruct the transforms as faithfully as possible from the associated publication.
We then extract features in response to each of the tensorized probe stimuli at each distinct layer of the network. Importantly, we define a layer here as a distinct computational (sub)module. This means, for example, that we treat convolution and the rectified nonlinearity that follows it as two distinct feature maps; crucially, for transformers, this also means we analyze the outputs not only of each attention head, but of the individual key-query-value modules used to compute them. At the end of our feature extraction procedure, for each model and each model layer, we arrive at a feature matrix of dimensionality number-of-images x number-of-features, the latter value of which represents the flattened dimensions of the original feature map. Beyond flattening, we perform no other transformation of the original features during extraction.
Classical RSA (cRSA)
To compute the classical representational similarity (cRSA) score31 for a single layer, we used the following procedure: First, we split the 1000 images into two sets of 500 (a training set, and a testing set). Using the training set of images, we compute the representational similarity matrices (RSMs) of each model layer (500 × 500 × number-of-layers) using Pearson correlation as the distance metric. We then compare each layer’s RSM to the brain RSM, also using Pearson similarity, and identify the layer with the highest correlation as the model’s most brain-predictive layer. Finally, using the held-out test set of 500 images, we compute that target layer’s RSM and correlate it with the brain RSM. This test score from the most predictive layer serves as the overall cRSA score for the target model.
Voxel-encoding RSA (veRSA)
To arrive at a voxel-encoding representational similarity (veRSA) score32–34 for a single model, the overall procedure was similar to that of cRSA, but with the addition of an intermediate encoding procedure wherein layerwise model features were fit to each individual voxel’s response profile across the image probe set.
The first step in the encoding procedure is the dimensionality reduction of model feature maps. We perform this step for two reasons: first, the features extracted from various deep neural networks can sometimes be massive (the first convolutional layer of VGG16, for example, yields a flattened feature matrix with ∼3.2 million dimensions per image); and second, the same dimensionality reduction procedure applied to all layers ensures that the explicit degrees of freedom across model layers is constant. To reduce dimensionality, we apply the SciKit-Learn implementation of sparse random projection124. This procedure relies on the Johnson-Lindenstrauss (JL) lemma125, which takes in a target number of samples and an epsilon distortion parameter, and returns the number of random projections necessary to preserve the euclidean distance between any two points up to a factor of 1 ± epsilion. (Note that this is a general formula; no brain data enter into this calculation). In our case, with the number of samples set to 1000 (the total number of images) and an epsilon distortion of 0.1, the JL lemma yields a target dimensionality of 5920 projections.
After computing this target dimensionality, we then proceed to compute the sparse random projection for each layer of our target DNN. The sparse random projection matrix consists of zeros and sparse ones, forming nearly orthogonal dimensions, which are then normalized by the density of the matrix (the inverse square root of the total number of features). The layerwise feature maps are then projected onto this matrix by taking the dot product between them. The output of the procedure is a reduced layerwise feature space of size of 1000 images × 5920 dimensions with a preserved representational geometry. Note that in cases where the number of features is less than the number of projections suggested by the JL lemma, the original feature map is effectively upsampled through the random projection matrix, again yielding a matrix of 1000 × 5920 dimensions.
We compute our encoding model for each voxel as a weighted combination of these 5920 dimensions, using brain data from our training set of 500 images. (We note that while the number of dimensions needed for only 500 images would be only D = 5326 according to the JL lemma, adding extra dimensions will only preserve the geometry with nominally less distortion than the epsilon provided, and does not meaningfully affect the results). The fitting procedure for each voxel leverages SciKit-Learn’s cross-validated ridge regression function (“RidgeCV”), a hyperefficient regression method that uses generalized cross-validation to provide a LOOCV prediction per image (per output). This fit was computed over a logarithmic range of alpha penalty parameters (1e−1 to 1e7), to identify each voxel’s optimal alpha parameter. We modified the RidgeCV function in order to select the best alpha using Pearson correlation as a score function (the same score function we use to evaluate the model at large), and to parallelize an internal for-loop for greater efficiency. This yielded a set of encoding weights for each voxel (number-of-voxels × 5920 reduced-feature-dimensions).
Next, with these encoding weights and the 500 training images, we compute the predicted response of every voxel to each image, and compute the corresponding predicted RSM using Pearson correlation. After computing each layer’s representational similarity via Pearson correlation between the layer-predicted RSM and the target brain RSM, we again select the most predictive layer on the basis of results from the training set and compute this layer’s RSA correspondence to the brain data using the held-out set of 500 test images. This test score from the most predictive layer serves as the final veRSA score for the target model.
We emphasize that this method contrasts with popular practices in primate and mouse benchmarking, which treat predictivity of unit-level univariate response profiles as the key outcome measure. Because fMRI affords more systematic spatial sampling over the cortex, rather than taking the aggregate of single voxel fits as our key measure, we choose to treat the population representational geometry over each ROI as the critical target for prediction. This multi-voxel similarity structure provides different kinds of information about the format of population-level coding than do individual units126. Computing the veRSA metric does, however, yield individual voxelwise-encoding models, the individual predictive accuracies of which we collect and have available in addition to the cRSA and veRSA scores for future analysis.
Statistical analysis
Opportunistic experiments
The statistical test for each targeted model set comparison consisted of a linear fixed effects model with brain-predictivity score (cRSA or veRSA, in units of rPearson) as the outcome variable, and two additive effects: that of the experimental manipulation and that of subject ID (to control for overall differences in predictivity across subjects). In the case of the ImagNet1K versus ImageNet21K comparison, a mixed effects model was used to capture the within-model structure of the comparison, with an additional random intercept for model ID. All linear fixed and mixed effects models were fit using R’s ‘stats’127 and ‘lme4’128 packages, respectively. Confidence intervals and p-values were estimated using R’s ‘parameters’ package129, which leverages a Wald t-distribution approximation (for fixed effects), and a Wald z-distribution approximation (for random effects).
We provide each of these linear models (in R-style formulaic pseudocode) below:
CNNs versus Transformers:
lm(Score ∼ ArchitectureClass + SubjectID, reference = CNN)
Taskonomy Encoders:
lm(Score ∼ Task + SubjectID, reference = Denoising)
Contrastive Self-Supervised Learning:
lm(Score ∼ TaskClass + SubjectID, reference = Non-Contrastive)
Language Alignment:
lm(Score ∼ TaskClass + SubjectID, reference = SimCLR)
ImageNet1K versus ImageNet21K:
lmer(Score ∼ DatasetSize + SubjectID + (1|ModelID), reference = ImageNet1K)
Objects, Faces, Places:
lm(Score ∼ Dataset + SubjectID, reference = ImageNet1K-Objects)
Notably, here we are treating subject-level variation as a fixed effect, as there are only 4 subjects whose data contain all available repetitions of the shared 1000 images. By implication, this means that the statistical inferences that are supported by these tests only apply within these 4 participants and not to the general population. On the other hand, we train and test these encoding models on an order of magnitude more brain data points than in prior datasets. Ultimately, our statistical methods were constrained by the particular structure of the NSD fMRI dataset, which prioritizes high-density within-subject stimulus sampling (e.g. 10000 images for 1 subject) at the expense of having a smaller overall number of subjects (e.g. the “Shared1000” images for 4 subjects or the “Special515” for 8 subjects). See130,131 for further discussion on the broader debate of fMRI experimental design.
Breakpoint analysis of model rankings
When analyzing variation across all models, we use a breakpoint analysis132 to quantify the distinct visual elbows that appear in the rank-order of these models. This piece-wise regression method estimates the origin and endpoint of distinct linear sub-segments across an otherwise nonlinear trend. Here, we perform this analysis using R’s ‘segmented’ package133, predicting score by model rank, setting the sole ψ hyperparameter (NBreakpoints) to 2, and computing confidence intervals with the ‘gradient’ method134 for breakpoint interval estimation. We use the first breakpoint yielded by this analysis (over the veRSA brain-predictivity scores) to divide the full set of models into the higher-ranking (‘top’) and lower-ranking (‘bottom’) model sets that we use in several subsequent analyses.
Model-to-Model analysis
To compare the representations of our surveyed models directly, we perform a model-to-model representational similarity analysis, comparing the RSMs from the most brain-predictive layer of each surveyed model directly, with (as in cRSA) or without (as in veRSA) the intermediate encoding of voxels. As in our main analyses, we compute these RSMs with a first-order Pearson correlation, then compare their flattened upper triangular portions with a second-order Pearson correlation.
We reduce the dimensionality (for visualization’s sake) of the resultant modelwise-RSM to 2 dimensions with classical multidimensional scaling (MDS135), as implemented in R’s ‘stats’ package127. To better visualize the representational similarities of distinct groups of models in this reduced space, we draw convex hulls around each group using R’s ‘ggforce’ package136, with the relative concavity of each hull set to 10.
Effective dimensionality
We compute the effective dimensionality (ED) of each of our target layers using the formula proposed by Del Giudice65: the squared sum of all eigenvalues, divided by the sum of the squared eigenvalues.
Note that we compute the ED of these representations both with and without sparse random projection. In a noteworthy validation of the JL lemma’s preservation of pointwise geometry, we find no analytically-relevant difference in the measured value of ED as a function of this dimensionality reduction (rSpearman = 0.993 between ED with and without SRP, across the layers we target here). Given this minimal difference, we use the effective dimensionality of the randomly-projected representations in all main analyses, as these were the feature spaces that were directly used in predicting the brain.
Our approach does differ from that of Elmoznino and Bonner66, as we perform no pooling or other forms of feature aggregation on our targeted layers before computing the ED of these layers. See Supplementary Information SI.2 for more detailed comparisons. We choose not to do a pooling operation before calculating effective dimensionality because (1) this is not a general operation that can be performed on other kinds of (non-convolutional) architectures, and (2) estimating the effective dimensionality over the same feature space used to fit the brain responses seems preferable from the theoretical standpoint of establishing a measure of model variation that meaningfully abstracts over the details of model implementation.
Supplementary information
Source data
Acknowledgements
This work was supported by a Hodgson’s Innovation Fund Grant (C.C.), an NDSEG fellowship (J.S.P.), and NSF-CAREER BCS-1942438 (T.K) and NIH grant R01EY034118 (K.K.). Collection of the NSD dataset was supported by NSF IIS-1822683 (K.K.) and NSF IIS-1822929.
Author contributions
All authors contributed substantially to the completion of this work, and were indispensable in its completion. Colin Conwell performed all model-based data collection and statistical analyses. Jacob Prince parsed the human fMRI data; performed multiple comprehensive reviews of the analytic pipeline and associated codebase; and contributed extensively to the final drafting of the manuscript. Kendrick Kay formulated the noise ceiling procedure, and edited all manuscript drafts. George Alvarez assisted with the formulation of the narrative, and edited all manuscript drafts. Talia Konkle constructed the overall narrative framing for the work, drafted large portions of the final manuscript, and was extensively involved in every step of the writing process.
Peer review
Peer review information
Nature Communications thanks Jeffrey Bowers, Heiko Schütt and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
All data generated in this study have been deposited in our Project GitHub: github.com/ColinConwell/DeepNSD/, and are available for use under a GNU General Public License 3.0. Source data (and versioned code) for the reproduction of all results in this publication (statistics, figures, tables) are available at github.com/ColinConwell/DeepNSD/publication. The brain data used in this analysis are a parse of the larger Natural Scenes Dataset, which is publicly available for download at: naturalscenesdataset.org/. Source data and code for reproducing all statistics, figures, and tables are also available in the accompanying source-data.zip file. Source data are provided with this paper.
Code availability
Code for the reproduction of these results is available at github.com/ColinConwell/DeepNSD/publication.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Colin Conwell, Email: conwell@g.harvard.edu.
Talia Konkle, Email: tkonkle@fas.harvard.edu.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-53147-y.
References
- 1.DiCarlo, J. J., Zoccolan, D. & Rust, N. C. How Does the Brain Solve Visual Object Recognition? Neuron73, 415–434 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hubel, D. H., & Wiesel, T. N. Receptive fields and functional architecture of monkey striate cortex. J. Physiol.195, 215–243 (1968). [DOI] [PMC free article] [PubMed]
- 3.Olshausen, B. A., Field, D. J. et al. Sparse coding of natural images produces localized, oriented, bandpass receptive fields. Submitted to Nature. Available electronically as ftp://redwood. psych. cornell. edu/pub/papers/sparse-coding. ps, 1995. Citeseer.
- 4.Krizhevsky, A., Sutskever, I. & Hinton, G. E. ImageNet classification with deep convolutional neural networks. In Advances in Neural Information Processing Systems, 1097–1105 (2012).
- 5.Yamins, D. L. et al. Performance-optimized hierarchical models predict neural responses in higher visual cortex. Proc. Natl Acad. Sci.111, 8619–8624, (2014). [DOI] [PMC free article] [PubMed]
- 6.Kriegeskorte, N. Deep neural networks: a new framework for modeling biological vision and brain information processing. Annu. Rev. Vis. Sci.1, 417–446, (2015). [DOI] [PubMed]
- 7.Eickenberg, M., Gramfort, A., Varoquaux, G. & Thirion, B. Seeing it all: Convolutional network layers map the function of the human visual system. NeuroImage, 152, 184–194, (2017). [DOI] [PubMed]
- 8.Kay, K. N. Principles for models of neural information processing. NeuroImage180, 101–109 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Kietzmann, T. C., McClure, P. & Kriegeskorte, N. Deep neural networks in computational neuroscience. 10.1093/acrefore/9780190264086.013.46 (2019).
- 10.Thomas, S. Deep learning: the good, the bad, and the ugly. Annu. Rev. Vis. Sci.5, 399–426. 10.1146/annurev-vision-091718-014951 (2019). [DOI] [PubMed]
- 11.Richards, B. A. et al. A deep learning framework for neuroscience. Nat. Neurosci.22, 1761–1770. 10.1038/s41593-019-0520-2 (2019). [DOI] [PMC free article] [PubMed]
- 12.Cao, R. & Yamins, D. Explanatory models in neuroscience, Part 1: Taking mechanistic abstraction seriously. Cognitive Systems Research, 101244 (2024).
- 13.Doerig, A. et al. The neuroconnectionist research programme. Nat. Rev. Neurosci.24, 431–450 (2023). [DOI] [PubMed]
- 14.Kanwisher, N., Khosla, M. & Dobs, K. Using artificial neural networks to ask ‘why’ questions of minds and brains. Trends Neurosci.46, 240–254 (2023). [DOI] [PubMed]
- 15.Güçlü, U. & van Gerven, M. A. Deep neural networks reveal a gradient in the complexity of neural representations across the ventral stream. J. Neurosci.35, 10005–10014 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamins, D. L. & DiCarlo, J. J. Using goal-driven deep learning models to understand sensory cortex. Nat. Neurosci.19, 356 (2016). [DOI] [PubMed]
- 17.Bashivan, P., Kar, K. & DiCarlo, J. J. Neural population control via deep image synthesis. Science, 364, 10.1126/science.aav9436 (2019). [DOI] [PubMed]
- 18.Xiao, W. & Kreiman, G. XDream: Finding preferred stimuli for visual neurons using generative networks and gradient-free optimization. PLoS Comput. Biol.16, e1007973 (2020). [DOI] [PMC free article] [PubMed]
- 19.Khaligh-Razavi, S. M. & Kriegeskorte, N. Deep supervised, but not unsupervised, models may explain IT cortical representation. PLoS Comput. Biol.10, e1003915 (2014). [DOI] [PMC free article] [PubMed]
- 20.Güçlü, U. & van Gerven, M. A. Deep neural networks reveal a gradient in the complexity of neural representations across the ventral stream. J. Neurosci.35, 10005–10014 (2015). [DOI] [PMC free article] [PubMed]
- 21.Cichy, R. M., Khosla, A., Pantazis, D., Torralba, A. & Oliva, A. Comparison of deep neural networks to spatiotemporal cortical dynamics of human visual object recognition reveals hierarchical correspondence. Sci. Rep.6, 10.1038/srep27755 (2016). [DOI] [PMC free article] [PubMed]
- 22.Long, B., Yu, C. P. & Konkle, T. Mid-level visual features underlie the high-level categorical organization of the ventral stream. Proc. Natl Acad. Sci.115, E9015–E9024 (2018). [DOI] [PMC free article] [PubMed]
- 23.Wen, H., Shi, J., Chen, W. & Liu, Z. Deep residual network predicts cortical representation and organization of visual features for rapid categorization. Sci. Rep.8, 1–17 (2018). [DOI] [PMC free article] [PubMed]
- 24.St-Yves, G. & Naselaris, T. The feature-weighted receptive field: an interpretable encoding model for complex feature spaces. NeuroImage, 180, 188–202 (2018). [DOI] [PMC free article] [PubMed]
- 25.Storrs, K. R., Kietzmann, T. C., Walther, A., Mehrer, J. & Kriegeskorte, N. Diverse Deep Neural Networks All Predict Human Inferior Temporal Cortex Well, After Training and Fitting. J. Cogn. Neurosci.33, 2044–2064 (2021). [DOI] [PubMed]
- 26.Martin S. et al. Brain-Score: Which Artificial Neural Network for Object Recognition is most Brain-Like? bioRxiv preprint, 10.1101/407007 (2018).
- 27.Martin S. et al. Integrative benchmarking to advance neurally mechanistic models of human intelligence. Neuron108, 413–423 (2020). [DOI] [PubMed]
- 28.Radoslaw, M. C. et al. The algonauts project: A platform for communication between the sciences of biological and artificial intelligence. arXiv Prepr. arXiv1905, 05675 (2019). [Google Scholar]
- 29.K. F. Willeke et al. The Sensorium competition on predicting large-scale mouse primary visual cortex activity. arXiv preprint arXiv:2206.08666, 2022.
- 30.Allen, E. J. et al. A massive 7T fMRI dataset to bridge cognitive neuroscience and artificial intelligence. Nature Neurosci.25, 116–126 (2022). [DOI] [PubMed]
- 31.Kriegeskorte, N., Mur, M. & Bandettini, P. Representational similarity analysis- connecting the branches of systems neuroscience. Fronti. Syst. Neurosci.2, 4 (2008). [DOI] [PMC free article] [PubMed]
- 32.Khaligh-Razavi, S. M., Henriksson, L., Kay, K. & Kriegeskorte, N. Fixed versus mixed rsa: Explaining visual representations by fixed and mixed feature sets from shallow and deep computational models. J. Math. Psychol.76, 184–197 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaniuth, P. & Hebart, M. N. Feature-reweighted representational similarity analysis: A method for improving the fit between computational models, brains, and behavior. NeuroImage257, 119294 (2022). [DOI] [PubMed] [Google Scholar]
- 34.Konkle, T. & Alvarez, G. A. A self-supervised domain-general learning framework for human ventral stream representation. Nat. Commun.13, 1–12 (2022). [DOI] [PMC free article] [PubMed]
- 35.Yann, L., Yoshua, B. & Geoffrey, H. Deep Learning. Nature521, 436–444 (2015).
- 36.Zhuang, L. et al. A convnet for the 2020s. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pages 11976–11986, 2022.
- 37.McGreivy, N. & Hakim A. Convolutional layers are not translation equivariant. arXiv preprint arXiv:2206.04979, 2022.
- 38.Maithra, R., Thomas, U., Simon, K., Chiyuan, Z. & Alexey, D. Do vision transformers see like convolutional neural networks? Adv. Neural Inf. Process. Syst.34, 12116–12128 (2021). [Google Scholar]
- 39.Muhammad Muzammal, N. et al. Intriguing properties of vision transformers. Adv. Neural Inf. Process. Syst.34, 23296–23308 (2021). [Google Scholar]
- 40.Hong-Yu, Z., Chixiang, L., Sibei, Y. & Yizhou, Y. ConvNets vs. Transformers: Whose visual representations are more transferable? In Proceedings of the IEEE/CVF International Conference on Computer Vision, 2230–2238 (2021).
- 41.Zamir, A. R. et al. Taskonomy: Disentangling task transfer learning. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 3712–3722 (2018).
- 42.Goyal, P. et al VISSL, https://github.com/facebookresearch/vissl 2021.
- 43.Mu, N., Kirillov, A., Wagner, D. & Xie, S. Slip: Self-supervision meets language-image pre-training. European conference on computer vision 529–544 (Springer Nature Switzerland, Cham, 2022).
- 44.Sax, A. et al. Learning to Navigate Using Mid-Level Visual Priors. arXiv:1912.11121 [cs], URL http://arxiv.org/abs/1912.11121. arXiv: 1912.11121 (2019).
- 45.Geirhos, R. et al. On the surprising similarities between supervised and self-supervised models. In: Proceedings of the Shared Visual Representations in Humans & Machines Workshop (NeurIPS) (2020).
- 46.Chen, T., Kornblith, S., Norouzi, M. & Hinton, G. A simple framework for contrastive learning of visual representations. In International conference on machine learning 1597–1607 (PMLR, 2020).
- 47.Conwell, C., Mayo, D., Barbu, A., Buice, M., Alvarez, G. & Katz, B. Neural regression, representational similarity, model zoology & neural taskonomy at scale in rodent visual cortex. Adv. Neural Inf. Process. Syst.34, 5590–5607 (2021). [Google Scholar]
- 48.Nayebi, A. et al. Unsupervised Models of Mouse Visual Cortex. bioRxiv. (Cold Spring Harbor Laboratory, 2021).
- 49.Zhuang, C. et al. Unsupervised neural network models of the ventral visual stream. Proc. Natl Acad. Sci.118, e2014196118 (2021). [DOI] [PMC free article] [PubMed]
- 50.Radford, A. et al. Learning transferable visual models from natural language supervision. In: Proceedings of the International Conference on Machine Learning, PMLR, 8748–8763 (2021).
- 51.Wortsman, M. et al. Robust fine- tuning of zero-shot models. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 7959–7971 (2022).
- 52.Wang, A. Y., Kay, K., Naselaris, T., Tarr, M. J., & Wehbe, L. Incorporating natural language into vision models improves prediction and understanding of higher visual cortex. BioRxiv, 2022–09 (Cold Spring Harbor Laboratory, 2022).
- 53.Kaplan, J. et al. Scaling laws for neural language models. arXiv preprint arXiv:2001.08361, 2020.
- 54.Goyal, P. et al. Vision models are more robust and fair when pretrained on uncurated images without supervision. arXiv preprint arXiv, (2022). 2202.08360.
- 55.Puigcerver, J. et al. Scalable transfer learning with expert models. arXiv preprint arXiv, (2020). 2009.13239.
- 56.Ridnik, T., Ben-Baruch, E., Noy, A. & Zelnik-Manor, L. Imagenet-21k pretraining for the masses. arXiv preprint arXiv, (2021). 2104.10972.
- 57.Wightman, R. Pytorch image models. https://github.com/rwightman/ pytorch-image-models, (2019).
- 58.Gallicchio, C. & Scardapane, S. Deep randomized neural networks. In Recent Trends in Learning From Data: Tutorials from the INNS Big Data and Deep Learning Conference (INNSB- DDL2019), 43–68. (Springer, 10.1007/978-3-030-43883-8_3 2020).
- 59.Cadena, S. A. et al. How well do deep neural networks trained on object recognition characterize the mouse visual system? In NeurIPS Neuro AI Workshop, (2019).
- 60.Hermann, K. & Lampinen, A. What shapes feature representations? exploring datasets, architectures, and training. Adv. Neural Inf. Process. Syst.33, 9995–10006 (2020). [Google Scholar]
- 61.Konkle, T. B. et al. Face detection in untrained deep neural networks. Nat. Commun.12, 7328 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Prince, J. S., Alvarez, G. A. & Konkle, T. Contrastive learning explains the emergence and function of visual category-selective regions. Science Advances, 10, (2024). [DOI] [PMC free article] [PubMed]
- 63.Nonaka, S., Majima, K., Aoki, S. C. & Kamitani, Y. Brain hierarchy score: Which deep neural networks are hierarchically brain-like? IScience24, 103013 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ratan Murty, N. A., Bashivan, P., Abate, A., DiCarlo, J. J. & Kanwisher, N. Compu- tational models of category-selective brain regions enable high-throughput tests of selectivity. Nat. Commun.12, 5540 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Marco Del Giudice. Effective dimensionality: A tutorial. Multivariate behavioral research, 56, 527–542. 10.1080/00273171.2020.1743631. (Taylor & Francis, 2021). [DOI] [PubMed]
- 66.Elmoznino, E. & Bonner, M. F. High-performing neural network models of visual cortex benefit from high latent dimensionality. bioRxiv, 2022–07, (Cold Spring Harbor Laboratory, 2022). [DOI] [PMC free article] [PubMed]
- 67.Garrido, Q., Balestriero, R., Najman, L. & Lecun, Y. Rankme: Assessing the downstream performance of pretrained self-supervised representations by their rank. In International conference on machine learning 10929–10974 (PMLR, 2023).
- 68.Yerxa, T., Kuang, Y., Simoncelli, E. & Chung, S. Learning efficient coding of natural images with maximum manifold capacity representations. Advances in Neural Information Processing Systems, 36, 24103–24128 (2023).
- 69.Linsley, D. et al. Performance-optimized deep neural networks are evolving into worse models of inferotemporal visual cortex. Advances in Neural Information Processing Systems, 36 (2024).
- 70.Geirhos, R. et al. Partial success in closing the gap between human and machine vision. Adv. Neural Inf. Process. Syst.34, 23885–23899 (2021). [Google Scholar]
- 71.Muttenthaler, L., Dippel, J., Linhardt, L., Vandermeulen, R. A. & Kornblith, S. Human alignment of neural network representations. In: Proceedings of the 11th International Conference on Learning Representations (ICLR), (2023).
- 72.Dehghani, M. et al. Scaling vision transformers to 22 billion parameters. arXiv preprint arXiv, (2023). 2302.05442.
- 73.Wood, J. N., Lee, D., Wood, B. & Wood, S. M. W. Reverse engineering the origins of visual intelligence. In CogSci, 2020. [DOI] [PubMed]
- 74.Vong, W. K., Wang, W., Orhan, A. E. & Lake, B. M. Grounded language acquisition through the eyes and ears of a single child. Science383, 504–511 (2024). [DOI] [PubMed] [Google Scholar]
- 75.Wang, A., Tarr, M. & Wehbe, L. Neural taskonomy: Inferring the similarity of task-derived representations from brain activity. Adv. Neural Inform. Proc. Syst.32, (2019).
- 76.Dwivedi, K., Bonner, M. F., Cichy, R. M. & Roig, G. Unveiling functions of the visual cortex using task-specific deep neural networks. PLoS Comput. Biol.17, e1009267 (2021). [DOI] [PMC free article] [PubMed]
- 77.Cadena, S. A. et al. Diverse task-driven modeling of macaque V4 reveals functional specialization towards semantic tasks. bioRxiv 2022–05. (Cold Spring Harbor Laboratory, 2022). [DOI] [PMC free article] [PubMed]
- 78.Wang, Z., Bovik, A. C., Sheikh, H. R. & Simoncelli, E. P. Image quality assessment: from error visibility to structural similarity. IEEE Trans. image Process.13, 600–612 (2004). [DOI] [PubMed] [Google Scholar]
- 79.Dosselmann, R. & Yang, X. D. A comprehensive assessment of the structural similarity index. Signal, Image Video Process.5, 81–91 (2011). [Google Scholar]
- 80.Gatys, L. A., Ecker, A. S., Bethge, M., Hertzmann, A. and Shechtman, E. Controlling perceptual factors in neural style transfer. In Proceedings of the IEEE conference on computer vision and pattern recognition, 3985–3993 (2017).
- 81.Jing, Y. et al. Neural style transfer: A review. IEEE Trans. Vis. computer Graph.26, 3365–3385 (2019). [DOI] [PubMed] [Google Scholar]
- 82.Abbas, A., Tirumala, K., Simig, D., Ganguli, S. & Morcos, A. S. Semdedup: Data-efficient learning at web-scale through semantic deduplication. In: Proceedings of the ICLR 2023: Multimodal Representation Learning Workshop (2023).
- 83.Gadre, S. Y. et al. Datacomp: In search of the next generation of multimodal datasets. Advances in Neural Information Processing Systems, vol. 36, (2024).
- 84.Sorscher, B., Geirhos, R., Shekhar, S., Ganguli, S. & Ari, M. Beyond neural scaling laws: beating power law scaling via data pruning. Adv. Neural Inf. Process. Syst.35, 19523–19536 (2022). [Google Scholar]
- 85.Wightman, R., Touvron, H. & Jégou, H. Resnet strikes back: An improved training procedure in timm. arxiv 2021. arXiv preprint arXiv:2110.00476, 2021.
- 86.Zhang, H., Cisse, M., Dauphin, Y. N. & Lopez-Paz, D. mixup: Beyond empirical risk minimization. In: Proceedings of the International Conference on Learning Representations (ICLR) (2018).
- 87.Yun, S. et al Cutmix: Regularization strategy to train strong classifiers with localizable features. In Proceedings of the IEEE/CVF international conference on computer vision, 6023–6032 (2019).
- 88.The Mosaic ML Team. composer. https://github.com/mosaicml/composer/, (2021).
- 89.Guillaume L., Andrew I., Logan E., Sung Min P., Hadi S. and Aleksander M. dry. Ffcv: Accelerating training by removing data bottlenecks. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 12011–12020 (2023).
- 90.Han, Y., Poggio, T. A. & Cheung, B. System Identification of Neural Systems: If We Got It Right, Would We Know? In: Proceedings of the 40th International Conference on Machine Learning, PMLR, vol. 202, 12430–12444 (2023).
- 91.Arend, L. et al. Single units in a deep neural network functionally correspond with neurons in the brain: preliminary results. Technical report, Center for Brains, Minds and Machines (CBMM), (2018).
- 92.Khosla, M. & Williams, A. H. Soft Matching Distance: A metric on neural representations that captures single-neuron tuning. Proceedings of UniReps: the First Workshop on Unifying Representations in Neural Models, (PMLR, 2024).
- 93.Lee, H. et al. Topographic deep artificial neural networks reproduce the hallmarks of the primate inferior temporal cortex face processing network. bioRxiv, 2020–07 (2020).
- 94.Nicholas, M. Blauch, Marlene Behrmann, and David C Plaut. A connectivity-constrained computational account of topographic organization in primate high-level visual cortex. Proc. Natl Acad. Sci.119, e2112566119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Margalit, E. et al. A unifying principle for the functional organization of visual cortex. bioRxiv, 2023–05, (2023).
- 96.Doshi, F. R. & Konkle, T. Cortical topographic motifs emerge in a self-organized map of object space. Sci. Adv.9, eade8187 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Williams, A. H., Kunz, E., Kornblith, S. & Linderman, S. W. Generalized shape metrics on neural representations. Adv. Neural Inf. Process. Syst.34, 4738–4750 (2021). [PMC free article] [PubMed] [Google Scholar]
- 98.Sucholutsky, I. et al. Getting aligned on representational alignment. arXiv preprint arXiv:2310.13018, 2023.
- 99.Geirhos, R. et al. ImageNet-trained CNNs are biased towards texture; increasing shape bias improves accuracy and robustness. Proceedings of the International Conference on Learning Representations (ICLR) (2019).
- 100.Xu, Y. & Vaziri-Pashkam, M. Limits to visual representational correspondence between convolutional neural networks and the human brain. Nat. Commun.12, 2065 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Feather, J., Leclerc, G., Mądry, A. & McDermott, J. H. Model metamers reveal divergent invariances between biological and artificial neural networks. Nat. Neurosci.26, 2017–2034 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rust, N. C. & Movshon, J. A. In praise of artifice. Nat. Neurosci.8, 1647–1650 (2005). [DOI] [PubMed] [Google Scholar]
- 103.Krakauer, J. W., Ghazanfar, A. A., Gomez-Marin, A., MacIver, M. A. & Poeppel, D. Neuroscience needs behavior: correcting a reductionist bias. Neuron93, 480–490 (2017). [DOI] [PubMed] [Google Scholar]
- 104.Bowers J. S. et al. Deep problems with neural network models of human vision. Behav. Brain Sci. 1–74, (2022). [DOI] [PubMed]
- 105.Golan, T., Raju, P. C. & Kriegeskorte, N. Controversial stimuli: Pitting neural networks against each other as models of human cognition. Proc. Natl Acad. Sci.117, 29330–29337 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Golan, T., Guo, W., Schütt, H. H. & Kriegeskorte, N. Distinguishing representational geometries with controversial stimuli: Bayesian experimental design and its application to face dissimilarity judgments. Proceedings of the SVRHM 2022 Workshop @ NeurIPS (2022).
- 107.Mehrer, J., Spoerer, C. J., Jones, E. C., Kriegeskorte, N. & Kietzmann, T. C. An ecologically motivated image dataset for deep learning yields better models of human vision. Proc. Natl Acad. Sci.118, e2011417118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Prasad A., Manor U. & Pereira T. Exploring the role of image domain in self-supervised dnn models of rodent brains. In SVRHM 2022 Workshop@ NeurIPS, 2022.
- 109.Cohen, U., Chung, S., Lee, D. D. & Sompolinsky, H. Separability and geometry of object manifolds in deep neural networks. Nat. Commun.11, 746 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sorscher, B., Ganguli, S. & Sompolinsky, H. Neural representational geometry underlies few-shot concept learning. Proc. Natl Acad. Sci.119, e2200800119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kuoch, M. et al. Probing biological and artificial neural networks with task-dependent neural manifolds. In Conference on Parsimony and Learning, 395–418. (PMLR, 2024).
- 112.Popham, S. F. et al. Visual and linguistic semantic representations are aligned at the border of human visual cortex. Nat. Neurosci.24, 1628–1636 (2021). [DOI] [PubMed]
- 113.Tang, J., Du, M., Vo, V., Lal, V. & Huth, A. Brain encoding models based on multimodal transformers can transfer across language and vision. Adv. Neural Inf. Process. Syst.36 (2024). [PMC free article] [PubMed]
- 114.Wang, A. Y., Kay, K., Naselaris, T., Tarr, M. J. & Wehbe, L. Better models of human high-level visual cortex emerge from natural language supervision with a large and diverse dataset. Nat. Mach. Intell.5, 1415–1426 (2023). [Google Scholar]
- 115.Prince, J. S. & Konkle, T. Computational evidence for integrated rather than specialized feature tuning in category-selective regions. J. Vis.20, 1577–1577 (2020). [Google Scholar]
- 116.Ren, Y. & Bashivan, P. How well do models of visual cortex generalize to out of distribution samples? bioRxiv, 2023–05 (2023). [DOI] [PMC free article] [PubMed]
- 117.Paszke, A. et al. PyTorch: An Imperative Style, High- Performance Deep Learning Library. In H. Wallach, H. Larochelle, A. Beygelzimer, F. d’ Alché- Buc, E. Fox, and R. Garnett, editors, Adv. Neural Inform. Proc. Syst.32, 8024–8035. Curran Associates, Inc., http://papers.neurips.cc/paper/9015-pytorch-an-imperative-style-high-performance-deep-learning-library.pdf (2019).
- 118.Sax, A. et al. Mid-Level Visual Representations Improve Generalization and Sample Efficiency for Learning Visuomotor Policies. (2018).
- 119.Wu, Y., Kirillov, A., Massa, F., Lo, W.-Y. & Girshick, R. Detectron2, https://github.com/facebookresearch/detectron2 2019.
- 120.Lin, T. -Y. et al. Microsoft coco: Common objects in context. In European conference on computer vision, 740–755 (Springer, 2014).
- 121.Prince, J. S. et al. Improving the accuracy of single-trial fMRI response estimates using GLMsingle. Elife, 11, e77599 (2022). [DOI] [PMC free article] [PubMed]
- 122.Tarhan, L. & Konkle T. Reliability-based voxel selection. NeuroImage, 207, 116350 (2020). [DOI] [PubMed]
- 123.Kay, K. et al. Disentangling signal and noise in neural responses through generative modeling. bioRxiv. (2024).
- 124.Pedregosa, F. et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 12, 2825–2830 (2011).
- 125.Achlioptas, D. Database-friendly random projections. In Proceedings of the twentieth ACM SIGMOD-SIGACT-SIGART symposium on Principles of database systems, 274–281 (2001).
- 126.Kriegeskorte, N. et al. Matching categorical object representations in inferior temporal cortex of man and monkey. Neuron60, 1126–1141 (2008). [DOI] [PMC free article] [PubMed]
- 127.R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria, http://www.R-project.org/.ISBN3-900051-07-0 (2013).
- 128.Bates, D., Mächler, M., Bolker, B. & Walker, S. Fitting linear mixed-effects models using lme4. J. Stat. Softw.67, 1–48 (2015). [Google Scholar]
- 129.Lüdecke, D., Ben-Shachar, M. S., Patil, I. & Makowski, D. Extracting, computing and exploring the parameters of statistical models using R. J. Open Source Softw.5, 2445 (2020). [Google Scholar]
- 130.Poldrack, R. A. et al. Scanning the horizon: towards transparent and reproducible neuroimaging research. Nat. Rev. Neurosci.18, 115–126 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Naselaris, T., Allen, E. & Kay, K. Extensive sampling for complete models of individual brains. Curr. Opin. Behav. Sci.40, 45–51 (2021). [Google Scholar]
- 132.Vito, M. R. Muggeo. Estimating regression models with unknown break-points. Stat. Med.22, 3055–3071 (2003). [DOI] [PubMed] [Google Scholar]
- 133.Vito, M. R. Muggeo. segmented: an r package to fit regression models with broken-line relationships. R. N.8, 20–25, https://cran.r-project.org/doc/Rnews/ (2008). [Google Scholar]
- 134.Vito, M. R. Muggeo. Interval estimation for the breakpoint in segmented regression: a smoothed score-based approach. Aust. N.Z. J. Stat.59, 311–322 (2017). [Google Scholar]
- 135.John, C. G. Some distance properties of latent root and vector methods used in multivariate analysis. Biometrika53, 325–338 (1966). [Google Scholar]
- 136.Pedersen, T. L. ggforce: Accelerating ’ggplot2’, https://ggforce.data-imaginist.com, https://github.com/thomasp85/ggforce (2022).
- 137.Richard, D. M. et al. Confidence intervals from normalized data: A correction to cousineau (2005). Tutorials in Quantitative Methods for Psychology4, 61–64 (2008).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated in this study have been deposited in our Project GitHub: github.com/ColinConwell/DeepNSD/, and are available for use under a GNU General Public License 3.0. Source data (and versioned code) for the reproduction of all results in this publication (statistics, figures, tables) are available at github.com/ColinConwell/DeepNSD/publication. The brain data used in this analysis are a parse of the larger Natural Scenes Dataset, which is publicly available for download at: naturalscenesdataset.org/. Source data and code for reproducing all statistics, figures, and tables are also available in the accompanying source-data.zip file. Source data are provided with this paper.
Code for the reproduction of these results is available at github.com/ColinConwell/DeepNSD/publication.