Abstract
Cis-regulatory elements (CREs), such as promoters and enhancers, are DNA sequences that regulate the expression of genes. The activity of a CRE is influenced by the order, composition, and spacing of sequence motifs that are bound by proteins called transcription factors (TFs). Synthetic CREs with specific properties are needed for biomanufacturing as well as for many therapeutic applications including cell and gene therapy. Here, we present regLM, a framework to design synthetic CREs with desired properties, such as high, low, or cell type–specific activity, using autoregressive language models in conjunction with supervised sequence-to-function models. We used our framework to design synthetic yeast promoters and cell type–specific human enhancers. We demonstrate that the synthetic CREs generated by our approach are not only predicted to have the desired functionality but also contain biological features similar to experimentally validated CREs. regLM thus facilitates the design of realistic regulatory DNA elements while providing insights into the cis-regulatory code.
Cis-regulatory elements (CREs), such as promoters and enhancers, are DNA sequences that regulate gene expression. Their activity is influenced by the presence, order, and spacing of sequence motifs (Wittkopp and Kalay 2012) that bind to proteins called transcription factors (TFs), similarly to how words and phrases define the meaning of a sentence. Synthetic CREs with specific properties are needed for biomanufacturing as well as numerous therapeutic applications including cell and gene therapy, for example, to maximize the activity of a therapeutic gene in the target cell type.
Such CREs are often designed manually based on prior knowledge (Fornes et al. 2023). Recent studies have used directed evolution (Taskiran et al. 2024) and gradient-based approaches (Schreiber and Lu 2020; Linder and Seelig 2021; Gosai et al. 2023) for CRE design, in which supervised “oracle” models are trained to predict the activity of a CRE from its sequence, and are then used to edit sequences iteratively until the desired prediction is achieved. However, such approaches are not truly generative and do not necessarily learn the overall sequence distribution of the desired CREs. Instead, they may only optimize specific features that have high predictive value. Consequently, the resulting CREs may be out-of-distribution and unrealistic, leading to unpredictable behavior when they are experimentally tested in a cell.
Autoregressive language models, such as generative pretrained transformer (GPT), can produce realistic content in natural languages (Brown et al. 2020). Here, we present regLM, a framework to design synthetic CREs with desired properties, such as high, low, or cell type–specific activity, using autoregressive language models in conjunction with supervised models. Although masked language models have been used to embed or classify DNA sequences (Ji et al. 2021; Benegas et al. 2023; Dalla-Torre et al. 2023; Fishman et al. 2023; Zhou et al. 2024), to our knowledge this is the first time language modeling has been used for DNA in a generative setting.
Results
regLM adapts the HyenaDNA framework for CRE generation
Several transformer-based foundation models for DNA have been developed (Ji et al. 2021; Benegas et al. 2023; Dalla-Torre et al. 2023; Fishman et al. 2023; Zhou et al. 2024). However, these methods are based on masked language modeling which is difficult to use for sequence generation. In contrast, the recent HyenaDNA foundation models (Nguyen et al. 2023) are single-nucleotide resolution autoregressive models trained on the human genome, and are hence suitable for regulatory element design. These models are based on the Hyena operator (Poli et al. 2023), which uses implicit convolutions to scale subquadratically with sequence length.
regLM builds on the HyenaDNA framework to perform generative modeling of CREs with desired properties using prompt tokens. This takes advantage of the resolution and computational efficiency of the HyenaDNA model. Further, the ability to fine-tune pretrained models which have already learned regulatory features assists in design tasks which lack sufficient labeled data for training.
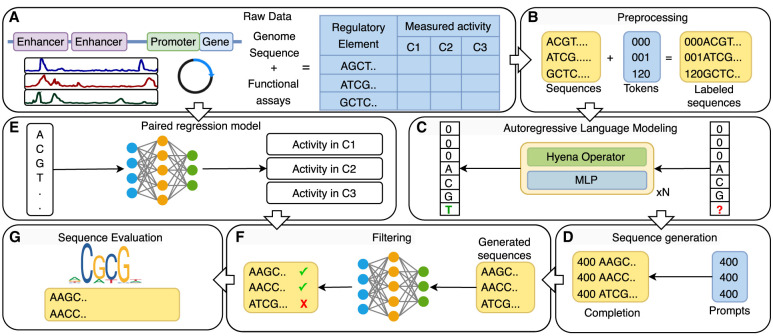
Given a data set of DNA sequences labeled with their measured activity (Fig. 1A), we encode the label in a sequence of categorical tokens (prompt tokens), which is prefixed to the beginning of the DNA sequence (Fig. 1B). We train or fine-tune a HyenaDNA model to take the processed sequences and perform next token prediction beginning with the prompt tokens (Fig. 1C). This formulation allows us to use any prior knowledge on sequences in the model explicitly.
Figure 1.
Schematic of regLM. (A,B) DNA sequences are prefixed with a sequence of prompt tokens representing functional labels. (C) A HyenaDNA model is trained or fine-tuned to perform next token prediction on the labeled sequences. (D) The trained model is prompted with a sequence of prompt tokens to generate sequences with desired properties. (E,F) A sequence-to-function regression model trained on the same data set is used to check and filter the generated sequences. (G) The regulatory content of generated sequences is evaluated.
Once trained, the language model can be prompted with the sequence of tokens representing any desired function. The model, now conditioned on the prompt tokens, generates a DNA sequence 1 nt at a time (Fig. 1D). In parallel, we train a supervised sequence-to-activity regression model on the same data set (Fig. 1E), and apply it to the generated sequences to select those that best match the desired activity (Fig. 1F). This combined approach allows us to use the regression model as an oracle like previous model-guided approaches, while the language model ensures that the generated sequences have realistic content. Finally, we provide several approaches to evaluate the generated sequences as well as the model itself (Fig. 1G).
regLM generates yeast promoters of varying strength
Training and evaluating a regLM model on yeast promoter sequences
We applied the regLM framework to a data set of randomly generated 80 bp DNA sequences and their measured promoter activities in yeast grown in complex and defined media (de Boer et al. 2020; Vaishnav et al. 2022). We prefixed each sequence with a two-token label, wherein each token ranges from 0 to 4 and represents the promoter activity in one of the media (Supplemental Fig. S1). For example, the label 00 indicates that the sequence has low activity in both media, whereas 04 indicates low activity in the complex medium and high activity in the defined medium (Fig. 2A).
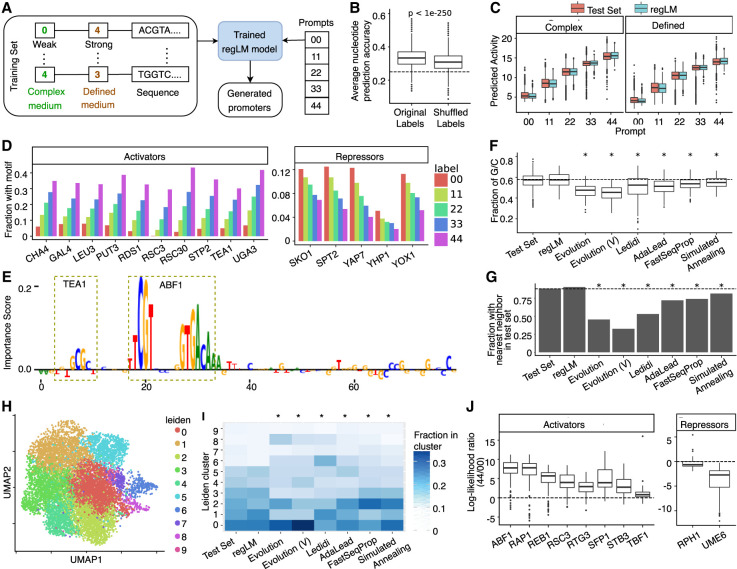
Figure 2.
regLM generates synthetic yeast promoters. (A) Schematic of the experiment. (B) Box plot showing the mean accuracy of the trained regLM model on test set sequences, before and after randomly shuffling the labels among sequences. The dashed line represents the accuracy of 0.25 expected by chance. (C) Predicted activity of regLM-generated promoters, compared to promoters from the test set with the same label. (D) Fraction of regLM promoters prompted with different labels that contain the TF motifs most strongly correlated with promoter activity in the test set. (E) Example of a regLM-generated strong promoter. Height represents the per-nucleotide importance score obtained from the paired regression model using in silico mutagenesis. Motifs with high importance are highlighted. (F) Fraction of G/C bases in strong promoters generated by different methods. (G) Fraction of generated promoters whose nearest neighbor based on k-mer content is a validated promoter from the test set, for different methods. (H) UMAP visualization of true (Test Set) and synthetic strong promoters, labeled by cluster membership. (I) Cluster distribution of strong promoters generated by different methods. (J) Box plots showing the log-ratio between the likelihood of the motif sequence given label 44 (high activity) versus label 00 (low activity) for activating or repressing TF motifs inserted in random sequences. Motifs were selected based on TF-MoDISco results. In F, G, and I, asterisks indicate significant (P < 0.05) differences from the test set, and Evolution (V) represents synthetic promoters generated by Vaishnav et al. (2022).
A regLM model trained to perform the next nucleotide prediction on this data set reached 31% mean accuracy on native yeast promoters and 33.8% accuracy on the test set (Supplemental Fig. S2). This test set performance exceeded the 25% accuracy expected by chance as well as the 31.3%–31.6% accuracy of n-gram models trained on the same data, and the 31.3%–32.7% accuracy of n-gram models trained on the test set itself (Supplemental Fig. S3). Accuracy reduced when we randomly shuffled the labels across sequences (Fig. 2B; Supplemental Fig. S4) (One-sided Mann–Whitney U test P-value = 8.8 × 10−37 for native promoters, P < 10−250 for test set), indicating that the model learned to use the information encoded in the prompt tokens.
Within the test set, we observed higher accuracy in motifs for known yeast TFs (Supplemental Fig. S5A) (One-sided Mann–Whitney U test P-value = 5.3 × 10−77). Accuracy increased with the abundance of the motif in the data set (Supplemental Fig. S5B) (Pearson's rho = 0.55, P-value = 2.7 × 10−12). As expected due to the autoregressive formulation of the model, accuracy increased along the length of the test sequences (Supplemental Fig. S6) (Pearson's rho = 0.21, P-value = 6 × 10−22). As a result, the model's mean accuracy within abundant motifs occurring in the last 15 nt of the promoter sequence was 40%, significantly higher than its accuracy across all nucleotides in the sequence (Supplemental Fig. S7) (Mann–Whitney U test P-value < 10−250).
Finally, we asked whether the model learned to associate specific motifs with categories of promoter activity. For each motif, we calculated the relative abundance of the motif in strong promoters (label 44) versus weak promoters (label 00). We also calculated the ratio between the model's accuracy within the motif when the motif was present in strong promoters versus weak promoters. The strong correlation between these two metrics (Supplemental Fig. S8) (Pearson's rho = 0.88, P-value = 5.6 × 10−45), indicates that the model has learned to associate the prompt tokens with motifs that are consistent with the corresponding promoter activity; for example, when it observes the prompt 44, the model is more accurate at predicting motifs that tend to occur in strong promoters.
Generating synthetic yeast promoters
We generated promoters of defined strength by prompting the trained regLM model with labels 00, 11, 22, 33, and 44 (Supplemental Table S1). Generated sequences were distinct from each other and from the training set, having a minimum edit distance of 25 bp from training sequences. Supervised regression models trained on the same data as the language model (Supplemental Fig. S9) were used to discard generated sequences whose predicted activity did not match the prompt. Only 1.1% of the generated sequences were discarded.
Independent regression models trained on separate data from the language model (Supplemental Fig. S10) predicted that regLM generates stronger promoters when prompted with higher labels, and that the activity of the generated promoters matches that of held-out test promoters with the same label (Fig. 2C). The abundance of TF motifs in the generated promoters was strongly correlated with their abundance in the test set; in other words, when regLM was prompted with the label 44 its generated sequences were more likely to contain motifs for activating yeast TFs that are often seen in strong promoters (Supplemental Fig. S11; Fig. 2D).
In addition to motif abundance, we also examined per-base importance using in silico mutagenesis (ISM). Using TF-MoDISco (Shrikumar et al. 2018), we identified motifs for known activator (ABF1, REB1, RAP1, RSC3, SFP1, STB3) and repressor (UME6) TFs with high importance in both the test set and the generated promoters, indicating that regLM generates motifs that contribute strongly to regulatory activity (Fig. 2E; Supplemental Figs. S12, S13; Supplemental Table S2).
regLM generates promoters with diverse and realistic sequence content
To assess the biological realism of regLM-generated promoters relative to CREs generated by other methods, we compared 200 putative strong promoters generated by regLM (prompted with label 44; Supplemental Table S3) to sequences of similar predicted activity generated by five approaches (Directed evolution, Ledidi [Schreiber and Lu 2020], AdaLead, FastSeqProp, and Simulated Annealing) as well as synthetic strong promoters generated in another study (Supplemental Fig. S14; Supplemental Table S4; Vaishnav et al. 2022). For a fair comparison, we performed all five model-guided methods using the regression model trained on the same data set as regLM as an oracle. All sets of synthetic promoters were compared to known strong promoters from the test set using Polygraph (Lal et al. 2023). Below, we use Evolution (V) to refer to synthetic promoters generated by Vaishnav et al. (2022) using an evolution-based approach.
GC content (the percentage of G or C nucleotides in a sequence) is a useful biological metric to evaluate the realism of synthetic sequences. regLM promoters were most similar to test set promoters in GC content, whereas other approaches produced sequences with lower GC content (Fig. 2F) (Kruskal–Wallis P-value 5.7 × 10−173; Dunn's post-hoc P-values 3.5 × 10−69 [Evolution vs. Test Set], 1.5 × 10−70 [Evolution (V) vs. Test Set], 2.3 × 10−15 [Ledidi vs. Test Set], 1.6 × 10−27 [AdaLead vs. Test Set], 4.2 × 10−11 [FastSeqProp vs. Test Set], 1.2 × 10−6 [Simulated Annealing vs. Test Set]).
We counted the frequency of all k-mers of length 4 in all promoters. No k-mers were differentially abundant (defined as having two-sided Mann–Whitney U test adjusted P-value < 0.05) in regLM promoters with respect to test set promoters, compared to 27–122 differentially abundant k-mers in the promoter sets generated by other methods (Supplemental Table S5). When we matched each sequence to its nearest neighbor based on their k-mer frequencies, over 90% of regLM promoters were matched to a test set promoter, unlike other methods (Fig. 2G; Supplemental Table S5). regLM-generated promoters were among the most difficult to distinguish from the test set using simple classifiers based on k-mer frequency (Supplemental Table S5).
We repeated the above analyses using the frequency of yeast TF binding motifs in all promoters (see Methods). regLM and Simulated Annealing were the only methods that returned no differentially abundant motifs (defined as having two-sided Mann–Whitney U test adjusted P-value < 0.05) with respect to the test set (Supplemental Table S5). regLM-generated promoters were the most likely to have a test set promoter as their nearest neighbor based on motif frequency (Supplemental Table S5). regLM-generated promoters were also among the most difficult to distinguish from the test set using simple classifiers based on motif frequency (Supplemental Table S5).
To assess realism at the level of regulatory syntax, we examined combinations of motifs present in the generated sequences. We first computed the frequencies of pairwise combinations of motifs. Out of 2321 motif pairs that were present in over 5% of any group of promoters, only one was differentially abundant (defined as having two-sided Fisher's exact test adjusted P-value < 0.01) in regLM promoters with respect to test set promoters. In contrast, 21–439 motif pairs were differentially abundant in the other sets of synthetic promoters (Supplemental Table S5). Motifs in regLM-generated promoters also did not occur in significantly different positions compared to their positions in the test set (Supplemental Table S5) (defined as two-sided Mann–Whitney U test P-value < 0.01).
We examined the distance and orientation between paired motifs in each group of promoters. For each motif pair, we counted the fraction of occurrences of the pair in which both motifs were in the same orientation, in each group of synthetic promoters as well as the test set. We found that the same-orientation fractions for motif pairs in regLM-generated promoters showed the highest Pearson's correlation with those in the test set (Supplemental Table S5). We also tested whether the distance between motifs in these pairs was significantly different in synthetic promoters relative to the test set. regLM-generated promoters were the third lowest in the number of motif pairs with significantly different distances (defined as having two-sided Mann–Whitney U test adjusted P-value < 0.01) (Supplemental Table S5).
To assess whether larger combinations of co-occurring motifs are shared between real and synthetic promoters, we performed graph-based clustering of real and synthetic strong promoters (Traag et al. 2019) based on their TF motif content. This revealed 10 clusters (Fig. 2H) corresponding to different combinations of co-occurring TF motifs (Supplemental Fig. S15; Supplemental Note S1). All 10 clusters were represented in regLM promoters in similar proportion to their abundance in the test set; in contrast, the sets of sequences generated by other methods had skewed cluster representation, suggesting a tendency to converge upon specific transcriptional programs (Fig. 2I; Supplemental Fig. S16) (χ2 P-values 2.6 × 10−14 [Evolution vs. Test Set], 3.6 × 10−54 [Evolution (V) vs. Test Set], 1.3 × 10−68 [Ledidi vs. Test Set], 1.0 × 10−11 [AdaLead vs. Test Set] 4.1 × 10−3 [FastSeqProp vs. Test Set] 2.8 × 10−3 [Simulated Annealing vs. Test Set]).
Finally, we embedded all the real and synthetic promoters in a latent space defined by the convolutional layers of the independent regression models. The distance between sequences in this latent space incorporates not only differences in the frequency of important motifs, but also more complex regulatory syntax learned by the regression model such as motif orientation and spacing. Within this latent space, regLM promoters were still the most likely to have a test set promoter as their nearest neighbor (Supplemental Table S5). Together, this evidence demonstrates comprehensively that regLM has learned many aspects of the yeast regulatory code.
Interrogating the trained regLM model reveals species-specific regulatory grammar
To learn whether interrogating the trained regLM model could reveal the regulatory rules of yeast cells, we selected motifs for activating and repressing yeast TFs based on TF-MoDISco results (see Methods) and inserted each motif into 100 random DNA sequences. We used the trained regLM model to compute the likelihoods of the resulting sequences (P(sequence|label)) given either label 44 (strong promoter) or 00 (weak promoter). For each synthetic promoter, we defined a log-likelihood ratio as follows:
A positive log-ratio indicates that the model has learned the motif is more likely to occur in sequences with label 44 than with label 00, whereas a negative log-ratio indicates the opposite. We observed that sequences containing activating motifs tend to have positive log-likelihood ratios, whereas sequences containing repressive motifs tend to have negative log-likelihood ratios (Fig. 2J). We also calculated the per-base log-likelihood ratios on all promoters in the test set and found a significant positive correlation with the ISM scores derived from regression models (Supplemental Fig. S17) further supporting our assertion that the language model has learned regulatory syntax, and suggesting that the log-likelihood ratio can be used as a nucleotide-level or region-level score to interpret these models.
regLM generates cell type–specific human enhancers
Training and evaluating a regLM model on human enhancer sequences
We trained a regLM model on a data set of 200 bp human enhancers and their measured activity in three cell lines (K562, HepG2, and SK-N-SH) (Gosai et al. 2023) with the aim of designing cell type–specific human enhancers. Each sequence was prefixed with a sequence of three prompt tokens, each ranging from 0 to 3 and representing the measured activity of the enhancer in one of the three lines (Supplemental Fig. S18). For example, label 031 indicates that the sequence has low activity in HepG2 cells, high activity in K562 cells, and weak activity in SK-N-SH cells (Fig. 3A).
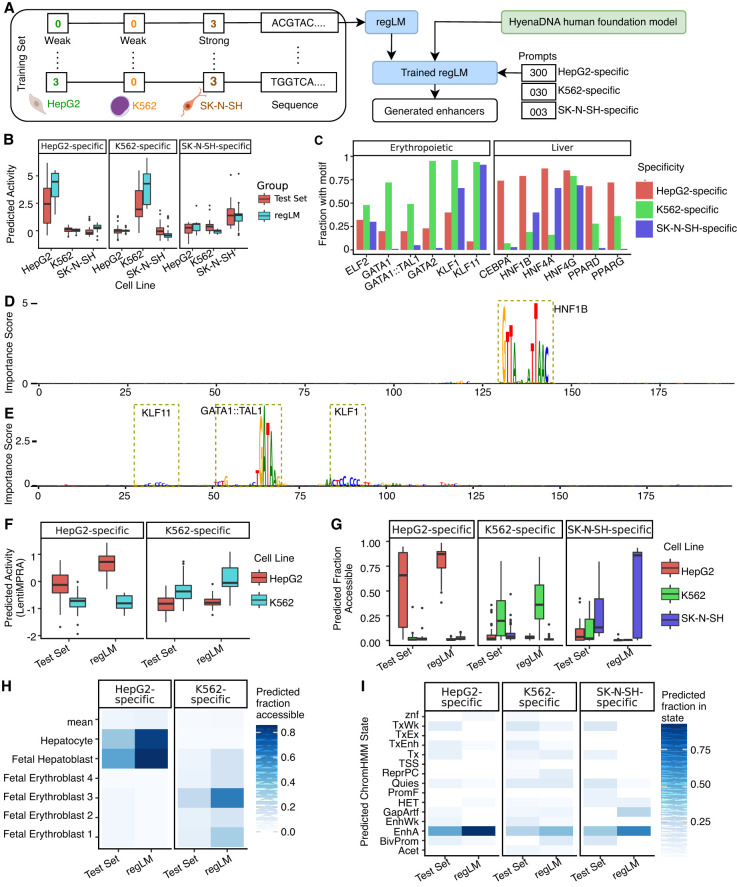
Figure 3.
regLM generates synthetic cell type–specific human enhancers. (A) Schematic of the experiment. (B) Predicted activity of cell type–specific human enhancers generated by regLM, compared to real cell line-specific human enhancers from the test set, in three cell lines. (C) Fraction of regLM-generated enhancers containing selected cell type–specific TF motifs. (D) Sequence of a HepG2-specific regLM-generated enhancer. (E) Sequence of a K562-specific regLM-generated enhancer. Height is proportional to per-nucleotide importance scores from the independent regression model using ISM. Motifs with high importance are highlighted. (F) Predicted activity of real and regLM-generated cell type–specific enhancers, using a model trained on LentiMPRA data. (G) Predictions of a binary classification model trained on ATAC-seq from human cell lines, on real and regLM-generated cell type–specific enhancers. (H) Predictions of a binary classification model trained on pseudobulk snATAC-seq from 204 cell types, on real and regLM-generated cell type–specific enhancers. Color intensity represents the fraction of sequences in the group that were predicted to be accessible. “Mean” represents the average of all remaining cell types. (I) Predictions of a classification model trained to classify genomic DNA into chromatin states defined by the full-stack ChromHMM annotation, on real and regLM-generated cell type–specific enhancers. Color intensity represents the fraction of sequences in the group that were predicted to belong to the given state. (Acet) acetylations, (BivProm) bivalent promoter, (EnhA) enhancers, (EnhWk) weak enhancers, (GapArtf) assembly gaps and artifacts, (HET) heterochromatin, (PromF) Flanking promoter, (ReprPC) polycomb repressed, (Quies) quiescent, (TSS) transcription start site, (Tx) transcription, (TxWk) weak transcription, (TxEnh) transcribed enhancer, (TxEx) exon and transcription, (znf) ZNF genes.
Here, instead of training a model from scratch, we could fine-tune a preexisting HyenaDNA model that had already learned regulatory information from the human genome (Nguyen et al. 2023). The pretrained model had a mean per-nucleotide accuracy of 34% on the test set before any fine-tuning, demonstrating that it learned relevant genomic properties. After fine-tuning, this accuracy increased to 45%. Despite the extreme rarity of cell type–specific enhancers in the training set (enhancers with labels 300, 030, or 003 comprised only 0.16% of the training set), accuracy remained high on cell type–specific enhancers (36.2%).
Generating and validating synthetic human enhancers
We used the trained regLM models to generate ∼4000 putative enhancers with activity specific to each cell line (Supplemental Table S6) by prompting them with the labels 300 (HepG2-specific), 030 (K562-specific), and 003 (SK-N-SH specific) and selecting generated sequences with a minimum edit distance of 20 nt with reference to the training set. We then used the paired regression models trained on the same data set (Supplemental Fig. S19) to select the 100 regLM-generated enhancers predicted to be most specific to each cell line (Supplemental Fig. S20; Supplemental Table S7).
Independent regression models (Supplemental Fig. S21) predicted that the regLM-generated elements have cell type–specific activity (higher predicted activity in the on-target cell type than in the off-target cell types). Although the full set of sequences generated by the language model display a wide range of activity (Supplemental Fig. S20), the top 100 HepG2- and K562-specific sequences generated by regLM are predicted to be more specific than the majority of enhancers with the corresponding label in the test set (Fig. 3B), and are even comparable to the predicted specificity of synthetic enhancers designed using model-guided approaches explicitly intended to maximize activity far beyond the range observed in the training set (Supplemental Fig. S22; Supplemental Table S8; Gosai et al. 2023). We focused on these 200 synthetic enhancers for the subsequent evaluation.
Because the test set contains very few enhancers with this level of cell type specificity, we did not evaluate the realism of the synthetic enhancers by quantitative comparisons of sequence content to the test set. However, we noted that motifs for known cell type–specific TFs occurred at higher frequency in regLM-generated enhancers of the appropriate specificity. For example, the motif for the erythropoietic GATA1 TF and the GATA1–TAL1 complex occur at higher frequency in regLM-generated K562-specific enhancers, whereas motifs for the liver-specific HNF4A and HNF1B factors occur at higher frequency in HepG2-specific synthetic enhancers (Fig. 3C). TF-MoDISco on ISM scores in the regLM-generated enhancers returned motifs for cell type–specific TFs (HNF1B and HNF4A for HepG2, and GATA2 for K562) (Supplemental Table S9). Examination of ISM scores for individual synthetic elements yielded these as well as additional cell type–specific motifs (FOXO3 for HepG2, and GATA1::TAL1 for K562) (Fig. 3D,E; Supplemental Figs. S23, S24). We did not observe motifs for neuron-specific TFs among the TF-MoDISco outputs for SK-N-SH cells but instead general enhancer-associated factors such as AP-1.
Compared to less specific regLM-generated enhancers, the top 100 regLM-generated enhancers specific for HepG2 are characterized by elevated abundance of the liver-related HNF1B and CEBPA motifs (FDR-adjusted Wilcoxon test P-value 1.2 × 10−11 for HNF1B and 1.2 × 10−5 for CEBPA). Among K562-specific enhancers, the top sequences are characterized by higher abundance of GATA1 and GATA1–TAL1 motifs and absence of the inhibitory SNAI3 motif as well as NFKB1 motifs (FDR-adjusted Wilcoxon test P-values 3.7 × 10−12 for GATA1, 4.7 × 10−6 for GATA1–TAL1, 1.1 × 10−8 for SNAI3 and 3.9 × 10−4 for NFKB1) (Supplemental Table S10). We also checked the abundance of these motifs in a set of synthetic enhancers designed by Gosai et al. (2023), selected to have similar predicted activity to the regLM-generated enhancers (Supplemental Fig. S25), and found similar trends (Supplemental Table S10).
We used several independent models to further validate the predicted cell type specificity of the HepG2- and K562-specific synthetic enhancers. First, we trained a regression model on lentiviral massively parallel reporter assay (MPRA) data from HepG2 and K562 cell lines (Agarwal et al. 2023), and applied it to our designed enhancers. The model predicted that the designed enhancers for K562 and HepG2 would have cell line-specific activity even in the context of lentiviral integration (Fig. 3F). Next, we trained binary classification models on chromatin accessibility data from cell lines (The ENCODE Project Consortium 2012) and predicted that the designed elements would have cell type–specific chromatin accessibility (Fig. 3G). In addition, a model trained on chromatin accessibility in numerous fetal and adult human cell types predicted that the designed elements would also maintain cell type–specific accessibility in related cell types (Fig. 3H). Finally, we trained a model to classify DNA elements into chromatin states defined by the ChromHMM full-stack annotation (Vu and Ernst 2022). This model predicted that most of the regLM-generated enhancers belong to enhancer-associated chromatin states (Fig. 3I). In all cases, these models supported our prediction that the cell type–specific enhancer activity of the regLM-generated set exceeds that of cell type–specific enhancers in the test set.
We also ran all of these models on the set of synthetic elements designed by Gosai et al. (2023) with similar predicted activity to ours (Supplemental Fig. S25), and found that the regLM-generated enhancers showed comparable cell type specificity based on all predictions (Supplemental Figs. S26–S29). Together, these diverse predictions greatly increase our confidence in the validity of regLM-generated enhancers.
Discussion
In recent years, many advances have been made in applying language modeling to genomic DNA sequences, demonstrating the ability of such models to learn genomic sequence composition and functional patterns. Most work in this field has focused on using language model-based sequence embeddings to predict biological activity (Ji et al. 2021; Dalla-Torre et al. 2023), as well as to predict variant effects (Benegas et al. 2023). Here, we ask whether autoregressive language models can generate novel DNA sequences with precisely controlled functions. We demonstrate that the regLM framework can learn the regulatory code of DNA in different species and cell types, and generate CREs with desired levels of activity in silico.
In contrast to predictive model-based approaches for regulatory DNA design (Vaishnav et al. 2022; Gosai et al. 2023; Taskiran et al. 2024), regLM not only generates sequences with controlled activity but also implicitly ensures that they have similar sequence content to the training set. Indeed, the evaluation of the synthetic sequences generated by regLM shows high concordance between the regulatory rules implemented in these sequences and known regulatory syntax. This increases our confidence in the generated sequences and may help ensure their predictable behavior in the genomic context. The fidelity of generated sequences to the training set can be tuned using sampling methods that have been shown to be effective in natural language (Koehn 2010; Holtzman et al. 2019). regLM also differs from predictive model-based methods in that it is not biased by user-provided initial sequences and instead generates a diverse set of sequences following different regulatory programs.
Although this demonstration focuses on short regulatory elements, it will be interesting to apply this framework to more complex tasks. The ability of the HyenaDNA architecture to scale to sequences of hundreds of thousands or even millions of bases (Nguyen et al. 2023), offers the possibility of generating novel genes as well as regulatory sequences and potentially even multigene genetic circuits and whole microbial genomes. Also, although we have used simple activity-based prompts in this study, our framework allows the user to define any number of arbitrary categories (Supplemental Note S2; Supplemental Fig. S30). Therefore, the prompts supplied to such models could become more complex in the future, including parameters such as species, cell type, GC content, or the presence of specific motifs.
Another interesting advantage of our approach is its interpretability based on base-level likelihood values, which allows us to understand the prompt-driven regulatory logic driving its decisions during generation. In this study, we analyzed a regLM model using likelihood-based approaches as well as per-nucleotide predictive accuracy, demonstrating that it learned species and cell type–specific cis-regulatory syntax. However, all existing tools for genomic model interpretation focus on supervised models (Shrikumar et al. 2018; Avsec et al. 2021b). As language models become more powerful and commonly used in genomics, it will be interesting to extend interpretation methods to these and compare the regulatory rules and biases learned by different modeling approaches.
On the other hand, a potential disadvantage is that language models may learn evolved features in natural genomes that are not actually necessary for regulatory function. This can reduce functionality and be a weakness for mechanistic understanding of gene regulation. Larger training sets including randomly generated, mutated and nongenomic sequences will help mitigate this problem (de Boer and Taipale 2024). In addition, models trained on data from multiple species and individuals may achieve better performance by learning signatures of evolutionarily conserved sequences (Karollus et al. 2024).
Methods
Data sources
Yeast promoter data from Vaishnav et al. (2022) was downloaded from Zenodo (https://zenodo.org/records/4436477). Measurements were available for 31,349,363 sequences in complex medium, and 21,037,407 sequences in defined medium. Human MPRA data was downloaded from the supplementary material of Gosai et al. (2023). This data set contains 798,064 enhancer sequences from the human genome with their measured enhancer activity in three cell lines.
Data processing
Yeast promoters
We removed the constant sequences flanking each promoter and selected promoter sequences that were 80 bp long and contained no N characters. This left measurements for 23,414,517 sequences in the complex medium and 16,799,784 sequences in the defined medium. We split the data set into 7,533,156 sequences whose activity was measured in both media, and sequences whose activity was measured in only one medium (15,881,361 sequences measured only in complex medium and 9,266,628 sequences measured only in defined medium). The sequences measured in both media were used to train regLM and its paired regression models, whereas the sequences measured only in one medium were used to train independent, medium-specific regression models.
Taking the sequences with measured activity in both media, we randomly split them into training (7,483,156 sequences), validation (50,000 sequences), and test (50,000 sequences) sets. We calculated the quintiles (five equally sized bins) of measured activity levels in complex medium and defined medium separately based on the training set. We assigned each sequence in the training set a token 0–4 based on its quintile of activity in the complex medium, and a second token 0–4 based on its quintile of activity in the defined medium; 0 indicates that the sequence belongs to the lowest quintile and 4 indicates that the sequence belongs to the highest quintile. For example, label 00 means a sequence in the lowest quintile of activity in both media. Label 40 means a sequence that is in the highest quintile of activity in complex medium but the lowest in defined medium. Sequences in the validation and test sets were assigned labels based on the quintiles calculated on the training set.
Human enhancers
We held out 94,451 sequences from Chromosomes 7, 13, 21, and 22 to train independent regression models, while the remaining 669,233 sequences were used to train the regLM model and its paired regression models. Out of these 669,233 sequences, we randomly sampled 50 sequences with cell type–specific activity to use as a validation set while the remaining sequences were used for training. The training set for regLM was used as a test set for the independent regression models, whereas the training set for the independent regression models was used as a test set for regLM and its paired regression models.
We assigned each sequence in the training set a token 0–3 based on its activity in HepG2 cells, a second token 0–3 based on its activity in K562 cells, and a third token 0–3 based on its activity in SK-N-SH cells. 0 indicates activity <0.2, 1 indicates activity between 0.2 and 0.75, 2 indicates activity between 0.75 and 2.5, and 3 indicates activity >2.5. These cutoff values roughly correspond to the 25th, 75th, and 95th percentiles of activity. For example, label 301 means an enhancer that is in the highest group of activity in HepG2 cells, the lowest group in K562 cells, and the second lowest group in SK-N-SH cells.
Training regLM models
Human enhancers
For human enhancers, we fine-tuned the pretrained foundation model “hyenadna-medium-160k-seqlen” from https://huggingface.co/LongSafari/hyenadna-medium-160k-seqlen/tree/main (Nguyen et al. 2023). This model has 6.55 million parameters and is trained to perform next token prediction on the human genome. The model was fine-tuned for 16 epochs on 1 NVIDIA A100 GPU using the AdamW optimizer with cross-entropy loss, learning rate of 10−4 and batch size of 1024. Validation loss and accuracy were computed every 100 steps and the model with the lowest validation loss was saved. During training, examples with each label were sampled from the training set with a weight inversely proportional to the frequency of the label, allowing the model to focus on cell type–specific enhancers that were extremely rare.
Yeast promoters
For yeast promoters, we trained from scratch a HyenaDNA model with the same architecture as “hyenadna-medium-160k-seqlen.” The model was trained for 100 epochs on 1 NVIDIA A100 GPU using the AdamW optimizer with cross-entropy loss, learning rate of 3 × 10−4, batch size of 2048, and a maximum context length of 84 (80 bp plus two label tokens as well as start and end tokens). Validation loss and accuracy were computed every 2000 steps and the model with the lowest validation loss was saved.
Training regression models
For both yeast and human data sets, we trained two sets of regression models. One set of models was trained on the same sequences as the regLM model, to use in conjunction with regLM to filter and prioritize generated sequences. A second set of models was trained on data that was held out from regLM and all generative methods and was used only for independent in silico validation of synthetic CREs. Within each set, we trained separate regression models for each medium or cell type. Hence, for yeast, we trained a total of four regression models (two each for the two media) and for humans, we trained a total of six (two each for three cell types). All regression models used the same architecture, based on the Enformer model (Avsec et al. 2021a) and the same hyperparameters. Further details are given in the Supplemental Methods.
Yeast promoters
The regLM-matched models were trained using the same training, validation, and test data used to train regLM. For the independent regression models, the 21,609,084 sequences whose activity was measured only in complex medium and 11,460,087 sequences whose activity was measured only in the defined medium were used. In each medium, 50,000 randomly chosen sequences were held out for validation and 50,000 were held out for testing. The remaining sequences were used for training.
Human enhancers
For human data, we used a reduced version of the pretrained Enformer model (Avsec et al. 2021a) and fine-tuned it separately for each cell type. For the regLM-matched regression models, we fine-tuned the model on the same sequences as regLM. For the independent models, sequences from Chromosome 21 were used for validation, while sequences from Chromosomes 7, 13, and 22 were used for training.
Generating synthetic CREs using regLM
Yeast promoters
We prompted the regLM model trained on yeast promoters to generate 1000 sequences each with labels 00, 11, 22, 33, and 44. During generation, we applied nucleus sampling (Holtzman et al. 2019) with a top-p cutoff of 0.85 to increase the reliability of generation. Generated promoters were filtered using the regression model trained on the same data as the language model. For each medium, we first used the paired regression model to predict the activity of all sequences in its training set, and computed the mean and standard deviation of predicted activity for training sequences with each class token (0, 1, 2, 3, and 4). We then used the same model to predict the activity of all generated promoters in both media. We discarded generated promoters whose predicted activity in either medium was more than two standard deviations from the mean predicted activity of promoters with the same token in the training set. We performed this procedure separately for complex and defined media. We then randomly selected 200 synthetic promoters of each generated class (00, 11, 22, 33, and 44) to compare with other methods.
Human enhancers
The regLM model trained on human enhancers was prompted to generate 5000 sequences each with tokens 300 (HepG2-specific), 030 (K562-specific), and 003 (SK-N-SH specific), using the beam search method (Koehn 2010) to increase reliability. We dropped sequences with an edit distance of <20 from the training set, leaving 3900–4000 sequences for each cell type (Supplemental Fig. S20).
Generated enhancers were then filtered using the regression models trained on the same data as regLM. We first filtered the generated sequences using absolute thresholds consistent with the prompted labels (predicted activity >3.5 in the target cell type and <0.2 in the off-target cell type). Next, we estimated the cell type specificity of each sequence as the difference between its predicted activity in the target cell type and its maximum predicted activity in off-target cell types. Based on this, we selected the 100 most specific regLM-generated enhancers for each cell type.
In silico evaluation of synthetic CREs
k-mer content
The frequency of all subsequences of length 4 (4-mers) was counted in each real or synthetic promoter. Each sequence was thus represented by a 256-dimensional vector. To calculate the fraction of real nearest neighbors, we matched each sequence to its nearest neighbor out of all real and synthetic sequences. For each group of synthetic CREs, we calculated the proportion of sequences whose nearest neighbor was an experimentally validated CRE from the test set. To compute classification performance, we trained a support vector machine (SVM) with fivefold cross-validation to distinguish each set of synthetic sequences from the reference set based on their k-mer frequencies. The area under the receiver operator curve (AUROC) for each SVM was reported as a measure of classification performance.
Transcription factor motif content
Position probability matrices (PPMs) were downloaded from the JASPAR 2024 database in MEME format. One hundred seventy PPMs for yeast were selected using the filters Species = “Saccharomyces cerevisiae” and Versions = “Latest version.” Seven hundred fifty-five PPMs were selected for humans using the filters Species = “Homo sapiens” and Versions = “Latest version.”
Pairwise correlations between motifs were also downloaded from the JASPAR 2024 database. Motifs were clustered based on their pairwise Pearson's correlations using agglomerative clustering with a distance threshold of 0.1. For clusters consisting of two motifs, the motif with higher information content was chosen as the cluster representative, and the other was discarded. For clusters containing more than two motifs, the motif that had the highest average Pearson's correlation to the other cluster members was selected as the representative and the others were discarded. This resulted in a filtered set of 140 motifs for yeast and 464 for human.
Reading the MEME files, conversion of PPMs to position weight matrices (PWMs) and sequence scanning was performed using the pymemesuite package, with a uniform background frequency, default pseudocount of 0.1, and P-value threshold of 0.001. Each sequence was represented by the 140-dimensional vector of its motif frequency for all motifs. Each sequence was matched to its nearest neighbors in this vector space, and the proportion of real nearest neighbors for each group of synthetic CREs was calculated as described above. Classification performance was calculated as described above.
Model-based embeddings
Real and synthetic CREs were embedded in a model-defined latent space by passing them as input to the model and taking the output of the convolutional tower. For yeast promoters, the embeddings from each regression model had 384 features. We concatenated the embeddings from the models trained on two media, resulting in an embedding vector of size 768 for each sequence. Values were clipped to the 1st and 99th percentiles of the distribution to remove extreme values. To compute nearest neighbors efficiently, we reduced the number of features to 50 using principal component analysis (PCA). Each sequence was matched to its nearest neighbors in PCA space and the proportion of real nearest neighbors for each group of synthetic CREs was calculated as described above.
Interpretation of the regLM model trained on yeast promoters
regLM is trained to perform next token prediction; that is, for each position in a DNA sequence, regLM predicts the probability of all possible bases (A, C, G, and T) conditioned on the previous bases as well as the initial label. Thus, we can obtain the likelihood of an observed sequence conditioned on its initial label (P(sequence|label)) as the product of probabilities of the base observed at each position.
To assess whether regLM has learned the function of a given motif, we generated 100 random DNA sequences and inserted the consensus sequence for the motif at the center of each. We prefixed each sequence with label 00 (low activity) and used the trained regLM model to predict the probability of each base in the motif. We calculated the likelihood of the motif conditioned on the sequence being labeled with 00 (P(sequence|label = 00)). We then prefixed all 100 sequences with the label 44 (high activity in both media) and repeated the procedure, calculating the likelihood of the motif conditioned on the sequence being labeled with 44 (P(sequence|label = 44)).
Additional models for human CRE validation
We validated synthetic human CREs using four additional models, trained on different data sets: one binary classification model trained on cell line ATAC-seq data from the ENCODE Project (The ENCODE Project Consortium 2012), one binary classification model trained on single-nucleus ATAC-seq from multiple human tissues (Zhang et al. 2021), one multiclass classification model trained on full-stack chromatin state annotations of the human genome (Vu and Ernst 2022) and one regression model trained on lentiviral MPRA data in human cell lines (Agarwal et al. 2023). All models were Enformer-based models, following the same structure as the regression models used to evaluate the generated yeast and human CREs described above.
We refer readers to the Supplemental Methods for more details on model training, model parameters, and running benchmark methods.
Software availability
regLM source code is available at GitHub (https://github.com/Genentech/regLM) under an MIT license, along with documentation and tutorials. The source code is also provided as Supplemental Code S1.
Code to perform the experiments in this paper is available at Zenodo (https://zenodo.org/records/12668907). All experiments were performed using Python v3.8, PyTorch v1.13.0, and PyTorch Lightning v1.8.2.
Model weights are available at Zenodo (https://zenodo.org/records/12668907). Additionally, the models used for human CRE validation are available on Weights and Biases at the following links:
Binary classification model trained on ATAC-seq from cell lines: https://wandb.ai/grelu/binary_atac_cell_lines/artifacts/model/model/v1
Regression model trained on Lentiviral MPRA: https://wandb.ai/grelu/human-mpra-agrawal-2023/artifacts/model/model/v0
Binary classification model trained on CATLAS snATAC-seq: https://wandb.ai/grelu/human-atac-catlas/artifacts/model/model/v3
Classification model trained on ChromHMM annotations: https://wandb.ai/grelu/human-chromhmm-fullstack/artifacts/model/model/v2
Supplemental Material
Acknowledgments
We thank the anonymous reviewers for their helpful and constructive comments.
Author contributions: A.L. and G.E. developed the regLM method. A.L. performed experiments and analyzed data. D.G. trained and applied the models used to validate synthetic human enhancers. T.B. provided mentorship and supervision. A.L., D.G., and G.E. wrote the manuscript. All authors read and approved the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.279142.124.
Freely available online through the Genome Research Open Access option.
Competing interest statement
All of the authors are employees of Genentech, Inc.
References
- Agarwal V, Inoue F, Schubach M, Martin BK, Dash PM, Zhang Z, Sohota A, Noble WS, Yardimci GG, Kircher M, et al. 2023. Massively parallel characterization of transcriptional regulatory elements in three diverse human cell types. bioRxiv 10.1101/2023.03.05.531189 [DOI]
- Avsec Ž, Agarwal V, Visentin D, Ledsam JR, Grabska-Barwinska A, Taylor KR, Assael Y, Jumper J, Kohli P, Kelley DR. 2021a. Effective gene expression prediction from sequence by integrating long-range interactions. Nat Methods 18: 1196–1203. 10.1038/s41592-021-01252-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avsec Ž, Weilert M, Shrikumar A, Krueger S, Alexandari A, Dalal K, Fropf R, McAnany C, Gagneur J, Kundaje A, et al. 2021b. Base-resolution models of transcription-factor binding reveal soft motif syntax. Nat Genet 53: 354–366. 10.1038/s41588-021-00782-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benegas G, Batra SS, Song YS. 2023. DNA language models are powerful predictors of genome-wide variant effects. Proc Natl Acad Sci 120: e2311219120. 10.1073/pnas.2311219120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown T, Mann B, Ryder N, Subbiah M, Kaplan JD, Dhariwal P, Neelakantan A, Shyam P, Sastry G, Askell A, et al. 2020. Language models are few-shot learners. In Advances in Neural Information Processing Systems 33: 34th Conference on Neural Information Processing Systems (NeurIPS 2020), Vancouver, Canada. [Google Scholar]
- Dalla-Torre H, Gonzalez L, Mendoza-Revilla J, Carranza NL, Grzywaczewski AH, Oteri F, Dallago C, Trop E, Sirelkhatim H, Richard G, et al. 2023. The nucleotide transformer: building and evaluating robust foundation models for human genomics. bioRxiv 10.1101/2023.01.11.523679 [DOI]
- de Boer CG, Taipale J. 2024. Hold out the genome: a roadmap to solving the cis-regulatory code. Nature 625: 41–50. 10.1038/s41586-023-06661-w [DOI] [PubMed] [Google Scholar]
- de Boer CG, Vaishnav ED, Sadeh R, Abeyta EL, Friedman N, Regev A. 2020. Deciphering eukaryotic gene-regulatory logic with 100 million random promoters. Nat Biotechnol 38: 56–65. 10.1038/s41587-019-0315-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishman V, Kuratov Y, Petrov M, Shmelev A, Shepelin D, Chekanov N, Kardymon O, Burtsev M. 2023. GENA-LM: a family of open-source foundational DNA language models for long DNA sequences. bioRxiv 10.1101/2023.06.12.544594 [DOI]
- Fornes O, Av-Shalom TV, Korecki AJ, Farkas RA, Arenillas DJ, Mathelier A, Simpson EM, Wasserman WW. 2023. Ontarget: in silico design of MiniPromoters for targeted delivery of expression. Nucleic Acids Res 51: W379–W386. 10.1093/nar/gkad375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosai SJ, Castro RI, Fuentes N, Butts JC, Kales S, Noche RR, Mouri K, Sabeti PC, Reilly SK, Tewhey R. 2023. Machine-guided design of synthetic cell type-specific cis-regulatory elements. bioRxiv 10.1101/2023.08.08.552077 [DOI] [PMC free article] [PubMed]
- Holtzman A, Buys J, Du L, Forbes M, Choi Y. 2019. The curious case of neural text degeneration. In 8th International Conference on Learning Representations (ICLR 2020).
- Ji Y, Zhou Z, Liu H, Davuluri RV. 2021. DNABERT: pre-trained bidirectional encoder representations from transformers model for DNA-language in genome. Bioinformatics 37: 2112–2120. 10.1093/bioinformatics/btab083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karollus A, Hingerl J, Gankin D, Grosshauser M, Klemon K, Gagneur J. 2024. Species-aware DNA language models capture regulatory elements and their evolution. Genome Biol 25: 83. 10.1186/s13059-024-03221-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehn P. 2010. Statistical machine translation. Cambridge University Press, Cambridge. [Google Scholar]
- Lal A, Gunsalus L, Gupta A, Biancalani T, Eraslan G. 2023. Polygraph: a software framework for the systematic assessment of synthetic regulatory DNA elements. bioRxiv 10.1101/2023.11.27.568764 [DOI]
- Linder J, Seelig G. 2021. Fast activation maximization for molecular sequence design. BMC Bioinformatics 22: 510. 10.1186/s12859-021-04437-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen E, Poli M, Faizi M, Thomas A, Wornow M, Birch-Sykes C, Massaroli S, Patel A, Rabideau C, Bengio Y, et al. 2023. HyenaDNA: long-range genomic sequence modeling at single nucleotide resolution. In Advances in Neural Information Processing Systems 36: Annual Conference on Neural Information Processing Systems 2023 (NeurIPS 2023), New Orleans. [Google Scholar]
- Poli M, Massaroli S, Nguyen E, Fu DY, Dao T, Baccus S, Bengio Y, Ermon S, Ré C. 2023. Hyena hierarchy: towards larger convolutional language models. In Proceedings of the 40th International Conference on Machine Learning (ICML 2023), Honolulu, HI. [Google Scholar]
- Schreiber J, Lu YY. 2020. Ledidi: designing genomic edits that induce functional activity. bioRxiv 10.1101/2020.05.21.109686 [DOI]
- Shrikumar A, Tian K, Avsec Ž, Shcherbina A, Banerjee A, Sharmin M, Nair S, Kundaje A. 2018. Technical note on transcription factor motif discovery from importance scores (TF-MoDISco) version 0.5.6.5. arXiv:1811.00416 [cs.LG]. 10.48550/arXiv.1811.00416 [DOI]
- Taskiran II, Spanier KI, Dickmänken H, Kempynck N, Pančíková A, Ekşi EC, Hulselmans G, Ismail JN, Theunis K, Vandepoel R, et al. 2024. Cell-type-directed design of synthetic enhancers. Nature 626: 212–220. 10.1038/s41586-023-06936-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traag VA, Waltman L, van Eck NJ. 2019. From Louvain to Leiden: guaranteeing well-connected communities. Sci Rep 9: 5233. 10.1038/s41598-019-41695-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaishnav ED, de Boer CG, Molinet J, Yassour M, Fan L, Adiconis X, Thompson DA, Levin JZ, Cubillos FA, Regev A. 2022. The evolution, evolvability and engineering of gene regulatory DNA. Nature 603: 455–463. 10.1038/s41586-022-04506-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu H, Ernst J. 2022. Universal annotation of the human genome through integration of over a thousand epigenomic datasets. Genome Biol 23: 9. 10.1186/s13059-021-02572-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittkopp PJ, Kalay G. 2012. Cis-regulatory elements: molecular mechanisms and evolutionary processes underlying divergence. Nat Rev Genet 13: 59–69. 10.1038/nrg3095 [DOI] [PubMed] [Google Scholar]
- Zhang K, Hocker JD, Miller M, Hou X, Chiou J, Poirion OB, Qiu Y, Li YE, Gaulton KJ, Wang A, et al. 2021. A single-cell atlas of chromatin accessibility in the human genome. Cell 184: 5985–6001.e19. 10.1016/j.cell.2021.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Ji Y, Li W, Dutta P, Davuluri R, Liu H. 2024. DNABERT-2: efficient foundation model and benchmark for multi-species genome. In The Twelfth International Conference on Learning Representations (ICLR 2024), Vienna. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.