Abstract
The spread of bacterial drug resistance has posed a severe threat to public health globally. Here, we cover bacterial resistance to current antibacterial drugs, including traditional herbal medicines, conventional antibiotics, and antimicrobial peptides. We summarize the influence of bacterial drug resistance on global health and its economic burden while highlighting the resistance mechanisms developed by bacteria. Based on the One Health concept, we propose 4A strategies to combat bacterial resistance, including prudent Application of antibacterial agents, Administration, Assays, and Alternatives to antibiotics. Finally, we identify several opportunities and unsolved questions warranting future exploration for combating bacterial resistance, such as predicting genetic bacterial resistance through the use of more effective techniques, surveying both genetic determinants of bacterial resistance and the transmission dynamics of antibiotic resistance genes (ARGs).
Keywords: Bacterial resistance, Antibiotics, Mechanism of action, Antimicrobial peptides, Strategies, Global health
1. Introduction
The use of antibiotics over 100 years has triggered the widespread and rapid transfer of antibiotic resistance genes (ARGs) among bacteria across the world (Andersson, 2003; Larsson and Flach, 2021). Clinical resistance of the spirochaete bacterium Treponema pallidum to the antibiotic arsphenamine (also known as Salvarsan) was first reported in 1924 (Silberstein, 1924; Stekel, 2018). This was followed by the detection of resistance to the commercial penicillin in Staphylococcus aureus as early as 1942, rapidly followed in the 1950s and 1960s by the acquisition of resistance by other bacteria such as Escherichia coli, Salmonella spp., Shigella spp., and other bacterial species (Aslam et al., 2018; Rammelkamp and Maxon, 1942). Bacterial antibiotic resistance emerges almost concurrently as new antibiotics enter the market and are widely implemented in our society (Levy, 1998). Today, nearly all known bacterial pathogens have developed resistance to antibiotics (Barber and Rozwadowska-Dowzenko, 1948). Resistance continues to spread not only in pathogenic bacteria of human or animal origin, but also in environmental microbes. Resistance can also develop to some probiotics under the long-term selective pressure exerted by antibiotics (Li et al., 2020a-c; Radimersky et al., 2010; Al-Zahrani et al., 2019; Ejaz et al., 2021; Mohapatra et al., 2018; Fluit et al., 2001; Hawkey, 1986; Wozniak et al., 2019; Peterson and Kaur, 2018; Sharma et al., 2014). Antibiotic-resistant bacteria (ARB) can survive and even grow in the presence of one or more antibacterial agents. Multidrug resistance (MDR) has been regarded as acquired resistance to at least one drug in three or more antimicrobial categories (Magiorakos et al., 2012). There is an increasing prevalence of MDR bacteria globally, which are exceedingly difficult to treat, constituting a growing global health problem (Zarfel et al., 2014; van Duin and Paterson, 2016; Breidenstein et al., 2011; Der Torossian and de la Fuente-Nunez, 2019), particularly for critical care practitioners. Thus, there is a tremendous interest in developing novel agents as alternative approaches for treating such infections (de la Fuente-Núñez et al., 2016; de la Fuente-Nunez et al., 2017; Torres et al., 2018; Torres et al., 2019; Mirghani et al., 2022).
Recently, increasing attention has focused on ribosomally synthesized antimicrobial peptides (AMPs) from animals, plants, and microorganisms (Magana et al., 2020). These peptides, initially described in the 1960s, are known as promising natural alternatives to classical antibiotics because of their broad-spectrum antimicrobial activity (Abdi et al., 2019; Torres et al., 2021; Melo et al., 2021; Torres et al., 2018; Silva et al., 2020; Wang et al., 2020b; Li et al., 2018). AMPs, also named host defense peptides, are critical components of the host innate defense system and have multi-functions, including anti-fungal, anti-viral, anti-parasitic, anti-tumor, and anti-inflammatory activity, which are different from conventional antibiotics (Hegedüs and Marx, 2013; Kurpe et al., 2020; Bakovic et al., 2021; Pérez-Cordero et al., 2011; Pereira et al., 2016; Arias et al., 2017; Yan et al., 2013; Mookherjee et al., 2006). AMPs can bind to the negatively charged bacterial surface and kill bacteria primarily through membrane disruption (Wang et al., 2020b; Nuri et al., 2015; Wang et al., 2017b). The development of bacterial resistance to AMPs is unlikely due to their action on multiple cellular targets (Zasloff, 2002; Duperthuy, 2020). However, a growing number of studies have demonstrated that some pathogenic bacteria, including Group A Streptococcus (GAS), Salmonella enterica, and Campylobacter jejuni, can develop significant resistance to various AMPs, such as defensins, LL-37, cecropins, melittin, and PR-39 (Duperthuy, 2020; Malik et al., 2017; Lofton et al., 2013; Nizet et al., 2001; Cullen and Trent, 2010; Gruenheid and Le Moual, 2012). Additionally, traditional herbal medicines are a previously untapped source of potential antibacterial drugs. Indeed, herbal medicines have been shown to contain antibacterial compounds including alkaloids, organic acids, flavonoids, terpenes, essential oils, anthraquinones, etc.; they are widely considered as antibiotic alternatives because of their antimicrobial activity and low cytotoxicity (Yang et al., 2018; Wang et al., 2013; Cushnie et al., 2014; Sieniawska et al., 2017; Tagousop et al., 2018; Adamczak et al., 2019; Parham et al., 2020; Soković et al., 2010; Manso et al., 2022; Kosalec et al., 2013). The use of naturally occurring herbal medicines to treat bacterial infections has a long history (over 5,000 years) in parts of Asia (such as China and India), Europe, North America, and Australia (Ekor, 2014; Majaz and Khurshid, 2016). For example, a combination of garlic, wine and other allium species described in a 1,000-year-old remedy from an Anglo-Saxon leechbook was able to kill methicillin-resistant S. aureus (MRSA) and suppress biofilm infection (Harrison et al., 2015). Ointments containing tea leaf extracts have been utilized to treat diseases caused by MDR S. aureus, and natural products containing alkaloids have been used to treat diarrheic infectious diseases (Orchard and van Vuuren, 2017; Romero et al., 2005; Chen et al., 2015; Chen et al., 2014; Rubinstein and Keynan, 2013). However, there is still little experimental evidence demonstrating that certain bacteria, including Enterobacteriaceae, Morganellaceae, Bacillaceae and staphylococci, develop resistance to extracts from herbal medicines (e.g., Cymbopogon citratus, Salvia officinalis, Artemisia vulgaris, etc.) (Singh et al., 2012; Singh et al., 2011; Singh et al., 2013).
In this review, we outline various mechanisms whereby bacteria become resistant to antibacterial agents (including traditional herbal medicines, conventional antibiotics, and AMPs), as well as highlight the impact and economic burden of bacterial resistance on global health. Fig. 1 delineates the evolution of bacterial resistance to antibacterial agents over time. According to the One Health concept (Kim and Cha, 2021), we propose 4A strategies to combat the spread of bacterial drug resistance, including prudent Application of antibacterial agents, scientific Administration, development of effective Assays and find novel Alternatives to antibiotics. While describing how bacteria develop resistance to antibiotics, we hope to obtain insights into the mechanisms and general physiological effects that antibacterial agents exert on bacteria. Collectively, these insights may enable the development of more effective antibacterial agents that minimize drug resistance.
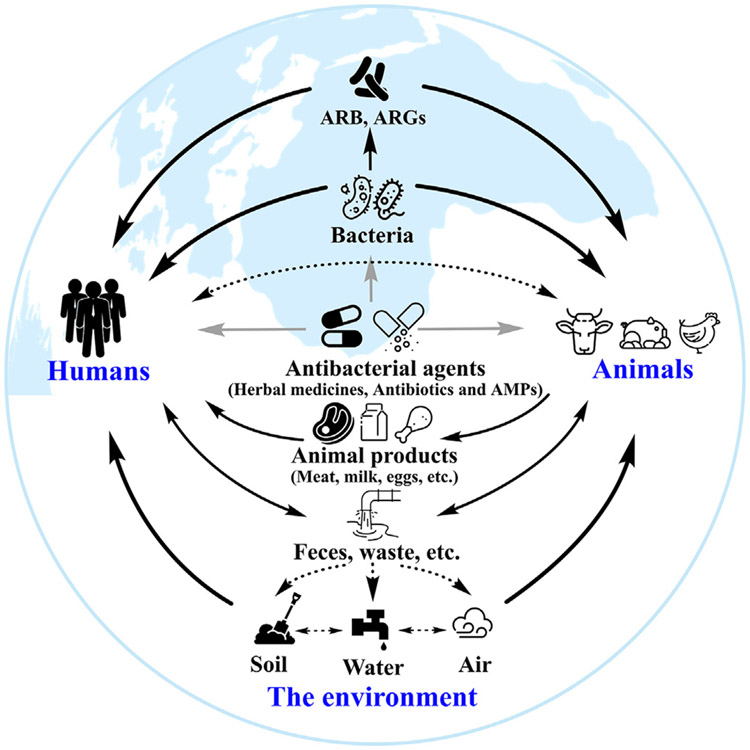
Fig. 1.
A brief profile of the evolution of bacterial resistance to current antibacterial agents. Of the large number of bacteria in the world, a fraction causes diseases in humans and animals, against which we need effective antibacterial agents. Some bacteria (such as the ESKAPE pathogens) are particularly resistant to antibiotics (Martin et al., 2015). These resistant bacteria and ARGs are rapidly disseminated in humans, animals, and the environment by direct or indirect contact with each other or with excreta, animal products, soil, and water contributing to the concept of One Health (Larsson and Flach, 2021; Rana et al., 1991). ARB: antibiotic-resistant bacteria. ARGs: antibiotic resistance genes. Black arrow: the spread of ARB and ARGs in humans, animals, and the environment. Gray arrow: effects of drugs on bacteria or bacterial infections in humans and animals.
2. Influence of bacterial drug resistance on health, food security, and economics
According to the WHO, drug resistance is currently one of the most severe threats to global health, food security, and economic burden, and can influence anyone at any age and any place. Drug resistance occurs naturally, but unfortunately, the misuse of conventional antibiotics in both human and animal health has greatly accelerated this process.
2.1. Global health
A growing number of pathogenic bacterial infections, such as tuberculosis, pneumonia, salmonellosis, and gonorrhea, are becoming harder to treat due to their increased resistance to conventional antibiotics. In the 2021 GLASS Report (involving 109 countries or territories), the proportions of E. coli and Klebsiella pneumoniae resistant to cotrimoxazole, a first-line drug, were 54.4 % and 43.1 % in urinary tract infections, 24.9 % for MRSA, and 65.48 % for Acinetobacter spp. against the third-generation cephalosporin in bloodstream infections (World Health Organization (WHO), 2021). Neisseria gonorrhoeae resistant to cefixime, the third-generation cephalosporin, has already been found in Thailand and the Philippines (World Health Organization (WHO), 2021). The prevalence of vancomycin-resistant S. aureus (VRSA) was 5 % in Asia, 16 % in Africa, 4 % in America, 3 % in South America, and 1 % in Europe (Shariati et al., 2020). The prevalence of colistin-resistant bacteria has increased to 43 % in Italy, 37.3–28.8 % in China, 31 % in Spain, and 20.8 % in Greece (Gogry et al., 2021; Que et al., 2022). In summary, bacterial antibiotic resistance has reached worrying levels in many countries worldwide.
In some cases, the available antibiotic treatment options for common bacterial infections are becoming ineffective. In countries such as Argentina, China, Australia, Chile, Lebanon, Colombia, the US, and Singapore, it has been shown that >50 % of patients infected with resistant K. pneumoniae have infections that do not respond to carbapenems (Wang et al., 2022a-c; Bassetti et al., 2021). Colistin and vancomycin, both last-resort drugs, even prove to be ineffective against MDR Enterobacteriaceae (e.g., Klebsiella and E. coli) and VRSA (Gogry et al., 2021). The lack of therapeutic solutions is resulting in increased mortality, longer hospital stays, and higher medical costs (Aslam et al., 2018). In 2019, approximately 1.27 million people globally died from infections caused by ARB, and it has been estimated that 10,000,000 people will die every year by 2050 in the absence of effective treatments (Murray et al., 2022; Nishanth and Palanichamy, 2020; WHO, 2019). In Thailand, around 38,000 people die every year and there are 3.2 million hospital days due to resistant bacterial infections. In Europe and the US, 33,000 and 35,000 deaths, respectively, occur annually amounting to 2–2.5 million extra hospital days, and in India resistance causes the death of 58,000 babies per year (WHO, 2019; Zhen et al., 2021). Notably, death rates of patients with bacteremia induced by resistant ESKAPE pathogens were up 20–70 % from 2006 to 2011 (De Oliveira et al., 2020). These statistics depict a dire scenario and point to drug-resistant bacterial infections as a global health threat.
2.2. Food security
Conventional antibiotics are widely used in food animals, including chickens, pigs, cattle, and fish (Van Boeckel et al., 2015). In some countries, such as Vietnam, over 70 % of the total antibiotics produced are applied to animal production, largely for growth promotion and disease prevention rather than for therapeutic means (Carrique-Mas et al., 2019; Martin et al., 2015; Carrique-Mas et al., 2020; Van et al., 2020). It is currently projected that the antibiotic use in the farming sector will rise to 67 % by 2030 in certain developing countries (Van Boeckel et al., 2015). This widespread application of antibiotics in livestock and other animals is considered to be partially responsible for the rapid emergence of ARB. If not tackled appropriately, agricultural uses of antibiotics may have significant public health implications. ARB from animals may be transferred to humans through a variety of means, such as direct contact with farmers, food products, animal excretions, and environmental dispersal (Fig. 1) (Graham et al., 2009; Price et al., 2005; Smith et al., 2013).
The transfer of resistant bacteria from farm animals to humans was first reported in the past 40 years, and a high incidence of antibiotic resistance has been shown to take place in the gut microbiota of farm animals and workers (Smith et al., 2013). Over 90 % of conventional antibiotics applied in animals are excreted in urine and feces and then widely transmitted by groundwater, fertilizer, and surface runoff, profoundly impacting the environmental microbiome while also transferring ARB and resistance genes to humans (Fig. 1) (Smith et al., 2013).
As a result, food products from animals may be contaminated with ARB and ARGs. These ARB and resistance genes are present in the soil, water and in excretions from animals (Verraes et al., 2013; Que et al., 2022). As shown in Fig. 1, food may be contaminated with ARB and ARGs present in the environment during food processing. In food, the competence development of transformation (see above) for the uptake of plasmid-borne erythromycin and kanamycin resistance genes including erm(B) and nptII has been shown for Enterococcus faecalis, L. lactis and Bacillus subtilis in milk (Kharazmi et al., 2002; Zenz et al., 1998; Toomey et al., 2009). Antibiotic resistance has also been transferred to Campylobacter jejuni via transformation (Verraes et al., 2013).
During the production of certain food products, probiotics, starter cultures, bacteriophages, and biopreserving microorganisms are intentionally used for technical reasons (Verraes et al., 2013). Starter cultures are added to foodstuffs to induce fermentation in drinks and food, e.g., yoghurt, sauerkraut, and fermented sausages (Verraes et al., 2013). Some probiotics, such as Lactobacillus, Lactococcus, Leuconostoc, and Pediococcus, are mostly used for fermentation and biopreservation (Li et al., 2020a-c). Almost all probiotics contain resistance genes that may be transmitted to other bacteria (Li et al., 2020a-c; Verraes et al., 2013). Many Lactobacillus and Enterococcus species possess transferable ARGs, such as tet, cat, blaZ, and ermB, suggesting a higher risk of transmission to humans, while a few studies have detected the transfer of erm and tet resistance genes of Bifidobacteria from humans (Aires et al., 2007; Gueimonde et al., 2010; Wang et al., 2017a).
2.3. Economic burden
The economic impact of antibiotic resistance is enormous and growing. In the US, medical costs associated with drug-resistant infections are in the range of $18,588–29,069 per patient (Aslam et al., 2018). Direct economic losses of about $20 and $42 billion resulting from such infections have been recorded in the US and China, respectively, which is equivalent to 0.11 % of the US' and 0.37 % of China's yearly gross domestic product (GDP), and amounting to a societal economic burden of $35 billion and $77 billion annually, respectively (Aslam et al., 2018; Ventola, 2015; Zhen et al., 2021). In Europe, bacterial resistance accounted for €1.5 billion in economic burden, with over €900 million in hospital costs; societal costs have been estimated to reach 569 million extra hospital days yearly by 2050 (Kobeissi et al., 2021). Moreover, Sub-Saharan Africa has lost 0.1–2.5 % of its GDP due to bacterial resistance (Taylor et al., 2014). In total, resulting from drug-resistance, the annual global GDP in 2050 would decline by 3.8 % and 1.0 % in high-income and low-income countries, respectively, with >5 % of GDP loss, and the global economic burden of antibiotic resistance is predicted to amount to about $120 trillion by 2050 (Aslam et al., 2018; Zhen et al., 2019; Prestinaci et al., 2015; Bank, 2017).
Additionally, bacterial infections caused by antibiotic-resistant pathogens also impose a burden on families and communities because of lost salaries and health care costs. In India, the average cost of treating a resistant bacterial infection is about $700, which is equivalent to 442 days of salary for a rural male worker. In Organization for Economic Co-operation and Development (OECD) countries, treatments for patients infected with antibiotic-resistant pathogens incur an additional cost of $10,000–$40,000 (Bank, 2017; Cecchini et al., 2015). It is predicted that approximately 24 million people across the globe would be pushed into extreme poverty due to bacterial resistance by 2030, especially in developing countries, and 28.3 million people would fall into poverty in 2050 (Zhen et al., 2019; Bank, 2017; Cecchini et al., 2015; Kotwani et al., 2021). In addition, annual global increases in health care costs due to drug-resistance infections may be up to $0.3–1 trillion per year by 2050, including $23 billion in Europe and the US, and $2.9 trillion in OECD countries (Bank, 2017; Cecchini et al., 2015). If effective strategies are not put into effect in the near future, resistant bacteria may have an even larger negative effect on global public health, food security, and the economy.
3. Bacterial resistance to antibacterial agents
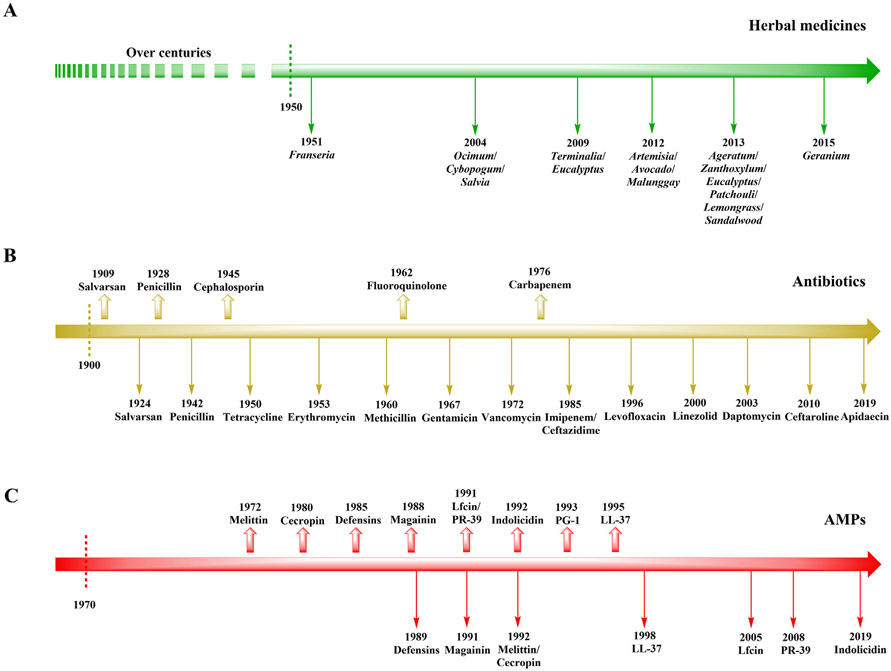
Various bacterial diseases may be treated with currently available antibacterial agents (including traditional herbal medicines and conventional antibiotics). Unfortunately, bacteria have developed resistance to many of these agents (Rubinstein and Keynan, 2013; Chromek et al., 2006; Kai-Larsen et al., 2010). Comparably, conventional antibiotics have contributed substantially to multidrug resistance due to their over prescription world-wide (Miranda et al., 2018; Remesh et al., 2013; Ozkurt et al., 2005). The timeline for bacterial resistance development toward antibacterial agents is provided in Fig. 2, which includes the discovery date of each drug (traditional herbal medicines, conventional antibiotics, and AMPs) along with the year in which bacterial resistance was first reported.
Fig. 2.
Timeline for bacterial resistance to develop against current antibacterial agents. (A) Traditional herbal medicines. (B) Conventional antibiotics. (C) Next-generation AMPs. Defensins: phase IV; magainin: phase III; PG-1: phase II; LL-37: phase I and II; other AMPs: no data. Rightward arrow: timeline; upward arrow: drug discovery date; downward arrow: Year when drug resistance was first reported.
3.1. Bacterial resistance to herbal medicines
For centuries, traditional herbal medicines have been used for the management of various bacterial diseases in many developed and developing countries, however the study of bacterial resistance to herbal drugs has remained understudied (Martin and Ernst, 2003; Bittner Fialová et al., 2021; Odongo et al., 2022). In recent decades, some bacteria (especially enteric bacteria) have been shown to develop resistance to herbal medicines (Fig. 2A; Table 1).
Table 1.
Bacterial resistance to traditional herbal medicines.
| Time | Bacteria | Resistance | Reference |
|---|---|---|---|
| 1951 | S. pullorum and D. pneumoniae | Extracts of Franseria ambrosioides leaves and stems in vitro and in vivo (chicks and mice) | (Parlett, 1951) |
| 1951 | K. pneumoniae | Extracts of Franseria ambrosioides in vitro | (Parlett, 1951) |
| 2004 | P. aeruginosa | Oils extracted from Ocimum gratissimum, Cybopogum citratus, and Salvia officinalis | (Pereira et al., 2004) |
| 2009 | P. aeruginosa, S. enterica Typhimurium, E. coli and K. pneumoniae | Extracts of Terminalia arjuna and Eucalyptus globulus | (Khan et al., 2009) |
| 2010 | P. aeruginosa | Oils extracted from Cymbopogon citratus | (Naik et al., 2010) |
| 2011 | Enterobacteriaceae (Hafnea alvei, Leclercia adecarboxylata, Salmonella, Escherichia, Citrobacter, Kluyvera cryocrescens, Erwinia ananas, Pragia fontium, and Klebsiella), Morganellaceae (Xenorhabdus luminescens and Providencia), Proteobacteria (Proteus), Yersiniaceae (Serratia), Bacillaceae (Bacillus) and staphylococci | Oils extracted from C. citratus | (Singh et al., 2011) |
| 2012 | Bordetella and Bacillus coagulans | Oils extracted from Artemisia vulgaris | (Singh et al., 2012) |
| 2012 | Pediococci (P. acidilactici and P. pentosaceous) and L. plantarum BS | Extracts of avocado (Persea americana Mill.) and malunggay (Moringa oleifera Lam.) | (Saguibo and Elegado, 2012) |
| 2013 | Citrobacter, Edwardsiella, Enterobacter, Klebsiella, Raoultella, Salmonella, etc. | Extracts of Ageratum conyzoides and Zanthoxylum rhetsa; essential oils from eucalyptus, patchouli, lemongrass, sandalwood, and Artemisia vulgaris | (Singh et al., 2013) |
| 2013 | Actinobacillus, Aeromonas, E. coli, Salmonella, Klebsiella, Proteus mirabilis, Pseudomonas, Staphylococcus, Vibrio minicus, Alcaligenes and Brucella | Oils extracted from Salvia officinalis L. sage | (Singh, 2013) |
| 2015 | E. faecium and S. thermophilus | Extracts of Geranium sanguineum and Salvia officinalis leaves | (Teneva-angelova and Beshkova, 2015) |
In the early 1950s, Parlett investigated the resistance of Salmonella pullorum, K. pneumoniae, and Diplococcus pneumoniae to plant extracts (Parlett, 1951). S. pullorum and D. pneumoniae were found to develop resistance to the extracts from the leaves and stems of Franseria ambrosioides in chicks and mice, and K. pneumoniae developed resistance in vitro: the inhibitory concentration of K. pneumoniae increased from 4.5 to 7.5 mg/ml over an 8-week period of subculturing. A few reports have demonstrated the prevalence of resistance to herbal drugs in clinical and environmental bacterial strains. Some clinical bacterial strains, such as Salmonella spp., Escherichia, Klebsiella spp., Pseudomonas aeruginosa, and staphylococci were also resistant to essential oils extracted from herbal plants, including Ocimum gratissimum, Cymbopogon citratus, Cybopogum citratus, Salvia officinalis, and A. vulgaris (Singh et al., 2012; Singh et al., 2011; Naik et al., 2010; Pereira et al., 2004). Singh et al. (2013) found that bacterial strains (including Actinobacillus spp., Aeromonas spp., Salmonella spp., E. coli, and Klebsiella spp.) isolated from sick or dead animals were resistant to S. officinalis L. sage oil (Singh, 2013). Moreover, MDR strains of E. coli, P. aeruginosa, S. enterica Typhimurium, and K. pneumoniae were resistant to extracts of Terminalia arjuna and Eucalyptus globulus (Khan et al., 2009). A total of 194 faecal samples were collected from house geckos and detected the antibiotic and herbal drug sensitivity of bacterial isolates from such samples (Singh et al., 2013). The results indicated that MDR bacteria (including Citrobacter, Edwardsiella, Enterobacter, Klebsiella, and Salmonella) accounted for 12.1 % of the isolates tested for antimicrobial drug sensitivity; among them, only 27.8 % and 13.9 % of the bacterial strains were inhibited by the extracts of Ageratum conyzoides and Zanthoxylum rhetsa; only 3.1–5.4 % of the bacterial strains were inhibited by essential oils from eucalyptus, patchouli, lemongrass, sandalwood, and Artemisia vulgaris. Of note, MDR strains of E. coli, Klebsiella and Raoultella developed resistance to extracts from herbal medicines (such as Ageratum conyzoides, Artemisia vulgaris, Zanthoxylum rhetsa, patchouli, etc.) (Singh et al., 2013). These results indicate that bacterial resistance to herbal drugs and antibiotics are likely to go hand in hand (Singh et al., 2013).
Additionally, Saguibo and Elegado (2012) detected resistance to leaf extracts of malunggay (Moringa oleifera Lam.) and avocado (Persea americana Mill.) leaves in probiotic lactic acid bacteria (LAB), including Lactobacillus plantarum BS, Pediococcus acidilactici strains (4E5, AA5a, 3G3 and K3A2–2) and P. pentosaceous K2A2–3 (Saguibo and Elegado, 2012). These results revealed that all LAB developed selective resistance to some plant extracts; the pediococci strains exhibited higher resistance than L. plantarum BS. Other probiotics, such as the Streptococcus thermophilus and Enterococcus faecium strains isolated from Geranium, were resistant to the extracts of Geranium sanguineum leaves at a concentration of 30 mg/ml; the E. faecium and S. thermophilus isolates from Salvia had increased resistance to S. officinalis leave extracts, at concentrations of 30–100 mg/ml (Table 1) (Teneva-angelova and Beshkova, 2015).
3.2. Bacterial resistance to antibiotics
The first antimicrobial, arsphenamine Salvarsan, was discovered in 1909 and clinical resistance to this drug was first reported in 1924 (Silberstein, 1924; Stekel, 2018). Penicillin was discovered in 1928, then commercialized and implemented in society to treat bacterial infections in the 1940s (Ventola, 2015). However, penicillin-resistant bacteria emerged as early as 1943, and this trend has continued with every antibiotic that has been deployed in our society (Fig. 2B) (Ventola, 2015). Some bacteria have intrinsic resistance and they may also develop resistance to antibiotics through mutation (i.e., adaptive resistance). Resistance can develop in the environment by the uptake of genetic elements encoding resistance genes or mutations in bacterial genomes (Kummerer, 2004; Méhi et al., 2014).
Recent years have seen a rapid rise in the prevalence of pathogenic MDR bacteria globally, representing a serious threat to global health (van Duin and Paterson, 2016). The most important types of ARB include E. faecium, S. aureus, K. pneumoniae, Acinetobacter baumannii, P. aeruginosa, and Enterobacter species (ESKAPE) (Santajit and Indrawattana, 2016). These resistant bacteria are endowed with various intrinsic resistance mechanisms to antibiotics and can also acquire new resistance genes or mutations (Rubinstein and Keynan, 2013).
(i). Enterococci.
Enterococci are Gram-positive facultative anaerobes often found in the gut of animals and humans. More than 20 Enterococcus species have been reported, which are resistant to aminoglycosides, vancomycin, and ampicillin (Rubinstein and Keynan, 2013). Among them, E. faecium and E. faecalis are the most common vancomycin-resistant pathogens (Santajit and Indrawattana, 2016). MDR enterococci have rapidly emerged in recent years; these strains exhibit high-level gentamicin and penicillin resistance due to β-lactamase production. Increasing numbers of vancomycin-resistant and ampicillin-resistant infections caused by enterococci have been detected in healthcare facilities, animals, and humans (Rubinstein and Keynan, 2013; Freitas et al., 2018; Correa-Martínez et al., 2022; Bunnell et al., 2022; Freitas et al., 2022; Samad et al., 2022).
Vancomycin-resistant enterococci (VRE) emerged in North America during the late 1980s and have disseminated across the world with an incidence of 20–30 % (Rubinstein and Keynan, 2013). For example, vancomycin-resistant MDR enterococci have emerged in Europe (Huycke and Gilmore, 1998; Murray et al., 1991; Leclercq et al., 1988). About 2–5 % of the European population and about 0.1–3.7 % of the population in China are intestinal carriers of vancomycin-resistant E. faecium acquired from food (Rubinstein and Keynan, 2013; Zhou et al., 2020). Individuals at the highest risk of infection by VRE are hospital workers and those working in agriculture, food industry, and waste and wastewater treatment, especially when in direct contact with patients and feces (Freitas et al., 2022; Samad et al., 2022). VRE outbreaks can also take place in transplant and intensive care units.
(ii). S. aureus.
The Gram-positive bacterium S. aureus is frequently present on the skin and in the upper respiratory tract, and it is the major cause of soft tissue and skin infectious diseases. Methicillin-resistant S. aureus (MRSA) was first identified in England in 1961 and disseminated globally, becoming a leading cause of bacterial infections in animals and humans (Lee et al., 2018a,b; Morgan, 2008; Romero and de Souza da Cunha, 2021; Argudín et al., 2021; Kejela and Dekosa, 2022). MRSA has also been found in livestock, poultry, and companion animals. New strains of MRSA emerging from animals, particularly pigs, are increasingly causing infections in humans (Morgan, 2008; Abdullahi et al., 2021). In the US, MRSA causes over 80,000 severe infections per year and was responsible for 100,000 deaths in 2019 (Murray et al., 2022). MRSA is regarded as a “superbug” as it is resistant to all β-lactams, among others, and constitutes a growing public health threat.
Vancomycin is one of the last-line antibacterial agents to treat MRSA infections and has been used for nearly 40 years since the 1980s (Werner and Witte, 2008). However, vancomycin-resistant S. aureus isolates have been found for over 20 years (Cong et al., 2020; Shariati et al., 2020; Kejela and Dekosa, 2022). Clinical cases of VRSA (with minimum inhibitory concentration (MIC) ≥ 16 μg/ml) and vancomycin-intermediate S. aureus (VISA) (with MIC of 4–8 μg/ml) are becoming increasingly common (Chambers and Deleo, 2009). The presence or absence of vanA or other van resistance genes is used to determine whether S. aureus isolates can be classified as VRSA (Werner and Witte, 2008). VRSA is of special concern because resistance genes from VRE are transmitted among species. VRSA isolates carry both the vanA and mecA resistance determinants, which confer resistance to vancomycin and methicillin, respectively (Appelbaum, 2007; Desta et al., 2022).
(iii). K. pneumoniae.
The Gram-negative bacterium K. pneumoniae is ubiquitously found on the mucosal surface in humans and animals, water and soil (Wang et al., 2020a). K. pneumoniae exists in the gastrointestinal tract and the nasopharynx of humans; it can also enter the blood circulation or other tissues, causing infection (Piperaki et al., 2017; Chang et al., 2021).
Data from the European Antimicrobial Resistance Surveillance Network for the years 2005–2015 identified K. pneumoniae resistant to all four main categories of antibiotics: carbapenems, aminoglycosides, fluoroquinolones, and third generation cephalosporins. Notably, carbapenem-resistant K. pneumoniae (CRKP) has been found in Italy, Greece, and Romania, with 40–60 % resistance rates, as well as other countries, highlighting the major role of CRKP in the burden of antibiotic resistance (Navon-Venezia et al., 2017). Infections caused by CRKP are highly recalcitrant and impose a tremendous problem particularly in severely ill patients.
Additionally, the production by K. pneumoniae of the enzyme known as New Delhi metallo-β-lactamase (NDM-1), which is encoded by the blaNDM-1 gene, has increased the incidence of CRKP isolates and may affect other antibiotics, including fluoroquinolones, aminoglycosides, and β-lactams (Kumarasamy et al., 2010; Yong et al., 2009; Zhang et al., 2021;Wang et al., 2022a-c ). Carbapenemase-mediated MDR K. pneumoniae cannot currently be completely eradicated with existing drugs.
(iv). A. baumannii.
The Gram-negative organism A. baumannii, a coccobacillus, is an important opportunistic pathogen (Joly-Guillou, 2005; Murray et al., 2022). A. baumannii, which is responsible for 2–10 % of all hospital diseases infected by Gram-negative bacteria, is regarded as one of six major MDR pathogens in hospitals by the Infectious Diseases Society of America (Talbot et al., 2006; Murray et al., 2022).
The WHO published in 2017 a list of global research priorities to treat MDR and broadly resistant bacteria and ranked A. baumannii first on its list (Isler et al., 2019; Kurihara et al., 2020). Treatment for each infection caused by MDR A. baumannii is estimated to cost $33,510–$129,917 (Zhou et al., 2019). Moreover, patients with bacteremia have high mortality rates (56.2 %) due to serious infections caused by MDR A. baumannii (Gramatniece et al., 2019). The attributed mortality rate of MDR A. baumannii varies from 8.4 to 89 % (Cornejo-Juarez et al., 2020; Falagas and Rafailidis, 2007).
(v). P. aeruginosa.
P. aeruginosa is a Gram-negative rod-shaped γ-Proteobacterium found in all habitats (Breidenstein et al., 2011; Gellatly and Hancock, 2013; Moradali et al., 2017). Although P. aeruginosa is considered a conditional pathogen, it is the bacterium most commonly related to hospital-acquired infectious diseases and ventilator-associated pneumonia (Barbier et al., 2013). This bacterium is responsible for high mortality and morbidity in immunocompromised individuals and in patients with cystic fibrosis (CF) (Sadikot et al., 2005). P. aeruginosa is the leading pathogen causing CF lung diseases; chronic infection with P. aeruginosa is recalcitrant to treatment with antibiotics and leads to decreased pulmonary function and finally to mortality in CF patients (Lyczak et al., 2002).
It is exceedingly difficult to completely eradicate P. aeruginosa as it possesses intrinsic, acquired or adaptive resistance, through its low membrane permeability, to a wide variety of antibiotics including fluoroquinolones, aminoglycosides, and β-lactams (Hancock and Speert, 2000; Nicas and Hancock, 1983; Pelegrin et al., 2021). Because most antibiotic molecules do not enter P. aeruginosa cells, they are ineffective. Some ARGs such as ftsI, mcr and oprD in P. aeruginosa may play a role in resistance to β-lactams, carbapenem, cetftolozan, tazobactam, among other antibiotics (Pang et al., 2019; Pelegrin et al., 2021).
(vi). Enterobacteriaceae.
Enterobacteriaceae (which includes the genera Escherichia, Citrobacter, Serratia, Salmonella, Enterobacter, Proteus, and others) are non-fastidious Gram-negative bacilli that can result in opportunistic infections in immunocompromised patients. Enterobacteriaceae have developed a variety of mechanisms against antibiotics (Santajit and Indrawattana, 2016). In 2020, the Global Antimicrobial Resistance and Use Surveillance System (GLASS) reported that the rate of resistance to ciprofloxacin commonly applied in the treatment of urinary tract infections was in the range of 4.1 % to 79.4 % for K. pneumoniae and of 8.4 % to 92.9 % for E. coli. Resistance to fluoroquinolone in E. coli and Salmonella, as well as other genera, is also widespread. In China, a distinct increase was detected in ARGs among E. coli isolates from livestock, leading to resistance to veterinary (norfloxacin and florfenicol) and human medicines (cephalosporins, meropenem and colistin) for 50 years (from the 1970s to 2019) (Yang et al., 2022). Enterobacteriaceae resistant to colistin, a last-line antibiotic, have also been detected in several countries (e.g., Italy, Spain, Belgium, UK, the US, Japan, Egypt, etc.) (Aghapour et al., 2019; Gharaibeh and Shatnawi, 2019; Qadi et al., 2021; Kawamoto et al., 2022).
A few studies investigating patients found that approximately 5.8 % of E. coli and 31.1 % of Enterobacter spp. displayed resistance to third- or fourth generation cephalosporins (Murray et al., 2022; Paterson, 2006; Shah et al., 2004). Genes encoding extended-spectrum β-lactamase (ESBL) are frequently found in Enterobacteriaceae, endowing high-level resistance to sulfonamides, aminoglycosides, and quinolones. ESBL-producing Enterobacteriaceae are MDR and highly resistant to all currently available antibiotics. Antibiotic resistance not only impedes the effective treatment and prevention of Enterobacteriaceae infections but can also be transferred from Enterobacteriaceae to a wide range of bacterial species (Paterson, 2006).
3.3. Bacterial resistance to next-generation AMPs
AMPs are vital components of the host native defense system targeting pathogenic bacteria. These peptides are typically short and positively charged (Koprivnjak and Peschel, 2011). Since bombinin (1962) and melittin (1972) were isolated from the skin secretions of Bombina variegate and honeybee venom, respectively, over 5,948 AMPs (containing natural and synthetic ones) have been found across the tree of life displaying broad-spectrum antimicrobial activity (https://dramp.cpu-bioinfor.org/) (Koprivnjak and Peschel, 2011; Habermann, 1972). However, most AMPs are still in preclinical and clinical development (Lazzaro et al., 2020; Boparai and Sharma, 2020). Bacteria take longer to initiate resistance to AMPs than to conventional antibiotics because AMPs exhibit multifunctional mechanisms of action (Zasloff, 2002). However, there is increasing data reporting that bacteria such as GAS, S. enterica, and Campylobacter jejuni eventually develop resistance to various AMPs (Nizet et al., 2001; Cullen and Trent, 2010; Gruenheid and Le Moual, 2012; Lofton et al., 2013; Andersson et al., 2016; Malik et al., 2017; Duperthuy, 2020; Wang et al., 2022a-c) (Fig. 2C; Table 2). Similarly to conventional antibiotics, the development of bacterial resistance to AMPs is eventually expected when these agents are deployed in clinical practice (Aslam et al., 2018).
Table 2.
Summary of bacterial resistance to new generation AMPs and its mechanisms.
| Types of AMPs |
Reported time |
Bacteria | AMPs | Genes involved in AMP resistance |
Mechanisms | References |
|---|---|---|---|---|---|---|
| Defensins | 1989 | S. enterica serovar Typhimurium | NP-1 | phoP | The phoP mutants play a role in resistance | (Fields et al., 1989) |
| 1996–2013 | B. pertussis | HNP-1, pBD-1 and pBD-2 | brkA and bvg | BrkA and bvg-activated genes and surface modification mediate resistance | (Elahi et al., 2006; Fernandez and Weiss, 1996; Taneja et al., 2013) | |
| 1998 | N. gonorrhoeae | rNP-2 and HNP-2 | mtrR | Energy-dependent mtr efflux systems contribute to resistance to defensins | (Shafer et al., 1998) | |
| 1999–2017 | S. aureus and MRSA | HNP-1, HNP-2, HNP-3, hBD-1, hBD-2, hBD-3 and hBD-4 | dltA/B/C, mprF, hemB, graR/S, qacA, srrAB, aps and mprF | The mutant teichoic acids lacking D-alanine, SCV phenotype and modification of lipid A by phosphoethanolamine transferase LptA endow S. aureus with resistance to defensin | (Habets and Brockhurst, 2012; Kubicek-Sutherland et al., 2017; Glaser et al., 2014; Li et al., 2007; Peschel et al., 1999; Ernst et al., 2009; Peschel et al., 1999; Peschel et al., 2001) | |
| 2003 | S. agalactiae | HNP-1, HNP-2 and HNP-3 | dtlA | D-alanylation of LTA contributes to resistance to AMPs | (Poyart et al., 2003) | |
| 2004 | P. aeruginosa | hBD-1 and rabbit NP1 | pmrAB | PhoPQ two-component system contributes to resistance to AMP | (Moskowitz et al., 2004) | |
| 2005 | Porphyromonas gingivalis | hBD-2 | NN | ND | (Shelburne et al., 2005) | |
| 2006 | B. bronchiseptica | pBD-1 and hBD-2 | NN | ND | (Elahi et al., 2006) | |
| 2007–2015 | H. ducreyi | α-Defensins (HNP-1, HNP-2 and HNP-3) and β-defensins (hBD-2, hBD-4 and hBD-5) | lptA, ptdA and ptdB | Modification of lipid A by phosphoethanolamine transferase LptA confer to resistance to AMPs | (Trombley et al., 2015; Mount et al., 2007; Mount et al., 2010) | |
| 1999–2017 | S. aureus and MRSA | HNP 1–3, hBD-1, hBD-2, hBD-3 and hBD-4 | dltA/B/C, mprF, graR/S, qacA, srrAB, aps and mprF | Reduced electron transport, SCV phenotype, modification of teichoic acids and resistance MprF protein are associated with resistance to defensins | (Kubicek-Sutherland et al., 2017; Li et al. 2007; Peschel et al., 1999; Ernst et al. 2009) | |
| 2011 | K. pneumoniae | hBD-1, hBD-2 and HNP-1 | cps, ugd, pmrF and pagP | Capsule expression and lipid A modification with aminoarabinose and palmitate contribute to resistance to AMPs | (Llobet et al., 2011) | |
| 2013 | B. pertussis | HNP-1 and HNP-2 | dra | dra contributes to AMP resistance by decreasing the electronegativity of the cell surface | (Taneja et al., 2013) | |
| 2015 | Bacteroides thetaiotaomicron | hAD5 | lpxF | LPS modification confers to resistance to AMPs | (Cullen et al., 2015) | |
| 2019 | E. coli | hBD-3 | folA, mlaD, malE, lpxA, acpS and ublA | Reduction in the net negative surface charge of the bacterial membrane by perturbing the Mla pathway confers to resistance to human membrane-targeting AMPs | (Kintses et al., 2019a,b) | |
| Cathelicidins | 1998–2016 | N. gonorrhoeae | LL-37 | mtrR, misR and lptA | The capsule and the endotoxin LOS, energy-dependent multiple transferable efflux systems (such as MtrCDE), MisR response regulator and MisS sensor kinase are involved in resistance to AMP | (Shafer et al., 1998; Kandler et al., 2016; Jones et al., 2009) |
| 2001–2005 | GAS | Cathelicidin and CRAMP | Tn917, crgR and dltA | Chromosomal location of the Tn917 insertion, crgR regulator and D-alanylation of teichoic acid endow with resistance to AMPs | (Nizet et al., 2001) | |
| 2004 | P. aeruginosa | LL-37, CAP18 and SMAP29 | pmrAB | PhoPQ two-component system contributes to resistance to AMP | (Moskowitz et al., 2004) | |
| 2005–2009 | N. meningitidis | LL-37 | mtrD, lptA, siaC, siaD and lipA | The endotoxin LOS and polysaccharide capsule contribute to resistance | (Spencer et al., 2018; Kristian et al., 2005; Tzeng et al., 2005) | |
| 2006–2019 | E. coli | LL-37 and PR-39 | folA, mlaD, malE, lpxA, acpS and ublA | Curli increased adherence to epithelial cells and resistance to AMPs | (Chromek et al., 2006; Kai-Larsen et al., 2010; Kintses et al., 2019a,b) | |
| 2010 | H. ducreyi | LL-37 | sapA | The SapA transporter confers to resistance to AMP | (Mount et al., 2010) | |
| 2007–2017 | S. aureus and MRSA | LL-37 and PR-39 | dltA/B/C, mprF, graR/S, qacA, hemB, aps and mprF | Reduced electron transport, SCV phenotype, the aps sensor system and resistance MprF protein are associated with resistance to AMPs | (Kubicek-Sutherland et al., 2017; Glaser et al., 2014; Sakoulas et al., 2014; Thedieck et al., 2006; Li et al. 2007; Ernst et al. 2009) | |
| 2013–2018 | A. baumannii | LL-37 | pmrB | Nonsynonymous point mutations within pmrB conferring resistance to AMP | (Spencer et al., 2018; Napier et al., 2013) | |
| 2013 | S. enterica Typhimurium | LL-37 and PR-39 | pmrB, phoP and waaY (or rfaY) | Mutations in two-component signal transduction pathways and in the lipopolysaccharide biosynthesis pathway confer to resistance to AMPs | (Lofton et al., 2013) | |
| 2015 | Bacteroides thetaiotaomicron | LL-37 and CRAMP | lpxF | LPS modification confers to resistance to AMPs | (Cullen et al., 2015) | |
| Others | 1991–2017 | S. enterica Typhimurium | Magainin 2, cecropin P1, mastoparan, melittin, CNY100HL, WGH, Lfcin and Cycloviolacin O2 | sbmA, hemA, hemB, hemC, hemL, pmrB, waaY and phoP | Inactivation and mutations in two-component signal transduction pathways and in the LPS biosynthesis pathway reduce the uptake of AMPs | (Malik et al., 2017; Lofton et al., 2013; Glaser et al., 2014; Pranting et al., 2008; Pranting and Andersson, 2010; Paterson et al., 2009; Groisman et al., 1992; Rana et al., 1991) |
| 1996 | B. pertussis | Cecropin P1 | brkA and bvg | BrkA protein and bvg-activated genes mediate resistance | (Fernandez and Weiss, 1996) | |
| 1998 | B. bronchiseptica and B. pertussis | Cecropin (P and B), magainine-II-amide, protamine and melittin | bvgS, wlbA and wlbL | The loss of the O-specific side chains and the prevalence of the LPS core structure confer to resistance to AMPs | (Banemann and Gross, 1998) | |
| 1998–2005 | N. gonorrhoeae | PG-1 | mtrR and lptA | Energy-dependent mtr efflux systems and lipid A modification contribute to resistance to AMPs | (Shafer et al., 1998; Tzeng et al. 2005) | |
| 1999–2020 | S. aureus | LfcinB, melittin, pexiganan, indolicidin, protegrins, tachyplesins and magainin II | hemB, pmtR (ytrA), vraG (bceB), atl, namA, menF, atl and aps | SCV phenotype and the aps sensor system endow S. aureus with resistance to AMPs | (Habets and Brockhurst, 2012; El Shazely et al., 2020; Samuelsen et al., 2005; Li et al. 2007; Peschel et al., 1999) | |
| 2000 | P. aeruginosa | CP28 and CP29 | phoQ | The PhoP-PhoQ system contributes to resistance to AMPs | (Macfarlane et al., 2000) | |
| 2003 | S. agalactiae | Indolicidin, magainin II, cecropin B, PG-1 and PG-3 | dtlA | D-alanylation of LTA contributes to resistance to AMPs | (Poyartet al. 2003) | |
| 2006 | P. fluorescens | Pexiganan | NN | ND | (Perron et al., 2006) | |
| 2006–2019 | E. coli | Membrane-targeting (pexiganan, CAP18, cecropin P1, etc.) and intracellular-targeting AMPs (apidaecin IB, bactenecin 5, indolicidin, etc.) | folA, mlaD, malE, lpxA, acpS and ublA | Reduction in the net negative surface charge of the bacterial membrane by perturbing the Mla pathway confers to resistance to human membrane-targeting AMPs | (Kintses et al., 2019a,b; Perron et al., 2006) | |
| 2011 | K. pneumoniae | Magainin 2 | ugd, pmrF and pagP | Capsule expression and lipid A modification with aminoarabinose and palmitate contribute to resistance to AMPs | (Llobet et al., 2011) | |
| 2017 | MRSA | CNY100HL and WGH | dltA/B/C, mprF, graR/S, qacA and srrAB | Reduced electron transport and SCV phenotype are associated with resistance to AMPs | (Kubicek-Sutherland et al., 2017) | |
| 2019 | Gut microbiota | Cecropin P1, indolicidin, PR-39, R8, tacyplesin II, omigannan, etc. | acr, alm, anr, pmr, arn, bra, col, cpr, dlt, epi, lpx, mcr1, mtr, par, pho, yej, etc. | Intrinsic resistant genes contribute to AMPs | (Kintses et al., 2019a,b) |
ND; no data.
(i). Defensins.
Defensins (18–45 amino acids) are small cationic defense peptides that are rich in cysteine and found in animals, plants, and fungi. Characterized by the linking pattern of highly conserved cysteines, the following three sub-families of defensins have been classified: α (Cys1–6, Cys2–4 and Cys3–5), β (Cys1–5, Cys2–4 and Cys3–6), and circular θ (no free N- and C-terminus), mainly secreted by neutrophils, epithelial cells, and leukocytes, respectively (Ganz, 2003). Defensins display potent antimicrobial and immune activities (Ganz, 2003; Arnett and Seveau, 2011; Contreras et al., 2020; Yu et al., 2022; Wu et al., 2022).
A few studies have shown that defensins can induce resistance in several bacterial pathogens (Table 2) (Elahi et al., 2006). For example, Bordetella pertussis displayed various levels of resistance to human α-defensin 1 (HNP-1) and α-defensin 2 (HNP-2) (Fernandez and Weiss, 1996; Taneja et al., 2013). The dra locus confers defensin resistance by reducing the cell surface electronegativity of B. pertussis (Taneja et al., 2013). Elahi et al. evaluated the protective effects of β-defensin against B. pertussis and Bordetella bronchiseptica in newborn piglets and found that B. bronchiseptica was fully resistant to porcine β-defensin 1 (pBD-1) and human β-defensin 2 (hBD-2), but B. pertussis was highly sensitive to pBD-1 and partially resistant to pBD-2 (Elahi et al., 2006). N. gonorrhoeae developed resistance to the rabbit defensin 2 (rNP-2) and HNP-2 by a mechanism related to energy-dependent mtr efflux systems (Shafer et al., 1998). Shelburne et al. investigated the effects of human β-defensin 1 (hBD-1), β-defensin 2 (hBD-2), β-defensin 3 (hBD-3), and β-defensin 4 (hBD-4) on the oral periodontal pathogen Porphyromonas gingivalis and found that these four hBDs can induce resistance in multiple P. gingivalis strains after pretreatment with sublethal levels of defensins or exposure to environmental conditions such as heat and peroxide stress (Shelburne et al., 2005).
H. ducreyi has high resistance to human α-defensins (including HNP-1, HNP-2 and HNP-3) and β-defensins (such as hBD-2, hBD-4 and hBD-5), respectively (Mount et al., 2007; Mount et al., 2010). This may be related to modification of pEtN transferase in Haemophilus ducreyi, conferring resistance to α-defensins and β-defensins (Trombley et al., 2015). Moreover, lipid A modification by phosphoethanolamine transferase LptA in H. ducreyi can confer resistance to human defensins (such as α-defensin HNP-3 and β-defensin hBD-5) (Trombley et al., 2015). Exposure of K. pneumoniae to polymyxin induces cross-resistance to hBD-1, hBD-2, and HNP-1 (Llobet et al., 2011). Treatment with polymyxin B led to upregulated expression of genes cps, pmrF, ugd, and pagP, conferring resistance to defensins (Llobet et al., 2011). A few studies have demonstrated that S. aureus rapidly became resistant to HNP-1, hBD-2 and hBD-3 (Habets and Brockhurst, 2012; Kubicek-Sutherland et al., 2017). MRSA mutants resistant to LL-37 exhibited increased cross-resistance to hBD-1, hBD-2 and hBD-3; PR-39-resistant S. aureus mutants displayed cross-resistance to hBD-1 and hBD-4; a wheat germ histone (WGH)-resistant S. aureus mutant showed resistance to hBD-1, hBD-3 and hBD-4. These observations indicate that resistant S. aureus mutants may evade host defensins (Kubicek-Sutherland et al., 2017); in fact, S. aureus resistance to defensins is linked to the formation of the small colony variant (SCV) phenotype and reduced electron transport (Habets and Brockhurst, 2012; Kubicek-Sutherland et al., 2017; Glaser et al., 2014).
(ii). Cathelicidins.
Cathelicidins are a third general class of epithelial AMPs that are expressed in the intestinal epithelium in humans and other animals and can kill bacteria by membrane disruption (Bals and Wilson, 2003; Batista Araujo et al., 2022). Recent data have demonstrated that some bacterial pathogens (including Neisseria, GAS, S. enterica, and E. coli) display resistance to cathelicidins (Table 2) (Lofton et al., 2013; Nizet et al., 2001; Chromek et al., 2006).
Wild-type N. gonorrhoeae strain FA19 is 4-fold to 8-fold more highly resistant to LL-37 than its isogenic misR-null mutant JK100, which is attributed to the MisR response regulator (Kandler et al., 2016). Moreover, the energy-dependent efflux systems-multiple transferable resistance (mtr), MtrCDE and resistance/nodulation/division (RND) in N. gonorrhoeae also contribute to resistance to LL-37 (Shafer et al., 1998; Kandler et al., 2016; Spencer et al., 2018). Another wild-type N. meningitidis serogroup C strain FAM20 was resistant to 10 μM of LL-37 after 1 h of attachment to the epithelial cell surface, which was attributed to its capsule and the endotoxin lipooligosaccharide (LOS) (Jones et al., 2009). Nizet et al. discovered that cathelicidin- and CRAMP-resistant GAS was more pathogenic than sensitive GAS in normal mice; chromosomal integration of transposon Tn917 and cathelicidin resistance gene regulator-crgR are associated with an inducible cathelicidin-resistant phenotype of GAS (Nizet et al., 2001). Meanwhile, the S. enterica Typhimurium LT2 strain was resistant to LL-37, resulting from to gene mutations in two-component signal transduction and in lipopolysaccharide (LPS) biosynthesis (Lofton et al., 2013). Clinical pyelonephritic E. coli strains from patients were more highly resistant to LL-37 than cystitic strains; these data suggested that E. coli invasiveness was related to bacterial resistance to cathelicidin (Chromek et al., 2006). Additionally, E. coli strains with mutant curli proteins or mutant cellulose synthase were more resistant to human LL-37 and mouse cathelicidin than their wild-type counterparts (Kai-Larsen et al., 2010).
In addition to the resistance of H. ducreyi to defensins as noted above, this species was found to be resistant to cathelicidin LL-37, which was linked to the SapA transporter (Mount et al., 2010). Additionally, previous studies showed that when MDR A. baumannii was treated with colistin, cross-resistance to host LL-37 increased, which was linked to mutations within pmrB (Spencer et al., 2018; Napier et al., 2013). Noticeably, community-associated (CA)-MRSA USA300 LAC JE2 and USA600 strains exhibited remarkably increased resistance to human LL-37 (Sakoulas et al., 2014) due to defects in electron transport (Kubicek-Sutherland et al., 2017). Additionally, the hemB S. aureus mutant displayed the SCV phenotype, conferring resistance to LL-37 (Glaser et al., 2014).
(iii). Other peptides.
Additional reports have demonstrated the development of resistance to other AMPs, such as magainin, melittin, and lactoferricin B (LfcinB) (Table 2) (Kintses et al., 2019a,b; Andersson et al., 2016). Fernandez and Banemann detected in B. bronchiseptica and B. pertussis resistance to various AMPs, including cecropin (P and B), melittin, magainine-II-amide, and protamine (Fernandez and Weiss, 1996; Banemann and Gross, 1998). These authors found that B. bronchiseptica displayed remarkably more resistance to all AMPs than B. pertussis, which may be due to structural changes in its LPS, specifically, the absence of highly charged O-specific sugar side chains and the presence of a crystalline porin structure mediated by wlbA, wlbL, and bvg gene products (Fernandez and Weiss, 1996; Banemann and Gross, 1998). Shafer et al. also found that the mtr efflux system of N. gonorrhoeae could enhance gonococcal resistance to LL-37 and protegrin-1 (PG-1) (Shafer et al., 1998).
Polymyxin-resistant P. aeruginosa developed cross-resistance to defensins, cathelicidins (including LL-37, CAP18 and SMAP29), and other peptides (such as CP28 and CP29) (Moskowitz et al., 2004; Macfarlane et al., 2000). S. aureus rapidly evolved cross-resistance to melittin, LfcinB, HNP-1, and pexiganan (magainin's derivative, phase III clinical trials) in vitro; four mutated genes — pmtR (ytrA), vraG (bceB), atl, and namA — were identified in each melittin-resistant strain (Habets and Brockhurst, 2012; El Shazely et al., 2020; Samuelsen et al., 2005). The S. enterica Typhimurium DA6192 strain developed cross-resistance to cycloviolacin O2, the cyclotide peptide isolated from Viola odorata after 100–150 passages (Malik et al., 2017). In S. enterica, some mutations (sbmA, waaY, and phoP) in two-component signal transduction pathways and LPS biosynthesis conferred resistance to CNY100HL, WGH, Lfcin, and PR-39; strains carrying these mutations were also cross-resistant to cycloviolacin O2 (Malik et al., 2017; Lofton et al., 2013; Glaser et al., 2014; Pranting et al., 2008; Pranting and Andersson, 2010). Exposure of K. pneumoniae to polymyxin induced cross-resistance to magainin 2 and human defensins (including hBD1, hBD2, and HNP1), which was associated with LPS modification and capsule expression (Llobet et al., 2011). The resistance of the CA-MRSA USA300 LAC JE2 strain to AMPs was assessed by serial passage; the results showed that this strain of S. aureus was highly resistant to CNY100HL, WGH, and PR-39, again resulting from defects in electron transport (Kubicek-Sutherland et al., 2017).
A study systematically assessing the resistance of E. coli to 15 AMPs revealed the prevalence of resistance or cross-resistance in E. coli to membrane-targeting peptides (pexiganan, CAP18, cecropin P1, HBD-3, LL-37, and others) and intracellular-targeting peptides with similar modes of action (apidaecin IB, bactenecin 5, indolicidin, PR-39, and others) (Kintses et al., 2019a,b; Perron et al., 2006). Perturbing the Mla pathway can decrease the net negative charge of the bacterial cell surface, leading bacteria to become resistant to AMPs that target bacterial membranes. Intracellular-targeting AMPs (PR39 and BAC5) can scarcely elicit bacterial cross-resistance to membrane-disruptive human AMPs (LL-37) (Kintses et al., 2019a,b). Similarly, P. fluorescens rapidly developed resistance to pexiganan, an analogue of magainin, after exposure to gradually increasing concentrations of this peptide (Perron et al., 2006).
4. Mechanisms of bacterial resistance to current antibacterial agents
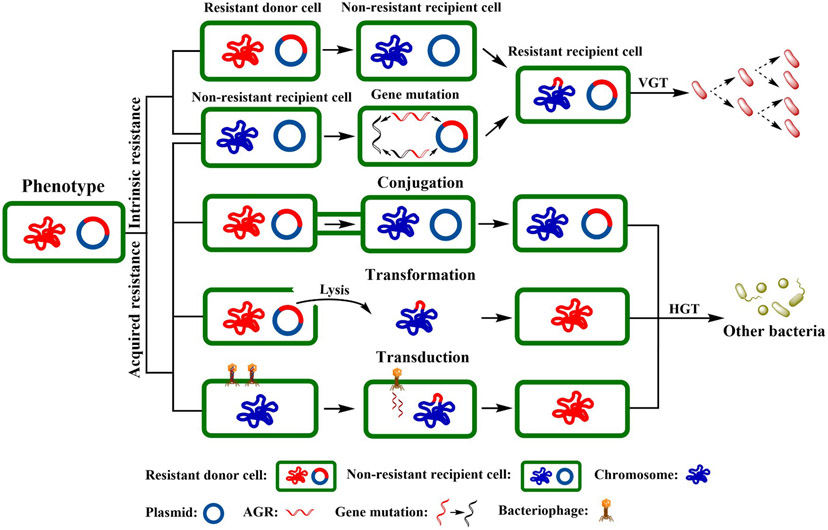
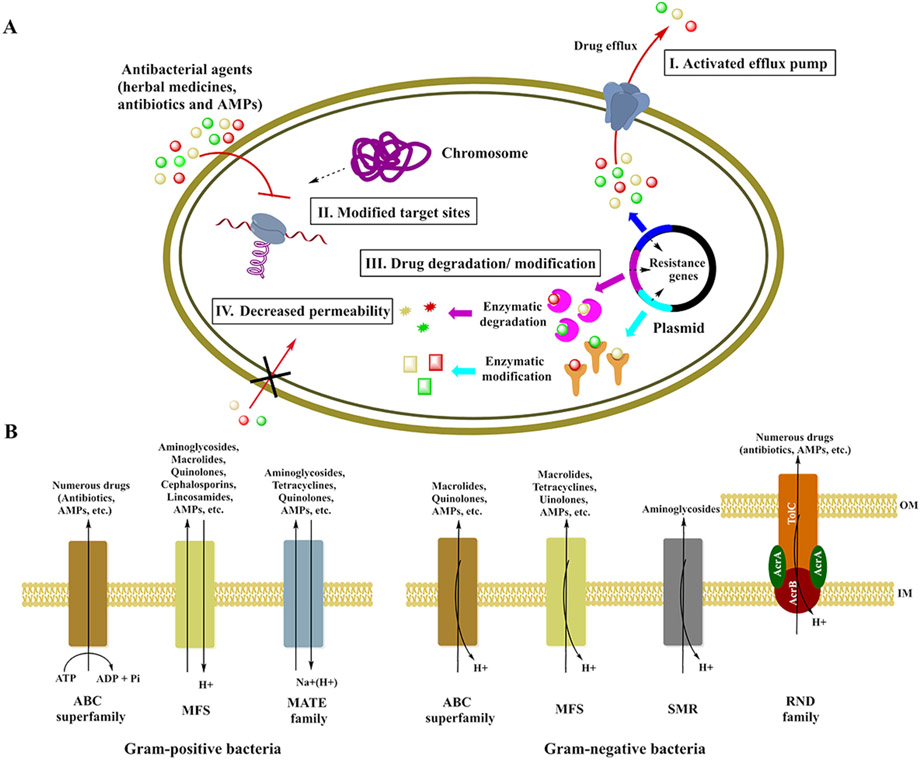
Bacterial resistance to antibacterial drugs can be classified as either intrinsic or acquired resistance. Table 3 describes instances of intrinsic resistance of both pathogenic bacteria and probiotics to conventional antibiotics, including aminoglycosides, β-lactams, carbapenems and quinolones, etc. Meanwhile, inherent ARGs vertically transfer during cell division to the next generation (Fig. 3) (Davies and Davies, 2010; Perry et al., 2014). The most common bacterial mechanisms involved in intrinsic resistance involve efflux pumps and modifying the bacterial outer membrane (Fig. 4A) (Reygaert, 2018). Comparably, acquired resistance occurs via DNA mutation or by horizontal transmission (Fig. 3) and may involve activation of efflux pumps, target modification, and drug modification or inactivation (Fig. 4A) (Regea, 2018; Reygaert, 2018).
Table 3.
Intrinsic resistance of bacteria to conventional antibiotics.
| Types | Bacteria | Antibiotics | References |
|---|---|---|---|
| Pathogens | Treponema pallidum | Arsphenamine (Salvarsan) | (Silberstein, 1924) |
| E. coli | Macrolides | (Reygaert, 2018; Liu et al., 2010; Blair et al., 2004) | |
| S. marcescens | Macrolides | (Reygaert, 2018; Blair et al., 2004; Stock et al., 2003) | |
| Bacteroides | Aminoglycosides, β-lactams and quinolones | (Reygaert, 2018) | |
| S. maltophilia | Aminoglycosides, β-lactams, carbapenems and quinolones | (Reygaert, 2018; Nicodemo and Paez, 2007; Alonso and Martínez, 2001; Alonso and Martínez, 2000) | |
| L. monocytogenes | β-Lactams and cephalosporins | (Reygaert, 2018) | |
| Klebsiella spp. | β-Lactams | (Reygaert, 2018; Fu et al., 2007) | |
| Acinetobacter spp. | β-Lactams and glycopeptides | (Reygaert, 2018; Vila et al., 1993; Seifert et al., 1993) | |
| P. aeruginosa | Sulfonamides, β-lactams, chloramphenicols and tetracyclines | (Reygaert, 2018; Deplano et al., 2005; Sellera et al., 2018) | |
| S. enterica Typhimurium | Melittin and cecropin | (Paterson et al., 2009) | |
| S. aureus | Indolicidin, hBD-3 and LL-37 | (Li et al., 2007) | |
| Probiotics | Enterococcus | Aminoglycosides, β-lactams, cephalosporins, lincosamides and glycopeptides | (Li et al., 2020a-c; Reygaert, 2018; Hollenbeck and Rice, 2012; Portillo et al., 2000; Courvalin et al., 1980; Zervos and Schaberg, 1985; Arias et al., 2007) |
| Lactobacillus | Aminoglycosides, glycopeptides, metronidazole and quinolones | (Guo et al., 2017; Courvalin et al., 1980; Jaimee and Halami, 2015) | |
| Bifiobacterium | Aminoglycosides, glycopeptides, metronidazole, quinolones and polypeptides | (Li et al., 2020a-c; Jose et al., 2014; Gueimonde et al., 2010) | |
| L. lactis | Aminoglycosides, quinolones, polypeptides and β-lactams | (Li et al., 2020a-c; Jose et al., 2014; Gueimonde et al., 2010; Hummel et al., 2007) | |
| S. thermophilus | Aminoglycosides and neomycin | (Li et al., 2020a-c; Jose et al., 2014; Gueimonde et al., 2010) |
Note: “R” in the table indicates resistance to the antibiotics.
Fig. 3.
Phenotypes of bacterial resistance to antibacterial agents. Bacterial resistance includes intrinsic resistance (only vertically transferred, taxa-specific) and acquired resistance (vertically or horizontally transferred, taxa-nonspecific). Bacteria can transfer ARGs by horizontal gene transfer (HGT), i.e., conjugation, transformation, or transduction, and by vertical gene transfer (VGT) during genome replication and cell division. HGT is regarded as one of the major mechanisms of ARG transfer and is responsible for the abundance of ARGs. VGT can determine the prevalence of ARGs within a bacterial community (Rana et al., 1991; Pereira et al., 2004; Cox and Wright, 2013).
Fig. 4.
Bacterial mechanisms of resistance to current antibacterial agents. (A) Resistance mechanisms. Bacterial resistance mechanisms include activated efflux pumps, modified targets, degradation or modification of drugs, and decreased permeability to drugs. (B) Bacterial efflux pumps. In Gram-positive bacteria, efflux pumps (such as the ABC superfamily, MFS, and the MATE family) are related to resistance to a few antibiotics and AMPs. Efflux pumps of Gram-negative bacteria include the ABC superfamily, MFS, SMR, and the RND family (Russell et al., 1986; Gunn et al., 1998; Shafer et al., 1998; Saar-Dover et al., 2012; Warner et al., 2008; Wang et al., 2017a; Uddin et al., 2021). IM: inner membrane; OM: outer membrane.
4.1. Activation of efflux pumps
Bacterial efflux pump systems, composed of membrane proteins, are present in bacterial cells and play a key role in resistance to antibacterial agents (including traditional herbal medicines, conventional antibiotics and AMPs); efflux pumps can also be involved in other functions in bacteria such as virulence and pathogenesis (Li and Nikaido, 2009; Alav et al., 2018; Toit, 2017). Efflux pumps can be encoded both within the bacterial chromosome and on plasmids. They involves five major families of efflux systems in bacteria: major facilitator superfamily (MFS), ATP binding cassettes (ABC), small multidrug resistance (SMR), resistance nodulation division (RND), and multidrug and toxic compound extrusion (MATE) (Li and Nikaido, 2009). The electrochemical potential of the membrane is an energy source for some efflux pumps. The ABC efflux pump, however, uses ATP hydrolysis for the export of toxic antibacterial agents or biological metabolites from bacterial cells out into the extracellular environment (Fig. 4A) (Mahamoud et al., 2007; Aires and Nikaido, 2005; Zgurskaya and Nikaido, 1999).
Several efflux pumps are constitutively produced at low levels and confer intrinsic resistance under any given condition, whereas others are transiently overexpressed in the presence of a mutation or an effector (i.e., acquired resistance) (Li and Nikaido, 2009). The most abundant efflux pumps are those of the MFS, which are encoded in both Gram-positive and Gram-negative bacteria, and RNDs, which exclusively exist in Gram-negative bacteria. Both types of efflux pumps contribute to resistance to fluoroquinolones, penicillins, macrolides, polymyxin B, tetracyclines, and other conventional antibiotics, as well as some AMPs (Table 4) (Alav et al., 2018; Aygul, 2015; Kabra et al., 2019; Bina et al., 2008; Mateus et al., 2021; Tian et al., 2010). The RND tripartite pump in Gram-negative bacteria is fixed in the inner membrane and comprised of an inner membrane transporter (such as AcrB, AcrD, or MexB), a periplasmic adaptor protein (such as AcrA, MexC, MexA, MexX, or MexE), and an outer membrane channel (such as TolC, OprJ, OprN, or OprM). The RND tripartite pump can catalyze the active efflux of various conventional antibiotics (Daury et al., 2016). For example, E. coli AcrABZ-TolC is composed of four distinct functional proteins: TolC, AcrA, AcrB, and AcrZ (Fig. 4B). TolC, which is similar to a porin, traverses the bacterial outer membrane. The inner membrane protein AcrB, which extends into the periplasmic region, is a permease that utilizes the potential energy of a proton motive force to drive the efflux process. AcrA connects AcrB and TolC and can transfer drug-triggered conformational changes in AcrB within the inner membrane to TolC in the outer membrane. The highly conserved AcrZ protein interacts with the AcrB transmembrane regions, influencing drug recognition (Nikaido, 2009). In the presence of antibiotics, the pump assumes a transport activated state, in which the AcrB trimer is either in a loose (L), tight (T), or open (O) conformation, representing different stages of the transport mechanism; the cycling of AcrB from the L to T to O conformation is driven by proton translocation through the transmembrane domain of AcrB. In the absence of antibiotics, this pump switches to a resting state, in which the AcrB trimer is in the LLL conformation and the TolC central channel is closed (Aron and Opperman, 2018; Wang et al., 2017a). The AcrABZ-TolC RND pump system can extrude most common antibiotics and detergents, thereby lowering their concentration inside the cell, and although AcrB cannot extrude aminoglycosides, a homologous protein, AcrD, can (Wang et al., 2017a; Russell et al., 1986).
Table 4.
Efflux pumps of ESKAPE and their resistance to antibacterial agents.
| ESKAPE | Transporter family | Efflux pump | Antibacterial agents | References |
|---|---|---|---|---|
| Enterococci | MFS and ABC | EmeA, EfmA, EfrAB, MsrC, ABC7 and ABC16 | Antibiotics: macrolides (azithromycin, erythromycin), aminoglycosides, tetracyclines, quinolones (ofloxacin, norfloxacin, ciprofloxacin) and lincosamides (lincomycin) | (Santajit and Indrawattana, 2016; Miller et al., 2014; Nishioka et al., 2009; Chouchani et al., 2012) |
| S. aureus | MFS, ABC, MATE and SMR | NorA, NorB, NorC, MsrA, MepA, SepA, MdeA, SdrM, Tet38, QacA and QacB | Antibiotics: quinolones (mupirocin, norfloxacin), macrolides, tetracyclines and β-lactams (nafcillin, penicillin G, methicillin and cefotaxime); AMPs: LL-37, HBD-3 and indolicidin | (Reygaert, 2018; Blair et al., 2004; Falord et al., 2012; Schrader-Fischer and Berger-Bachi, 2001) |
| K. pneumoniae | MFS, RND and SMR | KpnGH, KpnEF and AcrAB-TolC | Antibiotics: chloramphenicols (chloramphenicol), quinolones (nalidixic acid, ciprofloxacin, and levofloxacin) and cephalosporins (cefoxitin) AMPs: cathelicidin and defensins (HNP-1, hBD-1 and hBD-2) |
(Padilla et al., 2010; Srinivasan et al., 2014; Srinivasan and Rajamohan, 2013) |
| A. baumannii | MFS, MATE and RND | SmvA, AbeM, AdeIJK and AdeABC | Antibiotics: macrolides (erythromycin), quinolones and cephalosporins (cephalosporin) | (Vrancianu et al., 2020; Kyriakidis et al., 2021; Su et al., 2005) |
| P. aeruginosa | SMR, MATE and RND | MexAB-OprM, MexCD-OprJ, MexEF-OprN, MexXY-OprM and EmrE | Antibiotics: β-lactams (ampicillin), tetracyclines, chloramphenicols (chloramphenicol), quinolones and cephalosporins | (Nikaido, 2009; Li et al., 1995; Poole et al., 1996; Kohler et al., 1997; Rieg et al., 2009; Poole, 2001) |
| Enterobacter | MFS, RND, MATE and ABC | QepA, AcrAB-TolC, AcrEF-TolC, AcrAD-TolC, Mef(B), MacAB-Tolc, MsdAB-TolC, MdfA, MdtK, SmfY, SsmE and SmdAB | Antibiotics: quinolones (norfloxacin), β-lactams, tetracyclines, chloramphenicols and macrolides (erythromycin) AMPs: LL-37, HNP-1, HNP-2, HNP-3, HBD-1, HBD-2 and PG-1 |
(Russell et al., 1986; Padilla et al., 2010; Kohler et al., 1997; Rieg et al., 2009; Mine et al., 1999) |
Other orthologous RND pumps that extrude antibacterial drugs, such as MexAB-OprM and VexAB of K. pneumoniae, A. baumannii, P. aeruginosa, Enterobacter, N. meningitides, N. gonorrhoeae, and Vibrio cholerae, share the same architecture as AcrABZ-TolC; they are responsible for resistance to antibiotics as well as a few AMPs (including HNP-1, LL-37, HBD-1, HBD-2, PG-1, CRAMP-38, etc.) (Shafer et al., 1998; Padilla et al., 2010; Joo et al., 2016).
MFS efflux pumps are composed of 12 or 24 total membrane fractions and are associated with the transport of conventional antibiotics, metabolites, and other dyes (Reygaert, 2018). Most MFS efflux pumps are encoded in bacterial chromosomes. TetA, QepA, MefB, and NorA pumps in many Enterobacteriaceae and S. aureus actively extrude tetracyclines, fluoroquinolones, chloramphenicols, and macrolides; particularly in E. coli, approximately 50 % of efflux pumps belong to the MFS system (Reygaert, 2018; Blair et al., 2004; Falord et al., 2012).
ABC transporters confer resistance to various antibiotics and AMPs (including vancomycin, nisin, indolicidin, LL-37, bacitracin, and HBD-3) (Table 4) (Li and Nikaido, 2009; Falord et al., 2012). For example, the dual ABC and MFS efflux pumps found in S. pneumoniae play a key role in resistance to cathelicidin that can alter mefE and mel expression and this can confer cross-resistance to LL-37 and macrolides (Zahner et al., 2010). Notably, the multidrug-binding repressor TtgR can regulate the transcription of the RND family transporter TtgABC in Pseudomonas putida; the TtgR-TtgABC system recognizes and extrudes various conventional antibiotics (including chloramphenicol and tetracycline) and herbal flavonoids (such as quercetin, naringenin, and phloretin) (Alguel et al., 2007; Teran et al., 2006). The SapABCDF function in S. enterica may mediate resistance to melittin. AMPs bind to SapA in the periplasm and are transported to the cytoplasm, where they may be degraded by proteases or bind to a regulator, activating resistance determinants (Parra-Lopez et al., 1993).
Some efflux pumps also play an important role in bacterial biofilm resistance and undergo upregulation in biofilms in comparison with bacterial cells in the planktonic state (Gillis et al., 2005). ABC-, MFS- and RND-type efflux pumps were reported to be of great importance for the evolution of resistance to aminoglycosides, quinolones, β-lactams, and other antibiotics in a sub-population of bacterial biofilms, such as those produced by P. aeruginosa, E. coli, A. baumannii, S. enterica, and S. aureus (Li and Nikaido, 2009; Pamp et al., 2008). These resistance patterns are strongly associated with antibiotics actively extruded by the MexXY, MexCD-OprJ and MexAB-OprM pumps, thus linking these pumps to biofilm resistance to antibiotics (Gillis et al., 2005; Chiang et al., 2012).
4.2. Modification of target sites
A few cases of bacterial resistance attributable to target modification involve the mutation of target genes, leading to reduced binding to drugs (herbal medicines, antibiotics and AMPs) (Fig. 4A) (Nikaido, 2009; Su et al., 2018; Marshall et al., 2002). The chromosome or a plasmid may encode the target genes. Bacterial resistance to fluoroquinolones, for example, is elicited by mutations in their target enzymes-DNA gyrase and topoisomerase IV, which play a role in DNA replication (Hooper, 2000). Ince et al. determined the effects of mutations in topoisomerase IV and DNA gyrase in S. aureus on the MICs for gatifloxacin and ciprofloxacin. The MICs of these antibiotics against S. aureus with mutated grlA (A116E and S80F), grlB (R470N), or gyrA (S84L) increased by 2-, 4-, and 8-fold, respectively, indicating that mutations in these fluoroquinolone targets can result in bacterial resistance (Ince et al., 1999). Similarly, the gyrA S. pneumoniae mutants were cross-resistant to sparfloxacin and gatifloxacin; parC gene encodes subunits of topoisomerase IV and point mutations in this gene in S. pneumoniae led to increased MICs for ciprofloxacin, levofloxacin, trovafloxacin, and norfloxacin (Ince et al., 1999). Gootz et al. also demonstrated that the parC mutants of S. pneumoniae selected by ciprofloxacin are resistant to both ciprofloxacin and trovafloxacin (Gootz et al., 1996). These results support the notion that some target-altered mutants are resistant to fluoroquinolones.
Another example of bacterial high-level resistance ascribed to target mutation is streptomycin resistance conferred by mutations in rpsL, which encodes ribosomal proteins (e.g., RpsL) in E. coli (Pelchovich et al., 2013). Ribosomal mutations in target genes rpsL, rrs, and S12 are also found in Mycobacterium tuberculosis clinical strains, contributing to resistance to streptomycin, rifampicin, pyrazinamide, and ethambutol (Finken et al., 1993; Khosravi et al., 2017). The mechanism of action of antibiotics determines whether this resistance can be transferred between bacteria; in some cases, a plasmid carrying this mutated target gene will not make the recipient completely resistant due to the antibiotic targeting the ribosome (including chromosomally encoded intact RpsL), which can cause cell death (Nikaido, 2009). Additionally, the erm family of genes can confer clinical resistance to macrolides; mutation in 23S rRNA by altering A2058 or its neighboring sites and methylation of A2058 by an adenine-specific methylase-ErmC can cause resistance to lincosamides, macrolides, chloramphenicol, and streptogramins (the MLS antibiotics) in E. coli, S. aureus, and B. subtilis (Nikaido, 2009; Weisblum, 1995).
On the Gram-negative bacterial cell surface, the major modification target sites are located in LPS and LOS, particularly lipid A. These targets are modified by the removal of phosphate groups and the reduction of their net negative charge (Fleitas et al., 2016; Ding et al., 2008). The addition of L-Ara4N, glucosamine, galactosamine, palmitate, acylation, glycine or diglycine to the acyl chain of lipid A and to phosphatidylglycerol (PG) can confer resistance to peptide antibiotics (including colistin and polymyxin B) and AMPs (including indolicidin, LL-37, and defensins) in various bacteria, including S. enterica, P. aeruginosa, K. pneumoniae, B. pertussis, V. cholerae, and S. aureus (Fleitas et al., 2016; Ding et al., 2008; Hankins et al., 2012; Shah et al., 2014; Thaipisuttikul et al., 2014; Tamayo et al., 2005; Dalebroux et al., 2014). Meanwhile, modified lipoteichoic acid (LTA) in Gram-positive bacteria is regarded as a common cause of resistance to polymyxin B, colistin, and a subset of cationic AMPs (such as magainin 2, LL37, K9L6, and K6L9), especially through D-alanylation of LTA (Kristian et al., 2005; Saar-Dover et al., 2012). The dlt operon is responsible for encoding the D-alanyl carrier protein ligase, which adds D-alanine to the LTA Gro-P moiety; dlt mutants displayed enhanced sensitivity to AMPs, but overexpression of these genes conferred increased resistance (Neuhaus and Baddiley, 2003). Additionally, the capsular polysaccharides or exopolysaccharides produced by S. pneumoniae, K. pneumoniae, E. coli, and P. aeruginosa can directly trap drugs or inhibit their interaction with the cell membrane, leading to resistance to antibacterial agents including polymyxin B, LL-37, HNP-1, HBD-1, and HD-5 (Jones et al., 2009; Campos et al., 2004; Thomassin et al., 2013).
4.3. Modification or degradation of drugs
One general mechanism of resistance is inactivation of the conventional antibiotic or AMP (Nikaido, 2009; Fleitas et al., 2016). Aminoglycosides, such as kanamycin, amikacin, and tobramycin, are inactivated by phosphorylation, acetylation, or adenylation via plasmid- or chromosomeencoded enzymes; this enzymatic activity reduces the net positive charge of the aminoglycoside and leads to resistance in E. coli, Streptomyces fradiae, and other Gram-negative bacteria (Shaw et al., 1993; Bujnakova et al., 2014). Enzymes that hydrolyze β-lactam antibiotics contribute to resistance in Enterobacter, Serratia, and staphylococci (Reygaert, 2018; Vu and Nikaido, 1985).
Another common resistance mechanism is proteolytic degradation (Fig. 4A) (Nikaido, 2009; Fleitas et al., 2016). Some bacteria proteolytically degrade AMPs by expressing proteinases such as SpeB (Fleitas et al., 2016). The omptin family of aspartate proteases present in the outer membrane of enteric bacteria is the most thoroughly researched class of proteases in Gram-negative bacteria (Kukkonen and Korhonen, 2004). Metalloproteases also are involved in Gram-negative bacterial defense against AMPs (Kooi and Sokol, 2009). Some Gram-negative bacteria degrade AMPs, e.g., LL-37 and mCRAMP, inside the bacterial cell after transporting them with specific transporters (Parra-Lopez et al., 1993). Another bacterial resistance mechanism to AMPs involves neutralizing or binding to AMPs. Such a resistant mechanism can be achieved directly by interactions with the bacterial cell surface or released proteins or indirectly by AMP-binding molecules secreted from the host cell surface of pathogenic bacteria (Nizet, 2006).
4.4. Decreased permeability of bacterial cells to drugs
Resistant bacteria, particularly Gram-negative bacteria, can reduce the permeability of the cell outer membrane by changing porin channels located in the membrane; these changes make the membrane less permeable and so prevent the entry of hydrophilic drug molecules, such as tetracyclines, β-lactams, and fluroquinolones (Fig. 4A) (Munita and Arias, 2016; Hasdemir et al., 2004; Kiani et al., 2021). Mutations in certain porins, such as OmpK35, OmpK36 and OprD, have been found in K. pneumoniae, Enterobacter aerogenes, N. gonorrhoeae, and Pseudomonas isolates, conferring resistance to carbapenems, cephalosporins and imipenem (Kiani et al., 2021; Thiolas et al., 2004; Lee et al., 2018a,b; Hamzaoui et al., 2018). As fewer drug molecules enter the bacterial cell, the level of resistance to the drugs increases.
Sequestration, another mechanism of resistance, requires drug-binding proteins, which inhibit the antibiotic from approaching its target. In bacteria that produce the bleomycin (BLM) family of antibiotics, for example, the producing bacteria sequester the antibiotic by expressing the binding proteins BlmA, TlmA, and ZbmA (Sugiyama and Kumagai, 2002). Furthermore, some bacteria can circumvent an antibacterial response by downregulating AMP expression (Fleitas et al., 2016). Generally, most bacteria contain multiple cooperating mechanisms to enable response to the antibacterial agents, including antibiotics or active molecules that bacteria themselves produce (Peterson and Kaur, 2018).
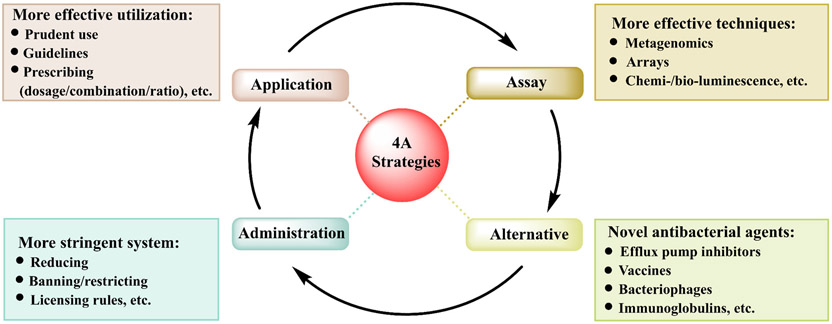
5. The 4A strategies to combat bacterial resistance
To counteract the increasing problem of bacterial drug resistance that then is transferred between humans, animals, and the environment, the following 4A strategies may be adopted based on the One Health concept (Fig. 5).
Fig. 5.
The 4A strategies to counter drug resistance. Effective strategies are urgently needed to combat bacterial resistance, including the prudent use of antibacterial agents, advanced molecular techniques, stringent administration and the need for novel alternatives to antibiotics, such as efflux pump inhibitors, vaccines, bacteriophages, immunoglobulins, AMPs, untapped herbal medicines, and probiotics (Kern and de With, 2012).
5.1. Application
First, we need to prudently manage the application of antibacterial agents under supervision, with a rational and appropriate drug prescribing approach to ultimately eradicate infection.
Extensive exposure to antibacterial agents can cause significant selective pressure favoring the occurrence and transfer of ARGs and ARB. It is critical to reduce drug consumption by prescribing a lower total amount of drugs [e.g., defined daily doses (DDDs)], thus reducing the likelihood of selecting for bacterial resistance. Identifying alternatives to the use of conventional antibiotics must also be pursued in agricultural applications. A classical five-year study focusing on MRSA and Clostridium difficile showed that a sustained reduction in the use of quinolones, cephalosporins, and penicillins lowered the incidence of MRSA infection (Cook et al., 2011). The prudent use of herbal medicines or antibiotics in both people and animals may slow down the evolution of resistance and prolong the useful lifetime of effective drugs (Yalew, 2020). It is expected that authorities such as the WHO, the World Organization for Animal Health (OIE), the Food and Drug Administration (FDA), the Food and Agricultural Organization (FAO) and the European Commission (EC) publish instructions and guidelines for the prudent use of antibacterial agents in humans and animals with the aim of reducing the use of these drugs to the lowest imperative level, ultimately minimizing the occurrence of bacterial resistance (Ungemach et al., 2006).
As stated by the Centers for Disease Control and Prevention (CDC), nearly 30 % of antibiotics prescribed by physicians are unnecessary. For example, antibiotics are incorrectly prescribed to treat viral infections, against which they are not effective. Regulating antibiotic prescription can help reduce the select pressure toward bacterial resistance and may consist of: determining the optimal ratio and dose of antibacterial agents to be used per treatment; starting at a low dose and increasing the dose slowly and progressively; using only one or two antibiotics from the same class; and taking a weekly “antibiotic holiday” or changing to another class of drugs (Allen and Brown, 2019; Lee et al., 2013a,b; Bhardwaj et al., 2016; Sun and Li, 2021). Formalized practical guidelines to be followed by formulary implementation should also be developed (Yalew, 2020; Lee et al., 2013a,b; Kern and de With, 2012).
Noticeably, synergistic combinations between two or more antibacterial agents, such as conventional antibiotics and AMPs, antibiotics and traditional herbal medicines, or AMPs and herbal medicines, could prove to be a more effective strategy against drug-resistant bacteria than using a single agent (Bhardwaj et al., 2016; Wu et al., 2020; Xu et al., 2018; Roque-Borda et al., 2021; Li et al., 2020a-c; Han et al., 2021).
5.2. Administration
It is essential to promote global law enforcement to prevent the illegal sale and use of drugs for humans and animals (Llor and Bjerrum, 2014; Alghadeer et al., 2018; Chen et al., 2018; Pokharel et al., 2020; Jacobs et al., 2019). Self-medication with drugs also contributes to the spread of resistance. Some antibacterial agents, particularly conventional antibiotics, are illegally sold and used without a prescription in Africa, Asia, and southern Europe (Llor and Bjerrum, 2014; Alghadeer et al., 2018). Approximately 70 % of the antibiotics produced in some developing countries (including Vietnam, China, India, etc.) and in the US are used in animals (Pokharel et al., 2020; Chen et al., 2018). Law enforcement prohibiting over-the-counter (OTC) sales of antibiotics has successfully reduced antibiotic sales in Brazil, Mexico, Chile, Venezuela, Colombia, Bosnia, Herzegovina, Azerbaijan, North Macedonia, and likely Vietnam and Thailand (Jacobs et al., 2019).
To decrease the antibiotic application in animals (Van et al., 2020), government officials may ban or restrict the use of certain antibiotics used for veterinary purposes (Li et al., 2020a-c; Fair and Tor, 2014; Abdallah et al., 2019; Lee et al., 2015). In 1986, Sweden was the first country in the world to prohibit adding conventional antibiotics to feed. Since then, over 128 countries have banned antibiotic use in animal feed, which previously used as growth promoters (Salim et al., 2018). In July 2020, China prohibited the use of growth-promoting antibiotics in feed, although herbal medicines are still allowed (Schoenmakers, 2020). National legislations and government action plans and policies can significantly reduce the overuse of antibiotics. Additionally, licensing rules for antibacterial drugs in the countries mentioned above should be made more strict and governments should impose heavy penalties when guidelines are not followed.
5.3. Assays and alternatives
Susceptibility testing of drugs can help determine effective drug dosage and may be used as a basis to formulate a profile of empirical therapy for the proper treatment of bacterial diseases. Advanced effective molecular techniques, including metagenomics, arrays, chemiluminescence and bioluminescence, should be further developed to rapidly identify ARB and ARGs, enabling the epidemiological surveillance and monitoring of bacterial outbreaks (Bai et al., 2022; Liang et al., 2021; Yadav and Kapley, 2021).
Novel strategies are also needed for the rational design and discovery of new classes of antibacterial agents. For example, nanotechnology can be applied to yield novel efflux pump inhibitors, helping to overcome bacterial drug resistance (Xu et al., 2019). Forward and reverse genetic approaches can be used to identify drug targets such as FrmB and GloB, which may help design targeted prodrugs to counter resistant bacteria (Miller et al., 2021). Furthermore, the clustered regularly interspaced short palindromic repeats (CRISPR)-Cas system has been exploited to develop smart antibacterial agents (Gholizadeh et al., 2020), and DNA-encoded chemical libraries are being used to effectively screen drugs (Wang et al., 2022a-c). Emerging advances in computational design are also helping to accelerate antibiotic design and discovery (Wan et al., 2022; Cesaro et al., 2022; Torres et al., 2022; Palmer et al., 2021; Torres et al., 2021; de la Fuente-Nunez, 2019; Torres and de la Fuente-Nunez, 2019; Stokes et al., 2020; Porto et al., 2018).
Additional alternative approaches to treat bacterial infections include vaccines, bacteriophages, immunoglobulins, new generation AMPs, traditional herbal medicines, and probiotics, which may help reduce the selection pressure for conventional antibiotic resistance and minimize drug resistance (Arqué et al., 2022; Torres et al., 2021; Silva et al., 2020; Wang et al., 2020b; Lee et al., 2013a,b; Micoli et al., 2021; Torres et al., 2022; Raj Singh et al., 2019).
Other strategies, including ongoing education, raising awareness of the threat of conventional antibiotic resistance, monitoring local resistance patterns, and consulting with physicians, pharmacists, and microbiologists, are also needed.
6. Conclusions and future perspectives
The widespread application of antibacterial agents has led to the spread of resistance in numerous bacteria. Genes endowing resistance to existing antibacterial agents can be easily transferred among bacteria. Here, we have reviewed the influence of drug-resistant bacteria on global health and the occurrence and mechanisms underlying bacterial resistance. We have also proposed the 4A strategies to control the “global resistome”. However, there are considerable gaps to fill in. Below, we list several areas that require further research to prevent a new pandemic, this time caused by drug-resistant bacteria:
Develop automated platforms to conduct phenotypic assays and use them in clinical spaces to rapidly identify resistant bacteria and evaluate their drug-resistance profiles.
Develop improved next-generation sequencing-based technologies (such as Nanopore and PacBio sequencing) and real-time computational methods to rapidly identify ARGs and to characterize a wide variety of MGEs that transfer antibiotic resistance.
Continuously monitor the global spread and distribution of ARGs and MDR bacteria, particularly the transmission dynamics of genes that contribute to resistance. It is critical to explore and track the genetic determinants that allow bacterial resistance to evolve and to understand the complex mechanisms involved in this evolution.
Pursue alternative assays, including the exploration of vaccines, efflux pump inhibitors, probiotics, bacteriophages, AMPs, and untapped herbal medicines, to cope with bacterial infections caused by resistant pathogens. Importantly, before the clinical application of a new antibacterial agent, it is crucial to assess their propensity to select for bacterial resistance. Of note, cross-resistance between antibacterial agents should be monitored carefully and synergistic interactions among different antibacterial molecules warrant further inquiry.
Study the influence of existing drug use on probiotics and other commensal bacteria, including the transfer and role of ARGs within the microbiota. More data are needed to assess the effects of resistance determinants on the microbiota.
Addressing these 4A strategies will enhance our ability to address the problems posed by bacterial resistance in the years to come.
HIGHLIGHTS.
The most important types of antibiotic resistant bacteria include ESKAPE species.
Bacteria develop resistance to antimicrobial peptides (AMPs) and herbal medicines.
Bacteria resist drugs by activated efflux pumps, modified drugs or targets, etc.
Bacterial resistance heavily threats global health, food security, and economic.
The 4A strategies may be adopted based on the One Health concept.
Acknowledgments
We want to thank all the scientists without their dedicated work this review would not have been possible. We also would like to thank the editor and the reviewers for their critical reading, thoughtful comments, and constructive suggestions. We also thank the National Natural Science Foundation of China (Grant No. 32072770) for their financial support. Cesar de la Fuente-Nunez holds a Presidential Professorship at the University of Pennsylvania, is a recipient of the Langer Prize by the AIChE Foundation and acknowledges funding from the IADR Innovation in Oral Care Award, the Procter & Gamble Company, United Therapeutics, a BBRF Young Investigator Grant, the Nemirovsky Prize, Penn Health-Tech Accelerator Award, the Dean’s Innovation Fund from the Perelman School of Medicine at the University of Pennsylvania, the National Institute of General Medical Sciences of the National Institutes of Health under award number R35GM138201, and the Defense Threat Reduction Agency (DTRA; HDTRA11810041 and HDTRA1-21-1-0014).
Footnotes
Declaration of competing interest
Cesar de la Fuente-Nunez provides consulting services to Invaio Sciences and is a member of the Scientific Advisory Boards of Nowture S.L. and Phare Bio. The de la Fuente Lab has received research funding or inkind donations from United Therapeutics, Strata Manufacturing PJSC, and Procter & Gamble, none of which were used in support of this work. All other authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this review.
CRediT authorship contributions statement
Ting Li wrote the manuscript and prepared the figures and tables. Zhenlong Wang investigated and wrote the manuscript. Jianhua Guo and Cesar de la Fuente-Nunez conceptualized and revised the manuscript. Jinquan Wang, Bin Han, Hui Tao and Jie Liu investigated and reviewed the manuscript. Xiumin Wang investigated, supervised and wrote the manuscript. All authors read and approved the final manuscript.
Data availability
No data was used for the research described in the article.
References
- Abdallah A, Zhang P, Zhong Q, et al. , 2019. Application of traditional Chinese herbal medicine by-products as dietary feed supplements and antibiotic replacements in animal production. Curr. Drug Metab 20, 54–64. [DOI] [PubMed] [Google Scholar]
- Abdi M, Mirkalantari S, Amirmozafari N, 2019. Bacterial resistance to antimicrobial peptides. J. Pept. Sci 25, e3210. [DOI] [PubMed] [Google Scholar]
- Abdullahi IN, Fernández-Fernández R, Juárez-Fernández G, et al. , 2021. Wild animals are reservoirs and sentinels of Staphylococcus aureus and MRSA clones: a problem with “One Health” concern. Antibiotics 10, 1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamczak A, Ożarowski M, Karpiński TM, 2019. Antibacterial activity of some flavonoids and organic acids widely distributed in plants. J. Clin. Med 9, 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aghapour Z, Gholizadeh P, Ganbarov K, et al. , 2019. Molecular mechanisms related to colistin resistance in Enterobacteriaceae. Infect. Drug Resist 12, 965–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aires J, Doucet-Populaire F, Butel MJ, 2007. Tetracycline resistance mediated by tet(W), tet(M), and tet(O) genes of Bifidobacterium isolates from humans. Appl. Environ. Microbiol 73, 2751–2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aires JR, Nikaido H, 2005. Aminoglycosides are captured from both periplasm and cytoplasm by the AcrD multidrug efflux transporter of Escherichia coli. J. Bacteriol 187, 1923–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alav I, Sutton JM, Rahman KM, 2018. Role of bacterial efflux pumps in biofilm formation. J. Antimicrob. Chemother 73, 2003–2020. [DOI] [PubMed] [Google Scholar]
- Alghadeer S, Aljuaydi K, Babelghaith S, et al. , 2018. Self-medication with antibiotics in Saudi Arabia. Saudi Pharm. J 26, 719–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alguel Y, Meng C, Teran W, et al. , 2007. Crystal structures of multidrug binding protein TtgR in complex with antibiotics and plant antimicrobials. J. Mol. Biol 369, 829–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen RC, Brown SP, 2019. Modified antibiotic adjuvant ratios can slow and steer the evolution of resistance: co-amoxiclav as a case study. MBio 10, e01831–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso A, Martínez JL, 2000. Cloning and characterization of SmeDEF, a novel multidrug efflux pump from Stenotrophomonas maltophilia. Antimicrob. Agents Chemother 44, 3079–3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso A, Martínez JL, 2001. Expression of multidrug efflux pump SmeDEF by clinical isolates of Stenotrophomonas maltophilia. Antimicrob. Agents Chemother 45, 1879–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Zahrani J, Al Dossari K, Gabr AH, et al. , 2019. Antimicrobial resistance patterns of uropathogens isolated from adult women with acute uncomplicated cystitis. BMC Microbiol. 19, 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson DI, 2003. Persistence of antibiotic resistant bacteria. Curr. Opin. Microbiol 6, 452–456. [DOI] [PubMed] [Google Scholar]
- Andersson DI, Hughes D, Kubicek-Sutherland JZ, 2016. Mechanisms and consequences of bacterial resistance to antimicrobial peptides. Drug Resist. Updat 26, 43–57. [DOI] [PubMed] [Google Scholar]
- Appelbaum PC, 2007. Reduced glycopeptide susceptibility in methicillin-resistant Staphylococcus aureus (MRSA). Int. J. Antimicrob. Agents 30, 398–408. [DOI] [PubMed] [Google Scholar]
- Argudín MA, Deplano A, Nonhoff C, et al. , 2021. Epidemiology of the Staphylococcus aureus CA-MRSA USA300 in Belgium. Eur. J. Clin. Microbiol. Infect. Dis 40, 2335–2347. [DOI] [PubMed] [Google Scholar]
- Arias CA, Torres HA, Singh KV, et al. , 2007. Failure of daptomycin monotherapy for endocarditis caused by an Enterococcus faecium strain with vancomycin-resistant and vancomycin-susceptible subpopulations and evidence of in vivo loss of the vanA gene cluster. Clin. Infect. Dis 45, 1343–1346. [DOI] [PubMed] [Google Scholar]
- Arias M, Hilchie AL, Haney EF, et al. , 2017. Anticancer activities of bovine and human lactoferricin-derived peptides. Biochem. Cell Biol 95, 91–98. [DOI] [PubMed] [Google Scholar]
- Arnett E, Seveau S, 2011. The multifaceted activities of mammalian defensins. Curr. Pharm. Des 17, 4254–4269. [DOI] [PubMed] [Google Scholar]
- Aron Z, Opperman TJ, 2018. The hydrophobic trap–the Achilles heel of RND efflux pumps. Res. Microbiol 169, 393–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arqué X, Torres MDT, Patiño T, et al. , 2022. Autonomous treatment of bacterial infections in vivo using antimicrobial micro- and nanomotors. ACS Nano 16, 7547–7558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslam B, Wang W, Arshad MI, et al. , 2018. Antibiotic resistance: a rundown of a global crisis. Infect. Drug Resist 11, 1645–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aygul A., 2015. The importance of efflux systems in antibiotic resistance and efflux pump inhibitors in the management of resistance. Mikrob. Bul 49, 278–291. [DOI] [PubMed] [Google Scholar]
- Bai Y, Ruan X, Li R, et al. , 2022. Metagenomics-based antibiotic resistance genes diversity and prevalence risk revealed by pathogenic bacterial host in Taihu Lake, China. Environ. Geochem. Health 44, 2531–2543. [DOI] [PubMed] [Google Scholar]
- Bakovic A, Risner K, Bhalla N, et al. , 2021. Brilacidin demonstrates inhibition of SARS-CoV-2 in cell culture. Viruses 13, 271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bals R, Wilson JM, 2003. Cathelicidins–a family of multifunctional antimicrobial peptides. Cell. Mol. Life Sci 60, 711–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banemann ADH, Gross R, 1998. The lipopolysaccharide of Bordetella bronchiseptica acts as a protective shield against antimicrobial peptides. Infect. Immun 66, 5607–5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bank W., 2017. Drug-resistant infections: a threat to our economic future. World Bank, Washington, DC. © World Bank. https://openknowledge.worldbank.org/handle/10986/26707 License:CCBY3.0IGO. [Google Scholar]
- Barber M, Rozwadowska-Dowzenko M, 1948. Infection by penicillin-resistant staphylococci. Lancet 2, 641–644. [DOI] [PubMed] [Google Scholar]
- Barbier F, Andremont A, Wolff M, et al. , 2013. Hospital-acquired pneumonia and ventilator-associated pneumonia: recent advances in epidemiology and management. Curr. Opin. Pulm. Med 19, 216–228. [DOI] [PubMed] [Google Scholar]
- Bassetti M, Echols R, Matsunaga Y, et al. , 2021. Efficacy and safety of cefiderocol or best available therapy for the treatment of serious infections caused by carbapenem-resistant Gram-negative bacteria (CREDIBLE-CR): a randomised, open-label, multicentre, pathogen-focused, descriptive, phase 3 trial. Lancet Infect. Dis 21, 226–240. [DOI] [PubMed] [Google Scholar]
- Batista Araujo J, Sastre de Souza G, Lorenzon EN, 2022. Indolicidin revisited: biological activity, potential applications and perspectives of an antimicrobial peptide not yet fully explored. World J. Microbiol. Biotechnol 38, 39. [DOI] [PubMed] [Google Scholar]
- Bhardwaj M, Singh BR, Sinha DK, et al. , 2016. Potential of herbal drug and antibiotic combination therapy: a new approach to treat multidrug resistant bacteria. Pharm. Anal. Acta 7, 1–4. [Google Scholar]
- Bina XR, Provenzano D, Nguyen N, et al. , 2008. Vibrio cholerae RND family efflux systems are required for antimicrobial resistance, optimal virulence factor production, and colonization of the infant mouse small intestine. Infect. Immun 76, 3595–3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittner Fialová S, Rendeková K, Mučaji P, et al. , 2021. Antibacterial activity of medicinalplants and their constituents in the context of skin and wound infections, considering European legislation and folk medicine-a review. Int. J. Mol. Sci 22, 10746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair JMA, Richmond GE, Piddock LJV, 2004. Multidrug efflux pumps in Gram-negative bacteria and their role in antibiotic resistance. Futur. Microbiol 9, 1165–1177. [DOI] [PubMed] [Google Scholar]
- Boparai JK, Sharma PK, 2020. Mini review on antimicrobial peptides, sources, mechanism and recent applications. Protein Pept. Lett 27, 4–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breidenstein EB, de la Fuente-Núñez C, Hancock REW, 2011. Pseudomonas aeruginosa: all roads lead to resistance. Trends Microbiol. 19, 419–426. [DOI] [PubMed] [Google Scholar]
- Bujnakova D, Strakova E, Kmet V, 2014. In vitro evaluation of the safety and probiotic properties of Lactobacilli isolated from chicken and calves. Anaerobe 29, 118–127. [DOI] [PubMed] [Google Scholar]
- Bunnell K, Duong Ringsred T., et al. , 2022. Aminopenicillins for treatment of ampicillin-resistant enterococcal urinary tract infections. Am. J. Health Syst. Pharm 79, 1056–1065. [DOI] [PubMed] [Google Scholar]
- Campos MA, Vargas MA, Regueiro V, et al. , 2004. Capsule polysaccharide mediates bacterial resistance to antimicrobial peptides. Infect. Immun 72, 7107–7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrique-Mas J, Van NTB, Cuong NV, et al. , 2019. Mortality, disease and associated antimicrobial use in commercial small-scale chicken flocks in the Mekong Delta of Vietnam. Prev. Vet. Med 165, 15–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrique-Mas JJ, Choisy M, Van Cuong N, et al. , 2020. An estimation of total antimicrobial usage in humans and animals in Vietnam. Antimicrob. Resist. Infect. Control 9, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecchini M, Langer J, Slawomirski L, 2015. Antimicrobial Resistance in G7 Countries and Beyond: Economic Issues, Policies and Options for Action. [Google Scholar]
- Cesaro A, Torres MDT, Gaglione R, et al. , 2022. Synthetic antibiotic derived from sequences encrypted in a protein from human plasma. ACS Nano 16, 1880–1895. [DOI] [PubMed] [Google Scholar]
- Chambers HF, Deleo FR, 2009. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat. Rev. Microbiol 7, 629–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang D, Sharma L, Dela Cruz CS, et al. , 2021. Clinical epidemiology, risk factors, and control strategies of Klebsiella pneumoniae infection. Front. Microbiol 12, 750662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Yu Z, Li Y, et al. , 2014. Effects of berberine in the gastrointestinal tract - a review of actions and therapeutic implications. Am. J. Chin. Med 42, 1053–1070. [DOI] [PubMed] [Google Scholar]
- Chen C, Tao C, Liu Z, et al. , 2015. A randomized clinical trial of berberine hydrochloride in patients with diarrhea-predominant irritable bowel syndrome. Phytother. Res 29, 1822–1827. [DOI] [PubMed] [Google Scholar]
- Chen X, Wu L, Xie X, 2018. Assessing the linkages between knowledge and use of veterinary antibiotics by pig farmers in rural China. Int. J. Environ. Res. Public Health 15, 1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang WC, Pamp SJ, Nilsson M, et al. , 2012. The metabolically active subpopulation in Pseudomonas aeruginosa biofilms survives exposure to membrane-targeting antimicrobials via distinct molecular mechanisms. FEMS Immunol. Med. Microbiol 65, 245–256. [DOI] [PubMed] [Google Scholar]
- Chouchani C, El Salabi A, Marrakchi R, et al. , 2012. First report of mefA and msrA/msrB multidrug efflux pumps associated with blaTEM-1 β-lactamase in Enterococcus faecalis. Int. J. Infect. Dis 16, e104–e109. [DOI] [PubMed] [Google Scholar]
- Chromek M, Slamova Z, Bergman P, et al. , 2006. The antimicrobial peptide cathelicidin protects the urinary tract against invasive bacterial infection. Nat. Med 12, 636–641. [DOI] [PubMed] [Google Scholar]
- Cong Y, Yang S, Rao X, 2020. Vancomycin resistant Staphylococcus aureus infections: a review of case updating and clinical features. J. Adv. Res 21, 169–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras G, Shirdel I, Braun MS, et al. , 2020. Defensins: transcriptional regulation and function beyond antimicrobial activity. Dev. Comp. Immunol 104, 103556. [DOI] [PubMed] [Google Scholar]
- Cook PP, Rizzo S, Gooch M, et al. , 2011. Sustained reduction in antimicrobial use and decrease in methicillin-resistant Staphylococcus aureus and Clostridium difficile infections following implementation of an electronic medical record at a tertiary-care teaching hospital. J. Antimicrob. Chemother 66, 205–209. [DOI] [PubMed] [Google Scholar]
- Cornejo-Juarez P, Cevallos MA, Castro-Jaimes S, et al. , 2020. High mortality in an outbreak of multidrug resistant Acinetobacter baumannii infection introduced to an oncological hospital by a patient transferred from a general hospital. PLoS One 15, e0234684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa-Martínez CL, Jurke A, Schmitz J, et al. , 2022. Molecular epidemiology of vancomycin-resistant enterococci bloodstream infections in Germany: a population-based prospective longitudinal study. Microorganisms 10, 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courvalin P, Carlier C, Collatz E, 1980. Plasmid-mediated resistance to aminocyclitol antibiotics in group D streptococci. J. Bacteriol 143, 541–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox G, Wright GD, 2013. Intrinsic antibiotic resistance: mechanisms, origins, challenges and solutions. Int. J. Med. Microbiol 303, 287–292. [DOI] [PubMed] [Google Scholar]
- Cullen TW, Schofield WB, Barry NA, et al. , 2015. Antimicrobial peptide resistance mediates resilience of prominent gut commensals during inflammation. Science 347, 170–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen TW, Trent MS, 2010. A link between the assembly of flagella and lipooligosaccharide of the Gram-negative bacterium Campylobacter jejuni. Proc. Natl. Acad. Sci. U. S. A 107, 5160–5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cushnie TPT, Cushnie B, Lamb AJ, 2014. Alkaloids: an overview of their antibacterial, antibiotic-enhancing and antivirulence activities. Int. J. Antimicrob. Agents 44, 377–386. [DOI] [PubMed] [Google Scholar]
- Dalebroux ZD, Matamouros S, Whittington D, et al. , 2014. PhoPQ regulates acidic glycerophospholipid content of the Salmonella Typhimurium outer membrane. Proc. Natl. Acad. Sci. U. S. A 111, 1868–1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daury L, Orange F, Taveau JC, et al. , 2016. Tripartite assembly of RND multidrug efflux pumps. Nat. Commun 7, 10731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies J, Davies D, 2010. Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev 74, 417–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Fuente-Nunez C., 2019. Toward autonomous antibiotic discovery. mSystems 4, e00151–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Fuente-Núñez C, Cardoso MH, de Souza Cândido E, et al. , 2016. Synthetic antibiofilm peptides. Biochim. Biophys. Acta 1858, 1061–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Fuente-Nunez C, Torres MD, Mojica FJ, et al. , 2017. Next-generation precision antimicrobials: towards personalized treatment of infectious diseases. Curr. Opin. Microbiol 37, 95–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Oliveira DMP, Forde BM, Kidd TJ, et al. , 2020. Antimicrobial resistance in ESKAPE pathogens. Clin. Microbiol. Rev 33, e00181–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deplano A, Denis O, Poirel L, et al. , 2005. Molecular characterization of an epidemic clone of panantibiotic-resistant Pseudomonas aeruginosa. J. Clin. Microbiol 43, 1198–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Der Torossian Torres M. de la Fuente-Nunez C, 2019. Reprogramming biological peptides to combat infectious diseases. Chem. Commun. (Camb.) 55, 15020–15032. [DOI] [PubMed] [Google Scholar]
- Desta K, Aklillu E, Gebrehiwot Y, et al. , 2022. High levels of methicillin-resistant Staphylococcus aureus carriage among healthcare workers at a teaching hospital in Addis Ababa Ethiopia: first evidence using mecA detection. Infect. Drug Resist 15, 3135–3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding JL, Li P, Ho B, 2008. The Sushi peptides: structural characterization and mode of action against Gram-negative bacteria. Cell. Mol. Life Sci 65, 1202–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duperthuy M., 2020. Antimicrobial peptides: virulence and resistance modulation in Gram-negative bacteria. Microorganisms 8, 280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ejaz H, Younas S, Abosalif KOA, et al. , 2021. Molecular analysis of blaSHV, blaTEM, and blaCTX-M in extended-spectrum beta-lactamase producing Enterobacteriaceae recovered from fecal specimens of animals. PLoS One 16, e0245126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekor M., 2014. The growing use of herbal medicines: issues relating to adverse reactions and challenges in monitoring safety. Front. Pharm 4, 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Shazely B, Yu G, Johnston PR, et al. , 2020. Resistance evolution against antimicrobial peptides in Staphylococcus aureus alters pharmacodynamics beyond the MIC. Front. Microbiol 11, 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elahi S, Buchanan RM, Attah-Poku S, et al. , 2006. The host defense peptide beta-defensin 1 confers protection against Bordetella pertussis in newborn piglets. Infect. Immun 74, 2338–2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst CM, Staubitz P, Mishra NN, et al. , 2009. The bacterial defensin resistance protein MprF consists of separable domains for lipid lysinylation and antimicrobial peptide repulsion. PLoS. Pathog 5, e1000660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fair RJ, Tor Y, 2014. Antibiotics and bacterial resistance in the 21st century. Perspect. Med. Chem 6, 25–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falagas ME, Rafailidis PI, 2007. Attributable mortality of Acinetobacter baumannii: no longer a controversial issue. Crit. Care 11, 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falord M, Karimova G, Hiron A, et al. , 2012. GraXSR proteins interact with the VraFG ABC transporter to form a five-component system required for cationic antimicrobial peptide sensing and resistance in Staphylococcus aureus. Antimicrob. Agents Chemother 56, 1047–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez RC, Weiss A, 1996. Susceptibilities of bordetella pertussis strains to antimicrobial peptides. Antimicrob. Agents Chemother 40, 1041–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields PI, Groisman EA, Heffron F, 1989. A Salmonella locus that controls resistance to microbicidal proteins from phagocytic cells. Science 243, 1059–1062. [DOI] [PubMed] [Google Scholar]
- Finken M, Kirschner P, Meier A, et al. , 1993. Molecular basis of streptomycin resistance in Mycobacterium tuberculosis: alterations of the ribosomal protein S12 gene and point mutations within a functional 16S ribosomal RNA pseudoknot. Mol. Microbiol 9, 1239–1246. [DOI] [PubMed] [Google Scholar]
- Fleitas O, Agbale CM, Franco OL, 2016. Bacterial resistance to antimicrobial peptides: an evolving phenomenon. Front. Biosci 21, 1013–1038. [DOI] [PubMed] [Google Scholar]
- Fluit AC, Schmitz FJ, Verhoef J, et al. , 2001. Multi-resistance to antimicrobial agents for the ten most frequently isolated bacterial pathogens. Int. J. Antimicrob. Agents 18, 147–160. [DOI] [PubMed] [Google Scholar]
- Freitas AR, Novais C, Duarte B, et al. , 2018. High rates of colonisation by ampicillin-resistant enterococci in residents of long-term care facilities in Porto, Portugal. Int. J. Antimicrob. Agents 51, 503–507. [DOI] [PubMed] [Google Scholar]
- Freitas AR, Tedim AP, Almeida-Santos AC, et al. , 2022. High-resolution genotyping unveils identical ampicillin-resistant Enterococcus faecium strains in different sources and countries: a One Health approach. Microorganisms 10, 632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Zhang F, Zhang W, et al. , 2007. Differential expression of blaSHV related to susceptibility to ampicillin in Klebsiella pneumoniae. Int. J. Antimicrob. Agents 29, 344–347. [DOI] [PubMed] [Google Scholar]
- Ganz T., 2003. Defensins: antimicrobial peptides of innate immunity. Nat. Rev. Immunol 3, 710–720. [DOI] [PubMed] [Google Scholar]
- Gellatly SL, Hancock RE, 2013. Pseudomonas aeruginosa: new insights into pathogenesis and host defenses. Pathog. Dis 67, 159–173. [DOI] [PubMed] [Google Scholar]
- Gharaibeh MH, Shatnawi SQ, 2019. An overview of colistin resistance, mobilized colistin resistance genes dissemination, global responses, and the alternatives to colistin: a review. Vet. World 12, 1735–1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gholizadeh P, Kose S, Dao S, et al. , 2020. How CRISPR-cas system could be used to combat antimicrobial resistance. Infect. Drug Resist 13, 1111–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillis RJ, White KG, Choi KH, et al. , 2005. Molecular basis of azithromycin-resistant Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother 49, 3858–3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser R, Becker K, von Eiff C, et al. , 2014. Decreased susceptibility of Staphylococcus aureus small-colony variants toward human antimicrobial peptides. J. Invest. Dermatol 134, 2347–2350. [DOI] [PubMed] [Google Scholar]
- Gogry FA, Siddiqui MT, Sultan I, et al. , 2021. Current update on intrinsic and acquired colistin resistance mechanisms in bacteria. Front. Med 8, 677720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gootz TD, Zaniewski R, Haskell S, et al. , 1996. Activity of the new fluoroquinolone trovafloxacin (CP-99,219) against DNA gyrase and topoisomerase IV mutants of S. pneumoniae selected in vitro. Antimicrob. Agents Chemother 40, 2691–2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham JP, Evans SL, Price LB, et al. , 2009. Fate of antimicrobial-resistant enterococci and staphylococci and resistance determinants in stored poultry litter. Environ. Res 109, 682–689. [DOI] [PubMed] [Google Scholar]
- Gramatniece A, Silamikelis I, Zahare I, et al. , 2019. Control of Acinetobacter baumannii outbreak in the neonatal intensive care unit in Latvia: whole-genome sequencing powered investigation and closure of the ward. Antimicrob. Resist. Infect. Control 8, 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groisman EA, Heffron F, Solomon F, 1992. Molecular genetic analysis of the Escherichia coli phoP locus. J. Bacteriol 174, 486–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenheid S, Le Moual H, 2012. Resistance to antimicrobial peptides in Gram-negative bacteria. FEMS Microbiol. Lett 330, 81–89. [DOI] [PubMed] [Google Scholar]
- Gueimonde M, Florez AB, van Hoek AH, et al. , 2010. Genetic basis of tetracycline resistance in Bifidobacterium animalis subsp. lactis. Appl. Environ. Microbiol 76, 3364–3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunn JS, Lim KB, Krueger J, et al. , 1998. PmrA-PmrB-regulated genes necessary for 4-aminoarabinose lipid A modification and polymyxin resistance. Mol. Microbiol 27, 1171–1182. [DOI] [PubMed] [Google Scholar]
- Guo H, Pan L, Li L, et al. , 2017. Characterization of antibiotic resistance genes from Lactobacillus isolated from traditional dairy products. J. Food. Sci 82, 724–730. [DOI] [PubMed] [Google Scholar]
- Habermann E., 1972. Bee and wasp venoms. Science 177, 314–322. [DOI] [PubMed] [Google Scholar]
- Habets MG, Brockhurst MA, 2012. Therapeutic antimicrobial peptides may compromise natural immunity. Biol. Lett 8, 416–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamzaoui Z, Ocampo-Sosa A, Martinez MF, et al. , 2018. Role of association of OmpK35 and OmpK36 alteration and blaESBL and/or blaAmpC genes in conferring carbapenem resistance among non-carbapenemase-producing Klebsiella pneumoniae. Int. J. Antimicrob. Agents 52, 898–905. [DOI] [PubMed] [Google Scholar]
- Han HH, Da Teng, Mao RY, et al. , 2021. Marine peptide-N6NH2 and its derivative-GUON6NH2 have potent antimicrobial activity against intracellular Edwardsiella tarda in vitro and in vivo. Front. Microbiol 12, 637427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock RE, Speert DP, 2000. Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and impact on treatment. Drug Resist. Updat 3, 247–255. [DOI] [PubMed] [Google Scholar]
- Hankins JV, Madsen JA, Giles DK, et al. , 2012. Amino acid addition to Vibrio cholerae LPS establishes a link between surface remodeling in Gram-positive and Gram-negative bacteria. Proc. Natl. Acad. Sci. U. S. A 109, 8722–8727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison F, Roberts AE, Gabrilska R, et al. , 2015. A 1,000-year-old antimicrobial remedy with antistaphylococcal activity. MBio 6, e01129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasdemir UO, Chevalier J, Nordmann P, et al. , 2004. Detection and prevalence of active drug efflux mechanism in various multidrug-resistant Klebsiella pneumoniae strains from Turkey. J. Clin. Microbiol 42, 2701–2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkey PM, 1986. Resistant bacteria in the normal human flora. J. Antimicrob. Chemoth 18, 133–139. [DOI] [PubMed] [Google Scholar]
- Hegedüs N, Marx F, 2013. Antifungal proteins: more than antimicrobials? Fungal Biol. Rev 26, 132–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenbeck BL, Rice LB, 2012. Intrinsic and acquired resistance mechanisms in Enterococcus. Virulence 3, 421–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper DC, 2000. Mechanisms of action and resistance of older and newer fluoroquinolones. Clin. Infect. Dis 31, S24–S28. [DOI] [PubMed] [Google Scholar]
- Hummel AS, Hertel C, Holzapfel WH, et al. , 2007. Antibiotic resistances of starter and probiotic strains of lactic acid bacteria. Appl. Environ. Microbiol 73, 730–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huycke MMSD, Gilmore MS, 1998. Multiple-drug resistant enterococci the nature of the problem and an agenda for the future. Emerg. Infect. Dis 4, 239–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ince D, Aras R, Hooper DC, 1999. Mechanisms and frequency of resistance to gatifloxacin in comparison with ciprofloxacin in Staphylococcus aureus. Drugs 58, 134–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isler B, Doi Y, Bonomo RA, et al. , 2019. New treatment options against carbapenem-resistant Acinetobacter baumannii infections. Antimicrob. Agents Chemother 63, e01110–e01118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs TG, Robertson J, van den Ham HA, et al. , 2019. Assessing the impact of law enforcement to reduce over-the-counter (OTC) sales of antibiotics in low- and middle-income countries; a systematic literature review. BMC Health Serv. Res 19, 536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaimee G, Halami PM, 2015. High level aminoglycoside resistance in Enterococcus, Pediococcus and Lactobacillus species from farm animals and commercial meat products. Annals Microbiol. 66, 101–110. [Google Scholar]
- Joly-Guillou ML, 2005. Clinical impact and pathogenicity of Acinetobacter. Clin. Microbiol. Infect 11, 868–873. [DOI] [PubMed] [Google Scholar]
- Jones A, Georg M, Maudsdotter L, et al. , 2009. Endotoxin, capsule, and bacterial attachment contribute to Neisseria meningitidis resistance to the human antimicrobial peptide LL-37. J. Bacteriol 191, 3861–3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo HS, Fu CI, Otto M, 2016. Bacterial strategies of resistance to antimicrobial peptides. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci 371, 20150292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jose NM, Bunt CR, Hussain MA, 2014. Implications of antibiotic resistance in probiotics. Food. Rev. Int 31, 52–62. [Google Scholar]
- Kabra R, Chauhan N, Kumar A, et al. , 2019. Efflux pumps and antimicrobial resistance: paradoxical components in systems genomics. Prog. Biophys. Mol. Biol 141, 15–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kai-Larsen Y, Luthje P, Chromek M, et al. , 2010. Uropathogenic Escherichia coli modulates immune responses and its curli fimbriae interact with the antimicrobial peptide LL-37. PLoS Pathog. 6, e1001010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandler JL, Holley CL, Reimche JL, et al. , 2016. The MisR response regulator is necessary for intrinsic cationic antimicrobial peptide and aminoglycoside resistance in Neisseria gonorrhoeae. Antimicrob. Agents Chemother 60, 4690–4700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamoto Y, Kaku N, Akamatsu N, et al. , 2022. The surveillance of colistin resistance and mobilized colistin resistance genes in multidrug-resistant Enterobacteriaceae isolated in Japan. Int. J. Antimicrob. Agents 59, 106480. [DOI] [PubMed] [Google Scholar]
- Kejela T, Dekosa F, 2022. High prevalence of MRSA and VRSA among inpatients of Mettu Karl Referral Hospital, Southwest Ethiopia. Trop. Med. Int. Health 27, 735–741. [DOI] [PubMed] [Google Scholar]
- Kern WV, de With K, 2012. Rational antibiotic prescribing. Challenges and successes. Bundesgesundheitsbl. Gesundheitsforsch. Gesundheitsschutz 55, 1418–1426. [DOI] [PubMed] [Google Scholar]
- Khan RIB, Akram M, Shakil S, et al. , 2009. Antimicrobial activity of five herbal extracts against multi drug resistant (MDR) strains of bacteria and fungus of clinical origin. Molecules 14, 586–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharazmi M, Hammes WP, Hertel C, 2002. Construction of a marker rescue system in Bacillus subtilis for detection of horizontal gene transfer in food. Syst. Appl. Microbiol 25, 471–477. [DOI] [PubMed] [Google Scholar]
- Khosravi AD, Etemad N, Hashemzadeh M, et al. , 2017. Frequency of rrs and rpsL mutations in streptomycin-resistant Mycobacterium tuberculosis isolates from Iranian patients. J. Glob. Antimicrob. Resist 9, 51–56. [DOI] [PubMed] [Google Scholar]
- Kiani M, Astani A, Eslami G, et al. , 2021. Upstream region of OprD mutations in imipenem-resistant and imipenem-sensitive Pseudomonas isolates. AMB Express 11, 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DW, Cha CJ, 2021. Antibiotic resistome from the One-Health perspective: understanding and controlling antimicrobial resistance transmission. Exp. Mol. Med 53, 301–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kintses B, Jangir PK, Fekete G, et al. , 2019. Chemical-genetic profiling reveals limited cross-resistance between antimicrobial peptides with different modes of action. Nat. Commun 10, 5731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kintses B, Mehi O, Ari E, et al. , 2019. Phylogenetic barriers to horizontal transfer of antimicrobial peptide resistance genes in the human gut microbiota. Nat. Microbiol 4, 447–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobeissi E, Menassa M, Moussally K, et al. , 2021. The socioeconomic burden of antibiotic resistance in conflict-affected settings and refugee hosting countries: a systematic scoping review. Confl Health 15, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler T, Michea-Hamzehpour M, Henze U, et al. , 1997. Characterization of iMexE-MexF-OprN, a positively regulated multidrug efflux system of Pseudomonas aeruginosa. Mol. Microbiol 23, 345–354. [DOI] [PubMed] [Google Scholar]
- Kooi C, Sokol PA, 2009. Burkholderia cenocepacia zinc metalloproteases influence resistance to antimicrobial peptides. Microbiology 155, 2818–2825. [DOI] [PubMed] [Google Scholar]
- Koprivnjak T, Peschel A, 2011. Bacterial resistance mechanisms against host defense peptides. Cell. Mol. Life Sci 68, 2243–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosalec I, Kremer D, Locatelli M, et al. , 2013. Anthraquinone profile, antioxidant and antimicrobial activity of bark extracts of Rhamnus alaternus, R. fallax, R. intermedia and R. pumila. Food Chem. 136, 335–341. [DOI] [PubMed] [Google Scholar]
- Kotwani A, Joshi J, Kaloni D, 2021. Pharmaceutical effluent: a critical link in the interconnected ecosystem promoting antimicrobial resistance. Environ. Sci. Pollut. Res. Int 28, 32111–32124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristian SA, Datta V, Weidenmaier C, et al. , 2005. D-alanylation of teichoic acids promotes Group A Streptococcus antimicrobial peptide resistance, neutrophil survival, and epithelial cell invasion. J. Bacteriol 187, 6719–6725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubicek-Sutherland JZ, Lofton H, Vestergaard M, et al. , 2017. Antimicrobial peptide exposure selects for Staphylococcus aureus resistance to human defence peptides. J. Antimicrob. Chemother 72, 115–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukkonen M, Korhonen TK, 2004. The omptin family of enterobacterial surface proteases/adhesins: from housekeeping in Escherichia coli to systemic spread of Yersinia pestis. Int. J. Med. Microbiol 294, 7–14. [DOI] [PubMed] [Google Scholar]
- Kumarasamy KK, Toleman MA, Walsh TR, et al. , 2010. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect. Dis 10, 597–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kummerer K., 2004. Resistance in the environment. J. Antimicrob. Chemother 54, 311–320. [DOI] [PubMed] [Google Scholar]
- Kurihara MNL, Sales RO, Silva KED, et al. , 2020. Multidrug-resistant Acinetobacter baumannii outbreaks: a global problem in healthcare settings. Rev. Soc. Bras. Med. Trop 53, e20200248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurpe SR, Grishin SY, Surin AK, et al. , 2020. Antimicrobial and amyloidogenic activity of peptides can antimicrobial peptides be used against SARS-Cov-2? Int. J. Mol. Sci 21, 9552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyriakidis I, Vasileiou E, Pana ZD, et al. , 2021. Acinetobacter baumannii antibiotic resistance mechanisms. Pathogens 10, 373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson DGJ, Flach CF, 2021. Antibiotic resistance in the environment. Nat. Rev. Microbiol 20, 257–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzaro BP, Zasloff M, Rolff J, 2020. Antimicrobial peptides: application informed by evolution. Science 368, eaau5480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclercq R, Derlot E, Courvalin P, 1988. Plasmid-mediated resistance to vancomycin and teicoplanin in Enterococcus faecium. N. Engl. J. Med 319, 157–161. [DOI] [PubMed] [Google Scholar]
- Lee CR, Cho IH, Jeong BC, et al. , 2013a. Strategies to minimize antibiotic resistance. Int. J. Environ. Res. Public Health 10, 4274–4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AS, de Lencastre H, Garau J, et al. , 2018a. Methicillin-resistant Staphylococcus aureus. Nat. Rev. Dis. Primers 4, 18033. [DOI] [PubMed] [Google Scholar]
- Lee J-J, Huang Y-C, Hsiao Y, et al. , 2018b. Insertion sequence-dependent ompK36 mutation associated ertapenem resistance in clinical Klebsiella pneumoniae. Adv. Microbiol 8, 253–269. [Google Scholar]
- Lee CR, Lee JH, Kang LW, et al. , 2015. Educational effectiveness, target, and content for prudent antibiotic use. Biomed. Res. Int 2015, 214021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee NH, Lee SM, Song DH, et al. , 2013b. Antimicrobial effect of emodin isolated from Cassia tora Linn. seeds against food-borne bacteria. J. Appl. Biol. Chem 56, 187–189. [Google Scholar]
- Levy SB, 1998. The challenge of antibiotic resistance. Sci. Am 278, 46–53. [DOI] [PubMed] [Google Scholar]
- Li Y, Cao W, Liang S, et al. , 2020c. Metagenomic characterization of bacterial community and antibiotic resistance genes in representative ready-to-eat food in southern China. Sci. Rep 10, 15175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Cha DJ, Lai Y, et al. , 2007. The antimicrobial peptide-sensing system aps of Staphylococcus aureus. Mol. Microbiol 66, 1136–1147. [DOI] [PubMed] [Google Scholar]
- Li XZ, Nikaido H, 2009. Efflux-mediated drug resistance in bacteria: an update. Drugs 69, 1555–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XZ, Nikaido H, Poole K, 1995. Role of MexA-MexB-OprM in antibiotic efflux in Pseudomonas aeruginosa. Antimicrob. Agents Chemother 39, 1948–1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ZZ, Teng D, Mao RY, et al. , 2018. Improved antibacterial activity of the marine peptide N6 against intracellular Salmonella Typhimurium by conjugating with the cell-penetrating peptide Tat11 via a cleavable linker. J. Med. Chem 61, 7991–8000. [DOI] [PubMed] [Google Scholar]
- Li T, Teng D, Mao R, et al. , 2020b. A critical review of antibiotic resistance in probiotic bacteria. Food Res. Int 136, 109571. [DOI] [PubMed] [Google Scholar]
- Li B, Yang N, Shan YX, et al. , 2020a. Therapeutic potential of a designed CSαβ peptide ID13 in Staphylococcus aureus-induced endometritis of mice. Appl. Microbiol. Biotechnol 104, 6693–6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H, Wang F, Mu R, et al. , 2021. Metagenomics analysis revealing the occurrence of antibiotic resistome in salt lakes. Sci. Total Environ 790, 148262. [DOI] [PubMed] [Google Scholar]
- Liu A, Tran L, Becket E, et al. , 2010. Antibiotic sensitivity profiles determined with an Escherichia coli gene knockout collection: generating an antibiotic bar code. Antimicrob. Agents. Chemother 54, 1393–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llobet E, Campos MA, Gimenez P, et al. , 2011. Analysis of the networks controlling the antimicrobial-peptide-dependent induction of Klebsiella pneumoniae virulence factors. Infect. Immun 79, 3718–3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llor C, Bjerrum L, 2014. Antimicrobial resistance: risk associated with antibiotic overuse and initiatives to reduce the problem. Ther. Adv. Drug Saf 5, 229–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lofton H, Pranting M, Thulin E, et al. , 2013. Mechanisms and fitness costs of resistance to antimicrobial peptides LL-37, CNY100HL and wheat germ histones. PLoS One 8, e68875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyczak JB, Cannon CL, Pier GB, 2002. Lung infections associated with cystic fibrosis. Clin. Microbiol. Rev 15, 194–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlane ELA, Kwasnicka A, Hancock REW, 2000. Role of Pseudomonas aeruginosa PhoP-PhoQ in resistance to antimicrobial cationic peptides and aminoglycosides. Microbiology 146, 2543–2554. [DOI] [PubMed] [Google Scholar]
- Magana M, Pushpanathan M, Santos AL, et al. , 2020. The value of antimicrobial peptides in the age of resistance. Lancet Infect. Dis 20, e216–e230. [DOI] [PubMed] [Google Scholar]
- Magiorakos AP, Srinivasan A, Carey RB, et al. , 2012. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect 18, 268–281. [DOI] [PubMed] [Google Scholar]
- Mahamoud A, Chevalier J, Alibert-Franco S, et al. , 2007. Antibiotic efflux pumps in Gram-negative bacteria: the inhibitor response strategy. J. Antimicrob. Chemother 59, 1223–1229. [DOI] [PubMed] [Google Scholar]
- Majaz AQ, Khurshid IM, 2016. Herbal medicine: a comprehensive review. Int. J. Pharm. Res 8, 1–5. [Google Scholar]
- Malik SZLM, Göransson U, Andersson DI, 2017. Resistance to the cyclotide cycloviolacin O2 in Salmonella enterica caused by different mutations that often confer cross-resistance or collateral sensitivity to other antimicrobial peptides. Antimicrob. Agents Chemother 61, e00684–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manso T, Marta L, de Miguel T, 2022. Antimicrobial activity of polyphenols and natural polyphenolic extracts on clinical isolates. Antibiotics 11, 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall SH, Donskey CJ, Hutton-Thomas R, et al. , 2002. Gene dosage and linezolid resistance in Enterococcus faecium and Enterococcus faecalis. Antimicrob. Agents Chemother 46, 3334–3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin KW, Ernst E, 2003. Herbal medicines for treatment of bacterial infections: a review of controlled clinical trials. J. Antimicrob. Chemother 51, 241–246. [DOI] [PubMed] [Google Scholar]
- Martin MJ, Thottathil SE, Newman TB, 2015. Antibiotics overuse in animal agriculture: a call to action for health care providers. Am. J. Public Health 105, 2409–2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateus C, Nunes AR, Oleastro M, et al. , 2021. RND efflux systems contribute to resistance and virulence of Aliarcobacter butzleri. Antibiotics 10, 823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méhi O, Bogos B, Csörgő B, et al. , 2014. Perturbation of iron homeostasis promotes the evolution of antibiotic resistance. Mol. Biol. Evol 31, 2793–2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo MCR, Maasch J, de la Fuente-Nunez C, 2021. Accelerating antibiotic discovery through artificial intelligence. Commun. Biol 4, 1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micoli F, Bagnoli F, Rappuoli R, et al. , 2021. The role of vaccines in combatting antimicrobial resistance. Nat. Rev. Microbiol 19, 287–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JJ, Shah IT, Hatten J, et al. , 2021. Structure-guided microbial targeting of antistaphylococcal prodrugs. eLife 10, e66657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller WR, Munita JM, Arias CA, 2014. Mechanisms of antibiotic resistance in enterococci. Expert Rev. Anti-Infect. Ther 12, 1221–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mine T, Morita Y, Kataoka A, et al. , 1999. Expression in Escherichia coli of a new multidrug efflux pump, MexXY, from Pseudomonas aeruginosa. Antimicrob. Agents Chemother 43, 415–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda CD, Godoy FA, Lee MR, 2018. Current status of the use of antibiotics and the antimicrobial resistance in the Chilean salmon farms. Front. Microbiol 9, 1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirghani R, Saba T, Khaliq H, et al. , 2022. Biofilms: formation, drug resistance and alternatives to conventional approaches. AIMS Microbiol. 8, 239–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohapatra DP, Debata NK, Singh SK, 2018. Extensively drug-resistant and pandrug-resistant Gram-negative bacteria in a tertiary-care hospital in Eastern India: a 4-year retrospective study. J. Glob. Antimicrob. Resist 15, 246–249. [DOI] [PubMed] [Google Scholar]
- Mookherjee N, Brown KL, Bowdish DM, et al. , 2006. Modulation of the TLR-mediated inflammatory response by the endogenous human host defense peptide LL-37. J. Immunol 176, 2455–2464. [DOI] [PubMed] [Google Scholar]
- Moradali MF, Ghods S, Rehm BH, 2017. Pseudomonas aeruginosa lifestyle: a paradigm for adaptation, survival, and persistence. Front. Cell. Infect. Microbiol 7, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan M., 2008. Methicillin-resistant Staphylococcus aureus and animals: zoonosis or humanosis? J. Antimicrob. Chemother 62, 1181–1187. [DOI] [PubMed] [Google Scholar]
- Moskowitz SM, Ernst RK, Miller SI, 2004. PmrAB, a two-component regulatory system of Pseudomonas aeruginosa that modulates resistance to cationic antimicrobial peptides and addition of aminoarabinose to lipid A. J. Bacteriol 186, 575–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount KL, Townsend CA, Bauer ME, 2007. Haemophilus ducreyi is resistant to human antimicrobial peptides. Antimicrob. Agents Chemother 51, 3391–3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount KL, Townsend CA, Rinker SD, et al. , 2010. Haemophilus ducreyi SapA contributes to cathelicidin resistance and virulence in humans. Infect. Immun 78, 1176–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munita JM, Arias CA, 2016. Mechanisms of antibiotic resistance. Microbiol. Spectr 4. 10.1128/microbiolspec.VMBF-0016-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray BE, Singh KV, Markowitz SM, et al. , 1991. Evidence for clonal spread of a single strain of beta-lactamase-producing Enterococcus (Streptococcus) faecalis to six hospitals in five states. J. Infect. Dis 163, 780–785. [DOI] [PubMed] [Google Scholar]
- Murray CJL, Ikuta KS, Sharara F, et al. , 2022. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 399, 629–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik MI, Fornda BA, Jaykumar E, et al. , 2010. Antibacterial activity of lemongrass (Cymbopogon citratus) oil against some selected pathogenic bacterias. Asian Pac. J. Trop. Med 3, 535–538. [Google Scholar]
- Napier BA, Burd EM, Satola SW, et al. , 2013. Clinical use of colistin induces cross-resistance to host antimicrobials in Acinetobacter baumannii. MBio 4, e00021–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navon-Venezia S, Kondratyeva K, Carattoli A, 2017. Klebsiella pneumoniae: a major world-wide source and shuttle for antibiotic resistance. FEMS Microbiol. Rev 41, 252–275. [DOI] [PubMed] [Google Scholar]
- Neuhaus FC, Baddiley J, 2003. A continuum of anionic charge: structures and functions of D-alanyl-teichoic acids in gram-positive bacteria. Microbiol. Mol. Biol. Rev 67, 686–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicas TI, Hancock RE, 1983. Pseudomonas aeruginosa outer membrane permeability: isolation of a porin protein F-deficient mutant. J. Bacteriol 153, 281–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicodemo AC, Paez JI, 2007. Antimicrobial therapy for Stenotrophomonas maltophilia infections. Eur. J. Clin. Microbiol. Infect. Dis 26, 229–237. [DOI] [PubMed] [Google Scholar]
- Nikaido H., 2009. Multidrug resistance in bacteria. Annu. Rev. Biochem 78, 119–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishanth MAD, Palanichamy P, 2020. Different classes of antimicrobial and their proneness to AMR. Vet Helpline E-magazine 8. http://www.vethelplineindia.co.in/antimicrobial-resistance/. [Google Scholar]
- Nishioka T, Ogawa W, Kuroda T, et al. , 2009. Gene cloning and characterization of EfmA, a multidrug efflux pump, from Enterococcus faecium. Biol. Pharm. Bull 32, 483–488. [DOI] [PubMed] [Google Scholar]
- Nizet V., 2006. Antimicrobial peptide resistance mechanisms of human bacterial pathogens. Curr. Issues Mol. Biol 8, 11–26. [PubMed] [Google Scholar]
- Nizet V, Ohtake T, Lauth X, et al. , 2001. Innate antimicrobial peptide protects the skin from invasive bacterial infection. Nature 414, 454–457. [DOI] [PubMed] [Google Scholar]
- Nuri R, Shprung T, Shai Y, 2015. Defensive remodeling: how bacterial surface properties and biofilm formation promote resistance to antimicrobial peptides. Biochim. Biophys. Acta 1848, 3089–3100. [DOI] [PubMed] [Google Scholar]
- Odongo EA, Mutai PC, Amugune BK, et al. , 2022. A systematic review of medicinal plants of Kenya used in the management of bacterial infections. Evid. Based Complement. Alternat. Med 2022, 9089360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orchard A, van Vuuren S, 2017. Commercial essential oils as potential antimicrobials to treat skin diseases. Evid. Based Complement. Alternat. Med 2017, 4517971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozkurt Z, Erol S, Kadanali A, et al. , 2005. Changes in antibiotic use, cost and consumption after an antibiotic restriction policy applied by infectious disease specialists. Jpn. J. Infect. Dis 58, 338–343. [PubMed] [Google Scholar]
- Padilla E, Llobet E, Domenech-Sanchez A, et al. , 2010. Klebsiella pneumoniae AcrAB efflux pump contributes to antimicrobial resistance and virulence. Antimicrob. Agents Chemother 54, 177–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer N, Maasch JRMA, Torres MDT, et al. , 2021. Molecular dynamics for antimicrobial peptide discovery. Infect. Immun 89, e00703–e00720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pamp SJ, Gjermansen M, Johansen HK, et al. , 2008. Tolerance to the antimicrobial peptide colistin in Pseudomonas aeruginosa biofilms is linked to metabolically active cells, and depends on the pmr and mexAB-oprM genes. Mol. Microbiol 68, 223–240. [DOI] [PubMed] [Google Scholar]
- Pang Z, Raudonis R, Glick BR, et al. , 2019. Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and alternative therapeutic strategies. Biotechnol. Adv 37, 177–192. [DOI] [PubMed] [Google Scholar]
- Parham S, Kharazi AZ, Bakhsheshi-Rad HR, et al. , 2020. Antioxidant, antimicrobial and antiviral properties of herbal materials. Antioxidants 9, 1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parlett RC, 1951. Studies of Induced Resistance of Certain Bacteria to Several Plant Extracts. The University of Arizona. [Google Scholar]
- Parra-Lopez C, Baer MT, Groisman EA, 1993. Molecular genetic analysis of a locus required for resistance to antimicrobial peptides in Salmonella typhimurium. EMBO J. 12, 4053–4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson DL, 2006. Resistance in Gram-negative bacteria: Enterobacteriaceae. Am. J. Infect. Control 34, S20–S28. [DOI] [PubMed] [Google Scholar]
- Paterson GK, Cone DB, Peters SE, et al. , 2009. The enzyme phosphoglucomutase (Pgm) is required by Salmonella enterica serovar Typhimurium for O-antigen production, resistance to antimicrobial peptides and in vivo fitness. Microbiology 155, 3403–3410. [DOI] [PubMed] [Google Scholar]
- Pelchovich G, Schreiber R, Zhuravlev A, et al. , 2013. The contribution of common rpsL mutations in Escherichia coli to sensitivity to ribosome targeting antibiotics. Int. J. Med. Microbiol 303, 558–562. [DOI] [PubMed] [Google Scholar]
- Pelegrin AC, Palmieri M, Mirande C, et al. , 2021. Pseudomonas aeruginosa: a clinical and genomics update. FEMS Microbiol. Rev 45, fuab026. [DOI] [PubMed] [Google Scholar]
- Pereira AV, de Barros G, Pinto EG, et al. , 2016. Melittin induces in vitro death of Leishmania (Leishmania) infantum by triggering the cellular innate immune response. J. Venom. Anim. Toxins. Incl. Trop. Dis 22, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira RS, Sumita TC, Furlan MR, et al. , 2004. Antibacterial activity of essential oils on microorganisms isolated from urinary tract infections. Rev. Saude Publ 38, 326–328. [DOI] [PubMed] [Google Scholar]
- Pérez-Cordero JJ, Lozano JM, Cortés J, et al. , 2011. Leishmanicidal activity of synthetic antimicrobial peptides in an infection model with human dendritic cells. Peptides 32, 683–690. [DOI] [PubMed] [Google Scholar]
- Perron GG, Zasloff M, Bell G, 2006. Experimental evolution of resistance to an antimicrobial peptide. Proc. Biol. Sci 273, 251–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschel A, Otto M, Jack RW, et al. , 1999. Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J. Biol. Chem 274, 8405–8410. [DOI] [PubMed] [Google Scholar]
- Perry JA, Westman EL, Wright GD., 2014. The antibiotic resistome: what’s new? Curr. Opin. Microbiol 21, 45–50. [DOI] [PubMed] [Google Scholar]
- Peschel A, Jack RW, Otto M, et al. , 2001. Staphylococcus aureus resistance to human defensins and evasion of neutrophil killing via the novel virulence factor MprF is based on modification of membrane lipids with L-lysine. J. Exp. Med 193, 1067–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson E, Kaur P, 2018. Antibiotic resistance mechanisms in bacteria: relationships between resistance determinants of antibiotic producers, environmental bacteria, and clinical pathogens. Front. Microbiol 9, 2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperaki ET, Syrogiannopoulos GA, Tzouvelekis LS, et al. , 2017. Klebsiella pneumoniae: virulence, biofilm and antimicrobial resistance. Pediatr. Infect. Dis. J 36, 1002–1005. [DOI] [PubMed] [Google Scholar]
- Pokharel S, Shrestha P, Adhikari B, 2020. Antimicrobial use in food animals and human health: time to implement 'One Health' approach. Antimicrob. Resist. Infect. Control 9, 181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole K., 2001. Multidrug efflux pumps and antimicrobial resistance in Pseudomonas aeruginosa and related organisms. J. Mol. Microbiol. Biotechnol 3, 255–264. [PubMed] [Google Scholar]
- Poole K, Gotoh N, Tsujimoto H, et al. , 1996. Overexpression of the mexC-mexD-oprJ efflux operon in nfxB-type multidrug-resistant strains of Pseudomonas aeruginosa. Mol. Microbiol 21, 224–713. [DOI] [PubMed] [Google Scholar]
- Portillo A, Ruiz-Larrea F, Zarazaga M, et al. , 2000. Macrolide resistance genes in Enterococcus spp. Antimicrob. Agents Chemother 44, 967–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porto WF, Irazazabal L, Alves ESF, et al. , 2018. In silico optimization of a guava antimicrobial peptide enables combinatorial exploration for peptide design. Nat. Commun 9, 1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poyart C, Pellegrini E, Marceau M, et al. , 2003. Attenuated virulence of Streptococcus agalactiae deficient in D-alanyl-lipoteichoic acid is due to an increased susceptibility to defensins and phagocytic cells. Mol. Microbiol 49, 1615–1625. [DOI] [PubMed] [Google Scholar]
- Pranting M, Andersson DI, 2010. Mechanisms and physiological effects of protamine resistance in Salmonella enterica serovar Typhimurium LT2. J. Antimicrob. Chemother 65, 876–887. [DOI] [PubMed] [Google Scholar]
- Pranting M, Negrea A, Rhen M, et al. , 2008. Mechanism and fitness costs of PR-39 resistance in Salmonella enterica serovar Typhimurium LT2. Antimicrob. Agents Chemother 52, 2734–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prestinaci F, Pezzotti P, Pantosti A, 2015. Antimicrobial resistance: a global multifaceted phenomenon. Pathog. Glob. Health 109, 309–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price LB, Johnson E, Vailes R, et al. , 2005. Fluoroquinolone-resistant Campylobacter isolates from conventional and antibiotic-free chicken products. Environ. Health Perspect 113, 557–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qadi M, Alhato S, Khayyat R, et al. , 2021. Colistin resistance among Enterobacteriaceae isolated from clinical samples in Gaza Strip. Can. J. Infect. Dis. Med. Microbiol 2021, 6634684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Que MY, Cao WW, Zhang H, et al. , 2022. The prevalence, antibiotic resistance and multilocus sequence typing of colistin-resistant bacteria isolated from Penaeus vannamei farms in earthen ponds and HDPE film-lined ponds in China. J. Fish Dis 45, 1289–1299. [DOI] [PubMed] [Google Scholar]
- Radimersky T, Frolkova P, Janoszowska D, et al. , 2010. Antibiotic resistance in faecal bacteria (Escherichia coli, Enterococcus spp.) in feral pigeons. J. Appl. Microbiol 109, 1687–1695. [DOI] [PubMed] [Google Scholar]
- Raj Singh B, Yadav A, Kumar Sinh D, et al. , 2019. Potential of herbal antibacterials as an alternative to antibiotics for multiple drug resistant bacteria: an analysis. Res. J. Vet. Sci 13, 1–8. [Google Scholar]
- Rammelkamp CH, Maxon T, 1942. Resistance of Staphylococcus aureus to the action of penicillin. Exp. Biol. Med 51, 386–389. [Google Scholar]
- Rana FR, Macias EA, Sultany CM, et al. , 1991. Interactions between magainin 2 and Salmonella typhimurium outer membranes: effect of lipopolysaccharide structure. Biochemistry 30, 5858–5866. [DOI] [PubMed] [Google Scholar]
- Regea G., 2018. Pharmacology & clinical research review on antibiotics resistance and its economic impacts. J. Pharmacol. Clin. Res 5, 555675. [Google Scholar]
- Remesh A, Gayathri AM, Singh R, et al. , 2013. The knowledge, attitude and the perception of prescribers on the rational use of antibiotics and the need for an antibiotic policy–a cross sectional survey in a tertiary care hospital. J. Clin. Diagn. Res 7, 675–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reygaert WC, 2018. An overview of the antimicrobial resistance mechanisms of bacteria. AIMS Microbiol. 4, 482–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieg S, Huth A, Kalbacher H, et al. , 2009. Resistance against antimicrobial peptides is independent of Escherichia coli AcrAB, Pseudomonas aeruginosa MexAB and Staphylococcus aureus NorA efflux pumps. Int. J. Antimicrob. Agents 33, 174–176. [DOI] [PubMed] [Google Scholar]
- Romero CD, Chopin SF, Buck G, et al. , 2005. Antibacterial properties of common herbal remedies of the southwest. J. Ethnopharmacol 99, 253–257. [DOI] [PubMed] [Google Scholar]
- Romero LC, de Souza da Cunha MLR, 2021. Insights into the epidemiology of community-associated methicillin-resistant Staphylococcus aureus in special populations and at the community-healthcare interface. Braz. J. Infect. Dis 25, 101636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roque-Borda CA, da Silva PB, Rodrigues MC, et al. , 2021. Challenge in the discovery of new drugs: antimicrobial peptides against WHO-list of critical and high-priority bacteria. Pharmaceutics 13, 773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein E, Keynan Y, 2013. Vancomycin-resistant enterococci. Crit. Care Clin 29, 841–852. [DOI] [PubMed] [Google Scholar]
- Russell AD, Hammond SA, Morgan JR, 1986. Bacterial resistance to antiseptics and disinfectants. J. Hosp. Infect 7, 213–225. [DOI] [PubMed] [Google Scholar]
- Saar-Dover R, Bitler A, Nezer R, et al. , 2012. D-alanylation of lipoteichoic acids confers resistance to cationic peptides in Group B Streptococcus by increasing the cell wall density. PLoS Pathog. 8, e1002891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadikot RT, Blackwell TS, Christman JW, et al. , 2005. Pathogen-host interactions in Pseudomonas aeruginosa pneumonia. Am. J. Respir. Crit. Care Med 171, 1209–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saguibo JD, Elegado FB, 2012. Resistance profile of probiotic lactic acid bacteria against inhibitory effects of selected plant extracts. Philipp. Agric. Sci 95, 22–32. [Google Scholar]
- Sakoulas G, Guram K, Reyes K, et al. , 2014. Human cathelicidin LL-37 resistance and increased daptomycin MIC in methicillin-resistant Staphylococcus aureus strain USA600 (ST45) are associated with increased mortality in a hospital setting. J. Clin. Microbiol 52, 2172–2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salim HM, Huque KS, Kamaruddin KM, et al. , 2018. Global restriction of using antibiotic growth promoters and alternative strategies in poultry production. Sci. Prog 101, 52–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samad MA, Sagor MS, Hossain MS, et al. , 2022. High prevalence of vancomycin non-susceptible and multi-drug resistant enterococci in farmed animals and fresh retail meats in Bangladesh. Vet. Res. Commun 46, 811–822. [DOI] [PubMed] [Google Scholar]
- Samuelsen O, Haukland HH, Kahl BC, et al. , 2005. Staphylococcus aureus small colony variants are resistant to the antimicrobial peptide lactoferricin B. J. Antimicrob. Chemother 56, 1126–1129. [DOI] [PubMed] [Google Scholar]
- Santajit S, Indrawattana N, 2016. Mechanisms of antimicrobial resistance in ESKAPE pathogens. Biomed. Res. Int 2016, 2475067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenmakers K., 2020. How China is getting its farmers to kick their antibiotics habit. Nature 586, S60–S62. [Google Scholar]
- Schrader-Fischer G, Berger-Bachi B, 2001. The AbcA transporter of Staphylococcus aureus affects cell autolysis. Antimicrob. Agents Chemother 45, 407–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert H, Baginski R, Schulze A, et al. , 1993. Antimicrobial susceptibility of Acinetobacter species. Antimicrob. Agents Chemother 37, 750–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellera FP, Fernandes MR, Moura Q, et al. , 2018. Draft genome sequence of an extensively drug-resistant Pseudomonas aeruginosa isolate belonging to ST644 isolated from a footpad infection in a Magellanic penguin (Spheniscus magellanicus). J. Glob. Antimicrob. Resist 12, 88–89. [DOI] [PubMed] [Google Scholar]
- Shafer WM, Qu X, Waring AJ, et al. , 1998. Modulation of neisseria gonorrhoeae susceptibility to vertebrate antibacterial peptides due to a member of the resistance/nodulation/division efflux pump family. Proc. Natl. Acad. Sci. U. S. A 95, 1829–1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah AA, Hasan F, Ahmed S, et al. , 2004. Extended-spectrum β-lactamases (ESbLs): characterization, epidemiology and detection. Crit. Rev. Microbiol 30, 25–32. [DOI] [PubMed] [Google Scholar]
- Shah NR, Hancock RE, Fernandez RC, 2014. Bordetella pertussis lipid a glucosamine modification confers resistance to cationic antimicrobial peptides and increases resistance to outer membrane perturbation. Antimicrob. Agents Chemother 58, 4931–4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shariati A, Dadashi M, Moghadam MT, et al. , 2020. Global prevalence and distribution of vancomycin resistant, vancomycin intermediate and heterogeneously vancomycin intermediate Staphylococcus aureus clinical isolates: a systematic review and meta-analysis. Sci. Rep 10, 12689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma P, Tomar SK, Goswami P, et al. , 2014. Antibiotic resistance among commercially available probiotics. Food Res. Int 57, 176–195. [Google Scholar]
- Shaw KJ, Rather PN, Hare RS, et al. , 1993. Molecular genetics of aminoglycoside resistance genes and familial relationships of the aminoglycoside-modifying enzymes. Microbiol. Rev 57, 138–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelburne CE, Coulter WA, Olguin D, et al. , 2005. Induction of β-defensin resistance in the oral anaerobe Porphyromonas gingivalis. Antimicrob. Agents Chemother 49, 183–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieniawska E, Swatko-Ossor M, Sawicki R, et al. , 2017. Natural terpenes influence the activity of antibiotics against isolated Mycobacterium tuberculosis. Med. Princ. Pract 26, 108–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silberstein S., 1924. Zur Frage der salvarsanresistenten Lues. Arch. Dermatol. Syph 147, 116–130. [Google Scholar]
- Silva ON, Torres MDT, Cao J, et al. , 2020. Repurposing a peptide toxin from wasp venom into antiinfectives with dual antimicrobial and immunomodulatory properties. Proc. Natl. Acad. Sci. U. S. A 117, 26936–26945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh BR, 2013. Evaluation of antibacterial activity of Salvia officinalis L. sage oil on veterinary clinical isolates of bacteria. Medicine 11, 27–30.23379695 [Google Scholar]
- Singh BR, Singh V, Singh RK, et al. , 2011. Antimicrobial activity of lemongrass (Cymbopogon citratus) oil against microbes of environmental, clinical and food origin. Int. Res. Pharm. Pharmacol 9, 228–236. [Google Scholar]
- Singh BR, Singh V, Singh RK, et al. , 2012. Comparative evaluation of antimicrobial effect of Artemisia vulgaris essential oils extracted from fresh and dried herb. Med. Plants 4, 76–82. [Google Scholar]
- Singh BR, Singh V, Ebibeni N, et al. , 2013. Antimicrobial and herbal drug resistance in enteric bacteria isolated from faecal droppings of common house lizard/gecko (Hemidactylus frenatus). Int. J. Microbiol 2013, 340848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith TC, Gebreyes WA, Abley MJ, et al. , 2013. Methicillin-resistant Staphylococcus aureus in pigs and farm workers on conventional and antibiotic-free swine farms in the USA. PLoS One 8, e63704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soković M, Glamočlija J, Marin PD, et al. , 2010. Antibacterial effects of the essential oils of commonly consumed medicinal herbs using an in vitro model. Molecules 15, 7532–7546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer JJ, Pitts RE, Pearson RA, et al. , 2018. The effects of antimicrobial peptides WAM-1 and LL-37 on multidrug-resistant Acinetobacter baumannii. Pathog. Dis 76. 10.1093/femspd/fty007. [DOI] [PubMed] [Google Scholar]
- Srinivasan VB, Rajamohan G, 2013. KpnEF, a new member of the Klebsiella pneumoniae cell envelope stress response regulon, is an SMR-type efflux pump involved in broad-spectrum antimicrobial resistance. Antimicrob. Agents Chemother 57, 4449–4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan VB, Singh BB, Priyadarshi N, et al. , 2014. Role of novel multidrug efflux pump involved in drug resistance in Klebsiella pneumoniae. PLoS One 9, e96288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stekel D., 2018. First report of antimicrobial resistance pre-dates penicillin. Nature 562, 192. [DOI] [PubMed] [Google Scholar]
- Stock I, Grueger T, Wiedemann B, 2003. Natural antibiotic susceptibility of strains of Serratia marcescens and the S. liquefaciens complex: S. liquefaciens sensu stricto, S. proteamaculans and S. grimesii. Int. J. Antimicrob. Agents 22, 35–47. [DOI] [PubMed] [Google Scholar]
- Stokes JM, Yang K, Swanson K, et al. , 2020. A deep learning approach to antibiotic discovery. Cell 180, 688–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su W, Kumar V, Ding Y, et al. , 2018. Ribosome protection by antibiotic resistance ATP-binding cassette protein. Proc. Natl. Acad. Sci. U. S. A 115, 5157–5162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su XZ, Chen J, Mizushima T, et al. , 2005. AbeM, an H+-coupled Acinetobacter baumannii multidrug efflux pump belonging to the MATE family of transporters. Antimicrob. Agents Chemother 49, 4362–4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama M, Kumagai T, 2002. Molecular and structural biology of bleomycin and its resistance determinants. J. Biosci. Bioeng 93, 105–116. [DOI] [PubMed] [Google Scholar]
- Sun J, Li Y, 2021. Long-term, low-dose macrolide antibiotic treatment in pediatric chronic airway diseases. Pediatr. Res 91, 1036–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagousop CN, Tamokou J, Ekom SE, et al. , 2018. Antimicrobial activities of flavonoid glycosides from Graptophyllum grandulosum and their mechanism of antibacterial action. BMC Complement. Altern. Med 18, 252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot GH, Bradley J, Edwards JE, et al. , 2006. Bad bugs need drugs: an update on the development pipeline from the antimicrobial availability task force of the infectious diseases society of America. Clin. Infect. Dis 42, 657–668. [DOI] [PubMed] [Google Scholar]
- Tamayo R, Choudhury B, Septer A, et al. , 2005. Identification of cptA, a PmrA-regulated locus required for phosphoethanolamine modification of the Salmonella enterica serovar Typhimurium lipopolysaccharide core. J. Bacteriol 187, 3391–3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taneja NK, Ganguly T, Bakaletz LO, et al. , 2013. D-alanine modification of a protease-susceptible outer membrane component by the Bordetella pertussis dra locus promotes resistance to antimicrobial peptides and polymorphonuclear leukocyte-mediated killing. J. Bacteriol 195, 5102–5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J, Hafner M, Yerushalmi E, et al. , 2014. Estimating the economic costs of antimicrobial resistance: model and results RAND Corporation. (Research Reports). [Google Scholar]
- Teneva-angelova T, Beshkova D, 2015. Resistance profile of plant-derived lactic acid bacteria against herb extracts. Sci. Bull. Ser. F Biotechnol 19, 109–116. [Google Scholar]
- Teran W, Krell T, Ramos JL, et al. , 2006. Effector-repressor interactions, binding of a single effector molecule to the operator-bound TtgR homodimer mediates derepression. J. Biol. Chem 281, 7102–7109. [DOI] [PubMed] [Google Scholar]
- Thaipisuttikul I, Hittle LE, Chandra R, et al. , 2014. A divergent Pseudomonas aeruginosa palmitoyltransferase essential for cystic fibrosis-specific lipid A. Mol. Microbiol 91, 158–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thedieck K, Hain T, Mohamed W, et al. , 2006. The MprF protein is required for lysinylation of phospholipids in Listerial membranes and confers resistance to cationic antimicrobial peptides (CAMPs) on Listeria monocytogenes. Mol. Microbiol 62, 1325–1339. [DOI] [PubMed] [Google Scholar]
- Thiolas A, Bornet C, Davin-Regli A, et al. , 2004. Resistance to imipenem, cefepime, and cefpirome associated with mutation in Omp36 osmoporin of Enterobacter aerogenes. Biochem. Biophys. Res. Commun 317, 851–856. [DOI] [PubMed] [Google Scholar]
- Thomassin JL, Lee MJ, Brannon JR, et al. , 2013. Both group 4 capsule and lipopolysaccharide O-antigen contribute to enteropathogenic Escherichia coli resistance to human α-defensin 5. PLoS One 8, e82475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian T, Wu XG, Duan HM, et al. , 2010. The resistance-nodulation-division efflux pump EmhABC influences the production of 2,4-diacetylphloroglucinol in Pseudomonas fluorescens 2P24. Microbiology 156, 39–48. [DOI] [PubMed] [Google Scholar]
- Toit AD, 2017. Bacterial physiology: efflux pumps, fitness and virulence. Nat. Rev. Microbiol 15, 512–513. [DOI] [PubMed] [Google Scholar]
- Toomey N, Monaghan A, Fanning S, et al. , 2009. Assessment of antimicrobial resistance transfer between lactic acid bacteria and potential foodborne pathogens using in vitro methods and mating in a food matrix. Foodborne Pathog. Dis 6, 925–933. [DOI] [PubMed] [Google Scholar]
- Torres MDT, Pedron CN, Higashikuni Y, et al. , 2018. Structure-function-guided exploration of the antimicrobial peptide polybia-CP identifies activity determinants and generates synthetic therapeutic candidates. Commun. Biol 1, 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres MDT, Sothiselvam S, Lu TK, et al. , 2019. Peptide design principles for antimicrobial applications. J. Mol. Biol 431, 3547–3567. [DOI] [PubMed] [Google Scholar]
- Torres MDT, Cao J, Franco OL, et al. , 2021. Synthetic biology and computer-based frameworks for antimicrobial peptide discovery. ACS Nano 15, 2143–2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres MDT, Melo MCR, Crescenzi O, et al. , 2022. Mining for encrypted peptide antibiotics in the human proteome. Nat. Biomed. Eng 6, 67–75. [DOI] [PubMed] [Google Scholar]
- Torres MT, de la Fuente-Nunez C, 2019. Toward computer-made artificial antibiotics. Curr. Opin. Microbiol 51, 30–38. [DOI] [PubMed] [Google Scholar]
- Trombley MP, Post DM, Rinker SD, et al. , 2015. Phosphoethanolamine transferase LptA in Haemophilus ducreyi modifies lipid A and contributes to human defensin resistance in vitro. PLoS One 10, e0124373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzeng YL, Ambrose KD, Zughaier S, et al. , 2005. Cationic antimicrobial peptide resistance in Neisseria meningitidis. J. Bacteriol 187, 5387–5396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin TM, Chakraborty AJ, Khusro A, et al. , 2021. Antibiotic resistance in microbes: history, mechanisms, therapeutic strategies and future prospects. J. Infect. Public Health 14, 1750–1766. [DOI] [PubMed] [Google Scholar]
- Ungemach FR, Muller-Bahrdt D, Abraham G, 2006. Guidelines for prudent use of antimicrobials and their implications on antibiotic usage in veterinary medicine. Int. J. Med. Microbiol 296, 33–38. [DOI] [PubMed] [Google Scholar]
- Van Boeckel TP, Brower C, Gilbert M, et al. , 2015. Global trends in antimicrobial use in food animals. Proc. Natl. Acad. Sci. U. S. A 112, 5649–5654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Duin D, Paterson DL, 2016. Multidrug-resistant bacteria in the community: trends and lessons learned. Infect. Dis. Clin. N. Am 30, 377–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van TTH, Yidana Z, Smooker PM, et al. , 2020. Antibiotic use in food animals worldwide, with a focus on Africa: pluses and minuses. J. Glob. Antimicrob. Resist 20, 170–177. [DOI] [PubMed] [Google Scholar]
- Ventola CL, 2015. The antibiotic resistance crisis: part 1: causes and threats. P T 40, 277–283. [PMC free article] [PubMed] [Google Scholar]
- Verraes C, Van Boxstael S, Van Meervenne E, et al. , 2013. Antimicrobial resistance in the food chain: a review. Int. J. Environ. Res. Public Health 10, 2643–2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vila JMA, Marco F, Abdalla S, et al. , 1993. In vitro antimicrobial production of β-lactamases, aminoglycoside-modifying enzymes, and chloramphenicol acetyltransferase by and susceptibility of clinical isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother 37, 138–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrancianu CO, Gheorghe I, Czobor IB, et al. , 2020. Antibiotic resistance profiles, molecular mechanisms and innovative treatment strategies of Acinetobacter baumannii. Microorganisms 8, 935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu H, Nikaido H, 1985. Role of beta-lactam hydrolysis in the mechanism of resistance of a beta-lactamase-constitutive Enterobacter cloacae strain to expanded-spectrum β-lactams. Antimicrob. Agents Chemother 27, 393–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan F, Kontogiorgos-Heintz D, de la Fuente-Nunez C, 2022. Deep generative models for peptide design. Digit Discov. 1, 195–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Huang L, Chen S, 2013. Senecio scandens Buch.-Ham: a review on its ethnopharmacology, phytochemistry, pharmacology, and toxicity. J. Ethnopharmacol 149, 1–23. [DOI] [PubMed] [Google Scholar]
- Wang N, Hang X, Zhang M, et al. , 2017a. Analysis of newly detected tetracycline resistance genes and their flanking sequences in human intestinal bifidobacteria. Sci. Rep 7, 6267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XM, Teng D, Mao RY, et al. , 2017b. Combined systems approaches reveal a multi-stage mode of action of a marine antimicrobial peptide against pathogenic Escherichia coli and its protective effect against bacterial peritonitis and endotoxemia. Antimicrob. Agents. Chemother 61, e01056–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Zhao G, Chao X, et al. , 2020a. The characteristic of virulence, biofilm and antibiotic resistance of Klebsiella pneumoniae. Int. J. Environ. Res. Public Health 17, 6278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZL, Liu XH, Teng D, et al. , 2020b. Development of chimeric peptides to facilitate the neutralisation of lipopolysaccharides during bactericidal targeting of multidrug-resistant Escherichia coli. Commun. Biol 3, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Earley M, Chen L, et al. , 2022a. Clinical outcomes and bacterial characteristics of carbapenem-resistant Klebsiella pneumoniae complex among patients from different global regions (CRACKLE-2): a prospective, multicentre, cohort study. Lancet Infect. Dis 22, 401–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Koirala B, Hernandez Y, et al. , 2022b. A naturally inspired antibiotic to target multidrug-resistant pathogens. Nature. 601, 606–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Yang N, Teng D, et al. , 2022c. Resistance response to arenicin derivatives in Escherichia coli. Appl. Microbiol. Biotechnol 106, 211–226. [DOI] [PubMed] [Google Scholar]
- Warner DM, Shafer WM, Jerse AE, 2008. Clinically relevant mutations that cause derepression of the Neisseria gonorrhoeae MtrC-MtrD-MtrE efflux pump system confer different levels of antimicrobial resistance and in vivo fitness. Mol. Microbiol 70, 462–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisblum B., 1995. Erythromycin resistance by ribosome modification. Antimicrob. Agents Chemother 39, 577–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner GSB, Witte W, 2008. Acquired vancomycin resistance in clinically relevant pathogens. Futur. Microbiol 3, 547–562. [DOI] [PubMed] [Google Scholar]
- WHO, 2019. New Report Calls for Urgent Action to Avert Antimicrobial Resistance Crisis. [Google Scholar]
- World Health Organization (WHO), 2021. Global Antimicrobial Resistance and Use Surveillance System (GLASS) Report. World Health Organization. [Google Scholar]
- Wozniak TM, Barnsbee L, Lee XJ, et al. , 2019. Using the best available data to estimate the cost of antimicrobial resistance: a systematic review. Antimicrob. Resist. Infect. Control 8, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CL, Hsueh JY, Yip BS, et al. , 2020. Antimicrobial peptides display strong synergy with vancomycin against vancomycin-resistant E. faecium, S. aureus, and wild-type E. coli. Int. J. Mol. Sci 21, 4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JH, Zhou X, Chen QQ, et al. , 2022. Defensins as a promising class of tick antimicrobial peptides: a scoping review. Infect. Dis. Poverty 11, 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Xu L, Yuan G, et al. , 2018. Synergistic combination of two antimicrobial agents closing each other's mutant selection windows to prevent antimicrobial resistance. Sci. Rep 8, 7237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu XL, Kang XQ, Qi J, et al. , 2019. Novel antibacterial strategies for combating bacterial multidrug resistance. Curr. Pharm. Des 25, 4717–4724. [DOI] [PubMed] [Google Scholar]
- Yadav S, Kapley A, 2021. Antibiotic resistance: global health crisis and metagenomics. Biotechnol. Rep 29, e00604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalew ST, 2020. Review on antibiotic resistance: resistance mechanisms, methods of detection and its controlling strategies. Bio. J. Sci. Tech. Res 24, 18651–18657. [Google Scholar]
- Yan D, Chen D, Shen J, et al. , 2013. Bovine lactoferricin is anti-inflammatory and anti-catabolic in human articular cartilage and synovium. J. Cell. Physiol 22, 447–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Xie Y, Guo M, et al. , 2018. Nephrotoxicity and Chinese herbal medicine. Clin. J. Am. Soc. Nephrol 13, 1605–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Shen Y, Jiang J, et al. , 2022. Distinct increase in antimicrobial resistance genes among Escherichia coli during 50 years of antimicrobial use in livestock production in China. Nat. Food 3, 197–205. [DOI] [PubMed] [Google Scholar]
- Yong D, Toleman MA, Giske CG, et al. , 2009. Characterization of a new metallo-beta-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother 53, 5046–5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu SS, Zhao ZH, Gong XF, et al. , 2022. Antimicrobial and immunomodulatory activity of beta-defensin from the Chinese spiny frog (Quasipaa spinosa). Dev. Comp. Immunol 126, 104264. [DOI] [PubMed] [Google Scholar]
- Zahner D, Zhou X, Chancey ST, et al. , 2010. Human antimicrobial peptide LL-37 induces MefE/Mel-mediated macrolide resistance in Streptococcus pneumoniae. Antimicrob. Agents Chemother 54, 3516–3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarfel G, Galler H, Luxner J, et al. , 2014. Multiresistant bacteria isolated from chicken meat in Austria. Int. J. Environ. Res. Public Health 11, 12582–12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zasloff M., 2002. Antimicrobial peptides of multicellular organisms. Nature 415, 389–395. [DOI] [PubMed] [Google Scholar]
- Zenz KI, Neve H, Geis A, et al. , 1998. Bacillus subtilis develops competence for uptake of plasmid DNA when growing in milk products. Syst. Appl. Microbio 21, 28–32. [DOI] [PubMed] [Google Scholar]
- Zervos MJ, Schaberg DR, 1985. Reversal of the in vitro susceptibility of enterococci to trimethoprim-sulfamethoxazole by folinic acid. Antimicrob. Agents Chemother 28, 446–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zgurskaya HI, Nikaido H, 1999. Bypassing the periplasm: reconstitution of the AcrAB multidrug efflux pump of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A 96, 7190–7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang WX, Chen HY, Chen C, et al. , 2021. Resistance phenotype and molecular epidemiology of carbapenem-resistant Klebsiella pneumoniae isolates in Shanghai. Microb. Drug Resist 27, 1312–1318. [DOI] [PubMed] [Google Scholar]
- Zhen X, Lundborg CS, Sun X, et al. , 2019. Economic burden of antibiotic resistance in ESKAPE organisms: a systematic review. Antimicrob. Resist. Infect. Control 8, 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen XM, Lundborg CS, Sun XS, et al. , 2021. Economic burden of antibiotic resistance in China: a national level estimate for inpatients. Antimicrob. Resist. Infect. Control 10, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Yao Y, Zhu B, et al. , 2019. Risk factors for acquisition and mortality of multidrug-resistant Acinetobacter baumannii bacteremia: a retrospective study from a chinese hospital. Medicine 98, e14937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Zhou H, Sun Y, et al. , 2020. Characterization of clinical enterococci isolates, focusing on the vancomycin-resistant enterococci in a tertiary hospital in China: based on the data from 2013 to 2018. BMC Infect. Dis 20, 356. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.