Abstract
A primary aim of computational psychiatry is to establish predictive models linking individual differences in brain functioning with symptoms. In particular, cognitive impairments are transdiagnostic, treatment resistant, and associated with poor outcomes. Recent work suggests that thousands of participants may be necessary for the accurate and reliable prediction of cognition, questioning the utility of most patient collection efforts. Here, using a transfer learning framework, we train a model on functional neuroimaging data from the UK Biobank to predict cognitive functioning in three transdiagnostic samples (ns = 101 to 224). We demonstrate prediction performance in all three samples comparable to that reported in larger prediction studies and a boost of up to 116% relative to classical models trained directly in the smaller samples. Critically, the model generalizes across datasets, maintaining performance when trained and tested across independent samples. This work establishes that predictive models derived in large population-level datasets can boost the prediction of cognition across clinical studies.
Neuroimaging reliably predicts cognition when models are translated from large datasets to smaller clinical studies.
INTRODUCTION
A key goal of computational psychiatry is the development of predictive models that provide personalized and robust estimates of clinically relevant phenotypes that can be used for prognostic and treatment decision-making. A primary barrier to progress in this area has been the historical use of small sample sizes, which has resulted in inflated prediction accuracies that largely fail to generalize across samples, populations, or collection sites (1–3). Here, we demonstrate a modeling strategy that uses measurements of brain function to robustly predict global cognitive function across multiple transdiagnostic samples, yielding models that generalize between independent cohorts despite modest sample sizes. The models also simultaneously provide interpretable insight into the neurobiology of global cognitive functioning in common psychiatric illness.
Impaired global cognitive functioning is a transdiagnostic characteristic of psychiatric illness (4–6). It is difficult to treat (7, 8), predicts social, occupational, and functional impairment (9–11), and is widely regarded by patients as a key priority for treatment (12, 13). Global cognitive functioning indexes the overall mental capacity and performance of an individual across multiple cognitive domains and is essential for everyday functioning and cognitive well-being. Impairments in global cognitive functioning have been implicated across all psychiatric diagnoses with evidence showing that it is a transdiagnostic phenomenon related to the presence of psychopathology and not to any specific disorder (4). The effect sizes related to underperformance range from small to medium in magnitude, with greater deficits found in mood and psychosis-spectrum disorders (4, 5). The overall performance across a broad range of cognitive tasks has repeatedly been linked to the structural and functional integrity of regions within transmodal association cortices. These regions are responsible for the integration of multiple sources of interoceptive and exteroceptive information and believed to underpin “higher-order” associative processes, which support cognition untethered from immediate sensory inputs (14–16), including adaptive goal-directed behavior (17), the application of complex rules (18), and the dynamic control of motor outputs (19). Across patient populations, converging evidence suggests the presence of altered functioning within the large-scale systems that comprise the association cortex (6, 20–24).
In particular, impaired connectivity within the default network, encompassing aspects of medial prefrontal, posterior/retrosplenial, and inferior parietal cortices, has been observed across diagnostic categories (14–18), while the level of dysconnectivity in the frontoparietal network, encompassing aspects of the dorsolateral prefrontal, dorsomedial prefrontal, lateral parietal, and posterior temporal cortices (19), often tracks the severity of diagnoses and observed cognitive deficits (20–22). However, despite the importance of establishing network-level predictors of symptom severity, the extent to which individual-specific profiles of brain functioning associate with clinically relevant cognitive impairments remains to be determined.
Inter-regional functional coupling of hemodynamic signals measured with functional magnetic resonance imaging (fMRI), here termed functional connectivity, has recently emerged as a powerful and robust predictor of global cognitive functioning in healthy populations (23–27). However, population neuroscience studies suggest that sample sizes exceeding thousands of participants may be required to develop accurate and stable brain-based predictive models of behavior (2, 28–30). This requirement far exceeds the vast majority of samples available to psychiatric research groups, calling into question both the utility and feasibility of developing clinically focused predictive models. Moreover, even brain-cognition predictive models derived from consortium-level samples can fail to generalize or show substantially reduced performance when applied to different datasets (2, 31–34), greatly diminishing the scope of their potential applications. This underscores a need for brain-based models that can reliably predict cognition using sample sizes that are feasible for psychiatric research groups to collect and that can generalize between independent datasets.
In large population-based cohorts, the functioning of specific brain systems can be leveraged to predict a broad variety of phenotypes, ranging from demographic factors to physical health–related and mental health–related variables (35–38). The associated brain-based models, which are derived from tens of thousands of healthy individuals, likely contain information that could be translated to smaller clinical cohorts, allowing for the prediction of illness-relevant and treatment-relevant phenotypes. In this regard, a recently developed framework called “meta-matching” (38) capitalizes on the fact that a limited set of overlapping functional circuits is associated with a wide variety of phenotypes and uses high-throughput population-based collection efforts to boost predictions of phenotypes in smaller cohorts. Using this framework, we have previously demonstrated a substantial increase in prediction accuracies for a broad range of variables in population-based healthy samples (38). However, the extent to which the meta-matching approach can improve the prediction of clinically relevant behaviors in small independent patient samples, yield cross-dataset generalizable predictions, and generate neurobiological insight remains to be determined.
Here, we use the meta-matching framework to develop an accurate, generalizable, and interpretable transdiagnostic model of global cognitive across a diverse set of datasets and psychiatric illnesses. We find that, across multiple distinct and reasonably sized datasets, the meta-matching model results in prediction accuracies that are statistically significant, superior to conventional models, and comparable to those regularly observed in much larger population-level studies. Moreover, suggesting the presence of shared brain-based features underlying cognitive impairments across patient populations, the derived models are generalizable, meaning that they maintain performance when trained and tested in independent datasets with differing diagnostic, imaging, and phenotypic characteristics. The brain features that drive prediction across the datasets become more similar at coarser spatial scales, with increased connectivity within transmodal association networks and decreased connectivity between transmodal and unimodal cortices being the most common transdiagnostic predictor of better global cognition.
RESULTS
Accurate and generalizable prediction of global cognitive functioning across psychiatric populations
Our overall aim was to develop a reliable and generalizable brain-based model that can predict global cognitive functioning across multiple samples of patients with psychiatric illness. To this end, we applied the recently developed meta-matching framework, which capitalizes on the fact that a limited set of overlapping functional circuits is associated with a wide variety of health, cognitive, and behavioral phenotypes (38). First, we used resting-state fMRI (rs-fMRI) data from 36,848 participants from the UK Biobank to derive functional connectivity estimates between 419 brain regions (39, 40). Next, we used these connectivity values to train a single fully connected feed-forward deep neural network (DNN) to predict 67 observed health, cognitive, and behavioral phenotypes in the UK Biobank.
Using the meta-matching approach (38), we then adapted this trained DNN (from the UK Biobank) to predict global cognitive function scores in three independent transdiagnostic clinical datasets: (i) the Human Connectome Project for Early Psychosis (HCP-EP; n = 145), which includes individuals diagnosed with affective and non-affective psychosis; (ii) the Transdiagnostic Connectome Project (TCP; n = 101), which largely includes individuals diagnosed with mood and anxiety disorders; and (iii) the Consortium for Neuropsychiatric Phenomics (CNP; n = 224), which is composed of individuals diagnosed with schizophrenia, bipolar disorder, or attention-deficit/hyperactivity disorder. All three samples also included a subset of healthy participants without psychiatric diagnoses. For a full demographic and diagnostic breakdown of the samples, see table S1. Global cognitive function scores were derived for each clinical dataset using principal components analysis (PCA) on a range of neuropsychological tests (see table S2). Of note, the test batteries varied across datasets, allowing for the assessment of model robustness to study design and associated phenotype selection. For details about the meta-matching approach, please see Materials and Methods.
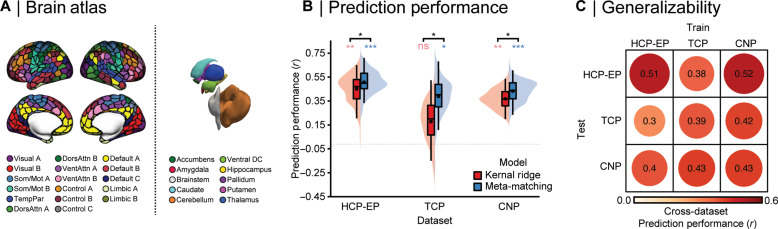
Our first aim was to determine if the meta-matching approach can make accurate and statistically significant predictions within clinical samples. For each dataset, we trained the meta-matching model using a nested cross-validation procedure, where each clinical sample was split into 100 unique training (70%) and test (30%) sets and the full meta-matching model was implemented for each train/test split. Performance was assessed as the mean Pearson correlation between the observed and predicted global cognition scores across the 100 test sets, and statistical significance was assessed using a permutation testing procedure (see Materials and Methods for details). As shown in Fig. 1B, the meta-matching approach yields statistically significant predictions (all ps < 0.05) across all three datasets, with mean prediction accuracies comparable to those found using much larger healthy samples (41). We find the same pattern of results when using the coefficient of determination to evaluate model performance (fig. S1). Furthermore, we establish that the meta-matching models systematically perform better than a standard prediction method, where a baseline comparison model was trained to predict cognition directly from the clinical sample functional connectivity values, with the difference between comparison and meta-matching models reaching statistical significance (all ps < 0.05).
Fig. 1. Accurate and generalizable prediction of global cognitive functioning across patient samples.
(A) Network organization of the human cortex. Colors reflect regions estimated to be within the same functional network according to the 17-network solution from Yeo et al. (47) across the 400-parcel atlas from Schaefer et al. (39), along with 19 subcortical regions (40). Cortical parcels and subcortical regions are used to extract blood-oxygen-level dependent time series and compute pair-wise functional connectivity estimates used for prediction. (B) Prediction performance (Pearson’s correlation between observed and predicted values) using KRR (red) and meta-matching (blue) across three transdiagnostic datasets: HCP-EP, TCP, and UCLA CNP. Colored asterisks denote above-chance prediction ( *P < 0.05; **P < 0.001; ***P < 0.0001; ns = P > 0.05), and black asterisks denotes the statistically significant difference between models. (C) Generalizability matrix for the meta-matching models, showing the prediction performance between the independent samples, where the meta-matching model is trained in one dataset and then used to make predictions in an independent dataset. The diagonal represents the mean prediction performance within each dataset, which is also represented by the black dots in (B).
Our second aim was to determine if the meta-matching model generalizes across independent clinical collection efforts. Generalizability was assessed as the Pearson correlation between the observed and predicted global cognition scores when a model was trained in one dataset and tested on another dataset. Here, we trained the meta-matching model on the full sample of one dataset and evaluated prediction performance on the other, resulting in six train-test prediction pairs between the three clinical datasets. Reflecting the presence of generalizable brain-behavior relationships across multiple independent clinical cohorts, we observed that the meta-matching model generalizes across datasets (Fig. 1C) and reached prediction accuracies both comparable to the mean in-sample accuracies shown in Fig. 1B and those reported in other studies that use in-sample validation and was statistically significant for all but one train/test pair (train/test: HCP-EP/TCP). In all cases except this same train/test pair, higher generalizability was found when using the meta-matching model, compared to a standard prediction model (fig. S2). Scatterplots of observed and predicted values are provided in fig. S3. We note that the meta-matching model generalized between datasets despite differences in diagnostic makeup, MRI scanners, acquisition parameters, and the fact that global cognition between each train-test pair was derived using different neurocognitive assessments, ranging from at-home online tests to gold-standard clinician-administered batteries.
Stable predictive network features across independent transdiagnostic datasets
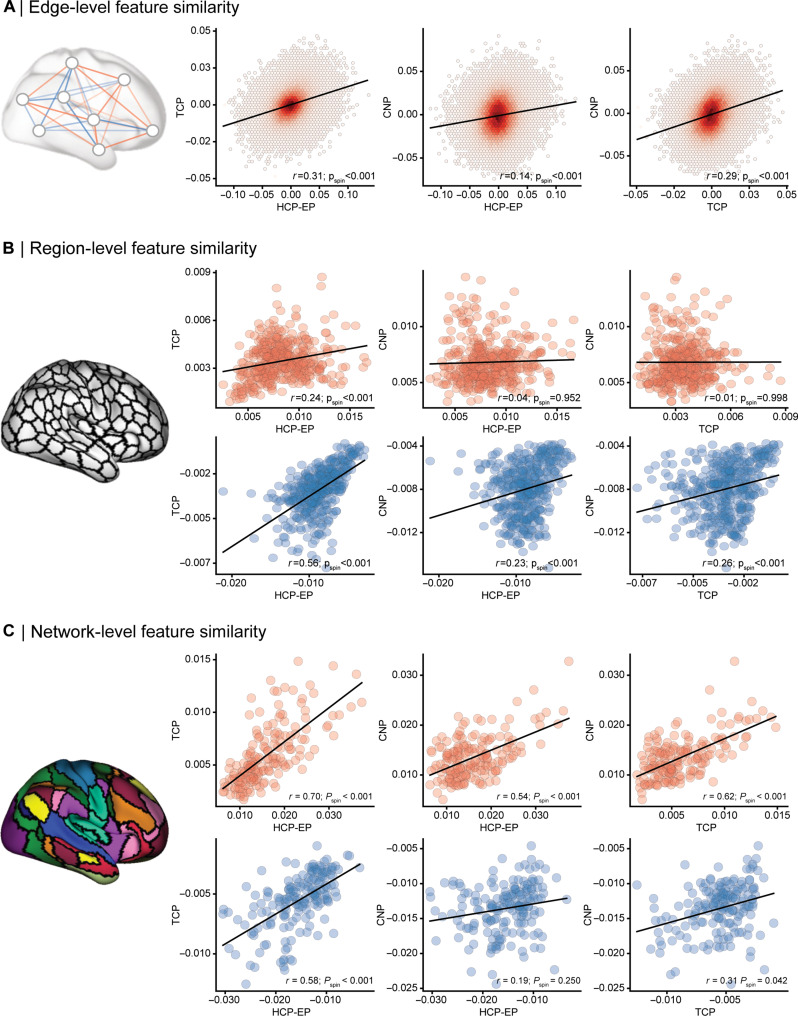
We next determined the extent to which the neurobiological features that drive the predictions are shared between datasets and if commonality is increased with coarser spatial scales. Predictive feature weights were derived using the Haufe transformation (31). This transformation considers the covariance between functional connectivity and global cognition scores and, unlike regression coefficients, ensures that feature weights are not statistically independent of global cognition. It also increases both the interpretability and reliability of predictive features (31, 42, 43). For each of the three datasets, we examined associations between average weights across the 100 cross-validation folds, at spatial scales of edges, regions, and networks. The edge-level spatial scale refers to the original 87,571 inter-regional pair-wise connections entered in the prediction models. By taking the mean of all edges attached to each of the 419 brain regions, edge-level connections can be aggregated into region-level predictive features. By taking the mean of all edges within and between 18 canonical functional networks including the subcortex [Fig. 1A; (39)], edge-level connections can also be aggregated into 171 network-level predictive features. For both aggregated scales (region-level and network-level), positive and negative feature weights were considered separately by zeroing negative or positive values before averaging, respectively.
When assessing associations between brain maps, spatial autocorrelation must be considered to ensure that any observed associations are not driven by low-level spatial properties of the brain (32). This same consideration extends to associations between edge-level network maps, where connectivity profiles of spatially adjacent regions demonstrate autocorrelation. To account for this property in the data, we implemented a spin test, which is a standardized procedure where the cortical regions of the atlas are rotated on an inflated sphere to generate configurations that preserve the spatial autocorrelation pattern of the cortex. We used these null atlas configurations to shuffle the rows and columns of the feature weight matrices to assess the statistical significance of feature weight correlations between datasets.
We find significant correlations across all three spatial scales (Fig. 2, A to C). At the edge level (Fig. 2A), we find low to moderate consistency between datasets, with the strongest correlation observed between the TCP and HCP-EP datasets (r = 0.31, Pspin < 0.001) and the TCP and CNP datasets (r = 0.29, Pspin < 0.001), with the CNP and HCP-EP datasets showing the weakest association (r = 0.14, Pspin < 0.001). At the region level (Fig. 2B), we again find low to moderate consistency between datasets, with the strongest associations between datasets when examining negative feature weights (rs = 0.23 to 0.56; psspin < 0.001), indicating that regions where lower functional connectivity predicts better cognition are more highly related between datasets, relative to regions where higher functional connectivity predicted better cognition (rs = 0.01 to 0.24; psspin < 0.001 to 0.998). The comparison between negative regional predictive features showing greater consistency between datasets than positive features was statistically significant for all three pairs of datasets (Zs =−2.60 to 5.97, psspin < 0.009). At the network level (Fig. 2C), we find the strongest overall consistency between datasets with moderate to high effect sizes observed when examining positive feature weights (r = 0.54 to 0.70; psspin < 0.001), indicating that network-level connections where lower functional connectivity predicts better cognition are strongly related between datasets, relative to network-level connections where higher functional connectivity predicted better cognition (r = 0.19 to 0.58; psspin = <0.001 to 0.250). The comparison between negative regional predictive features showing greater consistency between datasets than positive features was statistically significant for all three pairs of datasets (Zs = 2.88 to 5.75, ps < 0.004). Aggregating functional connectivity values at the canonical network-level capitalizes on the intrinsic functional architecture of the brain, with network-level brain function consistently being shown to have higher reliability (33, 34) compared to edge-level and region-level measures. Therefore, aggregating features at the network level may provide a more coherent signal than individual edge-level features, which may obscure associations between both individuals and datasets.
Fig. 2. Predictive features are correlated between independent transdiagnostic datasets across scales.
(A) Association between HCP-EP, TCP, and UCLA CNP prediction model feature weights at the edge level, which consist of 87,571 features per model. (B) Association between feature weights of the three datasets at the region level, where feature weights were averaged for all edges corresponding to a region, resulting in 419 regional features. Positive (red) and negative (blue) feature weights were considered separately by zeroing negative or positive values before averaging, respectively. All P values displayed account for spatial autocorrelation between edges, regions, and networks. (C) Association between feature weights of the three datasets at the network level, where feature weights were averaged within and between each network, resulting in 171 network features per dataset. Positive and negative feature weights were considered separately by zeroing negative or positive values before averaging, respectively.
Network-level predictors of better cognitive functioning
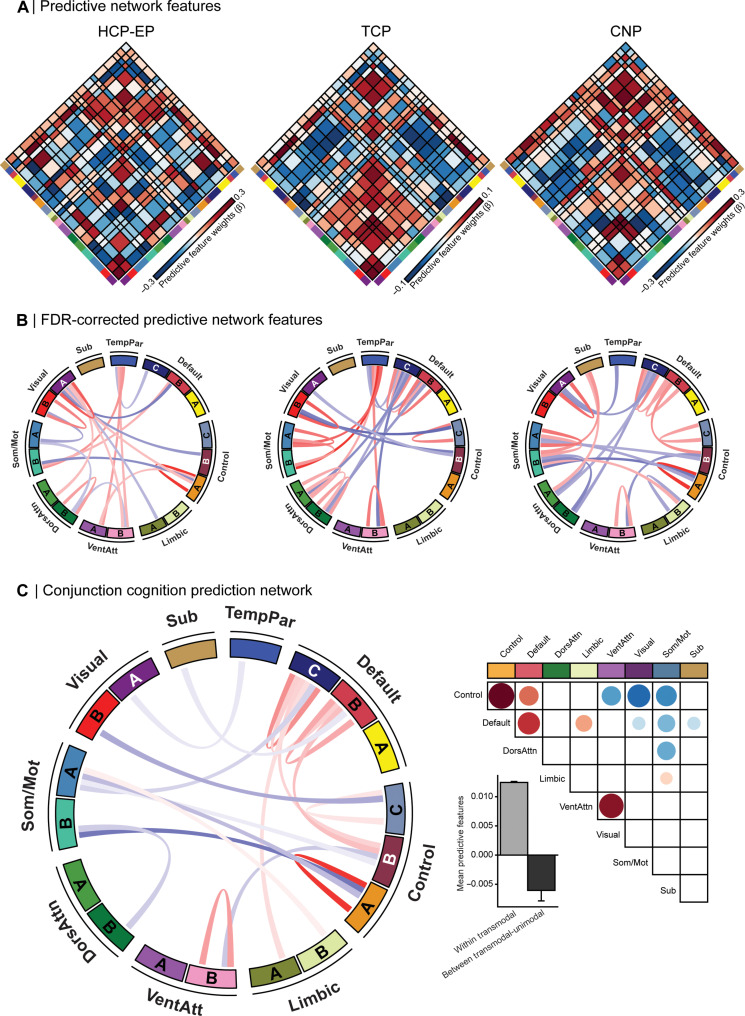
Given that predictive features were most stable between datasets at the network level, we examined the functional architecture of inter/intranetwork connections driving prediction performance (Fig. 3, A to C). In all three datasets, we observed a consistent, widespread, and complex pattern of network-level feature weights (Fig. 3B; PFDR < 0.05). In line with prior work that reliably links functional coupling in transmodal association networks with cognition (44–46), we find that brain-cognition relations converge on connections where higher functional connectivity within transmodal (default, frontoparietal, and ventral attention) networks and lower functional connectivity between transmodal and unimodal (visual and somatomotor) networks predict better cognition (Fig. 3C). We also find that connectivity within the frontoparietal subnetwork A, encompassing aspects of dorsolateral prefrontal, lateral parietal, medial cingulate, and posterior temporal cortices, was the strongest predictor of cognitive performance across datasets. More broadly, we find that, in each of the three datasets, increased connectivity within unimodal, transmodal, and all aggregated cortical networks was predictive of better cognition (Fig. 4).
Fig. 3. Increased within transmodal and reduced between network coupling is predictive of better cognitive functioning.
(A) Predictive feature matrices for each of the three datasets: HCP-EP, TCP, and UCLA CNP, averaged within and between network blocks. Non-averaged data are provided in the Supplementary Materials (fig. S5). Red, positive predictive feature weight (stronger coupling predicts better cognition); blue, negative predictive feature weight (weaker coupling predicts better cognition). (B) Top 10% of FDR-corrected predictive network connections for each dataset are displayed in Circos plots. See fig. S11 for all FDR-corrected predictive network connections for each dataset, displayed using Circos plots. (C) (Left) Circos plot showing the connections which survive multiple-comparison correction in a conjunction analysis across the three datasets. (Top right) Heat map of conjunction analysis results aggregated into a seven-network and subcortex atlas solution. (Bottom right) Mean feature weights from the conjunction analysis categorized into within and between transmodal and unimodal networks. Sub, subcortex; TempPar, temporoparietal; DorsAttn, dorsal attention; VentAttn, ventral attention; SomMot, somatomotor.
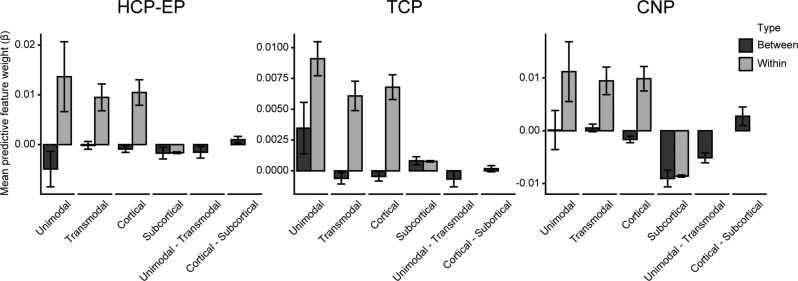
Fig. 4. Increased within and decreased between system coupling predicts better cognition across datasets.
Average predictive feature weights within (gray) and between (black) unimodal and transmodal cortical and subcortical regions across the three datasets: HCP-EP, TCP, and UCLA CNP. Error bars represent the SEM. Unimodal networks include all visual and somatomotor networks, and transmodal networks include default, control, ventral attention, dorsal attention, limbic, and temporoparietal networks.
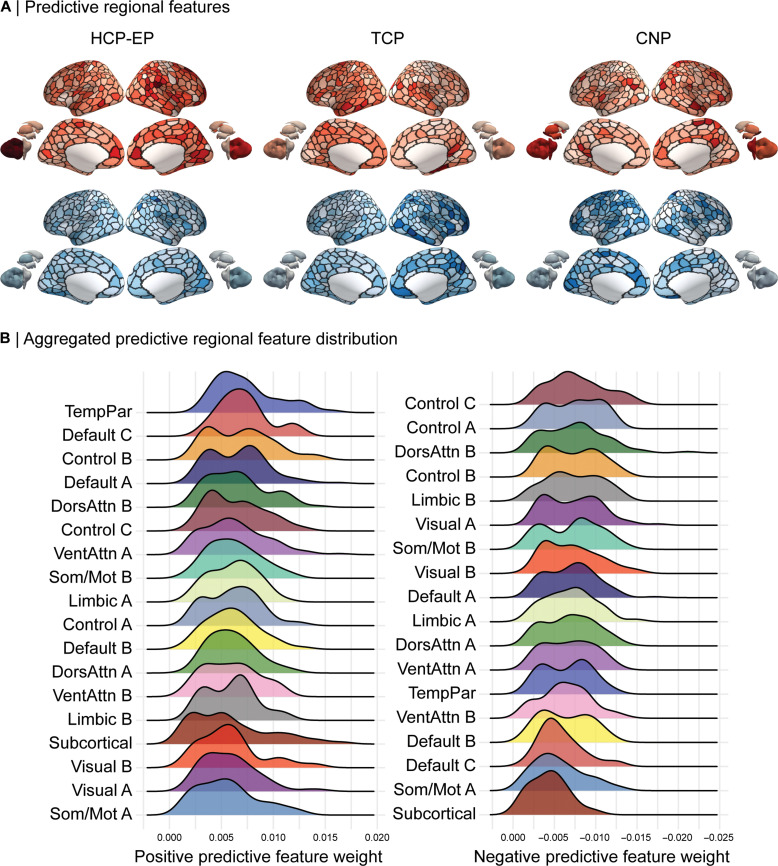
To provide an increasing level of granularity, we also examined the network-level architecture of regional predictive features (Fig. 5A). The strongest positive predictive regions for the HCP-EP dataset were the left cerebellar, right dorsal prefrontal, and temporoparietal cortices, and the negative predictive regions were right post-central and visual extrastriate cortices. For the TCP dataset, the strongest positive predictive regions included the right parahippocampal and left intraparietal cortices and negative predictive regions included the right intraparietal, anterior temporal, and precuneus regions. For the CNP dataset, positive predictive regions included the bilateral hippocampus, right temporoparietal, and dorsolateral prefrontal cortices and negative predictive regions were right post-central gyrus, somatomotor, and left visual extrastriate cortex. While there was some heterogeneity in region-level predictive features, when these were aggregated into canonical networks (Fig. 5B), across all datasets, the strongest positive drivers of prediction performance were regions in transmodal temporoparietal, default, and frontoparietal networks. The strongest negative drivers also included the frontoparietal, dorsal attention, limbic, and primary sensory regions, with the prominence of the frontoparietal network characterized by lower connectivity to sensory networks (Fig. 3C). We provide non-aggregated region-level distributions for each dataset individually, as well as distributions using a seven-network solution (47) in the Supplementary Materials (fig. S5).
Fig. 5. Predictive features at the regional level.
(A) Regional feature weights projected onto cortical and subcortical regions. Average positive (red) and negative (blue) feature weights are shown separately for each of the three datasets: HCP-EP, TCP, and UCLA CNP. (B) Positive (left) and negative (right) distributions of regional feature weights from all three datasets aggregated into 17 networks and subcortex and ordered by the strongest to weakest mean predictive feature weight.
DISCUSSION
Constructing robust and generalizable models that reliably predict clinical symptoms from brain markers has previously required sample sizes exceeding most current collection efforts. Here, we provide a proof of concept and define an associated roadmap for the generation of brain-based predictions in clinical populations. Critically, the models reported here are generalizable across independent datasets, maintaining prediction performance when trained in one dataset and tested on another, even when the datasets are independent and differ in their collection sites, demographic and diagnostic makeup, measures of global cognition, imaging acquisition sequences, and data processing methods. The neurobiological features that drive prediction performance are most consistent between datasets at the scale of canonical functional networks rather than individual brain regions or edges. In line with previous hypotheses concerning the neurobiological substrates of cognition (48–50), our findings converge on a global cognition predictive network where increased coupling within transmodal and the decreased coupling between transmodal and unimodal networks are linked with better cognition across transdiagnostic samples.
Widespread cognitive impairments are a core feature of common psychiatric illness, often presenting prior to illness onset (51, 52), and contribute to impaired social and occupational functioning (9–11). Here, we capture global cognitive impairments using PCA to extract the shared variance across multiple different submeasures of cognition such as processing speed, working memory, and executive function, which varied across the three cohorts used. Leveraging the meta-matching framework, we demonstrate that it is possible to achieve predictions of global cognitive functioning comparable to accuracies observed in the current state of the art for the field, using sample sizes that are much smaller than those that have recently been recommended for deriving stable and generalizable brain-based predictions (28, 30). A particular advantage of our approach is that it yields discoveries that generalize across both healthy controls and common psychiatric disorders. By combining multivariate predictive models with transfer learning approaches like meta-matching, we provide a framework to leverage high-throughput population-based cohorts to boost predictive power in smaller datasets.
Here, we demonstrate that the meta-matching model generalizes not only across diagnostic categories but also between independent datasets relying on different measures of cognition, neuroimaging protocols, and data processing strategies. Usually, models trained in one dataset lose much of their predictive capacity when applied to an independent dataset, even when the two datasets are diagnostically or demographically similar (2, 28, 53–55). The meta-matching approach likely achieves this high level of generalizability by exploiting correlations amongst phenotypes, relying on a common set of neurobiological features that predict a broad range of behaviors that underlie an individual’s global cognitive performance, independent of diagnosis or measurement methods.
We have previously demonstrated that a larger-sized test sample (i.e., n > 100) assists in boosting performance, but meta-matching outperforms baseline models even in samples as small as n = 10 (38). A critical factor affecting model performance is the correlations between the phenotypes being tested and those available in the larger dataset for initial model training. We previously showed that test phenotypes with stronger correlations with at least one training phenotype lead to greater prediction improvement with meta-matching (38).
While we find differences in the neurobiological features driving prediction performance between the independent datasets, we observe consistency across all spatial scales, with the strongest and consistently significant correspondence detected at the network level. Given the important methodological and phenotypic differences between datasets, feature weights from separate models are expected to show differences. Analogous to genetics, where broadening the spatial scale from single-nucleotide polymorphisms to gene pathways results in more consistent associations with complex behavior, we find that broadening the spatial scale from inter-regional edge-level connections to canonical networks results in more stable associations. The similarity of neurobiological features at the network level aligns with a large literature of explanatory studies implicating macroscale networks as the primary unit of analysis for complex behavioral traits (56, 57), as opposed to isolated regions or individual circuits, and evidences that the individual heterogeneity of patients assigned the same diagnosis is greatly attenuated when aggregating results across functional circuits and networks rather than brain regions (58). Moreover, the consistency we observed between datasets suggests that the meta-matching model is likely making predictions by indexing a common neurobiology closely associated with cognitive function. In line with this hypothesis, we find that the connectivity of transmodal association networks, including the default and frontoparietal networks, is the most prominent driver of prediction performance. Specifically, increased connectivity within association networks and decreased connectivity between these networks and visual and somatomotor sensory networks were consistently associated with better cognition. This finding converges with decades of empirical work demonstrating that the activation and integrity of association networks is a critical driver of complex cognition (44, 49, 50) and suggests that the prediction model is not relying on highly idiosyncratic characteristics or overfitting noise in the neuroimaging data to make predictions within each dataset. Suggesting the presence of shared neurobiological associates of impaired cognition across broad diagnostic categories, the observed set of brain-behavior relations was reliable and generalizable across a diverse set of patient populations.
While this pattern of connectivity represents the most consistent statistically significant network-level features, it is likely that a distributed range of shared and unique connections also contributes to the prediction of cognition within each dataset (36). Moreover, age-related functional alterations include desegregation of large-scale networks (59), which are often associated with poorer cognitive performance (60). Accordingly, we find that increased segregation of association networks from sensory networks is associated with better performance. Notably, our finding of increased connectivity within association networks predicting better cognitive function aligns with other large-scale investigations reporting a similar association with a general positive domain of behavior (35) but are likely more specific to cognitive functions as they represent overlapping, rather than district, brain features across multiple samples (36). Our current analyses establish the presence of shared brain-based predictive features of cognitive functioning between patient cohorts. Future work should examine the unique neurobiological contributors of illness-relevant shifts in cognition across broader symptom profiles both within and across diagnostic groups.
There are some limitations in the current work. While being able to make accurate and generalizable predictions of an observed phenotype such as cognition suggests the potential for clinical applications, future work should seek to develop models that can provide guidance on longitudinal outcomes. Such outcomes would include change in cognition over time, such as illness-related decline, response to medications, and transitions in illness severity. As large-scale population-based longitudinal data become available, the meta-matching framework can be adapted to predict symptom change and illness course. Moreover, in our analyses we focused on global cognition, which can be reliably measured (61–63), is consistently impaired across common psychiatric diagnoses (4–6), and is identified by patients as a key target for assessment and treatment (9, 10). Specific subdomains of cognition may be more or less impaired across populations, and future work should attempt to predict the results of specialized neurocognitive assessments targeting constructs like working memory, executive function, processing speed, or attention. This will likely require standardization of neurocognitive assessments between independent data collection efforts. While the current and previous findings (38) demonstrate a boost in prediction performance and generalizability even when there is a difference in age and imaging acquisition parameters between the UK Biobank and the clinical datasets, future work may find further improvements by training and testing the prediction model on age-matched and acquisition parameter–matched datasets.
The meta-matching framework leverages overlap in correlation structure between brain and behavioral phenotypes found in large population-level datasets and smaller clinical datasets. One notable advantage of this framework is that it allows the prediction of behavioral phenotypes that differ from those available in large population-level datasets, but it is likely that a closer match between the target and trained phenotypes will improve performance. We trained our meta-matching model on 67 variables from the UK Biobank, including various cognitive measures. These cognitive measures substantially contribute to the observed enhancements in prediction performance and generalizability (see Control analyses). While previous research has demonstrated that meta-matching results in an overall improvement in prediction performance across multiple broad categories of behavior, prediction targets diverging significantly from those used to train the meta-matching model may not benefit from this framework in terms of prediction performance or generalizability. Therefore, the effectiveness of the meta-matching method may vary depending on the similarity between the prediction target and the phenotypes used in training the model.
We initially trained the meta-matching model on the large UK Biobank sample (n = 36,848). Future work should examine if a similar boost in performance can be achieved with smaller samples. Future work should also examine if longer scan durations, which can improve both reliability and prediction performance (64, 65), can further enhance performance of meta-matching and other transfer learning models. Last, in determining the features that are most relevant for predicting cognition, we implemented the Haufe transformation, which enhances both reliability and interpretability of feature weights (36, 66). The Haufe transformation remains the best linear approximation of feature weights for nonlinear models (31), and we have previously demonstrated that the results of the transformation when using deep learning models in the prediction process are highly comparable to results using only linear models (38). However, future studies should compare the transformation to alternative and validated approaches for interpreting feature weights from nonlinear models, as they become available.
By translating predictive models derived in large community-based datasets, we can make an accurate and generalizable prediction of global cognition in transdiagnostic patient populations. The performance of these models is driven by increased coupling within transmodal networks and decreased coupling between transmodal and sensory networks.
MATERIALS AND METHODS
Overview
Our overall analysis strategy aimed to develop a robust and generalizable model that can accurately predict global cognitive function in transdiagnostic patient samples. Briefly, we first used meta-matching (38), which capitalizes on the correlation structure between phenotypes of clinical interest and those available in larger population-level datasets by (1) training a single generic fully connected feed-forward DNN to predict a set of 67 health, behavioral, and cognitive phenotypes using in vivo estimates of brain function from the UK Biobank dataset (67); (2) using this trained model to generate predictions of these phenotypes in smaller independent patient datasets; (3) and as a final step, training and validating a kernel ridge regression (KRR) model to predict global cognitive function using the predicted phenotypes generated from the DNN model in step 2. Global cognitive function was derived using PCA on a range of neuropsychological tests that varied between the patient datasets. The significance of prediction performance and the generalizability of models were assessed using permutation testing, and the feature weights from each model were correlated between datasets and mapped at differing spatial scales (edge, region, and network) to examine the consistency of neurobiological correlates. Please see Brain-based predictive modeling for a detailed overview of the modeling procedure.
Datasets
This study used data from four datasets: the UK Biobank (67), the HCP-EP (68), the TCP (69), and the CNP (70). Our analyses were approved by the Yale University Institutional Review Board, and the UK Biobank data were accessed under resource application 25163. The final number of included participants, demographic and diagnostic characteristics is described below with additional details provided in table S1.
UK Biobank
The UK Biobank (67) is a population epidemiology study of 500,000 adults aged 40 to 69 years and recruited between 2006 and 2010. A subset of 100,000 participants is being recruited for multimodal imaging, including brain structural MRI and rs-fMRI. A wide range of health, behavioral, and cognitive phenotypes was collected for each participant. Here, we used the January 2020 release of 37,848 participants with complete and usable structural MRI and rs-fMRI.
Human Connectome Project for Early Psychosis
The HCP-EP (68) study is acquiring high-quality brain MRI and behavioral and cognitive measures in a cohort of people aged 16 to 35 years, with either affective or non-affective early phase psychosis within the first 5 years of the onset of psychotic symptoms. The dataset also includes healthy control participants, and the data release used here (Release 1.1) comprises 140 patients and 63 controls. Inclusion and exclusion criteria are described elsewhere (68). In the current study, we used a subset of 145 participants who passed quality control and had complete and usable cognitive and rs-fMRI data. The included sample had a mean age of 23.41 (SD ± 3.68), was 38% female, and had a mean framewise displacement (head motion during rs-fMRI acquisition) of 0.06 mm (SD ± 0.04).
Transdiagnostic Connectome Project
The TCP is a publicly available data collection effort between Yale University and McLean Hospital, United States, to acquire brain MRI and behavioral and cognitive measures in a transdiagnostic cohort, including healthy controls and patients meeting the diagnostic criteria for an affective or psychotic illness. Recruitment details and inclusion and exclusion criteria can be found in the Supplementary Materials and elsewhere (69). The data included in the current study were composed of a subsample of 101 participants who passed quality control and had complete and usable cognitive and rs-fMRI data at the time of the study, including 60 patients and 41 healthy controls. The included sample had a mean age of 32.21 (SD ± 12.54), was 57% female, and had a mean framewise displacement of 0.09 mm (SD ± 0.05).
Consortium for Neuropsychiatric Phenomics
The CNP dataset is publicly available and composed of brain MRI and behavioral and cognitive measures from 272 participants, including 130 healthy individuals and 142 patients diagnosed with affective, neurodevelopmental, or psychotic illnesses. Details about participant recruitment can be found elsewhere (70). In the current study, we used a subset of 224 participants who passed quality control and had complete and usable cognitive and rs-fMRI data. The included sample had a mean age of 32.59 (SD ± 9.21), was 42% female, and had a mean framewise displacement of 0.08 mm (SD ± 0.03).
Quantifying brain function
MRI acquisition parameters
For the UK Biobank, a total of 490 functional volumes were acquired over 6 min at four imaging sites with harmonized Siemens 3T Skyra MRI scanners using the following parameters: repetition time = 735 ms, echo time = 42 ms, flip angle = 51°, resolution of 2.4 mm3, and a multiband acceleration factor of 8. For a T1-weighted image, an MPRAGE sequence with a total of 256 slices was acquired using the following parameters: repetition time (TR) = 2000 ms, inversion time (TI) = 880 ms, resolution of 1 mm3, and parallel imaging acceleration factor of 2.
For the HCP-EP, a total of four runs of 420 functional volumes were acquired over 5.6 min at three imaging sites with harmonized Siemens 3T Prisma MRI scanners using the following parameters: repetition time = 800 ms, echo time = 37 ms, flip angle = 52°, resolution of 2 mm3, and a multiband acceleration factor of 8. Spin echo field maps in the opposing acquisition direction were acquired to correct for susceptibility distortions. For a T1-weighted image, an MPRAGE sequence with a total of 208 slices was acquired using the following parameters: TR = 2400 ms, TI = 1000 ms, and resolution of 0.8 mm3.
For the TCP data, four runs of a total of 488 functional volumes were acquired over 5 min at two imaging sites with harmonized Siemens Magnetom 3T Prisma MRI scanners using the following parameters: repetition time = 800 ms, echo time = 37 ms, flip angle = 52°, resolution of 2 mm3, and a multiband acceleration factor of 8. Spin echo field maps in the opposing acquisition direction were acquired to correct for susceptibility distortions. For a T1-weighted image, an MPRAGE sequence with a total of 208 slices was acquired using the following parameters: TR = 2400 ms and resolution of 0.8 mm3.
For the CNP data, a total of 152 functional volumes were acquired over 5 min at 2 imaging sites with harmonized Siemens Trio 3T MRI scanners using the following parameters: repetition time = 2000 ms, echo time = 30 ms, flip angle = 90°, and a resolution of 4 mm3. For a T1-weighted image, an MPRAGE sequence with a total of 176 slices was acquired using the following parameters: TR = 1900 ms and resolution of 1 mm3.
MRI quality control
For all the clinical datasets, extensive quality control procedures were implemented, and the details can be found in the Supplementary Materials. Briefly, all raw images were first put through an automated quality control procedure (MRIQC), which resulted in the exclusion of scans with large artifacts. Recent studies have shown that multiband datasets (i.e., HCP-EP and TCP) with high temporal resolution contain additional respiratory artifacts that manifest in the six realignment parameters typically used to calculate summary statistics of head motion (71, 72). To mitigate this effect, framewise displacement traces were downsampled and bandpass filtering was applied on the realignment parameter between 0.2 and 0.5 Hz (73). Following this step, uniform motion exclusion criteria were applied to all clinical datasets, and scans with an established cutoff of mean framewise displacement greater than 0.55 mm, which has previously been shown to result in good control of motion artifacts (74), were excluded. Last, for all participants, functional connectivity matrices, carpet plots, and quality control–functional connectivity metrics were visualized and examined to ensure that the processing and denoising steps achieved the desired effects of reducing noise and associations between head motion and functional connectivity.
MRI processing
A detailed outline of the processing and denoising steps for each dataset is provided in the Supplementary Materials. Briefly, for each dataset, we used differing but widely accepted processing strategies, which all included nonlinear spatial normalization to Montreal Neurological Institute space, brain tissue segmentation, and independent component analysis (ICA)-based denoising. These strategies were tailored to address differences in fMRI acquisition parameters (i.e., single-band versus multiband) and to ensure that our models were robust to differences in preprocessing and denoising procedures.
Global signal regression was also applied to all scans, as we have previously demonstrated using multiple independent datasets that it improves behavioral prediction performance (75) and data denoising (73, 74). The final derivatives used for prediction were 419 × 419 matrices for each subject, which were computed using 400 cortical and 19 noncortical regions (for simplicity, noncortical regions are indicated as “subcortex”; Fig. 1A) by averaging the time series within each parcel and computing interregional pair-wise Pearson correlations. For each subject, the correlation values were z scored and the upper triangle of this matrix that consisted of 87,571 unique functional connectivity estimates was entered into the prediction models.
Quantifying global cognitive functioning
For each of the three clinical datasets, PCA was applied to all available cognitive and neuropsychological measures to derive a robust measure of global cognition. Each dataset had a distinct set of neuropsychological tests used to quantify cognitive functioning. A full list of measures for each dataset can be found in table S2. Briefly, for the HCP-EP, measures included the National Institutes of Health Toolbox (76) and Wechsler Abbreviated Scale of Intelligence (77). For the TCP, measures were administered online through the TestMyBrain platform (78), which included assessment of matrix reasoning, sustained attention, basic psychomotor speed, and processing speed, as well as Stroop and Hammer reaction time measures acquired during MRI acquisition. For the CNP, measures included subtests from the California Verbal Learning Test, Wechsler Memory Scale (79), and Wechsler Adult Intelligence Scale (80). To reduce the complexity of the prediction model, the PCA for each dataset was computed on the full sample, before any cross-validation, rather than computed on the training sample at each of the 100 splits. To ensure that data leakage between the train and test splits did not influence our results, we tested if prediction models generalized between the different and completely independent datasets, where the PCA was computed separately on full independent samples (see Evaluating model generalizability). For each dataset, the first principal component (PC) was retained. For the HCP-EP dataset, the first PC explained 57.2% of the variance, with the second and third PC examining 13.6 and 6% of the variance, respectively. For the TCP dataset, the first PC explained 25.9% of the variance, with the second and third PC examining 16.2 and 13.8% of the variance, respectively. For the CNP dataset, the first PC explained 32.5% of the variance, with the second and third PC examining 9.4 and 7% of the variance, respectively. For each dataset, a higher PC score indexed better global cognition. The full list of loadings for each item can be found in table S2.
Brain-based predictive modeling
Consistent with the approach outlined in (38), we trained a single fully connected feed-forward DNN using the UK Biobank dataset to predict 67 different cognitive, health, and behavioral phenotypes from resting-state functional connectivity matrices. This type of DNN has a generic architecture, where the connectivity values enter the model through an input layer, and each output layer is fully connected to the layer before it, meaning that values at each node are the weighted sum of node values from the previous layer. During the training process, these weights are optimized so that the output layer results in predictions that are close estimations of observed phenotypes. In practice, any multivariate prediction method can be used instead of the DNN, but the fully connected feed-forward DNN offers an effective and parsimonious method to predict 67 phenotypes using a single model (24, 38, 41, 81). The 67 cognitive, health, and behavioral variables were selected based on an initial list of 3937 phenotypes by a systematic procedure that excluded brain variables, categorical variables (except sex), repeated measures, and phenotypes that were not predictable using a held-out set of 1000 participants (38). A full list of selected phenotypes can be found in table S3, and further details of the DNN architecture and variable selection procedure within the UK Biobank can be found elsewhere (38). This trained DNN model is openly available and can be implemented in any sample with available resting-state functional connectivity data (see https://github.com/ThomasYeoLab/Meta_matching_models).
Following training of the DNN, it was applied to the clinical datasets using a nested cross-validation and stacking procedure. The procedure described below was implemented separately for each clinical dataset. First, the DNN was applied to the dataset, using resting-state functional connectivity matrices as inputs, resulting in 67 generated cognitive, health, and behavioral variables as outputs. These outputs and corresponding global cognitive scores were split into 100 distinct train (70%) and test (30%) sets without replacement. We then implemented a stacking procedure, where a KRR model using a linear kernel with L2 regularization was trained to predict global cognitive functioning scores using the generated 67 cognitive, health, and behavioral variables as inputs. KRR is a classical machine learning technique that makes a prediction of a given phenotype in an individual as a weighted version of similar individuals. Similarity was defined as the interindividual correlation of predicted phenotypes. KRR has one free parameter that controls the strength of regularization and was selected based on fivefold cross-validation within the training set. Once optimized, the model was evaluated on the held-out test set. This was repeated for the 100 distinct train-test splits to obtain a distribution of performance metrics. We have previously demonstrated that this stacking procedure improves the performance of the meta-matching framework in predicting behavioral phenotypes (38). In practice, any multivariable model can be used in place of KRR. However, KRR is a robust and flexible multivariable model for predicting behavioral phenotypes (24, 43, 75).
As a comparison to the meta-matching model described above, we also implemented a standard machine learning model to provide a baseline. Here, we used the standard implementation of KRR, where the model was trained to predict global cognitive function scores, using resting-state functional connectivity matrices as inputs. This is in contrast to the KRR model implemented during the meta-matching stacking process, which was trained using the DNN-generated cognitive, health, and behavioral as inputs. KRR was used as a baseline model as it is widely used and has repeatedly been shown to work well for functional connectivity–based behavioral and demographic prediction (24, 29, 38, 41, 81–83). In a recent work, we have further demonstrated that meta-matching with stacking is superior to a classic transfer learning approach (84). The nested cross-validation procedure used for the baseline comparison model was the same as the one used for the meta-matching model, where each dataset was split into 100 distinct train (70%) and test (30%) sets without replacement, followed by fivefold cross-validation within the training set to tune the model hyperparameters, and the model performance was evaluation on the held-out test set. All codes used for analysis and figure generation can be found online at https://github.com/sidchop/PredictingCognition.
Evaluating model performance
The performance of each model is defined as the Pearson correlation between the true and predicted behavioral scores for the test sample in each split. Average performance was computed by taking the mean across the 100 distinct splits. We also evaluated absolute, as opposed to relative, prediction performance using the coefficient of determination [fig. S2; (1)]. All models were evaluated on whether they performed better than chance using null distributions of performance metrics. For the meta-matching model, in each of the three clinical datasets, cognitive function scores were randomly permuted 10,000 times. Each permutation was used to train (70% of the sample) and test (30% of the sample) a null prediction model. The P value for the model’s significance was defined as the proportion of null prediction accuracies greater than the mean performance of the observed model. The same procedure was used to evaluate the statistical significance of the baseline comparison model.
Evaluating model generalizability
The generalizability of the model was evaluated by training the meta-matching model on all individuals from one dataset and testing on all individuals from another dataset. This results in six train-test pairs between the three clinical datasets (i.e., HCP-EP, TCP, and CNP). For each train-test pair, performance was again measured as the Pearson correlation between the predicted and actual scores on the test dataset. We also report absolute performance using the coefficient of determination [fig. S2; (1)]. Statistical significance was evaluated by permuting the training dataset cognitive function scores and computing a null meta-matching model 10,000 times. The P value corresponding to model significance was defined as the proportion null prediction accuracies greater than the performance of the observed model. To compare the within dataset prediction performance between standard baseline comparison and meta-matching models, we computed a paired-sampled t test for each of the three clinical datasets. This allowed us to evaluate if the difference in the cross-validated prediction performance of the models was significantly greater than zero.
Comparing neurobiological features between datasets and spatial scales
To increase the interpretability and reliability of feature weights from the prediction models, we used the Haufe transformation (31, 42, 66). To illustrate the need for the Haufe transformation, let us consider the prediction of a target variable, such as global cognition (y), based on the functional connectivity (FC) of two edges, denoted as FCA and FCB. In this example, let us assume that FCA = y−noise, and FCB = noise. Then, examining raw feature weights from a prediction model with 100% performance would erroneously show both edges as strongly related to and equally important for predicting global cognition. To address this issue, the Haufe transformation computes the covariance between the predicted target variable and the FC of the two edges. In this example, the Haufe transformation assigns a weight of zero to FCB, indicating that FCB is not related to global cognition, despite its contribution to the prediction performance. While originally developed for linear models, the Haufe transformation can also recover the best linear interpretation of nonlinear models such as DNNs (31). Moreover, the predictive features computed using Haufe transformation are more reliable and robust, further underscoring the importance of this inversion process (36, 66).
This procedure ensured that the feature weights index quantities that are statistically related to global cognition and results in a positive or negative predictive feature value for each edge of the functional connectivity matrix. A positive predictive feature value indicates that higher functional connectivity for the edge was associated with the greater predicted cognitive functioning, and a negative predictive feature value indicates that lower functional connectivity was associated with the greater predicted cognitive functioning. For each of the three models, the transformed feature weights were then averaged across the 100 splits to obtain mean feature weights, resulting in a single symmetric 419 × 419 predictive feature matrix for each dataset.
We assessed the association of neurobiological predictive features between each of the three predictive feature matrices at the edge, region, and network level. At the edge level, which comprises each of the 87,571 feature weights, similarity between the three samples was assessed using Pearson correlation. To account for spatial autocorrelation between each pair of feature weight matrices (32), we applied the spin test, where the cortical regions of the atlas are rotated on an inflated surface to generate 10,000 null atlas configurations, which preserve the spatial autocorrelation pattern of the cortex. These null atlas configurations are used to shuffle the rows and columns of the feature weight matrices, allowing the generation of a null distribution of Pearson correlation values between a pair of feature weight matrices at the edge, region, and network level. Statistical significance was assessed as the proportion of null values greater than the observed value (Pspin). As the spin test procedure can only be applied to cortical regions, the 19 noncortical regions were excluded when computing the P value. By taking the mean of all edges attached to each of the 419 brain regions, edge-level connections can be aggregated into region-level predictive features. By taking the mean of all edges within and between 18 canonical functional networks including the subcortex [Fig. 1A; (39)], edge-level connections can also be aggregated into 171 network-level predictive features. Pearson correlation was again used to assess association in region-level and network-level feature weights between the three samples. For both aggregated scales (region-level and network-level), positive and negative feature weights were considered separately by zeroing negative or positive values before averaging, respectively. This procedure allows examination of the relative contribution of the polarity of weights and is equivalent to summing the positive or negative feature weights. To compare the negative and positive feature weight correlations within each spatial scale, we used Fisher’s Z statistic modified for nonoverlapping correlations based on dependent groups (85, 86).
Evaluating neurobiological features
To evaluate the statistical significance of feature weights for each of the three datasets, we implemented a permutation testing procedure. To reduce the multiple comparison burden, we evaluated the significance of each model at the network level, where the observed feature weights for each model were averaged within and between 18 network blocks, resulting in 171 network-level features per model. This network averaging procedure was repeated for feature weights from 10,000 null models, where the cognitive function score had been randomly permuted. This results in null distribution of network-level feature weights for each of the 171 network connections, and the P value was computed as the proportion of the null network-level feature weights greater than the observed value. The P values were then false-discovery rate (FDR) corrected and evaluated at a P < 0.05 level. To uncover the network-level predictive features that drive performance across the three datasets, we implemented a conjunction analysis, where, at each network connection, the minimum FDR-corrected P value was retained and evaluated for significance at P < 0.016, accounting for the three contrasts.
Control analyses
Demographic characteristics such as age and sex as well as head motion during neuroimaging can bias the performance of prediction models (87). To ensure that model performance was not driven by these covariates, we repeated the primary models after adjusting for age, sex and mean framewise displacement. For each of the 100 train-test splits, the variables were first regressed out of the global cognition training data, and the resulting regression coefficients were used to residualize the global cognition test data (88), after which the entire prediction modeling procedure was repeated. The reported results were robust to covariate inclusion and all three meta-matching models remained statistically significant (figs. S6 and S7). Moreover, the edge-level feature weights from the original and covariate adjusted models were highly correlated at rs > 0.96 for all three datasets (fig. S7). Performance remained stable in the HCP-EP sample (r = 0.51) and decreased in the TCP dataset (r = 0.25) and CNP dataset (r = 0.28; fig. S6). The pattern of results showing the superior performance of the meta-matching compared to the conventional KRR model was maintained in all three datasets (fig. S6).
Meta-matching capitalizes on correlations between neurobiology associated with diverse demographic, health, and behavioral phenotypes. We implemented the meta-matching framework using a two-step stacking approach (see Brain-based predictive modeling) that allows us to examine the feature weights that drive the prediction of cognition associated with each of the 87,571 functional connections of the brain, as well as the feature weights associated with each of the 67 demographic, health, and behavioral phenotypes. By examining the feature weights associated with the 67 DNN-generated demographic, health, and behavioral variables, it is possible to evaluate which phenotypes are driving the prediction of cognitive outcomes. The generated variables driving performance were highly consistent across the three datasets (all rs > 0.95; fig. S8). The primary drivers of prediction were directly related to cognition (fluid intelligence, matrix pattern completion, and symbol digit substitution). However, across the three datasets, both age and the first genetic PC were strong predictors, with the latter indexing ancestry, which can also be a proxy for complex forms of societal and environmental bias, in turn affecting cognitive performance. To investigate if the observed improvements in behavior prediction performance extend beyond the functional connections associated with specific sociodemographic factors, we repeated the meta-matching stacking procedure for our primary analyses after removing the first genetic PC, age, and sex from the meta-matching model and found similar results to our original model (fig. S9). This analysis suggests that it is the functional connections associated with cognition-related variables in the UK Biobank that drives the boost in prediction performance in the three clinical datasets.
To assess if meta-matching prediction performance was dependent on sample characteristics such as age, sex, and diagnosis, we conducted a leave-one-out cross-validation. For each sample, this procedure resulted in a predicted score for each subject. The overall prediction performance (correlation between predicted and observed scores) remained comparable to the K-fold procedure used in the primary analyses (HCP-EP:r = 0.50; TCP:r = 0.35; CNP:r = 0.41; fig. S10). To evaluate differences in model performance between diagnostic and demographic subgroups, we fitted a general linear model to the observed and predicted scores separately for each subgroup. Age was converted to a binary variable using a mean split, sex was treated as a binary variable, and diagnostic group was treated as a categorical variable that included a healthy control group. For each sample, we compared the mean square error between subgroups using a nonparametric Kruskal-Wallis test. In this way, we were able to determine if, for each given characteristic (e.g., sex), there is a significant difference in relative prediction error between subgroups (fig. S10). We only find a significant effect within the CNP dataset for diagnosis, with post hoc tests demonstrating marginally worse prediction performance within patients diagnosed with bipolar disorder compared to the healthy control group (P = 0.024). These findings demonstrate that the meta-matching prediction model performance is largely robust to sample characteristics and diagnoses.
To ensure that the subgroup of patients diagnosed with psychosis were not the primary driver of the cross-dataset generalizability model performance, we repeated the analyses after removing all patients with psychosis (N = 37) from the CNP dataset. We find that the cross-dataset prediction performance between the CNP and both the TCP and HCP-EP remains comparable in magnitude and statistically significant (0.33 < r < 0.51; fig. S12). To ensure that performance and generalizability were maintained when applying the model to patients alone, we repeated the analyses after removing all healthy individuals. We find that both the performance (0.31 < r < 0.55; fig. S13A) and the cross-dataset generalizability (0.31 < r < 0.56; fig. S13, B and C) in all datasets are comparable in magnitude, statistically significant, and superior to the baseline model.
Acknowledgments
Funding: This work was supported by the National Institute of Mental Health (R01MH123245 to A.J.H. and R01MH120080 to A.J.H. and B.T.T.Y.). E.R.O. and S.C. are supported by an American Australian Association Graduate Education Fund Scholarship and the University of Melbourne McKenzie Fellowship. E.D. is supported by the Yale University Kavli Institute for Neuroscience Postdoctoral Fellowship for Academic Diversity, the Northwell Health/Feinstein Institutes for Medical Research Advancing Women in Science and Medicine Career Development Award, and the Feinstein Institutes for Medical Research Emerging Scientist Award. A.F. was supported by the Sylvia and Charles Viertel Foundation, National Health and Medical Research Council (IDs: 1197431 and 1146292), and Australian Research Council (IDs: DP200103509). B.T.T.Y. is supported by the Singapore National Research Foundation (NRF) Fellowship (Class of 2017), the NUS Yong Loo Lin School of Medicine (NUHSRO/2020/124/TMR/LOA), the Singapore National Medical Research Council (NMRC) LCG (OFLCG19May-0035), NMRC STaR (STaR20nov-0003), Singapore Ministry of Health (MOH) Centre Grant (CG21APR1009), and the US National Institutes of Health (R01MH120080). Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not reflect the views of the Singapore NRF, NMRC, or MOH. K.A. is a scientific advisor to and shareholder in BrainKey Inc., a medical image analysis software company. This research has been conducted using the UK Biobank Resource under application number 25163.
Author contributions: Conceptualization: S.C. and A.J.H. Data curation: S.C., C.L., J.A.R., J.M., L.C., P.L., A.H., K.A., A.F., I.H.-R., L.T.G., J.T.B., B.T.T.Y., and A.J.H. Formal analysis: S.C. Funding acquisition: J.T.B., B.T.T.Y., and A.J.H. Methodology: S.C., E.D., L.A., P.C., N.W., B.T.T.Y., and A.J.H. Supervision: A.J.H. Visualization: S.C. Writing—original draft: S.C. Writing—review and editing: S.C., E.D., C.L., E.R.O., L.A., P.C., N.W., P.K., A.R., J.M., L.C., P.L., A.H., K.A., A.F., I.H.-R., L.T.G., J.T.B., B.T.T.Y., and A.J.H.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. This study used four publicly available datasets: (i) the UK Biobank (https://ukbiobank.ac.uk/), (ii) the HCP-EP (https://humanconnectome.org/study/human-connectome-project-for-early-psychosis), (iii) the TCP (https://openneuro.org/datasets/ds005237), and (iv) the UCLA CNP project (https://openfmri.org/dataset/ds000030/). All codes used for analysis and figure generation can be found online at https://zenodo.org/records/7596453 and https://github.com/sidchop/PredictingCognition.
Supplementary Materials
This PDF file includes:
Figs. S1 to S13
Tables S1 to S3
Additional information on TCP dataset
Detailed information on functional MRI processing, denoising, and quality control
References
REFERENCES AND NOTES
- 1.Poldrack R. A., Huckins G., Varoquaux G., Establishment of best practices for evidence for prediction: A review. JAMA Psychiatry 77, 534–540 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Varoquaux G., Cross-validation failure: Small sample sizes lead to large error bars. Neuroimage 180, 68–77 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Whelan R., Garavan H., When optimism hurts: Inflated predictions in psychiatric neuroimaging. Biol. Psychiatry 75, 746–748 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Abramovitch A., Short T., Schweiger A., The C Factor: Cognitive dysfunction as a transdiagnostic dimension in psychopathology. Clin. Psychol. Rev. 86, 102007 (2021). [DOI] [PubMed] [Google Scholar]
- 5.East-Richard C., Mercier A. R., Nadeau D., Cellard C., Transdiagnostic neurocognitive deficits in psychiatry: A review of meta-analyses. Can. Psychol. 61, 190–214 (2020). [Google Scholar]
- 6.McTeague L. M., Goodkind M. S., Etkin A., Transdiagnostic impairment of cognitive control in mental illness. J. Psychiatr. Res. 83, 37–46 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Millan M. J., Agid Y., Brüne M., Bullmore E. T., Carter C. S., Clayton N. S., Connor R., Davis S., Deakin B., DeRubeis R. J., Dubois B., Geyer M. A., Goodwin G. M., Gorwood P., Jay T. M., Joëls M., Mansuy I. M., Meyer-Lindenberg A., Murphy D., Rolls E., Saletu B., Spedding M., Sweeney J., Whittington M., Young L. J., Cognitive dysfunction in psychiatric disorders: Characteristics, causes and the quest for improved therapy. Nat. Rev. Drug Discov. 11, 141–168 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Shilyansky C., Williams L. M., Gyurak A., Harris A., Usherwood T., Etkin A., Effect of antidepressant treatment on cognitive impairments associated with depression: A randomised longitudinal study. Lancet Psychiatry 3, 425–435 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mohamed S., Rosenheck R., Swartz M., Stroup S., Lieberman J. A., Keefe R. S., Relationship of cognition and psychopathology to functional impairment in schizophrenia. Am. J. Psychiatry 165, 978–987 (2008). [DOI] [PubMed] [Google Scholar]
- 10.Green M. F., Cognitive impairment and functional outcome in schizophrenia and bipolar disorder. J. Clin. Psychiatry 67, 3 (2006). [PubMed] [Google Scholar]
- 11.Diamond A., Executive functions. Annu. Rev. Psychol. 64, 135–168 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fitzgerald H. M., Shepherd J., Bailey H., Berry M., Wright J., Chen M., Treatment goals in schizophrenia: A real-world survey of patients, psychiatrists, and caregivers in the United States, with an analysis of current treatment (long-acting injectable vs oral antipsychotics) and goal selection. Neuropsychiatr. Dis. Treat. 17, 3215–3228 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McNaughton E. C., Curran C., Granskie J., Opler M., Sarkey S., Mucha L., Eramo A., Francois C., Webber-Lind B., McCue M., Patient attitudes toward and goals for MDD treatment: A survey study. Patient Prefer. Adherence 13, 959–967 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xia C. H., Ma Z., Ciric R., Gu S., Betzel R. F., Kaczkurkin A. N., Calkins M. E., Cook P. A., García de la Garza A., Vandekar S. N., Cui Z., Moore T. M., Roalf D. R., Ruparel K., Wolf D. H., Davatzikos C., Gur R. C., Gur R. E., Shinohara R. T., Bassett D. S., Satterthwaite T. D., Linked dimensions of psychopathology and connectivity in functional brain networks. Nat. Commun. 9, 3003 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elliott M. L., Romer A., Knodt A. R., Hariri A. R., A connectome-wide functional signature of transdiagnostic risk for mental illness. Biol. Psychiatry 84, 452–459 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vanes L. D., Dolan R. J., Transdiagnostic neuroimaging markers of psychiatric risk: A narrative review. Neuroimage 30, 102634 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anticevic A., Cole M. W., Murray J. D., Corlett P. R., Wang X.-J., Krystal J. H., The role of default network deactivation in cognition and disease. Trends Cogn. Sci. 16, 584–592 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chopra S., Francey S. M., O’Donoghue B., Sabaroedin K., Arnatkeviciute A., Cropley V., Nelson B., Graham J., Baldwin L., Tahtalian S., Yuen H. P., Allott K., Alvarez-Jimenez M., Harrigan S., Pantelis C., Wood S. J., McGorry P., Fornito A., Functional connectivity in antipsychotic-treated and antipsychotic-naive patients with first-episode psychosis and low risk of self-harm or aggression: A secondary analysis of a randomized clinical trial. JAMA Psychiatry 78, 994–1004 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vincent J. L., Kahn I., Snyder A. Z., Raichle M. E., Buckner R. L., Evidence for a frontoparietal control system revealed by intrinsic functional connectivity. J. Neurophysiol. 100, 3328–3342 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang C.-C., Luo Q., Palaniyappan L., Yang A. C., Hung C.-C., Chou K.-H., Lo C.-Y. Z., Liu M.-N., Tsai S.-J., Barch D. M., Feng J., Lin C. P., Robbins T. W., Transdiagnostic and illness-specific functional dysconnectivity across schizophrenia, bipolar disorder, and major depressive disorder. Biol. Psychiatry Cogn. Neurosci. Neuroimaging 5, 542–553 (2020). [DOI] [PubMed] [Google Scholar]
- 21.Fornito A., Yoon J., Zalesky A., Bullmore E. T., Carter C. S., General and specific functional connectivity disturbances in first-episode schizophrenia during cognitive control performance. Biol. Psychiatry 70, 64–72 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baker J. T., Dillon D. G., Patrick L. M., Roffman J. L., Brady R. O., Pizzagalli D. A., Öngür D., Holmes A. J., Functional connectomics of affective and psychotic pathology. Proc. Natl. Acad. Sci. U.S.A. 116, 9050–9059 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dhamala E., Jamison K. W., Jaywant A., Dennis S., Kuceyeski A., Distinct functional and structural connections predict crystallised and fluid cognition in healthy adults. Hum. Brain Mapp. 42, 3102–3118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He T., Kong R., Holmes A. J., Nguyen M., Sabuncu M. R., Eickhoff S. B., Bzdok D., Feng J., Yeo B. T., Deep neural networks and kernel regression achieve comparable accuracies for functional connectivity prediction of behavior and demographics. Neuroimage 206, 116276 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sripada C., Rutherford S., Angstadt M., Thompson W. K., Luciana M., Weigard A., Hyde L. H., Heitzeg M., Prediction of neurocognition in youth from resting state fMRI. Mol. Psychiatry 25, 3413–3421 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Finn E. S., Shen X., Scheinost D., Rosenberg M. D., Huang J., Chun M. M., Papademetris X., Constable R. T., Functional connectome fingerprinting: Identifying individuals using patterns of brain connectivity. Nat. Neurosci. 18, 1664–1671 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang R., Calhoun V. D., Fan L., Zuo N., Jung R., Qi S., Lin D., Li J., Zhuo C., Song M., Fu Z., Jiang T., Sui J., Gender differences in connectome-based predictions of individualized intelligence quotient and sub-domain scores. Cereb. Cortex 30, 888–900 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marek S., Tervo-Clemmens B., Calabro F. J., Montez D. F., Kay B. P., Hatoum A. S., Donohue M. R., Foran W., Miller R. L., Hendrickson T. J., Malone S. M., Kandala S., Feczko E., Miranda-Dominguez O., Graham A. M., Earl E. A., Perrone A. J., Cordova M., Doyle O., Moore L. A., Conan G. M., Uriarte J., Snider K., Lynch B. J., Wilgenbusch J. C., Pengo T., Tam A., Chen J., Newbold D. J., Zheng A., Seider N. A., Van A. N., Metoki A., Chauvin R. J., Laumann T. O., Greene D. J., Petersen S. E., Garavan H., Thompson W. K., Nichols T. E., Yeo B. T. T., Barch D. M., Luna B., Fair D. A., Dosenbach N. U. F., Reproducible brain-wide association studies require thousands of individuals. Nature 603, 654–660 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schulz M.-A., Yeo B., Vogelstein J. T., Mourao-Miranada J., Kather J. N., Kording K., Richards B., Bzdok D., Different scaling of linear models and deep learning in UKBiobank brain images versus machine-learning datasets. Nat. Commun. 11, 4238 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Helmer M., Warrington S., Mohammadi-Nejad A.-R., Ji J. L., Howell A., Rosand B., Anticevic A., Sotiropoulos S. N., Murray J. D., On the stability of canonical correlation analysis and partial least squares with application to brain-behavior associations. Commun. Biol. 7, 217 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haufe S., Meinecke F., Görgen K., Dähne S., Haynes J.-D., Blankertz B., Bießmann F., On the interpretation of weight vectors of linear models in multivariate neuroimaging. Neuroimage 87, 96–110 (2014). [DOI] [PubMed] [Google Scholar]
- 32.Váša F., Mišić B., Null models in network neuroscience. Nat. Rev. Neurosci. 23, 493–504 (2022). [DOI] [PubMed] [Google Scholar]
- 33.Tozzi L., Fleming S. L., Taylor Z. D., Raterink C. D., Williams L. M., Test-retest reliability of the human functional connectome over consecutive days: Identifying highly reliable portions and assessing the impact of methodological choices. Netw. Neurosci. 4, 925–945 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zuo X.-N., Xing X.-X., Test-retest reliabilities of resting-state FMRI measurements in human brain functional connectomics: A systems neuroscience perspective. Neurosci. Biobehav. Rev. 45, 100–118 (2014). [DOI] [PubMed] [Google Scholar]
- 35.Smith S. M., Nichols T. E., Vidaurre D., Winkler A. M., Behrens T. E., Glasser M. F., Ugurbil K., Barch D. M., Van Essen D. C., Miller K. L., A positive-negative mode of population covariation links brain connectivity, demographics and behavior. Nat. Neurosci. 18, 1565–1567 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen J., Tam A., Kebets V., Orban C., Ooi L. Q. R., Asplund C. L., Marek S., Dosenbach N. U., Eickhoff S. B., Bzdok D., Holmes A. J., Yeo B. T. T., Shared and unique brain network features predict cognitive, personality, and mental health scores in the ABCD study. Nat. Commun. 13, 1–17 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller K. L., Alfaro-Almagro F., Bangerter N. K., Thomas D. L., Yacoub E., Xu J., Bartsch A. J., Jbabdi S., Sotiropoulos S. N., Andersson J. L., Griffanti L., Douaud G., Okell T. W., Weale P., Dragonu I., Garratt S., Hudson S., Collins R., Jenkinson M., Matthews P. M., Smith S. M., Multimodal population brain imaging in the UK Biobank prospective epidemiological study. Nat. Neurosci. 19, 1523–1536 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He T., An L., Chen P., Chen J., Feng J., Bzdok D., Holmes A. J., Eickhoff S. B., Yeo B., Meta-matching as a simple framework to translate phenotypic predictive models from big to small data. Nat. Neurosci. 25, 795–804 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schaefer A., Kong R., Gordon E. M., Laumann T. O., Zuo X.-N., Holmes A. J., Eickhoff S. B., Yeo B. T. T., Local-global parcellation of the human cerebral cortex from intrinsic functional connectivity MRI. Cereb. Cortex 28, 3095–3114 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fischl B., Salat D. H., Busa E., Albert M., Dieterich M., Haselgrove C., Van Der Kouwe A., Killiany R., Kennedy D., Klaveness S., Montillo A., Makris N., Rosen B., Dale A. M., Whole brain segmentation: Automated labeling of neuroanatomical structures in the human brain. Neuron 33, 341–355 (2002). [DOI] [PubMed] [Google Scholar]
- 41.Ooi L. Q. R., Chen J., Zhang S., Kong R., Tam A., Li J., Dhamala E., Zhou J. H., Holmes A. J., Yeo B. T., Comparison of individualized behavioral predictions across anatomical, diffusion and functional connectivity MRI. Neuroimage 263, 119636 (2022). [DOI] [PubMed] [Google Scholar]
- 42.Tian Y., Zalesky A., Machine learning prediction of cognition from functional connectivity: Are feature weights reliable? Neuroimage 245, 118648 (2021). [DOI] [PubMed] [Google Scholar]
- 43.Chen J., Ooi L. Q. R., Kiat Tan T. W., Zhang S., Li J., Asplund C. L., Eickhoff S. B., Bzdok D., Holmes A. J., Yeo B. T., Relationship between prediction accuracy and feature importance reliability: An empirical and theoretical study. Neuroimage 274, 120115 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cole M. W., Repovš G., Anticevic A., The frontoparietal control system: A central role in mental health. Neuroscientist 20, 652–664 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zanto T. P., Gazzaley A., Fronto-parietal network: Flexible hub of cognitive control. Trends Cogn. Sci. 17, 602–603 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smallwood J., Brown K., Baird B., Schooler J. W., Cooperation between the default mode network and the frontal–parietal network in the production of an internal train of thought. Brain Res. 1428, 60–70 (2012). [DOI] [PubMed] [Google Scholar]
- 47.Yeo B. T., Krienen F. M., Sepulcre J., Sabuncu M. R., Lashkari D., Hollinshead M., Roffman J. L., Smoller J. W., Zöllei L., Polimeni J. R., Fischl B., Liu H., Buckner R. L., The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J. Neurophysiol. 106, 1125–1165 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.P. Flechsig, Die Localisation der geistigen Vorgänge insbesondere der Sinnesempfindungen des Menschen (De Gruyter, 1896). [Google Scholar]
- 49.Goldman-Rakic P. S., Topography of cognition: Parallel distributed networks in primate association cortex. Annu. Rev. Neurosci. 11, 137–156 (1988). [DOI] [PubMed] [Google Scholar]
- 50.Badre D., Nee D. E., Frontal cortex and the hierarchical control of behavior. Trends Cogn. Sci. 22, 170–188 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.K. Allott, A. Lin, “Cognitive risk factors for psychosis” in Risk Factors for Psychosis (Elsevier, 2020), pp. 269–287. [Google Scholar]
- 52.Catalan A., De Pablo G. S., Aymerich C., Damiani S., Sordi V., Radua J., Oliver D., McGuire P., Giuliano A. J., Stone W. S., Fusar-Poli P., Neurocognitive functioning in individuals at clinical high risk for psychosis: A systematic review and meta-analysis. JAMA Psychiatry 78, 859–867 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tong X., Xie H., Carlisle N., Fonzo G. A., Oathes D. J., Jiang J., Zhang Y., Transdiagnostic connectome signatures from resting-state fMRI predict individual-level intellectual capacity. Transl. Psychiatry 12, 367 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boeke E. A., Holmes A. J., Phelps E. A., Toward robust anxiety biomarkers: A machine learning approach in a large-scale sample. Biol. Psychiatry Cogn. Neurosci. Neuroimaging 5, 799–807 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dhamala E., Ooi L. Q. R., Chen J., Kong R., Anderson K., Chin R., Yeo B. T., Holmes A., Proportional intracranial volume correction differentially biases behavioral predictions across neuroanatomical features and populations. Neuroimage 260, 119485 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Park H.-J., Friston K., Structural and functional brain networks: From connections to cognition. Science 342, 1238411 (2013). [DOI] [PubMed] [Google Scholar]
- 57.Petersen S. E., Sporns O., Brain networks and cognitive architectures. Neuron 88, 207–219 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Segal A., Parkes L., Aquino K., Kia S. M., Wolfers T., Franke B., Hoogman M., Beckmann C. F., Westlye L. T., Andreassen O. A., Zalesky A., Harrison B. J., Davey C., Soriano-Mas C., Cardoner N., Tiego J., Yücel M., Braganza L., Suo C., Berk M., Cotton S., Bellgrove M. A., Marquand A. F., Fornito A., Regional, circuit, and network heterogeneity of brain abnormalities in psychiatric disorders. Nat. Neurosci. 26, 1613–1629 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.M. Y. Chan, “Age-related desegregation of functional systems in healthy adults: The underlying patterns of connections and protective life-course factors” thesis, The University of Texas at Dallas, Richardson, TX (2016). [Google Scholar]
- 60.Kong T. S., Gratton C., Low K. A., Tan C. H., Chiarelli A. M., Fletcher M. A., Zimmerman B., Maclin E. L., Sutton B. P., Gratton G., Fabiani M., Age-related differences in functional brain network segregation are consistent with a cascade of cerebrovascular, structural, and cognitive effects. Netw. Neurosci. 4, 89–114 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Johnson W., Bouchard T. J. Jr., Krueger R. F., McGue M., Gottesman I. I., Just one g: Consistent results from three test batteries. Intelligence 32, 95–107 (2004). [Google Scholar]
- 62.Fawns-Ritchie C., Deary I. J., Reliability and validity of the UK Biobank cognitive tests. PLOS ONE 15, e0231627 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Savage J. E., Jansen P. R., Stringer S., Watanabe K., Bryois J., De Leeuw C. A., Nagel M., Awasthi S., Barr P. B., Coleman J. R., Grasby K. L., Hammerschlag A. R., Kaminski J. A., Karlsson R., Krapohl E., Lam M., Nygaard M., Reynolds C. A., Trampush J. W., Young H., Zabaneh D., Hägg S., Hansell N. K., Karlsson I. K., Linnarsson S., Montgomery G. W., Muñoz-Manchado A. B., Quinlan E. B., Schumann G., Skene N. G., Webb B. T., White T., Arking D. E., Avramopoulos D., Bilder R. M., Bitsios P., Burdick K. E., Cannon T. D., Chiba-Falek O., Christoforou A., Cirulli E. T., Congdon E., Corvin A., Davies G., Deary I. J., DeRosse P., Dickinson D., Djurovic S., Donohoe G., Conley E. D., Eriksson J. G., Espeseth T., Freimer N. A., Giakoumaki S., Giegling I., Gill M., Glahn D. C., Hariri A. R., Hatzimanolis A., Keller M. C., Knowles E., Koltai D., Konte B., Lahti J., Le Hellard S., Lencz T., Liewald D. C., London E., Lundervold A. J., Malhotra A. K., Melle I., Morris D., Need A. C., Ollier W., Palotie A., Payton A., Pendleton N., Poldrack R. A., Räikkönen K., Reinvang I., Roussos P., Rujescu D., Sabb F. W., Scult M. A., Smeland O. B., Smyrnis N., Starr J. M., Steen V. M., Stefanis N. C., Straub R. E., Sundet K., Tiemeier H., Voineskos A. N., Weinberger D. R., Widen E., Yu J., Abecasis G., Andreassen O. A., Breen G., Christiansen L., Debrabant B., Dick D. M., Heinz A., Hjerling-Leffler J., Ikram M. A., Kendler K. S., Martin N. G., Medland S. E., Pedersen N. L., Plomin R., Polderman T. J. C., Ripke S., van der Sluis S., Sullivan P. F., Vrieze S. I., Wright M. J., Posthuma D., Genome-wide association meta-analysis in 269,867 individuals identifies new genetic and functional links to intelligence. Nat. Genet. 50, 912–919 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Noble S., Scheinost D., Constable R. T., A decade of test-retest reliability of functional connectivity: A systematic review and meta-analysis. Neuroimage 203, 116157 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.L. Q. R. Ooi, C. Orban, T. E. Nichols, S. Zhang, T. W. K. Tan, R. Kong, S. Marek, N. U. Dosenbach, T. Laumann, E. M. Gordon, J. H. Zhou, D. Bzdok, S. B. Eickhoff, A. J Holmes, B. T. T. Yeo; Alzheimer’s Disease Neuroimaging Initiative, MRI economics: Balancing sample size and scan duration in brain wide association studies. bioRxiv 580448 [Preprint] (2024). 10.1101/2024.02.16.580448. [DOI]
- 66.Chen J., Ooi L. Q. R., Tan T. W. K., Zhang S., Li J., Asplund C. L., Eickhoff S. B., Bzdok D., Holmes A. J., Yeo B. T. T., Relationship between prediction accuracy and feature importance reliability: An empirical and theoretical study. NeuroImage 274, 120115 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bycroft C., Freeman C., Petkova D., Band G., Elliott L. T., Sharp K., Motyer A., Vukcevic D., Delaneau O., O’Connell J., Cortes A., Welsh S., Young A., Effingham M., McVean G., Leslie S., Allen N., Donnelly P., Marchini J., The UK Biobank resource with deep phenotyping and genomic data. Nature 562, 203–209 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lewandowski K. E., Bouix S., Ongur D., Shenton M. E., Neuroprogression across the early course of psychosis. J. Psychiatr. Brain Sci. 5, e200002 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.S. Chopra, C. V. Cocuzza, C. Lawhead, J. A. Ricard, L. Labache, L. M. Patrick, P. Kumar, A. Rubenstein, J. Moses, L. Chen, C. Blankenbaker, B. Gillis, L. T. Germine, I. Harpaz-Rote, B. T. T. Yeo, J. T. Baker, A. J. Holmes, The Transdiagnostic Connectome Project: A richly phenotyped open dataset for advancing the study of brain-behavior relationships in psychiatry. medRxiv 24309054 [Preprint] (2024). 10.1101/2024.06.18.24309054. [DOI]
- 70.Poldrack R. A., Congdon E., Triplett W., Gorgolewski K., Karlsgodt K., Mumford J., Sabb F., Freimer N., London E., Cannon T., Bilder R. M., A phenome-wide examination of neural and cognitive function. Sci. Data 3, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Power J. D., Lynch C. J., Silver B. M., Dubin M. J., Martin A., Jones R. M., Distinctions among real and apparent respiratory motions in human fMRI data. Neuroimage 201, 116041 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fair D. A., Miranda-Dominguez O., Snyder A. Z., Perrone A., Earl E. A., Van A. N., Koller J. M., Feczko E., Tisdall M. D., van der Kouwe A., Klein R. L., Mirro A. E., Hampton J. M., Adeyemo B., Laumann T. O., Gratton C., Greene D. J., Schlaggar B. L., Hagler D. J. Jr., Watts R., Garavan H., Barch D. M., Nigg J. T., Petersen S. E., Dale A. M., Feldstein-Ewing S. W., Nagel B. J., Dosenbach N. U. F., Correction of respiratory artifacts in MRI head motion estimates. Neuroimage 208, 116400 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Power J. D., Mitra A., Laumann T. O., Snyder A. Z., Schlaggar B. L., Petersen S. E., Methods to detect, characterize, and remove motion artifact in resting state fMRI. Neuroimage 84, 320–341 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Parkes L., Fulcher B., Yücel M., Fornito A., An evaluation of the efficacy, reliability, and sensitivity of motion correction strategies for resting-state functional MRI. Neuroimage 171, 415–436 (2018). [DOI] [PubMed] [Google Scholar]
- 75.Li J., Kong R., Liégeois R., Orban C., Tan Y., Sun N., Holmes A. J., Sabuncu M. R., Ge T., Yeo B. T., Global signal regression strengthens association between resting-state functional connectivity and behavior. Neuroimage 196, 126–141 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Weintraub S., Dikmen S. S., Heaton R. K., Tulsky D. S., Zelazo P. D., Bauer P. J., Carlozzi N. E., Slotkin J., Blitz D., Wallner-Allen K., Fox N. A., Beaumont J. L., Mungas D., Nowinski C. J., Richler J., Deocampo J. A., Anderson J. E., Manly J. J., Borosh B., Havlik R., Conway K., Edwards E., Freund L., King J. W., Moy C., Witt E., Gershon R. C., Cognition assessment using the NIH Toolbox. Neurology 80, S54–S64 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.D. Wechsler, Wechsler Abbreviated Scale of Intelligence (Pearson Education, 1999). [Google Scholar]
- 78.Singh S., Strong R. W., Jung L., Li F. H., Grinspoon L., Scheuer L. S., Passell E. J., Martini P., Chaytor N., Soble J. R., The TestMyBrain digital neuropsychology toolkit: Development and psychometric characteristics. J. Clin. Exp. Neuropsychol. 43, 786–795 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.D. Wechsler, Wechsler Memory Scale (Psychological Corporation, 1945). [Google Scholar]
- 80.D. Wechsler, Wechsler Adult Intelligence Scale, Archives of Clinical Neuropsychology (Psychological Corporation, 1955). [Google Scholar]
- 81.Dhamala E., Ooi L. Q. R., Chen J., Ricard J. A., Berkeley E., Chopra S., Qu Y., Lawhead C., Yeo B. T. T., Holmes A. J., Brain-based predictions of psychiatric illness-linked behaviors across the sexes. Biol. Psychiatry. 94, 479–491 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.R. X. Rodriguez, S. Noble, C. C. Camp, D. Scheinost, Connectome caricatures: Removing large-amplitude co-activation patterns in resting-state fMRI emphasizes individual differences. bioRxiv 588578 [Preprint] (2024). 10.1101/2024.04.08.588578. [DOI]
- 83.A. Mihalik, M. Brudfors, M. Robu, F. S. Ferreira, H. Lin, A. Rau, T. Wu, S. B. Blumberg, B. Kanber, M. Tariq, “ABCD neurocognitive prediction challenge 2019: Predicting individual fluid intelligence scores from structural MRI using probabilistic segmentation and kernel ridge regression” in Challenge in Adolescent Brain Cognitive Development Neurocognitive Prediction (Springer, 2019), pp. 133–142. [Google Scholar]
- 84.Chen P., An L., Wulan N., Zhang C., Zhang S., Ooi L. Q. R., Kong R., Chen J., Wu J., Chopra S., Bzdok D., Eickhoff S. B., Holmes A. J., Yeo B. T. T., Multilayer meta-matching: translating phenotypic prediction models from multiple datasets to small data. Imaging Neurosci. 2, 1–22 (2024). [Google Scholar]
- 85.Raghunathan T. E., Rosenthal R., Rubin D. B., Comparing correlated but nonoverlapping correlations. Psychol. Methods 1, 178–183 (1996). [Google Scholar]
- 86.Diedenhofen B., Musch J., cocor: A comprehensive solution for the statistical comparison of correlations. PLOS ONE 10, e0121945 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dhamala E., Yeo B. T., Holmes A. J., One size does not fit all: Methodological considerations for brain-based predictive modeling in psychiatry. Biol. Psychiatry 93, 717–728 (2023). [DOI] [PubMed] [Google Scholar]
- 88.Chyzhyk D., Varoquaux G., Milham M., Thirion B., How to remove or control confounds in predictive models, with applications to brain biomarkers. Gigascience 11, giac014 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Alfaro-Almagro F., Jenkinson M., Bangerter N. K., Andersson J. L., Griffanti L., Douaud G., Sotiropoulos S. N., Jbabdi S., Hernandez-Fernandez M., Vallee E., Vidaurre D., Webster M., McCarthy P., Rorden C., Daducci A., Alexander D. C., Zhang H., Dragonu I., Matthews P. M., Miller K. L., Smith S. M., Image processing and Quality Control for the first 10,000 brain imaging datasets from UK Biobank. Neuroimage 166, 400–424 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Salimi-Khorshidi G., Douaud G., Beckmann C. F., Glasser M. F., Griffanti L., Smith S. M., Automatic denoising of functional MRI data: Combining independent component analysis and hierarchical fusion of classifiers. Neuroimage 90, 449–468 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Esteban O., Markiewicz C. J., Blair R. W., Moodie C. A., Isik A. I., Erramuzpe A., Kent J. D., Goncalves M., DuPre E., Snyder M., Oya H., Ghosh S. S., Wright J., Durnez J., Poldrack R. A., Gorgolewski K. J., fMRIPrep: A robust preprocessing pipeline for functional MRI. Nat. Methods 16, 111–116 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tustison N. J., Avants B. B., Cook P. A., Zheng Y., Egan A., Yushkevich P. A., Gee J. C., N4ITK: Improved N3 bias correction. IEEE Trans. Med. Imaging 29, 1310–1320 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Avants B. B., Tustison N., Song G., Advanced normalization tools (ANTS). Insight J. 2, 1–35 (2009). [Google Scholar]
- 94.Zhang Y., Brady M., Smith S., Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE Trans. Med. Imaging 20, 45–57 (2001). [DOI] [PubMed] [Google Scholar]
- 95.Cox R. W., AFNI: Software for analysis and visualization of functional magnetic resonance neuroimages. Comput. Biomed. Res. 29, 162–173 (1996). [DOI] [PubMed] [Google Scholar]
- 96.Jenkinson M., Bannister P., Brady M., Smith S., Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage 17, 825–841 (2002). [DOI] [PubMed] [Google Scholar]
- 97.Wang S., Peterson D. J., Gatenby J. C., Li W., Grabowski T. J., Madhyastha T. M., Evaluation of field map and nonlinear registration methods for correction of susceptibility artifacts in diffusion MRI. Front. Neuroinform. 11, 17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pruim R. H. R., Mennes M., van Rooij D., Llera A., Buitelaar J. K., Beckmann C. F., ICA-AROMA: A robust ICA-based strategy for removing motion artifacts from fMRI data. Neuroimage 112, 267–277 (2015). [DOI] [PubMed] [Google Scholar]
- 99.Aquino K. M., Fulcher B. D., Parkes L., Sabaroedin K., Fornito A., Identifying and removing widespread signal deflections from fMRI data: Rethinking the global signal regression problem. Neuroimage 212, 116614 (2020). [DOI] [PubMed] [Google Scholar]
- 100.Glasser M. F., Sotiropoulos S. N., Wilson J. A., Coalson T. S., Fischl B., Andersson J. L., Xu J., Jbabdi S., Webster M., Polimeni J. R., Van Essen D. C., Jenkinson M.; WU-Minn HCP Consortium , The minimal preprocessing pipelines for the Human Connectome Project. Neuroimage 80, 105–124 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jenkinson M., Beckmann C. F., Behrens T. E., Woolrich M. W., Smith S. M., FSL. Neuroimage 62, 782–790 (2012). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S13
Tables S1 to S3
Additional information on TCP dataset
Detailed information on functional MRI processing, denoising, and quality control
References