Abstract
A plausible, but largely unexplored reason for many weed biocontrol agents failing to establish or being ineffective, could involve abiotically induced changes to an invasive plants’ biochemical phenotype and consequent enhanced herbivore resistance. Considerable literature demonstrates that chemically altered plant phenotypes can impair insect life history performance. Heather beetle, (Lochmaea suturalis), introduced to control invasive heather (Calluna vulgaris) in New Zealand (NZ) was difficult to establish and displays variable effectiveness. Using UHPLC-MS non-targeted metabolomics, we analysed primary and secondary metabolites of C. vulgaris from its native range (Scotland) and it’s introduced range (NZ), between which, differences in soil nutrients and ultraviolet light exist. We also explored secondary metabolite variation between sites within each range. New Zealand samples had the highest number of amplified metabolites, most notably defensive phenylpropanoids, supporting the concept of abiotically induced upregulation of key biosynthetic pathways. Analysis of secondary metabolite variation within each range revealed differences between sites but found little correlation of phenylpropanoid levels being influenced by variable soil nutrients. These results validate questions about the possibility of abiotically altered biochemical phenotypes in invasive plants, influencing weed biocontrol agent establishment and effectiveness, and show the potential for metabolomics in assisting future, or retrospectively analysing biological control programmes.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-76228-w.
Subject terms: Chemical biology, Ecology
Introduction
Globally, invasive plants are a serious threat to the terrestrial habitats they invade, causing considerable economic loss in managed production systems such as agriculture, horticulture and forestry or ecological perturbations in environmentally sensitive natural systems including communal rangelands and conservation lands which provide essential ecosystem services1. In most instances such intrusions cumulatively result in losses of production and biodiversity at the species, community, or ecosystem level2.
For many of these ecosystems, the introduction of insect or pathogen biocontrol agents sourced from the plant’s native range (classical biocontrol) offers a sustainable long-term management alternative for invasive plants3,4. However, some biocontrol programmes do not achieve the desired goals due to agents either failing to establish3,5, being ineffective once they are established6,7 or display variable effectiveness between sites8. While evidence suggests biocontrol agent population establishment is likely influenced by many environmentally driven factors9–11, there is a paucity of literature considering target plant biochemistry and how changes in its genotype or biochemical phenotype might act as a component allee effect and impose limitations or extinctions on control agent establishment, particularly while at vulnerable low population numbers12. Likewise, literature addressing target plant biochemistry on agent ineffectiveness or variable inter-site effectiveness is sparse (but see8).
In natural habitats, plants as sessile organisms are simultaneously subjected to multiple stressors to which appropriate responses are required for establishment, growth, and survival13–15. Considerable evidence relating to abiotic and biotic stresses on plants exists16–18, demonstrating that plants are capable of responding to such stresses via a plastic and finely balanced response network involving activation of stress responsive genes which regulate phytohormone production, redox signalling pathways, growth, and calcium and protein kinase cascades, resulting in changes to both primary and secondary metabolism17,19,20, all of which are key endogenous components in mediating plant stress responses.
Plant responses resulting from adaptive changes driven by the genotype under changed environmental conditions can manifest either independently or as a combination of morphologically, physiologically, or biochemically altered phenotypes broadly termed phenotypic plasticity21–23. Phenotypic plasticity is thought to be a common characteristic of invasive plants24, but experimental evidence investigating this is equivocal25, citing it is often limited to the early stages of establishment26 and constrained within the ecological limits of the sites studied14,27,28.
Nevertheless, some recent publications have demonstrated abiotically induced alterations to the metabolome of invasive plants when compared with their conspecifics in the native range. Such alterations generally result in increased levels of various defensive secondary metabolites29–31, suggesting that this may not be an uncommon phenomenon.
Heather, (Calluna vulgaris) is an invasive shrub introduced from Europe now widely established on the North Islands Central Plateau (CP) of New Zealand (NZ), where it impacts this fragile sub-alpine environment32–34. The heather beetle (Lochmaea suturalis) (Coleoptera : Chrysomelidae) sourced and introduced from the United Kingdom (UK) in 1996 as a biocontrol agent, required multiple releases over several years before establishment, does not achieve the population densities of its home range in the UK and Europe and currently displays variable effectiveness within the CP region. A long-term research effort has investigated several factors to explain these scenarios with only low foliar nitrogen in CP heather providing a definitive explanation35–38, but potential changes in plant defensive biochemistry had not been explored. We hypothesize that the plastic nature of plant biochemical responses induced potentially by abiotic factors may cause introduced biocontrol agents, to encounter plants with altered defensive biochemistry compared to those plants from where the agent was sourced. We aim therefore to explore the biochemical phenotype of this plant in the invaded range (NZ) and its native range (UK), from where the biocontrol agent was sourced and we further aimed to explore such phenotypic variability between sites within each range.
In the CP, mean seasonal temperatures for this higher altitude region, while displaying slightly greater daily variation of maximums and minimums, are very closely matched to the higher latitudes of Scotland in the UK. The CP region however experiences a longer winter period (3–4 weeks) than that of northern UK latitudes39, but spring and early summer mean monthly temperatures, i.e., the growing period for C. vulgaris, are very closely matched. We assume therefore that C. vulgaris in this region experiences climatic conditions very similar to its northern hemisphere conspecifics. We hypothesize that two abiotic parameters of this region that may induce long term permanent changes to the biochemical profile of C. vulgaris are soil nutrients and light.
Soil nutrient availability (depending on the underlying geology) and long term increased chemical defences in plants (most notably phenylpropanoids such as polyphenolics, flavonoid glycosides, flavones and coumarins), are well documented40–45. Soils of heather dominated heathlands in the UK and Europe range from free draining to wet, are acidic and low in nutrients, especially phosphate and nitrogen46. Similarly, the young volcanic soils of the CP region are generally free draining but with levels even lower in phosphate and nitrogen38,47.
Light quantity (intensity) and quality (the balance of PAR, UV-A and UV-B), which is dependent on latitude and altitude48,49, also induce long term changes to the plant biochemical phenotype by modulating the jasmonate (JA) and abscisic acid (ABA) dependant pathways thus inducing Shikimate-phenylpropanoid derived flavonoids and phenolic acids. These metabolites play an important role in plant photoprotection but have also been shown to enhance plant defences against insect herbivores50–53 or provide systemically acquired resistance to biotrophic pathogens54,55. Light intensity and ambient UV are significantly different between the two regions, with peak summertime UV index figures of 12 to 13 at the North Island CP being approximately double that of higher latitude regions of the UK at 6 to 748,49. Furthermore, previous work demonstrating plasticity of secondary metabolites in C. vulgaris at varying altitudes56 and seasonally57 in Europe and in response to multiple abiotic factors in New Zealand58,59, indicates that this plant species readily responds to changing parameters in its environment, making it perhaps an ideal model species to explore.
In recent years, high throughput mass spectral chromatographic technology in conjunction with online cheminformatic platforms has markedly progressed metabolomic analytical capability. Metabolomics makes it feasible to now study plant biochemistry at the molecular level, providing a targeted or non-targeted characterisation and quantification of currently identifiable metabolites (the metabolome), in a particular plant tissue in response to its environment or treatment37,60–64.
In this study, we applied non-targeted metabolomics to investigate possible changes to both primary and secondary metabolites of C. vulgaris between its native range in Scotland (UK) and plants from the invaded range of the CP in New Zealand. Using unsupervised principal components analysis (PCA) our findings are exploratory only but do demonstrate clear differences in the plant metabolomes between the native and invaded ranges potentially linked to UV and soil nutrients.
It is beyond the scope of this study to independently assess the effects of these abiotic parameters (UV and soil nutrients) on the plant metabolome, as this requires controlled, experimental conditions. Testing the direct effects of altered foliar biochemistry on heather beetle performance is also beyond the scope of this study. These questions are now being addressed and will be presented in subsequent publications. However, we posit that our results validate the question of biochemically altered defences potentially exposing beetles at the CP sites to encounter host plants that are less assimilable than those in its native range. Furthermore, we propose that metabolomics is a powerful analytical tool that could be useful to assess biochemical changes in invasive plants due to differing abiotic influences, and such information could assist decision making in future biocontrol programmes or retrospectively elucidate unsuccessful ones.
Materials and methods
Sampling
Foliage
Five samples of mature heather (Calluna vulgaris) foliage were collected from each of four sites in Scotland (S Fig. 3a), during the Northern Hemisphere summer from 29th June to 3rd July 2018. The sites were, Glensaugh (GS) Lat. 56.910605° Lon. -2.569144°, alt. 323 m, soils - Strichen, peaty gleyed podzols; Ballogie Estate (BE) Lat. 56.998852° Lon. -2.743927°, alt. 316 m, soils - Countesswells, peaty gleyed podzols; Glenturret (GT) Lat. 56.415890° Lon. -3.912876°, alt 351 m, soils – Gourdie, noncalcareous gleys with peaty gleys; Creag Meagaidh (CM) Lat. 56.933362° Lon. -4.527912°, Alt 290 m, soils – Arkaig, peaty gleys with dystrophic semi-confined peat. During this period, most plants at GS, GT, and CM, were not flowering but at BE a small number had reached very early budburst. Plants with flowers were, where possible, avoided for sampling. Each sample comprised of equal quantities of 10–15 mm long fresh sprigs combined from three individual intertwined or adjacent mature plants. Each such sample was taken ≥ than 10 m apart and all were immediately cryo-frozen in nitrogen vapour then stored at -80 °C until freeze drying and grinding.
Mature C. vulgaris plants were sampled from four sites from the Central Plateau region of the North Island, New Zealand (S Fig. 3b), during January 17th − 18th 2019 (Southern Hemisphere summer) using the same sampling and storage protocol. The sites were Mangaturuturu (MU) Lat. -39.303293° Lon. 175.390239°, alt. 817 m, soil - Orthic Podzol; Waiouru (WU) Lat. -39.456172° Lon. 175.677246°, alt. 814 m, soil - Orthic Allophanic; Quarry (QU) Lat. -39.431120° Lon. 175.685689°, alt. 881 m, soil - Orthic Allophanic; Waihohunu (WH) Lat. -39.227263° Lon. 175.732654°, alt. 975 m, soil - Tephric Recent. At all sites, plants were at very early to early budburst with only a few flowers present, and again plants with flowers were avoided for sampling.
Soils and UV:
Five samples for soil nutrient analysis were collected from random positions in all sites. Each sample consisted of 3 soil cores each taken to a depth of 15 cm and air dried until no change in mass. Cores were sieved through a 1 mm precision sieve, then combined and analysed for Olsen P, Total N and pH. NZ samples were analysed by Hill Labs, Hamilton NZ and SC samples at the James Hutton Institute Laboratories, Aberdeen, UK. Phosphorus for Olsen P for both NZ and SC samples were measured on air dried soil using NaHCO3 (0.5 M; pH8.5) extractant. For Total N both NZ and SC samples were subjected to the Dumas combustion method then measured using a VarioMAX CN Macro Elementar analyser on NZ soils and a Thermo Flash EA 1112 elemental analyser (Thermo Fisher) on SC soils. For pH, both soils were slurried (1:2 v/v) soil:H2O and analysed using glass bulb pH probes. Parameters used for C. vulgaris’ exposure to ultra-violet for each range/region, are based on reviews of global summer noontime maxima of a standardised measurement of erythemal UV intensity known as the ultra-violet index (UVI) and with an adjustment for altitude (1000 masl) at the North Island Central Plateau sites58,59,65.
Invertebrates
To assess potential herbivore induced plant metabolite responses, using a beating tray and standardised beating protocol, we collected all invertebrates from the foliage of all plants at all sites and immediately preserved them in 70% ethanol. These were later sorted into family and their associated feeding guild, enumerated, then interrogated for Pearson correlation with phenylpropanoid compound intensities associated with their own site.
Foliage sample preparation for UHPLC-MS analysis
Foliage samples were freeze dried then stored for 2 weeks at -20 °C prior to grinding to ≈ 150–50 μm particle size before extraction. 50 ± 2.0 mg of ground sample were weighed into 2 mL microcentrifuge tubes and extracted in 800 µL of pre-chilled CHCl3:MeOH (1:1 v/v) with internal standards comprising 1.6 mg L-1 of d5-L-tryptophan, d4-citric acid, d10-leucine, d2-tyrosine, d35-stearic acid, d5-benzoic acid, 13C2-glucose, and d7-alanine. Samples were vortexed for 2 min and kept at -20 °C for 1 h after which 400 µL of H2O was added and again vortexed for 2 min. Samples were then centrifuged for 15 min (11000 RPM @ 4 °C) creating a biphasic layer. Two aliquots of 200 µL each of the upper, aqueous layer were transferred to 2 ml microcentrifuge tubes and evaporated to dryness, under a continuous stream of nitrogen (30 °C for 50 min) and stored @ -80 °C until reconstitution. For semi-polar compounds, reconstitution for C18-LC-MS analysis, was in 200 µL of C2H3N: H2O (1:9 v/v), vortexed for 1 min then transferred to a glass insert in an auto-sampler vial. For polar compounds reconstitution for HILIC-LC-MS analysis, was in 200 µL of C2H3N:H2O (1:1 v/v), vortexed for 1 min then similarly transferred to a glass insert in an auto-sampler vial. A pooled mix of all samples was similarly prepared (n = 7) and used as quality controls (QC) for each of the C18 and HILIC streams. These were evenly distributed (every 8th sample) to monitor any systematic effects on the corresponding analysis. Five extraction blanks were included at the beginning of and an amino acid standard (A9906; Sigma-Aldrich, NZ) at the beginning and end of the sampling sequences.
Chromatography and mass-spectrometry
Chromatography and tandem mass-spectrometry analysis of polar and semi-polar compounds were achieved using a Thermo LC–MS system (Thermo Fisher Scientific, Waltham, MA, USA) which consisted of an Accela 1250 quaternary UHPLC pump, a PAL auto-sampler fitted with a 15,000 psi injection valve (CTC Analytics AG., Zwingen, Switzerland) and 20 µl injection loop, and an Exactive Orbitrap mass spectrometer with electrospray ionisation run in both positive and negative modes.
For semi-polar compounds, samples were cooled in the auto-sampler at 4 °C and a 2 µL aliquot was injected into a 1.9 μm Thermo Hypersil Gold C18 column (UPLC, 100 mm × 2.1 mm, Thermo Fisher Scientific, USA) at 25 °C with a gradient elution programme and a flow rate of 400 µL/min. The mobile phase was water with 0.1% formic acid (solvent A), and acetonitrile with 0.1% formic acid (solvent B). Using the Xcalibur software package provided by the manufacturer the gradient elution programme was: held at 5% B (0–0.5 min),5–99% B (0.5–13 min), held at 99% B (13–15 min), returned to 5% B (15–16 min) and allowed to equilibrate for a further 4 min prior to the next injection. The first 1.5 min and the last 6 min of the chromatogram were diverted to waste. Mass spectral data were collected in profile mode over a mass range of m/z 60–1200, at a mass resolution setting of 25,000 with a maximum trap fill time of 100 ms. Samples were run in both positive and negative ionisation modes separately. Positive ion mode parameters were: spray voltage, 3.5 kV; capillary temperature, 325 °C; capillary voltage, 50 V, tube lens 120 V. Negative ion mode parameters were: spray voltage, − 3.5 kV; capillary temperature, 325 °C; capillary voltage, − 90 V, tube lens − 80 V. The nitrogen source gas desolvation settings were the same for both modes (arbitrary units): sheath gas, 40; auxiliary gas, 10; sweep gas, 566.
For polar compounds, samples were cooled in the auto-sampler at 4 °C and a 2 µL aliquot was injected into a 5 μm ZIC-pHILIC column (100 mm × 2.1 mm, Merck Darmstadt, Germany) at 25 °C with a gradient elution programme and a flow rate of 250 µL/min. The mobile phase was acetonitrile with 0.1% formic acid (solvent A) and 16 mM ammonium formate in water (solvent B). The gradient elution programme was: held at 97% A (0–1 min), 97–70% A (1–12 min), 70–10% A (12–14.5 min), held at 10% A (14.5–17 min), returned to 97% A (17–18.5 min) and allowed to equilibrate for a further 5.5 min prior to the next injection. The first 1.5 min and the last 5 min of the chromatogram were diverted to waste. Mass spectral data were collected in profile mode over a mass range of m/z 55-1100 at a mass resolution setting of 25,000 with a maximum trap fill time of 100 ms. Positive ion mode parameters were: spray voltage, 3.5 kV; capillary temperature, 325 °C; capillary voltage, 90 V, tube lens 120 V. Negative ion mode parameters were: spray voltage, − 3.0 kV; capillary temperature, 325 °C; capillary voltage, − 90 V, tube lens − 100 V. The nitrogen source gas desolvation settings were the same for both modes (arbitrary units): sheath gas, 40; auxiliary gas, 10; sweep gas, 567.
Data analysis
Thermo derived .raw files for each stream i.e., C18 and HILIC in both positive and negative modes, were converted to mzML format using MSConvertGUI68, uploaded into MZmine69 to determine the appropriate baseline noise threshold and then into XCMS online “xcmsonline.scripps.edu”, for feature detection, alignment and exploratory data analysis70. Feature detection parameters for C18 data were, m/z deviation 10 ppm, min and max peak width 5 and 20 respectively, mzdiff 0.001, s/n threshold 20, Prefilter intensity 1e4 and noise filter 4e4. For HILIC data the same parameters were 10ppm, 10 and 60, 0.001, 20, 1e4 and 3e4. After downloading the output, a series of procedures followed, converting raw mass spectrometry data into data matrices comprising m/z, retention time, and the corresponding ion intensity measurements suitable for statistical analysis.
Reduction of background variability in the full data matrix of each stream was performed using a QC vs. Blank t-test thus allowing subtraction of those features with p > 0.05 values or t.stat values corresponding to any features high in the blanks. These data matrices were each uploaded into MetaboAnalyst ver. 6.0 (MA 6.0)71 and data integrity checked to confirm the number of samples, number of peaks, missing values, and the number of treatment groups. No missing values were detected in any of the data sets. For filtration of variables showing low repeatability, the threshold to remove those with high percent relative standard deviation (RSD) was set at 30% to that of the QCs and the data normalised by auto-scaling (mean-centred and divided by the standard deviation of each variable) and Gaussian distribution confirmed so that feature mass intensities are comparable.
For each stream, we explored the entire data matrix at the site level for both the NZ and SC ranges by subjecting them to multiple principal component analysis (PCA) in MA 6.0. This indicated a degree of site cluster overlap within each range but a clear and consistent separation of clusters between the NZ and SC ranges (see S Fig. 2). We therefore considered it appropriate to apply paired PCA to each streams data matrix at the NZ and SC level for the purpose of demonstrating those differences between the metabolomes of each range.
Using the data matrices at the NZ and SC range level, final analyses were performed using the one factor statistical platform in MA 6.0. Unsupervised paired PCA’s were conducted on those p < 0.05 features remaining in each stream, to visualise and confirm the degree of separation between clusters (metabolomes) for each range. Then from these data matrices, features for annotation were achieved by applying a paired t-test with all features below the threshold value of FDR < 0.0572 being retained for that purpose.
Annotations were conducted and confidence levels confirmed73 for each metabolite by interrogating the original .raw files using Xcalibur Freestyle. Formula matches were confirmed, and mass accuracy parameters were set within +/– 10.0 ppm. For HILIC level 1 confidence, m/z and rt. results were matched against a Grasslands AgResearch in house spectral standards library (GL), based on authentic standards run under the same chromatographic conditions. For all C18 and the remaining HILIC features, level 2 and 3 (parent ion plus at least one fragment and parent ion only, respectively) confidence levels were confirmed using the MassBank.eu (https://massbank.eu/) spectral database. Where the same annotated confirmed compound (ion) appeared in both + and – modes the one with the highest intensity was included in the final data table. If a compound appeared in both C18 and HILIC streams the criteria for inclusion in the table was if they were a secondary or primary metabolite respectively.
Using the metabolites covering all three levels of confidence from Tables 1 and 2 and using the pathways analysis platform in MA 5.0 we conducted analyses to elucidate which pathways are enriched the most and provide the greatest impact on the data sets between each range.
Table 1.
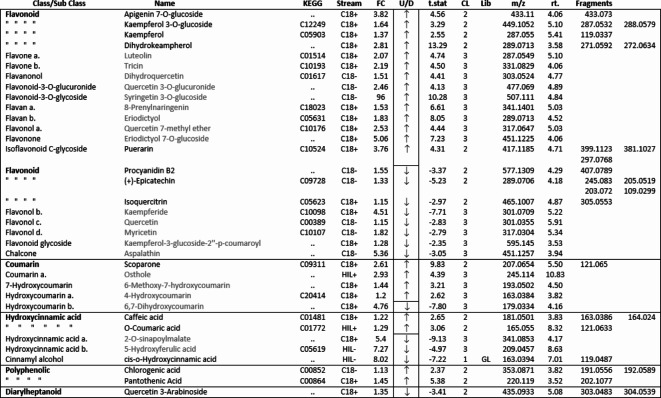
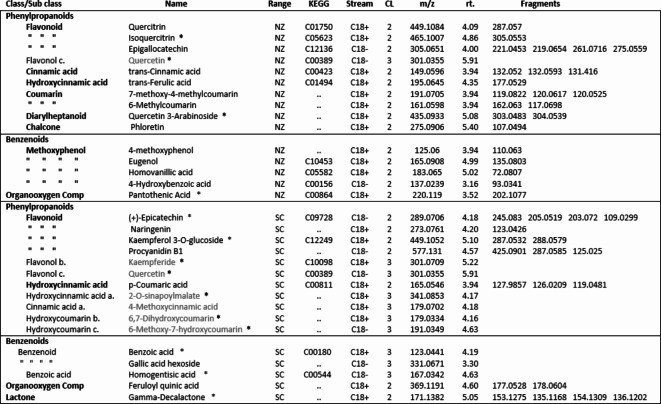
Phenylpropanoid compounds annotated from all four streams. level 1 and 2 confidence compound names are in black type. Level 3 confidence names are in grey type and identified to sub class only. FC = Fold Change, U/D = up or down, CL = confidence level, Lib = GL library.
Table 2.
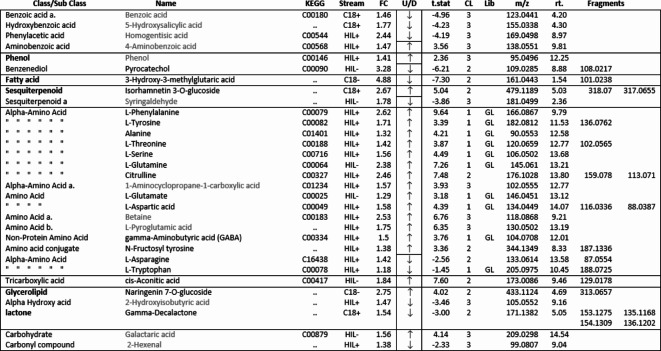
Primary and secondary compounds annotated from all four streams. level 1 and 2 confidence compound names are in black type Level 3 confidence names are in grey type and identified to sub class. FC = Fold Change, U/D = up or down, CL = confidence level, Lib = GL library.
To observe inter-site differences for secondary metabolites potentially due to soil nutrient status within each range, we ran multiple PCA on the C18 pos and C18 neg data matrices independently. Each feature in these data matrices was then subjected to one-way ANOVA in MA 6.0 to provide significant features for annotation. MA 6.0 also provides PERMANOVA analysis results to assess the significance of difference between PCA derived clusters. Annotated compounds were subjected to Minitab v 21.1.0 for Tukey post hoc allocation of significant differences. Phenylpropanoid compounds resulting from these analyses were graphed then individually subjected to Pearson correlation analysis to explore potential relationships with site soil nutrient status. The soil nutrient samples being randomly located within each site are therefore not paired with each foliage sample, so the mean site values for Olsen P and Total N were used.
Soil analysis results from the NZ range for Olsen P were converted from volumetric mg lt− 1 to gravimetric mg kg− 1 74 to match the SC result output and together with Total N and pH were for inter-site statistics subjected to one-way ANOVA with Tukey post hoc allocation analyses. Between NZ and SC range statistics for the same parameters were achieved using t-tests. Both analyses were achieved using Minitab v 21.1.0.
Results
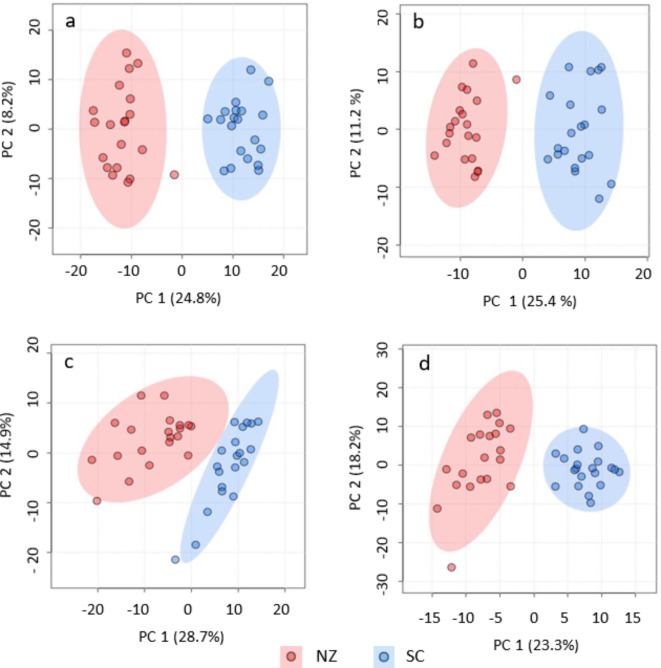
Paired PCA plots for both C18 and HILIC streams in both ionisation modes display clear unsupervised clustering of all samples from New Zealand’s Nth Is. Central Plateau (CP) and those of Scotland (SC) in the United Kingdom (UK), see Fig. 1. Separation between the NZ and SC clusters for C18 pos is explained by principal components 1 and 2 combined being 34.9% of the observed variance. These values are for C18 neg, 36.3%, HILpos, 44.6% and HILneg, 41.5% respectively. The MA 6.0 platform provides statistical testing to verify robustness of the clustering using PERMANOVA with 999 permutations. With (PERMANOVA); C18 pos, F = 27.17, p = 0.001; C18 neg F = 14.77, p = 0.001; HILIC pos F = 38.95, p = 0.001 and HILIC neg F = 38.73, p = 0.001 respectively, these results confirm the validity of the PCA analyses.
Fig. 1.
Paired PCA plots including 95% confidence ellipses for each stream showing significant separation between New Zealand (NZ) and Scotland (SC) sites for each ionization mode. PC 1 and PC 2 scores for each, explain the observed variance between clusters. a = C18 pos, b = C18 neg, c = HILIC pos and d = HILIC neg. PERMANOVA (999 permutations) provides verification of the robustness of the cluster formations in all four analyses. C18 pos, F = 27.17, p = 0.001; C18 neg F = 14.77, p = 0.001; HILIC pos F = 38.95, p = 0.001 and HILIC neg F = 38.73, p = 0.001.ara>
Interrogation of the .raw files resulted in 66 metabolites being annotated from the C18pos, C18neg, HILICpos and HILICneg streams combined (See S Fig. 1a and b). Twenty-two flavonoids, five coumarins, five hydroxycinnamic acids, two polyphenolics and one diarylheptanoid all phenylpropanoids, were revealed from both C18 pos and neg streams combined. Fourteen of these were confirmed at level 2 with the remainder being level 3 (Table 1). Of these 35 phenylpropanoids, 22 of them are amplified in the NZ samples compared to those of SC. Additionally, secondary metabolites including 4 benzoic acids, 2 phenols, 1 fatty acid plus 2 sesquiterpenoids, were identified from both streams (Table 2). For primary metabolites HILIC separation revealed 16 amino acids for which the Grasslands in house spectral standards library (GL) confirmed ten to be of confidence level 1, six to confidence level 2 and the remainder to level 3. Of these 16 amino acids, 14 are amplified in the NZ samples. Carboxylic acids, glycerolipids and organooxygen metabolites from both streams make up the remainder (Table 2).
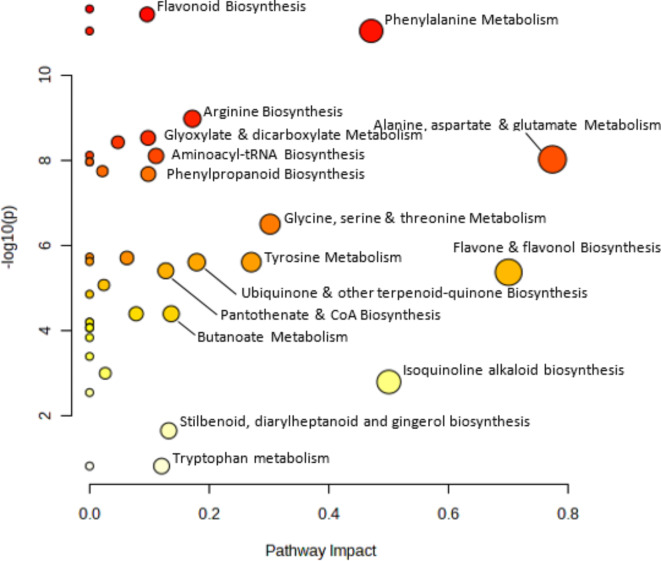
Pathway analysis identified those metabolic pathways to be enriched as indicated by the impact factor (x axis) and its p value of significance (y axis). Pathways with an impact factor ≥ 0.1 are labelled. The most significant of these were Alanine, aspartate and glutamate metabolism which revealed the greatest enrichment value of 0.77 followed by flavone and flavonol biosynthesis 0.7 and Phenylalanine metabolism 0.47 Note: The Isoquinoline alkaloid biosynthesis pathway is dependent on one compound only i.e., L-Tyrosine (Fig. 2).
Fig. 2.
Pathway enrichment analyses indicating the pathways having the greatest impact factor (x axis) and greatest p value of significance (y axis) between the two treatments (Ranges). Circle size indicates pathway impact value, and colour the p value. All pathways with an impact factor ≥ 0.1 are identified. Those in white to pale yellow are represented by one compound only.
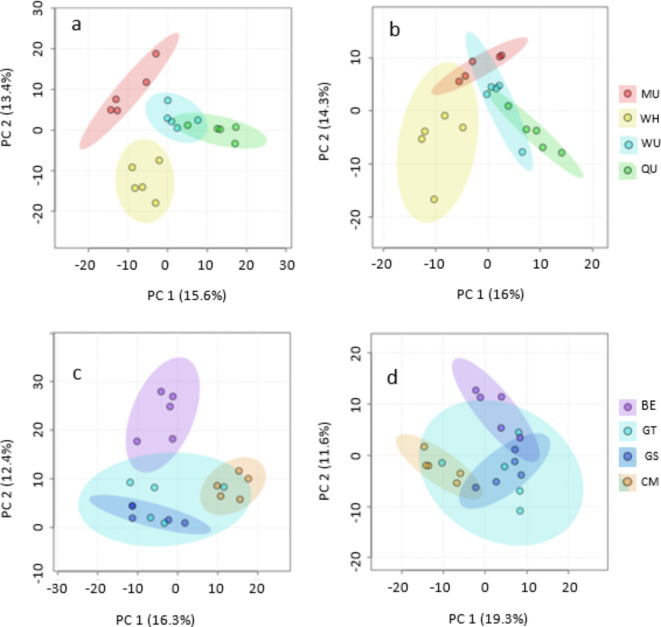
Multiple PCA using C18 pos and neg matrices revealed some overlap of clusters between sites in both ranges (Fig. 3). For the NZ sites, in C18 pos MU and WH are clearly separated but display a degree of overlap for QU and WU with a combined value for PC 1 and PC 2 of 29% explaining the observed variance between these groups. Separation is less well defined except for site WH in the C18 neg matrix however with a PC combined value of 30.3%. The SC sites generally display more overlap but BE displays the greatest separation in the C18 pos stream with a PC value of 28.7%. Separation of the SC C18 neg are the least well defined with a PC value of 30.9%. Statistical testing using PERMANOVA (999 permutations) however provides verification of the robustness of the cluster formations in all four of these PCA analyses. (PERMANOVA); NZ C18 pos, F = 28.23, p = 0.001; NZ C18 neg F = 15.87, p = 0.001; SC C18 pos F = 17.86, p = 0.001 and SC C18 neg F = 9.48, p = 0.001.
Fig. 3.
Multiple PCA’s of NZ and SC C18 pos and neg matrices showing clustering of sites in both ranges with 95% confidence ellipses. PC 1 and PC 2 scores for each, explain the observed variance between clusters. For NZ C18 pos (a) MU and WH clearly separate but with a degree of overlap for QU and WU. NZ C18 neg (b), separation is less well defined except for WH. SC C18 pos reveals BE, with the greatest separation but considerable overlap with the remaining three clusters. SC C18 neg separation is the least well defined. PERMANOVA (999 permutations) provides verification of the robustness of the cluster formations. For NZ C18 pos, F = 28.23, p = 0.001; NZ C18 neg F = 15.87, p = 0.001; SC C18 pos F = 17.86, p = 0.001 and SC C18 neg F = 9.48, p = 0.001. New Zealand CP sites are Mangaturuturu (MU), Waihohonu (WH), Waiouru (WU) and Quarry (QU). Scotland sites are, Ballogie Estate (BE), Glenturret (GT), Glensaugh (GS) and Creag Meagaidh (CM).
Of the one-way ANOVA identified features that differ significantly (all with p < 0.01) between the four sites in both NZ and SC, thirty-one compounds were annotated. Seventeen of these (see S Fig. 1c) are additional to those listed in Tables 1 and 2. The previously annotated compounds are marked with and asterisk, see Table 3. Of these additional seventeen, eleven are phenylpropanoids with the remainder comprising five benzenoids and one organooxygen compound.
Table 3.
Compounds annotated from the C18 pos and C18 neg streams within the New Zealand (NZ) and Scotland (SC) ranges. Level 2 confidence compound names are in black type . Level 3 confidence names are in grey type and identified to sub class only. CL = confidence level, Compounds in common with table 1a and 1b denoted by *.
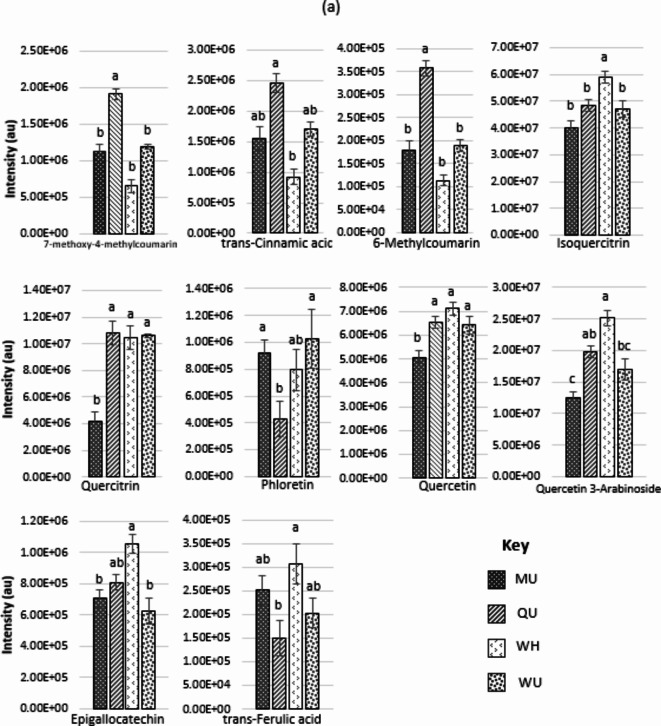
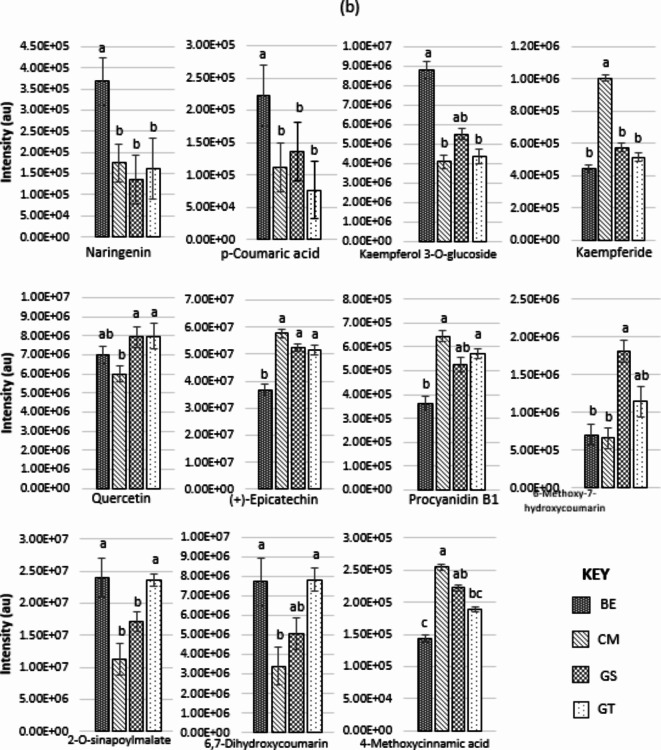
Ten phenylpropanoids for the NZ sites and eleven for the SC sites are graphically displayed in Fig. 4a and b respectively and were explored for Pearson correlation coefficient with site mean Olsen P and Total N values. There was little evidence either positive or negative for correlation between most of these phenylpropanoids. A moderate to weak negative but significant correlation does exist for the NZ compounds of trans-Ferulic acid (Olsen P, r(18) = − 0.43, p < 0.05 and Total N, r(18) = − 0.48, p < 0.05) and Epigallocatechin (Olsen P, r(18) = − 0.43, p < 0.05 and Total N, r(18) = − 0.58, p < 0.01). In the SC compounds slightly stronger moderate negative correlation with Total N only, exist for 2-O-Sinapoylmalate (Total N, r(18) = − 0.58, p < 0.01), 6,7-Dihydroxycoumarin (Total N, r(18) = − 0.61, p < 0.01) and a positive correlation for 4-Methoxycinnamic acid (Total N, r(18) = 0.59, p < 0.01).
Fig. 4.
(a) One-way ANOVA determined 10 phenylpropanoid compounds that differed significantly between the four New Zealand (CP) sites. Those sites sharing letters are not significant. All are p < 0.01. Sites are Mangaturuturu (MU), Quarry (QU), Waihohonu (WH) and Waiouru (WU). (b) One-way ANOVA determined 11 phenylpropanoid compounds that differed significantly between the four United Kingdom (SC) sites. Those sites sharing letters are not significant, all are p < 0.01. Sites are Ballogie Estate (BE), Creag Meagaidh (CM), Glensaugh (GS) and Glenturret (GT).
Phytophagous invertebrates were absent from the samples collected from the New Zealand CP sites. This was because the heather beetle biocontrol agent was not established at these sites and the only known New Zealand native spp. known to graze on C. vulgaris are the manuka beetle (Pyronota festiva)34 and an unidentified lepidopteran leaf tying caterpillar. These two native spp. are generally only present in very low numbers earlier in the season. From the SC sites, foliage dwelling invertebrates sorted into taxonomic groups and feeding guild resulted in three major taxa being identified. These were, piercing/sucking hemiptera, leaf chewing coleoptera and leaf chewing lepidoptera, (See S Table 1). Hemiptera were significantly different between sites with one-way ANOVA indicating GT significantly different from CM and BE but not GS, F(3, 16) = 4.58, p 0.017. Coleoptera with numbers approximately an order of magnitude higher at site GT differed significantly from CM, BE and GS, F(3, 16) = 10.72, p 0.001 and lepidoptera showed no significant difference between any site F(3, 16) = 3.04, p 0.115.
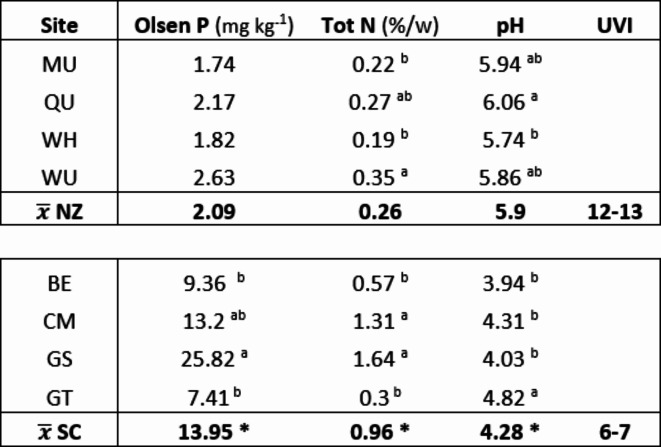
Soil nutrient analyses confirm soils of the 4 sites in SC to be relatively acidic and with very low levels of Olsen P and Total N. The volcanic soils of the CP in NZ however, while less acidic are even poorer in nutrient status with both Olsen P and Total N at extremely low levels, indeed Olsen P is at the limit of detection for this analytical method74. The paired t-test values between NZ and SC are highly significant for Olsen P t(1, 19) 5.45, p < 0.0001; Total N t(1, 19) 4.73, p < 0.0001, and pH t(1, 19) -18.4, p < 0.0001 respectively. One way ANOVA for Olsen P levels at the NZ sites show no significant difference F(3, 16) = 2.06, p = 0.1446 while for Total N F(3, 16) = 6.39, p = 0.0046 and pH F(3, 16) = 4.70, p = 0.015, do show a significant difference between these means. For the SC sites Olsen P F(3, 16) = 6.55, p = 0.0042, Total N F(3, 16) = 12.97, p = < 0.0001 and pH F(3, 16) = 16.01, p = < 0.0001, all show a significant difference between the means, Table 4.
Table 4.
Soil nutrient analysis values by site for Olsen P, Total N and pH with all Tukey post hoc levels of significance at p < 0.05. Those sites sharing letters are not significant. The New Zealand (NZ) and Scotland (SC) range values differ significantly by paired t-test and are indicated by * with p < 0.0001. Summer noontime average UV index for each country is also provided. The NZ Central Plateau UVI values are adjusted for 1000 masl). NZ sites are, Mangaturuturu (MU), Quarry (QU), Waihohonu (WH) and Waiouru (WU) and for SC are, Ballogie Estate (BE), Creag Meagaidh (CM), Glensaugh (GS) and Glenturret (GT).
Discussion
Our results show significant differences in the biochemical profile of Calluna vulgaris plants growing in its native range in the UK compared to its conspecifics in NZ. A notable feature of this difference is the greater number of phenylpropanoids and amino acid metabolites that are amplified in the NZ range (Tables 1 and 2). According to the literature, both soil nutrients and ultraviolet light influence and upregulate the same major shikimate-phenylpropanoid pathway75,76 and the subsequently dependent flavonoid and flavone/flavonol biosynthesis pathways. We provide good evidence for upregulation of all these pathways, and the resultant amplification of many primary and secondary metabolites which suggests that C. vulgaris is responding to differing environmental factors in these two ranges.
New Zealand CP sites have poorer soil nutrient status compared to the SC sites. Literature demonstrating depleted or low soil nitrogen and phosphate availability, increasing the level of phenylpropanoid chemical defences in plants, is provided by several studies40,41,43–45. Nitrogen deficiency can induce the accumulation of foliar phenylpropanoid compounds by increasing the activity of phenylalanine ammonia-lyase (PAL), a key enzyme of the shikimic acid – phenylpropanoid pathway41,44, resulting in increased levels of total phenolics77,78 as well as coumarins, anthocyanins, flavonoid glycosides, flavones, iso-flavones, and tannins41,45. Similarly, low levels of phosphate can increase foliar levels of caffeoylquinic acids, coumarins, anthocyanins and flavonoid glycosides41,44. Many of these compounds appear to be amplified in our NZ samples, supporting the existing evidence of low soil nutrients enhancing chemical defences.
In contrast to soils, NZ has considerably higher ambient ultra-violet intensity levels than in the UK48,49. Changes in the light environment, particularly increased levels of UV-B, induce signal transduction pathways to regulate plant physiological activity40,79 often resulting in soluble phenolic compounds, particularly phenolic acids, flavonoids, flavonol glycosides of kaempferol, quercetin and myricetin as well as hydroxy cinnamic esters to accumulate in plants, many of which play an important role in photoprotection and defence80,81.
Considerable literature exists demonstrating that such UV-radiation induced phenylpropanoids can not only increase plant resistance to insects and impair insect herbivore performance42,50,52 but also increase plant resistance to biotrophic pathogens54,55. With reference to insect herbivores, several flavonoids82, as well as chlorogenic acid83, several catechins84, caffeic and o-coumaric acid85 have all been demonstrated to impair growth and survival rates, fertility, fecundity and population growth rates in a range of insect families and we found amplified levels of many of these compounds at our NZ sites.
Of the primary metabolites, the non-protein amino acid γ-Aminobutyric acid (GABA) is also upregulated in our samples. This compound accumulates under a range of abiotic stresses including soil nutrients and light and is known to function directly in plant immune responses to biotrophic pathogens and fungi as well as being a powerful neuromuscular and growth inhibitor against insect herbivores, often causing ill thrift and death86,87. It’s therefore conceivable, that elevated levels of some of the metabolites we have recorded in C. vulgaris in this study could render the assimilability of the host plant more challenging to L. suturalis in its new environment. These amplified metabolites may contribute to a nutritional cascade, exacerbating the effects of low foliar N levels reported in another recent paper38 and may help explain the difficulties with the initial establishment of this control agent released to control C. vulgaris. in NZ.
When investigating differences between sites within the same range, we assume no variation in UV intensity exists and we know there is no influence of shade or aspect at either one, given all sites are in open shrubland communities. Therefore, we focused our analysis on response variables potentially linked to soil nutrients. While PCA revealed some overlap between sites (clusters) within each of the ranges, the one-way ANOVA revealed significant differences in phenylpropanoid compounds between some of those sites. For the CP sites we encountered significant differences in the intensities of ten phenylpropanoids with only trans-Ferulic acid and epigallocatechin however, having a negative correlation with both phosphate (Olsen P) and nitrogen (Total N) levels in those soils. Overall, there remains only weak evidence that soil nutrients in this region are influencing the intensities of the phenylpropanoids that we were able to positively annotate.
The SC sites revealed eleven phenylpropanoid compounds showing significant intensity differences between sites but similarly provide little evidence (notwithstanding the results for 2-O-Sinapoylmalate, 6,7-Dihydroxycoumarin and 4-Methoxycinnamic acid) from our Pearson correlation coefficients, that soil phosphate and nitrogen levels are greatly influencing phenylpropanoid intensities. We cannot therefore, claim a causal effect as other factors and their interaction may also contribute to the observed metabolite changes. Regarding the variable effectiveness of L. suturalis at some of the New Zealand sites then, it remains uncertain if varying intensity of any phenylpropanoids could be involved. We suggest this question could be addressed with controlled dietary experiments using some of the compounds that were amplified in the NZ range, that are known to impair insect performance.
While plant genetic variability is understood to influence the overall metabolite profile of plants, its effect on metabolite intensity appears equivocal. Literature indicates that for plants of the same species that vary genetically, metabolite intensity is influenced much more by abiotic parameters than genetic variability88–90. We acknowledge that genetics may be a contributor to the observed metabolite differences and are currently addressing the degree of genetic variability of C. vulgaris between ranges and within sites but posit that differences in soil nutrients and UV-radiation are likely the strongest drivers for these results. Support for both these abiotic influences altering the biochemical profile of C. vulgaris in NZ, is provided by recent field trials and a tunnel house experiment using UV attenuating screens58. From four sites on the CP region of the North Island, significant differences in the volatile organic compound (VOC) emissions of C. vulgaris were recorded between sites. Of the environmental variables collected i.e., soil nutrients, ambient daytime temperature, soil water content and soil temperature, the main contributing factor to these differences was soil nutrients59. Tunnel house manipulations of ultraviolet light using 20% and 95% attenuating screens and exposing mature and phenologically similar C. vulgaris plants for 75 days to this treatment, revealed significant differences between several VOC metabolites, demonstrating that this plant is also sensitive to differing levels of ultraviolet radiation. Which abiotic parameter may be driving the variances revealed in this study however requires controlled experiments manipulating nutrient availability and ultraviolet light both independently and combined, to provide quantitative data to confirm any direct effect of these parameters and further elucidate these results.
Average ambient temperatures vary little between our NZ and SC ranges and soil moisture contents (while we did not measure these) at the time of sampling were within normal summertime ranges, thus we expect little influence on metabolite profiles and intensities from those sources. Induced plant responses to differing insect herbivore communities however needed to be considered. The significantly high number of Hemiptera and Coleoptera at GT was the most likely to produce a positive correlation with high intensities of a given phenylpropanoid at that same site, but none were observed. Additionally, the PCA clusters for GS and CM show considerable overlap with GT, again suggesting that GT is not separating out as a result of any induced response to invertebrate herbivory. There was little evidence then of herbivory influencing the intensity measurements of secondary metabolites which may skew the NZ vs. SC range or inter-site soil nutrient PCA results. Plant ontogeny can influence secondary metabolites which has been reported in a recent paper for VOC compounds in C. vulgaris91, however all plants sampled at all sites in this study were mature and well developed, thus we expect any influence from that source to be negligible. For more in-depth understanding of constitutive metabolites of a successionally dynamic heather community displaying variable plant ontogeny, we also recommend further testing.
While we cannot ascribe direct causality from environmental factors, these results illuminate the potential use of metabolomic techniques for biological control of weeds. Once a potential agent has been selected, metabolomic assays may help determine sites with the closest matching target plant metabolomes, to potentially, both source and release the biocontrol agent. Such information may assist in reducing the chances of encountering difficult establishment or poor effectiveness scenarios. Additionally, it would be of interest to retrospectively compare plant metabolomes from both the source and release sites of already introduced control agents for both successful and unsuccessful programmes to explore to what extent the plant metabolome could be used as a predictor of biocontrol agent success. Further ideas and potential applications have been extensively discussed in a recent review37.
Conclusions
Our results clearly demonstrate significant differences between the metabolomes of C. vulgaris plants occurring in its native range in Scotland (UK) and those in its invaded range of the Central Plateau of the North Island of New Zealand and suggest that UV-radiation and soil nutrients could be driving the observed differences. However, to assess direct causality further experiments under controlled conditions are necessary.
It is also evident for sites within each of the ranges we tested, significant variation occurs for many phenylpropanoids. Bioassays exploring biocontrol agent behaviour, feeding preferences and life history performance are required to understand if both inter and intra site variance in this and potentially other classes of compounds could be a driver of poor agent establishment rates or variable effectiveness.
Our non-targeted metabolomics approach to these investigations revealed not just secondary defensive metabolites, but the majority of the primary amino acid metabolites identified (in addition to those key precursors of the shikimate-phenylpropanoid pathway i.e., phenylalanine and tyrosine) are also amplified suggesting overall increased biosynthetic pathway upregulation of this plant in the New Zealand CP environment. Our results therefore add to the very limited literature documenting biochemical phenotypic change to invasive plants that have established in a new, abiotically different region.
We conclude, this study has provided a primer, that abiotically induced biochemical change may not be uncommon in invasive plants. This validates our original question of the potential for biochemically modified phenotypes altering plant defensive capacity and/or nutritional assimilability which may compromise specialist control agents when reunited with their coevolutional host plant. The application of metabolomics therefore, may be a valuable tool to assist with determining such changes and help elucidate poor establishment and/or effectiveness scenarios in weed biocontrol.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We wish to acknowledge Prof. Glenn Iason for assistance with initial feasibility and Joan Beaton, Donald Barrie, Allan Sim, Gillian Green and Ailsa Johnson-Marshall of the James Hutton Institute in Aberdeen for technical assistance. Alex and Mary Seldon of Glenturret, Pete Littlejohn of Ballogie Estate, Kirsty North of the Scottish Natural Heritage, NZ Defence and NZ Dept. of Conservation for access to study sites. Evans Effah and Claire Zuchetta of Massey University for field assistance on the Central Plateau.
Author contributions
DPB. Conceived the primary questions and objectives of the investigations. Carried out all sampling, processing of samples, annotation and statistical analysis. Secured funding. Primary author and writer of the manuscript.AKS. Advised on appropriate chromatographic analysis, supervised and ran all UHPLC-MS technical processes, advised regarding metabolomic analysis techniques and revised the manuscript.RJP. Advised and assisted with site selection and sample collection carried out in Scotland UK. Facilitated hosting and access to laboratory facilities at the James Hutton Institute, Aberdeen and revised the manuscript.PP. Has provided 2 decades worth of background research information on the Heather biocontrol program. Now recently has provided expertise on foliar N impacts on heather beetle control agent performance and other invertebrates associated with this program and revised the manuscript.ACM. Principal investigator and project supervisor. Advised on concept, design and interpretation of investigations, supported fieldwork sampling, secured funding and revised the manuscript.
Funding
We wish to thank the following organisations for funding this project: The QE II technicians study award, awarded to Paul Barrett. Manaaki Whenua- Landcare Research, for MBIE - Strategic Scientific Investment Funding allocated to Paul Barrett. The Royal Society, Fast Start Marsden Grant “Plant Communication in times of rapid environmental change” awarded to ACM.
Data availability
All relevant .raw spectral data sets are available upon reasonable request.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Pimentel, D., Zuniga, R. & Morrison, D. Update on the environmental and economic costs associated with alien-invasive species in the United States. Ecol. Econ.52(3), 273–288 (2005). [Google Scholar]
- 2.Pyšek, P. et al. A global assessment of invasive plant impacts on resident species, communities, and ecosystems: The interaction of impact measures, invading species’ traits and environment. Glob. Change Biol.18(5), 1725–1737 (2012). [Google Scholar]
- 3.Hayes, L. et al. Biocontrol of weeds: Achievements to date and future outlook. In Ecosystem services in New Zealand—conditions and trends, 375–385 (Manaaki Whenua Press, Lincoln, 2013).
- 4.Schwarzländer, M., Hinz, H. L., Winston, R. & Day, M. Biological control of weeds: An analysis of introductions, rates of establishment and estimates of success, worldwide. BioControl63(3), 319–331 (2018). [Google Scholar]
- 5.McFadyen, R. C. Successes in biological control of weeds. Paper presented at the Proceedings of the X international symposium on biological control of weeds (2000).
- 6.McClay, A. S. & Balciunas, J. K. The role of pre-release efficacy assessment in selecting classical biological control agents for weeds—Applying the Anna Karenina principle. Biol. Control35(3), 197–207 (2005). [Google Scholar]
- 7.Raghu, S. & Dhileepan, K. The value of simulating herbivory in selecting effective weed biological control agents. Biol. Control34(3), 265–273 (2005). [Google Scholar]
- 8.Falla, C. et al. Effects of light intensity on Solanum mauritianum (Solanaceae) morphological and chemical traits and the performance of its biological control agent Gargaphia decoris (Hemiptera: Tingidae). Biol. Control181, 105218 (2023). [Google Scholar]
- 9.Grevstad, F. S. Factors influencing the chance of population establishment: Implications for release strategies in biocontrol. Ecol. Appl.9(4), 1439–1447 (1999). [Google Scholar]
- 10.Jonsen, I. D., Bourchier, R. S. & Roland, J. Influence of dispersal, stochasticity, and an Allee effect on the persistence of weed biocontrol introduction success failure. Ecol. Model.203(3–4), 521–526 (2007). [Google Scholar]
- 11.Harms, N. E., Cronin, J. T., Diaz, R. & Winston, R. L. A review of the causes and consequences of geographical variability in weed biological control successes. Biol. Control, 104398 (2020).
- 12.Tobin, P. C., Berec, L. & Liebhold, A. M. Exploiting Allee effects for managing biological invasions. Ecol. Lett.14(6), 615–624 (2011). [DOI] [PubMed] [Google Scholar]
- 13.Valladares, F., Gianoli, E. & Gómez, J. M. Plastic phenotypic response to light of 16 congeneric shrubs from a Panamanian rainforest. New Phytol.176(4), 749–763 (2007a). [DOI] [PubMed] [Google Scholar]
- 14.Valladares, F., Gianoli, E. & Gómez, J. M. Ecological limits to plant phenotypic plasticity. New Phytol.(176), 749–763. (2007). [DOI] [PubMed]
- 15.Clavijo McCormick, A. Can plant–natural enemy communication withstand disruption by biotic and abiotic factors? Ecol. Evol.6(23), 8569–8582 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Obata, T. & Fernie, A. R. The use of metabolomics to dissect plant responses to abiotic stresses. Cell. Mol. Life Sci.69(19), 3225–3243 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Foyer, C. H., Rasool, B., Davey, J. W. & Hancock, R. D. Cross-tolerance to biotic and abiotic stresses in plants: A focus on resistance to aphid infestation. J. Exp. Bot.67(7), 2025–2037 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Sharma, A. et al. Response of phenylpropanoid pathway and the role of polyphenols in plants under abiotic stress. Molecules24(13), 2452 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cramer, G. R., Urano, K., Delrot, S., Pezzotti, M. & Shinozaki, K. Effects of abiotic stress on plants a systems biology perspective. BMC Plant Biol.11(1), 163 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Verma, V., Ravindran, P. & Kumar, P. P. Plant hormone-mediated regulation of stress responses. BMC plant biology, 16, 1–10.New Phytologist(176), 749–763 (2016). [DOI] [PMC free article] [PubMed]
- 21.Pigliucci, M. Evolution of phenotypic plasticity: Where are we going now? Trends Ecol. Evol.20(9), 481–486 (2005). [DOI] [PubMed] [Google Scholar]
- 22.Van Kleunen, M., Weber, E. & Fischer, M. A meta-analysis of trait differences between invasive and non‐invasive plant species. Ecol. Lett.13(2), 235–245 (2010). [DOI] [PubMed] [Google Scholar]
- 23.Shaar-Moshe, L., Hayouka, R., Roessner, U. & Peleg, Z. Phenotypic and metabolic plasticity shapes life‐history strategies under combinations of abiotic stresses. Plant. Direct3(1), 1–13 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hauvermale, A. L. & Sanad, M. N. Phenological plasticity of wild and cultivated plants. In Plant Phenology: IntechOpen (2019).
- 25.Godoy, O., Valladares, F. & Castro-Díez, P. Multispecies comparison reveals that invasive and native plants differ in their traits but not in their plasticity. Funct. Ecol.25(6), 1248–1259 (2011). [Google Scholar]
- 26.Palacio-López, K. & Gianoli, E. Invasive plants do not display greater phenotypic plasticity than their native or non‐invasive counterparts: A meta‐analysis. Oikos120(9), 1393–1401 (2011). [Google Scholar]
- 27.Lázaro-Nogal, A. et al. Environmental heterogeneity leads to higher plasticity in dry‐edge populations of a semi‐arid Chilean shrub: Insights into climate change responses. J. Ecol.103(2), 338–350 (2015). [Google Scholar]
- 28.Bufford, J. L. & Hulme, P. E. Increased adaptive phenotypic plasticity in the introduced range in alien weeds under drought and flooding. Biol. Invasions23(8), 2675–2688 (2021). [Google Scholar]
- 29.Wolf, V. C., Berger, U., Gassmann, A. & Müller, C. High chemical diversity of a plant species is accompanied by increased chemical defence in invasive populations. Biol. Invasions13, 2091–2102 (2011). [Google Scholar]
- 30.Skubel, S. A. et al. Metabolomic differences between invasive alien plants from native and invaded habitats. Sci. Rep.10(1), 1–9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Medina, P. et al. Plant geographic distribution influences chemical defences in native and introduced Plantago lanceolata populations. bioRxiv, 2023.2006. 2005.543708 (2023).
- 32.Effah, E. et al. Effects of two invasive weeds on arthropod community structure on the Central Plateau of New Zealand. Plants9(7), 919 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Effah, E. et al. Seasonal and environmental variation in volatile emissions of the New Zealand native plant Leptospermum scoparium in weed-invaded and non-invaded sites. Sci. Rep.10(1), 1–11 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Effah, E., Svendsen, L., Barrett, D. P. & Clavijo McCormick, A. Exploring plant volatile-mediated interactions between native and introduced plants and insects. Sci. Rep.12(1), 15450 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peterson, P., Fowler, S. & Barrett, P. Is the poor establishment and performance of heather beetle in Tongariro National Park due to the impact of parasitoids predators or disease. New. Z. Plant. Prot.57, 89–93 (2004). [Google Scholar]
- 36.Fowler, S. V. et al. Investigating the poor performance of heather beetle, Lochmaea suturalis (Thompson)(Coleoptera: Chrysomelidae), as a weed biocontrol agent in New Zealand: Has genetic bottlenecking resulted in small body size and poor winter survival? Biol. Control87, 32–38 (2015). [Google Scholar]
- 37.Barrett, D. P., Fowler, S. V., Subbaraj, A. K., Groenteman, R. & Clavijo-McCormick, A. Metabolomic analysis of host plant biochemistry could improve the effectiveness and safety of classical weed biocontrol. Biol. Control, 104663. (2021).
- 38.Peterson, P., Fowler, S. & Barrett, D. P. Low host-plant nitrogen contributes to poor performance of heather beetle, an introduced weed biocontrol agent in New Zealand. Biol. Control197, 105589 (2024). [Google Scholar]
- 39.Peterson, P., Fowler, S. V., Harman, Barrett, D. P. & Merrett, M. Biological Control of heather. Technical Report: LCR0708/043 (2007).
- 40.Yang, L. et al. Response of plant secondary metabolites to environmental factors. Molecules23(4), 762 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gershenzon, J. Changes in the levels of plant secondary metabolites under water and nutrient stress. In Phytochemical Adaptations to Stress, 273–320. (Springer, 1984).
- 42.Mazid, M., Khan, T. & Mohammad, F. Role of secondary metabolites in defence mechanisms of plants. Biology Med.3(2), 232–249 (2011). [Google Scholar]
- 43.Wright, D. M. et al. Do leaves of plants on phosphorus-impoverished soils contain high concentrations of phenolic defence compounds? Funct. Ecol.24(1), 52–61 (2010). [Google Scholar]
- 44.Sampedro, L., Moreira, X. & Zas, R. Costs of constitutive and herbivore-induced chemical defences in pine trees emerge only under low nutrient availability. J. Ecol.99(3), 818–827 (2011). [Google Scholar]
- 45.Kováčik, J. & Klejdus, B. Induction of phenolic metabolites and physiological changes in chamomile plants in relation to nitrogen nutrition. Food Chem.142, 334–341 (2014). [DOI] [PubMed] [Google Scholar]
- 46.De Graaf, M. C., Bobbink, R., Smits, N. A., Van Diggelen, R. & Roelofs, J. G. Biodiversity, vegetation gradients and key biogeochemical processes in the heathland landscape. Biol. Conserv.142(10), 2191–2201 (2009). [Google Scholar]
- 47.Hewitt, A. E. New Zealand soil classification. Landcare Res. Sci. Ser.(1) (2010).
- 48.Liley, J. B. & McKenzie, R. L. Where on earth has the highest UV? RSNZ Misc Ser.68, 36–37 (2006). [Google Scholar]
- 49.Seckmeyer, G. et al. Europe’s darker atmosphere in the UV-B. Photochem. Photobiol. Sci.7(8), 925–930 (2008). [DOI] [PubMed] [Google Scholar]
- 50.Ballaré, C. L., Mazza, C. A., Austin, A. T. & Pierik, R. Canopy light and plant health. Plant Physiol.160(1), 145–155 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rousseaux, M. C. et al. Responses to solar ultraviolet-B radiation in a shrub‐dominated natural ecosystem of Tierra Del Fuego (southern Argentina). Glob. Change Biol.7(4), 467–478 (2001). [Google Scholar]
- 52.Izaguirre, M. M., Mazza, C. A., Biondini, M., Baldwin, I. T. & Ballaré, C. L. Remote sensing of future competitors: Impacts on plant defences. Proc. Natl. Acad. Sci.103(18), 7170–7174 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Qi, J. et al. Ultraviolet-B enhances the resistance of multiple plant species to lepidopteran insect herbivory through the jasmonic acid pathway. Sci. Rep.8(1), 1–9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fu, Z. Q. & Dong, X. Systemic acquired resistance: Turning local infection into global defense. Annu. Rev. Plant Biol.64, 839–863 (2013). [DOI] [PubMed] [Google Scholar]
- 55.Ballaré, C. L. Light regulation of plant defence. Annu. Rev. Plant Biol.65, 335–363 (2014). [DOI] [PubMed] [Google Scholar]
- 56.Monschein, M., Iglesias Neira, J., Kunert, O. & Bucar, F. Phytochemistry of heather (Calluna vulgaris (L.) Hull) and its altitudinal alteration. Phytochem. Rev.9, 205–215 (2010). [Google Scholar]
- 57.Jalal, M. A., Read, D. J. & Haslam, E. Phenolic composition and its seasonal variation in Calluna vulgaris. Phytochemistry21(6), 1397–1401 (1982). [Google Scholar]
- 58.Effah, E. et al. Herbivory and attenuated UV Radiation Affect Volatile emissions of the Invasive Weed Calluna vulgaris. Molecules25(14), 3200 (2020a). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Effah, E. et al. Natural variation in volatile emissions of the Invasive Weed Calluna vulgaris in New Zealand. Plants9(2), 283 (2020b). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hall, R. D. Plant metabolomics: From holistic hope, to hype, to hot topic. New Phytol.169(3), 453–468 (2006). [DOI] [PubMed] [Google Scholar]
- 61.Macel, M., van Dam, N. M. & Keurentjes, J. J. Metabolomics: The chemistry between ecology and genetics. Mol. Ecol. Resour.10(4), 583–593 (2010). [DOI] [PubMed] [Google Scholar]
- 62.Arbona, V., Manzi, M., Ollas, C. D. & Gómez-Cadenas, A. Metabolomics as a tool to investigate abiotic stress tolerance in plants. Int. J. Mol. Sci.14(3), 4885–4911 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jorge, T. F., Mata, A. T. & António, C. Mass spectrometry as a quantitative tool in plant metabolomics. Philos. Trans. R. Soc. A: Math. Phys. Eng. Sci.374(2079), 20150370 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sampaio, B. L., Edrada-Ebel, R. & Da Costa, F. B. Effect of the environment on the secondary metabolic profile of Tithonia diversifolia: A model for environmental metabolomics of plants. Sci. Rep.6, 29265 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vanicek, K., Frei, T., Litynska, Z. & Schmalwieser, A. UV-Index for the public. A guide for publication and interpretation of solar UV Index forecasts for the public prepared by the Working Group 4 of the COST-713 Action UV-B Forecasting. Eur. Coop. in the Field of Sci. and Tech. Res., Brussels (2000).
- 66.Fraser, K. et al. Analysis of metabolic markers of tea origin by UHPLC and high resolution mass spectrometry. Food Res. Int.53(2), 827–835 (2013). [Google Scholar]
- 67.Fraser, K. et al. Non-targeted analysis of tea by hydrophilic interaction liquid chromatography and high resolution mass spectrometry. Food Chem.134(3), 1616–1623 (2012). [DOI] [PubMed] [Google Scholar]
- 68.Adusumilli, R. & Mallick, P. Data conversion with ProteoWizard msConvert. Proteomics: Methods Protocols, 339–368 (2017). [DOI] [PubMed]
- 69.Pluskal, T. et al. (2020). Metabolomics data analysis using MZmine.
- 70.Domingo-Almenara, X. & Siuzdak, G. Metabolomics data processing using XCMS. In Computational Methods and Data Analysis for Metabolomics, 11–24 (Springer, 2020). [DOI] [PubMed]
- 71.Pang, Z. et al. MetaboAnalyst 5.0: Narrowing the gap between raw spectra and functional insights. Nucleic Acids Res.49(W1), W388–W396 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. Roy. Stat. Soc.: Ser. B (Methodol.)57(1), 289–300 (1995). [Google Scholar]
- 73.Sumner, L. W. et al. Proposed minimum reporting standards for chemical analysis. Metabolomics3(3), 211–221 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Taylor, M. et al. How closely do different Olsen P measures correlate? Occasional Report No. 31 Fertilizer and Lime Research Centre, Massey University, Palmerston North, New Zealand.(2018).
- 75.Bassman, J. H. Ecosystem consequences of enhanced solar Ultraviolet Radiation: Secondary plant metabolites as mediators of multiple trophic interactions in Terrestrial Plant communities. Photochem. Photobiol.79(5), 382–398 (2004). [DOI] [PubMed] [Google Scholar]
- 76.Kumar, K., Debnath, P., Singh, S. & Kumar, N. An overview of Plant Phenolics and their involvement in abiotic stress tolerance. Stresses3(3), 570–585 (2023). [Google Scholar]
- 77.Kováčik, J. & Bačkor, M. Changes of phenolic metabolism and oxidative status in nitrogen-deficient Matricaria chamomilla plants. Plant. soil.297(1–2), 255–265 (2007). [Google Scholar]
- 78.Borges, C. V., Minatel, I. O., Gomez-Gomez, H. A. & Lima, G. P. P. Medicinal plants: Influence of environmental factors on the content of secondary metabolites. In (eds Ghorbanpour, M. & Varma, A.) Medicinal Plants and Environmental Challenges(Springer, 2017).
- 79.Roberts, M. R. & Paul, N. D. Seduced by the dark side: Integrating molecular and ecological perspectives on the influence of light on plant defence against pests and pathogens. New Phytol.170(4), 677–699 (2006). [DOI] [PubMed] [Google Scholar]
- 80.Kotilainen, T. et al. Assessment of UV biological spectral weighting functions for phenolic metabolites and growth responses in silver birch seedlings. Photochem. Photobiol.85(6), 1346–1355 (2009). [DOI] [PubMed] [Google Scholar]
- 81.Kuhlmann, F. & Müller, C. Impacts of ultraviolet radiation on interactions between plants and herbivorous insects: A chemo-ecological perspective. In Progress in Botany, Vol. 72, 305–347 (Springer, 2010).
- 82.Singh, S., Kaur, I. & Kariyat, R. The multifunctional roles of polyphenols in plant-herbivore interactions. Int. J. Mol. Sci.22(3), 1442 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Leiss, K. A., Maltese, F., Choi, Y. H., Verpoorte, R. & Klinkhamer, P. G. Identification of chlorogenic acid as a resistance factor for thrips in chrysanthemum. Plant Physiol.150(3), 1567–1575 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li, X. et al. (+)-Catechin, epicatechin and epigallocatechin gallate are important inducible defensive compounds against Ectropis grisescens in tea plants. Plant. Cell. Environ.45(2), 496–511 (2022). [DOI] [PubMed] [Google Scholar]
- 85.Patton, C. A., Ranney, T. G., Burton, J. D. & Walgenbach, J. F. Feeding responses of Japanese beetle to naturally occurring metabolites found in rosaceous plants. J. Environ. Hortic.15(4), 222 (1997). [Google Scholar]
- 86.Ramos-Ruiz, R., Martinez, F. & Knauf-Beiter, G. The effects of GABA in plants. Cogent Food Agric.5(1), 1670553 (2019). [Google Scholar]
- 87.Tarkowski, Ł. P., Signorelli, S. & Höfte, M. γ-Aminobutyric acid and related amino acids in plant immune responses: Emerging mechanisms of action. Plant. Cell. Environ.43(5), 1103–1116 (2020). [DOI] [PubMed] [Google Scholar]
- 88.Robinson, A. R., Ukrainetz, N. K., Kang, K. Y. & Mansfield, S. D. Metabolite profiling of Douglas-fir (Pseudotsuga menziesii) field trials reveals strong environmental and weak genetic variation. New Phytol.174(4), 762–773 (2007). [DOI] [PubMed] [Google Scholar]
- 89.Frank, T., Röhlig, R. M., Davies, H. V., Barros, E. & Engel, K. H. Metabolite profiling of maize kernels—Genetic modification versus environmental influence. J. Agric. Food Chem.60(12), 3005–3012 (2012). [DOI] [PubMed] [Google Scholar]
- 90.Neugart, S. et al. The intrinsic quality of brassicaceous vegetables: How secondary plant metabolites are affected by genetic, environmental, and agronomic factors. Sci. Hort.233, 460–478 (2018). [Google Scholar]
- 91.Effah, E., Barret, D. P., Pakeman, R. J., Beaton, J. K. & Clavijo McCormick, A. Calluna vulgaris volatile emissions suggest varying anti-herbivore defence strategies with plant ontogeny. Plant Ecol. Divers. 1–10 (2024).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant .raw spectral data sets are available upon reasonable request.