Abstract
Accurate molecular representation of compounds is a fundamental challenge for prediction of drug targets and molecular properties. In this study, we present a molecular video-based foundation model, named VideoMol, pretrained on 120 million frames of 2 million unlabeled drug-like and bioactive molecules. VideoMol renders each molecule as a video with 60-frame and designs three self-supervised learning strategies on molecular videos to capture molecular representation. We show high performance of VideoMol in predicting molecular targets and properties across 43 drug discovery benchmark datasets. VideoMol achieves high accuracy in identifying antiviral molecules against common diverse disease-specific drug targets (i.e., BACE1 and EP4). Drugs screened by VideoMol show better binding affinity than molecular docking, revealing the effectiveness in understanding the three-dimensional structure of molecules. We further illustrate interpretability of VideoMol using key chemical substructures.
Subject terms: Cheminformatics, Drug development
Accurate molecular representation of compounds is a fundamental challenge for drug discovery. Here, the authors present a molecular video-based foundation model pretrained on 120 million frames of 2 million molecular videos, and apply it to molecular targets and properties prediction.
Introduction
Drug discovery is a complex and time-consuming process that involves the identification of potential drug targets, the design and synthesis of compounds, and the testing of compounds for efficacy and safety1,2. For example, in traditional drug discovery, medicinal chemists and pharmacologists select and optimize candidate compounds based on knowledge and experience and verify them by screening cellular or animal models3. Computational drug discovery that uses computational and artificial intelligence technologies to assist drug development offers a promising approach to speeding up this process4,5. By leveraging large datasets of biological and chemical information, these computational approaches, such as foundation models6,7, can rapidly identify new drug targets8, design candidate molecules9, and evaluate the efficacy and properties of those candidates10, which substantially reduces the time and cost of traditional drug discovery and development.
Accurate molecular representation of hundreds of millions of existing and novel compounds is a fundamental challenge for computational drug discovery communities11,12. Traditional approaches used hand-crafted fingerprints as molecular representations, such as physicochemical fingerprints13, and pharmacophore-based fingerprints14. Limited by domain knowledge, these traditional representation approaches are subjective and immutable, devoid of adequate generalizability. With the rise of deep learning and self-supervised learning, automated molecular representation learning approaches can extract representations from molecular sequences15,16, graphs17,18, and images19,20 by pre-training on large-scale molecular datasets19. These approaches showed a substantial performance improvement in the various tasks of drug discovery7,19,21. Based on recent advances in video representation learning and self-supervised learning in computer vision22–24, self-supervised video-based pre-trained models offer compelling opportunities to further improve the performance of drug discovery.
In this study, we present a molecular video-based foundation model (termed VideoMol) for molecular representation learning. Specifically, VideoMol utilizes dynamic awareness and physicochemical awareness to learn molecular representation from a vast quantity of molecular 3D dynamic videos in an unsupervised manner. We implemented a self-supervised pre-training framework to capture the physicochemical information of compounds from 120 million frames of 2 million molecular videos with diverse biological activities at the human proteome. We demonstrate that VideoMol outperforms existing state-of-the-art methods in drug discovery tasks, including drug target and multiple molecular property predictions.
Results
Framework of VideoMol
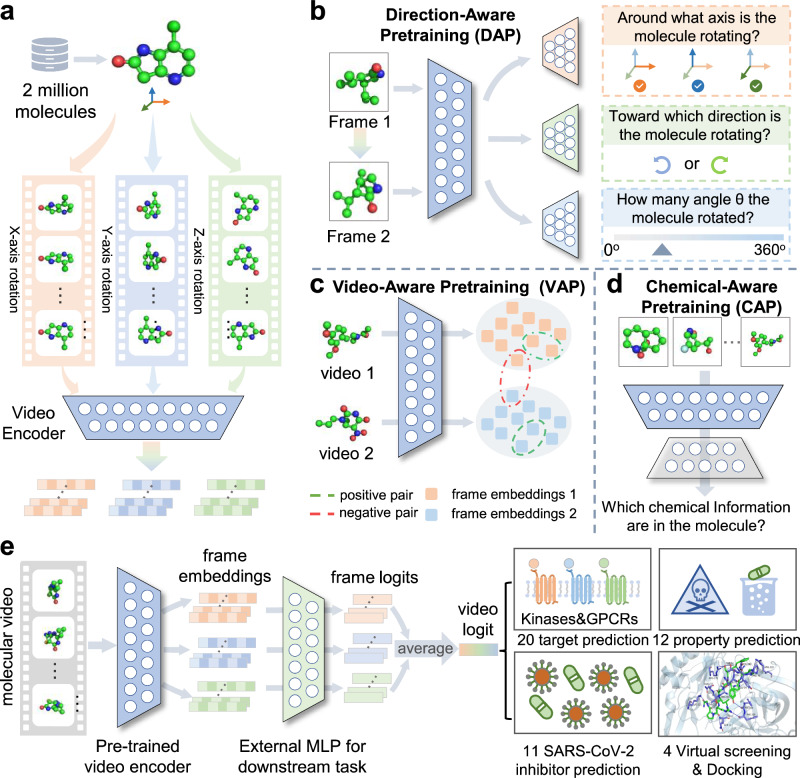
Molecules exist in nature and are constantly conformational dynamics, making video the most direct representation method. The molecular 3D information can be directly observed from the video without the help of manual feature extraction, such as the distance between pairs of atoms and the angle formed between multiple atoms and so on. In addition, we evaluated the advantages of different representations in feature extraction capabilities and found that our proposed video representation has obvious advantages over existing representations with a 39.8% improvement rate on 8 basic attributes (Supplementary Section C.2 and Supplementary Table 1). Therefore, these significant differences motivate us to develop VideoMol for accurately predicting the targets and properties of molecules in the form of videos derived from molecules. First, we generated conformations for 2 million drug-like and bioactive molecules and rendered a dynamic video with 60 frames for each 3D molecules (120 million frames in total). Then, we feed the molecular 3D videos into a video encoder to extract latent features (Fig. 1a) and implement three pretraining strategies to optimize the latent representation by considering changes of videos and physicochemical information of molecules (Fig. 1b–d). Finally, we fine-tune the pre-trained video encoder on downstream tasks (prediction of molecular targets and properties) to further improve the model performance (Fig. 1e). VideoMol achieves good interpretability through the use of Grad-CAM (Gradient-weighted Class Activation Mapping)25 to visualize the contribution of molecular videos to the prediction results with heatmaps (see below). To comprehensively evaluate the performance of VideoMol, we selected four types of tasks: (1) compound-kinase binding activity prediction, (2) ligand-GPCR (G Protein-Coupled Receptors) binding activity prediction, (3) anti-SARS-CoV-2 activity prediction, and (4) prediction of molecular properties. Details of these datasets are provided in the Methods section and the Supplementary Section A.1.
Fig. 1. Overview of the VideoMol foundational model.
a Feature extraction of molecular videos. First, we render 2 million molecules with conformers in 3D spatial structure. We then rotate the rendered molecule around the , , axes and generate snapshots for each frame of the molecule video. Finally, we feed the molecular frames into a video encoder to extract latent features. b–d Three self-supervised tasks for pre-training video encoder. The direction-aware pretraining (DAP) task is used to distinguish the relationship between pairs of molecular frames (such as the axis of rotation, the direction of rotation, and the angle of rotation) by using axis classifier (orange), rotation classifier (green) and angle classifier (blue). The video-aware pretraining (VAP) task is used to maximize intra-video similarity and minimize inter-video similarity. The chemical-aware pretraining (CAP) task is used to recognize information related to physicochemical structures in molecular videos by using chemical classifier (gray). e The finetuning of VideoMol on downstream benchmarks (such as binding activity prediction and molecular property prediction). A multi-layer perceptron (MLP) is added after the pre-trained video encoder for fine-tuning on four types of downstream drug discovery tasks (20 target prediction, 12 property prediction, 11 SARS-CoV-2 inhibitor prediction, and 4 virtual screening and docking). We assemble the results (logits) of each frame as the prediction result of molecular video (video logit).
Performance of VideoMol
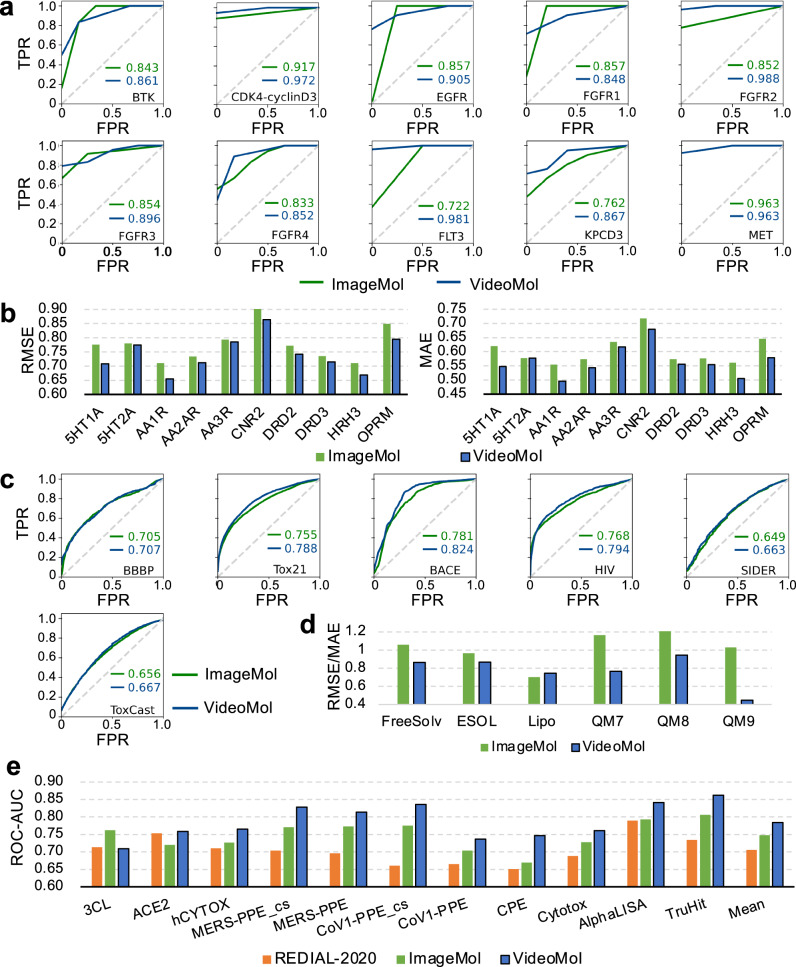
We first evaluated the performance of VideoMol with three types of state-of-the-art molecular representation learning methods on 10 compound-kinase (classification task) and 10 ligand-GPCR (regression task) binding activity prediction datasets with balanced scaffold split26. The state-of-the-art methods include: (1) sequence- (RNNLR, TRFMLR, RNNMLP, TRFMMLP, RNNRF, TRFMRF27, CHEM-BERT28), (2) graph- (MolCLRGIN, MolCLRGCN6), and (3) image-based models (ImageMol19) (Supplementary Table 2). For 10 compound-kinase interaction datasets, VideoMol achieves better AUC performance than other methods across BTK (AUC = 0.861 ± 0.023), CDK4-cyclinD3 (AUC = 0.972 ± 0.039), EGFR (AUC = 0.905 ± 0.017), FGFR1 (AUC = 0.848 ± 0.027), FGFR2 (AUC = 0.988 ± 0.017), FGFR3 (AUC = 0.896 ± 0.039), FGFR4 (AUC = 0.852 ± 0.080), FLT3 (AUC = 0.981 ± 0.026), KPCD3 (AUC = 0.867 ± 0.036), and MET (AUC = 0.963 ± 0.026) with an average performance improvement of 5.9% ranging from 1.8% to 20.3% (Fig. 2a and Supplementary Table 3). In particular, VideoMol outperforms the state-of-the-art methods of ImageMol and MolCLR with average improvements of 6.7% and 20.1%. For 10 ligand-GPCR binding datasets, VideoMol achieves the best results on all datasets with an average performance improvement by 4.5% on Root Mean Squared Error (RMSE) ranging from 0.60% to 8.8% and 5.9% on Mean Absolute Error (MAE) with a maximum of 11.4% (Fig. 2b and Supplementary Table 4). VideoMol achieves average performance improvement of 4.5% and 10.2% on RMSE, and 6.2% and 12.0% on MAE for ImageMol and MolCLR, respectively.
Fig. 2. Performance of the VideoMol framework on multiple drug discovery tasks.
a The ROC (Receiver Operating Characteristic) curves of ImageMol and VideoMol on 10 main types of biochemical kinases with balanced scaffold split. The x-axis and y-axis represent FPR (False Positive Rate) and TPR (True Positive Rate), respectively. b The RMSE (Root Mean Squared Error) and MAE (Mean Absolute Error) performance of ImageMol and VideoMol on 10 GPCR with balanced scaffold split. c The ROC curves of ImageMol and VideoMol on 6 molecular property prediction benchmarks with scaffold split. d The RMSE (FreeSolv, ESOL, Lipo) and MAE (QM7, QM8, QM9) performance of ImageMol and VideoMol with scaffold split. For each of presentation, the values of FreesSolv and QM7 are scaled down by a factor of 2 and 100, respectively, and the values of QM8 and QM9 are scaled up by a factor of 50 and 100, respectively. e The ROC-AUC (Area Under the Receiver Operating Characteristic Curve) performance of REDIAL-2020, ImageMol, and VideoMol on 11 SARS-CoV-2 datasets. Source data are provided as a Source Data file.
We next turned to evaluate the performance of VideoMol with 18 popular and competitive baselines on 12 types of molecular property prediction tasks with scaffold split: (1) molecular targets—beta-secretase (BACE, a key drug target in Alzheimer’s disease) and anti-viral activities in human immunodeficiency virus (HIV), (2) blood–brain barrier penetration (BBBP); (3) drug metabolism and side effect resource (SIDER); (4) molecular toxicities—toxicity using the Toxicology in the 21st Century (Tox21) and Toxicity Forecaster (ToxCast); (5) solubility—Free Solvation (FreeSolv) and Estimated Solubility (ESOL)—and lipophilicity (Lipo); (6) quantum—Quantum Machine 7 (QM7), QM8 and QM9. We compared VideoMol with 3 different types of state-of-the-art methods, including 2D-graph- (InfoGraph29, GPT-GNN30, ContextPred17, GraphLoG31, G-Contextual17, G-Motif18, AD-GCL32, JOAO33, SimGRACE34, GraphCL35, GraphMAE36, MGSSL33, AttrMask17, MolCLR6, Mole-BERT37), 3D-graph- (3D InfoMax38, GraphMVP39, Uni-Mol40), and image-based methods (ImageMol19) (Supplementary Table 2). In classification tasks, using the area under the receiver operating characteristic (ROC) curve (AUC), VideoMol achieves elevated performance across BBBP (AUC = 70.7% ± 1.5), Tox21 (AUC = 78.8% ± 0.4), HIV (AUC = 79.4% ± 0.5), BACE (AUC = 82.4% ± 0.9), SIDER (AUC = 66.3% ± 0.9), ToxCast (AUC = 66.7% ± 0.5), outperforming other methods (Fig. 2c and Supplementary Table 5). In regression task, VideoMol achieves better performance (low error values) than FreeSolv (RMSE = 1.725 ± 0.053), ESOL (RMSE = 0.866 ± 0.017), Lipo (RMSE = 0.743 ± 0.009), QM7 (MAE = 76.436 ± 1.561), QM8 (MAE = 0.01890 ± 0.0020), and QM9 (MAE = 0.00896 ± 0.00003), outperforming other methods (Fig. 2d and Supplementary Table 6).
We next turned to evaluate VideoMol on anti-SARS-CoV-2 viral activity prediction. Specifically, we evaluated 11 SARS-CoV-2 biological assays, which covers multiple therapeutic approaches such as viral replication, viral entry, counter-screening, in vitro infectivity, and live virus infectivity41. Compared with REDIAL-202041 (molecular fingerprint-based method) and ImageMol19 (image-based representation method), we found that VideoMol achieved better ROC-AUC performance (3CL = 0.709 ± 0.006, ACE2 = 0.759 ± 0.025, hCYTOX = 0.765 ± 0.003, MERS-PPE_cs=0.828 ± 0.027, MERS-PPE = 0.814 ± 0.004, CPE = 0.747 ± 0.013, CoV1-PPE_cs=0.836 ± 0.029, CoV1-PPE = 0.737 ± 0.007, Cytotox = 0.761 ± 0.002, AlphaLISA = 0.841 ± 0.004, TruHit=0.862 ± 0.002) with an average 3.9% improvement ranging from 3.3% to 7.8% compared with ImageMol and an average 8.1% improvement ranging from 0.6% to 17.5% compared with REDIAL-2020 (Fig. 2e and Supplementary Table 7).
We next turned to calculate the uncertainty intervals with 95% confidence intervals (CI) of ImageMol and VideoMol using 10 ligand-GPCR binding activity prediction datasets and 11 SARS-CoV-2 viral activity prediction datasets. In details, we used the popular bias-corrected and accelerated (BCa) bootstrap intervals42,43 to calculate 95% CI, which corrects for both bias and skewness of the bootstrap parameter estimated by incorporating a bias-correction factor and an acceleration factor. The results of the uncertainty interval show the effectiveness of VideoMol with an average improvement ranging from 5.44% to 10.07% compared to ImageMol (Supplementary Tables 8 and 9).
In summary, VideoMol is an effective molecular video-based representation learning method in multiple drug discovery tasks, outperforming state-of-the-art methods (Fig. 2 and Supplementary Tables 3–9).
Discovery of ligand-receptor interactions via VideoMol
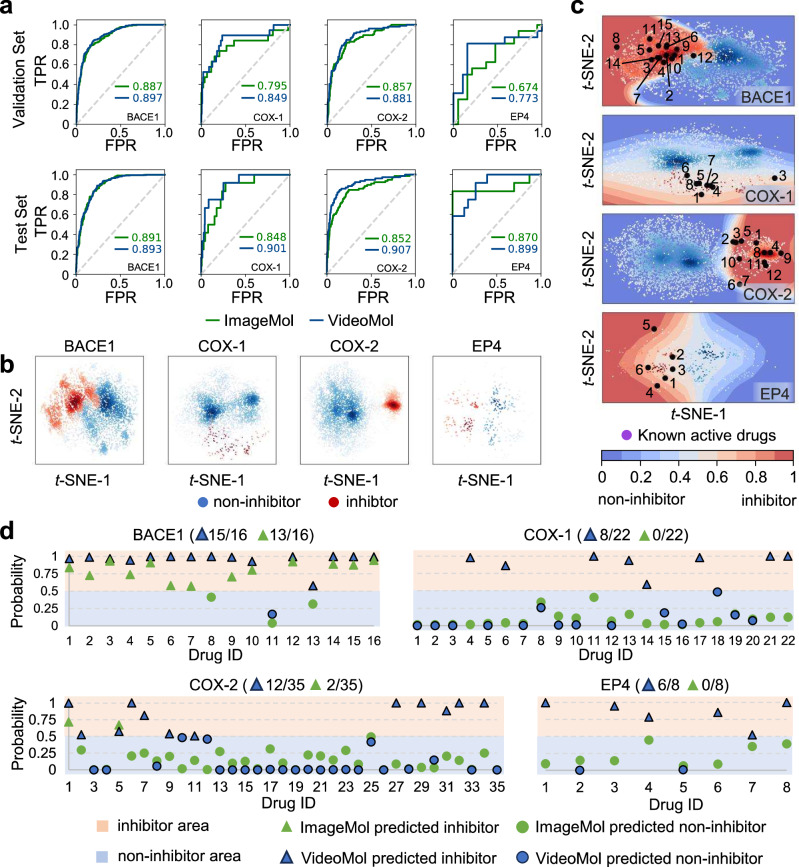
We next turned to identifying novel ligand-receptor interactions via VideoMol for 4 well-known human targets: beta-secretase 1 (BACE1), cyclooxygenase 1 (COX-1), COX-2, and prostaglandin E receptor 4 (EP4), in order to assess the generalizability of the model. We collected the training data of these 4 targets from the ChEMBL database44 and evaluated the performance of VideoMol on these targets with a random split of 8:1:1 (Supplementary Table 10). Using ROC-AUC metric evaluation, we found that VideoMol achieved high performance on both validation set (BACE1 = 0.897, COX-1 = 0.849, COX-2 = 0.881 and EP4 = 0.773) and test set (BACE1 = 0.893, COX-1 = 0.901, COX-2 = 0.907 and EP4 = 0.899), outperforming ImageMol with an average improvement rate of 6.4% in the validation set and an average improvement rate of 4.1% in the test set (Fig. 3a). The t-SNE (t-distributed Stochastic Neighbor Embedding)45 visualization in the latent space showed a clear boundary between inhibitors and non-inhibitors on all 4 targets, suggesting accurate representation of VideoMol to learn discriminative information (Fig. 3b).
Fig. 3. The virtual screening on four common drug targets (BACE1, COX-1, COX-2 and EP4).
a The ROC (Receiver Operating Characteristic) curves of ImageMol and VideoMol on validation set (the first row) and test set (the second row). b t-SNE (t-distributed Stochastic Neighbor Embedding) visualization of latent features extracted by VideoMol. c The drug discovery on BACE1, COX-1, COX-2, and EP4. Blue and red points represent non-inhibitors and inhibitors from the ChEMBL dataset, respectively. Black points indicate known inhibitors. The decision boundary is drawn by training an SVM using the ChEMBL dataset, where blue to red indicates that the probability of belonging to the inhibitor gradually increases. d Virtual screening on known inhibitors of BACE1 (16 drugs), COX-1 (22 drugs), COX2 (35 drugs), and EP4 (8 drugs). The x-axis and y-axis represent the number of the drug and the predicted probability (inhibitors), respectively. The orange and blue backgrounds represent inhibitor area and non-inhibitor area, respectively. The green and blue triangles represent inhibitors predicted by ImageMol and VideoMol respectively. The green and blue circles represent non-inhibitors predicted by ImageMol and VideoMol respectively. The numbers represent the precision of ImageMol and VideoMol. Source data are provided as a Source Data file.
We further collected 16 BACE1 inhibitors (Supplementary Table 11), 22 COX-1 inhibitors (Supplementary Table 12), 35 COX-2 inhibitors (Supplementary Table 13) and 8 EP4 inhibitors (Supplementary Table 14) from the MedChemExpress database (https://www.medchemexpress.com/, see Supplementary Section A.1). We found that VideoMol successfully re-identified 15 BACE1 inhibitors (93.8% success rate), 8 COX-1 inhibitors (36.4% success rate), 11 COX-2 inhibitors (34.3% success rate) and 6 EP4 inhibitors (75.0% success rate) (Fig. 3c, d and Supplementary Tables 15–18). Compared with ImageMol, VideoMol achieved significantly better generalizability on these four external validation targets with an average precision improvement of 38.1% ranging from 12.5% to 75.0%.
Discovery of BACE1 inhibitors from existing drugs via VideoMol
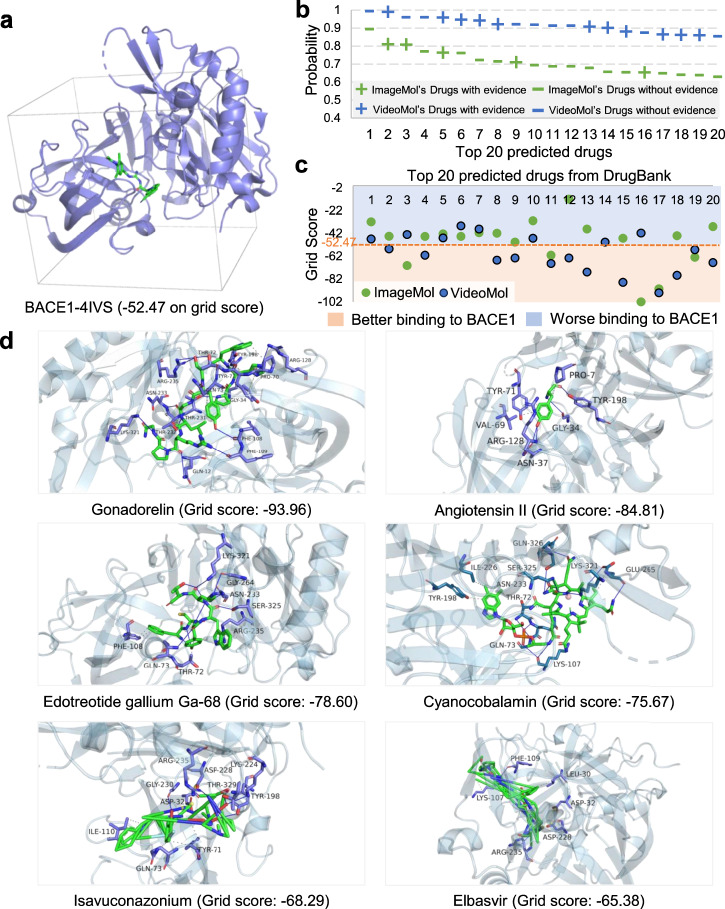
BACE1 (beta-site amyloid precursor protein cleaving enzyme 1), is a key drug target in Alzheimer’s disease (AD) and there is lack effective small molecular treatment for AD to date46. We next turned to screening potential inhibitors via specifically targeting BACE1 from 2,500 approved drugs from the DrugBank database47 using VideoMol (Supplementary Table 19). We downloaded the known X-ray crystal structure of BACE1 (PDB ID: 4IVS) with a co-crystallized inhibitor bearing the indole acylguanidine core structure (Ligand ID: VSI)48 from the PDB (Protein Data Bank)49 database. We evaluated grid score (a metric of binding ability and the smaller value denotes the better score) between ligand and receptor (PDB ID: 4IVS, Fig. 4a) by Dock6.1050. We illustrated the BACE1 inhibitor prediction results for both ImageMol and VideoMol in Supplementary Table 20 and Supplementary Table 21. We collected experimental evidence for the top 20 drugs predicted by VideoMol and ImageMol from the published literatures. We found that 11 of the 20 drugs predicted by VideoMol were validated as potential treatment for AD (55% success rate), which was higher than the 5 drugs of ImageMol (25% success rate). (Fig. 4b). These reflected that VideoMol can learn more 3D information and chemical information to ensure the high confidence of predictions. We further evaluated the grid scores of the top 20 predicted drugs to the 4IVS crystal structure by using Dock6.10. Using the grid score of 4IVS crystal structure (Fig. 4a) as a threshold, VideoMol prioritizes more drugs with better grid scores of −52.47 (60%, 12 out of 20 drugs) compared to ImageMol (20%, 4 out of 20 drugs) (Fig. 4c), revealing that VideoMol captures 3D information compared to ImageMol derived from static 1D and 2D image-based representation19. Finally, we selected 6 drugs with the best grid score from the top 20 drugs for molecular docking simulation (Fig. 4d). We found that 5 out of 6 drugs (83.3%) had been validated as potential treatment for AD based on existing published experimental data (Supplementary Table 22).
Fig. 4. Computational validation of VideoMol-predicted drugs on human beta-secretase 1 (BACE1).
a The 4IVS crystal structure of BACE1. The gray tetragon represents the area of the docking pocket. The grid score is calculated by Dock6.10 (the smaller, the better). b Top 20 drugs prioritized by VideoMol and ImageMol to be active against the BACE1 target. The x-axis and y-axis represent the drug and the predicted probability (inhibitors), respectively. Green and blue represent ImageMol’s drugs and VideoMol’s drugs respectively. Minus/plus signs indicate whether a predicted drug is supported by existing experimental data from publish literatures. c The docking results of the Top 20 drugs predicted by VideoMol. The x-axis and y-axis represent the index and grid scores of the drug respectively. Light blue and light orange areas indicate worse and better grid scores than the 4IVS (grid score = −52.47), respectively. d Docking examples of 6 drugs (Gonadorelin, Angiotensin II, Edotreotide gallium Ga-68, Cyanocobalamin, Isavuconazonium and Elbasvir) with the best grid scores. The numerical value in the bracket represents the grid score. The molecular structures shown here can be found in Supplementary Dataset 1. Source data are provided as a Source Data file.
Video visualization and model interpretability
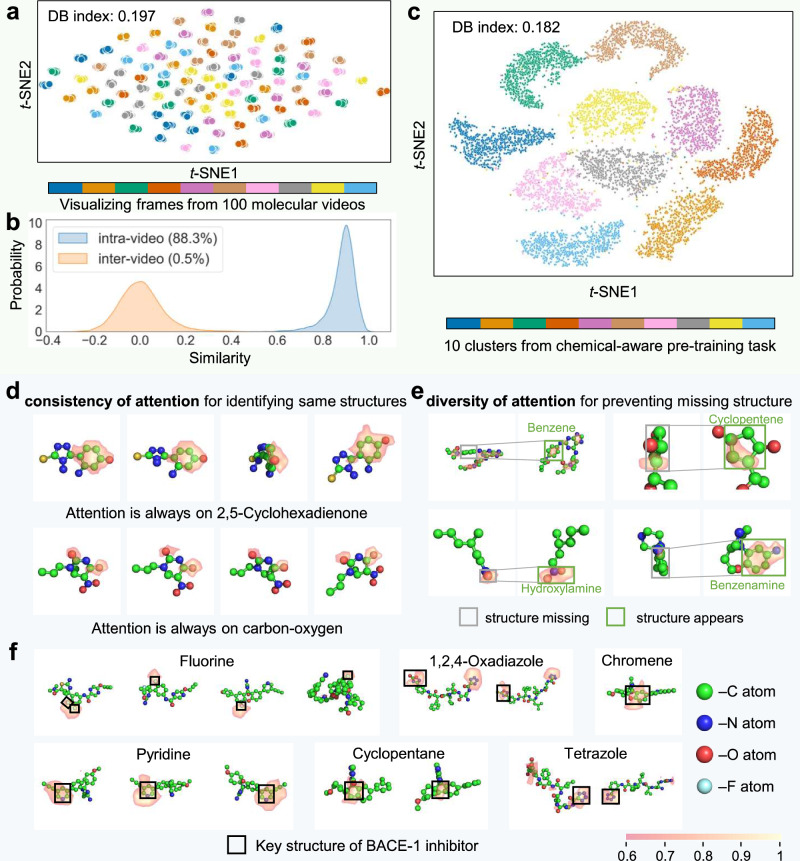
Since each frame in the molecular videos represents the same molecule, their projections in the feature space should be similar. To evaluate the discriminative power of VideoMol on molecular videos, we randomly selected 100 molecular videos and extract features for each frame in the videos. Subsequently, we used t-SNE to project each feature into a two-dimensional space (Fig. 5a and Supplementary Fig. 1). Frames from the same video are well clustered together, while frames from different videos are clearly separated. We also quantitatively evaluated the DB (Davies Bouldin) index51 of these clusters. VideoMol achieved a low DB index (the value is 0.197), indicating that VideoMol has the ability to recognize different frames of the same molecule. We randomly sample 10,000 pairs of molecular frames from the same and different molecular videos respectively and compute the cosine similarity between these paired samples. As expected, there is a high average similarity (88.3%) on intra-video and almost zero (0.5%) on inter-video, indicating that VideoMol is robust to different 3D views of the same molecule (Fig. 5b).
Fig. 5. Biological interpretation and feature distribution of VideoMol.
a Visualization of each frame in 100 molecular videos (60 frames for each video). Representations are extracted by VideoMol and dimensionally reduced by t-SNE. Different colors represent frames in different cluster videos. DB index is a metric to evaluate the clustering quality, and the larger the value, the better the clustering performance. b Similarity distribution (n = 20,000 samples) of intra-video and inter-video. Similarity is computed using a pair of frames from intra-video or inter-video. The content in brackets indicates the average similarity of the distribution. c t-SNE visualization (10,000 samples) of features extracted by VideoMol. Different colors represent different cluster labels (this cluster label is obtained in the chemical-aware pretraining task). d–f Grad-CAM visualization of VideoMol on molecular frames. We use 0.6 as the threshold for visualization, that is, set the importance lower than 0.6 to 0. In d, each row represents a molecular video. In e, pairs of molecular frames represent frames where structure is missing and frames where structure appears, respectively. In f each panel represents examples of key structures related to BACE-1 inhibitory activities from frames of different molecules. Source data are provided as a Source Data file.
To investigate how the physicochemical information contribute performance of VideoMol, we used t-SNE to visualize the representation of VideoMol with cluster labels from a chemical-aware pretraining task. We randomly selected 10 clusters (1000 samples for each cluster) for visualization. As shown in Fig. 5c, the representations extracted by VideoMol produce clusters with sharp boundaries with a low DB index = 0.182, indicating that VideoMol learned physicochemical knowledge well. We further visualized 30 additional clusters with 500 samples per cluster and found that different cluster labels still produce strong clustering effect (Supplementary Fig. 2).
To inspect how VideoMol performs the inference process, we used GradCAM25 to visualize attention heatmaps for molecular videos. Figure 5d, e respectively shows the consistency and diversity of VideoMol’s attention (green represents carbon atom C, blue represents nitrogen atom N, red represents oxygen atom O and cyan represents fluorine atom). We found that as the video played, VideoMol was always able to attend to the same molecular substructure (Fig. 5d), such as 2,5-Cyclohexadienone (composition of 6 carbon atoms and 1 oxygen) in the first row, and carbon-oxygen structure in the second row, which shows that VideoMol has consistency for different frames in the same video. In Fig. 5e, we see that VideoMol can pay attention to diverse structural information as the video plays to alleviate the missing structural information. For example, VideoMol cannot attend to benzene-, cyclopentene-, hydroxylamine- and benzenamine-structure through the use of Grad-CAM in the left frame, whereas VideoMol can attend to these structures in the right frame.
To test that VideoMol can provide chemists with meaningful knowledge related to predictive targets, we evaluate the interpretability of VideoMol in prediction of BACE-1 inhibitors. We found that VideoMol identified known chemistry knowledge related to BACE-1 inhibitors, such as fluorine52, 1,2,4-Oxadiazole53, chromene54, pyridine55, cyclopentane56, tetrazole53 (Fig. 5f), which was verified by wet experiments (all evidence can be found in Supplementary Table 23). For instance, VideoMol maintained high attention on fluorine when predicting a compound as an inhibitor of BACE-1, which was validated by a previous experimental study52.
Ablation study
To study the impact of the pre-training strategies on VideoMol, we train VideoMol with different pre-training tasks, including w/o pre-training, only video-aware strategy, only direction-aware strategy and only chemical-aware strategy, chemical&direction, chemical&video, and direction&video (Supplementary Table 24). We found that pre-training tasks can improve the performance of VideoMol on downstream tasks compared to VideoMol without pre-training with 27.1% average RMSE improvement and 31.0% average MAE improvement. Ablation experiments on single or double pre-training tasks show that chemical-awareness is important and the pre-training tasks guide VideoMol to learn meaningful chemical knowledge in the molecule. We can also see a trend that with the integration of pre-training tasks, the performance of VideoMol improves consistently in most evaluations, indicating that these pre-training tasks complement each other. Overall, the proposed pre-training tasks are effective for improving the performance of VideoMol.
To test the effectiveness of molecular features captured by VideoMol, we use VideoMol to extract the features of each frame in the molecular video and calculate their mean value as the molecular feature (called VideoMolFeat). For a fair comparison, we integrated 21 traditional molecular fingerprints, which are obtained by fingerprint stitching and PCA (Principal Component Analysis)57 dimensionality reduction (called EnsembleFP). We then evaluated the performance using the MLP (Multilayer Perceptron) implemented by scikit-learn58 and kept the same experimental settings. We found that VideoMolFeat achieved the best performance with 17.0% average RMSE improvement and 19.6% average MAE improvement compared with EnsembleFP on 10 GPCR datasets, which illustrates that a visual molecular feature is effective as a superior alternative to traditional molecular fingerprinting (Supplementary Table 25).
To explore the impact of different frame numbers on VideoMol, we sampled 5, 10, 20, 30, and 60 molecular frames from 5HT1A, AA1R, AA2AR, CNR2, DRD2, and HRH3 datasets at equal time intervals. We found that the performance of VideoMol is positively correlated with the number of frames with an average performance improvement of 6.6% (510 frames), 3.9% (1020 frames), 1.9% (2030 frames), 4.0% (3060 frames) on RMSE metric and 7.6% (510 frames), 4.6% (1020 frames), 2.3% (2030 frames), 4.5% (3060 frames) on MAE metric, which shows that the increase of frame number enriches the 3D information extracted by VideoMol and its performance may be expected to be further increased by expanding the frame number (Supplementary Table 26).
To verify the sensitivity of VideoMol to video generation sources, we used two additional platforms to generate molecular videos, which are OpenBabel59 and DeepChem60. We found that the video generation source of different platforms has no significant impact on VideoMol with an average performance of 0.755 ± 0.068 (Openbabel), 0.755 ± 0.072 (DeepChem), 0.742 ± 0.064 (RDKit) in RMSE metric and 0.581 ± 0.057 (Openbabel), 0.576 ± 0.060 (DeepChem), 0.566 ± 0.053 (RDKit) in MAE metric (Supplementary Table 27). Therefore, VideoMol has low sensitivity to video generation sources from different platforms.
To investigate whether VideoMol can identify conformational changes of molecules, we used pre-trained VideoMol to extract features of molecules with different conformers from 10 ligand-GPCR binding activity prediction datasets and compared the cosine similarities between different videos with different conformers. Since the similarity between conformers is related to their RMSD (Root-Mean-Square Deviation) distance, we also calculated the similarity of features in different RMSD intervals. We found that VideoMol is discriminative for videos from different conformers (Supplementary Table 28). Further, when the RMSD between two conformations is larger, the feature similarity extracted by VideoMol shows a decreasing trend. Especially in the 90-100 percentile range, the feature similarity extracted by VideoMol is always the lowest. These comprehensive observations show that VideoMol can identify conformational differences effectively.
Discussion
We have proposed a self-supervised video-processing-based pre-training framework, VideoMol, that learns molecular representations by utilizing dynamic awareness and physicochemical awareness. We showed the high performance of VideoMol on various drug discovery tasks, including predicting molecular target profiles (e.g., GPCRs, kinases, SARS-CoV-2) and molecular properties (e.g., pharmacology, biophysics, physical, and quantum chemistry). We evaluated the effectiveness of VideoMol on 4 common targets (BACE1, COX-1, COX-2, and EP4) from ChEMBL (Figs. 1a and 2b). We also verified the high precision of VideoMol on the virtual screening of 4 targets (BACE1, COX-1, COX-2 and EP4), which are consistent with ongoing clinical and experimental data (Figs. 3c and 4d). Compared with ImageMol, VideoMol achieved an average precision improvement of 38.1% on these 4 targets, which showed that VideoMol is able to generalize to external validation sets. Especially in the virtual screening of COX-1, COX-2 and EP4 inhibitors, VideoMol achieved significant advantages, demonstrating VideoMol can overcame data imbalance (imbalance rates of 0.043 and 0.253 in COX-1 and COX-2 from ChEMBL) and data scarcity (only 350 samples in EP4 from ChEMBL) scenarios.
On the interpretability of VideoMol, we found that the attention of VideoMol is different on different frames of the same video in Fig. 5e, which is due to occlusion of viewing angles problem that make useful information often scattered in different views61. This showed the advantage of molecular video, allowing VideoMol to learn more molecular information by scanning each frame. In addition, it is worth noting that VideoMol can perceive substructures in occlusion scenes (such as the third column of the first row in Fig. 5d).
We highlighted several improvements of VideoMol over state-of-the-art: (1) VideoMol achieves high performance on various benchmark datasets (including property prediction and target binding activity prediction), outperforming the state-of-the-art representation learning methods (Supplementary Tables 3–9); (2) VideoMol overcomes class imbalance and data scarcity scenarios and achieves high accuracy and strong generalization in virtual screening on 4 common targets (BACE1, COX-1, COX-2 and EP4) (Fig. 3, Supplementary Table 10 and Supplementary Tables 15–18); (3) VideoMol captures 3D information and is good at predicting ligands with high binding capacity to receptors (Supplementary Tables 21 and 22 and Fig. 4); (4) the representation of VideoMol is robust to inconsistent views of the molecule (Fig. 5a, b) and contains rich and meaningful physicochemical information (Fig. 5c); (5) VideoMol has good interpretability, which is intuitive and informative for identifying chemical structures or substructures related to molecular properties and target binding, and can solve the occlusion of viewing angles problem (Fig. 5d–f). Furthermore, to explore the effect of frame number on VideoMol, we extract frames in videos with equal spacing. We found that the performance of VideoMol can gradually improve with the number of frames, which shows that the combination of different frames can enrich the feature representation of molecules (Supplementary Table 26).
VideoMol is a novel molecular representation learning framework, which is significantly different from previous sequence-, graph- and image-based molecular representation learning methods. VideoMol treated molecules as dynamic videos and learned molecular representations in a video processing manner, which means that a large number of video representation learning technologies can be used for learning molecular representation23,62. Compared with our previous ImageMol, VideoMol had several substantial upgrades, including: (1) the content of molecular visual representation is upgraded from 2D pixel information to 3D pixel information; (2) molecular pre-training is upgraded from image-based learning to video-based learning; (3) the fingerprint information included is upgraded from the previous 1 fingerprint (MACCS key) to 21 fingerprints (Supplementary Table 29). Since VideoMol involves research fields such as image representation learning, video representation learning, and multi-view representation learning, it has greater research potential and motivates more researchers for greater performance improvement.
We acknowledged several potential limitations of VideoMol. While molecular video can achieve performance improvements, it will increase the computational complexity. Although the multi-view fine-tuning strategy can reduce the computational complexity, the choice of view is still a problem. Like other 3D-based molecular representation methods38,39,63, VideoMol does not take the diversity of conformers into account, but it can easily be improved by modeling consistency between different conformers. Several potential directions may further improve VideoMol: (1) Use of more biomedical data to train a larger version of VideoMol, noting that this would increase demand on computing resource; (2) Under resource constraints, use of pruning strategies (including data pruning and model pruning) to reduce the computational complexity of VideoMol; (3) Due to the rich physical and chemical information integrated in VideoMol, the distillation based on VideoMol is a meaningful research direction, which uses VideoMol as a teacher model to guide the learning of other student models (such as sequence-based models, graph-based models, etc.); (4) Use of better video processing methods and ensemble learning methods to integrate information between different frames is also an important direction to improve performance. Using a simple extension to VideoMol, we can allow the model to learn the correlations and variances between different conformations in the same molecule from videos of dynamic changes, thereby further playing an important role in molecular dynamics scenarios.
We believe that it is promising to represent molecules and perform inferences through videos as molecular imaging techniques continue to advance. In summary, the introduction of VideoMol on the one hand enriches the form of molecular representation in the field of computational drug discovery, and on the other hand inspires people to learn and understand the molecules from different perspectives.
Methods
Molecular conformer generation
When pre-training, we directly use the conformational information provided in PCQM4Mv2 database64. However, during fine-tuning, molecules in downstream tasks do not contain corresponding conformational information, so we obtain molecular conformers through a multi-stage generation method. We first remove the hydrogen atoms from the molecule and use MMFFOptimizeMolecule() in RDKit with MMFF94 (Merk Molecular Force Field 94) and a maximum number of iterations to generate conformers in a pre-determined coordinate system. Then, we judge whether the generated conformer has converged. If the conformer does not converge, we increase the maximum number of iterations by and repeat this process 10 times until convergence. Finally, if RDKit fails to generate a conformer or the conformer has not converged after 10 attempts, we directly use the 2D conformation instead. In addition, when the server resources are sufficient, we also use to directly generate conformers instead of multi-stage generation.
Molecular video generation
After obtaining molecular conformers, these conformers undergo counterclockwise rotations () about the positive , , and axes to generate snapshots (Fig. 1a), where is from 1 to 20 and snapshots are generated for all axes. Specifically, the matrix represents a counterclockwise rotation about the positive -axis by -th angle, which can be formalized as:
| 1 |
where . We generate a molecular frame (the -th frame of the -th molecule) for each snapshot using PyMOL (A software for visualization and rendering of molecular 3D structures)65 with stick-ball mode, where 3 represents the three channels (RGB or BGR) and 224 represents width and height. The key command used to render molecules in PyMOL is bg_color white;hide (hydro);set stick_ball,on;set stick_ball_ratio,3.5;set stick_radius,0.15;set sphere_scale,0.2;set valence,1;set valence_mode,0;set valence_size, 0.1. Since PyMol generates frames by default, we need to post-process them to get frames. Specifically, we expand the frame to by adding white pixels around it and resize it to . Finally, these 60 frames are stitched sequentially to generate molecular videos (where represents the number of molecules).
Strategies for pre-training VideoMol
Pretraining aims to improve the model’s ability to focus on crucial information in the molecular video, enabling more meaningful feature extraction.
In this paper, to obtain information in molecular videos from different perspectives, we consider three pre-training tasks (Fig. 1b–d): video-aware pre-training, direction-aware pre-training, and chemical-aware pre-training. Specifically, video-aware pre-training equips the model with the ability to distinguish different molecular videos, such as whether the two frames are from the same video. Direction-aware pre-training enables the model to discriminate the relationship between each frame, such as the angle of difference between two frames. Chemical-aware pre-training helps the model mine physiochemical-related information in videos.
Considering the efficiency and scalability of VideoMol, we perform independent feature extraction on each frame in molecular videos. Specifically, for the -th frame of the given -th video, we feed it into the video encoder to obtain the frame feature . In pre-training, given a batch of molecular videos , we randomly sample two frames and from each video, where and have the same axis of rotation. Then, we input and to video encoder to extract latent features and , where is the hidden dimension. Finally, the batch of data in the form of feature matrix is constructed for the following pre-training.
Identifying the differences between videos is important for the model to learn discriminative information between molecules because a video only describes one molecule. Meanwhile, different frames in a video describe different views of molecules in 3D space, which leads to unstable representation of the model when extracting different frames from the same video. Therefore, we propose a video-based pre-training task, called video-aware pretraining (VAP), to model the inter-video frame similarity and intra-video frame dissimilarity, i.e., frames from the same video should be close together, while frames from different videos should be far apart. Specifically, our approach uses contrastive learning to train molecular video representations, contrasting positive pairs of latent vectors against negative pairs. Given a batch of frame latent matrix , the is obtained by flattening . We define two frames from the same molecular video as positive pair and the others as negative. Therefore, the samples that can form positive pairs with the -th sample in are itself and the -th sample. Thus, our VAP objective is formalized based on InfoNCE loss as follows:
| 2 |
where is the -th latent vector in , is the temperature parameter, and is the parameters of the video encoder.
Correlating two snapshots from a continuously rotating object is trivial for humans, which benefits from the human ability to reason and imagine the 3D structure of objects based on prior knowledge. For example, humans can easily associate occluded regions when they only observe limited unoccluded local regions. Therefore, to equip the model with such ability, we propose direction-aware pretraining (DAP), which consists of three prediction tasks: i) axis, ii) rotation, and iii) angle prediction. First, a residual matrix is generated by substracting the first channel with the second, e.g. . Then, the features of each row in the residual matrix are passed separately through three classifiers (Multi-Layer Perceptrons), namely axis classifier , rotation classifier , and angle classifier . The classifiers are trained to predict the axis of rotation (-, -, or -axis), the direction of rotation (clockwise or counterclockwise), and the angle of rotation (an integer from 1 to 19) of with respect to , respectively. Cross-entropy loss is used for these MLPs, and the DAP losses are defined as follows:
| 3 |
where represents one of the prediction tasks (i.e., axis, rotation, angle), is the ground truth of corresponding prediction task in vector form, is the parameters of the classifier predicting , is the number of corresponding categories, and is the parameters of the video encoder.
The chemical knowledge is important for improving the performance of drug discovery19. Here, we introduced a Multi-Chemical Semantics Clustering (MCSC) with 20 additional fingerprint descriptors (details are provided in Supplementary Section B.1 and Supplementary Table 29), called Chemical-aware pretraining (CAP), to further extract more physicochemical information. In details, we first extract 21 molecular fingerprints for all molecules, and reduce the dimensionality of each fingerprint to 100 dimensions using PCA (Principal Components Analysis)66. We stitch together the reduced molecular fingerprints to get the MCSC fingerprint . Then, we use -Means to cluster MSCS fingerprints (molecules with similar MCSC distances were grouped together) and assign a corresponding pseudo-label to each cluster. We chose as the appropriate number of clusters. Finally, we employed a chemical classifier, which is MLP with as its parameters, to predict the pseudo-labels from the latent frame vectors and . The cost function of the CAP task can be formalized as follows:
| 4 |
where is the multinomial logistic loss or the negative log-softmax function, and is the parameters of the video encoder.
Pre-training process
To pre-train VideoMol, we sample 2 million unlabeled molecules and their corresponding conformers from the PCQM4Mv2 (a public access database on quantum chemistry, Supplementary Section A.1 for details), and generate molecular videos with 60 frames. We randomly sample 90% of molecular videos for training and the remaining 10% for evaluation in pre-training stage. The pre-training of VideoMol includes three important components, which are video encoder selection, data augmentation and pre-training process, respectively.
In video encoder selection, encouraged by the surprising performance of vision transformers (ViT)67 in computer vision, we use a 12-layer ViT as the video encoder of VideoMol. For each frame in the video, the video encoder splits it into patches as input and extracts 384-dimensional features. See Supplementary Table 30, Supplementary Section A.2 and Supplementary Section B.2 for more details of hyperparameters and model in pre-training stage.
Data augmentation is a simple and effective method to improve the generalization and robustness of model and is widely used in various artificial intelligence tasks. Because molecular videos contain structural and geometric information about compounds, augmentation methods that affect this information, such as RandomRotation and RandomFlip, cannot be used. Here, we have chosen four data augmentation methods: (1) CenterCrop with a size of 224; (2) RandomGrayscale with 30% probability of occurrence; (3) ColorJitter with brightness, contrast, saturation of and 30% probability of occurrence; (4) GaussianBlur with a kernel size of 3, sigma of (0.1, 2.0) and 30% probability of occurrence. Molecular videos are sequentially processed by these augmentations and normalized using Normalize with ImageNet default mean (0.485, 0.456, 0.406) and default standard deviation (0.229, 0.224, 0.225). These augmentation methods are provided by the PyTorch library68.
Since the pre-training process involves multiple optimization objectives (e.g. and ), we use a weighted multi-objective optimization algorithm to allow the model to benefit from each pre-training task, whose core idea is to use variable weights related to loss of task to control the pre-training tasks that the model focuses on. Specifically, we compute the loss weights by (), where represents any one of and and represents the number of tasks. The final weighted multi-task loss can be formalized as:
| 5 |
Finally, we use the weighted loss function to optimize the parameters of VideoMol by using mini-batch stochastic gradient descent. See Supplementary Section A.2, Supplementary Section B.2, and Supplementary Table 30 for more pre-training details and see Supplementary Section C.1 and Supplementary Figs. 3 and 4 for pre-training logging.
Fine-tuning process
After pre-training, we add an external multi-layer perceptron (MLP) after the video encoder for fine-tuning of downstream tasks. In the MLP, the number of output neurons in the last layer is equal to the number of downstream tasks . In details, given a batch of molecular videos with frames , we input each frame in molecular videos into video encoder to extract latent features , where is the hidden dimension. Then, we further forward-propagate latent features into the external MLP to obtain the logit (relevant to downstream tasks) of each frame. Since different frames of the same video describe the same molecule, we average the logit of frames from the same video as the final logit of the molecule . Finally, we use cross-entropy loss to optimize classification tasks and MSE (Mean Square Error) or Smooth L1 loss to optimize regression tasks.
Downstream details
We first describe datasets and splitting methods. In binding activity prediction task, we use 10 kinase targets in compound-kinase binding activity prediction and use 10 GPCR targets in ligand-GPCR binding activity prediction, which can be obtained from ImageMol19 (Supplementary Table 31 for statistical details). We use the same splitting method as ImageMol in kinase and GCPR targets, which uses a balanced scaffold split to divide the dataset into 80% training set, 10% validation set and 10% test set. In molecular property prediction task, we conduct experiments on 12 common benchmarks from the MoleculeNet69 (Supplementary Table 32). Following previous works6,63, we split all property prediction datasets using scaffold split, which splits molecules according to molecular substructure with 8:1:1. The scaffold split is a challenging splitting method for evaluating the generalization ability of models to out-of-distribution data samples. The balanced scaffold split ensures the balance of the scaffold size in the training set, validation set and test set. In anti-SARS-CoV-2 activity prediction task, we use the same data splitting as REDIAL-202041 and ImageMol on 11 SARS-CoV-2 activity prediction tasks (Supplementary Table 33).
We next describe metrics of evaluation. As suggested by MoleculeNet, we use ROC-AUC as the evaluation metric for the classification task, including 6 property predictions (BBBP, Tox21, HIV, BACE, SIDER and ToxCast). For the regression prediction of the remaining 6 molecular properties, we use RMSE (Root Mean Squared Error) and MAE (Mean Absolute Error) to evaluate FreeSolv, ESOL, lipophilicity and QM7, QM8, QM9, respectively. For compound-GPCR binding activity prediction, we report RMSE and MAE metrics. For compound-kinase binding activity prediction and anti-SARS-CoV-2 activity prediction task, we report ROC-AUC metric. Specifically, mean and standard deviations of MoleculeNet are reported with 10 different random seeds and mean and standard deviation of other datasets are reported with 3 different random seeds. See Supplementary Tables 34 and 35 and Supplementary Section A.3 for more details of fine-tuning stage. See Supplementary Section C.3 and Supplementary Table 36 for the details of computational requirements of VideoMol.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract HHSN261201500003I to R.N. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This Research was supported [in part] by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Author contributions
X.Z., F.C., and H.X. conceived and designed the study. H.X. and L.H. constructed the databases and drew the figures. H.X. and K.L. designed the framework. H.X. developed the codes and performed all experiments. H.X., L.Z., X.Z., and F.C. performed data analyses. H.X., X.Z., Z.F., F.C., Y.Q., R.N., J.H., and M.R.Z. discussed and interpreted all results. H.X., L.Z., X.Z., and F.C. wrote and critically revised the manuscript.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
The original datasets used in this project can be found at the following links: 2 million pre-training dataset: https://ogb.stanford.edu/docs/lsc/pcqm4mv2/; 10 GPCRs: https://figshare.com/articles/dataset/10_GPCRs/26941483 (Supplementary Table 31); 10 kinases: https://lincs.hms.harvard.edu/kinomescan/ (Supplementary Table 31); 12 molecular property prediction datasets: https://1drv.ms/f/s!Atau0ecyBQNTgRrf1iE-eogd17M-?e=m7so1Q (Replace the BBBP in the hyperlink with another dataset name to download other datasets) (Supplementary Table 32); 11 SARS-CoV-2 targets: https://opendata.ncats.nih.gov/covid19/assays (Supplementary Table 33); Source data are provided with this paper. All processed data are publicly available at https://github.com/HongxinXiang/VideoMol or https://github.com/ChengF-Lab/VideoMol. Source data are provided with this paper.
Code availability
All codes and the trained models are available at https://github.com/HongxinXiang/VideoMol or https://github.com/ChengF-Lab/VideoMol or Zenodo70.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Xiangxiang Zeng, Email: xzeng@hnu.edu.cn.
Feixiong Cheng, Email: chengf@ccf.org.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-53742-z.
References
- 1.Smith, A. Screening for drug discovery: The leading question. Nature418, 453–455 (2002). [DOI] [PubMed] [Google Scholar]
- 2.Gorgulla, C. et al. An open-source drug discovery platform enables ultra-large virtual screens. Nature580, 663–668 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schultz, D. C. et al. Pyrimidine inhibitors synergize with nucleoside analogues to block SARS-CoV-2. Nature604, 134–140 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lam, H. Y. I. et al. Application of variational graph encoders as an effective generalist algorithm in computer-aided drug design. Nat. Mach. Intell.5, 754–764 (2023). [Google Scholar]
- 5.Gentile, F. et al. Artificial intelligence–enabled virtual screening of ultra-large chemical libraries with deep docking. Nat. Protoc.17, 672–697 (2022). [DOI] [PubMed] [Google Scholar]
- 6.Wang, Y., Wang, J., Cao, Z. & Barati Farimani, A. Molecular contrastive learning of representations via graph neural networks. Nat. Mach. Intell.4, 279–287 (2022). [Google Scholar]
- 7.Xue, D. et al. X-MOL: large-scale pre-training for molecular understanding and diverse molecular analysis. Sci. Bull.67, 899–902 (2022). [DOI] [PubMed] [Google Scholar]
- 8.Liu, G. et al. GraphDTI: a robust deep learning predictor of drug-target interactions from multiple heterogeneous data. J. Cheminform.13, 1–17 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang, M. et al. Deep learning approaches for de novo drug design: an overview. Curr. Opin. Struct. Biol.72, 135–144 (2022). [DOI] [PubMed] [Google Scholar]
- 10.Wieder, O. et al. A compact review of molecular property prediction with graph neural networks. Drug Discov. Today.: Technol.37, 1–12 (2020). [DOI] [PubMed] [Google Scholar]
- 11.Wigh, D. S., Goodman, J. M. & Lapkin, A. A. A review of molecular representation in the age of machine learning. Wiley Interdiscip. Rev.: Comput. Mol. Sci.12, e1603 (2022). [Google Scholar]
- 12.Xiang, H. et al. An Image-enhanced Molecular Graph Representation Learning Framework. in Proceedings of the Thirty-Third International Joint Conference on Artificial Intelligence, IJCAI-24. (ed. K. Larson) 6107–6115, 10.24963/ijcai.2024/675 (International Joint Conferences on Artificial Intelligence Organization, 2024).
- 13.Raevsky, O. A. Physicochemical descriptors in property-based drug design. Mini Rev. Med. Chem.4, 1041–1052 (2004). [DOI] [PubMed] [Google Scholar]
- 14.Sun, H. Pharmacophore-based virtual screening. Curr. Med. Chem.15, 1018–1024 (2008). [DOI] [PubMed] [Google Scholar]
- 15.Weininger, D. SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules. J. Chem. Inf. Comp. Sci.28, 31–36 (1988). [Google Scholar]
- 16.Heller, S. R., McNaught, A., Pletnev, I., Stein, S. & Tchekhovskoi, D. InChI, the IUPAC international chemical identifier. J. Cheminform.7, 1–34 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu, W. et al. Strategies for pre-training graph neural networks. International Conference on Learning Representations (ICLR) (ICLR, 2020).
- 18.Rong, Y. et al. Self-supervised graph transformer on large-scale molecular data. Adv. Neural Inf. Process. Syst.33, 12559–12571 (2020). [Google Scholar]
- 19.Zeng, X. et al. Accurate prediction of molecular properties and drug targets using a self-supervised image representation learning framework. Nat. Mach. Intell.4, 1004–1016 (2022). [Google Scholar]
- 20.Xiang, H., Jin, S., Liu, X., Zeng, X. & Zeng, L. Chemical structure-aware molecular image representation learning. Brief. Bioinform.24, bbad404 (2023). [DOI] [PubMed] [Google Scholar]
- 21.Dai, H., Dai, B. & Song, L. Discriminative embeddings of latent variable models for structured data. International Conference On Machine Learning, p. 2702-2711 (PMLR, 2016).
- 22.Wang, J. et al. Self-supervised video representation learning by uncovering spatio-temporal statistics. IEEE Trans. Pattern Anal. Mach. Intell.44, 3791–3806 (2021). [DOI] [PubMed] [Google Scholar]
- 23.Wang, R. et al. Masked video distillation: rethinking masked feature modeling for self-supervised video representation learning. Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, p. 6312–6322 (IEEE, 2023).
- 24.Duan, H., Zhao, N., Chen, K. & Lin, D. Transrank: Self-supervised video representation learning via ranking-based transformation recognition. Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, p. 3000–3010 (IEEE, 2022).
- 25.Selvaraju, R.R. et al. Grad-cam: Visual explanations from deep networks via gradient-based localization. Proceedings of the IEEE International Conference On Computer Vision, p. 618–626 (IEEE, 2017).
- 26.Bemis, G. W. & Murcko, M. A. The properties of known drugs. 1. Molecular Frameworks. J. Med. Chem.39, 2887–2893 (1996). [DOI] [PubMed] [Google Scholar]
- 27.Honda, S., Shi, S. & Ueda, H.R.J.A.P.A. SMILES transformer: pre-trained molecular fingerprint for low data drug discovery. arXiv10.48550/arXiv.1911.04738 (2019).
- 28.Kim, H., Lee, J., Ahn, S. & Lee, J. R. A merged molecular representation learning for molecular properties prediction with a web-based service. Sci. Rep.11, 1–9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun, F.-Y., Hoffman, J., Verma, V. & Tang, J. Infograph: Unsupervised and semi-supervised graph-level representation learning via mutual information maximization. International Conference on Learning Representations (ICLR) (ICLR, 2020).
- 30.Hu, Z., Dong, Y., Wang, K., Chang, K.-W. & Sun, Y. Gpt-gnn: Generative pre-training of graph neural networks. Proceedings of the 26th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining, p. 1857–1867 (ACM, 2020).
- 31.Xu, M., Wang, H., Ni, B., Guo, H. & Tang, J. Self-supervised graph-level representation learning with local and global structure. International Conference on Machine Learning, 11548–11558 (PMLR, 2021).
- 32.Suresh, S., Li, P., Hao, C. & Neville, J. Adversarial graph augmentation to improve graph contrastive learning. Adv. Neural Inf. Process. Syst.34, 15920–15933 (2021). [Google Scholar]
- 33.Zhang, Z., Liu, Q., Wang, H., Lu, C. & Lee, C.-K. Motif-based Graph Self-Supervised Learning for Molecular Property Prediction. Advances in Neural Information Processing Systems34 (MIT Press, 2021).
- 34.Xia, J., Wu, L., Chen, J., Hu, B. & Li, S.Z. Simgrace: A simple framework for graph contrastive learning without data augmentation. Proceedings of the ACM Web Conference 2022, 1070–1079 (ACM, 2022).
- 35.You, Y. et al. Graph contrastive learning with augmentations. Adv. Neural Inf. Process. Syst.33, 5812–5823 (2020). [Google Scholar]
- 36.Hou, Z. et al. Graphmae: self-supervised masked graph autoencoders. Proceedings of the 28th ACM SIGKDD Conference on Knowledge Discovery and Data Mining, 594–604 (ACM, 2022).
- 37.Xia, J. et al. Mole-BERT: Rethinking Pre-training Graph Neural Networks For Molecules. Published as a conference paper at ICLR 2023 (2023).
- 38.Stärk, H. et al. 3D Infomax improves GNNs for Molecular Property Prediction. NeurIPS 2021 AI for Science Workshop (MIT Press, 2021).
- 39.Liu, S. et al. Pre-training Molecular Graph Representation with 3D Geometry. International Conference on Learning Representations (ICLR, 2021).
- 40.Zhou, G. et al. Uni-Mol: A UNIVersal 3D Molecular Representation Learning Framework. (2023).
- 41.Bocci, G. et al. A machine learning platform to estimate anti-SARS-CoV-2 activities. Nat. Mach. Intell. 3, 527–535 (2021).
- 42.Efron, B. Better bootstrap confidence intervals. J. Am. Stat. Assoc.82, 171–185 (1987). [Google Scholar]
- 43.Efron, B. & Tibshirani, R.J. An Introduction To The Bootstrap (Chapman and Hall/CRC, 1994).
- 44.Gaulton, A. et al. The ChEMBL database in 2017. Nucleic Acids Res.45, D945–D954 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hinton, G.E. & Roweis, S. Stochastic neighbor embedding. Advances In Neural Information Processing Systems. Vol. 15 (2002).
- 46.Hampel, H. et al. The β-secretase BACE1 in Alzheimer’s disease. Biol. Psychiatry89, 745–756 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wishart, D. S. et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res.46, D1074–D1082 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zou, Y. et al. Virtual screening and structure-based discovery of indole acylguanidines as potent β-secretase (BACE1) inhibitors. Molecules18, 5706–5722 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Berman, H. M. et al. The protein data bank. Nucleic Acids Res.28, 235–242 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Balius, T. E., Tan, Y. S. & Chakrabarti, M. DOCK 6: Incorporating hierarchical traversal through precomputed ligand conformations to enable large‐scale docking. J. Comput. Chem.45, 47–63 (2024). [DOI] [PubMed] [Google Scholar]
- 51.Davies, D.L. & Bouldin, D.W. A cluster separation measure. IEEE transactions on pattern analysis and machine intelligence, p. 224-227 (IEEE, 1979). [PubMed]
- 52.Gu, T. et al. Development and structural modification of BACE1 inhibitors. Molecules22, 4 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kimura, T. et al. Design and synthesis of potent β-secretase (BACE1) inhibitors with P1′ carboxylic acid bioisosteres. Bioorg. Med. Chem. Lett.16, 2380–2386 (2006). [DOI] [PubMed] [Google Scholar]
- 54.Garino, C. et al. BACE-1 inhibitory activities of new substituted phenyl-piperazine coupled to various heterocycles: chromene, coumarin and quinoline. Bioorg. Med. Chem. Lett.16, 1995–1999 (2006). [DOI] [PubMed] [Google Scholar]
- 55.Malamas, M. S. et al. Aminoimidazoles as potent and selective human β-secretase (BACE1) inhibitors. J. Med. Chem.52, 6314–6323 (2009). [DOI] [PubMed] [Google Scholar]
- 56.Hanessian, S., Hou, Y., Bayrakdarian, M. & Tintelnot-Blomley, M. Stereoselective synthesis of constrained oxacyclic hydroxyethylene isosteres of aspartic protease inhibitors: Aldol and Mukaiyama Aldol methodologies for branched tetrahydrofuran 2-carboxylic acids. J. Org. Chem.70, 6735–6745 (2005). [DOI] [PubMed] [Google Scholar]
- 57.Ringnér, M. What is principal component analysis? Nat. Biotechnol.26, 303–304 (2008). [DOI] [PubMed] [Google Scholar]
- 58.Pedregosa, F. et al. Scikit-learn: machine learning in Python. J. Mach. Learn. Res.12, 2825–2830 (2011). [Google Scholar]
- 59.O’Boyle, N.M. et al. Open Babel: an open chemical toolbox. J. Cheminform. 3, 1–14 (2011). [DOI] [PMC free article] [PubMed]
- 60.Altae-Tran, H., Ramsundar, B., Pappu, A. S. & Pande, V. Low data drug discovery with one-shot learning. ACS Cent. Sci.3, 283–293 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.He, Y., Yan, R., Fragkiadaki, K. & Yu, S.-I. Epipolar transformers. Proceedings of the IEEE/cvf Conference On Computer Vision And Pattern Recognition, 7779–7788 (IEEE, 2020).
- 62.Qian, R. et al. Spatiotemporal contrastive video representation learning. Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 6964–6974 (IEEE, 2021).
- 63.Fang, X. et al. Geometry-enhanced molecular representation learning for property prediction. Nat. Mach. Intell.4, 127–134 (2022). [Google Scholar]
- 64.Hu, W. et al. Ogb-lsc: a large-scale challenge for machine learning on graphs. Thirty-fifth Conference on Neural Information Processing Systems Datasets and Benchmarks Track (MIT Press, 2021).
- 65.DeLano, W. L. Pymol: an open-source molecular graphics tool. CCP4 Newsl. Protein Crystallogr40, 82–92 (2002). [Google Scholar]
- 66.Maćkiewicz, A. & Ratajczak, W. Principal components analysis (PCA). Comput. Geosci.19, 303–342 (1993). [Google Scholar]
- 67.Dosovitskiy, A. et al. An Image is Worth 16x16 Words: Transformers for Image Recognition at Scale. International Conference on Learning Representations (ICLR, 2020).
- 68.Paszke, A. et al. Pytorch: An imperative style, high-performance deep learning library. Advances in Neural Information Processing Systems32 (MIT Press, 2019).
- 69.Wu, Z. et al. MoleculeNet: a benchmark for molecular machine learning. Chem. Sci.9, 513–530 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xiang, H. et al. A Molecular Video-derived Foundation Model for Scientific Drug Discovery. VideoMol: v1.0, 10.5281/zenodo.13843803 (2024).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The original datasets used in this project can be found at the following links: 2 million pre-training dataset: https://ogb.stanford.edu/docs/lsc/pcqm4mv2/; 10 GPCRs: https://figshare.com/articles/dataset/10_GPCRs/26941483 (Supplementary Table 31); 10 kinases: https://lincs.hms.harvard.edu/kinomescan/ (Supplementary Table 31); 12 molecular property prediction datasets: https://1drv.ms/f/s!Atau0ecyBQNTgRrf1iE-eogd17M-?e=m7so1Q (Replace the BBBP in the hyperlink with another dataset name to download other datasets) (Supplementary Table 32); 11 SARS-CoV-2 targets: https://opendata.ncats.nih.gov/covid19/assays (Supplementary Table 33); Source data are provided with this paper. All processed data are publicly available at https://github.com/HongxinXiang/VideoMol or https://github.com/ChengF-Lab/VideoMol. Source data are provided with this paper.
All codes and the trained models are available at https://github.com/HongxinXiang/VideoMol or https://github.com/ChengF-Lab/VideoMol or Zenodo70.