Abstract
Long non-coding RNAs serve a crucial role in autophagy of vascular smooth muscle cells (VSMCs). The present study aimed to investigate the effect of small nucleolar RNA host gene 1 (SNHG1) on autophagy in VSMCs and the associated underlying mechanisms. Rapamycin was used to induce autophagy in VSMCs and the effects of SNHG1 on the proliferation and migration of VSMCs and the change in phenotype were tested following overexpression and silencing of SNHG1. The target gene of SNHG1 was predicted and validated. SNHG1-regulated autophagy of VSMCs via C-type lectin domain family 7 member A (CLEC7A) was determined by combined silencing of SNHG1 and overexpression of CLEC7A. Rapamycin-induced autophagy in VSMCs changed the cell phenotype from contractile to synthetic, with decreased expression of α-smooth muscle actin and smooth muscle protein 22a and increased expression of osteopontin. Overexpression of SNHG1 caused the same change in phenotype while the opposite change was observed following SNHG1 silencing. Overexpression of SNHG1 promoted the proliferation and migration of VSMCs. CLEC7A was identified as a target gene of SNHG1 and a direct binding relationship between them was confirmed by RNA immunoprecipitation and RNA pull-down assays. Overexpression of SNHG1 increased the expression of CLEC7A. The expression of both SNHG1 and CLEC7A was increased during autophagy of VSMCs. Overexpression of SNHG1 promoted autophagy of VSMCs and silencing of CLEC7A reduced this effect of SNHG1. In conclusion, SNHG1 and CLEC7A were increased in VSMCs following autophagy. SNHG1 promotes the conversion of VSMCs from a contractile phenotype to a synthetic phenotype by facilitating CLEC7A expression.
Keywords: vascular smooth muscle cells, small nucleolar RNA host gene 1, C-type lectin domain family 7 member A, autophagy, phenotype
Introduction
Abnormal changes in the phenotype and function of vascular smooth muscle cells (VSMCs) have been implicated in several vascular diseases, such as aneurysms and atherosclerosis (1). VSMCs can have two phenotypes: A contractile phenotype [markers: α-smooth muscle actin (SMA), smooth muscle protein 22a (SM22a), myosin heavy chain 11 (MYH11)] and a synthetic phenotype [markers: Matrix metalloproteinase (MMP) 2, tumor necrosis factor-α, osteopontin (OPN)] (2,3). During disease development, the phenotype of VSMCs can change from contractile to synthetic (4,5).
Autophagy is a process wherein cells use lysosomes to break down intracellular organelles and macromolecules (6). Alterations in the autophagy status of VSMCs due to various external factors can lead to abnormal cell proliferation, migration and matrix secretion (7). Cytokines activate signaling pathways that affect autophagy and thereby influence phenotypic and functional changes in VSMCs (8,9). The transition from the contractile to the synthetic phenotype occurs following autophagy of VSMCs (10). Therefore, identifying molecular targets that can effectively regulate the phenotype and function of VSMCs is crucial for the treatment of vascular diseases.
Long non-coding RNA (lncRNA) is a class of RNA molecules that are >200 nt in length and do not encode proteins. It has been discovered that lncRNAs play a crucial role in the development of various diseases (11–13). Small nucleolar RNA host gene 1 (SNHG1), a small ribosomal housekeeping gene, is a lncRNA that originates from the U22 host gene on chromosome 11 and is an important component of the 18 s ribosomal RNA. It consists of ~3,927 bases and contains eight small nucleotide RNAs (14,15). SNHG1 has been found to be abnormally expressed in various diseases and regulates the biological behavior of the disease via its target genes (16–20). C-type lectin domain family 7 member A (CLEC7A), also known as Dectin-1, is a member of the CLEC7 family and its gene encodes the C-lectin/C-lectin domain-like domain. The encoded glycoprotein is a small type II membrane receptor with an extracellular C-type lectin-like domain fold and a cytoplasmic domain with an immune receptor tyrosine activation motif (21). The functions of CLEC7A include phagocytosis, production of reactive oxygen species and the secretion of pro-inflammatory cytokines, which are essential for antifungal defense (22,23). In addition, CLEC7A recognizes other pathogens and endogenous ligands, suggesting a broader role in immunity (24).
Aneurysms are closely related to the abnormality of VSMCs. Overexpression of CLEC7A has been shown to be associated with aneurysm healing following embolization (25). CLEC7A can also distinguish intracranial aneurysms from normal samples (26).
Our previous study showed that the expression of CLEC7A is significantly higher in intracranial aneurysms compared with normal vascular tissue (27), indicating the association of CLEC7A with VMSCs.
Based on the existing literature and previous research, it was hypothesized that SNHG1 regulates CLEC7A to modulate the status of autophagy in VSMCs, which in turn leads to the development of vascular diseases. The present study used rapamycin to induce autophagy in VSMCs. Following manipulation of the expression of SNHG1 by silencing and overexpression, the occurrence of autophagy, phenotypic changes and migratory ability was assessed in VSMCs. VSMCs were then transduced with vectors containing silenced or overexpressed SNHG1 to examine the expression of CLEC7A. The interaction between SNHG1 and CLEC7A was determined using a RNA-binding protein immunoprecipitation (RIP) assay and a RNA pull-down assay. Finally, the phenotypic changes in VSMCs were investigated by combining overexpression of CLEC7A with silencing of SNHG1. The findings aimed to provide insights into the mechanism by which SNHG1 regulates the phenotypic transition of VSMCs by mediating autophagy via CLEC7A.
Materials and methods
Cell culture
Immortalized human aortic smooth muscle cells (VSMCs) were purchased from Meisen Chinese Tissue Culture Collections (cat. no. CTCC-001-0577) and seeded in complete medium (cat. no. CTCC-001-0577-CM, Meisen Cell Technology Co., Ltd), which consisted of F-12K medium containing 0.05 mg/ml ascorbic acid, 0.01 mg/ml insulin, 0.01 mg/ml transferrin, 10 ng/ml sodium selenite, 0.03 mg/ml endothelial cell growth supplement, 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, 10 mM TES, 10% fetal bovine serum and 1% antibiotic-antimycotic medium under humidified conditions (37°C; 5% CO2).
Cell transduction
Short hairpin RNAs (shRNAs) designed to target SNHG1 along with their corresponding empty vector (vector-sh) and the plasmids for overexpression (OE) of SNHG1 or CLEC7A and the empty vector (vector-OE) were procured from Hanbio Biotechnology Co., Ltd. The 2nd generation lentiviral system was used for transfection. All recombinant lentivirus sequences were generated as previously described (28). VSMCs were transduced with lentiviral vectors containing SNHG1 silencing sequences, designated sh-SNHG1. Similarly, lentiviral vectors containing human SNHG1 or CLEC7A overexpression sequences were introduced into VSMCs, designated OE-SNHG1 and OE-CLEC7A, respectively. The viral supernatant was used to infect target cells at an MOI of 10. The VSMCs underwent a 72-h incubation period with the recombinant lentiviruses and subsequently underwent puromycin selection for 72 h using 3 µg/ml puromycin for selection of transfected cells and 3 µg/ml for maintenance of the stable cell lines. Finally, transduction efficacy was validated by assessing mRNA expression levels using reverse transcription-quantitative (RT-q) PCR. Experiments were conducted 48 h after the completion of puromycin selection. The sequences of shRNA are detailed in Table SI, whereas the sequences utilized for overexpression are presented in Table SII.
RT-qPCR
RT-qPCR was used to quantitatively analyze gene expression as previously described (29). Total RNA was extracted from VSMCs with TRIzol® reagent (Thermo Fisher Scientific, Inc.). The cDNA was synthesized from the RNA using a reverse transcriptase kit (cat. no. RR036A; Takara Biotechnology Co., Ltd.) according to the manufacturer's protocol. qPCR was performed using SYBR Green qPCR Master Mix (cat. no. Q711-02; Nanjing Vazyme Biotech Co., Ltd) following the manufacturer's protocol. The qPCR thermocycling conditions were as follows: Initial denaturation at 95°C for 30 sec, followed by 40 cycles of denaturation at 95°C for 10 sec, annealing at 56°C for 30 sec, and extension at 72°C for 60 sec. GAPDH was used as an internal control. Relative expression levels were calculated using the 2−ΔΔCq method (30). The experiment was performed as three independent replicates. The primers used are listed in Table SIII.
Western blotting
Western blot procedures were in accordance with standard protocols as described in the earlier study by Yue et al (31). Briefly, the cellular proteins were extracted with radioimmunoprecipitation assay buffer (cat. no. R0010; Beijing Solarbio Science & Technology Co., Ltd.) and the protein concentration was determined using the BCA Protein Assay Kit (cat. no. P0012S; Beyotime Institute of Biotechnology) according to the manufacturer's protocol. Proteins were then separated by SDS-PAGE on 12.5% gels for LC3, SM22α and CLEC7A, and 10% gels for p62, BECN1, OPN, GAPDH and β-actin. Following electrophoresis, the proteins were transferred to a polyvinylidene membrane and subsequently blocked with 5% skimmed milk in Tris-buffered saline with 1% Tween 20 for 1 h. Primary antibodies were incubated overnight at 4°C, while treatment with secondary antibodies was performed at room temperature for ~1 h. The antibodies used in this work were as follows: Rabbit anti-LC3 (1:1,000; cat. no. 4108S; Cell Signaling Technology, Inc.); Rabbit anti-P62 (1:1,000; cat. no. 16177S; Cell Signaling Technology, Inc.); Rabbit anti-SM22a (1:1,000; cat. no. CSB-PA00804A0Rb; Cusabio Technology, LLC); rabbit anti-α-smooth muscle actin (α-SMA) (1:1,000; cat. no. R23450; Chengdu Zhengneng Biotechnology Co., Ltd.); rabbit anti-osteopontin (OPN) (1:1,000; cat. no. R26171; ZENBIO); rabbit anti-CLEC7A (1:1,000; cat. no. PA5-34382; Thermo Fisher Scientific, Inc.); rabbit anti-GAPDH (1:1,000; cat. no. GB15004-100; Wuhan Servicebio Technology Co., Ltd.); β-actin (1:1,000; cat. no. 380624; Chengdu Zhengneng Biotechnology Co., Ltd.) anti-rabbit immunoglobulin (Ig) G, horseradish peroxidase-linked antibody (1:10,000; cat. no. ab6721; Abcam). The proteins were visualized using the ECL Western Blotting Detection System (cat. no. 143237; Biosharp Life Sciences) according to the manufacturer's instructions. Relative target protein levels were quantified using ImageJ software (version 1.8.0; National Institutes of Health). GAPDH or β-actin served as the internal control for normalization of target protein levels. The experiment was performed in three independent replicates.
Cell counting kit-8 (CCK-8) assay
Rapamycin was first dissolved in a 100 mM dimethyl sulfoxide (DMSO) solution (cat. no. D8371-50 ml; Beijing Solarbio Science & Technology Co., Ltd.) and further diluted in fresh medium to reach a concentration of 100 nM before use. Cells in the logarithmic growth phase were dissociated using 0.25% trypsin to prepare cell suspension with a density of 3×104 cells/ml, subsequently seeded into 96-well plates at 100 µl per well. After a 48-h incubation period, the transduced cells were detached using trypsin and reseeded at a concentration of 1×104 cells/ml. Subsequently, 100 ml of the cell suspension was dispensed into the 96-well plates. Then three wells were randomly selected at 0, 12 and 24 h after cultivation in a controlled environment at 37°C. Each well was then treated with 10 ml of CCK-8 (cat. no. MA0218; MeilunBio) and further incubated for 2 h. Finally, the optical density (OD) was measured at 450 nm and a curve was plotted to represent the dynamics of cellular growth.
Wound healing assays
VSMCs were seeded in a six-well plate, transduced and subsequently treated. After a 24-h incubation period, a linear scratch was meticulously created across the plate with a 200-µl pipette tip and fresh serum-free medium was carefully added. Images were then captured with an inverted fluorescence microscope (Axio Vert.A1; Carl Zeiss AG) showing the progression of cell migration after 0, 12 and 24 h. The wound closure was quantified using ImageJ software (version 1.8.0). The width of the scratch was measured at multiple points along the scratch, and the average width was calculated. The wound closure rate was determined using the formula: Wound closure rate (%)=(initial scratch width-current scratch width)/initial scratch width ×100.
RIP assay
To investigate the interaction between the SNHG1 sequence and the CLEC7A protein, an RIP assay was performed according to the manufacturer's protocol (cat. no. Bes5101; Guangzhou BersinBio Biotechnology Co., Ltd.). Cells were washed with phosphate-buffered saline, lysed in a buffer containing protease and RNA enzyme inhibitors and divided into three groups: Input, IP and IgG. Anti-CLEC7A antibody (cat. no. 60128; Cell Signaling Technology, Inc.) and IgG antibody (cat. no. Bes5101; Guangzhou BersinBio Biotechnology Co., Ltd.) were used for incubation with the 0.8 ml lysates of the IP and IgG groups, respectively. RNA-protein complexes were isolated with 20 µl protein A/G magnetic beads and then digested with proteinase K. Proteins bound to RNA were extracted with phenol/chloroform/isoamyl alcohol (125:24:1) and analyzed by RT-qPCR. The percentage of input and fold enrichment were calculated to assess the changes in RNA-protein binding owing to CLEC7A stimulation.
RNA pull-down assay
Guangzhou BersinBio Biotechnology Co., Ltd. developed a precise probe targeting SNHG1 to uncover potential proteins that interact with SNHG1. Subsequently, an RNA pulldown kit (cat. no. Bes5102; Guangzhou BersinBio Biotechnology Co., Ltd.) was used to isolate proteins associated with SNHG1. For this purpose, 40 µl magnetic beads were conjugated with the 100 µl SNHG1 probe, forming a complex. This complex of probe and magnetic beads was subsequently introduced into the 0.8 ml cell lysate, where it underwent a 2-h incubation that allowed interaction between the SNHG1 probe and the specific proteins. Finally, 60 µl Protein Elution Buffer and 0.6 µl dithiothreitol were added, and the mixture was incubated at 37°C for 2 h with intermittent mixing. After incubation, the mixture was placed on a magnet for 1 min and the supernatant was collected. The eluted proteins then underwent SDS-PAGE followed by western blotting using specific antibodies against the proteins of interest. The probe sequence used for RNA pull-down used is listed in Table SIV.
RPI-Seq database prediction
We utilized the RPI-Seq database (Version 1.0, URL: http://pridb.gdcb.iastate.edu/RPISeq/), a computational tool designed to predict interactions between RNA and proteins. The RPI-Seq database was accessed through its online platform, and the sequences of SNHG1 and CLEC7A were submitted for analysis. The prediction algorithm within RPI-Seq evaluates the likelihood of interaction based on sequence and structural features of the RNA and protein molecules.
Statistical analysis
Data are presented as mean ± standard deviation. Statistical significance between two groups was determined using Student's t-test, while comparisons between multiple groups were performed using one-way analysis of variance followed by Tukey's post hoc test. Statistical analyses were performed using GraphPad Prism version 8 software (GraphPad; Dotmatics). P<0.05 was considered to indicate a statistically significant difference.
Results
Autophagy is induced by rapamycin in VSMCs
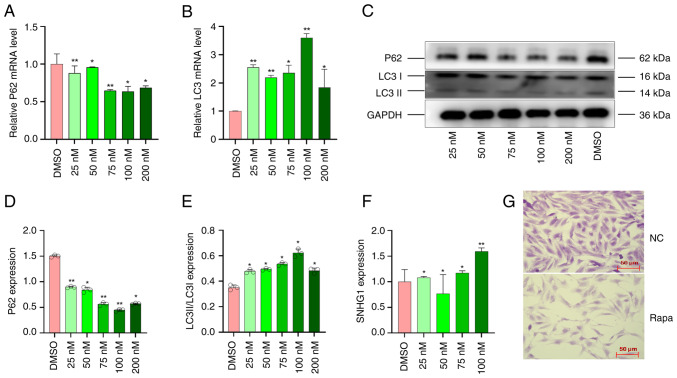
VSMCs were exposed to different concentrations of rapamycin (25, 50, 75, 100 and 200 nM) for 24 h to induce autophagy, while DMSO served as a control. The mRNA expression of autophagy markers (32) LC3 (LC3 I and LC3 II) and P62 was detected using RT-qPCR. Compared with the control group, the mRNA expression of P62 was lowest and LC3 was highest at the concentration of 100 nM (Fig. 1A and B). This indicated that the concentration of 100 nM was the most suitable for inducing autophagy. Western blot assay was used to measure the changes in autophagy markers and showed that the protein concentrations of P62 decreased while those of LC3 I and LC3 II increased (Fig. 1C and E). These results indicated that the optimal concentration of 100 nM can effectively induce autophagy in VSMCs.
Figure 1.
VSMCs autophagy was induced by rapamycin. Expression of (A) P62 mRNA and (B) LC3 mRNA in VSMCs following treatment with rapamycin at different concentrations. (C-E) Expression of P62 and LC3 protein in VSMCs following treatment with rapamycin at different concentrations. (F) Expression of SNHG1 following treatment with rapamycin at different concentrations; (G) Morphological changes of VCMC following rapamycin treatment at the concentrations of 100 nM, scale bar, 50 µm. Data were shown as mean ± standard deviation of a representative experiment performed in three independent replicates. Compared with DMSO treatment *P<0.05, **P<0.01. VSMCs, vascular smooth muscle cells; SNHG1, small ribosome housekeeping gene RNA1; DMSO, dimethyl sulfoxide.
In addition, the expression of SNHG1 was measured by RT-qPCR assay; SNHG1 expression increased significantly at a concentration of 100 nM compared with other concentrations (Fig. 1F). The morphological changes in VSMCs treated with 100 nM rapamycin are shown. Normally, VSMCs have a long spindle shape with clear boundaries and a smooth surface. Following autophagy is induced, the number of cells decreases and they become smaller, more irregular and lose their spindle shape, as shown in Fig. 1G.
Establishment of stable expression of SNHG1 in VSMCs
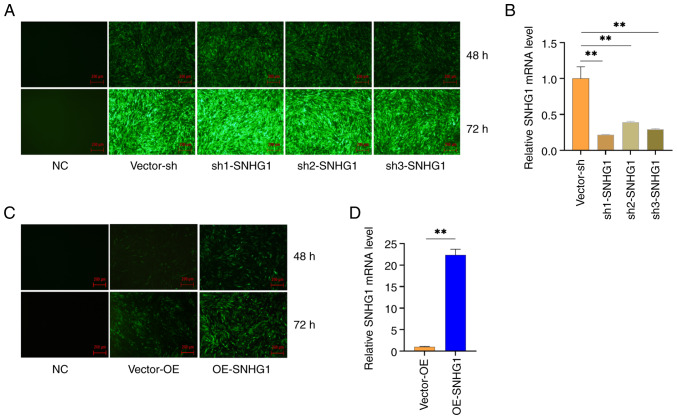
The present study introduced three interfering sequences into VSMCs using lentiviruses over a period of 48 and 72 h. Subsequently, a fluorescence microscope was used to observe the morphology and fluorescence expression of the cells. The results showed that all three interfering sequences led to fluorescence expression, with a fluorescence rate of >80%. By contrast, the blank group showed no fluorescence expression, confirming the successful transfer of the lentivirus into the cells (Fig. 2A). Therefore, a 72-h transduction was chosen for the following experiment. RT-qPCR was performed to measure the expression of SNHG1 in the cells. The results showed that all three silencing sequences significantly inhibited the expression of SNHG1 in the cells, with Sh1 showing the strongest inhibition (Fig. 2B). The overexpression sequence significantly increased the expression of SNHG1 in the cells, reaching a level 22 times higher than that in the control group (Fig. 2C and D). These results indicated that the present study was able to successfully manipulate SNHG1 levels in VSMCs.
Figure 2.
SNHG1 silencing and overexpression vector transduction of VSMCs. (A) Fluorescence map of lentivirus transduced SNHG1 silencing sequence into cells. (B) RT-qPCR detection of SNHG1 expression in silenced cells. (C) Fluorescence pattern of cells transduced with lentivirus following overexpression of SNHG1. (D) RT-qPCR detected the expression of SNHG1 in the cells following overexpression. Scale bar: 200 µm. Data were shown as mean ± standard deviation of a representative experiment performed in three independent replicates. **P<0.01. SNHG1, small ribosome housekeeping gene RNA1; VSMCs, vascular smooth muscle cells; RT-qPCR, reverse transcription-quantitative PCR; sh, short hairpin; NC, negative control; OE, overexpression.
SNHG1 facilitated transition of phenotype VSMCs
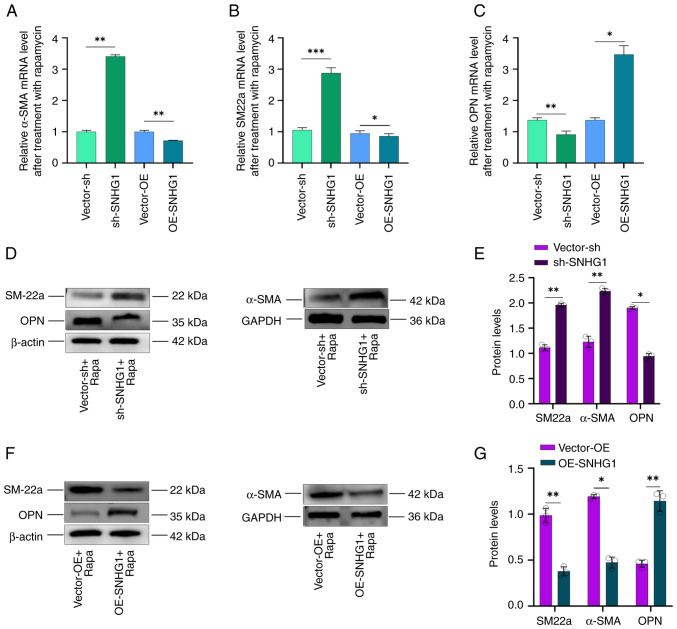
α-SMA, SM22a and OPN are important markers for phenotypic changes following autophagy. When the phenotype of VSMCs shifts from contractile to synthetic, the expression of α-SMA and SM22a decreases, while that of OPN increases (33–35). The present study investigated the effects of SNHG1 on phenotypic changes in VSMCs following autophagy induced by rapamycin at a concentration of 100 nM. It first overexpressed or silenced SNHG1 and then combined treatment with rapamycin in the cells and found that silencing SNHG1 led to an increase in SM22a and α-SMA expression, while OPN expression decreased. Conversely, overexpression of SNHG1 resulted in decreased SM22a and α-SMA expression but increased OPN expression (Fig. 3A-C). Western blotting examination of α-SMA, SM22a and OPN protein levels confirmed these results (Fig. 3D-G). These results indicated that silencing of SNHG1 in VSMCs can induce a shift from a synthetic to a contractile phenotype during autophagy.
Figure 3.
Changes in VSMCs phenotype following silencing and overexpression of SNHG1. (A) α-SMA, (B) SM22a and (C) OPN mRNA expression following overexpressing or silencing of SNHG1 combining rapamycin treatment in VSMCs; (D-G) SM22a, α-SMA and OPN protein levels following silencing or overexpression of SNHG1 combing with rapamycin treatment. Data were shown as mean ± standard deviation of a representative experiment performed in three independent replicates. *P<0.05, **P<0.01, ***P<0.001. VSMCs, vascular smooth muscle cells; SNHG1, small ribosome housekeeping gene RNA1; α-SMA, α-smooth muscle actin; SM22a, smooth muscle protein 22a; OPN, osteopontin; sh, short hairpin; NC, negative control; OE, overexpression; Rapa, rapamycin.
SNHG1 promotes migration and proliferation of VSMCs
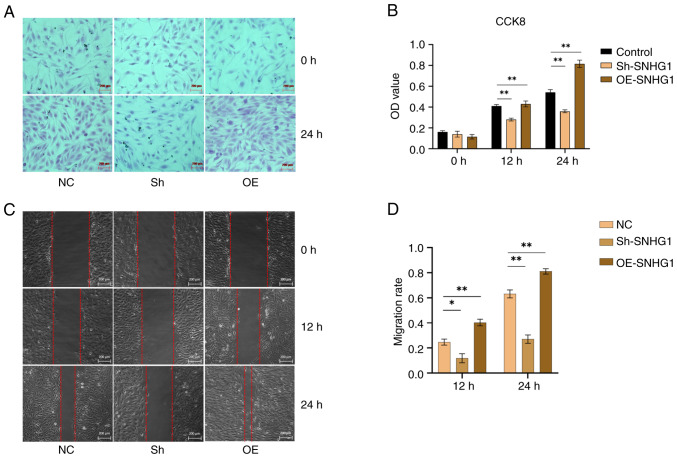
Following successful transduction of SNHG1, the cell viability was tested using the CCK8 assay. The results showed that overexpression of SNHG1 increased the proliferation of VSMCs. Conversely, this effect was suppressed when SNHG1 was silenced, leading to a decrease in the proliferation of VSMCs (Fig. 4A and B). Furthermore, wound healing assays showed that silencing SNHG1 reduced the migration of VSMCs, while overexpression of SNHG1 increasing their migration compared with the control group (Fig. 4C and D). Taken together, these results suggested that SNHG1 played a role in promoting the proliferation and migration ability of VSMCs.
Figure 4.
Changes in proliferation and migration of VSMCs following silencing and overexpression of SNHG1. (A) Images of VSMCs proliferation following silencing and overexpression of SNHG1. (B) OD value following silencing and overexpression of SNHG1 in VSMCs at different time by CCK-8 assay. (C) Images of VSMCs following silencing and overexpression of SNHG1 by wound healing assay. (D) Comparison of cells migration ability following silencing and overexpression of SNHG1. Scale bar: 200 µm. Data were shown as mean ± standard deviation of a representative experiment performed in three independent replicates. *P<0.05, **P<0.01. VSMCs, vascular smooth muscle cells; SNHG1, small ribosome housekeeping gene RNA1; OD, optical density; sh, short hairpin; NC, negative control; OE, overexpression.
CLEC7A is a regulatory target of SNHG1
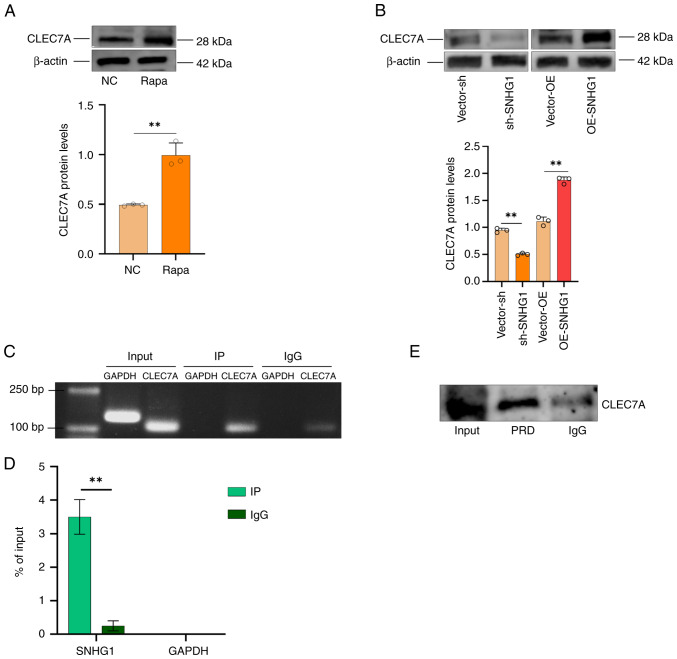
In our previous study, an increase in the expression of CLEC7A was observed in aneurysm tissues (27). In the present study, induction of autophagy in VSMCs was found to result in an increase in CLEC7A protein levels (Fig. 5A). This suggested that CLEC7A played a role in aneurysm formation by participating in autophagy in VSMCs. A possible relationship between SNHG1 and CLEC7A was predicted using the RPI-Seq database and the result indicated a strong association between them (Fig. S1). This suggested that CLEC7A is one of the targets regulated by SNHG1. To verify whether CLEC7A is a direct regulatory target of SNHG1, CLEC7A protein levels changed following silencing and overexpression of SNHG1 in VSMCs (Fig. 5B); RIP and RNA pull-down assays were used to verify the binding effect. As shown in Fig. 5C and D, SNHG1 was pulled down by an anti-CLEC7A antibody in the RIP assay. The RNA pull-down assay then showed that CLEC7A proteins were bound to SNHG1 in VSMCs (Fig. 5E). Taken together, these results demonstrated that SNHG1 directly binds to CLEC7A in VSMCs. SNGH1 and autophagy are interrelated and autophagy also regulated CLEC7A via SNGH1.
Figure 5.
CLEC7A is the regulatory target of SNHG1. (A) Change of CLEC7A protein levels in VSMCs following treatment with rapamycin. (B) CLEC7A protein levels changes following silence and overexpression of SNHG1 in VSMCs. (C and D) RIP assays together with RT-qPCR showed that SNHG1 was pulled down by an anti-CLEC7A antibody. (E) RNA pulldown and western-blotting revealed the direct binding of CLEC7A proteins to SNHG1. Data were shown as mean ± standard deviation of a representative experiment performed in three independent replicates. **P<0.01. Input, sample control group; PRD, SNHG1 probe pull down CLEC7A protein group; NC, Negative control group (LacZ); CLEC7A, C-type lectin domain family 7 member A; VSMCs, vascular smooth muscle cells; SNHG1, small ribosome housekeeping gene RNA1; RIP, RNA-binding protein immunoprecipitation; RT-qPCR, reverse transcription-quantitative PCR.
Effect of SNHG1 on the expression of CLEC7A on the phenotypic changes of VSMCs
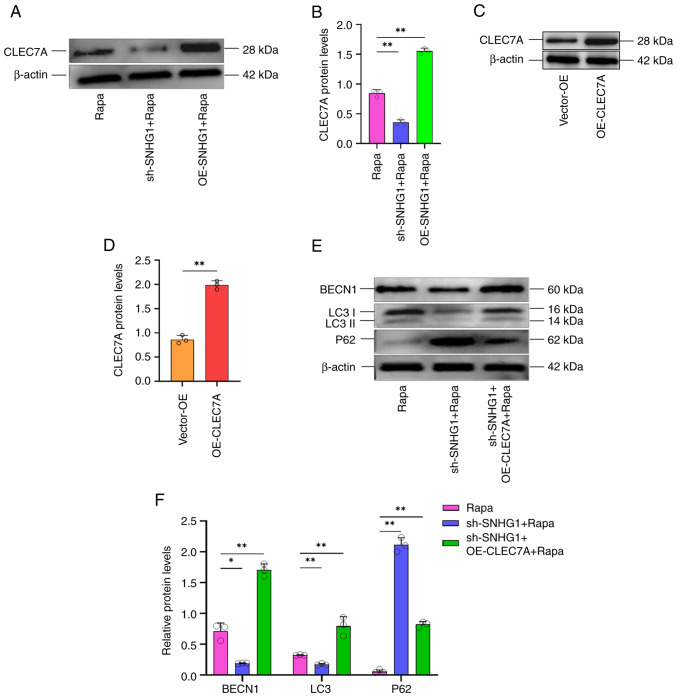
To investigate the effect of SNHG1, which regulates CLEC7A, on the phenotypic changes of VSMCs, autophagy was induced following silencing or overexpressing of SNHG1. Silencing of SNHG1 decreased CLEC7A protein levels following induction of autophagy, whereas overexpression of SNHG1 increased these levels (Fig. 6A and B). Next, an overexpression vector for CLEC7A was successfully constructed and transduced into VSMCs. Western blotting results confirmed the overexpression of CLEC7A (Fig. 6C and D). Subsequently, the silencing of SNHG1 was combined with the overexpression of CLEC7A in these cells and they were treated with rapamycin. As shown in Fig. 6E and F, western blotting showed that autophagy markers changed following silencing SNHG1, with LC 3II/LC 3I and Beclin1 decreasing and P62 increasing. This effect was offset by combing with overexpression of CLEC7A. Taken together, these results suggested that SNHG1 regulated autophagy in VSMCs by enhancing CLEC7A.
Figure 6.
SNHG1 regulation of CLEC7A expression affects phenotypic changes in VSMCs. (A and B) CLEC7A protein levels were altered following silencing and overexpression of SNHG1 combined with rapamycin treatment in VSMCs. (C and D) The expression of CLEC7A protein levels in VSMCs following overexpression. (E and F) Change of autophagy markers (BECN1, LC3II/LC3I and P62) following silencing of SNHG1 and overexpression of CLEC7A combined with rapamycin treatment in VSMCs. Data were shown as mean ± standard deviation of a representative experiment performed in three independent replicates. *P<0.05, **P<0.01. SNHG1, small ribosome housekeeping gene RNA1; CLEC7A, C-type lectin domain family 7 member A; VSMCs, vascular smooth muscle cells; BECN1, Beclin1.
Discussion
VSMCs undergo phenotypic transformation by switching from a contractile phenotype to a synthetic phenotype characterized by increased proliferation and migratory capacity. This transformation is accompanied by a downregulation of contractile proteins such as α-SMA and SM22a and an upregulation of MMPs and inflammatory mediators (5,36,37). This phenotypic transformation of VSMCs has been shown to contribute to the development and progression of vascular diseases (38). Autophagy is a cellular process responsible for the degradation and recycling of damaged or unnecessary cellular components, including proteins and organelles. It plays a crucial role in maintaining cellular homeostasis and is involved in various physiological and pathological processes. Autophagy can influence the processes in VSMCs that are associated with a change in phenotype and subsequently lead to the development of disease (39). Genes related to autophagy, such as ATG7 and ATG5, are implicated for VSMCs (40). SNHG1 is associated with the development of various diseases. In the case of ischemic stroke, Zhang et al (41) discovered that SNHG1, as a competitive endogenous RNA, can influence pathological changes in cerebral blood vessels via the hypoxia-inducible factor-1α/vascular endothelial growth factor signaling pathway. Wang et al (42) also found that increased expression of SNHG1 promotes angiogenesis in cerebral microvascular endothelial cells following oxygen-glucose deprivation therapy and this effect is achieved by targeting miR-199a. This suggests that SNHG1 plays a critical role in vascular endothelial cell proliferation and apoptosis. In addition, studies have shown that SNHG1 regulates vascular endothelial cell proliferation and angiogenesis through miR-196a (43), targeting the miR-340-5p/PIK3CA axis in diabetic retinopathy (44). Li et al (45) found that SNHG1 attenuates high glucose-induced calcification/senescence of VSMCs through post-transcriptional regulation of Bhlhe40 and autophagy via Atg10.
These results suggest a close association between SNHG1 and the development of vascular diseases. The present study found that overexpression of SNHG1 also enhanced the proliferation and migration of VSMCs. Furthermore, the results suggested that silencing of SNHG1 in VSMCs following autophagy could induce a transition from a synthetic phenotype to a contractile phenotype, demonstrating that SNHG1 mediates autophagy of VSMCs.
CLEC7A has been implicated in several diseases. Studies have shown that polymorphisms in CLEC7A could be promising biomarkers for susceptibility to ulcerative colitis (46–48). In addition, CLEC7A serves as a modifier gene in cytotoxic T-lymphocyte associated protein 4 to maintain immune homeostasis and tolerance (49). Another study reported that CLEC7A drives intestinal fungus-mediated host lipid deposition (50), involving diabetic cardiomyopathy (51) and Alzheimer's disease (52). Importantly, the CLEC7A pathway has been shown to activate robust autophagy-dependent unconventional protein secretion in human macrophages (53).
The present study investigated the relationship between SNHG1 and its target gene, CLEC7A, in regulating the biological function of VSMCs. Our previous research has shown that CLEC7A is involved in the development (27) and the present study showed that its expression in VSMCs increased when the cells transitioned from a contractile phenotype to a synthetic phenotype. The following mechanistic study confirmed that CLEC7A was a direct target gene regulated by SNHG1 in VSMCs by RIP and RNA pull-down assays. In addition, it was observed that the expression of both SNHG1 and CLEC7A increased following autophagy of VSMCs. When SNHG1 was overexpressed, the contractile phenotype of VSMCs decreased while the synthetic phenotype increased. Conversely, the trend was reversed when SNHG1 was silenced. Furthermore, when CLEC7A was overexpressed in VSMCs with silenced SNHG1 and induced autophagy, an increase in autophagy markers was observed. These results suggested that SNHG1 mediated the autophagy process in VSMCs by targeting CLEC7A, which in turn triggered the phenotypic transformation of VSMCs. Although the experiment was performed with rapamycin treatment, resulting in increases in both CLEC7A and SNHG1, an RIP assay for CLEC7A following rapamycin treatment would be helpful to consolidate the role of CLEC7A in autophagy. In addition, the subcellular localization of lncRNAs plays a crucial role in determining their functional capabilities within cells. These molecules exert an influence on different cellular compartments and thus enable a spectrum of biological activities. These activities range from the regulation of gene expression to the modulation of cell structure and the response to stress (54,55). For example, SNHG1 was previously identified primarily in the nuclei of bladder cancer cells (56); however, its distribution in VSMCs remains unexplored. Therefore, to decipher the mechanism of action of SNHG1 in the regulation of VSMCs, a fluorescence in situ hybridization experiment needs to be performed to determine precisely its subcellular localization in these cells. While the findings of the present study emphasized the crucial roles of SNHG1 and CLEC7A in regulating autophagy and VSMCs phenotypic transformation, it is noteworthy that incorporating MYH11 and Calponin as additional markers could have strengthened the evidence. Future research on the expression patterns of MYH11 and Calponin related to VSMCs phenotype may offer valuable insights for a comprehensive understanding of this process.
The present study demonstrated that SNHG1 promoted proliferation and migration of VSMCs and increased expression following autophagy of VSMCs. SNHG1 facilitated the transition of VSMCs from a contractile to a synthetic phenotype by promoting autophagy and this mechanism involves the regulation of CLEC7A.
Supplementary Material
Acknowledgements
Not applicable.
Glossary
Abbreviations
- VSMCs
vascular smooth muscle cells
- lncRNA
long non-coding RNA
- SNHG1
small ribosome housekeeping gene RNA1
- CLEC7A
C-type lectin domain containing 7A
Funding Statement
The present study was partly supported by research funding from the National Natural Science Foundation of China (grant nos. 81860222 and 82060226); The Basic Ability Enhancement Program for Young and Middle-aged Teachers of Guangxi (grant no. 2021KY0080); Scientific research team incubation project of Guangxi Minzu Hospital (grant no. FY202107) and Innovation Project of Guangxi Graduate Education (grant no. YCSW2023218).
Availability of data and materials
The data generated in the present study may be requested from the corresponding author.
Authors' contributions
CQ and RTH were responsible for study concept and design. HWD, ZMD, HWD and ZMY performed the literature review. CQ, RTH, HWD, WBT, SDZ, ZMD and ZMY were responsible for data analysis and interpretation. HWD, WBT, SDZ, ZMY, ZMD, RTH and CQ were responsible for manuscript writing and reviewing. CQ and RTH confirm the authenticity of all the raw data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no conflict of interest.
References
- 1.Penn DL, Witte SR, Komotar RJ, Connolly E., Jr The role of vascular remodeling and inflammation in the pathogenesis of intracranial aneurysms. J Clin Neurosci. 2014;21:28–32. doi: 10.1016/j.jocn.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 2.Liao XH, Wang N, Zhao DW, Zheng DL, Zheng L, Xing WJ, Ma WJ, Bao LY, Dong J, Zhang TC. STAT3 protein regulates vascular smooth muscle cell phenotypic switch by interaction with myocardin. J Biol Chem. 2015;290:19641–19652. doi: 10.1074/jbc.M114.630111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cao T, Zhang L, Yao LL, Zheng F, Wang L, Yang JY, Guo LY, Li XY, Yan YW, Pan YM, et al. S100B promotes injury-induced vascular remodeling through modulating smooth muscle phenotype. Biochim Biophys Acta Mol Basis Dis. 2017;1863:2772–2782. doi: 10.1016/j.bbadis.2017.07.002. [DOI] [PubMed] [Google Scholar]
- 4.Zhu LH, Huang L, Zhang X, Zhang P, Zhang SM, Guan H, Zhang Y, Zhu XY, Tian S, Deng K, Li H. Mindin regulates vascular smooth muscle cell phenotype and prevents neointima formation. Clin Sci (Lond) 2015;129:129–145. doi: 10.1042/CS20140679. [DOI] [PubMed] [Google Scholar]
- 5.Bennett MR, Sinha S, Owens GK. Vascular smooth muscle cells in atherosclerosis. Circ Res. 2016;118:692–702. doi: 10.1161/CIRCRESAHA.115.306361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhen Y, Stenmark H. Autophagosome biogenesis. Cells. 2023;12:668. doi: 10.3390/cells12040668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tai S, Hu XQ, Peng DQ, Zhou SH, Zheng XL. The roles of autophagy in vascular smooth muscle cells. Int J Cardiol. 2016;211:1–6. doi: 10.1016/j.ijcard.2016.02.128. [DOI] [PubMed] [Google Scholar]
- 8.Grootaert MOJ, Moulis M, Roth L, Martinet W, Vindis C, Bennett MR, De Meyer GRY. Vascular smooth muscle cell death, autophagy and senescence in atherosclerosis. Cardiovasc Res. 2018;114:622–634. doi: 10.1093/cvr/cvy007. [DOI] [PubMed] [Google Scholar]
- 9.Cheng CI, Lee YH, Chen PH, Lin YC, Chou MH, Kao YH. Free fatty acids induce autophagy and LOX-1 upregulation in cultured aortic vascular smooth muscle cells. J Cell Biochem. 2017;118:1249–1261. doi: 10.1002/jcb.25784. [DOI] [PubMed] [Google Scholar]
- 10.Li T, Tan X, Zhu S, Zhong W, Huang B, Sun J, Li F, Wang Y. SPARC induces phenotypic modulation of human brain vascular smooth muscle cells via AMPK/mTOR-mediated autophagy. Neurosci Lett. 2019;712:134485. doi: 10.1016/j.neulet.2019.134485. [DOI] [PubMed] [Google Scholar]
- 11.Sabeena S. Role of noncoding RNAs with emphasis on long noncoding RNAs as cervical cancer biomarkers. J Med Virol. 2023;95:e28525. doi: 10.1002/jmv.28525. [DOI] [PubMed] [Google Scholar]
- 12.Wang PS, Wang Z, Yang C. Dysregulations of long non-coding RNAs-The emerging ‘lnc’ in environmental carcinogenesis. Semin Cancer Biol. 2021;76:163–172. doi: 10.1016/j.semcancer.2021.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meybodi SM, Soleimani N, Yari A, Javadifar A, Tollabi M, Karimi B, Meybodi ME, Seyedhossaini S, Milan PB, Firoozabadi AD. Circulatory long noncoding RNAs (circulatory-LNC-RNAs) as novel biomarkers and therapeutic targets in cardiovascular diseases: Implications for cardiovascular diseases complications. Int J Biol Macromol. 2023;225:1049–1071. doi: 10.1016/j.ijbiomac.2022.11.167. [DOI] [PubMed] [Google Scholar]
- 14.Cui Y, Zhang F, Zhu C, Geng L, Tian T, Liu H. Upregulated lncRNA SNHG1 contributes to progression of non-small cell lung cancer through inhibition of miR-101-3p and activation of Wnt/β-catenin signaling pathway. Oncotarget. 2017;8:17785–17794. doi: 10.18632/oncotarget.14854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li J, Zhang Z, Xiong L, Guo C, Jiang T, Zeng L, Li G, Wang J. SNHG1 lncRNA negatively regulates miR-199a-3p to enhance CDK7 expression and promote cell proliferation in prostate cancer. Biochem Biophys Res Commun. 2017;487:146–152. doi: 10.1016/j.bbrc.2017.03.169. [DOI] [PubMed] [Google Scholar]
- 16.Lu Y, Xi J, Zhang Y, Chen W, Zhang F, Li C, Wang Z. SNHG1 inhibits ox-LDL-induced inflammatory response and apoptosis of HUVECs via up-regulating GNAI2 and PCBP1. Front Pharmacol. 2020;11:703. doi: 10.3389/fphar.2020.00703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li W, Dong X, He C, Tan G, Li Z, Zhai B, Feng J, Jiang X, Liu C, Jiang H, Sun X. Correction to: LncRNA SNHG1 contributes to sorafenib resistance by activating the Akt pathway and is positively regulated by miR-21 in hepatocellular carcinoma cells. J Exp Clin Cancer Res. 2021;40:377. doi: 10.1186/s13046-021-02183-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu J, Yang R, Hua X, Huang M, Tian Z, Li J, Lam HY, Jiang G, Cohen M, Huang C. lncRNA SNHG1 promotes basal bladder cancer invasion via interaction with PP2A catalytic subunit and induction of autophagy. Mol Ther Nucleic Acids. 2020;21:354–366. doi: 10.1016/j.omtn.2020.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao J, Geng L, Chen Y, Wu C. SNHG1 promotes MPP+-induced cytotoxicity by regulating PTEN/AKT/mTOR signaling pathway in SH-SY5Y cells via sponging miR-153-3p. Biol Res. 2020;53:1. doi: 10.1186/s40659-019-0267-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu Y, Zhu B, Yan Y, Bai S, Kang H, Zhang J, Ma W, Gao Y, Hui B, Li R, et al. Long non-coding RNA SNHG1 stimulates ovarian cancer progression by modulating expression of miR-454 and ZEB1. Mol Oncol. 2021;15:1584–1596. doi: 10.1002/1878-0261.12932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rizzetto L, De Filippo C, Rivero D, Riccadonna S, Beltrame L, Cavalieri D. Systems biology of host-mycobiota interactions: Dissecting Dectin-1 and Dectin-2 signalling in immune cells with DC-ATLAS. Immunobiology. 2013;218:1428–1437. doi: 10.1016/j.imbio.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y, Spatz M, Da Costa G, Michaudel C, Lapiere A, Danne C, Agus A, Michel ML, Netea MG, Langella P, et al. Deletion of both Dectin-1 and Dectin-2 affects the bacterial but not fungal gut microbiota and susceptibility to colitis in mice. Microbiome. 2022;10:91. doi: 10.1186/s40168-022-01273-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Al Madhoun A, Kochumon S, Al-Rashed F, Sindhu S, Thomas R, Miranda L, Al-Mulla F, Ahmad R. Dectin-1 as a potential inflammatory biomarker for metabolic inflammation in adipose tissue of individuals with obesity. Cells. 2022;11:2879. doi: 10.3390/cells11182879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choraghe RP, Kolodziej T, Buser A, Rajfur Z, Neumann AK. RHOA-mediated mechanical force generation through Dectin-1. J Cell Sci. 2020;133:jcs236166. doi: 10.1242/jcs.236166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rouchaud A, Johnson C, Thielen E, Schroeder D, Ding YH, Dai D, Brinjikji W, Cebral J, Kallmes DF, Kadirvel R. Differential gene expression in coiled versus flow-diverter-treated aneurysms: RNA sequencing analysis in a rabbit aneurysm model. AJNR Am J Neuroradiol. 2016;37:1114–1121. doi: 10.3174/ajnr.A4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Turhon M, Maimaiti A, Gheyret D, Axier A, Rexiati N, Kadeer K, Su R, Wang Z, Chen X, Cheng X, et al. An immunogenic cell death-related regulators classification patterns and immune microenvironment infiltration characterization in intracranial aneurysm based on machine learning. Front Immunol. 2022;13:1001320. doi: 10.3389/fimmu.2022.1001320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu R, Huang L, Zhou M, Zhou S, Hu R. Expression and clinical significance of CLEC7A in intracranial aneurysm tissues and serum. J Minim Invasive Surg. 2021;16:453–456. (In Chinese) [Google Scholar]
- 28.Su S, Shi YT, Chu Y, Jiang MZ, Wu N, Xu B, Zhou H, Lin JC, Jin YR, Li XF, Liang J. Sec62 promotes gastric cancer metastasis through mediating UPR-induced autophagy activation. Cell Mol Life Sci. 2022;79:133. doi: 10.1007/s00018-022-04143-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qiu WL, Zhang YW, Feng Y, Li LC, Yang L, Xu CR. Deciphering pancreatic islet β cell and α cell maturation pathways and characteristic features at the single-cell level. Cell Metab. 2017;25:1194–1205. e1194. doi: 10.1016/j.cmet.2017.04.003. [DOI] [PubMed] [Google Scholar]
- 30.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 31.Yue Y, Xu J, Li Y, Cheng K, Feng Q, Ma X, Ma N, Zhang T, Wang X, Zhao X, Nie G. Antigen-bearing outer membrane vesicles as tumour vaccines produced in situ by ingested genetically engineered bacteria. Nat Biomed Eng. 2022;6:898–909. doi: 10.1038/s41551-022-00886-2. [DOI] [PubMed] [Google Scholar]
- 32.Gomez-Sanchez R, Yakhine-Diop SM, Rodriguez-Arribas M, Bravo-San Pedro JM, Martínez-Chacón G, Uribe-Carretero E, Pinheiro de Castro DC, Pizarro-Estrella E, Fuentes JM, González-Polo RA. MRNA and protein dataset of autophagy markers (LC3 and p62) in several cell lines. Data Brief. 2016;7:641–647. doi: 10.1016/j.dib.2016.02.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ailawadi G, Moehle CW, Pei H, Walton SP, Yang Z, Kron IL, Lau CL, Owens GK. Smooth muscle phenotypic modulation is an early event in aortic aneurysms. J Thorac Cardiovasc Surg. 2009;138:1392–1399. doi: 10.1016/j.jtcvs.2009.07.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ding Y, Zhang M, Zhang W, Lu Q, Cai Z, Song P, Okon IS, Xiao L, Zou MH. AMP-activated protein kinase alpha 2 deletion induces VSMC phenotypic switching and reduces features of atherosclerotic plaque stability. Circ Res. 2016;119:718–730. doi: 10.1161/CIRCRESAHA.116.308689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Horita H, Wysoczynski CL, Walker LA, Moulton KS, Li M, Ostriker A, Tucker R, McKinsey TA, Churchill ME, Nemenoff RA, Weiser-Evans MC. Nuclear PTEN functions as an essential regulator of SRF-dependent transcription to control smooth muscle differentiation. Nat Commun. 2016;7:10830. doi: 10.1038/ncomms10830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Owens GK, Kumar MS, Wamhoff BR. Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol Rev. 2004;84:767–801. doi: 10.1152/physrev.00041.2003. [DOI] [PubMed] [Google Scholar]
- 37.Thyberg J. Phenotypic modulation of smooth muscle cells during formation of neointimal thickenings following vascular injury. Histol Histopathol. 1998;13:871–891. doi: 10.14670/HH-13.871. [DOI] [PubMed] [Google Scholar]
- 38.Sawyer DM, Pace LA, Pascale CL, Kutchin AC, O'Neill BE, Starke RM, Dumont AS. Lymphocytes influence intracranial aneurysm formation and rupture: Role of extracellular matrix remodeling and phenotypic modulation of vascular smooth muscle cells. J Neuroinflammation. 2016;13:185. doi: 10.1186/s12974-016-0654-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang B, Li X, Wang M, Li GX, Ren PW, Wang YQ, Xin SJ, Qin LF. Trehalose attenuates abdominal aortic aneurysm formation by inducing autophagy in smooth muscle cells. J Geriatr Cardiol. 2023;20:214–222. doi: 10.26599/1671-5411.2023.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fang ZM, Feng X, Chen Y, Luo H, Jiang DS, Yi X. Targeting autophagy in aortic aneurysm and dissection. Biomed Pharmacother. 2022;153:113547. doi: 10.1016/j.biopha.2022.113547. [DOI] [PubMed] [Google Scholar]
- 41.Zhang L, Luo X, Chen F, Yuan W, Xiao X, Zhang X, Dong Y, Zhang Y, Liu Y. LncRNA SNHG1 regulates cerebrovascular pathologies as a competing endogenous RNA through HIF-1alpha/VEGF signaling in ischemic stroke. J Cell Biochem. 2018;119:5460–5472. doi: 10.1002/jcb.26705. [DOI] [PubMed] [Google Scholar]
- 42.Wang Z, Wang R, Wang K, Liu X. Upregulated long noncoding RNA Snhg1 promotes the angiogenesis of brain microvascular endothelial cells after oxygen-glucose deprivation treatment by targeting miR-199a. Can J Physiol Pharmacol. 2018;96:909–915. doi: 10.1139/cjpp-2018-0107. [DOI] [PubMed] [Google Scholar]
- 43.Zhang L, Zhang Q, Lv L, Jianhua Z, Ting C, Wu Y. LncRNA SNHG1 regulates vascular endothelial cell proliferation and angiogenesis via miR-196a. J Mol Histol. 2020;51:117–124. doi: 10.1007/s10735-020-09862-z. [DOI] [PubMed] [Google Scholar]
- 44.He FT, Fu XL, Li MH, Fu CY, Chen JZ. LncRNA SNHG1 targets miR-340-5p/PIK3CA axis to regulate microvascular endothelial cell proliferation, migration, and angiogenesis in DR. Kaohsiung J Med Sci. 2023;39:16–25. doi: 10.1002/kjm2.12625. [DOI] [PubMed] [Google Scholar]
- 45.Li S, Ni Y, Li C, Xiang Q, Zhao Y, Xu H, Huang W, Wang Y, Wang Y, Zhan J, Liu Y. Long noncoding RNA SNHG1 alleviates high glucose-induced vascular smooth muscle cells calcification/senescence by post-transcriptionally regulating Bhlhe40 and autophagy via Atg10. J Physiol Biochem. 2023;79:83–105. doi: 10.1007/s13105-022-00924-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Legaki E, Koutouratsas T, Theocharopoulos C, Lagkada V, Gazouli M. Polymorphisms in CLEC5A and CLEC7A genes modify risk for inflammatory bowel disease. Ann Gastroenterol. 2024;37:64–70. doi: 10.20524/aog.2024.0843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Basson A, Trotter A, Rodriguez-Palacios A, Cominelli F. Mucosal interactions between genetics, diet, and microbiome in inflammatory bowel disease. Front Immunol. 2016;7:290. doi: 10.3389/fimmu.2016.00290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Iliev ID, Funari VA, Taylor KD, Nguyen Q, Reyes CN, Strom SP, Brown J, Becker CA, Fleshner PR, Dubinsky M, et al. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science. 2012;336:1314–1317. doi: 10.1126/science.1221789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Turnbull C, Bones J, Stanley M, Medhavy A, Wang H, Lorenzo AMD, Cappello J, Shanmuganandam S, Pandey A, Seneviratne S, et al. DECTIN-1: A modifier protein in CTLA-4 haploinsufficiency. Sci Adv. 2023;9:eadi9566. doi: 10.1126/sciadv.adi9566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ma J, Zhou M, Song Z, Deng Y, Xia S, Li Y, Huang X, Xiao D, Yin Y, Yin J. Clec7a drives gut fungus-mediated host lipid deposition. Microbiome. 2023;11:264. doi: 10.1186/s40168-023-01698-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang N, Wang M, Lin K, Wang M, Xu D, Han X, Zhao X, Wang Y, Wu G, Luo W, et al. Dectin-1 deficiency alleviates diabetic cardiomyopathy by attenuating macrophage-mediated inflammatory response. Biochim Biophys Acta Mol Basis Dis. 2023;1869:166710. doi: 10.1016/j.bbadis.2023.166710. [DOI] [PubMed] [Google Scholar]
- 52.Zhao X, Sun J, Xiong L, She L, Li L, Tang H, Zeng Y, Chen F, Han X, Ye S, et al. β-amyloid binds to microglia Dectin-1 to induce inflammatory response in the pathogenesis of Alzheimer's disease. Int J Biol Sci. 2023;19:3249–3265. doi: 10.7150/ijbs.81900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Öhman T, Teirila L, Lahesmaa-Korpinen AM, Cypryk W, Veckman V, Saijo S, Wolff H, Hautaniemi S, Nyman TA, Matikainen S. Dectin-1 pathway activates robust autophagy-dependent unconventional protein secretion in human macrophages. J Immunol. 2014;192:5952–5962. doi: 10.4049/jimmunol.1303213. [DOI] [PubMed] [Google Scholar]
- 54.Deng J, Xu W, Jie Y, Chong Y. Subcellular localization and relevant mechanisms of human cancer-related micropeptides. FASEB J. 2023;37:e23270. doi: 10.1096/fj.202301019RR. [DOI] [PubMed] [Google Scholar]
- 55.Wei C, Xu Y, Shen Q, Li R, Xiao X, Saw PE, Xu X. Role of long non-coding RNAs in cancer: From subcellular localization to nanoparticle-mediated targeted regulation. Mol Ther Nucleic Acids. 2023;33:774–793. doi: 10.1016/j.omtn.2023.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cai H, Xu H, Lu H, Xu W, Liu H, Wang X, Zhou G, Yang X. LncRNA SNHG1 facilitates tumor proliferation and represses apoptosis by regulating PPARgamma ubiquitination in bladder cancer. Cancers (Basel) 2022;14:4740. doi: 10.3390/cancers14194740. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated in the present study may be requested from the corresponding author.