Summary
Sciatica is characterized by radiating pain along the sciatic nerve, with a lifetime prevalence of up to 43%. This study explored blood biomarkers for sciatica using transcriptomic microarray data (GSE124272 and GSE150408). Differential gene expression analysis identified NADK2 as a potential diagnostic biomarker. A diagnostic model based on NADK2 showed strong validation performance in 200 clinical cases. Gene set enrichment analysis (GSEA) suggested a connection between NADK2 and the aminoacyl-tRNA biosynthesis pathway. In conclusion, NADK2 emerges as promising diagnostic and therapeutic targets for sciatica, significantly advancing our comprehension of potential pathogenic mechanisms and offering perspectives for early diagnosis and treatment.
Subject areas: Pathology, Transcriptomics
Graphical abstract
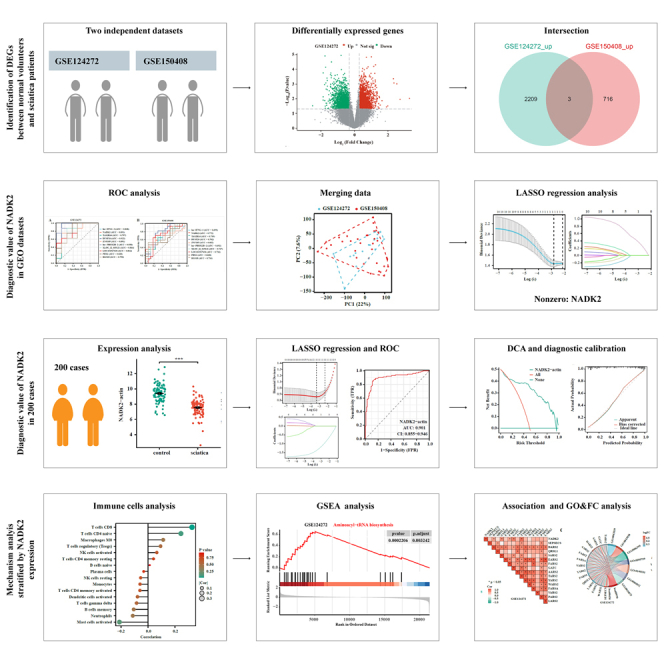
Highlights
-
•
Sciatica diagnosis relies on clinical criteria and imaging with known limitations
-
•
NADK2 expression is elevated in the blood of individuals with sciatica
-
•
High blood NADK2 expression is linked to the presence of sciatica
-
•
NADK2 expression correlates with the aminoacyl-tRNA biosynthesis pathway
Pathology; Transcriptomics
Introduction
Sciatica is a common disease with a lifetime prevalence of up to 43%.1Most sciatica is caused by lumbar disc herniation (LDH), and is mainly caused by single-level LDH at the L4/5 or L5/S1 level.2 The current treatment approach for sciatica involves a stepwise model, initially employing non-surgical methods such as exercise, medication, and interventional procedures. Surgical intervention is considered when non-surgical treatments prove ineffective or when there is persistent and progressive pain.3,4 However, the complications including dural tears, neurologic injury, hardware or bone graft migration, dysesthesia, loss of fixation, and infection, make patients prohibitive. The early diagnosis for sciatica cases is necessary since approximately 84% of LDH cases improve with conservative treatment such as medication and physical therapy.5
Sciatica is diagnosed mainly based on history and physical examination and there is no specific test for sciatica.6 A recent cohort study proposed clinical criteria for unilateral leg pain, mono radicular distribution of pain, positive straight leg raise test at <60° (or femoral stretch test), unilateral motor weakness, and asymmetric ankle reflex to predict sciatica caused by LDH. The patients excluded are those with serious pathology such as cancer, trauma, and infection.7 The sciatic nerve is generally visualized using magnetic resonance imaging (MRI).8 However, the long appointment and checkout time, and prohibition for patients with ferromagnetic objects, pregnancy, and claustrophobia make it challenging for many people to undergo MRI. Besides, sciatica lacks a specific and simple diagnostic method. A combination of various clinical examinations and auxiliary test results for confirmation is typically required.9
Identifying a biomarker in body fluids may provide a viable solution for diagnosing sciatica. In 2002, Brisby et al. reported that the proinflammatory cytokine interleukin (IL)-8 in cerebrospinal fluid could serve as a potential biomarker for disc herniation and sciatica.10 However, a study in 2019 involving 119 patients found that 17 inflammatory serum biomarkers failed to demonstrate diagnostic value for sciatica and referred leg pain.11 More recently, Jin et al.12 developed a diagnostic model using a five-gene immune-related signature to effectively distinguish sciatica patients. In this study, we analyzed the blood gene expression of sciatica patients and healthy individuals to explore diagnostic biomarkers for sciatica, providing insights for the early diagnosis and treatment of this condition.
Results
NADK2 is expressed higher in patients with sciatica than in volunteers
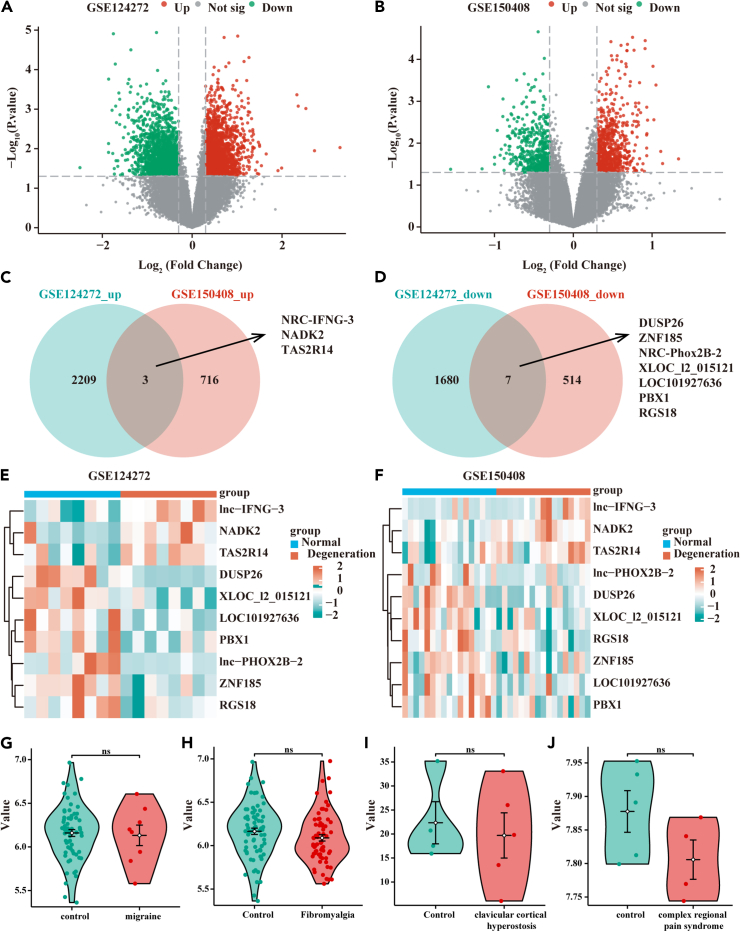
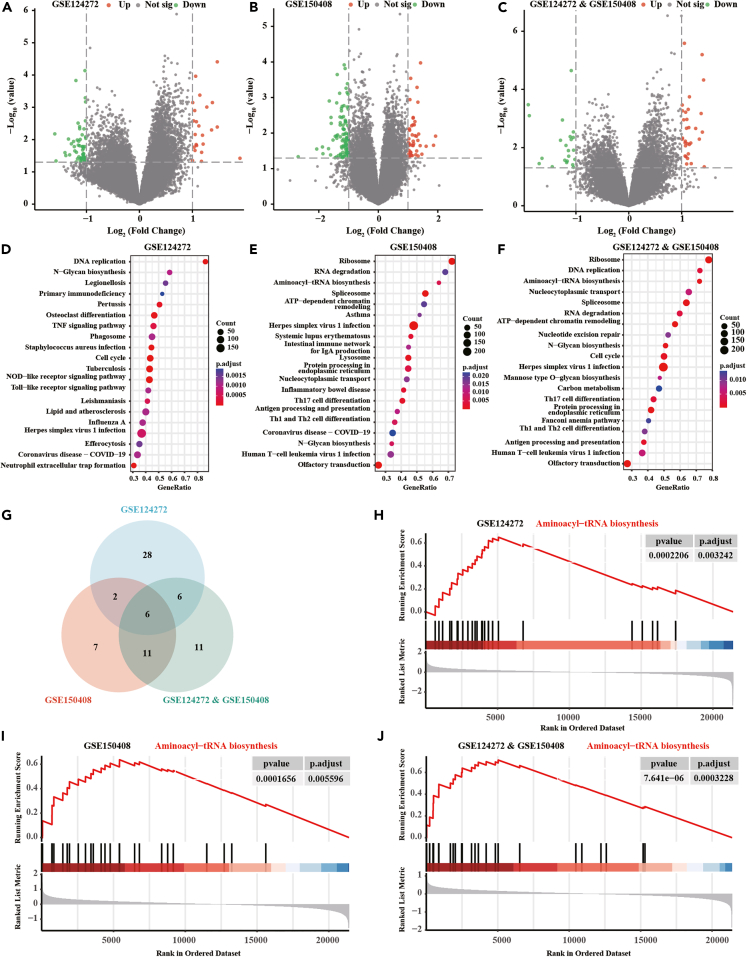
Two transcriptomic microarray data (GEO: GSE124272 and GEO: GSE150408) were sourced from NCBI’s Gene Expression Omnibus. Differentially expressed genes (DEGs) screening between volunteers and sciatica patients was conducted. Employing a threshold of |log fold change| > 0.3 and p < 0.05; 2,212 up-expressed and 1,687 down-expressed were identified in GEO: GSE124272. Additionally, 719 up-expressed and 521 DEGs were found in GEO: GSE150408. A Venn diagram illustrated the intersection of DEGs, revealing ten genes across the cohorts, including three upregulated (NRC-IFNG-3, NADK2, and TAS2R14) and seven downregulated genes (DUSP26, ZNF185, NRC-Phox2B-2, XLOC_l2_015121, LOC101927636, PBX1, and RGS18) (Figures 1C and 1D). The expression patterns of these ten DEGs in the two datasets were visually represented using heat maps (Figures 1E and 1F). Details were shown in Table S1. Additionally, NADK2 expression in the blood of patients with migraine (6.16 ± 0.31 vs. 6.13 ± 0.33, p > 0.05), fibromyalgia (6.17 ± 0.31 vs. 6.09 ± 0.30, p > 0.05), clavicular cortical hyperostosis (22.34 ± 8.79 vs. 19.70 ± 10.53, p > 0.05), and complex regional pain syndrome (7.88 ± 0.07 vs. 7.81 ± 0.06, p > 0.05) did not show an increase compared to healthy individuals (Figures 1G–1J), supporting the notion that NADK2 may be specifically upregulated in sciatica.
Figure 1.
Differential genes between sciatica patients and healthy individuals in GSE124272 and GSE150408 datasets
(A and B) Volcano plot displaying differential genes in GSE124272 and GSE150408 between sciatica patients and healthy individuals (independent-samples t test, mean ± SD).
(C) Venn diagram showing 3 commonly upregulated genes in both datasets.
(D) Venn diagram displaying 7 commonly downregulated genes in both datasets.
(E and F) Heatmap presenting the expression of intersecting genes in GSE124272 and GSE150408.
(G–J) Violin plot showing NADK2 expression in the blood of patients with migraine, fibromyalgia, clavicular cortical hyperostosis, and complex regional pain syndrome compared to healthy individuals (independent-samples t test, mean ± SD).
NADK2 expression in blood holds diagnostic relevance for sciatica
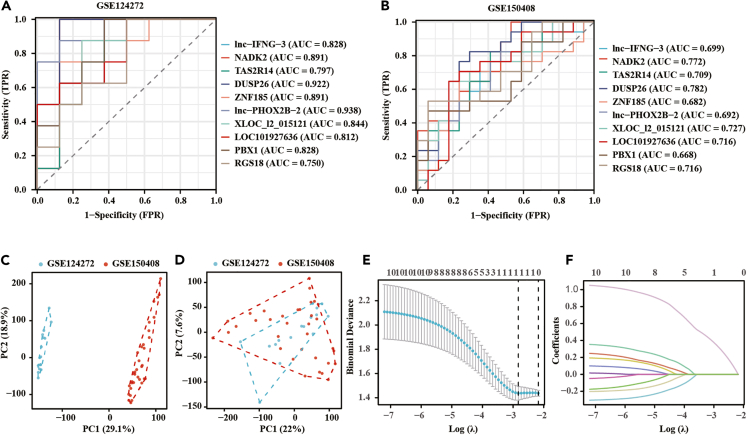
Exploring the diagnostic relevance of NADK2 expression in blood for sciatica, receiver operating characteristic (ROC) curves were employed after preliminary DEG screening. An area under the ROC curve (AUC) greater than 0.75 was considered a good diagnostic value. As shown in Figures 2A and 2B, all the ten DEGs had a high diagnostic value in GSE124272, while only DUSP26 and NADK2 exhibited a high diagnostic value in GSE150408. Considering the limited data volume in each dataset, the two datasets were merged to create a combined dataset. Initial principal-component analysis (PCA) analysis revealed distinct clusters for the two datasets, indicating a batch effect likely due to different study origins (Figure 2C). Post removal of the batch effect, PCA results showcased successful integration, as evidenced by unified clustering (Figure 2D). Subsequent analysis using LASSO regression with 10-fold cross-validation yielded an optimal lambda value (λ(min) = 0.032) (Figures 2E and 2F). Table S2 presented the LASSO coefficients of the ten DEGs. These results revealed that NADK2 exclusively held diagnostic relevance for sciatica.
Figure 2.
Establishment of NADK2 as a diagnostic gene for sciatica
(A and B) Diagnostic ROC curves displaying differential genes in each of GSE124272 and GSE150408.
(C and D) PCA analysis assessing batch effects in the two datasets before (C) and after (D) batch effect removal.
(E and F) LASSO regression analysis with 10-fold cross-validation on the merged dataset, revealing that only NADK2 holds diagnostic significance.
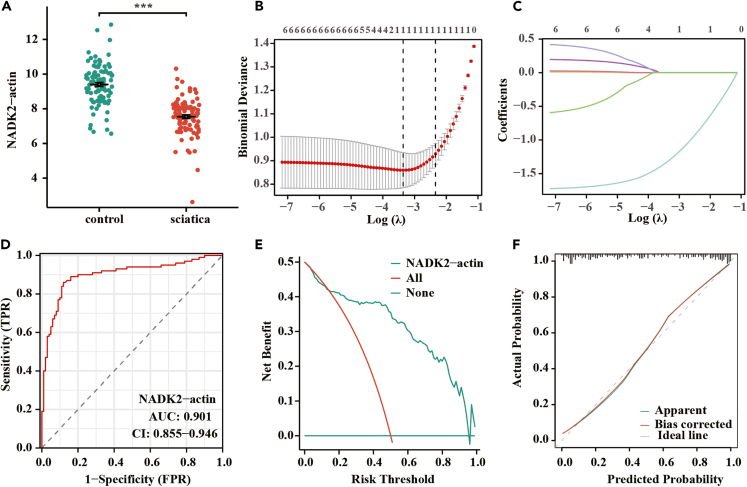
Verification of NADK2 expression and diagnostic value in clinical cases
The diagnostic potential of NADK2 for sciatica was externally validated using a cohort of 200 clinical cases. Demographic and clinicopathological characteristics were shown in Table 1. The expression level of NADK2 in sciatica patients was significantly lower than that in the control group (9.40 ± 1.14 vs. 7.55 ± 1.04, p < 0.001) (Figure 3A). Utilizing LASSO regression analysis with 10-fold cross-validation (λ(min) = 0.035) on this clinical dataset, NADK2 emerged as the sole significant marker (Figures 3B and 3C). Table S3 displayed the LASSO coefficients of the clinical factors. Reanalysis of the ROC curve reaffirmed the high diagnostic value of NADK2 for sciatica (AUC = 0.901) (Figure 3D). Further validation was provided by decision curve analysis (DCA) and a diagnostic calibration curve (Figures 3E and 3F).
Table 1.
Demographic and clinicopathological characteristics in participants
| Characteristics | No sciatica | Sciatica | p value |
|---|---|---|---|
| n | 100 | 100 | – |
| sex, n (%) | – | – | 0.12 |
| male | 40 (20%) | 51 (25.5%) | – |
| female | 60 (30%) | 49 (24.5%) | – |
| age, median (IQR) | 47 (32, 60) | 48 (31, 62) | 1 |
| BMI, median (IQR) | 22.55 (19.7, 25.0) | 26.15 (22.5, 28.5) | p < 0.05a |
| smoking, n (%) | – | – | 0.44 |
| yes | 27 (13.5%) | 32 (16%) | – |
| no | 73 (36.5%) | 68 (34%) | – |
| osteoarthritis pain, n (%) | – | – | 0.52 |
| yes | 29 (14.5%) | 25 (12.5%) | – |
| no | 71 (35.5%) | 75 (37.5%) | – |
BMI, body mass index; IQN, interquartile range.
p < 0.05.
Figure 3.
Validating the diagnostic efficacy of NADK2 in a clinical dataset
(A) Expression levels of NADK2 in the control group and discogenic sciatica (n = 100/group).
(B and C) LASSO regression analysis with 10-fold cross-validation on clinical data.
(D) Diagnostic ROC curve validating the diagnostic value of NADK2 expression for sciatica.
(E and F) Further validation of the diagnostic value of NADK2 using diagnostic curve analysis and diagnostic calibration curve. ∗∗∗p < 0.001.
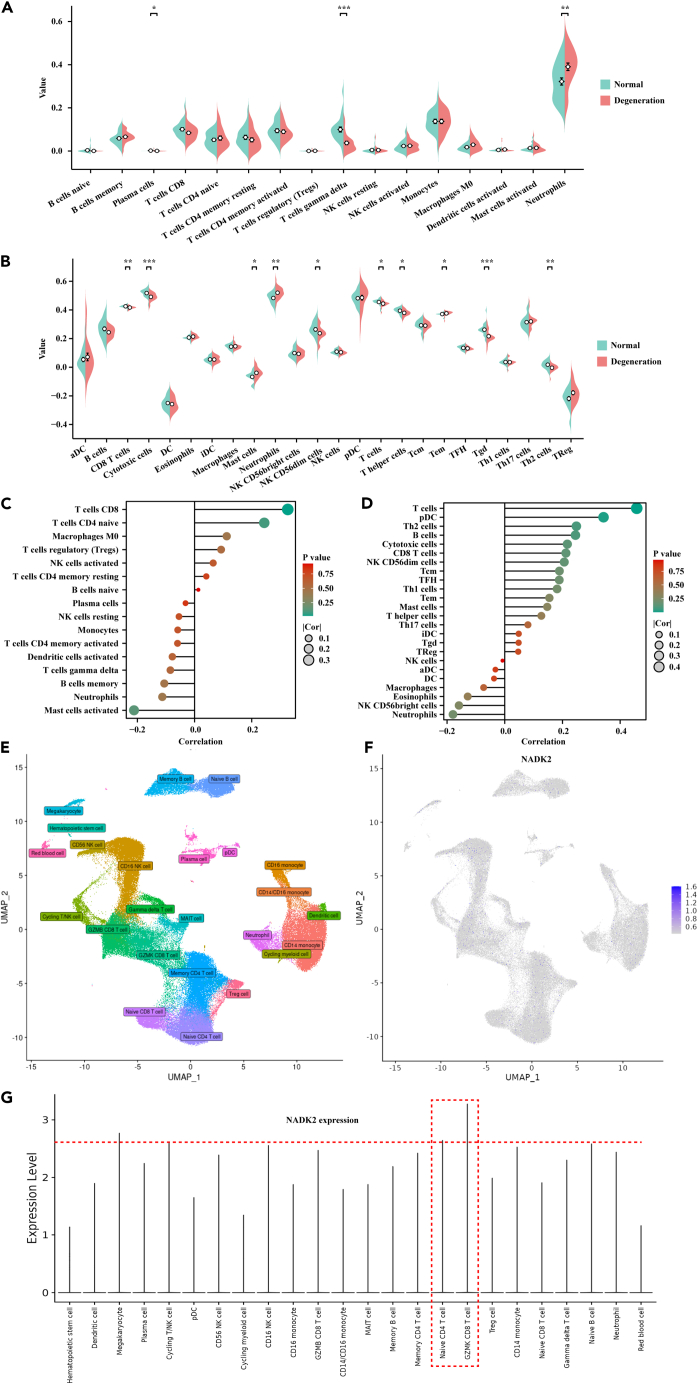
Immune cell infiltration analysis in GEO datasets
The distribution of blood immune cells in both volunteers and sciatica patients underwent analysis using the Cibersort and ssGSEA methods. Cibersort revealed a notable difference in plasma cells, T cells gamma delta, and neutrophils (Figure 4A), while CD8 T cells, cytotoxic cells, mast cells, neutrophils, natural killer (NK) CD56dim cells, pDC, T cells, T helper cells, Tem, Tgd, and Th2 cells exhibited a significant difference between volunteers and sciatica patients (Figure 4B). Subsequently, the correlation between various immune cells and NADK2 expression was explored, revealing significant differences in the expression of T cells CD8 (Cibersort), pDC, and T cells (ssGSEA) across the groups (Figures 4C and 4D). Besides, data analyzed and downloaded from the DISCO database indicate that NADK2 is primarily expressed in CD8 T cell in the blood (Figures 4E–4G). Therefore, the expression levels of NADK2 in CD8 T cells play a significant role in determining the overall NADK2 expression levels in the blood during disease.
Figure 4.
Distribution of immune cells in sciatica patients and healthy individuals
(A and B) Analysis of immune cell distribution in sciatica patients and healthy individuals using Cibersort (A) and ssGSEA algorithms (B).
(C and D) Comparison of the correlation between immune cells and NADK2 gene expression in the two methods.
(E) UMAP (uniform manifold approximation and projection) visualization of single-cell RNA sequencing dataset from blood in DISCO database.
(F and G) UMAP plot and histogram plot illustrating expression of NADK2 across all cell types in blood. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
The NADK2 expression was correlated with aminoacyl-tRNA biosynthesis signaling pathway
In conducting this analysis, three datasets were utilized: GSE124272, GSE150408, and integrated data. These datasets were stratified into high and low NADK2 expression groups, and a more stringent threshold for DEG analysis was performed (p < 0.05, |log fold change| > 1) to focus on genes with stronger differential expression when analyzing the correlation with NADK2 expression. In GSE124272, 135 DEGs were identified (43 upregulated and 92 downregulated), while in GSE150408, 75 DEGs were found (23 upregulated and 53 downregulated). The integrated data revealed 53 DEGs (31 upregulated and 22 downregulated) (Figures 5A–5C). Subsequent gene set enrichment analysis (GSEA) unveiled the top 20 enrichment pathways for both high-expression and low-expression groups (Figures 5D–5F). Cross-analysis highlighted that six signaling pathways were enriched in all the three datasets. Intersective GSEA results were shown in the Table S4-S6. Among them, the aminoacyl-tRNA biosynthesis signaling pathway draw our attention (Figures 5G–5J).
Figure 5.
Pathway analysis associated with NADK2 diagnosis
(A–C) DEG analysis was conducted on GSE124272, GSE150408, and integrated data, categorizing based on average expression of NADK2 (|logFC| > 1, p < 0.05).
(D–F) GSEA analysis obtained the top 20 enriched pathways for high and low expression groups in the three datasets.
(G–J) Cross-analysis revealed that the aminoacyl-tRNA biosynthesis signaling pathway had high enrichment scores in all three datasets.
Association between aminoacyl-tRNA biosynthesis pathway and NADK2
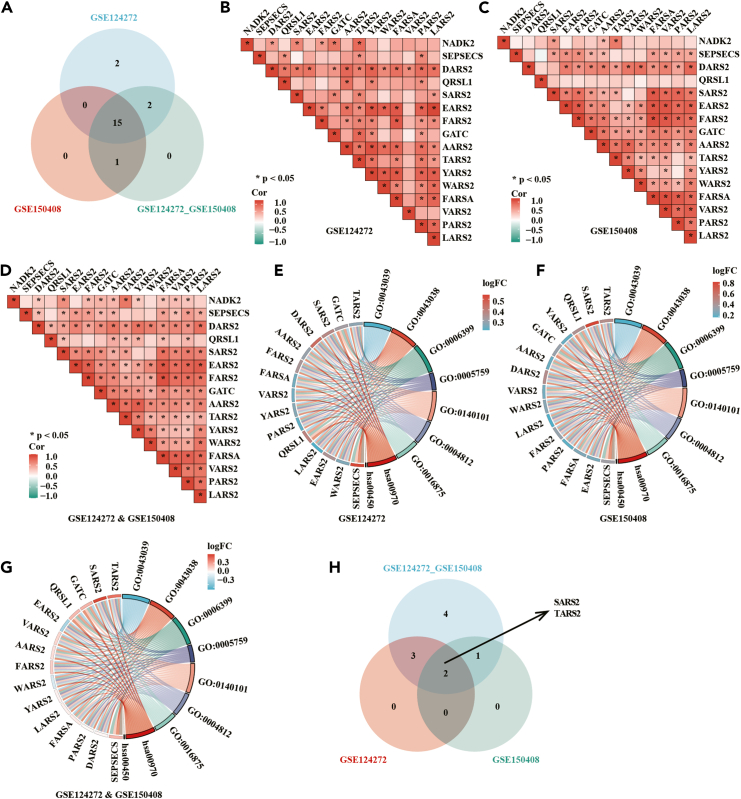
To unravel the link between the aminoacyl-tRNA biosynthesis pathway and NADK2, we pinpointed genes enriched in all three datasets, resulting in the identification of 15 genes (Figure 6A). An association analysis between these 15 intersection genes and NADK2 uncovered a significant correlation with the majority of pathway genes (Figures 6B–6D). Further scrutiny of these intersection genes was conducted through Gene Ontology (GO) and functional clustering analysis (Figures 6E–6G). Ultimately, among the screened genes, those intricately associated with NADK2 were discerned, indicating that seryl-tRNA synthetase 2 (SARS2) and threonyl-tRNA synthetase 2 (TARS2) might exhibit functional relevance to NADK2 (Figure 6H).
Figure 6.
Relationship between aminoacyl-tRNA biosynthesis pathway and NADK2
(A) Venn diagram illustrates 15 intersecting genes enriched in the pathway across three datasets.
(B–D) Association analysis between NADK2 and the 15 intersecting genes reveals that NADK2 is correlated with most genes within the pathway.
(E–G) GO co-enrichment and fold-change analysis for the intersecting genes.
(H) Intersection of genes associated with both NADK2 and the pathway enriched genes.
Discussion
Sciatica presents as a radiating pain along the sciatic nerve, exhibiting a considerable lifetime prevalence of up to 43%.9,13 Despite its prevalence, sciatica lacks a specific diagnostic method necessitating a comprehensive approach involving diverse clinical examinations and auxiliary tests for accurate diagnosis. Leveraging transcriptomic microarray data of whole blood from NCBI’s Gene Expression Omnibus, the RNA expression between normal volunteers and sciatica patients were analyzed. Firstly, the DEG analysis in the two datasets was performed. Then, after merging the two datasets and removing batch effects, a LASSO regression with 10-fold cross-validation was applied. Notably, this analysis identified NADK2 as the sole gene with substantial diagnostic implications for sciatica. To validate its diagnostic efficacy, NADK2 underwent scrutiny in a set of 200 clinical cases, affirming its effectiveness in diagnosing sciatica. GSEA analysis offered preliminary insights, suggesting a potential association between NADK2 and the aminoacyl-tRNA biosynthesis signaling pathway. Further correlation analysis between genes in this pathway and NADK2 elucidated the relevance of SARS2 and TARS2 to the diagnostic function of NADK2. Given their associations with mitochondrial function, these findings hint at a potential correlation between sciatica diagnosis involving NADK2 and mitochondrial function.
Nicotinamide adenine dinucleotide phosphate (NADPH) is the primary cellular currency found in both the cytoplasm and mitochondria, assumes a pivotal role in furnishing reducing equivalents crucial for reductive biosynthesis and antioxidant defenses, thereby facilitating cellular growth and proliferation.14 Its involvement extends to the biosynthesis of macromolecules, encompassing amino acids, fatty acids, tetrahydrofolate, and deoxyribonucleotides.15,16,17,18 Additionally, NADPH serves as a driving force for various redox systems, instigating the production of key antioxidants such as glutathione, essential for quenching reactive oxygen species (ROS) generated during cellular metabolism and growth processes.16,18,19,20 Thus, NAPDH-associated protein can be a biomarker for various disease. For example, increased NADPH oxidase (NOX) 5 and decreased levels of NOX4 were related with serum iron metabolism biomarkers in relapsing-remitting multiple sclerosis patients.21 Serum NOX2 levels are significantly associated with hemorrhage severity, poor 90-day prognosis and delayed cerebral ischemia (DCI) in aneurysmal subarachnoid hemorrhage patients.22 Nicotinamide phosphoribosyl-transferase, the rate-limiting enzyme in the nicotinamide adenine dinucleotide (NAD) salvage pathway, was highly elevated in serum of patients with gastrointestinal acute graft-versus-host disease.23 In this study, we found that the expression of blood NADK2, a mitochondrial NAD kinase, was associated with sciatica patients. NADK2 can mediate the mitochondrial NADPH generation which assumes a critical role in proline biosynthesis and glutamate reduction, where proline is utilized for cytoplasmic protein synthesis, thereby playing a crucial role in cell growth and proliferation.15,24 Deficiency or mutations in NADK2 can lead to mitochondrial dysfunction and cellular proline deficiency.24 Research has found that elevated levels of proline are associated with the neurophysiological pathologies of certain diseases, as patients with hyper-prolinemia may manifest neurological symptoms and brain abnormalities.25 Additionally, NADK2 deficiency was observed in four patients with complex neurological metabolic disorders.26 This suggests that mitochondrial dysfunction resulting from NADK2 deficiency may be associated with neurological functions. While these findings are promising, more research is needed to explore NADK2’s exact mechanistic role in sciatica and evaluate whether direct therapeutic targeting of NADK2 could provide clinical benefits. Future studies could investigate this possibility in preclinical models to determine if NADK2 modulation can alleviate sciatica symptoms.
Through the analysis of NADK2 and cellular metabolic pathways, a potential link between the aminoacyl-tRNA biosynthesis signaling pathway and the diagnosis of NADK2 has been revealed. Aminoacyl-tRNA synthetases (AARSs) play a pivotal role in aminoacyl-tRNA biosynthesis.27 As an indispensable and widely distributed enzyme family, AARSs catalyze the binding of specific amino acids to their corresponding transfer RNA (tRNA), playing a critical role in protein synthesis.28 Mitochondrial AARS mutations can disrupt the respiratory chain, contributing to various diseases such as encephalopathy and cardiomyopathy. However, their contribution to muscle and nerve diseases remains unclear.29 Moreover, AARSs may be linked to pain, as evidenced by a study on chronic pain highlighting the substantial impact of the aminoacyl-tRNA biosynthesis signaling pathway on chronic inflammatory pain,30 although the exact mechanisms remain elusive. Within this pathway, SARS2 and TARS2 are proposed to be potentially associated with NADK2 functioning.
SARS2 encodes the mitochondrial serine tRNA synthetase. Elevated SARS2 expression in various neurodegenerative diseases may lead to excessive mitochondrial protein production, resulting in protein accumulation. This disruption can compromise the protein homeostasis in the mitochondrial matrix, inducing mitochondrial stress.31,32,33 Furthermore, high levels of SARS2 may impede retrograde axonal transport in mitochondria,33,34,35,36 causing the accumulation of ROS and oxidative stress, ultimately inducing cell death.36 In summary, SARS2 overexpression renders neurons more susceptible to dysfunction in mitochondrial dynamics, transport, lipid metabolism, and other functions. TARS2 encodes the mitochondrial threonyl-tRNA synthetase.37 The pathogenicity of TARS2 was initially reported in patients with mitochondrial myopathy.38 Symptoms of TARS2-related mitochondrial diseases include developmental delay, intellectual impairment, degeneration, brain atrophy, changes in basal ganglia signaling, and mitochondrial features, including lactic acidosis.37 Additionally, TARS2 plays a crucial role in early central nervous system development, possibly influencing neuronal proliferation and apoptosis through mTORC1 signaling.37,39 In summary, both SARS2 and TARS2 can impact neuronal function, suggesting a potential association with sciatica to some extent. SARS2 and TARS2 are both associated with mitochondrial function, and enhancing mitochondrial function can alleviate sciatica associated pain.40 Additionally, NADK2 may be linked to the aminoacyl-tRNA biosynthesis function in mitochondria. Elevated expression of NADK2 leads to increased expression of SARS and TARS2, thereby inhibiting mitochondrial function and exacerbating sciatica.
In summary, our study involved comprehensive bioinformatics analysis and identified NADK2 as a potential diagnostic biomarker for sciatica. We emphasized the association of NADK2 with the aminoacyl-tRNA biosynthesis pathway in mitochondria, along with its correlation with the SARS2 and TARS2 genes. These identified genes and pathways may serve as valuable diagnostic and therapeutic targets for sciatica. This research contributes to a deeper understanding of the potential pathogenic mechanisms associated with sciatica, offering insights for the early diagnosis and treatment of this condition.
Limitations of the study
This study has certain limitations, primarily relying on public databases and limited clinical specimens. Larger-scale research and more direct evidence are needed to establish the relationship between sciatica and NADK2, SARS2, and TARS2.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Xin Zheng (thindy1980@163.com).
Materials availability
This study did not generate new unique reagents.
Data and code availability
This study utilized transcriptomic microarray data sourced from NCBI’s Gene Expression Omnibus with two datasets GSE124272 and GSE150408, encompassing 16 and 34 participants, respectively. The former included 8 individuals with sciatica and 8 healthy controls, while the latter comprised data from 17 patients with sciatica and 17 healthy volunteers. Additionally, transcriptomic sequencing of peripheral blood from patients with migraine, fibromyalgia, clavicular cortical hyperostosis, complex regional pain syndrome, and their respective healthy control groups was downloaded from datasets GSE172049 (migraine subgroup within the healthy controls), GSE47603, and GSE67311. Single-cell sequencing data of human blood were analyzed and downloaded from the DISCO database. The datasets used and/or analyzed in this study are available from the GEO database and the supplementary materials provided. This study did not generate any code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Acknowledgments
Funding supported by grants as follows: National Natural Science Foundation of China (grant no. 82272523 and 81902244]; President’s fund of Nanfang Hospital (grant no. 2022B013); Science and Technology Projects of Guangzhou (grant no. 2024A04J5097); The Guangdong Natural Science Foundation (grant no. 2023A1515012499); The Presidential Foundation of Zhujiang Hospital of Southern Medical University (grant no. yzjj2022ms17); China Postdoctoral Science Foundation Special Fund for Pneumonia Epidemic Prevention and Control (grant no. 212977); and The Guangzhou’s Key R&D Plan for agricultural and social development (grant no. 2024B03J0781).
Author contributions
X.W., B.W., Y.W., and X.Z. contributed reagents, protocols, samples, or experiments. X.W. and B.W. performed the experiments. X.W., Z.R., B.W., J.S., J.L., Y.W., and X.Z. analyzed, discussed, and interpreted the data. X.W., Z.R., B.W., Y.W., and X.Z. wrote the manuscript.
Declaration of interests
The authors declare no competing interests.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited data | ||
| Analyzed data | This paper | GEO: GSE124272 GEO: GSE150408 GEO: GSE172049 GEO: GSE47603 GEO: GSE67311 |
| Oligonucleotides | ||
| Primer: NADK2 forward, GCTCTACAGTCCGGAAGAACC; NADK2 reverse, GCATCCCAACAACGAGAACG, |
This paper | N/A |
| Primer: β-ACTIN forward, GATCATTGCTCCTCCTGAGC; β-ACTIN reverse, ACTCCTGCTTGCTGATCCAC. |
This paper | N/A |
| Software and algorithms | ||
| Limma package (version 3.54.2) | N/A | |
| ggplot2 package (version 3.4.4) | N/A | |
| ComplexHeatmap package (version 2.15.4) | N/A | |
| VennDiagram package (version 1.7.3) | N/A | |
| R software (version 4.2.2) | https://cloud.r-project.org/ | |
| SVA package (version 3.46.0) | N/A | |
| pROC package (version 1.18.0) | N/A | |
| rmda package (version 1.6) | N/A | |
| rms package (version 6.4.0) | N/A | |
| ResourceSelection packages (version 0.3–5) | N/A | |
| Cibersort/GSVA packages (version 1.44.5) | https://cibersortx.stanford.edu/ | |
| ClusterProfiler package (version 4.6.2) | N/A | |
Experimental model and study participant details
For validation, blood samples were collected from100 sciatica patients and 100 healthy volunteers in our hospital. Patients with cancer, severe diseases that restrict activity (such as cardiopulmonary failure, hepatic and renal dysfunction, serious cardiovascular and cerebrovascular diseases), and inflammatory conditions (including rheumatoid arthritis, septicopyemia, and severe pneumonia) were excluded. The samples were stored in liquid nitrogen until use, and tested for NADK2 by quantitative real-time PCR (RT-PCR). To ensure participant confidentiality, all personal identifying information was anonymized and replaced with coded identifiers during the data collection and analysis process. Only authorized research personnel had access to the data, which was securely stored in encrypted databases. In addition, all procedures were conducted in accordance with the Declaration of Helsinki and were approved by the Ethical Review Committee of our Hospital (Approval number: 2024-SYJS-164-01). These protective measures were implemented to safeguard the privacy and well-being of the participants throughout the study. Written informed consent was received.
Method details
Identification of differentially expressed genes
DEG identification between groups was executed using the 'limma' package (version 3.54.2), while the 'ggplot2′ package (version 3.4.4) facilitated the visualization via volcano plots. Variation in gene expression across datasets was depicted using heat maps, generated with the 'ComplexHeatmap' package (version 2.15.4). Shared genes under different conditions were analyzed and extracted employing the 'VennDiagram' package (version 1.7.3). All computational processes were conducted in R software (version 4.2.2).
Batch effect removal
PCA was employed to assess batch effects. Initial PCA analysis of the two data groups indicated the presence of batch effects. To address this, the ‘SVA’ package (version 3.46.0) in R software was employed, resulting in improved data integration post-removal of batch effects, thereby enabling more effective subsequent analyses.
Discovery and validation of diagnostic genes
The diagnostic efficacy of DEGs was evaluated using the ROC curve, analyzed via the ‘pROC’ package (version 1.18.0) and visualized using 'ggplot2'. The least absolute shrinkage and selection operator (LASSO) regression, incorporating 10-fold cross-validation, determined the optimal lambda value from the smallest partial likelihood deviation. The construction of a binary logistic model utilized the ‘glm’ function, and the diagnostic value of genes was assessed using DCA and diagnostic calibration curves calculated with the ‘rmda’ package (version 1.6) and visualized using 'ggplot2'; Diagnostic calibration curves analyzed using the ‘rms’ packages (version 6.4.0) and ‘ResourceSelection’ packages (version 0.3–5).
RNA extraction and quantitative RT-PCR
Total RNA was extracted from blood using TRIzol Reagent (Thermo Scientific, USA) according to the manufacturer’s protocol. A total of 2 μg RNA was reverse-transcribed to cDNA with Oligo(dT) (synthesized by Life Technologies), and RevertAid Reverse Transcriptase (Thermo Scientific). Expression of specific genes was calculated by the comparative cycle threshold method using SuperReal PreMix SYBR Green (FP204-02; TIANGEN) in an Applied Biosystem 7500 Fast RT-PCR system (Life Technologies). Primer sequences are list here: NADK2 forward, GCTCTACAGTCCGGAAGAACC; NADK2 reverse, GCATCCCAACAACGAGAACG, β-ACTIN forward, GATCATTGCTCCTCCTGAGC; β-ACTIN reverse, ACTCCTGCTTGCTGATCCAC.
Immune analysis
Immune cell infiltration in normal and sciatica patients was quantified using Cibersort and ssGSEA algorithms (R package: Cibersort/GSVA, version 1.44.5). The Cibersort algorithm, accessible via its website (https://cibersortx.stanford.edu/), applies markers of 22 immune cell types to analyze uploaded data. Similarly, the ssGSEA algorithm, based on Bindea, Gabriela et al.'s markers for 24 immune cells, was employed for assessing immune infiltration in the data.41
Enrichment analysis
GO and Kyoto Encyclopedia of Genes and Genomes enrichment analyses were conducted with the ‘ClusterProfiler’ R package (version 4.6.2). The analysis included all pathways from GSEA gene set. Statistical significance was established at a p-value of less than 0.05. The ‘ggplot2’ package (version 3.4.4) facilitated data visualization.
Quantification and statistical analysis
All statistical analyses, unless specified otherwise, were bilateral, with a significance threshold set at p < 0.05. These analyses were systematically performed using R software (version 4.2.2). Data were presented as the mean ± standard deviation. Quantitative data were compared using a 2-sided Student’s t test or Wilcoxon matched-pairs test. Categorical data compared with Fisher exact test. Details were shown in results or figure legends.
Published: October 18, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.111196.
Contributor Information
Yang Wang, Email: wangyang98289@smu.edu.cn.
Xin Zheng, Email: thindy1980@163.com.
Supplemental information
References
- 1.Liu C., Ferreira G.E., Abdel Shaheed C., Chen Q., Harris I.A., Bailey C.S., Peul W.C., Koes B., Lin C.W.C. Surgical versus non-surgical treatment for sciatica: systematic review and meta-analysis of randomised controlled trials. BMJ. 2023;381 doi: 10.1136/bmj-2022-070730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Valat J.P., Genevay S., Marty M., Rozenberg S., Koes B. Sciatica. Best Pract. Res. Clin. Rheumatol. 2010;24:241–252. doi: 10.1016/j.berh.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 3.Oliveira C.B., Maher C.G., Pinto R.Z., Traeger A.C., Lin C.W.C., Chenot J.F., van Tulder M., Koes B.W. Clinical practice guidelines for the management of non-specific low back pain in primary care: an updated overview. Eur. Spine J. 2018;27:2791–2803. doi: 10.1007/s00586-018-5673-2. [DOI] [PubMed] [Google Scholar]
- 4.Stochkendahl M.J., Kjaer P., Hartvigsen J., Kongsted A., Aaboe J., Andersen M., Andersen M.Ø., Fournier G., Højgaard B., Jensen M.B., et al. National Clinical Guidelines for non-surgical treatment of patients with recent onset low back pain or lumbar radiculopathy. Eur. Spine J. 2018;27:60–75. doi: 10.1007/s00586-017-5099-2. [DOI] [PubMed] [Google Scholar]
- 5.Ma Z., Yu P., Jiang H., Li X., Qian X., Yu Z., Zhu Y., Liu J. Conservative Treatment for Giant Lumbar Disc Herniation: Clinical Study in 409 Cases. Pain Physician. 2021;24:E639–E648. [PubMed] [Google Scholar]
- 6.Stynes S., Konstantinou K., Ogollah R., Hay E.M., Dunn K.M. Clinical diagnostic model for sciatica developed in primary care patients with low back-related leg pain. PLoS One. 2018;13 doi: 10.1371/journal.pone.0191852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Genevay S., Courvoisier D.S., Konstantinou K., Kovacs F.M., Marty M., Rainville J., Norberg M., Kaux J.F., Cha T.D., Katz J.N., Atlas S.J. Clinical classification criteria for radicular pain caused by lumbar disc herniation: the radicular pain caused by disc herniation (RAPIDH) criteria. Spine J. 2017;17:1464–1471. doi: 10.1016/j.spinee.2017.05.005. [DOI] [PubMed] [Google Scholar]
- 8.Ailianou A., Fitsiori A., Syrogiannopoulou A., Toso S., Viallon M., Merlini L., Beaulieu J.Y., Vargas M.I. Review of the principal extra spinal pathologies causing sciatica and new MRI approaches. Br. J. Radiol. 2012;85:672–681. doi: 10.1259/bjr/84443179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jensen R.K., Kongsted A., Kjaer P., Koes B. Diagnosis and treatment of sciatica. BMJ. 2019;367 doi: 10.1136/bmj.l6273. [DOI] [PubMed] [Google Scholar]
- 10.Brisby H., Olmarker K., Larsson K., Nutu M., Rydevik B. Proinflammatory cytokines in cerebrospinal fluid and serum in patients with disc herniation and sciatica. Eur. Spine J. 2002;11:62–66. doi: 10.1007/s005860100306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hider S.L., Konstantinou K., Hay E.M., Glossop J., Mattey D.L. Inflammatory biomarkers do not distinguish between patients with sciatica and referred leg pain within a primary care population: results from a nested study within the ATLAS cohort. BMC Muscoskel. Disord. 2019;20:202. doi: 10.1186/s12891-019-2604-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jin X., Wang J., Ge L., Hu Q. Identification of Immune-Related Biomarkers for Sciatica in Peripheral Blood. Front. Genet. 2021;12 doi: 10.3389/fgene.2021.781945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ropper A.H., Zafonte R.D. Sciatica. N. Engl. J. Med. 2015;372:1240–1248. doi: 10.1056/NEJMra1410151. [DOI] [PubMed] [Google Scholar]
- 14.Pollak N., Dölle C., Ziegler M. The power to reduce:: pyridine nucleotides -: small molecules with a multitude of functions. Biochem. J. 2007;402:205–218. doi: 10.1042/Bj20061638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tran D.H., Kesavan R., Rion H., Soflaee M.H., Solmonson A., Bezwada D., Vu H.S., Cai F., Phillips J.A., DeBerardinis R.J., Hoxhaj G. Mitochondrial NADP is essential for proline biosynthesis during cell growth. Nat. Metab. 2021;3:571. doi: 10.1038/s42255-021-00374-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hosios A.M., Vander Heiden M.G. The redox requirements of proliferating mammalian cells. J. Biol. Chem. 2018;293:7490–7498. doi: 10.1074/jbc.TM117.000239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoxhaj G., Manning B.D. The PI3K-AKT network at the interface of oncogenic signalling and cancer metabolism. Nat. Rev. Cancer. 2020;20:74–88. doi: 10.1038/s41568-019-0216-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiao W., Wang R.S., Handy D.E., Loscalzo J. NAD(H) and NADP(H) Redox Couples and Cellular Energy Metabolism. Antioxidants Redox Signal. 2018;28:251–272. doi: 10.1089/ars.2017.7216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeBerardinis R.J., Chandel N.S. Fundamentals of cancer metabolism. Sci. Adv. 2016;2 doi: 10.1126/sciadv.1600200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goodman R.P., Calvo S.E., Mootha V.K. Spatiotemporal compartmentalization of hepatic NADH and NADPH metabolism. J. Biol. Chem. 2018;293:7508–7516. doi: 10.1074/jbc.TM117.000258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doğan H.O., Yildiz Ö.K. Serum NADPH oxidase concentrations and the associations with iron metabolism in relapsing remitting multiple sclerosis. J. Trace Elem. Med. Biol. 2019;55:39–43. doi: 10.1016/j.jtemb.2019.05.011. [DOI] [PubMed] [Google Scholar]
- 22.Wu X., Ji D., Wang Z., Yu W., Du Q., Hu W., Zheng Y., Dong X., Chen F. Elevated Serum NOX2 Levels Contribute to Delayed Cerebral Ischemia and a Poor Prognosis After Aneurysmal Subarachnoid Hemorrhage: A Prospective Cohort Study. Neuropsychiatric Dis. Treat. 2023;19:1027–1042. doi: 10.2147/ndt.S407907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gerner R.R., Macheiner S., Reider S., Siegmund K., Grabherr F., Mayr L., Texler B., Moser P., Effenberger M., Schwaighofer H., et al. Targeting NAD immunometabolism limits severe graft-versus-host disease and has potent antileukemic activity. Leukemia. 2020;34:1885–1897. doi: 10.1038/s41375-020-0709-0. [DOI] [PubMed] [Google Scholar]
- 24.Zhu J., Schwörer S., Berisa M., Kyung Y.J., Ryu K.W., Yi J., Jiang X., Cross J.R., Thompson C.B. Mitochondrial NADP(H) generation is essential for proline biosynthesis. Science. 2021;372:968–972. doi: 10.1126/science.abd5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wyse A.T.S., Netto C.A. Behavioral and neurochemical effects of proline. Metab. Brain Dis. 2011;26:159–172. doi: 10.1007/s11011-011-9246-x. [DOI] [PubMed] [Google Scholar]
- 26.Pomerantz D.J., Ferdinandusse S., Cogan J., Cooper D.N., Reimschisel T., Robertson A., Bican A., McGregor T., Gauthier J., Millington D.S., et al. Clinical heterogeneity of mitochondrial NAD kinase deficiency caused by a NADK2 start loss variant. Am. J. Med. Genet. 2018;176:692–698. doi: 10.1002/ajmg.a.38602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zong B.B., Xiao Y., Li R., Li H.H., Wang P.Y., Yang X.P., Zhang Y.Y. Transcriptome and metabolome profiling to elucidate the mechanism underlying the poor growth of serotype 2 after orphan response regulator CovR deletion. Front. Vet. Sci. 2023;10 doi: 10.3389/fvets.2023.1280161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pang Y.L.J., Poruri K., Martinis S.A. tRNA synthetase: tRNA aminoacylation and beyond. Wires Rna. 2014;5:461–480. doi: 10.1002/wrna.1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rubio Gomez M.A., Ibba M. Aminoacyl-tRNA synthetases. RNA. 2020;26:910–936. doi: 10.1261/rna.071720.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang W., Lyu J., Xu J., Zhang P., Zhang S., Chen Y., Wang Y., Chen G. The related mechanism of complete Freund's adjuvant-induced chronic inflammation pain based on metabolomics analysis. Biomed. Chromatogr. 2021;35 doi: 10.1002/bmc.5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leuner K., Schütt T., Kurz C., Eckert S.H., Schiller C., Occhipinti A., Mai S., Jendrach M., Eckert G.P., Kruse S.E., et al. Mitochondrion-Derived Reactive Oxygen Species Lead to Enhanced Amyloid Beta Formation. Antioxidants Redox Signal. 2012;16:1421–1433. doi: 10.1089/ars.2011.4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Batlevi Y., La Spada A.R. Mitochondrial autophagy in neural function, neurodegenerative disease, neuron cell death, and aging. Neurobiol. Dis. 2011;43:46–51. doi: 10.1016/j.nbd.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parodi L., Barbier M., Jacoupy M., Pujol C., Lejeune F.X., Lallemant-Dudek P., Esteves T., Pennings M., Kamsteeg E.J., Guillaud-Bataille M., et al. The mitochondrial seryl-tRNA synthetase SARS2 modifies onset in spastic paraplegia type 4. Genet. Med. 2022;24:2308–2317. doi: 10.1016/j.gim.2022.07.023. [DOI] [PubMed] [Google Scholar]
- 34.Denton K.R., Lei L., Grenier J., Rodionov V., Blackstone C., Li X.J. Loss of Spastin Function Results in Disease-Specific Axonal Defects in Human Pluripotent Stem Cell-Based Models of Hereditary Spastic Paraplegia. Stem Cell. 2014;32:414–423. doi: 10.1002/stem.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Havlicek S., Kohl Z., Mishra H.K., Prots I., Eberhardt E., Denguir N., Wend H., Plötz S., Boyer L., Marchetto M.C.N., et al. Gene dosage-dependent rescue of HSP neurite defects in SPG4 patients neurons. Hum. Mol. Genet. 2014;23:2527–2541. doi: 10.1093/hmg/ddt644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grimm A., Eckert A. Brain aging and neurodegeneration: from a mitochondrial point of view. J. Neurochem. 2017;143:418–431. doi: 10.1111/jnc.14037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Accogli A., Lin S.J., Severino M., Kim S.H., Huang K., Rocca C., Landsverk M., Zaki M.S., Al-Maawali A., Srinivasan V.M., et al. Clinical, neuroradiological, and molecular characterization of mitochondrial threonyl-tRNA- synthetase (TARS2)-related disorder. Genet. Med. 2023;25 doi: 10.1016/j.gim.2023.100938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zheng W.Q., Pedersen S.V., Thompson K., Bellacchio E., French C.E., Munro B., Pearson T.S., Vogt J., Diodato D., Diemer T., et al. Elucidating the molecular mechanisms associated with-related mitochondrial disease. Hum. Mol. Genet. 2022;31:523–534. doi: 10.1093/hmg/ddab257. [DOI] [PubMed] [Google Scholar]
- 39.Chan W.Y., Lorke D.E., Tiu S.C., Yew D.T. Proliferation and apoptosis in the developing human neocortex. Anat. Rec. 2002;267:261–276. doi: 10.1002/ar.10100. [DOI] [PubMed] [Google Scholar]
- 40.Song Y., Lu S., Geng W., Feng X., Luo R., Li G., Yang C. Mitochondrial quality control in intervertebral disc degeneration. Exp. Mol. Med. 2021;53:1124–1133. doi: 10.1038/s12276-021-00650-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bindea G., Mlecnik B., Tosolini M., Kirilovsky A., Waldner M., Obenauf A.C., Angell H., Fredriksen T., Lafontaine L., Berger A., et al. Spatiotemporal Dynamics of Intratumoral Immune Cells Reveal the Immune Landscape in Human Cancer. Immunity. 2013;39:782–795. doi: 10.1016/j.immuni.2013.10.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study utilized transcriptomic microarray data sourced from NCBI’s Gene Expression Omnibus with two datasets GSE124272 and GSE150408, encompassing 16 and 34 participants, respectively. The former included 8 individuals with sciatica and 8 healthy controls, while the latter comprised data from 17 patients with sciatica and 17 healthy volunteers. Additionally, transcriptomic sequencing of peripheral blood from patients with migraine, fibromyalgia, clavicular cortical hyperostosis, complex regional pain syndrome, and their respective healthy control groups was downloaded from datasets GSE172049 (migraine subgroup within the healthy controls), GSE47603, and GSE67311. Single-cell sequencing data of human blood were analyzed and downloaded from the DISCO database. The datasets used and/or analyzed in this study are available from the GEO database and the supplementary materials provided. This study did not generate any code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.