Abstract
The p53 tumor suppressor protein is a sequence-specific transcription factor that modulates the response of cells to DNA damage. Recent studies suggest that full transcriptional activity of p53 requires the coactivators CREB binding protein (CBP)/p300 and PCAF. These coactivators interact with each other, and both possess intrinsic histone acetyltransferase activity. Furthermore, p300 acetylates p53 to activate its sequence-specific DNA binding activity in vitro. In this study, we demonstrate that PCAF also acetylates p53 in vitro at a lysine residue distinct from that acetylated by p300 and thereby increases p53’s ability to bind to its cognate DNA site. We have generated antibodies to acetylated p53 peptides at either of the two lysine residues that are targeted by PCAF or p300 and have demonstrated that these antibodies are highly specific for both acetylation and the particular site. Using these antibodies, we detect acetylation of these sites in vivo, and interestingly, acetylation at both sites increases in response to DNA-damaging agents. These data indicate that site-specific acetylation of p53 increases under physiological conditions that activate p53 and identify CBP/p300 and PCAF as the probable enzymes that modify p53 in vivo.
The tumor suppressor protein p53 responds to DNA damage to slow cell growth and promote programmed cell death (21, 31, 35). p53 achieves its antiproliferative properties through its action as a DNA-binding transcriptional activator, to induce expression of downstream target genes. These include p21/waf1/cip1 (16), GADD45 (30), cyclin G (46), bax (40), IGF-BP3 (11), and mdm2 (6, 60), whose gene products are involved in cell cycle arrest, apoptosis, and regulation of p53 function in cells exposed to DNA-damaging agents.
Three major functional domains have been identified in p53: an amino (N)-terminal transactivation domain (residues 1 to 80) (12, 17, 20, 49), a central sequence-specific DNA-binding domain (residues 94 to 293) (7, 24, 57), and a carboxyl (C)-terminal oligomerization domain (residues 325 to 355) (14, 28, 34, 50, 55). In addition to the oligomerization domain, the C terminus of p53 contains two regions (residues 290 to 325 [58] and residues 356 to 393 [26]) that negatively regulate its DNA-binding activity. Multiple posttranslational modifications to these regulatory domains, such as phosphorylation, affect p53 function through modulation of DNA binding (26, 56). In addition, the highly positively charged C-terminal regulatory region may interact with the core DNA-binding domain and lock p53 in an inactive conformation (42). Evidence that supports this idea is the activation of DNA binding by (i) deletion either of the C-terminal region or of the polyproline region at the N-terminal border of the core DNA-binding domain, (ii) binding of 14-3-3 proteins or the monoclonal antibody PAb421 to the C-terminal regulatory domain, and (iii) phosphorylation within the regulatory regions (24–26, 29, 42, 56, 58).
CREB binding protein (CBP) and p300 are structurally related transcriptional factors, involved in cell cycle control and differentiation, which coactivate numerous transactivators, including p53 (3, 23, 36, 52). CBP/p300 have extensive structural and functional similarity, including the capacity to bind both to the adenovirus oncoprotein E1A (1a) and to transactivators, such as CREB (1a, 33, 38), c-Jun/c-Fos (2, 5), c-Myb/v-Myb (15, 43), MyoD (62), and Stat1 (63) and Stat2 (9), and to p53. p300 and CBP associate with PCAF (p300 and CBP associated factor), which has been implicated as an important factor for cell cycle progression (61) and differentiation (48, 61). The complex formed between CBP and PCAF is disrupted by E1A (61), leading to suppression of p53 transactivation (36, 48, 52). These observations suggest that interaction of CBP and PCAF with p53 is critical for p53 function. Supportive evidence is provided by the finding that CBP/p300 and PCAF function as transcriptional coactivators for p53 to fully activate endogenous p21/waf1/cip1 gene expression (52).
An important feature common to coactivators CBP/p300 and PCAF is their intrinsic histone acetyltransferase (HAT) activity (45, 61). Acetylation of lysine residues in the N-terminal tails of histones facilitates gene activation, perhaps by reducing histone tail affinity for DNA and thereby promoting transcription factor binding to nucleosomal DNA (10, 37, 41, 54). The finding that coactivators are HATs has led to an appealing model that transactivators recruit these enzymes to provide promoter-specific chromatin remodeling. Although both are HATs, CBP/p300 and PCAF have little sequence similarity within their HAT domains (44, 45) and, accordingly, exhibit differences in substrate specificities: recombinant CBP/p300 equally acetylate all four histones (H3, H2A, H2B, and H4), even when incorporated into nucleosomes, while PCAF preferentially acetylates H3 and primarily while in a free, nonnucleosomal state. Other proteins, including components of the transcriptional machinery, such as TFIIEβ and TFIIF, are acetylated in vitro by CBP/p300 as well as by other HATs (27). Recently, p300 was shown to acetylate p53 on its C terminus and to enhance p53’s DNA-binding activity in vitro (22). This observation is consistent with the model discussed above, that p53’s C terminus regulates DNA binding. The observation that HATs acetylate substrates other than histones has generated increased interest in the role of acetylation in regulation of gene expression. However, the physiological significance of acetylation of targets other than histones remains an open question. Given the ability of p300 to acetylate p53 in vitro, we investigated whether PCAF is also able to acetylate p53. In this study, we demonstrate that PCAF does indeed acetylate p53 and that it does so at a specific lysine distinct from the residue acetylated by p300. Significantly, both of these lysine residues exhibit increased acetylation in cells in response to DNA damage.
MATERIALS AND METHODS
Plasmids and recombinant proteins.
Specific regions of p53 were amplified by PCR and subcloned into the pGEX-5X-1 vector in frame with the glutathione S-transferase (GST) coding region. The p53 substitution mutations were generated by two-step PCR within the p53 C-terminal region (nucleotides [nt] 900 to 1179) and subcloned into pGEX-5X-1. The acetyltransferases were expressed from pGEX-2T-p300 (amino acids [aa] 1195 to 1810), pGEX-5X-PCAF (aa 352–832), or pRSETA-PCAF containing only the acetyltransferase domain (13). For transfection assays, full-length p53 substitutions (lysine to arginine or alanine) were generated by PCR-directed mutagenesis and then subcloned into pCR3.1 vector at HindIII and PstI sites.
The GST fusion acetylation domains were expressed in Escherichia coli, bound to glutathione Sepharose beads, and eluted with buffer containing 25 mM glutathione, and purified proteins were dialyzed in buffer containing 20 mM Tris (pH 8), 0.5 mM EDTA, 100 mM KCl, 20% glycerol, 0.5 mM dithiothreitol (DTT), 1 mM phenylmethylsulfonyl fluoride (PMSF) before storage at −80°C, as previously described (8).
In vitro acetylation assays.
Substrate proteins (GST-p53 beads) were incubated with 0.25 μCi [3H]acetyl coenzyme A (CoA) (Amersham) and 0.2 μg of purified enzyme in 30 μl of acetylation buffer containing 50 mM Tris (pH 8.0), 5% glycerol, 0.1 mM EDTA, 50 mM KCl, 1 mM DTT, 1 mM PMSF, and 10 mM sodium butyrate. Reaction mixtures were incubated at 30°C for 30 min, stopped by addition of Laemmli buffer, and resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The reactions were analyzed first by Coomassie blue staining to verify the amounts of proteins used in each reaction, and the same gel was subsequently subjected to autoradiography to evaluate acetylation activity.
Electrophoretic mobility shift assay.
For DNA-binding assays, 30 ng of recombinant p53 was acetylated or mock acetylated and incubated with the p53 consensus binding oligonucleotide, 5′-TACAGAACATGTCTAAGCATGCTG-3′, which was labeled with [γ-32P]ATP. The binding reaction was done in 20 μl of DNA-binding buffer (20 mM Tris [pH 7.5], 50 mM KCl, 5 mM MgCl2, 1 mM DTT, 0.5 mM EDTA, 10% glycerol, 10 mM Na butyrate, 0.5 mg of bovine serum albumin, and 100 ng of carrier DNA) with or without 1 μl of PAb421 monoclonal anti-p53 antibody (100 μg/ml). PAb421 antibody was added to the reactions 20 min before DNA probe was added. After incubation at room temperature for another 30 min, the reactions were resolved on a 5% native polyacrylamide gel in Tris-glycine buffer (25 mM Tris base, 200 mM glycine, and 2 mM EDTA) and run at room temperature for 4 h, followed by autoradiography.
p53 Transcriptional activity.
Saos-2 cells were transfected by calcium phosphate precipitation with plasmids expressing wild-type p53 or mutant p53 (0.1 μg) and 29 μg of pEp21-TK-SEAP. Alkaline phosphatase activity was determined 48 h later, as previously described (59).
Generation of acetylation-specific antibodies.
The p53 peptides for acetyl-K320 and acetyl-K373 were SSPQKKKPLDGE and SHLKSKKGQSTSR, respectively, where the underlined lysine residues were acetylated. The peptides were injected into two rabbits each (Research Genetics, Inc.) and were boosted three times. For both peptides, we obtained high-titer and high-specificity antisera to detect p53 acetylated at the appropriate lysine.
Western blotting and immunoprecipitations with α-acetyl-K320 and a α-acetyl-K373 antibodies.
Thirty nanograms of highly purified recombinant wild-type p53 protein or the full-length p53 substitutions K320R and K373R was incubated with or without 0.2 μg of GST-PCAF or GST-p300 protein in the presence of 0.25 μCi of [3H]acetyl-CoA at 30°C for 30 min. The reactions were stopped by adding Laemmli buffer and loaded onto SDS–10% PAGE, transferred to a 0.2-μm-pore-size nitrocellulose membrane, and immunoblotted with the α-acetyl-K320 or α-acetyl-K373 antibodies.
To monitor p53 acetylation in vivo, whole cell extracts were prepared from U20S cells that were either untreated, UV irradiated at 50 J/m2, or γ irradiated at 10 Gray. Whole cell extracts were prepared 4 h after irradiation by lysing the cells in buffer containing 50 mM Tris (pH 8.0), 120 mM NaCl, 0.5% Nonidet P-40, 10 μM Tricostatin A, 1 mM DTT, 1 mM Pefabloc, and 1 mM Pepstatin A. The extracts were cleared by centrifugation and immunoprecipitated with the α-acetyl-K320 or α-acetyl-K373 antibodies or antibody DO1. The amounts of extracts used (1.3 mg of nonirradiated cell extracts and 0.65 mg of UV or γ-irradiated cell extracts) were predetermined by normalization of the amount of p53 by Western analysis with the anti-p53 monoclonal antibody, DO1. The amount of p53 in the immunoprecipitates was determined by immunoblotting with antibody DO1.
RESULTS
PCAF HAT domain acetylates p53 in vitro.
CBP/p300 and PCAF physically interact, and both acetylate histones in vitro. We have previously shown that CBP/p300 and PCAF synergistically enhance p53-mediated transcriptional activation of the endogenous p21 gene (52). The observation that CBP/p300 acetylates p53 raised the question of whether PCAF can also acetylate p53 and, if so, whether the substrate specificity would be distinct from that of CBP/p300. To address this question, in vitro acetyltransferase assays were performed. p53 was fused to GST to aid in purification and used as a substrate for PCAF or p300 acetylation domains (4, 13, 45, 61), which had comparable activity on histone H3, with core histones as substrates (data not shown).
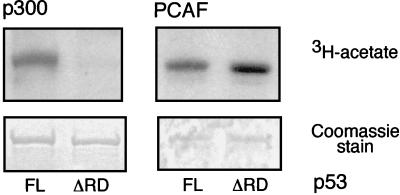
PCAF acetylated full-length p53 (p53FL), as did p300, and they had nearly equal acetylation activities (Fig. 1). Identical amounts of enzymes were then used to acetylate a C-terminal truncated p53 protein (p53ΔRD), lacking the last 25 residues, which had previously been shown to harbor the lysine residues acetylated by p300 (22). Consistent with previous observations, p300-mediated acetylation of p53ΔRD was completely lost. However, PCAF acetylated the truncation protein with no apparent reduction in efficiency compared to that of p53FL (Fig. 1). This result indicated that PCAF efficiently acetylates p53 in vitro and that PCAF and p300 target distinct residues within p53.
FIG. 1.
Acetylation of p53 by PCAF and p300 in vitro. GST fused to either full-length p53 (p53FL) or a p53 truncation mutant (p53ΔRD) lacking 25 amino acids from its C terminus was acetylated with either p300 HAT (left) or PCAF HAT (right). Reaction products were separated by SDS-PAGE. The gel was stained with Coomassie blue to demonstrate that equivalent substrate was used in each reaction (lower panels), and [3H]acetate reaction products were subsequently visualized by autoradiography of the identical gel (upper panels).
PCAF binds to and acetylates p53 within a region spanning residues 300 to 369.
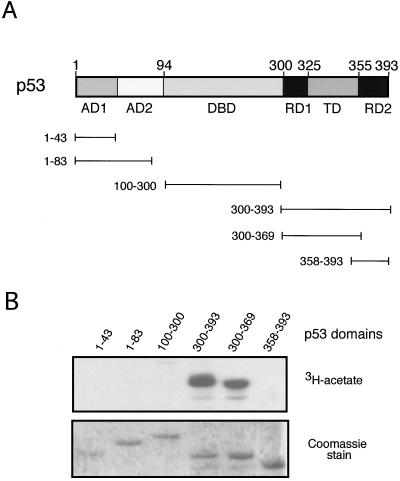
Next we sought to identify the domain of p53 acetylated by PCAF, using a series of deletions within p53 as acetylation substrates (Fig. 2A). The domains tested encompassed the transcriptional activation domain(s) (AD1 and AD2; aa 1 to 43 and 1 to 83), the DNA-binding domain (DBD; aa 100 to 300), the C-terminal region (aa 300 to 393), the C-terminal regulatory domain (RD2; residues 358 to 393), and the tetramerization domain plus a second regulatory domain (RD1 + TD; aa 300 to 369). The deletions were expressed as GST fusion proteins in E. coli, purified on glutathione Sepharose beads, and tested for acetylation by PCAF. Only GST fusion proteins containing the entire C-terminal domain (residues 300 to 393) of p53 or the RD1 + TD domains (aa 300 to 369) served as substrates for PCAF (Fig. 2B). Specifically, PCAF was not able to acetylate the RD2 domain (aa 358 to 393) of p53 (Fig. 2B), in contrast to p300 (22). In addition, the efficiency of PCAF acetylation of the 300 to 393 region was comparable to the RD1 + TD domains (aa 300 to 369), suggesting that the RD2 domain did not have an important role in the overall acetylation (Fig. 2B). These results indicate that PCAF acetylates target residues within one of the regulatory domains (RD1) and/or the tetramerization domain but not within the regulatory domain (RD2) acetylated by p300.
FIG. 2.
Identification of p53 domains acetylated by PCAF. (A) Schematic diagram of p53 domain structure. A series of GST-p53 constructs are depicted containing functional domains of p53. AD1 and AD2, activation domains; DBD, DNA-binding domain; TD, tetramerization domain; RD1 and RD2, regulatory domains. (B) Acetylation of p53 functional domains by PCAF. GST bead slurry of each domain indicated that p53 truncation protein was subjected to acetylation assays with PCAF HAT. Reactions were analyzed as described in the legend for Fig. 1.
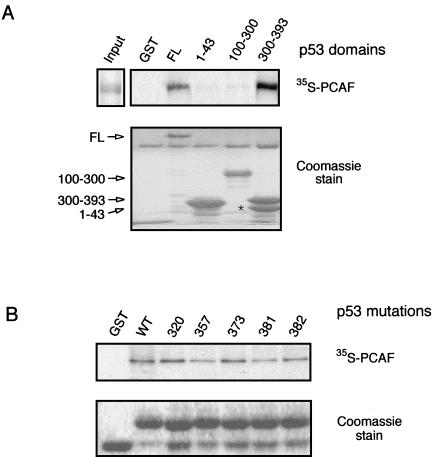
Since PCAF acetylates p53 within a specific domain, we wanted to determine whether a stable interaction could be detected between PCAF and p53. To accomplish this, the GST-p53 fusion proteins used in the acetylation assays (Fig. 2A) were incubated with in vitro-translated PCAF. Only full-length p53 and the C terminus (aa 300 to 393) interacted with PCAF (Fig. 3A). Importantly, this region of p53 was same as that acetylated by PCAF.
FIG. 3.
Interaction of p53 and PCAF in vitro. (A) Interaction of PCAF with p53 domains. GST-p53 truncation proteins, as shown in Fig. 1A, were incubated with in vitro-translated full-length [35S]methionine-labeled PCAF protein. Following incubation, the beads were washed and then subjected to SDS-PAGE, Coomassie staining, and autoradiography. PCAF input (2 μl) represents 10% of actual reaction volume. Unfused GST is present at the bottom of the gel in lane 1. The asterisk indicates a breakdown product of GST-p53(300–393). (B) Interaction of PCAF with GST-p53(300–393) bearing lysine substitutions, as indicated. Binding reactions were performed as described for panel A.
Lysine 320 in p53 is acetylated by PCAF.
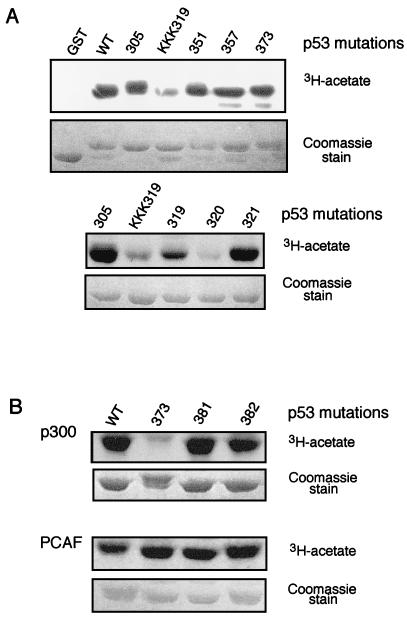
We next wished to define the lysine residue(s) acetylated by PCAF. Lysines within the aa 300 to 369 region of p53 were substituted with arginines either individually or in combination. The substitution mutations were generated within the GST-p53 (300–393) construct (Fig. 2A) and assayed for their ability to serve as substrates for PCAF acetylation. p53 protein containing substitutions at aa 305, 351, or 357 were acetylated by PCAF at the same level as the wild-type protein, whereas the triple substitution mutant K(319/320/321)R was a poor acetylation substrate for PCAF (Fig. 4A, upper panel). To precisely define the residue that is targeted by PCAF in K(319/320/321)R, the individual lysines were converted to arginine. The substitution mutants K319R and K321R were acetylated by PCAF with efficiency similar to that of wild-type p53 (Fig. 4A, lower panel), whereas the K320R substitution mutant was acetylated poorly (Fig. 4A, lower panel). Each of the lysine substitution mutants within GST-p53(300–393) was able to interact with in vitro-translated PCAF (Fig. 3B), indicating that the decreased acetylation of K320R did not result from inability of PCAF to interact with p53.
FIG. 4.
Determination of lysine residues acetylated by PCAF and p300. (A) PCAF acetylation of p53 lysine substitution mutants in RD1 and TD of p53. Lysine residues in the RD1 region (residues 300 to 324) and the tetramerization domain were mutated to arginine (aa 305, 319, 320, 321, 351, and 357). Each mutant was introduced within the 300 to 393 aa region of p53 fused to GST and named according to the position of the mutated amino acid(s). KKK319 stands for triple mutant at residues 319 to 321. Experimental details were the same as those described in the legend for Fig. 2. (B) Acetylation of p53 lysine substitution mutations in RD2 by PCAF and p300. GST fusion proteins containing lysine-to-arginine mutations at residues 373, 381, and 382 were analyzed for acetylation p300 (upper panel) or PCAF (lower panel). Experimental details were the same as those described in the legend for Fig. 2.
We also examined p300-mediated acetylation of GST-p53(300–393). In addition to the lysine substitution mutations within the aa 300 to 369 region described above, lysines in RD2 previously shown to be acetylated by p300 (aa 373, 381, and 382) were replaced by arginine. The K373R substitution significantly decreased the acetylation by p300 (Fig. 2B), whereas none of the lysine substitutions within RD2 affected acetylation by PCAF (Fig. 2B). Reduced acetylation by PCAF and p300 was also observed when the substitutions were made in the context of full-length p53 (data not shown). Thus, acetylation of p53 by PCAF and p300 is highly specific and, interestingly, is targeted to distinct lysines in the regulatory domains of p53.
PCAF acetylation activates p53’s sequence-specific DNA binding.
p53’s function as a tumor suppressor is tightly correlated with its DNA-binding activity (20, 47, 51), which is regulated in multiple ways. Both RD1 (56, 58) and RD2 (26, 31) are involved in allosteric negative regulation of DNA binding. For example, deletions within either region enhance DNA binding by p53, as does the p53-specific monoclonal antibody PAb421, which binds to an epitope in RD2 (aa 371 to 381), thereby relieving the inhibitory effect of the carboxyl-terminal region on DNA binding (25, 26). In addition, acetylation by p300 within RD2 stimulates p53 binding (22). Based on these findings and on our observation of acetylation by PCAF in RD1, we tested whether acetylation by PCAF alters p53’s DNA-binding properties. Using an electrophoretic mobility shift assay, we monitored the sequence-specific DNA-binding activity by p53 in the presence or absence of acetylation by PCAF. An oligonucleotide bearing a p53 consensus binding site was radiolabeled, and either the wild type (p53wt) or the K(319/320/321)R substitution mutant (p53mut) was tested for binding. The p53 proteins bound to DNA with similar affinity, and PAb421 equivalently stimulated the binding (Fig. 5, compare lanes 2 and 3 for p53wt to lanes 11 and 12 for p53mut). Therefore, the substitutions in RD1 did not affect the behavior of the DNA-binding domain of p53.
FIG. 5.
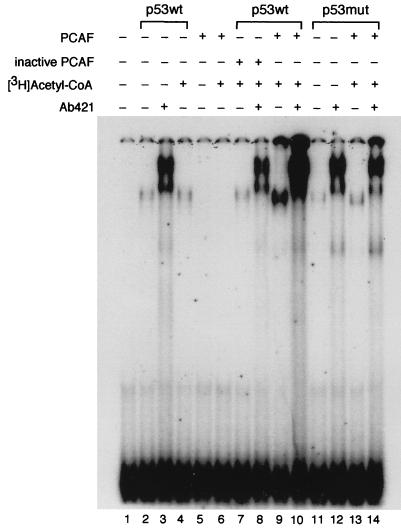
The effect of acetylation by PCAF on p53 sequence-specific binding to DNA. Purified recombinant human wild-type p53 (p53wt) or the triple lysine-to-arginine mutant (319/320/321; p53mut) was incubated with or without [3H]acetyl-CoA and either active PCAF or heat-treated PCAF (inactive). The reaction products were then incubated with a 32P-labeled p53 cognate site in binding buffer, and binding was determined by DNA mobility shift on a native polyacrylamide gel. The polyclonal antibody Ab421 was added as indicated. The mobility of the DNA probe alone is shown in lane 1. Multiple repetitions of the assay were performed, with quantitatively similar effects of K320 acetylation on DNA binding by p53.
We then determined the effect of acetylation of p53 by PCAF. The addition of acetyl-CoA and PCAF greatly stimulated the binding of p53, both in the presence and absence of PAb421 (Fig. 5, compare lanes 2 and 3 to lanes 9 and 10). Several negative controls were performed to determine whether this enhancement of DNA binding was due to acetylation of p53 by PCAF. First, PCAF protein alone or with acetyl-CoA did not possess DNA-binding activity (Fig. 5, lanes 5 and 6). Second, inactivation of PCAF by heat treatment eliminated the activation of p53 DNA binding (Fig. 5, lanes 7 and 8). Third, and most significantly, DNA binding of p53 bearing the K(319/320/321)R substitutions was not potentiated by PCAF and acetyl Co-A (Fig. 5, compare lanes 11 and 12 to 13 and 14).
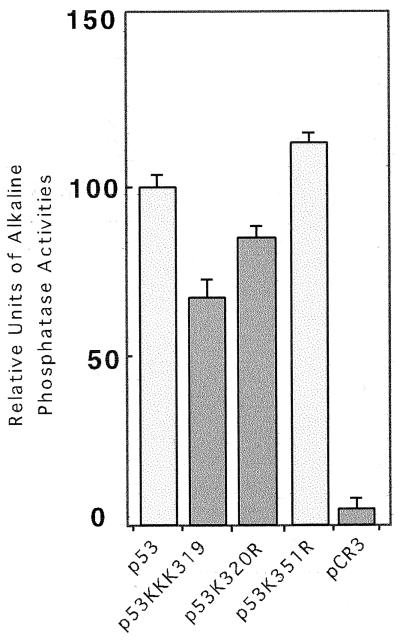
The effect of acetylation at K320 on p53’s DNA-binding activity in vitro suggested that transcriptional activation mediated by p53 may be altered by acetylation. To test this, wild-type p53 or p53 bearing substitutions at the PCAF-targeted acetylation site at K320 was cotransfected into Saos-2 cells with a p53-responsive reporter plasmid. The single substitution K320R exhibited a modest 15% lowering of transcriptional activity, and the triple substitution K(319/320/321)R was reduced 35% (Fig. 6). Thus, p53 bearing the triple substitution possesses lower activity than the single change, perhaps because the loss of the single K320 shifts acetylation to one of the adjacent lysine residues.
FIG. 6.
Transcriptional activity of p53 K320 substitutions in Saos-2 cells. Cells were transfected with 0.1 μg of p53 wild type or mutant expression plasmid and 29 μg of pEp21-TK-SEAP. The level of activity of each p53 construct was compared to that of wild-type p53.
Acetylation of K320 and K373 in p53 increases in vivo by UV or ionizing radiation treatment.
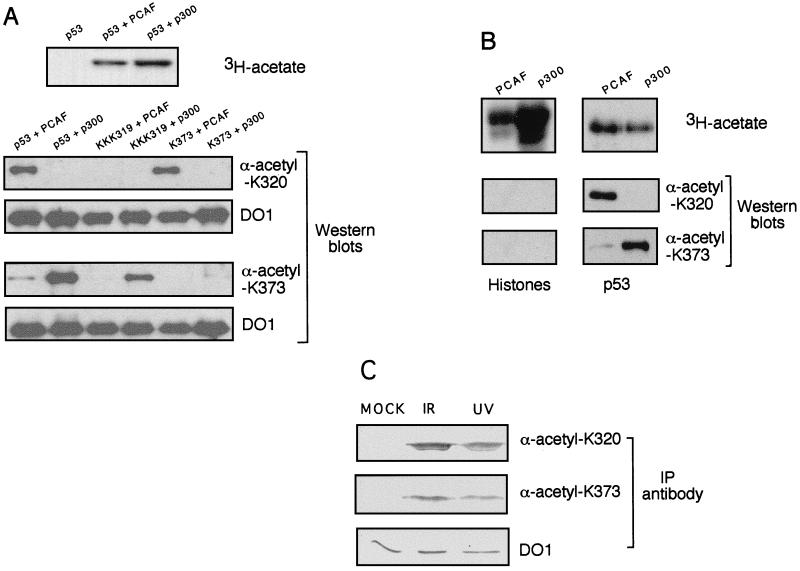
Our observations that PCAF specifically acetylates p53 at K320, that this acetylation stimulates p53’s DNA binding in vitro, and that alteration of this residue partially lowers p53 transcriptional activity in vivo raised the question of whether the acetylation occurs in vivo, and if so, whether it is regulated by previously known physiological stimuli of p53, such as DNA damage. To determine the acetylation state of p53 in vivo, we generated acetylation- and site-specific anti-p53 antibodies and used them to probe the acetylation status of p53. Peptides were synthesized comprising 13 amino acids, with the central residue containing either acetylated K320 or K373. These two peptides were then used as antigens to generate polyclonal antisera. The specificities of these antisera were determined by Western blot analysis of wild-type or substitution mutants of p53 that had been acetylated by either PCAF or p300 (Fig. 7A). As a first level of specificity, neither antibody detected unacetylated p53 (data not shown). Secondly, the anti-acetyl-K320 (α-ac-K320) antibody detected p53 acetylated by PCAF but not by p300, while the α-ac-K373 antibody behaved in the opposite fashion, detecting p53 acetylated by p300 but not by PCAF (Fig. 7A). Thirdly, detection of p53 by the α-ac-K320 was lost in the K(319/320/321)R substitution mutant, and detection by the α-ac-K373 was lost in the K373R mutant (Fig. 7A). Finally, neither antibody recognized core histones acetylated by PCAF (which acetylates primarily H3) or p300 (which acetylates all four core histones) (Fig. 7B). These results show that the α-ac-K320 and α-ac-K373 antibodies are highly specific for p53 acetylation patterns generated either by PCAF or by p300 in vitro.
FIG. 7.
Acetylation of p53 in response to DNA-damaging agents. (A) Specificity of antibodies raised to p53 peptides encoding acetylated lysines 320 or 373. Recombinant p53 was incubated with either PCAF or p300 in the presence of [3H]acetyl-CoA. A sample of each reaction was subjected to SDS-PAGE and analyzed by autoradiography to determine the level of acetylation (upper panel). Samples of each reaction were then tested by Western analysis with the anti-K320-acetyl antibody (lower left) and anti-K373-acetyl antibody (lower right). The Western blot membranes were then reprobed with the anti-p53 monoclonal antibody DO1 to compare the amounts of p53 proteins used in each reaction. (B) Specificity of antibodies raised to acetylated p53 peptides against acetylated core histones. Equal amounts of free core histones or p53 were acetylated by PCAF or p300. Samples were treated the same way as described for panel A. (C) Effect of DNA-damaging agents on the acetylation state of p53. Whole cell extracts were prepared from untreated cells or cells treated with either ionizing radiation (IR) or UV radiation. Extracts were immunoprecipitated (IP) with anti-ac-K320, anti-ac-K373, or anti-p53 monoclonal DO1 antibody. The amount of p53 in each immunoprecipitate was then analyzed by Western blotting with DO1 antibody.
Using the acetyl-specific antibodies, we then investigated the acetylation status of p53 in vivo. U20S osteosarcoma cells, which express wild-type p53, were untreated or exposed to either UV or ionizing radiation, since these DNA-damaging agents are known to induce p53 function in vivo (39). Because the amount of p53 increases in response to DNA damage, we first immunoprecipitated p53 with the DO1 monoclonal antibody to determine the amount of cell extract that contains an equivalent amount of p53 (Fig. 7C). Immunoprecipitation with the α-ac-K320 or α-ac-K373 antibodies revealed a very low amount of K320- or K373-acetylated p53 protein in mock-irradiated cells. However, following irradiation, both antibodies detected an increased amount of acetylated p53 protein (Fig. 7C, lower panel). Thus, p53 acetylation is induced at both K320 and K373 after DNA damage.
DISCUSSION
In this study we have identified a novel function for the acetyltransferase activity of the Gcn5 family member human PCAF. In addition to utilizing histones as a substrate, PCAF acetylates the transcription factor and tumor suppressor p53. The acetylation site in p53 is highly specific and, importantly, is distinct from the previously characterized sites acetylated by p300. The consequence of the acetylation by PCAF is greatly stimulated DNA binding by p53 in vitro. Most significant is that this study is the first to demonstrate that target lysines within a nonhistone substrate exhibit increased acetylation under physiological conditions that stimulate function. In particular, under in vivo conditions that cause DNA damage, which potentiate p53 activity, increased p53 acetylation is detected at lysines acetylated in vitro by PCAF and p300.
Previous studies have shown that CBP/p300 and PCAF form a coactivator complex to facilitate gene transcription by specific transcriptional activators (32). In particular, we have shown that CBP and PCAF function synergistically to activate p53-dependent transcription of the endogenous p21/waf1/cip1 gene (52). The finding that CBP and PCAF both acetylate histones in vitro has raised the question of why multiple acetyltransferases function in complexes to potentiate transcription of target genes. One possibility, suggested by the fact that each coactivator has distinct histone target specificity (27, 45), is that they acetylate distinct sites within nucleosomes to achieve a synergistic effect on chromatin remodeling, as previously suggested (45). Our results show that PCAF and CBP have clearly distinct specificities for acetylation of p53 and, hence, the potential for synergistic activation of p53 as well. Thus, at one level, CBP and PCAF acetylation of p53 enhances its binding affinity for promoters of target genes. At a second level, once bound to target genes, p53 recruitment of the CBP-PCAF coactivator complex acetylates nucleosomal histones, thereby promoting access to DNA of RNA polymerase II and other basal transcription factors. Another possible explanation for distinct acetylation by the different enzymes is that it allows p53 to respond to different activating signals.
We have compared the primary sequence of the PCAF-dependent acetylation site at K320 in p53 to the previously-determined yeast Gcn5-dependent acetylation sites in core histones H3 and H4 (32a). Interestingly, there is no apparent similarity between the sequences flanking K320 and the consensus histone acetylation site, suggesting great flexibility in the interaction of different acetylation substrates with the catalytic domain of the Gcn5 family. Structural studies of acetylation domains complexed with different substrates may reveal the basis of this sequence flexibility in the sites of acetylation.
The effects of p300 (22) and PCAF acetylation (Fig. 5) on p53 DNA binding in vitro are clear. In support of a similar effect in vivo, we observed a modest reduction in transcriptional activity of p53 in transfection assays, using the K320R single substitution or the K(319/320/321)R triple substitution. We also tested substitution at K373 and found that arginine at this position exhibited less than a 10% reduction in transcription activity, while alanine showed a decrease of approximately 65% lowering (52a). In general, these effects are qualitatively similar to reported results on mutation of other sites of posttranslational modification of p53. For example, phosphorylation at S15, S315, and S392 increases after irradiation, but substitutions at these residues have either a partial effect or no effect on p53 function, respectively (18, 19, 29, 53, 56). There are several explanations for these rather modest effects. First, p300 and PCAF are likely to both acetylate histones and p53 to activate p53-responsive genes, so mutation of the acetylation sites within p53 may lower p53-mediated activation only partially. Second, the amount of exogenous p53 introduced by transfection may exceed the regulatory capacity of cells. Third, as mentioned above, there is evidence of several or many functionally redundant posttranslational modifications within the regulatory region and the activation domains of p53. In further support of physiological significance, we have screened a database of p53 gene mutations found in human tumors and cell lines (maintained by the International Agency for Research on Cancer; www.iarc.fr/p53/homepage.htm) to determine the frequency of substitution mutations in RD1 and RD2. Among the reported potential regulatory sites (serine 315 [56], serines 376 and 378 [29], lysines 373, 381, and 382 [22], serine 389 [44], and serine 392 [44]), substitutions are found only at lysine 320.
Abundant evidence indicates that p53’s ability to bind to DNA is tightly linked to its physiological functions in tumor suppression (35). p53 is a tetramer (25) and is postulated to assume two dynamic states, a high-affinity state and a low-affinity state for DNA binding (24, 25). Activation of p53 is thought to require a conformational switch from the low-to-high affinity states. In addition, both C-terminal regulatory domains (RD1 and RD2) negatively regulate p53’s DNA-binding activity in vitro, perhaps by maintaining p53 in the low-affinity state (25, 26, 58). These results raise the possibility that RD1 and RD2 are targeted by agents, such as DNA damage, that regulate p53 function in vivo. Indeed, ionizing radiation leads to a specific dephosphorylation at serine 376 within RD2, which in turn leads to association of p53 with 14-3-3 proteins and enhanced sequence-specific DNA-binding activity (29). In addition, phosphorylation of serine 392 within RD2, which activates p53 DNA binding in vitro (26), has been observed in response to UV irradiation in vivo (18, 44).
The findings reported here and similar results from other laboratories (1) suggest that RD1 and RD2 are also targeted by acetyltransferases in vivo. Lysine 320 within RD1 and lysine 373 within RD2 become acetylated after exposure of cells to UV or ionizing radiation. In vitro, the acetylation site at K320 is targeted by PCAF and the acetylation at K373 is targeted by p300, and both p300 and PCAF increase the affinity of p53 to bind its cognate DNA site. This suggests that there are multiple and perhaps redundant pathways to increase p53’s capacity for DNA binding in response to DNA damage. It will be important to establish the conditions under which each of these pathways contributes to the p53 DNA damage response and whether these modifications can occur independently of each other or only in a specific sequence.
ACKNOWLEDGMENTS
We gratefully acknowledge the gifts of GST-PCAF and GST-p300 protein and expression plasmids from G. Blobel and E. Verdin, respectively. We thank H.-X. Zhang for assistance preparing GST-p53 fusion proteins, G. Simon for help using the p53 mutation database, and G. Moore, B. Lu, and members of the Berger lab for valuable discussions.
D.M.S. was supported by National Institutes of Health training grant to the Wistar Institute; R.C.T. was supported by a Howard Hughes Medical Institute predoctoral fellowship. The research was supported by grants from the National Cancer Institutes and the American Cancer Society (to T.D.H.) and from the National Institutes of General Medical Sciences, the National Science Foundation, the Council for Tobacco Research and the American Cancer Society (to S.L.B.).
REFERENCES
- 1.Anderson, C. Personal communication.
- 1a.Arany Z, Sellers W R, Livingston D M, Eckner R. E1A-associated p300 and CREB-associated CBP belong to a conserved family of coactivators. Cell. 1994;77:799–800. doi: 10.1016/0092-8674(94)90127-9. [DOI] [PubMed] [Google Scholar]
- 2.Arias J, Alberts A S, Brindle P, Claret F X, Smeal T, Karin M, Feramisco J, Montminy M. Activation of cAMP and mitogen responsive genes relies on a common nuclear factor. Nature. 1994;370:226–229. doi: 10.1038/370226a0. [DOI] [PubMed] [Google Scholar]
- 3.Avantaggiati M L, Ogryzko V, Gardner K, Giordano A, Levine A S, Kelly K. Recruitment of p300/CBP in p53-dependent signal pathways. Cell. 1997;89:1175–1184. doi: 10.1016/s0092-8674(00)80304-9. [DOI] [PubMed] [Google Scholar]
- 4.Bannister A, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 5.Bannister A J, Kouzarides T. CBP-induced stimulation of c-Fos activity is abrogated by E1A. EMBO J. 1995;14:4758–4762. doi: 10.1002/j.1460-2075.1995.tb00157.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barak Y, Juven T, Haffner R, Oren M. Mdm2 expression is induced by wild type p53 activity. EMBO J. 1993;12:461–468. doi: 10.1002/j.1460-2075.1993.tb05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bargonetti J, Manfredi J J, Chen X, Marshak D R, Prives C. A proteolytic fragment from the central region of p53 has marked sequence-specific DNA-binding activity when generated from wild-type but not from oncogenic mutant p53 protein. Genes Dev. 1993;7:2565–2574. doi: 10.1101/gad.7.12b.2565. [DOI] [PubMed] [Google Scholar]
- 8.Barlev N, Candau R, Wang L, Darpino P, Silverman N, Berger S. Characterization of physical interactions of the putative transcriptional adaptor, ADA2, with acidic activation domains and TATA-binding protein. J Biol Chem. 1995;270:19337–19344. doi: 10.1074/jbc.270.33.19337. [DOI] [PubMed] [Google Scholar]
- 9.Bhattacharya S, Eckner R, Grossman S, Oldread E, Arany Z, D’Andrea A, Livingston D. Cooperation of Stat2 and p300/CBP in signalling induced by interferon-alpha. Nature. 1996;383:344–347. doi: 10.1038/383344a0. [DOI] [PubMed] [Google Scholar]
- 10.Brownell J, Zhou J, Ranalli T, Kobayashi R, Edmondson D, Roth S, Allis C D. Tetrahymena histone acetyltransferase A: a transcriptional co-activator linking gene expression to histone acetylation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 11.Buckbinder L, Talbott R, Velasco-Miguel S, Takenaka I, Faha B, Seizinger B R, Kley N. Induction of the growth inhibitor IGF-binding protein 3 by p53. Nature. 1995;377:646–649. doi: 10.1038/377646a0. [DOI] [PubMed] [Google Scholar]
- 12.Candau R, Scolnick D M, Darpino P, Ying C Y, Halazonetis T, Berger S L. Two tandem and independent sub-activation domains in the amino terminus of p53 require the adaptor complex for activity. Oncogene. 1997;15:807–816. doi: 10.1038/sj.onc.1201244. [DOI] [PubMed] [Google Scholar]
- 13.Candau R, Zhou J, Allis C D, Berger S L. Histone acetyltransferase activity and interaction with ADA2 are critical for GCN5 function in vivo. EMBO J. 1997;16:555–565. doi: 10.1093/emboj/16.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clore G M, Omichinski J G, Sakaguchi K, Zambrano N, Sakamoto H, Appella E, Gronenborn A M. High-resolution structure of the oligomerization domain of p53 by multidimensional NMR. Science. 1994;265:386–391. doi: 10.1126/science.8023159. [DOI] [PubMed] [Google Scholar]
- 15.Dai P, Akimaru H, Tanaka Y, Hou D, Yasukawa T, Kanei-Ishii C, Takahashi T, Ishii S. CBP as a transcriptional coactivator of c-Myb. Genes Dev. 1996;10:528–540. doi: 10.1101/gad.10.5.528. [DOI] [PubMed] [Google Scholar]
- 16.el Deiry W, Tokino T, Velculescu V E, Levy D B, Parsons R, Trent J M, Lin D, Mercer W E, Kinzler K W, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 17.Fields S, Jang S K. Presence of a potent transcription activating sequence in the p53 protein. Science. 1990;249:1046–1049. doi: 10.1126/science.2144363. [DOI] [PubMed] [Google Scholar]
- 18.Fiscella M, Ullrich S J, Zambrano N, Shields M T, Lin D, Lees-Miller S P, Anderson C W, Mercer W E, Appella E. Mutation of the serine 15 phosphorylation site of human p53 reduces the ability of p53 to inhibit cell cycle progression. Oncogene. 1993;8:1519–1528. [PubMed] [Google Scholar]
- 19.Fiscella M, Zambrano N, Ullrich S J, Unger T, Lin D, Cho B, Mercer W E, Anderson C W, Appella E. The carboxy-terminal serine 392 phosphorylation site of human p53 is not required for wild-type activities. Oncogene. 1994;9:3249–3257. [PubMed] [Google Scholar]
- 20.Funk W D, Pak D T, Karas R H, Wright W E, Shay J W. A transcriptionally active DNA-binding site for human p53 protein complexes. Mol Cell Biol. 1992;12:2866–2871. doi: 10.1128/mcb.12.6.2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gottlieb T, Oren M. p53 in growth control and neoplasia. Biochim Biophys Acta. 1996;1287:77–102. doi: 10.1016/0304-419x(95)00019-c. [DOI] [PubMed] [Google Scholar]
- 22.Gu W, Roeder R G. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 23.Gu W, Shi X L, Roeder R G. Synergistic activation of transcription by CBP and p53. Nature. 1997;387:819–823. doi: 10.1038/42972. [DOI] [PubMed] [Google Scholar]
- 24.Halazonetis T D, Davis L J, Kandil A N. Wild-type p53 adopts a ’mutant’-like conformation when bound to DNA. EMBO J. 1993;12:1021–1028. doi: 10.1002/j.1460-2075.1993.tb05743.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Halazonetis T D, Kandil A N. Conformational shifts propagate from the oligomerization domain of p53 to its tetrameric DNA binding domain and restore DNA binding to select p53 mutants. EMBO J. 1993;12:5057–5064. doi: 10.1002/j.1460-2075.1993.tb06199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hupp T, Meek D, Midgley C, Lane D. Regulation of the specific DNA binding function of p53. Cell. 1992;71:875–886. doi: 10.1016/0092-8674(92)90562-q. [DOI] [PubMed] [Google Scholar]
- 27.Imhof A, Yang X, Ogryzko V, Nakatani Y, Wolffe A, Ge H. Acetylation of general transcription factors by histone acetyltransferases. Curr Biol. 1997;7:689–692. doi: 10.1016/s0960-9822(06)00296-x. [DOI] [PubMed] [Google Scholar]
- 28.Jeffrey P D, Gorina S, Pavletich N P. Crystal structure of the tetramerization domain of the p53 tumor suppressor at 1.7 angstroms. Science. 1995;267:1498–1502. doi: 10.1126/science.7878469. [DOI] [PubMed] [Google Scholar]
- 29.Kapoor M, Lozano G. Functional activation of p53 via phosphorylation following DNA damage by UV but not gamma radiation. Proc Natl Acad Sci USA. 1998;95:2834–2837. doi: 10.1073/pnas.95.6.2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kastan M B, Zhan Q, el Deiry W, Carrier F, Jacks T, Walsh W V, Plunkett B S, Vogelstein B, Fornace A J. A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell. 1992;71:587–597. doi: 10.1016/0092-8674(92)90593-2. [DOI] [PubMed] [Google Scholar]
- 31.Ko L J, Prives C. p53: puzzle and paradigm. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 32.Korzus E, Torchia J, Rose D, Xu L, Kurokawa R, McInerney E, Mullen T, Glass C, Rosenfeld M. Transcription factor-specific requirements for coactivators and their acetyltransferase functions. Science. 1998;279:703–707. doi: 10.1126/science.279.5351.703. [DOI] [PubMed] [Google Scholar]
- 32a.Kuo M-H, Brownell J E, Ranalli T A, Cook R G, Edmonson D G, Roth S Y, Allis C D. Transcription-linked acetylation by Gen5p of histones H3 and H4 at specific lysines. Nature. 1996;383:269–272. doi: 10.1038/383269a0. [DOI] [PubMed] [Google Scholar]
- 33.Kwok R P, Lundblad J R, Chrivia J C, Richards J P, Bachinger H P, Brennan R G, Roberts S G, Green M R, Goodman R H. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature. 1994;370:223–226. doi: 10.1038/370223a0. [DOI] [PubMed] [Google Scholar]
- 34.Lee W, Harvey T S, Yin Y, Yau P, Litchfield D, Arrowsmith C H. Solution structure of the tetrameric minimum transforming domain of p53. Nat Struct Biol. 1994;1:877–890. doi: 10.1038/nsb1294-877. [DOI] [PubMed] [Google Scholar]
- 35.Levine A J. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 36.Lill N L, Grossman S R, Ginsberg D, DeCaprio J, Livingston D M. Binding and modulation of p53 by p300/CBP coactivators. Nature. 1997;387:823–827. doi: 10.1038/42981. [DOI] [PubMed] [Google Scholar]
- 37.Luger K, Mader A, Richmond R, Sargent D, Richmond T. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 38.Lundblad J, Kwok R, Laurance M, Harter M, Goodman R. Adenoviral E1A-associated protein p300 as a functional homologue of the transcriptional coactivator CBP. Nature. 1995;374:85–88. doi: 10.1038/374085a0. [DOI] [PubMed] [Google Scholar]
- 39.Maltzman W, Czyzyk L. UV irradiation stimulates levels of p53 cellular tumor antigen in nontransformed mouse cells. Mol Cell Biol. 1984;4:1689–1694. doi: 10.1128/mcb.4.9.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miyashita T, Reed J C. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell. 1995;80:293–299. doi: 10.1016/0092-8674(95)90412-3. [DOI] [PubMed] [Google Scholar]
- 41.Mizzen C, Allis C. Linking histone acetylation to transcriptional regulation. Cell Mol Life Sci. 1998;54:6–20. doi: 10.1007/s000180050121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muller-Tiemann B, Halazonetis T, Elting J. Identification of an additional negative regulatory region for p53 sequence-specific DNA binding. Proc Natl Acad Sci USA. 1998;95:6079–6084. doi: 10.1073/pnas.95.11.6079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oelgeschlager M, Janknecht R, Krieg J, Schreek S, Luscher B. Interaction of the co-activator CBP with Myb proteins: effects on Myb-specific transactivation and on the cooperativity with NF-M. EMBO J. 1996;15:2771–2780. [PMC free article] [PubMed] [Google Scholar]
- 44.Ogryzko V, Kotani T, Zhang X, Schlitz R, Howard T, Yang X, Howard B, Qin J, Nakatani Y. Histone-like TAFs within the PCAF histone acetylase complex. Cell. 1998;94:35–44. doi: 10.1016/s0092-8674(00)81219-2. [DOI] [PubMed] [Google Scholar]
- 45.Ogryzko V, Schlitz R, Russanova V, Howard B, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 46.Okamoto K, Beach D. Cyclin G is a transcriptional target of the p53 tumor suppressor protein. EMBO J. 1994;13:4816–4822. doi: 10.1002/j.1460-2075.1994.tb06807.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pietenpol J A, Tokino T, Thiagalingam S, el Deiry W, Kinzler K W, Vogelstein B. Sequence-specific transcriptional activation is essential for growth suppression by p53. Proc Natl Acad Sci USA. 1994;91:1998–2002. doi: 10.1073/pnas.91.6.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Puri P L, Sartorelli V, Yang X J, Hamamori Y, Ogryzko V V, Howard B H, Kedes L, Wang J Y, Graessmann A, Nakatani Y, Levrero M. Differential roles of p300 and PCAF acetyltransferases in muscle differentiation. Mol Cell. 1997;1:35–45. doi: 10.1016/s1097-2765(00)80005-2. [DOI] [PubMed] [Google Scholar]
- 49.Raycroft L, Wu H Y, Lozano G. Transcriptional activation by wild-type but not transforming mutants of the p53 anti-oncogene. Science. 1990;249:1049–1051. doi: 10.1126/science.2144364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sakamoto H, Lewis M S, Kodama H, Apella E, Sakaguchi K. Specific sequences from the carboxyl terminus of human p53 gene product form anti-parallel tetramers in solution. Proc Natl Acad Sci USA. 1994;91:8974–8978. doi: 10.1073/pnas.91.19.8974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scharer E, Iggo R. Mammalian p53 can function as a transcription factor in yeast. Nucleic Acids Res. 1992;20:1539–1545. doi: 10.1093/nar/20.7.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Scolnick D, Chehab N, Stavridi E, Lien M, Caruso L, Moran E, Berger S, Halazonetis T. CREB-binding protein and p300/CBP-associated factor are transcriptional coactivators of the p53 tumor suppressor protein. Cancer Res. 1997;57:3693–3696. [PubMed] [Google Scholar]
- 52a.Scolnick, D. M., and L. Liu. Unpublished data.
- 53.Shieh S Y, Ikeda M, Taya Y, Prives C. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell. 1997;91:325–334. doi: 10.1016/s0092-8674(00)80416-x. [DOI] [PubMed] [Google Scholar]
- 54.Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 55.Sturzbecher H W, Brain R, Addison C, Rudge K, Remm M, Grimaldi M, Keenan E, Jenkins J R. A C-terminal α-helix plus basic region motif is the major structural determinant of p53 tetramerization. Oncogene. 1992;7:1513–1523. [PubMed] [Google Scholar]
- 56.Wang Y, Prives C. Increased and altered DNA binding of human p53 by S and G2/M but not G1 cyclin-dependent kinases. Nature. 1995;376:88–91. doi: 10.1038/376088a0. [DOI] [PubMed] [Google Scholar]
- 57.Wang Y, Reed M, Wang P, Stenger J E, Mayr G, Anderson M E, Schwedes J F, Tegtmeyer P. p53 domains: identification and characterization of two autonomous DNA-binding regions. Genes Dev. 1993;7:2575–2586. doi: 10.1101/gad.7.12b.2575. [DOI] [PubMed] [Google Scholar]
- 58.Waterman J, Shenk J, Halazonetis T. The dihedral symmetry of the p53 tetramerization domain mandates a conformational switch upon DNA binding. EMBO J. 1995;14:512–519. doi: 10.1002/j.1460-2075.1995.tb07027.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Waterman M, Waterman J, Halazonetis T. An engineered four-stranded coiled coil substitutes for the tetramerization domain of wild type p53 and alleviates transdominant inhibition by tumor-derived p53 mutants. Cancer Res. 1996;56:158–163. [PubMed] [Google Scholar]
- 60.Wu X, Bayle J H, Olson D, Levine A J. The p53-mdm-2 autoregulatory feedback loop. Genes Dev. 1993;7:1126–1132. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]
- 61.Yang X, Ogryzko V, Nishikawa J, Howard B, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral E1A oncoprotein. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 62.Yuan W, Condorelli G, Caruso M, Felsani A, Giordano A. Human p300 protein is a coactivator for the transcription factor MyoD. J Biol Chem. 1996;271:9003–9013. doi: 10.1074/jbc.271.15.9009. [DOI] [PubMed] [Google Scholar]
- 63.Zhang J, Vinkemeier U, Gu W, Chakravarti D, Horvath C, Darnell J J. Two contact regions between Stat1 and CBP/p300 in interferon gamma signaling. Proc Natl Acad Sci USA. 1996;93:15092–15096. doi: 10.1073/pnas.93.26.15092. [DOI] [PMC free article] [PubMed] [Google Scholar]