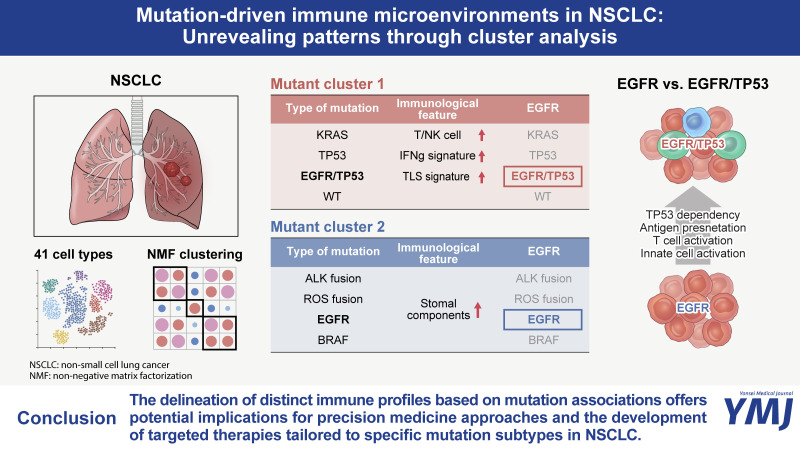
Abstract
Purpose
We aimed to comprehensively analyze the immune cell and stromal components of tumor microenvironment at the single-cell level and identify tumor heterogeneity among the major top-derived oncogene mutations in non-small cell lung cancer (NSCLC) using single-cell RNA sequencing (scRNA-seq) data.
Materials and Methods
The scRNA-seq dataset utilized in this study comprised 64369 primary tumor tissue cells from 21 NSCLC patients, focusing on mutations in EGFR, ALK, BRAF, KRAS, TP53, and the wild-type.
Results
Tumor immune microenvironment (TIM) analysis revealed differential immune responses across NSCLC mutation subtypes. TIM analysis revealed different immune responses across the mutation subtypes. Two mutation clusters emerged: KRAS, TP53, and EGFR+TP53 mutations (MC1); and EGFR, BRAF, and ALK mutations (MC2). MC1 showed higher tertiary lymphoid structures signature scores and enriched populations of C2-T-IL7R, C3-T/NK-CXCL4, C9-T/NK-NKG, and C1-B-MS4A1 clusters than cluster 2. Conversely, MC2 cells exhibited higher expression levels of TNF, IL1B, and chemokines linked to alternative immune pathways. Remarkably, co-occurring EGFR and TP53 mutations were grouped as MC1. EGFR+TP53 mutations showed upregulation of peptide synthesis and higher synthetic processes, as well as differences in myeloid and T/NK cells compared to EGFR mutations. In T/NK cells, EGFR+TP53 mutations showed a higher expression of features related to cell activity and differentiation, whereas EGFR mutations showed the opposite.
Conclusion
Our research indicates a close association between mutation types and tumor microenvironment in NSCLC, offering insights into personalized approaches for cancer diagnosis and treatment.
Keywords: Non-small cell lung cancer, ScRNA-seq, mutation, EGFR gene, TP53 gene
Graphical Abstract
INTRODUCTION
Lung cancer is a leading cause of cancer-related mortality worldwide.1,2 Non-small cell lung cancer (NSCLC) accounts for approximately 80% of all lung cancer cases.3 The advent of immune checkpoint inhibitors (ICIs) therapy has significantly improved the survival of NSCLC patients. However, only a subset of patients responds to ICI.4 Moreover, despite the initial response, majority of patients eventually develop disease progression or acquire resistance.5
NSCLC is characterized by a complex genetic landscape in which various somatic mutations significantly contribute to the initiation, progression, and response to therapy.6,7,8 Among these key genetic alterations are mutations in the epidermal growth factor receptor (EGFR), KRAS, and BRAF, and the translocation of ROS proto-oncogene 1 (ROS1) and anaplastic lymphoma kinase (ALK).9,10,11 STK11/LKB1 co-mutations can drive intrinsic resistance to ICI in KRAS mutant lung adenocarcinoma by decreasing PD-L1 expression and CD8+ T-cells infiltration in tumor microenvironment.12 In addition, NSCLC patients harboring genomic alterations in EGFR, ALK, and ROS1 exhibit extremely low response to ICI.13 Therefore, understanding the influence of key generic alterations on the tumor immune microenvironment and response to ICI is essential for the development of personalized immunotherapy.
Several studies have used single cell RNA-sequencing (scRNA-seq) to analyze mutations in NSCLC. However, as mentioned earlier, these studies focused on specific mutations or groups of mutations, rather than providing a comprehensive analysis of various NSCLC mutations.14,15,16 While it is feasible to merge multiple NSCLC scRNA-seq datasets to conduct a comprehensive study on mutations, the integration of scRNA-seq data presents challenges. Ensuring consistency across diverse datasets and seamlessly integrating them can pose challenges.17 To conduct such comprehensive studies, it is essential to explore various data sources and integrate and refine vast amounts of data.18 This process provides a crucial foundation for understanding the interactions between mutations and patient diversity. Therefore, exploring various databases to analyze the differences among NSCLC mutations comprehensively, we discovered a dataset that integrated 1283972 single cells from 556 samples and 318 patients across 29 datasets.19 We selected data from 48 patients, including EGFR, ALK, ROS1, BRAF, KRAS, and TP53 mutation information, as well as “not mutated” data. After performing data quality control (QC) , we selected scRNA-seq data from primary tumor samples of 21 of the 48 patients.
In this study, we meticulously analyzed tumor heterogeneity among mutations in primary NSCLC tumors. Specifically, we investigated the associations between various mutations and divided them into two groups based on their associations. Subsequently, we examined these groups’ differences, revealing different immune cell heterogeneities. Through this study, we present a multifaceted exploration of the interplay among mutations in patients with NSCLC and delineate distinct immune profiles based on mutation associations. Delineating the heterogeneous landscape of co-mutations, such as TP53, is important in EGFR mutant NSCLC. Through our investigation, we aim to uncover distinct characteristics associated with specific mutation profiles in order to facilitate the development of tailored treatment strategies for individual patients. By delving into these complexities, our purpose is to deepen our understanding of NSCLC biology and lay the groundwork for more personalized treatment approaches.
MATERIALS AND METHODS
scRNA-seq datasets
To obtain the scRNA-seq database, we utilized NSCLC data from the CZ Cell Gene Resource.20 This database, which incorporates clinical information, consists of 1283972 cells obtained from 318 patients with NSCLC, contains 17811 gene features, and serves as the basis for the analysis in this study.19 This database contains several public databases, among which we selected the one that contained mutation information. Mutation data were obtained and used by obtaining GSE123904 from the Gene Expression Omnibus database, CRA001963 from the National Genomic Data Center Genome Sequence Archive database, and PRJNA591860 from the Sequence Read Archive database.15,21,22 The data included information from 48 NSCLC patients, and after data QC, 133994 cells were used for analysis. Of these, 64369 cells from the primary tumor sample were used in the analysis.
QC and data processing
Our study initially filtered cells with unique feature counts of more than 5000. Subsequently, we normalized the dataset using the Seurat “NormalizedData” function, employing the global-scaling normalization method known as “LogNormalize,” and scaled it by a default scale factor of 10000. We then further scaled the data by applying the Seurat “ScaleData” function, considering the variation in “nCount_RNA” and “percent.mt.” We utilized the Seurat “ElbowPlot” function to determine the optimal dimensionality. Dimensionality reduction analysis was conducted using the Seurat “RunPCA” function and non-linear dimensional reduction with the Seurat “RunUMAP” function.
Software and statistical analysis
In this study, we employed the R programming language for data analysis, predominantly utilizing the Seurat package for preprocessing, visualization, and clustering of scRNA-seq. Alongside Seurat, we utilized various other R packages for analysis, including Seurat (version: 5.0.1), matrix (version: 1.6.5), dplyr (version: 1.0.8), patchwork (version: 1.2.0), ggplot2 (version: 3.4.4), ggpubr (version: 0.4.0), dittoSeq (version: 1.14.1), corrplot (version: 0.92), stats (version: 4.3.2), NMF (version: 0.27) , CellChat (version: 1.6.1), and EnhancedVolcano (version: 1.20.0). We used Student’s t-test to assess statistical significance, with p-values of 0.05. The findings that met this criterion were considered statistically significant. All data analyses were performed using R software (version 4.3.2) and Prism 10.0 software (GraphPad Software, Inc.; San Diego, CA, USA). Pearson’s correlation analysis was used for correlation analysis. Statistical significance analyses were performed using Student’s t-test (*p<0.05, **p<0.01, ***p<0.001, ****p<0.0001).
RESULTS
Differential proportions of the 41 clusters obtained from primary tumor samples of 21 NSCLC patients were observed across mutations following cell type annotation
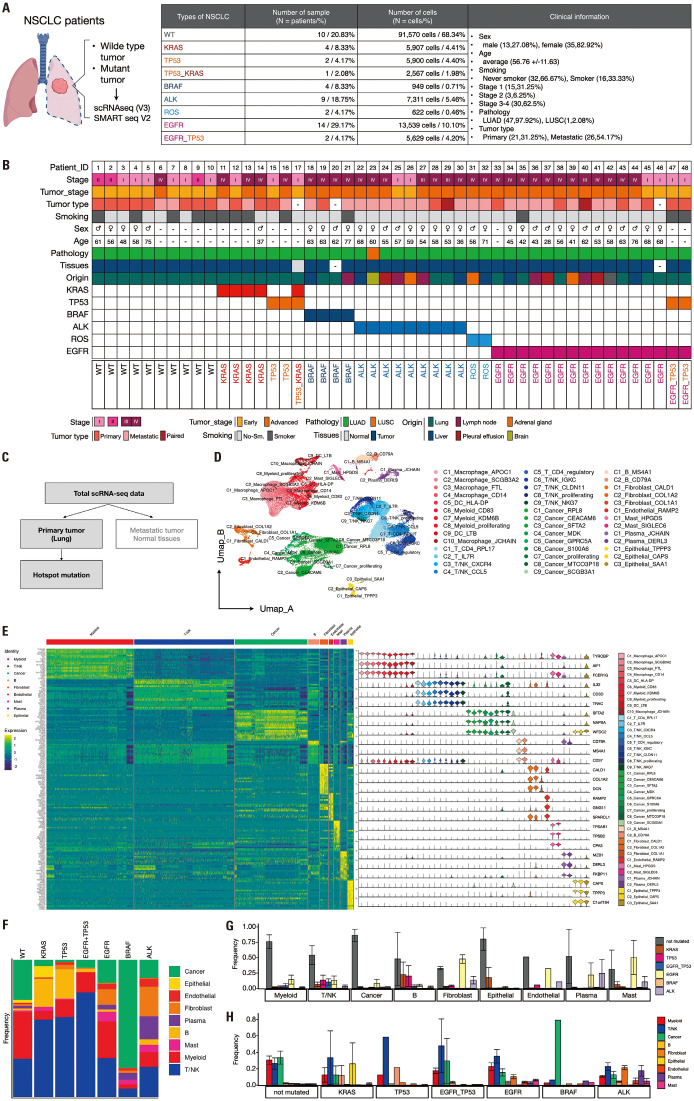
We utilized single-cell RNA sequencing (scRNA-seq) data from a cohort of 48 patients diagnosed with NSCLC. Our analysis incorporated 133994 cells, each containing information on somatic mutations. This dataset comprised detailed patient information, including demographic factors such as sex, age, and smoking history, as well as clinical parameters such as tumor stage, tumor type, pathology, tissue origin, and intricate mutation profiles for each individual (Fig. 1A and B). This extensive dataset offers a nuanced understanding of the molecular and cellular landscapes of NSCLC, enabling the exploration of the complex relationships between genetic alterations and various clinical parameters. We selected 21 of the 48 NSCLC patient datasets containing primary tumor data, totaling 64369 cells (Fig. 1C). Patients with TP53+KRAS and ROS1 mutations were excluded from the study. Subsequently, we analyzed primary data from NSCLC patients with mutations, including KRAS, TP53, EGFR, EGFR+TP53, BRAF, ALK, and wild-type (WT). The scRNA-seq dataset was preprocessed for further analysis. Initially, we applied log normalization, computed feature variances using the VST method, performed data scaling, and conducted a principal component analysis (PCA). Subsequently, we visualized an elbow plot to determine the cutoff point. This allowed us to proceed with the nearest neighbor calculation from one to eight dimensions, clustering with a resolution of 0.5, and dimensionality reduction using PCA. A total of 41 distinct clusters were identified within the dataset and visualized using a Uniform Manifold Approximation and Projection (UMAP) plot (Supplementary Fig. 1A, only online). The frequency of these clusters was examined according to the mutation type. KRAS, TP53, and EGFR+TP53 mutations exhibited similar cluster frequencies, whereas EGFR, ALK, and BRAF mutations exhibited different cluster frequencies (Supplementary Fig. 1B, only online). We examined the composition ratio of the predicted cell types and the top genes for each of the 41 clusters in the database. Based on this analysis, we assigned the cell types to each cluster as follows: myeloid, T/NK, cancer, B, fibroblast, endothelial, epithelial, and mast cells (Supplementary Fig. 1C, D, and E, only online). Subsequently, we conducted type annotation by confirming the expression of marker genes within each cluster. Proliferating cells were annotated using MKI67 and TOP2A, macrophages with CD68, dendritic cells (DC) with CD1C and LAMP3, and Tregs with FOXP3 and IL2RA. Subtype annotation was performed using the top marker genes for each cluster (Fig. 1D and Supplementary Fig. 2, only online). To assess the similarity among clusters designated as the same cell type, we selected 30000 cells and generated heat maps based on the top 20 genes for each cell type (Fig. 1E and Supplementary Fig. 3A and B, only online). Furthermore, we examined the expression of the top three genes per cell type across the 41 clusters (Fig. 1E). Additionally, when examining heat maps for the top 10 genes per cluster, we observed similarities among clusters assigned to the same cell type (Supplementary Fig. 3C and D, only online), validating our cell type annotations. Subsequently, we investigated the composition ratio of the cell types per mutation (Fig. 1F). We observed that KRAS, TP53, and EGFR+TP53 mutations exhibited similar cell-type composition ratios to those of EGFR, BRAF, and ALK mutations. Interestingly, EGFR+TP53 mutations showed a different cell type composition ratio than EGFR mutations, which resembled TP53 mutations. Therefore, we further confirmed the differences and similarities in the cell type composition ratios based on mutations by examining the frequency of mutations per cell type (Fig. 1G) and the frequency of cell types per mutation (Fig. 1H). We then proceeded with additional research on the association between these mutations.
Fig. 1. Comprehensive analysis of scRNA-seq data from NSCLC patients. (A) Schematic representation of the scRNA-seq dataset comprising 133,994 cells from 48 NSCLC patients. Each cell contains detailed somatic mutation information. (B) The rich patient details encompassed in the database including demographic factors (gender, age, smoking history) and clinical parameters (tumor stage, tumor type, pathology, tissue origin). The dataset provides intricate mutation profiles for individuals, facilitating a nuanced understanding of the molecular and cellular landscape within NSCLC and enabling the exploration of complex relationships between genetic alterations and clinical parameters. (C) Selection process of NSCLC patient datasets and exclusion criteria for specific mutations. (D) Uniform Manifold Approximation and Projection (UMAP) plot of the primary tumor data from NSCLC patients according to cluster assigned with cell type. (E) Visualization of top markers by cell type, visualized using heatmaps and violin plots. (F) Stacked bar plot visualizing the composition of cell types based on the types of mutations. (G) Bar plot visualizing the frequency of each mutation per cell type. The x-axis represents different cell types, while the y-axis represents the frequency of mutations. (H) Bar plot visualizing the frequency of each cell type per mutation. The x-axis represents different mutations, while the y-axis represents the frequency of cell types. NSCLC, non-small cell lung cancer; scRNA-seq, single-cell RNA sequencing.
Samples with distinct genomic alterations segregated into two statistically significant clusters, each characterized by differential cellular composition
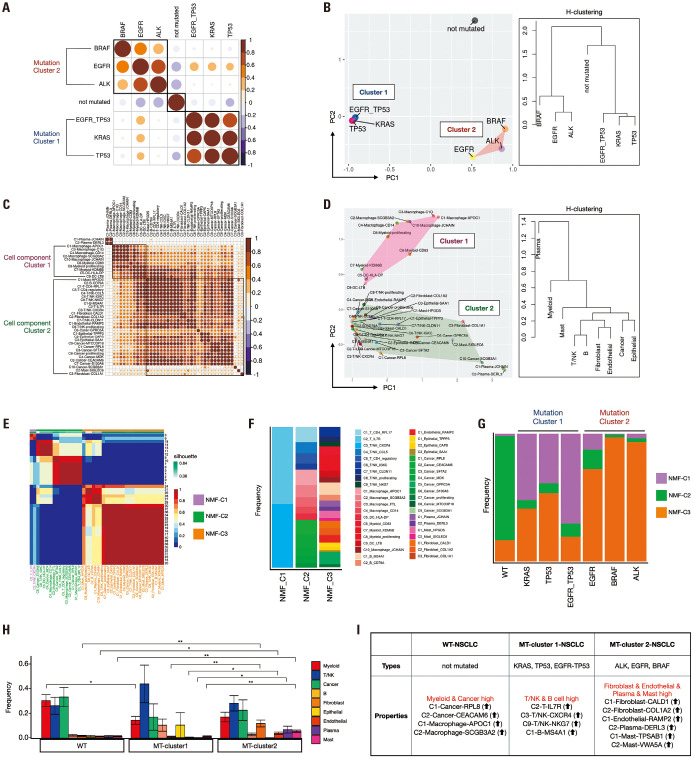
We investigated the correlations between these mutations and NSCLC. We evaluated the composition ratio of clusters per mutation and generated a correlation plot using hierarchical clustering (H-clustering) order. Notably, mutations were segregated into three groups: WT/KRAS, TP53, EGFR+TP53/EGFR, BRAF, and ALK. Therefore, we designated KRAS, TP53, and EGFR+TP53 mutations as mutation cluster 1 and EGFR, ALK, and BRAF mutations as mutation cluster 2 (Fig. 2A). This grouping was further supported by observations in the PCA plot, which revealed that the three identified groups exhibited similar characteristics (Fig. 2B). As seen earlier in the similarity of cell type composition ratios, this result confirmed that the WT, mutation cluster 1, and mutation cluster 2 were distinctly separated. We further investigated the correlation between the 41 cluster-assigned cell types using H-clustering. The 41 clusters were categorized into three groups: plasma, myeloid (cell component cluster 1), and the remaining cell types (cell component cluster 2) (Fig. 2C). Upon examining the features of each cluster using the PCA plot, we reaffirmed the resemblance in features among the clusters belonging to the plasma, myeloid, and remaining cell types (Fig. 2D and Supplementary Fig. 4, only online). We also performed non-negative matrix factorization (NMF) clustering using k-means (k-means=3) for 41 clusters. This analysis revealed three distinct NMF-clusters: C2-T-IL7R and C3-T/NK-CXCR4 in NMF-cluster 1; C1-T-CD4-RPL17, C4-T/NK-CCL5, C5-T-CD4-regulatory, C6-T/NK-IGKC, C1-Macrophage-APOC1, C2-Macrophage-SCGB3A2, C3-Macrophage-FTL, C4-Macrophage-CD14, C5-DC-HLA-DP, C1-Cancer-RPL8, C2-Cancer-CEACAM6, C3-Cancer-SFTA2, C4-Cancer-MDK, C6-Cancer-S100A6 in NMF-cluster 2; with the remaining clusters, C7-T/NK-CLDN11, C8-T/NK-proliferating, C9-T/NK-NKG7, C7-Myeloid-KDM6B, C8-Myeloid-proliferating, C9-DC-LTB, C10-Macrophage-JCHAIN, C8-Cancer-MTCO3P18, C9-Cancer-SCGB3A1, C1-B-MS4A1, C2-B-CD79A, C1-Endothelial-RAMP2, C1-Epithelial-TPPP3, C2-Epithelial-CAPS, C3-Epithelial-SAA1, C1-Plasma-JCHAIN, C2-Plasma-DERL3, C1-Mast-HPGDS, C2-Mast-SIGLEC6, C1-Fibroblast-CALD1, C2-Fibroblast-COL1A2, C3-Fibroblast-COL1A1, in NMF-cluster 3 (Fig. 2E and F). To investigate the distribution of NMF clusters according to the mutations, we examined the frequency of each NMF cluster permutation (Fig. 2G). Intriguingly, we observed similar patterns in the frequency distribution of NMF clusters across the three mutation groups. In the WT group, NMF-cluster 2 showed a higher frequency. At the same time, in mutation cluster 1 (KRAS, TP53, EGFR+TP53), NMF-cluster 1 exhibited a higher frequency, and in mutation cluster 2 (EGFR, BRAF, and ALK), NMF-cluster 3 displayed a higher frequency. We examined the frequency of different cell types per mutation cluster (Fig. 2H and Supplementary Fig. 5, only online). Based on these results, we could discern variations in the cellular composition within the mutation clusters. Myeloid and cancer cells in the WT showed higher frequencies, particularly C1-Cancer-RPL8, C2-Cancer-CEACAM6, C1-Macrophage-APOC1, and C2-Macrophage-SCGB3A2. In mutation cluster 1, the frequencies of T/NK and B were higher, whereas those of DCs/macrophages were lower, notably, with higher frequencies of C2-T-IL7R, C3-T/NK-CXCR4, C9-T/NK-NKG7, and C1-B-MS4A1. Conversely, fibroblasts, plasma, endothelial cells, and mast cells exhibited higher frequencies in mutation cluster 2 (Fig. 2I and Supplementary Fig. 5B, only online). Through these findings, we confirmed the similarity between specific mutations, such as KRAS, TP53, and EGFR+TP53, while also noting significant differences in features between EGFR and EGFR+TP53 mutations.
Fig. 2. Investigation of mutation correlations and cellular composition in NSCLC. (A) Correlation plot showing hierarchical clustering (H-clustering) order of mutations, segregating into three distinct groups. We designated mutation clusters 1 and 2 through this result. (B) Principal component analysis (PCA) plot and dendrogram using H-clustering confirming the grouping of mutations observed in (A), indicating similar features among the identified mutation groups. (C) The hierarchical clustering of the 21 clusters was based on the composition ratio of predicted cell types, categorizing clusters into three main groups. (D) PCA plot illustrating the resemblance in features among clusters belonging to plasma, DC/Macrophage, and the remaining cell types, as observed in (C). (E) Non-negative matrix factorization (NMF) clustering of the 41 clusters revealed three distinct NMF clusters. (F) Stacked bar plot visualizing the composition of 41 cluster assigned with cell type on each NMF-cluster. (G) Stacked bar plot visualizing the composition of NMF-clusters based on the types of mutations. (H) Bar plot visualizing the frequency of each cell type per mutation cluster. The x-axis represents different mutation clusters, while the y-axis represents the frequency of cell types. (I) Key results of cell proportions for the split clusters. Cluster 1 had a high proportion of T/NK and B cells, while cluster 2 had a high proportion of fibroblast, endothelial, plasma, and mast cells. NSCLC, non-small cell lung cancer; DC, dendritic cell.
Elevated ratios of C2-T-IL7R, C3-T/NK-CXCR4, and C9-T/NK-NKG7 clusters within the T/NK subset in mutation cluster 1 correlated with T cell migration and homeostasis
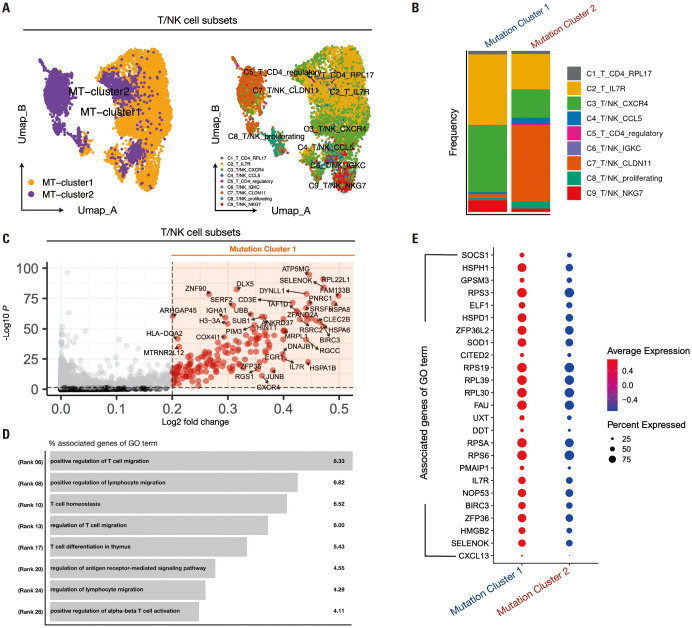
We previously investigated the frequency of cell types across mutation clusters (Fig. 2H and I). Differences in the composition ratio of cell types across mutation clusters were observed, suggesting potential effects on immune cell function. Therefore, we explored differences in the functionality of immune cells at varying frequencies across mutation clusters. Initially, to examine the differences in T cell and NK cell behavior between mutation clusters 1 and 2, we selected T/NK clusters along with mutation clusters 1 and 2. We conducted data preprocessing steps similar to those outlined in Fig. 2B. Nearest neighbors were computed up to the 15th dimension, followed by clustering with a resolution of 0.5. Subsequently, we visualized the distribution of the mutation clusters and previously obtained T/NK clusters using UMAP plots (Fig. 3A). Even when only T/NK cells were selected, mutation cluster 1 predominantly consisted of C2-T-IL7R, C3-T/NK-CXCR4, and C9-T/NK-NKG7 within the T/NK cluster composition ratio (Fig. 3B). Intriguing patterns emerged upon comparing cytokine expression between mutation clusters 1 and 2: IFNG exhibited higher expression in mutation cluster 1, whereas TNF, IL1B, IL2, and IL10 showed higher expression in mutation cluster 2 (Supplementary Fig. 6A, only online). Additionally, we examined chemokine expression in mutation clusters 1 and 2 and identified elevated expression of XCL1, CXCL13, and CCL3 in mutation cluster 1, whereas XCL2, CCL4, and CCL5 were highly expressed in mutation cluster 2 (Supplementary Fig. 6B, only online). Notably, CXCL13, highly expressed in T cells of mutation cluster 1, was identified as a signature gene for tertiary lymphoid structures (TLS). TLS are lymphoid structures that occur within tumors and are associated with B cell-mediated immune responses, which are known to affect ICI responses positively. Therefore, we compared the scores of the TLS signature genes between mutation clusters. The TLS signature score was significantly higher in cluster 1 than in cluster 2 (p<2.22e-16) (Supplementary Fig. 6E, only online). These findings highlight the distinct immune profiles associated with mutation clusters 1 and 2. We conducted a differential expression gene (DEG) analysis to explore these differences further. We performed DEG analysis using criteria of fold-change greater than 0.2 and a p-value below 0.05, resulting in the acquisition of 294 genes (Fig. 3C). We utilized gene ontology (GO) analysis to identify the biological pathways associated with the 294 genes. The identified pathways were related to T cell migration, homeostasis, differentiation, and activation (Fig. 3D), with a higher expression of GO term-associated genes observed in cluster 1 than in cluster 2 (Fig. 3E).
Fig. 3. Exploring T/NK cell function and differential gene expression in NSCLC mutant clusters. (A) UMAP representing the distribution of mutation clusters 1 and 2 in T/NK cells selected from primary tumor data of NSCLC patients, utilizing only the data selected from mutation clusters 1 and 2. (B) Stacked bar plot results show the distribution of T/NK cell subsets corresponding to mutation clusters 1 and 2. (C) Volcano plot results after DEG analysis of mutation clusters 1 and 2, visualizing the genes highly expressed in mutation cluster 1 (cutoff: logFC>0.2, p<0.05). (D) Result of the biological pathway list for the genes highly expressed in cluster 1 identified in (C). (E) The expression of the top marker genes for each of the biological pathways obtained earlier was compared in mutant clusters 1 and 2 and plotted in a dot plot. NSCLC, non-small cell lung cancer; UMAP, Uniform Manifold Approximation and Projection; DEG, differential expression gene.
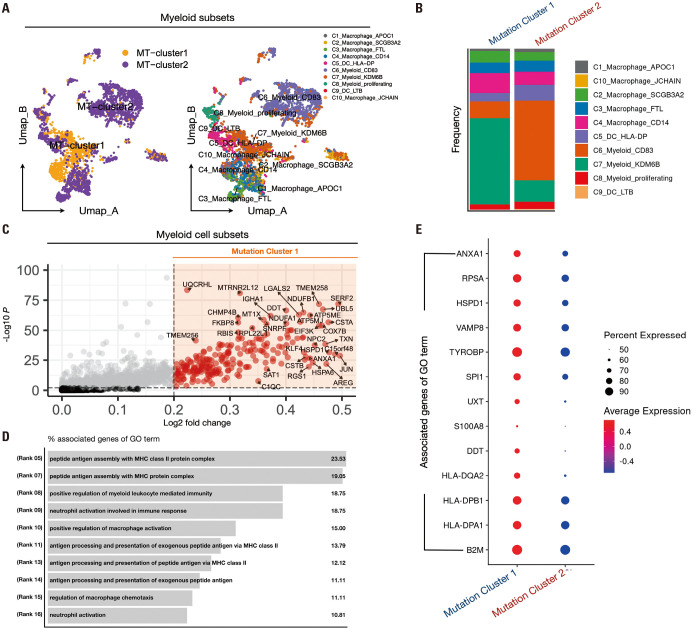
A higher proportion of the C7-Myeloid-KDM6B cluster within the myeloid subset in mutation cluster 1 was associated with antigen presentation
To investigate the differences in myeloid behavior between mutation clusters 1 and 2, we selected myeloid clusters and conducted data preprocessing steps identical to those previously performed on T/NK cells. We then visualized the distribution of mutation clusters and previously obtained DC/macrophage clusters using UMAP plots (Fig. 4A). When only myeloid cells were selected, mutation cluster 1 predominantly consisted of C7-Myeloid-KDM6B within the Myeloid cluster composition ratio (Fig. 4B). Upon comparing cytokine expression between mutation clusters 1 and 2, intriguingly similar expression patterns to those observed in T/NK cells emerged. IFNG exhibited a higher expression in mutation cluster 1, whereas TNF, IL1A, IL1B, IL6, and IL10 showed a higher expression in cluster 2 (Supplementary Fig. 6C, only online). Furthermore, we examined chemokine expression in mutation clusters 1 and 2 and found that CXCL5, CXCL8, and CXCL12 were highly expressed in mutation cluster 1, whereas CCL2, CCL3, CCL4, CCL8, CXCL1, CXCL2, CXCL3, CXCL9, CXCL10, and CXCL11 were highly expressed in mutation cluster 2 (Supplementary Fig. 6D, only online). We conducted a DEG analysis further to explore the dissimilarities between mutation clusters 1 and 2. We performed DEG analysis using the fold-change criteria greater than 0.2 and a p-value below 0.05, acquiring 353 genes (Fig. 4C). GO analysis revealed biological pathways associated with MHC-II-mediated antigen presentation, macrophage activation, and neutrophil activation (Fig. 4D), with higher expression of GO term-associated genes observed in mutation cluster 1 than in mutation cluster 2 (Fig. 4E).
Fig. 4. Exploring myeloid cell function and differential gene expression in NSCLC mutant clusters. (A) UMAP representing the distribution of mutation clusters 1 and 2 in myeloid cells selected from primary tumor data of NSCLC patients, utilizing only the data selected from mutation clusters 1 and 2. (B) Stacked bar plot results show the distribution of myeloid cell subsets corresponding to mutation clusters 1 and 2. (C) Volcano plot results after DEG analysis of mutation clusters 1 and 2, visualizing the genes highly expressed in mutation cluster 1 (cutoff: logFC>0.2, p<0.01). (D) Result of the biological pathway list for the genes highly expressed in cluster 1 identified in (C). (E) The expression of the top marker genes for each of the biological pathways obtained earlier was compared in mutant clusters 1 and 2 and plotted in a dot plot. NSCLC, non-small cell lung cancer; UMAP, Uniform Manifold Approximation and Projection; DEG, differential expression gene.
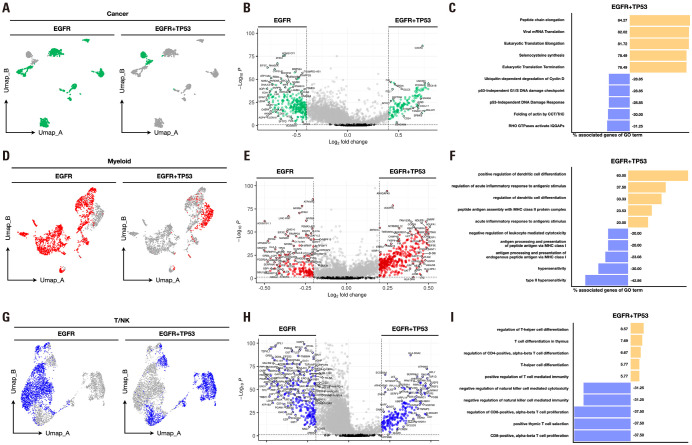
EGFR+TP53 mutation exhibited greater transcriptome alterations in tumors compared with EGFR mutation alone, with notable changes observed in the myeloid lineage
Previous studies confirmed that the double mutation of EGFR and TP53 exhibited distinct characteristics from mutations in EGFR alone. Therefore, we aimed to analyze the differences between EGFR mutations and EGFR+TP53 mutations in cancer, myeloid, and T/NK cell subsets. First, we selected EGFR and EGFR+TP53 mutations and cancer subsets. We then conducted data preprocessing steps identical to those outlined in Fig. 3A and 4A. Examination of the features of these two mutations revealed a clear distinction (Fig. 5A). We performed DEG analysis between the two mutations in the cancer subset to analyze the differences between distinct features. Using a cutoff of log2FC≥0.4 and p-value≤0.05, we obtained 302 genes associated with EGFR and 254 genes associated with EGFR+TP53 mutation (Fig. 5B). We conducted a reactome pathway analysis to identify the biological pathways associated with the DEGs. The pathways associated with EGFR included actin synthesis and p53-independent DNA damage response, whereas those associated with the EGFR+TP53 mutation included elongation, translation, and selenocysteine synthesis related to protein synthesis (Fig. 5C). Next, we selected EGFR and EGFR+TP53 mutations along with myeloid subsets. After conducting data pre-processing steps similar to those described above, we observed a clear distinction between the features of the two mutations (Fig. 5D). Consequently, we performed a DEG analysis of the two mutations in the myeloid subset. Using a cutoff of log2FC≥0.2 and p-value≤0.05, we obtained 187 EGFR and 520 genes associated with EGFR+TP53 mutation (Fig. 5E). We used the GO immune system process to identify the biological pathways associated with DEGs. The pathways associated with EGFR mutation included hypersensitivity, MHC-I-mediated antigen presentation, and negative regulation of leukocyte-mediated cytotoxicity, whereas those associated with the EGFR+TP53 mutation included DC differentiation (Fig. 5F). Finally, we selected EGFR and EGFR+TP53 mutations along with the T/NK cell subset. After conducting data pre-processing steps similar to those described above, we observed a clear distinction between the features of the two mutations (Fig. 5G). Consequently, we performed a DEG analysis of the two mutations in the T/NK subset. Using a cutoff of log2FC≥0.2 and p-value≤0.05, we obtained 292 genes associated with EGFR and 319 genes associated with EGFR+TP53 mutation (Fig. 5H). We used the GO immune system process to identify the biological pathways associated with DEGs. The pathways associated with EGFR included CD8+ T cell proliferation and negative regulation of NK cells. In contrast, those associated with the EGFR+TP53 mutation included CD4+ T cell differentiation and positive regulation of T cell-mediated immunity (Fig. 5I). These results, harboring both EGFR and TP53 mutations, demonstrate differences in EGFR and cell composition and cancer and immune cell function. This finding is predicted to be relevant to the previously described ICI responses, suggesting that further research is needed to consider these factors in future treatment strategies.
Fig. 5. Comparison of features seen in EGFR mutant and EGFR+TP53 mutant cells. (A) Differences in the distribution of T/NK cells in EGFR mutants and EGFR+TP53 mutants visualized by UMAP. (B) The genes highly expressed in T/NK cells of the EGFR mutant and EGFR+TP53 mutant were visualized in a volcano plot (DEG analysis cutoff: logFC>0.2, p<0.05). (C) Identified biological pathways associated with genes highly expressed in T/NK cells for each of the two mutations. (D) Differences in the distribution of myeloid cells in EGFR mutants and EGFR+TP53 mutants visualized by UMAP. (E) The genes highly expressed in EGFR mutant and EGFR+TP53 mutant myeloid cells were visualized in a volcano plot (DEG analysis cutoff: logFC>0.2, p<0.05). (F) Identified biological pathways associated with genes highly expressed in myeloid cells for each of the two mutations. (G) Differences in the distribution of cancer cells in EGFR mutants and EGFR+TP53 mutants visualized by UMAP. (H) The genes highly expressed in EGFR mutant and EGFR+TP53 mutant cancer cells were visualized in a volcano plot (DEG analysis cutoff: logFC>0.4, p<0.05). (I) Identified biological pathways associated with genes highly expressed in cancer cells for each of the two mutations.
DISCUSSION
Our study showed that the cellular-level components of the tumor microenvironment in NSCLC are heterogeneous according to major mutations, which provides important insights into the response to immunotherapeutic agents, such as ICIs. Harnessing the potential of scRNA-seq data the immune landscape that plays a pivotal role in tumor progression and response to therapy can be comprehensively explored.23,24,25,26 Given the analytical resolution of single-cell sequencing, we aimed to characterize the molecular and cellular features of NSCLC at the single-cell level and explore their complex relationships. Interestingly, the single-cell RNA database used in this study contained much mutational information. This allowed us to identify the molecular and cellular characteristics of mutations in NSCLC. Our results revealed interesting patterns and associations between the mutation and cell types in NSCLC.
First, we found similar patterns in the cellular composition across the major mutation types in NSCLC. KRAS, TP53, and EGFR+TP53 mutations had similar cellular compositions, suggesting similarities between mutation types. EGFR, BRAF, and ALK mutations were associated with different cellular compositions. These results indicate that mutation types can affect a tumor’s cellular composition and interactions. We grouped mutations with similar characteristics by examining the correlation between mutation types in NSCLC. KRAS, TP53, and EGFR+TP53 mutations appeared as a group with similar characteristics (mutation cluster 1), indicating that these mutations have similar biological properties in NSCLC. Mutation cluster 1 showed higher IFNG and CXCL13 expressions, its TLS signature score was also significantly higher than that of Mutant Cluster 2. For the immune cell cluster, the C2-T-IL7R, C3-T/NK-CXCL4, C9-T/NK-NKG, and C1-B-MS4A1 clusters were observed at higher levels than those in cluster 2.
In contrast, EGFR, ALK, and BRAF mutations exhibited characteristics similar to a separate group (mutation cluster 2). Mutation cluster 2 showed higher expression of TNF, IL1B, and chemokines associated with alternative immune pathways. Mutation cluster 2 also showed increased expression of most stromal cell subsets, including fibroblasts (C1-Fibroablst-CALD1, C2-Fibroblast-COL1A2), endothelial cells (C1-endothelial-RAMP2), and mast cells (C1-Mast-TPSAB1). This is consistent with previous findings and provides important insights into the similarities and differences among specific mutation types. In addition, previous studies have reported a 14% response rate to ICIs in patients with EGFR mutations.24 Although many studies have investigated the reasons for the lower responsiveness of these mutations compared to the WT, the causes are not clearly understood. Therefore, our results provide important insights into the causes of lower responsiveness of patients with mutations in EGFR, ALK, and ROS to ICIs.
A highlight of this study was that unlike mutation cluster 2, mutations co-occurring with EGFR and TP53 were observed in mutation cluster 1. Therefore, we focused on EGFR-mutant lung cancer and EGFR+TP53 co-occurring mutant lung cancer, and found that the most significant difference between EGFR-mutant lung cancer and EGFR+TP53 co-mutant lung cancer was the transcriptome difference in cancer cells. The transcriptome diversity of the EGFR+TP53 mutant was more regulated by higher synthetic processes, such as peptide synthesis and elongation. In addition, we found that the difference between the two mutations in myeloid cells was strongly characterized by MHC class 2 and DC activity and differentiation in EGFR+TP53 co-occurring mutant cancer. In contrast, MHC class 1 was strongly characterized in EGFR mutant cancer. The difference between the two mutations in T/NK cells was that EGFR+TP53 co-occurring mutant cancers had high expression of features associated with T/NK cell activity and differentiation. In contrast, EGFR mutant cancers had high expression of the opposite features. These results imply that the tumor microenvironment is characterized differently in the presence or absence of TP53 mutations. This may provide important insights into the poor response to ICIs in patients with EGFR mutations.
In our study, we conducted analysis using data from a total of 21 NSCLC patients, uncovering intriguing patterns and associations between mutations and cell types. However, due to the limited sample size utilized in our analysis, we encountered a limitation in not being able to conduct analysis dependent on NSCLC stage. Given that treatment strategies need to vary based on the stage of lung cancer, this is a critical factor that warrants further investigation.27 Recognizing this limitation underscores the importance of expanding our research to include a larger cohort, allowing for more robust analyses that account for the diverse stages of lung cancer.
In conclusion, these analyses allowed us to identify the characteristic differences in gene expression and biological pathways between KRAS, TP53, and EGFR+TP53 mutations and EGFR, BRAF, and ALK mutations. We also showed that these differences were linked to immune cell activity within the tumor. Our work and previous studies suggest a close relationship between mutation types and tumor microenvironment in NSCLC, and may help develop personalized approaches for cancer diagnosis and treatment.
ACKNOWLEDGEMENTS
This research was supported by the Bio & Medical Technology Development Program of the National Research Foundation (NRF) and funded by the Korean government (MSIT) (No. 2022M3E5F3081138). This work was also supported by the National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT) (No. 2022R1A2C3005817).
Footnotes
The authors have no potential conflicts of interest to disclose.
- Conceptualization: Youngtaek Kim, Kwangmin Na, Dong Kwon Kim, Seul Lee, Seong-san Kang, Sujeong Baek, and Kyoung-Ho Pyo.
- Data curation: Youngtaek Kim, Joon Yeon Hwang, Seong-san Kang, and Mi Hyun Kim.
- Formal analysis: Youngtaek Kim, Joon Yeon Hwang, Seul Lee, Seung Min Yang, Chai Young Lee, and Yu Jin Han.
- Funding acquisition: Youngtaek Kim, Joon Yeon Hwang, and Kyoung-Ho Pyo.
- Investigation: Youngtaek Kim, Kwangmin Na, Dong Kwon Kim, Sujeong Baek, Seung Min Yang, Mi Hyun Kim, Heekyung Han, Seong Su Jeong, Chai Young Lee, Yu Jin Han, Jie-Ohn Son, Sang-Kyu Ye, and Kyoung-Ho Pyo.
- Methodology: Youngtaek Kim.
- Project administration: Youngtaek Kim and Kyoung-Ho Pyo.
- Resources: Youngtaek Kim, Joon Yeon Hwang, Heekyung Han, Seong Su Jeong, Jie-Ohn Son, and Sang-Kyu Ye.
- Software: Youngtaek Kim.
- Supervision: Kyoung-Ho Pyo.
- Validation: Youngtaek Kim, Joon Yeon Hwang, Jie-Ohn Son, Sang-Kyu Ye, and Kyoung-Ho Pyo.
- Visualization: Youngtaek Kim.
- Writing—original draft: Youngtaek Kim.
- Writing—review & editing: Youngtaek Kim and Joon Yeon Hwang.
- Approval of final manuscript: all authors.
SUPPLEMENTARY MATERIALS
(A) UMAP plot depicting 41 clusters identified within the NSCLC patient dataset comprising 64369 cells. (B) Stacked bar plots representing the frequency of clusters 0 to 40 across the mutations in the dataset. (C) Markers and references for cell type assignment by cluster. Cell type annotation was performed using the following markers: Myeloid (CD14, LYZ), T cells (CD3E, CD3D), NK cells (NKG7, GNLY), cancer cells (NAPSA, NKX2-1), epithelial cells (CAPS, SNTN), B cells (MS4A1, LY9), fibroblast (COL1A2, DCN), endothelial cells (FLT1, PECAM1), plasma cells (MZB1, JCHAIN), and mast cells (KIT, CPA3). (D) Dot plot and violin plot representing the expression of cell type subset markers by 41 clusters. (E) Feature plot confirming the expression of cell type subset markers. NSCLC, non-small cell lung cancer; UMAP, Uniform Manifold Approximation and Projection.
Stacked bar plots showing the clinical information of the 21 patients with primary tumor data and the proportion of the 41 clusters assigned a cell type for each patient. The three stacked bar plots represent the total cell types, T/NK cell types, and immune cell types, respectively.
(A) UMAP plot of the primary tumor data from NSCLC patients according to nine cell types. (B) Heatmap visualizing the expression levels for each of the nine cell types and its top genes. (C) UMAP plot of the primary tumor data from NSCLC patients according to 41 cluster assigned with cell types. (D) Heatmap visualizing the expression levels for each of the 41 cluster assigned with cell types and their top genes. NSCLC, non-small cell lung cancer; UMAP, Uniform Manifold Approximation and Projection.
Dendrogram illustrating hierarchical clustering (H-clustering) of the 41 cluster assigned with cell types observed in Fig. 2C and D.
(A) Percentage of the composition of 41 cluster assigned with cell type by mutation, visualized using a stacked bar plot. (B) Percentage of the composition of 41 clusters with cell type assigned per cluster was visualized using a stacked bar plot.
(A) Expression of cytokines (IL1A, IL1B, IL2, IL4, IL6, IL10, TNF, IFNG, IL12A, IL12B) by cluster in T/NK subset visualized using a dot plot. (B) Expression of chemokines (CCL1, CCL2, CCL3, CCL4, CCL5, CCL7, CCL8, CCL11, CXCL1, CXCL2, CXCL3, CXCL5, CXCL6, CXCL8, CXCL9, CXCL10, CXCL11, CXCL12, CXCL13, CX3CL, XCL1, XCL2) by cluster in the T/NK subset visualized using a dot plot. (C) Expression of cytokines (IL1A, IL1B, IL2, IL4, IL6, IL10, TNF, IFNG, IL12A, IL12B) by cluster in the myeloid subset visualized using a dot plot. (D) Expression of chemokines (CCL1, CCL2, CCL3, CCL4, CCL5, CCL7, CCL8, CCL11, CXCL1, CXCL2, CXCL3, CXCL5, CXCL6, CXCL8, CXCL9, CXCL10, CXCL11, CXCL12, CXCL13, CX3CL, XCL1, XCL2) by cluster in the myeloid subset visualized using a dot plot. (E) Violin plot comparing the TLS signature scores between MT-cluster1 and MT-cluster2. The TLS signature scores were computed using the AddModuleScore function based on TLS-associated genes. The plot shows a significant difference in the TLS signature scores between the two clusters, with MT-cluster1 exhibiting a higher TLS signature score compared to MT-cluster2.
References
- 1.Thandra KC, Barsouk A, Saginala K, Aluru JS, Barsouk A. Epidemiology of lung cancer. Contemp Oncol (Pozn) 2021;25:45–52. doi: 10.5114/wo.2021.103829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leiter A, Veluswamy RR, Wisnivesky JP. The global burden of lung cancer: current status and future trends. Nat Rev Clin Oncol. 2023;20:624–639. doi: 10.1038/s41571-023-00798-3. [DOI] [PubMed] [Google Scholar]
- 3.Maione P, Rossi A, Sacco PC, Bareschino MA, Schettino C, Ferrara ML, et al. Treating advanced non-small cell lung cancer in the elderly. Ther Adv Med Oncol. 2010;2:251–260. doi: 10.1177/1758834010366707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou S, Yang H. Immunotherapy resistance in non-small-cell lung cancer: from mechanism to clinical strategies. Front Immunol. 2023;14:1129465. doi: 10.3389/fimmu.2023.1129465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yan X, Zhao L, Wu F, Shen B, Zhou G, Feng J, et al. Efficacy and safety analysis of immune checkpoint inhibitor rechallenge therapy in locally advanced and advanced non-small cell lung cancer: a retrospective study. J Thorac Dis. 2024;16:1787–1803. doi: 10.21037/jtd-23-1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blakely CM, Watkins TBK, Wu W, Gini B, Chabon JJ, McCoach CE, et al. Evolution and clinical impact of co-occurring genetic alterations in advanced-stage EGFR-mutant lung cancers. Nat Genet. 2017;49:1693–1704. doi: 10.1038/ng.3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang L, Diao M, Zhang Z, Jiang M, Chen S, Zhao D, et al. Comparison of the somatic genomic landscape between central- and peripheral-type non-small cell lung cancer. Lung Cancer. 2024;187:107439. doi: 10.1016/j.lungcan.2023.107439. [DOI] [PubMed] [Google Scholar]
- 8.Mansouri S, Heylmann D, Stiewe T, Kracht M, Savai R. Cancer genome and tumor microenvironment: reciprocal crosstalk shapes lung cancer plasticity. Elife. 2022;11:e79895. doi: 10.7554/eLife.79895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farago AF, Azzoli CG. Beyond ALK and ROS1: RET, NTRK, EGFR and BRAF gene rearrangements in non-small cell lung cancer. Transl Lung Cancer Res. 2017;6:550–559. doi: 10.21037/tlcr.2017.08.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lamberti G, Andrini E, Sisi M, Rizzo A, Parisi C, Di Federico A, et al. Beyond EGFR, ALK and ROS1: current evidence and future perspectives on newly targetable oncogenic drivers in lung adenocarcinoma. Crit Rev Oncol Hematol. 2020;156:103119. doi: 10.1016/j.critrevonc.2020.103119. [DOI] [PubMed] [Google Scholar]
- 11.Santarpia M, Ciappina G, Spagnolo CC, Squeri A, Passalacqua MI, Aguilar A, et al. Targeted therapies for KRAS-mutant non-small cell lung cancer: from preclinical studies to clinical development-a narrative review. Transl Lung Cancer Res. 2023;12:346–368. doi: 10.21037/tlcr-22-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Skoulidis F, Goldberg ME, Greenawalt DM, Hellmann MD, Awad MM, Gainor JF, et al. STK11/LKB1 mutations and PD-1 inhibitor resistance in KRAS-mutant lung adenocarcinoma. Cancer Discov. 2018;8:822–835. doi: 10.1158/2159-8290.CD-18-0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Calles A, Riess JW, Brahmer JR. Checkpoint blockade in lung cancer with driver mutation: choose the road wisely. Am Soc Clin Oncol Educ Book. 2020;40:372–384. doi: 10.1200/EDBK_280795. [DOI] [PubMed] [Google Scholar]
- 14.Wu F, Fan J, He Y, Xiong A, Yu J, Li Y, et al. Single-cell profiling of tumor heterogeneity and the microenvironment in advanced non-small cell lung cancer. Nat Commun. 2021;12:2540. doi: 10.1038/s41467-021-22801-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He D, Wang D, Lu P, Yang N, Xue Z, Zhu X, et al. Single-cell RNA sequencing reveals heterogeneous tumor and immune cell populations in early-stage lung adenocarcinomas harboring EGFR mutations. Oncogene. 2021;40:355–368. doi: 10.1038/s41388-020-01528-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moghal N, Li Q, Stewart EL, Navab R, Mikubo M, D’Arcangelo E, et al. Single-cell analysis reveals transcriptomic features of drug-tolerant persisters and stromal adaptation in a patient-derived EGFR-mutated lung adenocarcinoma xenograft model. J Thorac Oncol. 2023;18:499–515. doi: 10.1016/j.jtho.2022.12.003. [DOI] [PubMed] [Google Scholar]
- 17.Lee H, Kim C, Jeong J, Jung K, Han B. ScIntegral: a scalable and accurate cell-type identification method for scRNA-seq data with application to integration of multiple donors. [accessed on 2024 February 10];BioRxiv [Preprint] 2020 doi: 10.1101/2020.09.17.301911. Available at: [DOI] [Google Scholar]
- 18.Hwang B, Lee JH, Bang D. Single-cell RNA sequencing technologies and bioinformatics pipelines. Exp Mol Med. 2018;50:1–14. doi: 10.1038/s12276-018-0071-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salcher S, Sturm G, Horvath L, Untergasser G, Kuempers C, Fotakis G, et al. High-resolution single-cell atlas reveals diversity and plasticity of tissue-resident neutrophils in non-small cell lung cancer. Cancer Cell. 2022;40:1503–1520.e8. doi: 10.1016/j.ccell.2022.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.CZI Single-Cell Biology Program. Abdulla S, Aevermann B, Assis P, Badajoz S, Bell SM, et al. CZ CELL×GENE discover: a single-cell data platform for scalable exploration, analysis and modeling of aggregated data. [accessed on 2024 February 28];BioRxiv [Preprint] 2023 doi: 10.1101/2023.10.30.563174. Available at: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laughney AM, Hu J, Campbell NR, Bakhoum SF, Setty M, Lavallée VP, et al. Regenerative lineages and immune-mediated pruning in lung cancer metastasis. Nat Med. 2020;26:259–269. doi: 10.1038/s41591-019-0750-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maynard A, McCoach CE, Rotow JK, Harris L, Haderk F, Kerr DL, et al. Therapy-induced evolution of human lung cancer revealed by single-cell RNA sequencing. Cell. 2020;182:1232–1251.e22. doi: 10.1016/j.cell.2020.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tan Z, Xue H, Sun Y, Zhang C, Song Y, Qi Y. The role of tumor inflammatory microenvironment in lung cancer. Front Pharmacol. 2021;12:688625. doi: 10.3389/fphar.2021.688625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang R, Wang H, Tang M. Roles of tissue-resident immune cells in immunotherapy of non-small cell lung cancer. Front Immunol. 2023;14:1332814. doi: 10.3389/fimmu.2023.1332814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yeo AT, Rawal S, Delcuze B, Christofides A, Atayde A, Strauss L, et al. Single-cell RNA sequencing reveals evolution of immune landscape during glioblastoma progression. Nat Immunol. 2022;23:971–984. doi: 10.1038/s41590-022-01215-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li PH, Kong XY, He YZ, Liu Y, Peng X, Li ZH, et al. Recent developments in application of single-cell RNA sequencing in the tumour immune microenvironment and cancer therapy. Mil Med Res. 2022;9:52. doi: 10.1186/s40779-022-00414-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Araghi M, Mannani R, Heidarnejad Maleki A, Hamidi A, Rostami S, Safa SH, et al. Recent advances in non-small cell lung cancer targeted therapy; an update review. Cancer Cell Int. 2023;23:162. doi: 10.1186/s12935-023-02990-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) UMAP plot depicting 41 clusters identified within the NSCLC patient dataset comprising 64369 cells. (B) Stacked bar plots representing the frequency of clusters 0 to 40 across the mutations in the dataset. (C) Markers and references for cell type assignment by cluster. Cell type annotation was performed using the following markers: Myeloid (CD14, LYZ), T cells (CD3E, CD3D), NK cells (NKG7, GNLY), cancer cells (NAPSA, NKX2-1), epithelial cells (CAPS, SNTN), B cells (MS4A1, LY9), fibroblast (COL1A2, DCN), endothelial cells (FLT1, PECAM1), plasma cells (MZB1, JCHAIN), and mast cells (KIT, CPA3). (D) Dot plot and violin plot representing the expression of cell type subset markers by 41 clusters. (E) Feature plot confirming the expression of cell type subset markers. NSCLC, non-small cell lung cancer; UMAP, Uniform Manifold Approximation and Projection.
Stacked bar plots showing the clinical information of the 21 patients with primary tumor data and the proportion of the 41 clusters assigned a cell type for each patient. The three stacked bar plots represent the total cell types, T/NK cell types, and immune cell types, respectively.
(A) UMAP plot of the primary tumor data from NSCLC patients according to nine cell types. (B) Heatmap visualizing the expression levels for each of the nine cell types and its top genes. (C) UMAP plot of the primary tumor data from NSCLC patients according to 41 cluster assigned with cell types. (D) Heatmap visualizing the expression levels for each of the 41 cluster assigned with cell types and their top genes. NSCLC, non-small cell lung cancer; UMAP, Uniform Manifold Approximation and Projection.
Dendrogram illustrating hierarchical clustering (H-clustering) of the 41 cluster assigned with cell types observed in Fig. 2C and D.
(A) Percentage of the composition of 41 cluster assigned with cell type by mutation, visualized using a stacked bar plot. (B) Percentage of the composition of 41 clusters with cell type assigned per cluster was visualized using a stacked bar plot.
(A) Expression of cytokines (IL1A, IL1B, IL2, IL4, IL6, IL10, TNF, IFNG, IL12A, IL12B) by cluster in T/NK subset visualized using a dot plot. (B) Expression of chemokines (CCL1, CCL2, CCL3, CCL4, CCL5, CCL7, CCL8, CCL11, CXCL1, CXCL2, CXCL3, CXCL5, CXCL6, CXCL8, CXCL9, CXCL10, CXCL11, CXCL12, CXCL13, CX3CL, XCL1, XCL2) by cluster in the T/NK subset visualized using a dot plot. (C) Expression of cytokines (IL1A, IL1B, IL2, IL4, IL6, IL10, TNF, IFNG, IL12A, IL12B) by cluster in the myeloid subset visualized using a dot plot. (D) Expression of chemokines (CCL1, CCL2, CCL3, CCL4, CCL5, CCL7, CCL8, CCL11, CXCL1, CXCL2, CXCL3, CXCL5, CXCL6, CXCL8, CXCL9, CXCL10, CXCL11, CXCL12, CXCL13, CX3CL, XCL1, XCL2) by cluster in the myeloid subset visualized using a dot plot. (E) Violin plot comparing the TLS signature scores between MT-cluster1 and MT-cluster2. The TLS signature scores were computed using the AddModuleScore function based on TLS-associated genes. The plot shows a significant difference in the TLS signature scores between the two clusters, with MT-cluster1 exhibiting a higher TLS signature score compared to MT-cluster2.