Abstract
The maintenance of [PSI], a prion-like form of the yeast release factor Sup35, requires a specific concentration of the chaperone protein Hsp104: either deletion or overexpression of Hsp104 will cure cells of [PSI]. A major puzzle of these studies was that overexpression of Hsp104 alone, from a heterologous promoter, cures cells of [PSI] very efficiently, yet the natural induction of Hsp104 with heat shock, stationary-phase growth, or sporulation does not. These observations pointed to a mechanism for protecting the genetic information carried by the [PSI] element from vicissitudes of the environment. Here, we show that simultaneous overexpression of Ssa1, a protein of the Hsp70 family, protects [PSI] from curing by overexpression of Hsp104. Ssa1 protein belongs to the Ssa subfamily, members of which are normally induced with Hsp104 during heat shock, stationary-phase growth, and sporulation. At the molecular level, excess Ssa1 prevents a shift of Sup35 protein from the insoluble (prion) to the soluble (cellular) state in the presence of excess Hsp104. Overexpression of Ssa1 also increases nonsense suppression by [PSI] when Hsp104 is expressed at its normal level. In contrast, hsp104 deletion strains lose [PSI] even in the presence of overproduced Ssa1. Overproduction of the unrelated chaperone protein Hsp82 (Hsp90) neither cured [PSI] nor antagonized the [PSI]-curing effect of overproduced Hsp104. Our results suggest it is the interplay between Hsp104 and Hsp70 that allows the maintenance of [PSI] under natural growth conditions.
Prions are infectious proteins which are believed to cause transmissible neurodegenerative diseases in mammals, such as sheep scrapie, human kuru, and Creutzfeldt-Jacob disease, and bovine spongiform encephalopathy, or “mad cow” disease (see references 38 and 50 for reviews). According to the current model, the prion protein (PrPSc) is a conformational derivative of the normal protein (PrPC), which has acquired an ability to influence the normal protein, encoded by the same gene, to adopt the prion conformation. In the PrPSc conformation, PrP is characterized by increased proteinase resistance, high β-sheet content, and a capacity to form insoluble aggregates (38, 39).
Recent data suggest that prion-like phenomena, in which self-perpetuating changes in protein conformation alter the phenotype of the organism, are widespread in nature. It has been hypothesized that autocatalyzed misfolding and/or aggregation cause such a common human disease as Alzheimer’s disease (22, 26, 28). Even more remarkably, it has been noted that yeast non-Mendelian elements [URE3] and [PSI] (53) and Podospora non-Mendelian element [Het-S] (8) behave like prions. The propagation of these elements is believed to be due to the transmission of proteins with a self-perpetuating, functionally altered conformation, rather than to the transmission of a nucleic acid replicon. Therefore, the prion model provides an additional mechanism of genetic transmission, which is based on the information coded in the biopolymer’s three-dimensional structure rather than in its primary sequence. Existence of such systems of structural coding has revolutionary implications for our understanding of genetics and evolution.
[PSI] causes translational readthrough (nonsense suppression) in yeast (9, 10). Both genetic and biochemical data strongly suggest that [PSI] represents a functionally defective form of the evolutionary conserved release factor eRF3 (Sup35), which propagates through a self-perpetuating change in protein conformation (see references 29, 49, and 55 for reviews). Sup35 overproduction greatly increases the frequency of the spontaneous appearance of [PSI] (4, 14). Yeast Sup35 protein forms insoluble proteinase-resistant aggregates in [PSI+] cells (35, 36). The insoluble form of the Sup35 protein (Sup35PSI) stimulates aggregation of the soluble Sup35 protein in cell extracts, thus mimicking the prion-like propagation of the protein change in vitro (18, 37). The purified Sup35 protein has also been shown to undergo self-seeded polymerization in vitro, resulting in the formation of Congo red-staining amyloid-like fibers (18, 24), similar to those formed by the mammalian PrP (39).
Conformational switches and aggregation events, postulated by the prion model, make it reasonable to expect that chaperone proteins are involved in prion propagation. Indeed, we have previously described that [PSI] propagation requires an intermediate amount of the chaperone protein Hsp104 (5). Inactivation of Hsp104 cured cells of [PSI], suggesting that Hsp104 may be required for a partial unfolding of the normal protein that makes it susceptible to assuming a prion-like conformation. However, selective overproduction of Hsp104 also inhibited and eventually cured [PSI], with an efficiency that correlated with the level of Hsp104 produced. Since it has been shown that Hsp104 function in thermotolerance is to reverse heat-induced aggregation damage (34), one could suggest that excess Hsp104 can shift the balance from prion-like aggregates to partially unfolded intermediates and soluble forms, resulting in the loss of [PSI]. Biochemical experiments confirmed that both excess Hsp104 and hsp104 deletions lead to an increase in the proportion of the soluble Sup35 protein relative to insoluble Sup35 protein in [PSI+] cells (35, 36). In vitro experiments indicate that Hsp104 and Sup35 can physically interact with each other under certain circumstances (43). Recent in vitro data also suggest that Hsp104 influences the conformational transitions of mammalian PrP as well, promoting the acquisition of the structural characteristics of the disease form, PrPSc, with a high degree of specificity (13). This result indicates that folding intermediates formed by unrelated prion proteins possess similar features. Thus, investigating the nature of chaperone-Sup35 interaction not only is important for understanding the remarkable process that allows the transmission of genetic information by alternative protein conformations in yeast but also may lead to increased understanding of the general mechanisms of prion-like conformational switches.
No human homolog of Hsp104 has been identified to date, which raises the question of whether other chaperone proteins that are conserved in humans influence prion-like transitions. Indeed, in yeast cells, the relationship between the Hsp104 chaperone and the [PSI] prion-like element presents a paradox (see reference 54) that might be explained by interactions with other chaperones. Hsp104 levels increase during growth at high temperatures and upon heat shock (40, 41). When Hsp104 is selectively induced to similar levels by using a heterologous promoter, up to 90% of the culture is cured of [PSI] (5, 6, 31). Yet growth at 37 to 39°C causes neither inhibition nor curing of [PSI] (44, 48), while heat shock at 42 to 55°C causes only low-efficiency curing of [PSI] (6, 10, 31, 48). It might be postulated that other proteins, denatured by heat shock, compete with the Sup35 protein for Hsp104, preventing newly induced Hsp104 from curing cells of [PSI] under these circumstances. However, Hsp104 normally accumulates during heat shock at levels much higher than required to repair heat-induced damage (30). Even more strikingly, [PSI] is very stable in stationary-phase cells and spores (7, 10), when Hsp104 levels are also high (41) but protein denaturation is presumably low.
On the basis of these data, we (6) hypothesized that other heat- and stationary-phase-induced factors interfere with Hsp104’s capacity to cure cells of [PSI], allowing the stable propagation of [PSI] through diverse environmental conditions. Here, we report the effects of Ssa1 protein, a yeast member of the Hsp70 family, on Hsp104 mediated-curing of [PSI]. Ssa proteins are abundant chaperones of the yeast cytosol that are coinduced with Hsp104 during heat shock, stationary-phase growth, and sporulation (25, 51, 52). Moreover, they interact with Hsp104 in thermotolerance in vivo (42) and in protein disaggregation in vitro (17). Since Ssa proteins constitute a single essential complementation group with strongly overlapping functions, we have analyzed only Ssa1. To provide a broader range of circumstances in which to test the interactions between Hsp104, Hsp70, and [PSI], we took advantage of the existence of distinct types of [PSI] elements. Strong and weak [PSI] elements have stable, heritable differences in the efficiency of nonsense suppression as well as different mitotic transmission rates. Because these elements can be propagated in isogenic yeast strains, they are believed to result from somewhat different, self-perpetuating conformational states of Sup35 (14). Members of the yeast Hsp70 family modify Hsp104’s effect on both types of [PSI] element, providing an explanation for the maintenance of [PSI] under natural conditions.
MATERIALS AND METHODS
Yeast strains.
Genotypes of the Saccharomyces cerevisiae strains are shown in Table 1. [PSI+] strains OT55 (also called [PSI+]1-1-74-D694) and OT56 (also called [PSI+]7-74-D694) are independent derivatives of strain 74-D694, induced by overproduction of the wild-type Sup35 protein as described previously (14). The [psi−] ade1-14 strains are red on YPD medium. Presence of [PSI] leads to the suppression of the ade1-14UGA allele, detected as growth on adenine-deficient (−Ade) medium after 3 to 4 (OT56) or 7 to 8 (OT55) days of incubation and as white (OT56) or pink (OT55) color on YPD medium. OT56 exhibits higher mitotic stability of [PSI] in the presence of guanidine-HCl than OT55 (14). L-1607 is an hsp104::URA3 disruption obtained in strain OT55 as described previously (5). OT46 is a spontaneous Ura− derivative of L-1607 which, according to Southern and Western analysis, retains the hsp104 deletion (15). GT1-S31 and GT1-S13 were recovered from the meiotic progeny of the genetic cross between 35-D693 (15) and JN14. GT56-13B was recovered by A. Galkin (in Y. Chernoff’s lab) from the meiotic progeny of the cross between GT1-S13 and OT56.
TABLE 1.
S. cerevisiae strains used
| Name | Synonym(s) | Genotype | Reference(s) or source |
|---|---|---|---|
| OT55 | [PSI+]1-1-74-D694 | MATa ade1-14UGAhis3 leu2 trp1-289UAGura3 [PSI+] | 14, 15 |
| OT56 | [PSI+]7-74-D694 | MATa ade1-14UGAhis3 leu2 trp1-289UAGura3 [PSI+] | 14, 15 |
| L-1607 | 74-D694-hsp104del | MATa ade1-14UGAhis3 leu2 trp1-289UAGura3 hsp104::URA3 [psi−] | 5 |
| OT46 | L-1607 Ura− | MATa ade1-14UGAhis3 leu2 trp1-289UAGura3 hsp104Δ [psi−] | 15 |
| GT1-S31 | None | MATα ade1-14UGAhis3 leu2 lys2 trp1-289UAGura3 [PSI+] | This study |
| GT1-S13 | None | MATα ade1-14UGAhis3 leu2 trp1-289UAGura3 [PSI+] | This study |
| GT56-13B | None | MATα ade1-14UGAhis3 leu2 trp1-289UAGura3 [PSI+] | Unpublished data |
| JN14 | None | MATa his3 leu2 lys2 trp1-Δ1 ura3 ssa1::HIS3 ssa2::URA3 | Gift of E. Craig |
Plasmids.
Shuttle plasmids used in this study are shown in Table 2. Plasmid pLA1, which is a pRS313 (45) derivative containing the GAL1,10 promoter, was constructed by L. Arwood in S. Lindquist’s lab. Plasmid pSSA1-LEU2, also called pLH101, is a pRS425 derivative containing a wild-type SSA1 gene under its normal promoter and was constructed by L. Henninger in S. Lindquist’s lab. Plasmid pH28, constructed by E. Schirmer in S. Lindquist’s lab, is a pLA1 derivative which contains the entire promoterless HSP104 gene inserted into BamHI-SacI-cut polylinker immediately after GAL1,10 promoter. Plasmid pMC3, which contains a promoterless HSP82 gene fused into the BamHI site of the GAL1,10 promoter in centromeric URA3 vector pBM150 (46), was constructed by M. Fortin in S. Lindquist’s lab. Plasmids pUKC815 and pUKC819, kindly provided by M. F. Tuite, contain PGK-lacZ hybrid constructs from pUKC350 and pUKC353 (16), respectively, cloned in the centromeric URA3-containing vector YCp50.
TABLE 2.
Shuttle plasmids used
| Name | Other name(s) | Type | Copy no. | Yeast marker(s) | Reference |
|---|---|---|---|---|---|
| pRS316 | None | YCp | Low | URA3 | 45 |
| pRS425 | None | YEp | Moderate | LEU2 | 45 |
| YEp13 | None | YEp | Moderate | LEU2 | 3 |
| pLA1 | pRS313GAL | YCpGAL | Low | HIS3 | Unpublished data |
| pRS316GAL | None | YCpGAL | Low | URA3 | 32 |
| pYS104 | pRS316-HSP104 | YCp | Low | URA3, HSP104 | 40 |
| pSSA1-LEU2 | pRS425-SSA1, pLH101 | YEp | Moderate | LEU2, SSA1 | Unpublished data |
| pH28 | pLA1-HSP104 | YCpGAL | Moderate | HIS3, GAL::HSP104 | Unpublished data |
| pGAL::SSA1-URA3 | None | YCpGAL | Low | URA3, GAL::SSA1 | 42 |
| pMC3 | None | YCpGAL | Low | URA3, GAL::HSP82 | Unpublished data |
| pUKC815 | None | YCp | Low | URA3, PGK-lacZ | 16 |
| pUKC819 | None | YCp | Low | URA3, PGK-UGA-lacZ | 16 |
Antibodies.
Sup35 antipeptide antibodies specific to amino acid positions 137 to 151 of the Sup35 protein (35), Hsp104-specific antibody 8-1 (33), and Hsp82-specific antibodies (2) have been described previously. Ssa-specific polyclonal antibody SSA1 C1delB was kindly provided by E. Craig. Secondary anti-rabbit antibodies were purchased from Amersham. Western blotting, reaction to the primary and secondary antibodies, and detection were performed by the chemiluminescence method as described in the Amersham protocol. Densitometry assays were performed according to Image Tool (developed by Don Wilcox, Brent Dove, Doss McDavid, and David Greer; downloaded from http://www.uthscsa.edu).
Yeast media and growth conditions.
Standard yeast media and cultivation conditions (23) were used. Transformation was performed according to the modified Li+ procedure (20). Liquid cultures were grown on the shaker, normally at 200 to 250 rpm, with a liquid/flask volume ratio of 1:5 or more. Galactose induction on solid medium was performed by replica plating the yeast strains onto the corresponding synthetic medium containing galactose instead of glucose. To achieve galactose induction in liquid medium, yeast cultures were pregrown in the corresponding synthetic glucose medium up to 2 × 106 to 1 × 107 cells/ml, collected by centrifugation, washed twice with H2O, and inoculated into the corresponding synthetic medium, containing 2% galactose and 2% raffinose, at the starting concentration 2.5 × 105 to 5 × 105 cells/ml.
Standard procedures (23) were used for yeast sporulation, micromanipulation, and tetrad analysis. For isolation of random spores, sporulating cultures were suspended in H2O, vortexed with an equal volume of diethyl ether for 5 min, and plated on the synthetic medium, which was selective for the particular genotype. About 95 to 99% of the colonies are formed by the haploid ascospores, which are more resistant to ether treatment than the vegetative (diploid) cells.
Temperature resistance assays.
To measure temperature resistance of the exponential yeast cells, yeast precultures were grown overnight at 25°C with shaking in the corresponding synthetic medium selective for the plasmid(s), diluted to 106 cells/ml, and incubated for another 3 h at 25°C with shaking. After these incubations, 0.5-ml aliquots of each culture were either placed directly on ice (control samples) or incubated for 5 to 20 min in the 50°C water bath and then placed on ice (heat-shocked samples). Then, serial dilutions of both heat-shocked and control samples were prepared and either pipetted onto the corresponding synthetic medium, 5 μl per spot (semiquantitative assay), or plated onto the corresponding synthetic medium, 0.1 ml per plate (quantitative assay). Resulting plates were scored after 3 to 4 days of incubation at 30°C. Temperature resistance of the cultures preinduced at 37°C was determined in the same way, except that at 30 min before heat shock, 5-ml aliquots of each culture were placed into 37°C and incubated with shaking for another 30 min, as described previously (40, 41).
To measure temperature resistance of the galactose-induced yeast cells, yeast precultures were grown to 2.4 × 106 cells/ml at 25°C with shaking in 5 ml of the corresponding synthetic glucose medium, selective for the plasmids. Cells were collected by centrifugation, washed twice with H2O, resuspended in 50 ml of the same medium containing galactose and raffinose instead of glucose, and incubated with shaking at 25°C for about two cell divisions, resulting in the final concentration of about 106 cells/ml. Then 0.5-ml aliquots of each culture were taken and heat shocked as described above.
Protein isolation and analysis.
Hsp104, Hsp70 (Ssa1), and Hsp82 were identified by the reaction to the corresponding antibodies (see above) in the protein extracts, prepared from yeast cells which had been lysed on a vortex mixer with glass beads in 100 mM Tris-HCl (pH 7.5)–0.2 M NaCl–1 mM EDTA–0.5 mM dithiothreitol–2 mM phenylmethylsulfonyl fluoride–5% glycerol, or in the total-lysate fractions prepared by the centrifugation-fractionation (see below). Extracts were then separated by the standard sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) procedure. The centrifugation-fractionation performed as described previously (35), with slight modifications, was used for determination of the Sup35 protein distribution between the soluble and insoluble fractions. Yeast cotransformants were grown to 5 × 106 cells/ml in either synthetic glucose or synthetic galactose-raffinose medium, both selective for both plasmids. Cells were lysed on a vortex mixer with glass beads in lysis buffer containing 50 mM Tris-HCl (pH 7.5), 10 mM KCl, 5 mM MgCl2, 0.1 mM EDTA, 0.1 mM dithiothreitol, 100 μg of cycloheximide per ml, 2 mM phenylmethylsulfonyl fluoride, 1 mM benzamidine, 2 μg of pepstastin A per ml, 10 μg of leupeptin per ml, and 100 μg of RNase A per ml. High concentrations of proteinase inhibitors were required to keep proteins stable throughout the fractionation procedure. Cell debris was removed by centrifugation at 3,000 × g to produce a total-lysate fraction. Half of the total lysate was used as a control, while the remainder was fractionated by centrifugation at 8,300 × g for 15 min. The supernatant was placed into a fresh tube, and the pellet was resuspended in an equal amount of lysis buffer. SDS, glycerol, β-mercaptoethanol, and Tris-HCl (pH 6.8) were added to every sample up to final concentrations of 3%, 10%, 3%, and 0.15 M, respectively. Resulting samples were heated at 95°C for 10 min and run on the standard SDS-polyacrylamide gel. For the protein assays, gels were transferred onto Hybond ECL nitrocellulose membranes and reacted to the antibodies as described above.
The β-galactosidase activity assays.
For measuring β-galactosidase activity, yeast cultures were grown overnight at 30°C with shaking in liquid medium selective for the plasmids. Cell extracts were prepared by the standard procedure (23) and stored at −70°C. The β-galactosidase activity in extracts was measured by using a chemiluminescence assay (21), with modifications according to the most recent Tropix, Inc., protocol, using an Optocomp I luminometer (MGM Instruments, Inc.). The correspondence between relative light units and amounts of active β-galactosidase was determined by using standard solutions of pure β-galactosidase, purchased from Sigma. Galacton-Plus and Accelerator were purchased from Tropix; β-galactosidase activities were normalized per 1 mg of total cellular protein, the concentration of which was determined by the Bradford assay (Bio-Rad).
RESULTS
Ssa1 overproduction protects [PSI] from the effects of overproduced Hsp104.
As we have shown previously (5), a low-copy-number (centromeric) URA3 plasmid, containing the wild-type HSP104 gene (pYS104), inhibits the ability of [PSI] to suppress a nonsense mutation in the ADE1 gene, ade1-14UGA. Both a strong (OT56) and a weak (OT55) [PSI+] strain, both bearing the ade1-14 allele, became phenotypically Psi− when they carried this plasmid. That is, they did not grow on the synthetic medium which contained no adenine and was selective for the plasmid. To determine if the Hsp70 family protein Ssa1 altered the affect of Hsp104 on [PSI+], we cotransformed cells with both pYS104 and a multicopy SSA1 plasmid (pSSA1-LEU2). The cells containing both plasmids remained phenotypically Psi+: they grew in the absence of adenine (Fig. 1) and formed pink (OT55) or white (OT56) colonies on YPD, in contrast to more reddish colonies of the transformants bearing pYS104 alone (not shown). This result suggests that the Ssa protein interferes with Hsp104’s ability to inhibit [PSI]-mediated suppression.
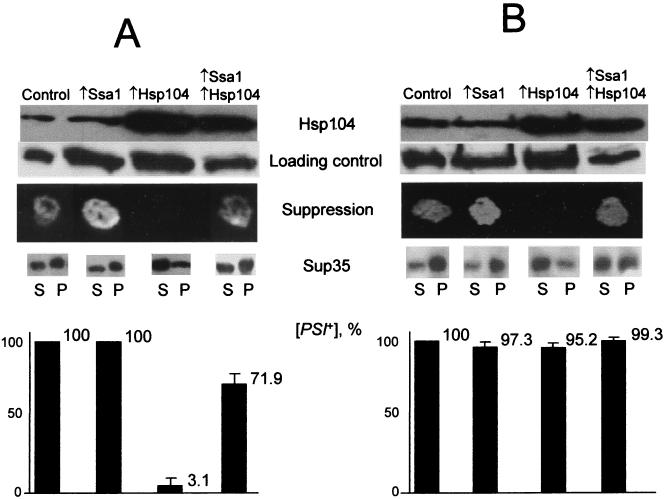
FIG. 1.
Effects of multicopy SSA1 plasmid on Hsp104-induced inhibition and curing of [PSI] in yeast strains OT55 (A) and OT56 (B). Plasmid combinations were as follows: control, YEp13 plus pRS316; Ssa1, pSSA1-LEU2 plus pRS316; Hsp104, YEp13 plus pYS104; Ssa1-Hsp104, pSSA1-LEU2 plus pYS104. (Plasmids YEp13 and pRS316, which contain no HSP genes, were used as matching controls.) To determine Hsp104 levels, proteins were isolated from the cultures growing in liquid −Ura-Leu/glucose medium at 25°C, separated by SDS-PAGE, and reacted to Hsp104-specific antibodies. Experiments were repeated at least two times with the same result and used at least two different transformants of each set. For the loading control, the same protein extracts were reacted to the Sup35 antibodies. In contrast to the previous findings by Paushkin et al. (36), levels of the Sup35 protein in these total lysates, which include both soluble and insoluble fractions (see description of Sup35 aggregation below), always remained constant in the strains used in this study regardless of the presence or absence of [PSI]. Relative protein amounts were also confirmed by Coomassie blue staining (not shown). To determine suppression, transformants grown on −Ura-Leu/glucose medium were replica plated onto −Ura-Leu-Ade/glucose medium and photographed after 6 (A) or 4 (B) days of incubation at 30°C. In each case, at least eight independent transformants for each plasmid combination were tested. No differences in growth between transformants were detected on the control −Ura-Leu/glucose (A and B) plates (not shown). To test for Sup35 aggregation, protein extracts, prepared from the cultures grown in liquid −Ura-His/glucose medium, were fractionated by centrifugation as described previously (35) (see Materials and Methods). For each strain, equal volumes of supernatant (S) and pellet (P) fractions were loaded onto the gel. Distributions of total cellular proteins between S and P fractions were similar for all strains, as verified by Coomassie blue staining (not shown). Experiments were repeated at least two times with at least two different transformants of each set. To examine [PSI] curing, cultures grown in liquid −Ura-Leu/glucose medium were diluted in H2O, plated onto the same medium (100 to 200 colonies per plate), grown for 3 to 4 days of incubation at 30°C, and replica plated onto 5-fluoroorotic acid medium to get rid of URA3-containing HSP104 plasmid; after 3 days of growth, cultures were replica plated onto −Ade medium and YPD medium, in order to identify [PSI+] and [psi−] colonies (see Materials and Methods). Ninety-five percent probability levels of variation are shown.
Next we examined whether the Ssa protein had prevented curing of [PSI] by Hsp104. To check whether the pYS104 plasmid had cured cells of [PSI] or had simply inhibited its phenotypic effect, transformants bearing HSP104 and SSA1 plasmids (or matching control plasmids) were incubated on −Ura-Leu medium, to select for the plasmids, and grown for 3 days at 30°C. Serial dilutions of each transformant were then plated onto −Ura-Leu medium to obtain single colonies, and these were then selected for loss of the URA3-containing pYS104 plasmid by replica plating colonies onto 5-fluoroorotic acid (23). Finally, colonies were replica plated on −Ade and YPD media to test for the presence of [PSI].
In the strong [PSI+] strain, OT56, the vast majority of colonies originating from the pYS104 transformants remained [PSI+] after plasmid was cured (Fig. 1B), and the suppressor efficiency of [PSI] had not been significantly changed, judging from both growth on −Ade and color on YPD. This finding indicates that expression of Hsp104 from this plasmid had temporarily inhibited the Psi+ phenotype, but had not efficiently cured cells of the element, as reported previously (5). In contrast, most pYS104 transformants of the weak [PSI+] strain, OT55, remained [psi−] after pYS104 was lost, indicating that Hsp104 had cured them of the [PSI] element. This correlation between the nature of the [PSI] element (strong versus weak) and the efficiency of Hsp104-induced curing is in agreement with our previous data (14). Notably, most of the OT55 transformants that had contained both pYS104 and pSSA1-LEU2 were [PSI+] when pYS104 was lost (Fig. 1A). This result indicates that Ssa1 can both negate the ability of Hsp104 to inhibit phenotypic expression of strong [PSI] elements and interfere with Hsp104-induced curing of weak [PSI] elements.
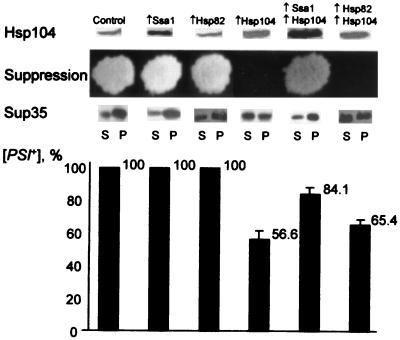
Strong [PSI+] strains such as OT56, which are [PSI+] after loss of the low-copy-number HSP104 plasmid, can be cured of [PSI] when HSP104 is expressed at a higher level from a strong inducible promoter (5). To determine if SSA1 overexpression is able to protect [PSI] against curing by high levels of Hsp104, we used the centromeric plasmids pGAL::SSA1-URA3 and pH28(HIS3), in which the GAL promoter was fused to the SSA1 and HSP104 genes, respectively. In strains carrying these constructs, SSA1 and HSP104 overexpression was induced by growth on medium containing galactose instead of glucose. Matching centromeric plasmids, pRS316GAL (URA3) and pLA1 (HIS3), without the Hsp104 and Ssa1 coding sequences were used as controls. All double-plasmid combinations were obtained in the strong [PSI+] strain OT56 and induced on galactose (see Materials and Methods). [PSI]-mediated suppression of ade1-14 on galactose medium and the proportion of cells that became [PSI+] and [psi−] after galactose induction were determined. GAL::HSP104-mediated inhibition of [PSI] suppression was efficient only in the absence of GAL::SSA1 (Fig. 2), and the curing of [PSI] by induced GAL::HSP104 was also reduced significantly in the presence of the induced GAL::SSA1 (Fig. 2). In the absence of GAL::HSP104, GAL::SSA1 did not cause [PSI] curing.
FIG. 2.
Hsp104 levels and effects on [PSI] in OT56 cells, which contain galactose-inducible SSA1 and HSP82 constructs. Plasmid combinations used were as follows: control, pRS316GAL plus pLA1; Ssa1, pGAL::SSA1-URA3 plus pLA1; Hsp82, pMC3 plus pLA1; Hsp104, pRS316GAL plus pH28; Ssa1 plus Hsp104, pGAL::SSA1-URA3 plus pH28; Hsp82 plus Hsp104, pMC3 plus pH28. (Plasmids pRS316GAL and pLA1, which contain no HSP genes, were used as matching controls.) Hsp104 levels were determined as described in the legend to Fig. 1 except that cultures were grown in the −Ura-His/galactose-raffinose medium at 25°C. All the samples shown contained the same amount of total protein loaded, as verified by Coomassie blue staining and/or by reaction to the Sup35-specific antibodies, as described for Fig. 1. Each experiment was repeated at least four times with at least two independent transformants per each plasmid combination. Background Hsp104 levels are higher than in Fig. 1 due to partial induction of the chromosomal HSP104 allele during growth on galactose, as described previously (41). To determine suppression, transformants grown on −Ura-His/glucose medium were replica plated onto −Ura-His-Ade/galactose medium and photographed after 6 days of incubation at 30°C. In each case, at least eight independent transformants for each plasmid combination were tested. No differences in growth between transformants were detected on the control −Ura-His/galactose plates (not shown). To test for Sup35 aggregation, protein extracts, prepared from the cultures grown in liquid −Ura-His/galactose-raffinose medium at 25°C, were fractionated by centrifugation as described elsewhere (35) (see Materials and Methods). For each strain, equal volumes of supernatant (S) and pellet (P) fractions were loaded onto the gel. Distributions of total cellular proteins between S and P fractions were similar for all strains, as verified by Coomassie blue staining (not shown). Experiments were repeated at least three times with at least two different transformants of each set. To examine [PSI] curing, cultures grown in liquid −Ura-His/galactose-raffinose medium at 25°C were washed twice with H2O, diluted in H2O, plated onto −Ura-His/glucose medium (100 to 200 colonies per plate), grown for 3 to 4 days at 30°C, and replica plated onto −Ura-His-Ade/glucose and YPD media in order to identify [PSI+] and [psi−] colonies. Ninety-five percent probability levels of variation are shown.
Ssa and Hsp104 protein levels in the overproducer strains.
The expression of Hsp70 is tightly controlled by autoregulatory mechanisms. Relatively low levels of Ssa induction in overproducer strains could be expected, due to both the presence of constitutively expressed members of the Ssa subfamily (52), which cross-react with the Ssa antibodies, and the ability of Ssa1 protein to down regulate the transcription of its own gene and that of other SSA genes (47). Densitometry analysis of the Western blots confirmed that exponentially growing cells, which bear multiple copies of the SSA1 gene, contain 1.53 ± 0.214-fold more Ssa protein than exponentially growing cells of isogenic transformants carrying the matching control vectors. Similar levels of Ssa overproduction (1.51 ± 0.127-fold) were achieved with the GAL::SSA1 construct on galactose medium. As previously described (42), Hsp104 overproduction had no effect on Ssa protein levels (not shown). Western blot results (Fig. 1 and 2), accompanied by densitometry measurements (not shown), also confirmed that the levels of overproduced Hsp104 in these strains were not affected by the overproduction of Ssa1. Both the centromeric pYS104 plasmid on glucose medium (Fig. 1) and the GAL::HSP104 plasmid on galactose medium (Fig. 2) caused an increase in Hsp104 protein levels independently of the Ssa1 plasmids. This result suggests that the ability of overproduced Ssa1 protein to protect [PSI] from excess Hsp104 is not due to a decrease in Hsp104 expression.
Excess Ssa1 prevents accumulation of soluble Sup35 protein in [PSI+] cells that overexpress Hsp104.
The Sup35PSI prion protein, found in [PSI+] cells, forms insoluble aggregates, in contrast to the normal soluble Sup35 protein in [psi−] cells (35, 36). It has been reported that when Hsp104 has cured cells of [PSI] aggregates, Sup35 protein is soluble. Quite remarkably, even low levels of Hsp104 overexpression (35, 36) led to a detectable shift in the ratio of soluble to insoluble Sup35 protein, although [PSI] curing was inefficient under these conditions.
Our results (Fig. 1 and 2) confirm that Hsp104 overproduction increases the amount of soluble Sup35 protein versus insoluble Sup35 protein in all strains and conditions tested. However, transformants which overexpress both SSA1 and HSP104 contained less soluble Sup35 protein and more insoluble Sup35 protein than transformants which overexpress HSP104 alone. Thus, at the molecular level, higher levels of Ssa1 interfere with the disappearance of Sup35PSI aggregates, observed during growth in the presence of increased levels of Hsp104. In cells expressing Hsp104 at wild-type levels, Ssa1 overproduction showed no reproducible effect on the ratio between soluble and insoluble Sup35 protein (Fig. 1 and 2).
Increased levels of Hsp82 protein do not interfere with Hsp104’s effect on [PSI].
The expression of Hsp82 is also increased under conditions that induce Hsp104 and Hsp70 (Ssa1): heat shock, stationary-phase growth, and sporulation (2). To determine whether Hsp82 modifies the effect of Hsp104 on [PSI], GAL::HSP82 and GAL::HSP104 constructs were induced individually and simultaneously in the [PSI+] strain OT56, as described for Ssa1. Hsp82 overproduction was verified by Western blotting (not shown). Normal [PSI] maintenance and [PSI]-mediated suppression were not affected by Hsp82 overproduction, and Hsp104-mediated inhibition and curing of [PSI] were not affected by excess Hsp82 (Fig. 2). We also examined whether excess Hsp82 affected the solubility of Sup35. There was no effect of excess Hsp82 on the distribution of Sup35 protein between the soluble and insoluble fractions in cells that expressed Hsp104 at normal levels or in cells that expressed it at high levels (Fig. 2). Similar results were obtained with the strain OT55 (not shown).
Excess Ssa1 increases the efficiency of nonsense suppression in [PSI+] strains.
We also examined the effect of Ssa1 on nonsense suppression in [PSI+] strains that do not carry Hsp104-overproducing plasmids. Our previous data (5, 31) suggested that multiple copies of the SSA1 gene increased the growth of some [PSI+] strains on media selective for nonsense suppression and inhibited the growth of other [PSI+] strains. In both OT55 and OT56 strains bearing the [PSI]-suppressible ade1-14UGA mutation, plasmid pSSA1-LEU2 increased growth on −Ade medium and decreased accumulation of the red pigment (not shown). A problem in interpreting this observation is that Ssa1 protein is involved in the general stress response, which helps cells survive stressful conditions. It requires 5 to 8 days to form an individual colony on −Ade medium through [PSI]-mediated suppression of the ade1-14 allele, whereas wild-type cells form colonies on this medium in 2 days. Thus, better growth of some strains on −Ade medium might be due to an effect of Ssa1 overexpression on growth under stressful conditions, rather than an effect on [PSI]-mediated suppression. On the other hand, Ssa1 overproduction is known to be harmful to some yeast strains (47), and this could explain the inhibition of growth in some Ssa1 overproducers on −Ade medium. To directly measure whether Ssa1 affects [PSI]-mediated suppression in OT55 and OT56, we used a quantitative assay for the readthrough of nonsense codons. Plasmid pUKC815 bears the promoter and N-terminal end of the yeast PGK gene fused in frame to the bacterial β-galactosidase gene (PGK-lacZ). In the matching vector for monitoring nonsense suppression, pUKC819, the PGK and lacZ open reading frames are interrupted by a UGA mutation (16) (see Materials and Methods). The β-galactosidase levels per 1 mg of total cellular protein were determined in OT56 derivatives transformed with either of these vectors, together with plasmid pSSA1-LEU2 or the YEp13 control plasmid. Efficiencies of suppression were calculated as ratios of β-galactosidase activities in matched strains carrying the two fusion constructs. Cells which contained the multicopy SSA1 plasmid exhibited three- to fourfold-higher efficiencies of suppression than cells carrying the vector control (Table 3). Therefore, results of all three assays used to measure nonsense suppression by [PSI] (i.e., growth on −Ade medium, color, and PGK-lacZ readthrough) correlated to each other in OT56 genetic background. This confirms that increased growth on −Ade medium and decreased accumulation of the red pigment, caused by excess Ssa1 protein in OT56 and isogenic OT55, result from a stimulating effect of excess Ssa1 on [PSI]-mediated nonsense-suppression rather than from the secondary effects on growth.
TABLE 3.
Effect of multicopy SSA1 plasmid on [PSI]-mediated nonsense suppression in strain OT56
| Plasmid | β-Galactosidase (ng)/mg of total protein
|
Suppression (%; mean ± SE) | |
|---|---|---|---|
| Readthrough (pUKC815) | UGA (pUKC819) | ||
| Control (YEp13) | 1,293 | 4.35 | 0.34 ± 0.16 |
| SSA1 (pSSA1-LEU2) | 835 | 12.5 | 1.41 ± 0.38 |
Determined as ratio UGA/readthrough. At least four independent transformants were analyzed per each strain-plasmid combination; at least two measurements were performed per each transformant.
Excess Ssa does not produce nonsense suppression in cells carrying a deletion of Hsp104.
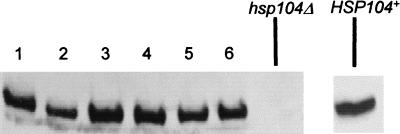
Deletion of HSP104 in a [PSI+] strain heritably cures cells of [PSI], eliminating nonsense suppression. Because Ssa1 increases the efficiency of nonsense suppression in cells that overexpress Hsp104 and in cells with wild-type levels of Hsp104, we next examined whether it would promote sufficient nonsense suppression to compensate for an hsp104 deletion. The hsp104::URA3 disruption strain L-1607, which bears the [PSI]-suppressible ade1-14 mutation, was transformed with the multicopy pSSA1-LEU2 plasmid and crossed to the [PSI+] strains GT1-S31 and GT56-13B. The resulting diploids, which are homozygous for ade1-14 and ura3 and heterozygous for the hsp104::URA3 disruption, were [PSI+]. Isogenic diploids bearing the matching vector without SSA1, YEp13, served as controls. The diploids were sporulated and dissected. Haploid progeny that grew on −Ura-Leu medium, and presumably carried the hsp104::URA3 deletion and plasmid pSSA1-LEU2 or YEp13, were tested for the ability to grow on −Ade medium. None did. To increase the number of colonies that could be tested, we used random spore platings to select cells that could grow on −Ura-Leu medium. (Rare surviving diploids were identified by their inability to mate and were discarded.) In both control and Ssa1-overproducing strains, most of the selected colonies were [psi−] and could not grow on −Ade medium (Table 4). To determine if the rare [PSI+] Ura+ haploids that carried plasmid pSSA1-LEU2 contained the hsp104::URA3 deletion, proteins were examined by Western blotting. All six colonies tested expressed full-length Hsp104 protein (Fig. 3). Presumably, these originated from meiotic gene conversion of the ura3 marker. Thus, a multicopy SSA1 plasmid cannot produce nonsense suppression in cells carrying an hsp104 deletion.
TABLE 4.
Analysis of the meiotic progeny of HSP104+/hsp104::URA3 diploids
| Plasmid | Diploid | Ura+ Leu+ spore clones
|
||
|---|---|---|---|---|
| [PSI+] | [psi−] | Total | ||
| pSSA1-LEU2 | GT1-S31 × L-1607 | 6 | 111 | 117 |
| GT56-13B × L-1607 | 1 | 109 | 110 | |
| Total (%) | 7 (3.1) | 220 | 227 | |
| YEp13 | GT1-S31 × L-1607 | 19 | 160 | 179 |
| GT56-13B × L-1607 | 17 | 151 | 168 | |
| Total | 36 (10.0) | 311 | 347 | |
FIG. 3.
Western analysis of the exceptional Ura+ [PSI+] spore clones from the diploids heterozygous by hsp104::URA3 disruption. Proteins were isolated from six exceptional Ura+ [PSI+] spore clones, obtained in the meiotic progeny of the diploids GT1-S31 × L-1607 and GT56-13B × L-1607, which bear plasmid pSSA1-LEU2 (lanes 1 to 6), and control hsp104 deletion strain L-1607. Extracts were subjected to SDS-PAGE, transferred onto a nitrocellulose filter, and reacted to Hsp104-specific antibodies. All extracts contained similar amounts of the total cellular protein as verified by Coomassie blue staining (not shown).
Effects of excess Ssa1 protein on Hsp104-mediated thermotolerance.
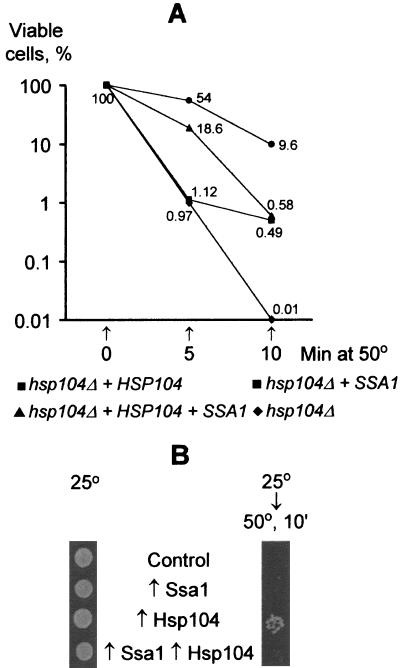
Next, we tested whether Ssa1 would interfere with the other known function of Hsp104. Hsp104 is a stress tolerance factor that greatly increases survival in yeast cells exposed to high temperatures (40, 41). This activity of Hsp104 correlates with its ability to promote the solubilization of aggregated proteins damaged by heat (34). In the absence of Hsp104, excess Ssa1 protein partially compensates for the temperature tolerance defect (42). Moreover, Ssa1 protein cooperates with Hsp104 in the reactivation of aggregated proteins in vitro (17). Ssa proteins appear to have at least two roles in stress tolerance: they bind unfolded proteins, reducing their tendency to aggregate, and they assist in the Hsp104-mediated reactivation of proteins that have already aggregated. We investigated the effects of Ssa1 on the basal and inducible thermotolerance of yeast cells that contained various levels of Hsp104. Exponentially growing cells were grown at 25°C, the conditions under which effects on [PSI]-mediated suppression were analyzed. The cells were then shifted directly to 50°C to measure basal thermotolerance. An hsp104 deletion derivative of OT46 carrying control vectors was extremely sensitive to a 50°C heat shock: only 0.01% of the cells were alive after a 10-min exposure (Fig. 4A). Under the same conditions, the viability of isogenic strains containing either the multicopy SSA1 plasmid or the low-copy-number HSP104 plasmid was higher, roughly 50- or 1,000-fold, respectively. Thus, as previously described (30, 40–42), both the HSP104 gene and multiple copies of the SSA1 gene are able to protect yeast cells against temperature-induced killing, and the effect of HSP104 is much stronger than that of SSA1. However, the OT46 transformant, which contained both multiple copies of SSA1 and a low-copy-number HSP104 plasmid, was about 15-fold less viable after 10 min at 50°C and about 3-fold less viable after 5 min at 50°C than the isogenic transformant containing HSP104 alone (Fig. 4A). Thus, multiple copies of SSA1 interfere with the temperature tolerance provided by Hsp104.
FIG. 4.
Effects of Ssa1 and Hsp104 on temperature tolerance of yeast cells. Temperature tolerance assays were performed as described in Materials and Methods. Plasmid designations are the same as in Fig. 1. Experiments were repeated twice (A) or four times (B) with similar results.
The same experiment was performed with strain OT56, which contains an intact chromosomal copy of the HSP104 gene (qualitative results in Fig. 4B; quantitative results not shown). Addition of the single extra copy of HSP104 increased the temperature resistance of OT56. Interestingly, our results indicate that strain OT56, carrying a single chromosomal copy of HSP104, is less resistant to 50°C treatment than strain OT46, carrying the HSP104 gene on the centromeric plasmid. This is likely because the plasmid-borne HSP104 gene yields higher constitutive expression than the chromosomal gene (reference 5 and Fig. 1). Temperature protection by multiple copies of SSA1 was not detected in OT56. However, the OT56 transformant containing both multiple copies of SSA1 and a single extra copy of HSP104 was less temperature resistant than one containing HSP104 alone, confirming an inhibitory effect of Ssa1 on basal thermotolerance in cells expressing Hsp104.
When OT46 and OT56 cultures were preincubated at 37°C to induce thermotolerance, the temperature resistance of all transformants containing HSP104 was greatly increased at 50°C, as reported previously for the other yeast strains (40, 41). Under these conditions, multiple copies of SSA1 did not affect temperature resistance of the strains containing the HSP104 plasmid as strongly as they did in the absence of the 37°C pretreatment. We detected an Hsp104-antagonizing effect of SSA1 only twice in four such experiments (data not shown). We also examined the temperature resistance of OT56 transformants bearing the galactose-inducible GAL::SSA1 and/or GAL::HSP104 plasmids. Again, GAL::HSP104 conferred temperature resistance to cells growing in galactose medium, independently of the GAL::SSA1 plasmid (data not shown). Apparently, excess Ssa1 protein interferes with Hsp104-mediated temperature tolerance only under some culture conditions. This may depend on both levels of Hsp104 induction and the effects of other factors induced by temperature pretreatment or growth on galactose.
DISCUSSION
We have shown that at least one of the proteins of the Hsp70 family, Ssa1, is able to protect the prion-like genetic element known as [PSI] from Hsp104-induced curing. This helps to resolve a major paradox concerning the inheritance of [PSI]. Transient selective overproduction of Hsp104 is sufficient to eliminate the [PSI] element (5), providing one of the strongest genetic arguments that [PSI] represents a new type of inheritance based upon the transmission of proteins with alternative, self-perpetuating structures. However, if Hsp104 is the only factor controlling [PSI], the [PSI] element should be lost during growth at high temperatures or in stationary phase, when Hsp104 levels increase. Since Ssa levels are also increased under these conditions, our results provide a likely explanation for ability of genetic information carried by [PSI] to be preserved in the face of environmental fluctuations.
The Ssa Hsp70 subfamily includes, in addition to Ssa1, the Ssa2, Ssa3, and Ssa4 proteins (47, 51, 52). These proteins exhibit different patterns of expression. For example, while Ssa2 is expressed at a high constitutive level in exponential cells, Ssa1 expression is increased at high temperatures, and Ssa3 and Ssa4 are normally expressed only at high temperatures. During stationary-phase growth, on the other hand, Ssa3 is the most strongly induced (51). These proteins are highly homologous and have strongly overlapping functions in other protein-folding processes. Thus, any or all of them is likely to play a role in the maintenance of [PSI], depending on the particular environmental conditions. Determining with certainty which (if any) members of the family preferentially effect [PSI+] will not be easy in vivo. We have constructed several [PSI+] strains bearing multiple ssa deletions (ssa1,3, ssa1,2,3, and ssa1,3,4) in various genetic backgrounds. In some (but not all) of these multiple-deletion strains, frequencies of [PSI] loss during growth at 37°C were increased markedly (e.g., loss in 5 to 20% of cells, compared to less than 0.5% at 25°C) (7). However, Western blots revealed that multiple ssa deletion strains still contain near wild-type levels of Ssa protein. This result is apparently due to compensatory induction of the remaining member(s) of SSA family in the cells bearing multiple ssa deletions, as described previously (1, 56). Simultaneous inactivation of all four SSA genes is lethal (52). Therefore, it is not yet possible to tell whether Ssa is the only factor protecting [PSI] from Hsp104 during growth at high temperature and whether Ssa is required for [PSI] maintenance in the normal conditions. Moreover, Hsp70 proteins regulate not only their own expression but also that of many other protein-folding agents, including other Hsps (12) and trehalose (19). We have shown that at least one of these, Hsp82 (a yeast homolog of the mammalian Hsp90) does not play an important role in [PSI] maintenance. However, we do not know whether Ssa1’s affects on [PSI] are an indirect consequence of its position in these regulatory circuits or direct consequence of interaction with Sup35, the protein determinant of [PSI].
Previous results suggest that Ssa protein does not influence Sup35 secondary structure in vitro (43). However, this does not rule out a possibility of direct interaction between Ssa and Sup35, which would not have a major effect on the secondary structure. In vivo interaction could also be assisted by other proteins, which are not present in vitro. It is also possible that Ssa could specifically recognize a prion isoform of the Sup35. Further experiments to determine whether there are specific in vivo interactions between Ssa and Sup35 are under way.
Ssa1 and Hsp104 functions also interface in another realm, providing cells with tolerance to heat stress. In vivo, Ssa1 overproduction partially compensates for the loss of temperature tolerance in the absence of Hsp104 (reference 42 and confirmed herein). Moreover, whereas Hsp104 is dispensable for growth at all temperatures in a wild-type background, it is essential for growth at high temperatures when Ssa protein levels are reduced (42). Hsp104 functions in stress tolerance as a “molecular crowbar,” promoting the resolubilization of heat-damaged proteins (34). In vitro work points to two different roles for Ssa in stress tolerance. First, it binds unfolded proteins and prevents them from aggregating, reducing the requirement for Hsp104’s disaggregating function. Second, together with another chaperone, Ydj1, Ssa1 helps previously aggregated proteins, targets of Hsp104’s disaggregating function, return to the folded state (17). Our finding that overexpressing Ssa1 during log-phase growth on glucose interferes with Hsp104’s thermotolerance functions when cells are shifted directly to high temperatures (Fig. 4) was, therefore, unexpected. There are two likely explanations.
First, at certain chaperone concentrations and/or with certain substrates, Ssa might interfere with Hsp104’s resolubilizing activity. It might do so either by binding directly to substrates and preventing Hsp104’s interaction with them or by titrating the free cellular concentration of Hsp104 cofactors, such as Ydj1. This same mechanism might account for Ssa1’s ability to interfere with Hsp104-mediated curing of [PSI]. That is, Hsp104 might cure cells of [PSI] simply by disaggregating previously aggregated Sup35, and Ssa1 might interfere with this disaggregation. Indeed, we find that a greater fraction of Sup35 remains in the pellet after lysate fractionation in cells that overexpress both Ssa1 and Hsp104 than in cells that overexpress Hsp104 alone (Fig. 1 and 2). However, we also find that under some conditions (for example, after a temperature pretreatment), Ssa1 overexpression does not significantly interfere with Hsp104-mediated thermotolerance. If it is Ssa’s interference with Hsp104 mediated [PSI] curing that saves the [PSI] elements during heat shock, some aspect of Ssa’s effects on Sup35 aggregates and heat-damaged aggregates would have to diverge under these conditions. An attractive possibility is that while Ssa is assisting Hsp104 in solubilizing the amorphous aggregates, it protects highly ordered protein complexes (e.g., cytoskeletal networks) from the disassembling effect of the stress-induced Hsp104. This would explain why Hsp104 normally has no effect on cytoskeletal structures. Prion polymers, which resemble some patterns of the highly ordered structures, could also be protected by Ssa to some extent. Indeed, mammalian Hsp70 proteins were shown to protect some cytoskeletal components (in particular, the centrosome and intermediate filaments) during heat shock (27).
A second explanation for Ssa1’s interference with Hsp104-mediated thermotolerance in log-phase cells shifted to high temperatures is that it acts indirectly, down regulating the basal expression of other thermotolerance or growth factors. Conditioning preheat treatments, by recruiting Ssa proteins away from regulatory factors (11), would derepress these factors and restore full thermotolerance. These other factors might also interact with Sup35 protein and influence its folding transitions during heat shock and stationary-phase growth, but the specific roles of these factors and the nature of their interaction with Sup35 is unclear. Indeed, the molecular mechanisms of Hsp104’s effects on [PSI] and Sup35 themselves remain to be uncovered. The surprising observation that both overexpression and inactivation of Hsp104 can cure cells of [PSI] has been explained by role of Hsp104 in forming of partially unfolded conversion intermediate (5), by stochiometric interaction between such an intermediate and Hsp104 hexamer (35), or by ability of Hsp104 to promote [PSI] segregation by breaking down huge aggregates into the small aggregation “seeds” (36). However, none of these models have been directly tested due to inherent difficulties of the analysis of aggregation-prone substrates in vitro.
The remarkable hypothesis that a heritable phenotypic change in yeast could be transmitted by a heritable change in protein structure, with no underlying change in a nucleic acid, was first proposed in 1994 (53). Since then a great deal of genetic, cell biological, and biochemical data has provided compelling support. We are still a long way from understanding the specific physical mechanisms that promote the underlying changes in protein state. However, we are now beginning to appreciate the complexity with which the [PSI] factor interfaces with the biology of yeast cells. We have now linked the expression of a second major yeast chaperone, a member of the evolutionarily conserved Hsp70 chaperone family whose expression changes in response to the environment, to the forces that control [PSI] inheritance. Therefore, one group of chaperone proteins plays a major role in both adaptation to the temperature stress and control of prion maintenance. This finding provides a framework for discovering how [PSI] is maintained in the face of environmental fluctuations. Moreover, further understanding of [PSI] propagation will serve as a powerful tool of investigating the molecular pathways by which cells respond to environmental changes. The question arises: Is [PSI] a beneficial factor that has prompted the evolution of mechanisms for its maintenance under diverse conditions? Or has [PSI], like viruses and transposable elements, learned to take advantage of complex regulatory mechanisms in the cell to promote its own propagation?
ACKNOWLEDGMENTS
We are grateful to A. D. Zink for help in performing the temperature resistance assays. We thank E. Craig for the Ssa-specific antibodies and M. Tuite for the gift of plasmids pUKC815 and pUKC819.
This work was supported by the grant 1R21GM55091 from the National Institute of General Medical Sciences to Y.O.C. and by Howard Hughes Medical Institute funds to S.L.L.
REFERENCES
- 1.Boorstein W R, Craig E A. Structure and regulation of the SSA4 HSP70 gene of Saccharomyces cerevisiae. J Biol Chem. 1991;265:18912–18921. [PubMed] [Google Scholar]
- 2.Borkovich K A, Farelly F W, Finkelstein D B, Taulien J, Lindquist S. Hsp82 is an essential protein that is required in higher concentrations for growth of cells at higher temperatures. Mol Cell Biol. 1989;9:3919–3930. doi: 10.1128/mcb.9.9.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Broach J R, Strathern J N, Hicks J B. Transformation in yeast: development of a hybrid cloning vector and isolation of the CAN1 gene. Gene. 1979;8:121–133. doi: 10.1016/0378-1119(79)90012-x. [DOI] [PubMed] [Google Scholar]
- 4.Chernoff Y O, Derkach I L, Inge-Vechtomov S G. Multicopy SUP35 gene induces de-novo appearance of psi-like factors in the yeast Saccharomyces cerevisiae. Curr Genet. 1993;24:268–270. doi: 10.1007/BF00351802. [DOI] [PubMed] [Google Scholar]
- 5.Chernoff Y O, Lindquist S L, Ono B, Inge-Vechtomov S G, Liebman S W. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+] Science. 1995;268:880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- 6.Chernoff Y O, Liebman S W, Patino M M, Lindquist S W. Response from Chernoff et al. [Re: R. B. Wickner, Prions of yeast and heat-shock protein 104: ‘coprion’ and cure.] Trends Microbiol. 1995;3:369. doi: 10.1016/s0966-842x(00)88978-7. [DOI] [PubMed] [Google Scholar]
- 7.Chernoff, Y. O., and G. P. Newnam. Unpublished data.
- 8.Coustou V, Deleu C, Saupe S, Begueret J. The protein product of the het-s heterokaryon incompatibility gene of the fungus Podospora anserina behaves as a prion analog. Proc Natl Acad Sci USA. 1997;94:9773–9778. doi: 10.1073/pnas.94.18.9773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cox B S. PSI, a cytoplasmic suppressor of super-suppressor in yeast. Heredity. 1965;20:505–521. [Google Scholar]
- 10.Cox B S, Tuite M F, McLaughlin C S. The PSI factor of yeast: a problem in inheritance. Yeast. 1988;4:159–178. doi: 10.1002/yea.320040302. [DOI] [PubMed] [Google Scholar]
- 11.Craig E A, Gross C A. Is hsp70 the cellular thermometer? Trends Biochem Sci. 1991;16:135–140. doi: 10.1016/0968-0004(91)90055-z. [DOI] [PubMed] [Google Scholar]
- 12.Craig E A, Jacobsen K. Mutations of the heat inducible 70 kilodalton genes of yeast confer temperature sensitive growth. Cell. 1984;39:841–849. doi: 10.1016/0092-8674(84)90279-4. [DOI] [PubMed] [Google Scholar]
- 13.DebBurman S K, Raymond G J, Caughey B, Lindquist S. Chaperone supervised conversion of prion protein to its proteinase resistant form. Proc Natl Acad Sci USA. 1997;94:13938–13943. doi: 10.1073/pnas.94.25.13938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Derkatch I L, Chernoff Y O, Kushnirov V V, Inge-Vechtomov S G, Liebman S W. Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae. Genetics. 1996;144:1375–1386. doi: 10.1093/genetics/144.4.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Derkatch I L, Bradley M, Zhou P, Chernoff Y O, Liebman S W. Genetic and environmental factors affecting the de novo appearance of the [PSI+] prion in Saccharomyces cerevisiae. Genetics. 1997;147:507–519. doi: 10.1093/genetics/147.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Firoozan M, Grant C, Duarte J A B, Tuite M F. Quantitation of readthrough of termination codons in yeast using a novel gene fusion assay. Yeast. 1991;7:173–183. doi: 10.1002/yea.320070211. [DOI] [PubMed] [Google Scholar]
- 17.Glover J, Lindquist S. Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell. 1998;94:1–20. doi: 10.1016/s0092-8674(00)81223-4. [DOI] [PubMed] [Google Scholar]
- 18.Glover J R, Kowal A S, Schirmer E C, Patino M M, Liu J-J, Lindquist S. Self-seeded fibers formed by Sup35, the protein determinant of [PSI+], a heritable prion-like factor of Saccharomyces cerevisiae. Cell. 1997;89:811–819. doi: 10.1016/s0092-8674(00)80264-0. [DOI] [PubMed] [Google Scholar]
- 19.Hottiger T, De Virgilio C, Bell W, Boller T, Wiemken A. The 70-kilodalton heat-shock proteins of the SSA subfamily negatively modulate heat-shock-induced accumulation of trehalose and promote recovery from heat stress in the yeast, S. cerevisiae. Eur J Biochem. 1992;210:125–132. doi: 10.1111/j.1432-1033.1992.tb17399.x. [DOI] [PubMed] [Google Scholar]
- 20.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jain V K, Magrath I T. A chemiluminescent assay for quantitation of β-galactosidase in the femtogram range: application to quantitation of β-galactosidase in lacZ-transfected cells. Anal Biochem. 1991;199:119–124. doi: 10.1016/0003-2697(91)90278-2. [DOI] [PubMed] [Google Scholar]
- 22.Jarrett J T, Lansbury P T. Seeding “one-dimensional crystallization” of amyloid: a pathogenic mechanism in Alzheimer’s disease and scrapie? Cell. 1993;73:1055–1058. doi: 10.1016/0092-8674(93)90635-4. [DOI] [PubMed] [Google Scholar]
- 23.Kaiser C, Michaelis S, Mitchell A. Methods in yeast genetics. A Cold Spring Harbor laboratory course manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 24.King C-Y, Tittmann P, Gross H, Gebert R, Aebi M, Wuthrich K. Prion-inducing domain 2-114 of yeast Sup35 protein transforms in vitro into amyloid-like filaments. Proc Natl Acad Sci USA. 1997;94:6618–6622. doi: 10.1073/pnas.94.13.6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kurtz S, Rossi J, Petko L, Lindquist S. An ancient developmental induction: heat-shock proteins induced in sporulation and oogenesis. Science. 1986;231:1154–1157. doi: 10.1126/science.3511530. [DOI] [PubMed] [Google Scholar]
- 26.Lansbury P T, Caughey B W. The chemistry of scrapie reaction: the “ice 9” metaphore. Chem Biol. 1995;2:1–5. doi: 10.1016/1074-5521(95)90074-8. [DOI] [PubMed] [Google Scholar]
- 27.Liang P, McRae T H. Molecular chaperones and the cytoskeleton. J Cell Sci. 1997;110:1431–1440. doi: 10.1242/jcs.110.13.1431. [DOI] [PubMed] [Google Scholar]
- 28.Liautard J-P. A hypothesis on the aetiology of Alzheimer’s disease: description of a model involving a misfolded chaperone. Med Hypotheses. 1994;43:372–380. doi: 10.1016/0306-9877(94)90012-4. [DOI] [PubMed] [Google Scholar]
- 29.Lindquist S. Mad cows meet psi-chotic yeast: the expansion of prion hypothesis. Cell. 1997;89:495–498. doi: 10.1016/s0092-8674(00)80231-7. [DOI] [PubMed] [Google Scholar]
- 30.Lindquist S, Kim G. Heat-shock protein 104 expression is sufficient for thermotolerance in yeast. Proc Natl Acad Sci USA. 1996;93:5301–5306. doi: 10.1073/pnas.93.11.5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lindquist S, Patino M M, Chernoff Y O, Kowal A S, Singer M A, Lee K-H, Blake T, Liebman S W. The role of Hsp104 in stress tolerance and [PSI+] propagation in S. cerevisiae. Cold Spring Harbor Symp Quant Biol. 1995;60:451–460. doi: 10.1101/sqb.1995.060.01.050. [DOI] [PubMed] [Google Scholar]
- 32.Liu H, Krizek J, Bretscher A. Construction of a GAL1-regulated yeast cDNA expression library and its application to the identification of genes whose overexpression causes lethality in yeast. Genetics. 1992;132:665–673. doi: 10.1093/genetics/132.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parsell D A, Sanchez Y, Stitzel J D, Lindquist S. Hsp104 is a highly conserved protein with two essential nucleotide-binding sites. Nature. 1991;353:270–273. doi: 10.1038/353270a0. [DOI] [PubMed] [Google Scholar]
- 34.Parsell D A, Kowal A S, Singer M A, Lindquist S. Protein disaggregation mediated by heat-shock protein Hsp104. Nature. 1994;372:475–478. doi: 10.1038/372475a0. [DOI] [PubMed] [Google Scholar]
- 35.Patino M M, Liu J-J, Glover J R, Lindquist S. Support for the prion hypothesis for inheritance of a phenotypic trait in yeast. Science. 1996;273:622–626. doi: 10.1126/science.273.5275.622. [DOI] [PubMed] [Google Scholar]
- 36.Paushkin S V, Kushnirov V V, Smirnov V N, Ter-Avanesyan M D. Propagation of the yeast prion-like [psi+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J. 1996;15:3127–3134. [PMC free article] [PubMed] [Google Scholar]
- 37.Paushkin S V, Kushnirov V V, Smirnov V N, Ter-Avanesyan M D. In vitro propagation of the prion-like state of yeast Sup35 protein. Science. 1997;277:381–383. doi: 10.1126/science.277.5324.381. [DOI] [PubMed] [Google Scholar]
- 38.Prusiner S B. Molecular biology and pathogenesis of prion diseases. Trends Biochem Sci. 1996;21:482–487. doi: 10.1016/s0968-0004(96)10063-3. [DOI] [PubMed] [Google Scholar]
- 39.Prusiner S B, McKinley M P, Bowman K A, Bolton D C, Bendheim P E, Groth D F, Glenner G G. Scrapie prions aggregate to form amyloid-like birefringent rods. Cell. 1983;35:349–358. doi: 10.1016/0092-8674(83)90168-x. [DOI] [PubMed] [Google Scholar]
- 40.Sanchez Y, Lindquist S L. HSP104 required for induced thermotolerance. Science. 1990;248:1112–1115. doi: 10.1126/science.2188365. [DOI] [PubMed] [Google Scholar]
- 41.Sanchez Y, Taulien J, Borkovich K A, Lindquist S. Hsp104 is required for tolerance to many forms of stress. EMBO J. 1992;11:2357–2364. doi: 10.1002/j.1460-2075.1992.tb05295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sanchez Y, Parsell D A, Taulien J, Vogel J L, Craig E A, Lindquist S. Genetic evidence for a functional relationship between Hsp104 and Hsp70. J Bacteriol. 1993;175:6484–6491. doi: 10.1128/jb.175.20.6484-6491.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schirmer E C, Lindquist S. Interactions of the chaperone Hsp104 with yeast Sup35 and mammalian PrP. Proc Natl Acad Sci USA. 1997;94:13932–13937. doi: 10.1073/pnas.94.25.13932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Singh A, Helms C, Sherman F. Mutation of the non-mendelian suppressor, psi+, in yeast by hypertonic media. Proc Natl Acad Sci USA. 1979;76:1952–1956. doi: 10.1073/pnas.76.4.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Srikantha T, Dhar B, Bustin M. Expression of human chromosomal proteins HMC-14 and HMC-17 in Saccharomyces cerevisiae. Exp Cell Res. 1990;191:71–75. doi: 10.1016/0014-4827(90)90037-b. [DOI] [PubMed] [Google Scholar]
- 47.Stone D E, Craig E A. Self-regulation of 70-kilodalton heat shock proteins in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:1622–1632. doi: 10.1128/mcb.10.4.1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tuite M F, Mundy C R, Cox B S. Agents that cause a high frequency of genetic change from [psi+] to [psi−] in Saccharomyces cerevisiae. Genetics. 1981;98:691–711. doi: 10.1093/genetics/98.4.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tuite M F, Lindquist S L. Maintenance and inheritance of yeast prions. Trends Genet. 1996;12:467–471. doi: 10.1016/0168-9525(96)10045-7. [DOI] [PubMed] [Google Scholar]
- 50.Weissmann C. The Ninth Datta Lecture. Molecular biology of transmissible spongiform encephalopathies. FEBS Lett. 1996;389:3–11. doi: 10.1016/0014-5793(96)00610-2. [DOI] [PubMed] [Google Scholar]
- 51.Werner-Washburne M, Craig E A. Expression of members of the Saccharomyces cerevisiae hsp70 multigene family. Genome. 1989;31:684–689. doi: 10.1139/g89-125. [DOI] [PubMed] [Google Scholar]
- 52.Werner-Washburne M, Stone D E, Craig E A. Complex interactions among members of an essential subfamily of hsp70 genes in Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:2568–2577. doi: 10.1128/mcb.7.7.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wickner R B. [URE3] as an altered URE2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science. 1994;264:566–569. doi: 10.1126/science.7909170. [DOI] [PubMed] [Google Scholar]
- 54.Wickner R B. Prions of yeast and heat-shock protein 104: ‘co-prion’ and cure. Trends Microbiol. 1995;3:367–369. doi: 10.1016/s0966-842x(00)88978-7. [DOI] [PubMed] [Google Scholar]
- 55.Wickner R B, Masison D C, Edskes H K. [PSI] and [URE3] as yeast prions. Yeast. 1995;11:1671–1685. doi: 10.1002/yea.320111609. [DOI] [PubMed] [Google Scholar]
- 56.Unno K, Kishido T, Hosaka M, Okada S. Role of Hsp70 subfamily, ssa, in protein folding in yeast cells, seen in luciferase-transformed ssa mutants. Biol Pharm Bull. 1997;20:1240–1244. doi: 10.1248/bpb.20.1240. [DOI] [PubMed] [Google Scholar]