Abstract
Human coronaviruses exhibit a spectrum of symptoms, ranging from mild seasonal colds to severe respiratory manifestations. Despite progress in understanding the host's innate defense mechanisms against coronaviruses, how these viruses manipulate the immune response to promote inflammation remains elusive. In this study, we unveil the role of the coronavirus nonstructural protein 14 (NSP14) in leveraging the host's linear ubiquitin chain assembly complex (LUBAC) to instigate NF-κB activation, thereby triggering proinflammatory responses. Our findings uncover that HOIL-1-interacting protein (HOIP) directly engages with NSP14, conferring linear polyubiquitin chains onto NSP14. Consequently, ubiquitinated NSP14 recruits NEMO and initiates the activation of the IKK complex. This NSP14-induced NF-κB activation stimulates the expression of proinflammatory factors but not type I interferon, leading to a skewed host innate immune response tilting to inflammation. Collectively, our study sheds light on a virus-initiated linear ubiquitination pathway that induces NF-κB signaling and provokes proinflammatory responses.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12964-024-01949-4.
Keywords: Coronavirus, NSP14, Linear ubiquitination, LUBAC, HOIP
Introduction
Coronaviruses are a group of single-stranded positive-sense RNA viruses with a broad spectrum of vertebrate hosts [1]. Within the human population, seven human coronaviruses (HCoVs) have been identified, among which is the pandemic strain, SARS-CoV-2. While HCoV-229E, HCoV-OC43, HCoV-HKU1, and HCoV-NL63 typically induce mild symptoms akin to the common cold, MERS-CoV, SARS-CoV, and SARS-CoV-2 are notably pathogenic, leading to severe lower respiratory tract damage, pneumonia, and bronchiolitis in humans [2, 3]. The COVID-19 pandemic has inflicted devastating consequences, resulting in significant loss of life. Despite the development of mRNA vaccines and antiviral drugs, the emergence of vaccine-resistant strains and viral mutations necessitates frequent updates of vaccine and antivirals. Moreover, the potential for future coronavirus pandemics underscores the urgent need to discover a universal target applicable to all coronaviruses.
Activation of the RIG-I-like receptor (RLR) signaling pathway triggers the expression of proinflammatory cytokines and interferon-stimulated genes (ISGs) through the IKK-NF-κB and TBK1-IRF signaling branches, respectively. Initially beneficial to the host during early infection by mobilizing immune cell infiltration to combat the virus, dysregulated proinflammatory cytokines can lead to harmful systemic hyperinflammation and tissue damage. While the extent of RLR signaling pathways' involvement in coronavirus-induced inflammation remains incompletely understood, coronaviruses, like many RNA viruses, counteract the RLR signaling pathway. For instance, the viral nucleoprotein (N) of SARS-CoV and MERS-CoV binds to the SPRY domain of TRIM25, inhibiting TRIM25-mediated K63-linked ubiquitination of RIG-I and thereby abrogating RIG-I activation [4]. Additionally, SARS-CoV-2 ORF9B disrupts RIG-I-MAVS antiviral signaling by interfering with the K63-linked ubiquitination of NEMO [5]. Intriguingly, recent transcriptome analyses have revealed that SARS-CoV-2 strongly suppresses the expression of type I IFN and ISGs but not proinflammatory factors [6–8]. Moreover, severe cases of COVID-19 often exhibit heightened levels of proinflammatory cytokines in the plasma, including interleukin-1 beta (IL-1β), interleukin-8 (IL-8), and tumor necrosis factor-alpha (TNFα) [9–11]. While current research concludes that coronavirus proteins inhibit host innate immunity, the preservation of proinflammatory factors remains not fully understood. Therefore, the outstanding question pertains to how coronaviruses skew the host's innate immune responses toward inflammation. A deeper understanding of how the innate immune system recognizes and reacts to HCoV infections will be crucial for guiding the development of potential therapeutic interventions.
Nonstructural protein 14 (NSP14) is a highly conserved viral bifunctional enzyme in the Coronaviridae family with two distinct domains: the exoribonuclease domain (ExoN) [12] and the S-adenosyl methionine (SAM)-dependent (guanine-N7) methyl transferase (N7-MTase) [13]. The ExoN domain plays a proofreading role associated with viral polymerase in averting lethal mutagenesis. Meanwhile, the N7-MTase domain plays a crucial role in orchestrating the assembly of the cap structure at the 5`-end of viral mRNA, thereby facilitating viral mRNA translation. Moreover, NSP14 sabotages the host’s innate immune defense by either cleaving viral RNA or camouflaging it through methylation, thus eluding recognition by RIG-I/MDA5 and hindering the expression of type I interferons [14–16].
Despite its role in suppressing innate immunity triggered by viral RNA, NSP14 also initiates NF-κB activation, as indicated by several recent studies [17–19]. Initially, a study revealed that NSP14 from the transmissible gastroenteritis virus (TGEV), a pig coronavirus, stimulated NF-κB reporter activity by interacting with DEAD-Box Helicase 1 (DDX1) [17]. However, DDX1 serves as a co-activator, augmenting NF-κB activity rather than directly activating it [20]. Subsequent studies proposed that NSP14 induces NF-κB activation through inosine-5'-monophosphate dehydrogenase 2 (IMPDH2), the rate-limiting enzyme in de novo guanine nucleotide biosynthesis [18, 19]. IMPDH2 inhibitors are known to impede DNA and RNA virus infection by depleting the GTP supply [21]. Nevertheless, the precise mechanism by which IMPDH2 activates the NF-κB signaling pathway remains unclear. In essence, these pioneering studies collectively affirm NSP14 as an NF-κB inducer, underscoring a distinctive mechanism by which coronaviruses activate NF-κB signaling. However, the exact mechanism through which NSP14 triggers NF-κB activation is not well elucidated.
Recently, it has been uncovered that head-to-tail-linked linear ubiquitination plays a crucial role in NF-κΒ-dependent inflammatory signaling and immune responses [22]. The formation of linear ubiquitin chains is orchestrated by the E2 conjugating enzyme UBE2L3 and the linear ubiquitin chain assembly complex (LUBAC) comprising the catalytic E3 ligase HOIP (also known as RNF31), an auxiliary E3 ligase HOIL-1L (also known as RBCK1), and the adaptor SHARPIN [23–28]. Linear polyubiquitin plays a critical role in regulating the canonical NF-κB pathway, such as TNFα, IL-1, and TLR signaling pathways [23–27, 29]. In addition, our previous study has demonstrated that linear ubiquitination facilitates NF-κB activation induced by the latent membrane protein 1 (LMP1) during Epstein-Barr virus (EBV) infection [30]. However, whether linear ubiquitination is involved in coronavirus-induced NF-κB activation is unknown.
In this study, we reveal that coronaviruses hijack the host LUBAC complex to attach linear polyubiquitin chains onto the viral protein NSP14. This linear ubiquitinated NSP14 then recruits and activates the IKK complex, initiating NF-κB activation and subsequent expression of proinflammatory factors. Notably, specific inhibition of linear ubiquitination effectively dampened coronavirus-induced mRNA expression of proinflammatory factors while leaving ISGs unaffected. Additionally, linear ubiquitination of NSP14 was found to enhance viral replication within cells. Taken together, our findings underscore how coronaviruses exploit host linear ubiquitination, a distinctive form of polyubiquitin, to drive NF-κB activation, manipulate the host immune response toward inflammation, and bolster viral replication.
Results
NSP14 induces NF-κB activity independent of its enzymatic activities
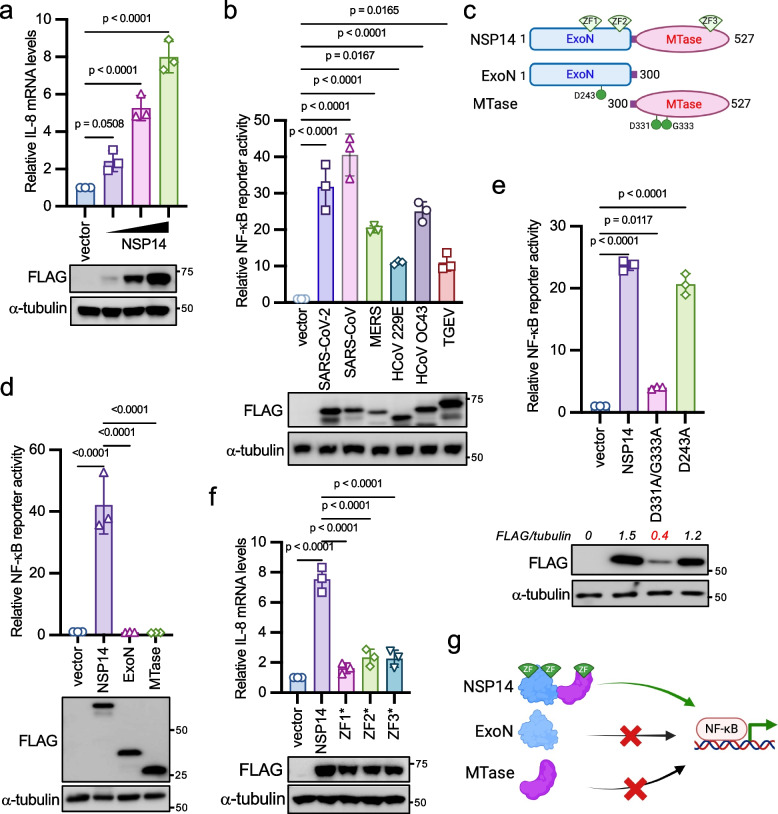
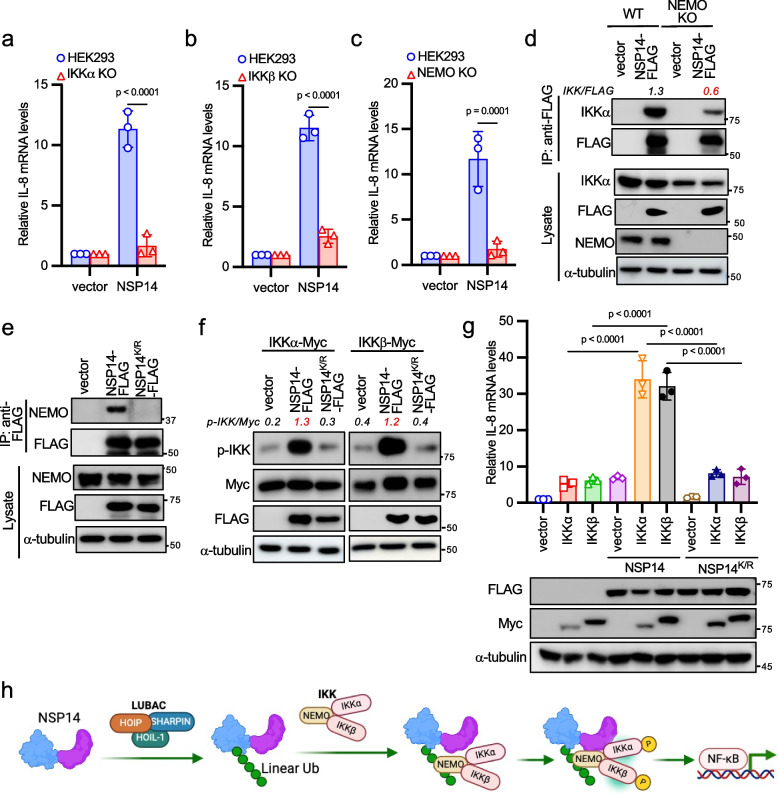
To investigate the impact of coronavirus proteins on the expression of proinflammatory factors, we adopted NF-κB reporter assays because most proinflammatory factors are induced by the NF-κB transcriptional factor. We transfected each SARS-CoV-2 gene along with the NF-κB firefly luciferase reporter and the Renilla luciferase construct as an internal control into HEK293 cells. We found that NSP14 and ORF7A significantly increased NF-κB reporter activity (Fig. S1a), aligning with recent similar screening studies [18, 19]. Some viral genes previously identified as NF-κB activators were not found in our screening, such as ORF3A [31], possibly due to the different screening system, varying levels of viral protein expression, or reporter assay readout windows. We aimed to investigate NSP14 because of its high activity in inducing NF-κB activation. First, we found that NSP14 activated NF-κB reporter (Fig. S1b) and IL-8 mRNA expression (Fig. 1a) in a dose-dependent manner. Next, we assessed whether NSP14-induced NF-κB activation is specific to viral species by evaluating NSP14 genes from other human coronaviruses (SARS-CoV, MERS-CoV, HCoV OC43, HCoV 229E) and the pig coronavirus TGEV. Remarkably, all NSP14 proteins induced NF-κB activity (Fig. 1b), suggesting a general and conserved mechanism across coronaviruses.
Fig. 1.
NSP14 activates the NF-κB signaling pathway. a 2 × 105 HEK293 cells were transfected with 0.05 µg, 0.1 µg, and 0.15 µg of FLAG-tagged SARS-CoV-2 NSP14 for 24 h. Real-time PCR analysis was conducted to assess the relative IL-8 mRNA levels. Lysates were blotted with anti-FLAG and anti-α-tubulin antibodies. b Co-transfection of FLAG-tagged NSP14 genes from indicated coronaviruses along with NF-κB firefly luciferase reporter and pRL-SV40 (Renilla luciferase as an internal control) into HEK293 cells. After 48 h, cells were harvested, and the relative reporter activity was determined by calculating the ratio of firefly luciferase to Renilla luciferase. Lysates were blotted with anti-FLAG and anti-α-tubulin antibodies. c Schematic of NSP14 domains and mutants. ExoN: 3′-to-5′ exoribonuclease; MTase: guanine-N7-methyltransferase; ZF: zinc finger. d FLAG-tagged SARS-CoV-2 wild-type NSP14 or the indicated mutant was co-transfected with the NF-κB reporter and pRL-SV40 into HEK293 cells. Lysates were blotted with anti-FLAG and anti-α-tubulin antibodies. e FLAG-tagged SARS-CoV-2 NSP14, the D243A mutant, or the D331A/G333A mutant was co-transfected with the NF-κB reporter and pRL-SV40 into HEK293 cells. Lysates were blotted with anti-FLAG and anti-α-tubulin antibodies. f HEK293 cells were transfected with FLAG-tagged SARS-CoV-2 NSP14 or the indicated zinc finger mutant for 24 h. ZF1*: C207S/C210S; ZF2*: C261S/H264A; ZF3*: C484S/H487A. Real-time PCR analysis was performed to determine the relative IL-8 mRNA levels. Lysates were blotted with anti-FLAG and anti-α-tubulin antibodies. The p-value was calculated by one-way ANOVA followed by Tukey's multiple comparisons test (a, b, d, e, f). g Model illustrating NSP14-mediated NF-κB activation
NSP14 is a bifunctional enzyme with distinct ExoN and N7-MTase domains (Fig. 1c). However, as reported previously [32], neither the ExoN nor the MTase domain alone triggered NF-κB activation (Fig. 1d), indicating that NSP14's enzymatic activities are not accountable for NF-κB activation. Moreover, while NSP10 is essential for NSP14's ExoN activity, it had minimal effect on NSP14-induced NF-κB activity (Fig. S1c), suggesting that ExoN activity is dispensable for NF-κB activation. This was further supported by the observation that mutants of NSP14 lacking ExoN activity (D243A) or MTase activity (D331A/G333A)[33] retained the ability to activate NF-κB (Fig. 1e). Of note, the low NF-κB activity induced by the D331A/G333A mutant could be due to its weak expression (Fig. 1e).
NSP14 harbors three zinc fingers (ZFs), two in the ExoN domain and one in the N7-MTase domain (Fig. 1c). Intriguingly, mutations in each ZF (ZF1*: C207S/C210S; ZF2*: C261S/H264A; ZF3*: C484S/H487A) impaired NSP14-induced NF-κB reporter activity (Fig. S1d) and IL-8 mRNA expression (Fig. 1f). ZFs are also known to mediate protein–protein interactions [34], implying that these ZFs might recruit other proteins to activate NF-κB. Hence, these results suggest that NF-κB activation induced by NSP14 is independent of its enzymatic activities but relies on its three zinc fingers (Fig. 1g).
NSP14 is linearly ubiquitinated to govern NF-κB activation
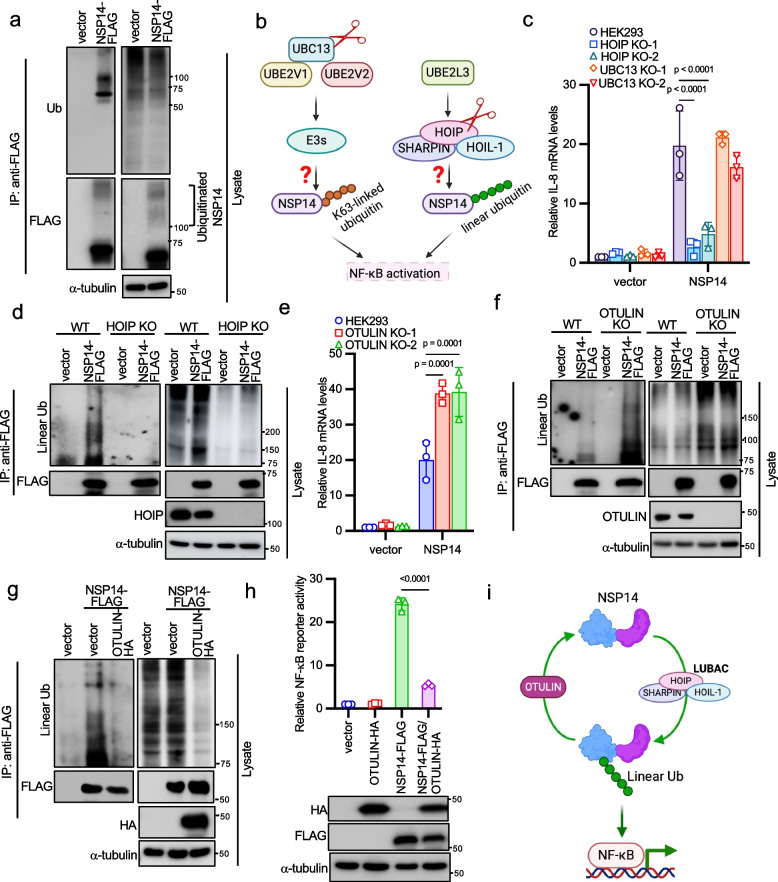
Upon examining SARS-CoV-2 NSP14 protein expression, we observed high molecular weight bands resembling polyubiquitination in the blot when exposing a longer time (Fig. 2a). Thus, we suspected that NSP14 might be subject to ubiquitination. However, the high molecular weight bands are not necessary ubiquitinated proteins. To ensure these bands are ubiquitinated NSP14 proteins, we performed immunoprecipitation to pull down NSP14 proteins and blotted it with an anti-ubiquitin antibody. Western blotting confirmed that these bands indeed represented polyubiquitinated NSP14 (Fig. 2a). Consistently, NSP14 proteins from all other five coronaviruses were also ubiquitinated (Fig. S2a). To determine the type of ubiquitination involved, we focused on K63-linked polyubiquitin and linear polyubiquitin, both known to regulate NF-κB signaling pathways [35]. UBC13 is associated with K63-linked ubiquitination, while HOIP is integral to the LUBAC complex responsible for linear ubiquitination (Fig. 2b). Therefore, we knocked out UBC13 and HOIP to abolish K63-linked ubiquitination and linear ubiquitination, respectively, and validated the functional loss in these cell lines in response to TNFα (Figs. S2b-S2e). Then, we transfected NSP14 together with NF-κB reporter into these knockout cells. Knockout of HOIP, but not UBC13, abolished NSP14-induced NF-κB reporter activity (Fig. S2f) and IL-8 mRNA expression (Fig. 2c), suggesting that linear ubiquitination is essential for NSP14-induced NF-κB activation. Furthermore, HOIP knockout abrogated NSP14 linear ubiquitination (Fig. 2d), further confirming that NSP14 undergoes linear ubiquitination.
Fig. 2.
NSP14 induces NF-κB activation through the linear ubiquitination pathway. a FLAG-tagged NSP14 or pCMV-3Tag-8 vector was transfected into HEK293 cells. Cell lysates were immunoprecipitated with the anti-FLAG antibody and blotted with anti-FLAG, anti-ubiquitin (Ub), and anti-α-tubulin antibodies. b Strategies targeting K63-linked ubiquitination and linear ubiquitination. c Wild-type HEK293 cells, two UBC13 knockout, and two HOIP knockout HEK293 cell lines were transfected with FLAG-tagged NSP14 for 24 h. Real-time PCR was performed to determine the relative IL-8 mRNA levels. d Vector or FLAG-tagged NSP14 (NSP14-FLAG) was transfected into HOIP wild-type and knockout HEK293 cells. After 48 h, cell lysates were immunoprecipitated with the anti-FLAG antibody and blotted with anti-FLAG, anti-linear ubiquitin, anti-HOIP, and anti-α-tubulin antibodies. e Wild-type and OTULIN knockout HEK293 cells were transfected with FLAG-tagged NSP14 for 24 h. Real-time PCR was performed to determine the relative IL-8 mRNA levels. f Wild-type and OTULIN knockout HEK293 cells were transfected with vector or NSP14-FLAG. After 48 h, cell lysates were immunoprecipitated with the anti-FLAG antibody and blotted with anti-FLAG, anti-linear ubiquitin, anti-OTULIN, and anti-α-tubulin antibodies. g NSP14-FLAG was transfected with vector or HA-tagged OTULIN (OTULIN-HA) into HEK293 cells. After 48 h, cell lysates were immunoprecipitated with the anti-FLAG antibody and blotted with anti-FLAG, anti-linear ubiquitin, anti-HA, and anti-α-tubulin antibodies. h HEK293 cells were transfected with NSP14-FLAG and OTULIN-HA, together with the NF-κB reporter and pRL-SV40. After 48 h, cells were harvested, and the relative reporter activity was determined by calculating the ratio of firefly luciferase to Renilla luciferase. Lysates were blotted with anti-FLAG, anti-HA and anti-α-tubulin antibodies. The p-value was calculated by one-way ANOVA followed by Tukey's multiple comparisons test (h) and two-way ANOVA followed by Sidak's multiple comparisons test (c, e). i Proposed model illustrating NSP14 linear ubiquitination regulated by LUBAC and OTULIN
Next, we investigated the role of OTULIN (OTU domain-containing deubiquitinase with linear linkage specificity), a deubiquitinase hydrolyzing linear polyubiquitin chains [36, 37], in NSP14-mediated NF-κB activity. Thus, we generated OTULIN knockout cell lines by CRISPR (Fig. S2g). Ablation of OTULIN enhanced TNFα-induced IκBα phosphorylation (Fig. S2h), confirming functional loss in this cell line. In OTULIN knockout cells, NSP14-induced NF-κB reporter activity (Fig. S2i) and IL-8 mRNA expression (Fig. 2e) were heightened. Consistently, OTULIN deficiency increased NSP14 linear ubiquitination (Fig. 2f), while OTULIN overexpression led to cleavage of NSP14's linear polyubiquitin (Fig. 2g) and suppression of NSP14-induced NF-κB reporter activity (Fig. 2h). Collectively, these findings suggest that HOIP mediates NSP14 linear ubiquitination, promoting NF-κB activation, while OTULIN counteracts this by cleaving NSP14's linear polyubiquitin, thus suppressing NF-κB activity (Fig. 2i).
NSP14 interacts with HOIP via zinc fingers
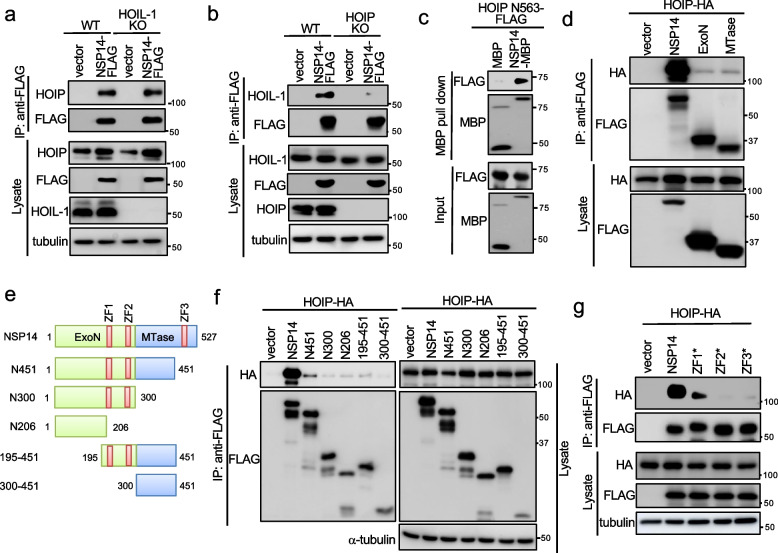
Given NSP14's linear ubiquitination, we hypothesized an interaction between NSP14 and one of the components in the LUBAC complex. To test this hypothesis, we first performed co-immunoprecipitation (co-IP) between NSP14 and each component of LUBAC. Co-IP revealed an interaction with HOIP, rather than with HOIL-1 or SHARPIN (Fig. S3a). Moreover, NSP14 interacted with endogenous HOIP (Fig. S3b) and co-localized with HOIP in the cytoplasm (Fig. S3c). To determine whether the NSP14-HOIP interaction is dependent on HOIL-1, we made HOIL-1 knockout A549 cells. Immunoprecipitation found that NSP14 still interacted with HOIP in HOIL-1 knockout cells (Fig. 3a), indicating that the NSP14-HOIP interaction is independent of HOIL-1. Conversely, NSP14 failed to interact with endogenous HOIL-1 in HOIP knockout cells (Fig. 3b), suggesting that NSP14 interacts with HOIL-1 through HOIP in the LUBAC complex.
Fig. 3.
HOIP interacts with and ubiquitinates NSP14. a NSP14-FLAG was transfected into HOIL-1 wild-type and knockout A549 cells. After 48 h, cell lysates were immunoprecipitated with the anti-FLAG antibody and blotted with anti-FLAG, anti-HOIP, anti-HOIL-1, and anti-α-tubulin antibodies. b NSP14-FLAG was transfected into HOIP wild-type and knockout A549 cells. After 48 h, cell lysates were immunoprecipitated with the anti-FLAG antibody and blotted with anti-FLAG, anti-HOIP, anti-HOIL-1, and anti-α-tubulin antibodies. c Purified bacterial recombinant FLAG-tagged HOIP N563 was incubated with either MBP or MBP-tagged NSP14 (NSP14-MBP) at 4 °C for 16 h. Subsequently, MBP pull-down assays were carried out, followed by Western blotting with anti-MBP and anti-FLAG antibodies. d FLAG-tagged NSP14 or the indicated domain was co-transfected with HA-tagged HOIP (HOIP-HA) into HEK293 cells. After 48 h, cell lysates were immunoprecipitated with the anti-FLAG antibody and blotted with anti-FLAG and anti-HA antibodies. e Schematics of NSP14 mutants. f Co-transfection of FLAG-tagged NSP14 or the indicated mutants with HOIP-HA into HEK293 cells. After 48 h, cell lysates were immunoprecipitated with the anti-FLAG antibody and blotted with anti-FLAG, anti-HA, and anti-α-tubulin antibodies. g FLAG-tagged NSP14 or the indicated ZF mutant was co-transfected with HOIP-HA into HEK293 cells. After 48 h, cell lysates were immunoprecipitated with the anti-FLAG antibody and blotted with anti-FLAG, anti-HA, and anti-α-tubulin antibodies
Next, we determined the binding domain of HOIP responsible for NSP14 interaction. We made a panel of HOIP mutants (Fig. S3d). Co-IP found that the region between amino acids 1 to 563 (N563) in HOIP was sufficient for NSP14 binding (Fig. S3e). To further validate the direct interaction between NSP14 and HOIP, pull-down assays were performed using MBP-tagged NSP14 and FLAG-tagged HOIP N563 protein purified from E. coli. In vitro MBP pull-down assays showed that NSP14, but not MBP, bound HOIP N563 (Fig. 3c). Similarly, we determined the domains of NSP14 responsible for HOIP interaction. ExoN or MTase domain alone barely bound HOIP (Fig. 3d), suggesting the requirement of both domains for interaction. To further dissect the binding region, we made a panel of NSP14 mutants (Fig. 3e). Co-IP revealed the deletion of any ZF in NSP14 dramatically reduced its interaction with HOIP (Fig. 3f). In line with this observation, disruption of individual ZF1, ZF2, or ZF3 in NSP14 reduced or abolished its interaction with HOIP (Fig. 3g). These findings suggest that ZFs mediate NSP14-HOIP interaction, aligning with the necessity of ZFs for NSP14-mediated NF-κB activation (Fig. 1f).
Multiple ubiquitination sites regulate NSP14-mediated NF-κB activation
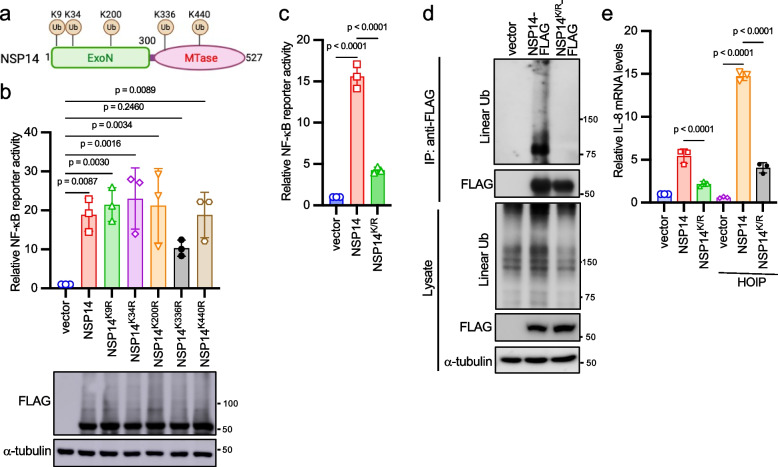
Given that NSP14 proteins from various coronaviruses undergo linear ubiquitination, it's reasonable to speculate that they might share conserved ubiquitination sites. Upon aligning their protein sequences, we identified five conserved lysines: K9, K34, K200, K336, and K440 (corresponding to sites in SARS-CoV-2 NSP14) (Fig. 4a). However, when point mutations were made for each lysine, the mutants still activated the NF-κB reporter (Fig. 4b), indicating potential redundancy in ubiquitination sites. To address this, all five lysines were mutated to arginines (designated as NSP14K/R). The NSP14K/R impaired NF-κB activity (Fig. 4c) and abolished the linear ubiquitination (Fig. 4d). In agreement with these results, wild-type NSP14, but not NSP14K/R, synergized with HOIP on IL-8 mRNA expression (Fig. 4e). Of note, our data cannot exclude that there might be only two, three, or four lysines from these five sites are responsible for NSP14 linear ubiquitination and NF-κB activity. Taken together, these findings suggest that LUBAC ubiquitinates NSP14 at multiple lysine sites for downstream NF-κB activation.
Fig. 4.
HOIP ubiquitinates NSP14 at multiple sites. a Conserved lysines in NSP14 proteins from six coronaviruses, including SARS-CoV, SARS-CoV-2, MERS, HCoV OC43, HCoV 229E, and TGEV. b HEK293 cells were transfected with FLAG-tagged NSP14 or the indicated mutant, along with NF-κB reporter and pRL-SV40. After 48 h, cells were harvested, and the relative reporter activity was determined by the ratio of firefly luciferase to Renilla luciferase. Lysates were blotted with anti-FLAG and anti-α-tubulin antibodies. c FLAG-tagged NSP14 or NSP14K/R was co-transfected with the NF-κB reporter and pRL-SV40 into HEK293 cells. After 48 h, cells were harvested, and the relative reporter activity was determined by the ratio of firefly luciferase to Renilla luciferase. d NSP14-FLAG or NSP14K/R-FLAG was transfected into HEK293 cells. After 48 h, cell lysates were immunoprecipitated with the anti-FLAG antibody and blotted with anti-FLAG, anti-linear ubiquitin, and anti-α-tubulin antibodies. e FLAG-tagged NSP14 or NSP14.K/R was co-transfected with vector or HA-tagged HOIP into HEK293 cells for 24 h. Real-time PCR analysis was performed to determine the relative IL-8 mRNA levels. The p-value was calculated by one-way ANOVA followed by Tukey's multiple comparisons test (b, c, e)
NSP14 activates NF-κB via the IKK complex
To ascertain NSP14's position in the NF-κB signaling cascade, we co-transfected IKKα, IKKβ, or NF-κB p65 alongside the NF-κB reporter into HEK293 cells. NSP14 synergistically activated the NF-κB reporter with IKKα and IKKβ but exhibited only a marginal effect on p65-induced NF-κB activity (Fig. S4a), indicating that NSP14 acts upstream of NF-κB p65.
The upstream NF-κB pathways mainly involve two distinct signaling branches: the canonical pathway and the non-canonical pathway, dependent on the IKK complex and IKKα, respectively [35, 38]. Additionally, TBK1, another member of the IKK kinase family, is also reported to activate NF-κB. To discern which signal cascade NSP14 activates, we assessed NSP14-mediated NF-κB activation in IKKα, IKKβ, NEMO knockout HEK293 cells, and TBK1 knockout mouse embryonic fibroblasts (MEFs). IKKα, IKKβ, and NEMO knockout HEK293 cells were generated via CRISPR and validated by reduced IκBα phosphorylation in response to TNFα (Figs. S4b-S4d). Deficiency of IKKα, IKKβ, or NEMO significantly diminished NSP14-induced NF-κB activity (Figs. S4e-S4g) and IL-8 mRNA expression (Figs. 5a-5c). In contrast, TBK1 knockout MEFs demonstrated a similar NF-κB activity as wild-type MEFs (Figs. S4h). These findings collectively suggest that NSP14 activates the canonical NF-κB signaling pathway through the IKK complex.
Fig. 5.
NSP14-mediated NF-κB activation through the IKK complex. a-c Transfection of either vector or NSP14-FLAG into wild-type, IKKα knockout (a), IKKβ knockout (b), or NEMO knockout (c) HEK293 cells for 24 h. Real-time PCR analysis was conducted to determine the relative IL-8 mRNA levels. d Vector or NSP14-FLAG was transfected into NEMO wild-type and knockout HEK293 cells. After 48 h, cell lysates were immunoprecipitated with the anti-FLAG antibody and blotted with anti-FLAG, anti-IKKα, anti-NEMO, and anti-α-tubulin antibodies. e Vector, NSP14-FLAG, or NSP14K/R-FLAG was transfected into HEK293 cells. After 48 h, cell lysates were immunoprecipitated with the anti-FLAG antibody and blotted with anti-FLAG, anti-NEMO, and anti-α-tubulin antibodies. f NSP14-FLAG or NSP14K/R-FLAG was transfected with Myc-tagged IKKα or IKKβ into HEK293 cells. After 48 h, cell lysates were blotted with anti-FLAG, anti-Myc, anti-p-IKK (Ser176/180), and anti-α-tubulin antibodies. g FLAG-tagged NSP14 or NSP14K/R was transfected with Myc-tagged IKKα or IKKβ into HEK293 cells for 24 h. Real-time PCR analysis was performed to determine the relative IL-8 mRNA levels. Lysates were blotted with anti-FLAG, anti-Myc, and anti-α-tubulin antibodies. The p-value was calculated by one-way ANOVA followed by Tukey's multiple comparisons test (g) or two-way ANOVA followed by Sidak's multiple comparisons test (a, b, c). h Model illustrating NSP14 linear polyubiquitin-mediated NEMO recruitment and IKK activation
NSP14 recruits and activates the IKK complex through linear polyubiquitin
In the TNFα-induced NF-κB pathway, linear polyubiquitin activates NF-κB by recruiting the IKK complex through the ubiquitin-binding domain in NEMO [39]. A recent study found that NSP14 interacted with NEMO [40]. Thus, it is plausible that the linear polyubiquitin of NSP14 recruits the IKK complex through binding NEMO, subsequently activating the IKK complex and NF-κB. To delineate the mechanism, we first examined the interaction between NSP14 and IKKα in NEMO knockout cells. The NSP14-IKKα interaction was reduced in NEMO knockout cells (Fig. 5d), suggesting that NSP14 binds the IKK complex through the regulatory subunit NEMO. Next, we examined the effect of NSP14 ubiquitination on NEMO binding. Wild-type NSP14, but not the ubiquitination-defective mutant NSP14K/R, interacted with endogenous NEMO (Fig. 5e), suggesting that linear ubiquitination is required for NSP14-NEMO interaction.
Next, we examined the effect of NSP14 on IKK phosphorylation, a hallmark of the activation of the IKK complex and NF-κB pathway. Wild-type NSP14 induced robust IKK phosphorylation whereas the NSP14K/R mutant had little impact on IKK phosphorylation (Fig. 5f). Consistently, wild-type NSP14 but not the NSP14K/R mutant activated NF-κB reporter and IL-8 mRNA expression synergistically with IKKα and IKKβ (Figs. S4i and 5 g). Taken together, these data suggest that the linear polyubiquitin of NSP14 recruits the IKK complex, leading to IKK activation and subsequent NF-κB pathway activation (Fig. 5h).
HCoV induces pro-inflammatory responses through linear ubiquitination
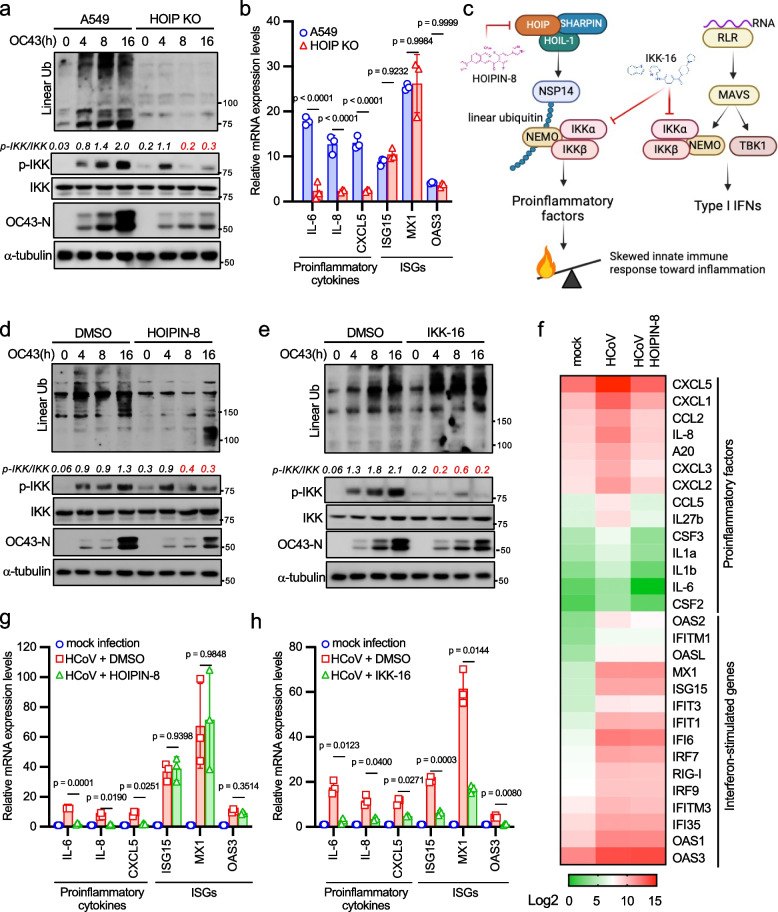
Our studies propose that NSP14 linear ubiquitination activates the NF-κB pathway. However, coronavirus RNA also triggers NF-κB activation through the RLR-MAVS signaling pathway. To determine the predominant or redundant role of these pathways in HCoV-induced inflammation, we knocked out HOIP and MAVS in A549 cells to deactivate these pathways individually. Subsequently, we evaluated the impact of HOIP and MAVS deficiency on HCoV OC43-induced innate immune responses. Upon infecting wild-type, HOIP knockout, and MAVS knockout A549 cells with HCoV OC43, we observed that HOIP ablation abolished HCoV-induced linear ubiquitination (Fig. 6a). Interestingly, HOIP deficiency impaired IKK phosphorylation at 8- and 16-h post-infection (Fig. 6a). Conversely, MAVS knockout had minimal effect on linear ubiquitination and reduced IKK phosphorylation at 4-h post-infection (Fig. S5a), suggesting distinct roles of HOIP and MAVS in controlling IKK activation at different infection stages.
Fig. 6.
HOIP deficiency impairs HCoV OC43-induced linear ubiquitination and proinflammatory cytokine expression. a HOIP wild-type and knockout A549 cells were infected with 1 MOI of HCoV OC43 for designated times. Lysates were blotted with anti-FLAG, anti-p-IKK (Ser176/180), anti-IKK, anti-linear ubiquitin, anti-OC43 N, and anti-α-tubulin antibodies. Band densitometry was calculated using Image J. The ratio of phosphorylated IKK to total IKK in each lane was indicated. b HOIP wild-type and knockout A549 cells were infected with 1 MOI of HCoV OC43 for 16 h. Real-time PCR was conducted to determine the relative mRNA levels of proinflammatory factors and ISGs. c Schematic representation of NSP14 and RLR pathways involved in coronavirus infection. Targets of HOIPIN-8 and IKK-16 are indicated. d A549 cells were treated with DMSO or 30 μM HOIPIN-8 for 2 h and then infected with 1 MOI of HCoV OC43 for designated times. Lysates were blotted with anti-FLAG, anti-p-IKK (Ser176/180), anti-IKK, anti-linear ubiquitin, anti-OC43 N, and anti-α-tubulin antibodies. e A549 cells were treated with DMSO or 10 μM IKK-16 for 2 h and then infected with HCoV OC43 for designated times. Lysates were blotted with anti-FLAG, anti-p-IKK (Ser176/180), anti-IKK, anti-linear ubiquitin, anti-OC43 N, and anti-α-tubulin antibodies. f A549 cells were treated with DMSO or 30 μM HOIPIN-8 for 2 h and then infected with 1 MOI of HCoV OC43 for 16 h. RNA sequencing was performed, and the normalized mean count of proinflammatory factors and ISGs is shown in the heatmap. g A549 cells were treated with DMSO or 30 μM HOIPIN-8 for 2 h and then infected with 1 MOI of HCoV OC43 for 16 h. Real-time PCR was performed to determine the relative mRNA levels of proinflammatory factors and ISGs. h A549 cells were treated with DMSO or 10 μM IKK-16 for 2 h and then infected with 1 MOI of HCoV OC43 for 16 h. Real-time PCR was performed to determine the relative mRNA levels of proinflammatory factors and ISGs. The p-value was calculated by two-way ANOVA followed by Sidak's multiple comparisons test (b, g, h)
Strikingly, knockout of HOIP in two lung epithelial cell lines, H1299 and A549, dramatically reduced the expression of proinflammatory factors but not ISGs (Figs. 6b and S5b). In contrast, MAVS deficiency caused an opposite effect by reducing mRNA expression of ISGs but not proinflammatory factors (Fig. S5c). The moderate effects on ISG15 and OAS3 mRNA expression in MAVS knockout cells suggest that other IFN induction pathways, such as TLRs, are also involved in ISG expression. These findings support a model wherein linear ubiquitination drives coronavirus-induced proinflammatory responses, while the RLR-MAVS pathway predominantly regulates type I IFN production (Fig. 6c).
To substantiate our model, we employed two inhibitors, HOIPIN-8 [41] and IKK-16 [42], to block NSP14 linear ubiquitination and IKK kinase activity, respectively (Fig. 6c). It's worth noting that HOIPIN-8 selectively targets the NSP14-mediated NF-κB signaling pathway, whereas IKK-16 affects both NF-κB and type I IFN signaling pathways (Fig. 6c). First, we tested the toxicity of HOIPIN-8 and IKK-16 in A549 cells using the previous reported dosage [41, 42]. Both inhibitors showed minimal toxic effect on A549 cells after 5 days (Fig. S5d). Then, we treated the A549 cells with either HOIPIN-8 or IKK-16 followed by HCoV OC43 infection. As anticipated, HOIPIN-8 effectively inhibited HCoV OC43-induced linear ubiquitination (Fig. 6d), akin to the effects observed with HOIP knockout. Furthermore, it inhibited IKK phosphorylation during the late stage of viral infection (Fig. 6d), consistent with our findings with HOIP knockout. Conversely, IKK-16 blocked IKK phosphorylation without affecting linear ubiquitination when compared to the DMSO control group (Fig. 6e).
Next, we examined mRNA expression in the cells treated with HOIPIN-8 using RNA sequencing. HCoV OC43 induced both proinflammatory factor and ISG mRNA expression (Fig. 6f). Mirroring the effects of HOIP knockout, HOIPIN-8 selectively suppressed the mRNA expression of proinflammatory factors but not ISGs (Fig. 6f-6g). In contrast, IKK-16 inhibited the expression of both proinflammatory factors and ISGs (Fig. 6h) due to its broad suppression of both NF-κB and type I IFN signaling pathways (Fig. 6c). Overall, these findings provide further evidence supporting the notion that coronaviruses induce proinflammatory responses by exploiting the host's linear ubiquitination system.
Linear ubiquitination facilitates HCoV viral replication
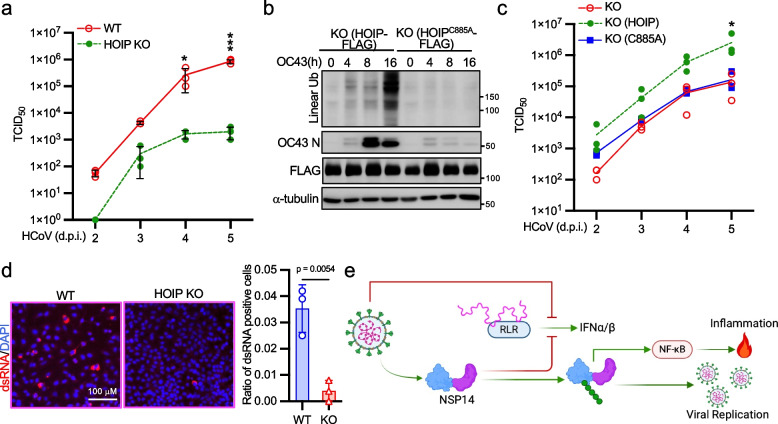
The impact of proinflammatory responses on HCoV infection prompts us to delve into the role of linear ubiquitination in coronavirus infection. TCID50 assays revealed a remarkable reduction in viral titer by more than 100-fold in HOIP knockout A549 cells at 4- and 5-days post-infection (Fig. 7a) with comparable cell viability as the wild-type cells (Fig. S6a). Similarly, the viral N protein expression level was reduced in HOIP knockout H1299 cells (Fig. S6b). To further elucidate the requirement of HOIP enzymatic activity in coronavirus replication, we reconstituted wild-type HOIP and the enzyme-dead mutant HOIPC885A in HOIP knockout A549 cells. Upon HCoV OC43 infection, wild-type HOIP but not the HOIPC885A rescued linear ubiquitination and viral N protein expression (Fig. 7b). Consistent with these observations, TCID50 assays showed that wild-type HOIP but not the HOIPC885A mutant rescued HCoV infection (Fig. 7c). Furthermore, HOIPIN-8 treatment resulted in a substantial reduction in viral titer by approximately 10- to 100-fold, while IKK-16 only had a minor effect (Fig. S6c), emphasizing the necessity of HOIP enzymatic activity for HCoV replication.
Fig. 7.
Linear ubiquitination facilitates coronavirus infection. a HOIP wild-type and knockout A549 cells were infected with 0.01 MOI of HCoV OC43 for the designated days. TCID50 of culture supernatants containing HCoV OC43 were determined on Vero cells. b HOIP knockout A549 cells reconstituted with HOIP or the C885A mutant were infected with 1 MOI of HCoV OC43 for the designated times. Lysates were blotted with anti-FLAG, anti-linear ubiquitin, anti-OC43 N, and anti-α-tubulin antibodies. c HOIP knockout A549 cells and knockout cells reconstituted with HOIP or the C885A mutant were infected with 0.01 MOI of HCoV OC43 for the designated days. TCID50 of culture supernatants containing HCoV OC43 were determined on Vero cells. d Wild-type and HOIP knockout A549 cells were infected with 0.1 MOI HCoV OC43 for 24 h. Cells were stained with anti-dsRNA (red) and DAPI (blue). The relative ratio of positively stained cells is summarized in the graph. The p-value was calculated by t-test. e Model depicting how HCoV skews host innate immune responses toward inflammation
The impairment of coronavirus titer and protein expression upon HOIP knockout suggests a potential impact of linear ubiquitination on coronavirus replication. Thus, we examined the quantities of viral dsRNA foci, a marker of viral replication. Remarkably, HOIP deficiency led to a significant reduction in dsRNA foci in infected cells (Fig. 7d). Consistently, HOIPIN-8 treatment also hindered dsRNA foci formation in HCoV-infected cells (Fig. S6d). Together, these findings underscore the essential role of linear ubiquitination in facilitating HCoV replication and the induction of viral infection-induced proinflammatory responses (Fig. 7e).
Discussion
A novel coronavirus-initiated linear ubiquitin signaling pathway
It is widely acknowledged that coronaviruses employ various mechanisms to hijack host ubiquitination processes, thus undermining the innate defense mechanisms. For instance, the viral N proteins of SARS-CoV and MERS-CoV disrupt TRIM25-mediated K63-linked ubiquitination of RIG-I, consequently suppressing type I IFN production [4]. Furthermore, K63-linked ubiquitination of SARS-CoV-2 ORF7A impedes the IFN response [43]. These findings, alongside many recent studies on viral IFN antagonisms, underscore the multifaceted evasion strategies employed by coronaviruses. However, whether human coronaviruses stimulate their own signaling pathways to enhance viral infection remains uncertain.
Our research reveals that coronaviruses exploit the host's linear ubiquitination machinery to initiate a novel NF-κB pathway. In this pathway, HOIP confers linear polyubiquitin onto NSP14. Subsequently, the linear polyubiquitin on NSP14 recruits the IKK complex through binding to NEMO, triggering IKK and downstream NF-κB activation. Activation of this pathway not only promotes viral replication but also skews the balance of the host's innate immune responses toward inflammation.
HCoV induces proinflammatory responses via NSP14 linear ubiquitination
Several SARS-CoV-2 proteins have been identified as activators of NF-κB signaling pathways. While one study indicated that the spike protein activates TLR4-NF-κB signaling [44], another suggested that hyper-inflammatory responses to the spike protein are limited to senescent cells [45]. Moreover, recent research has contested the involvement of TLR4 in SARS-CoV-2-induced proinflammatory factor expression, proposing instead that the E protein activates TLR2, leading to the expression of proinflammatory factors like IL-1β, IL-6, and MCP-1 [46]. The mechanism by which the E protein activates TLR2 remains unclear, particularly considering the typical topology of beta coronavirus E proteins, which position both the N- and C-termini towards the cytosol [47].
Our study proposes a model in which linear ubiquitination emerges as one of the drivers of NF-κB activation during coronavirus infection. Linear ubiquitination, prevalent in TNF and IL-1 signaling pathways, is well-known to facilitate NF-κB activation through NEMO binding to linear polyubiquitin chains [48]. Intriguingly, NSP14 interacts with the IKK complex via linear polyubiquitin-mediated interaction with NEMO, suggesting that coronaviruses employ a strategy akin to the TNF signaling pathway. Supporting this, recent evidence links transcription signatures in SARS-CoV-2 infection to TNFα signaling via NF-κB [49]. Furthermore, the deficiency of linear ubiquitination significantly diminishes NF-κB activity induced by HCoV infection, highlighting a role in HCoV-induced NF-κB activation. Of note, it is important to recognize that the 5 simultaneous K-to-R changes could have other effects. They could prevent proper protein folding of NSP14, alter its localization, or cause it to function as an inhibitor in some way. These warrants further investigations in the future.
HCoV skews host innate immune responses by differentially controlling NF-κB and IRF signaling pathways
The impact of HCoV infection on the innate immune response is well-documented, characterized by an imbalanced innate immune response tilting to inflammation with suppression of type I IFN. Given NF-κB's pivotal role in regulating both type I IFN production and proinflammatory factors, coronaviruses must simultaneously activate the NF-κB signaling pathway for proinflammatory factor expression while inhibiting the IFN signaling pathway. Indeed, many SARS-CoV-2 proteins, including NSP1, NSP3, NSP12, NSP13, NSP14, NSP15, NSP16, ORF3, ORF3B, ORF6, M, ORF7A, ORF7B, and ORF9B, have been reported to suppress type I IFN production [50–56] and STAT1/2 phosphorylation [57] by antagonizing the RLR and IFN signaling pathways. Similar strategies are adopted by other coronavirus proteins. For instance, the nucleoprotein of SARS-CoV interacts with TRIM25 to inhibit RIG-I via RIG-I ubiquitination [4], and liquid droplets formed by the nucleoprotein of SARS-CoV-2 inhibit MAVS K63-linked ubiquitination [58], thereby suppressing type I IFN expression. Additionally, ORF-9B of SARS-CoV promotes PCBP2-mediated degradation of MAVS [59], while the 4A protein of MERS-CoV represses MDA5 signaling [60]. This viral antagonism of the host IFN response is crucial for virus replication and the outcomes of coronavirus infection.
As both the RLR pathway and most TLR pathways bifurcate into NF-κB and IRF signaling branches, targeting these pathways would suppress the expression of both proinflammatory factors and type I IFN. Given the multitude of mechanisms through which coronaviruses inhibit these pathways, it is improbable that they activate the RLR and TLR pathways to induce proinflammatory responses. Our study suggests that coronaviruses differentially regulate NF-κB and IRF signaling pathways by suppressing the RLR pathways and activating the NSP14-mediated NF-κB signaling pathway. These mechanisms of differential control over NF-κB and IRF signaling pathways result in skewed innate immune responses (Fig. 7e).
Is NF-κB a friend or foe of coronavirus?
Our findings prompt this critical inquiry into why coronaviruses have evolved to employ their own NF-κB signaling pathway to activate proinflammatory factors. Two recent studies demonstrate the requirement of NF-κB for SARS-CoV-2 infection [40, 49]. Our study found that HOIP knockout and HOIP inhibitor impeded HCoV replication, indicating that linear ubiquitination-mediated NF-κB activation might facilitate viral infection. It is not clear whether NSP14 linear ubiquitination alone or along with other protein linear ubiquitination is responsible for this phenotype. To address this question, future investigation is warranted to generate an NSP14 linear ubiquitination defective mutant virus.
While it may seem counterintuitive, NF-κB's proviral role is not unprecedented, particularly among certain oncogenic DNA viruses and retroviruses. For instance, human T-cell leukemia virus type 1 and Epstein-Barr virus activate NF-κB to transform cells and inhibit apoptosis [61, 62]. HIV-1 and HIV-2 induce NF-κB activation via their Tat proteins, promoting p65 DNA binding and inhibiting the NF-κB repressor IκBα [63]. Interestingly, inhibition of NF-κB signaling has been shown to diminish influenza A virus infection, suggesting a potential proviral role for some RNA viruses [64, 65]. While it's unlikely that NF-κB's proviral activity is a universal mechanism among all viruses, there may be unique NF-κB target genes essential for HCoV infection and critical steps in the HCoV life cycle.
Several hypotheses have been proposed regarding HCoV-induced NF-κB activation. For instance, IKKα/β, essential kinases for almost all NF-κB signaling pathways, could mediate the phosphorylation of viral proteins. Consistent with this model, the knockout of TRAF2, an upstream signal molecule of the IKK complex, reduced SARS-CoV-2 infection [40]. However, depletion or inhibition of NF-κB, the transcription factor downstream of the IKK, still impaired SARS-CoV-2 infection [49], suggesting that SARS-CoV-2 replication requires the upregulation of one or more NF-κB target genes that may regulate the viral replication process. We have analyzed our dataset and published datasets based on several criteria. First, we searched RNA sequencing databases of cells infected with four different coronaviruses, including SARS-CoV-1, SARS-CoV-2, MERS, HCoV 229E [8]. As our data suggest that linear ubiquitination-mediated NF-κB activation is a general mechanism for coronaviruses, we collected the upregulated genes induced by all four coronaviruses. Second, we interrogated the collected upregulated gene to our RNA sequencing data. We found that 81 genes of the collection were induced by HCoV OC43, and their expression was also blocked by HOIPIN-8. Lastly, we interrogated several recent CRISPR screenings [66–69] on coronaviruses with these 81 genes and found that 13 of them are the hits in the CRISPR screenings (Supplementary Table 1). Future research will investigate whether and how these 13 genes regulate HCoV infection.
Enhanced comprehension of the common mechanisms shared by various coronaviruses will be indispensable in informing the development of potential therapeutic strategies. Our study's insights will help bridge gaps in understanding how coronaviruses modulate host innate immune responses towards inflammation and lay the groundwork for novel treatments against coronaviruses.
Materials and methods
Cells
HEK293 cells (ATCC, # CRL-1573), HRT-18G (ATCC, # CRL-11663) and Vero cells (ATCC, # CCL-81), were maintained in Dulbecco’s Modified Eagle Medium (Life Technologies, # 11,995–065) containing antibiotics (Life Technologies, # 15,140–122) and 10% fetal bovine serum (Life Technologies, # 26,140–079). A549 cells (ATCC, # CCL-185) were cultured in RPMI Medium 1640 (Life Technologies, # 11,875–093) plus 10% fetal bovine serum and 1 × MEM Non-Essential Amino Acids Solution (Life Technologies, # 11,140–050).
Viruses
HCoV OC43 was purchased from ATCC (#VR-1558). HCoV OC43 viral titration was performed as follows. Vero cells were infected with a serial diluted HCoV OC43. After 1 h, the medium was removed and replaced by the DMEM plus 2% FBS. Cells were examined for cytopathic effects to determine TCID50 for five days.
Plasmids and chemicals
NSP14 genes from SARS-CoV-2, SARS-CoV, MERS-CoV, HCoV OC43, HCoV 229E, TGEV, and the 5 K/5R mutant were synthesized by GenScript and then cloned into the pCMV-3Tag-8 vector (Stratagene). FLAG-tagged HOIP and OTULIN were reported before [28, 70]. Mutants of SARS-CoV-2 NSP14 were constructed by PCR or using a Q5® Site-Directed Mutagenesis Kit (New England Biolabs, Ipswich, MA, USA). Briefly, PCR was performed using the FLAG-tagged NSP14 in pCMV3Tag-8 vector (Stratagene, Santa Clara, CA, USA) as the template. After PCR, the PCR product is added to the Kinase-Ligase-DpnI (KLD) enzyme mix for circularization and template removal. The mixture was transformed into E. coli for generating plasmids. All mutants were validated by DNA sequencing. All primers used for cloning and mutagenesis are listed in Supplementary Table 2.
HOIPIN-8 was purchased from Axon Medchem (# 2972). IKK-16 was purchased from Selleck Chemicals (# S2882).
Antibodies
Primary antibodies: Anti-β-actin [Abcam, # ab8227, WB (1:1,000)], anti-FLAG [Sigma, # F3165, WB (1:1,000), IFA (1:100)], anti-linear ubiquitin [Millipore, # MABS451, WB (1:1,000)], anti-ubiquitin [Cell Signaling Technology, #3933S, WB (1:1,000)], anti-HA [Cell Signaling Technology, # 3724, WB (1:1,000), IFA (1:100)], anti-IκBα [Cell Signaling Technology, # 4814S, WB (1:1,000)], anti-phospho-IκBα (Ser32) [Cell Signaling Technology, # 2859S, WB (1:1,000)], anti-IKKα [Cell Signaling Technology, # 11930S, WB (1:1,000)], anti-IKKβ [Cell Signaling Technology, # 8943S, WB (1:1,000)], anti-phospho-IKKακβ (Ser176/180) [Cell Signaling Technology, # 2697S, WB (1:1,000)], anti-NEMO [Cell Signaling Technology, # 2695S, WB (1:1,000)], anti-TBK1 [Cell Signaling Technology, # 3504S, WB (1:1,000)], anti-phospho-TBK1 (Ser172) [Cell Signaling Technology, # 5483S, WB (1:1,000)], anti-human MAVS [Cell Signaling Technology, # 24930S, WB (1:1,000)], anti-Myc [Cell Signaling Technology, # 2276S, WB (1:1,000)], anti-Myc [Bethyl Laboratories, # A190-105A, WB (1:1,000)], anti-HOIP [R&D Systems, # MAB8039SP, WB (1:2,000), IFA (1:100)], anti-MyD88 [ABclonal, # A19082, WB (1:1,000)], anti-HCoV OC43 nucleoprotein [Sino Biological, # 40,643-T62, WB (1:1,000)].
Secondary antibodies: Goat anti-Mouse IgG-HRP [Bethyl Laboratories, # A90-116P, WB (1:10,000)], Goat anti-Rabbit IgG-HRP [Bethyl Laboratories, # A120-201P, WB (1:10,000)], Alexa Fluor 594 Goat Anti-Mouse IgG (H + L) [Life Technologies, # A11005, IFA (1:200)], Alexa Fluor 488 Goat Anti-Rabbit IgG (H + L) [Life Technologies, # A11034, IFA (1:200)].
Sample preparation, Western blotting, and immunoprecipitation
Approximately 1 X 106 cells were lysed in 500 µl of tandem affinity purification (TAP) lysis buffer [50 mM Tris–HCl (pH 7.5), 10 mM MgCl2, 100 mM NaCl, 0.5% Nonidet P40, 10% glycerol, the Complete EDTA-free protease inhibitor cocktail tablets (Roche, # 11,873,580,001)] for 30 min at 4 °C. The lysates were then centrifuged for 30 min at 15,000 rpm. Supernatants were collected and mixed with the Lane Marker Reducing Sample Buffer (Thermo Fisher Scientific, # 39,000).
Western blotting and immunoprecipitation were performed as described in a previous study [71]. Briefly, samples (10–15 μl) were loaded into Mini-Protean TGX Precast Gels, 15 well (Bio-Rad, # 456–103), and run in 1 × Tris/Glycine/SDS buffer (Bio-Rad, # 161–0732) for 35 min at 200 V. Protein samples were transferred to Immun-Blot PVDF Membranes (Bio-Rad, # 162–0177) in 1 × Tris/Glycine Buffer (Bio-Rad, # 161–0734) at 70 V for 60 min. PVDF membranes were blocked in 1 × TBS buffer (Bio-Rad, # 170–6435) containing 5% Blotting-Grade Blocker (Bio-Rad, # 170–6404) for 1 h. After washing with 1 × TBS buffer for 30 min, the membrane blot was incubated with the appropriately diluted primary antibody in antibody dilution buffer (1 × TBS, 5% BSA, 0.02% sodium azide) at 4 °C for 16 h. Then, the blot was washed three times with 1 × TBS (each time for 10 min) and incubated with secondary HRP-conjugated antibody in antibody dilution buffer (1:10,000 dilution) at room temperature for 1 h. After three washes with 1 × TBS (each time for 10 min), the blot was incubated with Clarity Western ECL Substrate (Bio-Rad, # 170–5060) for 1–2 min. The membrane was removed from the substrates and then exposed to the Amersham imager 600 (GE Healthcare Life Sciences, Marlborough, MA).
For immunoprecipitation, 2% of cell lysates were saved as an input control, and the remainder was incubated with 5–10 μl of the indicated antibody plus 20 μl of Pierce Protein A/G Plus Agarose (Thermo Fisher Scientific, # 20,423) or 10 μl of EZview Red Anti-FLAG M2 Affinity Gel (Sigma, # F2426). After mixing end-over-end at 4 °C overnight, the beads were washed 3 times (5 min each wash) with 500 μl of lysis buffer. For ubiquitin detection, all beads were washed with 1 M urea for 15 min, 3 times to exclude potential binding of unanchored polyubiquitin.
Immunofluorescence assay
Cells were cultured in the Lab-Tek II CC2 Chamber Slide System 4-well (Thermo Fisher Scientific, # 154,917). After the indicated treatment, the cells were fixed and permeabilized in cold methanol for 10 min at −20 °C. Then, the slides were washed with 1 × PBS for 10 min and blocked with Odyssey Blocking Buffer (LI-COR Biosciences, # 927–40,000) for 1 h. The slides were incubated in Odyssey Blocking Buffer with appropriately diluted primary antibodies at 4 °C for 16 h. After 3 washes (10 min per wash) with 1 × PBS, the cells were incubated with the corresponding Alexa Fluor conjugated secondary antibodies (Life Technologies) for 1 h at room temperature. The slides were washed three times (10 min each time) with 1 × PBS and counterstained with 300 nM DAPI for 1 min, followed by washing with 1 × PBS for 1 min. After air-drying, the slides were sealed with Gold Seal Cover Glass (Electron Microscopy Sciences, # 3223) using Fluoro-gel (Electron Microscopy Sciences, # 17,985–10). Images were captured and analyzed using a Revolve Microscope (Discover Echo Inc.).
Real-time PCR
Total RNA was prepared using the RNeasy Mini Kit (Qiagen, # 74,106). One µg quantity of RNA was reverse transcribed into cDNA using the QuantiTect reverse transcription kit (Qiagen, # 205,311). For one real-time reaction, 10 µl of SYBR Green PCR reaction mix (Eurogentec), including 100 ng of the synthesized cDNA plus an appropriate oligonucleotide primer pair, were analyzed on a CFX384 Touch Real-Time PCR Detection System (Bio-Rad). The comparative Ct method was used to determine the relative mRNA expression of genes normalized by the housekeeping gene GAPDH. The primer sequences: human GAPDH, forward primer 5`- AGGTGAAGGTCGGAGTCA-3`, reverse primer 5`-GGTCATTGATGGCAACAA-3`; human IL-6, forward primer 5`- ACTCACCTCTTCAGAACGAATTG −3`, reverse primer 5`- CCATCTTTGGAAGGTTCAGGTTG −3`; human IL-8, forward primer 5`- TTTTGCCAAGGAGTGCTAAAGA −3`, reverse primer 5`- AACCCTCTGCACCCAGTTTTC −3`; human IFNb1, forward primer 5`- TCATCCTGTCCTTGAGGCAGT −3`, reverse primer 5`- CAGCAATTTTCAGTGTCAGAAGC −3`; human CXCL5, forward primer 5`- AGCTGCGTTGCGTTTGTTTAC −3`, reverse primer 5`- TGGCGAACACTTGCAGATTAC −3`; human ISG15, forward primer 5`- CGCAGATCACCCAGAAGATCG −3`, reverse primer 5`- TTCGTCGCATTTGTCCACCA −3`; human MX1, forward primer 5`- GTTTCCGAAGTGGACATCGCA −3`, reverse primer 5`- CTGCACAGGTTGTTCTCAGC −3`; human OAS3, forward primer 5`- GCTTCAAGAGCTATGTGGACC −3`, reverse primer 5`- GGAAACGTGAGTCTCAGACCA −3`; human CXCL10 (IP10), forward primer 5`- TTCAAGGAGTACCTCTCTCTAG −3`, reverse primer 5`- CTGGATTCAGACATCTCTTCTC −3`; human CCL5 (RANTES) qPCR primers were purchased from Qiagen (# PPH00703B-200); HCoV OC43, forward primer 5`- ATGTTAGGCCGATAATTGAGGACTAT −3`, reverse primer 5`- AATGTAAAGATGGCCGCGTATT −3`.
RNA sequencing and data analysis
Total RNA was prepared using the RNeasy Mini Kit. RNA samples were sent to Novogene (Sacramento, CA) for sequencing. Each sample was sequenced to generate a minimum of 20 million reads. Raw sequencing reads were aligned to human genome (hg38; Genome Reference Consortium GRCH38). The alignments were conducted using the Spliced Transcripts Alignment to a Reference (STAR) aligner version 2.5.3 (–clip5pNbases 6, default options) [72] and were subjected to visual inspection using the Integrative Genomics Viewer (IGV) genome browser [73]. Transcript data from STAR were then analyzed using the RSEM software (version 1.3.0 [74]) for quantification of human gene expression. The EBSeq software [75] was utilized to call statistically differentially expressed genes using a false discovery rate (FDR) less than 0.05 as previous described [76].
Plasmid transfection
HEK293 cells were transfected using Lipofectamine 3000 (Life Technologies, # L3000015) Transfection Reagent according to the manufacturer’s protocol. For co-IP experiments, a total of 2.5 µg of plasmids was transfected into approximately 1.2 × 106 cells. For other experiments, a total of 0.5 µg of plasmids was transfected into approximately 2 × 105 cells.
Reporter assays
To perform NF-κB reporter assays, 0.2 µg of each SARS-CoV-2 plasmid was co-transfected with 0.1 µg of pNF-kB Luc reporter (Stratagene) and 0.02 µg of pRL-SV40 plasmid (Promega), containing the Renilla luciferase gene as an internal control, into 2 × 105 HEK293 cells. After 48 h, the activities of both firefly and Renilla luciferases were measured using the Dual Luciferase Reporter Assay Kit (Promega). The luciferase activities were normalized to the Renilla luciferase activity of the internal control. Each experiment was performed in triplicate.
MTT assay
Cells were seeded in a 96-well plate in the culture medium. After 24 h, cells were either infected with HCoV OC43 or treated with inhibitors for 5 days. Cell viability and growth were assessed every day for 5 days using the MTT Cell Proliferation Kit (Cayman Chemical, Ann Arbor, MI, USA).
Protein purification from E. coli
NSP14 and the 5 K/5R mutant were cloned into pMXB10 (New England Biolabs, # E6901S) to fuse an MBP tag. These constructs were transfected into BL21 (DE3) E. coli (New England Biolabs, # C2527I) and cultured in LB broth at 20 °C. IPTG (0.4 mM) was added to induce protein expression. MBP-tagged proteins were purified using the IMPACT kit (New England Biolabs, # E6901S), and the MBP pull-down assays were performed using the anti-MBP Magnetic Beads New England Biolabs, # E8037S).
CRISPR/Cas9
The single guide RNA (sgRNA) targeting sequences: control CRISPR sgRNA: CTGAAAAAGGAAGGAGTTGA; human HOIP sgRNA: 5`- CGTACGAGAACTTGCATTTG −3`; human MAVS sgRNA: 5`- ACAGGGTCAGTTGTATCTAC −3`; human UBC13 sgRNA: 5`- GGCGTTGCTCTCATCTGGTT −3`; human OTULIN sgRNA: 5`- GGCTCCGGATCGTTCGGAGC −3`; human IKKα sgRNA: 5`- GTCTGTACCAGCATCGGGTG −3`; human IKKβ sgRNA: 5`- GACTGTCACCCTCAGTTCGC −3`; human NEMO sgRNA: 5`- GGCAGCAGATCAGGACGTAC −3`. The sgRNA was cloned into lentiCRISPR v2 vector [77]. The lentiviral construct was transfected with psPAX2 and pMD2G into HEK293T cells using PEI. After 48 h, the media containing lentivirus were collected. The targeted cells were infected with the media containing the lentivirus supplemented with 10 μg/ml polybrene. Cells were selected with 10 μg/ml puromycin for 14 days. Single clones were expanded for knockout confirmation by Western blotting.
Statistics and reproducibility
The sample size was sufficient for data analyses. Data were statistically analyzed using the software GraphPad Prism 9.
Supplementary Information
Supplementary Material 3: Supplementary Fig. 1. NSP14 mediates NF-κB activation. (a) HEK293 cells were co-transfected with 0.2 µg of the indicated FLAG-tagged SARS-CoV-2 gene along with NF-κB reporter and pRL-SV40. After 48 h, cells were harvested, and the relative reporter activity was determined by calculating the ratio of firefly luciferase to Renilla luciferase. The p -value was calculated by one-way ANOVA followed by Dunnett's multiple comparisons test. Lysates were blotted with anti-FLAG and anti-α-tubulin antibodies. (b) HEK293 cells were co-transfected with 0.05 µg, 0.1 µg, and 0.15 µg of FLAG-tagged SARS-CoV-2 NSP14 along with NF-κB reporter and pRL-SV40. After 48 h, cells were harvested, and the relative reporter activity was determined by calculating the ratio of firefly luciferase to Renilla luciferase. Lysates were blotted with anti-FLAG and anti-α-tubulin antibodies. (c) 0.1 µg of FLAG-tagged SARS-CoV-2 NSP14 and 0.1 µg of FLAG-tagged SARS-CoV-2 NSP10 were co-transfected with the NF-κB reporter and pRL-SV40 into HEK293 cells. Western blotting demonstrates the expression levels of NSP14-FLAG and NSP10-FLAG. The position of NSP14 and NSP10 is denoted. (d) FLAG-tagged SARS-CoV-2 NSP14 or the indicated zinc finger mutant was co-transfected with the NF-κB reporter and pRL-SV40 into HEK293 cells. After 48 h, cells were harvested, and the relative reporter activity was determined by calculating the ratio of firefly luciferase to Renilla luciferase. Lysates were blotted with anti-FLAG and anti-α-tubulin antibodies. The p -value was calculated by one-way ANOVA followed by Tukey's multiple comparisons test (b, c, d). Supplementary Fig. 2. NSP14 activates NF-κB via linear ubiquitination. (a) HEK293 cells were transfected with FLAG-tagged NSP14 genes derived from various coronaviruses. After 48 h, cell lysates were subjected to immunoprecipitation and subsequent blotting using the indicated antibodies. (b) Lysates of HEK293 cells and two HOIP knockout cell lines were blotted as indicated. (c) Blotting analysis of lysates from HEK293 cells and two UBC13 knockout cell lines. (d-e) Wild-type, HOIP knockout (d), and UBC13 (e) knockout HEK293 cells were stimulated with 10 ng/mL TNFα for designated times. Subsequently, cell lysates were analyzed by Western blotting. (f) Wild-type HEK293 cells, along with two UBC13 knockout and two HOIP knockout HEK293 cell lines, were transfected with FLAG-tagged SARS-CoV-2 NSP14 along with NF-κB reporter and pRL-SV40. After 48 h, cells were collected, and the relative reporter activity was determined by calculating the ratio of firefly luciferase to Renilla luciferase. (g) Blotting analysis of lysates from OTULIN wild-type and knockout HEK293 cells. (h) OTULIN wild-type and knockout HEK293 cells were stimulated with 10 ng/mL TNFα for designated times. Cell lysates were blotted using the indicated antibodies. (i) Wild-type and two OTULIN knockout HEK293 cell lines were transfected with FLAG-tagged SARS-CoV-2 NSP14 with the NF-κB reporter and pRL-SV40. The p -value was calculated by two-way ANOVA followed by Sidak's multiple comparisons test (f, i). Supplementary Fig. 3. NSP14 interacts with HOIP. (a) NSP14-HA was co-transfected with FLAG-tagged HOIP, HOIL-1, or SHARPIN into HEK293 cells. After 48 h, cell lysates were subjected to immunoprecipitation with the anti-FLAG antibody and subsequent blotting with anti-FLAG and anti-HA antibodies. (b) NSP14-FLAG or GFP-FLAG was transfected into HEK293 cells. After 48 h, cell lysates were immunoprecipitated with the anti-FLAG antibody and blotted using the indicated antibodies. (c) Vector or NSP14-HA was transfected into A549 cells. After 48 h, cells were fixed and stained with anti-HOIP (red), anti-HA (green), and DAPI nuclear stain (blue). Scale bar = 10 μm. (d) Schematic of HOIP mutants. PUB: Peptide N-glycanase/UBA or UBX-containing proteins; ZnF: Zinc finger; NZF: Npl4 zinc finger; UBA: Ubiquitin-associated; RBR: RING between RING fingers. (e) FLAG-tagged HOIP or the indicated mutant was co-transfected with Myc-tagged (NSP14-Myc) into HEK293 cells. After 48 h, cell lysates were immunoprecipitated with the anti-FLAG antibody and blotted as indicated. Supplementary Fig. 4. The IKK complex is required for NSP14-mediated NF-κB activation. (a) Co-transfection of FLAG-tagged NSP14 with IKKα, IKKβ, or p65, along with NF-κB reporter and pRL-SV40, into HEK293 cells. After 48 h, cells were collected, and the relative reporter activity was determined by calculating the ratio of firefly luciferase to Renilla luciferase. (b-d) Wild type, IKKα knockout (b), IKKβ knockout (c), or NEMO knockout (d) HEK293 cells were stimulated with 10 ng/mL TNFα for designated times. Cell lysates were blotted as indicated. (e–g) Transfection of vector or FLAG-tagged NSP14 with NF-κB reporter and pRL-SV40 into wild-type, IKKα knockout (e), IKKβ knockout (f), and NEMO knockout (g) HEK293 cells. After 48 h, cells were collected, and the ratio of firefly luciferase to Renilla luciferase was calculated to determine the relative reporter activity. (h) Vector or FLAG-tagged NSP14 was transfected with the NF-κB reporter and pRL-SV40 into wild-type and TBK1 knockout MEFs. After 48 h, cells were harvested, and the relative reporter activity was determined by calculating the ratio of firefly luciferase to Renilla luciferase. (i) FLAG-tagged NSP14 or NSP14 K/R was transfected with Myc-tagged IKKα or IKKβ into HEK293 cells, along with NF-κB reporter and pRL-SV40. After 48 h, cells were harvested, and the relative reporter activity was determined by calculating the ratio of firefly luciferase to Renilla luciferase. Lysates were blotted with anti-FLAG, anti-Myc, and anti-α-tubulin antibodies. The p -value was calculated by two-way ANOVA followed by Sidak's multiple comparisons test (a, e, f, g, h) or one-way ANOVA followed by Tukey's multiple comparisons test (i). Supplementary Fig. 5. MAVS knockout impairs ISG but not proinflammatory factor expression. (a) MAVS wild-type and knockout A549 cells were infected with HCoV OC43 for the designated times. Cell lysates were blotted as indicated, and band densitometry was calculated using Image J. The ratio of phosphorylated IKK to total IKK in each lane was indicated. (b) HOIP wild-type and knockout H1299 cells were infected with 1 MOI of HCoV OC43 for 16 h. Real-time PCR was conducted to determine the relative mRNA levels of proinflammatory factors and ISGs. (c) MAVS wild-type and knockout A549 cells were infected with 1 MOI of HCoV OC43 for 16 h. Real-time PCR was conducted to determine the relative mRNA levels of proinflammatory factors and ISGs. (d) MTT assays of A549 cells treated with DMSO, 30 μM HOIPIN-8, or 10 μM IKK-16 for 5 days. The p- value was calculated by two-way ANOVA followed by Sidak's multiple comparisons test. Supplementary Fig. 6. HOIPIN-8 inhibits HCoV OC43 viral replication. (a) HOIP wild-type and knockout A549 cells were infected with 0.01 MOI of HCoV OC43 for the designated days. MTT assays were performed to determine cell viability. (b) HOIP wild-type and knockout H1299 cells were infected with 0.01 MOI of HCoV OC43 for the designated days. Lysates were blotted as indicated. (c) A549 cells were treated with DMSO, HOIPIN-8, or IKK-16 for 2 h and then infected with 0.01 MOI of HCoV OC43 for the designated days. TCID 50 of culture supernatants containing HCoV OC43 were determined on Vero cells. The p -value was calculated by two-way ANOVA followed by Sidak's multiple comparisons test. (d) A549 cells were treated with DMSO or HOIPIN-8 for 2 h and then infected with 0.1 MOI HCoV OC43 for 24 h. Cells were stained with anti-dsRNA (red) and DAPI (blue). The relative ratio of positively stained cells is summarized in the graph. The p -value was calculated by t -test.
Acknowledgements
This work was supported by the National Institutes of Health grants R01AI141399 (S.L.), R21AI167870 (S.L.), R21AI166043 (S.L.), R21AI137750 (S.L.), and R01CA261258 (Z.L.).
Authors’ contributions
L.W. and S.L. conceived and designed the project. F.H. performed experiments in Figs. 1a and d-e, 2c-h, 3d and f, 4c-e, 5b and g, 6a-b and d-h, S1a-b, S2a-i, S3b and e, S4b-d, and S5a and c-d. W.H. performed experiments in Figs. 1e, 3a and c, 3g, 5a and c-f, S1a and d, S3a and c, S4a and e, S4g, S5b, S6a and b. L.W. performed experiments in Figs. 7a-c, S4h, and S6c. K.S. performed experiments in Figs. 1b, 2a, 7d, S1a and c, and S6d. A.H. performed experiments in Fig. 4 b. Y.W. performed experiments in Figure S1a. F.H., W.H., K.S., A.H., S.L., and L.W. analyzed the data. F.H., W.H., K.S., K.L., Y.W., Y.S., A.H., and L.W. made the constructs. Z.L. assisted in the analysis of RNA-seq data. S.L. and L.W. wrote the draft. All authors participated in manuscript writing.
Data availability
The RNA-seq data that support the findings of this study have been deposited in NIH SRA with the accession number PRJNA729116.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Fang Hua and Wenzhuo Hao contributed equally to this work.
Contributor Information
Lingyan Wang, Email: lwang32@tulane.edu.
Shitao Li, Email: sli38@tulane.edu.
References
- 1.Ye ZW, et al. Zoonotic origins of human coronaviruses. Int J Biol Sci. 2020;16:1686–97. 10.7150/ijbs.45472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bucknall, R. A., King, L. M., Kapikian, A. Z. & Chanock, R. M. Studies with human coronaviruses. II. Some properties of strains 229E and OC43. Proc Soc Exp Biol Med. 1972;139:722-727. 10.3181/00379727-139-36224. [DOI] [PubMed]
- 3.Woo PC, et al. Clinical and molecular epidemiological features of coronavirus HKU1-associated community-acquired pneumonia. J Infect Dis. 2005;192:1898–907. 10.1086/497151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hu, Y. et al. The Severe Acute Respiratory Syndrome Coronavirus Nucleocapsid Inhibits Type I Interferon Production by Interfering with TRIM25-Mediated RIG-I Ubiquitination. J Virol. 2017;91. 10.1128/JVI.02143-16. [DOI] [PMC free article] [PubMed]
- 5.Wu J, et al. SARS-CoV-2 ORF9b inhibits RIG-I-MAVS antiviral signaling by interrupting K63-linked ubiquitination of NEMO. Cell Rep. 2021;34: 108761. 10.1016/j.celrep.2021.108761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ong, E. Z. et al. A Dynamic Immune Response Shapes COVID-19 Progression. Cell Host Microbe. 2020;27:879–882.e872. 10.1016/j.chom.2020.03.021. [DOI] [PMC free article] [PubMed]
- 7.Zhou, Z. et al. Heightened Innate Immune Responses in the Respiratory Tract of COVID-19 Patients. Cell Host Microbe. 2020;27:883–890. e882. 10.1016/j.chom.2020.04.017. [DOI] [PMC free article] [PubMed]
- 8.Blanco-Melo, D. et al. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell. 2020;181:1036–1045. 10.1016/j.cell.2020.04.026 [DOI] [PMC free article] [PubMed]
- 9.Huang C, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan. China Lancet. 2020;395:497–506. 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Merad M, Subramanian A, Wang TT. An aberrant inflammatory response in severe COVID-19. Cell Host Microbe. 2021;29:1043–7. 10.1016/j.chom.2021.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lowery SA, Sariol A, Perlman S. Innate immune and inflammatory responses to SARS-CoV-2: Implications for COVID-19. Cell Host Microbe. 2021;29:1052–62. 10.1016/j.chom.2021.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Minskaia E, et al. Discovery of an RNA virus 3’->5’ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc Natl Acad Sci U S A. 2006;103:5108–13. 10.1073/pnas.0508200103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Y, et al. Functional screen reveals SARS coronavirus nonstructural protein nsp14 as a novel cap N7 methyltransferase. Proc Natl Acad Sci U S A. 2009;106:3484–9. 10.1073/pnas.0808790106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Case JB, Ashbrook AW, Dermody TS, Denison MR. Mutagenesis of S-Adenosyl-l-Methionine-Binding Residues in Coronavirus nsp14 N7-Methyltransferase Demonstrates Differing Requirements for Genome Translation and Resistance to Innate Immunity. J Virol. 2016;90:7248–56. 10.1128/JVI.00542-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Becares M, et al. Mutagenesis of Coronavirus nsp14 Reveals Its Potential Role in Modulation of the Innate Immune Response. J Virol. 2016;90:5399–414. 10.1128/JVI.03259-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tahir M. Coronavirus genomic nsp14-ExoN, structure, role, mechanism, and potential application as a drug target. J Med Virol. 2021;93:4258–64. 10.1002/jmv.27009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou Y, et al. Cellular RNA Helicase DDX1 Is Involved in Transmissible Gastroenteritis Virus nsp14-Induced Interferon-Beta Production. Front Immunol. 2017;8:940. 10.3389/fimmu.2017.00940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li TW, et al. SARS-CoV-2 Nsp14 protein associates with IMPDH2 and activates NF-kappaB signaling. Front Immunol. 2022;13:1007089. 10.3389/fimmu.2022.1007089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zaffagni, M. et al. SARS-CoV-2 Nsp14 mediates the effects of viral infection on the host cell transcriptome. Elife. 2022;11. 10.7554/eLife.71945. [DOI] [PMC free article] [PubMed]
- 20.Ishaq M, et al. The DEAD-box RNA helicase DDX1 interacts with RelA and enhances nuclear factor kappaB-mediated transcription. J Cell Biochem. 2009;106:296–305. 10.1002/jcb.22004. [DOI] [PubMed] [Google Scholar]
- 21.Braun-Sand SB, Peetz M. Inosine monophosphate dehydrogenase as a target for antiviral, anticancer, antimicrobial and immunosuppressive therapeutics. Future Med Chem. 2010;2:81–92. 10.4155/fmc.09.147. [DOI] [PubMed] [Google Scholar]
- 22.Shimizu Y, Taraborrelli L, Walczak H. Linear ubiquitination in immunity. Immunol Rev. 2015;266:190–207. 10.1111/imr.12309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haas TL, et al. Recruitment of the linear ubiquitin chain assembly complex stabilizes the TNF-R1 signaling complex and is required for TNF-mediated gene induction. Mol Cell. 2009;36:831–44. 10.1016/j.molcel.2009.10.013. [DOI] [PubMed] [Google Scholar]
- 24.Tokunaga F, et al. Involvement of linear polyubiquitylation of NEMO in NF-kappaB activation. Nat Cell Biol. 2009;11:123–32. 10.1038/ncb1821. [DOI] [PubMed] [Google Scholar]
- 25.Tokunaga F, et al. SHARPIN is a component of the NF-kappaB-activating linear ubiquitin chain assembly complex. Nature. 2011;471:633–6. 10.1038/nature09815. [DOI] [PubMed] [Google Scholar]
- 26.Ikeda, F. et al. SHARPIN forms a linear ubiquitin ligase complex regulating NF-kappaB activity and apoptosis. Nature. 2011;471:637–641. https://doi.org/nature09814. [DOI] [PMC free article] [PubMed]
- 27.Gerlach, B. et al. Linear ubiquitination prevents inflammation and regulates immune signalling. Nature. 2011;471:591-596. https://doi.org/nature09816. [DOI] [PubMed]
- 28.Fu B, Li S, Wang L, Berman MA, Dorf ME. The ubiquitin conjugating enzyme UBE2L3 regulates TNFalpha-induced linear ubiquitination. Cell Res. 2014;24:376–9. 10.1038/cr.2013.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zak, D. E. et al. Systems analysis identifies an essential role for SHANK-associated RH domain-interacting protein (SHARPIN) in macrophage Toll-like receptor 2 (TLR2) responses. Proc Natl Acad Sci U S A. 2011;108:11536–11541. https://doi.org/1107577108. [DOI] [PMC free article] [PubMed]
- 30.Greenfeld H, et al. TRAF1 Coordinates Polyubiquitin Signaling to Enhance Epstein-Barr Virus LMP1-Mediated Growth and Survival Pathway Activation. PLoS Pathog. 2015;11: e1004890. 10.1371/journal.ppat.1004890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nie Y, et al. SARS-CoV-2 ORF3a positively regulates NF-kappaB activity by enhancing IKKbeta-NEMO interaction. Virus Res. 2023;328: 199086. 10.1016/j.virusres.2023.199086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tofaute, M. J. et al. SARS-CoV-2 NSP14 MTase activity is critical for inducing canonical NF-kappaB activation. Biosci Rep. 2024;44. 10.1042/BSR20231418. [DOI] [PMC free article] [PubMed]
- 33.Ma Y, et al. Structural basis and functional analysis of the SARS coronavirus nsp14-nsp10 complex. Proc Natl Acad Sci U S A. 2015;112:9436–41. 10.1073/pnas.1508686112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leon O, Roth M. Zinc fingers: DNA binding and protein-protein interactions. Biol Res. 2000;33:21–30. 10.4067/s0716-97602000000100009. [DOI] [PubMed] [Google Scholar]
- 35.Song, K. & Li, S. The Role of Ubiquitination in NF-kappaB Signaling during Virus Infection. Viruses.2021;13. 10.3390/v13020145. [DOI] [PMC free article] [PubMed]
- 36.Fiil BK, et al. OTULIN restricts Met1-linked ubiquitination to control innate immune signaling. Mol Cell. 2013;50:818–30. 10.1016/j.molcel.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Keusekotten K, et al. OTULIN antagonizes LUBAC signaling by specifically hydrolyzing Met1-linked polyubiquitin. Cell. 2013;153:1312–26. 10.1016/j.cell.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hayden MS, Ghosh S. NF-kappaB, the first quarter-century: remarkable progress and outstanding questions. Genes Dev. 2012;26:203–34. 10.1101/gad.183434.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dittmar G, Winklhofer KF. Linear Ubiquitin Chains: Cellular Functions and Strategies for Detection and Quantification. Front Chem. 2019;7:915. 10.3389/fchem.2019.00915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim DK, et al. A proteome-scale map of the SARS-CoV-2-human contactome. Nat Biotechnol. 2023;41:140–9. 10.1038/s41587-022-01475-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oikawa D, et al. Molecular bases for HOIPINs-mediated inhibition of LUBAC and innate immune responses. Commun Biol. 2020;3:163. 10.1038/s42003-020-0882-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Waelchli R, et al. Design and preparation of 2-benzamido-pyrimidines as inhibitors of IKK. Bioorg Med Chem Lett. 2006;16:108–12. 10.1016/j.bmcl.2005.09.035. [DOI] [PubMed] [Google Scholar]
- 43.Cao Z, et al. Ubiquitination of SARS-CoV-2 ORF7a promotes antagonism of interferon response. Cell Mol Immunol. 2021;18:746–8. 10.1038/s41423-020-00603-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao Y, et al. SARS-CoV-2 spike protein interacts with and activates TLR41. Cell Res. 2021. 10.1038/s41422-021-00495-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Camell CD, et al. Senolytics reduce coronavirus-related mortality in old mice. Science. 2021. 10.1126/science.abe4832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zheng M, et al. TLR2 senses the SARS-CoV-2 envelope protein to produce inflammatory cytokines. Nat Immunol. 2021;22:829–38. 10.1038/s41590-021-00937-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yuan Q, Liao Y, Torres J, Tam JP, Liu DX. Biochemical evidence for the presence of mixed membrane topologies of the severe acute respiratory syndrome coronavirus envelope protein expressed in mammalian cells. FEBS Lett. 2006;580:3192–200. 10.1016/j.febslet.2006.04.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rahighi S, et al. Specific recognition of linear ubiquitin chains by NEMO is important for NF-kappaB activation. Cell. 2009;136:1098–109. 10.1016/j.cell.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 49.Nilsson-Payant BE, et al. The NF-kappaB Transcriptional Footprint Is Essential for SARS-CoV-2 Replication. J Virol. 2021;95: e0125721. 10.1128/JVI.01257-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yuen CK, et al. SARS-CoV-2 nsp13, nsp14, nsp15 and orf6 function as potent interferon antagonists. Emerg Microbes Infect. 2020;9:1418–28. 10.1080/22221751.2020.1780953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lei X, et al. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat Commun. 2020;11:3810. 10.1038/s41467-020-17665-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zust R, et al. Ribose 2’-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat Immunol. 2011;12:137–43. 10.1038/ni.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jiang HW, et al. SARS-CoV-2 Orf9b suppresses type I interferon responses by targeting TOM70. Cell Mol Immunol. 2020;17:998–1000. 10.1038/s41423-020-0514-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Konno, Y. et al. SARS-CoV-2 ORF3b Is a Potent Interferon Antagonist Whose Activity Is Increased by a Naturally Occurring Elongation Variant. Cell Rep. 2020;32. https://doi.org/ARTN10.1016/j.celrep.2020.108185. [DOI] [PMC free article] [PubMed]
- 55.Fu YZ, et al. SARS-CoV-2 membrane glycoprotein M antagonizes the MAVS-mediated innate antiviral response. Cell Mol Immunol. 2021;18:613–20. 10.1038/s41423-020-00571-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zheng, Y. et al. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) membrane (M) protein inhibits type I and III interferon production by targeting RIG-I/MDA-5 signaling. Signal Transduct Tar. 2020;5. https://doi.org/ARTN2991038/s41392–020–00438–7. [DOI] [PMC free article] [PubMed]
- 57.Xia, H. J. et al. Evasion of Type I Interferon by SARS-CoV-2. Cell Rep. 2020;33. https://doi.org/ARTN1082341016/j.celrep.2020.108234. [DOI] [PMC free article] [PubMed]
- 58.Wang S, et al. Targeting liquid-liquid phase separation of SARS-CoV-2 nucleocapsid protein promotes innate antiviral immunity by elevating MAVS activity. Nat Cell Biol. 2021;23:718–32. 10.1038/s41556-021-00710-0. [DOI] [PubMed] [Google Scholar]
- 59.Shi CS, et al. SARS-coronavirus open reading frame-9b suppresses innate immunity by targeting mitochondria and the MAVS/TRAF3/TRAF6 signalosome. J Immunol. 2014;193:3080–9. 10.4049/jimmunol.1303196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Niemeyer D, et al. Middle East respiratory syndrome coronavirus accessory protein 4a is a type I interferon antagonist. J Virol. 2013;87:12489–95. 10.1128/JVI.01845-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cahir-McFarland ED, Davidson DM, Schauer SL, Duong J, Kieff E. NF-kappa B inhibition causes spontaneous apoptosis in Epstein-Barr virus-transformed lymphoblastoid cells. Proc Natl Acad Sci U S A. 2000;97:6055–60. 10.1073/pnas.100119497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hiscott J, Kwon H, Genin P. Hostile takeovers: viral appropriation of the NF-kappaB pathway. J Clin Invest. 2001;107:143–51. 10.1172/JCI11918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fiume G, et al. Human immunodeficiency virus-1 Tat activates NF-kappaB via physical interaction with IkappaB-alpha and p65. Nucleic Acids Res. 2012;40:3548–62. 10.1093/nar/gkr1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wurzer WJ, et al. NF-kappaB-dependent induction of tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) and Fas/FasL is crucial for efficient influenza virus propagation. J Biol Chem. 2004;279:30931–7. 10.1074/jbc.M403258200. [DOI] [PubMed] [Google Scholar]
- 65.Ehrhardt C, et al. The NF-kappaB inhibitor SC75741 efficiently blocks influenza virus propagation and confers a high barrier for development of viral resistance. Cell Microbiol. 2013;15:1198–211. 10.1111/cmi.12108. [DOI] [PubMed] [Google Scholar]
- 66.Kratzel A, et al. A genome-wide CRISPR screen identifies interactors of the autophagy pathway as conserved coronavirus targets. PLoS Biol. 2021;19: e3001490. 10.1371/journal.pbio.3001490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang, R. et al. Genetic Screens Identify Host Factors for SARS-CoV-2 and Common Cold Coronaviruses. Cell. 2021;184:106–119.e114. 10.1016/j.cell.2020.12.004 [DOI] [PMC free article] [PubMed]
- 68.Daniloski, Z. et al. Identification of Required Host Factors for SARS-CoV-2 Infection in Human Cells. Cell. 2021;184:92–105. e116. 10.1016/j.cell.2020.10.030 [DOI] [PMC free article] [PubMed]
- 69.Zhu S, et al. Genome-wide CRISPR activation screen identifies candidate receptors for SARS-CoV-2 entry. Sci China Life Sci. 2022;65:701–17. 10.1007/s11427-021-1990-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhao M, et al. Non-proteolytic ubiquitination of OTULIN regulates NF-kappaB signaling pathway. J Mol Cell Biol. 2020;12:163–75. 10.1093/jmcb/mjz081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhao M, et al. Non-proteolytic ubiquitination of OTULIN regulates NF-kappaB signaling pathway. J Mol Cell Biol. 2019. 10.1093/jmcb/mjz081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Robinson JT, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–6. 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Leng N, et al. EBSeq: an empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics. 2013;29:1035–43. 10.1093/bioinformatics/btt087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kheir, F. et al. Detection of Epstein-Barr Virus Infection in Non-Small Cell Lung Cancer. Cancers (Basel). 2019;11. 10.3390/cancers11060759. [DOI] [PMC free article] [PubMed]
- 77.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11:783–4. 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material 3: Supplementary Fig. 1. NSP14 mediates NF-κB activation. (a) HEK293 cells were co-transfected with 0.2 µg of the indicated FLAG-tagged SARS-CoV-2 gene along with NF-κB reporter and pRL-SV40. After 48 h, cells were harvested, and the relative reporter activity was determined by calculating the ratio of firefly luciferase to Renilla luciferase. The p -value was calculated by one-way ANOVA followed by Dunnett's multiple comparisons test. Lysates were blotted with anti-FLAG and anti-α-tubulin antibodies. (b) HEK293 cells were co-transfected with 0.05 µg, 0.1 µg, and 0.15 µg of FLAG-tagged SARS-CoV-2 NSP14 along with NF-κB reporter and pRL-SV40. After 48 h, cells were harvested, and the relative reporter activity was determined by calculating the ratio of firefly luciferase to Renilla luciferase. Lysates were blotted with anti-FLAG and anti-α-tubulin antibodies. (c) 0.1 µg of FLAG-tagged SARS-CoV-2 NSP14 and 0.1 µg of FLAG-tagged SARS-CoV-2 NSP10 were co-transfected with the NF-κB reporter and pRL-SV40 into HEK293 cells. Western blotting demonstrates the expression levels of NSP14-FLAG and NSP10-FLAG. The position of NSP14 and NSP10 is denoted. (d) FLAG-tagged SARS-CoV-2 NSP14 or the indicated zinc finger mutant was co-transfected with the NF-κB reporter and pRL-SV40 into HEK293 cells. After 48 h, cells were harvested, and the relative reporter activity was determined by calculating the ratio of firefly luciferase to Renilla luciferase. Lysates were blotted with anti-FLAG and anti-α-tubulin antibodies. The p -value was calculated by one-way ANOVA followed by Tukey's multiple comparisons test (b, c, d). Supplementary Fig. 2. NSP14 activates NF-κB via linear ubiquitination. (a) HEK293 cells were transfected with FLAG-tagged NSP14 genes derived from various coronaviruses. After 48 h, cell lysates were subjected to immunoprecipitation and subsequent blotting using the indicated antibodies. (b) Lysates of HEK293 cells and two HOIP knockout cell lines were blotted as indicated. (c) Blotting analysis of lysates from HEK293 cells and two UBC13 knockout cell lines. (d-e) Wild-type, HOIP knockout (d), and UBC13 (e) knockout HEK293 cells were stimulated with 10 ng/mL TNFα for designated times. Subsequently, cell lysates were analyzed by Western blotting. (f) Wild-type HEK293 cells, along with two UBC13 knockout and two HOIP knockout HEK293 cell lines, were transfected with FLAG-tagged SARS-CoV-2 NSP14 along with NF-κB reporter and pRL-SV40. After 48 h, cells were collected, and the relative reporter activity was determined by calculating the ratio of firefly luciferase to Renilla luciferase. (g) Blotting analysis of lysates from OTULIN wild-type and knockout HEK293 cells. (h) OTULIN wild-type and knockout HEK293 cells were stimulated with 10 ng/mL TNFα for designated times. Cell lysates were blotted using the indicated antibodies. (i) Wild-type and two OTULIN knockout HEK293 cell lines were transfected with FLAG-tagged SARS-CoV-2 NSP14 with the NF-κB reporter and pRL-SV40. The p -value was calculated by two-way ANOVA followed by Sidak's multiple comparisons test (f, i). Supplementary Fig. 3. NSP14 interacts with HOIP. (a) NSP14-HA was co-transfected with FLAG-tagged HOIP, HOIL-1, or SHARPIN into HEK293 cells. After 48 h, cell lysates were subjected to immunoprecipitation with the anti-FLAG antibody and subsequent blotting with anti-FLAG and anti-HA antibodies. (b) NSP14-FLAG or GFP-FLAG was transfected into HEK293 cells. After 48 h, cell lysates were immunoprecipitated with the anti-FLAG antibody and blotted using the indicated antibodies. (c) Vector or NSP14-HA was transfected into A549 cells. After 48 h, cells were fixed and stained with anti-HOIP (red), anti-HA (green), and DAPI nuclear stain (blue). Scale bar = 10 μm. (d) Schematic of HOIP mutants. PUB: Peptide N-glycanase/UBA or UBX-containing proteins; ZnF: Zinc finger; NZF: Npl4 zinc finger; UBA: Ubiquitin-associated; RBR: RING between RING fingers. (e) FLAG-tagged HOIP or the indicated mutant was co-transfected with Myc-tagged (NSP14-Myc) into HEK293 cells. After 48 h, cell lysates were immunoprecipitated with the anti-FLAG antibody and blotted as indicated. Supplementary Fig. 4. The IKK complex is required for NSP14-mediated NF-κB activation. (a) Co-transfection of FLAG-tagged NSP14 with IKKα, IKKβ, or p65, along with NF-κB reporter and pRL-SV40, into HEK293 cells. After 48 h, cells were collected, and the relative reporter activity was determined by calculating the ratio of firefly luciferase to Renilla luciferase. (b-d) Wild type, IKKα knockout (b), IKKβ knockout (c), or NEMO knockout (d) HEK293 cells were stimulated with 10 ng/mL TNFα for designated times. Cell lysates were blotted as indicated. (e–g) Transfection of vector or FLAG-tagged NSP14 with NF-κB reporter and pRL-SV40 into wild-type, IKKα knockout (e), IKKβ knockout (f), and NEMO knockout (g) HEK293 cells. After 48 h, cells were collected, and the ratio of firefly luciferase to Renilla luciferase was calculated to determine the relative reporter activity. (h) Vector or FLAG-tagged NSP14 was transfected with the NF-κB reporter and pRL-SV40 into wild-type and TBK1 knockout MEFs. After 48 h, cells were harvested, and the relative reporter activity was determined by calculating the ratio of firefly luciferase to Renilla luciferase. (i) FLAG-tagged NSP14 or NSP14 K/R was transfected with Myc-tagged IKKα or IKKβ into HEK293 cells, along with NF-κB reporter and pRL-SV40. After 48 h, cells were harvested, and the relative reporter activity was determined by calculating the ratio of firefly luciferase to Renilla luciferase. Lysates were blotted with anti-FLAG, anti-Myc, and anti-α-tubulin antibodies. The p -value was calculated by two-way ANOVA followed by Sidak's multiple comparisons test (a, e, f, g, h) or one-way ANOVA followed by Tukey's multiple comparisons test (i). Supplementary Fig. 5. MAVS knockout impairs ISG but not proinflammatory factor expression. (a) MAVS wild-type and knockout A549 cells were infected with HCoV OC43 for the designated times. Cell lysates were blotted as indicated, and band densitometry was calculated using Image J. The ratio of phosphorylated IKK to total IKK in each lane was indicated. (b) HOIP wild-type and knockout H1299 cells were infected with 1 MOI of HCoV OC43 for 16 h. Real-time PCR was conducted to determine the relative mRNA levels of proinflammatory factors and ISGs. (c) MAVS wild-type and knockout A549 cells were infected with 1 MOI of HCoV OC43 for 16 h. Real-time PCR was conducted to determine the relative mRNA levels of proinflammatory factors and ISGs. (d) MTT assays of A549 cells treated with DMSO, 30 μM HOIPIN-8, or 10 μM IKK-16 for 5 days. The p- value was calculated by two-way ANOVA followed by Sidak's multiple comparisons test. Supplementary Fig. 6. HOIPIN-8 inhibits HCoV OC43 viral replication. (a) HOIP wild-type and knockout A549 cells were infected with 0.01 MOI of HCoV OC43 for the designated days. MTT assays were performed to determine cell viability. (b) HOIP wild-type and knockout H1299 cells were infected with 0.01 MOI of HCoV OC43 for the designated days. Lysates were blotted as indicated. (c) A549 cells were treated with DMSO, HOIPIN-8, or IKK-16 for 2 h and then infected with 0.01 MOI of HCoV OC43 for the designated days. TCID 50 of culture supernatants containing HCoV OC43 were determined on Vero cells. The p -value was calculated by two-way ANOVA followed by Sidak's multiple comparisons test. (d) A549 cells were treated with DMSO or HOIPIN-8 for 2 h and then infected with 0.1 MOI HCoV OC43 for 24 h. Cells were stained with anti-dsRNA (red) and DAPI (blue). The relative ratio of positively stained cells is summarized in the graph. The p -value was calculated by t -test.
Data Availability Statement
The RNA-seq data that support the findings of this study have been deposited in NIH SRA with the accession number PRJNA729116.