ROPN1 is a highly expressed intracellular antigen in triple-negative breast cancer, and a T-cell receptor targeting ROPN1 enables sensitive and selective cancer cell recognition, supporting use as a safe and potent T-cell therapy.
Abstract
Triple-negative breast cancer (TNBC) has an urgent need for new therapies. We discovered Ropporin-1 (ROPN1) as a target to treat TNBC with T cells. ROPN1 showed high and homogenous expression in 90% of primary and metastatic TNBC but not in healthy tissues. Human leukocyte antigen-A2-binding peptides were detected via immunopeptidomics and predictions and used to retrieve T-cell receptors (TCR) from naïve repertoires. Following gene introduction into T cells and stringent selection, we retrieved a highly specific TCR directed against the epitope FLYTYIAKV that did not recognize noncognate epitopes from alternative source proteins. Notably, this TCR-mediated killing of three-dimensional (3D) tumoroids in vitro and tumor cells in vivo and outperformed standard-of-care drugs. Finally, the T-cell product expressing this TCR and manufactured using a clinical protocol fulfilled standard safety and efficacy assays. Collectively, we have identified and preclinically validated ROPN1 as a target and anti-ROPN1 TCR T cells as a treatment for the vast majority of patients with TNBC.
Significance: Metastatic TNBC has a dismal prognosis. This study discovers Ropporin-1 as a target for T-cell therapy for most patients. The selected TCR is highly specific and sensitive in advanced models, and preclinical testing shows that the T-cell product expressing this TCR, manufactured according to good manufacturing practice, has favorable safety and potency.
Introduction
Triple-negative breast cancer (TNBC) is an aggressive subtype of breast cancer (BC) that affects relatively young women at an age of 40 to 50 years at diagnosis and accounts for 15% to 20% of all breast cancer cases. The 5-year overall survival rate for metastatic TNBC is only 12% (American Cancer Society: http://www.cancer.org/), which reflects the lack of effective treatment options. Indeed, TNBC lacks targets for hormonal therapy and HER2 amplification, and patients with TNBC are therefore not eligible for standard therapies for breast cancer (1). The poly(ADP-ribose) polymerase (PARP) inhibitors olaparib and talazoparib have shown effectiveness in treating advanced TNBC with germline BRCA mutations [overall response rate (ORR) 55%–62%; refs. 2, 3], yet only 10% to 20% of patients with TNBC carry these mutations (4–6), and the relatively high ORR does not translate into overall survival benefit (7). The programmed cell death 1 (PD1) inhibitor pembrolizumab, when combined with chemotherapy, improves responses for a subgroup of patients with metastatic TNBC, whose tumors are positive for programmed cell death ligand 1 (PD-L1), resulting in an ORR of 53% compared with 41% for chemotherapy only, but this subgroup only accounts for 30% to 40% of all patients with TNBC (8). A more recently approved standard-of-care option for patients with patients with TNBC is the trophoblast cell surface antigen 2 (TROP2)-targeting antibody–drug conjugate sacituzumab govitecan, which has resulted in an ORR of 35% and showed a gain in ORR of 30% when compared with chemotherapy in a cohort of relapsed patients with TNBC. Treatment with sacituzumab govitecan, however, is accompanied with serious side effects, with more than 50% of patients experiencing adverse events of grade 3 or higher, such as neutropenia and leukopenia (9, 10). Despite new drugs having become available, there is still an unmet demand for effective and safe therapies for patients with TNBC.
A promising new treatment option for TNBC is adoptive T-cell therapy (ACT). ACT is a form of immune therapy that relies on the transfer of tumor-specific T cells that are generally generated by inserting a gene to express either a chimeric antigen receptor (CAR) or T-cell receptor (TCR) into the patients’ T cells ex vivo. CAR T cells constitute major breakthroughs for the treatment for different B-cell malignancies (11–15) as well as multiple myeloma (16–18) with six of these CAR T-cell products now implemented as standard-of-care (19). Unfortunately, the efficacy of CAR T cells to treat solid tumors lags behind significantly to that of hematologic tumors (20–25). Explanations may include that, in general, solid tumors lack tumor-specific targets available on the surface of solid tumors and harbor an immunosuppressive tumor microenvironment (26).
TCRs recognize targets through presentation via MHC molecules, not being restricted to cell surface antigens and broadening the spectrum of potential tumor target antigens. Notably, TCR-engineered T cells targeting intracellularly the cancer germline antigens New York esophageal squamous cell carcinoma 1 (NY-ESO1) or MAGE family member A4 (MAGE-A4) in the context of human leukocyte antigen (HLA-)A2 have shown clear clinical responses with ORRs up to 61% in patient with melanoma and synovial sarcoma. These two TCR T-cell products have a good safety profile, in which the latter has recently been granted accelerated FDA approval (27–29). However, high expression of NY-ESO-1 and MAGE-A4 is limited to the abovementioned solid tumor types, and expressions are often heterogeneous and variable among patients (30).
In the current study, we have identified and tested new target antigens and corresponding TCRs in a stepwise manner and according to stringent selection criteria for safety and efficacy to enable the development of ACT for TNBC. In our search, only targets were included that were absent in healthy tissues to reduce the risk of on-target toxicity. At the same time, target selection included high and homogenous expression in TNBC to facilitate effective antitumor T-cell responsiveness (31). TCRs were retrieved from naïve repertoires and selected when harboring a stringent epitope recognition motif to reduce the risk of off-target toxicity (32–34) and driving eradication of TNBC in advanced tumor models. Overall, our work reports the discovery of a novel target, the cancer germline antigen (CGA) Ropporin-1 (ROPN1), as well as the retrieval and extensive preclinical validation of an anti-ROPN1 TCR for the treatment of the majority of patients with TNBC.
Results
ROPN1 Is Absent in Healthy Tissues and Shows Abundant and Homogenous Expression in Primary and Metastatic TNBC
To discover a target suitable for ACT for TNBC, we have applied a stepwise and stringent selection process in which large gene expression datasets of normal healthy and tumor tissues were screened for intracellular antigens (Fig. 1A).
Figure 1.
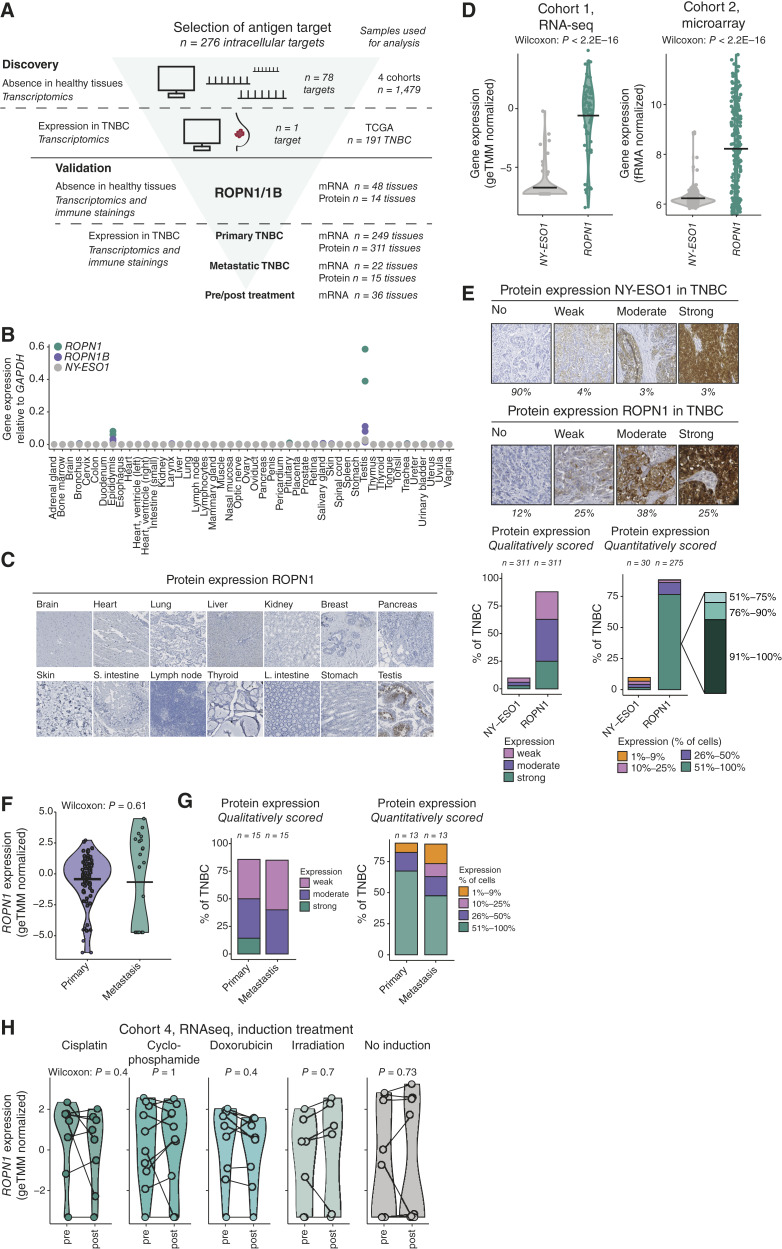
ROPN1/B is absent in healthy tissues and highly and homogenously present in primary and metastatic TNBC independent of pretreatment. A, Flowchart of the discovery of ROPN1/B as a target for T-cell treatment of TNBC and validation of its tumor-restricted expression using healthy tissues, as well as primary and metastatic TNBC tissues. Intracellular targets (CGAs: n = 276) were screened for absent expression in healthy tissues (n = 1,479 tissues) and expression in TNBC (n = 191 tissues). ROPN1/B mRNA and protein expression was further validated in multiple sets of healthy tissues (two additional cohorts) and four different cohorts of patient tumor tissues (see “Methods” for details). B, Dot plot shows gene expressions as fold changes relative to GAPDH (2−dCt) according to RT-qPCR using a cDNA library of 48 healthy tissue samples. NY-ESO1 was used as a reference. Green, ROPN1; purple, ROPN1B; gray, NY-ESO1 (CTAG1B), n = 2 to 3 per target antigen. C, Representative immune stainings of ROPN1/B using an array of 14 healthy tissues (2–6 donors per tissue, total n = 66). D, Violin plots show the distribution of gene expression of NY-ESO1 (CTAG1B, gray) and ROPN1 (green) in TNBC cohort 1 (n = 66, geTMM normalized) and cohort 2 (n = 183, fRMA-normalized; see “Methods” for details on cohorts). Data were analyzed using the Wilcoxon signed-rank test: cohort 1: P < 2.2E16; cohort 2: P < 2.2E16. E, Representative immune stainings of TNBC categorized according to staining intensity with different intensities for NY-ESO1 (top) and ROPN1/B (bottom). Stacked bar graphs (middle) show the fraction of TNBC tumors with weak, moderate, and strong immune staining of NY-ESO1 or ROPN1/B [tissue microarrays (TMA), n = 311]. Stacked bar graphs (right) show fractions of TNBC tumors with either 1% to 9%, 10% to 25%, 26% to 50%, or 51% to 100% of tumors cells positive for NY-ESO1 or ROPN1/B protein; the latter category of 51% to 100% ROPN1-stained cells is further subdivided into the fractions: 51% to 75%, 76% to 90%, and 91% to 100% (in zoomed-in stacked bar). F, Violin plot shows the distribution of gene expressions of ROPN1/B in primary and metastatic TNBC from TNBC cohort 1 (n = 66, geTMM normalized) and cohort 3 (n = 22) following batch correction. Data were analyzed using the Wilcoxon signed-rank test: P = 0.61. G, Stacked bar graphs (left) show the fraction of primary and metastatic TNBC with weak, moderate, and strong immune staining of ROPN1/B (whole tissue sections, n = 15 paired samples). Stacked bar graphs (right) show the fraction of primary and metastatic TNBC with either 1% to 9%, 10% to 25%, 26% to 50%, or 51% to 100% of tumor cells positive for ROPN1/B. H, Violin plots show the distribution of gene expression of ROPN1/B in pre- and post-induction treatment biopsies of metastatic TNBC retrieved from cohort 4 (n = 53 of which n = 44 are paired samples, geTMM normalized). Data of paired pre/postsamples was analyzed using the Wilcoxon signed-rank test: cisplatin: n = 8, P = 0.4; cyclophosphamide: n = 10, P = 1; doxorubicin: n = 9, P = 0.4; irradiation: n = 7, P = 0.7; no induction: n = 10, P = 0.73. ACT: L. intestine, Large intestine; S. intestine, small intestine.
In a first step, we observed that ROPN1 as well as its paralog ROPN1B (>95% amino acid sequence homology) was one of 78 intracellular proteins that was absent in large series of healthy organs according to multiple gene expression databases (Supplementary Fig. S1A). Gene and protein expression of ROPN1 in healthy tissues was confirmed to be limited to the testis and, to a lesser extent, the epididymis (Fig. 1B and C). It is noteworthy that both these tissues are immune privileged (35, 36), meaning these tissues lack MHC molecules and are generally ignored by T cells.
In a second step, the screening of tumor samples revealed that ROPN1 was expressed in more than 90% of patients with TNBC. In comparison, targets for ACT with clinical precedent, such as NY-ESO1 and MAGE-A4, were only expressed in 16% and 25% of patients with TNBC, respectively (Supplementary Fig. S1B). Furthermore, the levels of gene and protein expression were significantly higher for ROPN1 (Fig. 1D and E; Supplementary Fig. S1B) when compared with abovementioned reference targets. ROPN1 protein was detected in more than 50% of tumor cells in approximately 75% of primary TNBC (Fig. 1E). In fact, when zooming into ROPN1 expression in more than 90% of tumor cells, we observed this to be the case for approximately 60% of primary TNBC (Fig. 1E; right). These levels of abundance, extent of expression, and homogeneity are, to our knowledge, unique when compared with other targets of ACT. To assess the robustness of ROPN1 as a target for ACT, we also investigated gene and protein expressions in metastatic TNBC lesions (Fig. 1F and G) and observed that the majority of studied lymph node lesions expressed ROPN1 in more than 50% of tumor cells, highly concordant with the extent of expression observed in primary tumors. In addition, ROPN1 expression in distant TNBC lesions was not affected by preoperative treatments, such as cisplatin, cyclophosphamide, doxorubicin, or radiation (Fig. 1H). Notably, ROPN1 was also expressed in 90% of skin cutaneous melanoma (SKCM; Supplementary Fig. S1C), implying that these patient cohorts could also benefit from ACT targeting ROPN1.
HLA-A2-Restricted and Endogenously Processed Peptides Enable Retrieval of ROPN1-Specific TCRs from Naïve Repertoires
TCRs recognize small peptide fragments of the target antigen in the context of HLA molecules on the surface of cells. We set out to investigate which peptides originating from the ROPN1/B protein were recognized by TCRs, not shared with other source proteins, and restricted to the HLA-A2 subtype, the latter covering 30% to 50% of the EU and US population (37). For details on peptide selection, see Supplementary Table S1 and “Methods”. ROPN1/B proteins contain 212 amino acids and cover more than 200 theoretical HLA-A2-restricted peptides according to the prediction algorithm NetMHCpan 4.1. All these peptides were then filtered for in silico qualities that are considered relevant to antigen processing and presentation, such as location of protease cleavage sites and affinities for the transporter associated with antigen processing (TAP) and binding to HLA-A2, which shortlisted the number of peptides to 20 (Fig. 2A). This list of potential epitopes was supplemented with 17 peptides identified following mass spectrometry of HLA-bound peptides from ROPN1+ HLA-A2+ TNBC (2 out of 17), from ROPN1+ HLA-A2+ K562ABC cells (1 out of 17), and from datasets of primary tumor samples (14 out of 17; Supplementary Table S2; “Methods”; Supplementary Methods; refs. 38, 39). Notably, one peptide (FLYTIAKV) that was identified in primary tumor samples was also shortlisted based on in silico predictions, yielding a total set of 36 peptides. This set of peptides was then screened for lack of similarity with other source antigens using Expitope software (40). The peptides that were only present in ROPN1/B needed to fulfill minimal binding to HLA-A2 in vitro, after which 21 peptides survived that were further assessed for two parameters: the amplitude (maximal HLA-A2 binding at saturating peptide concentration) and the half-maximal effective concentration (EC50; Methods). Eleven peptides showed binding to HLA-A2 in a dose-dependent manner and were ranked based on amplitude (Fig. 2B).
Figure 2.
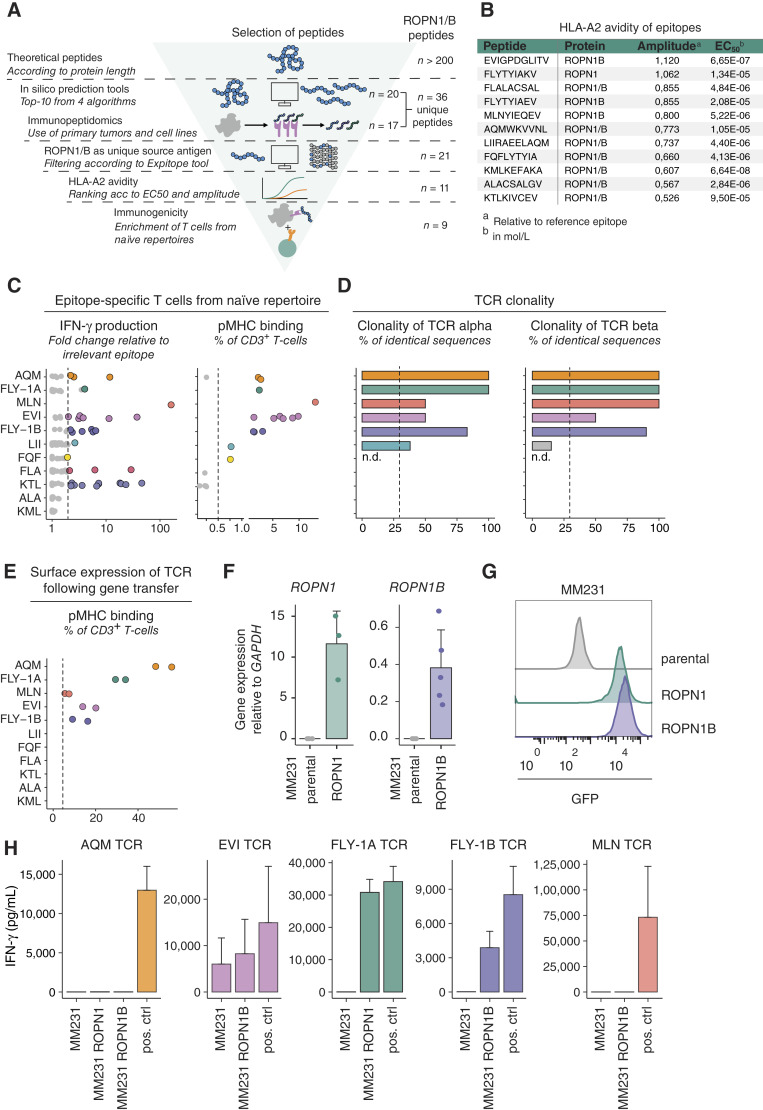
Retrieval and selection of ROPN1/B peptides according to uniqueness and HLA-A2 avidity, as well as natural TCRs against endogenously processed epitopes. A, Flowchart of retrieval and selection of ROPN1/B peptides according to in silico predictions (n = 20) and peptide elution (n = 17), resulting in n = 37 identified peptides, as well as non-cross-reactivity (n = 21, avidity for HLA-A2 (n = 11) and immunogenicity (n = 9; Methods for details on each tool/assay and Supplementary Table S1 for an overview of the results per tool). B, The table presents the overview of HLA-A2 binding characteristics for the 11 shortlisted peptides that went into T-cell enrichments. The gp100 epitope YLEPGPVTA (YLE) served as a reference epitope. Amplitudes are expressed as FC of median fluorescent intensity (MFI) of anti-HLA-A2-PE relative to YLE at highest peptide concentrations, and EC50 values (mean, calculated via GraphPad Prism 5.0) are listed in molarity (mol/L). C, Dots represent enriched T-cell populations (with peptides from B and each dot representing a single enrichment) according to FC of IFN-γ production compared with irrelevant epitope (RP; left). Epitope stimulated T cells which produced minimally 200 pg/mL, and 2× more IFN-γ than RP stimulated T cells were stained for pMHC binding, which is shown as percentage of CD3+ T cells (right). D, Clonality of TCRα and TCRβ sequences from epitope-specific T-cell populations (from C) are shown as percentage of total number of sequences. E, TCRαβ combinations derived from clonal populations (from D) were introduced into T cells and tested for pMHC binding, again shown as percentage of CD3+ T cells. Each epitope in C–E is shown with a unique color, and when dots are presented in gray, these epitopes did not meet the selection criteria for further characterization (Methods; Supplementary Fig. S8). F, Bar plots represent gene expression of ROPN1/B using MM231 transfectants depicted as FC relative to GAPDH (2−∆Ct) according to RT-qPCR. ROPN1-expressing MM231 cells are visualized in green (n = 3), ROPN1B-expressing MM231 cells in purple (n = 5), and parental MM231 cells (not expressing ROPN1/B) in gray (n = 5). Mean and SD are shown. G, Representative histograms show MFI of GFP expression of ROPN1-expressing MM231 (green), ROPN1B-expressing MM231 (purple), and parental MM231 (not expressing GFP, gray). H, Bar plots display the ability of TCR T cells (from E) to recognize endogenously processed and presented cognate epitope. IFN-γ levels (in pg/mL) were measured upon stimulation of TCR T cells with ROPN1/B-expressing or parental MM231 (F and G). Positive controls are BSM cells loaded with cognate epitope. Mean and SD are shown.
To retrieve ROPN1-specific T cells, these 11 peptides were individually fed to CD11c+ dendritic cells that were cocultured with autologous naïve CD8+ T cells from peripheral blood mononuclear cells (PBMC) derived from a set of 13 healthy donors. With our technique (41), which we improved for sensitive detection of epitope-specific T cells, we are able to isolate TCRs from precursor T cells present at low frequencies in healthy individuals, such as those reported for the CGAs MAGE-A3 and NY-ESO (42, 43). Nine out of 11 peptides yielded T cells that demonstrated significant epitope-specific IFN-γ secretion and peptide–MHC binding (Fig. 2C; Supplementary Fig. S2A). These epitope-specific T-cell populations were FACS sorted according to peptide–MHC binding or peptide-induced CD137 expression and subjected to bulk TCR sequencing, which revealed oligo or monoclonality for 13 TCRs (Fig. 2D; Supplementary Fig. S2B). Five sets of genes encoding TCRαβs directed against the epitopes FLYTYIAKV (FLY-1A), FLYTYIAEV (FLY-1B), EVIGPDGLITV (EVI), MLNYIEQEV (MLN), and AQMWKVVNL (AQM) showed surface expression by primary T cells upon retroviral transduction (Fig. 2E; Supplementary Fig. S2C). To test whether these TCRs recognize their cognate epitope following endogenous processing and HLA-A2 presentation, a critical go/no-go parameter for the respective epitope–TCR pair, we cocultured TCR T cells with the TNBC cell line MDA-MB-231 (MM231) that overexpresses either ROPN1 or ROPN1B (Fig. 2F and G). Transfectants provided robust models to test and select ROPN1 TCRs, which was necessitated by a lack of HLA-A2+ cancer cell lines that naturally express ROPN1. Patient-derived tumor models with endogenous levels of ROPN1 expression have been subsequently applied to validate these TCRs (see below). FLY-1A and 1B TCRs specifically recognized ROPN1+ or ROPN1B+ MM231 cells, respectively (Fig. 2H). In contrast, MLN- and AQM-specific TCRs did not recognize cancer cells expressing ROPN1/B, potentially indicating that these two epitopes are not naturally presented or these TCRs were of a too low affinity for their cognate epitopes (Fig. 2H). The EVI-specific TCR recognized the parental cancer cells that do not express ROPN1/B (Fig. 2H), potentially indicating cross-reactivity to other targets. TCRs specific for the EVI, MLN, and AQM epitopes were therefore excluded from further characterization.
TCRs Directed against FLY-1A and FLY-1B Are Highly Specific and Do Not Recognize Peptides from Non-ROPN1/B Source Proteins
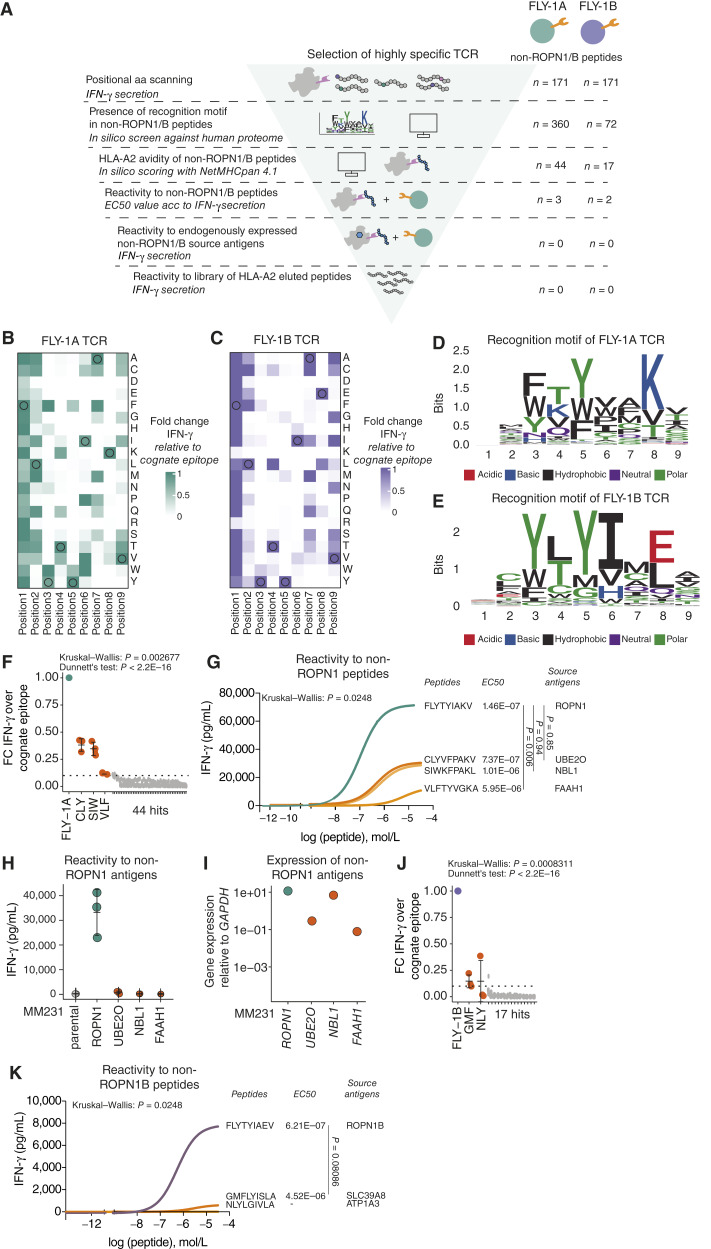
Cross-reactivity of TCR T cells toward other antigens is a potential safety concern, which when overlooked can result in severe side effects. For example, MAGE-A3 TCR T cells being cross-reactive to either Titin, a protein highly expressed in cardiomyocytes, or a protein expressed in brain cells, led to patient deaths when tested in clinical trials (32, 33). To assess the extent of cross-reactivity of the FLY-1A and FLY-1B TCRs (Fig. 3A), we positionally scanned amino acids of the cognate epitopes, replacing each amino acid by any other amino acid to determine which amino acids and which positions are critical for recognition by the corresponding TCR (see “Methods” for details). For both epitopes, we found that most amino acid replacements at positions 3 to 7 resulted in substantial reduction of IFN-γ production by FLY-1A (Fig. 3B) and FLY-1B (Fig. 3C) TCR T cells. With the resulting recognition motif of each TCR (Fig. 3D and E), an in silico search was performed against the complete human proteome to find non-ROPN1 peptides that could theoretically be recognized by either of these two TCRs. Potential peptides coming from this search were shortlisted according to binding to HLA-A2 and recognition by TCR T cells of peptide-pulsed target cells. These analyses revealed that the FLY-1A TCR recognized three non-ROPN1 peptides, namely, CLYVFPAKV (CLY), SIWKFPAKL (SIW), and VLFTYVGKA (VLF), coming from the source antigens (E3-independent) E2 ubiquitin-conjugating enzyme (UBE2O), neuroblastoma suppressor of tumorigenicity 1 (NBL1), and fatty-acid amide hydrolase 1 (FAAH1), respectively (Fig. 3F). Dose titrations demonstrated that half-maximal responses (i.e., sensitivities) of FLY-1A TCR T cells for the CLY and SIW peptides are comparable to the cognate FLY-1A epitope, in which the half-maximal response of this TCR for the VLF peptide is significantly lower compared with the FLY-1A epitope (Fig. 3G). The amplitudes (i.e., absolute quantities of IFN-γ) for all three non-ROPN1 peptides were significantly lower compared with the FLY-1A epitope (Fig. 3G). We subsequently assessed whether these alternative peptides can be processed and presented by TNBC cells transfected with either one of the three source antigens (Fig. 3H and I). Notably, the FLY-1A TCR was unable to recognize TNBC cells overexpressing such source antigens, suggesting that the CLY, SIW, and VLF peptides are not a product of the natural antigen processing and presentation machinery and therefore pose minimal risk to elicit FLY-1A-mediated off-target toxicity (Fig. 3H and I). The FLY-1B TCR could recognize two non-ROPN1B peptides, namely, GMFLYISLA (GMF) and NLYGIVLA (NLY), coming from the source antigens metal cation symporter ZIP8 (SLC39A8) and sodium/potassium-transporting ATPase subunit alpha-3 (ATP1A3), respectively (Fig. 3J). Dose titrations demonstrated that half-maximal responses as well as amplitudes of FLY-1B TCR T cells for the GMF and NLY peptides were significantly lower or nonexisting compared with the FLY-1B epitope (Fig. 3K). No further investigation of the natural processing and presentation of these peptides was needed.
Figure 3.
TCRs directed against FLY-1A and FLY-1B have a strict recognition motif and specifically recognize cognate but not alternative epitopes. A, Schematic overview of experiments performed to determine TCR specificity (see “Methods” for details). Heatmap shows the relative IFN-γ production by (B) FLY-1A TCR T cells or (C) FLY-1B TCR T cells upon positional amino acid scanning of the cognate epitope. TCR T cells were cocultured with BSM cells loaded with single amino acid variants that cover all amino acids at every position of the cognate epitope (n = 171). T-cell IFN-γ production is expressed as FC compared with the cognate FLY-1A epitope (n = 3). Original amino acids from the cognate epitope are circled. D, Sequence logo of the recognition motif of FLY-1A TCR T cells. E, Sequence logo of the recognition motif of FLY-1B TCR T cells. The height of each letter is scaled in bits using the R package ggseqlogo and represents the probability of that amino acid at that position. The colors of the amino acids represent the chemical properties explained below the logo. Motifs were queried against a human protein database using ScanProsite, which yielded 44 non-ROPN1 for FLY-1A TCR T cells and 17 non-ROPN1B peptides for FLY-1B TCR T cells, respectively, that harbored the recognition motif and were predicted to bind to HLA-A2 according to NetMHCpan 4.1. F, Dot plot shows IFN-γ production by FLY-1A TCR T cells upon stimulation with 44 peptides (from D, 10 μg/mL) expressed as FC compared with the cognate epitope (n = 3). Mean and SD are shown. The Kruskal–Wallis rank test was performed followed by Dunnett’s multiple comparisons test: P < 2.2E16. G, Representative dose response curves of FLY-1A TCR T cells following exposure to non-ROPN1 peptides from D with FC > 0.1. These peptides included CLYVFPAKV (CLY), SIWKFPAKL (SIW), and VLFTYVGKA (VLF; depicted in orange); the cognate FLY-1A epitope was included as a comparator (green; n = 3). Mean EC50 values (in mol/L, n = 3) and source antigens of peptides are shown on the right side of the plot. The Kruskal–Wallis rank test was performed followed by Dunnett’s multiple comparisons test: CLY vs. FLY-1A: P = 0.85; SIW vs. FLY-1A: P = 0.94; VLF vs. FLY-1A: P = 0.0060. H, Dot plots display the lack of ability of FLY-1A TCR T cells to recognize endogenously expressed non-ROPN1 source antigens. IFN-γ levels (in pg/mL) were measured upon stimulation of FLY-1A TCR T cells with MM231 expressing one of the three non-ROPN1 antigens (shown in orange) or ROPN1 (used as a comparator, shown in green; n = 3). I, Dot plot represents gene expression of the non-ROPN1 antigens in transfected MM231 cells. Gene expression of the source antigens is depicted as FC relative to GAPDH (2−∆Ct) according to RT-qPCR; colors are as in F (n = 1). J, Dot plot shows IFN-γ production by FLY-1B TCR T cells upon stimulation with 17 peptides (from in silico screen against human proteome using the recognition motif from E, 10 μg/mL), expressed as FC compared with the cognate epitope (n = 3). The Kruskal–Wallis rank test was performed followed by Dunnett’s multiple comparisons test: P < 2.2E16. Individual points, mean, and SD are shown. K, Representative dose response curves of FLY-1B TCR T cells following exposure to non-ROPN1B peptides from J with FC > 0.1. These peptides included GMFLYISLA (GMF) and NLYGIVLA (NLY; depicted in orange); the cognate FLY-1B epitope was included as a comparator (purple; n = 3). Mean EC50 values (in mol/L, n = 3) and source antigens of peptides are shown on the right side of the plot. The EC50 value could not be calculated for the NLY peptide. Data were analyzed using the Wilcoxon signed-rank test: FLY-1B vs. GMF: P = 0.08086.
FLY-1A TCR T Cells Effectively Kill Patient-Derived TNBC Organoids and Outperform Standard-of-Care Treatments
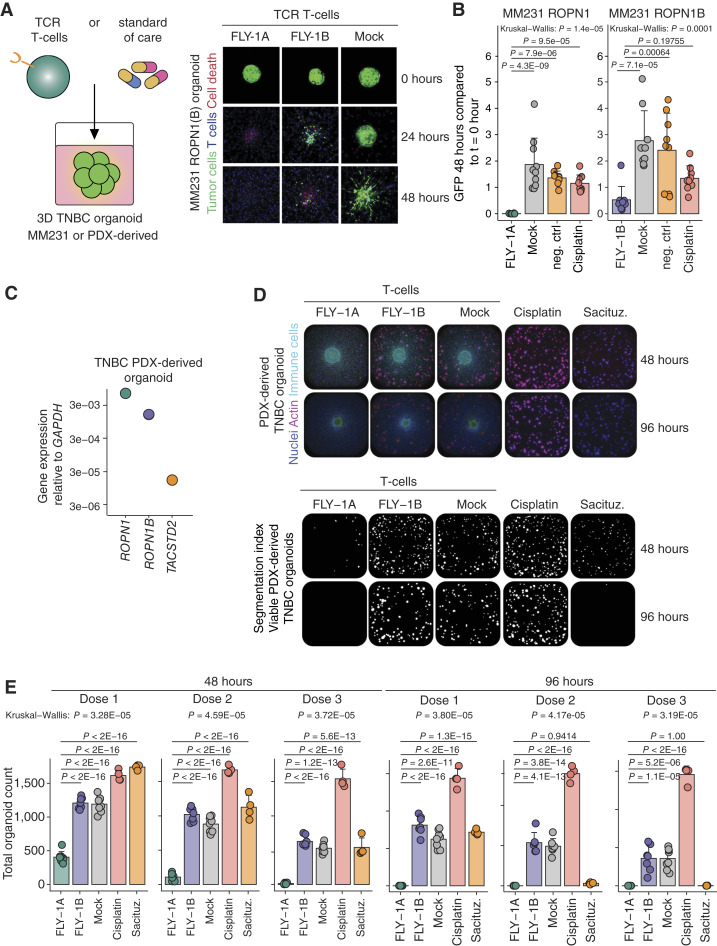
Following our studies into safety, we set out to investigate the killing capacity of ROPN1-specific TCR T cells using different TNBC models. In a first model, we used tumoroids that were derived from the ROPN1 or ROPN1B-overexpressing MM231 cell line (see “Methods” for details). We noted that FLY-1A and FLY-1B TCR T cells, when loaded on top of these collagen gels, migrated toward and mediated killing of tumoroids expressing ROPN1 and ROPN1B, respectively, over a period of 48 hours. Notably, FLY-1A TCR T cells caused 100% tumoroid killing and outperformed cisplatin (20 μmol/L), which was used as a comparator. FLY-1B TCR T cells could not completely eradicate the tumoroids within the same time frame and performed similarly to cisplatin (Fig. 4A and B).
Figure 4.
FLY-1A TCR T cells recognize 3D breast tumoroids and outperform standard-of-care in vitro. A, Cartoon depicting the assay setup to test TCR T cells or standard-of-care therapies for their reactivity to three-dimensional (3D) extracellular matrix (ECM)-embedded organoids (left). Representative confocal fluorescence microscopy images of organoids derived from ROPN1/B-overexpressing MM231 cells at t = 0 hour, t = 24 hours, and t = 48 hours after coculture with TCR T cells (n = 3 experiments; n = 4 replicates per experiment; right). FLY-1A and FLY-1B TCR T cells were tested against MM231 cells expressing ROPN1 or ROPN1/B, and mock T cells were included as a negative control. The green color indicates GFP-expressing organoid, the blue color represents TCR T cells, and the red color represents binding by PI. B, Bar graphs display differences in GFP signal from the MM231 ROPN1/B-derived organoids at 48 hours after addition of T cells relative to 0 hour. Cisplatin (20 μmol/L) or medium (negative control) was used as comparators. Individual points, mean, and SD are shown. C, Dot plot represents gene expression of ROPN1 (green), ROPN1B (purple), and TACSTD2 (TROP2, orange) in TNBC PDX-derived organoid. Gene expression of these targets is depicted as FC relative to GAPDH (2−∆Ct) according to RT-qPCR (n = 1). D, Representative images of TNBC PDX-derived organoids at 48 and 96 hours after the addition of T cells. Cisplatin (Dose 1: 0.1 μmol/L; Dose 2: 1.0 μmol/L; Dose 3: 10 μmol/L) or sacituzumab govitecan (Dose 1: 0.1 μmol/L; Dose 2: 1.0 μmol/L; Dose 3: 10 nmol/L) was used as a comparator. Left, actin shown in pink and nuclei visualized in blue represent living organoids, whereas immune cells are visualized in green. Right, segmented images show the viable organoids (white) from the images of the left at 48 and 96 hours. E, Bar plots represent total cell count of TNBC PDX-derived organoids at 48 hours (left) and 96 hours (right) after the addition of three different doses of T cells or drug compounds (n = 2 donors, 4 replicates per donor). Individual points, mean, and SD are shown. The Kruskal–Wallis rank test was performed followed by Dunnett’s multiple comparisons test. Sacituz., sacituzumab govitecan.
In a second series of experiments, we used 3D tumor organoids from patient with TNBC-derived xenograft (PDX) tissues that endogenously express ROPN1 and ROPN1B and are positive for HLA-A2. These organoids are also positive for TROP2, which is the target for the drug sacituzumab govitecan, a standard-of-care treatment for metastatic TNBC (Fig. 4C). Again, FLY-1A TCR T cells were highly efficient in killing the patient-derived organoids that endogenously expressed ROPN1 in a dose-dependent manner and more effectively when compared with the killing of organoids by FLY-1B TCR T cells after 48 hours (Fig. 4D and E). Importantly, FLY-1A TCR T cells significantly outperformed cisplatin as well as sacituzumab govitecan (0.1-10 nmol/L). Furthermore, ROPN1-expressing melanoma organoids were also efficiently killed by FLY-1A TCR T cells to a similar extent as the killing of NY-ESO1-expressing melanoids by NY-ESO1 TCR T cells (Supplementary Fig. S3A–S3C).
In line with the superiority of FLY-1A versus FLY-1B TCR T cells in both organoid models, there is at least a fivefold difference in effective concentration of cognate epitope that yields half-maximal production of IFN-γ (Fig. 3G and K), pointing to a significantly higher avidity of the FLY-1A TCR T cells. The TCR specific for FLY-1B was therefore excluded from further characterizations.
We extended the in vitro testing of FLY-1A TCR T cells, particularly about their specificity, and assessed T-cell recognition toward cancer cell lines (n = 10) or cell line-derived 3D tumoroids (n = 3) that lack ROPN1 and/or HLA-A2. FLY-1A TCR T cells did not recognize cell lines lacking expression of either ROPN1, HLA-A2, or both (Supplementary Fig. S4A) nor did they recognize tumoroids that lack either ROPN1, HLA-A2, or both (Supplementary Fig. S4B and S4C). Another line of evidence for lack of cross-reactivity comes from the testing of FLY-1A TCR T cells for their ability to recognize noncognate peptides eluted from HLA-A2. Coculturing of TCR T cells with target cells pulsed with a library of 114 such peptides confirmed the inability of the FLY-1A TCR to recognize noncognate epitopes (Supplementary Fig. S4D). In short, FLY-1A TCR T cells are highly specific for ROPN1+ HLA-A2+ tumor cells and are nonreactive toward tumor cells lacking expression of ROPN1 and/or HLA-A2 as well as noncognate epitopes.
FLY-1A TCR T Cells Infiltrate and Eradicate TNBC Tumors Effectively
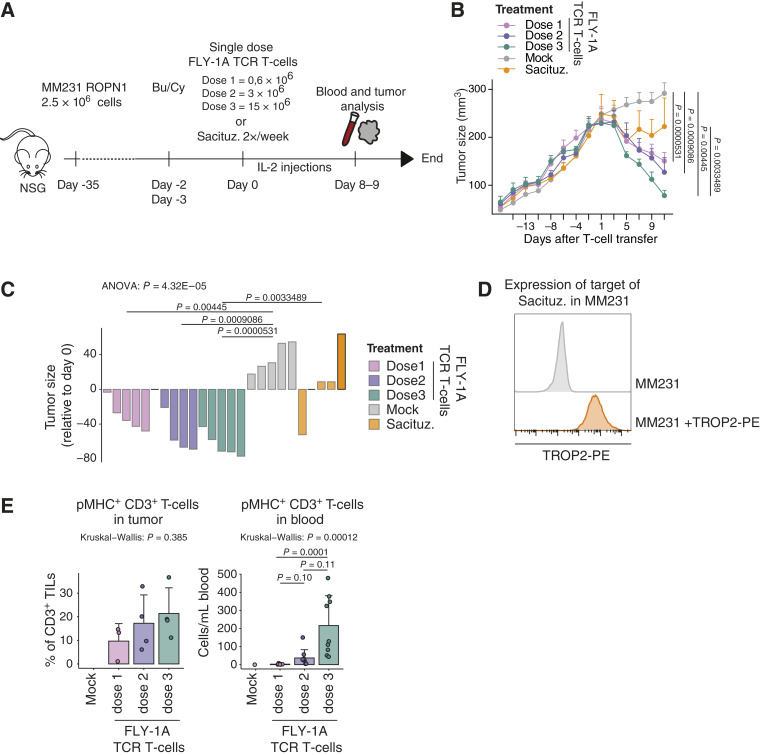
Next, we assessed tumor cell killing as well as pharmacokinetics of FLY-1A TCR T cells in mice engrafted human TNBC (see “Methods”; Fig. 5A). In these experiments, and in line with clinical trials, we preceded the transfer of TCR T cells with lymphodepleting chemotherapy and supported transferred TCR T cells with low-dose IL-2 (see “Methods” for details). In line with the organoid experiments, treatment of tumor-bearing mice with FLY-1A TCR T cells induced a clear and dose-dependent tumor regression, and a single infusion of TCR T cells significantly outperformed repetitive treatment with sacituzumab govitecan (2x/week 0.4 mg/kg) (Fig. 5B–D). In addition, FLY-1A TCR T cells were detected in dose-dependent quantities in tumors as well as in blood samples (Fig. 5E). Notably, mice treated with FLY-1A TCR T cells showed specific enrichment of pMHC+ CD8+ as well as pMHC+ CD4+ T cells within the regressing tumors (Supplementary Fig. S5A and S5B).
Figure 5.
FLY-1A TCR T cells lead to dose-dependent regression of large TNBC tumors and significantly outperform standard-of-care treatment in vivo. A, Scheme depicting the in vivo study design (see “Methods” for details). NSG mice bearing palpable subcutaneous tumors derived from MM231 ROPN1 cells were treated with either 1 transfer of FLY-1A TCR T cells (0.6, 3, or 15 × 106 TCR+ CD3+ T cells), mock T cells (equal to the no. of cells given for highest TCR T-cell dose), or sacituzumab govitecan (0.4 mg/kg) 2 times per week. Blood (n = 9 per group) and tumors (n = 4 per group) were collected at day 8/9. B, Line graph shows tumor size over time in mice treated with FLY-1A TCR T cells (dose 1: pink; dose 2: purple; dose 3: green), mock T cells (gray), or sacituzumab govitecan (orange; n = 5 per group). C, Waterfall plot represents tumor size at day 11 relative to day −1 per mouse per group (the same colors as in B). ANOVA test was performed followed by Tukey’s post hoc test. Only significant differences are shown. D, Flow cytometric determination of TROP2 protein expression in parental MM231 cells stained with antibody (depicted in orange) or not (negative control, depicted in gray). E, Presence of TCR T cells in tumor (left) and blood (right). T cells binding pMHC that were either present in single tumor cell suspensions or peripheral blood samples were detected by flow cytometry (see “Methods” for details). Individual points, mean, and SD are shown. The Kruskal–Wallis rank test was performed followed by Dunn’s multiple comparisons test. Significant differences between different treatments vs. mock T cells were not calculated because of low numbers. Bu/Cy, busulfan and cyclophosphamide; Sacituz., sacituzumab govitecan.
Small-Scale Clinical Product of FLY-1A TCR T Cells Passes Safety and Potency Test
In a final step, moving toward clinical development of the FLY-1A TCR, we questioned whether surface expression of this TCR could be enhanced as a function of preferential pairing between TCRα and TCRβ chains. Upon introduction of exogenous TCRα and TCRβ chains, there is a possibility of mispairing between transgenic chains and endogenous TCR chains. TCR mispairing could generate new TCR combinations with potentially self-reactive specificities (44, 45), but more importantly (as we have already ruled out the recognition of alternative peptides by TCR T cells, Fig. 3), it also dilutes the surface expression of transgenic TCRαβ and consequently limits anti-ROPN1 effectivity. To assess the proneness of the FLY-1A TCR to mispairing, we stained FLY-1A TCR T cells with an antibody specific for its TCR-Vβ13.1 chain as well as with its FLY-1A:HLA-A2 pMHC multimer. We observed equal fractions of TCR T cells being positive for either TCR-Vβ or pMHC [TCR-Vβ: 31% ± 19% (mean ± SD), pMHC: 31% ± 18% of five different donors, Fig. 6A and B], suggesting that all surface-expressed FLY-1A TCRβ chains are complexed with all surface-expressed FLY-1A TCRα chains, thereby enabling maximal binding of the cognate epitope coupled to HLA-A2. In fact, the introduction of a cysteine bridge (Cys) or substitutions of three amino acid residues (LRY), which are reported to reduce mispairing for some TCRs (46, 47), did not result in further improvement of either TCR-Vb expression or binding to pMHC (Fig. 6C).
Figure 6.
FLY-1A TCR is not prone to mispair, and small-scale clinical product of non-modified FLY-1A TCR T cells passes safety and potency assays. A, Schematic principle of mispairing between transgenic and endogenous TCRα and TCRβ chains following TCR engineering of T cells. B, Representative flow cytometry plot shows wild-type (wt) FLY-1A TCR expression following staining with pMHC (left) or anti-TCRVβ13.1 (middle) of CD3+ T cells. The percentages of pMHC (green) and TCR-Vβ (purple) binding T cells are not statistically different (right, n = 5 donors, individual data points are shown). Data were analyzed using the Wilcoxon signed-rank test: P = 1. C, Representative histogram shows MFI of pMHC+ CD3+ T cells transduced with wt FLY-1A TCR (green) compared with modified FLY-1A TCR that incorporates an extra cysteine bridge (Cys, purple) or the LRY motif (pink; right, n = 2 donors). D, Bar graph shows wt FLY-1A TCR expression according to % binding of pMHC within CD3+ (green), CD4+ (purple), and CD8+ (pink) T cells following an established GMP process performed at two different sites (Erasmus MC and NecstGen, n = 5 donors). Individual data points, mean, and SD are shown. E, Cell numbers of wt FLY-1A TCR T cells at manufacturing days 2, 4, 7, 9, and 11 of T-cell products from D. F, Wild-type FLY-1A TCR T-cell products do not recognize healthy tissue-derived primary cells. IFN-γ production by FLY-1A TCR was measured upon stimulation with cell cultures derived from 11 different HLA-A2+ healthy tissues without (green) or with (purple) preloading with FLY-1A peptide at 10 mmol/L (n = 3 donors). Individual data points and the mean of three biologic replicates are shown. Note that all healthy cell types tested were able to elicit a response by FLY-1A TCR T cells when preloaded with cognate epitope. Mock T cells (in gray) served as a negative control. The Wilcoxon signed-rank test was performed to test healthy tissue-derived cell cultures in the no peptide conditions: FLY-1A TCR vs. mock T cells for all cultures, P > 0.05. G, Wild-type FLY-1A TCR T-cell products recognize TNBC PDX. IFN-γ production by FLY-1A TCR was measured upon coculture with single-cell suspensions derived from TNBC PDX samples (n = 26 of 8 different ROPN1+ HLA-A2+ PDXs). Samples that were ROPN1+ HLA-A2− (n = 6 of 3 different PDX’s) or ROPN1− HLA-A2− (n = 4 of 2 different PDXs) served as additional specificity controls. Mock T cells (in gray) served as a negative control. Individual data points, mean, and SD are shown. The Wilcoxon signed-rank test was performed to test significance between FLY-1A TCR vs. mock T cells, and only significant differences are shown: ROPN1+ HLA-A2+, P = 8.58E05; ROPN1+ HLA-A2−, P = 0.589; ROPN1− HLA-A2−, P = 0.886. H, CD4+ FLY-1A TCR T cells recognize ROPN1 TNBC. IFN-γ and IL-2 productions by FLY-1A TCR T cells were measured following T-cell sorting into CD4+ and CD8+ T cells using magnetic beads and coculturing with ROPN1-expressing MM231 cells (n = 2 donors, 2 replicates per donor). Mock T cells served as negative controls. Individual data points and mean values are shown. I, Representative dose response curves of FLY-1A (green) and NY-ESO1 (pink) TCR T cells following exposure to titrated amounts of their cognate epitope. Mean EC50 values (in μmol/L, FLY-1A TCR: n = 6, NY-ESO1 TCR: n = 4) are shown above the plot. Data were analyzed using the Wilcoxon signed-rank test: FLY-1A vs. NY-ESO1 TCR T cells: P = 0.26. CD3, CD3+ T cells, CD14, CD14+ monocytes; CD19, CD19+ B cells; HBEPCs, human bronchial epithelial cells; HCAECs, human coronary artery endothelial cells; HCFs, human cardiac fibroblasts; HOBs, human osteoblasts; HPFs, human pulmonary fibroblasts; HWPs, human white preadipocytes (subcutaneous); MFI, median fluorescent intensity; NHDF, normal human dermal fibroblasts; NHEK, normal human epidermal keratinocytes.
After having determined that the FLY-1A TCR can be considered a so-called dominant TCR, not needing additional modifications to promote preferential pairing, we manufactured a small-scale FLY-1A TCR T-cell product up to 5 × 108 cells according to an optimized good manufacturing practice (GMP) process yielding highly effective and juvenile TCR T cells (48), currently used in a clinical trial conducted at Erasmus MC (clinicaltrial.gov NCT04729543). These TCR T cells expanded between 25- and 70-fold in 11 days and contained between 51% and 82% pMHC+ cells within the CD3+ T-cell population (Fig. 6D and E). Importantly, we subjected this clinical-like T-cell product to safety and potency assays that comply with current regulations for first inhuman ACT. First, we performed coculture experiments with the FLY-1A TCR T-cell product and HLA-A2+ primary cell lines that were derived from 11 different healthy organs and tissues (Supplementary Table S3). These experiments demonstrated no elevated IFN-γ levels of the TCR T-cell product when compared with the mock T-cell product (Fig. 6F), pointing to no or limited off-target toxicity toward healthy tissues. Note that exogenous loading of these primary cell lines with cognate epitope did evoke a TCR-mediated IFN-γ response (Fig. 6F, purple bars). Second, we performed coculture experiments of the FLY-1A TCR T-cell product with PDX tissues from patients with TNBC and SKCM with endogenous ROPN1 expression. This FLY-1A TCR T-cell product secreted significantly higher levels of IFN-γ upon coculture with ROPN1+ HLA-A2+ TNBC PDX (n = 26 of 8 different ROPN1+ HLA-A2+ PDX’s, Fig. 6G) or SKCM PDX single-cell suspensions (n = 2 of 1 ROPN1+ HLA-A2+ PDX’s, Supplementary Fig. S6A) compared with the mock T-cell product. In line with experiments with cell lines and tumoroids, the FLY-1A TCR T-cell product was nonreactive toward patient tumor samples lacking ROPN1 and/or HLA-A2 (n = 5, Fig. 6G), demonstrating that this TCR T-cell product is tumor reactive and specific in an ex vivo setting. Besides the abovementioned assays, we assessed whether the FLY-1A TCR is functionally expressed by CD4+ T cells, and we separated FLY-1A TCR CD4+ and CD8+ T cells following gene transduction and assessed cytokine production upon coculture with ROPN1+ TNBC cells. We observed that pMHC+ CD4+ T cells as well as their pMHC+ CD8+ counterpart specifically produced IFN-γ and IL-2, suggesting that for its performance the FLY-1A TCR does not require the CD8 coreceptor (Fig. 6H). Finally, we tested the sensitivity of the FLY-1A TCR T-cell product and demonstrated that these T cells recognize the cognate epitope with a functional avidity of 0.1 μmol/L. This avidity measure is comparable to the 0.7 μmol/L avidity of the NY-ESO1 TCR, the latter already being tested in clinical trials in which it induced regression in synovial sarcoma (Fig. 6I; ref. 28). In addition, we observed a positive correlation between ROPN1 mRNA expression in target cells and T-cell IFN-γ production (Spearman’s R = 0.74, n = 78 covering cell lines, PDXs and patient-derived organoids; Supplementary Fig. S7A). Notably, from this correlation, we extracted that the minimal ROPN1 expression required for T-cell recognition equals the lower limit of endogenous expression levels of patient-derived tumor samples. To exemplify the ability of TCR T cells to recognize tumor samples, we demonstrated that FLY-1A TCR T cells effectively kill patient-derived organoids with endogenous ROPN1 expression (Supplementary Fig. S7B).
Discussion
We have identified ROPN1, a novel target antigen for ACT with no detectable expression in healthy tissue and high and homogenous expression in more than 90% of TNBC, as well as an anti-ROPN1 TCR that demonstrates no cross-reactivity and significant effectiveness in preclinical models when compared with standard-of-care drugs. In this study, we have selected the target antigen, epitope, as well as the corresponding TCR using a stepwise approach and stringent filtering according to criteria for therapeutic safety as well as efficacy (Supplementary Fig. S8). It is noteworthy that these selections, when applying such criteria, are accompanied by a high number of dropouts. In fact, we started with nearly 300 antigens, 200 epitopes, and more than 25 TCRs, pointing to a selection stringency, at least for the TCRs, of 10% or less. These numbers may vary for different antigens and different tumor types but do point out that a high number of antigens, epitopes, and TCRs are not suited for clinical application and should be identified and dismissed early on during the preclinical development of ACT.
Currently, one of the critical challenges for ACT to treat solid tumors is the lack of suitable tumor-specific targets. Ideally, a suitable target is absent from healthy tissue, which would derisk the occurrence of autoreactivity following treatment with T cells. The search into suitable targets was triggered by our earlier study with CAR T cells that were directed against carbonic anhydrase IX (CAIX) antigen-positive renal cell carcinoma (49, 50). In this phase I trial, we observed serious adverse events likely because of recognition by CAR T cells of epithelial cells covering larger bile ducts and expressing low levels of CAIX antigen. To limit the chances of such on-target, off-tumor toxicity, we have stringently selected ROPN1 antigen for its absent expression in adult healthy tissues. In fact, we have extensively tested the absence of ROPN1 in healthy tissues using gene expressions from databases covering 1,479 individuals and 66 tissues, cDNA libraries covering 48 tissues, as well as immune stainings covering 14 major tissues. The only healthy tissues found to express ROPN1 are testis and epididymis, in which this protein is expressed in the fibrous sheath of sperm cells likely contributing to their motility (51). Not only are these tissues immune privileged, these tissues are also absent in female patients with TNBC, all in all indicative of a minimal risk for on-target off-tumor toxicity. The safety profile of ROPN1 extends that of other CGAs, such as MAGE-C2, NY-ESO1, and MAGE-A4, which are being targeted (clinicaltrial.gov NCT04729543) or already have been targeted to treat melanoma and synovial sarcoma (28, 29).
Besides its absence from adult healthy tissues, ROPN1 is highly and homogeneously present in tumor tissues from less than 300 patients with TNBC, even in the metastatic setting and independent of pretreatments. The striking abundance and homogeneity of ROPN1/B in TNBC likely limit the risk of tumor recurrence following ACT since it is reported that outgrowth of antigen-negative tumor cell clones is generally a consequence of low and heterogeneous expression of target antigens within tumors (27, 52, 53). Notably, a recent report highlighted that ROPN1 may act as an oncogene and drive metastasis formation in vivo and demonstrated an inverse correlation between ROPN1 expression and survival of patients with TNBC (54), further highlighting the therapeutic value of ROPN1 as a target for ACT.
Moving from target antigen to epitopes, we have applied in silico and laboratory tools to select potential T-cell epitopes according to predicted values for peptide processing and presentation, supplemented with peptides eluted from HLA, and actual binding to HLA-A2. Furthermore, ROPN1 peptides were selected that lack homology to non-ROPN1/B sequences in the human proteome to prevent potential off-target toxicities. Following shortlisting, 11 peptides remained, of which eight were predicted, two were eluted, and one peptide (FLY-1A) was predicted and eluted (Supplementary Tables S1 and S2). Interestingly, although numbers are limited, there is little concordance between in silico predictions and immunopeptidomics. To retrieve ROPN1-specific T cells, these 11 peptides were loaded onto dendritic cells that were cocultured with autologous CD8+ T cells from healthy donors. Using an optimized detection protocol (41), we were able to retrieve ROPN1/B-specific T-cell populations against nine out of 11 peptides, substantiating the reported notion that precursor T cells specific for CGAs are present, albeit at low frequencies, in healthy individuals (42, 43).
From these T-cell populations, we obtained 13 TCRαβs against five epitopes that showed oligo- or mono-clonality, of which one TCRαβ against each epitope was surface expressed upon gene transfer into primary human T cells. Of note, we have used bulk rather than single-cell methods to identify TCR sequences and, consequently, have not directly identified single-cell pairs of TCRα and β chains. This potentially implies that low frequency or subclonal TCRs may have been missed, for which reason we recommend single-cell sequencing for future retrievals of TCR sequences. When zooming in on T-cell avidity for these five TCRs, we observed a range between 0.1 and 4 μmol/L, which is comparable to the avidity observed for the reference TCR directed against NY-ESO1 (Fig. 6I), demonstrating the feasibility of retrieving highly specific TCRs with relatively high natural affinity from naïve repertoires. Next, in a critical “go/no-go step,” these TCRs directed against ROPN1/B were challenged for their ability to recognize processed and naturally presented peptides by TNBC cells. To this end, we have generated ROPN1/1B transfected cell lines enabling the selection of TCRs. Only the FLY-1A and FLY-1B TCRs survived this test, and the other three TCRs did not, after which the two former TCRs were subjected to extensive safety and efficacy analyses.
In recognition of the fact that transfectants are not truly representing tumor cells, we validated ROPN1/1B TCRs with the use of patient TNBC and melanoma-derived tissues as well as organoids. Importantly, these tumor models showed ROPN1 positivity that is concordant with what we observed with TNBC patient-derived tissues, namely, that ROPN1 is expressed in the vast majority of primary and metastatic patients with TNBC on gene and protein levels (Fig. 1). When testing FLY-1A and FLY-1B TCR T cells for their antitumor performance in these models, we demonstrated that FLY-1A TCR T cells outperformed FLY-1B TCR T cells with respect to the eradication of ROPN1(B)+ and HLA-A2+ 3D TNBC as well as melanoma organoids, which is likely linked to their higher functional avidity. In fact, the functional avidity of FLY-1A TCR T cells is slightly higher compared with NY-ESO1 TCR T cells (0.1 and 0.7 μmol/L, respectively), with the latter TCR (termed 1G4-α95:LY) already having shown clinical efficacy in treating melanoma and sarcoma patients (55, 56). Furthermore, the FLY-1A TCR is CD8 coreceptor independent and is expressed in CD4+ and CD8+ T cells, and FLY-1A TCR-expressing CD4+ T cells produce IFN-γ and IL2 upon stimulation with epitope, characteristics that may aid in engaging CD4+ T cells in an anti-ROPN1 response (57, 58), which are considered critical for inducing durable immune responses. The FLY-1A TCR T cells showed clear tumor infiltration and effective antitumor effects when tested in TNBC-bearing mice. Importantly, the FLY-1A TCR T cells significantly surpassed the antitumor effects seen with standard-of-care drugs, including cisplatin and sacituzumab govitecan, in tumoroid and mouse models.
Finally, in preparation of a clinical application, we assessed the FLY-1A TCR for its dominance about surface expression and demonstrated that the pairing of the transgenic TCRα and TCRβ chains could not be improved by cysteine or LRY modifications as described previously (46, 59, 60). With the nonmodified wild-type FLY-1A TCR, we manufactured a clinical-like T-cell product according to an optimized GMP protocol yielding T cells enriched for a young phenotype (according to CD45RO− and CCR7+; ref. 48) and which protocol is currently used in a clinical trial at Erasmus MC to treat MAGE-C2+ melanoma. Notably, this FLY-1A TCR T-cell product passed preclinical safety and potency assays. For example, this product was nonreactive to primary cell cultures derived from healthy organs. This, together with this TCR specifically recognizing cognate but not alternative epitopes, showing no proneness to mispair, and not mediating a response to ROPN1- and/or HLA-A2-negative tumor cells, is indicative of a minimal risk for off-target, off-tumor toxicity. Also, this FLY-1A TCR T-cell product showed a potent response toward patient tumor samples. In fact, T-cell IFN-γ production correlates with ROPN1 gene expression, in which the minimal ROPN1 expression required for T-cell recognition equals the lower limit of endogenous expression levels of patient-derived tumor samples.
Even though the preclinical data argue in favor of a highly specific and sensitive T-cell product to treat TNBC or melanoma, one cannot exclude that toxicities and/or lack of an antitumor response may occur in some patients. With respect to safety, the next step would be to test T cells gene engineered with this TCR in a clinical trial according to dose titration. To this end, the preparation of a clinical vector batch and its quality release is being scheduled. With respect to efficacy, we have previously reported that the immunosuppressive microenvironment of TNBC underlies resistance to anti-PD1 and can be grouped according to the presence and spatial phenotype of CD8+ T cells (61). For example, the excluded phenotype, with CD8+ T cells being confined to the tumor margins, harbors dense extracellular matrix and associates with activation of the TGFβ pathway; the ignored phenotype, with no CD8+ T cells, harbors CD163+ myeloid cells and associates with activation of the WNT pathway, whereas the inflamed phenotype, with CD8+ T cells present in tumor and stromal regions, reveals necrosis and enhanced expression of T-cell coinhibitory receptors. To study immune evasive mechanisms following treatment with FLY-1A TCR T cells, we are currently scheduling the testing of adoptive therapy in an immune-competent tumor model. Interestingly, high ROPN1 expression is observed independently of these spatial CD8 T-cell phenotypes (Supplementary Fig. S9A). Moreover, ROPN1 expression is also not related to response to anti-PD1 (Supplementary Fig. S9B, data from TONIC trial; ref. 62). Interestingly, however, patients with ROPN1+ HLA-A2+ metastatic TNBC who responded to anti-PD1 show an enrichment of the frequency of the FLY-1A TCRβ chain in blood (Supplementary Fig. S9C). These preliminary findings suggest that the targeting of ROPN1 may be agnostic for the type of immune suppression as well as the type of pretreatment and that the ROPN1 TCRβ may take part in an effective anti-TNBC response.
In summary, we have identified ROPN1 as novel and safe target present in the majority of TNBC patients for ACT and selected the FLY-1A TCR that is highly specific and shows high therapeutic value in advanced tumor models, in which it outperforms current treatment options. Currently, T cells gene engineered with this TCR are scheduled for use in a clinical trial to treat patients with TNBC or other ROPN1-positive cancers.
Methods
Patient Cohorts, Databases, and Code of Conduct
American Cancer Society: http://www.cancer.org/ (RRID:SCR_005756).
TNBC cohort 1: breast cancer with RNA-seq (n = 347 of which n = 66 TNBC, geTMM normalized) accessible through the European Genome-phenome Archive EGAS00001001178 (BASIS cohort; ref. 63).
TNBC cohort 2: Primary breast cancer with node-negative disease with microarray data (U133) not receiving adjuvant systemic treatment (n = 867 of which n = 183 TNBC). Data were retrieved from gene expression omnibus (GEO) GSE2034, GSE5327, GSE11121, GSE2990, and GSE7390. Data were normalized using fRMA and batch corrected with ComBat (RRID:SCR_010974; ref. 64). Details of combined cohorts have been described previously (65).
TNBC cohort 3: Metastatic TNBC of the Center for Personalized Cancer Treatment cohort obtained from the Hartwig Medical Foundation with RNA-seq (geTMM normalized; refs. 63, 66) combined and batch corrected with primary TNBC from BASIS using the R package ComBat. After batch correction, 22 metastatic TNBC and 66 primary TNBC were used for the analysis.
TNBC cohort 4: Metastatic TNBC from patients treated with anti-PD1 antibody in the TONIC trial (n = 53 of which n = 44 paired samples; ref. 62) with processed transcriptome data of pre- and post-induction treatment biopsies retrieved via controlled access (available through EGAS00001003535).
The Cancer Genome Atlas (TCGA): Pan-cancer RNA-seq data as well as sample annotation data were retrieved from the UCSC Xena Browser (n = 10,495 of which 1,211 breast cancer, 191 TNBC, and 245 SKCM, TPM normalized). The results shown are in part based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga.
Healthy tissues: RNA-seq data of four databases covering 66 healthy tissues [Uhlen: n = 122 individuals, n = 32 tissues (67); GTEx: n = 1,315 individuals, n = 53 tissues (68); Illumina body map: n = 32 individuals, n = 17 tissues; and Snyder Lab: n = 25 individuals, n = 13 tissues (69)] were downloaded on June 13, 2018, from Expression Atlas (TPM normalized).
This study was performed according to the Declaration of Helsinki and the “Code for Proper Secondary Use of Human Tissue in The Netherlands” (version 2002, update 2011) of the Federation of Medical Scientific Societies (FMSF), which aligns with authorized use of coded spare tissue for research. According to national guidelines, no informed consent was required for this study, and data as well as ex vivo analysis of spare TNBC tissues was approved by the Medical Ethical Committee of Erasmus MC (MEC.02.953 and MEC2020-0090, respectively). The collection of patient data and tissues for the generation and distribution of patient organoids was performed in accordance to the guidelines of the European Network of Research Ethics Committees, following European, national, and local laws.
Gene Expression: RNA-seq, Microarray, and qPCR
Expression of 276 CGAs (as in CTdatabase, Ludwig institute, http://www.cta.lncc.br/, RRID:SCR_007614; ref. 70) was analyzed in healthy and tumor tissues. Expression of ROPN1 and ROPN1B was evaluated in four different cohorts of healthy tissues and was considered expressed in a tissue when TPM values reached the threshold of more than 0.2 in at least two cohorts. Expression in tumors (TCGA) was classified as follows: TPM values between 1 and 9, between 10 and 100, and more than 100 were classified as low, moderate, and high expression, respectively. In case of RNA-seq data (TNBC cohorts 1 and 3) and microarray data (TNBC cohort 2), geTMM- and fRMA-normalized expression values were used, respectively. We used NY-ESO1 as a reference for expression studies.
Quantitative PCR (qPCR) was performed on cDNA panels of 48 healthy human tissues (OriGene Cat# HMRT104, Technologies, Rockville, MD) and TNBC cell lines using MX3000 [RRID:SCR_020526; Taqman probes ROPN1: Hs00250195_m1; ROPN1B: Hs00250195_m1; CTAG1B (NY-ESO1): Hs00265824_m1; GAPDH: Hs02758991_g1; NBL1: Hs01063631_m1; UBE2O: Hs00222904_m1; FAAH: Hs01038664_m1]. Ct values of genes of interest were normalized to GAPDH, and relative expression was expressed as 2−∆Ct.
Protein Expression: IHC Staining
Stainings were performed using large cores of healthy tissues (2 mm in diameter) covering 16 major tissues each from 2 to 6 individuals (derived from autopsy or resection, n = 62) as well as FFPE tissue microarrays of TNBC primary tumor tissue covering 311 patients (71). In addition, whole tissue sections of paired primary and metastatic TNBC tumors (n = 15) were stained (61). Staining with anti-ROPN1 antibody (Atlas Antibodies Cat# HPA052530, RRID:AB_2681861), which detects ROPN1 and ROPN1B protein or anti-NY-ESO1 antibody (Thermo Fisher Cat# 356200, RRID: AB_2533215), was performed following heat-induced antigen retrieval for 20 minutes at 95°C. After cooling to RT, staining was visualized by the antimouse EnVision+ System-HRP (DAB; DakoCytomation). Human testis tissue was used as a positive control tissue. Stainings were manually scored on intensity and percentage of positive tumor cells using Distiller (SlidePath, RRID:SCR_005597) software independently by three investigators (MT, DH, DK).
Generation and Culture of Cell Lines and T Cells
T cells were derived from PBMC from healthy human donors (obtained from Sanquin) by centrifugation via Ficoll-Isopaque (density = 1.077 g/cm3; Amersham Pharmacia Biotech) and following gene transduction (Methods) cultured in RPMI medium supplemented with 25 mmol/L HEPES, 6% human serum (Sanquin), 2 mmol/L L-glutamine, 1% antibiotics (T-cell medium), and 360 U/mL human rIL2 (Proleukin; Chiron). T cells were stimulated every 2 weeks with a mixture of irradiated allogeneic feeder cells, as described elsewhere (72).
The origin of TNBC cell lines has been described elsewhere (73, 74). These cell lines included BT549 (RRID:CVCL_1092), EVSA-T (RRID:CVCL_1207), HCC1569 (RRID:CVCL_1255), Hs578T (RRID:CVCL_0332), MDA-MB231 (MM231, RRID: CVCL_0062), MDA-MB435 s (RRID:CVCL_0622), SUM159PT (RRID:CVCL_5423), and SUM229PE (RRID:CVCL_5594) and were, together with the lymphoblastic K562ABC cell line (parental K562 cells RRID:CVCL_0004, kindly provided by Dr. B. Levine, University of Pennsylvania, Philadelphia, PA), cultured in RPMI medium supplemented with 10% FBS, 2 mmol/L L-glutamine, and 1% antibiotics. The packaging cell lines 293T (RRID:CVCL_0063, kindly provided by Dr. Y. Soneoka, Oxford University, Oxford, U.K.; ref. 75) and Phoenix-Ampho (RRID:CVCL_H716, kindly provided by Dr. G. Nolan, Stanford University; ref. 76) were cultured in DMEM supplemented with 10% FBS, 2 mmol/L L-glutamine, nonessential amino acids, and 1% antibiotics (DMEM complete). The TAP-deficient HLA-A2+ TxB hybrid cell line 174xCEM.T2 (T2, RRID:CVCL_2211, obtained from ATCC: CRL1992) and the HLA-A2+ lymphoblastoid cell line BSM (RRID:CVCL_E504, obtained from Sigma in May 2018, Cat #88052032) were cultured in RPMI medium supplemented with 10% FBS, 2 mmol/L L-glutamine, and 1% antibiotics. Cells were cultured at 37°C/5% CO2. Cells were used for experiments up to 10 passages (max. 5 weeks) and were monthly tested for Mycoplasma contamination by PCR. The latest date cells were tested on May 14, 2024.
Generation of ROPN1/B-Expressing TNBC Cell Lines
To generate ROPN1/B-overexpressing cancer cell lines, a ROPN1 or ROPN1B-GFP fragment [amino acid sequence accessible under UniProtKB (RRID:SCR_004426) Acc. No. Q9HAT0/Q9BZX4; ROPN1/B2A-GFP] was ordered via GeneArt and cloned into the PiggyBac vector PB510B1 (a kind gift from Dr. P.J. French, Erasmus MC) using Infusion Cloning kit (Takara, Cat# 638947). Subsequently, three TNBC cell lines MDA-MB231 cell line, Hs578T, and SUM159PT and the lymphoblastic cell line K562ABC were stably transfected with PiggyBac ROPN1/B-GFP DNA using Lipofectamine P3000 transfection reagent kit (Invitrogen Cat# L3000001) and Transposase Expression vector DNA (System Biosciences Cat# PB210PA1). Cells were cultured with 2 to 5 μg/mL puromycin (Life Technologies Cat# A1113803). Expression of ROPN1/B and GFP was confirmed with qPCR and flow cytometry, respectively.
Identification, Selection, and Ranking of ROPN1/B Peptides
ROPN1/B peptides were ranked according to multiple in silico methods to predict different aspects of immune reactivity (NetMHCpan, RRID:SCR_018182, NetCTLpan, SYFPEITHI, RRID:SCR_013182, and RANKPEP; refs. 77–80). For details on predicted features of these algorithms, see Hammerl and colleagues (81). For immunopeptidomics, the ROPN1/B-expressing HLA-A2+ MM231 cells (3 × 108) were treated with 50 pg/mL human recombinant IFN-γ (PreproTech, Cat# 300-02) for 24 hours and harvested using EDTA before immunoprecipitation of MHC class I molecules. In addition, 1 × 108 ROPN1B-expressing HLA-A2+ K562ABC cells were treated with epigenetic drugs (treatment regimen as described in ref. 82) prior to immunoprecipitation. K562ABC cells are K562 lymphoblastic cells that are engineered to express HLA-A, HLA-B, and HLA-C alleles and are typically used for antigen presentation research. Peptides were eluted and measured with mass spectrometry as described previously (Supplementary Methods; refs. 38, 83). Lastly, peptides were searched in available databases of peptides eluted from tumor tissues as well as cell lines (https://www.zhang-lab.org/caatlas/). All peptides identified through immunopeptidomics were used for below-described analysis, even when they not mapped to HLA-A2.
The top 10 predicted peptides per in silico tool as well as unique peptides (not overlapping with the predicted peptides) retrieved from immunopeptidomics were checked for cross-reactivity with Expitope (40). Peptides that overlapped with peptides from non-ROPN1/B human proteins with up to two amino acid mismatches (i.e., seven out of nine amino acids are identical) were excluded from further analysis. The final list of peptides was ordered at ThinkPeptides (ProImmune, Oxford, United Kingdom), dissolved in 50% to 75% DMSO, and stored at −20°C until use. The HLA-A2 stabilization assay was performed using T2 cells with a slightly adapted protocol (Supplementary Methods; ref. 84). In brief, T2 cells were exposed to titrated amounts of peptide (range from 31 nmol/L to 31 μmol/L, n = 3), and HLA-A2 expression using an HLA-A2-PE antibody (clone BB7.2, 1:20, Abcam Cat #ab79523, RRID:AB_1640177) was measured using flow cytometry (FACS Celesta, BD, RRID:SCR_019597) and analyzed using FlowJo software (version 10.7.1, TreeStar, RRID:SCR_008520).
We calculated two parameters of binding avidity to HLA-A2: (i) amplitude, which was the difference in fluorescence intensity between the highest concentration and baseline, and (ii) half-maximal effective concentration (EC50), which is the calculated concentration of peptide (using GraphPad Prism 5.0, RRID:SCR_002798) giving half-maximal response. Eleven peptides adhered to the following two criteria: (i) EC50 of less than1 × 10−4 mol/L and (ii) binding amplitude of more than 0.5 relative to the reference peptide gp100YLE; these peptides were ranked according to amplitude values.
Enrichment of ROPN1/B Epitope-Specific CD8+ T Cells
Enrichment of epitope-specific T cells was performed by coculturing naïve T cells with autologous CD11c+ cells that were loaded with the shortlisted peptides using PBMCs from two to seven healthy donors per tested peptide. Following the first coculture cycle, ROPN1-specific T cells were amplified over another three cycles using peptide-pulsed autologous PBMC supplemented with medium containing common g cytokines according to a protocol we have previously described (41). Following enrichment, T cells were tested for ROPN1 and/or ROPN1B epitope-specific IFN-γ production. To this end, a supernatant from overnight cocultures of T cells with T2 cells (1 × 106/mL) that were pulsed with epitope (10 ng/mL) was collected, and IFN-γ production was measured with an enzyme-linked immunosorbent assay (BioLegend, Cat #430115) according to the manufacturer’s protocol. T2 cells loaded with an irrelevant epitope were included as a negative control. T-cell IFN-γ was considered epitope-specific in case levels exceeded 200 pg/mL, and levels were minimally twice as high as for irrelevant epitope. T cells that fulfilled these criteria were stained with peptide:MHC (pMHC) tetramers (Tetramer Shop) to determine the frequency of epitope-specific T cells. Events were acquired with FACS Celesta and analyzed using FlowJo software. In case T-cell binding of pMHC was observed in more than 0.5% of CD3+ T cells, these cells were FACS sorted with pMHC multimers.
TCR Cloning and Sequence Identification
CD8+ T cells were exposed to the SMARTer RACE cDNA Amplification Kit (Clontech/Takara, Cat #634858) to identify and annotate ROPN1/B epitope-specific TCRα and TCRαβ chains based on Kunert and colleagues (82). Using the IMGT database and the HighV-QUEST tool (http://www.imgt.org, RRID:SCR_012780), the TCR-V, TCR-D, and TCR-J sequences were annotated according to the Lefranc nomenclature (85, 86). In case for a given T-cell population, TCRα sequences represented 30% or more of all functional sequences of TCRα and the same for TCRβ (i.e., >30% clonal sequences of TCRα and TCRβ), and then those TCR chains were matched with each other. These sets of TCRα and TCRβs were codon optimized (GeneArt) and cloned into the pMP71 vector (a kind gift of Prof. Wolfgang Uckert, MDC, Berlin, Germany; ref. 87) using a TCRβ2A-TCRα cassette.
TCR Gene Transfer
Upon activation with anti-CD3 Monoclonal Antibody (OKT3, Thermo Fisher Scientific Cat #16-0037-81, RRID:AB_468854), PBMCs from healthy donors were transduced with TCR-encoding retroviruses (pMP71) or empty vector that were produced by a coculture of 293T and Phoenix-Ampho packaging cells, as described previously (88, 89). Staining for surface-expressed TCR transgenes was performed using pMHC dextramers (Immudex; ref. 90). In case pMHC complexes were insensitive to detect TCR T cells, we used antibodies directed against TCR-Vβ (antihuman TCR-Vβ7.1, clone ZOE, Beckman Coulter Cat# IM2287, RRID:AB_131323) and CD137-APC (1:25, BD Biosciences Cat# 550890, RRID:AB_398477). The expression of CD137 was measured following 48-hour stimulation with epitope-loaded BSM melanoma cells. In case pMHC or CD137 expression in more than 5% of CD3+ T cells was observed in at least two donors, TCR T cells were then MACS sorted using pMHC complexes or FACS sorted according to upregulated CD137 expression.
Coculture Experiments of TCR T Cells with Target Cells
TCR-transduced T cells (6 × 104/well in a 96-well round-bottom tissue culture-treated plate) were cocultured with target cells (2 × 104/well) in a total volume of 200 μL RPMI medium supplemented with 25 mmol/L HEPES, 10% FBS, 2 mmol/L L-glutamine, and 1% antibiotics for 24 hours. Parental or ROPN1/B-expressing MM231 tumor cells were pretreated for 48 hours with IFN-γ prior to coculture. T-cell recognition of endogenously processed and presented epitopes was demonstrated in case IFN-γ levels exceeded 200 pg/mL, and levels were minimally twice as high as for mock T cells. To test the sensitivity of FLY-1A and FLY-1B TCR T cells, T cells were cocultured with BSM cells loaded with epitope concentrations ranging from 1 pmol/L to 30 μmol/L to determine EC50 values.
Positional Amino Acid Scanning of ROPN1/B Epitopes
Positional amino acid scanning of ROPN1/B epitopes was performed to assess the recognition motifs of TCRs. TCR T cells were cocultured with BSM cells that were pulsed with alternative peptides (i.e., 10 μg/mL) covering all individual amino acids as replacements at every single position in the cognate ROPN1/B epitope (n = 171). Critical amino acid substitutions were defined as those that resulted in at least a 90% drop in IFN-γ production when compared with the cognate epitope. Once having identified alternative peptides that contained such critical amino acid(s), we could assign a fold change value (FC) for every amino acid at every position relative to the cognate epitope. We calculated the frequency per amino acid at every position by dividing the FC of each amino acid by the sum of FCs of all amino acids per position. The resulting frequency matrix was used as input to create the sequence logo presented in bits with the R package ggseqlogo (91). The resulting recognition motif was scanned for its occurrence in the human proteome using the ScanProsite tool (RRID:SCR_024425; ref. 92). Non-ROPN1/B peptides that contained the recognition motif were scored for their in silico binding to HLA-A2 with the NetMHCpan 4.1 software (77). HLA-A2 binders (weak as well as strong binders) were subsequently synthesized and tested for their capability to induce responses by ROPN1/B TCR T cells. To this end, alternative peptides were loaded onto BSM cells and tested at a single dose of 10 μg/mL, and those that resulted in T-cell IFN-γ secretion (FC > 0.1 compared with the cognate epitope) were used to determine EC50 values according to dose titrations. Importantly, non-ROPN1/B source antigens that harbor such peptides were overexpressed in MM231 cells, after which these MM231 cells were cocultured with TCR T cells. Only in case there was no reactivity by TCR T cells toward alternative source antigens, the peptide was considered not to be a product of endogenous processing and presentation. Finally, TCR T cells were screened for a lack of reactivity toward 114 HLA-A2-eluted noncognate peptides (34).
Testing Reactivity of TCR T Cells toward 3D Organoid Models
TCR T cells were subjected to tracking and monitoring in three-dimensional (3D) tumoroid models of ROPN1/B-GFP-expressing MM231 cells and patient-derived organoids from breast and skin tumors.
To this end, we used image-guided injection of droplets with tumor cells into a collagen gel to form extracellular matrix (ECM)-embedded tumoroids as described previously (93–95). Overnight, gels were overlayed with a T-cell medium containing 0.4 μmol/L propidium iodide (PI). Subsequently, TCR or mock T cells were labeled with 0.1 μg/mL Hoechst 33242 (Invitrogen Cat# H1399) for 1 hour, and 50,000 labeled T cells were dispersed on top of the collagen gel. Some tumoroids were treated with 20 μmol/L cisplatin as a positive control for tumor cell killing. Tumoroids were imaged at indicated time points by confocal microscopy using either a Nikon TE2000 equipped with a prior stage controlled by NIS-Elements software (RRID:SCR_014329) or a confocal Opera Phenix High Content Screening System (PerkinElmer, RRID: SCR_021100) equipped with a ×10 air objective (NA 0.3) and a temperature- and CO2-controlled incubator. Approximately 10 to 15 μmol/L Z-stacks were captured spanning the entire tumoroid using a 10× or 20× objective. Images were stitched where necessary and analyzed by either CellProfiler (v 2.2.0, RRID:SCR_007358) or Harmony High Content Imaging and Analysis Software (RRID:SCR_018809). A maximum projection was generated through summing of Hoechst, GFP, and PI signals in each image.
Organoid models from PDX derived from breast and skin tumors were established as described (96). PDX organoids (BR5010B and ME12086B) were seeded in 3D hydrogel (Proprietary, Crown Bioscience Netherlands B.V.) in 384-well plates (Greiner Bio-One B.V.) and incubated for 48 hours. TCR T cells were stained with cell tracker (CellTracker Green CMFDA Dye, Invitrogen Cat# C2925), resuspended in T-cell medium, and added on top of the 3D hydrogel at three different ratios of organoids to T cells. Cisplatin and sacituzumab govitecan were added to tumor organoids at 0.1, 1, and 10 μmol/L and 0.1, 1, and 10 nmol/L, respectively. After 48 and 96 hours, plates were fixed and stained, and tumor volumes were analyzed. Imaging was performed using ImageXpress Micro XLS (Molecular Devices, RRID:SCR_025259) with a 4× NIKON objective. Image analysis was performed using Ominer software. The segmentation index provides a measure of viable cells in a given image. To this end, this index integrates multiple masking operations to isolate and analyze either DAPI (parameter: nuclei), TRITC (actin), or FITC (cell tracker, in this case immune cells). It integrates multiple masking operations to signal, ultimately identifying viable tumor organoids. Experiments and analyses were performed by personnel at the premises of Crown Bioscience, Netherlands, Leiden (Supplementary Methods).
In Vivo Performance of TCR T Cells in a TNBC Xenograft Model
ROPN1-expressing MM231 tumor cells (2.5 × 106) were suspended in Matrigel and subcutaneously (sc) transplanted in the right flank of 8- to 12-week-old female NSG mice (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ, Charles River Laboratories, RRID:IMSR_JAX:005557). Tumor engraftment and treatment regimen were carried out as described previously (97) with only minor changes. In short, mice (25–30 g) bearing established tumors (mean 217 mm3, range 120–318 mm3) were divided over five treatment groups for equal tumor size distribution (n = 9 per T-cell treatment group, in total n = 36 mice). Mice received a single intravenous (iv) transfer of 0.6, 3, or 15 × 106 human TCR or mock T cells followed by sc IL2 injections (1 × 105 IU) for eight consecutive days following T-cell transfer. Alternatively, mice (n = 5) received two iv injections per week of sacituzumab govitecan (Trodelvy, Gilead Sciences Ireland UC; 0.4 mg/kg according to the manufacturer’s instructions). At day 11, tumor regressions were measured relative to day 0, and tumor regression in mice treated with TCR T cells was compared with mock T cells and sacituzumab govitecan. In addition, blood (n = 9 mice) and tumor (n = 4 mice) samples were collected at day 8/9 to detect transferred T cells with flow cytometry. Single-cell suspensions were obtained from the tumors using collagenase treatment as described above. Cells (either from blood or tumor) were washed with PBS and incubated with pMHC and additional antibodies: CD3-BV421 (1:20, BD Biosciences Cat# 562877, RRID:AB_2737860), CD8-BV650 (1:100, BD Biosciences Cat# 563821, RRID:AB_2744462), CD4-V500 (1:50, BD Biosciences Cat# 560768, RRID:AB_1937323), CD137-APC (1:25, BD Biosciences Cat# 550890, RRID:AB_398477), CD279-APC-Cy7 (1:50, BioLegend Cat# 329922, RRID:AB_10933429), and CD366-BB515 (1:10, BD Biosciences Cat# 565568, RRID:AB_2744368). Events were acquired with FACS Celesta and analyzed using FlowJo and R packages ggplot2 (RRID:SCR_014601) and ComplexHeatmap (98, 99). Mouse experiments were approved by the Dutch Central Committee of Animal Experiments (project license numbers: EMCAVD10100202216075 and EMCAVD101002017867) in accordance with the Dutch Act on Animal Experimentation and executed in the animal facility of Erasmus Medical Center.
Assessing Preferential Pairing of TCR Chains
The mispairing between the transgenic and endogenous TCR genes was assessed by transducing T cells with the wild-type or modified TCRαβ chains or the TCRβ chain only. We applied two separate modifications of TCR chains, namely, the introduction of a cysteine bridge or the LRY amino acids, which have both been reported to enhance preferential pairing between the transgenic TCRα and TCRβ chains (46, 47). TCR expression was assessed by flow cytometry upon staining with pMHC multimers and antihuman TCR-Vβ13.1 (clone IMMU 222, Beckman Coulter Cat# IM2292, RRID:AB_131326).
GMP Manufacturing of T-cell Product
We made the TCR T-cell product using a GMP protocol that has been optimized previously (48) and implemented in a clinical trial (clinicaltrials.gov identifier: NCT04729543). To this end, frozen PBMCs were thawed and resuspended in a T-cell medium to a concentration of a 1 × 106 cells/mL. Cells were activated with soluble anti-CD3 (30 ng/mL, Miltenyi Biotec Cat# 130-093-377, RRID:AB_1036126) and anti-CD28 (30 ng/mL, Miltenyi Biotec Cat# 130-126-202, RRID:AB_2889536), hIL15 (110 IU/mL, Miltenyi Biotec Cat# 130-095-766), and hIL21 (0.1 IU/mL, Miltenyi Biotec Cat# 130-095-784) for 48 hours. RetroNectin-coated (overnight, 12 μg/mL, Takara Cat# T100A) nontissue culture flat-bottom plates (24 well) were blocked with 1-mL PBS/2%FBS, after which 0.3-mL retroviral supernatant supplemented with IL15/IL21 was added per well. Plates were centrifuged for 15 minutes at 1,000 g (slow brake). T cells (1 × 106/well) were resuspended in 0.3-mL retroviral supernatant and added to each well. Plates were centrifuged for 60 minutes at 1,000 g (slow brake) and incubated at 37°C/5% CO2. After 5 hours, 800 μL/well T-cell medium supplemented with IL15/IL21 was added to each well and incubated overnight, after which 1.2-mL supernatant was removed and 0.6-mL fresh retroviral supernatant supplemented with IL15/IL21 was added per well. Again, plates were centrifuged to prepare for a second hit with retroviral supernatant, as described above. T cells were then harvested and cultured at a concentration of 0.25 × 106/mL in T-cell medium supplemented with IL15/IL21 for 3 to 6 days. T cells were either directly used in coculture assays or frozen down. Manufacturing was performed at Erasmus MC, Rotterdam, and at the NecstGen facility, Leiden.
Reactivity of TCR T Cells toward Healthy Cells
Primary human healthy cells from HLA-A2+ donors (confirmed via genotyping and with flow cytometric staining for HLA-A2) were obtained via ScienCell or PromoCell (Supplementary Table S3) and cultured and frozen down according to the manufacturer’s instructions. On the day of assay, primary cells and TCR T cells were thawed and rested for 1 and 2 hours, respectively, after which coculture assays were performed as described above.
Reactivity of TCR T Cells toward Patient-Derived Xenografts In Vitro
We assessed T-cell reactivity toward TNBC and melanoma-derived PDX. PDXs were maintained using serial transplantation using NSG mice. Tumors, once isolated, were cut into smaller fragments in DMEM supplemented with 10% FBS, 2 mmol/L L-glutamine, nonessential amino acids, and 1% antibiotics and washed with PBS. Cells were resuspended in fresh collagenase A solution (Sigma Cat# 10103586001) in PBS (1 mg/mL) and incubated for 45 minutes at 37°C while being regularly mixed. EDTA (0.1 mmol/L) was added, and cells were pushed through a 70 μmol/L cell strainer. Single cells were either pretreated with recombinant IFN-γ for 24 hours or used directly in coculture assays. Mock or TCR T cells (1 × 105) were cocultured with single tumor cells in a 1:1 ratio overnight, after which the supernatant was harvested to detect IFN-γ.
Statistical Analyses
Statistical analysis was performed with GraphPad Prism (RRID: RRID:SCR_002798) and Rstudio (RRID:SCR_000432). Statistically significant differences between two groups were calculated with the Mann–Whitney U test for independent samples or the Wilcoxon signed-rank test for paired samples. To determine statistically significant differences between multiple groups, one-way ANOVA with adjustments for multiple comparisons with Tukey’s post hoc test was performed. If assumptions for ANOVA were not met, the Kruskal–Wallis rank test was performed followed by Dunnett’s or Dunn’s multiple comparisons test. Differences were considered statistically significant when P < 0.05.
Data Availability
Transcriptomic data of healthy tissues and primary tumors is available at the GEO (RRID:SCR_005012), the European Genome-phenome Archive, ArrayExpress (RRID:SCR_002964), and UCSC Xena Browser. RNA-seq data of cohort 1 is accessible through EGAS00001001178. Microarray data of cohort 2 is available using the following identifiers: GSE2034, GSE5327, GSE11121, GSE2990, and GSE7390. RNA-seq data of cohort 3 was obtained from the Hartwig Medical Foundation after data transfer agreement (no. DR026) and is accessible through the Hartwig Medical Foundation via https://www.hartwigmedicalfoundation.nl. RNA-seq data of cohort 4 is available at European Genome-phenome Archive (EGAS00001003535). Data generated by the TCGA Research Network is publicly available and can be retrieved from https://www.cancer.gov/tcga or https://xenabrowser.net/. RNA-seq data of healthy tissue cohorts were retrieved on June 13, 2018, from the Expression Atlas (www.ebi.ac.uk/gxa/home) and are available using the following ArrayExpress identifiers: Uhlen cohort, E-MTAB2836; GTEx cohort, E-MTAB5214; Illumina body map cohort, E-MTAB513; and Snyder cohort, E-MTAB4344. Peptide sequences are provided in Supplementary Table S1. Publicly available immunopeptidomics data can be retrieved using the PRIDE partner repository (http://www.ebi.ac.uk/pride) with identifiers PXD004894 and PXD013649. Immunopeptidomics data generated in this study have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the identifiers PXD054804 and PXD055042. The FLY-1A TCRα and TCRβ chain sequences have been deposited to the GenBank with accession numbers PQ177856 and PQ177857. The remaining data generated in this study are available upon request to the corresponding author.
Supplementary Material
This file contains supplementary methods and references.
Top-ranked ROPN1/B peptides and their scores for immunogenicity, non-cross reactivity, HLA-A2 binding and immunogenicity.
ROPN1/B peptides identified with immunopeptidomics.
Overview of healthy cell lines.
Expressions of ROPN1/B, NY-ESO1 and MAGE-A4 target antigens in healthy tissues, TNBC and other tumor types.
Retrieval of ROPN1/B-specific T-cells and TCR genes.
FLY-1A TCR T-cells recognize melanoma tumoroids.
FLY-1A TCR T-cells do not recognize ROPN1-negative and/or HLA-A2-negative tumor cells.
Extended phenotypes of intra-tumoral and peripheral T-cells following adoptive transfer of FLY-1A TCR T cells.
FLY-1A TCR T-cells recognize patient-derived melanoma cells.
The extent of recognition of the ROPN1 target by FLY-1A TCR T cells correlates with level of expression.
Stepwise approach from retrieval and identification of TCRs to the functional testing of their sensitivity, specificity and reactivity.
Presence of ROPN1 and FLY-1A TCRβ in metastatic TNBC in relation to endogenous T cell phenotype and anti-PD1 response.
Acknowledgments
We thank the FACS Sorting Shared Facility of Erasmus MC, especially Andrea Sacchetti, Harm de Wit, and Peter van Geelen, for their assistance. We thank Olga Isaeva (the Netherlands Cancer Institute) for assistance with data analysis of the TONIC trial and Michiel van Dijk (Leiden Academic Centre for Drug Research) for his contributions to experiments with breast cancer cell line-derived tumoroids. This work was supported by the Health∼Holland Public-Private Partnership award EMC-TKI LSH20020 awarded to R.J.M. Abbott and R. Debets, the Erasmus MC Daniel den Hoed Foundation awarded to R. Debets, and the Dutch Cancer Society (KWF; project no. 2014-7087) awarded to J.W.M. Martens and R. Debets.
Footnotes
Note: Supplementary data for this article are available at Cancer Discovery Online (http://cancerdiscovery.aacrjournals.org/).
Authors’ Disclosures
D. Hammerl reports grants from Dutch Cancer Society and Health Holland Public-private partnership during the conduct of the study and has a patent for P128827EP00 pending. M.T.A. de Beijer reports personal fees from the Dutch Ministry of Health, Welfare and Sport and personal fees from Erasmus University Medical Center outside the submitted work and has a patent for WO 2021/110919 pending to ISA Pharmaceuticals B.V. and Erasmus University Medical Center. S.I. Buschow reports grants and nonfinancial support from ISA Pharmaceuticals BV, and Merus BV, Pfizer Inc. and grants and nonfinancial support from Numab therapeutics AG outside the submitted work. M. Kok reports grants from Bristol Myers Squibb, Roche, and AZ, other support from MSD, AZ, and Gilead, and other support from BioNTech outside the submitted work. E.H.J. Danen reports grants from Genmab B.V. outside the submitted work. J.W.M. Martens reports grants from the Dutch Cancer Society during the conduct of the study and grants from the Dutch Cancer Society, Netherlands Organisation for Scientific Research (NWO), European Research Council, S. Komen Foundation, Erasmus University Medical Center Rotterdam, the Netherlands, breast cancer Now, MLDS NL, GSK, Pfizer, Philips, Cergentis, Therawis, Pamgene, Cytrotrack, and Menarini and personal fees from Novartis outside the submitted work. R.J.M. Abbott reports grants from Health∼Holland, other support from Swanbridge Capital, Van Herk Ventures, and Thuja Capital during the conduct of the study, and personal fees and other support from Enara Bio Ltd. and Adaptimmune Ltd. outside the submitted work. R. Debets reports grants from Health∼Holland Public-Private Partnership award EMC-TKI LSH20020, Erasmus MC Daniel den Hoed Foundation, and the Dutch Cancer Society KWF 2014-7087 during the conduct of the study and grants from MSD, Bayer, and Pan Cancer T outside the submitted work, as well as has patents for P128827EP00 and P136875EP00 pending and licensed to Pan Cancer T and a patent for P130556EP00 pending. No disclosures were reported by the other authors.
Authors’ Contributions
D. Kortleve: Conceptualization, data curation, formal analysis, investigation, visualization, methodology, writing–original draft, writing–review and editing. D. Hammerl: Conceptualization, data curation, formal analysis, supervision, investigation, visualization, methodology, writing–original draft, writing–review and editing. M. van Brakel: Investigation, methodology. R. Wijers: Investigation, methodology. D. Roelofs: Investigation. K. Kroese: Investigation. M.M. Timmermans: Data curation, investigation. C.-Y. Liao: Formal analysis, investigation, methodology. S. Huang: Formal analysis, investigation. A. Trapman-Jansen: Investigation. R. Foekens: Investigation. J. Michaux: Investigation. M.T.A. de Beijer: Formal analysis, investigation, writing–review and editing. S.I. Buschow: Data curation, formal analysis, writing–review and editing. J.A.A. Demmers: Formal analysis, investigation. M. Kok: Resources, data curation, writing–review and editing. E.H.J. Danen: Formal analysis, writing–review and editing. M. Bassani-Sternberg: Resources, data curation, formal analysis, writing–review and editing. J.W.M. Martens: Resources, data curation, funding acquisition, methodology, writing–review and editing. R.J.M. Abbott: Funding acquisition, methodology, writing–review and editing. R. Debets: Conceptualization, resources, supervision, funding acquisition, methodology, writing–review and editing.
References
- 1. Yao H, He G, Yan S, Chen C, Song L, Rosol TJ, et al. Triple-negative breast cancer: is there a treatment on the horizon? Oncotarget 2017;8:1913–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Robson M, Im S-A, Senkus E, Xu B, Domchek SM, Masuda N, et al. Olaparib for metastatic breast cancer in patients with a germline BRCA mutation. N Engl J Med 2017;377:523–33. [DOI] [PubMed] [Google Scholar]
- 3. Litton JK, Rugo HS, Ettl J, Hurvitz SA, Gonçalves A, Lee K-H, et al. Talazoparib in patients with advanced breast cancer and a germline BRCA mutation. N Engl J Med 2018;379:753–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kuchenbaecker KB, Hopper JL, Barnes DR, Phillips K-A, Mooij TM, Roos-Blom M-J, et al. Risks of breast, ovarian, and contralateral breast cancer for BRCA1 and BRCA2 mutation carriers. JAMA 2017;317:2402–16. [DOI] [PubMed] [Google Scholar]
- 5. Tung N, Lin NU, Kidd J, Allen BA, Singh N, Wenstrup RJ, et al. Frequency of germline mutations in 25 cancer susceptibility genes in a sequential series of patients with breast cancer. J Clin Oncol 2016;34:1460–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Winter C, Nilsson MP, Olsson E, George AM, Chen Y, Kvist A, et al. Targeted sequencing of BRCA1 and BRCA2 across a large unselected breast cancer cohort suggests that one-third of mutations are somatic. Ann Oncol 2016;27:1532–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Han Y, Yu X, Li S, Tian Y, Liu C. New perspectives for resistance to PARP inhibitors in triple-negative breast cancer. Front Oncol 2020;10:578095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cortes J, Rugo HS, Cescon DW, Im S-A, Yusof MM, Gallardo C, et al. Pembrolizumab plus chemotherapy in advanced triple-negative breast cancer. N Engl J Med 2022;387:217–26. [DOI] [PubMed] [Google Scholar]
- 9. Bardia A, Hurvitz SA, Tolaney SM, Loirat D, Punie K, Oliveira M, et al. Sacituzumab govitecan in metastatic triple-negative breast cancer. N Engl J Med 2021;384:1529–41. [DOI] [PubMed] [Google Scholar]
- 10. Carey LA, Loirat D, Punie K, Bardia A, Diéras V, Dalenc F, et al. Sacituzumab govitecan as second-line treatment for metastatic triple-negative breast cancer—phase 3 ASCENT study subanalysis. NPJ Breast Cancer 2022;8:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schuster SJ, Bishop MR, Tam CS, Waller EK, Borchmann P, McGuirk JP, et al. Tisagenlecleucel in adult relapsed or refractory diffuse large B-cell lymphoma. N Engl J Med 2019;380:45–56. [DOI] [PubMed] [Google Scholar]
- 12. Curran KJ, Margossian SP, Kernan NA, Silverman LB, Williams DA, Shukla N, et al. Toxicity and response after CD19-specific CAR T-cell therapy in pediatric/young adult relapsed/refractory B-ALL. Blood 2019;134:2361–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Neelapu SS, Locke FL, Bartlett NL, Lekakis LJ, Miklos DB, Jacobson CA, et al. Axicabtagene ciloleucel CAR T-cell therapy in refractory large B-cell lymphoma. N Engl J Med 2017;377:2531–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Locke FL, Ghobadi A, Jacobson CA, Miklos DB, Lekakis LJ, Oluwole OO, et al. Long-term safety and activity of axicabtagene ciloleucel in refractory large B-cell lymphoma (ZUMA-1): a single-arm, multicentre, phase 1–2 trial. Lancet Oncol 2019;20:31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Brentjens RJ, Davila ML, Riviere I, Park J, Wang X, Cowell LG, et al. CD19-Targeted T cells rapidly induce molecular remissions in adults with chemotherapy-refractory acute lymphoblastic leukemia. Sci Transl Med 2013;5:177ra38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Raje N, Berdeja J, Lin Y, Siegel D, Jagannath S, Madduri D, et al. Anti-BCMA CAR T-cell therapy bb2121 in relapsed or refractory multiple myeloma. N Engl J Med 2019;380:1726–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Munshi NC, Anderson LD Jr, Shah N, Madduri D, Berdeja J, Lonial S, et al. Idecabtagene vicleucel in relapsed and refractory multiple myeloma. N Engl J Med 2021;384:705–16. [DOI] [PubMed] [Google Scholar]
- 18. Brudno JN, Maric I, Hartman SD, Rose JJ, Wang M, Lam N, et al. T cells genetically modified to express an anti–B-cell maturation antigen chimeric antigen receptor cause remissions of poor-prognosis relapsed multiple myeloma. J Clin Oncol 2018;36:2267–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Moretti A, Ponzo M, Nicolette CA, Tcherepanova IY, Biondi A, Magnani CF. The past, present, and future of non-viral CAR T cells. Front Immunol 2022;13:867013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Haas AR, Tanyi JL, O’Hara MH, Gladney WL, Lacey SF, Torigian DA, et al. Phase I study of lentiviral-transduced chimeric antigen receptor-modified T cells recognizing mesothelin in advanced solid cancers. Mol Ther 2019;27:1919–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ahmed N, Brawley VS, Hegde M, Robertson C, Ghazi A, Gerken C, et al. Human epidermal growth factor receptor 2 (HER2)–specific chimeric antigen receptor–modified T cells for the immunotherapy of HER2-positive sarcoma. J Clin Oncol 2015;33:1688–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. O’Rourke DM, Nasrallah MP, Desai A, Melenhorst JJ, Mansfield K, Morrissette JJD, et al. A single dose of peripherally infused EGFRvIII-directed CAR T cells mediates antigen loss and induces adaptive resistance in patients with recurrent glioblastoma. Sci Transl Med 2017;9:eaaa0984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Heczey A, Louis CU, Savoldo B, Dakhova O, Durett A, Grilley B, et al. CAR T cells administered in combination with lymphodepletion and PD-1 inhibition to patients with neuroblastoma. Mol Ther 2017;25:2214–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Straathof K, Flutter B, Wallace R, Jain N, Loka T, Depani S, et al. Antitumor activity without on-target off-tumor toxicity of GD2–chimeric antigen receptor T cells in patients with neuroblastoma. Sci Transl Med 2020;12:eabd6169. [DOI] [PubMed] [Google Scholar]
- 25. Zhang C, Wang Z, Yang Z, Wang M, Li S, Li Y, et al. Phase I escalating-dose trial of CAR-T therapy targeting CEA+ metastatic colorectal cancers. Mol Ther 2017;25:1248–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Baulu E, Gardet C, Chuvin N, Depil S. TCR-engineered T cell therapy in solid tumors: state of the art and perspectives. Sci Adv 2023;9:eadf3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rapoport AP, Stadtmauer EA, Binder-Scholl GK, Goloubeva O, Vogl DT, Lacey SF, et al. NY-ESO-1-specific TCR–engineered T cells mediate sustained antigen-specific antitumor effects in myeloma. Nat Med 2015;21:914–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Robbins PF, Kassim SH, Tran TLN, Crystal JS, Morgan RA, Feldman SA, et al. A pilot trial using lymphocytes genetically engineered with an NY-ESO-1-reactive T-cell receptor: long-term follow-up and correlates with response. Clin Cancer Res 2015;21:1019–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hong DS, Van Tine BA, Biswas S, McAlpine C, Johnson ML, Olszanski AJ, et al. Autologous T cell therapy for MAGE-A4+ solid cancers in HLA-A*02+ patients: a phase 1 trial. Nat Med 2023;29:104–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kerkar SP, Wang Z-F, Lasota J, Park T, Patel K, Groh E, et al. MAGE-A is more highly expressed than NY-ESO-1 in a systematic immunohistochemical analysis of 3668 cases. J Immunother 2016;39:181–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Johnson LA, Morgan RA, Dudley ME, Cassard L, Yang JC, Hughes MS, et al. Gene therapy with human and mouse T-cell receptors mediates cancer regression and targets normal tissues expressing cognate antigen. Blood 2009;114:535–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Morgan RA, Chinnasamy N, Abate-Daga D, Gros A, Robbins PF, Zheng Z, et al. Cancer regression and neurological toxicity following anti-MAGE-A3 TCR gene therapy. J Immunother 2013;36:133–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Linette GP, Stadtmauer EA, Maus MV, Rapoport AP, Levine BL, Emery L, et al. Cardiovascular toxicity and titin cross-reactivity of affinity-enhanced T cells in myeloma and melanoma. Blood 2013;122:863–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kunert A, Obenaus M, Lamers CHJ, Blankenstein T, Debets R. T-cell receptors for clinical therapy: in vitro assessment of toxicity risk. Clin Cancer Res 2017;23:6012–20. [DOI] [PubMed] [Google Scholar]
- 35. Li N, Wang T, Han D. Structural, cellular and molecular aspects of immune privilege in the testis. Front Immunol 2012;3:152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhao S, Zhu W, Xue S, Han D. Testicular defense systems: immune privilege and innate immunity. Cell Mol Immunol 2014;11:428–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. González-Galarza FF, Takeshita LYC, Santos EJM, Kempson F, Maia MHT, da Silva ALS, et al. Allele frequency net 2015 update: new features for HLA epitopes, KIR and disease and HLA adverse drug reaction associations. Nucleic Acids Res 2015;43:D784–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chong C, Marino F, Pak H, Racle J, Daniel RT, Müller M, et al. High-throughput and sensitive immunopeptidomics platform reveals profound interferonγ-mediated remodeling of the human leukocyte antigen (HLA) ligandome. Mol Cell Proteomics 2018;17:533–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bassani-Sternberg M, Bräunlein E, Klar R, Engleitner T, Sinitcyn P, Audehm S, et al. Direct identification of clinically relevant neoepitopes presented on native human melanoma tissue by mass spectrometry. Nat Commun 2016;7:13404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jaravine V, Mösch A, Raffegerst S, Schendel DJ, Frishman D. Expitope 2.0: a tool to assess immunotherapeutic antigens for their potential cross-reactivity against naturally expressed proteins in human tissues. BMC Cancer 2017;17:892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mahajan S, Kortleve D, Debets R, Hammerl D. Detection of low-frequency epitope-specific T cells in blood of healthy individuals according to an optimized in vitro amplification system. J Immunol 2022;209:2239–47. [DOI] [PubMed] [Google Scholar]
- 42. Chaux P, Vantomme V, Coulie P, Boon T, van der Bruggen P. Estimation of the frequencies of anti-MAGE-3 cytolytic T-lymphocyte precursors in blood from individuals without cancer. Int J Cancer 1998;77:538–42. [DOI] [PubMed] [Google Scholar]
- 43. Alanio C, Lemaitre F, Law HKW, Hasan M, Albert ML. Enumeration of human antigen-specific naive CD8+ T cells reveals conserved precursor frequencies. Blood 2010;115:3718–25. [DOI] [PubMed] [Google Scholar]
- 44. Willemsen RA, Weijtens ME, Ronteltap C, Eshhar Z, Gratama JW, Chames P, et al. Grafting primary human T lymphocytes with cancer-specific chimeric single chain and two chain TCR. Gene Ther 2000;7:1369–77. [DOI] [PubMed] [Google Scholar]
- 45. Bendle GM, Linnemann C, Hooijkaas AI, Bies L, De Witte MA, Jorritsma A, et al. Lethal graft-versus-host disease in mouse models of T cell receptor gene therapy. Nat Med 2010;16:565–70. [DOI] [PubMed] [Google Scholar]
- 46. Kuball J, Dossett ML, Wolfl M, Ho WY, Voss R-H, Fowler C, et al. Facilitating matched pairing and expression of TCR chains introduced into human T cells. Blood 2007;109:2331–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Thomas S, Mohammed F, Reijmers RM, Woolston A, Stauss T, Kennedy A, et al. Framework engineering to produce dominant T cell receptors with enhanced antigen-specific function. Nat Commun 2019;10:4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lamers CHJ, van Steenbergen-Langeveld S, van Brakel M, Groot-van Ruijven CM, van Elzakker PMML, van Krimpen B, et al. T cell receptor-engineered T cells to treat solid tumors: T cell processing toward optimal T cell fitness. Hum Gene Ther Methods 2014;25:345–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lamers CHJ, Sleijfer S, Vulto AG, Kruit WHJ, Kliffen M, Debets R, et al. Treatment of metastatic renal cell carcinoma with autologous T-lymphocytes genetically retargeted against carbonic anhydrase IX: first clinical experience. J Clin Oncol 2006;24:e20–2. [DOI] [PubMed] [Google Scholar]
- 50. Lamers CH, Sleijfer S, Van Steenbergen S, Van Elzakker P, Van Krimpen B, Groot C, et al. Treatment of metastatic renal cell carcinoma with CAIX CAR-engineered T cells: clinical evaluation and management of on-target toxicity. Mol Ther 2013;21:904–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Fiedler SE, Dudiki T, Vijayaraghavan S, Carr DW. Loss of R2D2 proteins ROPN1 and ROPN1L causes defects in murine sperm motility, phosphorylation, and fibrous sheath integrity. Biol Reprod 2013;88:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Timm J, Walker CM. Mutational escape of CD8+ T cell epitopes: implications for prevention and therapy of persistent hepatitis virus infections. Med Microbiol Immunol 2015;204:29–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Verdegaal EME, De Miranda NFCC, Visser M, Harryvan T, Van Buuren MM, Andersen RS, et al. Neoantigen landscape dynamics during human melanoma–T cell interactions. Nature 2016;536:91–5. [DOI] [PubMed] [Google Scholar]
- 54. Liu Q, Huang X, Li Q, He L, Li S, Chen X, et al. Rhophilin-associated tail protein 1 promotes migration and metastasis in triple negative breast cancer via activation of RhoA. FASEB J 2020;34:9959–71. [DOI] [PubMed] [Google Scholar]
- 55. Robbins PF, Morgan RA, Feldman SA, Yang JC, Sherry RM, Dudley ME, et al. Tumor regression in patients with metastatic synovial cell sarcoma and melanoma using genetically engineered lymphocytes reactive with NY-ESO-1. J Clin Oncol 2011;29:917–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Robbins PF, Li YF, El-Gamil M, Zhao Y, Wargo JA, Zheng Z, et al. Single and dual amino acid substitutions in TCR CDRs can enhance antigen-specific T cell functions. J Immunol 2008;180:6116–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Willemsen R, Ronteltap C, Heuveling M, Debets R, Bolhuis R. Redirecting human CD4+ T lymphocytes to the MHC class I-restricted melanoma antigen MAGE-A1 by TCR alphabeta gene transfer requires CD8alpha. Gene Ther 2005;12:140–6. [DOI] [PubMed] [Google Scholar]
- 58. Willemsen RA, Sebestyén Z, Ronteltap C, Berrevoets C, Drexhage J, Debets R. CD8α coreceptor to improve TCR gene transfer to treat melanoma: down-regulation of tumor-specific production of IL-4, IL-5, and IL-10. J Immunol 2006;177:991–8. [DOI] [PubMed] [Google Scholar]
- 59. Cohen CJ, Li YF, El-Gamil M, Robbins PF, Rosenberg SA, Morgan RA. Enhanced antitumor activity of T cells engineered to express T-cell receptors with a second disulfide bond. Cancer Res 2007;67:3898–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Govers C, Sebestyén Z, Coccoris M, Willemsen RA, Debets R. T cell receptor gene therapy: strategies for optimizing transgenic TCR pairing. Trends Mol Med 2010;16:77–87. [DOI] [PubMed] [Google Scholar]
- 61. Hammerl D, Martens JWM, Timmermans M, Smid M, Trapman-Jansen AM, Foekens R, et al. Spatial immunophenotypes predict response to anti-PD1 treatment and capture distinct paths of T cell evasion in triple negative breast cancer. Nat Commun 2021;12:5668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Voorwerk L, Slagter M, Horlings HM, Sikorska K, van de Vijver KK, de Maaker M, et al. Immune induction strategies in metastatic triple-negative breast cancer to enhance the sensitivity to PD-1 blockade: the TONIC trial. Nat Med 2019;25:920–8. [DOI] [PubMed] [Google Scholar]
- 63. Smid M, Rodríguez-González FG, Sieuwerts AM, Salgado R, Prager-Van der Smissen WJC, Vlugt-Daane Mvd, et al. Breast cancer genome and transcriptome integration implicates specific mutational signatures with immune cell infiltration. Nat Commun 2016;7:12910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zhang Y, Parmigiani G, Johnson WE. ComBat-seq: batch effect adjustment for RNA-seq count data. NAR Genom Bioinform 2020;2:lqaa078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hammerl D, Massink MPG, Smid M, van Deurzen CHM, Meijers-Heijboer HEJ, Waisfisz Q, et al. Clonality, antigen recognition, and suppression of CD8+ T cells differentially affect prognosis of breast cancer subtypes. Clin Cancer Res 2020;26:505–17. [DOI] [PubMed] [Google Scholar]
- 66. Smid M, Coebergh van den Braak RRJ, van de Werken HJG, van Riet J, van Galen A, de Weerd V, et al. Gene length corrected trimmed mean of M-values (GeTMM) processing of RNA-seq data performs similarly in intersample analyses while improving intrasample comparisons. BMC Bioinformatics 2018;19:236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, et al. Proteomics. Tissue-based map of the human proteome. Science 2015;347:1260419. [DOI] [PubMed] [Google Scholar]
- 68. GTEx Consortium . Human genomics. The genotype-tissue expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 2015;348:648–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Lin S, Lin Y, Nery JR, Urich MA, Breschi A, Davis CA, et al. Comparison of the transcriptional landscapes between human and mouse tissues. Proc Natl Acad Sci U S A 2014;111:17224–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Almeida LG, Sakabe NJ, de Oliveira AR, Silva MCC, Mundstein AS, Cohen T, et al. CTdatabase: a knowledge-base of high-throughput and curated data on cancer-testis antigens. Nucleic Acids Res 2009;37:D816–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Liu NQ, De Marchi T, Timmermans A, Trapman-Jansen AMAC, Foekens R, Look MP, et al. Prognostic significance of nuclear expression of UMP-CMP kinase in triple negative breast cancer patients. Sci Rep 2016;6:32027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. van de Griend RJ, Bolhuis RLH. Rapid expansion of allospecific cytotoxic T cell clones using nonspecific feeder cell lines without further addition of exogenous IL2. Transplantation 1984;38:401–6. [DOI] [PubMed] [Google Scholar]
- 73. Riaz M, van Jaarsveld MTM, Hollestelle A, Prager-van der Smissen WJC, Heine AAJ, Boersma AWM, et al. miRNA expression profiling of 51 human breast cancer cell lines reveals subtype and driver mutation-specific miRNAs. Breast Cancer Res 2013;15:R33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Hollestelle A, Elstrodt F, Timmermans M, Sieuwerts AM, Klijn JGM, Foekens JA, et al. Four human breast cancer cell lines with biallelic inactivating α-catenin gene mutations. Breast Cancer Res Treat 2010;122:125–33. [DOI] [PubMed] [Google Scholar]
- 75. Grignani F, Kinsella T, Mencarelli A, Valtieri M, Riganelli D, Grignani F, et al. High-efficiency gene transfer and selection of human hematopoietic progenitor cells with a hybrid EBV/retroviral vector expressing the green fluorescence protein. Cancer Res 1998;58:14–9. [PubMed] [Google Scholar]
- 76. Weijtens ME, Willemsen RA, Hart EH, Bolhuis RL. A retroviral vector system ‘STITCH’ in combination with an optimized single chain antibody chimeric receptor gene structure allows efficient gene transduction and expression in human T lymphocytes. Gene Ther 1998;5:1195–203. [DOI] [PubMed] [Google Scholar]
- 77. Reynisson B, Alvarez B, Paul S, Peters B, Nielsen M. NetMHCpan-4.1 and NetMHCIIpan-4.0: improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Res 2020;48:W449–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Stranzl T, Larsen MV, Lundegaard C, Nielsen M. NetCTLpan: pan-specific MHC class I pathway epitope predictions. Immunogenetics 2010;62:357–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Rammensee H, Bachmann J, Emmerich NP, Bachor OA, Stevanović S. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics 1999;50:213–9. [DOI] [PubMed] [Google Scholar]
- 80. Reche PA, Glutting J-P, Reinherz EL. Prediction of MHC class I binding peptides using profile motifs. Hum Immunol 2002;63:701–9. [DOI] [PubMed] [Google Scholar]
- 81. Hammerl D, Rieder D, Martens JWM, Trajanoski Z, Debets R. Adoptive T cell therapy: new avenues leading to safe targets and powerful allies. Trends Immunol 2018;39:921–36. [DOI] [PubMed] [Google Scholar]
- 82. Kunert A, van Brakel M, van Steenbergen-Langeveld S, da Silva M, Coulie PG, Lamers C, et al. MAGE-C2–Specific TCRs combined with epigenetic drug-enhanced antigenicity yield robust and tumor-selective T cell responses. J Immunol 2016;197:2541–52. [DOI] [PubMed] [Google Scholar]
- 83. de Beijer MTA, Bezstarosti K, Luijten R, Doff WAS, Boor PPC, Pieterman RFA, et al. Immunopeptidome of hepatocytes isolated from patients with HBV infection and hepatocellular carcinoma. JHEP Rep 2022;4:100576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Miles KM, Miles JJ, Madura F, Sewell AK, Cole DK. Real time detection of peptide–MHC dissociation reveals that improvement of primary MHC-binding residues can have a minimal, or no, effect on stability. Mol Immunol 2011;48:728–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Brochet X, Lefranc M-P, Giudicelli V. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res 2008;36:W503–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Giudicelli V, Brochet X, Lefranc M-P. IMGT/V-QUEST: IMGT standardized analysis of the immunoglobulin (IG) and T cell receptor (TR) nucleotide sequences. Cold Spring Harb Protoc 2011;2011:695–715. [DOI] [PubMed] [Google Scholar]
- 87. Engels B, Cam H, Schüler T, Indraccolo S, Gladow M, Baum C, et al. Retroviral vectors for high-level transgene expression in T lymphocytes. Hum Gene Ther 2003;14:1155–68. [DOI] [PubMed] [Google Scholar]
- 88. Lamers CHJ, Willemsen RA, Van Elzakker P, Van Krimpen BA, Gratama JW, Debets R. Phoenix-ampho outperforms PG13 as retroviral packaging cells to transduce human T cells with tumor-specific receptors: implications for clinical immunogene therapy of cancer. Cancer Gene Ther 2006;13:503–9. [DOI] [PubMed] [Google Scholar]
- 89. Straetemans T, van Brakel M, van Steenbergen S, Broertjes M, Drexhage J, Hegmans J, et al. TCR gene transfer: MAGE-C2/HLA-A2 and MAGE-A3/HLA-DP4 epitopes as melanoma-specific immune targets. Clin Dev Immunol 2012;2012:586314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Kortleve D, van Brakel M, Wijers R, Debets R, Hammerl D. Gene engineering T cells with T-cell receptor for adoptive therapy. Methods Mol Biol 2022;2453:209–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Wagih O. ggseqlogo: a versatile R package for drawing sequence logos. Bioinformatics 2017;33:3645–7. [DOI] [PubMed] [Google Scholar]
- 92. Sigrist CJA, de Castro E, Cerutti L, Cuche BA, Hulo N, Bridge A, et al. New and continuing developments at PROSITE. Nucleic Acids Res 2013;41:D344–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Truong HH, de Sonneville J, Ghotra VPS, Xiong J, Price L, Hogendoorn PCW, et al. Automated microinjection of cell-polymer suspensions in 3D ECM scaffolds for high-throughput quantitative cancer invasion screens. Biomaterials 2012;33:181–8. [DOI] [PubMed] [Google Scholar]
- 94. Truong HH, Xiong J, Ghotra VPS, Nirmala E, Haazen L, Le Dévédec SE, et al. β1 integrin inhibition elicits a prometastatic switch through the TGFβ-miR-200-ZEB network in E-cadherin-positive triple-negative breast cancer. Sci Signal 2014;7:ra15. [DOI] [PubMed] [Google Scholar]
- 95. Balcioglu HE, van de Water B, Danen EHJ. Tumor-induced remote ECM network orientation steers angiogenesis. Sci Rep 2016;6:22580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Van De Wetering M, Francies HE, Francis JM, Bounova G, Iorio F, Pronk A, et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 2015;161:933–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Straetemans T, Coccoris M, Berrevoets C, Treffers-Westerlaken E, Scholten CEV, Schipper D, et al. T-cell receptor gene therapy in human melanoma-bearing immune-deficient mice: human but not mouse T cells recapitulate outcome of clinical studies. Hum Gene Ther 2012;23:187–201. [DOI] [PubMed] [Google Scholar]
- 98. Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016;32:2847–9. [DOI] [PubMed] [Google Scholar]
- 99. Gu Z. Complex heatmap visualization. Imeta 2022;1:e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This file contains supplementary methods and references.
Top-ranked ROPN1/B peptides and their scores for immunogenicity, non-cross reactivity, HLA-A2 binding and immunogenicity.
ROPN1/B peptides identified with immunopeptidomics.
Overview of healthy cell lines.
Expressions of ROPN1/B, NY-ESO1 and MAGE-A4 target antigens in healthy tissues, TNBC and other tumor types.
Retrieval of ROPN1/B-specific T-cells and TCR genes.
FLY-1A TCR T-cells recognize melanoma tumoroids.
FLY-1A TCR T-cells do not recognize ROPN1-negative and/or HLA-A2-negative tumor cells.
Extended phenotypes of intra-tumoral and peripheral T-cells following adoptive transfer of FLY-1A TCR T cells.
FLY-1A TCR T-cells recognize patient-derived melanoma cells.
The extent of recognition of the ROPN1 target by FLY-1A TCR T cells correlates with level of expression.
Stepwise approach from retrieval and identification of TCRs to the functional testing of their sensitivity, specificity and reactivity.
Presence of ROPN1 and FLY-1A TCRβ in metastatic TNBC in relation to endogenous T cell phenotype and anti-PD1 response.
Data Availability Statement
Transcriptomic data of healthy tissues and primary tumors is available at the GEO (RRID:SCR_005012), the European Genome-phenome Archive, ArrayExpress (RRID:SCR_002964), and UCSC Xena Browser. RNA-seq data of cohort 1 is accessible through EGAS00001001178. Microarray data of cohort 2 is available using the following identifiers: GSE2034, GSE5327, GSE11121, GSE2990, and GSE7390. RNA-seq data of cohort 3 was obtained from the Hartwig Medical Foundation after data transfer agreement (no. DR026) and is accessible through the Hartwig Medical Foundation via https://www.hartwigmedicalfoundation.nl. RNA-seq data of cohort 4 is available at European Genome-phenome Archive (EGAS00001003535). Data generated by the TCGA Research Network is publicly available and can be retrieved from https://www.cancer.gov/tcga or https://xenabrowser.net/. RNA-seq data of healthy tissue cohorts were retrieved on June 13, 2018, from the Expression Atlas (www.ebi.ac.uk/gxa/home) and are available using the following ArrayExpress identifiers: Uhlen cohort, E-MTAB2836; GTEx cohort, E-MTAB5214; Illumina body map cohort, E-MTAB513; and Snyder cohort, E-MTAB4344. Peptide sequences are provided in Supplementary Table S1. Publicly available immunopeptidomics data can be retrieved using the PRIDE partner repository (http://www.ebi.ac.uk/pride) with identifiers PXD004894 and PXD013649. Immunopeptidomics data generated in this study have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the identifiers PXD054804 and PXD055042. The FLY-1A TCRα and TCRβ chain sequences have been deposited to the GenBank with accession numbers PQ177856 and PQ177857. The remaining data generated in this study are available upon request to the corresponding author.