Abstract
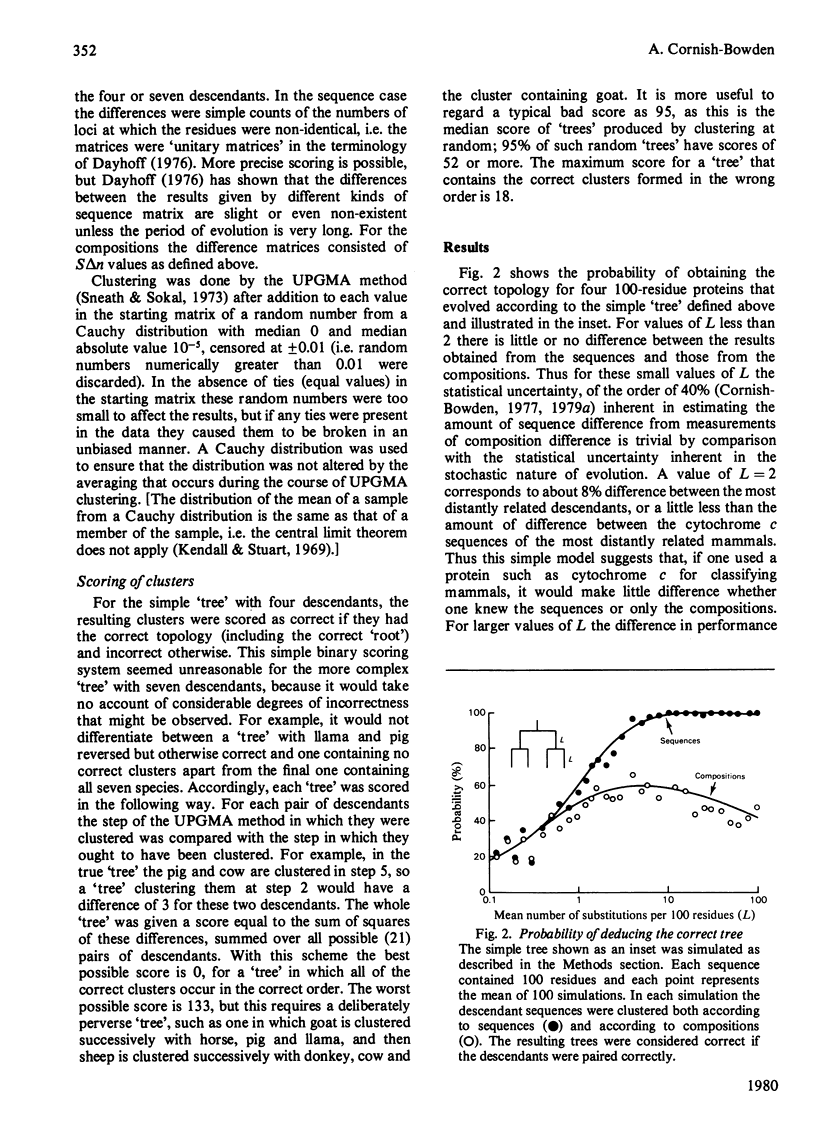
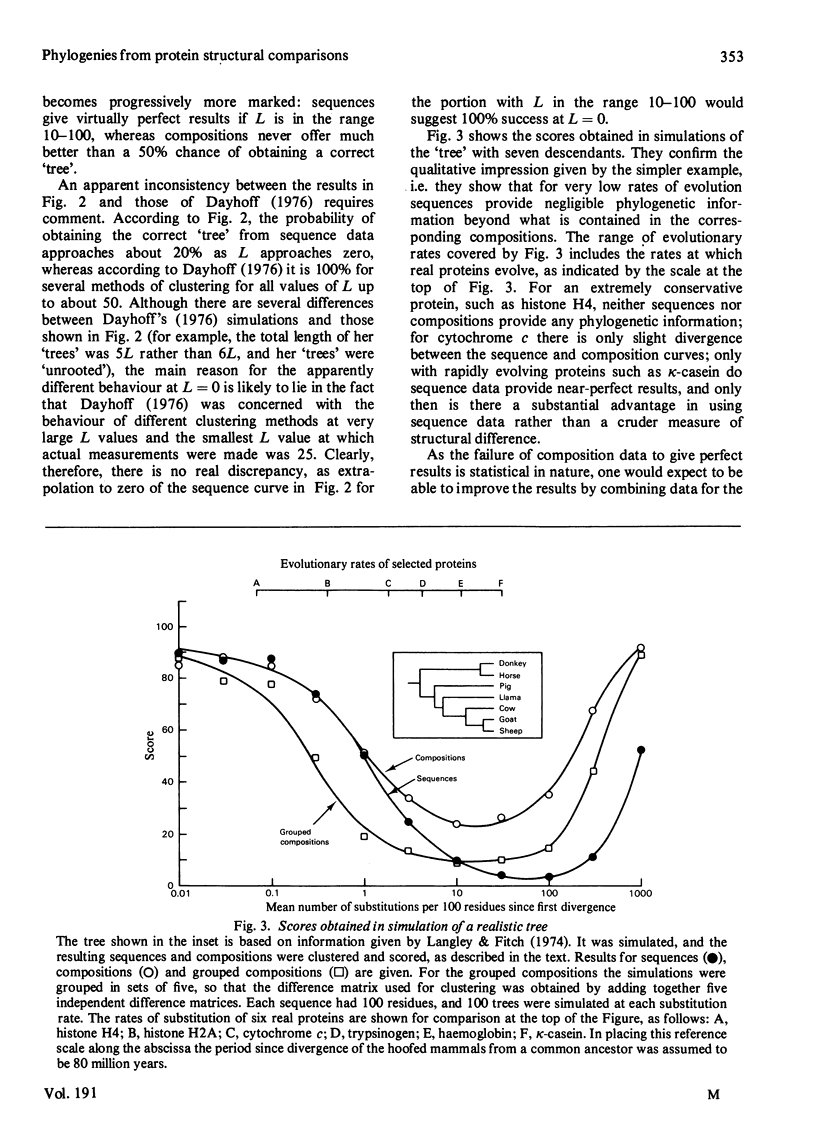
Because evolution occurs by random events, the actual number of substitutions that occur in any period is not exactly equal to the number expected from the mean rate of substitution, but is statistically distributed about it. In consequence, even if rates of evolution are constant in different lineages, 'trees' deduced from descendant protein sequences contain random errors. When there are fewer than about eight differences between the sequences of the most distantly related pair from a set of proteins, this random effect is very large. It can then render trivial the statistical disadvantage inherent in using a crude measure of protein difference, such as amino acid composition or immunological cross-reactivity, in preference to a measure based the sequences of the most distantly related pair from a set of proteins, this random effect is very large. It can then render trivial the statistical disadvantage inherent in using a crude measure of protein difference, such as amino acid composition or immunological cross-reactivity, in preference to a measure based the sequences of the most distantly related pair from a set of proteins, this random effect is very large. It can then render trivial the statistical disadvantage inherent in using a crude measure of protein difference, such as amino acid composition or immunological cross-reactivity, in preference to a measure based on amino acid sequence. In some cases, such as classification of mammals on the basis of cytochrome c structure, it appears to make little difference to the reliability of the results whether the sequences of the protein concerned are known or not. It may also be possible to obtain more reliable phylogenetic information from composition measurements on several kinds of protein than one could obtain from sequence measurements on a single kind of protein.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnheim N., Steller R. Multiple genes for lysozyme in birds. Arch Biochem Biophys. 1970 Dec;141(2):656–661. doi: 10.1016/0003-9861(70)90185-2. [DOI] [PubMed] [Google Scholar]
- Cornish-Bowden A. Assessment of protein sequence identity from amino acid composition data. J Theor Biol. 1977 Apr 21;65(4):735–742. doi: 10.1016/0022-5193(77)90019-4. [DOI] [PubMed] [Google Scholar]
- Cornish-Bowden A. How reliably do amino acid composition comparisons predict sequence similarities between proteins? J Theor Biol. 1979 Feb 21;76(4):369–386. doi: 10.1016/0022-5193(79)90007-9. [DOI] [PubMed] [Google Scholar]
- Dayhoff M. O. The origin and evolution of protein superfamilies. Fed Proc. 1976 Aug;35(10):2132–2138. [PubMed] [Google Scholar]
- Langley C. H., Fitch W. M. An examination of the constancy of the rate of molecular evolution. J Mol Evol. 1974;3(3):161–177. doi: 10.1007/BF01797451. [DOI] [PubMed] [Google Scholar]
- Nei M. Standard error of immunological dating of evolutionary time. J Mol Evol. 1977 May 13;9(3):203–211. doi: 10.1007/BF01796109. [DOI] [PubMed] [Google Scholar]
- Prager E. M., Wilson A. C. Construction of phylogenetic trees for proteins and nucleic acids: empirical evaluation of alternative matrix methods. J Mol Evol. 1978 Jun 20;11(2):129–142. doi: 10.1007/BF01733889. [DOI] [PubMed] [Google Scholar]
- Thompson R. B., Borden D., Tarr G. E., Margoliash E. Heterogeneity of amino acid sequence in hippopotamus cytochrome c. J Biol Chem. 1978 Dec 25;253(24):8957–8961. [PubMed] [Google Scholar]


