Abstract
Carbonic anhydrase (CA), an enzyme conserved across species, is pivotal in the interconversion of inorganic carbon (Ci; CO2, and HCO3−). Compared to the well-studied intracellular CA, the specific role of extracellular CA in photosynthetic organisms is still not well understood. In the green alga Chlamydomonas (Chlamydomonas reinhardtii), carbonic anhydrase 1 (CAH1), located at the periplasmic space, is strongly induced under CO2-limiting conditions by the Myb transcription factor LCR1. While the lcr1 mutant shows decreased Ci-affinity, the detailed mechanisms behind this phenomenon are yet to be elucidated. In this study, we aimed to unravel the LCR1-dependent genes essential for maintaining high Ci-affinity. To achieve this, we identified a total of 12 LCR1-dependent inducible genes under CO2-limiting conditions, focusing specifically on the most prominent ones—CAH1, LCI1, LCI6, and Cre10.g426800. We then created mutants of these genes using the CRISPR–Cas9 system, all from the same parental strain, and compared their Ci-affinity. Contrary to earlier findings that reported no reduction in Ci-affinity in the cah1 mutant, our cah1-1 mutant exhibited a decrease in Ci-affinity under high HCO3−/CO2-ratio conditions. Additionally, when we treated wild-type cells with a CA inhibitor with low membrane permeability, a similar reduction in Ci-affinity was observed. Moreover, the addition of exogenous CA to the cah1 mutant rescued the decreased Ci-affinity. These results, highlighting the crucial function of the periplasmic CAH1 in maintaining high Ci-affinity in Chlamydomonas cells, provide insights into the functions of periplasmic CA in algal carbon assimilation.
A periplasmic carbonic anhydrase in Chlamydomonas reinhardtii plays a crucial role in maintaining a high affinity for inorganic carbon, particularly under CO2-limiting conditions.
Introduction
Carbonic anhydrase (CA; EC 4.2.1.1) is a metalloenzyme that catalyzes the interconversion between CO2 and HCO3−. CA is among the enzymes that display the highest turnover rates (Chegwidden and Carter 2000), thereby fulfilling biological demands in diverse physiological processes such as pH homeostasis, inorganic carbon (Ci; CO2 and HCO3−) transport, and Ci assimilation. CA is classified into eight subclasses based on the primary structure (Aspatwar et al. 2022).
In land plants, CA is hypothesized to play a crucial role in carbon assimilation, although its function remains controversial. Historically, it has been posited that in C3 plants, abundant CA in the chloroplast stroma aids CO2 fixation by facilitating its diffusion (Jacobson et al. 1975). However, this understanding has been challenged by recent studies. For instance, Hines et al. (2021) found that the complete loss of stromal CA does not significantly alter carbon assimilation compared to wild-type (WT) plants. In contrast, CAs' role in aquatic organisms, such as microalgae and cyanobacteria, is more clearly defined within the operation of the CO2-concentrating mechanism (CCM) (Fukuzawa et al. 1992; Badger 2003). Notably, Rubisco, a key enzyme in photosynthetic CO2 fixation, exhibits a lower affinity for CO2 in microalgae and cyanobacteria than in its terrestrial counterparts (Jordan and Ogren 1981). In these aquatic organisms, the CCM helps overcome the disadvantage of Rubisco's lower CO2 affinity by actively transporting HCO3− into the chloroplast stroma through membrane transporters and channels. Once in proximity to Rubisco, HCO3− is converted to CO2 by intracellular CA, effectively concentrating CO2 (Raven et al. 2011).
In Chlamydomonas (Chlamydomonas reinhardtii), a freshwater green alga, CAs are crucial for driving CCM, and the compartmentalized CAs are integral to supplying CO2 specifically to the pyrenoid, where Rubisco is densely packed in the chloroplast (Moroney et al. 2011). Among them, carbonic anhydrase 3 (CAH3), an α-type CA, plays a unique role. It is localized in the lumen of the pyrenoid-invading thylakoid membrane, also known as the pyrenoid tubule (Sinetova et al. 2012). Here, CAH3 facilitates the conversion of HCO3− to CO2, a process enhanced by the lumen's acidic pH. CAH3-deficient mutant exhibits decreased Ci affinity with higher accumulation of internal Ci relative to WT cells, highlighting its role in the generation of CO2 from the stromal Ci pool (Funke et al. 1997; Karlsson et al. 1998). Additionally, the low-CO2 inducible protein B/C (LCIB/C) hexamer, a θ-type CA positioned around the pyrenoid, serves to reconvert CO2 leaking from the pyrenoid into HCO3−, maintaining optimal Ci concentration for photosynthesis (Wang and Spalding 2006; Yamano et al. 2010; Kasili et al. 2023). In addition to LCIB/LCIC and CAH3, Chlamydomonas has α-type CAs (CAH1 and CAH2), β-type CAs (CAH4 to CAH9), and γ-type CAs (CAG1 to CAG3), but their roles in CCM remain unresolved (Moroney et al. 2011).
CAH1, an α-type CA localized at the periplasmic space, was the first CA to be identified in Chlamydomonas (Coleman et al. 1984), but its importance in the CCM remains controversial. CAH1 is induced upon CO2-limitation, and its induction is dependent on a Myb transcription factor LCR1, whose expression is regulated by CCM1/CIA5, a master regulator of CCM (Fukuzawa et al. 1990, 2001; Xiang et al. 2001; Yoshioka et al. 2004). In addition to the abundant accumulation of CAH1 under CO2-limiting conditions, inhibition of periplasmic CA by weakly permeable CA inhibitors, such as acetazolamide (AZA), has been shown to decrease Ci-affinity (Moroney et al. 1985). While other CA isoforms, including the α-type CAH2 and the β-type CAH8, are also present in the periplasmic space, their expression levels under CO2-limiting conditions are lower compared to that of CAH1 (Moroney et al. 2011). These observations have led to the establishment of a well-known model in which periplasmic CA, particularly CAH1, facilitates diffusive CO2 entry by maintaining a CO2 gradient across the plasma membrane. This is achieved through the rapid equilibration of CO2 with bulk HCO3− at the cell surface. Because periplasmic CA activity was detected in diverse algae (Nimer et al. 1999; Elzenga et al. 2000; Tsuji et al. 2017, 2021), and its inhibition by AZA caused the decline of Ci-affinity, periplasmic CA-mediated CO2 uptake has been a widespread hypothesis. Conversely, it has also been suggested that the effects of AZA on photosynthetic kinetics may be due to the inhibition of intracellular CAs rather than extracellular ones (Williams and Turpin 1987). Moreover, a Chlamydomonas mutant lacking CAH1 showed no difference in growth or Ci-affinity difference under low-CO2 conditions (Van and Spalding 1999), challenging the hypothesis that periplasmic CA facilitates CO2 acquisition from bulk HCO3−. Another hypothesis based on the mathematical modeling is that periplasmic CA recaptures leaked CO2 through hydration reaction (Fridlyand 1997). Thus, while massive effort has been spent to elucidate the function of periplasmic CA, conclusive evidence to support either hypothesis has not been presented yet.
We previously demonstrated through macroarray analysis, which is limited to a specific number of genes, that the lcr1 mutant was unable to induce at least three low-CO2 (LC) inducible genes, namely CAH1, LCI1, and LCI6 (Yoshioka et al. 2004). Notably, LCI1, localized at the plasma membrane, is hypothesized to function as a CO2 channel due to its structural characteristics (Ohnishi et al. 2010; Kono et al. 2020). In addition, LCI1 interacts with high-light activated protein 3 (HLA3), an HCO3− transporter on the plasma membrane (Yamano et al. 2015; Mackinder et al. 2017). Although the lcr1 mutant shows a decrease of Ci-affinity under CO2-limiting conditions, the major contributor to this phenotype has not been determined yet. Among the three candidates, independent disruption of CAH1 and LCI1 do not show decreased Ci-affinities (Van and Spalding 1999; Kono and Spalding 2020), suggesting that cooperative functions of these three components or contribution of other unidentified factors for high-affinity photosynthesis for Ci. In this study, to gain further insight into LCR1-dependent CCM factors, we generated a lcr1 mutant and identified LCR1-dependent genes by RNA-seq analysis. Furthermore, by generating mutant strains of LCR1-dependent genes using the CRISPR–Cas9 method, we found that loss of CAH1 causes a decrease in Ci-affinity.
Results
Identification of LCR1-dependent inducible genes under CO2-limiting conditions
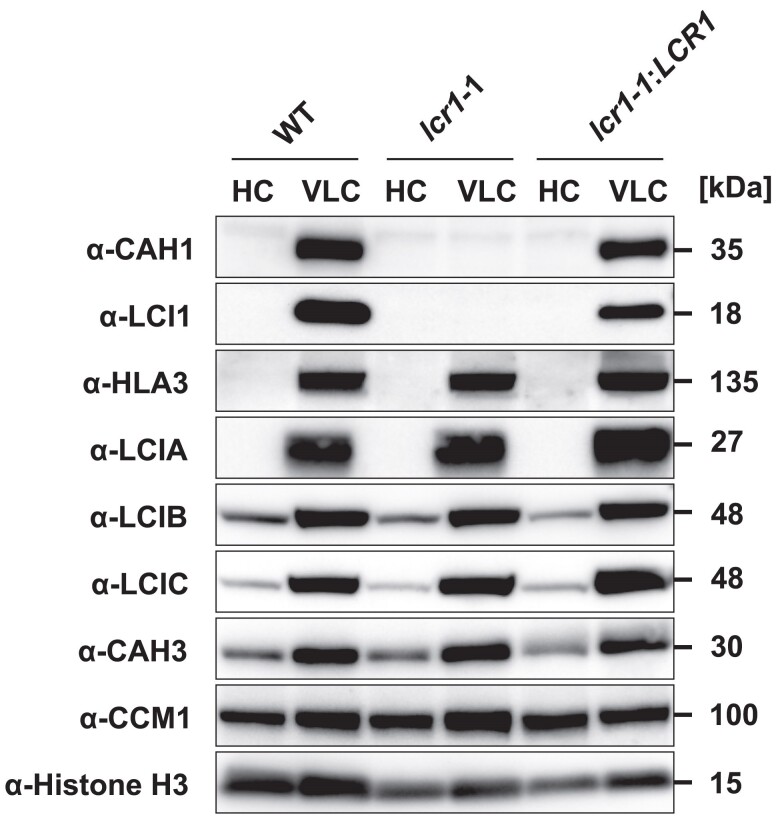
In our previous study (Yoshioka et al. 2004), we utilized the lcr1 insertion mutant derived from the parental strain Q304P3, where CAH1-promoter activity was monitored by arylsulfatase (Ars) enzyme activity (Kucho et al. 1999). However, due to the absence of a cell wall, this strain was unsuitable for physiological analysis. To address this, we generated a lcr1 mutant, named lcr1-1 in this study, derived from the WT strain C9 with a cell wall. To create the lcr1-1 mutant, we inserted the AphVII gene cassette, conferring hygromycin resistance, into the first exon of LCR1 using the CRISPR–Cas9 system (Supplementary Fig. S1A and B). The lcr1-1:LCR1 complemented strain was also created by reintroducing the LCR1 gene fragment, including its putative promoter, 5′-UTR, and 3′-UTR, into lcr1-1. In the lcr1-1:LCR1, the reduced accumulation levels of CAH1 and LCI1 observed in the lcr1-1 were rescued (Fig. 1). On the other hand, the accumulation levels of HLA3, LCIA, LCIB, LCIC, CAH3, and CCM1 did not change among the strains, consistent with previous findings that LCR1 specifically regulates CAH1 and LCI1 among CCM-related genes (Yoshioka et al. 2004).
Figure 1.
Accumulation of CCM-related proteins in the lcr1 mutant. Cells were first grown under 5% (v/v) CO2 condition for 24 h and shifted to 5% (v/v) CO2 (HC) or 0.04% (v/v) (VLC) CO2 conditions for 12 h at pH 7.0. Histone H3 was used as a loading control. WT, wild type.
Given the limitations of macroarray analysis in previous studies for quantifying all gene expression levels (Yoshioka et al. 2004), we explored whether LCR1 influences genes beyond CAH1, LCI1, and LCI6 under CO2-limiting conditions using RNA-seq analysis. We cultured WT, lcr1-1, and lcr1-1:LCR1 cells under 5% CO2 (high-CO2; HC) or 0.04% CO2 aerated (very-low CO2; VLC) conditions in MOPS-P liquid medium at pH 7.0 and quantified their transcriptome profiles. In WT cells, the expression levels of 1,647 genes were significantly increased at either 0.3 or 2 h after switching to VLC conditions compared to HC conditions [(FDR) < 0.01 and log2FC < −1] (Supplementary Data Set 1). Among them, under VLC conditions, the expression levels of 12 genes, including LCR1, CAH1, LCI1, and LCI6, were significantly decreased in lcr1-1 and recovered in lcr1-1:LCR1 (Table 1). Among these genes, Cre10.g426800, encoding a protein of unknown function with a transferase domain, was particularly notable as its expression level decreased more than 4-fold by LCR1 mutation both 0.3 and 2 h after VLC induction (Table 1). This led us to focus our subsequent analysis on CAH1, LCI1, LCI6, and Cre10.g426800.
Table 1.
Genes downregulated in the lcr1 mutant under VLC conditions
| Gene ID | Gene name | Description | VLC-0.3 h | VLC-2.0 h | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| lcr1-1/WT | lcr1-1/lcr1-1:LCR1 | lcr1-1/WT | lcr1-1/lcr1-1:LCR1 | |||||||
| log2FC | FDR | log2FC | FDR | log2FC | FDR | log2FC | FDR | |||
| Cre02.g095065 | 0.98 | 7.1.E–02 | −0.54 | 7.8.E–01 | −1.20 | 6.1.E–03 | −1.78 | 1.9.E–04 | ||
| Cre02.g095067 | 0.47 | 2.5.E–01 | −0.52 | 7.1.E–01 | −1.26 | 4.3.E–04 | −1.70 | 3.6.E–05 | ||
| Cre03.g162800 | LCI1 | Low-CO2-inducible membrane protein | −0.30 | 6.4.E–01 | −0.80 | 7.0.E–01 | −1.98 | 5.2.E–04 | −2.03 | 3.1.E–03 |
| Cre04.g223100 | CAH1 | Carbonic anhydrase | −3.49 | 5.9.E–27 | −2.24 | 1.9.E–10 | −2.86 | 3.0.E–19 | −2.30 | 3.8.E–11 |
| Cre06.g278137 | −1.90 | 2.4.E–06 | −0.05 | 9.9.E–01 | −3.41 | 8.9.E–21 | −1.85 | 1.0.E-04 | ||
| Cre08.g364050 | −0.10 | 7.6.E–01 | −1.01 | 1.6.E–02 | −1.04 | 9.2.E–05 | −1.75 | 2.8.E–10 | ||
| Cre08.g381450 | OPR35 | OctotricoPeptide Repeat Protein | −1.71 | 8.7.E–10 | −1.23 | 2.6.E–03 | −2.62 | 1.3.E–21 | −1.47 | 8.5.E–06 |
| Cre09.g399552 | LCR1 | Myb-like transcription factor | −2.54 | 4.4.E–05 | −1.74 | 1.9.E–01 | −2.55 | 6.3.E-05 | −2.20 | 4.3.E-03 |
| Cre10.g426800 | −2.37 | 2.9.E–20 | −2.12 | 2.3.E–14 | −2.79 | 1.6.E–27 | −2.20 | 7.4.E-16 | ||
| Cre10.g448200 | ARL9 | ARF-like GTPase | −1.84 | 1.3.E–09 | −0.45 | 7.3.E–01 | −2.19 | 1.3.E–13 | −1.14 | 3.0.E–03 |
| Cre12.g553350 | LCI6 | Low-CO2-inducible protein 6 | 0.69 | 5.9.E–02 | −0.96 | 1.9.E–01 | −1.64 | 9.5.E–07 | −1.43 | 4.9.E–04 |
| Cre16.g684022 | −2.68 | 2.8.E–20 | −1.86 | 6.4.E–08 | −2.34 | 1.1.E–15 | −1.20 | 1.6.E–03 | ||
Differentially expressed genes in lcr1-1 cells, with false discovery rate < 0.01 and log2FC < −1, in 0.04% CO2 aerated conditions for 0.3 or 2 h were indicated. WT, wild type; VLC, very low-CO2.
Impact of CAH1 and LCI1 mutations on Ci-affinity in Chlamydomonas cells
To clarify the contribution of LCR1-dependent genes to CCM, we employed the CRISPR–Cas9 method to generate mutants of CAH1, LCI1, LCI6, and Cre10.g426800. First, we created insertional mutants of LCI6 and Cre10.g426800 and measured their photosynthetic O2-evolution rates. Three strains with an insertional mutation in the first exon of LCI6 were isolated. Additionally, two strains of Cre10.g426800 were isolated: one with a mutation in the first exon and the other in the second exon (Supplementary Fig. S2, A and B). To evaluate Ci-affinity in these mutants, we measured their O2-evolving activity. The selected pH conditions of 6.2, 7.0, and 7.8 represent a range that encompasses typical environmental variations, allowing us to assess the mutants' responses under diverse but relevant scenarios. In these mutants, the K0.5 (Ci) values, the Ci concentrations required for half-maximal O2-evolving rate, were measured at pH 7.8, where CCM phenotypes are most pronounced. The mutants showed no increase in K0.5 (Ci) compared to the WT, unlike in the lcr1-1 strain (Supplementary Table S1). Furthermore, complementation of lcr1 with these genes rescued the Ci-affinity to WT levels. These results suggest that these two genes were not important for maintaining high Ci-affinity under CCM-inducing conditions.
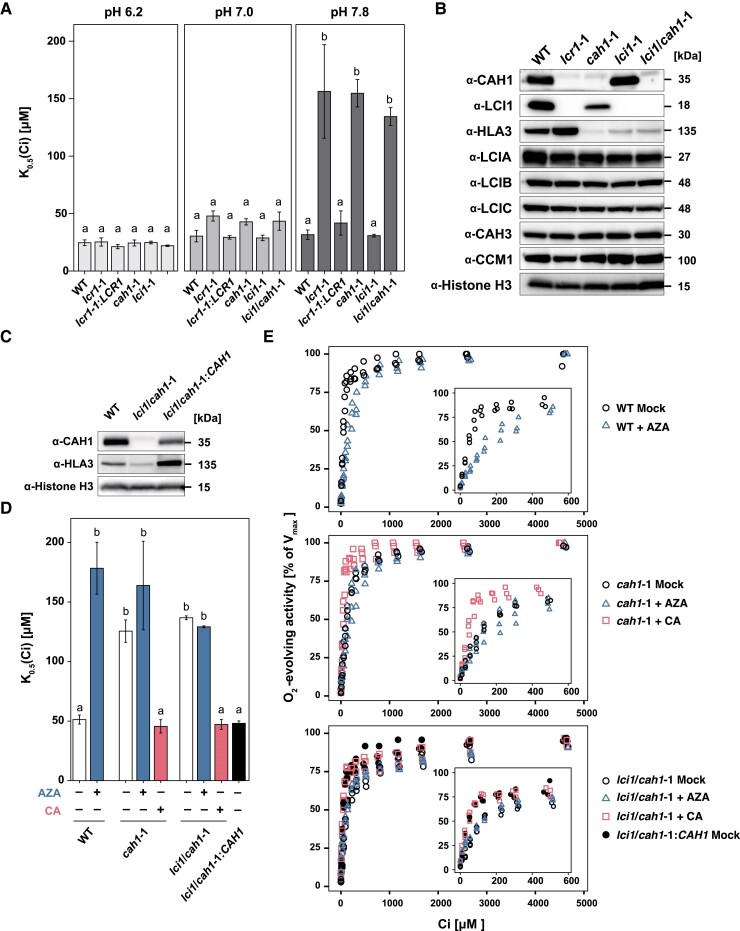
Next, we isolated mutants of CAH1 and LCI1, designated cah1-1 and lci1-1, respectively, and also produced a double mutant (lci1/cah1-1) by disrupting the CAH1 in the lci1-1 background (Supplementary Fig. S3, A and B). The K0.5 (Ci) values of the mutants were similar to those of the WT at pH 6.2 and 7.0, with significant differences emerging only at pH 7.8 (P < 0.05). At this pH, the K0.5 (Ci) value of lcr1-1 was notably higher than WT, more than 4-fold, indicating a reduced Ci-affinity (Fig. 2A; Supplementary Table S1). Interestingly, the cah1-1 mutant showed similar K0.5 (Ci) values to lcr1-1, while the K0.5 (Ci) value of lci1-1 did not significantly differ from WT (Supplementary Fig. S4). Additionally, the lci1/cah1-1 double mutant exhibited K0.5 (Ci) values comparable to cah1-1 and lcr1-1, highlighting a critical role for CAH1 in maintaining Ci affinity under a high HCO3−/CO2 ratio. Unexpectedly, we observed a reduction in HLA3 accumulation in cah1-1, lci1-1, and lci1/cah1-1 (Fig. 2B). This reduction in HLA3 accumulation complicates our understanding of the roles of CAH1 and HLA3 in maintaining Ci-affinity. It raises the question of whether the observed decrease in Ci-affinity in cah1-1 is solely due to CAH1 loss or if it might also involve a synergistic effect resulting from the simultaneous reduction of both CAH1 and HLA3.
Figure 2.
Physiological characteristics of lcr1, cah1, lci1, and lci1/cah1 mutants. A)K0.5 (Ci) values of cah1-1, lci1-1 and lci1/cah1-1 cells at pH 6.2, 7.0, and 7.8. Cells were grown in 0.04% (v/v) CO2 conditions for 12 h at pH 7.0. Data from all experiments show mean values ± standard error (Se) from three biological replicates. Statistical analysis was conducted using the Tukey–Kramer multiple comparison test, with different letters indicating significant differences (P < 0.05). B) Accumulation of CCM-related proteins in WT, lcr1-1, cah1-1, lci1-1 and lci1/cah1-1 cells grown under 0.04% (v/v) CO2 conditions for 12 h at pH 7.0. Histone H3 was used as a loading control. C) Accumulation of CAH1 and HLA3 in WT, lci1/cah1-1 and lci1/cah1-1:CAH1 cells under 0.04% (v/v) CO2 condition at pH 7.0. Histone H3 was used as a loading control. D)K0.5 (Ci) values in WT, cah1-1, lci1/cah1-1, and lci1/cah1-1:CAH1 cells at pH 7.8. Cells were grown in 0.04% (v/v) CO2 conditions for 12 h at pH 7.0. Data from all experiments show mean values ± Se from three biological replicates. Values for cells treated with AZA or bovine CA are shown in blue and red bars, respectively. The Tukey–Kramer multiple comparison test was utilized for statistical analysis, with differing letters indicating statistically significant variations (P < 0.05). E) Oxygen-evolving activity of WT, cah1-1, lci1/cah1-1, and lci1/cah1-1:CAH1 cells from three biological replicates treated with AZA or bovine CA in response to external dissolved Ci concentrations at pH 7.8 for the ranges of 0 to 5,000 µm Ci and 0 to 600 µm Ci (inset). Before measurements, cells were grown in the liquid culture aerated with 0.04% CO2 for 12 h. Values in each cell with AZA or bovine CA are shown as blue triangle and red square plots, respectively.
Acetazolamide's influence on CAH1-mediated Ci-affinity
To further elucidate CAH1's specific contribution to Ci-affinity and separate its effects from those of HLA3, we investigated the response of cells treated with AZA, a CA inhibitor with low membrane permeability, at pH 7.8. Additionally, we generated a lci1/cah1-1:CAH1 strain by introducing a gene fragment of CAH1, including its putative promoter, 5′-UTR and 3′-UTR, into lci1/cah1-1 for comparison. In lci1/cah1-1:CAH1, the accumulation levels of CAH1 and HLA3 were increased compared to lci1/cah1-1 (Fig. 2C). The addition of AZA to WT cells resulted in an increased K0.5 (Ci) value, aligning with the levels observed in cah1-1 and lci1/cah1-1 mutants (Fig. 2, D and E; Supplementary Table S2). Conversely, when cah1-1 and lci1/cah1-1 mutants were supplemented with bovine CA, their K0.5 (Ci) value was reduced to WT levels, suggesting that exogenous CA activity can compensate for the loss of CAH1 function by replenishing CO2 in the equilibrium. On the other hand, the addition of AZA to cah1-1 and lci1/cah1-1 cells did not cause a further significant increase in K0.5 (Ci) values. Furthermore, K0.5 (Ci) values in lci1-1/cah1:CAH1 were similar to WT. In combination with previous studies showing a very minor contribution of HLA3 to the maintenance of Ci affinity at pH 7.8 (Yamano et al. 2015), our results demonstrate that the alteration in Ci-affinity observed in cah1-1 and lci1/cah1-1 is primarily attributed to the loss of CAH1 activity. These results affirm the critical function of periplasmic CAH1 in CCM by maintaining high-Ci affinity under CO2-limiting conditions.
Effect of CAH1 mutation on growth rate
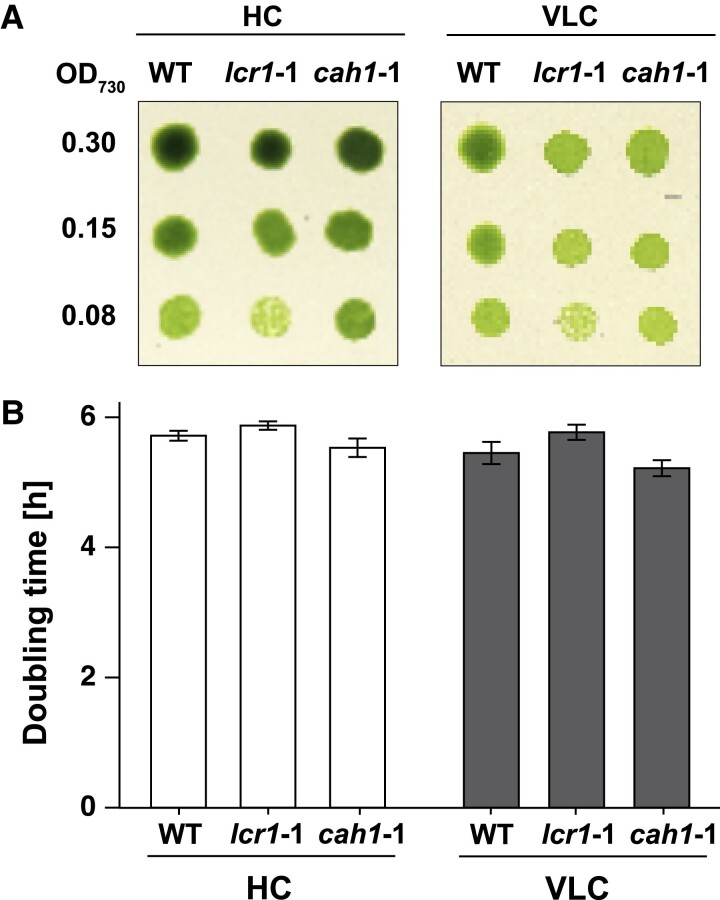
To further examine the impact of CAH1 deficiency, we evaluated the growth rates of WT, lcr1-1, and cah1-1 cells. Despite the significant role of CAH1 in maintaining Ci-affinity, no differences in growth rate among these strains were observed under both HC and VLC conditions, as evidenced by spot tests on agar plates at pH 7.8 (Fig. 3A). Additionally, the doubling times for these strains in a liquid medium were comparable (Fig. 3B). These findings suggest that, while CAH1 is crucial for maintaining Ci-affinity, its absence does not impede the overall growth rate under CO2-limiting conditions.
Figure 3.
The growth of lcr1 and cah1 mutants. A) Spot test of WT, lcr1-1, and cah1-1. Cells were diluted to the indicated optical density (OD700 = 0.30, 0.15, or 0.08). Subsequently, 3 μL of the cell suspensions were spotted on agar plates with pH 7.8. The plates were incubated for 4 d under 5% [v/v] CO2 (HC) or 0.01% [v/v] CO2 (VLC) conditions with continuous light at 120 μmol photons m−2 s−1. B) Doubling time of WT, lcr1-1 and cah1-1 cells were calculated from three independent experiments. Each cell was cultured in a 5% CO2 (HC) or 0.04 CO2 (VLC) aeration. Error bars represent standard error (Se).
Discussion
In this study, we assessed the Ci-affinity of LCR1-dependent gene mutants created using the CRISPR–Cas9 system across various pH conditions, ranging from acidic to alkaline, to understand their behavior under different environmental scenarios. Notably, at pH 7.8, a condition representative of high HCO3−/CO2 ratios, the cah1-1 mutant exhibited a significant decrease in Ci-affinity, highlighting the pivotal role of CAH1 in Chlamydomonas cells.
The function of LCR1 in various environmental stresses
LCR1 is instrumental in the activation of the CCM under CO2-limiting conditions, notably regulating the expression of CAH1 and LCI1 (Yoshioka et al. 2004). Conversely, under high-light conditions, LCR1 is critical for the expression of LHCSR3, essential for photoprotection (Arend et al. 2023). However, our study revealed that LCR1 did not regulate LHCSR3 expression under CO2-limiting conditions (Table 1), demonstrating that the genes controlled by LCR1 vary with environmental context. Our finding that LCR1 does not regulate LHCSR3 under CO2-limiting conditions builds upon our previous work (Yamano et al. 2008), which demonstrated the complex regulation of LHCSR3 (formerly known as Li818r-1 and Li818r-3). In that study, we showed that LHCSR3 is induced by high-light in a CCM1-independent manner, while under CO2-limiting conditions, its expression is CCM1-dependent. The current results, showing that LCR1 (a downstream factor of CCM1) does not regulate LHCSR3 under CO2-limiting conditions, suggest a more intricate regulatory network. This implies that while CCM1 is involved in LHCSR3 regulation under CO2-limiting conditions, it likely acts through factors other than LCR1.
To further elucidate the regulatory mechanism of LHCSR3 expression and the roles of CCM1 and LCR1 in this process, future studies should investigate the expression patterns of LHCSR3 under various combinations of light intensity and CO2 availability. Additionally, identifying transcription factors that mediate CCM1-dependent LHCSR3 expression under CO2-limiting conditions would be crucial. Exploring potential interactions between LCR1 and other transcription factors involved in LHCSR3 regulation could also provide valuable insights. These investigations could reveal how Chlamydomonas fine-tunes its gene expression in response to complex environmental changes, particularly in the context of carbon concentration mechanisms and photoprotection. Such findings underscore the importance of phenotypic analysis under various environmental conditions. Further insights into the diverse functions of transcription factors are expected from the recent large-scale systematic analysis (Fauser et al. 2022), which examines mutant phenotypes under various environmental growth conditions and chemical treatments.
CAH1 facilitates indirect HCO3− utilization under alkaline conditions
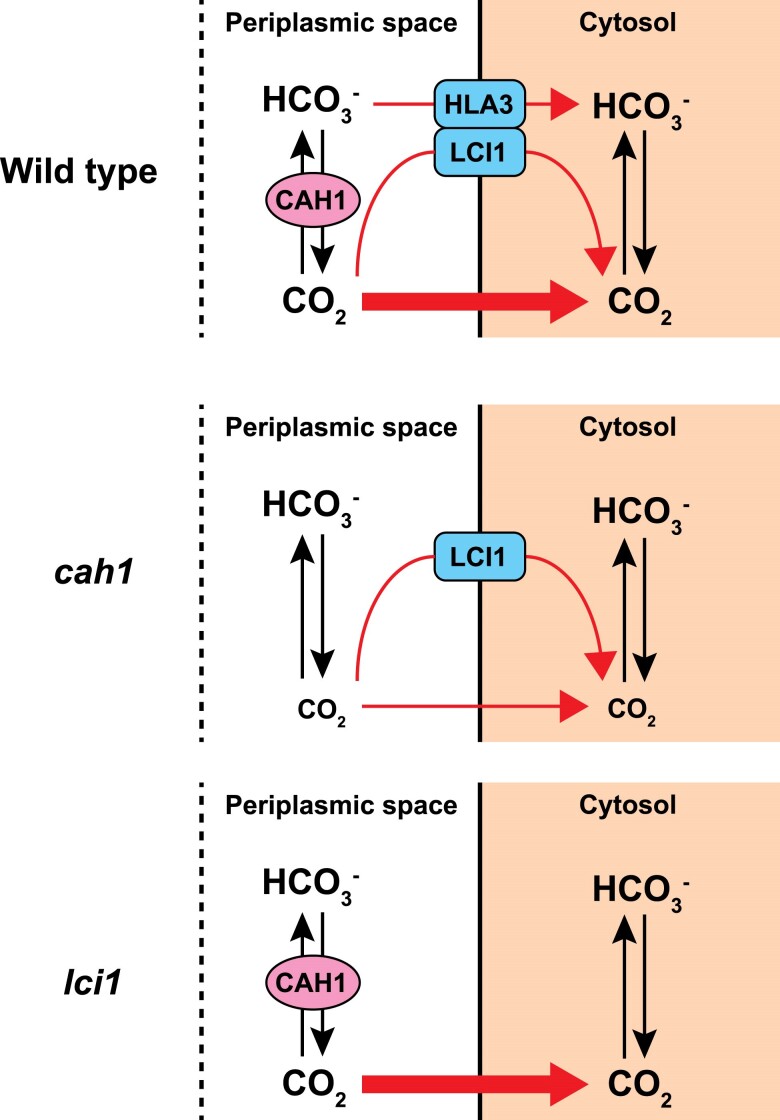
We demonstrated that CAH1 is crucial for maintaining high Ci-affinity in Chlamydomonas WT cells under alkaline conditions (pH 7.8), supporting the contribution of CAH1 to indirect utilization of abundant HCO3− (Fig. 4). This aligns with previous reports indicating enhanced transcription, protein accumulation, and CA activity of CAH1 at higher pH levels (Fett and Coleman 1994). An earlier study did not reveal significant differences in Ci-affinity between WT and cah1 mutants (Van and Spalding 1999), possibly due to the measurements performed at neutral pH. In addition, our research utilized a consistent parental strain for cah1 mutants, ensuring a more accurate evaluation of CAH1's impact.
Figure 4.
Models for Ci uptake pathway in WT, cah1 and lci1 mutants. Tentative models show how WT, cah1, and lci1 mutants uptake Ci across the plasma membrane under CO2-limiting conditions and at pH 7.8. Black arrows indicate the interconversion between CO2 and HCO3−. Red arrows show the Ci uptake pathway from the periplasmic space into the cytosol.
Whole genome sequencing of various Chlamydomonas laboratory strains has revealed genetic diversity among these strains (Gallaher et al. 2015). Furthermore, we have previously reported results suggesting that WT strains can acquire characteristics during long-term subculturing (Tsuji et al. 2023). These findings underscore the potential for genetic drift and the accumulation of spontaneous mutations in laboratory strains over time. As demonstrated in our previous studies (Toyokawa et al. 2020; Tsuji et al. 2023), this study reaffirms the importance of using mutants generated from the same parental strain for accurate phenotypic analysis in Chlamydomonas reverse genetics. By using the C9 strain as the common background for all our mutants, we minimize the confounding effects of strain-specific genetic variations, ensuring more reliable and reproducible results.
In Chlamydomonas, periplasmic CA was identified about four decades ago (Kimpel et al. 1983), and physiological experiments using weakly permeable sulfonamide inhibitors established the well-known model that periplasmic CA facilitates the indirect utilization of bulk HCO3− (Moroney et al. 1985, Aizawa and Miyachi 1986). Although the contradictory result in the previous analysis of cah1 mutant (Van and Spalding 1999) had raised controversy about the function of periplasmic CA, we demonstrated the importance of CAH1 at alkaline conditions, strengthening the original hypothesis that periplasmic CA supplies CO2 from HCO3−. Regarding catalytic direction (hydration or dehydration), there is an opposing hypothesis based on a mathematical modeling, in which periplasmic CA recaptures CO2 leaked from the cell through the hydration (Fridlyand 1997). However, this hypothesis is unlikely in Chlamydomonas because (i) analysis using membrane inlet mass spectrometry (MIMS) detected net CO2 uptake, but not CO2 efflux, by the cell when external CA was inhibited or removed (Shiraiwa et al. 1993; Sültemeyer et al. 1989), and (ii) light-dependent alkalization of medium was observed (Shiraiwa et al. 1993). These physiological measurements were performed at pH 8.0, which is similar to the conditions (pH 7.8) where our cah1 mutant displayed lower Ci-affinity than the parental strain (Fig. 2A). Importance of periplasmic CA was also suggested by the pronounced inhibitory effect of weakly permeable sulfonamide inhibitor at alkaline pH (pH 8.0) (Moroney et al. 1985). Thus, the long-standing discrepancy between physiological and genetic evidence has been solved, and both approaches provide consistent support for the original model that periplasmic CA enhances indirect HCO3− utilization by accelerating dehydration. Besides Chlamydomonas, the enhanced CO2 uptake by periplasmic CA-mediated dehydration is supported in various algal species. In the relatively distant green alga Chlorella, physiological studies have provided evidence for the role of periplasmic CA (Matsuda et al. 1999). Furthermore, the enhanced CO2 uptake by periplasmic CA-mediated dehydration is also supported in some marine diatoms such as T. pseudonana and O. sinensis by kinetic analysis of CO2 uptake using MIMS and direct measurement of cell surface pH changes, respectively (Hopkinson et al. 2013; Chrachri et al. 2018), suggesting the generality of the classical model in diverse algal groups. Notably, CAH1 in Chlamydomonas is α-type while diatoms have δ- and ζ-type in the periplasmic space (Samukawa et al. 2014), suggesting the convergent evolution of the CCM in different lineages as previously discussed (Matsuda et al. 2017).
Multiple strategies of Ci-uptake in Chlamydomonas
In Chlamydomonas, Ci uptake across the plasma membrane involves multiple transport strategies. These include direct pathways of CO2 through LCI1 and HCO3− through HLA3, respectively, along with indirect pathways involving CAH1 (Fig. 4). Despite no significant reduction in Ci affinity in lci1-1 mutants (Fig. 2A;Supplementary Table S1), LCI1's cooperative role with other channels cannot be ruled out. This unexpected result suggests a complex CO2 uptake mechanism in Chlamydomonas. It is possible that unidentified CO2 channels or transporters may compensate for the loss of LCI1. Additionally, functional redundancy in the Ci uptake system might allow other pathways to compensate for the deficiency of a single gene. Further analysis, such as creating multiple gene knockout mutants, could help elucidate the intricate nature of this Ci uptake system and the specific role of LCI1 within it. Additionally, post-translational modifications of LCI1 could play a crucial role in its function or regulation. Future studies investigating these aspects, including the identification of potential LCI1 interacting partners and analysis of its post-translational modifications, will be essential to fully understand the role of LCI1 in the CO2 uptake mechanism of Chlamydomonas.
Interestingly, the accumulation level of HLA3 was reduced in cah1-1 and lci1-1 (Fig. 4). The complete loss of CAH1 and LCI1 may have caused this phenotype, as HLA3 accumulation was not altered in lcr1-1, which still expresses low levels of CAH1 and LCI1 (Figs 1 and 2B; Supplementary Data Set 1). Since HLA3 and LCI1 interact and form a complex on the plasma membrane (Mackinder et al. 2017), it is possible that the formation of the HLA3–LCI1 complex was inhibited in lci1-1. Furthermore, the expression of HLA3 is closely tied to that of LCIA, a HCO3− transporter located on the chloroplast envelope (Yamano et al. 2015). This multitiered control of HLA3 expression by various factors, including CAH1, LCI1, and LCIA, suggests a complex regulatory network governing Ci uptake. To further unravel the complexities of this regulatory network, future studies should investigate the physical interaction between CAH1 and HLA3, as well as the impact of LCIA, LCI1, and CAH1 deficiency on HLA3 expression.
Notably, from our findings that cah1-1 mutants displayed a substantially higher K0.5 (Ci) value compared to WT under pH 7.8 conditions (Fig. 2A;Supplementary Table S1), emphasizing CAH1's primary role in Ci uptake into chloroplasts at this pH conditions (Fig. 4). Despite the reduced Ci-affinity, the absence of growth rate differences among lcr1-1, cah1-1, and WT under CO2-limiting conditions (Fig. 3, A and B) suggests that the CO2 concentrations used in our growth experiments were still sufficient to support normal growth in the mutants. This observation raises the possibility that the growth conditions used in this study may not have been optimal for detecting the effects of CAH1 loss on growth rate. Future studies exploring a wider range of CO2 concentrations and pH conditions are needed to fully elucidate the impact of CAH1 deficiency on growth under varying environmental conditions.
Diversity of periplasmic CA functions in Chlamydomonas
Our findings reveal that AZA significantly reduced Ci-affinity in cells, aligning with previous research (Moroney et al. 1985). Besides CAH1, CAH2 and CAH8 are also located in the periplasmic space (Moroney et al. 2011). Although CAH2 shares a similar amino acid sequence with CAH1, its expression is induced under high CO2 conditions, differing from CAH1 (Fujiwara et al. 1990). CAH8, a β-type CA with a transmembrane domain, is positioned closer to the plasma membrane than CAH1 under varying CO2 conditions (Ynalvez et al. 2008). The comparable Ci-affinity in AZA-treated WT cells and cah1 mutant underscores CAH1's greater role in Ci uptake under CO2-limiting conditions compared to CAH2 and CAH8. Future studies focusing on the regulation of these periplasmic CAs and their compensatory interactions are essential, potentially informing bioengineering approaches to enhance microalgae photosynthesis.
Materials and methods
Chlamydomonas (C. reinhardtii) strains and cultural conditions
The WT strain C9, obtained from the IAM Culture Collection at the University of Tokyo, was utilized for physiological and biochemical experiments. Strain C9 is now available from the Microbial Culture Collection at the National Institute for Environmental Studies, Japan, as strain NIES-2235 (alternatively named CC-5098 in the Chlamydomonas Resource Center). The cells were precultured in a TAP medium and subsequently resuspended in 50 mL of MOPS-P medium. They were grown under a 5% (v/v) CO2 atmosphere with a light intensity set at 120 μmol photons m−2 s−1, following the method described by Toyokawa et al. (2020), until they reached the mid-logarithmic phase of growth. For the induction of VLC conditions, cells acclimated to high-CO2 conditions were centrifuged, resuspended in fresh MOPS-P medium, and then cultured with air bubbling containing 0.04% (v/v) CO2 at the same light intensity.
Measurement of photosynthetic O2-evolving activity
Cells were harvested and resuspended in Ci-depleted 20 mm MES–NaOH (pH 6.2), MOPS–NaOH (pH 7.0), or HEPES–NaOH (pH 7.8) buffers, adjusting the density to 10 to 20 μg chlorophyll per milliliter. The photosynthetic oxygen evolution rate was then measured using a Clark-type oxygen electrode (Hansatech Instruments) as described previously (Yamano et al. 2008). AZA adjusted to a concentration of 5 mm and dissolved in DMSO, was added to the measuring buffer at a 1% [v/v]. Bovine CA was added into the buffer, achieving a concentration of 2.0 μg mL–1. For comparison, 1% DMSO was introduced to samples without AZA.
Generation of mutants by the CRISPR–Cas9 system
For CRISPR–Cas9-mediated genome editing, guide RNAs were designed using the CRISPOR tool (Concordet and Haeussler 2018), as detailed in Supplementary Figs. S1 to S3. The introduction of the ribonucleoprotein complex and the AphVII or AphVIII cassette into cells followed the method of Tsuji et al. (2023). Primer sets used for screening are shown in Supplementary Figs. S1 to S3, and their sequences are listed in Supplementary Table S3.
Immunoblotting analysis
Total protein extraction, SDS–polyacrylamide gel electrophoresis (SDS/PAGE), and immunoblotting analyses were carried out as previously described (Wang et al. 2016). Primary antibodies were utilized at the following indicated dilutions: anti-HLA3 at 1:1,250, anti-LCIA at 1:5,000, anti-LCI1 at 1:5,000, anti-LCIB at 1:5,000, anti-CAH1 at 1:2,500, anti-CAH3 at 1:2,000, anti-CCM1 at 1:2,500, and anti-Histone H3 at 1:10,000. A horseradish peroxidase-conjugated goat anti-rabbit IgG antibody from Life Technologies was employed as the secondary antibody at a dilution of 1:10,000 to detect the primary antibodies.
RNA-seq analysis
Total RNA was extracted from cells using the RNeasy Plant Mini Kit (QIAGEN), following the manufacturer's instructions. After RNA purification, the total RNA was analyzed using the Illumina Novaseq 6000 system. In each condition, sequencing data were obtained from two biological replicates. The resulting reads were aligned with version 5.6 of the C. reinhardtii genome annotation, which was downloaded from https://phytozome-next.jgi.doe.gov/. The alignment, counting of reads, and normalization of read counts were performed according to the methods previously described in Shimamura et al. (2023).
Accession numbers
The accession numbers of the Phytozome database for Chlamydomonas genes LCR1, CAH1, LCI1, and LCI6 are Cre09.g399552, Cre04.g223100, Cre03.g162800, and Cre12.g553350, respectively.
Supplementary Material
Acknowledgments
We thank Yoriko Matsuda for the technical assistance with mutant isolation.
Contributor Information
Daisuke Shimamura, Graduate School of Biostudies, Kyoto University, Kyoto 606-8502, Japan; RIKEN Center for Sustainable Resource Science, Yokohama 230-0045, Japan.
Tomoaki Ikeuchi, Graduate School of Biostudies, Kyoto University, Kyoto 606-8502, Japan.
Ami Matsuda, Graduate School of Biostudies, Kyoto University, Kyoto 606-8502, Japan.
Yoshinori Tsuji, Graduate School of Biostudies, Kyoto University, Kyoto 606-8502, Japan; Department of Bioscience, School of Biological and Environmental Sciences, Kwansei Gakuin University, Hyogo 669-1330, Japan.
Hideya Fukuzawa, Graduate School of Biostudies, Kyoto University, Kyoto 606-8502, Japan.
Keiichi Mochida, RIKEN Center for Sustainable Resource Science, Yokohama 230-0045, Japan.
Takashi Yamano, Graduate School of Biostudies, Kyoto University, Kyoto 606-8502, Japan; Center for Living Systems Information Science (CeLiSIS), Kyoto University, Kyoto 606-8501, Japan.
Author contribitions
T.Y. and H.F. conceived and designed the study; D.S. performed most of the experiments; T.I. and A.M. contributed to mutant isolation; K.M. provided additional supervision and resources; D.S., Y.T., and T.Y. wrote the article, and all authors approved it. T.Y. agreed to serve as the author responsible for contact and ensure communication.
Supplementary data
The following materials are available in the online version of this article.
Supplementary Figure S1. The lcr1 mutant generated by the CRISPR–Cas9 system.
Supplementary Figure S2. The mutants of lci6 and Cre10.g426800 generated by the CRISPR–Cas9 system.
Supplementary Figure S3. The mutants of cah1 and lci1 generated by the CRISPR–Cas9 system.
Supplementary Figure S4. Oxygen-evolving activity of WT and transformant cells in response to external dissolved Ci concentrations.
Supplementary Table S1. Photosynthetic parameters of WT and transformant cells.
Supplementary Table S2. Effect of AZA and bovine CA on photosynthetic parameters of WT and cah1-1 cells.
Supplementary Table S3. Sequences of primers used in this study.
Funding
This work was supported by the Japan Society for the Promotion of Science (Grants Numbers JP20H03073, JP21K19145, JP24K01851 to T.Y.), JST SPRING JPMJSP2110 (to D.S.), GteX Program Japan Grant Number JPMJGX23B0 (to T.Y.), JP16H06279 (PAGS), and the Asahi Glass Foundation (to T.Y.)
Data availability
Data deposition: The RNA-seq raw data in this paper have been deposited in the DNA Data Bank of Japan (DDBJ) Sequence Read Archive (DRA) (accession no. DRA017670).
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
References
- Aizawa K, Miyachi S. Carbonic anhydrase and CO2 concentrating mechanisms in microalgae and cyanobacteria. FEMS Microbiol Rev. 1986:2(3):215–233. 10.1111/j.1574-6968.1986.tb01860.x [DOI] [Google Scholar]
- Arend M, Yuan Y, Ruiz-Sola MÁ, Omranian N, Nikoloski Z, Petroutsos D. Widening the landscape of transcriptional regulation of green algal photoprotection. Nat Commun. 2023:14(1):2687. 10.1038/s41467-023-38183-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aspatwar A, Tolvanen MEE, Barker H, Syrjänen L, Valanne S, Purmonen S, Waheed A, Sly WS, Parkkila S. Carbonic anhydrases in metazoan model organisms: molecules, mechanisms, and physiology. Physiol Rev. 2022:102(3):1327–1383. 10.1152/physrev.00018.2021 [DOI] [PubMed] [Google Scholar]
- Badger M. The roles of carbonic anhydrases in photosynthetic CO2 concentrating mechanisms. Photosynth Res. 2003:77(2/3):83–94. 10.1023/A:1025821717773 [DOI] [PubMed] [Google Scholar]
- Chegwidden WR, Carter ND. Introduction to the carbonic anhydrases. In: Chegwidden WR, Carter ND, Edwards YH, editors. The carbonic anhydrases: new horizons. Basel: Birkhäuser; 2000. p. 13–28. [Google Scholar]
- Chrachri A, Hopkinson BM, Flynn K, Brownlee C, Wheeler GL. Dynamic changes in carbonate chemistry in the microenvironment around single marine phytoplankton cells. Nat Commun. 2018:9(1):74. 10.1038/s41467-017-02426-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman JR, Berry JA, Togasaki RK, Grossman AR. Identification of extracellular carbonic anhydrase of Chlamydomonas reinhardtii 1. Plant Physiol. 1984:76(2):472–477. 10.1104/pp.76.2.472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Concordet J-P, Haeussler M. CRISPOR: intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res. 2018:46(W1):W242–W245. 10.1093/nar/gky354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elzenga JTM, Prins HBA, Stefels J. The role of extracellular carbonic anhydrase activity in inorganic carbon utilization of Phaeocystis globosa (Prymnesiophyceae): a comparison with other marine algae using the isotopic disequilibrium technique. Limnol Oceanogr. 2000:45(2):372–380. 10.4319/lo.2000.45.2.0372 [DOI] [Google Scholar]
- Fauser F, Vilarrasa-Blasi J, Onishi M, Ramundo S, Patena W, Millican M, Osaki J, Philp C, Nemeth M, Salomé PA, et al. Systematic characterization of gene function in the photosynthetic alga Chlamydomonas reinhardtii. Nat Genet. 2022:54(5):705–714. 10.1038/s41588-022-01052-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fett JP, Coleman JR. Regulation of periplasmic carbonic anhydrase expression in Chlamydomonas reinhardtii by acetate and pH. Plant Physiol. 1994:106(1):103–108. 10.1104/pp.106.1.103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridlyand LE. Models of CO2 concentrating mechanisms in microalgae taking into account cell and chloroplast structure. Biosystems. 1997:44(1):41–57. 10.1016/S0303-2647(97)00042-7 [DOI] [PubMed] [Google Scholar]
- Fujiwara S, Fukuzawa H, Tachiki A, Miyachi S. Structure and differential expression of two genes encoding carbonic anhydrase in Chlamydomonas reinhardtii. Proc Natl Acad Sci U S A. 1990:87(24):9779–9783. 10.1073/pnas.87.24.9779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuzawa H, Fujiwara S, Yamamoto Y, Dionisio-Sese ML, Miyachi S. cDNA cloning, sequence, and expression of carbonic anhydrase in Chlamydomonas reinhardtii: regulation by environmental CO2 concentration. Proc Natl Acad Sci U S A. 1990:87(11):4383–4387. 10.1073/pnas.87.11.4383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuzawa H, Miura K, Ishizaki K, Kucho K, Saito T, Kohinata T, Ohyama K. Ccm1, a regulatory gene controlling the induction of a carbon-concentrating mechanism in Chlamydomonas reinhardtii by sensing CO2 availability. Proc Natl Acad Sci U S A. 2001:98(9):5347–5352. 10.1073/pnas.081593498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuzawa H, Suzuki E, Komukai Y, Miyachi S. A gene homologous to chloroplast carbonic anhydrase (icfA) is essential to photosynthetic carbon dioxide fixation by Synechococcus PCC7942. Proc Natl Acad Sci U S A. 1992:89(10):4437–4441. 10.1073/pnas.89.10.4437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funke RP, Kovar JL, Weeks DP. Intracellular carbonic anhydrase is essential to photosynthesis in Chlamydomonas reinhardtii at atmospheric levels of CO2. Demonstration via genomic complementation of the high-CO2-requiring mutant ca-1. Plant Physiol. 1997:114(1):237–244. 10.1104/pp.114.1.237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallaher SD, Fitz-Gibbon ST, Glaesener AG, Pellegrini M, Merchant SS. Chlamydomonas genome resource for laboratory strains reveals a mosaic of sequence variation, identifies true strain histories, and enables strain-specific studies. Plant Cell. 2015:27(9):2335–2352. 10.1105/tpc.15.00508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hines KM, Chaudhari V, Edgeworth KN, Owens TG, Hanson MR. Absence of carbonic anhydrase in chloroplasts affects C3 plant development but not photosynthesis. Proc Natl Acad Sci U S A. 2021:118(33):e2107425118. 10.1073/pnas.2107425118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkinson BM, Meile C, Shen C. Quantification of extracellular carbonic anhydrase activity in two marine diatoms and investigation of its role. Plant Physiol. 2013:162(2):1142–1152. 10.1104/pp.113.217737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson BS, Fong F, Heath RL. Carbonic anhydrase of spinach: studies on its location, inhibition, and physiological function. Plant Physiol. 1975:55(3):468–474. 10.1104/pp.55.3.468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan DB, Ogren WL. Species variation in the specificity of ribulose biphosphate carboxylase/oxygenase. Nature. 1981:291(5815):513–515. 10.1038/291513a0 [DOI] [Google Scholar]
- Karlsson J, Clarke AK, Chen ZY, Hugghins SY, Park YI, Husic HD, Moroney JV, Samuelsson G. A novel alpha-type carbonic anhydrase associated with the thylakoid membrane in Chlamydomonas reinhardtii is required for growth at ambient CO2. EMBO J. 1998:17(5):1208–1216. 10.1093/emboj/17.5.1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasili RW, Rai AK, Moroney JV. LCIB functions as a carbonic anhydrase: evidence from yeast and Arabidopsis carbonic anhydrase knockout mutants. Photosynth Res. 2023:156(2):193–204. 10.1007/s11120-023-01005-1 [DOI] [PubMed] [Google Scholar]
- Kimpel DL, Togasaki RK, Miyachi S. Carbonic anhydrase in Chlamydomonas reinhardtii I. Localization. Plant Cell Physiol. 1983:24(2):255–259. 10.1093/pcp/24.2.255 [DOI] [Google Scholar]
- Kono A, Chou T-H, Radhakrishnan A, Bolla JR, Sankar K, Shome S, Su C-C, Jernigan RL, Robinson CV, Yu EW, et al. Structure and function of LCI1: a plasma membrane CO2 channel in the Chlamydomonas CO2 concentrating mechanism. Plant J. 2020:102(6):1107–1126. 10.1111/tpj.14745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono A, Spalding MH. LCI1, a Chlamydomonas reinhardtii plasma membrane protein, functions in active CO2 uptake under low CO2. Plant J. 2020:102(6):1127–1141. 10.1111/tpj.14761 [DOI] [PubMed] [Google Scholar]
- Kucho K, Ohyama K, Fukuzawa H. CO2-responsive transcriptional regulation of CAH1 encoding carbonic anhydrase is mediated by enhancer and silencer regions in Chlamydomonas reinhardtii. Plant Physiol. 1999:121(4):1329–1338. 10.1104/pp.121.4.1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackinder LCM, Chen C, Leib RD, Patena W, Blum SR, Rodman M, Ramundo S, Adams CM, Jonikas MC. A spatial interactome reveals the protein organization of the algal CO2-concentrating mechanism. Cell. 2017:171(1):133–147.e14. 10.1016/j.cell.2017.08.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda Y, Hopkinson BM, Nakajima K, Dupont CL, Tsuji Y. Mechanisms of carbon dioxide acquisition and CO2 sensing in marine diatoms: a gateway to carbon metabolism. Philos Trans R Soc Lond B Biol Sci. 2017:372(1728):20160403. 10.1098/rstb.2016.0403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda Y, Williams TG, Colman B. Quantification of the rate of CO2 formation in the periplasmic space of microalgae during photosynthesis. A comparison of whole-cell rate constants for CO2 and HCO3– uptake among three species of the green alga Chlorella. Plant Cell Environ. 1999:22(4):397–405. 10.1046/j.1365-3040.1999.00399.x [DOI] [Google Scholar]
- Moroney JV, Husic HD, Tolbert NE. Effect of carbonic anhydrase inhibitors on inorganic carbon accumulation by Chlamydomonas reinhardtii. Plant Physiol. 1985:79(1):177–183. 10.1104/pp.79.1.177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroney JV, Ma Y, Frey WD, Fusilier KA, Pham TT, Simms TA, DiMario RJ, Yang J, Mukherjee B. The carbonic anhydrase isoforms of Chlamydomonas reinhardtii: intracellular location, expression, and physiological roles. Photosynth Res. 2011:109(1–3):133–149. 10.1007/s11120-011-9635-3 [DOI] [PubMed] [Google Scholar]
- Nimer NA, Brownlee C, Merrett MJ. Extracellular carbonic anhydrase facilitates carbon dioxide availability for photosynthesis in the marine dinoflagellate prorocentrum micans. Plant Physiol. 1999:120(1):105–112. 10.1104/pp.120.1.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi N, Mukherjee B, Tsujikawa T, Yanase M, Nakano H, Moroney JV, Fukuzawa H. Expression of a low CO₂-inducible protein, LCI1, increases inorganic carbon uptake in the green alga Chlamydomonas reinhardtii. Plant Cell. 2010:22(9):3105–3117. 10.1105/tpc.109.071811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raven JA, Giordano M, Beardall J, Maberly SC. Algal and aquatic plant carbon concentrating mechanisms in relation to environmental change. Photosynth Res. 2011:109(1–3):281–296. 10.1007/s11120-011-9632-6 [DOI] [PubMed] [Google Scholar]
- Samukawa M, Shen C, Hopkinson BM, Matsuda Y. Localization of putative carbonic anhydrases in the marine diatom, Thalassiosira pseudonana. Photosynth Res. 2014:121(2–3):235–249. 10.1007/s11120-014-9967-x [DOI] [PubMed] [Google Scholar]
- Shimamura D, Yamano T, Niikawa Y, Hu D, Fukuzawa H. A pyrenoid-localized protein SAGA1 is necessary for Ca2+-binding protein CAS-dependent expression of nuclear genes encoding inorganic carbon transporters in Chlamydomonas reinhardtii. Photosynth Res. 2023:156(2):181–192. 10.1007/s11120-022-00996-7 [DOI] [PubMed] [Google Scholar]
- Shiraiwa Y, Goyal A, Tolbert NE. Alkalization of the medium by unicellular green Algae during uptake dissolved inorganic carbon. Plant Cell Physiol. 1993:34(5):649–657. 10.1093/oxfordjournals.pcp.a078467 [DOI] [Google Scholar]
- Sinetova MA, Kupriyanova EV, Markelova AG, Allakhverdiev SI, Pronina NA. Identification and functional role of the carbonic anhydrase Cah3 in thylakoid membranes of pyrenoid of Chlamydomonas reinhardtii. Biochim Biophys Acta (BBA)—Bioenergetics. 2012:1817(8):1248–1255. 10.1016/j.bbabio.2012.02.014 [DOI] [PubMed] [Google Scholar]
- Sültemeyer DF, Miller AG, Espie GS, Fock HP, Canvin DT. Active CO2 transport by the green alga Chlamydomonas reinhardtii. Plant Physiol. 1989:89(4):1213–1219. 10.1104/pp.89.4.1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyokawa C, Yamano T, Fukuzawa H. Pyrenoid starch sheath is required for LCIB localization and the CO2-concentrating mechanism in green Algae. Plant Physiol. 2020:182(4):1883–1893. 10.1104/pp.19.01587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji Y, Kinoshita A, Tsukahara M, Ishikawa T, Shinkawa H, Yamano T, Fukuzawa H. A YAK1-type protein kinase, triacylglycerol accumulation regulator 1, in the green alga Chlamydomonas reinhardtii is a potential regulator of cell division and differentiation into gametes during photoautotrophic nitrogen deficiency. J Gen Appl Microbiol. 2023:69(1):1–10. 10.2323/jgam.2022.08.001 [DOI] [PubMed] [Google Scholar]
- Tsuji Y, Kusi-Appiah G, Kozai N, Fukuda Y, Yamano T, Fukuzawa H. Characterization of a CO2-concentrating mechanism with low sodium dependency in the centric diatom Chaetoceros gracilis. Mar Biotechnol (NY). 2021:23(3):456–462. 10.1007/s10126-021-10037-4 [DOI] [PubMed] [Google Scholar]
- Tsuji Y, Mahardika A, Matsuda Y. Evolutionarily distinct strategies for the acquisition of inorganic carbon from seawater in marine diatoms. J Exp Bot. 2017:68(14):3949–3958. 10.1093/jxb/erx102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van K, Spalding MH. Periplasmic carbonic anhydrase structural gene (Cah1) mutant in Chlamydomonas reinhardtii. Plant Physiol. 1999:120(3):757–764. 10.1104/pp.120.3.757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Yamano T, Takane S, Niikawa Y, Toyokawa C, Ozawa SI, Tokutsu R, Takahashi Y, Minagawa J, Kanesaki Y, et al. Chloroplast-mediated regulation of CO2-concentrating mechanism by Ca2+-binding protein CAS in the green alga Chlamydomonas reinhardtii. Proc Natl Acad Sci U S A. 2016:113(44):12586–12591. 10.1073/pnas.1606519113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Spalding MH. An inorganic carbon transport system responsible for acclimation specific to air levels of CO2 in Chlamydomonas reinhardtii. Proc Natl Acad Sci U S A. 2006:103(26):10110–10115. 10.1073/pnas.0603402103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams TG, Turpin DH. The role of external carbonic anhydrase in inorganic carbon acquisition by Chlamydomonas reinhardii at alkaline pH. Plant Physiol. 1987:83(1):92–96. 10.1104/pp.83.1.92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Zhang J, Weeks DP. The Cia5 gene controls formation of the carbon concentrating mechanism in Chlamydomonas reinhardtii. Proc Natl Acad Sci U S A. 2001:98(9):5341–5346. 10.1073/pnas.101534498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano T, Miura K, Fukuzawa H. Expression analysis of genes associated with the induction of the carbon-concentrating mechanism in Chlamydomonas reinhardtii. Plant Physiol. 2008:147(1):340–354. 10.1104/pp.107.114652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano T, Sato E, Iguchi H, Fukuda Y, Fukuzawa H. Characterization of cooperative bicarbonate uptake into chloroplast stroma in the green alga Chlamydomonas reinhardtii. Proc Natl Acad Sci U S A. 2015:112(23):7315–7320. 10.1073/pnas.1501659112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano T, Tsujikawa T, Hatano K, Ozawa S-I, Takahashi Y, Fukuzawa H. Light and low-CO2-dependent LCIB-LCIC complex localization in the chloroplast supports the carbon-concentrating mechanism in Chlamydomonas reinhardtii. Plant Cell Physiol. 2010:51(9):1453–1468. 10.1093/pcp/pcq105 [DOI] [PubMed] [Google Scholar]
- Ynalvez RA, Xiao Y, Ward AS, Cunnusamy K, Moroney JV. Identification and characterization of two closely related beta-carbonic anhydrases from Chlamydomonas reinhardtii. Physiol Plant. 2008:133(1):15–26. 10.1111/j.1399-3054.2007.01043.x [DOI] [PubMed] [Google Scholar]
- Yoshioka S, Taniguchi F, Miura K, Inoue T, Yamano T, Fukuzawa H. The novel Myb transcription factor LCR1 regulates the CO2-responsive gene Cah1, encoding a periplasmic carbonic anhydrase in Chlamydomonas reinhardtii. Plant Cell. 2004:16(6):1466–1477. 10.1105/tpc.021162 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data deposition: The RNA-seq raw data in this paper have been deposited in the DNA Data Bank of Japan (DDBJ) Sequence Read Archive (DRA) (accession no. DRA017670).