Abstract
Affinity cross-linking of the plasma membrane fraction to an 125I-labeled chitin oligosaccharide led to the identification and characterization of an 85-kD, chitin binding protein in plasma membrane-enriched fractions from both suspension-cultured soybean cells and root tissue. Inhibition analysis indicated a binding preference for larger (i.e. degrees of polymerization = 8) N-acetylated chitin molecules with a 50% inhibition of initial activity value of approximately 50 nm. N-Acetyl-glucosamine and chitobiose showed no inhibitory effects at concentrations as high as 250 μm. It is noteworthy that the major lipo-chitin oligosaccharide Nod signal produced by Bradyrhizobium japonicum was also shown to be a competitive inhibitor of ligand binding. However, the binding site appeared to recognize the chitin portion of the Nod signal, and it is unlikely that this binding activity represents a specific Nod signal receptor. Chitooligosaccharide specificity for induction of medium alkalinization and the generation of reactive oxygen in suspension-cultured cells paralleled the binding activity. Taken together, the presence of the chitin binding protein in the plasma membrane fraction and the specificity and induction of a biological response upon ligand binding suggest a role for the protein as an initial response mechanism for chitin perception in soybean (Glycine max).
Cell signaling and the perception of small phyto-active compounds comprise the basis for communication between plants and microbes in a number of widely studied systems. Small, diffusible compounds from both plants and microbes can initiate a wide range of biological responses in their respective counterpart. Chitin perception by plants in response to microbial invasion plays an integral role in cell signaling during pathogenesis (Wagner, 1994; Stacey and Shibuya, 1997). Modified chitin oligosaccharides, similarly, play a central role in the establishment of a host-specific symbiosis between legumes and their rhizobial symbionts (for review, see Cohn et al., 1998). Although much has been done to elucidate the numerous responses evoked upon ligand recognition, relatively little is known about how these signals are perceived by the host plant. However, it is apparent that chitin perception by diverse plant species shows some similarities (Stacey and Shibuya, 1997). For example, tomato, soybean (Glycine max), carrot, and rice have been shown to respond to the application of chitin oligosaccharides (De Jong et al., 1993; Ito et al., 1997; Felix et al., 1998). These responses include the induction of mRNA expression in soybean (Minami et al., 1996a), the depolarization of the rice plasma membrane (Kuchitsu et al., 1993), as well as an effect on carrot embryogenesis and re-initiation of meristematic cell division (De Jong et al., 1993). Thus, it is likely that most plants possess a common chitin recognition system.
Nodule formation in legumes involves the recognition of substituted lipo-chitin molecules produced by the invading rhizobium. Modified in a host-specific manner, these signal molecules, Nod signals, induce a number of physiological responses in the developing root of the legume host (Denarie et al., 1992; Long, 1996). In previous work (Minami et al., 1996a, 1996b), we examined the structure/function relationships of various chitin and lipo-chitin molecules on the expression of two soybean, early nodulin genes, ENOD40 and ENOD2, as a measure of chitin/lipo-chitin activity. It was surprising that our results showed that ENOD40 expression could be induced by chitopentaose. However, this expression appeared transient and was localized to the pericycle of the root stele (Minami et al., 1996a). In contrast, treatment of roots with the major lipo-chitin Nod signal produced by Bradyrhizobium japonicum, the soybean symbiont, resulted in sustained ENOD40 expression localized to both the pericycle and the developing nodule primordia. Minami et al. (1996b) subsequently showed that the induction of ENOD2 expression required the cooperative action of at least two structurally distinct signals, one of which had to be an active lipo-chitin molecule, but the other could be a chitin oligosaccharide. Taken together, these results argued for the involvement of two signal recognition events. One, somewhat non-specific, could recognize chitin oligomers resulting in the transient expression of ENOD40. However, the second recognition event required the specific B. japonicum lipo-chitin Nod signal. When both recognition systems were activated, their cooperative effect was evidenced by the induction of ENOD2 expression. Results from other laboratories, using different plants and experimental systems, support the idea of two chitin-signaling events involved in nodule initiation (Ardourel et al., 1994; Geurts and Franssen, 1996; Felle et al., 2000).
The impetus for the study described here was the identification of a chitin recognition site that could mediate the first response described above. Using an affinity cross-linking method, we identified an 85-kD protein in plasma membrane-enriched fractions from both soybean suspension-cultured cells and root tissue. The selectivity of chitin oligomer binding to membrane fractions mimicked that previously seen in rice (Shibuya et al., 1996) as well as the previously identified chitin binding site in tomato (Baureithel et al., 1994). The specificity of induction of medium alkalinization and the generation of reactive oxygen (ROX) paralleled chitin binding at the soybean plasma membrane surface. Therefore, the soybean chitin binding site may represent a member of a conserved family of chitin binding proteins responsible for the onset of certain plant-microbe interaction responses in plants. The common thread that unites all of the above-mentioned responses is that in each plant system, whether leguminous or non-leguminous, there appears to be a common perception mechanism for chitin oligosaccharides.
RESULTS
Preparation of Radiolabeled Chitin Oligosaccharide Ligands
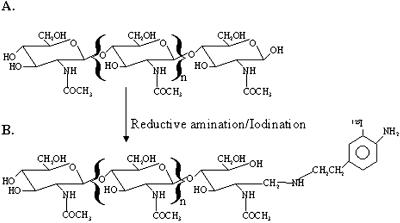
An aminophenyl group was conjugated to both N-acetylchito-pentaose and N-acetylchito-octaose by reductive amination with 2-(4-aminophenyl) ethylamine (APEA) according to the method of Ito et al. (1997). The structure of the synthesized ligands is shown in Figure 1. The ligands, 1-(2-(4-aminophenyl) ethyl) amino-1-(tetra-N-acetylchitotetraosyl)-N-acetyldeoxyglucosaminitol (GN5-APEA), and 1-(2-(4-aminophenyl) ethyl) amino-1-(hepta-N-acetylchitoheptaosyl)-N-acetyldeoxyglucosaminitol (GN8-APEA), were purified by gel-filtration, cation-exchange chromatography, and reverse-phase HPLC as previously described (Ito et al., 1997; data not shown).
Figure 1.
Structure of ligands used in saturation kinetic analysis, inhibition analysis, and affinity cross-linking studies. A, β-1,4-Linked N-acetylchitooligosaccharide. B, Chitin-APEA ligand. APEA-ligands were synthesized using either (GlcNAc)5 or (GlcNAc)8 as substrates for reductive amination. Iodination of the APEA-ligand was performed with carrier-free [Na+]-125I, using Iodogen as a solid phase catalyst.
Identification of a Chitin Binding Site in the Soybean Plasma Membrane
The perception and response of soybean to purified nodulation signals produced by B. japonicum, as well as to purified chitin oligosaccharides have been well documented (Carlson et al., 1993; Stokkermans et al., 1995; Minami et al., 1996a, 1996b). To further define the structural requirements of chitin recognition in soybean, we investigated the binding affinities of a number of chitin oligosaccharides for a high-affinity binding site localized in the soybean plasma membrane. We used two different modified chitin oligosaccharides, GN5-APEA and GN8-APEA. Binding of the APEA-ligands was monitored using plasma membrane enriched fractions from both suspension-cultured cells, as well as 5-d root tissue. As can be seen in Figure 2A, specific binding of GN8-APEA to plasma membrane-enriched fractions isolated from suspension-cultured soybean cells was saturable at a ligand concentration of approximately 100 nm. Ligand binding to plasma membrane purified from 5-d root tissue showed a similar pattern of saturation (data not shown). Saturation of ligand binding was determined in the presence (Fig. 2A, non-specific binding) and absence (Fig. 2A, total binding) of an unlabeled, competitive inhibitor, (GlcNAc)7, to determine non-specific and total binding, respectively. Specific binding was calculated as the difference in these two values. Binding of GN5-APEA to plasma membrane enriched fractions from both suspension-cultured cells and root tissue showed a similar pattern of saturable binding to that obtained with the GN8-APEA ligand (data not shown). To ensure that hydrolysis of the APEA-ligands had not occurred during the binding reactions, thin-layer chromatography of the ligands was performed following incubation with plasma membrane. Based on the relative mobility of the extracted ligands when compared with 125I-labeled APEA ligands that were not incubated with membrane fractions, we can concluded that significant hydrolysis (>1%) did not occur during the binding reaction (data not shown).
Figure 2.
Binding of 125I-(GlcNAc)8-APEA ligand to purified plasma membrane is saturable and of high affinity. A, Plasma membrane (approximately 20 μg of total protein) was incubated with an increasing amount of [125I]-(GlcNAc)8-APEA ligand in the presence (○, non-specific) or absence (♦, total binding, 4–5,000 cpm in each experiment) of unlabeled competitor, (GlcNAc)7. Specific binding (▪) was calculated as the difference between total and non-specific binding. For each concentration, the data are presented as the percentage of total binding (without unlabeled competitor) measured at saturation (i.e. 100 nm ligand). Similar data were obtained using [125I]-(GlcNAc)5-APEA as the ligand (data not shown). B, Scatchard analysis. Data obtained using plasma membrane from suspension-cultured soybean cells (I) or 5-d-old root tissue (II). r, Amount of bound ligand; c, concentration of free ligand.
Scatchard and Hill Plot Analysis
Scatchard analysis (Scatchard, 1949) of [125I]-GN5-APEA and [125I]-GN8-APEA binding to the plasma membrane preparations from both suspension-cultured cells, as well as root tissue, indicated the presence of a specific binding site for chitin oligosaccharides. Data obtained using [125I]-GN8-APEA yielded a Kd (dissociation constant) of 35 nm and a Bmax of 12.2 fmol μg−1 protein for suspension-cultured cells (Fig. 2B, I). Binding of GN8-APEA to plasma membrane preparations from root tissue indicated a Kd of 47 nm and a Bmax of 7.6 fmol μg−1 protein (Fig. 2B, II). In both cases, the data identify a high-affinity binding site for chitin at the plasma membrane surface. The slight difference in the Kd value may be due to differences in homogeneity of the preparations and, therefore, is not evidence for the presence of two distinct sites. The Hill coefficient for all Scatchard data, with an average value of approximately 0.93 (data not shown), indicated an absence of cooperative binding (Hill, 1913). Similar analysis of GN5-APEA binding to plasma membrane suspension-cultured cells yielded a Kd of 58 nm and a Bmax of 9.2 fmol μg−1 protein (data not shown). Using plasma membrane from root tissue, these values were 75 nm and 11.3 fmol μg−1 protein for Kd and Bmax, respectively (data not shown).
Inhibition Analysis of Chitin Binding
The biological action of the lipo-chitin Nod signal in the legume-rhizobium symbiosis exhibits a strict specificity with regard to the chemical structure of the signal molecule. To determine if chitin perception at the soybean plasma membrane surface demonstrated this same preference, we performed a analysis of chitin binding in the presence of competing chitin oligomers (degrees of polymerization [d.p.] = 1–8). As can be seen in Figure 3A, inhibition of chitin binding proceeded in a size-dependent fashion. The larger chitin oligosaccharides were better inhibitors of ligand binding to plasma membrane isolated from suspension-cultured soybean cells (Fig. 3A). In all cases, the N-acetylchitooctaose showed complete inhibition of ligand binding at a concentration of 25 μm (ID50 approximately 50 nm), indicating a high degree of preference for the chitooctaose. Figure 3B shows the result of [125I]-GN5-APEA inhibition analysis using soybean suspension cell plasma membrane. Again, inhibition of chitin binding was size-dependent with chitooctaose (ID50 approximately 80 nm) being the best inhibitor of ligand binding. In addition, GlcNAc and N-acetylchitobiose were shown to have no inhibitory effect on ligand binding at concentrations as high as 250 μm (data not shown), thereby, suggesting that there is a size minimum for binding inhibition. In addition to assessing the inhibitory effect of chitin binding by various chitin oligosaccharides, de-acetylated chitin oligosaccharides (chitosan) at concentrations as high as 250 μm were also tested for their ability to inhibit ligand binding. None of the chitosan oligomers (d.p. = 2–8) had an inhibitory effect on ligand binding (data not shown).
Figure 3.
Inhibition of chitin oligosaccharide binding to a plasma membrane-enriched fraction from soybean proceeds in a size-dependent manner. Ten picomoles of [125I]-(GlcNAc)8-APEA ligand (A) or [125I]-(GlcNAc)5-APEA (B) were incubated with plasma membrane (approximately 20 μg of total protein) isolated from soybean suspension-cultured cells in the presence of increasing amounts of inhibitor chitin oligosaccharide. ♦, Octamer; ▪, heptamer; ▴, hexamer; ×, pentamer; ○, tetramer; ●, trimer. Data from a single experiment (in triplicate) are shown (sd ± 5%). Numerous replicates gave identical results.
The Major Lipo-Chitin Nod Signal from B. japonicum Inhibits [125I]-GN5-APEA Binding to the Plasma Membrane of Soybean
The structural requirements for inhibition of chitin binding to the soybean plasma membrane suggest that Nod signals may also inhibit binding. Therefore, we tested the ability of the major B. japonicum Nod signal [i.e. NodBjV(C18:1Δ11, Me-Fuc)] to inhibit chitin binding to soybean plasma membrane. Figure 4A shows the pattern of inhibition of [125I]-GN5-APEA binding to root plasma membrane in the presence of unmodified chitin oligosaccharides. This pattern of inhibition was similar to that described in Figure 3 with oligosaccharides of a greater degree of polymerization (i.e. d.p. = 8) being better inhibitors of ligand binding. As shown in Figure 4B, inhibition of [125I]-GN5-APEA binding to soybean root plasma membrane was also observed using the Nod signal. However, the level of inhibition was less than that of the structurally similar N-chitopentaose. With an IC50 value of approximately 10 μm, the major Nod signal produced by B. japonicum was similar to chitotetraose in its ability to inhibit chitin binding to the plasma membrane of soybean.
Figure 4.
Inhibition of [125I]-(GlcNAc)5-APEA binding by the major lipo-chitin Nod signal produced by B. japonicum. Ten picomoles of [125I]-GN5-APEA was incubated with 5-d-old root plasma membrane (20 μg of total protein) in the presence of increasing amounts of chitin oligosaccharides or Nod signal, NodBjV(C18:1Δ11, Me-Fuc). A, Inhibition analysis of [125I]-GN5-APEA binding to plasma membrane isolated from 5-d-old G. soja roots. ♦, Octamer; ▪, heptamer; ▴, hexamer; ×, pentamer; ○, tetramer; ●, trimer. Data from a single experiment (in triplicate) are shown (sd ± 5%). Numerous replicates gave identical results. B, Inhibition of [125I]-GN5-APEA binding by the major lipo-chitin Nod signal produced by B. japonicum strain USDA110. ♦, Octamer; ×, pentamer; ●, trimer; ▵, Nod signal. Data from a single experiment (in triplicate) are shown (sd ± 5%). Numerous replicates gave identical results.
Affinity Cross-Linking of GN8-APEA to Plasma Membrane-Enriched Fractions from Soybean
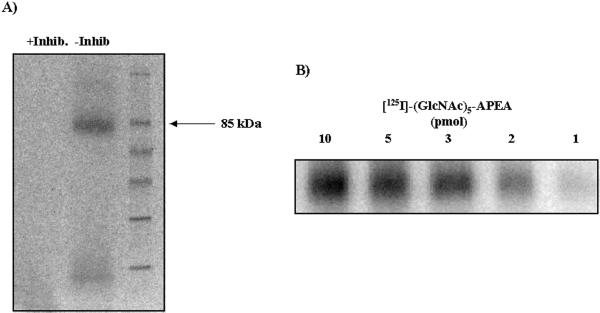
To identify the high-affinity chitin binding site, an affinity cross-linking method was used (Ito et al., 1997). SDS-PAGE analysis showed the presence of a major band of approximately 85 kD with minor bands at approximately 14 and approximately 150 kD (Fig. 5A). The major band at 85 kD is similar in size to a chitin binding protein previously described in rice (Ito et al., 1997). The largest band, 150 kD, may represent a dimer of the 85-kD protein. The smaller band may be a proteolytic fragment of the major band. In the presence of a 250-fold molar excess (25 μm) of competitive (GlcNAc)7 inhibitor, no signal was detected, indicating saturation of the binding site.
Figure 5.
Affinity cross-linking of 125I-(GlcNAc)-APEA ligand to the plasma membrane-localized 85-kD-binding protein. A, Plasma membrane was incubated in the presence of 10 pmol of 125I-(GlcNAc)8-ligand in the presence (+Inhib) or absence (−Inhib) of chitin oligosaccharide inhibitor (GlcNAc)7, (250 molar excess). Cross-linking was performed using 2% (v/v) glutaraldehyde/NaCNBH3 as the cross-linking agent. Fractions were separated on a 12% (v/v) SDS gel and visualized using a phosphor-imaging system. B, Ligand concentration dependence of affinity cross-linking. Plasma membrane (20 μg of total protein) from 5-d-old soybean roots was incubated in the presence of increasing concentrations of [125I]-GN5-APEA (1–10 pmol/100 μL). The membranes were subjected to a cross-linking reaction as described in A and the solubilized proteins were analyzed by SDS-PAGE. No labeling of the 85-kD binding site was observed when unlabeled competitive inhibitor was included as described in A (data not shown).
The binding affinity of [125I]-GN5-APEA was further evaluated by examining the concentration-dependence of affinity cross-linking to the 85-kD binding site. As shown in Figure 5B, binding of the radiolabeled ligand was reduced in the presence of decreasing ligand concentrations. Quantification of the relative intensities of the labeled ligands was performed by autoradiography using a phosphor screen, and further analyzed using image analysis software. Computer analysis indicated that the ligand concentration required for a half-maximal binding (approximately 33 nm) was in rough agreement with the Kd value obtained by Scatchard analysis (Kd = 47 nm; Fig. 2B) and the IC50 values shown in Figure 3. Therefore, this 85-kD protein may be responsible for the chitin binding activity measured in the soybean plasma membrane-enriched fractions.
Generation of ROX in Suspension-Cultured Soybean Cells by N-Acetylchitooligosaccharides
To correlate the presence of a chitin binding site with a biological response, the generation of ROX in response to chitin elicitation was monitored by the luminol-chemiluminescence assay (Anderson et al., 1991). As can be seen in Figure 6, ROX was generated in suspension-cultured soybean cells upon treatment with purified chitin oligosaccharides. The generation of ROX was directly dependent on the degree of polymerization of the chitin oligomers. The pattern of the response to chitin oligomers shown in Figure 6 is similar to the specificity of the chitin binding site in the plasma membrane (Fig. 3A) in that, in both cases, the larger chitin oligosaccharides (i.e. [GlcNAc]8) were preferred. Furthermore, (GlcNAc) and (GlcNAc)2, which lacked the ability to inhibit [125I]-GN8-APEA binding to plasma membrane preparations, were also incapable of eliciting production of ROX (Fig. 6). However, unlike the binding assays, the ROX response to (GlcNAc)4, (GlcNAc)5, and (GlcNAc)6 did not differ significantly. Differences in the ROX response to (GlcNAc)7 and (GlcNAc)8 was also not significant. These data suggest that a specific threshold must be exceeded for a ROX response and this differs depending on the chitin oligomer used. The levels of ROX production shown in Figure 6 are similar to the levels previously reported using rice suspension-cultured cells in response to chitin elicitation (Kuchitsu et al., 1993; Minami et al., 1996c; Shibuya et al., 1996).
Figure 6.
Generation of ROX in suspension-cultured soybean cells by N-acetylchitooligosaccharides. Suspension-cultured soybean cells were incubated in the presence or absence (dH2O) of 1 μm chitooligosaccharides of varied degrees of polymerization and assayed for the generation of ROX species. In the presence of both the (GlcNAc)7 and (GlcNAc)8 oligomers, a significant production of H2O2 was observed. The biological response to chitin was size-dependent with chitin oligomers of a lesser degree of polymerization being weak activators of an oxidative response. Data represent the average of three independent trails in triplicate.
N-Acetylchitooligosaccharides Induce Alkalinization of Culture Medium of Soybean Suspension Cells in a Size-Dependent Manner
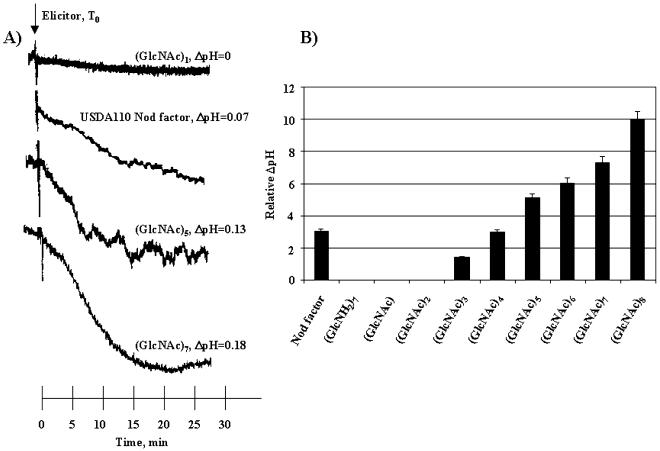
Alkalinization of the culture medium of suspension-cultured plant cells has been reported as an early response to microbial infection (He et al., 1998), as well as to the perception of purified chitin-oligosaccharide elicitors (Kuchitsu et al., 1993; Baureithel et al., 1994; Minami et al., 1996c). To determine if perception of chitin oligosaccharides by soybean suspension cells induces a rapid alkalinization of the culture medium, chitin oligosaccharides of varied degrees of polymerization were tested for their ability to induce a change in pH (ΔpH) of the medium upon elicitor treatment. As seen in Figure 7B, N-acetylchitooligosaccharides induced the alkalinization of the culture medium of soybean suspension cells in a size-dependent manner. To ensure a direct comparison of the different measurements, only the change in pH is given. Treatment of soybean cells with chitin oligosaccharides of a greater degree of polymerization (i.e. [GlcNAc]8) induced a greater ΔpH, as compared with N-acetylchitooligosaccharides of a lesser degree of polymerization (i.e. [GlcNAc]1). Further supporting the specificity of the chitin binding site, treatment of suspension cells with equal molar amounts of chitosan, the de-acetylated chitin oligosaccharide, resulted in no measurable ΔpH of the culture medium ([GlcNH2]7, Fig. 7B).
Figure 7.
Alkalinization of soybean suspension cell culture medium in response to chitin oligomers. Seven-hundred milligrams of suspension-cultured cells was incubated in the presence of 5 μm chitooligosaccharides of various degrees of polymerization or B. japonicum Nod signal, and the culture medium was monitored for a change in pH (ΔpH). The magnitude of ΔpH was shown to be directly dependent upon the degree of polymerization of the chitin backbone with larger oligomers (i.e. [GlcNAc]8) inducing the greatest ΔpH. All data were normalized for cell mass. ΔpH was calculated based upon an initial pH (T0) of 5.5. A, Representative tracing of the raw data of selected oligosaccharide and Nod signal treated cells. Cells were monitored for approximately 30 min following elicitor treatment at 25°C. pH changes were calculated based on standard pH shifts using K-phosphate buffers of varying pH. B, Normalized expression based on the magnitude of ΔpH based on fresh cell weight. Data are presented as the relative pH change compared with (GlcNAc)8, which elicited the largest ΔpH compared with all elicitors tested. The results shown are the average of three independent measurements. Sample sd ± 5%. T0, Time of elicitor treatment.
To determine if the major lipo-chitin Nod signal, a modified chitin pentamer, produced by B. japonicum was capable of inducing a rapid alkalinization of suspension-culture medium similar to that of chitin oligosaccharides, cells were treated with 5 μm of purified Nod signal. Treatment of soybean suspension cells with Nod signal resulted in rapid medium-alkalinization similar in degree (e.g. ΔpH) to that when cells were treated with (GlcNAc)4 or (GlcNAc)5 (Fig. 7A). These results suggest that chitin perception by the plasma membrane-localized chitin binding protein and the resultant alkalinization of the culture medium is dependent upon the degree of polymerization of the chitin backbone, and modifications to the oligosaccharide (e.g. fucosylation and acylation) do not appear to influence the specificity in terms of both binding (Fig. 4B) and the induction of a biological response (Fig. 7A).
DISCUSSION
The perception of chitin oligosaccharides has been shown to evoke a wide range of responses in a number of plant species (Cosio et al., 1988; De Jong et al., 1993; Minami et al., 1996a, 1996b, 1996c). In soybean, Minami et al. (1996a) showed that chitin oligomers could induce the transient expression of the early nodulin ENOD40. This suggested that a chitin binding protein could be involved in the nodulation response in soybean. We sought to identify such a binding site using an 125I-labeled chitin ligand. Two ligands were used, GN5-APEA and GN8-APEA. The first of these ligands matches in size the most active chitin oligomer involved in ENOD40 induction in soybean (Minami et al., 1996a). The second ligand is identical to that shown to be the most active in the binding to the rice plasma membrane (Ito et al., 1997). We analyzed the response of suspension-cultured cells to chitin since such cells are easily manipulated and have been widely used for studies of microbial elicitors. In addition, we analyzed tissue from 5-d-old roots since it is root tissue that responds to the Nod signals. In both cases, the binding of the [125I]-chitin ligands demonstrated relatively high affinities (e.g. Kd of 35 and 47 nm in suspension cells and root cells, respectively, using [125I]-GN8-APEA as a ligand). Binding of [125I]-GN5-APEA to plasma membrane-enriched fractions from both root tissue and suspension-cultured soybean cells also demonstrated a high affinity (i.e. 58–75 nm, data not shown). These Kd values compare favorably with those reported for chitin binding to rice (i.e. 29 nm; Shibuya et al., 1996) and tomato cells (i.e. 28 nm; Baureithel et al., 1994). These values also compare favorably with the lipo-chitin binding sites reported from the membrane preparations from Medicago (i.e. NFBS1, 86 nm and NFBS2, 2 nm, respectively; Bono et al., 1995; Gressent et al., 1999).
In soybean, the cellular responses of the developing root to modified lipo-chitin oligosaccharides result in the development of a new organ, the nodule (for review, see Cohn et al., 1998). Nodule initiation in legumes is triggered by a specific lipo-chitin Nod signal produced by the compatible rhizobium. The size-specificity for Nod signal perception follows a strict requirement for oligosaccharides with a degree of polymerization of 3 to 5 β-1,4-linked GlcNAc residues. In addition, chemical modifications to both the reducing and non-reducing ends of the Nod signal are important host-specificity determinants of the legume-Rhizobium interaction (van Rhijn and Vanderleyden, 1995; Pueppke, 1996). Bono et al. (1995), and more recently Gressent et al. (1999), used a 35S-labeled molecule to identify and characterize a Nod signal-binding activity in membrane fractions prepared from alfalfa roots. In the case of Nod signal binding to Medicago suspension-cultured cells, there appeared to be two distinct binding sites. The first site, termed NFSB1 (Nod factor binding site 1), exhibited a saturable and reversible binding of Nod signals, with an affinity (Kd) of 86 nm. The second site, termed NFSB2, displayed a high degree of specificity for modified lipo-chitin Nod signals, suggesting that the recognition was determined by the lipid and oligosaccharide structural elements. It is interesting that the presence of two binding sites, which exhibit differing affinities for ligand binding is consistent with the presence of two recognition events involved in nodule initiation (Ardourel et al., 1994; Minami et al., 1996a, 1996b). In the case of lipo-chitin perception in Medicago, NFBS1 may represent a non-specific site present in both legumes and non-legumes, whereas NFBS2 may be responsible for the perception of host-specific/modified lipo-chitin Nod signals.
In soybean, the situation may be different from alfalfa since Minami et al. (1996a) showed that chitin oligomers could induce the transient expression of the early nodulin ENOD40. This suggested that a chitin binding protein could be involved in the nodulation response in soybean. We sought to identify such a binding site using [125I]-labeled GN5-APEA and GN8-APEA ligands. Our data suggest that both ligands apparently interact with the same binding site on the soybean plasma membrane. Competition using chitin oligomers of increasing length indicate that the soybean chitin binding site prefers larger chitin oligomers. These results are very similar to what has been shown for chitin binding to rice membranes (Minami et al., 1996c; Shibuya et al., 1996; He et al., 1998).
Soybeans can be infected with fungal pathogens and respond with a defense response (e.g. Okinaka et al., 1995). Therefore, one might expect the presence of a chitin perception system that would help mediate such a response. As shown in Figure 6, chitin does elicit a potent oxidative burst response in soybean cells. Moreover, the specificity of this response generally matches the specificity of chitin binding in that larger chitin oligomers are more active. It is similar that perception of chitin oligosaccharides by suspension-cultured soybean cells induces a rapid alkalinization of the culture medium in a size-dependent fashion, which more closely matches the chitin binding (Fig. 7). The correlation between the structural specificity of chitin binding to the plasma membrane and the biological responses evoked upon perception of these elicitors suggests a role for chitin oligosaccharides in an elicitor-signaling pathway (He et al., 1998). By analogy to better studied systems in rice and other plants, the 85-kD chitin binding protein found in the plasma membrane is an excellent candidate for a chitin receptor involved in mediating the soybean response to chitin elicitation.
There have been reports suggesting that an incompatible Rhizobium (i.e. inoculated onto a heterologous host) might induce a defense response that could inhibit nodulation (Bhagwat et al., 1999). Such a mechanism could play an important role in determining the host specificity of rhizobia. Although preferring the larger oligomers, the plasma membrane chitin binding protein did recognize smaller chitin oligomers. Competition experiments using purified B. japonicum Nod signal as a competitive inhibitor of both GN5-APEA and GN8-APEA binding to plasma membrane fractions from soybean suggests that the B. japonicum Nod signal interacts with the high affinity chitin binding site localized in the plasma membrane of soybean (Fig. 4) and is capable of eliciting a phytoactive defense response in suspension-cultured cells (Fig. 7). In both cases, the response elicited by Nod signal treatment is similar to that elicited by the structurally similar chitotetraose and chitopentaose molecules. The ability of both chitin oligosaccharides and rhizobial Nod signals to interact with the same binding site suggests that the chitin backbone is the common element perceived by the soybean chitin binding protein. Thus, based on the structural similarities between the lipooligosaccharide Nod signals and chitooligosaccharides, it is theoretically possible that incompatible Nod signals could induce a defense response.
Our data indicate that chitin molecules of a lesser degree of polymerization (i.e. d.p. = 5) are relatively poor competitors for ligand binding. The data indicate that the Nod signal has low affinity for the chitin binding site and, therefore, it is unlikely that this site is the long sought after Nod signal receptor. It seems unlikely that the chitin binding protein found in the soybean plasma membrane mediates a nodulation-related response to Nod signals. However, the ability of the major lipo-chitin Nod signal from B. japonicum to inhibit chitin binding to the plasma membrane of soybean (Fig. 4), albeit at a relatively low affinity, suggests that there might be some structural elements that are common to both Nod signal and chitin perception. Diaz et al. (2000) recently reported that both modified rhizobial Nod signals as well as unmodified chitin oligosaccharides could induce cortical cell divisions in red clover (Trifolium pratensis) roots transformed with the Pisum sativum lectin (PSL). This study demonstrated that the oligochitin backbone was sufficient for the induction of cortical cell divisions in clover roots, arguing that Nod signal and chitin perception may share a common recognition event. It is interesting that this report also showed that chitin oligosaccharides of a lesser degree of polymerization (e.g. chitotriose), as well as de-N-acetylated chitin oligosaccharides (e.g. chitosan oligosaccharides) were less active than the larger N-acetylchitooligosaccharides in inducing cortical cell divisions. Application of GlcNAc failed to induce cortical cell divisions, suggesting the requirement for a minimum degree of polymerization of the chitin backbone. These results compare favorably with our data, which demonstrate the structural specificity for chitin binding to the soybean plasma membrane.
The characterization of a plasma membrane-localized binding site in soybean will provide us with a foundation from which we can further define the mode of action of chitin, as well as the perception of modified lipo-chitin oligosaccharides in soybean. The specificity of the chitin binding site identified in this study argues that it most likely does not play a specific role in lipo-chitin Nod signal recognition. However, binding specificity did match that of a chitin-elicited defense response (i.e. generation of ROX and medium alkalinization). Since these responses are well accepted as initial steps in a defense response to pathogen invasion, we propose that the chitin binding protein identified likely plays a role in eliciting a defense response against invading fungal pathogens (compare Cheong and Hahn, 1991; Cheong et al., 1993).
MATERIALS AND METHODS
Chemicals
Chitin and chitosan oligosaccharides were obtained from Seikagaku America (Falmouth, MA). Na-125Iodine was purchased from ICN (Costa Mesa, CA). [14C]Methylated proteins were purchased from Amersham Life Sciences (Arlington Heights, IL). 2-(4-Aminophenyl) ethylamine (APEA) was purchased from Aldrich Chemicals (Tokyo). Protease inhibitors, salicylhydroxamic acid, and phenylmethylsulfonyl flouride were obtained from Sigma (St. Louis). DNA primers were purchased from Integrated DNA Technologies (Coralville, IA). All other reagents were obtained from Fisher Scientific (Pittsburgh).
Purification of Plasma Membrane-Enriched Fractions from Both Suspension-Cultured Cells and Root Tissue
Soybean (Glycine max cv Essex) seeds were surface sterilized as described previously (Sanjuan et al., 1992) and germinated in the dark at 28°C for 5 d on moist, sterile Whatman filter paper. After 5 d, roots were excised, frozen in liquid nitrogen, and stored at −80°C. Approximately 12,000 roots were collected to yield 200 g (fresh weight) of plant material. Suspension-cultured soybean cells of G. max SB-1, derived from G. max (L.) Merr. cv Mandarin root tissue, were maintained in a modified B5 medium (1× B5 basal salt mixture (Sigma) + 2% [v/v] Suc, 1× B5 vitamins, pH 5.5) at 28°C, according to the method of Ho et al. (1988). Suspension cultured soybean cells of G. max cv Enrei derived from root tissue were maintained in a similar manner. Cells were harvested 10 d after transfer to fresh medium, frozen in liquid nitrogen, and stored at −80°C until use. Approximately 200 g (fresh weight) of either suspension-cultured cells or 5-d-old roots were used as starting material for the isolation of plasma membrane-enriched fractions. Tissue was homogenized in homogenization buffer (0.3 m Suc, 50 mm MES [2-(N-morpholino)-ethanesulfonic acid]-Tris, pH 7.6, 5 mm EDTA, 5 mm EGTA, 20 mm NaF, 1 mm dithiothreitol, 4 mm salicylhydroxamic acid, 2 mm phenylmethylsulfonyl flouride, 2.5 mm Na2S2O5, 0.5% [v/v] bovine serum albumin) on ice for 30 min. After homogenization, cells were lysed at 1,000 p.s.i. in a French pressure cell press (American Instrument Company, Silver Spring, MD). Following lysis, the homogenate was centrifuged at 4,000g for 10 min at 4°C. The supernatant was then subjected to an additional centrifugation at 4°C for 10 min at 15,000g. The resultant supernatant was then centrifuged at 100,000g for 1 h at 4°C in a Ti65 fixed angle rotor (Beckman Instruments, Fullerton, CA). After ultra-centrifugation, the microsomal fraction was resuspended in microsomal fraction buffer (0.25 m Suc + 10 mm sodium phosphate, pH 7.8) and homogenized using a hand-held Potter homogenizer. In both cases, plasma membrane-enriched fractions were prepared by aqueous two-phase partitioning using dextran T500 and polyethyleneglycol 3,500 according to the method of Shibuya et al. (1996). Further details on purification and assays for membrane purity can be found in Day et al. (2000). The plasma membrane-enriched fraction was resuspended in 1 mL of plasma membrane buffer {0.25 m Suc + 5 mm HEPES [4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid]-bistrispropane, pH 7.0 + 0.1 m dithiothreitol}. Typically, 3 to 5 mg of plasma membrane-enriched fractions were obtained from 200 g of cells (fresh weight). Protein concentrations were determined using the BCA Protein Assay (Pierce Chemical, Rockford, IL), using bovine serum albumin standards. In all assays, the quantity of plasma membrane is based on protein determination.
Synthesis of [125I]-Labeled Aminophenyl Derivatives of N-Acetylchitopentaose and N-Acetylchitooctaose
The chitin pentamer (GN5-APEA) and chitin octamer (GN8-APEA) aminophenyl conjugate ligands used in the binding assays were prepared and purified as previously described using a reductive amination reaction. (Ito et al., 1997). Iodination of the GN5-APEA and GN8-APEA ligands was performed using Iodogen (Pierce Chemical) as the oxidizing agent. Two microliters of either GN5-APEA or GN8-APEA (1-mm stock) and 10 μL of 0.5 m sodium phosphate buffer (pH 7.5) were mixed in a glass test tube, precoated with 50 μg of Iodogen. Five microliters of Na-[125 I] (16.3 × 105 Bq) (ICN Radiochemicals) was added to the mixture and incubated at room temperature for 1 h. After incubation, 5 μL of 0.1 m unlabeled NaI were added to the reaction tube and incubated for 5 min. The mixture was applied to a Poly Prep AG 1 × 8 column (Bio-Rad Laboratories, Hercules, CA) and eluted with distilled water. The radioactive, unbound fractions were stored at −20°C until use for affinity cross-linking and binding assays.
GN5-APEA and GN8-APEA were typically radio-iodinated to a specific activity of 1.8 × 109 Bq mmol−1 for use in binding assays, inhibition analysis, and affinity cross-linking of the plasma membrane enriched fraction.
Isolation of Lipo-Chitin Nod Signals Produced by Bradyrhizobium japonicum
The lipo-chitin Nod signal from B. japonicum strain USDA110 was purified according to the method of Sanjuan et al. (1992). The identity and purity of the Nod signal preparations was verified by mass spectrometry (data not shown). The B. japonicum Nod signal is a chitin pentamer, acylated at the non-reducing end with vaccenic acid, and substituted at the reducing end with 2-O-methyl-Fuc [i.e. BjNodV(C18:1; mefuc)].
Binding Assays and Inhibition Analysis
Binding assays were performed according to Shibuya et al. (1996) with slight modifications. In most cases, 20 μg of plasma membrane (equivalent to 20 μg of protein) was mixed with 10 pmol of either [125I]-GN5-APEA or [125I]-GN8-APEA, and the final volume was adjusted to 300 μL with binding buffer (25 mm Tris-HCl [pH 7.0] + 1 mm MgCl2, 1 m NaCl, 2 mm dithiothreitol). For inhibition analysis, an appropriate amount of unlabeled chitin oligosaccharide or chitosan oligosaccharide was added to the reaction mixture. The reaction mixture was directly applied to a Durapore membrane filter (Multiscreen-GV 96-well filtration plate, 0.22 μm, Millipore, Bedford MA). A vacuum was applied to the membrane, and the samples were washed twice with 200 μL of wash buffer (25 mm Tris-HCl [pH 7.0] + MgCl2, 2 mm dithiothreitol). Radioactivity retained on the membranes, which corresponds to bound ligand, was counted on a gamma counter (RiaGamma 1274 system, LKB Wallac Oy, Finland).
Thin Layer Chromatography of APEA Ligands following Binding Analysis
To ascertain the structural integrity of the APEA ligands following the binding assays, the ligands were extracted from the binding reactions and analyzed by thin-layer chromatography (TLC). Binding assays were performed as described above, using either GN5-APEA or GN8-APEA. The reaction mixture was centrifuged at 41,000 rpm for 30 min (TLA 100.2 fixed-angle rotor, Beckman Instruments). Following centrifugation, the supernatant was removed to a sterile Eppendorf tube and stored at room temperature until TLC analysis. The pellet was washed once with 500 μL of binding buffer and then resuspended in 200 μL N-butanol. The tube was vortexed vigorously for 20 s and then centrifuged at 41,000 rpm for an additional 30 min. After centrifugation, the supernatant was transferred to a sterile Eppendorf tube and dried under filtered N2 gas. The dried extract was resuspended in 50 μL of dH2O. Ten microliters of each extract was spot inoculated onto a normal-phase silica TLC plate (Sigma). The dried plate was developed in 100% (v/v) acetonitrile. After chromatography, the plate was air-dried and exposed to X-omat AR x-ray film (Kodak, Rochester, NY) for 7 d.
Affinity Cross-Linking
Affinity cross-linking of the [125I]-GN5-APEA and [125I]-GN8-APEA ligands to the plasma membrane or the microsomal fraction was performed in a manner similar to the binding assays. After 1-h incubation on ice, the reaction mixture was centrifuged at 41,000 rpm for 30 min (TLA 100.2 fixed-angle rotor, Beckman Instruments). The pellet was washed once with 1 mL of binding buffer, followed by 1 mL of distilled water. The pellet was then resuspended in 50 μL of phosphate-buffered saline (pH 7.4). Two-hundred microliters of 2.5% (v/v) glutaraldehyde, and 50 μL of Na- CNBH3 (1 mg mL−1) were added to the reaction tube. The reaction was incubated at room temperature (25°C) for 1 h. After incubation, the mixture was centrifuged at 41,000 rpm for 30 min. The pellet was washed twice with 1 mL of binding buffer, followed by two washes each with 1 mL of phosphate-buffered saline. The pellet was re-suspended in 50 μL of SDS sample buffer and analyzed by SDS-PAGE according to the method of Laemmli (1970) using 12% (v/v) acrylamide. Dried gels were analyzed by autoradiography using a phosphor screen processed on a Storm 840 Optical Scanner (Molecular Dynamics, Sunnyvale, CA). Images were analyzed using ImageQuant software (Molecular Dynamics). Quantification of the relative intensities of the labeled bands was done by autoradiography using an Instant Imager Electronic Autoradiograph (Packard Instrument Company, Meriden, CT).
ROX Detection and Quantification
The generation of ROX species induced by chitin oligomers was monitored as follows: Chitin oligosaccharides of various degrees of polymerization (d.p. = 1–8) were added to suspension-cultured SB-1 soybean cells (Ho et al., 1988). Fifty milligrams of suspension-cultured soybean cells was transferred to fresh modified B5 medium (Ho et al., 1988) and grown at 25°C. After 1 h, various chitin oligosaccharides were individually added to a final concentration of 1 μm. An equal amount of sterile dH2O was added to a control sample, instead of the chitin oligomers. The cells were incubated at 25°C for an additional 15 min, after which time they were transferred to an ice bath to quench the reaction. The cells were removed by low speed centrifugation, and the supernatant was used to determine the presence of ROX species by the luminol-dependent chemiluminescence assay (Anderson et al., 1991). In brief, 50 μL of the supernatant was added to 50 μL of 1.1 mm luminol, 100 μL of 14 mm potassium ferricyanide, and 300 μL of 50 mm potassium phosphate buffer (pH 7.9). Samples were immediately analyzed for chemiluminescence using a TD-20/20 luminometer (Turner Designs, Sunnyvale, CA).
Alkalinization of Soybean Suspension Cell Medium in Response to Chitin Oligosaccharides
To avoid wounding suspension-cultured soybean cells by mechanical stirring and thus preventing a slow but continuous alkalinization of the culture medium, 700 mg of cells were sequestered in a 15- × 50-mm polyester mesh bag (Kureha Chemical, Tokyo). Mesh bags containing the suspension cells were placed into Erlenmeyer flasks containing 30 mL of fresh, sterile, B5 medium (Ho et al., 1988), pH 5.5. After stirring the cells 300 rpm for 30 min at 25°C, chitin oligosaccharides of various degrees of polymerization (d.p. = 1–8), as well as the major lipo-chitin Nod signal produced by B. japonicum strain USDA110, were added individually to a final concentration of 5 μm. A HM-5S pH meter (TOA Electronics, Tokyo) equipped with a glass pH electrode (GS-5015C, TOA Electronics) was used to monitor the pH change (ΔpH) of the medium. After each replicate experiment, the cells were harvested from the mesh bags, and their fresh weight determined. Data were calculated as the magnitude of ΔpH normalized for cell mass (e.g. 700 mg). Values were expressed as the relative expression (e.g. ΔpH) based on the pH change observed when cells were treated with chitin octamer, GN8.
ACKNOWLEDGMENTS
The authors wish to thank the laboratory of Dr. Barry Bruce at the University of Tennessee for allowing the use of laboratory space for iodination experiments. We would also like to thank Dr. Donald K. Dougall (University of Tennessee) for providing laboratory space for tissue culture. Special thanks to Dr. Russell Carlson (University of Georgia, Athens) for MS analysis of the Nod signal preparations.
Footnotes
This work was supported by the Department of Energy (grant no. DE–FG02–97ER–20260 to G.S.) and by a research grant from Bio-oriented Technology Research Advancement Institute (PRO-BRAIN to N.S.).
LITERATURE CITED
- Anderson AJ, Rogers K, Tepper CS, Blee K, Cardon J. Timing of molecular events following elicitor treatment of plant cells. Plant Physiol Mol Plant Pathol. 1991;38:1–13. [Google Scholar]
- Ardourel M, Demont N, Debelle F, Maillet F, de Billy F, Prome JC, Denarie J, Truchet G. Rhizobium meliloti lipooligosaccharide nodulation factors: different structural requirements for bacterial entry into target root hair cells and induction of plant symbiotic developmental responses. Plant Cell. 1994;6:1357–1374. doi: 10.1105/tpc.6.10.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baureithel K, Felix G, Boller T. Specific, high affinity binding of chitin fragments to tomato cells and membranes: competitive inhibition of binding by derivatives of chitooligosaccharides and a Nod factor of Rhizobium. J Biol Chem. 1994;269:17931–17938. [PubMed] [Google Scholar]
- Bhagwat AA, Mithofer A, Pfeffer PE, Kraus C, Spickers N, Hotchkiss A, Ebel J, Keister DL. Further studies of the role of cyclic β-glucans in symbiosis: an NdvC mutant of Bradyrhizobium japonicum synthesizes cyclodecakis-(1–3)-β-glucosyl. Plant Physiol. 1999;119:1057–1064. doi: 10.1104/pp.119.3.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bono JJ, Riond J, Nicolaou KC, Bockovich NJ, Estevez VA, Cullimore JV, Ranjeva R. Characterization of a binding site for chemically synthesized lipo-oligosaccharidic NodRm factors in particulate fractions prepared from roots. Plant J. 1995;7:253–260. doi: 10.1046/j.1365-313x.1995.7020253.x. [DOI] [PubMed] [Google Scholar]
- Carlson RW, Sanjuan J, Bhat UR, Glushka J, Spaink HP, Wijfjes AW, van Brussel AA, Stokkermans TJ, Peters NK, Stacey G. The structures and biological activities of the lipo-oligosaccharide nodulation signals produced by type I and II strains of Bradyrhizobium japonicum. J Biol Chem. 1993;268:18372–18381. [PubMed] [Google Scholar]
- Cheong JJ, Alba R, Cote F, Enkerli J, Hahn MG. Solubilization of functional plasma membrane-localized Hepta-β-glucoside elicitor-binding proteins from soybean. Plant Physiol. 1993;103:1173–1182. doi: 10.1104/pp.103.4.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong JJ, Hahn MG. A specific, high-affinity binding site for the hepta-β-glucoside elicitor exists in soybean membranes. Plant Cell. 1991;3:137–147. doi: 10.1105/tpc.3.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn JR, Day RB, Stacey G. Legume nodule organogenesis. Trends Plant Sci. 1998;3:105–110. [Google Scholar]
- Cosio EG, Popperl H, Schmidt WE, Ebel J. High-affinity binding of fungal β-glucan fragments to soybean (Glycine max L.) microsomal fractions and protoplasts. Eur J Biochem. 1988;175:309–315. doi: 10.1111/j.1432-1033.1988.tb14198.x. [DOI] [PubMed] [Google Scholar]
- Day RB, McAlvin C, Loh J, Denny RL, Wood T, Young N, Stacey G. Differential expression of two soybean apyrases: one of which is an early nodulin. Mol Plant Microbe Interact. 2000;13:1053–1070. doi: 10.1094/MPMI.2000.13.10.1053. [DOI] [PubMed] [Google Scholar]
- De Jong AJ, Heidstra R, Spaink HP, Hartog MV, Meijer EA, Hendriks R, Schiavo FL, Terzi M, Bissling T, Kammen AV. A plant somatic embryo mutant is rescued by rhizobial lipo-oligosaccharides. Plant Cell. 1993;5:615–620. doi: 10.1105/tpc.5.6.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denarie J, Debelle F, Rosenberg C. Signaling and host range variation in nodulation. Annu Rev Microbiol. 1992;46:497–531. doi: 10.1146/annurev.mi.46.100192.002433. [DOI] [PubMed] [Google Scholar]
- Diaz C, Spaink HP, Kijne JW. Heterologous rhizobial lipochitin oligosaccharides and chitin oligomers induce cortical cell divisions in red clover roots, transformed with the pea lectin gene. Mol Plant Microbe Interact. 2000;13:268–276. doi: 10.1094/MPMI.2000.13.3.268. [DOI] [PubMed] [Google Scholar]
- Felix G, Baureithel K, Boller T. Desensitization of the perception system for chitin fragments in tomato cells. Plant Physiol. 1998;117:643–650. doi: 10.1104/pp.117.2.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felle HH, Kondorosi E, Kondorosi A, Schultze M. How alfalfa root hairs discriminate between nod factors and oligochitin elicitors. Plant Physiol. 2000;124:1373–1380. doi: 10.1104/pp.124.3.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geurts R, Franssen H. Signal transduction in Rhizobium-induced nodule formation. Plant Physiol. 1996;112:447–453. doi: 10.1104/pp.112.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gressent F, Drouillard S, Mantegazza N, Samain E, Geremia RA, Canut H, Niebel A, Driguez H, Ranjeva R, Cullimore J. Ligand specificity of a high-affinity binding site for lipo-chitooligosaccharidic nod factors in Medicago cell suspension cultures. Proc Natl Acad Sci USA. 1999;96:4704–4709. doi: 10.1073/pnas.96.8.4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He DY, Yazaki Y, Nishizawa Y, Takai R, Yamada K, Sakano K, Shibuya N, Minami E. Gene activation by cytoplasmic acidification in suspension-cultured rice cells in response to the potent elicitor, N-acetylchitoheptaose. Mol Plant Microbe Interact. 1998;11:1167–1174. [Google Scholar]
- Hill AV. The combination of hemoglobin with oxygen and with carbon monoxide. Biochem J. 1913;7:471–480. doi: 10.1042/bj0070471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho SC, Ye WZ, Schindler M, Wang JL. Quantitative assay for binding of Bradyrhizobium japonicum to cultured soybean cells. J Bacteriol. 1988;170:3882–3890. doi: 10.1128/jb.170.9.3882-3890.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y, Kaku H, Shibuya N. Identification of a high-affinity binding protein for N-acetylchitooligosaccharide elicitor in the plasma membrane of suspension-cultured rice cells by affinity labeling. Plant J. 1997;12:347–356. doi: 10.1046/j.1365-313x.1997.12020347.x. [DOI] [PubMed] [Google Scholar]
- Kuchitsu K, Kikuyama M, Shibuya N. N-Acetylchitooligosaccharides, biotic elicitor for phytoalexin production, induce transient membrane depolarization in suspension-cultured rice cells. Protoplasma. 1993;174:79–81. [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Long SR. Rhizobium symbiosis: Nod factors in perspective. Plant Cell. 1996;8:1885–1898. doi: 10.1105/tpc.8.10.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minami E, Kouchi H, Carlson RW, Cohn JR, Kolli VK, Day RB, Ogawa T, Stacey G. Cooperative action of lipo-chitin nodulation signals on the induction of the early nodulin, ENOD2, in soybean roots. Mol Plant Microbe Interact. 1996a;9:574–583. doi: 10.1094/mpmi-9-0574. [DOI] [PubMed] [Google Scholar]
- Minami E, Kouchi H, Cohn JR, Ogawa T, Stacey G. Expression of the early nodulin, ENOD40, in soybean roots in response to various lipo-chitin signal molecules. Plant J. 1996b;10:23–32. doi: 10.1046/j.1365-313x.1996.10010023.x. [DOI] [PubMed] [Google Scholar]
- Minami E, Kuchitsu K, He DY, Kouchi H, Midoh N, Ohtsuki Y, Shibuya N. Two novel genes rapidly and transiently activated in suspension-cultured rice cells by treatment with N-acetylchitoheptaose, a biotic elicitor for phytoalexin production. Plant Cell Physiol. 1996c;37:563–567. doi: 10.1093/oxfordjournals.pcp.a028981. [DOI] [PubMed] [Google Scholar]
- Okinaka Y, Mimori K, Takeo K, Kitamura S, Takeuchi Y, Yamaoka N, Yoshikawa M. A structural model for the mechanisms of elicitor release from fungal cell walls by plant β-1,3-endoglucanase. Plant Physiol. 1995;109:839–845. doi: 10.1104/pp.109.3.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pueppke SG. The genetic and biochemical basis for nodulation of legumes by rhizobia. Crit Rev Biotechnol. 1996;16:1–51. doi: 10.3109/07388559609146599. [DOI] [PubMed] [Google Scholar]
- Sanjuan J, Carlson RW, Spaink HP, Bhat UR, Barbour WM, Glushka J, Stacey G. A 2-O-methylfucose moiety is present in the lipo-oligosaccharide nodulation signal of Bradyrhizobium japonicum. Proc Natl Acad Sci USA. 1992;89:8789–8793. doi: 10.1073/pnas.89.18.8789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scatchard G. The attraction of proteins for small molecules and ions. Ann NY Acad Sci. 1949;51:660–672. [Google Scholar]
- Shibuya N, Ebisu N, Kamada Y, Kaku H, Cohn J, Ito Y. Localization and binding characteristics of a high-affinity binding site for N-acetylchitooligosaccharide elicitor in the plasma membrane from suspension-cultured rice cells suggest a role as a receptor for the elicitor signal at the cell surface. Plant Cell Physiol. 1996;37:894–898. [Google Scholar]
- Stacey G, Shibuya N. Chitin recognition in rice and legumes. Plant Soil. 1997;194:161–169. [Google Scholar]
- Stokkermans TJ, Ikeshita S, Cohn J, Carlson RW, Stacey G, Ogawa T, Peters NK. Structural requirements of synthetic and natural product lipo-chitin oligosaccharides for induction of nodule primordia on Glycine soja. Plant Physiol. 1995;108:1587–1595. doi: 10.1104/pp.108.4.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rhijn P, Vanderleyden J. The Rhizobium-plant symbiosis. Microbiol Rev. 1995;59:124–142. doi: 10.1128/mr.59.1.124-142.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner GP. Evolution and multi-functionality of the chitin system. EXS. 1994;69:559–577. doi: 10.1007/978-3-0348-7527-1_33. [DOI] [PubMed] [Google Scholar]