Summary
Background
Impaired mitochondrial protein import machinery leads to phenotypically heterogeneous diseases. Here, we report a recurrent homozygous missense variant in the gene that encodes the translocase of outer mitochondrial membrane 7 (TOMM7) in nine patients with microcephaly, short stature, facial dysmorphia, atrophic macular scarring, and moyamoya disease from seven unrelated families.
Methods
To prove the causality of the TOMM7 variant, mitochondrial morphology, proteomics, and respiration were investigated in CRISPR/Cas9-edited iPSCs-derived endothelial cells. Cerebrovascular defects and mitochondrial respiration were also examined in CRISPR/Cas9-edited zebrafish.
Findings
iPSC-derived endothelial cells with homozygous TOMM7 p.P29L showed increased TOM7 stability, enlarged mitochondria, increased senescence, and defective tube formation. In addition, proteomic analysis revealed a reduced abundance of mitochondrial proteins involved in ATP synthesis or coordinating TCA cycle and gluconeogenesis. Moreover, mitochondrial respiration was slightly decreased while ATP production from glycolysis was significantly increased. Furthermore, deletion of tomm7 in zebrafish caused craniofacial and cerebrovascular defects that recapitulated human phenotypes. Notably, homozygous iPSCs differentially expressed genes involved in glycolysis and response to hypoxia. Finally, the metabolic imbalance was evidenced by decreased oxygen consumption, increased level of hexokinase 2, and enhanced glycolysis in endothelial cells derived from the patient's iPSCs.
Interpretation
These results revealed the essential role of TOMM7 in balancing cellular sources of energy production at both proteomic and transcriptomic levels and provided the molecular mechanisms through which TOMM7 p.P29L variant leads to an autosomal recessive microcephalic osteodysplastic dwarfism with moyamoya disease.
Funding
This work is supported by National Science and Technology Council grants and National Cheng Kung University Hospital.
Keywords: TOM complex, iPSC-derived endothelial cells, Cerebrovascular disease, Mitochondrial protein import, Mitochondrial respiration, Glycolysis
Research in context.
Evidence before this study
The majority of mitochondrial proteins are synthesized as precursors in the cytosol and imported into mitochondria by the translocase of outer mitochondrial membrane (TOM) complex. Mitochondrial diseases caused by impaired mitochondrial import machinery are phenotypically heterogeneous. Mitochondrial dysfunction in endothelial cells was associated with moyamoya disease, although the mechanism remains largely unknown. Loss of TOM7, a small regulator of TOM complex, caused developmental and cerebrovascular defects in the mouse and zebrafish models. In humans, two studies of a single case carrying a homozygous TOMM7 variant with overlapping yet heterogeneous developmental phenotypes were recently reported. However, no cerebrovascular defect was detected in individuals with TOMM7 variant.
Added value of this study
In this study, a recurrent TOMM7 variant was identified in nine patients with postnatal growth failure, microcephaly, and facial dysmorphism from seven unrelated families. Notably, five of the patients had developed moyamoya disease. To prove the causative role of TOMM7 variant in moyamoya disease, CRISPR/Cas9-edited iPSCs-derived endothelial cell models were generated. Cells with homozygous TOMM7 variant showed decreased mitochondrial respiration and increased cellular glycolysis, indicating metabolic reprogramming. In addition, CRISPR/Cas9-edited tomm7 deficient zebrafish showed metabolic deficiencies and recapitulated the patient's developmental and cerebrovascular phenotypes. Finally, mitochondrial functional defects and metabolic reprogramming were detected in endothelial cells derived from patient's iPSCs and CRISPR/Cas9-corrected control iPSCs.
Implications of all the available evidence
This report showed the segregation of the variant allele in nine members from seven unrelated families and solidified that TOMM7 is the disease-causing gene of a syndrome with unique phenotypes. Differential diagnosis of TOMM7-associated syndrome and other genetic diseases with similar phenotypes can be made with characteristic dentition. In addition, early detection of TOMM7-associated syndrome can be made with atrophic macular scarring. Moreover, metabolic reprogramming may be distinct in different types of cells with TOMM7 variant. Therefore, extra caution is required when designing molecular treatments for patients with TOMM7 variant in the future.
Introduction
Mitochondria serve as the cellular powerhouse by conducting many bioenergetic processes.1, 2, 3 Of ∼1500 mammalian mitochondrial proteins, only 13 of them are encoded by mitochondrial DNA.4 The other mitochondrial proteins are encoded in the nuclear genome and must be imported into mitochondria after being synthesized as precursors in the cytosol.2,5 Entry of nearly all mitochondrial proteins is coordinated by the translocase of outer mitochondrial membrane (TOM) complex.6,7 TOM complex is composed of TOM22, TOM40, TOM70A, TOM20, and small regulators TOM5, TOM6, and TOM7. Small TOM proteins aid in maintaining the stability and function of the TOM complex.8 Continuous biogenesis, fusion, fission, and mitophagy contribute to mitochondrial quantity, quality, and morphology.9, 10, 11 TOM7 is essential for accumulating PTEN-induced kinase 1 (PINK1) on the outer mitochondrial membrane to initiate mitophagy in response to stress.12
Human mitochondrial disorders are a genetically and phenotypically heterogeneous group of diseases.13,14 Individuals with dysfunctional outer mitochondrial membrane (OMM) proteins showed developmental defects in multiple organs. For example, variants in TOMM70 (MIM: 606081) caused global developmental delay with microcephaly and neurological defects.15,16 In addition, variants in MTX2 (MIM: 608555), which encodes the OMM protein Metaxin-2, caused mandibuloacral dysplasia.17 Recently, two cases with TOMM7 (MIM: 607980) variants have been reported.18,19 The individual carrying homozygous TOMM7 p.W25R developed syndromic short stature, skeletal abnormalities, nystagmus, muscle hypotonia, microvesicular liver steatosis, and developmental delay.18 The other individual with Garg-Mishra syndrome carried a homozygous TOMM7 p.P29L showed progeria syndrome, severe dwarfism, mandibular hypoplasia, hyperopia, partial lipodystrophy, mild mitral regurgitation, and mild dilatation of the left ventricle.19
Knockdown of TOMM7 in human brain microvascular endothelial cells caused decreased activity of complex I and increased reactive oxygen species (ROS) levels.20 However, elevated mitochondrial respiration was detected in fibroblasts from the patient's fibroblasts and mouse embryonic fibroblasts (MEFs) with homozygous TOMM7 p.P29L variant.19 On the other hand, increased oxygen consumption rate (OCR) yet decreased ATP synthesis was observed in mutant mouse primary rib chondrocytes and MEFs with Tomm7 p.W25R variant.18 Hence, the role of TOM7 in mitochondrial respiration remains unclear.
In animal models, Tomm7 knockout and homozygous Tomm7 p.W25R knock-in mice recapitulated human phenotypes, such as growth defects and severe lipoatrophy, and died at first couple months of age.18 In another study, Tomm7 knockout mice showed developmental defects and, interestingly, sparse cerebral vascular tree.20 Cerebrovascular abnormalities were also observed in tomm7 knockdown zebrafish embryos.20 However, cerebrovascular phenotypes have not been reported in humans with TOMM7 variants.
Here, we identified the recurrent homozygous missense variant TOMM7 p.P29L in nine patients with microcephaly, short stature, facial dysmorphia, developmental delay, and macular scarring. Notably, five patients developed moyamoya disease, that correlates to cerebrovascular abnormalities found in animal studies. Moyamoya disease is characterized by progressive stenosis of internal carotid arteries and the development of fragile collateral vascular networks.21, 22, 23, 24 Transcriptome analyses in the middle cerebral artery from patients with moyamoya disease revealed downregulation of genes involved in mitochondrial function and mitochondrial oxidative phosphorylation (OXPHOS).25 Endothelial colony-forming cells from patients with moyamoya disease showed disrupted mitochondrial morphology, decreased OCR, and impaired angiogenic activities.26 These studies suggest the involvement of mitochondrial dysfunction in endothelial cells in the pathogenesis of moyamoya disease, although the mechanism remains largely unknown.25,26 Here, we investigate the molecular pathogenesis of TOMM7 in association with moyamoya disease in CRISPR/Cas9-edited induced pluripotent stem cells (iPSCs)-derived endothelial cells and zebrafish models.
Methods
Ethics
Informed written consent was obtained from the patients and their parents, including for the inclusion of patient images in publication. The entire procedure of this study was approved by the Institutional Review Board of the National Cheng Kung University Hospital, National Taiwan University Hospital, and Chung-Shan Medical University Hospital (IRB: A-BR-111-019 and B-BR-109-108). Animal studies have been reviewed and approved by the Institutional Animal Care and Use Committee of the National Cheng Kung University (IACUC: 111235 and 109321).
Whole exome sequencing
Genomic DNA was extracted from peripheral blood collected in EDTA-containing tubes. Exome libraries were built using SureSelect Human All Exon V6 (Agilent) or Hyperkappa (Roche) and sequenced with 2 × 75bp pair-end sequencing on the Illumina NextSeq 500 or NovaSeq6000 platform. Sequencing reads were aligned to human genome reference Hg38 using BWA-mem (bio-bwa.sourceforge.net). Single nucleotide variants and small insertions and deletions were identified with Genome Analysis Toolkit 3.5 (gatk.broadinstitute.org). Variants were annotated with ANNOVAR (annovar.openbioinformatics.org), and novel variants were filtered against 1000 Genomes, dbSNP, and Genome Aggregation Database (gnomAD). Candidate variants were prioritized by unbiased approaches. Variants are ranked by the allele frequency (AF) in the genome aggregation database (gnomAD) and by the prediction of deleteriousness, such as the score of combined annotation dependent deletion (CADD), according to the American College of Medical Genetics and Genomics guidelines.27,28 Segregation analysis with Sanger sequencing was used to validate the variants. Runs of homozygosity were analyzed with AutoMap.29 The fraction of the general population carrying homozygous missense or loss of function variants (MAF < 0.001) in TOMM7 was obtained by the sum of AF of homozygous variants in the global population in gnomAD (v4.1.0) is 0.000251. The significance of the rare variant identified in the current study being causal for the disease was calculated with the sorvastats program in the SORVA package (github.com/alizraksi/sorva; with parameters of -f 0.000251 -n [5, 2] -s [5, 2] -c [1, 0.25]) and then Bonferroni corrected by multiplying estimated 20,000 genes that were sequenced.30 Facial features were analyzed with GestaltMatcher.
Facial analysis
GestaltMatcher was performed to analyze the facial similarities among the five images of four patients who consented to the facial analysis.31 Photos of proband 1-III at different ages, i.e., image 1-III at 10 years old and 1-III-2 at 5 years old, were used in the analysis. Model ensemble and test-time augmentation for encoding each image to 12 512-dimensional vectors were first used. Cosine distance was used to quantify the similarity between two vectors, and the 12 cosine distances between the two images were further averaged to define the facial phenotypic similarity between the two images. When the cosine distance was smaller, indicating that the two images were close in the phenotype space, the two were more similar. Cohort-level analyses were then performed and the similarities on the image level were further analyzed.
To investigate whether the probands share a similar facial phenotype, mean pairwise distances were calculated, followed by random sampling 100 times. Because there were images (1-III and 1-III-2) from the same proband, sampling containing these two images was avoided. The distribution was compared to two distributions (same and random) built from the 1555 images from different subjects with 328 different syndromes from the GestaltMatcher Database (GMDB). To build the distribution resulting from the patients with the same syndrome, a sub-cohort was randomly selected, and the mean pairwise distance was calculated 100 times for every 328 syndromes. Next, the distribution was built by randomly sampling a sub-cohort, and the mean pairwise distance was calculated 100 times to build the “random” distribution. With the same random distribution, we performed the Receiver Operating Characteristic (ROC) analysis with five-fold cross-validation to obtain the threshold to distinguish these two distributions. In the fold with the highest Youden index, we obtained the threshold c = 0.909, corresponding to a sensitivity of 0.862 and a specificity of 0.792.
Pairwise comparison analysis was further performed to investigate the similarities among the five images on the image level. The five images from the patients were compared to 7459 images with 449 different disorders from GMDB by performing leave-one-out cross-validation to simulate the real-world scenario. For example, by testing the image 1-III, the rest four images were put in the space with the other 7459 images, and the ranks of the four images to the image 1-III were calculated. In Supplementary Figure S1a, we can visualize the similarity of each pair of individuals compared to the control cohort (7459 images). We further reported the pairwise distances in Supplementary Figure S1b to analyze the influence of age difference on the facial phenotypic similarities between images 1-III and 1-III-2.
Generation of inducible cell lines
Pooled cDNA was obtained from RNA extracted from Flp-In T-Rex 293 cells (R78007; ThermoFisher; RRID: CVCL_U427) with Trizol reagent (ThermoFisher) followed by reverse transcription by High-capacity cDNA archive kit (Applied Biosystems). Wild-type TOMM7 cDNA was obtained by PCR from the pooled cDNA. The TOMM7 p.P29L was generated by QuikChange Site-Directed Mutagenesis Kit (Agilent) with wild-type TOMM7 cDNA. N′-terminal myc-tagged TOMM7 WT or p.P29L cDNA was then constructed into pcDNA5/FRT vector (ThermoFisher) and co-transfected with pOG44 Flp-recombinase expression vector (ThermoFisher) in a 1:9 ratio into Flp-In T-Rex 293 cells using TransIT-293 transfection reagent (Mirus Bio). Flp-In T-Rex 293 cells were maintained in DMEM with 10% FBS and 1% Penicillin and Streptomycin, 100 μg/ml Zeocine (Invitrogen), and 15 μg/ml Blasticidin S HCl (Invitrogen) at 37 °C with 5% CO2. After 3 days, transfected cells were selected in 100 μg/ml hygromycin. Single colonies were isolated and sequenced with primer pairs targeting the FRT locus in the genome and the flipped-in TOMM7 cDNA. In all experiments, the Flp-In expressing cell lines were treated with 1 μg/ml tetracycline for 3 days to induce the expression of TOMM7. The sequences of primers used in this study are listed in Supplementary Table S1. Flp-In T-Rex 293 cells were regularly checked for mycoplasma infection with a PCR mycoplasma detection kit (APOLO biochemical).
Culture of induced pluripotent stem cells (iPSCs)
Human iPSCs (DF19-9-7T; WiCell Research Institute, RRID: CVCL_K055) were cultured following the procedures in the previous study.32 Briefly, iPSCs were seeded at 50,000 cells per well in 6-well plates pre-coated with 1250 ng/cm2 vitronectin (VTN, STEMCEL Technologies) and cultured in TeSR-E8 medium (STEMCEL Technologies). The culture medium was replaced every 24 h. To passage the cells, iPSCs were dissociated with ReLeSR (STEMCEL Technologies) and neutralized with an equal volume of poly-vinyl-alcohol solution (0.1% PVA in PBS). To increase cell survival, 10 μM of Y-27632 (BioVision) was added in the medium for the first 24 h after passage and removed the next day. iPSCs were authenticated by short tandem repeat region (STR) profiling33 and regularly checked for mycoplasma infection with a PCR mycoplasma detection kit (APOLO biochemical).
Generating iPSCs with patient-specific mutation through CRISPR/Cas9 gene editing
Guide RNA (gRNA) targeting TOMM7 were designed with CRISPOR (crispor.tefor.net) (Supplementary Table S2) and constructed into a Streptococcus pyogenes Cas9 (Cas9) and gRNA co-expressing plasmid eSpCas9 (1.1)-T2A-Puro (Addgene #101039). Anti-sense single-strand oligodeoxynucleotide (ssODN) with two phosphorothioate (PS) linkages at both 5′-ends and 3′-end was used to promote homology-directed repair. Donor ssODN was designed with asymmetric homology arms: the length of the right (5′) and left (3′) homology arms were 73-nt and 48-nt long, respectively. To knock in the patient-specific mutation (TOMM7 c.86C>T) in iPSCs, 1 × 106 of iPSCs were electroporated with 5 μg of plasmid and 6 μg of ssODN by NEPA21 (Nepa Gene). After electroporation, iPSCs were immediately transferred to pre-warmed TeSR-E8 medium with 10 μM Y-27632 (Biovision) for 24 h. The medium was replaced with the addition of 0.5 μg/ml puromycin every 24 h for colony selection. Successfully edited single colonies were isolated and genotyped by restriction enzyme-aided Sanger sequencing and targeted sequencing.
Reprogramming patient PBMC to iPSCs
Peripheral blood mononuclear cells (PBMC) from patient 1-III were reprogrammed to iPSC by the Cytotune™-iPS 2.0 Sendai Reprogramming kit (Thermo Fisher Scientific). The iPSCs were cultured in DMEM-F12 medium, 20% KnockOut serum (Gibco™), 0.1 mM Non-Essential Amino Acid, 2 mM l-glutamine, 0.1 mM 2-mercaptoethanol, 100 U/ml Penicillin, 100 μg/ml Streptomycin, and 10 ng/ml bFGF (Gibco™). The medium was changed daily, and iPSCs were mechanically passaged with ROCK inhibitor. iPSCs were maintained on mouse embryonic fibroblast (mMEF) pre-seeded plates for 7 passages, then in a feeder-free medium for additional 7 passages before culturing in E8 medium. Transgene-free confirmation was done with iPSCs IBMS-iPSC-091-01 P8. RNA extracted by Trizol was converted into cDNA by Superscript™ IV first strand synthesis system (Thermo Fisher Scientific). RT-PCR was performed with the positive control IBMS-iPSC-091-01 P0 and negative control DEPC water. STR of DNA from the patient's PBMC and the 13 IBMS-iPSC-091-01 P14/P7 (P14 represented the total passage number, including 7 passages cultured in feeder-dependent and 7 more passages after shifting to the feeder-free cultivation) were analyzed by GenePrint 10 PCR amplification system (Promega) (Supplementary Data 1). Pluripotency markers were evaluated by immunochemistry (ICC) in IBMS-iPSC-091-01 P12/P5 staining with SSEA4, SOX2, TRA-1-60, and OCT4, by flow cytometry of IBMS-iPSC-091-01 P16/P9 with SSEA4, TRA-1-60, TRA-1-81, OCT4, SOX2, and Nanog and by RT-PCR of IBMS-iPSC-091-01 P12/P5 with primers of Nanog, OCT4, and SOX2. In vivo and in vitro differentiation assays with embryoid body (EB) with accutase dissociated IBMS-iPSC-091-01 and Teratoma formation with 1 × 106 of accutase dissociated IBMS-iPSC-091-01 P12/P5 cells injected into 8 weeks old NOD/SCID mice's testes were harvested 8 weeks later and stained with Haematoxylin and Eosin (H&E) stain. Karyotyping was performed with 40 metaphase spreads of IBMS-iPSC-091-01 P12/P5. CRISPR/Cas9 gene correction was achieved with anti-sense ssODN with TOMM7 c.86T>C, two PS linkages at both 5′-ends and 3′-end and asymmetric homology arms of the length of 90-nt and 63-nt, respectively. Successfully edited single colonies were isolated and genotyped by restriction enzyme-aided Sanger sequencing and targeted sequencing.
Differentiation of iPSCs to endothelial cells
iPSCs were differentiated into endothelial cells following the previous protocol.32 Briefly, iPSCs were differentiated into mesodermal progenitor cells and then into endothelial cells. Initially, 5 × 105 of iPSCs were seeded in a 6-cm dish pre-coated with 250 ng/cm2 VTN and incubated in TeSR-E6 medium (STEMCEL Technologies) containing 10 μM Y-27632, 3 μM CHIR99021 (Selleckchem), and 2 ng/ml Activin A (ProSpec) for 2 days (Day 0–2). Cells were then dissociated by Accutase (Innovative Cell Technologies), re-seeded in a 10-cm dish, and cultured in TeSR-E6 medium with 2 mg/ml PVA and 20 ng/ml mVEGF-A (PeproTech) for an additional 3 days (Day 2–5). The iPSCs-derived endothelial (iEndo) cells were dissociated and harvested for subsequent experiments on day 6 of cell differentiation.
Differentiation of iPSCs to fibroblasts
iPSCs were differentiated into fibroblasts following the previous study.34 Briefly, iPSCs were cultured to form embryoid bodies and then differentiated into fibroblasts. Initially, iPSCs were incubated in ReLeSR to dissociate colonies into cell clumps and then seeded in a low-attachment dish containing 10 μM Y-27632 and 4 mg/ml PVA for 1 day to form embryoid bodies. Embryoid bodies were then spun down and transferred to dishes pre-coated with gelatin (Cell Biologics) and cultured in high glucose DMEM medium (HyClone) with 0.3 mM ascorbic acid (Sigma), 2 ng/ml Activin A (Sigma), and 20% fetal bovine serum (Gibco) for an additional 10 days. After fibronectin induction, cells were dissociated with Accutase (Innovative Cell Technologies) and cultured in DMEM with 10% FBS for at least 3 passages to obtain consistent spindle-like fibroblast cells.
Quantitative real-time PCR
iPSC markers (OCT4, SOX2, NANOG), mesodermal specific genes (HAND1, T, and PDGFRA), endothelial cell markers (CDH5 and PECAM1), and fibroblasts markers (COL1A1, CD10, CD73) are used in real-time quantitative PCR to confirm the cell identities. Total RNA was extracted from the cells using the RNeasy Plus Micro Kit (Qiagen) and reverse-transcribed into cDNA using a High-capacity cDNA Archive kit (Applied Biosystems). Quantitative real-time PCR (qRT-PCR) was performed with the SYBR Green master mix reagent (Applied Biosystems) on QuantStudio 3 Real-Time PCR machine (Applied Biosystems). The relative gene expression was calculated using the delta-delta Ct method and normalized to GAPDH mRNA levels.
Mitochondrial enrichment
Mitochondria were isolated with a Mammalian Mitochondria Isolation Kit for Tissue and Cultured Cells (BioVision, K288-50) following the manufacturer's protocol. For Flp-In cells, mitochondria were isolated after 3 days of tetracycline induction. For iEndo cells, mitochondria were isolated on day 6 of endothelial cell differentiation. Briefly, the cell pellet was resuspended with 1 ml of Mitochondria Isolation Buffer and incubated on ice for 2 min, followed by the addition of 10 μl of Reagent A and incubation on ice for 5 min, with vortex for 5 s every minute. Samples were then centrifuged at 600 ×g for 10 min at 4 °C, and the supernatant was collected for further centrifugation at 7000 ×g for 10 min at 4 °C. The supernatant was removed, and the pellet was washed with 1 ml of Mitochondria Isolation Buffer. Subsequently, the sample was centrifuged again to pellet the intact mitochondria. The supernatant was removed, and the mitochondria were resuspended with Storage Buffer for mitochondrial protein immunoprecipitation experiments, mitochondrial complex I activity assay, and Complex III activity assay. For immunoblotting, isolated mitochondria were lysed in RIPA lysis buffer. For proteomic analysis, isolated mitochondria were lysed in the reduction buffer. All buffers contain protease inhibitors. Samples were stored at −80 °C until used.
Mitochondria assays
Mitochondrial Complex I activity assay kit (Abcam), Mitochondrial Complex III activity assay kit (Abcam), and NAD+/NADH assay kit (Abcam) were used according to the manufacturer's protocol. For Complex I and III enzyme activity assays, 5 and 2 μg of intact mitochondria from iEndo cells were used as input, respectively. For NAD+/NADH assay, 2 × 106 of iEndo cells were used. To evaluate mitochondrial ROS, iEndo cells were stained with 2.5 μM MitoSOX (Invitrogen) for 20 min at 37 °C and analyzed using flow cytometry.
Mitochondrial protein immunoprecipitation
Eighty μg of isolated mitochondria were lysed in mitochondrial storage buffer with 1% digitonin and pre-cleared with 50 μl Protein A/G agarose beads (Santa Cruz) for 1 h at 4 °C. After spinning down the Protein A/G beads at 1000 ×g, the supernatant was collected and incubated with 5 μg anti-myc monoclonal antibody (Invitrogen) overnight at 4 °C. The next day, 50 μl Protein A/G beads were added and incubated for 1 h at 4 °C. The beads were then spun down at 1000 ×g and washed with mitochondrial storage buffer for 5 min at 4 °C 3 times. The beads were incubated with 4× protein loading dye (Invitrogen) for 10 min at 70 °C to elute the binding proteins. Samples were resolved by SDS-PAGE, followed by immunoblotting. The antibodies used in immunoblotting were mouse monoclonal anti-myc (Invitrogen), rabbit anti-TOM40 (ProteinTech), rabbit anti-TOM22 (Invitrogen), and anti-GAHDH (Santa Cruz). Details of antibodies are in Supplementary Table S2.
Immunoblotting
For SDS-PAGE, whole cells or isolated mitochondria were lysed in RIPA lysis buffer supplemented with cOmplete Mini Protease inhibitor Cocktail (Sigma). The proper amount of protein was prepared in 1× NuPAGE LDL sample Buffer and 1× NuPAGE Sample Reducing Agent (Invitrogen) with additional DTT (BioVision) at a final concentration of 100 mM. Samples were heated at 70 °C for 10 min before separating on NuPAGE 10% Bis-Tris Gel (Invitrogen) using 1× NuPAGE MES SDS Running buffer (Invitrogen). The gel was transferred to 0.22 μm PVDF membrane (Cytiva Amersham) by Trans-Blot Semi-Dry Transfer Cell (BIO-RAD). The membrane was then incubated with 5% skim milk in 0.01% TBS-T (0.01% Tween 20 in TBS) for at least 2 h at room temperature for blocking. To improve the detection of low molecular weight proteins, membranes were immersed in cold acetone for 30 min before blocking with 5% skim milk in 0.01% TBS-T. Primary antibodies were diluted in 5% skim milk in 0.01% TBS-T, and membranes were immersed with primary antibodies overnight at 4 °C. Secondary antibodies were diluted in 5% skim milk in TBS-T. Membranes were incubated with secondary antibodies for 2 h at room temperature. The antibodies used in immunoblotting were anti-Actin (Santa Cruz); rabbit anti-ATP5A (Abcam); rabbit anti-ATPH (ProteinTech); anti-GAPDH (Santa Cruz); mouse anti-Myc (Invitrogen); rabbit anti-PCK2 (ProteinTech); rabbit anti-PINK1 (Cell Signaling); rabbit anti-SDHA (ProteinTech); rabbit anti-TOMM7 (Invitrogen); rabbit anti-TOMM22 (Invitrogen); rabbit anti-TOMM40 (ProteinTech); rabbit anti-USMG5 (ProteinTech); rabbit anti-VDAC1 (Abcam). Details of the antibodies are in Supplementary Table S2. α-Tubulin was used as a negative control for mitochondria. Relative protein expression was measured using ImageJ software.
Protein half-life assay
iEndo cells were treated with 100 μg/ml cycloheximide (Sigma) at 37 °C for 0, 2, 4, 6, 8, 10, or 12 h. Cells were dissociated by Accutase, re-suspended with E6, washed with PBS, and counted with 0.4% trypan blue. Every 1 × 104 cell was lysed with 1 μl of RIPA buffer. Each well in SDS-PAGE was loaded with a sample containing 1.6 × 105 cells. Immunoblotting with an anti-TOM7 antibody (ThermoFisher) was used to detect the amount of TOM7. Images were quantified by ImageJ software and analyzed with GraphPad Prism 7. The half-life of TOM7 was calculated by nonlinear regression with exponential curve fit.
Immunofluorescent staining
For mitochondrial morphology, 8000 of iEndo cells were plated on chamber slides (ibidi) precoated with 1 μg/cm2 fibronectin for 24 h. For detecting the localization of myc-TOM7, Flp-in T-Rex cell lines were induced with tetracycline for 72 h before seeding at 8000 per well for 48 h on chambered slides precoated with 1 μg/cm2 poly-l-lysine (MilliporeSigma, A003E). Briefly, cells were fixed with 3.7% formaldehyde for 30 min at RT, washed three times with PBS, and permeabilized with 0.1% TritonX-100 in PBS for 30 min at RT. After blocking with 5% BSA and 20% FBS in PBS for 1 h at room temperature and three additional washes with PBS, cells were incubated overnight at 4 °C with primary antibody diluted in 1% BSA and 20% FBS in PBS. The cells were washed in PBS for 5 min three times. Secondary antibodies were then added to the cells and incubated for 1 h at room temperature. Slides were mounted with Fluoroshield Mounting Medium containing DAPI (Abcam, ab104139). The antibodies used in immunofluorescence were rabbit anti-TOM40 (ProteinTech), mouse anti-Calnexin (ProteinTech), mouse anti-myc (Invitrogen). Images were acquired with FluoView™ FV3000 Confocal Laser Scanning Microscope (Olympus). The z-stacks were acquired at 0.36 mm intervals for a total of approximately 9 mm. Mitochondrial structures were quantified by Mitochondrial Network Analysis (MiNA), an ImageJ macro (github.com/ScienceToolkit/MiNA). Mitochondrial footprint was calculated from the binarized image without skeletonizing and other parameters were calculated from the skeletonized image.
Transmission electron microscopy (TEM)
The iEndo cells were collected on day 6 of differentiation and fixed in a solution containing 2.5% glutaraldehyde and 3 mM CaCl₂ in 0.1 M cacodylate buffer for 1 h at 4 °C. After washing with 0.1 M cacodylate buffer containing 3 mM CaCl₂, the cells were post-fixed for 1 h at room temperature in 0.1 M cacodylate buffer containing 1% osmium tetroxide and 1.5% potassium ferricyanide. The samples were washed in ddH₂O and gradually dehydrated in a graded ethanol series of 70%, 80%, 90%, and 99.5% ethanol, each for 10 min, followed by three rinses in 100% ethanol, each for 20 min. The dehydrated samples were then infiltrated in stages with Spurr resin–ethanol solutions containing 25%, 50%, 75%, and 100% resin, with each stage lasting 1 h. The infiltrated samples were left in 100% Spurr resin overnight. Next, the samples were embedded in molds with fresh resin and polymerized at 70 °C for 48 h. The embedded cells were cut using an ultramicrotome (EM UC7, Leica) into 70 nm ultrathin sections with a diamond knife and post-stained with uranyl acetate and lead citrate. Transmission electron microscopy (TEM) was performed on a JEOL JEM-1400 at 120 keV, equipped with a CCD Camera System Model 895 (Ultrascan, Gatan).
Tube formation assay
Briefly, 2.5 × 104 iEndo cells were seeded in a 96-well plate precoated with 9.3 μg/ml Matrigel (Corning) and allowed to attach for 1 h at 37 °C with 5% CO2. Subsequently, images were acquired every 20 min and maintained at 37 °C with 5% CO2 using a Nikon TE2000-E inverted microscope.
Measurement of senescence
Senescence was examined in iEndo cells by a senescence-associated β-galactosidase assay following the manufacturer's protocol (Cell Signalling). Briefly, 5 × 104 iEndo cells were seeded on a 12-well plate precoated with 1 μg/cm2 fibronectin and cultured overnight. Then, cells were washed with PBS and fixed in 1× Fixative solution for 15 min at RT. Cells were washed in PBS and stained with SA-β-galactosidase staining solution for 3 days at 37 °C. Subsequently, cells were washed with PBS and replaced with 70% glycerol for long-term storage. Stained cells were visualized with an Olympus IX71 microscope.
Oxygen consumption rate measurements for iPSC-derived endothelial cells
For the Seahorse assay, 20 assay wells were divided into the desired number of samples (for example, 5 replicated assay wells per sample for 4 genotypes of iEndo cells). OCR was measured by Seahorse XF24 extracellular flux analyzer (Agilent) with at least three biological replications. OCR was measured by Seahorse XF24 extracellular flux analyzer (Agilent). For iEndo cells, 3 × 104 cells were seeded on an XFe-24 Seahorse plate (Seahorse Biosciences) precoated with 1 μg/cm2 fibronectin (ScienCell) and cultured in TeSR-E6 medium with 2 mg/ml PVA and 20 ng/ml mVEGF-A overnight at 37 °C with 5% CO2. The Seahorse XF24 Sensor Cartridge was hydrated in XF Calibration buffer (Seahorse Biosciences) overnight at 37 °C with 5% CO2. The next day, the culture medium of iEndo cells was exchanged for 675 μl of Seahorse XF DMEM medium (Seahorse Biosciences) freshly supplemented with 2 mM glutamine, 1 mM sodium pyruvate, and 5 mM glucose and equilibrated for 1 h at 37 °C without CO2. After equilibration, the plate with cells was inserted in the Seahorse analyzer, and the OCR was measured for each treatment condition at 37 °C with the following ETC inhibitors: Oligomycin (Sigma) at a final concentration of 3 μM, carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone (FCCP, Sigma) at a final concentration of 1 μM, Rotenone (Sigma) at a final concentration of 3 μM, and Antimycin A (Sigma) at a final concentration of 1.5 μM. For each condition (basal and after each inhibitor injection), cycles were performed with four repetitions of mixing for 3 min, waiting for 2 min, and measuring for 3 min. Data were normalized to the cell numbers that were determined by the hemocytometer when initially seeded on the plate.
Oxygen consumption rate and extracellular acidification rate measurements for iPSCs
OCR was measured by Seahorse XF24 extracellular flux analyzer (Agilent). For iPSCs, 3.5 × 104 cells were seeded on an XFe-24 Seahorse plate (Seahorse Biosciences) precoated with 1 μg/cm2 fibronectin (ScienCell) and cultured in TeSR-E8 medium with 10 μM Y-27632 overnight at 37 °C with 5% CO2. The Seahorse XF24 Sensor Cartridge was hydrated in XF Calibration buffer (Seahorse Biosciences) overnight at 37 °C with 5% CO2. The next day, the culture medium of iPSCs was exchanged for 675 μl of Seahorse XF DMEM medium (Seahorse Biosciences) freshly supplemented with 2 mM glutamine, 1 mM sodium pyruvate, and 5 mM glucose and equilibrated for 1 h at 37 °C without CO2. After equilibration, the plate with cells was inserted in the Seahorse analyser, and the OCR was measured for each treatment condition at 37 °C with the following ETC inhibitors: Oligomycin (Sigma) at a final concentration of 3 μM, carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone (FCCP, Sigma) at a final concentration of 0.5 μM, Rotenone (Sigma) at a final concentration of 3 μM, and Antimycin A (Sigma) at a final concentration of 1.5 μM. For each condition (basal and after each inhibitor injection), cycles were performed with four repetitions of mixing for 3 min, waiting for 2 min, and measuring for 3 min. Data were normalized to the cell numbers that were determined by the hemocytometer when initially seeded on the plate.
Extracellular acidification rate (ECAR) was measured by Seahorse XF24 extracellular flux analyser (Agilent). For iPSCs, 3.5 × 104 cells were seeded on an XFe-24 Seahorse plate (Seahorse Biosciences) coated with 1 μg/cm2 vitronectin (STEM CELL) and cultured in TeSR-E8 medium with 10 μM Y-27632 overnight at 37 °C with 5% CO2. The Seahorse XFe24 Sensor Cartridge was hydrated in XF Calibration buffer (Seahorse Biosciences) overnight at 37 °C with 5% CO2. The next day, the culture medium of iPSCs was changed to 675 μl of Seahorse XF DMEM medium (Seahorse Biosciences) freshly supplemented with 2 mM glutamine and equilibrated for 1 h at 37 °C without CO2. After equilibration, ECAR and OCR were measured with the following treatments: 10 mM d-Glucose (Sigma), 3 μM Oligomycin (Sigma), and 100 mM 2-Deoxy-d-glucose (Sigma). For each condition, 4 cycles of 3-min mixing, 2-min waiting, and 3-min measuring were performed. Data were normalized to the cell numbers that were determined by hemocytometer when initially seeded on the plate.
Oxygen consumption rate measurements for iPSC-derived fibroblasts
OCR was measured by Seahorse XF24 extracellular flux analyzer (Agilent). For iPSC-derived fibroblasts, 3 × 104 cells were seeded on an XFe-24 Seahorse plate (Seahorse Biosciences) and cultured in DMEM with 10% FBS overnight at 37 °C with 5% CO2. Seahorse XF24 Sensor Cartridge was hydrated in XF Calibration buffer (Seahorse Biosciences) overnight at 37 °C without CO2. The next day, the culture medium of iFibro cells was exchanged for 675 μl of XF DMEM Medium, pH 7.4 (Agilent) containing 2 mM glutamine, 1 mM sodium pyruvate, and 10 mM glucose, then equilibrated for 1 h at 37 °C without CO2. After equilibration, the plate with cells was inserted in the Seahorse analyzer, and the OCR was measured for each treatment condition at 37 °C with the following ETC inhibitors: Oligomycin (Sigma) at a final concentration of 2 μM, carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone (FCCP, Sigma) at a final concentration of 1 μM, Rotenone (Sigma) at a final concentration of 2 μM, and Antimycin A (Sigma) at a final concentration of 2 μM. For each condition (basal and after each inhibitor injection), cycles were performed with four repetitions of mixing for 3 min, waiting for 2 min, and measuring for 3 min. Data were normalized to the cell numbers that were determined by the hemocytometer when initially seeded on the plate.
Mass spectrometry and analysis
Mitochondria from iEndo were enriched on day 6 of endothelial differentiation as described above and dissolved in Reduction Buffer (8M urea (Sigma), 2 mM DTT, and 25 mM ammonium bicarbonate (NH4HCO3, Sigma)). Initially, 17.5 mg of mitochondrial proteins were prepared in 25 ml of Reduction Buffer and incubated for 1 h at 37 °C to reduce cysteine residues. Cysteines were then alkylated with 15 mM iodoacetamide (Sigma) for 1 h at RT in the dark. Additional DTT was added to a final concentration of 5 mM to quench alkylation. Subsequently, 0.36 μg of trypsin (Promega) was added for proteolytic digestion with a protein-to-trypsin ratio of 50:1 in 40 mM NH4HCO3 overnight at 37 °C. The next day, proteolysis was stopped by acidifying the samples with 0.2% formic acid (Sigma). The acidified peptides were desalted and concentrated using a 100-μl C18 tip, as described previously (Pierce). Peptides were dissolved in 50% acetonitrile (ACN, Sigma)/0.1% formic acid (FA, Sigma) and dried in a Speed Vac (Thermo) for 5–6 h at RT. The dried peptides were stored at −80 °C. The mass spectrum was performed with high-resolution Orbitrap CE/LC-MS in the core facility of the Instrument Center of National Cheng Kung University. The dried peptides were re-dissolved in 0.1% FA in ddH2O for LC-MS/MS analysis. Data-dependent acquisition (DDA) was acquired using the Ultimate 3000 RSLCnano system (Thermo Fisher Scientific) coupled with a Q Exactive Plus Hybrid Quadrupole-Orbitrap Mass Spectrometer (QE-plus, Thermo Fisher Scientific).
For liquid chromatography, 4 μl of peptides (0.1 μg/μl) were separated at a flow rate of 4 μl/min on a trap column (3 μm, 100 Å C18, 75 μm × 2 cm) for 60 min in 2% ACN and 0.1% FA in ddH2O and were then separated at a flow rate of 300 nl/min along a 60 min non-linear LC gradient on analytical column (2 μm, 100 Å C18, 75 μm × 25 cm). The LC gradient consisted of buffer A (0.1% formic acid in ddH2O) and buffer B (80% acetonitrile and 0.1% formic acid in ddH2O). Peptides were eluted using the following gradients: 5% of buffer B for 5 min (0–5 min), 5–16% buffer B for 3 min (5–8 min), 16–42.5% of buffer B for 27 min (8–35 min), 42.5–99% of buffer B for 2 min (35–37 min), wash out at 99% buffer B for 8 min (37–45 min), decreased to 5% of buffer B for 2 min (45–47 min), and 5% of buffer B for 13 min (47–60 min).
For mass spectrometer acquisition, eluted peptides were directly ionized at a potential of +1.9 kV into a QE-plus. MS1 full scans were acquired at a resolution of 70,000 at m/z 200 in the Orbitrap and ion funnel radio frequency (RF) setting of 50. The automatic gain control (AGC) target value was set to 3E6 charges and a maximum injection time (IT) of 200 ms. Dynamic exclusion was applied with a repeat count of 1 and an exclusion time of 10 s. The 10 most intense precursor ions (charge-state 2+ and higher) were selected using an AGC target value of 1E5 charges followed by fragmentation by higher-energy collision-induced dissociation (HCD) at normalized collision energy (NCE) of 35%. MS2 scans were acquired at a resolution of 17,500 at m/z 200 in the Orbitrap, a maximum IT of 50 ms, and an isolation window of 1.6 m/z. Raw files were converted into Mascot generic files (MGFs) using MSConvert. MGF files were searched using Mascot (version 2.8.0.1) against the Swiss-Prot part of the UniProt Knowledgebase (UniProtKB/Swiss-Prot) and decoy databases. Protein identification was performed by Mascot with the following settings: trypsin digestion, two allowed missed cleavages, variable modifications of Oxidation (M) and deamidated (NQ), peptide mass tolerance of 10 ppm, and fragment mass tolerance of 0.05 Da. Quantification analysis was done by LC-MS/MS with parallel reaction monitoring or selective ion monitoring. The exponentially modified protein abundance index (emPAI) of the identified protein was used for proDA (rdrr.io/bioc/proDA) to obtain the significance of differentially abundant proteins. The names of the proteins were converted to official gene names for gene set enrichment analysis analyses (GSEA).
RNA sequencing
Total RNA was extracted from iPSCs, iPSC-derived endothelial cells, or zebrafish brains using an Allprep DNA/RNA kit (QIAGEN). The mRNA libraries were prepared with TruSeq Standard mRNA Library Prep Kit (Illumina) and sequenced with 2 × 75 pair-end sequencing reaction with Illumina NextSeq500 system. For human samples, sequences were aligned to GRCh38 with HISAT2 (daehwankimlab.github.io/hisat2), and uniquely mapped reads were counted with HTSeq (www-huber.embl.de/HTSeq) and annotated with GTF file from ENSEMBL (Homo_sapiens.GRCh38.99). For zebrafish samples, sequences were aligned to GRCz10 with HISAT2, and uniquely mapped reads were counted with HTSeq, and annotated with GTF file from ENSEMBL (Danio_rerio.GRCz10.91) and converted to human orthologous genes. Differentially expressed genes were identified with DESeq2 (bioconductor.org/packages/release/bioc/html/DESeq2.html). Genes with less than 10 reads in all samples were not included in further analyses. Differential expression of genes and gene sets in probands when compared with control were analyzed with DESeq2. GSEA was performed with the enrichGO function from the clusterProfiler package (guangchuangyu.github.io/software/clusterProfiler). Enrichment in other ontologies or pathways in the Molecular Signatures Database v7.0 (software.broadinstitute.org/gsea/msigdb) was also analyzed with the GSEA function from the clusterProfiler package.
CRISPR/Cas gene editing in zebrafish
Adult AB line Zebrafish (Danio rerio) and embryos were maintained under standard conditions and kept on a light-dark cycle of 14 h in light and 10 h in dark at 27 °C in the Zebrafish Core Facility of National Cheng Kung University. Zebrafish carrying Tg (fli1:EGFP) was purchased from the Taiwan zebrafish core facility at the National Health Research Institute. The gene editing was done by injecting ribonucleotide protein (RNP) consisting of Alt-R HiFi S. pyogenes Cas9 nuclease (IDT) and crRNA:tracrRNA duplex into the 1-cell embryo.35 The potential gRNA target sites were identified with CRISPOR (crispor.tefor.net). The gRNA sequences used in this study are listed in Supplementary Table S1. To form RNP, chemically synthesized Alt-R-modified target-specific crRNAs and common tracrRNAs were synthesized (IDT) and dissolved in duplex buffer (IDT) to make 100 μM stock solution. To get 50 μM of crRNA:tracrRNA duplex guide RNA (dgRNA), equal volumes of 100 μM Alt-R crRNA and 100 μM Alt-R tracrRNA were mixed and annealed by the following conditions: 95 °C for 5 min, cool gradually at 0.1 °C/s to 25 °C, 25 °C for 5 min, and cool down to 4 °C rapidly. Then, 50 μM dgRNA was diluted into 25 μM with duplex buffer and stored at −80 °C. On the day of microinjection, 5 μM of Cas9 protein (Alt-R™ S.p. HiFi Cas9 Nuclease V3, IDT) was freshly complexed with 5 μM dgRNA for 10 min at 37 °C and then placed at room temperature. For injection, 2.3 nl of reaction mix containing 5 μM gRNA:Cas9 RNP complex was injected into the one-cell stage embryo from Tg (fli1a:EGFP) zebrafish. Injected mosaic zebrafish were crossed with wild-type (WT) fish to obtain heterozygous F0 generation. Genotyping was done by targeted sequencing. Knockout zebrafish was obtained by monohybrid intercross of heterozygous F1 generation.
Genotyping zebrafish
Genomic DNA was extracted from individual dechorionated embryos, larvae, or adult tail biopsies. Samples were incubated with 30 μl of 50 mM NaOH and heated for 20 min at 95 °C, then neutralized with 3 μl of 1M Tris–HCl (pH 8.0) and centrifuged at 2400 rpm for 15 min at 4 °C. The supernatant containing the genomic DNA was kept for genotyping. Amplicons with tomm7 gRNA targeting sites were amplified by Taq DNA polymerase (OnePCR, GeneDireX) and sequenced by next-generation sequencing. Primers for PCR are listed in Supplementary Table S1. Mosaic tomm7 zebrafish were crossed with wild-type fish to obtain heterozygous F0 fish. In all experiments, homozygous tomm7-deleted zebrafish were obtained by monohybrid intercross with heterozygous tomm7-deleted F1.
Visualizing vasculature in zebrafish embryos
Zebrafish embryos were anesthetized in 0.003% tricaine (3-amino benzoic acid ethyl ester, Sigma) and mounted in 0.5% agarose before observing under a fluorescence microscope. PTU (1-phenyl-2-thiourea, Sigma) was added in E3 water at 24 hpf to inhibit body pigmentation and facilitate observation of the vascular structure of fish embryos. Images were acquired with FluoView™ FV3000 Confocal Laser Scanning Microscope (Olympus). The z-stacks were acquired at 5-μm intervals for a total of approximately 200 μm. The diameter of the six intersegmental vessels from individual embryos was measured using ImageJ software.
Alcian blue staining
Zebrafish embryos were euthanized with an overdose of tricaine at 4 dpf. Embryonic tails were used for DNA extraction for genotyping, while the rest of the anterior tissues were fixed with 3.7% formaldehyde overnight at 4 °C. Fixed samples were washed three times with 0.1% PBS-T (0.1% Tween-20 in PBS) and stained with 0.1% Alcian blue solution (w/v) (Sigma) in 20% (v/v) acetic acid and 80% (v/v) ethanol for 3 h. After staining, the embryos were rinsed in a series of ethanol/PBS solutions, starting with 80% ethanol/PBS and then 50% ethanol/PBS and 30% ethanol/PBS. Soft tissues were then bleached with 3% H2O2 and 1% KOH for 3 h and cleared with 0.25% trypsin for 3 days at room temperature. Samples were washed in PBS for 15 min 3 times and rinsed with a series of PBS/glycerol solutions, starting from 70% PBS/30% glycerol, then 50% PBS/50% glycerol, and 30% PBS/70% glycerol. Stained samples were stored in 70% glycerol in PBS and visualized with an Olympus IX71 microscope. The width and length of the cranial cartilage were measured using ImageJ software.
Oxygen consumption rate measurements for zebrafish
OCR of 48-hpf embryos was measured by Seahorse XFe24 extracellular flux analyser (Agilent). Before assays, embryos were maintained under standard conditions until they reached 48 hpf. Seahorse XFe24 Sensor Cartridge was hydrated in XF Calibration buffer (Seahorse Biosciences) overnight at 28.5 °C. During the assay, embryos were incubated in E3 water with 0.003% tricaine (Sigma) to minimize embryo movement in the central position of the islet capture microplate (Seahorse Biosciences) in each well. On the day of the assay, a 200 μl blunt pipette tip was used to place 50 μl of E3 water with 0.003% tricaine containing a 48-hpf embryo in the central position of the islet capture microplate (Seahorse Biosciences) in each well. After that, the screens were inserted into the wells, and an additional 625 μl of E3 water with 0.003% tricaine was added for a total volume of 675 μl. After equilibration, the plate with embryos was inserted in the Seahorse analyzer, and OCR was measured for each treatment condition at 28.5 °C with the following ETC inhibitors: oligomycin (Sigma) at a final concentration of 40 μM, carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone (FCCP, Sigma) at a final concentration of 8 μM, Rotenone (Sigma) at a final concentration of 1.5 μM, and Antimycin A (Sigma) at a final concentration of 1.5 μM. A total of 30 cycles were conducted in the assays, with each cycle consisting of 1.5 min of mixing, followed by 2 min of waiting, and 3 min of measuring. Basal respiration was determined using 8 cycles. Subsequently, 4 cycles of measurements were conducted after the first injection of oligomycin, followed by an additional 4 cycles of measurements after the second oligomycin injection. Additionally, 7 cycles of measurements were conducted following the injection of FCCP, and finally, 7 cycles of measurements were conducted after the injection of Rotenone/Antimycin A.
Statistics
Statistical analyses were performed using GraphPad Prism v.8.4.3. Data are presented as mean ± SEM from at least three independent experiments. Statistical analyses used in the study include nonlinear regression with one-phase exponential decay fit and two-way ANOVA followed by Dunnett post hoc test for protein half-life assay; Kruskal–Wallis test with Dunn's post hoc test for multiple comparisons for quantification of tube formation assay, mitochondrial morphology, OCR measurements, as well as zebrafish brain size, body length, and cartilages measurement; Friedman test followed by Dunn's post hoc test for multiple comparisons for immune blotting, complex activity assay, qPCR, as well as zebrafish ISV diameter measurement; Mann–Whitney U-tests for PER measurements and ATP production rate measurements; chi-square goodness-of-fit test for zebrafish F1 intercross. For visual inspection of histograms and Q-Q plots with noticeable deviations from normality, non-parametric analyses were used to ensure accurate results (Supplementary Data 2a). Unless stated otherwise, all comparisons were shown between groups. Differences with P < 0.05 were considered significant. No statistical methods were used to predetermine the sample size. The experiments were not randomized, and the investigators were not blinded.
Role of funders
Funders had no role in study design, data collection, data analyses, interpretation, or writing of reports.
Results
Clinical features of patients
Previously, we identified three unrelated Taiwanese probands (1-III, 2-III, and 3-III) that developed hyperopia and atrophic macular scarring.36 The probands also have similar craniofacial features, including microcephaly and micrognathia, short stature and developmental delay (Fig. 1a, Table 1). Proband 1-III is a 10-year-old girl who, at 4 years old, presented to the pediatric emergency room with acute onset right upper limb weakness, showing a muscle power of 2 with decreased deep tendon reflex but without any sensory deficit. The right upper limb weakness improved gradually the next day, but a second episode occurred after 3 days with the manifestations of right central facial palsy, dysarthria, and dysphagia. Stroke was considered, and brain magnetic resonance imaging (MRI) revealed left parietal cytotoxic edema and bilaterally discontinued internal carotid to middle cerebral arteries, compatible with moyamoya disease (Fig. 1b). The neurological symptoms fully recovered after intravenous fluid supplement, and she was referred to a neurosurgeon for surgical intervention. Further neurocognitive examination showed that the proband had a mild intellectual disability requiring special education. Considering the phenotypes of failure to thrive, short stature, dysmorphic face, and moyamoya disease, microcephalic osteodysplastic primordial dwarfism type II (MOPD-II) was suspected initially.37, 38, 39 However, no pathogenic variants were identified in the PCNT gene. At the age of 10, she measured 110 cm (−4.5 standard deviation (SD)) in height and exhibited microcephaly (occipital frontal circumference (OFC) was 50 cm, −1.88 SD), micrognathia, alopecia partialis, and aged-looking skin on her hands (Table 1, Fig. 1a and c, and Supplementary Figure S2a). Protruding of maxillary central incisors and hair loss were more obvious when compared to those 5 years ago.36 Recently, her youngest sister (patient 1-V) was diagnosed with macular scarring at the age of 2 (Supplementary Figure S2b). Although patient 1-V has normal facial features and normal height (91 cm, −1.9 SD) at the age of 3, delayed neurodevelopment had been noticed (Table 1).
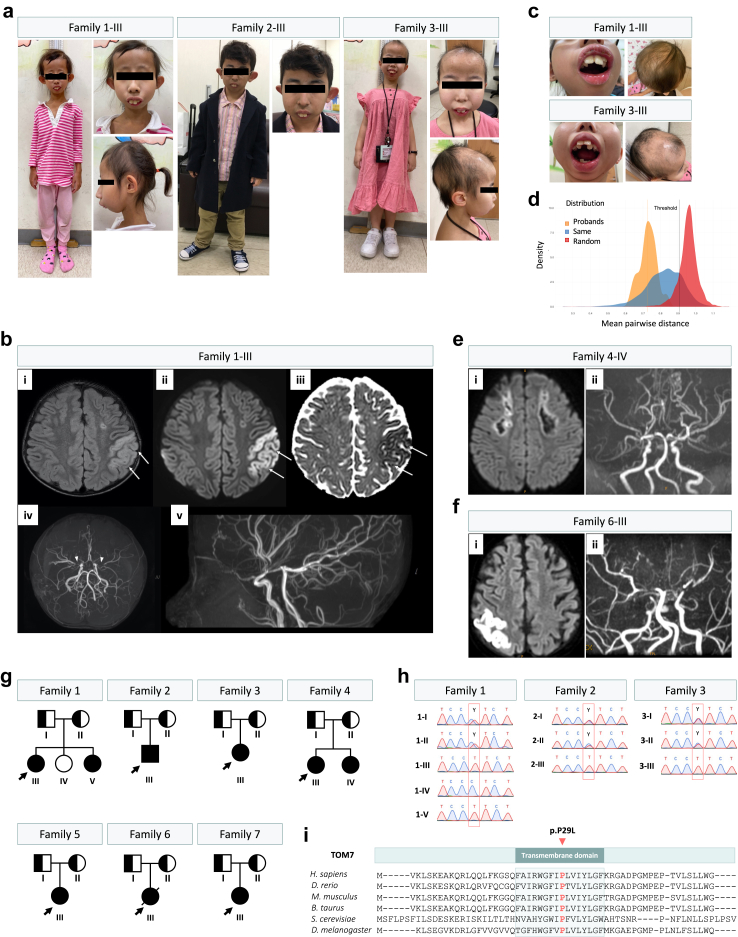
Fig. 1.
Recurrent TOMM7 p.P29L variant in four patients from three unrelated families. (a) Short stature and unique facial features, including low set and protruding ear, beaked nose, and micrognathia in four patients from three families. (b) The brain magnetic resonance imaging (MRI) study of patient 1-III revealed cytotoxic edema of the left parietal lobe (arrows), as shown by T2 hyperintensity (i), high signals on diffusion-weighted imaging (ii), and low apparent diffusion coefficient values (iii), Magnetic resonance arteriography (MRA) indicated bilateral occlusion of distal internal carotid arteries (ICA) and interrupted proximal middle cerebral arteries (MCA) ((iv), arrowheads) and abundance collateral vasculatures (v), compatible with Moyamoya disease. (c) Protruding maxillary central incisors, crowded and irregular teeth, hair thinning with obvious hair loss. (d) Analysis of facial similarity among the patients. Three distributions of mean pairwise distance were shown: our patients (orange), the random selection from the subjects with 328 disorders (red), and the selection from subjects with the same disorder (blue). The orange vertical line indicated the mean pairwise distance of the patients, and the black vertical line indicated the threshold that classifies whether it was from the same disorder or random selection. The entire (100%) distribution of patients was below the threshold. (e) The brain MRI of patient 4-IV showing confluent areas of encephalomalacia in bilateral frontal white matter and right anterior and superior frontal gray matter, suggesting an old border-zone infarction. The periphery of the lesions still showed mild diffusion restriction (i). Time-of-flight MRA revealed severe narrowing of distal ICA to proximal anterior cerebral artery (ACA) and MCA bilaterally with prominent basal collateral vessels, compatible with Moyamoya disease. The M2s and A2s can be seen. The posterior cerebral arteries (PCAs) were prominent with collaterals. Moyamoya disease with bilateral cerebral old infarctions and impaired cerebral perfusion, was more on the right side (ii). (f) The brain MRI of patient 6-III showing abnormal high signal intensity on T2WI at both parietal, both frontal cortical, subcortical areas, left temporal subcortical area, left parietal central parenchyma. Limited diffusion coefficients were evident at the right parietal cortical, left parietal central parenchyma, left temporal subcortical area. Recent ischemic changes were suspected at both parietal and left temporal areas (i). Remarkably decreased flow signals at both supraclinoid internal carotid artery, right petrous, cavernous internal carotid artery, both proximal MCAs and ACAs as well as left proximal PCA (ii). (g) Family pedigree of seven unrelated families in this study. (h) Confirmation of TOMM7 p.P29L variant by Sanger sequencing. (i) Proline 29 locates on the highly conserved transmembrane domain of TOM7 protein (NP_061932.1).
Table 1.
Clinical features.
| Patient | 1-III | 1-V | 2-III | 3-III | 4-III | 4-IV | 5-III | 6-III | 7-III | Summary |
|---|---|---|---|---|---|---|---|---|---|---|
| TOMM7 c.86T>C; p.P29L | Homozygous | Homozygous | Homozygous | Homozygous | Homozygous | Homozygous | Homozygous | Homozygous | Homozygous | − |
| Sex | Female | Female | Male | Female | Female | Female | Female | Female | Female | Male, 11% (1/9); Female, 89% (8/9) |
| Age (years) | 10 | 3 | 23 | 19 | 13 | 10 | 6 | 11 | 11 | |
| Age at presentation (years) | 4 | 2 | 5 | 5 | 6 | 6 | 2 | 3 | 4 | |
| Height (cm) | 110 (−4.5 SD) | 91 (−1.9SD) | 117.5 (−9 SD) | 106.3 (−6.7 SD) | 125 (−4.6 SD) | 95 (−7.3 SD) | 91 (−6.0 SD) | 115.5 (−4.1 SD) | 104 (−6.0 SD) | Short stature 100% (9/9) |
| Low body weight | + | + | + | + | + | + | + | − | NA | 89% (8/9) |
| Initial presenting symptoms | Right hemiparesis | Visual impairment | Nystagmus, exotropia | Nystagmus, exotropia | Blurred vision | Left hemiparesis | Left hemiparesis | Blurred vision | Short stature, visual impairment | − |
| Craniofacial features | ||||||||||
| OFC (cm) | 50 | 49 | 49 | 50 | NA | NA | NA | NA | NA | − |
| Micrognathia | + | + | + | + | + | + | + | + | + | 100% (9/9) |
| Sparse hair | + | + | − | + | − | + | + | − | + | 67% (6/9) |
| Microcephaly | + | − | + | + | + | + | − | + | + | 78% (7/9) |
| Dental crowding | + | − | − | + | − | + | − | − | + | 44% (4/9) |
| Prominent ears | − | − | + | − | − | − | − | + | − | 22% (2/9) |
| Hypertelorism | − | − | − | − | − | − | − | + | − | 11% (1/9) |
| Broad forehead | + | − | − | + | + | − | − | − | + | 44% (4/9) |
| Triangular face | + | + | − | − | − | − | − | − | − | 22% (2/9) |
| Bulbous nose | − | − | − | − | − | − | + | − | − | 11% (1/9) |
| Depressed nasal bridge | − | − | − | − | − | − | − | + | − | 11% (1/9) |
| Deformities | ||||||||||
| Narrow chest | + | − | − | + | − | + | + | + | + | 67% (6/9) |
| Pectus carinatum | − | − | − | − | − | + | − | − | − | 11% (1/9) |
| Short limbs | − | − | − | − | − | − | − | + | − | 11% (1/9) |
| Stubby fingers | + | + | + | + | − | − | − | − | − | 44% (4/9) |
| Dystrophic nails | + | − | + | − | − | + | + | − | − | 44% (4/9) |
| Acetabular dysplasia | − | − | − | − | − | + | + | + | − | 39% (3/9) |
| Genu valgum | − | − | − | − | − | + | − | − | − | 11% (1/9) |
| Coxa valga | − | − | − | + | + | + | + | + | + | 67% (6/9) |
| Kyphosis | + | − | − | − | − | + | + | + | − | 44% (4/9) |
| Scoliosis | + | − | − | + | − | − | − | + | + | 44% (4/9) |
| Rib deformities | − | − | − | − | − | − | − | + | + | 22% (2/9) |
| Other | NA | NA | NA | NA | Bone cyst at right femur | Prominent joint | Cafe-au-lait spot | Bilateral single transverse palmar crease | NA | − |
| Cognition | ||||||||||
| Cognition impairment | + | + | − | − | − | − | + | − | + | 44% (4/9) |
| Cognition/Development | Mild cognitive impairment | Delayed milestones | NA | Normal | Normal | Normal | Delayed milestones | Normal | Delayed language and cognitive skills | − |
| Brain MRI | ||||||||||
| Infarction | + | − | NA | − | − | + | + | + | − | 50% (4/8) |
| Moyamoya disease | + | − | NA | − | + | + | + | + | − | 63% (5/8) |
| Moyamoya diseaseonset age | 4 | NA | NA | NA | 13 | 6 (OP at age 7 years) | 2 | 12 (Died at age 12 years) | NA | − |
| Other intracranial lesions | NA | NA | NA | NA | Thin corpus callosum | Encephalo- malacia | NA | NA | Brain cyst was told by 2 years old | − |
| Ophthalmological presentation | ||||||||||
| Hyperopia | + | + | + | + | + | − | + | − | − | 67% (6/9) |
| Maculopathy | + | + | + | + | + | + | − | − | + | 78% (7/9) |
| Nystagmus | + | − | + | + | + | − | − | − | − | 44% (4/9) |
| Strabismus | + | − | + | + | − | − | − | + | − | 44% (4/9) |
| Amblyobia | + | + | + | + | + | − | + | − | − | 67% (6/9) |
| Optic atrophy | − | − | − | − | − | − | + | + | − | 22% (2/9) |
| Visceral abnormalities | ||||||||||
| Hepatosteatosis | − | NA | NA | NA | NA | NA | NA | − | NA | 0% (0/2) |
| Hepatomegaly | − | − | NA | NA | NA | NA | NA | − | NA | 0% (0/3) |
| Renal echo abnormalities | − | − | NA | NA | NA | NA | NA | − | NA | 0% (0/3) |
| Cardiomegaly | − | − | NA | NA | + | − | − | + | − | 29% (2/7) |
| Lipodystrophy-like appearance | + | − | NA | − | − | − | − | − | − | 13% (1/8) |
| Advanced bone age | − | + | NA | − | − | − | − | + | − | 25% (2/8) |
| Delayed bone age | − | − | NA | − | − | + | − | − | − | 13% (1/8) |
| Delayed puberty | NA | NA | NA | − | NA | NA | NA | NA | − | 0% (0/2) |
Proband 2-III was born with normal birth weight. At the age of 2, he showed failure to thrive despite normal endocrine levels. He developed nystagmus and deviated eyes at the age of 5. At the age of 23, he had an extremely short stature (117.5 cm, −9 SD) and microcephaly (OFC was 49 cm, −4.2 SD). He also had facial features of maxillary protrusion, receding chin, prominent ears, dystrophic nails, and stubby fingers (Table 1, Fig. 1a, and Supplementary Figure S2a).
Proband 3-III is a 19-year-old female who was born at full term to non-consanguineous parents with an uneventful antenatal history except intrauterine growth retardation with a body weight of 2.56 kg and a body length of 44 cm at birth. She had exodeviation and nystagmus at the age of 5. She presented hair loss and alopecia since she was 10 years old. Before visiting the endocrine clinic, she was karyotyped 46XX and tested for progeria or progeria-like syndromes. However, variant analyses were negative for LMNA, ERCC8, and ERCC6 genes. At the age of 19 years, she measured 106.3 cm (−6.7 SD) in height and exhibited microcephaly (OFC was 50 cm, −3.2 SD), micrognathia, alopecia partialis, a thin and upturned upper lip, and a small thorax (Table 1, Fig. 1a and c). Whole body bone X-ray showed bowed humeral bones and metaphyseal dysplastic changes (Supplementary Figure S2c). A provocative test showed an adequate growth hormone surge, and serum IGF-1 was high (589.58 ng/ml; reference range 140∼496 ng/ml; Supplementary Table S3). Further brain imaging revealed an enlarged pituitary gland.
Facial analysis showed a clear similar facial phenotype among patients from families 1, 2, and 3.31 All the distribution falls below the threshold, indicating that the distribution of the four patients differs from the random selection of images from 328 disorders (Fig. 1d). Moreover, the image of the 2-year-old patient 1-V is relatively not that similar to the other four images, indicating an age-related progression in facial features (Supplementary Figure S2a). Indeed, when we compared the distances of images of proband 1-III at 10 years old (images 1-III) and at 5 years old (image 1-III-2) to the other three images, pictures of 2-III and 3-III have smaller distances to 1-III than to 1-III-2. The picture of 1-V is more similar to that of 1-III-2 (distance of 0.69) than to 1-III (distance of 0.77) (Supplementary Figure S2b). These results indicate that the patients present a clearer and similar facial phenotype when they are older.
Recently, five more patients with similar clinical features, including moyamoya disease, were identified in multiple medical centers in Taiwan. Proband 4-III is a 13-year-old female who first presented at age 6 with blurred vision. She has a remarkable growth deficit, recorded at −4.6 SD below the norm, and continues to struggle with low body weight. Clinically, she exhibits micrognathia, microcephaly, and a broad forehead. She had coxa valga and a bone cyst at her right femur noted on the X-ray, and otherwise, no obvious skeletal deformities. Ophthalmological presentation includes hyperopia, maculopathy, and nystagmus. Cardiomegaly was depicted on a plain chest film (Table 1). Patient 4-IV, the younger sister of patient 4-III, is a 10-year-old girl who initially presented with left hemiparesis at age 6. Brain MRI depicted multiple infarctions, encephalomalacia of the previous infarcted sites, and severe narrowing of several intracranial main arteries, which is compatible with moyamoya disease (Fig. 1e). She underwent encephaloduroarteriosynangiosis (EDAS) at age of 7. Similar to her elder sister, she is significantly shorter than her peers, with a stature of −7.3 SD, and remains underweight. Her craniofacial features include micrognathia, sparse hair, microcephaly, and dental crowding, and she had skeletal deformities including a narrow chest, pectus carinatum, acetabular dysplasia, coxa valga, and kyphoscoliosis (Table 1).
Proband 5-III exhibited left hemiparesis at 2 years of age. Brain MRI demonstrated infarctions and arterial narrowing, suggesting moyamoya disease. Her craniofacial features include a beaked nose, relative micrognathia, and sparse hair. Her skeletal deformities include a narrow chest, acetabular dysplasia, coxa valga, and kyphoscoliosis. Nail dysplasia and some café-au-lait macules were also noticed. Her growth was severely affected, with a height of −6 SD by the age of 6 years, and she also had a global developmental delay (Table 1).
Proband 6-III was a girl who initially presented at 3 years old with blurred vision, at which ophthalmologic examination revealed optic atrophy, maculopathy, and strabismus. She was short for her age at −4.1 SD but maintained a normal weight. Further assessments showed micrognathia, microcephaly, depressed nasal bridge, hypertelorism, and prominent ears. She had skeletal deformities with a narrow chest, short limbs, acetabular dysplasia, coxa valga, rib deformities, and kyphoscoliosis. Cardiomegaly was visualized on a chest film (Table 1). At age 12, she presented left hemiparesis, and a brain MRI depicted multiple infarctions and severe vascular narrowing and occlusion, consistent with moyamoya disease (Fig. 1f). She received ERAD afterward, but unfortunately, she succumbed during the postoperative period due to complicated pneumonia, pneumothorax, and massive hemothorax complicated by cardiopulmonary failure.
Proband 7-III is an 11-year-old female, born prematurely at 32 weeks gestation, exhibiting stunted growth and developmental delays. At birth, she weighed 2200 g and measured 48 cm in length. Currently, she has a height of 104 cm (<6 SD) and a weight of 16 kg. Physical examination revealed dysmorphic features, including microcephaly, broad forehead, dental crowding, webbed neck, micrognathia, and spare hair, along with skeletal anomalies, such as a narrow chest and slight scoliosis. Ophthalmological examination confirmed congenital macular dystrophy, contributing to her visual impairment. MRI imaging of the brain identified cystic lesions. Developmentally, the patient exhibited delayed language and cognitive skills (Table 1).
Exome sequencing identifies recurrent homozygous missense variant in TOMM7
We performed trio-based exome sequencing to identify the disease-causing gene in the patients. No pathogenic variants in genes involved in MOPD-II or progeria syndrome were identified. No moyamoya-associated SNPs were identified, either.40 Instead, we identified a recurrent homozygous missense variant in the gene encoding for the translocase of outer mitochondrial membrane 7 (TOMM7) (NC_000007.14:g.22822694G>A; NM_019059.5:c.86C>T; NP_061932.1:p.P29L) in all 9 patients (Fig. 1g and h, Supplementary Figure S3a). This variant has been reported in a patient with some overlapped phenotypes.19 The parents of the probands are all heterozygotes, and the unaffected sibling of proband 1-III (1-IV) is homozygous for the common allele (Fig. 1g and h). The youngest sister of proband 1-III (1-V) also carries homozygous TOMM7 p.P29L variant (Fig. 1g and h), supporting the segregation and autosomal recessive inheritance of TOMM7 p.P29L variant among the families.
The allele frequency (AF) of TOMM7 c.86C>T is 0.000008055 in gnomAD (v4.1.0). The AF is 0.00132 in the Taiwan Biobank database. Analysis of single nucleotide variants from exome sequencing showed increases in homozygosity in probands compared to controls (Supplementary Figure S3b). TOMM7 is located in one of the two shared stretches of homozygous regions identified among the probands from families 1, 2, and 3 (Supplementary Figure S3c).29 In addition, increased numbers and length of runs of homozygosity in the probands also hint at the possibility of consanguinity despite none of the three families being aware of common ancestors (Supplementary Figure S3b).41 Rare homozygous missense or loss of function variants in TOMM7 were present in 0.0251% of individuals in all genetic ancestry groups, according to gnomAD. The identification of the homozygous TOMM7 c.86C>T variant in our patients, including 2 sibling pairs and 5 singletons, reached genome-wide significance when compared to general population (P = 2.5072 × 10−23; SORVA with Bonferroni correction).30 With the extremely low incidence reported worldwide, the fact that we were able to identify 9 patients in 7 unrelated families correlated with the higher AF of this TOMM7 variant in the Taiwanese population and indicated a founder effect.
Partial cellular mislocalization of TOM7P29L
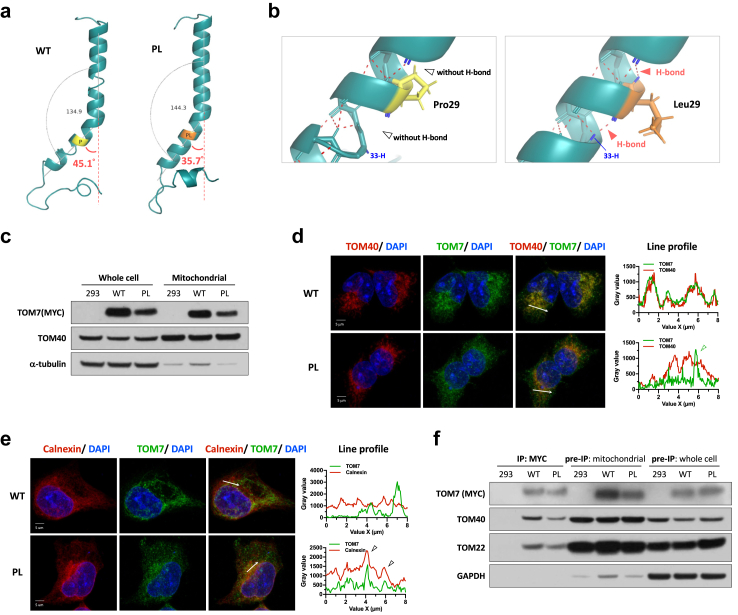
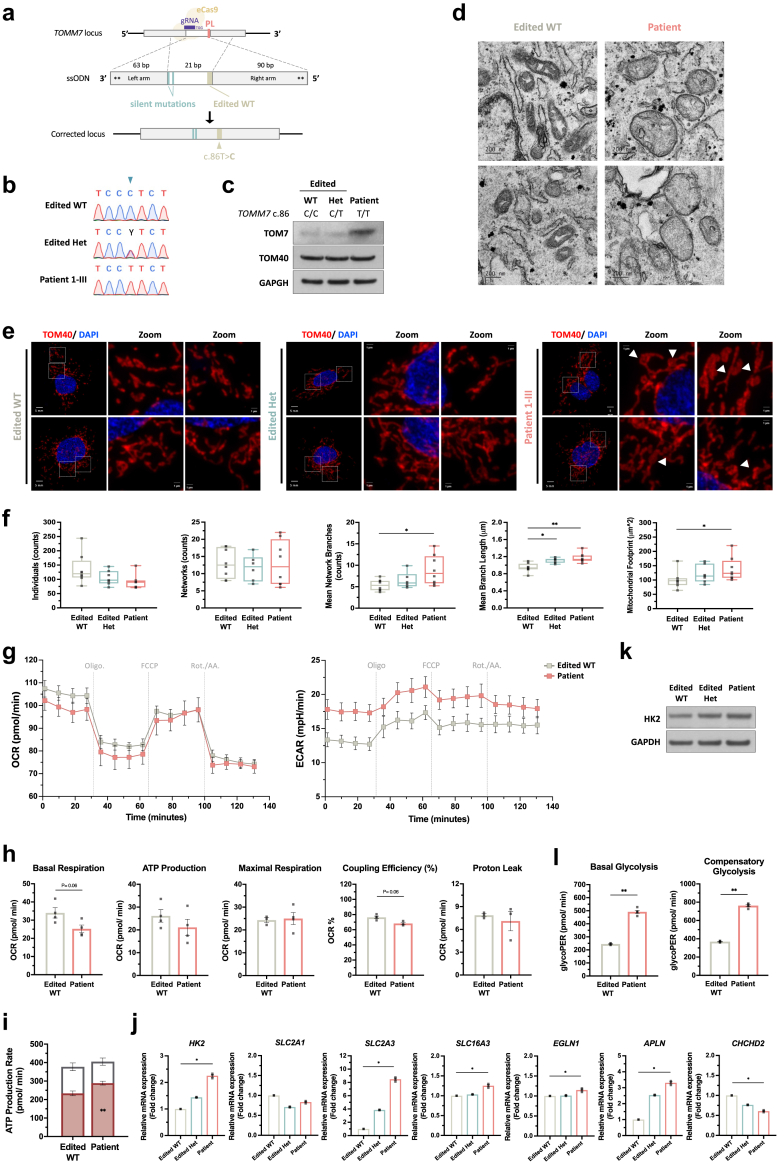
The proline residue29 in the TOM7 transmembrane domain is evolutionarily conserved (Fig. 1i) and is required to generate a kinked structure in the TOM7.42 In yeast, p.P29L led to the mislocalization of tom7 to ER.42 The predicted protein structure of TOM7 indicated that the p.P29L variant might cause conformational changes with a predicted smaller angle of the α-helix (Fig. 2a) and a predicted increased number of hydrogen bonds that may affect the stability of the α-helix of TOM7 (Fig. 2b). TOM7 p.P29L protein (TOM7P29L) was able to target mitochondria in the Flp-In TRex 293T cells expressing N-terminal myc-tagged TOM7P29L (Fig. 2c). However, a portion of TOM7P29L did not colocalize with mitochondria (Fig. 2d). Like yeast, we also observed partial colocalization of TOM7P29L and ER in human cells (Fig. 2e). Human TOM7 has been shown to form an import intermediate with TOM40 and then a stable TOM complex with TOM22.44 In mitochondria-enriched lysates, TOM7P29L can physically interact with TOM40 and TOM22 (Fig. 2f). These results suggest that TOM7P29L does not impair the TOM complex biogenesis on the OMM. Taken together, we showed that, although a portion of TOM7P29L did not target mitochondria, some TOM7P29L could form TOM complex with other TOM member proteins when localized to mitochondria.
Fig. 2.
Functional assays in inducible Flp-In TRex 293 cells with TOM7 variant. (a) Predicted structure of transmembrane domain of TOM7 by I-TASSER43 with full-length TOM7 (amino acids 1∼55) and annotated with PyMol. Angles of the kinks were determined by α-carbon of residue Glu7, Trp25, and Gly39 with PyMol. (b) Zoom-in view of the Pro29 residue (yellow) in the α-helix of wild-type TOM7. Red dotted lines represent the predicted hydrogen bonds. Zoom-in view of the Leu29 residue (orange) in the α-helix of TOM7 variant. Red dotted lines represent the predicted hydrogen bonds. (c) Immunoblotting of TOM7 with anti-myc antibody in whole-cell lysate or mitochondria-enriched lysate from Flp-In TRex293 cells expressing myc-tagged WT TOM7 or TOM7P29L. (d) Representative z-stack confocal images and line profile of immunofluorescent staining with anti-myc antibody (green; staining TOM7), anti-TOM40 antibody (Red; staining mitochondria) and DAPI (blue) in Flp-In TRex 293 cells carrying myc-tagged wild-type TOM7 or TOM7P29L. Zoom-in views were taken from a single z-plane. Tringle indicates non-mitochondrial TOM7P29L staining. (e) Representative z-stack confocal images and line profile of immunofluorescent staining with anti-myc antibody (green; staining TOM7), anti-Calnexin antibody (Red; staining ER) and DAPI (blue) in Flp-In TRex 293 cells carrying WT TOM7 or TOM7P29L. Zoom-in views were taken from a single z-plane. Triangles indicate co-localized ER and TOM7P29L staining. (f) Immunoprecipitation of TOM7 with anti-myc antibody pulled down the members of TOM complex, TOM22 and TOM40, in Flp-In TRex 293 cells expressing myc-tagged WT TOM7 or TOM7P29L. WT, wild-type TOM7; PL, TOM7p.P29L.
TOM7P29L leads to malfunction in iPSC-derived endothelial cells
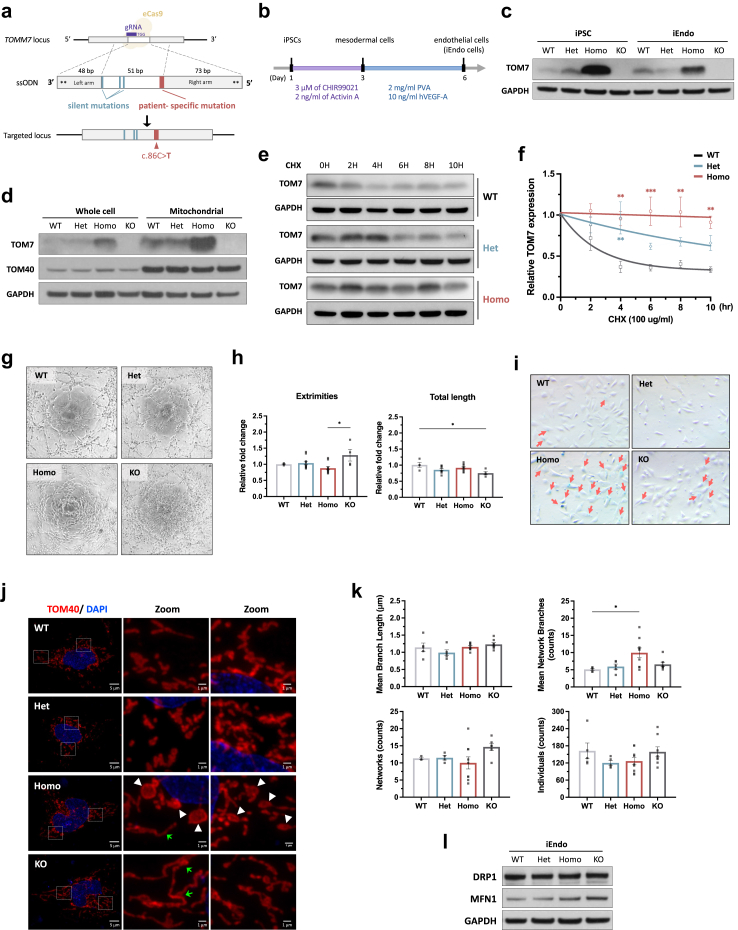
Moyamoya disease is a cerebrovascular disease associated with mitochondrial dysfunction in endothelial progenitor cells.26 To investigate the impact of TOM7P29L in endothelial cells, we chose to knock in TOMM7 p.P29L in iPSCs and then differentiate the iPSCs into endothelial cells (Fig. 3a and b, Supplementary Figure S4a). This cell model allowed us to select the correctly edited colonies without worrying about the limited passages of the cells. In addition, CRISPR/Cas9-edited iPSCs have an isogenic genetic background that avoids complications in data interpretation. At least two independent single colonies from each of heterozygous TOMM7 p.P29L (TOMM7+/PL, Het), homozygous TOMM7 p.P29L (TOMM7PL/PL, Homo), and TOMM7 knock-out (TOMM7−/−, KO) iPSCs were generated (Supplementary Figure S4b and c). For endothelial cell differentiation, iPSCs were first differentiated into mesodermal progenitor cells and then into endothelial cells32 (Fig. 3b, Supplementary Figure S5a and b). Flow cytometry with lineage-specific markers was performed for all genotypes to monitor the iEndo differentiation. Assays were performed with at least two colonies for every genotype. Quantitative real-time PCR (qRT-PCR) with cell markers confirmed the identities of the iPSC-derived endothelial (iEndo) cells, and no difference was found among the genotypes (Supplementary Figure S5c and d).
Fig. 3.
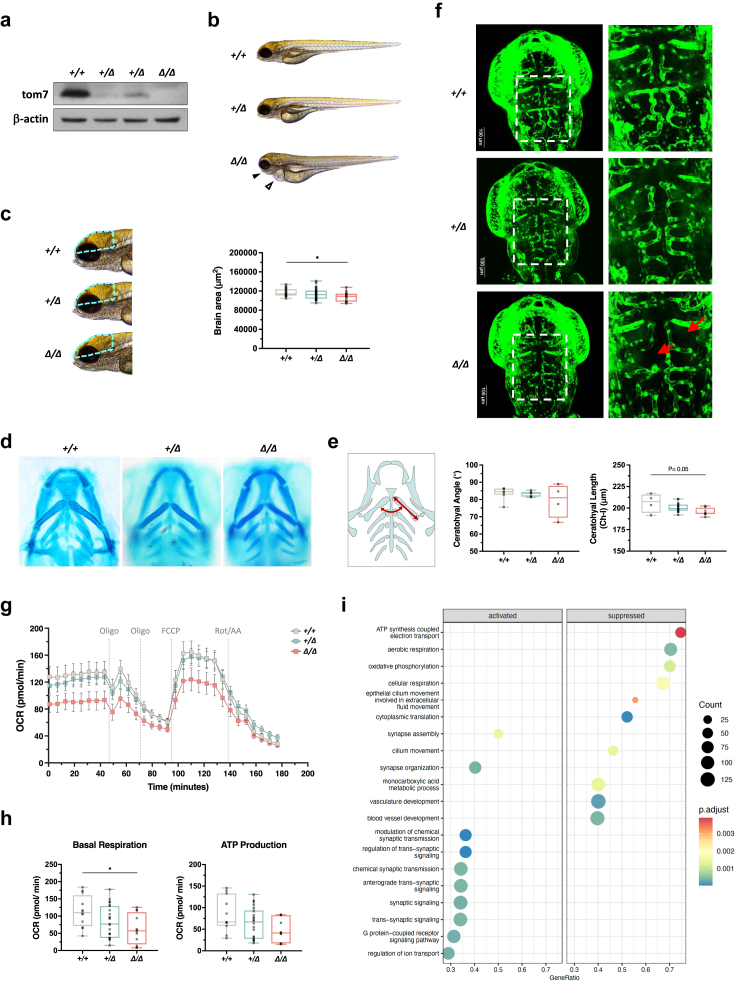
Functional assays in CRISPR-edited iPSC-derived endothelial cells with TOM7 variant. (a) Schematic presentation of CRISPR/Cas9-mediated homology-directed repair with a single-strand oligodeoxynucleotide (ssODN) template carrying TOMM7 p.P29L (red) and three silent mutations (cyan). Asterisks represent phosphorothioate modifications. eCas9, enhanced Cas9 protein containing K484A, K1003A, and R1060A mutations. (b) Workflow of deriving endothelial cells from iPSCs through mesodermal cells. (c) Immunoblotting with anti-TOM7 antibody in whole-cell lysate from iPSCs and iEndo cells. (d) Immunoblotting with anti-TOM7 and anti-TOM40 antibodies in whole-cell lysate and in mitochondria-enriched lysates from iPSCs. (e) Protein half-live was evaluated by immunoblotting with anti-TOM7 antibody in whole-cell lysates from iEndo cells treated with cycloheximide (CHX) for 0, 2, 4, 6, 8, and 10 h. (f) Quantitative analysis of the level of TOM7 from (f). Statistical analysis was done by nonlinear regression with one-phase exponential decay fit and two-way ANOVA followed by Dunnett post hoc test. (g) Representative pictures of tube formation of iEndo cells on Matrigel. (h) Quantitative analyses of the number of extremities and the total length of tubes from (h). Statistical analysis was performed using Kruskal–Wallis test with Dunn's post hoc test for multiple comparisons. (i) Senescence associated β-galactosidase staining in iEndo cells. Arrows indicate positively stained cells. (j) Representative confocal images of immunofluorescent staining with anti-TOM40 (Red; staining mitochondria) and DAPI in iEndo cells. White arrowheads indicate large, round mitochondria. Green arrows indicate elongated mitochondria. (k) Quantitative analyses of mitochondrial morphology in the images of immunofluorescent staining by Mitochondrial Network Analysis (MiNA) toolset showing mean branch length, mean network branches, individual mitochondrial counts, and mitochondrial networks per cell in iEndo cells from (k). Statistical analysis was performed using Kruskal–Wallis test with Dunn's multiple comparisons test. (l) Immunoblotting with anti-DRP1 and anti-MFN1 antibodies in whole-cell lysate from iEndo cells. WT, wild type; Het, heterozygous TOMM7 p.P29L; Homo, homozygous TOMM7 p.P29L; KO, TOMM7 knockout. Data were presented as mean ± S.E.M. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
When evaluating the expression of endogenous TOM7P29L, we found that the level of TOM7P29L was predominately higher in the whole-cell lysate and mitochondria-enriched lysates from TOMM7PL/PL iPSCs and iEndo cells (Fig. 3c and d, Supplementary Figure S5e). These results support that TOM7P29L can target mitochondria. In addition, we found TOM7P29L more stable than WT TOM7, with the half-life of TOM7 protein being 1.769 and >12 h in TOMM7PL/PL (Homo) and WT iEndo cells, respectively (Fig. 3e and f).
To examine the mitochondrial fitness of endothelial cells, we measured the angiogenesis ability, mitochondrial quality control in response to stress, mitochondrial morphology, and cell senescence. Decreased tube formation was observed in TOMM7PL/PL iEndo cells, although not as significant as that in TOMM7−/− iEndo cells (Fig. 3g and h). However, TOMM7PL/PL iEndo cells showed an increased percentage of senescent cells (Fig. 3i). When iEndo cells were induced with mitochondrial stress, a slightly lower level of PINK1 was stabilized in TOMM7PL/PL iEndo cells when compared with WT cells, suggesting a moderate effect of TOM7P29L on initiating mitophagy (Supplementary Figure S6a and b). In addition, round and enlarged mitochondria were present at the perinuclear area of TOMM7PL/PL iEndo cells in regular culture conditions without stress induction (Fig. 3j, Supplementary Figure S6c). In addition, elongated mitochondria were predominately seen in TOMM7−/− iEndo cells (Fig. 3j). Overall, the average number of mitochondrial branches was significantly increased in TOMM7PL/PL cells when compared with WT cells (P = 0.0105, Kruskal–Wallis test with Dunn's multiple comparisons test) (Fig. 3k). The level of mitofusin 1 (MFN1), which controls the fusion of mitochondrial outer membrane, was increased in both TOMM7PL/PL and TOMM7−/−iEndo cells (Fig. 3l). On the other hand, the level of dynamin-related protein 1 (DRP1), which regulates mitochondrial fission, remained unchanged in the iEndo cells (Fig. 3l). These results suggest that TOM7 is involved in maintaining proper function and mitochondrial dynamics in endothelial cells.
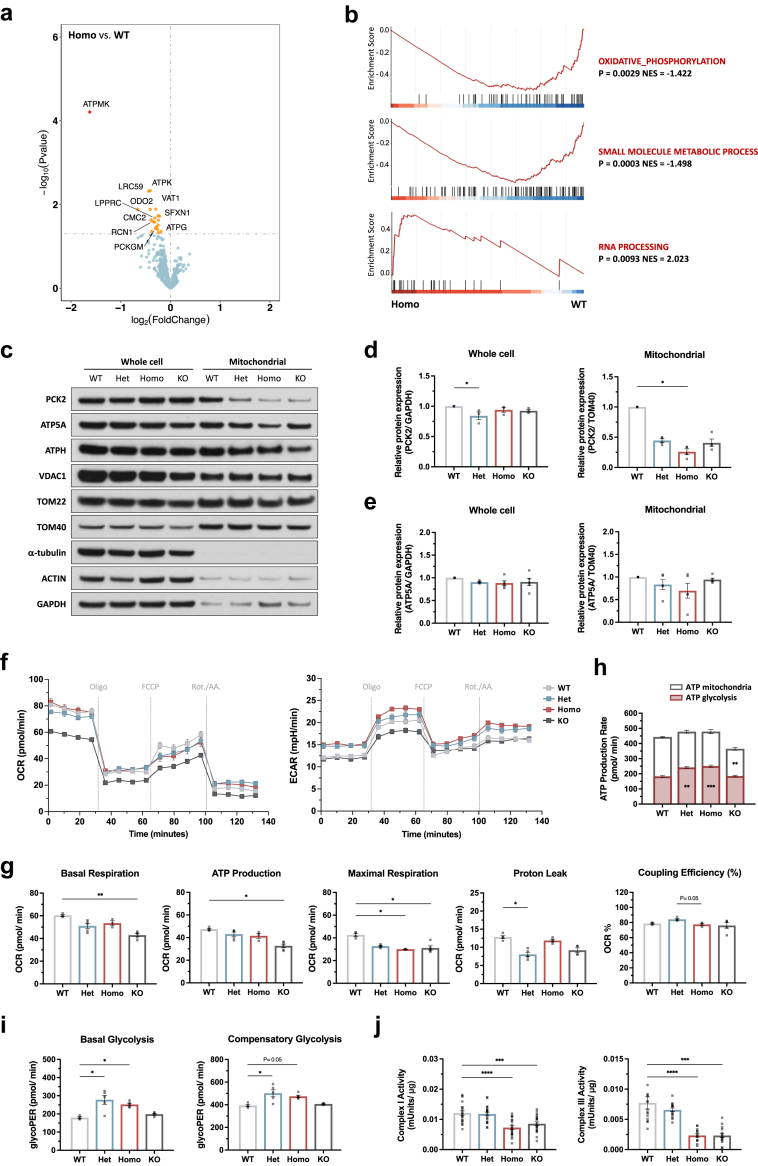
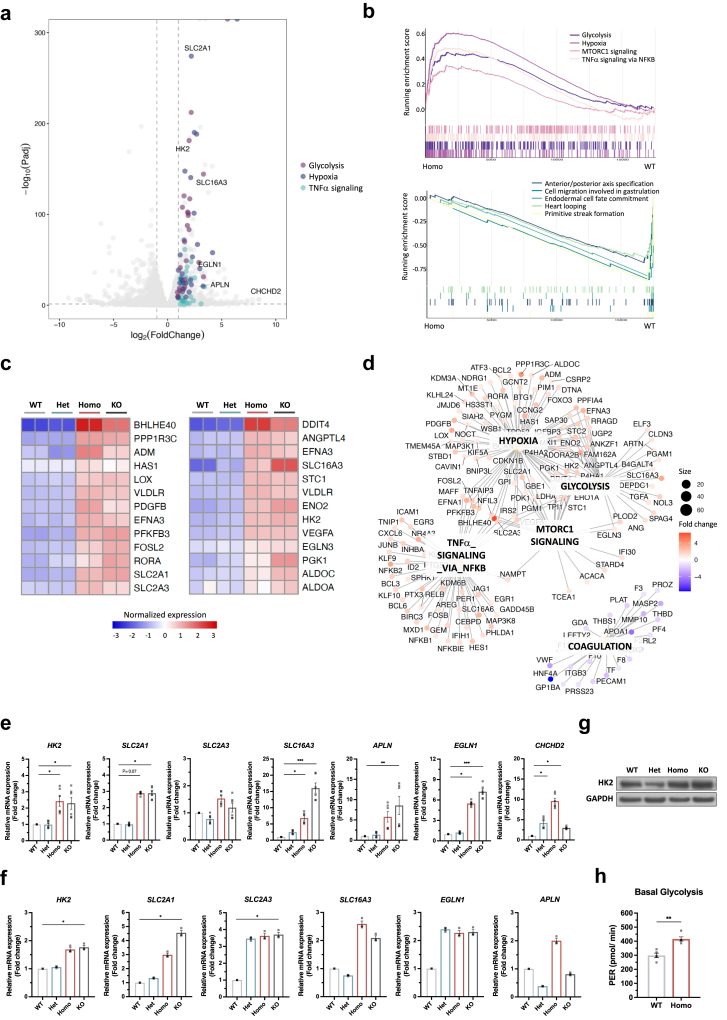
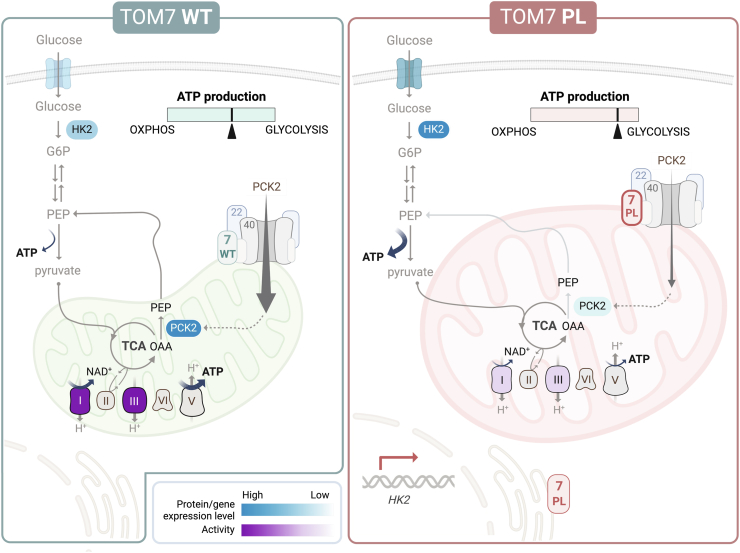
Altered mitochondrial proteome and defective OXPHOS in endothelial cells with TOM7P29L
Despite being able to form the TOM complex, TOM7P29L may affect protein import in the mitochondria. Indeed, mitochondrial proteome analyzed by mass spectrometry revealed, of 651 proteins identified, subunits of ATP synthase, such as ATPMK, ATPK, and ATPG, proteins involved in balancing glycolysis and gluconeogenesis, such as PCKGM (a phosphoenolpyruvate carboxykinase encoded by PCK2), were significantly decreased in TOMM7PL/PL iEndo cells when compared to WT cells (P = 0.00006, P = 0.0046, P = 0.0311, and P = 0.0372 for ATPMK, ATPK, ATPG, and PCKGM, respectively, proDA) (Fig. 4a). Overall, the abundance of proteins involved in mitochondrial oxidative phosphorylation (OXPHOS) and small molecule metabolic process was significantly decreased (P = 0.0029 and P = 0.0003, respectively, GSEA) while proteins involved in RNA processing were increased (P = 0.0093, GSEA) in TOMM7PL/PL iEndo cells (Fig. 4b). The reduced level of PCK2 and ATP5A was validated with immunoblotting in whole-cell lysate and mitochondrial fraction of iEndo cells (Fig. 4c–e). These results suggest TOM7PL alters the abundance of certain mitochondrial proteins in iEndo cells.
Fig. 4.
Mitochondrial protein and respiration in iPSC-derived endothelial cells with TOM7 variant. (a) Volcano plot showing mitochondrial proteomic analysis of the differential abundance of proteins in TOM7PL/PL iEndo cells when compared with WT iEndo cells. (b) Functional enrichment analysis of mitochondrial proteome revealed a down or up representation of proteins in GO biological process terms. (c) Immunoblotting of differentially abundant protein components in whole-cell lysate and mitochondria-enriched lysates from iEndo cells. (d) Quantitative analyses of the level of PCK2 in whole cell or mitochondria-enriched lysates from (c). Statistical analysis was performed using Friedman test followed by Dunn's post hoc test, with comparisons made to WT cells. (e) Quantitative analyses of the level of ATP5A in whole cell or mitochondria-enriched lysates. Statistical analysis was performed using Friedman test followed by Dunn's post hoc test, with comparisons made to WT cells. (f) Oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) measured by the Seahorse Mito Stress Assay with 3 × 104 iEndo cells. One representative result was shown. Oligo., oligomycin; FCCP, carbonyl cyanide-p-trifluoromethoxyphenylhydrazone; Rot, rotenone; AA, antimycin A. (g) Basal respiration, ATP-coupled respiration, percentage of respiration used for ATP synthesis, maximal mitochondrial respiratory capacity, and proton leak-linked respiration were calculated from one representative experiment (f). Statistical analysis was performed using Kruskal–Wallis test with Dunn's multiple comparisons test. (h) ATP production rates from mitochondrial respiration and glycolysis. Statistical analysis of mitochondrial ATP and glycolytic ATP was performed using the Kruskal–Wallis test followed by Dunn's post hoc test, with comparisons made to WT cells for each parameter. (i) Basal glycolysis and compensatory glycolysis in iEndo cells measured by glycolytic proton efflux rate (PER) in Seahorse assay. Statistical analysis was performed using Kruskal–Wallis test with Dunn's multiple comparisons test. (j) The activities of Complex I (NADH dehydrogenase) and Complex III (cytochrome c reductase) in iEndo cells were measured, with normalization to the input concentration of intact mitochondria. The Y-axis represents the activity levels (mUnits/μg) of Complex I and Complex III at each Δ[(t + 1) −t] 30-s interval. Statistical analysis was performed using Friedman test followed by Dunn's post hoc test, with comparisons made to WT cells. WT, wild-type; Het, heterozygous TOMM7+/PL; Homo, homozygous TOMM7PL/PL; KO, TOMM7 knock-out. Data were presented as mean ± S.E.M. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001.
The extracellular acidification rate (ECAR) was increased while the OCR was slightly decreased in TOMM7PL/PL iEndo cells when compared with WT or TOMM7+/PL cells (Homo vs. WT, P = 0.0389 for maximal respiration, Kruskal–Wallis test with Dunn's multiple comparisons test) (Fig. 4f and g). Interestingly, with the ATP production from glycolysis significantly increased, and the overall ATP production was not altered in TOMM7PL/PL iEndo cells (Homo vs. WT, P = 0.0006, Kruskal–Wallis test followed by Dunn's post hoc test) (Fig. 4h). Consistently, the measurement of proton efflux rate (PER) confirmed the significantly increased glycolysis in TOMM7PL/PL iEndo cells (P = 0.0206 for basal glycolysis and P = 0.0282 for compensatory glycolysis, Kruskal–Wallis test with Dunn's multiple comparisons test) (Fig. 4i). The activities of complex I (NADH dehydrogenase) and complex III (ubiquinol-cytochrome c reductase) were significantly lower in TOMM7PL/PL and TOMM7−/− iEndo cells (Homo vs. WT, P < 0.0001 for both complex I and complex III; KO vs. WT, P = 0.0006 for complex I and complex III, Friedman test followed by Dunn's post hoc test) (Fig. 4j, Supplementary Figure S6d). The NAD+/NADH ratio showed a decreasing trend (Supplementary Figure S6e). However, no difference in mitochondrial ROS was detected among the iEndo cells (Supplementary Figure S6f). These results suggest a metabolic reprogramming in the endothelial cells with TOM7P29L.
Deletion of tomm7 in zebrafish caused cerebrovascular and craniofacial defects
With 86% amino acids identical to human TOM7, zebrafish tom7 is a highly evolutionary conserved protein (Fig. 1i). Knock-down tomm7 in zebrafish with morpholino has been shown to cause intracranial haemorrhage with cerebral vessel instability.20 To create a genetic model, we generated zebrafish lines carrying tomm7 frameshift variant (tomm7Δ) through CRISPR/Cas9 genome editing (Supplementary Figure S7a–c). Immunoblotting confirmed the loss of tom7 in homozygous knockout (tomm7Δ/Δ) (Fig. 5a). Although tomm7Δ/Δ embryos survived (Supplementary Figure S7d), they had small dentary bones of the lower jaw, cardiac edema, and a curved body with regular body length (Fig. 5b, Supplementary Figure S7e and f). Brain area was significantly smaller in tomm7Δ/Δ embryos (P = 0.0404) (Fig. 5c, Kruskal–Wallis test with Dunn's multiple comparisons test). The tomm7Δ/Δ zebrafish also had shorter ceratohyal cartilages with normal angles (Fig. 5d and e). In tomm7Δ/Δ; Tg (fli1a:eGFP) zebrafish, which expresses endothelial cell-specific eGFP, missing central arteries in the hindbrain were noticed (Fig. 5f), while no abnormality was observed in the intersegmental vessels (Supplementary Figure S7g and h). Moreover, OCR measurement suggested decreased mitochondrial respiration in tomm7Δ/Δ embryos (P = 0.0439 for basal respiration, Kruskal–Wallis test with Dunn's multiple comparisons test) (Fig. 5g and h). Interestingly, transcriptome revealed through RNA sequencing in tomm7Δ/Δ zebrafish brain showed downregulated vasculature development and OXPHOS, as well as upregulated synaptic signaling, indicating loss of tom7 altered the nuclear gene expression (Fig. 5i). These data also suggest that loss of arteries in tomm7Δ/Δ may be the consequence of differential expression of genes required for vessel maintenance or morphogenesis (Fig. 5i). Taken together, these results suggest that tom7 deficiency in zebrafish leads to metabolic impairment as well as microcephaly, craniofacial dysmorphia, and cerebrovascular defects that recapitulate symptoms in the patients.
Fig. 5.
Deletion of tomm7 caused developmental defects and cerebral vessel dysmorphism in zebrafish. (a) Immunoblotting with anti-tom7 antibody in brain lysate from adult zebrafish. (b) Representative pictures of zebrafish embryos at 96 hpf after PTU treatment. The black triangle indicates a recessed chin, and the white triangle indicates cardiac edema in knockout zebrafish. (c) Quantification of the size of the brain (indicated by dash lines) at 96 hpf. +/+, N = 19; +/Δ, N = 34; Δ/Δ, N = 13. Statistical analysis was performed using Kruskal–Wallis test with Dunn's multiple comparisons test. (d) Representative pictures of craniofacial cartilage elements at 96 hpf with Alcian blue staining. (e) Illustration of cartilages and measurements of length and angle of ceratohyal cartilages in zebrafish at 96 hpf. For the angle of ceratohyal cartilages: +/+, N = 6; +/Δ, N = 7; Δ/Δ, N = 4. For the length of ceratohyal cartilages: WT, N = 4; +/Δ, N = 19; Δ/Δ, N = 8. Statistical analysis was performed using the Kruskal–Wallis test followed by Dunn's multiple comparisons test. (f) Maximum projections of head vasculature in Tg (fli1a:EGFP); tomm7+/+, Tg (fli1a:EGFP); tomm7+/Δ and Tg (fli1a:EGFP); tomm7Δ/Δ zebrafish embryo at 96 hpf. Arrows indicate missing central arteries. Scale bars, 100 μm. (g) Oxygen consumption rate (OCR) measured by the Seahorse Mito Stress Test with Tg (fli1a:EGFP); tomm7+/+, Tg (fli1a:EGFP); tomm7+/Δ and Tg (fli1a:EGFP); tomm7Δ/Δ zebrafish embryo at 48 hpf. +/+, N = 13; +/Δ, N = 24; Δ/Δ, N = 13. Oligo., oligomycin; FCCP, carbonyl cyanide-p-trifluoromethoxyphenylhydrazone; Rot, rotenone; AA, antimycin A. (h) Basal respiration and ATP-coupled respiration were calculated from (g) and analyzed using the Kruskal–Wallis test followed by Dunn's multiple comparisons test. (i) Dotplot showing the ratio of enriched genes in representing 20 significantly activated and suppressed GO biological process terms in tomm7Δ/Δ adult zebrafish brain when compared with WT zebrafish brain. +/+, wild type, tomm7+/+; +/Δ, tomm7+/Δ; Δ/Δ, tomm7Δ/Δ. ∗P < 0.05.
Altered transcriptome in iPSCs with TOM7P29L
To trace the molecular basis of the disease, we asked whether TOM7P29L-associated mitochondrial defects and differential gene expression can be detected at the early stage of cell differentiation, that is, in iPSCs. Pluripotent stem cells have been shown to offer prospects of gene regulation during early development, which is relevant to diseases that occur during fetal development.45 Through measuring the OCR, we found significantly decreased mitochondrial respiration in TOMM7PL/PL iPSCs when compared with WT or TOMM7+/PL iPSCs (Homo vs. WT, P = 0.0099 for basal respiration, P = 0.0044 for ATP production, and P = 0.0136 for coupling efficiency (%); Homo vs. Het P = 0.0161 for maximal respiration, Kruskal–Wallis test with Dunn's multiple comparisons test) (Supplementary Figure S8a and b). Intriguingly, RNA sequencing in TOMM7PL/PL iPSCs revealed upregulation of genes involved in glycolysis, hypoxia, and/or TNFα signaling when compared with WT (Fig. 6a–d) or TOMM7+/PL cells (Supplementary Figure S9a and b). In addition, genes involved in embryonic development and morphogenesis were downregulated in TOMM7PL/PL iPSCs (Fig. 6b, Supplementary Figure S10a). The upregulation of genes involved in glucose metabolism (HK2), glucose transmembrane transport (SLC2A1 and SLC2A3), lactate and pyruvate transport (SLC16A3), and hypoxia (EGLN1 and CHCHD2) was validated by qRT-PCR in both iPSCs and iEndo cells (Fig. 6e and f). Increased level of hexokinase 2 (encoded by HK2), which phosphorylates glucose in the first step of glucose metabolism, further supports the reliance on glycolysis in energy production in TOMM7PL/PL iPSCs (Fig. 6g). Indeed, basal glycolysis, measured by PER, was significantly increased in TOMM7PL/PL iPSCs (P = 0.0079, Mann–Whitney U-tests) (Fig. 6h). Taken together, these results suggest that TOMM7PL/PL iPSCs increase glycolysis to compensate for the decreased energy supply by mitochondria.
Fig. 6.
Mitochondrial respiration and transcriptome analysis of iPSCs with homozygous TOM7 variant. (a) Volcano plot showing RNAseq results of upregulated and downregulated genes in TOMM7PL/PL when compared with WT cells. Genes in indicated pathways with adjusted P value < 0.05 and log2 (fold change) > 1 were in color. (b) Gene set enrichment analysis (GSEA) showing significantly upregulated Hallmark datasets and downregulated GO biological process terms in TOMM7PL/PL iPSCs when compared with WT cells. (c) Heatmap showing expression of differentially expressed genes (identified by comparing TOMM7PL/PL iPSCs with WT cells) in all genotypes. (d) Category network plot showing the linkage of significantly upregulated Hallmark gene sets in TOMM7PL/PL iPSCs when compared with WT cells. (e) Quantitative RT-PCR confirmed the differentially expressed genes in iPSCs. Statistical analysis was performed using Friedman test followed by Dunn's post hoc test, with comparisons made to WT cells. (f) Quantitative RT-PCR confirmed the differentially expressed genes in iEndo cells. Statistical analysis was performed using Friedman test followed by Dunn's post hoc test, with comparisons made to WT cells. (g) Immunoblotting in whole-cell lysate from iPSCs with anti-HK2 antibody. (h) Glycolysis in iPSCs measured by glycolytic proton efflux rate (PER) in Seahorse assay. Statistical analysis was performed using Mann–Whitney U-tests. WT, wild-type; Het, heterozygous TOMM7+/PL; Homo, homozygous TOMM7PL/PL; KO, TOMM7 knock-out TOMM7−/−. Data were presented as mean ± S.E.M. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
While little difference in gene expression was observed between TOMM7+/PL and WT iPSCs, gene expression in TOMM7PL/PL or TOMM7−/− iEndo cells was distinct from that in WT iPSCs (Supplementary Figure S10b and c). Although TOMM7PL/PL and TOMM7−/− iPSCs shared only about half of the differentially expressed genes (Supplementary Figure S10c), many enriched pathways were overlapped in TOMM7PL/PL and TOMM7−/− iPSCs (Supplementary Figure S11). Taken together, these results suggest that TOM7P29L modulates glucose metabolism in iPSCs.
Metabolic reprogramming in endothelial cells and fibroblasts differentiated from patient's iPSCs
Finally, we established iPSCs reprogrammed from the patient's PBMC (Supplementary Figure S12). The iPSC colonies with WT TOMM7 and heterozygous TOMM7+/P29L were generated through CRISPR/Cas9 gene correction from the patient's iPSCs (Fig. 7a and b). Consistently, the level of TOM7P29L was higher than WT TOM7 in endothelial cells derived from the patient's iPSCs (Fig. 7c). Moreover, round and enlarged mitochondria were detected in the patient's iEndo cells under ultrastructural TEM, while normal mitochondria were evidenced in the variant-corrected WT iEndo cells (Fig. 7d). Round and enlarged mitochondria, with significantly increased network branches and branch length at the perinuclear area, were also detected with immunofluorescent staining in the patient's iEndo cells when compared with the variant-corrected WT iEndo cells (P = 0.0459 for network branches and P = 0.0018 for branch length, Kruskal–Wallis test with Dunn's multiple comparisons test) (Fig. 7e and f).
Fig. 7.
Functional assays in endothelial cells derived from CRISPR-edited patient's iPSC. (a) Schematic presentation of CRISPR/Cas9-mediated homology-directed repair of patient's TOMM7 p.P29L with a single-strand oligodeoxynucleotide (ssODN) template carrying WT TOMM7 and two silent mutations (cyan). Asterisks represent phosphorothioate modifications. eCas9, enhanced Cas9 protein containing K484A, K1003A, and R1060A mutations. (b) Sanger sequencing confirmed gene-edited iPSCs with heterozygous and homozygous TOMM7 p.P29L. (c) Immunoblotting in whole-cell lysate from iEndo cells derived from patient's iPSCs with anti-TOM7 and anti-TOM40 antibodies. (d) Representative transmission electron microscopy images of iEndo cells derived from patient's iPSCs and variant-corrected WT iPSCs (edited WT). Scale bars, 200 nm. (e) Representative confocal images of immunofluorescent staining with anti-TOM40 (Red; staining mitochondria) and DAPI (Blue) in patient's iEndo cells. White arrows indicate large, round mitochondria. Cyan arrows indicate elongated mitochondria. (f) Quantitative analyses of mitochondrial morphology in the images of immunofluorescent staining by Mitochondrial Network Analysis (MiNA) toolset showing counts of individual mitochondria, counts of network, mean mitochondrial branch length, mean network branches, and mitochondrial footprint in patient's iEndo cells from (e). Statistical analysis was performed using Kruskal–Wallis test with Dunn's multiple comparisons test. (g) Oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) measured by the Seahorse Mito Stress Test with 3 × 104 iEndo cells. One representative result was shown. Oligo., oligomycin; FCCP, carbonyl cyanide-p-trifluoromethoxyphenylhydrazone; Rot, rotenone; AA, antimycin A. (h) Basal respiration, ATP-coupled respiration, percentage of respiration used for ATP synthesis, maximal mitochondrial respiratory capacity, and proton leak-linked respiration in iEndo cells derived from patient's iPSCs were calculated from (g). Statistical analysis was performed using Mann–Whitney U-tests. (i) ATP production rates from mitochondrial respiration and glycolysis. Statistical analysis of mitochondrial ATP and glycolytic ATP was performed using Mann–Whitney U-tests, with comparisons made to WT cells for each parameter. (j) Quantitative RT-PCR confirmed the differentially expressed genes in patient's iEndos, and statistical analysis was performed using Friedman test followed by Dunn's post hoc test, with comparisons made to WT cells. (k) Immunoblotting in whole-cell lysate from iEndo cells derived from patient's iPSCs with anti-HK2 antibody. (l) Basal glycolysis and compensatory glycolysis in iEndo cells derived from patient's iPSCs measured by glycolytic proton efflux rate (PER) in Seahorse assay. Statistical analysis was performed using Mann–Whitney U-tests. Edited WT, wild type TOMM7 p.P29L; Edited Het, heterozygous TOMM7 p.P29L; Patient, patient 1-III, homozygous TOMM7 p.P29L; Edited KO, TOMM7 knockout. Data were presented as mean ± S.E.M. ∗P < 0.05, ∗∗P < 0.01.
The OCR measurement in patients' iEndo cells showed only a subtle decreasing trend in basal respiration and ATP-linked production when compared with variant-corrected WT iEndo cells (Fig. 7g and h). Consistently, ATP production from glycolysis was significantly increased in endothelial cells derived from the patient's iPSCs (P = 0.0028, Mann–Whitney U-tests) (Fig. 7i). Most importantly, the expression of genes involved in glucose metabolisms, such as HK2 and SLC2A3, was increased in the patient's iEndo cells (Fig. 7j). Indeed, the level of HK2 protein and glycolysis activity were significantly increased in the patient's iEndo cells when compared with variant-corrected iEndo cells (P = 0.0079 for basal glycolysis, P = 0.0079 for compensatory glycolysis, Mann–Whitney U-tests) (Fig. 7k and l).
Previous studies showed no change in the level of ATP5A in the liver and brain tissue lysates from Tomm7−/− mice18 or increased ATP5A in the fibroblasts from patients.19 In the current study, we found the level of ATP5A comparable in the mitochondrial lysate from TOMM7PL/PL and TOMM7−/− iEndo cells (Fig. 4c and e). In addition, previous studies showed that OCR was either unchanged in brain endothelial cells from Tomm7−/− mice20 or increased in human fibroblasts with TOMM7PL/PL and in Tomm7−/− mouse embryonic fibroblasts,18,19 yet we found decreased mitochondrial OCR in TOMM7PL/PL and TOMM7−/− iEndo cells. To test the possibility of cell type-specific effects on the differences among the studies, we differentiated the reprogrammed iPSCs into fibroblasts and investigated mitochondrial respiration in the TOMM7PL/PL iPSC-derived fibroblasts (iFibro) (Supplementary Figure S13a and b), which express specific markers such as COL1A1 and vimentin (Supplementary Figure S13c and d). We found that the level of ATP5A was comparable in whole-cell lysates and mitochondria from TOMM7PL/PL iFibro (Supplementary Figure S13e and f). We also detected lower OCR in TOMM7PL/PL and TOMM7 knockout iFibro, suggesting no phenotypic difference among iPSCs, iEndo cells, and iFibro (Supplementary Figure S13g–i). Together, these results indicate that correction of homozygous TOMM7 p.P29L alleles normalized mitochondrial morphology and cellular metabolism in endothelial cells derived from the patient's iPSCs.
Discussion
In seven unrelated families, we identified a homozygous missense variant in TOMM7 p.P29L in nine patients who had microcephaly, short stature, micrognathia, alopecia partialis, developmental delay, ophthalmic features, and, most importantly, moyamoya disease. We showed the autosomal recessive inheritance and segregation of TOMM7 p.P29L in these families. In addition, we found mitochondrial enlargement and alterations in mitochondrial proteomes in TOMM7PL/PL endothelial cells derived from iPSCs. We detected decreased complex I and III activities and mitochondrial oxygen consumption, as well as significantly increased glycolytic activity. Moreover, a metabolic change was evidenced by elevated gene expression and protein levels of HK2, along with reduced mitochondrial PCK2 protein levels (Fig. 8). Taken together, we provided molecular and cellular evidence of mitochondrial dysfunction and metabolic reprogramming and supported the causative role of TOMM7 in the disease.
Fig. 8.
Graphic summary of the study findings, highlighting the impact of TOMM7 p.P29L (PL) variant on metabolic reprogramming in iPSC-derived endothelial cells. The level of differentially expressed genes or abundant proteins are represented in blue gradient. Activities of complexes I and III are represented in purple gradient.
Together with previous studies of the cases with TOMM7 p.W25R and the TOMM7 p.P29L variants, it is now clear that TOMM7 is a disease-causing gene for a new mitochondrial-related syndrome with phenotypic heterogeneity.18,19 Compared with normal brain MRI findings in the previous case with Garg-Mishra syndrome carrying the same TOMM7 p.P29L variant, the most distinct phenotypic finding in our patients is that five of our patients (5 in 8 patients with brain MRI; 63%) developed moyamoya disease, which correlates to the cerebrovascular phenotypes observed in previous and current animal studies. In contrast to the previous case with Garg-Mishra syndrome who showed proportionate short stature with relative macrocephaly, our patients showed short stature with microcephaly. In addition, maculopathy, which was not detected in previous cases, was noticed in seven of our patients (7/9; 78%). Moreover, no hepatosteatosis was detected in two patients who received abdominal sonography, although patient 1-III had a lipodystrophy-like appearance. Compared with previous cases, our patients showed some overlapped symptoms, such as short stature, developmental delay, abnormal dentition, micrognathia, nystagmus, and hyperopia. Besides, two of our patients had alopecia partialis, and two had aged skin. The previous case with Garg-Mishra syndrome had mild mitral regurgitation and mild dilatation of the left ventricle; two of our patients also showed cardiomegaly. The phenotypic variability in individuals with the TOMM7 variant supports that human mitochondrial disorders are among the most phenotypically heterogeneous diseases.13,14 The genetic background of the patient may also contribute to phenotypic heterogeneity. However, despite various expressivity in TOMM7-associated syndrome, macular scarring is likely to be the most common symptom that can be diagnosed at a younger age, as we observed in patient 1-V during prospective follow-up, and thus a target to monitor as the first sign of symptom onset. In addition, our results also suggest that facial dysmorphism progressively appears as the patient ages; therefore, facial dysmorphism may not be a good indicator in young childhood.
Considering short stature in association with moyamoya disease, MOPD-II, a rare autosomal recessive disorder characterized by short stature, microcephaly, micrognathia, abnormal dentition, moyamoya disease and intracranial aneurysms (in 25%∼64% of patients), was the most favorable diagnosis.37,38,46 Previously, one of our patients with short stature who developed moyamoya disease was initially diagnosed as MOPD-II, illustrating the perplexity in the differential diagnosis between MOPD-II and TOMM7-associated syndrome.47 However, some distinctive features can provide clues for differential diagnosis. For example, dentition problems are distinct as MOPD-II is characterized by small teeth, while TOMM7-associated disorder is characterized by protruding maxillary central incisors with normal-size teeth. In addition, eight of the patients had average birth weight followed by age-dependent growth failure, which is not consistent with the prenatal growth retardation that usually presents with small-for-gestational-age in MOPD-II.
The molecular effects of TOM7 deficiency in mammalian cells were puzzling in recent studies. The earlier study showed that the brain endothelial cells from Tomm7−/− mice had comparable OCR and increased ROS compared to those from WT mice.20 However, the recent study in mouse primary rib chondrocytes with either homozygous Tomm7 p.W25R or null Tomm7 showed increased glycolysis, unchanged ROS, increased phospho-AMPK (an indication of inefficient ATP synthesis) yet increased OCR.18 The other recent study in fibroblasts from the patient and mouse embryonic fibroblasts with TOMM7 p.P29L showed increased OCR with no change in ROS.19 The current study detected increased glycolysis, unchanged ROS, and slightly decreased OCR in iPSC-derived endothelial cells and iPSC-derived fibroblasts carrying homozygous TOMM7 p.P29L. Moreover, the difference in OCR was more significant in iPSCs with homozygous TOMM7 p.P29L than in iEndo cells with homozygous TOMM7 p.P29L. On the other hand, the decrease of OCR in TOMM7 knockout iEndo cells was more significant than that in TOMM7 knockout iPSCs. The phenotypic variation is probably a result of complex regulatory events in different cell types, cell culture conditions (e.g., glucose concentration), or genetic backgrounds. Indeed, recent studies with single-cell RNA sequencing showed inter- and intra-tissue metabolic heterogeneity of the same cell type. For example, endothelial cells, which rely more on glycolysis or fatty acid oxidation for energy production, exhibit phenotypic plasticity and can reprogram their metabolism to fulfill physiological needs.48 Nevertheless, our results are consistent with previous findings of impaired OXPHOS and decreased ATP synthase components in fibroblasts from patients with Hutchinson–Gilford progeria syndrome,49 as well as reduced OCR in endothelial progenitor cells from patients with moyamoya disease.26 Importantly, we observed decreased OXPHOS in tomm7 knockout zebrafish associated with cerebrovascular defects, providing the correlation between mitochondrial respiration defects and vessel integrity in vivo. However, the impact of TOMM7 p.P29L in different cell lineages needs further investigation.
At the cellular level, our data suggest that angiogenesis, a process involved in collateral vessel development in moyamoya disease, can still be initiated in homozygous endothelial cells even though the tube-forming ability may be decreased. In addition, homozygous endothelial cells showed enlarged or elongated mitochondria, a morphology that has been seen in senescent cells,43 in agreement with the patient's progeria phenotypes. In fact, an imbalance of mitochondrial fission and fusion may contribute to aging.50,51 Endothelial cell senescence has been implicated in age-related vascular disease, including stroke.52 The mechanisms through which TOMM7 p.P29L affects mitochondrial fusion and fission and their correlation with cerebrovascular defects remain to be investigated.
The differential level of WT TOM7 vs. TOM7P29L highlighted the difference between ectopically expressed protein and endogenous protein. The level of endogenous TOM7P29L was higher when compared with WT TOM7 in iPSCs, iEndo cells, and endothelial cells derived from the patient's iPSCs. Future study is needed to investigate whether the degradation or turnover of TOM7 depends on the mitochondrial localization or enzymatic proteolysis in mitochondria. In addition, since RIPA lysis buffer used in immunoblotting only detects soluble proteins, whether TOM7P29L aggregates or not remains to be determined. In yeast, P29L led to the mislocalization of tom7 to ER.42 Previous study showed GFP-tagged TOMM7P29L localized to mitochondria.19 We observed partial mis-localization of TOM7P29L to ER in Flp-In TRex 293T cells. Although partial mislocalization of TOM7P29L probably affected the mitochondrial protein import in the cells, iEndo cells with homozygous TOM7PL and null-TOM7 showed overlapped phenotypes, suggesting that TOM7P29L is likely to be a hypomorphic variant in protein function.
Among the low-abundant mitochondrial proteins identified in iEndo cells with homozygous TOM7PL, PCK2 regulates mitochondrial respiration by reversing TCA cycle intermediate oxaloacetate to phosphoenolpyruvate, leading to decreased TCA cycle intermediates and enhanced gluconeogenesis in cancer cells.53 Hence, a low abundance of mitochondrial PCK2 may preserve the amount of TCA cycle intermediates, such as NADH and FADH2, and enhance ETC and OXPHOS in TOMM7PL/PL endothelial cells. However, because the activity of NADH dehydrogenase was also decreased, the NADH may not be oxidized efficiently, resulting in a slightly lower NAD+/NADH ratio in TOMM7PL/PL endothelial cells. How TOM7P29L affects the mitochondrial import of PCK2 is currently unknown. In addition, whether defects in mitochondrial protein import lead to the accumulation of precursor proteins in the cytosol and induce mitochondria-mediated proteostatic stress in cells with TOM7P29L, remains to be investigated.54,55
Recent studies highlight the communication between mitochondria and nuclear gene expression by retrograde signaling.56 Our data indicate that TOMP29L alters the transcriptome at the early stage of development. Homozygous iPSCs and iEndo cells may rely more on glycolysis for energy production by upregulating genes involved in glycolysis, including glucose transmembrane transporters GLUT1 (encoded by SLC2A1), which has been shown to promote postnatal central neuron system angiogenesis,57 GLUT3 (encoded by SLC2A3) and HK2, whose activation increased glucose metabolism in endothelial cells with hyperglycaemia,58 to help unburden the mitochondria.
Loss of the cerebral arteries and metabolic defects in tom7-deficient zebrafish can be detected early during embryonic vascular development in vivo. Both brain area and ceratohyal length were statistically significantly decreased in tom7-deficient zebrafish despite the moderate study power (Supplementary Data 2b). However, how tom7 regulates the genes involved in blood vessel development, especially in the cerebral vessels, remains to be determined. In conclusion, we provided evidence of TOMM7 p.P29L variant causing metabolic reprogramming and nominated TOMM7 as a disease-causing gene for an autosomal recessive microcephalic osteodysplastic dwarfism with moyamoya disease.
Contributors
Conceptualization, P.C.C.; Methodology, C.Y.L., P.X.L., T.C.H., P.M.K., and P.C.C.; Investigation, C.Y.L., L.W.C., M.C.T., Y.Y.C., Y.M.C., W.L.H., Y.H.C., J.L.L., H.A.C., N.C.L., P.H.S., P.X.L., T.C.H., H.K., Y.C.W., Y.T.H., M.H.S., and P.C.C.; Formal analysis, C.Y.L., P.X.L., T.C.H., H.K., S.H.L., and P.C.C.; Visualization, C.Y.L., L.W.C., P.X.L., T.C.H., H.K., and P.C.C.; Writing – original draft, P.C.C., C.Y.L., L.W.C., T.C.H., Y.M.C., N.C.L., and M.C.T.; Writing – review & editing, P.C.C., L.W.C., and Y.Y.C.; Funding acquisition, L.W.C., M.H.S., and P.C.C.; Resources, L.W.C., M.C.T., Y.Y.C., P.M.C., L.L.H.H., M.H.S., and P.C.C.; Accessed and verified the underlying data, C.Y.L., L.W.C., P.X.L., and P.C.C.; Supervision, P.C.C. All authors read and approved the final version of the manuscript.
Data sharing statement
Sequencing data have been deposited to the sequence read archive (SRA) (BioProject ID: PRJNA1173235). Uncropped western blots are provided in the Supplementary Information. Data collected for the study and data from sample analyses will be made available to the corresponding author upon reasonable request.
Declaration of interests
The authors declare no competing interests.
Acknowledgements
The authors gratefully acknowledge the use of Q-Orbitrap MS (QE Plus; CE/LC-MS) belonging to the Instrument Center of National Cheng Kung University. The authors thank the technical services provided by the “Bio-image Core Facility” and the “iMANI Center” of the National Core Facility for Biopharmaceuticals, Ministry of Science and Technology, Taiwan, as well as technical service from the Instrument Development Center of the National Cheng Kung University. The authors thank the Zebrafish Facility in the Animal Center of the School of Medicine, National Cheng Kung University, for maintaining the zebrafish. We thank the Human Disease iPSC Service Consortium for iPSC generation and technical support. The authors thank the Biostatistics Consulting Center, Clinical Medicine Research Center, and National Cheng Kung University Hospital for providing the statistical consulting services.
This work is supported by grants from National Science and Technology Council 111-2320-B-006-067-MY3 (to P.C.C.), National Cheng Kung University Hospital NCKUH-10802014 (to L.W.C. and P.C.C.) and NCKUH-11104014 (to M.H.S.).
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2024.105476.
Appendix A. Supplementary data
References
- 1.Dromparis P., Michelakis E.D. Mitochondria in vascular health and disease. Annu Rev Physiol. 2013;75:95–126. doi: 10.1146/annurev-physiol-030212-183804. [DOI] [PubMed] [Google Scholar]
- 2.Palmer C.S., Anderson A.J., Stojanovski D. Mitochondrial protein import dysfunction: mitochondrial disease, neurodegenerative disease and cancer. FEBS Lett. 2021;595(8):1107–1131. doi: 10.1002/1873-3468.14022. [DOI] [PubMed] [Google Scholar]
- 3.Spinelli J.B., Haigis M.C. The multifaceted contributions of mitochondria to cellular metabolism. Nat Cell Biol. 2018;20(7):745–754. doi: 10.1038/s41556-018-0124-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morgenstern M., Stiller S.B., Lubbert P., et al. Definition of a high-confidence mitochondrial proteome at quantitative scale. Cell Rep. 2017;19(13):2836–2852. doi: 10.1016/j.celrep.2017.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chacinska A., Koehler C.M., Milenkovic D., Lithgow T., Pfanner N. Importing mitochondrial proteins: machineries and mechanisms. Cell. 2009;138(4):628–644. doi: 10.1016/j.cell.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sokol A.M., Sztolsztener M.E., Wasilewski M., Heinz E., Chacinska A. Mitochondrial protein translocases for survival and wellbeing. FEBS Lett. 2014;588(15):2484–2495. doi: 10.1016/j.febslet.2014.05.028. [DOI] [PubMed] [Google Scholar]
- 7.Harbauer A.B., Zahedi R.P., Sickmann A., Pfanner N., Meisinger C. The protein import machinery of mitochondria-a regulatory hub in metabolism, stress, and disease. Cell Metab. 2014;19(3):357–372. doi: 10.1016/j.cmet.2014.01.010. [DOI] [PubMed] [Google Scholar]
- 8.Kato H., Mihara K. Identification of Tom5 and Tom6 in the preprotein translocase complex of human mitochondrial outer membrane. Biochem Biophys Res Commun. 2008;369(3):958–963. doi: 10.1016/j.bbrc.2008.02.150. [DOI] [PubMed] [Google Scholar]
- 9.Morciano G., Patergnani S., Bonora M., et al. Mitophagy in cardiovascular diseases. J Clin Med. 2020;9(3) doi: 10.3390/jcm9030892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Willems P.H., Rossignol R., Dieteren C.E., Murphy M.P., Koopman W.J. Redox homeostasis and mitochondrial dynamics. Cell Metab. 2015;22(2):207–218. doi: 10.1016/j.cmet.2015.06.006. [DOI] [PubMed] [Google Scholar]
- 11.Nunnari J., Suomalainen A. Mitochondria: in sickness and in health. Cell. 2012;148(6):1145–1159. doi: 10.1016/j.cell.2012.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sekine S., Wang C., Sideris D.P., Bunker E., Zhang Z., Youle R.J. Reciprocal roles of Tom7 and OMA1 during mitochondrial import and activation of PINK1. Mol Cell. 2019;73(5):1028–1043.e5. doi: 10.1016/j.molcel.2019.01.002. [DOI] [PubMed] [Google Scholar]
- 13.Russell O.M., Gorman G.S., Lightowlers R.N., Turnbull D.M. Mitochondrial diseases: hope for the future. Cell. 2020;181(1):168–188. doi: 10.1016/j.cell.2020.02.051. [DOI] [PubMed] [Google Scholar]
- 14.Stenton S.L., Prokisch H. Genetics of mitochondrial diseases: identifying mutations to help diagnosis. EBioMedicine. 2020;56 doi: 10.1016/j.ebiom.2020.102784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wei X., Du M., Xie J., et al. Mutations in TOMM70 lead to multi-OXPHOS deficiencies and cause severe anemia, lactic acidosis, and developmental delay. J Hum Genet. 2020;65(3):231–240. doi: 10.1038/s10038-019-0714-1. [DOI] [PubMed] [Google Scholar]
- 16.Dutta D., Briere L.C., Kanca O., et al. De novo mutations in TOMM70, a receptor of the mitochondrial import translocase, cause neurological impairment. Hum Mol Genet. 2020;29(9):1568–1579. doi: 10.1093/hmg/ddaa081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elouej S., Harhouri K., Le Mao M., et al. Loss of MTX2 causes mandibuloacral dysplasia and links mitochondrial dysfunction to altered nuclear morphology. Nat Commun. 2020;11(1):4589. doi: 10.1038/s41467-020-18146-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Young C., Batkovskyte D., Kitamura M., et al. A hypomorphic variant in the translocase of the outer mitochondrial membrane complex subunit TOMM7 causes short stature and developmental delay. HGG Adv. 2023;4(1) doi: 10.1016/j.xhgg.2022.100148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garg A., Keng W.T., Chen Z., et al. Autosomal recessive progeroid syndrome due to homozygosity for a TOMM7 variant. J Clin Invest. 2022;132(23) doi: 10.1172/JCI156864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi D., Qi M., Zhou L., et al. Endothelial mitochondrial preprotein translocase Tomm7-Rac1 signaling axis dominates cerebrovascular network homeostasis. Arterioscler Thromb Vasc Biol. 2018;38(11):2665–2677. doi: 10.1161/ATVBAHA.118.311538. [DOI] [PubMed] [Google Scholar]
- 21.Bang O.Y., Fujimura M., Kim S.K. The pathophysiology of moyamoya disease: an update. J Stroke. 2016;18(1):12–20. doi: 10.5853/jos.2015.01760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bersano A., Guey S., Bedini G., et al. Research progresses in understanding the pathophysiology of moyamoya disease. Cerebrovasc Dis. 2016;41(3–4):105–118. doi: 10.1159/000442298. [DOI] [PubMed] [Google Scholar]
- 23.Fox B.M., Dorschel K.B., Lawton M.T., Wanebo J.E. Pathophysiology of vascular stenosis and remodeling in moyamoya disease. Front Neurol. 2021;12 doi: 10.3389/fneur.2021.661578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dorschel K.B., Wanebo J.E. Genetic and proteomic contributions to the pathophysiology of moyamoya angiopathy and related vascular diseases. Appl Clin Genet. 2021;14:145–171. doi: 10.2147/TACG.S252736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu S., Wei W., Zhang F., et al. Transcriptomic profiling of intracranial arteries in adult patients with moyamoya disease reveals novel insights into its pathogenesis. Front Mol Neurosci. 2022;15 doi: 10.3389/fnmol.2022.881954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Choi J.W., Son S.M., Mook-Jung I., et al. Mitochondrial abnormalities related to the dysfunction of circulating endothelial colony-forming cells in moyamoya disease. J Neurosurg. 2018;129(5):1151–1159. doi: 10.3171/2017.5.JNS17147. [DOI] [PubMed] [Google Scholar]
- 27.MacArthur D.G., Manolio T.A., Dimmock D.P., et al. Guidelines for investigating causality of sequence variants in human disease. Nature. 2014;508(7497):469–476. doi: 10.1038/nature13127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Richards S., Aziz N., Bale S., et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quinodoz M., Peter V.G., Bedoni N., et al. AutoMap is a high performance homozygosity mapping tool using next-generation sequencing data. Nat Commun. 2021;12(1):518. doi: 10.1038/s41467-020-20584-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rao A.R., Nelson S.F. Calculating the statistical significance of rare variants causal for Mendelian and complex disorders. BMC Med Genomics. 2018;11(1):53. doi: 10.1186/s12920-018-0371-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hsieh T.C., Bar-Haim A., Moosa S., et al. GestaltMatcher facilitates rare disease matching using facial phenotype descriptors. Nat Genet. 2022;54(3):349–357. doi: 10.1038/s41588-021-01010-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu Y.T., Yu I.S., Tsai K.J., et al. Defining minimum essential factors to derive highly pure human endothelial cells from iPS/ES cells in an animal substance-free system. Sci Rep. 2015;5:9718. doi: 10.1038/srep09718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu J., Hu K., Smuga-Otto K., et al. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2009;324(5928):797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Itoh M., Umegaki-Arao N., Guo Z., Liu L., Higgins C.A., Christiano A.M. Generation of 3D skin equivalents fully reconstituted from human induced pluripotent stem cells (iPSCs) PLoS One. 2013;8(10) doi: 10.1371/journal.pone.0077673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu R.S., Lam I.I., Clay H., Duong D.N., Deo R.C., Coughlin S.R. A rapid method for directed gene knockout for screening in G0 zebrafish. Dev Cell. 2018;46(1):112–125.e4. doi: 10.1016/j.devcel.2018.06.003. [DOI] [PubMed] [Google Scholar]
- 36.Chen W.J., Huang F.C., Shih M.H. Ocular characteristics in a variant microcephalic primordial dwarfism type II. BMC Pediatr. 2019;19(1):329. doi: 10.1186/s12887-019-1685-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Waldron J.S., Hetts S.W., Armstrong-Wells J., et al. Multiple intracranial aneurysms and moyamoya disease associated with microcephalic osteodysplastic primordial dwarfism type II: surgical considerations. J Neurosurg Pediatr. 2009;4(5):439–444. doi: 10.3171/2009.6.PEDS08137. [DOI] [PubMed] [Google Scholar]
- 38.Bober M.B., Khan N., Kaplan J., et al. Majewski osteodysplastic primordial dwarfism type II (MOPD II): expanding the vascular phenotype. Am J Med Genet A. 2010;152A(4):960–965. doi: 10.1002/ajmg.a.33252. [DOI] [PubMed] [Google Scholar]
- 39.Hall J.G., Flora C., Scott C.I., Jr., Pauli R.M., Tanaka K.I. Majewski osteodysplastic primordial dwarfism type II (MOPD II): natural history and clinical findings. Am J Med Genet A. 2004;130A(1):55–72. doi: 10.1002/ajmg.a.30203. [DOI] [PubMed] [Google Scholar]
- 40.Wang X., Wang Y., Nie F., et al. Association of genetic variants with moyamoya disease in 13 000 individuals: a meta-analysis. Stroke. 2020;51(6):1647–1655. doi: 10.1161/STROKEAHA.120.029527. [DOI] [PubMed] [Google Scholar]
- 41.Ceballos F.C., Joshi P.K., Clark D.W., Ramsay M., Wilson J.F. Runs of homozygosity: windows into population history and trait architecture. Nat Rev Genet. 2018;19(4):220–234. doi: 10.1038/nrg.2017.109. [DOI] [PubMed] [Google Scholar]
- 42.Allen R., Egan B., Gabriel K., Beilharz T., Lithgow T. A conserved proline residue is present in the transmembrane-spanning domain of Tom7 and other tail-anchored protein subunits of the TOM translocase. FEBS Lett. 2002;514(2–3):347–350. doi: 10.1016/s0014-5793(02)02433-x. [DOI] [PubMed] [Google Scholar]
- 43.Yoon Y.S., Yoon D.S., Lim I.K., et al. Formation of elongated giant mitochondria in DFO-induced cellular senescence: involvement of enhanced fusion process through modulation of Fis1. J Cell Physiol. 2006;209(2):468–480. doi: 10.1002/jcp.20753. [DOI] [PubMed] [Google Scholar]
- 44.Johnston A.J., Hoogenraad J., Dougan D.A., et al. Insertion and assembly of human tom7 into the preprotein translocase complex of the outer mitochondrial membrane. J Biol Chem. 2002;277(44):42197–42204. doi: 10.1074/jbc.M205613200. [DOI] [PubMed] [Google Scholar]
- 45.Bonder M.J., Smail C., Gloudemans M.J., et al. Identification of rare and common regulatory variants in pluripotent cells using population-scale transcriptomics. Nat Genet. 2021;53(3):313–321. doi: 10.1038/s41588-021-00800-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perry L.D., Robertson F., Ganesan V. Screening for cerebrovascular disease in microcephalic osteodysplastic primordial dwarfism type II (MOPD II): an evidence-based proposal. Pediatr Neurol. 2013;48(4):294–298. doi: 10.1016/j.pediatrneurol.2012.12.010. [DOI] [PubMed] [Google Scholar]
- 47.Duker A.L., Kinderman D., Jordan C., et al. Microcephalic osteodysplastic primordial dwarfism type II is associated with global vascular disease. Orphanet J Rare Dis. 2021;16(1):231. doi: 10.1186/s13023-021-01852-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dumas S.J., Garcia-Caballero M., Carmeliet P. Metabolic Signatures of distinct endothelial phenotypes. Trends Endocrinol Metabol. 2020;31(8):580–595. doi: 10.1016/j.tem.2020.05.009. [DOI] [PubMed] [Google Scholar]
- 49.Rivera-Torres J., Acin-Perez R., Cabezas-Sanchez P., et al. Identification of mitochondrial dysfunction in Hutchinson-Gilford progeria syndrome through use of stable isotope labeling with amino acids in cell culture. J Proteonomics. 2013;91:466–477. doi: 10.1016/j.jprot.2013.08.008. [DOI] [PubMed] [Google Scholar]
- 50.Lima T., Li T.Y., Mottis A., Auwerx J. Pleiotropic effects of mitochondria in aging. Nat Aging. 2022;2(3):199–213. doi: 10.1038/s43587-022-00191-2. [DOI] [PubMed] [Google Scholar]
- 51.Sun N., Youle R.J., Finkel T. The mitochondrial basis of aging. Mol Cell. 2016;61(5):654–666. doi: 10.1016/j.molcel.2016.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Han Y., Kim S.Y. Endothelial senescence in vascular diseases: current understanding and future opportunities in senotherapeutics. Exp Mol Med. 2023;55(1):1–12. doi: 10.1038/s12276-022-00906-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bluemel G., Planque M., Madreiter-Sokolowski C.T., et al. PCK2 opposes mitochondrial respiration and maintains the redox balance in starved lung cancer cells. Free Radic Biol Med. 2021;176:34–45. doi: 10.1016/j.freeradbiomed.2021.09.007. [DOI] [PubMed] [Google Scholar]
- 54.Priesnitz C., Becker T. Pathways to balance mitochondrial translation and protein import. Genes Dev. 2018;32(19–20):1285–1296. doi: 10.1101/gad.316547.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maruszczak K.K., Ayyamperumal S., Chacinska A. Defective mitochondrial import as a challenge for cellular protein homeostasis. FEBS Lett. 2023;597(12):1555–1568. doi: 10.1002/1873-3468.14677. [DOI] [PubMed] [Google Scholar]
- 56.Martinez-Reyes I., Chandel N.S. Mitochondrial TCA cycle metabolites control physiology and disease. Nat Commun. 2020;11(1):102. doi: 10.1038/s41467-019-13668-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Veys K., Fan Z., Ghobrial M., et al. Role of the GLUT1 glucose transporter in postnatal CNS angiogenesis and blood-brain barrier integrity. Circ Res. 2020;127(4):466–482. doi: 10.1161/CIRCRESAHA.119.316463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rabbani N., Thornalley P.J. Hexokinase-2 glycolytic overload in diabetes and ischemia-reperfusion injury. Trends Endocrinol Metab. 2019;30(7):419–431. doi: 10.1016/j.tem.2019.04.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.