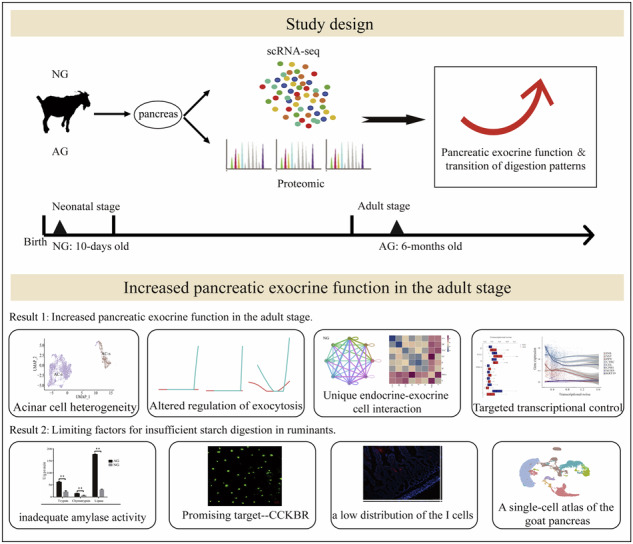
Abstract
Unlike monogastric animals, ruminants exhibit significantly lower starch digestibility in the small intestine. A better understanding of the physiological mechanisms that regulate digestion patterns in ruminants could lead to an increased use of starch concentrates. Here we show more robust pancreatic exocrine function in adult goats (AG) than in neonatal goats (NG) by combining scRNA-seq and proteomic analysis. Our findings suggest that inadequate amylase activity could be a limiting factor in starch digestion in ruminants. In addition, we show that insufficient starch hydrolysis in adult goats might be associated with low expression of a CCKBR receptor in the acinar cells. On top of that, the low expression of CCKBR in adult goats, in conjunction with a low distribution of the CCK-I cells in the duodenum, may jointly lead to a slow response of the intestinal-pancreatic reflex and induce an asynchronous process of food entering the small intestine and releasing of digestive enzymes, which ultimately limits the starch digestibility. Overall, the present findings generate a resource that can provide better insight into the mammalian pancreas.
Subject terms: Model vertebrates, Animal physiology
Multi-omics sheds light on the precise mechanisms of pancreatic functional transformation that underlie the two extreme digestion modes: non-ruminating mode (neonatal goats) and ruminating mode (adult goats).
Introduction
There are two primary modes of nutrient digestion in ruminants: rumen fermentation and enzymatic hydrolysis in the small intestine. Rumen fermentation is the dominant process, followed by enzymatic digestion1. The starch digestibility in the small intestinal lumen is generally lower in ruminants than in monogastric animals, and evidence suggests that ruminants have a limited capacity to enzymatically hydrolyze starch. Approximately 40% of starch that escapes rumen fermentation cannot be digested or utilized by ruminants2,3. Consistently, Harmon et al. reported that only 40% to 62% of rumen bypass starch is digested in dairy ruminants4. In contrast, the enzymatic digestion and utilization efficiency of starch in the small intestine is extremely high in monogastric animals, with the ileal starch digestibility exceeding 98%5–8. Notably, newborn ruminants exhibit the same digestive pattern as monogastric animals, characterized by the dominant enzymatic hydrolysis mode. This process depends on the secretion of trypsin, lipase, and α-amylase from the pancreas into the small intestine9. With the development of the rumen, ruminants possess a unique stomach structure, distinct from that of monogastric animals, comprising the rumen, reticulum, omasum, and abomasum. This stricture reflects the significant transition in digestion patterns. Thus, the pancreas function and regulation may differ between ruminants and monogastric animals, as the majority of dietary nutrients are fermented in the rumen. We hypothesized that the transition in digestion patterns following rumen development may lead to limited pancreatic function, ultimately inducing insufficient starch digestibility in the small intestine of ruminants.
Despite extensive research over the past decades, the exocrine function landscape had not been fully addressed and was challenging to construct because of the local concentration of hydrolytic enzymes that can rapidly degrade cells and RNA upon pancreatic resection10. Importantly, the physical location of the islets within the exocrine pancreas facilitates possible crosstalk, including positive and negative impacts on exocrine acinar cellular function, either by direct physical interactions or by secreted proteins and small molecules11. Emerging single-cell RNA sequencing (scRNA-seq) technology reinforced by advanced computational algorithms provides a powerful means for the systematic and genome-wide molecular identification of cell heterogeneity in pancreatic tissue12. Herein, we use 10x Genomics scRNA-seq and proteomic approaches to elucidate the precise mechanisms of pancreatic functional transformation that underlie the two extreme digestion modes: non-ruminating mode (neonatal goats) and ruminating mode (adult goats).
Results
The starch digestion capacity in ruminants is limited
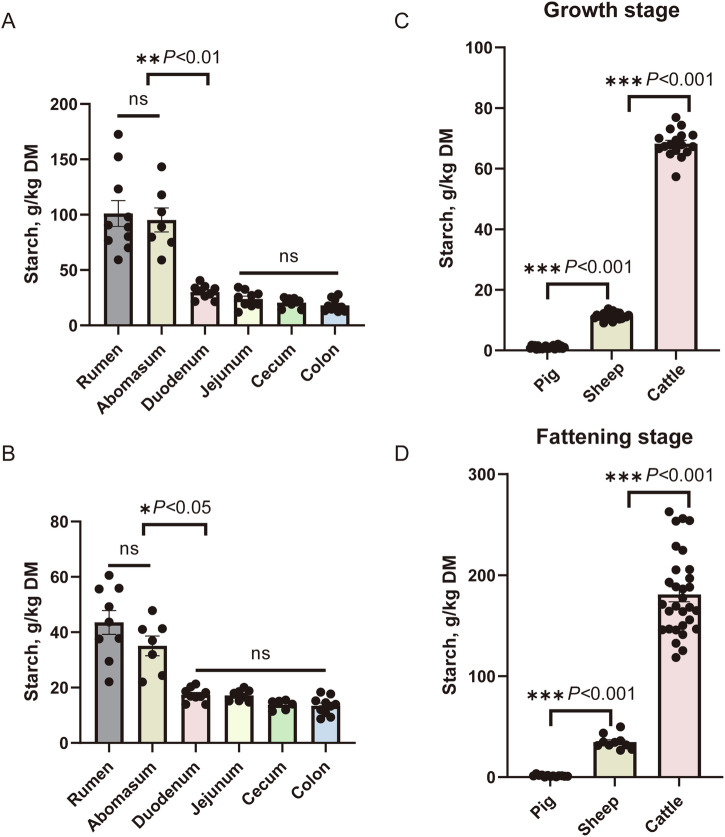
Our previous laboratory results showed that the main digestive site for rumen bypass starch was in the duodenum in sheep (Fig. 1A and B; Supplementary Data 1.1 and 1.2). The results also showed that starch content did not differ between the rumen and abomasum chyme. Furthermore, there was no significant difference in the concentration of starch between the small intestine (duodenum and jejunum) and the large intestine (cecum and colon). However, starch disappearance in the duodenum ranged from 50% to 68% of rumen bypass starch in sheep fed high-starch feed and high-fat diet. We compared the fecal starch residue levels in ruminants (cattle and sheep) and monogastric animals (pigs) during both the growth and fattening stages. The results showed that fecal starch residue was significantly higher in ruminants than in monogastric animals (sheep > pig, cattle > pig; p < 0.01) (Fig. 1C and D; Supplementary Data 1.3 and 1.4), suggesting that the limited starch digestibility in the small intestine of ruminants. Next, we studied the mechanisms underlying pancreatic functional transformation in non-ruminating (neonatal goats) and ruminating (adult goats) modes using scRNA-seq.
Fig. 1. Ruminant animals have limited starch digestion ability.
Fecal starch (g/kg dry matter) content was analyzed at various digestive parts (stomach, small intestine, and large intestine) of sheep fed a high-starch diet (A) or high-fat diet (B). Note: Due to improper sample storage and insufficient sample volume, the N values of each group are not completely consistent. For a high-starch diet, N = 10, 7, 9, 9, 9, 10 biologically independent samples per intestinal regions, respectively. For a high-fat diet, N = 9, 7, 9, 8, 6, 10 biologically independent samples per intestinal regions, respectively. C Fecal starch (g/kg dry matter) content analysis at various breeds (pig, sheep and cattle) at the growth stage and fattening stage. D Due to improper sample storage and insufficient sample volume, the N values of each group are not completely consistent. In the growth stage, N = 23, 24, and 17 biologically independent samples/breeds, respectively. In the fattening stage, N = 13, 10, and 30 biologically independent samples/breeds, respectively. The standard error of the mean (SEM) is represented by error bars.
Single-cell transcriptome for the pancreatic
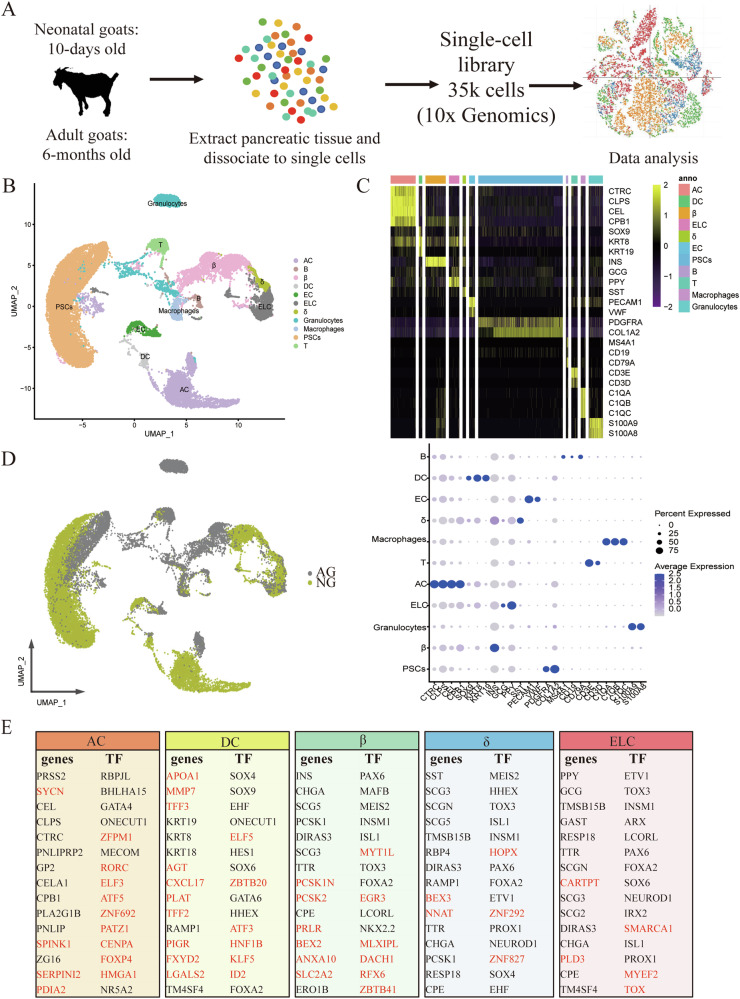
Here, we obtained more than 178 million sequence reads in total, with an average of more than 5000 reads per cell, which was sufficient for cell-type classification13 (Supplementary Data 2.1). After quality control, a total of 35,449 live cells, with a median of 1,240 genes detected per cell, were included in the downstream analysis (Supplementary Data 2.2) (Fig. 2A). We identified about 26 clusters that could be assigned into 11 major cell types (Fig. 2B, C; Fig. S1A, B; Table. S1), including acinar cells (AC), ductal cells (DC), beta cells (β), delta cells (δ), endocrine-like cells (ELC), endothelial cells (EC), activated pancreatic stellate cells (PSCs), B cells (B), T cells (T), macrophages and granulocytes. B, T, macrophages, and granulocytes cells are collectively referred to as immune cells (IC) hereafter. Interestingly, a group of cells (clusters 6 and 21, named ELC cells) was characterized by a high expression of multiple hormones (such as GCG and PPY) after the removal of the potential cell doublets (Fig. 2C). Indeed, such cells have been described by Segerstolpe et al.11 who have found that hormone co-expression was found in 0.9–1.8% of the remaining endocrine cells after the removal of the potential cell doublets. We notice that endocrine cells (β, δ and ELC) clustered together, and exocrine cells (AC and DC) clustered together (Fig. 2B), irrespective of digestion mode differences, showing the gene expression differences between exocrine and endocrine cell types. For each cell type, cells from the NG group were clustered separately from the AG group (Figs. 2D, S1A), indicating the transcriptional profile of each cell type changed with the digestive pattern transition.
Fig. 2. A transcriptomic map of the goat pancreas.
A Single-cell RNA-seq was carried out on the goat pancreas using the 10x Genomics Chromium Controller to generate data that quantify transcript abundance across cells and genes. B Uniform manifold approximation and projection (UAMP) clustering of 35,449 cells isolated from pancreas of neonatal and adult goats. Cell type was annotated by the expression of known marker genes. Each dot represents a single cell. C Heatmap summarizing mean expression (normalized and log-transformed) of selected canonical markers in each cluster. The top bar indicates the assigned cluster identity (upper). Dot plotting showing gene signatures among different clusters, the shadings denote average expression levels and the sizes of dots denote fractional expression (bottom). D UAMP clustering of cells isolated from all samples. Cells are colored based on the groups. E Tables denoting the top fifteen differentially expressed genes and transcription factors (TFs) when comparing one cell type to all other cells in the dataset. Genes whose cell-type specificity was previously unknown in the pancreas are marked in red.
We next generated the expression profiles of exocrine and endocrine cell types, and found their functional dedication to nutrient digestion and hormone secretion, respectively. In detail, a total of 21 highly expressed pancreatic digestive enzymes-related genes were identified in AC cells, accounting for a large fraction of the transcriptome (35%) in AC (Table S2, Supplementary Data 2.3 and 2.4). For endocrine cells, INS, SST, GCG and PPY transcripts accounting for 28%, 9%, and 29% of the total cellular transcripts in the β, δ, and ELC cells, respectively (Supplementary Data 2.4). The top 15 genes and transcription factors in each of β, δ, ELC, and exocrine cell types are shown in Fig. 2E. As expected, the classical transcription factors RBPJL, BHLHA15, GATA4, and NR5A2 found in AC of humans or mice14, were greatly expressed in AC of goats. Likewise, SOX9, HES1 and GATA6 were highly expressed in the DC cells of goats, consistent with the results from humans or mice14. Furthermore, our study in goats confirmed the well-known endocrine cell type-hallmark transcription factors, which were reported in humans: HHEX (δ), ARX (non-β/δ types), and PAX6 and INSM1 (all types)15. Interestingly, several genes and transcription factors found here have never been reported as markers for specific cell types of the pancreas (Fig. 2E). For example, ZFPM1(FOG1), initially discovered as a mediator of GATA4 function, was known to be co-expressed with GATA4 in intestinal epithelial cells in the mature mouse small intestine16. Our data provide evidence that ZFPM1 was co-expressed with GATA4 in AC cells of goats, opening up the question of whether ZFPM1 is a potential marker gene of AC cells. Consistent with our results in goats, the SYCN, SPINK1, SERPINI2, and PDIA genes were highly expressed in human pancreatic AC cells15. However, our data showed that MAFB and NKX2-2 were β-specific genes while NEUROD1 was restricted in non-β-specific genes in goats. These were inconsistent with the previous study in humans that showed that MAFB was restricted in α-and β cells while NEUROD1 and NKX2-2 were expressed in all endocrine cells17,18. Thus, although the overall functionality of the pancreas is conserved in evolutionary time, the specific expression across the cell types may be varied across species and deserve further investigation. Overall, we generated a high-quality cell-type-specific transcriptome of the pancreatic tissue of goats.
The mature acinar cell subtype was detected in the AG group, while the immature acinar cell subtype was detected in the NG group
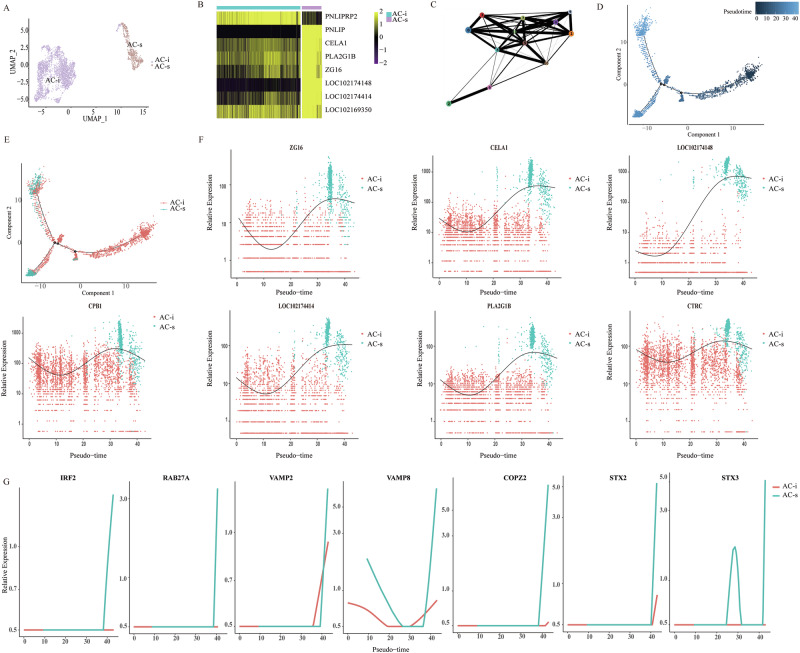
The exocrine acinar cells have attracted more attention due to the characteristic function of synthesizing multiple digestive enzymes, which are necessary for proteins, lipids, and starch digestion. We next performed differential gene expression analysis on acinar-specific genes between non-ruminating and ruminating goats and identified 13 pancreatic digestive enzyme-related genes with significantly elevated expression in acinar of the AG group (Supplementary Data 3; bimod, |log2 FC | ≥ 1 and q < 0.01). It was documented that the tight regulation of gene expression determines precise cell fates, suggesting molecular heterogeneity determines functional heterogeneity. To identify possible heterogeneity in AC cells, we merged the expression data from all AC cells and identified 12 clusters (Fig. S2A). Of interest, we found that the AC clusters 2 and 6 quantitatively consisted of cells derived from the AG samples. Conversely, the other clusters (clusters 0, 1, 3, 4, 5, 7, 8, 9, 10, 11) were quantitatively dominated by cells derived from the NG samples (Fig. S2B). Thus, we named this AC subtype with higher numbers of enzymes from the AG group as secretory acinar cells (hereafter, AC-s; clusters 2 and 6) (Fig. 3A, B, and S2C). In contrast, the expressions of digestive enzyme-related genes of AC cells from the NG group were markedly lower than those from AC-s. Thus, these AC appear less robust in their enzyme secretion and digestive processes (Fig. S2D), and we named this subset immature acinar cells (hereafter, AC-i; clusters 0, 1, 3, 4, 5, 7, 8, 9, 10, 11). As expected, GSEA analysis revealed enriched expression of genes related to the nutrient digestion process in AC-s cells of AG samples (Fig. S2D). Moreover, the difference in the AC subpopulation was further confirmed using computational algorithms. Firstly, we applied partition-based graph abstraction (PAGA) to elucidate relationships between the 12 clusters generated from all AC. The results also showed that clusters 2 and 6 were separate from the other clusters (Fig. 3C). Secondly, we used a Monocle219 to track the gene expression changes along the trajectory from the neonatal to adult stages and to identify further the key molecular events governing the AC-fate transition. We observed non-random expression patterns that ordered developmental ages along a trajectory (Fig. 3D). AC-s and AC-i were located at opposite ends of the developmental trajectory (Fig. 3E). Specifically, the expression of several AC-s candidate markers along pseudotime was elevated at the late developmental stage of the acinar cell (Figs. 3F and S2E). Overall, the mature acinar cell subtype (AC-s) with enhancing “typical” pancreatic digestive enzymes mRNA expression was found in the AG group, while the immature acinar cell subtype (AC-i) was found in the NG group.
Fig. 3. Uncovering subtypes of AC cells.
A UAMP clustering of 3483 AC cells isolated from the pancreas of neonatal and adult goats. Colored according to the assigned subtypes. B Heatmap shows the expression of the selected 8 candidates (rows) between the two AC subtypes (columns) by multi-omics analysis. The colors in the heatmap correspond to standardized log2 expression values, where each heatmap cell contains the distribution of values across the cells in each cluster. The top bar indicates the assigned subtype identity. C PAGA abstracted graph showing the most probable subgraph representing the AC data manifold. Each node corresponds to a cluster, the size of nodes is proportional to the number of cells in each cluster and edges indicate potential routes of cell transitions between them. Trajectory analysis of acinar cells colored according to pseudotime (D) and AC subtypes (E). F Expression of pancreatic digestive enzyme related genes along pseudotime. G Expression of exocytosis related genes along pseudotime.
Pancreatic enzymes are in the form of zymogen granules for transport and release into the acinus lumen via inducing the fusion of zymogen granules with the apical plasma membrane of acinar cells, namely exocytosis20. In line with our conception, IRF2, an integral transcription factor in the regulated exocytosis in the pancreas21, was elevated at the late developmental stage of AC-s (Fig. 3G). Similarly, expression of its downstream key proteins, such as soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) proteins21, RAS oncogene family member 27 A (RAB27A), vesicle-associated membranous protein 2 (VAMP2), VAMP8, coatomer protein complex subunit zeta 2 (COPZ2), syntaxin 2 (STX2) and STX3, was progressively increased along pseudotime (Fig. 3G). Digestive enzymes are separated from constitutively secreted proteins and packaged into zymogen granules (ZGs), and their fusion with the apical membrane and the process of exocytosis is physiologically controlled by well-characterized Ca2+-dependent signal transduction pathways22. Thus, pancreatic AC-s synthesize and store various enzymes, which may give rise to the intracellular electrical signal mechanisms in controlling enzyme secretion. Overall, the inferred exocytosis-related transcription factor/genes provide a valuable resource for understanding the enhancement of pancreatic exocrine function of AC-s from the AG group.
Predicted potential interactions between AC and endocrine cells correlate with reduced expression of NOTCH in the AG group
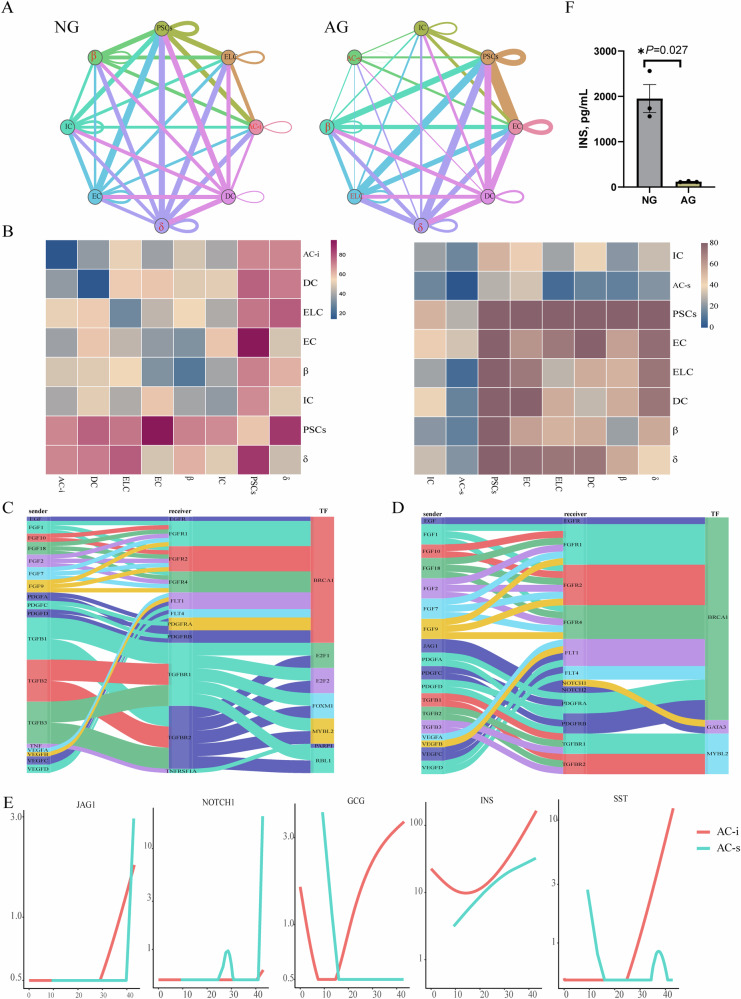
Understanding exocrine-endocrine crosstalk in the pre- and post-pancreatic development provides an opportunity to determine how these signals impact pancreatic cellular growth, differentiation, and viability, and/or contribute to endocrine or exocrine functions. We used a recently developed receptor/ligand database, CellPhoneDB23, to identify the potential of cell–cell interactions in the pancreas from the AG and NG groups to define the changes in interactions with digestion mode transition. While most interactions were unchanged, some were specific to the neonatal stage or the adult stage. Specifically, the number of predicted interactions between AC and endocrine cells was reduced in the AG group compared to the NG group (Figs. 4A, B and S3A-D). Analysis of the predicted cell–cell interactions between AC and ELC in both groups revealed a wealth of growth factor signaling pathways (Fig. 4C, D). However, AC–ELC interactions unique to the AG group included JAG1–NOTCH1 contacts (Fig. 4D), critical for pancreatic cell-fate determination, differentiation and many other biological processes24. Li et al.24 found that the activated NOTCH pathway prevents adjacent cells from adopting an endocrine fate. NOTCH signaling is also reported as a gatekeeper of acinar-to-β metaplasia in vitro25. Consistently, the expressions of JAG1 and NOTCH1 were elevated at the late developmental stage of AC-s, while the progressive increases of islet GCG, INS and SST expressions were only observed in AC-i during the late developmental stage in this study (Fig. 4E). Baeyens et al. reported that inhibition of Notch signaling promotes β-cell neogenesis via inhibiting expression of Ngn3, Pdx1, and insulin25. Consistent with this, serum insulin content in newborn goats was higher than that in adult goats (Fig. 4F; Supplementary Data 4). Thus, the above results were consistent with the possibility that the activated NOTCH pathway may modulate the enhanced pancreatic exocrine function of AC-s of AG by preventing acinar cells from adopting an endocrine fate.
Fig. 4. Pancreatic is characterized by unique cell-to-cell signaling networks at different developmental stages.
A Networks depicting cell types as nodes and interactions as edges. Edge thickness is proportional to the number of interactions between the connecting types. Interested cell communications are marked in red. B Heat map depicting the number of all possible interactions between the clusters analyzed. C Sankey diagram depicting selected AC–ELC interactions enriched in the NG group. D Sankey diagram depicting selected AC–ELC interactions enriched in the AG group. E Expression of selected genes along pseudotime. F Serum insulin content in the AG and NG groups. N = 3 biologically independent samples/group. The standard error of the mean (SEM) is represented by error bars.
Increased expression of atypical hormones and transcriptional noise in endocrine islet cells and decreased transcriptional noise in acinar cells of adult goats
Based on the results of the enhancement of pancreatic exocrine function in adult goats with changes in endocrine-to-exocrine cell signaling networks, we speculated that the endocrine function had changed in adult goats. At the next stage, we did a comparison of individual transcriptome profiles between NG and AG groups in β and δ cell types (|log2 FC | ≥ 1 and q < 0.01). We found that cellular insulin mRNA expression in β cells did not differ between the NG and AG groups. However, we observed a notable increase in the expression of “atypical” hormones, specifically SST and PPY, in the AG group (Fig. S3E). Exocrine AC in the pancreas displayed remarkable plasticity and could convert their phenotype in vitro to β cells when cultured in the presence of relevant factors26. A previous study showed two morphologically distinct types of pancreas intermediate cells (β-AC and α-AC)27. In line with these experiments, our data presented co-expression of insulin and pancreatic digestive enzymes in β cells, suggesting that AC-derived β cells may exist in vivo.
Similarly, cellular SST mRNA expression in the δ cells did not differ between the NG and AG groups; however, the expression of the marker gene PPY in γ cells was significantly increased (Fig. S3F). Analysis of single-cell transcriptomic data from mouse pancreas revealed that 95% of GHSR-expressing cells co-express PPY + SST and this expression was restricted to γ and δ cell clusters28. Analysis of several single-cell human pancreatic transcriptome data sets revealed that 57% of GHSR-expressing cells co-express PPY + SST, which was prominent in δ and β cell clusters28. Taken together, these findings indicated that a subset of δ cells co-expressed PPY and SST among goats, humans, and mouse. Overall, the fraction of β or δ co-expressing both typic and atypic hormone mRNA increased significantly in the AG group compared to the NG group (Fig. S3E, F). These findings indicate increased transcriptional instability in islet cells from the AG group relative to the NG group.
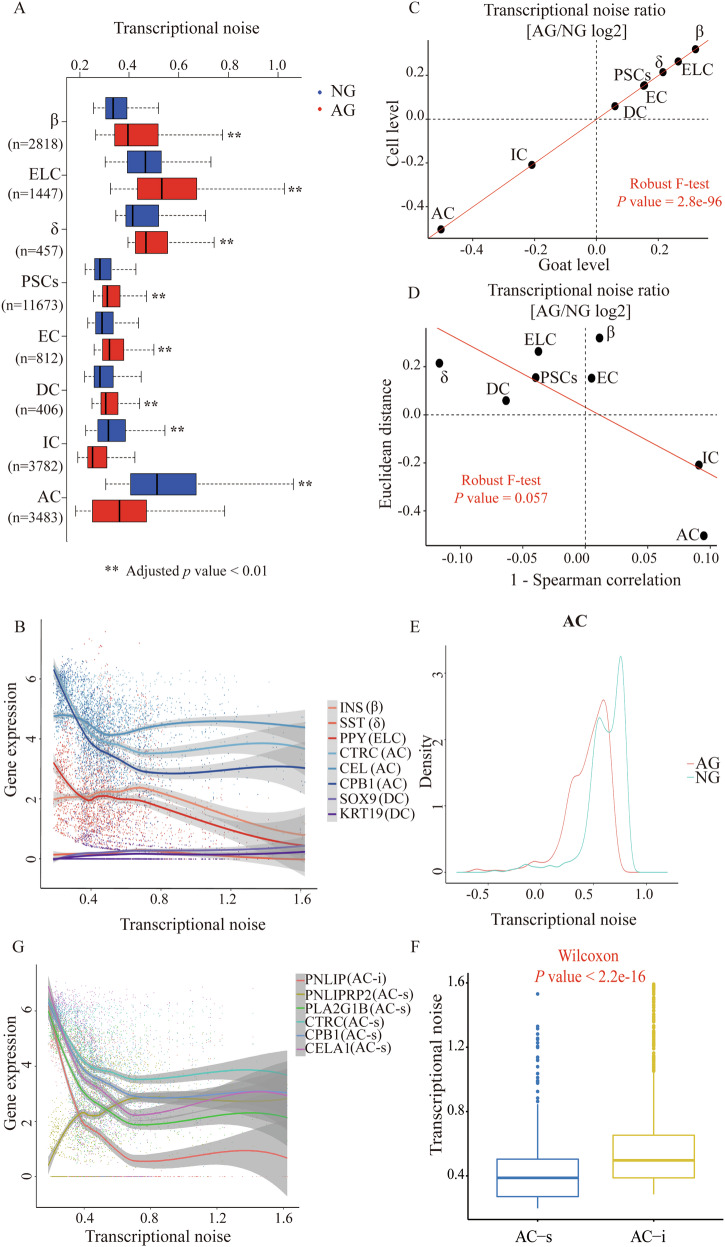
Transcriptional noise is defined as stochastic fluctuations in gene expression and it can be beneficial to population fitness by facilitating a bet-hedging strategy in fluctuating environments29. Aging-associated increases in transcriptional noise can lead to fate drifts and ambiguous cell type identities in humans30. However, it remains to be seen whether noise levels are expected to change globally in the neonatal and adult goat pancreas and in what direction to be. We quantified transcriptional noise following previous work30 and accounted for differences in total UMI counts and cell-type frequencies. As expected, we observed an increased transcriptional noise in most cell types in samples from the AG group compared to those from the NG group (Fig. 5A; Supplementary Data 5), especially for islet-associated cell types. Interestingly, the AC transcriptional noise results showed an inversion trend (Fig. 5A). We next performed linear regression on gene expression levels as a function of noise rank (batch corrected and within cell type) to investigate whether any systematic gene expression differences accompany a change in transcriptional noise. As shown in Fig. 5B, the expression of canonical hormone genes of islet cells decreased with increasing transcriptional noise. In contrast, marker genes of AC, including CTRC, CEL and CPB1, were highly expressed with decreasing transcriptional noise. To further exclude technical confounding, we averaged the transcriptional noise scores per goat and obtained highly concordant results (Fig. 5C). To further substantiate this finding, we quantified transcriptional noise in an alternative manner using Spearman’s correlations between cells. This analysis confirmed our finding that transcriptional noise in pancreatic endocrine cells was increased during the growth stage while the transcriptional noise of AC decreased (Fig. 5D, E). We further explored the difference in transcriptional noise between the AC-s from the AG group and AC-i from the NG group. Notably, the transcriptional noise in AC-s, accompanied by the decreasing transcriptional noise of its marker genes, was significantly lower than that in AC-i (Fig. 5F, G). Thus, the above results indicated that there might be a physiological basis characterized by the enhancement of exocrine function and inhibition of endocrine function by regulating the expression of “typical” enzymes and “atypical” hormones through transcriptional noise in adult goats compared to neonatal goats.
Fig. 5. Most cell types show increased transcriptional noise during the growth stage.
A Boxplot illustrates transcriptional noise by age and cell type for the indicated number of cells. For all boxplots, the box represents the interquartile range, the horizontal line in the box is the median, and the whiskers represent 1.5 times the interquartile range. Blue and red colors indicate neonatal and adult cells, respectively. Asterisk indicates significant changes (Wilcoxon’s rank sum test, adjusted p value < 0.01). Cell types are ordered by decreasing transcriptional noise ratio between neonatal and adult cells. B Expression of cell-typical markers in pancreatic endocrine and exocrine cells, ranked by transcriptional noise. Dots represent individual cells, line is running mean, with k = n / 5. C Scatterplot shows the log2 ratio of transcriptional noise between AG and NG samples as calculated using goat averages and single cells on the X and Y axes, respectively. D Scatterplot depicts the log2 ratio of transcriptional noise between AG and NG samples as calculated using 1–Spearman correlation and the Euclidean distance between cells on the X and Y axes, respectively. For both panels, the size of the dots corresponds to the negative log10 adjusted p value of the cell type-resolved differential transcriptional noise test and the red lines correspond to the robust linear model regression fit. E As an example, the distribution of 1–Spearman correlation coefficients between all pairs of neonatal and adult cells is shown for AC cells. Larger values represent increased transcriptional noise. Blue and red colors indicate NG and AG samples. F Boxplot illustrates transcriptional noise by acinar cell subtypes. G Expression of the selected markers in AC, ranked by transcriptional noise. Dots represent individual cells, line is running mean, with k = n / 5.
Increased pancreatic exocrine function was observed in the AG group compared to the NG group
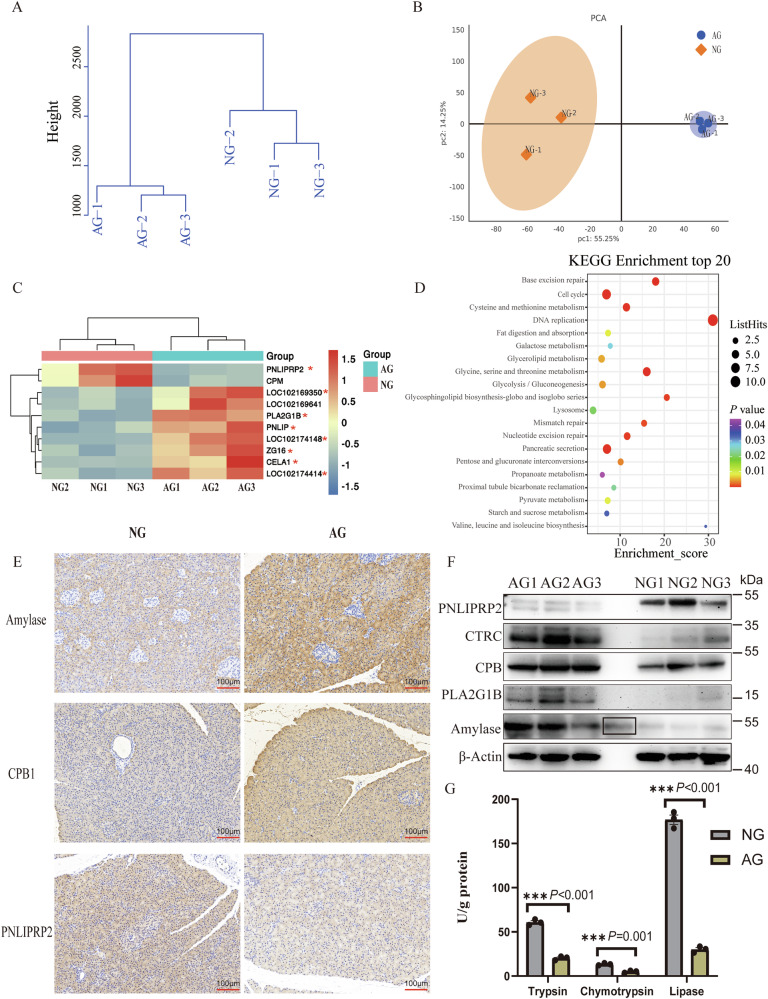
To validate the results of our single-cell RNA-sequencing (scRNA-seq) data, we performed proteomic sequencing assays. Our analysis showed a clear separation of the AG from the NG group (Fig. 6A, B). From the 5948 proteins quantified in this tissue, we identified 137 differentially expressed proteins, 47 proteins were significantly up-regulated and 90 proteins were significantly downregulated in the AG compared to the NG (t-test, |log2 FC | ≥ 1 and p < 0.05; Supplementary Data 6.1). Ten differentially expressed exocrine function-related proteins showed complementary expression patterns between the two groups (Fig. 6C). Among these, eight proteins (indicated by a red asterisk) were cross-referenced with the 13 DEGs listed in Supplementary File 5. KEGG analysis showed the up-regulated DEGs in the AG group were mainly involved in nutrient metabolism-related processes (Fig. 6D). Moreover, the results of immunohistochemistry (Fig. 6E) showed an increased expression of pancreatic digestive enzymes in the AG group that was also validated by WB analysis (Figs. 6F and S4). In detail, expression of amylase and CPB1 proteins, as well as CTRC and PLA2G1B, were increased in the AG group compared to the NG group. Expectedly, the PNLIPRP2, related to lipid digestion of neonatal animals31, was highly expressed in the NG group. Moreover, the AG cells expressed a higher level of pancreatic digestive enzymes, including trypsin, chymotrypsin, and pancreatic lipase (Fig. 6G; Supplementary Data 6.2). Notably, amylase was not detected in this study due to its low activity and/or the detection methods, which was in line with previous reports that indicated starch utilization in the small intestine of beef (55%)32 and dairy cattle (60%)33 are inefficient. Overall, we identified more robust pancreatic exocrine function in adults than in neonatal goats combine scRNA-seq and proteomic.
Fig. 6. Increased pancreatic exocrine function in adult goats.
A Hierarchical clustering dendrogram of sample Euclidean distance. B Principal component (PC) analysis of the pancreas transcriptomes for the AG (neonatal goats) and NG (adult goats (AG group) groups are shown by symbols of the different shapes. Circles represent the AG group, while diamonds represent the NG group. PC1 and PC2 represent the top two dimensions of detected proteins in the pancreatic tissue. C Heatmap of pancreatic digestive enzyme related differentially expressed proteins. Eight proteins were identified combined analysis of single-cell transcriptome and proteome and marked with red asterisks. D The top 20 KEGG enrichment pathways analysis of all DEPs in the AG group. E Immunohistochemical staining of Amylase, CPB1 and PNLIPRP2 in the pancreas from NG and AG group. Scale bar = 100 μm. F Western blot analysis of PNLIPRP2, CTRC, CPB1, PLA2G1B and Amylase in pancreatic tissue. β-Actin was used as the loading control. The black box represents the operation error during the spotting process and repeated spotting of AG3. G Pancreatic trypsin, chymotrypsin, and lipase activities in the AG and NG groups. N = 3 biologically independent samples/group. Standard error of the mean (SEM) represented error bars.
An insufficient abundance of a CCKBR receptor in the acinar cells of adult goats might limit pancreatic enzyme secretion
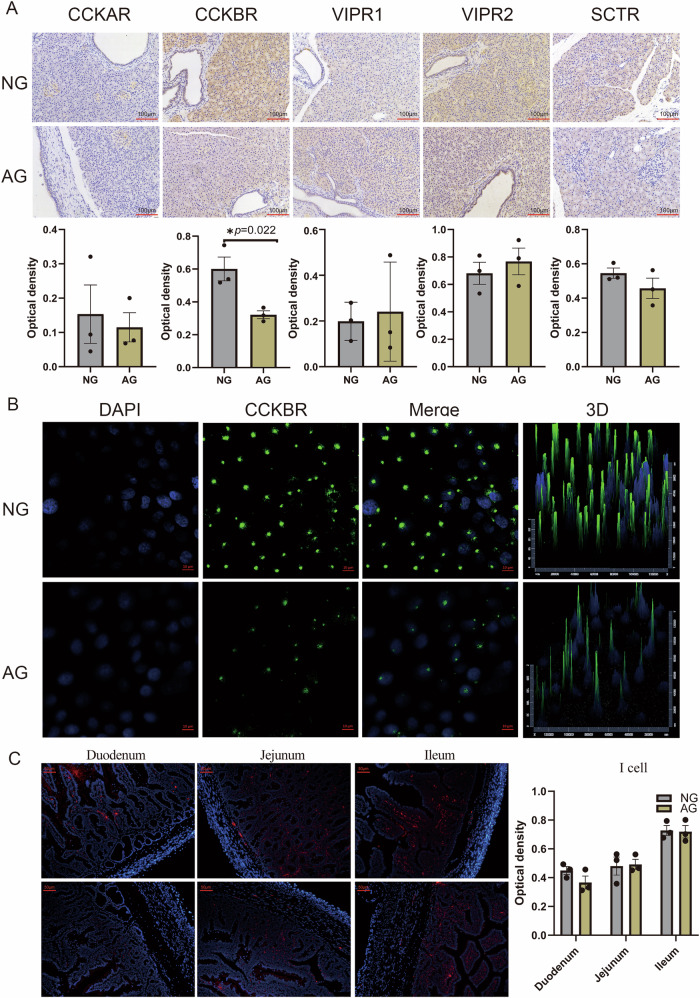
Functional receptors on acinar cells from different species that mediate pancreatic enzyme secretion have been identified for acetylcholine receptors, CCKR, GRPR, VIPR, and SCTR by measuring radio-labeled specific ligands-receptors binding responses34. In our work, only the expression of CCKBR on pancreatic acinar cells was significantly up-regulated in neonatal goats than that of adult goats (|log2 FC | ≥ 0.38 and P value ≤ 0.01) (Table 1). Consistently, immunohistochemistry with specific antibodies showed that the abundance of CCKBR, but not CCKAR, VIPR1, VIPR2, or SCTR, were higher in the acini of neonatal goats within the pancreas slice (Fig. 7A; Supplementary Data 7.1). To further prove the in vivo results, we performed the in vitro analysis at the cellular level, and the result confirmed the greater expression of CCKBR in pancreatic acinar cells of neonatal goats than that in adult goats (Fig. 7B), supporting the specificity of the changes observed. Furthermore, the distribution of CCKBR exhibited cellular polarity (Fig. 7B), which may be related to the secretion and exocytosis polarity of acinar cells. The CCK in the gut is a vital regulator of adult pancreatic growth and is essential and sufficient for the biosynthesis of pancreatic amylase, chymotrypsinogen, and trypsinogen35. In this study, we asked if the intestinal CCK-I cells, characterized by secreting CCK, has regional distribution characteristics during the development process of goats. We found that the number of CCK-immunoreactive I-cells did not vary with age in each intestinal region of goats. Of interest was the least distribution of CCK-I cells in the duodenum, which was the main site of pancreatic enzymes in the small intestine (Fig. 7C; Supplementary Data 7.2).
Table 1.
Expression of exocrine-related receptor genes on acinar cells of neonatal and adult goat pancreas using scRNA-seq
| Item | Treatment | log2FC | P-vlaue | significant | |
|---|---|---|---|---|---|
| NG | AG | ||||
| CCKAR | 0.034 | <0.001 | −0.033 | <0.001 | no |
| CCKBR | 0.703 | 0.109 | −0.430 | <0.001 | yes |
| VIPR1 | 0.004 | 0.008 | 0.004 | 0.523 | no |
| VIPR2 | 0.074 | 0.159 | 0.073 | <0.001 | no |
| SCTR | 0.032 | 0.015 | −0.017 | <0.001 | no |
| LOC102177821 | 0.004 | <0.001 | −0.017 | <0.001 | no |
LOC102177821: neuronal acetylcholine receptor subunit alpha-10.
NG neonatal goats (control group), AG adult goats (treatment group).
Fig. 7. An insufficient abundance of a CCKBR receptor in the acinar cells of adult goats compared to neonatal goats.
A Immunohistochemistry of the pancreatic CCKAR, CCKBR, VIPR1, VIPR2 and SCTR from neonatal (NG) and adult goats (AG). *P < .05 and **P < .01; values are shown as means ± SEM are shown. N = 3 biologically independent samples/group. Scale bar = 100 μm. B CCKBR staining on acinar cells from neonatal (NG) and adult goats (AG). Scale bar = 10 μm. C CCK staining on intestinal I-cells from neonatal (NG) and adult goats (AG). N = 3 biologically independent samples/group. Scale bar = 50 μm.The standard error of the mean (SEM) is represented by error bars.
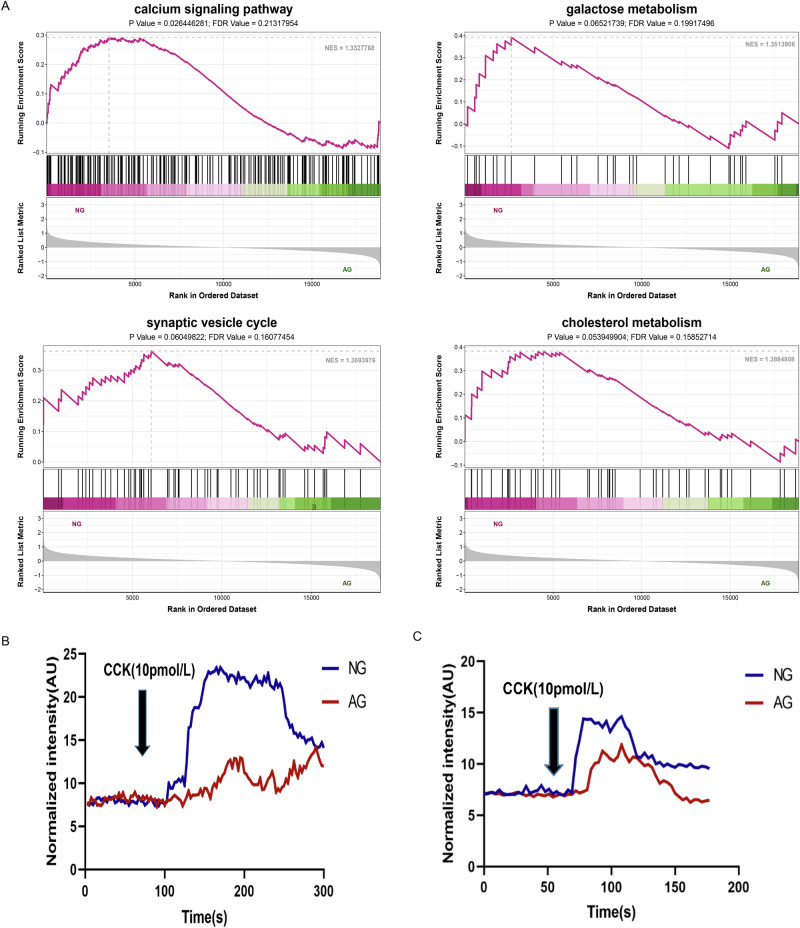
To investigate potential molecular mechanisms of how and whether CCKBR affects pancreatic exocrine function, we performed transcriptome analysis of the acinar cells from the NG and AG groups treated with a physiological concentration of CCK, respectively. Interestingly, our results showed that only CCKBR was significantly up-regulated in acini of neonatal goats compared with adults (Table 2). The GSEA showed the ‘calcium signaling pathway,’ ‘synaptic vesicle cycle,’ ‘galactose metabolism,’ and ‘cholesterol metabolism’ in the NG group were significantly enriched (Fig. 8A), indicating there may be a solid pro-pancreatic exocrine effect of CCK/CCKBR in neonatal goats by activating calcium ion signaling and exocytosis pathways. Thus, we used the calcium-sensitive fluorescence probe Fluo-4-am to detect the oscillatory Ca2+ of acinar cells under the stimulation of 10 pmol/L CCK. Firstly, the acini with good cell viability from the in vitro were imaged by confocal microscopy for oscillatory Ca2+ fluorescence for 3 min. Secondly, CCK was added to stimulate acinar cells at 60 seconds of exposure from a stable basal level. We observed a quickly CCK-stimulated Ca2+ response in acinar cells of neonatal goats at 72 s (Fig. 8B; Supplementary Movie 1). In comparison, it appeared in adult goats at 87 s (Fig. 7B; Supplementary Movie 2). Meanwhile, quantitative statistics of fluorescence signals showed that CCK initiated more robust global Ca release in the NG group than that in the AG group (Fig. 8B). Nonetheless, Ca2+ fluorescence traces showed a similar CCK-stimulated release with maximal Ca2+ fluorescence peaks at 108 s in the two groups and then returned to baseline when the stimulus was stopped (Fig. 8B). These results indicated that Ca2+ responses between the two group were relatively synchronized but uncoupled and CCK-elicited stronger increases of Ca2+ signals in the NG group than that in the AG group. CCK-elicited individual exocytotic events were measured as mentioned above. We observed exocytosis of a release-ready zymogen granule pool similar in rate and magnitude to that previously observed in Ca2+ response in such cells during CCK stimulation. Briefly, we observed an obvious exocytotic secretion in acinar cells of neonatal goats at 105 s (Fig. 8C; Supplementary Movie 3). In comparison, it appeared in adult goats at 167 s (Fig. 8C; Supplementary Movie 4). At the same time, the combined analysis of fluorescence signals with curve trend showed that CCK initiated a large amount of basolateral exocytosis in the NG group than that in the AG group (Fig. 8C).
Table 2.
Expression of exocrine-related receptor genes on acinar cells of neonatal and adult goat pancreas using RNA-seq
| Item | Treatment | log2FC | P-vlaue | significant | |
|---|---|---|---|---|---|
| NG | AG | ||||
| CCKBR | 1.397 | 0.731 | −0.934 | <0.001 | yes |
| VIPR1 | 0.331 | 0.309 | −0.097 | 0.958 | no |
| VIPR2 | 0.313 | 0.387 | −0.306 | 0.387 | no |
| SCTR | 19.587 | 20.903 | −0.094 | 0.698 | no |
| LOC102177821 | 0.255 | 0.358 | −0.489 | 0.140 | no |
LOC102177821: neuronal acetylcholine receptor subunit alpha-10.
NG neonatal goats (control group), AG adult goats (treatment group).
Fig. 8. CCKBR- mediated a strong pro-pancreatic exocrine effect by activating calcium ion signaling and exocytosis pathways.
A List of the significant GSEA terms. NG neonatal goats, AG adult goats. Statistical significance was declared at |NES | > 1, P-value < 0.05 and FDR value < 0.25. B Representative responses of Ca2+ in goat pancreatic acinar cells isolated from neonatal (NG) and adult goats (AG). A Cell viability of acinar cells from the two groups after treated with physiological concentration of CCK. B Representative responses of Ca2+ in the two groups after treated with physiological concentration of CCK. C Representative responses of exocytosis in goat pancreatic acinar cells isolated from neonatal (NG) and adult goats (AG).
Discussion
Improving nutrient digestion, especially starch in the small intestine of ruminants, is one of the prime goals for meat and milk producers. To better understand the changes in pancreatic function associated with digestive pattern transition, we present an atlas of goat pancreas and integrate scRNA-seq data with bulk proteomics to explore the underlying mechanisms. Here, our integrated scRNA-seq and proteomic analyses revealed that adult goats, which rely on rumen fermentation, exhibited stronger pancreatic exocrine function compared to neonates, which depend on pancreatic enzymes. The underlying reasons were as follows: 1) In this study, we identified AC-s subtype with higher expression of enzymes and exocytosis-related genes in adult goats. 2) There might be a physiological basis characterized by the enhancement of exocrine function and inhibition of endocrine function in adult goats. Firstly, we identified JAG1–NOTCH1 contacts in AC–ELC interactions, which were special to the AG group. Extensive theoretical and experimental research has shown that activated Notch signaling prevents acinar-to-endocrine metaplasia24,26. Consistently, serum insulin was significantly reduced in adult goats compared to neonatal goats. In the future, NOTCH pathway, its use for generating acinar metaplastic cells or mature acinar cells will help to precisely delineate pancreatic exocrine and endocrine function. Secondly, we found the increased expression of atypical hormones and transcriptional noise in endocrine islet cells of adult goats compared to neonatal goats. On the contrary, increased expression of typical enzymes and decreased transcriptional noise were found in the acinar cells of adult goats. Transcription is often stochastic, meaning it does not occur in all cells at the same time36. Thus, noise can result in significant transcript-level differences between cells37. Therefore, there may be targeted transcriptional control in adult goats that leads to declining endocrine function and increasing exocrine function. This regulation likely occurs through transcriptional noise affecting the fate of marker genes, distinguishing adult goats from neonatal goats. Our findings align with earlier studies of postnatal growth in rodents and humans, which show that the exocrine compartment accounts for approximately 90% of the pancreas in adults, compared to 50% in neonates10,38. In contrast, pancreatic endocrine cells account for 22% of detected cells in neonates compared to only 6% in adults10. 3) Increased pancreatic exocrine function in the AG group was further corroborated by IHC, WB, and enzyme activity assays.
In our work, the limitations in starch digestion and utilization in ruminants appear to be due to inadequate amylase activity rather than insufficient pancreatic enzyme secretion. Similarly, a previous review summarized that pancreatic α-amylase activity is a limiting factor for starch hydrolysis in ruminants39. Previous reports showed that the CCKAR subtype, which is presented on acinar cells, is involved in the pancreatic secretory response to feeding in humans and rodents40–42. On the contrary, our observation was that the CCKBR, not the CCKAR, was presented on acinar cells and involved in the pancreatic secretory response in goats. Furthermore, it has to be reminded that several lines of evidence indicate that the expression of CCKBR on acinar cells of newborn goats was stronger than that in adult goats. Firstly, its expression in the acinar cell clusters from neonatal goats was significantly higher than that of adult goats in our data on pancreatic scRNA-seq. Secondly, the specificity of the changes observed in scRNA-seq data was reproduced in vivo (Fig. 7A). Thirdly, as expected for the results in vivo, CCKBR increased further in the primary acinar cells of newborn goats in vitro (Fig. 7B), excluding the bias of experimental results caused by factors such as low immunohistochemical resolution and preference of the cell annotation of scRNA-seq. In addition, we validated the CCKBR-mediated a strong pro-pancreatic exocrine effect by activating calcium ion signaling and exocytosis pathways. Thus, our data provides two explanations for the insufficient starch hydrolysis in ruminants: (1) the lower proportion of starch disappearing in the small intestine of ruminants may be due to limited α-amylase activity. (2) The low expression of CCKBR in acinar cells of adult goats, combined with the sparse distribution of CCK-I cells in the duodenum, may lead to a delayed intestinal-pancreatic reflex. This delay can cause an asynchronous process where food enters the small intestine before the release of pancreatic digestive enzymes. Overall, CCKBR forms an interesting and promising target throughout the ruminants to influence important exocrine pancreas processes, which was ignored in other mammals, such as humans and rodents.
Materials and methods
Ethics approval and consent to participate
This study was approved by the Institutional Animal Care and the Use Committee of the Institute of Subtropical Agriculture, Chinese Academy of Sciences, Changsha, China (approval number 20200031). We have complied with all relevant ethical regulations regarding the use of animals.
Animals and sample collection
Six Liuyang black goats aged about 10 days and 6 months old were selected from commercial goat farms and divided into the NG (neonatal goats, n = 3) and AG (adult goats, n = 3) groups, respectively. Both groups consisted of a twin and a singleton female goat. The nutrient contents of the two groups are shown in Tables S3 and 4. Serum samples were collected before slaughter. Goats were humanely euthanized with an intravenous overdose of sodium pentobarbital, and the pancreas, mid segments of the duodenum, jejunum, and ileum tissues were freshly sampled.
Pancreatic single-cell RNA-sequencing
We performed scRNA-seq on all alive cells isolated from pancreatic tissue of neonatal and adult goats as reported previously43 (see Supplementary Materials for further details). Single-cell suspensions (700-1200 cells/μL) without cell debris and having greater than 90% viability were passed to the subsequent analysis. The scRNA-seq libraries were sequenced on the Illumina NovaSeq 6000 sequencing system in a 150-bp paired-ended manner by LC-Bio Technology Co., Ltd. (Hangzhou, China) at a minimum depth of 20,000 reads per cell. Raw data were demultiplexed, barcoded, and mapped to the goat reference genome (https://www.ncbi.nlm.nih.gov/assembly/GCF_001704415.1/). The process of clustering cells was performed using the “FindClusters” function with an appropriate resolution Two-dimensional visualization was obtained with a UMAP44. Cells were then subjected to cell trajectory analyses, transcriptional noise analyses, and functional enrichment analyses were conducted for the differentially expressed genes identified.
Determination of serum insulin
We employed a spectrophotometric method to detect serum insulin content as per instructions of the commercial kits (ELK Biotechnology Co., Ltd; ELK7816).
TMT quantitative proteomic analysis
A portion of pancreatic tissues from the AG (n = 3) and NG (n = 3) groups were used for protein extraction. The proteins were quantified by TMT 6 labeling and analyzed by Luming Biotechnology (Shanghai, China). For TMT labelling, the lyophilized samples after trypsin digestion were gently resuspended in 100 μL 200 mM TEAB, and about 40 μL of each sample was moved into new tubes. After adding 41 μL of the TMT label reagent and gently mixing, the mixture was subjected to incubation for 1 h at room temperature. To cease the reaction, we added 8 µL of 5% hydroxylamine and then incubated for 15 min. Finally, after lyophilizing the labelled peptide solutions, processed samples were placed at −80°C. A fold change (FC) > 2 or < 0.5 with a P < 0.05 was considered as significantly differential expression. Following the identification of the differentially expressed proteins (DEP), we performed KEGG enrichment analysis to describe the functions.
KEGG and GSEA analysis
The KEGG enrichment analysis, which was analyzed using DAVID, was employed to explore the potential functions and crucial pathways involved in pancreatic exocrine function. A pathway with the Benjamini–Hochberg adjusted P-value of < 0.05 was considered significant. To determine whether a wide range of previously defined gene sets were enriched in different phenotypes, we used the GSEA tool (v3.10.1) from the Broad Institute. The Molecular Signatures Database (MSigDB) was used to obtain a set of hallmark genes. Benjamini–Hochberg adjusted for P < 0.05 and false discovery rate <0.05 were used as significance cutoff criteria. The enrichment score indicated the degree to which a gene set was overrepresented at the top or bottom of a ranked list of genes.
Immunohistochemistry (IHC), WB and pancreatic digestive enzyme activities analysis
Fresh pancreatic tissue samples were formalin-fixed and paraffin-embedded for immunohistochemistry following standard protocols. For WB, total proteins were separated using 12% SDS-PAGE and subsequently electro-transferred onto a PVDF membrane, which was then sealed with QuickBlockTM Blocking Buffer (Beyotime, P0252) at room temperature for 20 min. After that, the PVDF membrane was cut according to the size of the target protein. Then, the membranes were incubated with primary antibodies in TBST, including 5% BSA, overnight at 4 °C, then incubated again with secondary antibodies. The antibody information is shown in Table S5.
Another portion of pancreatic samples (n = 3/group) were homogenized in 5 volumes v/w of DEPC water. The samples were centrifuged (3000 × g, 10 min at 4 °C), and then the supernatant was collected to determine the total protein and enzyme activities. We employed a spectrophotometric method to detect the activity levels of trypsin, chymotrypsin, lipase, and α-amylase as per instructions of the commercial kits (ZCIBIO Technology Co., Ltd).
Primary cell culture of pancreatic acinar cells and Immunofluorescence (IF)
A piece of the pancreas tissue was quickly transferred to the cell culture laboratory through transfer fluid, and fat and fascia were removed and trimmed. The tissue was chopped into small pieces and transferred into a 15-ml sterile enzyme-free centrifuge tube containing 10 ml collagenase type IV (1 mg/mL). The pancreas and enzyme solution were then incubated at 37 °C with reciprocal shaking at 80 r/min. After 20 minutes, we used a slender tip pipette to mix well and centrifuged at 1000 rpm for 5 min. The suspension was subjected to a filtration process using a 70-μm cell strainer, and then larger cell clumps were discarded. The filtered acini were rinsed thrice with DMEM/F12 medium containing 10x penicillin-streptomycin mixture, 2.5 μg/mL gentamicin, 2.5 μg/mL amphotericin, 10 μg/mL insulin, 10 ng/mL EGF and 10% FBS and resuspended at a cellular density of 3 × 105 cell/cm2 in the same media at 37 °C, 5% CO2.
For daily morphologic examination, we used an inverted phase-contrast photomicroscope to monitor and photograph cultures. The viability of acinar cells was tested using the CCK8 test method. The culture showing 91% viability was used for further studies. The purity of acinar cells was checked by differential digestion, adhesion, and cell scraping methods. To identify the acinar cells, we checked the expression of acinar-specific marker carboxypeptidase A1 (CPA1) using immunofluorescence staining (IF). A portion of primary pancreatic cells, duodenum, jejunum, and ileum tissues from twin female goats (n = 2/group) were fixed with a 4% buffered formalin solution for immunofluorescence staining assays following standard protocols as described in the study of Konno et al. 45. We used a well-characterized anti-CCK antibody, which has been validated in previous studies, to specifically label CCK-producing cells46. The antibody information is shown in Table S5.
RNA sequencing
The mRNA sequence was performed by the LC-Bio Technology Co.Ltd., (Hangzhou, China). Each NG and AG group has 4 culture plates, 10 pmol/L CCK was used to treat pancreatic acinar cells from the NG and AG groups for 30 min. Cell supernatant and cells were collected and centrifuged at 10,000 rpm at 4°C. Total RNA from the acinar cells was extracted using TRIzol (Thermofisher, 15596018) as per the manufacturer’s guidelines. After checking the quantity and purity of RNA, 1 μg of total RNA per sample (n = 4 / group) with good integrity (RIN number >7.0) was used for library preparation on the Illumina NovaseqTM 6000 sequence platform. TruSeq RNA Library Prep Kit v2 (Illumina, CA, USA) was used to construct the library for RNA-seq. The average size of paired-end library inserts was 300 bp (±50 bp). The sequencing data were mapped to the goat reference genome (capra hircus, GCF_001704415.2_ARS1.2) using the HISAT package47 and assembled using StringTie48. The expression data were estimated by calculating FPKM using StringTie. We also applied Ballgown with |log2 FC | > 0.38 and p-value < 0.05 to execute the differential expression analysis. We used the OmicStudio tools to perform the bioinformatic analysis (https://www.omicstudio.cn). Benjamini–Hochberg adjusted for P < 0.05 and false discovery rate <0.25 were used as significance cutoff criteria for GSEA analysis.
Ca2+ and exocytosis measurements in pancreatic acinar cell populations
Change in cytosolic Ca2+ ion concentration in response to CCK treatment or concentration was evaluated by using a Fluo-4 probe adapted from Murphy et al.49. Briefly, acinar cells from the NG and AG groups were loaded with Fluo-4 by preincubation in 5 μM Fluo-4-am (working fluids: 100 μg Fluo-4-am and 18.2325 ml PBS; MCE HY101896) for 30 min at 37 °C. After 1 minute of exposure, Fluo-4-acinar cells were loaded with a physiologically representative concentration of CCK (10 pmol/mL) for 2 min, and single-cell fluorescence was conducted at 488 nm and emitted at 512 ~ 520 nm. Confocal imaging of cells was recorded using a Zeiss LSM880 system (Carl Zeiss, Jena, Germany).
Exocytotic responses were measured using FM1-43 (5 μmol/L; Biorigin BN14014) dye, loaded at 37 °C for 15 minutes until a stable basal fluorescence level was attained (excitation, 480 nm; emission, >510 nm)50. Time-lapse imaging was performed with the above system with an oil immersion objective and a 491-nm laser at 5 min. After 1 min exposure time, FM1-43-acinar cells from the NG and AG groups were loaded with 10 pmol/mL CCK for 4 min, and imaging of the cells was performed using the imaging systems.
Statistics and reproducibility
The data, including residual fecal starch, INS, IHC, IF staining, and digestive enzyme profile, were subjected to statistical analysis using SPSS version 23. One-way ANOVA (with Multiple comparisons using Tamhane’s test) was used to compare multiple groups, while the independent-sample t-test was used to compare two groups. Statistical significance was determined at P < 0.05 (*P < 0.05, **P < 0.01, ***P < 0.001). The standard error of the mean (SEM) is represented by error bars; sample/group details are provided in Figure Captions.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
This research was funded by Chinese Academy of Sciences (Strategic Priority Research Program Grant NO. XDA26040304, XDA26050102), the Natural Science Foundation of Hunan Province of China (2022JJ10054) and Hunan Innovation Province Project (2019RS3021). We thank the LC-Bio Technology co.ltd., (Hangzhou, China) for providing single-cell transcriptomics services and the Shanghai Luming biological technology co., LTD (Shanghai, China) for proteomics services. We would like to thank Sanjie Jiang (from BGI Genomics, Shenzhen, China) for the assistance with the transcriptional noise analysis.
Author contributions
Yan Cheng: conceptualization, methodology, validation, formal analysis, investigation, writing—original draft, visualization. Tianxi Zhang: methodology, formal analysis, investigation, visualization. Chao Yang: methodology, validation, investigation. Kefyalew Gebeyew: validation, writing—review and editing, supervision. Chengyu Ye: methodology, software, data curation. Xinxin Zhou: software, data curation, visualization. Tianqi Zhang: conceptualization, methodology, writing—review and editing. Ganyi Feng: methodology, formal analysis. Rui Li: methodology, formal analysis. Zhixiong He: conceptualization, resources, writing—review and editing, supervision. Oren Parnas: conceptualization, writing—review and editing, supervision. Zhiliang Tan: conceptualization, resources, writing—review and editing, supervision.
Peer review
Peer review information
: Communications Biology thanks Shin Hamada, Stefan Pierzynowski and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Simona Chera and Laura Rodríguez Pérez. [A peer review file is available.]
Data availability
Raw data for TMT quantitative proteomic, scRNA-seq and RNA-seq have been submitted to the ProteomeXchange Consortium and National Center for Biotechnology Information with accession numbers: PXD034499 (https://www.iprox.cn/page/project.html?id=IPX0004563000), GSE207644 and PRJNA1007276, respectively. In addition, data for all graphs in the manuscript are provided in the Supplementary Data file.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Zhixiong He, Email: zxhe@isa.ac.cn.
Oren Parnas, Email: oren.parnas@gmail.com.
Zhiliang Tan, Email: zltan@isa.ac.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-024-07406-9.
References
- 1.Guo, L., Yao, J. H. & Cao, Y. C. Regulation of pancreatic exocrine in ruminants and the related mechanism: The signal transduction and more. Anim. Nutr.7, 1145–1151 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moharrery, A., Larsen, M. & Weisbjerg, M. R. Starch digestion in the rumen, small intestine, and hind gut of dairy cows - A meta-analysis. Anim. Feed Sci. Technol.192, 1–14 (2014). [Google Scholar]
- 3.Kreikemeier, K. K. & Harmon, D. L. ABOMASAL GLUCOSE, MAIZE STARCH AND MAIZE DEXTRIN INFUSIONS IN CATTLE - SMALL-INTESTINAL DISAPPEARANCE, NET PORTAL GLUCOSE FLUX AND ILEAL OLIGOSACCHARIDE FLOW. Br. J. Nutr.73, 763–772 (1995). [DOI] [PubMed] [Google Scholar]
- 4.Harmon, D. L., Yamka, R. M. & Elam, N. A. Factors affecting intestinal starch digestion in ruminants: A review. Can. J. Anim. Sci.84, 309–318 (2004). [Google Scholar]
- 5.Wang, J. F., Zhu, Y. H., Li, D. F., Jorgensen, H. & Jensen, B. B. The influence of different fiber and starch types on nutrient balance and energy metabolism in growing pigs, Asian-Australasian. J. Anim. Sci.17, 263–270 (2004). [Google Scholar]
- 6.Weurding, R. E., Veldman, A., Veen, W. A. G., van der Aar, P. J. & Verstegen, M. W. A. Starch digestion rate in the small intestine of broiler chickens differs among feedstuffs. J. Nutr.131, 2329–2335 (2001). [DOI] [PubMed] [Google Scholar]
- 7.Li, Y. et al. In vitro and in vivo digestibility of corn starch for weaned pigs: Effects of amylose: amylopectin ratio, extrusion, storage duration, and enzyme supplementation. J. Anim. Sci.93, 3512–3520 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Lin, F. D., Knabe, D. A. & Tanksley, T. D. APPARENT DIGESTIBILITY OF AMINO-ACIDS, GROSS ENERGY AND STARCH IN CORN, SORGHUM, WHEAT, BARLEY, OAT GROATS AND WHEAT MIDDLINGS FOR GROWING-PIGS. J. Anim. Sci.64, 1655–1663 (1987). [DOI] [PubMed] [Google Scholar]
- 9.Whitcomb, D. C. & Lowe, M. E. Human pancreatic digestive enzymes. Dig. Dis. Sci.52, 1–17 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Tosti, L. et al. Single-Nucleus and In Situ RNA-Sequencing Reveal Cell Topographies in the Human Pancreas. Gastroenterology160, 1330 (2021). [DOI] [PubMed] [Google Scholar]
- 11.Segerstolpe, A. et al. Single-Cell Transcriptome Profiling of Human Pancreatic Islets in Health and Type 2 Diabetes. Cell Metab.24, 593–607 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang, X. N. et al. Comparative Analysis of Droplet-Based Ultra-High-Throughput Single-Cell RNA-Seq Systems. Mol. Cell73, 130 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Pollen, A. A. et al. Low-coverage single-cell mRNA sequencing reveals cellular heterogeneity and activated signaling pathways in developing cerebral cortex. Nat. Biotechnol.32, 1053 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cleveland, M. H., Sawyer, J. M., Afelik, S., Jensen, J. & Leach, S. D. Exocrine ontogenies: On the development of pancreatic acinar, ductal and centroacinar cells. Semin. Cell Developmental Biol.23, 711–719 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li, J. et al. Single-cell transcriptomes reveal characteristic features of human pancreatic islet cell types. Embo Rep.17, 178–187 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beuling, E. et al. GATA4 mediates gene repression in the mature mouse small intestine through interactions with friend of GATA (FOG) cofactors. Developmental Biol.322, 179–189 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bonal, C. & Herrera, P. L. Genes controlling pancreas ontogeny. Int. J. Developmental Biol.52, 823–835 (2008). [DOI] [PubMed] [Google Scholar]
- 18.Artner, I. et al. MafB - An activator of the glucagon gene expressed in developing islet alpha- and beta-cells. Diabetes55, 297–304 (2006). [DOI] [PubMed] [Google Scholar]
- 19.Qiu, X. et al. Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods14, 979 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Palade, G. INTRACELLULAR ASPECTS OF PROCESS OF PROTEIN-SYNTHESIS. Science189, 347–358 (1975). [DOI] [PubMed] [Google Scholar]
- 21.Mashima, H. et al. Interferon Regulatory Factor-2 Regulates Exocytosis Mechanisms Mediated by SNAREs in Pancreatic Acinar Cells. Gastroenterology141, 1102–U459 (2011). [DOI] [PubMed] [Google Scholar]
- 22.Williams, J. A. Regulation of pancreatic acinar cell function. Curr. Opin. Gastroenterol.22, 498–504 (2006). [DOI] [PubMed] [Google Scholar]
- 23.Efremova, M., Vento-Tormo, M., Teichmann, S. A. & Vento-Tormo, R. CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat. Protoc.15, 1484–1506 (2020). [DOI] [PubMed] [Google Scholar]
- 24.X. Y. Li, W. J. Zhai, C. B. Teng, Notch Signaling in Pancreatic Development. Int. J. Mol. Sci. 17, 48 (2016). [DOI] [PMC free article] [PubMed]
- 25.Baeyens, L. et al. Notch Signaling as Gatekeeper of Rat Acinar-to-beta-Cell Conversion in Vitro,. Gastroenterology136, 1750–1760 (2009). [DOI] [PubMed] [Google Scholar]
- 26.Baeyens, L. et al. Notch Signaling as Gatekeeper of Rat Acinar-to-β-Cell Conversion in Vitro. Gastroenterology136, 1750–1760 (2009). [DOI] [PubMed] [Google Scholar]
- 27.Aughsteen, A. A. Immunofluorescence and electron-microscopic observations of intermediate cells in the pancreas of mice, rats and humans. Cells Tissues Organs170, 21–28 (2002). [DOI] [PubMed] [Google Scholar]
- 28.Gupta, D. et al. High Coexpression of the Ghrelin and LEAP2 Receptor GHSR With Pancreatic Polypeptide in Mouse and Human Islets. Endocrinology162, bqab148 (2021). [DOI] [PMC free article] [PubMed]
- 29.Acar, M., Mettetal, J. T. & van Oudenaarden, A. Stochastic switching as a survival strategy in fluctuating environments. Nat. Genet.40, 471–475 (2008). [DOI] [PubMed] [Google Scholar]
- 30.Angelidis, I. et al. An atlas of the aging lung mapped by single cell transcriptomics and deep tissue proteomics. Nat. Commun.10, 963 (2019). [DOI] [PMC free article] [PubMed]
- 31.Bakala N’Goma, J.-C., Amara, S., Dridi, K., Jannin, V. & Carriere, F. Understanding the lipid-digestion processes in the GI tract before designing lipid-based drug-delivery systems. Therapeutic Deliv.3, 105–124 (2012). [DOI] [PubMed] [Google Scholar]
- 32.Owens, F., Zinn, R. & Kim, Y. Limits to starch digestion in the ruminant small intestine. J. Anim. Sci.63, 1634–1648 (1986). [DOI] [PubMed] [Google Scholar]
- 33.Moharrery, A., Larsen, M. & Weisbjerg, M. R. Starch digestion in the rumen, small intestine, and hind gut of dairy cows–A meta-analysis. Anim. Feed Sci. Technol.192, 1–14 (2014). [Google Scholar]
- 34.Williams, J. A. Regulation of acinar cell function in the pancreas. Curr. Opin. Gastroenterol.26, 478–483 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Williams, J. A. Intracellular signaling mechanisms activated by cholecystokinin-regulating synthesis and secretion of digestive enzymes in pancreatic acinar cells. Annu. Rev. Physiol.63, 77–97 (2001). [DOI] [PubMed] [Google Scholar]
- 36.Raj, A. & van Oudenaarden, A. Nature, Nurture, or Chance: Stochastic Gene Expression and Its Consequences. Cell135, 216–226 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stapel, L. C., Zechner, C. & Vastenhouw, N. L. Uniform gene expression in embryos is achieved by temporal averaging of transcription noise. Genes Dev.31, 1635–1640 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kachar, B., Taga, R., Kniebel, G. A. & Sesso, A. MORPHOMETRIC EVALUATION OF THE NUMBER OF EXOCRINE PANCREATIC CELLS DURING EARLY POSTNATAL-GROWTH IN THE RAT. Acta Anatomica103, 11–15 (1979). [DOI] [PubMed] [Google Scholar]
- 39.Huntington, G. B. Starch utilization by ruminants: From basics to the bunk. J. Anim. Sci.75, 852–867 (1997). [DOI] [PubMed] [Google Scholar]
- 40.Liang, T. et al. Ex vivo human pancreatic slice preparations offer a valuable model for studying pancreatic exocrine biology. J. Biol. Chem.292, 5957–5969 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wank, S. A. et al. PURIFICATION, MOLECULAR-CLONING, AND FUNCTIONAL EXPRESSION OF THE CHOLECYSTOKININ RECEPTOR FROM RAT PANCREAS. Proc. Natl Acad. Sci. USA89, 3125–3129 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rehfeld, J.F. Cholecystokinin-From Local Gut Hormone to Ubiquitous Messenger. Front. Endocrinol. 8, 47 (2017). [DOI] [PMC free article] [PubMed]
- 43.Xu, Z. Y. et al. Single-cell RNA sequencing and lipidomics reveal cell and lipid dynamics of fat infiltration in skeletal muscle. J. Cachexia Sarcopenia Muscle12, 109–129 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Becht, E. et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol.37, 38 (2019). [DOI] [PubMed] [Google Scholar]
- 45.Konno, K. et al. Cellular and subcellular localization of cholecystokinin (CCK)-1 receptors in the pancreas, gallbladder, and stomach of mice. Histochemistry Cell Biol.143, 301–312 (2015). [DOI] [PubMed] [Google Scholar]
- 46.Zhang, W. X. et al. Micro/nanoplastics impair the feeding of goldfish by disrupting the complicated peripheral and central regulation of appetite. Sci. Total Environ. 946, 174112 (2024). [DOI] [PubMed]
- 47.Kim, D., Landmead, B. & Salzberg, S. L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods12, 357–U121 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pertea, M. et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol.33, 290 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Murphy, J. A. et al. Direct activation of cytosolic Ca2+ signaling and enzyme secretion by cholecystokinin in human pancreatic acinar cells. Gastroenterology135, 632–641 (2008). [DOI] [PubMed] [Google Scholar]
- 50.Gaisano, H. Y. et al. Supramaximal cholecystokinin displaces Munc18c from the pancreatic acinar basal surface, redirecting apical exocytosis to the basal membrane. J. Clin. Investig.108, 1597–1611 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
Raw data for TMT quantitative proteomic, scRNA-seq and RNA-seq have been submitted to the ProteomeXchange Consortium and National Center for Biotechnology Information with accession numbers: PXD034499 (https://www.iprox.cn/page/project.html?id=IPX0004563000), GSE207644 and PRJNA1007276, respectively. In addition, data for all graphs in the manuscript are provided in the Supplementary Data file.