Abstract
The application of traditional morphological and ecological species concepts to closely related, asexual fungal taxa is challenging due to the lack of distinctive morphological characters and frequent cosmopolitan and plurivorous behaviour. As a result, multilocus sequence analysis (MLSA) has become a powerful and widely used tool to recognise and delimit independent evolutionary lineages (IEL) in fungi. However, MLSA can mask discordances in individual gene trees and lead to misinterpretation of speciation events. This phenomenon has been extensively documented in Diaporthe, and species identifications in this genus remains an ongoing challenge. However, the accurate delimitation of Diaporthe species is critical as the genus encompasses several cosmopolitan pathogens that cause serious diseases on many economically important plant hosts. In this regard, following a survey of palm leaf spotting fungi in Lisbon, Portugal, Diaporthe species occurring on Arecaceae hosts were used as a case study to implement an integrative taxonomic approach for a reliable species identification in the genus. Molecular analyses based on the genealogical concordance phylogenetic species recognition (GCPSR) and DNA-based species delimitation methods revealed that speciation events in the genus have been highly overestimated. Most IEL identified by the GCPSR were also recognised by Poisson tree processes (PTP) coalescent-based methods, which indicated that phylogenetic lineages in Diaporthe are likely influenced by incomplete lineage sorting (ILS) and reticulation events. Furthermore, the recognition of genetic recombination signals and the evaluation of genetic variability based on sequence polymorphisms reinforced these hypotheses. New clues towards the intraspecific variation in the common loci used for phylogenetic inference of Diaporthe species are discussed. These results demonstrate that intraspecific variability has often been used as an indicator to introduce new species in Diaporthe, which has led to a proliferation of species names in the genus. Based on these data, 53 species are reduced to synonymy with 18 existing Diaporthe species, and a new species, D. pygmaeae, is introduced. Thirteen new plant host-fungus associations are reported, all of which represent new host family records for Arecaceae. This study has recognised and resolved a total of 14 valid Diaporthe species associated with Arecaceae hosts worldwide, some of which are associated with disease symptoms. This illustrates the need for more systematic research to examine the complex of Diaporthe taxa associated with palms and determine their potential pathogenicity. By implementing a more rational framework for future studies on species delimitation in Diaporthe, this study provides a solid foundation to stabilise the taxonomy of species in the genus. Guidelines for species recognition, definition and identification in Diaporthe are included.
Taxonomic novelties: New species: Diaporthe pygmaeae D.S. Pereira & A.J.L. Phillips. New synonyms: Diaporthe afzeliae Monkai & Lumyong, Diaporthe alangii C.M. Tian & Q. Yang, Diaporthe araliae-chinensis S.Y. Wang et al., Diaporthe australiana R.G. Shivas et al., Diaporthe australpacifica Y.P. Tan & R.G. Shivas, Diaporthe bombacis Monkai & Lumyong, Diaporthe caryae C.M. Tian & Q. Yang, Diaporthe chimonanthi (C.Q. Chang et al.) Y.H. Gao & L. Cai, Diaporthe conferta H. Dong et al., Diaporthe diospyrina Y.K. Bai & X.L. Fan, Diaporthe durionigena L.D. Thao et al., Diaporthe etinsideae Y.P. Tan & R.G. Shivas, Diaporthe eucalyptorum Crous & R.G. Shivas, Diaporthe fujianensis Jayaward. et al., Diaporthe fusiformis Jayaward. et al., Diaporthe globoostiolata Monkai & Lumyong, Diaporthe hainanensis Qin Yang, Diaporthe hongkongensis R.R. Gomes et al., Diaporthe hubeiensis Dissan. et al., Diaporthe infecunda R.R. Gomes et al., Diaporthe italiana Chethana et al., Diaporthe juglandigena S.Y. Wang et al., Diaporthe lagerstroemiae (C.Q. Chang et al.) Y.H. Gao & L. Cai, Diaporthe lithocarpi (Y.H. Gao et al.) Y.H. Gao & L. Cai, Diaporthe lutescens S.T. Huang et al., Diaporthe machili S.T. Huang et al., Diaporthe megabiguttulata M. Luo et al., Diaporthe middletonii R.G. Shivas et al., Diaporthe morindae M. Luo et al., Diaporthe nannuoshanensis S.T. Huang et al., Diaporthe nigra Brahman. & K.D. Hyde, Diaporthe orixae Q.T. Lu & Zhen Zhang, Diaporthe passifloricola Crous & M.J. Wingf., Diaporthe pimpinellae Abeywickrama et al., Diaporthe pseudoinconspicua T.G.L Oliveira et al., Diaporthe pungensis S.T. Huang et al., Diaporthe rhodomyrti C.M. Tian & Qin Yang, Diaporthe rosae M.C. Samar. & K.D. Hyde, Diaporthe rumicicola Manawas et al., Diaporthe salicicola R.G. Shivas et al., Diaporthe samaneae Monkai & Lumyong, Diaporthe subcylindrospora S.K. Huang et al., Diaporthe tectonae Doilom et al., Diaporthe tectonigena Doilom et al., Diaporthe theobromatis H. Dong et al., Diaporthe thunbergiicola Udayanga & K.D. Hyde, Diaporthe tuyouyouiae Y.P. Tan et al., Diaporthe unshiuensis F. Huang et al., Diaporthe vochysiae S.A. Noriler et al., Diaporthe xishuangbannaensis Hongsanan & K.D. Hyde, Diaporthe xylocarpi M.S. Calabon & E.B.G. Jones, Diaporthe zaobaisu Y.S. Guo & G.P. Wang, Diaporthe zhaoqingensis M. Luo et al.
Citation: Pereira DS, Phillips AJL (2024). Diaporthe species on palms – integrative taxonomic approach for species boundaries delimitation in the genus Diaporthe, with the description of D. pygmaeae sp. nov. Studies in Mycology 109: 487–594. doi: 10.3114/sim.2024.109.08
Keywords: coalescent-based methods, genetic distance-based methods, integrative taxonomy, new taxa, species limits
INTRODUCTION
Species are the fundamental units of biodiversity, ecology, evolution, and bioconservation. Yet, defining and recognising a species has been an ongoing challenge (De Queiroz 2007, Ellis 2011). Many species concepts and recognition criteria have been proposed over the years. However, whether it is based on morphological, biological, ecological or phylogenetic characters, there are numerous disagreements regarding the acceptable criteria to delineate a fungal species (Giraud et al. 2008, Lücking et al. 2020, Xu 2020, Stengel et al. 2022). The high degree of phenotypic plasticity, homologous sequence data and broad range of hosts, often lead to different conclusions about how to determine the variation and boundaries between fungal taxa (Jeewon & Hyde 2016, Steenkamp et al. 2018, Chethana et al. 2020, 2021a).
Since the early 1990s, the use of DNA sequence analyses increased the complexity of fungal taxonomy, giving rise to new difficulties in understanding evolutionary processes and relationships. Although phylogenetic-based studies have changed the perception of fungal diversity, relationships at species level and recognition of species remain controversial and subject to different interpretations (Hyde et al. 2013, Jeewon & Hyde 2016, Xu 2016, 2020, Maharachchikumbura et al. 2021). The concatenation of multilocus DNA sequence data represents a powerful and commonly used approach to recognise independent evolutionary lineages (IEL) in fungi (Dupuis et al. 2012). However, the process is not always straightforward as it can mask the discordances among loci and lead to misinterpretations of the speciation events (Kubatko & Degnan 2007).
The genealogical concordance phylogenetic species recognition (GCPSR) (Taylor et al. 2000), which relies on the comparison of more than one gene genealogy, has proven to be a suitable tool for species delimitation, especially in morphologically conserved fungi (Cai et al. 2011, Laurence et al. 2014, Udayanga et al. 2014a). According to the GCPSR, recombination within a lineage is likely to cause conflict between gene trees and the transition from conflict to congruence indicates the species limit (Taylor et al. 2000). Even so, genes can have significantly different evolutionary histories, which make the species boundaries difficult to ascertain in the early stages of speciation. Thus, processes such as incomplete lineage sorting (ILS) and other reticulation events can also cause gene-tree/species-tree discordance and ultimately mislead the evolutionary relationships among closely related taxa (Castresana 2007, Degnan & Rosenberg 2006, 2009, Schrempf & Szöllősi 2020).
For the above reasons, modern systematics rely on integrative taxonomic approaches that combine different sources of evidence for identifying, delimiting, and describing species. Subsequently, congruence between results from different methods and datasets is likely to lead to more reliably supported species limits (Padial et al. 2010, Carstens et al. 2013, Bustamante et al. 2019, Stengel et al. 2022). Recent studies have successfully applied coalescentand genetic distance-based methods to aid species delimitation in taxa where cryptic speciation processes have been reported. Coalescent-based methods, which stochastically connect sampled gene lineages backward in time, allow alternative hypotheses of evolutionary independence to be tested. These methods can infer the relationships among taxa and delimit IEL objectively even when there is incongruence between gene genealogies and lack of reciprocal monophyly among lineages. Thus, coalescent-based methods provide a more comprehensive view of speciation events than the GCPSR (Rannala & Yang 2003, 2020, Liu et al. 2009, Sukumaran & Knowles 2017). On the other hand, genetic distance-based methods rely on the close clustering of haplotypes as an indication of the species boundaries, which will therefore depend on the difference in variation of intra- and interspecific diversification rates. These methods can infer putative relationships among taxa by accessing pairwise nucleotide genetic distances in the sequences of a given dataset. These are grouped according to the assumption that the genetic variation within species is smaller than between species (DeSalle et al. 2005, Del-Prado et al. 2010, Zou et al. 2011, Krishnamurthy & Francis 2012).
Coalescent- and genetic distance-based methods have proved to be very useful to estimate species trees and support species boundaries for several animal (e.g., Waters et al. 2010, Satler et al. 2013, Yu et al. 2017, Guo & Kong 2022) and plant (e.g., Carston et al. 2009, Karbstein et al. 2020, Vieira et al. 2023) taxa. However, they have rarely been used in fungi. A few studies have examined the utility of these methods for closely related, morphologically conserved asexual fungal taxa, such as Alternaria (Stewart et al. 2014), Aspergillus (Sklenář et al. 2017, 2021), Beauveria (Bustamante et al. 2019), Colletotrichum (Liu et al. 2016), Fusarium (Achari et al. 2020) and, more recently, Diaporthe (Hilário et al. 2021a, b, Pereira et al. 2023).
Species of Diaporthe (syn. Phomopsis) have a diverse host range and a wide geographical distribution, occurring mostly as plant pathogens, endophytes and saprobes, but also as pathogens of humans and other mammals (Murali et al. 2006, Garcia-Reyne et al. 2011, Udayanga et al. 2011, Marin-Felix et al. 2019). Species recognition criteria in Diaporthe have evolved from morphology and host association (Uecker 1988) to the widespread use of DNA sequence data (Udayanga et al. 2012a, b, Tan et al. 2013, Dissanayake et al. 2017, Lambert et al. 2023). Modern systematic accounts of the genus rely on multilocus sequence analyses (MLSA) based on the nuclear ribosomal internal transcribed spacer (ITS) region, and the translation elongation factor 1-alpha (tef1), beta-tubulin (tub2), histone (his3), and calmodulin (cal) loci (Gomes et al. 2013, Santos et al. 2017a, Guo et al. 2020, Sun et al. 2021, Wang et al. 2021). Although Diaporthe has received much attention, and many phylogenetic studies have been conducted over the years, the taxonomy of the genus is still uncertain, and several lineages remain poorly understood. Cryptic diversification, phenotypic plasticity and extensive host associations, coupled with its paraphyletic nature, have long made it difficult to accurately identify Diaporthe spp. and ultimately impair a truthful analysis of their species boundaries (Gao et al. 2017, Norphanphoun et al. 2022).
In recent years, most studies have considered distinct well-supported Diaporthe clades recognised by MLSA as distinct species, disregarding key factors to determine species status, such as incongruences among gene genealogies and lack of gene flow between populations. However, gene concatenation has been shown to impair reliable tree topologies when there are high levels of ILS, recombination or other reticulation events, resulting in poor species discrimination (Kubatko & Degnan 2007, Mendes & Hahn 2018). Therefore, Diaporthe is an ideal case study for the application of quantitative species recognition methodologies, using methods that incorporate uncertainty in gene trees, and that assess intra- and interspecific diversification rates. Recent studies have shown significant incongruences among the gene genealogies and identified the presence of recombination events, indicating divergent evolutionary histories among the common loci used for phylogenetic inference in Diaporthe (Hilário et al. 2021a, b, Pereira et al. 2023). Hilário et al. (2021a, b) resolved the D. amygdali and D. eres species complexes through the application of the GCPSR principle, along with coalescent-based models, providing evidence that each complex constitutes a population with intraspecific variability rather than different lineages. Pereira et al. (2023) recognised three IEL in the D. arecae species complex (DASC) and reduced 52 species to synonym under D. arecae following an integrative taxonomic approach that included genealogical concordance and coalescent-based analyses. Hypotheses suggested by the authors to explain these results included recombination and ILS. Despite having been reported in previous studies, these phylogenetic incongruences are still overlooked in most Diaporthe spp.
Considering that the high intraspecific variability of Diaporthe has been erroneously used to delimit species, the true species diversity of the genus is likely to be highly overestimated. In this sense, in the present study, Diaporthe species occurring on Arecaceae hosts were used as a case study to gain new insights into the phylogenetic species delimitation in the genus. No intensive study supported by molecular data has yet been carried out to resolve the complex of Diaporthe species occurring on Arecaceae hosts. Although several Diaporthe species have been recorded on palms, most were mainly based on host association but without molecular data to confirm their phylogenetic position. As a result, most of these taxa have not been transferred to Diaporthe and remain in Phomopsis. Fröhlich et al. (1997) provided a synopsis of Diaporthe (as Phomopsis) species known from palms and several other species have been reported by Taylor & Hyde (2003).
Recently, Pereira et al. (2023) resolved the species boundaries delimitation in the DASC and provided new data on the Diaporthe species occurring on palms. The present study builds on this previous research and aims to: (1) implement an integrative taxonomic approach, comprising single and multilocus phylogenetic analyses, coalescent- and genetic distance-based species delimitation methods, phylogenetic networks and hierarchical cluster analysis of phenotypic data, to reliably identify species in the genus Diaporthe; (2) propose practical guidelines to provide a more rational framework, based on scientific data, on how to delineate species boundaries and establish novel taxa in the genus Diaporthe; (3) identify a set of potential phytopathogenic isolates of Diaporthe obtained from foliar lesions of ornamental palm trees in Lisbon, Portugal; and (4) resolve the complex of Diaporthe species occurring on Arecaceae hosts. A new synopsis of currently accepted and phylogenetically validated Diaporthe species reported from palms worldwide is also presented.
MATERIALS AND METHODS
Specimen collection, examination, and single-spore isolation
Between October 2018 and May 2021, diseased leaf segments and leaflets with foliar lesions were collected from different ornamental palm tree species in Lisbon, Portugal, including Chamaerops humilis, Chrysalidocarpus lutescens, Phoenix roebelenii, Trachycarpus fortunei, and Washingtonia filifera (detailed information of collection sites and respective hosts are referred to in the Taxonomy section). Plant material was transported to the laboratory in paper envelopes and examined with a Leica MZ9.5 stereo microscope (Leica Microsystems GmbH, Germany) for observations on lesion morphology and associated fungi.
Isolations were made directly from foliar lesions. Pieces of tissue 1–2 mm2 were cut from the edge of the lesion, surface disinfected in 5 % sodium hypochlorite for 1 min, rinsed in three changes of sterile water and blotted dry on sterile filter paper. The fragments were plated onto 1/2 strength Potato Dextrose Agar (1/2 PDA) (PDA; BIOKAR Diagnostics, France) containing 0.05 % chloramphenicol (CPDA) (Chloramphenicol; Sigma-Aldrich, Canada) and incubated at room temperature until colonies developed. Colonies were subcultured onto 1/2 PDA, and single-spore isolates were subsequently established. Sterilisation efficiency was controlled by sterilised leaf fragments impressions and plating of the last sterile water change onto CPDA.
Four of the isolates used in the present study, i.e., CDP 0022, CDP 0209, CDP 0315 (D. foeniculina) and CDP 0052 (D. pyracanthae), were previously reported as a preliminary study on Diaporthe occurring on palms published in Boonmee et al. (2021) and their morphomolecular characterisation was re-accessed here.
Morphological observation and characterisation
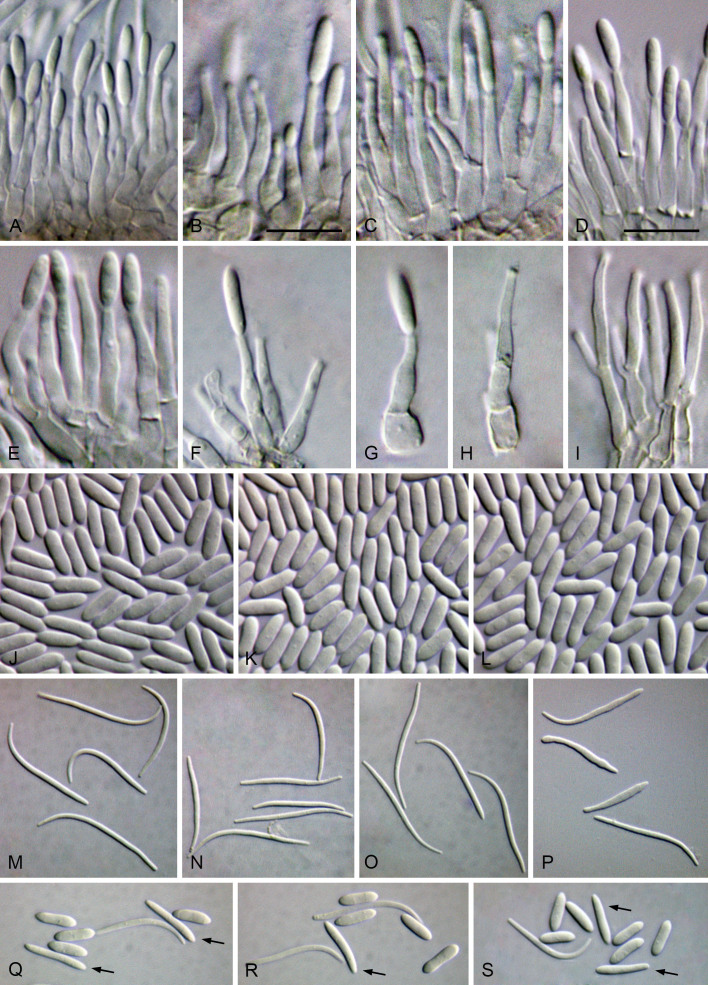
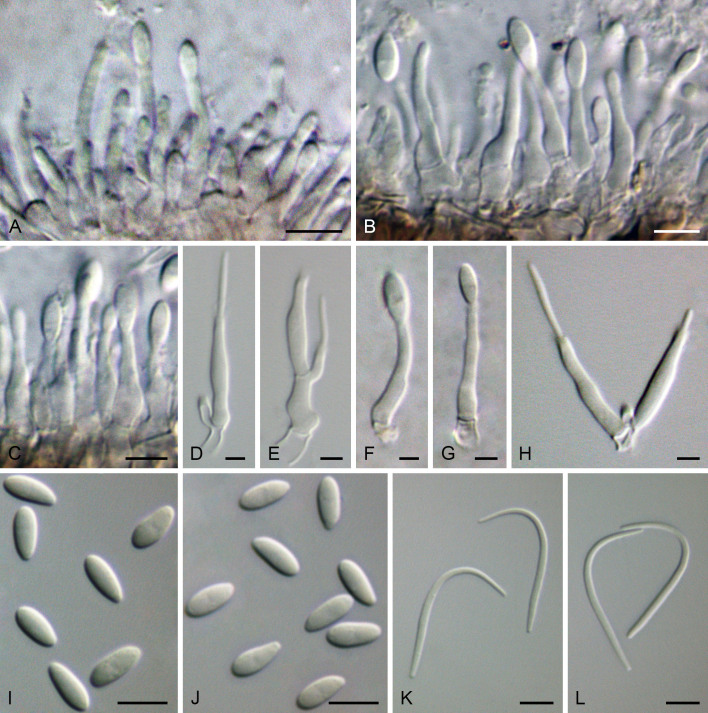
Cultures were induced to sporulate by culturing on 2 % water agar (WA) (Bacteriological Agar Type E; BIOKAR Diagnostics, France) bearing healthy doubled autoclaved (two cycles of 20 min, 121 °C and 1 bar with 48 h between each cycle) palm leaf pieces. After a suitable period of incubation at 28 °C under black light, ranging from 3 d to 1 wk, conidiomata were cut through vertically and the conidiogenous layer dissected. Microscopic structures (pycnidia, conidiophores, conidiogenous cells and conidia) were mounted in 100 % lactic acid and examined by differential interference contrast (DIC) microscopy. Observations on micromorphological features were made with Leica MZ9.5 and Leica DMR microscopes (Leica Microsystems GmbH, Germany), and digital images were recorded with Leica DFC300 and Leica DFC320 cameras (Leica Microsystems GmbH, Germany), respectively. Measurements were made with the measurement module of the Leica IM500 Image Management System (Leica Microsystems GmbH, Germany). Mean, standard deviation (SD) and 95 % confidence intervals were calculated from n = total of measured structures. Measurements are given as minimum and maximum dimensions with mean and SD in parenthesis. Photoplates were prepared with Adobe Photoshop CS6 Extended (Adobe, USA).
DNA extraction, PCR amplification and sequencing
Genomic DNA (gDNA) was extracted from mycelium of cultures grown on 1/2 PDA following a modified and optimized version of the guanidium thiocyanate method described by Pitcher et al. (1989). Amplification reactions were carried out with Taq polymerase, nucleotides, primers, PCR-grade water (ultrapure DNase/RNase-free distilled water) and buffers supplied by Invitrogen (USA). Primer details and respective amplification targets are listed in Table 1. Amplification reactions were performed in an UNO II Thermocycler (Biometra, Germany). The PCR products were checked on 1 % agarose electrophoresis gels stained with ethidium bromide and visualised on a UV transilluminator to assess PCR amplification. Amplified PCR products were purified and sequenced by Eurofins Genomics (Germany).
Table 1.
Details of primers used for amplification and sequencing.
| Locus1 | Primer name | Direction | Sequence (5’ → 3’) | Reference |
|---|---|---|---|---|
| cal | CAL-228F | Forward | GAGTTCAAGGAGGCCTTCTCCC | Carbone & Kohn (1999) |
| CAL-737R | Reverse | CATCTTTCTGGCCATCATGG | Carbone & Kohn (1999) | |
| ITS | ITS5 | Forward | GGAAGTAAAAGTCGTAACAAGG | White et al. (1990) |
| NL4 | Reverse | GGTCCGTGTTTCAAGACGG | O’Donnell (1993) | |
| tef1 | EF1-688F | Forward | CGGTCACTTGATCTACAAGTGC | Alves et al. (2008) |
| EF1-1251R | Reverse | CCTCGAACTCACCAGTACCG | Alves et al. (2008) | |
| tub2 | T1 | Forward | AACATGCGTGAGATTGTAAGT | O’Donnell & Cigelnik (1997) |
| Bt2b | Reverse | ACCCTCAGTGTAGTGACCCTTGGC | Glass & Donaldson (1995) |
1cal: partial calmodulin gene; ITS: partial cluster of nrRNA genes, including the nuclear 5.8S rRNA gene and its flanking internal transcribed spacer regions ITS1 and ITS2; tef1: partial translation elongation factor 1-alpha gene; tub2: partial beta-tubulin gene.
The PCR reaction mixtures and respective cycling conditions used to amplify the cluster of nrRNA genes (ITS) and part of the translation elongation factor 1-alpha gene (tef1) and the β-tubulin gene (tub2), were performed as described by Pereira & Phillips (2023). Primers CAL-228F and CAL-737R were used to amplify part of the calmodulin gene (cal). The PCR reaction mixture for each primer pair consisted of 50–100 ng of gDNA, 1× PCR buffer, 25 pmol of each primer, 200 μM of each dNTP, 3 mM MgCl2, 3 % of DMSO, 1 U Taq DNA polymerase and was made up to a total volume of 25 μL with PCR-water. Negative controls with PCR water instead of the template DNA were included in every set of amplification reactions. The following cycling conditions were used: initial denaturation at 95 °C for 5 min, followed by 40 cycles of denaturation at 94 °C for 30 s, annealing at 55 °C for 45 s and elongation at 72 °C for 1 min, and a final elongation step at 72 °C for 10 min.
ITS and tef1 were sequenced only in the forward direction using the primers ITS5 and EF1-688F, respectively, while tub2 and cal were sequenced in both directions using the same primers as used for the DNA amplification. Consensus sequences for tub2 and cal were produced using BioEdit v. 7.0.5.3 (Hall 1999). All newly generated sequences were deposited in GenBank and their accession numbers are listed in Table 2.
Table 2.
Collection details and GenBank accession numbers of taxa included in the phylogenetic analyses.
1 Taxon or strain previous name is noted in brackets if different from current name for taxa that have been synonymised (indicated by syn.) or resolved in the present study or in previous studies.
2 Acronyms of culture collections, AR, FAU: isolates in culture collection of Systematic Mycology and Microbiology Laboratory, USDA-ARS, Beltsville, Maryland, USA; ATCC: American Type Culture Collection, Virginia, USA; BRIP: Plant Pathology Herbarium, Department of Primary Industries, Dutton Park, Queensland, Australia; CAA: Collection of Artur Alves housed at Department of Biology, University of Aveiro, Portugal; CBS: CBS-KNAW Fungal Bio-diversity Centre, Utrecht, The Netherlands; CDP: culture collection of D.S. Pereira, housed at the Lab Bugworkers | M&B-BioISI | Tec Labs – Innovation Centre, Faculty of Sciences, University of Lisbon, Lisbon, Portugal; CECT: Spanish Type Culture Collection at University of Valencia, Valencia, Spain; CFCC: China Forestry Culture Collection Center, Beijing, China; CGMCC: China General Microbiological Culture Collection Center, China; CGMHD: Chang Gung Memorial Hospital, Taiwan; CPC: working collection of P.W. Crous, housed at CBS; DAL: strains deposited in fungal collection of the Instituto Agroforestal Mediterráneo, Universitat Politècnica de València, Valencia, Spain; GUCC: Guizhou University Culture Collection; GZCC: Guizhou Academy of Agricultural Sciences Culture Collection, Guizhou, China; JZB: culture collection of Institute of Plant and Environmental Protection, Beijing Academy of Agriculture and Forestry Sciences, Beijing 100097, China; KUMCC: Culture Collection of Kunming Institute of Botany, Kunming, China; KUNCC: Kunming Institute of Botany Culture Collection, Chinese Academy of Sciences, Kunming, China; LC: working collection of Lei Cai, housed at Laboratory State Key Laboratory of Mycology, Institute of Microbiology, Chinese Academy of Sciences, China; LGMF: Culture Collection of Laboratório de Genética de Microrganismos (LabGeM), Universidade Federal do Paraná, Curitiba, Brazil; MFLU: Herbarium of Mae Fah Luang University, Chiang Rai, Thailand; MFLUCC: Mae Fah Luang University Culture Collection, Chiang Rai, Thailand; MUM: Culture Collection of Micoteca da Universidade do Minho, Braga, Portugal; NCYUCC: National Chiayi University Culture Collection, Taiwan; NTUCC: Department of Plant Pathology and Microbiology, National Taiwan University Culture Collection, Taiwan; NTUPPMCC: Department of Plant Pathology and Microbiology, National Taiwan University Culture Collection; PSCG: personal culture collection of Y.S. Guo, China; SAUCC: Shandong Agricultural University Culture Collection, China; SCHM: Mycological Herbarium of South China Agricultural University, Guangzhou, China; SDBR-CMU: Culture Collection of Sustainable Development of Biological Resources Laboratory at Chiang Mai University, Chiang Mai, Thailand; STMA: culture collection of HZI, Helmholtz Center for Infection Research, Braunschweig, Germany; URM: Culture Collection of Universidade Federal de Pernambuco, Recife, Brazil; VPRI: Victorian Plant Pathology Herbarium, National Collection of Fungi, Knoxfield, Victoria, Australia; VTCC: Vietnam Type Culture Collection, Center of Biotechnology, Vietnam National University, Hanoi, Vietnam; ZHKUCC: University of Agriculture and Engineering Culture Collection, China.
3 Status of the strains or specimens are noted by bold superscript ET (ex-epitype), H (holotype), IT (ex-isotype), PT (ex-paratype) and T (ex-type).
4 Newly generated sequences are in bold; n/a: sequences not available; cal: partial calmodulin gene; his3: partial histone H3 gene; ITS: partial cluster of nrRNA genes, including the nuclear 5.8S rRNA gene and its flanking internal transcribed spacer regions ITS1 and ITS2; tef1: partial translation elongation factor 1-alpha gene; tub2: partial beta-tubulin gene.
Sequence alignment and phylogenetic analyses
A preliminary identification based on BLASTn searches with the ITS sequences of the isolates from the present study was carried out to determine the most closely related taxa, whose sequences were subsequently retrieved from GenBank. Isolates of Diaporthe obtained from palm tissues listed in recent literature (i.e., Gomes et al. 2013, Azuddin et al. 2021, Sun et al. 2021) or deposited in GenBank were also used. An initial phylogenetic analysis with all species currently accepted in the genus Diaporthe was conducted. The resulting tree was compared with recent literature on Diaporthe. Nine well-supported clades containing the Diaporthe isolates from palms examined in the present study were selected for further analyses. These clades were populated and the available ITS, tef1, tub2, cal and his3 sequences for the corresponding taxa were retrieved from GenBank and their accession numbers are listed in Table 2.
Sequences for each locus were aligned with ClustalX v. 2.1 (Thompson et al. 1997) using the following parameters: pairwise alignment parameters (gap opening = 10, gap extension = 0.1) and multiple alignment parameters (gap opening = 10, gap extension = 0.2, DNA transition weight = 0.5, delay divergent sequences = 25 %). Alignments were checked, and manual adjustments were made wherever necessary with BioEdit v. 7.0.5.3 (Hall 1999). Terminal regions with missing data and ambiguously aligned regions were visually checked and manually excluded from the analysis. Sequences were combined in concatenated matrices using MEGA X v. 10.2.6 (Kumar et al. 2018).
Maximum likelihood (ML), maximum parsimony (MP) and Bayesian analyses (BA) were used for phylogenetic inferences of the concatenated alignments and were implemented on the CIPRES Science Gateway portal v. 3.3 (Miller et al. 2010) using RAxML-NG v. 1.1.0 (Kozlov et al. 2019), PAUP v. 4.0a165 (Swofford 2002) and MrBayes v. 3.2.7a (Ronquist et al. 2012), respectively. The resulting trees were visualised with FigTree v. 1.4.4 (Rambaut 2018) and prepared with Adobe Illustrator CS2 v. 12.0.0 (Adobe, USA). All phylogenetic inferences included a set of outgroup taxa corresponding to Diaporthe species closely related to the Diaporthe clade under analysis as inferred by the initial phylogenetic analysis conducted.
The best-fit nucleotide substitution model for each locus was determined using MEGA X v. 10.2.6 (Kumar et al. 2018) under the Bayesian information criterion (BIC). Clade stability and robustness of the branches of the best scoring ML tree were estimated by conducting a rapid bootstrap (BS) analysis with iterations halted automatically by RAxML-NG.
Maximum parsimony analyses were performed using the heuristic search option with 1 000 random taxa additions and tree bisection and reconnection (TBR) as the branch-swapping algorithm. All characters were unordered and of equal weight, and alignment gaps were treated as missing data. Maxtrees was set to 10 000, branches of zero length were collapsed and all multiple, and equally parsimonious trees were saved. The first equally most parsimonious tree was used as reference when multiple equally most parsimonious trees were obtained. Clade stability and robustness of the most parsimonious trees were assessed using BS analysis with 1 000 pseudoreplicates, each with 10 replicates of random stepwise addition of taxa (Felsenstein 1985, Hillis & Bull 1993). Descriptive tree statistics for parsimony such as tree length (TL), homoplasy index (HI), consistency index (CI), retention index (RI) and rescaled consistency index (RC) were calculated.
The Bayesian analyses were computed with four simultaneous Markov Chain Monte Carlo (MCMC) chains for two runs, 10 000 000 generations and a sampling frequency of 10 generations, ending the run automatically when standard deviation of split frequencies (SDSF) fell below 0.01. The first 25 % of trees were discarded as the burn-in fraction, while the remaining 75 % were used to calculate the 50 % majority rule consensus tree and posterior probability (PP) values.
The resulting alignments and phylogenetic trees were deposited in figshare (www.figshare.com, DOI: 10.6084/m9.figshare.27144723, accessed during October 2024).
Phylogenetic species recognition
Individual gene trees were assessed to compare highly supported clades in order to detect clade conflicts between the individual phylogenies and to accordingly apply the GCPSR principle (Taylor et al. 2000) to determine the species boundaries of each Diaporthe clade. Subclades were ranked to independent evolutionary lineages (IEL) if they were a) well-delimited from other lineages as dichotomous branches with relevant relative length; and b) well-supported (ML-BS and MP-BS ≥ 70 %) in a single gene tree and not contradicted at or above this level of support in more than one other single gene tree. For these assessments, ML and MP analyses were conducted as described above for single gene sequence alignments. Species with missing data for a given gene were excluded from the analyses of that gene region. The IEL were determined conclusively if resolved with high support values (ML/MP-BS ≥ 70 % and PP ≥ 0.90) in most phylogenetic analyses of the combined datasets. For all potential synonyms and new species, their phylogenetic position, branch length to the closest sister species and support values for the clustering were evaluated.
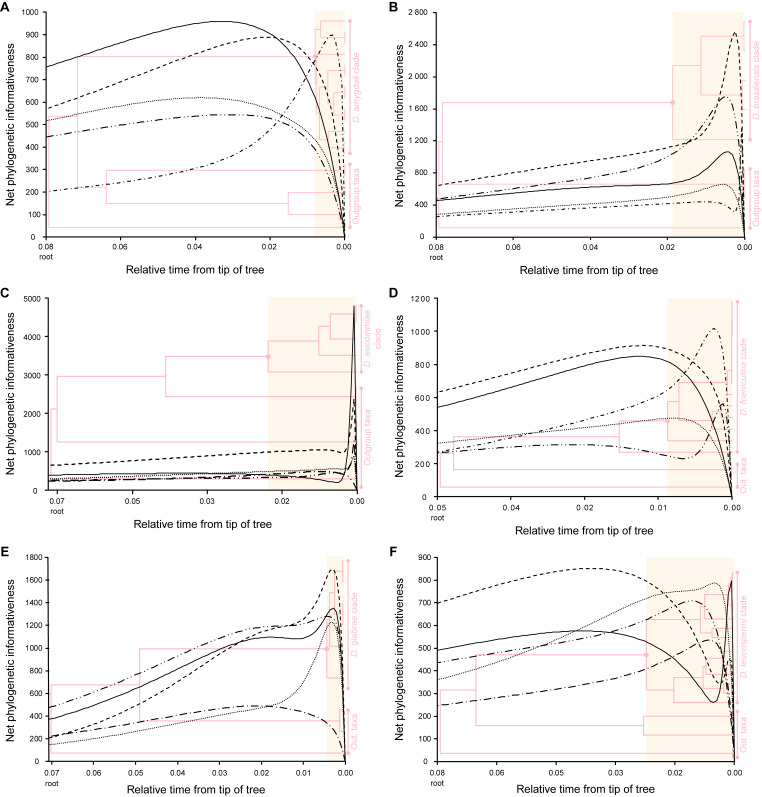
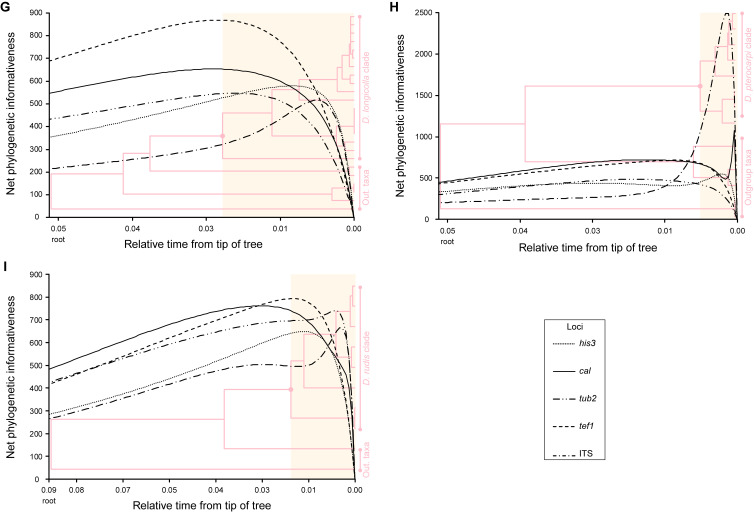
Phylogenetic informativeness analysis
To determine the loci most suitable for phylogenetic inference in each Diaporthe clade, the phylogenetic informativeness (PI) profiling method (Townsend 2007) was employed. The analysis was implemented in PhyDesign (López-Giráldez & Townsend 2011) web server (http://phydesign.townsend.yale.edu/, accessed during November 2023). The PI was measured from a partitioned combined dataset of the ITS, tef1, tub2, cal and his3 loci for the strains of each Diaporthe clade for which the five loci were available, including the set of closely related outgroup taxa. An ML inference from RAxML-NG analysis of the combined dataset was performed as described above, using a general time reversible (GTR) nucleotide substitution model including a discrete gamma distribution and estimation of proportion of invariable sites (GTR + G + I) to accommodate variable rates across sites. The ML inference was used to build a time tree using MEGA X v. 10.2.6 (Kumar et al. 2018) as described by Mello (2018). Relative divergence times were estimated for all branching points by applying the RelTime-ML method (Tamura et al. 2012, 2018) with no calibration constraints. Branch lengths were calculated using the same substitution model as previously used to estimate the ML inference. The PI for all five partitions were determined using the rates of change for each site under the HyPhy criteria (Pond et al. 2005).
Species delimitation analyses
Five DNA-based species delimitation analyses were carried out, including three genetic distance-based methods, i.e., statistical parsimony network (SPN) (Templeton et al. 1992), automatic barcode gap discovery (ABGD) (Puillandre et al. 2012) and assemble species by automatic partitioning (ASAP) (Puillandre et al. 2021), and two coalescent-based methods, i.e., Poisson tree processes (PTP) (Zhang et al. 2013) and multi-rate PTP (mPTP) (Kapli et al. 2017). For these assessments, single gene sequence alignments for the most phylogenetic informative locus (as determined by the PI profiling method) for ABGD and ASAP, and 5-loci combined datasets for all methods, were used to infer the species boundaries of each Diaporthe clade.
The species delimitation hypotheses inferred by each method were compared with the phylogenetic species recognised by the GCPSR principle. Subsequently, the taxa delimited by the DNA-based species delimitation analyses were considered as molecular operational taxonomic units (MOTU) and were used to calculate the delimitation efficiency ratio (DER) according to equation (1) to assess which method best infers the species limits recognised by the GCPSR principle. The delimitation of the closely related outgroup taxa included in each clade was not considered for the DER as the analyses conducted were not directed towards the delimitation of these species.
| (1) |
where, for a given Diaporthe clade, NMOTU stands for the number of MOTU inferred by a given species delimitation method, and Nspecies stands for the number of species recognised by the GCPSR principle.
The SPN analyses were performed in TCS v. 1.21 (Clement et al. 2000) with alignment gaps treated as missing data, and a maximum parsimony connection probability set at 95 % statistical confidence, as this value proved to be a useful general tool and simple quantitative standard for phylogenetic species recognition (Hart & Sunday 2007). Resulting networks were considered as MOTU.
The ABGD analyses were performed using the Kimura two-parameter (K2P) nucleotide substitution model (Kimura 1980) to compute the matrix of pairwise nucleotide distances; the remaining parameters were left as default, including a relative gap width (X) of 1.5, and a prior maximum divergence of intraspecific diversity (P) ranging from 0.001 (Pmin) to 0.1 (Pmax). The analyses were conducted on the web server for ABGD (https://bioinfo.mnhn.fr/abi/public/abgd/abgdweb.html, accessed during November 2023). Only the primary partitions produced by ABGD were considered to sort the aligned sequences into hypothetical species, since they are typically stable on a wider range of prior values and are usually closer to the number of groups described by taxonomists (Puillandre et al. 2012). However, recursive splits were checked and are reported if their results were considered significant given the species delimitation schemes produced by the other methods. Moreover, to avoid subjectivity in the analyses, the median number of ABGD partitions (i.e., closest to P = 0.01) were used as the basis for hypothetical species, as this has produced good correspondence with traditional species in empirical studies (Puillandre et al. 2012, Kekkonen & Herbert 2014).
The ASAP analyses were performed using the K2P nucleotide substitution model (Kimura 1980) to compute the matrix of pairwise nucleotide distances; the remaining parameters were left as default, including a recursive split probability of 0.01. The analyses were conducted on the web server for ASAP (https://bioinfo.mnhn.fr/abi/public/asap/, accessed during November 2023). Hypothetical species delimitation was defined evaluating both the partitions with best asap-score and with second best asap-score according to Puillandre et al. (2021). In highly discordant cases, the partition with the third best asap-score was also evaluated. The partition selected was the closest to the delimitation predicted based on phylogenetic analyses, as well as the species delimitation schemes produced by the other methods.
The PTP (Zhang et al. 2013) and mPTP (Kapli et al. 2017) analyses were performed using the ML inferences produced by RAxML-NG v. 1.1.0 (Kozlov et al. 2019). The PTP analyses were performed with 500 000 MCMC generations, thinning set to 100, burn-in of 10 % and conducted on the web server for PTP (http://species.h-its.org/ptp/, accessed during November 2023). Convergence of the MCMC iterations was assessed by visualising the log-likelihood trace plot. The mPTP analyses were conducted on the web server for mPTP (http://mptp.h-its.org, accessed during November 2023). The resulting trees were prepared with Adobe Illustrator CS2 v. 12.0.0 (Adobe, USA).
Coalescent-based species tree estimation methods have been shown to work reliably and produce accurate species trees even when there are substantial amounts of missing data (Nute et al. 2018), especially if they are randomly distributed (per gene and/or per taxa) and if a sufficiently large number of genes are sampled (Xi et al. 2016). To avoid possible erroneous species delimitations schemes, given the lack of cal and his3 partial sequences for several species of most Diaporthe clades under analysis, PTP and mPTP analyses were additionally applied to combined datasets that included those species whose five (ITS, tef1, tub2, cal and his3), four (ITS, tef1, tub2 and cal) and three (ITS, tef1 and tub2) loci were available, and were conducted using ML inferences of the 5-, 4- and 3-loci combined datasets, respectively. All coalescent-based species delimitation schemes obtained for each Diaporthe clade were qualitatively compared and discordant cases are noted and discussed. To aid conclusions on species excluded from these additional analyses due to lack of partial sequences for tef1 and/or tub2, their phylogenetic position within the established species boundaries of its respective clade was considered. A similar approach was followed for ABGD and ASAP analyses when there were a substantial number of species lacking tub2 and cal partial sequences for a given Diaporthe clade. Therefore, these analyses were conducted using 3-loci combined datasets.
Pairwise homoplasy index test and phylogenetic network analyses
The concatenated alignments were used to infer the occurrence of recombination events within each Diaporthe clade under analyses, and between potentially synonymous species within each Diaporthe clade through the pairwise homoplasy index (PHI, Φw) test (Bruen et al. 2006) implemented in SplitsTree4 v. 4.19.0 (Huson & Bryant 2006). For these assessments, when possible, the strains of a given species were tested for genetic interchange with a set of authentic strains, including those with type status, of the respective species under which the synonym is suggested. A similar approach was followed for potentially taxonomic novelties. Significant recombination was considered when the probably of the Φw-statistic was below 0.05 (P-value < 0.05).
To evaluate and visualise the impact of the potential recombination events, the relationships between taxa within each Diaporthe clade were visualised through phylogenetic networks based on the concatenated sequence alignments. The phylogenetic networks were constructed using the LogDet transformation (Steel 1994) for the distance matrix and the Neighbor-Net algorithm (Bryant & Moulton 2006) implemented in SplitsTree4 v. 4.19.0 (Huson & Bryant 2006). The resulting phylogenetic networks were prepared with Adobe Illustrator CS2 v. 12.0.0 (Adobe, USA).
Hierarchical cluster analysis of phenotypic data
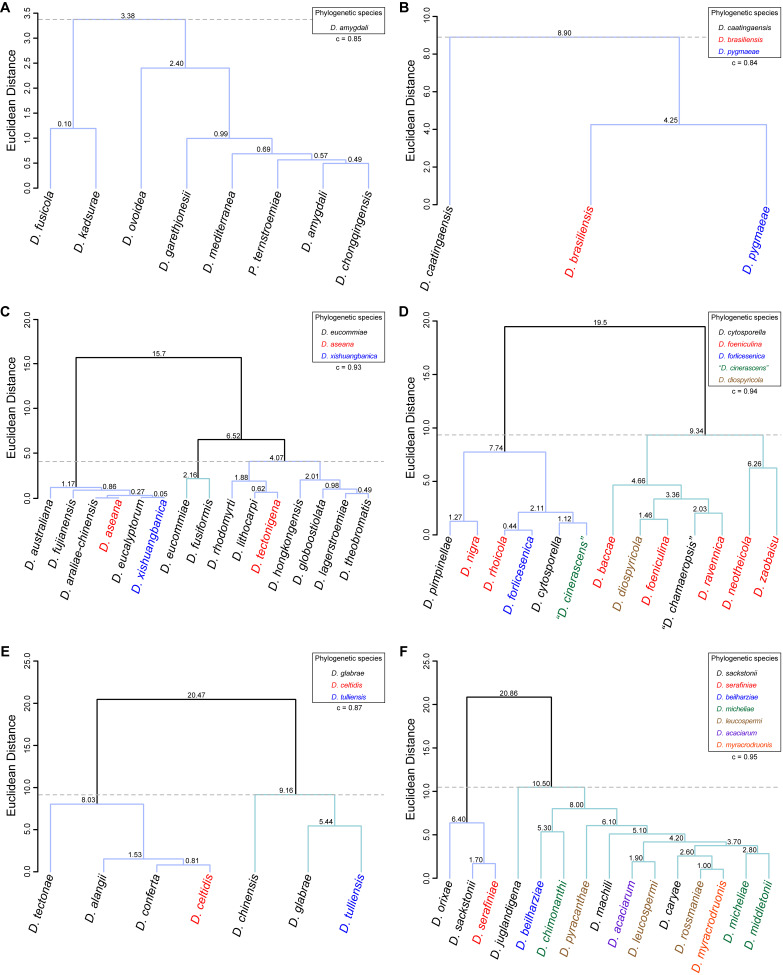
To assess the correlation between species phylogenetic boundaries and taxa morphology, measurements of the length and width of morphological features of all species belonging to each Diaporthe clade with published taxonomic descriptions were used. A hierarchical cluster analysis (HCA) was conducted using R Statistical Software v. 4.3.1 (R Core Team 2023). Pairwise distance among taxa were estimated with Euclidean distance index to generate the dissimilarity matrices and dendrograms were constructed by the unweighted pair group method with arithmetic mean (UPGMA) as the clustering algorithm. Dendrograms were generated using the following R packages: cluster v. 2.1.4 (Maechler et al. 2022), factoextra v. 1.0.7 (Kassambara et al. 2020) and dendextend v. 1.17.1 (Galili 2015). The number of clusters was determined by visual inspection of the dendrograms and by using the gap statistic method (Tibshirani et al. 2001). Goodness-of-fit of the dendrograms was evaluated by means of the cophenetic correlation coefficient (c) (Sokal & Rohlf 1962). Dendrograms were generated based on the length-to-width (L/W) ratios of alpha conidia, beta conidia and/or elements of the conidiogenous layer (i.e., conidiophores and/or conidiogenous cells). The species included in the analyses were only those with available dimensions for these features. When several taxa were lacking dimensions for a particular feature, it was excluded from the analyses or, in the case of beta conidia, coded with zeros. The L/W ratios were calculated for all taxa following equation (2) to standardise and make the data comparable between taxa.
| (2) |
where, for a given taxon and a given micromorphological structure, Lmin and Lmax stand for the length minimum and maximum dimensions, respectively, and Wmin and Wmax stand for the width minimum and maximum dimensions, respectively.
RESULTS
Phylogenetic analyses and phylogenetic species recognition
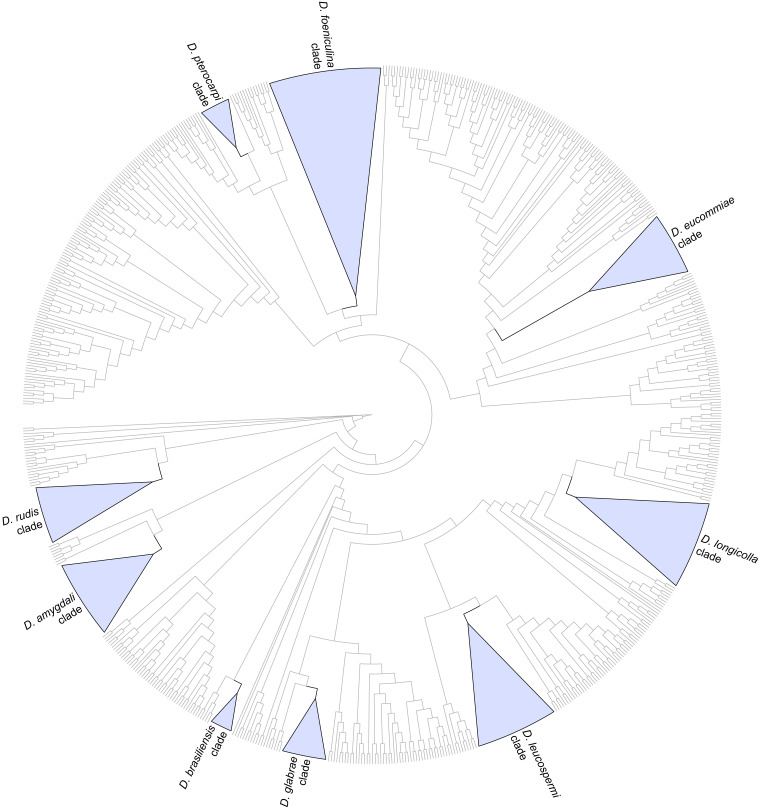
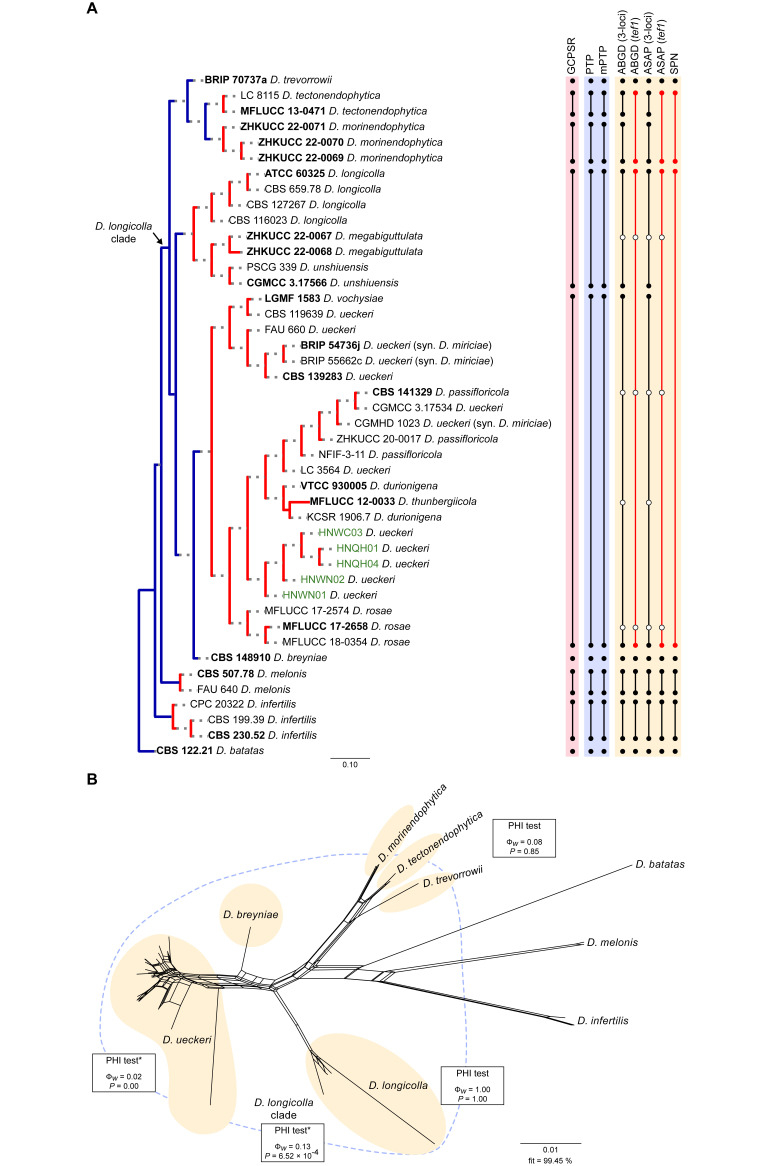
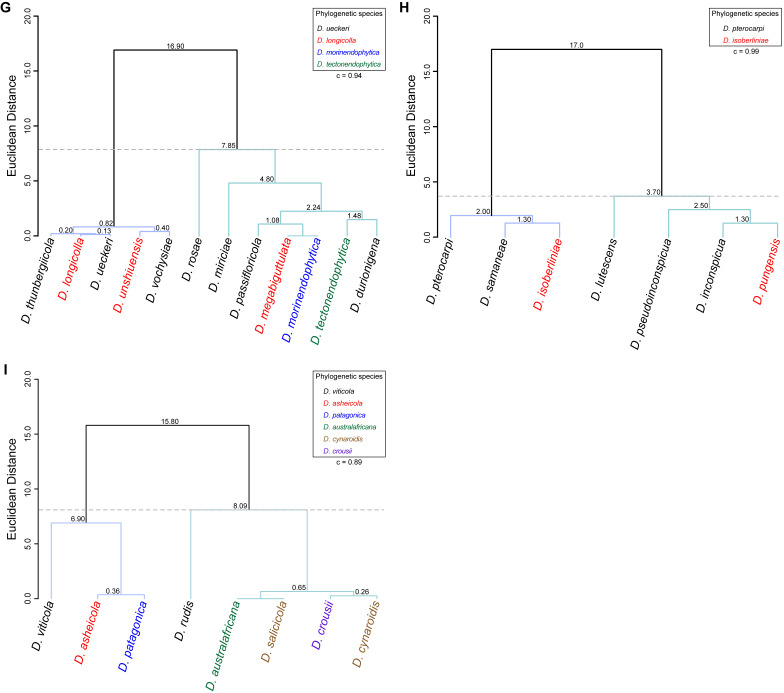
Twenty-six isolates obtained from foliar lesions of palms in Lisbon, Portugal were tentatively identified as diaporthe-like taxa based on macro- and micromorphological characters. These included the presence of fluffy, flattened, white or greyish aerial mycelium, often with scattered, solitary or aggregated, globose to subglobose, dark conidiomata, producing alpha and/or beta conidia. BLASTn searches in GenBank with the newly generated ITS sequences revealed about 99–100 % similarity to known Diaporthe species. Phylogenetic analyses of these isolates, along with 10 additional Diaporthe taxa isolated from palm tissues listed in recent literature or deposited in GenBank, revealed that they belong in nine well-supported (ML-BS ≥ 95 %) Diaporthe clades, which were assigned according to the oldest typified species (Figs 1–9). Fifteen isolates reside in the D. foeniculina clade (Fig. 5), five in the D. longicolla clade (Fig. 9), four in the D. leucospermi clade (Fig. 8), three in the D. amygdali clade (Fig. 2), two in each the D. brasiliensis (Fig. 3), D. glabrae (Fig. 6) and D. rudis (Fig. 10) clades, while the remaining two isolates reside in the D. eucommiae (Fig. 4) and D. inconspicua (Fig. 7) clades. All nine Diaporthe clades, along with a set of closely related outgroup taxa, were subsequently analysed to assess their species limits and correctly assign isolates from palm tissues to Diaporthe species.
Fig. 1.
Cladogram of the phylogenetic tree generated from a maximum likelihood analysis based on combined ITS, tef1, tub2, cal and his3 sequence data of the isolates from palms examined in this study and all species currently accepted in the genus Diaporthe. A total of 783 strains were included in the combined dataset that comprised 2 790 characters (including gaps) (576 characters for ITS, 507 for tef1, 560 for tub2, 614 for cal and 533 for his3) after alignment and manual adjustment. The analysis was conducted using the GTR+G+I nucleotide substitution model. Taxa names and phylogenetic support values have been removed for simplification purposes. The well-supported (ML-BS ≥ 95 %) clades containing the isolates from palms examined in this study have been collapsed and are highlighted with coloured triangles with their respective branches in black. Cladogram is rooted to Cytospora disciformis (CBS 116827 and CBS 118083).
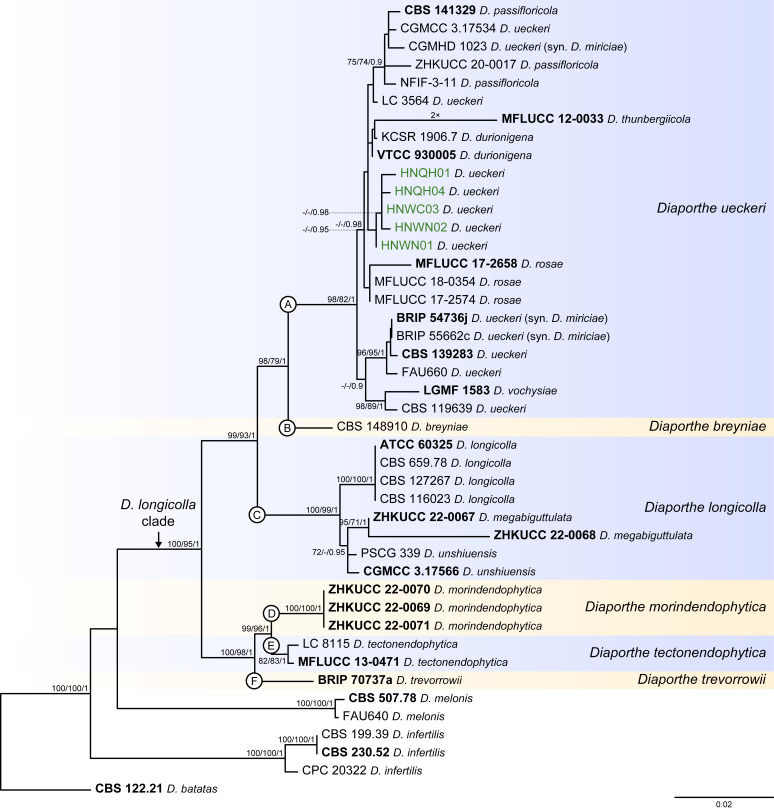
Fig. 9.
Phylogenetic tree generated from a maximum likelihood analysis based on combined ITS, tef1, tub2, cal and his3 sequence data of the Diaporthe longicolla clade and closely related species. Bootstrap support values for maximum likelihood, maximum parsimony (ML-BS/MP-BS ≥ 70 %) and Bayesian posterior probabilities (PP ≥ 0.90) are shown at the nodes. Strains with type status are indicated in bold font. The isolates from palm tissues included in the analyses are presented in green typeface. Species boundaries within the D. longicolla clade are delimited with coloured blocks and their respective branches are indicated by lettered circles (A–F). The scale bar represents the expected number of nucleotide changes per site. The tree is rooted to D. batatas (CBS 122.21).
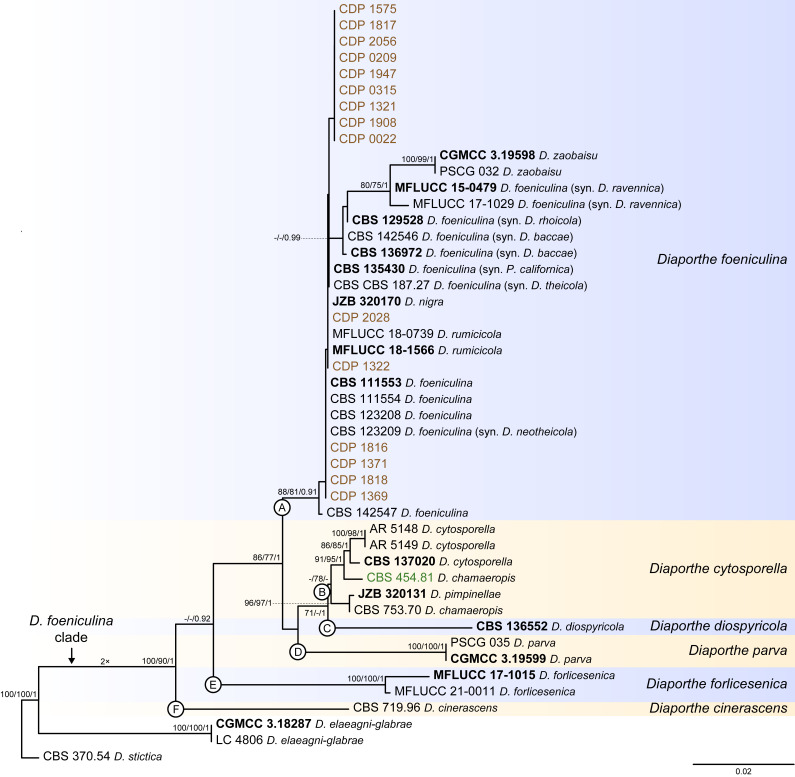
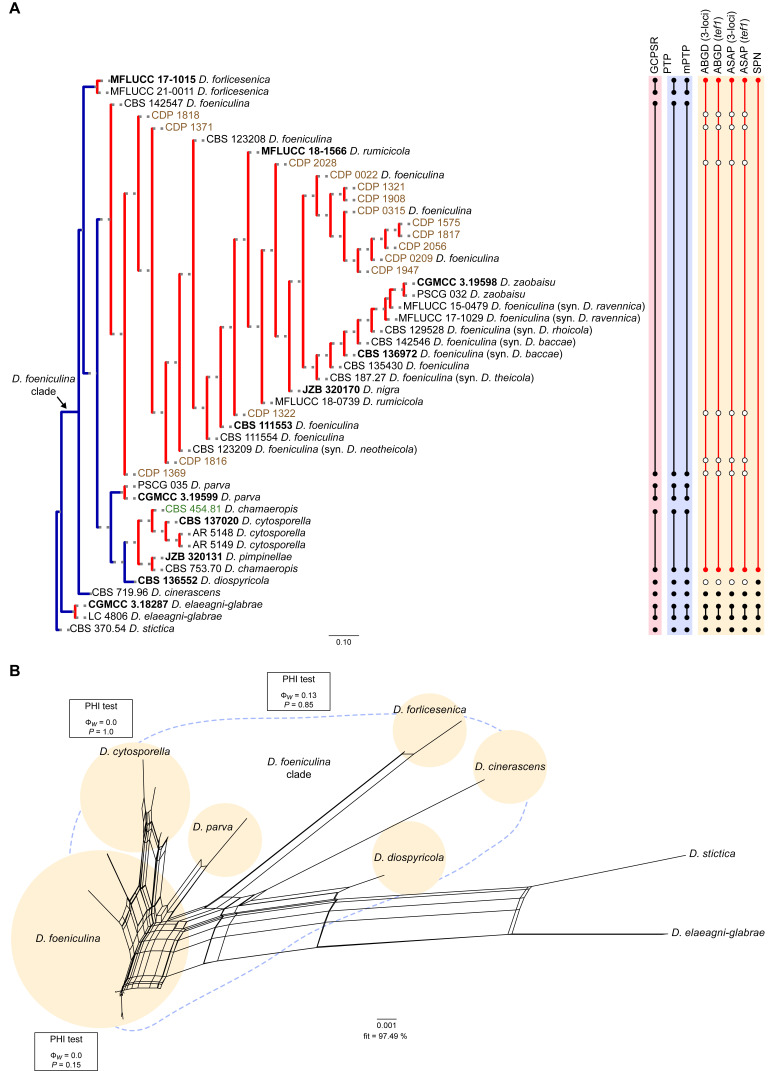
Fig. 5.
Phylogenetic tree generated from a maximum likelihood analysis based on combined ITS, tef1, tub2, cal and his3 sequence data of the Diaporthe foeniculina clade and closely related species. Bootstrap support values for maximum likelihood, maximum parsimony (ML-BS/MP-BS ≥ 70 %) and Bayesian posterior probabilities (PP ≥ 0.90) are shown at the nodes. Strains with type status are indicated in bold font. The isolates from this study are presented in brown typeface and the additional isolate from palm tissues included in the analyses is presented in green typeface. Species boundaries within the D. foeniculina clade are delimited with coloured blocks and their respective branches are indicated by lettered circles (A–F). The scale bar represents the expected number of nucleotide changes per site. The tree is rooted to D. stictica (CBS 370.54).
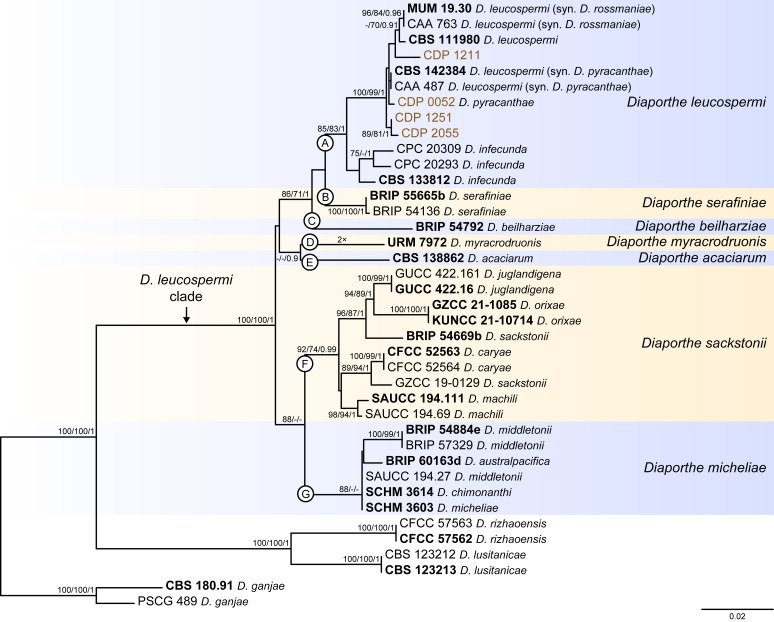
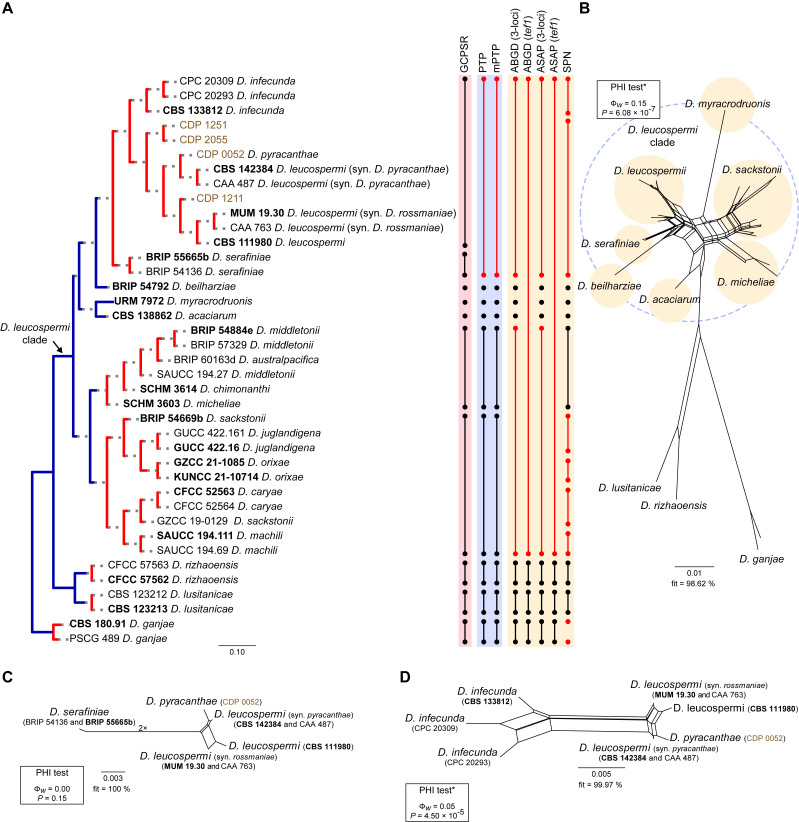
Fig. 8.
Phylogenetic tree generated from a maximum likelihood analysis based on combined ITS, tef1, tub2, cal and his3 sequence data of the Diaporthe leucospermi clade and closely related species. Bootstrap support values for maximum likelihood, maximum parsimony (ML-BS/MP-BS ≥ 70 %) and Bayesian posterior probabilities (PP ≥ 0.90) are shown at the nodes. Strains with type status are indicated in bold font. The isolates from this study are presented in brown typeface. Species boundaries within the D. leucospermi clade are delimited with coloured blocks and their respective branches are indicated by lettered circles (A–G). The scale bar represents the expected number of nucleotide changes per site. The tree is rooted to D. ganjae (CBS 180.91 and PSCG 489).
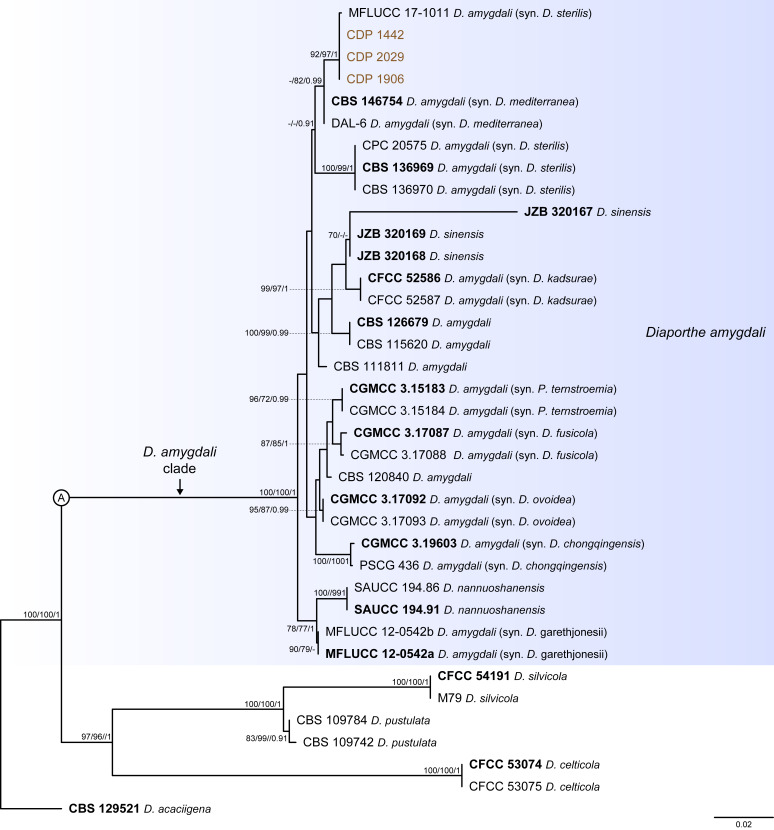
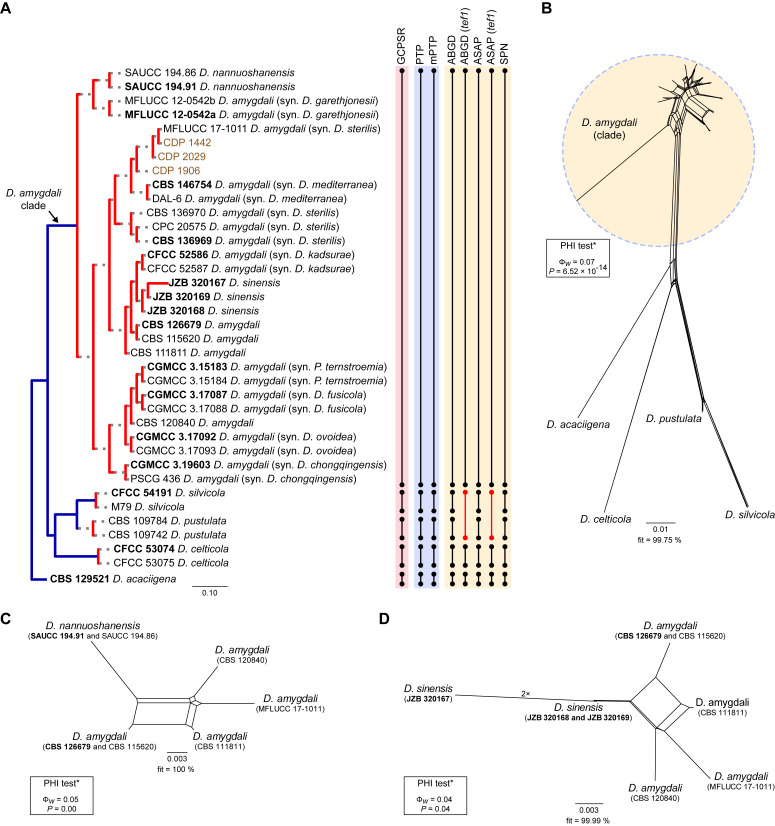
Fig. 2.
Phylogenetic tree generated from a maximum likelihood analysis based on combined ITS, tef1, tub2, cal and his3 sequence data of the Diaporthe amygdali clade and closely related species. Bootstrap support values for maximum likelihood, maximum parsimony (ML-BS/MP-BS ≥ 70 %) and Bayesian posterior probabilities (PP ≥ 0.90) are shown at the nodes. Strains with type status are indicated in bold font. The isolates from this study are presented in brown typeface. Species boundaries within the D. amygdali clade are delimited with a coloured block and its respective branch is indicated by a lettered circle (A). The scale bar represents the expected number of nucleotide changes per site. The tree is rooted to D. acaciigena (CBS 129521).
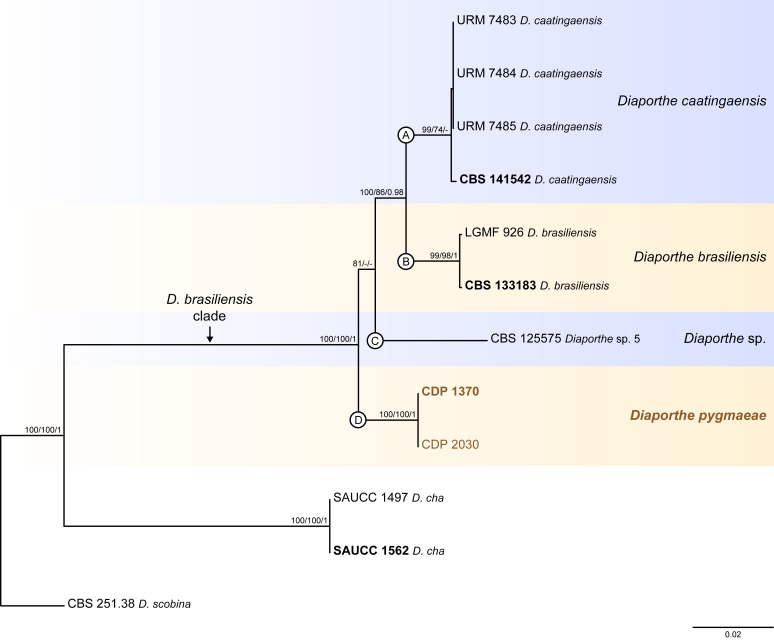
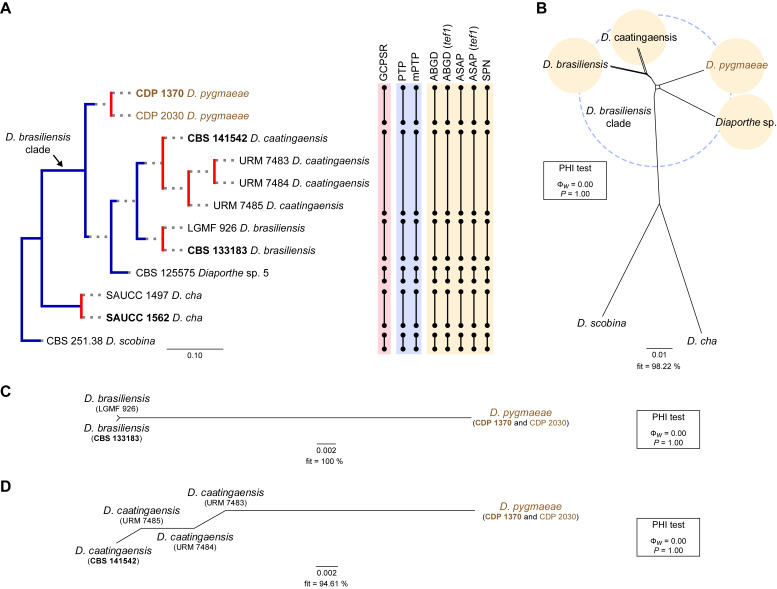
Fig. 3.
Phylogenetic tree generated from a maximum likelihood analysis based on combined ITS, tef1, tub2, cal and his3 sequence data of the Diaporthe brasiliensis clade and closely related species. Bootstrap support values for maximum likelihood, maximum parsimony (ML-BS/MP-BS ≥ 70 %) and Bayesian posterior probabilities (PP ≥ 0.90) are shown at the nodes. Strains with type status are indicated in bold font. The isolates from this study are presented in brown typeface. Species boundaries within the D. brasiliensis clade are delimited with coloured blocks and their respective branches are indicated by lettered circles (A–D). The scale bar represents the expected number of nucleotide changes per site. The tree is rooted to D. scobina (CBS 251.38).
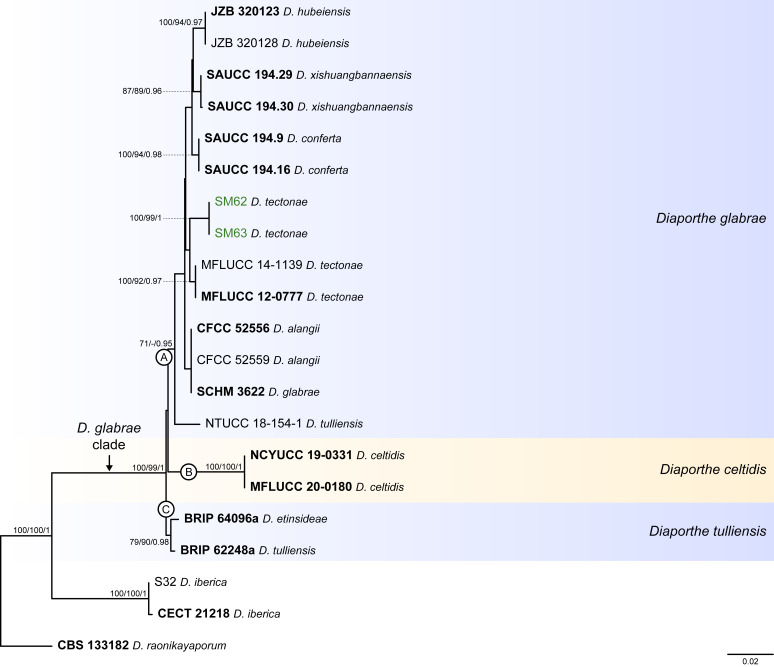
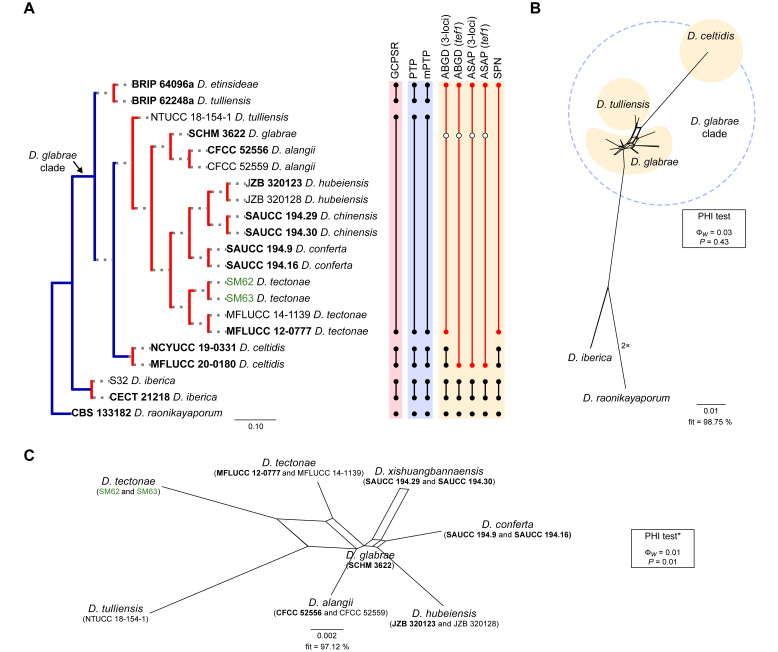
Fig. 6.
Phylogenetic tree generated from a maximum likelihood analysis based on combined ITS, tef1, tub2, cal and his3 sequence data of the Diaporthe glabrae clade and closely related species. Bootstrap support values for maximum likelihood, maximum parsimony (ML-BS/MP-BS ≥ 70 %) and Bayesian posterior probabilities (PP ≥ 0.90) are shown at the nodes. Strains with type status are indicated in bold font. The isolates from palm tissues included in the analyses are presented in green typeface. Species boundaries within the D. glabrae clade are delimited with coloured blocks and their respective branches are indicated by lettered circles (A–C). The scale bar represents the expected number of nucleotide changes per site. The tree is rooted to D. raonikayaporum (CBS 133182).
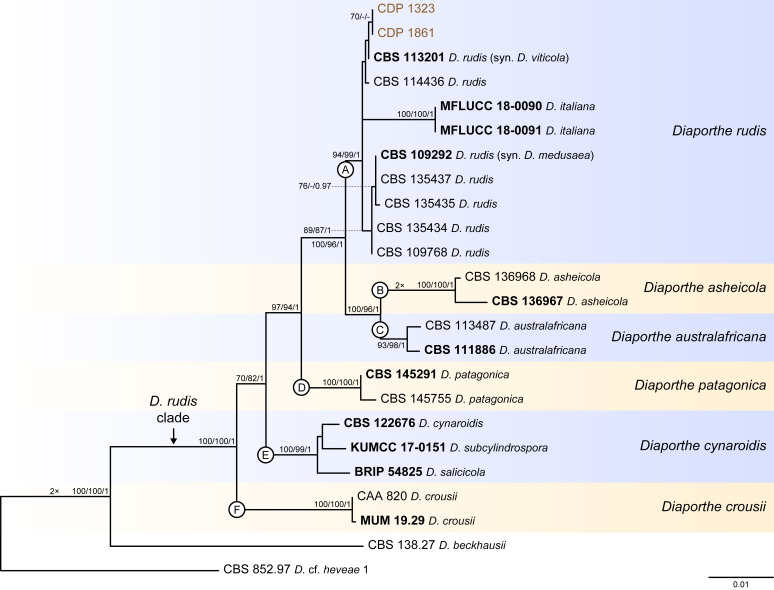
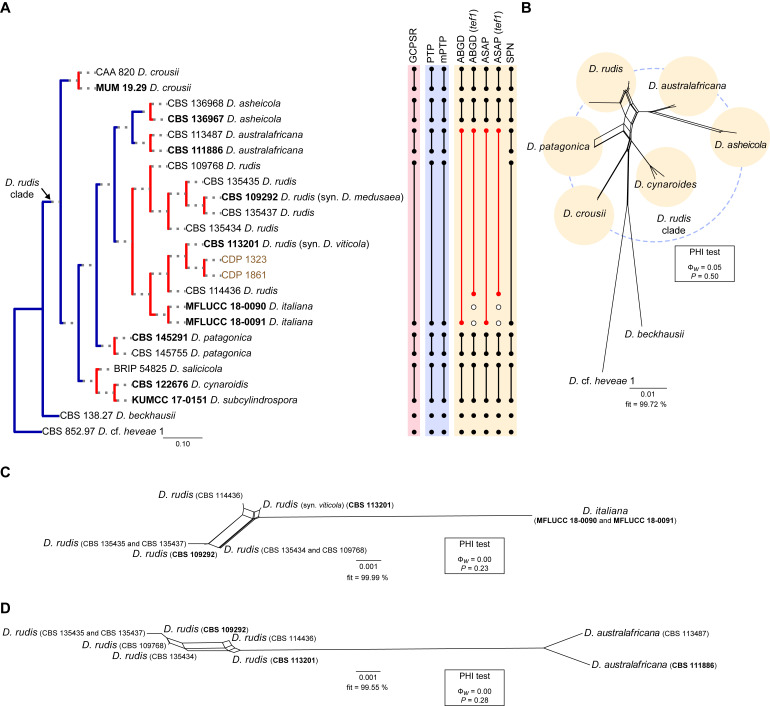
Fig. 10.
Phylogenetic tree generated from a maximum likelihood analysis based on combined ITS, tef1, tub2, cal and his3 sequence data of the Diaporthe rudis clade and closely related species. Bootstrap support values for maximum likelihood, maximum parsimony (ML-BS/MP-BS ≥ 70 %) and Bayesian posterior probabilities (PP ≥ 0.90) are shown at the nodes. Strains with type status are indicated in bold font. The isolates from this study are presented in brown typeface. Species boundaries within the D. rudis clade are delimited with coloured blocks and their respective branches are indicated by lettered circles (A–F). The scale bar represents the expected number of nucleotide changes per site. The tree is rooted to D. cf. heveae 1 (CBS 852.97).
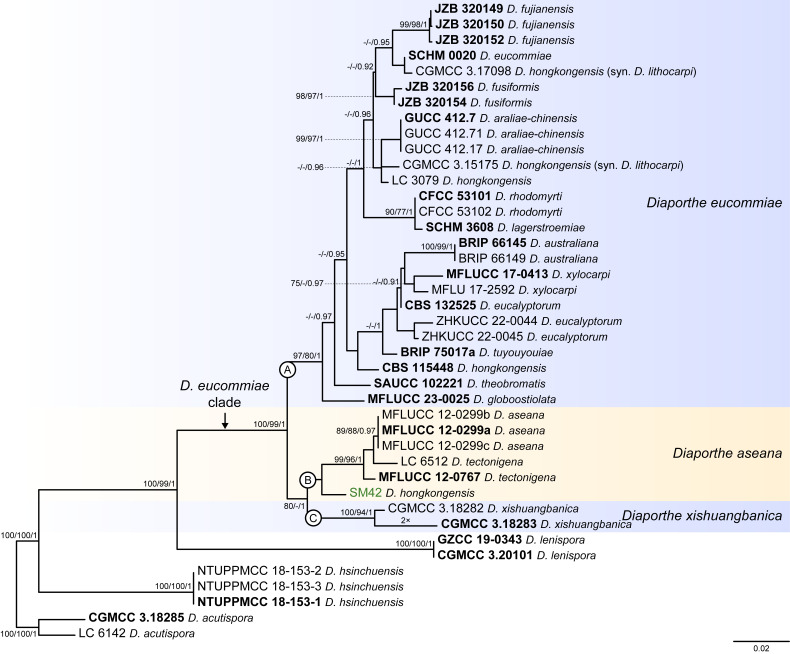
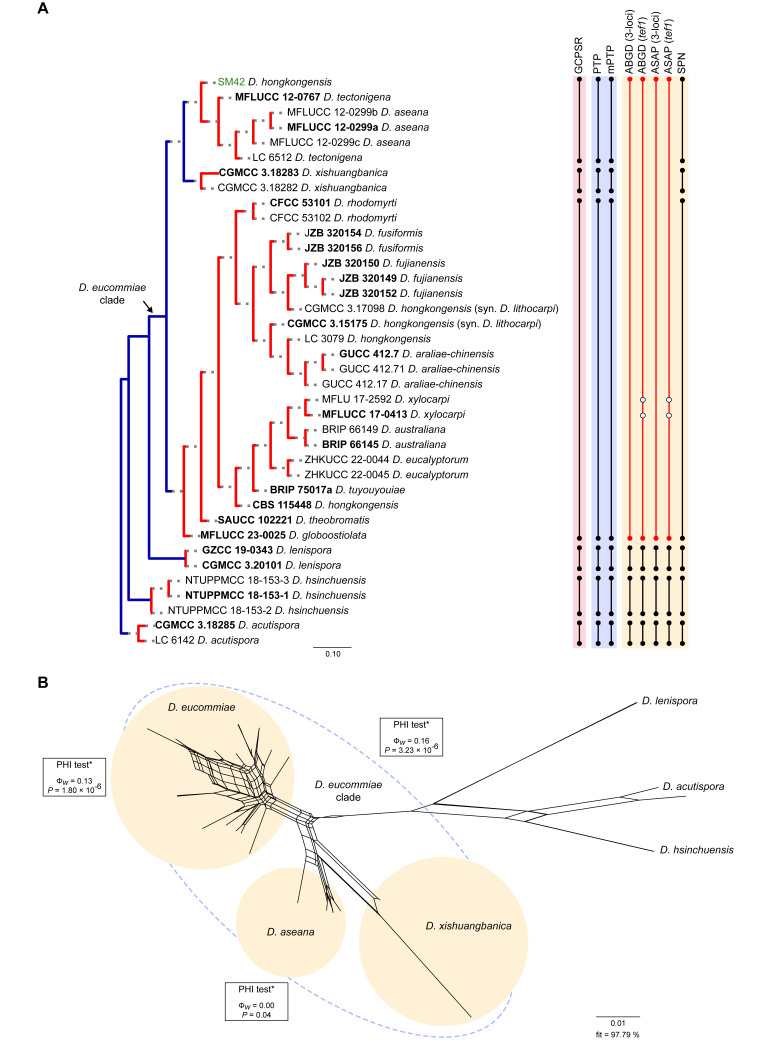
Fig. 4.
Phylogenetic tree generated from a maximum likelihood analysis based on combined ITS, tef1, tub2, cal and his3 sequence data of the Diaporthe eucommiae clade and closely related species. Bootstrap support values for maximum likelihood, maximum parsimony (ML-BS/MP-BS ≥ 70 %) and Bayesian posterior probabilities (PP ≥ 0.90) are shown at the nodes. Strains with type status are indicated in bold font. The isolate from palm tissues included in the analyses is presented in green typeface. Species boundaries within the D. eucommiae clade are delimited with coloured blocks and their respective branches are indicated by lettered circles (A–C). The scale bar represents the expected number of nucleotide changes per site. The tree is rooted to D. acutispora (CGMCC 3.18285 and LC 6142).
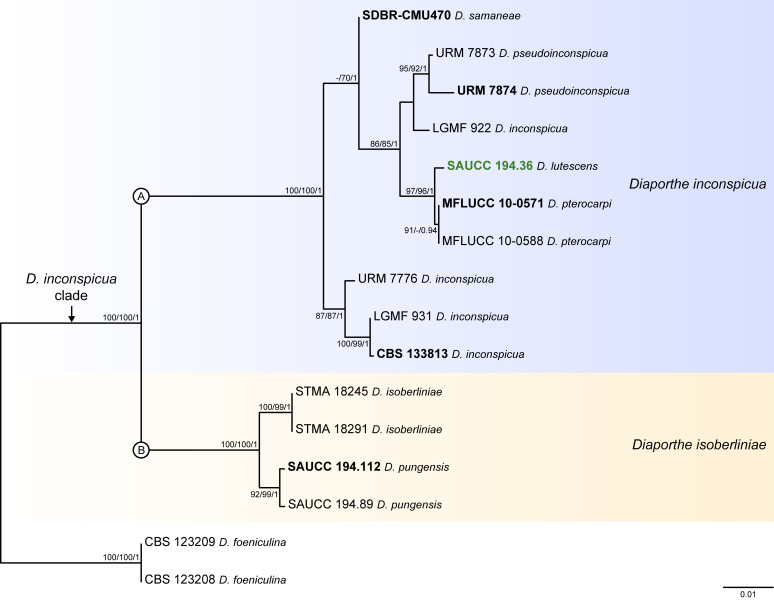
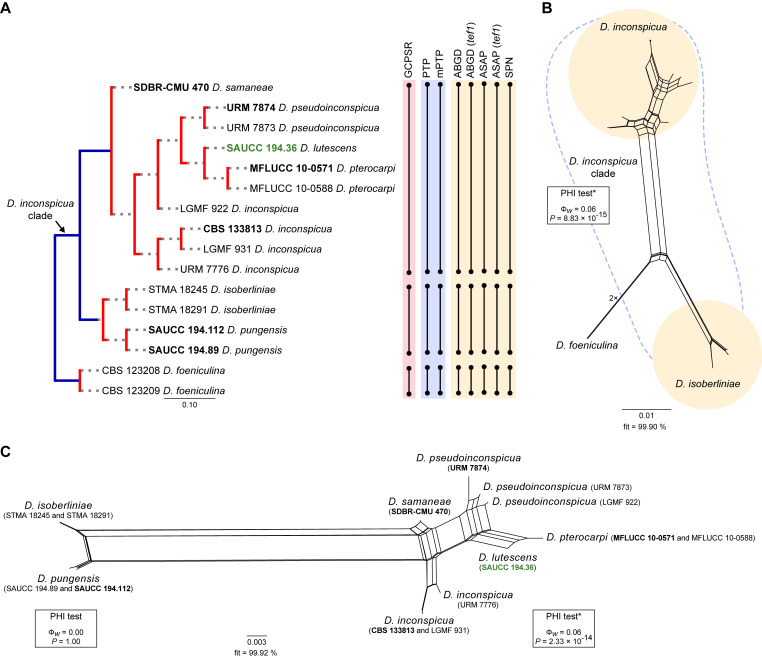
Fig. 7.
Phylogenetic tree generated from a maximum likelihood analysis based on combined ITS, tef1, tub2, cal and his3 sequence data of the Diaporthe inconspicua clade and closely related species. Bootstrap support values for maximum likelihood, maximum parsimony (ML-BS/MP-BS ≥ 70 %) and Bayesian posterior probabilities (PP ≥ 0.90) are shown at the nodes. Strains with type status are indicated in bold font. The isolate from palm tissues included in the analyses is presented in green typeface. Species boundaries within the D. inconspicua clade are delimited with coloured blocks and their respective branches are indicated by lettered circles (A and B). The scale bar represents the expected number of nucleotide changes per site. The tree is rooted to D. foeniculina (CBS 123208 and CBS 123209).
Diaporthe amygdali clade
Thirty-seven isolates were included in the phylogenetic analyses of the D. amygdali clade, namely 30 ingroup taxa (three obtained in this study, viz. CDP 1442, CDP 1906 and CDP 2029) and seven closely related outgroup taxa (Table 2). Statistics for the different datasets and respective phylogenetic trees are summarised in Table S1. Tree topologies resulting from ML, MP and BA inferences were similar, displaying roughly the same well-resolved terminal clades, mostly supported by high ML/MP-BS (≥ 70 %) and PP (≥ 0.90) values, whose relative position differed only in the MP inference. The ML tree is shown in Fig. 2 with ML-BS/MP-BS/PP values at the nodes. The final likelihood score for the best scoring ML tree was -6875.719343. The BA inference had an average standard deviation of split frequencies (SDSF) and an average potential scale reduction factor (PSRF) of 0.009856 and 1.001, respectively, after 570 000 generations, resulting in 85 502 trees sampled. The MP analysis of 434 parsimony-informative characters (21.5 %) resulted in 180 equally parsimonious trees of 808 steps with a low level of homoplasy as indicated by the measures of homoplasy and character fit (Table S1).
According to the phylogenetic analyses of the concatenated alignment, the three isolates from this study clustered in a highly supported clade (100 % ML-BS / 100 % MP-BS / 1 PP), which was designated here as the D. amygdali clade (clade A in Fig. 2). This clade contained 22 taxa previously recognised as D. amygdali, including its ex-type strain CBS 126679, as well as five taxa belonging to two other Diaporthe species, i.e., D. nannuoshanensis and D. sinensis. Both species appear to be intermingled with taxa previously recognised as D. amygdali, lacking a clear phylogenetic distinction. Moreover, the three strains of D. sinensis did not cluster in highly supported monophyletic lineage (Fig. 2).
Individual ML and MP gene trees were compared to identify concordant branches and accordingly apply the GCPSR principle. All individual ML and MP gene trees were topologically similar, presenting the same well-delimited clades. However, several conflicts were detected between the individual phylogenies of the different loci. Tree topologies between the individual gene genealogies varied substantially, with incongruent branches and most of the internal nodes receiving low or lacking bootstrap support values (Fig. S1). In general, tree topologies of the tef1-(Fig. S1B) and cal-phylograms (Fig. S1D) were more similar to the phylogenetic analyses of the combined dataset. The multilocus phylogenetic analyses showed a better delimitation of the D. amygdali clade when compared to the individual gene genealogies (Fig. 2).
Most individual gene genealogies failed to diagnose D. nannuoshanensis and D. sinensis as well-supported monophyletic lineages, except for tef1- and his3-phylograms (Fig. S1B, E), and ITS-phylograms (Fig. S1A), respectively. In most individual datasets, both species lack phylogenetic distinctiveness from several D. amygdali strains. Moreover, the relationships between both species and the remaining D. amygdali strains are highly discordant among the individual phylogenies (Fig. S1). Following the GCPSR principle, it was verified that the node that delimits the transition from concordant branches to incongruity among branches corresponds to the D. amygdali clade (Fig. 2), which was recovered with high ML/MP-BS values from almost all individual gene trees (-/88 %, 99/100 %, 100/100 %, 100/100 % and 100/100 % in ITS-, tef1-, tub2-, cal- and his3-phylograms, respectively) (Fig. S1). Subsequently, all taxa within the D. amygdali clade seem to be conspecific and were recognised as a single independent evolutionary lineage (IEL). Thus, considering the phylogenetic analyses of the combined dataset, as well as the GCPSR principle, D. nannuoshanensis and D. sinensis are regarded as potential synonyms of D. amygdali.
Diaporthe brasiliensis clade
Twelve isolates were included in the phylogenetic analyses of the D. brasiliensis clade, namely nine ingroup taxa (two obtained in this study, viz. CDP 1370 and CDP 2030) and three closely related outgroup taxa (Table 2). Statistics for the different datasets and respective phylogenetic trees are summarised in Table S2. Tree topologies resulting from ML, MP and BA inferences were similar, and presented the same well-resolved clades for each species included in the analyses, mostly supported by high ML/MP-BS and PP values. The ML tree is shown in Fig. 3 with ML-BS/MP-BS/PP values at the nodes. The final likelihood score for the best scoring ML tree was -5929.692434. The BA inference had an average SDSF and an average PSRF of 0.009819 and 1.001, respectively, after 135 000 generations, resulting in 20 252 trees sampled. The MP analysis of 289 parsimony-informative characters (11.7 %) resulted in one tree of 523 steps with a very low level of homoplasy as indicated by the measures of homoplasy and character fit (Table S2).
According to the phylogenetic analyses, CDP 1370 and CDP 2030 grouped together in a monophyletic lineage with maximum support values (100 % ML-BS / 100 % MP-BB / 1 PP; subclade D in Fig. 3). This formed a distinct subclade sister to D. caatingaensis (subclade A), D. brasiliensis (subclade B) and the strain CBS 125575 (Diaporthe sp. 5) (subclade C) in a well-supported clade (100 % ML-BS / 100 % MP-BS / 1 PP), which was designated here as the D. brasiliensis clade (Fig. 3). Subclade D is here considered to represent a new Diaporthe species, D. pygmaeae, which is introduced below in the Taxonomy section.
Individual ML and MP gene trees were compared to identify concordant branches and accordingly apply the GCPSR principle. All individual ML and MP gene trees were topologically similar, presenting the same well-resolved clades. In addition, almost no conflicts were detected between the individual phylogenies of the different loci, which were topologically similar to the analyses of the concatenated dataset (Figs S2, 3). The only exception was the ITS-phylograms, in which D. caatingaensis strains were paraphyletic (Fig. S2A). Thus, except for D. caatingaensis, all Diaporthe species were well-resolved and constant among phylogenetic trees, insomuch as all individual datasets had similar resolution, displaying the same well-supported groupings as resolved with the combined dataset phylogenies. All species, as well as the D. brasiliensis clade, were recovered with high ML/MP-BS (≥ 86 %) values from almost all individual gene trees (Fig. S2). Following the GCPSR principle, four different IEL were recognised within the D. brasiliensis clade, including an unnamed species, i.e., the strain CBS 125575. Therefore, all individual datasets supported a new subclade in the D. brasiliensis clade as representing a new Diaporthe species. Additionally, the support for D. pygmaeae sp. nov. was provided by base-pair sequence comparisons with the ex-type strains of D. brasiliensis (CBS 133183) and D. caatingaensis (CBS 141542) (detailed information of sequence comparisons are referred to in the Taxonomy section).
Diaporthe eucommiae clade
The phylogenetic analyses of the D. eucommiae clade included 34 ingroup taxa and seven closely related outgroup taxa. The ingroup taxa included one strain from palm substrata retrieved from recent literature, i.e., SM42 identified as D. hongkongensis (Table 2). Statistics for the different datasets and respective phylogenetic trees are summarised in Table S3. Tree topologies resulting from ML, MP and BA inferences were similar, displaying the same well-resolved terminal clades, whose relative position differed only in the MP inference. The ML tree is shown in Fig. 4 with ML-BS/MP-BS/PP values at the nodes. The final likelihood score for the best scoring ML tree was -6645.118752. The BA inference had an average SDSF and an average PSRF of 0.009519 and 1.002, respectively, after 425 000 generations, resulting in 63 752 trees sampled. The MP analysis of 358 parsimony-informative characters (19.1 %) resulted in 1 000 trees of 749 steps with a low level of homoplasy as indicated by the measures of homoplasy and character fit (Table S3).
According to the phylogenetic analyses, the ingroup taxa resolved into three subclades, which clustered in a monophyletic lineage with high support values (100 % ML-BS / 99 % MP-BS / 1 PP), which was designated here as the D. eucommiae clade (Fig. 4). Of these, subclade C represent a well-supported (100 % ML-BS / 94 % MP-BS / 1 PP) single lineage, while subclades A and, at less extent, B displayed substantial substructure (Fig. 4). The strain SM42 clustered with no relevant support values in subclade B, together with five strains belonging to D. aseana and D. tectonigena. These two species grouped in a well-supported lineage (99 % MLBS / 96 % MP-BS / 1 PP) without a clear phylogenetic distinction, since D. tectonigena strains (LC 6512 and MFLUCC 12-0767) did not group into a separate, dichotomous lineage from D. aseana (Fig. 4). Although identified as D. hongkongensis, SM42 did not group with strains previously recognised as D. hongkongensis, including its ex-type strain CBS 115448, and is phylogenetically closer to D. xishuangbanica (CGMCC 3.18282 and GCMCC 3.18283) (subclade C). The ex-type strain and other previously recognised strains of D. hongkongensis grouped together but dispersed among 22 strains belonging to 12 different Diaporthe species in a monophyletic lineage with support values of 97 % ML-BS, 80 % MP-BS and 1 PP (subclade A), namely D. araliaechinensis, D. australiana, D. eucalyptorum, D. eucommiae, D. fujianensis, D. fusiformis, D. globoostiolata, D. lagerstroemiae, D. rhodomyrti, D. theobromatis, D. tuyouyou and D. xylocarpi. Most of the evolutionary relationships between these species lack phylogenetic support and some species present phylogenetic indistinctiveness or even a paraphyletic nature (Fig. 4).
Considering the lack of phylogenetic distinction between the species included in the subclades A and B (Fig. 4), the multilocus phylogenetic inferences suggest that the species boundaries within the D. eucommiae clade are poorly defined, which makes it impossible to correctly classify the isolate from palm substrata included in the analyses. In this sense, the GCPSR principle was applied, and the individual ML and MP gene trees were compared to identify concordant branches. All individual ML and MP gene trees were topologically similar, presenting the same well-delimited clades. However, due to lack of partial sequences of cal and his3 loci for several taxa, IEL are difficult to define, and their boundaries become unclear due to the comparison of datasets with a substantially different number of taxa. Still, several conflicts were detected between the individual phylogenies of the different loci, most of which produced highly discordant branches (Fig. S3), which can be used to predict IEL within the D. eucommiae clade.
While tef1-phylograms presented a very similar topology to that observed with the combined dataset analyses (Figs S3B, 4), most individual phylograms presented a highly variable structure, with different groupings depending on the locus under analysis (Fig. S3). ITS- and tub2-phylograms appear to be the most divergent loci in depicting the evolutionary relationships within the D. eucommiae clade (Fig. S3A, C), although the substantially different number of taxa between gene trees makes it difficult to draw conclusions. In both analyses, the number of discordant branches and lack of phylogenetic support indicate that a single IEL can be recognised within the D. eucommiae subclade. In the ITS-phylograms, some species recognised within the D. eucommiae subclade in the multilocus phylogenetic inferences, clustered together with D. aseana and D. tectonigena (Fig. S3A), with no relevant support values, while in the tub2-phylograms some species clustered as distinct lineages from the D. eucommiae subclade (Fig. S3B). Thus, considering the phylogenetic analyses of the combined dataset and most of the conflicts detected through the GCPRS principle, three IEL are considered in the D. eucommiae clade. Therefore, D. araliae-chinensis, D. australiana, D. eucalyptorum, D. fujianensis, D. fusiformis, D. globoostiolata, D. hongkongensis, D. lagerstroemiae, D. rhodomyrti, D. theobromatis, D. tuyouyou and D. xylocarpi are regarded as potential synonyms of D. eucommiae, and D. tectonigena is regarded as a potential synonym of D. aseana. The third IEL recognised within the D. eucommiae clade, i.e., D. xishuangbanica, was found to represent a distinct well-supported lineage in all individual gene trees for which sequences are available, except for ITS-phylograms (Fig. S3).
Diaporthe foeniculina clade
The phylogenetic analyses of the D. foeniculina clade included 44 ingroup taxa and three closely related outgroup taxa. The ingroup taxa included 15 isolates from this study, namely CDP 0022, CDP 0209, CDP 0315, CDP 1321, CDP 1322, CDP 1369, CDP 1371, CDP 1575, CDP 1816, CDP 1817, CDP 1818, CDP 1908, CDP 1947, CDP 2028 and CDP 2056, and an additional strain from palm substrata retrieved from recent literature, i.e., CBS 454.81 identified as D. chamaeropis (Table 2). Statistics for the different datasets and respective phylogenetic trees are summarised in Table S4. Tree topologies resulting from ML, MP and BA inferences were similar, presenting generally the same well-resolved clades, although few terminal clades were different in the MP inference. The ML tree is shown in Fig. 5 with ML-BS/MP-BS/PP values at the nodes. The final likelihood score for the best scoring ML tree was -4865.036992. The BA inference had an average SDSF and an average PSRF of 0.009423 and 1.003, respectively, after 260 000 generations, resulting in 39 002 trees sampled. The MP analysis of 191 parsimony-informative characters (10.0 %) resulted in 1 000 trees of 400 steps with a low level of homoplasy as indicated by the measures of homoplasy and character fit (Table S4).
According to the phylogenetic analyses, the ingroup taxa resolved into six moderately to highly supported subclades. The 15 isolates from this study clustered in a well-supported subclade (88 % ML-BS / 81 % MP-BS / 0.91 PP), dispersed among 17 other Diaporthe strains (subclade A in Fig. 5). This subclade contains 11 taxa previously recognised as D. foeniculina, as well as five taxa belonging to three other Diaporthe species, i.e., D. nigra, D. rumicicola and D. zaobaisu. Diaporthe rumicicola and D. nigra cannot be distinguished from D. foeniculina strains considering the concatenated sequence analyses conducted, given that both species lack either support values and/or relevant branch length (Fig. 5). Although D. zaobaisu grouped in a well-supported lineage with a substantial relative branch length within the above-mentioned subclade, all three species appear to be intermingled among other strains previously recognised as D. foeniculina, lacking a clear phylogenetic distinction. The strain CBS 454.81 (D. chamaeropis) clustered in a sister subclade, supported by moderate ML-BS support value (78 %; subclade B), together with five Diaporthe strains, including two different species (D. cytosporella and D. pimpinellae) and another D. chamaeropis strain (CBS 753.70), which seems to be paraphyletic in relation to CBS 454.81. Both subclades clustered in a monophyletic lineage, together with D. diospyricola (subclade C), D. parva (subclade D), D. forlicesenica (subclade E) and D. cinerascens (subclade F), with high support values (100 % ML-BS / 90 % MP-BS / 1 PP), which was designated here as the D. foeniculina clade (Fig. 5).
To test the conflicts detected through the analyses of the concatenated dataset, the individual ML and MP gene trees were compared to identify concordant branches and define the IEL within the D. foeniculina clade, according to the GCPSR principle. All individual ML and MP gene trees were topologically similar, presenting the same well-delimited clades. As for the previous clade, the results of the GCPSR principle should be carefully interpreted due to lack of partial sequences of cal and his3 loci for several taxa and, subsequently, comparison of datasets with a substantially different number of taxa. Even so, several conflicts were detected between the individual phylogenies of the different loci, particularly among taxa composing the subclades that include D. foeniculina and D. chamaeropis strains (subclades A and B in Fig. 5), suggesting that strains in both lineages are conspecific. In both subclades, species are paraphyletic or produce polytomies in most individual gene genealogies, or even present phylogenetic indistinctiveness in different combinations of taxa depending on the locus under analysis (Fig. S4). In general, tree topologies of the tef1-phylograms (Fig. S4B) and cal-phylograms (Fig. S4D) were more similar to the phylogenetic analyses of the combined dataset. The multilocus phylogenetic analyses showed a better delimitation of the D. foeniculina clade when compared to the individual gene genealogies.
The phylogenetic position and distinction of D. zaobaisu is not clear, since it appears to be a distinct well-supported (ML-BS and/or MP-BS ≥ 82 %) lineage sister to D. foeniculina in tef1-, tub2- and his3-phylograms (Fig. S4B, C, E), but cannot be recognised as a distinct clade on ITS-phylograms (Fig. S4A) and on the multilocus phylogenies (Fig. 5). No cal partial sequences are available to confirm the evolutionary relationships of D. zaobaisu with D. foeniculina strains. Thus, considering the phylogenetic analyses of the combined dataset, as well as the GCPSR principle, D. nigra, D. rumicicola and D. zaobaisu are regarded as potential synonyms of D. foeniculina, and D. chamaeropis and D. pimpinellae are regarded as potential synonyms of D. cytosporella. The remaining IEL recognised within the D. foeniculina clade, i.e., D. cinerascens, D. diospyricola, D. forlicesenica and D. parva, were found to clear represent distinct well-supported lineages in most individual gene trees for which sequences are available. (Fig. S4).
Diaporthe glabrae clade
The phylogenetic analyses of the D. glabrae clade included 18 ingroup taxa and three closely related outgroup taxa. The ingroup taxa included two strains from palm substrata retrieved from recent literature, i.e., SM62 and SM63, both identified as D. tectonae (Table 2). Statistics for the different datasets and respective phylogenetic trees are summarised in Table S5. Tree topologies resulting from ML, MP and BA inferences were similar, and presented the same groupings with the same relative positions for each species included in the analyses, mostly supported by high ML/MP-BS and PP values. The ML tree is shown in Fig. 6 with ML-BS/MP-BS/PP values at the nodes. The final likelihood score for the best scoring ML tree was -4757.046661. The BA inference had an average SDSF and an average PSRF of 0.009705 and 1.001, respectively, after 410 000 generations, resulting in 30 751 trees sampled. The MP analysis of 184 parsimony-informative characters (9.6 %) resulted in 114 trees of 412 steps with a very low level of homoplasy as indicated by the measures of homoplasy and character fit (Table S5).
According to the phylogenetic analyses, SM62 and SM63 clustered in a well-supported subclade (71 % ML-BS / 0.97 PP; subclade A in Fig. 6), together with 11 strains belonging to six different Diaporthe species, i.e., D. alangii, D. conferta, D. glabrae, D. hubeiensis, D. chinensis and D. tectonae, as well as the strain NTUCC 18-154-1 identified as D. tulliensis. Most species in this subclade are well-resolved in monophyletic lineages with high support values, except for the lineage of the ex-type strains of D. alangii (CFCC 52556) and D. glabrae (SCHM 3622). This lineage did not present relevant support values, and both species are phylogenetically indistinct. Even so, the relative branch lengths in the entire clade are small and the strains from palm tissues (SM62 and SM53), which are identified as D. tectonae, did not cluster with the ex-type strain of D. tectonae (MFLUCC 12-0777) with relevant support values (Fig. 6). Similarly, the strain NTUCC 18-154-1 (D. tulliensis) did not cluster with the ex-type strain of D. tulliensis (BRIP 62248a), which grouped in a separate lineage, together with D. etinsideae, with moderate to high support values (79 % MLBS / 90 % MP-BS / 0.98 PP; subclade C), although with very small relative branch lengths (Fig. 6). Subclades A and C, together with D. celtidis (subclade B), clustered in a monophyletic lineage with support values of 100 % ML-BS, 99 % MP-BS and 1 PP, which was designated here as the D. glabrae clade (Fig. 6).
Individual ML and MP gene trees were compared to identify concordant branches and accordingly apply the GCPSR principle. All individual ML and MP gene trees were topologically similar, presenting the same well-delimited clades. However, some strains in the D. glabrae clade presented conflicts between the individual phylogenies of the different loci (Fig. S5). As previously mentioned, IEL should be carefully interpreted when applying the GCPSR principle due to lack of partial sequences of cal and his3 loci for several taxa. In general, tree topologies of the tub2-phylograms (Fig. S5C) were more similar to the phylogenetic analyses of the combined dataset. The multilocus phylogenetic analyses showed a better delimitation of the D. glabrae clade when compared to the individual gene genealogies (Fig. 6).
Considering the ITS-phylograms (Fig. S5A), most species from subclade A (Fig. 6) presented phylogenetic indistinctiveness. In tef1- and tub2-phylograms (Fig. S5B, C), although most species are apparently well-resolved, their groupings are highly discordant among the two loci. Most species with available sequences for cal and his3 loci, also presented phylogenetic indistinctiveness (Fig. S5D, E). Moreover, the ex-type strains of D. tulliensis (BRIP 62248a) and D. etinsideae (BRIP 64096a) are barely distinguishable in ITS-, tef1 and tub2-phylograms, with very inconspicuous relative branch lengths, suggesting that they are conspecific, while D. celtidis strains clustered as an independent lineage in all three individual gene genealogies (Fig. S5A–C). Thus, considering the phylogenetic analyses of the combined dataset, as well as the GCPSR principle, three IEL can be recognised within the D. glabrae clade, insomuch as D. alangii, D. conferta, D. hubeiensis, D. tectonae and D. xishuangbannaensis are regarded as potential synonyms of D. glabrae, and D. etinsideae is regarded as a potential synonym of D. tulliensis.
Diaporthe inconspicua clade
Sixteen isolates were included in the phylogenetic analyses of the D. inconspicua clade, namely 14 ingroup taxa and two closely related outgroup taxa. The ingroup taxa included one strain from palm substrata retrieved from recent literature, i.e., SAUCC 194.36, which corresponds to D. lutescens isolated from leaves of Chrysalidocarpus lutescens in Yunnan (Table 2). Statistics for the different datasets and respective phylogenetic trees are summarised in Table S6. Tree topologies resulting from ML, MP and BA inferences were similar, and presented the same well-resolved clades with the same relative positions for each species included in the analyses, supported by high ML-BS, MP-BS and PP values. The ML tree is shown in Fig. 7 with ML-BS/MP-BS/PP values at the nodes. The final likelihood score for the best scoring ML tree was -4017.269368. The BA inference had an average SDSF and an average PSRF of 0.008328 and 1.024, respectively, after 5 000 generations, resulting in 752 trees sampled. The MP analysis of 202 parsimony-informative characters (11.1 %) resulted in one tree of 264 steps with a low level of homoplasy as indicated by the measures of homoplasy and character fit (Table S6).
According to the phylogenetic analyses, D. lutescens (SAUCC 194.36) clustered in a monophyletic lineage with maximum support values (100 % ML-BS / 100 % MP-BS / 1 PP) together with nine strains belonging to four Diaporthe species, i.e., D. inconspicua, D. pseudoinconspicua, D. pterocarpi and D. samaneae (subclade A in Fig. 7). While most species in this lineage are well-resolved, strains identified as D. inconspicua are paraphyletic. Moreover, the relative length of the branches between D. lutescens and D. pterocarpi is very small and the two species are almost phylogenetically indistinguishable (Fig. 7). This lineage is sister to another subclade with maximum support values that includes two Diaporthe species, i.e., D. isoberliniae and D. pungensis, both apparently well-resolved (subclade B in Fig. 7). In a more general view, both subclades clustered in a monophyletic lineage with maximum support values, which was designated here as the D. inconspicua clade (Fig. 9). It is worth mentioning that the ex-type strain of D. isoberliniae (CBS 137981) was not included in the phylogenetic inferences conducted here. Only the sequences for ITS and tub2 loci are available for this strain, which results in a negative impact on the robustness and overall support of the phylogenetic inferences. However, ML phylogenetic inferences have been conducted to confirm that the D. isoberliniae strains used (STMA 18245 and STMA 18291) are related to its ex-type strain (data not shown).
Individual ML and MP gene trees were compared to identify concordant branches and accordingly apply the GCPSR principle. All individual ML and MP gene trees were topologically similar, presenting the same well-delimited clades. However, several conflicts were detected between the individual phylogenies of the different loci. None of the individual phylogenies displayed a similar topology to that observed in the multilocus phylogenetic analyses (Figd S6, 7). In most individual gene genealogies, all strains included in the subclade A presented phylogenetic indistinctiveness and the evolutionary relationships established are highly discordant (Fig. S6). For instance, D. lutescens is phylogenetically indistinct from D. pterocarpi in the cal-phylograms (Fig. S6D) and from D. samaneae in the his3-phylograms (Fig. S6E), while it lacks phylogenetic distinctiveness from most species in the subclade A in the tef1- and tub2-phylograms (Fig. S6B, C). Similarly, D. isoberliniae and D. pungensis are phylogenetically indistinct in tef1- and cal-phylograms, where the lineage is composed by polytomies for all or some of the strains (Fig. S6B, D). Only the ITS-phylograms displayed a roughly similar topology to that observed in the multilocus phylogenetic inferences, although some species remained unresolved in polytomies and/or in paraphyletic branches (Fig. S6A). Thus, considering the phylogenetic analyses of the combined dataset, as well as the GCPSR principle, two IEL can be recognised within the D. inconspicua clade, insomuch as D. lutescens, D. pseudoinconspicua, D. pterocarpi and D. samaneae are regarded as potential synonyms of D. inconspicua, and D. pungensis is regarded as a potential synonym of D. isoberliniae.
Diaporthe leucospermi clade
The phylogenetic analyses of the D. leucospermi clade included 33 ingroup taxa and six closely related outgroup taxa. The ingroup taxa included four isolates from this study, namely CDP 0052, CDP 1211, CDP 1251 and CDP 2055 (Table 2). Statistics for the different datasets and respective phylogenetic trees are summarised in Table S7. Tree topologies resulting from ML, MP and BA inferences were similar, displaying the same well-resolved clades with the same relative positions for each species included in the analyses, mostly supported by high ML/MP-BS and PP values. The ML tree is shown in Fig. 8 with ML-BS/MP-BS/PP values at the nodes. The final likelihood score for the best scoring ML tree was -7174.782693. The BA inference had an average SDSF and an average PSRF of 0.008864 and 1.003, respectively, after 235 000 generations, resulting in 35 252 trees sampled. The MP analysis of 423 parsimony-informative characters (20.9 %) resulted in 52 trees of 806 steps with a low level of homoplasy as indicated by the measures of homoplasy and character fit (Table S7).
According to the phylogenetic analyses, seven well-resolved subclades have been noted in a monophyletic lineage with high support values (100 % ML-BS / 100 % MP-BS / 1 PP), which was designated here as the D. leucospermi clade (Fig. 8). The four isolates from this study clustered in a well-supported subclade (85 % ML-BS / 83 % MP-BS / 1 PP; subclade A) together with eight strains belonging to two Diaporthe species. Five of these strains are identified as D. leucospermi, including its ex-type strain CBS 111980, and formed a well-resolved lineage with the isolates from this study with support values of 100 % ML-BS, 99 % MP-BS and 1 PP. The other three strains belong to D. infecunda and clustered in a sister lineage with no relevant support values (Fig. 8). Among the remaining six subclades noted, four are composed by single strains or species that represent well-resolved divergent lineages within the D. leucospermi clade, considering their phylogenetic support values and/or the relative size of their branches. These include D. serafiniae (subclade B), D. beilharziae (subclade C), D. myracrodruonis (subclade D) and D. acaciarum (subclade E) (Fig. 8). The other two subclades presented substantial substructure and are composed by several species, some of which comprise strains with a paraphyletic nature and lineages forming polytomies. In subclade F, D. caryae, D. juglandigena, D. machili and D. orixae grouped in well-resolved lineages, while the ex-type strain BRIP 54669b and GZCC 19-0129 of D. sackstonii did not group together. In subclade G, strains of D. middletonii, including its ex-type strain BRIP 54884e, clustered separately and intermingled with the ex-type strains of other three Diaporthe species, i.e., D. australpacifica, D. chimonanthi and D. micheliae. Moreover, D. chimonanthi, D. micheliae and the strain SAUCC 194.27 (D. middletonii) presented phylogenetic indistinctiveness and formed a polytomy basal to the remaining strains within subclade G, which is supported by 88 % ML-BS (Fig. 8).
To test and confirm the conflicts detected through the analyses of the concatenated dataset, the individual ML and MP gene trees were compared to identify concordant branches and define the IEL within the D. leucospermi clade, according to the GCPSR principle. All individual ML and MP gene trees were topologically similar, presenting the same well-delimited clades. As mentioned in previous clades, the partial sequences of cal and his3 loci were lacking for several taxa, and thus IEL are difficult to evaluate. Even so, several conflicts were detected between the individual phylogenies of the different loci (Fig. S7), especially among taxa composing the subclades that include D. leucospermi, D. sackstonii and D. micheliae (subclades A, F and G in Fig. 8). In general, tree topologies of the tef1-phylograms (Fig. S7B) were more similar to the phylogenetic analyses of the combined dataset. The multilocus phylogenetic analyses showed a better delimitation of the D. leucospermi clade when compared to the individual gene genealogies (Fig. 8).
Diaporthe infecunda was found to be paraphyletic in all individual gene genealogies, and to be intermingled with strains identified as D. leucospermi, which suggests that strains in subclade A are conspecific (Fig. S7). Species included in subclade F were well-resolved only in the tef1-phylograms (Fig. S7B). In the remaining individual gene genealogies several species revealed phylogenetic indistinctiveness in different combinations depending on the locus under analysis, while others presented a paraphyletic nature or grouped in lineages that lack phylogenetic support (Fig. S7A, C–E). Species in subclade G were also phylogenetically indistinguishable in most individual gene trees for which sequences are available. It is worth mentioning that only the ITS locus is available for the ex-type strains of D. chimonanthi and D. micheliae, which cannot distinguish both species from the strain SAUCC 194.27 (D. middletonii) (Fig. S7A). The same strain is phylogenetically indistinct from the ex-type strain of D. australpacifica in tef1-phylograms (Fig. S7B), while all D. middletonii strains are clearly conspecific with D. australpacifica in tub2-phylograms (Fig. S7C). Thus, considering the phylogenetic analyses of the combined dataset, as well as the GCPSR principle, subclades A, F and G have been recognised as three IEL and all strains within each subclade were found to be conspecific. Therefore, D. infecunda is regarded as a potential synonym of D. leucospermi, D. caryae, D. juglandigena, D. machili and D. orixae are regarded as potential synonyms of D. sackstonii, and D. australpacifica, D. chimonanthi and D. middletonii are regarded as potential synonyms of D. micheliae. The remaining IEL recognised within the D. leucospermi clade, i.e., D. acaciarum, D. beilharziae, D. myracrodruonis and D. serafiniae were found to clearly represent distinct well-supported lineages in most individual gene trees for which sequences are available (Fig. S7), although the distinction between D. beilharziae and D. serafiniae was not clear in both ITS- and tub2-phylograms.
Diaporthe longicolla clade
The phylogenetic analyses of the D. longicolla clade included 38 ingroup taxa and six closely related outgroup taxa. The ingroup taxa included five strains from palm substrata retrieved from recent literature, i.e., HNQH01, HNQH04, HNWC03, HNWN01 and HNWN02, identified as D. ueckeri (Table 2). Statistics for the different datasets and respective phylogenetic trees are summarised in Table S8. Tree topologies resulting from ML, MP and BA inferences were similar, displaying the same well-resolved terminal clades, whose relative position differed only in the MP inference for few strains. The ML tree is shown in Fig. 9 with MLBS/MP-BS/PP values at the nodes. The final likelihood score for the best scoring ML tree was -6100.067187. The BA inference had an average SDSF and an average PSRF of 0.009852 and 1.002, respectively, after 480 000 generations, resulting in 72 002 trees sampled. The MP analysis of 250 parsimony-informative characters (13.0 %) resulted in 324 trees of 617 steps with a low level of homoplasy as indicated by the measures of homoplasy and character fit (Table S8).
According to the phylogenetic analyses, six subclades were noted in a well-supported (100 % ML-BS / 95 % MP-BS / 1 PP; subclades A–F) monophyletic lineage, which was designated here as the D. longicolla clade (Fig. 8). Of these, subclades B (D. breyniae), D (D. morindendophytica), E (D. tectonendophytica) and F (D. trevorrowii) represent well-resolved (ML/MP-BS ≥ 82 % and 1 PP) single lineages, whereas subclades A and C displayed substantial to moderate substructure, respectively (Fig. 9). The five isolates from palm substrata clustered in a well-supported lineage (98 % ML-BS / 82 % MP-BS / 1 PP; subclade A) together with 18 strains belonging to six Diaporthe species, i.e., D. durionigena, D. passifloricola, D. rosae, D. thunbergiicola, D. ueckeri and D. vochysiae. Although these strains were identified as D. ueckeri, they did not cluster with its ex-type strain CBS 139283, which grouped in a well-supported lineage (96 % ML-BS / 95 % MPBS / 1 PP) with other strains of D. ueckeri. However, the taxa identified as D. ueckeri in previous studies were paraphyletic and scattered among other species included in subclade A. Moreover, most species in this subclade lack phylogenetic support or are unresolved in polytomies (Fig. 9). In subclade C, which is supported by 100 % ML/MP-BS and 1 PP, while D. longicolla and D. megabiguttulata grouped in distinct lineages with support values of 95 and 100 % ML-BS, 71 and 100 % MP-BS, respectively, and 1 PP, the phylogenetic resolution of D. unshiuensis is not clear, forming a polytomy instead of a dichotomic branch (Fig. 9).
To better diagnose the discordances observed in the multilocus phylogenetic inferences, the individual ML and MP gene trees were compared to identify concordant branches and define the IEL within the D. longicolla clade, according to the GCPSR principle. All individual ML and MP gene trees were topologically similar, presenting the same well-delimited clades. As mentioned in previous clades, this analysis should be applied with cautious due to lack of partial sequences of cal and his3 loci for several taxa. Several conflicts were observed between the taxa from subclades A and C (Figs S8, 9). Only tef1- (Fig. S8B) and cal-phylograms (Fig. S8D) displayed a similar topology to that observed with the multilocus phylogenetic inferences, which showed a better delimitation of the D. longicolla clade when compared to the individual gene genealogies (Fig. 9).
Most individual gene genealogies failed to diagnose species previously recognised in subclade A, insomuch as most of them were phylogenetically indistinct or presented a paraphyletic nature, which suggests that strains in subclade A are conspecific (Fig. S8). Similarly, subclade C was well-resolved only in tef1-phylograms (Fig. S8B), although branch lengths between D. longicolla and D. unshiuensis strains are very small. In the remaining individual gene trees, D. unshiuensis and D. megabiguttulata were unresolved, forming polytomies basal to D. longicolla, or lacking phylogenetic distinction (ITS-, tub2- and his3-phylograms; Fig. S8A, C, E). Thus, considering the phylogenetic analyses of the combined dataset, as well as the GCPSR principle, subclades A and C have been recognised as two IEL and all strains within each subclade were found to be conspecific. Therefore, D. durionigena, D. passifloricola, D. rosae, D. thunbergiicola and D. vochysiae are regarded as potential synonyms of D. ueckeri, and D. megabiguttulata and D. unshiuensis are regarded as potential synonyms of D. longicolla. The remaining IEL recognised within the D. longicolla clade, i.e., D. breyniae, D. morindendophytica, D. tectonendophytica and D. trevorrowii, were found to represent distinct well-supported lineages in most individual gene trees for which sequences are available (Fig. S8).
Diaporthe rudis clade
Twenty-four isolates were included in the phylogenetic analyses of the D. rudis clade, namely 22 ingroup taxa (two obtained in this study, viz. CDP 1323 and CDP 1861) and two closely related outgroup taxa (Table 2). Statistics for the different datasets and respective phylogenetic trees are summarised in Table S9. Tree topologies resulting from ML, MP and BA inferences were similar, and presented the same well-resolved clades for each species included in the analyses, mostly supported by high ML/MP-BS and PP values. The ML tree is shown in Fig. 10 with ML-BS/MP-BS/PP values at the nodes. The final likelihood score for the best scoring ML tree was -5750.998454. The BA inference had an average SDSF and an average PSRF of 0.007876 and 1.004, respectively, after 65 000 generations, resulting in 9 752 trees sampled. The MP analysis of 177 parsimony-informative characters (8.3 %) resulted in 36 trees of 529 steps with a very low level of homoplasy as indicated by the measures of homoplasy and character fit (Table S9).
According to the phylogenetic analyses, the ingroup taxa resolved into six well-supported subclades (ML/MP-BS ≥ 93 % and 1 PP; subclades A–F) from a monophyletic lineage with maximum support values, which was designated here as the D. rudis clade (Fig. 10). While subclades B (D. asheicola), C (D. australafricana), D (D. patagonica) and F (D. crousii) were composed by strains of a single species, subclades A and E included Diaporthe strains from more than one species. The isolates from this study grouped in subclade A, together with strains of D. rudis and two strains with type status of D. italiana. Although D. italiana strains grouped in a distinct lineage, they are nested in subclade A and mixed with strains previously recognised as D. rudis, including its ex-type strain CBS 109292 (Fig. 10). In subclade E, the ex-type strains of three Diaporthe species, i.e., D. cynaroidis (CBS 122676), D. salicicola (BRIP 54825) and D. subcylindrospora (KUMCC 17-0151), radiated with relatively small branch length distances, suggesting that they might be conspecific (Fig. 10).
Individual ML and MP gene trees were compared to identify concordant branches and accordingly apply the GCPSR principle. All individual ML and MP gene trees were topologically similar, presenting the same well-delimited clades. In general, tree topologies of the tef1- (Fig. S9B) and cal-phylograms (Fig. S9D) were more similar to the phylogenetic analyses of the combined dataset. The multilocus phylogenetic analyses showed a better delimitation of the D. rudis clade when compared to the individual gene genealogies (Fig. 2). Nonetheless, a very similar phylogenetic structure was observed between the individual phylogenies of the different loci, among which almost no conflicts were detected (Fig. S9), except for ITS-phylograms (Fig. S9A). However, D. italiana lacks phylogenetic distinctiveness from D. rudis in the tub2- and cal-phylograms (Fig. S9C, D) and appeared intermingled among them, along other species, in the ITS-phylograms (Fig. S9A). No his3 partial sequences are available for D. italiana, and tef1 partial sequences have been excluded from the present analyses (further details are given in the Taxonomy section). The low phylogenetic distinction between D. cynaroidis, D. salicicola and D. subcylindrospora was also observed in the individual gene genealogies for which sequences are available (Fig. S9B, C). Following the GCPSR principle, six different IEL were recognised in the D. rudis clade, corresponding to the six subclades observed in the multilocus phylogenetic inferences. Therefore, D. italiana is regarded as a potential synonym of D. rudis, and D. salicicola and D. subcylindrospora are regarded as potential synonyms of D. cynaroidis.
Phylogenetic informativeness and informative characters of each locus
To assess the relative power of the five markers to resolve a node in a given period of time in Diaporthe phylogenies, phylogenetic informativeness (PI) analyses were performed for each locus in each of the combined datasets. The phylogenetic informativeness profiles (PIP) generated for the nine Diaporthe clades treated in this study are shown in Fig. 11.
Fig. 11.
Net phylogenetic informativeness profiles (PIP) in arbitrary units of Diaporthe species through relative time scales for combined datasets of 5-loci (ITS, tef1, tub2, cal and his3). A. PIP of the D. amygdali clade and related species. B. PIP of the D. brasiliensis clade and related species. C. PIP of the D. eucommiae clade and related species. D. PIP of the D. foeniculina clade and related species. E. PIP of the D. glabrae clade and related species. F. PIP of the D. inconspicua clade and related species. G. PIP of the D. leucospermi clade and related species. H. PIP of the D. longicolla clade and related species. I. PIP of the D. rudis clade and related species. The lines represent individual loci profiles and are referred to in the chart legend. The respective time trees inferred by applying the RelTime-ML method are shown in the background of each panel, in corresponding time scales. All divergence times shown are relative times as no calibrations were used. Nodes of radiation of each Diaporthe clade are highlighted by dots and the corresponding graph areas are emphasised by coloured blocks.
The most informative markers for phylogenetic inference of Diaporthe species varied according to the clade under analysis. However, tef1 ranked as the most informative marker for phylogenetic inference in all the Diaporthe clades analysed, while ITS, tub2 and his3 ranked as the least informative (Fig. 11). Nonetheless, in most Diaporthe clades, different loci were coupled with tef1 as informative markers. For instance, in the D. amygdali (Fig. 11A), D. foeniculina (Fig. 11D), D. inconspicua (Fig. 11F), D. longicolla (Fig. 11H) and D. rudis (Fig. 11I) clades, cal also ranked as one of the most informative markers to resolve the node at which each clade radiates into different subclades (coloured blocks in Fig. 11). These results are congruent with the topologies of tef1- and cal-phylograms of most Diaporthe clades (Figs S1, S3, S4, S7–S9), which were in general more topologically similar to the respective multilocus phylogenetic inferences. On the other hand, tub2 was also one of the most informative markers to resolve species limits within the D. glabrae clade (Fig. 11E), which was also congruent with the respective individual gene genealogies obtained for this clade (Fig. S5).
In comparison with the percentage of parsimony informative characters and unique alignment patterns of each locus, tef1 and, at less extent, cal showed a congruent result with the PI profiles as the most informative loci for phylogenetic inference of most Diaporthe clades (Tables S1–S9). In most clades, the percentage of parsimony informative characters and unique alignment patterns of tef1 is almost twice or more than twice that of the other loci. For instance, in the D. eucommiae clade, while tef1 presented 39.1 % of parsimony informative characters and 48.8 % of unique alignment patterns, respectively, the remaining loci presented percentages of informative characters of less than or around 20 % (Table S3).
Integrating PI over specific periods of time provides information for ranking loci at specific nodes of a given phylogeny. The PI analyses for the D. amygdali (Fig. 11A), D. foeniculina (Fig. 11D) and D. inconspicua (Fig. 11F) clades showed a peak for the ITS curve corresponding to the relative period in which the respective clades radiate into several branches and subclades (coloured block in Fig. 11). Thus, for that specific relative period ITS ranks as the most informative marker. Nonetheless, ITS is the least informative locus as the trees approach theirs roots in all the Diaporthe clades analysed. A similar curve pattern is observed in the PIP of tub2 and his3 loci in some clades (Fig. 11). According to the informative characters provided by the phylogenetic analyses, ITS, together with tub2 and his3, displayed the least informative sequences, with the lowest percentage of parsimony informative characters and unique alignment patterns in most Diaporthe clades (Tables S1–S9). These results suggest that ITS and, to a lesser extent, tub2 and his3 loci may not be suitable for phylogenetic inference of Diaporthe species. These results are in line with the ITS- and tub2-phylograms, which were, in general terms, the most topologically discordant when compared to the corresponding multilocus phylogenetic inferences.
Species delimitation analyses, pairwise homoplasy test and phylogenetic network analyses
The IEL determined by the GCPSR principle and the multilocus phylogenetic analyses were tested through five different DNA-based species delimitation methods. The congruence of their results in relation to the GCPSR was evaluated by the delimitation efficiency ratio (DER), the results of which are presented in Table 3. It should be noted that the delimitation of the closely related outgroup taxa included in each Diaporthe clade was not considered here, regardless of the detection of conflicts in their delimitation. The present analyses were not directed at these taxa, which were only used to reinforce the delimitation of species within each of the Diaporthe clades treated.
Table 3.
Delimitation efficiency ratio (DER) results for the species delimitation analyses of the nine Diaporthe clades studied.
| Clade | Delimitation efficiency ratio (DER)1 | ||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| PTP | mPTP | ABGD2 | ASAP2 | SPN | |||
|
|
|
||||||
| Combined | tef1 | Combined | tef1 | ||||
| D. amygdali clade (Fig. 12A) | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| D. brasiliensis clade (Fig. 13A) | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| D. eucommiae clade (Fig. 14A) | 1 | 1 | 0.33 | 0.33 | 0.33 | 0.33 | 1 |
| D. foeniculina clade (Fig. 15A) | 1 | 1 | 0.25 | 0.25 | 0.25 | 0.25 | 0.4 |
| D. glabrae clade (Fig. 16A) | 1 | 1 | 0.67 | 0.33 | 0.33 | 0.33 | 0.67 |
| D. inconspicua clade (Fig. 17A) | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| D. leucospermi clade (Fig. 18A) | 0.86 | 0.86 | 0.71 | 0.14 | 0.71 | 0.14 | 1.43* |
| D. longicolla clade (Fig. 19A) | 1 | 1 | 1 | 0.67 | 1 | 0.67 | 0.67 |
| D. rudis clade (Fig. 20A) | 1 | 1 | 0.83 | 0.83 | 0.83 | 0.83 | 1 |
1 PTP: Poisson tree processes; mPTP: multi-rate PTP; ABGD: automatic barcode gap discovery; ASAP: assemble species by automatic partitioning; SPN: statistical parsimony network.
2 Combined: combined dataset based on 5- (ITS, tef1, tub2, cal and his3) or 3-loci (ITS, tef1 and tub2), as cal and his3 loci were excluded from certain clades due to lack of sequences for several taxa (for details see the corresponding figure).
DER > 1, the species delimitation analysis recognised a higher number of molecular operational taxonomic units (MOTU) when compared to the number of species recognised by the GCPSR principle.
Coalescent-based methods, i.e., Poisson tree processes (PTP) and multi-rate PTP (mPTP) produced, in general, species delimitation schemes with several molecular operational taxonomic units (MOTU) highly congruent with the number of putative phylogenetic species inferred by the GCPSR (DER = 1.00; Table 3). The only exception was observed in the D. leucospermi clade, for which both PTP and mPTP recognised two of the IEL as a single MOTU (DER = 0.86; Table 3). Moreover, both PTP and mPTP analyses gave congruent species delimitation results in all the Diaporthe clades treated.
As previously referred, missing data were very unevenly distributed among the different genes used, corresponding mostly to sequences of cal and his3 loci (Table 2). Given the lack of these data for several species of most of the Diaporthe clades, the PTP and mPTP analyses performed for the 5-loci combined datasets including all species in each clade, were compared with PTP and mPTP analyses of 5-, 4- and 3-loci combined datasets including only those species whose five, four or three loci were available, respectively. The species delimitation schemes produced by both analyses were, in general, highly congruent and missing data had little impact on the species hypotheses (data not shown). Nonetheless, in some clades, the decrease in the number of taxa and/or loci led to an over-lumping of species that were recognised as different IEL by the GCPSR principle, especially in the mPTP analyses (specific cases are mentioned below for the respective Diaporthe clade). However, the opposite scenario did not occur, i.e., species limits determined by the GCPSR principle were never over-divided by the coalescent-based methods, which provides further evidence for the potential synonyms suggested by the phylogenetic analyses.
Unlike the coalescent-based methods, the distance-based methods presented variable DER and produced in general a few MOTU high to moderately incongruent with the GCPSR principle (DER = 0.25–1.43; Table 3). Moreover, while the different coalescent-methods applied resulted in congruent results in each Diaporthe clade, the different distance-based methods often produced incongruent species delimitation hypotheses. In most of the Diaporthe clades, the distance-based methods showed lumping of the IEL recognised by the GCPSR (DER < 1.00; Table 3). Only three out of nine Diaporthe clades, i.e., D. amygdali, D. brasiliensis and D. inconspicua clades, were similarly resolved by all distance-based methods, which gave congruent results with the GCSPR (DER = 1.00; Table 3). Among the different distance-based methods tested, SPN analyses generated higher DER, which resulted in a greater number of delimitation hypotheses congruent with the GCPSR. Therefore, the over-lumping of IEL observed with distance-based methods were more prominent with ABGD and ASAP analyses. Over-splitting of IEL was only observed in the D. leucospermi clade when applying SPN analyses (DER = 1.43; Table 3).
It is worth mentioning that most of the incongruent ABGD and ASAP delimitation schemes corresponded to datasets from which cal and his3 loci have been excluded due to lack of sequence data for several taxa, which strongly disturbed the distance matrices generated and the corresponding barcode gaps. However, in the D. rudis clade, the same lumping of IEL of ABGD and ASAP analyses were observed, even though combined datasets of 5-loci were used (Table 3). Although tef1 ranked as the most informative loci for phylogenetic inference of all the Diaporthe clades treated (Fig. 11), distance-based analyses based on this single locus resulted in similar or worst species delimitation schemes, i.e., with more lumping of IEL, compared to the combined datasets (Table 3). These over-lumping results were also observed when tef1 phylogenies were tested with coalescent-based methods (data not shown). Thus, both coalescent- and distance-based methods produced results more congruent with the GCPSR principle when combined datasets were tested, instead of single locus. The over-lumping of IEL observed when applying distance-based analyses also provides further evidence for the potential synonyms suggested by the phylogenetic analyses.
The phylogenetic species recognised by the GCPSR principle were also tested by constructing phylogenetic networks to observe incompatible and ambiguous signals within each Diaporthe clade treated. All the phylogenetic networks built for each Diaporthe clade showed fit values greater than 90 %, indicating that the displayed networks represent well the LogDet distance matrices from which they were computed. The splits graphs for each Diaporthe clade are presented in Figs 12–20, along with the results of the DNA-based species delimitation analyses.
Fig. 12.
Species delimitation analyses of the Diaporthe amygdali clade. A. Comparison of species delimitation results for the D. amygdali clade based on a combined dataset of 5-loci (ITS, tef1, tub2, cal and his3), unless indicated otherwise (see Materials and Methods for further explanation). Schematic phylogenetic relationships are shown using the species delimitation scheme obtained from the PTP analysis, in which putative species clusters are represented as transitions from blue-coloured (speciation process) to red-coloured (population process) branches or as terminal, blue-coloured branches. Dot-bounded bars in the right-hand columns show the results of the genealogical concordance- (pink column; GCPSR), coalescence- (blue column; PTP and mPTP) and distance-based (yellow column; ABGD, ASAP and SPN) species delimitation analyses, which are referred to at the top. Incongruent results in relation to the GCPSR are highlighted in red. B–D. NeighborNet phylogenetic networks of D. amygdali and related species based on the LogDet transformation for a combined dataset of 5-loci (ITS, tef1, tub2, cal and his3). The D. amygdali clade and respective species are delimited by dashed and coloured shapes, respectively. The PHI test results are presented next to each set of species tested and if positive for recombination is indicated by an asterisk (*). Strains with type status are indicated in bold font. The isolates from this study are presented in brown typeface. The scale bars represent the expected number of nucleotide changes per site.
Fig. 20.
Species delimitation analyses of the Diaporthe rudis clade. A. Comparison of species delimitation results for the D. rudis clade based on a combined dataset of 5-loci (ITS, tef1, tub2, cal and his3), unless indicated otherwise (see Materials and Methods for further explanation). Schematic phylogenetic relationships are shown using the species delimitation scheme obtained from the PTP analysis, in which putative species clusters are represented as transitions from blue-coloured (speciation process) to red-coloured (population process) branches or as terminal, blue-coloured branches. Dots and dot-bounded bars in the right-hand columns show the results of the genealogical concordance- (pink column; GCPSR), coalescent- (blue column; PTP and mPTP) and distance-based (yellow column; ABGD, ASAP and SPN) species delimitation analyses, which are referred to at the top. B–D. NeighborNet phylogenetic networks of D. rudis and related species based on the LogDet transformation for a combined dataset of 5-loci (ITS, tef1, tub2, cal and his3). The D. rudis clade and respective species are delimited by dashed and coloured shapes, respectively. The PHI test results are presented next to each set of species tested and if positive for recombination is indicated by an asterisk (*). Strains with type status are indicated in bold font. The isolates from this study are presented in brown typeface. The scale bars represent the expected number of nucleotide changes per site.
The results of the five species delimitation methods in the D. amygdali clade (Fig. 12A) were in broad agreement and unequivocally supported the entire D. amygdali clade as conspecific. These results are in line with the networked relationships among the taxa composing this clade (Fig. 12B), which displayed many conflicting signals. The parallel edges and boxlike polygons between virtually all taxa within the D. amygdali clade suggest likelihood of recombination, which is congruent with the results obtained with the PHI test that found statistically significant evidence for recombination (P = 6.52 × 10−14; Fig. 12B). Moreover, both species regarded here as potential synonyms of D. amygdali – D. nannuoshanensis and D. sinensis – shown conflicting signals in their network relationships with a set of authentic strains of D. amygdali, including its ex-type strain, insomuch as the PHI test revealed statistically significant evidence for recombination (P = 0.00 and 0.04, respectively; Fig. 12C, D). Only one branch, which corresponds to D. sinensis (JZB 320167), appears to diverge from the intricate web of edges of the D. amygdali clade, which is related to the highly polymorphic cal sequence available for this strain (further details are referred to in the Taxonomy section). However, all analyses carried out were congruent and revealed that strains introduced as D. nannuoshanensis and D. sinensis represent intraspecific variation of D. amygdali.
All five species delimitation methods supported four species in the D. brasiliensis clade (Fig. 13A). Therefore, D. pygmaeae was recognised as a new Diaporthe species with the coalescent- and distance-based analyses applied to combined and single locus datasets. Similarly, the networked relationships among the taxa composing the D. brasiliensis clade showed that all four species are clearly placed apart from each other by an assemblage of relatively long branches and bifurcated evolutionary relationships (Fig. 13B). This treelike structure was congruent with the PHI test results, which found no statistically significant evidence for recombination within the D. brasiliensis clade (P = 1.00; Fig. 13B). The PHI test was also negative for recombination (P = 1.00) when the strains of D. pygmaeae sp. nov. were tested only against strains of D. brasiliensis or D. caatingaensis and no contradicting signals were observed in their networked relationships (Fig. 13C, D).
Fig. 13.
Species delimitation analyses of the Diaporthe brasiliensis clade. A. Comparison of species delimitation results for the D. brasiliensis clade based on a combined dataset of 5-loci (ITS, tef1, tub2, cal and his3), unless indicated otherwise (see Materials and Methods for further explanation). Schematic phylogenetic relationships are shown using the species delimitation scheme obtained from the PTP analysis, in which putative species clusters are represented as transitions from blue-coloured (speciation process) to red-coloured (population process) branches or as terminal, blue-coloured branches. Dot-bounded bars in the right-hand columns show the results of the genealogical concordance- (pink column; GCPSR), coalescence- (blue column; PTP and mPTP) and distance-based (yellow column; ABGD, ASAP and SPN) species delimitation analyses, which are referred to at the top. B–D. NeighborNet phylogenetic networks of D. brasiliensis and related species based on the LogDet transformation for a combined dataset of 5-loci (ITS, tef1, tub2, cal and his3). The D. brasiliensis clade and respective species are delimited by dashed and coloured shapes, respectively. The PHI test results are presented next to each set of species tested and if positive for recombination is indicated by an asterisk (*). Strains with type status are indicated in bold font. The isolates from this study are presented in brown typeface. The scale bars represent the expected number of nucleotide changes per site.
In the D. eucommiae clade, PTP, mPTP and SPN analyses were congruent in supporting three species (Fig. 14A). Additional PTP and mPTP analyses based on 3-, 4- and 5-loci combined datasets were also in broad agreement with the IEL recognised by the GCPSR principle and returned delimitation schemes of three species. The only exception was the mPTP analysis based on the 3-loci combined dataset, which indicated that the entire D. eucommiae clade is conspecific, similar to the results obtained with ABGD and ASAP analyses. Considering that SPN analyses, which are also distance-based, showed congruent results with most of the coalescent-based analyses, it is likely that the over-lumping results are be biased due to lack of sequence data.
Fig. 14.
Species delimitation analyses of the Diaporthe eucommiae clade. A. Comparison of species delimitation results for the D. eucommiae clade based on a combined dataset of 5-loci (ITS, tef1, tub2, cal and his3), unless indicated otherwise (see Materials and Methods for further explanation). Schematic phylogenetic relationships are shown using the species delimitation scheme obtained from the PTP analysis, in which putative species clusters are represented as transitions from blue-coloured (speciation process) to red-coloured (population process) branches. Dot-bounded bars in the right-hand columns show the results of the genealogical concordance- (pink column; GCPSR), coalescent- (blue column; PTP and mPTP) and distance-based (yellow column; ABGD, ASAP and SPN) species delimitation analyses, which are referred to at the top. Incongruent results in relation to the GCPSR are highlighted in red. Empty dots indicate taxa excluded from the analysis due to lack of sequence data. B, C. NeighborNet phylogenetic networks of D. eucommiae and related species based on the LogDet transformation for a combined dataset of 5-loci (ITS, tef1, tub2, cal and his3). The D. eucommiae clade and respective species are delimited by dashed and coloured shapes, respectively. The PHI test results are presented next to each set of species tested and if positive for recombination is indicated by an asterisk (*). Strains with type status are indicated in bold font. The isolate from palm tissues included in the analyses is presented in green typeface. The scale bars represent the expected number of nucleotide changes per site.
The phylogenetic network of the D. eucommiae clade revealed several conflicting signals, implying likelihood of recombination, which was shown to be statically significant (P = 3.23 × 10−6). Based on the relative distance of taxa and structure of the phylogenetic network, three subnetworks are observed in the D. eucommiae clade, which resemble bifurcated treelike evolutionary events and are congruent with the previous analyses that recognised three species (Fig. 14B). Moreover, D. aseana and D. eucommiae subnetworks tested positive for recombination (P = 1.80 × 10−6 and 0.04, respectively), which supports the potential synonyms proposed here for both species. However, statistically significant evidence for recombination was also detected between strains of D. aseana, D. eucommiae and D. xishuangbanica, as well as when the PHI test was performed on pairwise combinations of strains of these three species (P < 0.05).
The incongruent results observed between species delimitation analyses are well illustrated in the networked relationships among the taxa of the D. foeniculina clade. Several contradicting signals dominate the evolutionary relationships of these taxa, which are mostly connected by boxlike polygons and a set of branches with very irregular lengths that impair the clear definition of the boundaries between the different species (Fig. 15B). These contradicting signals support the potential synonyms noted here for D. foeniculina and D. cytosporella, whose networks appear to exhibit inherently non-treelike evolutionary events with several parallel edges. However, it is unclear if some of the IEL inferred by the GCPSR do represent the same species. The incongruences observed through phylogenetic networks analyses are congruent with the SPN analyses (Fig. 15A), which revealed that the haplotypes of D. foeniculina, D. cytosporella, D. diospyricola and D. parva are significantly connected in a single network and may be regarded as conspecific. However, this result may be biased due to the lack of sequence data or even insufficient sampling. For example, D. diospyricola only has the ITS sequence available for a single strain (Table 2). While D. diospyricola and D. parva seem to relatively distance themselves from the web of connections composing D. foeniculina, D. cytosporella subnetwork appear to exhibit inherently non-treelike evolutionary events with several parallel edges with D. foeniculina subnetwork (Fig. 15B). Nonetheless, the PHI test was negative for recombination (P > 0.05; Fig. 15B) for all the datasets tested within the D. foeniculina clade, and no statistically significant recombination was detected between strains of D. foeniculina and D. cytosporella (P = 0.40).
Fig. 15.
Species delimitation analyses of the Diaporthe foeniculina clade. A. Comparison of species delimitation results for the D. foeniculina clade based on a combined dataset of 5-loci (ITS, tef1, tub2, cal and his3), unless indicated otherwise (see Materials and Methods for further explanation). Schematic phylogenetic relationships are shown using the species delimitation scheme obtained from the PTP analysis, in which putative species clusters are represented as transitions from blue-coloured (speciation process) to red-coloured (population process) branches or as terminal, blue-coloured branches. Dots and dot-bounded bars in the right-hand columns show the results of the genealogical concordance- (pink column; GCPSR), coalescence- (blue column; PTP and mPTP) and distance-based (yellow column; ABGD, ASAP and SPN) species delimitation analyses, which are referred to at the top. Incongruent results in relation to the GCPSR are highlighted in red. Empty dots indicate taxa excluded from the analysis due to lack of sequence data. B. NeighborNet phylogenetic networks of D. foeniculina and related species based on the LogDet transformation for a combined dataset of 5-loci (ITS, tef1, tub2, cal and his3). The D. foeniculina clade and respective species are delimited by dashed and coloured shapes, respectively. The PHI test results are presented next to each set of species tested and if positive for recombination is indicated by an asterisk (*). Strains with type status are indicated in bold font. The isolates from this study are presented in brown typeface and the additional isolate from palm tissues included in the analyses is presented in green typeface. The scale bars represent the expected number of nucleotide changes per site.
As with the D. foeniculina clade, the coalescent- and distance-based analyses were not concordant regarding the number of MOTU inferred for the D. glabre clade. Moreover, only PTP and mPTP analyses were congruent in supporting three species as inferred by the GCPSR principle (Fig. 16A). As previously mentioned, the results of the different methods are likely to be influenced by datasets composed of a different number of loci due to lack of sequence data for several taxa. Nonetheless, PTP analyses on 3-loci datasets also generated species delimitation schemes of three species, although mPTP analyses on the same datasets provided similar lumping of species as observed with the distance-based methods.
Fig. 16.
Species delimitation analyses of the Diaporthe glabrae clade. A. Comparison of species delimitation results for the D. glabrae clade based on a combined dataset of 5-loci (ITS, tef1, tub2, cal and his3), unless indicated otherwise (see Materials and Methods for further explanation). Schematic phylogenetic relationships are shown using the species delimitation scheme obtained from the PTP analysis, in which putative species clusters are represented as transitions from blue-coloured (speciation process) to red-coloured (population process) branches or as terminal, blue-coloured branches. Dots and dot-bounded bars in the right-hand columns show the results of the genealogical concordance- (pink column; GCPSR), coalescent- (blue column; PTP and mPTP) and distance-based (yellow column; ABGD, ASAP and SPN) species delimitation analyses, which are referred to at the top. Incongruent results in relation to the GCPSR are highlighted in red. Empty dots indicate taxa excluded from the analysis due to lack of sequence data. B, C. NeighborNet phylogenetic networks of D. glabrae and related species based on the LogDet transformation for a combined dataset of 5-loci (ITS, tef1, tub2, cal and his3). The D. glabrae clade and respective species are delimited by dashed and coloured shapes, respectively. The PHI test results are presented next to each set of species tested and if positive for recombination is indicated by an asterisk (*). Strains with type status are indicated in bold font. The isolates from palm tissues included in the analyses are presented in green typeface. The scale bars represent the expected number of nucleotide changes per site.
The networked relationships among the taxa of the D. glabrae clade illustrate well the incongruencies of the DNA-based species delimitation analyses. While the branches leading to D. celtidis have a considerable relative size that separates and bifurcates them from the other taxa in the clade, strains recognised here as D. tulliensis and D. glabrae display many contradicting edges and boxes inferring a relationship among individual with likelihood of recombination (Fig. 16B). Moreover, the relative distance and structure of the phylogenetic network seem to indicate that both taxa are conspecific, which is in line with the haplotype networks inferred by the SPN analyses (Fig. 16A). However, the PHI test results are congruent with the species delimitation hypotheses inferred by the coalescent methods, which support the IEL recognised by the GCPSR principle. No statistically significant evidence for recombination was detected between strains of D. tulliensis and D. glabrae (P = 0.05), nor when the D. glabrae clade was tested (P = 0.43; Fig. 16B). Yet, significant recombination was detected when the PHI was tested only with strains of D. glabrae (P = 0.01; Fig. 16C), denoting lack of reproductive isolation and supporting the potential synonyms proposed here for this species.
The results of the five species delimitation methods in the D. inconspicua clade (Fig. 17A) were in broad agreement and unequivocally supported two species. These results are in line with the networked relationships among the taxa composing this clade, which relative position and distance resemble bifurcated treelike evolutionary events that resulted into two subnetworks (Fig. 17B). However, these subnetworks display many conflicting signals, with parallel edges and boxlike polygons present both within D. inconspicua subnetwork and between each subnetwork (Fig. 17C). The likelihood of recombination was proved to be statistically significant through the PHI test, which was positive (P = 8.83 × 10−15) for the entire D. inconspicua clade, as well as for the D. inconspicua subnetwork (P = 2.33 × 10−14), which supports the potential synonyms proposed here for this species. Strains of D. isoberliniae and D. pungensis, although clearly related, tested negative for recombination (P = 1.00; Fig. 17C). Moreover, pairwise combinations of set of strains in both subnetworks to test the presence of recombination between D. inconspicua and D. isoberliniae, also gave negative results and no statistically significant recombination was detected between both species (P > 0.05).
Fig. 17.
Species delimitation analyses of the Diaporthe inconspicua clade. A. Comparison of species delimitation results for the D. inconspicua clade based on a combined dataset of 5-loci (ITS, tef1, tub2, cal and his3), unless indicated otherwise (see Materials and Methods for further explanation). Schematic phylogenetic relationships are shown using the species delimitation scheme obtained from the PTP analysis, in which putative species clusters are represented as transitions from blue-coloured (speciation process) to red-coloured (population process) branches. Dot-bounded bars in the right-hand columns show the results of the genealogical concordance- (pink column; GCPSR), coalescent- (blue column; PTP and mPTP) and distance-based (yellow column; ABGD, ASAP and SPN) species delimitation analyses, which are referred to at the top. B, C. NeighborNet phylogenetic networks of D. inconspicua and related species based on the LogDet transformation for a combined dataset of 5-loci (ITS, tef1, tub2, cal and his3). The D. inconspicua clade and respective species are delimited by dashed and coloured shapes, respectively. The PHI test results are presented next to each set of species tested and if positive for recombination is indicated by an asterisk (*). Strains with type status are indicated in bold font. The isolate from palm tissues included in the analyses is presented in green typeface. The scale bars represent the expected number of nucleotide changes per site.
In the D. leucospermi clade, none of the five species delimitation methods agreed in depicting the number of species. While most of the analyses lumped IEL recognised by the GCPSR principle, the SPN analyses divided the taxa into a greater number of MOTU than the monophyletic lineages observed in the phylogenetic analyses (Fig. 18A). As referred in previous clades, the results of ABGD and ASAP should be interpreted with caution as they are based on datasets of three loci due to lack of sequence data. However, the lumping effect was similarly observed in additional analyses comprising only the species for which the four and five loci were available (data not shown). On the other hand, while the additional PTP analyses based on 4- and 5-loci datasets were congruent with the previous results, all mPTP analyses and the PTP analyses based on the 3-loci dataset resulted in over-lumping of IEL, producing species delimitation schemes like those obtained with ABGD and ASAP analyses (data not shown).
Fig. 18.
Species delimitation analyses of the Diaporthe leucospermi clade. A. Comparison of species delimitation results for the D. leucospermi clade based on a combined dataset of 5-loci (ITS, tef1, tub2, cal and his3), unless indicated otherwise (see Materials and Methods for further explanation). Schematic phylogenetic relationships are shown using the species delimitation scheme obtained from the PTP analysis, in which putative species clusters are represented as transitions from blue-coloured (speciation process) to red-coloured (population process) branches or as terminal, blue-coloured branches. Dots and dot-bounded bars in the right-hand columns show the results of the genealogical concordance- (pink column; GCPSR), coalescent-(blue column; PTP and mPTP) and distance-based (yellow column; ABGD, ASAP and SPN) species delimitation analyses, which are referred to at the top. B–D. NeighborNet phylogenetic networks of D. leucospermi and related species based on the LogDet transformation for a combined dataset of 5-loci (ITS, tef1, tub2, cal and his3). The D. leucospermi clade and respective species are delimited by dashed and coloured shapes, respectively. The PHI test results are presented next to each set of species tested and if positive for recombination is indicated by an asterisk (*). Strains with type status are indicated in bold font. The isolates from this study are presented in brown typeface. The scale bars represent the expected number of nucleotide changes per site.
The phylogenetic network of the D. leucospermi clade revealed several conflicting signals and a series of projected branches of different relative size that make it difficult to perceive the boundaries between the different taxa. The number of parallel edges and boxlike polygons are clearly implying likelihood of recombination among species, insomuch as the entire clade tested positive for recombination (P = 6.08 × 10−7; Fig. 18B).
The PTP and mPTP analyses were more congruent than distance-based methods with the IEL recognised by the GCPSR principle, except in the case of D. serafiniae, which was recognised as conspecific of D. leucospermi (Fig. 18A). The phylogenetic position and distinction of D. serafiniae was also unclear in the ITS- and tub2-phylograms (Fig. S7), but it was classified as an independent lineage, considering that its evolutionary relationship with the D. leucospermi subclade was not clear or well-supported by any of the available loci or by the multilocus phylogenetic analyses (Fig. 8). The relative position and distance of D. serafiniae and D. leucospermi strains in their networked relationships effectively support their distinction as different lineages (Fig. 18C). Moreover, no statistically significant evidence for recombination was detected between strains of both species (P = 0.15). However, the PHI tested positive (P = 4.50 × 10−5) when applied to strains of D. leucospermi and D. infecunda, supporting the recombination denoted in the conflicting signals observed in the network relationships between these two species (Fig. 18D). Thus, although the SPN analyses divided the haplotypes of D. leucospermi and D. infecunda into distinct networks, the remaining analyses were congruent in regarding both species to be synonymous. The SPN analyses also divided D. sackstonii into several independent MOTU, disconnecting its haplotypes in a more or less similar way to the number of species previously introduced in this lineage (Fig. 18A). However, the networked relationships between virtually all taxa within this lineage are dominated by boxes instead of a bifurcating evolutionary treelike structure (data not shown). Moreover, statistically significant recombination was detected within D. sackstonii lineage when applying the PHI test (P = 2.57 × 10−4), supporting the potential synonyms proposed here for this species.
Most of the species delimitation methods applied were congruent and supported six species in the D. longicolla clade (Fig. 19A). Only single-locus and SPN analyses were incongruent and over-lumped several of the IEL determined by the GCPSR principle. The results of all the additional coalescent- and distance-based analyses, including only the species for which the four and five loci were available, were also congruent with the GCPSR principle (data not shown), apart from the additional mPTP analyses, which also tend to produce over-lumped species delimitation schemes.
Fig. 19.
Species delimitation analyses of the Diaporthe longicolla clade. A. Comparison of species delimitation results for the D. longicolla clade based on a combined dataset of 5-loci (ITS, tef1, tub2, cal and his3), unless indicated otherwise (see Materials and Methods for further explanation). Schematic phylogenetic relationships are shown using the species delimitation scheme obtained from the PTP analysis, in which putative species clusters are represented as transitions from blue-coloured (speciation process) to red-coloured (population process) branches or as terminal, blue-coloured branches. Dots and dot-bounded bars in the right-hand columns show the results of the genealogical concordance- (pink column; GCPSR), coalescent- (blue column; PTP and mPTP) and distance-based (yellow column; ABGD, ASAP and SPN) species delimitation analyses, which are referred to at the top. B. NeighborNet phylogenetic networks of D. longicolla and related species based on the LogDet transformation for a combined dataset of 5-loci (ITS, tef1, tub2, cal and his3). The D. longicolla clade and respective species are delimited by dashed and coloured shapes, respectively. The PHI test results are presented next to each set of species tested and if positive for recombination is indicated by an asterisk (*). Strains with type status are indicated in bold font. The isolates from palm tissues included in the analyses are presented in green typeface. The scale bars represent the expected number of nucleotide changes per site.
The network relationships between the taxa of the D. longicolla clade displayed many conflicting signals, implying likelihood of recombination, which proved to be statistically significant (P = 6.52 × 10−4; Fig. 19B). However, most of these signals are concentrated in subnetworks that resemble a bifurcated treelike structure, which correspond to the previously determined species boundaries. Taxa in the D. ueckeri subnetwork were shown to be virtually all connected and most of them had relatively small branch sizes (Fig. 19B), which supports the previous analyses that regarded the entire subclade as conspecific. Moreover, the PHI test revealed statically significant evidence for recombination (P = 0.00).
No significant recombination was detected between strains of D. morindendophytica, D. tectonendophytica and D. trevorrowii (P = 0.85; Fig. 19B), which supports the previous analyses that regarded the three species as independent lineages. However, no significant recombination was detected between strains of the D. longicolla subnetwork (P = 1.00; Fig. 19B), although previous analyses regarded species in this subclade as conspecific.
The results of most of the species delimitation methods in the D. rudis clade (Fig. 20A) were in broad agreement and supported six species. Only the ABGD and ASAP analyses showed over-lumping between D. rudis and D. australafricana. The six species were also evident in the networked relationships among the taxa composing the D. rudis clade, as all species are clearly placed apart from each other by an assemblage of relatively long branches and bifurcated evolutionary relationships. Moreover, almost no contradicting signals were detected, except for few parallel edges between taxa previously recognised as D. rudis (Fig. 20B).
The treelike structure of the networked relationships in the D. rudis clade was congruent with the PHI test results, which found no statistically significant evidence for recombination within the clade (P = 0.50; Fig. 20B). Strains of D. rudis and D. australafricana tested negative for recombination (P = 0.28; Fig. 20C), which supports the previous analyses that indicated both lineages as different species. However, strains of D. italiana and D. rudis also tested negative for recombination (P = 0.23; Fig. 20D), although the remaining analyses were congruent in regarding both species to be synonymous.
Hierarchical cluster analysis of phenotypic data
Dendrograms were constructed by hierarchical cluster analyses based on published morphological descriptions of species of the Diaporthe clades treated in this study (Fig. 21). A synopsis of the morphological data is provided in Table 4, along with the host, country and ecological group of all specimens considered. The phenotypic data used to construct the dendrograms were the dimensions of the alpha conidia, beta conidia and/or elements of the conidiogenous layer, depending on the Diaporthe clade under analysis.
Fig. 21.
Dendrograms obtained by hierarchical cluster analysis of Diaporthe clades using Euclidean distance and UPGMA algorithm. A. Dendrogram of the D. amygdali clade based on the length-to-width ratio (L/W) of alpha conidia and elements of the conidiogenous layer. B. Dendrogram of the D. brasiliensis clade based on the L/W of alpha conidia and elements of the conidiogenous layer. C. Dendrogram of the D. eucommiae clade based on the L/W of alpha and beta conidia. D. Dendrogram of the D. foeniculina clade based on the L/W of alpha and beta conidia and elements of the conidiogenous layer. E. Dendrogram of the D. glabrae clade based on the L/W of alpha and beta conidia and elements of the conidiogenous layer. F. Dendrogram of the D. inconspicua clade based on the L/W of alpha and beta conidia and elements of the conidiogenous layer. G. Dendrogram of the D. leucospermi clade based on the L/W of alpha and beta conidia and elements of the conidiogenous layer. H. Dendrogram of the D. longicolla clade based on the L/W of alpha and beta conidia. I. Dendrogram of the D. rudis clade based on the L/W of alpha and beta conidia and elements of the conidiogenous layer. The taxa included in each analysis were those with available measurements for the micromorphological structure used to infer the respective dendrogram. The horizontal dashed lines indicate the putative distance cut-of level used to produce clusters. Euclidean distance values greater than 0.00 are shown at the nodes. Clusters are highlighted with coloured lines. Phylogenetic species are highlighted by different coloured typefaces and referred to in the chart legend. The cophenetic correlation coefficient (c) is noted below the chart legend.
Table 4.
Synopsis of the morphological data on asexual morphs of the species recognised in the Diaporthe clades treated in the present study, including a summary of host, country, and ecological group for the type or available specimens.
| Clade/Taxon1 | Conidiomata | Conidiogenous layer | Conidia | Origin2 | Reference |
|---|---|---|---|---|---|
| Diaporthe amygdali clade | |||||
| Diaporthe amygdali | |||||
|
Diaporthe amygdali (Delacr.) Udayanga, Crous & K.D. Hyde ≡Fusicoccum amygdali Delacr. CBS 126679ET |
Pycnidia subglobose to ampulliform, dark brown to black, exuding white to cream conidial droplets, 240–390 μm diam × 140–160 μm high (on host), 160–220 μm diam × 120–300 μm high (in culture) |
Conidiophores subcylindrical, hyaline, seldom branched, 7.4–36.3 × 1.5–3.2 μm (x̄= 14.5 × 2.3 μm, n = 380) Conidiogenous cells cylindrical, tapering towards apex, hyaline, periclinal thickening and collarette present, 5–20 × 1.5–3.2 μm, (x̄ = 9.8 × 2.3 μm, n = 380) |
Alpha conidia ovoid-ellipsoid, mostly with one end obtuse and the other acute, hyaline, aseptate, eguttulate to biguttulate, (4.18–)6.27–6.32(–9.64) × (1.63–)2.36–2.38(–3.31) μm, (x̄= 6.3 × 2.37 μm, n = 2100) Beta andgamma conidia not observed |
Host: on branches of Prunus amygdalus with symptoms of cankers or dieback Country: Portugal Ecological group: pathogen |
Diogo et al . (2010) |
|
Diaporthe chongqingensis Y.S. Guo & G.P. Wang. CGMCC 3.19603T |
Pycnidia globose, grey to black, exuding yellowish translucent conidial droplets from ostiole, 285–744 μm diam |
Conidiophores cylindrical, straight to sinuous, hyaline, septate, branched, 11–24.1 × 1.6–2.9 μm (x̄= 16 × 2.1 μm, n = 30) Conidiogenous cells cylindrical, straight or slightly curved, hyaline, rough, periclinal thickening present |
Alpha conidia fusiform, acutely rounded at one end, hyaline, aseptate, multiguttulate, 5.5–7.5 × 2–3 μm (x̄= 6.4 × 2.3 μm, n = 50; L/W = 2.8). Beta and gamma conidia not observed |
Host: on branches of Pyrus pyrifolia with symptoms of shoot canker Country: China (Chongqing) Ecological group: pathogen |
Guo et al . (2020) |
|
Diaporthe fusicola Y.H. Gao & L. Cai. HMAS 244837H |
Pycnidia subglobose to globose, with hairy necks, exuding yellowish translucent conidial droplets, aggregated in more or less cohesive clusters with variable number of pycnidia of variable morphology and dimensions, 175–500 μm diam |
Conidiophores ampulliform, hyaline, 1-septate, unbranched, densely aggregated, 6.5–12.5 × 2–6 μm Conidiogenous cells cylindrical, straight, terminal, tapering towards apex, hyaline, phialidic, 14–26 × 1.5–2.5 μm |
Alpha conidia fusoid, tapering towards ends, hyaline, aseptate, mostly biguttulate, 5.4–7.9 × 1.6–2.7 μm (x̄= 6.7 × 2 μm, n = 60) Beta and gamma conidia not observed |
Host: on diseased leaves of Lithocarpus glaber Country: China (Zhejiang) Ecological group: potential pathogen |
Gao et al . (2015) |
|
Diaporthe garethjonesii Dissanayake, Tangthirasunun & K.D. Hyde MFLU 13-0261H |
Pycnidia globose, unilocular, black, ostiolate, 85–125 μm diam× 80–100 μm high (x̄= 115 × 85 μm, n = 10) |
Conidiophores cylindrical, straight to sinuous, hyaline, smooth, 5–12 × 1–1.5 μm Conidiogenous cells cylindrical, terminal and lateral, slightly tapering towards apex, phialidic, 0.5–1 μm diam |
Alpha conidia fusoid to ellipsoid, straight, tapering towards ends, obtuse apex, hyaline, aseptate, guttulate, 5–6 × 2–3 μm Beta conidia curved or hamate, straight, hyaline, aseptate, smooth, less common than alpha conidia, 40–50 × 3–4 μm Gamma conidia not observed |
Host: on unknown dead leaves Country: Thailand Ecological group: saprophyte |
Hyde et al . (2016) |
|
Diaporthe kadsurae C.M. Tian & Q. Yang CFCC 52586T |
Pycnidia nearly flat, discoid, uniloculate and undivided, ectostromatic disc brown to dark, with one ostiole, 475–525 μm diam | Conidiophores cylindrical, straight or slightly curved, hyaline, unbranched, densely aggregated, tapering towards apex, 7–11 × 1.8–2.9 μm |
Alpha conidia oval or fusoid, tapering towards apex, hyaline, aseptate, biguttulate, 5.5–7.5 × 2.1–2.9 μm (x̄= 6.5 × 2.5 μm, n = 60) Beta and gamma conidia not observed |
Host: on symptomatic branches of Kadsura longipedunculata Country: China (Jiangxi) Ecological group: potential pathogen |
Yang et al . (2018) |
|
Diaporthe mediterranea M. León, Rodríguez-Reina & Armengol CBS 146754T |
Pycnidia globose or irregular, dark brown to black, exuding whitish translucent to creamy conidial droplets from ostiole (x̄ = 527 μm diam, n = 30) |
Conidiophores reduced to conidiogenous cells Conidiogenous cells cylindrical, straight, hyaline, smooth, densely aggregated (x̄ = 15.5 × 2.2 μm, n = 30) |
Alpha conidia fusiform, acute ends, hyaline, aseptate, multiguttulate (x̄= 6.6 × 2.4 μm, n = 30) Beta andgamma conidia not observed |
Host: on twig canker of Prunus amygdalus Country: Spain Ecological group: pathogen |
León et al . (2020) |
|
Diaporthe nannuoshanensis S.T. Huang, J.W. Xia, X.G. Zhang & Zhuang Li* SAUCC 194.91T |
Pycnidia globose to subglobose, black, brown to dark brown, coated with white and greyish hyphae, exuding cream conidial droplets from central ostiole, 275–530 μm diam × 205–435 μm high |
Conidiophores cylindrical, straight to sinuous, hyaline, smooth, eguttulate to biguttulate, densely aggregated, branched, 7.5–14 × 1.5–2.5 μm (x̄= 10.5 × 2.1 μm, n = 20) Conidiogenous cells cylindrical, ampulliform, straight or sinuous, terminal and lateral, tapering towards apex, phialidic, neck up to 4.0 μm long, 1–1.5 μm wide |
Beta conidia filiform, straight, bent or hamate, acutely rounded apex, truncate base, slightly narrowing towards apex, hyaline, aseptate, smooth, 20.5–24 × 1.5–2 μm (x̄= 22.3 × 1.7 μm, n = 50) Alpha andgamma conidia not observed |
Host: on leaf spots of Camellia sinensis Country: China (Yunnan) Ecological group: potential pathogen |
Huang et al . (2021a) |
|
Diaporthe ovoidea Y.H. Gao & L. Cai HMAS 244835H |
Pycnidia globose, exuding cream conidial droplets from ostiole, 140–250 μm diam |
Conidiophores reduced to conidiogenous cells Conidiogenous cells cylindrical, straight or slightly curved, tapering towards apex, single to multi-septate, rough, guttulate, phialidic, (12.3–)14.2–23.6(–26.5) × 1.6–2.3 μm (x̄= 18.7 × 1.9 μm, n = 30) |
Alpha conidia ovoid to fusoid, mostly with one end obtuse and the other acute, hyaline, aseptate, eguttulate to biguttulate, 5.3–8.3 × 1.7–3 μm (x̄= 6.7 × 2.3 μm, n = 60) Beta conidia filiform, curved, with rounded ends, hyaline, aseptate, eguttulate, 16.1–25 × (1–)1.2–1.5(–1.8) μm (x̄= 20.7 × 1.4 μm, n = 30) Gamma conidia not observed |
Host: on leaf spots of Lithocarpus glaber Country: China (Zhejiang) Ecological group: potential pathogen |
Gao et al . (2015) |
| “Diaporthe sinensis ” Jayaward.,Manawas., X.H. Li, J.Y. Yan & K.D. Hyde JZBH 3340167H |
Pycnidia globose, dark brown to black, 360–900 μm (x̄= 500 μm, n = 20) diam |
Conidiophores reduced to conidiogenous cells Conidiogenous cells simple, terminal, hyaline, smooth |
Alpha conidia oval, obtuse ends, hyaline, aseptate, 4–7 × 2–3 μm (x̄= 5 × 3 μm, n = 40) Beta and gamma conidia not observed |
Host: on dead leaves of Camellia sinensis Country: China (Fujian) Ecological group: saprophyte or potential pathogen |
Manawasinghe et al . (2021) |
|
Phomopsis ternstroemiae Y.H. Gao, W. Sun & L. Cai HMAS 244234T |
Pycnidia globose, white to dark brown, exuding yellowish translucent conidial droplets from ostiole, 180–390 μm diam | Conidiophores cylindrical, hyaline, branched, single to multi-septate, rough, 13.9–18.2 × 1.8–2.4 μm |
Alpha conidia fusiform to ellipsoidal, tapering towards ends or obtuse at one end, hyaline, aseptate, biguttulate, very often with several additional smaller guttules, 6.2–10.7 × 2.0–3.4 μm (x̄= 7.51 × 2.61 μm, n = 60) Beta andgamma conidia not observed |
Host: leaf spots of Ternstroemia gymnanthera Country: China (Zhejiang) Ecological group: potential pathogen |
Gao et al . (2014) |
| Diaporthe brasiliensis clade | |||||
| Diaporthe brasiliensis | |||||
|
Diaporthe brasiliensis R.R. Gomes, Glienke & Crous CBS 133183T |
Pycnidia globose to conical, brown to black, ostiolate, with globose conidial masses, white to pale-luteous, 70–160 μm diam × 60–140 μm high, with necks of 60–130 μm high. |
Conidiophores cylindrical, filiform, straight to curved, hyaline, 1–3-septate, (17–)20–27(–30) × 2(–4) μm Conidiogenous cells cylindrical, filiform, straight to curved, with flared collarette and slight periclinal thickening, (7–)8–12(–14) × 2(–3) μm |
Alpha conidia ellipsoid to irregular, bluntly rounded apex, obtuse to subtruncate base, hyaline, aseptate, bi- to multiguttulate, 6–7(–8) × 2–3 μm Beta andgamma conidia not observed |
Host: on a leaf of Aspidosperma tomentosum Country: Brazil (Rio de Janeiro) Ecological group: endophyte |
Gomes et al . (2013) |
| Diaporthe caatingaensis | |||||
|
Diaporthe caatingaensis J.D.P. Bezerra, L.M. Paiva, G.A. Silva, C.M. Souza-Motta & Crous CBS 141542T |
Pycnidia globose to subglobose-conical, dark brown to black, with hyaline to pale conidial masses at neck apex, up to 465 μm diam, with long black necks up to 510 μm tall |
Conidiophores cylindrical, straight to sinuous, hyaline, 3–5-septate, smooth, densely aggregated, sometimes branched, 30–37.5 × 2(–2.5) μm Conidiogenous cells cylindrical, terminal, with distinct collarette at apex and slight periclinal thickening, phialidic, 16–23.5 × 1–2(–2.5) μm |
Alpha conidia fusoid to ellipsoid, tapering towards ends, subobtuse to obtuse apex, subtruncate to truncate base, hyaline, aseptate, smooth, guttulate, (6.5–)8.5–9.5(–10.5) × (1.5–)2(–2.5) μm Beta andgamma conidia not observed |
Host: on Tacinga inamoena Country: Brazil (Pernambuco) Ecological group: endophyte |
Crous et al . (2006a) |
| Diaporthe pygmaeae sp. nov. | |||||
|
Diaporthe pygmaeae D.S. Pereira & A.J.L. Phillips CDP 1370T |
Pycnidia globose to subglobose, black, exuding white to pale conidial droplets, up to 210 μm diam |
Conidiophores mostly reduced to conidiogeneous cells, doliiform to subcylindrical, hyaline to subhyaline, aseptate to 1–3-septate, smooth, densely aggregated, unbranched Conidiogenous cells terminal, cylindrical, tapering towards apex, straight or slightly curved, hyaline, aseptate, rarely 1-septate, smooth, unbranched, rarely 1-branched, occasionally with minute, inconspicuous collarette, with periclinal thickenings,8.0–19.0(–31.6) × 1.9–3.9(–4.8) μm |
Alpha conidia fusoid to ellipsoid, acute ends, hyaline, aseptate, smooth, eguttulate, often with granular contents (7.8–)8.0–10.0(–10.7) × 2.4–3.5(–4.0) μm (x̄= 9.0 × 2.9 μm, n = 150) Beta andgamma conidia not observed |
Host: on foliar lesions of Phoenix roebelenii Country: Portugal Ecological group: potential pathogen |
Present study |
| Diaporthe eucommiae clade | |||||
| Diaporthe eucommiae | |||||
|
Diaporthe araliae-chinensis S.Y. Wang, Yong Wang bis & Y. Li* GUCC 412.7T |
Pycnidia globose or subglobose, deep green to black, up to 1 mm diam |
Conidiophores reduced to conidiogeneous cells Conidiogenous cells cylindrical, subulate, mostly straight, slightly tapering towards apex, hyaline, simple, smooth, densely aggregated, with inconspicuous periclinal thickening, 12–20 × 1.5–2.5 μm (x̄= 16.5 × 2 μm, n = 20) |
Alpha conidia fusoid to ellipsoidal, asymmetrical, mostly straight, tapering towards ends, hyaline, aseptate, smooth, 5.5–9.5 × 2–3 μm (x̄= 7.5 × 2.4 μm, n = 30) Beta andgamma conidia not observed |
Host: on leaves of Aralia chinensis Country: China (Guizhou) Ecological group: UN |
Hyde et al . (2023) |
|
Diaporthe australiana R.G. Shivas, Akinsanmi & Y.P. Tan* BRIP 66145T |
Pycnidia globose or irregular, dark brown to black, exuding whitish to pale yellow conidial droplets from central ostioles, up to 1 mm diam |
Conidiophores with an irregularly polygonal basal cell, hyaline, smooth, densely aggregated, 15 × 25 μm Paraphyses intermingled among conidiophores, tapering towards apex (1–2 μm wide), hyaline, 1–3-septate, smooth, up to 70 μm long Conidiogenous cells cylindrical, straight or flexuous, hyaline, phialidic, 10–20 × 1–2.5 μm |
Alpha conidia fusiform, acute ends, hyaline, aseptate, guttulate, 5–8.5 × 1.5–2 μm Beta andgamma conidia not observed |
Host: on husk rot of Macadamia sp. Country: Australia (New South Wales) Ecological group: pathogen |
Wrona et al . (2020) |
|
Diaporthe eucalyptorum Crous & R.G. Shivas* CBS 132525T |
Pycnidia subglobose, black, exuding white to cream conidial droplets from central ostioles, up to 350 μm |
Conidiophores r educed to conidiogeneous cells or straight to sinuous, hyaline, up to 4-septate, unbranched or branched below, smooth, densely aggregated, 15–60 × 3–4 μm Paraphyses c ylindrical, flexuous, hyaline, 1–3-septate, unbranched or branched below, smooth, up to 70 μm long, 2–3 μm wide at base Conidiogenous cells cylindrical, slightly tapering towards apex (1–1.5 μm diam), terminal and lateral, phialidic, with visible periclinal thickening and flared collarette (up to 2 μm long), surrounded by a prominent flaring mucoid sheath, 10–30 × 2–3 μm |
Alpha conidia fusoid, tapering towards ends, straight, subobtuse apex, subtruncate base, hyaline, aseptate, smooth, guttulate, (5.5–)6.5–7(–8) × (2–)2.5(–3) μm Beta and gamma conidia not observed |
Host: on leaf spots of Eucalyptus sp. Country: Australia (Queensland) Ecological group: potential pathogen |
Crous et al . (2012) |
|
Diaporthe eucommiae (F.X. Chao & P.K. Chi) Y.H. Gao & L. Cai* ≡ Phomopsis eucommiae F.X. Chao & P.K. Chi SCHM 0020H |
Pycnidia triangular to oblate, dark brown, ostiolate, uniloculate, 60–110 μm diam |
Conidiophores hyaline, branched Conidiogenous cells hyaline, phialidic |
Alpha conidia fusiform to ellipsoid, hyaline, aseptate, mono- to biguttulate, 3.9–4.7 × 1–1.7 μm Beta conidia filiform, straight to curved, hyaline, aseptate,6.6–14.9 × 0.33–0.66 μm Gamma conidia not observed |
Host: on leaf spots of Eucommia ulmoides Country: China (Guangdong) Ecological group: potential pathogen |
Cao & Chi (1990); Gao et al . (2017) |
|
Diaporthe fujianensis Jayaward., Manawas., X.H. Li, J.Y. Yan & K.D. Hyde* JZB 320149T |
Pycnidia globose, black | Conidiogenous cells terminal, hyaline, smooth |
Alpha conidia oval or ellipsoidal, obtuse ends, hyaline, aseptate, 4–6 × 2–3 μm (x̄= 5 × 2.5 μm, n = 30) Beta andgamma conidia not observed |
Host: on dead shoots of Camellia sinensis Country: China (Fujian) Ecological group: saprophyte or potential pathogen |
Manawasinghe et al . (2021) |
|
Diaporthe fusiformis Jayaward., Manawas., X.H. Li, J.Y. Yan & K.D. Hyde* JZB 320154T |
Pycnidia globose, black |
Conidiophores reduced to conidiogenous cells Conidiogenous cells hyaline, smooth, clustered |
Alpha conidia fusiform, angular ends, hyaline, aseptate, eguttulate, 8–5 × 2–3 μm (x̄= 7 × 2 μm, n = 40) Beta conidia filiform, hamate, tapering towards ends, hyaline, aseptate, 23–32 × 1.2–1.6 μm (x̄= 27 × 1.5 μm, n = 40) Gamma conidia not observed |
Host: on dead leaves of Camellia sinensis Country: China (Fujian) Ecological group: saprophyte or potential pathogen |
Manawasinghe et al . (2021) |
|
Diaporthe globoostiolata Monkai & S. Lumyong* MFLUCC 23-0025T |
Pycnidia subconical to subglobose, dark brown to black, with black ostiolar necks, uniloculate, 90–120 × 110–180 μm |
Conidiophores reduced to conidiogeneous cells Conidiogenous cells subcylindrical to ampulliform, slightly tapering towards apex, hyaline, terminal, monophialidic, with periclinal thickening and a prominent collarette, 3.5–11.4 × 1.4–3.7 μm (x̄= 6.5 × 2.2 μm, n = 30) |
Alpha conidia fusiform to ellipsoid, obtuse ends, hyaline, aseptate, smooth, mono- to biguttulate, 6–9.6 × 1.8–2.8 μm (x̄= 7.6 × 2.2 μm, n = 30) Beta conidia filiform, tapering towards apex, straight to slightly curved, truncate base, hyaline, smooth, eguttulate, 13.2–22 × 1–1.8 μm (x̄= 16.8 × 1.4 μm, n = 30) Gamma conidia not observed |
Host: on dead leaves of a member of Fagaceae Country: Thailand Ecological group: saprophyte |
Monkai et al . (2023) |
|
Diaporthe hongkongensis R.R. Gomes, Glienke & Crous* (CBS 115448T) |
Pycnidia g lobose, with central ostiole, exuding a creamy conidial cirrhus, up to 200 μm diam |
Conidiophores r educed to conidiogeneous cells Paraphyses hyaline, up to 4-septate, frequently branched below, smooth, with a clavate terminal cell, up to 80 μm long and apex of 2–8 μ diam Conidiogenous cells ampulliform to subcylindrical, with prominent apical tapper, hyaline, smooth, with periclinal thickening and minute collarette (1 μm long), phialidic, 5–12 × 2–4 μm |
Alpha conidia fusiform, tapering towards ends, mostly straight, acutely rounded apex, truncate base, hyaline, aseptate, smooth, granular to guttulate, (5–)6–7(–8) × (2–)2.5(–3) μm Beta conidia spindle-shaped, mostly curved in upper part, acutely rounded apex, truncate base, widest in mid region, hyaline, smooth, 18–22 × 1.5–2 μm Gamma conidia ellipsoid-fusoid, subobtuse apex, truncate base, hyaline, aseptate, smooth, 10–13 × 2 μm |
Host: on fruit of Dichroa febrifuga Country: Hong Kong Ecological group: UN |
Gomes et al . (2013) |
|
Diaporthe lithocarpi (Y.H. Gao, W. Sun & L. Cai) Y.H. Gao & L. Cai ≡ Phomopsis lithocarpi Y.H. Gao, W. Sun & L. Cai CGMCC 3.15175T |
Pycnidia globose to subglobose, dark brown to black, exuding cream conidial droplets from ostioles, 120–270 μm diam | Conidiophores cylindrical, frequently curved, tapering towards apex, hyaline, septate, rough, 12–15.4 × 1.9–2.6 μm |
Alpha conidia fusiform, tapering towards ends, hyaline, aseptate, biguttulate, 5.7–8.1 × 2.1–3.2 μm (x̄= 7 × 2.6 μm, n = 30) Beta conidia filiform, curved or hamate, obtuse ends, hyaline, aseptate, eguttulate, 17.6–28.1 × 0.92–1.81 μm (x̄= 23.6 × 1.4 μm, n = 30) Gamma conidia not observed |
Host: on leaf spots of Lithocarpus glaber Country: China (Zhejiang) Ecological group: potential pathogen |
Gao et al . (2014); Tan & Shivas (2023) |
|
Diaporthe lagerstroemiae (C.Q. Chang, M.M. Xiang & P.K. Chi) Y.H. Gao & L. Cai* ≡ Phomopsis lagerstroemiae C.Q. Chang, M.M. Xiang & P.K. Chi SCHM 3608H |
Pycnidia mostly triangular or ampullate, rarely compressed-triangular, eustromatic, unilocular, olivaceous brown to dark brown, wall becoming darker and thicker towards ostiole, 110–150 × 65–125 μm |
Conidiophores filiform, hyaline, septate, simple or branched, 11–35 × 1.4–2.1 μm Conidiogenous cells hyaline, phialidic, enteroblastic |
Alpha conidia fusiform, acute ends, hyaline, aseptate, biguttulate, 6–8.1 × 1.5–1.9 μm Beta conidia filiform, curved or hamate, rarely abnormal, clavate, straight or somewhat curved, hyaline, aseptate, 12–15 × 0.9–1.2 μm Gamma conidia not observed |
Host: on branches of Lagerstroemia indica Country: China (Hunan) Ecological group: UN |
Chang et al . (2005a); Gao et al . (2017) |
|
Diaporthe rhodomyrti C.M. Tian & Qin Yang* CFCC 53101T |
Pycnidia globose or rostrate, black, often exuding translucent conidial droplets from ostioles, 500–850 μm diam |
Conidiophores reduced to conidiogenous cells Conidiogenous cells cylindrical, straight, tapering towards apex, hyaline, septate, unbranched, (14.5–)15.5–23(–25.5) × 1.5–2 μm (n = 30; L/W = 8.5–13) |
Alpha conidia ellipsoidal, hyaline, aseptate, biguttulate, 6–7(–8.5) × 2–2.5(–3) μm (n = 30; L/W = 2.8–3.3) Beta conidia filiform, straight to sinuous, hyaline, aseptate, eguttulate, (15–)16.5–21.5(–23) × 1–1.5 μm (n = 30; L/W = 15.5–16.5) Gamma conidia not observed |
Host: On leaves of Rhodomyrtus tomentosa Country: China (Jiangxi) Ecological group: potential pathogen |
Cao et al . (2022) |
|
Diaporthe theobromatis H. Dong, J.W. Xia & X.G. Zhang* SAUCC 102221T |
Pycnidia globose, dark brown to black, covered with short hyphae, exuding whitish translucent to yellowish conidial droplets from ostioles |
Conidiophores cylindrical, hyaline, septate, branched, 14–21 × 1–2 μm Conidiogenous cells cylindrical, straight, tapering towards apex, hyaline, terminal, phialidic, 4.5–12 × 1–2 μm |
Alpha conidia ellipsoid or falcate, acute ends, hyaline, aseptate, mono- to biguttulate, 5–12 × 1.5–2.5 μm Beta conidia filiform, hamate or curved, hyaline, smooth, 15–25 × 1–2 μm Gamma conidia not observed |
Host: on leaf spot of Theobroma cacao Country: China (Yunnan) Ecological group: potential pathogen |
Dong et al . (2020) |
| Diaporthe aseana | |||||
|
Diaporthe aseana Dissan., Tangthir. & K.D. Hyde MFLUCC 12-0299aT |
Pycnidia globose, black, unilocular, ostiolate, 140–200 × 220–300 μm (x̄= 185 × 260 μm, n = 10) |
Conidiophores cylindrical, straight to sinuous, hyaline, smooth, densely aggregated, 8–15 × 2–3 μm Conidiogenous cells cylindrical, slightly tapering towards apex, terminal and lateral, phialidic, 6–10 × 2–3 μm |
Alpha conidia fusoid to ellipsoidal, subobtuse apex, tapering towards ends, straight, hyaline, aseptate, smooth, guttulate, 6–9 × 2–3 μm Beta and gamma conidia not observed |
Host: on an unknown dead leaf Country: Thailand Ecological group: saprophyte |
Hyde et al . (2016) |
|
Diaporthe tectonigena Doilom, Dissan. & K.D. Hyde* MFLUCC 12-0767T |
Pycnidia subglobose or variable, black, with an elongated black neck, multilocular, exuding hyaline to yellowish, reddish brown to umber, white to cream conidial droplets from central ostioles, (170–)270–310(–360) × (165–)280–295(–320) μm (x̄= 265 × 268 μm, n = 25) |
Conidiophores cylindrical, straight or slightly curved, rounded to obtuse apex, wider at base, hyaline, septate, branched or unbranched, smooth, in dense clusters, 9–32 × 1.8–3.7 μm Conidiogenous cells cylindrical, slightly tapering towards apex, terminal, phialidic, 2–5 × 1.3–3.9 μm |
Alpha conidia oblong to ellipsoid, bluntly rounded apex, obtuse to subtruncate base, hyaline, aseptate, smooth, multiguttulate, (5–)7.3–7.8(–8.3) × 2.5–3.5 μm (x̄= 7.2 × 2.9 μm, n = 30) Beta conidia filiform, curved or hamate, subtruncate base, hyaline, aseptate, (16–)29–30(–34) × 1.4–2 μm (x̄= 28 × 1.7 μm, n = 30) Gamma conidia ellipsoid-fusoid, acutely rounded apex, subtruncate base, hyaline, aseptate, smooth, multiguttulate, (9–)10–12(–14) × 1.7–3.2 μm (x̄= 11 × 2.3 μm, n = 30) |
Host: on a twig dieback of Tectona grandis Country: Thailand Ecological group: potential pathogen |
Doilom et al . (2016) |
| Diaporthe xishuangbanica | |||||
|
Diaporthe xishuangbanica Y.H. Gao & L. Cai CGMCC 3.18283T |
Pycnidia globose, 180–310 μm diam | Conidiophores cylindrical, straight, sometimes sinuous or lateral, branched, phialidic, 13–34.5 × 1.5–3 μm (x̄= 20.9 × 2.1 μm, n = 40) |
Alpha conidia fusiform, hyaline, aseptate, multiguttulate, 7–9.5 × 2.5–3.5 μm (x̄= 8.3 × 2.8 μm, n = 30) Beta and gamma conidia not observed |
Host: on diseased leaves of Camellia sinensis Country: China (Yunnan) Ecological group: potential pathogen |
Gao et al . (2017) |
| Diaporthe foeniculina clade | |||||
| Diaporthe cytosporella | |||||
| “Diaporthe chamaeropis ” (Cooke) R.R. Gomes, Glienke & Crous CBS 454.81 |
Pycnidia globose, black, exuding creamy conidial droplets from central ostioles, up to 400–600 μm diam |
Conidiophores cylindrical, straight to sinuous, hyaline, 1–5-septate, smooth, densely aggregated, branched, 10–50 × 2–2.5 μm Conidiogenous cells cylindrical, terminal and lateral, slightly tapering towards apex (1–1.5 μm diam), with visible periclinal thickening, phialidic, 10–20 × 1.5–2 μm |
Alpha conidia fusoid to ellipsoid, tapering towards ends, straight, subobtuse apex, subtruncate base, hyaline, aseptate, smooth, guttulate, (5–)6–8(–9) × 2(–2.5) μm Beta conidia spindle-shaped, curved, acutely rounded apex, truncate base, tapering from lower third towards apex, hyaline, aseptate, smooth, (20–)22–27(–30) × 1.5(–2) μm Gamma conidia not observed |
Host: on a dead part of leaf ofChamaerops humilis Country: Greece Ecological group: saprophyte |
Gomes et al . (2013) |
|
Diaporthe cytosporella (Penz. & Sacc.) Udayanga & Castl. ≡ Phoma cytosporella Penz. & Sacc. CBS 137020ET |
Pycnidia globose, with an elongated black neck, often exuding a yellowish conidial cirrus from ostiole, up to 450 μm diam, 65–100 μm high |
Conidiophores cylindrical, ampulliform, wider at base, hyaline, smooth, branched or unbranched, occurring in dense clusters, 7–18 × 1–2 μm Conidiogenous cells cylindrical, terminal, slightly tapering towards apex (0.5–1 μm), phialidic |
Alpha conidia ovate to ellipsoidal, subtruncate base, hyaline, aseptate, smooth, bi- to multiguttulate, (6.9–)8–9(–12.6) × (2.3–)2.6–3.2 μm (x̄= 8.8 × 3 μm, n = 30) Beta andgamma conidia not observed |
Host: on Citrus limon Country: Spain Ecological group: UN |
Udayanga et al . (2014b) |
|
Diaporthe pimpinellae Abeywickrama, Camporesi, Dissan. & K.D. Hyde* MFLU 19-0563H |
Pycnidia as black dots (on host), 150–250 μm diam × 150–200 μm high |
Conidiophores cylindrical, straight to sinuous, terminal, hyaline, aseptate, densely aggregated, slightly tapering towards apex Conidiogenous cells cylindrical, terminal and lateral, phialidic, 2–4 × 4–8 μm |
Alpha conidia fusiform or oval, hyaline, aseptate, single to biguttulate, 6–8 × 2–3 μm Beta andgamma conidia not observed |
Host: on aerial stems of Pimpinella peregrina Country: Thailand Ecological group: saprophyte |
Yuan et al . (2020) |
| “Diaporthe cinerascens ” | |||||
| “Diaporthe cinerascens ” Sacc. CBS 719.96 |
Pycnidia globose, black, exuding creamy-luteous conidial droplets from central ostioles, up to 300 μm diam |
Conidiophores cylindrical, straight to sinuous, hyaline, 1–3-septate, smooth, densely aggregated, branched, 17–30 × 2–3 μm Conidiogenous cells cylindrical, terminal and lateral, slightly tapering towards apex (1.5–2 μm diam), collarette mostly absent and slightly flared when present, with visible periclinal thickening, phialidic, 8–18 × 2–3 μm |
Alpha conidia fusoid to ellipsoid, tapering towards ends, subobtuse apex, subtruncate base, hyaline, aseptate, smooth, guttulate, 7–8(–9) × (2.5–)3 μm Gamma conidia ellipsoid to fusoid, acutely rounded apex, subtruncate base, hyaline, aseptate, smooth, 8–12 × 3 μm Beta conidia not observed |
Host: on branch of Ficus carica Country: Bulgaria Ecological group: UN |
Gomes et al . (2013) |
| Diaporthe diospyricola | |||||
|
Diaporthe diospyricola Crous CBS 136552T |
Pycnidia globose, black, exuding creamy conidial droplets from central ostioles, up to 400 μm diam |
Conidiophores c ylindrical, straight to sinuous, hyaline, 2–4-septate, smooth, densely aggregated, branched, 20–50 × 2.5–4 μm Conidiogenous cells cylindrical, terminal and lateral, slightly tapering towards apex (1–1.5 μm diam), with visible periclinal thickening, phialidic, 7–15 × 1.5–2.5 μm |
Alpha conidia fusoid to ellipsoid, tapering towards ends, straight, subobtuse apex, subtruncate base, hyaline, aseptate, smooth, guttulate, (5.5–)6–7(–7.5) × (2–)2.5(–3) μm Beta conidia spindle-shaped, curved, acutely rounded apex, truncate base, tapering from lower third towards apex, hyaline, aseptate, smooth, (18–)25–27(–30) × 1.5(–2) μm Gamma conidia not observed |
Host: on leaves of Diospyros whyteana Country: South Africa Ecological group: UN |
Crous et al . (2013) |
| Diaporthe foeniculina | |||||
|
Diaporthe foeniculina (Sacc.) Udayanga & Castl. ≡ Phoma foeniculina Sacc. CBS 111553ET |
Pycnidia globose to subglobose, with an elongated, black neck, often exuding a yellowish drop- like conidial cirrus from ostiole, 400–700 μm diam × (300–)500–800(–930) μm high |
Conidiophores cylindrical, straight to sinuous, hyaline, smooth, unbranched, 9–15(–18) × 1–2 μm Conidiogenous cells cylindrical, terminal, slightly tapering towards apex (0.5–1 μm diam) |
Alpha conidia ellipsoidal or fusiform, rarely with subtruncate base, hyaline, aseptate, smooth, eguttulate, bi- or multiguttulate, (7.5–)8.5–9(–9.2) × (2–)2.3–2.5(–2.7) μm (x̄= 8.8 × 2.4 μm, n = 30) Beta conidia hamate or slightly curved, acute apex, subtruncate base, hyaline, aseptate, eguttulate, (20–)22–28(–29) × (1.1–)1.4–1.6(–2) μm (x̄= 25.1 × 1.5 μm, n = 30) Gamma conidia not observed |
Host: on dead stems of Foeniculum vulgare Country: Portugal Ecological group: saprophyte |
Udayanga et al . (2014b) |
|
Diaporthe baccae L. Lombard, G. Polizzi & Crous CBS 136972T |
Pycnidia globose to conical, brown to black, eustromatic, multilocular, occasionally with ostiolate necks, surface covered with hyphae, exuding cream to pale conidial droplets from central ostioles, up to 650 μm diam |
Conidiophores cylindrical, straight to sinuous, hyaline, 1–3-septate, smooth, densely aggregated, branched, 20–57 × 2–3 μm Conidiogenous cells cylindrical, terminal and lateral, slightly tapering towards apex, with visible periclinal thickening, phialidic, 9–23 × 1–2 μm |
Alpha conidia fusoid to ellipsoid, tapering towards ends, straight, subobtuse apex, subtruncate base, hyaline, aseptate, smooth, guttulate, (6–)7–9 × 2–3 μm (x̄= 8 × 2 μm) Beta conidia spindle-shaped, acutely rounded apex, truncate base, tapering from lower third towards apex, hyaline, aseptate, smooth, (17–)20–24(–26) × 1–2 μm (x̄= 22 × 2 μm) Gamma conidia not observed |
Host: on Vaccinium corymbosum Country: Italy Ecological group: UN |
Lombard et al . (2014) |
|
Diaporthe neotheicola A.J.L. Phillips & J.M. Santos CBS 123208T |
Pycnidia of variable morphology and dimensions, globose to subglobose, cone-shaped, naked or covered with dense layer of hyphae, exuding translucent yellow conidial droplets, 420–730 μm diam × 280–1130 μm high |
Conidiophores cylindrical, hyaline, smooth, unbranched, single- to multi-septate, intermingled with long, septate and highly branched paraphyses, 6.7–23.5 × 1.6–3.1 μm Conidiogenous cells cylindrical to filiform, tapering towards apex, with periclinal thickening, phialidic, of two types, terminal (10.6–19.1 × 1.4–2.4 μm) and lateral (branched-like aspect of conidiophore) (2.6–9.8 × 1–1.6 μm) |
Alpha conidia fusoid, obtuse ends, hyaline, aseptate, mostly biguttulate, with basal conidiogenous scar, (6.6–)7.6–8(–9.5) × (1.9–)2.2–2.3(–2.6) μm (x̄= 7.8 × 2.2 μm, n = 50) Beta conidia filiform, curved, with rounded ends, hyaline, aseptate, eguttulate, (21.5–)25.7–26.7(–30.3) × (0.8–)1(–1.1) μm (x̄= 26.2 × 1 μm, n = 50) Gamma conidia not observed |
Host: on Foeniculum vulgare Country: Portugal Ecological group: UN |
Santos & Phillips (2009) |
|
Diaporthe ravennica Thambug., Camporesi & K.D. Hyde MFLU 16–0665H |
Stromata multiloculate, with subglobose or irregular locules of 140–275 μm diam × 65–175 μm high (x̄= 190 × 108 μm, n = 5), ostiolate or inostiolate, up to 500 μm diam |
Conidiophores cylindrical, straight to sinuous, hyaline, smooth, unbranched, 12–23 × 1.2–1.8 μm (x̄= 16.4 × 1.5 μm, n = 40) Conidiogenous cells cylindrical, terminal, slightly tapering towards apex, phialidic |
Alpha conidia ellipsoidal or fusiform, hyaline, aseptate, smooth, eguttulate, bi- or multiguttulate, (6.4–)7.2–10.5 × 1.7–2.8 μm (x̄= 8.6 × 2.3 μm, n = 40) Beta conidia filiform, slightly curved, acute apex, subtruncate base, hyaline, aseptate, eguttulate, 21–32(–38) × (1–)1.2–1.7 μm (x̄= 26.4 × 1.3 μm, n = 30) Gamma conidia not observed |
Host: on dead branches of Tamarix sp. Country: Italy Ecological group: saprophyte |
Thambugala et al . (2016) |
|
Diaporthe rhoicola Crous CBS 129528T |
Pycnidia flattened, multilocular, up to 600 μm diam |
Conidiophores subcylindrical, hyaline, 1–3-septate, smooth, unbranched or branched (below or above), 20–40 × 2–3 μm Conidiogenous cells subcylindrical, terminal, tapering towards a truncate apex (1–1.5 μm), with a flaring collarette (up to 5 μm wide and long), smooth, 15–25 × 2–3 μm Paraphyses intermingled among conidiophores, subcylindrical, hyaline, smooth, branched or unbranched, up to 80 μm long, 2–3 μm wide |
Alpha conidia subcylindrical to fusoid-ellipsoidal, obtuse apex, widest in middle, tapering to a truncate base (1 μm diam), hyaline, aseptate, smooth, guttulate, (7–)8–9(–10) × 3(–3.5) μm Beta and gamma conidia not observed | Host: on leaf spots of Rhus pendulina Country: South Africa Ecological group: potential pathogen |
Crous et al . (2011a) |
|
Diaporthe nigra Brahmanage & K.D. Hyde* JZBH 320170H |
Pycnidia globose or irregular, multiloculate, black |
Conidiophores reduced to conidiogeneous cells or rarely short, compressed cylindrical, straight to sinuous, hyaline, smooth, branched, densely aggregated, 8–25 × 1–3 μm (x̄= 20 × 1.5 μm) Conidiogenous cells subcylindrical to ampulliform, phialidic, with flared collarette and visible periclinal thickening, 190–295 × 120–175 μm (x̄= 270 × 160 μm) |
Alpha conidia ovate to ellipsoidal, obtuse to subtruncate base, straight, hyaline, aseptate, smooth, biguttulate, 17–28 × 7–7.5 μm (x̄= 25 × 7.2 μm) Beta andgamma conidia not observed |
Host: on dead aerial stem of Ballota nigra Country: Italy Ecological group: saprophyte |
Hyde et al . (2020a) |
|
Diaporthe rumicicola Manawasinghe, Camporesi & K.D. Hyde* MFLU 18-0739H |
Pycnidia ampulliform, ostiolate, with elongate black neck, 98–280 μm (x̄= 208 μm, n = 10) diam (on host) |
Conidiophores reduced to conidiogeneous cells on host, with paraphyses observed on culture Conidiogenous cells integrated, clustered, hyaline, smooth, enteroblastic with percurrent annelations |
Alpha conidia hyaline, smooth, mono- or bi- guttulate, 3–5 × 2–3 μm (x̄= 3.5 × 2.5 μm, n = 30) Beta and gamma conidia not observed |
Host: on dead aerial stem of Rumex sp. Country: Italy Ecological group: saprophyte |
Hyde et al . (2019) |
|
Diaporthe zaobaisu Y.S. Guo & G.P. Wang* CGMCC 3.19598T |
Pycnidia globose or irregular, dark brown to black, 235–445 μm diam |
Conidiophores cylindrical, straight, hyaline, 1-septate, densely aggregated, 6–13 × 2.5–4 μm Conidiogenous cells ampulliform, tapering towards apex, hyaline, terminal, phialidic, 8.5–12 × 2.5–3 μm |
Alpha conidia fusiform, hyaline, aseptate, biguttulate, 5.5–8.5 × 2–3 μm (x̄= 6.4 × 2.3 μm, n = 50; L/W = 2.8) Beta conidia filiform, curved, tapering towards ends, hyaline, aseptate, 21.5–28 × 1–1.4 μm (x̄= 24.5 × 1.1 μm, n = 41; L/W = 22.3) Gamma conidia not observed |
Host: on branches with shoot cankers symptoms of Pyrus bretschneideri Country: China (Yunnan) Ecological group: pathogen |
Guo et al . (2020) |
| Diaporthe forlicesenica | |||||
|
Diaporthe forlicesenica Bundhun, Camporesi & K.D. Hyde ≡ Diaporthe dorycnii Dissan., Camporesi & K.D. Hyde MFLUCC 17-1015T |
Pycnidia globose, brown to black, up to 420 μm diam × 380 μm high |
Conidiophores cylindrical, slightly tapering towards apex, straight to sinuous, hyaline, terminal, densely aggregated, 21–35 × 1.5–2.5 μm Conidiogenous cells subcylindrical, filiform, straight, tapering towards apex, 13–19 × 2–3 μm |
Alpha conidia fusiform or oval, obtuse ends, hyaline, biguttulate, 9–13.5 × 3–4 μm (x̄= 11 × 4 μm) Beta andgamma conidia not observed |
Host: on dead aerial stem of Dorycnium hirsutum Country: Italy Ecological group: saprophyte |
Bundhun et al . (2021); Dissanayake et al . (2017) |
| Diaporthe glabrae clade | |||||
| Diaporthe glabrae | |||||
|
Diaporthe alangii C.M. Tian & Qin Yang* CFCC 52556T |
Pycnidia discoid (ectostromatic disc), uniloculate and undivided, black, with one ostiole, 135–330 μm diam | Conidiophores cylindrical, straight, hyaline, unbranched, phialidic, 6–12 × 1.4–2 μm |
Alpha conidia ellipsoidal, mostly with one end obtuse and the other acute, hyaline, aseptate, occasionally submedian constriction, biguttulate, 6.5–8 × 2 μm (x̄= 7 × 2 μm, n = 30) Beta andgamma conidia not observed |
Host: on symptomatic branches of Alangium kurzii Country: China (Zhejiang) Ecological group: potential pathogen |
Yang et al . (2018) |
|
Diaporthe conferta H. Dong, J.W. Xia & X.G. Zhang* SAUCC 194.9T |
Pycnidia subglobose or irregular, black, covered with short hyphae, exuding yellowish translucent conidial droplets from ostioles |
Conidiophores cylindrical, straight to slightly curved, tapering towards apex, wider at base, hyaline, septate, branched or unbranched, smooth, in dense clusters, 5–25 × 2–3 μm Conidiogenous cells cylindrical, straight, tapering towards apex, terminal, phialidic, 5–15 × 2–3 μm |
Alpha conidia ellipsoidal or oval, bluntly rounded apex, obtuse base, hyaline, aseptate, smooth, biguttulate, 5–7 × 2–3 μm Beta andgamma conidia not observed |
Host: on leaf spots of Elaeagnus conferta Country: China (Yunnan) Ecological group: potential pathogen |
Dong et al . (2021) |
|
Diaporthe glabrae (C.Q. Chang, Z.D. Jiang & P.K. Chi) Y.H. Gao & L. Cai ≡ Phomopsis glabrae C.Q. Chang, Z.D. Jiang & P.K. Chi SCHM 3622H |
Pycnidia globose or triangular, eustromatic, unilocular, olivaceous to brown, 75–140 × 53–95 μm |
Conidiophores hyaline, septate, simple or branched, 7.5–20 × 1.6–2.5 μm Conidiogenous cells hyaline, phialidic, enteroblastic |
Alpha conidia oblong-ellipsoidal, obtuse ends, hyaline, aseptate, biguttulate, 5–7.4 × 1.6–2.1 μm Beta conidia filiform, mostly hamate, hyaline, aseptate, 13.5–22.5 × 0.6–1 μm Gamma conidia not observed |
Host: on branches of Bougainvillea glabra Country: China (Fujian) Ecological group: UN |
Chang et al . (2005b); Gao et al . (2017) |
|
Diaporthe hubeiensis Dissanayake, X.H. Li & K.D. Hyde* JZB 320123T |
Pycnidia subglobose, ostiolate, with up to 100 μm black cylindrical necks, up to 510 μm diam | Conidiophores reduced to conidiogeneous cells |
Alpha conidia ellipsoidal to cylindrical, blunt ends, hyaline, aseptate, smooth, biguttulate, 5.6–7.1 × 1–3.1 μm (x̄= 6.1 × 1.8 μm, n = 40) Beta conidia filiform, tapering towards ends, 17–27 × 1–1.5 μm (x̄= 24 × 1.5 μm, n = 40) Gamma conidia not observed |
Host: on diseased trunk of Vitis vinifera Country: China (Hubei) Ecological group: pathogen |
Manawasinghe et al . (2019) |
|
Diaporthe morindae M. Luo, W. Guo, M. P. Zhao, Manawas., K. D. Hyde & C. P. You* ZHKUCC 22-0072T |
Pycnidia oblate, subglobose, flask or irregularly shaped, uni or multilocular, 50–380 × 30–160 μm (x̄= 170 × 90 μm) | Conidiogenous cells hyaline, phialidic |
Alpha conidia ellipsoidal or torque circular, blunt ends, hyaline, aseptate, mono- or biguttulate, 6–7 × 2–4 μm (x̄= 6 × 3 μm) Beta andgamma conidia not observed |
Host: on healthy stems and roots of Morinda officinalis Country: China (Guangdong) Ecological group: endophyte |
Luo et al . (2022) |
|
Diaporthe tectonae Doilom, A.J. Dissanayake & K.D. Hyde* MFLUCC 12-0777T |
Pycnidia subglobose or irregular, black, with an elongated black neck, exuding hyaline to yellowish, white to cream conidial droplets from central ostioles, (460–)725–820(–1385) × (500–)900–1035(–2075) μm (x̄ = 773 × 918 μm, n = 25) |
Conidiophores cylindrical, straight or slightly curved, tapering towards apex, wider at base, hyaline, septate, branched or unbranched, smooth, in dense clusters, 11–18 × 1–2 μm Conidiogenous cells cylindrical, slightly tapering towards apex (0.5–1 μm), terminal, phialidic, 1.5–5.2 × 0.9–1.7 μm |
Alpha conidia oblong to ellipsoidal, bluntly rounded apex, obtuse to subtruncate base, hyaline, aseptate, smooth, mono- to mostly biguttulate, (4–)5.5–6(–6.8) × 2–2.9 μm (x̄= 5.5 × 2.6 μm, n = 30) Beta conidia rare, cylindrical, flexuous, hamate, rounded apex, subtruncate base, hyaline, aseptate, smooth, 10–13.5 × 1.3–1.7 μm (x̄= 12 × 1.5 μm, n = 10) Gamma conidia ellipsoid-fusoid, acutely rounded apex, subtruncate base, hyaline, aseptate, smooth, (7–)8–9(–10.6) × 1.3–2.1 μm (x̄= 8.5 × 1.8 μm, n = 30) |
Host: on dieback lesions of twigs of Tectona grandis Country: Thailand Ecological group: potential pathogen |
Doilom et al . (2016) |
|
Diaporthe xishuangbannaensis Hongsanan & K.D. Hyde* ≡ Diaporthe chinensis H. Dong, J.W. Xia & X.G. Zhang SAUCC 194.30T |
Pycnidia subglobose or irregular, black, covered with short hyphae, exuding whitish translucent to yellowish conidial droplets from ostioles |
Conidiophores cylindrical, straight to slightly curved, tapering towards apex, wider at base, hyaline, septate, branched or unbranched, smooth, in dense clusters, 4–20 × 1–3 μm Conidiogenous cells cylindrical, straight, tapering towards apex, terminal, phialidic, 5–7 × 2–3.5 μm |
Alpha conidia ellipsoidal or oval, bluntly rounded apex, obtuse base, hyaline, aseptate, smooth, biguttulate, 5–7 × 2–3.5 μm Beta conidia rare, filiform, hamate or curved, hyaline, smooth, 17–25 × 1–1.5 μm Gamma conidia rare, obovate to clavate, hyaline, 2–4-guttulate 7–10 × 1.5–2 μm |
Host: on leaf spots of Litchi chinensis Country: China (Yunnan) Ecological group: potential pathogen |
Dong et al . (2021); Hongsanan et al . (2023) |
| Diaporthe celtidis | |||||
|
Diaporthe celtidis Tennakoon, C.H. Kuo & K.D. Hyde MFLUCC 20-0180T |
Pycnidia globose to subglobose, dark brown to black, thin-walled, ostiolate, exuding light brown to pale yellow conidial droplets from ostioles, up to 500 μm diam |
Conidiophores sometimes reduced to conidiogenous cells, hyaline, 1-septate Conidiogenous cells subcylindrical, tapering towards apex, determinate, unbranched, smooth, phialidic, enteroblastic, 6–10 × 1.2–2.6 μm (x̄= 7.5 × 1.7 μm, n = 20) |
Alpha conidia oval to cylindrical, straight to slightly curved, rounded apex, hyaline, aseptate, smooth, biguttulate, 5–6 × 2–3 μm (x̄= 5.3 × 2.5 μm, n = 40) Beta andgamma conidia not observed |
Host: on dead leaves of Celtis formosana Country: Taiwan Ecological group: saprophyte |
Tennakoon et al . (2021) |
| Diaporthe tulliensis | |||||
|
Diaporthe tulliensis R.G. Shivas, Vawdrey & Y.P. Tan BRIP 62248aT |
Pycnidia subglobose, ostiolate, beaks absent or up to 1 mm long, exuding cream to pale yellow conidial droplets from ostioles, up to 500 μm diam |
Conidiophores reduced to conidiogeneous cells Conidiogenous cells cylindrical, tapering towards apex, hyaline, 15–20 × 1.5–2.5 μm |
Alpha conidia oval to cylindrical, rounded apex, obconically truncate base, hyaline, aseptate, (4.5–)5–7 × 2–2.5(–3) μm Beta conidia scarce, flexuous, hamate, hyaline, aseptate, 25–30 × 1(–1.5) μm Gamma conidia not observed |
Host: on rotted stem end of fruit of Theobroma cacao Country: Australia (Queensland) Ecological group: UN |
Crous et al . (2015) |
| Diaporthe inconspicua clade | |||||
| Diaporthe inconspicua | |||||
|
Diaporthe inconspicua R.R. Gomes, Glienke & Crous* URM 7775 |
Pycnidia globose, dark brown to black, ostiolate, exuding yellowish conidial droplets from ostioles, 424–954 × 371–742 μm |
Conidiophores cylindrical, straight to sinuous, hyaline, 0–2-septate, sometimes branched, in dense clusters, 11–21.5 × 2–2.5 μm Conidiogenous cells cylindrical, terminal, phialidic, (2–)5.5–8.5(–14.5) × (1.5–)2–2.5 μm |
Alpha conidia ellipsoidal to fusoid, acute ends, hyaline, aseptate, smooth, eguttulate, 5.5–6.5 × 1.5–2 μm Beta conidia filiform, straight to curved, tapering towards apex, truncate base, hyaline, aseptate, eguttulate, (17.5–)20–26(–28) × 1–1.5 μm Gamma conidia not observed |
Host: on Poincianella pyramidalis Country: Brazil Ecological group: endophyte |
Gomes et al . (2013); Bezerra et al . (2018) |
|
Diaporthe lutescens S.T. Huang, J.W. Xia, X.G. Zhang & Z. Li* SAUCC 194.36T |
Pycnidia subglobose, black, exuding white creamy conidial droplets from central ostioles |
Conidiophores subcylindrical, straight or slightly curved, obtuse apex, wider at base, hyaline, septate, unbranched, smooth, 10.2–17 × 1.8–3 μm Conidiogenous cells cylindrical, straight to sinuous, tapering towards apex, terminal, phialidic, 5.7–9.1 × 1.4–2.6 μm |
Beta conidia filiform, straight or slightly curved, subtruncate base, enlarged towards apex, hyaline, aseptate, 20.8–28.8 × 1.2–2 μm (x̄= 25.3 × 1.4 μm, n = 20) Alpha andgamma conidia not observed |
Host: on leaf spots of Chrysalidocarpus lutescens Country: China (Yunnan) Ecological group: potential pathogen |
Sun et al . (2021) |
|
Diaporthe pseudoinconspicua T.G.L. Oliveira, J.D.P. Bezerra, A.R. Machado, Souza-Motta & O.M.C. Magalh.* URM 7874T |
Pycnidia globose to subglobose, dark brown to black, exuding yellowish conidial droplets from ostioles, 200–320 × 160–190 μm |
Alpha conidiophores straight to sinuous, hyaline, branched, aggregated, 14.5–21.5(–23.5) × 2.5–3 μm Beta conidiophores straight to sinuous, hyaline, septate, branched, smooth, aggregated, 10.5–16(–18) × 2–2.5(–3) μm Conidiogenous cells straight to sinuous, bifurcate, hyaline, phialidic, (9–)10.5–13.5 × 2–2.5(–3) μm |
Alpha conidia fusoid, one end rounded and the other acute, hyaline, aseptate, bi- to multiguttulate, 5–7.5(–8.5) × 2–2.5(–3.5) μm Beta conidia filiform, straight to curved, one end obtuse and the other truncate, hyaline, aseptate, 18–21(–25.5) × 1–1.5(–2) μm Gamma conidia not observed |
Host: on branches of Poincianella pyramidalis Country: Brazil Ecological group: endophyte |
Crous et al . (2018) |
| “Diaporthe pterocarpi ” (S. Hughes) Udayanga, Xing Z. Liu & K.D. Hyde MFLUCC 10-0572ET |
Pycnidia globose, 100–120 μm diam, later conical, with a slightly elongated black neck, exuding dirty white, spiral conidial cirri from ostioles, up to 100 μm diam, 65–100 μm high |
Conidiophores ampulliform, straight to sinuous, hyaline, unbranched, smooth, 10–15 × 1–2 μm Conidiogenous cells cylindrical, slightly tapering towards apex (0.5–1 μm), terminal, phialidic |
Alpha conidia fusiform, subtruncate base, hyaline, aseptate, biguttulate, rarely 3-guttulate, smooth, (5–)6–7(–9) × (2–)2.5(–3) μm Beta andgamma conidia not observed |
Host: on leaf spot of Pterocarpus indicus Country: Thailand Ecological group: potential pathogen |
Udayanga et al . (2012b) |
|
Diaporthe samaneae Monkai & S. Lumyong* SDBR-CMU470T |
Pycnidia subglobose to ovoid, elongate, dark brown to brown, ostiolate, multiloculate, 300–480 × 290–740 μm |
Conidiophores subcylindrical, straight to sinuous, hyaline to pale brown, septate, branched, smooth, tightly aggregated, 7.5–31.7 × 1.5–2.7 μm (x̄= 19 × 2 μm, n = 30) Conidiogenous cells subcylindrical to ampulliform, tapering towards apex, hyaline, terminal, with periclinal thickening and a prominent collarette, 5.2–14.3 × 1.5–2.7 μm (x̄= 9.7 × 2 μm, n = 30) |
Alpha conidia ellipsoid to elongate fusiform, obtuse apex, subtruncate base, hyaline, aseptate, smooth, eguttulate, forming basipetal chains of two or more conidia on phialidic neck, 7–11 × 1.8–2.8 μm (x̄= 8.4 × 2.4 μm, n = 30) Beta andgamma conidia not observed |
Host: on dead wood of Samanea saman Country: Thailand Ecological group: saprophyte |
Monkai et al . (2023) |
|
Diaporthe inconspicua R.R. Gomes, Glienke & Crous* URM 7775 |
Pycnidia globose, dark brown to black, ostiolate, exuding yellowish conidial droplets from ostioles, 424–954 × 371–742 μm |
Conidiophores cylindrical, straight to sinuous, hyaline, 0–2-septate, sometimes branched, in dense clusters, 11–21.5 × 2–2.5 μm Conidiogenous cells cylindrical, terminal, phialidic, (2–)5.5–8.5(–14.5) × (1.5–)2–2.5 μm |
Alpha conidia ellipsoidal to fusoid, acute ends, hyaline, aseptate, smooth, eguttulate, 5.5–6.5 × 1.5–2 μm Beta conidia filiform, straight to curved, tapering towards apex, truncate base, hyaline, aseptate, eguttulate, (17.5–)20–26(–28) × 1–1.5 μm Gamma conidia not observed |
Host: on Poincianella pyramidalis Country: Brazil Ecological group: endophyte |
Gomes et al . (2013); Bezerra et al . (2018) |
| Diaporthe isoberliniae | |||||
|
Diaporthe isoberliniae Crous CBS 137981T |
Pycnidia g lobose, black, exuding creamy conidial droplets from central ostioles, up to 300 μm diam |
Conidiophores c ylindrical, straight to sinuous, hyaline, 2–3-septate, branched, smooth, densely aggregated, 15–40 × 3–4 μm Conidiogenous cells cylindrical, slightly tapering towards apex (1 μm diam), terminal and lateral, with periclinal thickening and flared collarette (up to 4 μm long), phialidic, 10–14 × 2.5–3 μm |
Alpha conidia fusoid-ellipsoid, tapering towards ends, subobtuse apex, subtruncate base, hyaline, aseptate, smooth, guttulate, (6.5–)8–9(–10) × (2.5–)3(–3.5) μm Beta and gamma conidia not observed |
Host: on Isoberlinia angolensis Country: Zambia Ecological group: UN |
Crous et al . (2014b) |
|
Diaporthe isoberliniae Crous STMA 18245, STMA 18291 |
Pycnidia g lobose or irregular, dark brown to black, exuding white to cream or yellow conidial droplets from ostioles, 200–460 μm diam | Conidiophores cylindrical to subcylindrical, subhyaline base to pale olivaceous, hyaline apex, 1–3-septate, smooth, densely aggregated, 13–42 × 1.5–4 μm Conidiogenous cells cylindrical to subcylindrical, tapering towards apex, terminal or lateral, hyaline, phialidic, (5.5–)6.5–14 × 1.5–3 μm |
Alpha conidia ellipsoid to obovoid, or fusoid-ellipsoid, rounded or subobtuse apex, acutate or subtruncate base, hyaline, aseptate, bi- to multiguttulate, 5.5–9(–10) × 2–3(–3.5) μm Beta conidia less frequent, filiform, curved, tapering towards apex, hyaline, aseptate, eguttulate, 11.5–27.5 × 1–2 μm Gamma conidia less frequent, broadly fusiform, straight to slightly curved, rarely sinuose, acutate or filiform apex, filiform base, hyaline, aseptate, multiguttulate, 10–18.5(–21) × 1.5–2.5 μm |
Host: on Pittosporum manii Country: Cameroon Ecological group: endophyte |
Lambert et al . (2023) |
|
Diaporthe pungensis S.T. Huang, J.W. Xia, X.G. Zhang & Z. Li* SAUCC 194.112T |
Pycnidia globose or subglobose, dark brown to black, thin-walled, exuding white creamy conidial droplets from ostioles |
Conidiophores cylindrical, straight to sinuous, hyaline, aseptate, unbranched, smooth, 11–14.5 × 1.5–2.3 μm Conidiogenous cells cylindrical, terminal, phialidic, 8–9.5 × 1–2.5 μm |
Alpha conidia ellipsoidal to fusoid, subobtuse apex, subtruncate base, hyaline, aseptate, smooth, 2–3-guttulate, 6–8.5 × 2–3.3 μm (x̄= 6.6 × 2.5 μm, n = 20) Beta conidia filiform, slightly curved, tapering towards apex, truncate base, hyaline, aseptate, eguttulate, 24–28.9 × 1–2 μm (x̄= 26.9 × 1.4 μm, n = 20) Gamma conidia not observed |
Host: on leaf spots of Elaeagnus pungens Country: China (Yunnan) Ecological group: potential pathogen |
Sun et al . (2021) |
| Diaporthe leucospermi clade | |||||
| Diaporthe acaciarum | |||||
|
Diaporthe acaciarum Crous & M.J. Wingf. CBS 138862T |
Pycnidia globose, black, exuding creamy conidial droplets from central ostioles, up to 300 μm diam |
Conidiophores cylindrical, straight to sinuous, hyaline, 2–3-septate, branched, smooth, densely aggregated, 20–30 × 2.5–4 μm Conidiogenous cells cylindrical, with a slight apical taper (1–1.5 μm diam), terminal and intercalary, with periclinal thickening and collarette (up to 2 μm long), phialidic, 15–25 × 2–3 μm |
Alpha conidia fusoid-ellipsoid, tapering towards ends, straight, subobtuse apex, subtruncate base, hyaline, aseptate, smooth, biguttulate, (6–)6.5–7(–7.5) × (2–)2.5(–3) μm Beta conidia spindle-shaped, acutely rounded apex, truncate base, tapering from lower third towards base, hyaline, aseptate, smooth, (20–)25–35(–40) × 1.5(–2) μm Gamma conidia not observed |
Host: on thorns of Acacia tortilis Country: Tanzania Ecological group: UN |
Crous et al . (2014a) |
| Diaporthe beilharziae | |||||
|
Diaporthe beilharziae R.G. Shivas, Jacq. Edwards & Y.P. Tan BRIP 54792IT |
Pycnidia subglobose, beaks absent or less than 300 μm, exuding pale yellow to salmon conidial droplets from ostioles, up to 250 μm diam |
Conidiophores reduced to conidiogenous cells or ampulliform to cylindrical, hyaline to pale yellowish brown, 1-septate, 5–15 × 1.5–3.5 μm Conidiogenous cells cylindrical, flexuous, tapering towards apex, hyaline, 5–20 × 1.5–3 μm |
Alpha conidia oval to cylindrical, rounded apex, obconically truncate base, hyaline, aseptate, mostly biguttulate, (5.5–)6.5–9(–10) × 2–2.5(–3) μm Beta conidia scarce, flexuous, hyaline, 15–25 × 1–1.5 μm Gamma conidia not observed |
Host: on leaf spot of Indigofera australis Country: Australia (New South Wales) Ecological group: potential pathogen |
Tan et al . (2013) |
| Diaporthe leucospermi | |||||
|
Diaporthe leucospermi Crous & Summerell CBS 111980T |
Pycnidia dark brown, exuding a creamy conidial cirrus from central ostiole, up to 300 μm diam |
Conidiophores reduced to conidiogeneous cells or subcylindrical, hyaline, pale brown at base, 1–3-septate, smooth, 15–30 × 2–3 μm Conidiogenous cells subcylindrical, with an apical taper, terminal or lateral, with periclinal thickening and flaring collarette (1 μm long), phialidic, 10–15 × 2–3 μm |
Alpha conidia ellipsoid, tapering to acutely rounded apex and obtuse to truncate base, hyaline, aseptate, smooth, biguttulate, flattened hilum (1 μm diam), (6–)7(–8) × (2.5–)3 μm
Beta conidia spindle-shaped, prominently hooked in apical part, acute apex, truncate base, hyaline, aseptate, smooth, (20–)25–30(–35) × (1–)1.5 μm Gamma conidia not observed |
Host: on leaves of Leucospermum sp. Country: Australia (New South Wales) Ecological group: UN |
Crous et al . (2011b) |
|
Diaporthe pyracanthae L. Santos & A. Alves CBS 142384T |
Pycnidia dark brown, exuding a creamy to white conidial cirrus from central ostiole |
Conidiophores reduced to conidiogeneous cells or subcylindrical, hyaline, smooth Conidiogenous cells subcylindrical, with an apical taper, hyaline, smooth, phialidic |
Alpha conidia fusiform to ellipsoid, rounded apex, obtuse to truncate base, hyaline, aseptate, smooth, frequently biguttulate, 6.7–6.8 × 2.2–2.4 μm (interpreted from means, n = 100) Beta conidia filiform, frequently hooked in apical part, acute apex, truncate base, hyaline, aseptate, smooth, 26.8–30 × 1.3 μm (interpreted from means, n = 100) Gamma conidia infrequent, fusoid, acutely rounded apex, subtruncate base hyaline, aseptate, smooth |
Host: on branch canker of Pyracantha coccinea Country: Portugal Ecological group: pathogen |
Santos et al . (2017b) |
|
Diaporthe rossmaniae S. Hilário, I. Amaral, L. Santos & A. Alves MUM 19.30T |
Pycnidia brown to black, exuding a creamy white to yellow conidial cirrus |
Conidiophores reduced to conidiogeneous cells Conidiogenous cells subcylindrical, hyaline to gold, smooth, x̄= 7.4 × 1.9 μm (n = 30) |
Alpha conidia ellipsoid, rounded apex, obtuse to truncate base, hyaline, aseptate, smooth, mono to biguttulate, x̄= 6.8 × 2.5 μm (n = 100) Beta conidia filiform, frequently hooked in apical part, acute apex, truncate base, hyaline, aseptate, smooth, x̄= 25.7 × 1.2 μm (n = 100) Gamma conidia not observed |
Host: on twig lesions of Vaccinium corymbosum Country: Portugal Ecological group: pathogen |
Hilário et al . (2020) |
| Diaporthe micheliae | |||||
|
Diaporthe chimonanthi (C.Q. Chang, M.M. Xiang & P.K. Chi) Y.H. Gao & L. Cai* ≡ Phomopsis chimonanthi C.Q. Chang, M.M. Xiang & P.K. Chi SCHM 3614H |
Pycnidia triangular or tuberous, eustromatic, unilocular, 150–238 × 130–230 μm |
Conidiophores slim, hyaline, septate, branched, 13–25 × 1.6–2.5 μm Conidiogenous cells hyaline, phialidic, enteroblastic |
Alpha conidia fusiform, somewhat obtuse base, hyaline, aseptate, biguttulate, 6.6–8.8 × 1.8–2.2 μm Beta conidia mostly filiform, curved or hamate, rarely abnormal, clavate, straight or somewhat curved, hyaline, aseptate, 15–18 × 1–1.5 μm Gamma conidia not observed |
Host: on branches of Chimonanthus praecox Country: China (Hunan) Ecological group: UN |
Chang et al . (2005a); Gao et al . (2017) |
|
Diaporthe micheliae Y.H. Gao & L. Cai ≡Phomopsis micheliae C.Q. Chang, Z.D. Jiang & P.K. Chi SCHM 3603H |
Pycnidia tuberous or irregular, eustromatic, unilocular, wall becoming darker and thicker towards ostiole, 85–188 × 55–118 μm |
Conidiophores hyaline, septate, simple, rarely branched, 8.5–19.5 × 1.6–2.6 μm Conidiogenous cells hyaline, phialidic, enteroblastic |
Alpha conidia fusiform, acute ends, hyaline, aseptate, biguttulate, 6.5–8.2 × 1.6–2 μm Beta conidia filiform, straight or hamate, hyaline, aseptate, 9–23 × 0.5–0.8 μm Gamma conidia not observed |
Host: on branches of Michelia alba Country: China (Fujian) Ecological group: UN |
Chang et al . (2005b); Gao et al . (2017) |
|
Diaporthe middletonii R.G. Shivas, L. Morin, S.M. Thomps. & Y.P. Tan* BRIP 54884eT |
Pycnidia subglobose, multilocular, necks absent or up to 200 μm, exuding cream conidial droplets from a few ostioles, up to 300 μm diam |
Conidiophores reduced to conidiogenous cells or cylindrical, hyaline to pale yellowish brown, 1-septate, 10–25 × 1.5–3.5 μm Conidiogenous cells cylindrical, hyaline, 5–20 × 1.5–2.5 μm |
Alpha conidia fusiform to cylindrical, rounded apex, obconically truncate base, hyaline, aseptate, mostly biguttulate, (5–)6–7.5(–8) × 2–2.5(–3) μm Beta conidia scarce, mostly J-shaped, flexuous, hyaline, aseptate, 20–35 × 1–1.5 μm Gamma conidia not observed |
Host: on stem of Rapistrum rugosum Country: Australia (Queensland) Ecological group: UN |
Thompson et al . (2015) |
| Diaporthe myracrodruonis | |||||
|
Diaporthe myracrodruonis A.P.S.L. Pádua, T.G.L. Oliveira, Souza-Motta, X.L. Fan & J.D.P. Bezerra URM 7972T |
Pycnidia globose to subglobose, dark brown to black, thin-walled, exuding whitish to light cream conidial droplets from ostioles, (424)636–954(–975) × (286–)318–795(–901) μm |
Conidiophores straight to sinuous, moderately curved, tapering towards apex, thick at base, hyaline, occasionally branched, smooth, phialidic, (5–)6–9(–11) × 1.5–2(–2.5) μm Conidiogenous cells cylindrical, terminal, phialidic, (2–)5.5–8.5(–14.5) × (1.5–)2–2.5 μm |
Alpha conidia fusoid or elongated ellipsoid, attenuating towards ends, hyaline, aseptate, smooth 2.5–3.5(–4) × 1–1.5 μm Beta conidia sickle-shaped, curved, curved and slightly tapered apex, truncate base, hyaline, aseptate, smooth, 18–20(–24) × 0.7–1 μm Gamma conidia not observed |
Host: on leaves of Myracrodruon urundeuva Country: Brazil Ecological group: endophyte |
da Silva et al . (2019) |
| Diaporthe sackstonii | |||||
|
Diaporthe caryae C.M. Tian & Qin Yang* CFCC 52563T |
Pycnidia n early flat, discoid, uniloculate and undivided, ectostromatic disc brown to dark, with one ostiole, 310–325 μm diam | Conidiophores cylindrical, sometimes inflated, hyaline, unbranched, phialidic, 7–11 × 1.4–2.2 μm |
Alpha conidia ellipsoidal or fusiform, obtuse ends, hyaline, aseptate, eguttulate, 7–8.5 × 2.1–2.5 μm (x̄= 8 × 2.3 μm, n = 30) Beta conidia filiform, straight or hamate, tapering towards apex, subtruncate base, hyaline, aseptate, eguttulate, 15.5–34 × 1.1–1.4 μm (x̄= 27.5 × 1.2 μm, n = 30) Gamma conidia not observed |
Host: on symptomatic twigs of Carya illinoensis Country: China (Beijing) Ecological group: potential pathogen |
Yang et al . (2018) |
|
Diaporthe juglandigena S.Y. Wang, Yong Wang bis* GUCC 422.16T |
Pycnidia irregular globose to subglobose, exuding black conidial masses surrounded by white mycelium and drops of water, up to 2 mm diam |
Conidiophores reduced to conidiogenous cells Conidiogenous cells subulate, cylindrical, straight to sinuous, tapering towards apex, hyaline, simple, rarely branched above, smooth, slightly thicker, densely aggregated, 19–34 × 1–2.5 μm (x̄= 27 × 1.7 μm, n = 20) |
Alpha conidia fusoid to ellipsoidal, asymmetrical, tapering towards ends, mostly straight, hyaline, aseptate or 1-septate, smooth, frequently guttulate, 5–8 × 2–3 μm (x̄= 6.4 × 2.3 μm, n = 30) Beta conidia infrequent, filiform, mostly curved, acute apex, hyaline, aseptate, eguttulate, smooth, 23–36 × 1–2 μm (x̄= 31 × 1.3 μm, n = 10) Gamma conidia not observed |
Host: on peel of Juglans regia Country: China (Guizhou) Ecological group: potential pathogen |
Wang et al . (2022) |
|
Diaporthe machili S.T. Huang, J.W. Xia, W.X. Sun & X.G. Zhang* SAUCC 194.111T |
Pycnidia globose to subglobose, black, exuding cream conidial droplets from central ostioles | Conidiophores mostly ampulliform, cylindrical, straight or slightly curved, swelling at base, tapering towards apex, hyaline, septate, unbranched, guttulate, densely aggregated, 7–11.4 × 1.8–2.8 μm |
Beta conidia J-shaped, filiform, mostly curved, swelling in middle, tapering towards ends, hyaline, aseptate, 29–39 × 1.3–1.5 μm (x̄= 32.5 × 1.4 μm, n = 20) Alpha and gamma conidia not observed |
Host: on diseased leaves of Machilus pingii Country: China (Yunnan) Ecological group: potential pathogen |
Huang et al . (2021b) |
|
Diaporthe orixae Q.T. Lu & Z. Zhang* KUNCC 21–10714T |
Pycnidia globose to conical, brown to dark brown, ostiolate, with short necks (36–88 × 49–73 μm), smooth or covered by sparce hyaline hyphae, 98–509 × 55–360 μm diam |
Conidiophores reduced to conidiogeneous cells or subcylindrical to cylindrical, hyaline, rarely septate Conidiogenous cells subcylindrical to cylindrical, rarely tapering towards apex, hyaline, with collarette and prominent periclinal thickening, 2–4.2 × 1–2.5 μm |
Alpha conidia oblong to ellipsoid, bluntly rounded apex, obtuse base, hyaline, aseptate, multiguttulate, 3.5–5.5 × 1.5–3 μm Beta andgamma conidia not observed |
Host: on stems of Orixa japonica Country: China (Guizhou) Ecological group: endophyte |
Lu et al . (2022) |
|
Diaporthe sackstonii R.G. Shivas, S.M. Thomps. & Y.P. Tan BRIP 54669bT |
Pycnidia multilocular, with necks up to 0.5 mm, exuding cream conidial droplets from ostioles, up to 1 mm diam |
Conidiophores reduced to conidiogenous cells or filiform, hyaline to pale yellowish brown, septate, 15–40 × 1.5–3 μm Conidiogenous cells cylindrical to lageniform, tapering toward apex, hyaline, 10–15 × 1.5–3 μm |
Alpha conidia fusiform, rounded apex, obconically truncate base, hyaline, aseptate, 6–7(–8) × 2–2.5 μm Beta and gamma conidia not observed |
Host: on petiole of Helianthus annuus Country: Australia (Queensland) Ecological group: UN |
Thompson et al . (2015) |
| Diaporthe serafiniae | |||||
|
Diaporthe serafiniae R.G. Shivas, S.M. Thomps. & Y.P. Tan BRIP 55665bT |
Pycnidia multilocular, with necks up to 1.5 mm, exuding cream conidial droplets from most ostioles, up to 2 mm diam |
Conidiophores fusiform, hyaline to pale yellowish brown, 1-septate, 15–25 × 1.5–3.5 μm Conidiogenous cells cylindrical, flexuous, tapering towards apex, hyaline, 5–20 × 1.5–2.5 μm |
Alpha conidia fusiform, rounded apex, narrowed towards base, hyaline, aseptate, biguttulate, 5.5–7 (–8) × 1.5–2.5(–3) μm Beta andgamma conidia not observed |
Host: on seed of an ornamental variety of Helianthus annuus Country: Australia (Queensland) Ecological group: UN |
Thompson et al . (2015) |
| Diaporthe longicolla clade | |||||
| Diaporthe longicolla | |||||
|
Diaporthe longicolla (Hobbs) J.M. Santos, Vrandečić & A.J.L. Phillips ≡ Phomopsis longicolla Hobbs ATCC 60325T |
Pycnidia globose, black, stromatic, uni- or multilocular, with prominent necks more than 200 μm long opening by an apical ostiole, locules uniostiolate or multiostiolate, up to 500 μm diam |
Conidiophores hyaline, septate, simple or usually branched, 3.5–24 × 1–4 μm Conidiogenous cells filiform, hyaline, phialidic |
Alpha conidia ellipsoid to fusiform, hyaline, aseptate, guttulate, 5–9.5 × 1.5–3.5 μm Beta conidia rare, filiform, hamate, hyaline Gamma conidia not observed |
Host: seeds, pods, and stems of Glycine max Country: USA (Ohio) Ecological group: pathogen |
Hobbs (1985); Santos et al . (2011) |
|
Diaporthe megabiguttulata M. Luo, W. Guo, Manawas., M.P. Zhao, K.D. Hyde & C.P. You* ZHKUCC 22-0067T |
Pycnidia oblate or hemispherical, uni- or multilocular, 50–440 × 40–270 μm (x̄= 180 × 120 μm) | Conidiogenous cells hyaline, phialidic |
Alpha conidia ellipsoid or lanceolate, one end obtuse and the other acute, hyaline, aseptate, biguttulate, 5–10 × 2–3 μm (x̄= 6 × 2 μm) Beta conidia hamate or curved, hyaline, truncate base, 20–30 × 1–2 μm (x̄= 30 × 1 μm) Gamma conidia not observed |
Host: on stem of Morinda officinalis Country: China (Guangdong) Ecological group: endophyte |
Luo et al . (2022) |
|
Diaporthe unshiuensis F. Huang, K.D. Hyde & Hong Y. Li* CGMCC 3.17566T |
Pycnidia globose, subglobose or irregular, dark brown | Conidiophores cylindrical, slightly tapering towards apex, hyaline, smooth, 14.3–24.2 × 1.4–2.6 μm (x̄= 19.5 × 2 μm) |
Alpha conidia ellipsoidal or clavate, truncate base, hyaline, aseptate, smooth, biguttulate, 5.2–7.5 × 2–3.9 μm (x̄= 6.5 × 2.8 μm; L/W = 2.4) Beta and gamma conidia not observed |
Host: on melanose fruit of Citrus unshiu Country: China (Zhejiang) Ecological group: potential pathogen |
Huang et al . (2015) |
| Diaporthe morindendophytica | |||||
|
Diaporthe morindendophytica M. Luo, W. Guo, Manawas., M.P. Zhao, K.D. Hyde & C.P. You ZHKUCC 22-0069T |
Pycnidia oblate, subglobose or irregular, uni- or multilocular, exuding black conidial droplets from ostioles, 40–200 × 30–130 μm (x̄= 120 × 70 μm) | Conidiogenous cells hyaline, phialidic |
Alpha conidia ellipsoid, torque circular or lanceolate, blunts ends, or one end slightly pointed, hyaline, aseptate, biguttulate, 5–10 × 2–3 μm (x̄= 6 × 3 μm) Beta conidia linear, curved, hyaline, 20–30 × 1–2 μm (x̄= 20 × 2 μm) Gamma conidia not observed |
Host: on stem of Morinda officinalis Country: China (Guangdong) Ecological group: endophyte |
Luo et al . (2022) |
| Diaporthe tectonendophytica | |||||
|
Diaporthe tectonendophytica Doilom, Dissan. & K.D. Hyde MFLUCC 13-0471T |
Pycnidia subglobose or irregular, black, with an elongated black neck, exuding hyaline to yellowish, reddish brown to umber, white to cream conidial droplets from central ostioles, (250–)550–675(–1048) × (300–)770–1000(–1490) μm (x̄= 542 × 660 μm, n = 25) |
Conidiophores cylindrical, straight or slightly curved, rounded to obtuse at apex, wider at base, hyaline, septate, branched or unbranched, smooth, in dense clusters, 15–36 × 1.5–2.5 μm Conidiogenous cells cylindrical, slightly tapering towards apex (0.5–1 μm), terminal, phialidic, 4–8 × 1.5–2 μm |
Alpha conidia ellipsoid, bluntly rounded apex, obtuse to subtruncate base, hyaline, aseptate, smooth, mono- to mostly biguttulate, (3.8–)4.8–5.4(–6) × 1.7–2.7 μm (x̄= 5 × 2.2 μm, n = 30) Beta conidia filiform, curved or hamate, subtruncate base, hyaline, aseptate, (14–)24.5–25.5(–29.3) × 1–1.6 μm (x̄= 23 × 1.3 μm, n = 30) Gamma conidia not observed |
Host: on branches of Tectona grandis Country: Thailand Ecological group: endophyte |
Doilom et al . (2016) |
| Diaporthe ueckeri | |||||
|
Diaporthe durionigena L.D. Thao, L.T. Hien, N.V. Liem, H.M. Thanh & T.N. Khanh* VTCC 930005T |
Pycnidia globose to subglobose, black, with up to six well-defined necks per conidioma, 200–400 μm diam |
Conidiophores reduced to conidiogeneous cells Conidiogenous cells cylindrical, hyaline, 10–18 × 2.5–3.5 μm |
Alpha conidia rare or absent, ellipsoidal, hyaline, aseptate, smooth, biguttulate, (5.6–)6.1–7.5(–7.9) × (1.8–)2.1–2.7(–3) μm Beta conidia hamate, hyaline, aseptate, (17.8–)20.3–26.1(–31.2) × 1.1–1.5(–1.7) μm Gamma conidia not observed |
Host: on branches of Durio zibethinus Country: Vietnam Ecological group: UN |
Crous et al . (2020, 2021a) |
|
Diaporthe miriciae R.G. Shivas, S.M. Thomps. & Y.P. Tan BRIP 54736jT |
Pycnidia multilocular, with necks up to 1 mm, exuding pale yellow conidial droplets from some ostioles |
Conidiophores reduced to conidiogeneous cells or cylindrical to obclavate, hyaline to subhyaline, 1–2-septate, 10–20 × 1.5–3 μm Conidiogenous cells cylindrical to obclavate, tapering towards apex, hyaline, 5–12 × 1.5–3 μm |
Alpha conidia fusiform to oval, rounded apex, narrowed base, hyaline, aseptate, 6–7.5(–9) × 2–2.5(–3) μm Beta conidia hamate, flexuous, hyaline, aseptate, 20–35 × 1–1.5 μm Gamma conidia not observed |
Host: on stubble of Helianthus annuus Country: Australia (New South Wales) Ecological group: UN |
Thompson et al . (2015) |
|
Diaporthe passifloricola Crous & M.J. Wingf.* CBS 141329T |
Pycnidia globose, black, with short black necks, exuding creamy conidial droplets from central ostioles, up to 250 μm diam |
Conidiophores cylindrical, straight to sinuous, hyaline, 2–3-septate, branched, smooth, densely aggregated, 20–50 × 3–4 μm Conidiogenous cells cylindrical, tapering towards apex (1–1.5 μm), terminal and lateral, phialidic, with periclinal thickening, 7–20 × 1.5–2.5 μm |
Alpha conidia fusoid-ellipsoid, tapering towards end, subobtuse apex, subtruncate base, hyaline, aseptate, smooth, guttulate, (5–)6–7(–9) × 2.5(–3) μm Beta conidia spindle-shaped, tapering from lower third towards apex, curved, acutely rounded apex, truncate base, hyaline, aseptate, (20–)22–25(–27) × 1.5(–2) μm Gamma conidia not observed |
Host: on leaf spots of Passiflora foetida Country: Malaysia Ecological group: potential pathogen |
Crous et al . (2016b) |
|
Diaporthe rosae Samarak. & K.D. Hyde* MFLUCC 17-2658 |
Pycnidia globose to pyriform, reddish brown, coriaceous, with elongate necks, 95–160 μm (x̄ = 123.9 μm, n = 22) long, 35–75 μm (x̄= 54.9 μm, n = 10) high, 80–140 μm (x̄= 110.9 μm, n = 10) diam at base |
Conidiophores reduced to conidiogenous cells or straight, hyaline, septate, branched, smooth, 10–19 × 1.9–3.3 μm (x̄= 15 × 2.6 μm, n = 20) Alpha conidiogenous cells ampulliform, slightly tapering towards apex, phialidic, with periclinal thickening, 13 × 1–2.5 μm (x̄= 9.9 × 2 μm, n = 25) Beta conidiogenous cells ampulliform, slightly tapering towards apex, phialidic, with periclinal thickening and flared collarette, 7.7–15 × 1.2–2.3 μm (x̄= 12.5 × 1.9 μm, n = 10) |
Alpha conidia ovate to ellipsoidal, subtruncate base, hyaline, aseptate, smooth, often biguttulate, 5.5–7.5 × 2–3 μm (x̄= 6.5 × 2.5 μm, n = 30; L/W = 2.6) Beta conidia scarce, fusiform to hooked, subtruncate base, hyaline, aseptate, smooth, 12.5–18 × 1–2 μm (x̄= 14.6 × 1.4 μm, n = 10; L/W = 10.4) Gamma conidia not observed |
Host: on dead pedicel of Rosa sp. Country: Thailand Ecological group: saprophyte |
Wanasinghe et al . (2018) |
|
Diaporthe thunbergiicola Udayanga & K.D. Hyde* MFLUCC 12-0033T |
Pycnidia subglobose to ovate, with an elongated black neck 100–200 μm long, often exuding a yellowish conidial cirrus from ostiole, 100–200 μm diam |
Conidiophores ampulliform, cylindrical to subcylindrical, with larger basal cell, hyaline, unbranched or branched at the basal cell, smooth, 6–14 × 1–2 μm (x̄= 11 × 1.5 μm, n = 30) Conidiogenous cells cylindrical, slightly tapering towards apex (0.5–1 μm), terminal, phialidic |
Alpha conidia ovate to ellipsoidal, subtruncate base, hyaline, aseptate, smooth, biguttulate, 5.7–7.5 × 2–3 μm (x̄= 6.6 × 2.8 μm, n = 30) Beta and gamma conidia not observed |
Host: on diseased leaves of Thunbergia laurifolia Country: Thailand Ecological group: potential pathogen |
Liu et al . (2015) |
|
Diaporthe ueckeri Udayanga & Castl. CBS 139283T |
Pycnidia g lobose, with an elongated black neck 200–300 μm long, often exuding a yellowish conidial cirrus from ostiole, 150–200 μm diam |
Conidiophores ampulliform, long, slender, hyaline, unbranched, smooth, (9–)12–28(–30) × 1.5–2.5 μm Conidiogenous cells cylindrical, slightly tapering towards apex (0.5–1 μm), terminal, phialidic |
Alpha conidia ellipsoidal, subtruncate base, hyaline, aseptate, smooth, often biguttulate, (6–)6.4–8.3(–8.6) × (2–)2.3–3 μm (x̄= 7.2 × 2.6 μm, n = 30) Beta andgamma conidia not observed |
Host: on crown of Cucumis melo Country: USA (Oklahoma) Ecological group: potential pathogen |
Udayanga et al . (2015) |
|
Diaporthe vochysiae S.A. Noriler, R.R. Gomes & C. Glienke LGMF 1583T |
Pycnidia globose to conical, brown to black, with necks 90 μm long, outer surface smooth, ostiolate, with globose, predominantly yellow and yellow to reddish brown conidial mass, 328 × 367 μm |
Conidiophores subcylindrical to cylindrical, tapering towards apex, hyaline, 1–3-septate, rarely branched above the septa, 21–29 × 2.1–3.3 μm Conidiogenous cells subcylindrical, tapering towards apex, hyaline, with slight periclinal thickening and flared collarette, (13.5–)12.2–14.8(–15) × 3(–5) μm |
Alpha conidia oblong to ellipsoid, bluntly rounded apex, obtuse base, hyaline, aseptate, biguttulate, (2.8–)3–4(–4.2) × (1.5–)2 μm Beta andgamma conidia not observed |
Host: on leaf of Vochysia divergens Country: Brazil (Mato Grosso do Sul) Ecological group: endophyte |
Noriler et al . (2019) |
| Diaporthe rudis clade | |||||
| Diaporthe asheicola | |||||
|
Diaporthe asheicola L. Lombard & Crous MFLU 15-2966 |
Pycnidia globose, flask-like to conical, brown to black, eustromatic, unilocular, with ostiolate necks, outer surface smooth, exuding a globose white, pale-yellow to yellow conidial mass or cirrus, up to 530 μm diam and 290–420 μm high |
Alpha conidiophores cylindrical, filiform, aseptate, rarely branched, 8–35 × 2–3 μm (x̄= 27 × 2 μm, n = 20) (alpha ), 10–27 × 1–2 μm (x̄= 16 × 1.5 μm, n = 20) (beta ) Conidiogenous cells subcylindrical, tapering towards apex, with periclinal thickening and collarette, phialidic, 3–15 × 1–2 μm (x̄= 10 × 1.5 μm,n = 20) (alpha ), 7–14 × 1–2 μm (x̄= 11 × 1.5 μm, n = 20) (beta ) |
Alpha conidia fusoid to ellipsoidal, acutely rounded apex, obtuse to subtruncate base, multiguttulate, rarely biguttulate, 6–9 × 2–3 μm (x̄ = 8 × 2 μm, n = 20) Beta conidia straight, curved or hamate, 20–25 × 1–2 μm (x̄= 23 × 1 μm, n = 20) Gamma conidia not observed |
Host: on a stem of Fraxinus pennsylvanica Country: Russia Ecological group: saprophyte |
Lombard et al . (2014); Chethana et al . (2021b) |
| Diaporthe australafricana | |||||
|
Diaporthe australafricana Crous & van Niekerk CBS 111886T |
N/A # | N/A # |
Alpha conidia fusoid, obtuse ends, biguttulate to eguttulate, 5–5.5(–6) × 1.5–2 μm Beta and gamma conidia not observed |
Host: on Vitis vinifera Country: South Africa Ecological group: UN |
van Niekerk et al . (2005) |
| Diaporthe crousii | |||||
|
Diaporthe crousii S. Hilário, L. Santos & A. Alves MUM 19.29T |
Pycnidia ellipsoid, brown to black, exuding yellow conidial mass from o stiole |
Conidiophores reduced to conidiogeneous cells Conidiogenous cells cylindrical, straight, hyaline, aggregated, x̄= 21.9 × 1.5 μm (n = 30) |
Alpha conidia ellipsoid, rounded apex, obtuse to truncate base, hyaline, aseptate, smooth, biguttulate, x̄= 5.7 × 2.3 μm (n = 100) Gamma conidia infrequent, hyaline, aseptate, smooth, x̄= 10.6 × 1.5 μm (n = 8) Beta conidia not observed |
Host: on dieback lesions of Vaccinium corymbosum Country: Portugal Ecological group: pathogen |
Hilário et al . (2020) |
| Diaporthe cynaroidis | |||||
|
Diaporthe cynaroidis Marinc., M.J. Wingf. & Crous MFLU 14-0847 |
Pycnidia globose to pyriform, black, coriaceous, with an elongated neck, often exuding a yellowish white conidial cirrus from ostiole, 265–300 μm diam at base × 125–140 μm high (x̄= 280 × 135 μm, n = 10) |
Conidiophores ampulliform, straight to sinuous, hyaline, smooth, unbranched, 4–6 × 4.5–8 μm (x̄= 4.6 × 6.6 μm, n = 20) Conidiogenous cells cylindrical, slightly tapering towards apex, terminal, phialidic, 8–14 × 1.5–3 μm (x̄= 11.2 × 2.2 μm, n = 20) |
Alpha conidia ovate to ellipsoidal, subtruncate base, hyaline, aseptate, smooth, often biguttulate, 5.8–7.5 × 2.5–3.5 μm (x̄= 6.4 × 2.8 μm, n = 10) Beta andgamma conidia not observed |
Host: on a dead aerial stem of Eupatorium cannabinum Country: Italy Ecological group: saprophyte |
Marincowitz et al . (2008); Hyde et al . (2020a) |
|
Diaporthe salicicola R.G. Shivas, Jacq. Edwards & Y.P. Tan* VPRI 32789T |
Pycnidia with ostiolar beaks mostly up to 400 μm long |
Conidiophores cylindrical, straight, hyaline, 1–3-septate, unbranched, 10–25 × 2–3 μm Conidiogenous cells cylindrical, tapering towards apex, terminal, hyaline, phialidic, 10–20 × 2–3 μm |
Alpha conidia cylindrical to oval, rounded apex, slightly attenuated base, hyaline, aseptate, (4–)5–7(–8) × 1.5–2.5 μm Beta and gamma conidia not observed |
Host: on leaf spot of Salix purpurea Country: Australia (Tasmania) Ecological group: potential pathogen |
Tan et al . (2013) |
| Diaporthe patagonica | |||||
|
Diaporthe patagonica M. Zapata, M.A. Palma, M.J. Anninat & E. Piontelli CBS 145291T |
Pycnidia globose, flask-shaped, eustromatic, convoluted to unilocular, brown to black, with black cylindrical ostiolate necks up to 1 mm, outer surface smooth, exuding light yellow to cream conidial mass, up to 550 μm diam |
Conidiophores reduced to conidiogenous cells or cylindrical, straight, hyaline, smooth, densely aggregated Conidiogenous cells cylindrical, slightly tapering towards apex, terminal and lateral, phialidic, with inconspicuous periclinal thickening, 10.5–22 × 1–2.5 μm |
Alpha conidia fusoid to ellipsoid, tapering towards ends, straight, hyaline, aseptate, smooth, mostly biguttulate, (5.5–)6–7.5(–10) × (2–)2.5(–3) μm (x̄= 7 × 2.4 μm) Beta conidia spindle shaped, some straight, mostly curved towards one end, subacute apex, lightly truncate base, hyaline, aseptate, smooth, (12–)16–22(–28.5) × 1–1.5 μm (x̄= 18.8 × 1.3 μm) Gamma conidia scarcely observed |
Host: on branches of Aristotelia chilensis with dieback symptoms Country: Chile Ecological group: potential pathogen |
Zapata et al . (2020) |
| Diaporthe rudis | |||||
|
Diaporthe rudis (Fr.) Nitschke ≡ Sphaeria rudis Fr. CBS 109292ET |
Pycnidia globose, 200–250 μm diam, and 400–500 μm diam at maturity |
Conidiophores cylindrical, ampulliform, straight to sinuous, hyaline, smooth, branched, 20–45 × 2–2.4 μm Paraphyses abundant among conidiophores, 20–40 × 1–2 μm Conidiogenous cells cylindrical, terminal, slightly tapering towards apex (0.5–1 μm), phialidic |
Alpha conidia ovate to ellipsoidal, subtruncate base, hyaline, aseptate, smooth, biguttulate, (6.3–)7–8(–8.7) × 2–2.5 μm (x̄= 7.5 × 2.2 μm, n = 30) Beta conidia fusiform or hooked, subtruncate base, hyaline, aseptate, smooth, 27–31(–35.2) × (3–)3.4–3.8(–4.2) μm (x̄= 29.5 × 3.6 μm, n = 30) Gamma conidia fusiform, subtruncate base, hyaline, aseptate, smooth, mostly biguttulate, (10–)14–15 × 1–2 μm (x̄= 14.4 × 1.7 μm, n = 30) |
Host: on stem of Laburnum anagyroides Country: Austria Ecological group: UN |
Udayanga et al . (2014b) |
|
Diaporthe viticola Nitschke CBS 113201ET |
Pycnidia black, exuding white or pale-yellow conidial mass |
Conidiophores tapering towards apex, septate, branched, branches arising from immediately below the septa, 17–34 × 1.5–2.5 μm (in culture), 18–24 × 2–3 μm (on host, some unbranched) Conidiogenous cells hyaline, aperture apical, channel minute, with funnel-shaped collarette up to 6 μm long, integrated, enteroblastic, phialidic, 10–14 × 1.5–2.5 μm (in culture), 10–14 × 2 μm (on host) |
Alpha conidia oblong to elliptical, hyaline, aseptate, smooth, biguttulate, 5–7 × 2.5 μm (in culture), 7–9 × 2–3 μm (on host) Beta conidia straight or curved or hamate, hyaline, smooth, 16–28 × 1 μm Gamma conidia not observed |
Host: on Vitis vinifera Country: Portugal Ecological group: UN |
Phillips (1999); van Niekerk et al . (2005); Udayanga et al . (2014b) |
1 Taxa synonymised in the present study are noted with a superscript asterisk (*); status of the strains or specimens on which the morphological data is based is noted by superscript ET (ex-epitype), H (holotype) and T (ex-type).
2 UN = unknown, i.e. information not mentioned by the respective authors; the ecological group “potential pathogen” stands for species recovered from symptomatic tissues, but for which pathogenicity tests were not conducted to prove their pathogenicity.
#N/A = not available, i.e. feature not mentioned by the respective authors in the taxonomic description of the species.
Note: several species of the Diaporthe clades treated (including some synonymised in the present study) were excluded from this synopsis due to lack of morphological data regarding their asexual morphs. Some details about their collections are given below.
Diaporthe amygdali clade: D. sterilis Lombard, Polizzi & Crous was introduced by Lombard et al. (2014) as a pathogen on Vaccinium corymbosum in Italy, based on the diagnosis of sequence data, since no sporulation was observed for five different isolates (including the ex-type strain CBS 136969) on different media or on sterilised organic material placed on the media; D. sterilis was synonymised under D. amygdali by Hilário et al. (2021a).
Diaporthe eucommiae clade: D. tuyouyouiae Y.P. Tan, Bishop-Hurley & R.G. Shivas was introduced by Tan & Shivas (2023) from a leaf spot on Decalobanthus peltatus in Queensland (Australia), based on the diagnosis of sequence data obtained apparently from the type specimen (BRIP 75017a), and no taxonomic description was provided. D. xylocarpi M.S. Calabon & E.B.G. Jones (as D. salinicola Dayar.) was introduced by Dayarathne et al. (2020) as a saprophyte on decaying submerged wood of Xylocarpus sp. in Thailand, based on morpho-molecular analyses [the species name was validly published by Calabon et al. (2023)]; however, only the sexual morph was observed on the host tissue, and no sporulation was observed in culture; both species are synonymised here under D. eucommiae.
Diaporthe foeniculina clade: D. parva Y.S. Guo & G.P. Wang was introduced by Guo et al. (2020) as a pathogen on branches with shoot cankers symptoms of Pyrus bretschneideri, based on the diagnosis of sequence data, since the isolates formed globose or irregular, dark brown to black conidiomata-like structure that remained sterile on different media or on sterilised organic material under different conditions.
Diaporthe glabrae clade: D. etinsideae Y.P. Tan, Grice & R.G. Shivas was introduced by Tan & Shivas (2022) on Annona muricata in Queensland (Australia), based on the diagnosis of sequence data obtained apparently from the type specimen (BRIP 64096a), and no taxonomic description was provided; D. etinsideae is synonymised here under D. tulliensis.
Diaporthe inconspicua clade: D. inconspicua R.R. Gomes, Glienke & Crous was introduced by Gomes et al. (2013) for three endophytic isolates from the medicinal plants Maytenus ilicifolia and Spondias mombin in Brazil, based on the diagnosis of sequence data, since the isolates (including the ex-type strain CBS 133813) were sterile in culture; however, the morphological description of D. inconspicua was later emended by Bezerra et al. (2018) based on a set of endophytic isolates from Poincianella pyramidalis in Brazil phylogenetically related to D. inconspicua, and thus the description provided by Bezerra et al. (2018) was considered here. Similarly, the morphological description of D. isoberliniae Crous on Isoberlinia angolensis in Zambia (Crous et al. 2014b) was emended by Lambert et al. (2023) based on two strains on Pittosporum manii in Cameroon, which produced beta and gamma conidia, features that were not observed by Crous et al. (2014b) for the type specimen; the emended taxonomic description of D. isoberliniae was considered in this synopsis, along with its original description.
Diaporthe leucospermi clade: D. infecunda R.R. Gomes, Glienke & Crous was introduced by Gomes et al. (2013) for a set of eight isolates from leaves of medicinal plants growing in Brazil, such as Schinus terebinthifolius and Maytenus ilicifolia, based on the diagnosis of sequence data, since the isolates (including the ex-type strain CBS 133812) were sterile in culture; D. infecunda is synonymised here under D. leucospermi. D. australpacifica Y.P. Tan & R.G. Shivas was introduced by Tan & Shivas (2022) on a stem of Amaranthus blitum in Norfolk Island (Australia), based on the diagnosis of sequence data obtained apparently from the type specimen (BRIP 60163d), and no taxonomic description was provided; D. australpacifica is synonymised here under D. micheliae.
Diaporthe longicolla clade: D. breyniae Y. Marín & C. Lamb. Was introduced by Matio Kemkuignou et al. (2022) as an endophyte on leaves of Breynia oblongifolia in Cameroon, based on the diagnosis of sequence data, since no sporulation was observed for the ex-type strain CBS 148910 on different media. D. trevorrowii Y.P. Tan, Grice, Czislowski & R.G. Shivas was introduced by Tan & Shivas (2022) on stems of Cucumis melo in Queensland (Australia), based on the diagnosis of sequence data obtained apparently from the type specimen (BRIP 70737a), and no taxonomic description was provided.
Diaporthe rudis clade: D. asheicola L. Lombard & Crous was introduced by Lombard et al. (2014) on Vaccinium ashei in Chile, based on the diagnosis of sequence data, since the two isolates representing the species (including the ex-type strain CBS 136967) could not be induced to sporulate on different media or on sterilised V. myrtillus tissue placed on the media; however, a morphological description for D. asheicola was provided by Chethana et al. (2021b) for a Russian specimen of Diaporthe on a stem of Fraxinus pennsylvanica, phylogenetically related to D. asheicola, and thus the description provided by Chethana et al. (2021b) was considered here. D. cynaroidis Marinc., M.J. Wingf. & Crous was introduced by Marincowitz et al. (2008) on leaf litter of Protea cynaroides in South Africa, but only the sexual morph was reported associated with its type collection, and the asexual morph was regarded as “Phomopsis sp.”; however, a morphological description of the asexual morph of D. cynaroidis was provided by Hyde et al. (2020a) for an Italian saprophyte specimen of Diaporthe on a dead aerial stem of Eupatorium cannabinum, phylogenetically related to D. cynaroidis, and thus the description provided by Hyde et al. (2020a) was considered here. D. subcylindrospora S.K. Huang & K.D. Hyde was introduced by Hyde et al. (2018) as a saprophyte on a dead branch of Salix sp. in China, based on morpho-molecular analyses, but only the sexual morph was observed on the host tissue; D. subcylindrospora is synonymised here under D. cynaroidis. Similarly, D italiana Chethana, Camporesi & K.D. Hyde was introduced by Hyde et al. (2019) as a saprophyte on a dead, aerial branch of Morus alba, based on morpho-molecular analyses, but only the sexual morph was observed on the host tissue; D. italiana is synonymised here under D. rudis.
All dendrograms presented cophenetic correlation coefficient (c) values greater than 0.80, revealing that the clustering obtained is reliable and well fit. Almost none of the dendrograms obtained were congruent with the phylogenetic analyses carried out for each
Diaporthe clade, as the phylogenetic species were not recognised when their morphological characters were compared (Fig. 21). An exception was observed in morphological comparisons between species previously recognised within the D. amygdali clade. The gap statistic grouped all taxa into a single cluster, revealing no statistical significance in the size differences between the morphological characters evaluated (Fig. 21A). This result is congruent with the previous molecular based approaches that regarded the entire clade as conspecific. However, a similar result was observed for the D. brasiliensis clade. Although the phylogenetic relationships between species of the D. brasiliensis clade clearly depicted them as well-delimited IEL, the gap statistic found no statistical evidence to group them in different clusters and all three species were considered as a single cluster (Fig. 21B). In the remaining dendrograms, taxa were grouped into three (for the D. eucommiae clade, Fig. 21C) or two (Fig. 21D–I) clusters, in which the phylogenetic species defined by the molecular approaches are mixed. The same phylogenetic species is often represented in different clusters or, when represented in a single cluster, is always mixed with different phylogenetic species. For instance, in the D. leucospermi clade, all taxa representing D. leucospermi (brown typeface in Fig. 21G) and D. micheliae (green typeface in Fig. 21G) are grouped in the same cluster. However, the morphological characters did not distinguish both species as distinct from each other, and they are further grouped together with D. acaciarum, D. beilharziae, D. myracrodruonis and three taxa representing D. sackstonii (black typeface in Fig. 21G).
In all dendrograms, the phylogenetic relationships between the different taxa are highly discordant with their morphological dissimilarities. This was highly evident in the clades in which the species boundaries were already well defined, such as in the D. brasiliensis clade (Fig. 3 vs Fig. 21B) and the D. rudis clade (Fig. 10 vs Fig. 21I). For instance, while D. brasiliensis is phylogenetically closer to D. caatingaensis, morphologically it seems to be more similar to D. pygmaeae sp. nov. (Fig. 21B). Therefore, the results indicate that there is no association between the morphology of the taxa and their phylogenetic position in the different Diaporthe clades analysed.
Taxonomy
A set of 36 isolates of Diaporthe taxa obtained from palm substrata, either isolated in this study or retrieved from recent literature or GenBank, were identified as belonging to nine well-supported Diaporthe clades. Phylogenetic analyses, coalescent- (PTP and mPTP) and genetic distance-based (ABGD, ASAP and SPN) methods, phylogenetic networks, recombination analyses and hierarchical cluster analyses of phenotypic data were conducted to determine the species boundaries of each of these clades and correctly assign the isolates from palms to Diaporthe species. Forty-eight Diaporthe species are reduced to synonymy and one species is delineated as new. Morphological descriptions and illustrations of the Diaporthe species isolated from foliar lesions of palms are provided. All the Diaporthe species that occur on palms and have been treated in this study are listed below and discussed in the respective taxonomic notes with justification for reducing previously accepted species to synonymy. In addition, new data is provided on D. arecae and D. chiangmaiensis, considering their occurrence on palms. Species that have not been recorded on palms, but which have been treated in this study, are also noted and briefly discussed (see the Other species treated section).
Diaporthe amygdali (Delacr.) Udayanga et al., Fungal Divers. 56: 166. 2012. MycoBank MB 800722. Fig. 22.
Fig. 22.
Diaporthe amygdali (CDP 1442). A, B. Conidiogenous layer. C–G. Conidiophores and conidiogenous cells. H, I. Alpha conidia (black arrow in panel H points at a gamma conidium). Scale bars: A–D, H, I = 5 μm, E–G = 2.5 μm.
Basionym: Fusicoccum amygdali Delacr., Bulletin de la SMF 21: 182. 1905. MycoBank MB 186607.
Synonyms: Phomopsis amygdali (Delacr.) J.J. Tuset & M.T. Portilla, Canad. J. Bot. 67: 1280. 1989. MycoBank MB 125252.
Phomopsis amygdaliana Canonaco, Riv. Patol. Veg.: 157. 1936. MycoBank MB 256738.
Phomopsis ternstroemiae Y.H. Gao et al. [as “ternstroemia”], Mycol. Prog. 13 (1): 119. 2014. MycoBank 818625.
Diaporthe ternstroemiae (Y.H. Gao et al.) Y.H. Gao & L. Cai [as “ternstroemia”], Fungal Biol. 119: 306. 2015. MycoBank MB 819774 [Nom. inval., Arts. 41.1, F.5.1 (Shenzhen)].
Diaporthe fusicola Y.H. Gao & L. Cai, Fungal Biol. 119: 300. 2014. MycoBank MB 805928.
Diaporthe ovoidea Y.H. Gao & L. Cai [as “ovoicicola”], Fungal Biol. 119: 298. 2014. MycoBank MB 827881.
Diaporthe sterilis L. Lombard et al., Phytopathol. Mediterr. 53: 94. 2014. MycoBank MB 807600.
Diaporthe garethjonesii Dissanayake et al., Fungal Divers. 80: 171. 2015. MycoBank MB 551403.
Diaporthe kadsurae C.M. Tian & Q. Yang, MycoKeys 39: 135. 2018. MycoBank MB 824713.
Diaporthe chongqingensis Y.S. Guo & G.P. Wang, Persoonia 45: 146. 2020. MycoBank MB 830656.
Diaporthe mediterranea M. León et al., Agronomy 10: 17. 2020. MycoBank MB 836048.
New synonym:
Diaporthe nannuoshanensis S.T. Huang, et al., Nova Hedwigia 112: 405. 2021. MycoBank MB 836439.
Typus: Portugal, Trás-os-Montes, Mirandela, on twigs of Prunus amygdalus (as P. dulcis) (Rosaceae), Sep. 2005, E. Diogo [epitype of Phomopsis amygdali CBS H-20420 designated by Diogo et al. (2010), exepitype culture CBS 126679].
See Diogo et al. (2010) for illustrations and descriptions of asexual morph. Sexual morph not reported.
Isolate CDP 1442. Sexual morph: Undetermined. Asexual morph: Conidiomata on palm leaflets in culture pycnidial, subglobose to conical or ampulliform, often tapering towards ostiole, nonstromatic, uniloculate, black, solitary, occasionally aggregated in small groups, superficial, slightly covered with whitish mycelium, exuding a white mucoid mass of conidia, up to 250 μm diam. Conidiophores formed from the cells lining the locule wall, often reduced to conidiogenous cells, hyaline to subhyaline, smooth- and thin-walled, doliiform to subcylindrical, straight, aseptate, rarely 1-septate and producing a lateral branch below the septum that becomes conidiogenous. Conidiogenous cells hyaline, smooth-and thin-walled, discrete, determinate, cylindrical, tapering towards the apex, occasionally with a broader, globose base, straight, rarely curved or slightly curved, aseptate, unbranched, rarely with minute and inconspicuous collarette, enteroblastic, proliferating at the same level giving rise to periclinal thickenings, (5.19–)7.05–11.70(–15.46) × 1.63–4.02 μm, 95 % confidence limits = 8.88–9.65 × 2.33–2.51 μm (mean ± SD = 9.27 ± 1.93 × 2.42 ± 0.45 μm, n = 95). Alpha conidia fusiform to oval, often ellipsoid, tapering towards both ends, acute to subacute base, often slightly subtruncate, subobtuse to obtuse apex, smooth- and thin-walled, hyaline, aseptate, eguttulate, often with few granular contents, 5.24–7.81(–8.11) × 1.73–2.53 μm, 95 % confidence limits = 6.35–6.52 × 2.10–2.16 μm (mean ± SD = 6.43 ± 0.52 × 2.13 ± 0.17 μm), mean ± SD conidium length/width ratio = 3.04 ± 0.39 (n = 150). Gamma conidia very infrequent, fusiform to ellipsoidal, smooth- and thin-walled, hyaline, aseptate, eguttulate. Beta conidia not observed.
Culture characteristics: Colonies on 1/2 PDA, reaching 75 mm diam after 7 d at 20 °C in darkness. Surface flat, slightly cottony towards centre, with irregular to undulate margin, circular to irregular shape, whitish, becoming ivory white with rare grey patches, opaque, slightly translucent towards margin. Reverse ivory to luteus white, becoming greyish to black. No diffusible pigment. Conidiomata black, sterile, sparse, developing in poorly defined circles on the surface in about 1 wk.
Materials examined: Portugal, Lisbon, Parque das Nações, on foliar lesions of segments of Chamaerops humilis (Arecaceae), 8 May 2021, D.S. Pereira (specimen HDP 089), living culture CDP 1442 (ITS sequence PP577989, tef1 sequence PP579314, tub2 sequence PP579329, cal sequence PP579345); on foliar lesions of leaflets of Washingtonia filifera (Arecaceae), 8 May 2021, D.S. Pereira (specimen HDP 097), living culture CDP 1906 (ITS sequence PP577990, tef1 sequence PP579315, tub2 sequence PP579330, cal sequence PP579346); on foliar lesions of segments of C. humilis (Arecaceae), 8 May 2021, D.S. Pereira (specimen HDP 099), living culture CDP 2029 (ITS sequence PP577991, tef1 sequence PP579316, tub2 sequence PP579331, cal sequence PP579347).
Hosts: Acer sp. (Sapindaceae), Amygdalus persica (Rosaceae) Camellia sinensis, Camellia sp. (Theaceae), Chamaerops humilis (Arecaceae), Citrus sp. (Rutaceae), Corylus avellana (Betulaceae), Cytisus sp. (Fabaceae), Juglans regia (Juglandaceae), Kadsura longipedunculata (Schisandraceae), Lithocarpus glaber (Fagaceae), Malus domestica (Rosaceae), Osmanthus fragrans (Oleaceae), Persea americana (Lauraceae), Pieris japonica (Ericaceae), Prunus amygdalus, P. armeniaca, P. armeniaca, P. persica, P. salicina, Prunus sp., Pyrus pyrifolia, Pyrus sp. (Rosaceae), Ternstroemia gymnanthera (Pentaphylacaceae), Vaccinium corymbosum (Ericaceae), Vitis vinifera, Vitis sp. (Vitaceae) and Washingtonia filifera (Arecaceae) (Farr & Rossman 2023, present study).
Distribution: China, France, Greece, Hungary, Italy, Japan, Portugal, South Africa, Spain, Thailand, Tunisia, Turkey, Uruguay and USA (Farr & Rossman 2023).
Notes: Diaporthe amygdali was introduced by Delacroix (1905) as Fusicoccum amygdali, causing cankers on the twigs of almond trees (Prunus amygdalus) in France. The same pathogen was found on almonds and peaches (P. persica) by Canonaco (1936), who named it Phomopsis amygdalina. Later, Tuset & Portilla (1989) re-examined the type specimen of F. amygdali and, based on morphology and symptomatology, they considered it to be better accommodated in the genus Phomopsis as P. amygdali. In addition, the authors also considered P. amygdalina to be a synonym of P. amygdali. Since there are no cultures unequivocally linked to any existing type specimen of P. amygdali, an epitype specimen was designated by Diogo et al. (2010), following a survey of Diaporthe (as Phomopsis) isolates obtained from almond trees showing symptoms of canker or dieback in Portugal. With the abolishment of the dual nomenclature for pleomorphic fungi, Udayanga et al. (2012a) transferred P. amygdali into Diaporthe as D. amygdali. Since the epitypification of D. amygdali (Diogo et al. 2010), several new species morphologically identical and phylogenetically closely related have been introduced. However, the phylogenetic relationships between some of those species were considered ambiguous, insomuch as the clade has been regarded as a putative species complex (Gao et al. 2016). Recently, Hilário et al. (2021a) resolved the D. amygdali species complex, providing evidence that it constitutes a single species and, subsequently, synonymised eight Diaporthe species under D. amygdali. The integrative taxonomic approach carried out in the present study confirmed the results of Hilário et al. (2021a) and revealed two new synonyms in the D. amygdali clade, namely D. nannuoshanensis (Huang et al. 2021a) and “D. sinensis” (Manawasinghe et al. 2021). Both “species” were shown to be intermingled in the clade of D. amygdali, lacking phylogenetic distinctiveness (Fig. 2). The exepitype culture of D. amygdali (CBS 126679) showed the following nucleotide similarities with the sequences of the ex-type culture of D. nannuoshanensis (SAUCC 194.91) and the ex-paratype culture of “D. sinensis” (JZB 320168). On ITS: 98.01 % (542/553, including 5 gaps) and 98.30 % (520/529, including 2 gaps), respectively. On tef1: 93.84 % (274/292, including 4 gaps) and 97.94 % (284/290, including 2 gaps), respectively. On tub2: 99.74 % (387/388) and 97.62 % (369/378, including 1 gap), respectively. On cal: 98.42 % (437/444, including 2 gaps) and 100 % (435/435), respectively. On his3: 99.40 % (418/423); no his3 sequence data is available for any of the ex-type cultures of “D. sinensis”. It should be noted that the above-mentioned nucleotide similarities were based on one of the ex-paratype cultures of “D. sinensis” rather than its ex-type culture (JZB 320167). This is because the cal partial sequence available for the latter is highly polymorphic and impairs nucleotide pairwise comparisons with the cal partial sequence available for the ex-epitype of D. amygdali. This highly polymorphic cal sequence causes “D. sinensis” to cluster on a longer branch in the D. amygdali clade (Fig. 2). The scientific name “Diaporthe sinensis” was preoccupied by an earlier fungus (Feng et al. 2019), and so the species was invalidly published (Nom. illegit., Art. 53.1). As a result, the strains of “D. sinensis” have been assigned here to D. amygdali, but the species epithet sinensis could not be made synonymous with D. amygdali. Similarly, although Hilário et al. (2021a) listed “D. ternstroemiae” as a synonym of D. amygdali, this species was invalidly introduced as a new combination for P. ternstroemiae without a recognised nomenclatural repository identifier [Nom. inval., Art. 41.1, F.5.1 (Shenzhen)]. Therefore, “D. ternstroemiae” has been excluded here from the list of synonyms of D. amygdali. Morphologically, all “species” previously described in the D. amygdali clade harbour ovoid/fusoid to ellipsoid alpha conidia of considerably overlapping dimensions, a common absence of beta and gamma conidia, as well as conidiomata, conidiophores and/or conidiogenous cells that lie within the same size ranges (Table 4). Considering the morphological data available for the type specimens of “species” in the D. amygdali clade, the mean dimensions of the alpha conidia produced by D. amygdali strains are 5.49–7.99 × 1.97–2.98 μm (L/W = 2.20–3.13), which clearly overlap with the dimensions reported for the ex-epitype culture of D. amygdali (CBS 126679; mean = 6.30 × 2.37 μm) (Table 4). Thus, except for the production of beta conidia observed in D. garethjonesii (Hyde et al. 2016) and D. ovoidea (Gao et al. 2015), the morphology of the asexual morph of all D. amygdali strains match the description provided by Diogo et al. (2010). Three isolates of D. amygdali were obtained from foliar lesions of palms in Lisbon, Portugal, namely CDP 1442 and CDP 2029 from Chamaerops humilis, and CDP 1906 from Washingtonia filifera. Isolates from distinct palm hosts were tentatively characterised to assess micromorphological variation. However, despite numerous attempts with different temperatures and with different organic material introduced on the agar surface, isolate CDP 1906 remained sterile even after long periods of incubation of about 2 mo. Isolates from C. humilis were morphologically similar to the ex-epitype of D. amygdali (Diogo et al. 2010) (Fig. 22). Considering the strain characterised here (CDP 1442) and the ex-epitype strain of D. amygdali (CBS 126679), both produce subglobose to ampulliform, black pycnidial conidiomata, exuding masses of ovoid-ellipsoid, hyaline and aseptate alpha conidia of remarkable similar dimensions (mean = 6.43 × 2.13 μm and 6.3 × 2.37 μm, respectively) (Diogo et al. 2010). Sequence comparisons with the ex-epitype of D. amygdali (CBS 126679) for ITS, tef1, tub2 and cal showed 98.35 %, 98.16 %, 97.35 % and 99.38 %, respectively, sequence similarity and differences are represented by few base pair changes and gaps. The nucleotide sequence similarity between the D. amygdali isolates obtained here was 100 % for the four loci. No his3 sequence data is available for these isolates. Diaporthe amygdali has not previously been recorded on Arecaceae and thus two new host records are reported here (Table 5). The isolates of D. amygdali studied here were recorded from foliar lesions of palms, but pathogenicity has not been tested. However, most of the “species” introduced in the D. amygdali clade have been described in association with plant diseases, including leaf spots (Table 4). Thus, it is likely that the isolates obtained from palms are pathogenic. Moreover, D. amygdali is considered the main pathogen associated with twig cankers and shoot blight symptoms of almond and peach trees in Mediterranean countries, as well as a damaging agent in other important fruit crops across the globe (León et al. 2020). Therefore, the geo-ecological data available for D. amygdali suggest that this species has a widespread distribution as a pathogen in a broad range of angiosperms.
Table 5.
List of accepted Diaporthe species associated with Arecaceae. The Diaporthe species considered were only those described with both morphological and molecular data. Currently valid Diaporthe species that were described on palms based solely on morphological data were disregarded.
| Species | Host1 | Country1 | Reference |
|---|---|---|---|
| Diaporthe amygdali | Chamaerops humilis | Portugal | Present study |
| Washingtonia filifera | Portugal | Present study | |
| D. arecae | Areca catechu | India | Srivastava et al. (1962) (as Subramanella arecae); Gomes et al. (2023) |
| China | Xu et al. (2020) (as D. limonicola); GenBank (as D. limonicola and D. pseudophoenicicola, see Pereira et al. 2023) | ||
| Arenga engleri | Hong Kong | Gomes et al. (2013) (as D. arengae) | |
| Calamus castaneus | Malaysia | Azuddin et al. (2021) (also as D. arengae) | |
| Caryota maxima | China (Xisha Islands) | GenBank (as D. melitensis) | |
| Chamaerops humilis | Portugal | Boonmee et al. (2021) (as D. chamaeropicola); Pereira et al. (2023) | |
| Chrysalidocarpus lutescens | China | Huang et al. (2021) (as D. chrysalidocarpi); Senanayake et al. (2023) | |
| Cocos nucifera | Brazil | GenBank | |
| China (Xisha Islands) | GenBank (as D. melitensis) | ||
| Elaeis guineensis | Indonesia | Yurnaliza et al. (2021) (as D. eucalyptorum) | |
| Livistona chinensis | China | Senanayake et al. (2023) | |
| Hong Kong | Guo et al. (2000) | ||
| Oncosperma sp. | Thailand | Saengket et al. (2021) | |
| Phoenix canariensis | Uruguay | Gao et al. (2017) (as D. pseudophoenicicola) | |
| Phoenix dactylifera | Portugal | Boonmee et al. (2021) (as D. pseudophoenicicola); Pereira et al. (2023) | |
| Spain | Gomes et al. (2013) (as D. pseudophoenicicola) | ||
| Phoenix paludosa | India (Andaman Islands) | Rajamani et al. (2018) (as D. hongkongensis) | |
| Trachycarpus fortunei | China | Senanayake et al. (2023) | |
| D. aseana | Calamus castaneus | Malaysia | Azuddin et al. (2021) (as D. hongkongensis) |
| D. chiangmaiensis | Calamus castaneus | Malaysia | Azuddin et al. (2021) (as “D. cf. heveae”) |
| D. cytosporella | Chamaerops humilis | Greece | Gomes et al. (2013) (as “D. chamaeropis”) |
| D. eres | Rhapis subtilis | China | Gao et al. (2016) |
| D. eucommiae | Nypa fruticans | India (Andaman Islands) | Thavamani et al. (2018) (as D. hongkongensis) |
| D. foeniculina | Chamaerops humilis | Portugal | Present study |
| Chrysalidocarpus lutescens | Portugal | Present study | |
| Phoenix roebelenii | Portugal | Present study | |
| Trachycarpus fortunei | Portugal | Present study | |
| Washingtonia filifera | Portugal | Present study | |
| D. glabrae | Calamus castaneus | Malaysia | Azuddin et al. (2021) (as D. tectonae) |
| Calamus thwaitesii | Sri Lanka | Dissanayake et al. 2016 | |
| D. inconspicua | Chrysalidocarpus lutescens | China | Sun et al. (2021) (as D. lutescens) |
| D. leucospermi | Chamaerops humilis | Portugal | Present study |
| Chrysalidocarpus lutescens | Portugal | Present study | |
| D. pygmaeae | Chamaerops humilis | Portugal | Present study |
| Phoenix roebelenii | Portugal | Present study | |
| D. rudis | Chrysalidocarpus lutescens | Portugal | Present study |
| Washingtonia filifera | Portugal | Present study | |
| D. ueckeri | Areca catechu | China | GenBank |
| Hyophorbe lagenicaulis | China | Guo et al. (2024) |
New host and new geographical records are noted in bold font.
Diaporthe arecae (H.C. Srivast. et al.) R.R. Gomes et al., Persoonia 31: 16. 2013. MycoBank MB 802924.
Basionym: Subramanella arecae H.C. Srivast. et al., Mycologia 54: 7. 1962. MycoBank MB 339830.
Synonyms: Phyllosticta nelumbonis Sawada, Special Publication College of Agriculture National Taiwan University 8: 140. 1959. MycoBank MB 336860 [Nom. inval., Art. 39.1 (Melbourne)].
Diaporthe nelumbonis Sawada ex R. Kirschner, Mycol. Prog. 17: 280. 2017. MycoBank MB 821926.
Phomopsis averrhoae C.Q. Chang et al., Mycosystema 24: 6. 2005. MycoBank MB 344467.
Diaporthe averrhoae (C.Q. Chang et al.) Y.H. Gao & L. Cai, IMA Fungus 8: 183. 2017. MycoBank MB 821437.
Phomopsis liquidambaris C.Q. Chang et al., Mycosystema 24: 9. 2005. MycoBank MB 344462.
Diaporthe liquidambaris (C.Q. Chang et al.) Udayanga & Castl., IMA Fungus 7: 291. 2016. MycoBank MB 819021.
Diaporthe liquidambaris (C.Q. Chang et al.) Y.H. Gao & L. Cai, IMA Fungus 8: 183. 2017. MycoBank MB 821446.
Phomopsis phyllanthicola C.Q. Chang et al., Mycosystema 24: 10. 2005. MycoBank MB 344466.
Diaporthe phyllanthicola (C.Q. Chang et al.) Y.H. Gao & L. Cai, IMA Fungus 8: 184. 2017. MycoBank MB 821461.
Phomopsis loropetali C.Q. Chang et al., Mycosystema 24: 148. 2005. MycoBank MB 344460.
Diaporthe loropetali (C.Q. Chang et al.) Y.H. Gao & L. Cai, IMA Fungus 8: 183. 2017. MycoBank MB 821448.
Diaporthe ceratozamiae Crous & R.G. Shivas, Persoonia 27: 133. 2011. MycoBank MB 560695.
Diaporthe musigena Crous & R.G. Shivas, Persoonia 26: 119. 2011. MycoBank MB 560160.
Diaporthe pterocarpicola Udayanga et al., Cryptogam. Mycol. 33: 303. 2012. MycoBank MB 801053.
Diaporthe arengae R.R. Gomes et al., Persoonia 31: 16. 2013. MycoBank MB 802925.
Diaporthe fraxini-angustifoliae R.G. Shivas et al., Fungal Divers. 61: 255. 2013. MycoBank MB 802384.
Diaporthe litchiicola R.G. Shivas et al. [as “litchicola”], Fungal Divers. 61: 256. 2013. MycoBank MB 545033.
Diaporthe pascoei R.G. Shivas, et al., Fungal Divers. 61: 258. 2013. MycoBank MB 802387.
Diaporthe pseudomangiferae R.R. Gomes et al., Persoonia 31: 30. 2013. MycoBank MB 802945.
Diaporthe pseudophoenicicola R.R. Gomes et al., Persoonia 31: 30. 2013. MycoBank MB 803839.
Diaporthe limonicola Guarnaccia & Crous, IMA Fungus 8: 328. 2017. MycoBank MB 821731.
Diaporthe melitensis Guarnaccia & Crous, IMA Fungus 8: 329. 2017. MycoBank MB 821732.
Diaporthe pescicola Dissanayake, J.Y. Yan et al., Mycosphere 8: 542. 2017. MycoBank MB 551988.
Diaporthe podocarpi-macrophylli Y.H. Gao & L. Cai, IMA Fungus 8: 176. 2017. MycoBank MB 820682.
Diaporthe sennae C.M. Tian & Qin Yang, Phytotaxa 302: 149. 2017. MycoBank MB 820452.
Diaporthe taoicola Dissanayake et al., Mycosphere 8: 543. 2017. MycoBank MB 551989.
Diaporthe cercidis C.M. Tian & Q. Yang, MycoKeys 39: 124. 2018. MycoBank MB 824707.
Diaporthe oculi Mochiz. & Kaz. Tanaka, J. Infect. Chemother. 25: 98. 2018. MycoBank MB 825540.
Diaporthe pandanicola Tibpromma & K.D. Hyde, MycoKeys 33: 44. 2018. MycoBank MB 823840.
Diaporthe pseudooculi Mochiz. & Kaz. Tanaka, J. Infect. Chemother. 25: 100. 2018. MycoBank MB 825541.
Diaporthe anhuiensis H. Zhou & C.L. Hou, Phytotaxa 422: 165. 2019. MycoBank MB 832081.
Diaporthe guangxiensis Dissanayake et al., Front. Microbiol. 10: 14. 2019. MycoBank MB 552578.
Diaporthe huangshanensis H. Zhou & C.L. Hou, Phytotaxa 422: 169. 2019. MycoBank MB 832082.
Diaporthe millettiae H. Long et al., MycoKeys 57: 119. 2019. MycoBank MB 829563.
Diaporthe osmanthi H. Long et al., MycoKeys 57: 120. 2019. MycoBank MB 829564.
Diaporthe viniferae Dissanayake, X.H. Li & K.D. Hyde, Front. Microbiol. 10: 21. 2019. MycoBank MB 552002.
Diaporthe acuta Y.S. Guo & G.P. Wang, Persoonia 45: 140. 2020. MycoBank MB 830655.
Diaporthe delonicis R.H. Perera et al., Mycosphere 11: 2129. 2020. MycoBank MB 556855.
Diaporthe drenthii Y.P. Tan et al., Plant Pathol. 69: 916. 2020. MycoBank MB 833828.
Diaporthe fulvicolor Y.S. Guo & G.P. Wang, Persoonia 45: 146. 2020. MycoBank MB 830657.
Diaporthe krabiensis Dayarathne, Mycosphere 11: 92. 2020. MycoBank MB 635831 [Nom. inval., Art. F.5.1 (Shenzhen)].
Diaporthe krabiensis Dayar. ex Hongsanan & K.D. Hyde, Mycosphere 14: 1030. 2023. MycoBank MB 900762 [isonym, Art. 6.3 Note 2].
Diaporthe krabiensis Dayar. ex M.S. Calabon & E.B.G. Jones, Bot. Mar. 66: 219. 2023. MycoBank MB 848522.
Diaporthe searlei R.G. Shivas et al., Plant Pathol. 69: 918. 2020. MycoBank MB 833830.
Diaporthe spinosa Y.S. Guo & G.P. Wang, Persoonia 45: 154. 2020. MycoBank MB 830659.
Diaporthe taiwanensis H.A. Ariyaw & I. Tsai, Phytotaxa 461: 161. 2020. MycoBank MB 835116.
Diaporthe camelliae-oleiferae Q. Yang, MycoKeys 84: 22. 2021. MycoBank MB 840451.
Diaporthe chamaeropicola D.S. Pereira & A.J.L. Phillips, Fungal Divers. 111: 166. 2021. MycoBank MB 557847.
Diaporthe chrysalidocarpi S.T. Huang et al., MycoKeys 78: 59. 2021. MycoBank MB 837812.
Diaporthe endocitricola Z.Y. Dong et al., Front. Microbiol. 11: 9. 2021. MycoBank MB 557628.
Diaporthe hunanensis Q. Yang, MycoKeys 84: 26. 2021. MycoBank MB 840452.
Diaporthe schimae C.M. Tian & Q. Yang, MycoKeys 77: 55. 2021. MycoBank MB 829526.
Diaporthe annellsiae Y.P. Tan & R.G. Shivas, Index Austral. Fungi 2: 1. 2022. MycoBank MB 559559.
Diaporthe bounty Y.P. Tan & R.G. Shivas, Index Austral. Fungi 2: 3. 2022. MycoBank MB 559562.
Diaporthe gossiae Y.P. Tan & R.G. Shivas, Index Austral. Fungi 2: 5. 2022. MycoBank MB 559565.
Diaporthe hongheensis E.F. Yang & Tibpromma, J. Fungi 8: 15. 2022. MycoBank MB 559411.
Diaporthe howardiae Y.P. Tan & R.G. Shivas, Index Austral. Fungi 2: 6. 2022. MycoBank MB 559570.
Diaporthe meliae C.M. Tian & Qin Yang, MycoKeys 91: 38. 2022. MycoBank MB 829523.
Diaporthe norfolkensis Y.P. Tan & R.G. Shivas, Index Austral. Fungi 2: 8. 2022. MycoBank MB 559574.
Diaporthe viciae W.S. Zhao et al., Mycosphere 14: 34. 2023. MycoBank MB 558423.
New synonyms:
Diaporthe afzeliae Monkai & Lumyong, J. Fungi 9 (no. 603): 7. 2023. MycoBank MB 900377.
Diaporthe bombacis Monkai & Lumyong, J. Fungi 9 (no. 603): 11. 2023. MycoBank MB 900378.
Diaporthe diospyrina Y.K. Bai & X.L. Fan, MycoKeys 98: 76. 2023. MycoBank MB 847473.
Diaporthe hainanensis Qin Yang, MycoKeys 102: 235. 2024. MycoBank MB 848328.
Typus: India, on fruit of Areca catechu (Arecaceae), during 1958–1959, H.C. Srivastava (holotype of Subramanella arecae IMI, anon. s. n., IARI, anon. s. n.); on fruit of Areca catechu, Feb. 1964, H.C. Srivastava (isotype of Subramanella arecae CBS H-7808, ex-isotype culture CBS 161.64).
See Srivastava et al. (1962) for illustrations and descriptions of asexual morph. Sexual morph was not reported for any of the type specimens but was reported under the species names D. hongheensis (Yang et al. 2022) and D. spinosa (Guo et al. 2020).
Hosts: Reported from more than 50 genera and 55 species in 38 families. Studies subsequent to Pereira et al. (2023) confirm the following hosts (see taxonomic notes for further details): in Arecaceae: Caryota maxima, Cocos nucifera, Elaeis guineensis, Livistona chinensis, Phoenix padulosa, Oncosperma sp. and Trachycarpus fortunei (Guo et al. 2000, Thavamani et al. 2018, Saengket et al. 2021, Yurnaliza et al. 2021, Senanayake et al. 2023, present study); in Bombacaceae: Bombax ceiba (Monkai et al. 2023); in Ebenaceae: Diospyros kaki (Bai et al. 2023); in Fabaceae: Afzelia xylocarpa (Monkai et al. 2023); in Rhizophoraceae: Rhizophora apiculata (Julia et al. 2022); in Rubiaceae: Morinda officinalis (Luo et al. 2022); and in Solanaceae: Solanum melongena (Aumentado & Balendres 2024).
Distribution: Studies subsequent to Pereira et al. (2023) confirm Philippines (Aumentado & Balendres 2024) as an additional country. Moreover, the analysis carried out in this study also confirms Brazil as an additional country to those listed by Pereira et al. (2023) (see taxonomic notes for further details).
Notes: Diaporthe arecae was introduced by Srivastava et al. (1962) as Subramanella arecae from Areca catechu in India and was later assigned to Diaporthe by Gomes et al. (2013). The phylogenetic position of D. arecae and its related species has been unclear for several years. As a result, it has been treated as a species complex with several poorly understood lineages. Pereira et al. (2023) carried out an integrative taxonomic approach that identified three well-delimited subclades within the D. arecae species complex (DASC). The authors reduced 52 Diaporthe spp. to synonymy under D. arecae, which were shown to be conspecific and sister to D. chiangmaiensis and D. smilacicola (Pereira et al. 2023). Since Pereira et al. (2023), four new species have been introduced in the DASC, namely D. afzeliae, D. bombacis (Monkai et al. 2023), D. diospyrina (Bai et al. 2023) and D. hainanensis (Liu et al. 2024). The phylogenetic position of these taxa has been reassessed in the present study (data not shown), which confirmed that they represent intraspecific variation of D. arecae. Therefore, the three species are herein treated as synonyms of D. arecae. Recently, the morphological description of D. arecae was emended by Senanayake et al. (2023), who isolated new strains of D. arecae from monocotyledons plants in China. Although additional data was provided for the taxonomic treatment of D. arecae, its species boundaries were incorrectly noted (fig. 99 in Senanayake et al. 2023) and the entire DASC [sensu Norphanphoun et al. (2022)] was considered conspecific. Consequently, two of the strains used to emend the taxonomic description of D. arecae, ZHKUCC 22-0182 and ZHKUCC 22-0192, did not cluster with strains previously assigned to D. arecae. Considering the current understanding of the DASC [sensu Pereira et al. (2023)], these strains and their respective host, along with the taxonomic emendation of D. arecae, were disregarded here. Further analyses are needed to correctly assign these strains to a Diaporthe species. Although D. arecae has primarily been described as a pathogen causing severe post-harvest fruit rot of A. catechu, it has a widespread distribution and a broad host range as a pathogen, endophyte and saprophyte. The palm hosts associated with D. arecae included Arenga engleri, Calamus castaneus, Chrysalidocarpus lutescens, Phoenix canariensis and P. dactylifera (Pereira et al. 2023). Senanayake et al. (2023) recorded new strains of D. arecae as saprophytes on the leaves of C. lutescens (syn. Dypsis lutescens), Livistona chinensis and Trachycarpus fortunei in China. Moreover, the present study has provided new insights regarding the range of palms hosts that are colonised by D. arecae through the analysis of unpublished reports, whose ITS sequences have been deposited in GenBank, as well as occasional reports of Diaporthe species in biodiversity or ecological studies (refer to Table 5 for a summary of the palm hosts from which D. arecae has been collected). The phylogenetic position of these strains has been (re)evaluated (data not shown) and is commented herein. Guo et al. (2000) identified a Diaporthe strain (Diaporthe sp. MS704; GenBank accession number: AF153737) as a foliar endophyte in the fronds of L. chinesis in Hong Kong. Due to limited availability of DNA sequence data at the time, the relationship of this strain with reference Diaporthe taxa was ambiguous. However, the present analysis confirmed that it represents an isolate of D. arecae. Thavamani et al. (2018) studied the fungal endophytes inhabiting the leaves of obligate mangrove hosts in the Andaman Islands, India and recorded two strains of Diaporthe on P. padulosa and Nypa fruticans. The strain from P. padulosa (GenBank accession number: MH371252) was found to be an isolate of D. arecae that was misidentified as D. hongkongensis. Saengket et al. (2021) isolated three endophytic strains of Diaporthe from tissues of healthy native palms of Oncosperma sp. in Thailand. Two of these strains, MFLUCC 20-0208 and MFLUCC 20-0214, clustered in the DASC. The ITS sequence for the strain MFLUCC 20-0208 has been deposited in GenBank (accession number: MW255349) and was confirmed to represent an isolate of D. arecae. Yurnaliza et al. (2021) identified an endophytic strain of Diaporthe in the petioles of Elaeis guineensis in Indonesia. This strain (GenBank accession number: MW488222) was found to be an isolate of D. arecae that was misidentified as D. eucalyptorum. In addition, five unpublished reports of D. arecae on arecaceous hosts were found in GenBank and their phylogenetic affiliations with D. arecae were confirmed. These include the strains XSZ-01, XSZ-02 and YXD-07 isolated from the leaves of Cocos nucifera and Caryota maxima in the Xisha Islands, China (GenBank accession numbers: OQ469324, OQ469325 and OQ469330, respectively), which were misidentified as D. melitensis; the endophytic strain Diaporthe sp. PB83 (GenBank accession number: MK508809) from leaves of C. nucifera in Brazil; and the strain Diaporthe sp. PV isolated as a leaf spot disease pathogen of A. catechu in India.
Diaporthe aseana Dissan. et al., Fungal Divers. 80: 167. 2015. MycoBank MB 551402.
New synonym: Diaporthe tectonigena Doilom et al., Fungal Divers. 82: 164. 2016. MycoBank MB 551977.
Typus: Thailand, Phayao, Jam Pa Thong Waterfall, on dead leaf, 12 Mar. 2012, N. Tangthirasunun (holotype MFLU 13-0256, ex-type culture MFLUCC 12-0299a).
See Hyde et al. (2016) for illustrations and descriptions of asexual morph. Sexual morph not reported.
Hosts: Abies beshanzuensis (Pinaceae) (Hyde et al. 2016), Camellia sinensis (Theaceae) (Gao et al. 2017), Cassia spectabilis (Fabaceae) (Udayanga et al. 2012a), Calamus castaneus (Arecaceae) (Azuddin et al. 2021), Cinchona calisaya (Rubiaceae) (Hyde et al. 2016), Tectona grandis (Lamiaceae) (Doilom et al. 2016).
Distribution: China (Hyde et al. 2016, Gao et al. 2017), Indonesia (Hyde et al. 2016), Malaysia (Azuddin et al. 2021) and Thailand (Udayanga et al. 2012a, Doilom et al. 2016, Hyde et al. 2016).
Notes: The subclade of D. aseana included the ex-type cultures of D. aseana and D. tectonigena that appear phylogenetically and morphologically indistinguishable and are thus made synonymous. Both species clustered in a well-supported lineage and are closely related (Fig. 4). The ex-type culture of D. aseana (MFLUCC 12-0299a) showed the following nucleotide similarities with the sequences of the ex-type culture of D. tectonigena (MFLUCC 12-0767). On ITS: 98.10 % (516/526, including 5 gaps). On tef1: 99.36 % (310/312, including 1 gap). On tub2: 99.20 % (372/375, including 1 gap). On cal: 100 % (443/443). No his3 sequence data is available for both ex-type cultures. Morphologically, both species produce fusoid to ellipsoidal, hyaline and aseptate alpha conidia of considerable overlapping dimensions (6–9 × 2–3 μm for D. aseana vs 7.3–7.8 × 2.5–3.5 μm for D. tectonigena) (Doilom et al. 2016, Hyde et al. 2016) (Table 4). Beta conidia were only observed in D. tectonigena (Doilom et al. 2016), but its production has been shown to be considerably influenced by growth conditions and thus tend to simply reflect intraspecific variability (Mostert et al. 2001). A closely related strain (SM42) to D. aseana has been isolated as an endophyte on Calamus castaneus spines and has been misidentified as D. hongkongensis (Azuddin et al. 2021) (Table 5). Most of the analyses carried out in this study have shown that this strain is conspecific with D. aseana. However, its phylogenetic position within the D. aseana subclade lacks relevant support values (Fig. 4) and coalescent-based analyses of 3-loci combined datasets suggested that this strain is a distinct lineage closely related to D. aseana (data not shown). The ex-type culture of D. aseana (MFLUCC 12-0299a) showed the following nucleotide similarities with the sequences of “D. hongkongensis” SM42. On ITS: 98.00 % (539/550, including 4 gaps). On tef1: 95.13 % (332/349, including 3 gaps). On tub2: 96.36 % (371/385, including 3 gaps). No cal sequence data is available for “D. hongkongensis” SM42. The low sequence similarity values on tef1and tub2 loci (p-distance = 0.05 and 0.04, respectively) also suggests that this strain represents a distinct lineage within the D. eucommiae clade. Therefore, future strains related to this isolate from C. castaneus may reveal that it constitutes a different taxon closely related to D. aseana. However, for taxonomic stability and clarity, the fungus from Malaysia was assigned to D. aseana. Although D. aseana (as D. tectonigena) has been isolated in association with twig dieback of Tectona grandis (Doilom et al. 2016), it is not known whether this fungus is a pathogen, an opportunistic saprobe or an endophyte. Most records of this species support its endophytic nature.
Diaporthe chiangmaiensis N.I. de Silva et al., Mycosphere 13: 1032. 2022. MycoBank MB 559527.
New synonym: Diaporthe zhaoqingensis M. Luo et al., J. Fungi 8 (no. 806): 22. 2022. MycoBank MB 553427.
Typus: Thailand, Chiang Rai Province, on dead attached twigs of Magnolia lilifera (Magnoliaceae), 11 Feb. 2019, N.I. de Silva (holotype MFLU 18-1305, ex-type culture MFLUCC 18-0544).
See de Silva et al. (2022) for illustrations and descriptions of sexual and asexual morphs.
Hosts: Alstonia scholaris (Apocynaceae) (de Silva et al. 2022), Calamus castaneus (Arecaceae) (Azuddin et al. 2021), Hevea brasiliensis (Euphorbiaceae) (Gomes et al. 2013), Magnolia lilifera (Magnoliaceae) (de Silva et al. 2022), Morinda officinalis (Rutaceae) (Luo et al. 2022) and Nephrolepis cordifolia (Nephrolepidaceae) (Seifollahi et al. 2023).
Distribution: China (Luo et al. 2022), India (Gomes et al. 2013), Malaysia (Azuddin et al. 2021) and Thailand (de Silva et al. 2022, Seifollahi et al. 2023).
Notes: De Silva et al. (2022) introduced D. chiangmaiensis as a saprophyte on attached dead twigs of Magnolia champaca in Thailand, with further collections of the same species as saprophyte and endophyte of Magnolia lilifera and Alstonia scholaris, respectively. The sister relationship of D. chiangmaiensis with the strain D. cf. heveae 2 has been noted by de Silva et al. (2022) and both taxa were found to be conspecific by Pereira et al. (2023), who recognised D. chiangmaiensis as separate lineage within the DASC. Later, Monkai et al. (2023) noted the close phylogenetic relationship between D. chiangmaiensis and D. zhaoqingensis, which was introduced as an endophyte on the root and stem of Morina officinalis in China by Luo et al. (2022). Monkai et al. (2023) considered both species to be distinct based on the evidence from the PHI test and DNA sequence comparison. However, this relationship was reevaluated here, and both taxa were found to be conspecific. Although the PHI tested negative for recombination between strains of D. chiangmaiensis and D. zhaoqingensis, both taxa were recognised as a single lineage following the GCPSR principle and DNA-based species delimitation analyses (data not shown). These results are congruent with the phylogenetic analyses in which both species grouped together in a well-supported monophyletic lineage (data not shown). Only the ITS and tef1 sequence data are available for the ex-type culture of D. chiangmaiensis (MFLUCC 18-0544), which showed the following nucleotide similarities with the sequences of the ex-type culture of D. zhaoqingensis (ZHKUCC 22-0056). On ITS: 98.42 % (501/509, including 1 gap). On tef1: 99.67 % (299/300). Morphological comparisons between the asexual morphs of both species are superfluous, since only gamma conidia were observed for D. zhaoqingensis (Luo et al. 2022), while only alpha conidia were observed for D. chiangmaiensis (de Silva et al. 2022). Sequence data of additional loci for the ex-type strain of D. chiangmaiensis are needed to clarify the taxonomic placement and relationship of both species. However, giving the available data, for taxonomic stability and clarity, D. chiangmaiensis and D. zhaoqingensis are made synonymous. Most records of D. chiangmaiensis support its endophytic nature. Regarding palms, the species has been recorded as an endophyte on the spines of Calamus castaneus in Malaysia (Azuddin et al. 2021) (Table 5).
Diaporthe cytosporella (Penz. & Sacc.) Udayanga & Castl., Persoonia 32: 95. 2014. MycoBank MB 803986.
Basionym: Phoma cytosporella Penz. & Sacc. 1887. MycoBank MB 220373.
Synonym: Phomopsis cytosporella (Penz. & Sacc.) H.S. Fawc. & H.A. Lee, Citrus diseases and their control: 407. 1926. MycoBank MB 251741.
New synonym: Diaporthe pimpinellae Abeywickrama et al., Fungal Divers. 104: 77. 2020. MycoBank MB 556363.
Typus: Italy, Rome, Modena, on Citrus limonia (Rutaceae), Jan. 1886 (holotype of Phoma cytosporella, BPI 798526). Spain, on Citrus limon (Rutaceae), M.E. Palm [epitype of Phoma cytosporella designated by Udayanga et al. (2014b), BPI 892459, dried culture, ex-type culture CBS 137020].
See Udayanga et al. (2014b) for illustrations and descriptions of asexual morph. The Latin description of the sexual morph of Phoma cytosporella has been cited by Saccardo (1892).
Hosts: Chamaerops humilis (Arecaceae) (Gomes et al. 2013), Citrus limon, C. sinensis (Rutaceae) (Udayanga et al. 2014b), Juglans regia (Juglandaceae) (Luna et al. 2022), Pimpinella peregrina (Apiaceae) (Yuan et al. 2020), Pistacia vera (Anacardiaceae) (Chen et al. 2014), Prunus amygdalus (Rosaceae) (Holland et al. 2021), Salix sp. (Salicaceae) (Lawrence et al. 2015), Spartium junceum (Fabaceae) (Gomes et al. 2013) and Vitis vinifera (Vitaceae) (Udayanga et al. 2014b, Lawrence et al. 2015).
Distribution: Croatia, Greece (Gomes et al. 2013), Italy (Udayanga et al. 2014b, Yuan et al. 2020), Spain and USA (Chen et al. 2014, Udayanga et al. 2014b, Lawrence et al. 2015, Holland et al. 2021, Luna et al. 2022).
Notes: Diaporthe cytosporella is phylogenetically closely related to D. foeniculina but clearly distinguished based on most of the DNA-based analyses carried out here. The status of this species has been unclear for many years. It was first described as Phoma cytosporella from Italy and later synonymised under D. citri by Fisher (1972), who considered Phomopsis cytosporella as the valid name for D. citri based on the chronology of names. Nonetheless, this interpretation was not adopted by plant pathologists or taxonomists, insomuch as P. cytosporella was determined to be a distinct taxon in Diaporthe by Udayanga et al. (2014b), who epitypified D. cytosporella based on fresh collections from Europe. According to the analyses conducted here, the strains “D. chamaeropis” CBS 454.81 and CBS 137020 were shown to be synonyms of D. cytosporella. However, the species D. chamaeropis was not considered in the synonyms proposed here, since no ex-type strain has been formally linked to this species. Diaporthe chamaeropis (as Phoma chamaeropis) was originally described on the petioles of Chamaerops humilis (as Chamaerops sp.) and other palms from Kew, England (Cooke 1885) and later transferred to Phomopsis (as P. chamaeropis) by Petrak (1919). Gomes et al. (2013) analysed the strains CBS 454.81 from the dead part of a leaf of C. humilis in Greece, and CBS 753.70 from a dead branch of Spartium junceum in Croatia. They considered that these cultures could be authentic for D. chamaeropis based on conidial dimensions, but no epitype was formally designated. As no ex-type cultures exist for D. chamaeropis, the strains “D. chamaeropis” CBS 454.81 and CBS 137020 were here assigned to D. cytosporella. In spite of this, since neither of these two strains are linked to the holotype, the species epithet chamaeropis could not be made a synonym of D. cytosporella. The analyses carried out here also revealed that D. cytosporella and D. pimpinellae (saprobic on aerial stems of Pimpinella peregrina in Italy) are conspecific. The ex-type cultures of both species clustered in a well-supported lineage, together with “D. chamaeropis” strains, and are closely related (Fig. 5). The ex-type culture of D. cytosporella (CBS 137020) showed the following nucleotide similarities with the sequences of the ex-type culture of D. pimpinellae (JZB 320131). On ITS: 98.44 % (506/514, including 2 gaps). On tef1: 98.63 % (289/293, including 2 gaps). On tub2: 99.15 % (464/468). On his3: 99.10 % (439/443, including 2 gaps). No cal sequence data is available for the ex-type culture of D. pimpinellae. Morphologically, both species produce similar fusoid to ellipsoidal, hyaline and aseptate alpha conidia, which are somewhat longer in D. cytosporella (8–9 × 2.6–3.2 μm) (Udayanga et al. 2014b) than D. pimpinellae (6–8 × 2–3 μm) (Yuan et al. 2020). These are also remarkably similar to the alpha conidia reported for “D. chamaeropis” (CBS 454.81) (6–8 × 2 μm) (Gomes et al. 2013) (Table 4). Although few records indicate a saprophytic nature for D. cytosporella, including its report on C. humilis, the species has historically been associated to melanose and stem-end rot diseases of Citrus spp. (Udayanga et al. 2014b). Moreover, D. cytosporella (as D. chamaeropis) has been reported as the causal agent of shoot blight and canker diseases of important fruit and nut crops in California, USA, such as pistachio (Chen et al. 2014), grape vine (Lawrence et al. 2015), almond (Holland et al. 2021) and walnut (Luna et al. 2022).
Diaporthe eucommiae (F.X. Chao & P.K. Chi) Y.H. Gao & L. Cai, IMA Fungus 8: 183. 2017. MycoBank MB 821440.
Basionym: Phomopsis eucommiae F.X. Chao & P.K. Chi [as “eucommi”], Journal of Central-South Forestry College 10: 41. 1990. MycoBank MB 363754.
New synonyms: Phomopsis lagerstroemiae C.Q. Chang et al., Mycosystema 24: 148. 2005. MycoBank MB 344476.
Diaporthe lagerstroemiae (C.Q. Chang et al.) Y.H. Gao & L. Cai, IMA Fungus 8: 183. 2017. MycoBank MB 821444.
Diaporthe eucalyptorum Crous & R.G. Shivas, Persoonia 28: 153. 2012. MycoBank MB 800374.
Diaporthe hongkongensis R.R. Gomes et al., Persoonia 31: 23. 2013. MycoBank MB 802934.
Phomopsis lithocarpi Y.H. Gao et al. [as “lithocarpus”], Mycol. Prog. 13: 115. 2014.
Diaporthe lithocarpi (Y.H. Gao et al.) Y.H. Gao & L. Cai [as “lithocarpus”], Fungal Biol. 119: 306. 2015. MycoBank MB 819872 [Nom. inval., Arts. 41.1, F.5.1 (Shenzhen)].
Diaporthe lithocarpi (Y.H. Gao et al.) Y.H. Gao & L. Cai, Index Austral. Fungi 6: 3. 2023. MycoBank MB 559481.
Diaporthe australiana R.G. Shivas et al., Plant Pathol. 69: 916. 2020. MycoBank MB 833827.
Diaporthe salinicola Dayar., Mycosphere 11: 91. 2020. MycoBank MB 556589 [Nom. illegit., Art. 53.1].
Diaporthe xylocarpi M.S. Calabon & E.B.G. Jones, Bot. Mar. 66: 219. 2023. MycoBank MB 848445.
Diaporthe theobromatis H. Dong et al., Mycosystema 39: 2247. 2020. Fungal Name MB 570757.
Diaporthe fujianensis Jayaward. et al., Mycosphere 12: 451. 2021. MycoBank MB 557997.
Diaporthe fusiformis Jayaward. et al., Mycosphere 12: 451. 2021. MycoBank MB 557998.
Diaporthe rhodomyrti C.M. Tian & Qin Yang, MycoKeys 91: 41. 2022. MycoBank MB 829525.
Diaporthe araliae-chinensis S.Y. Wang et al., Mycosphere 14: 680. 2023. MycoBank MB 845964.
Diaporthe globoostiolata Monkai & Lumyong, J. Fungi 9 (no. 603): 13. 2023. MycoBank MB 900380.
Diaporthe tuyouyouiae Y.P. Tan et al. [as “tuyouyou”], Index Austral. Fungi 6: 3. 2023. MycoBank MB 663080.
Typus: China, Nanxiang, Guangdong Province, on leaves of Eucommia ulmoides (Eucommiaceae), F.X. Cao [holotype as cited by Gao et al. (2017), SCHM 0020].
See Cao & Chi (1990) for illustrations and descriptions of asexual morph. A sexual morph was not reported for the type specimen but was reported under the species name D. xylocarpi (as D. salinicola) (Dayarathne et al. 2020).
Hosts: Reported from more than 25 genera and 30 species in 26 families, including Actinidiaceae (Actinidia deliciosa), Aquifoliaceae (Ilex latifolia), Araliaceae (Aralia chinensis), Arecaceae (Nypa fruticans), Caprifoliaceae (Patrinia sp.), Convolvulaceae (Decalobanthus peltatus), Cupressaceae (Cunninghamia lanceolata), Eucommiaceae (Eucommia ulmoides), Fabaceae (Wisteriopsis reticulata), Fagaceae (Castanopsis carlesii, C. eyrei, Lithocarpus glaber, Quercus glauca), Hamamelidaceae (Macadamia ternifolia), Hydrangeaceae (Dichroa febrifuga), Juglandaceae (Cyclocarya paliurus), Lythraceae (Lagerstroemia indica), Malvaceae (Theobroma cacao), Meliaceae (Xylocarpus sp.), Myrtaceae (Eucalyptus sp., Rhodomyrtus tomentosa), Pentaphylacaceae (Ternstroemia gymnanthera), Poaceae (Miscanthus sinensis), Proteaceae (Loropetalum chinense), Rhizophoraceae (Rhizophora apiculata), Rosaceae (Prunus persica, Pyrus pyrifolia), Rubiaceae (Morinda officinalis), Rutaceae (Citrus grandis, C. reticulata, C. sinensis, C. unshiu), Smilacaceae (Smilax china, S. glabra), Theaceae (Camellia sinensis, Schima superba) and Vitaceae (Vitis vinifera) (Farr & Rossman 2023).
Distribution: Australia, Hong Kong, India, Indonesia, Thailand and Turkey (Farr & Rossman 2023).
Notes: Diaporthe eucommiae was introduced by Cao & Chi (1990) as Phomopsis eucommiae on leaf spots of Eucommia ulmoides in China and was later accommodated in Diaporthe by Gao et al. (2017). Cao & Chi (1990) provided an ITS sequence derived from the holotype specimen of D. eucommiae, but it has been ignored in most studies and taxonomic revisions of Diaporthe. The phylogenetic analyses carried out in the present study revealed that D. eucommiae clustered in a well-supported clade, together with the types of 13 species of Diaporthe, which has been mainly associated with D. hongkongensis (Fig. 4). Diaporthe hongkongensis was introduced by Gomes et al. (2013) on a fruit of Dichroa febrifuga (Hydrangeaceae) in Hong Kong. In the following year, a closely related species to taxa with affiliations to D. arecae was introduced in the same clade by Gao et al. (2014), who isolated P. lithocarpi on leaf spots of Lithocarpus glaber and leaves of other plant host in China. This Phomopsis species was later accommodated in Diaporthe, as D. lithocarpi, by Gao et al. (2015), who established its closely phylogenetic relationship with D. hongkongensis [see fig. 1 in Gao et al. (2015)]. The first inconsistencies within this clade were observed by Gao et al. (2016), who isolated 40 isolates that clustered in a clade together with the types of D. lithocarpi and D. hongkongensis, which were found to be phylogenetically and morphologically indistinguishable. Although D. lithocarpi was made synonymous with D. hongkongensis, the first was invalidly introduced by Gao et al. (2015), who cited the identifier of the basionym (P. lithocarpi) in the protologue. Therefore, D. lithocarpi was introduced without a recognised nomenclatural repository identifier [Nom. inval., Arts. 41.1, F.5.1 (Shenzhen)] and the synonym provided by Gao et al. (2016) has become ineffective. Over the years, several species closely related to D. hongkongensis from various hosts have been introduced, mainly in China, regardless the inconsistencies observed by Gao et al. (2016). For instance, recently Manawasinghe et al. (2021) introduced the new species D. fujianensis and D. fusiformis as closely related to D. eucalyptorum, D. hongkongensis and D. lithocarpi based on a negative PHI test result, as they indicated that branch lengths and divergence times within the clade were indistinguishable. As a result, the clade became unmanageable, with poor species delimitation and several lineages lacking relevant nodal supports. The integrative taxonomic approach carried out here has shown that the entire clade is conspecific. Therefore, following the principle of priority, all 13 species have been assigned to the oldest species epithet, i.e., eucommiae, including D. lithocarpi that has been recently validly published by Tan & Shivas (2023). Diaporthe eucommiae strains tend to show great genetic variability, which has consequently led to an inflation of the speciation events recognised within the clade. As noted by Gao et al. (2016), internal nodes and sub-branches can be found in this clade, indicating that it is possibly undergoing active divergence and speciation. Sequence comparisons between the types of “species” introduced in the D. eucommiae clade for ITS, tef1, tub2, cal and his3 showed 96–99 %, 94–99 %, 95–99 %, 93–100 % and 96–98 % nucleotide similarities, respectively. Thus p-distances of up to 0.04, 0.06, 0.05, 0.06 and 0.04 in ITS, tef1, tub2, cal and his3 sequences, respectively, may be detected when future strains of D. eucommiae are isolated. Morphologically, all “species” in the D. eucommiae clade are virtually indistinguishable, with fusoid or ellipsoidal alpha conidia and filiform, curved or hamate beta conidia, often with considerably overlapping sizes. However, “species” can show morphological variability in the length of alpha and beta conidia, which are often longer than those observed for the type specimen of D. eucommiae. These “species” also present a common absence of gamma conidia (observed only in D. hongkongensis) (Table 4). Considering the morphological data available for the “species” synonymised here, the mean dimensions of the alpha and beta conidia produced by D. eucommiae strains are 5.63–7.71 × 1.87–2.55 μm (L/W = 2.00–4.25) and 15.24–22.56 × 0.98–1.57 μm (L/W = 11.43–21.72), respectively, which clearly express the morphological variability in conidial sizes when compared with the dimensions reported for the type specimen of D. eucommiae (SCHM 0020; alpha and beta conidia dimensions = 3.9–4.7 × 1–1.7 μm and 6.6–14.9 × 0.33–0.66 μm, respectively) (Cao & Chi 1990) (Table 4). Most “species” introduced in the D. eucommiae clade were found to be associated with leaf spots and other woody plant diseases, including branch dieback, cankers, shoot blight and stem-end and fruit rot (e.g., Chen et al. 2023). However, it is yet unresolved whether D. eucommiae is a primary or secondary pathogen and further research is required. Regarding palms, D. eucommiae (as D. hongkongensis) was recovered from the mangrove palm Nypa fruticans in the Andaman Islands, India (Thavamani et al. 2018) (Table 5). This record was excluded from the phylogenetic trees presented here, since only the ITS sequence is available, and it strongly affected the robustness of the trees. Interestingly, D. eucommiae (as D. xylocarpi and D. eucalyptorum) has also been recorded on decaying submerged mangrove wood of Xylocarpus sp. in Thailand (Dayarathne et al. 2020) and as an endophyte on leaves of Rhizophora apiculata (Julia et al. 2022), being one of the few currently known marine species of Diaporthe. Thus, the ecology of D. eucommiae appears to be complex, as the species can colonise both terrestrial and marine habitats, as well as a wide range of plant hosts, which may be related to the high genetic variability detected in the clade. Geographically, D. eucommiae seems to be particularly common in China.
Diaporthe foeniculina (Sacc.) Udayanga & Castl., Persoonia 32: 95. 2014. MycoBank MB 803929. Fig. 23.
Fig. 23.
Diaporthe foeniculina. A–C, F–J, M, Q–S: CDP 1369; D, E, P: CDP 0022; K, N: CDP 1818; L, O: CDP 1947. A–E. Conidiogenous layer. F–I. Conidiophores and conidiogenous cells. J–L. Alpha conidia. M–P. Beta conidia. Q–S. Alpha, beta and gamma (black arrows) conidia. Scale bars: A–F, J–S = 5 μm, G–I = 2.5 μm.
Basionym: Phoma foeniculina Sacc., Michelia 2: 95. 1880. MycoBank MB 234536.
Synonyms: Phomopsis theicola Curzi, Atti Ist. Bot. Lab. Crittog. Univ. Pavia 3: 60. 1927. MycoBank MB 446039.
Diaporthe theicola Curzi, Atti Ist. Bot. Lab. Crittog. Univ. Pavia 3: 64. 1927. MycoBank MB 255097.
Phomopsis foeniculina (Sacc.) Sousa da Câmara, Agron. Lusit. 9: 104. 1947. MycoBank MB 492792.
Diaporthe neotheicola A.J.L. Phillips & J.M. Santos, Fungal Divers. 34: 120. 2009. MycoBank MB 512257.
Diaporthe rhoicola Crous [as “rhusicola”], Persoonia 26: 135. 2011. MycoBank MB 827844.
Diaporthe baccae L. Lombard et al., Phytopathol. Mediterr. 53: 93. 2014. MycoBank MB 807599.
Diaporthe ravennica Thambug. et al., Fungal Divers. 82: 296. 2016. MycoBank MB 552100.
New synonyms: Diaporthe rumicicola Manawas. et al., Fungal Divers. 96: 138. 2019. MycoBank MB 555379.
Diaporthe nigra Brahman. & K.D. Hyde, Fungal Divers. 100: 185. 2020. MycoBank MB 556927.
Diaporthe zaobaisu Y.S. Guo & G.P. Wang, Persoonia 45: 156. 2020. MycoBank MB 830660.
Typus: Italy, on Camellia sinensis (Theaceae), M. Curzi (epitype of Diaporthe theicola, dried culture specimen, designated by Udayanga et al. (2014b), BPI 892462, ex-epitype culture CBS 187.27, same as ex-isotype culture of Phomopsis theicola; lectotype of Phomopsis theicola designated by Udayanga et al. (2014b), MBT 175964). France, on stems of Foeniculum “arvensis” (Apiaceae), Brunaud (holotype of Phoma foeniculina cited in Phillips (2003) with illustration, PAD 281). Portugal, Évora, on Foeniculum vulgare (Apiaceae), Nov. 2007, A.J.L. Phillips (holotype of Diaporthe neotheicola CBS-H 20131, ex-type culture CBS 123208); Madeira, Serra da Agua, at base of 2-yr-old stem of Foeniculum vulgare (Apiaceae), Aug. 2001, A.J.L. Phillips (epitype of Phoma foeniculina designated by Udayanga et al. (2014b), LISE 94791, ex-epitype culture CBS 111553). USA, California, Santa Barbara County, on dead outer bark and decaying fruit of Citrus limonia (Rutaceae), 3 Mar. 1922, H.S. Fawcett (holotype of Phomopsis californica, BPI 0358313); California, San Diego, on branch of Citrus limonia (Rutaceae), 16 Nov. 2012, A. Eskalen (epitype of Phomopsis californica designated by Udayanga et al. (2014b), BPI 892460, ex-epitype culture CBS 135430).
See Udayanga et al. (2014b) for illustrations and description of asexual morph (ex-epitype culture of P. foeniculina). See Santos & Phillips (2009) for illustrations and description of asexual and sexual morphs (ex-type culture of D. neotheicola).
Isolate CDP 1369. Sexual morph: Undetermined. Asexual morph: Conidiomata on palm leaflets in culture pycnidial, subglobose to pulvinate, non-stromatic, uniloculate, black, solitary or aggregated, semi-immersed, covered with whitish and yellowish mycelium, exuding a yellowish mucoid mass of conidia, up to 360 μm diam. Conidiophores formed from the cells lining the locule wall, rarely reduced to conidiogenous cells, hyaline to subhyaline, smooth- and thin-walled, doliiform to subcylindrical, straight, sometimes curved, aseptate or 1–2-septate, unbranched or sometimes producing a lateral branch that becomes conidiogenous. Conidiogenous cells rising from conidiophores, sometimes in groups of 2 conidiogenous cells, hyaline, smooth- and thin-walled, discrete, determinate, cylindrical, tapering towards the apex, occasionally with a broader, globose or irregular base, straight, rarely curved or slightly curved, occasionally with small guttules, enteroblastic, proliferating at the same level giving rise to periclinal thickenings, (9.70–)10.10–20.90(–27.06) × 1.48–3.36 μm, 95 % confidence limits = 14.62–16.98 × 2.03–2.26 μm (mean ± SD = 15.80 ± 4.46 × 2.14 ± 0.43 μm, n = 55). Alpha conidia fusiform, tapering towards the base, acute to subacute base, often subtruncate, obtuse to subobtuse apex, smooth- and thin-walled, hyaline, aseptate, eguttulate, often with few granular contents, 6.41–8.89(–9.46) × 1.72–2.65 μm, 95 % confidence limits = 7.61–7.77 × 2.14–2.19 μm (mean ± SD = 7.69 ± 0.51 × 2.16 ± 0.18 μm), mean ± SD conidium length/width ratio = 3.58 ± 0.41 (n = 150). Beta conidia filiform, hamate, sometimes slightly sigmoid, rarely cylindrical and wider from the upper third towards the base, base rounded or truncate, apex subacute and tapered, smooth- and thin-walled, hyaline, aseptate, eguttulate, (17.26–)20.88–27.44(–29.23) × 1.05–1.99 μm, 95 % confidence limits = 23.65–24.98 × 1.29–1.40 μm (mean ± SD = 24.32 ± 2.39 × 1.35 ± 0.18 μm), mean ± SD conidium length/width ratio = 18.44 ± 3.25 (n = 50). Gamma conidia infrequent, fusiform, acute at both ends, slightly truncate base, hyaline, smooth- and thin-walled, aseptate, eguttulate, 9.91–14.82 × 1.42–2.04 μm, 95 % confidence limits = 12.09–13.04 × 1.73–1.84 μm (mean ± SD = 12.57 ± 1.35 × 1.79 ± 0.15 μm), mean ± SD conidium length/width ratio = 7.12 ± 1.21 (n = 30).
Culture characteristics: Colonies on 1/2 PDA, reaching 55 mm diam after 7 d at 20 °C in darkness. Surface flat, with sparse aerial mycelium, with filamentous to filiform margin, irregular shape, white, becoming luteous white, with white tufts, opaque. Reverse ivory to luteus white. No diffusible pigment. Conidiomata black, abundant, of variable shapes and dimensions, solitary or aggregated in numerous groups, developing scattered on the surface in about 1 wk.
Materials examined: Portugal, Lisbon, Parque das Nações, Jardins Garcia d’Orta, Talhão do Colante, on foliar lesions of segments of Trachycarpus fortunei (Arecaceae), 5 Oct. 2018, D.S. Pereira (specimen HDP 013/02), living culture CDP 0022 (ITS sequence MT000992, tef1 sequence MN990208, tub2 sequence MN990209); Marvila, Ferreira de Castro Street, near Casa dos Direitos Sociais, on foliar lesions of segments of Chamaerops humilis (Arecaceae), 13 Oct. 2018, D.S. Pereira (specimen HDP 025/02), living culture CDP 0209 (ITS sequence MT004913, tef1 sequence MT011071, tub2 sequence MT011077); Parque das Nações, Jardins da Água, near Oceanário de Lisboa, on foliar lesions of segments of C. humilis (Arecaceae), 16 Oct. 2018, D.S. Pereira (specimen HDP 035/02), living culture CDP 0315 (ITS sequence MT004914, tef1 sequence MT011072, tub2 sequence MT011078); Parque das Nações, on foliar lesions of leaflets of Chrysalidocarpus lutescens, 8 May 2021, D.S. Pereira (specimen HDP 087), living cultures CDP 1321 (ITS sequence PP577994, tef1 sequence PP579319, tub2 sequence PP579334), CDP 1322 (ITS sequence PP577995); Parque das Nações, on foliar lesions of leaflets of Phoenix roebelenii, 8 May 2021, D.S. Pereira (specimen HDP 088), living cultures CDP 1369 (ITS sequence PP577996), CDP 1371 (ITS sequence PP577997); Parque das Nações, on foliar lesions of segments of C. humilis, 8 May 2021, D.S. Pereira (specimen HDP 091), living culture CDP 1575 (ITS sequence PP577998, tef1 sequence PP579320, tub2 sequence PP579335); Parque das Nações, on foliar lesions of segments of Washingtonia filifera, 8 May 2021, D.S. Pereira (specimen HDP 095), living cultures CDP 1816 (ITS sequence PP577999), CDP 1817 (ITS sequence PP578000, tef1 sequence PP579321, tub2 sequence PP579336), CDP 1818 (ITS sequence PP578001); Parque das Nações, on foliar lesions of segments of W. filifera, 8 May 2021, D.S. Pereira (specimen HDP 097), living culture CDP 1908 (ITS sequence PP578002, tef1 sequence PP579322, tub2 sequence PP579337); Parque das Nações, on foliar lesions of segments of C. humilis, 8 May 2021, D.S. Pereira (specimen HDP 098), living culture CDP 1947 (ITS sequence PP578003, tef1 sequence PP579323, tub2 sequence PP579338); Parque das Nações, on foliar lesions of segments of C. humilis, 8 May 2021, D.S. Pereira (specimen HDP 099), living culture CDP 2028 (ITS sequence PP578004); Parque das Nações, on foliar lesions of segments of C. humilis, 8 May 2021, D.S. Pereira (specimen HDP 100), living culture CDP 2056 (ITS sequence PP578005, tef1 sequence PP579324, tub2 sequence PP579339).
Hosts: Reported from more than 55 genera and 80 species in 37 families, including Actinidiaceae (Actinidia deliciosa), Amaranthaceae (Chenopodium sp.), Anacardiaceae (Pistacia vera, Searsia pendulina), Apiaceae (Angelica sylvestris, Foeniculum vulgare), Arecaceae (Chamaerops humilis, Chrysalidocarpus lutescens, Phoenix roebelenii, Trachycarpus fortunei, Washingtonia filifera), Asparagaceae (Asparagus sp.), Asphodelaceae (Hemerocallis fulva), Asteraceae (Achillea millefolium, Arctium minus), Betulaceae (Corylus avellana), Brassicaceae (Lunaria rediviva), Cupressaceae (Cupressus sempervirens), Ebenaceae (Diospyros kaki), Ericaceae (Vaccinium corymbosum), Euphorbiaceae (Euphorbia pulcherrima), Fabaceae (Acacia sp., Amorpha fruticosa, Aspalathus linearis, Cyclopia genistoides, C. longifolia, C. subternata, Glycine max, Melilotus officinalis, Paraserianthes lophantha, Robinia pseudoacacia, Wisteria sinensis), Fagaceae (Castanea sativa, Quercus robur, Q. suber), Grossulariaceae (Ribes nigrum), Hydrangeaceae (Hydrangea macrophylla), Juglandaceae (Juglans regia), Lamiaceae (Ballota nigra, Salvia sp.), Lauraceae (Persea americana), Moraceae (Ficus benjamina, F. carica), Myrtaceae (Eucalyptus camaldulensis), Nyctaginaceae (Bougainvillea spectabilis), Oleaceae (Olea europaea), Onagraceae (Fuchsia excorticata), Platanaceae (Platanus hispanica, P. orientalis), Polygonaceae (Rumex sp.), Proteaceae (Protea sp.), Rosaceae (Malus domestica, Prunus amygdalus, P. armeniaca, P. avium, P. persica, Pyrus bretschneideri, P. communis, P. pyrifolia, Rosa canina, Rubus sp.), Rutaceae (Citrus aurantiifolia, C. australasica, C. bergamia, C. japonica, C. latifolia, C. limon, C. limonia, C. maxima, C. medica, C. mitis, C. paradisi, C. reticulata, C. sinensis), Salicaceae (Salix sp.), Sapindaceae (Acer negundo), Simaroubaceae (Ailanthus altissima), Tamaricaceae (Tamarix sp.), Theaceae (Camellia sinensis) and Vitaceae (Vitis vinifera) (Farr & Rossman 2023, present study).
Distribution: Australia, Chile, China, Croatia, Cyprus, France, Germany, Greece, Iran, Italy, Lebanon, Malta, Netherlands, New Zealand, Portugal, Serbia, South Africa, Spain, Turkey, Uruguay and USA (Farr & Rossman 2023).
Notes: Diaporthe foeniculina was introduced by Saccardo (1880) as Phoma foeniculina on stems of Foeniculum “arvensis” in France and was later transferred to Phomopsis by Sousa da Câmara (1947). Udayanga et al. (2014b) resolved the status of the species by accommodating it in Diaporthe and providing an epitype specimen for Phoma foeniculina. They recognised its wide host range and its importance as a pathogen of Citrus spp. Furthermore, molecular data derived from the epitype of Phoma foeniculina showed that this taxon was conspecific with the ex-type isolates of D. neotheicola and P. theicola, names that have been widely used to refer to D. foeniculina (Udayanga et al., 2014b). Many other possible synonyms have been reviewed and discussed by Udayanga et al. (2014b) based on available molecular data, living cultures and type specimens. However, several lineages closely related to D. foeniculina have been introduced over the years, impairing the interpretation of its species limits. Recently, Hilário et al. (2021c) synonymised D. baccae and D. ravennica under D. foeniculina. The integrative taxonomic approach carried out in the present study confirmed the synonyms provided by Hilário et al. (2021c) and revealed three new synonyms in the D. foeniculina clade, namely D. rumicicola (Hyde et al. 2019), D. nigra (Hyde et al. 2020a) and D. zaobaisu (Guo et al. 2020). The three species have been introduced as taxa closely related but phylogenetically distinct from D. foeniculina based on minor morphological and molecular differences. However, the phylogenetic analyses carried out here showed that they are intermingled in the D. foeniculina clade, lacking relevant phylogenetic distinctiveness (Fig. 5). The ex-epitype culture of D. foeniculina (CBS 111553) showed the following nucleotide similarities with the sequences of the ex-type cultures of D. rumicicola (MFLUCC 18-1566), D. nigra (JZB 320170) and D. zaobaisu (CGMCC 3.19598). On ITS: 99.81 % (534/535, including 1 gap), 99.62 % (523/525, including 1 gap) and 97.82 % (493/504, including 2 gaps), respectively. On tef1: 99.64 % (274/275, including 1 gap), 98.97 % (288/291, including 3 gaps) and 88.28 % (256/290, including 31 gaps), respectively. On tub2: 100 % (413/413, including 1 gap), 99.57 % (465/467) and 99.40 % (499/502, including 1 gap), respectively. No cal sequence data is available for the ex-type cultures of the three species. The his3 sequence data is available only for the ex-type cultures of D. zaobaisu and have the following nucleotide similarities with the his3 sequence of the ex-epitype culture of D. foeniculina: 98.64 % (435/441). Only tef1 sequence of D. zaobaisu shows a substantial number of nucleotide differences to that of D. foeniculina (p-distance = 0.12), which implies its clustering on longer branches in the D. foeniculina clade (Fig. 5). However, most of these differences correspond to two long gap regions in the beginning and end of the tef1 partial sequence (equivalent to 31 of the 34 nucleotide differences between the two sequences) and only three nucleotides appear to represent fixed alleles. Morphologically, all “species” previously described in the D. foeniculina clade are virtually indistinguishable with highly overlapping micromorphological characters: fusoid to ellipsoidal alpha conidia with a frequently subtruncate base and a common presence of filiform, hamate or curved beta conidia with dimensions lying within the same ranges (Table 4). The only exception is D. nigra, whose alpha conidia are considerably larger (17–28 × 7–7.5 μm) (Hyde et al. 2020a) than those reported for other “species” in the D. foeniculina clade (Table 4). Nonetheless, the size of the alpha conidia of D. nigra are atypical when compared to the size of alpha conidia usually reported for Diaporthe spp. Moreover, the size ranges reported for the alpha conidia of D. nigra are not in line with what can be observed on the photoplate presented along with its taxonomic description (fig. 115e,f in Hyde et al. 2020a, given the scale bars shown), so the size ranges reported appear to be unreliable. Considering the morphological data available for the type specimens of “species” in the D. foeniculina clade (excluding D. nigra for the reasons detailed above), the mean dimensions of the alpha and beta conidia produced by D. foeniculina strains are 6.69–8.43 × 2.17–2.80 μm (L/W = 1.60–3.06) and 22.04–27.74 × 1.12–1.54 μm (L/W = 14.67–26.20), respectively, which clearly overlap with the dimensions reported for the ex-epitype culture of D. foeniculina (CBS 111553; mean = 8.8 × 2.4 μm for alpha conidia and 25.1 × 1.5 μm for beta conidia) (Table 4). Thus, except for absence of the production of beta conidia in D. rhoicola (Crous et al. 2011a), D. nigra (Hyde et al. 2020a) and D. rumicicola (Hyde et al. 2019), the morphology of the asexual morph of all D. foeniculina strains match the description provided by Udayanga et al. (2014b). Fifteen isolates of D. foeniculina were obtained from foliar lesions of five different species of palms in Lisbon, Portugal, including six isolates from Chamaerops humilis, four from Washingtonia filifera, two from Chrysalidocarpus lutescens and Phoenix roebelenii, and one from Trachycarpus fortunei. Isolates from distinct palm hosts were tentatively characterised to assess micromorphological variation. However, despite numerous attempts with different temperatures and with different organic material introduced on the agar surface, isolates from C. lutescens remained sterile even after long periods of incubation of about 2 mo. Isolates from the other palm hosts were morphologically similar to the ex-epitype of D. foeniculina (Udayanga et al. 2014b) (Fig. 23). Considering the strain characterised here (CDP 1369) and the ex-epitype strain of D. foeniculina (CBS 111553), both produce globose to subglobose, black pycnidial conidiomata, exuding yellowish masses of fusiform, hyaline and aseptate alpha conidia and hamate, hyaline and aseptate beta conidia of similar dimensions (mean = 7.69 × 2.16 μm and 24.32 × 1.35 μm, respectively, for CDP 1369, and 8.8 × 2.4 μm and 25.1 × 1.5, respectively, for CBS 111553) (Udayanga et al. 2014b). Sequence comparisons with the ex-epitype of D. foeniculina (CBS 111553) for ITS, tef1 and tub2 showed 99.24–100 %, 99.35 % and 99.41–100 %, respectively, sequence similarity and differences are represented by few base pair changes and gaps. The nucleotide sequence similarity between the D. foeniculina isolates obtained here was either 100 % or nearly so for all three loci. No cal and his3 sequence data are available for these isolates. Diaporthe foeniculina has not previously been recorded on Arecaceae and thus five new host records are reported here (Table 5). The isolates of D. foeniculina studied here were recorded from foliar lesions of palms, but pathogenicity has not been tested. Despite being commonly recorded as a saprophyte on dead plant material from different hosts, D. foeniculina has also been reported as the causal agent of cankers, dieback and fruit rot in numerous crops, including almond, avocado, citrus, chestnut and grapevine (e.g., Makris et al. 2022). Therefore, the geo-ecological data available for D. foeniculina suggest that this species has a widespread distribution in both temperate and tropical regions as a pathogen and saprophyte in a wide range of herbaceous weeds and woody fruit crops.
Diaporthe glabrae (C.Q. Chang et al.) Y.H. Gao & L. Cai, IMA Fungus 8: 183. 2017. MycoBank MB 821443.
Basionym: Phomopsis glabrae C.Q. Chang et al., Mycosystema 24: 8. 2005. MycoBank MB 344454.
New synonyms: Diaporthe tectonae Doilom et al., Fungal Divers. 82: 163. 2016. MycoBank MB 551976.
Diaporthe alangii C.M. Tian & Q. Yang, MycoKeys 39: 118. 2018. MycoBank MB 824704.
Diaporthe hubeiensis Dissan. et al., Front. Microbiol. 10 (no. 1936): 20. 2019. MycoBank MB 552579.
Diaporthe chinensis N.I. de Silva et al., Mycosphere 12: 188. 2021. MycoBank MB 556135 [Nom. illegit., Art. 53.1].
Diaporthe xishuangbannaensis Hongsanan & K.D. Hyde, Mycosphere 14: 1140. 2023. MycoBank MB 900765.
Diaporthe conferta H. Dong et al., Mycosystema 40: 442. 2021. MycoBank MB 570778.
Diaporthe morindae M. Luo et al., J. Fungi 8(no. 806): 17. 2022. MycoBank MB 553409.
Typus: China, Putian, Fujian Province, on living branches of Bougainvillea glabra (Nyctaginaceae), Sep. 2001, Y-H. Cheng (holotype of Phomopsis glabrae, SCHM 3622).
See Chang et al. (2005b) for illustrations and descriptions of asexual morph. Sexual morph not reported.
Hosts: Actinidia chinensis, A. deliciosa, Actinidia sp. (Actinidiaceae), Alangium kurzii (Cornaceae), Bougainvillea glabra (Nyctaginaceae), Calamus castaneus, C. thwaitesii (Arecaceae), Camellia oleifera (Theaceae), Elaeagnus conferta (Elaeagnaceae), Litchi chinensis (Sapindaceae), Morinda officinalis (Rubiaceae), Rhizophora apiculata (Rhizophoraceae), Tectona grandis (Lamiaceae), Vigna unguiculata (Fabaceae) and Vitis vinifera (Vitaceae) (Farr & Rossman 2023, present study).
Distribution: China, India, Indonesia, Korea, Malaysia, Sri Lanka, Taiwan, Thailand (Farr & Rossman 2023, present study).
Notes: Diaporthe glabrae was introduced by Chang et al. (2005b) as P. glabrae on branches of Bougainvillea glabra in China and was later accommodated in Diaporthe by Gao et al. (2017). Chang et al. (2005b) provided an ITS sequence derived from the holotype specimen of D. glabrae, but it has been ignored in most studies and taxonomic revisions of Diaporthe. The phylogenetic analyses carried out in the present study revealed that D. glabrae clustered in a well-supported clade, together with the types of five species of Diaporthe (Fig. 6). Branch lengths and divergence times within the clade are virtually indistinguishable and all species were introduced based on minor morphological differences and small nucleotide dissimilarities. The integrative taxonomic approach carried out here has shown that the entire clade is conspecific. Therefore, following the principle of priority, all five species have been assigned to the oldest species epithet, i.e., glabrae. Sequence comparisons between the types of “species” introduced in the D. glabrae clade for ITS, tef1, tub2, cal and his3 showed 99–100 %, 99–100 %, 97–99 %, 98–100 % and 99–100 % nucleotide similarities, respectively. Morphologically, all “species” in the D. glabrae clade are virtually indistinguishable, producing oval to ellipsoidal alpha conidia, mostly with bluntly rounded apex and obtuse base, and filiform, hamate beta conidia of overlapping sizes. These “species” also present a common absence of gamma conidia (observed only in D. tectonae) (Table 4). Considering the morphological data available for the “species” synonymised here, the mean dimensions of the alpha and beta conidia produced by D. glabrae strains are 5.43–7.08 × 1.77–2.77 μm (L/W = 2.18–3.63) and 14.38–22.00 × 0.98–1.43 μm (L/W = 7.83–22.50), respectively, which clearly overlap with the dimensions reported for the type of D. glabrae (SCHM 3622; mean = 5–7.4 × 1.6–2.1 μm for alpha conidia and 13.5–22.5 × 0.6–1 μm for beta conidia) (Table 4). Thus, except for absence of the production of beta conidia in D. alangii (Yang et al. 2018) and D. conferta (Dong et al. 2021a), the morphology of the asexual morph of all D. glabrae strains match the description provided by Chang et al. (2005b). Another species, D. morindae (Luo et al. 2022), was confirmed to belong to the D. glabrae clade but was not included in the primary phylogenetic analyses carried out here. Further analyses have shown, however, that it is conspecific with taxa in the D. glabrae clade (data not shown) and are, thus, treated as synonym. Most “species” introduced in the D. glabrae clade were found to be associated with leaf spots and other plant diseases, including dieback, shoot and leaf blight (e.g., Du et al. 2021, Nair et al. 2021). Few reports have also noted D. glabrae as an endophyte (e.g., Julia et al. 2022), so it is yet unresolved whether this species is a primary or a latent or secondary pathogen and further research is required. Regarding palms, D. glabrae (as D. tectonae) has been isolated as an endophyte on Calamus castaneus spines in Malaysia (Azuddin et al. 2021), as well as an endophyte (strain RDWW-10) on C. thwaitesii in Sri Lanka (Dissanayake et al. 2016). The later record was excluded from the phylogenetic trees presented here, since only the ITS sequence is available, and it strongly affected the robustness of the trees. Geographically, D. glabrae seems to be particularly common in China and their records are apparently restricted to Asian countries.
Diaporthe inconspicua R.R. Gomes et al., Persoonia 31: 23. 2013. MycoBank MB 802936.
New synonyms: Diaporthe pseudoinconspicua T.G.L. Oliveira et al., Persoonia 40: 275. 2018. MycoBank MB 824820.
Diaporthe lutescens S.T. Huang et al., MycoKeys 77: 81. 2021. MycoBank MB 837597.
Diaporthe samaneae Monkai & Lumyong, J. Fungi 9 (no. 603): 14. 2023. MycoBank MB 900381.
Typus: Brazil, on petiole of Maytenus truncata (as Maytenus ilicifolia) (Celastraceae), Jul. 2007, R.R. Gomes (holotype CBS H-21102, ex-type culture CBS 133813).
No sporulation has been observed for the type specimen of D. inconspicua and the species was introduced based solely on phylogenetic analyses (Gomes et al. 2013). However, its morphological description has since been emended by Bezerra et al. (2018), who provided illustrations and descriptions of the asexual morph.
Hosts: Anacardium occidentale (Anacardiaceae), Atriplex nummularia (Amaranthaceae), Brosimum gaudichaudii (Moraceae), Cenostigma pyramidale (Fabaceae), Chrysalidocarpus lutescens (Arecaceae), Copaifera langsdorffii (Fabaceae), Cucumis melo (Cucurbitaceae), Cyclopia longifolia (Fabaceae), Magnolia sp. (Magnoliaceae), Monteverdia truncata (Celastraceae), Pterocarpus erinaceus, P. indicus (Fabaceae), Pyrus communis (Rosaceae), Samanea saman (Fabaceae), Spondias mombin (Anacardiaceae) and Vellozia gigantea (Velloziaceae) (Farr & Rossman 2023).
Distribution: Brazil, China, Costa Rica, South Africa, Thailand, Togo and Uruguay (Farr & Rossman 2023).
Notes: Subclade of D. inconspicua included the types of D. inconspicua, D. lutescens, D. pseudoinconspicua and D. samaneae, as well as two strains recognised as D. pterocarpi, which grouped with relevant nodal support and are phylogenetically closely related (Fig. 8). Although some substructure and well-supported groupings can be observed, the integrative taxonomic approach carried out has shown that the entire clade is conspecific and therefore all the corresponding species are treated as synonyms. However, it is likely that the clade is undergoing active divergence and speciation. Diaporthe pterocarpi was introduced by Hughes (1953) as Phomopsis pterocarpi on leaves of Pterocarpus erinaceus in Togo (West Africa). This species was later placed in Diaporthe by Udayanga et al. (2012b), who selected a fresh collection from Pterocarpus indicus in Thailand to serve as an epitype for P. pterocarpi based on comparisons with an isotype specimen (PDD 14878). Udayanga et al. (2012b) examined both specimens for symptoms on the host and micromorphology and considered them to be the same species. However, the epitypification provided by the authors was not Code compliant as the supported holotype, lectotype or neotype was not explicitly cited as required by Art. 9.9 (Shenzhen). In addition, the isolate from Thailand is morphologically distinct from the holotype of P. pterocarpi from Togo (conidiomata 65–100 × 100 μm, conidiophores 10–15 × 1–2 μm, and conidia (5–)6–7(–9) × (2–)2.5(–3) μm vs conidiomata 100 × 200 μm, conidiophores 10 × 2–3 μm, and conidia 6–9 × 2.5–3 μm, respectively) (Hughes 1953, Udayanga et al. 2012). Thus, the decision to select the material from Thailand as the epitype of P. pterocarpi is questionable, given that the only morphological similarities are in their conidial dimensions, and the countries are also geographically distant from one another. Therefore, following the principle of priority, the entire clade has been assigned to the oldest valid species epithet, i.e., inconspicua. Despite the strains “D. pterocarpi” MFLUCC 10-0572 and MFLUCC 10-0575 being assigned to D. inconspicua, the species epithet pterocarpi could not be made a synonym, as the epitypification provided by Udayanga et al. (2012b) for D. pterocarpi has become ineffective. Similar results have been recently proposed by Ferro et al. (2024), who isolated new endophytic strains of D. inconspicua in Brazil. The ex-type culture of D. inconspicua (CBS 133813) showed the following nucleotide similarities with the sequences of the ex-type cultures of D. lutescens (SAUCC 194.36), D. pseudoinconspicua (URM 7874), D. samaneae (SDBR-CMU470) and the strain “D. pterocarpi” MFLUCC 10-0571. On ITS: 94.41 % (507/537, including 7 gaps), 93.06 % (429/461, including 11 gaps), 96.34 % (553/574, including 9 gaps) and 94.63 % (476/503, including 6 gaps), respectively. On tef1: 95.74 % (270/282, including 7 gaps), 94.89 % (223/235, including 5 gaps), 96.70 % (293/303, including 5 gaps) and 96.70 % (293/303, including 5 gaps), respectively. On tub2: 99.49 % (390/392, including 2 gaps), 99.47 % (377/379), 100 % (423/423) and 100 % (349/349), respectively. On cal: 98.03 % (399/407, including 3 gaps), 98.31 % (408/415, including 4 gaps), 98.24 % (390/397, including 1 gap) and 94.90 % (409/431, including 3 gaps), respectively. On his3: 99.05 % (419/423), 99.38 % (478/481) and 99.16 % (475/479), respectively. No his3 sequence data is available for “D. pterocarpi” MFLUCC 10-0571. Most relevant nucleotide dissimilarities correspond to polymorphisms in the ITS1 region or sets of gaps in the other loci. Diaporthe inconspicua was introduced for endophytic isolates based on the diagnosis of sequence data, since the isolates (including the ex-type strain CBS 133813) were sterile in culture (Gomes et al. 2013). However, the morphological description of D. inconspicua has been emended by Bezerra et al. (2018) based on a set of endophytic isolates phylogenetically related to D. inconspicua. Therefore, the description provided by Bezerra et al. (2018) was considered here as representative of the morphology of D. inconspicua (Table 4). Morphologically, all “species” in the D. inconspicua clade are similar, producing fusoid to ellipsoidal alpha conidia with some overlap in size and showing a common absence of beta conidia (Table 4). Considering the morphological data available for the “species” synonymised here, the mean dimensions of the alpha and beta conidia produced by D. inconspicua strains are 5.88–8.00 × 1.95–2.45 μm (L/W = 2.60–3.91) and 19.60–25.27 × 1.07–1.67 μm (L/W = 15.50–18.40), respectively. Alpha conidia mean dimensions clearly overlap with the dimensions reported by Bezerra et al. (2018) for D. inconspicua (URM 7775; 5.5–6.5 × 1.5–2 μm) (Table 4). Diaporthe inconspicua has been mostly isolated as an endophyte of medicinal and other plants in Brazil, as well as a leaf spotting fungus (e.g., Gomes et al. 2013, Ferreira et al. 2017, Crous et al. 2018, de Carvalho et al. 2021, Ferro et al. 2024). Therefore, the ecological data available for D. inconspicua suggests that it is either an endophyte or a secondary pathogen. Recently, D. inconspicua was isolated from pear trees in Uruguay by Sessa et al. (2018), who considered the species to possibly represent a true endophyte based on laboratory pathogenicity tests. However, D. inconspicua has also been associated with shoot blight of Atriplex nummularia in Brazil (dos Santos Freire 2017) and dieback of Cyclopia longifolia in South Africa (Smit et al. 2021), so data regarding its pathogenicity, host range and aetiology remains preliminary and unknown. Diaporthe inconspicua is an uncommon species with few scattered reports from different regions of the world, which may explain the genetic variability observed in some strains. In relation to palms, D. inconspicua (as D. lutescens) has been isolated in association with leaf spots of Chrysalidocarpus lutescens in China (Sun et al. 2021) (Table 5).
Diaporthe leucospermi Crous & Summerell, Persoonia 27: 32. 2011. MycoBank MB 560561. Fig. 24.
Fig. 24.
Diaporthe leucospermi (CDP 0052). A–C. Conidiogenous layer. D–H. Conidiophores and conidiogenous cells. I, J. Alpha conidia. K, L. Beta conidia. Scale bars: A–C, I–L = 5 μm, D–H = 2.5 μm.
Synonyms: Diaporthe pyracanthae L. Santos & A. Alves, Mycosphere 8: 493. 2017. MycoBank MB 820224.
Diaporthe rossmaniae S. Hilário et al., Mycologia 112: 302. 2020. MycoBank MB 831452.
New synonym: Diaporthe infecunda R.R. Gomes et al., Persoonia 31: 24. 2013. MycoBank MB 802937.
Typus: Australia, New South Wales, the Blue Mountains Botanic Gardens, Mount Tomah, on leaves of Leucospermum sp. (Proteaceae), Aug 1999, P.W. Crous & B. Summerell (holotype CBS H-20674, culture ex-type CBS 111980).
See Crous et al. (2011b) for illustrations and descriptions of asexual morph. Sexual morph not reported.
Isolate CDP 0052. Sexual morph: Undetermined. Asexual morph: Conidiomata on palm leaflets in culture pycnidial, subglobose to pyriform, stromatic, uniloculate, dark-brown to black, solitary, superficial, mostly glabrous, often covered with hyphal outgrows, exuding a pearl mucoid mass of conidia, up to 600 μm diam. Conidiophores formed from the cells lining the locule wall, often reduced to conidiogenous cells, hyaline to subhyaline, smooth-and thin-walled, doliiform to subcylindrical, straight, aseptate or 1-septate, unbranched or sometimes producing a lateral branch below the septum that becomes conidiogenous. Conidiogenous cells hyaline, smooth- and thin-walled, discrete, determinate, cylindrical, occasionally ampulliform, tapering towards the apex, straight or curved, aseptate, unbranched,, with minute, sometimes conspicuous, collarette, enteroblastic, proliferating at the same level giving rise to periclinal thickenings, 5.9–22.6 × 1.5–3.4 μm, 95 % confidence limits = 11.58–13.50 × 1.97–2.20 μm (mean ± SD = 12.54 ± 3.47 × 2.09 ± 0.42 μm, n = 50). Alpha conidia fusiform to ellipsoidal, tapering towards the base, apex rounded, base subobtuse, sometimes with both ends rounded, smooth- and thin-walled, hyaline, aseptate, mostly biguttulate, with a big, conspicuous guttule at each end, 5–6.4 × 1.8–2.9 μm, 95 % confidence limits = 5.67–5.86 × 2.33–2.45 μm (mean ± SD = 5.77 ± 0.34 × 2.39 ± 0.22 μm), mean ± SD conidium length/width ratio = 2.43 ± 0.26 (n = 50). Beta conidia spindle-shaped, mostly hooked at the apex, base truncate, apex acute and tapered, smooth- and thin-walled, hyaline, aseptate, eguttulate, 21.6–32.1 × 0.9–1.8 μm, 95 % confidence limits = 26.94–28.00 × 1.23–1.33 μm (mean ± SD = 27.47 ± 1.91 × 1.28 ± 0.17 μm), mean ± SD conidium length/width ratio = 21.82 ± 3.34 (n = 50). Gamma conidia very infrequent, fusiform, smooth- and thin-walled, hyaline, aseptate, biguttulate.
Culture characteristics: Colonies on 1/2 PDA, reaching 55 mm diam after 7 d at 20 °C in darkness. Surface flat, with sparse aerial mycelium in raised concentric circles, with filamentous to filiform margin, circular to irregular shape, white, pale brown to yellowish towards the centre, opaque. Reverse luteus, buff to marron towards the centre. No diffusible pigment. Conidiomata black, developing scattered on the surface in about 1 wk.
Materials examined: Portugal, Lisbon, Parque das Nações, Jardins Garcia d’Orta, Talhão do Coloane, on foliar lesions of segments of Chamaerops humilis (Arecaceae), 16 Oct. 2018, D.S. Pereira (specimen HDP 039/02), living culture CDP 0052 (ITS sequence MT002843, tef1 sequence MT011070, tub2 sequence MT011076, cal sequence MT011066); Alvalade, on foliar lesions of leaflets of Chrysalidocarpus lutescens (Arecaceae), 9 Apr. 2021, D.S. Pereira (specimen HDP 082), living culture CDP 1211 (ITS sequence PP578006, tub2 sequence PP579340, cal sequence PP579350); Alvalade, on foliar lesions of leaflets of C. lutescens (Arecaceae), 9 Apr. 2021, D.S. Pereira (specimen HDP 084), living culture CDP 1251 (ITS sequence PP578007, tef1 sequence PP579325, tub2 sequence PP579341, cal sequence PP579351); Parque das Nações, on foliar lesions of segments of C. humilis (Arecaceae), 8 May 2021, D.S. Pereira (specimen HDP 0100), living culture CDP 2055 (ITS sequence PP578008, tef1 sequence PP579326, tub2 sequence PP579342).
Hosts: Acer negundo (Aceraceae), Astragalus mongholicus (Fabaceae), Chamaerops humilis, Chrysalidocarpus lutescens (Arecaceae), Coffea arabica (Rubiaceae), Diospyros kaki (Ebenaceae), Eucalyptus globulus, Eugenia uruguayensis (Myrtaceae), Hydrangea macrophylla (Hydrangeaceae), Justicia brandegeeana (Acanthaceae), Leucospermum sp. (Proteaceae), Maytenus truncata (Celastraceae), Passiflora edulis (Passifloraceae), Phaseolus vulgaris (Fabaceae), Pyracantha coccinea, Pyrus communis (Rosaceae), Schinus terebinthifolia (Anacardiaceae) and Vaccinium corymbosum (Ericaceae) (Farr & Rossman 2023, present study).
Distribution: Australia, Brazil, Portugal, South Africa, South Korea, Uruguay and USA (Farr & Rossman 2023).
Notes: Diaporthe leucospermi was introduced by Crous et al. (2011b) on leaves of Leucospermum sp. in Australia and, since then, no records of this species have been documented. However, according to Hilário et al. (2021c), few isolates collected from Hydrangea macrophylla and Acer negundo in Portugal by Santos et al. (2010) fall under the concept of D. leucospermi based on the available ITS and tef1 sequences. They also showed that D. pyracanthae (Santos et al. 2017b) and D. rossmaniae (Hilário et al. 2020) are indistinguishable and synonymous with D. leucospermi. The integrative taxonomic approach carried out in the present study confirmed the synonyms provided by Hilário et al. (2021c) and revealed that the subclade of D. leucospermi also included the ex-type culture of D. infecunda, which appears phylogenetically indistinguishable and are thus made synonymous with D. leucospermi. Both species clustered in a well-supported lineage and are closely related (Fig. 8). The ex-type culture of D. leucospermi (CBS 142384) showed the following nucleotide similarities with the sequences of the ex-type culture of D. infecunda (CBS 133812). On ITS: 97.55 % (558/572, including 3 gaps). On tef1: 98.49 % (327/332). On tub2: 98.34 % (415/422). On cal: 98.78 % (487/493). On his3: 97.59 % (446/458, including 1 gap). Morphological comparisons between the two “species” cannot be made because D. infecunda was introduced by Gomes et al. (2013) for a set of sterile isolates from leaves of medicinal plants in Brazil. Four isolates of D. leucospermi were obtained from foliar lesions of palms in Lisbon, Portugal, namely CDP 0052 and CDP 2055 from Chamaerops humilis, and CDP 1211 and CDP 1251 from Chrysalidocarpus lutescens. Isolates from distinct palm hosts were tentatively characterised to assess micromorphological variation. However, despite numerous attempts with different temperatures and with different organic material introduced on the agar surface, isolates from C. lutescens remained sterile even after long periods of incubation of about 2 mo. Isolates from C. humilis were morphologically similar to the ex-epitype of D. leucospermi (Crous et al. 2011b) (Fig. 23). Considering the strain characterised here (CDP 0052) and the ex-epitype strain of D. leucospermi (CBS 142384), both produce ellipsoid, hyaline and aseptate alpha conidia and spindle-shaped, hooked at apex, truncated at base, hyaline and aseptate beta conidia. While beta conidia of both strains showed overlapping dimensions (21.6–32.1 × 0.9–1.8 μm for CDP 0052 and 25–30 × 1.5 μm for CBS 142384), alpha conidia of CDP 0052 are slightly smaller (6.43 × 2.13 μm) than those reported for the type of D. leucospermi (7 × 3 μm) (Crous et al. 2011b). Sequence comparisons with the ex-epitype of D. leucospermi (CBS 142384) for ITS, tef1, tub2 and cal showed 97.74–100 %, 99.70–100 %, 98.19–98.79 % and 99.19–100 %, respectively, sequence similarity and differences are represented by few base pair changes and gaps, which were more substantial in the ITS1 region of CDP 2055. The nucleotide sequence similarity between the D. leucospermi isolates obtained here was 99–100 % for the four loci, except for the ITS sequence of CDP 2055, which is more polymorphic and showed sequence similarities of around 98 % with the other isolates. No his3 sequence data is available for these isolates. Diaporthe leucospermi has not previously been recorded on Arecaceae and thus two new host records are reported here (Table 5). The isolates of D. leucospermi studied here were recorded from foliar lesions of palms, but pathogenicity has not been tested. Diaporthe leucospermi has been recorded as an endophyte and weak pathogen on few plant hosts (Moreira et al. 2020). Furthermore, its nature as a latent pathogen and saprobe on fruit trees in Uruguay has recently been discussed (as D. infecunda) (Sessa et al. 2017, 2018).
Diaporthe pygmaeae D.S. Pereira & A.J.L. Phillips, sp. nov. MycoBank MB 851790. Fig. 25.
Fig. 25.
Diaporthe pygmaeae. A–F, H–K, N: CDP 1370, ex-type; G, L, M: CDP 2030. A–D. Conidiogenous layer. E–J. Conidiophores and conidiogenous cells. K–N. Alpha conidia. M. Pigmented alpha conidia. N. Microcyclic conidiogenesis (black arrow). Scale bars: A–C, K, L = 5 μm, D–J = 2.5 μm.
Etymology: Named after the common name, pygmy date palm, of the host species from which it was first isolated, Phoenix roebelenii.
Typus: Portugal, Lisbon, Parque das Nações, on foliar lesions of leaflets of Phoenix roebelenii (Arecaceae), 8 May 2021, D.S. Pereira, specimen HDP 088 (holotype AVE-F-17, ex-type culture CDP 1370 = CBS 151619, ITS sequence PP577992, tef1 sequence PP579317, tub2 sequence PP579332, cal sequence PP579348).
Sexual morph: Undetermined. Asexual morph: Conidiomata on palm leaflets in culture pycnidial, globose to subglobose, nonstromatic, uniloculate, black, solitary, immersed to semi-immersed, often covered with whitish mycelium, exuding a globose, white to pale mucoid mass of conidia, up to 210 μm diam. Conidiophores formed from the cells lining the locule wall, densely aggregated, hyaline to subhyaline, smooth- and thin-walled, doliiform to subcylindrical, aseptate or 1–3-septate, unbranched or branched, producing a lateral branch just below the septum that becomes conidiogenous. Conidiogenous cells hyaline, smooth- and thin-walled, terminal, rarely lateral, discrete, determinate, cylindrical, tapering towards the apex, straight or slightly curved, aseptate, unbranched, occasionally with minute and inconspicuous collarette, enteroblastic, proliferating at the same level giving rise to periclinal thickenings, 7.99–19.17(–31.55) × 1.92–3.91(–4.79) μm, 95 % confidence limits = 13.37–14.80 × 2.76–2.90 μm (mean ± SD = 14.08 ± 4.47 × 2.83 ± 0.44 μm, n = 150). Alpha conidia fusoid to ellipsoid, acute at both ends, smooth- and thin-walled, hyaline, aseptate, eguttulate, often with granular contents, rarely becoming light brown to goldish, (7.75–)8.01–9.97(–10.69) × 2.37–3.45(–4.01) μm, 95 % confidence limits = 8.90–9.09 × 2.84–2.93 μm (mean ± SD = 9.00 ± 0.61 × 2.89 ± 0.26 μm), mean ± SD conidium length/width ratio = 3.14 ± 0.38 (n = 150), microcyclic conidiogenesis occasionally observed. Beta and gamma conidia not observed.
Culture characteristics: Colonies on 1/2 PDA, reaching 52 mm diam after 7 d at 20 °C in darkness. Surface flat, with entire margin, circular shape, whitish, greyish white, opaque. Reverse luteus with patches of orange to brown. No diffusible pigment. Conidiomata black, sparse and developing scattered on the surface in about 1 wk.
Additional material examined: Portugal, Lisbon, Parque das Nações, on foliar lesions of segments of Chamaerops humilis (Arecaceae), 8 May 2021, D.S. Pereira (specimen HDP 089), living culture CDP 2030 = CBS 151618 (ITS sequence PP577993, tef1 sequence PP579318, tub2 sequence PP579333, cal sequence PP579349).
Hosts: Chamaerops humilis, Phoenix roebelenii (Arecaceae).
Distribution: Portugal.
Notes: Based on single and multilocus phylogenetic inferences, D. pygmaeae resolved as a highly supported basal lineage in the D. brasiliensis clade and is sister to D. brasiliensis, D. caatingaensis and the strain CBS 125575 (Diaporthe sp.) (Fig. 3). The new lineage was also supported by both coalescent- and genetic distance-based analyses, which depicted D. pygmaeae as a distinct MOTU. Diaporthe pygmaeae (CDP 1370) can be distinguished from D. brasiliensis (CBS 133183) based on 75 fixed unique single nucleotide polymorphisms (SNP) and 23 deletion/insertion polymorphisms (DIP) in the partial sequences of four gene regions (8, 31, 15 and 21, and 8, 2, 9 and 4 in ITS, tef1, tub2 and cal loci, respectively). On the other hand, it can be distinguished from D. caatingaensis (CBS 141542) based on 71 SNP and 15 DIP (8, 33, 13 and 17, and 2, 6, 4 and 3 in ITS, tef1, tub2 and cal loci, respectively). In short, the ex-type strain of D. pygmaeae (CDP 1370) have the following sequence similarities with the sequences of the ex-type strains of D. brasiliensis (CBS 110864) and D. caatingaensis. On ITS: 97.06 % (528/544, including 8 gaps) and 98.14 % (528/538, including 2 gaps), respectively. On tef1: 90.43 % (312/345, including 2 gaps) and 88.79 % (309/348, including 6 gaps), respectively. On tub2: 96.61 % (685/709, including 9 gaps) and 97.57 % (684/701, including 4 gaps), respectively. On cal: 95.04 % (479/504, including 4 gaps) and 96.02 % (483/503, including 3 gaps), respectively. No his3 sequence data is available for strains of D. pygmaeae. Morphologically, D. pygmaeae (CDP 1370), D. brasiliensis (CBS 133183) and D. caatingaensis (CBS 141542) are highly similar, producing globose, black pycnidial conidiomata, exuding masses of fusoid to ellipsoid, hyaline and aseptate conidia of considerable overlapping dimensions, except for D. brasiliensis (6–7 × 2–3 μm) (Gomes et al. 2013), whose conidial length is somewhat shorter when compared to D. pygmaeae and D. caatingaensis (8.0–10. × 2.4–3.5 μm and 8.5–9.5 × 2 μm, respectively) (Crous et al. 2016a) (Fig. 25, Table 4). Two strains of D. pygmaeae were isolated, namely CDP 1370 (ex-type) and CDP 2030. They exhibited a minute degree of variation in colony morphology when cultured on 1/2 PDA, which was expressed by darker, greyish surface mycelium and slightly higher growth rate in CDP 2030 (63 mm vs 52 mm after 7 d at 20 °C in darkness). No relevant variation in micromorphology was observed between these strains, which displayed very similar conidiogenous cells (mean = 14.08 × 2.83 μm for CDP 1370 and 8.77 × 3.05 μm for CDP 2030) and conidial (mean = 9.00 × 2.89 μm for CDP 1370, with L/W = 3.14, and 9.47 × 3.09 μm for CDP 2030, with L/W = 3.09) dimensions. However, the alpha conidia of CDP 2030 occasionally became pigmented with age, while this feature was apparently absent in the alpha conidia of CDP 1370. The nucleotide sequence similarity between them was 99.81 % for ITS, which result from an additional gap in ITS1 in CDP 2030 and 100 % for tef1, tub2 and cal loci. The isolates of D. pygmaeae studied were recorded from foliar lesions of palms, including Chamaerops humilis and Phoenix roebelenii (Table 5), but pathogenicity has not been tested. It is unknown if D. pygmaeae is host specific to Arecaceae members. However, species in the D. brasiliensis clade have uncommonly been found, so they have restricted host and geographical ranges. D. brasiliensis and D. caatingaensis have been virtually only isolated as endophytes from medicinal plants in Brazil (Gomes et al. 2013, Crous et al. 2016a), while D. pygmaeae has been isolated as a potential phytopathogen from ornamental palms in Portugal. Therefore, the ecological role of species in the D. brasiliensis clade as either endophytes or latent pathogens should be clarified as more strains of each species are isolated.
Diaporthe rudis (Fr.) Nitschke, Pyrenomycetes Germanici 2: 282. 1870. MycoBank MB 139900. Fig. 26.
Fig. 26.
Diaporthe rudis (CDP 1323). A–D. Conidiogenous layer. E–I. Conidiophores and conidiogenous cells (black arrows point to collarettes). J, K. Alpha conidia. Scale bars: A–D, J, K = 5 μm, E–I = 2.5 μm.
Basionym: Sphaeria rudis Fr., Elenchus Fungorum 2: 98. 1828. MycoBank MB 184043.
Synonyms: Rabenhorstia rudis (Fr.) Fr., Summa Veg. Scand. 2: 410. 1849. MycoBank MB 160642.
Diplodia rudis (Fr.) Desm., Mem. Acad. R. Sci. Lett. Belg 23: 27. 1849. MycoBank MB 247966.
Aglaospora rudis (Fr.) Tul. & C. Tul., Selecta Fungorum Carpologia 2: 165. 1863. MycoBank MB 168475.
Hercospora rudis (Fr.) Auersw., Hedwigia 7: 185. 1868. MycoBank MB 195007.
Phomopsis rudis (Fr.) Höhn., Fungi Saxonici Exsiccati XLIV: no. 2183. 1912. MycoBank MB 655685.
Aplosporella rudis (Fr.) Petr. & Syd. [as “Haplosporella”], Ann. Mycol. 21: 356. 1923. MycoBank MB 277015.
Valsa faginea Berk. & Broome, Ann. Mag. Nat. Hist. 3: 368. 1859. MycoBank MB 240431.
Sphaeria faginea Curr., Trans. Linn. Soc. Lond. 22: 281. 1859. MycoBank MB 465748.
Diaporthe faginea (Berk. & Broome) Sacc., Syll. Fung. 1: 619. 1882. MycoBank MB 174356.
Diaporthe macrostoma Nitschke, Pyrenomycetes Germanici 2: 284. 1870. MycoBank MB 172265.
Diaporthe medusaea Nitschke, Pyrenomycetes Germanici 2: 251. 1870. MycoBank MB 166990.
Diaporthe viticola Nitschke, Pyrenomycetes Germanici 2: 264. 1870. MycoBank MB 234198.
Phoma rudis Sacc., Michelia 1: 257. 1878. MycoBank MB 176930.
Phomopsis rudis (Sacc.) Höhn., Sitz.-Ber. K. Akad. Wiss., Math.-Naturwiss. Kl. Abt. I, 115: 680. 1906. MycoBank MB 121326.
Diaporthe silvestris Sacc. & Berl., Atti dell′Istituto Veneto Scienze 3: 737. 1885. MycoBank MB 148577.
New synonym: Diaporthe italiana Chethana et al., Fungal Divers. 96: 136. 2019. MycoBank MB 555376.
Typus: Australia, Vienna, 19.7763/2, Reisenbergbach-Weg, on stem of Laburnum anagyroides (Fabaceae), 8 Apr. 2000, W. Jaklitsch (epitype of Sphaeria rudis designated by Udayanga et al. (2014b), BPI 748231, exepitype culture CBS 109292); Vienna, 19.7763/2, Reisenbergbach-Weg, on stem of Laburnum anagyroides (Fabaceae), 8 Apr. 2000, W. Jaklitsch (epitype of Diaporthe medusaea designated by Udayanga et al. (2014b), BPI 748231, ex-epitype culture CBS 109292). France, on a dead branch of Laburnum anagyroides (as Cytisus laburnum) (Fabaceae), ex herb. Guépin no. 163 (holotype of Sphaeria rudis UPS F-004948). Germany, Nordrhein-Westfalen, Landkreis Unna, Cappenberg, Schloßgarten zu Cappenberg, on twigs of Laburnum anagyroides (as Cytisus laburnum) (Fabaceae), 18 Aug. 1866, T. Nitschke (holotype of Diaporthe medusaea B 70 0009168); Nordrhein-Westfalen, Munsterland, Munster Botanischer Garten, on thin branch of Fagus sylvatica (Fagaceae), 18 May 1866, T. Nitschke (holotype of Diaporthe macrostoma B 70 0009167); Westfalen, Munster, bei der Wienburg, on Vitis vinifera (Vitaceae), Feb. 1866, T. Nitschke (holotype of Diaporthe viticola B, anon. s. n.); Westfalen, Munster, bei der Wienburg, on Vitis vinifera (Vitaceae), Feb. 1866, T. Nitschke (isotype of Diaporthe viticola BPI 797316). Italy, Sardinia, on Vitis vinifera (Vitaceae), “in sarmentis Vitis viniferae silvestris, Cervarese” (Saccardo & Berlese 1885) (holotype of Diaporthe silvestris cited in van Niekerk et al. (2005), PAD 228). Portugal, Santo Tirso, Burgaes, on Vitis vinifera (Vitaceae), 16 Feb. 1998, A.J.L. Phillips (epitype of Diaporthe viticola designated by van Niekerk et al. (2005), CBS-H 7950, ex-epitype culture CBS 113201).
See Udayanga et al. (2014b) for illustrations and description of asexual and sexual morphs.
Isolate CDP 1323. Sexual morph: Undetermined. Asexual morph: Conidiomata on palm leaflets in culture pycnidial, globose, non-stromatic, uniloculate, black, solitary, immersed, becoming erumpent through the ostiolar region, covered with whitish mycelium. Conidiophores formed from the cells lining the locule wall, hyaline to subhyaline, smooth- and thin-walled, doliiform to subcylindrical, straight or curved, aseptate, often 1-septate, rarely 2-septate, unbranched or branched. Conidiogenous cells hyaline, smooth- and thin-walled, discrete, determinate, cylindrical, tapering towards the apex, straight or curved, aseptate, with a prominent collarette, up to 2.5 μm, enteroblastic, proliferating at the same level giving rise to periclinal thickenings, (5.79–)11.02–26.14 × 1.28–2.63 μm, 95 % confidence limits = 14.80–16.70 × 1.73–1.88 μm (mean ± SD = 15.75 ± 3.69 × 1.80 ± 0.29 μm, n = 55). Alpha conidia oval to ellipsoidal, tapering towards the ends, both ends acute, base slightly truncated, smooth- and thin-walled, hyaline, aseptate, eguttulate, often with granular contents, 5.34–7.65 × 1.77–2.65 μm, 95 % confidence limits = 6.24–6.37 × 2.08–2.13 μm (mean ± SD = 6.31 ± 0.42 × 2.11 ± 0.14 μm), mean ± SD conidium length/width ratio = 3.01 ± 0.29 (n = 150). Beta and gamma conidia not observed.
Culture characteristics: Colonies on 1/2 PDA, reaching 85 mm diam after 7 d at 20 °C in darkness. Surface flat, with sparce aerial mycelium, with entire margin, circular shape, white luteous to pale, opaque. Reverse luteus with patches of orangish. No diffusible pigment.
Materials examined: Portugal, Lisbon, Parque das Nações, on foliar lesions of segments of Chrysalidocarpus lutescens (Arecaceae), 8 May 2021, D.S. Pereira (specimen HDP 087), living culture CDP 1323 (ITS sequence PP578009, tef1 sequence PP579327, tub2 sequence PP579343, cal sequence PP579352); Parque das Nações, on foliar lesions of leaflets of Washingtonia filifera (Arecaceae), 8 May 2021, D.S. Pereira (specimen HDP 096), living culture CDP 1861 (ITS sequence PP578010, tef1 sequence PP579328, tub2 sequence PP579344, cal sequence PP579353).
Hosts: Reported from more than 40 genera and 55 species in 32 families, including Actinidiaceae (Actinidia deliciosa), Adoxaceae (Sambucus racemosa), Anacardiaceae (Pistacia vera), Aquifoliaceae (Ilex aquifolium), Arecaceae (Chrysalidocarpus lutescens, Washingtonia filifera), Asphodelaceae (Asphodelus albus), Asteraceae (Carlina vulgaris), Betulaceae (Alnus sp., Carpinus betulus, Corylus avellana), Cistaceae (Cistus tauricus), Cornaceae (Cornus kousa), Dioscoreaceae (Dioscorea communis), Dipsacaceae (Dipsacus fullonum), Ericaceae (Vaccinium corymbosum, V. myrtillus), Fabaceae (Acacia sp., Glycine max, Laburnum anagyroides, Lupinus arboreus), Fagaceae (Castanea sativa, Fagus sylvatica, Quercus robur), Garryaceae (Aucuba japonica), Geraniaceae (Pelargonium sp.), Hydrangeaceae (Hydrangea macrophylla), Lauraceae (Persea americana), Loranthaceae (Ileostylis micranthus), Moraceae (Morus alba), Myrtaceae (Eucalyptus globulus), Oleaceae (Fraxinus excelsior, Olea europaea), Onagraceae (Epilobium angustifolium), Pinaceae (Pinus pinaster), Poaceae (Anthoxanthum odoratum, Holcus lanatus), Rosaceae (Malus domestica, Pyrus communis, Rosa canina, R. rugosa), Rutaceae (Citrus × paradisi, C. aurantiacum, C. aurantiifolia, C. aurantium, C. latifolia, C. limon, C. maxima, C. reticulata, C. reticulata × paradisi, C. sinensis), Salicaceae (Salix sp.), Sapindaceae (Acer campestre, A. opalus, A. pseudoplatanus), Solanaceae (Brugmansia sp.) and Vitaceae (Vitis vinifera) (Farr & Rossman 2023, present study).
Distribution: Australia, Austria, Canada, Chile, China, Czech Republic, France, Germany, Greece, Italy, Latvia, Netherlands, New Zealand, Poland, Portugal, Serbia, South Africa, Spain, Sweden, Switzerland, UK, Ukraine and USA (Farr & Rossman 2023).
Notes: The name D. rudis is based on the oldest epithet of the many synonyms that have been proposed for this species over the years. Udayanga et al. (2014b) resolved the status and boundaries of D. rudis by providing an epitype specimen and associated molecular data. Thus, based on current understanding, D. rudis was originally described by Fries (1828) as Sphaeria rudis on a dead branch of Laburnum anagyroides. Udayanga et al. (2014b) also recognised that D. medusaea and D. viticola, names that have been used for important pathogens of Citrus and Vitis, respectively, were synonyms of D. rudis. The integrative taxonomic approach carried out in the present study revealed that D. italiana is a new synonym of D. rudis. Diaporthe italiana was introduced by Hyde et al. (2019) as saprophyte on a dead aerial branch of Morus alba as a closely related but phylogenetically distinct taxon from D. rudis based on minor morphomolecular differences. However, the phylogenetic analyses carried out here showed that D. italiana is nested within the D. rudis subclade, lacking relevant phylogenetic distinctiveness (Fig. 10). The ex-epitype culture of D. rudis (CBS 109292) showed the following nucleotide similarities with the sequences of the ex-type and ex-isotype cultures of D. italiana (MFLUCC 18-0090 and MFLUCC 18-0091, respectively). On ITS: 97.44 % (494/507, including 1 gap). On tef1: 98.04 % (300/306, including 3 gaps) and 98.65 % (293/297, including 3 gaps), respectively. On tub2: 99.60 % (497/499). On cal: 98.95 % (469/474, including 5 gaps) and 98.94 % (467/472, including 5 gaps), respectively. No his3 sequence data is available for the type cultures of D. italiana and D. rudis. The tef1 sequences available for the type cultures of D. italiana were excluded from the phylogenetic trees presented here, as they strongly affected the robustness of the trees and the resolution of the subclades within the D. rudis clade. Sequence comparisons between the ex-type and ex-isotype cultures of D. italiana showed 97.07 % (331/341, including 10 gaps) nucleotide similarity, evidencing that there are more dissimilarities in tef1 between the types of D. italiana than between them and the ex-epitype culture of D. rudis. This is due to a region of seven gaps that highly impacts the phylogenetic placement of both strains within the D. rudis clade. Morphological comparisons between the asexual morphs of the two “species” cannot be made because only the sexual morph of D. italiana was observed by Hyde et al. (2019). However, the sexual morphs of D. italiana and D. rudis are similar, producing ascospores that overlap in size (mean = 10.5–13 × 2.7–4.5 μm vs 12–14.2 × 3.5–3.7 μm, respectively) (Udayanga et al. 2014b, Hyde et al. 2019). Two isolates of D. rudis were obtained from foliar lesions of two different species of palms in Lisbon, Portugal, namely CDP 1323 from Chrysalidocarpus lutescens and CDP 1861 from Washingtonia filifera. The two isolates were tentatively characterised to assess micromorphological variation related to host colonisation. However, despite numerous attempts with different temperatures and with different organic material introduced on the agar surface, the isolate from W. filifera remained sterile even after long periods of incubation of about 2 mo. CDP 1323 was morphologically similar to the ex-epitype of D. rudis (Udayanga et al. 2014b) (Fig. 26). Both produce oval to ellipsoidal, hyaline and aseptate alpha conidia, with a subtruncate base. Nevertheless, the conidial length of CDP 1323 was slightly smaller than that reported for the exepitype of D. rudis (mean = 6.31 × 2.11 μm vs 7.5 × 2.2 μm, respectively). Furthermore, beta conidia have not been observed for the isolate from C. lutescens, although they have been reported for the ex-epitype of D. rudis (Udayanga et al. 2014b). Sequence comparisons with the ex-epitype culture of D. rudis (CBS 109292) for ITS, tef1, tub2 and cal showed 98.90 %, 99.70 %, 99.80 % and 99.79–100 %, respectively, sequence similarity and differences are represented by few base pair changes and gaps. The nucleotide sequence similarity between the D. rudis isolates obtained here was 100 % for all four loci, except for cal, where there was a difference of three gaps between the two isolates. No his3 sequence data is available for these isolates. Diaporthe rudis has not previously been recorded on Arecaceae and thus two new host records are reported here (Table 5). The isolates of D rudis studied here were recorded from foliar lesions of palms, but pathogenicity has not been tested. However, D. rudis is a well-known, widespread pathogen causing stem and fruit rot, seed decay, cankers, and dieback on agro-economically valuable hosts (González & Ciordia 2023). In Europe, D. rudis (as D. viticola) has mostly been associated with diseases of Vitis vinifera, being widely recognised as an important pathogen of grapevine (Guarnaccia et al. 2018).
Diaporthe ueckeri Udayanga & Castl. [as “ueckerae”], Fungal Biol. 119: 401. 2014. MycoBank MB 827136.
Synonym: Diaporthe miriciae R.G. Shivas et al., Persoonia 35: 46. 2015. MycoBank MB 808673.
New synonyms: Diaporthe thunbergiicola Udayanga & K.D. Hyde, Fungal Divers. 72: 22. 2015. MycoBank MB 551072.
Diaporthe passifloricola Crous & M.J. Wingf., Persoonia 36: 395. 2016. MycoBank MB 817057.
Diaporthe rosae M.C. Samar. & K.D. Hyde, Fungal Divers. 89: 185. 2018. MycoBank MB 554072.
Diaporthe vochysiae S.A. Noriler et al., Fitoterapia 138 (no. 104273): 273. 2019. MycoBank MB 827590.
Diaporthe durionigena L.D. Thao et al., Persoonia 44: 385. 2020. MycoBank MB 835458 [Nom. inval., Art. 40.8 (Shenzhen)].
Diaporthe durionigena L.D. Thao et al., Fung. Syst. Evol. 7: 288. 2021. MycoBank MB 839285.
Typus: USA, Oklahoma, on crown of Cucumis melo (Cucurbitaceae), F.A. Uecker (holotype BPI 748011, ex-type culture CBS 139283).
See Udayanga et al. (2015) for illustrations and descriptions of asexual morph. Sexual morph not reported.
Hosts: Anacardium occidentale (Anacardiaceae), Arachis hypogaea (Fabaceae), Areca catechu (Arecaceae), Brosimum gaudichaudii (Moraceae), Camellia sinensis (Theaceae), Citrus grandis, C. reticulata (Rutaceae), Copaifera pubiflora (Fabaceae), Costus spiralis (Costaceae), Cucumis melo (Cucurbitaceae), Durio zibethinus (Malvaceae), Eucalyptus citriodora (Myrtaceae), Glycine max (Fabaceae), Gossypium hirsutum (Malvaceae), Helianthus annuus (Asteraceae), Hyophorbe lagenicaulis (Arecaceae), Magnolia champaca, M. shiluensis (Magnoliaceae), Mangifera indica (Anacardiaceae), Melocactus ernestii (Cactaceae), Miconia sp. (Melastomataceae), Nelumbo nucifera (Nelumbonaceae), Nepenthes ampullaria (Nepenthaceae), Nephelium lappaceum (Sapindaceae), Passiflora foetida (Passifloraceae), Rosa sp. (Rosaceae), Selenicereus monacanthus, S. undatus (Cactaceae), Senna siamea (Fabaceae), Solanum melongena (Solanaceae), Stryphnodendron adstringens (Fabaceae), Thunbergia laurifolia (Acanthaceae), Vigna radiata (Fabaceae) and Vochysia divergens (Vochysiaceae) (Farr & Rossman 2023).
Distribution: Australia, Brazil, China, Colombia, Malaysia, Philippines, Taiwan, Thailand, USA and Vietnam (Farr & Rossman 2023).
Notes: Diaporthe ueckeri was introduced by Udayanga et al. (2015) on the crown of Cucumis melo in USA as member of the D. sojae species complex. The phylogenetic analyses carried out in the present study revealed that D. ueckeri clustered in a well-supported clade, together with the types of five species of Diaporthe (Fig. 9). However, although some substructure can be observed within the clade, most lineages lack phylogenetic support and are intermingled with several strains previously recognised as D. ueckeri. Gao et al. (2016) first observed inconsistencies within this clade, noting that the types of D. ueckeri and D. miriciae were phylogenetically and morphologically indistinguishable. Although D. miriciae was made synonymous with D. ueckeri, subsequent studies have largely overlooked the phylogenetic evidence provided by Gao et al. (2016). As a result, several lineages closely related to D. ueckeri and D. miriciae have been introduced over the years mostly based on minor morphological differences and small nucleotide dissimilarities. The integrative taxonomic approach carried out here has shown that the entire clade is conspecific and, therefore, all five species have been assigned to the oldest species name, i.e., D. ueckeri. Sequence comparisons between the types of “species” in the D. ueckeri clade for ITS, tef1, tub2, cal and his3 showed 95–100 %, 92–99 %, 98–100 %, 98–99 % and 97–99 % nucleotide similarities, respectively. The only “species” that appears to have relevant nucleotide dissimilarities from D. ueckeri is D. thunbergiicola. However, only ITS and tef1 sequence data are available for comparison, and several of the nucleotide differences observed in ITS correspond to gaps and polymorphic regions in ITS1. Considering the phylogenetic analyses conducted here, D. thunbergiicola clustered as a very long branch that could not segregate from strains in the D. ueckeri clade (Fig. 9). Hence, for taxonomic stability, D. thunbergiicola is treated here as a synonym of D. ueckeri until sequence data from other loci of its ex-type strain or additional strains can resolve this lineage. Morphologically, all “species” in the D. ueckeri clade are virtually indistinguishable. They have oval/oblong to ellipsoidal alpha conidia and filiform, hooked beta conidia, which are frequently absent, and their sizes fall within the same ranges, especially in alpha conidia (Table 4). Considering the morphological data available for the “species” synonymised here, the mean dimensions of the alpha and beta conidia produced by D. ueckeri strains are 5.53–7.04 × 2.13–2.67 μm (L/W = 1.75–3.00) and 18.70–26.03 × 1.15–1.63 μm (L/W = 10.17–22.00), respectively. The mean dimensions of alpha conidia partially overlap with the dimensions reported for the ex-type culture of D. ueckeri (CBS 139283; mean = 7.2 × 2.6 μm) (Table 4) (Udayanga et al. 2015). However, the deviation is mainly related to D. vochysiae, whose alpha conidia dimensions are considerably different when compared to other species in the clade. Specifically, their length is particularly shorter (3–4 × 2 μm) (Noriler et al. 2019) (Table 4). Thus, except for D. vochysiae, and considering that the production of beta conidia tends to simply reflect intraspecific variability associated with growth conditions (Mostert et al. 2001), the morphology of the asexual morph of all D. ueckeri strains match the description provided by Udayanga et al. (2015). Diaporthe ueckeri was first introduced as a pathogen on Cucumis melo, whose disease symptoms were unknown (Udayanga et al. 2015). However, subsequent reports have confirmed that D. ueckeri has a broad host range as a pathogen, endophyte and saprophyte. As a pathogen, it has been associated with various plant diseases, including leaf spots, cankers, dieback, and stem rot of fruit hosts (e.g., Lim et al. 2019, Lopez-Cardona et al. 2021, Aumentado & Balendres 2024). This is not surprising given the close affiliation of this species with taxa of the D. sojae species complex, which includes several important phytopathogenic species of Diaporthe associated with herbaceous field crops and weeds. Therefore, D. ueckeri may represent a serious threat to soybean and cucurbits, hosts on which its pathogenic nature has been confirmed. Moreover, D. ueckeri has also been associated to human diseases, so this may be considered as a ubiquitous pathogen that can opportunistically infect both humans and plant hosts (Udayanga et al. 2015). Regarding palms, D. ueckeri has been recently reported as the causal agent of brown blotch disease on Hyophorbe lagenicaulis in China (Guo et al. 2024). In addition, five unpublished reports of D. ueckeri causing leaf spots on Areca catechu in Hainan, China were found in GenBank and their phylogenetic affiliations with D. ueckeri were confirmed in the present study (Table 5).
Other species treated
Diaporthe cynaroidis Marinc. et al., CBS Biodiversity Series 7: 39. 2008. MycoBank MB 506209.
New synonyms: Diaporthe salicicola R.G. Shivas et al., Fungal Divers. 61: 258. 2013. MycoBank MB 803338.
Diaporthe subcylindrospora S.K. Huang et al., Mycosphere 9: 381. 2018. MycoBank MB 554077.
Notes: Subclade of D. cynaroidis included the types of D. cynaroidis, D. salicicola and D. subcylindrospora, which grouped together with relevant nodal support values and appear phylogenetically indistinguishable (Fig. 10). Therefore, the three “species” are treated as synonyms. The ex-type culture of D. cynaroidis (CBS 122676) showed the following nucleotide similarities with the sequences of the ex-type cultures of D. salicicola (VPRI 32789) and D. subcylindrospora (KUMCC 17-0151). On ITS: 99.31 % (576/580) and 99.28 % (554/558, including 2 gaps), respectively. On tef1: 99.39 % (326/328) and 98.48 % (323/328), respectively. On tub2: 98.25 % (672/684, including 4 gap) and 98.58 % (416/422), respectively. According to the current understanding of Diaporthe species diversity, these are considered small, insignificant nucleotide differences. No cal or his3 sequence data are available for the ex-type cultures of D. salicicola and D. subcylindrospora. Morphological comparisons between the types of the three “species” cannot be made because D. cynaroidis and D. subcylindrospora were introduced based on morpho-molecular analyses, but only the sexual morph was reported (Marincowitz et al. 2008, Hyde et al. 2020a), while only the asexual morph has been observed for D. salicicola (Tan et al. 2013) (Table 4). Nonetheless, the sexual morphs of D. salicicola and D. subcylindrospora are virtually indistinguishable, producing fusiform, hyaline and 1-septate ascospores of remarkably similar sizes (mean = 12.3 × 3.1 μm vs 12 × 3 μm, respectively) (Marincowitz et al. 2008, Hyde et al. 2020a). Regarding the eco-geographical data, all three “species” have been introduced based on collections from different countries/continents, i.e., D. cynaroidis was introduced as a saprophyte on leaf litter of Protea cynaroides in South Africa (Marincowitz et al. 2008), D. salicicola was introduced on a leaf spot of Salix purpurea in Tasmania, Australia (Tan et al. 2013), and D. subcylindrospora was introduced as a saprophyte on a dead branch of Salix sp. in China (Hyde et al. 2020a).
Diaporthe isoberliniae Crous, Persoonia 32: 221. 2014. MycoBank MB 808909.
New synonym: Diaporthe pungensis S.T. Huang et al., MycoKeys 77: 85. 2021. MycoBank MB 837599.
Notes: The subclade of D. isoberliniae include the types of D. isoberliniae and D. pungensis. The ex-type culture of D. isoberliniae was excluded from the phylogenetic trees presented here, since only the ITS and tub2 sequences are available and strongly affected the robustness of the trees. However, two strains of D. isoberliniae were isolated in Cameroon and used to amend the taxonomic description of the species (Lambert et al. 2023). These were considered here to represent authentic strains and were used in the phylogenetic analyses. Diaporthe isoberliniae and D. pungensis grouped together with relevant nodal support values and were found to be phylogenetically indistinguishable (Fig. 8). Therefore, the two species are treated as synonyms. Sequence comparisons between a representative strain of D. isoberliniae (STMA 18291) and the ex-type culture of D. pungensis (SAUCC 194.112) for ITS, tef1, tub2, cal and his3 showed 97.03 % (522/538, including 9 gaps), 98.96 % (286/289, including 3 gaps), 99.15 % (464/468, including 2 gaps), 99.34 % (452/455, including 1 gap), 97.24 % (422/434, including 7 gaps) respectively, nucleotide similarities; similar comparisons using the ex-type culture of D. isoberliniae (CBS 137981) showed 96.11 % (519/540, including 12 gaps) and 98.62 % (429/435, including 2 gaps) nucleotide similarities for ITS and tub2, respectively. According to the current understanding of Diaporthe species diversity, these are considered small, insignificant nucleotide differences, and, therefore, D. pungensis has been introduced primarily due to a polymorphic ITS sequence. Morphologically both species are indistinguishable, with fusoid to ellipsoidal alpha conidia, with subobtuse apex and subtruncate base, and filiform, curved and tapered towards apex beta conidia of considerable overlapping dimensions (5.5–9 × 2–3 μm and 11.5–27.5 × 1–2 μm, respectively, for D. isoberliniae, and 6–8.5 × 2–3.3 μm and 24–28.9 × 1–2 μm, respectively, for D. pungensis) (Sun et al. 2021, Lambert et al. 2023) (Table 4). Regarding the eco-geographical data, while D. isoberliniae was introduced on Isoberlinia angolensis in Zambia (Africa) (Crous et al. 2014b), D. pungensis was introduced on diseased leaves of Elaeagnus pungens in China (Asia) (Sun et al. 2021). Diaporthe isoberliniae is an uncommon Diaporthe species that has only been recollected once in Cameroon (Africa) as an endophyte on Pittosporum manii (Lambert et al. 2023). Further data is required to determine its ecological group.
Diaporthe longicolla (Hobbs) J.M. Santos et al., Persoonia 27: 13. 2011. MycoBank MB 563213.
Basionym: Phomopsis longicolla Hobbs, Mycologia 77: 542. 1985. MycoBank MB 105685.
New synonyms: Diaporthe unshiuensis F. Huang et al., Fungal Biol. 119: 344. 2015. MycoBank MB 810845.
Diaporthe megabiguttulata M. Luo et al., J. Fungi 8 (no. 806): 16. 2022. MycoBank MB 553404.
Notes: The subclade of D. longicolla include the types of D. megabiguttulata and D. unshiuensis, which grouped together with relevant nodal support values and were found to be phylogenetically and morphologically indistinguishable (Fig. 9). Therefore, the three species are treated as synonyms. Sequence comparisons between the ex-type culture of D. longicolla (ATCC 60325) and the type cultures of D. megabiguttulata (ZHKUCC 22-0067, type, and ZHKUCC 22-0068, paratype) and D. unshiuensis (CGMCC 3.17566) showed the following nucleotide similarities. On ITS: 98.36 % (541/550, including 2 gaps) and 98.17 % (484/493, including 2 gaps), respectively. On tef1: 98.63 % (216/219, including 1 gap) and 97.08 % (332/342, including 6 gaps), respectively. On tub2: 99.10 % (442/446) and 96.78 % (316/373, including 6 gaps), respectively. On cal: 99.03 % (511/516, including 1 gap); not available for D. unshiuensis. On his3: 98.24 % (446/454, including 4 gaps) and 96.70 % (469/485, including 10 gaps), respectively. Although D. unshiuensis presents some relevant nucleotide dissimilarities, most are related to the presence of sets of sequence gaps. Morphologically, the three species are highly similar and harbour ellipsoidal to clavate alpha conidia of considerably overlapping dimensions (5–9.5 × 1.5–3.5 μm for D. longicolla, 5–10 × 2–3 μm for D. megabiguttulata and 5.2–7.5 × 2–3.9 μm for D. unshiuensis) (Santos et al. 2011, Huang et al. 2015, Luo et al. 2022). Beta conidia have only been observed in D. megabiguttulata (Luo et al. 2022) (Table 4). Regarding the eco-geographical data, D. longicolla has a widespread distribution as a pathogen and endophyte in a broad range of plant hosts. This species is regarded as the main cause of seed decay of soybean (Santos et al. 2011) but has been associated with several other plant diseases (particularly as D. unshiuensis) (e.g., Guo et al. 2020, Du et al. 2021, Liao et al. 2022).
Diaporthe micheliae Y.H. Gao & L. Cai [as “michelina”], IMA Fungus 8: 184. 2017. MycoBank MB 823925.
Basionym: Phomopsis micheliae C.Q. Chang et al., Mycosystema 24: 9. 2005. MycoBank MB 335171 [Nom. illegit., Art. 53.1].
New synonyms: Phomopsis chimonanthi C.Q. Chang et al., Mycosystema 24: 146. 2005. MycoBank MB 344473.
Diaporthe chimonanthi (C.Q. Chang et al.) Y.H. Gao & L. Cai, IMA Fungus 8: 183. 2017. MycoBank MB 821439.
Diaporthe middletonii R.G. Shivas et al., Persoonia 35: 45. 2015. MycoBank MB 808672.
Diaporthe australpacifica Y.P. Tan & R.G. Shivas, Index Austral. Fungi 2: 2. 2022. MycoBank MB 559560.
Notes: The subclade of D. micheliae included the types of D. australpacifica, D. chimonanthi, D. micheliae and D. middletonii, which grouped with relevant nodal support values and are phylogenetically closely related (Fig. 8). The integrative taxonomic approach carried out has shown that all four species are conspecific and are therefore treated as synonyms. Although only ITS sequence data is available for the type specimens of D. chimonanthi and D. micheliae, both species are undoubtedly related to D. middletonii and D. australpacifica. Sequence comparisons between the types of the four species for ITS revealed 97–100 % nucleotide similarity. Moreover, sequence comparison between the types of D. middletonii and D. australpacifica for tef1 and tub2 showed 98.77 % (562/569, including 2 gaps) and 100 % (709/709). No cal or his3 sequence data are available for the types of both species. According to the current understanding of Diaporthe species diversity, these are considered small, insignificant nucleotide differences. Therefore, following the principle of priority, all three species have been assigned to the oldest species epithet, i.e., micheliae. Morphologically, these taxa are similar and harbour fusiform alpha conidia of overlapping dimensions and filiform, curved or hamate beta conidia that appear to vary in length. No taxonomic description is available for D. australpacifica (Table 4). Regarding the eco-geographical data, while D. micheliae and D. chimonanthi were introduced on branches of Michelia alba (Chang et al. 2005b) and Chimonanthus praecox (Chang et al. 2005a), respectively, in China, D. australpacifica and D. middletonii were introduced on a stem of Amaranthus blitum (Tan & Shivas 2022), and Rapistrum rugosum and Chrysanthemoides monilifera (Thompson et al. 2015), respectively, in Australia. The ecological groups of this species are virtually unknown. Few reports in China have isolated D. micheliae (as D. middletonii) on diseased leaves of Litchi chinensis, Lithocarpus glaber and L. craibianus (Huang et al. 2021b).
Diaporthe sackstonii R.G. Shivas et al., Persoonia 35: 46. 2015. MycoBank MB 808674.
New synonyms: Diaporthe caryae C.M. Tian & Q. Yang, MycoKeys 39: 124. 2018. MycoBank MB 824706.
Diaporthe machili S.T. Huang et al., MycoKeys 78: 63. 2021. MycoBank MB 837814.
Diaporthe juglandigena S.Y. Wang et al., J. Fungi 8 (no. 1301): 20. 2022. MycoBank MB 446039.
Diaporthe orixae Q.T. Lu & Zhen Zhang, Phytotaxa 544: 45. 2022. MycoBank MB 558801.
Notes: The subclade of D. sackstonii included the types of D. caryae, D. juglandigena, D. machili and D. orixae, which grouped with relevant nodal support values and are phylogenetically closely related (Fig. 8). Although some substructure and well-supported groupings can be observed within this clade, the integrative taxonomic approach carried out has shown that all five species are conspecific and are therefore treated as synonyms. However, it is likely that the clade is undergoing active divergence and speciation. The ex-type culture of D. sackstonii (BRIP 54669b) showed the following nucleotide similarities with the sequences of the ex-type cultures of D. caryae (CFCC 52563), D. juglandigena (GUCC 422.16), D. machili (SAUCC 194.111) and D. orixae (KUNCC 21–10714). On ITS: 99.28 % (555/559), 98.95 % (564/570), 98.89 % (534/540, including 3 gaps) and 95.89 % (466/486, including 5 gaps), respectively. On tef1: 96.01 % (337/351, including 5 gaps), 95.99 % (335/349, including 5 gaps), 95.57 % (302/316, including 6 gaps) and 95.83 % (345/360, including 5 gaps), respectively. On tub2: 96.43 % (676/701, including 6 gaps), 99.60 % (492/499), 96.62 % (458/474, including 3 gaps) and 98.86 % (612/619), respectively. No cal or his3 sequence data are available for the ex-type culture of D. sackstonii. Although some relevant nucleotide dissimilarities are observed between the sequences of the three loci of these “species”, most are related to the presence of sets of sequence gaps. Morphologically, these taxa are highly similar and harbour ellipsoidal or fusiform alpha conidia of considerably overlapping dimensions (alpha conidia have not been observed in D. machili). Beta conidia have only been observed in D. caryae, D. juglandigena and D. machili and in all three “species” they are produced with a similar shape (filiform) and overlapping sizes (reaching more than 30 μm in length) (Table 4). Regarding the eco-geographical data, while D. sackstonii was introduced on the petiole of Helianthus annuus in Queensland, Australia (Thompson et al. 2015), the other taxa have been introduced as potential pathogens or endophytes on collections of different woody plant hosts in China, including Carya illinoensis (Yang et al. 2018), Juglans regia (Wang et al. 2022), Machilus pingii (Huang et al. 2021b) and Orixa japonica (Lu et al. 2022).
Diaporthe tulliensis R.G. Shivas et al., Persoonia 35: 301. 2015. MycoBank MB 812896.
New synonym: Diaporthe etinsideae Y.P. Tan & R.G. Shivas, Index Austral. Fungi 2: 4. 2022. MycoBank MB 559564.
Notes: The subclade of D. tulliensis included the types of D. etinsideae and D. tulliensis, which grouped with relevant nodal support values and appear phylogenetically indistinguishable (Fig. 6). Therefore, D. etinsideae is made synonymous with D. tulliensis. Sequence comparisons between the types of both species for ITS, tef1 and tub2 showed 99.54 % (647/650, including 1 gap), 99.65 % (566/568) and 99.35 % (764/769), respectively, nucleotide similarities, which represent small, insignificant nucleotide differences. No cal or his3 sequence data are available for the types of both species. Morphological comparisons between the two species cannot be made because D. etinsideae was introduced based on the diagnosis of sequence data and no taxonomic description was provided (Tan & Shivas 2022). Both species were introduced based on collections from Queensland, Australia, but from different hosts. While D. tulliensis was isolated from the rotten end of a fruit of Theobroma cacao (Crous et al. 2015), D. etinsideae was isolated from Annona muricata (Tan & Shivas 2022).
Review of Diaporthe species reported from Arecaceae
A search of the US National Fungus Collections Fungus-Host Database (Farr & Rossman 2023) was conducted to reveal the associations reported between Diaporthe and members of the family Arecaceae. These were supplemented with additional reports from available literature on Diaporthe species reported from palms not covered by Farr & Rossman (2023), as well as with additional unpublished reports whose sequences have been deposited in GenBank. About 40 species of Diaporthe/Phomopsis were found to be associated with hosts in the family Arecaceae, along with several unnamed reports. These species names were verified against MycoBank and Index Fungorum databases (Crous et al. 2004, Index Fungorum Partnership 2023), as well as the available literature, including the most recent overviews of the taxonomy of Diaporthe (e.g., Norphanphoun et al. 2022, Hongsanan et al. 2023). Moreover, all reports with associated and available sequence data were analysed following an integrative taxonomic approach to correctly assign isolates from palms to Diaporthe species. Taking into account that some species have been regarded as synonyms, and based on the data from this study, it can be accepted that 14 valid Diaporthe species are associated with Arecaceae worldwide.
Most species of Diaporthe/Phomopsis reported from arecaceous hosts were identified based on morphological data and host association (Fröhlich et al. 1997, Taylor & Hyde 2003), which have historically been the species recognition criteria used to delimit and identify species in Diaporthe (Rehner & Uecker 1994, Udayanga et al. 2011). Nonetheless, Diaporthe tends to be morphologically conserved, with overlapping morphological characters and extensive phenotypic plasticity. Moreover, it is now widely accepted that multiple species of Diaporthe can often be found on a single host and a single species of Diaporthe can be associated with many different hosts (Mostert et al. 2001, Gomes et al. 2013, Gao et al. 2017, Marin-Felix et al. 2019). Thus, both morphology and host association are of limited value to reliably discriminate and identify species in Diaporthe, insomuch as many of the published names based on such characters are likely to be synonyms. Therefore, the older species names in Diaporthe or Phomopsis for which cultures and/or DNA sequence data are not available, cannot be considered reliable, and should be disregarded until they are re-collected and epitypified. About 15 species of Diaporthe or Phomopsis have been introduced based on their association with Arecaceae hosts and lack associated cultures and/or DNA sequence data. Fröhlich et al. (1997) summarised most Phomopsis species, which have not yet been transferred to Diaporthe. Few Diaporthe species have been later introduced or reported by Taylor & Hyde (2003). Diaporthe arctii was recorded from Trachycarpus fortunei in Switzerland and the United Kingdom. This species has been epitypified by Udayanga et al. (2015) who provided an ex-epitype culture and associated DNA sequence data. However, the validity of this name for the isolates recovered from T. fortunei cannot be confirmed, as they were identified based solely on morphology. Thus, these reports and respective palm hosts associations could be misidentifications. For the above reasons, all reports of Diaporthe taxa collected from Arecaceae hosts lacking associated molecular data were considered unreliable and were disregarded from this review. Furthermore, some reports with associated molecular data have also been excluded and comments on their exclusion are provided below. Table 5 lists all current accepted names of Diaporthe species associated with Arecaceae, their respective hosts and countries from which they were recorded.
The most recorded species of Diaporthe on palm hosts is D. arecae (Table 5). The present study has provided new insights into the range of palm hosts colonised by D. arecae, revealing five new plant host-fungus associations between this species and members of the Arecaceae (see the Taxonomy section for further details). New collections of Diaporthe taxa from palms will likely broaden the range of arecaceous hosts for D. arecae. The remaining Diaporthe spp. reported from palms have only been collected occasionally and are mostly associated with a single Arecaceae host (Table 5). These species have been analysed and treated in the present study. Therefore, almost all Arecaceae hosts-Diaporthe spp. associations listed have been validated here. In addition, five species of Diaporthe isolated from foliar lesions in Lisbon, Portugal, namely D. amygdali, D. foeniculina, D. leucospermi, D. pygmaeae sp. nov. and D. rudis, are reported from palm hosts for the first time, representing new host records of Diaporthe spp. for Arecaceae.
Diaporthe eres was recorded on diseased leaves of Rhapis subtilis by Gao et al. (2016). The authors regarded its phylogenetic position as a species complex, since many of the isolates evaluated revealed ambiguous clades with short branches and moderate bootstrap support. DNA sequence data for this strain, LC 4331, is not available, and thus it was not possible to confirm its phylogenetic position. However, given that the D. eres species complex was shown to constitute a single species by Hilário et al. (2021b), this record was considered in the present listing. Another strain with affiliations to the D. eres species complex, identified as D. cf. nobilis BR67, was isolated from pines of Calamus castaneus in Malaysia by Azuddin et al. (2021). The phylogenetic position of this strain was reassessed here but remained unclear. Thus, the correct name of this species needs to be clarified and the record was disregarded. Similarly, D. angustiapiculata was also disregarded from the present listing. Diaporthe angustiapiculata was introduced as a saprobe on the petioles of Phoenix roebelenii in China. Although it is supported by morphomolecular data, its phylogenetic position needs to be clarified, as it clustered in a basal clade adjacent to the D. eres species complex [sensu Hilário et al. (2021b)]. The species boundaries of this clade remain unclear, with several poorly understood lineages. Thus, future analyses are needed to clarify the status of this new Diaporthe species from palms.
Udayanga et al. (2012a, b) recorded another Diaporthe species from Rhapis sp. in Thailand. Although this strain, MFLUCC 10-0581, is supported with molecular data, no morphological information was included, and the species remained unnamed and was simply regarded as Diaporthe sp. The phylogenetic position of this strain was reassessed here, and it clustered in a clade inside the D. sojae species complex [sensu Norphanphoun et al. (2022)], closely related to D. yunnanensis. Other Diaporthe records from arecaceous hosts retrieved from GenBank or from recent literature have been found to cluster in the same clade, including D. phaseolorum C3349P (from Elaeis guineensis in Malaysia; GenBank accession number: OQ132539), Diaporthe sp. PB60 (from Cocos nucifera in Brazil; GenBank accession number: MK508811), the endophytic strains Phomopsis sp. RDWW-21 and RDWW-23 isolated from leaves of C. thwaitesii in Sri Lanka (Dissanayake et al. 2016) and the endophytic strains Diaporthe sp. BR103, SM59 and SM46 isolated from pines of C. castaneus in Malaysia (Azuddin et al. 2021). As this clade was out of the scope of the present study, these records have been excluded from the present listing. However, this may represent a common species of Diaporthe on palm trees, considering that several isolates collected from palm tissues clustered in this clade, and thus further studies are required to correctly assign them to known Diaporthe species.
Several other Diaporthe records from arecaceous hosts have been deposited in GenBank or noted in recent literature. Their phylogenetic positions were found to be scattered across several different Diaporthe clades. These records were not included in the present analysis, as the species boundaries of most of these clades are unclear and need to be resolved in future studies. Nevertheless, some notes are provided here to aid the revision of Diaporthe species on palms in the future. Saengket et al. (2021) isolated three endophytic strains of Diaporthe from healthy tissues of native Oncosperma palms in Thailand. While two of these strains were confirmed here to be D. arecae (see the Taxonomy section for further details), the third strain (MFLUCC 20-0206) clustered closely to the D. glabrae clade. The strain Diaporthe sp. AA71 (from leaves of C. nucifera in Brazil; GenBank accession number: MK508808) presents affiliation with the D. inconspicua clade. The strains Diaporthe sp. YXD-11 and ZSD-34 (from leaves of Chrysalidocarpus lutescens in the Xisha Islands, China; GenBank accession numbers: OQ469334 and OQ469357, respectively) and the strain Diaporthe sp. PB80 (from leaves of C. nucifera in Brazil) present affiliation with D. sojae. The strains D. biconispora PPR and D. pseudomangiferae NFR (from Phoenix padulosa and Nypa fruticans, respectively, in the Andaman Islands, India; GenBank accession numbers: ON974974 and ON974971, respectively) clustered closely to strains of D. biconispora.
DISCUSSION
Accurate identification of fungal phytopathogens is crucial for understanding the epidemiology and control of plant diseases and is the cornerstone for implementing international phytosanitary measures (Rossman & Palm-Hernández 2008, Wingfield et al. 2012). The criteria used to delimit and name species of phytopathogenic fungi have changed over time and the process has become more challenging by the increasing availability of DNA sequence data (Cai et al. 2011, Steenkamp et al. 2018). Molecular approaches have undoubtedly altered the perception of fungal diversity. However, they have also introduced new hurdles in understanding its evolutionary processes, particularly due to cryptic speciation (Lücking & Hawksworth et al. 2018, Lücking et al. 2020). Cryptic taxa have been increasingly documented in many genera of phytopathogenic fungi and have become an underlying problem in understanding evolutionary relationships in the genus Diaporthe (Udayanga et al. 2012a, Gao et al. 2017). Diaporthe has undergone numerous taxonomic revisions and updates over the years and is currently considered one of the most speciose fungal genera (Bhunjun et al. 2022). New species are continuously being introduced, mostly based on multilocus phylogenies due to the lack of diagnostic morphological characters (e.g., Guo et al. 2020, Huang et al. 2021b, Lambert et al. 2023). Yet, most researchers recognise distinct, well-supported clades in multilocus phylogenies as separate species without carefully examining their boundaries. Therefore, speciation events in Diaporthe remain poorly understood and are often overestimated. In this regard, the present study aimed to implement an integrative taxonomic approach to reliably identify species boundaries and introduce new species in Diaporthe. For this purpose, Diaporthe species occurring on Arecaceae hosts have been used as a case study, following a survey of leaf spotting fungi associated with palm trees in Lisbon, Portugal.
This study introduced a new species and reduced 48 species to synonymy with 16 existing Diaporthe taxa distributed across nine clades, whose species boundaries have been duly resolved. In addition, five species were reduced to synonymy with two existing species previously recognised in the D. arecae species complex (DASC). Overall, the combination of GCPSR and coalescent-based methods, along with a thorough analysis of recombination signals and genetic variability based on sequence polymorphisms, has proven to be a reliable methodology to accurately identify species in Diaporthe.
Multilocus phylogenetic analyses and locus prioritisation in Diaporthe systematics
Multilocus sequence analyses (MLSA) is the current golden standard methodology to understand the evolutionary relationships in Diaporthe. Even so, a gene tree does not necessarily correspond to a species tree, so simply recognising different clades in gene trees as species is likely to misrepresent the true evolutionary history of taxa. As Gatesy & Baker (2005) observed, combining multiple loci that do not individually support a clade, can often reveal emergent support for or conflict within that clade. This phenomenon was clearly in evidence in the present study. Phylogenetic analyses of combined datasets revealed several well-supported lineages within each Diaporthe clade, which have previously been interpreted as distinct species. However, several of these lineages showed phylogenetic discordance in individual phylograms, revealing incongruent nodes, conflicting branches and lack of phylogenetic support. Furthermore, strains of the same species frequently manifested as polyphyletic or paraphyletic in multiple individual phylograms. This, in turn, often resulted in poorly resolved multilocus phylogenetic trees, lacking definitive support values at the internodes between distinct species. For instance, most of the taxa introduced in the D. eucommiae subclade have diverged into well-supported terminal clades, but their phylogenetic relationships remain apparently unclear. A similar result was observed in the subclades of D. amygdali and D. ueckeri. Such phylogenetic discordance is commonly observed in Diaporthe (e.g., Baumgartner et al. 2013, Udayanga et al. 2014a, Hilário et al. 2021a, b, Pereira et al. 2023), but it is often overlooked when new species are introduced. For this reason, it has long been recommended that fungal species circumscription should be based on the simultaneous and rigorous application of multilocus phylogenetic analyses and genealogical concordance (Taylor et al. 2000, 2006, Cai et al. 2011).
According to the Genealogical Concordance Phylogenetic Species Recognition (GCPSR), the lack of genealogical congruence among individual gene trees suggests that the diversity sampled is below the species level. On the other hand, phylogenetic concordance can provide compelling evidence for the lack of genetic exchange and, therefore, evolutionary independence of lineages (Avise & Ball 1990, Taylor et al. 2000). Previous studies have already shown that the GCPSR has profound implications for accurate species recognition in several genera of cryptic phytopathogenic fungi (e.g., Dettman et al. 2003, Coetzee et al. 2005, Cai et al. 2009, Laurence et al. 2014), including Diaporthe (e.g., Udayanga et al. 2014a, b, 2015, Hilário et al. 2021a, b, Pereira et al. 2023). This was corroborated in the present analyses, which shown that the GCPSR is an essential and primary tool for precisely identifying species in Diaporthe. Once the genealogical concordant nodes were determined, the resulting phylogenies for all nine Diaporthe clades became more stable and coherent, making interspecific boundaries well supported and easily identifiable. In addition, the GCPSR strongly suggested a significant taxonomic inflation in the genus due to the incorrect use of highly polymorphic loci to interpret speciation events. As observed in most clades, populations of a given species may separate into distinct subclades when using tree reconstruction methods if there is a strong signal for lineage radiation in the concatenated dataset (Degnan & Rosenberg 2006, Rokas & Carrol 2006, Edwards et al. 2007, Liu et al. 2009). However, this signal can be tracked and properly identified as intraspecific variability when individual loci are gauged for genealogical concordance. This was evident when comparing individual and multilocus phylograms. While tef1-phylograms proved to be largely congruent with the species boundaries ascertained by the GCPSR, ITS- and, at less extent, tub2-phylograms were generally more congruent with the subclades of populations within these boundaries. For instance, the tef1-phylograms clearly delimited the existence of three independent evolutionary lineages (IEL) within the D. eucommiae clade, with minor substructure evidencing evolutionary differences between strains of the same IEL. However, the ITS- and tub2-phylograms resolved into highly variable subclades within each IEL, whose evolutionary relationships are unclear, mostly lacking relevant nodal support. It is possible that these loci are evolving more rapidly and have a higher sequence variability, thus dominating the topology of the multilocus phylograms. A similar hypothesis was suggested by Santos et al. (2010), who noted that the ITS region in Diaporthe appears to be evolving at a much faster rate than tef1 or even mating-type (mat) genes, thus resulting in many potential phylogenetic species.
The uncertainty regarding which set of loci is best suited to infer evolutionary relationships remains an underlying issue in Diaporthe systematics and contrasting results are often reported. Gomes et al. (2013) analysed five loci from 95 Diaporthe species and stated that tef1 poorly distinguished between them, insomuch as they recommended the use of his3 or tub2, along with ITS, as the most appropriate markers for phylogenetic inference in the genus. Udayanga et al. (2014a) recognised tef1, along with apn2 and his3, as the best markers for defining species in the D. eres species complex and excluded ITS due to sequence heterogeneity and resulting taxonomic confusion. Later, Santos et al. (2017a) regarded tef1 as the most informative out of the five common loci used for molecular identification in Diaporthe. More recently, Pereira et al. (2023) ranked cal as the most appropriate locus to infer species limits in the DASC. To access this issue, phylogenetic informativeness (PI) analyses were carried out here to compare each locus with respect to the species boundaries inferred by the multilocus phylogenetic analyses and the GCPSR. The most informative markers for phylogenetic inference of Diaporthe species varied according to the clade being analysed. However, tef1 showed the highest net PI, which was congruent with the percentage of informative characters that also suggested tef1 as the most informative locus for phylogenetic inference of most Diaporthe clades. These results are in line with the similarities observed between the topology of the combined gene analyses and the tef1-phylograms, which showed highly congruent terminal nodes. The approximately 300 bp complete intron sequence of tef1 has long been recognised as a powerful marker to resolve cryptic speciation in Diaporthe (Castlebury et al. 2001, Santos et al. 2010, Udayanga et al. 2012a, b, Santos et al. 2017a). As suggested by Santos et al. (2017a), using tef1 for species identification in Diaporthe may be a better option when only one locus barcoding is possible. The present results further suggest that future studies on Diaporthe should use tef1 as the primary marker to provide an estimate of the species boundaries, instead of ITS, which is currently used as the guide barcode for identifying Diaporthe isolates.
The ITS region has been proposed as the standard fungal DNA barcode and is widely used in fungal systematics to delimit species and understand evolutionary relationships (Schoch et al. 2012, Vu et al. 2019). However, there are several known issues with the effectiveness of this region, including the overestimation and underestimation of fungal diversity (e.g., Crouch et al. 2009, Gaziz et al. 2011, Maharachchikumbura et al. 2012). Several authors have argued that ITS is uninformative for accurate species identification in Diaporthe due mainly to its high intraspecific and intragenomic variability (e.g., Santos et al. 2010, Udayanga et al. 2014a, Hilário et al. 2021b). Hilário et al. (2022) showed that ITS sequences from individuals of the same Diaporthe species may differ by 3.0–9.4 %, and they also detected ITS paralogs with a variation of up to 14 %. This intraspecific variability of ITS was evident in each Diaporthe clade treated in the present study. ITS showed the lowest informative characters to resolve each Diaporthe clade, indicating that this locus might not be suitable for species delimitation in Diaporthe. However, while ITS was the least informative locus as the trees approached their roots, a substantial peak in its net PI profiles (PIP) was observed in several Diaporthe clades. This apparently informative signal of ITS corresponds to the relative period in which the respective clades radiate into several branches and subclades, reflecting the high intraspecific variability of this locus. Integrating PI over specific periods provides a metric for ranking loci, as the integration values will be higher for the loci that have the greatest probability of substitution during that period (Townsend 2007, López-Giráldez & Townsend 2011). Therefore, the peak on the PIP of ITS indicates that this locus is the most informative marker to infer intraspecific variation within several Diaporthe clades. This has also been previously documented for the DASC (Pereira et al. 2023). Although ITS may not be relevant in studies that aim to determine the species limits in Diaporthe, it may be a suitable locus in studies that aim to investigate the genetic diversity and population structure of Diaporthe species in relation to their host and geographical ranges. This will further allow testing of evolutionary hypotheses in the genus, such as the occurrence of recombination.
It should be noted that the resolution of some clades in this study has led to the recognition of species whose oldest type specimens do not have associated ex-type cultures. Examples of such species include D. eucommiae, D. glabrae, and D. micheliae. As a result, the type of these species only has ITS sequences available and lacks additional loci to support their phylogenetic positions. However, the available data for these taxa cannot be disregarded, and thus, they have been used in the taxonomic decisions made here. Although the ITS region has been deemed uninformative for molecular identification in Diaporthe, this is primarily due to its high intraspecific variability, which tends to split the same Diaporthe species into several hypothetical phylogenetic species (Santos et al. 2010). Therefore, the results obtained in the present study provide convincing evidence that the ITS sequences available for D. eucommiae, D. glabrae, and D. micheliae are efficient in determining their phylogenetic position in a certain Diaporthe clade. However, it is important not to overlook the need for further resolution of these species, and future studies should aim to epitypify them or provide reference specimens with associated cultures and molecular data. Epitypification is a useful tool to resolve taxonomic issues and to stabilise the systematics of poorly circumscribed species (Hyde & Zhang 2008, Ariyawansa et al. 2014). The epitypification of the aforementioned Diaporthe species is necessary to fix the precise application of their names and confirm their phylogenetic position.
The considerations previously made for tef1 and ITS are not exclusive to these loci. In most Diaporthe clades, cal was paired with tef1 as one of the most informative loci. Moreover, tub2 and his3 frequently displayed a weak discriminatory signal with a tree-tip associated peak similar to that of ITS, reflecting their potential high intraspecific variability. However, evaluating the effectiveness of these loci is not an easy task, as they seem to vary substantially depending on the clade under analysis. For instance, tub2 ranks, together with tef1, as one of the most informative loci to resolve the D. brasiliensis and D. glabrae clades. The tub2 alignment used in the D. brasiliensis dataset was much longer, which clearly affected the informativeness of this locus. Nonetheless, the length of the tub2 alignment used in the D. glabrae dataset was similar to that used in the phylogenetic analyses conducted for the remaining clades. Therefore, the relative power of the common loci used in Diaporthe systematics is variable and seems to be case-specific. Phylogenetic studies on Diaporthe have been using loci that have been chosen in a more or less ad hoc manner, by considering how conserved these loci were shown to be in other fungal genera (e.g., Gomes et al. 2013, Udayanga et al. 2012a, 2014a, b). Yet, the phylogenetic resolution of these markers is still poorly understood, and their evolutionary nuances in Diaporthe are virtually unknown. This can be easily observed when accessing the nucleotide sequence similarities between different Diaporthe species, or between different strains of a given species. For instance, the present study detected nucleotide dissimilarities of up to 7 % in the loci available for the strains of D. amygdali and D. eucommiae studied here. Interestingly, these dissimilarities were more prominent in the partial sequences of tef1 and cal for few strains. These results do not necessarily indicate that tef1 partial sequences exhibit more intraspecific variability than ITS or tub2. In fact, generally, the length of the available tef1 and cal partial sequences is considerably smaller than that of ITS or tub2. Therefore, only a few nucleotide differences can strongly impact the p-distances evaluated for both loci. Moreover, the intraspecific variation in these loci may be greater than the interspecific variation, which further complicates the identification of Diaporthe species. This has already been documented for ITS sequences (Hilário et al. 2022) and was also occasionally observed for the remaining loci in the present study.
Jeewon & Hyde (2016) suggested that, for practical purposes, a minimum of more than 1.5 % nucleotide dissimilarities in the ITS region, and higher percentages for fast-evolving introns of protein-coding genes, may be indicative of a new species. Later, Vu et al. (2019) predicted an even more conservative optimal identity threshold of 99.6 % to discriminate between filamentous fungal species. However, there is no consensus on the percentage of matching base pairs between a reference and query sequences required for a reliable identification, and it is evident that a single threshold will not apply to all fungi (Crous et al. 2021b). This is true for Diaporthe, whose species appear to be evolving rapidly and at a variable rate depending on the clade, consequently making it challenging to interpret genetic variability on the loci used to understand their evolutionary relationships. Although tef1 is considered here as the most suitable locus for phylogenetic inference in Diaporthe, it is not yet possible to determine a single, standard locus to resolve species boundaries in the genus. Similarly, there is no fixed threshold of nucleotide similarity to determine whether a lineage represents a new species, and this may vary according to the set of Diaporthe species or strains. For instance, Guo et al. (2020) introduced D. zaobaisu as a closely related taxa to the D. foeniculina subclade based on multilocus phylogenetic analyses of a combined ITS-tef1-tub2-his3 dataset. Nevertheless, only tef1 showed relevant sequence dissimilarity when compared to the sequences available for the ex-epitype culture of D. foeniculina. This included 34 nucleotide differences, consisting of 3 substitutions and 31 indels, spanning two regions located at the beginning and the end of the tef1 intron. Although these sequences have a considerable number of differences that are shared between the three strains representing D. zaobaisu, they cannot fully separate D. zaobaisu from D. foeniculina in the phylogenetic analyses. According to the GCPSR, D. zaobaisu strains are considered to represent intraspecific variation of D. foeniculina, as only one locus, out of four loci, separates the two species. If only tef1 sequence data were available for D. zaobaisu, it could be erroneously concluded that the species belongs to a new lineage. This example emphasises the importance of carefully and objectively interpreting the phylogenetic analyses to avoid overestimating species diversity within the genus.
The above discussion on the hints observed in the DNA barcoding of Diaporthe species, highlights the impact that multilocus DNA sequence data have on improving the appropriate identification of Diaporthe, and fungi in general. Indeed, while data from a single locus tends to infer information about the evolutionary history of a particular gene, sequence data from multiple markers can infer the evolutionary history of taxa (Boluda et al. 2019). The multilocus phylograms constructed here provided a clearer view of both interspecific and intraspecific diversity in all Diaporthe clades compared to any individual gene tree. Thus, even though the evaluation of individual gene trees is fundamental for a better understanding of evolutionary patterns and independence of lineages, the use of MLSA is necessary to establish robust species boundaries between taxa (Dupuis et al. 2012). Although recent taxonomic accounts of Diaporthe suggest that MLSA, including ITS, tef1, tub2, cal and his3 loci, should always be considered, several authors continue to introduce new species based solely on ITS (e.g., Crous et al. 2013, Dayarathne et al. 2020), or on the combination of ITS and tef1 and/or tub2 loci (e.g., Tan et al. 2013, Gao et al. 2014, Gao et al. 2017, Liu et al. 2015, Thompson et al. 2015, Wrona et al. 2020). Giving the high intraspecific variability observed in these loci, several of the species introduced in recent years are likely to represent an overestimation of speciation events, which was corroborated by the GCPSR results. However, this issue becomes increasingly problematic as more species are introduced based on a single or a few barcodes. This is because many researchers rely on ITS due to its high availability in public databases, coupled with the limited number of sequences for additional markers for several taxa (Hongsanan et al. 2018, Crous et al. 2021b). For example, Liu et al. (2015) introduced D. thunbergiicola as a sister species to D. ueckeri based on multilocus phylogenetic analyses of a combined ITS-tef1 dataset. Although some relevant nucleotide differences were observed between the sequences of the types of D. thunbergiicola and D. ueckeri, they cannot fully separate both species in the phylogenetic analyses. Diaporthe thunbergiicola clustered as a very long branch intermingled with D. ueckeri strains, suggesting that it is likely to represent intraspecific variability of D. ueckeri. For taxonomic stability, D. thunbergiicola was made synonym with D. ueckeri and only sequence data from other loci will be able to fully resolve this lineage. This example highlights the crucial role of MLSA in preventing the unwarranted proliferation of Diaporthe species names, as well as the establishment of lineages with ambiguous phylogenetic affiliations.
The notable disagreement regarding the optimal set of loci for species identification in Diaporthe reflects the lack of knowledge about its speciation process and evolutionary history. Speciation of fungal phytopathogens has been linked to reproductive isolation resulting from host jumps, host domestication, clonal divergence, and hybridisation (Stukenbrock 2013, Steenkamp et al. 2018). Although little is still known about the genetics underlying these processes, comparative genomics studies suggest that genome plasticity is a mechanism for rapid adaptive evolution and potentially speciation (Stukenbrock 2013). The phylogenetic incongruences detected in this and other studies on Diaporthe are probably a typical example of within-species genome plasticity. Such plasticity could have been driven by host and/or environmental adaptation or specialisation. Indeed, most Diaporthe species present a cosmopolitan behaviour, with the ability to infect a wide range of hosts, with different lifestyles and often with a worldwide distribution (Gomes et al. 2013, Gao et al. 2017). Thus, it is reasonable to speculate that the great genetic variability of Diaporthe species is a consequence of evolutionary adaption and specialisation to different ecological niches. As observed in the present study, Diaporthe species with a broad host range and/or a wide and extensive geographical distribution tend to present a greater genetic variability. For instance, genetic variability at some loci was more expressive between strains of D. eucommiae than between strains of D. brasiliensis and D. caatingaensis. While D. eucommiae has been reported as an endophyte, saprophyte and/or pathogen on more than 25 genera and 30 species in 26 plant families and six countries, D. brasiliensis and D. caatingaensis have virtually been only described as endophytes from medicinal plants in Brazil or occasional hosts in surrounding countries. This genetic variability has also been widely documented for D. eres, whose recurrent worldwide identification often resulted in taxonomic instability due to the generation of ambiguous and weakly supported clades (Udayanga et al. 2014b, Gao et al. 2016, Chaisiri et al. 2021b, Hilário et al. 2021b). The phylogenetic inference of Diaporthe clades with high intraspecific diversity also displays a considerable and somewhat uncertain substructure. This includes unsupported internal nodes and sub-branches of varying length, suggesting that these species may be undergoing active divergence and speciation. This has first been recognised by Gao et al. (2016) for the D. eucommiae (as D. hongkongensis) clade but has been extensively observed in several of the clades treated in the present study. These results are in line with the previously formulated hypothesis of within-species genome plasticity as a function of adaptive radiation to distinct ecological niches explored by populations of a given Diaporthe species. The reduction of gene flow between groups of individuals, caused by host and/or geographical barriers, results in the loss of ancestral polymorphisms or the emergence of new, unshared genomic variants. Therefore, alleles are sorted due to genetic drift or other stochastic evolutionary forces, leading to a decrease in the proportion of shared polymorphisms within a population, as genetic isolation is resulting in new divergent species. Loci from divergent species pass through polyphyly and paraphyly stages before becoming reciprocally monophyletic (Avise & Ball 1990, Rosenberg 2003, Sakamoto & Innan 2019). It is likely that the phylogenetic incongruences observed in Diaporthe are a consequence of an adaptive radiation process. As a result, future studies may recognise divergent lineages as new species due to the recurrent isolation of new Diaporthe strains. Therefore, it is increasingly important to investigate speciation events in the genus using an integrative taxonomic approach that includes methods capable of distinguish incomplete lineage sorting (ILS) from reticulation processes associated with intraspecific diversity.
Integrative taxonomy for species delimitation in Diaporthe
Recent studies have already advocated that in addition to multilocus phylogenies, other methodologies and datasets should be used to establish well-supported boundaries among taxa where cryptic speciation processes have been reported (Carstens et al. 2013), which is commonly known as integrative taxonomy (Dayrat 2005, Padial et al. 2010, Stengel et al. 2022). This approach has already been shown to have a considerable impact on Diaporthe systematics (e.g., Pereira et al. 2023) and its use has been formalised in this study considering a larger number of clades. In this regard, DNA-based species delimitation methods, along with phylogenetic networks, recombination and morphological analyses, were used to test the species boundaries ascertained by the GCPSR. The incongruences detected between individual gene genealogies in all Diaporthe clades indicate that the respective loci have different evolutionary histories, impairing a reliable analysis of their concatenation. Evolutionary phenomena such as ILS, horizontal gene transfer, gene duplication and loss, hybridisation, and recombination, are well-known sources of such phylogenetic incongruences (Degnan & Rosenberg 2009, Som 2015). For this reason, it has recently been recommended the use of methods that can account for these phenomena and accordingly make quantitative predictions about the likelihood of species delimitation hypotheses (Degnan & Rosenberg 2009, Rannala & Yang 2020). Due to their objectivity and precision, DNA-based species delimitation methods are becoming frequently used as powerful tools in fungal taxonomic studies (Maharachchikumbura et al. 2021, Chethana et al. 2021a).
The present analyses combined five quantitative species recognition methods based on two different approaches, namely coalescent (viz. Poisson tree processes, PTP; multi-rate PTP, mPTP) and genetic distance (viz. statistical parsimony network, SPN; automatic barcode gap discovery, ABGD; assemble species by automatic partitioning, ASAP). All methods yielded largely congruent results regarding the taxonomic decisions made according to the GCPSR. Nonetheless, different approaches often produced different species delimitation scenarios, which is congruent with the findings of previous studies (Bustamante et al. 2019, Maharachchikumbura et al. 2021). Coalescent- and genetic distance-based methods have already been successfully applied to delimit boundaries of cryptic species complexes in different fungal genera, including Alternaria (Stewart et al. 2014), Beauveria (Bustamante et al. 2019), Colletotrichum (Liu et al. 2016), Chloridium (Réblová et al. 2022), Fusarium (Glässnerová et al. 2022) and, more recently, Diaporthe (Pereira et al. 2023). These methods are particularly useful in cases where there is a lack of distinctive morphological characters, which limits polyphasic approaches that end up relying almost exclusively on multilocus phylogenies. Besides avoiding arbitrary cut-offs, these methods can also overcome the common over-supporting poorly resolved clades in MLSA (Rannala & Yang 2003, Belfiore et al. 2008, and references therein). Stewart et al. (2014) questioned the reliability of concatenation methods in the presence of divergent evolutionary histories and, using quantitative species recognition approaches, provided strong evidence for an overestimation of Alternaria species associated with brown spot of citrus worldwide. A similar result was reported by Liu et al. (2016), who showed that species introduced based on gene concatenation in Colletotrichum siamense species complex were recognised as a single entity by quantitative species recognition approaches. Their results are similar to that of the present study on all the Diaporthe clades treated. Most of the well-supported lineages depicted by concatenated phylograms were not supported by the quantitative species recognition methods implemented. Both coalescent- and genetic distance-based methods showed that Diaporthe species were overestimated in all nine clades. Therefore, these findings reinforced the existence of a significant taxonomic inflation in the interpretation of speciation events in the genus. However, while the coalescent-based methods were highly in agreement with the GCPSR results, exhibiting high values of delimitation efficiency ratio (DER), the distance-based methods tended to produce false negatives. Indeed, several IEL recognised by the GCPSR were over-lumped by the genetic distance approaches. Variability of results from species recognition methods is relatively common between loci and settings (Maharachchikumbura et al. 2021), which may be related either to the fundamentals of the methodology or to the inherent characteristics of the datasets. Therefore, to understand these conflicting results can help to maintain some consistency in the decision-making process about species limits.
Overall, the present results suggested that coalescent-based PTP and mPTP methods outperformed the distance-based analyses for delineating Diaporthe species. Nonetheless, although the over-lumping effect was observed in all the distance-based analyses and for most of the Diaporthe clades treated, they were found to be more prominent with ABGD and ASAP than with SPN analyses. SPN associates sequences of a dataset by calculating the maximum number of mutational steps that constitutes a parsimonious connection between two haplotypes at 95 % statistical confidence, and then joins haplotypes into networks. Haplotypes separated by more mutational steps, i.e., for which the probability of secondary mutations exceeds 5 %, remain disconnected in different networks, with are taken as hypothetical species (Templeton et al. 1992). Hart & Sunday (2007) found empirical evidence that subnetworks in SPN analyses were significantly consistent with Linnaean names, insomuch as they could be used to differentiate species in DNA sequence datasets. Although most of the Diaporthe clades in the present study corroborate the empirical evidence of Hart & Sunday (2007), some of them showed inconsistent results with the IEL determined by the GCPSR. In the D. foeniculina, D. glabrae and D. longicolla clades, several IEL were grouped together in a single subnetwork, indicating that they may be the same species. However, the consistency observed between the GCPSR and the coalescent-based approaches suggests that the lumping effect of this genetic distance-based approach is likely to represent an artefact of sequence divergence between Diaporthe taxa. As previously discussed, loci used in Diaporthe present some evolutionary conflicts that are difficult to interpret. The results of the SPN analyses could be indicative of the presence of reticulation or ILS processes, which are in line with the phylogenetic incongruences widely documented in Diaporthe. Due to a variable degree of sequence divergence between different clades of Diaporthe, it may not be sufficient to unambiguously delimit species. This is because of a high likelihood of shared alleles between distinct Diaporthe species resulting from the retention of ancestral polymorphisms or lack of gene flow barriers between populations (e.g., Hilário et al. 2021a, b). Likewise, the high intraspecific diversity of Diaporthe taxa is expected to highly influence the results of the SPN analyses. For instance, undersampling of intraspecific haplotype diversity has been shown to produce false positives and accordingly over-split taxa by the presence of few strains with highly polymorphic haplotypes (Chen et al. 2010). Indeed, over-splitting of IEL were only observed by SPN analyses on the subclades of D. leucospermi and D. sackstonii (belonging to the D. leucospermi clade). These subclades were found to harbour high intraspecific diversity, which is likely to be caused by some degree of host or geographical isolation. The genotypic variant of D. infecunda (syn. D. leucospermi) has only been virtually recorded as an endophyte in South American countries, which is probably leading to a substantial genetic divergence of these strains. Similarly, most taxa in the D. sackstonii subclade have been introduced based on a few strains isolated from different woody hosts in China, which has probably reduced the gene flow between populations leading to a substantial genetic divergence. These conflicting effects caused by the patterns of genetic diversity of Diaporthe taxa were even more expressive in the ABGD and ASAP analyses.
The ABGD and ASAP analyses detect a barcode gap between intra- and interspecific diversity in the distribution of pairwise differences among all sequences of a dataset. These sequences are then partitioned into the maximum number of groups (i.e., hypothetical species) such that the distance between two sequences from distinct groups will always be larger than a given threshold distance (i.e., barcode gap) (Puillandre et al. 2012, Puillandre et al. 2021). As these methods rely exclusively on discontinuities in DNA sequence variation, they may not be effective in species with strong population genetics structures or species that are less divergent (Yu et al. 2017, Bhunjun et al. 2020). Both scenarios are plausible in the evolutionary history of Diaporthe. In fact, the previous analyses regarding the common loci used in phylogenetic inference of Diaporthe species have suggested that clades of a given species harbour substantial substructure. Moreover, some species seem to reveal patterns of ILS in some loci, which are consistent with a relatively recent speciation process (e.g., Pereira et al. 2023). Therefore, it is not surprising that distance-based ABGD and ASAP methods were found to be inadequate for species recognition in Diaporthe. Several studies have already criticised the lack of phylogenetic content and evolutionary justification of distance-based methods, which do not rely on an underlying genealogical tree (Moritz & Cicero 2004, Taylor & Harris 2012). As shown by previous studies, the gap between intra- and interspecific divergences may vary among taxonomic groups or it may not even exist (e.g., Kvist et al. 2014, Wilson et al. 2023). Intra- and interspecific diversity in Diaporthe varied according to the clade of taxa considered. As a result, distance-based methods are expected to produce unstable results depending on the set of Diaporthe species being analysed, which was clearly in evidence in the present results. Both ABGD and ASAP calculate the pairwise distances without considering species affinities and infer a model-based confidence limit for the empirical divergences, using an a priori determined range of intraspecific divergence (Puillandre et al. 2012, 2021). Consequently, relatively high intraspecific variation values can easily be confused with low interspecific divergence values, resulting in pairwise distances distributions that are difficult to interpret. The inconsistent intra- and interspecific diversity among Diaporthe clades justify why these methods gave such variable and conflicting results when compared to the GCPSR. A correct interpretation of the results from ABGD or ASAP presupposes a reasonable range of intra- and interspecific divergences, which is not clearly the case in Diaporthe. Species assigned to different IEL by the GCPSR presented slightly overlapping intra- and interspecific diversity. This resulted in a potential wrong evaluation of the barcode gaps computed by ABGD and ASAP analyses, which frequently lead to an underestimation of species diversity, as corroborated by previous studies (e.g., Yang & Rannala 2016, Yu et al. 2017).
Considering that ABGD and ASAP were developed for single locus datasets and have yet not been previously tested in Diaporthe, the tef1 alignments were also used in this study to understand the impact of multilocus datasets on the species delimitation schemes. This locus was chosen considering that it was shown to be the most informative according to the PI profiling method. However, in most Diaporthe clades, no variation was detected between the ABGD and ASAP species delimitation schemes produced using tef1 or multilocus datasets. In the clades where variation was detected, such as the D. glabrae, D. leucospermi and D. longicolla clades, the ABGD and ASAP results with the tef1 datasets were even more conflicting than the results based on the multilocus datasets, with lower DER values. Similar results were obtained when coalescent-based methods were applied to tef1-phylograms. These outcomes endorse the discussion regarding the importance of MLSA in Diaporthe systematics. Although tef1 is, generally, the most informative locus to infer Diaporthe phylogenies, its evolutionary history may still reveal poor species delimitation. Importantly, most of the incongruent ABGD and ASAP species delimitation schemes were based on datasets that excluded the cal and his3 loci due to insufficient sequence data for several taxa. Their inclusion strongly disturbed the distance matrices generated and the corresponding barcode gaps. Therefore, the accuracy of these analyses seems to be more influenced by missing data than the coalescent-based methods, whose delimitation schemes were more stable when tested on datasets of 3- and 4-loci.
The PTP methods are based on the distinct branching patterns between divergence (Poisson model) and intraspecific diversification (coalescent model). These patterns are used to distinguish between speciation and population processes, which are measured in terms of the number of nucleotide substitutions per site. Considering that within-species branching events will be substantially more frequent than between species, the transition between different branching patterns is the threshold used to predict species boundaries (Zhang et al. 2013, Kapli et al. 2017). Contrarily to the genetic distance-based methods, PTP and mPTP rely on underlying evolutionary relationships and can accommodate genealogical ambiguities into species recognition (Fujita et al. 2012). This was evident on the PTP and mPTP species delimitation schemes obtained in this study. Despite the various phylogenetic incongruences detected among individual gene genealogies, in almost all clades, the species delimitation schemes produced by both methods were identical to the IEL depicted by the GCPSR. These results provide convincing evidence for the taxonomic decisions made in this study regarding the proposed synonyms in each Diaporthe clade. Moreover, they strongly suggest that PTP and mPTP coalescent-based methods are a precise and accurate approach, together with the GCPSR, to define species boundaries in Diaporthe, as suggested by previous studies (Hilário et al. 2021a, b, Pereira et al. 2023). Although PTP and mPTP analyses were highly congruent, mPTP analyses frequently over-lumped IEL when tested on 5-, 4-and 3-loci datasets to evaluate the impact of missing data and/or number of taxa. This was verified, for instance, in the D. foeniculina and D. glabrae clades. According to Kapli et al. (2017), mPTP accommodates different degrees of intraspecific genetic diversity within a phylogeny and has an improved delimitation accuracy compared to PTP. However, considering the limited understanding of the speciation process and evolutionary history of Diaporthe, this study suggests that PTP may be less susceptible to false negatives or false positives. Its delimitation schemes were found to be less affected by a decrease in the number of taxa or loci. Therefore, a conservative approach was taken here to estimate the boundaries of species within each clade, taking into consideration a majority rule consensus between the different approaches (Carstens et al. 2013).
The generalised over-lumping of IEL observed with distance-based approaches, or in some mPTP results, provided further support for the potential synonyms suggested by the phylogenetic analyses, as an over-splitting scenario has rarely been observed. To ensure a meaningful and cautious taxonomic treatment, IEL suggested by the GCPSR have not been made synonymous, even if considered as a single entity by other methodologies. This conservative approach avoids merging distinct taxonomic entities under the same species name. However, it may also result in retaining taxa that correspond to the same species as separate entities. For example, D. serafiniae was regarded as conspecific with D. leucospermi by PTP and mPTP analyses. Yet, due to lack of cal and his3 sequence data, the phylogenetic position of this species within the D. leucospermi clade remained unclear following the GCPSR. Evolutionary relationships in the D. leucospermi clade were highly variable among the different species recognition analyses conducted. This suggests that interpreting intra- and interspecific variation in this clade is particularly challenging, as observed in the haplotype connections inferred by the SPN analyses. Due to the limited available loci and the small sample size for D. serafiniae, this study refrained from synonymising it under D. leucospermi, since the presence of ILS cannot be disregarded as a potential source of phylogenetic discordance. Incomplete lineage sorting occurs when recently diverged lineages retain ancestral polymorphisms due to insufficient time to achieve reciprocal monophyly. Typically, the lack of complete lineage sorting cannot be determined without using multiple individuals per taxon (Maddison & Knowles 2006). Future studies, with a larger sample size, may conclude that D. serafiniae and D. leucospermi species are conspecific. This rationale is applicable to all studied Diaporthe clades and highlights the importance of a large sampling size in species delimitation analyses (Ahrens et al. 2016).
The phylogenetic species concept assumes that the fixation of a particular character state in a population indicates a long history of reproductive isolation. However, species recognition in phylogenetic studies is typically based on the characters of a small sample of individuals rather than the entire population of a species (Walsh et al. 2000). Consequently, individuals from a small sample size sharing a few unique characters can often be easily identified as distinct lineages from the population of a given polymorphic species (Davis & Nixon 1992, Goldstein et al. 2000). In species with a widespread distribution, such as most Diaporthe species, a few individuals often fail to represent the true diversity of the species, insomuch as new taxa can be mistakenly introduced when sampled from divergent populations. This phenomenon has been often detected in the present study and might be the main reason for the ambiguous species boundaries in most of the Diaporthe clades treated. For instance, Gao et al. (2016) showed that the phylogenetic distinctiveness between D. hongkongensis and D. lithocarpi (in the D. eucommiae clade) and between D. ueckeri and D. miriciae (in the D. longicolla clade) was completely erased by the addition of a number of new strains in both groups of species. Despite the incongruencies observed by Gao et al. (2016) in both clades, subsequent studies have introduced several closely related taxa that were found here to represent intraspecific variation, rather than distinct lineages, when a larger number of strains were used in their phylogenetic inferences (e.g., Wanasinghe et al. 2018, Crous et al. 2020, Manawasinghe et al. 2021, Cao et al. 2022). A similar outcome was also observed in the D. foeniculina clade. Although Hyde et al. (2019, 2020a) have introduced D. rumicicola and D. nigra, respectively, as distinct lineages sister to D. foeniculina, their distinctiveness was not supported in the analyses carried out here when additional strains were included in the clade. Therefore, to accurately delimit species or introduce new taxa in Diaporthe, it is essential to assess intraspecific diversity using a large number of strains, ideally from diverse origins, as suggested by previous studies (e.g., Gao et al. 2016, Hilário et al. 2021a, 2022).
It should be emphasised that while the coalescent-based results were deemed superior to those of the distance-based approaches, the latter still provided valuable insights into the phenomena underlying the evolutionary history of Diaporthe. The previously hypothesised highly rapid speciation for Diaporthe taxa has also been hinted by the distance-based approaches. Indeed, their over-lumping results seem to indicate an evolutionary history with patterns of ILS and reticulation processes, such as recombination. This is likely the reason why coalescent-based methods outperformed distance-based ones, as the former provide a more comprehensive view of the speciation process through the recognition and accommodation of gene tree discordant events (Edwards 2009, Jiao et al. 2021, Mirarab et al. 2021). This hypothesis was also corroborated by the topology of the phylogenetic networks built for each Diaporthe clade.
Phylogenetic networks are a generalisation of phylogenetic trees into diagrams that represent ancestral relationships between taxa whose evolutionary history includes non-treelike events (reticulations), such as recombination, hybridisation, and horizontal gene transfer (Huson & Bryant 2006, Bapteste et al. 2013). In the present study, the phylogenetic networks further evidenced the conflicting phylogenetic signals implied by the species recognition approaches. Most of the clades examined show that parallel edges and boxlike polygons are dominant in the evolutionary relationships among Diaporthe species. Furthermore, clades with well-resolved phylogenetic relationships did not generally exhibit contradictory phylogenetic information in the networked relationships of their taxa, as observed in the D. brasiliensis and D. rudis clades. The remaining clades exhibited extensive conflicting signals, indicating a lack of barriers to gene flow between populations and implying likelihood of recombination. In this regard, the pairwise homoplasy index (PHI) test was used to further analyse these results and complement the GCPSR criteria. Thus, all clades, subclades and proposed synonyms were tested for genetic exchange to indicate their evolutionary independence. Remarkably, the PHI test results confirmed most of the predicted evolutionary relationships, suggesting new hints to support the decision-making process about species limits and a few examples are discussed herein. The PHI test yielded a positive result between D. infecunda and D. leucospermi, but a negative result between D. serafiniae and D. leucospermi. Although the limited sample size should not be ignored, these results support the hypothesised ILS as a potential source of discordance for the phylogenetic resolution of D. serafiniae, as well as the conspecificity of D. infecunda and D. leucospermi. Recombination was shown to be statistically significant between strains of D. eucommiae, D. glabrae, D. ueckeri and D. sackstonii and, therefore, the proposed species boundaries in each subclade were reinforced by the presence of genetic exchange. Interestingly, none of the subclades in the D. foeniculina clade tested positive for recombination, despite the extensive conflicting signals in their networked relationships. Considering that D. foeniculina is commonly recorded worldwide and in association with a wide range of hosts, it is likely that recombination among these taxa has been minimised over time due to their adaptive divergence and ecological allopatry. This may also explain why D. zaobaisu (syn. D. foeniculina) has highly polymorphic loci. While most strains of D. foeniculina have been recorded in European countries, only a few have been recorded in two provinces of China, including the genotypic variant “D. zaobaisu”, which is likely a result of adaptive divergence due to geographical isolation. A similar result was observed in the subclades of D. isoberliniae, D. longicolla and D. rudis. For instance, although D. rudis has a widespread distribution and has been recorded from more than 40 genera and 55 species across 32 plant families, in Europe it has mostly been associated with Vitis vinifera. The adaptive divergence of D. italiana (syn. D. rudis) to Morus alba in Italy, a host on which D. rudis has not previously been recorded, is likely to explain the negative PHI test result between these taxa. Statistically significant evidence for recombination was found between D. aseana, D. eucommiae and D. xishuangbanica, which is consistent with the over-lumping of these taxa in a few DNA-based species delimitation analyses. Given the high intraspecific variation in the D. eucommiae clade, the detection of significant recombination between these lineages may be the result of a recent speciation process and, consequently, a lack of complete lineage sorting. This hypothesis is supported by the conflicting phylogenetic signals and relative branch distances observed in the phylogenetic network built for these taxa. A similar hypothesis has been suggested for the evolutionary patterns observed in the DASC and for the radiation of D. smilacicola and D. chiangmaiensis from D. arecae (Pereira et al. 2023). However, the existence of a putative hybrid in Diaporthe was recently reported (Hilário et al. 2022). Therefore, these taxa may also be linked by occasional hybridisation, which can be also a potential source of the phylogenetic incongruences detected. However, this hypothesis could only be tested with genome-scale data and phylogenomic analyses.
Considering that morphological characters have historically been used to define species in Diaporthe, the present study also investigated a possible correlation between phylogenetic boundaries and taxa morphology as part of the integrative taxonomic approach. However, the hierarchical cluster analysis (HCA) performed based on the available phenotypic data for taxa of each Diaporthe clade revealed that species recognised in all clades lack morphological distinctiveness. Moreover, the groupings displayed on the dendrograms of phenotypic data do not support any of the previously identified species in each Diaporthe clade, nor the IEL defined according to the species recognition analyses conducted in this study. Is has been long established that the morphological species concept is unreliable in reflecting the evolutionary history of Diaporthe, as morphological characters in the genus are highly conserved and display great plasticity depending on environmental conditions (e.g., Mostert et al. 2001). The HCA of phenotypic data carried out here reinforces the existence of cryptic speciation in Diaporthe, indicating no correlation between its morphology and phylogenetics. This complex diversification of Diaporthe, together with its phenotypic plasticity under different conditions, has limited the comprehension of the emergence of novel species and genotypic variants in the genus. As a result, the ecology of most Diaporthe lineages is poorly understood, including their host ranges and lifestyles. For many hosts, the complex of Diaporthe species remains relatively unknown or poorly annotated due to the erroneous introduction of genotypic variants as speciation events. This study has uncovered, for the first time, the complex of Diaporthe species on Arecaceae hosts.
Global distribution, ecology, and significance of Diaporthe on palms
The imprecise delimitation of Diaporthe species has practical implications for the study of the genus, as they are often associated with plant diseases (Hilário & Gonçalves 2023). Indeed, a reliable framework for the identification of fungal phytopathogens is essential for disease prevention and control, and can avoid economic losses in the production, import and export of agricultural and forestry products (McCartney et al. 2003, Rossman & Palm-Hernández 2008, Bhunjun et al. 2021). The present study significantly improves the understanding of the taxonomy and host diversity of various lineages in Diaporthe by clarifying species boundaries in different clades. Moreover, it illustrates the lack of knowledge about the association of Diaporthe taxa with Arecaceae hosts. This is not surprising considering that most records of Diaporthe from palms were identified based on morphology and host association (Fröhlich et al. 1997, Taylor & Hyde 2003), which are both inadequate to delineate species in the genus. Although a relatively small collection of Diaporthe strains was obtained from foliar lesions of ornamental palms in Lisbon, Portugal, five species were identified, all representing new host family records for Arecaceae. A total of 13 new plant host-fungus associations were ascertained, mostly of widespread and well-known Diaporthe pathogens in European countries, including Portugal.
In contrast to the other palm species sampled in this study, which are exclusively grown as ornamentals in Europe, Chamaerops humilis is one of only two native European palms and the sole species found in the Iberian Peninsula (Guzmán et al. 2017). However, only one Diaporthe species has previously been reported in association with C. humilis in Greece, and its identity was resolved in the present study as D. cytosporella. Although this record has been linked to D. chamaeropis (Gomes et al. 2013), which was first reported as Phoma chamaeropis on C. humilis and other palms from Kew, England (Cooke 1885), no epitype specimen has been formally designated for this species. Therefore, its phylogenetic position among extant Diaporthe species remains undetermined. In this study, four Diaporthe species were isolated from foliar lesions of C. humilis, including D. amygdali, D. foeniculina, D. leucospermi and the new species D. pygmaeae. Diaporthe pygmaeae was tentatively compared with the morphological description of D. chamaeropis (Petrak 1919). Nonetheless, substantial morphological differences, such as the size of alpha conidia and the absence of beta conidia in D. pygmaeae, refrained a possible epitypification. Phylogenetically, D. pygmaeae is sister to species in the D. brasiliensis clade, which includes uncommonly found endophytes that apparently have restricted host and geographical ranges. It is uncertain whether D. pygmaeae is an endophyte or a latent pathogen, as species in the D. brasiliensis clade have virtually only been isolated as endophytes and no pathogenicity tests were conducted in this study. Sessa et al. (2018) suggested, based on pathogenicity tests, that D. brasiliensis obtained from Pyrus communis in Uruguay could be considered as a true endophyte. Some plant pathogenic and endophytic Diaporthe species appear to be host-specific, which may explain why species in the D. brasiliensis clade are uncommonly reported in Diaporthe studies worldwide, despite their introduction over 10 years ago (Gomes et al. 2013). Thus, future studies should aim to understand the ecological role of D. pygmaeae as an endophyte and its potential host specificity towards Arecaceae hosts.
Like species in the D. brasiliensis clade, D. leucospermi is also an uncommonly found Diaporthe species. It was first introduced by Crous et al. (2011b) on Leucospermum sp. in Australia, and only a few strains have since been isolated from four different plant hosts in Portugal, mostly representing weak pathogens (Hilário et al. 2021c). However, this study has shown that D. infecunda, a sister lineage to that including the type of D. leucospermi, represents an intraspecific variation of the latter, which has broadened its host range. Moreover, D. leucospermi has not previously been recorded on palms and, therefore, two new plant host records, namely C. humilis and Chrysalidocarpus lutescens, have been established for D. leucospermi. Interestingly, while strains in the D. leucospermi lineage are mostly associated with plant diseases (Hilário et al. 2021c), strains in the D. infecunda lineage are primarily reported as endophytes (Gomes et al. 2013). The ecological role of D. leucospermi was first questioned by Crous et al. (2011b). Based on BLASTn similarity results, the authors suggested that the species may represent an endophyte with a broader host range, as the species showed affinities to Phomopsis sp. (GenBank accession number: EU002916), isolated as a fruit endophyte from Coffea arabica in Hawaii (Vega et al. 2010). Diaporthe infecunda has been isolated as an endophyte of medicinal plants in Brazil (Gomes et al. 2013) and South Korea (Kim et al. 2017), as well as common bean trees (Phaseolus vulgaris) in Brazil (dos Santos et al. 2016). However, it has also been identified as a pathogen causing trunk diseases of persimmon trees (Diospyros kaki) in South Africa (Moyo et al. 2016) and as the cause of flower and fruit rot of Passiflora edulis in Brazil (Moreira et al. 2020). Recently, Sessa et al. (2017, 2018) also discussed the nature of D. infecunda as a latent pathogen and saprobe on fruit trees in Uruguay. The ecological role of D. leucospermi has been well established as both a pathogen and an endophyte. Therefore, the strains recovered from foliar lesions of palms may represent either endophytic or latent pathogenic fungi, and further studies are needed to test their pathogenicity. However, given the affiliation of these isolates with strains in the D. leucospermi lineage, which have often been isolated from other plant hosts in Portugal, it is likely that the strains from palms also represent weak pathogens.
Unlike D. leucospermi, the other Diaporthe species obtained from palm foliar lesions, such as D. amygdali, D. foeniculina and D. rudis, are well-known pathogens with a worldwide distribution. It is noteworthy that they have never been detected in Arecaceae hosts. However, these pathogens have mostly been associated with plant hosts in European countries, including Portugal (Guarnaccia et al. 2022). Therefore, due to the scarcity of indigenous palms in Europe and temperate countries in general, it is likely that the representativeness of these Diaporthe species in Arecaceae is linked to their preferential distribution in hosts and countries located in temperate regions.
Diaporthe amygdali is considered the primary pathogen of twig cankers and shoot blight of almond (Prunus amygdalus), peach and nectarine (P. persica) in Mediterranean countries where these hosts are cultivated, such as France (Delacroix 1905), Greece (Michailides & Thomidis 2006), Hungary (Varjas et al. 2017), Italy (Gusella et al. 2023), Portugal (Diogo et al. 2010), Spain (León et al. 2020, Beluzán et al. 2021) and Tunisia (Rhouma et al. 2008). This species has also been reported as a pathogen in other fruit and nut crops in Mediterranean countries, such as apricot (P. armeniaca) (Garofalo 1973), blueberry (Vaccinium corymbosum) (Lombard et al. 2014, Hilário et al. 2021c) and walnut (Juglans regia) (López-Moral et al. 2020). Furthermore, it has also been found to affect diverse hosts in other continents, including almond and peach in the USA (Adaskaveg et al. 1999, Farr et al. 1999), grapevine (Vitis viniferae) in South Africa (Mostert et al. 2001), peach in Japan (Kanematsu et al. 1999), peach and nectarine in Uruguay (Sessa et al. 2017) and peach, pear (Pyrus spp.) and walnut in China (Dai et al. 2012, Bai et al. 2015, Meng et al. 2018). In this study, D. amygdali has been identified associated with Arecaceae hosts for the first time, having been isolated from C. humilis and Washingtonia filifera.
Diaporthe foeniculina is a commonly found opportunistic pathogen in a wide range of herbaceous plants, ornamentals, and fruit trees in Europe (Guarnaccia & Crous 2017). It is also frequently reported as an endophyte, saprobe, or pathogen in many woody hosts across the globe (e.g., Úrbez-Torres et al. 2013, Udayanga et al. 2014b, Makris et al. 2022). Despite its worldwide distribution in both tropical and temperate regions, D. foeniculina has not been previously recorded on Arecaceae hosts. Notably, D. foeniculina was the most frequently isolated Diaporthe species in this study, representing 15 out of the 29 strains (ca. 58 %). Moreover, D. foeniculina was the only Diaporthe species that has been isolated from all of the sampled palm hosts, including C. humilis, C. lutescens, Phoenix roebelenii, Trachycarpus fortunei and W. filifera. This suggests that this species has colonised palm trees, which are widely planted as ornamentals in Portuguese cities, due to its common occurrence on various plant hosts in Portugal (e.g., Lopes et al. 2021). It remains undetermined whether these strains are primary pathogens or endophytes acting as weak secondary pathogens or saprophytes, and pathogenicity studies are necessary to determine their ecological role. However, several studies have shown that D. foeniculina causes dieback, cankers, and fruit rot in many economically important crops, although it is often regarded as weakly aggressive. Diaporthe foeniculina has been most commonly found to cause various disease symptoms in Citrus spp., such as stem and shoot cankers, blight and dieback, in Europe (e.g., Guarnaccia & Crous 2017, Vakalounakis et al. 2019, Tekiner et al. 2020), USA (e.g., Udayanga et al. 2014b) and other countries (e.g., Habib et al. 2015). As D. neotheicola, this species has been reported to cause diseases in temperate and tropical fruit trees mostly from Australia, Europe and South Africa (e.g., Golzar et al. 2012, Kaliterna et al. 2012, van Dyk et al. 2021). Indeed, there are many reports of diseases caused by D. foeniculina worldwide, including shoot blight and trunk diseases of persimmon in Australia and South Africa (Golzar et al. 2012, Moyo et al. 2016), shoot blight of kiwi (Actinidia deliciosa) in Greece (Thomidis et al. 2013), stem and shoot cankers and dieback of sweet chestnut (Castanea sativa) in Italy (Annesi et al. 2016), branch cankers and stem-end rot of avocado (Persea americana) in Italy and Greece (Guarnaccia et al. 2016, Mathioudakis et al. 2020), reddish sunken cankers on apple trees (Malus domestica) in Uruguay (Sessa et al. 2017), black tip and necrotic spots on hazelnut (Corylus avellana) and stem cankers on blueberries (Vaccinium spp.) in Chile (Elfar et al. 2013, Guerrero et al. 2020), yellowing and dieback on Ficus benjamina in Iran (Esmaeilzadeh et al. 2020), woody canker and shoot blight of mango (Mangifera indica) and lychee (Litchi chinensis) in Italy (Aiello et al. 2022) and trunk diseases of grapevine in Cyprus (Makris et al. 2022). In Portugal, D. foeniculina has been reported as a weak pathogen associated with apple, pear (Pyrus communis) (Santos et al. 2017), almond (Diogo et al. 2010) and blueberry (Hilário et al. 2020, 2021c), as well as a saprophyte on economically important herbaceous plants, such as wild fennel (Foeniculum vulgare) (Santos & Phillips 2009).
Diaporthe rudis, usually referred to as D. viticola, is a well-known pathogen in Europe and New Zealand that has been mostly associated with disease symptoms in grapevines (Udayanga et al. 2014b, Guarnaccia et al. 2018). However, this species has also been found on a variety of other plant hosts (Gomes et al. 2013). In recent years, it has increasingly been reported as a postharvest pathogen in hosts of high agro-economic value, causing fruit rots in apples, avocado, blackberries (Rubus sp.), blueberries, grapes, kiwi and pears (Torres et al. 2016, Diaz et al. 2017, Cardinaals et al. 2018, KC & Rasmussen 2019, Lorenzini & Zapparoli 2019, Guarnaccia et al. 2020 Vučković et al. 2022, González & Ciordia 2023), as well as seed decays of hazelnut and soybean (Petrović et al. 2016, Pscheidt et al. 2019). In Portugal, D. rudis has only been occasionally isolated in association with grapevines (van Niekerk et al. 2015). It has also been identified as a minor pathogen of blueberries (Hilário et al. 2020, 2021c) and found on both symptomatic and asymptomatic wood of Eucalyptus globulus and Pinus pinaster, respectively (Lopes et al. 2021). In this study, D. rudis has been identified with Arecaceae hosts for the first time, having been isolated from C. lutescens and W. filifera.
Although the isolates of D. amygdali, D. foeniculina and D. rudis in this study were obtained from foliar lesions of palms, their pathogenicity has not been proven. Therefore, future studies should aim to better understand the phytopathogenic potential of these genotypic variants. This is important as they may pose a threat to important Portuguese crops, such as Citrus spp., Prunus spp. and Vitis vinifera, for which these species have been shown to be main pathogens with significant impact (Udayanga et al. 2014b). While palm trees are undoubtedly a key resource in the ornamentation of parks and gardens in Portuguese cities, they can also act as alternative hosts for pathogens that affect other economically important crops. Indeed, the isolation of Diaporthe species with a cosmopolitan behaviour in Arecaceae hosts for the first time, highlights the potential of ornamental palms, and ornamentals in general, to act as a reservoir for the dissemination of potential fungal pathogens. Furthermore, the broad lack of host specificity indicates the ability of D. amygdali, D. foeniculina and D. rudis to spread widely among different plant hosts before establishing themselves as potential emerging fungal pathogens in their main hosts, as suggested by previous studies (e.g., Sessa et al. 2018, Vakalounakis et al. 2019). Therefore, to detect and characterise new genotypic variants of these species is of the utmost importance, as the dissemination of fungal pathogens is a threat that causes severe economic losses in agriculture and forestry (Baldi & La Porta 2020).
Diaporthe is one of the most frequently isolated genera of endophytic fungi, with a wide host range and worldwide distribution (Gomes et al. 2013). However, it is often overlooked whether these endophytes represent true endophytes or latent pathogens. Recent studies have shown that healthy shoots of apple, pear, blueberry, and peach can host many true endophytic Diaporthe species, along with potential wood disease-causing strains that should be considered as latent pathogens (Sessa et al. 2017). Thus, the presence of certain genotypic variants of common Diaporthe species in certain hosts can lead to outbreaks of major infections. This is especially relevant in the current scenario of global climate change, which stresses plant communities and may facilitate the emergence of more aggressive Diaporthe strains capable of colonising new hosts (Desprez-Loustau et al. 2006, 2007). Furthermore, in response to changing environments, fungi may switch from an endophytic or saprophytic lifestyle to a pathogenic one (Manawasinghe et al. 2019). This would not be unexpected in species such as D. amygdali, D. foeniculina and D. rudis, which have been recorded as pathogens, saprobes, and endophytes on various plant hosts (Udayanga et al. 2014b). This has recently been discussed by Pereira et al. (2023) for D. arecae, which also exhibits a cosmopolitan behaviour ranging from minor pathogens (e.g., Ariyawansa et al. 2020, as D. taiwanensis) to causal agents of devastating dieback diseases of Citrus plants in European countries (Guarnaccia & Crous 2017, as D. limonicola and D. melitensis).
Although Diaporthe species on palms are scattered throughout the phylogeny of the genus, D. arecae is the most frequently recorded species of Diaporthe on Arecaceae. It has been reported from 14 different palm hosts, five of which, namely Caryota maxima, Cocos nucifera, Elaeis guineensis, Oncosperma sp. and Phoenix paludosa, have been resolved in this study based on sequences deposited in GenBank and occasional reports in the literature. It is likely that new collections of Diaporthe taxa from palms will broaden the range of arecaceous hosts for D. arecae. For example, three new palm hosts were recently established by Senanayake et al. (2023), who isolated new strains of D. arecae as saprophytes on the leaves of C. lutescens, Livistona chinensis and T. fortunei in China. The remaining Diaporthe spp. reported from palms have only been collected occasionally and are mostly associated with a single Arecaceae host. However, there are still many undiscovered Diaporthe species on palm trees, as evidenced by the number of Phomopsis and Diaporthe species introduced based on palm collections that have yet to be recollected and epitypified (Fröhlich et al. 1997, Taylor & Hyde 2003). Moreover, the taxonomic status of several records of Diaporthe taxa on arecaceous hosts remains unresolved due to their phylogenetic position in clades whose species boundaries are poorly understood. For instance, the present analyses revealed that some Diaporthe taxa found on palm tissues are phylogenetically related to the D. sojae species complex [sensu Norphanphoun et al. (2022)], which might represent another common clade of Diaporthe species on palm trees. Additional research is needed to correctly assign them to known Diaporthe species and understand their significance for Arecaceae hosts. Indeed, the D. sojae species complex includes several important phytopathogenic species of Diaporthe that cause diseases in herbaceous field crops and weeds, such as soybean and cucurbits (Udayanga et al. 2015). Currently, the only Diaporthe species associated with palm trees and with affiliations to the D. sojae species complex is D. ueckeri. This species has recently been identified as the causal agent of brown blotch disease, which has had a significant impact on the growth and ornamental value of Hyophorbe lagenicaulis palms in China (Guo et al. 2024).
Given the complex ecology and potential role of Diaporthe species as plant pathogens, the importance of accurately identifying species in the genus cannot be understated, as it is crucial for disease surveillance, control, and trade. Therefore, the integrative taxonomic approach carried out in this study is an essential tool to resolve species boundaries and, consequently, uncover and fine-tune the ecological role, host range, and epidemiology of Diaporthe species.
Guidelines for accurate species recognition, definition, and identification in Diaporthe
Based on the integrative taxonomic approach conducted, this study proposes new recommendations for accurately delimiting and introducing species in the genus Diaporthe, which build upon those previously provided by Hilário et al. (2022). The following guidelines also take into account the broad recommendations provided by Jeewon & Hyde (2016) for appropriate use of phylogenetic and morphological data in fungal species delineation, as well as the practical guide on how to name phytopathogenic fungi by Crous et al. (2021b), and the best practices for the description of new fungal species and publication of new names by Aime et al. (2021).
Before assigning any Diaporthe strain to a new or existing species, it is recommended to conduct a general phylogenetic analysis with all currently accepted species in the genus. This initial analysis will help determine the diversity of new strains in Diaporthe clades and assess which Diaporthe species are in their phylogenetic vicinity. This information is essential in subsequent analyses aimed at assigning a correct name to Diaporthe taxa. Although ITS is currently the guide barcode in these initial assessments, tef1 may provide a more accurate estimate of species boundaries and could be chosen as the primary marker (Udayanga et al. 2012a, Santos et al. 2017a).
The Diaporthe clades selected for further analyses should be as populated as possible, including all taxa previously synonymised in a given clade, as well as strains of diverse origins for each of them whenever available. Adopting this strategy of a large sampling size will help determine intraspecific variation and thus avoid the spurious introduction of new Diaporthe names for more polymorphic genotypic variants of a given species (Walsh et al. 2000, Ahrens et al. 2016). It is worth noting that BLASTn search results may also be included, but their annotations must be carefully evaluated as they can lead to misinterpretations. Recent studies have been showing that sequences deposited in public sequence databases may harbour technical errors and atypical chimeras that make reliable species identification problematic (e.g., Nilsson et al. 2006, 2012, 2014, Baturo-Cieśniewska 2020).
To prevent overestimation or misidentification of Diaporthe taxa, it is essential to always apply the GCPSR principle (Taylor et al. 2000). Although there are variations in the criteria used to apply the GCPSR principle (e.g., Dettman et al. 2003), in its strict sense, it recognises phylogenetic species as monophyletic, statistically supported and genealogically concordant lineages (Taylor et al. 2000). For practical purposes, it is generally recommended that terminal clades should produce consistent topologies when using different phylogenetic tree reconstruction methods, with bootstrap and Bayesian posterior probability values greater than or equal to 70 % and 0.90, respectively (e.g., Gazis et al. 2011).
In addition to critically examining the phylogenetic inferences, it is important to carefully analyse and interpret the underlying nucleotide diversity to stabilise the taxonomy of Diaporthe species. Previous recommendations have primarily focused on ITS sequence data, whose validity for molecular identification in Diaporthe has been questioned due to its intraspecific and intragenomic variability (e.g., Santos et al. 2010, Hilário et al. 2022). However, this study has shown that the intra and interspecific variation of the common loci used in Diaporthe systematics is variable and dependent on the clade being analysed. It is not uncommon to find that intraspecific variation is greater than interspecific variation in some loci. Therefore, different clades may present varying evolutionary patterns that require evaluation. For example, tef1 and cal partial sequences from strains of the same species have been shown to exhibit a p-distance of up to 0.07, particularly in lineages with extensive intraspecific adaptive radiation. Additionally, certain instances have demonstrated even greater intraspecific diversity in a single locus, which may not be associated with a speciation event. Although Jeewon & Hyde (2016) have suggested that a minimum of more than 1.5 % nucleotide dissimilarities may indicate a new species, this fixed threshold may not be applicable to any of the loci used in Diaporthe.
The introduction of novel Diaporthe species lacking molecular data from at least three loci (excluding ITS) is strongly discouraged. The MLSA including tef1, tub2, cal and his3 has shown to be a promising methodology for defining species boundaries in the genus (e.g., Santos et al. 2017a). Given that the relative power of the common loci used in Diaporthe systematics may vary depending on the clade being analysed, it is recommended that PI analyses be used for locus prioritisation when there are limitations to sequencing multiple loci due to lack of resources or similar reasons.
Taxonomic notes on novel Diaporthe species should include a detailed comparison of its phylogenetic relationships with closely related taxa. Therefore, nucleotide dissimilarity of the available loci should be used to support and justify the evolution of a novel clade/lineage. In addition, molecular information should be combined with morphological analyses to provide a more robust framework for establishing novel lineages, with particular reference to any significant apomorphs (Hyde et al. 2010, Jeewon & Hyde 2016). However, considering the great phenotypic plasticity of Diaporthe taxa (Mostert et al. 2001), minor morphological differences in size and shape, or even the presence or absence of dimorphic conidia, should never be used as the main delimiting criteria for establishing a novel Diaporthe species.
It is recommended that morphological characters should also be provided for new geographical and/or plant host records to provide insights into intraspecific phenotypic diversity, which may strengthen future studies of species boundaries in the genus. This is particularly relevant for Diaporthe clades with extensive intraspecific variability and whose phylogenetic analyses are frequently obscured by the presence of highly polymorphic DNA barcodes. Furthermore, as several Diaporthe species are known to be cryptic (Gomes et al. 2013), for taxa that have been incorrectly synonymised or identified it will be easier to resurrect a (new) lineage, using the available morphomolecular information.
Species identification should be complemented by an integrative approach that includes DNA-based species delimitation analyses to test the independence of lineages identified by multilocus phylogenetic analyses. Coalescent-based methods have been shown to allow for more robust and reliable species delimitation in Diaporthe (Hilário et al. 2021a, b, Pereira et al. 2023) due to their ability to detect and accommodate gene tree discordant events (Fujita et al. 2012). Likewise, in the present study, coalescent approaches, particularly the PTP method, provided consistent support for species limits in clades where speciation events are unclear. Additionally, genetic distance approaches can offer insights into the evolutionary processes that cause phylogenetic incongruences, which can provide additional support for taxonomic decisions when identifying Diaporthe strains.
To better understand phylogenetic incongruences, it is recommended to use phylogenetic networks and analyses to detect recombination. These methodologies will enable the evaluation of potential reticulate events between taxa and inference of more precise evolutionary relationships (Huson & Scornavacca 2011). It should be noted that the results of these methodologies constitute additional evidence to test evolutionary hypotheses and not absolute truths for taxonomic decisions. For instance, the PHI test (Bruen et al. 2006) has been used to determine the recombination level between taxa and, accordingly, warrant the GCPSR criteria by evaluating the significance of gene flow between them (e.g., Quaedvlieg et al. 2014). Recently, Hyde et al. (2020b) advocated that the detection of significant recombination within closely related taxa should be considered as an important method to justify a species. However, a statistically significant positive or negative PHI test result for recombination alone does not necessarily indicate that taxa represent the same or distinct species, respectively, as has been assumed in some Diaporthe studies (e.g., Monkai et al. 2023). As hypothesised in this study, the detection of statistically significant recombination may result from occasional hybridisation between distinct taxa. Likewise, the absence of recombination between strains of a given species may result from adaptive divergence, as well as insufficient sampling of populations polymorphisms.
Epitypification, through the provision of modern interpretative types, is recognised as the best approach to resolve taxonomic and phylogenetic issues of poorly circumscribed taxa (Hyde & Zhang 2008). As noted in this study, the resolution of species boundaries in Diaporthe may result in the delimitation of taxa whose types lack ex-type cultures and/or are supported by a limited number of DNA barcodes, or by old and poorly informative morphological descriptions. To ensure taxonomic stability, it is recommended that future studies dealing with clades in which these problems arise should provide epitypes or reference specimens whenever possible, following the appropriate guidelines previously recommended (e.g., Ariyawansa et al. 2014). Moreover, many Diaporthe (and Phomopsis) names are not linked to DNA sequence data or ex-type cultures, so the epitypification of these species is necessary to provide clarification and knowledge about the evolutionary history, phylogeny and species boundaries of the genus (Udayanga et al. 2011, Hongsanan et al. 2023). Efforts should therefore be made to recollect these taxa and fully characterise them with DNA sequences and modern morphological descriptions.
Conclusions
Although Diaporthe is considered one of the most speciose fungal genera, this claim is strongly challenged by the present study. Many of the recently introduced species into Diaporthe are the result of a profound lack of understanding of the phenomena that drive intra- and interspecific diversification in the genus. This study demonstrates that intraspecific variability has often been used to introduce new Diaporthe species. The interpretation of speciation events has often been obscured by divergent evolutionary histories among the common loci used for molecular identification in the genus. As a result, concatenated phylogenetic inferences, coupled with small sample sizes, have led to an inflation of species names. Therefore, the present study also highlights the need to critically examine phylogenies and especially the underlying nucleotide diversity to avoid overestimating species diversity in Diaporthe. This approach sets an important precedent for understanding conflicting phylogenetic signals and their potential sources.
Rapid adaptive divergence, linked to recent or ongoing speciation events, has been hypothesised to explain the detected phylogenetic incongruencies. These may be due to either the retention of ancestral polymorphisms, resulting in lack of complete lineage sorting, or to the absence of barriers to gene flow in nature, resulting in recombination or occasional hybridisation. Therefore, upcoming studies should adopt a polyphasic approach that integrates coalescent-based methods to stabilise the taxonomy of Diaporthe. These methods can incorporate phylogenetic incongruences into species recognition, providing a more precise and objective species delimitation. The present results indicate that this approach will be important to restructure the phylogenetic relationships between Diaporthe species, potentially revolutionising the current understanding of their boundaries. This study has not aimed to determine which evolutionary processes are occurring in the Diaporthe clades treated. However, it serves as a baseline for investigating the causes of phylogenetic incongruencies, which can be identified by further phylogenomic analyses. Large-scale whole genome sequencing should therefore be considered in the future to provide insights into the validity of the current loci used in Diaporthe and to identify new suitable markers for species delimitation in the genus. Although full genome sequences are still forthcoming for Diaporthe species, the development of a genome-based taxonomy approach in the future will certainly help to resolve the current taxonomic confusion that characterises most phylogenetic relationships between Diaporthe taxa.
Accurately delimiting Diaporthe species is critical for understanding the evolution of their pathogenicity and the emergence of new infectious diseases. The presence of unknown Diaporthe taxa, or even certain genotypic variants of extant species not previously identified on imported tropical hosts such as palms, could lead to outbreaks of infections. This study identified new genotypic variants of commonly reported pathogenic Diaporthe species on foliar lesions of ornamental palms in Lisbon, Portugal. The isolation of these species on palms for the first time highlights the potential threat that Diaporthe taxa may pose to woody hosts in both urban and natural environments, as well as to commercially important Portuguese crops. Therefore, the delimitation of Diaporthe species has significant socioeconomic implications and plays a critical role in global biosecurity, as their correct identification is essential for the implementation of measures to restrict the movement of potential plant pathogens.
ACKNOWLEDGEMENTS
The authors acknowledge the support from UIDB/04046/2020 (DOI: 10.54499/UIDB/04046/2020) and UIDP/04046/2020 (DOI: 10.54499/UIDP/04046/2020) Centre grants from FCT, Portugal (to BioISI), and from the PhD grant to D.S. Pereira (SFRH/BD/09742/2020) from FCT, Portugal.
DECLARATION ON CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
Supplementary Material: https://studiesinmycology.org/
Phylogenetic trees generated from a maximum likelihood analysis of the Diaporthe amygdali clade and related species. A. Based on ITS sequence data. B. Based on tef1 sequence data. C. Based on tub2 sequence data. D. Based on cal sequence data. E. Based on his3 sequence data. Bootstrap support values for maximum likelihood and maximum parsimony (ML-BS/MP-BS ≥ 70 %) are shown at the nodes. Taxa numbers were generated, and the corresponding strains and species are shown in the ITS-phylogram (panel A). Strains with type status are indicated in bold font. The isolates from this study are presented in brown typeface. The scale bars represent the expected number of nucleotide changes per site. The trees are rooted to D. acaciigena (CBS 129521).
Phylogenetic trees generated from a maximum likelihood analysis of the Diaporthe brasiliensis clade and related species. A. Based on ITS sequence data. B. Based on tef1 sequence data. C. Based on tub2 sequence data. D. Based on cal sequence data. E. Based on his3 sequence data. Bootstrap support values for maximum likelihood and maximum parsimony (ML-BS/MP-BS ≥ 70 %) are shown at the nodes. Taxa numbers were generated, and the corresponding strains and species are shown in the ITS-phylogram (panel A). Strains with type status are indicated in bold font. The isolates from this study are presented in brown typeface. The scale bars represent the expected number of nucleotide changes per site. The trees are rooted to D. scobina (CBS 129521).
Phylogenetic trees generated from a maximum likelihood analysis of the Diaporthe eucommiae clade and related species. A. Based on ITS sequence data. B. Based on tef1 sequence data. C. Based on tub2 sequence data. D. Based on cal sequence data. E. Based on his3 sequence data. Bootstrap support values for maximum likelihood and maximum parsimony (ML-BS/MP-BS ≥ 70 %) are shown at the nodes. Taxa numbers were generated, and the corresponding strains and species are shown in the ITS-phylogram (panel A). Strains with type status are indicated in bold font. The isolate from palm tissues included in the analyses is presented in green typeface. The scale bars represent the expected number of nucleotide changes per site. The trees are rooted to D. acutispora (CGMCC 3.18285 and LC 6142).
Phylogenetic trees generated from a maximum likelihood analysis of the Diaporthe foeniculina clade and related species. A. Based on ITS sequence data. B. Based on tef1 sequence data. C. Based on tub2 sequence data. D. Based on cal sequence data. E. Based on his3 sequence data. Bootstrap support values for maximum likelihood and maximum parsimony (ML-BS/MP-BS ≥ 70 %) are shown at the nodes. Taxa numbers were generated, and the corresponding strains and species are shown in the ITS-phylogram (panel A). Strains with type status are indicated in bold font. The isolates from this study are presented in brown typeface and the additional isolate from palm tissues included in the analyses is presented in green typeface. The scale bars represent the expected number of nucleotide changes per site. The trees are rooted to D. stictica (CBS 370.54).
Phylogenetic trees generated from a maximum likelihood analysis of the Diaporthe glabrae clade and related species. A. Based on ITS sequence data. B. Based on tef1 sequence data. C. Based on tub2 sequence data. D. Based on cal sequence data. E. Based on his3 sequence data. Bootstrap support values for maximum likelihood and maximum parsimony (ML-BS/MP-BS ≥ 70 %) are shown at the nodes. Taxa numbers were generated, and the corresponding strains and species are shown in the ITS-phylogram (panel A). Strains with type status are indicated in bold font. The isolates from palm tissues included in the analyses are presented in green typeface. The scale bars represent the expected number of nucleotide changes per site. The trees are rooted to D. raonikayaporum (CBS 133182).
Phylogenetic trees generated from a maximum likelihood analysis of the Diaporthe inconspicua clade and related species. A. Based on ITS sequence data. B. Based on tef1 sequence data. C. Based on tub2 sequence data. D. Based on cal sequence data. E. Based on his3 sequence data. Bootstrap support values for maximum likelihood and maximum parsimony (ML-BS/MP-BS ≥ 70 %) are shown at the nodes. Taxa numbers were generated, and the corresponding strains and species are shown in the ITS-phylogram (panel A). Strains with type status are indicated in bold font. The isolate from palm tissues included in the analyses is presented in green typeface. The scale bars represent the expected number of nucleotide changes per site. The trees are rooted to D. foeniculina (CBS 123208 and CBS 123209).
Phylogenetic trees generated from a maximum likelihood analysis of the Diaporthe leucospermi clade and related species. A. Based on ITS sequence data. B. Based on tef1 sequence data. C. Based on tub2 sequence data. D. Based on cal sequence data. E. Based on his3 sequence data. Bootstrap support values for maximum likelihood and maximum parsimony (ML-BS/MP-BS ≥ 70 %) are shown at the nodes. Taxa numbers were generated, and the corresponding strains and species are shown in the ITS-phylogram (panel A). Strains with type status are indicated in bold font. The isolates from this study are presented in brown typeface. The scale bars represent the expected number of nucleotide changes per site. The trees are rooted to D. ganjae (CBS 180.91 and PSCG 489).
Phylogenetic trees generated from a maximum likelihood analysis of the Diaporthe longicolla clade and related species. A. Based on ITS sequence data. B. Based on tef1 sequence data. C. Based on tub2 sequence data. D. Based on cal sequence data. E. Based on his3 sequence data. Bootstrap support values for maximum likelihood and maximum parsimony (ML-BS/MP-BS ≥ 70 %) are shown at the nodes. Taxa numbers were generated, and the corresponding strains and species are shown in the ITS-phylogram (panel A). Strains with type status are indicated in bold font. The isolates from palm tissues included in the analyses are presented in green typeface. The scale bars represent the expected number of nucleotide changes per site. The trees are rooted to D. batatas (CBS 122.21).
Phylogenetic trees generated from a maximum likelihood analysis of the Diaporthe rudis clade and related species. A. Based on ITS sequence data. B. Based on tef1 sequence data. C. Based on tub2 sequence data. D. Based on cal sequence data. E. Based on his3 sequence data. Bootstrap support values for maximum likelihood and maximum parsimony (ML-BS/MP-BS ≥ 70 %) are shown at the nodes. Taxa numbers were generated, and the corresponding strains and species are shown in the ITS-phylogram (panel A). Strains with type status are indicated in bold font. The isolates from this study are presented in brown typeface. The scale bars represent the expected number of nucleotide changes per site. The trees are rooted to D. cf. heveae 1 (CBS 852.97).
Synopsis of the alignment properties, statistics results and nucleotide substitution models used for the phylogenetic analyses of the Diaporthe amygdali clade.
Synopsis of the alignment properties, statistics results and nucleotide substitution models used for the phylogenetic analyses of the Diaporthe brasiliensis clade.
Synopsis of the alignment properties, statistics results and nucleotide substitution models used for the phylogenetic analyses of the Diaporthe eucommiae clade.
Synopsis of the alignment properties, statistics results and nucleotide substitution models used for the phylogenetic analyses of the Diaporthe foeniculina clade.
Synopsis of the alignment properties, statistics results and nucleotide substitution models used for the phylogenetic analyses of the Diaporthe glabrae clade.
Synopsis of the alignment properties, statistics results and nucleotide substitution models used for the phylogenetic analyses of the Diaporthe inconspicua clade.
Synopsis of the alignment properties, statistics results and nucleotide substitution models used for the phylogenetic analyses of the Diaporthe leucospermi clade.
Synopsis of the alignment properties, statistics results and nucleotide substitution models used for the phylogenetic analyses of the Diaporthe longicolla clade.
Synopsis of the alignment properties, statistics results and nucleotide substitution models used for the phylogenetic analyses of the Diaporthe rudis clade.
REFERENCES
- Achari SR, Kaur J, Dinh Q. et al. (2020). Phylogenetic relationship between Australian Fusarium oxysporum isolates and resolving the species complex using the multispecies coalescent model. BMC Genomics 21: 248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adaskaveg JE, Förster H, Connell JH. (1999). First report of fruit rot and associated branch dieback of almond in California caused by a Phomopsis species tentatively identified as P. amygdali. Plant Disease 83: 1073. [DOI] [PubMed] [Google Scholar]
- Ahrens D, Fujisawa T, Krammer H-J, et al. (2016). Rarity and incomplete sampling in DNA-based species delimitation. Systematic Biology 65: 478–494. [DOI] [PubMed] [Google Scholar]
- Aiello D, Guarnaccia V, Costanzo MB. et al. (2022). Woody canker and shoot blight caused by Botryosphaeriaceae and Diaporthaceae on mango and litchi in Italy. Horticulturae 8: 330. [Google Scholar]
- Aime MC, Miller AN, Aoki T. et al. (2021). How to publish a new fungal species, or name, version 3.0. IMA Fungus 12: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves A, Crous PW, Correia A. et al. (2008). Morphological and molecular data reveal cryptic speciation in Lasiodiplodia theobromae. Fungal Diversity 28: 1–13. [Google Scholar]
- Annesi T, Luongo L, Vitale S. et al. (2016). Characterization and pathogenicity of Phomopsis theicola anamorph of Diaporthe foeniculina causing stem and shoot cankers on sweet chestnut in Italy. Journal of Phytopathology 164: 412–416. [Google Scholar]
- Ariyawansa HA, Hawksworth DL, Hyde KD. et al. (2014). Epitypification and neotypification: guidelines with appropriate and inappropriate examples. Fungal Diversity 69: 57–91. [Google Scholar]
- Ariyawansa HA, Tsai I, Wang J-Y, et al. (2021). Molecular phylogenetic diversity and biological characterization of Diaporthe species associated with leaf spots of Camellia sinensis in Taiwan. Plants 10: 1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariyawansa HA, Tsai I, Withee P. et al. (2020). Diaporthe taiwanensis: a new taxon causing leaf spots and necrosis on Ixora chinensis in Taiwan. Phytotaxa 461: 155–165. [Google Scholar]
- Aumentado HD, Balendres MA. (2024). Novel species and new records of Diaporthe causing eggplant leaf and fruit blight in the Philippines. Mycological Progress 23: 23. [Google Scholar]
- Avise JC, Ball RM. (1990). Principles of genealogical concordance in species concepts and biological taxonomy. Oxford Surveys in Evolutionary Biology 7: 45–67. [Google Scholar]
- Azuddin NF, Mohd MH, Rosely NFN. et al. (2021). Molecular phylogeny of endophytic fungi from rattan (Calamus castaneus Griff.) spines and their antagonistic activities against plant pathogenic fungi. Journal of Fungi 7: 301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Q, Zhai L, Chen X. et al. (2015). Biological and molecular characterization of five Phomopsis species associated with pear shoot canker in China. Plant Disease 99: 1704–1712. [DOI] [PubMed] [Google Scholar]
- Bai YK, Lin L, Pan M. et al. (2023). Studies of Diaporthe (Diaporthaceae, Diaporthales) species associated with plant cankers in Beijing, China, with three new species described. MycoKeys 98: 59–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldi P, La Porta N. (2020). Molecular approaches for low-cost point-of-care pathogen detection in agriculture and forestry. Frontiers in Plant Science 11: 570862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bapteste E, van Iersel L, Janke A. et al. (2013). Networks: expanding evolutionary thinking. Trends in Genetics 29: 439–441. [DOI] [PubMed] [Google Scholar]
- Baturo-Cieśniewska A, Pusz W, Patejuk K. (2020). Problems, limitations, and challenges in species identification of Ascomycota members on the basis of ITS regions. Acta Mycologica 55: 5512. [Google Scholar]
- Baumgartner K, Fujiyoshi PT, Travadon R. et al. (2013). Characterization of species of Diaporthe from wood cankers of grape in eastern North American vineyards. Plant Disease 97: 912–920. [DOI] [PubMed] [Google Scholar]
- Belfiore NM, Liu L, Moritz C. (2008). Multilocus phylogenetics of a rapid radiation in the genus Thomomys (Rodentia: Geomyidae). Systematic Biology 57: 294–310. [DOI] [PubMed] [Google Scholar]
- Beluzán F, Olmo D, León M. et al. (2021) First report of Diaporthe amygdali associated with twig canker and shoot blight of nectarine in Spain. Plant Disease 105: 3300. [DOI] [PubMed] [Google Scholar]
- Bezerra JDP, Machado AR, Firmino AL. et al. (2018). Mycological Diversity Description I. Acta Botanica Brasilica 32: 656–666. [Google Scholar]
- Bhunjun CS, Dong Y, Jayawardena RS. et al. (2020). A polyphasic approach to delineate species in Bipolaris. Fungal Diversity 102: 225–256. [Google Scholar]
- Bhunjun CS, Niskanen T, Suwannarach N. et al. (2022). The numbers of fungi: are the most speciose genera truly diverse? Fungal Diversity 114: 387–462. [Google Scholar]
- Bhunjun CS, Phillips AJL, Jayawardena RS. et al. (2021). Importance of molecular data to identify fungal plant pathogens and guidelines for pathogenicity testing based on Koch’s postulates. Pathogens 10: 1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boluda CG, Rico VJ, Divakar PK. et al. (2019). Evaluating methodologies for species delimitation: the mismatch between phenotypes and genotypes in lichenized fungi (Bryoria sect. Implexae, Parmeliaceae). Persoonia 42: 75–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonmee S, Wanasinghe DN, Calabon MS. et al. (2021). Fungal diversity notes 1387–1511: taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Diversity 111: 1–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruen TC, Philippe H, Bryant D. (2006). A simple and robust statistical test for detecting the presence of recombination. Genetics 172: 2665–2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant D, Moulton V. (2004). Neighbor-Net: An agglomerative method for the construction of phylogenetic networks. Molecular Biology and Evolution 21: 255–265. [DOI] [PubMed] [Google Scholar]
- Bundhun D, Senanayake IC, Jayawardena RS. et al. (2021). First reports of the sexual morphs of Diaporthe forlicesenica nom. nov. and Diaporthe goulteri (Diaporthaceae, Diaporthales) revealed by molecular phylogenetics. Phytotaxa 516: 1–27. [Google Scholar]
- Bustamante DE, Oliva M, Leiva S. et al. (2019). Phylogeny and species delimitations in the entomopathogenic genus Beauveria (Hypocreales, Ascomycota), including the description of B. peruviensis sp. nov. MycoKeys 58: 47–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L, Giraud T, Zhang N. et al. (2011). The evolution of species concepts and species recognition criteria in plant pathogenic fungi. Fungal Diversity 50: 121–133. [Google Scholar]
- Cai L, Hyde KD, Taylor PWJ. et al. (2009). A polyphasic approach for studying Colletotrichum. Fungal Diversity 39: 183–204. [Google Scholar]
- Calabon MS, Jones EBG, Pang KL. et al. (2023). Updates on the classification and numbers of marine fungi. Botanica Marina 66: 213–238. [Google Scholar]
- Canonaco A. (1936). Il seccume dei rameti di mandorlo in relazione ad alcuni micromiceti. Rivista di Patologia Vegetale 26: 145–164. [Google Scholar]
- Cao FX, Chi PK. (1990). Identifications on fungal diseases of Phellodendron chinese, Magnolia officinalis var. bioba and Eucominis ulmoides. Journal of Central-South Forestry College 10: 34–43. [Google Scholar]
- Cao L, Luo D, Lin W. et al. (2022). Four new species of Diaporthe (Diaporthaceae, Diaporthales) from forest plants in China. MycoKeys 91: 25–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone I, Kohn LM. (1999). A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91: 553–556. [Google Scholar]
- Cardinaals J, Wenneker M, Voogd JGB. et al. (2018). Pathogenicity of Diaporthe spp. on two blueberry cultivars (Vaccinium corymbosum). EPPO Bulletin 48: 128–134. [Google Scholar]
- Carstens BC, Pelletier TA, Reid NM. et al. (2013). How to fail at species delimitation. Molecular Ecology 22: 4369–4383. [DOI] [PubMed] [Google Scholar]
- Castlebury LA, Farr DF, Rossman AY. (2001). Phylogenetic distinction of Phomopsis isolates from cucurbits. Inoculum 52: 25. [Google Scholar]
- Castresana J. (2007). Topological variation in single-gene phylogenetic trees. Genome Biology 8: 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaisiri C, Liu X, Lin Y. et al. (2021b). Phylogenetic and haplotype network analyses of Diaporthe eres species in China based on sequences of multiple loci. Biology 10: 179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaisiri C, Liu X-Y, Yin W-X. et al. (2021a). Morphology characterization, molecular phylogeny and pathogenicity of Diaporthe passifloricola on Citrus reticulata cv. Nanfengmiju in Jiangxi Province, China. Plants 10: 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CQ, Cheng Y-H, Xiang MH. et al. (2005b). New species of Phomopsis on woody plants in Fujian province. Mycosystema 24: 6–11. [Google Scholar]
- Chang CQ, Xi PG, Xiang MM. et al. (2005a). New species of Phomopsis on woody plants in Hunan Province. Mycosystema 24: 145–154. [Google Scholar]
- Chen H, Strand M, Norenburg JL. et al. (2010). Statistical parsimony networks and species assemblages in cephalotrichid nemerteans (Nemertea). PLoS ONE 5: e12885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P, Abeywickrama PD, Ji S. et al. (2023). Molecular identification and pathogenicity of Diaporthe eres and D. hongkongensis (Diaporthales, Ascomycota) associated with cherry trunk diseases in China. Microorganisms 11: 2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SF, Morgan DP, Michailides TJ. (2014). Botryosphaeriaceae and Diaporthaceae associated with panicle and shoot blight of pistachio in California, USA. Fungal Diversity 67: 157–179. [Google Scholar]
- Chethana KWT, Jayawardena RS, Hyde KD. (2020). Hurdles in fungal taxonomy: effectiveness of recent methods in discriminating taxa. Megataxa 1: 114–122. [Google Scholar]
- Chethana KWT, Manawasinghe IS, Hurdeal VG. et al. (2021a). What are fungal species and how to delineate them? Fungal Diversity 109: 1–25. [Google Scholar]
- Chethana KWT, Niranjan M, Dong W. et al. (2021b). AJOM new records and collections of fungi: 101–150. Asian Journal of Mycology 4: 113–260. [Google Scholar]
- Clement M, Posada D, Crandall KA. (2000). TCS: a computer program to estimate gene genealogies. Molecular Ecology 9: 1657–1659. [DOI] [PubMed] [Google Scholar]
- Coetzee MPA, Wingfield BD, Bloomer P. et al. (2005). Phylogenetic analyses of DNA sequences reveal species partitions amongst isolates of Armillaria from Africa. Mycological Research 109: 1223–1234. [DOI] [PubMed] [Google Scholar]
- Cooke MC. (1885). New British fungi. Grevillea 13: 89–100. [Google Scholar]
- Cranston KA, Hurwitz B, Ware D. et al. (2009). Species trees from highly incongruent gene trees in rice. Systematic Biology 58: 489–500. [DOI] [PubMed] [Google Scholar]
- Crouch JA, Clarke BB, Hillman BI. (2009). What is the value of ITS sequence data in Colletotrichum systematics and species diagnosis? A case study using the falcate-spored graminicolous Colletotrichum group. Mycologia 101: 648–656. [DOI] [PubMed] [Google Scholar]
- Crous PW, Gams W, Stalpers JA. et al. (2004). MycoBank: an online initiative to launch mycology into the 21st century. Studies in Mycology 50: 19–22. [Google Scholar]
- Crous PW, Groenewald JZ, Shivas RG. et al. (2011b). Fungal Planet description sheets: 69–91. Persoonia 26: 108–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Hernández-Restrepo M, Schumacher RK. et al. (2021a). New and Interesting Fungi. 4. Fungal Systematics and Evolution 7: 255–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Rossman AY, Aime C. et al. (2021a). Names of phytopathogenic fungi: a practical guide. Phytopathology 111: 1500–1508. [DOI] [PubMed] [Google Scholar]
- Crous PW, Shivas RG, Quaedvlieg W. et al. (2014b). Fungal Planet description sheets: 214–280. Persoonia 32: 184–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Summerell BA, Shivas RG. et al. (2012). Fungal Planet description sheets: 107–127. Persoonia 28: 138–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Summerell BA, Swart L. et al. (2011b). Fungal pathogens of Proteaceae. Persoonia 27: 20–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Burgess TI. et al. (2016a). Fungal Planet description sheets: 469–557. Persoonia 37: 218–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Burgess TI. et al. (2018). Fungal Planet description sheets: 716–784. Persoonia 40: 240–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Chooi YH. et al. (2020). Fungal Planet description sheets: 1042–1111. Persoonia 44: 301–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Guarro J. et al. (2013). Fungal Planet description sheets: 154–213. Persoonia 31: 188–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Le Roux JJ. et al. (2015). Fungal Planet description sheets: 371–399. Persoonia 35: 264–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Richardson DM. et al. (2016b). Fungal Planet description sheets: 400–468. Persoonia 36: 316–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Schumacher RK. et al. (2014a). Fungal Planet description sheets: 281–319. Persoonia 33: 212–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva RMF, Soares AM, de Pádua APSL. et al. (2019). Mycological Diversity Description II. Acta Botanica Brasilica 33: 163–173. [Google Scholar]
- Dai FM, Zeng R, Lu JP. (2012). First report of twig canker on peach caused by Phomopsis amygdali in China. Plant Disease 96: 288–289. [DOI] [PubMed] [Google Scholar]
- Davis JI, Nixon KC. (1992). Populations, genetic variation, and the delimitation of phylogenetic species. Systematic Biology 41: 421–435. [Google Scholar]
- Dayarathne MC, Jones EBG, Maharachchikumbura SSN. et al. (2020). Morpho-molecular characterization of microfungi associated with marine based habitats. Mycosphere 11: 1–188. [Google Scholar]
- Dayrat B. (2005). Towards integrative taxonomy. Biological Journal of the Linnean Society 85: 407–417. [Google Scholar]
- de Carvalho CR, Maia MQ, Sobral M. et al. (2021). Diversity and antimicrobial activity of culturable endophytic fungi associated with the neotropical ethnomedicinal plants Copaifera langsdorffii and Copaifera pubiflora. South African Journal of Botany 142: 305–315. [Google Scholar]
- De Queiroz K. (2007). Species concepts and species delimitation. Systematic Biology 56: 879–886. [DOI] [PubMed] [Google Scholar]
- de Silva NI, Hyde KD, Lumyong S. et al. (2022). Morphology, phylogeny, host association and geography of fungi associated with plants of Annonaceae, Apocynaceae and Magnoliaceae. Mycosphere 13: 955–1076. [Google Scholar]
- Degnan JH, Rosenberg NA. (2006). Discordance of species trees with their most likely gene trees. PLoS Genetics 2: e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnan JH, Rosenberg NA. (2009). Gene tree discordance, phylogenetic inference and the multispecies coalescent. Trends in Ecology and Evolution 24: 332–340. [DOI] [PubMed] [Google Scholar]
- Delacroix G. (1905). Sur une maladie des Amendiers en Provence. Bulletin de la Société Mycologique de France 21: 180–185. [Google Scholar]
- Del-Prado R, Cubas P, Lumbsch HT. et al. (2010). Genetic distances within and among species in monophyletic lineages of Parmeliaceae (Ascomycota) as a tool for taxon delimitation. Molecular Phylogenetics and Evolution 56: 125–133. [DOI] [PubMed] [Google Scholar]
- DeSalle R, Egan MG, Siddall M. (2005). The unholy trinity: taxonomy, species delimitation and DNA barcoding. Philosophical transactions of the Royal Society of London. Series B Biological Sciences 360: 1905–1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desprez-Loustau M-L, Marçais B, Nageleisen L-M, et al. (2006). Interactive effects of drought and pathogens in forest trees. Annals of Forest Science 63: 597–612. [Google Scholar]
- Desprez-Loustau M-L, Robin C, Reynaud G. et al. (2007). Simulating the effects of a climate-change scenario on the geographical range and activity of forest-pathogenic fungi. Canadian Journal of Plant Pathology 29: 101–120. [Google Scholar]
- Dettman JR, Jacobson DJ, Taylor JW. (2003). A multilocus genealogical approach to phylogenetic species recognition in the model eukaryote Neurospora. Evolution 57: 2703–2720. [DOI] [PubMed] [Google Scholar]
- Diaz GA, Latorre BA, Lolas M. et al. (2017). Identification and characterization of Diaporthe ambigua, D. australafricana, D. novem, and D. rudis causing a postharvest fruit rot in kiwifruit. Plant Disease 101: 1402–1410. [DOI] [PubMed] [Google Scholar]
- Diogo ELF, Santos JM, Phillips AJL. (2010). Phylogeny, morphology and pathogenicity of Diaporthe and Phomopsis species on almond in Portugal. Fungal Diversity 44: 107–115. [Google Scholar]
- Dissanayake AJ, Camporesi E, Hyde KD. et al. (2017). Molecular phylogenetic analysis reveals seven new Diaporthe species from Italy. Mycosphere 8: 853–877. [Google Scholar]
- Dissanayake AJ, Chen YY, Liu JJ. (2020). Unravelling Diaporthe species associated with woody hosts from Karst Formations (Guizhou) in China. Journal of Fungi 6: 251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dissanayake RK, Ratnaweera PB, Williams DE. et al. (2016). Antimicrobial activities of mycoleptodiscin B isolated from endophytic fungus Mycoleptodiscus sp. of Calamus thwaitesii Becc. Journal of Applied Pharmaceutical Science 6: 1–6. [Google Scholar]
- Doilom M, Dissanayake AJ, Wanasinghe DN. et al. (2016). Microfungi on Tectona grandis (teak) in Northern Thailand. Fungal Diversity 82: 107–182. [Google Scholar]
- Dong H, Mu T-C, Zhang Z-X, et al. (2020). Two new species of Diaporthe from Yunnan Province, southwestern China. Mycosystema 39: 2241–2250. [Google Scholar]
- Dong H, Mu T-C, Zhang Z-X, et al. (2021a). Molecular phylogenetic analysis reveals two new species of Diaporthe from Yunnan Province, southwestern China. Mycosystema 40: 436–446. [Google Scholar]
- Dong Z, Manawasinghe IS, Huang Y. et al (2021b). Endophytic Diaporthe associated with Citrus grandis cv. Tomentosa in China. Frontiers in Microbiology 11: 609387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Santos Freire MBG. (2017). First report of Diaporthe inconspicua associated with shoot blight of Atriplex nummularia in Brazil Journal of Plant Pathology 99: 542. [Google Scholar]
- dos Santos TT, de Souza LT, de Queiroz CB. et al. (2016). High genetic variability in endophytic fungi from the genus Diaporthe isolated from common bean (Phaseolus vulgaris L.) in Brazil. Journal of Applied Microbiology 120: 388–401. [DOI] [PubMed] [Google Scholar]
- Du Y, Wang X, Guo Y. et al. (2021). Biological and molecular characterization of seven Diaporthe species associated with kiwifruit shoot blight and leaf spot in China. Phytopathologia Mediterranea 60: 177–198. [Google Scholar]
- Dupuis JR, Roe AD, Sperling FA. (2012). Multilocus species delimitation in closely related animals and fungi: one marker is not enough. Molecular Ecology 18: 4422–4436. [DOI] [PubMed] [Google Scholar]
- Edwards SV. (2009). Is a new and general theory of molecular systematics emerging? Evolution 63: 1–19. [DOI] [PubMed] [Google Scholar]
- Edwards SV, Liu L, Pearl DK. (2007). High-resolution species trees without concatenation. Proceedings of the National Academy of Sciences of the USA 104: 5936–5941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfar K, Torres R, Díaz GA. et al. (2013). Characterization of Diaporthe australafricana and Diaporthe spp. associated with stem canker of blueberry in Chile. Plant Disease 97: 1042–1050. [DOI] [PubMed] [Google Scholar]
- Ellis MW. (2011). The problem with the species problem. History and Philosophy of the Life Sciences 33: 343–363. [PubMed] [Google Scholar]
- Esmaeilzadeh A, Zafari D, Bagherabadi S. (2020). First report of Diaporthe foeniculina causing yellowing and dieback on Ficus benjamina. New Disease Reports 41: 16. [Google Scholar]
- Farr DF, Castlebury LA, Pardo-Schultheiss RA. (1999). Phomopsis amygdali causes peach shoot blight of cultivated peach trees in the southeastern United States. Mycologia 91: 1008–1015. [Google Scholar]
- Farr DF, Rossman AY. 2023. Fungal Databases, US National Fungus Collections, ARS, USDA. Available at: https://nt.ars-grin.gov/fungaldatabases/. Accessed on 15 August 2023. [Google Scholar]
- Felsenstein J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39: 783–791. [DOI] [PubMed] [Google Scholar]
- Feng XX, Chen JJ, Wang GR. et al. (2019). Diaporthe sinensis, a new fungus from Amaranthus sp. in China. Phytotaxa 425: 259–268. [Google Scholar]
- Ferreira MC, Cantrell CL, Wedge DE. et al. (2017). Diversity of the endophytic fungi associated with the ancient and narrowly endemic neotropical plant Vellozia gigantea from the endangered Brazilian rupestrian grasslands. Biochemical Systematics and Ecology 71: 163–169. [Google Scholar]
- Ferro LO, Bezerra JDP, da Silva TM. et al. (2024). Endophytic Diaporthe species from Brazil. Fungal Systematics and Evolution 14: 251–269. [Google Scholar]
- Fisher FE. (1972). Diaporthe citri and Phomopsis citri: a correction. Mycologia 64: 422. [Google Scholar]
- Fries EM. (1828). Elenchus Fungorum, sistens Commentarium in Systema Mycologicum 2. Gryphiswaldiae: Sumptibus Ernesti Mauritii, Germany. [Google Scholar]
- Fröhlich J, Hyde KD, Guest DI. (1997). Fungi associated with leaf spots of palms in north Queensland, Australia. Mycological Research 101: 721–732. [Google Scholar]
- Fujita MK, Leaché AD, Burbrink FT. et al. (2012). Coalescent-based species delimitation in an integrative taxonomy. Trends in Ecology & Evolution 27: 480–488. [DOI] [PubMed] [Google Scholar]
- Galili T. (2015). Dendextend: An R package for visualizing, adjusting, and comparing trees of hierarchical clustering. Bioinformatics 31: 3718–3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Liu F, Cai L. (2016). Unravelling Diaporthe species associated with Camellia. Systematics and Biodiversity 14: 102–117. [Google Scholar]
- Gao Y, Liu F, Duan W. et al. (2017). Diaporthe is paraphyletic. IMA Fungus 8: 153–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Su Y, Sun W. et al. (2015). Diaporthe species occurring on Lithocarpus glabra in China, with descriptions of five new species. Fungal Biology 119: 295–309. [DOI] [PubMed] [Google Scholar]
- Gao Y-H, Sun W, Su YY. et al. (2014). Three new species of Phomopsis in Gutianshan Nature Reserve in China. Mycological Progress 13: 111–121. [Google Scholar]
- Garcia-Reyne A, López-Medrano F, Morales JM. et al. (2011). Cutaneous infection by Phomopsis longicolla in a renal transplant recipient from Guinea: first report of human infection by this fungus. Transplant Infectious Disease 13: 204–207. [DOI] [PubMed] [Google Scholar]
- Garofalo F. (1973). L’Albicocco “Tonda di Costigliole”, nuovo ospite di Fusicoccum amygdali Del. Informatore Fitopatologico 23: 13–15. [Google Scholar]
- Gatesy J, Baker RH. (2005). Hidden likelihood support in genomic data: can forty-five wrongs make a right? Systematic Biology 54: 483–492. [DOI] [PubMed] [Google Scholar]
- Gazis R, Rehner S, Chaverri P. (2011). Species delimitation in fungal endophyte diversity studies and its implications in ecological and biogeographic inferences. Molecular Ecology 20: 3001–3013. [DOI] [PubMed] [Google Scholar]
- Gaziz S, Rehner SA, Chaverri P. (2011). Species delimitation in fungal endophyte diversity studies and its implications in ecological and biogeographic inferences. Molecular Ecology 20: 3001–3013. [DOI] [PubMed] [Google Scholar]
- Giraud T, Refrégier G, Le Gac M. et al. (2008). Speciation in fungi. Fungal Genetics and Biology 45: 791–802. [DOI] [PubMed] [Google Scholar]
- Glass NL, Donaldson GC. (1995). Development of primer sets designed for use with PCR to amplify conserved genes from filamentous ascomycetes. Applied and Environmental Microbiology 61: 1323–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glässnerová K, Sklenář F, Jurjević Ž, et al. (2022). A monograph of Aspergillus section Candidi. Studies in Mycology 102: 1–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein PZ, Desalle R, Amato G. et al. (2000). Conservation genetics at the species boundary. Conservation Biology 14: 120–131. [Google Scholar]
- Golzar H, Tan YP, Shivas RG. et al. (2012). First report of shoot blight of persimmon caused by Diaporthe neotheicola in Australia. Australasian Plant Disease Notes 7: 115–117. [Google Scholar]
- Gomes RR, Glienke C, Videira SI. et al. (2013). Diaporthe: a genus of endophytic, saprobic and plant pathogenic fungi. Persoonia 31: 1–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González AJ, Ciordia M. (2023). Shoot dieback in thornless blackberries in Northern Spain caused by Diaporthe rudis and Gnomoniopsis idaeicola. Horticulturae 9: 965. [Google Scholar]
- Guarnaccia V, Crous PW. (2017). Emerging citrus diseases in Europe caused by species of Diaporthe. IMA Fungus 8: 317–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarnaccia V, Groenewald JZ, Woodhall J. et al. (2018). Diaporthe diversity and pathogenicity revealed from a broad survey of grapevine diseases in Europe. Persoonia 40: 135–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarnaccia V, Kraus C, Markakis E. et al. (2022). Fungal trunk diseases of fruit trees in Europe: pathogens, spread and future directions. Phytopathologia Mediterranea 61: 563–599. [Google Scholar]
- Guarnaccia V, Martino I, Tabone G. et al. (2020). Fungal pathogens associated with stem blight and dieback of blueberry in northern Italy. Phytopathologia Mediterranea 59: 229–245. [Google Scholar]
- Guarnaccia V, Vitale A, Cirvilleri G. et al. (2016). Characterisation and pathogenicity of fungal species associated with branch cankers and stem-end rot of avocado in Italy. European Journal of Plant Pathology 146: 963–976. [Google Scholar]
- Guerrero JC, Galdames RG, Ogass KC. et al. (2020). First report of Diaporthe foeniculina causing blacktip and necrotic spot on hazelnut kernel in Chile. Plant Disease 104: 975. [Google Scholar]
- Guo B, Kong L. (2022). Comparing the efficiency of single-locus species delimitation methods within Trochoidea (Gastropoda: Vetigastropoda). Genes 13: 2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JM, Liang JJ, Li KY. et al. (2024). First report of brown blotch disease caused by Diaporthe ueckeri on Hyophorbe lagenicaulis in China. Plant Disease 108: 224. [DOI] [PubMed] [Google Scholar]
- Guo LD, Hyde KD, Liew ECY. (2000). Identification of endophytic fungi from Livistona chinensis based on morphology and rDNA sequences. The New Phytologist 147: 617–630. [DOI] [PubMed] [Google Scholar]
- Guo YS, Crous PW, Bai Q. et al. (2020). High diversity of Diaporthe species associated with pear shoot canker in China. Persoonia 45: 132–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusella G, La Quatra G, Agustí-Brisach C. et al. (2023). Elucidating the almond constriction canker caused by Diaporthe amygdali in Sicily (South Italy). Journal of Plant Pathology 105: 987–1000. [Google Scholar]
- Guzmán B, Fedriani JM, Delibes M. et al. (2017). The colonization history of the Mediterranean dwarf palm (Chamaerops humilis L., Palmae). Tree Genetics & Genomes 13: 24. [Google Scholar]
- Habib W, Gerges E, Antelmi I. et al. (2015). Diaporthe foeniculina associated with severe shoot blight of lemon in Lebanon. Phytopathologia Mediterranea 54: 149–150. [Google Scholar]
- Hall TA. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41: 95–98. [Google Scholar]
- Hart MW, Sunday J. (2007). Things fall apart: biological species form unconnected parsimony networks. Biology Letters 3: 509–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilário S, Amaral IA, Gonçalves MFM. et al. (2020). Diaporthe species associated with twig blight and dieback of Vaccinium corymbosum in Portugal, with description of four new species. Mycologia 112: 293–308. [DOI] [PubMed] [Google Scholar]
- Hilário S, Gonçalves MFM. (2023). Mechanisms underlying the pathogenic and endophytic lifestyles in Diaporthe: an omics-based approach. Horticulturae 9: 423. [Google Scholar]
- Hilário S, Gonçalves MFM, Alves A. (2021b). Using genealogical concordance and coalescent-based species delimitation to assess species boundaries in the Diaporthe eres complex. Journal of Fungi 25: 507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilário S, Santos L, Alves A. (2021a). Diaporthe amygdali, a species complex or a complex species? Fungal Biology 125: 505–518. [DOI] [PubMed] [Google Scholar]
- Hilário S, Santos L, Alves A. (2021c). Diversity and pathogenicity of Diaporthe species revealed from a survey of blueberry orchards in Portugal. Agriculture 11: 1271. [Google Scholar]
- Hilário S, Santos L, Phillips AJL. et al. (2022). Caveats of the internal transcribed spacer region as a barcode to resolve species boundaries in Diaporthe. Fungal Biology 126: 54–74. [DOI] [PubMed] [Google Scholar]
- Hillis DM, Bull JJ. (1993). An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Systematic Biology 42: 182–192. [Google Scholar]
- Hobbs TW, Schmitthenner AF, Kuter GA. (1985). A new Phomopsis species from soybean. Mycologia 77: 535–544. [Google Scholar]
- Holland LA, Trouillas FP, Nouri MT. et al. (2021). Fungal pathogens associated with canker diseases of almond in California. Plant Disease 105: 346–360. [DOI] [PubMed] [Google Scholar]
- Hongsanan S, Jeewon R, Purahong W. et al. (2018). Can we use environmental DNA as holotypes? Fungal Diversity 92: 1–30. [Google Scholar]
- Hongsanan S, Norphanphoun C, Senanayake IC. et al. (2023). Annotated notes on Diaporthe species. Mycosphere 14: 918–1189. [Google Scholar]
- Huang F, Udayanga D, Wang XH. et al. (2015). Endophytic Diaporthe associated with Citrus: a phylogenetic reassessment with seven new species from China. Fungal Biology 119: 331–347. [DOI] [PubMed] [Google Scholar]
- Huang ST, Xia JW, Pan Y. et al. (2021a). Morphological and molecular description of Diaporthe nannuoshanensis sp. nov. (Diaporthaceae, Diaporthales) in south-western China. Nova Hedwigia 112: 399–411. [Google Scholar]
- Huang ST, Xia JW, Zhang XG. et al. (2021b). Morphological and phylogenetic analyses reveal three new species of Diaporthe from Yunnan, China. MycoKeys 78: 49–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes SJ. (1953). Fungi from the Gold Coast. II. Mycological Papers 50: 1–104. [Google Scholar]
- Huson DH, Bryant D. (2006). Application of phylogenetic networks in evolutionary studies. Molecular Biology and Evolution 23: 254–267. [DOI] [PubMed] [Google Scholar]
- Huson DH, Scornavacca C. (2011). A survey of combinatorial methods for phylogenetic networks. Genome Biology and Evolution 3: 23–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde KD, Abd-Elsalam K, Cai L. (2010). Morphology: still essential in a molecular world. Mycotaxon 114: 439–451. [Google Scholar]
- Hyde KD, Chaiwan N, Norphanphoun C. et al. (2018). Mycosphere notes 169–224. Mycosphere 9: 271–430. [Google Scholar]
- Hyde KD, Dong Y, Phookamsak R. et al. (2020a). Fungal diversity notes 1151–1276: taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Diversity 100: 5–277. [Google Scholar]
- Hyde KD, Hongsanan S, Jeewon R. et al. (2016). Fungal diversity notes 367–491: taxonomic and phylogenetic contributions to fungal taxa. Fungal Diversity 80: 1–270. [Google Scholar]
- Hyde KD, Jeewon R, Chen YJ. et al. (2020b). The numbers of fungi: is the descriptive curve flattening? Fungal Diversity 103: 219–271. [Google Scholar]
- Hyde KD, Norphanphoun C, Ma J. et al. (2023). Mycosphere notes 387– 412 – novel species of fungal taxa from around the world. Mycosphere 14: 663–744. [Google Scholar]
- Hyde KD, Tennakoon DS, Jeewon R. et al. (2019). Fungal diversity notes 1036–1150: taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Diversity 96: 1–242. [Google Scholar]
- Hyde KD, Udayanga D, Manamgoda DS. et al. (2013). Incorporating molecular data in fungal systematics: a guide for aspiring researchers. Current Research in Environmental & Applied Mycology 3: 1–32. [Google Scholar]
- Hyde KD, Zhang Y. (2008). Epitypification: should we epitypify? Journal of Zhejiang University Science B 9: 842–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Index Fungorum Partnership (2023). Index Fungorum. Available at: http://www.indexfungorum.org/. Accessed on 15 August 2023. [Google Scholar]
- Jeewon R, Hyde KD. (2016). Establishing species boundaries and new taxa among fungi: recommendations to resolve taxonomic ambiguities. Mycosphere 7: 1669–1677. [Google Scholar]
- Jiang N, Voglmayr H, Piao C-G, et al. (2021). Two new species of Diaporthe (Diaporthaceae, Diaporthales) associated with tree cankers in the Netherlands. MycoKeys 85: 31–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao X, Flouri T, Yang Z. (2021). Multispecies coalescent and its applications to infer species phylogenies and cross-species gene flow. HAYATI Journal of Biosciences 8: nwab127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julia ZR, Sukarno N, Ardie SW. et al. (2022). Endophytic fungi isolated from the mangrove species Rhizophora apiculata and their efficacy as herbicides. HAYATI Journal of Biosciences 29: 605–620. [Google Scholar]
- Kaliterna J, Miličević T, Cvjetković B. (2012). Grapevine trunk diseases associated with fungi from the Diaporthaceae family in Croatian vineyards. Archives of Industrial Hygiene and Toxicology 63: 471–479. [DOI] [PubMed] [Google Scholar]
- Kanematsu S, Yokoyama Y, Kobayashi T. et al. (1999). Taxonomic reassessment of the causal fungus of peach Fusicoccum canker in Japan. Annals of the Phytopathological Society of Japan 65: 531–536. [Google Scholar]
- Kapli P, Lutteropp S, Zhang J. et al. (2017). Multi-rate Poisson tree processes for single-locus species delimitation under maximum likelihood and Markov chain Monte Carlo. Bioinformatics 33: 1630–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbstein K, Tomasello S, Hodač L, et al. (2020). Phylogenomics supported by geometric morphometrics reveals delimitation of sexual species within the polyploid apomictic Ranunculus auricomus complex (Ranunculaceae). Taxon 69: 1191–1220. [Google Scholar]
- Kassambara A, Mundt F. (2020). Factoextra: extract and visualize the results of multivariate data analyses. R package version 1.0.7. Available at: https://CRAN.R-project.org/package=factoextra (accessed on 15 November 2023). [Google Scholar]
- KC A, Rasmussen L. (2019). First report of Diaporthe rudis causing fruit rot of European pears in the United States. Plant Disease 103: 2132. [Google Scholar]
- Kekkonen M, Hebert PD. (2014). DNA barcode-based delineation of putative species: efficient start for taxonomic workflows. Molecular Ecology Resources 14: 706–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Kim DY, Park H. et al. (2017). Two endophytic Diaporthe species isolated from the leaves of Astragalus membranaceus in Korea. Mycobiology 45: 430–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. (1980). A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. Journal of Molecular Evolution 16: 111–120. [DOI] [PubMed] [Google Scholar]
- Kozlov AM, Darriba D, Flouri T. et al. (2019). RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 35: 4453–4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamurthy PK, Francis RA. (2012). A critical review on the utility of DNA barcoding in biodiversity conservation. Biodiversity and Conservation 21: 1901–1919. [Google Scholar]
- Kubatko LS, Degnan JH. (2007). Inconsistency of phylogenetic estimates from concatenated data under coalescence. Systematic Biology 56: 17–24. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M. et al. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution 35: 1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvist S, Laumer CE, Junoy J. et al. (2014). New insights into the phylogeny, systematics and DNA barcoding of Nemertea. Invertebrate Systematics 28: 287–308. [Google Scholar]
- Lambert C, Schweizer L, Matio Kemkuignou B. et al. (2023). Four new endophytic species of Diaporthe (Diaporthaceae, Diaporthales) isolated from Cameroon. MycoKeys 99: 319–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurence MH, Summerell BA, Burgess LW. et al. (2014). Genealogical concordance phylogenetic species recognition in the Fusarium oxysporum species complex. Fungal Biology 118: 374–384. [DOI] [PubMed] [Google Scholar]
- Lawrence DP, Travadon R, Baumgartner K. (2015). Diversity of Diaporthe species associated with wood cankers of fruit and nut crops in northern California. Mycologia 107: 926–940. [DOI] [PubMed] [Google Scholar]
- León M, Berbegal M, Rodríguez-Reina JM. et al. (2020). Identification and characterization of Diaporthe spp. associated with twig cankers and shoot blight of almonds in Spain. Agronomy 10: 1062. [Google Scholar]
- Liao YC, Sun JW, Li DW. et al. (2022). First report of top blight of Cunninghamia lanceolata caused by Diaporthe unshiuensis and Diaporthe hongkongensis in China. Plant Disease 107: 962. [DOI] [PubMed] [Google Scholar]
- Lim L, Mohd MH, Zakaria L. (2019). Identification and pathogenicity of Diaporthe species associated with stem-end rot of mango (Mangifera indica L.). European Journal of Plant Pathology 155: 687–696. [Google Scholar]
- Liu F, Wang M, Damm U. et al. (2016). Species boundaries in plant pathogenic fungi: a Colletotrichum case study. BMC Evolutionary Biology 16: 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HY, Luo D, Huang HL. et al. (2024). Two new species of Diaporthe (Diaporthaceae, Diaporthales) associated with Camellia oleifera leaf spot disease in Hainan Province, China. MycoKeys 102: 225–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JK, Hyde KD, Jones EBG. et al. (2015). Fungal Diversity notes 1–100: Taxonomic and phylogenetic contributions to fungal species. Fungal Diversity 72: 1–197. [Google Scholar]
- Liu L, Yu L, Kubatko L. et al. (2009). Coalescent methods for estimating phylogenetic trees. Molecular Phylogenetics and Evolution 53: 320–328. [DOI] [PubMed] [Google Scholar]
- Liu L, Yu L, Pearl DK. et al. (2009). Estimating species phylogenies using coalescence times among sequences. Systematic Biology 58: 468–477. [DOI] [PubMed] [Google Scholar]
- Lombard L, Leeuwen GC, Guarnaccia V. et al. (2014). Diaporthe species associated with Vaccinium, with specific reference to Europe. Phytopathologia Mediterranea 53: 287–299. [Google Scholar]
- Lopes AF, Batista E, Hilário S. et al. (2021). Occurrence of Diaporthe species in Eucalyptus globulus, Pinus pinaster and Quercus suber in Portugal. Forest Pathology 51: e12674. [Google Scholar]
- Lopez-Cardona N, Guevara-Castro A, Ganan-Betancur L. et al. (2021). First report of Diaporthe ueckerae causing stem canker on soybean (Glycine max) in Colombia. Plant Disease 105: 4162. [DOI] [PubMed] [Google Scholar]
- López-Giráldez F, Townsend JP. (2011). PhyDesign: an online application for profiling phylogenetic informativeness. BMC Evolutionary Biology 11: 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Moral A, Lovera M, Raya MDC. et al. (2020). Etiology of branch dieback and shoot blight of English walnut caused by Botryosphaeriaceae and Diaporthe species in Southern Spain. Plant Disease 104: 533–550. [DOI] [PubMed] [Google Scholar]
- Lorenzini M, Zapparoli G. (2019). Diaporthe rudis associated with berry rot of postharvest grapes in Italy. Plant Disease 103: 1030. [Google Scholar]
- Lu QT, Zhang JY, Sun YR. et al. (2022). Diaporthe orixae sp. nov., an endophytic species isolated from Orixa japonica in southern China. Phytotaxa 544: 37–51. [Google Scholar]
- Lücking R, Aime MC, Robbertse B. et al. (2020). Unambiguous identification of fungi: where do we stand and how accurate and precise is fungal DNA barcoding? IMA Fungus 11: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lücking R, Hawksworth DL. (2018). Formal description of sequence-based voucherless Fungi: promises and pitfalls, and how to resolve them. IMA Fungus 9: 143–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna IJ, Doll D, Ashworth VETM. et al. (2022). Comparative profiling of wood canker pathogens from spore traps and symptomatic plant samples within California almond and walnut orchards. Plant Disease 106: 2182–2190. [DOI] [PubMed] [Google Scholar]
- Luo M, Guo W, Zhao MP. et al. (2022). Endophytic Diaporthe associated with Morinda officinalis in China. Journal of Fungi 8: 806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddison WP, Knowles LL. (2006). Inferring phylogeny despite incomplete lineage sorting. Systematic Biology 55: 21–30. [DOI] [PubMed] [Google Scholar]
- Maechler M, Rousseeuw P, Struyf A. et al. (2022). Cluster: cluster analysis basics and extensions. R package version 2.1.4. Available at: https://CRAN.R-project.org/package=cluster (accessed on 15 November 2023).
- Maharachchikumbura SS, Chen Y, Ariyawansa HA. et al. (2021). Integrative approaches for species delimitation in Ascomycota. Fungal Diversity 109: 155–179. [Google Scholar]
- Maharachchikumbura SS, Guo LD, Cai L. et al. (2012). A multi-locus backbone tree for Pestalotiopsis, with a polyphasic characterization of 14 new species. Fungal Diversity 56: 95–129. [Google Scholar]
- Makris G, Solonos S, Christodoulou M. et al. (2022). First report of Diaporthe foeniculina associated with grapevine trunk diseases on Vitis vinifera in Cyprus. Plant Disease 106: 1294. [DOI] [PubMed] [Google Scholar]
- Manawasinghe IS, Dissanayake AJ, Li X. et al. (2019). High genetic diversity and species complexity of Diaporthe associated with grapevine dieback in China. Frontiers in Microbiology 10: 1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manawasinghe IS, Jayawardena RS, Li HL. et al. (2021). Microfungi associated with Camellia sinensis: a case study of leaf and shoot necrosis on Tea in Fujian, China. Mycosphere 12: 430–518. [Google Scholar]
- Marincowitz S, Crous PW, Groenewald JZ. et al. (2008). Microfungi occurring on the Proteaceae in the fynbos. CBS Biodiversity Series No. 7. CBS Fungal Biodiversity Centre, the Netherlands. [Google Scholar]
- Marin-Felix Y, Hernández-Restrepo M, Wingfield MJ. et al. (2019). Genera of phytopathogenic fungi: GOPHY 2. Studies in Mycology 92: 47–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathioudakis MM, Tziros GT, Kavroulakis N. (2020). First report of Diaporthe foeniculina associated with branch canker of avocado in Greece. Plant Disease 104: 3057. [Google Scholar]
- Matio Kemkuignou B, Schweizer L, Lambert C. et al. (2022). New polyketides from the liquid culture of Diaporthe breyniae sp. nov. (Diaporthales, Diaporthaceae). MycoKeys 90: 85–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCartney HA, Foster SJ, Fraaije BA. et al. (2003). Molecular diagnostics for fungal plant pathogens. Pest Management Science 59: 129–142. [DOI] [PubMed] [Google Scholar]
- Mello B. (2018). Estimating TimeTrees with MEGA and the TimeTree Resource. Molecular Biology and Evolution 35: 2334–2342. [DOI] [PubMed] [Google Scholar]
- Mendes FK, Hahn MW. (2018). Why concatenation fails near the anomaly zone. Systematic Biology 67: 158–169. [DOI] [PubMed] [Google Scholar]
- Meng L, Yu C, Wang C. et al. (2018). First report of Diaporthe amygdali causing walnut twig canker in Shandong province of China. Plant Disease 102: 1859.30125178 [Google Scholar]
- Michailides TJ, Thomidis T. (2006). First report of Phomopsis amygdali causing fruit rot on peaches in Greece. Plant Disease 90: 1551. [DOI] [PubMed] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. (2010). Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: 2010 Gateway Computing Environments Workshop (GCE), 14 November 2010, New Orleans, LA, USA: 1–8. [Google Scholar]
- Mirarab S, Nakhleh L, Warnow T. (2021). Multispecies coalescent: theory and applications in phylogenetics. Annual Review of Ecology, Evolution, and Systematics 52: 247–268. [Google Scholar]
- Monkai J, Hongsanan S, Bhat DJ. et al. (2023). Integrative taxonomy of novel Diaporthe species associated with medicinal plants in Thailand. Journal of Fungi 9: 603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira RR, Caus G, Gomes Figueiredo JA. et al. (2020). Phomopsis rot caused by Diaporthe infecunda on fruit and flowers of Passiflora edulis in Brazil. Australasian Plant Pathology 49: 141–145. [Google Scholar]
- Moritz C, Cicero C. (2004). DNA barcoding: promise and pitfalls. PLoS Biology 2: 1529–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostert L, Crous PW, Kang JC. et al. (2001). Species of Phomopsis and a Libertella sp. occurring on grapevines with specific reference to South Africa: morphological, cultural, molecular and pathological characterization. Mycologia 93: 146–167. [Google Scholar]
- Moyo P, Mostert L, Bester M. et al. (2016). Trunk disease fungi associated with Diospyros kaki in South Africa. Plant Disease 100: 2383–2393. [DOI] [PubMed] [Google Scholar]
- Mu TC, Liu RY, Zhang XG, Li Z, Xia JW. (2022). Diaporthe cha sp. nov. on Camellia sinensis in China. Nova Hedwigia 115: 473–485. [Google Scholar]
- Murali TS, Suryanarayanan TS, Geeta R. (2006). Endophytic Phomopsis species: host range and implications for diversity estimates. Canadian Journal of Microbiology 52: 673–680. [DOI] [PubMed] [Google Scholar]
- Nair DS, Sajeena A, Johnson JM. et al. (2021). First report of leaf blight of yardlong bean caused by Diaporthe tectonae in India. Journal of Plant Pathology 103: 1069–1070. [Google Scholar]
- Nilsson R, Tedersoo L, Abarenkov K. et al. (2012). Five simple guidelines for establishing basic authenticity and reliability of newly generated fungal ITS sequences. MycoKeys 4: 37–63. [Google Scholar]
- Nilsson RH, Hyde KD, Pawłowska J. et al. (2014). Improving ITS sequence data for identification of plant pathogenic fungi. Fungal Diversity 67: 11–19. [Google Scholar]
- Nilsson RH, Ryberg M, Kristiansson E. et al. (2006). Taxonomic reliability of DNA sequences in public sequence databases: a fungal perspective. PLoS ONE 1: e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noriler SA, Savi DC, Ponomareva L. et al. (2019). Vochysiamides A and B: two new bioactive carboxamides produced by the new species Diaporthe vochysiae. Fitoterapia 138: 104273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norphanphoun C, Gentekaki E, Hongsanan S. et al. (2022). Diaporthe: formalizing the species-group concept. Mycosphere 13: 752–819. [Google Scholar]
- Nute M, Chou J, Molloy EK. et al. (2018). The performance of coalescent-based species tree estimation methods under models of missing data. BMC Genomics 19: 286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell K. (1993). Fusarium and its near relatives. In: The fungal holomorph: mitotic, meiotic and pleomorphic speciation in fungal systematics (Reynolds DR, Taylor JW. eds). CAB International, UK: 225–233. [Google Scholar]
- O’Donnell K, Cigelnik E. (1997). Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Molecular Phylogenetics and Evolution 7: 103–116. [DOI] [PubMed] [Google Scholar]
- Padial JM, Miralles A, De la Riva I. et al. (2010). The integrative future of taxonomy. Frontiers in Zoology 7: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira DS, Hilário S, Gonçalves MFM. et al. (2023). Diaporthe species on palms: molecular re-assessment and species boundaries delimitation in the D. arecae species complex. Microorganisms 11: 2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira DS, Phillips AJL. (2023). Botryosphaeriaceae on palms – a new species of Neodeightonia, N. chamaeropicola, and new records from diseased foliage of ornamental palms in Portugal. Phytotaxa 627: 1–64. [Google Scholar]
- Petrak F. (1919). Mykologische Notizen. I. Annales Mycologici 17: 59–100. [Google Scholar]
- Petrović K, Riccioni L, Vidić M. et al. (2016). First report of Diaporthe novem, D. foeniculina, and D. rudis associated with soybean seed decay in Serbia. Plant Disease 100: 2324–2324. [Google Scholar]
- Phillips AJL. (1999). The relationship between Diaporthe perjuncta and Phomopsis viticola on grapevines. Mycologia 91: 1001–1007. [Google Scholar]
- Phillips AJL. (2003). Morphological characterization of Diaporthe foeniculacea and its Phomopsis anamorph on Foeniculum vulgare. Sydowia 55: 274–285. [Google Scholar]
- Pitcher DG, Saunders NA, Owen RJ. (1989). Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Letters in Applied Microbiology 8: 151–156. [Google Scholar]
- Pond SLK, Frost SDW, Muse SV. (2015). HyPhy: hypothesis testing using phylogenies. Bioinformatics 21: 676–679. [DOI] [PubMed] [Google Scholar]
- Pscheidt JW, Heckert S, Wiseman M. et al. (2019). Fungi associated with and influence of moisture on development of kernel mold of hazelnut. Plant Disease 103: 922–928. [DOI] [PubMed] [Google Scholar]
- Puillandre N, Brouillet S, Achaz G. (2021). ASAP: assemble species by automatic partitioning. Molecular Ecology Resources 21: 609–620. [DOI] [PubMed] [Google Scholar]
- Puillandre N, Lambert A, Brouillet S. et al. (2012). ABGD, Automatic Barcode Gap Discovery for primary species delimitation. Molecular Ecology 21: 1864–1877. [DOI] [PubMed] [Google Scholar]
- Quaedvlieg W, Binder M, Groenewald JZ. et al. (2014). Introducing the Consolidated Species Concept to resolve species in the Teratosphaeriaceae. Persoonia 33: 1–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2021). R: a language and environment for statistical computing. R Foundation for Statistical Computing, Austria. Available at: https://www.R-project.org/. Accessed on 15 November 2023. [Google Scholar]
- Rambaut A. (2018). FigTree v1.4.4. Available at: http://tree.bio.ed.ac.uk/software/figtree/. Accessed on 15 August 2023.
- Rannala B, Yang Z. (2003). Bayes estimation of species divergence times and ancestral population sizes using DNA sequences from multiple loci. Genetics 164: 1645–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rannala B, Yang Z. (2020). Species delimitation. In: Phylogenetics in the Genomic Era (Scornavacca C, Delsuc F, Galtier N. eds). No commercial publisher: 5.5:1–5.5:18. [Google Scholar]
- Rannala B, Yang ZH. (2003). Bayes estimation of species divergence times and ancestral population sizes using DNA sequences from multiple loci. Genetics 164: 1645–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Réblová M, Hernández-Restrepo M, Sklenář F, et al. (2022). Consolidation of Chloridium: new classification into eight sections with 37 species and reinstatement of the genera Gongromeriza and Psilobotrys. Studies in Mycology 103: 87–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehner SA, Uecker FA. (1994). Nuclear ribosomal internal transcribed spacer phylogeny and host diversity in the coelomycetes Phomopsis. Canadian Journal of Botany 72: 1666–1674. [Google Scholar]
- Rhouma A, Triki MA, Ouertani K. et al. (2008). Chemical and biological control of Phomopsis amygdali the causal agent of constriction canker of Almond in Tunisia. Tunisian Journal of Plant Protection 3: 69–77. [Google Scholar]
- Rokas A, Carroll SB. (2006). Bushes in the tree of life. PLoS Biology 4: e352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P. et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg NA. (2003). The shapes of neutral gene genealogies in two species: probabilities of monophyly, paraphyly, and polyphyly in a coalescent model. Evolution 57: 1465–1477. [DOI] [PubMed] [Google Scholar]
- Rossman AY, Palm-Hernández ME. (2008). Systematics of plant pathogenetic fungi: why it matters? Plant Disease 92: 1376–1386. [DOI] [PubMed] [Google Scholar]
- Saccardo PA. (1880). Fungi Gallici lecti a cl. viris P. Brunaud, Abb. Letendre, A. Malbranche, J. Therry, vel editi in Mycotheca Gallica C. Roumeguèri. Series II. Michelia 2: 39–135. [Google Scholar]
- Saccardo PA. (1892). Supplementum Universale, Pars II. Discomyceteae-Hyphomyceteae. Sylloge Fungorum 10: 1–964. [Google Scholar]
- Saccardo PA, Berlese AN. (1885). Miscellania mycologica. Series II. Atti dell’Istituto Veneto Scienze 3: 711–742. [Google Scholar]
- Saengket M, Hyde KD, Kumar V. et al. (2021). Endophytic fungi from Oncosperma sp. with promising in vitro plant growth promotion and antagonistic activities. Chiang Mai Journal of Science 48: 837–852. [Google Scholar]
- Sakamoto T, Innan H. (2019). The evolutionary dynamics of a genetic barrier to gene flow: from the establishment to the emergence of a peak of divergence. Genetics 212: 1383–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos JM, Correia VG, Phillips AJL. (2010). Primers for mating-type diagnosis in Diaporthe and Phomopsis: their use in teleomorph induction in vitro and biological species definition. Fungal Biology 114: 255–270. [DOI] [PubMed] [Google Scholar]
- Santos JM, Phillips AJL. (2009). Resolving the complex of Phomopsis species and their Diaporthe teleomorphs on Foeniculum vulgare. Fungal Diversity 34: 111–125. [Google Scholar]
- Santos JM, Vrandecic K, Cosic J. et al. (2011). Resolving the Diaporthe species occurring on soybean in Croatia. Persoonia 27: 9–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos L, Alves A, Alves R. (2017a). Evaluating multilocus phylogenies for species boundaries determination in the genus Diaporthe. PeerJ 5: e3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos L, Phillips AJL, Crous PW. et al. (2017b). Diaporthe species on Rosaceae with descriptions of D. pyracanthae sp. nov. and D. malorum sp. nov. Mycosphere 8: 485–511. [Google Scholar]
- Satler JD, Carstens BC, Hedin M. (2013). Multilocus species delimitation in a complex of morphologically conserved trapdoor spiders (Mygalomorphae, Antrodiaetidae, Aliatypus). Systematic Biology 62: 805–823. [DOI] [PubMed] [Google Scholar]
- Schoch CL, Seifert KA, Huhndorf S. et al. (2012). Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proceedings of the National Academy of Sciences of the USA 109: 6241–6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrempf D, Szöllősi G. (2020). The sources of phylogenetic conflicts. In: Phylogenetics in the Genomic Era (Scornavacca C, Delsuc F, Galtier N. eds). No commercial publisher: 3.1–3.23. [Google Scholar]
- Seifollahi E, de Farias ARG, Jayawardena RS. et al (2023). Taxonomic advances from fungal flora associated with ferns and fern-like hosts in Northern Thailand. Plants 12: 683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senanayake IC, Rossi W, Leonardi M. et al. (2023). Fungal diversity notes 1611–1716: taxonomic and phylogenetic contributions on fungal genera and species emphasis in south China. Fungal Diversity 122: 161–403. [Google Scholar]
- Sessa L, Abreo E, Bettucci L. et al. (2017). Diversity and virulence of Diaporthe species associated with wood disease symptoms in deciduous fruit trees in Uruguay. Phytopathologia Mediterranea 56: 431–444. [Google Scholar]
- Sessa L, Abreo E, Lupo S. (2018). Diversity of fungal latent pathogens and true endophytes associated with fruit trees in Uruguay. Journal of Phytopathology 166: 633–647. [Google Scholar]
- Sklenář F, Jurjević Ž, Houbraken J. et al. (2021). Re-examination of species limits in Aspergillus section Flavipedes using advanced species delimitation methods and description of four new species. Studies in Mycology 99: 100120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sklenář F, Jurjević Ž, Zalar P. et al. (2017). Phylogeny of xerophilic aspergilli (subgenus Aspergillus) and taxonomic revision of section Restricti. Studies in Mycology 88: 161–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit L, Langenhoven WE, Petersen Y. (2021). Diaporthe species associated with dieback on Cyclopia (honeybush). European Journal of Plant Pathology 161: 565–578. [Google Scholar]
- Sokal RR, Rohlf FJ. (1962). The comparison of dendrograms by objective methods. Taxon 11: 33–40. [Google Scholar]
- Som A. (2015). Causes, consequences and solutions of phylogenetic incongruence. Briefings in Bioinformatics 16: 536–548. [DOI] [PubMed] [Google Scholar]
- Sousa da Câmara M. (1947). Mycetes aliquot Lusitaniae VII. Agronomia Lusitana 9: 85–128. [Google Scholar]
- Srivastava HC, Banu Z, Govindarajan VS. (1962). Fruit rot of areca nut caused by a new fungus. Mycologia 54: 5–11. [Google Scholar]
- Steel MA. (1994). Recovering a tree from the leaf colorations it generates under a Markov model. Applied Mathematics Letters 7: 19–24. [Google Scholar]
- Steenkamp ET, Wingfield MJ, McTaggart AR. et al. (2018). Fungal species and their boundaries matter – definitions, mechanisms and practical implications. Fungal Biology Reviews 32: 104–116. [Google Scholar]
- Stengel A, Stanke KM, Quattrone AC. et al. (2022). Improving taxonomic delimitation of fungal species in the age of genomics and phenomics. Frontiers in Microbiology 13: 847067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart JE, Timmer LW, Lawrence CB. et al. (2014). Discord between morphological and phylogenetic species boundaries: incomplete lineage sorting and recombination results in fuzzy species boundaries in an asexual fungal pathogen. BMC Evolutionary Biology 14: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stukenbrock EH. (2013). Evolution, selection and isolation: a genomic view of speciation in fungal plant pathogens. The New Phytologist 199: 895–907. [DOI] [PubMed] [Google Scholar]
- Sukumaran J, Knowles LL. (2017). Multispecies coalescent delimits structure, not species. Proceedings of the National Academy of Sciences of the USA 114: 1607–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Huang S, Xia J. et al. (2021). Morphological and molecular identification of Diaporthe species in south-western China, with description of eight new species. MycoKeys 77: 65–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL. (2002). PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4.0a165. Sinauer Associates, USA. [Google Scholar]
- Tamura K, Battistuzzi FU, Billing-Ross P. et al. (2012). Estimating divergence times in large molecular phylogenies. Proceedings of the National Academy of Sciences of the USA 109: 19333–19338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Qiqing T, Kumar S. (2018). Theoretical foundation of the RelTime method for estimating divergence times from variable evolutionary rates. Molecular Biology and Evolution 35: 1770–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan YP, Edwards J, Grice KRE. et al. (2013). Molecular phylogenetic analysis reveals six new Diaporthe species from Australia. Fungal Diversity 61: 251–260. [Google Scholar]
- Tan YP, Shivas RG. (2022). Nomenclatural novelties. Index of Australian Fungi 2: 1–12. [Google Scholar]
- Tan YP, Shivas RG. (2023). Nomenclatural novelties. Index of Australian Fungi 6: 1–17. [Google Scholar]
- Taylor HR, Harris WE. (2012). An emergent science on the brink of irrelevance: a review of the past 8 years of DNA barcoding. Molecular Ecology Resources 12: 377–388. [DOI] [PubMed] [Google Scholar]
- Taylor JE, Hyde KD. (2003). Microfungi of tropical and temperate palms. Fungal Diversity Research Series No. 12. Fungal Diversity Press, China. [Google Scholar]
- Taylor JW, Jacobson DJ, Kroken S. et al. (2000). Phylogenetic species recognition and species concepts in fungi. Fungal Genetics and Biology 31: 21–32. [DOI] [PubMed] [Google Scholar]
- Taylor JW, Turner E, Townsend JP. et al. (2006). Eukaryotic microbes, species recognition and the geographic limits of species: examples from the kingdom Fungi. Philosophical Transactions of the Royal Society B: Biological Sciences 361: 1947–1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tekiner N, Tozlu E, Guarnaccia V. (2020). First report of Diaporthe foeniculina causing fruit rot of lemon in Turkey. Journal of Plant Pathology 102: 277. [Google Scholar]
- Templeton AR, Crandall KA, Sing CF. (1992). A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and DNA sequence data. III. Cladogram estimation. Genetics 132: 619–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennakoon DS, Kuo CH, Maharachchikumbura SSN. et al. (2021). Taxonomic and phylogenetic contributions to Celtis formosana, Ficus ampelas, F. septica, Macaranga tanarius and Morus australis leaf litter inhabiting microfungi. Fungal Diversity 108: 1–215. [Google Scholar]
- Thambugala KM, Daranagama DA, Phillips AJL. et al. (2017). Microfungi on Tamarix. Fungal Diversity 82: 239–306. [Google Scholar]
- Thavamani R, Suryanarayanan TS, Murali TS. et al. (2018). Distribution and diversity of foliar endophytic fungi in the mangroves of Andaman Islands, India. Fungal Ecology 36: 109–116. [Google Scholar]
- Thomidis T, Exadaktylou E, Chen SF. (2013). Diaporthe neotheicola, a new threat for kiwifruit in Greece. Crop Protection 47: 35–40. [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F. et al. (1997). The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research 25: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson SM, Tan YP, Shivas RG. et al. (2015). Green and brown bridges between weed and crop hosts reveal novel Diaporthe spp. in Australia. Persoonia 35: 39–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibshirani R, Walther G, Hastie T. (2001). Estimating the number of clusters in a data set via the gap statistic. Journal of the Royal Statistical Society Series B: Statistical Methodology 63: 411–423. [Google Scholar]
- Toghueo RMK, Vázquez de Aldana BR, Zabalgogeazcoa I. (2023). Diaporthe species associated with the maritime grass Festuca rubra subsp. pruinosa. Frontiers in Microbiology 14: 1105299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres C, Camps R, Aguirre R. et al. (2016). First report of Diaporthe rudis in Chile causing stem-end rot on ‘Hass’ avocado fruit imported from California. Plant Disease 100: 1951. [Google Scholar]
- Townsend JP. (2007). Profiling phylogenetic informativeness. Systematic Biology 56: 222–231. [DOI] [PubMed] [Google Scholar]
- Tuset JJ, Portilla MT. (1989). Taxonomic status of Fusicoccum amygdali and Phomopsis amygdalina. Canadian Journal of Botany 67: 1275–1280. [Google Scholar]
- Udayanga D, Castlebury LA, Rossman AY. et al. (2014a). Insights into the genus Diaporthe: phylogenetic species delimitation in the D. eres species complex. Fungal Diversity 67: 203–229. [Google Scholar]
- Udayanga D, Castlebury LA, Rossman AY. et al. (2014b). Species limits in Diaporthe: molecular re-assessment of D. citri, D. cytosporella, D. foeniculina and D. rudis. Persoonia 32: 83–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udayanga D, Castlebury LA, Rossman AY. et al. (2015). The Diaporthe sojae species complex: phylogenetic re-assessment of pathogens associated with soybean, cucurbits and other field crops. Fungal Biology 119: 383–407. [DOI] [PubMed] [Google Scholar]
- Udayanga D, Liu X, McKenzie EHC. et al. (2011). The genus Phomopsis: biology, applications, species concepts and names of common phytopathogens. Fungal Diversity 50: 189–225. [Google Scholar]
- Udayanga D, Liu XX, Crous PW. et al. (2012b). Multi-locus phylogeny reveals three new species of Diaporthe from Thailand. Cryptogamie, Mycologie 33: 295–309. [Google Scholar]
- Udayanga D, Liu XZ, Crous PW. et al. (2012a). A multilocus phylogenetic evaluation of Diaporthe (Phomopsis). Fungal Diversity 56: 157–171. [Google Scholar]
- Uecker FA. (1988). A World List of Phomopsis Names with Notes on Nomenclature, Morphology and Biology. Contributions from the U.S. National Fungus Collection No. 3. Mycologia Memoir, Volume 13. Cramer Publisher, Germany. [Google Scholar]
- Úrbez-Torres JR, Peduto F, Smith RJ. et al. (2013). Phomopsis dieback: a grapevine trunk disease caused by Phomopsis viticola in California. Plant Disease 97: 1571–1579. [DOI] [PubMed] [Google Scholar]
- Vakalounakis DJ, Ntougias S, Kavroulakis N. et al. (2019). Neofusicoccum parvum and Diaporthe foeniculina associated with twig and shoot blight and branch canker of citrus in Greece. Journal of Phytopathology 167: 527–537. [Google Scholar]
- van Dyk M, Spies CFJ, Mostert L. et al. (2021). Pathogenicity testing of fungal isolates associated with olive trunk diseases in South Africa. Plant Disease 105: 4060–4073. [DOI] [PubMed] [Google Scholar]
- van Niekerk JM, Groenewald JZ, Farr DF. et al. (2005). Reassessment of Phomopsis species on grapevines. Australasian Plant Pathology 34: 27–39. [Google Scholar]
- Varjas V, Vajna L, Izsépi F. et al. (2017). First report of Phomopsis amygdali causing twig canker on almond in Hungary. Plant Disease 101: 1674. [Google Scholar]
- Vega FE, Simpkins A, Aime MC. et al. (2010). Fungal endophyte diversity in coffee plants from Colombia, Hawai’i, Mexico and Puerto Rico. Fungal Ecology 3: 122–138. [Google Scholar]
- Vieira JPS, Selbach-Schnadelbach A, Braz M. et al. (2023). Coalescent-based species delimitation in herbaceous bamboos (Bambusoideae, Olyreae) from Eastern Brazil: implications for taxonomy and conservation in a group with weak morphological divergence coupled with low genetic diversity. Plants 12: 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu D, Groenewald M, de Vries M. et al. (2019). Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Studies in Mycology 92: 135–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vučković N, Vico I, Duduk B. et al. (2022). Diversity of Botryosphaeriaceae and Diaporthe species associated with postharvest apple fruit decay in Serbia. Phytopathology 112: 929–943. [DOI] [PubMed] [Google Scholar]
- Walsh PD. (2000). Sample size for the diagnosis of conservation units. Conservation Biology 14: 1533–1537. [Google Scholar]
- Wanasinghe DN, Phukhamsakda C, Hyde KD. et al. (2018). Fungal diversity notes 709–839: taxonomic and phylogenetic contributions to fungal taxa with an emphasis on fungi on Rosaceae. Fungal Diversity 89: 1–236. [Google Scholar]
- Wang S-Y, McKenzie EHC, Phillips AJL. et al. (2022). Taxonomy and multigene phylogeny of Diaporthales in Guizhou Province, China. Journal of Fungi 8: 1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Guo Y, Du Y. et al. (2021). Characterization of Diaporthe species associated with peach constriction canker, with two novel species from China. MycoKeys 80: 77–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters JM, Rowe DL, Burridge CP. et al. (2010). Gene trees versus species trees: reassessing life-history evolution in a freshwater fish radiation. Systematic Biology 59: 504–517. [DOI] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee S. et al. (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: PCR protocols: a guide to methods and applications (Innis MA, Gelfand DH, Sninsky JJ. et al., eds). Academic Press, USA: 315–322. [Google Scholar]
- Wilson AW, Eberhardt U, Nguyen N. et al. (2023). Does one size fit all? Variations in the DNA barcode gaps of macrofungal genera. Journal of Fungi 9: 788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingfield MJ, De Beer ZW, Slippers B. et al. (2012). One fungus, one name promotes progressive plant pathology. Molecular Plant Pathology 13: 604–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrona CJ, Mohankumar V, Schoeman MH. et al. (2020). Phomopsis husk rot of macadamia in Australia and South Africa caused by novel Diaporthe species. Plant Pathology 69: 911–921. [Google Scholar]
- Xi Z, Liu L, Davis CC. (2016). The impact of missing data on species tree estimation. Molecular Biology and Evolution 33: 838–860. [DOI] [PubMed] [Google Scholar]
- Xu G, Qiu F, Li X. et al. (2020). Diaporthe limonicola causing leaf spot disease on Areca catechu in China. Plant Disease 104: 1869. [Google Scholar]
- Xu J. (2016). Fungal DNA barcoding. Genome 59: 913–932. [DOI] [PubMed] [Google Scholar]
- Xu J. (2020). Fungal species concepts in the genomics era. Genome 63: 459–468. [DOI] [PubMed] [Google Scholar]
- Yang EF, Karunarathna SC, Dai DQ. et al. (2022). Taxonomy and phylogeny of fungi associated with Mangifera indica from Yunnan, China. Journal of Fungi 8: 1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Fan X-L, Guarnaccia V. et al. (2018). High diversity of Diaporthe species associated with dieback diseases in China, with twelve new species described. MycoKeys 39: 97–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Rannala B. (2016). Species identifcation by Bayesian fngerprinting: a powerful alternative to DNA barcoding. bioRxiv: 041608. [Google Scholar]
- Yu G, Rao D, Matsui M. et al. (2017). Coalescent-based delimitation outperforms distance-based methods for delineating less divergent species: the case of Kurixalus odontotarsus species group. Scientific Reports 7: 16124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan HS, Lu X, Dai YC. et al. (2020). Fungal diversity notes 1277–1386: taxonomic and phylogenetic contributions to fungal taxa. Fungal Diversity 104: 1–266. [Google Scholar]
- Yurnaliza Y, Jamilah ID, Hartanto A. et al. (2021). Screening of endophytic fungi from oil palm (Elaeis guineensis) in producing exopolysaccharides. Biodiversitas 22: 1467–1473. [Google Scholar]
- Zapata M, Palma MA, Aninat MJ. et al. (2020). Polyphasic studies of new species of Diaporthe from native forest in Chile, with descriptions of Diaporthe araucanorum sp. nov., Diaporthe foikelawen sp. nov. and Diaporthe patagonica sp. nov. International Journal of Systematic and Evolutionary Microbiology 70: 3379–3390. [DOI] [PubMed] [Google Scholar]
- Zhang J, Kapli P, Pavlidis P. et al. (2013). A general species delimitation method with applications to phylogenetic placements. Bioinformatics 29: 2869–2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y-Q, Ma C-Y, Xue H. et al. (2023). Two new species of Diaporthe (Diaporthaceae, Diaporthales) in China. MycoKeys 95: 209–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou S, Li Q, Kong L, Yu H. et al. (2011). Comparing the usefulness of distance, monophyly and character-based DNA barcoding methods in species identification: a case study of Neogastropoda. PLoS ONE 6: e26619. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phylogenetic trees generated from a maximum likelihood analysis of the Diaporthe amygdali clade and related species. A. Based on ITS sequence data. B. Based on tef1 sequence data. C. Based on tub2 sequence data. D. Based on cal sequence data. E. Based on his3 sequence data. Bootstrap support values for maximum likelihood and maximum parsimony (ML-BS/MP-BS ≥ 70 %) are shown at the nodes. Taxa numbers were generated, and the corresponding strains and species are shown in the ITS-phylogram (panel A). Strains with type status are indicated in bold font. The isolates from this study are presented in brown typeface. The scale bars represent the expected number of nucleotide changes per site. The trees are rooted to D. acaciigena (CBS 129521).
Phylogenetic trees generated from a maximum likelihood analysis of the Diaporthe brasiliensis clade and related species. A. Based on ITS sequence data. B. Based on tef1 sequence data. C. Based on tub2 sequence data. D. Based on cal sequence data. E. Based on his3 sequence data. Bootstrap support values for maximum likelihood and maximum parsimony (ML-BS/MP-BS ≥ 70 %) are shown at the nodes. Taxa numbers were generated, and the corresponding strains and species are shown in the ITS-phylogram (panel A). Strains with type status are indicated in bold font. The isolates from this study are presented in brown typeface. The scale bars represent the expected number of nucleotide changes per site. The trees are rooted to D. scobina (CBS 129521).
Phylogenetic trees generated from a maximum likelihood analysis of the Diaporthe eucommiae clade and related species. A. Based on ITS sequence data. B. Based on tef1 sequence data. C. Based on tub2 sequence data. D. Based on cal sequence data. E. Based on his3 sequence data. Bootstrap support values for maximum likelihood and maximum parsimony (ML-BS/MP-BS ≥ 70 %) are shown at the nodes. Taxa numbers were generated, and the corresponding strains and species are shown in the ITS-phylogram (panel A). Strains with type status are indicated in bold font. The isolate from palm tissues included in the analyses is presented in green typeface. The scale bars represent the expected number of nucleotide changes per site. The trees are rooted to D. acutispora (CGMCC 3.18285 and LC 6142).
Phylogenetic trees generated from a maximum likelihood analysis of the Diaporthe foeniculina clade and related species. A. Based on ITS sequence data. B. Based on tef1 sequence data. C. Based on tub2 sequence data. D. Based on cal sequence data. E. Based on his3 sequence data. Bootstrap support values for maximum likelihood and maximum parsimony (ML-BS/MP-BS ≥ 70 %) are shown at the nodes. Taxa numbers were generated, and the corresponding strains and species are shown in the ITS-phylogram (panel A). Strains with type status are indicated in bold font. The isolates from this study are presented in brown typeface and the additional isolate from palm tissues included in the analyses is presented in green typeface. The scale bars represent the expected number of nucleotide changes per site. The trees are rooted to D. stictica (CBS 370.54).
Phylogenetic trees generated from a maximum likelihood analysis of the Diaporthe glabrae clade and related species. A. Based on ITS sequence data. B. Based on tef1 sequence data. C. Based on tub2 sequence data. D. Based on cal sequence data. E. Based on his3 sequence data. Bootstrap support values for maximum likelihood and maximum parsimony (ML-BS/MP-BS ≥ 70 %) are shown at the nodes. Taxa numbers were generated, and the corresponding strains and species are shown in the ITS-phylogram (panel A). Strains with type status are indicated in bold font. The isolates from palm tissues included in the analyses are presented in green typeface. The scale bars represent the expected number of nucleotide changes per site. The trees are rooted to D. raonikayaporum (CBS 133182).
Phylogenetic trees generated from a maximum likelihood analysis of the Diaporthe inconspicua clade and related species. A. Based on ITS sequence data. B. Based on tef1 sequence data. C. Based on tub2 sequence data. D. Based on cal sequence data. E. Based on his3 sequence data. Bootstrap support values for maximum likelihood and maximum parsimony (ML-BS/MP-BS ≥ 70 %) are shown at the nodes. Taxa numbers were generated, and the corresponding strains and species are shown in the ITS-phylogram (panel A). Strains with type status are indicated in bold font. The isolate from palm tissues included in the analyses is presented in green typeface. The scale bars represent the expected number of nucleotide changes per site. The trees are rooted to D. foeniculina (CBS 123208 and CBS 123209).
Phylogenetic trees generated from a maximum likelihood analysis of the Diaporthe leucospermi clade and related species. A. Based on ITS sequence data. B. Based on tef1 sequence data. C. Based on tub2 sequence data. D. Based on cal sequence data. E. Based on his3 sequence data. Bootstrap support values for maximum likelihood and maximum parsimony (ML-BS/MP-BS ≥ 70 %) are shown at the nodes. Taxa numbers were generated, and the corresponding strains and species are shown in the ITS-phylogram (panel A). Strains with type status are indicated in bold font. The isolates from this study are presented in brown typeface. The scale bars represent the expected number of nucleotide changes per site. The trees are rooted to D. ganjae (CBS 180.91 and PSCG 489).
Phylogenetic trees generated from a maximum likelihood analysis of the Diaporthe longicolla clade and related species. A. Based on ITS sequence data. B. Based on tef1 sequence data. C. Based on tub2 sequence data. D. Based on cal sequence data. E. Based on his3 sequence data. Bootstrap support values for maximum likelihood and maximum parsimony (ML-BS/MP-BS ≥ 70 %) are shown at the nodes. Taxa numbers were generated, and the corresponding strains and species are shown in the ITS-phylogram (panel A). Strains with type status are indicated in bold font. The isolates from palm tissues included in the analyses are presented in green typeface. The scale bars represent the expected number of nucleotide changes per site. The trees are rooted to D. batatas (CBS 122.21).
Phylogenetic trees generated from a maximum likelihood analysis of the Diaporthe rudis clade and related species. A. Based on ITS sequence data. B. Based on tef1 sequence data. C. Based on tub2 sequence data. D. Based on cal sequence data. E. Based on his3 sequence data. Bootstrap support values for maximum likelihood and maximum parsimony (ML-BS/MP-BS ≥ 70 %) are shown at the nodes. Taxa numbers were generated, and the corresponding strains and species are shown in the ITS-phylogram (panel A). Strains with type status are indicated in bold font. The isolates from this study are presented in brown typeface. The scale bars represent the expected number of nucleotide changes per site. The trees are rooted to D. cf. heveae 1 (CBS 852.97).
Synopsis of the alignment properties, statistics results and nucleotide substitution models used for the phylogenetic analyses of the Diaporthe amygdali clade.
Synopsis of the alignment properties, statistics results and nucleotide substitution models used for the phylogenetic analyses of the Diaporthe brasiliensis clade.
Synopsis of the alignment properties, statistics results and nucleotide substitution models used for the phylogenetic analyses of the Diaporthe eucommiae clade.
Synopsis of the alignment properties, statistics results and nucleotide substitution models used for the phylogenetic analyses of the Diaporthe foeniculina clade.
Synopsis of the alignment properties, statistics results and nucleotide substitution models used for the phylogenetic analyses of the Diaporthe glabrae clade.
Synopsis of the alignment properties, statistics results and nucleotide substitution models used for the phylogenetic analyses of the Diaporthe inconspicua clade.
Synopsis of the alignment properties, statistics results and nucleotide substitution models used for the phylogenetic analyses of the Diaporthe leucospermi clade.
Synopsis of the alignment properties, statistics results and nucleotide substitution models used for the phylogenetic analyses of the Diaporthe longicolla clade.
Synopsis of the alignment properties, statistics results and nucleotide substitution models used for the phylogenetic analyses of the Diaporthe rudis clade.