Abstract
The Helicobacter pylori immunodominant protein, CagA, is associated with severe gastritis and carcinoma. Injection of CagA into gastric epithelial cells by type IV secretion leads to actin-cytoskeletal rearrangements and cell scattering. CagA has been reported to have no role in the induction of transcription factor NF-κB and IL-8, which are crucial determinants for chronic inflammation. Here, we provide several lines of evidence showing that CagA is able to induce IL-8 in a time- and strain-dependent manner. We also show that by exchanging specific cagA genes, high IL-8-inducing H. pylori strains could be converted into low inducing strains and vice versa. Our results suggest that IL-8 release induced by CagA occurs via a Ras→Raf→Mek→Erk→NF-κB signaling pathway in a Shp-2- and c-Met-independent manner. Thus, CagA is a multifunctional protein capable of effecting both actin remodeling and potentiation of chemokine release.
Keywords: molecular pathogenesis, pathogenicity island, type IV secretion, virulence
The innate immune system responds to microbial pathogens in multiple organisms by recognition of specific pathogenic components based on interactions between pattern-recognition receptors (PRRs) and bacterial-associated molecules, such as LPS, peptidoglycan, DNA-containing unmethylated CpG motifs, flagellin, and lipoteichoic acid absent from self tissues. The largest and best studied family of PRRs comprises the Toll-like receptors (TLRs) (1-3). In addition to TLRs, a second class of PRRs consisting of the Nod1 and Nod2 cytoplasmic proteins is essential in intracellular bacterial sensing (4-6). A long-standing question is how the innate immunity discriminates noninvasive and extracellular bacterial pathogens from commensals.
Although some intracellular Helicobacter pylori have been reported (7, 8), this bacterium is generally considered as an extracellular pathogen, which has an important role in the pathogenesis of chronic gastritis, peptic ulcers, gastric adenocarcinoma, and gastric mucosa-associated lymphoid tissue (MALT) lymphoma (9-12). Persistent colonization of H. pylori in the human stomach results in release of chemoattractants, such as IL-8, which stimulate significant infiltration of neutrophils into the gastric mucosa, leading to chronic gastritis. IL-8 induction was shown to depend on the outer-membrane protein OipA (13) but predominantly on a functional type IV secretion system (T4SS) encoded by the cag pathogenicity island (cagPAI) present in virulent H. pylori strains (13-17). However, the mechanism by which cagPAI+ H. pylori strains induce proinflammatory responses in gastric epithelial cells has long remained a mystery. CagA, the only H. pylori T4SS effector protein known to date (18), has been shown to have an important role in H. pylori-induced actin-cytoskeletal rearrangements and cell scattering (19-22). However, it is a common belief that CagA has no role in the proinflammatory responses such as IL-8 release. Based on the observations that cagA mutants often still induce a considerable amount of IL-8, CagA is generally considered dispensable in the induction of proinflammatory responses (14-17).
Viala et al. recently reported that the T4SS of H. pylori delivers peptidoglycan into the host cells, which can then be sensed by the intracellular receptor Nod1 (also known as CARD4) leading to NF-κB activation (5). Here, we present lines of evidence demonstrating that transfected and translocated CagA from a subset of H. pylori strains are also able to induce IL-8 release through NF-κB activation. Our data strongly suggest that CagA represents another important mediator for cagPAI-dependent induction of potent proinflammatory responses during H. pylori infection.
Materials and Methods
Bacterial Strains and Cell Lines. All H. pylori strains used in this study have been described (20-24). AGS cells (human gastric adenocarcinoma epithelial cell line, CRL 1739c, American Type Culture Collection) were cultured according to standard procedures (22). Fibroblasts derived from Shp-2-/- mice (expressing a nonfunctional Shp-2 with an internal deletion of residues 46-110 in the N-terminal SH2 domain) or those from the Shp-2+/+ control expressing WT Shp-2 were cultured as described (25).
Mutagenesis, Cloning of cagA, Genetic Complementation, and Sequencing. Mutagenesis of cagA and virB11 genes has been described (22, 23). For complementation of cagA, cagA genes (of strains 26695, P310, and 2808) containing their own promoters were cloned in the Escherichia coli/H. pylori shuttle vector pSB19 containing the oriT of RP4 and a kanamycin resistance gene cassette (aph-A3) as selectable marker (23). Sequences of CagA from the various strains were determined by standard procedures and deposited in the GenBank database (see Fig. 7, which is published as supporting information on the PNAS web site).
Synchronized Infection and Bacterial Adherence Assays. H. pylori (2 × 108 colony forming units, CFU) were suspended in 0.5 ml of PBS and added to 2 × 106 AGS cells at a multiplicity of infection of 50 or 100 for the indicated periods of time. To determine the adherence of WT and mutant H. pylori to AGS cells, the infected cells were rigorously washed twice with PBS to remove unbound bacteria. Subsequently, the cells were harvested, and the CFU was determined on agar plates.
Expression Constructs, Transfection Assay, and Inhibitors. For transient expression of CagA, the cagA gene of H. pylori strain NCTC11637 was cloned into pSP65SRα vector containing a hemagglutinin (HA) tag (19). The phosphorylation-deficient CagA mutants were constructed by substituting all of the tyrosine residues in the five EPIYA sequence repeats (Y-893, Y-912, Y-965, Y-999, and Y-1033) by either alanines (CagAY→A) or phenylalanines (CagAY→F) (19, 20). The CagAWT, CagAY→A, Shp-2, and H-Ras constructs (19, 26) were kindly provided by Masanori Hatakeyama (Hokkaido University, Sapporo, Japan). Chihiro Sasakawa (University of Tokyo, Tokyo) kindly provided the CagAY→F mutant (20), which was then subcloned into pSP65SRα. The c-Met construct (27) was a gift from Craig P. Webb (Van Andel Institute, Bostwick, MI). To investigate the dynamics of NF-κB, we used pNF-κB-d2EGFP, a construct harboring a fusion of the NF-κB p65 subunit with GFP (p65-GFP, Clontech). The pIL-8-GFP construct comprises the proximal IL-8 promoter (nucleotides +420 to +102) in fusion to GFP cDNA (28). The Mek1 and Raf constructs were gifts of Petra Dersch (Robert Koch Institute, Berlin). Each of these expression constructs (10 μg) was transfected into 0.8 × 106 AGS by using Lipofectamine (Invitrogen). All pharmacological inhibitors were obtained from Calbiochem-Merck.
Antibodies and Immunoblotting Analysis. The following primary antibodies were used: mouse monoclonal α-phosphotyrosine PY99, mouse monoclonal α-Shp-2, goat α-GAPDH antibodies (all obtained from Santa Cruz Biotechnology), and rabbit polyclonal α-CagA antibody (Austral Biological). Rabbit polyclonal α-OipA antibody was kindly provided by Stefan Odenbreit (Ludwig Maximilian University, Munich). Horseradish peroxidase-conjugated α-mouse or α-rabbit polyvalent sheep Ig was used as secondary antibody (DAKO). Blots were developed with the ECL Plus Western blot reagents (Amersham Pharmacia).
IL-8 ELISA. The amount of IL-8 secreted into the cell culture supernatant was determined by ELISA using the OptEIA human IL-8 kit II (BD Biosciences) or the mouse KC IL-8 kit (R & D Systems). Statistical evaluation was performed by using Student's t test with sigmastat statistical software (version 2.0). (*, P < 0.05; **, P < 0.005; P ≥ 0.05 was considered not significant).
Immunofluorescence Microscopy. Immunostaining was carried out by standard procedures described in ref. 22. Transfected CagA was stained with a rabbit α-HA antibody (Sigma) and revealed with tetramethylrhodamine B isothiocyanate (TRITC)-conjugated goat α-rabbit (Sigma). Images were acquired on a DMIRE 2 fluorescence microscope (Diagnostic Instruments) by using spot advanced software.
Results
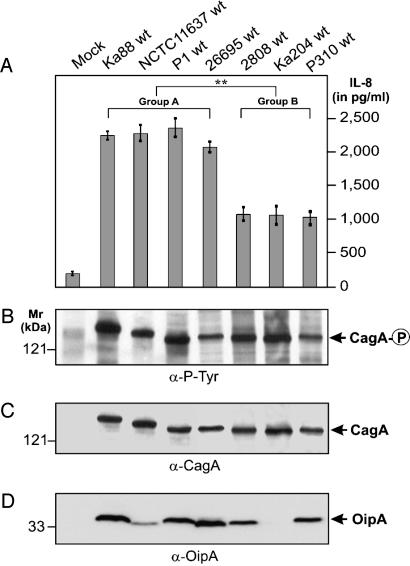
Identification of High and Low IL-8 Inducers Among H. pylori Strains Carrying a Functional cagPAI. To reexamine the ability of CagA to stimulate IL-8 induction in AGS gastric epithelial cells, we first analyzed clinical H. pylori isolates with respect to their capabilities in triggering IL-8 release. In agreement with our recent study on clinical isolates from gastritis, peptic ulcer, and gastric cancer patients (24), most cagPAI+ strains were able to induce high amounts of IL-8 (>2,000 pg/ml), whereas cagPAI- or T4SS-defective strains induced only a low level of IL-8 (<500 pg/ml), even after 9 h of infection. However, among the H. pylori strains that carry a functional cagPAI, we detected a distinct variation in their abilities to induce IL-8 (Fig. 1A). The strains that are capable of inducing >2,000 pg/ml of IL-8 are referred to as group A strains (or high IL-8 inducers), whereas strains that induce a lower level of IL-8 (≈1,000 pg/ml) are referred to as group B strains (or low IL-8 inducers). Interestingly, among the 62 cagPAI+ clinical H. pylori strains, 58 belong to group A (94%) and only 4 belong to group B, indicating that group A strains are significantly more prevalent in patients with gastric disease. Both group A and B strains were able to give rise to phosphorylated CagA in the host cells (Fig. 1 B and C), indicating that the T4SS is functional in both groups of strains (29, 30). Thus, the significantly reduced ability of group B strains to stimulate IL-8 appears to be due to reason(s) other than a nonfunctional T4SS. OipA is also unlikely to be a determining factor because the levels of OipA expression did not correlate with the amounts of IL-8 induced by the group A and B strains (Fig. 1D).
Fig. 1.
Identification of high and low IL-8-inducers among H. pylori strains that carry a functional cagPAI. (A) IL-8 release into the culture supernatant of AGS cells infected with various H. pylori strains was measured by standard ELISA. (B) Translocation and phosphorylation of CagA was analyzed by Western blotting with the phosphotyrosine-specific antibody PY-99. Stripping and reprobing of the blot with an α-CagA (C) or an α-OipA (D) antibody is shown. Arrows indicate the positions of the different proteins on the gel. Infection was for 9 h at a multiplicity of infection of 100. For the mock control, PBS was added to AGS. The data are mean values ± SD from at least three independent experiments.
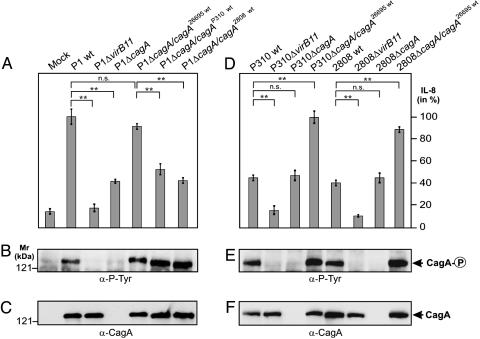
CagA Is Involved in IL-8 Induction. To reveal the molecular basis underlying the variation in IL-8-inducing abilities, we generated isogenic T4SS mutants (ΔvirB11 and ΔcagA) of both the high and low IL-8-inducing strains (23). Consistent with previous findings (5, 13-17), we observed an overall reduction in the level of IL-8 induction among all of the ΔvirB11 mutants, confirming that IL-8 induction by H. pylori is T4SS-dependent (Fig. 2 and Fig. 8A, which is published as supporting information on the PNAS web site). In contrast, we found that mutation of cagA had different effects on group A and group B strains. The ΔcagA mutants of group A strains induced significantly less IL-8 as compared with their WT H. pylori, whereas mutation of cagA in the group B strains (low IL-8 inducers) had no effect on their IL-8-inducing ability. Because a similar level of adherence to AGS cells was observed with all WT and mutant strains (Fig. 8B), our results strongly suggest that cagA encoded by the group A strains (or high IL-8 inducers) might have a role in the induction of IL-8.
Fig. 2.
CagA is involved in IL-8 induction. Genetic exchange of cagA genes reveals that low IL-8 inducers can be converted into high IL-8 inducers and vice versa. (A and D) ELISA reveals that exchange of cagA genes could alter the ability to induce IL-8 release. The cagA genes were exchanged between high and low IL-8-inducing H. pylori strains with the use of the shuttle plasmid pSB19. (B and E) CagA translocation and tyrosine phosphorylation in infected AGS cells was analyzed by Western blotting with antibody PY-99. (C and F) Stripping and reprobing of the blots with an α-CagA antibody. Arrows indictate the position of the different CagA protein species on the gel. Infection was for 9 h at a multiplicity of infection of 100. The data are mean values ± SD from at least three independent experiments.
To test this hypothesis, we exchanged the cagA genes between low- and high-IL-8-inducing strains. The cagA genes from strains 26695 (high inducer), 2808 (low inducer), and P310 (low inducer) are designated cagA26695, cagA2808, and cagAP310, respectively. Each of these genes were cloned into the E. coli/H. pylori shuttle vector pSB19, which were then used for complementation in the various high inducer and low inducer strains. As shown in Fig. 2A, expression of cagA26695 in P1ΔcagA restored the capability of P1ΔcagA to induce high amounts of IL-8, whereas complementation of P1ΔcagA with cagA2808 or cagAP310 led to a substantial drop in IL-8 induction as compared with that seen with WT P1. Conversely, after substitution of the cagA genes in the low IL-8 inducing group B strains (2808 and P310) with cagA26695 of the high inducing strain 26695, the capability of the low inducer strains 2808 and P310 to stimulate IL-8 release was restored to a level seen with the high IL-8 inducing strains 26695 or P1 (Fig. 2D). Expression, translocation, and phosphorylation of the CagA protein species during infection were confirmed by immunoblotting (Fig. 2 B, C, E, and F). Together, our data indicate that (i) the CagA protein of the group A H. pylori strains is involved in IL-8 induction, and (ii) genetic exchange of cagA genes with different IL-8-inducing capabilities among the clinical H. pylori strains could convert low IL-8 inducers into high IL-8 inducers and vice versa. Thus, this set of data provides direct proof that CagA is a determining factor in the induction of chemokine release.
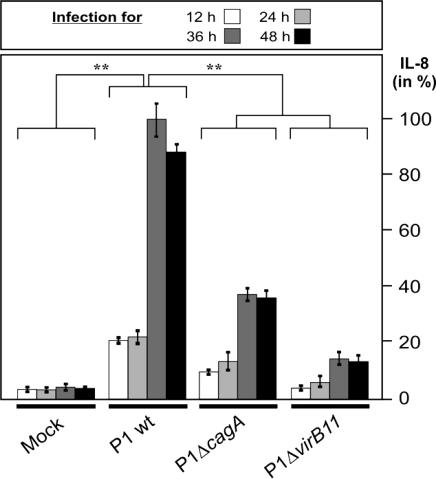
CagA-Mediated Induction of IL-8 Release Is Time-Dependent and Potentiates During the Course of Infection. Most of the previous studies investigated the H. pylori-induced IL-8 release at only one time point over a short course of infection (5, 13-17). Interestingly, by monitoring the IL-8 induction profile in a time course over 48 h, we observed that the amount of IL-8 induced by H. pylori in fact increased steadily with time until it reached a plateau by 36 h (Fig. 3). Most notably, the level of IL-8 induced by WT H. pylori was strongly enhanced at 24-36 h after infection as compared with that induced by the ΔcagA and ΔvirB11 mutants. A comparison of the levels of IL-8 release induced by WT and mutant bacteria at 36 h enables one to deduce that ≈64% and 86% of the IL-8 release stimulated by group A H. pylori strains were CagA- and cagPAI-dependent, respectively.
Fig. 3.
CagA-mediated induction of IL-8 release is time-dependent and potentiates during the course of infection. ELISA reveals that the amount of IL-8 induced by WT H. pylori enhances in a cagPAI- and CagA-dependent manner during the course of infection at a multiplicity of infection of 50.
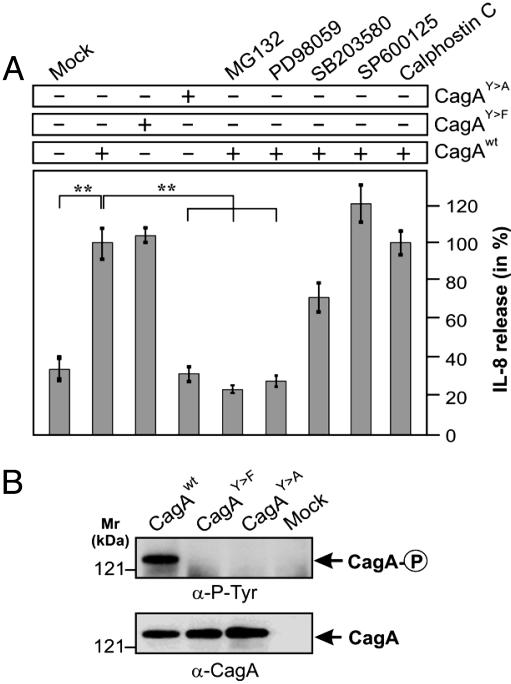
CagA Is Sufficient for Induction of IL-8 by an Erk- and NF-κB-Dependent Pathway. To test the hypothesis of whether CagA alone is able to stimulate IL-8 release by AGS cells, we transiently expressed CagA in AGS cells (19) and measured the level of IL-8 released by the transfected cells. Transient expression of WT CagA in AGS induced secretion of IL-8 ≈3-fold (up to 400-600 pg/ml IL-8) as compared with mock control (Fig. 4), demonstrating that transfected CagA is sufficient for the induction of a proinflammatory response. Furthermore, the phosphorylation-deficient CagAY→F mutant induced a similar level of IL-8 release, whereas another phosphorylation-deficient CagAY→A mutant failed to induce IL-8 (Fig. 4). This observation suggests that (i) the EPIYA motifs are important for the signaling and (ii) tyrosine phosphorylation appears to be nonessential or involved only indirectly.
Fig. 4.
Induction of IL-8 by transfection of CagA. (A) WT and mutant CagA from the high-inducing H. pylori strain NCTC11637 were transiently expressed in AGS in the presence or absence of the proteasome inhibitor MG132 (30 μM), Erk inhibitor PD98059 (25 μM), MAP kinase p38 inhibitor SB203580 (25 μM), c-Jun N-terminal kinase inhibitor SP600125 (25 μM), and PKC inhibitor calphostin C (100 nM) as indicated. At 48 h after infection, IL-8 release into the culture supernatant was measured by standard ELISA. For the mock control, empty vector was transfected. The data are mean values ± SD from at least three independent experiments. (B) Expression and tyrosine phosphorylation of CagA were verified by Western blot analysis.
Several recent studies have indicated that activation of the transcription factor NF-κB has a dominant role in H. pylori-induced IL-8 production in gastric epithelial cells (13-17). NF-κB can be activated by phosphorylation via different signaling pathways leading to subsequent proteolytic degradation of IκB. Activated NF-κB translocates to the nucleus where it up-regulates IL-8 gene transcription (15, 31). To unravel the signaling pathway involved in CagA-mediated IL-8 induction, we transfected AGS cells with CagA in the presence and absence of specific cell-permeable pharmacological inhibitors. As shown in Fig. 4, inhibitors for the mitogen-activated protein (MAP) kinases Erk1/2 (PD98059) and NF-κB (MG132) abolished the ability of CagA to induce IL-8, whereas the inhibitor of MAP kinase p38 (SB203580) or that of PKC (calphostin C) showed no inhibitory effect. The inhibitor for c-Jun N-terminal kinase (SP600125) even slightly enhanced the CagA-stimulated IL-8 release. Thus, these results suggest that CagA can induce IL-8 release through activation of an Erk→NF-κB-mediated signaling pathway.
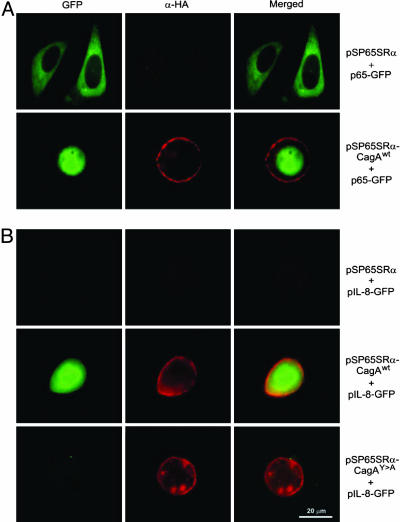
To reveal whether CagA expression indeed leads to activation of NF-κB, we examined the nuclear localization of NF-κB in AGS cells cotransfected with WT CagA and the p65 subunit of NF-κB in fusion to GFP (p65-GFP). WT CagA but not the empty vector control induced the recruitment of p65-GFP into the nucleus (Fig. 5A). These data suggest that expression of CagA results in translocation of NF-κB into the nucleus and, hence, activation of IL-8 transcription. To obtain further evidence for IL-8 gene activation, we cotransfected AGS cells with CagA constructs and GFP as a reporter under the control of the IL-8 promoter, pIL-8-GFP (28). Transient expression of WT CagA resulted in expression of GFP from the IL-8 promoter but not in transfections with the CagAY→A mutant or empty vector control (Fig. 5B).
Fig. 5.
Immunofluorescence of AGS cells transfected with (A) NF-κB p65 subunit (p65-GFP, green) or (B) pIL-8-GFP (green) in the presence of HA-tagged CagA constructs expressed from vector pSP65SRα (α-HA-staining, red). Transient expression of CagAWT triggered both translocation of p65-GFP into the nucleus as well as induction of pIL-8-GFP, neither of which was seen with the empty pSP65SRα vector control. The CagAY→A mutant also did not induce pIL-8-GFP (B) or nuclear translocation of p65-GFP (data not shown).
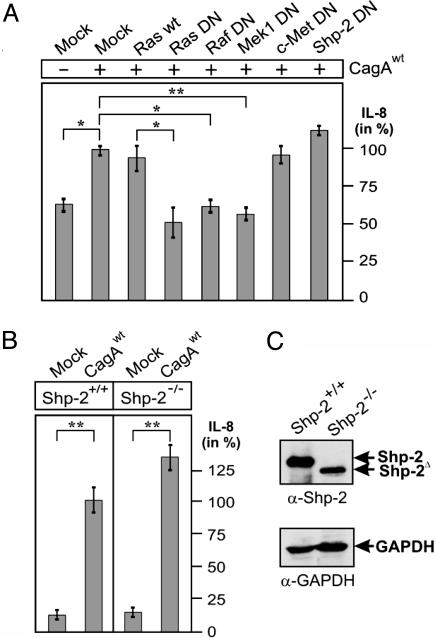
Induction of IL-8 by CagA Involves Ras and Raf but Not Shp-2 or c-Met. Last, we aimed to investigate which pathway upstream of Erk is involved in the CagA-induced NF-κB activation and subsequent IL-8 release. Ras is a well known regulator upstream of Erk (20, 32). Thus, we cotransfected WT CagA with dominant-negative (DN) Ras into AGS cells. Transient expression of DN Ras significantly inhibited the CagA-induced IL-8 secretion (Fig. 6A). Similar inhibition was observed by the Ras-specific farnesyltransferase inhibitor III (data not shown). In addition, transient expression of the DN construct of either Raf or Mek1, two downstream kinases in the Ras signaling pathway, also exhibited a significant blocking effect, whereas expression of DN c-Met or DN Shp-2 showed no inhibition (Fig. 6A). The tyrosine protein-tyrosine phosphatase (PTPase) Shp-2 has been described as a major binding partner of CagA (19, 26, 33). To ascertain that Shp-2 is indeed dispensable for CagA-induced IL-8 release, we transiently expressed WT CagA in fibroblasts derived from mice containing a nonfunctional Shp-2 (Shp-2-/-) or fibroblasts from their littermate controls containing WT Shp-2 (Shp-2+/+). The results indicate that the level of IL-8 release did not vary significantly between the Shp-2-/- and Shp-2+/+ cells (Fig. 6 B and C), supporting the notion that Shp-2 is not involved in the CagA-induced IL-8 release. Also, the CagA-induced IL-8 release in the fibroblasts was blocked in the presence of the PD98059 inhibitor (data not shown), suggesting that CagA induces IL-8 by a similar pathway as in AGS. Together, these results suggest that the CagA-induced IL-8-release proceeds via activation of the Ras→Raf→Mek→Erk→NF-κB signaling pathway in a Shp-2- and c-Met-independent manner.
Fig. 6.
Induction of IL-8 by CagA involves Ras, Raf, and Mek, but not Shp-2 or c-Met. (A) IL-8 release in AGS cells transfected with WT CagA in the presence of either WT or DN constructs. (B) IL-8 release in Shp-2-/- and Shp2+/+ cells transfected with CagAWT. As mock control, empty vector was transfected into the cells. The data are mean values ± SD from at least three independent experiments. (C) Expression of Shp-2 was verified by Western blot analysis with GAPDH as loading control.
Sequence Analysis of CagA from the Strong and Weak IL-8-Inducing H. pylori Strains. Alignment of the CagA sequences from both the high inducer and low inducer H. pylori strains shows overall strong homology except at the C terminus (Fig. 7A). The EPIYA motifs exist in various copies, depending on the H. pylori strain: either three or five copies per CagA in the group A strains or two or three copies in the group B strains (Fig. 7B). The fact that both the high IL-8 inducing CagA26695 and the low IL-8 inducing CagAP310 have three EPIYA motifs indicates that the number of EPIYA motifs is not sufficient for determining the ability to induce IL-8 release. Apart from the different number of EPIYA motifs, distinct single amino acid mutations or deletions are found in the sequences upstream of EPIYA repeats 2 and 3. Thus, it is tempting to speculate that both the number and sequence of the EPIYA motifs, as well as the amino acid residues upstream of the motifs may all be important for triggering IL-8 release. This postulation is in good agreement with the notion that differences in the arrangement of the EPIYA motifs in CagA proteins may underlie the different prevalence of gastric carcinoma in different geographic areas (33). Further studies are underway to elucidate the structural basis for the varying abilities of different CagA species to activate NF-κB signaling and, hence, induction of IL-8.
Discussion
The transcription factor NF-κB in human intestinal epithelial cells has a central role in regulating genes that govern the onset of mucosal inflammatory responses following microbial infections (1-6). H. pylori colonizes the extracellular surfaces of gastric epithelial cells and induces a strong inflammatory response that can progress to diseases such as peptic ulcer or even carcinoma. However, surface-expressed TLR2, TLR4, and TLR5 as typical pathogen-recognition molecules are not involved in the recognition of H. pylori (see refs. 5 and 12 for reviews). Viala et al. (5) have recently shown that the H. pylori-induced NF-κB activation in human embryonic kidney (HEK) 293 (cells deficient in TLR2 and TLR4) at 1 h after infection proceeds through translocation of peptidoglycan by the T4SS encoded in the cagPAI. They clearly demonstrated that the translocated peptidoglycan then activates Nod1 and, subsequently, NF-κB, leading to IL-8 release. Here, we describe a mechanism by which H. pylori is able to mediate proinflammatory responses from the outside of epithelial cells occurring at later time points when CagA is delivered. Our data support the notion that the peptidoglycan→Nod1→NF-κB pathway is not the exclusive cascade activated by H. pylori in the course of proinflammatory induction in infected gastric epithelial cells. Instead, H. pylori-induced NF-κB-mediated IL-8 production was shown in this study to be triggered predominantly by CagA, making CagA a bacterial T4SS effector protein capable of activating NF-κB.
Our current findings are in apparent contradiction to the previous notion that CagA is crucial for H. pylori-induced pathogenesis but does not have a role in triggering proinflammatory responses (9-18). However, paradoxically, they also provide clues to a possible explanation of this dilemma. It is now evident that the role of CagA in NF-κB and IL-8 induction might have previously been obscured by the following three major factors. (i) Depending on the H. pylori strain used, CagA mutants were shown to be capable of inducing IL-8 release, albeit often at a reduced level; (ii) the effect of sequence heterogeneity in CagA among clinical isolates on IL-8-inducing ability; and (iii) a possible time dependency had not been considered (13-17). First, we demonstrated here that the induction of IL-8 by H. pylori is time- and strain-dependent. High IL-8 inducing strains were shown to significantly potentiate this response over 48 h of infection. The potentiating effect of CagA on the induction of IL-8 was particularly obvious at the late time points, at 24-36 h after infection. Studies that have monitored the induction of IL-8 at much earlier time points could have underestimated the influence of CagA on the induction of IL-8 release. Second, by means of genetic exchange and site-directed mutagenesis, we were able to demonstrate a significant gain of proinflammatory function by CagA proteins. Our data showed that, by substituting specific cagA genes, high IL-8 inducing H. pylori strains could be converted into low IL-8-inducing strains and vice versa. Third, we have sequenced two cagA genes from low IL-8 inducers and the sequence data suggest that the EPIYA regions are likely to be critical. In agreement with this observation, we showed by transient expression experiments that WT CagA alone but not the CagAY→A EPIYA mutant is sufficient for triggering NF-κB activation, translocation of NF-κB into the nucleus and subsequent IL-8 release. Thus, CagA, the T4SS effector protein previously believed not to be involved in inflammatory responses, is now found to be capable not only of inducing actin-cytoskeletal rearrangements but also potentiating the induction of IL-8, thereby contributing directly to inflammation in H. pylori-induced gastric disease.
To dissect the signaling cascade involved in the CagA-induced IL-8 release, we examined in detail the effects of specific inhibitors, DN constructs as well as Shp-2 knockout cells, on the induction of IL-8 by transfected CagA. First, our data show that neither Shp-2, a tyrosine phosphatase and known binding partner of CagA (19, 26, 33), nor c-Met is required for triggering the signaling leading to IL-8 release. Second, findings from our pharmacological studies argue against the involvement of the MAP kinases p38 and c-Jun N-terminal kinase as well as PKC in the signaling cascade. Third, we demonstrated with the use of specific inhibitors that Erk kinase and NF-κB have a significant role in the signaling involved in the induction of IL-8. Also, we showed that expression of CagA in the presence of the DN constructs of Ras, Raf, and Mek1 inhibited the CagA-induced IL-8 release. Together, the data obtained allowed us to reveal that CagA induces IL-8 induction via the Ras→Raf→Mek→Erk→NF-κB signaling pathway. This conclusion is in good agreement with previous reports showing that MAP kinases play an important role in H. pylori-induced IL-8 production (34-37) and that nonphosphorylated CagA can activate the Ras→Erk pathway (20, 38). The binding of CagA to the adaptor protein Grb-2, which has been shown to activate Sos and Ras (20), could be the initial event in CagA-induced NF-κB activation.
In the light of our current findings, the functional role of CagA appears not to be restricted to the modulation of the host actin cytoskeleton and cell scattering (18-24, 26, 29, 30). Our data support a virulence model suggesting that CagA, after being translocated by T4SS in the host cell, acts as a multifunctional protein capable of triggering not only actin-cytoskeletal rearrangements and cell scattering but also NF-κB activation and the production of IL-8. The IL-8 release leads to a marked recruitment and infiltration of neutrophils into the gastric mucosa, contributing directly to the pronounced inflammatory response that is regarded as a hallmark of H. pylori infections. Our current data also reveal that different clinical cagPAI+ H. pylori strains express different CagA sequence variants, most of which have significant potentials for triggering IL-8 release. Thus, given the genetic plasticity of clinical H. pylori strains, it is tempting to hypothesize that cocolonizing H. pylori strains with different CagA species might be able to modulate their own proinflammatory potentials by genetic exchange during lifelong colonization of the human stomach. The elucidation of the structure-function of CagA remains one of the key challenges toward understanding how this important virulent factor of H. pylori exerts its wide spectrum of pathological effects on the host cell.
Supplementary Material
Acknowledgments
We thank Drs. M. Hatakeyama, C. Sasakawa, C. P. Webb, P. Dersch, and M. Husmann for providing plasmid constructs; G.-S. Feng for the Shp-2 knockout cells; M. Gerhard (Hubrecht Labs, Utrecht, The Netherlands) for H. pylori strain 2808; and S. Odenbreit for the α-OipA antibody. We also thank two anonymous reviewers for their critical comments. The work of S.B. is supported by NBL3-Magdeburger Forschungsverbund PFG4 and Priority Program SPP1150 of the Deutsche Forschungsgemeinschaft (Ba1671-3). T.K. is supported by an NBL3 fellowship.
Author contributions: S. Brandt, T.K., R.H., and S. Backert performed research; S. Brandt, T.K., R.H., and S. Backert analyzed data; W.K. contributed new reagents/analytic tools; S. Backert designed research; and S. Backert wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: TLR, Toll-like receptor; cagPAI, cag pathogenicity island; T4SS, type IV secretion system; DN, dominant negative; HA, hemagglutinin; MAP, mitogen-activated protein.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. DQ011619 and DQ011620).
References
- 1.Medzhitov, R. (2001) Nat. Rev. Immunol. 1, 135-145. [DOI] [PubMed] [Google Scholar]
- 2.Akira, S. (2003) Curr. Opin. Immunol. 15, 5-11. [DOI] [PubMed] [Google Scholar]
- 3.Beutler, B. (2004) Nature 430, 257-263. [DOI] [PubMed] [Google Scholar]
- 4.Girardin, S. E., Sansonetti, P. J. & Philpott, D. J. (2002) Trends Microbiol. 10, 193-199. [DOI] [PubMed] [Google Scholar]
- 5.Viala, J., Chaput, C., Boneca, I. G., Cardona, A., Girardin, S. E., Moran, A. P., Athman, R., Memet, S., Huerre, M. R., Coyle, A. J., et al. (2004) Nat. Immunol. 5, 1166-1174. [DOI] [PubMed] [Google Scholar]
- 6.Kim, J. G., Lee, S. J. & Kagnoff, M. F. (2004) Infect Immun. 72, 1487-1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amieva, M. R., Salama, N. R., Tompkins, L. S. & Falkow, S. (2002) Cell. Microbiol. 4, 677-690. [DOI] [PubMed] [Google Scholar]
- 8.Kwok, T., Backert, S., Schwarz, H., Berger, J. & Meyer, T. F. (2002) Infect. Immun. 70, 2108-2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Covacci, A., Telford, J. L., Del Giudice, G., Parsonnet, J. & Rappuoli, R. (1999) Science 284, 1328-1333. [DOI] [PubMed] [Google Scholar]
- 10.Montecucco, C. & Rappuoli, R. (2001) Nat. Rev. Mol. Cell Biol. 2, 457-466. [DOI] [PubMed] [Google Scholar]
- 11.Peek, R. M., Jr., & Blaser, M. J. (2002) Nat. Rev. Cancer 2, 28-37. [DOI] [PubMed] [Google Scholar]
- 12.Monack, D. M., Mueller, A. & Falkow, S. (2004) Nat. Rev. Microbiol. 2, 747-765. [DOI] [PubMed] [Google Scholar]
- 13.Yamaoka, Y., Kwon, D. H. & Graham, D. Y. (2000) Proc. Natl. Acad. Sci. USA 97, 7533-7538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crabtree, J. E., Xiang, Z., Lindley, I. J., Tompkins, D. S., Rappuoli, R. & Covacci, A. (1995) J. Clin. Pathol. 48, 967-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sharma, S. A., Tummuru, M. K., Blaser, M. J. & Kerr, L. D. (1998) J. Immunol. 160, 2401-2407. [PubMed] [Google Scholar]
- 16.Censini, S., Lange, C., Xiang, Z., Crabtree, J. E., Ghiara, P., Borodovsky, M., Rappuoli, R. & Covacci, A. (1996) Proc. Natl. Acad. Sci. USA 93, 14648-14653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fischer, W., Puls, J., Buhrdorf, R., Gebert, B., Odenbreit, S. & Haas, R. (2001) Mol. Microbiol. 42, 1337-1348. [DOI] [PubMed] [Google Scholar]
- 18.Censini, S., Stein, M. & Covacci, A. (2002) Curr. Opin. Microbiol. 4, 41-46. [DOI] [PubMed] [Google Scholar]
- 19.Higashi, H., Tsutsumi, R., Muto, S., Sugiyama, T., Azuma, T., Asaka, M. & Hatakeyama, M. (2001) Science 295, 683-686. [DOI] [PubMed] [Google Scholar]
- 20.Mimuro, H., Suzuki, T., Tanaka, J., Asahi, M., Haas, R. & Sasakawa, C. (2002) Mol. Cell 10, 745-755. [DOI] [PubMed] [Google Scholar]
- 21.Amieva, M. R., Vogelmann, R., Covacci, A., Tompkins, L. S., Nelson, W. J. & Falkow, S. (2003) Science 300, 1430-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Selbach, M., Moese, S., Hurwitz, R., Hauck, C. R., Meyer, T. F. & Backert, S. (2003) EMBO J. 22, 515-528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Backert, S., Moese, S., Selbach, M., Brinkmann, V. & Meyer, T. F. (2001) Mol. Microbiol. 42, 631-644. [DOI] [PubMed] [Google Scholar]
- 24.Backert, S., Schwarz, T., Miehlke, S., Kirsch, C., Sommer, C., Kwok, T., Gerhard, M., Goebel, U. B., Lehn, N., Koenig, W. & Meyer, T. F. (2004) Infect. Immun. 72, 1043-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi, Z. Q., Lu, W. & Feng, G. S. (1998) J. Biol. Chem. 27, 4904-4908. [DOI] [PubMed] [Google Scholar]
- 26.Higashi, H., Nakaya, A., Tsutsumi, R., Yokoyama, K., Fujii, Y., Ishikawa, S., Higuchi, M., Takahashi, A., Kurashima, Y., Teishikata, Y., et al. (2004) J. Biol. Chem. 279, 17205-17216. [DOI] [PubMed] [Google Scholar]
- 27.Furge, K. A., Kiewlich, D., Le, P., Vo, M. N., Faure, M., Howlett, A. R., Lipson, K. E., Woude, G. F. & Webb, C. P. (2001) Proc. Natl. Acad. Sci. USA 98, 10722-10727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dragneva, Y., Anuradha, C. D., Valeva, A., Hoffmann, A., Bhakdi, S. & Husmann, M. (2001) Infect. Immun. 69, 2630-2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stein, M., Bagnoli, F., Halenbeck, R., Rappuoli, R., Fantl, W. J. & Covacci, A. (2002) Mol. Microbiol. 43, 971-980. [DOI] [PubMed] [Google Scholar]
- 30.Selbach, M., Moese, S., Hauck, C. R., Meyer, T. F. & Backert, S. (2002) J. Biol. Chem. 277, 6775-6778. [DOI] [PubMed] [Google Scholar]
- 31.Muenzenmaier, A., Lange, C., Glocker, E., Covacci, A., Moran, A., Bereswill, S., Baeuerle, P. A., Kist, M. & Pahl, H. L. (1997) J. Immunol. 159, 6140-6147. [PubMed] [Google Scholar]
- 32.Kolch W. (2000) Biochem. J. 351, 289-305. [PMC free article] [PubMed] [Google Scholar]
- 33.Higashi, H., Tsutsumi, R., Fujita, A., Yamazaki, S., Asaka, M., Azuma, T. & Hatakeyama, M. (2002) Proc. Natl. Acad. Sci. USA 99, 14428-14433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Covacci, A. & Rappuoli, R. (2000) J. Exp. Med. 191, 587-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aihara, M., Tsuchimoto, D., Takizawa, H., Azuma, A., Wakebe, H., Ohmoto, Y., Imagawa, K., Kikuchi, M., Mukaida, N. & Matsushima, K. (1997) Infect. Immun. 65, 3218-3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Keates, S., Sougioultzis, S., Keates, A. C., Zhao, D., Peek, R. M., Jr., Shaw, L. M. & Kelly, C.P. (2001) J. Biol. Chem. 276, 48127-48134. [DOI] [PubMed] [Google Scholar]
- 37.Nozawa, Y., Nishihara, K., Peek, R. M., Nakano, M., Uji, T., Ajioka, H., Matsuura, N. & Miyake, H. (2002) Biochem. Pharmacol. 64, 21-30. [DOI] [PubMed] [Google Scholar]
- 38.Hirata, Y., Maeda, S., Mitsuno, Y., Tateishi, K., Yanai, A., Akanuma, M., Yoshida, H., Kawabe, T., Shiratori, Y. & Omata, M. (2002) Gastroenterology 23, 1962-1971. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.